Abstract
Translational initiation of the human BiP mRNA is directed by an internal ribosomal entry site (IRES) located in the 5′-untranslated region (5′-UTR). In order to understand the mechanism of the IRES-dependent translation of BiP mRNA, cellular proteins interacting with the BiP IRES were investigated. La autoantigen, which augments the translation of polioviral mRNA and hepatitis C viral mRNA, bound specifically to the second half of the 5′-UTR of the BiP IRES and enhanced translation of BiP mRNA in both in vitro and in vivo assays. This finding suggests that cellular and viral IRESs containing very different RNA sequences may share a common mechanism of translation.
INTRODUCTION
Translation of the majority of eukaryotic mRNAs is initiated through a cap-dependent mechanism termed ‘ribosome scanning’ (1). Binding of the small ribosomal subunit is mediated by interaction of a cap-binding initiation factor complex (eIF4F) to the 7-methylguanosine cap structure at the 5′-end of a mRNA. In contrast, translation of some mRNAs occurs in a cap- and 5′-end-independent manner through an internal segment in the 5′-untranslated region (5′-UTR). This RNA element has been termed the ‘internal ribosomal entry site’ (IRES) (2,3). IRES elements were first discovered in picornavirus mRNAs, which contain long, highly structured 5′-UTRs but lack a cap structure at the 5′-end of their mRNAs (3,4). Since then, many IRES structures have been discovered in related picornaviruses (5) and unrelated viruses such as hepatitis C virus (6), simian immunodeficiency virus (7) and Plautia stali intestine virus (8). The human immunoglobulin heavy chain-binding protein (BiP/GRP78) was the first cellular mRNA reported to harbor an IRES (9). The latest list of cellular IRESs includes the proto-oncogene c-myc (10,11), platelet-derived growth factor 2 (PDGF2/c-sis) (12), the eukaryotic translation initiation factor eIF4G (13), vascular endothelial growth factor (VEGF) (14), ornithine decarboxylase (15), p58 (PITSLRE) (16), X-linked inhibitor of apoptosis protein (XIAP) (17), NF-κB repressing factor (NRF) (18), AML1/RUNX1 (CBFA/PEBP2α) (19) and apoptotic protease activating factor (Apaf-1) (20).
BiP protein has been identified as an immunoglobulin heavy chain-binding protein (21). BiP protein binds transiently to a variety of nascent secretory and transmembrane proteins and it binds to misfolded proteins that accumulate in the endoplasmic reticulum (ER). Suggested functions for the BiP protein include mediation of proper folding, assembly of nascent proteins in the ER and scavenging of misfolded proteins in the ER (22). BiP protein is also known as glucose-regulated protein 78 (GRP78). It is homologous to heat-shock protein 70 (HSP70) (23). Expression of BiP is regulated at the transcriptional level where synthesis of BiP mRNA is induced under a variety of stress conditions such as glucose starvation and treatment with calcium ionophores, calcium-chelating agents such as EGTA and compounds that block cellular glycosylation, such as tunicamycin and glucosamine (24,25). Furthermore, several reports have suggested that expression of BiP protein is also regulated at the translational level (26–29). For example, translation of BiP mRNA becomes enhanced when translation of most host cellular mRNAs is inhibited upon poliovirus infection (27).
Although a number of cellular and viral IRES elements have been identified, little is known about the details of the mechanism of IRES-dependent translation except that some primary sequences and secondary structures of certain IRES elements are crucial for efficient translation. It is generally believed that IRES-dependent translation requires some of the canonical initiation factors functioning in the cap-dependent translation process (30). Certain IRES-dependent mRNAs require other specific cellular factors, which specifically interact with the mRNA for efficient translation. La autoantigen (31), polypyrimidine tract-binding protein (PTB) (32), upstream of N-ras (33) and poly(C)-binding protein (PCBP) (34) are included among the cellular factors enhancing IRES-dependent translation.
Human La autoantigen (SS-B) has been shown to stimulate IRES-dependent translation of polioviral mRNA and HCV mRNA (31,35). La protein was originally identified as an autoantigen recognized by sera from patients with systemic lupus erythematosus and Sjögren’s syndrome (36). La protein belongs to the RNA-binding proteins containing a RNA recognition motif (RRM) (37). It is involved in regulation of the initiation and termination of transcription by RNA polymerase III (38,39). In addition, La protein is associated with polymerase II RNA transcripts, such as U1 RNA (40), as well as viral RNAs, including Epstein–Barr virus encoded RNAs (41), adenovirus VA RNAs (42), vesicular stomatitis virus leader RNAs (43), the 5′-UTR of picornaviruses (31) and the HIV TAR sequence (44). In the case of poliovirus and hepatitis C virus, addition of purified La to rabbit reticulocyte lysates (RRL) stimulates viral IRES-dependent translation (31,35). Furthermore, the binding of La protein to the HIV TAR sequence alleviates the translational repression exerted by the TAR sequence on a downstream reporter gene (45). In the case of EMCV, translational suppression by surplus PTB was relieved by addition of La protein (46).
Polypyrimidine tract-binding protein/heterogeneous nuclear ribonucleoprotein I (PTB/hnRNP I), which was originally proposed to be a splicing factor, specifically interacts with IRESs of encephalomyocarditis virus (EMCV) (47,48), poliovirus (49) and foot-and-mouth disease virus (FMDV) (50). Binding of PTB often results in an enhancement of IRES-dependent translation (33,48,51–53). On the other hand, binding of PTB to the BiP IRES was shown to inhibit translation of BiP mRNA (26).
A few cellular proteins have been reported to interact with cellular IRESs. For instance, hnRNP C binds in a differentiation-induced manner to the c-sis IRES (54) and La modulates internal initiation directed by the XIAP IRES (17). The mechanisms of modulation of IRES-dependent translation by RNA-binding proteins remains to be elucidated.
