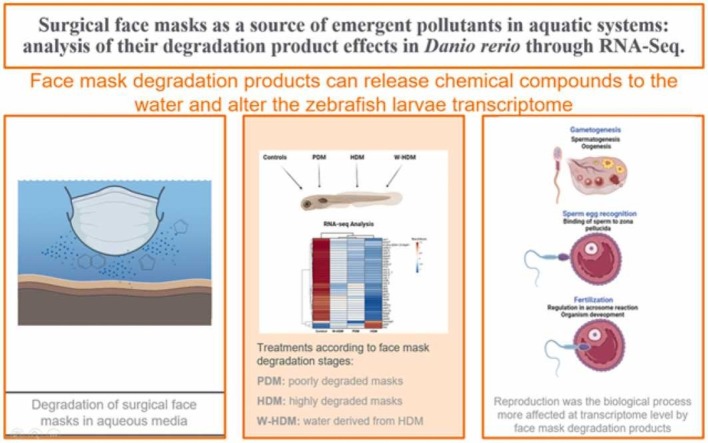
Abstract
Surgical face masks are the most popularised and effective personal equipment for protecting public health during the COVID-19 pandemic. They are composed of plastic polymer fibres with a large amount of inorganic and organic compounds that can be released into aquatic environments through degradation processes. This source of microplastics and inorganic and organic substances could potentially impact aquatic organisms. In this study, the toxicogenomic effects of face masks at different stages of degradation in water were analysed in zebrafish larvae (Danio rerio) through RNA-Seq. Larvae were exposed for 10 days to three treatments: 1) face mask fragments in an initial stage of degradation (poorly degraded masks -PDM- products) with the corresponding water; 2) face mask fragments in an advanced stage of degradation (highly degraded masks -HDM- products) with the corresponding water; and 3) water derived from HDM (W-HDM). Transcriptome analyses revealed that the three treatments provoked the down-regulation of genes related to reproduction, especially the HDM products, suggesting that degradation products derived from face masks could act as endocrine disruptors. The affected genes are involved in different steps of reproduction, including gametogenesis, sperm-egg recognition and binding or fertilisation. Immune-related genes and metabolic processes were also differentially affected by the treatments.
Keywords: Face mask, Degradation, Danio rerio, Transcriptome, Reproductive processes, Endocrine disruptors
Graphical Abstract
1. Introduction
The concept of "One Health” proposed by the World Health Organization (WHO) with the COVID-19 pandemic is currently more applicable than ever. One health is the intersection of human, animal and environmental health. Therefore, the synergies and harmony of these three blocks is the cornerstone of wellness and global health. Environmental degradation due to human behaviours and waste mismanagement shows an unsure future for the biosphere and humanity itself.
During the COVID-19 pandemic, the use of personal protective equipment (PPE), such as face masks, has supported significant advancements in public health (Howard, 2020). Currently, face masks are a legal requirement in public spaces in many countries. The reported monthly consumption of face masks has been approximately 129 billion for 7.8 billion people across the globe (Kalina et al., 2020). However, the management of PPE residues as well as uncivil behaviours of the population have already revealed face masks in terrestrial and aquatic compartments. A recent study in Toronto, Canada, showed that 95% of face masks from six survey locations in urban zones were surgical with respect to reusable face masks, with an incidence of approximately 0.00475 items·m2 around large grocery store parking areas (Ammendolia et al., 2021). Furthermore, 105,000 tonnes of face masks per month are disposed into the environment by Africans (Benson et al., 2021a, Benson et al., 2021b). The Annacis Island Wastewater Treatment Plant of Metro Vancouver has observed an increased number of discarded surgical masks since the beginning of the COVID-19 pandemic. This wastewater plant estimates that due to these emergent pollutants, Canada-wide maintenance costs could exceed $250 million per year (Rasmussen, 2020). From February 2020, face masks have already been found in natural compartments, reaching more than 70 surgical face masks along 100 m of shoreline, as reported by Oceans Asia Organization in Hong Kong. In the Mediterranean Sea, face masks have been seen floating like jellyfish and in the seabed off the coast of Antibes town, as reported by French environmental NGOs. Once again, the sink of waste products due to inappropriate human behaviours and waste management is the aquatic system. As a reflection, to protect human health from COVID-19, are we putting aside the other two fundamental pillars of the One Health concept? What should the balance be among them when one is truly committed?
The daily use of surgical face masks increases difficulties related to an environmental issue that is far from solved: plastic consumption and therefore plastic pollution. Before the COVID-19 pandemic, different directives and regulations (such as the Basel Convention, United Nations Convention on the Law of the Sea, International Convention for the Prevention of Pollution from Ships, Joint Group of Experts on the Scientific Aspects of Marine Environmental Protection and United Nations Global Partnership on Marine Litter) were advanced to alleviate plastic pollution and therefore the health of the planet. However, surgical face masks are a and large source of plastic pollution and nanofibre delivery due to their fabrication process (Leung and Sun, 2020). Moreover, when face masks reach the natural environment, environmental agents initiate their degradation and fragmentation processes to degrade the mask into smaller pieces called microplastics (MPs < 5 mm; (Thompson et al., 2004)) and nanoplastics (NPs < 100 nm; (Jambeck et al., 2015)).
Wastewater treatment plants (WWTPs) are the sink of many emerging pollutants and MPs (de Oliveira et al., 2020, Raju et al., 2020). Nonspecific treatments for the depuration of pollutants of emerging concern make WWTPs a source of pollutants in their surrounding environment, which in most cases is aquatic ecosystems. Due to their small size, MPs are available for a vast group of aquatic animals (Ferreira et al., 2019). Fish are one of the most sensitive groups to MP ingestion, with a frequency of accumulated MPs of approximately 72% (Provencher et al., 2019) and, since ingestion is the main way for MPs accumulation, fish are an animal group of special concern in this sense. The ingestion of MPs by fish is mainly due to their attractive coloration, low density, buoyancy, and resemblance to food (Güven et al., 2017). The effects of MPs are varied, from abrasions and ulcers in the digestive tract (Kim et al., 2019, Limonta et al., 2019, Wright et al., 2013), dysbiosis (Jin et al., 2018), inflammation in the gut tissues (Jeong and Choi, 2019, Qiao et al., 2019b), impairments in the triglyceride:cholesterol ratio in blood serum (Cedervall et al., 2012) and immune responses (Sendra et al., 2021a) to metabolome alterations (Qiao et al., 2019a). However, although surgical face masks are composed of fibres of plastic polymers and can be a source of MPs, they could also release other compounds, such as resins, which are used for their hydrophobic capacity, and different dyes used in their manufacture (Sullivan et al., 2021). Therefore, the impact of MPs and face mask degradation products are not equivalent, and MP studies are not completely shed unequivocable light on the effects caused by face masks in aquatic organisms (Gigault et al., 2018).
To investigate if surgical face masks at different degradation stages could release different quantity and types of organic and inorganic chemical compounds, we conducted a characterisation of the substances leached into water from face masks. Due to the potential risk posed by face masks and their degradation compounds to the aquatic environment, we selected zebrafish (Danio rerio) larvae as a model organism in toxicological studies (Dai et al., 2014) to determine the acute effects of the exposure to surgical face mask fragments under different stages of degradation as well as to the water in which face masks had degraded. To evaluate if different degradation stages can provoke different biological effects in zebrafish larvae, a transcriptomic approach using RNA-Seq analysis has been addressed in this work. RNA-Seq analysis allowed us to identify different processes that could be meaningfully affected by surgical face mask degradation products by the significant and powerful modulation of genes involved in those processes. This was the case for the reproduction process, which seemed to be highly affected by face mask degradation products, especially those in a more advanced stage of degradation.
2. Material and methods
2.1. Fragmentation and degradation of surgical face masks
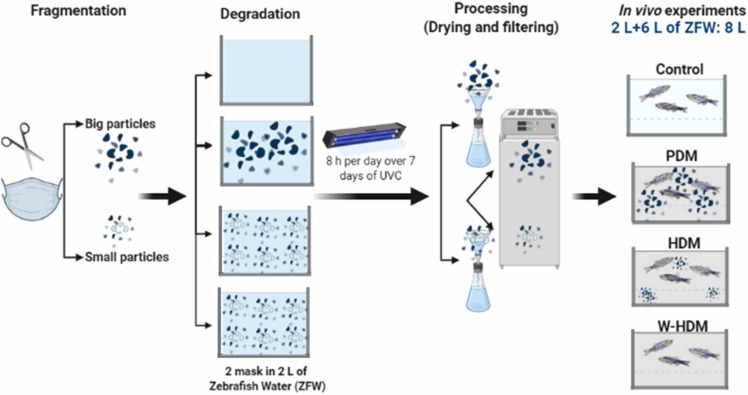
The experimental fragmentation and degradation of face masks is shown in Fig. 1. The surgical face masks used in this study were commercial face masks (LyncMed, Beijing, China) sold in a local pharmacy. Aluminium and elastic subjections were removed from the face masks prior to fragmentation.
Fig. 1.
Procedures and experimental design for PDM, HDM and W-HDM treatments and exposure to zebrafish.
The first fragmentation was performed with a scissor previously cleaned with ethanol. Six face masks were cut into pieces of approximately 2 mm, and the pieces corresponding to two face masks were reserved for poorly degraded mask (PDM) product treatment. The pieces from the other four masks were milled to obtain high fragmentation and were used for highly degraded mask (HDM) product treatments. After fragmentation, the degradation process consisted of placing the pieces of fragmented face masks in water: 1 tank of 2 L of zebrafish water for PDM products and 2 tanks for HDM products with two fragmented face masks each. One control tank with zebrafish water but without face mask fragments was also included. The tanks were exposed to ultraviolet C light (UVC; 0.62 µW·cm−2) for 8 h per day for 7 days to simulate a UV degradation process. After this period, the PDM and HDM fragments were collected from the tanks and dried at 40 °C. The water from each of the tanks (2 L) was filtered through a 0.4-µm-pore-size hydrophilic polycarbonate filter (Merck, HTTP04700) to avoid the release of fibres from face masks. A volume of 40 mL of the water from each tank was kept for posterior analysis of inorganic and organic compounds (water samples used in 2.2.2, 2.2.3). The remaining water (1960 mL) was diluted in 6 L of fresh zebrafish water (final dilution: 24.6%) for the experiments and water renovations of zebrafish larvae. The treated zebrafish water was oxygenated for use in the experiment and the water parameters remained practically unaltered during the experiment (pH: 6.9 -7; dissolved oxygen: 6.5-7 mg/L; conductivity: ~ 690 µS/cm).
2.2. Surgical face mask characterisation and its behaviour in freshwater systems
2.2.1. Size distribution and behaviour
The size distribution of PDM and HDM pieces was analysed by optical microscopy (n:200). In addition, the three layers of the surgical face masks, as well as different zones of face masks, were analysed by Fourier transform infra-red spectroscopy (FTIR) through the Bruker Alpha System.
Secondary characteristics of PDM and HDM fragments were analysed by static light scattering (SLS, Malvern Master Sizer 2000, with software version 5.61). The behaviour of the face mask fragments was studied in zebrafish water over time: 0, 24, 48 and 72 h (time between water tank changes). The protocol for zebrafish water preparation is included in the Supplementary materials.
2.2.2. Characterisation of organic compounds
For organic compound characterisation, the elastics of the face masks and aluminium nose clips were also removed. Between 1 and 1.3 g was used to extract the organic compounds with 35 mL of chloroform as the polar dissolvent. The extractions were performed in 100-mL Erlenmeyer flasks and under bath sonication. The liquid obtained from these face masks and the water from the PDM and HDM stored in Section 2.1 were concentrated by a rotavapor to reach a liquid volume of 1 mL. The samples were transferred to vials for organic compound analysis, which was conducted by gas chromatography–mass spectrometry (GC-MS). Details about the GS-MS analysis are provided in the Supplementary materials (Table S1)).
2.2.3. Characterisation of inorganic compounds
The elastics of the face masks and aluminium nose clip were also removed, and the face masks were cut with clean scissors. Approximately 0.2 g of face masks was digested by acid digestion in a microwave following the procedures of Rodríguez-Seijo et al. (2015). After digestion, the samples were evaporated and diluted in 10 mL of ultrapure water. The water samples were acidified with 2% of HNO3. The analysis of the samples was performed by inductively coupled plasma mass spectrometry (ICP-MS; Thermo elemental) in semiquantitative mode to detect the concentration of all the elements of the periodic table.
2.3. Zebrafish larvae treatments
A total of 240 wild-type zebrafish (120 larvae for mortality analysis and 120 for transcriptome experiments) at an age of 15 days postfertilisation (dpf) were obtained from the facilities of the Instituto de Investigaciones Marinas (Vigo, Spain), where zebrafish are maintained following established protocols (Nusslein-Volhard and Dahm, 2002, Westerfield, 2000). At this stage zebrafish larvae are not sexually differentiated, which allows to avoid the gender bias of the data. Fish care and challenge experiments were conducted according to the guidelines of the CSIC National Committee on Bioethics under approval number ES360570202001/17/FUN.01/INM06/BNG. Zebrafish larvae were maintained under a 12:12-h light-dark cycle at pH 7.0 and temperature 28–29 °C and, from 5 dpf, were fed daily with commercial food (Sera Micron, North Rhine-Westphalia, Germany).
