ABSTRACT
Background
Periconceptional folate intake is associated with the establishment of DNA methylation in offspring; however, variations in this relation by food sources compared with folic acid supplements are not described. Also, maternal folate intake is associated with decreased risk of pediatric acute lymphoblastic leukemia (ALL), but the mechanism is not known.
Objectives
We evaluated the relation between periconceptional folate intake by source and DNA methylation at birth in a cohort of pediatric ALL cases and controls in an epigenome-wide association study.
Methods
Genome-wide DNA methylation status obtained from archived neonatal blood spots from pediatric ALL cases (n = 189) and controls (n = 205) in the California Childhood Leukemia Study (CCLS) from 1995–2008 was compared with periconceptional folate from total, food, and supplemental sources using multivariable linear regression. Further stratification was performed by income, education, ethnicity, and total folate intake. We evaluated variable DNA methylation response to periconceptional folate by ALL case status through an interaction term.
Results
Two significant differentially methylated probes (DMPs) were associated with food and supplemental periconceptional folate intake in all subjects (n = 394). The top differentially methylated region at the promoter region of DUSP22(dual specificity phosphatase 22) demonstrated DNA hypermethylation in ALL cases but not in controls in response to total and food folate intake. We further identified 8 interaction term DMPs with variable DNA methylation response to folate intake by ALL case status. Further stratification of the cohort by education and ethnicity revealed a substantially higher number of DMPs associated with supplemental folic acid intake in Hispanic subjects with lower income and educational level.
Conclusions
We identified modest associations between periconceptional folate intake and DNA methylation differing by source, including variation by ALL case status. Hispanic subjects of lower income and education appear uniquely responsive to periconceptional folate supplementation.
Keywords: DNA methylation, folate, acute lymphoblastic leukemia, periconceptional nutrition, epigenetics, pediatric oncology
Introduction
The etiology of pediatric acute lymphoblastic leukemia (ALL) is likely multifactorial (1) with a prenatal origin (2), identifying the significance of the intrauterine period in future leukemia risk. This includes an association between maternal diet during pregnancy and development of childhood ALL. Consumption of vegetables, fruits, proteins (3), and legumes (4) during pregnancy is associated with a reduced risk of ALL in offspring after accounting for socioeconomic status. To explain these findings, focus has been placed on micronutrients involved in one-carbon metabolic processes, including folate (vitamin B-9), cobalamin (vitamin B-12), vitamin B-6, riboflavin (vitamin B-2) and methionine (5), which regulate processes including nucleotide synthesis and de novo DNA methylation (6).
Maternal diets high in folate decrease the risk of pediatric ALL (7, 8). Combined case-control data from 12 studies in the Childhood Leukemia International Consortium, including 6963 pediatric ALL subjects, demonstrated a reduction in ALL risk with periconceptional folate supplementation (9), a finding since confirmed independently (10). Neonatal serum folate concentrations are not associated with the development of ALL (11), indicating that exposure in the periconceptional and early gestational period has the strongest impact on leukemia risk. Folate source may additionally play an important role in regulating ALL risk. Natural folate exists in a wide array of plant and animal sources, whereas folate-fortified foods and dietary supplements occur as synthetic folic acid. While total maternal folate intake appears to reduce ALL risk across all populations, children of Hispanic background (5) and mothers of low educational levels (9) demonstrate particular benefit from higher folate intake, including folic acid supplementation.
DNA methylation patterns are largely established during early embryogenesis under genetic, stochastic, and environmental influences, including maternal folate consumption (12–14). This suggests that DNA methylation may mediate the association of maternal dietary folate and pediatric ALL (15). While aberrant DNA methylation typifies pediatric ALL at diagnosis (16), its role as an early or predisposing contributor to leukemia development is unknown. We hypothesized that periconceptional maternal folate intake influences pediatric ALL risk by modifying DNA methylation, and that this effect varies by food versus supplemental sources. We assessed periconceptional folate intake through a retrospective maternal dietary survey collected by the California Childhood Leukemia Study (CCLS) in association with DNA methylation at birth in pediatric ALL cases, prior to diagnosis, and matched healthy controls through an epigenome-wide association study (EWAS).
Methods
Study population
The CCLS is a population-based, case-control study that enrolled subjects with newly diagnosed pediatric (0–14 y) leukemia at California hospitals from 1995 to 2008 (17). Controls were randomly selected from birth certificates from the California Department of Public Health Office of Vital Records at a 1:1 or 2:1 ratio matched by birth date (i.e., matched to date of diagnosis for cases), sex, maternal race, and Hispanic ethnicity. A subset of pediatric ALL cases (B-cell, T-cell, or unknown lineage) and matched controls were assigned for whole-genome DNA methylation analysis using the Illumina Infinium Methylation EPIC BeadChip platform (Illumina) (14, 18). Participant dried blood spots (DBSs) obtained from peripheral blood heel-stick samples at birth were obtained from archived samples maintained by the California Department of Public Health. DNA was isolated from DBS samples, as described elsewhere (14), prior to being sent for methylation analysis. Written informed consent was obtained from mothers of participants, with assent from subjects aged 7 y or older. The institutional review boards of participating institutions and the California Committee for the Protection of Human Subjects reviewed and approved the study protocol.
Periconceptional folate estimation
Retrospective maternal dietary history spanning the year prior to pregnancy was obtained through a modified Block food-frequency questionnaire (mBFFQ) administered at diagnosis for cases or at the time of enrollment for controls (19). Specifications of the mBFFQ are reviewed elsewhere (4, 5). The mBFFQ surveyed frequency of intake of common foods and supplements, including 7 Hispanic-specific foods for Spanish-speaking patients. Folate intake was quantified as dietary folate equivalents (DFEs) and accounted for folic acid fortification in grain products in 1998. DFE measurements were categorized as total folate (from all sources), food folate (including natural and fortified food sources), and supplemental folic acid. Supplemental DFEs were based on exact labeled content of folic acid in multivitamin products and dietary supplements assessed on the mBFFQ. A total of 411 subjects were evaluated using the mBFFQ, including 198 ALL cases and 213 controls. One subject with incomplete folate intake estimation data was omitted.
DNA methylation analysis
Extracted DNA samples were modified by sodium bisulfite for conversion of unmethylated cytosines to uracil using the EZ DNA Methylation Kit (Zymo Research) for use in genome-wide methylation assessment. Samples were subsequently sent for methylation assessment using the Illumina Infinium Methylation EPIC BeadChip platform (Illumina) (14, 18). Raw IDAT (intensity data file)files were imported into R (Version 4.0.0, The R Foundation) for data preprocessing and normalization using the openSesame pipeline from the SeSAMe package (20) in minfi (21), with the distribution of signal background calibrated by type I probe out-of-band signal. Probes with a detection P-value threshold > 0.05 were masked from further analysis. NOOB background subtraction was performed, followed by removal of residual background. Nonlinear scaling was used to correct for dye balance. Probes and subjects with >5% missing values were removed. Probes previously identified as cross-hybridizing or overlapping genetic variants were additionally omitted (22). A total of 16 subjects were removed due to excessive missingness. Missing values were imputed using the impute.knn function in the R package impute. Chromosome X and Y probes were included in analysis. A total of 697,560 probes passed quality-control measures for inclusion in the study.
