Abbreviations
- CESC
cervical squamous cell carcinoma
- scRNA‐seq
single‐cell RNA sequencing
- UMAP
uniform manifold approximation and projection
- M‐MDSC
monocytic myeloid‐derived suppressor cells
- APOE
apolipoprotein E
- CXCL8
C‐X‐C Motif Chemokine Ligand 8
- CCR7
C‐C Motif Chemokine Receptor 7
- GZMK
Granzyme K
- NK
natural killer
- Treg
regulatory T
Dear Editor,
Increasing numbers of studies have revealed the immunomodulating effects of radiotherapy when combined with immune checkpoint inhibitors, as radioimmunotherapy has proven to be a promising treatment [1]. Radioimmunotherapy has shown significantly improved tumor responses than radiotherapy or immunotherapy alone in various malignant tumors [2, 3, 4]. It has also been applied to cervical cancer in multiple ongoing clinical trials (NCT03612791 [5] and NCT02635360 [6]). However, tumor recurrence and metastasis are often unavoidable. As such, investigations into radioimmunotherapy‐induced tumor ecosystem evolution are essential for guiding improvements in treatment strategies that achieve better long‐term disease control. To date, several studies have investigated radiochemotherapy‐induced tumor ecosystem evolution using bulk RNA‐sequencing and immune staining [7, 8]. However, these findings were limited owing to the cellular heterogeneity in cancers. Single‐cell RNA‐sequencing (scRNA‐seq) enables the characterization of cell compositions and transcriptomic states in the tumor at single‐cell resolution.
To investigate radioimmunotherapy‐induced intratumoral changes, we performed scRNA‐seq on a pair of cervical squamous cell carcinoma (CESC) samples before and during radioimmunotherapy. The study protocols are found in the Supplementary Materials. A total of 17,769 cells were obtained, with an average gene number of 1,875 after quality control. Using uniform manifold approximation and projection (UMAP) analysis, we identified nine main cell types: T cells (CD3D +, CD3E +), plasma cells (IGHG1 +, IGHG3 +), macrophages (CD14 +, CD68 +), monocytic myeloid‐derived suppressor cells (M‐MDSCs) (CD14 +, S100A12 +), mast cells (CPA3 +, KIT +), two types of cancer‐associated fibroblasts (ACTA2 +, DCN +), endothelial cells (VWF +, CDH5 +), and epithelial cells (KRT19 +, EPCAM +) (Supplementary Figure S1). The epithelial and immune cells accounted for 82% of all cells we obtained; therefore, we focused on the evolution of these cells at single‐cell resolution.
In this study, five epithelial cell subclusters (Epi1‐5) were identified by UMAP with distinct transcriptional features, such as Epi3 with high expression of squamous carcinoma cell markers KRT5, KRT6A, and KRT13, and Epi4 with high expression of normal cervical columnar cell markers MUC5AC and MUC5B (Figure 1A, B, Supplementary Figure S2A, Supplementary Table S1).
FIGURE 1.
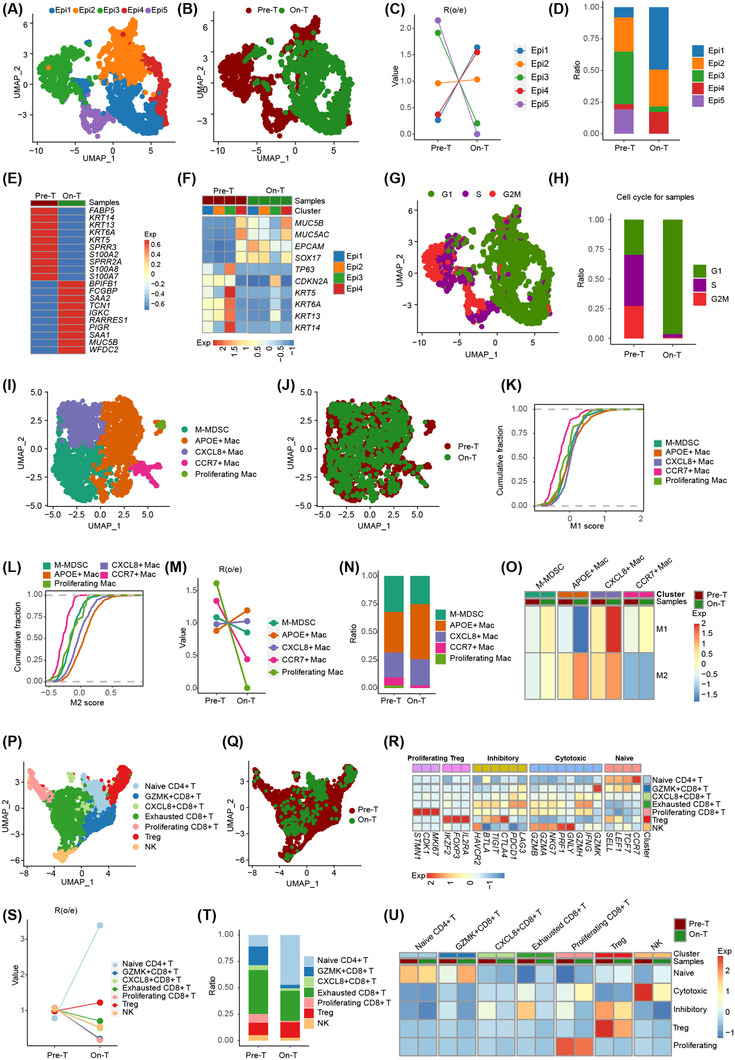
Single‐cell transcriptomics identifies tumor and immune microenvironment evolution induced by radioimmunotherapy in human CESC. (A, B) UMAP of epithelial cells showing five epithelial cell subclusters. Each dot represents a single cell, color‐coded by cell clusters (A) and treatment status (B). (C) Changes in the composition of the five epithelial cell subclusters before and during radioimmunotherapy. (D) Bar graph showing the proportion of each of the five epithelial cell subclusters quantified in tissue before and during radioimmunotherapy. (E) The top