ABSTRACT
Precision nutrition is an emerging concept that aims to develop nutrition recommendations tailored to different people's circumstances and biological characteristics. Responses to dietary change and the resulting health outcomes from consuming different diets may vary significantly between people based on interactions between their genetic backgrounds, physiology, microbiome, underlying health status, behaviors, social influences, and environmental exposures. On 11–12 January 2021, the National Institutes of Health convened a workshop entitled “Precision Nutrition: Research Gaps and Opportunities” to bring together experts to discuss the issues involved in better understanding and addressing precision nutrition. The workshop proceeded in 3 parts: part I covered many aspects of genetics and physiology that mediate the links between nutrient intake and health conditions such as cardiovascular disease, Alzheimer disease, and cancer; part II reviewed potential contributors to interindividual variability in dietary exposures and responses such as baseline nutritional status, circadian rhythm/sleep, environmental exposures, sensory properties of food, stress, inflammation, and the social determinants of health; part III presented the need for systems approaches, with new methods and technologies that can facilitate the study and implementation of precision nutrition, and workforce development needed to create a new generation of researchers. The workshop concluded that much research will be needed before more precise nutrition recommendations can be achieved. This includes better understanding and accounting for variables such as age, sex, ethnicity, medical history, genetics, and social and environmental factors. The advent of new methods and technologies and the availability of considerably more data bring tremendous opportunity. However, the field must proceed with appropriate levels of caution and make sure the factors listed above are all considered, and systems approaches and methods are incorporated. It will be important to develop and train an expanded workforce with the goal of reducing health disparities and improving precision nutritional advice for all Americans.
Keywords: precision nutrition, data science, food, genomics, nutrigenomics
Introduction
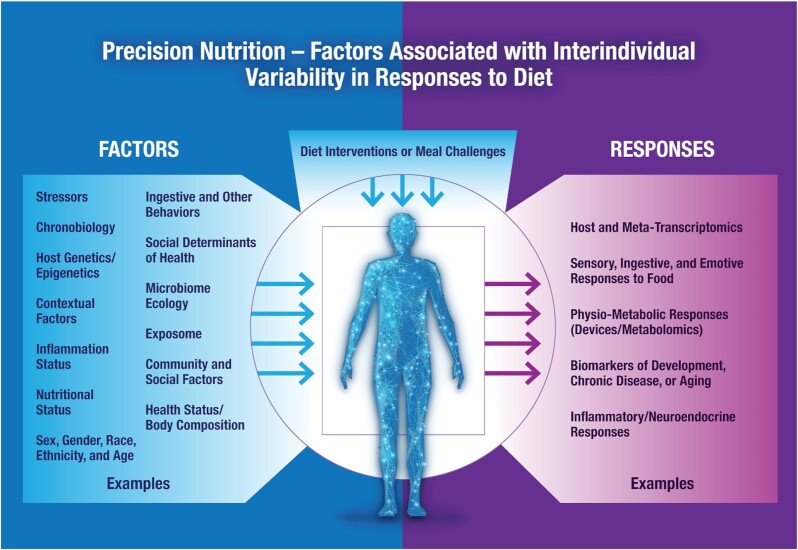
The importance of nutrition for health and disease prevention is well established (1), and global dietary guidelines are established to guide impactful public health policy (2). However, the practical question of what, when, and how to eat to stay healthy and aid individuals in their quest to optimize health is much more complex. As shown in Figure 1, many factors require consideration, including individual differences in disease risk, socio-environmental and cultural factors, and biological, behavioral, physiological, and psychosocial responses to dietary interventions. As defined by Maruvada et al. (3), “Precision nutrition is defined as nutrition or dietary guidance designed to optimize health, facilitate disease prevention, and enhance therapeutic benefit through molecular (metabolomic, genomic, proteomic, metagenomic) profiling at the level of the individual.” Precision nutrition aims to understand these complex interrelations to optimize metabolic responses to diet and ultimately make sustainable and targeted individual nutritional recommendations to prevent and treat diseases and improve overall health and well-being. This paper is not intended to be a comprehensive or systematic review of the current status of precision nutrition research. Rather, its objective is to summarize the specific topics, concepts, and issues raised during presentations and discussions during the NIH-sponsored “Research gaps and opportunities in precision nutrition” workshop on 11–12 January 2021. Given the many recent developments in this field, definitions of nutritional genomics, nutrigenomics/nutrigenetics, personalized, and precision nutrition are evolving, and different definitions have been presented (Supplemental Table 1).
FIGURE 1.
Precision nutrition. Factors associated with interindividual variability in responses to diet.
Precision Nutrition in Diet-Related Chronic Diseases
Chronic diseases [e.g., cardiovascular diseases (CVDs), cancers, diabetes, chronic respiratory diseases, and neurological diseases], also known as noncommunicable diseases, result from genetic, physiological, environmental, and behavioral factors. In the words of Richard Lewontin (4), “There are no genetic factors that can be studied independently of the environment, and there are no environmental factors that function independently of the genome.” Among the environmental risk factors, unhealthy diets and sedentary lifestyles are associated with several metabolic risk factors, such as increased blood glucose and lipids and increased risk of hypertension and obesity, leading to CVDs. These cardiometabolic risk factors can lead to CVD, the most burdensome global noncommunicable disease.
A fundamental approach to controlling the increasing prevalence of noncommunicable diseases is to reduce their risk factors. While the Dietary Guidelines for Americans (DGA) are intended as a foundation for a healthy population and guide government food and agricultural policy, the National Academies of Science, Engineering, and Medicine (NASEM) report from 2017 entitled Redesigning the Process for Establishing the Dietary Guidelines for Americans did indicate that, “Diet constitutes an extremely complex system of exposure that is known to influence health, and these modeling exercises can help make sense of that complex system,” and the following about the food pattern modeling used to inform the DGA: “the heterogeneity of the population is largely not accounted for, such as the distribution of requirements for energy and all nutrients, widely varying food choices by numerous demographic factors, and some food groups not being consumed by all Americans” (5). During the first session of the workshop, participants discussed precision nutrition concerning major chronic diseases, including cardiometabolic diseases [e.g., type 2 diabetes (T2D) and impaired glucose regulation] and CVD, cognitive decline, and diet-related cancers. These topics were presented as examples of the many areas of disease that are impacted by nutrition.
Nutrition and CVD phenotypes
Gene–diet interactions and CVD
Nutrigenetics, or gene–diet interactions, are the forerunners to precision nutrition initially defined for CVD-related traits. Many studies have been reported showing gene–diet interactions for intermediate conditions (e.g., hyperlipidemia, diabetes, obesity, and hypertension) and CVD events. These studies have been summarized in several reviews for different types of experimental designs (observation and intervention) and outcomes (intermediate CVD phenotypes and CVD outcomes) (6). Hundreds of gene–diet studies support (7) the notion that diet can modify genetic susceptibility to CVD. However, the level of specific scientific evidence to achieve precision desired remains too low to apply it in practice to individualize the dietary recommendations needed to prevent CVD. Limitations in the existing body of evidence include the small number of studies that replicate the evidence in different populations and the paucity of large phase III dietary intervention trials testing gene–diet interactions, specifically those having CVD incidence as the outcome. For those randomized controlled trials (RCTs) focusing on cardiometabolic traits (e.g., weight loss) no evidence was found that genotype–diet interaction is a main determinant of obesity treatment success (8, 9). There is some evidence from the Food4Me trial conducted in Europe that personalized dietary interventions using biomarkers could improve overall diet quality (10, 11). Several RCTs and other experimental approaches have tested whether genetic testing or other personalized dietary intervention approaches are useful in improving compliance and improving diet quality (12–21). However, the results are inconsistent, even at the level of systematic reviews and meta-analysis (22–24). Therefore, beyond the need to further conduct such interventions, additional meta-analyses of gene–diet interactions of existing randomized intervention trials and prospective cohorts involving similar dietary interventions and genetic markers are needed.
Epigenetics, dietary response, and CVD-related traits
Epigenetic responses (DNA methylation, histone modifications, and non-protein coding RNAs) to dietary factors and environmental conditions complement genetic variability in contributing to health and to the development and progression of chronic diseases. Diet and lifestyle influence the epigenetic regulation of key products of energy metabolism. The role of genes such as leptin (LEP), insulin receptor (INSR), TNF-α (TNF), and fatty acid synthase (FAS) (25) in the development of several chronic disorders can be traced back to epigenetic mechanisms during fetal development. This concept is further supported by various non-Mendelian features of metabolic diseases and cancer and the clinical differences between men and women or monozygotic twins. Unlike genetic factors that remain constant throughout life, epigenetics is malleable. Thus, epigenetic signals regulate genes of nutrient metabolism, but nutrient metabolism modifies epigenetic signaling (26). Moreover, DNA methylation at specific loci can be influenced by sequence variations, such that individual genotypes at a given locus may result in different patterns of DNA methylation due to allele-specific methylation. These methylation quantitative trait loci (mQTLs) can influence the methylation pattern across an extended genomic region.
Few studies have examined the contribution of epigenetic markers to variability in dietary response. As an illustrative example and using data from the Genetics of Lipid-Lowering Drugs and Diet Network Study (GOLDN) Study, Lai et al. (27) conducted an epigenome-wide association study (EWAS) on 979 subjects challenged with a high-fat diet. DNA methylation was measured in CD4+ T cells. Eight methylation sites encompassing 5 genes—lipoma-preferred partner (LPP), carnitine palmitoyltransferase 1A (CPT1A), apolipoprotein A5 (APOA5), sterol regulatory element binding transcription factor 1 (SREBF1), and ATP binding cassette subfamily G member 1 (ABCG1)—were significantly associated with postprandial lipemia (PPL). Higher methylation at LPP, APOA5, SREBF1, and ABCG1 and lower methylation at CPT1Awere correlated with increased plasma triacylglycerol (TG) concentrations that contributed to the PPL response. These PPL-associated methylation sites also correlated with fasting TG and accounted for the substantially higher phenotypic variance (∼15%) in PPL and fasting TG (∼16%) when compared with the genetic contribution of loci identified by a previous genome-wide association study (GWAS) (4.5%) in the same participants (28). However, such findings must be validated by larger studies, and similar approaches applied to studies of other nutrients. The cross-talk between epigenetic markers and genetic variability in relation to dietary response has been demonstrated in several studies (29). Thus, the epigenetic status of the APOA2 regulatory region was associated with saturated fat intake, and the APOA2 − 265T > C genotype promoted an APOA2 expression difference between APOA2 genotypes on a high-SFA diet (30). Along those lines, Ma et al. (31) demonstrated that higher n–3 PUFAs were associated with lower methylation at the IL6 promoter, but single nucleotide polymorphisms modified this association at the IL6 locus.
An important caveat related to measuring epigenetic changes in humans is that epigenetic markers are usually tissue specific. Therefore, the usual epigenetic analyses in human lymphocytes may not be representative of the changes in liver, muscle, and brain DNA. New methodological developments, such as the use of circulating cell-free DNA released from such tissues, may more precisely assess epigenetic markers within those tissues (32).
Dietary response, microbiome, and cardiometabolic traits
Research targeting precision nutrition approaches to prevent and treat cardiometabolic diseases has increasingly implicated a complex interactive role for the host microbiome (33–35). Some investigations have contrasted a Western and a Mediterranean-style dietary pattern (MedDiet) (36) with evidence of associated changes in the gut microbiome structure and function (37). The Health Professionals Follow-Up Study findings further supported protective associations between adherence to the MedDiet and cardiometabolic disease risk. This study demonstrated that such cardiometabolic disease as determined by measurements of blood biomarkers of glucose homeostasis, lipid metabolism, and inflammation was significantly stronger among participants with decreased abundance of Prevotella copri (38) bacteria. However, most existing studies are observational and cross-sectional, while mechanisms and causal factors remain largely unexplored. In this regard, studies both published and underway take a multi-omic approach and demonstrate relations between diet, the microbiome, and the circulating metabolome. The Personalized Responses to Dietary Composition Trial-1 (PREDICT-1) study (34) reported many significant associations between microbes and specific nutrients, foods, food groups, and general dietary indices, which were driven especially by the presence and diversity of healthy and plant-based foods. Moreover, microbial biomarkers of obesity were reproducible across external publicly available cohorts and in agreement with circulating blood metabolites that are indicators of cardiovascular disease risk. While some specific microbes [e.g., Prevotella copri reported by Asnicar et al. (34) and Blastocystis spp.] were indicators of favorable postprandial glucose metabolism, a microbiome fingerprint was predictive of cardiometabolic biomarkers (e.g., fasting and postprandial glycemic, lipemic, and inflammatory indices) and was also associated with healthy dietary habits. However, in acute experiments, measures of colonic fermentation and abiotic factors were not shown to be significantly associated with variability in postprandial responses, perhaps because of resilience in healthy adults (39).
It has been suggested that the habitual long-term diet can benefit the gut microbiome and the metabolome synergistically. For example, consistent intake of a predominantly plant-based diet appears to favorably influence circulating metabolites, especially bile acids. Thus, it is likely that such studies will increasingly identify dietary compounds and phytochemicals that may modulate bacterial abundance and quality within the gut that interact with the microbiome composition and thereby influence host metabolism (40, 41). It is important to emphasize that most of the current studies are still using a “global” population approach and we need to evolve towards more “personalized”/individual studies.
Type 2 diabetes
Predicting and controlling glycemic response to diet in T2D
The dysglycemia observed in T2D can be thought of as having 3 separate components: glycemic variability, ambient hyperglycemia, and hypoglycemic episodes (42). Glycemic variability refers to glucose fluctuations from peaks to nadirs, with hyperglycemic peaks reflecting postprandial glycemic response (PPGR). Ambient hyperglycemia is elevated glycemia that results from the loss of insulin-secreting β cells and their replacement by glucagon-secreting ɑ cells. Hypoglycemic episodes are adverse consequences generally driven by medication management. Glycemic control is an important management target and PPGR appears to be more damaging than ambient hyperglycemia (43). Glycemic variability increases oxidative stress and epidemiologic studies have shown postprandial hyperglycemic peaks to be a powerful predictor of cardiovascular risk (44). Further, some clinical trials have shown that lowering postprandial hyperglycemic peaks with medication reduces the risk of progression to diabetes, hypertension, and cardiovascular events in those with impaired glucose tolerance (45, 46). There is universal recognition that dietary management is key to successful T2D treatment. To date, most studies have used one-size-fits-all dietary interventions (47–57) with mixed results. Uniform dietary interventions may fail to manage postprandial hyperglycemia because individuals vary greatly in their glycemic response to the same food (58). Given the importance of interindividual variability in dietary response, new approaches are being used to investigate the range of responses and the variability in response due to measurement error or the consequence of behavior compensation, with the goal of understanding how dietary macronutrient manipulation may be optimized for individuals.
T2D and the microbiome
Recent studies have shown that the composition and function of the intestinal microbiota are critical factors in glucose homeostasis (59). In a series of mouse gut microbiota studies, Turnbaugh et al. (60) demonstrated that the obese microbiome has an increased capacity to harvest energy from the diet. Others have shown the composition and function of gut microbiota to be associated with glucose intolerance (61, 62), insulin resistance (63), and T2D (64, 65). Vrieze et al. (66) showed that transferring intestinal microbiota from lean humans to those with metabolic syndrome increased insulin sensitivity. In 2015, Zeevi et al. reported on the development of a machine-learning algorithm for predicting PPGR to specific foods consumed by the participant (35). In the model discovery phase, a glycemic profiling training dataset was compiled on 800 Israeli participants, with data including the gut microbiome, various blood tests, questionnaires, a date- and time-stamped food record, sleep, physical activity, and interstitial glucose measured with continuous glucose monitoring. The model demonstrated high between-subject variability in PPGR to the same foods, suggesting that universal dietary recommendations are of limited utility for limiting PPGR. The derived model was validated in a new cohort of 100 participants, with model predictions for meals being highly correlated with the measured glycemic response (R = 0.6) and substantially higher than naive predictions based only on the meal's carbohydrate content (R = 0.28) (35). The model was recently validated in a large US sample (58, 67) but, to date, has not been evaluated for limiting PPGR in patients with T2D. Validation is important as computer algorithms have not been usually demonstrated to be effective in improving health outcomes in different populations, and different computer algorithms may offer inconsistent or even contradictory nutrition recommendations.
Cancer
Nutrients, nonnutrient bioactives, energy balance, and dietary patterns are key determinants of cancer risk (68) and established guidelines for prevention can significantly impact the global burden of cancer (69). Such efforts also highlight the potential for personalized interventions to serve as important and cost-effective, nonpharmaceutical strategies for primary cancer prevention and suppressing cancer progression. In the past, diet and cancer interactions have traditionally focused on primary prevention. Despite solid preclinical evidence, nutrition intervention studies with specific dietary patterns, foods, or nutrients, and the impact on cancer outcomes are extremely limited. Studies such as the Selenium and Vitamin E Cancer Prevention Trial (SELECT) for prostate cancer (70) and the Alpha-Tocopherol Beta-Carotene Cancer Prevention Study (ATBC) with β-carotene focusing on lung cancer (71) have shown limited success, and were perhaps suboptimally designed (72), and reduced enthusiasm for additional studies. A major gap in these studies was a lack of understanding of the duration of intervention that may be necessary, as well as the form, bioavailability, and mechanistic targets, coupled with knowledge of the baseline nutrient status of the host, and an understanding of the significant individual variability in responses. One challenge in the field of cancer research relates to the limited number of intermediate biomarkers for risk, compared with those that are available for CVD risk. Definitive phase III trials should be designed based on knowledge derived from phase I/II data that clearly defines the pharmacodynamics of the intervention and the relevant factors impacting human heterogeneity.