Dry PDM and HDM fragments were mixed with their corresponding filtered water (described in Section 2.1) to obtain the same concentration in the treatments. Three replicates (10 larvae per replicate) were used for each treatment: a) control, b) PDM (10 mg·L−1 of the PDM fragments with the corresponding water), c) HDM (10 mg·L-1 of the HDM fragments with the corresponding water) and d) water derived from HDM (W-HDM); Fig. 1. The water volume of each tank was 0.5 L, and it was renewed every 3 days with the corresponding treatment. After 10 days of treatment exposure, seven zebrafish larvae were collected from each tank and pooled for RNA-Seq analysis. The larvae were euthanized by thermal shock on ice, and RNA isolation was immediately carried out after zebrafish collection. A parallel identical experiment was conducted to analyse whether the treatments had some impact on the survival of the zebrafish larvae.
2.4. RNA isolation and Illumina sequencing
Total RNA was isolated using the Maxwell 16LEV Simply RNA Tissue Kit (Promega) according to the manufacturer`s instructions. The quantity of RNA was measured in a NanoDrop ND-1000 (NanoDrop Technologies, Inc.), and RNA integrity was analysed in an Agilent 2100 Bioanalyzer (Agilent Technologies Inc.) according to the manufacturer's instructions. All the samples passed the quality control tests and were used for Illumina library preparation.
Three biological replicates per treatment (control, PDM, HDM and W-HDM), composed of each replicate by 7 pooled larvae, were used for mRNA sequencing. Double-stranded cDNA libraries were constricted using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, USA), and sequencing was performed using Illumina NovaSeq 6000 technology at Macrogen Inc. (Seoul, Republic of Korea). The raw read files (in FASTQ format) were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under BioProject accession number PRJNA770826.
2.5. Trimming, mapping, RNA-Seq and differential expression analysis of mRNAs
CLC Genomics Workbench v. 20.0.4 software (CLC Bio, Qiagen, Denmark) was used to filter and trim raw reads and perform the RNA-Seq and differential expression analyses. Raw reads were trimmed to remove adaptor sequences and low-quality reads (Phred score limit of 0.05). RNA-Seq analyses were carried out by mapping the filtered reads against the last version of the zebrafish genome (GRCz11) with the following parameters: length fraction = 0.8, similarity fraction 0.8, mismatch cost = 2, insertion cost = 3 and deletion cost = 3. The expression values were set as transcripts per million (TPMs). Finally, a differential expression analysis test was used to compare gene expression levels and to identify differentially expressed genes (DEGs) among the treatments. Transcripts with absolute fold change (FC) values ≥ 2 and p values ≤ 0.05 were retained for further analyses.
To identify and quantify the directions of variability in the data, a principal component analysis (PCA) plot was constructed using the TPM expression values with the ClustVis tool (https://biit.cs.ut.ee/clustvis/; Metsalu and Vilo, 2015). Heatmaps were created to show a hierarchical clustering of gene expression (TPM values) using the average linkage method with the Euclidean distance with the ClustVis tool.
To illustrate the common and exclusive DEGs among the treatments compared to the control larvae, Venn diagrams were performed using the Venny 2.1 tool (https://bioinfogp.cnb.csic.es/tools/venny/).
2.6. Gene Ontology (GO) terms, Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways and protein domain enrichment analyses
GO enrichment analysis of biological processes (BP), KEGG pathway analysis and protein domain (PD) enrichment analysis were conducted using DAVID software (https://david.ncifcrf.gov/; (Huang et al., 2007)). In all cases, a Bonferroni correction was applied (p < 0.05), and a minimal count of 5 was established. For domain enrichment, the InterPro database (Apweiler et al., 2001) was used. The representation of the different categories was based on the fold-enrichment value.
2.7. Protein–protein interaction networks
The interactions of the proteins encoded by the DEGs in each treatment were analysed with STRING v11.0 software (https://string-db.org); (Szklarczyk et al., 2021). The clusters formed in the STRING software indicated that the expressed proteins had more interactions among themselves than would be expected for a random set of proteins of similar size drawn from the genome. Such an enrichment indicated that the proteins were at least partially biologically connected as a group.
3. Results
3.1. Primary chemical characterisation of surgical face masks and secondary chemical characterisation of face mask degradation products in freshwater
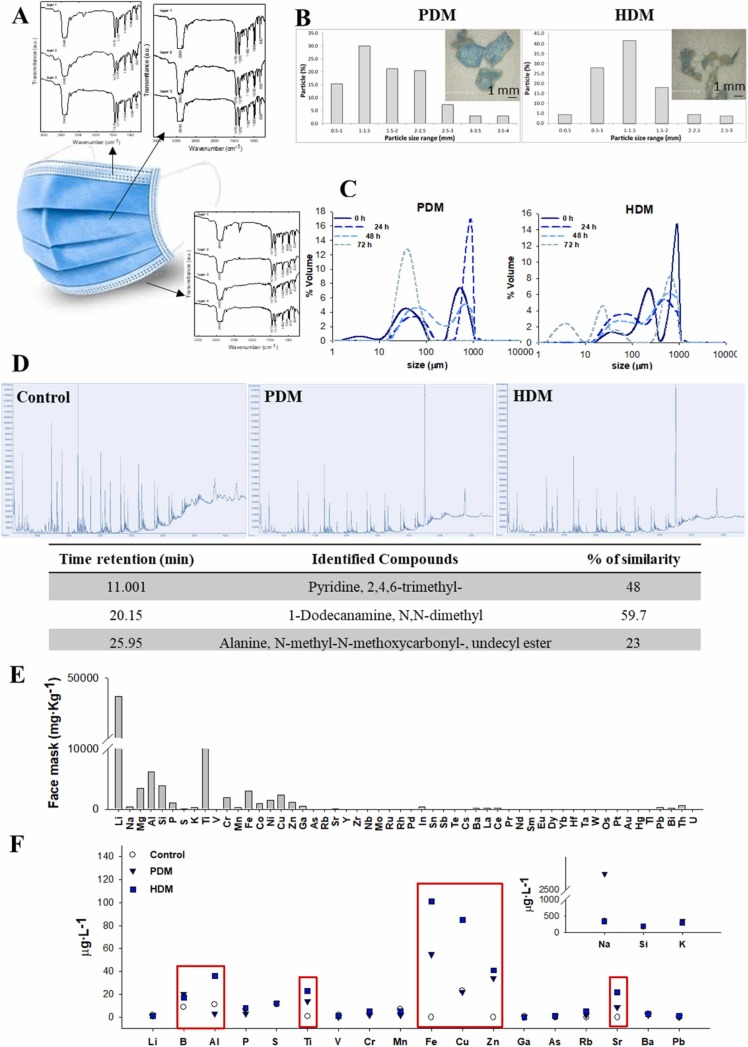
The commercial face masks used in this study were composed of three layers, with polypropylene as a plastic polymer ( Fig. 2A). The nominal sizes of the fragments used for the PDM and HDM treatments were 3.10 ± 1.6 mm and 1.25 ± 0.52 mm (mean ± standard deviation), respectively (Fig. 2B). Both PDM and HDM fragments showed continuous fragmentation/degradation behaviour after 72 h in zebrafish water (Fig. 2C). After this period, the face mask fragments reached a homogeneous population with only a peak between 17.37 and 12.22 µm for the PDM and a heterogeneous population with four peaks between 1.44 and 954.90 µm for the HDM (Fig. 2C).
Fig. 2.
Surgical face mask characterisation and its behaviour in the aquatic matrix. A) FTIR spectra of the different parts of the surgical face mask and different layers. B) Nominal sizes of fragments from PDM and HDM treatments. C) Behaviour of both size fragments of face mask in zebrafish water over 72 h (time between zebrafish water changes). D) Spectrum of the organic substances released into the water from the PDM and HDM treatments. E) Concentration of different elements present in the surgical face masks. For some elements the concentration was zero. F) Inorganic elements released into water from PDM and HDM treatments.
The organic and inorganic compounds were analysed in the face masks and in the water derived from PDM and HDM. The chromatography analysis of the face masks showed a profile of waxes and hydrocarbons (Fig. 2A). With respect to inorganic compounds, the face masks showed high concentrations of Li (49,695 mg kg−1), Ti (10,475.19 mg kg−1), Al (6197.64 mg kg−1), Si (3904.01 mg kg−1) and Mg (340.88 mg kg−1). Values of Cr, Mn, Fe, Co, Ni Cu Zn and Ga were also present, and low values were found for In, Ba, La, Ce, Pb, Bi and Th (Fig. 2E). However, these profiles of organic and inorganic compounds found in the surgical face masks were not the same as those found in the zebrafish water after a week of UV-C irradiation (degradation conditions). Waxes and hydrocarbons found in the face masks were not released into the water. However, other compounds, such as pyridine 2,4,6-trimethyl, 1-dodecanamine N,N-dimethyl and alanine N-methyl-N-methoxycarbonyl-undecyl ester, were found in the water derived from PDM and HDM degradation (Fig. 2D). No peaks for these compounds were found in the control zebrafish water under the same exposure conditions; therefore, they could be byproducts formed under surgical face mask degradation conditions.
Low concentrations of inorganic compounds, such as Na, B, Al, Ti, Fe and Cu, were only released into the water of HDM. Conversely, Zn and Sr were found in both the PDM and HDM water samples (Fig. 2F).
3.2. Transcriptome modulation of zebrafish larvae exposed to the different face mask degradation products
The survival of the larvae was higher than 91% over the experiment for all the tanks. Although no significant mortalities were observed in the larvae treated with the different face mask products (data not shown), RNA-Seq analyses were conducted to evaluate the impact of the different face mask degradation products on the overall transcriptome of zebrafish larvae.
A total of 321,832,678 million reads were obtained from the twelve sequenced samples, with a mean of 26,819,390 million reads per sample. From these, 99.9% of the raw reads passed the quality control, and these high-quality reads were mapped against the zebrafish genome. For the different samples, between 95% and 97% of the reads were successfully mapped against the genome (Table S2).
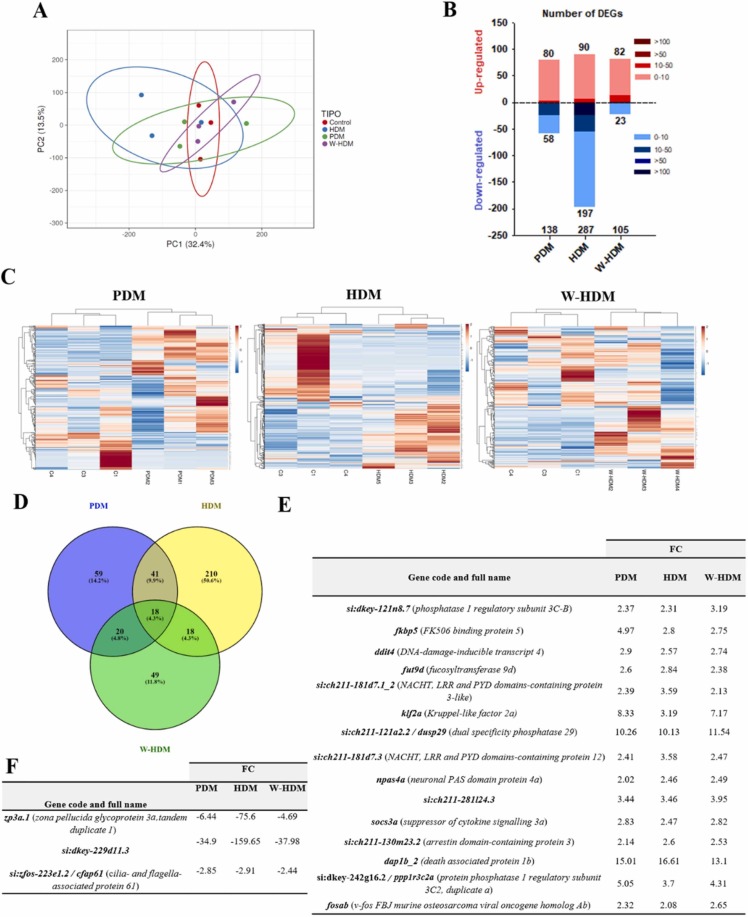
PCA of the twelve samples revealed a poor separation of the samples belonging to the four different treatments ( Fig. 3A). However, despite the low differentiation of the overall transcriptomes, an important group of genes were significantly modulated after exposure to the treatments. RNA-Seq and further differential expression analyses revealed 138, 287 and 105 differentially expressed genes (DEGs) in zebrafish larvae exposed to PDM, HDM and W-HDM products, respectively, compared with the control larvae (Fig. 3B; Tables S3–S5). Whereas a similar number of up-regulated DEGs was observed for the three treatments (80, 90 and 82 for the larvae exposed to PDM, HDM and W-HDM products, respectively), the number of down-regulated genes was much higher for those larvae treated with the HDM products (58, 197 and 23 for the larvae exposed to PDM, HDM and W-HDM products, respectively) (Fig. 3B). The expression profiles of the DEGs in TPM values were represented in heatmaps where the three biological replicates of each treatment clustered together, showing good consistency among the replicates corresponding to the same treatment (Fig. 3C).
Fig. 3.
