Abstract
To begin to understand the role of the lipooligosaccharide (LOS) molecule in chancroid infections, we constructed mutants defective in expression of glycosyltransferase genes. Pyocin lysis and immunoscreening was used to identify a LOS mutant of Haemophilus ducreyi 35000. This mutant, HD35000R, produced a LOS molecule that lacked the monoclonal antibody 3F11 epitope and migrated with an increased mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Structural studies indicated that the principal LOS glycoform contains lipid A, Kdo, and two of the three core heptose residues. HD35000R was transformed with a plasmid library of H. ducreyi 35000 DNA, and a clone producing the wild-type LOS was identified. Sequence analysis of the plasmid insert revealed one open reading frame (ORF) that encodes a protein with homology to the WaaQ (heptosyltransferase III) of Escherichia coli. A second ORF had homology to the LgtF (glucosyltransferase) of Neisseria meningitidis. Individual isogenic mutants lacking expression of the putative H. ducreyi heptosyltransferase III, the putative glucosyltransferase, and both glycosyltransferases were constructed and characterized. Each mutant was complemented with the representative wild-type genes in trans to restore expression of parental LOS and confirm the function of each enzyme. Matrix-assisted laser desorption ionization mass spectrometry and SDS-PAGE analysis identified several unique LOS glycoforms containing di-, tri-, and poly-N-acetyllactosamine repeats added to the terminal region of the main LOS branch synthesized by the heptosyltransferase III mutant. These novel H. ducreyi mutants provide important tools for studying the regulation of LOS assembly and biosynthesis.
Haemophilus ducreyi is a gram-negative bacterium which causes chancroid, a sexually transmitted disease (STD). Although this infection is uncommon in the United States, it is a major cause of genital ulcer disease in developing countries worldwide (40). Recently, it was reported that ulcerative sexually transmitted diseases, such as chancroid, serve as cofactors for human immunodeficiency virus (HIV) transmission, inreasing the risk of acquiring HIV infection two- to fivefold (11). Histological analyses of genital ulcers resulting from H. ducreyi infection have increased numbers of CD4+ lymphocytes and therefore may increase a person's susceptibility to HIV infection (39). Another disturbing fact regarding H. ducreyi infections is the emergence of antibiotic-resistant strains in areas where this disease is more prevalent (22, 40). These factors have stimulated increasing research efforts designed to understand the mechanisms and virulence factors involved in the pathogenesis of chancroid.
Although little is known about the bacterial components of H. ducreyi that contribute to colonization and subsequent ulcer formation, several putative virulence factors have been described. Two cytotoxins, a hemolysin with cytotoxic activity and a diffusible cytotoxin, were shown to be toxic to human foreskin fibroblasts and epithelial cells in vitro (2, 12, 32–34). Also, unique pili have been described which may function in attachment (38). Another putative virulence factor is the lipooligosaccharide (LOS) molecule, which has been a primary focus of our research (29). Structural analysis has demonstrated that the principal LOS glycoform of H. ducreyi shares common epitopes with the LOS of other mucosal pathogens, such as Neisseria gonorrhoeae, Neisseria meningitidis, and Haemophilus influenzae (9, 29). More importantly, these LOS epitopes have been implicated as important virulence factors for these latter human pathogens.
There are several lines of evidence which suggest that H. ducreyi LOS plays a role in the pathogenesis of chancroid. Our previous data demonstrated that injection of purified LOS causes intradermal inflammation in experimental animal models (10). Also, H. ducreyi mutants expressing truncated LOS molecules exhibit reduced virulence in the temperature-dependent rabbit model of infection (4, 5). Zaretzky and Kawula reported that purified LOS induced interleukin-8 expression from HaCaT cells in vitro, which could stimulate an inflammatory response that may indirectly lead to lesion formation (45). Alfa and DeGagne showed that a high concentration of pure LOS could inhibit H. ducreyi adherence to human foreskin fibroblasts in vitro (1). In addition, a Tn916 H. ducreyi mutant, with a disruption in a d-glycero-d-manno-heptose heptosyltransferase gene, exhibited reduced adherence and invasion of human keratinocytes in vitro (18).
In this study, we used pyocin lysis to initially identify LOS mutants of H. ducreyi 35000. We have previously shown that this method can select organisms that express truncated LOS molecules (8), suggesting that these bacteria contain defects in the biosynthetic pathway or assembly of this principal glycolipid. One LOS mutant, termed HD35000R, was isolated and used to clone and sequence two separate genes involved in LOS biosynthesis. The first gene codes for a protein which has homology to heptosyltransferase III of Escherichia coli (20, 44), while the second gene codes for a protein with homology to β1,4-glucosyltransferase of N. meningitidis (24). Individual isogenic mutants with disruptions in the glucosyltransferase, the heptosyltransferase, and both glycosyltransferase genes were constructed, and structures of the resulting LOS glycoforms were determined. Restoration of the wild-type LOS was accomplished by providing the wild-type H. ducreyi genes in trans in each isogenic mutant. These mutants will be important tools in providing a better understanding of the regulation and assembly of H. ducreyi LOS, which may lead to insight into the role of this molecule in pathogenesis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strains were grown at 37°C on Luria-Bertani (LB) agar plates or in LB broth. When needed, LB medium was supplemented with kanamycin, ampicillin, or chloramphenicol at a final concentration of 20, 50, or 30 μg/ml, respectively. H. ducreyi strains were cultured at 35°C in 5% CO2 on chocolate agar plates or in brain heart infusion broth as previously described (10). When needed, chocolate agar plates were supplemented with kanamycin (20 μg/ml), chloramphenicol (1 μg/ml), and/or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml). Pseudomonas aeruginosa strain C was grown in Pseudomonas broth (14).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Host strain used for sequencing | Life Technologies |
| XL1-Blue | Host strain used for cloning | Stratagene |
| H. ducreyi strains | ||
| 35000 | Wild-type strain isolated in Winnipeg; binds MAb 3F11 | 19 |
| HD35000R | LOS mutant derived from strain 35000 by pyocin selection; does not bind MAb 3F11 | This study |
| 35000hep− | Isogenic LOS mutant derived from strain 35000 by insertion of a cat cartridge into the XbaI site of the waaQ gene; binds MAb 3F11 | This study |
| 35000glu− | Isogenic LOS mutant derived from strain 35000 by insertion of a cat cartridge into the BglII site of the lgtF gene; does not bind MAb 3F11 | This study |
| 35000hepglu− | Isogenic LOS mutant derived from strain 35000 by insertion of a cat cartridge into the XbaI and BglII sites of the waaQ and lgtF genes: does not bind MAb 3F11 | This study |
| HD35000R (pLS88.8) | Strain derived from the electroporation of the pLS88 plasmid library of H. ducreyi into HD35000R; binds MAb 3F11 | This study |
| 35000hep−(pHEP) | Strain derived from the electroporation of pHEP into strain 35000hep− to complement the inactivated waaQ gene; binds MAb 3F11 | This study |
| 35000glu−(pGLU) | Strain derived from the electroporation of pGLU into strain 35000glu− to complement the inactivated lgtF gene; binds MAb 3F11 | This study |
| 35000hepglu−(pHEPGLU) | Strain derived from the electroporation of pHEPGLU into strain 35000hepglu− to complement the inactivated waaQ and lgtF genes; binds MAb 3F11 | This study |
| Plasmids | ||
| pLS88 | E. coli-H. ducreyi shuttle vector; Kanr, Smr, Sulr | 13 |
| pLS88.8 | pLS88 containing a 3.2-kb insert of H. ducreyi chromosomal DNA which complements HD35000R; contains 4 ORFs | This study |
| pRSM2072 | pRSM1791 suicide vector used for allele replacement with an improved multiple cloning site; trp′-′lacZ, Ampr | Robert Munson |
| pUCΔECAT | Plasmid that contains cat cartridge | Bruce Green |
| pCR2.1 | TA cloning vector | Invitrogen |
| pLS88.8CAThep | pLS88.8 with a cat cartridge inserted into the XbaI site within the waaQ gene | This study |
| pLS88.8CATglu | pLS88.8 with a cat cartridge inserted into the BglII site within the lgtF gene | This study |
| pLS88.8CAThepglu | pLS88.8 with a cat cartridge inserted into the XbaI and BglII sites within the waaQ and lgtF genes | This study |
| pGLU | pLS88 vector with a 1.2-kb DNA fragment containing the lgtF gene | This study |
| pHEP | pLS88 vector with a 1.2 kb DNA fragment containing the waaQ gene | This study |
| pHEPGLU | pLS88 vector with a 2.2-kb DNA fragment containing the waaQ and lgtF genes | This study |
| pMJFhep | pRSM2072 with the 4.4-kb NotI fragment from pLS88.8CAThep cloned into pRSM2072 | This study |
| pMJFglu | pRSM2072 with the 4.4-kb NotI fragment from pLS88.8CATglu cloned into pRSM2072 | This study |
| pMJFhepglu | pRSM2072 with the 4.3-kb NotI fragment from pLS88.8CAThepglu cloned into pRSM2072 | This study |
Pyocin isolation.
