Abstract
We previously established that a unidirectional site-specific recombinase, the phage φC31 integrase, can mediate integration into mammalian chromosomes. The enzyme directs integration of plasmids bearing the phage attB recognition site into pseudo attP sites, a set of native sequences related to the phage attP recognition site. Here we use two cycles of DNA shuffling and screening in Escherichia coli to obtain evolved integrases that possess significant improvements in integration frequency and sequence specificity at a pseudo attP sequence located on human chromosome 8, when measured in the native genomic environment of living human cells. Such integrases represent custom integration tools that will be useful for modifying the genomes of higher eukaryotic cells.
INTRODUCTION
Genetics still lacks tools for efficient site-specific integration of DNA sequences into the genomes of living higher cells. We have been investigating site-specific recombinases that may have the potential to provide such tools. This class of enzymes performs efficient recombination at recognition sites having the correct length to be present at low frequencies in large genomes (1). Phage integrases are site-specific recombinases that perform a unidirectional integration reaction with no competing reverse reaction, and may therefore be optimal for achieving integration at high frequency. The integrase from the Streptomyces phage φC31 (2,3) requires no cofactors (4) and performs efficient recombination between its attB and attP recognition sites on extrachromosomal plasmids in the mammalian cell environment (5).
Furthermore, the φC31 integrase recognizes native sequences in the human and mouse genomes that possess partial sequence identity to attP, called pseudo attP sites, and mediates the integration of plasmids bearing an attB site into such pseudo attP sequences (6). This reaction occurs at a frequency at least 5–10-fold above that of random integration and involves a hierarchy of endogenous sequences bearing varying degrees of identity to attP (6). The wild-type φC31 integrase thus carries out the general type of reaction we are seeking, directing relatively efficient and site-specific integration, compared with random integration. However, there appeared to be an opportunity to create integration reagents with even greater utility by using the wild-type enzyme as the starting material for a directed evolution study in which we might generate enzymes with greater sequence specificity and higher integration frequencies.
The requirements for this outcome would be expected to include alteration of the DNA recognition domain of the enzyme so that it avidly recognizes a particular native pseudo attP sequence, while losing ability to react with related sequences. Because the integrase recognizes both attP and attB, although not addressed in the present work, an optimal result might require parallel changes in attB. As well, the overall catalytic efficiency of the enzyme must increase. Gain of these features might require alterations involving both the DNA binding domain of the protein and the active site. We lack detailed structural information about the φC31 integrase. Even if we had such data, it would be difficult to predict how to engineer the enzyme. In this situation, non-rational methods are valuable to generate large libraries of variants from which enzymes having the desired properties can be found by screening (7,8).
A strikingly effective directed evolution strategy that includes mutagenesis and combinatorial exchanges is the DNA shuffling protocol (9–11). We have applied DNA shuffling in combination with a genetic screen that is capable of identifying mutant enzymes that possess an increased ability to perform integration at the desired sequence. We report here on such a screen and how we have used it to isolate new integrases that now display improved specificity for an endogenous sequence in the context of the native genome of a living human cell. This type of technology, in conjunction with more powerful screens, is likely to be effective for generating integration reagents customized to act at a wide variety of target sequences in a broad range of species. Such integration tools would have extensive applications across genetics, because they can be used to insert DNA site specifically and efficiently in processes such as functional genomics analysis, gene therapy, modification of the genomes of stem cells and construction of transgenic organisms.
MATERIALS AND METHODS
Plasmids
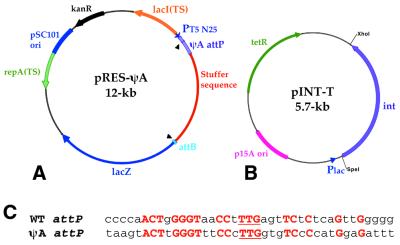
The screening assay uses two plasmids, called the resident plasmid, pRES-ψA, and the cloning plasmid, pINT-T. Both plasmids have compatible origins and can be propagated together in the same bacterial cell. The plasmid pTSK30 (12), which is temperature sensitive for replication, was used as the backbone for pRES-ψA (Fig. 1A). pTSK30 was cut with DrdI and SmaI to remove the lacZ alpha gene, which was replaced with the components below through a series of cloning steps involving several custom linkers (details available upon request). The phage T5 promoter (13) was added to provide a strong promoter for lacZ. The pseudo attP site ψA (Fig. 1C) from the human genome (6) (GenBank accession number AF333429) was added downstream of the T5 promoter. The ∼470-bp ψA sequence was copied from human genomic DNA by PCR with the primers 5′-ATTTGTAGAACTATTATGGG and 5′-AAGTCTTCTGGCTATACAGG. A ‘stuffer sequence’ was then inserted to act as a spacer between the two recombining att sites and to prevent transcriptional read-through. The stuffer sequence was obtained by PCR from human genomic DNA and encompassed a 2.3-kb GC-rich fragment from an intron of the human FGFR3 gene. A 45-bp φC31 attB site made from the oligonucleotides 5′-CGCGCCTGCGGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGTACTCCGG and 5′-CGCGCCGGAGTACGCGCCCGGGGAGCCCAAGGGCACGCCCTGGCACCCGCAGG was cloned downstream of the stuffer. The full-length lacZ gene from pCMVSPORTβgal (Life Technologies) was added. A temperature-sensitive muNStant of the lac repressor gene (lacI TS) was introduced from the plasmid pNH40lacIqTS (14).
