Abstract
Cyclohexene nucleic acid (CeNA) forms a duplex with RNA that is more stable than a DNA–RNA duplex (ΔTm per modification: +2°C). A cyclohexenyl A nucleotide adopts a 3′-endo conformation when introduced in dsDNA. The neighbouring deoxynucleotide adopts an O4′-endo conformation. The CeNA:RNA duplex is cleaved by RNase H. The Vmax and Km of the cleavage reaction for CeNA:RNA and DNA:RNA is in the same range, although the kcat value is about 600 times lower in the case of CeNA:RNA.
INTRODUCTION
RNase H is an ubiquitous enzyme that hydrolyses the RNA strand of a RNA/DNA hybrid. This enzyme plays an important role in the mode of action of antisense oligonucleotides. RNase H degrades RNA in DNA:RNA heteroduplexes in the 3′→5′ direction (1). The RNase H cleavage rate is significantly reduced when the RNA substrate is structured and the selection of the target sequence is of utmost importance for antisense activity of oligodeoxynucleotides (2). Phosphorothioates are, likewise, known to serve as substrates for RNase H when bound to their complementary RNA targets (3). The RNA target is cleaved with a slightly lower Km than with phosphodiester oligonucleotides (1). At higher concentrations, phosphorothioate oligonucleotides inhibit RNase H (1). Oligodeoxynucleotide boranophosphate induces RNase H with a cleavage rate that is enhanced in comparison with the cleavage rate of phosphodiester and phosphorothioate oligonucleotides. These results were obtained using a poly(A):oligo(dT) model (4). The gapmer approach was developed with the aim of obtaining oligonucleotides that have higher affinities for RNA targets combined with RNase H activity. Chimeric oligonucleotides with modified wings and with gaps of less than four deoxynucleotides do not allow RNase H to cleave of the target RNA; those with gaps of five deoxynucleotides are cleaved at 3-fold slower rate than unmodified substrates (1). More than five deoxynucleotides in the gap are needed for effective antisense activity.
Recently, we synthesised cyclohexene nucleic acids (CeNA) (5). When introducing one to three cyclohexene nucleotides into the DNA strand of a DNA/RNA hybrid, the duplex becomes thermodynamically more stable. A fully modified CeNA is more stable against enzymatic degradation in serum than DNA. An interesting observation is that a window of six cyclohexene nucleotides with adenine base moieties incorporated into a DNA oligonucleotide is sufficient to observe RNase H cleavage of a uridine hexamer window in a 2′-OMe oligonucleotide (5). This was attributed to the flexibility of the CeNA and the absence of steric hindrance in the minor groove for the approach of the enzyme and for promoting catalytic activity.
As the cleavage rate of the complementary RNA target by RNase H is important with respect to the practical use of oligonucleotides in antisense technology, we investigated the cleavage kinetics of CeNA in the aforementioned window approach.
MATERIALS AND METHODS
Oligonucleotides
The synthesis and characterisation of all oligonucleotides that have been used in the experiments have been described previously (5).
Thermal stability studies
Oligonucleotide concentrations were determined by measuring the absorbance in pure RNase-free water at 260 nm at 80°C and the following extinction coefficients were used: adenine nucleotide ɛ = 15 000; thymine nucleotides ɛ = 8500; guanine nucleotides ɛ = 12 500; cytosine nucleotides ɛ = 7500; uridine nucleotides ɛ = 10 000. Tm values were determined in a buffer containing 0.1 M NaCl, 0.02 M potassium phosphate pH 7.5 and 0.1 mM EDTA with a 4 µM concentration for each strand. Melting curves were obtained with a Cary 100 Bio spectrophotometer. Cuvettes were maintained at a precise temperature by means of water circulation through the cuvette holder. The temperature of the solution was measured with a thermistor directly immersed in one of the cuvettes. Temperature control and data acquisition was carried out with an IBM compatible computer using Cary WinUV thermal application software. The samples were heated at a rate of 0.2°C min–1 starting at 10°C up to 80°C. Melting temperatures were determined by plotting the first derivative of the absorbance versus temperature curve. Up and down curves showed identical Tm values.
