Abstract
The potential of lipid nanoparticles (LNPs) as nucleic acid delivery vehicles has been demonstrated in recent years, culminating in the emergency use approval of LNP-based mRNA SARS-CoV-2 vaccines in late 2020. The determination of RNA content relative to LNP size can be important to the understanding of efficacy and adverse effects. This work presents the first description of a facile and rapid analytical method for online, size-dependent RNA payload distribution measurement using data from multi-angle light scattering, ultraviolet and refractive index detectors following separation of the LNPs by size-exclusion chromatography. The analysis was validated by size-based fractionation of the LNPs with subsequent offline analysis of the fractions. Four LNPs formulated with different PEG-lipids and different lipid compositions were tested. Good agreement was observed between the online and offline size-based RNA distributions among all four LNPs, demonstrating the utility of the online method for LNP-encapsulated RNA in general, and suggesting a means for simplified biophysical quantitation of a dosing-related critical quality attribute.
Keywords: Lipid nanoparticle, RNA, Multi-angle light scattering, Size-dependent, Online analysis, Size exclusion chromatography
1. Introduction
Lipid nanoparticle (LNP) is a promising delivery platform for nucleic acids, for example mRNA or siRNA, in the areas of infectious disease [1], [2], [3], [4], [5], [6], [7], oncology [8], [9], [10], [11] and SARS-CoV-2 vaccine [12], [13]. LNPs encapsulating RNA (LNP-RNA) are formed through electrostatic capture and rapid precipitation by mixing of the payload and lipid packets comprised of cationic and neutral lipids. Various efforts have been made to study the physicochemical characteristic of the particles, such as size [14], [15] and compositional heterogeneity [14], morphology [14], [16], [17], macromolecular structure [18] and RNA encapsulation efficiency [17], [19]. Deep understanding of LNPs at the molecular level can help elucidate correlation of structure and in vitro/ in vivo performance, and develop effective manufacturing and control strategies.
Research has shown that the in vivo potency and tissue-penetrating ability of LNPs are related to particle size [20], [21]. Through fractionation by semi-preparative SEC and subsequent offline size measurement of the fractionated samples by hydrodynamic light scattering (DLS), Zhang et al. studied the size dependence of LNP composition, RNA encapsulation and in vitro gene-silencing activities [15]. Their data showed correlation between the cationic polymer/RNA ratio and RNA gene-silencing potency, which highlighted the importance of size-based compositional assessment of the LNP-RNAs.
Although semi-preparative fractionation by size with offline fraction analysis provides a viable approach to analysis of size-based compositional heterogeneity, the experiment is lengthy and laborious, and not suitable for use as a routine analytical procedure. Online analysis of the RNA payload versus size, in a single automated run, would be highly desirable. A candidate method for this type of characterization is SEC-MALS-UV-dRI, where size-exclusion chromatography (SEC) is coupled to a multi-angle light scattering (MALS) detector and concentration detectors such as a UV photometer and a differential refractometer (dRI).
SEC-MALS-UV-dRI is often used for the characterization of large molecules or complex macro-entities such as LNPs. This technique enables scientists to obtain size and sample concentration across the SEC peak without the need of a reference standard. For binary conjugates, and with the knowledge of the UV extinction coefficient and dn/dc values (or specific refractive index increment) of each constituent in the conjugate, such a multi-detector system enables immediate quantification of the two constituent components in the conjugate at each eluting fraction. However, for particles larger than roughly 50 nm in diameter such as LNP-RNA, the standard conjugate analysis is confounded by size-dependent UV light scattering in the UV detector. Instead of providing purely absorptive quantification of the RNA concentration, the total measured UV extinction at 260 nm (a characteristic absorption wavelength of RNA) consists of absorption by the RNA payload and scattering by the complete particles, which makes the direct analysis of RNA concentration and correlation with size information by MALS, RI and UV detectors highly challenging.
Methods for correction of the scattering contribution have been reported [22], [23], [24]. Porterfield and Zlotnick used the Rayleigh theory of scattering, wherein the extinction due to scattering is proportional to R6/λ4 (where R is the particle radius and λ the illuminating wavelength) to correct for the scattering effect by measuring extinction at the non-absorbed wavelengths of 340 nm and 360 nm, and obtained improved measurements of protein and nucleic acid content in viruses [24]. This method can be relatively successful if the particles are uniform in size and small compared to the UV wavelengths. However, it does not take into account a variety of effects such as the size-dependent variation of scattering efficiency (especially significant when the particle size is not small compared to the UV wavelengths and therefore the dependence is not strictly proportional to R6/λ4) and the variation of the particle’s refractive index with wavelength (the simple R6/λ4 correction assumes a wavelength-independent refractive index). Consequently, the application of this approach is limited.
Herein we report extended characterization of LNP-RNA wherein the quantity of LNP-associated RNA is determined as a function of LNP size. The novel method utilizes data from online MALS, UV, and dRI detectors following size-based separation by SEC. The approach is validated through comparison with RNA quantitation using a reverse-phase liquid chromatography (RPLC) assay performed on individual size fractions. Our study concludes that the novel online method provides accurate measurement of size-dependent RNA content in LNPs.
2. Materials and methods
2.1. Materials and lipid nanoparticle formulation
Chemically modified dsRNA were synthesized at Merck & Co., Inc., Kenilworth, NJ, USA. Ionizable amino lipid was synthesized at Merck & Co., Inc., Kenilworth, NJ, USA, as previously described [25]. Distearoylphosphatidylcholine (DSPC) and cholesterol were obtained from Sigma-Aldrich (St. Louis, MO). Poly(ethylene glycol) lipid (PEG-lipid) 1 (Sunbright GM-020) [26] and PEG-lipid 2 (Sunbright GM-020CN) [18] were manufactured by NOF Corporation (White Plains, NY). PEG-lipid 3, with similar structure as PEG-lipid 2 but different PEG repeating unit, was synthesized at Merck & Co., Inc., Kenilworth, NJ, USA. Phosphate-buffered saline (PBS) and Tris G buffers were purchased from HyClone (Logan, UT, USA). Triton X-100, ammonium bicarbonate and ethanol were obtained from Sigma-Aldrich (St. Louis, MO, USA). SYBR gold was obtained from Fisher Scientific (Walthm, MA, USA). Acetonitrile was from Fisher Scientific (Waltham, MA, USA). HPLC grade Milli-Q water was generated by Milli-Q purification system (Millipore-Sigma, Burlington, MA, USA).
