Abstract
The photooxidative damage of DNA, specifically guanine oxidation and strand-break formation, by sidechain-oxyfunctionalized acetophenones (hydroxy, methoxy, tert-butoxy and acetoxy derivatives), has been examined. The involvement of triplet-excited ketones and their reactivity towards DNA has been determined by time-resolved laser-flash spectroscopy. The generation of carbon-centered radical species upon Norrish-type I cleavage has been assessed by spin-trapping experiments with 5,5-dimethyl-1-pyrroline N-oxide, coupled with electron paramagnetic resonance spectroscopy. The observed DNA-base oxidation and strand-break formation is discussed in terms of the peroxyl radicals derived from the triplet-excited ketones by α cleavage and molecular oxygen trapping, as well as direct interaction of the excited states by electron transfer and hydrogen-atom abstraction. It is concluded that acetophenone derivatives, which produce radicals upon photolysis, in particular the hydroxy (AP-OH) and tert-butoxy (AP-OtBu) derivatives, are more effective in oxidizing DNA.
INTRODUCTION
The importance of oxidative DNA damage in mutagenesis, carcinogenesis and aging has motivated the current intensive activity on the elucidation of the interaction of DNA with reactive oxygen species (ROS) like hydroxyl, alkoxyl and peroxyl radicals and singlet oxygen, which are involved in oxidative stress (1–4). All these species are formed in cellular systems on exposure to the UV radiation of sunlight and during the oxygen-based metabolism. In the latter case, lipid peroxidation is an effective source of peroxyl radicals; alternatively, these oxidants are formed by O2 trapping of carbon-centered radicals produced in cellular H-abstraction processes (5).
As for the oxidizing power of the peroxyl radicals, they are of lower reactivity than alkoxyl and hydroxyl radicals and, thus, more selective oxidants (6). In the case of the DNA damage by peroxyl radicals, several methods have been employed for these reactive oxygen species. For example, the enzymatic oxidation of hydroperoxides by peroxidases constitutes an effective method of oxidizing the guanine in DNA by peroxyl radicals (7,8). A commonly used peroxyl-radical source is the thermal decomposition of azoalkanes in the presence of molecular oxygen. For such studies, 2,2′-azobis(2-methylpropanimidamide)hydrochloride (AAPH) has been employed (9–14), but recently it has been questioned whether AAPH serves as a reliable source of peroxyl radical (15–17).
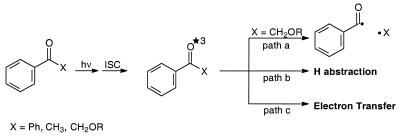
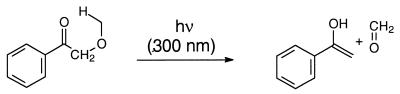
A third method of generating peroxyl radicals concerns the photolysis of ketones capable of undergoing α cleavage (Norrish-type I photoreaction), in which the released carbon-centered radicals are trapped by molecular oxygen. However, a potential problem of this photochemical method of peroxyl-radical generation in regard to the oxidation of DNA is the difficulty of distinguishing between alternative oxidative mechanisms by the intermediary excited ketone (Scheme 1). Besides α cleavage (path a), the excited state may interact with DNA directly by H abstraction (path b) or by electron transfer (path c) (18–21), as evidenced by the oxidation of guanine (22–25), the most vulnerable base towards oxidative damage (26).
Despite the shortcomings of the photochemical method, the incentive for the present study was the utilization of ketones prone to undergo α cleavage on photolysis for the generation of peroxyl radicals through trapping by molecular oxygen. The efficacy of DNA damage by the peroxyl radicals generated by this photochemical route was to be assessed in terms of guanine oxidation and strand-break formation. Previous work for this purpose has employed mainly substituted acetone derivatives (24,27–31). Several disadvantages of such acetone derivatives for photocleavage are evident. The low extinction coefficient of the acetone chromophore obliges the use of high ketone concentrations in the DNA oxidation experiments. Thus, quenching of the excited ketone species by the ground state ketone (32–34) should compete more effectively with the desired α cleavage of the ketone (Scheme 1, path a) to afford the required carbon-centered radicals for the generation of peroxyl radicals by trapping with molecular oxygen. Further, the excited states of acetone derivatives possess unfavorable electronic absorption characteristics, which encumber the direct determination of the α-cleavage kinetics by time-resolved laser-flash spectroscopy (30).
