Abstract
Collagens are the most abundant proteins in the body and among the most biosynthetically complex. A molecular ensemble of over 20 endoplasmic reticulum resident proteins participates in collagen biosynthesis and contributes to heterogeneous post-translational modifications. Pathogenic variants in genes encoding collagens cause connective tissue disorders, including osteogenesis imperfecta, Ehlers–Danlos syndrome, and Gould syndrome (caused by mutations in COL4A1 and COL4A2), and pathogenic variants in genes encoding proteins required for collagen biosynthesis can cause similar but overlapping clinical phenotypes. Notably, pathogenic variants in lysyl hydroxylase 3 (LH3) cause a multisystem connective tissue disorder that exhibits pathophysiological features of collagen-related disorders. LH3 is a multifunctional collagen-modifying enzyme; however, its precise role(s) and substrate specificity during collagen biosynthesis has not been defined. To address this critical gap in knowledge, we generated LH3 KO cells and performed detailed quantitative and molecular analyses of collagen substrates. We found that LH3 deficiency severely impaired secretion of collagen α1α1α2(IV) but not collagens α1α1α2(I) or α1α1α1(III). Amino acid analysis revealed that LH3 is a selective LH for collagen α1α1α2(IV) but a general glucosyltransferase for collagens α1α1α2(IV), α1α1α2(I), and α1α1α1(III). Importantly, we identified rare variants that are predicted to be pathogenic in the gene encoding LH3 in two of 113 fetuses with intracranial hemorrhage—a cardinal feature of Gould syndrome. Collectively, our findings highlight a critical role of LH3 in α1α1α2(IV) biosynthesis and suggest that LH3 pathogenic variants might contribute to Gould syndrome.
Keywords: collagen, post-translational modification, hydroxylase, glycosylation, basement membrane
Abbreviations: AAA, amino acid analyses; AHA, l-azidohomoalanine; DMEM, Dulbecco's modified Eagle's medium; EDS, Ehlers–Danlos syndrome; ER, endoplasmic reticulum; gRNA, guide RNA; HSP47, heat shock protein 47; Hyl, hydroxylysine; 4Hyp, 4-hydroxyproline; ICH, intracerebral hemorrhage; LH3, lysyl hydroxylase 3; MEF, mouse embryonic fibroblast; MS, mass spectrometry; OI, osteogenesis imperfecta; P4H, prolyl 4-hydroxylase; PTM, post-translational modification; PVDF, polyvinylidene difluoride; SI-collagen, stable isotope–labeled collagen; SPR, surface plasmon resonance
Collagens are the most abundant proteins in the human body and comprise 28 members encoded by 46 genes (1, 2). Pathogenic variants in genes encoding collagens cause a wide range of disorders affecting almost all tissues and organs (3, 4). Type I, type III, and type V collagens form fibrils contributing to structural frameworks and pathogenic variants in these collagens cause connective tissue disorders—osteogenesis imperfecta (OI) and Ehlers–Danlos syndrome (EDS)—that affect fibril-rich tissues, such as bone, skin, and vascular structures (5, 6, 7, 8, 9, 10). In contrast, type IV collagens form flexible networks that are present in the basement membranes of all tissues. Pathogenic variants in type IV collagens cause Alport syndrome (COL4A3, COL4A4, and COL4A5), which primarily affects the glomerular basement membrane (11, 12) and Gould syndrome (COL4A1 and COL4A2), a congenital multisystem disorder that includes highly variable cerebrovascular disease, pediatric epilepsy, ocular anterior segment dysgenesis, skeletal myopathy, and nephropathy (13, 14, 15, 16, 17, 18).
Collagen biosynthesis is a complex and highly orchestrated process that occurs in the endoplasmic reticulum (ER) and is facilitated by a molecular ensemble (19) of over 20 proteins that participate in four major steps: post-translational modifications (PTMs) of individual collagen chains, trimer assembly, triple helix formation, and secretion (19, 20, 21). Although most components of the molecular ensemble are shared among collagens, the collagen-modifying enzymes differ between collagens (22, 23, 24), suggesting subtype specialization of the molecular ensemble (24, 25). Moreover, the molecular ensemble and PTMs for type I collagen, the most extensively characterized collagen, show that the PTM profile is also tissue specific (24, 26). PTMs on the collagen triple helix are classified into three types of primary modifications to prolyl or lysyl residues and two types of secondary modifications to hydroxylysines. Prolyl modifications include 4-hydroxyproline (4Hyp) and 3-hydroxyproline that are generated by prolyl 4 hydroxylases and prolyl 3 hydroxylases, respectively (27). Approximately 40% or more of the prolines are modified to 4Hyp (22, 24, 28), which provides thermal stability to the collagen triple helix (29, 30, 31). In contrast, less than 5% of the prolines are modified to 3-hydroxyproline (28, 32), which is proposed to mediate protein–protein interactions (32, 33). Primary modification of lysine to hydroxylysine (Hyl) is attributed to lysyl hydroxylases (LHs), (27) and secondary modifications of Hyl include two successive and coordinated O-glycosylation steps. Hyl is required for subsequent galactose attachment to make galactosyl Hyls, which are further modified by glucose attachment to make glucosyl galactosyl Hyls. This sequence of modifications from lysine, to Hyl, to galactosyl Hyl and glucosyl galactosyl Hyl occurs in the ER during collagen biosynthesis (34, 35).
In the case of OI and EDS, it is well documented that pathogenic variations in genes encoding collagens and genes encoding proteins required for collagen biosynthesis lead to similar clinical outcomes (5, 6, 7, 8, 9, 10). A similar paradigm may also exist for Gould syndrome as pathogenic variants in glycosyltransferase 25 domain containing 1 (GLT25D1), encoded by collagen β (1-O) galactosyltransferase 1 (COLGALT1) cause musculoskeletal (36) and cerebrovascular (37, 38) manifestations similar to those observed in individuals with COL4A1 and COL4A2 pathogenic variants. Notably, because molecular ensemble proteins can be multifunctional and employed across diverse collagen types, pathogenic variants in encoding genes often cause phenotypes that blend across different clinical categories. An example being LH3, encoded by procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (PLOD3), whereby pathogenic variants cause connective tissue phenotypes that overlap with Stickler-like syndrome and epidermolysis bullosa (39, 40, 41, 42). Indeed, clinical features of patients with biallelic loss of function PLOD3 variants include craniofacial dysmorphisms, skeletal and ocular disorders, sensorineural hearing loss, and variable skin manifestations. Interestingly, in addition to clinical manifestations overlapping Stickler-like syndrome and epidermolysis bullosa, cerebral hemorrhages have been reported in three cases with PLOD3 variants (39, 43). LH3 is proposed to have substrate preference for heavily glycosylated collagens (44, 45, 46) such as type IV collagen (28, 32), raising the possibility that PLOD3 pathogenic variants could also cause Gould syndrome. Supporting this, Plod3 deficiency in the mouse lead to basement membrane disruptions caused by intracellular accumulation and impaired secretion of type IV collagen that had lower molecular mass suggesting incomplete PTMs (45). Importantly, according to the peptide-based in vitro enzymatic analyses, LH3 is striking in that it is a multifunctional enzyme that can execute all the enzymatic roles required for fully modifying lysine to glucosyl galactosyl Hyl—LH, galactosyltransferase, and glucosyltransferase (39, 44, 47, 48, 49, 50, 51). However, detailed quantitative PTM analysis has never been conducted for any type of collagen in the context of LH3 deficiency. Thus, in this study, we used CRISPR-mediated gene editing to knockout LH3 both in PFHR9 cells and mouse embryonic fibroblasts (MEFs) and performed a multifaceted investigation including cellular biochemistry, structural assessment, and quantitative PTM characterization for collagen α1α1α2(I), α1α1α1(III), and α1α1α2(IV) to elucidate the role of LH3 in the collagen molecular ensemble. Furthermore, since perinatal cerebral hemorrhage is a cardinal feature of Gould syndrome, we performed a genetic screen for PLOD3 variants in a fetal stroke cohort that was negative for COL4A1 and COL4A2 pathogenic variants.
Results
LH3 is required for secretion of collagen α1α1α2(IV) but not collagens α1α1α2(I) or α1α1α1(III)
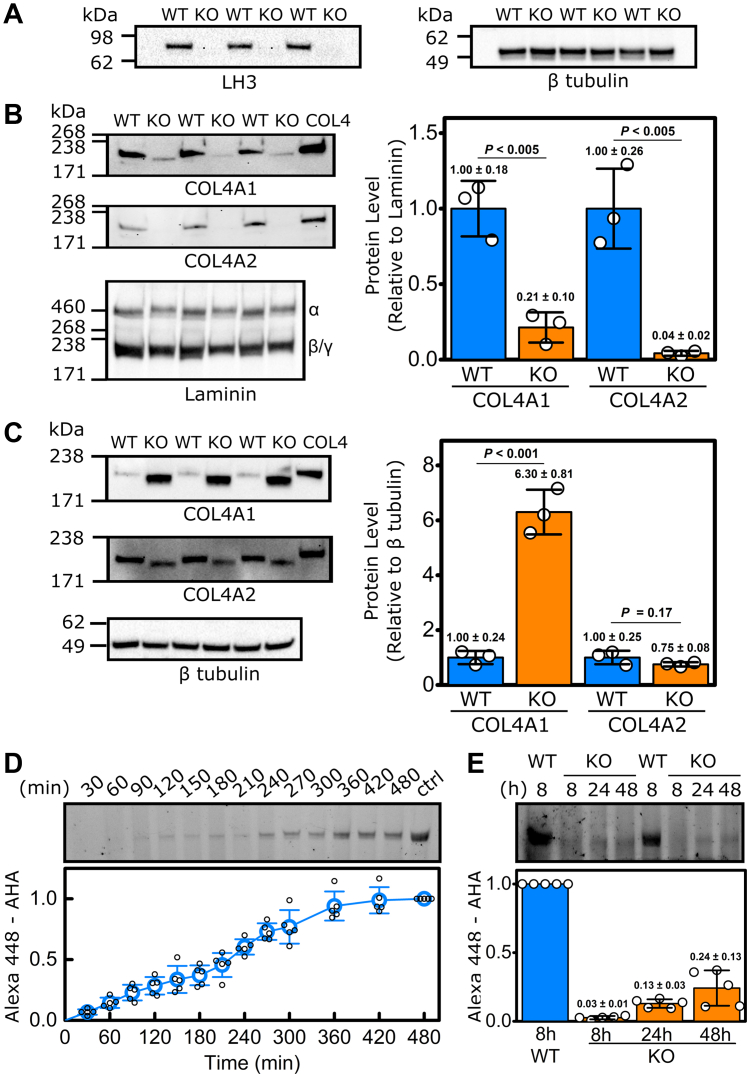
To explore the role of LH3 in the molecular ensemble for type IV collagen, we used CRISPR-mediated gene editing to knockout LH3 in PFHR9 cells (Figs. 1A, S1 and S2A). We selected PFHR9 cells because they are well characterized and secrete large amounts of collagen α1α1α2(IV) because of a duplication of the Col4a1/Col4a2 locus (28, 32, 52). Western blot analysis revealed a significant reduction in COL4A1 and COL4A2 protein levels in the conditioned culture medium of LH3 KO PFHR9 cells (Figs. 1B and S2B) and a concomitant increase in intracellular COL4A1 protein levels (Figs. 1C and S2C), indicating that LH3 deficiency impairs collagen α1α1α2(IV) secretion. To quantitively evaluate the effect of LH3 KO on collagen α1α1α2(IV) biosynthesis, we tested the rate of secretion using pulse-chase labeling with the methionine analog l-azidohomoalanine (AHA) (53, 54). Although this approach is well established for collagen α1α1α2(I), precise measurements of collagen α1α1α2(IV) secretion have not been carefully determined (55, 56, 57, 58). AHA pulse-chase analysis in WT PFHR9 cells showed that newly synthesized collagen α1α1α2(IV) takes approximately 7 to 8 h to be secreted (Fig. 1D, Table S1, and Fig. S3) compared with approximately 2 h for collagen α1α1α2(I) (59, 60). However, in LH3 KO PFHR9 cells, collagen α1α1α2(IV) secretion was significantly delayed, and only one-fourth of newly synthesized collagen α1α1α2(IV) was secreted even after 48 h in culture (Fig. 1E and Table S1).
