Abstract
Brain-derived neurotrophic factor (BDNF) is a widely expressed neurotrophin that supports the survival, differentiation, and signaling of various neuronal populations. Although it has been well described that expression of BDNF is strongly regulated by neuronal activity, little is known whether regulation of BDNF expression is similar in different brain regions. Here, we focused on this fundamental question using neuronal populations obtained from rat cerebral cortices and hippocampi of both sexes. First, we thoroughly characterized the role of the best-described regulators of BDNF gene - cAMP response element binding protein (CREB) family transcription factors, and show that activity-dependent BDNF expression depends more on CREB and the coactivators CREB binding protein (CBP) and CREB-regulated transcriptional coactivator 1 (CRTC1) in cortical than in hippocampal neurons. Our data also reveal an important role of CREB in the early induction of BDNF mRNA expression after neuronal activity and only modest contribution after prolonged neuronal activity. We further corroborated our findings at BDNF protein level. To determine the transcription factors regulating BDNF expression in these rat brain regions in addition to CREB family, we used in vitro DNA pulldown assay coupled with mass spectrometry, chromatin immunoprecipitation (ChIP), and bioinformatics, and propose a number of neurodevelopmentally important transcription factors, such as FOXP1, SATB2, RAI1, BCL11A, and TCF4 as brain region-specific regulators of BDNF expression. Together, our data reveal complicated brain region-specific fine-tuning of BDNF expression.
SIGNIFICANCE STATEMENT To date, majority of the research has focused on the regulation of brain-derived neurotrophic factor (BDNF) in the brain but much less is known whether the regulation of BDNF expression is universal in different brain regions and neuronal populations. Here, we report that the best described regulators of BDNF gene from the cAMP-response element binding protein (CREB) transcription factor family have a more profound role in the activity-dependent regulation of BDNF in cortex than in hippocampus. Our results indicate a brain region-specific fine tuning of BDNF expression. Moreover, we have used unbiased determination of novel regulators of the BDNF gene and report a number of neurodevelopmentally important transcription factors as novel potential regulators of the BDNF expression.
Keywords: BDNF cortex, CREB, hippocampus, neuronal activity
Introduction
Brain-derived neurotrophic factor (BDNF) is a neurotrophin that contributes to neuronal survival, differentiation and synaptic plasticity (Chao, 2003; Park and Poo, 2013). The expression of BDNF gene, consisting of numerous 5′ noncoding exons that are spliced together with the common protein-coding 3′ exon, is controlled by distinct promoter regions (Timmusk et al., 1993; Aid et al., 2007; Pruunsild et al., 2007) that are used for stimulus-specific, cell type-specific and brain region-specific BDNF expression. For example, in rodents BDNF transcripts containing BDNF exon II or exon IXa are highly expressed in the hippocampus, but at much lower levels in the cortex (Aid et al., 2007). Transgenic mice encompassing the whole rat or human BDNF locus revealed that the transgene could not recapitulate BDNF expression in hippocampal dentate granule cells (Koppel et al., 2009, 2010), implying differences in BDNF expression between cortex and hippocampus. The expression of different BDNF transcripts is dysregulated in a brain region-specific manner in patients with schizophrenia and in response to antidepressants (Wong et al., 2010), and in mice after chronic mild stress (P. Huang et al., 2018). Overall, the expression of BDNF varies in different brain regions in both health and disease.
It is acknowledged that BDNF has critical and distinguished roles in the cortex and hippocampus (Bergami et al., 2008; Hong et al., 2008; Sakata et al., 2009; Bambah-Mukku et al., 2014; Wang et al., 2015; Ortiz-López et al., 2017; Mudd et al., 2019). Understanding cell type-specific and brain region-specific stimulus-dependent regulation of BDNF expression serves as the basis for discerning how BDNF affects brain development and function. Therefore, we set out to compare the regulation of neuronal activity-dependent BDNF expression in cortical and hippocampal neurons.
To date, a number of transcription factors, including calcium-response factor (CaRF; Tao et al., 2002), Neuronal PAS Domain Protein 4 (NPAS4; Lin et al., 2008; Pruunsild et al., 2011), cAMP-response element binding protein (CREB; Shieh et al., 1998; Tao et al., 1998; Tabuchi et al., 2002; Hong et al., 2008; Benito et al., 2011; Pruunsild et al., 2011; Palomer et al., 2016; Tai et al., 2016), nuclear factor of activated T-cells (NFAT; Vashishta et al., 2009), and myocyte enhancer factor 2 (MEF2; Flavell et al., 2008; Lyons et al., 2012) families have been described in regulating neuronal activity-dependent BDNF expression (for review, see West et al., 2014). Notably, CaRF has been shown to regulate the levels of BDNF exon IV in cortex but not in hippocampus (McDowell et al., 2010) and MEF2 family transcription factors have been suggested to participate in the regulation of BDNF exon I levels in hippocampal but not in cortical neurons (Flavell et al., 2008; Lyons et al., 2012). Among the transcription factors that regulate activity-dependent BDNF expression, CREB transcription factor family is the best described. Notably, CREB binding to CRE-elements in the genome is cell-type specific (Cha-Molstad et al., 2004), and CREB target genes vary in different brain regions (Tanis et al., 2008). Although the role of CREB in the regulation of BDNF gene expression is well-established in both cortical and hippocampal neurons, it is not known whether the role of CREB in activity-dependent expression of BDNF is the same in both of these neuronal populations.
Here, focusing on the CREB transcription factor family we describe that the regulation of BDNF transcription by these factors and their coactivators CBP and CRTC1 is more prominent in cortical than in hippocampal neurons. Furthermore, our results indicate the role of CREB family in regulating the basal and early induced levels of BDNF mRNA after neuronal activity, whereas CREB family has a minor role in the late induction. We also show that CREB and coactivators CBP and CRTC1 are important for proper induction of BDNF protein expression after membrane depolarization in cortical neurons. We describe the component of BDNF-TrkB signaling in membrane-depolarization induced expression of BDNF in cortical but not in hippocampal neurons. Additionally, we used in vitro promoter pulldown coupled with mass spectrometric detection of the bound transcription factors and report novel regulators of BDNF, namely FOXP1, SATB2, RAI1, TCF4, BCL11A. Our results highlight the brain region-specific fine-tuning of BDNF expression.
Materials and Methods
Rat primary neuron culture and treatments
All animal procedures were performed in compliance with the local ethics committee. Cortical and hippocampal neuron cultures were generated from Sprague Dawley rat male and female pups of the same litter at embryonic day 20–21 as described in Esvald et al. (2020). For lentivirus-mediated overexpression, neurons were transduced at 0 days in vitro (DIV). All lentiviruses encoding EGFP, A-CREB, negative guide RNA (gRNA), CREB1 gRNA, CRTC1 gRNAs, and dCas9-KRAB-T2A-GFP were used as detailed by Esvald et al. (2020). At 2 DIV, half of the growth medium (for experiments with low molecular-weight inhibitors) or the whole media (in experiments where neurons were transduced with lentiviral particles) was changed and a final concentration of 10 μm mitotic inhibitor 5-fluoro-2′-deoxyuridine (Sigma) was added. For the Western blotting experiments, the neurons were plated in DMEM (Invitrogen) and 10% FBS (Pan Biotech) and the media was changed to supplemented NBA media after ∼1.5 h. Half of the media was changed at 2 DIV and 5 DIV. In all the experiments, at 7 DIV (∼24 h before the experiment), 1 μm tetrodotoxin (Tocris Bioscience) was added to the media to inhibit spontaneous neuronal activity. For experiments with low molecular-weight inhibitors, 5 μm CBP-CREB interaction inhibitor (CCII; in DMSO, Merck Millipore, catalog #217505, lot 2758191) or 25 nm Az23 (in DMSO, Axon Medchem) was added 1 h or 30 min, respectively, before KCl treatment at 8 DIV. Control cells were treated with the corresponding volume of DMSO. At 8 DIV, 25 mm KCl (Applichem; with 5 μm D-(2R)-amino-5-phosphonovaleric acid (D-APV, Cayman Chemical Company) to reduce excitotoxicity) was used to elicit membrane depolarization for 1, 3, or 6 h.
RNA extraction and RT-qPCR
After treatment, cultured neurons were lysed at 8 DIV in RLT lysis buffer (containing 1% β-mercaptoethanol (VWR Life Sciences) in experiments with lentivirus transduction) and RNA was purified with RNeasy Mini kit (QIAGEN) according to the manufacturer's recommendations with on-column digestion of genomic DNA using RNase-Free DNase set (QIAGEN). RNA concentration was measured with BioSpec-Nano (Shimadzu) or with Nanodrop 2000c (Thermo Scientific) spectrophomoteter.
To compare mRNA levels from cortical and hippocampal tissues, the lysates were prepared from the same pup and biological replicates were obtained from pups of different litters. The cortex and hippocampus of 8-day-old Sprague Dawley rat pups were dissected and frozen in 80°C. Tissue was weighed and homogenized in QIAzol lysis reagent (QIAGEN) at a ratio of 1 ml/100 mg of tissue (but not <500 µl). The amount of cortex was normalized to the amount of hippocampus by taking mass equivalent of homogenous lysate for RNA purification. Next, Qiazol was added to all lysates to achieve a volume of 1 ml and incubated for 5 min at room temperature. Then, 200 µl of chloroform was added, the mixture was shaken vigorously, and incubated at room temperature for 3 min. Finally, the solutions were centrifuged at 12,000 × g at 4°C for 15 min and the aqueous phase was transferred to a new tube. Lastly, equal volume of 70% ethanol was added, vortexed, and transferred to a Mini Spin column (EconoSpin). RNA purification was then performed as for cultured neurons using RNeasy Mini kit (QIAGEN) solutions and on-column digestion of DNA. The concentrations of RNA were measured with Nanodrop 2000c (Thermo Scientific) spectrophomoteter.
Equal amounts of RNA were taken to synthesize complementary DNA (cDNA) using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) and oligo(dT)20 primer. qPCR was performed using HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne) and LightCycler 480 Instrument II (Roche). Primers used in qPCR are listed in Tuvikene, 2021. All mRNA expression levels were normalized to HPRT1 expression levels.
Western blotting
The neurons were grown on 24-well plates, treated at 8 DIV with KCl, and lysed in 1× Laemmli buffer (without bromophenol blue and β-mercaptoethanol) followed by heating at 95°C for 5 min. The protein concentration was measured with Pierce BCA Protein Assay kit (Thermo Scientific) according to the manufacturer's protocol. After measuring the concentration, bromophenol blue and β-mercaptoethanol (VWR Life Sciences, 5% final concentration) were added, lysates were heated at 95°C for 5 min, and centrifuged at 16,000 × g for 1 min.
Equal amount of total protein (16–20 µg) per experiment was loaded along with 100–200 pg of recombinant mature BDNF protein (Icosagen, catalog #P-105-100) and separated on 15% gel using SDS-PAGE. The proteins were transferred to a PVDF membrane using Trans-Blot Turbo Transfer system (Bio-Rad) high MW program (constant 1.3 A, 10 min). Next, the membranes were blocked for at least 1 h at room temperature in 5% skimmed milk in TBST buffer (1× Tris-buffered saline (pH 7.4) and 0.1% Tween 20), incubated overnight at 4°C with primary anti-BDNF antibody (Icosagen, catalog #327-100, clone 3C11, 1 mg/ml, 1:1000) in 2% milk in TBST, and then overnight at 4°C with secondary antibody anti-mouse IgG conjugated with horseradish peroxidase (HRP; Thermo Scientific, 1:5000) in 2% milk. After both incubations the membrane was washed three times with TBST for 5 min at room temperature. Chemiluminescence signal was produced with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) or SuperSignal West Atto Maximum Sensitivity Substrate (Thermo Scientific). The signal was measured using ImageQuant Las 4000 (GE Healthcare Life Sciences) imaging system.
To measure CREB protein levels, the anti-mouse IgG-HRP signal was quenched with 30% H2O2 for 20 min at 37°C, followed by three washes with TBST buffer. The membrane was blocked again for at least 1 h at room temperature in 5% skimmed milk in TBST buffer. Anti-CREB antibody (Cell Signaling, D76D11, rabbit mAb #4820, 1:2500) was incubated overnight at 4°C in 2% milk in TBST and secondary antibody anti-rabbit IgG-HRP (Thermo Scientific, 1:5000) in 2% milk for at least 1 h at room temperature. To measure FLAG-A-CREB levels, the previous signal was again quenched with H2O2, the membrane was washed and blocked as described above, and then reprobed with mouse monoclonal anti-FLAG M2-HRP antibody (Sigma-Aldrich, #A8592, 1:5000) overnight at 4°C in 2% milk.
CRTC1 protein levels were analyzed from a separate 10% gel. The proteins were transferred to PVDF membrane using Trans-Blot Turbo Transfer system (Bio-Rad) with modified high MW program (constant 1.3 A, 15 min). The membrane was blocked and washed as described for other membranes. Primary anti-CRTC1 antibody (Cell Signaling, C71D11, rabbit mAb #2587, 1:1000) was incubated overnight at 4°C in 2% milk and secondary antibody anti-rabbit IgG-HRP (Thermo Scientific, 1:5000) in 2% milk for at least 1 h at room temperature.
For loading control, the membranes were stained with Coomassie solution (0.1% Coomassie Brilliant Blue R-250 Dye, 25% ethanol, 7% acetic acid), followed by washes with destaining solution (30% ethanol, 10% acetic acid) and rinsing with tap water. The membranes were imaged using ImageQuant Las 4000 (GE Healthcare Life Sciences) system.
BDNF, CREB, CRTC1, and FLAG-A-CREB protein levels were quantified using densitometric analysis with ImageQuant TL software (GE Healthcare Life Sciences, version 7.0). A marginally small constant number was added to each densitometric value (to account for undetectable signal of pro-BDNF in untreated cortical neurons in some experiments), and the specific protein levels were then normalized using Coomassie staining values. Statistical analysis was performed as described below (Experimental design and statistical analysis).
In vitro DNA pulldown and mass spectrometry
Rat BDNF promoter I and BDNF promoter IV regions were amplified by PCR from luciferase reporter plasmids (published in Esvald et al., 2020). The primers used are listed in Tuvikene, 2021. The biotinylated PCR products were purified using DNA Clean and Concentrator-100 kit (Zymo Research).
Nine hippocampi and three cortices from 8-day-old Sprague Dawley rat pups of both sexes were dissected and snap-frozen in liquid nitrogen. Nuclear lysates were prepared and in vitro DNA pulldown was performed as described previously (Tuvikene et al., 2021). Here, 350 µg of nuclear proteins were bound to 75 pmol of biotinylated DNA in the pulldown assay. The subsequent mass spectrometric analysis and custom R-script are also described previously (Tuvikene et al., 2021). At least a 1.5-fold enrichment to one promoter respective to the other was considered as specific binding and tissue-specificity was calculated only for transcription factors passing the specific binding criteria. All proteins detected in mass spectrometry are listed in Extended Data Figure 7-1.
Analysis of RNA-sequencing (RNA-seq) data
Raw RNA-seq data of previously published RNA-seq experiments [SRA accession numbers ERP016406 (Dias et al., 2016), SRP049264 (Araujo et al., 2015), SRP058362 (McKenna et al., 2015), SRP068801 (Jaitner et al., 2016), SRP074487 (W.H. Huang et al., 2016), SRP102696 (Araujo et al., 2017), SRP166779 (Garay et al., 2020), SRP261474 (Carullo et al., 2020)] were downloaded directly as fastq files from European Nucleotide Archive with the assistance of SRA-explorer (www.sra-explorer.info). Adapter and quality trimming was done using BBDuk (part of bbmap version 38.79) using the following parameters: ktrim = r k = 23 mink = 11 hdist = 1 tbo qtrim = lr trimq = 10, and minlen = 50 (for SRP261474), minlen = 45 (for ERP016406, SRP102696), minlen = 30 (for SRP049264), minlen = 25 (for SRP166779, SRP068801, SRP058362) or minlen = 65 (for SRP074487). Reads were mapped to rn6 (annotation from ENSEMBL, release 101, for SRP261474), hg19 genome (annotation from Gencode release 34, for SRP049264), or mm10 (annotation from Gencode M25 primary assembly, for the rest of the datasets) using STAR aligner (version 2.7.3a). Detected splice junctions were combined, junctions on mitochondrial DNA and junctions with noncanonical intron motifs were removed. Novel splice sites with at least 10 reads (for ERP016406, SRP261474 and SRP074487), or at least five reads (for the rest of the datasets) in the whole dataset were added for 2nd pass alignment using STAR. Aligned reads were assigned to genes (to determine total BDNF expression levels) or exons (to determine the expression levels of BDNF exon I and exon IV) using FeatureCounts (version 2.0.0), with additional -B -C arguments for paired-end reads. Counts were normalized using DESeq2 R package (version 1.28.1) and visualized using ggplot2 R package (version 3.3.2). Statistical analysis was performed on log-transformed normalized counts using two-tailed equal variance t test in Microsoft Excel 365.
