Abstract
Magnetization transfer contrast MR fingerprinting (MTC-MRF) is a novel quantitative imaging method that simultaneously quantifies free bulk water and semisolid macromolecule parameters using pseudo-randomized scan parameters. To improve acquisition efficiency and reconstruction accuracy, the optimization of MRF sequence design has been of recent interest in the MRF field, but has been challenging due to a large number of degrees of freedom to be optimized in the sequence. Herein, we propose a framework for learning-based optimization of the acquisition schedule (LOAS), which optimizes RF saturation-encoded MRF acquisitions with a minimal number of scan parameters for tissue parameter determination. In a supervised-learning framework, scan parameters were subsequently updated to minimize a pre-defined loss function that can directly represent tissue quantification errors. We evaluated the performance of the proposed approach with a numerical phantom and in in vivo experiments. For validation, MRF images were synthesized using the tissue parameters estimated from a fully connected neural network (FCNN) framework and compared with references. Our results showed that LOAS outperformed existing indirect optimization methods with regard to quantification accuracy and acquisition efficiency. The proposed LOAS method could be a powerful optimization tool in the design of MRF pulse sequences.
Keywords: Deep-learning, Optimization, MR fingerprinting, MT, CEST, NOE
1. Introduction
Magnetization transfer contrast (MTC) experiments measure the transfer (or exchange) of magnetization from semisolid macromolecular protons to surrounding free bulk water molecules1–5. While the effect of low-concentration macromolecular protons on the water resonance is not detectable after a single exchange, the cumulative effect of repeated saturation and exchange allows indirect assessment of the semisolid macromolecular protons through the free bulk water signal. MTC imaging offers great potential for the characterization of comprehensive tissue composition (e.g., myelin and mobile protein concentrations), and hence, could act as a biomarker for the clinical diagnosis of tissue disorders6–9. Typically, the MTC effect has been measured by calculating the MT ratio (MTR), which reflects the difference between two images obtained with and without RF saturation of macromolecular protons; however, the ratio is dependent on the choice of scan parameters and tissue relaxation effects. The most promising MTC quantification currently requires fitting of MTC-weighted signals acquired from repeated and serial image acquisitions with various saturation powers and frequency offsets using the analytical solution of the Bloch equations2,10–12. Although the model-based quantification approach can largely remove the dependence of experimental settings and separate MTC contributions from relaxation effects, it typically requires a long scan time and the post-processing for MTC quantification is computationally complex. In addition, the quantification accuracy is sensitive to fitting parameters, thus limiting its clinical utility.
Recently, a strategy called “MR fingerprinting (MRF)” was introduced as a novel acquisition and reconstruction approach to overcome time constraints13. Imaging scan parameters are intentionally changed for every acquisition to generate transient-state signal evolutions. Then, tissue parameters are quantified by comparing experimentally measured signal profiles with a simulated library of signal variation patterns based on the sequence used14–17. However, the size of the library is dramatically increased when a multiple-exchange model is taken into account, and thus, the construction of a huge library and an exhaustive search are unavoidable18–21. Moreover, a conventional MRF library with discrete values could lead to quantization errors due to the large dictionary step size. Recent advances in deep neural networks have opened a new possibility to solve general inverse problems in MRF reconstruction in an efficient manner and to produce reliable estimates of tissue parameters22–24. A deep-learning architecture can be designed to quantify multiple tissue parameters by learning the mapping relation between fingerprints and tissue parameter spaces, which bypasses the exhaustive dictionary search. Recent studies have proposed combining MTC- and chemical exchange saturation transfer (CEST)-MRF with deep-learning techniques to improve the quantification accuracy and accelerate the imaging time25–29.
To further accelerate data acquisition and improve accuracy, an optimal design of the MRF acquisition schedule is critical for efficient and accurate tissue parameter-mapping20,30,31. However, optimization of the MRF schedule has been challenging due to a large number of degrees of freedom available in an MRF sequence. Most of the current methods are based on indirect measurements, such as maximizing the signal discrimination between tissue types30, or maximizing the signal-to-noise ratio (SNR) efficiency using the constrained Cramer-Rao lower bound (CRLB)31. However, MRF schedules optimized by the aforementioned indirect metrics cannot guarantee the low reconstruction errors for each tissue parameter. In this study, to optimize RF saturation-encoded MRF acquisitions with a minimal number of saturation parameters for tissue parameter determination, we proposed a learning-based optimization framework. Unlike the optimization methods based on indirect measurements, the proposed approach can optimize scan parameters by directly computing quantitative errors in tissue parameters.
2. Theory
Transient-state signal model
The MTC-MRF signal model is based on modified Bloch equations for a two-pool exchange system (w: free bulk water pool, m: semisolid macromolecule pool). In the presence of MT exchange, the time evolution for longitudinal magnetization in the two pools during an RF saturation can be described with tissue parameters and scan parameters, such as RF saturation power (B1), frequency offset (Ω), saturation time (Ts), and delay time (Td)2,32:
| [1] |
| [2] |
| [3] |
| [4] |
| [5] |
| [6] |
where T1i and T2i are the longitudinal and transverse relaxation times of pool i, respectively; kij is the proton exchange rate from pool i to pool j; M0i is the equilibrium magnetization of pool i; ω1 is the RF saturation amplitude (= 2πγB1); and γ is the gyromagnetic ratio. The analytical solution of time-dependent longitudinal magnetization of the free bulk water pool from Eqs. [1] – [6] can be written as:
| [7] |
where
| [8] |
| [9] |
| [10] |
| [11] |
and where Mssw is the longitudinal magnetization of the free bulk water pool under steady-state (dM/dt = 0), and the saturation of the magnetization for the free bulk water and semisolid pools is governed by the RF absorption rates as follows:
| [12] |
During the relaxation delay time period, the magnetization vector was computed solely by the relaxation recovery process in the absence of B1, and the magnetization vector at the endpoint of each dynamic scan served as an initial condition for the next dynamic scan. Finally, the transient-state MTC-MRF signal evolution (SMTC-MRF) can be described by25,26:
| [13] |
The analytical solution for the transient-state signal model was incorporated with the deep-learning framework to optimize an MRF acquisition schedule with respect to the scan parameters (B1, Ω, Ts, and Td).
3. Methods
3.1. MTC-MRF acquisition schedules
3.1.1. Linear acquisition schedule
A linear acquisition schedule was generated by linearly increasing values from the minimum to maximum points of each scan parameter range. Specifically, the parameter ranges were: 8 – 50 ppm for frequency offsets; 0.9 – 2 μT for RF saturation powers; 0.4 – 2 s for RF saturation times; and 3.5 – 4.5 s for relaxation delay times. The range of scan parameters was determined by specific absorption rate (SAR) constraints. In particular, the limited range of the RF saturation power was used to stay within the clinically permitted SAR, mainly due to the use of the SAR-intensive, time-interleaved parallel transmission (pTX)-based RF saturation and 3D TSE readout with multiple inversion RF pulses. The same range of scan parameters was applied to the other acquisition schedules.
3.1.2. Interior-Point (IP)-based acquisition schedule
An acquisition schedule was generated using an IP algorithm by maximizing signal discrimination between different tissue types30. The optimization problem was solved by minimizing the correlation coefficient between MRF signals of the pre-defined dictionary, which could be formulated as follows:
| [14] |
where (d: length of the schedule) is the RF saturation and acquisition schedule, is the identity matrix, and D is the dictionary of MTC-MRF signals for all possible tissue parameter values. A total of ten thousand tissue parameter sets (N = 10,000) were contained in the dictionary, which were randomly sampled from each tissue parameter range. The signals in the dictionary were normalized to power one for accurate calculation of the correlation coefficients between MTC-MRF profiles. The IP algorithm was initialized with a pseudo-random (PR) schedule (see 3.1.4).
3.1.3. Cramer-Rao Lower Bound (CRLB)-based acquisition schedule
An acquisition schedule was optimized by minimizing CRLB values for the MTC-MRF signal model (Eq. 13)31. The CRLB with respect to tissue parameters , where q is the number of the parameters, can be defined as:
| [15] |
where represents the CRLB matrix and I(p) is the Fischer Information Matrix (FIM) defined as:
| [16] |
where is the log-likelihood function, SMTC-MRF is the MTC-MRF signal, and x is the RF saturation and acquisition schedule. With a white Gaussian noise, the Fisher information matrix can be expressed as:
| [17] |
where is the Jacobian matrix of the MTC-MRF signal at nth repetition time (TR), and σ is the standard deviation of the noise. The CRLB optimization focuses on minimizing the variance of tissue parameter estimates, thereby maximizing SNR efficiency. For any given tissue parameter dataset, the objective function of the CRLB optimization can be written as follows:
| [18] |
where x is the RF saturation schedule, N is the number of tissue parameter sets used to calculate the objective function, and W is the weighting function, which was heuristically determined. The tissue parameter dataset used for CRLB optimization was the same as that used for IP optimization.
3.1.4. Pseudo-random (PR) acquisition schedule
A pseudo-random acquisition schedule was generated by increasing spectral and temporal incoherence between dynamic scans, which could reduce information redundancy in multiple image acquisitions. The PR schedule was the same as that used in our previous studies25,26.
