Abstract
Cell membranes are the initial site of stimulus perception from environment and phospholipids are the basic and important components of cell membranes. Phospholipases hydrolyze membrane lipids to generate various cellular mediators. These phospholipase-derived products, such as diacylglycerol, phosphatidic acid, inositol phosphates, lysophopsholipids, and free fatty acids, act as second messengers, playing vital roles in signal transduction during plant growth, development, and stress responses. This review focuses on the structure, substrate specificities, reaction requirements, and acting mechanism of several phospholipase families. It will discuss their functional significance in plant growth, development, and stress responses. In addition, it will highlight some critical knowledge gaps in the action mechanism, metabolic and signaling roles of these phospholipases and their products in the context of plant growth, development and stress responses.
Keywords: Phospholipases, Phospholipid, Diacylglycerol, Free fatty acids, Phosphatidic acid, Signaling, Stress response
1. Introduction
Phospholipids are the backbone of cell membranes and their hydrolysis generates various cellular signals, such as free fatty acids (FFAs), phosphatidic acid (PA), diacylglycerol (DAG), lysophospholipids, and soluble head groups. Phospholipases catalyze the hydrolysis of membrane lipids , and they can be grouped into acyl-hydrolysing phospholipase A (PLA) and head group-hydrolyzing phospholipase C (PLC) and phospholipase D (PLD) [1,2]. PLAs cleave sn-1 and/or sn-2 position of glycerophospholipids to release free fatty acids and lysolipids [3]. PLCs hydrolyze the phosphodiester bond of the glycerol side to produce DAG and phosphorylated head groups, whereas PLDs hydrolyze the phosphodiester bond at the head group side to produce PA and free head groups [1,2] (Fig. 1). The PLA, PLC, and PLD are classified into different families and subfamilies, according to the DNA/protein sequences and conserved domains [1-3]. The diverse phospholipases indicate that they may play key roles in various cellular processes in plants. This review will focus on the structures, mechanism of action, substrate specificity, and reaction requirements, and physiological functions of several phospholipase families, including patatin-related PLA (pPLA), non-specific PLC (NPC), phosphoinostide-hydrolyzing PLC (PI-PLC), and PLD.
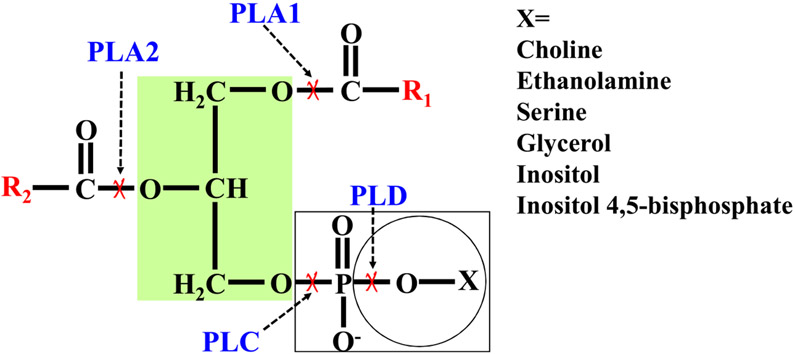
Fig. 1.
Phospholipid structure and hydrolysis site by phospholipase. The red cross shows the cleavage site. PLA1, phospholipase A1. PLA2, phospholipase A2. PLC, phospholipase C. PLD, phospholipase D. R1 and R2 are fatty acids. X represents the head group of phospholipids.
2. Patatin-related phospholipase As (pPLAs) in plants
2.1. Classification and domain structure of pPLAs
pPLAs are related structurally to patatins that are the major storage proteins in potato tubers and have acyl-hydrolyzing activity. pPLAs hydrolyze glycerolipids to generate lysolipids and free fatty acids [3]. In Arabidopsis, the core pPLA family consists of ten members that are classified into three groups, pPLA?, pPLA?? (α, β, γ, δ, ε), and pPLAIII (α, β, γ, δ) [3]. In addition, three triacylglycerol (TAG)-hydrolyzing lipases also contain the patatin domain [3-5]. pPLAI is the only gene in group I with 18 exons, which is much larger than other pPLAs in gene sizes. pPLAI has a leucine-rich repeat domain in C-terminal and a weak ankyrin-like homology domain in N-terminal [3]. pPLAI is considered as the most similar to ancestral gene among all pPLAs in evolution, because it keeps the recognizable homology to animal iPLA2s [3]. pPLAIIs have five to six introns and pPLAIIIs only have one intron. pPLAIIs are more closely related to potato tuber patatin and the catalytic domain in pPLAIIs is similar to that in animal iPLA2s [3].
The catalytic region of the pPLA family includes the esterase box, the phosphate or anion binding element and a catalytic dyad-containing motif. The esterase box GxSxG is conserved in pPLAI and pPLAIIs, but the middle Ser in pPLAIIIs is substituted with Gly, having the non-canonical esterase motif GxGxG [3,6]. The DGGGxxG represents phosphate or anion binding site and in Arabidopsis. The catalytic dyad-containing motif is represented by DGG sequence in pPLAIIα, pPLAIIβ, pPLAIIδ, pPLAIIε, pPLAIIIα and pPLAIIIδ, while DGA in pPLA1 and pPLAIIγ, and GGG in pPLAIIIβ and pPLAIIIγ [3,6].
2.2. Enzymatic properties of pPLAs
pPLAI, pPLAIIs, and pPLAIIIs hydrolyze both phospholipids and galactolipids in vitro [7,8]. pPLAI prefers galactolipids to phospholipids, displaying much higher enzyme activity on monogalactosyldiacylglycerol (MGDG) than PC or PE [7]. In comparison, pPLAIIγ, δ, and ε have a similar enzyme activity towards galactolipids and phospholipids [9]. The specific activity of pPLAIIα is higher than that of the GxGxG motif-containing pPLAIIIβ [6]. The pPLAIIIs have been shown to have thioesterase activity, hydrolyzing acyl-CoAs [6,10]. The calcium independent phospholipase A2 (iPLA2) enzyme from macrophage-like cell line P388D1 was shown to utilize different molecular species of PC [11]. These differences in the substrate preferences indicate that pPLAs have the capability to hydrolyze different membrane lipids.
In animals, some PLA2s have TAG lipase activity [3]. While none of 10 Arabidopsis pPLAs with TAG lipase activity was reported, three others, such as SUGAR-DEPENDENT1 (SDP1), are TAG lipases. Some pPLAs display much higher enzyme activity when the concentration of calcium raises to 1 mM, but they are still active in the absence of calcium ions [12]. pPLAs lack a functionally characterized signal peptide. Hence, it is hypothesized that pPLAs might be localized in the cytosol or attached to the outer surface of the intracellular membranes [8,12,13]. Previous studies have shown that pPLAs could also be activated by the heterotrimeric G proteins and phosphorylation of calcium-dependent protein kinases [14]. In vitro, calcium-dependent protein kinases can phosphorylate pPLAIIδ and pPLAIIε, enhancing their activity towards phosphatidylcholine (PC) and phosphatidylglycerol (PG) [9]. These activation patterns are analogous to the activation of animal cPLAs, which also can be activated by the receptor-dependent elevation in calcium ion concentration and the heterotrimeric G protein [15,16]. Further knowledge of the reaction conditions, such as the cofactors requirement of pPLAs, will expand the insights in the role of pPLA-driven lipid metabolism and signal transduction.
2.3. The functions of pPLAs in plant growth, development, and stress responses
2.3.1. Role of pPLAs in jasmonic acid (JA) biosynthesis and defense response
JA is an oxylipin and its biosynthesis is initiated in the chloroplast where linolenic acid is the substrate [7,17]. Most linolenic acid is esterified in monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), which are the main galactolipids in plastid membrane. In vivo, the release of linolenic acid from complex membrane lipids is accomplished through lipolytic activities, such as PLA enzymes (Fig. 2) [18-20]. The subcellular localization of pPLAI in Arabidopsis suggests that the enzyme might exert its function through hydrolysis of chloroplast membrane lipids [8]. Knockout of pPLAI impeded basal JA accumulation but not pathogen-triggered production of JA or galactolipid and phospholipid hydrolysis. These findings were further supported by the decreased level of free linolenic acid and repressed transcription of VSP2, a JA responsive gene [21,22]. However, it is still not clear whether JA biosynthesis is the result of hydrolysis of complex membrane lipids or the change of free fatty acids in vivo. The level of linolenic acid-containing DGDG-34:3 and DGDG-36:6 decreased after Botrytis cinerea infection, and the extra-plastidic lipids PC, PE, PI and PS also exhibited various patterns of hydrolysis during pathogenesis [7]. It is possible that the increase in the extra-plastidic phospholipid hydrolysis is linked with disease damage rather than controlling the initial step of JA biosynthesis [7]. These findings also suggest the presence of other acyl-hydrolyzing enzymes in plants that might contribute to membrane lipid degradation. Therefore, further study of other acyl-hydrolyzing activities in the JA production in the pPLAI-knockout background may provide valuable insights into the other enzymes involved.
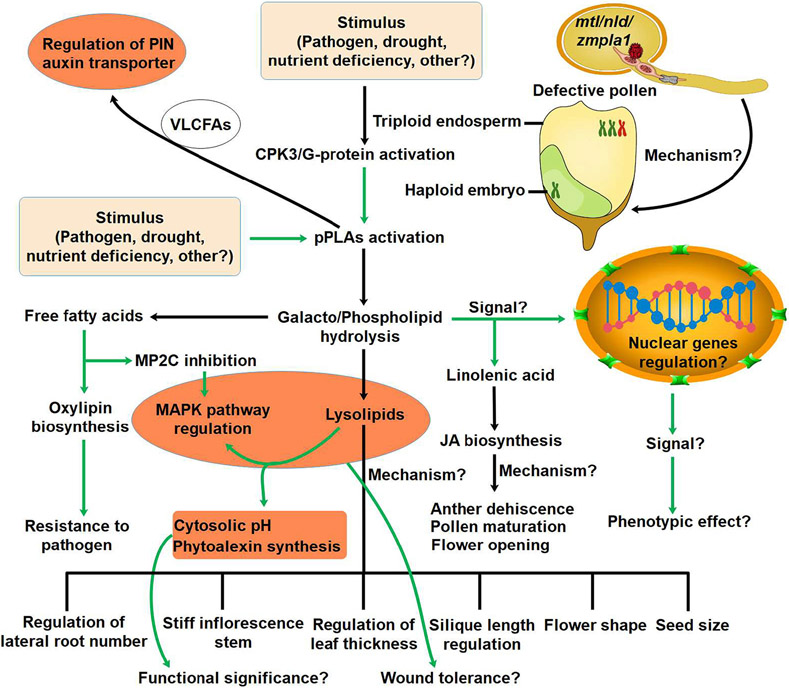
Fig. 2.
Activation and signaling functions of pPLAs. The black arrow indicates established links while the green arrow represents putative links. pPLAs are activated by heterotrimeric G proteins and phosphorylation of calcium dependent protein kinases. Phosphorylation of pPLAs by CPK3 may cause both galactolipids and phospholipids hydrolysis. pPLAs activity results in the generation of free fatty acids and lysolipids. Linolenic acid acts as precursor for JA involved in host specific resistance. The lysolipids may have link with cytosolic pH and phytoalexin synthesis to combat pathogen attack. Mutation in MTL/NLD/ZmPLA1 gene results in haploid embryo formation, resulting in the generation of haploid plants in maize. Oscillations of VLCFAs due to pPLA activity is assumed to be associated with the regulation of PIN auxin transporter.
pPLAI plays a positive role in plant defense against B. cinerea infection probably by maintaining the homeostasis of free fatty acids and basal JA pools. In contrast, knockouts of some pPLAIIs confer Arabidopsis resistance to B. cinerea infection and their overexpression renders plants more sensitive to the infection [8]. Arabidopsis leaves infected with P. syringae displayed faster and larger necroses in lines overexpressing pPLAIIα, pPLAIIγ and pPLAIIIα [23]. However, the JA-responsive PR-6 gene was not induced significantly. The overexpression of pPLAIIα, pPLAIIγ, or pPLAIIIα also did not produce significantly higher levels of JA [23]. These results indicate that the fatty acid produced by pPLAIIα, pPLAIIγ, or pPLAIIIα may not be involved in JA accumulation. It is possible that the basal JA production and pathogen-induced JA biosynthesis are controlled by different mechanisms.
2.3.2. pPLAs in hormone signaling and its impact on plant phenotypes
One of the major effects of some pPLAIIs is related to the regulation of root development [9,24]. pPLAIIε is highly expressed in roots except root tips. The root length increased and the lateral root density decreased in pPLAIIε-knockout mutants under phosphate deficiency, but these phenotypes disappeared after an abscisic acid (ABA) treatment. These results indicate that pPLAIIs might impact auxin signaling [9]. Alterations of root architecture under phosphate deficiency were also linked to changes in endogenous cytokinin and gibberellin [24-26]. However, the evidence linking pPLAIIε and cytokinin or gibberellin signaling are not strong. Rather, auxin promoting the expression of pPLAs is proposed [9]. Some studies suggest a possible mechanism about the linkage between phosphate deficiency and auxin signaling. Under phosphate deficiency, the concentration of auxin changes in lateral roots, and the auxin-responsive promoter DR5 and the auxin receptor promoter TIR1 are upregulated [24,27,28]. The sensitivity of auxin under phosphate starvation is considered to modulate the root structures [24]. Auxin promotes lateral root growth, and pplaIIγ mutants were also observed to keep the primary root growth while restricting the lateral root density under phosphate deficiency. These results suggest that pPLAIIs contribute positively to the auxin modulation of root growth.
In addition, the expression of auxin-response genes was delayed in the knockout mutants of pPLAIIIα, β, γ, and δ [29]. However, pPLAIIIδ-knockout plants displayed increased lateral root density while delayed the expression of early auxin-inducible genes under an auxin treatment [29]. pPLAIIIδ is also associated with longitudinal and transverse growth of different organs in Arabidopsis and oilseed rape [30]. Previous study suggests that pPLAIIIδ controls cell expansion and elongation by activating endogenous auxin distribution machinery [30]. Similarly, some auxin-related phenotypes such as longer hypocotyls and roots are shown in early seedling stages in pPLAIIIβ knockout lines, and the opposite phenomenon was observed in pPLAIIIβ overexpression lines [6]. However, how pPLAIIIs impact the distribution or expression of auxin is still unknown.
2.3.3. Role of pPLAs in uniparental genome elimination and seed germination
A major discovery in haploid plant generations with one set of chromosomes eliminated from the genome is the identification of a specific pPLAII in maize. This key gene named as MATRILINEAL(MTL)/NOT LIKE DAD(NLD)/ZmPHOSPHOLIPASE-A1(ZmPLA1) is represented by the GRMZM2G471240 locus in maize and its corresponding protein is predominantly localized in sperm cells [31-34]. A survey of wide range of maize haploid inducer lines identified a 4-bp insertion in the MTL/NLD/ZmPLA1 gene. This insertion causes a frameshift giving rise to a new unstable truncated variant which loses its intracellular membrane association [31,33]. The knockout of MTL/NLD/ZmPLA1 gene results in haploid induction in maize [31-33]. The molecular events perturbed in mtl/nld/zmpla1 mutant sperms are not known. Nonetheless, the synchronicity between the abnormal chromosome number in the inducer lines and transcriptional activation of MTL/NLD/ZmPLA1 during sperm cell formation suggest a role of MTL/NLD/ZmPLA1 in unknown metabolic and/or signaling processes that probably operate during the formation of sperm cells rather than the initial stages of normal pollen development [33,35]. How the loss of MTL/NLD/ZmPLA1 affects membrane lipid dynamics and intracellular signaling cascades leading to haploid formation is yet to be elucidated.
The rice plants overexpressing pPLAIIIα and camelina plants overexpressing Arabidopsis pPLAIIIδ yielded shorter and round seeds [36,37]. When a segment of rice pPLAIIIδ gene is deleted, the rice plants produced dense and erected panicles with smaller and round-shaped seeds [38]. The loss of pPLAIIIα rendered Arabidopsis more sensitive to ABA inhibition of germination and the mutants had more endogenous ABA contents and a higher transcript level of the GA repressor, GA2ox1, a GA oxidase. The overexpression of pPLAIIIα resulted in a higher expression level of GA2ox1. pPLAIIIα may regulate seed germination phenotype probably by maintaining the homeostasis of active and non-active forms of GA, as well as ABA/GA balances [39].