Chromatin immunoprecipitation (ChIP) assay
For ChIP assay from tissue, 10–12 hippocampi and six cortices from 8-day-old Sprague Dawley rat pups of both sexes from the same litter were dissected and snap-frozen in liquid nitrogen. Pups from different litters were considered biological replicates. Tissue pieces were weighed and homogenized in 1% formaldehyde (Cell Signaling, catalog #12606, diluted in PBS; the volume of the fixing solution was 20× the mass of the tissue) with Dounce tissue grinder (Wheaton) using loose pestle 20 times. After that, the homogenate was transferred to a 15-ml tube and incubated for 10 min at room temperature with rotation. Final concentration of 0.125 m glycine was added to quench formaldehyde and the mixture was rotated for another 10 min at room temperature. Next, the mixture was centrifuged at 2600 × g at 4°C for 5 min and the pellet was washed two times with ice-cold PBS (20× volume of the tissue weight). A protocol to obtain nuclear lysates was developed based on Vierbuchen et al. (2017). Briefly, the pellet was resuspended in 20× volume of ice-cold L1 [50 mm HEPES (pH 7.5), 140 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.25% Triton X-100, 0.5% NP-40, 10% glycerol, 1× cOmplete protease inhibitor cocktail (Roche)], rotated for 10 min at 4°C and centrifuged at 2600 × g at 4°C for 5 min. The pellet was next resuspended in L2 [10 mm Tris-HCl (pH 8.0), 200 mm NaCl, 1× cOmplete protease inhibitor cocktail (Roche)] in a volume of 20× the weight of the initial tissue piece, rotated for 10 min at 4°C and centrifuged at 2600 × g at 4°C for 5 min. Finally, the nuclear pellet was resuspended in 10× volume of L3 [10 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine, 1× cOmplete protease inhibitor cocktail (Roche)], incubated on ice for a couple of minutes, and transferred to Diagenode 15 ml tubes onto prewashed sonication beads (0.2 g of beads per 0.5 ml of nuclear lysate was prepared by washing three times with 1 ml PBS, vortexing and spinning the beads down). After transferring the nuclear lysates onto the beads, the mixture was vortexed vigorously to improve sonication efficiency. Chromatin was sonicated using the BioRuptor Pico device (Diagenode) for five cycles 30 s on and 30 s off, vortexed vigorously and sonicated for another five cycles. Next, the lysate was transferred to a new tube, final concentration of 1% Triton X-100 was added, and centrifuged at 4500 × g at 8°C for 5 min in a swing-out rotor. The protein concentration was measured with BCA kit, and 200–275 µg of nuclear protein was used per IP; 10 µg of CREB (Cell Signaling, D76D11, rabbit mAb #4820) or TCF4 (CeMines) antibody or 4.8 µg of BCL11A (Developmental Studies Hybridoma Bank, PCRP-BCL11A-1G10) antibody was used for overnight incubation at 4°C. At the same time, 50 µl Dynabeads Protein G (Invitrogen) per IP were prewashed 2 times with 0.02% Tween 20-PBS and blocked overnight with 200 µg/ml BSA (Thermo Scientific) at 4°C.
The next day, the Dynabeads were washed twice with 0.02% Tween 20-PBS and diluted in L3, containing Triton X-100 and protease inhibitor solution. Dynabeads were added to the lysates and incubated on a rotator for 5–7 h at 4°C. Finally, the beads were washed four times in 1 ml wash buffer [1% Triton X-100, 0.1% SDS, 150 mm NaCl, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), 1× cOmplete protease inhibitor cocktail (Roche)] and once with final wash buffer [1% Triton X-100, 0.1% SDS, 500 mm NaCl, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), 1× cOmplete protease inhibitor cocktail (Roche)]. DNA-protein complexes were eluted with 50 μl elution buffer (1% SDS, 100 mm NaHCO3, 1 mm EDTA) at 37°C for 10 min for three times with the final elution performed at additional 65°C for 5 min. At the same time, 10% of input samples were also diluted with elution buffer to a final volume of 150 μl. Final concentration of 250 mm NaCl was added to IP and input samples and samples were decrosslinked overnight at 65°C.
Next day, 125 μg/ml RNase A (PanReac Applichem) was added and incubated at 37°C for 1–2 h. Then, final concentrations of 6 mm EDTA and 240 μg/ml proteinase K (Fisher Bioreagent) were added and incubated at 45°C for 2 h. The genomic DNA was purified using QIAquick PCR Purification kit (QIAGEN).
DNA enrichment was measured with qPCR using primers listed in Tuvikene, 2021 and LightCycler 480 SYBR Green I Master kit (Roche). The DNA enrichment was calculated as percent of input relative to the levels of unrelated region (URR).
Experimental design and statistical analysis
Sample size estimation was not performed, and randomization and blinding were not used. Statistical analysis was performed on log-transformed, mean-centered and autoscaled data (where applicable). For data in Figures 3 and 5 that show the direct comparison of the role of the effector in different cultures, the data were only log-transformed. All tested hypotheses were specified before conducting the experiments. Two-tailed paired t tests were performed using R 4.1.1 programming language or Microsoft Excel 365 and p-values were adjusted for multiple comparisons with Holm or Holm–Šídák family-wise error rate correction method using R p.adjust function or GraphPad Prism 7.03 (GraphPad Software), respectively. Exact p-values are shown in extended data files. All experiments were performed using three or four independent biological replicates (cultures obtained from different litters, n = 3–4). For graphical representation, transformed and scaled means and mean ± SEM were back-transformed to linear scale with error bars representing upper and lower limits of back-transformed mean ± SEM. For RNA-seq data from (Carullo et al., 2020), statistical analysis was performed using DESeq2 package in R.
Figure 3.
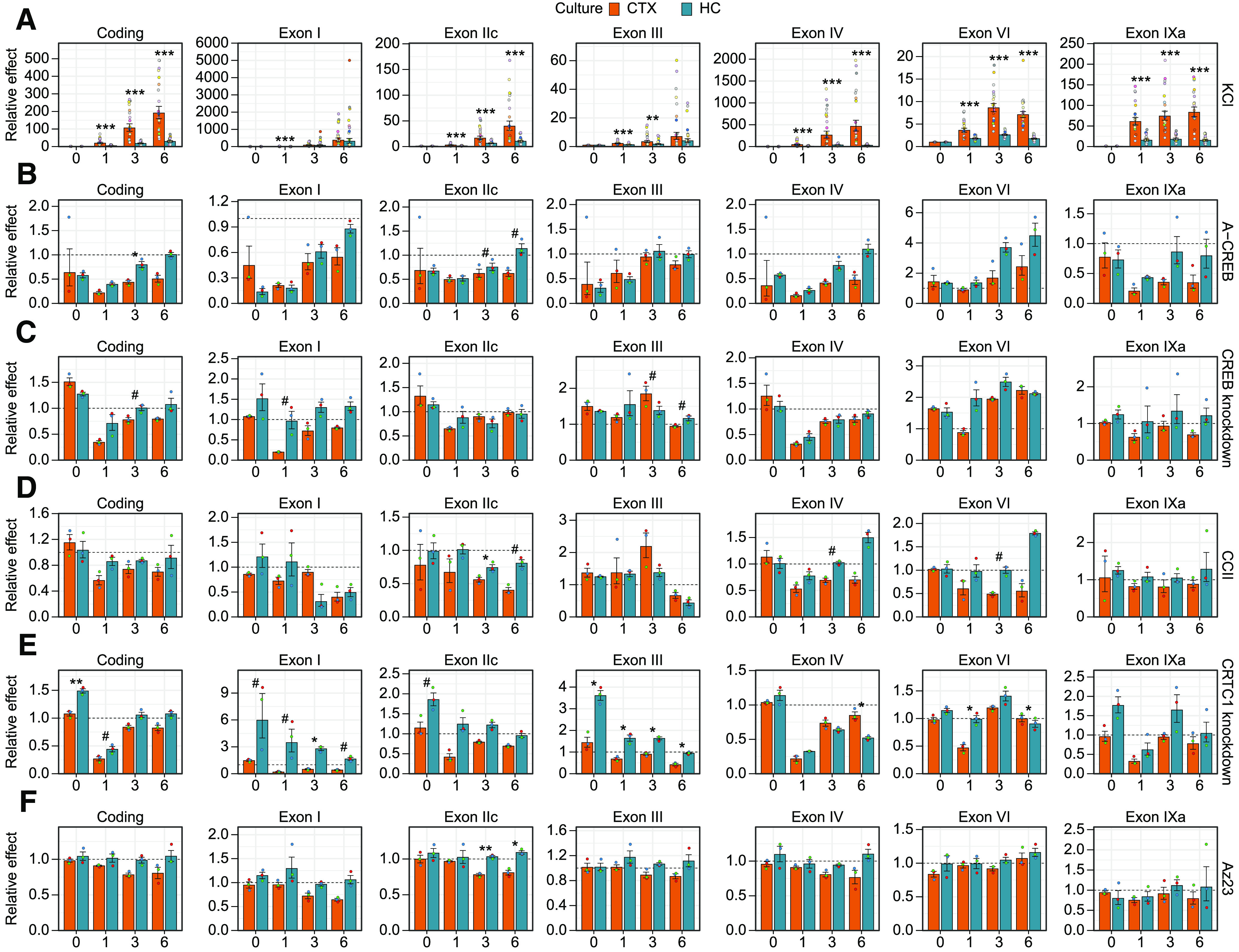
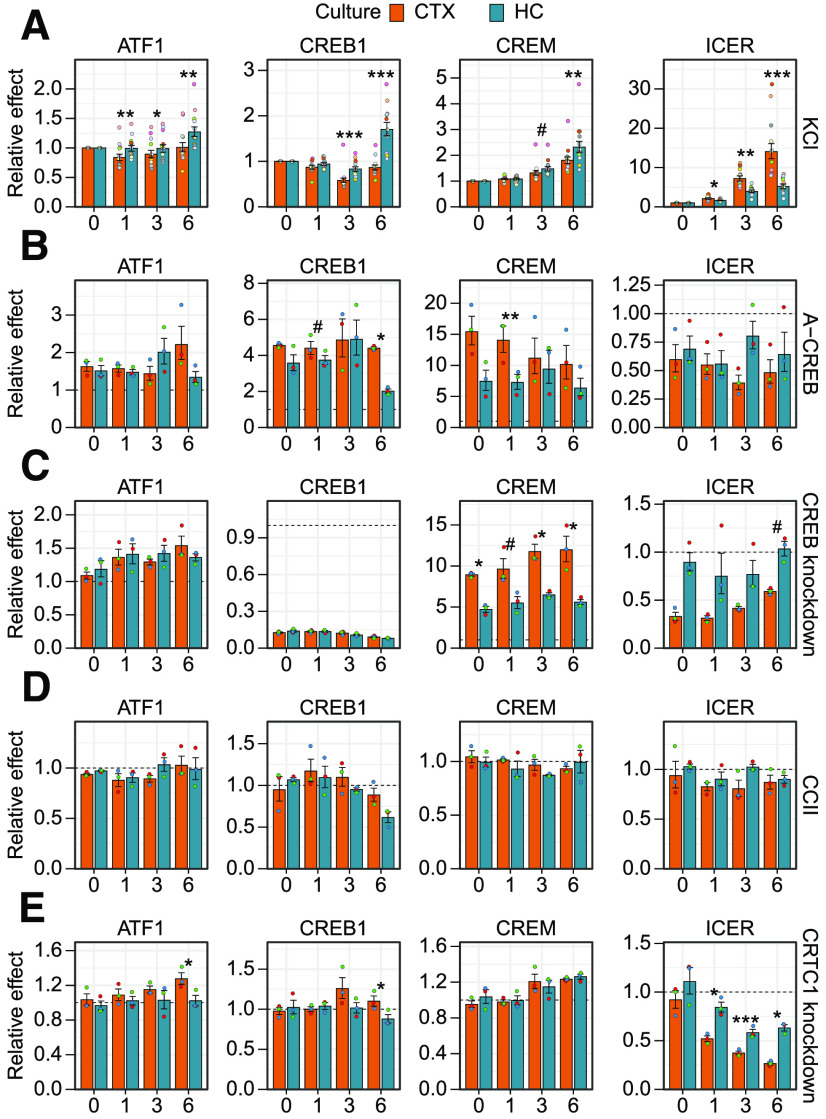
Comparison of the effect of impairing CREB family and coactivators in cortical and hippocampal neurons on activity-dependent BDNF gene expression. Cultured rat cortical (CTX) or hippocampal (HC) neurons were left untreated (0 h) or treated with 25 mm KCl at 8 DIV for the indicated time (1, 3, or 6 h). Total BDNF levels (Coding) or different BDNF transcripts (indicated by the respective 5′ exon name) were measured using RT-qPCR. Data from Figure 2 are shown relative to the transcripts' levels in the control cells [cells expressing EGFP, or negative guide RNA (gRNA) and dCas9-KRAB, or treated with DMSO] at the respective treatment time point. The bar graphs show the average fold changes of the effector, all data points are shown as dots, and each biological replicate [i.e., cultures derived from animals of different litters, n = 18 (A) or n = 3 (B–F)] is denoted with the same color. Error bars represent mean ± SEM. Statistical significance is shown compared with the effector fold change between cortical and hippocampal neurons at the respective time point of KCl treatment. Group averages and exact p-values are shown in Extended Data Figure 3-1. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm method.
Figure 5.
Comparison of impairing CREB family and coactivators in cortical and hippocampal neurons on the gene expression of CREB family members. Cultured rat cortical (CTX) or hippocampal (HC) neurons were left untreated (0 h) or treated with 25 mm KCl at 8 DIV for the indicated time (1, 3, or 6 h). Levels of different CREB family members [CREB1, ATF1, CREM activator forms containing the KID domain (CREM) or CREM negative regulator ICER] were measured using RT-qPCR. Data from Figure 4 are shown relative to the transcripts' levels in the control cells [expressing EGFP, or negative guide RNA (gRNA) and dCas9-KRAB, or treated with DMSO] at the respective treatment time point. The bar graphs show the average fold changes of the effector, all data points are shown as dots and each biological replicate [i.e., cultures derived from animals of different litters, n = 12 (A) or n = 3 (B–E)] is denoted with the same color. Error bars represent mean ± SEM. Statistical significance is shown compared with the effector fold change between cortical and hippocampal neurons at the respective time point of KCl treatment. Group averages and exact p-values are shown in Extended Data Figure 5-1. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm method.