Personalized medicine and targeted therapy approaches are the foundation of oncology therapeutics. Gene profiling of the cancer, to define relevant mutations and targets, has allowed for the development of “gene prognosis signatures”—knowledge that can improve the selection of higher risk individuals for novel adjuvant/neoadjuvant therapy trials. In addition, molecular signatures of individual cancers increasingly can define responders and nonresponding subgroups for specific treatments while also providing a specific target for defining mutation-specific agents (73, 74). In parallel, host genetics may define pharmacokinetic and pharmacodynamic responses in an individual, thereby impacting response to therapy and toxicity. Few studies address how dietary variables interact to impact these processes and improve individual outcomes.
Finally, early detection and improved therapy are greatly increasing the number of cancer survivors, including many with long-term cures. Providing evidence-based guidelines, specific for individuals experiencing unique combinations of therapeutics, are needed to improve long-term outcomes. Opportunities for precision nutrition trials are abundant and much needed. Critically, clinical trials in cancer must address issues of dose of an exposure and the duration of intervention necessary to achieve an optimal impact. Greater mechanistic insight into how genetic, epigenetic, microbiome, and other factors contribute to individual variability in response to dietary patterns, nutrients, and bioactive phytochemicals will enhance the design of impactful intervention studies for cancer prevention, therapy, and survivorship.
Gene–diet interactions and cancer
Precision nutrition research related to cancer started with the study of gene–diet interactions, similar to cardiometabolic traits and CVD risk. Theodoratou et al. (75) critically and comprehensively evaluated the evidence across 13 meta-analyses of observational studies of gene–diet interactions for the 5 most common cancers (breast, lung, prostate, colorectal, and stomach). The authors focused on gene–diet interactions for food and nutrient associations that were classified as convincing (class I), highly suggestive (class II), or suggestive (class III) and classified them as strong, moderate, weak, or no evidence. Among all the evaluated gene–diet interactions with prior weak, moderate, or high scores, only the interaction between the 10p14 locus near GATA binding protein 3 (GATA3) and processed meat in relation to colorectal cancer risk (76) was classified as moderate. The following interactions were classified as weak: interactions between rs17468277 [caspase 8 (CASP8)] and alcohol in relation to breast cancer risk as well as interactions between rs1805087 [methyltransferase (MTR)] and alcohol (77), rs16892766 (8q23.3) and vegetables (78), and glutathione S-transferase theta 1 (GSTT1) deletion polymorphism and cruciferous vegetables (79) in relation to colorectal cancer risk. The remaining studied associations did not show any evidence of an interaction. A more recent review focused on gene–diet interactions related only to breast cancer (80). The authors listed 18 genes investigated in 25 studies, and most of them reported significant gene–diet interactions. However, the strength of the evidence was not assessed. Additional strategies, perhaps in carefully controlled clinical trials of specific nutrients, phytochemicals, or dietary patterns examining the specific gene variants relevant to the intervention, can provide greater insight (81). Genetically engineered animal models can also provide critical preclinical evidence for relevant gene–nutrient interactions (82, 83).
Diet response, epigenetics, and cancer
Animal models support associations between diet and epigenetic alterations and between epigenetic alterations and cancer (84–87). However, data in human populations are sparse and inconsistent, both for observation studies and randomized clinical trials. These studies have failed to show clear, consistent, and predictable effects of diet or supplements on cancer risk because of high phenotypic variability in response. Therefore, the conclusion that diet is linked directly to epigenetic alterations and that these epigenetic alterations directly increase or decrease the risk of human cancer remained speculative based on the current evidence (88), suggesting opportunities for future research.
Dietary response, microbiome, and cancer
Rapidly emerging data implicate the gut microbiome's responsiveness to diet, in terms of both structure and function, impacting many bioprocesses related to carcinogenesis. We are learning that dietary patterns characterized by ultra-processed foods, refined sugar and grains, and meat compared with intake patterns rich in whole grains, legumes, vegetables, fruits, and fiber can significantly alter the microbial composition in association with improved host metabolism and health outcomes (89–92). There is a critical need to appreciate the potential of the overall dietary pattern, defined by multiple components of a healthy diet, to define a healthy and stable microbiome that is also optimized for cancer prevention.
The diet may impact the colonic microbiome in ways that alter the propensity for developing obesity and, conversely, obesity may subsequently lead to changes in colonic composition and function, impacting the host and perhaps risk of cancer (60, 93–98). Bacteria in various sites may alter the host response to bioactive phytochemicals through their metabolic conversion to metabolites that are subsequently absorbed and may have more or less bioactivity as modulators of carcinogenesis (99–103). For example, thioglucosidase myrosinase activity, which releases the bioactive sulforaphane from glucosinolates found in cruciferous vegetables, often depends upon gut bacterial thioglycosinodases (104), indicating that the gut microbiome influences sulforaphane bioavailability (105). However, at the present time, there is a large gap in knowledge regarding our understanding of the interplay between the gut microbial flora at various sites and the host in regard to the metabolism and anti-cancer activity of the diverse array of dietary bioactives. Studies that examine this interplay are needed to ultimately aid with personalized stratification-based prevention strategies to improve anti-cancer efficacy.
The growing evidence that the host microbiome influences the therapeutic response to cancer immunotherapy is driving new initiatives that may have a significant impact. The recognition that the microbiome is a modifiable target for dietary interventions and can influence efficacy and safety of novel immunotherapeutics, such as immune checkpoint blockade, raises the question of defining the optimal dietary interventions that are specific for the rapidly emerging array of treatment regimens (106, 107).
Thus, research is rapidly expanding regarding how dietary patterns, nutrients, and phytochemicals impact the microbiome at various sites as a central mediator of the diet and cancer relation.
Alzheimer disease
Optimal brain function results from highly complex interactions between genetic and environmental factors, including food intake, physical activity, age, and stress. Specifically, nutrition affects the brain throughout life, with profound implications for cognitive decline and dementia. These cognitive effects are mediated by changes in the expression of multiple genes, and responses to nutrition are, in turn, affected by individual genetic variability. Alzheimer disease (AD), the most common cause of dementia, develops decades before any clinical symptoms manifest. Lifestyle and genetic factors contribute to AD risk. The best-known genetic risk factor is the presence of the apolipoprotein E (ApoE4) allele at the APOE gene (108). The ApoE4 allele codes for an alternative form of ApoE, which can disrupt its ability to perform essential functions in lipid transport and metabolism in the brain. ApoE4 allele frequency in the general US population is ∼20%; however, carriers of this allele account for ∼40–65% of AD cases. Still, most people with the ApoE4 allele will not express the disease. Therefore, gene–environment interactions are thought to play a mediating role (109).
Gene–diet interactions, AD, and cognitive decline
Migration studies provide a clear example of the importance of gene–environment interactions in disease expression (110). For example, the ApoE4 allele at the APOE gene is common in persons of West African ancestry but does not meaningfully contribute to AD risk in those living in West Africa (111, 112). However, incidence of AD is much higher in Africans living in the United States (113). Likewise, southern Italians carrying ApoE4 and who live in Italy can reach old age, apparently unaffected by the genetic risk. However, southern Italians living in the United States carrying ApoE4 exhibit a highly reduced chance of living into late old age (114). Findings suggest that environment and lifestyle, including diet, may mediate ApoE4 effects on development of AD.
Longitudinal observational studies further demonstrate the potential interaction between dietary factors, cognitive decline, and the APOE locus. These studies highlight the importance of dietary components, as well as timing, duration, and “dose” of dietary and nutrient changes along the spectrum of preclinical AD, mild cognitive impairment due to AD, and AD dementia (110, 115). Trials of DHA supplements for AD dementia treatment failed, possibly due to starting supplementation too late in the disease process. Other trials that have focused on AD prevention only were largely negative possibly due to a low-dose supplementation. However, recent studies have shown that larger DHA doses are necessary for adequate brain bioavailability and that ApoE4 is associated with reduced DHA and EPA delivery to the brain prior to the onset of cognitive decline (116). Further long-term randomized studies are warranted at the earliest phases of the AD pathophysiological process to inform more definitive conclusions.
Dietary response, epigenetics, AD, and cognitive function
Epigenetic mechanisms are central to brain development, structure, and function. Studies linking nutrition with advances in neuroscience, genomics, and epigenomics should provide novel approaches to preventing cognitive decline, as well as treating dementia and AD. For example, curcumin consumption has been associated with better cognitive performance and lower prevalence of AD. These associations may be explained by curcumin's ability to downregulate DNA acetylation expression in specific cell lineages (117, 118). Similarly, flavonoids, which are polyphenolic compounds found in fruits, vegetables, and other natural sources, have been shown to reduce the expression of proinflammatory cytokines and prevent neural damage through epigenetic modulation (119).
Overall, recent studies have shown that personalized clinical recommendations based on genetics, in combination with modifiable risk factors (e.g., nutrition, physical activity, stress management), can help improve cognition and reduce calculated AD risk in patients at risk for AD (108, 120).
Dietary response, microbiome, AD, and cognitive function
The action of the gut microbiome on the ApoE4 allele (CNS) has been of increasing interest. A bidirectional relation has been reported between the gut microbiome and the CNS and has been referred to as the “brain-gut-microbiota axis.” Briefly, the brain-gut-microbiota axis means that the CNS can regulate the digestive tract by acting upon the enteric system and, vice versa, the intestinal microbiome can influence the CNS via afferent signaling pathways and secretion of active substances. Studies have implicated the role of the brain-gut-microbiota axis in AD pathology (121). Preclinical animal studies have shown that microbiota obtained from the intestines of AD mice possessing the human APP gene can increase amyloid-B deposition in the intestines of normal mice (122–124). Furthermore, studies in older populations suggest an association between increased levels of proinflammatory bacteria in gut microbiota and amyloid deposition in the brain and cognitive deficits (125).
The “Western diet” has been shown to contribute to changes in gut microbiota and the development of dementia. This higher-fat diet may change the bacterial composition in both the colon and cecum, resulting in a higher abundance of bacterial species associated with cognitive impairment and cerebral hypometabolism. Conversely, a healthier dietary plan such as a modified Mediterranean-ketogenic diet has been shown to improve microbial diversity in subjects with mild cognitive impairment due to AD compared with a more conventional Western diet (126). This suggests that diet may modulate the gut microbiota in a way that potentially reduces their risk of developing AD.
Individual Characteristics Influencing Response to Dietary Intakes
During the second session of the workshop, participants discussed many potential variables that contribute to interindividual variability in dietary response. Recent research has highlighted the importance of the rigorous methodology needed to assess nutritional physiology and postprandial metabolism.
A past focus on limited subsets of individual nutrients
The role of diet in health is well documented by decades of research in nutrition (127). However, understanding how diet affects human health (128, 129) and the methodologies utilized to acquire this knowledge are the subject of constant self-reflection and occasionally of fierce debate (130, 131). From a dietary perspective, what is known is mostly focused on the role of ∼150 dietary components, including most of the defined nutrients, systemically tracked by food-composition databases (e.g., by USDA), which represent only 0.5% of the biochemicals present in food (132). The need to enhance the USDA and other databases with rigorously collated, synthesized, and disseminated food-composition data is currently a major roadblock in the scientific community's ability to advance the field and assess the exceptional biochemical diversity of food. Arguably, accurate information on the biochemical composition of foods is just as essential to nutrition and health science as the genome project was to biology, revolutionizing the understanding of long-ignored environmental factors and their impact on the molecular roots of human disease. Indeed, one can validly argue by analogy that current coverage of food composition is where genetics was before the Human Genome Project. Quantifying the full biochemical palette of the diet could offer the possibility of linking these patterns to molecular processes with ultimate accuracy, leverage high-throughput approaches common in genomics, pave the way to personalized dietary recommendations, and improve the understanding of how diet modulates the efficacy of drugs and treatments (133). For example, a better understanding of how individuals differ in their absorption of nutrients can lead to the discovery of molecular and physiological targets for drug development that may improve postprandial handling of nutrients (134). Therefore, a Big Data strategy is needed to create and experimentally validate a high-resolution compendium of the biochemical composition of food and to make this new resource widely available in an actionable form to the research community. Deep mining of the chemical and biological literature for information on biochemical composition of food can be complemented with machine-learning tools to infer missing knowledge, and with systematic experimental validation to estimate the precision and the completeness of our current knowledge. The resulting platform would transform health science by catalyzing dramatic leaps in scientific and health insights and opening up novel avenues by which to understand, avoid, and control disease.
Impact of race, ethnicity, and culture on diet-related health outcomes
It is well established that individual nutritional needs and responses to diets vary across populations according to biological, demographic, and environmental features (135, 136). Although differences in populations and ethnicities are being increasingly acknowledged, these factors are often ignored in nutrition-based studies. For example, diverse groups are combined under the heading of Asian Americans, although there are striking social, cultural, and genetic differences among the subgroups within this population (137, 138). Similar criteria are applied to people of African-American and Hispanic heritage, although genotype and allele frequencies differ within these populations/ethnicities. Changes in diet or epidemiological transition have not equally affected all ethnic groups or subgroups (139, 140) and a significant gap exists in the understanding of genetic variability on nutrient requirements, as well as how genetics impacts responsiveness to nutrient change. Inclusion of diverse populations and their subgroups in studies is critical to understanding the physiological response to diet and characterization of responder versus nonresponder phenotypes. These phenotypes should be further evaluated in relation to their exposure to social and environmental factors such as physical activity and mobility; socioeconomic status; geography, including rural/urban settings; food security/availability; living situation; and exposure to pollutants. Identification of genetic variants that make specific subgroups susceptible to diseases and an understanding of dietary response in the context of exposure to the environmental factors will support the development of nutritional interventions targeted to these groups, which is a key step towards personalized nutrition.
Nutritional status as a source of interindividual variability
As described above, participants in nutrition research studies exhibit a large variability in response to dietary interventions. For many years, it was assumed that the variation in response was due to incomplete adherence to protocols. An individual's ability to adhere to dietary change is, in itself, an important factor to understand. Being able to make behavior changes to comply with dietary protocols may be different between participants due to factors such as social support, mental health, stress, financial, and environmental stability. Data on these outcomes should be collected in precision nutrition studies. Furthermore, in addition to factors that reduce an individual's ability to adhere to a research protocol, participants can have acceptable levels of adherence to dietary intervention protocols, particularly in the context of well-designed and highly controlled intervention studies, yet exhibit a wide array of biological responses to a specific intervention (34, 141). Study participants are not uniform in terms of the characteristics they bring into a dietary intervention—they differ in age, body habitus, microbiome, metabolome, epigenetic characteristics, usual diet prior to the intervention, dietary supplement use, medical conditions, and other lifestyle habits that influence metabolism and intervention response. Statistical designs that include block randomization can help distribute some of these characteristics across intervention groups. However, this strategy is typically only practical for a limited number of categorical variables, such as age groups, BMI groups, and sex. Importantly, stratification across more groups requires larger sample sizes to fill the blocks. Moreover, it is not feasible to block the randomization scheme to include complex, multidimensional exposures such as baseline microbiome and metabolome features. It is also important to recognize that the diet itself is complex. Shifting 1 dietary component, whether it is the macronutrient distribution or specific food components (e.g., cruciferous vegetables), will often produce changes in the metabolome, proteome, and microbiome. For comparisons of isocaloric diets, an increase in 1 of the 3 macronutrients (carbohydrate, fat, protein) will necessitate the reduction in another of the macronutrients as a percentage of total energy and may induce shifts in downstream metabolic by-products. Thus, both larger sample sizes and advanced computational strategies are needed to handle high multidimensional pathways to understand the metabolic effects of macronutrient change.
Measuring physiologic responses to eating
Fasting blood concentrations of metabolites are used to determine disease risk and treatment outcomes in health care. However, daily excursions of metabolites and hormones—exemplified by the rise and fall of blood concentrations of glucose, lipids, insulin between meals—are better predictors of disease risk compared with fasting blood values. For example, elevated concentrations of blood TGs after meals have a stronger association with CVD risk compared with fasting measurements of TGs (142, 143). Similarly, the use of glycated hemoglobin (HbA1c) levels—a read-out of plasma glucose concentrations over months—is well established to predict diabetes risk. Over the past 40 y, studies of meal carbohydrate metabolism have focused on utilizing an oral-glucose-tolerance test (OGTT) to assess fed-state glucose metabolism. The use of an OGTT simplifies test procedures and multiple indices of insulin sensitivity can be calculated (144). However, people rarely consume glucose in isolation and interactions exist between the metabolism of many components in mixed meals. Thus, although the OGTT is an important tool to understand metabolism, it should not be referred to as a meal test. Similarly, to understand TG metabolism, tests should contain mixed amounts of fat, representing more traditional daily food intake. New methods have been developed recently using metabolomics to track the processing and fate of dietary nutrients (35, 145).
Currently, state-of-the-art meal tolerance tests (MTTs) are performed in the morning after a fast of at least 10 h. Blood samples are drawn before and after the MTT as well as intermittently over 4–9 h. Longer times are needed to capture the concentrations of metabolites as they return to baseline levels. The total energy of the MTT can vary between subjects, so that it is scaled to be a set proportion of the subject's total daily energy needs (145). Several factors have been identified that influence the postmeal metabolism of nutrients (Table 1). These factors should be considered during the development of study designs to decide whether to intentionally control for these variables or not. Moving the field of precision nutrition forward will require an understanding of the biology of interindividual variability in response to dietary interventions. Careful planning and execution of metabolic tests will be required to accomplish this goal (141).
TABLE 1.
