Abstract
The Neurospora Varkud Satellite (VS) RNA is capable of promoting a reversible self-cleavage reaction important for its replication pathway. In vivo the VS RNA performs a cis-cleavage reaction to generate monomeric length transcripts that are subsequently ligated to produce circular VS RNA. The predominant form of VS RNA observed in vivo is the closed circular form, though minimal VS ribozyme self-cleavage constructs lack detectable ligation activity. MFOLD analysis of the entire VS RNA sequence revealed an extended region 5′ and 3′ of the minimal self-cleaving region that could anneal to form a complementary helix, which we have termed helix 7. In full-length VS RNA, this helix appears to span over 40 bp of sequence and brings the 5′- and 3′-ends of the RNA into proximity for the ligation reaction. Here we report a variant of the VS ribozyme with an extended 5′- and 3′-terminus capable of forming a truncated helix 7 that promotes the ligation reaction in vitro. Through mutation and selection of this RNA we have identified a ribozyme containing two point mutations in the truncated helix 7 that ligates with >70% efficiency. These results show that an additional helical element absent in current VS ribozyme constructs is likely to be important for the ligation activity of VS RNA.
INTRODUCTION
The Neurospora Varkud Satellite (VS) RNA is an abundant non-coding RNA found in the mitochondria of certain natural isolates of the species Neurospora crassa (1). In vivo VS RNA exists as a satellite RNA of the V plasmid, a large mitochondrial DNA that encodes a reverse transcriptase necessary for replication of the VS RNA (2). The VS RNA contains a catalytic element capable of a reversible self-cleavage reaction. Like most small satellite RNA molecules, VS is thought to replicate by rolling circle replication (3). Initially, VS RNA is synthesized as a multimeric transcript transcribed from the VS plasmid DNA. A site-specific cleavage event produces linear monomeric and some multimeric length transcripts that are ligated to produce circular VS RNA, the predominant form of VS observed in vivo (4).
The VS ribozyme, encoded within the VS RNA, is a small catalytic motif that catalyzes a site-specific cleavage reaction generating products containing 2′,3′-cyclic phosphate and 5′-OH termini (4). The reaction is mechanistically similar to other small catalytic RNAs, including the hammerhead, hairpin and hepatitis delta virus ribozymes (5). VS RNA itself is 881 nt in length, however, the minimal contiguous sequence required for self-cleavage consists of only 154 nt, a single nucleotide upstream and 153 nt downstream of the cleavage site (6). The secondary structure of this minimal self-cleaving construct (clone G11), based on RNA folding programs and biochemical data, is arranged as a set of six helical elements (7). A 3 bp long-range pseudoknot formed between the loops of helices 1 and 5 is also required for folding and activity of the RNA (7). While clone G11 represents the minimal self-cleaving domain, it is not capable of the reverse (ligation) reaction in vitro (8). Therefore, we reasoned that sequence elements essential for ligation may be absent from this construct. In an attempt to define elements necessary for ligation, the sequence at the 5′- and 3′-ends of the RNA were analyzed, revealing a long region of base pair complementarity. Based upon biochemical analysis of several ribozyme constructs we demonstrate that formation of this seventh helical element is critical for the ligation activity of the VS ribozyme.
MATERIALS AND METHODS
DNA templates and RNA synthesis
RNAs were synthesized in vitro by transcription of linearized plasmids with T7 RNA polymerase, purified by 8% denaturing PAGE, and extracted by the crush-and-soak method at 4°C in 10 mM Tris–HCl, 0.1 mM EDTA, 250 mM NaCl, pH 8.0. Transcripts were concentrated by ethanol precipitation and resuspended in an appropriate volume of TE (10 mM Tris–HCl, 0.1 mM EDTA, pH 8.0). Transcription reactions contained 40 mM Tris–HCl pH 8.0, 10 mM MgCl2, 2 mM spermidine, 5 mM DTT, 0.005% Triton X-100 and 1 mM each NTP.