3.1.5. Learning-based optimization of acquisition schedule (LOAS)
A fully-connected neural network (FCNN) was designed to optimize the RF saturation schedule for tissue parameter quantification. MTC-MRF signal profiles were generated using initial scan parameters and tissue parameters through a forward two-pool Bloch transform. The simulated MTC-MRF signals were fed to the FCNN architecture as an input, which outputs tissue parameter estimates (Fig. 1). The estimated tissue parameters were then compared to the input (ground-truth) for a loss calculation. The loss function was a mean square difference between the ground-truth and the estimated tissue parameters. The calculated loss was back-propagated to update scan parameters to minimize the loss via the adaptive moment estimation (ADAM) optimizer. Thus, the scan parameters were updated iteratively through epochs toward decreasing quantification errors. The objective function of the architecture was formulated as:
| [19] |
where p represents the ground-truth tissue parameters (input), x is the RF saturation and acquisition schedule, SMTC-MRF represents the MTC-MRF signal, η is the noise, T2w is the transverse relaxation time of the free bulk water, θ represents the parameters (weights) of the neural network, and represents the estimated tissue parameters (output). The FCNN architecture (f) consisted of FC (ds+1,256) – ReLU – FC (256,256) – ReLU – FC (256,4)– Sig, where FC(n,m) is a fully connected layer of input size n and output size m, ReLU is the rectified linear unit activation function, Sig is the sigmoid activation function, and ds is the number of dynamic scans. To normalize the MTC parameters, the sigmoid function was adopted at the last layer. The learning-based optimization architecture was trained by minimizing the mean square difference (loss function, Lθ) between p and , as follows:
| [20] |
Figure 1.
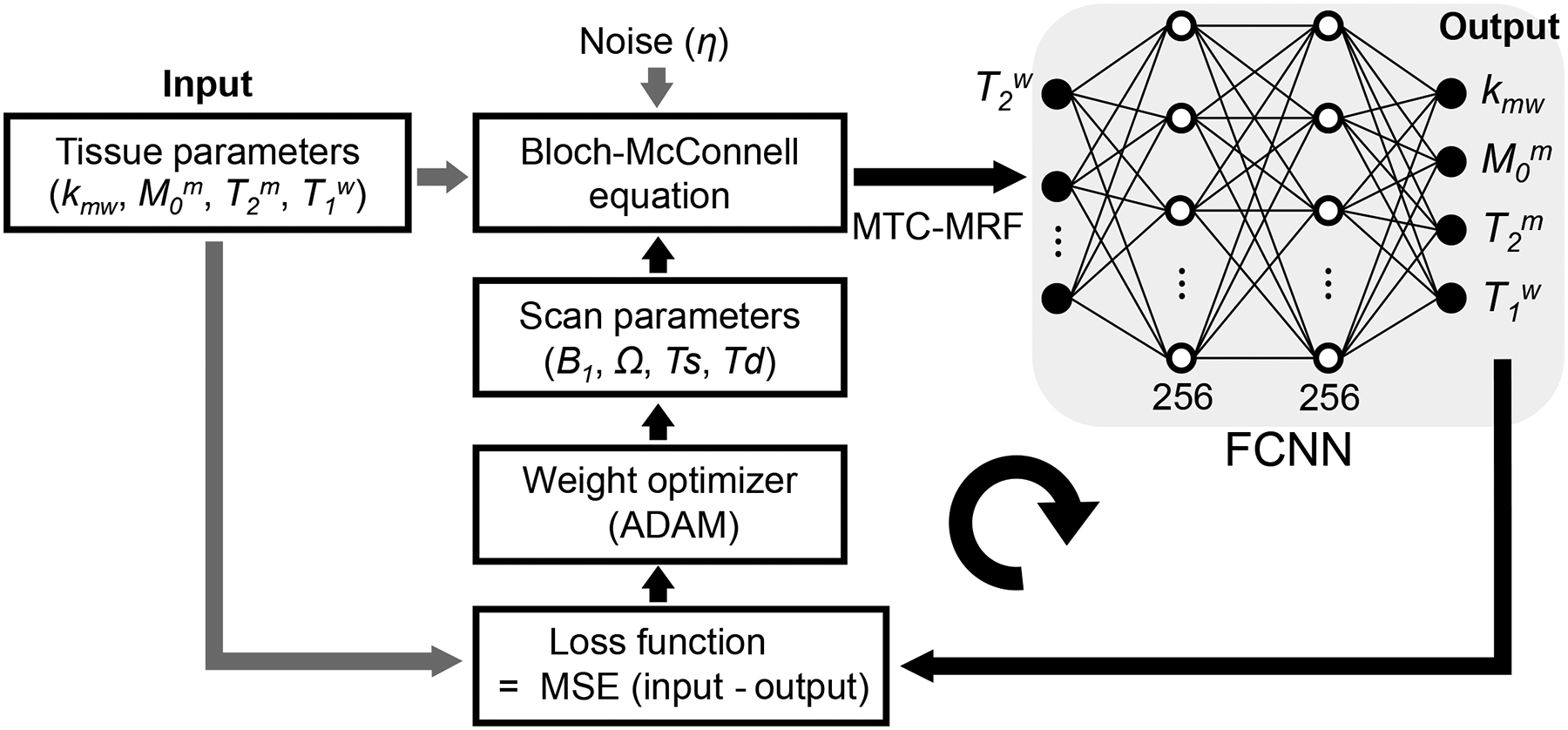
A scheme of the learning-based optimization of the acquisition schedule (LOAS). MRF signals are synthesized using initialized scan parameters, noise, and tissue parameters (Input) and fed to the fully connected neural network (FCNN). The FCNN outputs tissue parameter estimates (Output). A loss function is a mean square error between the ground-truths and estimated tissue parameters. The calculated loss was back-propagated with an ADAM optimizer to update scan parameters.
For the training dataset, forty million sets of tissue parameters were randomly sampled from the pre-defined range of each parameter. Specifically, the ranges of tissue parameters were: 5 – 100 Hz for Kmw; 2 – 17 % for M0m; 1 – 100 μs for T2m; and 0.2 – 3.0 s for T1w. In addition, T1 of macromolecular pool was set to a constant value of 1 s because it was not well determined2,33. The networks were trained using Tensorflow with ADAM as the optimizer34. The initial learning rate was set to 10−3 with a batch size of 200. Training was implemented on an NVIDIA TITAN RTX GPU (Santa Clara, CA).
3.2. MTC-MRF Bloch simulations
Two Bloch-McConnell equation-based simulation studies were performed to evaluate the reconstruction accuracy and efficiency of MTC-MRF schedules. For the first simulation study, four digital phantoms were constructed to evaluate the accuracy of tissue parameters (kmw, M0m, T2m, and T1w) with various schedules (Linear, IP, CRLB, PR, and LOAS). For each phantom with a matrix size of 100 × 500, one tissue parameter had five uniformly sampled values (5, 25, 50, 75, and 100 Hz for Kmw, 2, 6, 10, 14, and 17 % for M0m, 1, 25, 50, 75, and 100 μs for T2m, and 0.2, 0.9, 1.6, 2.3, and 3.0 s for T1w), while other three tissue parameter values were randomly selected. The parameter ranges were given as 5 to 100 Hz for kmw, 2 to 17% for M0m, 1 to 100 μs for T2m, and 0.2 to 3.0 s for T1w. Longitudinal magnetization evolutions of the free bulk water protons were simulated with the tissue parameters and scan parameters, and fed to the newly trained 6-layer FCNN architecture for tissue quantifications. Quantification results of the FCNN method were compared with those of the Bloch-fitting approach.
In the second digital phantom study, the efficiency of the MRF schedule optimized by the LOAS method was evaluated with various numbers of dynamic scans (#5, #10, #20, #30, and #40). The optimization was performed for each dynamic scan number schedule. The quantification accuracy was evaluated by calculating normalized root mean square errors (nRMSE) between ground-truths and tissue parameters estimated by the FCNN-based quantification method. The Bloch simulations were performed on a 64-bit Windows operating system (12-CORE, 3.8-GHz AMD processor and 32 GB of memory) using MATLAB (The MathWorks, Natick, MA).
3.3. In vivo MRI experiments
MRI experiments were performed on a 3T MRI system (Achieva dStream, Philips Healthcare, Best, The Netherlands), using a body coil for transmission and a 32-channel head coil for reception. Six subjects were included in the in vivo experiments, two females and four males (mean age: 36 years; range: 27 to 41 years of age). All subjects were examined with the approval of the institutional ethics committee, and written informed consent was obtained prior to the study. 3D MTC-MRF images were obtained from a fat-suppressed (spectral pre-saturation with inversion recovery, SPIR) multi-shot TSE pulse sequence with a four-fold (2 × 2) compressed sensing (CS) acceleration in the Ky-Kz plane35,36. The imaging parameters of MTC-MRF were: TE = 6 ms; FOV = 212 × 186 × 60 mm3; spatial resolution = 1.8 × 1.8 × 4 mm3; slice-selective 120° refocusing pulses; turbo factor = 104; and slice oversampling factor = 1.4. The pTX technique was used to achieve continuous RF saturation (100% duty-cycle saturation over 2 s, BW = 24.5 Hz), which is capable of reducing amplifier limitations while detecting highly sensitive saturation effects on clinical scanners35,37. In addition, the pTX-based pseudo-continuous-wave RF irradiation allowed for a larger degree of freedom for the RF saturation schedule and a simple analytical solution of the two-pool exchange model. Forty dynamic scans were acquired with varied frequency offsets, RF saturation powers, RF saturation times, and relaxation delay times. For amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging, another series of saturated images was obtained with a saturation power of 1.2 μT, a saturation time of 2 s, a delay time of 4 s, and six frequency offsets (±3, ±3.5, and ±4 ppm), following the MTC-MRF scans. For normalization, an unsaturated image (S0) was acquired by turning off the saturation pulse. In addition, water saturation shift-referencing (WASSR) images (26 frequency offsets range of ±1.2 ppm at intervals of 0.125 ppm, B1 = 0.5 μT)38 and multi-echo gradient-spin-echo (GRASE)39,40 images (TR = 3 s, echoes = 20, 40, 60, 80, and 100 ms, TSE factor = 5) were acquired for B0 and T2 maps, respectively. Note that water T2 maps were separately measured due to lack of water spin information from MTC-MRF images with far off-resonance frequency offsets (Ω >8ppm).
