Abstract
The cationic liposome is well-known as an efficient nucleic acid delivery tool; however, the stress responses induced by liposome per se have been rarely revealed. In this study, we found that Lipofectamine™ 2000 (lipo2000), a commonly used commercial cationic liposome transfection, could upregulate EphA2 mRNA expression in multiple cells at transfection dose. Furthermore, lipo2000 treatment could increase the level of EphA2 hnRNA (heterogeneous nuclear RNA). Lipo2000-induced EphA2 upregulation could be depleted upon global transcription inhibition, proving that lipo2000 upregulates EphA2 expression via activating its transcription. Moreover, HDAC4 depletion, a known EphA2 trans-acting regulatory factor, could eliminate the lipo2000-induced EphA2 upregulation, demonstrating that lipo2000 promotes EphA2 transcription in an HDAC4 dependent manner. Functionally, EphA2 knockdown did not affect GFP expression level and the interfering efficacy of siGAPDH, suggesting that EphA2 is unrelated to the nucleic acid delivery capacity of lipo2000. Nevertheless, EphA2 depletion significantly activated autophagy and apoptosis, increasing the cytotoxic effects of lipo2000, which could be rescued by EphA2 restoration, indicating that EphA2 is essential to overcome liposome-related cytotoxicity. Finally, we found that lipo2000 could activate EphA2 transcription in an HDAC4-dependent manner. EphA2 is not associated with the transfection efficiency of lipo2000, but it is vital to reduce lipo2000 cytotoxicity, suggesting that when conducting liposome-mediated gene function studies, especially for EphA2, the stress response of liposomes should be considered to obtain objective results.
Keywords: Lipofectamine™ 2000, EphA2, HDAC4, Liposome cytotoxicity
Lipofectamine™ 2000; EphA2; HDAC4; Liposome cytotoxicity.
1. Introduction
Transfection, a tool for delivering foreign nucleic acids into living cells, plays a critical role in biological function studies of target genes/proteins [1, 2]. Transfection methods are generally classified into biological, chemical, and physical approaches [1]. Chemical transfection, mediated by liposome and non-liposome chemicals, is widely applied for the merits such as easy use, lower cost, high performance, and wide applicability [1, 2]. Lipofectamine 2000™ (referred to as lipo2000 hereafter), one of the commercial cationic liposome transfection reagents, has been broadly used to efficiently introduce synthetic DNAs and RNAs into various mammalian cell types [3, 4]. In addition to being a transfection reagent, it can trigger stress responses in treated cells, also known as off-target effects [1, 3, 5, 6, 7]. More attention and efforts should be taken to reveal these off-target effects and the corresponding mechanisms for preventing mis-interpretation results obtained through transfection-mediated methods.
For instance, when exploring the functions of rat stress-inducible Hspa1b gene, researchers found that cationic liposomes like lipo2000 could activate Hspa1b promoter activity in B16F10 mouse melanoma cells [8]. Importantly, the microarray-based gene profile analysis revealed that lipo2000 altered many genes involved in stress response, apoptotic signaling, cell cycle regulation, metabolism, etc. [8]. Furthermore, Lipo2000/siNC (negative control of small interfering RNAs) complexes could induce autophagosome formation and autophagy in hepatoma cells in a dose and time-dependent manner [6]. Lipofectamine 2000/siRNA complexes could also induce protective autophagy, and endoplasmic reticulum unfolded protein response in human endothelial cells [3]. Moreover, proteomic analysis indicates that transfection reagent and DNA complexes could remarkably evoke interferon-response at mRNA and protein levels in HeLa cells [9]. These reporters strongly emphasize the urgency and significance of studies focusing on the off-target effects of transfect reagents.
During exploring the function of EphA2 in hepatocellular carcinoma (HCC), upregulation of EphA2 was always observed in the control group treated with lipo2000/siNC and lipo2000/vector plasmid complexes compared to the untreated blank group (unpublished data). In this study, we systematically investigated the functions and mechanisms of this phenotype. Lipo2000, at transfection dose, was found to promote EphA2 transcription in an HDAC4-dependent manner significantly. Functionally, EphA2 knockdown did not affect the transfection efficiency of lipo2000 but significantly increased its cytotoxicity, suggesting that EphA2 upregulation serves as a protective response to liposome treatment to maintain cell viability.
2. Methods and materials
2.1. Cell lines and reagents
All cells, including HepG2 (HCC), Huh7 (HCC), HEK-293T (Human embryonic kidney cell), AGS, DLD1, HeLa, and H460, were purchased from Procell Life Science&Technology Co., Ltd (Wuhan, China). HepG2, Huh7, 293T, and HeLa cells were cultured in DMEM (BI, Jerusalem, Israel) medium supplemented with 10% fetal bovine serum (FBS, BI, Jerusalem, Israel). AGS, DLD1, and H460 cells were maintained in RPMI-1640 (BI, Jerusalem, Israel) with 10% fetal bovine serum (BI, Jerusalem, Israel). All cells were grown in a humidified cell incubator at 37 °C with 5% CO2.