Determination of the kinetic parameters
The oligonucleotide 5′-CmGmGmCmGMrUrUrUrUrUrUCmAmGmGmAm-3′ was radiolabelled (32P) at the 5′-end using T4 polynucleotide kinase (Gibco BRL) and [γ-32P]ATP (4500 Cu/mmol) by standard procedures and purified on a NAP-5 column (Pharmacia). At various concentrations, ranging from 30 to 2000 nM, the radiolabelled RNA containing oligonucleotide was annealed in reaction buffer (10 mM Tris–HCl pH 7.5, 1 mM MgCl2, 50 mM KCl), with a 2-fold excess of the complementary oligonucleotide with deoxy, cyclohexene or phosphorothioate building blocks opposite the RNA gap and deoxynucleotides in the wings. Substrates were incubated with RNase H (Escherichia coli RNase H; Epicentre) at 37°C in a total volume of 25 µl, 0.125 U for the deoxy and phosphorothioate analogues and 25 U for the cyclohexene containing substrate. Aliquots (3 µl) were taken at specific time intervals (once every 2–10 min) and quenched in 5 µl stop mix (EDTA 50 mM, xylene cyanol FF 0.1% and bromophenol blue 0.1% in formamide 90%) and chilled on ice. Samples were analysed by denaturing 20% polyacrylamide gel electrophoresis (PAGE) containing urea (50%) with TBE buffer at 1000 V for 1.25 h. The gels were analysed and quantified by phosphorimaging (Packard Cyclone, Optiquant software). All assays were performed in the linear range of the phosphorimager. The amount of substrate hydrolysed was converted in concentrations and initial rate velocities and Michaelis–Menten parameters (Km and kcat) were calculated using GraphPad Prism software. Data reported were based on three independent experiments.
Structural investigations using NMR spectroscopy: resonance assignment
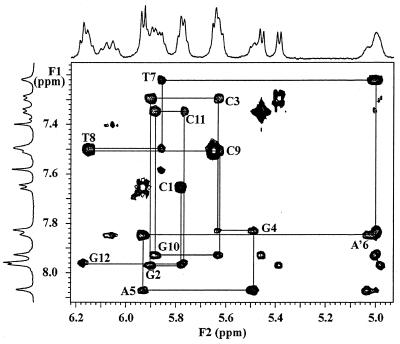
The structure of a dodecamer d(CGCGAA′TTCGCG) with a cyclohexenyl A introduced at the sixth position was investigated. Non-exchangeable protons in the DNA part of the modified oligonucleotide duplex were assigned starting from a standard anomeric to aromatic proton walk (6) in a NOESY (7) spectrum (mixing time 250 ms) in D2O. The modified nucleoside A′6, however, does not have an anomeric proton, since there is no oxygen in the cyclohexene ring. The H1′ of the cyclohexene ring is therefore upfield shifted (resonance at 5.00 p.p.m.) compared to the H1′ anomeric proton in ribose sugars. The intraresidue A′6:H1′ to A′6:H8 and the interresidue A′6:H1′ to T7:H6 and T6:H5 were clearly visible. In this way, sequential connectivities for the entire strand could be identified, thereby assigning all H1′and H8/H6 resonances (Fig. 1). Most of the other DNA sugar resonances of the DNA strand were identified by their TOCSY (8) signals to the well-dispersed anomeric proton signals. Many of these were confirmed by a DQF-COSY (9) spectrum. In A′6, assignment was started from H1′, which was identified through the sequential walk, and showed COSY crosspeaks to the highfield H2′1 and H2′2 (at 2.45 and 2.53 p.p.m., respectively) and the lowfield H6′ (6.06 p.p.m.). The same spectrum was used to assign H3′ (crosspeak to H2′1 and H2′2) and H4′ (crosspeak H3′). A TOCSY spectrum (mixing time 65 ms) allowed for assignment of H5′ and H7′/H7″. Sequential H3′-P-H5′/5″ (H7′/7″ in A′6) contacts observed in the two-dimensional [1H,31P] correlation spectrum (10) (HETCOR) confirmed assignments of these resonances (results not shown).
Figure 1.
(H8/H6/H2)–H1′/H5 region of the 150 ms NOESY at 20°C in 100% D2O. Sequential H8/H6(n)–H1′(n)–H8/H6(n+1) connectivities starting from the 5′-end (C1) are shown in solid lines. Resonances at 7.42 and 7.61 p.p.m. are from A5H2 and A6H2, respectively.
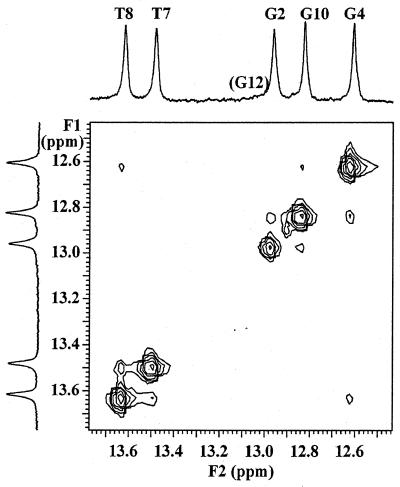
One-dimensional jump-return spectra (11) in H2O/D2O (9/1) had five sharp signals from guanine and thymine imino protons, indicative of strong hydrogen bonding in the base pairs of the duplex. The G1:H1 was broadened, due to fraying at the helix end. Those exchangeable protons were assigned in a two-dimensional watergate NOESY experiment (12). The five sharp signals could be identified via imino–imino contacts and to H1′ resonances. Strong NOE from adenine H2 and the thymine iminosignals in the same base pair, and weaker ones to iminoprotons of neighbouring base pairs, confirmed their assignment.