2.3.4. The role of pPLAIIIs in cell wall composition, lipid biosynthesis and seed oil accumulation
Previous studies reported that the overexpression of pPLAIIIβ and pPLAIIIδ in Arabidopsis and pPLAIIIα in rice led to a decrease in cellulose contents [6,36]. Meanwhile, overexpression of pPLAIIIβ from ginseng or pPLAIIIα from Arabidopsis decreased lignin content but not cellulose content [40,41]. These results imply that different pPLAIIIs exert their functions differently in altering cell wall composition. pPLAIIIδ was shown to regulate cellulose and oil contents in camelina through regulation of carbon partitioning [37]. Therefore, pPLAIIIs may have a role to regulate the central carbon flux involved in cell wall biosynthesis [40,41]. The effect of pPLAIIIs on cellulose deposition raises interesting questions about their function in lipid metabolism and oil accumulation in seeds. Subsequent study showed that pPLAIIIδ had higher expression in the developing radicle and cotyledons in Arabidopsis seeds. The seed oil content was decreased in pPLAIIIδ knockout lines, but increased in overexpression lines [10]. These results suggest that pPLAIIIδ hydrolyzes PC to generate free fatty acids and lysophosphatidycholine (LPC), which plays a role in channelling fatty acyl chains from plastids to ER [10]. Kinetic labelling indicates that fatty acids in the plastid are first converted into PC and later into TAG in the embryo of soybean [42-44]. In higher plants, fatty acids are synthesized in plastids and then exported to ER where the oil biosynthesis takes place [42]. Acyl-CoA:lysoPC acyltransferases (LPCAT1 and 2) in Arabidopsis seeds were shown to catalyze the incorporation of fatty acids into PC [45,46]. pPLAIIIδ hydrolyzes PC to release fatty acids in ER and generate LPC, which could be utilized by LPCAT to accept fatty acids from plastids. Lin et al. (2019) reported that overexpression of pPLAIIIβ from P. fendleri in Arabidopsis resulted in decreased levels of hydroxy fatty acids (HFA) both in PC and TAG, indicating that released HFAs from PC were not incorporated into TAG [47]. These results imply that LPCAT-pPLA cycle might facilitate fatty acid export from plastids to ER.
2.3.5. Enthralling questions regarding acting mechanism of pPLAs and their products
The forgone studies suggest that individual pPLAs have distinctive and yet overlapping functions in plant growth, development, and stress responses. Hence, one key to understanding those enzymes is to elucidate and distinguish the mechanism of actions for various pPLAs. For instance, what are the key downstream signals generated by pPLAs under specific stress conditions? FFAs and lysolipids as the products of pPLAs are the cellular mediators, but how they regulate plant functions are largely unknown. For example, the pPLA activity is triggered upon treatment with elicitor and this induction involves the transcriptional regulation of heterotrimeric G protein α-subunit [14]. This phenomenon causes the efflux of vacuolar protons and thereby lowers the pH in the cytosol. This integrated signal then initiates phytoalexin biosynthesis (Fig. 2) [14,48]. It is speculated that the production of LPC from pPLA hydrolysis of lipids could possibly trigger H+/Na+ exchange transporter and impact the pH [48]. Further, the reacylation of the produced LPC prevents itself to become a toxic compound [49]. Elevation of LPC physiological levels upon pPLA induction was also observed in response to other stimuli including mycorrhiza and peptide 13 elicitor [50,51]. However, the experiment of fluorescent labelling showed the accumulation of FFAs instead of LPC, suggesting the existence of unknown mechanism in the subsequent step [49]. The phosphorylation of pPLAs by CPKs is another potential mechanism reported to activate pPLAs [9]. In a recent study, pPLAIIIγ was found to confer tolerance against osmotic and salt stress in Arabidopsis during seed germination and seedling growth by regulating the levels of lysolipids and FFAs [52]. How the equilibrium of lysolipids and FFAs interfere with MAPK, SOS, and other cascades in modulating abiotic stress responses is also unknown.
Previous studies found that pPLAI-produced unsaturated fatty acids could stimulate an unknown protein kinase to downregulate the expression of MP2C, which is a protein phosphatase involved in the MAP kinase pathway under wounding conditions (Fig. 2) [53-55]. Furthermore, studies reported that polyunsaturated fatty acids could control the stomatal aperture and potassium ion channels [55]. Oleic acid could trigger the activity of PLDδ and lysophosphatidyletholamine (LPE) inhibited the activity of PLD to retard fruit senescence [56-59]. Whether the products of pPLA participate in these processes are not clear. It is worth noting that many other acylating or lipolytic enzymes also influence the content of lysolipids and FFAs intracellularly. Therefore, distinguishing FFAs coming from which route is the key step to study whether pPLA and its products regulate cellular processes as signaling mediator.
Another important question is how specific pPLAs are activated. Do stressors induce the transcriptional activation of pPLAs directly or through intermediate signals? The identification of variety of signals and environmental challenges inducing the transcriptional activation of various pPLAs will greatly facilitate the cross talk of pPLAs-mediated signaling with other signaling networks. The identification of novel specific activators, inhibitors, and elicitors as upstream regulators of pPLA may broaden our knowledge to understand the activation mechanism of a particular pPLA isoform in response to a particular stimulus. Furthermore, extensive functional characterization of identified pPLAs in plants is needed to elucidate which pPLAs are responsible for the generation of lysolipids and FFA species that could promote plant cell signaling under stress conditions.
3. Phospholipase Cs (PLCs) in plants
PLC hydrolyzes the phosphodiester bond close to the side of glycerol to produce phosphorylated head group and DAG. Depending on the substrate preferences, PLCs are classified into nonspecific phospholipase C (NPC) and phosphatidylinositol-specific PLC (PI-PLC) in plants [60].
3.1. NPCs in plants
3.1.1. Classification and domain structure of NPCs
This class of phospholipase is found only bacteria and higher plants and were originally identified as toxins in certain bacteria and now identified as having potential roles in plants [61,62]. Six NPC homologs named as NPC1 to NPC6 are identified in Arabidopsis [61 and references therein]. The orthologs of NPC have also been identified in other plants such as rice [63] and Brassica napus [64]. The protein structure of Arabidopsis NPCs comprises of phosphoesterase domain, which is essential for esterase activity and other three unannotated domains that are highly conserved with PC-PLC in Mycobacterium tuberculosis bacteria [60]. No transmembrane domain was found in NPCs and signal peptides were predicated in some Arabidopsis NPCs including NPC1, NPC2 and NPC6 [60]. Besides, the S-acylation site exists at the C-terminal Cys-533 of NPC4, conferring NPC4 the ability to anchor into lipid rafts on the plasma membrane [62,65]. It is proposed that the C-terminal domain of NPCs comprises of highly diverse protein sequences which might be responsible for the functional diversity of different NPCs [65,66].
3.1.2. Substrate preferences and catalytic properties of NPCs
NPCs hydrolyze common membrane phospholipids such as PC and PE. In addition to PC and PE, NPC4 also hydrolyzes PA, PS, PG and PI (4,5)P2 to a lower extent [67]. These results were based on the mass spectrometry-based profiling of DAG and its acyl chain specificity was not directly measured [67]. In Arabidopsis, the preferred substrates of both NPC5 and NPC4 are PC and PE. However, NPC5 activity is much lower than NPC4 [68]. Recently, NPC4 has been reported to prefer the major sphingophospholipid glycosyl inositol phosphoceramides (GIPC) to PC as substrate, especially under the condition of phosphorus deficiency in plants [62,65]. NPC3 exhibited phosphatase activity using LPA as substrate to produce monoacylglycerol [69]. Although NPC3 could use all the molecular species of LPA, the affinity was higher towards LPA-18:0 than other LPA species [69,70]. The other members like NPC1, NPC2, and NPC6 have the similar enzymatic characteristics to NPC4, preferring PC and PE [71,72]. Recently, NPC6 is found to hydrolyze galactolipids to produce DAG [73]. Likewise, rice NPC1 apart from phospholipid-hydrolyzing activity, was also active towards the galactolipids MGDG and DGDG, and its activity against MGDG was roughly half of DGDG [74]. The enzymatic properties of NPCs were also characterized in other plants such as cultured parsley, tobacco cells and petunia extracts [51,75].
Unlike PI-PLCs that require Ca2+, NPC activity is independent of Ca2+. The activity of NPC4 slightly increased when EGTA was added because of chelation of inhibitory divalent cations such as Co2+, Mn2+, or Zn2+ [60]. NPC3 was non-responsive to the divalent cations such as Mg2+, Ca2+, and Mn2+. Some detergents like Triton X-100, CHAPS, and NP-40 could reduce the activity of NPC3 in a dose-dependent manner [66]. Studies also showed that the activity of NPCs was impacted by AlCl3, glycoprotein and small protein cryptogein [51,76].
3.1.3. Subcellular localization and expression patterns
In Arabidopsis, NPCs exhibit highly unique subcellular distribution probably due to the presence of signal sequences in N-terminal in NPC1, NPC2 and NPC6, but not in the remaining NPCs [60]. The subcellular localization of NPC1 is confined to the compartments of the secretary pathway including endoplasmic reticulum (ER), trans-Golgi network and Golgi apparatus. NPC2 and NPC6 are localized in the plastid of mesophyll cells in leaves [71,73,77]. Moreover, NPC2 is predominantly localized in Golgi apparatus in roots and weak GFP signals GFP-tagged NPC2 have also been detected in ER [78]. The GFP imaging revealed that NPC6 was localized in microsomal and chloroplast membranes when GFP-tagged NPC6 was transiently expressed in tobacco mesophyll cells [73]. NPC4 was found to be localized at the plasma membrane and NPC5 was detected in both soluble and microsomal fractions [60,65,68]. There is no transmembrane domain in both NPC4 and NPC5. The membrane localization of NPC4 is determined by the acylation at the C-terminal Cys-533. The difference in C-terminal sequences endows NPCs with various subcellular localization and function [65]. The expression patterns of NPCs also have their own characteristics. For instance, Arabidopsis NPC3 and NPC4 are mainly expressed in vegetative tissues including root tip, leaf margin and cotyledon [61]. NPC2 and NPC6 exhibit preferential expression in petioles, leaf vasculature and trichome [71,77,79]. Besides, NPC2 and NPC6 are highly expressed in cotyledon and hypocotyl in germinating seeds, respectively [71,77,79]. Moreover, NPC2 also has high transcript abundance in anther, filament and stigma, while NPC6 is highly expressed in ovules and style [71,77,79].
3.1.4. Mechanism of NPCs action with reference to DAG functions
The regulatory function of NPC-generated DAG is intriguing in plants because the generation of DAG involves several metabolic pathways including the Kennedy pathway and membrane lipid hydrolysis [80]. The DAG production by hydrolysis involves three main routes: NPC of membrane phospholipids, PI-PLC hydrolysis of phosphoinositides, and PA phosphatase (PAP)- or lipid phosphate phosphatase (LPP)-catalyzed dephosphorylation of PLD-derived PA [81,82]. The NPC-produced DAG can also serve as backbone for the biosynthesis of sulpholipids, phospholipids, glycolipids and TAGs. NPCs play a role both in both plant growth and development by supplying DAG backbone for the production of a variety of glycerolipid species, as well as stress-induced signal transduction by delivering DAG as a lipid second messenger [83]. However, unlike well-documented DAG target protein kinase C (PKC) in animal cells, the molecular mechanism of DAG as a second messenger is not well defined in plants [66,84].
The application of medium-chain (8:0/8:0) DAG fully restored the lateral root number in salt-treated npc5 mutants, indicating that NPC-produced DAG has a role in regulating the development of lateral roots under saline conditions [85]. Another study reported that reduced NPC activity and DAG levels upon treatment of BY-2 cells of tobacco with aluminium resulting in retarded growth of pollen tube [76]. Later study revealed that the growth of the pollen tube was partially rescued upon overexpression of NPC4 [86]. The phenotype was restored by exogenous DAG application providing evidence that DAG promoted a cellular response to stress [86]. In another study, NPC4-generated DAG was found to be involved in regulating stomatal aperture under well-watered conditions [67]. Under drought environment, npc4 mutants exhibited low water use efficiency [67]. It is suggested that synthetic DAG triggers the ion pump in the plasma membrane of guard cell during stomatal opening and inhibits the K+ ion efflux leading to stomatal closure [87]. To unveil the cellular functions of NPCs, it would be worthwhile to characterize the DAG-binding domain-containing proteins for their affinities towards DAG and identify downstream targets along with the pathways having core DAGs signaling in plants.
3.1.5. The functions of NPCs in plant growth, development, and stress responses
3.1.5.1. Role of NPCs in gametophyte development and root growth.
NPC2 and NPC6 have been characterized in Arabidopsis playing potential roles in the development of gametophyte and root growth [71,77,79]. NPC2 is expressed highly in anther filament and stigma, while NPC6 is highly specific to ovules and style. A study showed that the npc2 npc6 double mutant was defective in the development of male and female gametophytes [71]. Both NPC2 and NPC6 preferably hydrolyze PC and PE and lipidome analysis revealed an increase in PC and PE levels and a decrease in MGDG in the floral buds of double mutants, suggesting the catabolism of phospholipids by NPC2 and NPC6 at a specific developmental stage of flowers [60,71]. The double mutants were unable to produce viable seeds because the gametogenesis event was arrested. Subsequently, the leaky knockdown double mutants were generated, such as suppressing the NPC6 expression in npc2-1 mutant or NPC2 in npc6-2. The lipid content and gametophyte development had no apparent changes between the leaky knockdown double mutants and the correspondent single mutant, suggesting the functional redundancy of NPC2 and NPC6 [77]. However, these mutants were impaired in root architecture, and the defective root phenotype could be recovered partially after phosphocholine (PCho) was supplied in the growth media [77,88]. Gene expression analysis showed that the transcript level of phospho-base N-methyltransferase (PMT1) is high in both mutants. PMT1 catalyzes the formation of PCho and hence the interaction of NPC2 and NPC6 with PMTs pathway was proposed during promotion of root growth [77]. It is suggested that PC as the secondary product after an NPC-catalyzed reaction played a role in root architecture [88]. Moreover, NPC4 distinctively plays a unique role in regulating the density and growth of root hair under phosphate deficiency in Arabidopsis [89].
3.1.5.2. Role of NPCs in combating heat and salt stress.
The role of NPC1 in plant thermotolerance was recently revealed. Knockout mutants of NPC1 are sensitive to heat stress while the overexpression lines are tolerant [72]. In addition, NPCs are also involved in plant response to high salinity. The number of lateral roots and the DAG content of the root were reduced in npc5 mutant as compared to that in WT, but the content of DAG was higher in overexpression lines [85]. The root phenotype was rescued in npc5 mutants after application of exogenous DAG [85]. NPC4 is also reported to regulate plant response to salt stress. The npc4 mutants were observed defective in seed germination, plant biomass and root length under salt stress [67,90]. It remains to explore how these two NPCs function synergistically in plant response to mild and severe concentration of salt stress.
3.1.5.3. Role of NPCs in plant response to hormones.
ABA is a well-known phytohormone that induces the expression of many genes under stress conditions [91]. NPCs have been shown to play important roles in regulation of plant response to various stresses. For example, the expression of ABA responsive genes was suppressed in npc4 mutants under salt stress, suggesting that loss of the function of NPC4 affects ABA signaling [67,90]. Compared to WT, the overexpression lines of NPC4 exhibited hypersensitivity to ABA in seed germination, root formation and stomatal closure, while knockout mutants displayed opposite effects [67]. NPC5 affects auxin signaling. The number of lateral roots of npc5 mutants showed no change when supplemented with IAA [85]. It seems that the mutation of NPC5 disrupts the signal transmission of IAA. However, the lateral root density was reduced after treatment with salt, while the root morphology of the mutants was not affected upon sorbitol- or mannitol-induced osmotic stress [85]. These findings suggest that NPC4 may take part in osmotic stress through ABA signaling while NPC5 may be involved in salt stress response by regulating auxin signaling. The NPC6 in rice is also involved in the mesocotyl elongation mediated by GA in rice [92]. Mutation of NPC3 or NPC4 affects the root architecture through brassinolide (BL) response as evident from the expression of GUS induced by the promoter from either NPC3 or NPC4 [70]. BL may induce NPC activity through transcriptional activation of cell expansion genes, like TCH4 encoding xyloglucan endotransglycosylase and LRX2 encoding leucine-rich extensin. The cell cycle events and cell expansion might be controlled by NPC3/NPC4 produced DAG through BL. These cell cycle events regulate root growth (Fig. 3).