The pVS6 plasmid contains VS nucleotides 599–791 (numbering based upon that of Saville and Collins; 4). VS6 RNA was prepared by transcription of EcoRI-cut pVS6. The pVSE plasmid also contains VS nucleotides 599–791, with mutations A782U and U785C incorporated via site-directed mutagenesis. The VS6 clone contains some vector sequence at the 3′-end, however, the VSE clone was designed with an EarI restriction site at the 3′-end that can be used to generate full-length VSE RNA. VSE RNA was prepared by transcription of pVSE cut with EarI. The G11 plasmid contains VS nucleotides 617–881 (6). G11 RNA was prepared by transcription of G11 linearized with SspI. Substrate RNA, SUB13, was prepared by transcription from a G11-AvaI template, and 5′-end-labeled as described below. SUB24 RNA was prepared by transcription of either EcoRI-cut pVS6 or AvaI-cut pVS6 (for 5′-end-labeling). The compensatory substrate, SUB24M, was prepared by transcription of plasmid 5A linearized with AvaI (plasmid 5A contains VS nucleotides 599–791, with mutations G604C, A605U and G606C). Transcription of pVS6-EcoRI followed by self-cleavage yielded the SUB24 substrate with a 2′,3′-cyclic phosphate terminus. Transcription of pVS6-AvaI, pG11-AvaI or p5A-AvaI yielded stem–loop I substrates which could be 5′-end-labeled and then cleaved by the VS trans-cleaving ribozyme to generate the oligonucleotide substrates (9). Concentrations of unlabeled transcripts were determined by UV absorption assuming an extinction coefficient at 260 nm of 2.2 × 106 for VS6 or VSE RNA, and 2.6 × 105 for substrate RNAs.
The AvaI substrate RNA was 5′-end-labeled using T4 polynucleotide kinase (T4 PNK) and [γ-32P]ATP. After labeling the RNA at 37°C for 45 min, the kinase was heat inactivated at 70°C for 20 min. The 32P-5′-end-labeled AvaI RNA was then cleaved with VS trans-cleaving ribozyme at 30°C for 1 h to generate the 2′,3′-cyclic phosphate end. Products were resolved on a 20% native polyacrylamide gel, visualized by autoradiography and eluted by the crush-and-soak method at 4°C in 10 mM Tris–HCl and 0.1 mM EDTA. This procedure enabled efficient 5′-end-labeling of the substrate RNA while avoiding the 3′-phosphatase activity of T4 PNK.
Ligation reactions
Ligation reactions were performed at 30°C in 50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 25 mM KCl and 2 mM spermidine. The ribozyme (0.5 µM) was preincubated at 50°C for 30 min and slow cooled, followed by an additional 10 min incubation at 30°C. Ligation reactions were initiated by addition of a pre-warmed solution of labeled substrate RNA and incubated at 30°C. The reaction was quenched by addition of 2 vol formamide loading buffer. Products were resolved by denaturing gel electrophoresis and visualized by autoradiography. For characterization of VSE and mutant RNAs, ligation reactions were performed as described using 0.5 µM ribozyme and trace amounts of labeled substrate (∼10 nM). Aliquots were removed at various times and quenched in 2 vol formamide loading buffer, and reaction products were resolved by denaturing gel electrophoresis. The amounts of radioactivity in the substrate and product bands were quantitated using a Molecular Dynamics PhosphorImager and ImageQuant software. Data were plotted and analyzed using KaleidaGraph software. Initial rates were determined based on a linear fit of the data.
Pool construction and selection assay
The initial randomized pool was constructed from two degenerate oligonucleotides, VS-3D and VS-5D, and their complements, VS-3 and VS-5, respectively, which span the full length of the VS6 ribozyme sequence. The pool was designed to give a degeneracy level of 7.5%, yielding an expected mutation rate of 12–14 mutations per molecule. The oligonucleotides were purified by 8% denaturing PAGE, extracted by the crush-and-soak method, concentrated with ethanol and resuspended in an appropriate volume of TE. Degenerate oligonucleotides, VS-3D and VS-5D, were annealed at 90°C to their complements, VS-3 and VS-5, respectively, and slow cooled to room temperature. The double-stranded templates, each comprising the 5′- and 3′-ends of the pool, were then digested with BsaI and ligated with T4 DNA ligase at 16°C to give full-length Pool 0 DNA. Pool 0 DNA was engineered with a T7 RNA polymerase promoter at the 5′-end and a primer binding site (PBS 1B) at the 3′-end. Pool 0 RNA was transcribed in vitro using T7 RNA polymerase and self-cleavage products purified by 8% denaturing PAGE. The RNA was extracted by the crush-and-soak method at 4°C, concentrated by ethanol precipitation and resuspended in an appropriate volume of TE.