RESULTS
RNase H is found in all organisms. The ability of RNase H to recognise and cleave the RNA strand of a DNA/RNA hybrid has found its major application in antisense technology. The successful use of phosphorothioate oligonucleotides as antisense constructs proves the central role of RNase H in this strategy. For the increase in potency of antisense oligonucleotides the strategy to increase the affinity of the antisense oligomer for its target while fully retaining RNase H activity has been followed. Increase in affinity for the target RNA, without inducing RNase H activity, does not result in an equal increase in potency (2).
The structural requirements of oligonucleotide hybrids to recognise and be cleaved by RNase H is well studied. Although the antisense technology makes use of mammalian RNase H, most studies have been performed by E.coli RNase H1 because it is more easy available. The substrate requirements for E.coli and mammalian RNase H are similar (13). Lima and Crooke (13) described that E.coli RNase H bind A-form duplexes 60-fold higher than B-form duplexes. It has become clear that, for an RNA/modified-DNA duplex to become a substrate for RNase H, the functional groups present in the minor groove of the duplex should not inhibit binding of the enzyme, the 2′-OH group of the target RNA should stay accessible to the enzyme and the duplex should have enough flexibility to allow optimal RNase H cleavage activity (14). In agreement with this analysis is the presence of 2′-OMe modifications in the DNA strand abolishing RNase H cleavage (15) and conformational rigid oligonucleotides such as HNA (16) and LNA (M.D.Sørensen, L.Kvaernø, T.Bryld, A.E.Håkansson, G.Gaubert, B.Verbeure, P.Herdewijn and J.Wengel, manuscript in preparation) being very poor RNase H inducers. As mentioned before, CeNA might fulfill these requirements.
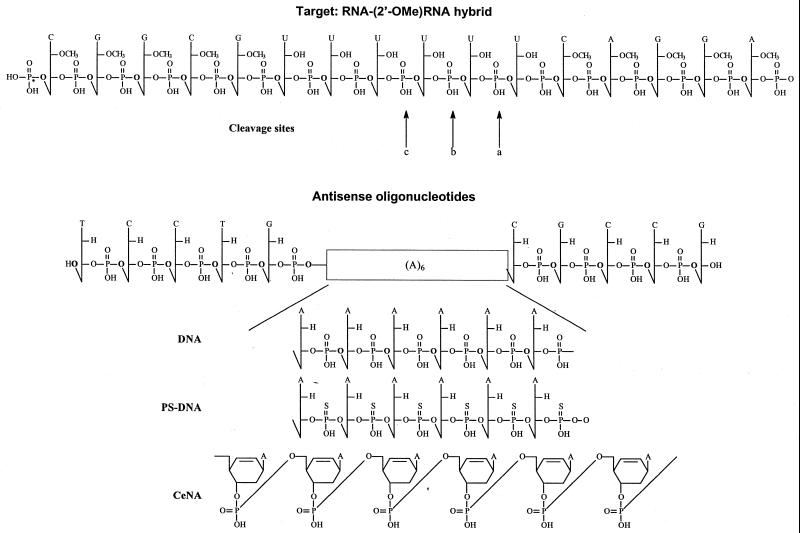
We studied the cleavage kinetics of hexadecamers comprised of a centred six-base ‘ribo U-gap’ flanked with 2′-OMe modified sequences as target (Fig. 2). The hexamer gap was selected to provide RNase H with a binding and cleavage site and to limit the number of initial cleavage sites (1).
Figure 2.
Structure of the RNA target and of the modified antisense oligonucleotides. The cleavage sites of RNase H are indicated with arrows.
The antisense strand is comprised of 2′-deoxynucleotides with a (A)6 gap where A is either a 2′-deoxynucleotide, a 2′-deoxynucleotide phosphorothioate or a cyclohexenyl nucleotide (Fig. 2).
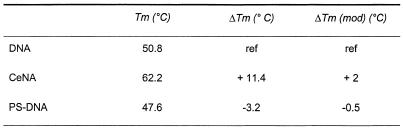
To avoid results being influenced by the different thermal stabilities of the oligonucleotide duplexes at working temperature (37°C), we first determined the Tm of the three complexes (Table 1). The Tm of the modified RNA/DNA duplex is 50.8°C. As expected, the Tm of the modified RNA/PS-DNA duplex is ∼3°C lower. Incorporation of six cyclohexenyl nucleotides into the DNA sequence gives a Tm of 62.2°C, which means a ΔTm of about +2°C per modification. The stabilisation effect of cyclohexenyl nucleotides on RNA/DNA duplexes has been described before (5). In all cases Tm values of the duplexes are much higher than working temperature for RNase H assay.
Table 1. Tm of duplexes shown in Figure 2.
