Abstract
Transforming growth factor β‐induced factor homeobox 1 (TGIF1) reportedly promotes the pathological processes of various malignant tumors. However, few studies have investigated the role of TGIF1 in gliomas. We aimed to explore the relationship between TGIF1 expression and the clinical characteristics of patients with glioma, including their overall survival. A total of thousands transcriptome datapoints were downloaded from public databases to determine the correlations between TGIF1 and various clinicopathological features using the Wilcoxon or Kruskal–Wallis tests. The Kaplan–Meier and Cox statistical methods were used to explore the prognostic significance of TGIF1. Gene set enrichment analysis (GSEA) was used to indirectly identify the pathological mechanisms modulated by TGIF1, and compounds that inhibit its expression were determined using a connectivity map (CMap). TGIF1 was significantly overexpressed in gliomas and was correlated with unfavorable prognostic factors and shorter overall survival. Cox analysis confirmed that TGIF1 expression was a significant predictor of poor prognosis in patients with glioma. GSEA revealed that the signaling pathways associated with TGIF1 expression in glioma included extracellular matrix receptor‐ and cell cycle‐modulating proteins. CMap analysis showed that the small molecules scriptaid, torasemide, dexpropranolol, ipratropium bromide, and harmine were potential negative regulators of TGIF1. Finally, in vitro experiments demonstrated that knockdown of TGIF1 significantly inhibited the proliferation and invasion of glioma cell. Taken together, our study, which is the first to comprehensively analyze TGIF1 in gliomas, revealed it to be a novel oncogene in terms of its association with this disease. As such, TGIF1 may be a potential therapeutic target for individualized treatment of patients with glioma.
Keywords: biomarker, glioma, prognosis, TGIF1
This study is the first to comprehensively analyze and reveal TGIF1, as a new oncogene, is closely related to the malignancy‐related phenotype of glioma, and that TGIF1 could be an independent risk factor for poor prognosis of glioma, which is expected to become a new target for individualized treatment of glioma patients.
1. INTRODUCTION
Glioma is a malignant tumor of the central nervous system that exhibits high recurrence and fatality rates. 1 Surgical resection followed by adjuvant chemoradiotherapy is widely prescribed by clinicians and has become the most effective treatment; moreover, novel therapies such as immunotherapy, photodynamic therapy, and transcranial magnetic stimulation adjuvant therapy continue to be developed. 2 , 3 , 4 Nevertheless, the prognoses of patients with glioma have not substantially improved; this is largely owing to the aggressive growth of the tumor and its unclear boundaries, which results in low total resection and high recurrence rates. To improve treatment outcomes, the World Health Organization (WHO) incorporated molecular criteria into the revised classification system of glioma in 2016. 5 This laid the foundation for enhancing the personalized treatment of patients with this disease, and emphasized the significance of molecular markers in their diagnosis, treatment, and prognosis.
Significant breakthroughs have been made in the basic and clinical research of glioma, and new diagnostic identifiers and treatment targets have been discovered and applied in clinical practice. Some patients who carry an IDH1 mutation and 1p/19q co‐deletion have been found to have a better prognosis 6 , 7 ; moreover, anaplastic gliomas with MGMT promoter methylations are sensitive to radiotherapy and chemotherapy, 8 , 9 while mutations in the PTEN gene are useful for evaluating the prognoses of patients with anaplastic astrocytoma. 10 Additionally, the mutation statuses of BRAF and TP53, as well as the expression of miR‐181d, have been useful for devising glioma treatment regimens and determining prognoses. 11 , 12 However, given that the outcomes of patients with this disease remain poor, screening for—and identifying—additional molecular markers of glioma is of far‐reaching significance for purposes of further understanding the biomolecular characteristics of this disease.
Transforming growth factor‐beta (TGF‐β) plays a key role in carcinogenesis 13 ; it is produced and secreted by tumor cells and promotes their invasive and metastatic potentials. 14 To date, a large number of studies found that TGF‐β‐induced factor homeobox 1 (TGIF1), a transcriptional repressor, plays an important biological role in various cancers. For example, silencing the TGIF1 gene can inhibit the pathological progression of papillary thyroid cancer. 15 Moreover, TGIF1 can significantly promote the proliferation and invasion of gastric cells by participating in the XTP8/TGIF1 signaling pathway. 16 Another study suggested that the increased expression of TGIF1 can promote the malignant progression of triple‐negative breast cancer cells and shorten the survival of patients with that disease. 17 Although Shaw et al. found that the expression of TGIF1 is downregulated in oligodendrogliomas with a 1p/19q co‐deletion, 18 no comprehensive analyses of the relationship between TGIF1 and the molecular and clinical characteristics of glioma, including the relevant cellular mechanisms, have been performed.
Hence, we performed this first study of its kind to systematically analyze TGIF1 in glioma and elucidate its biological function as well as its role in patient prognosis using a large sample. Our findings suggest that TGIF1 is a potentially novel molecular marker of glioma, and provide an additional understanding of the biomolecular characteristics of this disease, including its pathogenesis.
2. METHODS
2.1. Data collection and tissue sampling
Genomic data from thousands of gliomas were collected from the Chinese Glioma Genome Atlas (CGGA; http://www.cgga.org.cn/) database. We also downloaded RNA sequencing (RNA‐seq) and microarray data of 1018 and 301 gliomas, respectively, from the CGGA database. After excluding samples with incomplete clinical information, RNA‐seq, and microarray data from 748 to 268 gliomas, respectively, were used to investigate the role of TGIF1 in this disease. We also acquired data pertaining to 698 glioma tissues from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/), which is a multi‐tumor cancer gene‐mapping program; we analyzed 653 of these samples for which complete clinical information was available to validate our findings using the CGGA.
Additionally, we analyzed the expression of TGIF1 in 163 glioblastoma multiforme samples, 518 low‐grade gliomas, and 207 normal brain tissues from the Gene Expression Profiling Interactive Analysis (GEPIA2: http://gepia2.cancer‐pku.cn/), a large public online analysis tool that contains a large amount of gene expression data from tumor and corresponding normal tissues. We also used Oncomine (https://www.oncomine.org/resource/), a database for differential gene analysis that collects a large amount of tumor‐related microarray data, as a validation dataset for the differential expression of TGIF1 in gliomas. Moreover, the microarray gene expression databases GSE4290 and GSE50161, which together contain 111 glioma and 36 normal brain tissues, were downloaded from the Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) database. All supported data we used can be reached on the GitHub page: https://github.com/Ligang111/2022‐03‐24.
2.2. Tissues and cell lines
Twenty‐three glioma and 10 paracancerous brain tissue samples were collected via surgical resection performed at Henan Provincial People's Hospital and immediately stored in liquid nitrogen. Histopathological diagnoses were independently assessed by two physicians, who provided consistent results. The samples used in this study were approved by the hospital ethics committee, and each participant signed an informed consent form before surgery. Human astrocytes (HA) and the glioma cell lines LN229, A172, and U251 were purchased from GenePharma (Suzhou, China). Cells were cultured in Dulbecco's Modified Eagle's Medium with 10% fetal bovine serum and 1% penicillin and were cultured in a sterile incubator containing 5% CO2 at 37°C.