Statistical modeling
B-Values ranging from 0 (unmethylated) to 1 (fully methylated) were logit-transformed to M-values to normalize probe distribution for statistical modeling. Multivariable linear regression was conducted independently for the 3 folate variables (total, food, and supplemental). Supplemental DFEs were treated categorically (0,194, 360, and 540 DFEs), whereas the remainder were treated as continuous variables. The model controlled for array batch effect (plate), ALL case status, sex, the top 10 principal components (PCs) for nucleated cell proportions and top 10 PCs for genetic ancestry. Reference-Free Adjustment for Cell Type Composition (ReFACTor) was used to estimate nucleated cell proportions (23), whereas genetic ancestry was estimated using the EPISTRUCTURE (24) approach in GLINT (25). No significant variation in nucleated cell proportions (B cell, CD4 and CD8 T cell, granulocytes, monocytes, NK cells, and nucleated RBCs) was identified between ALL cases and controls using results of a separate reference-based deconvolution method (26–28). Differentially methylated probes (DMPs) were defined by false discovery rate (FDR) <0.05 based on the Benjamini and Hochberg method to correct for multiple comparisons. Differentially methylated regions (DMRs) were identified through Comb-p (29) using a seed P value < 0.05, maximum probe distance of 1000 base pairs, a minimum of 2 probes per region, and a significance threshold of Šidák-corrected P value < 0.05. All probes were annotated using the Bioconductor package IlluminaHumanMethylationEPICanno.ilm10b4.hg19 (30).
Subgroup analysis
Given prior evidence of variation in the association between folate consumption and ALL risk by ethnicity and socioeconomic factors (5, 9, 31), we conducted a subgroup analysis stratified by Hispanic compared with non-Hispanic White ethnicity, annual household income, the highest level of household education, and overall folate consumption. Income stratifications were defined as low (<$75,000) compared with high (≥$75,000) annual income. Household education stratifications were defined as lower educational level (high school or less) compared with higher educational level (some college or more). Median total folate intake for all participants was used to define low (<530.6 total DFEs) compared with high (≥530.6 total DFEs) overall folate consumption.
Interaction term analysis
To identify DNA methylation sites with variable response to periconceptional folate intake by ALL case status, an interaction term (ALL case status multiplied by folate DFEs) was incorporated in the linear regression model as the main covariate of interest while controlling sex, batch effect (plate), ALL case status, folate DFEs by source, the top 10 EPISTRUCTURE PCs, and top 10 ReFACTor PCs. After evaluating B-value distributions of ALL cases and controls, extreme outliers were adjusted through winsorization. Specifically, for cg08267698, cg00277198, cg04656615, and cg19839822, the 2 most extreme outliers among ALL cases were adjusted to the remaining most extreme value; for cg14946192 and cg11943916, the most extreme outlier was adjusted; and for cg14541523, the 3 most extreme outliers were adjusted. Interaction term CpGs with FDR <0.05 were subsequently assessed separately in ALL-case–stratified populations using winsorized data to confirm an opposing direction of effect in cases compared with controls.
Validation testing
To validate findings and test consistency with prior reports of maternal folate and DNA methylation at birth, results from the probe-specific EWAS were compared with folate-responsive CpGs previously published (13). The prior study evaluated 2 separate mother–offspring linked cohorts, the Norwegian Mother, Father, and Child Cohort Study (MoBa) and the Generation R Study (GenR), as well as a meta-analysis of the 2 cohorts revealing a total of 443 folate-responsive CpGs. A total of 359 CpGs overlapped with sites investigated in the current study. Spearman correlation coefficients were calculated between EWAS coefficients for total folate intake from this study and coefficients from the previously published meta-analysis, as well as the MoBa and GenR cohorts independently.
Data availability
The data underlying this article are available in the article and within Supplemental Tables 1–15. Results of the EWAS analysis presented in this study are provided as a downloadable file for all subjects, Hispanic subjects only, and non-Hispanic White subjects (32). This study used biospecimens from the California Biobank Program, which prohibits uploading of genomic data (including genome-wide DNA methylation) and/or sharing of individual-level data obtained from these biospecimens under the statutory scheme of the California Health and Safety Code sections 124980(j), 124991(b) and (h), and 103850(a) and (d).
Results
Cohort description and folate distribution
A total of 394 subjects were evaluated, including 189 ALL cases and 205 controls (Supplemental Figure 1). Cases and controls did not differ by sex, birth weight, race, Hispanic ethnicity, income, or highest household educational level (Table 1). Age at diagnosis ranged from 0.2 to 15.0 y for ALL cases (median: 4.2; IQR: 3.9), whereas age at enrollment for controls ranged from 0.3 to 14.7 y (median: 4.9; IQR: 5.3; P = 0.071). Median total folate intake was 530.6 DFEs. Median dietary intake was 419.0 DFEs from all food sources. For supplemental folic acid, 280 subjects reported no intake, 27 reported 194 DFEs, 14 reported 360 DFEs, and 73 reported 540 DFEs (Table 1, Supplemental Figure 2A). In this CCLS subset, cases and controls did not differ in folate intake for any source (Table 1, Supplemental Figure 2B). Of the 394 subjects included in the study, 201 (51.0%) had total DFEs equal to or greater than 520, the Estimated Average Requirement during pregnancy in the United States. Of the 189 cases, 99 (52.4%) had total DFEs equal to or greater than 520. Of the 205 controls, 102 (49.8%) had total DFEs equal to or greater than 520.
TABLE 1.