Tm values were taken in 0.1 M NaCl, phosphate buffer pH 7.5 at oligonucleotide concentration of 4 µM. ΔTm is given with respect to the modified RNA/DNA duplex.
It is well documented that the activity of RNase H is dependent on the conformation of the duplex (14). To study the structural influence of incorporation of a CeNA in a natural DNA duplex, a high resolution NMR study was performed on the self-complementary dodecamer d(CGCGAA′TTCGCG), where the sixth nucleoside A′ (A′6) is a d-cyclohexenyl-adenine residue. The potential for induction of local conformational changes in a duplex by a 3′-endo preorganised nucleotide can be revealed more clearly when incorporated into dsDNA than when incorporated into a DNA:RNA or dsRNA duplex. The obtained results were compared to the well known DNA Dickerson dodecamer with the same nucleotide sequence and a comparable chimeric duplex [d(CGC)r(AAA)d(TTTGCG)]2.
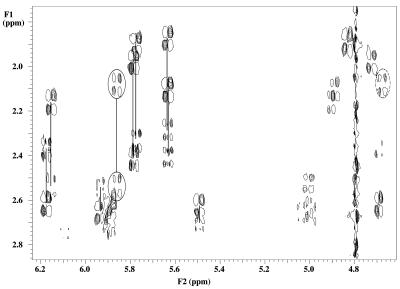
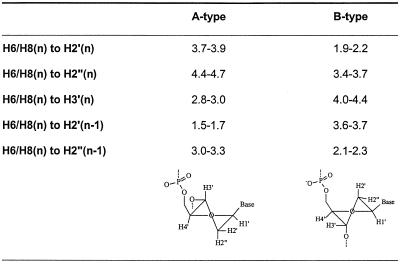
The sugar conformation of DNA residues can be determined from the values of J1′-2′, J2′′-3′ and J3′-4′, which are good indicators for the deoxyribose sugar puckering. For S-type sugars J1′-2′ is large (∼10 Hz) while for N-type sugars it is small (∼1 Hz); the opposite is true for J3′-4′ and J2′′-3′. Intermediate values of the coupling constants in flexible regions such as helix ends and hairpin loops are considered as an average that reflects a rapid interconversion of the furanose ring between S- and N-type conformation. Intermediate values of coupling constants in more rigid parts such as junctions in DNA–RNA chimeric duplexes (17) and DNA:RNA hybrids (18) are explained as an intermediate conformation of the deoxyribose ring that is neither N- or S-type but O4′-endo (E-type). All DNA residues, except for T7, showed the typical pattern for an S-type sugar in the DQF-COSY spectrum (residues 2 and 10 could not be evaluated due to overlap). In residue T7, both H1′ to H2′ and to H2″ have a comparable crosspeak pattern with an observed active coupling of ∼6.5 Hz (Fig. 3). This corresponds to a 40–50% N-type conformer in a fast equilibrium between S- and N-type puckering. The sum of coupling constants over H1′ (∼13 Hz) and H3′ (∼16 Hz) are also in agreement with 40–50% N-type conformer (6). However, there is no reason to assume ‘high flexibility’ in this central part of the duplex where strong hydrogen bonding between the base pairs occurs, as indicated by the strong and sharp iminoproton resonances of T7 and T8 depicted in Figure 4. The similar values of JH1′-H2′ and JH1′-H2′′ in combination with the intermediate value of JH3′-H4′ and strong JH2′-H3′ are most likely caused by an O4′-endo conformation as described for deoxyriboses at the junction DNA–RNA chimeric duplexes and DNA:RNA hybrids.
Figure 3.
Expansion of the DQF-COSY spectrum at 20°C showing the H2′ and H2″ to H1′ and H3′ crosspeaks in deoxyribose sugars of the studied duplex. the resolved H2′ and H2″ to H1′ crosspeaks from each sugar ring are connected with lines. Cross signals from residue T7 are encircled with solid and dotted lines for H1′ and H3′, respectively.
Figure 4.
Expansion of the iminoproton region in a two-dimensional watergate NOESY spectrum at 20°C, showing the imino–imino contacts in the central part of the duplex.
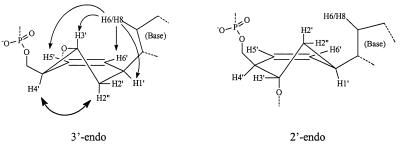
The conformation of the modified nucleotide itself (A′6) was mainly determined by intraresidue NOE contacts. Strong NOE from the base H8 to H3′ and H5′/H6′ cyclohexene protons in combination with NOE interaction of H2″ with H4′ are only possible when the cyclohexene ring is predominantly in a 3′-endo-like conformation as depicted in Figure 5. The weak J couplings of H1′ to both H2′ and H2″ are also in agreement with such a conformation, since in a 2′-endo conformation H1′ and H2′ have an axial position, which should result in strong J coupling between both protons. Coupling constants of cyclohexene H3′ to H4′ and H2″ are expected to be significantly different for both conformational types. However, this H3′ proton resonates exactly under the water signal in the experimental conditions used, hampering a reliable measurement on any of its crosspeaks in the COSY spectra.