2.3. Real‐time quantitative polymerase chain reaction
Total RNA from tissue samples and cells was extracted using Trizol (ThermoFisher Scientific), and the quality and quantity of total RNA were determined using Novoscript plus (catalog #E047; Novoprotein Scientific Inc.). Finally, we used 2× RealStar Green (catalog #A303‐05; GenStar) to conduct Real‐time quantitative polymerase chain reaction (RT‐qPCR). GAPDH was used as a housekeeping gene (forward: 5‐AAGAAGGTGGTGAAGCAGG‐3; reverse: 5‐GTCAAAGGTGG AGGAGTGG‐3). The primer sequences for TGIF1 were forward: 5′‐GGATTGGCTGTATG AGCACCGT‐3; reverse: 5‐GCCATCCTTTCTCAGCATGT CAG‐3′.
2.4. Gene set enrichment analysis of TGIF1
After downloading CGGA RNA‐seq, CGGA microarray, and TCGA RNA‐seq data from their respective databases, the downloaded data were batch‐corrected and normalized using the ‘limma’ package of the R statistical software. Data from all three datasets were divided into two groups according to the median expression level of TGIF1. Gene set enrichment analysis (GSEA; version 4.0) was used to complete the functional enrichment analysis of TGIF1; after the data were uploaded, Kyoto Encyclopedia of Genes and Genomes cell signaling pathways were selected as the enrichment target path to analyze the three datasets. All enrichment analysis results were considered significantly enriched if p‐values were <0.05 and q‐values were <0.25.
2.5. Connectivity map analysis
Connectivity map (CMap; https://clue.io/) is a well‐known database in the field of pharmacogenomics. Based on CGGA RNA‐seq data, we used Pearson correlation analysis to identify genes that were associated with TGIF1. Two lists of genes, one positive and the other negative correlations with TGIF1, were uploaded to the CMap database; parameters with p < 0.01 and enrichment <−0.8 were used to identify associated small molecules.
2.6. Cell transfection
Based on the expression level of TGIF1 in the cell lines, of which high expressed glioblastoma‐derived LN229 was used for in vitro experiments. The siRNA targeted on TGIF1 was purchased from GenePharma. LN229 cells transfected with negative control siRNA (S: UUCUCCGAACGUGUCACGUTT, AS: ACGUGACACGUUCGGAGAATT) were set as the control group (siNC), and LN229 transfected with siRNA targeting TGIF1 (S: GGAUGGCAAAGAUCCAAAUTT, AS: AUUUGGAUCUUUGCCAUCCTT) were set as the experimental group (siTGIF1). Then the mRNA level of TGIF1 in both groups was tested by RT‐qPCR.
2.7. Cell proliferation assay
To test the effect of knocking down TGIF1 on the proliferative capacity of glioma cells, the expression levels of the proliferation‐associated antigen Ki67 in different subgroups were first verified. After LN229 transfection of siRNA for 36 h, the cells were fixed and permeabilized and then blocked using 10% serum, followed by incubation using Ki67‐specific antibodies (Abcam, 1:200). After incubation with the specific secondary antibody, DAPI staining was performed. Furthermore, transfected LN229 cells were planted in 96‐well plates at a density of 2000 cells per well, and CCK8 solution was added at the set detection time points, and the absorbance value at 450 nm was detected after continued incubation for 4 h. Meanwhile, transfected LN229 cells were planted in a six‐well plate at a density of 500 cells per well. After continuing the culture for 14 days, the cell colonies were stained using crystal violet solution.
2.8. Cell invasion assay
Wound healing assay and Transwell assay were used to examine the effect of TGIF1 on the invasive ability of glioma cells. After the full growth of transfected cells in the six‐well plate, a wound was made using a sterile pipette tip, and the culture was continued using serum‐free DMEM medium after PBS washing, and relative distances of the wound at the same location were photographed at the set time points. In addition, a certain amount of transfected cells were added to Transwell chambers and cultured using 5% serum medium. The lower chamber was added to the medium with 20% serum and continued to incubate for 48 h, and then the cells invading to the lower wall of Transwell chambers were stained using crystal violet solution.
2.9. Meta‐analysis of TGIF1
In order to collect more evidence to verify the impact of TGIF1 on the prognosis of glioma patients, the study collected seven datasets including 1914 glioma patients (TCGA RNA‐seq: 653; CGGA microarray: 268; CGGA RNA‐seq: 748; GSE43378: 50; GSE4412: 85; GSE74187: 60; GSE83300: 50). These data were used for a meta‐analysis to verify the effect of TGIF1 on the prognosis of glioma. First of all, the above seven datasets were subjected to a Cox analysis one by one to verify the impact of TGIF1 on the prognosis of glioma patients. The Q‐test was used to verify the heterogeneity among the seven datasets, and the random‐effect model was used to complete the meta‐analysis based on I 2 = 92% and p < 0.01.
2.10. Statistical analysis
Statistical analyses were performed using the R software (version 3.6.3). The relationships between patient characteristics and TGIF1 expression were evaluated using the Wilcoxon or Kruskal–Wallis tests. Kaplan–Meier and Cox regression analyses were used to analyze the effect of TGIF1 overexpression on the survival of patients with glioma. The experimental data were analyzed by one‐way ANOVA, and all statistical analyses were processed by SPSS 22.0. Two‐sided p‐values <0.05 were considered statistically significant.
3. RESULTS
3.1. TGIF1 was significantly increased in gliomas
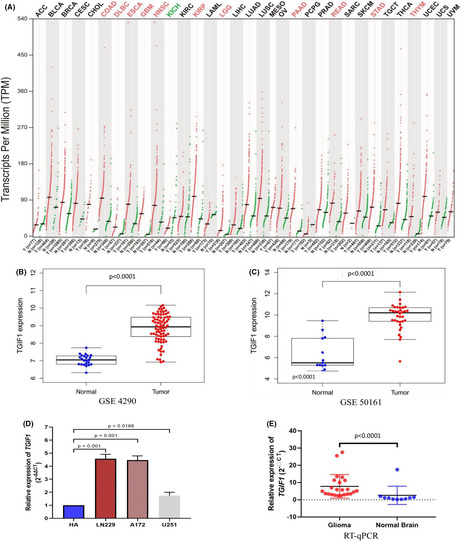
We first analyzed the expression of TGIF1 in tumors using the GEPIA and Oncomine databases and found that this gene's expression was higher in central nervous system tumors than in normal brain tissues in both databases (Figure 1A and Figure S1). Further exploration of different GEO and Oncomine datasets revealed that TGIF1 was significantly overexpressed in gliomas compared to normal tissue (Figure 1B,C and Figure S1). RT‐qPCR confirmed that the mRNA expression levels of TGIF1 in glioma cell lines and tissues were markedly increased over controls (Figure 1D,E). These results laid the foundation for further studying the role of TGIF1 in glioma.
FIGURE 1.