Results
Activity-dependent BDNF gene regulation by CREB family transcription factors
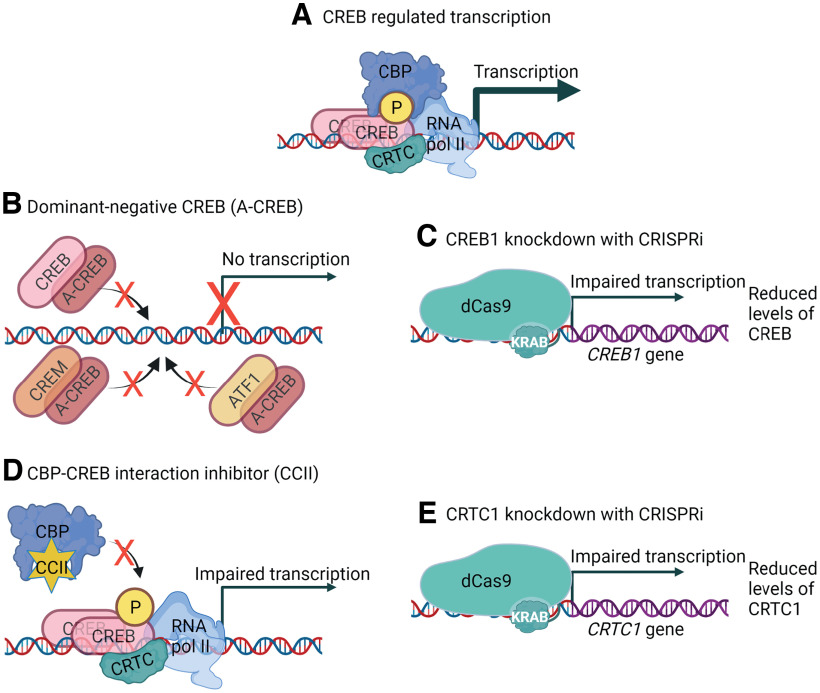
The expression of BDNF is induced in response to various stimuli, of which induction by neuronal activity is one of the most well-studied (for review, see West et al., 2014). We cultured rat cortical and hippocampal neurons from pups of the same litter to reduce the role of biological variability and ensure the comparability of the two neuronal populations, and investigated the role of CREB family transcription factors in membrane depolarization-induced BDNF gene expression (Fig. 1A). CREB family consists of three basic-leucine zipper transcription factors – activating transcription factor 1 (ATF1), cAMP-response element-binding protein (CREB), and cAMP response element modulator (CREM; Mayr and Montminy, 2001). To investigate the involvement of CREB family transcription factors in membrane depolarization-dependent BDNF induction, we overexpressed a dominant-negative form of CREB, named A-CREB. The mechanism of function for A-CREB is based on dimerization with CREB basic leucine zipper (bZip) domain (Ahn et al., 1998). Since the bZip domain in ATF1, CREB, and CREM is highly similar, A-CREB can impair the DNA binding of all the CREB family members (Fig. 1B).
Figure 1.
Schematical depiction of the tools used in the current study to elucidate the role of CREB family in BDNF gene regulation. A, CREB regulates transcription of the target genes together with coactivators CBP (or P300) and CRTCs. B, Overexpression of a dominant-negative form of CREB, named A-CREB, which dimerizes with CREB family members based on their basic leucine zipper domain (bZip) and inhibits binding to the DNA, therefore inhibiting CREB family-regulated transcription. C, CRISPR interference (CRISPRi) system targeting dCas9-KRAB to the CREB1 gene promoter impairs the transcription of CREB1 gene, reducing the levels of CREB. D, A low molecular weight compound CCII binds to the c-Myb-site on CBP and prevents CBP binding to CREB family members. E, CRISPRi system targeting dCas9-KRAB to the CRTC1 gene promoter impairs the transcription of CRTC1, reducing the levels of CRTC1. Schematics were created with BioRender.com.
CREB family transcription factors have two main types of coactivators that potentiate CREB-regulated transcription: CREB-binding protein (CBP) and P300 (for review, see Mayr and Montminy, 2001) and CREB-regulated transcriptional coactivators or CRTCs (for review, see Saura and Cardinaux, 2017). Of these, CBP interacts with phosphorylated kinase-inducible domain (KID; Parker et al., 1996; Radhakrishnan et al., 1997; Shaywitz et al., 2000; Dahal et al., 2017a,b) and CRTCs bind the bZip domain of CREB family members (Luo et al., 2012; Y. Song et al., 2018). To determine the relevance of interaction between CREB family and its coactivator CBP, we used a low molecular weight CBP-CREB interaction inhibitor (CCII; Best et al., 2004; Li and Xiao, 2009; Fig. 1D). To further elucidate the specific role of CREB transcription factor and CREB family coactivator CRTC1 in BDNF gene regulation, we silenced their expression with CRISPR interference (Fig. 1C,D, respectively).
Finally, we have previously described a BDNF-TrkB signaling-dependent positive feedback loop in rat cortical neurons where BDNF-TrkB signaling induces the expression of all major BDNF transcripts in a CREB-family and CBP-dependent manner (Esvald et al., 2020). As membrane depolarization induces both BDNF expression and BDNF secretion (for review, see Sasi et al., 2017; M. Song et al., 2017), it is plausible that at least part of the induction of BDNF expression after membrane depolarization depends on TrkB signaling. Therefore, to determine whether the BDNF-TrkB signaling component participates in depolarization-dependent BDNF gene regulation, we treated the cells with Trk receptor inhibitor Az23 (Thress et al., 2009) before KCl treatment. We measured the expression levels of total BDNF, major BDNF transcripts (exon I, IIc, III, IV, VI, IXa), and CREB family members using RT-qPCR to elucidate gene expression regulation after membrane depolarization in cortical and hippocampal neurons.
First, we determined the extent and dynamics of neuronal activity-dependent BDNF induction. Our results collectively showed that total BDNF expression is more inducible in response to membrane depolarization in cortical neurons than in hippocampal neurons (Figs. 2A, 3A). Our data also showed that all major BDNF transcripts are induced faster in cortical neurons than in hippocampal neurons as the inductions in cortical neurons after 1 h treatment were higher than in hippocampal neurons (Figs. 2A, 3A). Similarly, after prolonged depolarization all major transcripts except BDNF exon I transcripts showed higher inducibility in cortical neurons than in hippocampal neurons, whereas BDNF exon I transcripts showed similar induction between the two cultures (Figs. 2A, 3A).
Figure 2.
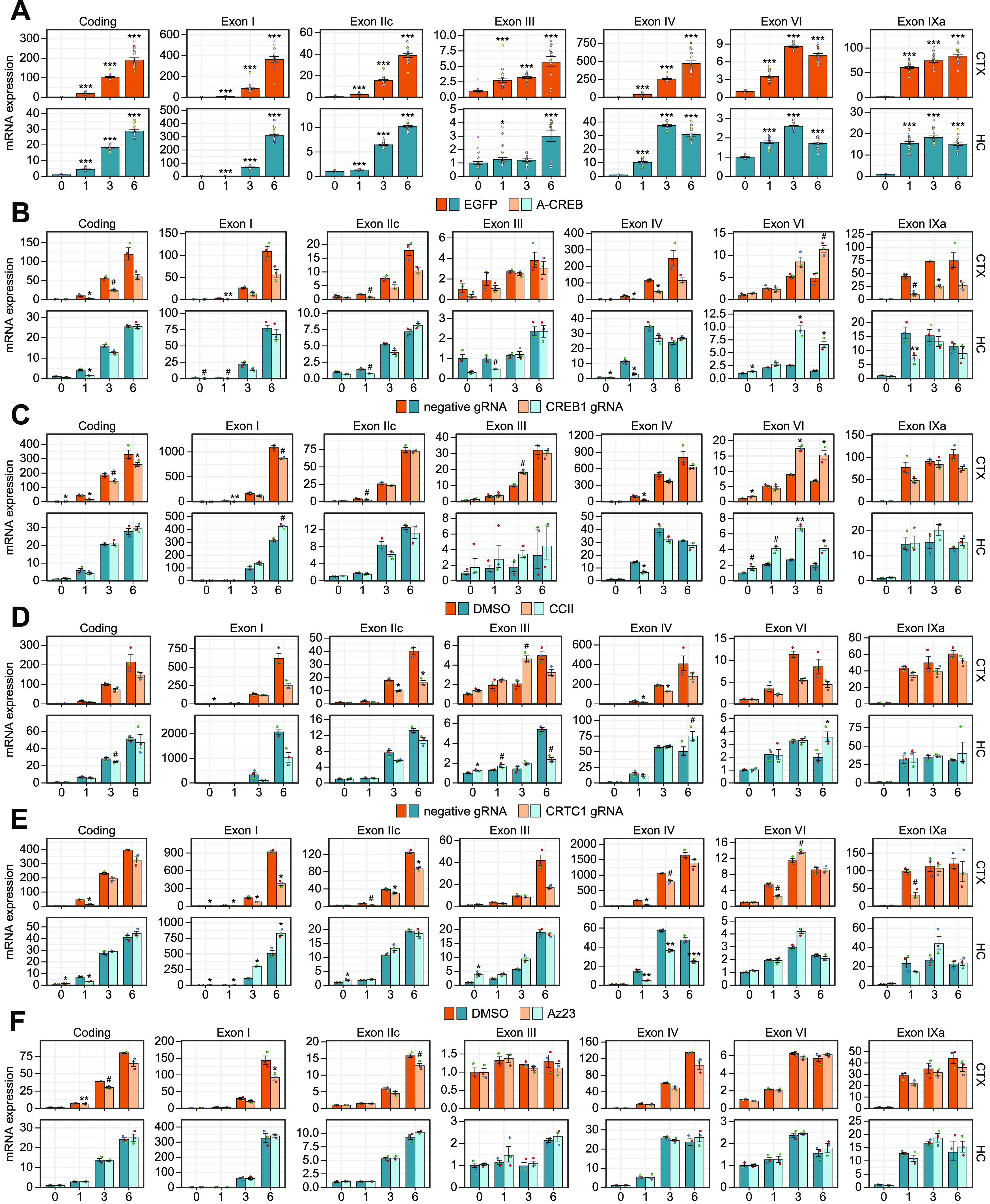
Regulation of activity-dependent BDNF gene expression by CREB family transcription factors and their coactivators in cortical and hippocampal neurons. Cultured rat cortical (CTX) or hippocampal (HC) neurons were left untreated (0 h) or treated with 25 mm KCl at 8 DIV for the indicated time (1, 3, or 6 h). Total BDNF levels (Coding) or different BDNF transcripts (indicated by the respective 5′ exon name) were measured using RT-qPCR. A, Induction of BDNF in response to membrane depolarization with KCl. Regulation of activity-dependent BDNF expression by various effectors: (B) overexpression of dominant-negative for the CREB family (A-CREB), (C) knock-down of CREB1 gene expression using CRISPRi, (D) disruption of the interaction between CBP and CREB (CCII), (E) knock-down of coactivator CRTC1 with CRISPRi, and (F) inhibition of Trk receptor signaling with Az23. Data are shown relative to the transcripts' levels in untreated cells in the respective culture (A) or in the control cells [expressing EGFP, or negative guide RNA (gRNA) and dCas9-KRAB, or treated with DMSO] at the indicated time point in the respective culture (B–F). The bar graphs show the average fold changes for n = 18 (A, 3 independent experiments plus the combined data from all control cells shown on panels B–F) or n = 3 (B–F) independent biological experiments. All data points are shown as dots, and each biological replicate (i.e., cultures derived from animals of different litters) is denoted with the same color. Error bars represent mean ± SEM. Statistical significance is shown compared with the expression levels in untreated cells in the respective culture (A) or in control cells at the respective time point of KCl treatment in the respective culture (B–F). Group averages and exact p-values are shown in Extended Data Figure 2-1. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm method.
Underlying data and exact p-values of the statistical analysis for Figure 2. Download Figure 2-1, XLSX file (76.5KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 3. Download Figure 3-1, XLSX file (43.6KB, xlsx) .
Next, we focused on the role of CREB family in the regulation of BDNF gene expression. In cortical neurons, overexpression of a dominant-negative form of the CREB family, A-CREB, decreased the basal expression and both the early and late induction of the major BDNF transcripts (Figs. 2B, 3B). Meanwhile, in hippocampal neurons overexpression of A-CREB diminished mainly basal expression and early induction of the major BDNF transcripts, while the late induction was largely unaffected (Figs. 2B, 3B). In contrast to other BDNF transcripts, the levels of BDNF exon VI transcripts were increased in response to A-CREB overexpression both in unstimulated neurons and after depolarization in both cortical and hippocampal neurons (Figs. 2B, 3B). Collectively our results show that the neuronal activity-induced levels of BDNF are decreased by A-CREB overexpression more in cortical than in hippocampal neurons (Fig. 3B).
The specific knock-down of CREB1 gene expression (knock-down ∼7- to 8-fold in both cortical and hippocampal untreated neurons, see Fig. 4C) revealed that CREB transcription factor participates in the early induction of BDNF exon I and exon IV transcripts in cortical neurons (Figs. 2C, 3C). In contrast, knock-down of CREB1 had only mild effect on BDNF expression in hippocampal neurons with the strongest effect on the early induction of BDNF exon IV (Figs. 2C, 3C). Notably, in both neuronal cultures we again detected increase in the expression of BDNF exon VI transcripts (Figs. 2C, 3C), showing an opposite regulation compared with other BDNF transcripts by CREB transcription factor.
Figure 4.
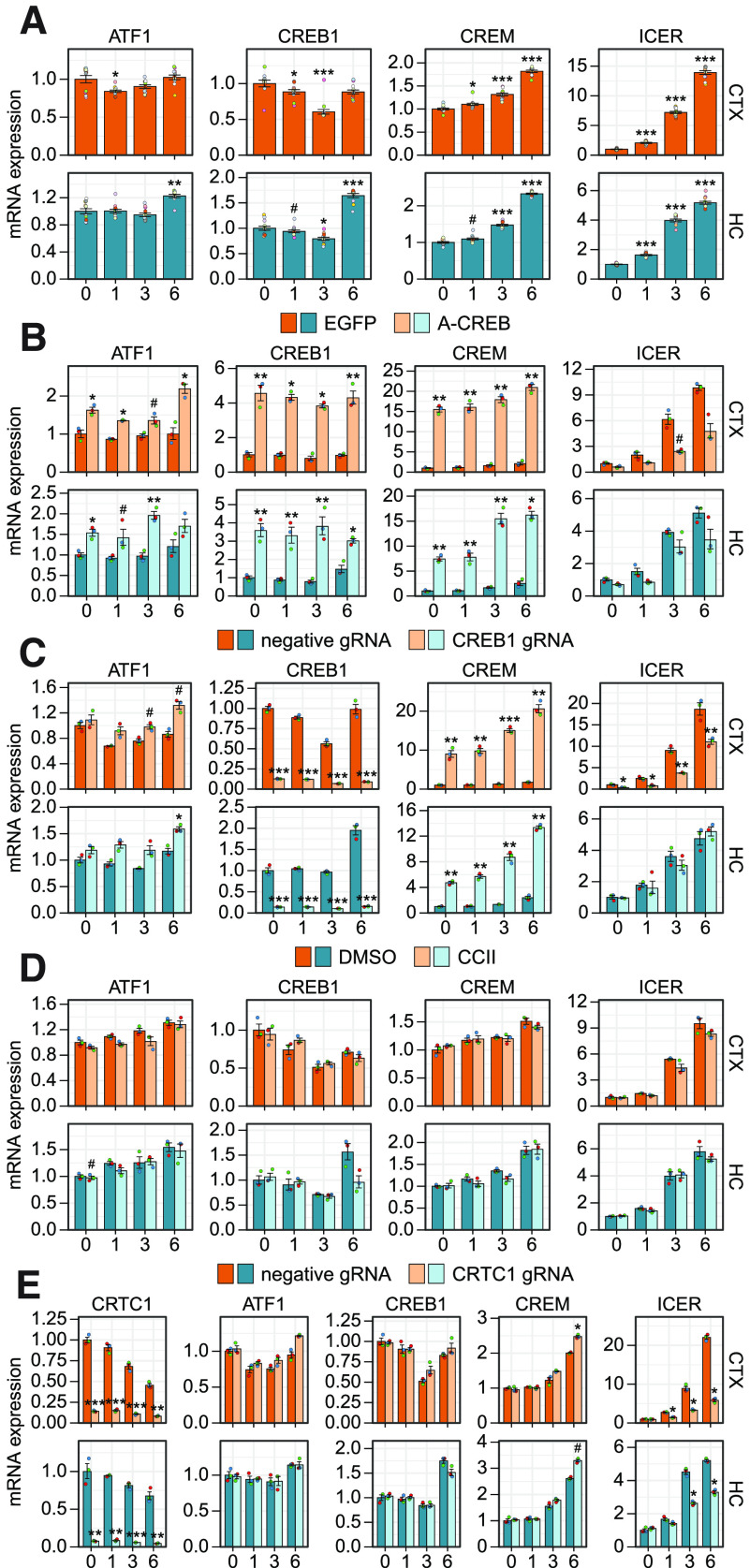
Regulation of ATF1, CREB1, and CREM by CREB family transcription factors and their coactivators in cortical and hippocampal neurons. Cultured rat cortical (CTX) or hippocampal (HC) neurons were left untreated (0 h) or treated with 25 mm KCl at 8 DIV for the indicated time (1, 3, or 6 h). Levels of different CREB family members (ATF1, CREB1, and CREM activator forms containing the KID domain (CREM) or CREM negative regulator ICER) were measured using RT-qPCR. A, Regulation of CREB family members in response to membrane depolarization with KCl. Regulation of activity-dependent CREB family expression by various effectors: (B) overexpression of dominant-negative for the CREB family (A-CREB), (C) knock-down of CREB1 gene expression using CRISPRi, (D) disruption of the interaction between CBP and CREB (CCII), (E) knock-down of coactivator CRTC1 with CRISPRi. Data are shown relative to the transcripts' levels in untreated cells in the respective culture (A) or in the control cells [expressing EGFP, or negative guide RNA (gRNA) and dCas9-KRAB, or treated with DMSO] at the indicated time point in the respective culture (B–E). The bar graphs show the average fold changes for n = 12 (A, combined data from all control cells shown on panels B–E) or n = 3 (B–E) independent biological experiments. All data points are shown as dots, and each biological replicate (i.e., cultures derived from animals of different litters) is denoted with the same color. Error bars represent mean ± SEM. Statistical significance is shown compared with the expression levels in untreated cells in the respective culture (A) or in control cells at the respective time point of KCl treatment in respective culture (B–E). Group averages and exact p-values are shown in Extended Data Figure 4-1. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm method.