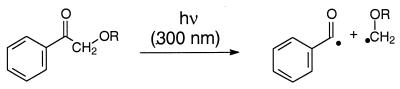
In view of these photophysical disadvantages of acetone derivatives, for this study we chose the benzoyl chromophore as shown in the hydroxy (AP-OH), methoxy (AP-OMe) tert-butoxy (AP-OtBu) and acetoxy (AP-OAc) derivatives of acetophenone (AP). Except for AP itself, which does not α-cleave (35), the other derivatives are expected to generate the required carbon-centered radicals through α cleavage to afford the desired peroxyl radicals by 3O2 trapping. For comparison, to assess the non-radical DNA oxidation directly by the triplet-excited ketones, it was necessary to examine the photolytically non-cleavable AP and benzophenone (BP) as typical type I sensitizers (27). The advantage of the acetophenone derivatives is their higher extinction coefficient, which permits short irradiation times and low ketone concentrations to minimize undesirable side reactions. Moreover, such phenyl-substituted ketone triplets possess appropriate absorption characteristics (high ɛ, near UV) to assess directly the kinetics of interaction with DNA by means of time-resolved laser-flash spectroscopy. As photobiological monitors of the DNA oxidative damage by peroxyl radicals, guanine oxidation in terms of the 8-oxo-7,8-dihydroguanine (8-oxoGua) and the guanidine-releasing products (GRP; in particular oxazolone, guanidinohydantoin and oxaluric acid) and strand-break formation were to be employed. To confirm that radicals are responsible for the DNA damage, 5,5-dimethyl-1-pyrroline N-oxide (DMPO) was to be employed as radical scavenger. Detection of the expected carbon-centered radicals in the α cleavage of the triplet ketone was, thus, to be achieved through spin trapping by DMPO, coupled with EPR spectroscopy.
MATERIALS AND METHODS
Synthesis of 2-tert-butoxyacetophenone
To a solution of 1.00 g (7.34 mmol) 2-hydroxyacetophenone and 1.01 g (7.34 mmol) tert-butyl bromide in 10 ml dry ethyl ether, 850 mg (3.67 mmol) silver oxide was added and stirred for 4 days at 20°C. The solids were removed by filtration and the organic phase washed with 10 ml distilled water, dried over magnesium sulfate and the solvent evaporated (40°C, 10 mbar). The crude product was purified by flash-column chromatography [silica gel, with 2:1 petroleum ether (30–50) : ethyl ether as eluent] to afford 643 mg (46%) of a colorless oil. 1H-NMR (CDCl3, 250 MHz) δ 1.21 (s, 9 H), 4.59 (s, 2 H), 7.3–7.5 (m, 3 H), 7.9 (m, 2 H); 13C-NMR (CDCl3, 63 MHz) δ 27.8, 66.6 (CH2), 74.9, 128.5, 128.6, 133.6, 135.7, 197.5.
Synthesis of 2-acetoxyacetophenone
To a solution of 1.27 g (1.16 ml, 16.2 mmol) acetyl chloride in 10 ml dichloromethane at 0°C, 1.28 g (1.31 ml, 16.2 mmol) pyridine and 2.00 g (14.7 mmol) AP-OH were added and the mixture was stirred for 10 min. The solution was washed once with 10 ml saturated sodium bicarbonate solution and twice with 10 ml of distilled water, dried over magnesium sulfate and the solvent evaporated (40°C, 20 mbar). The crude product was purified by flash-column chromatography [silica gel, with petroleum ether (30–50) : ethyl ether (gradient 10:1 to 5:1) as the eluent] to afford 2.22 g (12.4 mmol, 77%) colorless needles. 1H-NMR (CDCl3, 250 MHz) δ 2.21 (s, 3 H), 5.32 (s, 2 H), 7.4–7.7 (m, 3 H), 7.9 (m, 2 H); 13C-NMR (CDCl3, 63 MHz) δ 21.0, 66.4 (CH2), 128.2, 129.3, 134.3, 134.6, 170.8, 192.6.
Other materials
8-oxoGua and hydrofluoric acid (HF)/pyridine (70% HF), the sodium salt of 1,2-naphthoquinone-4-sulfonic acid (NQS), tris(hydroxylmethyl)aminomethane (trisbase), AP-OH, AP-OMe and bromophenol blue ‘gel loading solution’ were acquired from Sigma-Aldrich (Deisenhofen, Germany). Guanine, guanidine hydrochloride, sodium acetate solution (3 M in water), AP, BP, ammonium formate (97%), DMPO and tert-butyl bromide were obtained from Fluka (Buchs, Switzerland). The calf thymus DNA, ethidium bromide, boric acid and disodium hydrogen phosphate were purchased from Merck (Darmstadt, Germany). Methanol and acetonitrile (HPLC grade) were acquired from Fisher Scientific (Loughborough, UK). Citric acid (analytical grade) was purchased from Riedel de Haën (Seelze, Germany), pyridine from Grüssing (Filcum, Germany) and peracetic acid (40% in acetic acid) from Kesla Pharma (Wolfen, Germany). pBR 322 DNA was obtained from Amersham Pharmacia Biotech (Piscataway, NJ). All aqueous solutions were prepared with purified water (Milli-Q; Millipore, Bedford, MA). Agarose was supplied by Serva Feinbiochemica (Heidelberg, Germany).
Instrumentation
Apparatus. 1H-NMR spectra were measured on a Bruker AC 250 (250 MHz) and 13C-NMR spectra on a Bruker AC 250 (63 MHz), both with chloroform-d as the internal standard. Column chromatography was conducted on silica gel (32–64 µm) from Woelm (Erlangen, Germany) as stationary phase with a 100:1 adsorbent/substrate ratio.