3.4. Image processing for APT and NOE imaging
MTC images at ±3.5 ppm, synthesized using scan parameters, estimated tissue parameters from the FCNN framework, and acquired T2 relaxation time, were used as reference baseline images for APT and NOE imaging. APT and NOE images were calculated based on the subtraction method41:
| [21] |
| [22] |
where Zlab is the normalized label signal intensity (from free water + semisolid macromolecule + multiple CEST pools), Zref is the normalized reference signal intensity containing only MTC and direct saturation effects (from free water + semisolid macromolecules), ΔΩ is the frequency offset of a specific labile group, Ssat is the signal intensity measured with RF saturation, and S0 is the signal intensity measured without RF saturation. Here, ΔΩ of +3.5 ppm and −3.5 ppm were selected for APT and NOE images, respectively42,43. Zref (±3.5ppm) images were synthesized whereas Zlab (±3.5ppm) images were experimentally acquired.
3.5. Validation of MTC-MRF with synthetic MRI
Due to the lack of in-vivo ground truth information, a synthetic MRI technique was adopted to validate the proposed LOAS method using in-vivo images. The in-vivo tissue parameter maps reconstructed from the FCNN framework using 40 MTC-MRF images acquired with the PR schedule were defined as the reference (namely, Reference). Then, synthetic 3D MTC-MRF images were generated using the reference tissue parameters and the MRF schedules (LOAS, PR, CRLB, IP, and Linear) via a two-pool Bloch transform, and fed to 6-layer FCNN architectures newly trained with the MRF schedules. The reference tissue parameters were used to validate water and MTC quantification, comparing tissue parameters estimated with the various MRF schedules to the explicitly known ground-truth, and to gauge how tissue parameter estimation errors depend on the particular choice of scan parameters (e.g., MRF schedule and the number of scans). To assess the efficiency of the LOAS schedule, the quantification accuracy was evaluated by calculating nRMSE between the reference and tissue parameters estimated with various numbers of scans from 5 to 40 in an increment of 5. Eight FCNN architectures that corresponded to the different dynamic scan numbers were separately trained and used to estimate tissue parameters. In total, 176 different FCNN architectures were separately trained and tested with the various MRF schedules: 78 for CLRB; 58 for IP; and 40 for LOAS, PR, CRLB, IP, and Linear, with eight different dynamic scan numbers. Moreover, the accuracy of estimated MTC at 3.5 ppm, APT, and NOE signals were evaluated. A Bland-Altman analysis was performed to assess the agreement of tissue parameters estimated from the different dynamic scan numbers.
4. Results
4.1. Reconstruction error vs. optimization cost
Reconstruction errors of LOAS and optimization costs of CRLB and IP were monitored through epochs or iterations, as shown in Fig. 2. Overall training loss of LOAS (Fig. 2A), CRLB (Fig. 2C), and correlation (Fig. 2E) values decreased as the iteration and epoch proceeded. In the test process, reconstruction errors (converted to nRMSE) of the LOAS for each tissue parameter decreased with the epoch, as shown in Fig. 2B. However, the reconstruction errors of CRLB and IP were invariant even the optimization costs decreased with iterations, as shown in Figs. 2D and 2F. Note that the CRLB algorithm ran for 58 iterations to minimize the objective function, and thus, the corresponding 58 MRF schedules were used to train each FCNN architecture for water and MTC quantification. The reconstruction errors for each parameter were calculated from the test dataset, with 58 respectively trained FCNN models (Fig. 2D). The IP algorithm ran for 78 iterations and the reconstruction errors for each parameter (Fig. 2F) were evaluated in the same way as with the CRLB. The RF saturation schedules optimized by LOAS, CRLB, and IP strategies, and PR and Linear schedules are shown in Fig. 3 (also see Supporting Information Table S1).
Figure 2.
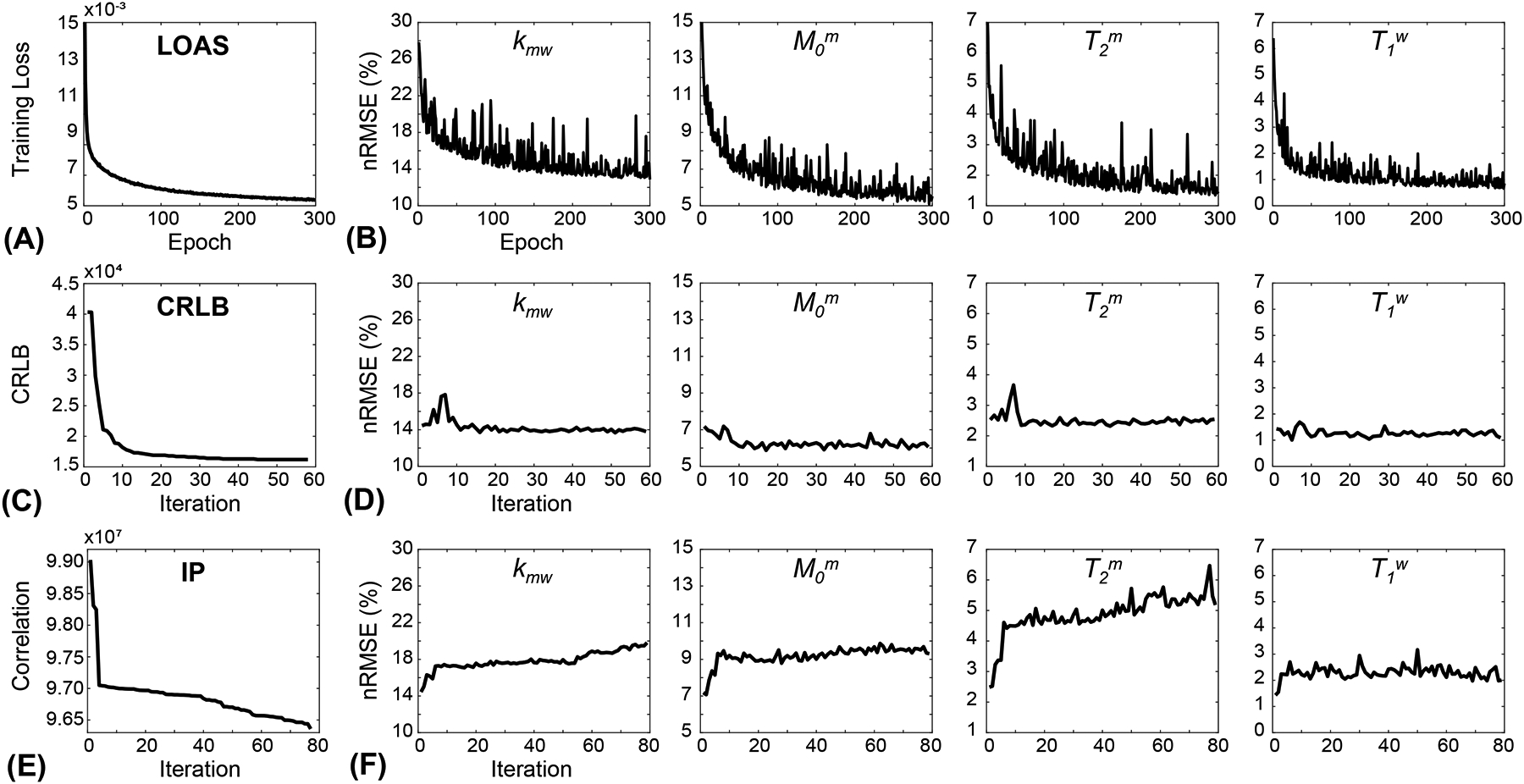
(A) A training loss with respect to epoch numbers from the LOAS architecture. (B) Test losses (converted to nRMSE values) of each tissue parameter. (C) Optimization cost as a function of iteration numbers from CRLB. Fifty-eight FCNN architectures were trained with corresponding MRF schedules obtained at each iteration. (D) nRMSE values for the estimated tissue parameters with respect to iteration numbers. (E) Optimization cost with respect to iteration numbers from IP. Seventy-eight FCNN architectures were trained with corresponding MRF schedules obtained at each iteration. (F) nRMSE values for the estimated tissue parameters with respect to iteration numbers.
Figure 3.