Pyocin was isolated from cultures of P. aeruginosa strain C by the method described by Morse et al. (31).
Pyocin lysis assay.
The pyocin selection assay was performed as described previously (8, 14). Clones resistant to lysis were tested for loss of reactivity with monoclonal antibody (MAb) 3F11. This antibody reacts to the terminal Gal-GlcNAc moiety conserved on the LOS of most gonococcal strains and also reacts with the LOS of most H. ducreyi strains (9, 28).
Complementation of HD35000R.
A plasmid library of H. ducreyi chromosomal DNA was previously constructed in the pLS88 shuttle vector which had been modified by the addition of NotI restriction sites (39a). This plasmid library was electroporated into HD35000R by a previously described procedure (6). Transformants were selected on chocolate agar containing kanamycin (20 μg/ml) and immunoscreened by probing nitrocellulose lifts with MAb 3F11.
Recombinant DNA techniques.
Plasmid isolations were performed using Qiagen purification kits (Qiagen, Chatsworth, Calif.). Restriction enzymes were purchased from New England Biolabs Inc., Beverly, Mass. T4 DNA ligase was purchased from Promega (Madison, Wis.). Restriction enzyme digests, ligations, transformations, and electroporation of E. coli were performed using standard methods (37). Electroporation of H. ducreyi was performed as previously described (6). PCR was used to produce double-stranded DNA probes and evaluate isogenic mutants. Approximately 100 ng of chromosomal DNA was used for the template. Annealing temperatures varied with primers used. Taq DNA polymerase was purchased from Fisher Scientific (Pittsburgh, Pa.).
Nucleotide sequence analysis.
Both strands of the 3.2-kb NotI insert of pLS88.8 were sequenced. Sequence analysis of the cloned genes was performed using MacVector 6.0 and the Wisconsin sequence analysis packages (Genetics Computer Group, Madison, Wis.).
Construction of isogenic mutants.
To mutate the H. ducreyi waaQ gene located on pLS88.8, pUCΔECAT was digested with EcoRI to remove the cat cartridge, treated with Klenow fragment, and ligated with pLS88.8, which had been digested with XbaI, and treated with Klenow fragment. To mutate the lgtF gene on pLS88.8, the cat cartridge from pUCΔCAT was isolated as a BamHI fragment and ligated to pLS88.8 which had been digested with BglII. For construction of mutations in both the waaQ and lgtF genes, pLS88.8 was digested with XbaI and BglII, liberating a 1.3-kb fragment containing portions of the waaQ and lgtF genes. The digested plasmid was then treated with Klenow fragment and ligated to the cat cartridge, which had also been blunt ended. Ligation mixtures were used to electroporate E. coli XL1-Blue, and transformants containing the cat cartridge in derivatives of pLS88.8 were selected on chloramphenicol and kanamycin. Plasmids with the correct restriction map were saved as pLS88.8CAThep, pLS88.8CATglu, and pLS88.8CAThepglu. Isogenic mutants of H. ducreyi 35000 were constructed as previously described by Bozue et al. (6). The NotI fragments from pLS88.8CAThep, pLS88.8CATglu, and pLS88.8CAThepglu were individually cloned into pRSM2072 (a derivative of pRSM1791 with an improved cloning site (L. Taratino and R. S. Munson, Jr., unpublished data) which had been digested with NotI. After transformation into E. coli, plasmids with the correct restriction map were saved as pMJFhep, pMJFglu, and pMJFhepglu. pMJFhep, pMJFglu, and pMJFhepglu were electroporated into H. ducreyi 35000, and cointegrates were selected on chocolate agar containing chloramphenicol. Isogenic mutants were selected by their ability to grow normally (large white colonies) on chocolate agar containing chloramphenicol and X-Gal. The isogenic mutants were designated 35000hep−, 35000glu−, and 35000hepglu−.
Southern blot analysis.
Chromosomal DNA was isolated from H. ducreyi strains using a modification of a previously described procedure (36). H. ducreyi strains were grown overnight on chocolate agar. Bacteria were harvested and resuspended in brain heart infusion broth, and chromosomal DNA was isolated. Chromosomal DNA was then digested to completion with HindIII or NcoI, electrophoresed on a 0.8% agarose gel, and then transferred to Immobilon-Ny+ membranes (Millipore, Bedford, Mass.) by capillary blotting overnight. Probes for the waaQ, lgtF, and cat genes were generated by PCR with the following primers. Primers P1 (5′-GATGCCTGTTGAGCCTCAGATTC-3′) and P2 (5′-TTGTTTACCGCTAGGGGGACAG-3′) were used to amplify a 505-bp internal probe to the H. ducreyi waaQ gene. A 571-bp internal probe to the lgtF gene was generated using primers P3 (5′-ACTCCGTGTACGATCCCATAAGTC-3′) and P4 (5′-TGGTCCACAGAGACAATTTGCTC-3′). To analyze 35000hep-glu, a 320-bp probe to a region upstream of waaQ was prepared by PCR using primers P5 (5′-TCCAACGATAATGAAAAAACTGCTC-3′) and P6 (5′-GATAGCGAATACCACTTTGCCAAG-3′). A 550-bp probe to the cat gene was also used in the Southern blot analysis. The templates for generation of the probes were pLS88.8 and p1710. DNA probes were biotinylated using a NEBlot Phototope labeling kit (New England Biolabs) as recommended by the manufacturer. Hybridizations were performed overnight at 64°C in 50 ng of denatured biotinylated probe per ml of 6× SSC–5× Denhardt's reagent, 0.5% sodium dodecyl sulfate (SDS) (20× SSC is 3 M NaCl plus 0.3 M sodium citrate [pH 7.0]). Washes were performed according to the NEBlot Phototope kit instruction manual. Detection was performed with a Phototope-Star detection kit (New England Biolabs).