RNA-Seq analysis results of zebrafish larvae exposed to the three face mask treatments. A) Principal component analysis (PCA) of the different samples. B) Stacked column chart showing the number and intensity of the up- and down-regulated DEGs after PDM, HDM and W-HDM treatments. C) Heatmaps representing the DEGs for the three treatments. D) Venn diagram reflecting the exclusive and common DEGs among the three. E) Common up- and F) down-regulated DEGs among the three treatments.
A Venn diagram of the DEGs for each treatment compared with the control revealed 18 common DEGs among the three treatments (Fig. 3D), 15 of which were up-regulated and the other 3 down-regulated. The common up- and down-regulated genes are shown in Fig. 3E and F, respectively. No significant biological processes or KEGG pathways were found to be enriched for these common DEGs. However, for the common down-regulated genes among the three treatments, two of them (zp3a.1 and si:zfos-223e1.2) were linked to the reproductive process, since zp3a.1 (zona pellucida glycoprotein 3a, tandem duplicate 1) encodes a component of the oocyte’s zona pellucida and si:zfos-223e1.2 (cfap61; cilia- and flagella-associated protein 61) is required for sperm flagellum formation and male fertility (Huang et al., 2020). Conversely, the common up-regulated DEGs seemed to be involved in a variety of processes, with the most affected genes across the three treatments being si:dkey-242g16.2 (ppp1r3c2a; protein phosphatase 1 regulatory subunit 3C2, duplicate a) and si:ch211–121a2.2 (dusp29; dual specificity phosphatase 29), which are involved in glucose metabolism, among other functions.
3.3. Main processes and pathways affected by the different treatments
3.3.1. Zebrafish larvae exposed to PDM products
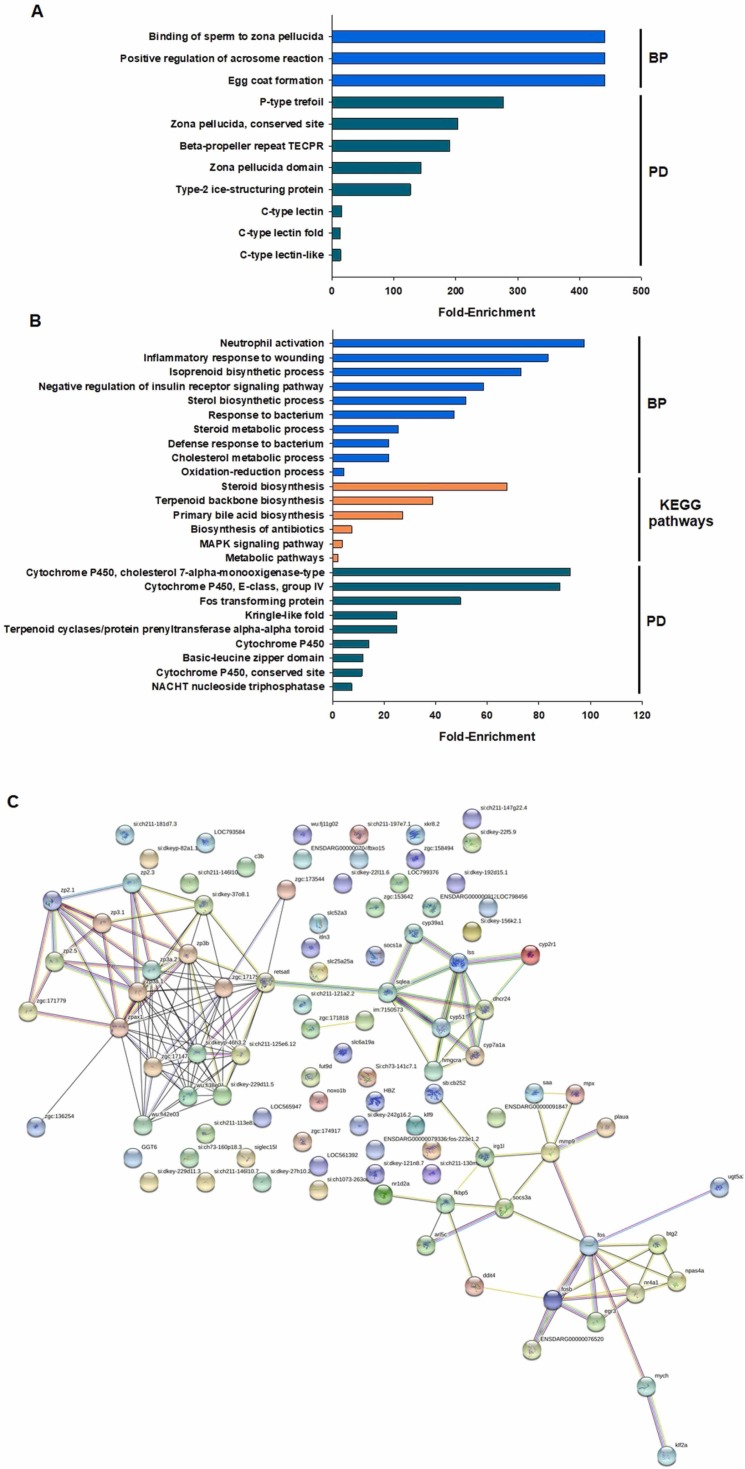
The top 25 most up- and down-regulated DEGs in zebrafish larvae exposed to PDM products are summarised in Table 1. Among the top 25 down-regulated DEGs, genes involved in reproductive processes, such as several genes encoding zona pellucida glycoproteins (zp2.1, zp3, zp2.5, zp2.2, zp3e, zp3.2_1 and zp2.3), were found. These down-regulated genes were inhibited more than 10-fold compared with the controls. GO biological process enrichment analysis of the down-regulated DEGs in zebrafish larvae exposed to PDM products revealed three enriched terms, all of which were related to reproduction: “binding of sperm to zona pellucida”, “positive regulation of acrosome reaction” and “egg coat formation” ( Fig. 4A). Significantly enriched KEGG pathways were not observed for these genes. However, protein domain enrichment analysis revealed different domains linked to the reproductive process, such as “P-type trefoil”, “zona pellucida, conserved site”, “zona pellucida domain”, “C-type lectin”, “C-type lectin fold” and “C-type lectin-like” (Fig. 4A).
Table 1.
Top 25 most up- and down-regulated DEGs in zebrafish larvae exposed to PDM for 10 days.
| Up-regulated genes |
Down-regulated genes |
||||
|---|---|---|---|---|---|
| Blast2 Go UnitProt/SwissProt Description |
Blast2 Go UnitProt/SwissProt Description |
||||
| Name |
Complete name |
FC |
Name |
Complete name or Predicted description |
FC |
| PDM | PDM | ||||
| dap1b_2 | death associated protein 1b | 15.01 | zp2.1 | zona pellucida glycoprotein 2, tandem duplicate 1 | -40.75 |
| itln3 | intelectin 3 | 12.74 | si:dkey-229d11.3 | si:dkey-229d11.3 | -37.98 |
| si:ch211–121a2.2 | dual specificity phosphatase 29 | 10.26 | zgc:171445 | serine-type endopeptidase inhibitor activity | -33.53 |
| itln2_1 | intelectin 2 | 8.61 | si:ch211–146l10.7 | si:ch211–146l10.7 | -27.78 |
| klf2a | kruppel-like factor 2a | 8.33 | zgc:173856 | zgc:173856 | -21.26 |
| si:ch211–113d11.8 | si:ch211–113d11.8 | 6.72 | wu:fi42e03 | wu:fi42e03 | -20 |
| si:dkey-22f5.9 | liver-expressed antimicrobial peptide 2 | 5.1 | zp3 | zona pellucida glycoprotein 3b | -19.92 |
| si:dkey-242g16.2 | protein phosphatase 1 regulatory subunit 3 C-B | 5.05 | zgc:173544 | zgc:173544 | -19.9 |
| cyp2r1 | cytochrome P450, family 2, subfamily R, | 5.03 | si:ch73–160p18.3 | flagellar attachment zone protein 1 | -19.65 |
| fkbp5 | FK506 binding protein 5 | 4.97 | si:ch211–146l10.8 | si:ch211–146l10.8 | -18.44 |
| zgc:153642 | GTPase IMAP family member 4 | 4.31 | zp2.5 | zona pellucida glycoprotein 2, tandem duplicate 5 | -17.75 |
| zgc:174917 | phytanoyl-CoA dioxygenase domain-containing protein 1 | 3.97 | zp2.2 | zona pellucida glycoprotein 2, tandemduplicate 2 | -17.7 |
| si:ch211–113e8.5 | GTP binding activity | 3.67 | zgc:136254 | fish-egg lectin | -16.87 |
| gene:ENSDARG00000104576 | glycogen binding activity and protein phosphatase 1 binding activity | 3.63 | si:dkey-4j21.2_1 | stonustoxin subunit beta | -14.56 |
| si:ch1073–286c18.5 | collagen alpha-1(XI) chain | 3.51 | zgc:171474 | zgc:171474 | -14.29 |
| rgs2 | regulator of G protein signalling 2 | 3.47 | zgc:171446 | ovostatin homologue | -14.18 |
| si:ch211–281l24.3 | oestrogen-responsive finger protein | 3.44 | si:ch211–147g22.4 | major histocompatibility complex class I-related gene protein | -14.1 |
| sqlea | squalene epoxidase a | 3.33 | si:dkey-229d11.5 | Endo-1,4-beta-xylanase B | -13.46 |
| slc52a3 | solute carrier family 52 (riboflavin | 3.31 | si:dkeyp-98a7.9 | adhesion G protein-coupled receptor L2 | -12.49 |
| cyp7a1 | cytochrome P450, family 7, subfamily A, | 3.25 | zp3e | zona pellucida glycoprotein 3e | -12.46 |
| zgc:163057 | haemoglobin subunit zeta | 3.19 | si:ch211–125e6.12 | galactose-specific lectin nattectin | -11.34 |
| si:dkey-202l22.6 | interferon-induced very large GTPase 1 | 3.15 | zp3.2_1 | zona pellucida glycoprotein 3, tandem duplicate 2 | -11.02 |
| c3a.2 | complement component c3a, duplicate 2 | 3.09 | zp2.3 | zona pellucida glycoprotein 2, tandem duplicate 3 | -10.67 |
| nr4a1 | nuclear receptor subfamily 4, group A, | 3.03 | si:dkey-37o8.1 | Elongation factor 1-alpha | -10.06 |
| si:zfos-741a10.3 | si:zfos-741a10.3 | 2.92 | si:ch1073–263o8.2 | si:ch1073–263o8.2 | -8.9 |
Fig. 4.
Biological processes (BP), KEGG pathways, protein domains (PD) and protein–protein interactions enriched for the genes modulated by the PDM treatment. A) Enrichment analyses of the down-regulated genes. B) Enrichment analyses of the up-regulated genes. C) STRING analysis (protein–protein interaction) of the DEGs under PDM treatment.
Conversely, the top 25 up-regulated genes after PDM treatment were related to i) innate immune response, such as dap1b_2 (death associated protein 1b), itln3 (intelectin 3), itln2_1 (intelectin 2), si:dkey-22f5.9 (liver-expressed antimicrobial peptide 2), si:dkey-202l22.6 (interferon-induced very large GTPase 1) and c3a.2 (complement component c3a, duplicate 2), and ii) isoprenoid/sterol biosynthesis, such as cyp2r1 (cytochrome P450, family 2, subfamily r), sqlea (squalene epoxidase a) and cyp7a1 (cytochrome P450, family 7, subfamily a) (Table 1). GO biological process enrichment analysis of the up-regulated DEGs in larvae exposed to PDM products revealed several terms linked to the immune response (“neutrophil activation”, “inflammatory response to wounding”, “response to bacterium” and “defence response to bacterium”) but also to the isoprenoid/sterol biosynthesis and redox process (“isoprenoid biosynthetic process”, “sterol biosynthetic process”, “steroid metabolic process”, “cholesterol metabolic process” and “oxidation-reduction process”) (Fig. 4B). Among the KEGG pathways, the most enriched pathways were also related to isoprenoid/sterol biosynthesis (“steroid biosynthesis”, “terpenoid backbone biosynthesis” and “primary bile acid biosynthesis”), and the protein domains also reflected these processes (“cytochrome P450, cholesterol 7-alpha-monooxigenase-type”, “cytochrome P450, E-class, group IV”, “terpenoid cyclases/protein prenyltransferase alpha-alpha toroid”, “cytochrome P450″ or “cytochrome P450, conserved site”), among others (Fig. 4B).
To obtain an overview of the function of the genes regulated by the PDM products, a protein–protein interaction network was constructed with STRING software (Fig. 4C). Treatment with the PDM products affected three main clusters of genes. The first two clusters were composed of different up-regulated DEGs; one of them included genes related to metabolic processes such as bile acid biosynthesis, cholesterol biosynthesis, sterol biosynthesis and steroid metabolism processes (cyp7a1a, shcr24, hmcra, sqlea, hmcs1, mvda, cyp51, lss and cyp39a1), and this cluster was connected through the steroid receptor nr4a1 with some DEGs involved in the immune response and inflammation (fbxo15, socs3a, socs1a, c3b, p2ry2, saa, ddit4, mmp9, irg1l, mpx, fos, fosb, btg2 and erg3). Conversely, the third cluster (the largest cluster) was not interconnected with the previous clusters and corresponded to the main down-regulated DEGs, which are related to different reproductive processes, such as cumulus cells and zona pellucida (zp2.1, zp2.3, zp2.5, zp3b, zp3a.1, zp3b) and chromosome condensation in primordial germ cells (h1m).
