Graphical abstract
Keywords: Covid-19, Temporal query, EHR, Big data
Abstract
Fast temporal query on large EHR-derived data sources presents an emerging big data challenge, as this query modality is intractable using conventional strategies that have not focused on addressing Covid-19-related research needs at scale. We introduce a novel approach called Event-level Inverted Index (ELII) to optimize time trade-offs between one-time batch preprocessing and subsequent open-ended, user-specified temporal queries. An experimental temporal query engine has been implemented in a NoSQL database using our new ELII strategy. Near-real-time performance was achieved on a large Covid-19 EHR dataset, with 1.3 million unique patients and 3.76 billion records. We evaluated the performance of ELII on several types of queries: classical (non-temporal), absolute temporal, and relative temporal. Our experimental results indicate that ELII accomplished these queries in seconds, achieving average speed accelerations of 26.8 times on relative temporal query, 88.6 times on absolute temporal query, and 1037.6 times on classical query compared to a baseline approach without using ELII. Our study suggests that ELII is a promising approach supporting fast temporal query, an important mode of cohort development for Covid-19 studies.
1. Introduction
Covid-19 is an unfolding global pandemic calling for urgent and accelerated efforts in identifying appropriate treatment strategies, developing accurate and rapid testing methods, and producing effective vaccines. Due to this urgency, clinical trials for treatment and prevention of Covid-19 must be complemented by population-based approaches. Clinical data about patients in electronic health records (EHR) provide an important source of information for Covid-19 research [1]. Benefits include those of traditional retrospective analyses such as identifying risk profiles, revealing health disparities, and understanding long-term health implications [2], [3]. They also enable machine learning approaches for outcome predication, drug repurposing, and poly-pharmacy (combinational drug effects) investigation [4], [5].
However, population-based Covid-19 study brings into sharp focus two unique query requirements in the context of EHR-derived big data. One is temporal query, particularly on medical events around a patient’s Covid-19 diagnosis. The second is interface for interactive cohort exploration, which requires near real-time responses to user-specified queries to facilitate study design and data access.
1.1. Temporal query
Covid-19 studies often involve temporal relationships on patient phenotype and healthcare events (e.g. diagnosis, medication, lab test and procedure) before and after Covid-19 diagnosis [6], [7], [8]. Here are some sample cohorts specifications that involve temporal relationships:
-
1.
All patients who received polymerase chain reaction (PCR) test in May 2020;
-
2.
All patients who developed neurologic complications after extracorporeal membrane oxygenation (ECMO) for Covid;
-
3.
All patients who had stroke within a month after Covid diagnosis;
-
4.
All patients who did not have any cardiovascular condition before positive Covid diagnosis.
Example 1 involves absolute temporal query, while the rest involves relative temporal queries. Example 1 is an instance of query with negation.
Temporal query, an important query modality for population-based Covid-19 research, has not been a traditional focus of clinical query systems [9] which were mostly focused on patient-recruitment for clinical trials [10], [11]. Fast temporal query on large EHR-derived datasets presents an emerging big data challenge, since temporal query is computationally expensive and takes too long to execute using a brute-force approach. Specifically, relative temporal query involves pairwise comparison of dates between clinical events, so a new data structure and query execution strategy is required to achieve a suitable level of response speed. Near real-time interface response is a critical factor to achieve a sense of “interactivity” for cohort exploration.
1.2. Interactive cohort exploration
Two general types of data processing pipeline for population-based studies exist. One is ad-hoc data processing, which involves the development of study-specific data extraction programming scripts to run directly on source data to obtain specific patient cohorts of interest [12]. The second is cohort discovery based on a clinical query interface, allowing an investigator to interactively explore and formulate patient cohorts of interest [13], [14], [15]. Such interfaces are often built on top of databases constructed using common data models such as those populated through i2b2 [16], PCORnet [17], and OHDSI/MOP [18].
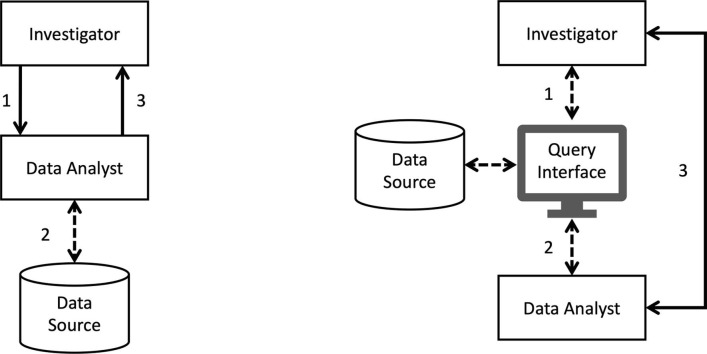
As pointed out in [15], the main distinction between the two pipelines can be seen in Fig. 1 . The ad-hoc approach (Fig. 1, left) requires an investigator to communicate data request to a data analyst (1), who in turn implements the request as a data extraction script to run on the data source (which can be in database or file-based format) (2), and then obtains requested data and finally returns results for further analysis (3). The time span between steps 1 and 3 can be weeks if not months, and steps 1–3 often need to be iterated as study criteria are refined. The second approach (Fig. 1: right) supports a paradigm which allows investigators and data analysts (1–2) to construct and issue query directly through a web interface without requiring the knowledge about how the backend data are structured and stored, whereby shortening the data-access life-cycle and facilitate collaborative data exploration (Fig. 1, right).
Fig. 1.
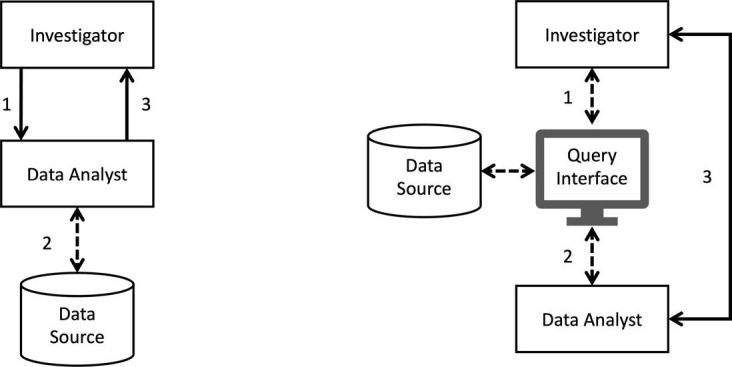
Typical data processing pipelines for population-based studies (adapted from [15]). Left: ad-hoc scripting. Right: data exploration using a web-based interface.
Many existing EHR data warehouses require the use of a command-line query language to extract data. Even for those that are equipped with a graphical user interface, there is a general lack of systematic and dedicated support for temporal queries in both the query language and the user interface [19]. Near real-time response to temporal query is one of the most computationally challenging aspects for interactive cohort exploration [20]. Methods for exploring, querying and interacting with data need to be improved to cope with the size and complexity of data. Indeed, an estimated 40% of study respondents reported that they sometimes gave up because the task was too time-consuming [19]. This query response latency challenge is amplified when using larger Covid-19 EHR dataset for population-based research.
To address these challenges, we introduce a novel approach called Event-level Inverted Index (ELII) to optimize time trade-offs between one-time batch preprocessing and open-ended, user-specified temporal queries. To demonstrate the feasibility of ELII, we developed an experimental query engine in a NoSQL (not only SQL) database using the ELII strategy to support the temporal query modality. Near-real-time performance was achieved on a large Covid-19 EHR dataset, with 1.3 million unique patients and 3.76 billion records. We evaluated the performance of ELII on five types of queries: non-temporal (classical), absolute temporal, relative temporal, query with negation, and patient-level event sequence look-up. Our experimental results show that ELII handled all five types of queries in seconds, achieving average speed accelerations of 26.8 times on relative temporal query, 88.6 times on absolute temporal query, and 1037.6 times on classical query compared to a baseline approach without using ELII. To summarize, our main contributions are:
-
•
A novel inverted index, ELII, to support fast temporal query on clinical events;
-
•
A data preprocessing pipeline for a document-based data model for ELII implementation;
-
•
An experimental query engine to support common queries and related evaluation using a real-world, large Covid-19 EHR dataset to demonstrate the enhanced performance using ELII.
2. Background
2.1. Database structure
Relational data store. Relational data store is a commonly used method for storing and managing clinical data [21]. EHR data is represented as tables, where each row in a table represents a record, and each column represents an attribute. Table 1 illustrates a clip of Lab table from an EHR dataset, where the first column captures patient identifier (ID).
Table 1.