Figure 1.
Plasmids used in genetic screen for detection of improved integrases. (A) pRES-ψA is established in E.coli cells and carries the intramolecular integration substrate. Recombination between attB and the human ψA attP sequence deletes the stuffer sequence and permits expression of lacZ from the T5 promoter. (B) pINT-T is the recipient for the shuffled integrase library, which is cloned into the plasmid in place of the wild-type φC31 integrase gene. The shuffled library is then introduced into cells carrying pRES-ψA. (C) Wild-type (WT) and ψA attP target sites. The top line shows the DNA sequence of the wild-type minimal 39-bp attP site (5) recognized by the φC31 integrase. The lower line shows the corresponding region of the human pseudo attP site ψA, identified as the most prevalent insertion site used by the φC31 integrase in human 293 cells (6). Uppercase letters denote perfect identity between the sequences, which totals 44%. The 3-bp core in which the recombination cross-over occurs is underlined.
To make pINT-T (Fig. 1B), the pInt plasmid (5) was modified. To first make the vector tetracycline resistant (TcR), pINT was cut with DraIII and PflMI and made blunt with T4 polymerase. This step provided a position for the TcR gene and also removed the kanamycin resistance gene from the pInt vector. From pBR322, the TcR gene was removed with EcoRI and PflMI, made blunt, and used to replace the kanamycin resistance gene, resulting in the plasmid pINT-Tc 2nd(+). pINT-Tc 2nd(+) was cut with BstEII and SpeI, which removed the integrase gene, producing pREC. A linker created with the oligonucleotides 5′-GTCACGCTCGAGAGATCTGA and 5′-CTAGTCAGATCTCTCGAGC was placed into these sites, which introduced unique BglII and XhoI restriction enzyme sites to pREC. The integrase gene was re-introduced into pREC by removing integrase from pINT-Tc 2nd(+) with BamHI and SpeI. pREC was cut with BglII and SpeI to accept the integrase gene (BglII and BamHI ends are compatible), producing pINT-T. XhoI and SpeI sites are unique to the pINT-T vector and can be used to shuttle an integrase library into and out of the vector. These same enzymes were later used to clone the shuffled integrases in place of wild-type integrase in pCMVInt (5) for their expression in human cells.
DNA shuffling
DNA shuffling of the integrase gene for cycles 1 and 2 was performed similarly to published protocols (9,10). Briefly, the integrase gene was copied from the pINT-T vector by PCR with the primers 5′-CTAAAGGGAACAAAAGCTGGAG and 5′-TGATATGGGGCAAATGGTGGTC. These primers lie directly adjacent to the unique XhoI and SpeI restriction sites used to clone the shuffled library back into the vector. An aliquot of 5–6 µg of integrase gene was treated with 0.15 U of DNase I in a 100 µl reaction volume for 20 min at room temperature. Fragments of the integrase gene were run out on a 1.6% NuSieve (BioWhittaker Molecular Applications) gel in 1× TAE. Fragments 50–250 bp long were cut out of the gel. DNA fragments were removed from the low-melt gel with β-agarase (New England Biolabs). Forty-five cycles of primerless extensions were performed as described (9,10) in a Bio-Rad iCycler Model PCR machine with Ready-To-Go PCR Beads (Amersham Pharmacia Biotech, Piscataway, NJ) supplemented with magnesium to 2.2 mM. To amplify the shuffled integrase library, a portion of the primerless reaction was added to the primers shown above, and further PCR was performed. The resulting PCR product of 1.9 kb was gel isolated. The integrase gene library was cut with XhoI and SpeI and ligated into pINT-T backbone devoid of the integrase gene. Ligation reactions used to produce the plasmid library were cleaned with Micropure-EZ columns (Millipore) followed by additional purification/concentration using Microcon YM-100 spin dialysis columns (Millipore). From a sampling of the library, we estimate ∼50% of colonies contained the integrase insert.