Ligation reactions were conducted as described above. The initial round of selection contained 1.0 µM Pool 0 RNA (∼1012 molecules) and a 2 µM solution of labeled and unlabeled SUB24 RNA, although in subsequent rounds the amount of substrate RNA was dropped to 0.5 µM. Ligated RNA was purified by 8% denaturing PAGE, visualized by autoradiography and extracted by the crush-and-soak method. The RNA was concentrated by ethanol precipitation, resuspended in an appropriate volume of TE and reverse transcribed using AMV reverse transcriptase (AMV-RT) with primer SLX1B, d(CGTGCGGATCCCTCTTCG), complementary to the 3′-end of the RNA. The AMV-RT was then heat inactivated at 90°C for 5 min and the reaction products PCR amplified using primers SLX1B and SLX3A, d(GCAAGCTTATTAATACGACTCACTATAGGTAGTAGAGTGTCGCAATCTGCG), to regenerate the pool (Fig. 2). RNA was prepared as described for Pool 0 RNA. The protocol was repeated for subsequent rounds of selection. The ligation efficiency of individual pools was determined using the ligation assay described above. Pool 3 DNA, representing the most active pool, was digested with EcoRI and HindIII and cloned into vector pUC19. RNA was prepared by in vitro transcription of plasmids linearized with EarI. Self-cleavage products were purified by 8% denaturing PAGE, eluted by the crush-and-soak method, ethanol precipitated and resuspended in TE. Individual clones were tested for ligation activity using the ligation assay described above and sequenced by dideoxy sequencing.
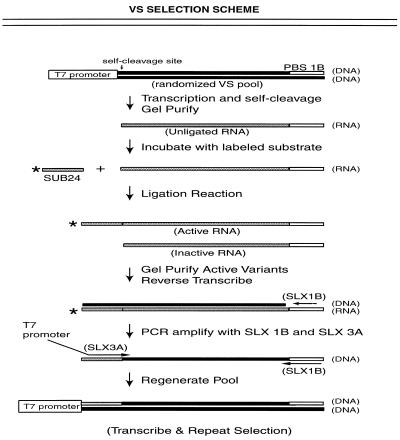
Figure 2.
Schematic representation of the selection scheme. Randomized Pool 0 DNA was designed with a primer binding site (PBS 1B) at its 3′-end and a T7 promoter at the 5′-end. Unligated RNA from the randomized pool was incubated with an oligonucleotide substrate (SUB24) containing a 2′,3′-cyclic phosphate terminus. Active RNAs ligate the oligonucleotide substrate and become sequence tagged at their 5′-end. Active variants were size selected from inactive variants by denaturing gel electrophoresis, reverse transcribed and PCR amplified with selective primers (SLX 1B and SLX 3A) to reintroduce the T7 promoter and regenerate the pool. The scheme was repeated for subsequent rounds of selection.
RESULTS
The majority of VS RNA isolated in vivo is in the closed circular form, suggesting that the internal equilibrium of this RNA favors ligation over cleavage. The minimal self-cleaving VS ribozyme construct (G11) provides only a short 3 bp helix for substrate binding interactions and this may explain why it is unable to efficiently ligate a complementary oligonucleotide substrate. The fact that full-length VS RNA can perform the reverse (ligation) reaction to generate circular products suggests that this minimal self-cleaving ribozyme may lack essential elements required for ligation activity.