LNP encapsulating RNA were manufactured by Merck & Co., Inc., Kenilworth, NJ, USA using a rapid precipitation process as previously described [27]. Briefly, LNPs were assembled by mixing an organic solution of lipids with an aqueous RNA solution. RNA duplexes were prepared in an aqueous sodium citrate buffer (20 mM, pH 5) at a concentration targeting a molar ratio of the amine in the amino lipid to the phosphate group in the RNA (N:P) of 3:1. The lipid components of the LNP comprised an ionizable amino lipid, DSPC, cholesterol, and PEG-lipid in a molar ratio of 58:10:30:2 (LNP-1, LNP-2, LNP-3) and 58:10:28:4 (LNP-4). Three PEG-lipids were used in the formulation with PEG-lipid 1 for LNP-1, PEG-lipid 2 for LNP-2 and PEG-lipid 3 for LNP-3 and LNP-4. The resulting LNP-RNAs were finally concentrated, sterilized via filtration through 0.45 and 0.2 μm sterile filters (Pall Corp., Port Washington, NY), and dispensed into sterile vials under aseptic conditions. Empty LNPs were prepared using the same procedure, but excluding RNA from the aqueous solution in LNP assembly. Eight LNP variants, four empty LNPs (LNP-1E, LNP-2E, LNP-3E, LNP-4E) and four LNP-RNAs (LNP-1F, LNP-2F, LNP-3F, LNP-4F) were prepared, with lipid molar compositions as defined per LNP-1, LNP-2, LNP-3 and LNP-4.
2.2. Cryo-transmission electron microscopy and RNA encapsulation efficiency assay
An aliquot of 3 µL of the resulting LNP sample was applied to C-flat (Protochips, NC, USA) holey carbon grids, plasma cleaned by Solarus plasma cleaner (Gatan, CA, USA). The grids were blotted with filter paper and then plunge-frozen into liquid ethane at −170 °C using a manual plunger. Grids were imaged on a Thermo Fisher Scientific (Hillsboro, OR, USA) Glacios Cryo Transmission Electron Microscope (Cryo-TEM) operated at 200 kV and equipped with a Falcon 3 direct electron detector. Automated data-collection was carried out using Leginon software (PMID: 15890530) where high magnification images were acquired by selecting targets at a lower magnification. For each sample condition, >50x high magnification images were collected for morphological studies of the LNPs and diameter measurements for size distribution.
The RNA encapsulation into the LNPs were measured by following the previously published procedure [15].
2.3. SEC analysis of LNP and fractionation
LNPs were analyzed by SEC-MALS-UV-dRI on an Agilent 1200 HPLC system equipped with a UV/Vis photodiode array (PDA) detector (Agilent Technologies, Santa Clara, CA, USA) which was further connected to a DAWN HELEOS II MALS detector and an Optilab T-rEX dRI detector (both from Wyatt Technology Corporation, Goleta, CA, USA). In order to prevent photodiode saturation by the strongly scattering LNP particles, the laser power of the DAWN detector was set to 20 %. A DynaPro NanoStar dynamic light scattering (DLS) detector (Wyatt Technology Corporation) was connected by optical fiber to the 16th angle of MALS detector for online Rh analysis. Data were acquired and analyzed with ASTRA software version 6.1.7.15 (Wyatt Technology).
SEC separation was performed on a TSKgel-G6000PWxl-CP SEC column of dimensions 7.8 mm × 300 mm and 13 µm particle size (TOSOH Bioscience, Tokyo, Japan), with 1x PBS as mobile phase at 0.5 mL/min flow rate and column temperature 35 °C. UV extinction was monitored at 260 nm. The delay volumes and band broadening between the UV, MALS and RI detectors were determined and corrected using the Pierce bovine serum albumin (BSA) standard (Thermo Fisher Scientific, Waltham, MA).
For online LNP-RNA characterization by SEC-MALS, ∼20 μg of sample were loaded onto the column. For offline RNA payload profiling, ∼300 µg of LNP were loaded onto the column and eluted fractions were collected at 0.5 min intervals. To avoid extensive peak broadening, fractions were collected directly after the PDA detector into an Agilent G1364C 54-vial-tray fraction collector (Agilent Technologies). Additional measurements with ∼ 300 µg loads were performed with the full set of UV, MALS and dRI detectors for the purpose of determining the total mass in each fraction collected by the fraction collector, as described in the next section.
2.4. Determination of total mass in collected fractions
The specific refractive index increment dn/dc of the LNPs was measured in batch mode on the Optilab T-rEX dRI detector. A series of LNP solutions diluted with the sample formulation matrix to a known concentration was infused into the RI detector through a syringe pump (Fusion 200, Chemyx Inc., Stafford, TX, USA). The dn/dc value determined for LNPs manufactured with either PEG-lipid 1 or PEG-lipid 2 and 3 was 0.16 mL/g. This value was utilized to calculate the total mass in each fraction with dRI measurements.
The UV peak of the fractionated ∼ 300 µg injection (PDA only, referred as ‘300UV-frac’) was compared to the UV peak of the ∼ 300 µg injection measured with all three detectors in line (‘300UV-MD’) to determine the retention time difference. The latter was combined with the UV – RI delay volume and band broadening corrections, determined with an injection of BSA mentioned above, in order to align 300UV-frac with the RI peak of the multi-detector injection (‘300RI-MD’). 300RI-MD was then sliced to correlate with the collected fractions of 300UV-frac. The total mass of the fractions was calculated from the sliced 300RI-MD peaks by integrating the concentration over the fraction collection interval with ASTRA software, using the measured dn/dc value.
2.5. Batch dynamic light scattering
Batch-mode measurements of hydrodynamic radius (Rh) were performed using the above DynaPro NanoStar, with the optical fiber disconnected from the DAWN MALS detector. In this mode aliquots were pipetted into 4 µL disposable NanoStar microcuvettes which were subsequently inserted into the DLS detector. Measurements were performed at a temperature of 25 °C and solvent refractive index of 1.333 was used.
2.6. Online quantification of the size-dependent RNA distribution
Four LNP-RNA samples and their corresponding empty LNPs were measured by the SEC-MALS-UV-dRI system for online quantification of their size-dependent RNA distribution. The SEC peak was divided into seven or eight regions. Using data from MALS and the two concentration detectors (UV at 260 nm and dRI), weight-average molecular weights (Mw) of the complete LNP-RNA and just the encapsulated RNA across each region were calculated using the Nanoconjugate Analysis module in ASTRA software. A sphere model was used in the Nanoconjugate Analysis module. The dn/dc value of 0.17 mL/g and 0.16 mL/g for RNA and lipids, respectively, and UV extinction coefficient of 22.5 mL/(mg com) were used for of RNA analysis. The enabled MALS angles were detectors 4 to 17. The RNA weight percentage (wt %), defined as the percent of RNA Mw relative to LNP-RNA Mw, was then analyzed for each region and plotted against the radius determined from MALS using ASTRA’s sphere model.