Complementation of H. ducreyi waaQ and lgtF mutants.
pLS88.8 was digested with both MunI and EcoRI to remove the entire waaQ gene. This digestion was subjected to agarose gel electrophoresis followed by purification with a GeneClean II kit (Bio 101 Inc., La Jolla, Calif.). The 1.2-kb DNA fragment containing the waaQ gene was ligated to the pLS88 vector which had been previously digested with EcoRI. To construct a plasmid containing the lgtF gene, pLS88.8 was used as a template in a PCR to amplify a DNA fragment containing the lgtF gene, using primers P7 (5′-TAAAGGTGAACGGGAACGAGCG-3′) and P8 (5′-AATAGCACAAAAGGGGCGG-3′). The PCR product (1.2 kb) was then cloned into the pCR2.1 vector (Invitrogen). This fragment was excised by digestion with EcoRI, subjected to agarose gel electrophoresis followed by purification with a GeneClean II kit, and then ligated to EcoRI-digested pLS88. To construct a plasmid containing the waaQ and lgtF genes, pLS88.8 was digested with both MlsI and Eco1471 to remove open reading frames 3 and 4 (ORF3 and ORF4). This digestion was subjected to agarose gel electrophoresis followed by purification with a GeneClean II kit. The 6.7-kb DNA fragment containing the pLS88 vector and the two transferase genes was religated. Ligation mixtures were used to transform E. coli XL1-Blue. Plasmids were purified (Qiagen), and restriction analyses were performed to verify the constructs. The plasmids, pHEP, pGLU, and pHEPGLU, were used to electroporate 35000hep−, 35000glu−, and 35000hepglu−, respectively. Selection of complemented H. ducreyi mutants was accomplished using chocolate agar containing kanamycin.
Preparation and analysis of H. ducreyi LOS.
LOS from proteinase K-treated whole-cell lysates was resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on a 14% acrylamide gel and visualized by silver staining (10, 41). Western blot analysis was performed by transferring LOS to a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) using a previously described procedure (26). LOS for structural analysis was extracted by the modified hot phenol-water procedure from bacteria that were grown overnight in 1600 ml of broth and dried (3, 23, 43).
Analysis of outer membranes proteins.
Outer membrane proteins of H. ducreyi 35000 and the isogenic mutants were prepared by a previously described method (15, 25). Proteins were resolved by SDS-PAGE and stained with Coomassie blue (25).
Mass spectrometric analysis of H. ducreyi LOS.
LOS structures from H. ducreyi strains 35000 and the three knockout mutants 35000hep−, 35000glu−, and 35000hepglu−, along with the corresponding complemented strains, were analyzed by mass spectrometry. In each case, approximately 0.1 to 1.0 mg of LOS was converted to the corresponding water-soluble O-deacylated LOS glycoforms by treatment with hydrazine (37°C, 30 min) (21). Samples were then analyzed by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) using a PE Biosystems (Framingham, Mass.) Voyager DE time-of-flight mass spectrometer or a Voyager DESTR time-of-flight mass spectrometer as previously described (17). Both instruments were operated with a nitrogen laser (337 nm) in the negative-ion mode under delayed extraction conditions (42); delay time was 100 to 175 ns, and grid voltage was 92 to 94% of full acceleration voltage (20 to 30 kV). Samples were purified and desalted by drop dialysis using a 0.025-μm-diameter nitrocellulose membrane and/or by anion-exchange Zip TipsAX (Millipore). Approximately 0.1 to 0.2 μg of O-deacylated LOS was mixed with 1 μl of a 320 mM 2,5-dihydroxybenzoic acid in 4:1 acetone-water (vol/vol) containing 175 mM 1-hydroxyisoquinoline (30), desalted with cation-exchange resin beads (DOWEX, 50×, NH4+) and then air dried on a stainless steel target. Spectra were acquired and averaged (typically 20 to 50 laser shots), and mass was calibrated with an external calibrant consisting of an equimolar mixture of angiotensin II, bradykinin, luteinizing hormone-releasing hormone, bombesin, α-melanocyte-stimulating hormone (CZE mixture; Bio-Rad) and ACTH 1-24 (Sigma, St. Louis, Mo.).
To determine the identity and linkages of terminal sugars in several of these LOS preparations, O-deacylated LOS was subjected to treatment with specific glycosidases at 37°C and reanalyzed by MALDI-MS. Glycosidases used were neuraminidase type VI-A from Clostridium perfringens (Sigma), β-galactosidase from jack bean meal, and/or β-n-acetylhexosaminidase from jack bean meal (Oxford GlycoScience, Oxford, United Kingdom).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited with GenBank and assigned accession no. AF215936.
RESULTS
Identification of a LOS mutant using pyocin selection.
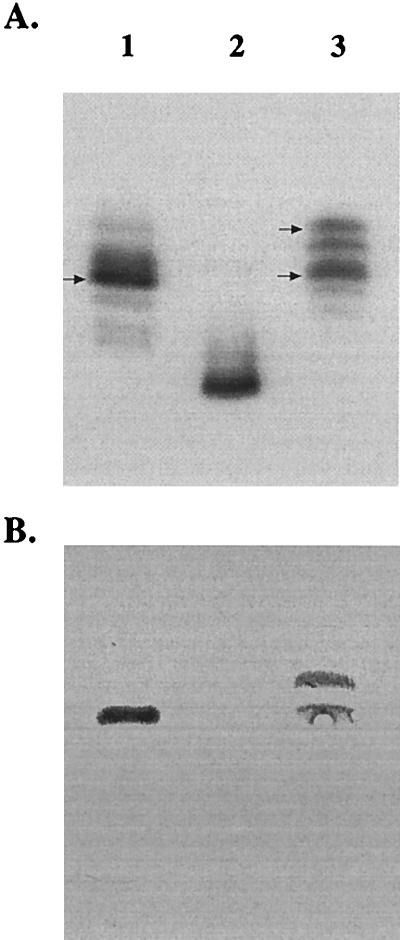
To identify genes involved in LOS biosynthesis, pyocin C was used to select for LOS mutants of H. ducreyi 35000 as described previously (8, 14). A single resistant colony was isolated and named HD35000R. Colony lift assay showed that this isolate did not react to MAb 3F11 (data not shown). MAb 3F11, developed to the LOS of N. gonorrhoeae strain 1291, reacts to an LOS epitope containing a terminal N-acetyllactosamine moiety conserved on over 90% of the H. ducreyi strains tested (9, 28). LOS from HD35000R was compared to the wild-type LOS by SDS-PAGE and Western blot analysis. The LOS from HD35000R (Fig. 1A, lane 2) migrated as a single band with a more rapid mobility than the LOS glycoforms produced by the parental strain (Fig. 1A, lane 1), which is indicative of a truncated LOS molecule. Western blot analysis demonstrated that the LOS of HD35000R lost reactivity to MAb 3F11 (Fig. 1B, lane 2), suggesting this structure lacks all or part of the terminal N-acetyllactosamine.
FIG. 1.
Composite of a silver-stained SDS-polyacrylamide gel (A) and the corresponding Western blot (B). The gel contains LOS from H. ducreyi 35000 (lane 1), HD35000R (lane 2), and the complemented pyocin mutant, HD35000R(pLS88.8) (lane 3). The blot was probed with MAb 3F11, demonstrating reactivity to wild-type LOS (lane 1) and with two glycoforms expressed by the complemented mutant HD35000R(pLS88.8) (lane 3). The LOS glycoforms that react with the MAb are marked with arrows.
MALDI-MS analysis of O-deacylated LOS from strain HD35000R identified two major deprotonated molecular ions, (M − H)− at m/z 1,759.6 and m/z 1,636.8, and a minor (M − H)− peak at m/z 1,882.1. These masses are consistent with an LOS structure that terminated with a core consisting of only two of the three heptoses with no additional branch structures, Hep2-KdoP(PEA)1,0-lipid A, and containing a variable number of phosphoethanolamine (PEA; 123 Da) (data not shown). These results suggest that H. ducreyi 35000R contains a defect which has affected the biosynthesis and/or assembly of LOS.
Complementation of H. ducreyi 35000R.