Examples of factors that can influence the metabolic response to eating1
| Factor | Examples of the possible effects |
|---|---|
| Age | Meal glucose and TG metabolism slows with aging (146). |
| Sex | Compared with women, men are more susceptible to dietary-induced hypertriglyceridemia (147). |
| Time of day | Compared with the first meal of the day, the processing of meal CHO from a second meal exhibits a pattern of insulin resistance; CHO and fat consumed at night clear from the plasma more slowly (148). |
| Exercise | Exercise the night before, or the morning before a meal test, will increase apparent glucose and fat clearance rates (149, 150). |
| Alcohol | Alcohol consumed with meals raises postmeal blood TG concentrations (151–153). |
| Fasting values | The fasting concentration of TG is the strongest predictor of postprandial TG excursions and fasting hyperglycemia is associated with greater glucose excursions after a CHO bolus (154). |
| Cross-macronutrient effects | A high-CHO evening meal raises fasting blood TG the next morning and slows postmeal TG clearance; conversely, a high-fat evening meal slows CHO clearance following the next morning's meal (155). Fiber slows glucose absorption (156). |
CHO, carbohydrate; TG, triacylglycerol.
The influence of sensory nutrition
Consumers report that the “taste” of foods drives what they like and choose to consume. “Taste” and flavor are complex chemosensory experiences with multiple peripheral inputs (true taste, smell, and chemesthesis) carried centrally for perceptual integration and coupling with hedonic, reward, decisional, and satiety responses. Multiple factors that drive variation in flavor perception and food preference influence dietary behaviors, chronic diet-related diseases, and the compliance with dietary interventions. These factors include conditions that impede chemicals reaching chemoreceptors and influence the function of chemosensory-related nerves [including coronavirus disease 2019 (COVID-19)], genetic variation in chemoreceptors, interactions between internal and external environments to influence the plasticity of chemosensory systems, and age-related changes. Genetic variation in taste and flavor perception frames the development of dietary behaviors in early life, in interaction with food environments. Aging, with changing experiences and environmental exposures, can modify flavor perception, with or without changes in food preferences and behaviors (157, 158). Interdisciplinary research should leverage sensory nutrition as markers of usual dietary behaviors and responses to improve understanding of variability in dietary interventions (159). Clinical nutrition research needs to account for chemosensation in measures that reflect sensory function and are relevant to food perception, including self-report and measures of food preference. Even the perception of the energy content of food can influence research results (160). Population-based research should incorporate feasible and informative measures of sensory nutrition and food preference with new study designs to enhance understanding of diet–disease relations (161). Last, the formulations of the diets and test meals being fed can impact research findings because palatability and acceptability can influence the postingestive processing of nutrients.
Immune system status and inflammatory response to diet
Optimal immune system status is paramount for health, as evidenced by the current COVID-19 pandemic (162). In summarizing the current state-of-the-art, there is relatively good prospective evidence demonstrating the relation between diet, nutritional status, the immune system, and inflammation (163). Also, there is well-developed knowledge with respect to the impact of different dietary elements on various aspects of immunity and inflammation, although this knowledge is yet to be fully elucidated. A key area for development is in the translation to human clinical prospective interventions and defining the impact on health outcomes mediated by the immune system.
Immune system status is determined by factors ranging from age, gender, circadian biology, infection history, and vaccination status to diet, alcohol intake, physical fitness, and the microbiome. Recent research relating to the adaptive and innate immune response has greatly advanced our knowledge (164). First, the adaptive immune response has memory (i.e., prior exposure to a pathogen heightens the future responses to the same pathogen). This is critical because the cells of the adaptive immune system such as T cells are specific to each pathogen and, without the propagation upon exposure, they would not be able to mount an effective defense. On the other hand, the innate immune response does not have a memory and can respond to any pathogen. Second, the metabolic configuration of an immune cell determines the nature of the immune response. Obesity-induced innate and adaptive immune dysregulation is characterized by heightened proinflammatory reactivity of myeloid cells and predominant glycolytic metabolic reconfiguration supporting a proinflammatory phenotype as well as suppressed adaptive immune response, including ability to produce an effective antibody response to vaccines. The profound impact of obesity on innate and adaptive immunity plays a crucial role in the severity of COVID-19 disease in this population. Nevertheless, knowledge in this field needs to deepen the understanding of the impact of different dietary elements on immune modulation beyond obesity (163). The challenge in the future will be to determine if and how different nutrients and nonnutrient food components train and re-configure immune response. The real challenge will be to understand the extent to which these paradigms, with co-regulation of both metabolic and inflammatory processes (165), translates to humans.
A key stumbling block is the lack of sensitive and specific biomarkers that accurately reflect the dynamic and circadian rhythm of the immune response. MTTs are 1 potential tool, with the prospect to capture both the metabolic and inflammatory dynamics in response to food intake (166, 167). This response varies substantially with age, gender, and metabolic phenotype. Nevertheless, there remains a significant body of work required to understand the impact of different dietary constituents on the acute postprandial immuno-metabolic response in a temporal, cell-specific manner. In this realm, although the adverse effects of some dietary elements are well known, greater focus needs to be paid to the extent to which immuno-metabolic responses can be attenuated or resolved with dietary change and how these effects can be robustly measured or quantified. To this end, precise nutrition approaches should add greater clarity with respect to optimal opportunities to maximize knowledge concerning the interactions between diet-related metabolism, immune function, and health.
Circadian rhythm, food intake, and sleep
Food intake
“When should I eat?” has emerged as a compelling research question. That food-intake timing matters is supported by an established body of evidence linking metabolism to the circadian system, a system composed of a network of internal biological clocks (141, 168, 169). As such, when we eat intimates a dimension of time, although there are several aspects of time that warrant consideration. Time may be social and determined by clock time (e.g., work time, school time); it may be biological and determined by internal body clocks (e.g., chronotype, age); or it may also be environmental and determined by sunrise and sunset, which vary by season and geographic latitude (e.g., solar time). Research to date has focused on clock time with regard to eating behavior. Early appreciation for the influence of the timing of food intake (170) on total daily energy intake has advanced to more sophisticated analyses of eating occasions across the 24-h day (171). Earlier versus later eating occasions, as well as intermittent fasting and restricted time windows for eating, have been associated with better metabolic health outcomes in ecologically valid settings (172–175). However, some evidence suggests that the benefits of earlier versus later eating occasions may be more relevant for the metabolic health of some individuals, such as MTNR1B risk-carriers or sedentary individuals, than others (176–178).
Data indicate that eating later in the day, later relative to the sleep episode and later compared with the central circadian clock (using the gold-standard dim-light melatonin onset), is associated with increased body mass, increased adiposity, increased odds of being obese or overweight, and decreased success of weight loss during dietary weight-loss interventions and in the years following bariatric surgery, both in children and adults and without apparent differences in energy intake (173, 175, 179–185). One of the possible mechanisms for this effect is if the magnitude of the increase in energy expenditure following a meal (known as diet-induced thermogenesis, or the thermic effect of food) is dependent on the time at which the meal is consumed. Diet-induced thermogenesis (DIT) is substantially higher when a test meal is consumed in the morning as compared with the evening. This variation in DIT may be primarily driven by the circadian timing system (186). Glucose tolerance also is relatively impaired in the evening compared with the morning (187), and the endogenous circadian system plays a key role in this modulation via regulation of B-cell function (188–190). With regard to personalized chrono-nutrition, it should also be pointed out that the effect of dinner timing on glucose control can depend importantly on individual genotype (176, 191). Progress in this area is stymied by the lack of an accurate and feasible assessment for biological time in ecologically valid settings. The complexity of the internal body clock system introduces further challenges. With clocks existing in every tissue, it remains uncertain which tissue would best represent biological time relevant to nutrition. This is especially important for the important segment of the population working night shifts, which has been associated with altered circadian rhythms, lifestyle habits, and cardiometabolic risks, and should be factored in when developing effective workplace wellness and precision nutrition programs (192). This is a promising and active area of ongoing research that may have implications for dietary interventions.
Sleep restriction and timing
The association between sleep and cardiometabolic function and disease, including the interaction with nutrition, has received increasing recognition. Controlled experimental studies have shown reduced insulin sensitivity to a morning OGTT in healthy adults during short-sleep, early-waking conditions (193). Short sleep, poor sleep quality, and sleep disorders are associated with increased risk for T2D (194–196), obesity, subclinical atherosclerosis, and CVD (197–199). Sleep deficiency affects hunger, appetite, food choice, and intake; however, this effect varies across individuals. Sleep loss exerts these effects via alterations in homeostatically regulated pathways, assessed by changes in circulating concentrations of leptin and ghrelin. A positive correlation has been reported between the increase in ghrelin-to-leptin ratio and increase in hunger following sleep restriction (200). Interindividual variability can also be observed within hedonic systems following sleep loss, as seen within the endocannabinoid system, which is known to regulate hedonically driven food intake (200). Sleep loss facilitates a robust increase in ratings for sweet, salty, and starchy foods as well as an actual increase in snack food intake specifically (201–204). There is also developing evidence for the reverse relation: the influence of nutrition on sleep (204, 205). Finally, the disturbances in daily routines due to the COVID-19 confinement and remote work have impacted circadian rhythms, energy balance, and body weight (206). More research is needed in this burgeoning field.
Social determinants influencing food and health
Social determinants of health refer to the economic and political structures, social and physical environments, and access to health services that shape a person's health and well-being. Social factors contribute significantly to health and may account for up to 20–50% of the disease burden of society (207). They also expose—and explain—nutrition and health disparities, as the social and environmental factors that influence a person's dietary choices are unequally distributed within a population. For example, structural barriers to healthy food choices (including low income, unavailability of food stores that sell healthy foods, neighborhood characteristics like walkability and crime, lack of transportation, literacy, discriminatory practices) exist for susceptible groups, such as racial/ethnic minorities. The concentration of structural barriers within communities can also give rise to unhealthy social and cultural food norms, which further reinforce unhealthy eating patterns. As such, diet quality is consistently poorer among populations with low incomes and among some minority populations like African-Americans (208, 209). Additionally, exposures to environmental stressors like discrimination, social isolation, and pollution affect not only food access and choice but also biological processes including inflammatory responses and gene expression (210). A precision nutrition approach should use all relevant information on an individual's characteristics, including their social context, to drive implementation strategies to achieve optimal compliance with nutritional guidelines and recommendations for better health. Inequitable social and environmental experiences may explain disparities in health outcomes mediated by epigenetics or the microbiome (211, 212). Unfortunately, past work in precision nutrition has often failed to integrate social determinants of health and, even when it does, investment in research to optimize translation to vulnerable communities is scarce. Current research opportunities to promote a wider integration of social determinant data in precision nutrition are presented in Table 2. To help close critical knowledge gaps, researchers may leverage and augment existing cutting-edge tools and databases to collect, share, and harmonize data and resources. Active inclusion of social and environmental factors should be done in novel research lines, training, diverse and inclusive interventions, and policies for optimal precision nutrition.
TABLE 2.
Current research limitations to understand the influence of social determinants in precision nutrition
| 1. Lack of consensus on the definition of social determinants (213) and their metrics |
| 2. Complexity of interactions across social factors requiring advanced analytical technologies |
| 3. Limited data from diverse and vulnerable populations |
| 4. Lack of understanding of mechanistic pathways and of the individual responses or embodiment of social factors |
| 5. Inattention to protective social factors such as social support, resilience, and optimism |
| 6. Unknown potential for sustainability, outreach, and implementation of interventions that account for social determinants |
| 7. Lack of approaches and techniques that fill data gaps and rank the most influential measurements in ways that are transparent, fair, and free from errors |
| 8. Dearth of technologies and methods that link different scales that affect precision nutrition (e.g., biology, behavior, social networks, environment, policy, economics, culture) |
| 9. Lack of studies that use systems approaches and methods to better elucidate how social determinates may affect and be affected by nutrition |
Differences in body weight as a source of individual response to diet
One important aspect of precision nutrition is to understand the factors that govern an individual's response to a weight-loss intervention. This is particularly relevant given the challenge of overweight and obesity in the United States. In addition to genetic and metabolic characteristics, tailoring specific dietary patterns to individual needs may help increase long-term adherence to weight-loss programs. This will, in turn, produce lasting weight management and aid in the prevention and treatment of obesity-related comorbidities, such as insulin resistance and dyslipidemia. Specific characteristics that should be considered include genetics, race, ethnicity, sex, menopausal status, level of glycemic control, disease history, gut microbial composition, food and taste preferences, eating behaviors (e.g., dietary restraint), and level of physical activity. Well-designed trials, such as the PREDICT-1 study (214), will be needed to clarify individual differences in responses to diet. When sufficient data are available in this area, it may be possible to create an online tool to help clinicians quickly identify what diets would work for certain individuals based on their unique characteristics. These tools could greatly augment a patient's success with their prescribed weight-loss intervention and other health outcomes (215).
Opportunities exist to close these gaps, including utilizing cutting-edge tools for data collection and developing databases to collect, share, and harmonize data. Recommendations and resources are available from several agencies on how to conduct research and intervene on social factors for optimal precision nutrition strategies. It is thus essential for social determinants of health to be prioritized in research, education, and policies relevant to precision nutrition.
The Need for New Technology and Computer-Aided Approaches to Better Understand the Complex Systems Involved
The third session of the workshop focused on analytical approaches in precision nutrition; application of knowledge to benefit individuals; ethical and legal challenges in study design, data collection, and implementation; and the need to train scientists in computational methodology, data science, systems science, machine learning, artificial intelligence (AI), and data infrastructure. As described earlier, achieving precision nutrition entails understanding and addressing the complex systems of factors that affect and may be affected by a person's diet and nutritional health (216). Progress will depend upon the development and integration of novel computational methods, tools, and approaches. While direct cause-and-effect relations may be easier to elucidate, unaided scientists can struggle with following secondary, tertiary, and other indirect effects, especially when time elapses between the initiation of the cause and the surfacing of the effects (217, 218). Often, the impact of a factor may not manifest for years, and the specific importance of such factors can differ between individuals. In addition, numerous feedback loops and dynamic factors that change over time can make relations even more challenging to disentangle.
Our society is at a key inflection point when it comes to the use of computer-aided analytics
Computer-aided methods, approaches, and tools have transformed a number of other disciplines, industries, and sectors (219–222). For example, meteorology relied considerably on direct observation and inaccurate conjecture before the advent of such methods. Imagine how difficult it was to anticipate what the weather might be beyond the next several hours. Nowadays, weather maps help bring together very disparate data streams that allow decision makers to better understand and address complex weather streams and to prepare better for more tailored responses to the weather. These weather maps are essentially visualizations of computer-generated information (223). People across the United States do not all have to remain prepared for rain, tornados, hurricanes, snow, and other inclement weather all the time. Instead, there are more precise preparations for and responses to the weather in different parts of the country.
Similarly, computer-aided methods, approaches, and tools can transform and advance the field of precision nutrition. Computer-aided methods include AI methods. The Encyclopedia Britannica (224) defines AI as “the ability of a digital computer or computer-controlled robot to perform tasks commonly associated with intelligent beings,” and Merriam-Webster (225) defines AI as “1) A branch of computer science dealing with intelligent behavior in computers and 2) The capability of a machine to imitate intelligent human behavior.” As can be seen by these definitions, AI is a very broad umbrella term encompassing any use of computers or computer-driven technology to perform tasks that intelligent beings would typically perform. This can range from sorting information in ways that are easier to digest, to performing tasks that human beings usually do to make decisions based on available information, to deriving insights from data. A computer-aided method becomes an AI method when it goes beyond the simple, straightforward, “mindless,” and predictable execution of tasks.
Included under AI are a range of different methods such as computer simulation modeling and machine learning (226). A computer simulation model is a computer-generated representation of a real-world situation. Machine learning is the use of algorithms to get computers to identify patterns in data and “learn” as humans do. This includes both traditional machine-learning algorithms that learn patterns and identify new relations from the data and thereby make predictions as well as AI capable of learning in new ways that mimic additional aspects of human intelligence. Machine learning can proceed with varying degrees of guidance from humans (227). Supervised learning is when an initial labeled training dataset is shown to the computer algorithm. A labeled training dataset is one that is organized and labeled in a way that shows the algorithm what, specifically, it should look for and assess, predetermining both the input and the output. Once the algorithm has learned from the training dataset, it can then be applied to other datasets. Unsupervised learning is when the algorithms train on unlabeled data, giving the algorithms the opportunity to figure things out. Computer-aided methods and technology such as machine learning can also facilitate precision nutrition by further elucidating the key factors and processes involved with precision nutrition. Since many of these may not be obvious and have clear, simple, and direct connections, and the clean and easily analyzable data may not be readily available, computer-aided methods such as machine learning can help organize and dig through available data to identify both the existence and the contribution of sometimes hidden factors that impact precision nutrition and diet-related diseases.
For example, Zeevi et al. (35) monitored glucose concentrations over the course of 1 wk in a cohort of 800 adults after measuring responsiveness to identical meals, and found high individual variability in response to the same meal and developed and validated a machine-learning algorithm integrating blood parameters, dietary habits, anthropometrics, physical activity, and gut microbiota in an attempt to predict individual responses to meals. Last, they conducted a dietary intervention to control glucose response, based on what they were learning from the algorithm, which resulted in significantly lower glucose responses after the meal as well as consistent changes to gut microbiota. In another study of 1002 healthy UK adults, Berry et al. (141) found that postprandial response to identical meals had large interindividual variability in both a clinical setting and at home. The research team found that factors such as the gut microbiome had a greater influence on postprandial lipidemia than meal macronutrients, but not on postprandial glycemia. In addition, genetic variation also had a modest impact on predictions. Additional studies have developed simulation models of metabolic events such as the glucose-insulin system following a meal, which can be used to understand and address how the body responds to meals among both healthy adults and individuals with diet-related health conditions (e.g., T2D) (228).