The electron paramagnetic resonance (EPR) experiments were carried out at room temperature (∼20°C) on a Bruker EPR 300 spectrometer, operated at 9.76 GHz with 100 kHz modulation frequency by using a flat quartz cell (10 × 1 mm). The EPR spectrometer settings were 20 mW microwave power, 0.52 G modulation amplitude, 1.28 ms time constant, 100 G/41.9 s scan rate and 2 × 104 gain.
The irradiation of the ketones was carried out in glass vessels by employing a Rayonet photoreactor (RPR-100, Southern New England Ultraviolet Company, Branfort, USA), equipped with sixteen 300-nm lamps (RPR 3000, 24 W each). All samples were degassed before irradiation (three ‘freeze–pump–thaw’ cycles). Time-resolved laser-flash experiments were performed in a 4 × 1 × 1 cm glass cell by irradiation with a 308-nm Lambda Physik EMG 101 MSC XeCl Laser (36).
Detection of DNA base oxidation
Oxidation of calf thymus DNA (CT DNA). The reaction mixture (400 µl) contained 0.100 g/l CT DNA (corresponds to 62.5 µM guanine) and (if no other value is stated) 1.00 mM ketone, in a 9:1 mixture of phosphate buffer (5.00 mM, pH 7.0) and acetonitrile. After photolysis under air at 300 nm and 0°C for 45 min, a 100- and a 200-µl aliquot were used for the analysis of 8-oxoGua and GRP. The unreacted ketone was extracted with ethyl acetate (2 × 200 and 2 × 400 µl) and the DNA was precipitated in the cold (–50°C) upon addition of sodium acetate (300 mM final concentration, pH 5.0) and addition of 3 vol ethanol (–50°C).
Acidic hydrolysis of DNA. The precipitated DNA was centrifuged for 3 min at 15 000 r.p.m., and after removal of the ethanol solution the resulting pellet was dried under vacuum (20°C, 0.01 torr). For the analysis of the GRP, the pellet was dissolved in 100 µl water and the solution was stored at ambient temperature for at least 24 h, followed by the procedure for the analysis of GRP described subsequently. For the 8-oxoGua analysis, treatment of the pellet with 13.0 µl HF/pyridine (70% HF) for 30 min at 37°C afforded a brown solution, which was neutralized by addition of 15.0 mg calcium carbonate, suspended in 200 µl water, and stirred vigorously for 30 min. After centrifugation (15 min at 15 000 r.p.m.) and washing of the residue with 200 µl water, the combined aqueous solutions were lyophilized (20°C, 0.01 torr) and dissolved in 100 µl water prior to the quantitative determination of 8-oxoGua by HPLC/EC analysis and guanine by high performance liquid chromatography/UV analysis.
HPLC analysis
The HPLC analytical system consisted of Bischoff model 2200 analytical pumps (Bischoff GmbH, Leonberg, Germany) equipped with a Rheodyne model 7125 loop injector (Rheodyne, Berkeley, CA) and a SpectraFlow 600 photodiode array detector (SunChrom, Friedrichsdorf, Germany). The latter was connected in series with an ESA Coulochem model 5100A electrochemical detector, supplied with a model 5011 high sensitivity analytical cell. Alternatively, a Shimadzu model RF-551 spectrofluorometric detector (Shimadzu, Kyoto, Japan) was employed for the quantification of the GRP.
The separation of 8-oxoGua was achieved on a 250 × 4.6 mm (inner diameter) Eurospher 100-C18 7-µm column (Knauer, Berlin, Germany) by using a mixture of 50.0 mM sodium citrate buffer (pH 5.0) and methanol (90:10, v/v; 1.00 ml/min) as eluent. For the detection of 8-oxoGua, the oxidation potential of the electrode was set at Eox = + 350 mV (37). The DNA bases were monitored at 254 nm by UV spectroscopy. The GRP were detected by an indirect fluorescence labeling assay, after release of the guanidine on alkaline treatment and its condensation with NQS. A 200-µl sample of the crude reaction mixture was washed twice with 400 µl ethyl acetate, freeze dried (20°C, 0.01 torr), and redissolved in 100 µl water. After addition of 38.0 µl aqueous NaOH (1.00 M) and 20.0 µl of an aqueous solution of NQS (5.00 mg/ml), the mixture was kept at 65°C for 10 min in the dark. The resulting solution was acidified by addition of 42.0 µl HCl [1.00 normal (N)] and diluted with 400 µl water. The products were separated on a 250 × 4.6 mm (i.d.) Eurospher 100-C18 5-µm column (Knauer GmbH, Berlin, Germany) with a mixture of 25.0 mM ammonium formate and methanol (75:25, v/v; 1.00 ml/min) as eluent and detected spectrofluorometrically (λex = 355 nm, λem = 405 nm) (38–40). The injection of the samples for the quantification of GRP was performed by an autosampler (Spark Basic-Marathon-Plus, Emmen, The Netherlands). All HPLC eluents were filtered under suction through a 0.45 µM Sartorius cellulose filter before use for the removal of small particles and degassing purposes.
