Letter to the editor
In the context of the COVID-19 pandemic, shortage of material forced laboratories to turn to diverse alternative solutions. Confronted to this situation for virus transport media (VTM), we conducted a study to evaluate the influence of pre-analytical parameters on respiratory virus molecular detection [1, 2]. Fifteen specimens were identified through Allplex™ Respiratory panel-1 assay (Seegene Eurobio), 5 influenza A virus (H3:FA1–3 and H1:FA4–5 subtypes) positive, 5 influenza B virus positive (FB1–5), 3 RSV-A (VA1–3) and 2 RSV-B (VB1–2) positive. Samples were selected within a defined Ct range of 21–34 Ct. Three VTM were evaluated: ESwab®, Virocult® and Cepheid® according to the study design (Fig. 1 ). Samples were kept under different temperature conditions for up to 72 h and tested at each time point using Xpert Xpress Flu / RSV assay (Cepheid). For each VTM, the reference Ct value is the one obtained at initial testing (T0). The dCt was calculated by comparing the obtained values under each condition compared to the T0 values: H4-T0; H24 [+4 °C]-T0; H72 [+4 °C]-T0 and H72 [AT]-T0 (Fig. 2 and table 1 ). A dCt over 2 was considered significant [3].
Fig. 1.
Experimental design for each tested VTM. Fifteen positive clinical samples were analyzed: influenza A virus (n = 5), influenza B virus (n = 5), RSV-A (n = 3) or RSV-B (n = 2). Three VTM were compared: Cepheid®, Virocult® and E swab® under several temperature storage (+4 °C and ambient temperature (TA) and delay (T0, H4, H24 and H72).
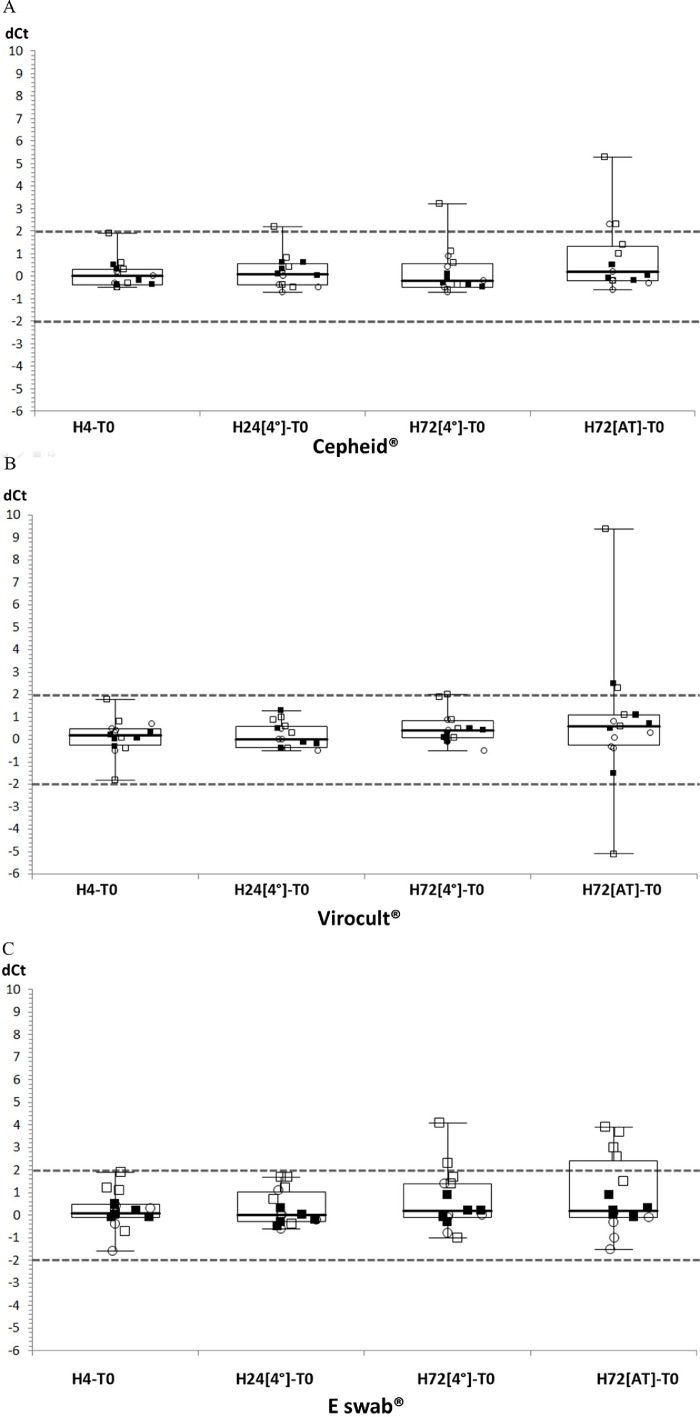
Fig. 2.