2.7. Offline RNA quantitation of SEC fractions
2.7.1. Reverse-phase HPLC RNA assay
RNA content in the collected fractions was analyzed on a Classic Acquity ultra-performance liquid chromatography (UPLC) system (Waters Corp., Milford, MA). A CSH C18 column with dimensions of 2.1 X 100 mm and 1.7 um particle size was used for separation (Waters Corp.). Mobile phase A consisted of 80 % of 50 mM ammonium bicarbonate in water and 20 % acetonitrile, while mobile phase B was mixture of ethanol and methanol at 1:1 v/v ratio. Mobile phase flow rate was maintained at 0.3 mL/min, column temperature was set to 30 °C, autosampler tray temperature was 10 °C, and UV signal was monitored at 260 nm. The chromatographic gradient began with 100 % A from 0 to 2 min, ramping up to 90 % B in 3 min.
2.7.2. Sample preparation
A 2 % solution (w/v) of Triton X-100 was prepared by dissolving 2 g of Triton X-100 into 100 mL of Milli-Q water. To prepare the fractionated sample for RNA analysis, the LNP was first disrupted by mixing 200 μL of the sample solution with 10 μL of 2 % Triton X-100. The disrupted solution was then further diluted 2x to 5x with a diluent consisting 50 mM ammonium bicarbonate and 0.1 % Triton X-100, to match with the mobile phase buffer and adjust the RNA concentration of fractions at the apex portion of the peak.
2.7.3. RNA assay linearity and accuracy
Approximately 1 mg/mL RNA stock standard solution was prepared by accurately weighing the RNA standard, dissolving and diluting with HyPure Water for Injection (WFI). The exact concentration of the RNA stock solution was calculated by UV absorption at 260 nm (measured by Spectramax M5 Plate Reader, Molecular Devices, San Jose, CA) with the accepted extinction coefficient of the RNA (21.51 L/g). The empty LNP matrix solution was made by mixing 300 μL of empty LNP solution with 45 mL diluent.
Samples for assessing the linearity of the RNA assay method were prepared by serial dilution of the 1 mg/mL stock standard with the diluent. Samples to evaluate spike recovery or lipid matrix effect were prepared by spiking RNA standard into disrupted empty LNP matrix solution.
3. Results and discussion
3.1. Characterization of LNPs by cryo-TEM, batch DLS and SEC-MALS-DLS
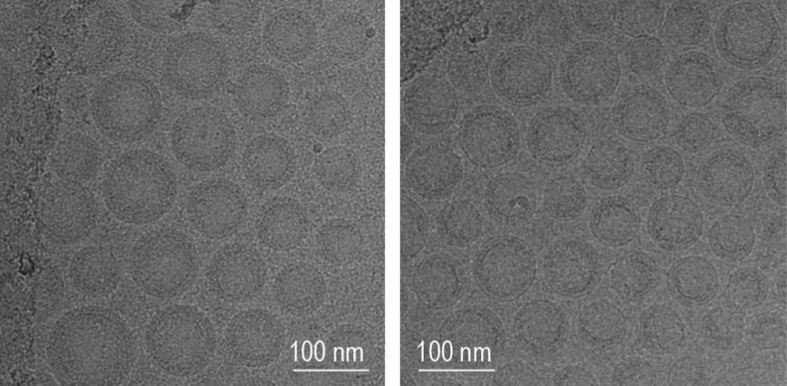
Fig. 1a and 1b show representative cryo-TEM images of LNP-1F and LNP-2F. Both images indicate spherical particles with diameters less than 120 nm. LNP-3F also appeared spherical (Supplementary Fig. 1). The z-average R h values of all LNPs, as determined by batch DLS, are below 50 nm irrespective of RNA content (Table 1 ).
Fig. 1.
Representative Cryo-TEM images of LNPs made of (a) LNP-1F and (b) LNP-2F.
Table 1.
Comparison of empty LNP and LNP-RNA particle attributes. Rh was measured by batch mode DLS and SEC-DLS. Particle radius (Rz) and Mw were measured by SEC-MALS-dRI.
| Rh(batch DLS) [nm] |
Rh(SEC-DLS) [nm] |
Rz (online MALS) [nm] |
Mw (online MALS) [MDa] |
|
|---|---|---|---|---|
| LNP-1E | 43.5 ± 0.9 | 42.6 | 41.7 | 84 |
| LNP-1F | 41.0 ± 0.2 | 41.2 | 40.6 | 111 |
| LNP-2E | 44.8 ± 1.8 | 43.6 | 42.8 | 113 |
| LNP-2F | 39.0 ± 0.5 | 38.6 | 36.0 | 77 |
| LNP-3E | 46.3 ± 0.7 | 46.4 | 46.6 | 144 |
| LNP-3F | 43.4 ± 0.1 | 42.6 | 41.6 | 112 |
| LNP-4E | 25.7 ± 0.3 | 26.8 | 27.9 | 20.3 |
| LNP-4F | 38.7 ± 0.5 | 44.4 | 44.4 | 58 |
The four LNP-RNA samples and their corresponding empty LNP samples were analyzed by SEC-MALS with online DLS (SEC-MALS-DLS). Zhang and others [14] showed that the TOSOH TSKgel G6000PWXL-CP column, designed for SEC of water-soluble cationic polymers, is also suitable for the analytical separation of cationic LNPs with average Rh between 33 and 70 nm. In our LNP formulations, cationic lipid is the most abundant component and an imaged capillary isoelectric focusing (IC-IEF) study demonstrated that LNPs with similar composition carry positive charge [28]. The average R h values of all LNPs except LNP-4E (25.7 nm) are within the range studied by Zhang and others. All LNP-4E particles were eluted on the Tosoh TSKgel G6000PWXL-CP column before the inclusion volume about 13.2 mL (Supplementary Fig. 2), indicating that this column is also suitable for LNP-4E. Thus, the Tosoh TSKgel G6000PWXL-CP column was selected for SEC separation.