Induction and detection of single strand breaks (SSB) in pBR 322 DNA (41)
Treatment. The samples were prepared in a final volume of 10.0 µl, placed in Eppendorf tubes and photolyzed at 300 nm and 0°C for 20 min. The concentration of supercoiled pBR 322 DNA was 10.0 mg/l in a 9:1 mixture of phosphate buffer (50.0 mM, pH 7.0) and acetonitrile. The concentration of the ketone was 10.0 µM.
Agarose-gel electrophoresis. After photolysis, 2.50 µl of a buffer solution (Sigma), which contained 0.05% (w/v) bromophenol blue, 40% (w/v) sucrose, 0.5% (w/v) sodium lauryl sulfate and 0.100 M EDTA, were added to the above samples and a 10.0-µl aliquot of each was loaded onto a 1% agarose gel, which contained 0.500 mg/l ethidium bromide. The gel electrophoresis was carried out in Tris buffer (pH 8.0; 18.0 mM trisbase, 18.0 mM boric acid and 10.0 mM EDTA) by running the gels on a Pharmacia horizontal apparatus (GNA 100), with the power supply set at 78 V for 2 h at room temperature (∼20°C). The DNA spots were detected by ethidium bromide excitation (fluorescence) with a UV transilluminator (366 nm) and recorded by photography with a Herolab camera E.A.S.Y. 429K, which was connected to a personal computer equipped with a Herolab E.A.S.Y. software program. The ratio of open-circular (OC) DNA relative to the total amount of DNA was determined from the light intensities of the spots. The greater binding constant of ethidium bromide with OC DNA rather than super-coiled (SC) DNA was corrected by applying a factor of 1.22 (42). The extent of strand-break formation was corrected for the blank value, which was set to zero.
RESULTS
Laser-flash photolysis
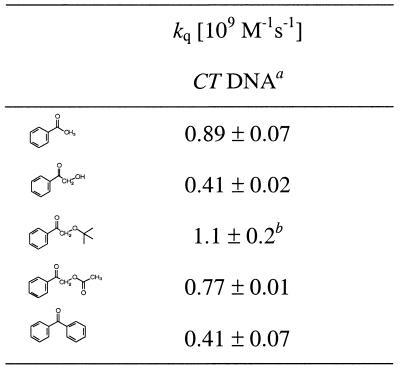
The quenching rate data of the triplet-ketone transients by CT DNA were determined through time-resolved absorption spectroscopy (excitation at 308 nm with a Lambda Physik EMG 101 MSC XeCl Excimer Laser). The triplet–triplet absorption of AP and its derivatives AP-R (Fig. 1) was detected at 340 nm. The BP triplet was measured at 600 nm because of interfering overlap of its absorption maximum (525 nm) (43) with the absorption of its ketyl radical. The BP ketyl radical was observed as a weak long-lived transient, whose intensity increased with the DNA concentration. For the determination of the quenching rate constants of the triplet ketones by DNA, the time dependence of the absorption decay curve at 340 nm was monitored at various DNA concentrations. For each kinetic run, the rate was calculated from a monoexponential fit of the decay curves. Second-order kinetics was applied for the ketyl radicals formed during the quenching process. The rate constants of the monoexponential fits were plotted against the DNA concentration (kobs = k0 + kq[DNA]) and the rate constant for the DNA quenching was obtained from the slope of such a plot (Table 1). For AP and AP-OAc, values of 0.89 × 109 and 0.77 × 109 M–1s–1 have been determined; for AP-OH and BP quenching, constants of ∼0.4 × 109 M–1s–1 were obtained. The quenching rate constants were calculated from the apparent guanine concentration in the CT DNA.
Figure 1.
Structures.
Table 1. Quenching rate constants for CT DNA of the triplet states formed during the ketone photolysis.
aThe concentration of guanine (0.100 mg/ml CT DNA = 62.5 µM Gua) has been used to calculate the rate constants. The mean values and their standard errors have been calculated from two experiments.
bOnly one run, error estimated ±20% due to short triplet lifetime.
DNA-base oxidation products
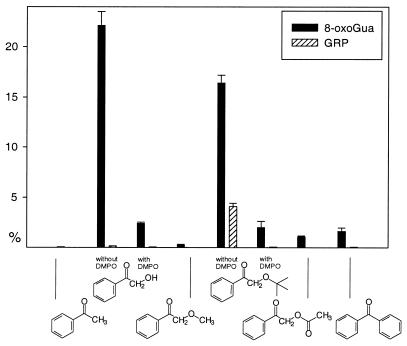
The guanine oxidation products 8-oxoGua and GRP were determined quantitatively in the ketone-mediated photooxidation of calf thymus DNA by standard HPLC methods (Fig. 2) (44). The ketones AP, AP-OMe, AP-OAc and BP only led to minor amounts (<2%) of 8-oxoGua, whereas the photolysis of AP-OH and AP-OtBu afforded large quantities (23 and 17%, respectively) of 8-oxoGua. Significant amounts (4%) of GRP were only detected in the AP-OtBu photolysis.
Figure 2.