Lipofectamine™ 2000 (Lip2000) and Lipofectamine™ 3000 (Lipo3000) were purchased from Thermo Fisher (CA, USA). PEI 40000 was purchased from BIOHUB (Shanghai, China). Actinomycin D and puromycin were obtained from MCE (Shanghai, China). Hoechst 33342 Staining Solution was obtained from Beyotime (Shanghai, China). siRNAs (small interfering RNAs), including siNC and siGAPDH, were synthesized by Sangon Biotech (Shanghai, China). pEGFP-N1 plasmids were purchased from HonorGene (Changsha, Hunan).
2.2. Transfection performance detection
pEGFP-N1 plasmids and siGAPDH were introduced into HCC cells via lipo2000 to detect lipo2000 performance after EphA2 depletion, as previously described [10]. The rate of positive fluorescent GFP was calculated by GFP expressing cells (green fluorescence)/total cells (Hoechst positive, blue fluorescence). The interfering efficacy was demonstrated by the relative expression of GAPDH with the siNC group as normalization.
2.3. Western blot
Western blot was performed as previously described [10]. Briefly, total proteins were isolated from RIPA (NCM, Suzhou, China) treated cell lysis via centrifugation at 4 °C. The denatured proteins were then separated through SDS-PAGE, incubated with antibodies, and visualized. The antibodies used in the study were listed as follows: rabbit anti-EphA2 (CST, CA, USA); rabbit anti-HDAC4, mouse anti-GAPDH, and mouse anti-GFP-tag (Abclonal, Wuhan, China); mouse anti-α-Tubulin (Abmart, Shanghai, China); rabbit anti-Cleaved-PARP-1 (D214) (Immunoway, Suzhou, China); mouse anti-LC3B (MBL, MA, USA); anti-rabbit or anti-mouse IgG HRP-conjugated secondary antibodies (BBI, Shanghai, China).
2.4. Quantitative reverse transcription polymerase chain reaction (qRTPCR) assay
Total RNA extraction and the qPCR assay were performed as previously described [10]. Total RNAs were extracted from HCC cells using Trizol reagent (Simgen, Hangzhou, China) and transcribed into cDNA by Hifair® Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) (Yeasen, Shanghai, China). The level of targeted genes in HCC cells was then tested by qPCR using Hieff® qPCR SYBR Green Master Mix (Yeasen, Shanghai, China) with GAPDH as normalized control. The primer sequences are listed in Supplementary able 1.
2.5. Lentivirus package and stable cell line establishment
Lentivirus interfering plasmids pLKO.1-shEphA2 and pLKO.1-shHDAC4 were constructed from pLKO.1-TRC-puro plasmid as described previously [11]. Lentivirus expression plasmid, pCDH-CMV-EphA2 (with hygromycin resistance), and control plasmid, were purchased from HonorGene (Changsha, China). The primer sequences are listed in Supplementary able 1. The shEphA2 and EphA2 lentivirus particles and control lentivirus were packaged as previously described [10]. Then, HepG2 and Huh7 cells were infected twice by shEphA2 lentivirus particles for 48 h. The cells were then cultured in media containing puromycin (2 μg/mL). The HepG2 and Huh7 cells with EphA2 depletion were re-infected with EphA2 lentivirus particles and cultured with media containing hygromycin (500 μg/mL) to obtain EphA2 expression rescue HCC cells. The living cells were harvested for interfering efficacy validation by PCR and Western blot. The control cell lines were parallelly produced.
2.6. CCK-8 assay
CCK-8 assay was performed as previously described [10]. The cells were seeded at a concentration of 5×103 cells per well in a 96-well plate. Lipo2000 was added to the cells the following day. After 6 h, the medium was replaced with fresh medium without lipo2000. After 48 h, CCK-8 reagent was added to the cell medium for 1 h, and the absorbance value was measured at 450 nm by a spectrophotometer (BioTek, WI, USA). The inhibitory rate was calculated with the shNC + vehicle group as normalized reference (OD (experimental group)/OD (shNC + vehicle group)). Three paralleled wells were set up, and the experiments were independently performed in triplicate.
2.7. Plate clone formation assay
The plate clone assay was performed as previously described [12]. HCC cells with EphA2 depletion or restoration were digested into single cell suspension enzymatically. Then, the cells were counted and seeded at a density of 1000 cells per well in a 6-well plate. After treatment with lipo2000 (2.5 μg/mL) for 6 h, the culture media was freshly replaced, and the cells were cultured for seven days. The cells were then fixed with methanol and stained with 0.1% crystal violet for 15 min, respectively. Then, the pictures of 6-well plates were photographed and the clone containing more than 50 cells was calculated under an inverted microscope (Leica, Solms, Germany).
2.8. Transwell invasion assay
Transwell assay was also performed as per our previous description [10]. HCC cells were treated with lipo2000 for 6 h. The transwell chamber (8 μm pore size) (Costar, ME, USA), pre-coated with Matrigel (Corning, NY, USA), was then filled with 300 μL basal DMEM medium containing 2.5×104 treated cells. The lower well (24 well plate) was filled with 700 μL DMEM medium containing 5% FBS to form a chemoattractant. The chambers were treated with methanol and stained with 0.5% crystal violet after 24 h. Finally, the cells on the upper side of the membrane were swabbed, and the invasive cells were visualized with an inverted microscope (Leica, Solms, Germany).