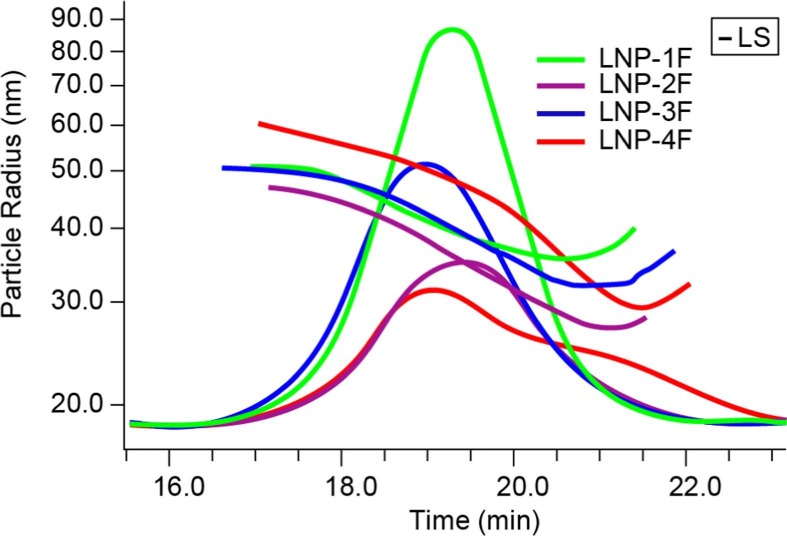
Fig. 2 presents the radius (R) values of the four LNP-RNA samples at each eluting data slice as determined by fitting the MALS data to a sphere model, overlaid on the corresponding 90° light scattering chromatogram. For the majority portion of each chromatogram the radius decreases with retention time, suggesting that good size-based SEC separation was achieved using the conditions described above. At the tail of each peak a slight increase in radius was observed, opposite to the expected trend. This phenomenon, often referred to as “non-ideal SEC effect”, is consistent with co-elution of a small number of large particles along with the main population of small particles. The non-ideal SEC effect is likely caused by the so-called “restricted diffusion” and, to a lesser degree, column interaction [29]. Field-flow fractionation (FFF) has been demonstrated to ameliorate this limitation of SEC [29], [30]. Due to the likely heterogeneity at the peak tail region, the region where radius increases with time was excluded from the analysis of size-dependent RNA wt % distribution in this study. The overlay of molar mass distribution with the corresponding 90° light scattering chromatogram of the four LNP-RNA samples showed that LNP-4 is much highly dispersed when compared with the other three LNPs (supplementary Fig. 3).
Fig. 2.
Particle radius plots overlaid on the corresponding 90° light scattering chromatogram of the four LNP-RNA samples.
Additionally, offline fluorescence assay of RNA encapsulation of the four LNPs showed that the encapsulation efficiency were higher than 90% for LNP-1, LNP-2 and LNP-3, while only 57% for LNP-4.
Table 1 summarizes the results of SEC-MALS-DLS measurements and batch DLS. Analysis of MALS and dRI signals determined: 1) z-average radius Rz, 2) weight-average molar mass Mw. Z-average Rh was determined by both online DLS and batch DLS. The latter was calculated as the average of 10 separate measurements and is reported along with the standard deviation. The results from online MALS and DLS are the average of two injections, and the differences between the duplicates are all less than 2 % for each quantity.
The good agreement between Rh from SEC-DLS and Rz by SEC-MALS (Table 1) is consistent with the hypothesis that these LNPs are spherical in shape, which is supported by the Cryo-TEM images. Non-spherical particles would result in discrepancies between Rh and Rz. Furthermore, batch Rh values are in excellent agreement with online DLS and radius by MALS for all samples except LNP-4F. The generally good agreement between batch and online DLS data indicate that the SEC separation did not alter these LNP samples during analysis.
Compared to online DLS, a slightly smaller size by batch DLS was observed for LNP-4E, but the values are still very close. The significant discrepancy between batch and online DLS sizes for LNP-4F is very likely attributed to their bimodal distribution characteristic evidenced in Fig. 2, as inaccuracy is expected in batch DLS when the size distribution of a sample is not monomodal. While it might be conjectured that the discrepancy is a result of shear stress on the SEC column, separation by field-flow fractionation—a technique that, much like size-exclusion chromatography, separates particles by hydrodynamic size but does so in an open channel with very little shear stress—produces a similar bimodal distribution (data not shown) and thus appears to refute that explanation. We integrated the bimodal peaks of LNP-4E and LNP-4F separately (Supplementary Fig. 4) and estimated the mass percent of each peak from the online dRI signal. For LNP-4E, approximately 98 % of the particles fall into peak 2, while Peak 1, which contains mostly larger particles, incorporates only 1.6 % of total particle mass. This might explain the minor difference between batch and online DLS size results for slightly bimodal LNP-4E. However, for LNP-4F, the larger particles in peak 1 make up about 19 % of total mass. From further analysis of the sizes of the particles of LNP-4F in peaks 1 and 2 by online DLS and MALS, and comparison to batch DLS (Supplementary Table 1), it seems that the size by batch DLS is close to the 81 % smaller particles group, while size by online MALS or DLS is the average of peaks 1 and 2.
3.2. Online analysis of size-based RNA distribution
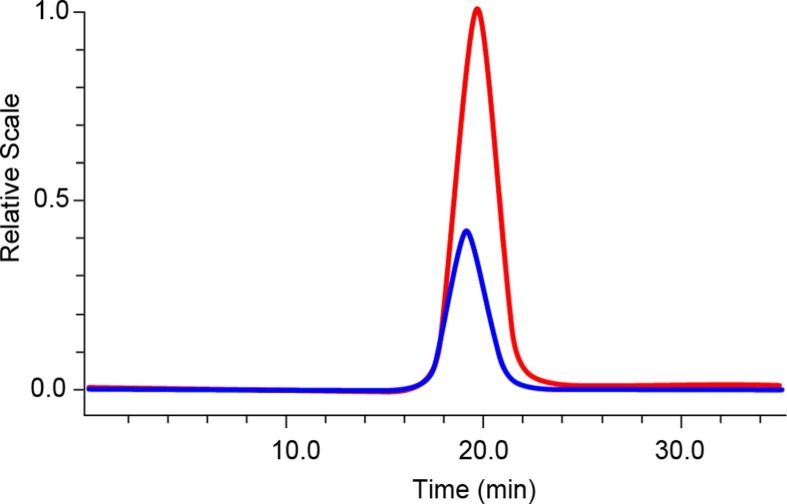
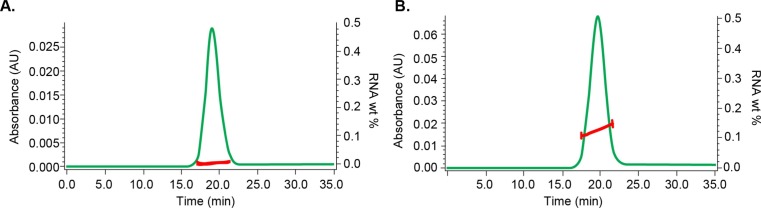
MALS, dRI, and UV absorption data from online SEC have been used to quantify the composition of various biological conjugates of two different types of molecules, such as protein and lipid, protein and polysaccharide, or protein and DNA [31], [32], [33]. However, the same conjugate analysis protocol cannot be applied to LNP-RNA, because the UV extinction signal from the LNP particles contains a significant contribution from the scattering phenomenon when the particle radius is>25 nm [34]. Fig. 3 shows the overlay of UV traces at 260 nm of LNP-2E (empty) and LNP-2F (RNA-filled) from the SEC-MALS-UV-dRI analysis. Though all the lipids used to form LNP particles, as well as the disrupted empty LNP-2E particles, do not exhibit UV absorption at 260 nm when measured on a UV spectrometer (data not shown), the empty LNP produced a sizeable UV extinction peak (Fig. 3), which is due to UV scattering (rather than absorption). Consequently, the UV signal from the LNP particles must be corrected to remove the scattering contribution when applying the standard conjugate analysis. A proprietary correction algorithm devised by Wyatt Technology establishes the correlation between particle size and scattering contribution to the UV absorption at 260 nm [35]. This correction algorithm is implemented in the Nanoconjugate Analysis module of ASTRA software. Using this algorithm, the respective total MW of LNP-RNA and MW of RNA are calculated for each eluting fraction, resulting in determination of the MW ratio of RNA and LNP-RNA, or the RNA wt %. Fig. 4 shows the RNA wt % of LNP-2E and LNP-2F calculated by the online analysis method overlaid with the UV chromatogram at 260 nm. The results from the online analysis of RNA wt % of all the empty LNPs and LNP-RNAs, and comparison with the corresponding offline measurements, will be discussed in 3.3.3.
