Abstract
Pancreatic cancer is one of the most serious health issues in developed and developing countries, with a 5‐year overall survival rate currently <9%. Patients typically present with advanced disease due to vague symptoms or lack of screening for early cancer detection. Surgical resection represents the only chance for cure, but treatment options are limited for advanced diseases, such as distant metastatic or locally progressive tumors. Although adjuvant chemotherapy has improved long‐term outcomes in advanced cancer patients, its response rate is low. So, exploring other new treatments is urgent. In recent years, increasing evidence has shown that lipid metabolism can support tumorigenesis and disease progression as well as treatment resistance through enhanced lipid synthesis, storage, and catabolism. Therefore, a better understanding of lipid metabolism networks may provide novel and promising strategies for early diagnosis, prognosis estimation, and targeted therapy for pancreatic cancer patients. In this review, we first enumerate and discuss current knowledge about the advances made in understanding the regulation of lipid metabolism in pancreatic cancer. In addition, we summarize preclinical studies and clinical trials with drugs targeting lipid metabolic systems in pancreatic cancer. Finally, we highlight the challenges and opportunities for targeting lipid metabolism pathways through precision therapies in pancreatic cancer.
Keywords: clinical trials, lipid metabolism, pancreatic cancer, targeting therapy
Abbreviations
- ABCA1
ATP‐binding cassette transporter A1
- ABCG1
ATP‐binding cassette transporter G1
- ACAT
acetyl‐CoA acetyltransferase
- ACC
Acetyl‐CoA carboxylase
- ACLY
ATP–citrate lyase
- ACSS
acetate by acetyl‐CoA synthetase
- ADM
acinar‐to‐ductal metaplasia
- AKT
the serine/threonine kinase
- AMPK
5′AMP‐activated protein kinase
- ASCT2
also named SLC1A5, solute carrier family 1 member 5
- BCL‐2
B‐cell lymphoma‐2
- BCL‐XL
B‐cell lymphoma extra‐large
- BET
bromo and extra terminal domain
- CACT
carnitine/acylcarnitine translocase
- ccRCC
clear cell renal cell carcinoma
- CD36
fatty acid translocase
- CDK4/6
cyclin‐dependent kinase 4/6
- CE
cholesteryl ester
- CPT1/2
carnitine palmitoyltransferase 1/2
- CSC
cancer stem cell
- EGCG
Epigallocatechin‐3 gallate
- EGFR
epidermal growth factor receptor
- ER
endoplasmic reticulum
- ERK
extracellular signal‐regulated kinase
- FA
fatty acid
- FABP
fatty acid‐binding protein
- FAO
fatty acid oxidation
- FASN
fatty acid synthase
- FATP
fatty acid transport protein
- GLS1/2
glutaminase 1/2
- GLUT1
glucose transporter 1
- GNAS
G‐protein αs
- GOT1
cytosolic aspartate aminotransaminase
- GPX4
glutathione peroxidase 4
- GSK‐3
glycogen synthase kinase 3
- HCC
hepatocellular carcinoma
- HDL
high‐density lipoprotein
- HDL‐C
high‐density lipoprotein‐cholesterol
- HER2
epidermal growth factor receptor 2
- HMGCR
3‐hydroxy‐3‐methyl‐glutaryl coenzyme A reductase
- IDH1/2
isocitrate dehydrogenase 1/2
- IKKβ
IkB kinase β
- IPMN
intraductal papillary mucinous neoplasia
- KRAS
Kirsten rat sarcoma viral oncogene
- LD
lipid droplet
- LDL
low‐density lipoprotein
- LDLR
low‐density lipoprotein receptor
- LN
lymph node
- LOX
lipoxygenase
- LXRs
liver X receptors
- MBOAT
membrane‐bound O‐acyltransferase
- MCL‐1
myeloid cell leukemia‐1
- MCT
monocarboxylate transporter
- MEK
mitogen‐activated protein kinase
- MUFA
monounsaturated fatty acid
- MVA
mevalonate
- NF‐κB
nuclear factor‐kappaB
- NSCLC
non‐small‐cell lung cancer
- PARP
poly (ADP‐ribose) polymerase
- PC
pancreatic cancer
- PDH
pyruvate dehydrogenase
- PI3K
phosphatidylinositol 3‐kinase
- PKA
protein kinase A
- PNKP
polynucleotide kinase/phosphatase
- PPAR
peroxisome proliferator‐activated receptor
- PUFA
polyunsaturated fatty acid
- ROS
reactive oxygen species
- SCD
stearoyl‐CoA desaturase
- SIK
salt‐inducible kinase
- SM
Squalene monooxygenase
- SOAT
Sterol‐o‐acyltransferase
- SP1
specificity protein 1
- SR‐B1/2
scavenger receptor class B type 1/2
- SREBP
sterol regulatory element‐binding protein
- STAT3
signal transducer and activator of transcription 3
- TAG
triacylglycerol
- TCA
tricarboxylic acid
- TFEB
transcription factor EB
- USP30
ubiquitin‐specific peptidase 30
- WNT signaling
Wingless / Integrated signaling
- Yarrow SFE
Yarrow CO2 supercritical extract
1. BACKGROUND
Pancreatic cancer (PC) is one of the most serious diseases, and its 5‐year survival rate remains less than 10% in the US and 7.2% in China. Patients with PC always typically present advanced diseases principally owing to a lack of early detection and effective treatment [1, 2]. Besides, extensive research has shown that PC also ranks 4th and 6th leading cause of cancer‐related deaths in the US and China, respectively [3, 4]. Additionally, PC is projected to become the second leading cause of cancer‐related deaths in the US by 2030 [5, 6].
Although progress has been made in multimodality treatment, surgery remains the only way to cure PC. Unfortunately, since the lack of symptoms in the early stage, over 80% of patients with PC are diagnosed when the lesion is no longer primarily resectable, resulting in a continuous increase in PC‐related deaths throughout the years [7, 8, 9]. Like many other types of cancers, PC is suggested to be a systematic disease that can benefit from comprehensive treatment following multidisciplinary management [10]. Additionally, recent research has established that targeted therapy is emerging as an anticancer strategy and plays an important role in treating a myriad of cancer types [11]. According to recent research, although targeted therapy has not demonstrated remarkable success in treating PC, a few clinical trials have reported encouraging outcomes [12]. Thus, it is fundamental to learn more about the molecular mechanisms of PC so that researchers can find novel hallmarks of signaling pathways.
Over the past two decades, changes in cell metabolism contributing to tumorigenicity and tumor progression have aroused researchers’ interest [13, 14]. These alterations mainly include aerobic glycolysis and glutamine metabolism [15, 16, 17]. Besides the well‐characterized reprogramming of glucose and glutamine metabolism, lipid metabolism is being increasingly recognized as an important pathway in cancer cells in the past few years [18, 19]. Lipids are a large and diverse set of nutrients composed of fat and lipoid, widely distributed in cellular organelles, and are critical components of all membranes [20]. Additionally, lipids are stored in lipid droplets (LDs) by cells when energy supplies are sufficient, but nutrient scarcity, on the other hand, can trigger homeostatic mechanisms of lipids which could function as second messengers to transduce signals within cells and serve as important energy sources [21]. Dysregulation of lipid metabolism contributes to the occurrence and progression of various metabolic diseases, including cardiovascular diseases, diabetes, and malignant diseases [22, 23, 24]. Currently, extensive evidence has revealed lipid metabolic abnormality, including membrane formation, lipid synthesis and degradation, and cellular signaling driven by lipids in various cancers [25, 26, 27]. Although several high‐quality reviews discuss the lipid metabolism of other cancer types [28, 29, 30], there is still no comprehensive review summarizing recent advances in the context of lipid metabolism reprogramming in PC. Therefore, considering the importance of lipid metabolism in malignant tumors, a complete understanding of reprogrammed lipid metabolism has a pivotal role in finding novel hallmarks and providing new insights for targeting cellular metabolic networks in PC. In this review, we summarize and discuss how lipid metabolic dysregulations contribute to the development and progression of PC and introduce therapeutic strategies for targeting lipid metabolism in PC.
2. ALTERATIONS OF LIPID METABOLISM IN PC
It is widely appreciated that lipids are essential to pancreatic cancer cells, primarily because they sustain membrane biosynthesis during rapid proliferation, are used in energy storage under conditions of metabolic stress, and have important roles as signaling molecules for many cellular activities. Although multifaceted and diverse biological functions of lipids and their by‐products are beginning to be uncovered gradually, there are still some unclear aspects in dire need of exploring.
2.1. Aberrant lipid uptake contributes to tumorigenesis and cancer progression
Lipid uptake from the exogenous environment is an important route through which cells can acquire fatty acids (FAs) and cholesterol [31]. FA uptake is conducted through membrane‐associated transport proteins, including fatty acid transport proteins (FATPs), fatty acid translocase (CD36), and fatty acid‐binding proteins (FABPs) [32]. CD36, a scavenger receptor class B type 2 (SR‐B2), is a transmembrane glycoprotein expressed on the cell surface in multiple cell types. In tumor tissues, CD36 is expressed in tumor cells, stromal cells, and immune cells, but the expression level varies in distinct cell types and tumor stages [33]. It is wildly accepted that CD36 is involved in metastasis initiation and proliferation of metastatic cells [34]. For example, a study has shown that the highly aggressive breast tumor MDA‐MB‐231 cell line expresses much less CD36 than the less aggressive MCF‐7 and T47‐D cells [35]. Additionally, CD36 was demonstrated to promote metastasis by activating the serine/threonine kinase (AKT) phosphorylation and inhibiting the degradation of glycogen synthase kinase 3 (GSK‐3)/β‐catenin in gastric cancer [36]. In parallel, there is evidence suggesting that FA uptake through CD36 may promote tumor metastasis and distant proliferation of cancer cells in hepatocellular carcinoma [37]. In the case of PC, the low expression of CD36 was associated with large tumor size and reduced survival rate [38], possibly due to decreased CD36 reduced tumor cell adhesion to the extracellular matrix and increased cell mobility, thereby speeding up the metastasis [36]. Besides, a large number of FAs via CD36 contributes to β‐oxidation, supporting cancer cell proliferation [39]. In addition to promoting tumor metastasis and cancer proliferation, CD36 can regulate chemoresistance in PC. The CD36 strong expression was correlated with gemcitabine resistance and the poor outcome in PC patients since CD36 enhances anti‐apoptosis protein expression, including B‐cell lymphoma‐2 (BCL‐2), B‐cell lymphoma extra‐large (BCL‐XL), and myeloid cell leukemia‐1 (MCL‐1), which protect cancer cells from drug‐induced cell death [40]. Moreover, it has been proved that CD36 can function as a mediator of the engulfment of PC microvesicles [41], but it is relatively less studied. In summary, the expression of CD36 is discrepant in different PC patients, and according to this we can develop individual treatment plans and predict prognosis.
Cholesterol, an essential lipid component of the mammalian cell membrane, plays a crucial role in maintaining membrane integrity and fluidity and forming membrane microstructures [42]. Intracellular cholesterol metabolism is maintained by a complex network, including cholesterol biosynthesis, uptake, export, and esterification [43, 44]. Cholesterol comes from diet and endogenous synthesis in the body. Specifically, cholesterol can be synthesized by cells de novo and through internalizing low‐density lipoprotein (LDL) [45]. In the case of cholesterol and cholesterol ester delivery, it occurs through a receptor‐mediated mechanism implicating the LDL receptor (LDLR) (Figure 1) [46]. According to recent evidence, genetically raised LDL cholesterol levels are associated with lower risks of endometrial cancer, including all endometrioid and non‐endometrioid subtypes [47]. Besides, another mendelian randomization analysis found that genetically predicted LDL cholesterol and total cholesterol were suggestively associated with an increased risk of colorectal cancer [48]. In addition, LDLR expression was positively correlated with poor prognosis in patients with small‐cell lung cancer and breast cancer, and LDLR depletion in epidermal growth factor receptor 2 (HER2)‐overexpressed breast cancer cells showed reduced cholesterol uptake and limited tumor growth in mice with hyperlipidemia [49, 50]. In regard to PC, several lines of evidence suggested that dysregulation of cholesterol uptake contributed to PC carcinogenesis. A retrospective study has found an unusual pattern of decreasing total cholesterol and LDL levels from 18 months to 6 months before PC diagnosis [51]. In addition, several meta‐analyses have shown a significant association between dietary cholesterol and the risk of PC in North America, Europe, and Japan, respectively [52]. Moreover, other studies revealed that genetically higher levels of LDL‐cholesterol were associated with PC progression. For example, recent evidence suggested that LDL‐cholesterol promotes PC cell proliferation, migration, and invasion by activating the signal transducer and activator of transcription (STAT)‐3 phosphorylation which contributes to both tumor survival and progression through the regulation of various established hallmarks of cancer [53]. Disruption of LDLR could impair proliferation in PC cell lines and tumorigenic capacities in mouse models and also inhibit the extracellular signal‐regulated kinase (ERK)‐dependent survival pathway, which is also overactive in numerous cancers by downregulating cholesterol uptake [54, 55]. Clinically, LDLR expression was positively correlated with decreased survival and a high risk of recurrence in patients with PC [55, 56, 57]. Although some recent research has revealed partial bio‐function of LDL cholesterol in PC, the exact correlation between LDL‐cholesterol levels and PC has not been completely deciphered (Figure 1).
FIGURE 1.
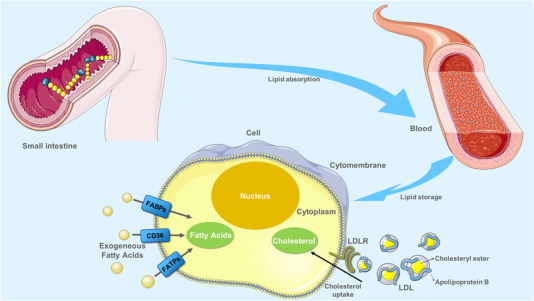
Process of lipid uptake. Digestion of lipids in food mainly occurs in the small intestine, where they are absorbed into the blood. The exogenous uptake of FAs from the surrounding microenvironment is facilitated by specialized transporters, including CD36, FATPs, and FABPs. Cholesterol carried by LDL particles in the blood can be taken up by LDLR at the basal surface of cells. Abbreviations: CD36, fatty acid translocase; FATPs, fatty acid transport proteins; FABPs, fatty acid‐binding proteins. LDL, low‐density lipoprotein; LDLR, low‐density lipoprotein receptor
2.2. De novo lipid synthesis is a metabolic source for tumor growth
In addition to lipid uptake, mammalian cells can acquire lipids through de novo synthesis. Under physiological conditions, lipogenesis is primarily restricted to hepatocytes and adipocytes, but cancer cells can activate lipogenesis in response to their high metabolic demand even in the presence of exogenous lipid sources [58, 59]. In contrast to non‐cancerous cells, approximately 93% of triacylglycerol FAs in tumor cells are de novo synthesized from mitochondrial citrate [60]. In addition, various enzymes that mediate lipid synthesis are transcriptionally upregulated in tumors [61, 62]. Since both FA and cholesterol are synthesized from acetyl‐CoA (acetyl coenzyme A; a key node in metabolism due to its intersection with many metabolic pathways and transformations) through a series of reactions, acetyl‐CoA levels are a key element for lipid production. In matters of acetyl‐CoA, it can either be derived from citrate by ATP‐citrate lyase (ACLY) or acetate by acetyl‐CoA synthetase (ACSS). Besides, glucose and glutamine contribute to citrate production from mitochondrial pyruvate oxidation in the tricarboxylic acid (TCA) cycle and reductive carboxylation, respectively. Glucose‐derived pyruvate is converted to acetyl‐CoA by pyruvate dehydrogenase, followed by citrate synthase‐mediated production of citrate, which is then exported to the cytosol from mitochondria by mitochondrial citrate transport proteins. Glutamine is converted to α‐ketoglutarate mediated by cytosolic glutaminase 1 (GLS1) or mitochondrial GLS2, which is followed by cytosolic isocitrate dehydrogenase 1 (IDH1)‐ and mitochondrial IDH2‐dependent isocitrate and subsequent citrate production (Figure 2) [63, 64, 65, 66]. Also, under conditions of metabolic stress such as hypoxia or lipid depletion, cancer cells can generate acetyl‐CoA from acetate by upregulating acetyl‐CoA synthetase 2 (ACSS2) [67].