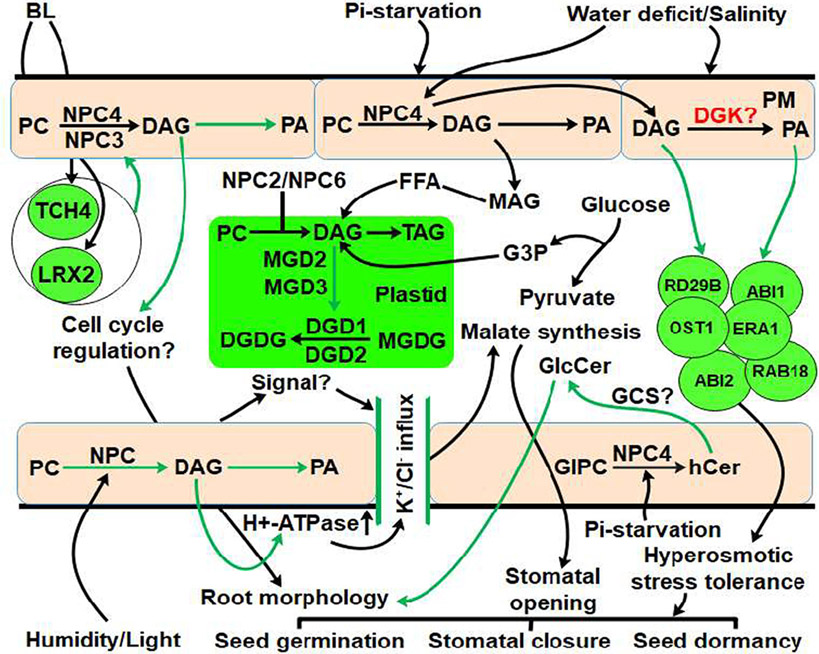
Fig. 3.
Signaling functions of NPCs under stress conditions. The black arrow indicates established links while the green arrow represents putative links. BL may induce NPC activity through transcriptional activation of cell expansion genes, TCH4 and LRX2. The cell cycle events and cell expansion might be controlled by NPC3/NPC4-produced DAG through BL signaling. In response to humidity and light, NPC generated-DAG probably activates H + -ATPase pump through an unknown intracellular signal leading to malate synthesis. Salt stress modulates NPC4 and it is speculated that NPC4's product DAG may undergo lipid-protein interaction to mediate the signaling events. In addition, NPC4 also accomplishes phosphosphingolipid hydrolysis under phosphate deficit condition to regulate root growth. Phospholipid to galactolipid conversion is also accomplished through NPC4/NPC5 activity under phosphate deficit conditions.
3.1.6. Critical knowledge gaps in understanding metabolic and signaling roles of NPCs
Phospholipases in plants have been investigated for their roles in lipid signaling. The alteration in the membrane lipid composition by phospholipases themselves is an indicator for a cell to initiate downstream signaling events. However, the regulatory signaling events that act downstream of NPC activation are poorly understood in plants. The NPC generated-DAG probably activates H+-ATPase pump through an unknown intracellular signal leading to malate synthesis in response to humidity and light (Fig. 3) [87]. This phenomenon has been shown to regulate stomatal opening and the involvement of exact NPC isoform or other DAG-generators is not known. The role of NPC-derived DAG in root growth promotion provided new insights that the polar head groups may also play a downstream regulatory role upon NPC activation [77]. Earlier studies found that knockout of NPC4 apparently does not alter membrane lipid contents but a mutant deficient in JA signaling exhibited higher transcription of NPC4 [60,93,94]. This observation led researchers to the hypothesis that NPC4 may have role in signaling networks instead of the primary metabolism of glycerolipids. However, recent studies reveal that NPC4 hydrolyzes GIPC to release phosphorus from membrane lipids to maintain cell membrane homeostasis and dynamics under phosphorus deficiency [62]. The galactolipid biosynthesis is governed by both prokaryotic and eukaryotic pathways. It was hypothesized that eukaryotic pathway-derived DAG for galactolipid biosynthesis through prokaryotic pathway initiates from PC [95]. The hydrolysis of PC to produce DAG is an important metabolism step to synthesize TAG. Higher transcription rate of NPC4 and NPC5 has been reported during TAG accumulation [96,97]. Apart from this, NPC2 and NPC6 have recently been reported to take part in TAG biosynthesis [73,79]. The DAG production from PC also involves different PLDs. Hence, this conversion of phospholipids to TAG and galactolipid biosynthesis seems to be more complex than that presented above. Therefore, the contribution of individual NPCs to the synthesis of lipids is still an open question to be addressed.
3.2. Phosphoinositide-hydrolyzing PLCs (PI-PLCs) in plants
PI-PLCs use PI(4,5)P2 as a substrate to yield DAG and inositol 1,4,5-phosphate (IP3) (Fig. 4) [98-101]. Those enzymes play important roles in plant growth, development, and stress responses, but the mechanism of action of PI-PLCs remains largely elusive in plants [83].
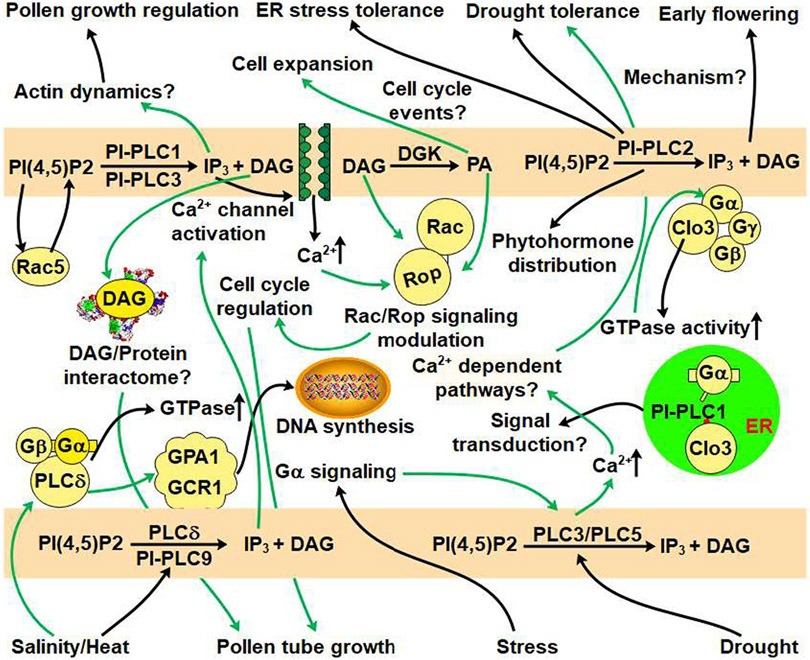
Fig. 4.
PI-PLC-mediated signaling under stress conditions. The black arrow indicates established links while the green arrow represents putative links. PI-PLCs-induced signaling events or phenotypic effects should include PI-PLC-generated phosphoinositides phosphates. PI-PLC3/PI-PLC5 confer drought tolerance possibly through calcium-dependent pathways or α-subunit of heterotrimeric G protein. PI-PLC2 is involved in early flowering. Heat stress induces PI-PLC9 activity and produced IP3 which may cause calcium ions oscillations. PLCδ interaction with Gα and Gβ subunits of heterotrimeric G-protein is already known but whether this interaction exists in response to salinity and heat stress is not known yet. The PI-PLC generated IP3 regulates pollen growth probably by regulating the actin dynamics, while the same phenotype might be regulated by PI-PLC produced DAG through DAG-protein interaction. Gα subunit of heterotrimeric G protein and Clo3 also interact with PI-PLC1 and this interaction is further strengthened by high Ca2+ levels in cell. Distinct functions of DAG and PA in PI-PLC signaling pathway are not clearly known but the Rac/Rop signaling could be modulated by DAG and PA, leading to cell cycle regulation which controls pollen tube growth.
3.2.1. Classification and domain structure of PI-PLCs
In mammals, thirteen PI-PLCs have been reported and are classified into six subgroups including PI-PLCβ, γ, δ, ε, η and ζ, depending on the structure of the conserved domain and biochemical properties [102-104]. PI-PLCs are identified in several plants [105-109]. Arabidopsis has nine PI-PLCs named as AtPLC1-AtPLC9 [106]. Plant PI-PLCs contain several conserved structural domains, including the X and Y catalytic domains that have the TIM barrel-like shape important for phosphoesterase activity. Another structure located in the C-terminus is activated by Ca2+ ions whereas the N-terminal protein sequences varies in different PI-PLCs in plants [102,110-112]. In animals, the X/Y domain of PI-PLCζ was found to bind PI(4,5)P2 [105]. The regulatory domain of EF-hand in animal PI-PLC was responsible for binding of the substrate lipids and Ca2+ [113]. The same domain is found in the N-terminus in majority of plant PI-PLCs and plays vital roles in the enzymes, tethering them to the plasma membrane. A study showed that EF-hand in PLC2 in Arabidopsis is essential for catalytic activity [114]. On the other hand, the binding of Ca2+ to the C2 domain changes the hydrophobicity of PI-PLCs, resulting in lipid tethering to the cell membrane [115]. There are several other factors that can affect the lipid tethering to plasma membrane. For example, the interaction of transmembrane protein NtC7 with the C2 domain of PI-PLC results in lipid trafficking to plasma membrane [75]. Also, the EF-hand present in plant PI-PLCs plays a vital role in PI-PLCs translocation to the plasma membrane [114].
3.2.2. Substrate preferences and reaction requirements of PI-PLCs
The mode of action and substrate preferences of PI-PLC is dependent on Ca2+ which modulates its activity, subcellular localization, and substrate affinities. Usually, PI-PLCs prefer using PI(4,5)P2, PI(4)P and PI as substrates, whereas they do not use PI(3)P, PI(3,4)P2, PI(3,5)P2 and PI(3,4,5)P3 [116]. Under millimolar Ca2+, soluble PI-PLC prefers PI to PI (4,5)P2 and PI(4)P. However, PI-PLC has higher affinity towards PI(4,5)P2 and PI(4)P under micromolar concentrations of Ca2+ [117,118]. Additionally, different PI-PLCs show diverse biochemical properties even under the same concentration of Ca2+ [109]. For instance, under micromolar concentrations of Ca2+, PpPLC1 of Physcomitrella patens prefers to utilize PI(4,5)P2 as the substrate, but PpPLC2 does not. However, PpPLC2 prefers using PI but PpPLC1 does not when Ca2+ is increased to millimolar concentrations [109]. Meanwhile, the type of divalent cations impacts the affinity of PI-PLC towards the substrate. Using Mn2+ and Co2+ instead of millimolar concentrations of Ca2+, wheat PI-PLC showed less affinity towards PI(4,5)P2 but higher towards PI(4)P [119]. The pH gradient also affects the optimal activity of PI-PLC towards its substrate; the optimal pH for PI(4)P hydrolysis is 6.0-7.0, and 6-6.5 for PI(4,5)P2 [119]. Furthermore, calmodulin (CaM), G-protein and phosphorylation also affect the activity of PI-PLC. For example, the activity of PI-PLC in Lily (Lilium daviddi) could be stimulated by CaM or G protein (active in cholera toxin), while inhibition of this activity occurred after adding CaM antibody or G-protein antagonist pertussis toxin, respectively [120]. Plant PI-PLCs lack the conserved motifs that are found in PLCβ or PLCε from animals reported to be involved in G-proteins interaction [121]. However, the C2 domain of PI-PLC of Pisum sativum was found to bind to Gα1 [122]. These results imply that the C2 domain of PI-PLC in plants may have diverse roles. The phosphorylation of tyrosine between the X and Y domains of PI-PLCγ in animals was reported to regulate the activity of PI-PLCγ [123]. Several phosphorylation sites have been identified in the N-terminal EF-hand, X or Y domains of the PI-PLCs in Arabidopsis [124]. It will be interesting to understand which and how phosphorylation at those sites affects the activity of PI-PLCs and whether and how the phosphorylation alters the function of PI-PLCs in plants.
3.2.3. Expression pattern and subcellular localization
Plant PI-PLCs are expressed in various vegetative and reproductive organs, including roots, stems, leaves, flowers, and fruits and at different developmental stages [125-127]. PI-PLC1 and PI-PLC2 have high expression levels in rosette and immature seeds. PI-PLC3 and PI-PLC7 are highly expressed in phloem of roots, leaves and flowers. The PI-PLC7 is also transcriptionally active in trichomes and hydathodes. PI-PLC4, PI-PLC5 and PI-PLC8 are specifically expressed in pollen [126-128]. Most PI-PLCs are induced under several types of stressors, such as ABA, salt, low temperature, SA, and drought in rapeseed, mung bean, maize, potato, wheat, lily, tomato, rice, and Arabidopsis [106,107,110-112,129,130]. Under cold, drought and salt conditions, rice PI-PLC4 exhibited a reduction in its transcriptional level while PI-PLC1 and PI-PLC3 showed an increase [63]. It was documented that mung bean PI-PLC3 and Arabidopsis PI-PLC2, PI-PLC3 and PI-PLC9 were associated with the plasma membrane, whereas PI-PLCs of soybean and rice were localized in the cytoplasm and membrane [63,106,110,114,131,132]. Wheat PI-PLCs was localized in ER and plasma membrane in root while they were detected in plasma membrane fractions in mature and germinating seeds of rapeseed [119,133]. It seems that the subcellular localization of PI-PLCs is different in different plants. In addition, PI-PLCs can be translocated between cytoplasm and membrane depending on under the concentration of Ca2+ [110,113].
3.2.4. Mechanism of PI-PLCs in plants
The induction of PI-PLCs change the physiological levels of PI(4,5)P2, PI(4)P, IP3, IP6, DAG and PA which are vital cellular mediators affecting various cellular processes, such as signal transduction, cytoskeleton framework, vesicular trafficking and membrane structural properties [83]. The activity of PI-PLCs could also be induced by salt, temperature and dehydration to cause elevation in the physiological levels of IP3 followed by increases in cytosolic calcium ions [105,106,134,135]. However, the receptors of IP3 are yet to be identified in plants. It is possible that plants may possess IP3 to IP6 phosphorylation mechanism instead of IP3 receptors as the IP6-triggered release of Ca2+ is significantly more than that of IP3 [136]. Moreover, the spatial distribution and physiological levels of the cellular mediators PI(4,5)P2 and conceivably PI(4)P and other polyphosphoinositides are altered as a result of PI-PLC activity. Polyphosphoinositides contribute to <1% of the total phospholipid content of a cell. The level of PI(4,5)P2 in plant cells is dynamic and lower than PI(4)P [116,137]. In spite of the extremely low physiological level in plant cells, PI(4,5)P2 still modulates tip growth, membrane asymmetry, vesicular transport and PLD activities [135,136,138]. PI(4)P also plays dynamic roles: it acts as a substrate for PI-PLC, precursor for the synthesis of PI(4,5)P2 and signaling molecule [116]. The other product of PI-PLC activation is DAG [83]. Unlike animal cells, plant cells lack protein kinase C (PKC) that is activated by DAG in the membranes [103,104,139]. A study reported that DAG derived from PI-PLC activity mediated lipid metabolism occurring at ER, mitochondria and plastids [140]. PI-PLC-derived DAG can be phosphorylated to PA by DAG kinase (DGK), but PA produced by the PI-PLC-DGK pathway may function differently than PLD-produced PA [141]. Therefore, the type of downstream targets of PI-PLCs in plants could be different from their mammalian counterparts, and the product DAG may have a role in lipid remodeling and metabolism.
3.2.5. The functions of PI-PLCs in plant growth, development, and stress responses
3.2.5.1. Role of phosphoinositides in determining organelle identity and tissue differentiation.
The physiological level of phosphoinositides is less than 1% of the overall phospholipid content [142]. Each compartment of a cell has a unique pattern of phosphoinositides accumulation determining their subcellular identity [143,144]. Phosphoinositides determine the physiochemical attributes of the plasma membrane and other endomembrane systems because of the shape and nature of charge on them [143]. Some of these parameters remain the same among different tissues: for instance, the plasma membrane of all types of cells is highly electronegative due to the accumulation of most predominant PI species PI4P in the membrane [145]. PI4P accumulation at the equatorial plate during cell division and the localization of PI4Kβ1 controls cytoplasmic division and establishment of phragmoplast [146,147].
Unlike PI4P, the PI(4,5)P2 patterning is not limited to the cell type, but to the tissue and organ level. The quantitative imaging of PI(4,5)P2 sensor lines provided evidence for PI(4,5)P2 patterning at the tissue scale [148]. The indirect way to localize PI(4,5)P2 by a sensor is to introduce an expression cassette containing fluorescent proteins to which a PI(4,5)P2-binding domain is fused in transgenic lines [142,149-152]. This fusion protein is recruited to the membranes due to the accumulation of PI(4,5)P2. In the cell with no or low levels of PI(4,5)P2, the localization of such fusion protein that interact with PI(4,5)P2 is disrupted, resulting in strong signals in the cytoplasm [142,149-152]. The PI(4,5)P2-binding Plekstrin Homology (pH) domain abundantly accumulates at the margins instead of centre of the SAM cells of Arabidopsis [148]. Therefore, it was hypothesized that PI(4,5)P2 could be important for self-maintenance of stem cell and organogenesis. In addition, it was hypothesized that low levels of PI(4,5)P2 rendered the PH domain unstable according to the finding that Lee et al., 2019 found a four-fold accumulation of the PH in the cotyledon under a high salt concentrations known to stimulate the PI(4,5)P2 synthesis [150,153]. In the perspective of this hypothesis, the clv3-17 mutants were imaged and indicated that the PH biosensors underwent degradation because of the absence of their binding partner PI(4,5)P2, suggesting a role of PI(4,5)P2 in the maintenance of meristem [148]. Alteration in the PI(4,5)P2/PI4P ratios is linked to the enhanced trafficking across the vacuole and differentiation of protophloem, indicating that the phosphoinositide-mediated biogenesis of vacuole and related transport could be linked to the development of phloem [154]. Also, the dynamics of phosphoinositides as a result of PI-PLC activity are involved in tolerance to ER stress induced by tunicamycin in Arabidopsis [126]. Further research should be conducted to reveal how the individual PI-PLCs contribute to PI(4,5)P2/PI4P ratios to regulate organelle identity and tissue differentiation in plants. Simultaneously, the use of integrative biology approaches will greatly assist to blend the intensive biochemical knowledge of PI-PLCs and their substrate and products into their cellular and developmental functions.