A plasmid library of H. ducreyi 35000 DNA, constructed in a derivative of the shuttle vector pLS88, was electroporated into HD35000R (Sun et al., submitted). Kanamycin-resistant transformants were screened for reactivity to MAb 3F11. Plasmid DNA was isolated from a positive transformant and electroporated back into HD35000R to confirm that this plasmid was responsible for the phenotype. The LOS from the complemented mutant, HD35000R(pLS88.8), was analyzed by SDS-PAGE and Western blotting. Figure 1A shows that the LOS isolated from HD35000R(pLS88.8) (Fig. 1A, lane 3) had an SDS-PAGE profile similar to that of the LOS of H. ducreyi 35000 (lane 1). However, Fig. 1A also shows that the complemented strain synthesized other larger glycoforms (lane 3) that were not readily apparent in the wild-type LOS (lane 1). Figure 1B shows that the LOS of HD35000R (pLS88.8) (lane 3) has reacquired the epitope recognized by MAb 3F11, indicating that the gene(s) present in the insert of plasmid pLS88.8 is sufficient to complement the LOS defect. Unexpectedly, MAb 3F11 also reacted with a slower-migrating LOS glycoform (Fig. 1B, lane 3) which appeared to contain di-N-acetyllactosamine, based on subsequent MALDI-MS analysis presented below.
Nucleotide sequence analysis of pLS88.8.
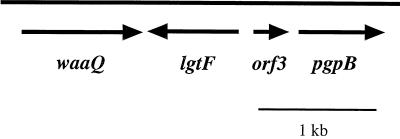
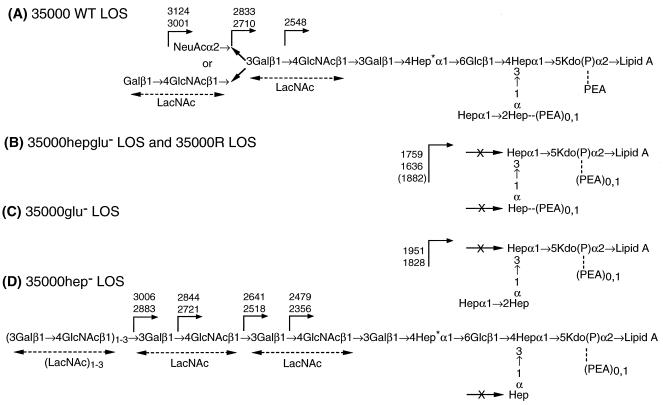
The 3.2-kb DNA insert in pLS88.8 was sequenced and found to contain four complete ORFs (Fig. 2). ORF1 encodes a polypeptide of 344 amino acids, with a predicted molecular mass of 38.5 kDa, and shares 20% identity and 37% similarity to the waaQ gene product of E. coli (20, 44). The E. coli waaQ gene encodes heptosyltransferase III. On the basis of sequence homologies and LOS structural analysis (see below), we conclude that ORF1 encodes a heptosyltransferase III which adds the third heptose of the triheptose core of H. ducreyi 35000 LOS.
FIG. 2.
ORF map of the sequenced portion of pLS88.8. The 3.2-kb DNA insert was found to contain four ORFs. ORF1 is the putative heptosyltransferase III (waaQ); ORF2 is the putative glucosyltransferase (lgtF); ORF3 has homology to hypothetical protein HI1333 of H. influenzae; ORF 4 shares homology with a phosphatidylglycerophosphate phosphatase B (pgpB) of E. coli. Arrows represent the direction of transcription.
A second ORF (ORF2), located immediately downstream of the waaQ homologue, is transcribed from the opposite strand. The derived amino acid sequence of ORF2 is 32% identical and 49% similar to a glucosyltransferase (LgtF) of N. meningitidis (24). Again, this sequence homology and the structural analysis of HD35000R LOS indicate that ORF2 encodes the enzyme responsible for the addition of the first glucose to the heptose core of the oligosaccharide branch via a β1,4 linkage. The next ORF (ORF3) was 82% identical and 91% similar to a hypothetical protein (HI1333) of H. influenzae, with no known function (16). Seventy-six base pairs downstream of ORF3 is ORF4, which encodes a homologue of phosphatidylglycerophosphatase B of H. influenzae (16). These last two ORFs were not analyzed further.
Construction of isogenic mutants.
Isogenic mutants were constructed to confirm that the genes we identified encode the proteins responsible for each of the proposed functions. The putative H. ducreyi waaQ and lgtF homologues were independently inactivated by insertion of a cat cartridge into a unique restriction site within each of the genes. In addition, both genes were simultaneously inactivated by removal of a 1.3-kb DNA fragment containing portions of the heptosyl- and glucosyltransferase genes followed by insertion of the cat cartridge into this region. The resultant plasmids bearing insertions into waaQ (pLS88.8CAThep), lgtF (pLS88.8CATglu), and both genes (pLS88.8CAThepglu) were each digested with NotI, and the 4.4-kb (4.3 kb for the double mutant) DNA fragments containing the inactivated genes were then ligated into NotI-digested pRSM2072. Each construct was individually electroporated into H. ducreyi 35000, and clones were selected on chocolate agar supplemented with chloramphenicol. The isogenic strains were designated 35000hep−, 35000glu−, and 35000hepglu−.
To confirm that the proper allelic exchange had occurred, Southern blot analysis was performed for each of the isogenic H. ducreyi mutants. Internal probes to the waaQ and lgtF genes were generated and used to probe HindIII-digested chromosomal DNA from 35000, 35000hep−, and 35000glu−. Each of the probes hybridized to a fragment approximately 6 kb in size from the wild-type chromosomal DNA and a fragment of approximately 7 kb from the mutants. A cat gene probe hybridized with a DNA fragment of approximately 7 kb from 35000glu− and 35000hep− but did not hybridize with wild-type chromosomal DNA (data not shown).
Because of the size similarity of the deleted XbaI-BglII fragment (1.3 kb) and the inserted cat cartridge (1.2 kb), the correct allelic exchange in the double-mutant strain 35000hepglu− was confirmed by digesting chromosomal DNA from the wild type and 35000hepglu− with NcoI. There is a single NcoI restriction site within the cat cartridge, and no cleavage sites exist in the sequenced insert of pLS88.8. A probe generated to the upstream region of waaQ, including a portion of the 5′ end of this gene, hybridized to a fragment of approximately 15 kb from the wild-type chromosomal DNA. This probe also hybridized to a 10-kb fragment from the mutant strain 35000hepglu−. Hybridization to this smaller fragment is consistent with the presence of the NcoI site within the cat cartridge. As expected, the cat gene probe hybridized to a 10-kb and a 5-kb fragment from the double mutant, confirming that a single allelic exchange had occurred at the predicted region of the chromosome. The cat gene probe did not hybridize to wild-type chromosomal DNA (data not shown). These results demonstrate that we have replaced the wild-type gene with the disrupted gene(s) in all three mutant strains.
Characterization of the isogenic mutants.