Mammalian PCR assay
293 cells that had reached 50–80% confluency in a 60-mm diameter dish were transfected with 50 ng of plasmid pHZ-attB (6) and 5 µg of a plasmid expressing either lacZ (pCMVSPORTβGal), the wild-type φC31 integrase pCMV-Int (5), or a shuffled integrase replacing wild-type Int in pCMV-Int, by using Lipofectamine (Life Technologies, Rockville, MD). Forty-eight hours after transfection, genomic DNA was harvested from 70% of the transfected cells. Genomic DNA was prepared using the DNEasy tissue kit (Qiagen). This DNA was subjected to quantitative PCR analysis (15,16) to detect the frequency of recombination at ψA . A forward primer (5′-CGATGTAGGTCACGGTCTCGA), a reverse primer (5′-TTGCGTCATGGCTTA) and a Taqman probe (5′-6FAM-CCAGGGCGTGCCCTTGGTGT-TAMRA) were designed so that a product would be detected only if a site-specific reaction had occurred at ψA. 6FAM is the fluorophore at the 5′ end of the Taqman probe, and TAMRA is the quencher at the 3′ end. The underlined TTG represents the att site core. A second set of primers to a nearby genomic sequence (forward primer, 5′-TTTCTCCTGTGCTATCGCAGAA; reverse primer, 5′-CTCCCGCAACATTGGCTT) was designed to normalize for the amount of DNA used in PCR. Quantification for this PCR was done using the SYBR Green kit (Applied Biosystems). Amplification reactions were performed in an ABI 7700 machine and the results analyzed with SDS v1.7 software (Applied Biosystems). Each sample was tested in triplicate, and each transfection was performed at least three times. The number of recombination junctions and the amount of DNA used were calculated using genomic DNA from a 293-derived cell line containing a single integration event at ψA (6) as a standard.
Mammalian selection assay and analysis of recombinant clones
293 cells were transfected as described above. Forty-eight hours after transfection, selection with 200 µg/ml of hygromycin B was carried out on 30% of the transfected cells for 14 days, and individual colonies were counted. At least three replicates of each experiment were performed, and the average number of colonies obtained was calculated. Single hygromycin-resistant colonies obtained from experiments performed with the 1C2 and 11C2 integrases were picked and expanded. Genomic DNA was prepared from 20 such clones generated by each integrase by using the DNEasy tissue kit (Qiagen). This DNA was then analyzed by PCR amplification for the presence of the specific attL recombination junction that would result from integration at ψA. The forward and reverse primers used were 5′-CGATGTAGGTCACGGTCTCGA and 5′-TTGCATGGCCTCATTTCCGTC, respectively. The products of PCR amplification were analyzed by electrophoresis on an agarose gel stained with ethidium bromide.
RESULTS
Genetic screen for improved integrases
We previously identified a DNA sequence on human chromosome 8 as the most prevalent genomic integration site for φC31 integrase-mediated genomic integration of a plasmid bearing attB (6). On this basis, the ψA site appeared to be relatively well recognized by the φC31 integrase and was chosen as the DNA substrate for this directed evolution study. As shown in Figure 1C, ψA shares the TTG common core and possesses identity at 44% of the positions in the 39-bp attP site that we previously determined to be the minimal site for obtaining full reaction in an Escherichia coli assay (5).
The primary property we were seeking to improve was the ability of the integrase to perform a recombination reaction between attB and the ψA pseudo attP site. For convenience, we developed a screen in E.coli, with the expectation that some of the improved integrases would also show enhanced function in the human cell environment. In order to identify improved integrases from a shuffled library of integrase mutants, we devised a genetic screen in which ability of the enzyme to complete a reaction at ψA could be read off Xgal indicator plates (17) as blue colony color. As shown in Figure 1A, we constructed an assay plasmid pRes-ψA in which the attB and ψA sites were separated by a stuffer sequence that would block transcription by a promoter upstream of ψA into the lacZ gene positioned downstream of attB. Only after an intramolecular integration event between attB and ψA would the transcription occur.
A 1.9-kb fragment containing the φC31 integrase gene was subjected to DNA shuffling (9). The shuffled products were cloned into the pINT-T plasmid, and the shuffled library transformed into E.coli cells carrying pRES-ψA. Colonies were allowed to form at 30°C on Xgal plates. Under these conditions, the pRES-ψA plasmid replicated and lacI was expressed, keeping most transcription of the integrase gene repressed. Once a moderately large colony size was attained, induction of the integrase was started. Induction was achieved simply by moving the plates to 37°C. At this temperature, most of the lac repressor product of lacI was inactive, enabling transcription and expression of the library of mutant integrases. In addition, replication of pRES-ψA was inhibited due to the temperature sensitive replication origin on the plasmid. Thus, during the growth period, colonies underwent little expansion, but expression of integrase and any consequent intramolecular integration reactions proceeded. When recombination occurred between attB and the ψA site, transcription of lacZ was activated, resulting in hydrolysis of Xgal to indigo, read as blue color on Xgal agar plates.
The timing, pattern and degree of blue color gave a measure of the integration activity characteristic of the integrase gene present in that bacterial colony. With an induction period of 24 h, we found that the wild-type integrase produced essentially no blue color in the colonies (Fig. 2). Therefore, any mutant integrase producing blue color in 24 h or less probably had increased ability to perform recombination between attB and ψA. By reducing the time allowed for induction, we increased the stringency of the screen.
Figure 2.

Colony color screen for improved integrases. Color photographs indicate the appearance of bacterial colonies carrying wild-type (WT INT) and shuffled integrases. Blue color on plates containing Xgal reflects the occurrence of intramolecular integration. The time of incubation at 37°C indicates the period of integrase expression induction. Even the low level of the 11C2 integrase present at time 0 in uninduced cells is sufficient to give significant blue color.