Figure 5.
The 3′-endo form and 2′-endo form of a cyclohexenyl nucleotide. A′6 in the self-complementary sequence d(CGCGAA′6TTCGCG) occurs in the 3′-endo conformation.
Relative intensities of the sequential and intraresidue H6/H8 to H1′ protons differ in A- and B-type helices (Table 2). However, low dispersion of A5:H2″, A′6:H2′, T7:H2′ and T8:H2′ prevented the use of this difference in the modified region of the duplex. In the unmodified part of the duplex (C1→G4 and C9→G12), the H6/H8(n) to H2′(n) were the most intense and stronger than the sequential H6/H8(n) to H2′(n–1), which were stronger than the H6/H8(n) to H2″(n–1). These data correspond to a B-type conformation in the unmodified part of the duplex. Intensities of intraresidue H3′ to aromatic H6/H8 are also characteristic for the sugar puckering in A- and B-type helices (Table 2) (19). Only in T7 and A′6 is this NOE crosspeak intense, indicative of an N-type character of the ‘sugar’ ring in those residues.
Table 2. Intraresidue and interresidue, interproton distances Å for canonical A- and B-DNA.
The sugar phosphate backbone clearly undergoes a conformational change in the central part of the duplex. In the [1H,31P]-HETCOR, the 31P resonance is downfield shifted at the A5 to A′6 as well as at the A′6 to T7 nucleotide linkage, indicative of altered α and/or ζ dihedral angles that are not directly measurable by NMR but are considered to influence the 31P chemical shift (20). The crosspeak pattern of T7:H5′ to T7:P is different from that occuring in regular B-type DNA. A strong passive T7:H5′ to T7:H4′ is present as typical for a rather exotic γ t conformation.
The effect of incorporation of a ribonucleotide into an oligodeoxynucleotide is sequence dependent. Incorporation of a single RNA residue in d(GCGTATACGC)2 (21) or d(CCGGCGCCGG)2 (22) drive the conformational equilibrium to the A-form (as determined by X-ray diffraction), while a single RNA insertion into d(CGCTAGCG)2 has no effect on the overall conformation (which remains a B-form) and only the sugar pucker of the ribonucleotide is C3′-endo (as determined by NMR spectrometry) (23). It has been observed that incorporation of a conformational preorganized N-type nucleotide into an oligodeoxynucleotide increases the thermal stability of the DNA/RNA duplex. The local conformational change is transmitted to the neighbouring nucleotides (24,25). A similar, but less pronounced, effect is seen with cyclohexenyl A. The observation that a single cyclohexene nucleotide adopts an N-type conformation when incorporated in dsDNA is an indication that a CeNA:RNA duplex might be of the A-type.
Next, we determined the cleavage rate of the modified RNA target by the three antisense oligonucleotides. The RNA target has 2′-OMe wings. It was demonstrated before that 2′-OMe wings increase the stability of duplexes, but that cleavage of the RNA by RNase H is less efficient using chimeric duplexes (1). However, the lower initial rate of cleavage facilitates experimental setup, and all data were obtained at 37°C. The cleavage rate for the full substrate is predominantly the product of two cleavage sites (Fig. 6). The first cleavage occurred between the first and second nucleotide of the RNA substrate closest to the 3′-end of the U6 gap (Fig. 2, arrow ‘a’). A second cleavage site is situated between the second and third nucleotide (using DNA and PS-DNA antisense) and between the third and fourth nucleotide (using CeNA, DNA and PS-DNA antisense) (Fig. 2, arrows ‘b’ and ‘c’). Other cleavage sites are apparently of minor importance.
Figure 6.
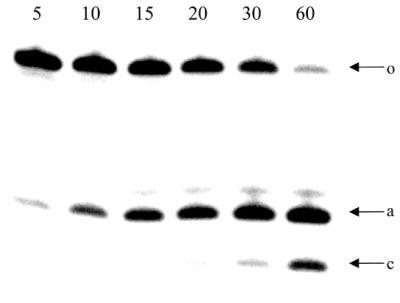
RNase H induced cleavage pattern of RNA–(2′-OMe)RNA hybrid in the presence of the CeNA antisense oligonucleotide: samples were taken at different time intervals (5, 10, 15, 20, 30 and 60 min); Arrows: o, full substrate; a, cleavage between rU1 and rU2; c, cleavage between rU3 and rU4.
It has been reported previously that the RNase H of the HIV RT induces an endonucleolytic cleavage, followed by exonucleolytic degradation of the RNA strand in the DNA–RNA duplex (26). It is not clear if the second product formed is the result of initial endonucleolytic cleavage or the result of the subsequent exonucleolytical degradation.