2.9. Statistical analysis
Graphpad Prism 9.0 software was used to perform statistical analysis and plot charts. Analysis of variance followed by multiple comparison tests and non-paired Student t-test were applied for comparison between multiple and two groups. P < 0.05 was considered statistically significant.
3. Results
3.1. Lipo2000 treatment induces EphA2 levels to increase in multiple cells
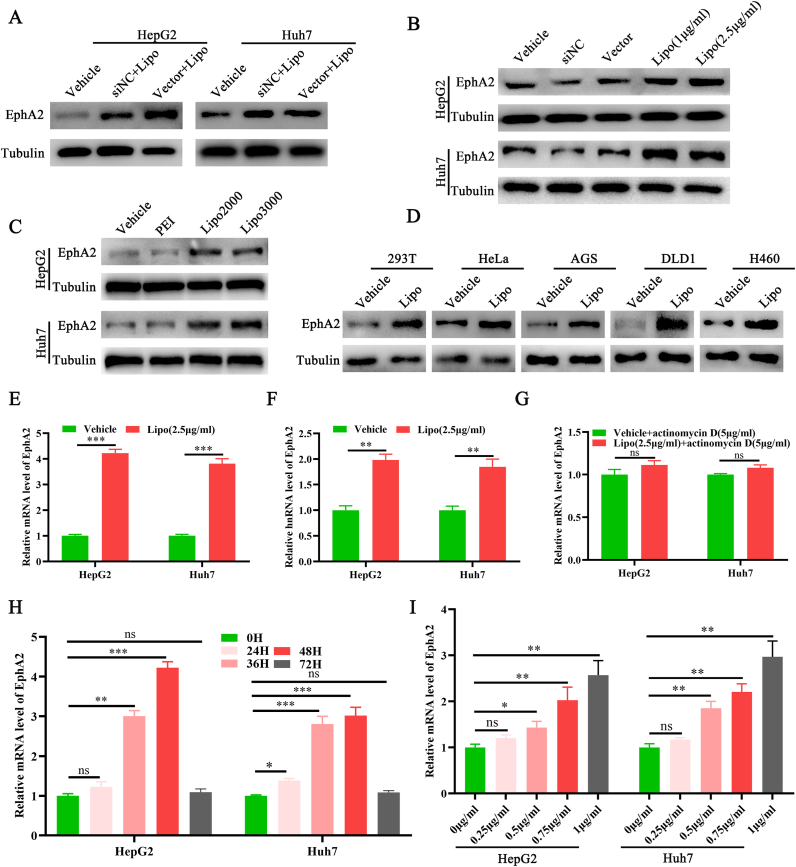
The level of EphA2 in HCC cells transfected with siNC or vector plasmids was found to be higher than the blank cells during lipo2000 mediated cell transfection (Figure 1A), suggesting that lipo2000 or lipo2000/siRNA or plasmids complexes could upregulate EphA2 in HCC cells. To determine which components caused EphA2 upregulation, we evaluated the effects of siNC, vector plasmid, and lipo2000 treatments on EphA2 levels in HCC cells, respectively. The results indicated that only Lipo2000 could significantly upregulate EphA2 in HCC cells in a dose-dependent manner (Figure 1B), suggesting that lipo2000 but not nucleic acid treatment could upregulate EphA2 in HCC cells. Moreover, we found that other cationic liposomes and cationic polymer transfection reagents might upregulate EphA2. The results showed that lipo3000, another cationic liposome, but not PEI, one of the widely used cationic polymer transfection reagents, could similarly upregulate EphA2 in HCC cells like lipo2000 (Figure 1C), demonstrating that cationic liposomes share the same capacity in EphA2 upregulation. Furthermore, we validated this effect in several other cell lines, including 293T, HeLa, AGS, DLD1, and H460 cells (Figure 1D), demonstrating that these effects may be common in human cells. Therefore, these findings indicated that the cationic liposome lipo2000 could upregulate EphA2 in multiple cells.
Figure 1.
Lipo2000 upregulates EphA2 expression by activating its transcription. Western blot results indicating the level of EphA2 in HCC cells with different treatments: (A) Vehicle (basal DMEM medium), lipo2000 (2.5 μg/mL)/siNC (5 nM) complexes and lipo2000 (2.5 μg/mL)/pEGFP-N plasmids (2.5 μg/mL) complexes. (B) Vehicle, siNC (5 nM), pEGFP-N plasmids (2.5 μg/mL), lipo2000 (1 μg/mL) and lipo2000 (2.5 μg/mL). (C) Vehicle, PEI (3 μg/mL), lipo2000 (2.5 μg/mL), and lipo3000 (3.75 μL). (D) Western blot results indicating the level of EphA2 in cells including 293T, HeLa, AGS, DLD1, and H460, treated with lipo2000 (2.5 μg/mL). (E) and (F) qPCR results demonstrating the relative expression of EphA2 mRNA and hnRNA in HCC cells treated by lipo2000. (G) qPCR results showing the relative expression of EphA2 mRNA co-treated with lipo2000 and actinomycin. (H) and (I) qPCR results showing the relative expression of EphA2 mRNA treated by lipo2000 at different times and doses. “Vehicle” means equal volume of pure DMEM medium. “Lipo” means lipo2000. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, “ns” means no significant difference.