Figure 1.
LH3 deficiency in PFHR9 cells impairs collagen α1α1α2(IV) secretion.A, semiquantitative Western blots showing successful ablation of LH3 in PFHR9 cells following CRISPR/Cas9-mediated LH3 inactivation (LH3 KO PFHR9 cells). β-tubulin was used as a loading control. B, semiquantitative Western blots and quantification of COL4A1 and COL4A2 levels in the conditioned cell culture medium showing reduced secretion of COL4A1 and COL4A2 in LH3 KO PFHR9 cells (KO) compared with their WT controls. Laminin α/β/γ was used as a loading control. C, semiquantitative Western blots and quantification of COL4A1 and COL4A2 levels in cell lysates showing intracellular COL4A1 accumulation in LH3 KO PFHR9 cells. β-tubulin was used as a loading control. COL4 indicates control-purified collagen α1α1α2(IV). The quantification data in B and C are shown as fold relative to WT levels and presented as means ± SD with individual data points representing independent preparations of culture medium and cell lysate and (n = 3). D, representative SDS-PAGE gel (top) and quantification (bottom) of time-dependent extracellular collagen α1α1α2(IV) levels in WT PFHR9 cells measured by AHA-Alexa Fluor pulse-chase labeling. “ctrl” indicates purified AHA-incorporated collagen α1α1α2(IV). The quantification of AHA-incorporated collagen α1α1α2(IV) was set to 1.0 at 480 min (n = 5). E, representative SDS-PAGE gel (top) and quantification (bottom) of time-dependent extracellular collagen α1α1α2(IV) levels for LH3 KO PFHR9 cells measured by AHA-Alexa Fluor pulse-chase labeling, demonstrating significantly impaired collagen α1α1α2(IV) secretion in LH3 KO PFHR9 cells compared with their WT AHA-incorporated collagen α1α1α2(IV) controls at 8 h (n = 5). Source data files for the figure: The uncropped images of Western blots used in the figure are presented in Fig. S2; generation of the purified AHA-incorporated collagen α1α1α2(IV) used as a control in the figure is presented in Fig. S3; and the value of AHA-incorporated collagen α1α1α2(IV) at each time points is shown in Table S1. AHA, l-azidohomoalanine; LH3, lysyl hydroxylase 3.
Because PFHR9 cells produce very low levels of fibrillar collagens, we also used CRISPR-mediated gene editing to knockout LH3 in MEFs to test the effect of LH3 deficiency on collagen α1α1α2(I) and collagen α1α1α1(III) biosynthesis (Figs. 2A, S4 and S5A). Notably, Western blot analysis showed that the levels of extracellular collagen α1α1α2(I) were increased in LH3 KO MEFs compared with their WT controls (Figs. 2B and S5B). Since the signals of α2(I) chain were stronger than that of α1(I) chain in Western blot (Figs. 2B and S5B), we quantitively tested the effect of LH3 KO on collagen secretion rates in MEFs. AHA pulse-chase labeling showed that the secretion rates of fibrillar collagens α1α1α2(I) and α1α1α1(III) were comparable between WT and LH3 KO MEFs, although delays were observed at early time points for collagen α1α1α2(I) (Fig. 2, C and D, Table S2 and Fig. S6). Collectively, these data suggest that LH3 is critical for biosynthesis of collagen α1α1α2(IV) but not collagens α1α1α2(I) and α1α1α1(III).
Figure 2.
LH3 deficiency in MEFs does not impair biosynthesis of collagen α1α1α2(I) or collagen α1α1α1(III).A, semiquantitative Western blots showing successful ablation of LH3 in MEFs following CRISPR/Cas9-mediated LH3 inactivation (LH3 KO MEFs). β-actin was used as a loading control. B, semiquantitative Western blots and quantification of collagen α1α1α2(I) in conditioned cell culture medium showing increased secretion of ProCOL1A1 and ProCOL1A2 in LH3 KO MEFs (KO) compared with their WT controls. Fibronectin was used as a loading control. COL1 indicates purified pepsin-treated collagen α1α1α2(I). The quantification data in B are shown as fold relative to WT levels and presented as means ± SD with individual data points representing independent preparations of culture medium (n = 3). C, representative SDS-PAGE gel. D, quantification of time-dependent extracellular levels of collagens α1α1α2(I) and α1α1α1(III) measured by AHA-Alexa Fluor pulse-chase labeling, demonstrating comparable secretion rate between LH3 KO and WT MEFs. “ctrl” indicates purified AHA-incorporated collagens α1α1α2 (I) and α1α1α1(III). WT AHA incorporation signal was set to 1.0 at 180 min (n = 5). Source data files for the figure: The uncropped images of Western blots used in the figure are presented in Fig. S5; generation of the purified AHA-incorporated pepsin-treated collagens α1α1α2(I) and α1α1α1(III) mixture used as a control in C is presented in Fig. S6; the value of AHA-incorporated collagens α1α1α2(I) and α1α1α1(III) at each time points is shown in Table S2. AHA, l-azidohomoalanine; LH3, lysyl hydroxylase 3; MEF, mouse embryonic fibroblast.
LH3 is required for collagen α1α1α2(IV) to form triple helices and higher order oligomeric structures
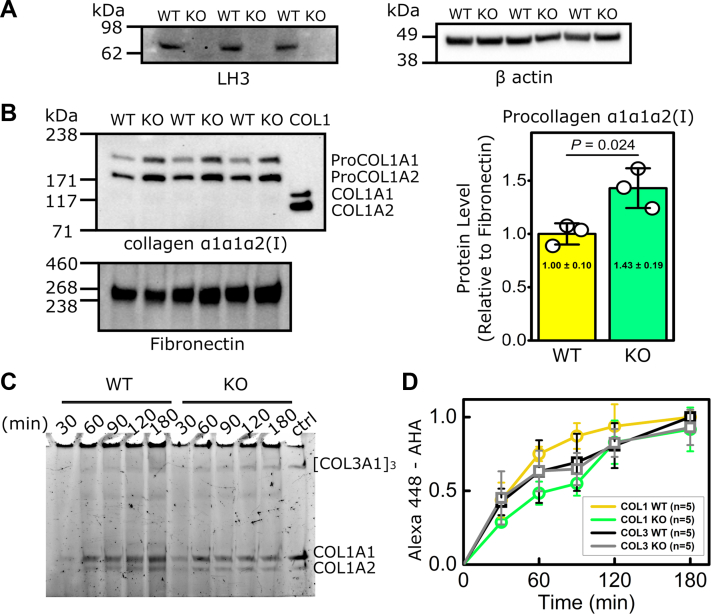
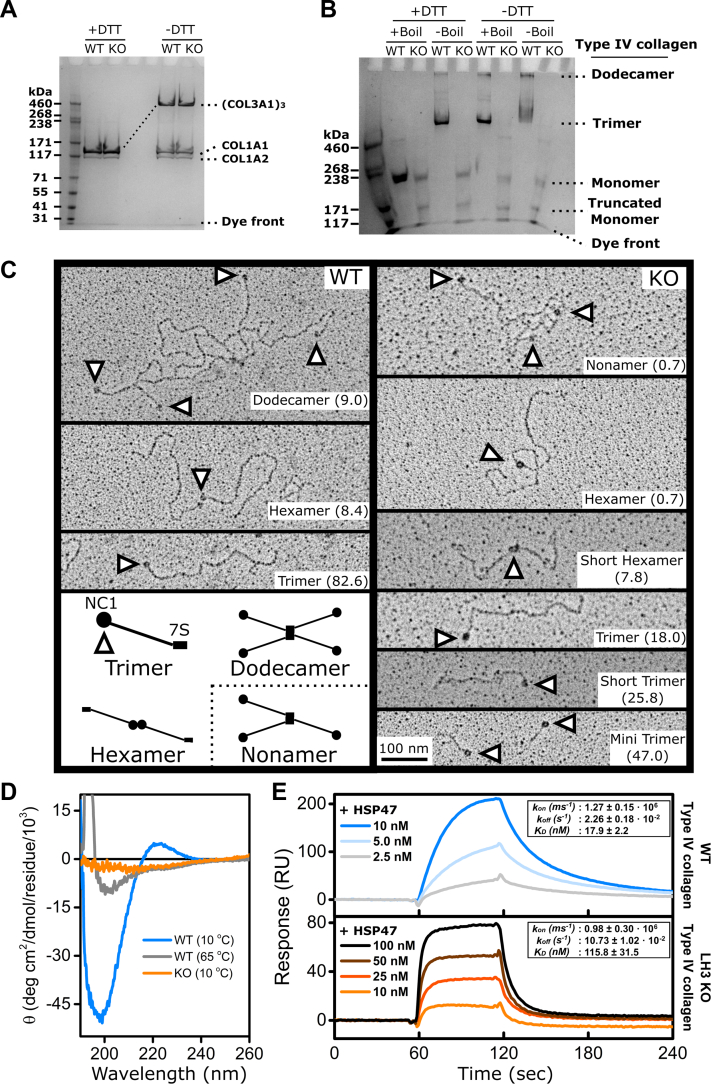
To molecularly characterize the effects of LH3 deficiency on collagens, we purified collagens α1α1α2(I), α1α1α1(III), and collagen α1α1α2(IV) from the conditioned culture medium of MEFs and PFHR9 cells, respectively (Fig. S7, A and B). Consistent with the Western blot and AHA pulse-chase data, there was no difference between collagens α1α1α2(I) and α1α1α1(III) produced by WT and LH3 KO MEFs (Fig. 3A). In contrast, collagen α1α1α2(IV) from LH3 KO PFHR9 cells did not show the higher order collagen α1α1α2(IV) structures that were observed in collagen α1α1α2(IV) from WT PFHR9 cells suggesting an absence of collagen α1α1α2(IV) disulfide crosslinking (Fig. S7B). We next used SDS-agarose gel electrophoresis to resolve the oligomeric state of collagen α1α1α2(IV). Consistent with previous reports (52, 56), the protein banding pattern observed in WT PFHR9 cells showed that most collagen α1α1α2(IV) was present as trimers or dodecamers unless denatured under reducing conditions when they were resolved as monomers (Fig. 3B). In contrast, most of the collagen α1α1α2(IV) isolated from LH3 KO PFHR9 cells was monomeric and failed to form higher order structures (Fig. 3B). Interestingly, we detected a lower molecular weight band in the samples isolated from LH3 KO PFHR9 cells on both SDS-agarose and SDS-PAGE gels (Figs. 3B and S7C), which was identified as truncated COL4A1 and COL4A2 proteins by LC–MS analysis (Fig. S7D). To better characterize collagen α1α1α2(IV) oligomeric structures and network formation, we performed rotary shadow electron microscopy on samples isolated from WT and LH3 KO PFHR9 cells. During type IV collagen network formation, two collagen α1α1α2(IV) heterotrimers associate with each other by way of head-to-head interactions of their carboxy terminal NC1 domains (hexamers) (61) and four collagen α1α1α2(IV) heterotrimers interact by way of lateral and antiparallel interactions of their amino-terminal 7S domains (dodecamers) (62). Electron micrographs from WT PFHR9 cells showed that over 80% of the collagen α1α1α2(IV) was trimeric, but we also found frequent examples of hexamers (∼8.5%) and dodecamers (∼9.0%; Fig. 3C). In contrast, we found reduced proportions of normal trimers and hexamers and did not observe dodecamers in samples from LH3 KO cells. Instead, we detected shortened versions of collagen α1α1α2(IV) hexamers (7.8%) and trimers (72.8%) that appeared truncated at the amino termini and corroborate the SDS-gel electrophoreses and LC–MS data (Figs. 3, B, C, and S7, B–D).