Computers have theoretical advantages over humans. They have the potential to perform calculations very quickly, holding, retrieving, and considering large amounts of information at a time, and remaining relatively “objective” when completing their tasks. At present, computers cannot do everything humans do, such as come up with original thoughts. Moreover, their execution of tasks depends heavily on how humans programmed and structured the tasks. Computer-aided approaches can help with all stages of achieving precision nutrition. This includes the following:
Determining what data are needed
Designing studies and data-collection activities
Helping collect and gather data and other information
Organizing, managing, and making data readily available
Analyzing and interpreting data and information
Communicating and disseminating insights
Implementing policies and interventions
Our society is at an inflection point. The past several decades have seen major growth in the capabilities and use of computers. Computational power has grown to the point where calculations can be done very rapidly, and a large amount of data can be readily stored and processed. Available infrastructure and platforms allow tools and information to be easily shared. There has been increasing acceptance and use of computer-driven approaches in health-related areas (229, 230). Additionally, the amount of data available has grown exponentially, rapidly exceeding the capacity of traditional methods to process and analyze the data (216).
The need for systems approaches when using computer-aided analytics for precision nutrition
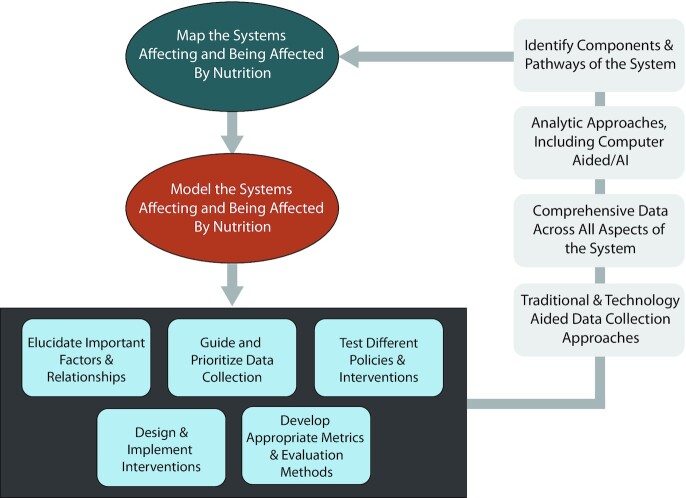
When computer-aided analytics do not account for or represent the actual systems it can result in “band aids” rather than sustainable solutions, overlooking indirect (e.g., secondary, tertiary, and beyond) effects of any situation or change, skewing choices among alternatives, unintended consequences, not collecting needed information and/or collecting superfluous or even misleading information, and wasting time, effort, and resources that, in many cases, end up not working, at best, and having negative effects at worst. Traditional approaches can overlook the complexities of the systems involved. For example, large-cohort studies that use multivariate regression to identify broad associations between a given nutritional factor and health outcomes may overlook differences in physiology, behaviors, and social determinants. Such approaches are also insufficient for understanding dietary factors or influences that are dynamic, such as exposures to and use of food environments. Just using computer-aided approaches to make such calculations faster may not really advance precision nutrition but rather deepen the current drawbacks of nutrition research. This is why computer-based approaches need to incorporate systems science and more systems approaches to nutrition research. It is important to remember that computer-aided approaches in and of themselves are neither bad nor good. Incorporating systems approaches means designing and implementing computer-based and other technology approaches in ways that account for and help characterize the complex systems involved (216, 231). Figure 2 shows how a systems approach can utilize technology to iteratively better understand these complex systems.
FIGURE 2.
How a systems approach can utilize technology to iteratively better understand complex systems. AI, artificial intelligence.
Systems-oriented computer-aided methods and technology can also facilitate precision nutrition by bringing different factors, processes, and components together to better understand and address the systems involved. Computer models can attempt to represent all of the key components and processes of a system and serve as virtual laboratories to test the effects of different changes in circumstances and interventions (232–237). These virtual laboratories have the ability to represent systems and complex interactions between dynamic variables that influence each other in complex ways, which can enable deeper understanding into processes and causal pathways that affect diet and related diseases (238). A “Virtual Infant” agent-based model representing infant–caregiver pairs has been developed that exemplifies this type of virtual laboratory. This agent-based model allowed virtual caregivers to feed virtual infants each day according to major feeding guidelines. The model simulated the development of the infants from birth to 6 mo. These simulations identified several scenarios where caregivers followed the guidelines, but infants still became overweight/with obesity by 6 mo even when caregivers adjusted feeding based on the infant's weight. This study exemplifies how one-size-fits-all nutritional guidelines might not result in the best health outcomes for all (184).
Similarly, 3 agent-based models of Baltimore, San Francisco, and Philadelphia were developed to evaluate the potential impact of implementing sugar-sweetened beverage warning labels in the different cities (236). Detailed representations of each city's layouts, including household, school, grocery, corner store, and restaurant locations, as well as detailed representations for each person, including behaviors, movements, clinical status, and physiology, were brought together using agent-based models. New technology can help with the implementation and adoption of prescribed precision nutrition behaviors as well. AI approaches can help identify barriers and facilitators of precision nutrition adoption. Systems modeling can show what adoption may be under different circumstances and test various policies and interventions. And sensors such as apps and wearables that monitor eating, movement, and mobility can provide real-time prompts, deliver “just in time adaptive interventions,” and track the adoption and long-term maintenance of dietary changes. In a longer-term vision, approaches can be designed to leverage all of these technologies in an end-to-end AI-enabled systems approach that facilitates improved adoption of precision nutrition interventions.
The need for a new systems-oriented data ecosystem
One way that new computer-aided methods and technology can bring the systems approaches needed to precision nutrition is by gathering and assessing data on different and often neglected parts of the system. For example, eating is a social practice, and what people eat is influenced by the people they eat with, as well as the family, friends, and community members who make up their social network (239, 240). These social ties influence eating through many socially influenced mechanisms, such as mimicry and normative influence, and by providing social support and social capital that can impact food access and consumption. Such information is rarely captured in nutrition research. New opportunities exist for using wearable devices, ecological momentary assessments, and digital traces of social phenomena to more precisely monitor social exposures and influences in context, and in real time, and to better understand the unique and varied situations that influence a person's food intake and nutritional health. These insights may inform intervention strategies that leverage, or even alter, social network phenomena (e.g., group-based interventions, targeting key opinion leaders and influencers) as part of efforts that promote the adoption of tailored dietary recommendations.
To realize the promise of precision medicine, it is also necessary that the data is AI/machine learning ready. AI/machine learning–ready data include large repositories of patient/human data, which are privacy protected, available to all, and serve as a standard dataset for comparison across studies. They also need to contain data pertaining to a wide variety of health and disease states, from a wide variety of sources (multi-omics, clinical, imaging, wearables) over the life course and before, during, and after episodes of care. Further, data ought to provide linkage across many types of data within the same subjects, utilize common data formats, and be sourced ethically and with input from communities participating in research (i.e., those whose data are being utilized for research purposes). Historically, data have been collected in a more ad hoc way without first determining what data need to be collected to account for all aspects of the system. A systems approach to data collection starts by mapping out all components, processes, relations, and mechanisms in a system, and using the map to determine where data collection is actually needed.
The need to include and incorporate more diverse populations
New technology can also facilitate a better understanding of neglected, disadvantaged, and vulnerable populations. For example, exposure to multiple risky social determinants of health, such as poverty and food insecurity, can limit individuals’ ability to eat recommended healthy foods (241, 242), as well as encompass exposures to stressors that have “under the skin” effects, such as increased inflammation, which may play an important role in physiological processes relevant to nutritional health (243). Conducting and implementing precision nutrition science without a central focus on the translation of this science, by considering the socio-ecological and economic challenges that vulnerable populations face, could further exacerbate health disparities. Specifically, people with few barriers to eating well will be better able to adopt new and tailored information to improve their diet and health. On the other hand, people facing socio-ecological barriers—including low incomes, food insecurity, and discrimination—and community divestment may not be able to change their diet, unless these specific barriers are addressed.
For example, recent research has begun to use population-scale mobility data passively collected from smartphones and novel data analytic techniques to study the food environments that large, diverse urban populations are exposed to throughout their day-to-day experiences (244). Early findings have uncovered that lower-income populations not only are more likely to live in areas surrounded by low nutritional quality food environments than richer populations, such as those with much more fast food than other types of prepared or fresh food, but are also more likely to be surrounded by these types of low-quality food environments when at work or conducting other daily activities. Achievable personalized nutrition interventions will need to take these types of barriers to access of food of higher nutritional quality into account.
The need to translate precision nutrition and the use of computer-aided analytics to the “real world”
Translating precision nutrition findings and recommendations to the “real world” is important yet challenging. Determining how nutritional recommendations can be better tailored to individuals or different groups of people is not the same as implementing such recommendations. Translating scientific findings into policies and practice is not trivial. Historically, even the translation of fairly simple dietary recommendations (e.g., consuming a variety of fruits and vegetables) has not been successful, with the majority of the population not following this nutritional guidance, and vulnerable groups having even lower adoption. We present several considerations and approaches that may help move precision nutrition from research to practice.
Scalability and sustainability are 2 closely related concepts that are critical for practitioners (245). Likelihood scenarios need to be translated into practical solutions that can be applied to real-world challenges and implemented with scalability and sustainability. Translation of likelihood scenarios into action addresses bridging the gap between the availability of systems models and their uses (246). Bridging research, education, policy, and practice can be accomplished using a variety of approaches. For example, awareness and understanding of how systems work can be accomplished through training and education or through the use of participatory approaches to model building where the users or decision makers can see the impact of certain strategies on outcomes or identify where unintended consequences may surface (229). From a translation perspective, moving evidence into practice demands a framework or model that leverages system drivers, deep knowledge, and expertise that bridges research and practice, and the a priori inclusion of user and stakeholder perspectives (229, 246, 247).
Computing platforms and tools need to provide access to conducting research on AI/machine learning datasets’ computing power, analytic tools, algorithms, visualizations, and secure electronic storage for data, data derivatives, visualizations, results, and other related research products. The platforms should foster collaborative research among interdisciplinary teams through enhanced discovery of one another's work and through direct support for shared pipelines, workflows, shared standards, etc. In addition, all tools need to facilitate transparent, reproducible, shareable research practices, such as through virtual machines that save workflows and analytic plans as executed. To foster transparent collaboration they should provide access to algorithms for reuse/adaptation, reproducibility, and transparency and facilitate dissemination of research findings.
Translation and dissemination frameworks need to go beyond publications of research findings into initial implementation efforts, include pragmatic evaluation, iterate, and adapt to local needs and demands, and scale-up to broader implementations that include scaling-up to different audiences, settings, and population sizes. To date, much of translation, dissemination, and implementation research has focused on initial uptake by early adopters of 1 health intervention at a time. Efforts to successfully scale and sustain must optimize context. Translation and dissemination of evidence-based interventions must be able to vary by context because few such efforts can be implemented according to identical protocols, resources, or levels of expertise (248–250). In fact, the need to recognize that things vary by context is so important that explicit efforts have focused on evidence-based principles that can be used in the context of evidence-informed decision making (251), a concept that appreciates that decisions to be made are not only based on research but also need to consider other variables such as political, organizational, or community health factors (251, 252).
Experts are required to interpret, understand, and disseminate the knowledge generated by research. Such expertise comes from multiple disciplines, including the fields of medicine, health, technology, and engineering, and provides deep insights into systems science, AI, and machine learning, as well as areas critical to successful translation into practice, such as human-centered design and health communications. Stakeholders’ perspectives need to be surfaced, considered, valued, and acted upon for translation to occur so that dissemination and, ultimately, implementation may successfully bring evidence-based solutions into practice. Stakeholders’ perspectives may, at times, be similar or vastly different because different people perceive and experience the world in different ways. Furthermore, any given stakeholder may hold various perspectives on the same system (253). Perspectives related to the implementation of systems epidemiology models should be gleaned from patients, providers, health care providers, care teams, payers, policymakers, caregivers, and community agencies. In addition, points of view should consider the biological, social, financial, behavioral, and political context.
The interpretation and translation of systems science approaches to precision nutrition call for an appreciation of the physical, social, and economic environments that modify the outcomes. There is a need to observe and summarize the strength of evidence of effectiveness and adapt such insights to the marketplace using scalable and sustainable solutions, including business plans that allow for sustained implementation. Engagement of leadership from across multiple sectors in an ongoing dialogue could facilitate the creation of models that reflect an alternative way of thinking designed to simultaneously benefit society and business while mobilizing resources and activating funding mechanisms from federal and private sources (254–259).
Based on the above, and in alignment with previously identified considerations (253), several public health–related recommendations for moving research findings toward practical implementation may be applied to precision nutrition. First, the ability to leverage existing system drivers will provide opportunities to advance translation, dissemination, and implementation efforts. This includes understanding the processes of adoption, implementation, and sustainability of new initiatives within organizations and institutions. Second is the application of pragmatic evaluation. Newly implemented programs or interventions should be assessed according to principles of evaluability that allow for conclusions to be made about their performance in the field and generate options for improvement (260). Furthermore, it is recommended to use existing measures to document performance as a cornerstone of an iterative learning approach. Third is the use of an explanatory, process, and outcome framework for translation and implementation. This will allow for moving beyond reporting on effectiveness and include descriptions and explanations of why things worked the way they did and how it was done. This approach should include both qualitative and quantitative observations, thereby providing insights on outcomes, mechanisms of action, and processes deployed.
The need to address key ethical and legal challenges with new technology and precision nutrition
Precision nutrition presents an opportunity to shift health care from being reactive to being proactive by providing insight into the specific dietary needs of individuals and populations. But the field's promise depends on adequate attention to issues of informed consent, representative datasets, socio-culturally sensitive design, and equitable access to products. These considerations must be built into research and implementation, as opposed to retrofitted. Building these considerations into research will ensure that personal information is not used for unapproved or malicious purposes, that tailored dietary recommendations are accurate and helpful, and that such recommendations benefit the people in greatest need.
The capacity to assess risks and benefits and authorize action in accordance with one's values is a critical consideration for various aspects of precision nutrition. Basic respect for autonomy requires that we do not impose risk on individuals without their understanding and agreement. The AI systems on which precision nutrition will increasingly rely must be fed significant amounts of protected health information (PHI) and personally identifiable information (PII). It can be argued on privacy grounds that one's PHI/PII should not be accessed by others without permission, regardless of actual harm incurred. Data providers must also be aware of tangible harms—entities can use PHI/PII to raise insurance premiums and hackers can use it to obtain ransom or steal an identity. For those receiving precision nutrition recommendations, disclosure of limitations and risks is critical for informed decision making. As precision nutrition hangs adjacent to health research and health care, there will be ambiguity as to when practices fall under the auspices of the federal “common rule” and Health Insurance Portability and Accountability Act (HIPAA) privacy rule and are thus subject to legal informed consent and security requirements. There will also be ambiguity as to whether precision nutrition applications will be regulated by the US FDA through the Software as a Medical Device pathway, meeting the same approval obstacles faced by medical AI applications, or whether a different regulatory process will be required. One challenge will be striking a balance between having enough regulatory procedures and oversight in place to protect the population to maintain and not unduly impede the adoption of such computational approaches. Regardless of legal requirements, the actual risks should inform the development of measures necessary to protect and respect individuals.
Data used to train algorithms for precision nutrition must be adequately representative of diverse populations, and recommendations must adequately recognize socio-cultural elements of the diet. Findings of 1 precision nutrition study using a specific cohort may not necessarily be applicable to predict responsiveness in other cohorts. If training datasets are inaccurate or incomplete, regardless of algorithm quality, the output will be inapplicable or harmful to populations missing from or inaccurately represented by the training set. Poorly designed algorithm systems and services that conflate race with genetics, inappropriately narrow user options, or under- or over-account for nonbiological socio-cultural correlations can also lead to bias and harmful recommendations. Research and development teams have begun developing strategies and toolkits for assessing and mitigating bias in AI algorithms (261–264). Precision nutrition developers should build off these strategies, with significant attention to testing before deployment and monitoring after deployment.
Precision nutrition has the potential to help address social determinants of health that lead to racially and socioeconomically disparate health outcomes. However, without subsidization or sponsorship, the whole field risks becoming a “boutique” service available only to those able to pay. To ensure populations in greatest need benefit, precision nutrition must be 1) accurately grounded in scientific research and sound algorithm development, 2) actionable based on available resources, and 3) supported by appropriately trained experts. Diverse populations must be involved in development, and design must keep diverse populations in mind.
The need to train a new generation familiar with computer-aided analytics and precision nutrition
The precipitous rise in the volume, velocity, and variety of available data and the corresponding arrival of Big Data analytics have created a new landscape for health research (265, 266) and make the vision of precision nutrition possible. Yet, precision nutrition can only be realized if the complex interplay of factors that influence nutrition and health outcomes are understood. These factors are present at multiple levels of influence: 1) external context, such as environment (including but not limited to food environment and opportunities for physical activity), policy, culture; 2) individual behavioral context such as dietary habits, physical activity; 3) clinical context, including current health status, treatments, and health history; and 4) below the skin contexts, such as genetic make-up, microbiome composition, metabolic factors. Understanding the joint impact of these factors is challenging because the sheer number of factors is large (i.e., high dimensionality) and compounded by the presence of bidirectional and nonlinear relations and moderated by the temporality of contexts (critical periods of exposure, timing, periodicity, duration, frequency).
Understanding how the entire web of interacting factors would play out over all health and disease topics at once is far too ambitious of a goal. A more feasible approach would be assembling teams of interdisciplinary scientists, as indicated by the specialized questions within the field. These teams will require a wide breadth and depth of collective domain expertise spanning across nutrition science, biomedical science, and behavioral science, but also core competencies in computational methodology, data science, systems science, machine learning, AI, and data infrastructure.