3.2. Lipo2000 upregulates EphA2 via promoting its transcription
Next, we further explored the underlying mechanism of how lipo2000 upregulated EphA2 in HCC cells. Firstly, the qPCR results revealed that lipo2000 treatment could significantly increase the level of EphA2 mRNA in HCC cells (Figure 1E). To determine whether lipo2000 regulates EphA2 at the transcriptional or post-transcriptional level, the level of EphA2 hnRNA (heterogeneous nuclear RNA) directly demonstrates the level of primary transcription products, was further tested using primers targeting the EphA2 transcript intron region [12]. Lipo2000 treatment consistently increased the level of EphA2 hnRNA (Figure 1F). Furthermore, actinomycin D, a gene transcription inhibitor, could eliminate the pro-transcription effects of lipo2000 on EphA2 (Figure 1G). Moreover, the qPCR results demonstrated that lipo2000 induced EphA2 upregulation in a dose-dependent manner, with the highest at 48 h (Fig. 1H, I). These results revealed that lipo2000 upregulated EphA2 via promoting its transcription.
3.3. Lipo2000-induced EphA2 upregulation depends on HDAC4
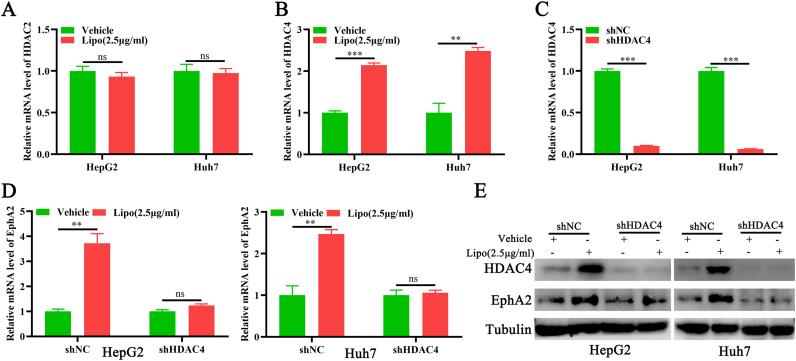
A recent study found that HDAC2 and HDAC4 activity is necessary for EphA2 transcription, implying that HDAC2 and HDAC4 may be involved in the Lip2000-induced EphA2 upregulation [13]. Initially, the effects of lipo2000 treatment on HDAC2 and HDAC4 expression were observed. The qPCR results indicated that lipo2000 had no regulatory influence on HDAC2 (Figure 2A) but significantly increased the level of HDAC4 (Figure 2B), suggesting that HDAC4 may be involved in the lipo2000 related EphA2 upregulation. Subsequently, the stable cell lines with HDAC4 depletion were successfully constructed to detect its effects on lipo2000-induced EphA2 upregulation (Figure 2C). Indeed, HDAC4 depletion stopped the lipo2000-related EphA2 overexpression, as demonstrated by qPCR and Western blot (Figure 2D, E). Therefore, these results suggest that Lipo2000-induced EphA2 upregulation was dependent on HDAC4.
Figure 2.
Lipo2000 upregulates EphA2 via HDAC4. (A) and (B) qPCR results indicating the relative mRNA level of HDAC2 and HDCA4 in HCC cells treated with lipo2000, respectively. (C) qPCR results indicating the relative expression of HDAC4 in HCC cells with HDAC4 knockdown. (D) and (E) qPCR and Western blot results indicating the relative expression of EphA2 in lipo2000 treated HCC cells with HDAC4 knockdown. ∗∗P < 0.01, ∗∗∗P < 0.001, “ns” means no significant difference.
3.4. EphA2 depletion does not affect the performance of lipo2000
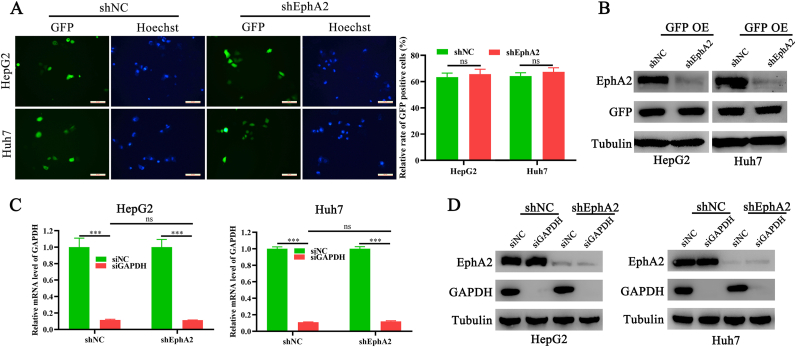
Subsequently, the study further investigated whether EphA2 depletion affects the transfection performance of lipo2000. First, HCC cells with EphA2 knockdown and corresponding control cells were equally transfected with pEGFP-N1 plasmids using lipo2000, and the relative transfection efficacy was determined by comparing the positive rate of fluorescence and GFP expression level. Both rates of GFP positive cells and GFP levels were comparable between HCC control cells and cells with EphA2 knockdown, suggesting that EphA2 depletion did not change the DNA transfection efficacy of lipo2000 (Figure 3A, B). Moreover, siRNAs targeting GAPDH were introduced into HCC control cells and cells with EphA2 knockdown. The interfering rate was then determined by qPCR and Western blot. As a result, siGAPDH significantly decreased GAPDH expression in both HCC control cells and cells with EphA2 knockdown (Figure 3C, D), indicating that EphA2 depletion did not affect the RNA delivery efficacy of lipo2000. Therefore, these results revealed that EphA2 alteration does not affect the transfection performance of lipo2000.