Figure 3.
LH3 deficiency impairs collagen α1α1α2(IV) oligomerization and triple helix formation.A, representative Coomassie blue–stained 3 to 8% Tris–acetate gel showing pepsin-treated collagens α1α1α2(I) and α1α1α1(III) purified from conditioned culture medium of WT and LH3 KO MEFs. B, representative Coomassie blue–stained 3.5% SDS-agarose gel showing collagen α1α1α2(IV) purified from conditioned culture medium of WT and LH3 KO PFHR9 cells. C, representative transmission electron rotary shadow microscopy images showing structural alterations of collagen α1α1α2(IV) purified from conditioned culture medium of WT and LH3 KO PFHR9 cells. Arrowheads point to globular NC1 domains. Diagram at the bottom left illustrates normal collagen α1α1α2(IV) network formation. The frequency of each molecular species is indicated in parentheses (%). D, CD spectra of purified collagen α1α1α2(IV) from conditioned culture medium of WT and LH3 KO PFHR9 cells under native (10 °C) and denaturing (65 °C) conditions. E, surface plasmon resonance analysis showing concentration-dependent binding kinetics between HSP47 and collagen α1α1α2(IV) purified from conditioned culture medium of WT and LH3 KO PFHR9 cells. Each curve represents the average of a minimum of three measurements. The binding constants and affinities are shown as mean ± SD in the inset. LH3, lysyl hydroxylase 3; MEF, mouse embryonic fibroblast.
To evaluate the secondary structure of the collagen α1α1α2(IV) triple helix, we measured CD spectra of collagen purified from WT or LH3 KO PFHR9 cells (Fig. 3D). At 10 °C, collagen α1α1α2(IV) isolated from WT cells showed the characteristic triple helical spectra with negative and positive peaks at 200 and 220 nm, respectively (63), and these peaks were lost following heat denaturation at 65 °C. In contrast, even at 10 °C, collagen α1α1α2(IV) isolated from LH3 KO PFHR9 cells resembled denatured WT collagen α1α1α2(IV) indicating that collagen α1α1α2(IV) produced by LH3 KO cells did not form stable triple helical helices. The triple helical structure is required for binding of the collagen molecular chaperone heat shock protein 47 (HSP47) (64), which plays a critical role in collagen trafficking by anchoring collagens to TANGO1 at ER exit sites to continue along the secretory pathway to the Golgi (65). To test whether HSP47 could bind purified collagen α1α1α2(IV), we performed surface plasmon resonance (SPR) analysis. HSP47 interacted with immobilized collagen α1α1α2(IV) from WT PFRH9 cells in a concentration-dependent manner (Fig. 3E). Although HSP47 also interacted with collagen α1α1α2(IV) from LH3 KO PFRH9 cells, the binding kinetics were altered with lower binding affinity and increased dissociation constant.
LH3 is a selective LH but a general collagen glucosyltransferase
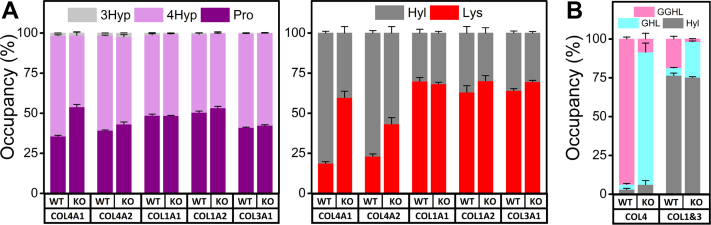
PTMs are important for the structural integrity of collagen triple helices. To understand how LH3 deficiency impacted collagen PTMs, we quantified prolyl hydroxylation, lysyl hydroxylation, and O-glycosylated Hyl using amino acid analyses (AAA) combined with LC–MS. First, we performed acid hydrolysis AAA (which removes sugar groups) of individual collagen α chains from WT or LH3 KO cells. We found that prolyl 3 hydroxylation, which occurs infrequently, was largely unchanged by LH3 deficiency across COL1A1, COL1A2, COL3A1, COL4A1, and COL4A2 proteins and that prolyl 4 hydroxylation was reduced for COL4A1 but not for the other α chains (Fig. 4A and Table S3). In contrast, we found more unmodified lysines (significantly fewer hydroxylysines), on COL4A1 and COL4A2 proteins isolated from LH3 KO PFHR9 cells compared with WT PFHR9 cells. The levels of lysine/Hyl for COL1A1, COL1A2, and COL3A1 isolated from MEFs were largely unchanged, suggesting that the LH function of LH3 is selective for collagen α1α1α2(IV) (Fig. 4A and Table S3). Next, we used alkaline hydrolysis AAA (which preserves sugar attachments) combined with LC–MS to compare the extent of galactosyl Hyl and glucosyl galactosyl Hyl on collagens between WT and LH3 KO cells. Type IV collagen is highly glycosylated, and Hyl residues from collagen α1α1α2(IV) produced by WT PFHR9 cells were almost fully modified to glucosyl galactosyl Hyl (Figs. 4B and S8). In contrast, collagen α1α1α2(IV) derived from LH3 KO PFHR9 cells had high occupancy of galactosyl Hyl but not glucosyl galactosyl Hyl suggesting that in addition to its role as an LH, LH3 also participates in glucosyl, but not galactosyl, modifications. As reported previously (44, 66), glucosyl galactosyl Hyl was also reduced in collagen α1α1α2(I) and collagen α1α1α1(III) suggesting that LH3 is also involved in glucosyl modifications of fibrillar collagens (Figs. 4B and S8). Taken together, these data suggest that LH3 has selective LH activity but general collagen glucosyltransferase activity.
Figure 4.
LH3 is a selective lysyl hydroxylase and general collagen glycosyltransferase.A, acid hydrolysis amino acid analysis. Histograms showing the occupancy of post-translational modifications in proline (3Hyp + 4Hyp + Pro = 100%) and lysine (Hyl + Lys = 100%) in individual alpha chains of collagens α1α1α2(IV), α1α1α2(I), and α1α1α1(III). B, alkaline hydrolysis amino acid analysis. Histogram showing the occupancy of O-glycosylation attached to Hyl (GGHL + GHL + Hyl = 100) in purified collagens α1α1α2(IV) and mixture of collagens α1α1α2(I) and α1α1α1(III). Source data files for the figure: The value of each amino acid, the number of biological replicates, and detailed statistical analyses for data shown in the figure are presented in Table S3 and Fig. S8, respectively. GGHL, glucosyl galactosyl Hyl; GHL, galactosyl Hyl; Hyl, hydroxylysine; 3Hyp, 3-hydroxyproline; 4Hyp, 4-hydroxyproline; LH3, lysyl hydroxylase 3; Lys, lysine; Pro, proline.
Rare PLOD3 variants in fetuses with intracerebral hemorrhage
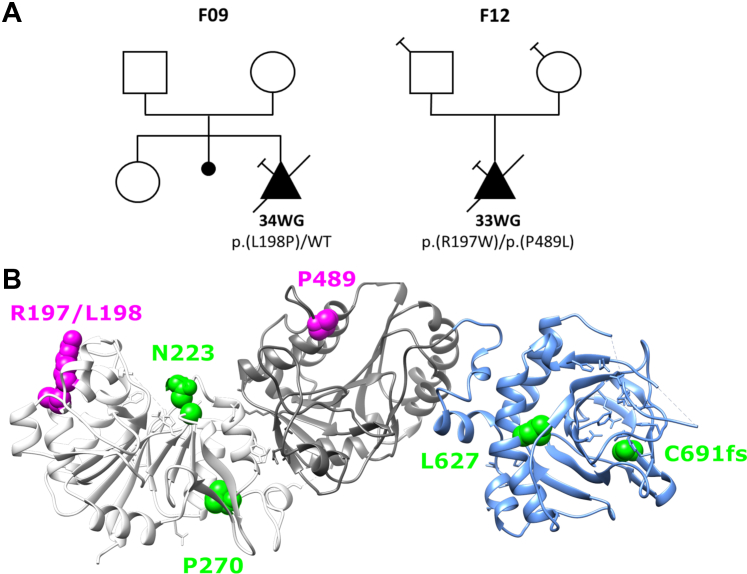
The role for LH3 in collagen α1α1α2(I) and collagen α1α1α1(III) glycosylation is consistent with the identification of PLOD3 pathogenic variants in individuals with connective tissue disorders (39, 41). However, our data demonstrate that LH3 plays an even greater role in collagen α1α1α2(IV) biosynthesis. Consistent with this finding, Plod3 deficiency in the mouse caused basement membrane defects (44, 45), suggesting that PLOD3 pathogenic variants may also contribute to intracerebral hemorrhages (ICHs), which constitute a central pathophysiological hallmark of Gould syndrome caused by COL4A1 and COL4A2 pathogenic variants (13, 14, 15, 67, 68, 69, 70, 71). In addition, cerebrovascular hemorrhages have been reported in three patients with a connective tissue disorder caused by biallelic pathogenic variants of PLOD3 (39, 43). To investigate the potential involvement of PLOD3 in ICH, we specifically evaluated the presence of PLOD3 variants in exome sequencing data from a fetal ICH cohort consisting of 113 probands who did not have COL4A1 or COL4A2 pathogenic variants. We did not find an enrichment of rare and predicted damaging PLOD3 qualifying variants between this cohort and the gnomAD control database (Table 1). However, in two unrelated fetuses, we identified three rare (<1/1000) PLOD3 missense variants—all three were predicted to be damaging by PolyPhen-2 (Fig. 5A and Table 2). Fetus F09 was heterozygous for p.L198P variant, and fetus F12 was compound heterozygous for p.R197W and p.P489L. Variants p.R197W and p.L198P are located in the catalytic glycosyltransferase domain close to a poly Asp sequence and predicted to affect the overall structure of active site in the transferase domain (50) (Fig. 5B). The p.P489L variant is located in the accessory glycosyltransferase domain and predicted to affect the substrate recognition (Fig. 5B). The two fetuses with predicted pathogenic variants had ICH identified on ultrasound follow up during the third trimester of pregnancy. Upon cerebral imaging, they presented a bilateral ventriculomegaly and intraparenchymal lesions (Table 3). Because of the severity of these lesions, the parents opted for termination of pregnancy. The lesions were confirmed by postmortem examination, and the fetuses showed some phenotypic differences. The fetus F12 had intraparenchymal hemorrhagic and ischemic lesions, whereas the fetus F09 had a microcephaly, a very thin corpus callosum, and a white matter atrophy. Neither had bone abnormalities or signs of external visceral malformation.