Here we report that La specifically binds to the second half of the BiP 5′-UTR (nt 115–207). Purified La protein augments translation of BiP mRNA in an in vitro translation system and overexpression of La protein in Cos-7 cells enhances BiP IRES-dependent translation.
MATERIALS AND METHODS
Plasmid construction
Enzymes used for cloning and modifying DNA were purchased from New England Biolabs and Boehringer Mannheim. Construction of the plasmids containing BiP IRES [pSK(+)/CATΔ-BCAT, pSK(+)/BiP-CAT, pSK(+)/BiP-CAT(50–117) and pSK(+)/BiP-CAT(115–221)] has been described by Kim et al. (26). In plasmid pSK(+)/CATΔ-BCAT the T7 promoter is followed by a dicistronic mRNA sequence (CATΔ represents a truncated CAT gene).
Dicistronic constructs were generated by assembling a cassette vector containing the Renilla (RLuc) and firefly (FLuc) luciferase genes as cistrons 1 and 2, respectively. Plasmid pEGFP-N1 (Promega) was digested with PinAI and SspB1 to remove the EGFP gene and then self-ligated to construct pΔEGFP-N1. Plasmid pELuc containing a FLuc gene was generated by ligating a PstI–NotI fragment of plasmid pΔEGFP-N1 to a PstI–NotI fragment of PCR-amplified FLuc gene. The PCR reaction was carried out with pGL2-control (Promega) and the oligonucleotides 5′-AATTTGCGGCCGCAAGCTTACATTTTACAATTTGGAC-3′ and 5′-AAACTGCAGATGGAAGACGCCAAAAAC-3′ as template and primers, respectively. To construct the dicistronic plasmid pRF, which contains the RLuc and FLuc reporter genes, a XbaI–NotI fragment of pRL-CMV (Promega) containing a RLuc gene was ligated to a NheI–NotI fragment of pELuc. Plasmids pR/BiP/F and pR/EMCV/F were constructed by inserting the BiP IRES and EMCV IRES sequences into the intercistronic site of plasmid pRF, respectively.
For construction of plasmid pT7-7/La, La cDNA was amplified by PCR using pGEX-KG/La as template (46). The oligonucleotides 5′-GGGAATTCACATGGCTGAAAATGGTGATAATG-3′ and 5′-ACTCTGCAGCTACTGGTCTCCAGCACC-3′ were used as primers. The PCR product was digested with PstI and EcoRI and then ligated into the PstI–EcoRI fragment of pT7-7. Plasmid pEGFP-La was generated by ligating a HindIII–Klenow-filled EcoRI fragment of plasmid pT7-7/La to a SalI–Klenow-filled EcoRI fragment of pEGFP-C1/PTB (46). Plasmid pH(1–402) was constructed by ligating a fragment of pSK(–)/CAT (55) digested with KpnI and BamHI to a PCR amplified fragment digested with KpnI and BamHI. The PCR reaction was carried out using pKI5 and the oligonucleotides 5′-CGCGGATCCCTGTGGGCGGCGGTTG-3′ and 5′-CGGGGTACCGCCAGCCCCCGATTGGGGGCGACACTCCACCATAGATC-3′ as template and primers, respectively.
In vitro transcription and translation
Plasmid DNAs were purified using the polyethyleneglycol precipitation method (56) and linearized with appropriate restriction enzymes. The linearized DNAs were then treated with phenol/chloroform and ethanol precipitated. Transcription reactions were performed with T7 RNA polymerase (Boehringer Mannheim) at 37°C for 90 min as described by the manufacturer. To produce capped mRNA, 1 mM m7GpppG (Pharmacia Biotech) was included in the transcription reaction mixture. The concentration of the RNA transcripts was determined by UV spectrophotometry. Plasmid pSK(+)/CATΔ-BCAT digested with BamHI was used to generate the dicistronic CATΔ-BCAT mRNA. To produce mRNAs with serial deletions in the BiP 5′-UTR [BiP UTR(1–232), UTR(1–53), UTR(1–207), UTR(50–117), UTR(115–207) and UTR(115–232)], EcoRI-digested pSK(+)/BiP-CAT, NruI-digested pSK(+)/BiP-CAT, BglI-digested pSK(+)/BiP-CAT, NarI-digested pSK(+)/BiP-CAT(50–117), BglI-digested pSK(+)/BiP-CAT(115–221) and EcoRI-digested pSK(+)/BiP-CAT(115–221) were used in transcription reactions. The BalI-digested pBS-ECAT (48), MunI-digested pMPS1-ECAT (48) and BamHI-digested pH(1–402) plasmids were used to generate the full-length EMCV IRES, poliovirus IRES and HCV IRES, respectively. SalI-digested pTM1-PTB1 (57) and HindIII-digested pT7-7/La were used as templates for in vitro transcription of PTB1 and La mRNA, respectively. Luciferase mRNA was purchased from Promega. Biotinylated RNAs were synthesized by including biotin-labeled UTP (Clontech) in the transcription reaction.
In vitro translations in micrococcal nuclease-treated RRL (Promega) were performed in 20 µl reaction mixtures containing 6 or 30 nM mRNA. The translation reactions were carried out at 30°C for 1 h in the presence of [35S]methionine (NEN). Translation products were analyzed by 15% SDS–PAGE. The intensity of the autoradiographic images was enhanced by fluorography using salicylic acid. The gels were dried and exposed to Kodak XAR-5 or Agfa Curix RP1 film for 12–18 h. The efficiencies of the translations were quantified using a densitometer (Bioimage 50S Series; BioImage System Corp.).
The purification procedure for recombinant human La protein has been described previously (46). All purified proteins were dialyzed against LD buffer (16.2 mM HEPES–KOH pH 7.5, 50 mM KCl, 0.5 mM DTT, 0.1 mM EDTA, 10% glycerol).