In an effort to identify sequence elements that may stabilize substrate–ribozyme interactions and promote ligation activity in vitro we conducted an MFOLD analysis of the entire VS RNA sequence (10). MFOLD (v.2.1) revealed an extended region of complementarity 5′ and 3′ to the minimal self-cleaving domain. This extended helix, termed helix 7, appears to extend over 40 bp of VS RNA sequence (Fig. 1). The putative helix consists of a long region of complementarity between 5′ nucleotides G564–A614 and 3′ nucleotides U774–U828. The helix 7 extension includes a few short mispaired regions and several non-canonical G-U base pairs, but as many as 34 standard Watson–Crick base pairs. These features appear to make helix 7 very stable, with a calculated Tm of 75°C (10). We reasoned that this element may be important for ligation activity by aligning the substrate on the ribozyme and thus may play an important biological role.
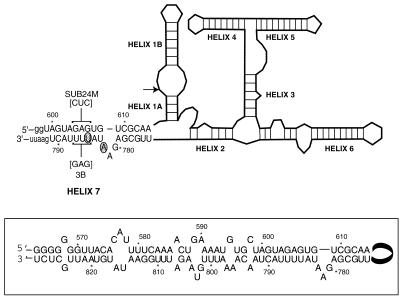
Figure 1.
Secondary structure of the VS6 clone predicted by MFOLD (v.2.1). The site of cleavage and ligation is indicated by an arrow. The RNA consists of seven helical regions (helices 1A–7). Base paired regions in the ribozyme are indicated by horizontal and vertical lines, while base pairing interactions in helix 7 are shown. The VS6 ribozyme contains a truncated version of helix 7, however, in full-length VS RNA helix 7 may extend over 40 bp of sequence (boxed region). Wild-type sequence is denoted in upper case and vector sequence in lower case. The VSE ribozyme contains two helix 7 mutations, U785C and A782U (shaded circles), that are observed to enhance ligation activity. Mutations in ribozyme 3B and compensatory mutations in SUB24M (used to demonstrate the helix 7 specificity switch) are indicated. In addition to the mutations indicated, ribozyme 3B also contains the A782U mutation.
To examine the potential contribution of this helix to VS ribozyme ligation activity several constructs were designed, each containing a truncated version of helix 7. One construct (VS6, Fig. 1), which contained an 8 nt extension to the 3′-end of the original VS ribozyme, was capable of ligating in trans a complementary oligonucleotide substrate (SUB24) that contained a 22 nt extension to the 5′-end of the cleavage site. Ligation of VS6 to SUB24, however, proceeded with an efficiency of only 2%. Gel shift analysis revealed that VS6 RNA bound the oligonucleotide substrate very poorly (<5%; data not shown). Further analysis by native polyacrylamide gel electrophoresis revealed that VS6 adopted multiple folding conformations (data not shown). This suggested that the low level ligation activity of this RNA may have resulted from structural heterogeneity and that the 3′ extension necessary for substrate binding may have been inaccessible, thus inhibiting ligation.
In order to identify an RNA construct with a truncated helix 7 that contained a minimal number of mutations yet could fold more readily and ligate a complementary oligonucleotide substrate with greater efficiency, we employed the technique of in vitro selection. Similar methods have been adapted to isolate ribozymes from random sequence, as well as to define essential sequence elements of naturally occurring ribozymes (11,12).
A randomized pool of VS6 DNA was designed with a degeneracy level of 7.5%, expected to yield an average mutation rate of 12–14 mutations per molecule. The randomized pool was constructed from two degenerate oligonucleotides, VS-3D and VS-5D, comprising the 5′- and 3′-ends of the VS6 sequence. This enabled us to randomize the entire region of VS6 sequence (nucleotides 621–791), with the exception of nucleotides 691–694 in helix 5, which were used to anneal the respective oligonucleotides together during generation of the initial pool. A T7 RNA polymerase promoter was engineered at the 5′-end of the sequence to facilitate in vitro transcription, as well as a selective primer binding site (PBS 1B) at the 3′-end.