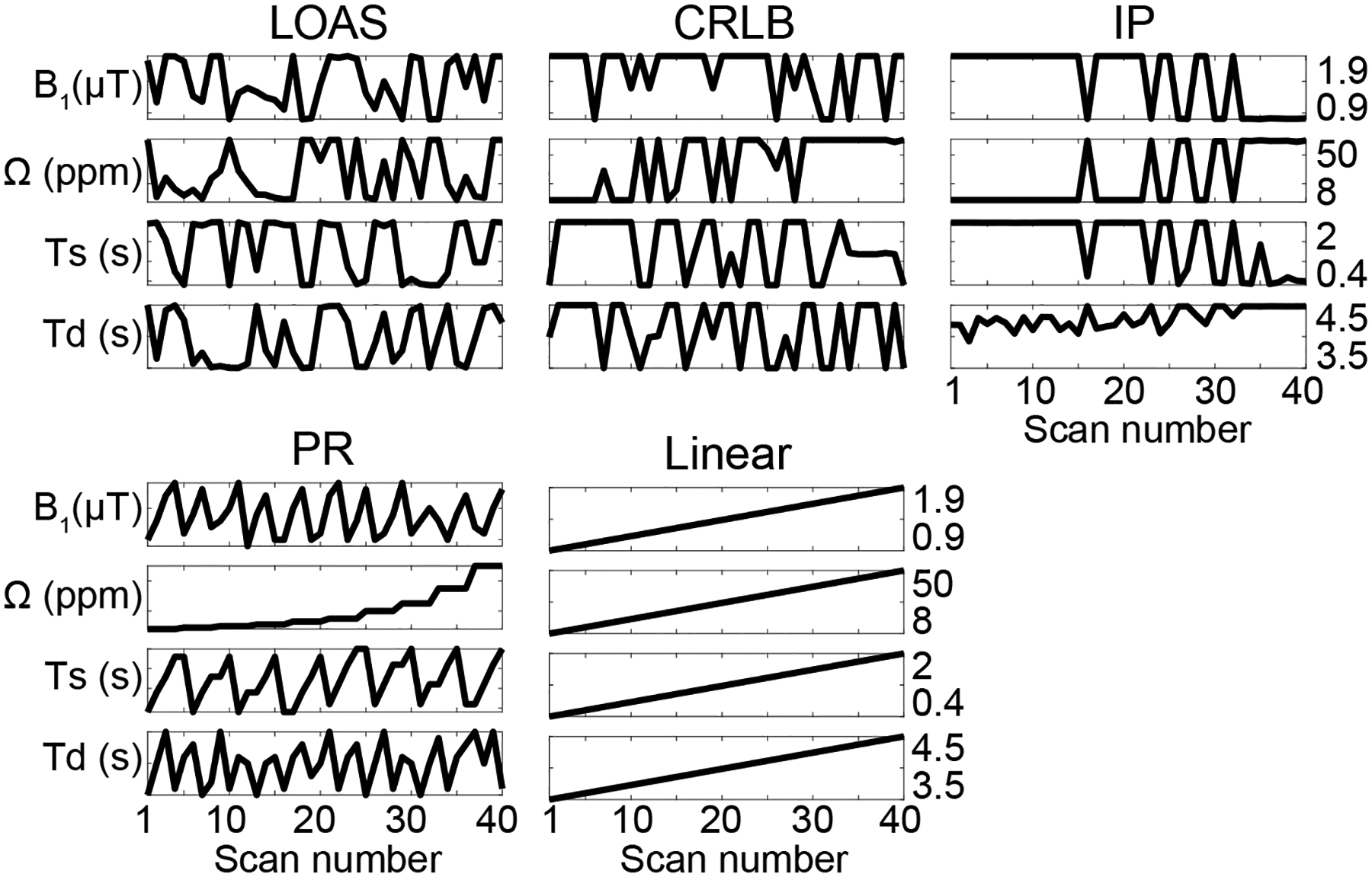
Optimized MRF schedules (B1, Ω, Ts, and Td) with 40 dynamic scans from the LOAS, CRLB, IP, PR, and Linear methods.
4.2. Digital phantom studies
Optimized MRF schedules were evaluated using digital phantoms encoded by the two-pool transient-state MTC model. Water and MTC parameters estimated from the deep-learning FCNN and Bloch fitting-based quantification methods were compared to the ground-truths, as shown in Fig. 4. Overall, the nRMSE values of the FCNN method were lower than those of the Bloch fitting method except for T1w estimated using LOAS, PR, and CRLB schedules. Nevertheless, both quantification methods still showed a high degree of accuracy for T1w estimation. The FCNN-based quantification approach with the LOAS schedule showed the highest accuracy in the quantification of tissue parameters. The average nRMSE values of kmw, M0m, T2m, and T1w between the FCNN and the ground-truth were 15.2%, 6.6%, 3.5%, and 1.3% for LOAS, 15.9%, 7.4%, 3.8%, and 1.6% for PR, 15.3%, 6.6%, 4.0%, and 1.3% for CRLB, 21.1%, 9.8%, 7.4%, and 2.0% for IP, and 32.4%, 16.1%, 14.1%, and 8.8% for Linear, respectively. Tissue parameters estimated from the FCNN with different numbers of dynamic scans (e.g., #40, #30, #20, #10, and #5 dynamic scans) were compared to the ground-truth and nRMSE values were calculated to evaluate the scan efficiency of the MTC-MRF (Fig. 5). A reduction of the number of dynamic scans resulted in increased nRMSE values. However, the parameters estimated from the LOAS schedule with 10 dynamic scans were not significantly different from the ground-truths. Moreover, significant correlations were observed between all estimated parameters and the ground-truth in all dynamic scan numbers (p< 0.05 for all parameters) and the correlation coefficients were all over 0.8, even for #5 dynamic scans (see Supporting Information Table S2). The nRMSE values with the dynamic scan numbers of #40, #30, #20, #10, and #5 were 15.2%, 15.8%, 16.9%, 18.8%, and 20.7% for kmw, 6.6%, 6.9%, 7.7%, 8.8%, and 11.0% for M0m, 3.5%, 3.9%, 4.5%, 6.0%, and 8.7% for T2m, and 1.3%, 1.4%, 1.7%, 2.2%, and 2.9% for T1w, respectively.
Figure 4.
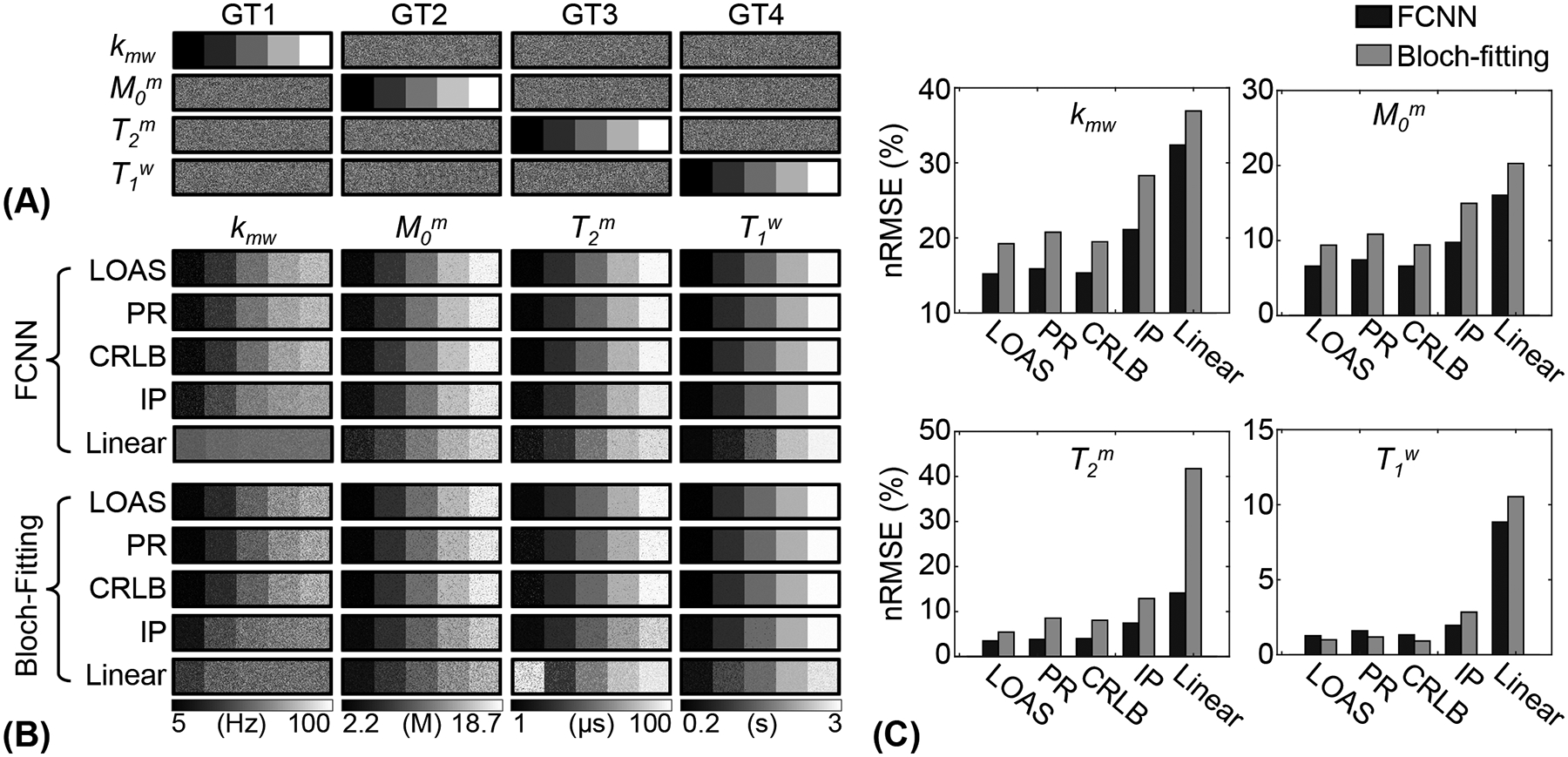
Bloch equation-based digital phantom experiments with the optimized MRF schedules. (A) Four ground-truth (GT1, GT2, GT3, and GT4) digital phantoms. (B) Water and MTC parameter maps estimated with various MRF schedules. (C) The resulting parameter maps from the Bloch-fitting and FCNN methods were compared to the ground-truths by calculating nRMSE.
Figure 5.