Streptavidin–acrylamide precipitation of La protein with biotinylated RNA
Co-precipitation of La with BiP RNA was investigated as described by Hahm et al. (55), with slight modifications. 35S-labeled La, PTB or luciferase, generated by in vitro translation in RRL, was mixed with 1.25 µg biotinylated RNA and 10 µg yeast tRNA (Boehringer Mannheim) in 15 µl of KHN buffer (150 mM KCl, 20 mM HEPES pH 7.9, 0.05% Nonidet P-40, 0.2 mM DTT). The binding reaction was carried out at 4°C for 1 h. The reaction mixture was further incubated at 4°C for 1 h after addition of 1 ml of KHN buffer and 70 µl of streptavidin–acrylamide beads (Pierce). The beads were collected by centrifugation, washed three times with 1 ml of KHN buffer, resuspended in 20 µl of protein sample buffer and then boiled for 4 min. After centrifugation, 20 µl of the supernatant was analyzed by 12% SDS–PAGE followed by autoradiography. For the binding assay using western blot analysis, 100 µg HeLa cell cytoplasmic extract was mixed with 10 µg or 0.9 µM biotinylated RNA and 30 µg yeast tRNA (Boehringer Mannheim) in 52 µl of KHN buffer. The other steps were as described above.
UV crosslinking of labeled RNAs and proteins
UV crosslinking reactions were performed as described previously (26). RNA probes (2 × 105–4 × 105 c.p.m.) purified on push columns (Stratagene) were incubated at 30°C for 20 min with 0.1 µg purified La in 30 µl of reaction mixture. After RNA binding, the reaction mixtures were irradiated with UV light on ice for 15 min using a UV-Stratalinker (Stratagene). Unbound RNA was removed by digestion with 20 µg RNase A, 200 U RNase T1 and 1 U RNase V1 (cobra venom nuclease; Pharmacia Biotech) at 37°C for 30 min. The RNA–protein complexes were analyzed by 12% SDS–PAGE followed by autoradiography.
Cell culture and transfection
Cos-7 cells were transfected using the electroporation procedure. Cells were co-transfected with a dicistronic reporter plasmid and a vector expressing the effector protein. About 48 h after transfection the cells were harvested and lysed by freezing and thawing. Renilla luciferase and firefly luciferase activities of cell extracts were measured using the dual-luciferase reporter assay system (Promega).
Western blot analysis
Cell extracts or RNA-bound samples were resolved by 12% SDS–PAGE and transferred to a nitrocellulose membrane (Amersham). The membrane was blocked overnight with 5% skimmed milk in TBS buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% Tween-20) and then incubated with anti-human La monoclonal antibody (46) or anti-GFP monoclonal antibody (Clontech) for 2 h. Horseradish peroxidase-conjugated anti-mouse IgG was used as secondary antibody. To visualize bands, the membrane was developed employing the ECL method according to the supplier’s instructions (Amersham).
RESULTS
La autoantigen binds to the BiP 5′-UTR
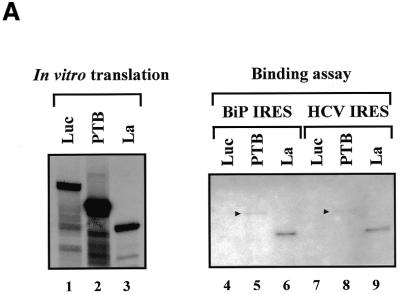
In order to investigate whether La protein interacts with the BiP 5′-UTR, we performed an RNA affinity resin binding assay (Fig. 1A). RNAs corresponding to the full-length BiP and HCV IRESs were synthesized by in vitro transcription in the presence of biotinylated UTP. 35S-labeled luciferase, PTB and La proteins, synthesized in an in vitro RRL translation system (Fig. 1A, lanes 1–3, respectively), were used to test the interactions with the biotinylated RNAs. After incubation of the labeled proteins with the biotinylated RNAs, the RNAs were precipitated with streptavidin–acrylamide and then the co-precipitated proteins were analyzed by SDS–PAGE. 35S-labeled luciferase, a negative control protein, did not show any interaction with the BiP or HCV IRES (Fig. 1A, lanes 4 and 7). As had been demonstrated in a previous report (26), PTB bound weakly to the BiP and HCV IRESs (Fig. 1A, lanes 5 and 8). On the other hand, 35S-labeled La was co-precipitated with both the BiP and HCV IRESs, as seen in lanes 6 and 9 of Figure 1A. These data therefore indicate that the La autoantigen can interact with the BiP IRES.
Figure 1.
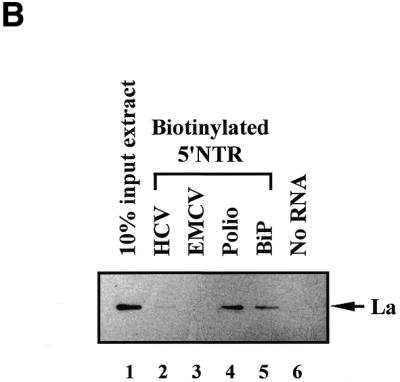
La protein binds to the BiP IRES. (A) In vitro translation products of luciferase, PTB and La are shown in lanes 1–3, respectively. Proteins co-precipitated with biotinylated RNAs corresponding to the full-length BiP IRES and HCV IRES are shown in lanes 4 and 7 (luciferase), 5 and 8 (PTB) and 6 and 9 (La), respectively. The positions of PTB are indicated by arrowheads. (B) Comparison of the RNA-binding activity of La protein to different IRESs. Biotinylated RNAs (72 pmol each) corresponding to the HCV IRES (lane 2), the EMCV IRES (lane 3), the polioviral IRES (lane 4) and the BiP IRES (lane 5) were incubated with a cytoplasmic extract of HeLa cells and then precipitated with streptavidin–acrylamide beads. The bead-bound proteins were analyzed by western blot analysis using a monoclonal antibody against the human La protein. Proteins bound to empty beads (lane 6) and 10% of input HeLa cell extract (lane 1) were included in the western blot analysis as negative and positive controls.