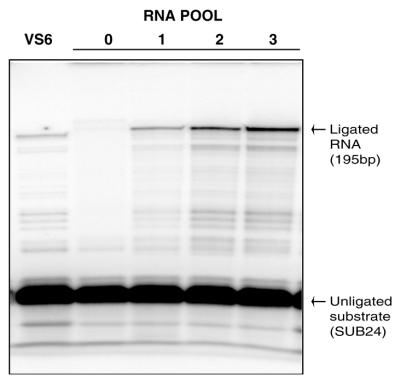
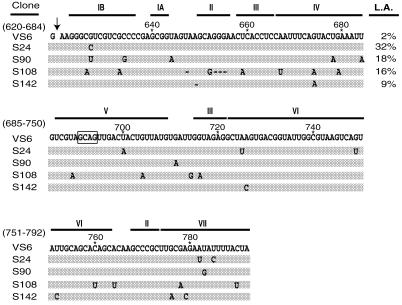
The selection and amplification scheme is outlined in Figure 2. Active variants were selected based upon their ability to ligate in trans an oligonucleotide substrate (SUB24). The initial RNA pool (Pool 0) had a ligation efficiency of 0.1%, however, after three rounds of selection the overall ligation efficiency of the RNA pool improved to 10% (Table 1 and Fig. 3). This corresponds to a 100-fold enhancement compared to the original Pool 0 RNA and a 5-fold enhancement compared to wild-type VS6 RNA. Several members of this pool were cloned and sequenced. Individual ligation efficiencies were determined based on the ability of the RNA to ligate 5′-end-labeled SUB24 in 30 min at 30°C. Several active clones were identified, the two most active of which, S24 and S90, had ligation efficiencies of 32 and 18% in 30 min, respectively (Fig. 4). S24 and S90 both had a mutation at position G627 in helix 1B, mutations in the 3′-segment of helix 7 and several unrelated mutations scattered throughout the RNA (Fig. 4). Many other active selected RNAs also contained mutations in the 3′-segment of helix 7, which suggested that these mutations may be contributing to the increased ligation efficiency of these constructs (Fig. 4).
Table 1. Results of the in vitro selection assay.
| RNA pool | Ligation activity (%) after 30 min (SUB24) |
|---|---|
| 0 | 0.1 |
| 1 | 1.4 |
| 2 | 5.1 |
| 3 | 10 |
Figure 3.
Ligation activity of RNA pools (Pools 0–3) compared to wild-type VS6 RNA. Pool RNA was incubated with 5′-end-labeled SUB24 at 30°C for 30 min under the conditions described in Materials and Methods. The ligation efficiency of the pool increased with each round of selection, as those sequences that promote ligation were selectively enriched.
Figure 4.
Sequence alignment of the most active selected clones. Individual mutations are shown with respect to their location in the VS6 ribozyme. –, deleted bases. Ligation activity (L.A.), reported after 30 min at 30°C, are shown to the right of the first row. Values do not reflect end points of reactions. The entire region of VS6 was randomized, with the exception of nucleotides 691–694 in helix 5 (boxed region).
To determine which mutations were responsible for the improved VS ribozyme ligation activity, three additional RNA constructs were prepared containing mutations commonly observed among the active population. Although a G627 mutation was present in both variants S24 and S90, mutating G627 to C showed only a modest improvement in the total ligation activity (<4%) compared to VS6. The G627U mutation had a reduced ligation efficiency of 1%. To assess a potential role for mutations observed in helix 7, an additional construct, termed VSE, was created that contained the two helix 7 mutations, A782U and U785C, observed in the most active clone, S24. VSE RNA was able to ligate a complementary substrate (SUB24) with an efficiency of 30% in 30 min, compared to 32% for S24. Further characterization found that VSE RNA ligates with an efficiency exceeding 70% in 4 h, with an initial rate constant of 1.6 min–1. Ribozymes containing each of the single helix 7 mutations were also prepared. The U785C mutant (1A) had a maximal ligation efficiency of 35% after 6 h, with an initial rate constant of 0.64 min–1 (Table 2). The ligation efficiency of the A782U mutant (2B) did not exceed 10% efficiency and had an initial rate constant of only 0.09 min–1 (Table 2). Thus, while the majority of the improved ligation activity appears to derive from the U785C mutation, both mutations are required for optimal efficiency of the ribozyme.