Lab test in EHR represented as a conventional relational data table.
| ID | CODE | NAME | TYPE | RES | UNIT | DATE |
|---|---|---|---|---|---|---|
| PT001 | 26464-8 | White blood cell | HEMAT | 9.4 | 2018-06-04 | |
| PT001 | 59408-5 | Oxygen saturation | BLOOD GAS | 99.0 | 2019-08-05 | |
| PT001 | N/A | Covid19 pcr | N/A | negative | N/A | 2020-05-21 |
| PT002 | 59408-5 | Oxygen saturation | BLOOD GAS | 96.0 | 2019-08-05 | |
| PT002 | N/A | Covid19 pcr | N/A | positive | N/A | 2020-06-23 |
Entity-attribute-value (EAV) data store. One limitation of the relational data store is the inflexible of attributes [22]. An attribute may have different types of data and some records may have missing data or without applicable attributes. EVA data model is an alternative row-based design. In general, an EAV table stores the attribute-value pairs of an entity. An entity may have multiple tables separated by data types. In Table 2 , for instance, a Lab record may have text, numeric and date-time data, and they are stored in three different EAV tables. In each table, the “ID” (Table 2, first column) is the lab record ID which serves as a linkage between the EVA tables. i2b2, a popular clinical query engine, uses EAV data store in the so-called “star-schema” design [16], [10].
Table 2.
Lab test in EHR represented as EAV data tables. ID - lab record ID; A - Attribute; V - Value.
| Value type: text |
Value type: numeric |
Value type: date |
||||||
|---|---|---|---|---|---|---|---|---|
| ID | A | V | ID | A | V | ID | A | V |
| PT001-1 | TEST CODE | 26464-8 | PT001-1 | TEST RESULT | 9.4 | PT001-1 | TEST DATE | 2018-06-04 |
| PT001-1 | TEST NAME | White blood cell | PT001-2 | TEST RESULT | 99.0 | PT001-2 | TEST DATE | 2019-08-05 |
| PT001-1 | TEST TYPE | HEMAT | PT002-1 | TEST RESULT | 96.0 | PT001-3 | TEST DATE | 2020-05-21 |
| PT001-1 | TEST UNIT | PT002-1 | TEST DATE | 2019-08-05 | ||||
2.2. Document-oriented data store and MongoDB
Document-oriented data store. Document-oriented data store, or document-oriented database, is a type of NoSQL database [23]. Different from traditional relational databases, document-oriented databases are designed to store, retrieve and manage information [24] represented as a collection of “documents” in JSON (JavaScript Object Notation) or XML (Extensible Markup Language) format [25]. A document can be large, complex, and semi-structured. It serves as the basic unit of data processing and conceptually is equivalent to a record in a relational data table. MongoDB is a document-oriented database system using JSON-type documents with optional schemas [26]. A group of documents in MongoDB is called a “collection.” Each element or entry in a document is represented as a key-value pair. Key-value pairs can be nested to provide tremendous expressive power for capturing complex information. MongoDB’s collections do not enforce document structure; therefore, it is “schema-less,” providing flexibility for data type and data-modeling choices to match application requirements. Table 3 shows three lab-related documents for the same patient (PT001) in previous examples using a document-oriented data store.
Table 3.
Lab test in EHR represented as a document-based data store.
| {“DOCUMENT ID”:“1” | {“DOCUMENT ID”:“2” | {“DOCUMENT ID”:“3” |
| “PATIENT ID”: “PT001” | “PATIENT ID”: “PT001” | “PATIENT ID”: “PT001” |
| “TEST CODE”: “26464-8” | “TEST CODE”: “59408-5” | “TEST NAME”: “Covid19 pcr” |
| “TEST NAME”: “White blood cell” | “TEST NAME”: “Oxygen saturation” | “TEST RESULT”: “negative” |
| “TEST TYPE”:“HEMATOLOGY” | “TEST TYPE”:“BLOOD GAS” | “TEST DATE”: 2020-05-21} |
| “TEST RESULT”: 9.4 | “TEST RESULT”: 99.0 | |
| “TEST UNIT”: “” | “TEST UNIT”: “” | |
| “TEST DATE”: 2018-06-04} | “TEST DATE”: 2019-08-05} |
For this study, we use document-oriented data store implemented in MongoDB for the following reasons:
-
1.
Rapid response to the Covid-19 pandemic requires data from multiple sites and sources to be pooled together in a short timeframe. This entails that not all source data can be mandated to follow the same data model, data format, or coding standard [27]. Document-oriented data store provides the flexibility for managing data format and coding variability.
-
2.
MongoDB provides querying facility which can take advantage of customized indices, nested/embedded objects and arrays, and support for on-the-fly regular expression processing during query. MongoDB is also scalable in database size, with a built-in constraint that the maximum BSON (a binary representation of MongoDB’s JSON document data records) document size is 16 MB. The number of levels of nesting for BSON documents is limited at 100. These generous constraints provide enough flexibility for most applications (but see Discussion on document size).
-
3.
We have had prior successful experiences using MongoDB [28], so this database environment is familiar to our team, which can help facilitate rapid development through code reuse and software architecture repurposing.
Column-oriented data store. Column-oriented data store is an alternative data structure used for NoSQL databases. In a column-oriented database, each column is stored in a single table. Table 4 is an example of five lab records in the column-oriented data store. Two of the five records do not have “TYPE” data so only other three records were stored in the colum “TYPE.” For a query on “TYPE,” three records with “TYPE” column will be fetched instead of all five records with entire columns. Such flexibility can help accelerate query execution on sparse dataset, or when not all the columns are involved in the execution of a query.
Table 4.
Lab test in EHR represented as column-oriented data tables. ID - Lab record ID.
| Column: NAME |
Column: TYPE |
Column: DATE |
|||
|---|---|---|---|---|---|
| ID | NAME | ID | TYPE | ID | DATE |
| PT001-1 | White blood cell | PT001-1 | HEMATOLOGY | PT001-1 | 2018-06-04 |
| PT001-2 | Oxygen saturation | PT001-2 | BLOOD GAS | PT001-2 | 2019-08-05 |
| PT001-3 | Covid19 pcr | PT002-1 | BLOOD GAS | PT001-3 | 2020-05-21 |
| PT002-1 | Oxygen saturation | PT002-1 | 2019-08-05 | ||
| PT002-2 | Covid19 pcr | PT002-2 | 2020-06-23 | ||
Though not used in this study, we mention graph-based data store for completeness. Graph-based data store is used in graph databases (GDB), which uses graph structure for semantic query and uses nodes, edges and attributes to represent and store data [29]. A node in the GDB represent an instance, similar to a record or a row in a relational database (or a document in a document-store database), such as person, business, and account. An edge represent a relationship connecting two nodes, an abstraction not directly implemented in a relational model or a document-store model. Resource Description Framework (RDF) is special type of GDB that uses XML syntax to describe the characteristics of web resources and the relationship between resources [30]. The main idea of RDF is to create statements about resources (in particular web resources) in expressions of the form (subject, predicate, object), known as triples [31]. This model provides an infrastructure for metadata of different web applications [32]. A collection of RDF statements intrinsically represents a labeled, directed multi-graph, and this makes an RDF data model better suited to certain types of knowledge representation [33], [34].
2.3. Inverted index
Originating from the field of information retrieval, a forward index, , associates each document D (or its identifier-ID) to the list of words that the document contains. For example, to answer the query “Which documents contain word X,” the forward index requires exhaustive iteration through each document and each word to locate a hit.
Inverted index, , is a common technique used for enhancing query performance. For text-based search, an inverted index consists of a list of all the unique words appearing in a document collection, and for each word, a list of identifiers for those documents that contain the word. An inverted index is usually implemented as a hash map: the key is the word and the value is an array of document identifiers. If the array only contains the document identifiers, it is called a “record-level inverted index.” If the array also contains the location of each word, then it is called a “word-level inverted index” [35].
In the context of document-oriented EHR dataset, document identifiers can be patient IDs and document content can be a list of records for each patient. Inverted index for lab test consists of, for each lab test, a list of patients (IDs) who had the corresponding lab test. For example, if patients had Covid-19 PCR test, then this would be captured in the inverted index as a corresponding entry:
This comes handy when querying for patients who had Covid-19 PCR test: it is merely a lookup (for Covid-19 PCR) to retrieve the patient list. In contrast, forward index requires searching the entire patient list, as well as each patient’s lab records to look for Covid-19 PCR test, which becomes extremely time-consuming for large EHR dataset.
2.4. Temporal information in EHR data
There are two conceptual types of temporal information in EHR. The first is event-type, where each occurrence is associated with a single timestamp. Multiple events can be aligned in a one dimensional timeline [36]. In an EHR dataset, lab test and medication are event-type data. The second is interval-type, where each occurrence is associated with a start timestamp and an end timestamp. Basic interval relations are often captured in Allen’s interval algebra [37]. Encounters are typical interval-type occurrences in a patient’s medical record. For this study, we are interested in the representation and query of event-type information in EHR dataset.
