Abstract
Ensemble learning uses multiple algorithms to obtain better predictive performance than any single one of its constituent algorithms could. With the growing popularity of deep learning technologies, researchers have started to ensemble these technologies for various purposes. Few, if any, however, have used the deep learning approach as a means to ensemble Alzheimer’s disease classification algorithms. This paper presents a deep ensemble learning framework that aims to harness deep learning algorithms to integrate multisource data and tap the ‘wisdom of experts.’ At the voting layer, two sparse autoencoders are trained for feature learning to reduce the correlation of attributes and diversify the base classifiers ultimately. At the stacking layer, a nonlinear feature-weighted method based on a deep belief network is proposed to rank the base classifiers, which may violate the conditional independence. The neural network is used as a meta classifier. At the optimizing layer, over-sampling and threshold-moving are used to cope with the cost-sensitive problem. Optimized predictions are obtained based on an ensemble of probabilistic predictions by similarity calculation. The proposed deep ensemble learning framework is used for Alzheimer’s disease classification. Experiments with the clinical dataset from National Alzheimer’s Coordinating Center demonstrate that the classification accuracy of our proposed framework is 4% better than six well-known ensemble approaches, including the standard stacking algorithm as well. Adequate coverage of more accurate diagnostic services can be provided by utilizing the wisdom of averaged physicians. This paper points out a new way to boost the primary care of Alzheimer’s disease from the view of machine learning.
Keywords: Deep learning, Ensemble learning, Stacking, Classification, Alzheimer’s disease
1. INTRODUCTION
Ensemble learning utilizes a group of decision-making systems that apply various strategies to combine classifiers to improve prediction on new data. Stacking is a well-known approach among the ensembles in which the predictions of a collection of models are given as inputs to a second-level learning algorithm. It has been employed successfully on a wide range of problems, such as chemometrics [1], spam filtering [2], signal processing [3, 4], and healthcare [5].
Nevertheless, the correlation of base classifiers is hard to eliminate. Currently, most methods are focusing on diversity among the members of a team of classifiers. For example, different learning algorithms and training data sets have been used for this purpose [6, 7]. However, few efforts have been made to reduce the correlation of base classifiers in the second-level algorithm of stacking.
The Restricted Boltzmann Machine (RBM) is a representative example of deep learning, which has become popular in several applications over the last decades, including image recognition [8], bioinformatics [9] and natural language processing [10]. It is a probabilistic model that uses a layer of hidden binary variables or units to model the distribution of a visible layer of variables. As a generative model, it has been used for analyzing different types of data, including labeled or unlabeled images [11], and acoustic data [12]. RBM does not require the independent of input components [11]. It is indeed an advantage to fuse the predictions of base classifiers even they might depend on each other.
Alzheimer’s disease (AD) is a chronic neurodegenerative disorder, which makes up more than 60% of all dementia cases [13, 14]. Age is the major risk factor for AD. With a rapidly aging world population, diagnosis services in many middle-income countries strive to meet actual demand and are mostly confined to tertiary care hospitals in major population centers [15]. Deep learning with some variants has been used for AD prediction in previous works [16, 17], but lacked the generalization capability needed for application by medical practitioners owing to insufficient data and the inherent physicians’ bias clinical judgments. Making full use of limited resources to improve AD diagnostic accuracy poses a severe challenge in improving healthcare. Hence there is an increasing need for new methods that can enhance the primary care of AD.
The diagnosis of AD is generally based on history-taking, clinical presentation, and behavioral observations. Specialists working in memory clinics sometimes show surprisingly low levels of diagnostic agreement with each other [18], making it hard to obtain objective and reproducible diagnose. Alternatively, more opinions should be sought from primary care services because of the lack of AD specialists in many parts of the world. Therefore, it is vital to find ways to leverage the wisdom of experts better [19]. Our framework is an effective strategy to assist existing or new health professionals, who have insufficient AD-related training, in making a clinical diagnosis.
We regard the clinical decision making of physicians as a learning algorithm that searches a hypothesis space about AD outcome for the best one. Without sufficient data or expertise, the learning algorithms or physicians may derive different AD outcome hypotheses in hypothesis space that would result in the same level of predictive accuracy. By constructing an ensemble of these classifiers or physicians, the algorithm can average decisions and reduce the risk of reliance on the wrong classifier or physician. Many learning algorithms perform local searches for outcome hypotheses that are constrained in local optima. Similarly, physicians may have more expertise in a specific disease, and their diagnoses are often leaned toward what they are most familiar with. An ensemble may provide a better approximation to the real unknown outcome than any individual classifier. Wu et al. combine three different classifiers using weighted and unweighted schemes to improve AD prediction [20]. They use the 11C-PIB PET image data, but the diversity of base classifiers can be further considered. In other words, the base classifiers may depend on each other. There have been recent works on how to combine ensemble learning with deep learning systems to achieve greater prediction accuracy [21, 22].
Most of the existing frameworks for AD prediction tend to achieve lower error rates by assuming the same loss for any misclassification. A computer-aided diagnosis system is developed that uses feature ranking and genetic algorithms to analyze structural magnetic resonance imaging data [23]. The conversion of mild cognitive impairment (MCI) to AD is predicted with this system. However, different mistakes may lead to significantly different clinical consequences. For example, failing to detect AD has a more potentially significant consequence than a false positive prediction. Cost-sensitive learning provides a solution to this problem by considering misclassification costs in the learning process [24].
Although using automated computer tools to facilitate medical analysis and prediction is a promising and essential area [25–27], most existing classification methods only use one individual modality of biomarkers for AD prediction, and the data collection process is subject to variability, which may affect the overall classification performance. Voxel wise tissue probability, cortical thickness, and hippocampal volumes are all neuroimaging features often used for AD classification [28–30]. There are, however, also several biological and/or genetic biomarkers that have been identified as well as being significantly related to increased risk of AD. Different measures provide complementary information, which in combination may significantly increase AD prediction performance. The Uniform Data Set (UDS) stored by the National Alzheimer’s Coordinating Center (NACC) includes detailed clinical information of participants, such as cognition outcome, neuropsychological test results, and family history, as well as neuroimaging indices of neurodegeneration [31]. It is a valuable resource that has promoted a wealth of Alzheimer’s disease research findings [32–34].
Based on this multi-dimensional data, we propose a deep ensemble learning framework (DELearning) to leverage the clinical expertise of averaged physicians to obtain more accurate AD prediction. It could be used in primary care settings in which there are limited accesses to specialists. DELearning is a three-layer framework with five stages. Firstly, to fuse multi-source data and reduce the correlation of original features, sparse auto-encoder (SAE) is used for feature learning to construct three feature spaces. Secondly, base classifiers are built by using different learning algorithms and feature spaces. Multiple hypotheses that can be likened to different physician opinions are generated through this kind of manipulation of training data. Thirdly, a new dataset composed of prediction values of classifiers is fed to a Deep Belief Network (DBN), which is used as a stacking method to tackle violations of conditional independence of the base classifiers. Through a contrastive divergence learning procedure [35], DELearning can evaluate different experts and integrate their diagnosis decisions. Fourthly, three Neural Networks (NNs) are constructed based on a back-propagation algorithm, and cost-sensitive methods such as over-sampling and threshold moving. Finally, probabilistic predictions of these models are mapped to a three-dimensional space. Prototypes of different categories are extracted based on mean values. Discrimination is carried out based on the similarity between individuals and prototypes.
The contributions of this paper are as follows:
We propose a novel ensemble learning method for AD classification with 4% better than six well-known ensemble approaches, which points out a new way to boost the primary care of Alzheimer’s disease.
A stacking method is proposed that uses DBN to combine predictions of base classifiers and cope with their dependencies.
In our framework, base classifiers are served as surrogates to physicians with different clinical expertise, which can leverage the wisdom of experts and multisource data to make a sound outcome that could be referenced in clinical settings.
The remainder of this paper is organized as follows. Section 2 presents the learning methods of DELearning. Section 3 discusses the empirical results and some observations. Section 4 presents the conclusion of the paper.