Underlying data and exact p-values of the statistical analysis for Figure 4. Download Figure 3-1, XLSX file (47.4KB, xlsx) .
Next, we focused on the coactivators of CREB family in the regulation of BDNF expression. Disrupting the interaction between coactivator CBP and CREB with low molecular weight inhibitor, CCII, showed that the late induction of BDNF exon I containing transcripts depends on CBP in both cortical and hippocampal neurons (Figs. 2D, 3D). Interestingly, in cortical neurons impairing CBP recruitment decreased both the early and late induction of BDNF exon IV and exon VI transcripts, whereas in hippocampal neurons the late induction of both transcripts was increased (Figs. 2D, 3D).
Knock-down of the coactivator CRTC1 in cortical neurons reduced CRTC1 levels ∼7-fold (see Fig. 4E) and mostly decreased the early induction of major BDNF transcripts (Figs. 2E, 3E). In hippocampal neurons the CRTC1 knock-down was slightly more efficient and reduced the CRTC1 levels ∼13-fold (see Fig. 4E) and this increased both the basal expression of BDNF transcripts of the first cluster of exons (exons I, IIc, and III) and induced levels of exon I and III transcripts (Figs. 2E, 3E). In contrast to the opposite effect on BDNF exon I transcripts in cortical and hippocampal neurons (Fig. 3E), CRTC1 knock-down decreased the induction of BDNF exon IV transcripts in both neuron cultures, although the effect was more prominent in hippocampal neurons (Figs. 2E, 3E). CRTC1 knock-down also decreased the early induction of BDNF exon VI transcripts in cortical neurons, whereas such effect was not seen in hippocampal neurons (Figs. 2E, 3E).
Lastly, we found that BDNF-TrkB signaling slightly contributed to the membrane depolarization-induced BDNF expression at later time points in cortical neurons, whereas no effect of inhibiting BDNF-TrkB signaling was seen in hippocampal neurons (Figs. 2F, 3F). The difference between cultures was consistently seen for all transcripts after prolonged membrane depolarization, although the difference was mostly not statistically significant (Fig. 3F).
Collectively our results show that BDNF exon I and IV transcripts, the most inducible BDNF transcripts, were consistently dependent on CREB family and their coactivators both in cortical and hippocampal neurons. Our results indicate that the induction of BDNF exon I in cortical neurons depends on CREB, CBP, and CRTC1, with CRTC1 being important throughout the measured timepoints of depolarization and CBP rather at later timepoints. The induction of BDNF exon IV in cortical neurons was also regulated by CREB, CBP, and CRTC1, whereas the induction in hippocampal neurons was regulated by CREB and CRTC1. The opposite effect of CRTC1 knock-down on BDNF exon I levels in cortical and hippocampal neurons and the opposite regulation of BDNF exon VI compared with other BDNF transcripts need further investigation. Overall, our results suggest that the activity-dependent regulation of BDNF transcripts in cortical neurons relies more on the CREB family and coactivators than in hippocampal neurons. Our results also suggest that the faster inducibility of BDNF in cortical neurons could be a result of the increased regulation by CREB family.
Activity-dependent expression and regulation of CREB family members
To further elucidate the mechanism of CREB family-dependent gene expression in different neuronal populations, we measured the activity-dependent expression levels of all the CREB family members, CREB1, ATF1, and CREM, in cortical and hippocampal neurons. CREB1 is known to be a constitutively expressed gene and CREB protein activity is mainly regulated by posttranslational mechanisms (for review, see Johannessen et al., 2004), as phosphorylation of CREB in the kinase-inducible domain (KID) stabilizes its interaction with the transcriptional coactivator CBP (Dahal et al., 2017a, b). Our results showed that membrane depolarization slightly decreases CREB1 levels after 3 h membrane depolarization in both cortical and hippocampal neurons (Fig. 4A). However, in hippocampal but not in cortical neurons there was a slight but consistent upregulation of both CREB1 and ATF1 mRNA levels at the 6-h time point (Figs. 4A, 5A).
Underlying data and exact p-values of the statistical analysis for Figure 5. Download Figure 5-1, XLSX file (31.7KB, xlsx) .
Next, we focused on the expression of CREM and inducible cAMP early repressor (ICER). The expression of ICER inside the CREM gene is controlled by an intronic promoter with four tandemly placed CRE elements, making it highly inducible by CREB family transcription factors (Molina et al., 1993). ICER contains only the CREM DNA binding domain and competes with the CREB family members in binding to DNA, therefore attenuating CREB family-regulated gene expression (Molina et al., 1993; Walker et al., 1998; Mioduszewska et al., 2003). Since CREM gene can also produce various repressor and activator forms of CREM via alternative splicing and exon shuffling (Foulkes et al., 1991; Laoide et al., 1993), we specifically measured the expression levels of CREM activator forms that contain the KID domain. Our data showed that the levels of the activator forms of CREM were slightly and the negative regulator ICER were remarkably increased in response to membrane depolarization (Fig. 4A). Notably, while the induction of CREM activator forms was relatively similar in both cortical and hippocampal neurons, the induction of ICER was faster and reached a higher induced level in cortical neurons than in hippocampal neurons (Figs. 4A, 5A).
Next, we analyzed the expression of CREB family members after overexpression of A-CREB, knock-down of CREB1 and CRTC1 and inhibition of CBP recruitment by CCII. Notably, overexpression of A-CREB strongly induced the expression of CREB1 and CREM, with the effect being stronger in cortical neurons, and also slightly induced ATF1 expression in both cultures (Figs. 4B, 5B), indicating that CREB family members try to normalize their functional levels in response to impairing CREB family-dependent transcription. In agreement with Esvald et al. (2020), knock-down of CREB1 expression remarkably induced the expression of CREM in both neuronal cultures (Fig. 4C), but more strongly in cortical neurons (Fig. 5C), indicating the existence of a compensatory mechanism between CREB and CREM, and also providing a possible explanation why A-CREB, which inhibits all the members of the CREB family, tends to have a more profound effect on BDNF gene expression than specific silencing of CREB1. Neither inhibition of CBP recruitment nor knock-down of CRTC1 affected the expression of CREB1 and ATF1 in cortical neurons (Fig. 4D, E). Interestingly, the late induction of CREB1 in hippocampal neurons seems to slightly depend on CBP and CRTC1 (Fig. 4D, E). The late induction of CREM was increased upon CRTC1 knock-down similarly in both cultures (Figs. 4E, 5E).
The regulation of ICER by CREB family members was different in cortical and hippocampal neurons. The expression of ICER was more inducible in cortical neurons (Figs. 4A, 5A) and this activity-dependent induction of ICER was impaired by CREB1 knock-down in cortical neurons but not in hippocampal neurons (Figs. 4C, 5C). The knock-down of CRTC1 decreased the induced levels of ICER in both cultures (Fig. 4E), but the effect on ICER levels was stronger and also apparent earlier after membrane depolarization in cortical neurons compared with hippocampal neurons (Fig. 5E). It appears that the induction of ICER is lower and less dependent on CREB in hippocampal neurons, suggesting that the ICER-dependent CREB family negative feedback loop is also less potent in hippocampal neurons than in cortical neurons.
Activity-dependent regulation of BDNF protein levels
Next, we investigated whether BDNF protein levels are also affected by impairing CREB family transcription factors and coactivators. For that we used the same tools as for qPCR analysis (Fig. 1), treated cortical and hippocampal neurons at 8 DIV with KCl and analyzed the protein levels using Western blotting. To detect BDNF we used a highly sensitive BDNF antibody that has been validated on BDNF knock-out animal brain tissue (Wosnitzka et al., 2020) and can detect even 15 pg of recombinant BDNF in Western blotting (our unpublished data).
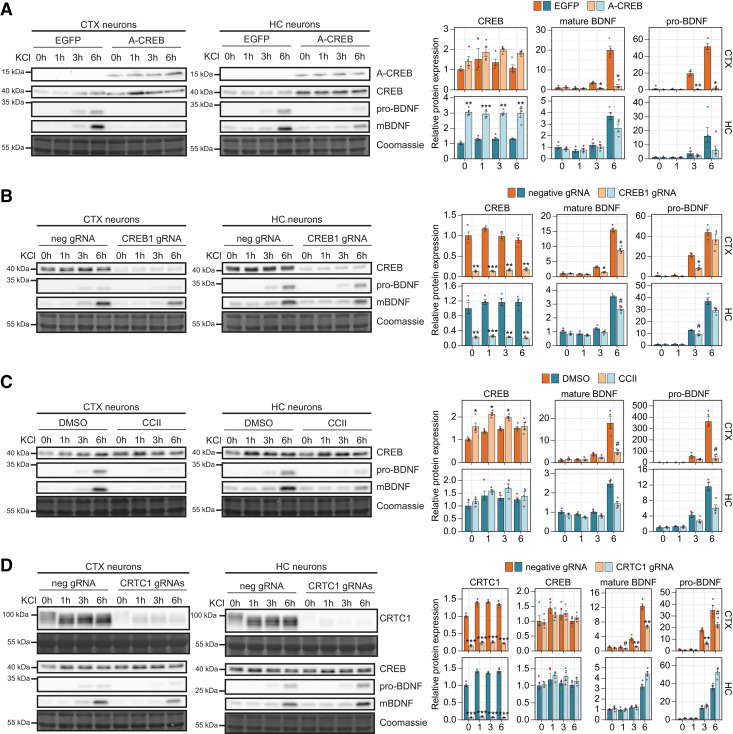
In agreement with the BDNF mRNA levels, our data show that both pro-BDNF and mature BDNF protein levels are more inducible in cortical neurons than in hippocampal neurons (Fig. 6A–D). Our results also indicate that the BDNF protein induction is faster in cortical than in hippocampal neurons, and that an increase in pro-BDNF levels can be detected before mature BDNF levels rise (Fig. 6A–D). The overexpression of A-CREB abolished the activity-dependent expression of mature BDNF protein in cortical neurons, while in hippocampal neurons only a minor effect on mature BDNF protein levels was seen (Fig. 6A). Similarly, the overexpression of A-CREB almost completely abolished the induction of pro-BDNF in cortical neurons, and decreased the induction in hippocampal neurons (Fig. 6A). In accordance with the CREB1 mRNA levels, the overexpression of A-CREB increased CREB protein levels in both cortical and hippocampal neurons, but the effect on protein levels was more prominent in hippocampal neurons (Fig. 6A).
Figure 6.
Regulation of activity-dependent BDNF protein levels by CREB family transcription factors and coactivators in cortical and hippocampal neurons. Cultured rat cortical (CTX) or hippocampal (HC) neurons were left untreated (0 h) or treated with 25 mm KCl at 8 DIV for the indicated time (1, 3, or 6 h). Regulation of activity-dependent BDNF protein expression was analyzed using lentiviruses encoding EGFP- or A-CREB (A), dCas9-KRAB along with negative control guide RNA (neg gRNA) or gRNA targeting CREB1 promoter (CREB1 gRNA; B), or preteated with CBP-CREB interaction inhibitor (CCII; C), or with dCas9-KRAB along with gRNA targeting CRTC1 promoter (CRTC1 gRNA; D). A-CREB, CREB, CRTC1, pro-BDNF, and mature BDNF (mBDNF) protein levels were measured using Western blotting and representative Western blotting images in cortical and hippocampal neurons are shown on the left, with Coomassie staining as a loading control. The pro-BDNF and mature BDNF levels were measured from the same membrane and their protein levels are shown using the same exposition time. The bar graphs on the right show densitometric analysis of the expression levels of CREB, CRTC1, pro-BDNF, and mature BDNF normalized with Coomassie signal. The protein levels in untreated control neurons (EGFP- or neg gRNA-expressing neurons or neurons treated with DMSO) were taken as 1. Error bars represent mean ± SEM of four biological replicates (n = 4), and individual data points of each biological replicate are shown with dots. Statistical significance is shown relative to the respective protein levels in control cells at the respective time point of KCl treatment in respective culture. Group averages and exact p-values are shown in Extended Data Figure 6-1. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm method.
Underlying data and exact p-values of the statistical analysis for Figure 6. Download Figure 6-1, XLSX file (36.3KB, xlsx) .
We next focused on the specific role of CREB transcription factor. The knock-down of CREB1 gene decreased CREB protein levels 5- to 8-fold in both cortical and hippocampal neurons, and decreased the protein induction of mature BDNF in cortical neurons at both 3 and 6 h of KCl treatment, and in hippocampal neurons only at the 6 h time point (Fig. 6B). A decrease in the levels of pro-BDNF was detected in both cortical and hippocampal neurons, with the effect being smaller in hippocampal neurons (Fig. 6B).
Next, we focused on the coactivators CBP and CRTC1 in the regulation of BDNF expression levels. By impairing CBP recruitment the induction of pro-BDNF and mature BDNF induction was greatly inhibited in cortical neurons, and was again mostly evident in hippocampal neurons only at the 6 h time point (Fig. 6C). Interestingly, CCII treatment slightly increased CREB protein levels in cortical neurons (Fig. 6C), although we did not detect any effect on the CREB1 mRNA levels (Fig. 4D).
Finally, CRTC1 knock-down decreased CRTC1 protein levels ∼7- and ∼15-fold in cortical and hippocampal neurons, respectively (Fig. 6D). We also detected depolarization-dependent dephosphorylation of CRTC1 as a shift in the mobility in Western blot analysis. Interestingly, CRTC1 knock-down had an opposing effect on the BDNF protein levels in cortical and hippocampal neurons – CRTC1 knock-down decreased the depolarization-induced levels of pro-BDNF and mature BDNF in cortical neurons but increased their levels in hippocampal neurons (Fig. 6D). Although the CRTC1 knock-down did not increase total BDNF mRNA levels, it increased BDNF exon I mRNA levels in hippocampal neurons (Fig. 2E), and thus the increased BDNF protein levels in hippocampal neurons could be explained by more efficient translation of BDNF exon I-containing transcripts as shown previously (Koppel et al., 2015). The lower levels of CRTC1 did not affect the CREB protein levels (Fig. 6D).
Collectively, our data show that CREB family and coactivators CBP and CRTC1 are the main regulators of activity-dependent induction of BDNF in cortical neurons, whereas they play a less prominent role in the induction of BDNF levels in hippocampal neurons.
Identification of tissue-specific transcription factors binding to BDNF promoters I and IV in vitro
Since, to our surprise, the role of CREB family in BDNF gene regulation was smaller than we expected, especially for the late induction and in the hippocampus, we next aimed to decipher which transcription factors additionally regulate BDNF gene expression in the cortex and hippocampus. For that we employed in vitro DNA pulldown assay with promoter I and IV fragments using 8-day-old rat cortical and hippocampal tissue nuclear lysates and identified the bound proteins using label-free liquid chromatography-coupled mass spectrometry (LC-MS; Fig. 7A). A similar approach has previously been used to determine transcription factors that bind to an obesity-related SNP region within the BDNF gene (Mou et al., 2015) and to an intronic enhancer region inside the BDNF gene (Tuvikene et al., 2021). Here, we selected transcription factors from mass spectrometric analysis according to Gene Ontology classification and compared their signal between pulldowns using either BDNF promoter I or IV. We set a ratio of 1.5 as a threshold for specific binding between the two promoter regions and then determined the brain region preference of the identified promoter-specific transcription factors (Fig. 7B).
Figure 7.