DNA photooxidation [(Gua) = 62.5 µM, 300 nm, 20 min, 0°C] during the ketone (200 µM) photolysis in 5.00 mM phosphate buffer (pH 7.0) : CH3CN (9:1) with and without DMPO (500 µM). The mean values and their standard erors have been calculated from two experiments.
Time profile and relative efficiency of the DNA photooxidation by ketones
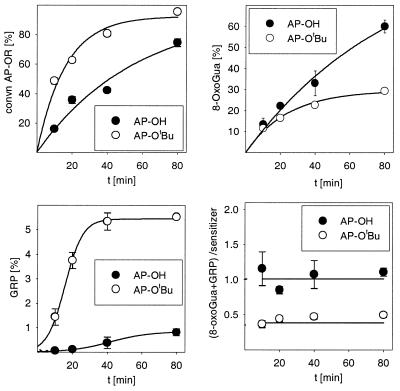
The time profiles were recorded for the DNA photooxidation by the oxyfunctionalized ketones AP-OH and AP-OtBu (Fig. 3). For this purpose, the ketone conversion (determined by HPLC) and the formation of 8-oxoGua and of GRP were monitored with time. The time profiles for the ketone conversion (Fig. 3, top left) reveal that the AP-OtBu is photolyzed approximately two times more rapidly than AP-OH and is consumed >80% after 1 h of irradiation. This photochemical behavior parallels that of the ketones in the absence of DNA (data not shown); moreover, the time profiles for the formation of 8-oxoGua (Fig. 3, top right) parallel the decomposition of the sensitizer, however, the more photostable AP-OH induces more 8-oxoGua than AP-OtBu. GRP (Fig. 3, bottom left) is not formed in the beginning, the time profile shows sigmoidal behavior. While AP-OH affords only traces of GRP, AP-OtBu is more efficient (6%). The efficiency of the DNA photooxidation is displayed in the time plot (Fig. 3, bottom right) for the ratio of the amount of oxidation products (8-oxoGua + GRP) per ketone sensitizer (AP-OH or AP-OtBu) consumed.
Figure 3.
Time profile for the DNA oxidation during the photolysis of the ketones AP-OH and AP-OtBu ([Gua] = 62.5 µM, 300 nm, 20 min, 0°C, (ketone) = 200 µM) in 5.00 mM phosphate buffer (pH 7.0) : CH3CN (9:1). The mean values and their standard errors have been calculated from two experiments.
Formation of single strand breaks
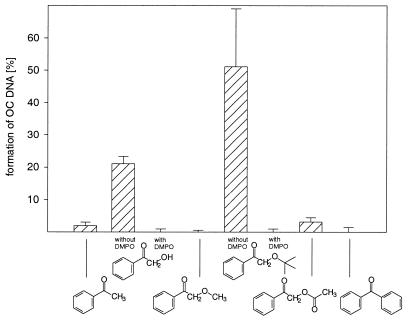
The photolysis of AP-OH and AP-OtBu induces SSBs in supercoiled pBR 322 DNA, as manifested by gel-electrophoretic measurements (Fig. 4). The ketones AP, AP-OMe, AP-OAc and BP were ineffective in causing strand breaks (<5%) under the applied conditions, whereas AP-OH (∼25%) and AP-OtBu (∼50%) were quite efficient. Addition of DMPO quenched completely the SSB formation by AP-OH and AP-OtBu.
Figure 4.
Formation of the OC form of pBR 322 DNA (10 mg/l) during the ketone photolysis (10 µM, 300 nm, 0°C, 20 min) in 0.500 mM phosphate buffer (pH 7.4) : CH3CN (9:1) with and without DMPO (5.00 µM). The mean values and their standard errors have been calculated from two experiments.
Base oxidation (guanine) versus hydrogen abstraction (sugar backbone)
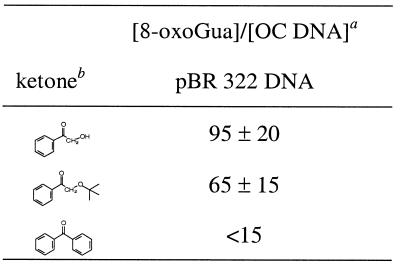
To determine the ratio of base versus sugar damage, conditions were chosen that, for AP-OH, AP-OtBu and BP, led to ∼50% SSB in pBR 322 DNA; this is equivalent to 0.023% SSB per guanine base. For AP-OH and AP-OtBu, these experiments were performed with 0.010 mg/ml DNA, whereas 0.100 mg/ml DNA was employed for BP. The extent of SSB formation was assessed by gel electrophoresis directly after photolysis, the amount of 8-oxoGua was determined by HPLC/EC after hydrolysis (HF/pyridine, 37°C, 40 min) of the oxidized pBR 322 DNA (see Table 2 for ratios). For AP-OH and AP-OtBu the ratios are 85 ± 15 and 60 ± 15, respectively, and within the experimental error are approximately the same, which indicates that base oxidation dominates strand cleavage; a value <15 was estimated for BP. Although in the latter case a considerable number of strand breaks have been induced, the amount of 8-oxoGua for BP falls within the error limit of detection and, thus, only an upper limit could be assessed for the ratio. The same situation also applies for the ketones AP and AP-OAc, for which no 8-oxoGua formation could be detected in the error limit, although the strand-break induction was appreciable (8 and 18%).