3.3.2. Zebrafish larvae exposed to HDM treatment
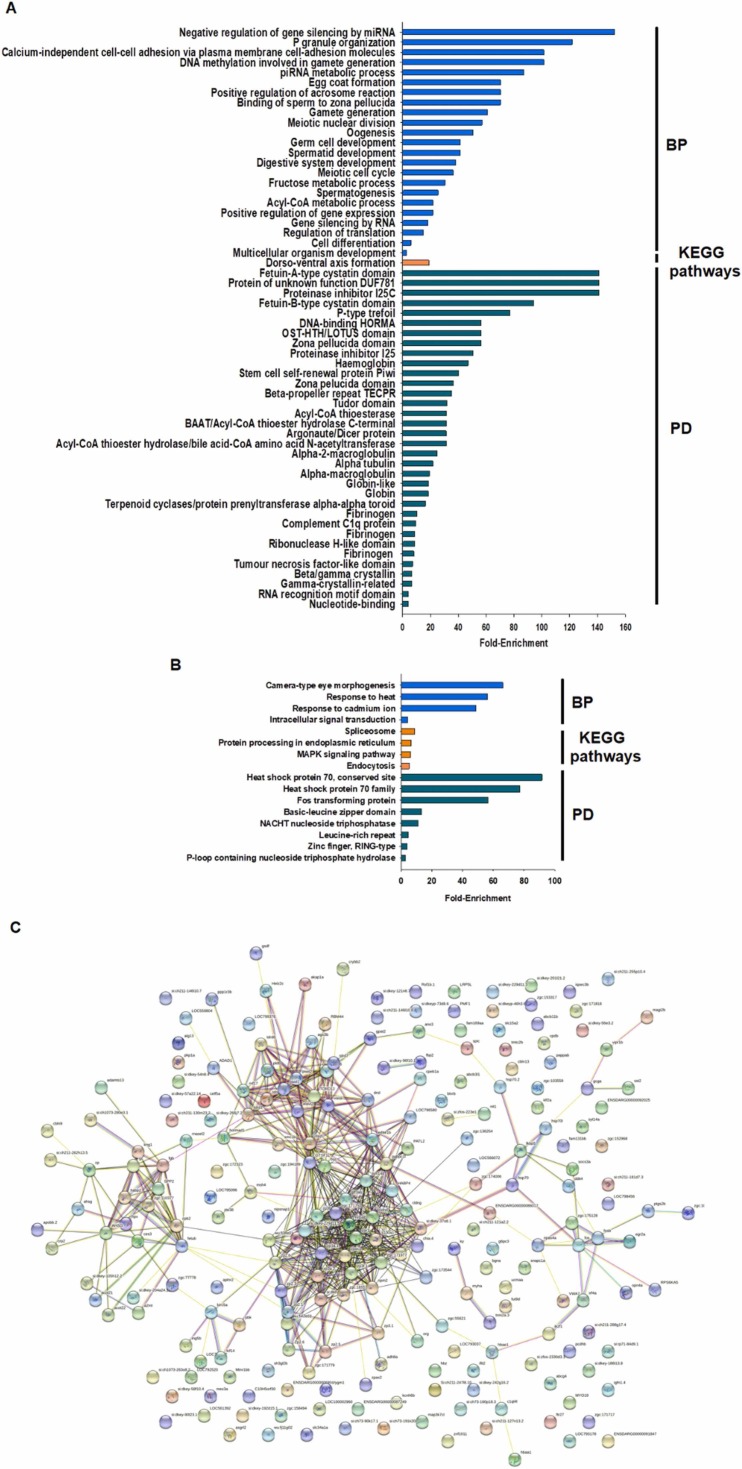
The top 25 most up- and down-regulated DEGs in zebrafish larvae exposed to HDM products are shown in Table 2. Among the top 25 down-regulated genes were also genes related to reproductive processes, such as the zona pellucida glycoproteins zp2.1, zp3, zp2.3, zp2.5, zp2.2, zp2.6, zpax4, zp3a.1 and zp3.2_1, which were inhibited more than 50-fold compared with the control larvae. Although not restricted therein, GO biological process enrichment analyses of the down-regulated DEGs revealed numerous terms related to reproductive processes (“P granule organization”, “DNA methylation involved in gamete generation”, “egg coat formation”, “positive regulation of acrosome reaction”, “binding of sperm to zona pellucida”, “gamete generation”, “meiotic nuclear division”, “oogenesis”, “germ cell development”, “spermatid development”, “meiotic cell cycle” and “spermatogenesis”) and RNA silencing (“negative regulation of gene silencing by miRNA”, “piRNA metabolic process”, “regulation of translation” and “gene silencing by RNA”) ( Fig. 5A). Similar to the results obtained for the GO terms, the enriched protein domains showed a high representation of terms linked to reproduction (“Fetuin-A-type cystatin domain”, “Fetuin-B-type cystatin domain”, “P-type trefoil”, “DNA binding HORMA”, “zona pellucida domain”) or RNA silencing during gametogenesis (“OST-HTH/LOTUS domain”, “stem cell self-renewal protein Piwi”, “argonaute/Dicer protein”, “Tudor domain”) (Fig. 5A). Interestingly, only one KEGG pathway was significantly enriched, which corresponded to “dorsoventral axis formation” (Fig. 5A).
Table 2.
Top 25 most up- and down-regulated DEGs in zebrafish larvae exposed to HDM for 10 days.
| Up-regulated genes |
Down-regulated genes |
||||
|---|---|---|---|---|---|
| Blast2 Go UnitProt/SwissProt Description |
Blast2 Go UnitProt/SwissProt Description |
||||
| Name |
Complete name or Predicted description |
FC |
Name |
Complete name or Predicted description |
FC |
| HDM | HDM | ||||
| dap1b_2 | death associated protein 1b | 16.61 | zp2.1 | zona pellucida glycoprotein 2, tandem | -562.51 |
| rsf1b.1_2 | remodelling and spacing factor 1b | 16.56 | zp3 | zona pellucida glycoprotein 3 | -394.92 |
| BX005305.1 | legumain-like | 12.71 | zp2.3 | zona pellucida glycoprotein 2, tandem duplicate 3 | -321.14 |
| zgc:101699 | cytosolic phospholipase A2 gamma | 11.04 | zp2.5 | zona pellucida glycoprotein 2, tandem duplicate 5 | -290.81 |
| znf1026_1 | zinc finger protein 1026 | 10.32 | zgc:173856 | zgc:173856 | -282.11 |
| si:dkey-9l20.3 | si:dkey-9l20.3 | 10.15 | si:ch211–146l10.7 | si:ch211–146l10.7 | -203.1 |
| si:ch211–121a2.2 | dual specificity phosphatase 29 | 10.13 | si:ch1073–263o8.2 | si:ch1073–263o8.2 | -199.07 |
| zgc:171534 | zgc:171534 | 8.7 | zgc:171446 | Ovostatin homologue | -195.34 |
| opn4a | opsin 4a (melanopsin) | 6.19 | si:ch211–146l10.8 | si:ch211–146l10.8 | -193.35 |
| hsp70.1 | heat shock cognate 70-kd protein, tandem | 6.01 | si:dkey-241l7.5 | Ladderlectin | -178.41 |
| nit1 | nitrilase 1 [Source:ZFIN;Acc:ZDB-GENE-040912–65] | 5.73 | zp2.2 | zona pellucida glycoprotein 2, tandem duplicate 2 | -167.84 |
| msh4 | mutS homologue 4 | 5.24 | zp2.6 | zona pellucida glycoprotein 2, tandem duplicate 6 | -167.48 |
| dnah11_2 | dynein, axonemal, heavy chain 11 | 4.96 | si:dkey-208k4.2 | P43 5 S RNA-binding protein | -160.94 |
| cyp2k6_2 | cytochrome P450, family 2, subfamily K, | 4.77 | pabpn1l | poly(A) binding protein, nuclear 1-like | -159.65 |
| hsp70.2 | heat shock cognate 70-kd protein, tandem | 4.33 | si:dkey-229d11.3 | si:dkey-229d11.3 | -144.39 |
| kcnh6b | potassium voltage-gated channel, subfamily | 4.08 | si:dkey-229d11.5 | endo-1,4-beta-xylanase B | -135.25 |
| efna5b | ephrin-A5b | 4.03 | si:dkeyp-98a7.3_2 | adhesion G protein-coupled receptor L2 | -134.93 |
| si:dkey-242g16.2 | Protein phosphatase 1 regulatory subunit 3 C-B | 3.7 | si:zfos-1505d6.3 | orthologous to human ZP4 (zona pellucida glycoprotein 4) | -130.69 |
| hsp70l | heat shock cognate 70-kd protein, like | 3.66 | zgc:171445 | Ovostatin homologue | -130.07 |
| fosb | FBJ murine osteosarcoma viral oncogene | 3.65 | zpax4 | zona pellucida protein AX 4 | -123.95 |
| si:ch73–191k20.3 | si:ch73–191k20.3 | 3.63 | si:dkeyp-46h3.2 | Rhamnose-binding lectin | -115.89 |
| si:ch211–181d7.1_2 | NACHT, LRR and PYD domains-containing protein 3 | 3.59 | zgc:171977 | zgc:171977 | -115.74 |
| si:ch211–181d7.3 | NLR family CARD domain-containing protein 3 | 3.58 | zp3a.1 | zona pellucida glycoprotein 3a, tándem duplicate 1 | -75.6 |
| si:dkey-204a24.11 | interferon-induced very large GTPase 1 | 3.53 | retsatl | retinol saturase (all-trans-retinol | -75.59 |
| MACIR | macrophage immunometabolism regulator | 3.48 | zp3.2_1 | zona pellucida glycoprotein 3, tandem duplicate 2 | -54.1 |
Fig. 5.
Biological processes (BP), KEGG pathways, protein domains (PD) and protein–protein interactions enriched for the genes modulated by HDM treatment. A) Enrichment analyses of the down-regulated genes. B) Enrichment analyses of the up-regulated genes. C) STRING analysis (protein–protein interaction) of the DEGs under HDM treatment.
The top 25 up-regulated DEGs in zebrafish larvae exposed to HDM products are shown in Table 2. Among them, three members of the heat shock protein 70 family were observed (hsp70.1, hsp70.2 and hsp70l); these genes can be induced by different factors, such as ischaemia, inflammation, osmotic and oxidative stress (Franzellitti and Fabbri, 2005). Up-regulated genes involved in the immune response were recognised, such as dap1b_2 (death-associated protein 1b), efna5b (ephrin-A5b), fosb (FBJ murine osteosarcoma viral oncogene), si:ch211–181d7.1_2 (nlrp3; NACHT, LRR and PYD domain-containing protein 3), si:ch211–181d7.3 (nlrp12; NACHT, LRR and PYD domain-containing protein 12), si:dkey-204a24.11 (gvin1; interferon-induced very large GTPase 1) and macir (macrophage immunometabolism regulator). GO biological process enrichment analysis of the up-regulated DEGs revealed terms involved in the response to stimulus (“response to heat”, “response to cadmium ion” and “intracellular signal transduction”) and developmental process (“camera-type eye morphogenesis”) (Fig. 5B). The KEGG pathways significantly enriched for the up-regulated genes were “spliceosome”, “protein processing in endoplasmic reticulum”, “MAPK signalling pathway” and “endocytosis” (Fig. 5B). Due to the high representation of HSP70-encoding genes, the most enriched protein domains corresponded to “heat shock protein 70, conserved site” and “heat shock protein 70 family” (Fig. 5B).
For the HDM product treatment, the protein-protein interconnections were higher than those observed for PDM products (Fig. 5C). In this case, the main cluster formed by the DEGs corresponded to genes that were also involved in cumulus cells and zona pellucida (zp2.1, zp2.3, zp2.5, zp2.6, zp3.1, zp3a.1, zp3a.2, zp3.b), but some additional genes related to gametogenesis and embryogenesis (patl2, zar1, buc, gtsf1l, dazl, kpna7) were also observed, as well as DEGs involved in cell adhesion and tight junctions (cldnd, cldng), cell proliferation (s100a1, npm2), RNA transcription and protein translation (pabpc1l, lsm14b, gtf3ab, habp4, eif4e1b), among others (Fig. 5C). This cluster was connected with a second cluster, also mainly composed of genes encoding proteins with a role in germ cell development, proliferation and integrity (smc1b, vasa, fkbp6, rnf17, tdrd1, tdrd5, tdrd7, dnd1, piwil1, ago3b, pld6, hormad1). The third cluster included several genes with a role in coagulation (cpb2, habp2, ahsg, kng1, spp2, fgb) but also with iron (hpx, cp), vitamin D (gc) and lipids (apobb.2), transport, and hormones and receptors linked to glucose metabolism (gcga, sst2, vipr1b) (Fig. 5C).