Table 1.
Enrichment analysis of rare predicted damaging PLOD3 variants in fetus cohort versus gnomAD cohorts
| gnomAD | Number of loss-of-function and missense variants with an MAF ≤1/1000 in PLOD3 gene | p |
|---|---|---|
| v2.1 (N = 141,456 individuals) | 2160 | 0.69 |
| v3.1 (N = 76,156 individuals) | 1225 | 0.7 |
MAF, minor allele frequency.
Figure 5.
Identification of four rare PLOD3 variants in fetal ICH cases.A, genealogical trees of the three families with PLOD3 variants. Square = male, circle = female, triangle = pregnancy not carried to term, black filled symbol = affected individual, empty symbol = clinically healthy relative, diagonal black line = deceased fetuses, and syringe symbol = blood sampled individual. The age of termination of pregnancy is indicated under each case. B, the crystal structure of the LH3 shows three different domains: catalytic glycosyltransferase domain (white), accessory glycosyltransferase domain (gray), and lysine dioxygenase domain (light blue). Missense variants identified in humans are indicated in different colors on the LH3 crystal structure. Green indicates previously reported variants have connective disorders resembling Stickler syndrome-like and epidermolysis bullosa. Magenta indicates variants identified in this study. ICH, intracerebral hemorrhage; LH3, lysyl hydroxylase 3.
Table 2.
Rare variants identified in PLOD3 gene in the cohort of 113 ICH fetuses
| Fetus | HGVSc position (NM_001084.5) | Protein change | Variant class | Status | gnomAD, v.2.1 |
PolyPhen-2 prediction | ACMG class |
|---|---|---|---|---|---|---|---|
| AF (%) | |||||||
| F09 | c.593T>C | p.(L198P) | Missense | HTZ | 0.0004 | Probably damaging (0.994) | 3 |
| F12a | c.589C>T | p.(R197W) | Missense | HTZ | 0.0012 | Possibly damaging (0.735) | 3 |
| F12a | c.1466C>T | p.(P489L) | Missense | HTZ | 0.051 | Possibly damaging (0.769) | 3 |
Abbreviations: ACMG, American College of Medical Genetics and Genomics; AF, allele frequency; HTZ, heterozygous.
ClinVar accession numbers: SCV002558784 (p.L198P), SCV002558785 (p.R197W), and SCV002558786 (p.P489L).
Biallelic variants (inherited from each parent).
Table 3.
Main clinical and pathological features of ICH-affected fetuses with PLOD3 variants
| Fetus | F09 | F12 |
|---|---|---|
| Sex | Female | Female |
| Gestation at diagnosis (weeks) | 31 | 30 |
| Brian imaging findings | Microcephaly, ventriculomegaly, interhemispheric cyst, porencephalic cavities, thin corpus callosum | Asymmetric bilateral ventriculomegaly, ischemic–hemorrhagic lesions in the right hemisphere |
| Outcome | TOP | TOP |
| Fetal brain examination | Important ventriculomegaly, white matter atrophy, severe microcephaly with diffuse bilateral lesions, absent or very thin corpus callosum | Asymmetric bilateral ventriculomegaly, intraparenchymal hemorrhages with cortical and subcortical ischemic foci |
| Skeletal X-ray examination | No sign of bone abnormality except for the absence of the 12th right rib | No sign of bone abnormality |
| Other autopsy anomalies | No sign of external or visceral malformation | No sign of external or visceral malformation |
Abbreviation: TOP, termination of pregnancy.
Discussion
To clarify the role(s) for LH3 in collagen biosynthesis, we generated LH3 KO cells using CRIPSR-mediated gene editing to conduct detailed quantitative and molecular analyses of collagens α1α1α2(I), α1α1α1(III), and α1α1α2(IV) implicated in OI, EDS, and Gould syndrome, respectively. We discovered that LH3 deficiency severely impaired biosynthesis of collagen α1α1α2(IV) but not collagen α1α1α2(I) or collagen α1α1α1(III). Our findings revealed altered PTMs in collagen α1α1α2(IV) produced by LH3 KO PFHR9 cells characterized by reduced and impaired modification of lysine to Hyl and galactosyl Hyl to glucosyl galactosyl Hyl. These PTM alterations prevented proper heterotrimer formation and HSP47 binding, which impaired collagen α1α1α2(IV) secretion. Notably, collagen α1α1α2(IV) that was secreted from LH3 KO PFHR9 cells showed impaired collagen α1α1α2(IV) network formation and failed to make higher order oligomers. In contrast, LH3 deficiency did not impact primary modifications to prolyl and lysyl residues for collagens α1α1α2(I) and α1α1α1(III) but specifically reduced modification of galactosyl Hyl to glucosyl galactosyl Hyl. Thus, we suggest that LH3 may be a selective LH at least for collagen α1α1α2(IV) and that network-forming and fibrillar collagens are both important substrates for LH3 glucosyltransferase activity.
Our results indicate that LH3 plays a critical role for collagen α1α1α2(IV) biosynthesis in PFHR9 cells but not for collagens α1α1α2(I) and α1α1α1(III) in MEFs. Surprisingly, Western blot analysis for PFHR9 cells showed that protein levels of intracellular COL4A2 in KO cells were similar to WT control cells; however, the protein migrated slightly faster and its secretion was significantly impaired (Fig. 1, B and C), indicating there could be a different quality control mechanism between COL4A1 and COL4A2 proteins. Although the majority of synthesized collagen α1α1α2(IV) was accumulated intracellularly in LH3 KO PFHR9 cells, some collagen α1α1α2(IV) was successfully secreted but did not have stable triple helical structures. Based on the current understanding of fibrillar collagen biosynthesis, this finding is intriguing for two reasons. First, the amount of 4Hyp is essential to the formation of triple helix rigidity (29). Type II collagen containing approximately 30% 4Hyp from P4ha1+/−, P4ha2−/− mice (P4ha1 and P4ha2 encoded prolyl 4-hydroxylase 1 and 2, respectively) decreased its thermal stability compared with a WT control; however, this type II collagen remained capable of forming a triple helical structure (72). Notably, the COL4A1 produced by LH3 KO PFHR cells showed a reduction of 4Hyp (Fig. 4A) but still contained around total 45% 4Hyp in the collagen α1α1α2(IV) heterotrimer isolated from LH3 KO PFHR9 cells, which is calculated by the amount of 4Hyp in COL4A1 and COL4A2 (Table S3). Second, the amounts of Hyl do not affect triple helix formation. Collagen α1α1α2(I) containing only 1.5% Hyl can form fully rigid triple helices and secrete into the extracellular matrix properly (24). Also, increased hydroxylysines in collagen α1α1α2(I) do not improve collagen thermal stability (73). Based on these observations, it is tempting to speculate that lysyl hydroxylation and/or the subsequent O-glycosylation critically contributes triple helix stability in collagen α1α1α2(IV).
Interestingly, the AHA pulse-chase experiments showed that the secretion rate of collagen α1α1α2(IV) was remarkably slower than that of collagens α1α1α2(I) and α1α1α1(III) (Figs. 1D and 2D). A significant difference between collagen α1α1α2(IV) and fibrillar collagens (type I, II, III, and V) is the presence of multiple interruptions of the collagenous repeats, which occur more frequently nearer the amino terminus (74). Repeat interruptions, which would be pathogenic if they occurred in fibrillar collagens, may interfere with propagation of triple helix formation, requiring renucleation of the three alpha chains, and likely contribute to the much longer secretion time for collagen α1α1α2(IV) compared with collagens α1α1α2(I) and α1α1α1(III). We hypothesize that the complement of PTMs near repeat interruptions may provide thermal or structural stability required for renucleation and propagation of the collagen α1α1α2(IV) triple helix. Supporting this notion, collagen α1α1α2(IV) from LH3 KO cells appeared shorted on rotary shadow images, which may suggest that the amino termini are disordered and cannot be resolved by electron microscopy. Alternatively, they may be more susceptible to cleavage, leading to a truncated protein of lower molecular weight as demonstrated by SDS gel electrophoreses and LC–MS analysis (Figs. 3, B, C and S7, B–D). Structural alterations of the 7S domain at the amino termini could be responsible for the inability to form higher order dodecamer structures in collagen α1α1α2(IV) isolated from LH3 KO PFHR9 cells (Figs. 3, B, C and S7, B–D). While the CD spectra showed that the overall secondary structure of collagen α1α1α2(IV) produced by LH3 KO was not fully triple helical (Fig. 3D), HSP47 was still able to bind with this collagen α1α1α2(IV). We speculate that this binding occurs on the triple helical carboxyl terminus region and weakly associate with the immature amino-terminal gelatin-like structure (Fig. 3E). Interestingly, small amounts of collagen α1α1α2(IV) were secreted from LH3 KO cells without a robust triple helical structure, which raises the question: what are the essential chemical or structural factors for collagen α1α1α2(IV) triple helix formation and for passing through quality control checkpoints in the ER and secretory pathway? Further experiments are required to provide additional mechanistic insights into the role of PTMs in collagen α1α1α2(IV) biosynthesis, secretion, and function(s).
Since our findings establish a critical role for LH3 in collagen α1α1α2(IV) biosynthesis, we predicted that PLOD3 might lead to clinical manifestations similar to those observed in individuals with COL4A1 and COL4A2 pathogenic variants causing Gould syndrome, a multisystem disorder frequently manifesting as fetal and perinatal ICH (13, 14, 15, 67, 68, 69, 70). Previously identified pathogenic PLOD3 missense variants cause phenotypically overlapping connective disorders including Stickler-like syndrome and epidermolysis bullosa (39, 40, 41, 42). In addition to the clinical features encountered in these two conditions, spontaneous ICH has been reported by Salo et al. (39) in two siblings. One of them was a female child with a connective tissue disorder, consisting of craniofacial dysmorphism, sensorineural deafness, and skeletal features, who presented a spontaneous large brain hematoma. Her male sibling was stillborn at 28 weeks of gestation; in addition to skeletal anomalies, autopsy of this fetus showed porencephaly. DNA analysis showed biallelic PLOD3 variants (p.N223S/p.C691AfsX9) located in a conserved region of the LH3 amino acid sequence responsible for collagen glycosyltransferase and LH activities. Recently, a homozygous PLOD3 small inframe deletion was shown to cause a cerebral small vessel disease characterized by multiple white matter hypersignals and bleeding foci in an infant with a few dysmorphic features (43). To evaluate the contribution of PLOD3 pathogenic variants to cerebrovascular manifestations, we analyzed a series of 113 cases of fetal ICH that were negative for COL4A1 and COL4A2 pathogenic variants. We identified a rare and predicted damaging heterozygous variant in one fetus (F09) and two distinct biallelic candidate variants in another fetus (F12) (Fig. 5 and Table 2). When comparing the whole cohort of fetuses to control databases, we did not detect an enrichment in rare and predicted damaging PLOD3 variants; however, this lack of enrichment could be attributable to the high genetic heterogeneity of fetal ICH (71). Additional work is required to establish the causality of the heterozygous PLOD3 variant present in fetus F09. Indeed, connective tissue disorders associated with PLOD3 are typically autosomal recessive, and heterozygous parents of affected cases are reported to be asymptomatic. We cannot exclude however that some heterozygous PLOD3 variants could have dominant negative effects or that they could act as risk factors acting in concert with other triggering events to cause fetal ICH. For fetus F12, who is a compound heterozygote with both variants being predicted to be possibly pathogenic, no tissue was available to evaluate the consequences of the two variants at the protein level. Autopsy data including skeletal X-ray data were normal. Additional in vitro experiments are required to determine the causality of these variants, and larger cohorts of fetuses and children with ICH should be analyzed to identify additional cases.