TGIF1 is highly expressed in gliomas. (A) Analysis of the GEPIA database revealed that TGIF1 is expressed in various tumors. Red represents an increased expression of TGIF1 relative to the corresponding control, while green represents decreased TGIF1 expression. (B) TGIF1 expression in normal brain tissues and glioma tissues according to the GSE4290 dataset. (C) TGIF1 expression in normal brain tissues and glioma tissues according to the GSE50161 dataset. (D) The expression levels of TGIF1 mRNA in human astrocytes and glioblastoma cell line LN229, A172, and U251. (E) TGIF1 expression in normal brain and glioma tissues collected from clinical patients. p value less than 0.05 is statistically significant
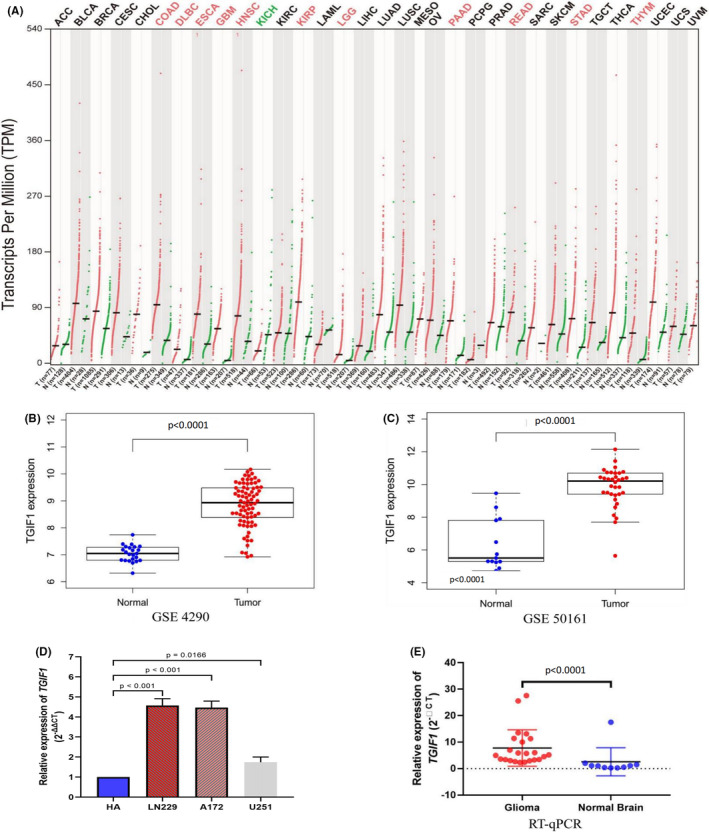
3.2. TGIF1 is correlated with the clinical characteristics of glioma
TCGA‐RNA‐seq, CGGA‐RNA‐seq, and CGGA‐microarray data were grouped according to the different grades of gliomas. As shown in Figure 2A, the expression level of TGIF1 was highest in WHO grade IV gliomas and was lower in WHO grade III tumors and lowest in WHO grade II gliomas. Additionally, recurrent gliomas showed higher expression levels of TGIF1 than primary gliomas (Figure 2G); this phenomenon was observed in several histological subtypes (Figure 2C). These findings suggest that TGIF1 may be involved in the malignant progression of glioma. We also found that the expression levels of TGIF1 in elderly patients, patients with IDH wildtype, and patients with 1p/19q non‐codeletion were higher than in the corresponding control groups (Figure 2B,D,E). IDH mutation and 1p/19q co‐deletion are known to be favorable prognostic indicators in patients with glioma. In contrast, our data indicated that TGIF1 promotes the malignant progression of gliomas and is associated with poor prognosis.
FIGURE 2.
Correlation between TGIF1 expression and molecular/clinical features associated with glioma. (A) TGIF1 expression in gliomas of various World Health Organization grades. (B) The relationship between TGIF1 expression and age in patients with glioma. (C) TGIF1 expression in gliomas of different histological subtypes. (D) The relationship between TGIF1 expression and IDH mutation status. (E) The relationship between TGIF1 expression and 1p/19q co‐expression status. (F) Differential analysis of TGIF1 expression in gliomas treated with chemotherapy after surgery. (G) TGIF1 expression in primary, recurrent, and secondary gliomas. p value less than 0.05 is statistically significant. Chemo, chemotherapy; IDH, isocitrate dehydrogenase; PRS, primary‐recurrent‐secondary; WHO, World Health Organization
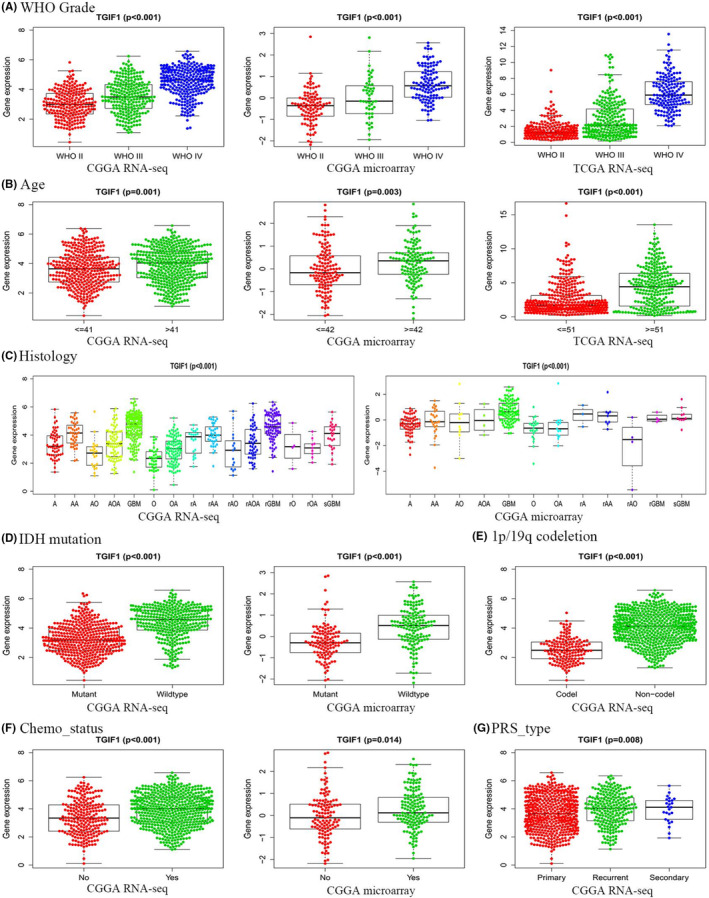
3.3. Effect of high expression of TGIF1 on overall survival and diagnostic value of prognosis of glioma patients
To understand the role of TGIF1 expression in the survival of patients with glioma, we divided the samples derived from the aforementioned databases into high‐ and low‐expression groups based on the median expression level of TGIF1 in the samples and used the Kaplan–Meier method to explore the relationship between this gene's expression and patient survival. When considering all grades of gliomas collectively, higher expression levels of TGIF1 correlated with shorter overall survival (Figure 3A–C). We then divided the patients into high‐grade (III and IV) and low‐grade (II) gliomas to evaluate the effect of TGIF1 on their prognoses; a favorable effect of low TGIF1 expression on the prognosis of patients with low‐grade glioma was only observed when analyzing the RNA‐seq data from the CGGA and TCGA (Figure S2A–C). However, three different datasets consistently showed that high expression of TGIF1 was associated with a reduced overall survival time among patients with high‐grade glioma (Figure S2D–F).
FIGURE 3.
The relationship between TGIF1 expression and overall survival of patients with glioma. (A–C) Correlations between TGIF1 expression levels and overall survival of glioma patients based on the CGGA RNA‐seq dataset, CGGA microarray dataset, and TCGA RNA‐seq dataset, respectively. (D–F) Receiver operating characteristic curves based on the CGGA RNA‐seq, CGGA microarray, and TCGA RNA‐seq datasets. p < 0.05 is statistically significant, AUC > 0.7 is considered credible
Given the influence of IDH mutations on prognosis, we also investigated the impact of TGIF1 on the prognosis of patients with varying IDH mutation statuses. According to the CGGA data, the high expression of TGIF1 significantly shortened the overall survival of patients with IDH mutations regardless of glioma grade and 1p19q codeletion (Figure S2H–I and K). Finally, the high expression of TGIF1 did affect the prognoses of patients of any glioma grade who had wildtype IDH regardless of 1p19q codeletion (Figure S2G and J).