3.3.3. Zebrafish larvae exposed to W-HDM treatment
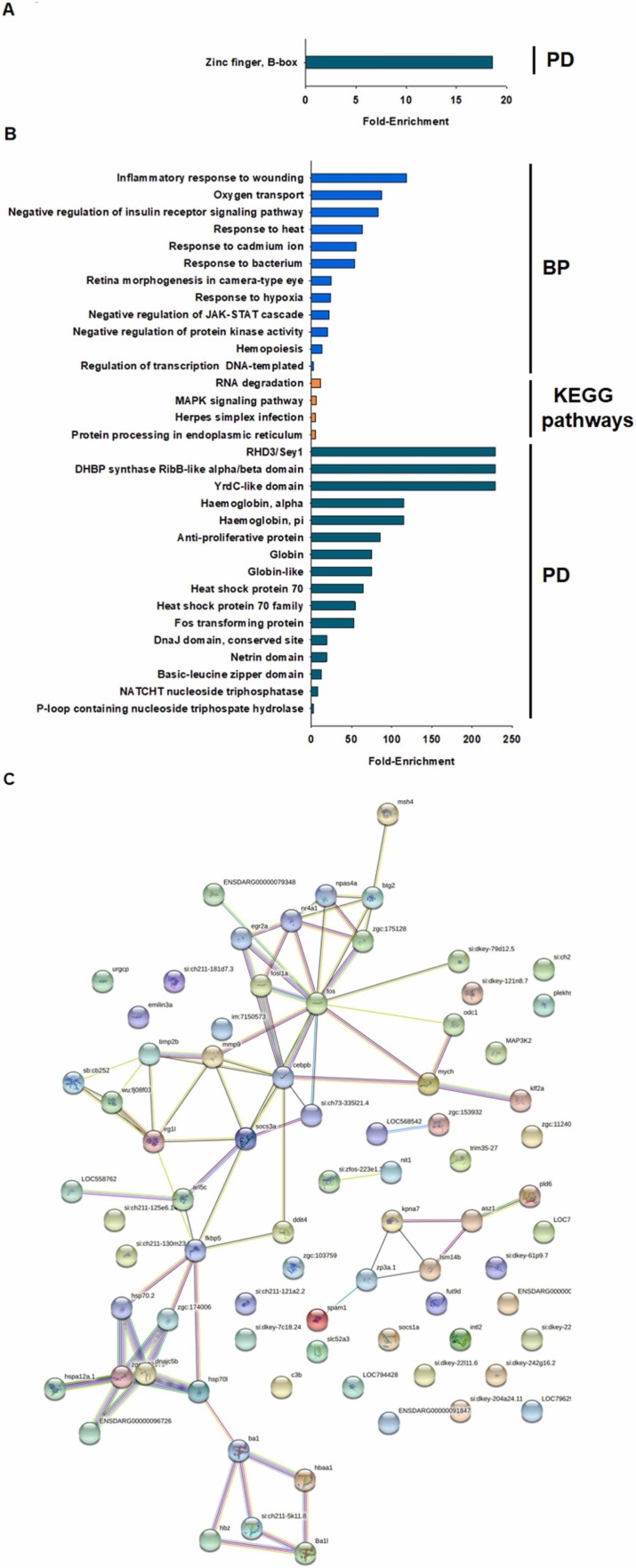
The top 25 most up- and down-regulated DEGs in zebrafish larvae exposed to W-HDM treatment are shown in Table 3. Among the top 25 down-regulated genes were also DEGs related to reproductive processes, such as spam1 (sperm adhesion molecule 1), zp3a.1 and si:zfos-223e1.2 (cfap61), and two members of the HSP70 family, but these DEGs were different from those differentially expressed with the HDM products hspa12a.1 and hspa12a.2. For the down-regulated genes after W-HDM treatment, no GO biological processes or KEGG pathways were significantly enriched, and only the protein domain “Zinc finger, B-box” was overrepresented ( Fig. 6A).
Table 3.
Top 25 most up- and down-regulated DEGs in zebrafish larvae exposed to W-HDM for 10 days.
| Up-regulated genes |
Down-regulated genes |
||||
|---|---|---|---|---|---|
| Blast2 Go UnitProt/SwissProt Description |
Blast2 Go UnitProt/SwissProt Description |
||||
| Name |
Complete name or Predicted description |
FC |
Name |
Complete name or Predicted description |
FC |
| W-HDM | W-HDM | ||||
| si:ch211–170p16.1_1 | SH3 domain and nuclear localisation signals | 83.14 | si:dkey-229d11.3 | AAA - ATPases associated with a variety of cellular activities | -54.27 |
| EIF3J | eukaryotic translation initiation factor 3 subunit J | 70.51 | si:dkey-240e12.6 | rhamnose-binding lectin | -34.9 |
| hbba1_2 | haemoglobin, beta adult 1 | 32.21 | si:ch211–189a21.1 | cytohesin-3 | -16.82 |
| hbba1_1 | haemoglobin, beta adult 2 | 31.58 | kpna7 | karyopherin alpha 7 | -9.09 |
| hbaa1 | haemoglobin, alpha adult 1 | 21.35 | hspa12a.1 | heat shock protein family A (Hsp70) member 12 A.1 | -7.35 |
| si:ch211–125e6.14 | ladderlectin | 20.51 | hspa12a.2 | heat shock protein family A (Hsp70) member 12 A.2 | -4.99 |
| BX548011.4 | BX548011.4 | 20.37 | spam1 | sperm adhesion molecule 1 | -4.91 |
| si:ch211–5k11.8 | haemoglobin subunit alpha | 15.69 | gpr20 | G protein-coupled receptor 20 | -4.72 |
| dap1b_2 | death-associated protein 1b | 13.1 | zp3a.1 | zona pellucida glycoprotein 3a, tándem duplicate 1 | -4.69 |
| itln2_1 | intelectin 2 | 12.51 | lsm14b | LSM family member 14B | -4.54 |
| si:ch211–121a2.2 | dual specificity phosphatase 29 | 11.54 | si:dkey-57c15.1 | acid-sensing ion channel 1 | -4.17 |
| si:ch73–129a22.11 | si:ch73–129a22.11 | 11.33 | si:ch211–194m7.8 | olfactomedin-like domain | -3.94 |
| CR847870.4 | gamma-tubulin binding | 9.45 | ly6m6 | lymphocyte antigen 6 family member M6 | -3.38 |
| zgc:112408 | Stomatin | 8.04 | asz1 | ankyrin repeat, SAM and basic leucine | -3.31 |
| klf2a | kruppel-like factor 2a | 7.17 | si:dkey-22l11.6 | si:dkey-22l11.6 | -3.08 |
| BX088712.5 | BX088712.5 | 6.92 | pld6 | phospholipase D family, member 6 | -2.77 |
| nit1 | nitrilase 1 | 6.37 | map3k2 | mitogen-activated protein kinase kinase | -2.61 |
| hbae5 | haemoglobin, alpha embryonic 5 | 6.21 | si:dkey-61p9.7 | si:dkey-61p9.7 | -2.47 |
| emilin3a | elastin microfibril interfacer 3a | 5.81 | si:zfos-223e1.2 | cilia- and flagella-associated protein 61 | -2.44 |
| msh4 | mutS homologue 4 | 5.32 | trim35–27 | tripartite motif containing 35–27 | -2.28 |
| cyp2k6_2 | cytochrome P450, family 2, subfamily K, | 5.22 | zgc:173581 | zgc:173581 | -2.28 |
| fosl1a | FOS-like antigen 1a | 4.89 | GRIN2B | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | -2.22 |
| si:dkey-242g16.2 | protein phosphatase 1 regulatory subunit 3 C-B | 4.31 | |||
| si:dkey-204a24.11 | interferon-induced very large GTPase 1 | 4.21 | |||
| si:ch211–281l24.3 | oestrogen-responsive finger protein | 3.95 |
Fig. 6.
Biological processes (BP), KEGG pathways, protein domains (PD) and protein–protein interactions enriched for genes modulated by the W-HDM treatment. A) Enrichment analyses of the down-regulated genes. B) Enrichment analyses of the up-regulated genes. C) STRING analysis (protein–protein interaction) of the DEGs under W-HDM treatment.
With regard to the top 25 up-regulated DEGs after W-HDM treatment (Table 3), an important insight is the number of DEGs encoding different haemoglobin types: hbba1_2 (haemoglobin, beta adult 1), hbba1_1 (haemoglobin, beta adult 2), hbbaa1 (haemoglobin, alpha adult 1) and hbae5 (haemoglobin, alpha embryonic 5). The high representation of these haem-binding proteins explains the GO biological process enriched terms “oxygen transport”, “response to hypoxia” and “haemopoiesis” for the up-regulated DEGs (Fig. 6B); different immune, response to stimulus, metabolism and development processes were also significantly enriched (Fig. 6B). The KEGG pathways enriched under H-HDM exposure were “RNA degradation”, “MAPK signalling pathway”, “herpes simplex infection” and “protein processing in endoplasmic reticulum” (Fig. 6B). As expected, among the most enriched protein domains were those related to the haemoglobin proteins (“haemoglobin, alpha”, “haemoglobin, pi”, “globin”, “globin-like”) but also, as observed for the up-regulated genes after HDM treatment, protein domains linked to the HSP70 family (“heat shock protein 70″ and “heat shock protein 70 family”) (Fig. 6B).
In relation to the STRING analysis, the DEGs predominantly grouped into a main cluster composed of genes encoding proteins related to cell proliferation (fos, fosl1a, btg2), immunity and inflammation (irg1l, socs1, socs3a, cebpb, mmp9), extracellular matrix degradation (mmp9, timp2b) and haem binding (hbba1, hbaa1, hbz), among other functions (Fig. 6C).
3.4. Face mask degradation products differentially impact biological processes
Based on the enrichment analyses, we wanted to examine in a more detailed manner different processes that were significantly affected by at least one of the treatments. To achieve this goal, we constructed heatmaps representing the TPM values of the genes across the different treatments. Although the initial face mask material was the same for the three treatments, the stage of degradation of the face masks provoked significant differences in the zebrafish transcriptome response.
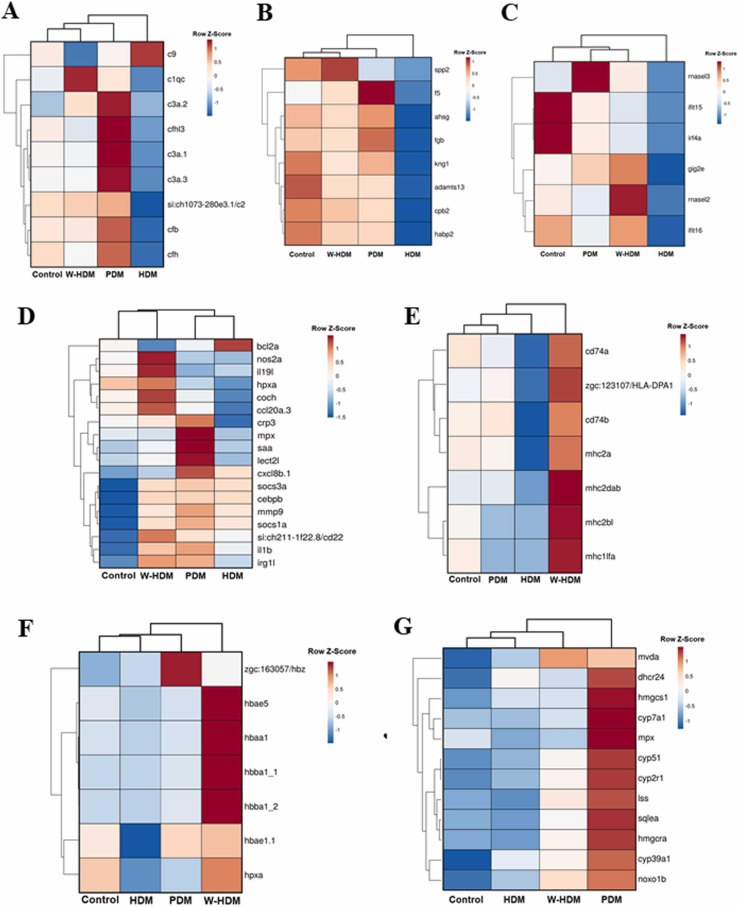
Since several enriched terms linked to the immune response were found to be significantly enriched after the face mask treatments, we separately analysed groups of genes that were well-defined for their main immune function (complement pathway, coagulation, type I interferon system, antigen presentation and inflammation). As expected, due to complement-coagulation cross-talk (Kenawy et al., 2015), both processes followed a similar tendency, with most of the genes involved in these pathways being strongly inhibited in zebrafish larvae exposed to the HDM products and with the lower impact caused by W-HDM products ( Fig. 7A and B). Similar to that observed for the complement and coagulation genes, HDM treatment inhibited certain antiviral genes involved in the type I interferon system (Fig. 7C). In relation to the inflammation process, some genes were up- and down-regulated according to the face mask treatment, and a clear pattern was not observed; however, for the selected genes, the W-HDM treatment clustered together with the control, reflecting an overall lower impact of this treatment on inflammation than the other treatments (Fig. 7D). However, the analysis of the DEGs involved in antigen presentation and modulation at least in one of the treatments revealed that, in this case, W-HDM products had the highest effect, with an up-regulation of these genes after the treatment (Fig. 7E). An unexpected result was also the high representation of haem-binding protein overexpression in the zebrafish larvae treated with the W-HDM products, which were minimally affected by the HDM treatment (Fig. 7F).