A challenge of temporal query using the conventional data model involves comparison of timestamps between two events with a specified temporal relation. Such a query involves quadratic time complexity of , where n is the number of events for a single patient. In extreme cases, a patient may have thousands of records, and the number of comparison will be millions. For millions of patients, this operation quickly becomes intractable. Therefore, optimization is necessary for temporal query on large datasets. Indexing time (on the timestamp column) is the most commonly used strategy to speed up temporal query [38]. Preprocessing query before execution is another approach to reduce condition evaluation effort [39]. Approach also has been proposed [40] to optimize the data model by pre-computing the relation between two events and cache such information for future use. Another study [41] demonstrated that complete in-memory processing can improve query execution time on a dataset with millions of rows, although memory remains a relatively expensive commodity today. Our ELII approach is different from those proposed in such studies in that we address the temporal query performance challenge using an innovative collection of forward and inverted indices specifically designed to handle EHR data and clinical events (see Discussion for general applicability).
3. Methods
Typical EHR data contain four types of source files about a patient: demographics (), diagnosis (), medication (), and lab test (). We use such file types to introduce our concept of event-level inverted index.
3.1. Event-level inverted index
An event-level inverted index consists of four main components:
-
1.
Patient Timeline, which contains all the clinical events and related information (e.g., date and time) for each patient;
-
2.
Conventional Inverted Index, which includes the inverted indices of time-invariant variables, especially for demographic data, such as “gender” and “race;”
-
3.
Timeline Inverted Index (tiII), which consists of inverted indices of time-dependent variables (i.e., event labels with timestamps), such as “diagnosis code” and “lab test;” and
-
4.
Global Lookup Table, which is a forward index of all variables and associated inverted indices.
Patient Timeline. We use multi-level nested documents (supported by MongoDB) to store patients’ events where “Patient ID” serves as the “primary key” of documents. Each timeline document consists of all clinical events of a patient. Each patient may have multiple events and each event may have multiple attributes. The result follows a structure of JSON-like nested key-value pairs:
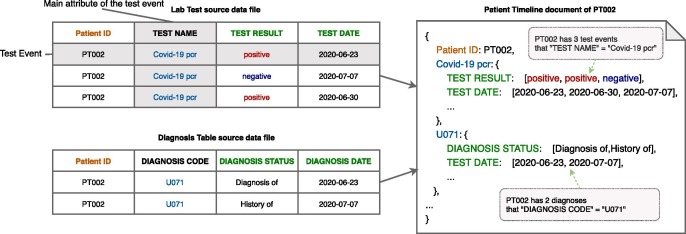
Fig. 2 illustrates the conversion to Patient Timeline document from a lab test file and a diagnosis file . Each record in the source file with the same “Patient ID” (PT002 in this example) is stored in the same document. For each table on left of Fig. 2, we selected the values of a column (marked in blue, “TEST NAME” for lab test table in Fig. 2 and “DIAGNOSIS CODE” for diagnosis table in Fig. 2) as the event keys. The rest of the columns (in green) in the source file are the attributes for the events. In Patient Timeline, the record values of each attribute are sorted by record date in the structure of array. The length of the array represents the number of occurrence of a certain type of events for a patient.
Fig. 2.
Illustrative conversion of row-oriented document data to Patient Timeline.
We use a column-oriented store for the values of each event attribute. In Patient Timeline for PT002 (Fig. 2), for example, the “TEST RESULT” field of “Covid-19 pcr” only contains the value in the column “TEST RESULT” with “Patient ID” equals to “PT002.” Query performance will benefit from a column-oriented data store because we only need to access the values of the query attributes instead of loading the entire document for event-specific queries.
For time-related event, we sorted the “Date” attribute for each event, which is another key idea for speeding up temporal query. In Fig. 2, for example, “TEST DATE” field of “Covid-19 pcr” is sorted from the oldest to the latest, and the values of “TEST RESULT” are sorted with the same order of “TEST DATE.” Some temporal queries only involve the first or the latest event, which can be retrieved in constant time. Using sorted array, searching for a specific date has worst time-complexity , where n is the number of elements in the array.
Conventional Inverted Index. Conventional inverted indices are used for time-invariant variables, such as those involved in file type . In the demographic source file (Table 5 ), each column represents a different demographic variable as an attribute, such as gender and race, and each row represents a patient. This inverted index has a key-value pair structure: {}, which the key () is the value of attribute and value () is the list of patient IDs. For Table 5, the corresponding inverted indices for “GENDER” looks like:
Table 5.
Demographics data in table format.
| ID | GENDER | RACE | AGE |
|---|---|---|---|
| PT001 | male | Caucasian | 35 |
| PT002 | female | Asian | 62 |
| PT003 | female | Caucasian | 19 |
| PT004 | male | African American | 47 |
| PT005 | unknown | other | 35 |
= [(male: [PT001, PT004]), (female: [PT002, PT003]), (other: [PT005])].
Timeline Inverted Index (tiII). There are two types of tiII: 1) single-attribute tiII; and 2) multi-attribute tiII.
For each event, the single-attribute tiII is structured as an attribute column (e.g., “TEST NAME” or “DIAGNOSIS CODE”), stored as key-value based documents. Each document consists of event name, the first date of event for all patients, and the last date of event for all patients. A 2-dimensional array (named ) is used to capture all patients who have had this event. This 2-dimensional array divides and sorts patient by event date chronologically:
where is an array consisting of patients (IDs) who had this event i-number of days since the first date. For example, represents patients who had this event on the first date of event and represents patients who had this event on the last date of event. This way, one can query and access the data by date in constant time by looking up the array-index of the date instead of executing an iteration of temporal comparisons between event time and query-specified time.
Multi-attribute tiII has a similar data structure to that of single-attribute tiII. The difference is that it handles combinations of events, such as
[“TEST NAME” and “TEST RESULT’]
or
[“DIAGNOSIS CODE” and “DIAGNOSIS STATUS”].
Therefore, multi-attribute tiII has additional fields for multiple event labels. Multi-attribute tiII provides a mechanism for pre-computed multi-event join using inverted index.
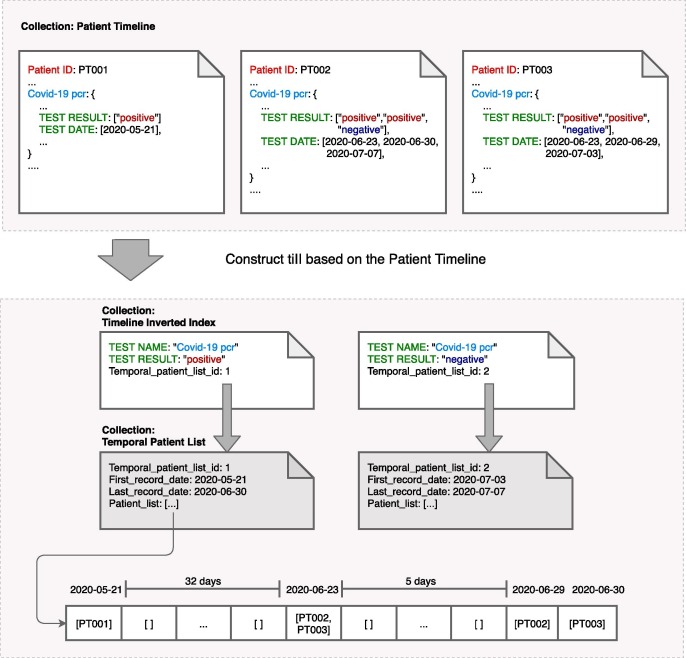
Fig. 3 shows an example of multi-attribute tiII for “Covid-19 pcr” test combined with test result (“positive” or “negative”). The dotted box on top is an example of Patient Timeline containing three patients who had “Covid-19 pcr” test. The sample event has “TEST NAME” being “Covid-19 pcr” and “TEST RESULT” being “positive,” with the corresponding multi-attribute tiII in the second dotted box. is shown in the last dotted box, which represents consecutive dates of all occurrences from the first date of the event to the last date of the event. In this example, with (Fig. 3, below) stores the identifiers of patients who had “positive” in “Covid-19 pcr” test from “2020-05-21” to “2020-06-30.” Each item of the array is a list, which stores the identifiers of all patients who had the event on this particular date. For example, the item of the array indicates PT002 and PT003 were tested “positive” for “Covid-19 pcr” in 2020-06-23 (34 days after 2020-05-21). If there is no event record of any patient on a specific day, this array item will be empty (a place holder).
Fig. 3.
Construct Multi-attribute tiII based on the Patient Timeline.
Global Lookup Table. This is a global forward index designed for inverted index management. This global forward index makes it straightforward to look up and access all inverted indices using all the original variable in the EHR data. For example, the documents for inverted index of “gender” can be looked up in the collection of demographic inverted indices.
3.2. Query execution
The template for temporal query can be structured as:
“Find patients who had [events] with [temporal constraints].”
User input parameters are “events” and “temporal constraints” (optional). “Events” is one or multiple attributes and value pairs such as diagnosis code: U071, diagnosis status: diagnosis of.