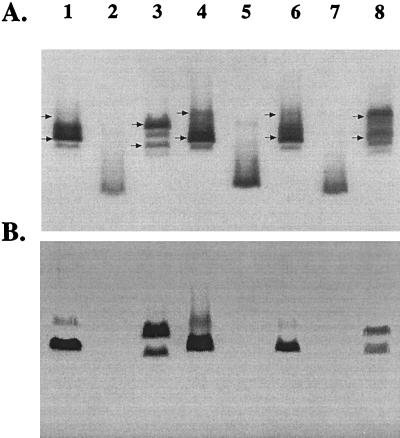
Outer membrane protein profiles and in vitro growth characteristics for all mutants did not reveal any significant differences in comparison to the wild type (data not shown). The LOS from the isogenic mutants, along with the wild-type strain 35000 and HD35000R, were analyzed by SDS-PAGE and Western blotting. Figure 3A shows that 35000hep− synthesized multiple LOS glycoforms (lane 3), with a migration pattern similar to that of the wild-type LOS (lane 1). The LOS of 35000glu− (lane 5) migrated more rapidly than the wild-type LOS (lane 1) but slower than HD35000R (lane 2). This observation suggests that 35000glu− LOS contains the complete triheptose core of the wild-type LOS. 35000hepglu− synthesized a highly truncated LOS molecule which migrated identically to HD35000R (lane 7). The corresponding Western blot probed with MAb 3F11 (Fig. 3B) revealed prominent reactivity to a LOS band which is consistent with the principal LOS glycoform synthesized by strain 35000 (lane 1). In addition, the antibody also reacted to a larger glycoform which appeared to represent a minor component of the wild-type LOS. In comparison, MAb 3F11 reacted with similar intensity to two prominent bands in the heptosyltransferase mutant (lane 3). Closer inspection reveals a slight downward shift in the migration pattern of these two glycoforms (lane 3) compared to the bands detected in the wild-type LOS (lane 1). This minor shift was thought to be the result of the absence of the third heptose of the triheptose core in both LOS species detected. Subsequent MALDI-MS analysis confirmed this hypothesis and provided detailed structures for each LOS species synthesized by all the mutants (see below). MAb 3F11 did not react to LOS from 35000glu− and 35000hepglu− (Fig. 3B, lanes 5 and 7).
FIG. 3.
Composite of a silver-stained SDS-polyacrylamide gel (A) and Western blot (B) probed with MAb 3F11. Shown are LOS from 35000 (lane 1), HD35000R (lane 2), 35000hep− (lane 3), 35000hep−(pHEP) (lane 4), 35000glu− (lane 5), 35000glu−(pGLU) (lane 6), 35000hepglu− (lane 7), and 35000hepglu−(pHEPGLU) (lane 8). The LOS glycoforms that react with the MAb are marked with arrows.
Structural analysis.
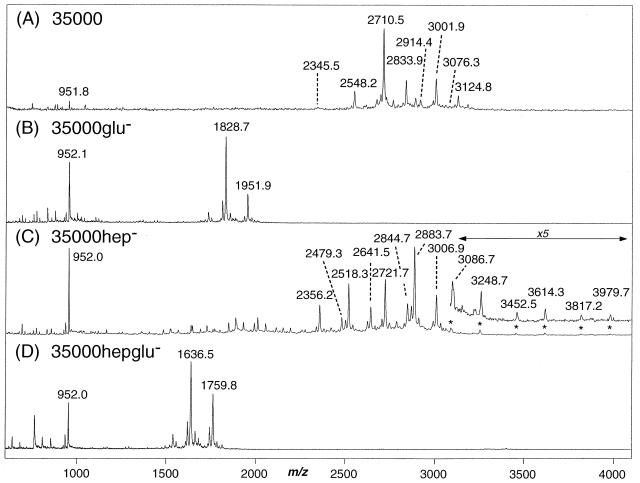
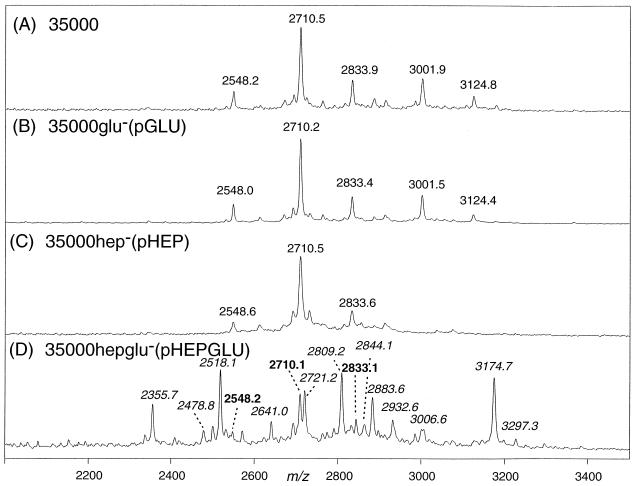
Mass spectrometric analysis of the O-deacylated LOS prepared from the three isogenic mutants confirmed the expected structures (Fig. 4B to D) and were clearly shifted in mass compared to the parental strain 35000 (Fig. 4A). The O-deacylated LOS obtained from wild-type strain 35000 gave a series of peaks, the four most abundant corresponding to (M − H)− ions for LOS glycoforms terminating in N-acetyllactosamine (m/z 2,710.5), previously referred to as A5 (7), and sialyl-N-acetyllactostamine (m/z 3,001.9, A5 + 291 Da), both of which were additionally substituted with a single PEA group (m/z 2,833.9 and 3,124.8, respectively). An additional molecular ion at m/z 2,548.2 appears to arise from a loss of galactose from the terminal N-acetyllactosamine unit (A5 − 162 Da). These masses and their assignments were previously reported for the human-passaged wild-type strain (7).
FIG. 4.
Negative-ion MALDI-MS spectra of O-deacylated LOS from wild-type strain 35000 (A) 35000glu− (B), 35000hep− (C), and 35000hepglu− (and HD35000R) (D). MS spectra for HD35000R and 35000hepglu− were essentially identical.
In contrast, O-deacylated LOS from the three isogenic mutants showed abundant peaks at lower masses, (M − H)−, or in the case of the 35000hep− mutant, peaks at both higher and lower mass. Mass spectra for mutant 35000hepglu− (Fig. 4D) and strain HD35000R were essentially identical. Molecular ion peaks were observed for strain 35000hepglu− at m/z 1,636.5 and 1,759.8 and could be assigned as Hep2-KdoP(PEA)0,1-lipid A, differing in the presence or absence of a single PEA (123 Da). Strain 35000glu− (Fig. 4B) showed two major glycoforms at m/z 1,828.7 and m/z 1,951.9 that are consistent with a complete triheptose core structure Hep3-KdoP(PEA)0,1-lipid A but lacking an oligosaccharide branch structure. The mass difference between O-deacylated LOS molecular ions from strain 35000glu− and strain 35000hepglu− corresponds to a single heptose residue, i.e., m/z 1,636 → 1,828 and m/z 1759 → 1951 (Δm = 192 Da), as would be expected for the loss of the terminal heptose from the core region. In the case of the 35000hep− strain, a complex mixture of LOS glycoforms was observed (Fig. 4C). The (M − H)− peaks at m/z 2,518.3 and 2,641.5 correspond to the major wild-type glycoforms terminating in N-acetyllactosamine, Gal-GlcNAc-Gal-Hep-Glc-Hep2-KdoP(PEA)0,1-lipid A, but lacking one of the core heptose residues. Similarly, the peaks at m/z 2,356.2 and 2,479.3 appear to arise from the additional loss of the terminal galactose, a minor branch structure also seen in the wild type. However, in addition to these expected LOS glycoforms, we observed several additional species that one would not expect from simple loss of heptose from the core. For example, LOS species terminating in N-acetyllactosamine did not appear to be partially sialylated (NeuAcα2→3Galβ1→4GlcNAc), as is the case in the parental strain. Rather, LOS glycoforms with extended polylactosamine structures were present, with the major glycoforms consisting of two N-acetyllactosamine repeats at m/z 2,883.7 and 3,006.9. Further addition of N-acetylglucosamine and galactose to this di-N-acetyllactosamine structures led to the formation of LOS showing relatively weak molecular ions peaks that corresponded to structures containing as many as five N-acetyllactosamine repeats, i.e., m/z 3,086.7, m/z 3,248.7, m/z 3,452.5, m/z 3,614.3, m/z 3,817.2, and m/z 3,979.7. Although the di-N-acetyllactosamine glycoform was detectable on SDS-PAGE (Fig. 3, lane 3), the polylactosamine structures were not readily apparent. This was likely due to a combination of the lack of sensitivity of SDS-PAGE coupled with the low abundance of these glycoforms. To visualize these minor polylactosamine glycoforms by SDS-PAGE, it was necessary to significantly overload the LOS (data not shown).