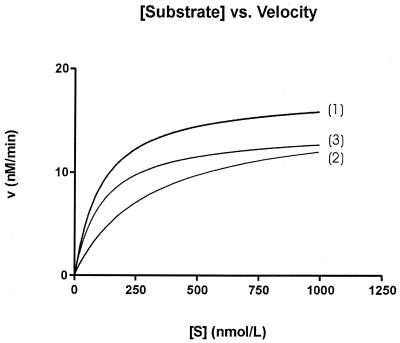
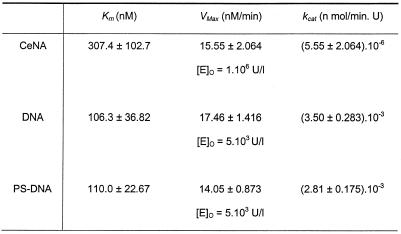
The plot of the reaction velocity as a function of the substrate concentration for the three complexes is given in Figure 7. Table 3 gives the different parameters. The differences in Km, Vmax and kcat for phosphorothioate and phosphodiester gapmers are small. The Km, Vmax and kcat are 106.3 nM, 17.5 nM/min and 3.5 × 10–3 nM/min.U (for phosphorothioate gapmer) and 110.0 nM, 14.15 nM/min and 2.8 × 10–3 nM/min.U (for phosphodiester gapmer), respectively. Both reaction kinetics were determined using the same enzyme concentrations.
Figure 7.
RNase H mediated cleavage kinetics of an RNA window flanked by (2′-OMe)RNA by DNA (1), CeNa (2) and PS-DNA (3). The structures of the oligonucleotides are depicted in Figure 2.
Table 3. Kinetic parameters of the cleavage of the three double-stranded gapmers, depicted in Figure 2, by E.coli RNase H.
The CeNA gapmer shows the same Vmax (15.6 nM/min) and an increased Km (307.4 nM) than the other antisense oligonucleotides. However, enzyme concentrations used to obtain these data were 200 times higher, which is reflected in a kcat that is three orders of magnitude lower than the kcat of the reference oligomers. Thus, the maximal catalytic rate of CeNA is significantly lower than that of phosphodiester and phosphorothioate oligonucleotides.
DISCUSSION
Permanent inactivation of the RNA target by an antisense oligonucleotide using the RNase H degradation mechanism is a very attractive approach for inhibition of gene expression. The success of this strategy, however, is dependent on several factors, which we do not have in hand and which are difficult to predict. The RNase H contribution to antisense effect varies from one cell type to another and from one target site to another and its contribution is dependent on the way the oligonucleotide is administered to the cell (27). The modified oligonucleotides that are enzymatically stable and are able to activate RNase H are limited.
Phosphorothioate oligonucleotides, showing fast cleavage kinetics, have three disadvantages. The affinity for the RNA target is reduced when compared with phosphodiester oligonucleotides. Excess phosphorothioates are detrimental to antisense activity when RNase H degradation is the terminating mechanism because large amounts of single-stranded phosphorothioates inhibit the enzyme (1). Phosphorothiate oligomers may hybridise with regions of partial complementarity and, due to their fast kinetics, induce RNase H mediated cleavage of non-targeted mRNA resulting in severe side effect or in loss of selectivity (in case the antisense drug is directed at a point mutation) (27). 2′-Fluoro-2′-deoxy-arabinofuranosyl nucleic acid (2′F-ANA) is the first example of a fully sugar-modified nucleic acid that increases duplex stability and activates RNase H (28). This has been explained by the O4-endo sugar conformation of the 2′-fluoro-2′-deoxy-arabinofuranosyl nucleotides in the 2′F-ANA strand, which is assumed to be an energetically favourable conformation allowing the necessary conformational changes of the duplex with minimal energy penalty for RNase H interaction and function (14). Indeed, cleavage of the phosphodiester function is expected to occur via an axial in-line displacement of the 5′-phosphoryl leaving group and conformational changes of the local helical structure are, most probably, needed for this process to occur. The cleavage kinetics (kcat) for 2′F-ANA are, at present, not published. The potential liver toxicity of C-5 propynyl-modified phosphorothioates has hampered further development of these oligomers (29).