FIGURE 2.
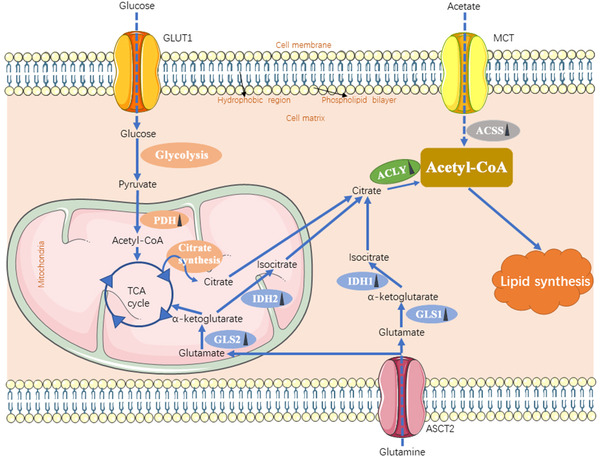
The source of acetyl‐CoA in cancer cells. Cancer cells obtain acetyl‐CoA from ACLY‐catalyzed citrate and ACSS‐catalyzed acetate, which is used for lipid synthesis. Additionally, glutamine and glucose can contribute to citrate production through the TCA cycle. Each solid blue arrow represents a specific biological process respectively in the figure. The dotted arrow means the process of transporting mediated by transporters. Dark triangles following the enzymes represent the alteration in cancer cells: upward means upregulation; downward means downregulation. Abbreviations: GLUT1, glucose transporter 1; MCT, monocarboxylate transporter; ASCT2, also named SLC1A5, solute carrier family 1 member 5; PDH, pyruvate dehydrogenase; GLS, glutaminase; IDH, isocitrate dehydrogenase; ACLY, ATP‐citrate lyase; ACSS, acyl‐CoA synthetase short‐chain family; TCA, tricarboxylic acid
2.2.1. Enzymes of producing acetyl‐CoA in PC: ACLY and ACSS
Human ACLY is a 480‐k‐DA tetramer with a total of 1101 amino acids and is principally located in the cytosol and nucleus [68]. ACLY catalyzes the conversion of citrate and CoA to oxaloacetate and acetyl‐CoA, playing a critical role in lipogenesis [69]. The upregulation of ACLY is correlated with tumor progression in various cancer types, including glioblastoma, colorectal cancer, breast cancer, lung cancer, and hepatocellular carcinoma (HCC) [70]. Inhibiting ACLY at the genetic level or pharmacologically significantly reduces tumor cell viability and suppresses cancer cell proliferation, invasion, and migration [71, 72]. For example, a recent study revealed that the IkB kinase β (IKKβ, which plays an important role in mediating inflammation and oncogenesis) phosphorylated ubiquitin‐specific peptidase 30 (USP30, which is involved in antagonizing mitophagy, regulating mitochondrial morphology, and restricting cell apoptosis) and ACLY, which in turn promoted ACLY deubiquitination in HCC. Besides, the inhibition of ACLY dramatically suppressed hepatocarcinogenesis [73]. Incidentally, increased ACLY expression was associated with poor outcomes for patients [74]. About PC, ACLY supports pancreatic tumorigenesis and facilitates tumor progression. It has been found that ACLY is required for efficient in vitro KRAS‐driven acinar‐to‐ductal metaplasia (ADM) and pancreatic tumorigenesis [75]. Both cell proliferation and tumor growth can be suppressed by targeting ACLY via concurrent Bromo and extra terminal domain (BET) inhibition and statin treatment [75]. Further, a novel study found that berberine, an isoquinoline alkaloid characterized by a diversity of pharmacological effects, could significantly decrease the expression of ACLY in the cytosol, leading to the disruption of lipid metabolism and inhibiting PC cells proliferation and migration [76]. In addition to participating in lipogenesis in the cytosol, nucleus‐translocated ACLY generates acetyl‐CoA for histone acetylation and gene transcription regulation [77]. For instance, the AKT‐ACLY signaling pathway could be activated by growth factors or oncogenic KRAS, which then promotes global histone acetylation in cancer cells which is correlated with poor prognosis, cell proliferation, and tumor growth [75, 77]. In the view of ACLY‐mediated acetyl‐CoA production plays important roles in both the cytosol and nucleus, targeting ACLY may serve as an attractive anti‐cancer strategy.
ACSS, an acetyl‐CoA synthetase short‐chain family, produces acetyl‐CoA via the ligation of acetate and CoA. ACSS family contains three members, ACSS1, ACSS2, and ACSS3. ACSS1 and ACSS3 are mitochondrial proteins, and ACSS2 is localized in both the cytoplasm and nucleus [78, 79]. In general, acetyl‐CoA synthetase was involved in the tumor progression. Specifically, ACSS1 was originally found to be required for acetate uptake and cell survival in HCC [80, 81]. ACSS3, a relatively new ACSS member, has been proposed as an important prognosis biomarker in gastric cancer [82]. Besides, recent studies have suggested that ACSS3 serves as a biomarker to stratify subtypes of HCC and promotes bladder cancer cell growth under metabolic stress [83, 84]. However, the role of ACSS1 and ACSS3 in PC has hardly been reported. Among the three ACSSs, ACSS2 was the most extensively studied one. It was transcriptionally upregulated by sterol regulatory element‐binding protein (SREBP) and highly expressed in many human tumors, especially under metabolic stress [67, 85]. In the glucose‐deprived condition, 5′AMP‐activated protein kinase (AMPK)‐mediated phosphorylation of ACSS2 at S659 was activated, which exposed the nuclear localization signal of ACSS2 for importin α5 binding and nuclear translocation [78, 86]. In the nucleus, ACSS2 formed complexes with transcription factor EB (TFEB). These complexes were translocated to lysosomal and autophagy gene promoter regions, leading to lysosomal biogenesis, autophagy, cell survival, and tumorigenesis [87, 88]. Knocking down ACSS2 largely inhibits tumor growth, which also signifies the pivotal role of acetate consumption in tumor growth [89, 90, 91]. In terms of PC, high expression of ACSS2 promotes acetate uptake for lipid synthesis and membrane phospholipids under metabolic stress such as hypoxia and lack of nutrition [92]. Nevertheless, information regarding the relationship between the ACSS family and PC is still limited. Thus, considering the critical role ACSS plays in lipid synthesis, it is necessary to figure out the upstream regulators and downstream signaling pathways of ACSSs in PC.
Many other enzymes are involved in the production of acetyl‐CoA, including pyruvate dehydrogenase (PDH), IDH1/2, and GLS1/2, which are closely linked with glucose and amino acids acid metabolism. Since we focus on lipid metabolism in PC, we just enumerate these enzymes in Figure 2 and they are not covered in detail.
2.2.2. FA biosynthesis enzymes in PC: ACC, FASN, and SCD
Acetyl‐CoA carboxylase (ACC), the rate‐limiting enzyme for FA synthesis, catalyzes the carboxylation of acetyl‐CoA to malonyl‐CoA. Mammalian acetyl‐CoA carboxylase occurs in two isoforms: ACC1 and ACC2 [93, 94]. ACC1, encoded by ACACA, is a cytosolic enzyme primarily expressed in lipogenic tissues and is critical for FA synthesis. On the one hand, ACC1 is genetically regulated by SREBP at the transcriptional level [95]. On the other hand, its expression is affected by a complex interplay of phosphorylation, the binding of allosteric regulators, and protein‐protein interactions at the protein level [93]. Previous research has established that ACC1 is highly expressed in various human cancers, including breast [94], prostate [96, 97], liver [98], gastric [99], and non‐small‐cell lung cancer (NSCLC) [100]. ACC1 depletion decreases FA synthesis and suppresses tumor growth. ACC2, encoded by ACACB, is anchored to the outer mitochondrial membrane, which controls FA β‐oxidation and FA uptake [101]. Recent evidence suggested that a high ACC2 expression level was positively associated with poor clinical cancer stage and a decreased 5‐year survival rate in laryngocarcinoma [102]. For PC, it has been found that higher plasma triglyceride levels and intratumor expression of ACC1 were associated with lower benefits from everolimus‐based treatment [103]. Besides, recent research found that ACC inhibitors could attenuate the Wingless / Integrated (WNT) and Hedgehog signaling, thus suppressing pancreatic tumor growth [104]. In addition, a recent report has shown that CPI‐613, a novel lipoate analog inhibiting mitochondrial metabolism, exhibits strong anticancer activity in PC cells via reactive oxygen species (ROS)‐associated apoptosis, which is coupled with AMPK activation. The upregulated AMPK‐ACC signaling rewires lipid metabolism, promoting the progression of apoptosis in PC cells [105]. Similarly, it was also revealed that berberine inhibited ACC activity by activating AMPK, which suppressed intracellular FA synthesis, decreased the biogenesis of extracellular vesicles, and finally inhibited the proliferation capacity of cancer cells [106]. Intriguingly, a lower level of ACC1 expression has been tested in PANC‐1 cells compared with other PC cell lines, which may indicate that PANC‐1 cells are independent, or only minimally dependent, on de novo FA synthesis for survival [107]. In summary, these findings suggest the critical roles of ACC in lipid metabolism, and part of its molecular mechanisms are relatively clear. However, whether inhibition of ACC has a bright future in treating PC depends on the results of reliable clinical studies.
Fatty acid synthase (FASN), a key lipogenic enzyme in the de novo biogenesis of FAs, condenses one molecule of acetyl‐CoA and seven malonyl‐CoA molecules into the 16‐carbon palmitate, which is used for the synthesis of more complex FAs, plasma membrane structures, and post‐translational protein palmitoylation [108]. FASN has been studied extensively in various cancers, including breast, prostate, colorectal, bladder, and lung carcinoma, and its overexpression and hyperactivity predict poor prognosis [109]. In regard to PC, it has been shown that serum FASN levels are higher in patients with PC and intraductal papillary mucinous neoplasia (IPMN) than in healthy controls [110]. Besides, FASN is highly up‐regulated in PC stem cells compared to parental cells [111]. Moreover, reports showed an increased expression level of FASN during proliferation of PANC‐1, and suppression of FASN by orlistat which is a FASN inhibitor resulted in a significant reduction of PANC‐1 proliferation and enhanced apoptosis of these cells [112, 113]. In addition, PC patients with a high level of FASN showed a shorter overall survival than patients with low FASN expression, and its overexpression is also associated with poor response to gemcitabine therapy which is the foundation of PC chemotherapy through upregulating the expression of pyruvate kinase M2 [114]. Additionally, novel paclitaxel nano‐formulation not only reduces the expression of FASN but also enhances gemcitabine efficacy [115, 116]. in vitro, it has been reported that FASN overexpression causes resistance to genotoxic treatments by increasing poly (ADP‐ribose) polymerase (PARP)‐1 expression and DNA repair activity via the nuclear factor‐kappaB (NF‐κB) and specificity protein 1 (SP1) in PC cells [117]. In addition, palbociclib, the cyclin‐dependent kinase 4 and 6 (CDK4/6) inhibitor, could negatively regulate FASN due to the upregulation of AMPKα and MiR‐33a levels to increase apoptosis in PANC‐1 and MIA PaCa‐2 cells [118]. Tumor‐associated FASN overexpression is preferentially regulated at the transcription level by SREBP1, which is downstream of several signaling pathways and factors such as the phosphatidylinositol 3‐kinase (PI3K)/AKT and mitogen‐activated protein kinase (MEK)/ERK pathways [119]. Being required for oncogenic KRAS‐induced pancreatic tumorigenesis, the activation of the epidermal growth factor receptor (EGFR) signaling can promote the expression of FASN in PC cells via an ERK‐dependent manner [120, 121]. Therefore, PC patients with overexpression of SREBP1 have shorter overall survival than patients with low SREBP1 expression, and knockdown of SREBP1 inhibits the growth capacity of PC cells [122].
Stearoyl‐CoA desaturase (SCD), an endoplasmic reticulum‐resident integral membrane protein, catalyzes the formation of a double bond at position Δ9 in stearic acid (C18:0) and palmitic acid (C16:0), to generate the monounsaturated FA (MUFA) oleic acid (C18:1) and palmitoleic acid (C16:1), respectively. SREBP controls the expression of SCD. There are 5 SCD genes (SCD1‐5), and humans contain the SCD homologs SCD1 and SCD5 [123, 124]. SCD1 overexpression was found in multiple tumors, and SCD1 inhibition could reduce the formation of tumors derived from human gastric, colon, lung, and prostate cancer cells in xenograft mouse models [125]. SCD5 is unique to primates and highly expressed in the brain and pancreas. Beyond that, SCD5 expression has also been observed in several human oncogene‐transformed and cancer cells [126]. In PC cells and patients, it has been observed an increased expression of SCD1 [127], and extensive research has shown that SCD1 prevents the death of PC cells by anti‐ferroptosis [128, 129]. Moreover, recent data suggest that the SCD enzyme is required for early pancreatic tumor growth, and inhibition of SCD induces the unfolded protein response, which in turn suppresses the growth of pancreatic tumors [130]. Compared to SCD1, the bio‐function of SCD5 in cancers remains relatively mysterious, particularly in PC. Hence, further research on the relationship between the SCD family and the malignant biological behavior of pancreatic tumors is worthy.
In summary, substantial evidence already showed that ACC, FASN, and SCD are closely correlated with tumorigenesis and progression, implying that targeting them might offer an innovative approach to improve the therapeutic outcome of PC patients.
2.2.3. Cholesterol and cholesterol ester biosynthesis enzymes in PC: acetoacetyl‐CoA thiolase, HMGCR, SM, and SOAT
Acetoacetyl‐CoA thiolase, also known as acetyl‐CoA acetyltransferase (ACAT), starts cholesterol biosynthesis by condensing two molecules of acetyl‐CoA into acetoacetyl‐CoA. ACAT corresponds to two ubiquitous metabolic enzymes, which are respectively localized in the mitochondria and cytoplasm [131]. In recent years, mitochondrial acetoacetyl‐CoA thiolase (T2) has been reported to be upregulated in diverse human cancer cells and play roles in anti‐cancer drug resistance, cancer cell proliferation, and tumor growth [132, 133, 134]. Additionally, it has also been suggested that ACAT participated in tumorigenesis of hypopharynx cancer and clear cell renal cell carcinoma(ccRCC) [135]. As for the bio‐function of ACAT in PC, it has been reported that T2 showed a down‐regulated trend in PC and was associated with radioresistance in the cell lines [136, 137].