ten scaled differentially expressed genes of epithelial cells before and during radioimmunotherapy. (F) Changes in gene expression of typical markers for normal and malignant cervical epithelial cells induced by radioimmunotherapy, colored by scaled mean expression levels. (G) Cell cycle stage of the five epithelial cell subclusters presented in the UMAP plot, colored by cell cycle phases. (H) Bar graph showing the proportion of cells in different cell cycle stages in tissues before and during radioimmunotherapy. (I, J) UMAP of macrophages showing five subtypes. Each dot represents a single cell, color‐coded by cell clusters (I) and treatment status (J). (K, L) Cumulative distribution function showing the distribution of M1 (K) and M2 (L) signature scores in each macrophage subtype. A rightward shift of the curve indicates an increased score. (M) Changes in the composition of the five macrophage subtypes induced by radioimmunotherapy. (N) Bar graph showing the proportion of the five macrophage subtypes in tissues before and during radioimmunotherapy. (O) Radioimmunotherapy‐induced M1/M2 signature changes in each macrophage subtype. (P, Q) UMAP of T and NK cells showing seven subclusters. Each dot represents a single cell, color‐coded by cell clusters (P) and treatment status (Q). (R) Expression of signature gene sets in the seven T and NK cell subclusters. (S) Changes in the composition of the seven T and NK cell subclusters before and during radioimmunotherapy. (T) Bar graph showing the proportion of the seven T and NK cell subclusters in tissues before and during radioimmunotherapy. (U) Radioimmunotherapy‐induced signature gene sets changes in each T and NK cell subclusters. Note: “Pre‐T” denotes cells taken before radioimmunotherapy, and “On‐T” denotes cells taken during radioimmunotherapy treatment. Abbreviations: CESC, cervical squamous cell carcinoma; UMAP, uniform manifold approximation and projection; M‐MDSC, monocytic myeloid‐derived suppressor cells; APOE, apolipoprotein E; CXCL8, C‐X‐C Motif Chemokine Ligand 8; CCR7, C‐C Motif Chemokine Receptor 7; GZMK, Granzyme K; Epi, epithelial cell; NK, natural killer cell; Mac, macrophage; Treg, regulatory T cell; Pre‐T, pre‐treatment; On‐T, on‐treatment; Exp, expression
Next, we investigated the change in epithelial cell subclusters before and during radioimmunotherapy. The proportions of Epi4 and Epi1 were elevated, while Epi3 decreased and Epi5 disappeared during radioimmunotherapy (Figure 1C, D). Differentially expressed genes in epithelial cells showed that the expression levels of squamous carcinoma cell markers, such as KRT5 and KRT6A, were decreased, while normal cervical columnar cell markers, such as MUC5B, were increased during radioimmunotherapy (Figure 1E, Supplementary Table S2). Interestingly, we compared the expression profiles of normal (MUC5B, MUC5AC, EPCAM, and SOX17) and malignant (TP63, CDKN2A, KRT5, KRT6A, KRT13, and KRT14) marker genes in epithelial cells before and during radioimmunotherapy and found that malignant marker gene expression of Epi1‐3 was enriched before radioimmunotherapy and significantly decreased during radioimmunotherapy, while normal epithelial marker genes showed contrary results (Figure 1F). Only Epi3 cells still showed enrichment of malignant cervical epithelial cell marker genes, but the Epi3 cells were at a low fraction of all epithelial cells during radioimmunotherapy.