Figure 3.
EphA2 depletion does not affect the transfection capacity of lipo2000. pEGFP-N plasmids (2.5 μg) were transfected into HCC shEphA2 and shNC cells using lipo2000. (A) The representative pictures of fluorescent GFP and Hoechst of HCC cells, Magnification 200×, scale bar 50μm. (B) Western blot indicating the level of GFP in HCC cells. SiNC (10 nm) and siGAPDH (10 nM) were transfected into HCC shEphA2 cells and shNC cells using lipo2000. (C) and (D) qPCR and Western blot indicating the level of EphA2 in HCC cells. ∗∗∗P < 0.001, “ns” means no significant difference.
Lipo2000-induced EphA2 upregulation enhances liposome-related cytotoxicity tolerance of cells, possibly by suppressing autophagic apoptosis.
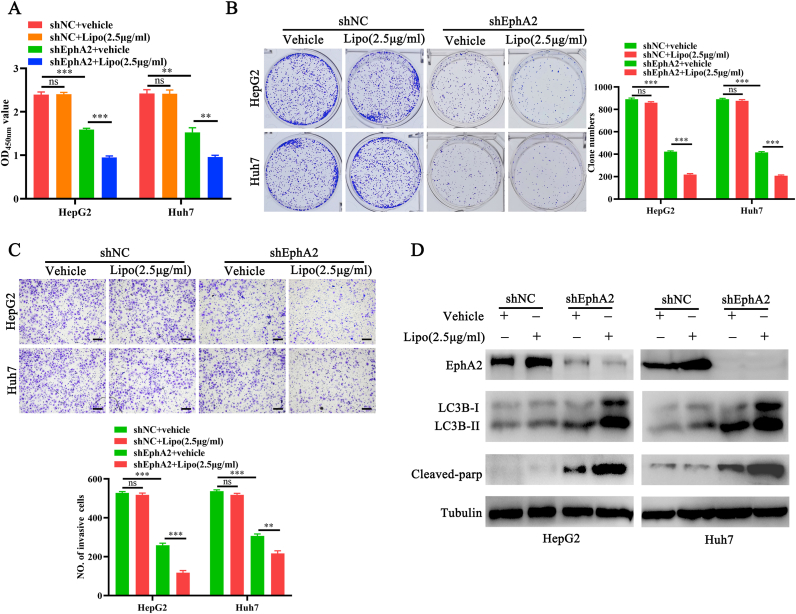
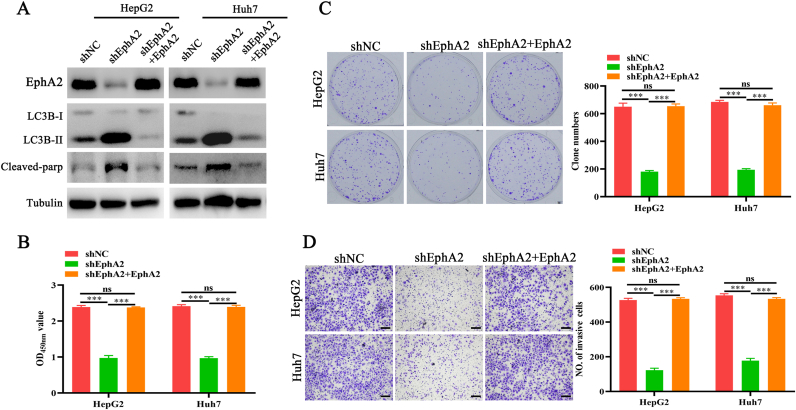
Because EphA2 is a potent pro-survival regulator in many cells, the effects of EphA2 depletion on lipo2000-induced cytotoxicity were explored [14]. Lipo2000 had no significant inhibition on control cells in CCK-8, plate clone formation, and Transwell invasion assays but significantly suppressed the proliferation and invasion of cells with EphA2 depletion, as shown by lower OD450nm values (Figure 4A), fewer cell clones (Figure 4B), and invasive cells (Figure 4C). A previous study found that lipo2000/siNC complexes and EphA2 depletion were involved in autophagy and apoptosis regulation [3, 6, 15, 16]. Thus, we measured autophagy and apoptosis level in control cells and cells with EphA2 depletion after lipo2000 treatment. Lipo2000 and EphA2 depletion could promote LC3B-II accumulation in control cells, respectively (Figure 4D), and lipo2000 exhibited remarkably synergistic effects in the promotion of LC3-II accumulation in cells with EphA2 knockdown. Furthermore, lipo2000 significantly increased the level of cleaved-parp (Figure 4D), a critical apoptotic marker [17] in HCC cells with EphA2 depletion, suggesting that EphA2 knockdown aggravated lipo2000-induced autophagy and subsequently activated autophagic apoptosis in HCC cells.