Subject characteristics1
| All subjects (n = 394) | Cases (n = 189) | Controls (n = 205) | P | |
|---|---|---|---|---|
| Female sex, n (%) | 174 (44.2%) | 86 (45.5%) | 88 (43.1%) | 0.612 |
| Median (IQR) gestational age, wk (IQR) | 40 (2) | 40 (3) | 40 (1) | 0.043 |
| Mean (SD) birth weight, g | 3411.1 (563.2) | 3361.7 (549.7) | 3456.6 (572.9) | 0.094 |
| Race, n (%) | 0.415 | |||
| White/Caucasian | 216 (54.8%) | 96 (50.8%) | 120 (58.5%) | |
| Black/African American | 9 (2.3%) | 4 (2.1%) | 5 (2.4%) | |
| Native American | 6 (1.5%) | 4 (2.1%) | 2 (1.0%) | |
| Asian or Pacific Islander | 26 (6.6%) | 11 (5.8%) | 15 (7.3%) | |
| Mixed or others | 135 (34.3%) | 72 (38.1%) | 63 (30.7%) | |
| Ethnicity, n (%) | 0.962 | |||
| Hispanic | 175 (44.4%) | 83 (43.9%) | 92 (44.9%) | |
| Non-Hispanic | 219 (55.6%) | 106 (56.1%) | 113 (55.1%) | |
| Annual income, n (%) | 0.442 | |||
| <$15,000 | 46 (11.7%) | 24 (12.7%) | 22 (10.7%) | |
| $15,000–29,999 | 68 (17.3%) | 39 (20.6%) | 29 (14.1%) | |
| $30,000–44,999 | 45 (11.4%) | 22 (11.6%) | 25 (12.2%) | |
| $45,000–59,999 | 54 (13.7%) | 27 (14.3%) | 27 (13.2%) | |
| $60,000–74,999 | 42 (10.7%) | 18 (9.5%) | 23 (11.2%) | |
| ≥$75,000 | 137 (34.8%) | 58 (30.7%) | 79 (38.5%) | |
| Household education, n (%) | 0.212 | |||
| None or elementary school | 15 (3.8%) | 9 (4.8%) | 6 (2.9%) | |
| High school or similar | 117 (29.7%) | 57 (30.2%) | 60 (29.3%) | |
| Some college or similar | 120 (30.5%) | 49 (25.9%) | 71 (34.6%) | |
| Bachelor's degree or higher | 142 (36.0%) | 74 (39.2%) | 68 (33.2%) | |
| Median (IQR) maternal folate DFEs | ||||
| Total | 530.6 (532.1) | 536.4 (533.9) | 519.8 (516.9) | 0.853 |
| Food | 419.0 (308.3) | 439.6 (316.1) | 410.0 (299.8) | 0.833 |
| Supplemental, n subjects per category | ||||
| 0 DFEs | 280 | 134 | 146 | 0.912 |
| 194 DFEs | 27 | 12 | 15 | |
| 360 DFEs | 14 | 8 | 6 | |
| 540 DFEs | 73 | 35 | 38 |
Characteristics of the n = 394 subjects assessed in this study from the California Childhood Leukemia Study (CCLS) with genome-wide DNA-methylation data available are shown. Characteristics of the n = 189 pediatric acute lymphoblastic leukemia cases and n = 205 matched controls included in this group are presented separately. No notable differences between case and control subjects were identified by sex, birth weight, reported race and ethnicity, annual income, or highest level of household education. Maternal periconceptional folate intake was calculated as DFEs and assessed by total intake, intake from food sources, and intake from supplemental sources. Total and food DFEs were assessed on a continuous scale, whereas supplemental folic acid was assessed categorically. DFE, dietary folate equivalent.
Chi-square test,
Wilcoxon test,
Welch's t-test,
Fisher's exact test.
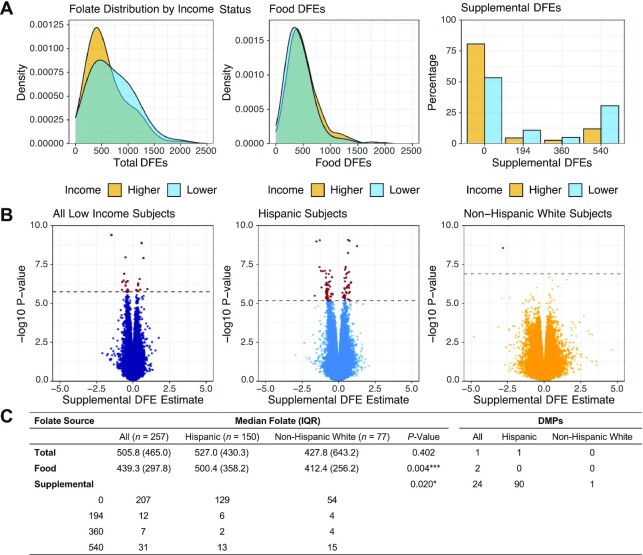
Subjects were further stratified by self-reported Hispanic ethnicity, annual income, household education, and total folate intake (Supplemental Figure 2C–F). Hispanic subjects reported higher folate intake from food sources, whereas non-Hispanic White subjects reported higher supplemental folic acid intake (Supplemental Figure 2C). Subjects with low annual income reported lower total folate intake compared with those with high income due to reduced supplemental folic acid intake, despite reporting higher intake from food sources (Supplemental Figure 2D), a trend similarly reflected in subjects with low household education (Supplemental Figure 2E).
Probe-specific analysis by folate source
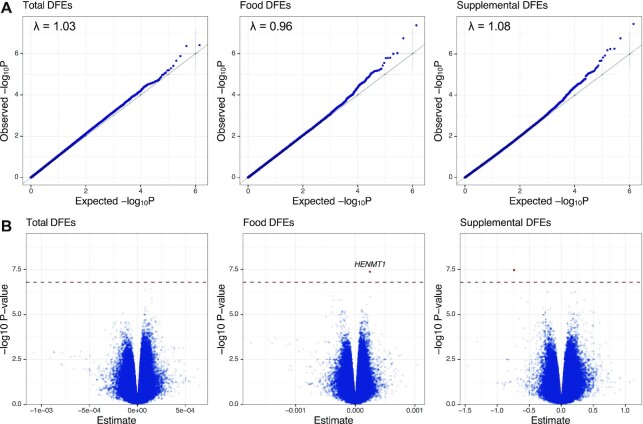
A total of 697,560 EPIC array CpG probes passed quality control measures for use in multivariable linear regression. Genomic inflation (λ) values by source were λ = 1.08 for total, 1.03 for food, and 0.96 for supplemental DFEs (Figure 1A). No significant DMPs were identified for total DFEs, 1 was identified for food DFEs, and 1 for supplemental DFEs (Supplemental Table 1, Figure 1B). Full EWAS results are available as a downloadable file (32). A DMP at HENMT1(HEN methyltransferase 1) (cg16977637) on chromosome 1 had a positive association (regression coefficient: 2.45 × 10−04; FDR = 0.030) with food DFEs, whereas an intergenic probe (cg17582160) was inversely associated with supplemental DFEs (coefficient: –0.724; FDR = 0.032). The top 10 probe-specific results for each folate variable are shown in Supplemental Table 1. Gene set enrichment analysis for the top 1000 probes was unremarkable for all folate sources. Correlation between regression coefficients was evaluated to characterize consistency of effect between folate sources (Supplemental Figure 3). The strength of correlation was strongest between total folate and food DFEs (Spearman's R = 0.75, P < 2.2 × 10−16). Correlation remained significant, although weaker, between total and supplemental DFEs (R = 0.16, P < 2.2 × 10−16) and food and supplemental DFEs (R = 0.05, P < 2.2 × 10−16).
FIGURE 1.
CpG-specific epigenome-wide association study results by maternal periconceptional folate source. A total 697,560 CpGs from the EPIC DNA-methylation array (Illumina) were included in multivariable linear regression analysis. (A) Quantile-quantile (QQ) plots demonstrating distribution of observed versus expected –log(10) raw P values for total, food, and supplemental periconceptional DFEs. Lambda values (λ) on plots represent estimation of genomic inflation, which was minimal across the 3 folate sources. (B) Volcano plot demonstrating –log(10) raw P value by regression coefficient. One significant DMP was identified in association with food folate and a second in association with supplemental folic acid intake. The hatched line represents FDR <0.05. Significant DMPs are positioned above the hatched line and are bolded, with associated gene names presented on the plot; unnamed DMPs are intergenic. DFE, dietary folate equivalent; DMP, differentially methylated probe; FDR, false discovery rate.