To properly examine interdisciplinary questions with complex and voluminous data, the members of scientific teams must possess core knowledge and competencies in data analytics; data infrastructure; data sharing; Findable, Accessible, Interoperable, and Reusable (FAIR) data principles (267); and algorithmic bias [e.g., detection, mitigation (268)], with additional understanding of statistical analysis, database design, technical coding skills, and data visualization along with relevant biomedical or behavioral science domain knowledge. Moreover, these competencies must be taught through a lens of equity, inclusion, and human rights (269), such as the Framework on Integrating Health Equity and Racial Justice into the AI development lifecycle (270).
To meet this charge, at least 2 types of training are needed. On the one hand, we need to retrain the existing investigative workforce not only to work in interdisciplinary teams but also to learn core competencies in data science. On the other hand, we need to reimagine how we currently train the next generation of scientists to incorporate interdisciplinary and data science competencies into their main disciplinary degree programs. To be successful, this cross-disciplinary training must include experiential, project-based learning. The authors offer 2 models of training from their own experience that incorporate this approach. The Institute for Systems Science in Health (ISSH) was an intensive, week-long summer course sponsored by the NIH Office of Behavioral and Social Sciences Research (OBSSR) from 2009 to 2012 (271). The ISSH was intentionally designed to provide an incubator space for luminaries in systems science to interact with public health researchers with applied research questions that could be addressed with systems science methodologies. Each year, ∼45 early-career and established investigators received hands-on training in 1 of 3 methods (agent-based modeling, system dynamics modeling, and network analysis) through a week-long in-residence training course. This type of intensive short-term, face-to-face approach could easily be adapted to the training needs of precision nutrition.
A second example of experiential project-based training is a predoctoral training program established in 2020 by OBSSR, the Training in Advanced Data Analytics for Behavioral and Social Sciences Research (TADA-BSSR) Program [NIH, 2019: RFA-OD-19–011 (272)]. The aim was to create new predoctoral programs to integrate innovative computational and data science analytic approaches directly into doctoral training. The program is offered at 8 universities (273). While this program was focused solely for OBSSR doctoral students, the principles and approach should be readily translatable to other disciplines involved in biomedical and health research.
Providing training in the core competencies in modern data science and opportunities for researchers at all career levels to apply their biomedical domain knowledge in the context of large interdisciplinary scientific teams will be key for the future precision nutrition scientific workforce.
Summary
Table 3 presents the many research opportunities that were identified from the workshop's presentations, discussions, and participant comments.
TABLE 3.
Research needs and future directions1
| Study design needs |
| Well-controlled intervention studies are needed to address individual differences in response to dietary exposures, food bioactives, and dietary patterns, including timing, duration, and dose-response. Studies specifically designed to target high-risk populations for prevalent health conditions (obesity, CVD, T2D, cancer, cognitive decline, and AD) are needed. Moreover, it is essential to determine the applicability of the findings to real-world settings. |
| Develop and test novel intervention strategies to help people change their dietary intake patterns over the long term and test if changing one's diet significantly affects disease risk and outcome. Considerable efforts are needed to help people 1) know what their dietary pattern is, 2) effectively modify their dietary pattern and be more “adherent” to a healthier diet, and 3) maintain these dietary and behavioral changes over time, including over a lifetime. |
| Studies that account for factors that cross all the different relevant scales (e.g., genetics, physiology, behavior, social networks, environment, economics). |
| Studies that utilize systems approaches and methods (e.g., maps and models) that can help better elucidate and bring together different components, factors, and mechanisms. |
| Hybrid approaches that combine different types of study design approaches (e.g., integrating systems models with intervention studies) to work synergistically. |
| Technologies and methods |
| Develop and validate accurate and precise objective measures of dietary intake, including real-time monitoring of food intake, postprandial response, and noninvasive biological responses. |
| Develop tools and methods to standardize, harmonize and improve interoperability of nutrition and food data. |
| Develop robust methods to integrate data from the genome, epigenome, microbiome, metabolome, and the exposome (i.e., single or multi-nutrient diet components, dose and timing of dietary modulations, and health behaviors (e.g., physical activity and sleep) into the precision nutrition framework. |
| Develop standardized and harmonized study procedures and data collection to control for and/or at least assess factors that can influence precision nutrition outcomes, including sleep and circadian biology (274). |
| Identify biomarkers for diet-related cancers and CVD is prioritized to more quickly elucidate the underlying basis of interindividual variability in diet and disease risks. |
| New methods and technologies to extract insights from existing data and sources (e.g., natural language-processing techniques to mine text for information). |
| Develop methods to extract and collapse larger data sources, including Big Data sources, into more refined datasets in ways that do not introduce bias. |
| Develop methods and tools to fill in missing data in ways that do not introduce bias. |
| Develop new AI/ML algorithms that can draw insights from datasets in ways that do not introduce bias. |
| Develop systems modeling methods that can better represent the actual mechanisms that affect and are affected by nutrition. |
| Develop mathematical and computational methods that help cross different scales (e.g., genetics, physiology, behavior, social networks, environment, economics). |
| Knowledge gaps |
| Develop more in-depth and precise knowledge of foods, food composition and groups and eating patterns, and related biomarkers. |
| Identify individual nutrigenomic/behavioral/lifestyle differences in chronic diet-related disease (e.g., CVD, neurodegenerative disease, cancer, T2D) and risk factors in order to personalize approaches for primary and/or secondary prevention of disease over the life course. |
| Collect data using objective measures of dietary intake episodically and prospectively over longer periods of time to learn if dietary patterns among the same individuals are reliable/repeatable and to what extent changes in dietary patterns affect disease risk and outcome. |
| Determine the predictive role of metabolomics and microbiome data in precision nutrition and chronic disease interrelations. |
| Determine the contribution and mechanisms of sleep and circadian effects in precision nutrition research and interventions based on chronobiological insights. |
| Quantify the effect of food policy, the food environment, socioeconomic and other personal factors, and industry on peoples’ dietary intake. Identify ways to change policy, the food environment, and industry to improve people's diets and presumably their health. |
| Better understand how complex systems are involved, affect, and are affected by nutrition. |
| Needs related to training in precision nutrition |
| Fill gaps in the implementation and dissemination of scientific research for evidence-based precision nutrition strategies and medical nutrition to reduce chronic diseases. |
| Develop a diverse workforce that has training in AI/nutrition science. |
| Develop a new generation of truly interdisciplinary researchers able to cross different content areas of nutrition and different new methodological areas such as mathematical and computer modeling and other types of AI. |
| Train more people well versed in systems, mathematical, and computational methods. |
| Train people to better recognize and address bias. |
| Train people to be better versed in social determinants. |
AD, Alzheimer disease; AI, artificial intelligence; CVD, cardiovascular disease; ML, machine learning; T2D, type 2 diabetes.
Advancements in precision nutrition will mean better tailoring of dietary interventions to different people's circumstances and situations. It is not yet clear how specific this tailoring may need to be, as precision nutrition does not necessarily mean personalized nutrition when each person should get dietary recommendations specific to himself or herself. As discussed above, much consideration and research will be needed before more precise nutrition recommendations can be achieved. This includes better understanding and accounting for variables such as age, sex, ethnicity, medical history, genetics, and social and environmental factors. The advent of new methods and technologies and the availability of considerably more data bring tremendous opportunity. However, we must proceed with appropriate levels of caution and make sure that the variables listed above are all appropriately considered, and systems approaches, and methods, are incorporated. Last, it will be important to develop and train an expanded workforce with the goal toward reducing health disparities and improving the precision of nutritional advice for all Americans.
Supplementary Material
Acknowledgements
The authors extend their appreciation to the members of the NIH organizing committee for the workshop: Kimberly Barch [Office of Nutrition Research (ONR)], Christopher Lynch, PhD (ONR), Charlotte A Pratt, PhD, RD [National Heart, Lung, and Blood Institute (NHLBI)]; Andrew Bremer, MD, PhD [Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)]; Allison Brown, PhD, MS (NHLBI); Jill Reedy, PhD, MPH, RD [National Cancer Institute (NCI)]; Karen Regan, MS, RD (ONR); Scarlet Shi, PhD (NHLBI); Pothur Srinivas, PhD, MPH (NCI) and Ashley Vargas, PhD, MPH, RDN (NICHD). The following members of the Public Health Informatics, Computational, and Operations Research (PHICOR) and Center for Advanced Technology and Communication in Health (CATCH) assisted with the preparation of this manuscript: Marie C Ferguson, MSPH; Kelly J O'Shea, BSFS; Sarah M Bartsch, MPH; and Patrick T Wedlock, MSPH.
The authors’ responsibilities were as follows—All authors: are responsible for the design, writing, and final content of the manuscript, and read and approved the final manuscript. The authors report no conflicts of interest.
Notes
The workshop was co-sponsored by the National Institutes of Health (NIH) Office of Nutrition Research (ONR); the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); the National Heart, Lung, and Blood Institute (NHLBI); and the NIH Office of Disease Prevention (ODP). This was also supported by the Stanford BSSR Pre-Doctoral Training Program at the Intersection of Data Sciences with Behavioral, Social, and Population Health Research, funded by NHLBI via grant 5T32HL151323-02 and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) via grant U01HD08686. This work was also supported by the NIH Common Fund and the National Center for Advancing Translational Sciences of the National Institutes of Health via award U54TR004279.
Supplemental Table 1 is available from the “Supplementary data” link in the online posting of the article at https://academic.oup.com/ajcn/.
EJP is a member of the Journal's Editorial Board.
Abbreviations used: AD, Alzheimer disease; AI, artificial intelligence; CNS, central nervous system; COVID-19, coronavirus disease 2019; CVD, cardiovascular disease; DGA, Dietary Guidelines for Americans; DIT, diet-induced thermogenesis; ISSH, Institute for Systems Science in Health; MedDiet, Mediterranean-style dietary pattern; MTT, meal tolerance test; OBSSR, NIH Office of Behavioral and Social Sciences Research; OGTT, oral-glucose-tolerance test; PHI, protected health information; PII, personally identifiable information; PPGR, postprandial glycemic response; PPL, postprandial lipemia; RCT, randomized controlled trial; TG, triacylglycerol; T2D, type 2 diabetes.
Contributor Information
Bruce Y Lee, Health Policy and Management, City University of New York Graduate School of Public Health and Health Policy, New York, NY, USA.
José M Ordovás, USDA-Human Nutrition Research Center on Aging at Tufts University, Boston, MA, USA.
Elizabeth J Parks, Nutrition and Exercise Physiology, University of Missouri School of Medicine, MO, USA.
Cheryl A M Anderson, Public Health, University of California, San Diego, San Diego, CA, USA.
Albert-László Barabási, Network Science Institute and Department of Physics, Northeastern University, Boston, MA, USA.
Steven K Clinton, The Ohio State University, Columbus, OH, USA.
Kayla de la Haye, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Valerie B Duffy, Allied Health Sciences, University of Connecticut, Storrs, CT, USA.
Paul W Franks, Novo Nordisk Foundation, Hellerup, Denmark, Copenhagen, Denmark, and Lund University Diabetes Center, Sweden; The Lund University Diabetes Center, Malmo, SwedenInsert Affiliation Text Here.
Elizabeth M Ginexi, National Institutes of Health, Office of Behavioral and Social Sciences Research, Bethesda, MD, USA.
Kristian J Hammond, Computer Science, Northwestern University McCormick School of Engineering, IL, USA.
Erin C Hanlon, Department of Medicine, The University of Chicago, Chicago, IL, USA.
Michael Hittle, Epidemiology and Clinical Research, Stanford University, Stanford, CA, USA.
Emily Ho, Public Health and Human Sciences, Linus Pauling Institute, Oregon State University, Corvallis, OR, USA.
Abigail L Horn, Information Sciences Institute, Viterbi School of Engineering, University of Southern California, Los Angeles, CA, USA.
Richard S Isaacson, Neurology, Weill Cornell Medical College, New York, NY, USA.
Patricia L Mabry, HealthPartners Institute, Bloomington, MN, USA.
Susan Malone, Rory Meyers College of Nursing, New York University, New York, NY, USA.
Corby K Martin, Ingestive Behavior Laboratory, Pennington Biomedical Research Center, Baton Rouge, LA, USA.
Josiemer Mattei, Nutrition, Harvard TH Chan School of Public Health, Boston, MA, USA.
Simin Nikbin Meydani, USDA-Human Nutrition Research Center on Aging at Tufts University, Boston, MA, USA.
Lorene M Nelson, Epidemiology and Population Health, Stanford University, Stanford, CA, USA.
Marian L Neuhouser, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Brendan Parent, Grossman School of Medicine, New York University, New York, NY, USA.
Nicolaas P Pronk, HealthPartners Institute, Bloomington, MN, USA.
Helen M Roche, UCD Conway Institute, School of Public Health, Physiotherapy, and Sports Science, University College Dublin, Dublin, Ireland.
Suchi Saria, Johns Hopkins University, Baltimore, MD, USA.
Frank A J L Scheer, Brigham and Women's Hospital, Boston, MA, USA; Medicine and Neurology, Harvard Medical School, Boston, MA, USA.
Eran Segal, Computer Science and Applied Math, Weizmann Institute of Science, Rehovot, Israel.
Mary Ann Sevick, Grossman School of Medicine, New York University, New York, NY, USA.
Tim D Spector, Twin Research and Genetic Epidemiology, King's College London, London, United Kingdom.
Linda Van Horn, Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Krista A Varady, Kinesiology and Nutrition, University of Illinois at Chicago, Chicago, IL, USA.
Venkata Saroja Voruganti, Nutrition and Nutrition Research Institute, Gillings School of Public Health, The University of North Carolina, Chapel Hill, NC, USA.
Marie F Martinez, Health Policy and Management, City University of New York Graduate School of Public Health and Health Policy, New York, NY, USA.