3.2.5.2. Role of PI-PLC2 in modulating male and female gametogenesis.
PI-PLC2 is reported to have a role in gametophyte development in Arabidopsis through the auxin signaling pathway probably by controlling the cell division events [155,156]. The male gametes produced by the pollen of pi-plc2 mutant were defective and even entirely infertile in homozygote. The segregation pattern was against the Mendel law of segregation (0.64: 1 compared to 1:1) after pollinating the heterozygous pollen to WT (Col) stigma [156]. Furthermore, the homozygous pi-plc2 mutants had defective female gametophyte and their megaspores exhibited abnormalities in mitotic division in ovules. A Mendelian ratio of 1.01: 1 was obtained after pollinating the WT pollen to the stigma of heterozygous pi-plc2 [152]. The results of a further study showed that the inflorescence of homozygous pi-plc2 mutants had higher expression levels of auxin biosynthesis genes including YUCCA1, YUCCA2, YUCCA4, YUCCA6 and YUCCA8 than those of WT and the IAA content in the mutants was higher than that in WT [156]. It was suggested that the homozygous mutant plants had an abnormal distribution of auxins, which may have a role in the embryo sac deformation [156]. These findings suggest that PI-PLC2 may maintain a strict equilibrium of auxin homeostasis by controlling transcriptional activation of auxin biosynthesis genes during gametogenesis. However, it remains to be determined whether IP3 or DAG as products of PI-PLC2 independently modulate the transcription of YUCCA genes or they act in a coordinated fashion to modulate auxin signaling during gametophyte development. Therefore, genetic complementation of the gametophyte-lethal phenotype of pi-plc2 knockout might shed light in future to conclude whether PI-PLC2 indeed plays a role in gametogenesis. In addition to this, Rac/Rop signaling is also involved in pollen tube growth [157,158]. The Rac component of the signaling is influenced by the PI-PLC substrate, PI(4,5)P2 in tobacco [157]. Therefore, it is possible that the gametophyte development in plants could be modulated by PI-PLC produced IP3 or DAG by regulating Rac/Rop signaling pathway. However, it is too early to give a mechanism how these players interact.
3.2.5.3. Roles of PI-PLCs in gravitropic response.
Previous reports suggest that IP3 produced by the action of PL-PLCs played a vital role in plant gravitropic response [134,159]. However, the mechanism by which gravity signals are perceived in plant cells through the activation of PI-PLCs is unknown. Studies are available highlighting that the involvement of auxin trafficking in gravitropism induced higher levels of IP3 [134,160]. In stark contrast to this, okadaic acid and lanthanum ions that are the inhibitors of protein phosphatases 1 and phosphatase 2A, and calcium channels, respectively, inhibited the accumulation of IP3 upon gravistimulation [160]. It is also suggested that the activation of PI-PLCs is influenced by amyloplast sedimentation, calcium and protein dephosphorylation during gravity signaling [161]. In contrast to long term fluctuations, the transient changes in IP3 contents are not affected by PI-PLCs inhibitors [134].
Suppression of PI-PLC leaded to inhibition of the gravitropic response in plants [159-161,162]. Further studies indicate that this phenomenon might involve PI3 mediated Ca2+ to change the activity of calcium/calmodulin-dependent protein kinase (MCK1), or interceded by inositol hexakisphosphate (IP6) in modifying auxin receptor transport inhibitor response 1 (TIR1) [163,164]. The mutant of PpPLC1 led to reduced cytokinin sensitivity and also reduced gravitropic response of P. patens [162]. It would be worthy to dissect the distinct mechanisms of action of individual PI-PLCs during gravity signaling in plants.
3.2.5.4. Role of PI-PLCs in regulating different phenotypic attributes and drought tolerance.
PI-PLC3 has been characterized for its role in regulating root architecture, seed germination and stomatal aperture in Arabidopsis. The loss of function of PI-PLC3 resulted in slight changes in the root architecture including shorter primary roots, reduced lateral root number and density [127]. Auxins are an important determinant of lateral root formation and IP6 has been previously shown to bind to auxin receptor TIR1 probably controlling its function [140,164]. However, it is still not known whether TIR1 binding IP6 is specifically produced by PI-PLC3 [127]. The reduced content of IP6 in the pi-plc3 mutants could be the cause of a reduced response of the plant to auxin. Furthermore, under normal conditions, the mutant seeds germinated more slowly than WT. However, the phenotype of mutant seed germination was not impacted when ABA was added, indicating that the loss of PI-PLC3 could reduce the seed sensitivity to ABA [127]. Another study showed that the seed germination event required rapid cleavage of IP6 to produce IP3. The pi-plc3 mutant maintained high PIP2 level in germinating seeds, indicating that the hydrolysis of PIP2 might impact the seed germination of pi-plc3 mutant. However, it is difficult to prove whether the product IP3 impacts the seed germination in this case, because the content of IP3 is too low to be detected. Meanwhile, the mutants were also insensitive to ABA-induced stomatal closure. Application of exogenous ABA induced more PIP2 in germinating mutant seeds, seedlings and guard cells of stomata than that in WT. On the contrary, overexpression of PI-PLC3 conferred plant drought tolerance by reducing the stomatal aperture [165].
Recently, PI-PLC5 has also been reported to regulate plant root growth, stomatal aperture and drought tolerance. The result of phospholipid analysis revealed that the content of PIP and PIP2 was reduced while PA content was increased [128]. It suggests that PI-PLC5 hydrolyzes PIP and PIP2 to generate DAG that can be phosphorylated to produce PA. PI-PLC7 apparently did not affect plant root architecture and drought tolerance like PI-PC3 and PI-PLC5, but it played roles in leaf serration and seed mucilage attachment. The double mutant of pi-plc3/7 was lethal, but pi-plc5/7 was viable and exhibited leaf serration, more seed mucilage attachment and enhanced drought tolerance [166]. Mucilage is characterized by the presence of pectin containing predominantly rhamnogalacturonan I (RGI) and polygalacturonic acid (PGA). Therefore, additional research is required to discover the relationship between pectin composition and PI-PLC7 activity. The above studies indicate PI-PLCs may play various roles in plant development and drought stress response.
3.2.5.5. Involvement of PI-PLCs in salt stress tolerance through Ca2+ signaling.
The transcriptional regulation of PI-PLC1,3,4-7 in Arabidopsis, PLCδ1 from Nicotiana tabacum, PLC3 from Vigna radiate, PLC1 from Populus tomentosa, PLC1 from Triticum aestivum and PI-PLC7 from Glycine max has been reported [129,167-172]. Their activation mechanism and the downstream signaling consequences under salt stress are still unknown. Previous studies highlighted PI-PLCs in modulating plant salt stress tolerance through changes of Ca2+ concentration in cells. However, the isoform was not identified. It was shown that PI-PLC4 negatively modulated the seedling growth of Arabidopsis [173]. Mutation in PLC4 resulted in hyposensitivity while its overexpression rendered the seedlings hypersensitive to the salt stress. PI-PLC4 also affected the expression of salt responsive genes including RD29B, MYB15 and ZAT10 [173]. Since IP3 receptors are yet to be discovered in plants, a phosphorylation mechanism involving unknown kinases responsible for IP3 to IP6 conversion is suggested. This unknown mechanism involving unknown multi-kinases causes the release of Ca2+. Oscillation in the frequency, amplitude and waveform of Ca2+ generates stimulus-specific signals [174]. The calcium signal generated by IP3 as a result of PI-PLC activity during salt stress accounts for ~30% of the total Ca2+ signal [175,176]. The overexpression of PI-PLC4 caused an increase of Ca2+ while its knockout repressed Ca2+ signals in response to salt stress [173]. Treatment of seedlings with EGTA did not affect their sensitivity to salt stress [173]. Furthermore, PLCδ interaction with Gα and Gβ subunits of heterotrimeric G-protein was reported but whether this interaction exists in response to salinity stress is not known (Fig. 4). These results highlight that PI-PLCs are involved in the Arabidopsis response to salt stress through modulating Ca2+ signals.
3.2.5.6. PI-PLCs-mediated signaling in plant innate immunity.
Plants respond to pathogen attack by activating both early and late signaling events. The early signaling events include protein phosphorylation, alteration in the composition of membrane phospholipids, increase in cytosolic Ca2+, NO and ROS generation. Subsequently, the synthesis of phytoalexin, induction of glucanases and chitinases, activation of the phenylpropanoid metabolism pathway and accumulation of phenolic compounds are the later defense-related responses [177,178]. PI-PLCs have been shown to act downstream of the immune receptors and are involved in both early signaling and late defense responses [179]. The pharmacological inhibitor U73122 of PI-PLC significantly suppressed the internalization of immune receptor Flagellin Sensing 2 (FLS2) that was induced by flg22, indicating that PI-PLC activity might have a role in affecting the localization of FLS2 immune receptor. This phenomenon seemed also to occur with another immune receptor Cf-4 [179]. Additionally, fluctuation in IP3 or IP6 levels results in cytosolic Ca2+ oscillations modulating the activities of several cellular proteins such as transporters, protein kinases and transcription factors etc. [174]. The PI-PLC activity inhibition blocks the defense-mediated medium alkalization response, followed by a repression of effector-modulated endocytosis of the immune receptors thereby inhibiting their signaling role [179-181]. In these processes, the change of Ca2+ concentration and pH in cytoplasm altering the activity of PI-PLC suggests that the activity of PI-PLC is indirectly mediated by receptor [179]. Furthermore, the substrate PIP2 of PI-PLC is reported to be involved in plant defense against pathogens.
Depletion of PIP2 resulted in blockage of K+ channels, leading to decreased concentration of potassium ions. Additionally, it also caused stomatal closure and arrested the cell cycle at equatorial plate resulting in programmed cell death (PCD), thereby avoiding pathogen spread [182]. The DAG produced by PI-PLC may also participate in recruiting and activating signaling proteins involved in initial defense responses [174]. However, it is unknown which PI-PLCs are activated upon stimulation of cell-surface immune receptors by pathogen. Further investigations concerning PI-PLCs signaling during plant defense and disease resistance may help in the identification of key processes/interactions facilitating plants to combat microbial infection.
4. PLDs in plants
4.1. Classification and domain structure of PLDs
PLD hydrolyzes the phosphodiester bond in phospholipids to generate PA and soluble head groups [183]. In Arabidopsis, twelve PLD genes including PLDα(1-3), β(1,2), γ(1-3), δ, ε and ζ(1, 2) are classified into six groups depending on protein structures and enzyme properties [184]. In B. napus, 32 PLDs were identified, such as PLDα1(4), α2(2), α3(4), β1(2), β2(2), γ(6), δ(7), ε(2), ζ1(2) and ζ1(1) [185]. Based on protein structures, these PLDs are classified as C2-PLDs and PHPX-PLDs due to the presence of the C2 or PH and Phox homology (PX) domains. Most PLDs belong to C2-PLDs except PLDζs which belong to PHPX-PLDs [186,187]. The C2 domain mediates Ca2+-dependent phospholipid binding. All PLDs have two HKD (HxKxxxxD) domains [188]. Recently, it was shown that the N-terminal C2 domain of Arabidopsis PLDα1 exhibited hydrophobic interaction with the C-terminal catalytic domain that features two HKD motifs [189]. In addition, some PLDs have specific motifs and domains. For instance, PLDβ1 has a PI(4,5)P2 binding region (PBR1) following the first HKD domain. The PBR1 binding to PI(4,5)P2 is critical for PLDβ1 activity [190]. Furthermore, PLDβ1 also has two polybasic motifs (K/RxxxxK/RxK/RK/R) responsible for PI(4,5)P2 binding close to the second HKD domain, while PLDα, γ, δ and ε lack some key residues in the corresponding region [187,190]. PLDα1 contains a motif located between 562 and 586 amino acid residues with a high resemblance to DRY motif in proteins, which is required to interact with the heterotrimeric G protein subunit Gα [191]. In PLDδ, there is an oleate binding motif responsible for oleate-dependent stimulation, which is localized in front of the first HKD domain [56]. The presence of these domains or motifs in the PLDs underlies a structural basis for the different biochemical properties and diverse functions of PLDs.
4.2. Substrate selectivity, catalytic and regulatory properties of PLDs
Earlier studies indicated that the presence of detergents such as SDS, and millimolar Ca2+ stimulated PLDα1 activity in vitro. All C2-PLDs require Ca2+ for activity; whereas PLDγ1, PLDγ2, PLDδ and PLDβ1 are most active at micromolar Ca2+, PLDα1 and PLDα3 are most active at millimolar Ca2+ [192-194]. Ca2+ binding to the C2 domain can promote the ability of PLD binding to substrates such as PC, PE, PG and PS [195,196]. PIP2 as a cofactor is required for the activity of PLDγ, β and ζ [190]. The production of N-acylethanolamine by PLDβ and PLDγ inhibits the activity of PLDα1 [197]. The activities of PLDα and PLDε are not affected by PIP2 because they lack some key residues in the PIP2 binding domain [187]. However, the role of PIP2 is not only a cofactor but an enhancer for the activity of PLDδ which is the PLD induced by oleate [56]. Furthermore, PLDα1, PLDα3 and PLDε show strong affinity towards PC. PLDα3 and PLDε can also cleave PS with a relatively slower rate [198,199]. By comparison, PLDδ and PLDγ1 preferentially hydrolyze PE whereas PLDζs are specific to use PC [187]. The different cofactor requirements and lipid preferences suggests that individual PLDs are activated differently and hydrolyze different lipid species to generate PA in specific biological processes.
4.3. Tissue distribution and subcellular location of PLDs
Different PLDs have different subcellular associations in plant cells [200,201]. In Arabidopsis, PLDα1 was reported to be present in both soluble and membrane fractions. It tends to move towards the membranes to hydrolyze membrane lipids when plants encounter stress [202,203]. PLDγ has multi-subcellular locations and is present in the plasma membrane, nuclei, intracellular membrane and mitochondria [200]. PLDβ1 was unsuccessfully detected in different subcellular fractions in Arabidopsis, probably due to its low abundance. PLDζ2 was localized to tonoplasts [204]. PLDα3, δ and ε showed strong signals in the plasma membrane [198,205,206].
4.4. The functions of PLDs in plant growth, development, and stress responses
Various stress conditions, such as high salinity, drought, diseases, and pest attacks, induce PLD expression. Extensive studies involving genetic manipulations, phospholipid profiling and physiological analyses have resulted in identifications of different PLD genes in specific physiological processes, and elucidation of the modes of action in specific signaling cascades [83,207].
4.4.1. Unique roles of individual PLDs in plant responses to different stressors
In Arabidopsis, PLDα1 is highly active and abundant in many tissues. pldα1 mutants were observed to have impairments in stomatal closure under drought, reactive oxygen species production, and plant response to salt and ABA treatments. PLDδ is the second abundant PLD in Arabidopsis, playing significant roles in freezing tolerance, dehydration, H2O2-induced cell death, salt stress and the ABA responses [57,208-212]. PLDα3 mediates plant response to hyperosmotic stress and PLDε has a significant effect in nitrogen signaling, leading to enhanced root hair development and biomass production [198,205]. In addition, PLDs are also involved in pathogen defense in Arabidopsis. PLDβ1 confers resistance against necrotrophic fungal pathogens and negatively impacts the plant resistance to the biotrophic Pst DC3000 [213]. PLDδ plays a positive role in powdery mildew tolerance [214]. PLDγs are involved in aluminium stress, pldγ1 and pldγ2 knockdown plants display increased tolerance to aluminium stress, suggesting PLDγs are involved in plant responses to heavy metal stress [215]. The expression of PLDζs, particularly PLDζ2, increased when phosphate was deprived, and the single mutant pldζ1, pldζ2 or the double mutant pldζ1pldζ2 showed slower elongation of primary roots than did WT under phosphate limitation [216-219]. In addition, the loss of PLDζ function caused a reduction in PC and an increase in DGDG levels under phosphorous deficiency. The results indicate that PLDζs hydrolyze PC to provide phosphorus for other cellular needs and DAG for the biosynthesis of galactolipids under phosphorus starvation [218]. These reports suggest that PLDζs are positive regulators helping plants to adapt to phosphate deficiency. These studies show that different PLDs can play unique roles in mediating plant responses to different stressors, suggesting that PLDs are important to coordinate plant adaptation to adverse conditions (Fig. 5).