Table 2. 8-oxoGua : OC DNA ratio in the ketone photooxidation of pBR 322 DNA.
aApproximately 50% SSB, which is equivalent to 0.023% SSB per guanine base. The mean values and their standard errors have been calculated from two experiments.
bFor AP-OH and AP-OtBu, 0.010 mg/ml DNA; for BP, 0.100 mg/ml DNA.
DISCUSSION
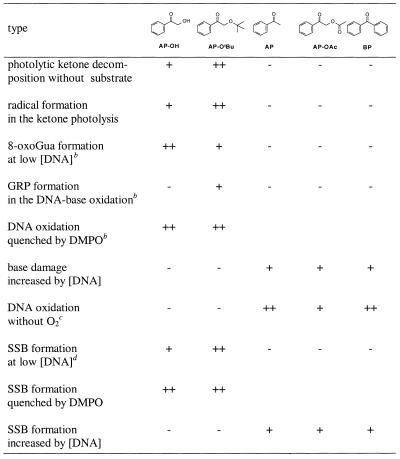
In order to rationalize the present DNA oxidation mechanistically, the pertinent qualitative trends in the data are summarized in Table 3, together with the essential experimental facts on the photochemical behavior of the ketones (first two entries). The data for AP-OMe are not shown because of the fast deactivation of its triplet state by Norrish-type II cleavage (Scheme 2) and, thus, this acetophenone derivative exhibits a low oxidative reactivity toward DNA. The other five ketones may be classified into two groups as to their efficacy of photooxidative damage of DNA on the basis of their ability of producing radicals on photolysis: the first group consists of AP-OH and AP-OtBu, from which radicals are efficiently released to induce base oxidation and SSB; the second group consists of AP, AP-OAc and BP, which do not produce radicals and show only a low oxidative reactivity toward DNA at the employed reaction conditions. However, at DNA concentrations higher than used herein, increased oxidation by the second group of excited ketones is observed, for both guanine oxidation and DNA cleavage (data not shown).
Table 3. Qualitative reactivity trendsa in the photochemical and photobiological activity of triplet-excited ketones.
aQualitative order: high (++), appreciable (+), none (-).
bSee Figure 2.
cAs a monitor for DNA oxidation, 8-oxoGua has been used.
dSee Figure 4.
The rate constants kq (Table 1) for the quenching of triplet ketones by the guanine base in the DNA are relatively high. However, direct DNA damage (Scheme 1, paths b and c) through excited-state quenching cannot account for the observed reactivity because the experiments were performed under air, conditions at which the triplet lifetimes are short {the lifetimes of the triplet ketones in the presence of air [1.00 mM ketone, 0.300 mM O2, in H2O:CH3CN (9:1)] are 710 µs for AP, 410 µs for AP-OH, 390 µs for AP-OtBu, 910 µs for AP-OAc and 500 µs for BP}, and because the concentration of DNA was low (0.100 mg/ml = 62.5 µM Gua). This is the reason for the low amount of 8-oxoGua and GRP formed in the photooxidation of CT DNA by the non-radical-generating ketones AP, AP-OAc and BP (Fig. 2). In contrast, much more of these DNA oxidation products is found in the photolysis of AP-OH and AP-OtBu. Both compounds are known to undergo efficient Norrish-type I cleavage and, thus, readily form radicals upon excitation. This has been shown by spin-trapping experiments with DMPO [with 50.0 µM DMPO in H2O:CH3CN (9:1) the DMPO adducts of the benzoyl radical (g = 2.0056, αN = 15.3 G, αH = 17.9 G) and the hydroxymethyl radical (g = 2.0055, αN = 16.0, αH = 22.6 G) were formed upon photolysis (300 nm) of AP-OH (2.00 mM), and the DMPO adducts of the benzoyl and the tert-butoxymethyl radical (g = 2.0055, αN = 16.0 G, αH = 21.1 G) were formed upon photolysis (300 nm) of AP-OtBu (2.00 mM)] and is also manifested in the shorter triplet lifetimes compared to AP, AP-OAc and BP {The triplet lifetimes in the absence of molecular oxygen [1.00 mM ketone solution in H2O:CH3CN (9:1)] are 6.6 µs for AP, 1.1 µs for AP-OH, 0.54 µs for AP-OtBu, 6.7 µs for AP-OAc and 9.2 µs for BP}. Consequently, upon α cleavage of the triplet-excited AP-OH and AP-OtBu, the benzoyl radical and an oxysubstituted methyl radical are formed (Scheme 3). Since the carbon-centered radicals react with molecular oxygen at nearly diffusion-controlled rates (5,6), both PhCO· and ROCH2· will form peroxyl radicals efficiently with O2, which are expected to oxidize the DNA. Indeed, a significantly higher quantity of oxidation products has been observed under aerobic conditions for the radical-generating ketones AP-OH and AP-OtBu than for the photo-resistent AP, AP-OAc and BP (Fig. 2). Thus, the peroxyl radicals formed upon photolysis are held responsible for the efficient oxidation of CT DNA. That radicals are involved in the present DNA photooxidation has been confirmed by the addition of DMPO as spin trap, which inhibited the formation of the oxidation products almost completely (Fig. 2).