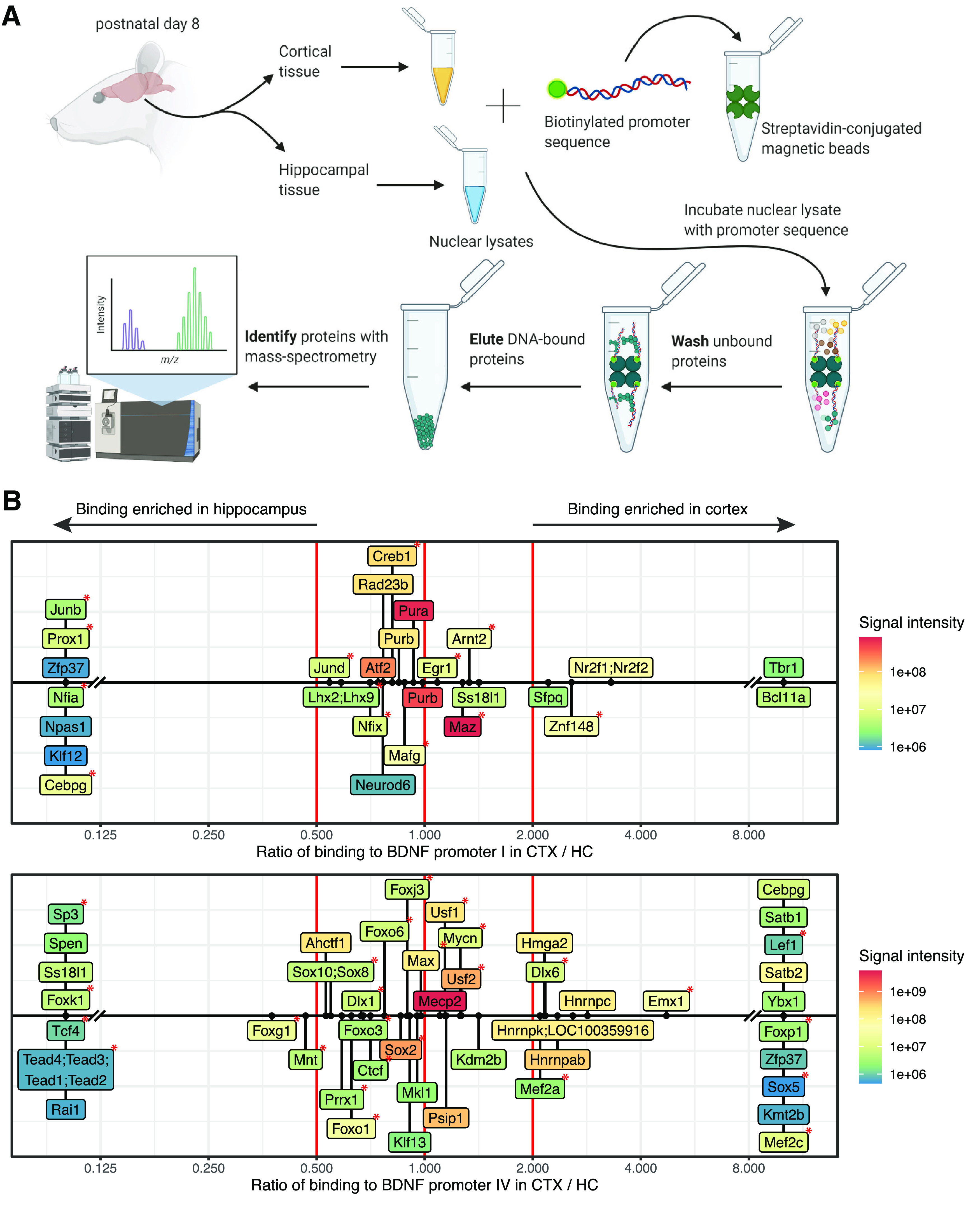
Numerous novel and previously reported transcription factors bind to BDNF promoters I and IV in vitro. A, Schematic of the setup of the in vitro DNA pulldown experiment (schematics created with BioRender). Cortical and hippocampal tissues were isolated from 8-day-old Sprague Dawley rat pups, nondenaturing nuclear lysates were prepared and subjected to in vitro promoter pulldown. Transcription factors bound to BDNF promoters I and IV were determined by LC-MS/MS. All peptides, proteins and transcription factors detected in mass spectrometry are listed in Extended Data Figure 7-1. B, Transcription factors showing promoter-specific binding to BDNF promoters I (upper panel) and IV (lower panel). Specific binding was determined as ≥1.5 ratio of binding signal to one promoter relative to the other promoter in the respective tissue sample. The figure depicts only transcription factors that were found to bind specifically to the respective promoter in either cortical or hippocampal sample. List of transcription factors with semicolons between gene names mark uncertainty in assigning the detected peptides to proteins. The red lines represent zero (no tissue-specific difference) and borders of 2-fold difference in label free quantification (LFQ) intensity between the two tissues. Colors indicate the maximal LFQ signal intensity for the indicated transcription factor in either cortical or hippocampal samples. The red asterisks mark transcription factors that have a binding site in the respective promoter according to Jaspar database (Fornes et al., 2020).
Lists of all peptides, proteins and transcription factors detected in mass-spectrometric analysis in the in vitro DNA pulldown experiment. Download Figure 7-1, XLSX file (794.7KB, xlsx) .
For BDNF promoter I, we detected binding of several factors that are widely acknowledged to regulate the activity of the promoter, i.e., cAMP-response element binding protein (CREB; Tabuchi et al., 2002; Pruunsild et al., 2011), aryl hydrocarbon receptor nuclear translocator 2 (ARNT2; Pruunsild et al., 2011; Bloodgood et al., 2013) and activator protein 1 (AP1) family transcription factors JunB and JunD (Tuvikene et al., 2016). For BDNF promoter IV, our assay also revealed numerous transcription factors previously reported to regulate the activity of the promoter (Fig. 7B), including myocyte enhancer factor 2 (MEF2) family members MEF2a and MEF2c (Flavell et al., 2008; Lyons et al., 2012; Chen et al., 2020), USF1 and USF2, the members of upstream stimulatory factor (USF) family of basic helix-loop-helix leucine zipper proteins (Chen et al., 2003b; Pruunsild et al., 2011), and methyl CpG binding protein 2 (MeCP2; Chen et al., 2003a; Martinowich et al., 2003; Yazdani et al., 2012; Tai et al., 2016). Moreover, several of the factors have a predicted binding site (Fig. 7B) according to Jaspar in silico prediction (Fornes et al., 2020). Based on the rediscovery of many known regulators of BDNF, we conclude that our in vitro pulldown assay is reliable and could therefore possibly also determine novel regulators of BDNF. We discovered numerous potential regulators of BDNF promoter I that might confer brain region-specific expression. For example, nuclear factor 1 protein NFIA, homeobox protein PROX1, and zinc finger proteins ZFP37 and KLF12 showed enriched binding to BDNF promoter I in the hippocampal lysate and zinc finger protein BCL11A and T-box family transcription factor TBR1 in the cortical lysate (Fig. 7B). Meanwhile, homeobox proteins SATB1 and SATB2, and forkhead box protein FOXP1, identified by us as novel regulators of BDNF promoter IV, showed cortex-specific binding and zinc finger protein RAI1 and helix-loop-helix protein TCF4 showed hippocampus-specific binding to BDNF promoter IV (Fig. 7B).
Differential expression of the in vitro bound factors in different neuronal populations
Next, we asked whether the transcription factors that showed brain region-preference in the in vitro promoter pulldown assay are expressed in a brain region-specific manner which could possibly explain their binding to BDNF promoters in certain brain regions. For that we used a previously published RNA-sequencing dataset (Carullo et al., 2020) to determine whether the identified transcription factors are differentially expressed in cultured neurons originating from different brain regions. First, we analyzed the levels of BDNF and determined that the levels were slightly higher in the cultured cortical neurons than in hippocampal neurons (Fig. 8A). Next, of the 88 transcription factors that we determined by in vitro pulldown assay, 17 showed at least 2-fold difference in expression levels between cultured cortical and hippocampal neurons (Fig. 8B; Extended Data Fig. 8-1). Of the novel factors that showed specific binding to BDNF promoter I, Prox1 has remarkably higher expression levels in hippocampal neurons, and Tbr1 is more highly expressed in cortical neurons (Fig. 8B), in agreement with our in vitro pulldown assay. Foxp1, Mef2c, Satb1, Satb2, and the high-mobility group (HMG) domain-containing Sox5, which showed preferential binding to BDNF promoter IV in cortical lysates, also have higher expression levels in cortical neurons than in hippocampal neurons (Fig. 8B).
Figure 8.
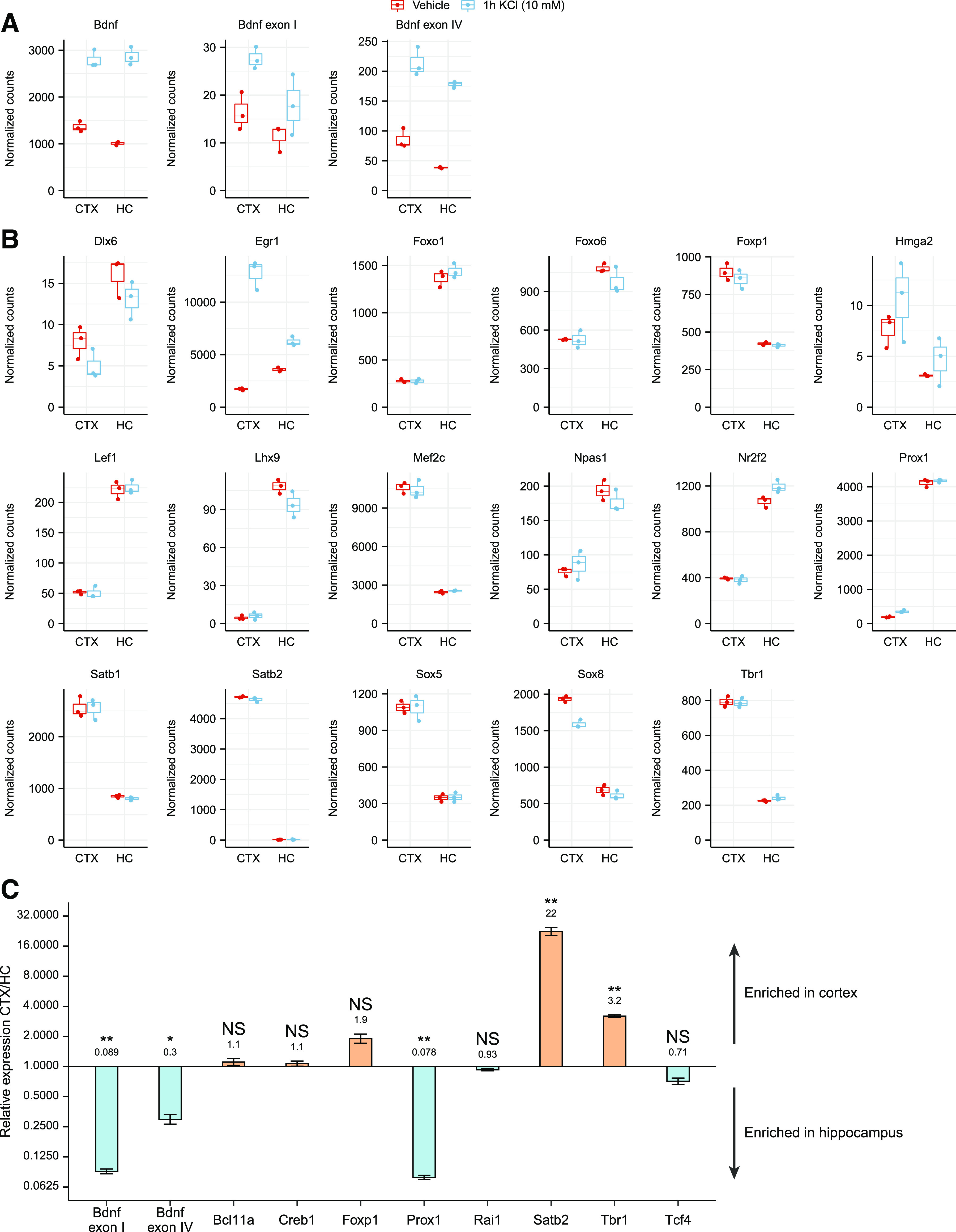
Various transcription factors that bind BDNF promoters in vitro are differentially expressed in cortical and hippocampal neurons and in cortex and hippocampus in vivo. A, B, RNA-sequencing (RNA-seq) data from embryonic day 18-derived rat cultured cortical (CTX) or hippocampal (HC) neurons treated with 10 mm KCl or vehicle for 1 h at 11 DIV (from Carullo et al., 2020) was used to analyze the expression levels of BDNF (A) and the transcription factors that we determined as specifically bound to BDNF promoter regions in the in vitro promoter pulldown experiment (B). Only transcription factors showing at least 2-fold expression difference between cultured cortical and hippocampal neurons in either untreated or KCl-treated neurons are shown. RNA-seq counts were normalized with DESeq2 and are shown as box plots, where the hinges show 25% and 75% quartiles, the horizontal line shows the median value, the upper whisker extends from the hinge to the largest value no further than 1.5× interquartile range from the hinge, the lower whisker extends from the hinge to the smallest value at most 1.5× interquartile range of the hinge. All data points are shown with dots. Expression data and statistical analysis for all the transcription factors detected to bind BDNF promoters in the in vitro pulldown assay are listed in Extended Data Figure 8-1. C, mRNA levels of the selected transcription factors in cortex and hippocampus of P8 Sprague Dawley pups shown as relative to the expression levels in cortex measured by RT-qPCR (group averages and exact p-values are listed in Extended Data Fig. 8-2). The expression levels of the determined transcription factors in the human cortex based on the GTEx portal are shown in Extended Data Figure 8-3. Error bars represent upper and lower limits of back-transformed mean ± SEM of three different litters (n = 3). Statistical significance is shown compared with the expression levels in cortex. *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm–Šídák method.
The expression levels and statistical analysis of the Carullo et al., 2020 RNA-sequencing data of the transcription factors detected to bind BDNF promoters in our in vitro promoter pulldown experiment. Download Figure 8-1, XLSX file (27.3KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 8C. Download Figure 8-2, XLSX file (18.2KB, xlsx) .
Comparison of the expression levels of the determined transcription factors in the human cortex and hippocampus based on the GTEx portal. Download Figure 8-3, XLSX file (14.4KB, xlsx) .
Next, we decided to validate the expression levels of a selection of transcription factors found in our in vitro pulldown assay in the cerebral cortex and hippocampus of 8-day-old rat pups using RT-qPCR (Fig. 8C; Extended Data Fig. 8-2). Interestingly, we discovered that in vivo, both BDNF exon I and exon IV-containing transcripts are substantially more expressed in the hippocampus than in the cortex (Fig. 8C). Of the transcription factors, we determined that Prox1 was markedly more expressed in the hippocampus and Satb2 and Tbr1 were more expressed in the cortex (Fig. 8C), which is in agreement with the preference shown in the pulldown assay. Meanwhile, in agreement with no preference in binding in the pulldown assay, the expression of Creb1 was similar in both brain regions. Interestingly, Rai1 and Tcf4, that showed a strong preference in the in vitro pulldown assay, did not show remarkable differences in their expression levels between different brain regions. Finally, we verified the expression of the identified transcription factors in the human cortex and hippocampus using the data from the Genotype-Tissue Expression (GTEx) portal (Extended Data Fig. 8-3). Collectively, we determined that many of the novel region-specific transcription factors binding to BDNF promoter regions, e.g., TBR1, FOXP1, SATB1, SATB2, and PROX1, are differentially expressed, possibly explaining their brain region-specific binding to BDNF promoters and suggesting their potential role in tissue-specific regulation of BDNF expression.
Validation of brain region-specific regulators of BDNF promoters I and IV
Next, to study whether the determined transcription factors identified by us are brain region-specific regulators of BDNF, we searched Gene Expression Omnibus (GEO) database for previously published RNA-seq datasets investigating the factors that were determined by us to bind BDNF promoters in the in vitro pulldown assay. First, in Foxp1 conditional knock-out (cKO) animals (Araujo et al., 2017), total BDNF levels were slightly increased in the cortex but not in hippocampus (Fig. 9A). When analyzing BDNF expression at a transcript level, we determined no change in the levels of BDNF exon I-containing transcripts, however, in agreement with our in vitro DNA pulldown experiment, we determined a slight increase of BDNF exon IV-containing transcripts in the FOXP1 cKO animals in cortex but not in hippocampus. Furthermore, overexpressing FOXP1 in human neural progenitor cells (Araujo et al., 2015) increased the levels of both total and BDNF exon IV-containing transcripts, whereas the levels of BDNF exon I-containing transcripts were undetectable in both control and FOXP1 overexpressing cells (Fig. 9B). These results suggest that FOXP1 is a novel regulator of BDNF exon IV-containing transcripts, with the exact outcome depending on the brain region and/or cell type.