The rate‐limiting enzyme of the mevalonate (MVA) metabolic pathway, 3‐hydroxy‐3‐methyl‐glutaryl coenzyme A reductase (HMGCR), converts HMG‐CoA to mevalonate [138]. HMGCR is commonly perceived as one of the pathogenesis for cardiovascular diseases, but recently increased data have shown that HMGCR is also involved in oncogenesis and metastasis [139, 140]. Sterols and isoprenoids, the production of the MVA pathway, are integral to tumor growth and progression [141]. It has been observed previously that HMGCR expression is upregulated in cancers derived from the adrenal gland, blood, lymph, brain, breast, colon, connective tissue, embryo, esophagus, liver, lung, ovary, pancreas, prostate, skin, and stomach [142]. Additionally, increased HMGCR expression promoted the growth and metastasis of some types of cancer, while HMGCR knockdown inhibited their growth, metastasis, and tumorigenesis [143, 144, 145]. Furthermore, it was observed that the HMGCR inhibitor statins reduced cancer risk, the grade and stage of patients at diagnosis, recurrence, and cancer‐related mortality [146, 147, 148]. Concerning PC, expression of HMGCR was shown to be elevated in PC in an oncogenic KRAS mouse model, and the MVA pathway gene expression was upregulated in PC [57, 75]. Suppression of HMGCR has been reported to decrease the risk of PC and had significant chemopreventive efficacy in PC cells [149]. Furthermore, inhibition of HMGCR sensitizes PC cells to gemcitabine and is effective in treating chemotherapy‐resistant PC cases [150, 151]. Intriguingly, according to two large prospective cohort studies in the US, regular statin use was not associated with PC risk [152]. Collectively, although inhibition of HMGCR could reduce the risk of PC remains controversial, it was proved that HMGCR contributed to tumorigenesis, and suppression of HMGCR could improve the therapeutic effect of PC.
Squalene monooxygenase (SM), another rate‐limiting cholesterol biosynthesis enzyme downstream of HMGCR, converts nonsterol intermediate squalene to 2,3(S)‐oxidosqualene. It is encoded by the squalene epoxidase gene and regulated by SREBP2 [153]. An early study showed that cancer cell proliferation was indeed highly dependent on sterol biosynthesis [154]. To date, robust scientific evidence shows that SM plays a vital role in multiple types of cancer [155]. The overexpression of SM was detected in hepatocellular cancer, breast cancer, prostate cancer, colorectal cancer, and squamous lung cancer [156, 157, 158, 159, 160]. Moreover, increasing evidence suggests a positive correlation between SM overexpression and poor prognosis in various tumors [155]. Of note, the loss of SM expression also contributes to tumor growth or promotes tumor aggressiveness. In ALK+ anaplastic large cell lymphoma cell lines, the loss of SM resulted in the accumulation of the squalene, which in turn altered the cellular lipid profile and prevented cancer cells from ferroptosis, contributing to a growth advantage under conditions of oxidative stress and in tumor xenografts [161, 162]. Taken together, SM might act as a double‐edged sword in various cancer. According to bioinformatic analyses of gene expression data in the ONCOMINE database, SM was significantly highly expressed in PC [158]. Furthermore, SM was considered a novel ferroptosis‐related risk signature, and a ferroptosis‐related high‐risk level can adversely affect the anti‐tumor immune process [163]. Additionally, SM has also been associated with radioresistance in PC [136]. However, there are still many unanswered questions about the role of SM in pathogenesis, progression, treatment, and prognosis in PC. Given the importance of SM in other cancers, focusing on the molecular mechanism of SM is expected to provide a novel idea for treating PC.
Sterol‐o‐acyltransferase (SOAT, also known as acyl‐CoA cholesterol acyltransferase), the founding member of the membrane‐bound O‐acyltransferase (MBOAT) family, catalyzes the transfer of an acyl group from acyl‐coenzyme A to cholesterol to generate cholesteryl ester (CE) at endoplasmic reticulum (ER) membrane [164]. Two mammalian isoforms of SOAT have been identified, SOAT1 and SOAT2, and they have gained attention as potential drug targets for treating diseases such as atherosclerosis, Alzheimer's disease, and cancers [165]. SOAT1 is ubiquitously expressed in most cell types and tissues, and high levels of SOAT1 expression have also been previously observed in diverse cancers [44, 166]. In addition, pharmacological inhibition of SOAT1 has been shown to reduce the size of HCC, inhibit the growth of prostate cancer cells, prolong survival in glioblastoma xenograft models, and enhance the antitumor response of CD8+ T cells and efficacy of immunotherapy in vitro [167, 168, 169, 170]. Human SOAT2 is mainly expressed in the intestine and fetal liver [171]. The previous research showed the induction of SOAT2 is linked to HCC development and demonstrated that HCC‐linked promoter hypomethylation played a conclusive role in the induction of human SOAT2 gene expression [172]. Besides, SOAT2 is also involved in the migration and invasion processes of breast cancer cells [173]. Further, an in vitro study revealed that the inhibitor of SOAT2 exhibited moderate cytotoxicity in the HeLa cell line [174]. The role of SOAT in PC is gradually being elucidated. An early study showed that SOAT was activated significantly in tumor models compared to normal mice or rats [175]. Moreover, a case report described an undifferentiated pancreatic carcinoma with rhabdoid features which strongly expressed SOAT1 [176]. Based on current work, SOAT1 expression dependent on p53 status was upregulated during PC progression. SOAT1 loss significantly impairs PC progression, and heterozygous loss of TP53 sensitizes tumor cells to SOAT1 deficiency [177]. Moreover, the overexpression of SOAT1 has been reported to be associated with a poor prognosis in patients with PC, and inhibitors of SOAT1 could suppress the growth and metastasis of tumors [178, 179]. In contrast, the mechanism of SOAT2 act in PC remains almost unknown, which needs further exploration (Figure 3).
FIGURE 3.
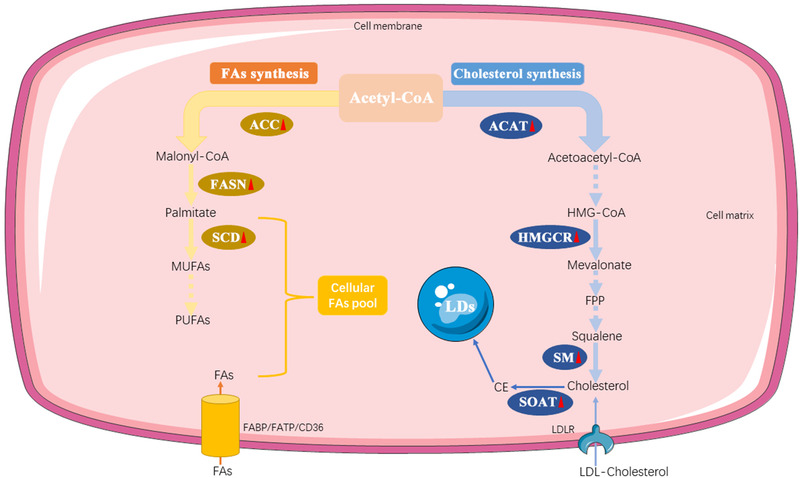
The synthesis of FAs and cholesterol. FA and cholesterol biosynthesis both starts with acetyl‐CoA. Acetyl‐CoA is catalyzed into a series of unsaturated FAs by ACC, FSAN, and SCD in sequence. The production of cholesterol is mediated by several key enzymes, including ACAT, HMGCR, and SM. Furthermore, cholesterol can be converted to CE by SOAT, which is stored in LDs. The solid arrow represents a one‐step specific biological process, whereas the dotted arrow represents a multi‐step specific biological process. Red triangles following the enzymes represent the alteration in PC cells: upward means upregulation; downward means downregulation. Abbreviations: FAs, fatty acids; ACC, Acetyl‐CoA carboxylase; FASN, fatty acid synthase; SCD, stearoyl‐CoA desaturase; MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids; ACAT, Acetyl‐CoA acetyltransferase; HMGCR, 3‐hydroxy‐3‐methyl‐glutaryl coenzyme A reductase; SM, Squalene monooxygenase; SOAT, Sterol‐o‐acyltransferase; CE, cholesterol ester; LDs, lipid droplets. CD36, fatty acid translocase; FATP, fatty acid transport protein; FABP, fatty acid‐binding protein. LDLR, low‐density lipoprotein receptor
2.3. Multiple roles of abnormal lipid catabolism to malignancy
2.3.1. Fatty acid oxidation
Fatty acid oxidation (FAO), also called β‐oxidation, allows the mitochondrial conversion of long‐chain FAs into acetyl‐CoA (which enters the TCA cycle), NADH, and FADH2 (which are coenzymes used in the electron transport chain) [180]. FAO mainly occurs in mitochondria and involves a series of reactions. The first step of FAO is fatty acid activation, producing fatty acyl‐CoA catalyzed by acyl‐CoA synthetase. Then, on the outer mitochondrial membrane, fatty acyl‐CoA is converted to fatty acylcarnitine by carnitine palmitoyltransferase I (CPT1), which comprises three subtypes, CPT1A, CPT1B, and CPT1C. After that, carnitine/acylcarnitine translocase (CACT) on the inner mitochondrial membrane shuttles acylcarnitine into the mitochondrial matrix. Next, carnitine palmitoyltransferase II (CPT2) on the matrix side of the inner membrane reconverts acylcarnitine to acyl‐CoA. Finally, in the mitochondrion, acyl‐CoA is cleaved into acetyl‐CoA by a repeated 4‐step cycle, and the breakdown product acetyl‐CoA enters the TCA cycle to generate ATP [181, 182, 183].
Studies over the past two decades have highlighted the importance of FAO for cancer metabolism, and many types of cancer exhibit high FAO activity [184, 185, 186]. Cancer cells frequently use FAs as catabolic substrates under conditions of ATP depletion, and the NADH and/or NADPH generated supports ATP production, redox homeostasis, and biosynthesis reactions, which in turn ensure cell survival and proliferation, and even some cancer cells preferentially use FAO to fuel growth when nutrients are abundant [187]. Instead, FAO was reported to be involved in the metastasis of some tumors [188, 189]. One of the main mechanisms of tumor metastasis is that FAO promotes the proliferation of lymphatic endothelial cells and lymphangiogenesis [190]. Another study also found that tumor metastasis to lymph node (LN) in mice required cancer cells to undergo a metabolic shift toward FAO, and the FAO signaling pathway was upregulated [191]. In addition, current evidence has revealed that FAO participates in the occurrence of chemoresistance [192]. Moreover, excessive FAO leads to oxidative stress, p38 activation, and impaired muscle growth in cancer cachexia [193].
Increasingly more studies have recognized the critical role of FAO in PC. Recent research showed that FA, not glucose, was a major source of electrons for ATP production through FAO and oxidative phosphorylation in cancer cells. Besides, dysregulated FAO contributed to the drug resistance of cancer stem cells (CSCs) [194, 195]. Similarly, another study has suggested that in PC with G‐protein αs (GNAS) mutations, cancer cells acquire acetyl‐CoA through FAO, which is activated by the protein kinase A (PKA)‐salt‐inducible kinase (SIK) axis, which leads to pancreatic tumorigenesis [196]. Besides, recent research has reported that FAO supports cell viability and invasion of PC in vitro under acidic extracellular conditions [197]. At the molecular level, it has been observed that CPT1A and CPT1B levels are higher in drug‐resistant pancreatic CSCs, and the oxidative phosphorylation in drug‐resistant cells was due to FAO [195]. Knockdown of CPT1C inhibited the tumorigenesis of PANC‐1 cells in vivo and further suppressed xenograft tumor growth in situ [198]. In conclusion, the dynamic FAO regulation exerts a critical role in cancer progression, and targeting the FAO signaling pathway might be a promising therapeutic strategy for PC.
2.3.2. Lipid peroxidation and cell death
Lipids, particularly polyunsaturated fatty acids (PUFAs), are susceptible to oxidation, leading to lipid peroxidation that is harmful to cells and tissues [199]. Lipid peroxides are formed through non‐enzymatic and enzymatic lipid peroxidation. Non‐enzymatic lipid peroxidation (also named auto‐oxidation of lipids) is a free radical‐driven chain reaction in which ROS initiates the oxidation of PUFAs. On the other hand, enzymatic lipid peroxidation is mediated by the lipoxygenase (LOX) family that catalyzes the deoxygenation of free and esterified PUFAs to generate various lipid hydroperoxides [200]. Lipid peroxidation has been shown to play a role in several types of cell death, including apoptosis, necroptosis, ferroptosis, pyroptosis, and alkaliptosis, which are associated with inflammation, neurodegenerative disease, and cancers [201]. In recent years, there has been an increasing interest in lipid peroxidation, especially ferroptosis, which was originally described as a type of cell death specifically occurring in cancer cells with mutant RAS and characterized by iron‐dependent lipid peroxidation [202]. Generally speaking, ferroptosis can be induced in two ways. On the one hand, the extrinsic pathway is mainly induced by the suppression of system XC − (for example, with erastin, sorafenib, or sulfasalazine). On the other hand, the intrinsic pathway of ferroptosis can be directly activated by reducing the expression or activity of glutathione peroxidase 4 (GPX4) [203].
It has been repeatedly reported that ferroptosis works as a tumor‐suppressive mechanism as well as apoptosis, and has been attracting attention as a novel therapeutic method against drug‐tolerant cancer cells [204]. The way of cell death like ferroptosis caused by lipid peroxidation plays a crucial part in the development and progression of PC. Recent evidence has suggested that ferroptosis acts as an inhibitor of pancreatic tumorigenesis in mice, and autophagy‐dependent ferroptotic cancer cell death may be required for ROS‐induced KRASG12D release from PC cells [205, 206]. A novel study also has reported that inhibition of the cytosolic aspartate aminotransaminase (GOT1) used to maintain redox balance in PC can promote ferroptosis and suppress the growth of numerous PC cell lines, primary cancer models, and xenograft tumors [207]. Moreover, inducing ferroptosis at the pharmacological or genetic level inhibits the formation and development of PC cells, particularly in subtypes of drug resistance. For example, activated ferroptosis potentiated the cytotoxic effect of gemcitabine and mitigated gemcitabine resistance in PC cells [208, 209]. Additionally, a predictive model based on ferroptosis regulators accurately predicted PC patient survival time, and elevated sensitivity to ferroptosis was correlated with enhanced immune activity [210]. However, recent studies have revealed the harmful impact of ferroptosis on tumor biology. For instance, new preclinical animal studies and clinical retrospective analyses documented that ferroptotic damage promoted KRAS‐driven pancreatic tumorigenesis [211], implicating that induction or inhibition of ferroptosis may depend on specific tumor subtypes. In regard to other kinds of cell death, such as necroptosis, previous evidence showed that inducing necroptosis could dramatically increase the survival times of mice with orthotopic PC and reduce tumor growth, stroma, and metastasis [212]. Overall, cell death associated with lipid peroxidation has emerged as an active area of research that may lead to new anti‐cancer approaches, particularly against metabolically active tumors.