It has been reported that La protein binds to certain IRES elements, such as those of poliovirus and HCV (31,35). To compare the interactions of La with different IRES elements, RNA affinity resin binding assays were performed using HeLa cell extracts and different IRES RNAs (Fig. 1B). Biotinylated RNAs spanning the HCV, EMCV, poliovirus and BiP IRESs were synthesized using T7 RNA polymerase. After incubating HeLa cell extracts with the biotinylated RNAs at the same molarity, the RNA-bound proteins were precipitated with streptavidin–acrylamide beads, resolved by SDS–PAGE and then subjected to western blot analysis with anti-human La antibody. Whereas La protein bound strongly to the poliovirus or BiP IRES (Fig. 1B, lanes 4 and 5), it bound weakly to the HCV IRES (Fig. 1B, lane 2). On the other hand, La protein was not detected among the proteins bound to the EMCV IRES under these conditions (Fig. 1B, lane 3). All of the tested RNAs (HCV, EMCV, poliovirus and BiP) showed similar stabilities under the binding assay conditions (data not shown). This indicates that the differences in intensity of the La protein bands in Figure 1B reflect differences in RNA affinity rather than RNA stability.
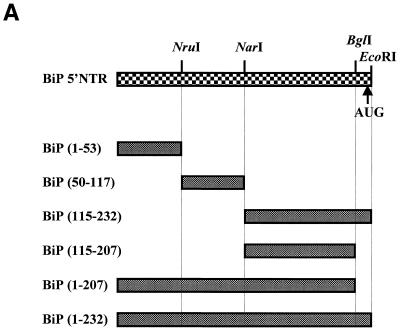
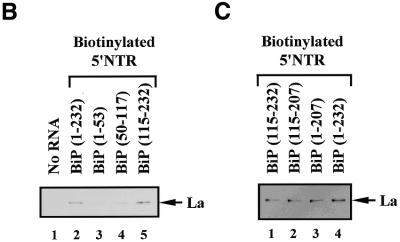
In order to determine the domain of BiP IRES required for interaction with La protein, we performed RNA–protein binding assays with RNAs corresponding to various parts of the BiP 5′-UTR as depicted in Figure 2A. La protein bound preferentially to the full-length BiP IRES and BiP UTR(115–232) (Fig. 2B, lanes 2 and 5, respectively). La protein bound weakly to the BiP UTR(50–117) fragment (Fig. 2B, lane 4). On the other hand, binding of La to the BiP UTR(1–53) RNA fragment was not detected (Fig. 2B, lane 3).
Figure 2.
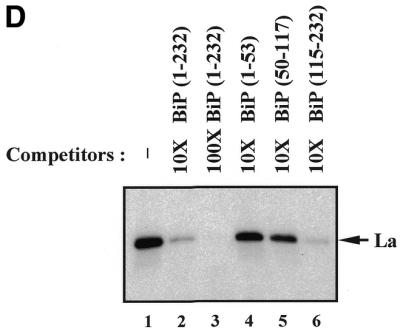
Determination of La-binding sites in the BiP IRES. (A) Schematic diagram of the BiP 5′-UTR and its deletion mutants used in La-binding assays. AUG denotes the initiator AUG codon. (B) Protein-binding assays were carried out using biotinylated RNAs corresponding to the full-length BiP IRES (lane 2) or to its deletion mutants (lanes 3–5). Lane 1 is the negative control showing proteins interacting with the beads themselves. (C) Effect of the initiator AUG on binding of La protein to the BiP IRES. The protein-binding assays were performed using biotinylated RNAs with (lanes 1 and 4) and without (lanes 2 and 3) the initiator AUG and the sequence immediately preceding the AUG. (D) The IRES-binding efficiency of purified La was monitored by UV crosslinking/competition assay. The 32P-labeled full-length BiP IRES and 0.1 µg purified La protein were used as probe and protein source, respectively. Competition experiments were carried out by adding cold competitor RNAs in 10- (lanes 2 and 4–6) and 100-fold (lane 2) molar excess. The competitor RNAs used were BiP UTR(1–232), UTR(1–53), UTR(50–117) and UTR(115–232), as indicated at the top of the panel. Lane 1 shows a control reaction with no competitor.
The effect of the initiator AUG codon on La binding was also investigated, since the initiator AUG codon of HCV had been reported to affect binding of La to the IRES element (35). The RNA-binding activity of La was tested using RNAs corresponding to the BiP 5′-UTR sequences (115–232), (115–207), (1–207) and (1–232) (Fig. 2C). We found that La protein bound efficiently to BiP UTRs containing nt 115–207, such as UTR(1–232), UTR(1–207), UTR(115–232) and UTR(115–207), regardless of the presence of the initiator AUG codon (Fig. 2C, compare lanes 1 and 3 with 2 and 4). This indicates that the initiator AUG codon is not involved in binding of La protein. Instead, nucleotides between 115 and 207 of the BiP IRES are required for the interaction with the La autoantigen.
To further confirm the direct interaction of La protein and the BiP IRES, UV crosslinking experiments using 32P-labeled full-length BiP IRES and purified La were performed in the presence of cold competitor RNAs corresponding to different portions of the BiP 5′-UTR (Fig. 2D). Addition of a 10- and 100-fold molar excess of cold full-length BiP IRES strongly inhibited binding of La protein to the probe (Fig. 2D, lanes 2 and 3). Moreover, BiP UTR(115–232) also strongly competed for binding of La to the BiP IRES, similarly to the full-length BiP IRES (Fig. 2D, compare lane 6 with 2). BiP UTR(50–117) competed slightly for La binding (Fig. 2D, lane 5), whereas BiP UTR(1–53) showed no competition (Fig. 2D, lane 4). These observations coincide well with the RNA–protein co-precipitation results shown in Figure 2B and C. This indicates that La preferentially binds to the second half of the BiP 5′-UTR (nt 115–207).