“Temporal constraints” provide the specification for the time interval in which the events take place. For example, a temporal constraint can be a period or temporal relations of two events such as “any stroke diagnosis AFTER first Covid-19 diagnosis BETWEEN 2020-02-01 and 2020-05-01.”
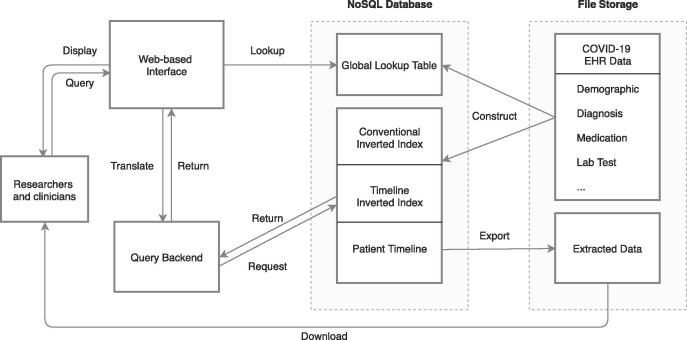
Fig. 4 presents the architecture of our query engine. For database content (Fig. 4, right), a NoSQL database with ELII is constructed from a collection of EHR source files. On the frontend (Fig. 4, left), a user builds a query using a web-based graphic interface. The query engine translates a user’s query as a group of database statements for the “Query Backend.” The Query Backend then executes the database statements according to statement type by consulting the Global Lookup Table and combines the results according to query logic. The final result is then presented back to the user in the web-interface. Query results can be exported and downloaded for further analysis. Three basic types of query are available:
-
1.
Classical (non-temporal),
-
2.
Absolute temporal, and
-
3.
Relative temporal.
Fig. 4.
Query engine architecture.
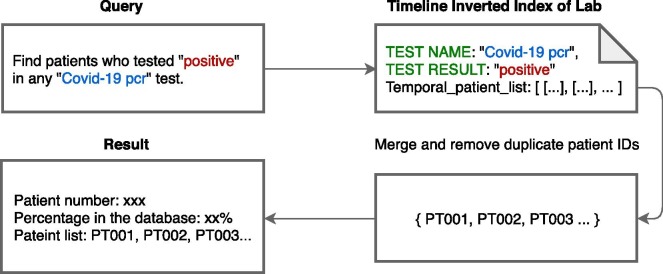
Classical. Classical query involves only non-temporal attributes, such as demographics and clinical events without time constraint. Conventional inverted index are used to support this kind of query. Pseudocode for classical query appears in Appendix A, Algorithm 1. Fig. 5 demonstrates the main steps involved in executing the classical query “Find patients who tested positive in any Covid-19 pcr test.” These steps are instantiated in the following statements in Algorithm 1:
-
1.
Look up Algorithm 1 line 1
-
2.
Merge resulting patient IDs Algorithm 1 line 2–5
-
3.
Find unique patient IDs Algorithm 1 line 6
-
4.
Return results Algorithm 1 line 7
Fig. 5.
Query without temporal constraints.
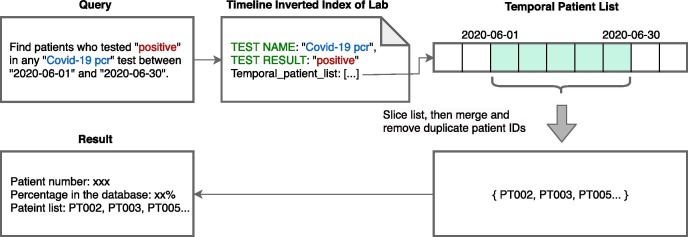
Absolute temporal. Absolute temporal query contains input parameters that restrict the times for events, executable using tiII. The pseudocode for absolute temporal query execution appears as Appendix A, Algorithm 2. Fig. 6 demonstrates the main steps involved in executing absolute temporal query “Find patients who tested positive in any Covid-19 pcr test between 2020-06-01 and 2020-06-30.” These steps are instantiated in the following statements in Algorithm 2:
-
1.
Look up within interval Algorithm 2 line 1–19
-
2.
Merge resulting patient IDs Algorithm 2 line 20
-
3.
Find unique patient IDs Algorithm 2 line 22
-
4.
Return results Algorithm 2 line 23
Fig. 6.
Query with absolute temporal condition.
Relative temporal. A relative temporal query contains two events: A and B, instead of a single event in the previous two query types. The query also specifies a temporal relation between A and B. In EHR, events of the same type may have multiple occurrences. Therefore, further elaboration of the temporal relation, as shown in Table 6 , is needed. In total, there are six possible relations between two clinical events with multiple occurrences (and corresponding timestamps).
Table 6.
Possible relations between two clinical events with multiple timestamps.
| Relation | Operator | Interpretation |
|---|---|---|
| A takes place before B | All | Last A occurs before first B |
| A takes place before B | Any | First A occurs before last B |
| A takes place before B | First | First A occurs before first B |
| A takes place before B | Last | Last A occurs before last B |
| A meets B | All | Every A occurs on the same day that B occurs |
| A meets B | Any | At least one A occurs on the same day that B occurs |
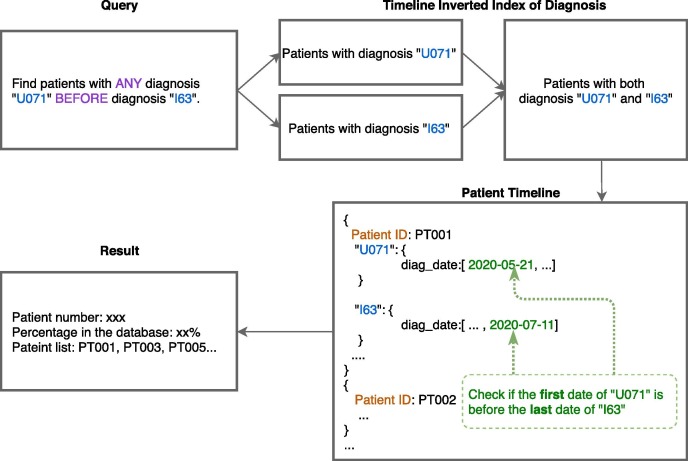
The pseudocode for query with relative temporal constraints appears as Appendix A, Algorithm 3. Fig. 7 shows the main steps involved in executing relative temporal query “Find patients with any diagnosis U071 (Covid-19) before diagnosis I63 (stroke).” These steps are instantiated in the following statements in Algorithm 3:
-
1.
Algorithm 3 line 5
-
2.
Algorithm 3 line 6
-
3.
U = unique IDs in Algorithm 3 line 8
-
4.
Find with , where
= first date for U071 and = last date for I63 Algorithm 3 line 9–49
Fig. 7.
Query with relative temporal condition.
In addition to these basic types of query, the query engine also supports full boolean queries and individual patient lookup.
Boolean query. Boolean query involves a combination of multiple sub-queries using logical operators (AND and OR). Sub-queries are executed independently first. Then, set intersection is performed on the resulting patient identifiers resulting from sub-queries for AND and set union (with unique patient identifiers) is performed on the resulting patient identifiers resulting from sub-queries for OR. For example, result for the following OR query
“Find patients who had a Covid-19 diagnosis OR a Covid-19 test with a positive result”
will be the union of patient identifiers from sub-queries “patients with any Covid-19 diagnosis” and “patients with any positive Covid-19 test.”
Negation. Negation is a useful operation for clinical studies to find patients without certain conditions. For EHR data, negation is handled by appropriately interpreting “lack of data or information.” Three typical scenarios are possible with the lack of information: 1. using “Closed-World Assumption,” lack of information entails negation. For example, if a patient did not have cancer diagnosis in medical record, it is safe to infer that the patient did not have cancer. 2. No information. For example, if a patient did not have HIV test, we have no way of knowing if the person has HIV or not. 3. No available result. For example, a patient may be coded as a smoker, but we have no “packs per day” information at all.
With these possible interpretations in mind, “lack of data or information” query can be executed by forming two groups of patients: a group Z of patients to exclude, and the group of all patients U. For example, the query “Find patients who did NOT have any diagnosis U071 (Covid-19) before diagnosis I63 (stroke)” can be performed by following these steps:
-
1.
-
2.
-
3.
Individual patient data look up. ELII is also designed for fast individual patient history data retrieval, another important query modality. It uses the Patient Timeline forward index . The query input is a patient ID, with other options including specific events the user wants to show in the output, and a period of time the events fall within.
3.3. Evaluation method
Data source. We used OPTUM® de-identified Covid-19 EHR dataset with Aug 6, 2020 release date. This dataset consisted 1.3 million patients who had either Covid-19 related diagnoses (U071, U072 and U073) or had Covid-19 specific lab tests (positive or negative). 16 individual source files were contained in the release, for different types of EHR records, such as patient demographics, diagnosis, medication, and lab. Each source file comes with several types of attributes (i.e., the “columns”). For instance, the PATIENT source file contains demographic attributes such as gender, age, and race. The DIAGNOSIS source file contains attributes such as diagnosis code, diagnosis code type, and diagnosis status. The MED ADMINISTRATIONS source file contains attributes such as drug name, National Drug Code (NDC), quantity of dose, and dose frequency. Definition of these attribute types are given in the accompanying data dictionary provided by OPTUM®. In total, 132 such attribute types are suitable for query construction. Table 7 lists the numbers of attributes and lines of records in the main source files, status of relevant date fields, and file size.