Regional analysis by folate source
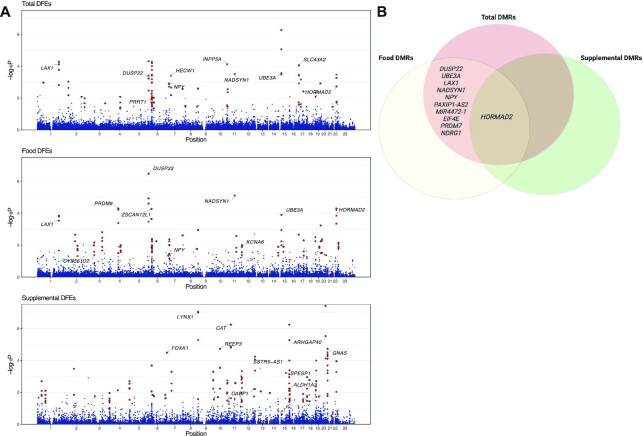
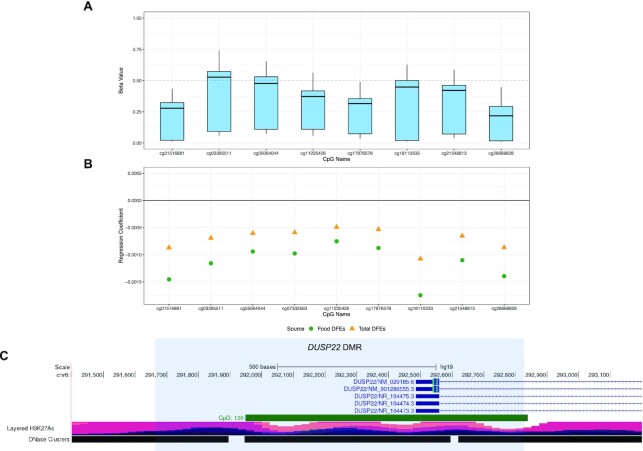
DMRs were analyzed using Comb-p (29). A total of 32 DMRs were identified in association with total folate DFEs [18 (56.3%) predominantly hypermethylated and 14 (43.8%) hypomethylated in response to folate intake], 35 were associated with food DFEs [14 (40.0%) hypermethylated, 21 (60.0%) hypomethylated], and 48 were associated with supplemental DFEs [41 (85.4%) hypermethylated, 7 (14.6%) hypomethylated] (Figure 2, Table 2, Supplemental Tables 3 and 4). Regional gene set enrichment analysis was unremarkable. A region spanning 1137 bp on chromosome 6 within the promoter region of DUSP22(dual specificity phosphatase 22) was the top DMR for total and food DFEs, although it was not associated with supplemental DFEs (Figure 3). Regression coefficients for the 2 folate sources were negative for all 9 probes in the DUSP22 region, indicating an inverse relation with DNA methylation (Supplemental Figure 4A). Probes in the region showed P < 0.05 only in ALL cases and not controls when analyzed independently (Supplemental Figure 4B).
FIGURE 2.
Differentially methylated regions by folate source. DMRs by folate source were identified based on spatial correlation of P values using Comb-p Šidák P values, which are corrected for multiple comparisons across regions according to the Comb-p algorithm. A Šidák P-value threshold <0.05 was used to define significance. (A) Manhattan plots shown demonstrate –log(10) region P values from Comb-p analysis by chromosomal position. Probes within DMRs are bolded, with labels for the top 10 DMRs by P value. A total of 32 DMRs were identified in association with total folate intake, 35 associated with food folate intake, and 48 with supplemental folic acid. (B) Venn diagrams of probes overlapping between folate sources demonstrate a single DMR (at HORMAD2,HORMA domain containing 2) common to all sources, along with 10 additional DMRs overlapping between total and food sources. No additional DMRs overlapped between total and supplemental sources or food and supplemental sources. Panel B was created with BioRender.com. DFE, dietary folate equivalent; DMR, differentially methylated region.
TABLE 2.
Top differentially methylated regions by folate source1
| Folate source and chromosome | Position (Hg19) | Width, bp | No. of probes | Gene association | Šidák P | DNA methylation response to folate | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| Total | |||||||
| 6 | 291687 | 292823 | 1137 | 9 | DUSP22 | 8.18 × 10–13 | Negative |
| 10 | 134331790 | 134332442 | 653 | 6 | INPP5A | 9.76 × 10–10 | Positive |
| 15 | 25684578 | 25684849 | 272 | 5 | UBE3A | 4.00 × 10–9 | Positive |
| 1 | 203734167 | 203734559 | 393 | 7 | LAX1 | 3.49 × 10–06 | Negative |
| 17 | 1508261 | 1508723 | 463 | 8 | SLC43A2 | 4.52 × 10–06 | Positive |
| 6 | 32120773 | 32121368 | 596 | 20 | PRRT1 | 9.85 × 10–06 | Negative |
| 22 | 30476089 | 30476525 | 437 | 11 | HORMAD2 | 6.18 × 10–05 | Negative |
| 11 | 71210210 | 71210295 | 86 | 2 | NADSYN1 | 9.61 × 10–05 | Positive |
| 7 | 43288635 | 43288940 | 306 | 5 | HECW1 | 1.34 × 10–04 | Positive |
| 7 | 24323559 | 24323939 | 381 | 8 | NPY | 2.28 × 10–04 | Positive |
| Food | |||||||
| 6 | 291687 | 292823 | 1137 | 9 | DUSP22 | 1.25 × 10–16 | Negative |
| 7 | 24323559 | 24323939 | 381 | 8 | NPY | 1.30 × 10–11 | Positive |
| 4 | 81118343 | 81119473 | 1131 | 10 | PRDM8 | 6.96 × 10–10 | Positive |
| 12 | 4918075 | 4919230 | 1156 | 10 | KCNA6 | 4.68 × 10–07 | Positive |
| 11 | 71210210 | 71210295 | 86 | 2 | NADSYN1 | 1.08 × 10–06 | Negative |
| 22 | 30476089 | 30476525 | 437 | 11 | HORMAD2 | 3.97 × 10–06 | Negative |
| 6 | 28058724 | 28059208 | 485 | 9 | ZSCAN12P1 | 4.53 × 10–06 | Negative |
| 1 | 203734167 | 203734559 | 393 | 7 | LAX1 | 1.68 × 10–05 | Negative |
| 15 | 25684578 | 25684849 | 272 | 5 | UBE3A | 1.72 × 10–05 | Positive |
| 3 | 50388823 | 50388924 | 102 | 5 | CYB561D2 | 6.14 × 10–05 | Positive |
| Supplemental | |||||||
| 15 | 69222592 | 69223018 | 427 | 6 | SPESP1 | 3.05 × 10–14 | Positive |
| 8 | 143859669 | 143860090 | 422 | 9 | LYNX1 | 1.06 × 10–10 | Positive |
| 20 | 37230326 | 37230741 | 416 | 6 | ARHGAP40 | 1.64 × 10–09 | Positive |
| 11 | 34460107 | 34460789 | 683 | 12 | CAT | 1.14 × 10–08 | Positive |
| 7 | 4762236 | 4762371 | 136 | 2 | FOXK1 | 3.20 × 10–07 | Negative |
| 16 | 1121693 | 1122047 | 355 | 5 | SSTR5-AS1 | 5.77 × 10–07 | Positive |
| 10 | 65733092 | 65733575 | 484 | 5 | REEP3 | 1.92 × 10–06 | Positive |
| 15 | 101389272 | 101390023 | 752 | 10 | ALDH1A3 | 2.66 × 10–06 | Positive |
| 20 | 57427412 | 57427977 | 566 | 18 | GNAS | 3.30 × 10–06 | Positive |
| 12 | 121087689 | 121088408 | 720 | 3 | CABP1 | 6.57 × 10–06 | Positive |
The top 10 DMRs by folate source identified based on spatial correlation of P values using Comb-p. Šidák P values are corrected for multiple comparisons across regions according to the Comb-P algorithm. The top region associated with total and food folate intake located on chromosome 22 and associated with the promoter region of DUSP22. A total of 32 DMRs were identified in association with total, 35 associated with food, and 48 with supplemental folic acid intake. ALDH1A3, aldehyde dehydrogenase 1 family member A3; ARHGAP40, Rho GTPase activating protein 40; CABP1, calcium binding protein 1; CAT, catalase; CYB561D2, cytochrome b561 family member D2; DMR, differentially methylated region; FOXK1, forkhead box K1; GNAS, GNAS complex locus; Hg19, human genome reference assembly (GRCh37); HECW1, HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1; HORMAD2, HORMA domain containing 2; INPP5A, inositol polyphosphate-5-phosphatase A; KCNA6, potassium voltage-gated channel subfamily A member 6; LAX1, lymphocyte transmembrane adaptor 1; LYNX1, Ly6/neurotoxin 1; NADSYN1, NAD synthetase 1; NPY, neuropeptide Y; PRDM8, PR/SET domain 8; PRRT1, proline rich transmembrane protein 1; REEP3, receptor accessory protein 3; SLC43A2, solute carrier family 43 member 2; SPESP1, sperm equatorial segment protein 1; SSTR5-AS1, SSTR5 antisense RNA 1; UBE3A, ubiquitin protein ligase E3A; ZSCAN12P1, zinc finger and SCAN domain containing 12 pseudogene 1.