Fig. 7.
Heatmaps representing the mean TPM expression values of the DEGs involved in different biological processes. A) Complement pathway. B) Coagulation system. C) Type I interferon-related genes. D) Inflammation; E) Antigen-presentation. F) Haem-binding proteins. G) Isoprenoid/sterol biosynthesis and redox process.
Because terms related to isoprenoid/sterol biosynthesis and redox processes were highly enriched for the PDM treatment, a heatmap representing the genes included in those terms was constructed. In this case, the genes were also almost unaffected by the HDM treatment, whereas some of them were induced by the W-HDM product, and a strong induction was observed in those larvae exposed to PDM products (Fig. 7G).
3.5. Face mask degradation products clearly impair the expression of genes involved in the reproductive process
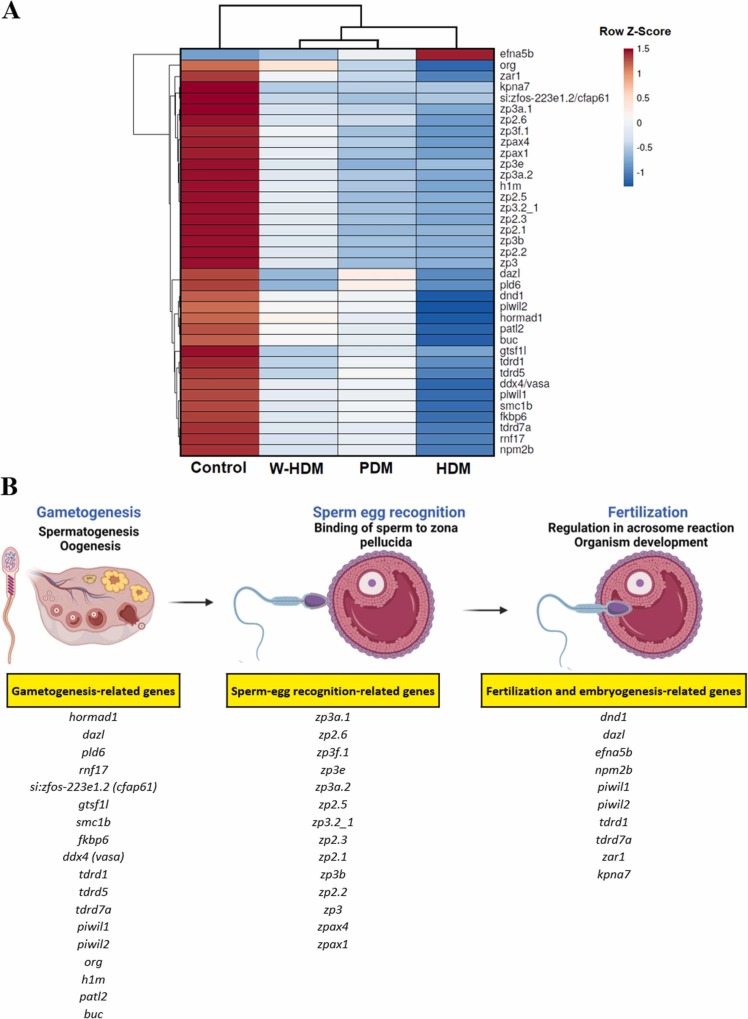
Although only zebrafish larvae exposed to face mask fragments (PDM and HDM) showed an enrichment of reproductive processes for the down-regulated genes in the GO enrichment analyses, genes associated with reproduction were significantly regulated by the three treatments (PDM, HDM and W-HDM); ( Fig. 8A)). The magnitude of the impact on the reproductive process showed an increasing tendency from the W-HDM, followed by PDM, to the HDM treatment, with more than 30 genes significantly affected by the HDM products (Fig. 8A).
Fig. 8.
Analysis of the DEGs involved in reproduction. A) Heatmap representing the mean TPM expression values of the DEGs with a role in reproduction across the different treatments. B) Schematic representation of the different reproduction steps with genes affected by the face mask treatments.
The exposure of zebrafish larvae to the different face mask degradation products induced a generalised inhibition of a multitude of genes with a pivotal role in different stages of reproduction (gametogenesis, sperm-egg recognition, fertilisation and embryo development) (Fig. 8B). Only efna5b (ephrin-a5), a gene involved in fertilisation and embryonic development, was significantly up-regulated by HDM treatment, although the protein encoded by this gene exerts a variety of functions independent of embryogenesis (Worku et al., 2018).
Regarding gametogenesis, genes involved in both male (spermatogenesis) and female (oogenesis) gamete formation were affected; this included genes with a direct role in meiosis (horma1 and smc1b), specific translational regulation in gametocytes (dazl and patl2), oocyte axis specification (buc) or sperm flagella development (cfap61), but most of the affected genes were involved in PIWI-interacting RNA (piRNA) biogenesis (piwil1, piwil2, rnf17, gtsf1l, fkbp6, ddx4, tdrd1, tdrd5, tdrd7a), which protect the germline from invasive transposable elements (Ishizu et al., 2012). Conversely, the DEGs belonging to the sperm-egg recognition process encode a variety of zona pellucida glycoproteins (zp2.1, zp2.2, zp2.3, zp2.5, zp2.6, zp3, zp3a.1, zp3a.2, zp3b, zp3f.1, zp3e, zp3.2_1, zpax1, zpax4), also known as sperm-binding proteins, since they mediate sperm binding to oocytes. Finally, genes with a role in fertilisation and embryogenesis were also down-regulated by the face mask degradation products.
4. Discussion
4.1. Surgical face masks are a material of emerging concern in aquatic environments
After face mask use, wastes are mismanaged on land (Aragaw, 2020, Dharmaraj et al., 2021), and their degradation can occur by means of different processes, such as weathering, ageing, and microbial degradation (Hasan et al., 2021). After fragmentation and degradation, small pieces of face masks are released into aquatic systems and can release microplastics and other substances, such as B, Al, Ti, Fe, Cu and Sr. The concentration of these compounds after 10 days of degradation with UVC in water did not increase much with respect to the controls, although the concentrations of the cited inorganic elements were dependent on the face mask degradation stage. Although the leaching of inorganic compounds is characteristic of the type of face mask (Sullivan et al., 2021), the degradation processes, the time in the aquatic media and the size of the face mask fragments are relevant in the leaching of chemical compounds from face masks into aquatic systems. According to the leached organic substances, only three compounds were identified as secondary metabolites of face mask degradation, which were not found in intact face masks. Therefore, these compounds are probably byproducts of the degradation of face masks in water. Two of the identified substances corresponded to heterocyclic aromatic compounds (2,4,6-trimethyl-pyridine and N,N-dimethyl 1-dodecanamine) that can be found in creosote-contaminated sites formed by thermal processes related to coal and fossil fuel (Johansen et al., 1997, Zemanek et al., 1997). In the case of 2,4,6-trimethyl-pyridine, this compound is found in various man-made compounds, such as dyes, industrial solvents, herbicides, pesticides, polymers and many natural metabolites (Stankevičiūtė et al., 2016). Most likely, 2,4,6-trimethyl-pyridine occurs from the degradation of olefin polymers. Polyolefins are commercial polymers that are more common and cheaper than others on the market. The trimethylpyridine and dimethylpyridine mixture represents an important part of the composition of polyolefins (Zhang et al., 2020). The use of similar chemical substances, such as 2,4,6-trimethyl-triazine, in olefins offers highly crystalline honeycomb-like structures and displays well-defined nanofibrillar morphologies with uniform diameters (Zhang et al., 2020). Therefore, this structure could be perfect for an efficient face mask design (Ullah et al., 2020). In the case of the second organic compound identified, N,N-dimethyl 1-dodecanamine is identified in alkanes and olefins (Zhang et al., 2020). Luryldimethylamine oxide (lauryl amine oxide), also known as dodecyl dimethylamine oxide (DDAO), is an amine oxide with a C12 alkyl tail and is one of the most frequently used surfactants (Varade et al., 2005). The third compound found was alanine, N-methyl-N-methoxycarbonyl-, undecyl ester, which is probably a byproduct of degradation or a prophase compound, since its industrial application has not been assigned but could include antibacterial compounds or dyes. A recent paper showed that single-use surgical face masks can act as dye carriers (methylene blue, crystal violet and malachite green) in aquatic environments (Anastopoulos and Pashalidis, 2021).
4.2. Toxicogenomic impacts of face mask degradation products in zebrafish larvae
Although fragments of face masks do not seem to be a high source of inorganic and nonpolar pollutants leaching into aquatic systems, the face mask fragments and fibres (mainly composed of plastic polymers) released from their degradation could be a source of emerging pollutants. Although face masks have not yet been of special concern, they could have environmental and health impacts on aquatic organisms exposed to these materials in specific locations. Currently, the concentration of face mask degradation products in aquatic ecosystems is not known; therefore, this study is based on the microplastic concentration already known in natural compartments (Koutnik et al., 2021) and used in previous works (Sendra et al., 2019a, Sendra et al., 2019b, Sendra et al., 2021b).
The degradation stage of the surgical face masks influences their effects, since smaller fragments are more bioavailable for organisms (Wang et al., 2020). This finding could explain the higher gene modulation induced by HDM products on zebrafish larvae as determined by RNA-Seq analysis. However, differential responses were observed for a variety of processes, with certain gene groups remaining unaffected by HDM treatment but being strongly regulated by the PDM and W-HDM treatments. This result could probably be a consequence of the presence of different byproducts generated during the degradation process of the face mask fragments, which could undergo modifications over time. These modifications are probably accelerated with the HDM and W-HDM treatments compared with the PDM due to the higher initial fragmentation. However, the presence of face mask fragments in the HDM and PDM treatments compared with the W-HDM treatment could allow an accumulation of microfibres and probably a higher variety of pollutants directly ingested by larvae. Each treatment in this study showed a particular profile of biological processes and pathways affected, offering a specific footprint in gene regulation. However, a common effect among the three treatments was the down-regulation of genes involved in reproductive processes. Under environmental conditions, the three stages of face mask degradation could be found in nature; therefore, more complex biological processes could be found compared with the results obtained in this work.
In this work it was revealed that under the selected conditions, exposure of zebrafish larvae to face mask degradation products could provoke effects on the immune response since, as determined by RNA-Seq analysis, several immune-related genes were affected by the three treatments. Xenobiotics frequently found in natural compartments can significantly suppress or enhance immune responsiveness, provoking immunotoxicity (Kreitinger et al., 2016). While W-HDM and PDM were able to induce the expression of genes involved in certain aspects of the immune response, such as antigen presentation or inflammation-related genes, HDM treatment suppressed the expression of genes involved in complement, coagulation and antiviral responses and dysregulated the expression of several genes with a role in inflammation. Moreover, numerous xenobiotics can also bind haemoglobin members via noncovalent interactions and can affect their function through alteration of the equilibrium between oxygenated and deoxygenated haemoglobin (Gaurav et al., 2021), which could explain the up-regulation of different haemoglobin members in the W-HDM treatment.
A mechanism to fight against xenobiotics is the activation of the KEAP1-NRF2 system, which is involved in the antioxidant enzyme activity of phase II transformation of xenobiotics. In the larvae treated with PDM products, induction of certain genes related to the immune response was observed, which could be in accordance with an up-regulation of genes involved in redox metabolism. The anti-inflammatory gene irg1l (immunoresponsive gene 1-like), which transforms cis-aconitate into aconitate, is involved in the activation of the KEAP1-NRF2 system, and this gene showed a higher expression level in larvae treated with PDM, especially in larvae exposed to W-HDM products. Therefore, the higher expression of redox-related genes and sterol biosynthesis (which requires oxygen radicals for their correct functioning) in those treatments could be related to the modulation of the immune genes observed in those groups of treatments (Spann and Glass, 2013). Moreover, some of the enzymes involved in sterol biosynthesis (cytochrome P450 enzymes) have a role in xenobiotic metabolism (Esteves et al., 2021, Lepesheva et al., 2008, Stravoravdis et al., 2021). Previous works have reported specific plastic additives as plasticizers, flame retardants and biocides as endocrine disrupters that mimic natural steroid hormones (Howdeshell et al., 2007, Sendra et al., 2020), which could also be a reasonable explanation for the modulation of genes involved in steroid biosynthesis by PDM treatment. Interestingly, although endocrine disruptors can interfere with the synthesis and activity of sex steroid hormones and, consequently, lead to reproductive disorders at different levels (Tyler et al., 1998), the treatment showing the lowest alteration of genes involved in steroid biosynthesis (HDM) was the same, inducing the highest effect on reproduction-related genes. Therefore, other routes, not only sterol pathways, could induce impairments in the expression of genes related to reproductive processes.