2. METHODS
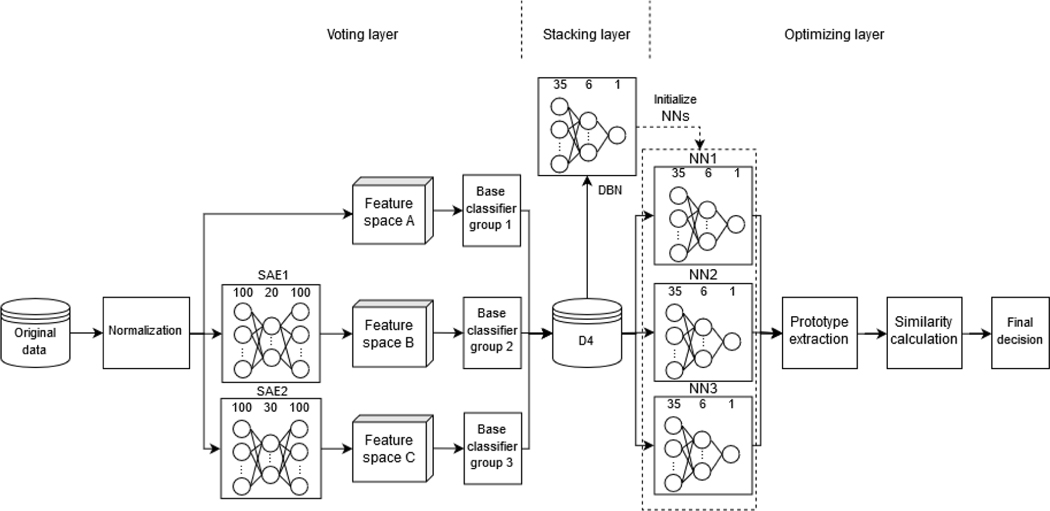
This paper interests in two outcomes: probable and possible AD (AD) and non-demented control (NDC). Probable and possible AD is the terminology used in all clinical settings [36]. Suppose we are given a group of samples defined on R, where 1≤ i≤ N the sample size is denoted by N. Each sample has the label representing AD and NDC, respectively. We refer to are the predictions of L physicians or base classifiers for the sample xi, where . Each physician or base classifier Li gets training samples to predict. All physicians will predict samples. In this paper, we consider the case where there are physicians (simulated by different base classifiers) with different expertise and try to address the question of how to obtain high prediction accuracy based on these circumstances. Refer to Fig. 1 for the framework of DELearning, which composes of three ensemble layers.
Fig. 1.
The framework of DELearning.
Voting layer
First, SAE [37] is used for feature learning to fuse normalized data and construct two feature spaces with a reduced correlation of attributes. Three data spaces are created by adding the normalization of the original data. Next, base classifiers are built by different learning algorithms on these spaces to generate multiple preliminary diagnoses. Thus, the samples are quantified by the predictions of these classifiers.
Stacking layer
DBN is used as a meta-classifier to combine the predictions of base classifiers in a weighted manner. It is a probabilistic generative model comprised of multiple Restricted Boltzmann Machines. In some cases, while the predictions of some base classifiers may be correlated with each other, the learned features in the hidden layer of RBM can be almost entirely uncorrelated [38]. Therefore, as a probabilistic generative model comprised of multiple RBMs, DBN is trained on the quantized samples as an ensemble method to tackle the dependence of base classifiers.
Optimizing layer
Three back-propagation NNs are built by the cost-sensitive method and assembled to optimize the predictions. NN1 is trained on the predictions of base classifiers with threshold moving. NN2 is trained on the dataset whose distribution is adjusted by over-sampling. NN3 is initialized as NN1 and NN2 by the parameters of the trained DBN model and trained on the same dataset. Then, we map the probabilistic diagnosis of these NNs in a 3-dimensional space and choose the mean values vector as the prototypes of AD and NDC. The similarities between a sample and the two prototypes are calculated by Euclidean distances. Finally, the outcome of the prototype, which is closest to the sample, is selected as the final decision.
2.1. Datasets
The data comes from the National Alzheimer’s Coordinating Center [31], which founded in 1999 and has maintained a cumulative database consisting of various types of clinical data such as clinical evaluations, brain MRI imaging, and neuropathology. Many researchers have been making use of this resource to get valuable findings [39]. We extracted 23,165 samples with 100 measures (attributes) from NACC UDS [40]. There are seven groups of measures selected including medical history (MH), Hachinski ischemic score (HIS), cerebrovascular disease (CVD), Unified Parkinson’s Disease Rating Scale (UPDRS), Neuropsychiatric Inventory Questionnaire (NPIQ), Geriatric Depression Scale (GDS) and Functional Activities Questionnaire (FAQ). HIS and CVD are separated into two subsets here. Refer to Table 1 for the details.
Table 1.
Seven groups of measures selected from NACC UDS.
| Groups | Measures |
|---|---|
|
| |
| MH | CVHATT, CVAFIB, CVANGIO, CVBYPASS, CVPACE, CVCHF, CVOTHR, CBSTROKE, CBTIA, CBOTHR, PD, SEIZURES, TRAUMBRF, HYPERTEN, HYPERCHO, DIABETES, B12DEF, THYROID, INCONTU, INCONTF |
|
| |
| HIS | ABRUPT, STEPWISE, SOMATIC, EMOT, HXHYPER, HXSTROKE, FOCLSYM, FOCLSIGN, HACHIN |
|
| |
| CVD | CVDVOG, STROKCOG, CVDIMAG, CVDIMAG1, CVDIMAG2, CVDIMAG3, CVDIMAG4 |
|
| |
| UPDRS | SPEECH, FACEXP, TRESTFAC, TRESTRHD, TRESTLHD, TRESTRFT, TRESTLFT, TRACTRHD, TRACTLHD, RIGDNECK, RIGDUPRT, RIGDUPLF, RIGDLORT, RIGDLOLF, TAPSRT, TAPSLF, HANDMOVR, HANDMOVL, HANDALTR, HANDALTL, LEGRT, LEGLF, ARISING, POSTURE, GAIT, POSSTAB, BRADYKIN |
|
| |
| NPIQ | DEL, HALL, AGIT, DEPD, ANX, ELAT, APA, DISN, IRR, MOT, NITE, APP |
|
| |
| GDS | SATIS, DROPACT, EMPTY, BORED, SPIRITS, AFRAID, HAPPY, HELPLESS, STAYHOME, MEMPROB, WONDRFUL, WRTHLESS, ENERGY, HOPELESS, BETTER |
|
| |
| FAQ | BILLS, TAXES, SHOPPING, GAMES, STOVE, MEALPREP, EVENTS, PAYATTN, REMDATES, TRAVEL |
|
| |
| Total number | samples: 23165; outcomes: 2; measures: 100 |
2.2. Voting layer
Due to the heterogeneous nature of 7 groups of attributes, a single classifier has difficulties in leveraging multisource information sufficiently to obtain a satisfying performance on AD classification no matter the amount of available data. More specifically, the clinical decision boundary that discriminates participants from different outcomes may be linear for some attributes while non-linear for another part. It may lie outside the space of functions that can be implemented by the chosen classifier. Even though a single classifier could achieve satisfying classification performance on the available data, it might not generalize for other data sources.
Three feature spaces are constructed by normalizing original data and two sparse autoencoders. Sixteen classification algorithms are trained on them to forming three base classifier groups to increase the generalization ability and diversify the decision boundaries. These classification algorithms consist of Bayes Network, Naive Bayes, J48, Hoeffding tree, REPTree, Filtered Classifier, Iterative Classifier Optimizer, Logistic Regression, LogitBoost, Multilayer Perception, Stacking, Random Committee, Random Forest, Random Subspace, AdaBoostM1, and Voted Perceptron. The prediction results from thirty-five classifiers selected with better performance are combined and fed into the next layer.
This section briefly illustrates three classification algorithms. Logistic Regression is a well-behaved classification algorithm, specially when the features to be studied can be treated as roughly linear, or the problem is linearly separable [41]. As a commonly used classifier, it can also deal with nonlinear problems through discretization and mapping of features. This discriminative model is also robust to noise and can avoid overfitting by using L1 or L2 regularization.
Tree ensembles like Random Forests are a combination of a bunch of decision trees [42]. One dominant advantage of tree ensemble is that they do not presume linear features for data. So, they are quite suitable for handling certain features.
Naive Bayes classifier is a probabilistic model based on Bayes theorem that can simplify learning by assuming that features are independent given class [43]. In the case of conditional independence, less data is needed in training a naive Bayes classifier due to that often converge faster than discriminative models such as Logistic Regression. Although this assumption is often impractical in the real world, Naive Bayes classifier still performs better in practice.