FIGURE 3.
Regional description of the top DMR at DUSP22(dual specificity phosphatase 22). (A) Box plot diagram of DNA methylation B-values for all subjects (n = 394) for the 9 CpG probes located within the 1137-bp region on chromosome 6 associated with the promoter region of DUSP22 identified in Comb-p in association with maternal total and food folate intake. (B) Regression coefficients for the same 9 CpG probes within the region for maternal total and food folate intake. (C) Diagram of local region surrounding the DUSP22 DMR (shaded region) on chromosome 6 with annotations from the UCSC Genome Browser including CpG island regions and H3K27 acetylation sites. DMR, differentially methylated region; DUSP22,dual specificity phosphatase 22.
Subgroup-stratified EWAS analysis
Regression analysis was repeated in subgroups stratified by Hispanic ethnicity, household education, annual income, and total folate intake (Supplemental Table 2 and Supplemental Tables 5–9; Supplemental Figure 2B–F). Two DMPs were identified among Hispanic subjects (n = 175, n exposed to supplemental folic acid = 33), both associated with supplemental DFEs. One DMP was identified in association with supplemental folic acid intake in non-Hispanic White subjects (n = 159, n exposed = 59) (Supplemental Table 9). Subjects with low annual income (n = 257, n exposed to supplemental folic acid = 50) had 1 DMP for total, 2 for food, and 21 for supplemental DFEs, whereas high-income subjects (n = 137, n exposed = 64) had no significant DMPs for any folate category. Subjects with a lower educational level (n = 132, n exposed to supplemental folic acid = 15) had 241 significant DMPs for supplemental DFEs and no significant DMPs for total and food DFEs, whereas those with a higher education (n = 262, n exposed = 99) had 22 significant DMPs for food DFEs but none for total and supplemental DFEs. Subjects with low total folate intake (n = 197, n exposed to supplemental folic acid = 10) had 3 DMPs associated with supplemental DFEs and none for total and food DFEs, whereas high-total-folate subjects (n = 197, n exposed = 104) had no significant DMPs for any folate category.
To assess ethnicity-related differences within these subgroups, we next stratified Hispanic and non-Hispanic subjects by annual income, household education, and total folate intake separately for analysis. Hispanic subjects with low household education (n = 102, n exposed to supplemental folic acid = 12), low annual income (n = 150, n exposed = 21), and low total folate intake (n = 83, n exposed = 2) demonstrated 215, 90, and 387 DMPs associated with supplemental DFEs, respectively (Figure 4, Supplemental Figure 5A–C). Hispanic subjects with low household education additionally had 1 DMP associated with total DFEs and 2 associated with food DFEs. In Hispanic subjects with high household education (n = 73) and high total folate intake (n = 92) there were no significant DMPs for any folate category, whereas those with high income levels (n = 25) could not be assessed independently due to the small sample size. The sample size for non-Hispanic White subjects with a low educational level (n = 23) precluded regression analysis. Non-Hispanic White subjects with low income (n = 77, n exposed to supplemental folic acid = 23) had 1 significant DMP associated with supplemental DFEs (Supplemental Figure 6A–C). Non-Hispanic White subjects with lower total folate intake (n = 84) had 2 significant DMPs for food DFEs and 14 DMPs for supplemental DFEs (n exposed = 5). Non-Hispanic White subjects with high household education (n = 136) had 1 DMP associated with food DFEs, whereas those with high income (n = 82) and high total folate intake (n = 75) had no significant findings. Subgroup-specific DMRs are provided in Supplemental Table 3 and Supplemental Tables 10–14.
FIGURE 4.
Hispanic subjects within the low-income stratification show a stronger association with supplemental folic acid intake compared with non-Hispanic White subjects. (A) Folate distribution by source within lower-income subjects (annual income <$75,000/y) stratified by Hispanic (n = 150) and non-Hispanic White (n = 77) subjects. Hispanic subjects were found to take in higher concentrations of folate from food sources, whereas non-Hispanic White subjects take in higher concentrations of folate from supplemental sources. (B) Regression results were associated with supplemental folic acid intake in all lower-income subjects (n = 257; left panel) compared with regression results further stratified by Hispanic subjects only (middle panel) and non-Hispanic White subjects (right panel). The hatched line represents FDR <0.05. Bolded points positioned above the hatched line represent significant DMPs. (C) Table demonstrating median folate values in lower-income subjects further stratified by Hispanic ethnicity. DFE, dietary folate equivalent; DMP, differentially methylated probe; FDR, false discovery rate.
Comparison to prior maternal folate-responsive DNA methylation sites
To evaluate consistency with prior studies, we compared the coefficient direction of effect for 359 CpG sites that overlapped with 443 folate-responsive sites (13) identified in 2 European cohorts: MoBa and GenR (Supplemental Table 15). Total folate intake coefficients were associated with published meta-analysis results from the 2 cohorts (Spearman's R = 0.15, P = 0.003; Supplemental Figure 7A–C). Independently, our results showed a stronger correlation with the GenR cohort (R = 0.25, P = 1.10 × 10−06) than the MoBA cohort (R = 0.065, P = 0.220).
Leukemia-specific associations
A total of 8 CpGs were found to have an interaction between ALL case status and food folate intake (FDR = 0.003–0.026; Table 3). No significant interactions were identified for total or supplemental DFEs. Significant probes were subsequently analyzed in subgroups stratified by ALL case status (Supplemental Figure 8) following winsorization of extreme outliers (Supplemental Figure 9). Coefficients for all 8 CpGs were opposed in direction of effect between ALL cases and controls. P values ranged from 3.03 × 10−05to 0.014 in cases, whereas just 1 CpG (cg24425686, associated with CERK, ceramide kinase; P = 0.037) had P < 0.05 in controls (remaining control P values ranged from 0.162 to 0.844).