2.3.3. Lipid droplets
Ubiquitous in cells, LDs, which mainly consist of triacylglycerol (TAG) and CE, are storage organelles at the center of lipid and energy homeostasis. Lipids stored during conditions of nutrient surplus are mobilized for energy production during starvation or phospholipid synthesis during high demand for membranes [213]. Recently, alterations in LD metabolism are emerging as important parts of cancer metabolic reprogramming [214]. The biosynthesis and breakdown of LDs play different roles in tumor progression. Extensive evidence has revealed that an increased abundance of LDs is a feature of many aggressive cancers, and their accumulation has been associated with neoplastic processes and tumor invasiveness [26, 215]. Moreover, compromising the ability of cancer cells to form LDs could also impair their chemoresistance and immune evasion [216]. Analogously, the activation of LD breakdown could be a beneficial strategy. For example, stimulation of lipolysis is detrimental for cancer cells under certain conditions since it may increase the levels of oxidative and ER stress, elevate lipid peroxidation and even lead to ferroptotic cell death [217]. Whereas caution should be exerted because LDs do not always play a cancer‐friendly role. LDs can promote drug accumulation and activation to kill certain types of cancer cells, such as the MCF7, H1650, A549, and RKO cell lines [218]. In addition, LD breakdown promotes the resistance of cancer cells to stress [219]. Thus, to sum up, the feasibility of targeting LDs should be carefully examined in different tumor types and particular contexts.
Based on the double‐edged roles of LDs in multiple cancers, the influence of LDs on tumor growth and metastasis needs to be spelled out in the context of PC. Previous data showed that LDs were drastically increased in PC [57], and that LD accumulation was correlated with the tumor growth and aggressiveness [220, 221]. Another study showed that the number of LDs in pancreatic CSCs was higher than that in non‐CSCs, and the peroxisome proliferator‐activated receptor α (PPARα) activation, which is considered a downstream factor of LD‐derived signaling, was observed in CSCs, implicating that LD‐PPARα axis is possibly involved in the maintenance of CSC properties [222]. Furthermore, depletion of LDs inhibited invasion of KRAS‐mutant PC [223]. Of note, instead of the amount of LDs, high LD movement speed, especially in a more acidic tumor microenvironment, promoted tumor invasion regulated by V‐ATPase, a key membrane proton (H+) pump which is associated with multidrug resistance of PC [224].
2.3.4. Cholesterol efflux
Instead of de novo synthesis, uptake, storage, and cholesterol efflux are also critical for cholesterol homeostasis. The rate of cholesterol efflux is dependent on cholesterol load in the cell, composition, size, and concentration of high‐density lipoprotein (HDL), and the expression of the efflux transporters. Four main pathways of cholesterol efflux have been identified: 1. passive diffusion of cholesterol to mature HDL particles; 2. facilitated diffusion via Scavenger receptor type B, class‐I (SR‐B1); 3. efflux to lipid‐poor apolipoprotein, apo A1 (pre β1 HDL) via ATP‐binding cassette transporter A1 (ABCA1); 4. ATP‐binding cassette transporter G1 (ABCG1) mediated efflux to lipid‐rich, mature HDL [225, 226]. Increasing evidence has suggested that cholesterol efflux contributes to diverse types of tumors. For example, HDL‐cholesterol levels are a potential biomarker for some cancers, and epidemiological data have also shown levels of HDL‐cholesterol are significantly inversely correlated with risk for multiple cancers [227]. In addition, SR‐B1, a multiligand membrane receptor protein that functions as a physiologically relevant HDL receptor, is reported to be overexpressed in many tumors [228]. ABCA1, a plasma membrane‐bound transporter that promotes cholesterol export and thereby reduces intracellular cholesterol levels, is well accounted for its anticancer activity [229]. Moreover, the expression of ABCG1 was also reported to be enhanced in cancer cell lines and was inversely related to overall survival, and depletion of ABCG1 triggered tumor regression [230].
The role of cholesterol efflux in PC is becoming increasingly clear. A previous study found serum HDL‐cholesterol levels were significantly lower in PC postmenopausal women [52]. Similarly, a two‐center retrospective study showed that PC patients exhibited a lower serum HDL level on admission versus the non‐PC tumor group [231]. Additionally, recent data comparing PC samples with those of normal tissues demonstrated considerably increased expression of SR‐B1 [232]. Meanwhile, a study showed an upregulation in transcript levels of ABCA1 and ABCG1 in PC compared to non‐neoplastic tissues [233]. Further, ABCA1 and ABCG1 were correlated with tumor progression and survival. Besides, ATP‐binding cassette (ABC) transporters are known to play a pivotal role in the development of PC chemoresistance due to their ability to pump anticancer drugs out of cancer cells [234], but relatively little is known about the roles of ABCA1 and ABCG1 in drug resistance. Collectively, the abnormal cholesterol efflux is evident in PC, and its underlying molecular mechanisms urgently need to be explored, which could provide a new hematologic index for diagnosis and prognosis combined with cholesterol uptake and synthesis, and also may point to a potentially novel anti‐tumor therapeutic strategy against PC.
2.4. Transcriptional regulation of lipid metabolism
2.4.1. SREBPs
Cellular lipid metabolism and homeostasis are controlled by SREBPs, a family of helix‐loop‐helix leucine zipper transcription factors consisting of three isoforms: SREBP1a and SREBP1c encoded by the SREBF1 gene, and SREBP2 encoded by the SREBF2 gene. SREBP1 mainly regulates the expression of FA synthesis genes and LDLR, while SREBP2 preferentially controls cholesterol biosynthesis gene expression [235]. The importance of SREBPs in cancer has begun to be recognized, which are frequently activated in cancer cells. Cleavage of SREBPs is stimulated by the PI3K‐AKT‐mTOR pathway, which is the most frequently activated oncogenic signaling pathway found in various cancers [236]. Additionally, the above‐mentioned ACLY is also a downstream target of SREBPs, which has an inseparable relationship with the occurrence and progress of PC.
2.4.2. LXRs
Liver X receptors (LXRs), including LXRα and LXRβ, are nuclear receptors with pivotal roles in the transcriptional control of lipid metabolism. LXRs regulate FA metabolism by controlling SREBP1c, and LXRs also participate in cholesterol absorption, transport, efflux, and excretion by binding to and regulating the expression of relevant genes [237]. Besides, LXRs have important effects on the metabolism of phospholipids [238]. LXRs play a divergent role in tumorigenesis, functioning as either tumor‐suppressors or tumor‐promoter dependent on the cell types and animal species [239, 240]. Thus, the roles of LXRs deserve further elaboration in PC. A study analyzed transcriptomic data of PC clinical samples and found overexpression of LXRs in tumors as compared to normal tissue controls [241]. In contrast, another research showed that the expression of LXRs and sterol response element‐binding factor‐1 (SREBF1), which controls the transcription of a key DNA repair gene polynucleotide kinase/phosphatase (PNKP), was significantly reduced in the tumor tissues from human PC patients compared with the adjacent normal tissues, contributing to the DNA repair deficiency in cancer by the newly identified LXR‐SREBF1‐PNKP signaling pathway [242]. Therefore, keeping LXRs in a dynamic balance seems capable of preventing cancer cell proliferation.
3. THERAPEUTICALLY EXPLORING LIPID METABOLISM IN PC TREATMENT
It has now been increasingly accepted that the integrated and mutual regulation between oncogenic signaling and lipid metabolism promotes cancer cell growth, invasion, and metastasis. Thus, developing therapeutic drugs to intervene in lipid metabolism at different levels is an emerging strategy against cancer (Figure 4). Here, we discussed several drugs or inhibitors targeting lipid metabolism not only in PC but also in other tumor types potentially relevant for future PC treatment (Table 1).
FIGURE 4.
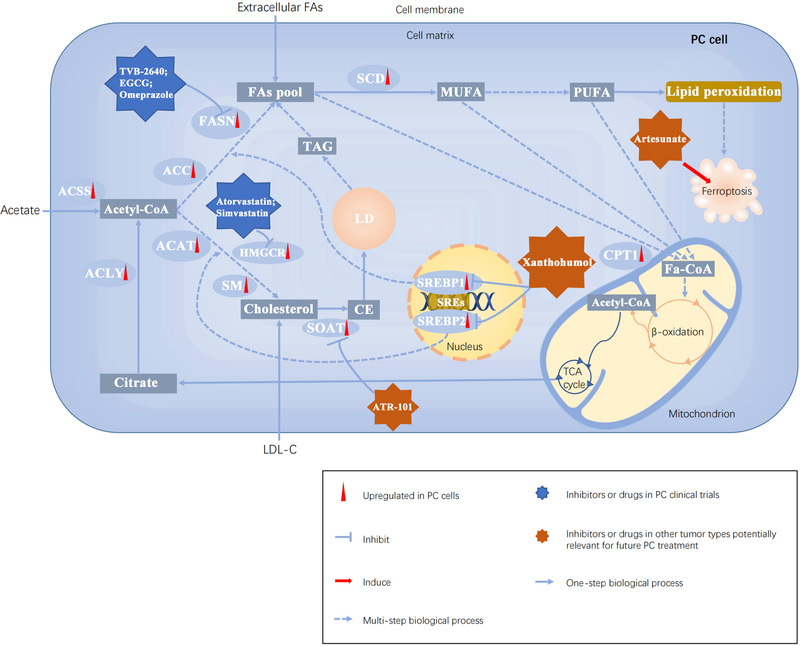
The landscape of lipid metabolism in PC cells and its potential targets in PC treatment. Abbreviations: ACLY, ATP‐citrate lyase; ACSS, acyl‐CoA synthetase short‐chain family; FAs, fatty acids; ACC, Acetyl‐CoA carboxylase; FASN, fatty acid synthase; SCD, stearoyl‐CoA desaturase; MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids; ACAT, Acetyl‐CoA acetyltransferase; HMGCR, 3‐hydroxy‐3‐methyl‐glutaryl coenzyme A reductase; SM, Squalene monooxygenase; SOAT, Sterol‐o‐acyltransferase; CE, cholesterol ester; LD, lipid droplet; LDL‐C, low‐density lipoprotein cholesterol; SREs, sterol regulatory elements; SREBP1/2, sterol regulatory element (SRE)‐binding protein 1/2; TCA, tricarboxylic acid; TAG, triacylglycerol; CPT1, carnitine palmitoyltransferase 1; EGCG, epigallocatechin gallate
TABLE 1.
Representative completed and ongoing clinical trials with inhibitors or drugs modulating lipid metabolism
| NCT number | Main target | Inhibitors/drugs | Tumor types | Stage of clinical trial | Addendum |
|---|---|---|---|---|---|
| NCT02223247 | FASN | TVB‐2640 | Solid tumors | Phase Ⅰ | Combination treatment |
| NCT02891538 | FASN | EGCG | Colon cancer | Early phase Ⅰ | Monotherapy |
| NCT02336087 | FASN | EGCG | Pancreatic cancer | Phase Ⅰ | Combination treatment |
| NCT02580279 | FASN | EGCG | Breast neoplasms | Phase Ⅱ | Combination treatment |
| NCT01340599 | FASN | EGCG | Prostate cancer | Phase III | Combination treatment |
| NCT00666562 | FASN | EGCG | Bladder cancer | Phase Ⅱ | Monotherapy |
| NCT01317953 | FASN | EGCG | Lung carcinoma | Phase Ⅰ | Monotherapy |
| NCT01116336 | FASN | EGCG | Neoplasms of head and neck | Phase Ⅰ | Combination treatment |
| NCT00303823 | FASN | EGCG | Cervical cancer | Phase Ⅱ | Combination treatment |
| NCT04930991 | FASN | Omeprazole | Pancreatic cancer | Early phase Ⅰ | Monotherapy |
| NCT04862260 | HMGCR | Atorvastatin | Pancreatic cancer | Early phase Ⅰ | Combination treatment |
| NCT02201381 | HMGCR | Atorvastatin | Pancreatic cancer and other cancers | Phase III | Combination treatment |
| NCT00944463 | HMGCR | Simvastatin | Pancreatic cancer | Phase Ⅱ | Combination treatment |
| NCT03889795 | HMGCR | Simvastatin | Pancreatic cancer | Phase Ⅰ | Combination treatment |
| NCT00416403 | HMGCR | Fluvastatin | Breast neoplasms | Phase Ⅱ | Monotherapy |
| NCT01992042 | HMGCR | Fluvastatin | Prostate cancer | Phase Ⅱ | Monotherapy |
| NCT02115074 | HMGCR | Fluvastatin | Glioma | Phase Ⅰ | Combination treatment |
| NCT00584012 | HMGCR | Lovastatin | Solid tumors | Phase Ⅰ | Combination treatment |
| NCT01898715 | SOAT1 | ATR‐101 | Adrenocortical carcinoma | Phase Ⅰ | Monotherapy |
| NCT02432651 | SREBPs | Xanthohumol | Oxidative DNA damage | Phase Ⅰ | Monotherapy |
| NCT02353026 | Ferroptosis | Artesunate | Solid tumors | Phase Ⅰ | Monotherapy |
| NCT02304289 | Ferroptosis | Artesunate | Hepatocellular carcinoma | Phase Ⅰ | Monotherapy |
| NCT02633098 | Ferroptosis | Artesunate | Colorectal cancer | Phase Ⅱ | Combination treatment |
Abbreviations: FASN, fatty acid synthase; EGCG, epigallocatechin gallate; HMGCR, 3‐hydroxy‐3‐methyl‐glutaryl coenzyme A reductase; SOAT1, Sterol‐o‐acyltransferase 1; SREBPs, sterol regulatory element (SRE)‐binding proteins.
3.1. Targeting FA synthesis
Several lines of evidence have suggested that targeting FA synthesis might be effective in treating PC. A preclinical study showed that intraperitoneally administered SB‐204990, an ACLY inhibitor, reduces tumor xerograph development in mice [243]. For inhibiting ACC, BAY ACC002 blocked tumor growth and converted the poorly differentiated histological phenotype to epithelial phenotype in multiple cell line‐based and patient‐derived PC xenograft models [104]. Besides, targeting FASN can be performed by several different inhibitors since FASN is a multi‐enzyme protein complex. Epigallocatechin‐3 gallate (EGCG) can inhibit the growth of pancreatic tumors orthotopically implanted in mice by blocking the β‐ketoacyl‐ACP synthase domain of FASN [244]. Orlistat blocked the thioesterase‐domain of FASN, and it has been shown that orlistat reduced human PC cell proliferation [112]. Beyond that, lansoprazole, rabeprazole, omeprazole, and pantoprazole, which are proton pump inhibitors, can induce PC cell death by suppressing thioesterase activity [245]. Moreover, targeting SCD has also been considered an anti‐cancer strategy, in which SCD inhibitor A939572 induced cell death in early pancreatic tumors [130]. In terms of clinical trials, although targeting FASN, which is overexpressed by many solid and hematopoietic tumors, including non‐small cell lung, breast, ovarian, prostate, colon, pancreatic cancers, and lymphoma, arouses the interest of most investigators, there is no direct and solid evidence to prove that inhibiting FASN is effective and safe for patients with PC (both NCT02336087 and NCT04930991 are recruiting). Nevertheless, we may draw inspiration from another completed clinical trial (NCT02223247) [246], demonstrating that responses to TVB‐2640 in combination with paclitaxel were seen across multiple tumor types, including in patients with KRASMUT NSCLC, ovarian, and breast cancer, and the primary adverse events are non‐serious and reversible like skin and subcutaneous tissue disorders [246]. Therefore, the efficacy and safety of targeting FASN in PC patients are promising and need to be warranted urgently.