DNA shuffling cycle 1
For the first cycle of shuffling, we subjected the wild-type φC31 integrase gene to the DNA shuffling protocol, then transformed the shuffled library into E.coli carrying pRES-ψA. After colony formation at 30°C, the plates were incubated for 24 h at 37°C to allow integrase expression and subsequent recombination. The plates were scored for blue color, with the result that 12 of ∼10 000 colonies screened showed specks of blue in the colony. Plasmid DNA was purified from these colonies and retested in the assay to confirm the color phenotype. Mini-prep DNA from blue colonies was analyzed by restriction mapping to verify that the expected intramolecular integration reaction had occurred in each case. For the most promising mutants, i.e. the ones that generated most blue color (1C1, 12C1, 17C1), the attR junction generated by recombination from several blue colonies was sequenced. The attR junction generated by recombination between attB and ψA was perfect to the base in each case. This result confirmed that the mutant integrases were performing the appropriate integration reaction, were precise, and were not simply mediating promiscuous integration at a variety of sequences. Instead, the mutant integrases appeared to have improved recognition of and activity at the ψA attP site.
The best candidates from cycle 1 were arranged in a tentative order of ability to perform recombination at ψA, based on the degree of blueness developed by the bacterial colony at a particular time of induction. By this measure, 17C1 appeared to be the most effective, since it formed colonies that turned uniformly pale blue after only 6 h of integrase expression (Fig. 2). The other cycle 1 mutants required longer induction times to develop blue color.
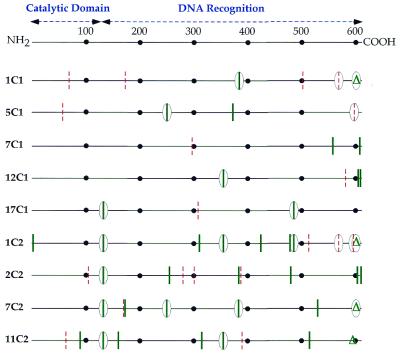
Five of the cycle 1 integrases were completely analyzed at the DNA sequence level in order to characterize what mutations were present. This information is depicted schematically in Figure 3 and reported completely in Table S1 (see Supplementary Material). In each case, three to six mutations were present. About half of the mutations were silent with respect to amino acid change, while the others led to amino acid substitutions. Most of the mutations were single base changes, although a single base deletion in 1C1 led to a frameshift near the C-terminus of the protein (Table S2). Each of the mutations was distinct. This outcome is typical of cycle 1 shuffling (7).
Figure 3.
Schematic diagram of mutational changes present in shuffled integrases. The 613 amino acid length of the φC31 integrase is demarcated by dots at 100 amino acid intervals. The approximate location of the catalytic domain, in analogy with other serine recombinase family members, is indicated, and the balance of the protein may carry segments involved in DNA recognition. Base changes that do not lead to amino acid changes are indicated by vertical red dotted lines. Amino acid changes are indicated by vertical green lines. Single base deletions that result in frameshifts are shown by green triangles. The symbol is circled if the identical change occurred in both cycle 1 and cycle 2 mutants.
DNA shuffling cycle 2
Plasmid pINT-T DNAs purified from the best 12 candidates from cycle 1 were mixed together in equal proportion and subjected to DNA shuffling to produce cycle 2. In this way, the distinct mutations present in the cycle 1 mutants could be mixed in a combinatorial way to produce new configurations in which favorable features could be combined to produce integrases with additional benefits. Because the cycle 1 mutants all gave evidence of blue color after 24 h of induction at 37°C, to increase the stringency of the cycle 2 assay the induction period was reduced to 6 h. None of the cycle 1 mutations showed more than a pale degree of blueness at this time point, so appearance of deep blue colonies at 6 h would indicate a further gain in integration efficiency.
The cycle 2 library was transformed into E.coli, and from a screen of ∼10 000 colonies we obtained 11 candidates that produced blue colonies after 6 h of integrase induction (Fig. 2). Based on the amount of blue color present, 1C2, 2C2 and 11C2 appeared to be the most efficient integrases. The mutant integrases were re-tested and the DNA of pRES-ψA from blue colonies was examined by restriction mapping and DNA sequencing. Again, the attR recombination junction was found to be perfect to the base in each case, indicating that the mutant integrases mediated a precise recombination reaction, had not become promiscuous and appeared to have an elevated ability to perform intramolecular integration between attB and ψA.
The complete DNA sequences of four of the best cycle 2 integrases were determined and are reported schematically in Figure 3 and in detail in Table S1. We found evidence that efficient DNA shuffling had occurred, because many of the mutations present in the cycle 2 mutants had already been seen in cycle 1 and were now present in new combinations. Since not all of the cycle 1 mutants were sequenced, we could not determine whether new mutations appeared in cycle 2. The Gln to Pro mutation at amino acid 134, derived from 17C1, appeared in all four cycle 2 mutants analyzed at the sequence level (Fig. 3 and Table S1). A single base deletion distinct from the one in 1C1 was present in 11C2 and caused a different frameshift that changed the amino acid sequence of the C-terminus of the protein (Fig. 3 and Table S2).