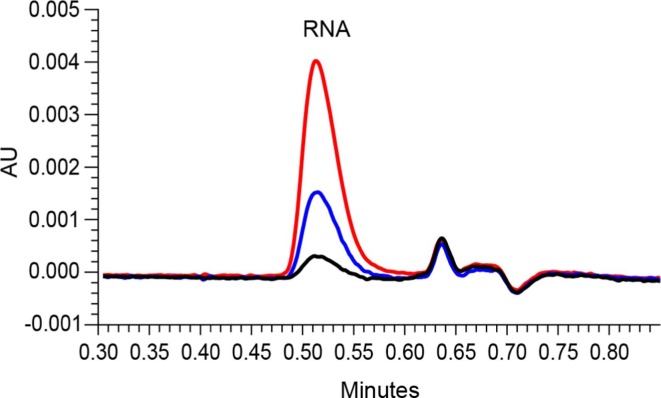
Fig. 3.
UV overlay of empty LNP-2E (blue) and RNA filled LNP-2F (red) from the SEC-MALS-UV-dRI analysis showing the significant UV signal from LNP-2E due to the particle scattering phenomenon in the UV detector. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
The UV chromatogram at 260 nm overlaid with RNA wt% (right Y-axis) for (A) LNP-2E and (B) LNP-2F.
3.3. Validation of online size-dependent LNP-RNA content analysis
3.3.1. RNA assay method development
Given the small fraction mass and large numbers of samples to be analyzed, a fast and sensitive offline assay method is required to quantify RNA in the collected fractions. SEC-UV, ion-pairing (IP)-RPLC and ion-exchange (IEX) have been used for analysis of RNA in LNP [36]. We developed a fast and sensitive RPLC method using a Waters CSH C18 (100 mm × 2.1 mm, 1.7 um) column with 20 % acetonitrile in 50 mM ammonium bicarbonate aqueous solution as mobile phase A and methanol/ethanol 1:1 as mobile phase B. Since the LNP samples also contain hydrophobic lipids, a gradient elution chromatographic condition was developed to ensure that the lipids were washed out. The mobile phase is held at 100 % A from 0 to 2 min for the elution of RNA, then increased to 90 % B from 2.01 min to 5 min to elute lipid components.
Under this condition, the RNA peak elutes earlier than the solvent front (Fig. 5 ). This chromatographic behavior can be explained by the following rational: at the pH of the 50 mM ammonium bicarbonate (∼7.6), RNA molecules acquire a strong negative charge, so there is almost no hydrophobic interaction between the RNA molecule and the stationary phase. Furthermore, under the mobile phase pH, the pores of the CSH C18 particles are negatively charged, and this charge architecture would prevent the RNA from entering the pores; rather they are expelled to the extra-particle interstitial space.
Fig. 5.
Overlay of RNA peak of 47.5 ppb (black), 190 ppb (blue) and 475 ppb(red) RNA standard solutions analyzed by the fast RNA assay method. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The method demonstrated good linearity from 47.5 ppb to 9.5 ppm with R2 = 0.9999 (data not shown). The limit of detection was determined to be 19 ppb with S/N = 7, and limit of quantitation 47.5 ppb with S/N = 16. The accuracy of the method was demonstrated by recovery of an RNA standard, which was spiked into the empty LNP matrix at concentrations from 47.5 ppb to 9.5 ppm. Within the calibrated linearity range (47.5 ppb to 9.5 ppm), recovery was found to be between 98 % and 104 % (Table 2 ), demonstrating a good measurement accuracy.
Table 2.
RNA spike recovery showing the accuracy of the method.
| RNA (ppb) | Recovery (%) |
|---|---|
| 47.5 | 97.8 |
| 95 | 104.4 |
| 190 | 100.6 |
| 475 | 100.7 |
| 950 | 99.2 |
| 1900 | 103.9 |
| 4750 | 103.6 |
| 9500 | 99.8 |
3.3.2. Offline measurement of size-dependent LNP-RNA content
LNPs were fractionated by size and the fractions subsequently measured to determine RNA concentration. To ensure reliable correlation of RNA wt % between size fractions analyzed by online SEC-MALS and those from offline measurements of fractions, we performed the separations for fraction collection on the same analytical-scale chromatographic column used for online analysis, injecting the maximum feasible load–300 μg–considering the loading capacity of a 7.8 × 300 mm SEC column [37]. Fractions were collected at 0.5 min intervals and RNA concentrations were analyzed by the fast RPLC assay discussed in Section 3.3.1.