Figure 4.
EphA2 knockdown significantly enhances the cytotoxicity of lipo2000 and activates autophagy and apoptosis. HCC shEphA2 and shNC cells are parallelly treated by lipo2000 (2.5 μg/mL). (A) CCK-8 assay indicating the growth of HCC cells. (B) Plate clone formation indicating the proliferation of HCC cells. (C) Transwell assay indicating the invasion of HCC cells. Magnification 100×, scale bar 200μm (D) Western blot indicating the level of EphA2, LC3B-I, LC3B-II and cleaved-parp in HCC cells. ∗∗P < 0.01, ∗∗∗P < 0.001, “ns” means no significant difference.
To further validated the protective role of EphA2 in respond to lipo2000, the expression of EphA2 was exogenously restored (Figure 5A). Consistently, the results demonstrated that EphA2 rescue recovered the cell viability indicated by the level of LC3B-II and cleaved-parp (Figure 5A), the OD450nm values (Figure 5B), cell clones (Figure 5C), and invasive cells (Figure 5D).
Figure 5.
EphA2 restoration recovers tolerance of HCC cells with EphA2 depletion to lipo2000 related cytotoxicity. HCC shNC, shEphA2 and shEphA2 + EphA2 cells are parallelly treated by lipo2000 (2.5 μg/mL). (A) Western blot indicating the level of EphA2, LC3B-I, LC3B-II and cleaved-parp in HCC cells. (B) CCK-8 assay indicating the growth of HCC cells. (C) Plate clone formation indicating the proliferation of HCC cells. (D) Transwell assay indicating the invasion of HCC cells. Magnification 100×, scale bar 200μm. ∗∗∗P < 0.001, “ns” means no significant difference.
Thus, these results reveal that lipo2000-induced EphA2 overexpression improves cell tolerance to liposome-related cytotoxicity, potentially by suppressing autophagic apoptosis.
4. Discussion
Liposome-mediated transfection, an efficient tool for delivering nucleic acids into cells, serves as a stress stimulus for treated cells, evoking a series of cellular responses. For instance, lipo2000/siNC RNA complexes significantly suppress cholesterol biosynthesis and induce autophagy in hepatoma cells without affecting cell viability [6, 18]. Lipo2000/siNC RNA complexes treatment induces cell apoptosis in human endothelial cells via synchronously modifying protective autophagy, and endoplasmic reticulum unfolded protein response [3]. Lipo2000/vector plasmid complexes could alter genes involved in interferon type I/II response in HeLa cells [9]. At physiological temperature, lipo2000-mediated transfection could activate the promoter of Hspa1b, one of the heat shock/stress genes, and modify the expression of several genes involved in the regulation of cell survival and stress responses in mouse B16F10 cells [8]. Lipo2000 alone could promote EphA2 transcription and expression in an HDAC4-dependent manner. Functionally, EphA2 depletion effect lipo2000 transfection efficiency but significantly enhances lipo2000 cytotoxicity, which may be related to autophagic apoptosis induced by EphA2 knockdown.
EphA2, a receptor tyrosine kinases (RTK) from Eph family, is a key regulator in multiple physiological and pathological processes, especially carcinogenesis [14]. EphA2 is upregulated and acts as an oncogene in multiple cancers, as previously demonstrated [14, 19, 20]. EphA2 expression regulation has been revealed at both the transcriptional and post-transcriptional levels. Non-coding regulatory RNAs, such as miRNAs, circRNAs, and lncRNAs, are the main post-transcriptional regulators in EphA2 mRNA level regulation targeting EphA2 mRNA for degradation [21, 22, 23, 24]. Moreover, several transcript factors, such as c-Myc, p53, and Sp1, and trans-acting factors like HDAC2 and HDAC4, are involved in the regulation of EphA2 transcription [13, 14, 25, 26]. In this study, lipo2000 induces overexpression of EphA2 hnRNA, and global transcription arrest abolishes EphA2 upregulation, confirming the transcriptional mechanism accounting for lipo2000 induced EphA2 upregulation, leading us to investigate and validate the function of HDAC4 in EphA2 regulation.
EphA2 activation and upregulation are essential for cancer cells to maintain malignant phenotypes, such as proliferative capacity, apoptosis resistance, high metastasis and stemness priority, and therapeutic tolerance [14, 19]. Downregulation and inactivation of EphA2 could effectively induce cell death and limit cell growth, migration, and invasion in tumors such as HCC [14, 19, 20, 27]. Thus, in our study, decreased survival capacity 1 is mostly responsible for the significant toxicity and inhibitory effects of lipo2000 on HCC cells with EphA2 depletion, as shown by excessive autophagy and apoptosis.