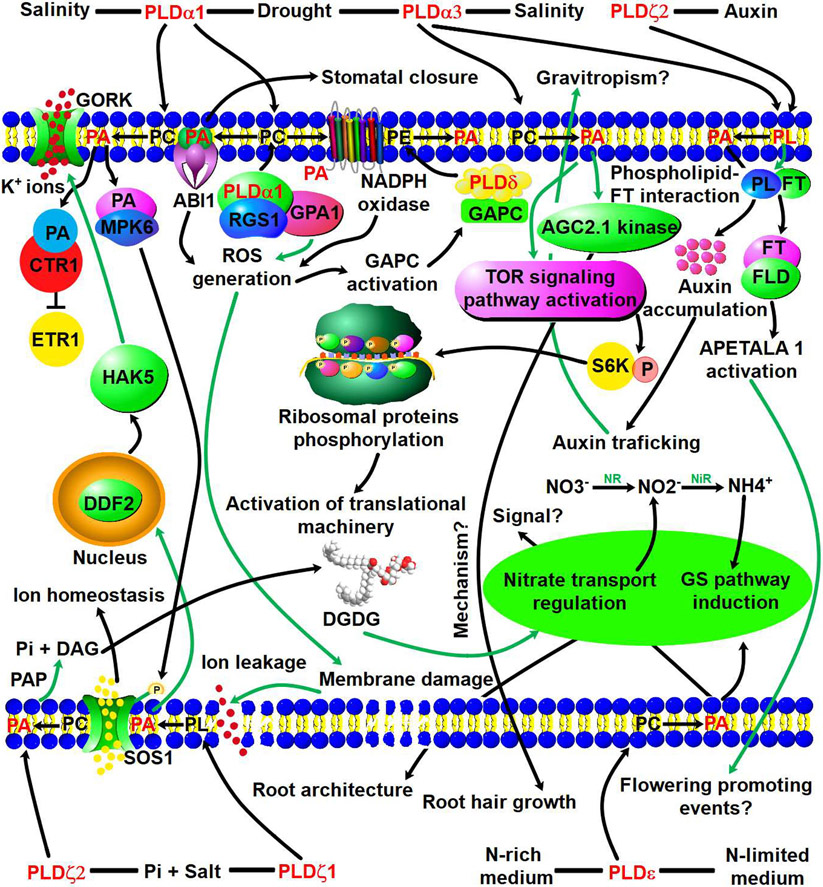
Fig. 5.
Signaling roles of PLD under stress conditions. The black arrow indicates established links while the green arrow represents putative links. PLDα1 produces PA under drought stress which interacts with ABI1 and NADPH oxidase. PLDα1 also plays a role in ABA regulated stomatal responses in G-protein dependent manner. GAPC interacts with PLDδ and mediates response to ROS in ABA signaling pathway to regulate stomatal aperture. Under salt stress, the PLDα1-produced PA activates MAPK cascade leading to the activation of Na+/H+ antiporter. Moreover, salt stress triggers PA production by PLDα1 which binds and inhibits CTR1 and blocks the interaction between ETR1 and CTR1. PLDα3 controls root growth probably through AGC2.1 kinase TOR signaling pathway. PLDζ2 responds to exogenous auxin and probably regulates gravitropism by impacting the auxin accumulation and transport. PA produced by PLDε under nutrient stress regulates nitrate transport and GS pathway but the intimate signal produced and its phenotypic consequence to plants is not known. Under salt stress, PLDζ1 produces PA which might regulate K+ ion concentration through HAK5 transporter. PA generated by PLDζ1/2 is hydrolyzed to DAG and inorganic phosphate under phosphorous deficit condition.
4.4.2. Different PLDs with unique modes of action in the same physiological processes in plants
Multiple PLDs can be involved in the same plant process but they occupy different steps with different mechanisms. The loss of the function of either PLDα1 or PLDδ rendered Arabidopsis plants insensitive to ABA-induced stomatal closure [206,210,220,221]. During plant response to ABA, PLDα1 interacts with Gα to promote stomatal opening, and the PLDα1-derived PA promotes stomatal closure through interacting with ABA INSENSITIVE 1 (ABI1). Moreover, PLDα1's product PA activates NADPH oxidase to mediate the production of ROS that promotes stomatal closure [208,210,222] (Fig. 5). However, the activity of PLDδ is activated by interacting with the cytosolic glyceraldehyde-3-phosphate dehydrogenases (GAPCs) which is oxidized via H2O2. Activated PLDδ then produces PA to control stomatal aperture (Fig. 5). It has been proposed that PA produced from PLDδ modulates the downstream target in ABA and H2O2 signaling during stomatal closure [223]. Thus, in plant response to ABA, PLDα1 enhances the production of H2O2 that induces the activity of PLDδ to produce PA in regulation of stomatal aperture. Thus, PLDδ acts downstream of PLDα1 in plant response to ABA [223].
PA produced by different PLDs are involved in plant response to salt stress. In Arabidopsis, high salinity induced an increase in PA in WT plants, but the increase was attenuated in pld pldα1, pldα3 and pldδ mutants [209,212,224-226]. Further studies showed that PLDα1-derived PA bound to mitogen-activated protein kinase 6 (MPK6), resulting in the regulation of MAPK cascade that includes MAP65-1 to enhance the rate of microtubule polymerization under the presence of NaCl [209,225]. The mutants of PLDα1 or PLDδ both have low levels of PA and exhibit increased sensitivity to salt stress while their overexpression lines elevate the tolerance to salt stress [226]. It suggests both PLDα1 and PLDδ play an important role in plant response to salt stress. The PLDα3 is also reported to modulate the plant tolerance to salt stress via enhancing root growth [205]. However, the mechanism of PLDα3 and PLDδ in regulation of the plant response to salt stress remains unclear. Together, PLDs have multifaceted mechanism of action in plants, and understanding the mode of action is important to distinguish the function of different PLDs.
4.4.3. PLDs' functions in lipid remodeling under cold stress
PLDs are the major enzymes responsible for membrane lipid hydrolysis in plant tissues under stress and tissue damages [201]. Cold stress results in a significant decrease of the main phospholipids including PC, PE and PG with increases in PA, LPC and LPE. PLD increased PA levels up to 5 folds upon exposure to −8 °C whereas the PA levels rose only half as much in the PLDα1-deficient plants compared to WT under freezing [227]. The PLDα1-deficient plants were more tolerant to freezing than WT [227]. These results suggest that PLDα1 plays an important role in the membrane integrity and negatively affects plant tolerance to freezing stress [227]. However, overexpression of PLDδ was reported to increase plant freezing tolerance whereas PLDδ knockout plants were more sensitive to freezing compared to WT [211]. This suggests PLDδ plays a positive role in plant freezing tolerance. A further study shows the WT plant produces more PA than that in pldδ mutant during freezing [211]. It is suggested that PA produced by PLDs in different patterns such as the amounts, the location and the timing may impact its function in plant response to freezing [211].
4.4.4. PLDs in plant immunity: plant-bacterial fungal, pathogen and viral interactions
Cell membranes are the initial defensive barrier against the invasion of pathogens. Phospholipids are the dominant components in cell membrane, and play important roles in defending exogenous biological invasion. PLDs-catalyzed hydrolysis of phospholipids is considered to have regulatory roles in plant response to diverse biotic infections. It is suggested that PLDs are an important mediator in the plant immunity. A previous study showed that the PA content increased in Arabidopsis upon infection with either necrotrophic fungus Botrytis cinerea or the hemibiotrophic bacteria Pseudomonas syringae [213]. PLDβ1 was identified as the major generator of PA involved in the plant response to biotic infection. In Arabidopsis, the loss of PLDβ1 increases plant sensitivity to Botrytis cinerea, but resistance to P. syringae by elevating the levels of SA and ROS [213]. The function of other PLDs in resistance to pathogen penetration has also been reported. The powdery fungi Blumeria graminis f. sp. hordei and Erysiphe pisi can easily penetrate into cells in the absence of PLDδ. PLDδ accumulated at the entry site of the fungal pathogen or the place of treatment with chitin [228]. The pldδ mutants exhibited a delay in the expression of chitin-induced-defense genes, indicating PLDδ has positive relation to plant innate immunity [229]. Additionally, PLDγ1 is found to be translocated to the plasma membrane induced by the intracellular immune receptor RPS2, implying a role of PLDγ1 in regulating plant immunity [230]. Further research indicated that the resistance towards bacterial and fungal infections was enhanced after the pldγ1 mutants were treated with microbe-associated molecular pattern (MAMP) but no difference in PA levels was observed between pldγ1 mutants and WT plants [231]. Moreover, PLDγ1 was found to interact with BAK1-INTERACTING RECEPTOR-LIKE KINASES (BIR2 and BIR3), which are the potent negative regulators of pattern-triggered immunity [231]. These findings indicate that PLDγ1 regulates plant innate immunity not by changing the levels of PA but through association with BIR2 and BIR3, highlighting a novel function of PLDs in plant immunity-related responses [231]. In future, the role of other PLDs in plant immunity needs to be investigated.
4.5. Mechanism of PLD-derived PA in cellular and physiological processes
4.5.1. Modulation of enzymatic activity and tethering proteins to the membrane
It is now evident that PLD plays critical roles in stomatal closure mediated by ABA. Thus, manipulating PLD gene expression results in decreasing water loss in plants [184,210,232]. In this process, PLDα1-derived PA played the role by binding to ABI1 and NADPH oxidase (Fig. 5) [208,210]. ABI1 is a negative regulator in ABA signaling, PA binding to ABI1 improves the release of ABA signaling and then enhances the stomatal closure [210]. NADPH oxidase is regulated by PA and this regulation was reported to be involved in stomatal closure through improving the production of H2O2 in Arabidopsis [208]. PLD-derived PA can also interact with sphingosine kinase (SPHK), which participates in the ABA signaling pathway [206,233]. In this process, PA interacts with SPHK directly and promotes the activity of SPHK [233]. In addition, under salt stress, the expression of PLDα1 and PLDδ was induced significantly to produce PA. Further studies certified that PLDα1 and PLDδ could enhance the salt tolerance in Arabidopsis [212,226]. In the pldα1 mutant, deactivation of MPK6 suggests that PA produced from PLDα1 can interact with and promote the activity of MPK6 [209]. Moreover, PLDα1-derived PA can also interact with MAP65-1 which enhances the bundling of microtubule-polymerization [225]. The experimental results indicate that PLDs and the product PA could regulate MPK6 and MAP65-1 in the plant response to salt stress [209,225].
Another important function of PA is to direct proteins towards membranes involved in modulation of the intracellular location and of protein-protein interaction. PA interacts with and binds ABI1, resulting in tethering AB11 to the plasma membrane and limiting its translocation to the nucleus [210]. PA binds to GAPC under salt stress and pushes it towards membrane [234,235]. PA induces the production ROS in plants by interacting with the cytoplasmic region of NADPH oxidase [208,210]. These results proposed that apart from the translocation of intracellular soluble proteins to the membranes, PA tethering also modulates the membrane association of cytosolic regions of important membrane proteins [83]. Some studies identified that PA had affinity towards additional putative proteins in plants [235,236]. However, the role of PA binding to these proteins needs to be verified. PA has also been shown to interacts with a MYB transcription factor, WEREWOLF (WER) at its R2 subdomain [237]. Deletion of PA-binding motif from WER interfered with its nuclear localization and role in epidermal cell fate determination. It is also shown that the suppression of PLDζ driven PA production also inhibits the nuclear location of WER and impedes root hair formation, and elongation [237]. These findings indicate that PLD-derived PA can bind to the specific site in a target protein, leading to change the localization or the activity of the proteins, thus impacting their function.
4.5.2. Effect of PA on membrane structural properties
PA has the hexagonal type ?? structural configuration and induces structural changes in the membrane. PA has the tendency to form a slackened structure around lipids thereby rendering the water repelling zone of membrane lipids exposed to effector proteins [238,239]. For example, PA binding proteins having hydrophobic residues can be positioned into the lipid bilayers of membrane [238,239]. In addition, an increase in cone-shaped PA in membrane lipid bilayers triggers negative curvatures that are normally found around the vesicle neck at the time of fusing acceptor from membrane donor [240]. Thus, the structural effects of PA interfere with membrane-protein interaction, and membrane budding and fusion [238,239].
4.5.3. PA as lipid mediators
PA is a class of minor phospholipids comprising around 1% of the total glycerophospholipids. The physiological level of PA is altered in response to various stimuli [184]. Stimulus-promoted PA synthesis can be induced either by hydrolysis of phospholipids by PLD or by DAG phosphorylation by DAG kinase [184,241]. Studies show that PA can bind to different classes of proteins, such as transcription factors, protein kinases and cytoskeletal proteins. A theory of “electrostatic/hydrogen bond switch mechanism” effectively explains that PA has more affinity to effector proteins than the other anionic phospholipids [242]. This theory suggests that electrostatic/hydrogen bond and the existence of hydrophobic region in the adjacent PA binding point in the particular amino acids in proteins may specify the affinity of PA-protein binding [184,243]. The interaction of PA with proteins may have two major consequences: One is regulating the catalytic activities and the intracellular distribution of the effector proteins. The other one is that local PA building up influences the membrane structure [184,243]. Moreover, PA is the core intermediate for the biosynthesis of glycerolipids including phospholipids, galactolipids, DAG and TAG [184,243]. PA plays a direct role in modulating the metabolism of lipids and their transport to the destination membranes [184,243]. Further, PA as lipid mediator produced by ZmPLD3 is probably involved in haploid induction (HI) as the zmpld3 mutant triggered HI in maize [244]. Therefore, distinct PA mediated pathways need to be identified for their direct or indirect effects on plant growth and development as PA can be produced by several other reactions, in addition to different PLDs. Distinguishing the sources of PA will benefit to better understand the roles of PA in plants.
5. Future perspectives
Although phospholipases are extensively studied for their roles in plant stress-related signaling events, there are still critical knowledge gaps in terms of their acting mechanism, metabolic, cellular, and physiological functions. Much remains to be learned to fully understand the role of the products of pPLA in signaling pathways. Extensive functional characterization of the identified pPLAs in plants is needed to figure out which pPLA is responsible for the generation of particular lysolipids and FFA species to promote plant cell signaling under stress conditions. Novel protein kinases, phosphatases and other signaling enzymes as FFA- and lysolipid-interacting partners and the receptors of lysolipids and FFAs are completely unknown in plants. Hence, thorough dissection of the physiological and biochemical events behind the phenotypes caused by genetic manipulation of pPLAs is needed.
DAG can be produced by several reactions, and the contribution of NPCs to the total physiological levels of DAG species produced remains unclear. It makes the NPC-specific regulatory functions intriguing. Therefore, identification and characterization of DAG-domain containing proteins for their affinities towards DAG and downstream targets along with the pathways having core DAGs signaling in plants will greatly unveil the cellular functions of NPCs. Furthermore, the concentration of IP3 in germinating seeds is extremely low, hence advance fluorescence techniques and mass spectrometric tools are needed in future to detect such minute concentrations. The IP3 generation causes Ca2+ oscillations which brings the question about their signaling potential. An established link determining the existence of IP3 receptors or of phosphorylation mechanism converting IP3 to IP6 is still missing.
Both NPC and PI-PLC generate DAG that undergoes phosphorylation to produce PA while PLDs are also the cellular generators for PA production. Meanwhile, lipid phosphate phosphatases convert PA to DAG. This complex conversion makes it challenging to delineate the molecular targets and biological roles of DAG and PA. Thus, precise elucidation of the dynamic changes of DAG and PA controlled by different routes under different conditions will greatly assist to understand the biological functions of phospholipases and their products as second messengers or mediators in plant growth, development, and stress responses.
Acknowledgements
The research in LG's lab was supported by the National Natural Science Foundation of China (31871658), Hubei Hongshan Laboratory (2021HSZD004) and Higher Education Discipline Innovation Project (B20051). UA, TF, and SI are the recipients of scholarship from the China Scholarship Council (CSC). XW acknowledges the support by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM141374.