Table 7.
OPTUM® Covid-19 dataset summary statistics.
| Table | # Attribute | # Record | Temporal | File Size (GB) |
|---|---|---|---|---|
| PATIENT | 18 | 1,317,565 | No | 0.15 |
| DIAGNOSIS | 13 | 567,441,527 | Yes | 49.94 |
| MED ADMINISTRATIONS | 21 | 170,517,390 | Yes | 42.32 |
| PRESCRIPTIONS WRITTEN | 21 | 78,359,556 | Yes | 15.02 |
| PATIENT REPORTED MEDS | 15 | 115,972,976 | Yes | 13.60 |
| LAB | 18 | 761,370,078 | Yes | 106.19 |
| Other | 93 | 2,067,229,913 | N/A | 131.22 |
| Total | 199 | 3,762,209,005 | N/A | 358.44 |
Query performance was evaluated using MongoDB with the entire dataset imported (1.3 million total patients). To test the performance of ELII’s four components (patient timeline, inverted index, timeline inverted index and global lookup table), three types of query (described in Section 3.2) were tested:
-
1.
Classical: Find patients who diagnosed/tested with [xxx];
-
2.
Absolute temporal: Find patients who diagnosed/tested with [xxx] after 2020-02-20;
-
3.
Relative temporal: Find patients who diagnosed/tested with [xxx] after 2020-02-20 and before first Covid-19 diagnosis.
For these three types of queries, we selected test events from Lab and Diagnosis for two reasons: 1) they are two commonly used record types for temporal query; 2) they are the top three largest record types in data size and the record number of the OPTUM® Covid-19 dataset version 20200806. As listed in Table 7, “Lab” has the largest data size and the second largest record number, and the data size and record number of “Diagnosis” are both the third largest.
The “[xxx]” in each test query template is the place holder for query parameter. Since the output of the temporal query is a list of patients, we selected 19 query parameters with different number of patient counts. We first picked 5 events with the number of patients ranging from 10 to 100,000 in base 10 logarithmic scale.
For events involving a number of patients greater than 100,000, we picked additional four events with a number of patients from 100,000 to 500,000 in linear scale (every 100,000 patients). We also tested an extreme case of “Lab” records: “test name” = “Oxygen saturation (SpO2).pulse oximetry,” which has the largest number of patients in this data release. The last event is Covid-19 diagnosis (“diagnosis code” = “U071”), used as Event B for relative temporal query testing. Summary statistics for each event tested are shown in Table 8 . For each event, we executed all three query testing templates and recorded the execution times for MongoDB statement translation, MongoDB running time, and result array conversion. All queries were repeated 10 times consecutively and the average time were reported.
Table 8.
19 events used for query evaluation.
| ID | Attribute | Orignial Table | # Record | # Patient |
|---|---|---|---|---|
| D1 | diagnosis code | DIAGNOSIS | 89 | 10 |
| D2 | diagnosis code | DIAGNOSIS | 247 | 100 |
| D3 | diagnosis code | DIAGNOSIS | 1,718 | 1,000 |
| D4 | diagnosis code | DIAGNOSIS | 52,863 | 10,015 |
| D5 | diagnosis code | DIAGNOSIS | 295,752 | 100,680 |
| D6 | diagnosis code | DIAGNOSIS | 2,053,756 | 203,739 |
| D7 | diagnosis code | DIAGNOSIS | 994,277 | 328,912 |
| D8 | diagnosis code | DIAGNOSIS | 3,987,244 | 452,383 |
| D9 | diagnosis code | DIAGNOSIS | 12,954,359 | 515,517 |
| L1 | test name | LAB | 19 | 10 |
| L2 | test name | LAB | 147 | 100 |
| L3 | test name | LAB | 1,035 | 1,004 |
| L4 | test name | LAB | 11,941 | 10,327 |
| L5 | test name | LAB | 305,057 | 102,405 |
| L6 | test name | LAB | 655,291 | 206,620 |
| L7 | test name | LAB | 1,151,535 | 300,434 |
| L8 | test name | LAB | 2,970,457 | 405,081 |
| L9 | test name | LAB | 3,034,999 | 531,467 |
| L10 | test name | LAB | 120,677,164 | 1,141,175 |
| Reference | diagnosis code | DIAGNOSIS | 555,733 | 100,686 |
Notes: D1: T25321S, D2: 236439005, D3: 3739, D4: I878, D5: 110033, D6: E559, D7: Z1159, D8: Z79899, D9: I10, L1: Urate, L2: angle, L3: rv psp, L4: 2019 novel coronavirus naat, L5: Human chorionic gonadotropin (HCG).quantitative, L6: Iron binding capacity.total (TIBC), L7: Thyroxine.free (FT4), L8: Prothrombin time (PT), L9: Hemoglobin A1C, L10: Oxygen saturation (SpO2).pulse oximetry. The last query value “U071” was only used in relative temporal query as a reference event.
Baseline database. For baseline performance, we imported the OPTUM® Covid-19 EHR dataset into MongoDB and kept its original row-oriented data format. Each row in the source files is stored as a document (the document keys are contained in the first column of each source file). To optimize query performance for the baseline database, we use MongoDB’s standard built-in indexing for all query experiments for a fair comparison. In this setup, classical query shows how the conventional inverted index in ELII improves performance. Temporal query shows how “timeline inverted index” and “patient timeline” in ELII improves performance, at a different scale.
4. Results
4.1. Data preprocessing
We developed an automated script for preprocessing OPTUM® Covid-19 EHR dataset version 20200806 into MongoDB. 3.76 billion records from 360 GB of text files were processed, creating event timelines for 1.3 million patients. The global lookup table contained 132 attribute types (diagnosis code, test code, test name, etc.), based on which we built single-attribute tiII for all 1.94 million attribute-value pairs. Single-attribute tiII is indexed for only one data field, and multi-attribute tiII is for multiple-field queries. For example, to query the patients with “Covid-19 pcr” test and “positive” result, we needed two-attribute tiII for the “TEST NAME” and the “TEST RESULT.” Creating multi-attribute tiII will increase the performance for multi-field queries but it requires extra pre-processing time.
4.2. Query performance
Table 9 shows the performance result for queries without temporal constraints. Conventional inverted index worked well: all queries were completed in less than 0.1 s for classical query.
Table 9.
Execution time (in seconds) for classical query “Find patients with [query event].”
| ID | Baseline | ELII | Saved | X-faster |
|---|---|---|---|---|
| D1 | 0.001 | 0.0004 | 0.0006 | 2.5 |
| D2 | 0.0016 | 0.0003 | 0.0013 | 5.3 |
| D3 | 0.0078 | 0.0005 | 0.0073 | 15.6 |
| D4 | 0.2574 | 0.0009 | 0.2565 | 286.0 |
| D5 | 1.3608 | 0.0056 | 1.3552 | 243.0 |
| D6 | 8.0205 | 0.015 | 8.0055 | 534.7 |
| D7 | 4.3322 | 0.0611 | 4.2711 | 70.9 |
| D8 | 14.5332 | 0.0499 | 14.4833 | 291.2 |
| D9 | 36.1807 | 0.0607 | 36.12 | 596.1 |
| L1 | 0.0007 | 0.0003 | 0.0004 | 2.3 |
| L2 | 0.0013 | 0.0004 | 0.0009 | 3.3 |
| L3 | 0.0077 | 0.0005 | 0.0072 | 15.4 |
| L4 | 0.0883 | 0.0009 | 0.0874 | 98.1 |
| L5 | 1.8039 | 0.0057 | 1.7982 | 316.5 |
| L6 | 3.2602 | 0.0135 | 3.2467 | 241.5 |
| L7 | 5.4892 | 0.0215 | 5.4677 | 255.3 |
| L8 | 17.2616 | 0.0545 | 17.2071 | 316.7 |
| L9 | 16.8289 | 0.0481 | 16.7808 | 349.9 |
| L10 | 345.8775 | 0.099 | 345.7785 | 3493.7 |
| Avg | 23.9639 | 0.0231 | 23.9408 | 1037.6356 |
Notes: Baseline: baseline model using row-oriented data store, ELII: our method using document-oriented data store with ELII, Saved: ELII Time - Baseline Time, X-faster: Baseline Time ELII Time.
Table 10 shows the performance result for absolute temporal query. ELII performed an average 88 times faster than the baseline setup. It was not as dramatic as in the case of classical query because query execution time for the baseline setting is reduced by filtering out more records with the time constraints. Meanwhile, query time for ELII was slightly increased since an additional step was involved to slice the patient list according to time period.