3.2. Targeting cholesterol synthesis
Targeting cholesterol synthesis is considered an emerging treatment, especially by blockage of the rate‐limiting enzyme HMGCR. According to in vitro studies, statins exert anti‐cancer activities against PC cells resistant to gemcitabine [151]. Additionally, several meta‐analyses suggested that the improved survival of PC patients was associated with statin use [247, 248]. Furthermore, a pooled analysis of two‐phase III studies showed that statin use was associated with better overall survival among patients with metastatic PC treated with first‐line chemotherapy [249]. However, according to another randomized controlled trial (NCT00944463), adding low‐dose simvastatin to gemcitabine did not provide clinical benefits in patients histologically or cytologically confirmed with metastatic or unresectable pancreatic adenocarcinoma [250]. Given sophisticated pharmacologic effects and the complexity of tumor molecular biology, a scrap of clinical findings does not conclusively exclude the use of simvastatin for advanced PC. For instance, inhibiting cholesterol synthesis endogenously by HMGCR inhibitors may cause an increased rate of cholesterol uptake so that cancer cells can remain alive and proliferate. This hypothesis needs to be confirmed by further clinical trials combining anti‐HMGCR drugs like atorvastatin, simvastatin, and fluvastatin combined with exogenous inhibitors of cholesterol uptake.
3.3. Targeting lipid catabolism
FAs are important energy resources through FAO, which are required for cancer cell growth and survival. Accordingly, FAO is considered a potential target for cancer therapy. A recent study found that inhibiting FAO with etomoxir, which acts as a potent inhibitor of CPT1, effectively suppressed low pH‐induced invasion in PC [197]. Besides, the knockdown of CPT1C inhibited the growth and tumorigenesis of PC [198]. Regarding the role of cell death related to lipid catabolism, targeting ferroptosis may be another attractive treatment for cancer. Great efforts have been made to develop therapeutic drugs to induce ferroptosis in PC cells [251]. Particularly, the antimalarial drug artesunate and the antiviral drug zalcitabine, which are two clinically used drugs, have been demonstrated to effectively suppress PC cells as ferroptosis inducers [252, 253]. Further, the combined application of gemcitabine and ferroptotic inducers could effectively attenuate gemcitabine resistance in vitro [129]. Additionally, targeting LDs is also a new strategy for treating PC by inhibiting cholesterol esterification. For example, avasimin, a potent SOAT1 inhibitor, caused apoptosis and suppression of proliferation in PC cell lines [179]. For now, the safety of SOAT1 inhibitor ATR‐101 has been demonstrated in a phase I study (NCT01898715), but the current formulation of ATR‐101 had limited efficacy in patients with advanced adrenocortical carcinoma [254]. There is a lack of clinical trial findings in PC patients.
3.4. Targeting transcriptional regulators of lipid metabolism
The transcription of genes required for FA and cholesterol synthesis is controlled by membrane‐bound transcription factor SREBPs, which are potential targets for cancer therapy. Betulin and fatostatin have been proposed as SREBP inhibitors. For example, the combined application of betulinic acid and mithramycin A effectively suppressed the development of PC xenografts in mice [255]. Besides, fatostatin has been reported to possess high antitumor activity against PC [256]. In addition to these, recent research has found that Yarrow CO2 supercritical extract (Yarrow SFE) diminished the tumor growth in a xenograft mouse model of PC, which suggests that Yarrow SFE could be a complementary adjuvant or nutritional supplement in PC therapy [257]. LXRs are nuclear receptors that regulate the transcription of genes involved in lipid metabolism. Targeting LXRs with the novel LXR inverse agonist and degrader (GAC0001E5) inhibited PC cell proliferation, supporting their potential application as treatments for advanced PC and other recalcitrant malignancies [258].
4. CONCLUSIONS AND PERSPECTIVES
It is now widely appreciated that cancer cells display significant rewiring in their lipid metabolism. Malignant cells enhance lipid metabolism to satisfy their demand for proliferation and progression under a nutrient‐ and oxygen‐scarce microenvironment. Various alterations in FA and cholesterol uptake have been reported and can contribute to cancer cells' aggressiveness. Furthermore, elevated expression of the key enzymes in lipogenic synthesis is closely related to the malignant biological behaviors of hematological and solid tumors. Aberrant lipid catabolism, such as high FAO activity, enhanced lipid peroxidation and abnormal cholesterol efflux, also appears to play an essential role in tumor growth and poor prognosis. Thus, the complexity of the lipid metabolism is embodied in functioning as a network of pathways with flexibility, feedback loops, and crosstalk. FA and cholesterol intake, synthesis, and degradation, as well as the involved key proteins and transcriptional regulators, constitute the core of this framework. Accordingly, most drugs or inhibitors in preclinical cancer models or clinical trials also target lipid metabolism. In addition, targeted drugs combined with classic chemotherapeutic agents have shown some promising results, especially in treating intractable PC. However, some results from completed clinical trials in PC or other solid tumors are not encouraging. Give an illustrative instance, solely inhibiting FA or cholesterol de novo synthesis does not affect cancer cells remarkably, which is probably associated with upregulated feedback of exogenous lipid uptake. Therefore, further clinical studies on the combination of exogenous lipid uptake inhibition and de novo synthesis might be promising. It is also worth noting that lipid homeostasis in cancer cells is controlled by lipid anabolism and catabolism. Compared with lipid anabolism, the roles of lipid catabolism and its related factors remain relatively unclear in PC, while reprogramming lipid catabolism is crucial in supporting cancer cell proliferation and migration. Importantly, focusing on the specific molecular mechanism of lipid catabolism and drugs targeting catabolic processes in PC may yield promising results.
In summary, lipid metabolism‐targeted therapy may be a novel and potentially effective strategy for PC patients. Thus, further understanding of the lipid metabolic network is required.
AUTHOR CONTRIBUTIONS
All authors contributed to the writing and editing of this manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
The authors declare no potential conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
All authors agree to publish the article.
ACKNOWLEDGMENTS
Not applicable.
Yin X, Xu R, Song J, Ruze R, Chen Y, Wang C, et al. Lipid metabolism in pancreatic cancer: emerging roles and potential targets. Cancer Commun. 2022;42:1234–1256. 10.1002/cac2.12360
Xinpeng Yin, Ruiyuan Xu and Jianlu Song contributed equally to this work.
Contributor Information
Xinpeng Yin, Email: yinxinpeng0222@163.com.
Ruiyuan Xu, Email: xry970124@163.com.
Jianlu Song, Email: SongJianlu2017@hotmail.com.
Rexiati Ruze, Email: rishatruzi@hotmail.com.
Yuan Chen, Email: chenyuan19961023@hotmail.com.
Chengcheng Wang, Email: wangchengcheng3638@163.com.
Qiang Xu, Email: xuqiang@pumch.cn.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐32. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209‐49. [DOI] [PubMed] [Google Scholar]
- 4. Wei W, Zeng H, Zheng R, Zhang S, An L, Chen R, et al. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol. 2020;21(7):e342‐e9. [DOI] [PubMed] [Google Scholar]
- 5. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913‐21. [DOI] [PubMed] [Google Scholar]
- 6. Park W, Chawla A, O'Reilly EM. Pancreatic Cancer: A Review. JAMA. 2021;326(9):851‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang L, Jansen L, Balavarca Y, Molina‐Montes E, Babaei M, van der Geest L, et al. Resection of pancreatic cancer in Europe and USA: an international large‐scale study highlighting large variations. Gut. 2019;68(1):130‐9. [DOI] [PubMed] [Google Scholar]
- 8. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73‐85. [DOI] [PubMed] [Google Scholar]
- 9. Yang J, Xu R, Wang C, Qiu J, Ren B, You L. Early screening and diagnosis strategies of pancreatic cancer: a comprehensive review. Cancer Commun (Lond). 2021;41(12):1257‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brunner M, Wu Z, Krautz C, Pilarsky C, Grutzmann R, Weber GF. Current Clinical Strategies of Pancreatic Cancer Treatment and Open Molecular Questions. Int J Mol Sci. 2019;20(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: Treating cancer with specificity. Eur J Pharmacol. 2018;834:188‐96. [DOI] [PubMed] [Google Scholar]
- 12. Qian Y, Gong Y, Fan Z, Luo G, Huang Q, Deng S, et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2020;13(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168(4):657‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Snaebjornsson MT, Janaki‐Raman S, Schulze A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020;31(1):62‐76. [DOI] [PubMed] [Google Scholar]
- 15. Bader JE, Voss K, Rathmell JC. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol Cell. 2020;78(6):1019‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao Y. Adipocyte and lipid metabolism in cancer drug resistance. J Clin Invest. 2019;129(8):3006‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang WH, Qiu Y, Stamatatos O, Janowitz T, Lukey MJ. Enhancing the Efficacy of Glutamine Metabolism Inhibitors in Cancer Therapy. Trends Cancer. 2021;7(8):790‐804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond). 2018;38(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bacci M, Lorito N, Smiriglia A, Morandi A. Fat and Furious: Lipid Metabolism in Antitumoral Therapy Response and Resistance. Trends Cancer. 2021;7(3):198‐213. [DOI] [PubMed] [Google Scholar]
- 20. Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017;16(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Efeyan A, Comb WC, Sabatini DM. Nutrient‐sensing mechanisms and pathways. Nature. 2015;517(7534):302‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 2015;161(1):161‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poggiogalle E, Jamshed H, Peterson CM. Circadian regulation of glucose, lipid, and energy metabolism in humans. Metabolism. 2018;84:11‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Broadfield LA, Pane AA, Talebi A, Swinnen JV, Fendt SM. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev Cell. 2021;56(10):1363‐93. [DOI] [PubMed] [Google Scholar]
- 25. Byrne A, Savas P, Sant S, Li R, Virassamy B, Luen SJ, et al. Tissue‐resident memory T cells in breast cancer control and immunotherapy responses. Nat Rev Clin Oncol. 2020;17(6):341‐8. [DOI] [PubMed] [Google Scholar]
- 26. Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, et al. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev. 2020;159:245‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomaraschi M. Role of Lipoproteins in the Microenvironment of Hormone‐Dependent Cancers. Trends Endocrinol Metab. 2020;31(3):256‐68. [DOI] [PubMed] [Google Scholar]
- 28. Filali‐Mouncef Y, Hunter C, Roccio F, Zagkou S, Dupont N, Primard C, et al. The menage a trois of autophagy, lipid droplets and liver disease. Autophagy. 2022;18(1):50‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Satriano L, Lewinska M, Rodrigues PM, Banales JM, Andersen JB. Metabolic rearrangements in primary liver cancers: cause and consequences. Nat Rev Gastroenterol Hepatol. 2019;16(12):748‐66. [DOI] [PubMed] [Google Scholar]
- 30. Garcia‐Estevez L, Moreno‐Bueno G. Updating the role of obesity and cholesterol in breast cancer. Breast Cancer Res. 2019;21(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med. 2021;218(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsushita Y, Nakagawa H, Koike K. Lipid Metabolism in Oncology: Why It Matters, How to Research, and How to Treat. Cancers (Basel). 2021;13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J, Li Y. CD36 tango in cancer: signaling pathways and functions. Theranostics. 2019;9(17):4893‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, et al. Targeting metastasis‐initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41‐5. [DOI] [PubMed] [Google Scholar]
- 35. Yu X, Guo C, Fisher PB, Subjeck JR, Wang XY. Scavenger Receptors: Emerging Roles in Cancer Biology and Immunology. Adv Cancer Res. 2015;128:309‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanase C, Gheorghisan‐Galateanu AA, Popescu ID, Mihai S, Codrici E, Albulescu R, et al. CD36 and CD97 in Pancreatic Cancer versus Other Malignancies. Int J Mol Sci. 2020;21(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nath A, Li I, Roberts LR, Chan C. Elevated free fatty acid uptake via CD36 promotes epithelial‐mesenchymal transition in hepatocellular carcinoma. Sci Rep. 2015;5:14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia S, Zhou L, Shen T, Zhou S, Ding G, Cao L. Down‐expression of CD36 in pancreatic adenocarcinoma and its correlation with clinicopathological features and prognosis. J Cancer. 2018;9(3):578‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Z, Kang Y. Lipid Metabolism Fuels Cancer's Spread. Cell Metab. 2017;25(2):228‐30. [DOI] [PubMed] [Google Scholar]
- 40. Kubo M, Gotoh K, Eguchi H, Kobayashi S, Iwagami Y, Tomimaru Y, et al. Impact of CD36 on Chemoresistance in Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2020;27(2):610‐9. [DOI] [PubMed] [Google Scholar]
- 41. Soung YH, Ford S, Zhang V, Chung J. Exosomes in Cancer Diagnostics. Cancers (Basel). 2017;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pellerin L, Carrie L, Dufau C, Nieto L, Segui B, Levade T, et al. Lipid metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives. Cancers (Basel). 2020;12(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang B, Song BL, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab. 2020;2(2):132‐41. [DOI] [PubMed] [Google Scholar]
- 44. Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer. 2020;1874(1):188394. [DOI] [PubMed] [Google Scholar]
- 45. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21(4):225‐45. [DOI] [PubMed] [Google Scholar]