Various indicators such as Q-statistic and “difficulty” θ have been discussed for quantitative assessment of diversity [44]. Q-statistic is a measure to evaluate the similarity of two classifiers’ predictions. It is formulated as
| (1) |
of which Nab is the number of samples where the classifier i has outcome a, and the classifier j has outcome b, where a and b could have value 0 (when the classifier predicts the wrong class) or 1 (when the classifier predicts the correct class for the sample).
As to multiple classifiers’ diversity assessment, the difficulty θ is used and defined as the variance of a discrete random variable X, which denotes the proportion of base classifiers that correctly classify a participant drawn randomly from the original training data.
We can treat the trained base classifiers as physicians from different fields with different clinical expertise because of these diversities. For a participant, the prediction of a classifier is equivalent to the diagnosis result of the corresponding physician.
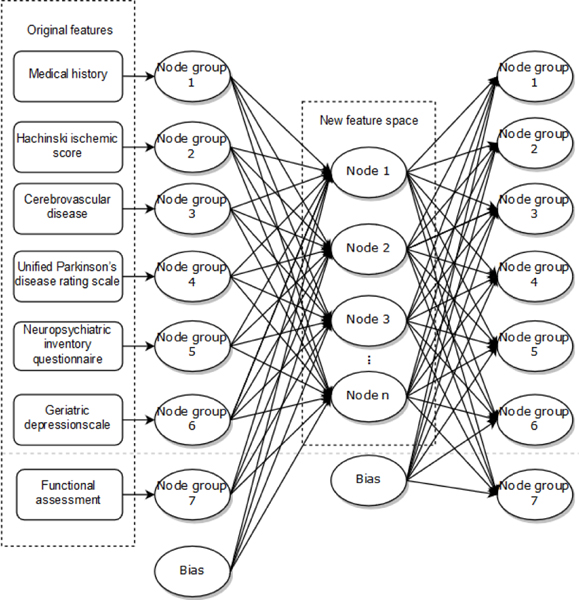
In addition to different learning algorithms, resampling of the training data is another way to increase the diversity of base classifiers. The clinical measures extracted from NACC are sparse, which affects the performance of some classifiers. To improve diversity and obtain high-level feature representation, two SAEs are used in the voting layer to automatically learn different feature spaces defined by the activations of their hidden nodes. We take SAE with a three-layer symmetrical structure including an input layer, a hidden layer, and an output layer. Seven node groups in the input layer consists of 100 nodes corresponding to measures in Table 1. The suitable number of nodes in the hidden layer can be selected according to reconstruction error to determine the dimensions of feature spaces. The activation values of hidden nodes are extracted as high-level features. Refer to Fig. 2 for the details of the SAE structure.
Fig. 2.
The structure of SAE.
Suppose the activation function of hidden units is the sigmoid function, the average activation of hidden unit j over n samples is denoted as
| (2) |
where is the activation function of the hidden unit j given a sample . A sparse representation is used to represent the input data better. The constraint is enforced to make hidden unit’s activation mostly near the sparsity parameter whose typical value close to 0.
To satisfy the constraint, we apply Kullback-Leibler (KL) divergence to optimization objective J (reconstruction error), serving as a sparsity penalty term. It is formulated as follows [45].
| (3) |
where W,b is weight and bias, respectively, , n is the number of hidden units.
The most crucial advantage of feature learning by SAE is that the correlation of transformed features is significantly reduced. Thereby, trained on these feature spaces, the diversity of base classifiers can be further improved.
2.3. Stacking layer
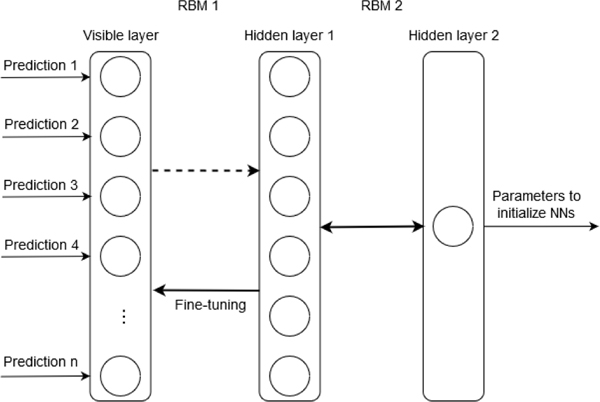
Stacking is an ensemble technique in which the predictions of a group of predictive models are combined through other learning models to generate a final output. In our stacking layer, the predictions of base classifiers from the voting layer are combined as inputs to the DBN that composed of two RBMs. RBM is a probabilistic model that uses variables of a hidden layer to model the distribution of observed data. Typically, an RBM is trained in an unsupervised manner to model the distribution of the inputs. The most outstanding strength of RBM is that the hidden units are conditionally independent, given the visible units [11]. It can learn the abstract representation from the predictions of base classifiers. As shown in Fig. 3, we train the DBN that consists of 35 nodes in the input layer, which corresponding to base classifiers with better performance selected, six nodes in the first hidden layer, and one node in the second hidden layer on the predictions of base classifiers. After the pre-training of RBMs in a layer-wise manner, the back-propagation learning algorithm is adopted for fine-tuning based on mean square error (MSE). The parameters of the trained DBN are used to initialize three neural networks with the same structure in the next layer, which reduces the effect of correlation from base classifiers’ predictions.
Fig. 3.
Stacking layer of DELearning with DBN.
2.4. Optimizing layer
One challenge in AD diagnosis is that the number of patients is fewer than healthy people in primary care settings. The cost of a missed diagnosis is higher than that of misdiagnosis under this circumstance. Cost-sensitive learning is a suitable tool for learning both from imbalanced datasets and unequal costs [24]. Based on Ref. [6], DELearning adapts over-sampling, threshold moving as cost-sensitive strategies to train NNs and assemble their probabilistic predictions.
In DELearning, there are two diagnosing outcomes for a participant, i.e., . The participant number corresponding to these two outcomes is Ni and Nj respectively. denotes the cost of a participant that belongs to the ith outcome misdiagnosing as the jth outcome (if the diagnosis is right then ). Under binary classification circumstances, it is clear that the cost of the ith outcome Cost[i] is equal to The cost matrix can be represented as
| (4) |
In order to solve the problem of unbalanced data and categories, we consider adopting oversampling for categories with fewer data. Suppose Ni< Nj, the ith outcome will have individuals after resampling. It can be computed as follows.
| (5) |
Where is called the coefficient of cost. The ranges of this parameter should fall into , and the category not be overfitted. Thus participants can be resampled for the ith category with random replacement.
Since under-sampling discards potentially useful training samples, the performance of the resulting classifier may be degraded. DELearning uses over-sampling for training NN in the optimizing layer.
Besides over-sampling, we also use threshold moving to adjust the output unit in NN toward an inexpensive outcome so that participants with higher misclassification costs will be easily identified.
Suppose Oi and are denoted as the output of NN in optimizing layer with or without threshold moving, the normalized real value can be adjusted in DELearning as follows.
| (6) |
After training DBN in the stacking layer, the parameters and structure are used to initialize three neural networks in the optimizing layer. NN1 is trained with threshold-moving and NN2 with over-sampling, respectively, on the predictions of base classifiers. NN3 is also trained directly on these predictions. The real-value outputs of these three NNs are combined to form prototypes for each category. The Euclidean distances are computed between samples and the two prototypes. The category with the smallest distance is selected as the final prediction result for the samples.
The DELearning algorithm is shown in Table 2.
Table 2.