In the photooxidation of DNA by AP-OtBu, the formation of minor amounts of GRP have been detected; however, the GRP is not formed right from the beginning, as displayed by the sigmoidal curve for the concentration profile (Fig. 3, bottom left), with most of the GRP observed after ∼30 min. This experimental fact implies that the GRP is not a primary photoproduct, but is formed by the subsequent oxidation of another photoproduct, presumably 8-oxoGua. For AP-OH, a similar behavior is noted in Figure 3 (bottom left), although the level of GRP generation is by an order of magnitude less.
Since both AP-OH and AP-OtBu release benzoyl radicals on photolysis, the difference in the photooxidative behavior toward the guanine base in DNA of these two oxyfunctionalized ketones has to be due to the other radical fragment, namely the tert-butoxymethyl peroxyl versus the hydroxymethyl peroxyl radical. In view of the fact that AP-OtBu is more effective in producing GRP than AP-OH (Fig. 2), of the two peroxyl species, evidently, the tert-butoxymethyl peroxyl radical is the more effective in oxidizing the 8-oxoGua. Control experiments confirm that AP-OH is quite inefficient in the photooxidation of 8-oxodG compared to AP-OtBu (data not shown).
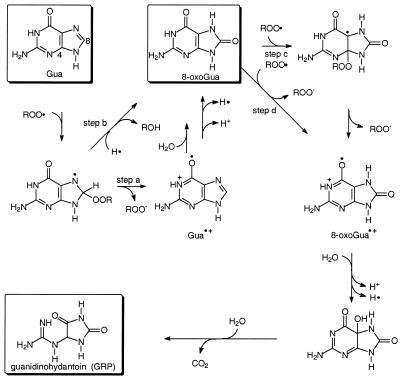
The mechanism for the formation of 8-oxoGua and GRP in the DNA photooxidation of the AP-OR (R = H, tBu) photolysis is proposed in Scheme 4. The formation of 8-oxoGua as the major product may proceed through the guanine radical cation (Gua·+) as an intermediate, which is known to produce 8-oxoGua by addition of water (45). Thus, addition of the peroxyl radical at the C-8 position of guanine in the DNA, followed by elimination of the peroxy anion or electron transfer to the peroxyl radical (step a), is proposed as the favored pathway. However, we cannot rule out the direct formation of 8-oxoGua from the peroxyl radical adduct to Gua (step b). The formation of the substantial quantities of GRP in the photooxidation by AP-OtBu (Fig. 2, bottom left) is explained by further one-electron oxidation of the more easily oxidized 8-oxoGua in the DNA to guanidinohydantoin by the peroxyl radical, as described for other one-electron oxidants in the literature (46). Alternatively, addition of the peroxyl radical to the C-4 position of 8-oxoGua (step c) and subsequent elimination of the peroxy anion or electron transfer (step d) would generate the radical cation 8-oxoGua·+. Upon alkaline hydrolysis, the guanidine is released from the GRP precursor and detected.
Although AP-OtBu is consumed approximately two times faster than AP-OH (Fig. 3, top left), the latter produces more 8-oxoGua (Fig. 3, top right). This phenomenon is only observed in DNA, since the oxidation of 2′-deoxyguanosine (dG) by AP-OtBu is more efficient than by AP-OH [for the photooxidation (300 nm, 45 min) of dG (200 µM) by AP-OtBu (400 µM) in H2O:CH3CN (9:1) a conversion of 52 ± 3% was observed, whereas under the same experimental conditions only 41 ± 1% was detected for AP-OH]. To account for the higher oxidative power of AP-OH towards DNA compared with AP-OtBu, we speculate that hydrogen bonding between the hydroxyl group of AP-OH with the sugar backbone of the DNA increases the effective AP-OH concentration near the DNA.
Strand-break experiments performed with supercoiled pBR 322 DNA showed a similar behavior as observed for the base oxidation. No significant number of strand breaks was found with the photo-resistent AP, AP-OAc and BP, whereas the radical-generating ketones AP-OH and AP-OtBu induced substantial strand breaks (Fig. 4). Since the SSB formation was efficiently inhibited by the addition of DMPO, radical species, presumably peroxyl radicals, are proposed as the oxidizing species. AP-OtBu is more efficient than AP-OH, which may be explained in terms of the faster decomposition of the former (Fig. 3, top left).
In summary, from the data in Table 2 we conclude that the major photooxidative damage in DNA is the base damage caused by the radical-releasing ketones, whereas the formation of SSB only plays a minor role. To rationalize this mechanistically, we propose that the intermediary peroxyl radicals, derived from O2 trapping of the carbon-centered radicals generated in the ketone photocleavage, react with the guanine base in the DNA to afford base oxidation products. Induction of strand breaks by the relatively sluggish peroxyl radicals through H abstraction from the sugar backbone is of subordinate efficacy, but damage transfer between the base and the sugar backbone may additionally operate (47,48).