- 46. Alberts A, Klingberg A, Hoffmeister L, Wessig AK, Brand K, Pich A, et al. Binding of Macrophage Receptor MARCO, LDL, and LDLR to Disease‐Associated Crystalline Structures. Front Immunol. 2020;11:596103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kho PF, Amant F, Annibali D, Ashton K, Attia J, Auer PL, et al. Mendelian randomization analyses suggest a role for cholesterol in the development of endometrial cancer. Int J Cancer. 2021;148(2):307‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cornish AJ, Law PJ, Timofeeva M, Palin K, Farrington SM, Palles C, et al. Modifiable pathways for colorectal cancer: a mendelian randomisation analysis. Lancet Gastroenterol Hepatol. 2020;5(1):55‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gallagher EJ, Zelenko Z, Neel BA, Antoniou IM, Rajan L, Kase N, et al. Elevated tumor LDLR expression accelerates LDL cholesterol‐mediated breast cancer growth in mouse models of hyperlipidemia. Oncogene. 2017;36(46):6462‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou T, Zhan J, Fang W, Zhao Y, Yang Y, Hou X, et al. Serum low‐density lipoprotein and low‐density lipoprotein expression level at diagnosis are favorable prognostic factors in patients with small‐cell lung cancer (SCLC). BMC Cancer. 2017;17(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sah RP, Sharma A, Nagpal S, Patlolla SH, Sharma A, Kandlakunta H, et al. Phases of Metabolic and Soft Tissue Changes in Months Preceding a Diagnosis of Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2019;156(6):1742‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Revilla G, Cedo L, Tondo M, Moral A, Perez JI, Corcoy R, et al. LDL, HDL and endocrine‐related cancer: From pathogenic mechanisms to therapies. Semin Cancer Biol. 2021;73:134‐57. [DOI] [PubMed] [Google Scholar]
- 53. Jung YY, Ko JH, Um JY, Chinnathambi A, Alharbi SA, Sethi G, et al. LDL cholesterol promotes the proliferation of prostate and pancreatic cancer cells by activating the STAT3 pathway. J Cell Physiol. 2021;236(7):5253‐64. [DOI] [PubMed] [Google Scholar]
- 54. Acier A, Godard M, Gassiot F, Finetti P, Rubis M, Nowak J, et al. LDL receptor‐peptide conjugate as in vivo tool for specific targeting of pancreatic ductal adenocarcinoma. Commun Biol. 2021;4(1):987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gonias SL, Karimi‐Mostowfi N, Murray SS, Mantuano E, Gilder AS. Expression of LDL receptor‐related proteins (LRPs) in common solid malignancies correlates with patient survival. PLoS One. 2017;12(10):e0186649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu Y, Gentiluomo M, Lorenzo‐Bermejo J, Morelli L, Obazee O, Campa D, et al. Mendelian randomisation study of the effects of known and putative risk factors on pancreatic cancer. J Med Genet. 2020;57(12):820‐8. [DOI] [PubMed] [Google Scholar]
- 57. Guillaumond F, Bidaut G, Ouaissi M, Servais S, Gouirand V, Olivares O, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2015;112(8):2473‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16(11):732‐49. [DOI] [PubMed] [Google Scholar]
- 59. Corn KC, Windham MA, Rafat M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog Lipid Res. 2020;80:101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763‐77. [DOI] [PubMed] [Google Scholar]
- 61. Guri Y, Colombi M, Dazert E, Hindupur SK, Roszik J, Moes S, et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell. 2017;32(6):807‐23 e12. [DOI] [PubMed] [Google Scholar]
- 62. Munir R, Lisec J, Swinnen JV, Zaidi N. Lipid metabolism in cancer cells under metabolic stress. Br J Cancer. 2019;120(12):1090‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Feng X, Zhang L, Xu S, Shen AZ. ATP‐citrate lyase (ACLY) in lipid metabolism and atherosclerosis: An updated review. Prog Lipid Res. 2020;77:101006. [DOI] [PubMed] [Google Scholar]
- 64. Granchi C. ATP citrate lyase (ACLY) inhibitors: An anti‐cancer strategy at the crossroads of glucose and lipid metabolism. Eur J Med Chem. 2018;157:1276‐91. [DOI] [PubMed] [Google Scholar]
- 65. Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, et al. Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature. 2019;568(7753):571‐5. [DOI] [PubMed] [Google Scholar]
- 66. Masisi BK, El Ansari R, Alfarsi L, Rakha EA, Green AR, Craze ML. The role of glutaminase in cancer. Histopathology. 2020;76(4):498‐508. [DOI] [PubMed] [Google Scholar]
- 67. Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl‐CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27(1):57‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP‐citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bempedoic Acid to Lower LDL Cholesterol — Safety and Efficacy. New England Journal of Medicine. 2020;383(7). [DOI] [PubMed] [Google Scholar]
- 70. Khwairakpam AD, Banik K, Girisa S, Shabnam B, Shakibaei M, Fan L, et al. The vital role of ATP citrate lyase in chronic diseases. J Mol Med (Berl). 2020;98(1):71‐95. [DOI] [PubMed] [Google Scholar]
- 71. Wen J, Min X, Shen M, Hua Q, Han Y, Zhao L, et al. ACLY facilitates colon cancer cell metastasis by CTNNB1. J Exp Clin Cancer Res. 2019;38(1):401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kumari R, Deshmukh RS, Das S. Caspase‐10 inhibits ATP‐citrate lyase‐mediated metabolic and epigenetic reprogramming to suppress tumorigenesis. Nat Commun. 2019;10(1):4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gu L, Zhu Y, Lin X, Lu B, Zhou X, Zhou F, et al. The IKKbeta‐USP30‐ACLY Axis Controls Lipogenesis and Tumorigenesis. Hepatology. 2021;73(1):160‐74. [DOI] [PubMed] [Google Scholar]
- 74. Guo W, Ma J, Yang Y, Guo S, Zhang W, Zhao T, et al. ATP‐Citrate Lyase Epigenetically Potentiates Oxidative Phosphorylation to Promote Melanoma Growth and Adaptive Resistance to MAPK Inhibition. Clin Cancer Res. 2020;26(11):2725‐39. [DOI] [PubMed] [Google Scholar]
- 75. Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, et al. Acetyl‐CoA Metabolism Supports Multistep Pancreatic Tumorigenesis. Cancer Discov. 2019;9(3):416‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu J, Luo X, Guo R, Jing W, Lu H. Cell Metabolomics Reveals Berberine‐Inhibited Pancreatic Cancer Cell Viability and Metastasis by Regulating Citrate Metabolism. J Proteome Res. 2020;19(9):3825‐36. [DOI] [PubMed] [Google Scholar]
- 77. Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, et al. Akt‐dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20(2):306‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee JH, et al. Nucleus‐Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol Cell. 2017;66(5):684‐97 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li Z, Liu H, He J, Wang Z, Yin Z, You G, et al. Acetyl‐CoA Synthetase 2: A Critical Linkage in Obesity‐Induced Tumorigenesis in Myeloma. Cell Metab. 2021;33(1):78‐93 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lakhter AJ, Hamilton J, Konger RL, Brustovetsky N, Broxmeyer HE, Naidu SR. Glucose‐independent Acetate Metabolism Promotes Melanoma Cell Survival and Tumor Growth. J Biol Chem. 2016;291(42):21869‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yun M, Bang SH, Kim JW, Park JY, Kim KS, Lee JD. The importance of acetyl coenzyme A synthetase for 11C‐acetate uptake and cell survival in hepatocellular carcinoma. J Nucl Med. 2009;50(8):1222‐8. [DOI] [PubMed] [Google Scholar]
- 82. Chang WC, Cheng WC, Cheng BH, Chen L, Ju LJ, Ou YJ, et al. Mitochondrial Acetyl‐CoA Synthetase 3 is Biosignature of Gastric Cancer Progression. Cancer Med. 2018;7(4):1240‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bidkhori G, Benfeitas R, Klevstig M, Zhang C, Nielsen J, Uhlen M, et al. Metabolic network‐based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc Natl Acad Sci U S A. 2018;115(50):E11874‐E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang J, Duan H, Feng Z, Han X, Gu C. Acetyl‐CoA synthetase 3 promotes bladder cancer cell growth under metabolic stress. Oncogenesis. 2020;9(5):46. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85. Miller KD, Pniewski K, Perry CE, Papp SB, Shaffer JD, Velasco‐Silva JN, et al. Targeting ACSS2 with a Transition‐State Mimetic Inhibits Triple‐Negative Breast Cancer Growth. Cancer Res. 2021;81(5):1252‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang X, Shao F, Shi S, Feng X, Wang W, Wang Y, et al. Prognostic Impact of Metabolism Reprogramming Markers Acetyl‐CoA Synthetase 2 Phosphorylation and Ketohexokinase‐A Expression in Non‐Small‐Cell Lung Carcinoma. Front Oncol. 2019;9:1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu Y, Wan W. Acetylation in the regulation of autophagy. Autophagy. 2022:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li X, Qian X, Lu Z. Local histone acetylation by ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Autophagy. 2017;13(10):1790‐1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moffett JR, Puthillathu N, Vengilote R, Jaworski DM, Namboodiri AM. Acetate Revisited: A Key Biomolecule at the Nexus of Metabolism, Epigenetics and Oncogenesis‐Part 1: Acetyl‐CoA, Acetogenesis and Acyl‐CoA Short‐Chain Synthetases. Front Physiol. 2020;11:580167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Moffett JR, Puthillathu N, Vengilote R, Jaworski DM, Namboodiri AM. Acetate Revisited: A Key Biomolecule at the Nexus of Metabolism, Epigenetics, and Oncogenesis ‐ Part 2: Acetate and ACSS2 in Health and Disease. Front Physiol. 2020;11:580171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu H, Luo J, Ma G, Zhang X, Yao D, Li M, et al. Acyl‐CoA synthetase short‐chain family member 2 (ACSS2) is regulated by SREBP‐1 and plays a role in fatty acid synthesis in caprine mammary epithelial cells. J Cell Physiol. 2018;233(2):1005‐16. [DOI] [PubMed] [Google Scholar]
- 92. Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, et al. Acetate Recapturing by Nuclear Acetyl‐CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017;18(3):647‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, et al. Structural basis for regulation of human acetyl‐CoA carboxylase. Nature. 2018;558(7710):470‐4. [DOI] [PubMed] [Google Scholar]
- 94. Rios Garcia M, Steinbauer B, Srivastava K, Singhal M, Mattijssen F, Maida A, et al. Acetyl‐CoA Carboxylase 1‐Dependent Protein Acetylation Controls Breast Cancer Metastasis and Recurrence. Cell Metab. 2017;26(6):842‐55 e5. [DOI] [PubMed] [Google Scholar]
- 95. Chen L, Duan Y, Wei H, Ning H, Bi C, Zhao Y, et al. Acetyl‐CoA carboxylase (ACC) as a therapeutic target for metabolic syndrome and recent developments in ACC1/2 inhibitors. Expert Opin Investig Drugs. 2019;28(10):917‐30. [DOI] [PubMed] [Google Scholar]
- 96. Ueda K, Nakatsu Y, Yamamotoya T, Ono H, Inoue Y, Inoue MK, et al. Prolyl isomerase Pin1 binds to and stabilizes acetyl CoA carboxylase 1 protein, thereby supporting cancer cell proliferation. Oncotarget. 2019;10(17):1637‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Penfold L, Woods A, Muckett P, Nikitin AY, Kent TR, Zhang S, et al. CAMKK2 Promotes Prostate Cancer Independently of AMPK via Increased Lipogenesis. Cancer Res. 2018;78(24):6747‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lally JSV, Ghoshal S, DePeralta DK, Moaven O, Wei L, Masia R, et al. Inhibition of Acetyl‐CoA Carboxylase by Phosphorylation or the Inhibitor ND‐654 Suppresses Lipogenesis and Hepatocellular Carcinoma. Cell Metab. 2019;29(1):174‐82 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fang W, Cui H, Yu D, Chen Y, Wang J, Yu G. Increased expression of phospho‐acetyl‐CoA carboxylase protein is an independent prognostic factor for human gastric cancer without lymph node metastasis. Med Oncol. 2014;31(7):15. [DOI] [PubMed] [Google Scholar]
- 100. Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, et al. Inhibition of acetyl‐CoA carboxylase suppresses fatty acid synthesis and tumor growth of non‐small‐cell lung cancer in preclinical models. Nat Med. 2016;22(10):1108‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang Y, Yu W, Li S, Guo D, He J, Wang Y. Acetyl‐CoA Carboxylases and Diseases. Front Oncol. 2022;12:836058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Li K, Zhang C, Chen L, Wang P, Fang Y, Zhu J, et al. The role of acetyl‐coA carboxylase2 in head and neck squamous cell carcinoma. PeerJ. 2019;7:e7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Vernieri C, Pusceddu S, Fuca G, Indelicato P, Centonze G, Castagnoli L, et al. Impact of systemic and tumor lipid metabolism on everolimus efficacy in advanced pancreatic neuroendocrine tumors (pNETs). Int J Cancer. 2019;144(7):1704‐12. [DOI] [PubMed] [Google Scholar]
- 104. Petrova E, Scholz A, Paul J, Sturz A, Haike K, Siegel F, et al. Acetyl‐CoA carboxylase inhibitors attenuate WNT and Hedgehog signaling and suppress pancreatic tumor growth. Oncotarget. 2017;8(30):48660‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gao L, Xu Z, Huang Z, Tang Y, Yang D, Huang J, et al. CPI‐613 rewires lipid metabolism to enhance pancreatic cancer apoptosis via the AMPK‐ACC signaling. J Exp Clin Cancer Res. 2020;39(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gu S, Song X, Xie R, Ouyang C, Xie L, Li Q, et al. Berberine inhibits cancer cells growth by suppressing fatty acid synthesis and biogenesis of extracellular vesicles. Life Sci. 2020;257:118122. [DOI] [PubMed] [Google Scholar]
- 107. Nishi K, Suzuki M, Yamamoto N, Matsumoto A, Iwase Y, Yamasaki K, et al. Glutamine Deprivation Enhances Acetyl‐CoA Carboxylase Inhibitor‐induced Death of Human Pancreatic Cancer Cells. Anticancer Res. 2018;38(12):6683‐9. [DOI] [PubMed] [Google Scholar]
- 108. Bruning U, Morales‐Rodriguez F, Kalucka J, Goveia J, Taverna F, Queiroz KCS, et al. Impairment of Angiogenesis by Fatty Acid Synthase Inhibition Involves mTOR Malonylation. Cell Metabolism. 2018;28(6):866‐80.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Buckley D, Duke G, Heuer TS, O'Farrell M, Wagman AS, McCulloch W, et al. Fatty acid synthase ‐ Modern tumor cell biology insights into a classical oncology target. Pharmacol Ther. 2017;177:23‐31. [DOI] [PubMed] [Google Scholar]
- 110. Walter K, Hong SM, Nyhan S, Canto M, Fedarko N, Klein A, et al. Serum fatty acid synthase as a marker of pancreatic neoplasia. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2380‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Brandi J, Dando I, Pozza ED, Biondani G, Jenkins R, Elliott V, et al. Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. J Proteomics. 2017;150:310‐22. [DOI] [PubMed] [Google Scholar]
- 112. Sokolowska E, Presler M, Goyke E, Milczarek R, Swierczynski J, Sledzinski T. Orlistat Reduces Proliferation and Enhances Apoptosis in Human Pancreatic Cancer Cells (PANC‐1). Anticancer Res. 2017;37(11):6321‐7. [DOI] [PubMed] [Google Scholar]
- 113. Saleh A, ElFayoumi HM, Youns M, Barakat W. Rutin and orlistat produce antitumor effects via antioxidant and apoptotic actions. Naunyn Schmiedebergs Arch Pharmacol. 2019;392(2):165‐75. [DOI] [PubMed] [Google Scholar]
- 114. Tadros S, Shukla SK, King RJ, Gunda V, Vernucci E, Abrego J, et al. De Novo Lipid Synthesis Facilitates Gemcitabine Resistance through Endoplasmic Reticulum Stress in Pancreatic Cancer. Cancer Res. 2017;77(20):5503‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tian S, Li P, Sheng S, Jin X. Upregulation of pyruvate kinase M2 expression by fatty acid synthase contributes to gemcitabine resistance in pancreatic cancer. Oncol Lett. 2018;15(2):2211‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shetty A, Nagesh PKB, Setua S, Hafeez BB, Jaggi M, Yallapu MM, et al. Novel Paclitaxel Nanoformulation Impairs De Novo Lipid Synthesis in Pancreatic Cancer Cells and Enhances Gemcitabine Efficacy. ACS Omega. 2020;5(15):8982‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wu X, Dong Z, Wang CJ, Barlow LJ, Fako V, Serrano MA, et al. FASN regulates cellular response to genotoxic treatments by increasing PARP‐1 expression and DNA repair activity via NF‐kappaB and SP1. Proc Natl Acad Sci U S A. 2016;113(45):E6965‐E73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Rencuzogullari O, Yerlikaya PO, Gurkan AC, Arisan ED, Telci D. Palbociclib negatively regulates fatty acid synthesis due to upregulation of AMPKalpha and miR‐33a levels to increase apoptosis in Panc‐1 and MiaPaCa‐2 cells. Biotechnol Appl Biochem. 2022;69(1):342‐354. [DOI] [PubMed] [Google Scholar]