Proposed DELearning algorithm.
| Input: Samples D with clinical measures and outcomes; Output: Prediction of AD or NDC |
|---|
|
|
| Voting Layer: |
| 1 Normalize the original data set D, get D1. |
| 2 Train two SAEs on D1. |
| 3 Derive the optimal structure of SAEs based on MSE. |
| 4 Construct data sets D2 and D3 by extracting activation values of hidden layers in SAEs. |
| 5 Train base classifiers on D1, D2, and D3. |
| 6 Generate data set D4 consisting of the predictions of all base classifiers. |
| Stacking Layer: |
| 7 Train DBN on D4 to stack the predictions of base classifiers. |
| 8 Generate the parameters and structures of the trained DBN. |
| Optimizing Layer: |
| 9 Initialize three neural networks NN1, NN2, and NN3 with the parameters and structures of DBN. |
| 10 Train NN1 with threshold-moving on D4. |
| 11 Train NN2 with over-sampling on D4. |
| 12 Train NN3 on D4. |
| 13 Generate predictions of NN1, NN2, NN3. |
| 14 Generate prototypes for AD and NDC. |
| 15 Calculate the similarity between samples and |
| prototypes. |
| 16 Make a final decision according to similarity. |
2.5. Performance measures
A confusion matrix that contains the actual outcome and predicted outcome is used to evaluate the performance of AD classification. Table 3 presents the confusion matrix for AD classification with two outcomes. TP is the number of AD patients that are correctly classified as AD. FP is the number of NDC participants that are diagnosed as AD. FN is the number of AD patients that are incorrectly classified as NDC. TN is the number of NDC participants that are classified correctly. We use the following four measures to evaluate DELearning as well as all the base classifiers. The statistically significant comparison of performances of DELearning and benchmarks are performed by using McNemar’s test [46]. The adjustment for multiple comparisons is performed using Bonferroni correction [47].
Table 3.
Confusion matrix for AD prediction.
| Predicted outcome | |||
|---|---|---|---|
|
|
|||
| AD | NDC | ||
|
| |||
| Actual outcome | AD | TP | FN |
|
| |||
| NDC | FP | TN | |
Accuracy is the proportion of all participants that are correctly classified as either AD or NDC. It is formulated as the following:
| (7) |
Precision denotes the proportion of predicted AD cases that are real AD patients.
| (8) |
The recall is the proportion of AD patients that are correctly classified. It reflects the ability of a classifier to recognize positive examples. In a medical context, recall is regarded as a more primary measure than precision [48], as the aim is to identify all real positive cases.
| (9) |
F1-measure provides a way to combine precision and recall into a single measure with no imbalanced manner, which can be formulated as follows:
| (10) |
3. RESULTS AND DISCUSSION
The original dataset is randomly split into three subsets, namely subset1 (50%), subset2 (30%), and subset3 (20%). Subset1 (50%) is used to select the base classifiers. Subset1 and subset2 are used as the training set (80%). Subset3 is used as the testing set (20%). In the training process, subset1 is used to train and further select the base classifiers. Table 4–7 shows the performance of classifiers on subset1 by 5-fold cross-validation. Based on it, the classifiers with better performance are incorporated into our model as the base classifiers. Then, the base classifiers make predictions on the training set (subset1 and subset2). These predictions are used to train DBN as well as three NNs that initialized by the parameters of DBN. The prototypes of NDC and AD are extracted based on the predictions of three NNs. In the testing phase, it is an end-to-end evaluation which is carried out on the testing set (subset3), which successively experienced SAEs, base classifiers, NNs, and similarity calculation to generate the final prediction result.
Table 4.
Accuracy of base classifiers in three feature spaces. (A) normalized original space, (B) 20-dimensional space learned by SAE, (C) 30-dimensional space learned by SAE.
| Classifier | Feature space | ||
|---|---|---|---|
|
| |||
| A | B | C | |
|
| |||
| Bayes Nets | 75.6% | 71.4% | 72.6% |
| Filtered Classifier | 79.2% | 75.4% | 75.3% |
| Hoeffding Tree | 78.5% | 76.0% | 74.7% |
| Iterative Classifier Optimizer | 79.9% | 75.3% | 76.3% |
| J48 | 79.1% | 75.1% | 75.8% |
| Logistic Regression | 80.4% | 77.2% | 78.0% |
| LogitBoost | 79.9% | 75.3% | 76.3% |
| Random Committee | 82.7% | 75.6% | 75.4% |
| Random Forest | 81.7% | 83.9% | 78.4% |
| Random SubSpace | 81.8% | 76.1% | 77.4% |
| REPTree | 80.4% | 76.0% | 75.9% |
| AdaBoostM1 | 76.2% | 74.4% | 75.5% |
| Multilayer Perception | 80.5% | 79.7% | 80.1% |
| Naive Bayes | 72.1% | 71.5% | 73.6% |
| Stacking | 63.1% | 62.6% | 63.1% |
| Voted Perceptron | 78.5% | 77.4% | 76.4% |
Table 7.
F1-measure of base classifiers in three feature spaces. (A) normalized original space, (B) 20-dimensional space learned by SAE, (C) 30-dimensional space learned by SAE.
| Classifier | Feature space | ||
|---|---|---|---|
|
| |||
| A | B | C | |
|
| |||
| Bayes Nets | 75.8% | 71.9% | 72.1% |
| Filtered Classifier | 79.2% | 75.4% | 75.2% |
| Hoeffding Tree | 78.7% | 76.0% | 73.5% |
| Iterative Classifier Optimizer | 79.9% | 75.6% | 76.4% |
| J48 | 79.0% | 74.7% | 76.0% |
| Logistic Regression | 80.2% | 77.3% | 78.0% |
| LogitBoost | 79.9% | 75.6% | 76.4% |
| Random Committee | 82.7% | 75.3% | 75.1% |
| Random Forest | 82.7% | 82.9% | 78.4% |
| Random SubSpace | 81.8% | 75.8% | 77.3% |
| REPTree | 80.4% | 76.1% | 75.9% |
| AdaBoostM1 | 76.1% | 74.9% | 75.6% |
| Multilayer Perception | 80.3% | 79.5% | 79.8% |
| Naive Bayes | 71.4% | 72.0% | 74.0% |
| Stacking | 62.2% | 60.3% | 60.6% |
| Voted Perceptron | 78.1% | 77.5% | 77.8% |
3.1. Feature learning
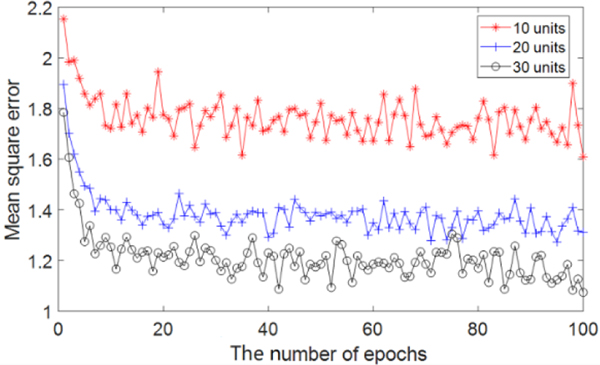
DELearning utilizes SAE as a feature learning method to obtain more discriminative features compared with the original set. In order to determine the optimal dimension of transformed space and reduce data correlation, we trained three-layer SAE models with 100 input units and 10, 20, 30 hidden units, respectively. The activation function was sigmoid. Obvious biases and weights were initialized to zero and random numbers sampled from a normal distribution with zero mean and standard deviation 1. Momentum was set to 0.5. The model was trained for 100 epochs. As shown in Fig. 4, the MSE of models with 20, 30 hidden units are much smaller than the model with 10 hidden units. Considering lower dimensions of data selection in situations of limited reduction of MSE, the number of hidden units of SAEs is set to 20 and 30 for the transformation of feature space.
Fig. 4.
The performance comparison of SAEs with 10, 20, 30 hidden units. The x-axis indicates the number of epochs, and the y-axis indicates the MSE with the corresponding epoch.
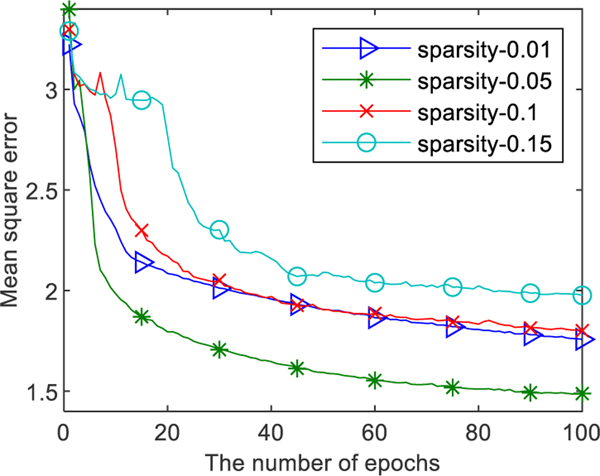
As shown in Fig. 5, we compared the performance of SAE with commonly used sparsity parameter ρ= 0.01, 0.05, 0.1, and 0.15, respectively. It can be seen that the best performance at ρ= 0.05. So, we set ρ= 0.05 in DELearning.