Further, we evaluated the cell cycle activation of the five epithelial cell subclusters and found that most Epi3 and Epi5 cells experienced S and G2M phases, while most Epi1‐2 and Epi4 cells experienced the G1 phase (Figure 1G, Supplementary Figure S2B). Radioimmunotherapy changed the cell cycle statuses of epithelial cells from the S and G2M phases to the G1 phase (Figure 1H). Taken together, radioimmunotherapy reprogramed epithelial cell subclusters from a malignant phenotype to a normal phenotype with a low fraction of residual Epi3 cells maintaining a malignant phenotype, implying a heterogeneous intratumoral response to radioimmunotherapy.
Macrophages have diverse roles, with complicated pro‐inflammatory and anti‐inflammatory characteristics in the tumor immune microenvironment. They were categorized into five subclusters with distinct transcriptional profiles in our study, consisting of M‐MDSCs (FCN1 +, S100A12 +), APOE+ macrophages, CXCL8+ macrophages (CXCL8 +), CCR7+ macrophages (CCR7 +), and proliferating macrophages (MKI67 +) (Figure 1I, J, Supplementary Figure S3, Supplementary Table S3). Evaluation of the M1 and M2 features of these macrophage subclusters revealed that APOE+ macrophages had the highest M2 score, while all subclusters showed similar M1 scores except CCR7+ macrophages (Figure 1K, L, Supplementary Figure S4). We also found that the proportion of APOE+ macrophages was increased and occupied the largest proportion of macrophages, while the proportion of proliferating macrophages was decreased during radioimmunotherapy (Figure 1M, N). Evaluation of the M1 and M2 scores before and during radioimmunotherapy showed that APOE+ macrophages had a decreased M1 score and an increased M2 score, while CXCL8+ macrophages had an increased M1 score (Figure 1O). Given the synergy between macrophage reprogramming strategies and immunotherapy [9], a strategy based on targeting tumor‐associated macrophages might reverse the immunosuppressive microenvironment during radioimmunotherapy and achieve a better immune response, such as targeting APOE macrophages.
Tumor‐infiltrating lymphocytes also have an important role in eliminating cancer cells, which become exhausted in tumors characterized by high expression of inhibitory molecules. For lymphocytes in our study, we identified seven subclusters with distinct transcriptional profiles, consisting of naïve CD4+ T (CD4 +, CCR7 +), regulatory T cell (Treg) (CD4 +, FOXP3 +), GZMK+CD8+ T (CD8A+, GZMK +), CXCL8+CD8+ T (CD8A +, CXCL8 +), exhausted CD8+ T (CD8A +, LAG3 +), proliferating CD8+ T (MKI67 +), and natural killer (NK) (NCAM1 +) cells (Figure 1P, Q, Supplementary Figure S5, Supplementary Table S4). Then, we evaluated naïve, cytotoxic, inhibitory, Treg, and proliferating scores for the seven subclusters and found that naïve CD4+ T cells had the highest naïve scores, NK and exhausted CD8+ T cells had high cytotoxic scores, Treg and exhausted CD8+ T cells had high inhibitory scores, Treg cells had the highest Treg scores, and proliferating CD8+ T cells had the highest proliferating scores (Figure 1R, Supplementary Figure S6). Next, among the T and NK cells during radioimmunotherapy, we found that the proportion of naïve CD4+ T cells was increased, while GZMK+CD8+ T and proliferating CD8+ T cells were decreased; naïve CD4+ T, Treg, and exhausted CD8+ T cells accounted for the majority of these cells (Figure 1S, T). These findings differ from those recently reported in another study that focused on the tumor immune response after low‐dose radiotherapy [10]; here, CD4+ T cells with features of exhausted effector cytotoxic cells were predominantly elicited. Treg and exhausted CD8+ T cells showed decreased inhibitory scores; NK and exhausted CD8+ T cells showed decreased cytotoxic scores during radioimmunotherapy (Figure 1U). Together, these data show that radioimmunotherapy induced a decrease in the inhibitory scores of residual exhausted CD8+ T and Treg cells in CESC.
It is important to consider several limitations in our study. First, this was a preliminary study with a single patient dataset that does not necessarily represent the cohort of patients with CESC who received radioimmunotherapy because of the intertumoral heterogeneity. A large‐scale study is thus needed to confirm and extend our findings. Second, spatial information is needed to understand the location of cells and their interactions to better understand the basis of radioimmunotherapy‐induced CESC evolution.