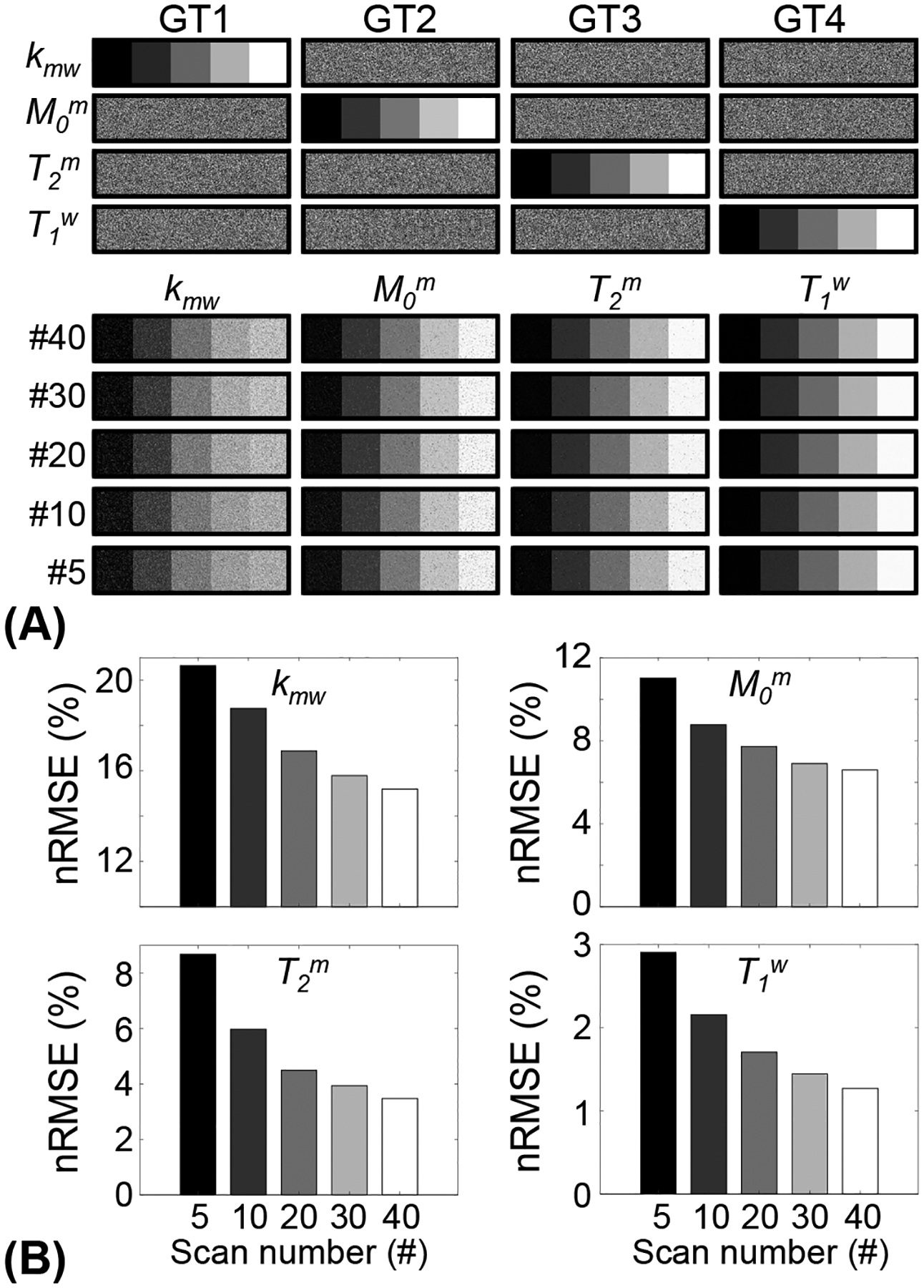
Bloch equation-based digital phantom experiments with different numbers of dynamic scans. (A) Parameter maps reconstructed from FCNN with respect to number of dynamic scans with LOAS schedules. (B) Reconstruction accuracy was evaluated for each tissue parameter with various dynamic scan numbers.
4.3. In vivo MRI studies
To evaluate the LOAS method with in-vivo data, synthetic MTC-MRF images were generated using the five MRF schedules and reference tissue parameters obtained from the PR schedule with 40 dynamic scans (Fig. 6). Then, synthetic images, as a test dataset, were fed into the newly-trained FCNN with each MRF schedule. The estimation accuracy was evaluated by calculating the mean absolute error (MAE) between the reference and tissue parameter estimates with the different MRF schedules. The lowest MAE values (0.95 for kmw, 0.36 for M0m, 0.97 for T2m, and 0.02 for T1w) were obtained with the LOAS method. In addition, good agreements were still observed for the PR and CRLB-based schedules. However, the IP and Linear schedules showed poor quantification quality.
Figure 6.
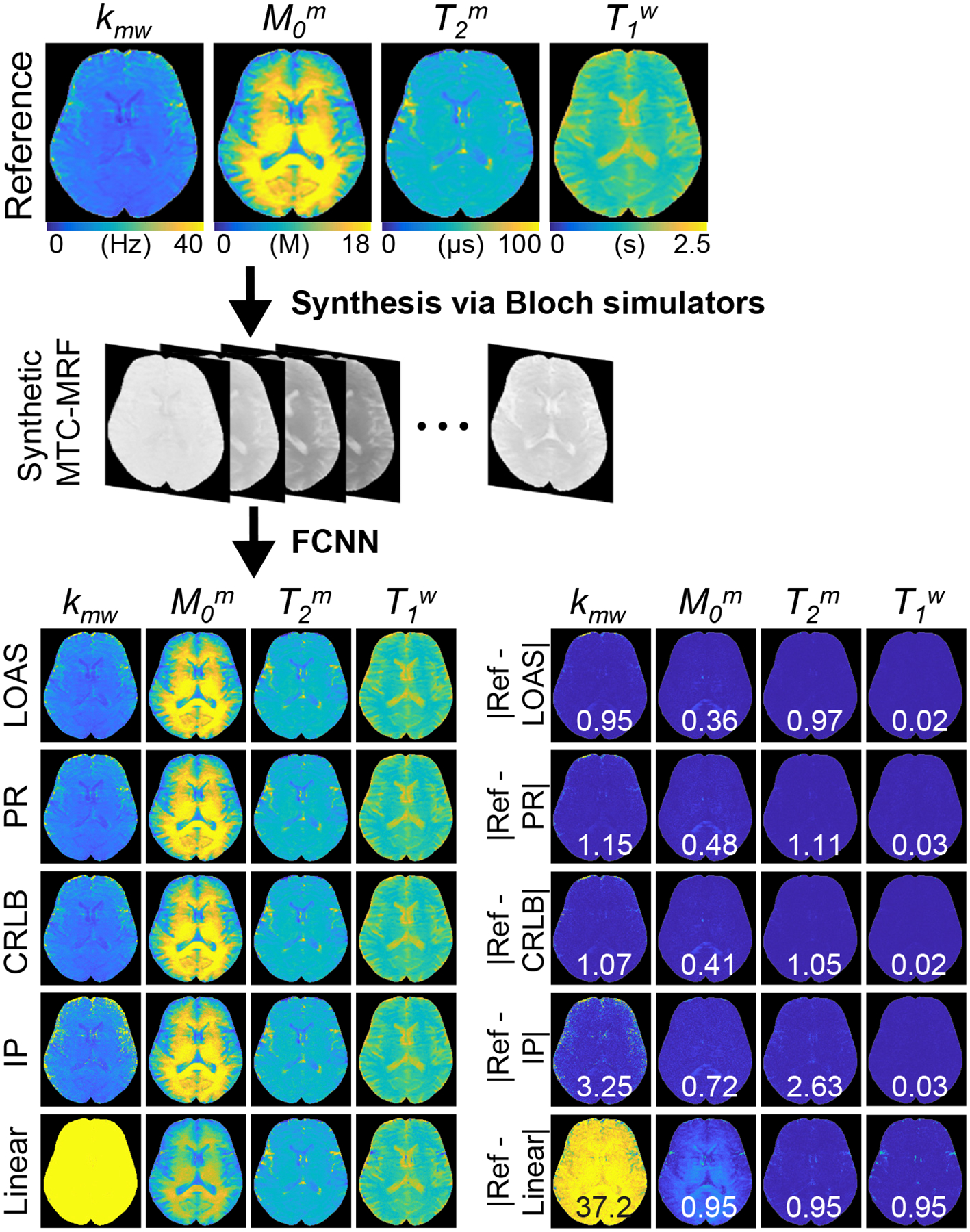
A schematic of the validation process using a synthetic MRI technique. Synthetic MTC-MRF images were generated using the reference maps and MRF schedules optimized with the LOAS, PR, CRLB, IP, and Linear methods and fed to newly-trained FCNN frameworks corresponding to the optimized schedules. Difference images between the reference and the estimated maps are shown. The mean difference value of each map is shown in the insert (white). Note that the MTC proton concentration was converted to absolute units using the water proton concentration (110 M).
Figure 7 shows the accuracy of tissue parameters, MTC at 3.5 ppm, APT, and NOE signals estimated using LOAS with different numbers of data sampling points. When the number of dynamic scans decreased, nRMSE values increased for water and MTC parameters. However, MTC, APT, and NOE contrast-weighted signals were relatively insensitive to the number of dynamic scans. Compared to the reference values obtained from 40 dynamic scans, parameter quantification with 10 dynamic scans was accurate, where nRMSE values were 3.44%, 3.23%, 1.84%, 2.03%, 0.49%, 0.49%, and 0.49% for kmw, M0m, T2m, T1w, Zref(3.5ppm), APT, and NOE, respectively. The estimated tissue parameter maps and contrast-weighted images (Zref(3.5ppm), APT, and NOE) are shown in Fig. 8.
Figure 7.