Table 2. Comparison of initial rates and percentage ligation for the VSE ribozyme and ribozymes 1A, 2B and 3B.
| Ribozyme | Mutation(s) | Substrate | Initial rate (min–1) | Ligation activity (%) after 6 h |
|---|---|---|---|---|
| VSE | A782U, U785C | SUB24 | 1.6 | 70 |
| 1A | U785C | SUB24 | 0.64 | 35 |
| 2B | A782U | SUB24 | 0.09 | 10 |
| 3B | A782U, U785G, U786A, U787G | SUB24 | 0.07 | 10 |
| 3B | A782U, U785G, U786A, U787G | SUB24M | 1.8a | 40 |
aInitial rate of ligation of ribozyme 3B with SUB24M which restores helix 7 base pairing interactions; SUB24M, 5′-ggUAGUACUCUGUCGCAAUCUGCG.
The importance of helix 7 in VS RNA ligation activity was further examined by looking at the effect on ligation activity of RNA constructs that lacked either the 5′ or 3′ extension required to form helix 7. To this end we utilized the G11 RNA, i.e. the original VS ribozyme that lacks the 3′ extension of helix 7, and a shorter substrate (SUB13) which lacks the 5′ extension of helix 7. SUB13 contains only 4 nt of wild-type VS sequence and is capable of forming only a short 3 bp helix with the ribozyme. In the absence of either the 5′ or 3′ extension, ligation activity was reduced to <1% efficiency, suggesting that neither the 3′ nor the 5′ extension alone are sufficient to confer ligation activity (Table 3).
Table 3. Both the 5′- and 3′-extensions of helix 7 are required for ligation activity.
| Ribozyme | Ligation activity (%) after 6 h |
|
|---|---|---|
| SUB13 | SUB24 | |
| SUB13, 5′-gggaaagcuUGCG; SUB24, 5′-ggUAGUAGAGUGUCGCAAUCUGCG; lower case letters denote vector sequence. | ||
| G11 | <1 | <1 |
| VS6 | <1 | 2 |
| VSE | <1 | 70 |
To investigate the importance of the alignment of helix 7 proposed in Figure 1, we designed a ribozyme (3B, Fig. 1) containing mutations in the 3′ extension of helix 7 expected to disrupt predicted base pairing interactions. The mutant ribozyme 3B ligates substrate SUB24 with reduced efficiency (<10%), at a rate >20-fold slower than the initial rate of VSE (0.07 min–1, Table 2). However, if the compensatory mutations are made in the substrate (SUB24M, Fig. 1) then the mutant ribozyme is capable of a ligation efficiency very close to that of the wild-type construct (40%, initial rate 1.8 min–1). This provides direct evidence for base pairing interactions between at least this segment of the helix 7 region.
DISCUSSION
We have described the identification and initial characterization of a putative helical element in the VS RNA secondary structure that appears to serve an important role in VS ligation activity. Helix 7 may play a biological role in VS RNA ligation activity by properly aligning the substrate on the ribozyme to facilitate ligation. Self-cleaving constructs lacking this helix were incapable of ligation activity (<1%), although recent circular permutation constructs of VS RNA have provided alternative means to achieve ligation activity in vitro (13). These circular permutated constructs, in which stem 1 is covalently linked to the 3′-end of G11 RNA, exploit the fact that VS RNA exists in vivo as head-to-tail multimers to align the substrate and ribozyme in close proximity.
Based on MFOLD analysis helix 7 appears to span a significant amount of the total VS RNA sequence. The presence of this extended region of complementarity may contribute binding energy to stabilize substrate–ribozyme interactions and maintain closed circular products in vivo. The VS6 ribozyme identified in this study contains only a truncated portion of the wild-type helix 7 and is capable of ligation activity. Through mutation and selection we have identified a highly efficient VS ligation construct with a significantly shorter helix than is likely present in the full-length VS RNA. VSE RNA contains only two mutations in the helix 7 region and ligates with an efficiency exceeding 70%. These results suggest that the internal equilibrium of VSE ribozyme favors ligation over cleavage, as is the case for the VS RNA.
The two helix 7 mutations, A782U and U785C, could either play a role in promoting the properly folded RNA conformation or they may directly stabilize interactions with the substrate in order to promote greater ligation efficiency. Both mutations appear to be important for ligation activity, though the U785C mutation makes the greatest contribution. The U785C mutation converts a G-U base pair to a G-C base pair, which may contribute additional stability to the helix. It is unclear how the A782U mutation makes its modest contribution to the ligation activity of the RNA. These mutations do not increase ligation activity in the absence of their complementary sequence at the 5′-end, demonstrating that the 3′ extension of helix 7 alone is not sufficient for ligation activity.