Figure 9.
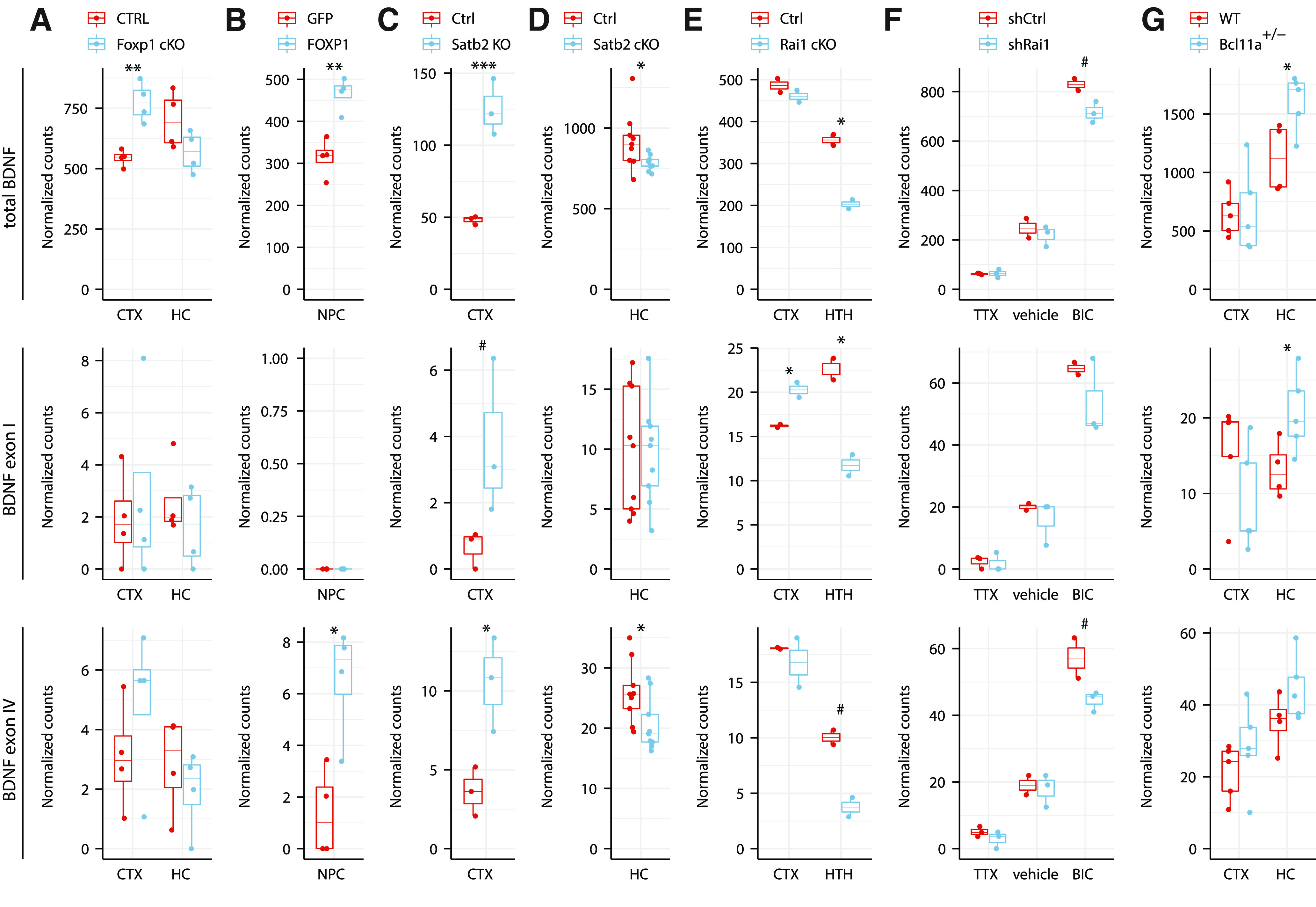
Transcription factors FOXP1, SATB2, RAI1, BCL11A regulate BDNF expression in a brain region-specific manner. Total BDNF levels (upper panels), levels of BDNF exon I (middle panels), and BDNF exon IV (lower panels) were analyzed from previously published RNA-seq experiments: (A) postnatal day 47 adult male mice cortex (CTX) and hippocampus (HC) in pyramidal neuron-specific Foxp1 conditional knock-out (cKO) animals using Emx1-Cre driver (Araujo et al., 2017); (B) human neural progenitor cells (NPC) overexpressing GFP or FOXP1 (Araujo et al., 2015); (C) postnatal day 0 CTX of Satb2 knock-out animals (McKenna et al., 2015); (D) adult mice HC CA1 region in Satb2 conditional knock-out in adult neurons using Camk2a-Cre driver (Jaitner et al., 2016); (E) three-week-old Rai1 flox/flox (Ctrl) and Rai1 cKO using Nestin-Cre driver in CTX and eight-week-old Rai1 cKO using Vglut2-Cre driver in hypothalamus (HTH; W.H. Huang et al., 2016); (F) Bru-seq to determine nascent transcripts in cultured cortical and hippocampal neurons derived from embryonic day 18 mice, grown 17 DIV and infected with lentiviruses expressing shRNA against Rai1 (shRai1) and treated with either tetrodotoxin (TTX), vehicle, or bicuculline (BIC) for 4 h (Garay et al., 2020); (G) CTX and HC of 16-week-old male wild-type (WT) or Bcl11a heterozygous knock-out mice (Dias et al., 2016). RNA-seq counts were normalized with DESeq2 and are shown as box plots, where the hinges show 25% and 75% quartiles, the horizontal line shows the median value, the upper whisker extends from the hinge to the largest value no further than 1.5× interquartile range from the hinge, the lower whisker extends from the hinge to the smallest value at most 1.5× interquartile range of the hinge. All data points are shown with dots. Statistical analysis was performed on log-transformed data. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, unpaired equal variance two tailed t test.
Second, we investigated the role of Satb family transcription factors in the regulation of tissue-specific BDNF expression. It has been reported that total BDNF mRNA levels are lower in Satb1 knock-out mice and that SATB1 binds to the BDNF locus in vivo in cerebral cortex (Balamotis et al., 2012). Here, we found that in the cortex of postnatal day 0 Satb2 knock-out mice (McKenna et al., 2015), total BDNF levels were strongly increased (Fig. 9C). Similarly, the levels of BDNF exon I and exon IV-containing transcripts were both increased (Fig. 9C). In contrast, in the hippocampus of adult conditional knock-out of Satb2 mice (Jaitner et al., 2016) the BDNF expression levels were the same as in wild type (Fig. 9D). While these results are in agreement with our in vitro pulldown assay (Fig. 7B) and higher Satb2 expression levels in cortex (Fig. 8B,C), we acknowledge that different effects in different brain regions could also be because of the use of different genetic models of Satb2 knock-out or because of the difference in the age of the animals at the time of the analysis. Additional experiments are needed to confirm the role of SATB2 as a tissue-specific regulator of BDNF expression.
Third, we focused on RAI1, which showed binding to BDNF promoter IV preferably in the hippocampus in our in vitro pulldown experiment (Fig. 7B), and which has previously been shown to regulate BDNF expression in the hypothalamus (Burns et al., 2010; W.H. Huang et al., 2016). To decipher the role of RAI1 in the regulation of BDNF, we analyzed BDNF expression levels in various Rai1 conditional knock-out (cKO) animals (W.H. Huang et al., 2016). Our results indicate that cKO of Rai1 had minor or no effect on total BDNF expression in cortex of three-week-old animals, whereas a strong decrease in the levels of total BDNF, exon I and exon IV-containing transcripts was observed in the hypothalamus of eight-week-old animals (Fig. 9E). Although no data were available on the effect of Rai1 knock-out in the hippocampus, these results indicate that RAI1 regulates BDNF expression in a brain region-specific manner, although it is not possible to rule out developmental stage-specific effects. As RAI1 has been described to regulate activity-dependent gene expression (Garay et al., 2020), we also analyzed BDNF expression levels in cultured neurons where the expression of Rai1 was silenced (Garay et al., 2020), and noted a slight decrease in the BDNF expression levels in bicuculline-treated neurons on silencing Rai1 expression (Fig. 9F). However, this effect was similar for total BDNF levels and BDNF exon I and exon IV-containing transcripts (Fig. 9F). Our results suggest that RAI1 could be a stimulus-specific regulator of BDNF gene expression.
Finally, we investigated the role of BCL11A (also known as CTIP1) that bound to BDNF promoter I and showed stronger binding in the cortex in our promoter pulldown assay (Fig. 7B). In the heterozygous Bcl11a knock-out mice (Dias et al., 2016) total BDNF levels were increased in the hippocampus, but remained the same in the cortex (Fig. 9G). At the transcript level, Bcl11a knock-out slightly reduced BDNF exon I levels in the cortex, but increased the levels in the hippocampus (Fig. 9G). However, the levels of BDNF exon IV were increased in both cortex and hippocampus of Bcl11a knock-out mice (Fig. 9G). Collectively, these results indicate that BCL11A has different effect on BDNF gene expression in different brain regions, however, the exact mechanism needs further clarification.
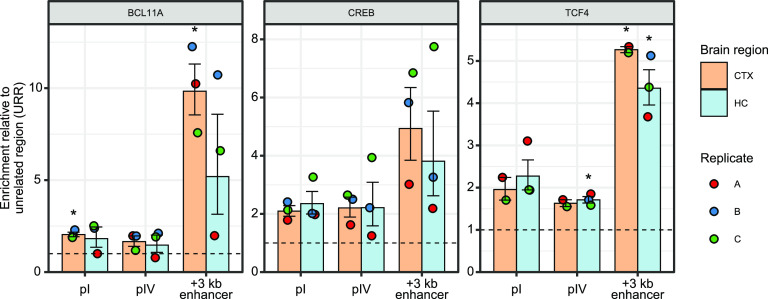
Finally, we focused on the promising candidates of region-specific regulators of BDNF and investigated whether they bind in vivo to BDNF promoters using ChIP assay. We focused on the binding of CREB, BCL11A, and TCF4 and our results show that all these factors bind in vivo to a BDNF intronic enhancer that is located 3 kb downstream from BDNF promoter I (Fig. 10) as was previously described for CREB and TCF4 in cultured neurons (Tuvikene et al., 2021). Furthermore, all the factors bind to BDNF promoters, however, not as efficiently as to the BDNF +3 kb enhancer region (Fig. 10). First, while BCL11A does bind to BDNF promoters, it only displays brain region-preference for the +3-kb region. Next, CREB binding to both BDNF promoters (and +3 kb region) was very stable in both cortex and hippocampus, indicating a lack of brain region-dependent binding specificity that is in agreement with our in vitro pulldown assay. Finally, TCF4 showed a slight preference in binding to BDNF promoter I and strong binding to the +3-kb enhancer region. Unfortunately, as we could not find antibodies suitable for ChIP assay for FOXP1, SATB2, and RAI1, we could not analyze their binding in vivo. Collectively, our ChIP experiment suggests that BCL11A, CREB, and TCF4 bind to BDNF promoters in vivo, however, with no notable differences in binding between cortex and hippocampus.
Figure 10.
BCL11A, CREB, and TCF4 bind to BDNF promoters in vivo. ChIP-qPCR analysis of BCL11A, CREB, and TCF4 binding to BDNF intronic enhancer located +3 kb downstream of BDNF promoter I (+3 kb enhancer), BDNF promoters I (pI) and IV (pIV) in the cortex and hippocampus of 8-d-old Sprague Dawley pups. Data are shown as percent of input relative to the binding of respective factor to unrelated region (URR). All data points are shown as dots, and each biological replicate (i.e., animals from different litters) is denoted with the same color. Error bars represent mean ± SEM of two or three different litters (n = 2–3). Statistical significance is shown compared with the binding of the respective factor to URR in the respective brain region. *p < 0.05, **p < 0.01, ***p < 0.001, paired two tailed t test, corrected for multiple comparisons using Holm–Šídák method.
Discussion
In this study, we investigated comparatively BDNF gene expression in response to neuronal signaling in cortical and hippocampal neurons and compared the role of CREB transcription factor family. Our results show that, as expected, CREB family transcription factors and specifically CREB in combination with its coactivators CBP and CRTC1 regulate the early induction of BDNF exon I and IV transcripts after neuronal signaling in cortical neurons. To our surprise, impairing the activity of CREB family or its coactivators had remarkably smaller effects on BDNF mRNA levels after prolonged stimulation than during the early time points of membrane depolarization. The rapid activation of CREB via posttranslational mechanisms and the lower levels of ICER, the endogenous negative regulator of the CREB family, could explain the faster inducibility of BDNF mRNA in cortical neurons. Although the early induction of BDNF mRNA seems to be strongly regulated by CREB family, the much higher expression levels of BDNF that are reached after prolonged neuronal signaling, seem to be mediated by other potent transcription factors that are synthesized as a second wave in response to neuronal activity.
Notably, the involvement of CREB family in the regulation of BDNF gene expression in hippocampal neurons appears to be complex. We made several observations on the regulation of BDNF exon I transcripts in hippocampal neurons: (1) overexpression of dominant-negative CREB and CREB knock-down had opposing effects on the basal levels of BDNF exon I transcripts, (2) inhibiting CBP recruitment had a profound effect on the late induction of BDNF exon I transcript, although neither overexpression of A-CREB nor CREB knock-down had such strong effect, and (3) expression of BDNF exon I transcripts was increased after CREB or CRTC1 knock-down, indicating their potential (probably indirect) repressor activities. Notably, the effect of CRTC1 knock-down on increased BDNF exon I mRNA levels also seems to result in higher BDNF protein levels in hippocampal neurons. Compared with cortical neurons where both CRTC1 and CBP participated in the induction of BDNF exon IV transcripts, CRTC1 was clearly the dominant coactivator in hippocampal cells. Collectively, this data show that in cortical and hippocampal neurons BDNF gene regulation is different at the level of CREB transcription factor family.
An interesting implication of our results is that the feedback loop regulating CREB family-dependent gene expression is different between cortical and hippocampal neurons. While the regulation of CREM activator forms by CREB family transcription factors was similar in both cortical and hippocampal neurons, we noted differences in the regulation of the endogenous negative regulator of CREB family, ICER, between the two neuronal populations. Namely, in cortical neurons neuronal activity-dependent induction of ICER was stronger and CREB knock-down had a more profound effect on its neuronal activity-dependent expression than in hippocampal neurons. Furthermore, knock-down of CRTC1 had stronger effect on the induction of ICER in cortical than in hippocampal neurons although the knock-down itself was stronger in hippocampal neurons. It would be of interest to investigate how the differences in the CREB family negative feedback loop affect CREB-dependent signaling.
Our results indicate that CREB family is a positive regulator of the expression of BDNF exon IV, but is a negative regulator of BDNF exon VI in neurons after membrane depolarization. In contrast, CREB family is a positive regulator of both transcripts after BDNF-TrkB signaling (Esvald et al., 2020). It could be explained by suppression of the expression of BDNF exon VI by transcriptional interference arising from transcription starting from BDNF promoter IV. Also, as CBP is highly associated with enhancer regions and impairing CBP recruitment decreased the induced levels of BDNF exon VI, it is also plausible that BDNF gene expression relies on a specific 3D chromatin structure and enhancer region(s) that are regulated by CBP recruitment. Competitive looping between specific promoter and enhancer regions would explain the opposite effects seen on BDNF exon IV and VI expression levels. We have previously shown that an intronic enhancer region governs the regulation of first cluster of BDNF exons, but not exons IV or VI (Tuvikene et al., 2021), showing that enhancers could potentiate only specific BDNF promoter regions. Further work will delineate the opposite effects of CREB family on BDNF exon IV and VI levels.