Table 10.
Execution time (in seconds) for absolute temporal query “Find patients with [Query Event] after Feb 20, 2020.”
| ID | Baseline | ELII | Saved | X-faster |
|---|---|---|---|---|
| D1 | 0.001 | 0.0008 | 0.0002 | 1.3 |
| D2 | 0.0014 | 0.0007 | 0.0007 | 2.0 |
| D3 | 0.0069 | 0.0009 | 0.006 | 7.7 |
| D4 | 0.2112 | 0.0024 | 0.2088 | 88.0 |
| D5 | 0.8956 | 0.0078 | 0.8878 | 114.8 |
| D6 | 5.4155 | 0.0702 | 5.3453 | 77.1 |
| D7 | 4.2298 | 0.3043 | 3.9255 | 13.9 |
| D8 | 10.5411 | 0.3058 | 10.2353 | 34.5 |
| D9 | 24.707 | 0.9459 | 23.7611 | 26.1 |
| L1 | 0.0007 | 0.0009 | -0.0002 | 0.8 |
| L2 | 0.0014 | 0.001 | 0.0004 | 1.4 |
| L3 | 0.0063 | 0.0012 | 0.0051 | 5.3 |
| L4 | 0.0942 | 0.0036 | 0.0906 | 26.2 |
| L5 | 1.3034 | 0.1885 | 1.1149 | 6.9 |
| L6 | 2.3589 | 0.051 | 2.3079 | 46.3 |
| L7 | 4.3796 | 0.0694 | 4.3102 | 63.1 |
| L8 | 14.7904 | 0.2539 | 14.5365 | 58.3 |
| L9 | 14.9931 | 0.2764 | 14.7167 | 54.2 |
| L10 | 319.2168 | 2.0664 | 317.1504 | 154.5 |
| Avg | 21.2186 | 0.2395 | 20.9791 | 88.5839 |
Notes: Baseline: baseline model using row-oriented data store, ELII: our method using document-oriented data store with ELII, Saved: ELII Time - Baseline Time, X-faster: Baseline Time ELII Time.
Table 11 shows the performance result for relative temporal query. The baseline result shows relative temporal query was the most time-consuming in our experiments. ELII achieved even better performance than the other two query types. For the large scale query L10, it had a significantly improvement over the baseline, achieving an average 26 times faster and reduced query execution time by 35 s on average.
Table 11.
Execution time (in seconds) for relative temporal query “Find patients had any [Query Event A] before first Covid-19 diagnosis.”
| ID | Baseline | ELII | Saved | X-faster |
|---|---|---|---|---|
| D1 | 2.652 | 0.1338 | 2.5182 | 19.8 |
| D2 | 2.6504 | 0.1325 | 2.5179 | 20.0 |
| D3 | 2.6716 | 0.1382 | 2.5334 | 19.3 |
| D4 | 2.9773 | 0.2031 | 2.7742 | 14.7 |
| D5 | 4.4099 | 0.7081 | 3.7018 | 6.2 |
| D6 | 11.9138 | 1.3572 | 10.5566 | 8.8 |
| D7 | 7.8546 | 1.2973 | 6.5573 | 6.1 |
| D8 | 17.8092 | 2.5436 | 15.2656 | 7.0 |
| D9 | 39.6454 | 3.2416 | 36.4038 | 12.2 |
| L1 | 2.4523 | 0.1175 | 2.3348 | 20.9 |
| L2 | 2.4235 | 0.0865 | 2.337 | 28.0 |
| L3 | 2.4848 | 0.0888 | 2.396 | 28.0 |
| L4 | 2.5785 | 0.1044 | 2.4741 | 24.7 |
| L5 | 4.5704 | 0.7681 | 3.8023 | 6.0 |
| L6 | 7.144 | 1.3152 | 5.8288 | 5.4 |
| L7 | 10.5915 | 1.6565 | 8.935 | 6.4 |
| L8 | 41.8886 | 2.4278 | 39.4608 | 17.3 |
| L9 | 38.694 | 3.2004 | 35.4936 | 12.1 |
| L10 | 493.3144 | 6.5753 | 486.7391 | 75.0 |
| Avg | 36.7751 | 1.3735 | 35.4016 | 26.7747 |
Notes: Baseline: baseline model using row-oriented data store, ELII: our method using document-oriented data store with ELII, Saved: ELII Time - Baseline Time, X-faster: Baseline Time ELII Time.
Our ELII-based query engine surpassed the baseline in almost every single testing query by execution time except for L1 (negligible difference for a small query). Our approach demonstrated more significant time reduction as the number of records became larger. The most time-consuming query was “test name” = “Oxygen saturation (SpO2).pulse oximetry,” which had 120 million records and 1.1 million patients. For all three tested query types, baseline setup for SpO2 took 345.9, 319.2, and 493.3s respectively, while ELII reduced the time to 0.1, 2.1, and 6.6s, demonstrating dramatic performance improvements for larger queries.
We performed validation of ELII result for query accuracy by comparing the results with the same query using the baseline setup. For all 57 tested queries (19 query events in Table 8 tested on three query types), the results of the two approaches are exactly the same in both number of patients and sets of patient IDs. We also randomly selected and manually validated query results using the patient timeline lookup function. By reviewing the records of a set of randomly selected patients, we were able to independently confirm that the query results met our query conditions.
4.3. Query interface
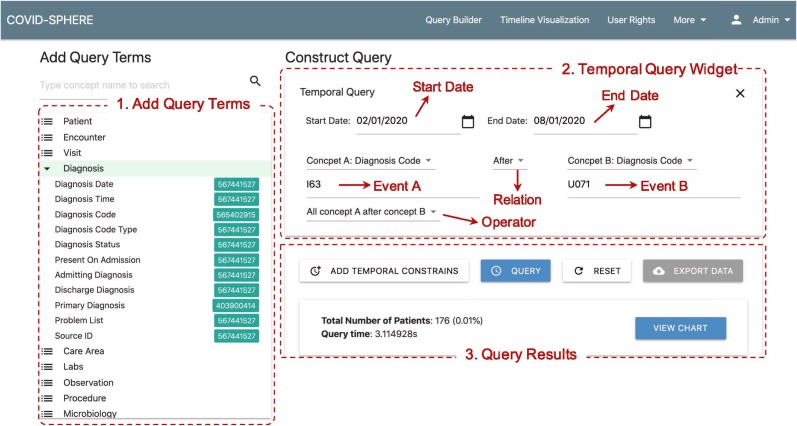
To demonstrate the feasibility of ELII for interactive cohort exploration, we developed a web-based user interface called CovidSphere with which to build temporal queries for cohort exploration. The main idea and design of CovidSphere followed the best practice of our previous established query interface design experiences. These include those reported in publications such as “MEDCIS’ [42], “X-search” [13], as well as “DataSphere” [28]. Fig. 8 shows the query builder interface layout with three areas annotated. In Fig. 8 area 1, a user may select an attribute type among “query terms,” such as diagnosis code or test name, to construct classic and absolute temporal queries. In Fig. 8 area 2, a query template for relative temporal query is provided, where a user may select different settings within four components: i) time period (the start date and the end date), ii) events that include attribute type and attribute value (i.e., event A and event B), iii) temporal relation between two events (BEFORE or AFTER), and iv) operator mode for the temporal relation as specified in Table 6. The example shown in Fig. 8 area 2 represents a query to find all patients who have ALL diagnosis code: I63 AFTER diagnosis code: U071 between Feb. 1, 2020 and Aug. 1, 2020. By clicking the “QUERY” button, the query engine will translate the user input into a backend query statement and obtain the corresponding list of patient identifiers. Query result information is displayed in area 3 of Fig. 8, which includes the number of patients meeting the query criteria. A user may download the resulting list of patients with demographics information in comma separated value format by clicking “EXPORT DATA.”
Fig. 8.
Layout of our temporal query interface.
Our experimental Covid-19 EHR database was implemented using MongoDB 4.4 Community Edition on MacOS. The query library for performance evaluation was written in Python (version 3.8.3). The ELII query engine was built and evaluated on a Mac Pro 2019 with 2.7 GHz 24-Core Intel Xeon processor and 768 GB 2933 MHz DDR4 memory.
5. Discussion
5.1. Preprocessing effort
We developed ELII to enhance query performance and demonstrate this for a large Covid-19 EHR dataset. To achieve this goal, we needed to pre-compute ELII indices as part of data conversion to MongoDB data. Our approach for building ELII involved two steps: (1) building a hash map in such a way that the hash key is an existing event and the hash value is a list of patient IDs with the associated records containing the corresponding event. The time complexity for this step is , where n is the total number of data elements in the dataset. (2) sorting the events associated with each patient, which has time complexity , where m is the total number of events in the database and is the total number of records that contain the event. Additional preprocessing time of 22 h was spent in building all single-attribute tiII of the 360 GB text files. This one time, preprocessing can be speeded up by using parallel computing.