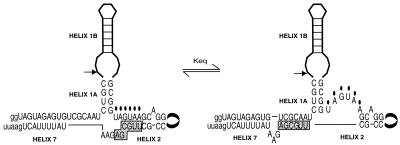
Based upon sequence alignments and chemical probing data we propose that helix 7 exists in equilibrium between two alternative conformations. In one conformation a weak helix 7 is formed that would exhibit poor substrate interactions (Fig. 5, left). In the alternative conformation a stronger helix 7 is formed that would allow better substrate binding interactions and thus increased ligation activity (Fig. 5, right). In switching between these two conformations, base pairing interactions in helix 2 are also disrupted (Fig. 5, right). This is consistent with modification interference experiments that previously demonstrated that disruptions in helix 2, achieved through reaction of base functional groups with hydrazine or DEPC, enhanced self-cleavage activity of the ribozyme (8). This region of secondary structure has been implicated as part of a VS attenuator structure evolved to affect the cleavage and ligation equilibrium of VS RNA and to promote the formation of closed circular products in vivo (8). While the precise base pairs that are formed within helix 7 and disrupted in helix 2 remain to be determined, these results clearly demonstrate that RNA sequence to the 5′- and 3′-sides of the traditionally defined boundaries of the VS ribozyme are important for its RNA ligation activity.
Figure 5.
Potential conformational switch between helices 2 and 7. Helix 7 can exist in equilibrium between two possible conformations, a weak helix 7 (left) and a more stable helix 7 (right). Formation of a stable helix 7 results in disruption of base pairing interactions in helix 2. Disruption of these base pairs has been observed to enhance self-cleavage activity in vitro. Sites of modified bases in helix 2 observed to enhance self-cleavage of G11 RNA are indicated by ovals above the nucleotides.
Acknowledgments
ACKNOWLEDGEMENTS
An NIH Individual Research Award to F.D.J and an NSF grant CHE 0100057 to S.A.S supported this work.
REFERENCES
- 1.Griffiths A.J.F. (1995) Natural plasmids of filamentous fungi. Microbiol. Rev., 59, 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennell J.C., Saville,B.J. and Collins,R.A. (1995) The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev., 9, 294–303. [DOI] [PubMed] [Google Scholar]
- 3.Symons R.H. (1992) Small catalytic RNAs. Annu. Rev. Biochem., 61, 641–671. [DOI] [PubMed] [Google Scholar]
- 4.Saville B.J. and Collins,R.A. (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell, 61, 685–696. [DOI] [PubMed] [Google Scholar]
- 5.McKay D.B. and Wedekind,J.E. (1999) Small ribozymes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 265–286.
- 6.Guo H.C., De Abreu,D.M., Tillier,E.R., Saville,B.J., Olive,J.E. and Collins,R.A. (1993) Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol., 232, 351–361. [DOI] [PubMed] [Google Scholar]
- 7.Beattie T.L., Olive,J.E. and Collins,R.A. (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA, 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beattie T.L. and Collins,R.A. (1997) Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol., 267, 830–840. [DOI] [PubMed] [Google Scholar]
- 9.Guo H.C.T. and Collins,R.A. (1995) Efficient trans-cleavage of a stem–loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J., 14, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. Kluwer Academic Publishers, The Netherlands, pp. 11–43.
- 11.Bartel D.P. and Szostak,J.W. (1993) Isolation of new ribozymes from a large pool of random sequences. Science, 261, 1411–1417. [DOI] [PubMed] [Google Scholar]
- 12.Berzal-Herranz A., Joseph,S., Chowrira,B.M., Butcher,S.E. and Burke,J.M. (1993) Essential nucleotide sequences and secondary structure elements of the hairpin ribozyme. EMBO J., 2, 2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen A.A. and Collins,R.A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell, 5, 469–478. [DOI] [PubMed] [Google Scholar]