EphA2 is a key host receptor for regulating virus infection and entry than an oncogenic RTK. Studies implying functional RNAi kinase screening strategies reveal that EphA2 is the major receptor to bind virus proteins and mediate the fusion and entry processes of viruses, including human herpesvirus [28], hepatitis C virus [29], Epstein-Barr Virus, and Kaposi's Sarcoma-Associated Herpesvirus [30, 31]. Virus-mediated transfection is also an efficient gene delivery method [2]. Although EphA2 is vital for virus infection and entry, EphA2 depletion does not affect the nucleic acid delivery performance of liposomes, suggesting that EphA2 is not essential for liposome entry.
5. Conclusion
Finally, we report that lipo2000 induces EphA2 transcription and expression in an HDAC4-dependent manner. EphA2 is not associated with lipo2000 transfection efficiency; however, it is vital for lipo2000 cytotoxicity reduction. The results emphasize that when conducting liposome-mediated gene function studies, especially for EphA2, the stress response of the liposome should be considered to obtain objective results.
Declarations
Author contribution statement
Zhiguo Huang: Performed the experiments; Analyzed and interpreted the data.
Jie Liu, Doctor: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Canzhi Zhang: Analyzed and interpreted the data.
Xin Yang, M.D: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Jie Liu was supported by Natural Science Foundation of Hunan Province [2021JJ40623], Changsha Municipal Natural Science Foundation [kq2014024], the Scientific Research Project of Hunan Provincial Health Commission [202102060121].
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix. ASupplementary data
The following is the supplementary data related to this article:
References
- 1.Stepanenko A.A., Heng H.H. Transient and stable vector transfection: pitfalls, off-target effects, artifacts. Mutat. Res. Rev. Mutat. Res. 2017;773:91–103. doi: 10.1016/j.mrrev.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Kim T.K., Eberwine J.H. Mammalian cell transfection: the present and the future. Anal. Bioanal. Chem. 2010;397:3173–3178. doi: 10.1007/s00216-010-3821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z., Zhang C., Wang Z., Shen J., Xiang P., Chen X., Nan J., Lin Y. Lipofectamine 2000/siRNA complexes cause endoplasmic reticulum unfolded protein response in human endothelial cells. J. Cell. Physiol. 2019;234:21166–21181. doi: 10.1002/jcp.28719. [DOI] [PubMed] [Google Scholar]
- 4.Lin C.W., Jan M.S., Kuo J.S. The vector-related influences of autophagic microRNA delivery by Lipofectamine 2000 and polyethylenimine 25K on mouse embryonic fibroblast cells. Eur. J. Pharmaceut. Sci. 2017;101:11–21. doi: 10.1016/j.ejps.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Guo X., Wang H., Li Y., Leng X., Huang W., Ma Y., Xu T., Qi X. Transfection reagent Lipofectamine triggers type I interferon signaling activation in macrophages. Immunol. Cell Biol. 2019;97:92–96. doi: 10.1111/imcb.12194. [DOI] [PubMed] [Google Scholar]
- 6.Mo R.H., Zaro J.L., Ou J.H., Shen W.C. Effects of Lipofectamine 2000/siRNA complexes on autophagy in hepatoma cells. Mol. Biotechnol. 2012;51:1–8. doi: 10.1007/s12033-011-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damodaran A.P., Courtheoux T., Watrin E., Prigent C. Alteration of SC35 localization by transfection reagents. Biochim. Biophys. Acta Mol. Cell Res. 2020;1867 doi: 10.1016/j.bbamcr.2020.118650. [DOI] [PubMed] [Google Scholar]
- 8.Fiszer-Kierzkowska A., Vydra N., Wysocka-Wycisk A., Kronekova Z., Jarzab M., Lisowska K.M., Krawczyk Z. Liposome-based DNA carriers may induce cellular stress response and change gene expression pattern in transfected cells. BMC Mol. Biol. 2011;12:27. doi: 10.1186/1471-2199-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagen L., Sharma A., Aas P.A., Slupphaug G. Off-target responses in the HeLa proteome subsequent to transient plasmid-mediated transfection. Biochim. Biophys. Acta. 2015;1854:84–90. doi: 10.1016/j.bbapap.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Song T., Hu Z., Liu J., Huang W. FLOT2 upregulation promotes growth and invasion by interacting and stabilizing EphA2 in gliomas. Biochem. Biophys. Res. Commun. 2021;548:67–73. doi: 10.1016/j.bbrc.2021.02.062. [DOI] [PubMed] [Google Scholar]