La autoantigen selectively enhances BiP IRES-dependent translation in vitro
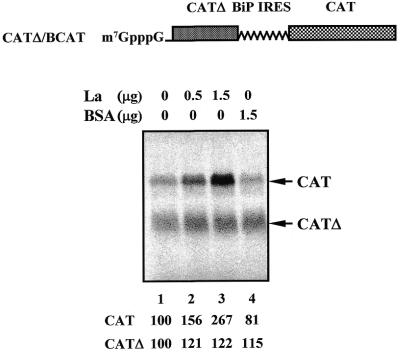
The role of the La protein in BiP IRES-dependent translation was examined in a RRL translation system, which contains less endogenous La protein compared with HeLa cell extract (31,46). In the case of capped dicistronic CATΔ-BCAT mRNA, the relative ratio of translation directed by the BiP IRES to translation by ribosome scanning was inversely correlated with the mRNA concentration used in the in vitro translation reaction (data not shown). In vitro translation reactions were performed with 6 nM capped CATΔ-BCAT mRNA, shown in the top panel of Figure 3. Addition of increasing amounts of purified La protein gradually enhanced BiP IRES-dependent translation up to 2.7-fold, whereas the first cistron (CATΔ) was unaffected (Fig. 3, lanes 1–3). On the other hand, BSA did not affect either internal initiation directed by the BiP IRES or cap-dependent translation (Fig. 3, lane 4).
Figure 3.
Effect of La protein on in vitro translation directed by the BiP IRES. (Top) Schematic diagram of capped mRNA CATΔ-BCAT. In vitro translations were performed using 6 nM capped CATΔ-BCAT mRNAs. Purified La (lanes 2 and 3) or BSA (lane 4) was added to reaction mixtures. The amounts of protein added to the reactions are indicated at the top of the panel. No protein was added to the sample loaded in lane 1. The intensities of the bands were measured with a densitometer. The relative efficiencies of the translation reactions were normalized by setting the products of the control translation reaction without additional protein as 100%.
Overexpression of La autoantigen enhances BiP IRES-dependent translation in vivo
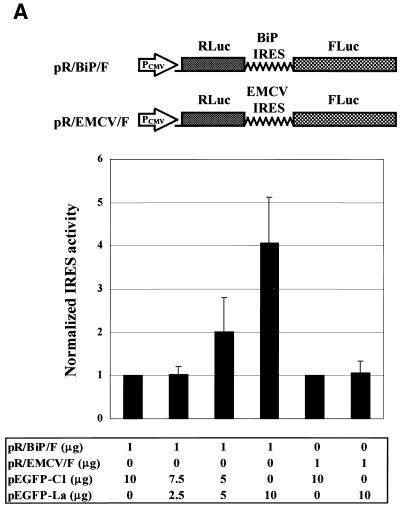
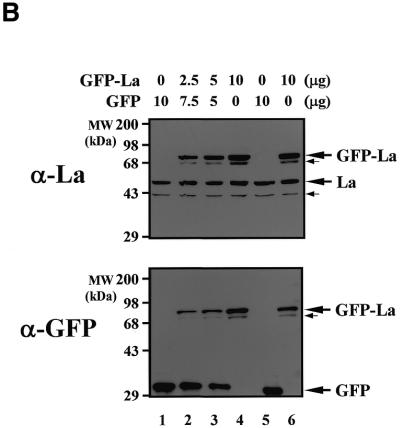
The enhancing effect of La autoantigen on BiP IRES-dependent translation in vivo was examined using a transient expression system. A dicistronic vector pR/BiP/F, comprising RLuc–BiP IRES–FLuc, was constructed for use as a reporter (Fig. 4A). The reporter plasmid was co-transfected into Cos-7 cells with effector plasmid pEGFP or pEGFP-La. Renilla luciferase and firefly luciferase activities were measured to monitor translation of the reporter genes (see Materials and Methods). The expression levels of endogenous La protein or the effector proteins in the transfected cells were monitored by western blot analysis using anti-human La monoclonal antibody or anti-GFP monoclonal antibody (Fig. 4B). The densitometric measurement of total La protein indicated that the level of GFP–La was ∼3-fold higher than that of endogenous La protein. Taking the transfection efficiency of the cells (∼10–15%) into account, the level of GFP–La in each transfected cell was much higher than that of endogenous La protein (∼20-fold). Interestingly, western blot analyses revealed some La-related proteins (indicated by arrows). These proteins are likely to be C-terminally truncated La and GFP-fused La proteins that could have been generated by caspase-3 (58).
Figure 4.
Effect of co-expression of La protein on translation directed by the BiP IRES in Cos-7 cells. (A) (Top) Schematic diagrams of dicistronic vectors pR/BiP/F and pR/EMCV/F. Reporter plasmid (pR/BiP/F or pR/EMCV/F) and effector plasmid (pEGFP-C1 or pEGFP-La) were co-transfected into Cos-7 cells. (Bottom) The numbers of transfected plasmids. (Middle) Relative activities of Renilla luciferase (RLuc) and firefly luciferase (FLuc). The relative values FLuc/RLuc of the samples are compared to that of the control. The columns and bars represent the means ± SD of triplicate experiments. (B) Expression level of La protein in the transfected cells. Western blot analyses were performed using a monoclonal antibody against human La autoantigen (upper) or a monoclonal antibody against GFP (lower). The GFP-fused La, endogenous La, and GFP are indicated by bold arrows. Small arrows depict La-related proteins.
The expression of FLuc directed by BiP IRES was enhanced as the GFP–La level increased in a dose-dependent manner (Fig. 4A). The relative ratio of Fluc to Rluc was increased up to 4.1-fold when GFP–La was co-expressed (Fig. 4A, compare lanes 1 and 4). In contrast, expression of FLuc via the EMCV IRES was unaffected by co-expression of 10 µg GFP–La (Fig. 4A, compare lanes 5 and 6). These results, along with the in vitro data, indicate that La autoantigen enhances translation directed by the BiP IRES.