To further investigate the nature of these higher-molecular-weight structures, we performed a series of alternating enzymatic digestions using β-galactosidase followed by β-N-acetylhexosaminidase. These experiments supported the linear nature of these higher-mass species and were consistent with the presence of a poly-N-acetyllactosamine structure (data not shown). Further in-depth structural analysis of these “polylactosamine” LOS glycoforms from strain 35000hep− is ongoing and will be presented in detail elsewhere. The proposed structures of strain 35000hep− and the two other mutants (35000glu− and 35000hepglu−) are shown in Fig. 6 along with the previously determined structure of the parental wild-type strain.
FIG. 6.
Structure of LOS from isogenic strains 35000 (wild type [WT]) (A), 35000hepglu− (identical to strain 35000R) (B), 35000glu− (C), and 35000hep− (D). Expected masses for deprotonated molecular ions, (M − H)−, for O-deacylated LOS are indicated. All core heptoses are l-glycero-d-manno-heptose with the exception of the branch heptose (labeled with an asterisk) which is d-glycero-manno-heptose.
Complementation of isogenic mutants.
To confirm that the LOS phenotypes observed for the isogenic mutants were not the result of secondary mutations, each of the mutants was complemented with the corresponding wild-type genes. Plasmids containing the individual genes (pHEP and pGLU) or both genes (pHEPGLU) were electroporated into each of the respective H. ducreyi mutants and immunoscreened using MAb 3F11. As a control, the pLS88 vector was used to electroporate each of the mutants. The presence of vector alone in the mutants had no effect on reactivity to MAb 3F11 or migration of LOS by SDS-PAGE (data not shown). All kanamycin-resistant transformants tested from the 35000glu− and 35000hepglu− complementations reacquired reactivity with MAb 3F11, indicating that expression of the principal LOS glycoform had been regained at the bacterial surface (data not shown). As shown in Fig. 3A, there was a major LOS glycoform, from the complemented mutant strains 35000glu(pGLU) (lane 6) and 35000hepglu(pHEPGLU) (lane 8), with an apparent mass equivalent to the principal LOS glycoform expressed by the wild type (lane 1). In addition, there is a larger glycoform observed in the LOS of 35000hepglu(pHEPGLU) (lane 8). The Western blot, probed with MAb 3F11 (Fig. 3B), demonstrated reactivity to a 4.5-kDa band in the complemented mutant LOS (lanes 6 and 8) and parental LOS (lane 1), as predicted. There is also reactivity to a larger glycoform detected in both the wild-type LOS (lane 1) and the LOS of 35000hepglu(pHEPGLU) (lane 8). This reactivity was consistent with the reactivity observed in the original complementation of HD35000R (lane 3), suggesting that this larger LOS glycoform contains di-N-acetyllactosamine.
Restoration of the parental LOS phenotype for 35000hep− was also confirmed by SDS-PAGE and Western blot analysis. Figure 3A shows that the LOS glycoforms from the complemented 35000hep− mutant (lane 4) exhibited a migration pattern that was consistent with the wild-type LOS (lane 1). The corresponding blot also shows a slight upward shift in the LOS glycoforms which react with MAb 3F11 (Fig. 3B, lane 4). These results are consistent with the addition of the third heptose to the core, which has apparently resulted in the expression of wild-type LOS (lane 1).
Structural analysis of the LOS from complemented mutants.
MALDI-MS analysis of the O-deacylated LOS prepared from three complemented strains revealed partial to complete complementation of the corresponding glycosyltransferase genes (Fig. 5B to D). The MALDI-MS spectrum of the O-deacylated LOS from the complemented isogenic mutant 35000glu−(pGLU) (Fig. 5B) showed peaks at m/z 2,548.0, 2,710.2, 2,833.4, 3,001.5, and 3,124.4, identical to molecular ion masses observed in the parental LOS phenotype (Fig. 5A, wild-type strain 35000). The complemented isogenic mutant 35000hep−(pHEP) also regained expression of several parental LOS glycoforms with (M − H)− peaks at m/z 2,548.6, 2,710.5, and 2,833.6 (Fig. 5C). However, the sialylated glycoforms containing terminal NeuAcα2→3Galβ1→4GlcNAc were not observed.
FIG. 5.
Negative-ion MALDI-MS spectra of the molecular ion region of O-deacylated LOS from wild-type strain 35000 (A), complemented mutant strain 35000glu−(pGLU) (B), complemented mutant strain 35000hep−(pHEP) (C), and double-complemented mutant strain 35000hepglu−(pHEPGLU) (D) containing masses indicative of complete complementation (bold) and incomplete (italics). See text for details.
Lastly, the MALDI-MS spectrum of the complemented isogenic mutant 35000hepglu−(pHEPGLU) revealed the presence of several LOS glycoforms at m/z 2,710.1, 2,833.1, and 2,548.2 (Fig. 5D) which are consistent with the wild-type LOS (Fig. 5A). As was the case for the 35000hep(pHEP) complemented strain, no sialylated glycoforms were observed on LOS structures that contained the completed triheptose core.
However, several additional molecular ion peaks were observed that appeared to result from incomplete complementation of the 35000hepglu−. These LOS glycoforms (Fig. 5D), i.e., m/z 2,518.1, 2,641.0, 2,355.7, 2,478.8, 2,721.2, 2,844.1, 2,883.6, and 3,006.6, were consistent with glycoforms present in the 35000hep− mutant (Fig. 4D). In addition, some sialylated glycoforms were detected in the LOS of the complemented double mutant, i.e., m/z 2,809.2, 2,932.6, 3,174.7, and 3,297.3 (Fig. 5D). These peaks were assigned as monosialylated counterparts to the LOS glycoforms containing terminal mono- and di-N-acetyllactosamine. The presence of terminal sialic acid in these latter glycoforms was confirmed by enzymatic digestion with neuraminidase followed by repeat mass spectrometry analysis, which selectively eliminated the masses for these four sialylated LOS species (data not shown; mass shifted by 291 Da in all cases).
DISCUSSION
Pyocin lysis has previously been used as a strategy to identify LOS mutants of H. ducreyi and N. gonorrhoeae (8, 14). Although the actual mechanism of lysis is poorly understood, a recent report by Lee et al. suggests that these particles contain nucleic acid (27). In this study, we used pyocin to select for LOS mutants of H. ducreyi strain 35000. One such mutant, HD35000R, produced a LOS molecule that lacked the MAb 3F11 epitope and migrated with an increased mobility on SDS-PAGE. Complementation of this mutant with a plasmid library containing H. ducreyi 35000 chromosomal DNA resulted in the identification of a clone expressing wild-type LOS. Western blot analysis confirmed that this transformant, HD35000R(pLS88.8), expressed the LOS epitope reactive with MAb 3F11. The sequence analysis of the complementing plasmid revealed a 3.2-kb DNA insert containing four complete ORFs. The putative protein product of ORF1 shared similarity (37%) with the E. coli WaaQ, the heptosyltransferase responsible for addition of the third heptose residue to the inner core of E. coli lipopolysaccharide (LPS) (44). The H. ducreyi WaaQ homologue also exhibited similarity to the WaaF homologue of H. ducreyi, which may function to attach the HepII via an α1,3 linkage to the HepI of the LOS core (5). However, MALDI-MS analysis confirmed that the core of HD35000R lacked the third heptose; therefore, we concluded that the waaQ encodes the H. ducreyi heptosyltransferase III.