The area under the receiver operating characteristic curve suggested that TGIF1 expression carries a high sensitivity for predicting the prognoses of these patients (Figure 3D–F). Additionally, according to the grouping of all subtypes in survival analysis, TGIF1 in all subtypes has diagnostic value for the prognosis of patients (Figure S3). The area under the curve (AUC) was greater than 0.7 in multiple points in time. Therefore, it can be concluded that the increased expression of TGIF1 has a significant impact on the overall survival of patients with gliomas and has diagnostic value, especially in high‐grade glioma.
3.4. TGIF1 is an independent predictor of poor prognosis in patients with glioma
To determine whether TGIF1 expression is an independent predictor of patient survival, we established Cox proportional hazards models based on the different datasets. Univariate and multivariate analyses suggested that TGIF1 expression is an independent negative prognostic risk factor for patients with all grades of gliomas (hazard ratio [HR] >1) (Figure 4). We then divided the patients into high‐grade (III and IV) and low‐grade (II) gliomas to evaluate the risk of TGIF1 on the prognosis of patients; the expression of TGIF1 has a significant risk for the prognosis of patients with low‐grade glioma was only observed when analyzing the RNA‐seq data from the CGGA and TCGA (HR >1) (Figures S4A–C and S5A–C). However, three different datasets consistently showed that the expression of TGIF1 has a significant risk for the prognosis of patients with high‐grade glioma (HR>1) (Figures S4D–F and S5D–F). More importantly, no matter which group TGIF1 is in, it shows that the high expression of TGIF1 is a risk to the prognosis of glioma patients according to the molecular subtypes of gliomas (HR >1) (Figures S4G–K and S5G–K). Mutual validation among the above databases confirmed that TGIF1 was an independent risk factor for the prognosis of glioma patients, especially high‐grade glioma.
FIGURE 4.
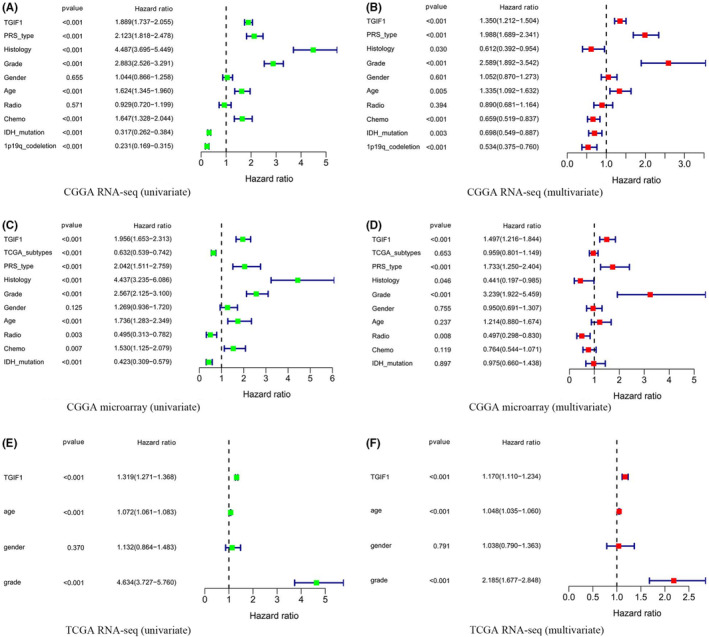
Prognostic value of various factors of patients with glioma. (A, B) Univariate and multivariate analyses based on the CGGA RNA‐seq dataset. (C, D) Univariate and multivariate analyses based on the CGGA microarray dataset. (E, F) Univariate and multivariate analyses based on TCGA RNA‐seq dataset. The range of hazard ratio less than 1 is considered as a protective factor; The range of HR more than 1 is considered as a risk factor. p < 0.05 considered statistically significant. PRS, primary‐recurrent‐secondary; radio, Radiotherapy; chemo, Chemotherapy; IDH, isocitrate dehydrogenase
3.5. Biological function of TGIF1 in glioma
Given our evidence that TGIF1 may be involved in the pathological processes of glioma, we further explored the cellular mechanisms potentially involved using GSEA. Several signaling pathways involved in cancer progression were identified, including those related to actin cytoskeleton rearrangement, extracellular matrix (ECM)‐receptor interaction, cell cycle regulation, and focal adhesion formation (Figure 5, Table 1).
FIGURE 5.
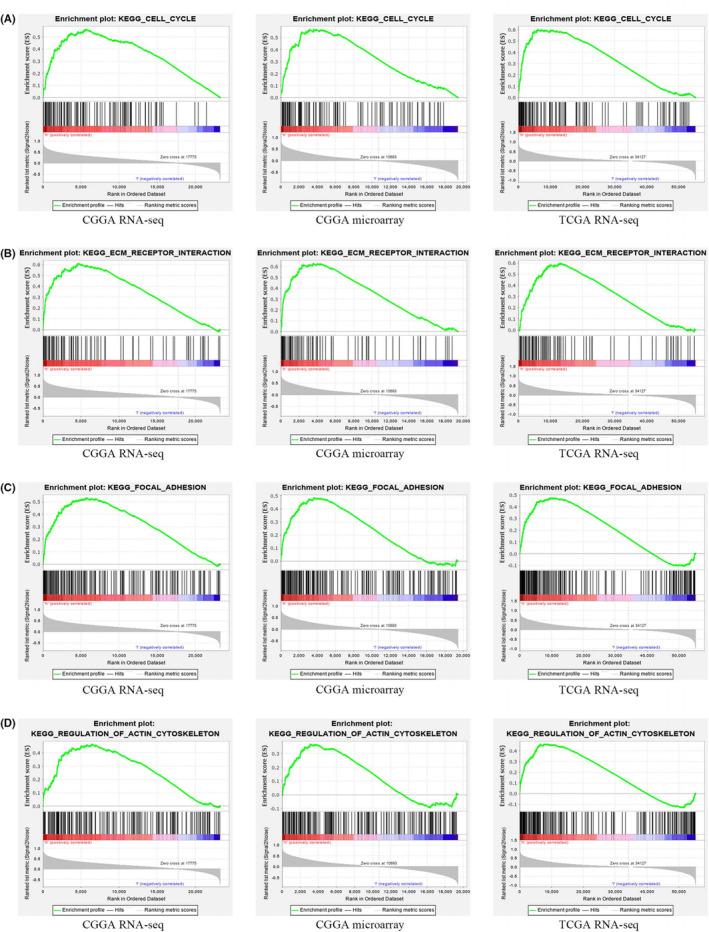
The enrichment of TGIF1 gene in signaling pathways based on gene set enrichment analyses of the CGGA RNA‐seq, CGGA microarray, and TCGA RNA‐seq datasets. (A) Enrichment in cell cycle signaling pathway; (B) Enrichment in extracellular matrix receptor interaction signaling pathway. (C) Enrichment in focal adhesion signaling pathway. (D) Enrichment in actin cytoskeleton signaling regulatory pathway. p < 0.05 and FDR q‐value <0.25 were considered as significantly enriched. See Table 1 for more details
TABLE 1.