In conclusion, our results suggest that LH3 plays a critical and multifunctional role in collagen α1α1α2(IV) biosynthesis, and that PLOD3 variants could lead to clinical manifestations associated with COL4A1 and COL4A2 pathogenic variants.
Experimental procedures
Cell culture
PFHR9 cells (CRL-2423) and MEFs (CRL-1503) were purchased from American Type Culture Collection. These cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/high glucose/pyruvate (Gibco; catalog no.: 11995065) supplemented with 10% (v/v) fetal bovine serum (Atlanta Biologicals), pencillin streptomycin glutamine 100× (Gibco; catalog no.: 10378016), and 5 mM Hepes in the presence of ascorbic acid phosphate (100 μg/ml; Wako Chemicals). After this section, standard DMEM indicates the aforementioned DMEM.
CRISPR-mediated KO of LH3
LH3 was knocked out in PFHR9 cells and MEFs using two CRISPR/Cas9 All-in-One plasmid of pSpCas9(BB)-2A-Puro (PX459) v2.0 and pSpCas9(BB)-2A-GFP (PX458) including the LH3 guide RNA (gRNA) sequence for Plod3 exon 5 (5′-TCCACTGGCGGACAATCTGATGG-3′) and exon 7 (5′-TCGTGTGCGCATCCGGAATGTGG-3′), which were designed and purchased from GenScript, respectively. Two plasmids were transfected into the cells using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions. After 24 h, transfected cells were treated with 10 μg/ml puromycin (Gibco) to select cells containing the PX459 plasmid. After puromycin selection, the GFP-positive cells containing the PX458 plasmid were isolated using a corning ring and cultured them with standard DMEM.
DNA sequencing
To confirm LH3 gRNA edited Plod3 exon 5 and 7, PCR products including the region targeted with gRNA were sequenced. DNA was prepared from WT and LH3 KO cells. Cells were plated onto 6 cm dish and grown to 100% confluency. Cells were collected by centrifugation after treating with trypsin (Promega). This cell pellet was digested by 3 mg/ml proteinase K in 20 μl of 50 mM Tris–HCl buffer containing 20 mM NaCl, 1% SDS, and 1 mM EDTA, pH 8.0, at 95 °C for 5 min after preincubating at 60 °C for 60 min. After the digestion, 200 μl of water was added to this solution as a template for PCR. PCR was performed with primer set A (forward: 5′-AAGCGCTTCCTCAACTCTG-3′ and reverse: 5′-TCCCTCATTACCCACCTCTAA-3′) and B (forward: 5′-ACTCCTGAGCATGTGTGTTAG-3′ and reverse 5′-ACAGGGTTGAGTGGCTAAAG-3′) for exon 5 and 7, respectively. Sequencing samples including the forward primer were sent to Quintara Biosciences. Sequencing results were analyzed by GeneStudio Professional Edition, version 2.2.0.0 (GeneStudio, Inc).
Protein analyses in cell lysate and conditioned culture medium
WT and LH3 KO cells were plated and grown to 80 to 90% confluency. Following stimulation of procollagen biosynthesis with ascorbic acid for 1 day, the medium was replaced to the fresh standard DMEM, and the cells were cultured for 2 days. The medium containing secreted proteins was dissolved in Bolt LDS sample buffer 4× (Life Technologies) with reducing agents. After the cells were washed with PBS once, cell lysates were extracted using M-PER (Thermo Fisher Scientific) containing Halt Protease Inhibitor Cocktail, EDTA-Free (Thermo Fisher Scientific) at 4 °C according to the manufacturer’s instructions. After centrifugation, soluble proteins in the supernatant were mixed with the same sample buffer with reducing agents. These protein solutions were separated on SDS-PAGE gels and then transferred to polyvinylidene difluoride (PVDF) membranes, and Western blots were performed. Gels, buffers, transfer condition, and antibodies are listed in Table S4. Blots were developed with horseradish peroxidase–enhanced Super-Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and detected by ChemiDoc MP imaging system (Bio-Rad) using the software Image Lab, version 4.0.1 (Bio-Rad). The intensities of protein signals were measured by ImageJ (National Institutes of Health).
Secretion rate assay for PFHR9 cells
WT and LH3 KO cells were cultured in a 16 cm dish grown to 100% confluency and equally plated to three 6-well tissue culture plates after the cells were trypsinized. Following stimulation of procollagen biosynthesis with ascorbic acid overnight, the cells were preincubated in pulse medium (Met- and Cys-free DMEM with 10% fetal bovine serum and 100 μg/ml ascorbic acid 2-phosphate) without AHA (Click Chemistry Tools) at 37 °C for 30 min to deplete methionine. Pulse labeling was performed with 1 mM AHA in pulse medium at 37 °C for 15 min, and cells were incubated in 1.5 ml chase medium (standard DMEM) at 37 °C for different time intervals (i.e., every 30 min by 360 min, 420 min 480 min, 24 h and 48 h). The conditioned culture medium was collected at each time point and frozen at −20 °C. After thawing and centrifuging at high speed to remove residual cells and large aggregates, 1.4 ml of medium was mixed with 1.4 ml of 0.1 M Tris–HCl containing 1.0 M urea, pH 7.5. This solution was incubated with 0.4 ml of Q Sepharose Fast Flow (GE Healthcare) to remove AHA-incorporated laminins at room temperature for an hour. After centrifugation at high speed for 10 min, 2.8 ml supernatant was mixed with 0.2 ml 10 M acetic acid and 1 ml of 5 M NaCl at 4 °C overnight. Collagen α1α1α2(IV) was precipitated at the highest speed by a tabletop centrifuge. The pellet containing collagen α1α1α2(IV) was dissolved in 27 μl of Tris-buffered saline and labeled with 3 μl of Click-iT Alexa Fluor 488 sDIBO Alkyne (Thermo Scientific) at room temperature for an hour. In LH3 KO secretion rate assay, the pellets from two wells in the 6-well plate were combined to detect adequate signals. This labeled solution was mixed with 10 μl of 4× Bolt LDS sample buffer with reducing agent and separated on precast 6% Tris–glycine gel (Invitrogen). The signals from AHA-Alexa Fluor 488 were detected by the ChemiDoc MP imaging system using the software Image Lab, version 4.0.1. The intensities of fluorescent signals were measured by ImageJ. The AHA-incorporated collagen α1α1α2(IV) as a loading control was purified from conditioned culture medium after an hour pulse and the overnight chase. The purification method is described in the section “Collagen α1α1α2(IV) purification” later. This assay was performed five times, and representative images are shown.
Secretion rate assay for MEFs
WT and LH3 KO cells were cultured in a 16 cm dish grown to 100% confluency and equally plated to five 10 cm dishes plate after the cells were trypsinized. Following stimulation of procollagen biosynthesis with ascorbic acid overnight, cells were preincubated in pulse medium without AHA at 37 °C for 30 min to deplete methionine. Pulse labeling was performed with 1 mM AHA in pulse medium at 37 °C for 15 min, and cells were incubated in 3.7 ml chase medium (standard DMEM) at 37 °C for different time intervals (i.e., every 30, 60, 90, 120, and 180 min). The conditioned culture medium was collected at each time point and frozen at −20 °C. After thawing and centrifuging at high speed to remove residual cells and large aggregates, 3.5 ml of medium was mixed with 0.5 ml 10 M acetic acid and 20 μl of 100 mg/ml pepsin at 4 °C overnight. After adding 1.0 ml of 5 M NaCl to the pepsin-treated solution and incubating at 4 °C overnight, collagens α1α1α2(I) and α1α1α1(III) were precipitated at the highest speed by a tabletop centrifuge. The pellet containing collagens α1α1α2(I) and α1α1α1(III) was dissolved in 27 μl of Tris-buffered saline and labeled with 3 μl of Click-iT Alexa Fluor 488 sDIBO Alkyne at room temperature for an hour. This labeled solution was mixed with 10 μl of 4× Bolt LDS sample buffer without reducing agent and separated on precast 6% Tris–glycine gel. The signals from AHA-Alexa Fluor 488 were detected by the ChemiDoc MP imaging system using the software Image Lab, version 4.0.1. The intensities of fluorescent signals were measured by ImageJ. The AHA-incorporated type I and type III collagen as a loading control was purified from conditioned culture medium after an hour pulse and the overnight chase. The purification method is described in the section”Collagens α1α1α2(I) and α1α1α1(III) purification” later. This assay was performed five times, and representative images are shown.
Collagen α1α1α2(IV) purification
Extraction of collagen α1α1α2(IV) was performed using the standard DMEM with PFHR9 cells. DMEM was prepared by the same procedures as described in the aforementioned “Protein level analyses” section, except using serum-free DMEM instead of the standard DMEM. The DMEM was dialyzed with 0.2 M acetic acid at 4 °C overnight. After spinning down to remove residual cells or aggregates, the DMEM was dialyzed with 0.2 M acetic acid containing 1.2 M NaCl at 4 °C overnight. The precipitates including collagen α1α1α2(IV) were collected by centrifugation at 13,000 rpm for 20 min using JA-14 rotor (Beckman) and resuspended with 50 mM Tris–HCl buffer containing 25 mM NaCl and 0.5 M urea, pH 7.5. After dialysis to the same buffer at 4 °C overnight and centrifugation at 13,000 rpm for 10 min using JA-20 rotor (Beckman), the solution was applied to HiTrap Q FF column (cytiva), and the flow through fraction was collected. The flow through was dialyzed to 50 mM Mes buffer containing 25 mM NaCl and 0.5 M urea, pH 6.0, at 4 °C overnight and applied to HiTrap SP HP column (cytiva). After washing with the same Mes buffer at least five column volumes, collagen α1α1α2(IV) was eluted by 50 mM Mes buffer containing 250 mM NaCl and 0.5 M urea, pH 6.0. The fractions containing collagen α1α1α2(IV) were concentrated using Amicon Ultra centrifugal filters Ultracel—100 K (Millipore) at the speed × 100g up to 1.5 ml as a total volume. The concentrated fraction was dialyzed to 0.2 M acetic acid at 4 °C overnight, and 0.5 ml of 5 M NaCl was added (final concentration of 1.25 M NaCl) to the dialyzed solution. After mixing at 4 °C overnight, collagen α1α1α2(IV) was precipitated by centrifugation at 14,000 rpm for 20 min using a tabletop centrifuge. The pellet was resuspended with 0.2 M acetic acid dialyzed to 0.2 M acetic acid at 4 °C overnight to remove residual NaCl for experiments.
Collagens α1α1α2(I) and α1α1α1(III) purification
Extraction of collagens α1α1α2(I) and α1α1α1(III) was performed using the cultured standard DMEM with MEFs. The DMEM was prepared by the same procedures as described in aforementioned “Protein level analyses” section. The DMEM was dialyzed with 0.5 M acetic acid at 4 °C overnight. After spinning down to remove residual cells or aggregates, the DMEM was incubated with pepsin added to a final concentration of 0.25 mg/ml at 4 °C overnight. The pepsin-treated DMEM was dialyzed with 0.5 M acetic acid containing 0.7 M NaCl at 4 °C overnight. The precipitates including collagens α1α1α2(I) and α1α1α1(III) were collected by centrifugation at 13,000 rpm for 20 min using JA-14 rotor. The pellet was resuspended in 0.2 M acetic acid, and, this solution contained enriched collagens. α1α1α2(I) and α1α1α1(III) were dialyzed to 0.2 M acetic acid at 4 °C overnight to remove residual NaCl for experiments.