Cyclohexenyl nucleic acid is a second example of an oligonucleotide with fully modified backbone that shows enhanced affinity for its RNA target while retaining RNase H activity. Structurally, it is unique because the furanose ring is replaced here by a six-membered unsaturated ring. The antisense activity and potential toxicity of these oligomers has not been fully evaluated yet. Structural studies, using NMR spectrometry, of a cyclohexenyl A incorporated at the sixth position in d(CGCGAA′6TTCGCG) demonstrates that cyclohexenyl A occurs in a 3′-endo mimicking conformation. The unmodified C/G part of the duplex remains in the B-type conformation while the T nucleotides flanking A′6 adopt an O4′-endo conformation. Cyclohexenyl A is, thus, able to induce conformational changes in an oligodeoxynucleotide. This, together with its presumed flexibility (5), argues for its potential to activate RNase H when incorporated in an antisense oligonucleotide. The kinetic data obtained from a gapmer approach using CeNA in the antisense window demonstrate that Vmax and Km are similar to those of phosphodiester and phosphorothioate oligonucleotides. The Km values are comparable with those found for ribonuclease A but 104-fold less favourable when compared with a ribozyme (30). The kcat of CeNA, however, is 630 and 500 times lower than the kcat of the phosphodiester and phosphothioate analogues, respectively. The RNase H activity has been studied with an (A/rU)6 sequence. It is known that duplex stability, conformation and susceptibility to RNase H varies with the sequences considered and the overall distribution of pyrimidines and purines between the two strands (31,32). A definitive conclusion about their potential to promote RNase H activity on RNA target in antisense experiments should wait for more studies with hybrids of mixed sequences and the use of mammalian RNase H.
It is difficult to draw definitive conclusions from these results with respect to the potency of CeNA as antisense oligonucleotides. One can argue that stronger binding of modified oligonucleotide to the RNA target may also imply a stronger binding to sites of partial complementarity. The slower cleavage kinetics, however, may result in fewer side effects due to cleavage of the non-targeted sequences. The eventual advantage of CeNA as an antisense drug will also be dependent on the terminating mechanism and thus on the selected target. Increasing affinity for the target with a reduced efficiency of cleavage might result in a better antisense molecule when binding of the oligonucleotide to its target RNA is rate limiting (1). However, when the rate limiting step in the antisense experiment is RNase H cleavage, CeNA will fail to result in increased potency when compared with phosphothioate oligonucleotides.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Chantal Biernaux for editorial help. This research was supported by a grant from the Katholieke Universiteit Leuven (GOA 97/11)
REFERENCES
- 1.Crooke S.T., Lemonidis,K.M., Neilson,L., Griffey,R., Lesnik,E.A. and Monia,B.P. (1995) Kinetics characteristics of Escherichia coli RNase H1: cleavage of various antisense oligonucleotide-RNA duplexes. Biochem. J., 312, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima W.F., Mohan,V. and Crooke,S.T. (1997) The influence of antisense olignucleotide-induced RNA structure on Escherichia coli RNase H1 activity. J. Biol. Chem., 272, 18191–18199. [DOI] [PubMed] [Google Scholar]
- 3.Gao W.Y., Han,F.S., Storm,C., Egan,W. and Cheng,Y. (1992) Phosphorothioate oligonucleotides are inhibitors of human DNA polymerases and RNase H: implications for antisense technology. Mol. Pharmacol., 41, 223–229. [PubMed] [Google Scholar]
- 4.Rait V.K. and Ramsay Shaw,B. (1999) Boranophosphates support the RNase H cleavage of polyribonucleotides. Antisense Nucleic Acid Drug Dev., 9, 53–60. [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Verbeure,B., Luyten,I., Lescrinier,E., Froeyen,M., Hendrix,C., Rosemeyer,H., Seela,F., Van Aerschot,A. and Herdewijn,P. (2000) Cyclohexene nucleic acids (CeNA): serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA. J. Am. Chem. Soc., 122, 8595–8602. [Google Scholar]
- 6.Wijmenga S.S., Mooren,M.W. and Hilbers,C.W. (1993) NMR of nucleic acids; from spectrum to structure. In Roberts,G. (ed.), NMR of Macromolecules: A Practical Approach. Oxford University Press, Oxford, UK, pp. 217–288.