The gene set enriches the high TGIF1 in three databases
| Gene set name | CGGA RNA‐seq | CGGA microarray | TCGA RNA‐seq | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NES | NOM p‐val | FDR q‐val | NES | NOM p‐val | FDR q‐val | NES | NOM p‐val | FDR q‐val | |
| Regulation of actin cytoskeleton | 1.72 | 0.00 | 0.14 | 1.52 | 0.04 | 0.14 | 1.77 | 0.01 | 0.04 |
| ECM receptor interaction | 1.86 | 0.00 | 0.11 | 1.78 | 0.01 | 0.08 | 1.86 | 0.00 | 0.04 |
| Cell cycle | 1.68 | 0.04 | 0.10 | 1.73 | 0.04 | 0.08 | 1.86 | 0.01 | 0.04 |
| Focal adhesion | 1.88 | 0.00 | 0.13 | 1.76 | 0.02 | 0.09 | 1.67 | 0.01 | 0.06 |
Note: Gene sets with NOM p < 0.05 and FDR q‐value <0.25 were considered as significantly enriched.
Abbreviations: FDR: false discovery rate; NES, normalized enrichment score; NOM, nominal.
3.6. Analysis of genes co‐expressed with TGIF1
Owing to the complexity of the mechanisms governing the malignant progression of glioma, other factors may be involved in its development. Pearson's correlation analysis was used to analyze the CGGA RNA‐seq data to identify other genes associated with TGIF1 in glioma tissues. Circle plots revealed the interconnections among the 20 genes most related to TGIF1 (Figure 6A,B); VIM, PDIA5, HDAC1, CASP6, TNFRSF12A, GPX7, PDIA4, ANXA1, SERPINH1, and SYDE1 were positively correlated with TGIF1 (Figure 6C–E and Figure S6A–G), while HRH3, SCN3B, CHGA, NSG2, RP1‐293 L6.1, REPS2, CPLX2, SVOP, RP5‐1119A7.17, and AMER3 were negatively correlated (Figure 6B,F–H and Figure S6H–N). These results further suggest that TGIF1 does not exert its tumorigenic effects in gliomas via a single signaling pathway; rather, they highlight the multifaceted pathways of this process.
FIGURE 6.
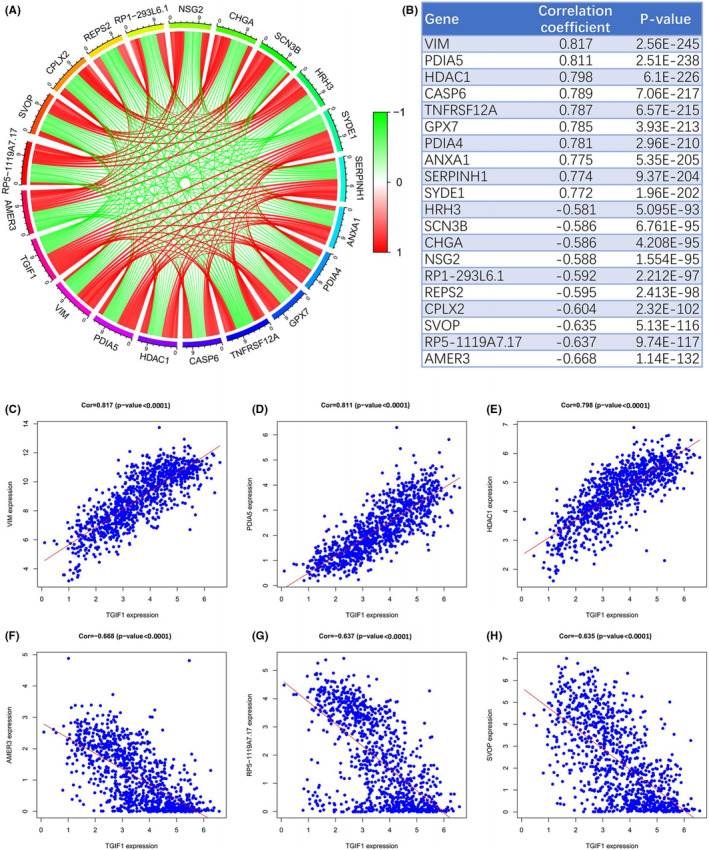
Analyses of TGIF1 co‐expression with other genes. (A) Circle plot shows 10 upregulated and 10 downregulated genes whose expression levels were significantly associated with TGIF1. (B) Enrichment parameters of the 20 genes whose expression levels are significantly associated with TGIF1. (C–E) Correlation analysis between VIM, PDIA5, HDAC1, and TGIF1. (F–H) Correlation analysis between AMER3, RP5‐1119A7.17, SVOP, and TGIF1. p < 0.05 is statistically significant
3.7. Identification of small molecules targeting TGIF1
We identified five small molecule compounds with potential value in treating glioma using the CMap online analysis tool; their corresponding parameters are shown in Table 2. Moreover, their two‐ and three‐dimensional structures were determined using the PubChem online tool (https://pubchem.ncbi.nlm.nih.gov) (Figure 7).
TABLE 2.
Small molecule drugs targeting TGIF1 against glioma
| CMap name | Enrichment | p value |
|---|---|---|
| Scriptaid | −0.920 | 0.0009 |
| Torasemide | −0.903 | 0.0002 |
| Dexpropranolol | −0.865 | 0.0050 |
| Ipratropium bromide | −0.846 | 0.0073 |
| Harmine | −0.828 | 0.0017 |
FIGURE 7.
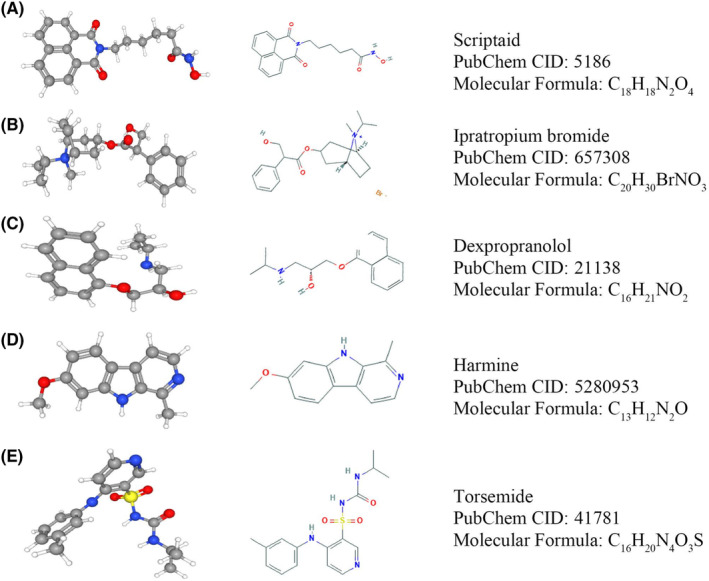
Two‐ and three‐dimensional structures of anti‐glioma small molecule drugs targeting TGIF1. (A) Scriptaid, (B) Ipratropium bromide, (C) Dexpropranolol, (D) Harmine, and (E) Torasemide
3.8. To verify the role of TGIF1 as a pathogenic molecule in the prognosis of glioma
In the cells of the experimental group, the expression level of TGIF1 was significantly less than that of the control group (p < 0.0001) (Figure 8A). The proportion of Ki67‐stained positive cells was significantly lower in the siTGIF1 group than that in the siNC group (p < 0.01) (Figure 8B). Besides, CCK8 assay showed that the cell viability of LN229 cells in the experimental group was lower than that of the control group at 24 h (p < 0.05), 48 h (p < 0.05), 72 h (p < 0.01), and 96 h (p < 0.01) (Figure 8C). Meanwhile, the number of cell colonies formed in the experimental group was significantly less than that in the control group (p < 0.01) (Figure 8D). Furthermore, compared to the control group, the invasive ability of LN229 cells in the experimental group was significantly reduced as confirmed by wound healing assay (p < 0.01) (Figure 8E) and Transwell assay (p < 0.0001) (Figure 8F). Therefore, it can be confirmed that the high expression of TGIF1 can promote the malignant behavior of glioma cells. in order to try to find more evidence to confirm that TGIF1 is a pathogenic molecule for the prognosis of patients with glioma, the study used a meta‐analysis to find that each data showed that TGIF1 was a risk for the prognosis of patients in seven independent datasets and the pooled HR for the association between TGIF1 expression and OS in patients was 1.61 (1.32–1.96). Thus, TGIF1 is a reliable pathogenic factor for glioma (Figure S7).