In conclusion, we revealed that radioimmunotherapy induced tumor and immune microenvironment evolution in human CESC at single‐cell resolution. Radioimmunotherapy changed epithelial cell subclusters from a malignant to a normal phenotype with a few residual malignant cells, increased APOE+ macrophages with high levels of M2 features, and induced a decrease in the inhibitory scores of residual exhausted CD8+ T and Treg cells. These results provide deep insights into cancer radioimmunotherapy and identify potential therapeutic targets that could be combined with radioimmunotherapy.
DECLARATIONS
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
AUTHORSHIP
JMY and LGX designed this study; CL, HY, RH, TYL, ML, XHL, QYH, and QLD collected the samples, performed the experiments, analyzed the data, and drafted the manuscript. All authors participated in writing, have read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethical Committee of Shandong Cancer Hospital and Institute (SDTHEC201906009). The patient provided written informed consent.
CONSENT FOR PUBLICATION
Not applicable.
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
This study was supported by funds from the National Natural Science Foundation of China (82030082), the Academic Promotion Program of Shandong First Medical University (2019ZL002), the Radiation Oncology Innovate Unit, Chinese Academy of Medical Sciences (2019RU071), Key Research and Development Project of Shandong Province of China (2021CXGC011102), Shandong Provincial Natural Science Foundation (ZR2021QH006) and Beijing Bethune Charitable Foundation (flzh202118).
Contributor Information
Ligang Xing, Email: xinglg@medmail.com.cn.
Jinming Yu, Email: sdyujinming@126.com.
DATA AVAILABILITY STATEMENT
All data in the study are available from corresponding authors, upon reasonable request.
REFERENCES
- 1. Deutsch E, Chargari C, Galluzzi L, Kroemer G. Optimising efficacy and reducing toxicity of anticancer radioimmunotherapy. Lancet Oncol. 2019;20(8):e452‐63. [DOI] [PubMed] [Google Scholar]
- 2. Altorki NK, McGraw TE, Borczuk AC, Saxena A, Port JL, Stiles BM, et al. Neoadjuvant durvalumab with or without stereotactic body radiotherapy in patients with early‐stage non‐small‐cell lung cancer: a single‐centre, randomised phase 2 trial. Lancet Oncol. 2021;22(6):824–35. [DOI] [PubMed] [Google Scholar]
- 3. Fizazi K, Drake CG, Beer TM, Kwon ED, Scher HI, Gerritsen WR, et al. Final Analysis of the Ipilimumab Versus Placebo Following Radiotherapy Phase III Trial in Postdocetaxel Metastatic Castration‐resistant Prostate Cancer Identifies an Excess of Long‐term Survivors. Eur Urol. 2020;78(6):822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sasaki A, Nakamura Y, Togashi Y, Kuno H, Hojo H, Kageyama S, et al. Enhanced tumor response to radiotherapy after PD‐1 blockade in metastatic gastric cancer. Gastric Cancer. 2020;23(5):893–903. [DOI] [PubMed] [Google Scholar]
- 5. Dyer BA, Feng CH, Eskander R, Sharabi AB, Mell LK, McHale M, et al. Current Status of Clinical Trials for Cervical and Uterine Cancer Using Immunotherapy Combined With Radiation. Int J Radiat Oncol Biol Phys. 2021;109(2):396–412. [DOI] [PubMed] [Google Scholar]
- 6. Feng CH, Mell LK, Sharabi AB, McHale M, Mayadev JS. Immunotherapy With Radiotherapy and Chemoradiotherapy for Cervical Cancer. Semin Radiat Oncol. 2020;30(4):273–80. [DOI] [PubMed] [Google Scholar]
- 7. Cosper PF, McNair C, Gonzalez I, Wong N, Knudsen KE, Chen JJ, et al. Decreased local immune response and retained HPV gene expression during chemoradiotherapy are associated with treatment resistance and death from cervical cancer. Int J Cancer. 2020;146(7):2047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J, Chen C, Zhan Y, Zhou L, Chen J, Cai Q, et al. Heterogeneity of IFN‐Mediated Responses and Tumor Immunogenicity in Patients with Cervical Cancer Receiving Concurrent Chemoradiotherapy. Clin Cancer Res. 2021;27(14):3990–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiang X, Wang J, Lu D, Xu X. Targeting tumor‐associated macrophages to synergize tumor immunotherapy. Signal Transduction and Targeted Therapy. 2021;6(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herrera FG, Ronet C, Ochoa de Olza M, Barras D, Crespo I, Andreatta M, et al. Low‐Dose Radiotherapy Reverses Tumor Immune Desertification and Resistance to Immunotherapy. Cancer Discov. 2022;12(1):108–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Data Availability Statement
All data in the study are available from corresponding authors, upon reasonable request.