Despite all the effects provoked by face mask products at transcriptome level, reproductive processes showed the most remarkable changes, as determined by RNA-Seq analysis of zebrafish larvae exposed to the three treatments (PDM, HDM and W-HDM) and compared with the control larvae. Although enriched GO terms related to reproduction were only observed for the down-regulated genes in the PDM and HDM treatments, some of the genes included in those categories were also down-regulated in the W-HDM treatment. A clear increasing effect on reproduction was observed with the different treatments as follows: W-HDM<PDM<HDM. In addition to gametogenesis (with representative genes for both oogenesis and spermatogenesis) and embryogenesis, genes involved in fertility and reproductive processes were significantly down-regulated in the three steps of fertilisation Step 1; species-specific binding of sperm to eggs, Step 2; zona pellucida proteins, Ca2+, G proteins and the acrosome reaction, and Step 3; penetration of the egg zona pellucida by sperm (Wassarman et al., 2001).
Gametogenesis-related genes seem to be highly impacted by face mask treatments, with down-regulated genes involved in meiosis, translational regulation in gametocytes, oocyte axis specification or sperm flagella development. However, most gametogenesis-affected genes are related to PIWI-interacting RNA (piRNA) biogenesis. piRNAs are a class of small noncoding RNAs that form the piRNA-induced silencing complex (piRISC) in the animal germ line (Siomi et al., 2011), which protects the integrity of the genome from invasive transposable elements (Ishizu et al., 2012, Siomi et al., 2011). piRNAs are tightly bound to PIWI proteins of the Argonaute family, and these can interact with many other proteins required for PIWI function, regulating piRNA-mediated silencing (Siomi et al., 2011). This phenomenon is observed for Tudor-domain-containing proteins (TDRDs) (Ishizu et al., 2012), but several non-PIWI, non-TDRD proteins are also involved in piRNA biogenesis, such as DDX4 (VASA), FKBP6 or GTSF1 (Dönertas et al., 2013, Ishizu et al., 2012). In this work, we observed a drastic down-regulation of PIWI-like genes (piwil1, piwil2), TDRD genes (tdrd1, tdrd5, tdrd7a, rnf17 -tdrd4-), ddx4, fkbp6 and gtsf1l genes, which suggested a severe impairment of piRNA-mediated silencing function. Consequently, based on gene expression analyses, gametogenesis disorders could be expected in zebrafish exposed to face mask degradation products. Nevertheless, further investigations are needed to validate this finding.
Among the genes involved in the reproductive process and affected by the treatments, we found a multitude of zona pellucida glycoproteins. Whereas mammals possess four genes encoding different types of zona pellucida proteins (ZP1, ZP2, ZP3 and ZP4), the copy number of these genes has suffered an expansion in teleosts. This evolutionary process, called concerted evolution, is probably a consequence of the reproductive strategy of fish, which need to produce eggs with high frequency and in large numbers (Mold et al., 2009). ZP proteins have been conserved in mammalian and nonmammalian eggs over the course of several hundred million years of evolution. They play vital roles during oogenesis, fertilisation and preimplantation development (Wassarman, 2008), but they also participate in several steps in the fertilisation pathway, such as receptors for sperm, inducers of sperm exocytosis and the acrosome reaction (Florman et al., 2010, Wassarman et al., 2001). Although some of the genes mentioned above also have a role in fertilisation, certain genes with a specific function in embryogenesis were also affected by the treatments, including zar1 (zygote arrest protein 1), an oocyte-specific gene that encodes a protein that is thought to function in the initiation of embryogenesis (Wu et al., 2003).
In mammals, most of the abovementioned proteins are produced in reproductive organs; however, in teleosts and in many other oviparous vertebrates, the major constituents of the eggs are synthesised in the liver and transported to the oocyte for uptake (Arukwe and Goksøyr, 2003). The liver is the main organ for detoxification of xenobiotics; therefore, damage to this organ due to face masks with plastic additives as endocrine disruptors (Sendra, et al., 2020) could provoke impairments in reproductive, metabolic and signalling processes (Qiao et al., 2016), as observed in the transcriptome of zebrafish larvae exposed to PDM and HDM treatments. The potential impairment of reproduction by face mask products could include effects at the individual but also at the population level, since fertility or reproductive success can compromise the population (Mathieu-Denoncourt et al., 2015). Furthermore, these changes could be transferred to offspring. From the beginning of this century, there is evidence that epigenetic processes play a pivotal role in regulating many genes involved in the endocrine system. There were modulated genes in this work belonging to the cytochrome P450 family, which are regulated by DNA methylation (an epigenetic mark); consequently, they could impact the major endocrine pathways in which they are involved (Jacobs et al., 2017, Rosenfeld, 2019). Disturbances in meiotic epigenetic inheritance were observed in enriched biological processes in zebrafish exposed to HDM, which could impact further generations owing to the involvement of the germ line. Environmentally induced alterations in the germline, whether genetic or epigenetic, will be transmitted to subsequent generations and could induce altered phenotypes in progeny and therefore exert effects at the population level (Jacobs et al., 2017). Alterations in the reproduction process and fitness were observed in natural populations of different animals exposed to plastic waste (Patrício Silva et al., 2021), which could be an indicative of similar effects for face mask degradation products. Indeed, reproduction is one of the sublethal effects induced by toxic chemicals more commonly extrapolated to a population level, since reproduction integrates the outcome of many interacting biological process (Martin et al., 2014).
This work only shows the complexity and effects of face mask products as emergent pollutants in aquatic organisms at transcriptome level. However, face mask fragments and their leaching products coexist with other environmental xenobiotics, such as metals, antibiotics, nanoparticles, pesticides, veterinary drugs, chemical compounds used in industry and personal care products. Therefore, the additive and synergic effects of all these pollutants have been unknown until now. Knowing the behaviour and biological effects of face mask products in combination with other xenobiotics could provide more real insight into the processes occurring in natural environments.
5. Conclusions
This study reveals, at the transcriptome level, the impact of the most commonly used PPE over the COVID-19 pandemic (face mask products) in the model organism zebrafish. As hypothesised, surgical face masks can release inorganic and organic compounds in aquatic systems, and the release of these compounds could be related to the degradation stage of face masks. The degradation stages are associated with the size of face mask fragments, showing different toxicogenomic effects according to each treatment. Despite face mask fragments coming from degradation, water that has been in contact with face masks could leave a footprint in aqueous media with unwanted effects in aquatic organisms. Although W-HDM showed significant toxicogenomic effects in zebrafish larvae, RNA-Seq analysis confirmed that those treatments containing face mask fragments (PDM and HDM) induced a more severe effect. Although not restricted to, reproductive process was the most remarkable function affected in zebrafish larvae exposed to face mask degradation products. Among the different treatments, HDM products induced the most powerful effect, with the down-regulation of a multitude of genes with a pivotal role in different steps of the reproductive process, from gametogenesis to embryogenesis.
The results suggest that the biological processes affected by face mask fragments could not only occur at the individual but also at the population level through fertility and reproductive impairments. However, these effects will depend on the stage of degradation of the surgical face masks.
Funding sources
Our laboratory is funded by projects PID2020-119532RB-I00 from Ministerio de Ciencia e Innovación), 0474_BLUEBIOLAB from EU FEDER Programa Interreg España-Portugal and IN607B 2019/01 from Consellería de Economía, Emprego e Industria (GAIN), Xunta de Galicia.
CRediT authorship contribution statement
Marta Sendra: Investigation, Writing – original draft. Patricia Pereiro: Investigation, Writing – review & editing. Pilar Yeste: Investigation, Writing – review & editing. Beatriz Novoa: Conceptualization, Supervision, Writing – review & editing, Funding acquisition. Antonio Figueras: Conceptualization, Supervision, Methodology, Software, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Marta Sendra wishes to acknowledge the Spanish Ministerio de Ciencia, Innovación y Universidades for her Juan de la Cierva contract (TJFI-2017-32493) and CEI·MAR for the funding of young researcher projects. Patricia Pereiro wishes to thank the Axencia Galega de Innovación (GAIN, Xunta de Galicia) for her postdoctoral contract (IN606B-2018/010).
Editor: Dr. R Teresa
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jhazmat.2021.128186.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
References
- Ammendolia J., Saturno J., Brooks A.L., Jacobs S., Jambeck J.R. An emerging source of plastic pollution: environmental presence of plastic personal protective equipment (PPE) debris related to COVID-19 in a metropolitan city. Environ. Pollut. 2021;269 doi: 10.1016/j.envpol.2020.116160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastopoulos I., Pashalidis I. Single-use surgical face masks, as a potential source of microplastics: do they act as pollutant carriers? J. Mol. Liq. 2021;326 doi: 10.1016/j.molliq.2020.115247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D.R., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N.J., Oinn T.M., Pagni M., Servant F., Sigrist C.J.A., Zdobnov E.M. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragaw T.A. Surgical face masks as a potential source for microplastic pollution in the COVID-19 scenario. Mar. Pollut. Bull. 2020;159 doi: 10.1016/j.marpolbul.2020.111517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arukwe A., Goksøyr A. Eggshell and egg yolk proteins in fish: hepatic proteins for the next generation: Oogenetic, population, and evolutionary implications of endocrine disruption. Comp. Hepatol. 2003;2:1–21. doi: 10.1186/1476-5926-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson N.U., Bassey D.E., Palanisami T. COVID pollution: impact of COVID-19 pandemic on global plastic waste footprint. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson N.U., Fred-Ahmadu O.H., Bassey D.E., Atayero A.A. COVID-19 pandemic and emerging plastic-based personal protective equipment waste pollution and management in Africa. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/j.jece.2021.105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedervall T., Hansson L.A., Lard M., Frohm B., Linse S. Food chain transport of nanoparticles affects behaviour and fat metabolism in fish. PLoS One. 2012;7:1–6. doi: 10.1371/journal.pone.0032254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y.J., Jia Y.F., Chen N., Bian W.P., Li Q.K., Ma Y.B., Chen Y.L., Pei D.S. Zebrafish as a model system to study toxicology. Environ. Toxicol. Chem. 2014;33:11–17. doi: 10.1002/etc.2406. [DOI] [PubMed] [Google Scholar]
- Dharmaraj S., Ashokkumar V., Hariharan S., Manibharathi A., Show P.L., Chong C.T., Ngamcharussrivichai C. The COVID-19 pandemic face mask waste: a blooming threat to the marine environment. Chemosphere. 2021;272 doi: 10.1016/j.chemosphere.2021.129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönertas D., Sienski G., Brennecke J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013;27:1693–1705. doi: 10.1101/gad.221150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves F., Rueff J., Kranendonk M. The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family. J. Xenobiotics. 2021;11:94–114. doi: 10.3390/jox11030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira I., Venâncio C., Lopes I., Oliveira M. Nanoplastics and marine organisms: what has been studied? Environ. Toxicol. Pharmacol. 2019;67:1–7. doi: 10.1016/j.etap.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Florman H.M., Jungnickel M.K., Sutton K.A. Shedding light on sperm pHertility. Cell. 2010;140:310–312. doi: 10.1016/j.cell.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Franzellitti S., Fabbri E. Differential HSP70 gene expression in the Mediterranean mussel exposed to various stressors. Biochem. Biophys. Res. Commun. 2005;336:1157–1163. doi: 10.1016/j.bbrc.2005.08.244. [DOI] [PubMed] [Google Scholar]
- Gaurav M., Natesh A., Arundhati A., Mariam D. Biochemical aspects of hemoglobin-xenobiotic interactions and their implications in drug discovery. Biochimie. 2021 doi: 10.1016/j.biochi.2021.08.006. [DOI] [PubMed] [Google Scholar]
- Gigault J., Halle A., ter Baudrimont M., Pascal P.Y., Gauffre F., Phi T.L., El Hadri H., Grassl B., Reynaud S. Current opinion: what is a nanoplastic? Environ. Pollut. 2018;235:1030–1034. doi: 10.1016/j.envpol.2018.01.024. [DOI] [PubMed] [Google Scholar]
- Güven O., Gökdağ K., Jovanović B., Kıdeyş A.E. Microplastic litter composition of the Turkish territorial waters of the Mediterranean Sea, and its occurrence in the gastrointestinal tract of fish. Environ. Pollut. 2017;223:286–294. doi: 10.1016/j.envpol.2017.01.025. [DOI] [PubMed] [Google Scholar]
- Hasan N.A., Heal R.D., Bashar A., Haque M.M. Face masks: protecting the wearer but neglecting the aquatic environment? A perspective from Bangladesh. Environ. Chall. 2021;4 doi: 10.1016/j.envc.2021.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.C. Understanding face mask use to prevent coronavirus and other illnesses: development of a multidimensional face mask perceptions scale. Br. J. Health Psychol. 2020;25:912–924. doi: 10.1111/bjhp.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howdeshell K.L., Furr J., Lambright C.R., Rider C.V., Wilson V.S., Gray L.E. Cumulative effects of dibutyl phthalate and diethylhexyl phthalate on male rat reproductive tract development: altered fetal steroid hormones and genes. Toxicol. Sci. 2007;99:190–202. doi: 10.1093/toxsci/kfm069. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Yin Y., Liu C., Li M., Yu X., Wang X., Zhang H., Muhammad T., Gao F., Li W., Chen Z.J., Liu H., Ma J. Absence of murine CFAP61 causes male infertility due to multiple morphological abnormalities of the flagella. Sci. Bull. 2020;65:854–864. doi: 10.1016/j.scib.2020.01.023. [DOI] [PubMed] [Google Scholar]
- Ishizu H., Siomi H., Siomi M.C. Biology of Piwi-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.N., Marczylo E.L., Guerrero-Bosagna C., Rüegg J. Marked for life: epigenetic effects of endocrine disrupting chemicals. Annu. Rev. Environ. Resour. 2017;42:105–160. doi: 10.1146/annurev-environ-102016-061111. [DOI] [Google Scholar]
- Jambeck J.R., Ji Q., Zhang Y.-G., Liu D., Grossnickle D.M., Luo Z.-X. Plastic waste inputs from land into the ocean. Science. 2015;347:764–768. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- Jeong J., Choi J. Adverse outcome pathways potentially related to hazard identification of microplastics based on toxicity mechanisms. Chemosphere. 2019;231:249–255. doi: 10.1016/j.chemosphere.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Jin Y., Xia J., Pan Z., Yang J., Wang W., Fu Z. Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish. Environ. Pollut. 2018;235:322–329. doi: 10.1016/j.envpol.2017.12.088. [DOI] [PubMed] [Google Scholar]
- Johansen, S.S., Hansen, A.B., Mosbœk, H., Arvin, E., 1997. Identification of Heteroaromatic and Other Organic Compounds in Ground Water at Creosote‐contaminated Sites in Denmark.