Mammalian integration frequency analyzed by quantitative PCR
Our genetic screen in E.coli effectively identified mutant enzymes that could perform more efficient recombination between attB and ψA in bacterial cells. Our goal was to obtain mutants that were more effective at performing integration at the ψA target site in its native context in the human genome. Because this reaction probably has requirements that would not be optimized in E.coli, not all of the mutant integrases we isolated in E.coli were expected to perform well in the mammalian context. The most direct measure of the ability of the mutants to mediate the desired integration event in human cells was to monitor the frequency of recombination at the ψA chromosomal position using quantitative PCR (15,16). This assay directly monitored the creation of the desired recombination junction and was free of artifacts, such as silencing of the integrated gene, which might be present in a genetic assay that relied on selection. To perform quantitative PCR, we used primers that flanked the expected recombination junction. One primer was located in the integrated plasmid and the other in flanking genomic sequences downstream of attL. Detection of this PCR band could only occur if the integration event we desired had occurred.
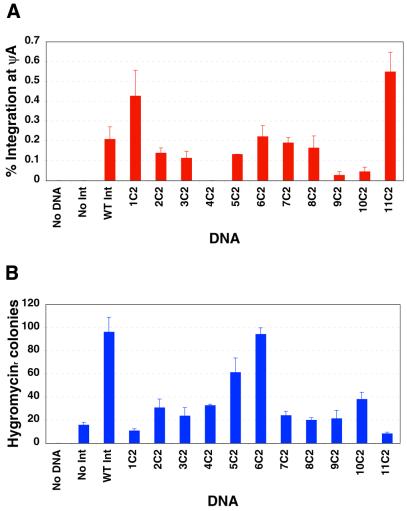
To perform the quantitative PCR assay, human 293 cells were transfected with plasmids carrying the mutant integrase gene to be tested and an attB plasmid, pHZ-attB. Forty-eight hours after transfection, integration was assayed. Cellular DNA was purified, and the PCR primers and Taqman probe were added and PCR performed. Each of the cycle 1 and 2 integrase mutants were tested in the quantitative PCR assay. None of the cycle 1 mutants showed an improvement in absolute integration frequency in human cells, despite their improved performance in E.coli. However, among the cycle 2 mutants, two of the eleven mutants, 1C2 and 11C2, showed a significant increase of 2–3-fold in absolute integration efficiency at ψA (Fig. 4A). Wild-type integrase mediated site-specific integration at ψA at a frequency of 0.21%, whereas the frequency mediated by 1C2 was 0.43% (P < 0.05) and that mediated by 11C2 was 0.55% (P < 0.01). These frequencies are uncorrected for transfection efficiency.
Figure 4.
Integration frequency assays for shuffled integrases in human cells. (A) Quantitative PCR assay for integration frequency at ψA. Human 293 cells were transfected with pHZ-attB and either no integrase, the wild-type φC31 integrase, or one of 11 cycle 2 shuffled integrases, 1C2–11C2. After 2 days, genomic DNA was prepared and analyzed with probes specific for the ψA site with quantitative PCR. The absolute frequency of integration in the sample is shown, uncorrected for transfection efficiency. (B) Selection assay for total integration frequency. The number of hygromycin-resistant colonies generated by each integrase after co-transfection with pHZ-attB into 293 cells. Error bars in (A) and (B) represent the standard error.
Overall genomic integration frequency and specificity
The above quantitative PCR data showed that the 1C2 and 11C2 integrases directed an increased absolute integration frequency at ψA. However, the PCR data did not reveal whether these evolved integrases also displayed an elevated integration frequency at other genomic sites. To get a sense of the relative specificity of the evolved integrases, we followed overall integration frequency by using a genetic assay, then determined what fraction of the integrants were located at ψA.
To determine overall integration frequency, we transfected the pHZ-attB plasmid carrying attB and the hygromycin resistance gene, along with an expression plasmid encoding the integrase to be tested, into human 293 cells. After 3 weeks of hygromycin selection, colonies were counted. This analysis (Fig. 4B) revealed that the shuffled integrases did not mediate an increase in overall integration frequency. Rather, one mutant was indistinguishable from wild-type integrase and the other 10 had significantly lower overall integration frequencies. In particular, the 1C2 and 11C2 mutants, which had higher integration frequencies specifically at ψA, had the lowest overall integration frequencies, as reflected by numbers of hygromycin-resistant colonies ∼10-fold below that of the wild-type integrase.
This result could be explained if the shuffled integrases now possessed an elevated specificity for ψA that decreased the background of integration into other genomic sequences, such as the other ∼100–1000 pseudo attP sequences thought to be present in the human genome (6). To measure the integration specificity, 20 randomly chosen hygromycin-resistant colonies generated by the 1C2 and 11C2 integrases were expanded. Genomic DNA was prepared and analyzed by PCR using primers that would detect integration at ψA. The results of this PCR analysis (Fig. S1) demonstrated that 30% (6/20) of integration events mediated by these two shuffled integrases now occurred at ψA. This result was in contrast to the previously determined figure of 5.2% (6) for the wild-type φC31 integrase. These data suggested that an increase of ∼6-fold in specificity for ψA accompanied the 2–3-fold increase in absolute integration frequency at ψA.