- 119. Rawson RB. The SREBP pathway–insights from Insigs and insects. Nat Rev Mol Cell Biol. 2003;4(8):631‐40. [DOI] [PubMed] [Google Scholar]
- 120. Bian Y, Yu Y, Wang S, Li L. Up‐regulation of fatty acid synthase induced by EGFR/ERK activation promotes tumor growth in pancreatic cancer. Biochem Biophys Res Commun. 2015;463(4):612‐7. [DOI] [PubMed] [Google Scholar]
- 121. Karmakar S, Kaushik G, Nimmakayala R, Rachagani S, Ponnusamy MP, Batra SK. MicroRNA regulation of K‐Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin Cancer Biol. 2019;54:63‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sun Y, He W, Luo M, Zhou Y, Chang G, Ren W, et al. SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol. 2015;36(6):4133‐41. [DOI] [PubMed] [Google Scholar]
- 123. Kubota CS, Espenshade PJ. Targeting Stearoyl‐CoA Desaturase in Solid Tumors. Cancer Res. 2022;82(9):1682‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bogie JFJ, Grajchen E, Wouters E, Corrales AG, Dierckx T, Vanherle S, et al. Stearoyl‐CoA desaturase‐1 impairs the reparative properties of macrophages and microglia in the brain. J Exp Med. 2020;217(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. AL AM, Syed DN, Ntambi JM. Insights into Stearoyl‐CoA Desaturase‐1 Regulation of Systemic Metabolism. Trends Endocrinol Metab. 2017;28(12):831‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Igal RA, Sinner DI. Stearoyl‐CoA desaturase 5 (SCD5), a Delta‐9 fatty acyl desaturase in search of a function. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866(1):158840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Macasek J, Vecka M, Zak A, Urbanek M, Krechler T, Petruzelka L, et al. Plasma fatty acid composition in patients with pancreatic cancer: correlations to clinical parameters. Nutr Cancer. 2012;64(7):946‐55. [DOI] [PubMed] [Google Scholar]
- 128. Gao J, Zhang Z, Liu Y, Zhang Z, Wang M, Gong A, et al. Stearoyl‐CoA Desaturase 1 Potentiates Hypoxic plus Nutrient‐Deprived Pancreatic Cancer Cell Ferroptosis Resistance. Oxid Med Cell Longev. 2021;2021:6629804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 129. Ye Z, Zhuo Q, Hu Q, Xu X, Mengqi L, Zhang Z, et al. FBW7‐NRA41‐SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biol. 2021;38:101807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Skrypek K, Balog S, Eriguchi Y, Asahina K. Inhibition of Stearoyl‐CoA Desaturase Induces the Unfolded Protein Response in Pancreatic Tumors and Suppresses Their Growth. Pancreas. 2021;50(2):219‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Abdelkreem E, Harijan RK, Yamaguchi S, Wierenga RK, Fukao T. Mutation update on ACAT1 variants associated with mitochondrial acetoacetyl‐CoA thiolase (T2) deficiency. Hum Mutat. 2019;40(10):1641‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Goudarzi A. The recent insights into the function of ACAT1: A possible anti‐cancer therapeutic target. Life Sci. 2019;232:116592. [DOI] [PubMed] [Google Scholar]
- 133. Garcia‐Bermudez J, Birsoy K. Drugging ACAT1 for Cancer Therapy. Mol Cell. 2016;64(5):856‐7. [DOI] [PubMed] [Google Scholar]
- 134. Fan J, Lin R, Xia S, Chen D, Elf SE, Liu S, et al. Tetrameric Acetyl‐CoA Acetyltransferase 1 Is Important for Tumor Growth. Mol Cell. 2016;64(5):859‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhao Z, Lu J, Han L, Wang X, Man Q, Liu S. Prognostic significance of two lipid metabolism enzymes, HADHA and ACAT2, in clear cell renal cell carcinoma. Tumour Biol. 2016;37(6):8121‐30. [DOI] [PubMed] [Google Scholar]
- 136. Souchek JJ, Baine MJ, Lin C, Rachagani S, Gupta S, Kaur S, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J Cancer. 2014;111(6):1139‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tao L, Zhong L, Li Y, Li D, Xiu D, Zhou J. Integrated proteomics and phosphoproteomics reveal perturbed regulative pathways in pancreatic ductal adenocarcinoma. Mol Omics. 2021;17(2):230‐40. [DOI] [PubMed] [Google Scholar]
- 138. Lu XY, Shi XJ, Hu A, Wang JQ, Ding Y, Jiang W, et al. Feeding induces cholesterol biosynthesis via the mTORC1‐USP20‐HMGCR axis. Nature. 2020;588(7838):479‐84. [DOI] [PubMed] [Google Scholar]
- 139. Ference BA, Kastelein JJP, Ginsberg HN, Chapman MJ, Nicholls SJ, Ray KK, et al. Association of Genetic Variants Related to CETP Inhibitors and Statins With Lipoprotein Levels and Cardiovascular Risk. JAMA. 2017;318(10):947‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yarmolinsky J, Bull CJ, Vincent EE, Robinson J, Walther A, Smith GD, et al. Association Between Genetically Proxied Inhibition of HMG‐CoA Reductase and Epithelial Ovarian Cancer. JAMA. 2020;323(7):646‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Mullen PJ, Yu R, Longo J, Archer MC, Penn LZ. The interplay between cell signalling and the mevalonate pathway in cancer. Nat Rev Cancer. 2016;16(11):718‐31. [DOI] [PubMed] [Google Scholar]
- 142. Mo H, Jeter R, Bachmann A, Yount ST, Shen CL, Yeganehjoo H. The Potential of Isoprenoids in Adjuvant Cancer Therapy to Reduce Adverse Effects of Statins. Front Pharmacol. 2018;9:1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Qiu Z, Yuan W, Chen T, Zhou C, Liu C, Huang Y, et al. HMGCR positively regulated the growth and migration of glioblastoma cells. Gene. 2016;576(1 Pt 1):22‐7. [DOI] [PubMed] [Google Scholar]
- 144. Ashida S, Kawada C, Inoue K. Stromal regulation of prostate cancer cell growth by mevalonate pathway enzymes HMGCS1 and HMGCR. Oncol Lett. 2017;14(6):6533‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chushi L, Wei W, Kangkang X, Yongzeng F, Ning X, Xiaolei C. HMGCR is up‐regulated in gastric cancer and promotes the growth and migration of the cancer cells. Gene. 2016;587(1):42‐7. [DOI] [PubMed] [Google Scholar]
- 146. Rosch PJ, McCully K. Statin use and reduced cancer‐related mortality. N Engl J Med. 2013;368(6):576. [DOI] [PubMed] [Google Scholar]
- 147. Singh S, Singh PP, Singh AG, Murad MH, Sanchez W. Statins are associated with a reduced risk of hepatocellular cancer: a systematic review and meta‐analysis. Gastroenterology. 2013;144(2):323‐32. [DOI] [PubMed] [Google Scholar]
- 148. Longo J, van Leeuwen JE, Elbaz M, Branchard E, Penn LZ. Statins as Anticancer Agents in the Era of Precision Medicine. Clin Cancer Res. 2020;26(22):5791‐800. [DOI] [PubMed] [Google Scholar]
- 149. Bathaie SZ, Ashrafi M, Azizian M, Tamanoi F. Mevalonate Pathway and Human Cancers. Curr Mol Pharmacol. 2017;10(2):77‐85. [DOI] [PubMed] [Google Scholar]
- 150. Zhou L, Wang Q, Zhang H, Li Y, Xie S, Xu M. YAP Inhibition by Nuciferine via AMPK‐Mediated Downregulation of HMGCR Sensitizes Pancreatic Cancer Cells to Gemcitabine. Biomolecules. 2019;9(10):620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kawashiri T, Tokunaga A, Kobayashi D, Shimazoe T. Anti‐tumor Activities of 3‐Hydroxy‐3‐methylglutaryl Coenzyme A (HMG‐CoA) Reductase Inhibitors and Bisphosphonates in Pancreatic Cell Lines Which Show Poor Responses to Gemcitabine. Biol Pharm Bull. 2020;43(1):49‐52. [DOI] [PubMed] [Google Scholar]
- 152. Hamada T, Khalaf N, Yuan C, Babic A, Morales‐Oyarvide V, Qian ZR, et al. Statin use and pancreatic cancer risk in two prospective cohort studies. J Gastroenterol. 2018;53(8):959‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Chua NK, Coates HW, Brown AJ. Squalene monooxygenase: a journey to the heart of cholesterol synthesis. Prog Lipid Res. 2020;79:101033. [DOI] [PubMed] [Google Scholar]
- 154. Kuzu OF, Noory MA, Robertson GP. The Role of Cholesterol in Cancer. Cancer Res. 2016;76(8):2063‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Cirmena G, Franceschelli P, Isnaldi E, Ferrando L, De Mariano M, Ballestrero A, et al. Squalene epoxidase as a promising metabolic target in cancer treatment. Cancer Lett. 2018;425:13‐20. [DOI] [PubMed] [Google Scholar]
- 156. Polycarpou‐Schwarz M, Gross M, Mestdagh P, Schott J, Grund SE, Hildenbrand C, et al. The cancer‐associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene. 2018;37(34):4750‐68. [DOI] [PubMed] [Google Scholar]
- 157. Sui Z, Zhou J, Cheng Z, Lu P. Squalene epoxidase (SQLE) promotes the growth and migration of the hepatocellular carcinoma cells. Tumour Biol. 2015;36(8):6173‐9. [DOI] [PubMed] [Google Scholar]
- 158. Ge H, Zhao Y, Shi X, Tan Z, Chi X, He M, et al. Squalene epoxidase promotes the proliferation and metastasis of lung squamous cell carcinoma cells though extracellular signal‐regulated kinase signaling. Thorac Cancer. 2019;10(3):428‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Stopsack KH, Gerke TA, Andren O, Andersson SO, Giovannucci EL, Mucci LA, et al. Cholesterol uptake and regulation in high‐grade and lethal prostate cancers. Carcinogenesis. 2017;38(8):806‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kim JH, Kim CN, Kang DW. Squalene Epoxidase Correlates E‐Cadherin Expression and Overall Survival in Colorectal Cancer Patients: The Impact on Prognosis and Correlation to Clinicopathologic Features. J Clin Med. 2019; 8(5):632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Garcia‐Bermudez J, Baudrier L, Bayraktar EC, Shen Y, La K, Guarecuco R, et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature. 2019;567(7746):118‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Jun SY, Brown AJ, Chua NK, Yoon JY, Lee JJ, Yang JO, et al. Reduction of Squalene Epoxidase by Cholesterol Accumulation Accelerates Colorectal Cancer Progression and Metastasis. Gastroenterology. 2021;160(4):1194‐207 e28. [DOI] [PubMed] [Google Scholar]
- 163. Xu F, Zhang Z, Zhao Y, Zhou Y, Pei H, Bai L. Bioinformatic mining and validation of the effects of ferroptosis regulators on the prognosis and progression of pancreatic adenocarcinoma. Gene. 2021;795:145804. [DOI] [PubMed] [Google Scholar]
- 164. Long T, Sun Y, Hassan A, Qi X, Li X. Structure of nevanimibe‐bound tetrameric human ACAT1. Nature. 2020;581(7808):339‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, et al. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020;581(7808):333‐8. [DOI] [PubMed] [Google Scholar]
- 166. Cavdarli S, Delannoy P, Groux‐Degroote S. O‐acetylated Gangliosides as Targets for Cancer Immunotherapy. Cells. 2020; 9(3):741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531(7596):651‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19(3):393‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, et al. Proteomics identifies new therapeutic targets of early‐stage hepatocellular carcinoma. Nature. 2019;567(7747):257‐61. [DOI] [PubMed] [Google Scholar]
- 170. Geng F, Cheng X, Wu X, Yoo JY, Cheng C, Guo JY, et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP‐1–Mediated Lipogenesis. Clinical Cancer Research. 2016;22(21):5337‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Chang CC, Sakashita N, Ornvold K, Lee O, Chang ET, Dong R, et al. Immunological quantitation and localization of ACAT‐1 and ACAT‐2 in human liver and small intestine. J Biol Chem. 2000;275(36):28083‐92. [DOI] [PubMed] [Google Scholar]
- 172. Lu M, Hu XH, Li Q, Xiong Y, Hu GJ, Xu JJ, et al. A specific cholesterol metabolic pathway is established in a subset of HCCs for tumor growth. J Mol Cell Biol. 2013;5(6):404‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Huang Y, Jin Q, Su M, Ji F, Wang N, Zhong C, et al. Leptin promotes the migration and invasion of breast cancer cells by upregulating ACAT2. Cell Oncol (Dordr). 2017;40(6):537‐47. [DOI] [PubMed] [Google Scholar]
- 174. Iwasaki A, Tadenuma T, Sumimoto S, Ohshiro T, Ozaki K, Kobayashi K, et al. Biseokeaniamides A, B, and C, Sterol O‐Acyltransferase Inhibitors from an Okeania sp. Marine Cyanobacterium. J Nat Prod. 2017;80(4):1161‐6. [DOI] [PubMed] [Google Scholar]
- 175. Rao KN, Kottapally S, Eskander ED, Shinozuka H, Dessi S, Pani P. Acinar cell carcinoma of rat pancreas: regulation of cholesterol esterification. Br J Cancer. 1986;54(2):305‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Ohmoto T, Yoshitani N, Nishitsuji K, Takayama T, Yanagisawa Y, Takeya M, et al. CD44‐expressing undifferentiated carcinoma with rhabdoid features of the pancreas: molecular analysis of aggressive invasion and metastasis. Pathol Int. 2015;65(5):264‐70. [DOI] [PubMed] [Google Scholar]
- 177. Oni TE, Biffi G, Baker LA, Hao Y, Tonelli C, Somerville TDD, et al. SOAT1 promotes mevalonate pathway dependency in pancreatic cancer. J Exp Med. 2020; 217(9):e20192389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Li J, Gu D, Lee SS, Song B, Bandyopadhyay S, Chen S, et al. Abrogating cholesterol esterification suppresses growth and metastasis of pancreatic cancer. Oncogene. 2016;35(50):6378‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lee SS, Li J, Tai JN, Ratliff TL, Park K, Cheng JX. Avasimibe encapsulated in human serum albumin blocks cholesterol esterification for selective cancer treatment. ACS Nano. 2015;9(3):2420‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Lim GB. Inhibiting fatty acid oxidation promotes cardiomyocyte proliferation. Nat Rev Cardiol. 2020;17(5):266‐7. [DOI] [PubMed] [Google Scholar]
- 181. Houten SM, Violante S, Ventura FV, Wanders RJ. The Biochemistry and Physiology of Mitochondrial Fatty Acid beta‐Oxidation and Its Genetic Disorders. Annu Rev Physiol. 2016;78:23‐44. [DOI] [PubMed] [Google Scholar]
- 182. Knottnerus SJG, Bleeker JC, Wust RCI, Ferdinandusse S, IJ L, Wijburg FA, et al. Disorders of mitochondrial long‐chain fatty acid oxidation and the carnitine shuttle. Rev Endocr Metab Disord. 2018;19(1):93‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Adeva‐Andany MM, Carneiro‐Freire N, Seco‐Filgueira M, Fernandez‐Fernandez C, Mourino‐Bayolo D. Mitochondrial beta‐oxidation of saturated fatty acids in humans. Mitochondrion. 2019;46:73‐90. [DOI] [PubMed] [Google Scholar]
- 184. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ma Y, Temkin SM, Hawkridge AM, Guo C, Wang W, Wang XY, et al. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 2018;435:92‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhelev Z, Aoki I, Lazarova D, Vlaykova T, Higashi T, Bakalova R. A "Weird" Mitochondrial Fatty Acid Oxidation as a Metabolic "Secret" of Cancer. Oxid Med Cell Longev. 2022;2022:2339584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Martinez‐Outschoorn UE, Peiris‐Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14(1):11‐31. [DOI] [PubMed] [Google Scholar]