References
- 1. US Burden of Disease Collaborators, Mokdad AH, Ballestros K, Echko M, Glenn S, Olsen HE, Mullany Eet al. The state of US health, 1990–2016: burden of diseases, injuries, and risk factors among US states. JAMA. 2018;319(14):1444–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Phillips JA. Dietary guidelines for Americans, 2020–2025. Workplace Health Safety. 2021;69(8):395. [DOI] [PubMed] [Google Scholar]
- 3. Maruvada P, Lampe JW, Wishart DS, Barupal D, Chester DN, Dodd Det al. Perspective: dietary biomarkers of intake and exposure—exploration with omics approaches. Adv Nutr. 2020;11(2):200–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lewontin R. The genetics of human diversity. New York (NY): Freeman Press; 1980. [Google Scholar]
- 5. National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Food and Nutrition Board; Committee to Review the Process to Update the Dietary Guidelines for Americans . Redesigning the process for establishing the Dietary Guidelines for Americans. Washington (DC): National Academies Press (US); 2017. Available from: https://www.ncbi.nlm.nih.gov/books/NBK469837/ [Google Scholar]
- 6. Corella D, Coltell O, Mattingley G, Sorli JV, Ordovas JM. Utilizing nutritional genomics to tailor diets for the prevention of cardiovascular disease: a guide for upcoming studies and implementations. Expert Rev Mol Diagn. 2017;17(5):495–513. [DOI] [PubMed] [Google Scholar]
- 7. Parnell LD, Blokker BA, Dashti HS, Nesbeth PD, Cooper BE, Ma Yet al. CardioGxE, a catalog of gene-environment interactions for cardiometabolic traits. BioData Mining. 2014;7(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bayer S, Winkler V, Hauner H, Holzapfel C. Associations between genotype–diet interactions and weight loss—a systematic review. Nutrients. 2020;12(9):2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holzapfel C, Sag S, Graf-Schindler J, Fischer M, Drabsch T, Illig Tet al. Association between single nucleotide polymorphisms and weight reduction in behavioural interventions—a pooled analysis. Nutrients. 2021;13(3):819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Celis-Morales C, Livingstone KM, Marsaux CF, Macready AL, Fallaize R, O'Donovan CBet al. Effect of personalized nutrition on health-related behaviour change: evidence from the Food4Me European randomized controlled trial. Int J Epidemiol. 2017;46(2):578–88. [DOI] [PubMed] [Google Scholar]
- 11. Livingstone KM, Celis-Morales C, Navas-Carretero S, San-Cristobal R, Forster H, Woolhead Cet al. Personalised nutrition advice reduces intake of discretionary foods and beverages: findings from the Food4Me randomised controlled trial. Int J Behav Nutr Phys Activity. 2021;18(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arkadianos I, Valdes AM, Marinos E, Florou A, Gill RD, Grimaldi KA. Improved weight management using genetic information to personalize a calorie controlled diet. Nutr J. 2007;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horne J, Gilliland J, O'Connor C, Seabrook J, Hannaberg P, Madill J. Study protocol of a pragmatic randomized controlled trial incorporated into the Group Lifestyle BalanceTM program: the nutrigenomics, overweight/obesity and weight management trial (the NOW trial). BMC Public Health. 2019;19(1):310. doi: 10.1186/s12889-019-6621-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nishigaki M, Tokunaga-Nakawatase Y, Nishida J, Taru C, Miyawaki I, Sanada Het al. Randomized controlled trial of the effectiveness of genetic counseling and a distance, computer-based, lifestyle intervention program for adult offspring of patients with type 2 diabetes: background, study protocol, and baseline patient characteristics. J Nutr Metab. 2012;2012:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nielsen D E, El-Sohemy A. A randomized trial of genetic information for personalized nutrition. Genes Nutr. 2012;7(4):559–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Hoogh IM, Winters BL, Nieman KM, Bijlsma S, Krone T, van den Broek TJet al. A novel personalized systems nutrition program improves dietary patterns, lifestyle behaviors and health-related outcomes: results from the Habit Study. Nutrients. 2021;13(6):1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rollo ME, Haslam RL, Collins CE. Impact on dietary intake of two levels of technology-assisted personalized nutrition: a randomized trial. Nutrients. 2020;12(11):3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoevenaars FPM, Berendsen CMM, Pasman WJ, van den Broek TJ, Barrat E, de Hoogh IMet al. Evaluation of food-intake behavior in a healthy population: personalized vs. one-size-fits-all. Nutrients. 2020;12(9):2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Al-Awadhi B, Fallaize R, Zenun Franco R, Hwang F, Lovegrove JA. Insights into the delivery of personalized nutrition: evidence from face-to-face and web-based dietary interventions. Front Nutr. 2020;7:570531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McBride CM, Bryan AD, Bray MS, Swan GE, Green ED. Health behavior change: can genomics improve behavioral adherence?. Am J Public Health. 2012;102(3):401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mustapa MAC, Amin L, Frewer LJ. Predictors of stakeholders' intention to adopt nutrigenomics. Genes Nutr. 2020;15(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li SX, Ye Z, Whelan K, Truby H. The effect of communicating the genetic risk of cardiometabolic disorders on motivation and actual engagement in preventative lifestyle modification and clinical outcome: a systematic review and meta-analysis of randomised controlled trials. Br J Nutr. 2016;116(5):924–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jinnette R, Narita A, Manning B, McNaughton SA, Mathers JC, Livingstone KM. Does personalized nutrition advice improve dietary intake in healthy adults? A systematic review of randomized controlled trials. Adv Nutr. 2021;12(3):657–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teasdale N, Elhussein A, Butcher F, Piernas C, Cowburn G, Hartmann-Boyce Jet al. Systematic review and meta-analysis of remotely delivered interventions using self-monitoring or tailored feedback to change dietary behavior. Am J Clin Nutr. 2018;107(2):247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milagro FI, Martínez JA. Epigenetics of obesity and weight loss. Endocrinol Nutr. 2013;60(Suppl 1):12–14. [DOI] [PubMed] [Google Scholar]
- 26. Ideraabdullah FY, Zeisel SH. Dietary modulation of the epigenome. Physiol Rev. 2018;98(2):667–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lai CQ, Wojczynski MK, Parnell LD, Hidalgo BA, Irvin MR, Aslibekyan Set al. Epigenome-wide association study of triglyceride postprandial responses to a high-fat dietary challenge. J Lipid Res. 2016;57(12):2200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wojczynski MK, Parnell LD, Pollin TI, Lai CQ, Feitosa MF, O'Connell JRet al. Genome-wide association study of triglyceride response to a high-fat meal among participants of the NHLBI Genetics of Lipid Lowering Drugs and Diet Network (GOLDN). Metabolism. 2015;64(10):1359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Y, Ordovas JM. The integration of epigenetics and genetics in nutrition research for CVD risk factors. Proc Nutr Soc. 2017;76(3):333–46. [DOI] [PubMed] [Google Scholar]
- 30. Lai CQ, Smith CE, Parnell LD, Lee YC, Corella D, Hopkins Pet al. Epigenomics and metabolomics reveal the mechanism of the APOA2-saturated fat intake interaction affecting obesity. Am J Clin Nutr. 2018;108(1):188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma Y, Smith CE, Lai CQ, Irvin MR, Parnell LD, Lee YCet al. The effects of omega-3 polyunsaturated fatty acids and genetic variants on methylation levels of the interleukin-6 gene promoter. Mol Nutr Food Res. 2016;60(2):410–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard Ret al. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang ZZ, Chen G, Hong Q, Huang S, Smith HM, Shah RDet al. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front Genet. 2019;10:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DAet al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27(2):321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger Aet al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]
- 36. Estruch R, Ros E, Salas-Salvadó J, Covas MI, Corella D, Arós Fet al. Primary prevention of cardiovascular disease with a Mediterranean diet supplemented with extra-virgin olive oil or nuts. N Engl J Med. 2018;378(25):e34. [DOI] [PubMed] [Google Scholar]
- 37. Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri Met al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69(7):1218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang DD, Nguyen LH, Li Y, Yan Y, Ma W, Rinott Eet al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat Med. 2021;27(2):333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nestel N, Hvass JD, Bahl MI, Hansen LH, Krych L, Nielsen DSet al. The gut microbiome and abiotic factors as potential determinants of postprandial glucose responses: a single-arm meal study. Front Nutr. 2021;7:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Othaim AA, Voreades N, Goodwin N, Curley J, Marasini D, Weir Tet al. Amounts and botanical diversity of dietary fruits and vegetables affect distinctly the human gut microbiome. Curr Dev Nutr. 2020;4(Suppl 2):1545. [Google Scholar]
- 41. van der Merwe M. Gut microbiome changes induced by a diet rich in fruits and vegetables. Int J Food Sci Nutr. 2021;72(5):665–9. [DOI] [PubMed] [Google Scholar]
- 42. Monnier L, Colette C, Owens D. The glycemic triumvirate and diabetic complications: is the whole greater than the sum of its component parts?. Diabetes Res Clin Pract. 2012;95(3):303–11. [DOI] [PubMed] [Google Scholar]
- 43.; Diabetes Control and Complications Trial Research Group, Nathan DM, Genuth S, Lachin J, Cleary P, Crofford Oet al. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993;329(14):977–86. [DOI] [PubMed] [Google Scholar]
- 44. Coutinho M, Gerstein HC, Wang Y, Yusuf S. The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes Care. 1999;22(2):233–40. [DOI] [PubMed] [Google Scholar]
- 45. Chiasson JL, Josse RG, Gomis R, Hanefeld M, Karasik A, Laakso M. Acarbose for prevention of type 2 diabetes mellitus: the STOP-NIDDM randomised trial. Lancet North Am Ed. 2002;359(9323):2072–7. [DOI] [PubMed] [Google Scholar]
- 46. Chiasson JL, Josse RG, Gomis R, Hanefeld M, Karasik A, Laakso M. Acarbose treatment and the risk of cardiovascular disease and hypertension in patients with impaired glucose tolerance: the STOP-NIDDM trial. JAMA. 2003;290(4):486–94. [DOI] [PubMed] [Google Scholar]
- 47. Heilbronn LK, Noakes M, Clifton PM. The effect of high- and low-glycemic index energy restricted diets on plasma lipid and glucose profiles in type 2 diabetic subjects with varying glycemic control. J Am Coll Nutr. 2002;21(2):120–7. [DOI] [PubMed] [Google Scholar]
- 48. Wolever TM, Gibbs AL, Mehling C, Chiasson JL, Connelly PW, Josse RGet al. The Canadian Trial of Carbohydrates in Diabetes (CCD), a 1-y controlled trial of low-glycemic-index dietary carbohydrate in type 2 diabetes: no effect on glycated hemoglobin but reduction in C-reactive protein. Am J Clin Nutr. 2008;87(1):114–25. [DOI] [PubMed] [Google Scholar]
- 49. Ma Y, Olendzki BC, Merriam PA, Chiriboga D E, Culver AL, Li Wet al. A randomized clinical trial comparing low-glycemic index versus ADA dietary education among individuals with type 2 diabetes. Nutrition. 2008;24(1):45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sacks FM, Carey VJ, Anderson CA, Miller ER 3rd, Copeland T, Charleston Jet al. Effects of high vs low glycemic index of dietary carbohydrate on cardiovascular disease risk factors and insulin sensitivity: the Omnicarb randomized clinical trial. JAMA. 2014;312(23):2531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jenkins DJ, Kendall CW, McKeown-Eyssen G, Josse RG, Silverberg J, Booth GLet al. Effect of a low-glycemic index or a high-cereal fiber diet on type 2 diabetes: a randomized trial. JAMA. 2008;300(23):2742–53. [DOI] [PubMed] [Google Scholar]
- 52. Reynolds A, Tekinkaya H, Venn B. The effect on day-long glycemia of consuming lower and higher glycemic index diets in people with type 2 diabetes: a randomized crossover study. J Diabetes Metab. 2014;5(9):1000436. [Google Scholar]
- 53. Silva FM, Kramer CK, Crispim D, Azevedo MJ. A high-glycemic index, low-fiber breakfast affects the postprandial plasma glucose, insulin, and ghrelin responses of patients with type 2 diabetes in a randomized clinical trial. J Nutr. 2015;145(4):736–41. [DOI] [PubMed] [Google Scholar]
- 54. Tay J, Luscombe-Marsh ND, Thompson CH, Noakes M, Buckley JD, Wittert GAet al. A very low-carbohydrate, low-saturated fat diet for type 2 diabetes management: a randomized trial. Diabetes Care. 2014;37(11):2909–18. [DOI] [PubMed] [Google Scholar]
- 55. Saslow LR, Kim S, Daubenmier JJ, Moskowitz JT, Phinney SD, Goldman Vet al. A randomized pilot trial of a moderate carbohydrate diet compared to a very low carbohydrate diet in overweight or obese individuals with type 2 diabetes mellitus or prediabetes. PLoS One. 2014;9(4):e91027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Andrews RC, Cooper AR, Montgomery AA, Norcross AJ, Peters TJ, Sharp DJet al. Diet or diet plus physical activity versus usual care in patients with newly diagnosed type 2 diabetes: the Early ACTID randomised controlled trial. Lancet North Am Ed. 2011;378(9786):129–39. [DOI] [PubMed] [Google Scholar]
- 57. Coppell K, Kataoka M, Williams S, Chisholm A, Vorgers S, Mann J. Nutritional intervention in patients with type 2 diabetes who are hyperglycaemic despite optimised drug treatment—Lifestyle Over and Above Drugs in Diabetes (LOADD) study: randomised controlled trial. BMJ. 2010;341:c3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mendes-Soares H, Raveh-Sadka T, Azulay S, Edens K, Ben-Shlomo Y, Cohen Yet al. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes. JAMA Network Open. 2019;2(2):e188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy Aet al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci. 2004;101(44):15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. [DOI] [PubMed] [Google Scholar]
- 61. Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza Oet al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514(7521):181–6. [DOI] [PubMed] [Google Scholar]
- 62. Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang Cet al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8(8):e71108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony Get al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. [DOI] [PubMed] [Google Scholar]
- 64. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg Bet al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. [DOI] [PubMed] [Google Scholar]
- 65. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang Fet al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- 66. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFet al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916.e7. e7. [DOI] [PubMed] [Google Scholar]
- 67. Mendes-Soares H, Raveh-Sadka T, Azulay S, Ben-Shlomo Y, Cohen Y, Ofek Tet al. Model of personalized postprandial glycemic response to food developed for an Israeli cohort predicts responses in Midwestern American individuals. Am J Clin Nutr. 2019;110(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Milner JA. Diet and cancer: facts and controversies. Nutr Cancer. 2006;56(2):216–24. [DOI] [PubMed] [Google Scholar]
- 69. Clinton SK, Giovannucci EL, Hursting SD. The World Cancer Research Fund/American Institute for Cancer Research third expert report on diet, nutrition, physical activity, and cancer: impact and future directions. J Nutr. 2020;150(4):663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Albanes D, Till C, Klein EA, Goodman PJ, Mondul AM, Weinstein SJet al. Plasma tocopherols and risk of prostate cancer in the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Cancer Prev Res. 2014;7(9):886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Albanes D, Heinonen OP, Huttunen JK, Taylor PR, Virtamo J, Edwards Bet al. Effects of alpha-tocopherol and beta-carotene supplements on cancer incidence in the Alpha-Tocopherol Beta-Carotene Cancer Prevention Study. Am J Clin Nutr. 1995;62(6):1427S–30S. [DOI] [PubMed] [Google Scholar]
- 72. Mayne ST, Playdon MC, Rock CL. Diet, nutrition, and cancer: past, present and future. Nat Rev Clin Oncol. 2016;13(8):504–15. [DOI] [PubMed] [Google Scholar]
- 73. Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17(3):297–303. [DOI] [PubMed] [Google Scholar]
- 74. Roychowdhury S, Chinnaiyan AM. Translating genomics for precision cancer medicine. Annu Rev Genomics Hum Genet. 2014;15:395–415. [DOI] [PubMed] [Google Scholar]
- 75. Theodoratou E, Timofeeva M, Li X, Meng X, Ioannidis JPA. Nature, nurture, and cancer risks: genetic and nutritional contributions to cancer. Annu Rev Nutr. 2017;37:(1):293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Figueiredo JC, Hsu L, Hutter CM, Lin Y, Campbell PT, Baron JAet al. Genome-wide diet-gene interaction analyses for risk of colorectal cancer. PLos Genet. 2014;10(4):e1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ding W, Zhou DL, Jiang X, Lu LS. Methionine synthase A2756G polymorphism and risk of colorectal adenoma and cancer: evidence based on 27 studies. PLoS One. 2013;8(4):e60508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan Det al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72(8):2036–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tse G, Eslick GD. Cruciferous vegetables and risk of colorectal neoplasms: a systematic review and meta-analysis. Nutr Cancer. 2014;66(1):128–39. [DOI] [PubMed] [Google Scholar]
- 80. Sellami M, Bragazzi NL. Nutrigenomics and breast cancer: state-of-art, future perspectives and insights for prevention. Nutrients. 2020;12(2):512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moran NE, Thomas-Ahner JM, Fleming JL, McElroy JP, Mehl R, Grainger EMet al. Single nucleotide polymorphisms in β-carotene oxygenase 1 are associated with plasma lycopene responses to a tomato-soy juice intervention in men with prostate cancer. J Nutr. 2019;149(3):381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kovalenko PL, Zhang Z, Yu J-G, Li Y, Clinton SK, Fleet JC. Dietary vitamin D and vitamin D receptor level modulate epithelial cell proliferation and apoptosis in the prostate. Cancer Prev Res. 2011;4(10):1617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tan H-L, Thomas-Ahner JM, Moran NE, Cooperstone JL, Erdman JW, Young GSet al. β-Carotene 9′,10′ oxygenase modulates the anticancer activity of dietary tomato or lycopene on prostate carcinogenesis in the TRAMP model. Cancer Prev Res. 2017;10(2):161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3(4):503–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Meeran SM, Ahmed A, Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenet. 2010;1(3-4):101–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17(5):363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr. 2009;139(12):2393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sapienza C, Issa JP. Diet, nutrition, and cancer epigenetics. Annu Rev Nutr. 2016;36:(1):665–81. [DOI] [PubMed] [Google Scholar]
- 89. Beaumont M, Portune KJ, Steuer N, Lan A, Cerrudo V, Audebert Met al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: a randomized, parallel, double-blind trial in overweight humans. Am J Clin Nutr. 2017;106(4):1005–19. [DOI] [PubMed] [Google Scholar]
- 90. O'Keefe S, Li J, Lahti L, Ou J, Carbonero F, Mohammed Ket al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6(1):6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shively CA, Register TC, Appt SE, Clarkson TB, Uberseder B, Clear KYet al. Consumption of Mediterranean versus Western diet leads to distinct mammary gland microbiome populations. Cell Rep. 2018;25(1):47–56, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang Xet al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–6. [DOI] [PubMed] [Google Scholar]
- 93. Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow STEet al. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018;2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sze MA, Schloss PD. Looking for a signal in the noise: revisiting obesity and the microbiome. Mbio. 2016;7(4):e01018–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Thingholm LB, Rühlemann MC, Koch M, Fuqua B, Laucke G, Boehm Ret al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019;26(2):252–264.e10. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman Ret al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau ALet al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chapkin RS, Navarro SL, Hullar MAJ, Lampe JW. Diet and gut microbes act coordinately to enhance programmed cell death and reduce colorectal cancer risk. Dig Dis Sci. 2020;65(3):840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hullar MA, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer Treat Res. 2014;159:377–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gerhauser C. Impact of dietary gut microbial metabolites on the epigenome. Philos Trans R Soc B Biol Sci. 2018;373(1748):20170359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Paul B, Barnes S, Demark-Wahnefried W, Morrow C, Salvador C, Skibola Cet al. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenet. 2015;7:(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Paul B, Royston KJ, Li Y, Stoll ML, Skibola CF, Wilson LSet al. Impact of genistein on the gut microbiome of humanized mice and its role in breast tumor inhibition. PLoS One. 2017;12(12):e0189756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Conaway CC, Getahun SM, Liebes LL, Pusateri DJ, Topham DK, Botero-Omary Met al. Disposition of glucosinolates and sulforaphane in humans after ingestion of steamed and fresh broccoli. Nutr Cancer. 2000;38(2):168–78. [DOI] [PubMed] [Google Scholar]
- 105. Tian S, Liu X, Lei P, Zhang X, Shan Y. Microbiota: a mediator to transform glucosinolate precursors in cruciferous vegetables to the active isothiocyanates. J Sci Food Agric. 2018;98(4):1255–60. [DOI] [PubMed] [Google Scholar]
- 106. Russo E, Nannini G, Dinu M, Pagliai G, Sofi F, Amedei A. Exploring the food-gut axis in immunotherapy response of cancer patients. World J Gastroenterol. 2020;26(33):4919–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77–91. [DOI] [PubMed] [Google Scholar]