The 2–3-fold higher integration frequency at ψA measured for 1C2 and 11C2 in Figure 4A would have predicted that for these mutants we would have seen approximately 10 colonies with integrations at ψA per 100 total integrations by the wild-type integrase in Figure 4B, rather than the three we observed. We do not have a full explanation for this result, but note that the assays measured two different things. The quantitative PCR assay directly measured recombination events at ψA at an early time, with no selection. The selection assay measured sustained gene expression at the composite of all integrated loci. It is possible that integration events were more poorly recovered in the selection assay in the case of the shuffled integrases simply due to the low colony numbers, which give rise to depressed growth on the plates. The lower colony numbers obtained with 1C2 and 11C2 suggested that the mutants did not confer hyper recombination or relaxed specificity, but rather represented altered specificity mutants.
In order to analyze the precision of the integration events mediated by the shuffled recombinases, we performed DNA sequence analysis on fragments containing the attL recombination junctions resulting after genomic integration. The attL junction was obtained by PCR from individual colonies representing several independent integration events at ψA. The sequence analysis revealed small deletions of from 6 to 17 bp in each case, overlapping the 3-bp cross-over region. This result is similar to results reported for the wild-type φC31 integrase, where most recombination junctions contained small deletions (6). The small deletions may indicate an impaired ability of both the wild-type and shuffled integrases to complete the recombination process on pseudo attP sites and possible involvement of host repair enzymes to complete the reaction.
DISCUSSION
This study demonstrates the feasibility of using DNA shuffling to evolve integration tools with improved site specificity for modifying large genomes. Starting with the phage φC31 integrase, which already displayed the ability to integrate into a set of sites in the human genome, we used directed evolution to create enzyme mutants with a stronger preference for one DNA sequence target site on chromosome 8. We isolated mutants that displayed higher absolute integration frequencies at this site and also a higher degree of preference for this site versus other locations in the genome.
Integration frequency
The screen we employed to detect integrases with improved function at the ψA sequence was carried out in E.coli for convenience. Integration mediated by the wild-type enzyme proceeds poorly at ψA in the bacterial environment. It seems likely that the screen primarily revealed integrases that had increased DNA sequence recognition of ψA rather than an increase in non-sequence-specific activity. This interpretation is supported by the DNA sequences of integration junctions, which were precise to the base in E.coli and not promiscuous, for both wild-type and shuffled integrases. The reaction evidently proceeds slightly differently in mammalian cells, where both the wild-type integrase and the shuffled 1C2 integrase often created small deletions of a few base pairs at the integration junction. This result may reflect the involvement of mammalian repair enzymes in the integration process.
Would the enzymes optimized for intramolecular integration in E.coli also be improved in the mammalian context? Of the 11 cycle 2 shuffled integrases, the top two performers in E.coli did show significant (2–3-fold) increases in absolute integration frequency at ψA, when carrying out an intermolecular integration reaction in the context of chromatin and other features of a living mammalian cell. Quantitative PCR showed that, in the presence of the 1C2 or 11C2 integrases, ∼0.5% of the cells in the population had an integration event at ψA. Because only transfected cells can have an integration event, these figures must be corrected by the transfection frequency. Since the transfection efficiency in these conditions is ∼5–10% (6), we infer that the true integration frequency is 10–20-fold higher, well into the percent range. These integration frequencies are unprecedented for a specific integration event in mammalian cells and indicate that our goal of obtaining enzymes that mediate integration at a frequency of ∼100% may be feasible.
Increase in specificity
We also monitored what fraction of the integration events mediated by the shuffled integrases were occurring at the desired ψA target site. From a random sample of integration events mediated by the 1C2 and 11C2 shuffled integrases, the two mutants that showed an increase in absolute integration efficiency at ψA, 30% (6/20) of integrations took place at ψA for each integrase. When a sample of integration events mediated by the wild-type integrase was analyzed, we found that 5.2% (5/96) were located at ψA (6). This result suggests that the specificity of the shuffled integrases has improved significantly, by 5–6-fold. In a 293 cell line containing an artificially placed wild-type attP site, 14/96 or 14.6% of the integrations occurred at this site (6). Since the shuffled integrases have a specificity of 30% for ψA, it appears that these evolved integrases are seeing the ψA pseudo attP site as well or better than the wild-type φC31 enzyme sees its own wild-type attP site.
To obtain a measure of whether the evolved integrases still recognize attP, we transferred the wild-type, 1C2 and 11C2 integrases into an E.coli strain carrying the pRES plasmid bearing the wild-type attP site and measured the level of recombination, as indicated by the degree of blue color on Xgal plates. The colonies carrying the wild-type integrase were bluer than those in which the 1C2 and 11C2 integrases had been introduced (data not shown). These results are consistent with the two shuffled integrases recognizing attP less well than the wild-type integrase does, verifying that they are likely to be altered specificity rather than relaxed specificity mutants.