Abbreviation:
- FFA
free fatty acid
- PA
phosphatidic acid
- DAG
diacylglycerol
- PLA
phospholipase A
- PLC
phospholipase C
- PLD
phospholipase D
- pPLA
patatin-related PLA
- sPLA2
secretory PLA2
- iPLA2
calcium-independent PLA2
- cPLAs
calcium dependent PLAs
- PAF-AHS
platelet-activating factor-acetyl hydrolases
- AdPLA
adipose PLA
- NPC
nonspecific phospholipase C
- PI-PLC
phosphatidylinositol-specific PLC
- TAG
triacylglycerol
- PC
phosphatidylcholine
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- PG
phosphatidyl glycerol
- GIPC
glucosylinositolphosphorylceramide
- JA
jasmonic acid
- ABA
abscisic acid
- MTL
matrilineal
- NLD
not like dad
- GA
gibberellic acid
- LPA
lysophosphatidic acid
- LPC
lysophosphatidycholine
- LPCAT1
lysophosphatidylcholine acyltransferase 1
- HFA
hydroxy fatty acids
- CPK
calcium dependent protein kinase
- MAPK
mitogen activated protein kinase
- SOS
salt overly sensitive
- LPE
lysophosphatidyletholamine
- CHAPS
3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate
- PAP
phosphatidic acid phosphatase
- PKC
protein kinase C
- LPP
lipid phosphate phosphatase
- PMT1
phospho-base N-methyltransferase 1
- IAA
indole acetic acid
- BL
brassinolide
- LRX2
leucine-rich repeat/extensin 2
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- IP3
inositol 1,4,5-trisphosphate
- PI(4)P
phosphatidylinositol-4-phosphate
- PI
phosphatidylinositol
- CaM
calmodulin
- PH
plekstrin homology
- PBR1
phosphatidylinositol 4,5-bisphosphate PI(4,5)P2 binding region
- MCK1
calcium/calmodulin-dependent protein kinase
- TIR1
transport inhibitor response
- RG1
rhamnogalacturonan I
- PGA
polygalacturonic acid
- RD29B
responsive to desiccation 29B
- ZAT10
salt tolerance zinc finger 10
- FLS2
flagellin sensing 2
- ABI1
ABA insensitive 1
- GAPC
glyceraldehyde-3-phosphate dehydrogenases
- BIR2
bak1-interacting receptor-like kinase 2
- NADPH
nicotinamide adenine dinucleotide phosphate
- SPHK
sphingosine kinase
- WER
werewolf
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- [1].Wang G, Ryu S, Wang X. Plant phospholipases: an overview. Lipases and Phospholipases 2012:123–37. [DOI] [PubMed] [Google Scholar]
- [2].Wang X. Plant phospholipases. Annu Rev Plant Biol 2001;52(1):211–31. [DOI] [PubMed] [Google Scholar]
- [3].Scherer GF, Ryu SB, Wang X, Matos AR, Heitz T. Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci 2010;15(12):693–700. [DOI] [PubMed] [Google Scholar]
- [4].Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 2006;18(3):665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kelly AA, Quettier A-L, Shaw E, Eastmond PJ. Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 2011;157(2):866–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li M, Bahn SC, Guo L, Musgrave W, Berg H, Welti R, et al. Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 2011;23(3):1107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang W, Devaiah SP, Pan X, Isaac G, Welti R, Wang X. AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J Biol Biochem 2007;282(25):18116–28. [DOI] [PubMed] [Google Scholar]
- [8].La Camera S, Geoffroy P, Samaha H, Ndiaye A, Rahim G, Legrand M, et al. A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J 2005;44(5):810–25. [DOI] [PubMed] [Google Scholar]
- [9].Rietz S, Dermendjiev G, Oppermann E, Tafesse FG, Effendi Y, Holk A, et al. Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Mol Plant 2010;3(3):524–38. [DOI] [PubMed] [Google Scholar]
- [10].Li M, Bahn SC, Fan C, Li J, Phan T, Ortiz M, et al. Patatin-related phospholipase pPLAIIIδ increases seed oil content with long-chain fatty acids in Arabidopsis. Plant Physiol 2013;162(1):39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ackermann EJ, Kempner E, Dennis E. Ca (2+)-independent cytosolic phospholipase A2 from macrophage-like P388D1 cells. Isolation and characterization. J Biol Biochem 1994;269(12):9227–33. [PubMed] [Google Scholar]
- [12].Holk A, Rietz S, Zahn M, Quader H, Scherer GF. Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol 2002;130(1):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Camera SL, Balagué C, Göbel C, Geoffroy P, Legrand M, Feussner I, et al. The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol Plant Microbe Interact 2009;22(4):469–81. [DOI] [PubMed] [Google Scholar]
- [14].Heinze M, Steighardt J, Gesell A, Schwartze W, Roos W. Regulatory interaction of the Gα protein with phospholipase A2 in the plasma membrane of Eschscholzia californica. Plant J 2007;52(6):1041–51. [DOI] [PubMed] [Google Scholar]
- [15].Evans JH, Spencer DM, Zweifach A, Leslie CC. Intracellular calcium signals regulating cytosolic phospholipase A2 translocation to internal membranes. J Biol Biochem 2001;276(32):30150–60. [DOI] [PubMed] [Google Scholar]
- [16].Handlogten ME, Huang C, Shiraishi N, Awata H, Miller RT. The Ca2+–sensing receptor activates cytosolic phospholipase A2 via a Gqα-dependent ERK-independent pathway. J Biol Biochem 2001;276(17):13941–8. [DOI] [PubMed] [Google Scholar]
- [17].Wang K, Guo Q, Froehlich JE, Hersh HL, Zienkiewicz A, Howe GA, et al. Two abscisic acid-responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell 2018;30(5):1006–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Blée E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci 2002;7(7):315–22. [DOI] [PubMed] [Google Scholar]
- [19].Howe GA, Schilmiller AL. Oxylipin metabolism in response to stress. Curr Opin Plant Biol 2002;5(3):230–6. [DOI] [PubMed] [Google Scholar]
- [20].Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci 1990;87(19):7713–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mason HS, Mullet JE. Expression of two soybean vegetative storage protein genes during development and in response to water deficit, wounding, and jasmonic acid. Plant Cell 1990;2(6):569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mason HS, DeWald DB, Creelman RA, Mullet JE. Coregulation of soybean vegetative storage protein gene expression by methyl jasmonate and soluble sugars. Plant Physiol 1992;98(3):859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zahn M, Wimalasekara R, Göbel C, Feussner I, Holk A, Scherer GF. Expression of Arabidopis phospholipase A genes in Petunia x hybrida. Increased hypersensitive-like response after infection with Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 demonstrates a function for phospholipase A in pathogen defence. Physiol Mol Plant Pathol 2005;67(1):2–14. [Google Scholar]
- [24].Pérez-Torres C-A, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 2008;20(12):3258–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang X, Yi K, Tao Y, Wang F, Wu Z, Jiang D, et al. Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant Cell Environ 2006;29(10):1924–35. [DOI] [PubMed] [Google Scholar]
- [26].Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2009;2(1):43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 2005;138(4):2061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, et al. Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 2007;144(1):232–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Labusch C, Shishova M, Effendi Y, Li M, Wang X, Scherer GF. Patterns and timing in expression of early auxin-induced genes imply involvement of phospholipases A (pPLAs) in the regulation of auxin responses. Mol Plant 2013;6(5):1473–86. [DOI] [PubMed] [Google Scholar]
- [30].Dong Y, Li M, Zhang P, Wang X, Fan C, Zhou Y. Patatin-related phospholipase pPLAIIIδ influences auxin-responsive cell morphology and organ size in Arabidopsis and Brassica napus. BMC Plant Biol 2014;14(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kelliher T, Starr D, Richbourg L, Chintamanani S, Delzer B, Nuccio ML, et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017;542(7639):105–9. [DOI] [PubMed] [Google Scholar]
- [32].Liu C, Li X, Meng D, Zhong Y, Chen C, Dong X, et al. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol Plant 2017;10(3):520–2. [DOI] [PubMed] [Google Scholar]
- [33].Gilles LM, Khaled A, Laffaire JB, Chaignon S, Gendrot G, Laplaige J, et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J 2017;36(6):707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang S, Jin W, Wang K. Centromere histone H3-and phospholipase-mediated haploid induction in plants. Plant Methods 2019;15(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li X, Meng D, Chen S, Luo H, Zhang Q, Jin W, et al. Single nucleus sequencing reveals spermatid chromosome fragmentation as a possible cause of maize haploid induction. Nat Commun 2017;8(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu G, Zhang K, Ai J, Deng X, Hong Y, Wang X. Patatin-related phospholipase A, pPLAIIIα, modulates the longitudinal growth of vegetative tissues and seeds in rice. J Exp Bot 2015;66(21):6945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li M, Wei F, Tawfall A, Tang M, Saettele A, Wang X. Overexpression of patatin-related phospholipase AIII δ altered plant growth and increased seed oil content in camelina. Plant Biotechnol J 2015;13(6):766–78. [DOI] [PubMed] [Google Scholar]
- [38].Qiao Y, Piao R, Shi J, Lee S-I, Jiang W, Kim B-K, et al. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor Appl Genet 2011;122(7):1439–49. [DOI] [PubMed] [Google Scholar]
- [39].Jang JH, Nguyen NQ, Légeret B, Beisson F, Kim Y-J, Sim H-J, et al. Phospholipase pPLAIIIα increases germination rate and resistance to turnip crinkle virus when overexpressed. Plant Physiol 2020;184(3):1482–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jang JH, Lee OR. Patatin-related phospholipase AtpPLAIIIα affects lignification of xylem in Arabidopsis and hybrid poplars. Plants (Basel, Switzerland) 2020;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jang JH, Bae EK, Choi YI, Lee OR. Ginseng-derived patatin-related phospholipase PgpPLAIIIβ alters plant growth and lignification of xylem in hybrid poplars. Plant Sci 2019;288:110224. [DOI] [PubMed] [Google Scholar]
- [42].Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J, Lu C. Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol 2012;160(3):1530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bates PD, Durrett TP, Ohlrogge JB, Pollard M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 2009;150(1):55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bates PD, Browse J. The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 2011;68(3):387–99. [DOI] [PubMed] [Google Scholar]
- [45].Tjellström H, Yang Z, Allen DK, Ohlrogge JB. Rapid kinetic labeling of Arabidopsis cell suspension cultures: implications for models of lipid export from plastids. Plant Physiol 2012;158(2):601–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, et al. Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 2012;24(11):4652–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lin Y, Chen G, Mietkiewska E, Song Z, Caldo KMP, Singer SD, et al. Castor patatin-like phospholipase A IIIβ facilitates removal of hydroxy fatty acids from phosphatidylcholine in transgenic Arabidopsis seeds. Plant Mol Biol 2019;101(6):521–36. [DOI] [PubMed] [Google Scholar]
- [48].Viehweger K, Dordschbal B, Roos W. Elicitor-activated phospholipase A2 generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signaling via the activation of Na+–dependent proton fluxes. Plant Cell 2002;14(7):1509–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Schwartze W, Roos W. The signal molecule lysophosphatidylcholine in Eschscholzia californica is rapidly metabolized by reacylation. Planta 2008;229(1):183–91. [DOI] [PubMed] [Google Scholar]
- [50].Scherer GF, Paul RU, Holk A. Phospholipase A2 in auxin and elicitor signal transduction in cultured parsley cells (Petroselinum crispum L.). Plant Growth Regul 2000;32(2):123–8. [Google Scholar]
- [51].Scherer GF, Paul RU, Holk A, Martinec J. Down-regulation by elicitors of phosphatidylcholine-hydrolyzing phospholipase C and up-regulation of phospholipase A in plant cells. Biochem Biophys Res Commun 2002;293(2):766–70. [DOI] [PubMed] [Google Scholar]
- [52].Li J, Li M, Yao S, Cai G, Wang X. Patatin-related phospholipase pPLAIIIγ involved in osmotic and salt tolerance in Arabidopsis. Plants 2020;9(5):650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Klucis E, Polya G. Calcium-independent activation of two plant leaf calcium-regulated protein kinases by unsaturated fatty acids. Biochem Biophys Res Commun 1987;147(3):1041–7. [DOI] [PubMed] [Google Scholar]
- [54].Baudouin E, Meskiene I, Hirt H. Unsaturated fatty acids inhibit MP2C, a protein phosphatase 2C involved in the wound-induced MAP kinase pathway regulation. Plant J 1999;20(3):343–8. [DOI] [PubMed] [Google Scholar]
- [55].Lee Y, Lee HJ, Crain RC, Lee A, Korn SJ. Polyunsaturated fatty acids modulates stomatal aperture and two distinct K+ channel currents in guard cells. Cell Signal 1994;6(2):181–6. [DOI] [PubMed] [Google Scholar]
- [56].Wang C, Wang X. A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol 2001;127(3):1102–12. [PMC free article] [PubMed] [Google Scholar]
- [57].Zhang W, Wang C, Qin C, Wood T, Olafsdottir G, Welti R, et al. The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 2003;15(10):2285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Farag KM, Palta JP. Use of lysophosphatidylethanolamine, a natural lipid, to retard tomato leaf and fruit senescence. Physiol Plant 1993;87(4):515–21. [Google Scholar]
- [59].Ryu SB, Karlsson BH, Özgen M, Palta JP. Inhibition of phospholipase D by lysophosphatidylethanolamine, a lipid-derived senescence retardant. Proc Natl Acad Sci 1997;94(23):12717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K-i, Ohta H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Biochem 2005;280(9):7469–76. [DOI] [PubMed] [Google Scholar]
- [61].Nakamura Y, Ngo AH. Non-specific phospholipase C (NPC): an emerging class of phospholipase C in plant growth and development. J Plant Res 2020;133(4):489–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang B, Li M, Phillips A, Li L, Ali U, Li Q, et al. Nonspecific phospholipase C4 hydrolyzes phosphosphingolipids and sustains plant root growth during phosphate deficiency. Plant Cell 2021;33(3):766–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Singh A, Kanwar P, Pandey A, Tyagi AK, Sopory SK, Kapoor S, et al. Comprehensive genomic analysis and expression profiling of phospholipase C gene family during abiotic stresses and development in rice. PLoS One 2013;8(4):e62494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Iqbal S, Ali U, Fadlalla T, Li Q, Liu H, Lu S, et al. Genome wide characterization of phospholipase A & C families and pattern of lysolipids and diacylglycerol changes under abiotic stresses in Brassica napus L. Plant Physiol Biochem 2020;147:101–12. [DOI] [PubMed] [Google Scholar]
- [65].Yang B, Zhang K, Jin X, Yan J, Lu S, Shen Q, et al. Acylation of non-specific phospholipase C4 determines its function in plant response to phosphate deficiency. Plant J 2021;106(6):1647–59. [DOI] [PubMed] [Google Scholar]
- [66].Pokotylo I, Pejchar P, Potocký M, Kocourková D, Krčková Z, Ruelland E, et al. The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog Lipid Res 2013;52(1):62–79. [DOI] [PubMed] [Google Scholar]
- [67].Peters C, Li M, Narasimhan R, Roth M, Welti R, Wang X. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 2010;22(8):2642–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 2008;56(1):28–39. [DOI] [PubMed] [Google Scholar]
- [69].Reddy VS, Rao DV, Rajasekharan R. Functional characterization of lysophosphatidic acid phosphatase from Arabidopsis thaliana. Biochim Biophys Acta 2010;1801(4):455–61. [DOI] [PubMed] [Google Scholar]
- [70].Wimalasekera R, Pejchar P, Holk A, Martinec J, Scherer GF. Plant phosphatidylcholine-hydrolyzing phospholipases C NPC3 and NPC4 with roles in root development and brassinolide signaling in Arabidopsis thaliana. Mol Plant 2010;3(3):610–25. [DOI] [PubMed] [Google Scholar]
- [71].Ngo AH, Lin YC, Liu Yc, Gutbrod K, Peisker H, Dörmann P, et al. A pair of nonspecific phospholipases C, NPC 2 and NPC 6, are involved in gametophyte development and glycerolipid metabolism in Arabidopsis. New Phytol 2018;219(1):163–75. [DOI] [PubMed] [Google Scholar]
- [72].Krčková Z, Brouzdová J, Daněk M, Kocourková D, Rainteau D, Ruelland E, et al. Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress. Front Plant Sci 2015;6:928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cai G, Fan C, Liu S, Yang Q, Liu D, Wu J, et al. Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants. New Phytol 2020;226(4):1055–73. [DOI] [PubMed] [Google Scholar]
- [74].Cao H, Zhuo L, Su Y, Sun L, Wang X. Non-specific phospholipase C1 affects silicon distribution and mechanical strength in stem nodes of rice. Plant J 2016;86(4):308–21. [DOI] [PubMed] [Google Scholar]