FIGURE 8.
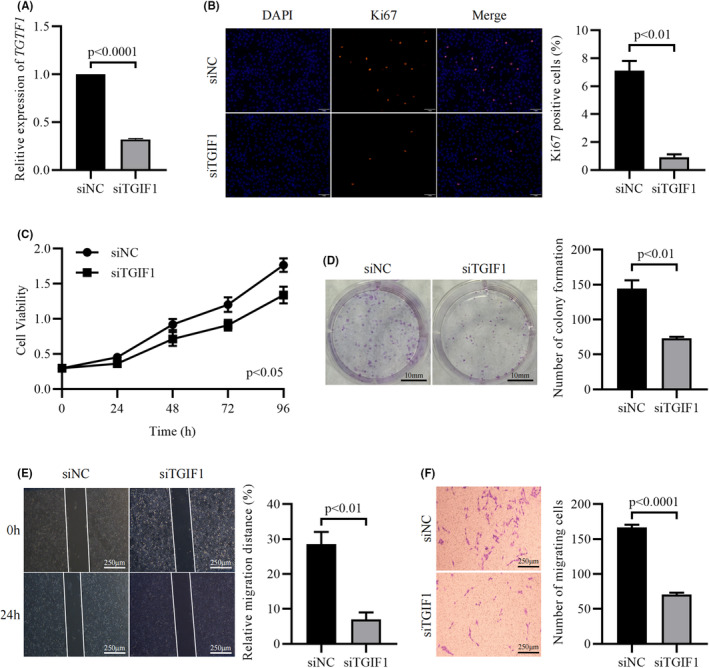
Knockdown of TGIF1 significantly inhibits the proliferation and invasion of LN229 cells. (A) The expression of TGIF1 in both siNC and siTGIF1 group. (B) Staining of Ki67 positive cells in both groups. (C) Cell viability of LN229 cells in siNC and siTGIF1 group at different time points. (D) Staining of cell colonies in both groups. (E) Relative distances of the wound healing assay. (F) Staining of invasive LN229 cells in the Transwell assay. p < 0.05 is statistically significant
4. DISCUSSION
A large number of studies have found that TGIF1 plays a critical biological function in a variety of cancers and that its expression is associated with the enhanced invasion, migration, and metastasis of tumor cells. However, systematic studies of TGIF1 in gliomas have not been performed to date. In our present analysis of glioma samples using public databases, we found an association between TGIF1 expression levels and unfavorable clinical features. We also clarified biological mechanisms of TGIF1 involvement in glioma using GSEA.
Using the online GEPIA tool, we discovered that the expression levels of TGIF1 in numerous tumor tissues such as glioblastoma multiforme and low‐grade glioma were higher than those in the matched normal tissues; this was also verified using two GEO datasets (GSE4290 and GSE50161). Previous studies have shown that elevated TGIF1 in triple‐negative breast cancer can promote malignant progression of the tumor and shorten the patients' survival. 17 Our analysis of the CGGA RNA‐seq data showed that increased expression of TGIF1 can shorten the survival of patients with glioma regardless of disease grade; this was verified when examining the CGGA microarray and TCGA RNA‐seq data. WHO grade IV gliomas are the most malignant and are associated with shorter median and overall survival times, and patients with gliomas carrying IDH mutations and 1p/19q co‐deletions achieve better survival than do those with IDH wildtype and no 1p/19q co‐deletions. 6 , 7 To investigate the impact of TGIF1 expression on the survival of patients with glioma, we analyzed the correlation between TGIF1 mRNA levels and clinical features associated with glioma prognosis. TGIF1 mRNA was markedly elevated in highly malignant gliomas compared to low‐grade counterparts; moreover, the expression of TGIF1 in recurrent gliomas was higher than that in primary gliomas. We also found that the expression level of TGIF1 in IDH‐mutated and 1p/19q co‐deleted tissues were lower than that in the matched control group. These results suggest that TGIF1, which is increasingly expressed commensurate with higher stage disease, may play an active role in glioma tumorigenesis and progression while being a negative prognostic factor. Our Cox analysis further revealed that TGIF1 is an independent predictor of the prognosis of patients with glioma.
GSEA revealed that actin cytoskeleton regulation, ECM receptor interaction, cell cycle regulation, and focal adhesion signaling pathways (all of which play important roles in tumor proliferation, migration, and invasion) were remarkably enriched in patient samples with higher TGIF1 mRNA; this was validated in different datasets. Previous studies have found that actin cytoskeleton regulation is critical for the proliferation and invasion of glioma cells. 19 Jiang et al. also found that cellular ECM receptors regulate glioma invasion and metastasis using transcriptome integration analysis, 20 while other investigators found that cell cycle protein alterations are involved in glioma cell proliferation and apoptosis 21 , 22 and that the dysregulation of focal adhesion assembly can promote cancer cell metastasis resistance to chemoradiotherapy. 23 , 24 As such, TGIF1 (as a transcriptional regulator) appears to have a multifaceted role in the proliferation, differentiation, metastasis, invasion, and apoptosis of glioma cells, thereby illustrating the complexity of glioma tumorigenesis.
We also found that there are many other genes co‐expressed with TGIF1 that may contribute to the malignant progression of glioma. For example, protein disulfide isomerase family A member 4, which is encoded by a gene (PDIA4) whose expression is directly correlated with TGIF1, can disrupt the DNA repair mechanism, thereby promoting the proliferation and metastasis of cancer cells. 25 , 26 ‘RALBP1‐associated Eps domain containing 2’, whose encoding gene REPS2’s expression levels are inversely correlated with those of TGIF1, blocks the proliferation and migration of tumor cells and inhibits their apoptosis, and is therefore an indicator of favorable prognosis as related to prostate, breast, and esophageal cancers. 27 , 28 , 29 These findings strongly indicate that TGIF1 is an oncogene that promotes tumor progression and shortens patient survival by participating in different tumorigenic biological processes. Moreover, our results provide a rationale for developing therapeutic agents that target TGIF1.