The predicted amino acid sequence of ORF2 was similar (49%) to the sequence of LgtF of N. meningitidis, a β1,4-glucosyltransferase (24). This is the first carbohydrate added to the heptose core on the main oligosaccharide branch of the principal glycoform synthesized by wild-type H. ducreyi 35000. Again, based on this sequence homology and subsequent structural analysis of the LOS of HD35000R, we designated ORF2 as the lgtF homologue in H. ducreyi. To confirm that the cloned genes were responsible for the proposed functions, isogenic mutants were constructed and then complemented with the corresponding wild-type genes.
Inactivation of the H. ducreyi lgtF gene resulted in the expression of a truncated LOS molecule lacking the first glucose of the oligosaccharide branch and all subsequent sugars distal to this carbohydrate. However, this disruption did not affect the assembly of the triheptose inner core. This result confirms that the H. ducreyi lgtF is the homologue of the β1,4 glucosyltransferase of N. meningitidis (24). Furthermore, our data confirm that the expression of this protein is required for chain elongation extending from the HepI of the core of H. ducreyi LOS.
Recently it was reported that a mutation in E. coli waaQ (heptosyltransferase III) resulted in synthesis of a LPS molecule which migrated similarly to wild-type LPS, lacking only the third heptose of the inner core (44). Inactivation of H. ducreyi waaQ also resulted in the expression of LOS molecules with electrophoretic mobility patterns that were similar, but not identical, to those of the LOS glycoforms produced by the wild-type strain. Structural analysis confirmed that these LOS molecules lacked only the third heptose of the core. The full-length LOS structure produced by the waaQ mutant demonstrates that the glucosyltransferase (LgtF) functions in the absence of a complete triheptose core. In addition, the reactivity to MAb 3F11 provided evidence that the LOS glycoforms of the waaQ mutant, lacking the third heptose of the inner core, retained the proper conformation of the terminal lactosamine epitope recognized by this antibody.
Interestingly, the H. ducreyi waaQ mutant also synthesized additional LOS glycoforms, albeit in minor abundance, that were not detected in the LOS of the wild-type strain. MALDI-MS analysis determined that these larger glycoforms were primarily the result of the addition of di-, tri-, and polylactosamines to the terminal portion of the main LOS branch. Previously Melaugh et al. detected that the dilactosamine glycoform existed in LOS isolated from H. ducreyi 35000, and these investigators speculated that this structure may have arisen from an alternative, as yet undefined, biosynthetic pathway (29). Complementation of 35000hep− mutant resulted in production of wild-type LOS and the disappearance of the slower-migrating LOS glycoforms. This result suggests that the synthesis and addition of terminal lactosamine may be directly or indirectly linked to the amount of WaaQ expressed during the synthesis of H. ducreyi LOS. This hypothesis is further supported by the fact that complementation of the 35000hepglu− mutant, with the plasmid containing only the lgtF gene, resulted in appearance of LOS structures consistent with the multiple glycoforms detected in the 35000hep− strain (data not shown). While it is intriguing to speculate as to possible regulatory mechanisms involved in the biosynthetic pathway of H. ducreyi LOS, more detailed studies are clearly needed before any conclusions can be made.
The mutant disrupted in both waaQ and lgtF (35000hepglu−) produced a LOS molecule that migrated with a mobility identical to the mobility of the principal glycoform produced by HD35000R. This result suggests that HD35000R contains disruptions in both LOS biosynthesis genes, and we are currently attempting to amplify the waaQ and lgtF genes from HD35000R to determine if both of these genes are disrupted by insertions, deletions, or point mutations. This information could provide more insight into possible regulation of these LOS genes. We also considered the possibility that HD35000R has a mutation in a regulatory gene that controls expression of both transferase genes. However, this scenario is highly unlikely, because these genes are found to be adjacent to one another and are transcribed convergently. While genes involved in LPS biosynthesis have been shown to be in contiguous clusters, LOS genes for many gram-negative human pathogens have been shown to be scattered throughout the chromosome (35). In addition to the downstream sequence, the sequence 5′ of the waaQ gene was determined to contain a homologue to the argininosuccinate synthase of H. influenzae (data not shown) (16). Therefore, our sequence analysis upstream and downstream of the waaQ and lgtF confirmed that these genes were not arranged in a contiguous cluster with other LOS synthesis genes, which is consistent with findings for other gram-negative bacteria such as N. meningitidis, N. gonorrhoeae, and H. influenzae (35).
It is also interesting that sialic acid is absent among LOS glycoforms terminating in N-acetyllactosamine in the 35000hep− mutant and in some of the complemented strains (Fig. 6). Although this phenomenon is unexplained, there are multiple factors that could be involved in the synthesis and addition of sialic acid. The gene which encodes for the H. ducreyi sialyltransferase has been cloned and sequenced (7). This enzyme has been reported to be unique in comparison to other known sialyltransferases, and there are currently no data describing the regulation of sialic acid addition to H. ducreyi LOS. It is possible that factors such as growth conditions and media may effect this mechanism. The presence of a complete LOS core or the presence of the proper accepting terminal region of the LOS molecule could also be critical. Most likely a combination of multiple factors is involved in this complex process. Although the mechanism of sialylation of H. ducreyi LOS is beyond the scope of this study, we now have the constructs that are essential to further investigations designed to understand the sialylation of H. ducreyi LOS.
In conclusion, we have identified, cloned, and sequenced two genes involved in expression and biosynthesis of the principal LOS glycoform of H. ducreyi 35000. Isogenic mutants deficient in expression of the heptosyltransferase III, the β1,4-glucosyltransferase and both glycosyltransferase genes were constructed and characterized. Future studies will be aimed at comparing these LOS mutants with the parental strain in biological assays such as adherence and invasion of human keratinocytes. In addition, these mutants can be compared with the wild type in the human challenge model of infection. Such studies may increase our knowledge of the mechanisms that control the regulation of expression of these glycosyltransferases and the role of LOS in pathogenesis of this organism.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants R01 AI30006 (to A.A.C.), R01 AI38444 (to R.S.M.), and R01 AI31254 (to B.W.G.) and by PE Biosystems, Framingham, Mass., which kindly provided instrumentation to B.W.G. DNA sequence was determined by the Core Facility at Children's Research Institute, which was supported in part by NIH grant HD34615.
We thank Huachun Zhong for excellent technical assistance.