Neuronal signaling is known to cause BDNF secretion (Sasi et al., 2017; M. Song et al., 2017) and the released BDNF can signal the cells in an autocrine or paracrine manner by binding to TrkB receptor to induce BDNF mRNA production (Tuvikene et al., 2016; Esvald et al., 2020). Previous reports have shown that infusion of BDNF to the adult rat hippocampus can elicit the expression of different BDNF transcripts (Wibrand et al., 2006; Esvald et al., 2020), implying that BDNF-TrkB signaling is also relevant for BDNF expression in the hippocampus. Our previous work has suggested that BDNF transcriptional autoregulation and membrane depolarization-induced expression of BDNF are regulated via distinct mechanisms in cortical neurons (Esvald et al., 2020). Here, we note that TrkB signaling slightly contributes to neuronal activity-dependent expression of BDNF in cortical neurons, but not in hippocampal neurons. Further work is necessary to understand why the TrkB signaling participates in cortical but not in hippocampal neurons in activity-regulated expression of BDNF, and what is the biological implication of this difference.
Finally, we focused on deciphering brain region-specific regulators of BDNF and determined numerous transcription factors that are involved in BDNF transcriptional regulation. Interestingly, we detected transcription factors from all the transcriptional waves of neuronal maturation during cortical neurogenesis (Telley et al., 2016) and many of the determined transcription factors are important for neuronal differentiation and formation of specific brain regions. For example, FOXP1 is needed for the differentiation of neural stem cells (Braccioli et al., 2017), SATB2 for the differentiation of subcerebral projection neurons (McKenna et al., 2015), BCL11A for brain development (Dias et al., 2016) and the migration and formation of polarity of cortical neurons (Wiegreffe et al., 2015), TCF4 for neuronal migration and cortical layering (Page et al., 2018; Li et al., 2019; Wittmann et al., 2021) and other processes of brain development (Kennedy et al., 2016; Thaxton et al., 2018; Phan et al., 2020; Schoof et al., 2020), TBR1 for synapse formation in cortical layer 6 (Fazel Darbandi et al., 2018), and PROX1 for suppression of neurite outgrowth (Kaltezioti et al., 2020) and determining granular cell identity in the dentate gyrus (Iwano et al., 2012). Collectively, this array of different transcription factors implies that BDNF has a complex spatiotemporal regulation depending on the developmental stage and brain region. In support of this, it has also been shown that NURR1 transcription factor regulates BDNF expression in midbrain (Volpicelli et al., 2007) but not in cortical neurons (Abdollahi and Fahnestock, 2022). It is plausible that BDNF is an effector molecule guiding the development of the nervous system and formation of neuronal circuits. Moreover, many of the transcription factors we identified as novel regulators of BDNF gene expression have been linked to various neurodevelopmental diseases. For example, in humans mutations in Foxp1 gene cause severe forms of autism spectrum disorder and intellectual disability (Hamdan et al., 2010; Horn et al., 2010) and in mice FOXP1 regulates genes implicated in autism (Araujo et al., 2015) and Foxp1 deletion causes anxiety-like behavior, impaired social communication and spatial learning (Bacon and Rappold, 2012; Araujo et al., 2017), alterations of Satb2 gene cause SATB2-associated syndrome characterized by delayed development and intellectual disability (Zarate and Fish, 2017), mutations of Bcl11a gene lead to intellectual disability and autism spectrum disorders (Simon et al., 2020), TCF4 haploinsufficiency causes syndromal mental retardation – Pitt-Hopkins syndrome (Amiel et al., 2007; Brockschmidt et al., 2007; Zweier et al., 2007), Tbr1 is one of the highly associated genes in autism (O'Roak et al., 2012) and its knock-out causes increased anxiety and aggression in adult mice (Fazel Darbandi et al., 2018) as well as behavioral abnormalities that resemble autism (T.N. Huang et al., 2014). Notably, many of the phenotypes caused by defects in these transcription factors resemble those obtained with altered levels of BDNF, e.g., hyperactivity, aggressiveness, obesity, memory problems, lower intellect, and autism (Chao, 2003; Gray et al., 2006; Han et al., 2013; Zheng et al., 2016; Skogstrand et al., 2019). It is tempting to speculate that some of the phenotypes observed when these transcription factors are perturbed could arise from the insufficiency of BDNF. For example, haploinsufficiency of RAI1, which is required for motor functions and learning (Bi et al., 2007; W.H. Huang et al., 2016), causes Smith-Magenis syndrome (SMS; Slager et al., 2003; Bi et al., 2004). Some phenotypic effects of SMS, including obesity, have been explained by decreased levels of BDNF in the hypothalamus (Burns et al., 2010; W.H. Huang et al., 2016) and overexpression of BDNF has been shown to be an effective therapeutic approach for SMS (Javed et al., 2021). However, further work is needed to confirm this hypothesis for other novel BDNF regulators.
Our results deepen the understanding of BDNF gene expression, highlighting the complex regulation of BDNF in different brain regions. This data could give insight for understanding various neurodevelopmental disorders but also for developing therapeutics to modulate the expression of BDNF in a brain region and cell type-specific manner. However, a substantial amount of work is still required to understand the exact mechanisms that govern BDNF gene expression, including chromatin conformation, the set of transcription factors expressed in a given cell, their posttranslational modifications and the different participating signaling pathways in different neuronal populations.
Footnotes
This work was supported by the Estonian Research Council Grant PRG805 and the European Union through the European Regional Development Fund Project No. 2014-2020.4.01.15-0012 and H2020-MSCA-RISE-2016 (EU734791). We thank the TUT Institutional Development Program for 2016–2022 Graduate School in Clinical Medicine, which received funding from the European Regional Development Fund under Program ASTRA 2014-2020.4.01.16-0032 in Estonia. We thank Epp Väli for technical assistance and Priit Pruunsild, Indrek Koppel, Florencia Cabrera-Cabrera, and Annela Avarlaid for critical reading of the manuscript.
The authors declare no competing financial interests.
References
- Abdollahi M, Fahnestock M (2022) Nurr1 is not an essential regulator of BDNF in mouse cortical neurons. Int J Mol Sci 23:6853. 10.3390/ijms23126853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C (1998) A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol 18:967–977. 10.1128/MCB.18.2.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T (2007) Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res 85:525–535. 10.1002/jnr.21139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J, Rio M, de Pontual L, Redon R, Malan V, Boddaert N, Plouin P, Carter NP, Lyonnet S, Munnich A, Colleaux L (2007) Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet 80:988–993. 10.1086/515582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo DJ, Anderson AG, Berto S, Runnels W, Harper M, Ammanuel S, Rieger MA, Huang HC, Rajkovich K, Loerwald KW, Dekker JD, Tucker HO, Dougherty JD, Gibson JR, Konopka G (2015) FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev 29:2081–2096. 10.1101/gad.267989.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo DJ, Toriumi K, Escamilla CO, Kulkarni A, Anderson AG, Harper M, Usui N, Ellegood J, Lerch JP, Birnbaum SG, Tucker HO, Powell CM, Konopka G (2017) Foxp1 in forebrain pyramidal neurons controls gene expression required for spatial learning and synaptic plasticity. J Neurosci 37:10917–10931. 10.1523/JNEUROSCI.1005-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C, Rappold GA (2012) The distinct and overlapping phenotypic spectra of FOXP1 and FOXP2 in cognitive disorders. Hum Genet 131:1687–1698. 10.1007/s00439-012-1193-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamotis MA, Tamberg N, Woo YJ, Li J, Davy B, Kohwi-Shigematsu T, Kohwi Y (2012) Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol Cell Biol 32:333–347. 10.1128/MCB.05917-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambah-Mukku D, Travaglia A, Chen DY, Pollonini G, Alberini CM (2014) A positive autoregulatory BDNF feedback loop via C/EBPβ mediates hippocampal memory consolidation. J Neurosci 34:12547–12559. 10.1523/JNEUROSCI.0324-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Valor LM, Jimenez-Minchan M, Huber W, Barco A (2011) cAMP response element-binding protein is a primary hub of activity-driven neuronal gene expression. J Neurosci 31:18237–18250. 10.1523/JNEUROSCI.4554-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergami M, Rimondini R, Santi S, Blum R, Götz M, Canossa M (2008) Deletion of TrkB in adult progenitors alters newborn neuron integration into hippocampal circuits and increases anxiety-like behavior. Proc Natl Acad Sci U S A 105:15570–15575. 10.1073/pnas.0803702105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M (2004) Identification of small-molecule antagonists that inhibit an activator:coactivator interaction. Proc Natl Acad Sci U S A 101:17622–17627. 10.1073/pnas.0406374101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Saifi GM, Shaw CJ, Walz K, Fonseca P, Wilson M, Potocki L, Lupski JR (2004) Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum Genet 115:515–524. 10.1007/s00439-004-1187-6 [DOI] [PubMed] [Google Scholar]
- Bi W, Yan J, Shi X, Yuva-Paylor LA, Antalffy BA, Goldman A, Yoo JW, Noebels JL, Armstrong DL, Paylor R, Lupski JR (2007) Rai1 deficiency in mice causes learning impairment and motor dysfunction, whereas Rai1 heterozygous mice display minimal behavioral phenotypes. Hum Mol Genet 16:1802–1813. 10.1093/hmg/ddm128 [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME (2013) The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503:121–125. 10.1038/nature12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braccioli L, Vervoort SJ, Adolfs Y, Heijnen CJ, Basak O, Pasterkamp RJ, Nijboer CH, Coffer PJ (2017) FOXP1 promotes embryonic neural stem cell differentiation by repressing Jagged1 expression. Stem Cell Reports 9:1530–1545. 10.1016/j.stemcr.2017.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockschmidt A, Todt U, Ryu S, Hoischen A, Landwehr C, Birnbaum S, Frenck W, Radlwimmer B, Lichter P, Engels H, Driever W, Kubisch C, Weber RG (2007) Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum Mol Genet 16:1488–1494. 10.1093/hmg/ddm099 [DOI] [PubMed] [Google Scholar]
- Burns B, Schmidt K, Williams SR, Kim S, Girirajan S, Elsea SH (2010) Rai1 haploinsufficiency causes reduced Bdnf expression resulting in hyperphagia, obesity and altered fat distribution in mice and humans with no evidence of metabolic syndrome. Hum Mol Genet 19:4026–4042. 10.1093/hmg/ddq317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carullo NVN, Phillips RA, IIISimon RC, Soto SAR, Hinds JE, Salisbury AJ, Revanna JS, Bunner KD, Ianov L, Sultan FA, Savell KE, Gersbach CA, Day JJ (2020) Enhancer RNAs predict enhancer–gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res 48:9550–9570. 10.1093/nar/gkaa671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH (2004) Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc Natl Acad Sci U S A 101:13572–13577. 10.1073/pnas.0405587101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV (2003) Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 4:299–309. 10.1038/nrn1078 [DOI] [PubMed] [Google Scholar]
- Chen LF, Lyons MR, Liu F, Green MV, Hedrick NG, Williams AB, Narayanan A, Yasuda R, West AE (2020) The NMDA receptor subunit GluN3A regulates synaptic activity-induced and myocyte enhancer factor 2C (MEF2C)-dependent transcription. J Biol Chem 295:8613–8627. 10.1074/jbc.RA119.010266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME (2003a) Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885–889. 10.1126/science.1086446 [DOI] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME (2003b) Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci 23:2572–2581. 10.1523/JNEUROSCI.23-07-02572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal L, Kwan TOC, Shammas SL, Clarke J (2017a) pKID binds to KIX via an unstructured transition state with nonnative interactions. Biophys J 113:2713–2722. 10.1016/j.bpj.2017.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal L, Shammas SL, Clarke J (2017b) Phosphorylation of the IDP KID modulates affinity for KIX by increasing the lifetime of the complex. Biophys J 113:2706–2712. 10.1016/j.bpj.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, et al. (2016) BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am J Hum Genet 99:253–274. 10.1016/j.ajhg.2016.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvald E-E, Tuvikene J, Sirp A, Patil S, Bramham CR, Timmusk T (2020) CREB family transcription factors are major mediators of BDNF transcriptional autoregulation in cortical neurons. J Neurosci 40:1405–1426. 10.1523/JNEUROSCI.0367-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazel Darbandi S, Robinson Schwartz SE, Qi Q, Catta-Preta R, Pai EL-L, Mandell JD, Everitt A, Rubin A, Krasnoff RA, Katzman S, Tastad D, Nord AS, Willsey AJ, Chen B, State MW, Sohal VS, Rubenstein JLR (2018) Neonatal Tbr1 dosage controls cortical layer 6 connectivity. Neuron 100:831–845.e7. 10.1016/j.neuron.2018.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim T-K, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60:1022–1038. 10.1016/j.neuron.2008.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A (2020) JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48:D87–D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes NS, Borrelli E, Sassone-Corsi P (1991) CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell 64:739–749. 10.1016/0092-8674(91)90503-q [DOI] [PubMed] [Google Scholar]
- Garay PM, Chen A, Tsukahara T, Rodríguez Díaz JC, Kohen R, Althaus JC, Wallner MA, Giger RJ, Jones KS, Sutton MA, Iwase S (2020) RAI1 regulates activity-dependent nascent transcription and synaptic scaling. Cell Rep 32:108002. 10.1016/j.celrep.2020.108002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Yeo GSH, Cox JJ, Morton J, Adlam A-LR, Keogh JM, Yanovski JA, Gharbawy AE, Han JC, Tung YCL, Hodges JR, Raymond FL, O'Rahilly S, Farooqi IS (2006) Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) Gene. Diabetes 55:3366–3371. 10.2337/db06-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Daoud H, Rochefort D, Piton A, Gauthier J, Langlois M, Foomani G, Dobrzeniecka S, Krebs MO, Joober R, Lafrenière RG, Lacaille JC, Mottron L, Drapeau P, Beauchamp MH, Phillips MS, Fombonne E, Rouleau GA, Michaud JL (2010) De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet 87:671–678. 10.1016/j.ajhg.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JC, Thurm A, Golden Williams C, Joseph LA, Zein WM, Brooks BP, Butman JA, Brady SM, Fuhr SR, Hicks MD, Huey AE, Hanish AE, Danley KM, Raygada MJ, Rennert OM, Martinowich K, Sharp SJ, Tsao JW, Swedo SE (2013) Association of brain-derived neurotrophic factor (BDNF) haploinsufficiency with lower adaptive behaviour and reduced cognitive functioning in WAGR/11p13 deletion syndrome. Cortex 49:2700–2710. 10.1016/j.cortex.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME (2008) A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron 60:610–624. 10.1016/j.neuron.2008.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D, et al. (2010) Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum Mutat 31:E1851–E1860. 10.1002/humu.21362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Gao T, Dong Z, Zhou C, Lai Y, Pan T, Liu Y, Zhao X, Sun X, Hua H, Wen G, Gao L, Lv Z (2018) Neural circuitry among connecting the hippocampus, prefrontal cortex and basolateral amygdala in a mouse depression model: associations correlations between BDNF levels and BOLD – fMRI signals. Brain Res Bull 142:107–115. 10.1016/j.brainresbull.2018.06.019 [DOI] [PubMed] [Google Scholar]
- Huang TN, Chuang H-C, Chou W-H, Chen C-Y, Wang H-F, Chou S-J, Hsueh Y-P (2014) Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat Neurosci 17:240–247. 10.1038/nn.3626 [DOI] [PubMed] [Google Scholar]
- Huang WH, Guenthner CJ, Xu J, Nguyen T, Schwarz LA, Wilkinson AW, Gozani O, Chang HY, Shamloo M, Luo L (2016) Molecular and neural functions of Rai1, the causal gene for Smith-Magenis syndrome. Neuron 92:392–406. 10.1016/j.neuron.2016.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano T, Masuda A, Kiyonari H, Enomoto H, Matsuzaki F (2012) Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development 139:3051–3062. 10.1242/dev.080002 [DOI] [PubMed] [Google Scholar]