The CMap online analysis tool identified five small molecules that are potentially potent against TGIF1. The anticancer properties of these drugs have already been demonstrated in previous studies. For example, scriptaid, which is an inhibitor of histone deacetylase, can induce apoptosis of glioma cells by activating Jun N‐terminal kinase. 30 , 31 Dexpropranolol is a well‐characterized blocker of non‐selective beta‐adrenergic receptors, and several studies have shown that it exhibits anti‐tumor properties in gastric, prostate, and breast cancers. 32 , 33 , 34 Harmine, a natural plant extract that has been shown to have anticancer characteristics in numerous tumor types, depletes stem‐like cells in glioblastoma by inhibiting Akt phosphorylation. 35 , 36 , 37 The repurposing of traditional medicines continues to gain attention, and some previously released drugs have demonstrated good clinical efficacy in the treatment of new diseases. For example, aspirin, which is widely prescribed to treat antipyretic analgesia, has been used to prevent transient cerebral ischemia attacks and myocardial infarction because of its inhibitory effect on platelet aggregation. 38 Moreover, the lipid‐lowering agent atorvastatin has recently been used for the conservative treatment of chronic subdural hemorrhage. 39 The global outbreak of the novel coronavirus (2019‐nCoV) in 2019 brought about a catastrophic pandemic owing to the lack of drugs or vaccines; however, the fact that old drugs such as ribavirin, chloroquine, and hydroxychloroquine have played important roles in saving patients' lives illustrates the importance of novel uses of traditional medicines for combating diseases. 40 , 41 , 42
Furthermore, this study confirmed the effect of TGIF1 on glioblastoma cells through in vitro experiments. As a hallmark feature of malignant tumors, unlimited malignant proliferation is a fundamental factor in the rapid development of glioma and tumor recurrence. 43 Many of the researches on glioma have focused on the proliferative capacity of tumor cells, such as overexpression of miR‐129‐5p can inhibit proliferation of LN229 cells and blocked the cell cycle in G1 Phase. 44 In the present study, it is similarly demonstrated that TGIF1 can significantly affect the proliferative capacity of gliomas, suggesting that TGIF1 serves as a clearly therapeutic target to inhibit glioma proliferation. In addition, the fact that tumors invade into normal tissues and cause indistinct tumor margins, thus inducing tumor recurrence recognizes invasion as another feature of malignant glioma. 45 It is reported that miR‐128 can significantly decrease the stability of COX‐2 mRNA and inhibit proliferation and invasion of LN229 cells. 46 Also, neural‐specific ECM molecule BEHAB/Brevican affects the invasion of glioma cells through the mediation of the extracellular matrix. 47 The present study also confirmed that TGIF1 can significantly affect the invasive ability of glioma by wound healing assay and transwell assay. At this point, the critical role of TGIF1 in the malignant progression of glioma has been clearly demonstrated. Combined with the above bioinformatics results, a conclusion is reached that TGIF1 can serve as a diagnostic and therapeutic target for glioma.
There were some limitations to this study. First, because various datasets have different categories of clinical information of patients, the clinical features of some patients with glioma could not be fully verified. These included a lack of data on the original recurrence status of tumors in the CGGA microarray and TCGA datasets, as well as incomplete data on glioma tissue subtypes in TCGA datasets. Additionally, the lack of information on gene expression profiles after chemoradiotherapy precluded further studies of the effect of chemoradiotherapy on TGIF1 expression. However, the correlation observed between chemotherapy and TGIF1 expression in this study further illustrated that tissues of greater malignant potentials have higher levels of TGIF1. Patients who receive additional chemotherapy owing to residual postoperative pathological findings generally have tumors with a higher degree of malignancy.
5. CONCLUSION
We found that the high expression of TGIF1 in gliomas is closely correlated with clinical characteristics associated with poor prognosis. Moreover, TGIF1 was found to be an independent predictor of patient prognosis. Finally, knockdown of TGIF1 significantly inhibits the proliferation and invasion of glioma cell. Therefore, TGIF1 appears to be a novel prognostic indicator and therapeutic target in patients with gliomas.
CONFLICT OF INTEREST
All authors have no conflict of interest in the ownership of the study.
AUTHOR CONTRIBUTIONS
Gang Li, Zhendong Liu, and Baoya Wang conceived and designed the experiment. Qiong Ma and Xuelin Wang analyzed and checked the data, Zhendong Liu, Baoya Wang, and Kunshan Guo wrote the manuscript. The final manuscript has been approved by the authors.
ETHICAL APPROVAL
The study protocol was approved by the Ethics Committee of Henan Provincial People‘s Hospital (Zhengzhou, China).
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
ACKNOWLEDGMENTS
Thanks to the staff of this study.
Wang B, Ma Q, Wang X, Guo K, Liu Z, Li G. TGIF1 overexpression promotes glioma progression and worsens patient prognosis. Cancer Med. 2022;11:5113‐5128. doi: 10.1002/cam4.4822
Contributor Information
Zhendong Liu, Email: superliuyisheng@outlook.com.
Gang Li, Email: lglabmed@hotmail.com.
DATA AVAILABILITY STATEMENT
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Davis M. Epidemiology and overview of gliomas. Semin Oncol Nurs. 2018;34(5):420‐429. [DOI] [PubMed] [Google Scholar]
- 2. Brown C, Alizadeh D, Starr R, et al. Regression of glioblastoma after chimeric antigen receptor T‐cell therapy. N Engl J Med. 2016;375(26):2561‐2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu J, Yu S, Wang X, et al. High affinity of chlorin e6 to immunoglobulin G for intraoperative fluorescence image‐guided cancer photodynamic and checkpoint blockade therapy. ACS Nano. 2019;13(9):10242‐10260. [DOI] [PubMed] [Google Scholar]
- 4. Diehl C, Schwendner M, Sollmann N, et al. Application of presurgical navigated transcranial magnetic stimulation motor mapping for adjuvant radiotherapy planning in patients with high‐grade gliomas. Radiother Oncol. 2019;138:30‐37. [DOI] [PubMed] [Google Scholar]
- 5. Louis D, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803‐820. [DOI] [PubMed] [Google Scholar]
- 6. Morshed R, Young J, Hervey‐Jumper S, Berger M. The management of low‐grade gliomas in adults. J Neurosurg Sci. 2019;63(4):450‐457. [DOI] [PubMed] [Google Scholar]
- 7. Dimitrov L, Hong C, Yang C, Zhuang Z, Heiss J. New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma. Int J Med Sci. 2015;12(3):201‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Rhun E, Taillibert S, Chamberlain M. Anaplastic glioma: current treatment and management. Expert Rev Neurother. 2015;15(6):601‐620. [DOI] [PubMed] [Google Scholar]
- 9. Binabaj M, Bahrami A, ShahidSales S, et al. The prognostic value of MGMT promoter methylation in glioblastoma: a meta‐analysis of clinical trials. J Cell Physiol. 2018;233(1):378‐386. [DOI] [PubMed] [Google Scholar]