TABLE 3.
Differences in DNA methylation responsiveness to maternal periconceptional food folate by pediatric leukemia case status1
| Interaction Term | Cases (n = 189) | Controls (n = 205) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CpG | Chr | Position (Hg19) | Gene | Coef. | SE | FDR | Coef. | P | Coef. | P |
| Food | ||||||||||
| cg24425686 | 22 | 47100062 | CERK | −5.44 × 10−04 | 9.13 × 10−05 | 0.003 | −1.65 × 10−04 | 0.002 | 9.97 × 10−05 | 0.037 |
| cg08267698 | 6 | 33386073 | CUTA | 4.66 × 10−04 | 7.99 × 10−05 | 0.003 | 2.02 × 10−04 | 1.19 × 10−04 | −5.23 × 10−05 | 0.162 |
| cg00277198 | 10 | 49973686 | WDFY4 | −5.04 × 10−04 | 8.70 × 10−05 | 0.003 | −1.66 × 10−04 | 2.61 × 10−04 | 6.04 × 10−05 | 0.112 |
| cg14946192 | 11 | 65729164 | SART1 | 3.00 × 10−04 | 5.27 × 10−05 | 0.005 | 7.82 × 10−05 | 0.002 | −6.07 × 10−06 | 0.844 |
| cg04656615 | 20 | 43711547 | — | −4.77 × 10−04 | 8.47 × 10−05 | 0.005 | −1.36 × 10−04 | 0.003 | 3.51 × 10−05 | 0.354 |
| cg19839822 | 22 | 42579537 | TCF20 | −6.26 × 10−04 | 1.12 × 10−04 | 0.005 | −1.48 × 10−04 | 3.92 × 10−04 | 2.37 × 10−05 | 0.523 |
| cg11943916 | 5 | 172282611 | ERGIC1 | −5.19 × 10−04 | 9.81 × 10−05 | 0.021 | −1.37 × 10−04 | 0.014 | 3.65 × 10−05 | 0.442 |
| cg14541523 | 12 | 132089933 | — | −6.29 × 10−04 | 1.21 × 10−04 | 0.026 | −3.12 × 10−04 | 3.03 × 10−05 | 2.40 × 10−05 | 0.767 |
To identify CpG sites with variation in effect by leukemia case status, an interaction term incorporating source-specific folate DFEs and leukemia case status was analyzed in the linear regression model. Interaction term CpGs with FDR <0.05 were then assessed using multivariable linear regression in leukemia case- and control-stratified subgroups to assess variation in the effect of maternal periconceptional folate on DNA methylation. A total of 8 interaction CpGs with FDR <0.05 were identified in association with maternal food folate intake. Following winsorization of extreme outliers, case-stratified regression analysis at these 8 probes demonstrated opposing direction of effect in ALL cases and controls. All probes were significant (P < 0.05) in ALL cases, whereas just one (cg24425686) was significant in controls. ALL, acute lymphoblastic leukemia; CERK, ceramide kinase; Chr, chromosome; Coef., coefficient; CUTA, cutA divalent cation tolerance homolog; ERGIC1, endoplasmic reticulum-golgi intermediate compartment 1; DFE, dietary folate equivalent; FDR, false discovery rate; Hg19, human genome build version 19; SART1, spliceosome associated factor 1;TCF20, transcription factor 20; WDFY4, WDFY family member 4.
Discussion
These results identify site-specific and regional associations between periconceptional folate intake and DNA methylation in offspring varying by dietary source. We identified DMPs for food and supplemental sources, but none when assessing these sources as combined total folate intake. Although 11 DMRs overlapped between total and food folate sources, just 1 supplemental DMR overlapped with the other folate sources. This divergence in results between total, food, and supplemental sources implies a distinct relation for each folate source in contributing to the early establishment of DNA methylation.
We additionally show that the relation between maternal dietary folate intake and DNA methylation at birth varies by Hispanic ethnicity, household education, annual income, and the overall amount of folate consumed. For individuals with lower educational and income levels, the strongest relation with DNA methylation variation resulted from supplemental folic acid intake. This finding was likely driven by Hispanic subjects, who showed substantially greater association with supplemental folic acid intake compared with non-Hispanic White subjects in the low-education and low-income subgroups. This finding agrees with prior studies that identified ALL risk reduction in Hispanic patients with the highest concentrations of maternal supplemental folic acid (5) and those with mothers of low education (9), indicating a potential additional benefit to targeted folate supplementation in these groups. All subjects in the low-overall-folate subgroup failed to meet US national guidelines for folate intake during pregnancy (>600 DFEs). Although the identified association with supplemental folic acid intake remained consistent across Hispanic compared with non-Hispanic White subgroups, the low number of subjects exposed to supplemental folic acid within these stratifications may have contributed to the large number of significant findings.
We identified a DMR linked to total and food periconceptional folate intake at the promoter region of DUSP22, a protein phosphatase and known tumor suppressor involved in mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription 3 (STAT3) regulation (33). DUSP22 rearrangements constitute 30% of ALK (anaplastic lymphoma kinase)-negative anaplastic large-cell lymphoma (34) and are found dysregulated in chronic lymphocytic leukemia (35). The gene body region of DUSP22 has additionally been identified in multiple studies to be differentially methylated in pediatric ALL at diagnosis (16, 36), and has been shown to be hypermethylated in response to maternal folate depletion in mouse models (15). This study is the first to confirm a similar direction of effect in response to maternal folate in humans. Although the direction of effect was the same between ALL cases and controls in this study, the strength of association was greater in ALL cases.
We identified unique relations between periconceptional folate and DNA methylation by ALL case status, including DMPs associated with CERK(ceramide kinase), CUTA(cutA divalent cation tolerance homolog), WDFY4 (WDFY family member 4), SART1 (spliceosome associated factor 1), TCF20 (transcription factor 20), and ERGIC1 (endoplasmic reticulum-golgi intermediate compartment 1). Each of these genes are previously identified as differentially methylated at diagnosis in B-cell precursor ALL (36). All sites demonstrated an opposing direction of effect in ALL cases and controls. We did not directly test for an underlying mechanism to explain variation in folate responsiveness at these sites, including mediation by genetic variation. However, evidence of a variable response in ALL cases lends support for folate to act as a driver of early changes in DNA methylation, which impact future pediatric ALL development.