- 188. Wright HJ, Hou J, Xu B, Cortez M, Potma EO, Tromberg BJ, et al. CDCP1 drives triple‐negative breast cancer metastasis through reduction of lipid‐droplet abundance and stimulation of fatty acid oxidation. Proc Natl Acad Sci U S A. 2017;114(32):E6556‐E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Wang YN, Zeng ZL, Lu J, Wang Y, Liu ZX, He MM, et al. CPT1A‐mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene. 2018;37(46):6025‐40. [DOI] [PubMed] [Google Scholar]
- 190. Wong BW, Wang X, Zecchin A, Thienpont B, Cornelissen I, Kalucka J, et al. The role of fatty acid beta‐oxidation in lymphangiogenesis. Nature. 2017;542(7639):49‐54. [DOI] [PubMed] [Google Scholar]
- 191. Lee CK, Jeong SH, Jang C, Bae H, Kim YH, Park I, et al. Tumor metastasis to lymph nodes requires YAP‐dependent metabolic adaptation. Science. 2019;363(6427):644‐9. [DOI] [PubMed] [Google Scholar]
- 192. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, et al. MSC‐regulated lncRNA MACC1‐AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38(23):4637‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Fukawa T, Yan‐Jiang BC, Min‐Wen JC, Jun‐Hao ET, Huang D, Qian CN, et al. Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia. Nat Med. 2016;22(6):666‐71. [DOI] [PubMed] [Google Scholar]
- 194. Lee JS, Oh SJ, Choi HJ, Kang JH, Lee SH, Ha JS, et al. ATP Production Relies on Fatty Acid Oxidation Rather than Glycolysis in Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2020;12(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Nimmakayala RK, Leon F, Rachagani S, Rauth S, Nallasamy P, Marimuthu S, et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene. 2021;40(1):215‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Patra KC, Kato Y, Mizukami Y, Widholz S, Boukhali M, Revenco I, et al. Mutant GNAS drives pancreatic tumourigenesis by inducing PKA‐mediated SIK suppression and reprogramming lipid metabolism. Nat Cell Biol. 2018;20(7):811‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Shin SC, Thomas D, Radhakrishnan P, Hollingsworth MA. Invasive phenotype induced by low extracellular pH requires mitochondria dependent metabolic flexibility. Biochem Biophys Res Commun. 2020:S0006‐291X(20)30284‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Wang Y, Yu T, Zhou Y, Wang S, Zhou X, Wang L, et al. Carnitine palmitoyltransferase 1C contributes to progressive cellular senescence. Aging (Albany NY). 2020;12(8):6733‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zielinski ZA, Pratt DA. Lipid Peroxidation: Kinetics, Mechanisms, and Products. J Org Chem. 2017;82(6):2817‐25. [DOI] [PubMed] [Google Scholar]
- 200. Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell. 2019;35(6):830‐49. [DOI] [PubMed] [Google Scholar]
- 201. Nishizawa H, Matsumoto M, Chen G, Ishii Y, Tada K, Onodera M, et al. Lipid peroxidation and the subsequent cell death transmitting from ferroptotic cells to neighboring cells. Cell Death Dis. 2021;12(4):332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron‐dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Tang D, Kroemer G. Ferroptosis. Curr Biol. 2020;30(21):R1292‐R7. [DOI] [PubMed] [Google Scholar]
- 204. Liang C, Zhang X, Yang M, Dong X. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv Mater. 2019;31(51):e1904197. [DOI] [PubMed] [Google Scholar]
- 205. Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee H‐J, Purohit V, et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. 2020;368(6486):85‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Dai E, Han L, Liu J, Xie Y, Kroemer G, Klionsky DJ, et al. Autophagy‐dependent ferroptosis drives tumor‐associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy. 2020;16(11):2069‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Kremer DM, Nelson BS, Lin L, Yarosz EL, Halbrook CJ, Kerk SA, et al. GOT1 inhibition promotes pancreatic cancer cell death by ferroptosis. Nat Commun. 2021;12(1):4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Kasukabe T, Honma Y, Okabe‐Kado J, Higuchi Y, Kato N, Kumakura S. Combined treatment with cotylenin A and phenethyl isothiocyanate induces strong antitumor activity mainly through the induction of ferroptotic cell death in human pancreatic cancer cells. Oncol Rep. 2016;36(2):968‐76. [DOI] [PubMed] [Google Scholar]
- 209. Ye Z, Hu Q, Zhuo Q, Zhu Y, Fan G, Liu M, et al. Abrogation of ARF6 promotes RSL3‐induced ferroptosis and mitigates gemcitabine resistance in pancreatic cancer cells. Am J Cancer Res. 2020;10(4):1182‐93. [PMC free article] [PubMed] [Google Scholar]
- 210. Tang R, Hua J, Xu J, Liang C, Meng Q, Liu J, et al. The role of ferroptosis regulators in the prognosis, immune activity and gemcitabine resistance of pancreatic cancer. Ann Transl Med. 2020;8(21):1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Dai E, Han L, Liu J, Xie Y, Zeh HJ, Kang R, et al. Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING‐dependent DNA sensor pathway. Nature Communications. 2020;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Xie Y, Zhu S, Zhong M, Yang M, Sun X, Liu J, et al. Inhibition of Aurora Kinase A Induces Necroptosis in Pancreatic Carcinoma. Gastroenterology. 2017;153(5):1429‐43 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20(3):137‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Li Z, Liu H, Luo X. Lipid droplet and its implication in cancer progression. Am J Cancer Res. 2020;10(12):4112‐22. [PMC free article] [PubMed] [Google Scholar]
- 215. Sunami Y, Rebelo A, Kleeff J. Lipid Droplet‐Associated Factors, PNPLA3, TM6SF2, and HSD17B Proteins in Hepatopancreatobiliary Cancer. Cancers (Basel). 2021;13(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Cotte AK, Aires V, Fredon M, Limagne E, Derangere V, Thibaudin M, et al. Lysophosphatidylcholine acyltransferase 2‐mediated lipid droplet production supports colorectal cancer chemoresistance. Nat Commun. 2018;9(1):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Cruz ALS, Barreto EdA, Fazolini NPB, Viola JPB, Bozza PT. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death & Disease. 2020;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Dubey R, Stivala CE, Nguyen HQ, Goo YH, Paul A, Carette JE, et al. Lipid droplets can promote drug accumulation and activation. Nat Chem Biol. 2020;16(2):206‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Petan T. Lipid Droplets in Cancer. Rev Physiol Biochem Pharmacol. 2020. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 220. Grachan JJ, Kery M, Giaccia AJ, Denko NC, Papandreou I. Lipid droplet storage promotes murine pancreatic tumor growth. Oncol Rep. 2021;45(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS Controls Pancreatic Cancer Cell Lipid Metabolism and Invasive Potential through the Lipase HSL. Cancer Res. 2020;80(22):4932‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Kuramoto K, Yamamoto M, Suzuki S, Togashi K, Sanomachi T, Kitanaka C, et al. Inhibition of the Lipid Droplet‐Peroxisome Proliferator‐Activated Receptor alpha Axis Suppresses Cancer Stem Cell Properties. Genes (Basel). 2021;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Man J, Pajic M, Joshua AM. Fats and Mets, KRAS‐Driven Lipid Dysregulation Affects Metastatic Potential in Pancreatic Cancer. Cancer Res. 2020;80(22):4886‐7. [DOI] [PubMed] [Google Scholar]
- 224. Nardi F, Fitchev P, Brooks KM, Franco OE, Cheng K, Hayward SW, et al. Lipid droplet velocity is a microenvironmental sensor of aggressive tumors regulated by V‐ATPase and PEDF. Lab Invest. 2019;99(12):1822‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Sharma B, Agnihotri N. Role of cholesterol homeostasis and its efflux pathways in cancer progression. J Steroid Biochem Mol Biol. 2019;191:105377. [DOI] [PubMed] [Google Scholar]
- 226. Ouimet M, Barrett TJ, Fisher EA. HDL and Reverse Cholesterol Transport. Circ Res. 2019;124(10):1505‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Delk SC, Chattopadhyay A, Escola‐Gil JC, Fogelman AM, Reddy ST. Apolipoprotein mimetics in cancer. Seminars in Cancer Biology. 2020;73:158‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Rajora MA, Zheng G. Targeting SR‐BI for Cancer Diagnostics, Imaging and Therapy. Front Pharmacol. 2016;7:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Moon SH, Huang CH, Houlihan SL, Regunath K, Freed‐Pastor WA, Morris JPt, et al. p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression. Cell. 2019;176(3):564‐80 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Namba Y, Sogawa C, Okusha Y, Kawai H, Itagaki M, Ono K, et al. Depletion of Lipid Efflux Pump ABCG1 Triggers the Intracellular Accumulation of Extracellular Vesicles and Reduces Aggregation and Tumorigenesis of Metastatic Cancer Cells. Front Oncol. 2018;8:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Wang F, Huang L, Zhang J, Fan J, Wu H, Xu J. Dyslipidemia in Chinese Pancreatic Cancer Patients: A Two‐Center Retrospective Study. J Cancer. 2021;12(17):5338‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Shen WJ, Azhar S, Kraemer FB. SR‐B1: A Unique Multifunctional Receptor for Cholesterol Influx and Efflux. Annu Rev Physiol. 2018;80:95‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Mohelnikova‐Duchonova B, Brynychova V, Oliverius M, Honsova E, Kala Z, Muckova K, et al. Differences in transcript levels of ABC transporters between pancreatic adenocarcinoma and nonneoplastic tissues. Pancreas. 2013;42(4):707‐16. [DOI] [PubMed] [Google Scholar]
- 234. Adamska A, Falasca M. ATP‐binding cassette transporters in progression and clinical outcome of pancreatic cancer: What is the way forward? World J Gastroenterol. 2018;24(29):3222‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Shimano H, Sato R. SREBP‐regulated lipid metabolism: convergent physiology ‐ divergent pathophysiology. Nat Rev Endocrinol. 2017;13(12):710‐30. [DOI] [PubMed] [Google Scholar]
- 236. Pons‐Tostivint E, Thibault B, Guillermet‐Guibert J. Targeting PI3K Signaling in Combination Cancer Therapy. Trends Cancer. 2017;3(6):454‐69. [DOI] [PubMed] [Google Scholar]
- 237. Wang Y, Viscarra J, Kim SJ, Sul HS. Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol. 2015;16(11):678‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Wang B, Tontonoz P. Liver X receptors in lipid signalling and membrane homeostasis. Nat Rev Endocrinol. 2018;14(8):452‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Wang Q, Wang J, Wang J, Zhang H. Molecular mechanism of liver X receptors in cancer therapeutics. Life Sci. 2021;273:119287. [DOI] [PubMed] [Google Scholar]
- 240. Zhao Y, Li H, Zhang Y, Li L, Fang R, Li Y, et al. Oncoprotein HBXIP Modulates Abnormal Lipid Metabolism and Growth of Breast Cancer Cells by Activating the LXRs/SREBP‐1c/FAS Signaling Cascade. Cancer Res. 2016;76(16):4696‐707. [DOI] [PubMed] [Google Scholar]
- 241. Srivastava S, Widmann S, Ho C, Nguyen D, Nguyen A, Premaratne A, et al. Novel Liver X Receptor Ligand GAC0001E5 Disrupts Glutamine Metabolism and Induces Oxidative Stress in Pancreatic Cancer Cells. Int J Mol Sci. 2020;21(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Yang B, Zhang B, Cao Z, Xu X, Huo Z, Zhang P, et al. The lipogenic LXR‐SREBF1 signaling pathway controls cancer cell DNA repair and apoptosis and is a vulnerable point of malignant tumors for cancer therapy. Cell Death Differ. 2020;27(8):2433‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8(4):311‐21. [DOI] [PubMed] [Google Scholar]
- 244. Shankar S, Marsh L, Srivastava RK. EGCG inhibits growth of human pancreatic tumors orthotopically implanted in Balb C nude mice through modulation of FKHRL1/FOXO3a and neuropilin. Mol Cell Biochem. 2013;372(1‐2):83‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Fako VE, Wu X, Pflug B, Liu JY, Zhang JT. Repositioning proton pump inhibitors as anticancer drugs by targeting the thioesterase domain of human fatty acid synthase. J Med Chem. 2015;58(2):778‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Falchook G, Infante J, Arkenau HT, Patel MR, Dean E, Borazanci E, et al. First‐in‐human study of the safety, pharmacokinetics, and pharmacodynamics of first‐in‐class fatty acid synthase inhibitor TVB‐2640 alone and with a taxane in advanced tumors. EClinicalMedicine. 2021;34:100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Tamburrino D, Crippa S, Partelli S, Archibugi L, Arcidiacono PG, Falconi M, et al. Statin use improves survival in patients with pancreatic ductal adenocarcinoma: A meta‐analysis. Dig Liver Dis. 2020;52(4):392‐9. [DOI] [PubMed] [Google Scholar]
- 248. Wang D, Rodriguez EA, Barkin JS, Donath EM, Pakravan AS. Statin Use Shows Increased Overall Survival in Patients Diagnosed With Pancreatic Cancer: A Meta‐Analysis. Pancreas. 2019;48(4):e22‐e3. [DOI] [PubMed] [Google Scholar]
- 249. Abdel‐Rahman O. Statin treatment and outcomes of metastatic pancreatic cancer: a pooled analysis of two phase III studies. Clin Transl Oncol. 2019;21(6):810‐6. [DOI] [PubMed] [Google Scholar]
- 250. Hong JY, Nam EM, Lee J, Park JO, Lee SC, Song SY, et al. Randomized double‐blinded, placebo‐controlled phase II trial of simvastatin and gemcitabine in advanced pancreatic cancer patients. Cancer Chemother Pharmacol. 2014;73(1):125‐30. [DOI] [PubMed] [Google Scholar]
- 251. Chen X, Kang R, Kroemer G, Tang D. Targeting ferroptosis in pancreatic cancer: a double‐edged sword. Trends Cancer. 2021;7(10):891‐901. [DOI] [PubMed] [Google Scholar]
- 252. Wang K, Zhang Z, Wang M, Cao X, Qi J, Wang D, et al. Role of GRP78 inhibiting artesunate‐induced ferroptosis in KRAS mutant pancreatic cancer cells. Drug Des Devel Ther. 2019;13:2135‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Li C, Zhang Y, Liu J, Kang R, Klionsky DJ, Tang D. Mitochondrial DNA stress triggers autophagy‐dependent ferroptotic death. Autophagy. 2021;17(4):948‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Smith DC, Kroiss M, Kebebew E, Habra MA, Chugh R, Schneider BJ, et al. A phase 1 study of nevanimibe HCl, a novel adrenal‐specific sterol O‐acyltransferase 1 (SOAT1) inhibitor, in adrenocortical carcinoma. Invest New Drugs. 2020;38(5):1421‐9. [DOI] [PubMed] [Google Scholar]
- 255. Gao Y, Jia Z, Kong X, Li Q, Chang DZ, Wei D, et al. Combining betulinic acid and mithramycin a effectively suppresses pancreatic cancer by inhibiting proliferation, invasion, and angiogenesis. Cancer Res. 2011;71(15):5182‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Siqingaowa, SS , Gopalakrishnan V, Taghibiglou C. Sterol regulatory element‐binding protein 1 inhibitors decrease pancreatic cancer cell viability and proliferation. Biochem Biophys Res Commun. 2017;488(1):136‐40. [DOI] [PubMed] [Google Scholar]
- 257. Mouhid L, Gomez de Cedron M, Garcia‐Carrascosa E, Reglero G, Fornari T, Ramirez de Molina A. Yarrow supercritical extract exerts antitumoral properties by targeting lipid metabolism in pancreatic cancer. PLoS One. 2019;14(3):e0214294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Karaboga H, Huang W, Srivastava S, Widmann S, Addanki S, Gamage KT, et al. Screening of Focused Compound Library Targeting Liver X Receptors in Pancreatic Cancer Identified Ligands with Inverse Agonist and Degrader Activity. ACS Chem Biol. 2020;15(11):2916‐28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