- 10. Smith J, Tachibana I, Passe S, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93(16):1246‐1256. [DOI] [PubMed] [Google Scholar]
- 11. Ludwig K, Kornblum H. Molecular markers in glioma. J Neurooncol. 2017;134(3):505‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang W, Zhang J, Hoadley K, et al. miR‐181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14(6):712‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derynck R, Akhurst R, Balmain A. TGF‐beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117‐129. [DOI] [PubMed] [Google Scholar]
- 14. Batlle E, Massagué J. Transforming growth factor‐β signaling in immunity and cancer. Immunity. 2019;50(4):924‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pu Y, Xiang J, Zhang J. KDM5B‐mediated microRNA‐448 up‐regulation restrains papillary thyroid cancer cell progression and slows down tumor growth via TGIF1 repression. Life Sci. 2020;250:117519. [DOI] [PubMed] [Google Scholar]
- 16. Jia K, Wen Q, Zhao X, Cheng J, Cheng L, Xi M. XTP8 stimulates migration and invasion of gastric carcinoma through interacting with TGIF1 . Eur Rev Med Pharmacol Sci. 2020;24(5):2412‐2420. [DOI] [PubMed] [Google Scholar]
- 17. Zhang M, Ferrigno O, Wang Z, et al. TGIF governs a feed‐forward network that empowers Wnt signaling to drive mammary tumorigenesis. Cancer Cell. 2015;27(4):547‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaw E, Haylock B, Husband D, et al. Gene expression in oligodendroglial tumors. Cell Oncol (Dordr). 2011;34(4):355‐367. [DOI] [PubMed] [Google Scholar]
- 19. Hua S, Li H, Liu Y, Zhang J, Cheng Y, Dai C. High expression of GALNT7 promotes invasion and proliferation of glioma cells. Oncol Lett. 2018;16(5):6307‐6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang Y, He J, Guo Y, Tao H, Pu F, Li Y. Identification of genes related to low‐grade glioma progression and prognosis based on integrated transcriptome analysis. J Cell Biochem. 2020;121:3099‐3111. [DOI] [PubMed] [Google Scholar]
- 21. Wu W, Chien C, Liu K, Chen Y, Chiu W. Evodiamine prevents glioma growth, induces glioblastoma cell apoptosis and cell cycle arrest through JNK activation. Am J Chin Med. 2017;45(4):879‐899. [DOI] [PubMed] [Google Scholar]
- 22. Song D, Liang H, Qu B, et al. Moxidectin inhibits glioma cell viability by inducing G0/G1 cell cycle arrest and apoptosis. Oncol Rep. 2018;40(3):1348‐1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gu C, Wang X, Long T, et al. FSTL1 interacts with VIM and promotes colorectal cancer metastasis via activating the focal adhesion signalling pathway. Cell Death Dis. 2018;9(6):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65‐75. [DOI] [PubMed] [Google Scholar]
- 25. Kuo T, Chen T, Jiang S, et al. Protein disulfide isomerase a4 acts as a novel regulator of cancer growth through the procaspase pathway. Oncogene. 2017;36(39):5484‐5496. [DOI] [PubMed] [Google Scholar]
- 26. Wang Z, Zhang H, Cheng Q. PDIA4: The basic characteristics, functions and its potential connection with cancer. Biomed Pharmacother. 2020;122:109688. [DOI] [PubMed] [Google Scholar]
- 27. Zhang H, Duan C, Zhang H, Cheng Y, Zhang C. Expression and clinical significance of REPS2 in human esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2013;14(5):2851‐2857. [DOI] [PubMed] [Google Scholar]
- 28. Oosterhoff J, Kühne L, Grootegoed J, Blok L. EGF signalling in prostate cancer cell lines is inhibited by a high expression level of the endocytosis protein REPS2. Int J Cancer. 2005;113(4):561‐567. [DOI] [PubMed] [Google Scholar]
- 29. Doolan P, Clynes M, Kennedy S, et al. TMEM25, REPS2 and Meis 1: favourable prognostic and predictive biomarkers for breast cancer. Tumour Biol. 2009;30(4):200‐209. [DOI] [PubMed] [Google Scholar]
- 30. Sharma V, Koul N, Joseph C, Dixit D, Ghosh S, Sen E. HDAC inhibitor, scriptaid, induces glioma cell apoptosis through JNK activation and inhibits telomerase activity. J Cell Mol Med. 2010;14(8):2151‐2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pont L, Naipal K, Kloezeman J, et al. DNA damage response and anti‐apoptotic proteins predict radiosensitization efficacy of HDAC inhibitors SAHA and LBH589 in patient‐derived glioblastoma cells. Cancer Lett. 2015;356:525‐535. [DOI] [PubMed] [Google Scholar]
- 32. Liao X, Chaudhary P, Qiu G, Che X, Fan L. The role of propranolol as a radiosensitizer in gastric cancer treatment. Drug Des Devel Ther. 2018;12:639‐645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brohée L, Peulen O, Nusgens B, et al. Propranolol sensitizes prostate cancer cells to glucose metabolism inhibition and prevents cancer progression. Sci Rep. 2018;8(1):7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Montoya A, Varela‐Ramirez A, Dickerson E, et al. The beta adrenergic receptor antagonist propranolol alters mitogenic and apoptotic signaling in late stage breast cancer. Biom J. 2019;42(3):155‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu L, Zhang J, Rao M, Zhang Z, Zhu H, Zhang C. Harmine suppresses the proliferation of pancreatic cancer cells and sensitizes pancreatic cancer to gemcitabine treatment. Onco Targets Ther. 2019;12:4585‐4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao J, Zhu H, Wan H, Zou X, Ma X, Gao G. Harmine suppresses the proliferation and migration of human ovarian cancer cells through inhibiting ERK/CREB pathway. Oncol Rep. 2017;38(5):2927‐2934. [DOI] [PubMed] [Google Scholar]
- 37. Liu H, Han D, Liu Y, et al. Harmine hydrochloride inhibits Akt phosphorylation and depletes the pool of cancer stem‐like cells of glioblastoma. J Neurooncol. 2013;112(1):39‐48. [DOI] [PubMed] [Google Scholar]
- 38. McFadyen J, Schaff M, Peter K. Current and future antiplatelet therapies: emphasis on preserving haemostasis. Nat Rev Cardiol. 2018;15(3):181‐191. [DOI] [PubMed] [Google Scholar]
- 39. Jiang R, Zhao S, Wang R, et al. Safety and efficacy of atorvastatin for chronic subdural hematoma in Chinese patients: a randomized ClinicalTrial. JAMA Neurol. 2018;75(11):1338‐1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019‐nCoV). Nat Rev Drug Discov. 2020;19(3):149‐150. [DOI] [PubMed] [Google Scholar]
- 41. Tang W, Cao Z, Han M, et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369:m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferner R, Aronson J. Chloroquine and hydroxychloroquine in covid‐19. BMJ. 2020;369:m1432. [DOI] [PubMed] [Google Scholar]
- 43. Li J, Liao T, Liu H, et al. Hypoxic glioma stem cell‐derived exosomes containing Linc01060 promote progression of glioma by regulating the MZF1/c‐Myc/HIF1α Axis. Cancer Res. 2021;81(1):114‐128. [DOI] [PubMed] [Google Scholar]
- 44. Gu X, Gong H, Shen L, Gu Q. MicroRNA‐129‐5p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting DNMT3A. Am J Translat Res. 2018;10(9):2834‐2847. [PMC free article] [PubMed] [Google Scholar]
- 45. Vollmann‐Zwerenz A, Leidgens V, Feliciello G, Klein C, Hau P. Tumor cell invasion in glioblastoma. Int J Mol Sci. 2020;21(6):1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin Y, Wu Z. MicroRNA‐128 inhibits proliferation and invasion of glioma cells by targeting COX‐2. Gene. 2018;658:63‐69. [DOI] [PubMed] [Google Scholar]
- 47. Giamanco K, Matthews R. The role of BEHAB/Brevican in the tumor microenvironment: mediating glioma cell invasion and motility. Adv Exp Med Biol. 2020;1272:117‐132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.