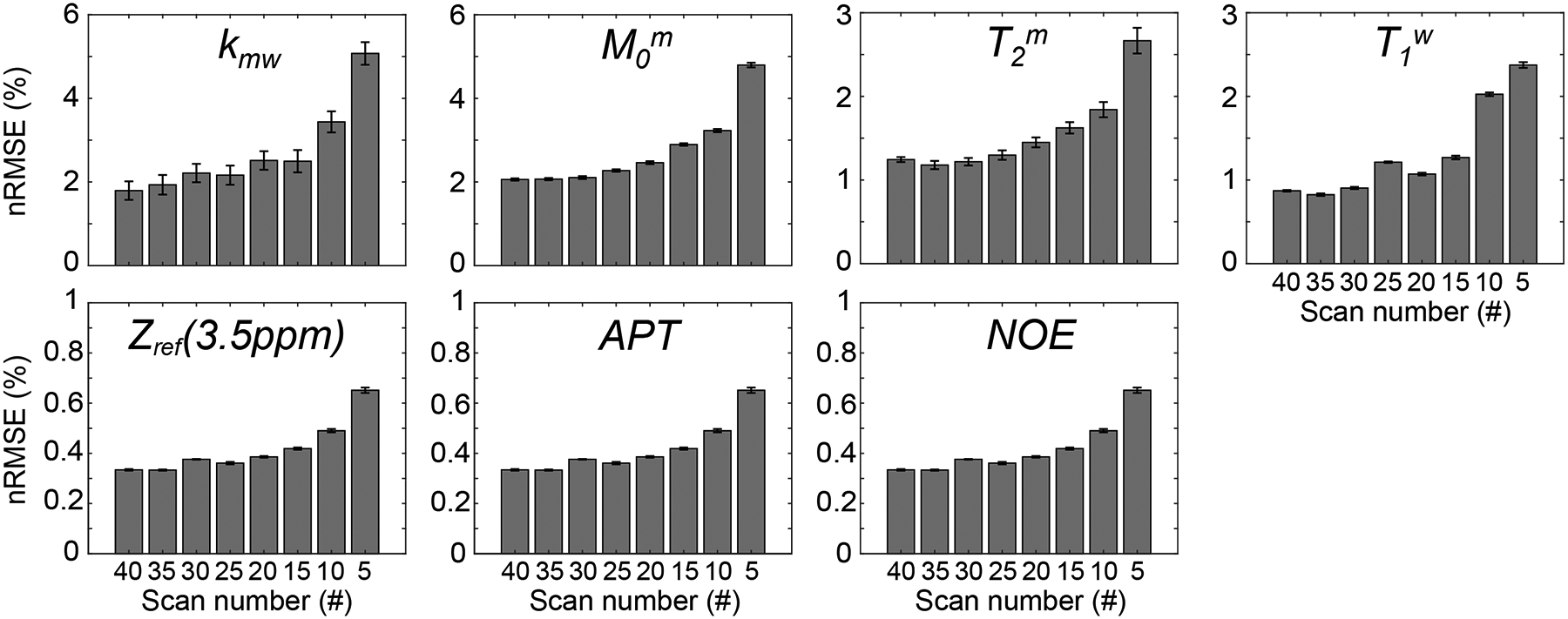
Quantification accuracy of LOAS with respect to the number of dynamic scans (#40, #35, #30, #25, #20, #15, #10, and #5) for water and MTC parameters, Zref (3.5ppm), APT, and NOE signal intensities from six healthy volunteers.
Figure 8.
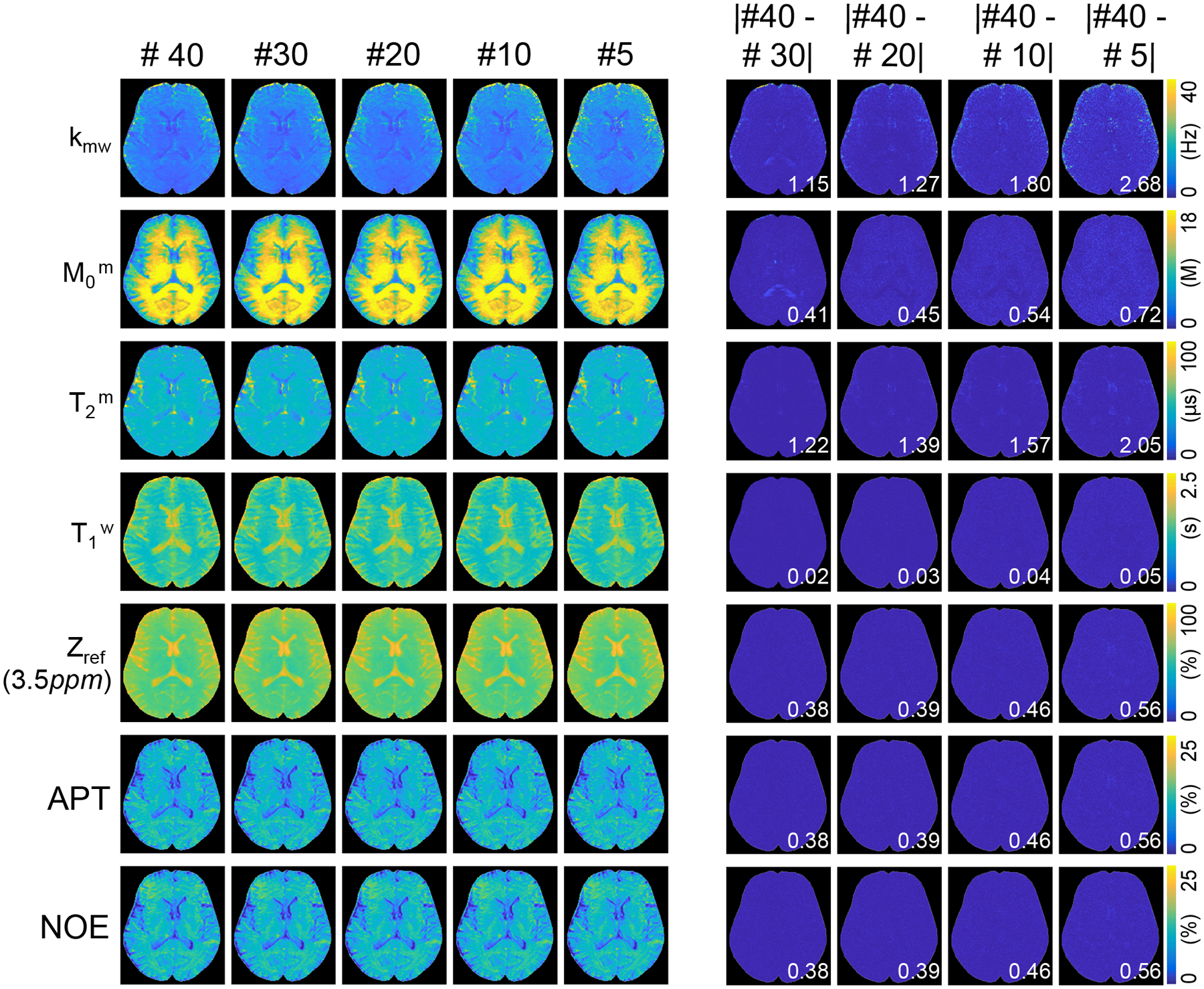
Quantitative MTC parameter maps, water T1 map, Zref (3.5ppm), APT, and NOE images of a representative human brain of a healthy volunteer. Difference images of the estimated maps with #40 dynamic scan vs. #30, #20, #15, #10, and #5 dynamic scans. The mean difference value of each map is shown in the insert (white).
A Bland Altman analysis was performed to evaluate the agreement among tissue parameter estimates for different numbers of dynamic scans, as shown in Fig. 9. No significant bias for the measurement of kmw, M0m, T2m, and T1w was observed over all dynamic scan numbers, except the dynamic scan number of #5 in kmw. The analysis revealed that the 95% confidence interval (CI) of the agreement became wider and more outliers were presented at a smaller number of dynamic scans. Particularly, when reducing the dynamic scan number from 10 to 5, a huge expansion of the 95% CI was observed for all tissue parameters. The Pearson correlation coefficient values between the dynamic scan number of #40 and the dynamic scan numbers of #30, #20, #10, and #5, respectively were 0.93, 0.88, 0.85, 0.75 for kmw, 0.99, 0.99, 0.99, and 0.98 for M0m, 0.98, 0.97, 0.97, and 0.94 for T2m, and 0.99, 0.99, 0.98, and 0.97 for T1w (p < 0.01 for all). The structural similarity index measure (SSIM) values between the dynamic scan number of #40 and the dynamic scan numbers of #30, #20, #10, and #5, respectively, were 0.97, 0.96, 0.94, 0.90 for kmw, 0.98, 0.98, 0.97, and 0.95 for M0m, 0.99, 0.98, 0.98, and 0.97 for T2m, and 0.99, 0.99, 0.98, and 0.97 for T1w (p < 0.01 for all).
Figure 9.
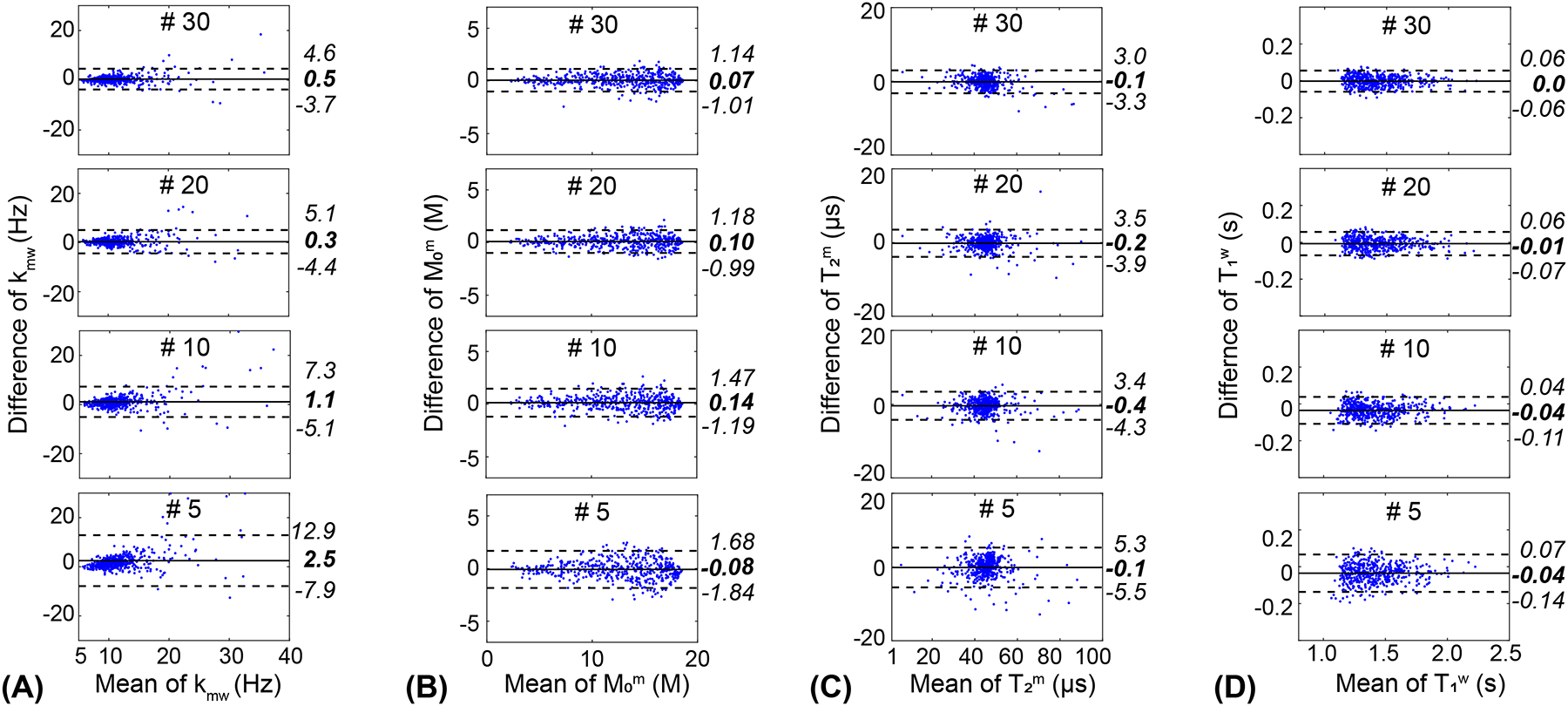
Bland-Altman plots comparing MTC (A) exchange rate, (B) concentration, (C) T2 relaxation time, and (D) water T1 relaxation time between #40 dynamic scans and #30, #20, and #10, respectively. The central solid lines show the mean bias, and the upper and lower dashed lines show the variation limits (±1.96 standard deviation).