DISCUSSION
La protein has been suggested to stimulate IRES-dependent translation of several viral mRNAs, such as from HCV and poliovirus (31,35). However, the role of La protein in cellular IRES-dependent translation has not as yet been described. Here we report that La protein interacts with a cellular IRES (BiP) and enhances IRES-dependent translation.
Rather large quantities of purified La protein were required for the enhancement of BiP IRES-dependent translation (Fig. 3). This might be due to a low specific activity of the La protein expressed in Escherichia coli, which produces modification-deficient proteins. Fan et al. (59) demonstrated that La is a phosphoprotein and that phosphorylation of the La protein is involved in the regulation of recycling of the RNA polymerase III transcription complex. Therefore, it is possible that phosphorylation of La protein may be required for the full activity, although both phosphorylated and dephosphorylated La protein bind to RNA equally well (59).
Previously, Yang and Sarnow (60) tried to identify cellular proteins interacting with the BiP IRES. The authors found 60 and 90 kDa proteins specifically interacting with the 3′-half (nt 129–220) of the BiP IRES. Immunochemical studies indicated that the 60 kDa protein is neither PTB nor La protein. The authors, however, did not perform experiments that could show the interaction between the the BiP IRES and La protein described in this report. Therefore, the discrepancy between these results is likely due to the differences in biochemical approaches used in the experiments. In the previous report the authors used a nuclear extract, while we used a cytoplasmic extract as protein source. Moreover, the authors did not investigate the effect of La protein on translation of BiP mRNA.
La antigen is localized mainly in the nucleus, but it may shuttle between the nucleus and the cytoplasm (61). Moreover, intracellular redistribution (cytoplasmic accumulation) of La protein occurs under certain stress conditions, such as UV irradiation (62), virus infection (31,63), apoptosis (58) and inhibition of RNA synthesis (61). Although the level of GFP–La in each transfected cell was much higher than that of endogenous La protein (∼20-fold; Fig 4B), most of the GFP–La was localized in the nucleus (data not shown). Only a small portion of GFP–La seems to stay in the cytoplasm and participate in IRES-dependent translation. This subcellular localization pattern of La protein likely contributes to the requirement for a large quantity of La protein for translational activation.
It is not clear how La protein enhances translation of BiP mRNA. It is possible that La may recruit the translational machinery, such as translational initiation factors and/or the 40S ribosomal subunit, near the initiation codon. In this respect, interaction of La protein with the 40S ribosomal subunit, perhaps by direct association with 18S rRNA, may give a clue to this kind of mechanism (64). Alternatively, La may induce a conformational change in the IRES element to facilitate interaction between RNA and translational machinery. The RNA helicase (65) and molecular chaperone (66,67) activities of La protein may play a role in this process.
What would be the physiological implications of translational regulation of BiP mRNA by La protein? La protein binds to the cellular IRES of XIAP (17), which is up-regulated by low dose ionizing irradiation (68). A dominant-negative deletion mutant of La (amino acids 226–348) reduced translation of XIAP mRNA in vitro and in vivo. This suggests that La protein is also involved in cellular mRNA translation. Intriguingly, La mRNAs that are composed of at least two isoforms (La1 and La1′) through alternative splicing, themselves contain IRES elements in the 5′-UTRs (69). This may be a survival strategy of cells under stress conditions that suppresses cap-dependent translation. Even under stress conditions, La protein may be translated continuously through the IRES elements. La protein, in turn, may assist translation of some mRNAs, such as XIAP and BiP, that are required for down-regulation of the stress response when environmental conditions become normal.
BiP protein, which is also known as GRP78, is homologous to HSP70 (23). La protein is one of the autoantigens recognized by sera from autoimmune patients with systemic lupus erythematosus and Sjögren’s syndrome (36). Overexpression of some heat-shock proteins has been detected in human autoimmune diseases such as systemic lupus erythematosus (70,71). Moreover, Ku protein, which was discovered as an autoantigen in a patient with scleroderma-polymyositis overlap syndrome, has been shown to modulate expression of hsp70 at the transcription level (72–74). Here we report that the autoantigen La enhances translation of a heat-shock protein (BiP) at the translation level. The relationship between heat-shock proteins and autoantigens, therefore, is an intriguing aspect of autoimmune diseases.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Dr M. Bachmann (Institut für Physiologische Chemie, Johannes-Gutenberg Universität, Germany) for kindly providing us with the monoclonal antibody against human La (3B9). The present study was supported in part by the G7, NRL and Molecular Medicine Research Group Programs of MOST.