Fig. 5.
The MSE of SAE with specific parameter ρ.
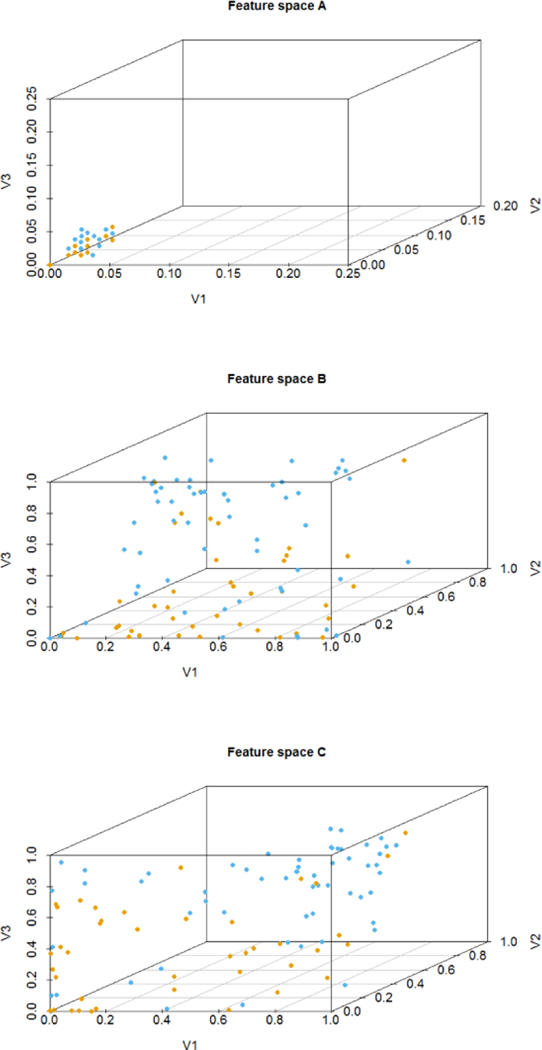
The samples after feature transformation are easier to distinguish from their categories. Fig. 6 presents 100 participants randomly selected from three feature spaces. In the original space, most features of participants with different outcomes are overlap with each other. With the transformed features, the boundary between AD and NDC is relatively obvious, which intuitively indicates that the dimension of features in transformed spaces is not only reduced but also more clearly representing the two outcome groups.
Fig. 6.
Visualization of 100 participants in 3 dimensions. (a) original space with some points overlapped, (b) feature space learned by SAE with 20 hidden units, (c) feature space learned by SAE with 30 hidden units. Each dot represents a participant. The color of the dot indicates the outcomes, orange for AD, and blue for NDC, respectively.
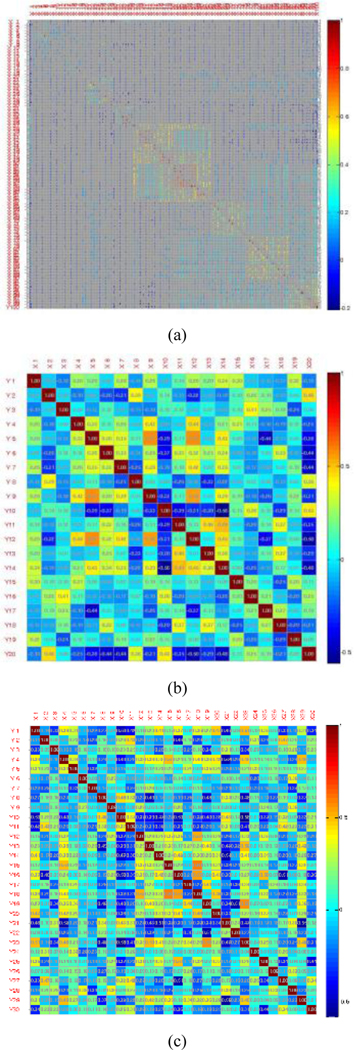
Fig. 7 shows the correlation matrices of features in different spaces constructed. The shaded rectangular tiling represents the correlation value of the corresponding two attributes. It uses a color scale ranging from blue (low correlation value) to red (high correlation value). It indicates that most features in the original space are not conditional independent of each other. The features in 20- and 30-dimensional spaces are approximately uncorrelated. It demonstrates that SAE reduces the dependence of samples’ attributes.
Fig. 7.
Correlation matrix of three feature spaces. (a) original space, (b) 20-dimensional space transformed by SAE, (c) 30-dimensional space transformed by SAE.
3.2. Construction of base classifiers
This section evaluates the performance of the classifiers constructed by multiple classification algorithms on samples of subset1 in three feature spaces. Here are some details of the classifiers. The structure learning method of Bayes network is a hill climbing search for optimal Bayes score. The maximum likelihood estimates method is used for the parameter learning with the obtained structure. The filtered classifier is a combination of the J48 decision tree and the MDL discretization method [49]. The minimum number of participants in the leaf is set to 2. 3-fold of participants is used for pruning with the confidence factor is 0.25. The rest is used for growing the tree. Hoeffding tree is an incremental anytime decision tree induction algorithm, which splitting criterion is the Gini index. The leaf prediction strategy is set to the majority class. REPTree is a decision tree using information gain and prunes with reducederror pruning with backfitting. Random subspace consists of multiple REPTree constructed systematically by randomly selecting 50% components of all features. The activation function of Multilayer Perception is a sigmoid function. The number of hidden units is set to 51, 11, and 16 for the models in 3 different feature spaces. The learning rate is set to 0.3. Momentum is 0.2. The others adopt the standard parameter settings.
Table 4 to Table 7 shows the performance of classifiers in the reduced feature spaces, namely 20- and 30-dimensional spaces learned by SAE, which are comparable with the original feature space. It indicates that our feature learning method not only dramatically reduces the correlation of clinical measures but also retains the useful information. The classifiers with better performance are selected as the base classifiers. At the same time, physicians in different fields can be imitated by these classifiers.
Through these different learning algorithms and feature spaces, the diversity of these base classifiers or physicians has been improved. Here is an example to validate it. Table 8 shows the classification results of Logistic Regression (LR) and REPTree (RT) in the 20-dimensional feature space. N11 and N00 are the numbers of participants that are correctly and wrongly recognized by these two classifiers, respectively. N10 is the number of participants who were correctly diagnosed by LR but wrongly diagnosed by RT, and N01 vice versa. The Q statistics between LR and RT is 0.86, which is smaller than 1. It shows that these two classifiers tend to recognize the same individuals correctly.
Table 8.
The relationship between Logistic Regression and REPTree in classification on 20-dimensional feature space.
| RT correct(1) | RT wrong(0) | |
|---|---|---|
|
| ||
| LR correct(1) | N11(7535) | N10(1102) |
| LR wrong(0) | N01(958) | N00(1905) |
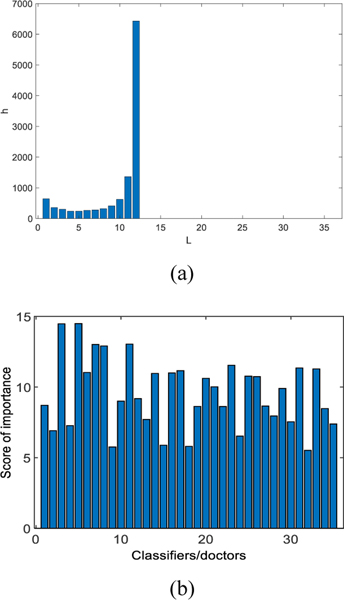
Let X be the number of base classifiers that correctly identify a participant. The variance of X is a measure of diversity based on the distribution of difficulty. Fig. 8(a) shows that all classifiers in the group made the correct predictions for around 4500 samples in the test dataset. All base classifiers which are trained by different learning algorithms and feature spaces achieved relatively high performance. Diverse groups of classifiers will have a smaller variance of X. Here, X= 1.1751e+006. As shown in Fig. 8(b), after using our classifier ranking method in the stacking layer, we observed that the base classifiers No. 3 and 5, have the highest score whose prediction accuracy are concurrently higher. Therefore, DELearning can evaluate classifiers or physicians automatically.