- 108. Berkowitz CL, Mosconi L, Rahman A, Scheyer O, Hristov H, Isaacson RS. Clinical application of APOE in Alzheimer's prevention: a precision medicine approach. J Prev Alzheimers Dis. 2018;5(4):245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Norwitz NG, Saif N, Ariza IE, Isaacson RS. Precision nutrition for Alzheimer's prevention in apoE4 carriers. Nutrients. 2021;13(4):1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yassine HN, Cordova I, He X, Solomon V, Mack WJ, Harrington MGet al. Refining omega-3 supplementation trials in APOE4 carriers for dementia prevention. Alzheimers Dement. 2020;16(S5):e039029. [Google Scholar]
- 111. Osuntokun BO, Sahota A, Ogunniyi AO, Gureje O, Baiyewu O, Adeyinka Aet al. Lack of an association between apolipoprotein E epsilon 4 and Alzheimer's disease in elderly Nigerians. Ann Neurol. 1995;38(3):463–5. [DOI] [PubMed] [Google Scholar]
- 112. Kalaria RN, Ogeng'o JA, Patel NB, Sayi JG, Kitinya JN, Chande HMet al. Evaluation of risk factors for Alzheimer's disease in elderly East Africans. Brain Res Bull. 1997;44(5):573–7. [DOI] [PubMed] [Google Scholar]
- 113. Hendrie HC, Ogunniyi A, Hall KS, Baiyewu O, Unverzagt FW, Gureje Oet al. Incidence of dementia and Alzheimer disease in 2 communities: Yoruba residing in Ibadan, Nigeria, and African Americans residing in Indianapolis, Indiana. JAMA. 2001;285(6):739–47. [DOI] [PubMed] [Google Scholar]
- 114. Gurinovich A, Andersen SL, Puca A, Atzmon G, Barzilai N, Sebastiani P. Varying effects of APOE alleles on extreme longevity in European ethnicities. J Gerontol A. 2019;74(Suppl 1):S45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang Y, Jin X, Lutz MW, Ju SY, Liu K, Guo Get al. Interaction between APOE ε4 and dietary protein intake on cognitive decline: a longitudinal cohort study. Clin Nutr. 2021;40(5):2716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Arellanes IC, Choe N, Solomon V, He X, Kavin B, Martinez AEet al. Brain delivery of supplemental docosahexaenoic acid (DHA): a randomized placebo-controlled clinical trial. EBioMedicine. 2020;59:102883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Reddy PH, Manczak M, Yin X, Grady MC, Mitchell A, Tonk Set al. Protective effects of Indian spice curcumin against amyloid-β in Alzheimer's disease. J Alzheimers Dis. 2018;61(3):843–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vahid F, Zand H, Nosrat-Mirshekarlou E, Najafi R, Hekmatdoost A. The role dietary of bioactive compounds on the regulation of histone acetylases and deacetylases: a review. Gene. 2015;562(1):8–15. [DOI] [PubMed] [Google Scholar]
- 119. Spagnuolo C, Moccia S, Russo GL. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur J Med Chem. 2018;153:105–15. [DOI] [PubMed] [Google Scholar]
- 120. Isaacson RS, Hristov H, Saif N, Hackett K, Hendrix S, Melendez Jet al. Individualized clinical management of patients at risk for Alzheimer's dementia. Alzheimers Dement. 2019;15(12):1588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kowalski K, Mulak A. Brain-gut-microbiota axis in Alzheimer's disease. J Neurogastroenterol Motil. 2019;25(1):48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. 2015;28(2):203–9. [PMC free article] [PubMed] [Google Scholar]
- 123. Burokas A, Moloney RD, Dinan TG, Cryan JF. Microbiota regulation of the mammalian gut-brain axis. Adv Appl Microbiol. 2015;91:1–62. [DOI] [PubMed] [Google Scholar]
- 124. Petra AI, Panagiotidou S, Hatziagelaki E, Stewart JM, Conti P, Theoharides TC. Gut-microbiota-brain axis and its effect on neuropsychiatric disorders with suspected immune dysregulation. Clin Ther. 2015;37(5):984–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari Cet al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging. 2017;49:60–8. [DOI] [PubMed] [Google Scholar]
- 126. Nagpal R, Neth BJ, Wang S, Craft S, Yadav H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer's disease markers in subjects with mild cognitive impairment. EBioMedicine. 2019;47:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Willett W. Nutritional epidemiology. Oxford (UK): Oxford University Press; 2012. [Google Scholar]
- 128. Badimon L, Vilahur G, Padro T. Systems biology approaches to understand the effects of nutrition and promote health. Br J Clin Pharmacol. 2017;83(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kussmann M, Affolter M, Nagy K, Holst B, Fay LB. Mass spectrometry in nutrition: understanding dietary health effects at the molecular level. Mass Spectrom Rev. 2007;26(6):727–50. [DOI] [PubMed] [Google Scholar]
- 130. Ioannidis JPA. The challenge of reforming nutritional epidemiologic research. JAMA. 2018;320(10):969–70. [DOI] [PubMed] [Google Scholar]
- 131. Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Barabási A-L, Menichetti G, Loscalzo J. The unmapped chemical complexity of our diet. Nat Food. 2020;1(1):33–7. [Google Scholar]
- 133. Hopkins BD, Pauli C, Du X, Wang DG, Li X, Wu Det al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature. 2018;560(7719):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jacome-Sosa M, Hu Q, Manrique-Acevedo CM, Phair RD, Parks EJ. Human intestinal lipid storage through sequential meals reveals faster dinner appearance is associated with hyperlipidemia. JCI Insight. 2021;6(15):e148378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Voruganti VS. Nutritional genomics of cardiovascular disease. Curr Genet Med Rep. 2018;6(2):98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hou R, Cole SA, Graff M, Haack K, Laston S, Comuzzie AGet al. Genetic variants affecting bone mineral density and bone mineral content at multiple skeletal sites in Hispanic children. Bone. 2020;132:115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Davey Smith G. Use of genetic markers and gene-diet interactions for interrogating population-level causal influences of diet on health. Genes Nutr. 2011;6(1):27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Satia JA. Diet-related disparities: understanding the problem and accelerating solutions. J Am Diet Assoc. 2009;109(4):610–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Fenech M, El-Sohemy A, Cahill L, Ferguson LR, French TA, Tai ESet al. Nutrigenetics and nutrigenomics: viewpoints on the current status and applications in nutrition research and practice. J Nutrigenet Nutrigenomics. 2011;4(2):69–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mathers JC. Nutrigenomics in the modern era. Proc Nutr Soc. 2017;76(3):265–75. [DOI] [PubMed] [Google Scholar]
- 141. Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf Jet al. Human postprandial responses to food and potential for precision nutrition. Nat Med. 2020;26(6):964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kolovou GD, Mikhailidis DP, Kovar J, Lairon D, Nordestgaard BG, Ooi TCet al. Assessment and clinical relevance of non-fasting and postprandial triglycerides: an expert panel statement. Curr Vasc Pharmacol. 2011;9(3):258–70. [DOI] [PubMed] [Google Scholar]
- 143. Higgins V, Adeli K. Postprandial dyslipidemia: pathophysiology and cardiovascular disease risk assessment. EJIFCC. 2017;28(3):168–84. [PMC free article] [PubMed] [Google Scholar]
- 144. Monzillo LU, Hamdy O. Evaluation of insulin sensitivity in clinical practice and in research settings. Nutr Rev. 2003;61(12):397–412. [DOI] [PubMed] [Google Scholar]
- 145. Mucinski JM, Vena JE, Ramos-Roman MA, Lassman ME, Szuszkiewicz-Garcia M, McLaren DGet al. High-throughput LC-MS method to investigate postprandial lipemia: considerations for future precision nutrition research. Am J Physiol Endocrinol Metab. 2021;320(4):E702–15. [DOI] [PubMed] [Google Scholar]
- 146. Spitler KM, Davies BS. Aging and plasma triglyceride metabolism. J Lipid Res. 2020;61(8):1161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Beveridge J, Jagannathan S, Connell WF. The effect of the type and amount of dietary fat on the level of plasma triglycerides in human subjects in the postabsorptive state. Can J Biochem. 1964;42(7):999–1003. [DOI] [PubMed] [Google Scholar]
- 148. Hadjadj S, Paul J-L, Meyer L, Durlach V, Verges B, Ziegler Oet al. Delayed changes in postprandial lipid in young normolipidemic men after a nocturnal vitamin A oral fat load test. J Nutr. 1999;129:(9):1649–55. [DOI] [PubMed] [Google Scholar]
- 149. Gill JMR, Hardman AE. Postprandial lipemia: effects of exercise and restriction of energy intake compared. Am J Clin Nutr. 2000;71:(2):465–71. [DOI] [PubMed] [Google Scholar]
- 150. Kurti SP, Frick H, Wisseman WS, Malin SK, Edwards DA, Emerson SRet al. Acute exercise improves glucose and TAG metabolism in young and older adults following high-fat, high-carbohydrate meal intake. Br J Nutr. 2022:127(5):687–95. [DOI] [PubMed] [Google Scholar]
- 151. Lee YJ, Yoo MG, Kim HK, Jang HB, Park KJ, Lee HJet al. The association between alcohol metabolism and genetic variants of ADH1A, SRPRB, and PGM1 in Korea. Alcohol. 2019;79:137–45. [DOI] [PubMed] [Google Scholar]
- 152. Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- 153. Fielding BA, Reid G, Grady M, Humphreys SM, Evans K, Frayn KN. Ethanol with a mixed meal increases postprandial triacylglycerol but decreases postprandial non-esterified fatty acid concentrations. Br J Nutr. 2000;83(6):597–604. [DOI] [PubMed] [Google Scholar]
- 154. Cohn JS, McNamara J, Cohn S, Ordovas J, Schaefer E. Postprandial plasma lipoprotein changes in human subjects of different ages. J Lipid Res. 1988;29(4):469–79. [PubMed] [Google Scholar]
- 155. Robertson MD, Henderson RA, Vist GE, Rumsey RD. Extended effects of evening meal carbohydrate-to-fat ratio on fasting and postprandial substrate metabolism. Am J Clin Nutr. 2002;75(3):505–10. [DOI] [PubMed] [Google Scholar]
- 156. Ellis PR, Rayment P, Wang Q. A physico-chemical perspective of plant polysaccharides in relation to glucose absorption, insulin secretion and the entero-insular axis. Proc Nutr Soc. 1996;55(3):881–98. [DOI] [PubMed] [Google Scholar]
- 157. Sergi G, Bano G, Pizzato S, Veronese N, Manzato E. Taste loss in the elderly: possible implications for dietary habits. Crit Rev Food Sci Nutr. 2017;57(17):3684–9. [DOI] [PubMed] [Google Scholar]
- 158. Spence C, Youssef J. Aging and the (chemical) senses: implications for food behaviour amongst elderly consumers. Foods. 2021;10(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Reed DR, Alhadeff AL, Beauchamp GK, Chaudhari N, Duffy VB, Dus Met al. NIH workshop report: sensory nutrition and disease. Am J Clin Nutr. 2021;113(1):232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Park C, Pagnini F, Langer E. Glucose metabolism responds to perceived sugar intake more than actual sugar intake. Sci Rep. 2020;10(1):15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Barragán R, Coltell O, Portolés O, Asensio EM, Sorlí JV, Ortega-Azorín Cet al. Bitter, sweet, salty, sour and umami taste perception decreases with age: sex-specific analysis, modulation by genetic variants and taste-preference associations in 18 to 80 year-old subjects. Nutrients. 2018;10(10):1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Calder PC. Nutrition and immunity: lessons for COVID-19. Eur J Clin Nutr. 2021;11(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Charles-Messance H, Mitchelson KAJ, De M, Castro E, Sheedy FJ, Roche HM. Regulating metabolic inflammation by nutritional modulation. J Allergy Clin Immunol. 2020;146(4):706–20. [DOI] [PubMed] [Google Scholar]
- 164. Wu D, Lewis ED, Pae M, Meydani SN. Nutritional modulation of immune function: analysis of evidence, mechanisms, and clinical relevance. Front Immunol. 2019;9:3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ralston JC, Lyons CL, Kennedy EB, Kirwan AM, Roche HM. Fatty acids and NLRP3 inflammasome-mediated inflammation in metabolic tissues. Annu Rev Nutr. 2017;37:(1):77–102. [DOI] [PubMed] [Google Scholar]
- 166. Matone A, O'Grada CM, Dillon ET, Morris C, Ryan MF, Walsh Met al. Body mass index mediates inflammatory response to acute dietary challenges. Mol Nutr Food Res. 2015;59(11):2279–92. [DOI] [PubMed] [Google Scholar]
- 167. Mazidi M, Valdes AM, Ordovas JM, Hall WL, Pujol JC, Wolf Jet al. Meal-induced inflammation: postprandial insights from the Personalised REsponses to DIetary composition trial (PREDICT) study in 1000 participants. Am J Clin Nutr. 2021;114(3):1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Dibner C, Schibler U. Circadian timing of metabolism in animal models and humans. J Intern Med. 2015;277(5):513–27. [DOI] [PubMed] [Google Scholar]
- 169. Kohsaka A, Bass J. A sense of time: how molecular clocks organize metabolism. Trends Endocrinol Metab. 2007;18(1):4–11. [DOI] [PubMed] [Google Scholar]
- 170. de Castro JM. The time of day of food intake influences overall intake in humans. J Nutr. 2004;134(1):104–11. [DOI] [PubMed] [Google Scholar]
- 171. Aqeel MM, Guo J, Lin L, Gelfand SB, Delp EJ, Bhadra Aet al. Temporal dietary patterns are associated with obesity in US adults. J Nutr. 2020;150(12):3259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wilkinson MJ, Manoogian ENC, Zadourian A, Lo H, Fakhouri S, Shoghi Aet al. Ten-hour time-restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab. 2020;31(1):92–104, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ruiz-Lozano T, Vidal J, de Hollanda A, Scheer F, Garaulet M, Izquierdo-Pulido M. Timing of food intake is associated with weight loss evolution in severe obese patients after bariatric surgery. Clin Nutr. 2016;35(6):1308–14. [DOI] [PubMed] [Google Scholar]
- 174. Sutton EF, Beyl R, Early KS, Cefalu WT, Ravussin E, Peterson CM. Early time-restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018;27(6):1212–21, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Garaulet M, Gómez-Abellán P, Alburquerque-Béjar JJ, Lee YC, Ordovás JM, Scheer FA. Timing of food intake predicts weight loss effectiveness. Int J Obesity. 2013;37(4):604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lopez-Minguez J, Saxena R, Bandin C, Scheer FA, Garaulet M. Late dinner impairs glucose tolerance in MTNR1B risk allele carriers: a randomized, cross-over study. Clin Nutr. 2018;37(4):1133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Colberg SR, Zarrabi L, Bennington L, Nakave A, Somma CT, Swain DPet al. Postprandial walking is better for lowering the glycemic effect of dinner than pre-dinner exercise in type 2 diabetic individuals. J Am Med Dir Assoc. 2009;10(6):394–7. [DOI] [PubMed] [Google Scholar]
- 178. Reynolds AN, Mann JI, Williams S, Venn BJ. Advice to walk after meals is more effective for lowering postprandial glycaemia in type 2 diabetes mellitus than advice that does not specify timing: a randomised crossover study. Diabetologia. 2016;59(12):2572–8. [DOI] [PubMed] [Google Scholar]
- 179. Jakubowicz D, Barnea M, Wainstein J, Froy O. High caloric intake at breakfast vs. dinner differentially influences weight loss of overweight and obese women. Obesity (Silver Spring). 2013;21(12):2504–12. [DOI] [PubMed] [Google Scholar]
- 180. Bandín C, Scheer FA, Luque AJ, Ávila-Gandía V, Zamora S, Madrid JAet al. Meal timing affects glucose tolerance, substrate oxidation and circadian-related variables: a randomized, crossover trial. Int J Obesity. 2015;39(5):828–33. [DOI] [PubMed] [Google Scholar]
- 181. McHill AW, Phillips AJ, Czeisler CA, Keating L, Yee K, Barger LKet al. Later circadian timing of food intake is associated with increased body fat. Am J Clin Nutr. 2017;106(5):1213–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Xiao Q, Garaulet M, Scheer F. Meal timing and obesity: interactions with macronutrient intake and chronotype. Int J Obesity. 2019;43(9):1701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Martínez-Lozano N, Tvarijonaviciute A, Ríos R, Barón I, Scheer F, Garaulet M. Late eating is associated with obesity, inflammatory markers and circadian-related disturbances in school-aged children. Nutrients. 2020;12(9):2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Dashti HS, Gómez-Abellán P, Qian J, Esteban A, Morales E, Scheer Fet al. Late eating is associated with cardiometabolic risk traits, obesogenic behaviors, and impaired weight loss. Am J Clin Nutr. 2021;113(1):154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Romon M, Edme JL, Boulenguez C, Lescroart JL, Frimat P. Circadian variation of diet-induced thermogenesis. Am J Clin Nutr. 1993;57(4):476–80. [DOI] [PubMed] [Google Scholar]
- 186. Morris CJ, Garcia JI, Myers S, Yang JN, Trienekens N, Scheer FA. The human circadian system has a dominating role in causing the morning/evening difference in diet-induced thermogenesis. Obesity (Silver Spring). 2015;23(10):2053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Van Cauter E, Shapiro ET, Tillil H, Polonsky KS. Circadian modulation of glucose and insulin responses to meals: relationship to cortisol rhythm. Am J Physiol. 1992;262(4 Pt 1):E467–E75. [DOI] [PubMed] [Google Scholar]
- 188. Morris CJ, Yang JN, Garcia JI, Myers S, Bozzi I, Wang Wet al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci. 2015;112(17):E2225–E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Qian J, Dalla Man C, Morris CJ, Cobelli C, Scheer F. Differential effects of the circadian system and circadian misalignment on insulin sensitivity and insulin secretion in humans. Diabetes Obesity Metab. 2018;20(10):2481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Mason IC, Qian J, Adler GK, Scheer F. Impact of circadian disruption on glucose metabolism: implications for type 2 diabetes. Diabetologia. 2020;63(3):462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Garaulet M, Qian J, Florez JC, Arendt J, Saxena R, Scheer F. Melatonin effects on glucose metabolism: time to unlock the controversy. Trends Endocrinol Metab. 2020;31(3):192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Peñalvo JL, Mertens E, Muñoz-Cabrejas A, León-Latre M, Jarauta E, Laclaustra Met al. Work shift, lifestyle factors, and subclinical atherosclerosis in Spanish male workers: a mediation analysis. Nutrients. 2021;13(4):1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Eckel RH, Depner CM, Perreault L, Markwald RR, Smith MR, McHill AWet al. Morning circadian misalignment during short sleep duration impacts insulin sensitivity. Curr Biol. 2015;25(22):3004–10. [DOI] [PubMed] [Google Scholar]
- 194. Nedeltcheva AV, Scheer FA. Metabolic effects of sleep disruption, links to obesity and diabetes. Curr Opin Endocrinol Diabetes Obesity. 2014;21(4):293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Reutrakul S, Van Cauter E. Sleep influences on obesity, insulin resistance, and risk of type 2 diabetes. Metabolism. 2018;84:56–66. [DOI] [PubMed] [Google Scholar]
- 196. Rosique-Esteban N, Papandreou C, Romaguera D, Warnberg J, Corella D, Martínez-González MÁet al. Cross-sectional associations of objectively-measured sleep characteristics with obesity and type 2 diabetes in the PREDIMED-Plus trial. Sleep. 2018;41(12):zsy190. [DOI] [PubMed] [Google Scholar]
- 197. Zhou T, Yuan Y, Xue Q, Li X, Wang M, Ma Het al. Adherence to a healthy sleep pattern is associated with lower risks of all-cause, cardiovascular and cancer-specific mortality. J Intern Med. 2022;291(1):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Dashti HS, Ordovás JM. Genetics of sleep and insights into its relationship with obesity. Annu Rev Nutr. 2021;41(1):223–52. [DOI] [PubMed] [Google Scholar]
- 199. Domínguez F, Fuster V, Fernández-Alvira JM, Fernández-Friera L, López-Melgar B, Blanco-Rojo Ret al. Association of sleep duration and quality with subclinical atherosclerosis. J Am Coll Cardiol. 2019;73(2):134–44. [DOI] [PubMed] [Google Scholar]
- 200. Hanlon EC, Tasali E, Leproult R, Stuhr KL, Doncheck E, de Wit Het al. Sleep restriction enhances the daily rhythm of circulating levels of endocannabinoid 2-arachidonoylglycerol. Sleep. 2016;39(3):653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. St-Onge MP, Roberts AL, Chen J, Kelleman M, O'Keeffe M, RoyChoudhury Aet al. Short sleep duration increases energy intakes but does not change energy expenditure in normal-weight individuals. Am J Clin Nutr. 2011;94(2):410–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Nedeltcheva AV, Kilkus JM, Imperial J, Kasza K, Schoeller DA, Penev PD. Sleep curtailment is accompanied by increased intake of calories from snacks. Am J Clin Nutr. 2009;89(1):126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Dashti HS, Zuurbier LA, de Jonge E, Voortman T, Jacques PF, Lamon-Fava Set al. Actigraphic sleep fragmentation, efficiency and duration associate with dietary intake in the Rotterdam Study. J Sleep Res. 2016;25(4):404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Dashti HS, Scheer FA, Jacques PF, Lamon-Fava S, Ordovás JM. Short sleep duration and dietary intake: epidemiologic evidence, mechanisms, and health implications. Adv Nutr. 2015;6(6):648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. St-Onge MP, Mikic A, Pietrolungo CE. Effects of diet on sleep quality. Adv Nutr. 2016;7(5):938–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Baquerizo-Sedano L, Chaquila JA, Aguilar L, Ordovás JM, González-Muniesa P, Garaulet M. Anti-COVID-19 measures threaten our healthy body weight: changes in sleep and external synchronizers of circadian clocks during confinement. Clin Nutr. 2021; Jun 25: S0261-5614(21)00315-0. doi: 10.1016/j.clnu.2021.06.019. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Magnan S. NAM perspectives. Washington (DC: ): National Academy of Medicine; 2017. [Google Scholar]
- 208. Wang DD, Leung CW, Li Y, Ding EL, Chiuve SE, Hu FBet al. Trends in dietary quality among adults in the United States, 1999 through 2010. JAMA Intern Med. 2014;174(10):1587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Liu J, Rehm CD, Onopa J, Mozaffarian D. Trends in diet quality among youth in the United States, 1999–2016. JAMA. 2020;323(12):1161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci. 2011;108(7):3080–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Notterman DA, Mitchell C. Epigenetics and understanding the impact of social determinants of health. Pediatr Clin North Am. 2015;62(5):1227–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Amato KR, Arrieta MC, Azad MB, Bailey MT, Broussard JL, Bruggeling CEet al. The human gut microbiome and health inequities. Proc Natl Acad Sci. 2021;118(25):e2017947118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Office of Disease Prevention and Health Promotion . Social determinants of health [Internet]. Available from: https://health.gov/healthypeople/objectives-and-data/social-determinants-health (accessed 9 September2021).