REFERENCES
- 1.Alfa M J, DeGagne P. Attachment of Haemophilus ducreyi to human foreskin fibroblasts involves LOS and fibronectin. Microb Pathog. 1997;22:39–46. doi: 10.1006/mpat.1996.0089. [DOI] [PubMed] [Google Scholar]
- 2.Alfa M J, DeGagne P, Totten P A. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect Immun. 1996;64:2349–2352. doi: 10.1128/iai.64.6.2349-2352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apicella M A, Griffiss J M, Schneider H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 1994;235:242–252. doi: 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- 4.Bauer B A, Lumbley S R, Hansen E J. Characterization of a WaaF (RfaF) homolog expressed by Haemophilus ducreyi. Infect Immun. 1999;67:899–907. doi: 10.1128/iai.67.2.899-907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer B A, Stevens M K, Hansen E J. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect Immun. 1998;66:4290–4298. doi: 10.1128/iai.66.9.4290-4298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozue J A, Tarantino L, Munson R S., Jr Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol Lett. 1998;164:269–273. doi: 10.1111/j.1574-6968.1998.tb13097.x. [DOI] [PubMed] [Google Scholar]
- 7.Bozue J A, Tullius M V, Wang J, Gibson B W, Munson R S., Jr Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J Biol Chem. 1999;274:4106–4114. doi: 10.1074/jbc.274.7.4106. [DOI] [PubMed] [Google Scholar]
- 8.Campagnari A A, Karalus R, Apicella M, Melaugh W, Lesse A J, Gibson B W. Use of pyocin to select a Haemophilus ducreyi variant defective in lipooligosaccharide biosynthesis. Infect Immun. 1994;62:2379–2386. doi: 10.1128/iai.62.6.2379-2386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campagnari A A, Spinola S M, Lesse A J, Kwaik Y A, Mandrell R E, Apicella M A. Lipooligosaccharide epitopes shared among gram-negative non-enteric mucosal pathogens. Microb Pathog. 1990;8:353–362. doi: 10.1016/0882-4010(90)90094-7. [DOI] [PubMed] [Google Scholar]
- 10.Campagnari A A, Wild L M, Griffiths G E, Karalus R J, Wirth M A, Spinola S M. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect Immun. 1991;59:2601–2608. doi: 10.1128/iai.59.8.2601-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. HIV prevention through early detection and treatment of other sexually transmitted diseases—United States. Recommendations of the Advisory Committee for HIV and STD prevention. Morbid Mortal Weekly Rep. 1998;47:1–24. [PubMed] [Google Scholar]
- 12.Cope L D, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–61. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon L G, Albritton W L, Willson P J. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid. 1994;32:228–232. doi: 10.1006/plas.1994.1060. [DOI] [PubMed] [Google Scholar]
- 14.Dudas K C, Apicella M A. Selection and immunochemical analysis of lipooligosaccharide mutants of Neisseria gonorrhoeae. Infect Immun. 1988;56:499–504. doi: 10.1128/iai.56.2.499-504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 17.Gibson B W, Engstrom J J, John C M, Hines W, Falick A M. Characterization of bacterial lipooligosaccharides by delayed extraction matrix-assisted laser desorption ionization time-flight mass spectrometry. J Am Soc Mass Spectrosc. 1997;8:645–658. [Google Scholar]
- 18.Gibson B W, Campagnari A A, Melaugh W, Phillips N J, Apicella M A, Grass S, Wang J, Palmer K L, Munson R S., Jr Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J Bacteriol. 1997;179:5062–5071. doi: 10.1128/jb.179.16.5062-5071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond G W, Lian C J, Wilt J C, Ronald A R. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob Agents Chemother. 1978;13:608–12. doi: 10.1128/aac.13.4.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 21.Helander I M, Nummila K, Kilpelainen I, Vaara M. Increased substitution of phosphate groups in lipooligosaccharides and lipid A of polymyxin-resistant mutants of Salmonella typhimurium and Escherichia coli. Prog Clin Biol Res. 1995;392:15–23. [PubMed] [Google Scholar]
- 22.Ison C A, Dillon J A, Tapsall J W. The epidemiology of global antibiotic resistance among Neisseria gonorrhoeae and Haemophilus ducreyi. Lancet. 1998;351(Suppl. 3):8–11. doi: 10.1016/s0140-6736(98)90003-4. . (Erratum, 352:1316.) [DOI] [PubMed] [Google Scholar]
- 23.Johnson K G, Perry M B. Improved techniques for the preparation of bacterial lipopolysaccharides. Can J Microbiol. 1976;22:29–34. doi: 10.1139/m76-004. [DOI] [PubMed] [Google Scholar]
- 24.Kahler C M, Carlson R W, Rahman M M, Martin L E, Stephens D S. Two glycosyltransferase genes, lgtF and rfaK, constitute the lipooligosaccharide ice (inner core extension) biosynthesis operon of Neisseria meningitidis. J Bacteriol. 1996;178:6677–6684. doi: 10.1128/jb.178.23.6677-6684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klesney-Tait J, Hiltke T J, Maciver I, Spinola S M, Radolf J D, Hansen E J. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179:1764–1773. doi: 10.1128/jb.179.5.1764-1773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knecht D A, Dimond R L. Visualization of antigenic proteins on Western blots. Anal Biochem. 1984;136:180–184. doi: 10.1016/0003-2697(84)90321-x. [DOI] [PubMed] [Google Scholar]
- 27.Lee F K, Dudas K C, Hanson J A, Nelson M B, LoVerde P T, Apicella M A. The R-type pyocin of Pseudomonas aeruginosa C is a bacteriophage tail-like particle that contains single-stranded DNA. Infect Immun. 1999;67:717–25. doi: 10.1128/iai.67.2.717-725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandrell R, Schneider H, Apicella M, Zollinger W, Rice P A, Griffiss J M. Antigenic and physical diversity of Neisseria gonorrhoeae lipooligosaccharides. Infect Immun. 1986;54:63–69. doi: 10.1128/iai.54.1.63-69.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melaugh W, Phillips N J, Campagnari A A, Tullius M V, Gibson B W. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry. 1994;33:13070–13078. doi: 10.1021/bi00248a016. [DOI] [PubMed] [Google Scholar]
- 30.Mohr M D, Bornsen K O, Widmer H M. Matrix-assisted laser desorption ionization mass spectrometry—improved matrix for oligosaccharides. Rapid Commun Mass Spectrom. 1995;9:809–914. doi: 10.1002/rcm.1290090919. [DOI] [PubMed] [Google Scholar]
- 31.Morse S A, Vaughan P, Johnson D, Iglewski B H. Inhibition of Neisseria gonorrhoeae by a bacteriocin from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1976;10:354–362. doi: 10.1128/aac.10.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer K L, Goldman W E, Munson R S., Jr An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol Microbiol. 1996;21:13–19. doi: 10.1046/j.1365-2958.1996.00615.x. [DOI] [PubMed] [Google Scholar]
- 33.Palmer K L, Grass S, Munson R S., Jr Identification of a hemolytic activity elaborated by Haemophilus ducreyi. Infect Immun. 1994;62:3041–3043. doi: 10.1128/iai.62.7.3041-3043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer K L, Thornton A C, Fortney K R, Hood A F, Munson R S, Jr, Spinola S M. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J Infect Dis. 1998;178:191–199. doi: 10.1086/515617. [DOI] [PubMed] [Google Scholar]
- 35.Preston A, Mandrell R E, Gibson B W, Apicella M A. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit Rev Microbiol. 1996;22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 36.Russo T A, Guenther J E, Wenderoth S, Frank M M. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol Microbiol. 1993;9:357–364. doi: 10.1111/j.1365-2958.1993.tb01696.x. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Spinola S M, Castellazzo A, Shero M, Apicella M A. Characterization of pili expressed by Haemophilus ducreyi. Microb Pathog. 1990;9:417–426. doi: 10.1016/0882-4010(90)90060-4. [DOI] [PubMed] [Google Scholar]
- 39.Spinola S M, Orazi A, Arno J N, Fortney K, Kotylo P, Chen C Y, Campagnari A A, Hood A F. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J Infect Dis. 1996;173:394–402. doi: 10.1093/infdis/173.2.394. [DOI] [PubMed] [Google Scholar]
- 39a.Sun S, Schilling B, Tarantino L, Tullius M V, Gibson B W, Munson R S., Jr Cloning and characterization of the lipooligosaccharide galactosyltransferase II gene of Haemophilus ducreyi. J Bacteriol. 2000;182:2292–2298. doi: 10.1128/jb.182.8.2292-2298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 42.Vestal M L, Juhasz P, Martin S A. Delayed extraction matrix-assisted laser desorption time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1995;9:1044–1050. [Google Scholar]
- 43.Westphal O, Jann K. Bacterial lipopolysaccharide extraction with phenol-water and further application of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 44.Yethon J A, Heinrichs D E, Monteiro M A, Perry M B, Whitfield C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem. 1998;273:26310–26316. doi: 10.1074/jbc.273.41.26310. [DOI] [PubMed] [Google Scholar]
- 45.Zaretzky F R, Kawula T H. Examination of early interactions between Haemophilus ducreyi and host cells by using cocultured HaCaT keratinocytes and foreskin fibroblasts. Infect Immun. 1999;67:5352–5360. doi: 10.1128/iai.67.10.5352-5360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]