Although our experiments were carried out in a typical high-end desktop machine, the query engine can also be hosted in a more powerful environment, such as a computer cluster. One of our contributions is that, using commonly available computer hardware configurations, ELII can already achieve near-realtime query performance for a large OPTUM® Covid-19 EHR dataset.
5.2. MongoDB Cold-start
MongoDB uses Memory Mapped Storage Engine (MMAP) which maps files in the disk to memory for faster process. If the documents of a query are not in memory, MongoDB will perform the mapping first and then execute the query, which is called a cold-start. Experiments for this study were performed with warm-start by making sure data was already loaded in memory, in order for the results to only reflect the execution time for queries. Even with cold-start, the ELII approach remains speedy because it covers most queries without prompting MongoDB to load all documents in memory.
5.3. MongoDB limit on document size
The maximum document size for MongoDB is 16 megabytes. This restriction was intended for managing RAM and I/O bandwidth. In extreme cases of the Covid-19 EHR dataset, a patient may have an extremely large number of events or an event may have taken place for an extremely large number of patients. If the corresponding document is larger than the allowable size, such a document can be split into several. Splitting will reduce query performance, but only for queries that triggered the splitting scenario. For example, {“diagnosis code”:“I10”}(D9 in Table 10) had 12,954,359 records that were split into 34 documents using our automated script. As a consequence, this absolute temporal query (D9) took much longer than other events involving fewer patients.
5.4. Patient timeline export
Query result consists of a list of patient IDs. A users can retrieve all the records from any individual patient in the result immediately after a query. The export function helps users to not only validate whether patients match their search criteria, but also inspect and make sense of the clinical event sequence in the specific cohort.
5.5. Time complexity estimation
MongoDB used + Tree for indexing, so a “find” statement is in time complexity, where x is the number of records in a document. Let n be the total number of patients, and m be the total number of unique events. Then the time complexity for classical and absolute temporal query is , and the time complexity of individual patient timeline lookup is . Relative temporal query is , where x is the number of resulting patients, and y is the maximum number of an individual patient’s events.
5.6. General applicability
Although we primarily designed ELII for Covid-19 EHR data to improve the performance of temporal query, our approach can be applied to other standard and typical EHR data for chronic diseases and conditions, with or without temporal query. In fact, our earlier work [28] demonstrated that a NoSQL (e.g., MongoDB) approach can cut query construction time by half while improving the speed of representative queries by a magnitude.
ELII also is applicable to other data models such as those for i2b2, PCORNet, and OMOP. Query engines built for these widely used data models have not provided sufficient support for temporal query. ELII offers a pathway to creating MongoDB-like query engines for databases using i2b2, PCORNet, and OMOP data models. The general steps involved would consist of exporting data from i2b2, PCORNet, and OMOP into text files for patient demographics, diagnosis, medication, and labs, with appropriately linkable identifies for each record line in the file. Then these source text files can be processed using our pre-processing and ELII-construction scripts, and imported into a MongoDB environment, with a CovidSphere-like query interface. Limitations and topics for further study include the need to reduce pre-processing time using parallel computing, incremental database updates without the need for full database reconstruction when updated source data become available, and inefficiency in space utilization that is typically associated with a document-oriented data store.
5.7. User interface usability evaluation
This is a substantial topic beyond the scope of the current paper. However, the design of our CovidSphere user interface followed the best practice from our previous established query interface design experiences. These include interfaces reported in publications such as MEDCIS [42], X-search [13], as well as DataSphere [28], and we expect to have similar outcomes. We recognize that some additional query functionalities would be desirable, such as a query manager to save and share queries, interfaces supporting case-control design and exploration, built-in tools for visualization, and more flexibility for customized data export. Using a similar strategy as our previously developed cohort query and exploration tools, we plan to develop a comprehensive interactive cohort exploration interface based on this new ELII data model and our preliminary CovidSphere interface for OPTUM® Covid-19 data. We plan to report the design, implementation, and user evaluation of the system in a separate future paper.
5.8. Research utility
We have not performed a formal research utility assessment to provide publishable evidence supporting our original design motivation in enabling population-based Covid-19 research. However, the temporal query functionality in CovidSphere using ELII is already benefiting several ongoing studies locally on the impact and consequence of Covid-19 on topics such as outcome differences between sex groups, and long-term neurological impacts. Our approach provided an intuitive way to communicate and refine study requirements for clinical investigators. It provided live feedback on “what if” questions. Results on such topics will be reported in disease-specific venues.
6. Conclusion
To better leveraging large EHR data for Covid-19 research, we developed an innovative inverted index system to support fast temporal query. Experimental results showed that a set of temporal queries tested on 1.3 million patients resulted in average execution time of seconds or less. Our study suggests that ELII is a promising approach supporting fast temporal query, an important mode of cohort development for Covid-19 and other population-based research.
Contributors
GQZ developed the ELII concept. YH and XJL designed and refined the ELII system and evaluation. YH wrote code for data preprocessing and ELII backend. XJL implemented the ELII interface. All authors contributed to writing and editing the manuscript.
Dataset availability
The dataset used for experiments in this study is provided by OPTUM®, a third-party vendor. The University of Texas Health Science Center at Houston licensed this dataset.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We sincerely thank our colleagues Shiqiang Tao and Licong Cui for valuable technical discussions. We also thank colleague Wei-Chun Chou for her assistance in enhancing the rendering of the diagrams used in this paper.
Appendix A. Pseudocode for temporal query
Algorithm 1
No temporal constraints
Procedure = EventQuery() InputinvertedIndexCollection: The client for accessing ELII collection in MongoDB event: The query event OutputpatentIDList: The list of patient IDs after query 1: ; 2: ; 3: fordocumentinqueryDocumentsdo 4: ; 5: end for 6: patentIDList = ; 7: returnpatentIDList
Algorithm 2
Absolute temporal condition
Procedure = AbsoluteTemporalEventQuery() InputinvertedIndexCollection: The client for accessing ELII collection in MongoDB event: The query event period: The query period OutputpatentIDList: The list of patient IDs after query 1: ; 2: ; 3: fordocumentinqueryDocumentsdo 4: ; 5: ; 6: 7: 8: ifis notNonethen 9: ; 10: ifthen 11: 12: end if 13: end if 14: ifis notNonethen 15: ; 16: ifthen 17: 18: end if 19: end if 20: ; 21: end for 22: patentIDList = ; 23: returnpatentIDList
Algorithm 3
Relative temporal condition
Procedure = RelativeTemporalEventQuery(, ) InputinvertedIndex: The client for accessing inverted index collection in MongoDB patientTimeline: The client for accessing patient timeline collection in MongoDB eventA: The first event A eventB: The second event B period: The query period cond: The relation between event A and event B, as shown in Table 6 op: The operator of temporal query, as shown in Table 6 OutputpatentIDList: The list of patient IDs after query 1: ifperiodisNonethen 2: ; 3: ; 4: else 5: ; 6: ; 7: end if 8: ; 9: ; 10: ; 11: patentIDList = ; 12: forpatientIDinpatientListAandBdo 13: if (isNone) or (isNone) then 14: Continue; 15: end if 16: ; 17: ; 18: ifcondisthen 19: switchopdo 20: case 21: ifthen 22: ; 23: end if 24: case 25: ifthen 26: ; 27: end if 28: case 29: ifthen 30: ; 31: end if 32: case 33: ifthen 34: ; 35: end if 36: else ifcondisthen 37: switchopdo 38: case 39: ifthen 40: ; 41: end if 42: case 43: if 0 then 44: ; 45: end if 46: end if 47: end for 48: patentIDList = ; 49: returnpatentIDList
References
- 1.National covid cohort collaborative (N3C), https://ncats.nih.gov/n3c (accessed: Oct 12, 2020).