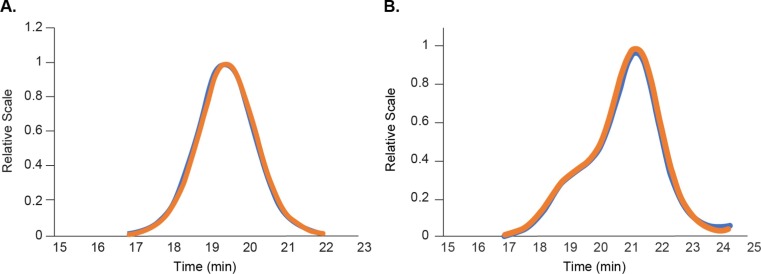
Due to strong scattering by the LNP particles, the MALS detector was saturated when separating 300 µg of sample, and size information could not be obtained. A lower loading of 20 μg was performed under the same conditions for size determination by MALS, under the assumption that identical size fractions elute at the same time regardless of sample load. In order to confirm this assumption, the RI peaks of the 20 μg and 300 μg injections were normalized to the same apex values and the consistency of peak shapes and retention times were examined. Fig. 6 shows the overlay of the 20 μg and 300 μg RI profiles for LNP-1F (Fig. 6A) and LNP-4F (Fig. 6B). For both samples, the two different loadings showed nearly identical peak shapes and retention times. The consistent agreement of the normalized RI peaks of the 20 μg and 300 μg injections suggests that the resolution within the peak is unaffected by the overall sample load, and that the offline fraction analysis with 300 μg loading is directly comparable to the online SEC-MALS analysis with 20 μg injection: no adjustments are required to account for different elution behavior under disparate sample loads. LNP-2F and LNP-3F also exhibited nearly identical peak shapes and retention times under the two loading conditions, confirming that the eluting size fractions are directly comparable for these samples as well (data not shown).
Fig. 6.
Overlay of normalized RI peaks of the 20 µg (orange) and 300 µg (blue) injections of (A) LNP-1F and (B) LNP-4F. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Finally, the RNA content in each fraction was analyzed by the fast RPLC method. The weight percent of RNA (RNA wt %) in each fraction was calculated as the ratio of the measured RNA concentration to the total mass of that fraction measured by RI. The RNA wt % measured on duplicate injections and the average radius measured by online MALS are tabulated for each fraction in Table 3 for all four LNP-RNA samples. The RNA wt % from duplicate experimental data showed good reproducibility for all four LNP-RNA samples.
Table 3.
Size-based RNA wt% in LNP-1F, LNP-2F, LNP-3F and LNP-4F from fractionation experiment (excluding the particle size curve up region of the peak).
| LNP-1F | |||||||
|---|---|---|---|---|---|---|---|
| R (nm) | 50.1 | 48.5 | 45.3 | 41.9 | 39.1 | 37.1 | 35.7 |
| duplicate-1 (%RNA) | 10.3 | 10.5 | 11.3 | 12.3 | 12.1 | 11.4 | 11.4 |
| duplicate-2 (%RNA) | 10.0 | 10.1 | 11.2 | 11.2 | 11.9 | 11.8 | 11.7 |
| avg (%RNA) | 10.1 | 10.3 | 11.3 | 11.7 | 12.0 | 11.6 | 11.5 |
| LNP-2F | ||||||||
|---|---|---|---|---|---|---|---|---|
| R (nm) | 46.7 | 45.4 | 42.9 | 39.7 | 36.3 | 33.1 | 30.5 | 29.1 |
| duplicate-1 (%RNA) | 10.4 | 10.5 | 10.0 | 10.7 | 11.5 | 12.0 | 12.5 | 12.8 |
| duplicate-2 (%RNA) | 9.7 | 10.2 | 10.1 | 10.8 | 11.4 | 12.1 | 12.8 | 12.9 |
| avg (%RNA) | 10.1 | 10.3 | 10.0 | 10.7 | 11.5 | 12.1 | 12.6 | 12.9 |
| LNP-3F | |||||||
|---|---|---|---|---|---|---|---|
| R (nm) | 49.9 | 48.1 | 45.3 | 42.1 | 38.6 | 35.4 | 33.2 |
| duplicate-1 (%RNA) | 9.6 | 10.2 | 11.1 | 11.7 | 12.0 | 12.7 | 13.3 |
| duplicate-2 (%RNA) | 9.2 | 9.4 | 10.3 | 11.2 | 12.1 | 13.1 | 14.2 |
| avg (%RNA) | 9.4 | 9.8 | 10.7 | 11.4 | 12.1 | 12.9 | 13.7 |
| LNP-4F | ||||||||
|---|---|---|---|---|---|---|---|---|
| R (nm) | 55.9 | 53.4 | 50.6 | 47.4 | 43.4 | 38.2 | 33.5 | 31.8 |
| duplicate-1 (%RNA) | 0.8 | 1.2 | 1.7 | 2.7 | 4.3 | 6.1 | 7.5 | 8.1 |
| duplicate-2 (%RNA) | 0.8 | 1.2 | 1.7 | 2.7 | 4.3 | 6.2 | 7.8 | 8.3 |
| avg (%RNA) | 0.8 | 1.2 | 1.7 | 2.7 | 4.3 | 6.2 | 7.7 | 8.2 |
3.3.3. Comparison of online and offline size-dependent RNA distribution
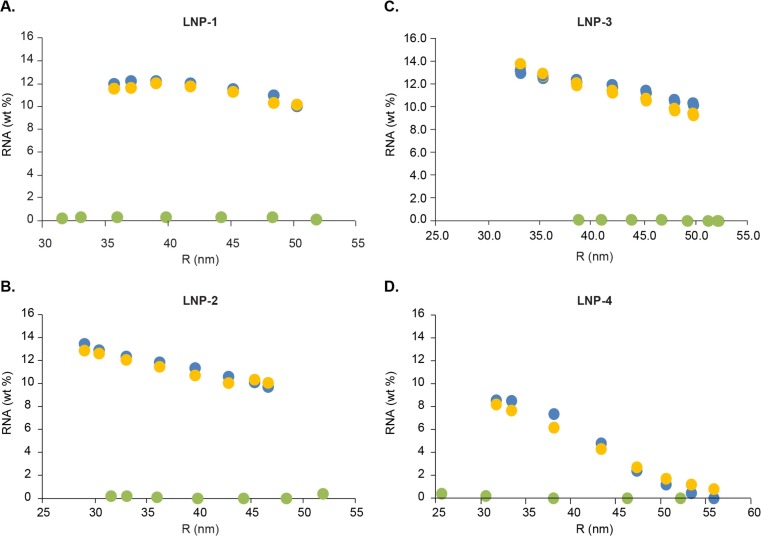
The RNA wt % values obtained from the online and offline methods are plotted against the average radius of each fraction in Fig. 7 . Both offline and online data showed that the encapsulated RNA profile was affected by the type of PEG-lipid and the PEG-lipid mol % in the formulation. LNP-1F showed relatively consistent RNA wt % across the size ranges, with slightly lower RNA wt % on the very large particle side. LNP-2F and −3F (made with the same molar ratio of PEG-Lipid 2 and PEG-lipid 3, respectively), exhibited a monotonic decrease of RNA wt % with increasing particle size.