SDS-gel analyses
To check purity, the isolated collagens were run on a Bolt 4 to 12% Bis–Tris Plus gel (Invitrogen) in the presence or the absence of DTT with Mes running buffer (Novex) after boiling denaturation. To compare differences in migration, the purified collagens α1α1α2(I) and α1α1α1(III) mixture were run on a 3 to 8% Tris–acetate gel (Invitrogen) in the presence or the absence of DTT with boiling denaturation. To demonstrate higher order collagen α1α1α2(IV) assemblies, SDS-agarose gel electrophoresis was performed for purified WT and LH3 KO collagen α1α1α2(IV). About 3.5% agarose was dissolved in 0.2 M Tris–HCl, pH 8.8. After measuring the total weight of the beaker and agarose solution, the beaker was heated in a microwave oven and gently swirled to thoroughly mix the agarose solution. After adding 20% SDS up to 0.25% and sufficient hot distilled water to obtain the initial weight, the SDS-agarose solution was mixed thoroughly again. Bio-Rad Mini-PROTEAN Tetra Handcast Systems was adapted to cast the gel, and hot SDS-agarose solution was loaded between preheated glass plates of a vertical electrophoresis apparatus. Four different sample conditions were prepared using purified WT and LH3 KO collagen α1α1α2(IV) in the presence or the absence of DTT with and without boiling denaturation. The samples were separated on a SDS—3.5% agarose gels with Tris–glycine SDS running buffer using 75 V constant voltage at 4 °C for about 2 h. All gels were stained with GelCode Blue Stain Reagent (Thermo Scientific). For PTM analysis, we used 6% Tris–glycine SDS-PAGE and 3.0% SDS-agarose gel to separate individual collagen chains of collagens α1α1α2(I) and α1α1α1(III) and collagen α1α1α2(IV) under reducing condition, respectively. After gels were electrophoretically transferred to PVDF membranes with 0.05% SDS, the membranes were stained with Ponceau S.
Protein identification by LC–MS
Gel bands were reduced with 10 mM DTT at 56 °C for 30 min followed by alkylation with 50 mM iodoacetamide at room temperature for 30 min. The samples were digested with trypsin at 37 °C for 16 h, and the generated tryptic peptides were sequentially extracted from the gels with 5% formic acid, 5% formic acid/50% acetonitrile, and 5% formic acid/95% acetonitrile. The extracted solutions were concentrated by a centrifugal evaporator CVE-3100 (EYELA) and then analyzed by LC–MS on a maXis II quadrupole time-of-flight mass spectrometer (Bruker Daltonics) coupled to a Shimadzu Prominence UFLC-XR system (Shimadzu) with chromatographic separation using an Ascentis Express Peptide ES-C18 column (2.7 μm particle size, L × I.D. 150 mm × 2.1 mm; Supelco) (75). The mass spectrometry (MS) scan and MS/MS acquisition were performed over the m/z ranges of 50 to 2500 with a frequency of 5 Hz. The acquired MS/MS spectra were searched against the UniProtKB/Swiss-Prot database (release 2018_05) for Mus musculus species using ProteinPilot software 4.5 (AB Sciex), as described previously (24).
Rotary shadow electron microscopy
Rotary shadowing was performed by methods described previously (76). In brief, samples were prepared to mix WT and LH3 KO collagen α1α1α2(IV) (approximate concentrations of 100 μg/ml) in 0.2 M acetic acid with glycerol to a final concentration of 70% glycerol. Each protein was nebulized onto freshly cleaved mica chips using an airbrush. The samples were then rotary shadowed with carbon/platinum using an electron beam gun within a vacuum evaporator. Images were acquired using an FEI G20 transmission electron microscope. Proportions of different heterotrimer structures were obtained from images of WT (n = 690) and LH3 KO (n = 295) molecules, and representative images were used in Figure 4C.
CD
CD spectra were recorded on an AVIV 202 spectropolarimeter (AVIV Biomedical, Inc) using a Peltier thermostat–controlled cell holder and a 1 mm path length rectangular quartz cell (Starna Cells, Inc). Protein concentrations were determined by acid hydrolysis amino acid analysis described in the section ”PTM analysis by LC–MS” and using collagen solution instead of PVDF membrane. Both WT and LH3 KO collagen α1α1α2(IV) were measured in 0.2 M acetic acid at 10 °C. The denatured WT collagen α1α1α2(IV) was measured using the cuvette after the measurements at 10 °C with heating up to 65 °C for 1 h. The spectra represent the average of at least three scans recorded at a wavelength resolution of 0.1 nm.
SPR analysis
SPR experiments were carried out using a BIAcore X instrument (GE Healthcare). Purified WT and LH3 KO collagen α1α1α2(IV) were immobilized on a CM5 sensor chip by amide coupling. The approximate coupled protein concentration was 1.2 ng/mm2 (1200 response units) and 1.4 ng/mm2 (1400 response units) of WT and LH3 KO collagen α1α1α2(IV), respectively. The experiments were performed at 20 °C in 10 mM Hepes, pH 7.4, containing 150 mM NaCl and 0.005% Surfactant P20 using a flow rate of 10 μl/min. All curves are the average of at least three replicates, and three independent measurements were performed. For the analysis of the binding affinity, the curves were fitted with the Langmuir binding model (BIAevaluation software; GE Healthcare). Hsp47 were purified from 17-day-old chicken embryos using methods described previously (77).
PTM analysis by LC–MS
The hydroxylation rate of Lys (Lys + Hyl = 100%) and Pro (Pro + 4-Hyp + 3-Hyp = 100%) in each α chain of collagens α1α1α2(IV), α1α1α2(I), and α1α1α1(III) was evaluated by LC–MS after acid hydrolysis, as described previously (78). In brief, after respective α chains were transferred to PVDF membranes as described in aforementioned SDS-gel analysis, the membrane bands were excised and subjected to acid hydrolysis (6 N HCl/1% phenol, 110 °C for 20 h in the gas phase under N2) after adding stable isotope–labeled collagen (SI-collagen) (79) as an internal standard. The acid hydrolysates were analyzed by LC–MS in multiple reaction monitoring mode on a QTRAP 5500+ hybrid triple quadrupole/linear ion trap mass spectrometer (AB Sciex) coupled to an ExionLC AD HPLC system (AB Sciex) with a ZIC-HILIC column (3.5 μm particle size, L × I.D. 150 mm × 2.1 mm; Merck Millipore). The content of Pro, 3-Hyp, 4-Hyp, Lys, and Hyl was quantitated by the peak area ratio of the analytes relative to the corresponding stable isotopically heavy analytes derived from SI-collagen (79). The occupancy of O-glycosylations attached to Hyl (free Hyl + galactosyl Hyl + glucosyl galactosyl Hyl = 100) in the collagens α1α1α2(I) and α1α1α1(III) mixture and purified collagen α1α1α2(IV) was evaluated by LC–MS after alkaline hydrolysis, as described previously (79). In brief, the purified collagen samples were subjected to alkaline hydrolysis (2 N NaOH, 110 °C for 20 h under N2) after adding SI-collagen as the internal standard. The content of free Hyl, galactosyl Hyl, and glucosyl galactosyl Hyl was quantitated by LC–MS analysis of the alkaline hydrolysates as described previously.
Human material and ethics statement
About 113 unrelated fetuses affected by an ICH grade III or IV were included following termination of pregnancy or intrauterine fetal death. They were referred for COL4A1/COL4A2 screening to the French national molecular genetics reference center for inherited cerebrovascular disorders (Saint Louis Hospital, Paris) and shown to be negative using a targeted high-throughput sequencing (conditions available upon request). Fetal ICH was in most cases detected at systematic second and third trimester ultrasound examinations and in some cases confirmed by fetal MRI and/or pathological examination. Systematic review of medical charts was performed in order to exclude fetuses with an identifiable cause or known risk factor for ICH, including evidence of maternal trauma during pregnancy, cocaine or maternal drug use, maternal or neonatal infections, and fetal alloimmune thrombocytopenia. Written informed consent for genetic investigation and research was provided by parents in accordance with the declaration of Helsinki and the French law. This study has been approved by the Inserm Ethics Committee (INSERM IRB00003888). Genomic DNA was isolated from postmortem fetal tissue and peripheral blood leukocytes of both parents and relatives when available. Prior to whole exome sequencing, we excluded the presence of a mutation in COL4A1 and COL4A2 genes by performing a targeted high-throughput sequencing (conditions available upon request).
Exome sequencing
Exon capture was performed at the IntegraGen platform (Evry) for fetus probands and relatives using the SureSelect Human All Exon V5-UTR (Agilent Technologies) or the Twist Human Core Exome Enrichment System (Twist Bioscience). Followed by 100 base pair paired-end sequencing using an Illumina NovaSeq platform. Data analysis was performed with the IntegraGen in house bioinformatic pipeline. Sequence reads were aligned to the human genome reference GRCh38/hg38 using BWA. Variant calling for the identification of single nucleotide variations, and small insertions/deletions were performed via the Broad Institute’s GATK Haplotype Caller GVCF tool (GATK 3.8.1). Ensembl’s VEP (Variant Effect Predictor, release VEP 95.1) program was used to process variants for further annotations. Allele frequency annotations was based on gnomAD (version 2.1.1) and 1000Genomes datasets. PolyPhen-2 algorithm was used to predict deleteriousness of missense variants. Finally, we used CANOES for the detection of copy number variation in whole exome sequencing data (80).
Qualifying variants in the PLOD3 gene
Several criteria were used for the selection of qualifying variants in the PLOD3 gene: (1) quality control: a minimum of 10 reads was required for depth coverage and a minimum of 25% for allelic balance, (2) variants’ nature: all coding and splice site variants were retained, (3) minor allele frequency ≤1/1000 in external databases (gnomAD, version 2.1.1 and 1000Genomes phase 3), and (4) for missense variants, predicted possibly or probably damaging with PolyPhen-2. Investigation for variant enrichment in the PLOD3 gene was performed by comparing our fetus cohort (N = 113) to cohorts of control individuals in gnomAD v2.1 and v3.1 with the same filter criteria. Burden tests/p values were used for statistical analyses.
Mapping of the amino acid residues in protein structure
The human LH3 crystal structure was retrieved from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (www.rcsb.org, accession number: 6FXK) and used for variant modeling using the UCSF Chimera software (version 1.14; build 42094).
Statistical analyses
For comparisons between two groups, one-way ANOVA was performed to determine whether differences between groups are significant using OriginPro, version 9.1 (OriginLab Corp). A p value of less than 0.05 was considered statistically significant.
Data availability
All data are contained within the article and supporting information. All source data are available from the corresponding author upon reasonable request. The MS datasets for protein identification of collagen α1α1α2(IV) samples have been deposited to the ProteomeXchange consortium via the jPOST partner repository with the dataset identifier PXD035051 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD035051).
(References: https://www.nature.com/articles/nbt.2839 and https://academic.oup.com/nar/article/45/D1/D1107/2605695?login=false).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We gratefully acknowledge the research department of Shriners Hospitals for Children in Portland for using CD instrument. We thank Hans Peter Bächinger and Cassandre Labelle-Dumais for useful discussions and suggestions on this work. Research reported in this publication was supported in part by the UCSF Vision Core shared resource of the National Institutes of Health/National Eye Institute P30 EY002162 and by an unrestricted grant from Research to Prevent Blindness, New York, NY.