REFERENCES
- 1.Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol., 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang S.K., Davies,M.V., Kaufman,R.J. and Wimmer,E. (1989) Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J. Virol., 63, 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang S.K., Krausslich,H.G., Nicklin,M.J., Duke,G.M., Palmenberg,A.C. and Wimmer,E. (1988) A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol., 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J. and Sonenberg,N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- 5.Jackson R.J., Hunt,S.L., Gibbs,C.L. and Kaminski,A. (1994) Internal initiation of translation of picornavirus RNAs. Mol. Biol. Rep., 19, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama-Kohara K., Iizuka,N., Kohara,M. and Nomoto,A. (1992) Internal ribosome entry site within hepatitis C virus RNA. J. Virol., 66, 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohlmann T., Lopez-Lastra,M. and Darlix,J.L. (2000) An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem., 275, 11899–11906. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki J. and Nakashima,N. (1999) Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol., 73, 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macejak D.G. and Sarnow,P. (1991) Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature, 353, 90–94. [DOI] [PubMed] [Google Scholar]
- 10.Nanbru C., Lafon,I., Audigier,S., Gensac,M.C., Vagner,S., Huez,G. and Prats,A.C. (1997) Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem., 272, 32061–32066. [DOI] [PubMed] [Google Scholar]
- 11.Stoneley M., Paulin,F.E., Le Quesne,J.P., Chappell,S.A. and Willis,A.E. (1998) C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene, 16, 423–428. [DOI] [PubMed] [Google Scholar]
- 12.Jeanne B., Sella,O., Le,S. and Elroy-Stein,O. (1997) PDGF2/c-sis mRNA Leader contains a differentiation-linked internal ribosomal entry site (D-IRES). J. Biol. Chem., 272, 9356–9362. [DOI] [PubMed] [Google Scholar]
- 13.Gan W. and Rhoads,R.E. (1996) Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J. Biol. Chem., 271, 623–626. [DOI] [PubMed] [Google Scholar]
- 14.Huez I., Creancier,L., Audigier,S., Gensac,M.C., Prats,A.C. and Prats,H. (1998) Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol., 18, 6178–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pyronnet S., Pradayrol,L. and Sonenberg,N. (2000) A cell cycle-dependent internal ribosome entry site. Mol. Cell, 5, 607–616. [DOI] [PubMed] [Google Scholar]
- 16.Cornelis S., Bruynooghe,Y., Denecker,G., Van Huffel,S., Tinton,S. and Beyaert,R. (2000) Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell, 5, 597–605. [DOI] [PubMed] [Google Scholar]
- 17.Holcik M. and Korneluk,R.G. (2000) Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol., 20, 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oumard A., Hennecke,M., Hauser,H. and Nourbakhsh,M. (2000) Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol. Cell. Biol., 20, 2755–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pozner A., Goldenberg,D., Negreanu,V., Le,S.Y., Elroy-Stein,O., Levanon,D. and Groner,Y. (2000) Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol., 20, 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coldwell M.J., Mitchell,S.A., Stoneley,M., MacFarlane,M. and Willis,A.E. (2000) Initiation of Apaf-1 translation by internal ribosome entry. Oncogene, 19, 899–905. [DOI] [PubMed] [Google Scholar]
- 21.Haas I.G. and Wabl,M. (1983) Immunoglobulin heavy chain binding protein. Nature, 306, 387–389. [DOI] [PubMed] [Google Scholar]
- 22.Pelham H.R.B. (1986) Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell, 46, 959–961. [DOI] [PubMed] [Google Scholar]
- 23.Munro S. and Pelham,H.R.B. (1986) An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell, 46, 291–300. [DOI] [PubMed] [Google Scholar]
- 24.Lee A.S. (1987) Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem. Sci., 12, 20–23. [Google Scholar]
- 25.Lee A.S. (1992) Mammalian stress response: induction of the glucose-regulated protein family. Curr. Opin. Cell Biol., 4, 267–273. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y.K., Hahm,B. and Jang,S.K. (2000) Polypyrimidine tract-binding protein inhibits translation of BiP mRNA. J. Mol. Biol., 304, 119–133. [DOI] [PubMed] [Google Scholar]
- 27.Sarnow P. (1989) Translation of glucose-regulated protein 78/immunoglobulin heavy-chain binding protein mRNA is increased in poliovirus-infected cells at a time when cap-dependent translation of cellular mRNAs is inhibited. Proc. Natl Acad. Sci. USA, 86, 5795–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prostko C.R., Brostrom,M.A., Galuska-Malara,E.M. and Brostrom,C.O. (1991) Stimulation of GRP78 gene transcription by phorbol ester and cAMP in GH3 pituitary cells. The accommodation of protein synthesis to chronic deprivation of intracellular sequestered calcium. J. Biol. Chem., 266, 19790–19795. [PubMed] [Google Scholar]
- 29.Ulatowski L.M., Lam,M., Vanderburg,G., Stallcup,M.R. and Distelhorst,C.W. (1993) Relationship between defective mouse mammary tumor virus envelope glycoprotein synthesis and GRP78 synthesis in glucocorticoid-treated mouse lymphoma cells. Evidence for translational control of GRP78 synthesis. J. Biol. Chem., 268, 7482–7488. [PubMed] [Google Scholar]
- 30.Scheper G.C., Voorma,H.O. and Thomas,A.A.M. (1992) Eukaryotic initiation factors-4E and -4F stimulate 5′ cap-dependent as well as internal initiation of protein synthesis. J. Biol. Chem., 267, 7269–7274. [PubMed] [Google Scholar]
- 31.Meerovitch K., Svitkin,Y.V., Lee,H.S., Lejbkowicz,F., Kenan,D.J., Chan,E.K.L., Agol,V.I., Keene,J.D. and Sonenberg,N. (1993) La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol., 67, 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellen C.U.T., Witherell,G.W., Schmid,M., Shin,S.H., Pestova,T.V., Gil,A. and Wimmer,E. (1993) A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA, 90, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell S.A., Brown,E.C., Coldwell,M.J., Jackson,R.J. and Willis,A.E. (2001) Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol., 21, 3364–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blyn L.B., Towner,J.S., Semler,B.L. and Ehrenfeld,E. (1997) Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol., 71, 6243–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali N. and Siddiqui,A. (1997) The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl Acad. Sci. USA, 94, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan E.M. (1989) Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv. Immunol., 44, 93–151. [DOI] [PubMed] [Google Scholar]
- 37.Kenan D.J., Query,C.C. and Keene,J.D. (1991) RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci., 16, 214–220. [DOI] [PubMed] [Google Scholar]