An interesting aspect of the behavior of the shuffled integrases in human cells is that the best integrases actually give the lowest numbers of hygromycin-resistant colonies upon co-transfection with an attB-hygromycin resistant plasmid (Fig. 4B). Since the 1C2 and 11C2 integrases have higher absolute integration frequencies and an elevated specificity of 30%, the lower colony numbers suggest that activity toward alternative pseudo attP sites is being suppressed. We took no steps to move the enzyme away from recognition of other sequences, but it seems likely that such an outcome occurs automatically as a side-effect of evolving toward recognition of one particular sequence. The remaining 70% of colonies generated by 1C2 and 11C2 that were not at ψA could be accounted for by random integration. Presumably, with further rounds of shuffling and further increases in specificity, the colony numbers would rise and an even higher fraction of these colonies would be the result of integration at ψA.
Insights into protein structure
The three-dimensional structure of the φC31 integrase is not currently available. The X-ray structure of another serine recombinase family member, the γδ resolvase, has been determined (18). While only distantly related to the φC31 integrase, it provides guidance, notably on location of the catalytic domain, which is likely to occupy the first ∼130 amino acids (19). The catalytic serine that is the most prominent hallmark of this family of site-specific recombinases (20) is located at amino acid 12 in the φC31 integrase (4). The pattern of mutations that we observed in the integrase gene during the first and second cycles of shuffling begins to suggest further features of the protein’s structure.
The two to three sense mutations per integrase mutant seen in the first round of shuffling were primarily scattered in the DNA recognition portion of the protein, currently loosely defined as the balance of the protein that is not in the catalytic domain, with the most striking clustering of mutations occurring at the C-terminal end of the protein (Table S2). Two areas of the protein were universally affected by the six to eight sense changes in cycle 2 mutants. The first was the mutation at amino acid 134 that was derived from 17C1. This glutamine to proline change adjacent to the catalytic domain of the protein has gone to fixation in the sequenced sample (4/4) of cycle 2 integrases and may lead to an overall increase in activity. The most active cycle 2 mutants, 1C2 and 11C2, each have additional changes in the catalytic domain, at amino acids 2 and 89, respectively, that may also increase catalytic activity. The other area universally affected in cycle 2 is the C-terminus of the protein. All four cycle 2 mutants sequenced bear changes in the final 20 amino acids of the protein (Fig. 3). Most of the mutations result in a more positive charge (Table S2), which may affect the enzyme’s binding to DNA.
There is little literature on changing the DNA recognition specificity of proteins. Sequence-specific binding of transcription factors to their DNA recognition sites has been altered through amino acid changes at specific positions (for example see 21). The restriction enzyme EcoRV was altered to begin to extend the length of its recognition site by undertaking random mutation of 22 amino acids thought to be involved in DNA recognition (22). Directed evolution including DNA shuffling was performed on the FLP recombinase to change its temperature optimum, but not its DNA recognition preference (23). A study of Cre recombinase by amino acid changes in its DNA binding region produced mutants with either loss of lox target binding or broadening of lox target recognition specificity (24). The recognition specificity of the phage λ integrase was altered by mutation to recombine the similar att sites of the closely related integrase of phage HK022, by screening random and oligonucleotide-directed mutations (25) or recombinant chimeras between the two integrases (26). A λ integrase mutant having five amino acid changes had nearly complete HK022 specificity (25). Mutants of the related serine recombinase, γδ resolvase, enable it to function in mammalian cells (27). Altered DNA-binding mutants have also been described for Mu transposase (28) and Tn5 transposase (29).
Utility
It seems likely that additional improvements can be realized with further rounds of shuffling and more stringent screens. This integrase technology has widespread areas of potential utility for applications requiring site-specific genome manipulation of cells, tissues and organs. Enzymes that have high integration efficiencies toward DNA sequences of choice would be valuable, for example in gene therapy, where they could be used to bring about site-specific integration of therapeutic genes into patient tissue. The higher the integration efficiency, the more effective the gene therapy, which is often now limited by inadequate levels of gene expression. High site specificity for a well-expressed location that does not disrupt a gene is beneficial for ensuring safety and guaranteeing stable gene expression. The technology would similarly be valuable for efficiently altering stem cells and other cells for transplantation.
The availability of a powerful technology for efficient site-specific integration would also improve the efficiency of production and the quality of transgenic animals and plants and open up the possibility of creating custom integration tools for functional genomics that could be used to specifically disrupt or alter genes of interest. In addition, the system will be a valuable tool for bringing about desired recombination events in tissue culture studies, such as obtaining multiple cell lines, each carrying integrations of different cassettes at the same location and manipulation of virus genomes, large plasmids such as BACs, and artificial chromosomes. The success achieved in this study suggests that creation of tools for all of these uses will be feasible.
SUPPLEMENTARY MATERIAL
Supplementary material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank N. Hasan and W. Szybalski for providing placIts and A. McCaffrey for advice on quantitative PCR. C.R.S. and B.T. were supported by PHS grant CA09302 from the National Cancer Institute. This work was supported by NIH grant DK58187 awarded to M.P.C.