Comparison of dCt for each sample stored for different periods of time, under different temperature conditions and gathered according to VTM: A: Cepheid®; B: Virocult®; C: E-swab®. Each virus family is represented by a different symbol: FluA, open squares; FluB, closed squares; RSV, open circles. The dotted horizontal lines represent the 2 Ct cut-off.
Table 1.
Delta Ct calculated against T0 values and obtained for each virus and storage condition (dCt values above 2 are represented on gray background).
| Cepheid® | Virocult® | E-swab® | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H4-T0 | H24 | H72 | H72 | H4-T0 | H24 | H72 | H72 | H4-T0 | H24 | H72 | H72 | |
| [4°]-T0 | [4°]-T0 | [AT]-T0 | [4°]-T0 | [4°]-T0 | [AT]-T0 | [4°]-T0 | [4°]-T0 | [AT]-T0 | ||||
| FluA (n = 5) | ||||||||||||
| Minimum | −0.5 | −0.5 | −0.6 | −0.2 | −1.8 | −0.4 | 0.1 | −5.1 | −0.7 | −0.4 | −1 | 1.5 |
| Median | 0.3 | 0.4 | 0.6 | 1.4 | 0.1 | 0.6 | 0.9 | 1.1 | 1.1 | 1.2 | 1.7 | 3 |
| Maximum | 1.9 | 2.2 | 3.2 | 5.3 | 1.8 | 1 | 2 | 9.4 | 1.9 | 1.7 | 4.1 | 3.9 |
| Values over 2 dCt (n) | 0 | 1 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 4 |
| FluB (n = 5) | ||||||||||||
| Minimum | −0.4 | 0 | −0.5 | −0.2 | −0.3 | −0.4 | −0.1 | −1.5 | −0.1 | −0.5 | −0.3 | −0.1 |
| Median | −0.20 | 0.3 | −0.3 | 0 | 0.1 | −0.1 | 0.2 | 0.7 | 0 | −0.2 | 0.2 | 0.2 |
| Maximum | 0.5 | 0.6 | 0.1 | 0.5 | 0.3 | 1.3 | 0.5 | 2.5 | 0.5 | 0.3 | 0.9 | 0.9 |
| Values over 2 dCt (n) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| RSV (n = 5) | ||||||||||||
| Minimum | −0.5 | −0.7 | −0.7 | −0.6 | −0.5 | −0.5 | −0.5 | −0.4 | −1.6 | −0.6 | −0.8 | −1.5 |
| Median | 0 | −0.4 | −0.2 | −0.2 | 0.4 | 0 | 0.1 | 0.1 | 0.1 | 0 | 0 | −0.3 |
| Maximum | 0.2 | 0.2 | 0.9 | 2.3 | 0.7 | 0.5 | 0.9 | 0.8 | 0.4 | 1.1 | 1.4 | 0.1 |
| Values over 2 dCt (n) | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| All viruses (n = 15) | ||||||||||||
| Values over 2 dCt (n) | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 3 | 0 | 0 | 2 | 4 |
For FluA, the most divergent Ct were observed with Eswab (72h-AT) with 5 values over the defined cut-off of 2 Ct. Less divergent values were observed for shorter time points and lower temperatures. For FluB and RSV, the dispersion was less striking than for FluA and only one divergent point (FluB/Virocult and RSV/Cepheid) was observed for each virus, both after storage at AT for 72 h. When comparing results for all detected viruses tested at H4, storage for 72 h at 4 °C or AT led to more results exceeding the defined cut-off (n = 4 p = 0.03 or 10 p<0.001).