Fig. 8.
(a) Patterns of difficulty θ for classifiers group with L = 35, N = 11500 and p > 0.7. The histograms show the number of samples that are correctly diagnosed by classifiers. The x-axis is the number of classifiers. (b) The important score of base classifiers calculated by DELearning. The x-axis indicates the classifier number. The y-axis is the score of the corresponding classifier.
3.3. Stacking by DBN
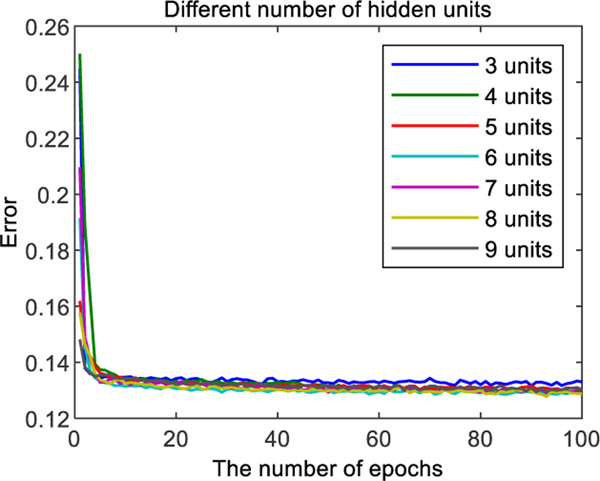
We formulate each participant as a vector composing of the predictions of these base classifiers. DBN is trained on the training set to ensemble these opinions from base classifiers with a greedy layer-wise unsupervised method in a mini-batch size of 100 cases [50]. Obvious biases and weights are initialized to 0, and random numbers obeyed a zero-mean normal distribution with a standard deviation 0.01. Momentum is set as 0.5. We run 100 epochs with a learning rate of 0.002. Here, we compare different settings of the number of hidden units from 3 to 9. Refer to Fig. 9 for the details. The DBN with 6 hidden units has the lowest error, and so it is selected. The trained DBN is used to initialize artificial neural networks.
Fig. 9.
Errors with different sizes of hidden layers.
3.4. Optimization for the final decision
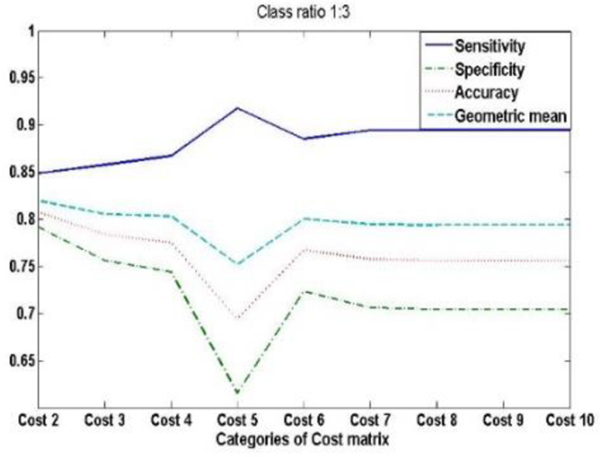
We compare 9 cost matrixes with integer values between 2.0 and 10.0. Each matrix is required to have at least one non-diagonal element to be equal to 1. From Fig. 10, we can see that the Geometric mean of cost 2 is highest; the cost matrix we used are shown in Table 9.
Fig. 10.
The performance of NNs with different cost matrixes.
Table 9.
Cost matrix used in DELearning.
| AD | NDC | |
|---|---|---|
|
| ||
| AD | 0 | 2 |
| NDC | 1 | 0 |
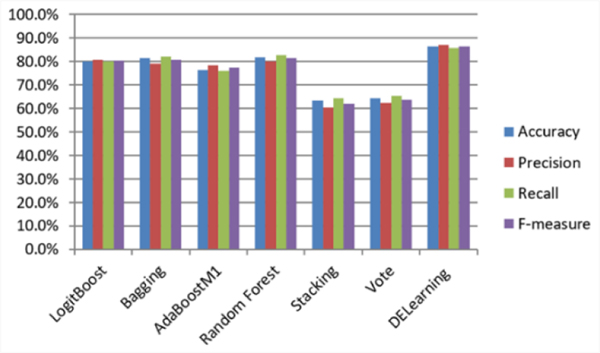
Specifically, after all the parameters are determined by training on training set in which the validation set is included in it, we compared DELearning with six ensemble learning methods, including LogitBoost [51], Bagging [52], Random Forest [53], AdaBoostM1 [54], Stacking [55], Vote [56]. As shown in Fig. 11, our method outperforms other ensemble methods in terms of precision, recall, accuracy, and F1-measure, with increases in the recall and accuracy of more than 3% and 4%, respectively. The differences between the performances of DELearning and benchmarks are statistically significant, with P<0.001.
Fig. 11.
The performance comparison of DELearning with six representative ensemble learning methods on NACC UDS.
Two cost-sensitive learning strategies, including over-sampling and threshold-moving, are adopted in optimizing layer to improve the AD recognition ability of our model by respectively manipulating data and model. By modifying the distribution of the training data, over-sampling technology creates a balanced dataset that provides more AD cases for training NN. By moving the decision threshold of NN toward AD outcome, threshold-moving makes AD cases harder to be misclassified by the NN. The higher recall of our method indicates that more AD cases can be correctly detected. Our method has a low probability of a missed diagnosis of AD whose cost is higher than that of misdiagnosis in the primary care settings. For the application scenario of this paper, high recall is more important than high precision since averaged physicians in primary care settings are interested in screening cases that may be AD. Then, these cases can be advised to go to a superior hospital for more medical resources to confirm the diagnosis. This approach could alleviate the pressure on superior hospitals with more and more demands on the service of AD diagnosis. From another perspective, through combining opinions of multiple primary care physicians simulated by base classifiers, the higher accuracy and recall can be achieved by our method. It could help boost the primary diagnosis of AD. The scarce medical resources can then be well allocated, specially for the areas where diagnostic coverage is low. It could help boost the primary diagnosis of AD, especially for the areas where medical resources are scarce.
4. CONCLUSIONS
The challenge of AD classification leads us to leverage the wisdom of experts and integrate multisource data to come up with better outcome prediction modality that could be used in primary care. This paper proposes DELearning, a three-layer framework, for AD classification that uses the deep learning approach to ensemble at each layer.
Using the clinical data from NACC UDS, we compared the performance of DELearning with six representative ensemble learning methods. The experimental results show that DELearning outperforms the others in terms of AD classification accuracy. It provides a data-driven solution to aid AD primary care, especially where access to AD expertise is limited.
DELearning can also be applied to other scenarios, including medical image tagging, where it may be not feasible or too expensive to obtain objective and reliable labels. DELearning can collect subjective labels from multiple experts or annotators and find meaningful yet hidden labels.
Table 5.
The precision of base classifiers in three feature spaces. (A) normalized original space, (B) 20-dimensional space learned by SAE, (C) 30-dimensional space learned by SAE.
| Classifier | Feature space | ||
|---|---|---|---|
|
| |||
| A | B | C | |
|
| |||
| Bayes Nets | 76.1% | 73.9% | 73.1% |
| Filtered Classifier | 79.1% | 75.5% | 75.1% |
| Hoeffding Tree | 79.1% | 76.1% | 74.6% |
| Iterative Classifier Optimizer | 79.8% | 76.1% | 76.4% |
| J48 | 78.9% | 74.7% | 76.2% |
| Logistic Regression | 80.2% | 77.3% | 78.0% |
| LogitBoost | 80.1% | 76.1% | 76.4% |
| Random Committee | 82.7% | 75.2% | 75.1% |
| Random Forest | 80.1% | 80.9% | 78.1% |
| Random SubSpace | 81.8% | 75.8% | 77.2% |
| REPTree | 80.4% | 76.3% | 76.0% |
| AdaBoostM1 | 78.5% | 77.1% | 75.8% |
| Multilayer Perception | 80.3% | 79.5% | 78.3% |
| Naive Bayes | 71.5% | 73.8% | 75.4% |
| Stacking | 60.2% | 61.3% | 60.5% |
| Voted Perceptron | 78.3% | 77.5% | 78.1% |
Table 6.