- 11.Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., Carpenter A.E., Foo S.Y., Stewart S.A., Stockwell B.R., Hacohen N., Hahn W.C., Lander E.S., Sabatini D.M., Root D.E. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 12.Huang W., Zeng C., Hu S., Wang L., Liu J. ATG3, a target of miR-431-5p, promotes proliferation and invasion of colon cancer via promoting autophagy. Cancer Manag. Res. 2019;11:10275–10285. doi: 10.2147/CMAR.S226828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T., Li J., Ma X., Yang Y., Sun W., Jin W., Wang L., He Y., Yang F., Yi Z., Hua Y., Liu M., Chen Y., Cai Z. Inhibition of HDACs-EphA2 signaling Axis with WW437 demonstrates promising preclinical antitumor activity in breast cancer. EBioMedicine. 2018;31:276–286. doi: 10.1016/j.ebiom.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao T., Xiao Y., Wang W., Tang Y.Y., Xiao Z., Su M. Targeting EphA2 in cancer. J. Hematol. Oncol. 2020;13:114. doi: 10.1186/s13045-020-00944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J., Wang L., Lv H., Liu J., Dong Y., Shi L., Ji Q. EphA2 inhibits SRA01/04 cells apoptosis by suppressing autophagy via activating PI3K/Akt/mTOR pathway. Arch. Biochem. Biophys. 2021;711 doi: 10.1016/j.abb.2021.109024. [DOI] [PubMed] [Google Scholar]
- 16.Amato K.R., Wang S., Hastings A.K., Youngblood V.M., Santapuram P.R., Chen H., Cates J.M., Colvin D.C., Ye F., Brantley-Sieders D.M., Cook R.S., Tan L., Gray N.S., Chen J. Genetic and pharmacologic inhibition of EPHA2 promotes apoptosis in NSCLC. J. Clin. Invest. 2014;124:2037–2049. doi: 10.1172/JCI72522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pistritto G., Trisciuoglio D., Ceci C., Garufi A., D'Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danielli M., Marinelli R.A. Lipid-based transfection reagents can interfere with cholesterol biosynthesis. Anal. Biochem. 2016;495:1–2. doi: 10.1016/j.ab.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Wilson K., Shiuan E., Brantley-Sieders D.M. Oncogenic functions and therapeutic targeting of EphA2 in cancer. Oncogene. 2021;40:2483–2495. doi: 10.1038/s41388-021-01714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao P., Jiang D., Huang Y., Chen C. EphA2: a promising therapeutic target in breast cancer. J. Genet. Genom. 2021;48:261–267. doi: 10.1016/j.jgg.2021.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Peng G., Meng H., Pan H., Wang W. CircRNA 001418 promoted cell growth and metastasis of bladder carcinoma via EphA2 by miR-1297. Curr. Mol. Pharmacol. 2021;14:68–78. doi: 10.2174/1874467213666200505093815. [DOI] [PubMed] [Google Scholar]
- 22.Niu X., Sun H., Qiu F., Liu J., Yang T., Han W. miR-10b-5p suppresses the proliferation and invasion of primary hepatic carcinoma cells by downregulating EphA2. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/1382061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Xiang Y., Huang Y., Sun H., Pan Y., Wu M., Zhang J. Deregulation of miR-520d-3p promotes hepatocellular carcinoma development via lncRNA MIAT regulation and EPHA2 signaling activation. Biomed. Pharmacother. 2019;109:1630–1639. doi: 10.1016/j.biopha.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Li Q.G., Xiao T., Zhu W., Yu Z.Z., Huang X.P., Yi H., Lu S.S., Tang Y.Y., Huang W., Xiao Z.Q. HDAC7 promotes the oncogenicity of nasopharyngeal carcinoma cells by miR-4465-EphA2 signaling axis. Cell Death Dis. 2020;11:322. doi: 10.1038/s41419-020-2521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G., Njauw C.N., Park J.M., Naruse C., Asano M., Tsao H. EphA2 is an essential mediator of UV radiation-induced apoptosis. Cancer Res. 2008;68:1691–1696. doi: 10.1158/0008-5472.CAN-07-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelinski D.P., Zantek N.D., Walker-Daniels J., Peters M.A., Taparowsky E.J., Kinch M.S. Estrogen and Myc negatively regulate expression of the EphA2 tyrosine kinase. J. Cell. Biochem. 2002;85:714–720. doi: 10.1002/jcb.10186. [DOI] [PubMed] [Google Scholar]
- 27.Wang H., Hou W., Perera A., Bettler C., Beach J.R., Ding X., Li J., Denning M.F., Dhanarajan A., Cotler S.J., Joyce C., Yin J., Ahmed F., Roberts L.R., Qiu W. Targeting EphA2 suppresses hepatocellular carcinoma initiation and progression by dual inhibition of JAK1/STAT3 and AKT signaling. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Light T.P., Brun D., Guardado-Calvo P., Pederzoli R., Haouz A., Neipel F., Rey F.A., Hristova K., Backovic M. Human herpesvirus 8 molecular mimicry of ephrin ligands facilitates cell entry and triggers EphA2 signaling. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupberger J., Zeisel M.B., Xiao F., Thumann C., Fofana I., Zona L., Davis C., Mee C.J., Turek M., Gorke S., Royer C., Fischer B., Zahid M.N., Lavillette D., Fresquet J., Cosset F.L., Rothenberg S.M., Pietschmann T., Patel A.H., Pessaux P., Doffoel M., Raffelsberger W., Poch O., McKeating J.A., Brino L., Baumert T.F. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J., Schaller S., Jardetzky T.S., Longnecker R. Epstein-barr virus gH/gL and Kaposi's sarcoma-associated herpesvirus gH/gL bind to different sites on EphA2 to trigger fusion. J. Virol. 2020;94 doi: 10.1128/JVI.01454-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su C., Wu L., Chai Y., Qi J., Tan S., Gao G.F., Song H., Yan J. Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses. Nat. Commun. 2020;11:5964. doi: 10.1038/s41467-020-19617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.