- 38.Maraia R.J. (1996) Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc. Natl Acad. Sci. USA, 93, 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maraia R.J., Kenan,D.J. and Keene,J.D. (1994) Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol., 14, 2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madore S.J., Wieben,E.D. and Pederson,T. (1984) Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J. Biol. Chem., 259, 1929–1933. [PubMed] [Google Scholar]
- 41.Toczyski D.P. and Steitz,J.A. (1991) EAP, a highly conserved cellular protein associated with Epstein–Barr virus small RNAs (EBERs). EMBO J., 10, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francoeur A.M. and Mathews,M.B. (1982) Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc. Natl Acad. Sci. USA, 79, 6772–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurilla M.G. and Keene,J.D. (1983) The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell, 34, 837–845. [DOI] [PubMed] [Google Scholar]
- 44.Chang Y.N., Kenan,D.J., Keene,J.D., Gatignol,A. and Jeang,K.T. (1994) Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol., 68, 7008–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svitkin Y.V., Pause,A. and Sonenberg,N. (1994) La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol., 68, 7001–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y.K. and Jang,S.K. (1999) La protein is required for efficient translation driven by encephalomyocarditis virus internal ribosomal entry site. J. Gen. Virol., 80, 3159–3166. [DOI] [PubMed] [Google Scholar]
- 47.Borovjagin A.V., Ezrokhi,M.V., Rostapshov,V.M., Ugarova,T.Y., Bystrova,T.F. and Shatsky,I.N. (1991) RNA–protein interactions within the internal translation initiation region of encephalomyocarditis virus RNA. Nucleic Acids Res., 19, 4999–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang S.K. and Wimmer,E. (1990) Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev., 4, 1560–1572. [DOI] [PubMed] [Google Scholar]
- 49.Hellen C.U.T., Pestova,T.V., Litterst,M. and Wimmer,E. (1994) The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J. Virol., 68, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luz N. and Beck,E. (1991) Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol., 65, 6486–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borovjagin A., Pestova,T. and Shatsky,I. (1994) Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett., 351, 299–302. [DOI] [PubMed] [Google Scholar]
- 52.Hunt S.L. and Jackson,R.J. (1999) Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA, 5, 344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaminski A., Hunt,S.L., Patton,J.G. and Jackson,R.J. (1995) Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA, 1, 924–938. [PMC free article] [PubMed] [Google Scholar]
- 54.Sella O., Gerlitz,G., Le,S.Y. and Elroy-Stein,O. (1999) Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell. Biol., 19, 5429–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahm B., Kim,Y.K., Kim,J.H., Kim,T.Y. and Jang,S.K. (1998) Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol., 72, 8782–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Oh Y.L., Hahm,B., Kim,Y.K., Lee,H.K., Lee,J.W., Song,O.-K., Tsukiyama-Kohara,K., Kohara,M., Nomoto,A. and Jang,S.K. (1998) Determination of functional domains in polypyrimidine tract-binding protein. Biochem. J., 331, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ayukawa K., Taniguchi,S., Masumoto,J., Hashimoto,S., Sarvotham,H., Hara,A., Aoyama,T. and Sagara,J. (2000) La autoantigen is cleaved in the COOH terminus and loses the nuclear localization signal during apoptosis. J. Biol. Chem., 275, 34465–34470. [DOI] [PubMed] [Google Scholar]
- 59.Fan H., Sakulich,A.L., Goodier,J.L., Zhang,X., Qin,J. and Maraia,R.J. (1997) Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell, 88, 707–715. [DOI] [PubMed] [Google Scholar]
- 60.Yang Q. and Sarnow,P. (1997) Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA–protein interactions. Nucleic Acids Res., 25, 2800–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bachmann M., Pfeifer,K., Schröder,H.C. and Müller,W.E.G. (1989) The La antigen shuttles between the nucleus and the cytoplasm in CV-1 cells. Mol. Cell. Biochem., 85, 103–114. [DOI] [PubMed] [Google Scholar]
- 62.Bachmann M., Chang,S., Slor,H., Kukulies,J. and Müller,W.E.G. (1990) Shuttling of the autoantigen La between nucleus and cell surface after uv irradiation of human keratinocytes. Exp. Cell Res., 191, 171–180. [DOI] [PubMed] [Google Scholar]
- 63.Shiroki K., Isoyama,T., Kuge,S., Ishii,T., Ohmi,S., Hata,S., Suzuki,K., Takasaki,Y. and Nomoto,A. (1999) Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol., 73, 2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peek R., Pruijn,G.J. and Van Venrooij,W.J. (1996) Interaction of the La (SS-B) autoantigen with small ribosomal subunits. Eur. J. Biochem., 236, 649–655. [DOI] [PubMed] [Google Scholar]
- 65.Huhn P., Pruijn,G.J.M., van Venrooij,W.J. and Bachmann,M. (1997) Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res., 25, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pannone B.K., Xue,D. and Wolin,S.L. (1998) A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J., 17, 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoo C.J. and Wolin,S.L. (1997) The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]
- 68.Holcik M., Lefebvre,C., Yeh,C., Chow,T. and Korneluk,R.G. (1999) A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nature Cell Biol., 1, 190–192. [DOI] [PubMed] [Google Scholar]
- 69.Carter M.S. and Sarnow,P. (2000) Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J. Biol. Chem., 275, 28301–28307. [DOI] [PubMed] [Google Scholar]
- 70.Latchman D.S. and Isenberg,D.A. (1994) The role of hsp90 in SLE. Autoimmunity, 19, 211–218. [DOI] [PubMed] [Google Scholar]
- 71.Stephanou A., Latchman,D.S. and Isenberg,D.A. (1998) The regulation of heat shock proteins and their role in systemic lupus erythematosus. Semin. Arthritis Rheum., 28, 155–162. [DOI] [PubMed] [Google Scholar]
- 72.Li G.C., Yang,S.H., Kim,D., Nussenzweig,A., Ouyang,H., Wei,J., Burgman,P. and Li,L. (1995) Suppression of heat-induced hsp70 expression by the 70-kDa subunit of the human Ku autoantigen.Proc. Natl Acad. Sci. USA, 92, 4512–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mimori T., Ohosone,Y., Hama,N., Suwa,A., Akizuki,M., Homma,M., Griffith,A.J. and Hardin,J.A. (1990) Isolation and characterization of cDNA encoding the 80-kDa subunit protein of the human autoantigen (p70/p80) recognized by autoantibodies from patients with scleroderma-polymyositis overlap syndrome. Proc. Natl Acad. Sci. USA, 87, 1777–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang S.H., Nussenzweig,A., Li,L., Kim,D., Ouyang,H., Burgman,P. and Li,G.C. (1996) Modulation of thermal induction of hsp70 expression by Ku autoantigen or its individual subunits. Mol. Cell. Biol., 16, 3799–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]