Recall rate of base classifiers in three feature spaces. (A) normalized original space, (B) 20-dimensional space learned by SAE, (C) 30-dimensional space learned by SAE.
| Classifier | Feature space | ||
|---|---|---|---|
|
| |||
| A | B | C | |
|
| |||
| Bayes Nets | 75.6% | 71.4% | 72.3% |
| Filtered Classifier | 79.3% | 75.4% | 75.4% |
| Hoeffding Tree | 78.5% | 76.0% | 74.7% |
| Iterative Classifier Optimizer | 79.9% | 75.4% | 76.3% |
| J48 | 79.1% | 75.1% | 75.8% |
| Logistic Regression | 80.5% | 77.3% | 78.1% |
| LogitBoost | 79.9% | 75.4% | 76.3% |
| Random Committee | 82.8% | 75.6% | 75.5% |
| Random Forest | 82.8% | 84.9% | 77.5% |
| Random SubSpace | 81.9% | 76.2% | 77.4% |
| REPTree | 80.4% | 76.0% | 75.9% |
| AdaBoostM1 | 76.1% | 74.5% | 75.5% |
| Multilayer Perception | 80.4% | 79.8% | 79.9% |
| Naive Bayes | 72.2% | 71.6% | 73.6% |
| Stacking | 64.2% | 62.7% | 62.8% |
| Voted Perceptron | 78.0% | 77.4% | 77.9% |
ACKNOWLEDGMENTS
This work was supported partially by the National Key R&D Program of China (No. 2018YFB1003204), Anhui Provincial Key Technologies R&D Program (No. 1804b06020378, No. 1704e1002221), CAMS Initiative for Innovative Medicine (CAMS-I2M, No. 2016-I2M-1-004). The NACC database is funded by NIA/NIH Grant U01 AG016976. NACC data are contributed by the NIA-funded ADCs: P30 AG019610 (PI Eric Reiman, MD), P30 AG013846 (PI Neil Kowall, MD), P50 AG008702 (PI Scott Small, MD), P50 AG025688 (PI Allan Levey, MD, PhD), P50 AG047266 (PI Todd Golde, MD, PhD), P30 AG010133 (PI Andrew Saykin, PsyD), P50 AG005146 (PI Marilyn Albert, PhD), P50 AG005134 (PI Bradley Hyman, MD, PhD), P50 AG016574 (PI Ronald Petersen, MD, PhD), P50 AG005138 (PI Mary Sano, PhD), P30 AG008051 (PI Thomas Wisniewski, MD), P30 AG013854 (PI M. Marsel Mesulam, MD), P30 AG008017 (PI Jeffrey Kaye, MD), P30 AG010161 (PI David Bennett, MD), P50 AG047366 (PI Victor Henderson, MD, MS), P30 AG010129 (PI Charles DeCarli, MD), P50 AG016573 (PI Frank LaFerla, PhD), P50 AG005131 (PI James Brewer, MD, PhD), P50 AG023501 (PI Bruce Miller, MD), P30 AG035982 (PI Russell Swerdlow, MD), P30 AG028383 (PI Linda Van Eldik, PhD), P30 AG053760 (PI Henry Paulson, MD, PhD), P30 AG010124 (PI John Trojanowski, MD, PhD), P50 AG005133 (PI Oscar Lopez, MD), P50 AG005142 (PI Helena Chui, MD), P30 AG012300 (PI Roger Rosenberg, MD), P30 AG049638 (PI Suzanne Craft, PhD), P50 AG005136 (PI Thomas Grabowski, MD), P50 AG033514 (PI Sanjay Asthana, MD, FRCP), P50 AG005681 (PI John Morris, MD), P50 AG047270 (PI Stephen Strittmatter, MD, PhD).
REFERENCES
- [1].Xu L, Jiang JH, Zhou YP, Wu HL, Shen GL, and Yu RQ, “MCCV stacked regression for model combination and fast spectral interval selection in multivariate calibration,” Chemometrics and Intelligent Laboratory Systems, vol. 87, pp. 226–230, Jun 15 2007. [Google Scholar]
- [2].Sakkis G, Androutsopoulos I, Paliouras G, Karkaletsis V, Spyropoulos CD, and Stamatopoulos P, “Stacking classifiers for anti-spam filtering of e-mail,” Proceedings of the 2001 Conference on Empirical Methods in Natural Language Processing, pp. 44–50, 2001. [Google Scholar]
- [3].Xiao L, Wan XY, Lu XZ, Zhang YY, and Wu D, “IoT Security Techniques Based on Machine Learning How do IoT devices use AI to enhance security?,” Ieee Signal Processing Magazine, vol. 35, pp. 41–49, Sep 2018. [Google Scholar]
- [4].Xiao L, Wan XY, and Han Z, “PHY-Layer Authentication With Multiple Landmarks With Reduced Overhead,” Ieee Transactions on Wireless Communications, vol. 17, pp. 1676–1687, Mar 2018. [Google Scholar]
- [5].Hsieh NC, Hung LP, Shih CC, Keh HC, and Chan CH, “Intelligent Postoperative Morbidity Prediction of Heart Disease Using Artificial Intelligence Techniques,” Journal of Medical Systems, vol. 36, pp. 1809–1820, Jun 2012. [DOI] [PubMed] [Google Scholar]
- [6].Zhou ZH, Wu JX, and Tang W, “Ensembling neural networks: Many could be better than all (vol 137, pg 239, 2002),” Artificial Intelligence, vol. 174, pp. 1570–1570, Dec 2010. [Google Scholar]
- [7].Melville P. and Mooney RJ, “Constructing diverse classifier ensembles using artificial training examples,” in IJCAI, 2003, pp. 505–510. [Google Scholar]
- [8].Schmidt U. and Roth S, “Shrinkage fields for effective image restoration,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2014, pp. 2774–2781. [Google Scholar]
- [9].Movahedi F, Coyle JL, and Sejdić E, “Deep belief networks for electroencephalography: A review of recent contributions and future outlooks,” IEEE journal of biomedical and health informatics, vol. 22, pp. 642–652, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mesnil G, Dauphin Y, Yao KS, Bengio Y, Deng L, Hakkani-Tur D, et al. , “Using Recurrent Neural Networks for Slot Filling in Spoken Language Understanding,” Ieee-Acm Transactions on Audio Speech and Language Processing, vol. 23, pp. 530–539, Mar 2015. [Google Scholar]
- [11].Hinton GE, Osindero S, and Teh YW, “A fast learning algorithm for deep belief nets,” Neural Computation, vol. 18, pp. 1527–1554, Jul 2006. [DOI] [PubMed] [Google Scholar]
- [12].Dahl G, Mohamed A.-r., and Hinton GE, “Phone recognition with the mean-covariance restricted Boltzmann machine,” in Advances in neural information processing systems, 2010, pp. 469–477. [Google Scholar]
- [13].Burns A. and Iliffe S, “Alzheimer’s disease,” BMJ, vol. 338, p. b158, 2009. [DOI] [PubMed] [Google Scholar]
- [14].Lam B, Masellis M, Freedman M, Stuss DT, and Black SE, “Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome,” Alzheimers Res Ther, vol. 5, p. 1, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Prince M, Comas-Herrera A, Knapp M, Guerchet M, and Karagiannidou M, “World Alzheimer report 2016: improving healthcare for people living with dementia: coverage, quality and costs now and in the future,” 2016. [Google Scholar]
- [16].Liu SQ, Liu SD, Cai WD, Pujol S, Kikinis R, and Feng DG, “Early Diagnosis of Alzheimer’s Disease with Deep Learning,” 2014. IEEE 11th; International Symposium on Biomedical Imaging (ISBI), pp. 10151018, 2014. [Google Scholar]
- [17].Suk HI and Shen DG, “Deep Learning-Based Feature Representation for AD/MCI Classification,” Medical Image Computing and Computer-Assisted Intervention - Miccai 2013, Pt Ii, vol. 8150, pp. 583–590, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Martin-Khan M, Flicker L, Wootton R, Loh PK, Edwards H, Varghese P, et al. , “The Diagnostic Accuracy of Telegeriatrics for the Diagnosis of Dementia via Video Conferencing,” Journal of the American Medical Directors Association, vol. 13, Jun 2012. [DOI] [PubMed] [Google Scholar]
- [19].Duboff RS, “The wisdom of (expert) crowds,” Harvard Business Review, vol. 85, pp. 28–0, Sep 2007. [Google Scholar]
- [20].Wu W, Venugopalan J, and Wang MD, “11C-PIB PET image analysis for Alzheimer’s diagnosis using weighted voting ensembles,” in 2017. 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2017, pp. 3914–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deng L. and Platt JC, “Ensemble deep learning for speech recognition,” in Fifteenth Annual Conference of the International Speech Communication Association, 2014. [Google Scholar]