- 214. Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf Jet al. Human postprandial responses to food and potential for precision nutrition. Nat Med. 2020;26(6):964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Gabel K, Cienfuegos S, Kalam F, Ezpeleta M, Varady KA. Time-restricted eating to improve cardiovascular health. Curr Atheroscler Rep. 2021;23(5):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Lee BY. Precision Nutrition: Research Gaps and Opportunities Workshop. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); 2021. [Google Scholar]
- 217. Green LW. Public health asks of systems science: to advance our evidence-based practice, can you help us get more practice-based evidence?. Am J Public Health. 2006;96(3):406–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Flood RL, Carson ER. Dealing with complexity: an introduction to the theory and application of systems science. New York (NY): Springer Science & Business Media; 2013. [Google Scholar]
- 219. Marrink SJ, Corradi V, Souza PC, Ingólfsson HI, Tieleman DP, Sansom MS. Computational modeling of realistic cell membranes. Chem Rev. 2019;119(9):6184–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Bower JM, Bolouri H. Computational modeling of genetic and biochemical networks. Cambridge (MA): MIT Press; 2001. [Google Scholar]
- 221. Schutter ED. Computational modeling methods for neuroscientists. Cambridge (MA): MIT Press; 2010. [Google Scholar]
- 222. Nolan J, Parker D, Van Kooten GC, Berger T. Malden (MA): Blackwell Publishing, Inc; 2009. [Google Scholar]
- 223. Wilks DS, Wilby RL. The weather generation game: a review of stochastic weather models. Prog Phys Geog Earth Environ. 1999;23(3):329–57. [Google Scholar]
- 224. Copeland BJ. Artificial intelligence [Internet]. Available from: https://www.britannica.com/technology/artificial-intelligence (accessed October 2, 2022).
- 225. Artificial intelligence [Internet]. Available from: https://www.merriam-webster.com/dictionary/artificial%20intelligence (accessed 23 March 2022).
- 226. Burch TK. Computer modeling of theory: explanation for the twenty-first century. In: Model-Based Demography Essays on Integrating Data, Technique and Theory. Series title: Demographic Research Monologues [Internet]. New York (NY): Springer Cham; 2018. p. 43–65. Available from: 10.1007/978-3-319-65433-1(accessed September 14, 2021). [DOI] [Google Scholar]
- 227. Das K, Behera RN. A survey on machine learning: concept, algorithms and applications. Int J Innovat Res Computer Comm Eng. 2017;5(2):1301–9. [Google Scholar]
- 228. Dalla Man C, Rizza RA, Cobelli C. Meal simulation model of the glucose-insulin system. IEEE Trans Biomed Eng. 2007;54(10):1740–9. [DOI] [PubMed] [Google Scholar]
- 229. Lee BY, Bartsch SM, Mui Y, Haidari LA, Spiker ML, Gittelsohn J. A systems approach to obesity. Nutr Rev. 2017;75(Suppl 1):94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Lee BY, Mueller LE, Tilchin CG. A systems approach to vaccine decision making. Vaccine. 2017;35(Suppl 1):A36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Mabry PL, Pronk NP, Amos CI, Witte JS, Wedlock PT, Bartsch SMet al. Cancer systems epidemiology: Overcoming misconceptions and integrating systems approaches into cancer research. PLoS Med. 2022;19:e1004027. doi: 10.1371/journal.pmed.1004027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Fallah-Fini S, Adam A, Cheskin LJ, Bartsch SM, Lee BY. The additional costs and health effects of a patient having overweight or obesity: a computational model. Obesity (Silver Spring). 2017;25(10):1809–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Ferguson MC, O'Shea KJ, Hammer LD, Hertenstein DL, Schwartz NJ, Winch LEet al. The impact of following solid food feeding guides on BMI among infants: a simulation study. Am J Prev Med. 2019;57(3):355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Ferguson MC, O'Shea KJ, Hammer LD, Hertenstein DL, Syed RM, Nyathi Set al. Can following formula-feeding recommendations still result in infants who are overweight or have obesity?. Pediatr Res. 2020;88(4):661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Ferguson MC, O'Shea KJ, Hammer LD, Hertenstein DL, Syed RM, Nyathi Set al. Can following formula-feeding recommendations still result in infants who are overweight or have obesity?. Pediatr Res. 2020:88(4):661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Lee BY, Ferguson MC, Hertenstein DL, Adam A, Zenkov E, Wang PIet al. Simulating the impact of sugar-sweetened beverage warning labels in three cities. Am J Prev Med. 2018;54(2):197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Seifu L, Ruggiero C, Ferguson M, Mui Y, Lee BY, Gittelsohn J. Simulation modeling to assist with childhood obesity control: perceptions of Baltimore City policymakers. J Public Health Policy. 2018;39(2):173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Ackley SF, Lessler J, Glymour MM. Dynamical modeling as a tool for inferring causation. Am J Epidemiol. 2022;191(1):1–6. doi: 10.1093/aje/kwab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Higgs S, Ruddock H. Social influences on eating. In: Handbook of Eating and Drinking: Interdisciplinary Perspectives, Meiselman HL, editor. Cham (Switzerland): Springer; 2020. p. 277–91. [Google Scholar]
- 240. Higgs S. Social norms and their influence on eating behaviours. Appetite. 2015;86:38–44. [DOI] [PubMed] [Google Scholar]
- 241. Patrick H, Nicklas TA. A review of family and social determinants of children's eating patterns and diet quality. J Am Coll Nutr. 2005;24(2):83–92. [DOI] [PubMed] [Google Scholar]
- 242. Shepherd R. Social determinants of food choice. Proc Nutr Soc. 1999;58(4):807–12. [DOI] [PubMed] [Google Scholar]
- 243. Solar O, Irwin A. A conceptual framework for action on the social determinants of health. Geneva (Switzerland): World Health Organization; 2010. Available from: https://apps.who.int/iris/handle/10665/44489(accessed October 1, 2022). [Google Scholar]
- 244. Horn AL, Bell BM, Bueno BGB, Bahrami M, Bozkaya B, Cui Yet al. , Investigating mobility-based fast food outlet visits as indicators of dietary intake and diet-related disease. 2021. medRxiv 2021.10.28.21265634; doi.org/10.1101/2021.10.28.21265634.
- 245. Pronk NP. Designing and evaluating health promotion programs. Dis Manage Health Outcomes. 2003;11(3):149–57. [Google Scholar]
- 246. Pronk NP, Dehmer SP, Hammond R, Halverson PK, Lee BY. Complex systems science and modeling. Submitted to the Secretary of Health and Human Services. U.S. Department of Health and Human Services, Washington, D.C. March, 2019.
- 247. Kern ML, Williams P, Spong C, Colla R, Sharma K, Downie Aet al. Systems informed positive psychology. J Posit Psychol. 2020;15(6):705–15. [Google Scholar]
- 248. Chambers DA, Norton WE. The adaptome: advancing the science of intervention adaptation. Am J Prev Med. 2016;51(4):S124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Yost J, Dobbins M, Traynor R, DeCorby K, Workentine S, Greco L. Tools to support evidence-informed public health decision making. BMC Public Health. 2014;14(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Pronk N. Practice and research connected: a synergistic process of translation through knowledge. transfer. In: ACSM's worksite health handbook. 2nd ed.Champaign (IL): Human Kinetics; 2009. p. 92–100. [Google Scholar]
- 251. Armstrong R, Pettman T, Waters E. Shifting sands—from descriptions to solutions. Public Health. 2014;128(6):525–32. [DOI] [PubMed] [Google Scholar]
- 252. Downey SM, Wages J, Jackson SF, Estabrooks PA. Adoption decisions and implementation of a community-based physical activity program: a mixed methods study. Health Promot Pract. 2012;13(2):175–82. [DOI] [PubMed] [Google Scholar]
- 253. Estabrooks PA, Brownson RC, Pronk NP. Dissemination and implementation science for public health professionals: an overview and call to action. Prev Chron Dis. 2018;15:E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Aarons GA, Ehrhart MG, Farahnak LR, Sklar M. Aligning leadership across systems and organizations to develop a strategic climate for evidence-based practice implementation. Annu Rev Public Health. 2014;35:(1):255–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Meadows DH. Thinking in systems: a primer. White River Junction (VT): Chelsea Green Publishing; 2008. [Google Scholar]
- 256. Scharmer C. Theory U: leading from the future as it emerges. Oakland (CA): Berrett-Koehler Publishers; 2008. [Google Scholar]
- 257. Uhl-Bien M, Marion R. A framework for leadership in the twenty-first century. In: Complexity Leadership, Part 1, Conceptual Foundations. Silver Spring (MD): International Leadership Association, Inc. 2008; p. 6–24. [Google Scholar]
- 258. Porter ME, Kramer MR. Creating shared value. Harvard Business Review. 2011;89:(1-2):62–77. [Google Scholar]
- 259. Pronk NP. Public health, business, and the shared value of workforce health and wellbeing. Lancet Public Health. 2019;4(7):e323. [DOI] [PubMed] [Google Scholar]
- 260. Leviton LC, Khan LK, Rog D, Dawkins N, Cotton D. Evaluability assessment to improve public health policies, programs, and practices. Annu Rev Public Health. 2010;31:(1):213–33. [DOI] [PubMed] [Google Scholar]
- 261. Noseworthy PA, Attia ZI, Brewer LC, Hayes SN, Yao X, Kapa Set al. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circulation Arrhythmia Electrophysiology. 2020;13(3):e007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Straw I. The automation of bias in medical artificial intelligence (AI): decoding the past to create a better future. Artificial Intelligence in Medicine. 2020;110:101965. [DOI] [PubMed] [Google Scholar]
- 263. Bellamy RK, Dey K, Hind M, Hoffman SC, Houde S, Kannan Ket al. AI Fairness 360: an extensible toolkit for detecting and mitigating algorithmic bias. IBM J Res Dev. 2019;63(4/5):4:1–4:15. [Google Scholar]
- 264. Faal F, Yu JY, Schmitt K. Domain adaptation multi-task deep neural network for mitigating unintended bias in toxic language detection. In: Proceedings of the 13th International Conference on Agents and Artificial Intelligence (ICAART 2021) - Volume 2. 2021; p. 932–40.
- 265. Kaplan RM, Riley WT, Mabry PL. News from the NIH: leveraging big data in the behavioral sciences. Transl Behav Med. 2014;4(3):229–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Riley WT. Chapter 18 - A new era of clinical research methods in a data-rich environment. In: Oncology Informatics, Using Health Information Technology to Improve Processes and Outcomes in Cancer. Hesse BW, Ahearn DK, Beckjord E, editors. New York (NY): Elsevier Inc; 2016. p. 343–55. [Google Scholar]
- 267. Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak Aet al. The FAIR guiding principles for scientific data management and stewardship. Scientific Data. 2016;3:(1):160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Fu R, Huang Y, Singh PV. Pushing the boundaries: frontiers in impactful OR/OM research. Catonsville (MD): INFORMS; 2020. p. 39–63. [Google Scholar]
- 269. Matheny M, Israni ST, Ahmed M, Whicher D. Artificial intelligence in health care: the hope, the hype, the promise, the peril. NAM Special Publication. Washington (DC): National Academy of Medicine; 2019. p. 154. [PubMed] [Google Scholar]
- 270. Dankwa-Mullan I, Scheufele EL, Matheny ME, Quintana Y, Chapman WW, Jackson Get al. A proposed framework on integrating health equity and racial justice into the artificial intelligence development lifecycle. J Health Care Poor Underserved. 2021;32(2S):300–17. [Google Scholar]
- 271. Mabry PL, Kaplan RM. Systems science: a good investment for the public's health. Health Educ Behav. 2013;40(1 Suppl):9S–12S. [DOI] [PubMed] [Google Scholar]
- 272. National Institutes of Health . Request for Application (RFA-OD-19-011) Predoctoral Training in Advanced Data Analytics for Behavioral and Social Sciences Research (BSSR) - Institutional Research Training Program [T32]. Bethesda (MD): National Institutes of Health; 2019. Available from: https://grants.nih.gov/grants/guide/rfa-files/RFA-OD-19-011.html(accessed September 14, 2021). [Google Scholar]
- 273. National Institutes of Health . OBSSR launches the Training in Advanced Data and Analytics for Behavioral and Social Sciences Research (TADA-BSSR) Program [Internet]. Updated 11 August 2020. Available from: https://obssr.od.nih.gov/news-and-events/news/director-voice/obssr-launches-training-advanced-data-and-analytics-behavioral (accessed October 4, 2021).
- 274. Dashti HS, Scheer F, Saxena R, Garaulet M. Timing of food intake: identifying contributing factors to design effective interventions. Adv Nutr. 2019;10(4):606–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.