5. Discussion
In this study, the MRF optimization problem was solved using a deep-learning model trained with a large amount of simulated MTC-MRF signals. In the supervised-learning framework, RF saturation parameters were updated toward minimizing a loss function that could directly calculate tissue quantification errors. Therefore, the LOAS method improved parametric reconstruction quality compared to existing indirect optimization methods19,31, enabling a further four-fold temporal acceleration by reducing the number of dynamic scans, in addition to the CS acceleration in two phase encoding directions (eight-fold acceleration in total).
A high degree of freedom and flexibility of MRF scan parameters pose challenges for sequence optimization13,44,45. Current MRF optimization methods are based on indirect optimization metrics. The CRLB-based MRF optimization technique minimizes the Cramer-Rao lower bound of tissue parameter estimates, which is equivalent to maximizing SNR efficiency31. The IP algorithm-based optimization method minimizes the signal correlation, or maximizes the discrimination between MRF signals of different tissue properties20,30. The indirect optimization metrics, however, cannot guarantee an optimal solution for tissue parameter quantification. In this study, the uncertainty of the objective functions in the CRLB and IP methods were evaluated by calculating direct tissue parameter errors for each iteration. Interestingly, the CRLB and correlation metrics were inconsistent with the tissue parameter errors, although the optimization cost values decreased with the number of iterations. In contrast, the LOAS updated the MRF scan parameters based on the gradient of the estimated tissue parameter error, and thus, the training loss of the LOAS directly reflected the tissue parameter error. It is interesting to note that Bang-Bang behavior was observed in the CRLB and IP optimization results, in which optimal values were often found at either the lower bound or the upper bound of the pre-defined scan parameter ranges (Fig. 3). Such a structural behavior was also observed in a previous study31. We speculate that this could be attributed, in part, to the limited ranges of scan parameters, thus limiting the maximum encoding efficiency. In our case, particularly, RF saturation powers and durations, as well as relaxation delay times, were highly constrained by SAR.
The proposed method was validated using simulated digital phantoms, and in vivo experiments against other sub-optimal MTC-MRF schedules. The performances of the various optimization methods were retrospectively evaluated by solving Bloch equations since it would not be feasible to prospectively acquire many images with different MRF schedules and dynamic scan numbers in a limited scan time. Our results showed that the LOAS outperformed other state-of-the-art methods30,31 in terms of quantification accuracy and acquisition efficiency. Moreover, the LOAS takes advantage of compatibility with various types of noise distributions, while the CRLB optimization requires the inherent assumption of a Gaussian noise distribution. In addition, the LOAS method could be integrated with the deep-learning-based tissue quantification or Bloch-fitting approaches. Importantly, the LOAS framework can be extended to include any other analytical signal models, tissue properties, sequences, and target scan parameters, which makes it extremely versatile and suitable for a wide range of multi-parameter MRF applications. A super-Lorentzian lineshape has been known to provide a good approximation for RF absorption rate by a semisolid MTC pool2,46,47. However, the calculation of numerical integration in the super-Lorentzian lineshape is significantly time-consuming. Instead, the Lorentzian lineshape was used in this study to reduce the computational burden on generating massive amounts of training dataset for 176 different FCNN architectures.
For water and MTC quantification, a deep-learning neural network was used to learn a mapping relation between a high-dimensional MR fingerprint space and a low-dimensional tissue parameter space. Although the FCNN required a long training time to optimize the weights of the network (20 hours for 40 million datasets), the neural network had low computational complexity in the test phase. A conventional Bloch equation-based fitting approach enabled tissue parameter estimation at a high computational cost (two hours for an image matrix of 256 × 256 × 9 × 40). In comparison, the FCNN framework enabled almost instantaneous estimation of tissue parameters (four seconds for an image matrix of 256 × 256 × 9 × 40). Furthermore, we exploited flexibility and degrees of freedom in MRF sequence parameters to improve quantification accuracy and efficiency. With the LOAS method, ten dynamic scans provided the best trade-off between quantification accuracy and acquisition efficiency, possibly by four-fold reduction in scan time. Corresponding tissue parameter maps are shown in Supporting Information Figure S1. Notably, the optimized MTC-MRF framework can provide baseline MTC reference signals for APT and NOE imaging and potentially be extended to quantitative CEST and NOE parameter mappings (e.g., proton exchange rate and concentration) within a clinically feasible scan time.
6. Conclusion
We proposed a learning-based optimization framework to accelerate data acquisition and improve quantification accuracy for magnetization transfer contrast MR fingerprinting. Unlike optimization methods based on indirect measurements, the proposed approach optimized scan parameters by directly calculating quantitative errors of the tissue parameters and significantly improved the accuracy and also reduced data acquisition time. The flexible LOAS framework could be a powerful optimization tool for MRF pulse sequence design.
Supplementary Material
Supporting Information Table S1. LOAS-based MTC-MRF schedules for dynamic scan numbers 40 and 10.
Supporting Information Figure S1. Quantitative water and MTC parameter maps from the LOAS, PR, CRLB, IP, and Linear schedules, and their corresponding difference images. The reference images (Ref) were obtained from deep-learning reconstruction with 40 MTC-MRF images acquired from the PR schedule. The mean difference value of each map is shown in the insert (white).
Supporting Information Table S2. The correlation coefficients between ground-truth values of digital phantoms and estimated parameters for free bulk water and semisolid MTC parameters for five different dynamic scan numbers.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Rad BriteStar Award from the Department of Radiology Johns Hopkins University School of Medicine, the National Institutes of Health (R01NS112242 and P41EB031771), and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1135).