- 2.J.H. Moore, I. Barnett, M.R. Boland, Y. Chen, G. Demiris, G. Gonzalez-Hernandez, D.S. Herman, B.E. Himes, R.A. Hubbard, D. Kim, et al., Ideas for how informaticians can get involved with Covid-19 research, 2020. [DOI] [PMC free article] [PubMed]
- 3.J. Wang, H. Anh, F. Manion, M. Rouhizadeh, Y. Zhang, Covid-19 signsym–a fast adaptation of general clinical nlp tools to identify and normalize Covid-19 signs and symptoms to omop common data model, ArXiv. [DOI] [PMC free article] [PubMed]
- 4.Randhawa G.S., Soltysiak M.P., El Roz H., de Souza C.P., Hill K.A., Kari L. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: Covid-19 case study. Plos One. 2020;15(4):e0232391. doi: 10.1371/journal.pone.0232391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. Alimadadi, S. Aryal, I. Manandhar, P.B. Munroe, B. Joe, X. Cheng, Artificial intelligence and machine learning to fight Covid-19, 2020. [DOI] [PMC free article] [PubMed]
- 6.J. Toubiana, C. Poirault, A. Corsia, F. Bajolle, J. Fourgeaud, F. Angoulvant, A. Debray, R. Basmaci, E. Salvador, S. Biscardi, et al., Kawasaki-like multisystem inflammatory syndrome in children during the Covid-19 pandemic in paris, France: prospective observational study, bmj 369. [DOI] [PMC free article] [PubMed]
- 7.W. Guo, M. Li, Y. Dong, H. Zhou, Z. Zhang, C. Tian, R. Qin, H. Wang, Y. Shen, K. Du, et al., Diabetes is a risk factor for the progression and prognosis of Covid-19, Diabetes/metabolism research and reviews (2020) e3319. [DOI] [PMC free article] [PubMed]
- 8.Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in Covid-19: A single center experience. J. Med. Virol. 2020;92(7):814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganslandt T., Mate S., Helbing K., Sax U., Prokosch H. Unlocking data for clinical research–the German i2b2 experience. Appl. Clin. Informatics. 2011;2(1):116. doi: 10.4338/ACI-2010-09-CR-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maier C., Christoph J., Schmidt D., Ganslandt T., Prokosch H., Kraus S., Sedlmayr M. Experiences of transforming a complex nephrologic care and research database into i2b2 using the idrt tools. J. Healthcare Eng. 2019 doi: 10.1155/2019/5640685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshmukh V.G., Meystre S.M., Mitchell J.A. Evaluating the informatics for integrating biology and the bedside system for clinical research. BMC Med. Res. Methodol. 2009;9(1):70. doi: 10.1186/1471-2288-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iavindrasana J., Cohen G., Depeursinge A., Müller H., Meyer R., Geissbuhler A. Clinical data mining: a review. Yearbook Med. Informatics. 2009;18(01):121–133. [PubMed] [Google Scholar]
- 13.Cui L., Zeng N., Kim M., Mueller R., Hankosky E.R., Redline S., Zhang G.-Q. X-search: an open access interface for cross-cohort exploration of the national sleep research resource. BMC Med. Informatics Decision Making. 2018;18(1):99. doi: 10.1186/s12911-018-0682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobbins N.J., Spital C.H., Black R.A., Morrison J.M., de Veer B., Zampino E., Harrington R.D., Britt B.D., Stephens K.A., Wilcox A.B., et al. Leaf: an open-source, model-agnostic, data-driven web application for cohort discovery and translational biomedical research. J. Am. Med. Inform. Assoc. 2020;27(1):109–118. doi: 10.1093/jamia/ocz165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G.-Q., Siegler T., Saxman P., Sandberg N., Mueller R., Johnson N., Hunscher D., Arabandi S. Visage: a query interface for clinical research. Summit Translat. Bioinformatics. 2010;2010:76. [PMC free article] [PubMed] [Google Scholar]
- 16.González L., Pérez-Rey D., Alonso E., Hernández G., Serrano P., Pedrera M., Gómez A., Schepper K.D., Crepain T., Claerhout B. Building an i2b2-based population repository for clinical research. Digital Personalized Health and Medicine: Proceedings of MIE. 2020;2020(270):78. doi: 10.3233/SHTI200126. [DOI] [PubMed] [Google Scholar]
- 17.C.B. Forrest, K.M. McTigue, A.F. Hernandez, L.W. Cohen, H. Cruz, K. Haynes, R. Kaushal, A.N. Kho, K.A. Marsolo, V.P. Nair, et al., PCORnet 2020: Current state, accomplishments, and future directions, J. Clin. Epidemiol. [DOI] [PMC free article] [PubMed]
- 18.Klann J.G., Joss M.A., Embree K., Murphy S.N. Data model harmonization for the all of us research program: Transforming i2b2 data into the OMOP common data model. PloS One. 2019;14(2):e0212463. doi: 10.1371/journal.pone.0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rind A., Wang T.D., Aigner W., Miksch S., Wongsuphasawat K., Plaisant C., Shneiderman B. Interactive information visualization to explore and query electronic health records. Found. Trends Human-Comput. Interact. 2013;5(3):207–298. [Google Scholar]
- 20.Binnig C., Basık F., Buratti B., Cetintemel U., Chung Y., Crotty A., Cousins C., Ebert D., Eichmann P., Galakatos A., Hättasch B. Real-Time Business Intelligence and Analytics 2015 Aug 31. Springer; Cham: 2015. Towards interactive data exploration; pp. 177–190. [Google Scholar]
- 21.C. Friedman, G. Hripcsak, S.B. Johnson, J.J. Cimino, P.D. Clayton, A generalized relational schema for an integrated clinical patient database, in: Proceedings of the Annual Symposium on Computer Application in Medical Care, American Medical Informatics Association, 1990, p. 335.
- 22.Dinu V., Nadkarni P. Guidelines for the effective use of entity–attribute–value modeling for biomedical databases. Int. J. Med. Informatics. 2007;76(11–12):769–779. doi: 10.1016/j.ijmedinf.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. Han, E. Haihong, G. Le, J. Du, Survey on nosql database, in: 2011 6th international conference on pervasive computing and applications, IEEE, 2011, pp. 363–366.
- 24.O. Tezer, A comparison of nosql database management systems and models, DigitalOcean. Np 21.
- 25.H. Vera, W. Boaventura, M. Holanda, V. Guimaraes, F. Hondo, Data modeling for nosql document-oriented databases, in: CEUR Workshop Proceedings, vol. 1478, 2015, pp. 129–135.
- 26.Banker K. Manning Publications Co.; 2011. MongoDB in action. [Google Scholar]
- 27.Dong X., Li J., Soysal E., Bian J., et al. Covid-19 TestNorm: A tool to normalize Covid-19 testing names to LOINC codes. J. Am. Med. Informat. Assoc. 2020;27(9):1437–1442. doi: 10.1093/jamia/ocaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Tao, L. Cui, X. Wu, G.-Q. Zhang. Facilitating cohort discovery by enhancing ontology exploration, query management and query sharing for large clinical data repositories, in: InAMIA Annual Symposium Proceedings 2017, vol. 2017, American Medical Informatics Association, p. 1685. [PMC free article] [PubMed]
- 29.A. Silvescu, D. Caragea, A. Atramentov, Graph databases, Artificial Intelligence Research Laboratory Department of Computer Science, Iowa State University.
- 30.O. Lassila, R.R. Swick, et al., Resource description framework (rdf) model and syntax specification.
- 31.Miller E. An introduction to the resource description framework. Bull. Am. Soc. Inform. Sci. Technol. 1998;25(1):15–19. [Google Scholar]
- 32.Jevsikova T., Berniukevicius A., Kurilovas E. Application of resource description framework to personalise learning: Systematic review and methodology., Informatics. Education. 2017;16(1):61–82. [Google Scholar]
- 33.Modoni G.E., Sacco M., Terkaj W. 2014 International Conference on Engineering, Technology and Innovation (ICE) IEEE; 2014. A survey of rdf store solutions; pp. 1–7. [Google Scholar]
- 34.Powers S. O’Reilly Media, Inc; 2003. Practical RDF: solving problems with the resource description framework. [Google Scholar]
- 35.R. Baeza-Yates, B. Ribeiro-Neto, et al., Modern information retrieval, vol. 463, ACM Press, New York, 1999.
- 36.W.-N. Lee, A.K. Das, Local alignment tool for clinical history: temporal semantic search of clinical databases, in: AMIA Annual Symposium Proceedings, vol. 2010, American Medical Informatics Association, 2010, p. 437. [PMC free article] [PubMed]
- 37.Allen J.F. Maintaining knowledge about temporal intervals. Commun. ACM. 1983;26(11):832–843. [Google Scholar]
- 38.Johnston T. Newnes; 2014. Bitemporal data: theory and practice. [Google Scholar]
- 39.Kvet M., Matiasko K. Temporal data performance optimization using preprocessing layer. J. Inform. Syst. Eng. Manage. 2018;3(2):13. [Google Scholar]
- 40.El-Sappagh S.H., El-Masri S., Riad A.M., Elmogy M. Electronic health record data model optimized for knowledge discovery. Int. J. Comput. Sci. Issues (IJCSI) 2012;9(5):329. [Google Scholar]
- 41.S. Lam, Patternfinder in microsoft amalga: Temporal query formulation and result visualization in action. unpublished, 2008. http://www.cs.umd.edu/hcil/patternFinderInAmalga/PatternFinderS-HonorsPaper.pdf.
- 42.G.Q. Zhang, L. Cui, S. Lhatoo, S.U. Schuele, S.S. Sahoo, MEDCIS: multi-modality epilepsy data capture and integration system, in: AMIA Annual Symposium Proceedings 2014, vol. 2014, American Medical Informatics Association, p. 1248. [PMC free article] [PubMed]