- [22].Qiu XH, Zhang L, Ren Y, Suganthan PN, and Amaratunga G, “Ensemble Deep Learning for Regression and Time Series Forecasting,” 2014. Ieee Symposium on Computational Intelligence in Ensemble Learning (Ciel), pp. 21–26, 2014. [Google Scholar]
- [23].Beheshti I, Demirel H, Matsuda H, and A s. D. N. Initi, “Classification of Alzheimer’s disease and prediction of mild cognitive impairment-to-Alzheimer’s conversion from structural magnetic resource imaging using feature ranking and a genetic algorithm,” Computers in Biology and Medicine, vol. 83, pp. 109–119, Apr 1 2017. [DOI] [PubMed] [Google Scholar]
- [24].Ting KM, “An instance-weighting method to induce cost-sensitive trees,” Ieee Transactions on Knowledge and Data Engineering, vol. 14, pp. 659–665, May-Jun 2002. [Google Scholar]
- [25].An N, Jin L, Yang J, Yin Y, Jiang S, Jing B, et al. , “Data Platform for the Research and Prevention of Alzheimer’s Disease,” in Healthcare and Big Data Management, ed: Springer, 2017, pp. 55–78. [DOI] [PubMed] [Google Scholar]
- [26].Ang TF, An N, Ding H, Devine S, Auerbach SH, Massaro J, et al. , “Using data science to diagnose and characterize heterogeneity of Alzheimer’s disease,” Alzheimer’s & Dementia: Translational Research & Clinical Interventions, vol. 5, pp. 264–271, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ding H, An N, Au R, Devine S, Auerbach SH, Massaro J, et al. , “Exploring the Hierarchical Influence of Cognitive Functions for Alzheimer’s Disease in a Cohort Study,” Journal of Medical Internet Research, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Magnin B, Mesrob L, Kinkingnehun S, Pelegrini-Issac M, Colliot O, Sarazin M, et al. , “Support vector machine-based classification of Alzheimer’s disease from whole-brain anatomical MRI,” Neuroradiology, vol. 51, pp. 73–83, Feb 2009. [DOI] [PubMed] [Google Scholar]
- [29].Oliveira PPD, Nitrini R, Busatto G, Buchpiguel C, Sato JR, and Amaro E, “Use of SVM Methods with Surface-Based Cortical and Volumetric Subcortical Measurements to Detect Alzheimer’s Disease,” Journal of Alzheimers Disease, vol. 19, pp. 1263–1272, 2010. [DOI] [PubMed] [Google Scholar]
- [30].Gerardin E, Chetelat G, Chupin M, Cuingnet R, Desgranges B, Kim HS, et al. , “Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging,” Neuroimage, vol. 47, pp. 1476–1486, Oct 1 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Beekly DL, Ramos EM, Lee WW, Deitrich WD, Jacka ME, Wu J, et al. , “The National Alzheimer’s Coordinating Center (NACC) database: the uniform data set,” Alzheimer Disease & Associated Disorders, vol. 21, pp. 249–258, 2007. [DOI] [PubMed] [Google Scholar]
- [32].Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I, et al. , “Primary age-related tauopathy (PART): a common pathology associated with human aging,” Acta Neuropathologica, vol. 128, pp. 755–766, Dec 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Toledo JB, Arnold SE, Raible K, Brettschneider J, Xie SX, Grossman M, et al. , “Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer’s Coordinating Centre,” Brain, vol. 136, pp. 2697–2706, Sep 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nelson PT, Braak H, and Markesbery WR, “Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship (vol 68, pg 1, 2009),” Journal of Neuropathology and Experimental Neurology, vol. 68, pp. 339–339, Mar 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hinton GE, “Training products of experts by minimizing contrastive divergence,” Neural Computation, vol. 14, pp. 1771–1800, Aug 2002. [DOI] [PubMed] [Google Scholar]
- [36].McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al. , “The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease,” Alzheimers & Dementia, vol. 7, pp. 263–269, May 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ng A, “Sparse autoencoder,” CS294A Lecture notes, vol. 72, pp. 1–19, 2011. [Google Scholar]
- [38].Shaham U, Cheng X, Dror O, Jaffe A, Nadler B, Chang J, et al. , “A deep learning approach to unsupervised ensemble learning,” in International conference on machine learning, 2016, pp. 30–39. [Google Scholar]
- [39].Rosenberg PB, Mielke MM, Appleby BS, Oh ES, Geda YE, and Lyketsos CG, “The Association of Neuropsychiatric Symptoms in MCI with Incident Dementia and Alzheimer Disease,” American Journal of Geriatric Psychiatry, vol. 21, pp. 685–695, Jul 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bradt J, Shim M, and Goodill SW, “Dance/movement therapy for improving psychological and physical outcomes in cancer patients,” Cochrane Database Syst Rev, vol. 1, p. CD007103, Jan 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Peng CYJ, Lee KL, and Ingersoll GM, “An introduction to logistic regression analysis and reporting,” Journal of Educational Research, vol. 96, pp. 3–14, Sep-Oct 2002. [Google Scholar]
- [42].Kocev D, Vens C, Struyf J, and Dzeroski S, “Tree ensembles for predicting structured outputs,” Pattern Recognition, vol. 46, pp. 817–833, Mar 2013. [Google Scholar]
- [43].Rish I, “An empirical study of the naive Bayes classifier,” in IJCAI 2001. workshop on empirical methods in artificial intelligence, 2001, pp. 41–46. [Google Scholar]
- [44].Kuncheva LI and Whitaker CJ, “Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy,” Machine Learning, vol. 51, pp. 181–207, May 2003. [Google Scholar]
- [45].Jia F, Lei YG, Guo L, Lin J, and Xing SB, “A neural network constructed by deep learning technique and its application to intelligent fault diagnosis of machines,” Neurocomputing, vol. 272, pp. 619–628, Jan 10 2018. [Google Scholar]
- [46].Casella G. and Berger R, Statistical inference. Duxbury Press, Belmont, CA, 1990. [Google Scholar]
- [47].Napierala MA, “What is the Bonferroni correction,” AAOS Now, vol. 6, p. 40, 2012. [Google Scholar]
- [48].Lee J, Oh J, Shah SK, Yuan XH, and Tang SJ, “Automatic Classification of Digestive Organs in Wireless Capsule Endoscopy Videos,” Applied Computing 2007, Vol 1 and 2, pp. 1041–0, 2007. [Google Scholar]
- [49].Fayyad U. and Irani K, “Multi-interval discretization of continuous-valued attributes for classification learning,” 1993. [Google Scholar]
- [50].Bengio Y, Lamblin P, Popovici D, and Larochelle H, “Greedy layer-wise training of deep networks,” in Advances in neural information processing systems, 2007, pp. 153–160. [Google Scholar]
- [51].Ridgeway G, “Additive logistic regression: A statistical view of boosting - Discussion,” Annals of Statistics, vol. 28, pp. 393–400, Apr 2000. [Google Scholar]
- [52].Breiman L, “Bagging predictors,” Machine Learning, vol. 24, pp. 123–140, Aug 1996. [Google Scholar]
- [53].Liaw A. and Wiener M, “Classification and regression by randomForest,” R news, vol. 2, pp. 18–22, 2002. [Google Scholar]
- [54].Freund Y. and Schapire RE, “Experiments with a new boosting algorithm,” in icml, 1996, pp. 148–156. [Google Scholar]
- [55].Nagi S. and Bhattacharyya DK, “Classification of microarray cancer data using ensemble approach,” Network Modeling Analysis in Health Informatics and Bioinformatics, vol. 2, pp. 159–173, 2013. [Google Scholar]
- [56].Dietterich TG, “Ensemble methods in machine learning,” Multiple Classifier Systems, vol. 1857, pp. 115, 2000. [Google Scholar]