Abbreviations:
- MRF
MR Fingerprinting
- MTC
Magnetization Transfer Contrast
- APT
Amide Proton Transfer
- CEST
Chemical Exchange Saturation Transfer
- LOAS
Learning-based Optimization of Acquisition Schedule
- FCNN
Fully Connected Neural Network
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
References
- 1.Balaban RS, Ceckler TL. Magnetization transfer contrast in magnetic resonance imaging. Magn Reson Q 1992;8:116–137. [PubMed] [Google Scholar]
- 2.Henkelman RM, Huang X, Xiang QS, Stanisz GJ, Swanson SD, Bronskill MJ. Quantitative interpretation of magnetization transfer. Magn Reson Med 1993;29(6):759–766. [DOI] [PubMed] [Google Scholar]
- 3.Pike GB. Pulsed magnetization transfer contrast in gradient echo imaging: a two-pool analytic description of signal response. Magn Reson Med 1996;36(1):95–103. [DOI] [PubMed] [Google Scholar]
- 4.van Zijl PCM, Lam WW, Xu J, Knutsson L, Stanisz GJ. Magnetization Transfer Contrast and Chemical Exchange Saturation Transfer MRI. Features and analysis of the field-dependent saturation spectrum. Neuroimage 2018;168:222–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanisz GJ, Kecojevic A, Bronskill MJ, Henkelman RM. Characterizing white matter with magnetization transfer and T(2). Magn Reson Med 1999;42(6):1128–1136. [DOI] [PubMed] [Google Scholar]
- 6.Filippi M, Rocca MA, Martino G, Horsfield MA, Comi G. Magnetization transfer changes in the normal appearing white matter precede the appearance of enhancing lesions in patients with multiple sclerosis. Ann Neurol 1998;43(6):809–814. [DOI] [PubMed] [Google Scholar]
- 7.Vavasour IM, Laule C, Li DK, Traboulsee AL, MacKay AL. Is the magnetization transfer ratio a marker for myelin in multiple sclerosis? J Magn Reson Imaging 2011;33(3):713–718. [DOI] [PubMed] [Google Scholar]
- 8.Smith SA, Golay X, Fatemi A, Mahmood A, Raymond GV, Moser HW, van Zijl PC, Stanisz GJ. Quantitative magnetization transfer characteristics of the human cervical spinal cord in vivo: application to adrenomyeloneuropathy. Magn Reson Med 2009;61(1):22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ropele S, Seewann A, Gouw AA, van der Flier WM, Schmidt R, Pantoni L, Inzitari D, Erkinjuntti T, Scheltens P, Wahlund LO, Waldemar G, Chabriat H, Ferro J, Hennerici M, O’Brien J, Wallin A, Langhorne P, Visser MC, Barkhof F, Fazekas F, group Ls. Quantitation of brain tissue changes associated with white matter hyperintensities by diffusion-weighted and magnetization transfer imaging: the LADIS (Leukoaraiosis and Disability in the Elderly) study. J Magn Reson Imaging 2009;29(2):268–274. [DOI] [PubMed] [Google Scholar]
- 10.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PC, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med 2007;58(4):786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heo HY, Jones CK, Hua J, Yadav N, Agarwal S, Zhou J, van Zijl PC, Pillai JJ. Whole-brain amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging in glioma patients using low-power steady-state pulsed chemical exchange saturation transfer (CEST) imaging at 7T. J Magn Reson Imaging 2016;44(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heo HY, Zhang Y, Jiang S, Lee DH, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semisolid magnetization transfer reference (EMR) signals: II. Comparison of three EMR models and application to human brain glioma at 3 Tesla. Magn Reson Med 2016;75(4):1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton JI, Jiang Y, Chen Y, Ma D, Lo WC, Griswold M, Seiberlich N. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med 2017;77(4):1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma D, Jones SE, Deshmane A, Sakaie K, Pierre EY, Larvie M, McGivney D, Blumcke I, Krishnan B, Lowe M, Gulani V, Najm I, Griswold MA, Wang ZI. Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions. J Magn Reson Imaging 2019;49(5):1333–1346. [DOI] [PubMed] [Google Scholar]
- 16.Ma D, Pierre EY, Jiang Y, Schluchter MD, Setsompop K, Gulani V, Griswold MA. Music-based magnetic resonance fingerprinting to improve patient comfort during MRI examinations. Magn Reson Med 2016;75(6):2303–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cauley SF, Setsompop K, Ma D, Jiang Y, Ye H, Adalsteinsson E, Griswold MA, Wald LL. Fast group matching for MR fingerprinting reconstruction. Magn Reson Med 2015;74(2):523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heo HY, Han Z, Jiang S, Schar M, van Zijl PCM, Zhou J. Quantifying amide proton exchange rate and concentration in chemical exchange saturation transfer imaging of the human brain. Neuroimage 2019;189(1):202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen O, Huang S, McMahon MT, Rosen MS, Farrar CT. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magn Reson Med 2018;80(6):2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlman O, Herz K, Zaiss M, Cohen O, Rosen MS, Farrar CT. CEST MR-Fingerprinting: Practical considerations and insights for acquisition schedule design and improved reconstruction. Magn Reson Med 2020;83(2):462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Han P, Zhou B, Christodoulou AG, Shaw JL, Deng Z, Li D. Chemical exchange saturation transfer fingerprinting for exchange rate quantification. Magn Reson Med 2018;80(4):1352–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Z, Chen Y, Liu M, Xiang L, Zhang Q, Wang Q, Lin W, Shen D. Deep Learning for Fast and Spatially Constrained Tissue Quantification From Highly Accelerated Data in Magnetic Resonance Fingerprinting. IEEE Trans Med Imaging 2019;38(10):2364–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoppe E, Korzdorfer G, Wurfl T, Wetzl J, Lugauer F, Pfeuffer J, Maier A. Deep Learning for Magnetic Resonance Fingerprinting: A New Approach for Predicting Quantitative Parameter Values from Time Series. Stud Health Technol Inform 2017;243:202–206. [PubMed] [Google Scholar]
- 24.Cohen O, Zhu B, Rosen MS. MR fingerprinting Deep RecOnstruction NEtwork (DRONE). Magn Reson Med 2018;80(3):885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang B, Kim B, Schar M, Park H, Heo HY. Unsupervised Learning for Magnetization Transfer Contrast MR Fingerprinting (MTC-MRF): Application to Chemical Exchange Saturation Transfer (CEST) and Nuclear Overhauser Enhancement (NOE) Imaging. Magn Reson Med 2020:DOI: 10.1002/mrm.28573. [DOI] [PubMed] [Google Scholar]
- 26.Kim B, Schar M, Park H, Heo HY. A deep learning approach for magnetization transfer contrast MR fingerprinting and chemical exchange saturation transfer imaging. Neuroimage 2020;221:117165. [DOI] [PubMed] [Google Scholar]
- 27.Glang F, Deshmane A, Prokudin S, Martin F, Herz K, Lindig T, Bender B, Scheffler K, Zaiss M. DeepCEST 3T: Robust MRI parameter determination and uncertainty quantification with neural networks-application to CEST imaging of the human brain at 3T. Magn Reson Med 2020;84(1):450–466. [DOI] [PubMed] [Google Scholar]
- 28.Zaiss M, Deshmane A, Schuppert M, Herz K, Glang F, Ehses P, Lindig T, Bender B, Ernemann U, Scheffler K. DeepCEST: 9.4 T Chemical exchange saturation transfer MRI contrast predicted from 3 T data - a proof of concept study. Magn Reson Med 2019;81(6):3901–3914. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Schar M, Chan KWY, Huang J, Wei Z, Lu H, Qin Q, Weiss RG, van Zijl PCM, Xu J. In vivo imaging of phosphocreatine with artificial neural networks. Nat Commun 2020;11(1):1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen O, Rosen MS. Algorithm comparison for schedule optimization in MR fingerprinting. Magn Reson Imaging 2017;41:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bo Z, Haldar JP, Congyu L, Dan M, Yun J, Griswold MA, Setsompop K, Wald LL. Optimal Experiment Design for Magnetic Resonance Fingerprinting: Cramer-Rao Bound Meets Spin Dynamics. IEEE Trans Med Imaging 2019;38(3):844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quesson B, Thiaudiere E, Delalande C, Chateil JF, Moonen CT, Canioni P. Magnetization transfer imaging of rat brain under non-steady-state conditions. Contrast prediction using a binary spin-bath model and a super-lorentzian lineshape. J Magn Reson 1998;130(2):321–328. [DOI] [PubMed] [Google Scholar]
- 33.Ramani A, Dalton C, Miller DH, Tofts PS, Barker GJ. Precise estimate of fundamental in-vivo MT parameters in human brain in clinically feasible times. Magn Reson Imaging 2002;20(10):721–731. [DOI] [PubMed] [Google Scholar]
- 34.Abadi M, Barham P, Chen JM, Chen ZF, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng XQ. TensorFlow: A system for large-scale machine learning. Proceedings of Osdi’16: 12th Usenix Symposium on Operating Systems Design and Implementation 2016:265–283. [Google Scholar]
- 35.Heo HY, Xu X, Jiang S, Zhao Y, Keupp J, Redmond KJ, Laterra J, van Zijl PCM, Zhou J. Prospective acceleration of parallel RF transmission-based 3D chemical exchange saturation transfer imaging with compressed sensing. Magn Reson Med 2019;82(5):1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heo HY, Zhang Y, Lee DH, Jiang S, Zhao X, Zhou J. Accelerating chemical exchange saturation transfer (CEST) MRI by combining compressed sensing and sensitivity encoding techniques. Magn Reson Med 2017;77(2):779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Togao O, Hiwatashi A, Keupp J, Yamashita K, Kikuchi K, Yoshiura T, Yoneyama M, Kruiskamp MJ, Sagiyama K, Takahashi M, Honda H. Amide Proton Transfer Imaging of Diffuse Gliomas: Effect of Saturation Pulse Length in Parallel Transmission-Based Technique. PLoS One 2016;11(5):e0155925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med 2009;61(6):1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu ML, Chang HC, Oshio K, Chen NK. A single-shot T2 mapping protocol based on echo-split gradient-spin-echo acquisition and parametric multiplexed sensitivity encoding based on projection onto convex sets reconstruction. Magn Reson Med 2018;79(1):383–393. [DOI] [PubMed] [Google Scholar]
- 40.Baessler B, Schaarschmidt F, Stehning C, Schnackenburg B, Giolda A, Maintz D, Bunck AC. Reproducibility of three different cardiac T2 -mapping sequences at 1.5T. J Magn Reson Imaging 2016;44(5):1168–1178. [DOI] [PubMed] [Google Scholar]
- 41.Heo HY, Lee DH, Zhang Y, Zhao X, Jiang S, Chen M, Zhou J. Insight into the quantitative metrics of chemical exchange saturation transfer (CEST) imaging. Magn Reson Med 2017;77(5):1853–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Med 2003;9:1085–1090. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Heo HY, Knutsson L, van Zijl PCM, Jiang S. APT-weighted MRI: Techniques, current neuro applications, and challenging issues. J Magn Reson Imaging 2019;50(2):347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panda A, Mehta BB, Coppo S, Jiang Y, Ma D, Seiberlich N, Griswold MA, Gulani V. Magnetic Resonance Fingerprinting-An Overview. Curr Opin Biomed Eng 2017;3:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee PK, Watkins LE, Anderson TI, Buonincontri G, Hargreaves BA. Flexible and efficient optimization of quantitative sequences using automatic differentiation of Bloch simulations. Magn Reson Med 2019;82(4):1438–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrison C, Henkelman RM. A model for magnetization transfer in tissues. Magn Reson Med 1995;33:475–482. [DOI] [PubMed] [Google Scholar]
- 47.Heo HY, Zhang Y, Lee DH, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 tesla. Magn Reson Med 2016;75(1):137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table S1. LOAS-based MTC-MRF schedules for dynamic scan numbers 40 and 10.
Supporting Information Figure S1. Quantitative water and MTC parameter maps from the LOAS, PR, CRLB, IP, and Linear schedules, and their corresponding difference images. The reference images (Ref) were obtained from deep-learning reconstruction with 40 MTC-MRF images acquired from the PR schedule. The mean difference value of each map is shown in the insert (white).
Supporting Information Table S2. The correlation coefficients between ground-truth values of digital phantoms and estimated parameters for free bulk water and semisolid MTC parameters for five different dynamic scan numbers.