The strength of this study is the large sampling of subjects with both perinatal DNA methylation data, prior to onset of pediatric ALL, and detailed periconceptional dietary folate calculations for mothers. We also show that our results are correlated with a prior study investigating maternal folate-responsive DNA methylation sites in offspring (13). This correlation was stronger for the GenR cohort than the MoBa cohort when assessed independently. We did not identify a clear explanation for this difference, as both cohorts are based on European populations (Norway and the Netherlands) and did not differ obviously in subject characteristics (13). In part, this difference may be attributable to variation in the timing and method of folate measurement, as the GenR cohort evaluated maternal blood folate concentrations at 12 wk of gestational age and the MoBa cohort measured this concentration at 16 wk. This study, in contrast, evaluated DFEs calculated through retrospectively obtained maternal dietary history, creating a potential for inaccuracies in data acquisition and recall bias, in particular given that no formal assessment of the reliability of the mBFFQ exists (31). However, although knowledge of a general benefit for folate intake in pregnancy is prevalent, its specific association with ALL risk is less widely known, which may serve to limit variation in recall between ALL cases and controls included in this study (37). Dietary folate calculations may also fail to account for variation in folate bioavailability resulting from potential differences in dietary absorption, drug interactions, and variants in genes involved in folate metabolism, which also are associated with risk of pediatric ALL (38–40). However, direct measurement of serum folate may vary temporally by recency of food intake and thus may not be fully reflective of maternal folate stores (41) available to the developing fetus. Finally, as ALL-defining chromosomal translocations have been identified at birth, we cannot fully rule out the influence of a subpopulation of pre-leukemic cells on the reported results of this study.
In summary, we identified a source-specific relation between dietary periconceptional folate and DNA methylation at birth. We additionally identified a particular impact for supplemental folic acid in Hispanic subjects, specifically those with lower educational and lower income levels. Further investigations into the relation of maternal folate on DNA methylation at birth should account for these varying folate sources. We also identified DNA methylation variation by ALL case status, including a notable region of interest at DUSP22. Further investigation into a potential functional role for this early molecular alteration in ALL predisposition in children is warranted.
Supplementary Material
Acknowledgements
The authors thank Hong Quach and Diana Quach for support on DNA isolation and execution of the Illumina arrays. They thank Robin Cooley and Steve Graham of the California Department of Public Health for advice and logistical support. For recruitment of subjects enrolled in the CCLS replication set, the authors gratefully acknowledge the clinical investigators at the following collaborating hospitals: University of California Davis Medical Center (Dr. Jonathan Ducore), University of California San Francisco (Drs. Mignon Loh and Katherine Matthay), Children's Hospital of Central California (Dr. Vonda Crouse), Lucile Packard Children's Hospital (Dr. Gary Dahl), Children's Hospital Oakland (Dr. James Feusner), Kaiser Permanente Roseville (formerly Sacramento) (Drs. Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs. Carolyn Russo, Alan Wong, and Denah Taggart), Kaiser Permanente San Francisco (Dr. Kenneth Leung), and Kaiser Permanente Oakland (Drs. Daniel Kronish and Stacy Month). The authors additionally thank the families for their participation in the California Childhood Leukemia Study (formerly known as the Northern California Childhood Leukemia Study).
The authors’ responsibilities were as follows—EMN, JLW, CM, and AJdS: designed the study; AYK, LM, CM, EMN, and SL: collected data and prepared data for analysis; EMN: analyzed the data and statistical approach and prepared the manuscript; AYK, LM, and CM: organized the provision of study samples; and all authors: read and approved the final manuscript. The authors report no conflicts of interest.
Notes
Supported by research grants from the National Institutes of Health [National Institute of Environmental Health Sciences (NIEHS); R01ES009137, P42ES004705, P01ES018172, P42ES0470518 and R24ES028524], the Environmental Protection Agency (EPA; RD83451101), United States of America, and the UK-based Children with Cancer Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIEHS and the EPA. The biospecimens and/or data used in this study were obtained from the California Biobank Program (CBP request #1531), section 6555(b), 17 CCR. The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication.
Supplemental Tables 1–15 and Supplemental Figures 1–9 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: ALL, acute lymphoblastic leukemia; CCLS, California Childhood Leukemia Study; CERK, ceramide kinase; DFE, dietary folate equivalent; DMP, differentially methylated probe; DMR, differentially methylated region; DUSP22,dual specificity phosphatase 22; EWAS, epigenome-wide association study; FDR, false discovery rate; GenR, Generation R Study; mBFFQ, modified Block food-frequency questionnaire; MoBa, Norwegian Mother, Father, and Child Cohort Study; PC, principal component; ReFACTor, Reference-Free Adjustment for Cell Type Composition.
Contributor Information
Eric M Nickels, Children's Hospital Los Angeles, Center for Blood Disease Institute, Los Angeles, CA, USA; University of Southern California Keck School of Medicine, Center for Genetic Epidemiology, Los Angeles, CA, USA.
Shaobo Li, University of Southern California Keck School of Medicine, Center for Genetic Epidemiology, Los Angeles, CA, USA.
Libby Morimoto, School of Public Health, University of California, Berkeley, Berkeley, CA, USA.
Alice Y Kang, School of Public Health, University of California, Berkeley, Berkeley, CA, USA.
Adam J de Smith, University of Southern California Keck School of Medicine, Center for Genetic Epidemiology, Los Angeles, CA, USA.
Catherine Metayer, School of Public Health, University of California, Berkeley, Berkeley, CA, USA.
Joseph L Wiemels, University of Southern California Keck School of Medicine, Center for Genetic Epidemiology, Los Angeles, CA, USA.
Data Availability
Data described in the manuscript, code book, and analytic code will be made available upon reasonable request pending approval from authors. The data underlying this article are available in the article and in its online supplementary material. This study used biospecimens from the California Biobank Program, which prohibits uploading of genomic data (including genome-wide DNA-methylation) and/or sharing of individual level data obtained from these biospecimens under the statutory scheme of the California Health and Safety Code Sections 124980(j), 124991(b), (h), and 103850(a) and (d). Results of the EWAS analysis presented in this study are provided as a downloadable file for all subjects, Hispanic subjects only, and non-Hispanic white subjects (32).