- 7.Jeener J., Meier,B.H., Bachmann,P. and Ernst,R.R. (1979) Investigation of exchange processes by two dimensional NMR spectroscopy. J. Chem. Phys., 71, 4546–4553. [Google Scholar]
- 8.Griesinger C., Otting,G. and Wüthrich,K. (1988) Clean TOCSY for 1H spin system identification in macromolecules. J. Am. Chem. Soc., 110, 7870–7872. [Google Scholar]
- 9.Rance M., Sørensen,O.W., Bodenhausen,G., Wagner,G., Ernst,R.R. and Wüthrich,K. (1983) Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun., 117, 479–485. [DOI] [PubMed] [Google Scholar]
- 10.Sklenar V., Miyashiro,H., Zon,G., Miles,H.T. and Bax,A. (1986) Assignment of the 31P and 1H resonances in oligonucleotides by two dimensional spectroscopy. FEBS Lett., 208, 94–98. [DOI] [PubMed] [Google Scholar]
- 11.Plateau P. and Guéron,M. (1982) Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc., 104, 7310–7311. [Google Scholar]
- 12.Piotto M., Saudek,V. and Sklenar,V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. V. J. Biomol. NMR, 2, 661–665. [DOI] [PubMed] [Google Scholar]
- 13.Lima W.F. and Crooke,S.T. (1997) Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide-RNA hybrids. Biochemistry, 36, 390–398. [DOI] [PubMed] [Google Scholar]
- 14.Zamaratski E., Pradeepkumar,P.I. and Chattopadhyaya,J. (2001) A critical survey of the structure-function of the antisense oligo.RNA hybrid duplex as substrate for RNase H. J. Biochem. Biophys. Meth., 48, 187–203. [DOI] [PubMed] [Google Scholar]
- 15.Inoue H., Hayase,Y., Iwai,S. and Ohtsuka,E. (1988) Sequence-specific cleavage of RNA using chimeric DNA splints and RNase H. Nucleic Acids Symp. Ser., 19, 135–138. [PubMed] [Google Scholar]
- 16.Hendrix C., Rosemeyer,H., De Bouvere,B., Van Aerschot,A., Seela,F. and Herdewijn,P. (1997) 1′,5′-Anhydrohexitol oligonucleotides: hybridization and strand displacement with oligoribonucleotides, interaction with RNase H and HIV reverse transcriptase. Chem. Eur. J., 3, 1513–1520. [Google Scholar]
- 17.Hsu S.T., Chou,M.T. and Cheng,J.W. (2000) The solution structure of [d(CGC)r(aaa)d(TTTGCG)]2: hybrid junctions flanked by DNA duplexes. Nucleic Acids Res., 28, 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar M., Fedoroff,J.M.M., Ribiero,N.S. and Reid,B.R. (1993) The DNA strand in DNA.RNA hybrid duplexes is neither B-form nor A-form in solution. Biochemistry, 32, 4207–4215. [DOI] [PubMed] [Google Scholar]
- 19.Goljer I. and Bolton,P.H. (1994) Studies of nucleic acids structures based on NMR results. In Croasmun,W.R. and Carlson,R.M.K. (eds), Two Dimensional NMR Spectroscopy: Applications for Chemists and Biochemists. VCH Publishers, Inc., New York, NY, 699–740.
- 20.Gorenstein D.G. (1992) 31P NMR of DNA. Methods Enzymol., 211, 254–286. [DOI] [PubMed] [Google Scholar]
- 21.Egli M., Usman,N. and Rich,A. (1993) Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA–RNA chimeric duplexes. Biochemistry, 32, 3221–3237. [PubMed] [Google Scholar]
- 22.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1994) A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA–RNA chimeric decamer duplex d(CCGGC)r(G)d(CCGG) with a novel intermolecular GC base-paired quadruplet. J. Mol. Biol., 236, 275–285. [DOI] [PubMed] [Google Scholar]
- 23.Jaishree T.N., van der Marel,G.A., van Boom,J.H. and Wang,A.H.J. (1993) Structural influence of RNA incorporation in DNA: quantitative nuclear magnetic resonance refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG). Biochemistry, 32, 4903–4911. [DOI] [PubMed] [Google Scholar]
- 24.Pradeepkumar P.I., Zamaratski,E., Földesi,A. and Chattopadhyaya,J. (2001) Transmission of the conformational information in the antisense/RNA hybrid duplex influences the pattern of the RNase H cleavage reaction. Tetrahedron Lett., 41, 8601–8607. [Google Scholar]
- 25.Bondensgaard K., Petersen,M., Singh,S.K., Rajwanshi,V.K., Kumar,R., Wengel,J. and Jacobsen,J.P. (2000) Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chem. Eur. J., 6, 2687–2695. [DOI] [PubMed] [Google Scholar]
- 26.Schatz O., Mous,J. and Le Grice,S.F. (1990) HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′–5′ exonuclease activity. EMBO J., 9, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toulmé J.J. and Tidd,D. (1998) Rate of ribonuclease H in antisense oligonucleotide-mediated effects. In Crouch,R.J. and Toulmé,J.J. (eds), Research in Ribonucleases H. INSERM, Paris, France, pp. 225–250.
- 28.Damha M.J., Wilds,C.J., Noronha,A., Brukner,I., Borkow,G., Arion,D. and Parniak,M.A. (1998) Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of ribonuclease H. J. Am. Chem. Soc., 120, 12976–12977. [Google Scholar]
- 29.Flanagan W.M., Wolf,J.J., Olson,P., Grant,D., Lin,K.-Y., Wagner,R.W. and Matteucci,M.D. (1999) A cytosine analog that confers enhanced potency to antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 96, 3513–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratmeyer L., Vinayak,R., Zhong,Y.-Y., Zon,G. and Wilson,W.D. (1994) Sequence specific thermodynamic and structural properties of DNA cntdot RNA duplexes. Biochemistry, 33, 5298–5304. [DOI] [PubMed] [Google Scholar]
- 32.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Solution structures of DNA·RNA hybrids with purine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]