- Jaitner C, Reddy C, Abentung A, Whittle N, Rieder D, Delekate A, Korte M, Jain G, Fischer A, Sananbenesi F, Cera I, Singewald N, Dechant G, Apostolova G (2016) Satb2 determines miRNA expression and long-term memory in the adult central nervous system. Elife 5:e17361. 10.7554/eLife.17361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed S, Lee YJ, Xu J, Huang W-H (2021) Temporal dissection of Rai1 function reveals brain-derived neurotrophic factor as a potential therapeutic target for Smith-Magenis syndrome. Hum Mol Genet 31:275–288. 10.1093/hmg/ddab245 [DOI] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U (2004) What turns CREB on? Cell Signal 16:1211–1227. 10.1016/j.cellsig.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Kaltezioti V, Foskolou IP, Lavigne MD, Ninou E, Tsampoula M, Fousteri M, Margarity M, Politis PK (2020) Prox1 inhibits neurite outgrowth during central nervous system development. Cell Mol Life Sci 78:3443–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J, Lewis JW, Posey J, Strange SK, Guzman-Karlsson MC, Phillips SE, Decker K, Motley ST, Swayze EE, Ecker DJ, Michael TP, Day JJ, Sweatt JD (2016) Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep 16:2666–2685. 10.1016/j.celrep.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel I, Aid-Pavlidis T, Jaanson K, Sepp M, Pruunsild P, Palm K, Timmusk T (2009) Tissue-specific and neural activity-regulated expression of human BDNF gene in BAC transgenic mice. BMC Neurosci 10:68. 10.1186/1471-2202-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel I, Aid-Pavlidis T, Jaanson K, Sepp M, Palm K, Timmusk T (2010) BAC transgenic mice reveal distal cis-regulatory elements governing BDNF gene expression. Genesis 48:214–219. 10.1002/dvg.20606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel I, Tuvikene J, Lekk I, Timmusk T (2015) Efficient use of a translation start codon in BDNF exon I. J Neurochem 134:1015–1025. 10.1111/jnc.13124 [DOI] [PubMed] [Google Scholar]
- Laoide BM, Foulkes NS, Schlotter F, Sassone-Corsi P (1993) The functional versatility of CREM is determined by its modular structure. EMBO J 12:1179–1191. 10.1002/j.1460-2075.1993.tb05759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BX, Xiao X (2009) Discovery of a small-molecule inhibitor of the KIX–KID interaction. Chembiochem 10:2721–2724. 10.1002/cbic.200900552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu Y, Morozov YM, Chen X, Page SC, Rannals MD, Maher BJ, Rakic P (2019) Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol Psychiatry 24:1235–1246. 10.1038/s41380-019-0353-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME (2008) Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455:1198–1204. 10.1038/nature07319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Viste K, Urday-Zaa JC, Senthil Kumar G, Tsai W-W, Talai A, Mayo KE, Montminy M, Radhakrishnan I (2012) Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc Natl Acad Sci U S A 109:20865–20870. 10.1073/pnas.1219028109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MR, Schwarz CM, West AE (2012) Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J Neurosci 32:12780–12785. 10.1523/JNEUROSCI.0534-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890–893. 10.1126/science.1090842 [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2:599–609. 10.1038/35085068 [DOI] [PubMed] [Google Scholar]
- McDowell KA, Hutchinson AN, Wong-Goodrich SJE, Presby MM, Su D, Rodriguiz RM, Law KC, Williams CL, Wetsel WC, West AE (2010) Reduced cortical BDNF expression and aberrant memory in Carf knock-out mice. J Neurosci 30:7453–7465. 10.1523/JNEUROSCI.3997-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Ortiz-Londono CF, Mathew TK, Hoang K, Katzman S, Chen B (2015) Mutual regulation between Satb2 and Fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proc Natl Acad Sci U S A 112:11702–11707. 10.1073/pnas.1504144112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioduszewska B, Jaworski J, Kaczmarek L (2003) Inducible cAMP early repressor (ICER) in the nervous system–a transcriptional regulator of neuronal plasticity and programmed cell death. J Neurochem 87:1313–1320. 10.1046/j.1471-4159.2003.02116.x [DOI] [PubMed] [Google Scholar]
- Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P (1993) Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell 75:875–886. 10.1016/0092-8674(93)90532-u [DOI] [PubMed] [Google Scholar]
- Mou Z, et al. (2015) Human obesity associated with an intronic SNP in the brain-derived neurotrophic factor locus. Cell Rep 13:1073–1080. 10.1016/j.celrep.2015.09.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd DB, Balmer TS, Kim SY, Machhour N, Pallas SL (2019) TrkB activation during a critical period mimics the protective effects of early visual experience on perception and the stability of receptive fields in adult superior colliculus. J Neurosci 39:4475–4488. 10.1523/JNEUROSCI.2598-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, et al. (2012) Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338:1619–1622. 10.1126/science.1227764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-López L, Vega-Rivera NM, Babu H, Ramírez-Rodríguez GB (2017) Brain-derived neurotrophic factor induces cell survival and the migration of murine adult hippocampal precursor cells during differentiation in vitro. Neurotox Res 31:122–135. 10.1007/s12640-016-9673-x [DOI] [PubMed] [Google Scholar]
- Page SC, Hamersky GR, Gallo RA, Rannals MD, Calcaterra NE, Campbell MN, Mayfield B, Briley A, Phan BN, Jaffe AE, Maher BJ (2018) The schizophrenia- and autism-associated gene, transcription factor 4 regulates the columnar distribution of layer 2/3 prefrontal pyramidal neurons in an activity-dependent manner. Mol Psychiatry 23:304–315. 10.1038/mp.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomer E, Carretero J, Benvegnù S, Dotti CG, Martin MG (2016) Neuronal activity controls Bdnf expression via Polycomb de-repression and CREB/CBP/JMJD3 activation in mature neurons. Nat Commun 7:11081. 10.1038/ncomms11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Poo M (2013) Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci 14:7–23. 10.1038/nrn3379 [DOI] [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR (1996) Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol 16:694–703. 10.1128/MCB.16.2.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan BN, et al. (2020) A myelin-related transcriptomic profile is shared by Pitt-Hopkins syndrome models and human autism spectrum disorder. Nat Neurosci 23:375–385. 10.1038/s41593-019-0578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T (2007) Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90:397–406. 10.1016/j.ygeno.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T (2011) Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J Neurosci 31:3295–3308. 10.1523/JNEUROSCI.4540-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I, Pérez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE (1997) Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91:741–752. 10.1016/S0092-8674(00)80463-8 [DOI] [PubMed] [Google Scholar]
- Sakata K, Woo NH, Martinowich K, Greene JS, Schloesser RJ, Shen L, Lu B (2009) Critical role of promoter IV-driven BDNF transcription in GABAergic transmission and synaptic plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A 106:5942–5947. 10.1073/pnas.0811431106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasi M, Vignoli B, Canossa M, Blum R (2017) Neurobiology of local and intercellular BDNF signaling. Pflugers Arch 469:593–610. 10.1007/s00424-017-1964-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Cardinaux J-R (2017) Emerging roles of CREB-regulated transcription coactivators in brain physiology and pathology. Trends Neurosci 40:720–733. 10.1016/j.tins.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Schoof M, Hellwig M, Harrison L, Holdhof D, Lauffer MC, Niesen J, Virdi S, Indenbirken D, Schüller U (2020) The basic helix-loop-helix transcription factor TCF4 impacts brain architecture as well as neuronal morphology and differentiation. Eur J Neurosci 51:2219–2235. 10.1111/ejn.14674 [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Dove SL, Kornhauser JM, Hochschild A, Greenberg ME (2000) Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein. Mol Cell Biol 20:9409–9422. 10.1128/MCB.20.24.9409-9422.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A (1998) Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron 20:727–740. 10.1016/s0896-6273(00)81011-9 [DOI] [PubMed] [Google Scholar]
- Simon R, Wiegreffe C, Britsch S (2020) Bcl11 transcription factors regulate cortical development and function. Front Mol Neurosci 13:51. 10.3389/fnmol.2020.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogstrand K, Hagen CM, Borbye-Lorenzen N, Christiansen M, Bybjerg-Grauholm J, Bækvad-Hansen M, Werge T, Børglum A, Mors O, Nordentoft M, Mortensen PB, Hougaard DM (2019) Reduced neonatal brain-derived neurotrophic factor is associated with autism spectrum disorders. Transl Psychiatry 9:252. 10.1038/s41398-019-0587-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH (2003) Mutations in RAI1 associated with Smith–Magenis syndrome. Nat Genet 33:466–468. 10.1038/ng1126 [DOI] [PubMed] [Google Scholar]
- Song M, Martinowich K, Lee FS (2017) BDNF at the synapse: why location matters. Mol Psychiatry 22:1370–1375. 10.1038/mp.2017.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Zhai L, Valencia Swain J, Chen Y, Wang P, Chen L, Liu Y, Xiang S (2018) Structural insights into the CRTC2-CREB complex assembly on CRE. J Mol Biol 430:1926–1939. 10.1016/j.jmb.2018.04.038 [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Sakaya H, Kisukeda T, Fushiki H, Tsuda M (2002) Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J Biol Chem 277:35920–35931. 10.1074/jbc.M204784200 [DOI] [PubMed] [Google Scholar]
- Tai DJC, Liu YC, Hsu WL, Ma YL, Cheng SJ, Liu SY, Lee EHY (2016) MeCP2 SUMOylation rescues Mecp2-mutant-induced behavioural deficits in a mouse model of Rett syndrome. Nat Commun 7:10552. 10.1038/ncomms10552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis KQ, Duman RS, Newton SS (2008) CREB binding and activity in brain: regional specificity and induction by electroconvulsive seizure. Biol Psychiatry 63:710–720. 10.1016/j.biopsych.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME (1998) Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20:709–726. 10.1016/s0896-6273(00)81010-7 [DOI] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME (2002) A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron 33:383–395. 10.1016/s0896-6273(01)00561-x [DOI] [PubMed] [Google Scholar]
- Telley L, Govindan S, Prados J, Stevant I, Nef S, Dermitzakis E, Dayer A, Jabaudon D (2016) Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351:1443–1446. 10.1126/science.aad8361 [DOI] [PubMed] [Google Scholar]
- Thaxton C, Kloth AD, Clark EP, Moy SS, Chitwood RA, Philpot BD (2018) Common pathophysiology in multiple mouse models of Pitt-Hopkins syndrome. J Neurosci 38:918–936. 10.1523/JNEUROSCI.1305-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K, MacIntyre T, Wang H, Whitston D, Liu Z-Y, Hoffmann E, Wang T, Brown JL, Webster K, Omer C, Zage PE, Zeng L, Zweidler-McKay PA (2009) Identification and preclinical characterization of AZ-23, a novel, selective, and orally bioavailable inhibitor of the Trk kinase pathway. Mol Cancer Ther 8:1818–1827. 10.1158/1535-7163.MCT-09-0036 [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H (1993) Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron 10:475–489. 10.1016/0896-6273(93)90335-o [DOI] [PubMed] [Google Scholar]
- Tuvikene J (2021) Differential regulation of the BDNF gene in cortical and hippocampal neurons - additional materials. Available at https://osf.io/vbdxg/. [DOI] [PMC free article] [PubMed]
- Tuvikene J, Pruunsild P, Orav E, Esvald E-E, Timmusk T (2016) AP-1 transcription factors mediate BDNF-positive feedback loop in cortical neurons. J Neurosci 36:1290–1305. 10.1523/JNEUROSCI.3360-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuvikene J, Esvald E-E, Rähni A, Uustalu K, Zhuravskaya A, Avarlaid A, Makeyev EV, Timmusk T (2021) Intronic enhancer region governs transcript-specific Bdnf expression in rodent neurons. Elife 10:e65161. 10.7554/eLife.65161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishta A, Habas A, Pruunsild P, Zheng J-J, Timmusk T, Hetman M (2009) Nuclear factor of activated T-cells isoform c4 (NFATc4/NFAT3) as a mediator of antiapoptotic transcription in NMDA receptor-stimulated cortical neurons. J Neurosci 29:15331–15340. 10.1523/JNEUROSCI.4873-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, Greenberg ME (2017) AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol Cell 68:1067–1082.e12. 10.1016/j.molcel.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli F, Caiazzo M, Greco D, Consales C, Leone L, Perrone-Capano C, D'Amato LC, Porzio UD (2007) Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J Neurochem 102:441–453. 10.1111/j.1471-4159.2007.04494.x [DOI] [PubMed] [Google Scholar]
- Walker WH, Daniel PB, Habener JF (1998) Inducible cAMP early repressor ICER down-regulation of CREB gene expression in Sertoli cells. Mol Cell Endocrinol 143:167–178. 10.1016/s0303-7207(98)00082-3 [DOI] [PubMed] [Google Scholar]
- Wang L, Chang X, She L, Xu D, Huang W, Poo M (2015) Autocrine action of BDNF on dendrite development of adult-born hippocampal neurons. J Neurosci 35:8384–8393. 10.1523/JNEUROSCI.4682-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Pruunsild P, Timmusk T (2014) Neurotrophins: transcription and translation. Handb Exp Pharmacol 220:67–100. 10.1007/978-3-642-45106-5_4 [DOI] [PubMed] [Google Scholar]
- Wibrand K, Messaoudi E, Håvik B, Steenslid V, Løvlie R, Steen VM, Bramham CR (2006) Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur J Neurosci 23:1501–1511. 10.1111/j.1460-9568.2006.04687.x [DOI] [PubMed] [Google Scholar]
- Wiegreffe C, Simon R, Peschkes K, Kling C, Strehle M, Cheng J, Srivatsa S, Liu P, Jenkins NA, Copeland NG, Tarabykin V, Britsch S (2015) Bcl11a (Ctip1) controls migration of cortical projection neurons through regulation of Sema3c. Neuron 87:311–325. 10.1016/j.neuron.2015.06.023 [DOI] [PubMed] [Google Scholar]
- Wittmann MT, Katada S, Sock E, Kirchner P, Ekici AB, Wegner M, Nakashima K, Lie DC, Reis A (2021) scRNA sequencing uncovers a TCF4-dependent transcription factor network regulating commissure development in mouse. Development 148:dev196022. 10.1242/dev.196022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Hyde TM, Cassano HL, Deep-Soboslay A, Kleinman JE, Weickert CS (2010) Promoter specific alterations of brain-derived neurotrophic factor mRNA in schizophrenia. Neuroscience 169:1071–1084. 10.1016/j.neuroscience.2010.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosnitzka E, Nan X, Nan J, Chacón-Fernández P, Kussmaul L, Schuler M, Hengerer B, Barde Y-A (2020) A new mouse line reporting the translation of brain-derived neurotrophic factor using green fluorescent protein. eNeuro 7:ENEURO.0462-19.2019. 10.1523/ENEURO.0462-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani M, Deogracias R, Guy J, Poot RA, Bird A, Barde Y-A (2012) Disease modeling using embryonic stem cells: meCP2 regulates nuclear size and RNA synthesis in neurons. Stem Cells 30:2128–2139. 10.1002/stem.1180 [DOI] [PubMed] [Google Scholar]
- Zarate YA, Fish JL (2017) SATB2-associated syndrome: mechanisms, phenotype, and practical recommendations. Am J Med Genet A 173:327–337. 10.1002/ajmg.a.38022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Zhang L, Zhu T, Huang J, Qu Y, Mu D (2016) Peripheral brain-derived neurotrophic factor in autism spectrum disorder: a systematic review and meta-analysis. Sci Rep 6:31241. 10.1038/srep31241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J, Reardon W, Saraiva J, Cabral A, Gohring I, Devriendt K, de Ravel T, Bijlsma EK, Hennekam RC, Orrico A, Cohen M, Dreweke A, Reis A, Nurnberg P, Rauch A (2007) Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). Am J Hum Genet 80:994–1001. 10.1086/515583 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Underlying data and exact p-values of the statistical analysis for Figure 2. Download Figure 2-1, XLSX file (76.5KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 3. Download Figure 3-1, XLSX file (43.6KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 4. Download Figure 3-1, XLSX file (47.4KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 5. Download Figure 5-1, XLSX file (31.7KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 6. Download Figure 6-1, XLSX file (36.3KB, xlsx) .
Lists of all peptides, proteins and transcription factors detected in mass-spectrometric analysis in the in vitro DNA pulldown experiment. Download Figure 7-1, XLSX file (794.7KB, xlsx) .
The expression levels and statistical analysis of the Carullo et al., 2020 RNA-sequencing data of the transcription factors detected to bind BDNF promoters in our in vitro promoter pulldown experiment. Download Figure 8-1, XLSX file (27.3KB, xlsx) .
Underlying data and exact p-values of the statistical analysis for Figure 8C. Download Figure 8-2, XLSX file (18.2KB, xlsx) .
Comparison of the expression levels of the determined transcription factors in the human cortex and hippocampus based on the GTEx portal. Download Figure 8-3, XLSX file (14.4KB, xlsx) .