Qualitatively, the concordance was 100% whatever the VTM, the storage period and temperature, and no false negative was observed. While the sample Ct suffered only little variations over time for each VTM, significant differences were mostly observed after storage for 72 h. It is therefore advisable to respect the supplier's recommendations and to analyze the sample before 72 h. Noteworthy, the signal obtained for influenza A virus was the most sensitive to duration or temperature of storage but no evident explanation could be raised for this observation.
To our knowledge, this proof of concept study is the first to evaluate Xpert Flu/RSV performance to detect influenza virus and RSV under several temperatures and delay storage using E-swab® and Virocult® as alternative VTM for Xpert Xpress Flu/RSV testing. However, storage of samples for 72 h could possibly lead to false negative results.
Due to the limited number of samples, covering a narrow range of viral loads, this study should be considered exploratory and be reinforced by complementary studies including virus inactivating VTM and SARS-CoV-2 detection [4]. Indeed, the use of a VTM capable of viral inactivation, such as Copan eNat, constitutes a real asset [5]. Preliminary data obtained using the inactivating media Sun-Trine Red with the Xpert Xpress ® Flu/RSV/SARS-CoV-2 assay are encouraging.
Ethics approval and consent to participate
Not applicable
Consent to publish
The corresponding author has obtained all author's approvals to publish this work.
Availability of data and material
Yes, upon request
Funding source
All reagents were provided by Cepheid, Inc.
Authors' contributions
ZM, CH, CG performed the experiments. CG, CP and VT performed the literature review, and wrote the first draft of the article. All authors approved the submitted version of the manuscript.
Declaration of competing interest
The authors report no conflict of interest related to this work.All reagents were provided by Cepheid. This work was conducted independently to the manufacturer.
Acknowledgements
Not applicable
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.105063.
Appendix. Supplementary materials
References
- 1.Garnett L., Bello A., Tran K.N., Audet J., Leung A., Schiffman Z., Griffin B.D., Tailor N., Kobasa D., Strong J.E. Comparison analysis of different swabs and transport mediums suitable for SARS-CoV-2 testing following shortages. J. Virol. Methods. 2020;285 doi: 10.1016/j.jviromet.2020.113947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlton C.L., Babady E., Ginocchio C.C., Hatchette T.F., Jerris R.C., Li Y., Loeffelholz M., McCarter Y.S., Miller M.B., Novak-Weekley S., Schuetz A.N., Tang Y.-.W., Widen R., Drews S.J. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin. Microbiol. Rev. 2018;32 doi: 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferroni A., Izopet J., Miedougé M. Vérification d'une méthode qualitative d'amplification génique. QUAMIC. 2019:273–282. th ed. Société Française de Microbiologie. [Google Scholar]
- 4.Wolters F., Grünberg M., Huber M., Kessler H.H., Prüller F., Saleh L., Fébreau C., Rahamat-Langendoen J., Thibault V., Melchers W.J.G. European multicenter evaluation of Xpert® Xpress SARS-CoV-2/Flu/RSV test. J. Med. Virol. 2021;93:5798–5804. doi: 10.1002/jmv.27111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard-Greenblatt M., Comar C.E., Flevaud L., Berti M., Harris R.M., Weiss S.R., Glaser L. Copan eNAT transport system to address challenges in COVID-19 diagnostics in regions with limited testing access. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00110-21. e00110-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yes, upon request