- [75].Nakamura K, Sano H. A plasma-membrane linker for the phosphoinositide-specific phospholipase C in tobacco plants. Plant Signal Behav 2009;4(1):26–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pejchar P, Potocký M, Novotná Z, Veselková Š, Kocourková D, Valentová O, et al. Aluminium ions inhibit the formation of diacylglycerol generated by phosphatidylcholine-hydrolysing phospholipase C in tobacco cells. New Phytol 2010;188(1):150–60. [DOI] [PubMed] [Google Scholar]
- [77].Ngo AH, Kanehara K, Nakamura Y. Non-specific phospholipases C, NPC2 and NPC6, are required for root growth in Arabidopsis. Plant J 2019;100(4):825–35. [DOI] [PubMed] [Google Scholar]
- [78].Krčková Z, Kocourková D, Daněk M, Brouzdová J, Pejchar P, Janda M, et al. The Arabidopsis thaliana non-specific phospholipase C2 is involved in the response to pseudomonas syringae attack. Ann Bot 2018;121(2):297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bose D, Ngo AH, Nguyen VC, Nakamura Y. Non-specific phospholipases C2 and C6 redundantly function in pollen tube growth via triacylglycerol production in Arabidopsis. Plant J 2021;106(2):409–18. [DOI] [PubMed] [Google Scholar]
- [80].Eichmann TO, Lass A. DAG tales. The multiple faces of diacylglycerol-stereochemistry, metabolism, and signaling. Cell Mol Life Sci 2015;72(20):3931–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nakamura Y, Ohta H. The diacylglycerol forming pathways differ among floral organs of Petunia hybrida. FEBS Lett 2007;581(28):5475–9. [DOI] [PubMed] [Google Scholar]
- [82].Douce R, Joyard J. Chloroplast envelope lipids: detection and biosynthesis. Methods Enzymol 1980;1980(69):290–301. [Google Scholar]
- [83].Hong Y, Zhao J, Guo L, Kim S-C, Deng X, Wang G, et al. Plant phospholipases D and C and their diverse functions in stress responses. Prog Lipid Res 2016;62:55–74. [DOI] [PubMed] [Google Scholar]
- [84].Dong W, Lv H, Xia G, Wang M. Does diacylglycerol serve as a signaling molecule in plants? Plant Signal Behav 2012;7(4):472–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Peters C, Kim SC, Devaiah S, Li M, Wang X. Non-specific phospholipase C 5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ 2014;37(9):2002–13. [DOI] [PubMed] [Google Scholar]
- [86].Pejchar P, Potocký M, Krčková Z, Brouzdová J, Daněk M, Martinec J. Non-specific phospholipase C4 mediates response to aluminum toxicity in Arabidopsis thaliana. Front Plant Sci 2015;6:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lee Y, Assmann SM. Diacylglycerols induce both ion pumping in patch-clamped guard-cell protoplasts and opening of intact stomata. Proc Natl Acad Sci 1991;88(6):2127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, Zurita-Silva A, Sánchez-Calderon L, Ramírez-Chávez E, et al. The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 2004;16(8):2020–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Su Y, Li M, Guo L, Wang X. Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. Plant J 2018;94(2):315–26. [DOI] [PubMed] [Google Scholar]
- [90].Kocourkova D, Krčková Z, Pejchar P, Veselková Š, Valentova O, Wimalasekera R, et al. The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J Exp Bot 2011;62(11):3753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zhu J-K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 2002;53(1):247–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Yang D, Liu X, Yin X, Dong T, Yu M, Wu Y. Rice non-specific phospholipase C6 is involved in mesocotyl elongation. Plant Cell Physiol 2021;62(6):985–1000. [DOI] [PubMed] [Google Scholar]
- [93].Chevalier F, Cuyas L, Jouhet J, Gros V, Chiarenza S, Secco D, et al. Interplay between jasmonic acid, phosphate signaling and the regulation of glycerolipid homeostasis in Arabidopsis. Plant Cell Physiol 2019;60(6):1260–73. [DOI] [PubMed] [Google Scholar]
- [94].Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1. An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998;280(5366):1091–4. [DOI] [PubMed] [Google Scholar]
- [95].Roughan P. Turnover of the glycerolipids of pumpkin leaves. The importance of phosphatidylcholine. Biochem J 1970;117(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Meï CE, Cussac M, Haslam RP, Beaudoin F, Wong Y-S, Maréchal E, et al. C1 metabolism inhibition and nitrogen deprivation trigger triacylglycerol accumulation in Arabidopsis thaliana cell cultures and highlight a role of NPC in phosphatidylcholine-to-triacylglycerol pathway. Front Plant Sci 2017;7:2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sagar S, Singh A. Emerging role of phospholipase C mediated lipid signaling in abiotic stress tolerance and development in plants. Plant Cell Rep 2021:1–11. [DOI] [PubMed] [Google Scholar]
- [98].Wang X. Lipid signaling. Curr Opin Plant Biol 2004;7(3):329–36. [DOI] [PubMed] [Google Scholar]
- [99].Berridge MJ. Inositol trisphosphate and calcium signalling. Nature 1993;361(6410):315–25. [DOI] [PubMed] [Google Scholar]
- [100].Wang X, Liu Y, Li Z, Gao X, Dong J, Zhang J, et al. Genome-wide identification and expression profile analysis of the phospholipase C gene family in wheat (Triticum aestivum L.). Plants 2020;9(7):885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang K, Li Y-l, Chen S. Genome-wide identification of phospholipase C related to chilling injury in peach fruit. J Plant Biochem Biotechnol 2020:1–10. [Google Scholar]
- [102].Chen G, Snyder CL, Greer MS, Weselake RJ. Biology and biochemistry of plant phospholipases. Crit Rev Plant Sci 2011;30(3):239–58. [Google Scholar]
- [103].Bunney TD, Katan M. PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem Sci 2011;36(2):88–96. [DOI] [PubMed] [Google Scholar]
- [104].Munnik T, Testerink C. Plant phospholipid signaling:“in a nutshell”. J Lipid Res 2009;50:S260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Georges F, Das S, Ray H, Bock C, Nokhrina K, Kolla VA, et al. Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant Cell Environ 2009;32(12):1664–81. [DOI] [PubMed] [Google Scholar]
- [106].Zheng SZ, Liu YL, Li B, Shang Zl, Zhou RG, Sun DY. Phosphoinositide-specific phospholipase C9 is involved in the thermotolerance of Arabidopsis. Plant J 2012;69(4):689–700. [DOI] [PubMed] [Google Scholar]
- [107].Vossen JH, Abd-El-Haliem A, Fradin EF, Van Den Berg GC, Ekengren SK, Meijer HJ, et al. Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J 2010;62(2):224–39. [DOI] [PubMed] [Google Scholar]
- [108].Khalil HB, Wang Z, Wright JA, Ralevski A, Donayo AO, Gulick PJ. Heterotrimeric Gα subunit from wheat (Triticum aestivum), GA3, interacts with the calcium-binding protein, Clo3, and the phosphoinositide-specific phospholipase C, PI-PLC1. Plant Mol Biol 2011;77(1–2):145. [DOI] [PubMed] [Google Scholar]
- [109].Mikami K, Repp A, Graebe-Abts E, Hartmann E. Isolation of cDNAs encoding typical and novel types of phosphoinositide-specific phospholipase C from the moss Physcomitrella patens. J Exp Bot 2004;55(401):1437–9. [DOI] [PubMed] [Google Scholar]
- [110].Kim YJ, Kim JE, Lee J-H, Lee MH, Jung HW, Bahk YY, et al. The Vr-PLC3 gene encodes a putative plasma membrane-localized phosphoinositide-specific phospholipase C whose expression is induced by abiotic stress in mung bean (Vigna radiata L.). FEBS Lett 2004;556(1–3):127–36. [DOI] [PubMed] [Google Scholar]
- [111].Liu H-T, Huang W-D, Pan Q-H, Weng F-H, Zhan J-C, Liu Y, et al. Contributions of PIP2-specific-phospholipase C and free salicylic acid to heat acclimation-induced thermotolerance in pea leaves. J Plant Physiol 2006;163(4):405–16. [DOI] [PubMed] [Google Scholar]
- [112].Zhai S, Sui Z, Yang A, Zhang J. Characterization of a novel phosphoinositide-specific phospholipase C from Zea mays and its expression in Escherichia coli. Biotechnol Lett 2005;27(11):799–804. [DOI] [PubMed] [Google Scholar]
- [113].Kouchi Z, Shikano T, Nakamura Y, Shirakawa H, Fukami K, Miyazaki S. The role of EF-hand domains and C2 domain in regulation of enzymatic activity of phospholipase Cζ. J Biol Biochem 2005;280(22):21015–21. [DOI] [PubMed] [Google Scholar]
- [114].Otterhag L, Sommarin M, Pical C. N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett 2001;497(2–3):165–70. [DOI] [PubMed] [Google Scholar]
- [115].Rupwate SD, Rajasekharan R. Plant phosphoinositide-specific phospholipase C: an insight. Plant Signal Behav 2012;7(10):1281–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Munnik T, Irvine R, Musgrave A. Phospholipid signalling in plants. Biochim Biophys Acta 1998;1389(3):222–72. [DOI] [PubMed] [Google Scholar]
- [117].McMURRAY WC, Irvine R. Phosphatidylinositol 4, 5-bisphosphate phosphodiesterase in higher plants. Biochem J 1988;249(3):877–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Drøbak BK. The plant phosphoinositide system. Biochem J 1992;288(Pt 3):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Melin P-M, Pical C, Jergil B, Sommarin M. Polyphosphoinositide phospholipase C in wheat root plasma membranes. Partial purification and characterization. Biochim Biophys Acta 1992;1123(2):163–9. [DOI] [PubMed] [Google Scholar]
- [120].Pan Y-Y, Wang X, Ma L-G, Sun D-Y. Characterization of phosphatidylinositol-specific phospholipase C (PI-PLC) from Lilium daviddi pollen. Plant Cell Physiol 2005;46(10):1657–65. [DOI] [PubMed] [Google Scholar]
- [121].Friedman EJ, Temple BR, Hicks SN, Sondek J, Jones CD, Jones AM. Prediction of protein–protein interfaces on G-protein β subunits reveals a novel phospholipase C β2 binding domain. J Mol Biol 2009;392(4):1044–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Misra S, Wu Y, Venkataraman G, Sopory SK, Tuteja N. Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant J 2007;51(4):656–69. [DOI] [PubMed] [Google Scholar]
- [123].Sekiya F, Poulin B, Kim YJ, Rhee SG. Mechanism of tyrosine phosphorylation and activation of phospholipase C-γ1: tyrosine 783 phosphorylation is not sufficient for lipase activation. J Biol Biochem 2004;279(31):32181–90. [DOI] [PubMed] [Google Scholar]
- [124].Nühse TS, Stensballe A, Jensen ON, Peck SC. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 2004;16(9):2394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Takáč T, Novák D, Šamaj J. Recent advances in the cellular and developmental biology of phospholipases in plants. Front Plant Sci 2019;10:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kanehara K, Yu C-Y, Cho Y, Cheong W-F, Torta F, Shui G, et al. Arabidopsis AtPLC2 is a primary phosphoinositide-specific phospholipase C in phosphoinositide metabolism and the endoplasmic reticulum stress response. PLoS Genet 2015;11(9):e1005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Zhang Q, Van Wijk R, Shahbaz M, Roels W, Schooten Bv, Vermeer JE, et al. Arabidopsis phospholipase C3 is involved in lateral root initiation and ABA responses in seed germination and stomatal closure. Plant Cell Physiol 2018;59(3):469–86. [DOI] [PubMed] [Google Scholar]
- [128].Zhang Q, Van Wijk R, Zarza X, Shahbaz M, Van Hooren M, Guardia A, et al. Knock-down of Arabidopsis PLC5 reduces primary root growth and secondary root formation while overexpression improves drought tolerance and causes stunted root hair growth. Plant Cell Physiol 2018;59(10):2004–19. [DOI] [PubMed] [Google Scholar]
- [129].Tasma IM, Brendel V, Whitham SA, Bhattacharyya MK. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol Biochem 2008;46(7):627–37. [DOI] [PubMed] [Google Scholar]
- [130].Zhu J, Zhou Y, Li J, Li H. Genome-wide investigation of the phospholipase C gene family in Zea mays. Front Genet 2020;11:1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Shi J, Gonzales RA, Bhattacharyya MK. Characterization of a plasma membrane-associated phosphoinositide-specific phospholipase C from soybean. Plant J 1995;8(3):381–90. [DOI] [PubMed] [Google Scholar]
- [132].Rupwate SD, Rajasekharan R. C2 domain is responsible for targeting rice phosphoinositide specific phospholipase C. Plant Mol Biol 2012;78(3):247–58. [DOI] [PubMed] [Google Scholar]
- [133].Novotna Z, Valentova O, Martinec J, Feltl T, Nokhrina K. Study of phospholipases D and C in maturing and germinating seeds of Brassica napus. Portland Press Ltd; 2000. [PubMed] [Google Scholar]
- [134].Perera IY, Heilmann I, Chang SC, Boss WF, Kaufman PB. A role for inositol 1, 4, 5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol 2001;125(3):1499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Parre E, Ghars MA, Leprince A-S, Thiery L, Lefebvre D, Bordenave M, et al. Calcium signaling via phospholipase C is essential for proline accumulation upon ionic but not nonionic hyperosmotic stresses in Arabidopsis. Plant Physiol 2007;144(1):503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lemtiri-Chlieh F, MacRobbie EA, Brearley CA. Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci 2000;97(15):8687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Munnik T, Irvine R, Musgrave A. Rapid turnover of phosphatidylinositol 3-phosphate in the green alga Chlamydomonas eugametos: signs of a phosphatidylinositide 3-kinase signalling pathway in lower plants? Biochem J 1994;298(2):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Gonorazky G, Ramirez L, Abd-El-Haliem A, Vossen JH, Lamattina L, Ten Have A, et al. The tomato phosphatidylinositol-phospholipase C2 (SlPLC2) is required for defense gene induction by the fungal elicitor xylanase. J Plant Physiol 2014;171(11):959–65. [DOI] [PubMed] [Google Scholar]
- [139].Pokotylo I, Kolesnikov Y, Kravets V, Zachowski A, Ruelland E. Plant phosphoinositide-dependent phospholipases C: variations around a canonical theme. Biochimie 2014;96:144–57. [DOI] [PubMed] [Google Scholar]
- [140].Munnik T. PI-PLC: Phosphoinositide-phospholipase C in plant signaling. In: Phospholipases in plant signaling. Springer; 2014. p. 27–54. [Google Scholar]
- [141].Rodas-Junco BA, Racagni-Di-Palma GE, Canul-Chan M, Usorach J, Hernández-Sotomayor S. Link between lipid second messengers and osmotic stress in plants. Int J Mol Sci 2021;22(5):2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Colin LA, Jaillais Y. Phospholipids across scales: lipid patterns and plant development. Curr Opin Plant Biol 2020;53:1–9. [DOI] [PubMed] [Google Scholar]
- [143].Noack LC, Jaillais Y. Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr Opin Plant Biol 2017;40:22–33. [DOI] [PubMed] [Google Scholar]
- [144].Boutté Y, Moreau P. Modulation of endomembranes morphodynamics in the secretory/retrograde pathways depends on lipid diversity. Curr Opin Plant Biol 2014;22:22–9. [DOI] [PubMed] [Google Scholar]
- [145].Simon MLA, Platre MP, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, et al. A PtdIns (4) P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2016;2(7):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Lin F, Krishnamoorthy P, Schubert V, Hause G, Heilmann M, Heilmann I. A dual role for cell plate-associated PI4Kβ in endocytosis and phragmoplast dynamics during plant somatic cytokinesis. EMBO J 2019;38(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA. Electron tomography of RabA4b-and PI-4Kβ1-labeled trans golgi network compartments in Arabidopsis. Traffic 2011;12(3):313–29. [DOI] [PubMed] [Google Scholar]
- [148].Stanislas T, Platre MP, Liu M, Rambaud-Lavigne LE, Jaillais Y, Hamant O. A phosphoinositide map at the shoot apical meristem in Arabidopsis thaliana. BMC Biol 2018;16(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Simon MLA, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, et al. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J 2014;77(2):322–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Van Leeuwen W, Vermeer JE, Gadella TW Jr, Munnik T. Visualization of phosphatidylinositol 4, 5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J 2007;52(6):1014–26. [DOI] [PubMed] [Google Scholar]
- [151].Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, et al. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol 2005;168(5):801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Platre MP, Jaillais Y. Guidelines for the use of protein domains in acidic phospholipid imaging. In: Lipid signaling protocols. Springer; 2016. p. 175–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lee E, Vanneste S, Pérez-Sancho J, Benitez-Fuente F, Strelau M, Macho AP, et al. Ionic stress enhances ER–PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc Natl Acad Sci 2019;116(4):1420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Gujas B, Cruz TM, Kastanaki E, Vermeer JE, Munnik T, Rodriguez-Villalon A. Perturbing phosphoinositide homeostasis oppositely affects vascular differentiation in Arabidopsis thaliana roots. Development 2017;144(19):3578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Di Fino LM, D’Ambrosio JM, Tejos R, van Wijk R, Lamattina L, Munnik T, et al. Arabidopsis phosphatidylinositol-phospholipase C2 (PLC2) is required for female gametogenesis and embryo development. Planta 2017;245(4):717–28. [DOI] [PubMed] [Google Scholar]
- [156].Li L, He Y, Wang Y, Zhao S, Chen X, Ye T, et al. Arabidopsis PLC 2 is involved in auxin-modulated reproductive development. Plant J 2015;84(3):504–15. [DOI] [PubMed] [Google Scholar]