REFERENCES
- 1.Thyagarajan B., Guimaraes,M.J., Groth,A.C. and Calos,M.P. (2000) Mammalian genomes contain active recombinase recognition sites. Gene, 244, 47–54. [DOI] [PubMed] [Google Scholar]
- 2.Kuhstoss S. and Rao,R.N. (1991) Analysis of the integration function of the Streptomycete bacteriophage ΦC31. J. Mol. Biol., 222, 897–908. [DOI] [PubMed] [Google Scholar]
- 3.Rausch H. and Lehmann,M. (1991) Structural analysis of the actinophage ΦC31 attachment site. Nucleic Acids Res., 19, 5187–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorpe H.M. and Smith,M.C.M. (1998) In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl Acad. Sci. USA, 95, 5505–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groth A.C., Olivares,E.C., Thyagarajan,B. and Calos,M.P. (2000) A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA, 97, 5995–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thyagarajan B., Olivares,E.C., Hollis,R.P., Ginsburg,D.S. and Calos,M.P. (2001) Site-specific genomic integration in mammalian cells mediated by phage φC31 integrase. Mol. Cell. Biol., 21, 3926–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore J.C. and Arnold,F.H. (1996) Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents.Nat. Biotechnol., 14, 458–467. [DOI] [PubMed] [Google Scholar]
- 8.Minshull J. and Stemmer,W.P.C. (1999) Protein evolution by molecular breeding. Curr. Opin. Chem. Biol., 3, 284–290. [DOI] [PubMed] [Google Scholar]
- 9.Stemmer W.P.C. (1994) DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA, 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stemmer W.P.C. (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature, 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 11.Crameri A., Raillard,S., Bermudez,E. and Stemmer,W.P.C. (1998) DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature, 391, 288–291. [DOI] [PubMed] [Google Scholar]
- 12.Phillips G.J. (1999) New cloning vectors with temperature-sensitive replication. Plasmid, 41, 78–81. [DOI] [PubMed] [Google Scholar]
- 13.Deuschle U., Kammerer,W., Gentz,R. and Bujard,H. (1986) Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J., 5, 2987–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan N. and Szybalski,W. (1995) Construction of lacIts and lacIqts expression plasmids and evaluation of the thermosensitive lac repressor. Gene, 163, 35–40. [DOI] [PubMed] [Google Scholar]
- 15.Gibson U.E., Heid,C.A. and Williams,P.M. (1996) A novel method for real time quantitative RT-PCR. Genome Res., 6, 995–1001. [DOI] [PubMed] [Google Scholar]
- 16.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 17.Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 18.Yang W. and Steitz,T.A. (1995) Crystal-structure of the site-specific recombinase gamma-delta resolvase complexed with a 34 bp cleavage site. Cell, 82, 193–207. [DOI] [PubMed] [Google Scholar]
- 19.Leschziner A.E., Boocock,M.R. and Grindley,N.D.F. (1995) The tyrosine-6 hydroxyl of gamma delta resolvase is not required for the DNA cleavage and rejoining reactions. Mol. Microbiol., 15, 865–870. [DOI] [PubMed] [Google Scholar]
- 20.Stark W.M., Boocock,M.R. and Sherratt,D.J. (1992) Catalysis by site-specific recombinases. Trends Genet., 8, 432–439. [PubMed] [Google Scholar]
- 21.Bulyk M., Huang,X., Choo,Y. and Church,G.M. (2001) Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl Acad. Sci. USA, 98, 7158–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanio T., Jeltsch,A. and Pingoud,A. (1998) Towards the design of rare cutting restriction endonucleases: Using directed evolution to generate variants of EcoRV differing in their substrate specificity by two orders of magnitude. J. Mol. Biol., 283, 59–69. [DOI] [PubMed] [Google Scholar]
- 23.Buchholz F., Angrand,P.O. and Stewart,A.F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol., 16, 657–662. [DOI] [PubMed] [Google Scholar]
- 24.Hartung M. and Kisters-Woike,B. (1998) Cre mutants with altered DNA binding properties. J. Biol. Chem., 273, 22884–22891. [DOI] [PubMed] [Google Scholar]
- 25.Dorgai L., Yagil,E. and Weisberg,R. (1995) Identifying determinants of recombination specificity: construction and characterization of mutant bacteriophage integrases. J. Mol. Biol., 252, 178–188. [DOI] [PubMed] [Google Scholar]
- 26.Yagil E., Dorgai,L. and Weisberg,R.A. (1995) Identifying determinants of recombination specificity: Construction and characterization of chimeric bacteriophage integrases. J. Mol. Biol., 252, 163–177. [DOI] [PubMed] [Google Scholar]
- 27.Schwikardi M. and Droge,P. (2000) Site-specific recombination in mammalian cells catalyzed by γδ resolvase mutants: implications for the topology of episomal DNA. FEBS Lett., 471, 147–150. [DOI] [PubMed] [Google Scholar]
- 28.Namgoong S.-Y., Sankaralingam,S. and Harshey,R.M. (1998) Altering the DNA-binding specificity of Mu transposase in vitro. Nucleic Acids Res., 26, 3521–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M., Bhasin,A. and Reznikoff,W.S. (1998) Molecular genetic analysis of transposase-end DNA sequence recognition: Cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 276, 913–925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.