Author contributions
Y. I., K. M., E. T.-L., and D. B. G. conceptualization; Y. I., Y. T., S. F. T., and D. R. K. methodology; Y. I., Y. T., T. C., S. F. T., and D. R. K. validation; Y. I., Y. T., T. C., S. F. T., and D. R. K. formal analysis; Y. I., Y. T., T. C., S. F. T., and D. R. K. investigation; Y. I. and T. C. resources; Y. I., Y. T., T. C., S. F. T., D. R. K., K. M., E. T.-L., and D. B. G. data curation; Y. I. and D. B. G. writing–original draft; Y. I., Y. T., T. C., S. F. T., D. R. K., K. M., E. T.-L., and D. B. G. writing–review & editing; Y. I. visualization; Y. I., E. T.-L., and D. B. G. supervision; Y. I., E. T.-L., and D. B. G. project administration; Y. I. and D. B. G. funding acquisition.
Funding and additional information
Research reported in this publication was supported by All May See Foundation award 7031182 (to Y. I.) and the National Institutes of Health under award number R01NS096173 (to D. B. G.). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agencies.
Edited by Ursula Jakob
Contributor Information
Yoshihiro Ishikawa, Email: yoshihiro.ishikawa@uscf.edu.
Douglas B. Gould, Email: douglas.gould@ucsf.edu.
Supporting information
References
- 1.Bella J., Hulmes D.J. Fibrillar collagens. Subcell Biochem. 2017;82:457–490. doi: 10.1007/978-3-319-49674-0_14. [DOI] [PubMed] [Google Scholar]
- 2.Ricard-Blum S., Ruggiero F. The collagen superfamily: from the extracellular matrix to the cell membrane. Pathol. Biol. 2005;53:430–442. doi: 10.1016/j.patbio.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Lamande S.R., Bateman J.F. Genetic disorders of the extracellular matrix. Anat. Rec. (Hoboken) 2020;303:1527–1542. doi: 10.1002/ar.24086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman J.F., Boot-Handford R.P., Lamande S.R. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 5.Marini J.C., Forlino A., Bachinger H.P., Bishop N.J., Byers P.H., Paepe A., et al. Osteogenesis imperfecta. Nat. Rev. Dis. Primers. 2017;3 doi: 10.1038/nrdp.2017.52. [DOI] [PubMed] [Google Scholar]
- 6.Malfait F., Castori M., Francomano C.A., Giunta C., Kosho T., Byers P.H. The Ehlers-Danlos syndromes. Nat. Rev. Dis. Primers. 2020;6:64. doi: 10.1038/s41572-020-0194-9. [DOI] [PubMed] [Google Scholar]
- 7.Besio R., Chow C.W., Tonelli F., Marini J.C., Forlino A. Bone biology: insights from osteogenesis imperfecta and related rare fragility syndromes. FEBS J. 2019;286:3033–3056. doi: 10.1111/febs.14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claeys L., Storoni S., Eekhoff M., Elting M., Wisse L., Pals G., et al. Collagen transport and related pathways in Osteogenesis Imperfecta. Hum. Genet. 2021;140:1121–1141. doi: 10.1007/s00439-021-02302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omar R., Malfait F., Van Agtmael T. Four decades in the making: collagen III and mechanisms of vascular Ehlers danlos syndrome. Matrix Biol. Plus. 2021;12 doi: 10.1016/j.mbplus.2021.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Damme T., Colman M., Syx D., Malfait F. The ehlers-danlos syndromes against the backdrop of inborn errors of metabolism. Genes (Basel) 2022;13:265. doi: 10.3390/genes13020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson B.G., Tryggvason K., Sundaramoorthy M., Neilson E.G. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N. Engl. J. Med. 2003;348:2543–2556. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]
- 12.Boudko S.P., Pokidysheva E., Hudson B.G. Prospective collagen IValpha345 therapies for Alport syndrome. Curr. Opin. Nephrol. Hypertens. 2022;31:213–220. doi: 10.1097/MNH.0000000000000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeanne M., Gould D.B. Genotype-phenotype correlations in pathology caused by collagen type IV alpha 1 and 2 mutations. Matrix Biol. 2017;57-58:29–44. doi: 10.1016/j.matbio.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao M., Alavi M.V., Labelle-Dumais C., Gould D.B. Type IV collagens and basement membrane diseases: cell biology and pathogenic mechanisms. Curr. Top Membr. 2015;76:61–116. doi: 10.1016/bs.ctm.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Kuo D.S., Labelle-Dumais C., Gould D.B. COL4A1 and COL4A2 mutations and disease: Insights into pathogenic mechanisms and potential therapeutic targets. Hum. Mol. Genet. 2012;21:R97–110. doi: 10.1093/hmg/dds346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyce D., McGee S., Shank L., Pathak S., Gould D. Epilepsy and related challenges in children with COL4A1 and COL4A2 mutations: a Gould syndrome patient registry. Epilepsy Behav. 2021;125 doi: 10.1016/j.yebeh.2021.108365. [DOI] [PubMed] [Google Scholar]
- 17.Mao M., Kiss M., Ou Y., Gould D.B. Genetic dissection of anterior segment dysgenesis caused by a Col4a1 mutation in mouse. Dis. Model Mech. 2017;10:475–485. doi: 10.1242/dmm.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao M., Smith R.S., Alavi M.V., Marchant J.K., Cosma M., Libby R.T., et al. Strain-dependent anterior segment dysgenesis and progression to glaucoma in Col4a1 mutant mice. Invest. Ophthalmol. Vis. Sci. 2015;56:6823–6831. doi: 10.1167/iovs.15-17527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa Y., Bachinger H.P. A molecular ensemble in the rER for procollagen maturation. Biochim. Biophys. Acta. 2013;1833:2479–2491. doi: 10.1016/j.bbamcr.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Ito S., Nagata K. Quality control of procollagen in cells. Annu. Rev. Biochem. 2021;90:631–658. doi: 10.1146/annurev-biochem-013118-111603. [DOI] [PubMed] [Google Scholar]
- 21.Onursal C., Dick E., Angelidis I., Schiller H.B., Staab-Weijnitz C.A. Collagen biosynthesis, processing, and maturation in lung ageing. Front. Med. (Lausanne) 2021;8 doi: 10.3389/fmed.2021.593874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ihme A., Krieg T., Nerlich A., Feldmann U., Rauterberg J., Glanville R.W., et al. Ehlers-danlos syndrome type VI: collagen type specificity of defective lysyl hydroxylation in various tissues. J. Invest. Dermatol. 1984;83:161–165. doi: 10.1111/1523-1747.ep12263502. [DOI] [PubMed] [Google Scholar]
- 23.Weis M.A., Hudson D.M., Kim L., Scott M., Wu J.J., Eyre D.R. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J. Biol. Chem. 2010;285:2580–2590. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa Y., Taga Y., Zientek K., Mizuno N., Salo A.M., Semenova O., et al. Type I and type V procollagen triple helix uses different subsets of the molecular ensemble for lysine posttranslational modifications in the rER. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa Y., Bächinger H.P. A substrate preference for the rough endoplasmic reticulum resident protein FKBP22 during collagen biosynthesis. J. Biol. Chem. 2014;289:18189–18201. doi: 10.1074/jbc.M114.561944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. Developmental stage-dependent regulation of prolyl 3-hydroxylation in tendon type I collagen. J. Biol. Chem. 2016;291:837–847. doi: 10.1074/jbc.M115.686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salo A.M., Myllyharju J. Prolyl and lysyl hydroxylases in collagen synthesis. Exp. Dermatol. 2021;30:38–49. doi: 10.1111/exd.14197. [DOI] [PubMed] [Google Scholar]
- 28.Basak T., Vega-Montoto L., Zimmerman L.J., Tabb D.L., Hudson B.G., Vanacore R.M. Comprehensive characterization of glycosylation and hydroxylation of basement membrane collagen IV by high-resolution mass spectrometry. J. Proteome Res. 2016;15:245–258. doi: 10.1021/acs.jproteome.5b00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg R.A., Prockop D.J. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem. Biophys. Res. Commun. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 30.Mizuno K., Hayashi T., Peyton D.H., Bachinger H.P. Hydroxylation-induced stabilization of the collagen triple helix. Acetyl-(glycyl-4(R)-hydroxyprolyl-4(R)-hydroxyprolyl)(10)-NH(2) forms a highly stable triple helix. J. Biol. Chem. 2004;279:38072–38078. doi: 10.1074/jbc.M402953200. [DOI] [PubMed] [Google Scholar]
- 31.Taga Y., Tanaka K., Hattori S., Mizuno K. In-depth correlation analysis demonstrates that 4-hydroxyproline at the Yaa position of Gly-Xaa-Yaa repeats dominantly stabilizes collagen triple helix. Matrix Biol. Plus. 2021;10 doi: 10.1016/j.mbplus.2021.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montgomery N.T., Zientek K.D., Pokidysheva E.N., Bachinger H.P. Post-translational modification of type IV collagen with 3-hydroxyproline affects its interactions with glycoprotein VI and nidogens 1 and 2. J. Biol. Chem. 2018;293:5987–5999. doi: 10.1074/jbc.RA117.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grafe I., Yang T., Alexander S., Homan E.P., Lietman C., Jiang M.M., et al. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 2014;20:670–675. doi: 10.1038/nm.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hennet T. Collagen glycosylation. Curr. Opin. Struct. Biol. 2019;56:131–138. doi: 10.1016/j.sbi.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 35.De Giorgi F., Fumagalli M., Scietti L., Forneris F. Collagen hydroxylysine glycosylation: non-conventional substrates for atypical glycosyltransferase enzymes. Biochem. Soc. Trans. 2021;49:855–866. doi: 10.1042/BST20200767. [DOI] [PubMed] [Google Scholar]
- 36.Geister K.A., Lopez-Jimenez A.J., Houghtaling S., Ho T.H., Vanacore R., Beier D.R. Loss of function of Colgalt1 disrupts collagen post-translational modification and causes musculoskeletal defects. Dis. Model Mech. 2019;12 doi: 10.1242/dmm.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyatake S., Schneeberger S., Koyama N., Yokochi K., Ohmura K., Shiina M., et al. Biallelic COLGALT1 variants are associated with cerebral small vessel disease. Ann. Neurol. 2018;84:843–853. doi: 10.1002/ana.25367. [DOI] [PubMed] [Google Scholar]
- 38.Teunissen M.W.A., Kamsteeg E.J., Sallevelt S., Pennings M., Bauer N.J.C., Vermeulen R.J., et al. Biallelic variants in the COLGALT1 gene causes severe congenital porencephaly: a case report. Neurol. Genet. 2021;7:e564. doi: 10.1212/NXG.0000000000000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salo A.M., Cox H., Farndon P., Moss C., Grindulis H., Risteli M., et al. A connective tissue disorder caused by mutations of the lysyl hydroxylase 3 gene. Am. J. Hum. Genet. 2008;83:495–503. doi: 10.1016/j.ajhg.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewans L.J., Colley A., Gaston-Massuet C., Gualtieri A., Cowley M.J., McCabe M.J., et al. Pathogenic variants in PLOD3 result in a Stickler syndrome-like connective tissue disorder with vascular complications. J. Med. Genet. 2019;56:629–638. doi: 10.1136/jmedgenet-2019-106019. [DOI] [PubMed] [Google Scholar]