Fig. 7.
Size-dependent RNA distribution in LNPs. Average of duplicate fractionation and offline RPLC analyses (orange) vs data from online analysis (blue for RNA-LNP and green for empty LNP). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
LNP-4F (made with 4 mol % of PEG-lipid 3) produced a sigmoidal distribution of RNA wt % versus size. Correlating this behavior with the bimodal LS and dRI profiles, i.e. the presence of two groups of particles containing distinct sizes (Supplementary Fig. 4 and Supplementary Table 1), we found that a very low wt % of RNA was detected in the large particle population, while the majority of RNA was encapsulated in the smaller particle group.
In general, excellent correlation of RNA wt % between the online and offline methods is seen for all four RNA-LNPs studied. The difference between the offline and online results is less than 1 % for all the data points of LNP-1F, −2F and −3F (relative difference less than 10%). For LNP-4F, the RNA wt % is generally lower than for the other samples – less than 10 % at all data points – and especially the larger particles, R > 50 nm, contain less than 2 % RNA. Therefore, the larger difference compared to the other samples (up to 18 % relative difference, or 1.3% in absolute terms) between online and offline values for LNP-4F may be considered an artifact of the low overall RNA wt % values. As expected, online analysis of all four empty LNPs resulted in 0 or close to 0 % RNA, as plotted in Fig. 7A–D.
Although LNP-4F produced the largest difference between online and offline methods, we expect this has limited impact in the application of the online method since only monomodal LNPs are acceptable for therapeutic development.
4. Conclusion
We evaluated a new online method for determining the distribution of RNA content versus LNP-RNA size, enabling advanced LNP-RNA characterization. The method utilizes a standard SEC-MALS-UV-dRI system and is implemented in the Nanoconjugate Analysis module of ASTRA software. The calculated RNA wt % results from four LNPs, made with different types of PEG-lipids and compositional ratios, were found to be in excellent agreement with the experimental data obtained by and fractionation with offline RPLC analysis. This work indicates that the correction of the scattering contribution to the UV signal implemented in the Nanoconjugate Analysis algorithm was adequate for reliable size-dependent RNA content analysis of lipid nanoparticles. Based on these results, we conclude that the online method is, in all likelihood, suitable for rapid and facile determination of the sized-dependent payload distribution of other LNPs encapsulating nucleic acids, or other carriers constructed from excipients that do not absorb UV at the characteristic absorption wavelength of the payload. The analytical advancement reported here provides additional characterization information, and potentially sheds light on understanding the therapeutic outcome of the RNA/DNA filled LNPs. We foresee the potential for use in quality control procedures, to evaluate the total RNA content in LNPs within a specified size range, which would constitute a critical quality attribute of a therapeutic LNP-RNA product.
CRediT authorship contribution statement
Xiujuan Jia: Conceptualization, Methodology, Validation, Investigation, Writing – original draft. Yong Liu: Writing – review & editing. Angela M. Wagner: Resources, Writing – review & editing. Michelle Chen: Methodology, Software, Formal analysis, Writing – original draft. Yuejie Zhao: Resources. Katelyn J. Smith: Resources. Dan Some: Methodology, Software, Writing – review & editing. Andreas M. Abend: Writing – review & editing. Justin Pennington: Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to thank Amy Doty from Discovery Pharmaceutical Science and Tyler Corts from Sterile and Specialty Product, Pharmaceutical Sciences, Merck & Co., Inc., Kenilworth, NJ, USA. for their help on LNP manufacture process. The authors would also like to thank Marian Gindy from Preclinical Development, Pharmaceutical Sciences, Merck & Co., Inc., Kenilworth, NJ, USA. for her reviewing of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jchromb.2021.123015.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.M.N. Ramasamy, A.M. Minassian, K.J. Ewer, A.L. Flaxman, P.M. Folegatti, D.R. Owens, M. Voysey, P.K. Aley, B. Angus, G. Babbage, S. Belij-Rammerstorfer, L. Berry, S. Bibi, M. Bittaye, K. Cathie, H. Chappell, S. Charlton, P. Cicconi, E.A. Clutterbuck, R. Colin-Jones, C. Dold, K.R.W. Emary, S. Fedosyuk, M. Fuskova, D. Gbesemete, C. Green, B. Hallis, M.M. Hou, D. Jenkin, C.C.D. Joe, E.J. Kelly, S. Kerridge, A.M. Lawrie, A. Lelliott, M.N. Lwin, R. Makinson, N.G. Marchevsky, Y. Mujadidi, A.P.S. Munro, M. Pacurar, E. Plested, J. Rand, T. Rawlinson, S. Rhead, H. Robinson, A.J. Ritchie, A.L. Ross-Russell, S. Saich, N. Singh, C.C. Smith, M.D. Snape, R. Song, R. Tarrant, Y. Themistocleous, K.M. Thomas, T.L. Villafana, S.C. Warren, M.E.E. Watson, A.D. Douglas, A.V.S. Hill, T. Lambe, S.C. Gilbert, S.N. Faust, A.J. Pollard, Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial, Lancet Lond. Engl. 396 (2021) 1979–1993. Doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., Cipolla M., Viant C., Barnes C.O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K.G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Robbiani D.F., Zhao Z., Gazumyan A., Schwartz R.E., Hatziioannou T., Bjorkman P.J., Mehandru S., Bieniasz P.D., Caskey M., Nussenzweig M.C. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le T.K., Paris C., Khan K.S., Robson F., Ng W.-L., Rocchi P. Nucleic Acid-Based Technologies Targeting Coronaviruses. Trends Biochem. Sci. 2021;46(5):351–365. doi: 10.1016/j.tibs.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S., Ploegh H.L., Anderson D.G. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113(29):E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.-J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30(12):1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 7.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D., Hope M.J., Krammer F., Hensley S.E., Weissman D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papachristofilou A., Hipp M.M., Klinkhardt U., Früh M., Sebastian M., Weiss C., Pless M., Cathomas R., Hilbe W., Pall G., Wehler T., Alt J., Bischoff H., Geißler M., Griesinger F., Kallen K.-J., Fotin-Mleczek M., Schröder A., Scheel B., Muth A., Seibel T., Stosnach C., Doener F., Hong H.S., Koch S.D., Gnad-Vogt U., Zippelius A. Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J. Immunother. Cancer. 2019;7:38. doi: 10.1186/s40425-019-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kallen K.-J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., Baumhof P., Scheel B., Koch S.D., Fotin-Mleczek M. A novel, disruptive vaccination technology: self-adjuvanted RNActive(®) vaccines. Hum. Vaccines Immunother. 2013;9(10):2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida S., Kinoh H., Ishii T., Matsui A., Tockary T.A., Takeda K.M., Uchida H., Osada K., Itaka K., Kataoka K. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials. 2016;82:221–228. doi: 10.1016/j.biomaterials.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy. Nano Lett. 2017;17(3):1326–1335. doi: 10.1021/acs.nanolett.6b0332910.1021/acs.nanolett.6b03329.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines. 2021;9:65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Haas R.M., Leone A.M. Polydispersity characterization of lipid nanoparticles for siRNA delivery using multiple detection size-exclusion chromatography. Anal. Chem. 2012;84(14):6088–6096. doi: 10.1021/ac3007768. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Pei Y.i., Zhang H., Wang L., Arrington L., Zhang Y.e., Glass A., Leone A.M. Assessing the heterogeneity level in lipid nanoparticles for siRNA delivery: size-based separation, compositional heterogeneity, and impact on bioperformance. Mol. Pharm. 2013;10(1):397–405. doi: 10.1021/mp3005337. [DOI] [PubMed] [Google Scholar]
- 16.Leung A.K.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid Nanoparticles Containing siRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C. 2012;116(34):18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26(9):363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M.E. Gindy, K. DiFelice, V. Kumar, R.K. Prud’homme, R. Celano, R.M. Haas, J.S. Smith, D. Boardman, Mechanism of Macromolecular Structure Evolution in Self-Assembled Lipid Nanoparticles for siRNA Delivery, (2014). Doi: 10.1021/la500630h. [DOI] [PubMed]
- 19.Belliveau N.M., Huft J., Lin P.JC., Chen S., Leung A.KK., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura T., Kawai M., Sato Y., Maeki M., Tokeshi M., Harashima H. The Effect of Size and Charge of Lipid Nanoparticles Prepared by Microfluidic Mixing on Their Lymph Node Transitivity and Distribution. Mol. Pharm. 2020;17(3):944–953. doi: 10.1021/acs.molpharmaceut.9b0118210.1021/acs.molpharmaceut.9b01182.s001. [DOI] [PubMed] [Google Scholar]
- 21.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Controlled Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 22.Cox A.J., DeWeerd A.J., Linden J. An experiment to measure Mie and Rayleigh total scattering cross sections. Am. J. Phys. 2002;70(6):620–625. doi: 10.1119/1.1466815. [DOI] [Google Scholar]
- 23.Sweeney J.A., Hennessey J.P. Evaluation of accuracy and precision of adenovirus absorptivity at 260 nm under conditions of complete DNA disruption. Virology. 2002;295(2):284–288. doi: 10.1006/viro.2002.1406. [DOI] [PubMed] [Google Scholar]
- 24.Porterfield J.Z., Zlotnick A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology. 2010;407(2):281–288. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D. Brown, J. Cunningham, M. Gindy, V. Pickering, M. Stanton, S. Stirdivant, W. Strapps, RNA INTERFERENCE MEDIATED INHIBITION OF CATENIN (CADHERIN-ASSOCIATED PROTEIN), BETA 1 (CTNNB1) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA), 2012. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2012018754&_cid=P10-KP5UVP-36366-1 (accessed May 26, 2021).
- 26.SUNBRIGHT® GM-020(DMG-PEG) | Phospholipids, (n.d.). https://www.nofamerica.com/store/index.php?dispatch=products.view&product_id=329 (accessed October 12, 2021).
- 27.Gindy M.E., Feuston B., Glass A., Arrington L., Haas R.M., Schariter J., Stirdivant S.M. Stabilization of Ostwald Ripening in Low Molecular Weight Amino Lipid Nanoparticles for Systemic Delivery of siRNA Therapeutics. Mol. Pharm. 2014;11(11):4143–4153. doi: 10.1021/mp500367k. [DOI] [PubMed] [Google Scholar]
- 28.Loughney J.W., Minsker K., Ha S., Rustandi R.R. Development of an imaged capillary isoelectric focusing method for characterizing the surface charge of mRNA lipid nanoparticle vaccines. Electrophoresis. 2019;40:2602–2609. doi: 10.1002/elps.201900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Light Scattering, Size Exclusion Chromatography and Asymmetric Flow Field Flow Fractionation: Powerful Tools for the Characterization of Polymers, Proteins and Nanoparticles | Wiley, Wiley.Com. (n.d.). https://www.wiley.com/en-us/Light+Scattering%2C+Size+Exclusion+Chromatography+and+Asymmetric+Flow+Field+Flow+Fractionation%3A+Powerful+Tools+for+the+Characterization+of+Polymers%2C+Proteins+and+Nanoparticles-p-9780470386170 (accessed April 13, 2021).
- 30.Mildner R., Hak S., Parot J., Hyldbakk A., Borgos S.E., Some D., Johann C., Caputo F. Improved multidetector asymmetrical-flow field-flow fractionation method for particle sizing and concentration measurements of lipid-based nanocarriers for RNA delivery. Eur. J. Pharm. Biopharm. Off. J. Arbeitsgemeinschaft Pharm. Verfahrenstechnik EV. 2021;163:252–265. doi: 10.1016/j.ejpb.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Gimpl K., Klement J., Keller S. Characterising protein/detergent complexes by triple-detection size-exclusion chromatography. Biol. Proced. Online. 2016;18(1) doi: 10.1186/s12575-015-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176(5):1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Characterizing Protein-Nucleic Acid Conjugates with Light Scattering, Chromatogr. Online. (n.d.). https://www.chromatographyonline.com/view/characterizing-protein-nucleic-acid-conjugates-light-scattering (accessed April 13, 2021).
- 34.Uitz J., Stramski D., Baudoux A.-C., Reynolds R.A., Wright V.M., Dubranna J., Azam F. Variations in the optical properties of a particle suspension associated with viral infection of marine bacteria. Limnol. Oceanogr. 2010;55(6):2317–2330. doi: 10.4319/lo.2010.55.6.2317. [DOI] [Google Scholar]
- 35.M. Chen, D. Some, Calculating molar mass values of components of and molar mass concentration values of conjugate molecules/particles, n.d.
- 36.Liu Y., Zhao Y., Socia A., Abend A. Application of liquid chromatography in characterisation of lipid nanoparticle-based oligonucleotides. Eur. Pharm. Rev. 2020;25:24–28. [Google Scholar]
- 37.TOSOH, TOSOH product overview: TSKgel PWXL-CP columns., (n.d.). https://www.separations.us.tosohbioscience.com/landing/PO17.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.