- Kalina, M., Ali, F., Tilley, E., 2020. Since January 2020 Elsevier Has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID- 19. The COVID-19 Resource Centre Is Hosted on Elsevier Connect, the Company's Public News and Information.
- Kenawy H.I., Boral I., Bevington A. Complement-coagulation cross-talk: a potential mediator of the physiological activation of complement by low pH. Front. Immunol. 2015;6:1–10. doi: 10.3389/fimmu.2015.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.W., Chae Y., Kim D., An Y.J. Zebrafish can recognize microplastics as inedible materials: quantitative evidence of ingestion behavior. Sci. Total Environ. 2019;649:156–162. doi: 10.1016/j.scitotenv.2018.08.310. [DOI] [PubMed] [Google Scholar]
- Koutnik V.S., Leonard J., Alkidim S., DePrima F.J., Ravi S., Hoek E.M.V., Mohanty S.K. Distribution of microplastics in soil and freshwater environments: global analysis and framework for transport modeling. Environ. Pollut. 2021;274 doi: 10.1016/j.envpol.2021.116552. [DOI] [PubMed] [Google Scholar]
- Kreitinger J.M., Beamer C.A., Shepherd D.M. Environmental immunology: lessons learned from exposure to a select panel of immunotoxicants. J. Immunol. 2016;196:3217–3225. doi: 10.4049/jimmunol.1502149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva G.I., Hargrove T.Y., Kleshchenko Y., Nes W.D., Villalta F., Waterman M.R. CYP51: a major drug target in the cytochrome P450 superfamily. Lipids. 2008;43:1117–1125. doi: 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.W.F., Sun Q. Electrostatic charged nanofiber filter for filtering airborne novel coronavirus (COVID-19) and nano-aerosols. Sep. Purif. Technol. 2020;250 doi: 10.1016/j.seppur.2020.116886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limonta G., Mancia A., Benkhalqui A., Bertolucci C., Abelli L., Fossi M.C., Panti C. Microplastics induce transcriptional changes, immune response and behavioral alterations in adult zebrafish. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-52292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B., Jager T., Nisbet R.M., Preuss T.G., Grimm V. Limitations of extrapolating toxic effects on reproduction to the population level. Ecol. Appl. 2014;24:1972–1983. doi: 10.1890/14-0656.1. [DOI] [PubMed] [Google Scholar]
- Mathieu-Denoncourt J., Wallace S.J., de Solla S.R., Langlois V.S. Plasticizer endocrine disruption: highlighting developmental and reproductive effects in mammals and non-mammalian aquatic species. Gen. Comp. Endocrinol. 2015;219:74–88. doi: 10.1016/j.ygcen.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mold D.E., Dinitz A.E., Sambandan D.R. Regulation of zebrafish zona pellucida gene activity in developing oocytes. Biol. Reprod. 2009;81:101–110. doi: 10.1095/biolreprod.108.071720. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Dahm R. Oxford University Press; 2002. Zebrafish. [Google Scholar]
- de Oliveira M., Frihling B.E.F., Velasques J., Filho F.J.C.M., Cavalheri P.S., Migliolo L. Pharmaceuticals residues and xenobiotics contaminants: occurrence, analytical techniques and sustainable alternatives for wastewater treatment. Sci. Total Environ. 2020;705 doi: 10.1016/j.scitotenv.2019.135568. [DOI] [PubMed] [Google Scholar]
- Patrício Silva A.L., Prata J.C., Mouneyrac C., Barcel D., Duarte A.C., Rocha-Santos T. Risks of Covid-19 face masks to wildlife: present and future research needs. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher J.F., Ammendolia J., Rochman C.M., Mallory M.L. Assessing plastic debris in aquatic food webs: what we know and don’t know about uptake and trophic transfer. Environ. Rev. 2019;27:304–317. doi: 10.1139/er-2018-0079. [DOI] [Google Scholar]
- Qiao Q., Le Manach S., Huet H., Duvernois-Berthet E., Chaouch S., Duval C., Sotton B., Ponger L., Marie A., Mathéron L., Lennon S., Bolbach G., Djediat C., Bernard C., Edery M., Marie B. An integrated omic analysis of hepatic alteration in medaka fish chronically exposed to cyanotoxins with possible mechanisms of reproductive toxicity. Environ. Pollut. 2016;219:119–131. doi: 10.1016/j.envpol.2016.10.029. [DOI] [PubMed] [Google Scholar]
- Qiao R., Lu K., Deng Y., Ren H., Zhang Y. Combined effects of polystyrene microplastics and natural organic matter on the accumulation and toxicity of copper in zebrafish. Sci. Total Environ. 2019;682:128–137. doi: 10.1016/j.scitotenv.2019.05.163. [DOI] [PubMed] [Google Scholar]
- Qiao R., Sheng C., Lu Y., Zhang Y., Ren H., Lemos B. Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci. Total Environ. 2019;662:246–253. doi: 10.1016/j.scitotenv.2019.01.245. [DOI] [PubMed] [Google Scholar]
- Raju S., Carbery M., Kuttykattil A., Senthirajah K., Lundmark A., Rogers Z., SCB S., Evans G., Palanisami T. Improved methodology to determine the fate and transport of microplastics in a secondary wastewater treatment plant. Water Res. 2020 doi: 10.1016/j.watres.2020.115549. [DOI] [PubMed] [Google Scholar]
- Rasmussen G. CBC; 2020. Those COVID-19 Masks, Gloves and Wipes We’re All Using Are Polluting Land and Sea. (n.d.) [Google Scholar]
- Rodríguez-Seijo A., Arenas-Lago D., Andrade M.L., Vega F.A. Identifying sources of Pb pollution in urban soils by means of MC-ICP-MS and TOF-SIMS. Environ. Sci. Pollut. Res. 2015;22:7859–7872. doi: 10.1007/s11356-014-4027-9. [DOI] [PubMed] [Google Scholar]
- Rosenfeld C.S. Effects of phytoestrogens on the developing brain, gut microbiota, and risk for neurobehavioral disorders. Front. Nutr. 2019;6 doi: 10.3389/fnut.2019.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendra M., Saco A., Yeste M.P., Romero A., Novoa B., Figueras A. Nanoplastics: from tissue accumulation to cell translocation into Mytilus galloprovincialis hemocytes. resilience of immune cells exposed to nanoplastics and nanoplastics plus Vibrio splendidus combination. J. Hazard. Mater. 2019 doi: 10.1016/j.jhazmat.2019.121788. [DOI] [PubMed] [Google Scholar]
- Sendra M., Staffieri E., Yeste M.P., Moreno-Garrido I., Gatica J.M., Corsi I., Blasco J. Are the primary characteristics of polystyrene nanoplastics responsible for toxicity and ad/absorption in the marine diatom Phaeodactylum tricornutum? Environ. Pollut. 2019;249:610–619. doi: 10.1016/J.ENVPOL.2019.03.047. [DOI] [PubMed] [Google Scholar]
- Sendra M., Pereiro P., Figueras A., Novoa B. An integrative toxicogenomic analysis of plastic additives. J. Hazard. Mater. 2020 doi: 10.1016/j.jhazmat.2020.124975. [DOI] [PubMed] [Google Scholar]
- Sendra M., Pereiro P., Yeste M.P., Mercado L., Figueras A., Novoa B. Size matters: zebrafish (Danio rerio) as a model to study toxicity of nanoplastics from cells to the whole organism. Environ. Pollut. 2021;268 doi: 10.1016/j.envpol.2020.115769. [DOI] [PubMed] [Google Scholar]
- Sendra M., Sparaventi E., Novoa B., Figueras A. An overview of the internalization and effects of microplastics and nanoplastics as pollutants of emerging concern in bivalves. Sci. Total Environ. 2021;753 doi: 10.1016/j.scitotenv.2020.142024. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Spann N.J., Glass C.K. Sterols and oxysterols in immune cell function. Nat. Immunol. 2013;14:893–900. doi: 10.1038/ni.2681. [DOI] [PubMed] [Google Scholar]
- Stankevičiūtė J., Vaitekūnas J., Petkevičius V., Gasparavičiūtė R., Tauraitė D., Meškys R. Oxyfunctionalization of pyridine derivatives using whole cells of Burkholderia sp. MAK1. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep39129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stravoravdis, S., Marra, R.E., Leblanc, N.R., Crouch, J.A., Hulvey, J.P., 2021. Evidence for the Role of CYP51A and Xenobiotic Detoxification in Differential Sensitivity to Azole Fungicides in Boxwood Blight Pathogens. [DOI] [PMC free article] [PubMed]
- Sullivan G.L., Delgado-Gallardo J., Watson T.M., Sarp S. An investigation into the leaching of micro and nano particles and chemical pollutants from disposable face masks – linked to the COVID-19 pandemic. Water Res. 2021;196 doi: 10.1016/j.watres.2021.117033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., Jensen L.J., von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.C., Olson Y., Mitchell R.P., Davis A., Rowland S.J., John A.W.G., McGonigle D., Russell A.E. Lost at sea: where is all the plastic? Science. 2004;304:838. doi: 10.1126/science.1094559. [DOI] [PubMed] [Google Scholar]
- Tyler C.R., Jobling S., Sumpter J.P. Endocrine disruption in wildlife: a critical review of the evidence. Crit. Rev. Toxicol. 1998;28:319–361. doi: 10.1080/10408449891344236. [DOI] [PubMed] [Google Scholar]
- Ullah S., Ullah A., Lee J., Jeong Y., Hashmi M., Zhu C., Joo K., Il, Cha H.J., Kim I.S. Reusability comparison of melt-blown vs nanofiber face mask filters for use in the coronavirus pandemic. ACS Appl. Nano Mater. 2020;3:7231–7241. doi: 10.1021/acsanm.0c01562. [DOI] [PubMed] [Google Scholar]
- Varade D., Joshi T., Aswal V.K., Goyal P.S., Hassan P.A., Bahadur P. Micellar behavior of mixtures of sodium dodecyl sulfate and dodecyldimethylamine oxide in aqueous solutions. Colloids Surf. A Physicochem. Eng. Asp. 2005;259:103–109. doi: 10.1016/j.colsurfa.2005.02.019. [DOI] [Google Scholar]
- Wang W., Ge J., Yu X. Bioavailability and toxicity of microplastics to fish species: a review. Ecotoxicol. Environ. Saf. 2020;189 doi: 10.1016/j.ecoenv.2019.109913. [DOI] [PubMed] [Google Scholar]
- Wassarman P.M. Zona pellucida glycoproteins. J. Biol. Chem. 2008;283:24285–24289. doi: 10.1074/jbc.R800027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman P.M., Jovine L., Litscher E.S. A profile of fertilization in mammals. Review. Nat. Cell Biol. 2001;3:E59–E64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- Westerfield, M. (2000). The zebrafish book: a guide for the laboratory use of zebrafish. http://zfin.org/zf_info/zfbook/zfbk.html.
- Worku T., Wang K., Ayers D., Wu D., Ur Rehman Z., Zhou H., Yang L. Regulatory roles of ephrinA5 and its novel signaling pathway in mouse primary granulosa cell apoptosis and proliferation. Cell Cycle. 2018;17:892–902. doi: 10.1080/15384101.2018.1456297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.L., Thompson R.C., Galloway T.S. The physical impacts of microplastics on marine organisms: a review. Environ. Pollut. 2013;178:483–492. doi: 10.1016/j.envpol.2013.02.031. [DOI] [PubMed] [Google Scholar]
- Wu X., Viveiros M.M., Eppig J.J., Bai Y., Fitzpatrick S.L., Matzuk M.M. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- Zemanek M.G., Pollard S.J., Kenefick S.L., Hrudey S.E. Toxicity and mutagenicity of component classes of oils isolated from soils at petroleum- and creosote-contaminated sites. J. Air Waste Manag. Assoc. 1997;47:1250–1258. doi: 10.1080/10473289.1997.10464076. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wei S., Wei W., Zou J., Gu G., Wu D., Bi S. Trimethyltriazine-derived olefin-linked covalent organic framework with ultralong nanofibers. Sci. Bull. 2020;65:1659–1666. doi: 10.1016/j.scib.2020.05.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.