- [157].Ischebeck T, Stenzel I, Hempel F, Jin X, Mosblech A, Heilmann I. Phosphatidylinositol-4, 5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J 2011;65(3):453–68. [DOI] [PubMed] [Google Scholar]
- [158].Nibau C, Wu H-m, Cheung AY. RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci 2006;11(6):309–15. [DOI] [PubMed] [Google Scholar]
- [159].Perera IY, Heilmann I, Boss WF. Transient and sustained increases in inositol 1, 4, 5-trisphosphate precede the differential growth response in gravistimulated maize pulvini. Proc Natl Acad Sci 1999;96(10):5838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Yun HS, Joo SH, Kaufman PB, Kim TW, Kirakosyan A, Philosoph-Hadas S, et al. Changes in starch and inositol 1, 4, 5-trisphosphate levels and auxin transport are interrelated in graviresponding oat (Avena sativa) shoots. Plant Cell Environ 2006;29(11):2100–11. [DOI] [PubMed] [Google Scholar]
- [161].Kolesnikov YS, Kretynin SV, Volotovsky ID, Kordyum EL, Ruelland E, Kravets VS. Molecular mechanisms of gravity perception and signal transduction in plants. Protoplasma 2016;253(4):987–1004. [DOI] [PubMed] [Google Scholar]
- [162].Repp A, Mikami K, Mittmann F, Hartmann E. Phosphoinositide-specific phospholipase C is involved in cytokinin and gravity responses in the moss Physcomitrella patens. Plant J 2004;40(2):250–9. [DOI] [PubMed] [Google Scholar]
- [163].Lu Y-T, Hidaka H, Feldman LJ. Characterization of a calcium/calmodulin-dependent protein kinase homolog from maize roots showing light-regulated gravitropism. Planta 1996;199(1):18–24. [DOI] [PubMed] [Google Scholar]
- [164].Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007;446(7136):640–5. [DOI] [PubMed] [Google Scholar]
- [165].Stevenson-Paulik J, Bastidas RJ, Chiou S-T, Frye RA, York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci 2005;102(35):12612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Van Wijk R, Zhang Q, Zarza X, Lamers M, Marquez FR, Guardia A, et al. Role for Arabidopsis PLC7 in stomatal movement, seed mucilage attachment, and leaf serration. Front Plant Sci 2018;9:1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc Natl Acad Sci 1995;92(9):3903–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kim YJ, Kim JE, Lee J-H, Lee MH, Jung HW, Bahk YY, et al. The V-PLC3 gene encodes a putative plasma membrane-localized phosphoinositide-specific phospholipase C whose expression is induced by abiotic stress in mung bean (Vigna radiata L.) 1. FEBS Lett 2004;556(1–3):127–36. [DOI] [PubMed] [Google Scholar]
- [169].Tripathy MK, Tyagi W, Goswami M, Kaul T, Singla-Pareek SL, Deswal R, et al. Characterization and functional validation of tobacco PLC delta for abiotic stress tolerance. Plant Mol Biol Rep 2012;30(2):488–97. [Google Scholar]
- [170].Zhang K, Jin C, Wu L, Hou M, Dou S, Pan Y. Expression analysis of a stress-related phosphoinositide-specific phospholipase C gene in wheat (Triticum aestivum L.). PLoS One 2014;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Zhang J, Zhang Z, Zhu D, Guan Y, Shi D, Chen Y, et al. Expression and initial characterization of a phosphoinositide-specific phospholipase C from Populus tomentosa. J Plant Biochem Biotechnol 2015;24(3):338–46. [Google Scholar]
- [172].Chen Z-F, Ru J-N, Sun G-Z, Du Y, Chen J, Zhou Y-B, et al. Genomic-wide analysis of the PLC family and detection of GmPI-PLC7 responses to drought and salt stresses in soybean. Front Plant Sci 2021;12:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Xia K, Wang B, Zhang J, Li Y, Yang H, Ren D. Arabidopsis phosphoinositide-specific phospholipase C 4 negatively regulates seedling salt tolerance. Plant Cell Environ 2017;40(8):1317–31. [DOI] [PubMed] [Google Scholar]
- [174].Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol 2010;61:593–620. [DOI] [PubMed] [Google Scholar]
- [175].Perera IY, Hung C-Y, Moore CD, Stevenson-Paulik J, Boss WF. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 2008;20(10):2876–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Deng X, Yuan S, Cao H, Lam SM, Shui G, Hong Y, et al. Phosphatidylinositol-hydrolyzing phospholipase C4 modulates rice response to salt and drought. Plant Cell Environ 2019;42(2):536–48. [DOI] [PubMed] [Google Scholar]
- [177].Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B, et al. Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe Interact 2006;19(7):711–24. [DOI] [PubMed] [Google Scholar]
- [178].Abd-El-Haliem AM, Joosten MH. Plant phosphatidylinositol-specific phospholipase C at the center of plant innate immunity. J Integr Plant Biol 2017;59(3):164–79. [DOI] [PubMed] [Google Scholar]
- [179].Abd-El-Haliem AM, Vossen JH, van Zeijl A, Dezhsetan S, Testerink C, Seidl MF, et al. Biochemical characterization of the tomato phosphatidylinositol-specific phospholipase C (PI-PLC) family and its role in plant immunity. Biochim Biophys Acta 2016;1861(9):1365–78. [DOI] [PubMed] [Google Scholar]
- [180].Mbengue M, Bourdais G, Gervasi F, Beck M, Zhou J, Spallek T, et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci 2016;113(39):11034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Postma J, Liebrand TW, Bi G, Evrard A, Bye RR, Mbengue M, et al. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol 2016;210(2):627–42. [DOI] [PubMed] [Google Scholar]
- [182].Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell 2006;126(5):969–80. [DOI] [PubMed] [Google Scholar]
- [183].Wang X. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol 2005;139(2):566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res 2006;45(3):250–78. [DOI] [PubMed] [Google Scholar]
- [185].Lu S, Fadlalla T, Tang S, Li L, Ali U, Li Q, et al. Genome-wide analysis of Phospholipase D gene family and profiling of phospholipids under abiotic stresses in Brassica napus. Plant Cell Physiol 2019;60(7):1556–66. [DOI] [PubMed] [Google Scholar]
- [186].Eliáš M, Potocký M, Cvrčková F, Žárský V. Molecular diversity of phospholipase D in angiosperms. BMC Genomics 2002;3(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Qin C, Wang X. The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 2002;128(3):1057–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Arhab Y, Abousalham A, Noiriel A. Plant phospholipase D mining unravels new conserved residues important for catalytic activity. Biochim Biophys Acta 2019;1864(5):688–703. [DOI] [PubMed] [Google Scholar]
- [189].Li J, Yu F, Guo H, Xiong R, Zhang W, He F, et al. Crystal structure of plant PLDα1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D. Cell Res 2020;30(1):61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Zheng L, Shan J, Krishnamoorthi R, Wang X. Activation of plant phospholipase Dβ by phosphatidylinositol 4, 5-bisphosphate: characterization of binding site and mode of action. Biochemistry 2002;41(14):4546–53. [DOI] [PubMed] [Google Scholar]
- [191].Zhao J, Wang X. Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Biochem 2004;279(3):1794–800. [DOI] [PubMed] [Google Scholar]
- [192].Pappan K, Zheng S, Wang X. Identification and characterization of a novel plant phospholipase D that requires polyphosphoinositides and submicromolar calcium for activity in Arabidopsis. J Biol Biochem 1997;272(11):7048–54. [DOI] [PubMed] [Google Scholar]
- [193].Pappan K, Qin W, Dyer JH, Zheng L, Wang X. Molecular cloning and functional analysis of polyphosphoinositide-dependent phospholipase D, PLDβ, from Arabidopsis. J Biol Biochem 1997;272(11):7055–61. [DOI] [PubMed] [Google Scholar]
- [194].Arisz S. Plant phosphatidic acid metabolism in response to environmental stress. Universiteit van Amsterdam; 2010. [Host]. [Google Scholar]
- [195].Zheng L, Krishnamoorthi R, Zolkiewski M, Wang X. Distinct Ca2+ binding properties of novel C2 domains of plant phospholipase Dα and β. J Biol Biochem 2000;275(26):19700–6. [DOI] [PubMed] [Google Scholar]
- [196].Qin C, Wang C, Wang X. Kinetic analysis of Arabidopsis phospholipase Dδ: substrate preference and mechanism of activation by Ca2+ and phosphatidylinositol 4, 5-bisphosphate. J Biol Biochem 2002;277(51):49685–90. [DOI] [PubMed] [Google Scholar]
- [197].Austin-Brown SL, Chapman KD. Inhibition of Phospholipase Dα byN-Acylethanolamines. Plant Physiol 2002;129(4):1892–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, et al. Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J 2009;58(3):376–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Hong Y, Pan X, Welti R, Wang X. Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 2008;20(3):803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Fan L, Zheng S, Cui D, Wang X. Subcellular distribution and tissue expression of phospholipase Dα, Dβ, and Dγ in Arabidopsis. Plant Physiol 1999;119(4):1371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Li M, Hong Y, Wang X. Phospholipase D-and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 2009;1791(9):927–35. [DOI] [PubMed] [Google Scholar]
- [202].Wang C, Zien CA, Afitlhile M, Welti R, Hildebrand DF, Wang X. Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 2000;12(11):2237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Zien CA, Wang C, Wang X, Welti R. In vivo substrates and the contribution of the common phospholipase D, PLDα, to wound-induced metabolism of lipids in Arabidopsis. Biochim Biophys Acta 2001;1530(2–3):236–48. [DOI] [PubMed] [Google Scholar]
- [204].Yamaryo Y, Dubots E, Albrieux C, Baldan B, Block MA. Phosphate availability affects the tonoplast localization of PLDζ2, an Arabidopsis thaliana phospholipase D. FEBS Lett 2008;582(5):685–90. [DOI] [PubMed] [Google Scholar]
- [205].Hong Y, Pan X, Welti R, Wang X. The effect of phospholipase Dα3 in Arabidopsis response to hyperosmotic stress and glucose. Plant Signal Behav 2008;3(12):1099–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Guo L, Mishra G, Markham JE, Li M, Tawfall A, Welti R, et al. Connections between sphingosine kinase and phospholipase D in the abscisic acid signaling pathway in Arabidopsis. J Biol Biochem 2012;287(11):8286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Deepika D, Singh A. Plant phospholipase D: novel structure, regulatory mechanism, and multifaceted functions with biotechnological application. Crit Rev Biotechnol 2021:1–19. [DOI] [PubMed] [Google Scholar]
- [208].Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, et al. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 2009;21(8):2357–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, et al. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 2010;188(3):762–73. [DOI] [PubMed] [Google Scholar]
- [210].Zhang W, Qin C, Zhao J, Wang X. Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci 2004;101(25):9508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Li W, Li M, Zhang W, Welti R, Wang X. The plasma membrane–bound phospholipase Dδ enhances freezing tolerance in Arabidopsis thaliana. Nat Biotechnol 2004;22(4):427–33. [DOI] [PubMed] [Google Scholar]
- [212].Katagiri T, Takahashi S, Shinozaki K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J 2001;26(6):595–605. [DOI] [PubMed] [Google Scholar]
- [213].Zhao J, Devaiah SP, Wang C, Li M, Welti R, Wang X. Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytol 2013;199(1):228–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Zhang Q, Berkey R, Blakeslee JJ, Lin J, Ma X, King H, et al. Arabidopsis phospholipase Dα1 and Dδ oppositely modulate EDS1-and SA-independent basal resistance against adapted powdery mildew. J Exp Bot 2018;69(15):3675–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Zhao J, Wang C, Bedair M, Welti R, Sumner LW, Baxter I, et al. Suppression of phospholipase Dγs confers increased aluminum resistance in Arabidopsis thaliana. PLoS One 2011;6(12):e28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, et al. Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 2003;300(5624):1427–30. [DOI] [PubMed] [Google Scholar]
- [217].Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci 2006;103(17):6765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Li M, Qin C, Welti R, Wang X. Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 2006;140(2):761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Li G, Xue H-W. Arabidopsis PLDζ2 regulates vesicle trafficking and is required for auxin response. Plant Cell 2007;19(1):281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Uraji M, Katagiri T, Okuma E, Ye W, Hossain MA, Masuda C, et al. Cooperative function of PLDδ and PLDα1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol 2012;159(1):450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Distéfano AM, Scuffi D, García-Mata C, Lamattina L, Laxalt AM. Phospholipase Dδ is involved in nitric oxide-induced stomatal closure. Planta 2012;236(6):1899–907. [DOI] [PubMed] [Google Scholar]
- [222].Mishra G, Zhang W, Deng F, Zhao J, Wang X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 2006;312(5771):264–6. [DOI] [PubMed] [Google Scholar]
- [223].Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, et al. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 2012;24(5):2200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Hong Y, Zhang W, Wang X. Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ 2010;33(4):627–35. [DOI] [PubMed] [Google Scholar]
- [225].Zhang Q, Lin F, Mao T, Nie J, Yan M, Yuan M, et al. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 2012;24(11):4555–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Bargmann BO, Laxalt AM, Riet Bt, Van Schooten B, Merquiol E, Testerink C, et al. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol 2009;50(1):78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, et al. Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Biochem 2002;277(35):31994–2002. [DOI] [PubMed] [Google Scholar]
- [228].Xing J, Li X, Wang X, Lv X, Wang L, Zhang L, et al. Secretion of phospholipase Dδ functions as a regulatory mechanism in plant innate immunity. Plant Cell 2019;31(12):3015–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Pinosa F, Buhot N, Kwaaitaal M, Fahlberg P, Thordal-Christensen H, Ellerström M, et al. Arabidopsis phospholipase Dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol 2013;163(2):896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Elmore JM, Liu J, Smith B, Phinney B, Coaker G. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics 2012;11(4). M111. 014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Schlöffel MA, Salzer A, Wan W-L, van Wijk R, Del Corvo R, Šemanjski M, et al. The BIR2/BIR3-associated phospholipase Dγ1 negatively regulates plant immunity. Plant Physiol 2020;183(1):371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Jacob T, Ritchie S, Assmann SM, Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc Natl Acad Sci 1999;96(21):12192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Guo L, Mishra G, Taylor K, Wang X. Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J Biol Biochem 2011:286(15):13336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].McLoughlin F, Arisz SA, Dekker HL, Kramer G, De Koster CG, Haring MA, et al. Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem J 2013;450(3):573–81. [DOI] [PubMed] [Google Scholar]
- [235].Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, De Koster CG, et al. Isolation and identification of phosphatidic acid targets from plants. Plant J 2004;39(4):527–36. [DOI] [PubMed] [Google Scholar]
- [236].Takáč T, Pechan T, Šamajová O, Šamaj J. Proteomic analysis of Arabidopsis pldα1 mutants revealed an important role of phospholipase D alpha 1 in chloroplast biogenesis. Front Plant Sci 2019;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Yao H, Wang G, Guo L, Wang X. Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell 2013;25(12):5030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Killian JA, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta 2004;1666(1–2):275–88. [DOI] [PubMed] [Google Scholar]
- [239].Kooijman EE, Chupin V, de Kruijff B, Burger KN. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 2003;4(3):162–74. [DOI] [PubMed] [Google Scholar]
- [240].Roth MG. Molecular mechanisms of PLD function in membrane traffic. Traffic 2008;9(8):1233–9. [DOI] [PubMed] [Google Scholar]
- [241].Arisz SA, Testerink C, Munnik T. Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 2009;1791(9):869–75. [DOI] [PubMed] [Google Scholar]
- [242].Kooijman EE, Tieleman DP, Testerink C, Munnik T, Rijkers DT, Burger KN, et al. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J Biol Biochem 2007;282(15):11356–64. [DOI] [PubMed] [Google Scholar]
- [243].Shin JJ, Loewen CJ. Putting the pH into phosphatidic acid signaling. BMC Biol 2011;9(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Li Y, Lin Z, Yue Y, Zhao H, Fei X, Liu C, et al. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat Plants 2021;7(12):1579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]