- 41.Vahidnezhad H., Youssefian L., Saeidian A.H., Touati A., Pajouhanfar S., Baghdadi T., et al. Mutations in PLOD3, encoding lysyl hydroxylase 3, cause a complex connective tissue disorder including recessive dystrophic epidermolysis bullosa-like blistering phenotype with abnormal anchoring fibrils and type VII collagen deficiency. Matrix Biol. 2019;81:91–106. doi: 10.1016/j.matbio.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Maddirevula S., Alzahrani F., Al-Owain M., Al Muhaizea M.A., Kayyali H.R., AlHashem A., et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet. Med. 2019;21:736–742. doi: 10.1038/s41436-018-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J., Feng W., Zhuo X., Lu W., Wang J., Fang F., et al. Cerebral small vessel disease caused by PLOD3 mutation: expanding the phenotypic spectrum of lysyl hydroxylase-3 deficiency. Pediatric Investig. 2022;6:219–223. doi: 10.1002/ped4.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruotsalainen H., Sipila L., Vapola M., Sormunen R., Salo A.M., Uitto L., et al. Glycosylation catalyzed by lysyl hydroxylase 3 is essential for basement membranes. J. Cell Sci. 2006;119:625–635. doi: 10.1242/jcs.02780. [DOI] [PubMed] [Google Scholar]
- 45.Rautavuoma K., Takaluoma K., Sormunen R., Myllyharju J., Kivirikko K.I., Soininen R. Premature aggregation of type IV collagen and early lethality in lysyl hydroxylase 3 null mice. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14120–14125. doi: 10.1073/pnas.0404966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sipila L., Ruotsalainen H., Sormunen R., Baker N.L., Lamande S.R., Vapola M., et al. Secretion and assembly of type IV and VI collagens depend on glycosylation of hydroxylysines. J. Biol. Chem. 2007;282:33381–33388. doi: 10.1074/jbc.M704198200. [DOI] [PubMed] [Google Scholar]
- 47.Heikkinen J., Risteli M., Wang C., Latvala J., Rossi M., Valtavaara M., et al. Lysyl hydroxylase 3 is a multifunctional protein possessing collagen glucosyltransferase activity. J. Biol. Chem. 2000;275:36158–36163. doi: 10.1074/jbc.M006203200. [DOI] [PubMed] [Google Scholar]
- 48.Rautavuoma K., Takaluoma K., Passoja K., Pirskanen A., Kvist A.P., Kivirikko K.I., et al. Characterization of three fragments that constitute the monomers of the human lysyl hydroxylase isoenzymes 1-3. The 30-kDa N-terminal fragment is not required for lysyl hydroxylase activity. J. Biol. Chem. 2002;277:23084–23091. doi: 10.1074/jbc.M112077200. [DOI] [PubMed] [Google Scholar]
- 49.Wang C., Luosujarvi H., Heikkinen J., Risteli M., Uitto L., Myllyla R. The third activity for lysyl hydroxylase 3: Galactosylation of hydroxylysyl residues in collagens in vitro. Matrix Biol. 2002;21:559–566. doi: 10.1016/s0945-053x(02)00071-9. [DOI] [PubMed] [Google Scholar]
- 50.Scietti L., Chiapparino A., De Giorgi F., Fumagalli M., Khoriauli L., Nergadze S., et al. Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3. Nat. Commun. 2018;9:3163. doi: 10.1038/s41467-018-05631-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo H.F., Bota-Rabassedas N., Terajima M., Leticia Rodriguez B., Gibbons D.L., Chen Y., et al. A collagen glucosyltransferase drives lung adenocarcinoma progression in mice. Commun. Biol. 2021;4:482. doi: 10.1038/s42003-021-01982-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duncan K.G., Fessler L.I., Bachinger H.P., Fessler J.H. Procollagen IV. Association to tetramers. J. Biol. Chem. 1983;258:5869–5877. [PubMed] [Google Scholar]
- 53.Mirigian L.S., Makareeva E., Leikin S. Pulse-chase analysis of procollagen biosynthesis by azidohomoalanine labeling. Connect Tissue Res. 2014;55:403–410. doi: 10.3109/03008207.2014.959120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Syx D., Ishikawa Y., Gebauer J., Boudko S.P., Guillemyn B., Van Damme T., et al. Aberrant binding of mutant HSP47 affects posttranslational modification of type I collagen and leads to osteogenesis imperfecta. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fessler L.I., Fessler J.H. Identification of the carboxyl peptides of mouse procollagen IV and its implications for the assembly and structure of basement membrane procollagen. J. Biol. Chem. 1982;257:9804–9810. [PubMed] [Google Scholar]
- 56.Lunstrum G.P., Bachinger H.P., Fessler L.I., Duncan K.G., Nelson R.E., Fessler J.H. Drosophila basement membrane procollagen IV. I. Protein characterization and distribution. J. Biol. Chem. 1988;263:18318–18327. [PubMed] [Google Scholar]
- 57.Toth M., Sado Y., Ninomiya Y., Fridman R. Biosynthesis of alpha2(IV) and alpha1(IV) chains of collagen IV and interactions with matrix metalloproteinase-9. J. Cell Physiol. 1999;180:131–139. doi: 10.1002/(SICI)1097-4652(199907)180:1<131::AID-JCP15>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 58.Matsuoka Y., Kubota H., Adachi E., Nagai N., Marutani T., Hosokawa N., et al. Insufficient folding of type IV collagen and formation of abnormal basement membrane-like structure in embryoid bodies derived from Hsp47-null embryonic stem cells. Mol. Biol. Cell. 2004;15:4467–4475. doi: 10.1091/mbc.E04-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishida Y., Kubota H., Yamamoto A., Kitamura A., Bachinger H.P., Nagata K. Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol. Biol. Cell. 2006;17:2346–2355. doi: 10.1091/mbc.E05-11-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vranka J.A., Pokidysheva E., Hayashi L., Zientek K., Mizuno K., Ishikawa Y., et al. Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin, and bones. J. Biol. Chem. 2010;285:17253–17262. doi: 10.1074/jbc.M110.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedchenko V., Bauer R., Pokidysheva E.N., Al-Shaer A., Forde N.R., Fidler A.L., et al. A chloride ring is an ancient evolutionary innovation mediating the assembly of the collagen IV scaffold of basement membranes. J. Biol. Chem. 2019;294:7968–7981. doi: 10.1074/jbc.RA119.007426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanov S.V., Bauer R., Pokidysheva E.N., Boudko S.P. Collagen IV exploits a Cl- step gradient for scaffold assembly. Adv. Exp. Med. Biol. 2021;21:129–141. doi: 10.1007/5584_2020_582. [DOI] [PubMed] [Google Scholar]
- 63.Bhatnagar R.S., Gough C.A. In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman G.D., editor. Springer US; Boston, MA: 1996. Circular dichroism of collagen and related polypeptides; pp. 183–199. [Google Scholar]
- 64.Ono T., Miyazaki T., Ishida Y., Uehata M., Nagata K. Direct in vitro and in vivo evidence for interaction between Hsp47 protein and collagen triple helix. J. Biol. Chem. 2012;287:6810–6818. doi: 10.1074/jbc.M111.280248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa Y., Ito S., Nagata K., Sakai L.Y., Bachinger H.P. Intracellular mechanisms of molecular recognition and sorting for transport of large extracellular matrix molecules. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E6036–E6044. doi: 10.1073/pnas.1609571113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sricholpech M., Perdivara I., Yokoyama M., Nagaoka H., Terajima M., Tomer K.B., et al. Lysyl hydroxylase 3-mediated glucosylation in type I collagen: molecular loci and biological significance. J. Biol. Chem. 2012;287:22998–23009. doi: 10.1074/jbc.M112.343954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gould D.B., Phalan F.C., Breedveld G.J., van Mil S.E., Smith R.S., Schimenti J.C., et al. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science. 2005;308:1167–1171. doi: 10.1126/science.1109418. [DOI] [PubMed] [Google Scholar]
- 68.Gould D.B., Phalan F.C., van Mil S.E., Sundberg J.P., Vahedi K., Massin P., et al. Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N. Engl. J. Med. 2006;354:1489–1496. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- 69.Breedveld G., de Coo I.F., Lequin M.H., Arts W.F., Heutink P., Gould D.B., et al. Novel mutations in three families confirm a major role of COL4A1 in hereditary porencephaly. J. Med. Genet. 2006;43:490–495. doi: 10.1136/jmg.2005.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vahedi K., Boukobza M., Massin P., Gould D.B., Tournier-Lasserve E., Bousser M.G. Clinical and brain MRI follow-up study of a family with COL4A1 mutation. Neurology. 2007;69:1564–1568. doi: 10.1212/01.wnl.0000295994.46586.e7. [DOI] [PubMed] [Google Scholar]
- 71.Coste T., Vincent-Delorme C., Stichelbout M., Devisme L., Gelot A., Deryabin I., et al. COL4A1/COL4A2 and inherited platelet disorder gene variants in fetuses showing intracranial hemorrhage. Prenat Diagn. 2022;42:601–610. doi: 10.1002/pd.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aro E., Salo A.M., Khatri R., Finnila M., Miinalainen I., Sormunen R., et al. Severe extracellular matrix abnormalities and chondrodysplasia in mice lacking collagen prolyl 4-hydroxylase isoenzyme II in combination with a reduced amount of isoenzyme I. J. Biol. Chem. 2015;290:16964–16978. doi: 10.1074/jbc.M115.662635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou Y., Donkervoort S., Salo A.M., Foley A.R., Barnes A.M., Hu Y., et al. P4HA1 mutations cause a unique congenital disorder of connective tissue involving tendon, bone, muscle and the eye. Hum. Mol. Genet. 2017;26:2207–2217. doi: 10.1093/hmg/ddx110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Al-Shaer A., Lyons A., Ishikawa Y., Hudson B.G., Boudko S.P., Forde N.R. Sequence-dependent mechanics of collagen reflect its structural and functional organization. Biophys. J. 2021;120:4013–4028. doi: 10.1016/j.bpj.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010397. M111 010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris N.P., Keene D.R., Glanville R.W., Bentz H., Burgeson R.E. The tissue form of type VII collagen is an antiparallel dimer. J. Biol. Chem. 1986;261:5638–5644. [PubMed] [Google Scholar]
- 77.Macdonald J.R., Bachinger H.P. HSP47 binds cooperatively to triple helical type I collagen but has little effect on the thermal stability or rate of refolding. J. Biol. Chem. 2001;276:25399–25403. doi: 10.1074/jbc.M102471200. [DOI] [PubMed] [Google Scholar]
- 78.Fujii K.K., Taga Y., Sakai T., Ito S., Hattori S., Nagata K., et al. Lowering the culture temperature corrects collagen abnormalities caused by HSP47 gene knockout. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-53962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. Stable isotope-labeled collagen: A novel and versatile tool for quantitative collagen analyses using mass spectrometry. J. Proteome Res. 2014;13:3671–3678. doi: 10.1021/pr500213a. [DOI] [PubMed] [Google Scholar]
- 80.Backenroth D., Homsy J., Murillo L.R., Glessner J., Lin E., Brueckner M., et al. CANOES: detecting rare copy number variants from whole exome sequencing data. Nucleic Acids Res. 2014;42:e97. doi: 10.1093/nar/gku345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information. All source data are available from the corresponding author upon reasonable request. The MS datasets for protein identification of collagen α1α1α2(IV) samples have been deposited to the ProteomeXchange consortium via the jPOST partner repository with the dataset identifier PXD035051 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD035051).
(References: https://www.nature.com/articles/nbt.2839 and https://academic.oup.com/nar/article/45/D1/D1107/2605695?login=false).