References
- 1. Whitehead TP, Metayer C, Wiemels JL, Singer AW, Miller MD. Childhood leukemia and primary prevention. Curr Probl Pediatr Adolesc Health Care. 2016;46(10):317–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera Get al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet North Am Ed. 1999;354(9189):1499–503. [DOI] [PubMed] [Google Scholar]
- 3. Jensen CD, Block G, Buffler P, Ma X, Selvin S, Month S. Maternal dietary risk factors in childhood acute lymphoblastic leukemia (United States). Cancer Causes Control. 2004;15(6):559–70. [DOI] [PubMed] [Google Scholar]
- 4. Kwan ML, Jensen CD, Block G, Hudes ML, Chu LW, Buffler PA. Maternal diet and risk of childhood acute lymphoblastic leukemia. Public Health Rep. 2009;124(4):503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singer AW, Selvin S, Block G, Golden C, Carmichael SL, Metayer C. Maternal prenatal intake of one-carbon metabolism nutrients and risk of childhood leukemia. Cancer Causes Control. 2016;27(7):929–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu JJ, Ward RL. Folate and one-carbon metabolism and its impact on aberrant DNA methylation in cancer. Adv Genet. 2010;71:79–121. [DOI] [PubMed] [Google Scholar]
- 7. Thompson JR, Gerald PF, Willoughby ML, Armstrong BK. Maternal folate supplementation in pregnancy and protection against acute lymphoblastic leukaemia in childhood: a case-control study. Lancet North Am Ed. 2001;358(9297):1935–40. [DOI] [PubMed] [Google Scholar]
- 8. Bailey HD, Miller M, Langridge A, de Klerk NH, van Bockxmeer FM, Attia Jet al. Maternal dietary intake of folate and vitamins B6 and B12 during pregnancy and the risk of childhood acute lymphoblastic leukemia. Nutr Cancer. 2012;64(7):1122–30. [DOI] [PubMed] [Google Scholar]
- 9. Metayer C, Milne E, Dockerty JD, Clavel J, Pombo-de-Oliveira MS, Wesseling Cet al. Maternal supplementation with folic acid and other vitamins and risk of leukemia in offspring: a Childhood Leukemia International Consortium study. Epidemiology. 2014;25(6):811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ajrouche R, Rudant J, Orsi L, Petit A, Baruchel A, Nelken Bet al. Maternal reproductive history, fertility treatments and folic acid supplementation in the risk of childhood acute leukemia: the ESTELLE study. Cancer Causes Control. 2014;25(10):1283–93. [DOI] [PubMed] [Google Scholar]
- 11. Chokkalingam AP, Chun DS, Noonan EJ, Pfeiffer CM, Zhang M, Month SRet al. Blood levels of folate at birth and risk of childhood leukemia. Cancer Epidemiol Biomarkers Prev. 2013;22(6):1088–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKay JA, Waltham KJ, Williams EA, Mathers JC. Folate depletion during pregnancy and lactation reduces genomic DNA methylation in murine adult offspring. Genes Nutr. 2011;6(2):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett Eet al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 2016;7(1):10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gonseth S, Roy R, Houseman EA, de Smith AJ, Zhou M, Lee STet al. Periconceptional folate consumption is associated with neonatal DNA methylation modifications in neural crest regulatory and cancer development genes. Epigenetics. 2015;10(12):1166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Potter C, Moorman AV, Relton CL, Ford D, Mathers JC, Strathdee Get al. Maternal red blood cell folate and infant vitamin B. Mol Nutr Food Res. 2018;62(22):1800411. [DOI] [PubMed] [Google Scholar]
- 16. Nordlund J, Bäcklin CL, Wahlberg P, Busche S, Berglund EC, Eloranta MLet al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14(9):r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma X, Buffler PA, Layefsky M, Does MB, Reynolds P. Control selection strategies in case-control studies of childhood diseases. Am J Epidemiol. 2004;159(10):915–21. [DOI] [PubMed] [Google Scholar]
- 18. Lee ST, Muench MO, Fomin ME, Xiao J, Zhou M, de Smith Aet al. Epigenetic remodeling in B-cell acute lymphoblastic leukemia occurs in two tracks and employs embryonic stem cell-like signatures. Nucleic Acids Res. 2015;43(5):2590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Block G, Hartman AM, Naughton D. A reduced dietary questionnaire: development and validation. Epidemiology. 1990;1(1):58–64. [DOI] [PubMed] [Google Scholar]
- 20. Zhou W, Triche TJ, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium beadchips in genomic deletions. Nucleic Acids Res. 2018;46(20):e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KDet al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy Pet al. Critical evaluation of the Illumina MethylationEPIC Beadchip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rahmani E, Zaitlen N, Baran Y, Eng C, Hu D, Galanter Jet al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods. 2016;13(5):443–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rahmani E, Shenhav L, Schweiger R, Yousefi P, Huen K, Eskenazi Bet al. Genome-wide methylation data mirror ancestry information. Epigenetics Chromatin. 2017;10(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahmani E, Yedidim R, Shenhav L, Schweiger R, Weissbrod O, Zaitlen Net al. GLINT: a user-friendly toolset for the analysis of high-throughput DNA-methylation array data. Bioinformatics. 2017;33(12):1870–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KTet al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC Beadarray. Genome Biol. 2018;19(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gervin K, Salas LA, Bakulski KM, van Zelm MC, Koestler DC, Wiencke JKet al. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenet. 2019;11(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koestler DC, Jones MJ, Usset J, Christensen BC, Butler RA, Kobor MSet al. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL). BMC Bioinf. 2016;17(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28(22):2986–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord Ret al. De novo identification of differentially methylated regions in the human genome. Epigenet Chromatin. 2015;8(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singer AW, Carmichael SL, Selvin S, Fu C, Block G, Metayer C. Maternal diet quality before pregnancy and risk of childhood leukaemia. Br J Nutr. 2016;116(8):1469–78. [DOI] [PubMed] [Google Scholar]
- 32. Nickels E. Periconceptional folate intake influences DNA methylation at birth based on dietary source in an analysis of pediatric acute lymphoblastic leukemia cases and controls: Datasets. Harvard Dataverse, V1, 2022. 10.7910/DVN/ADBAIS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sekine Y, Ikeda O, Hayakawa Y, Tsuji S, Imoto S, Aoki Net al. DUSP22/LMW-DSP2 regulates estrogen receptor-alpha-mediated signaling through dephosphorylation of Ser-118. Oncogene. 2007;26(41):6038–49. [DOI] [PubMed] [Google Scholar]
- 34. Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood. 2017;130(4):410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arruga F, Gizdic B, Bologna C, Cignetto S, Buonincontri R, Serra Set al. Mutations in NOTCH1 PEST domain orchestrate CCL19-driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. Leukemia. 2017;31(9):1882–93. [DOI] [PubMed] [Google Scholar]
- 36. Chaber R, Gurgul A, Wróbel G, Haus O, Tomoń A, Kowalczyk Jet al. Whole-genome DNA methylation characteristics in pediatric precursor B cell acute lymphoblastic leukemia (BCP ALL). PLoS One. 2017;12(11):e0187422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Infante-Rivard C, Jacques L. Empirical study of parental recall bias. Am J Epidemiol. 2000;152(5):480–6. [DOI] [PubMed] [Google Scholar]
- 38. Metayer C, Scélo G, Chokkalingam AP, Barcellos LF, Aldrich MC, Chang JSet al. Genetic variants in the folate pathway and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2011;22(9):1243–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wiemels JL, Smith RN, Taylor GM, Eden OB, Alexander FE, Greaves MFet al. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc Natl Acad Sci. 2001;98(7):4004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schraw JM, Yiu TT, Lupo PJ, Tsavachidis S, Rau R, Bondy MLet al. Maternal folate genes and aberrant DNA hypermethylation in pediatric acute lymphoblastic leukemia. PLoS One. 2018;13(5):e0197408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sobczyńska-Malefora A, Harrington DJ. Laboratory assessment of folate (vitamin B). J Clin Pathol. 2018;71(11):949–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and within Supplemental Tables 1–15. Results of the EWAS analysis presented in this study are provided as a downloadable file for all subjects, Hispanic subjects only, and non-Hispanic White subjects (32). This study used biospecimens from the California Biobank Program, which prohibits uploading of genomic data (including genome-wide DNA methylation) and/or sharing of individual-level data obtained from these biospecimens under the statutory scheme of the California Health and Safety Code sections 124980(j), 124991(b) and (h), and 103850(a) and (d).
Data described in the manuscript, code book, and analytic code will be made available upon reasonable request pending approval from authors. The data underlying this article are available in the article and in its online supplementary material. This study used biospecimens from the California Biobank Program, which prohibits uploading of genomic data (including genome-wide DNA-methylation) and/or sharing of individual level data obtained from these biospecimens under the statutory scheme of the California Health and Safety Code Sections 124980(j), 124991(b), (h), and 103850(a) and (d). Results of the EWAS analysis presented in this study are provided as a downloadable file for all subjects, Hispanic subjects only, and non-Hispanic white subjects (32).