Scheme 1. Photochemical pathways of triplet-excited ketones.
Scheme 2. Norrish-type II reaction in the photolysis of AP-OMe.
Scheme 3. Radical formation in the photolysis of the AP-OR ketones by α cleavage.
Scheme 4. Proposed mechanism for the photooxidation of guanine (Gua) in DNA by peroxyl radicals generated in the photolysis of AP-OH and AP-OtBu.
Acknowledgments
ACKNOWLEDGEMENTS
Generous financial support by the Deutsche Forschungsgemeinschaft (SFB 172 ‘Molekulare Mechanismen kanzerogener Primärveränderungen’) and by the Fonds der Chemischen Industrie is gratefully appreciated.
REFERENCES
- 1.Ames B.N. (1983) Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science, 221, 1256–1264. [DOI] [PubMed] [Google Scholar]
- 2. von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor & Francis, London, UK.
- 3.Meneghini R. (1988) Genotoxicity of active oxygen species in mammalian cells. Mutat. Res., 195, 215–230. [PubMed] [Google Scholar]
- 4.Cadenas E. (1989) Biochemistry of oxygen toxicity. Annu. Rev. Biochem., 58, 79–110. [DOI] [PubMed] [Google Scholar]
- 5.Marchaj A., Kelley,D.G., Bakac,A. and Espenson,H. (1991) Kinetics of the reactions between alkyl radicals and molecular oxygen in aqueous solution. J. Phys. Chem., 95, 4440–4441. [Google Scholar]
- 6.Ingold K.U. (1969) Peroxy radicals. Acc. Chem. Res., 2, 1–9. [Google Scholar]
- 7.Adam W., Kurz,A. and Saha-Möller,C.R. (2000) Peroxidase-catalyzed oxidative damage of DNA and 2′-deoxyguanosine by model compounds of lipid hydroperoxides: involvement of peroxyl radicals. Chem. Res. Toxicol., 13, 1199–1207. [DOI] [PubMed] [Google Scholar]
- 8.Adam W., Kurz,A. and Saha-Möller,C.R. (1999) DNA and 2′-deoxyguanosine damage in the horseradish-peroxidase-catalyzed autoxidation of aldehydes: the search for the oxidizing species. Free Radic. Biol. Med., 26, 566–579. [DOI] [PubMed] [Google Scholar]
- 9.Martini M. and Termini,J. (1997) Peroxy radical oxidation of thymidine. Chem. Res. Toxicol., 10, 234–241. [DOI] [PubMed] [Google Scholar]
- 10.Valentine M.R., Rodriguez,H. and Termini,J. (1998) Mutagenesis by peroxy radical is dominated by transversions at deoxyguanosine: evidence for the lack of involvement of 8-oxo-dG and/or abasic site formation. Biochemistry, 37, 7030–7038. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez H., Valentine,M.R., Holmquist,G.P., Akman,S.A. and Termini,J. (1999) Mapping of peroxyl radical induced damage on genomic DNA. Biochemistry, 38, 16578–16588. [DOI] [PubMed] [Google Scholar]
- 12.Simandan T., Sun,J. and Dix,D.A. (1998) Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochem. J., 335, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harkin L.A. and Burcham,P.C. (1997) Formation of novel C1-oxidised abasic sites in alkylperoxyl radical-damaged plasmid DNA. Biochem. Biophys. Res. Commun., 237, 1–5. [DOI] [PubMed] [Google Scholar]
- 14.Burcham P.C. and Harkin,L.A. (1999) Mutations at G:C Base pairs predominate after replication of peroxyl radical-damaged pSP189 plasmids in human cells. Mutagenesis, 14, 135–140. [DOI] [PubMed] [Google Scholar]
- 15.Wahl R.U.R., Zeng,L., Madison,S.A., DePinto,R.L. and Shay,B.J. (1998) Mechanistic studies on the decomposition of water-soluble azo-radical initiators. J. Chem. Soc. [Perkin], 2, 2009–2017. [Google Scholar]
- 16.Krainev A.G. and Bigelow,D.J. (1996) Comparison of 2,2′-azobis (2-amidinopropane) hydrochloride (AAPH) and 2,2′-azobis (2,4-dimethylvaleronitrile) (AMVN) as free radical initiators: a spin-trapping study. J. Chem. Soc. [Perkin], 2, 747–754. [Google Scholar]
- 17.Paul T., Young,M.J., Hill,I.E. and Ingold,K.U. (2000) Strand cleavage of supercoiled DNA by water-soluble peroxyl radicals. the overlooked importance of peroxyl radical charge. Biochemistry, 39, 4129–4135. [DOI] [PubMed] [Google Scholar]
- 18.Lamola A.A. (1970) Triplet photosensitization and the photobiology of thymine dimers in DNA. Pure Appl. Chem., 24, 599–610. [DOI] [PubMed] [Google Scholar]
- 19.Rahn R.O. (1983) Sensitized photoinduction of pyrimidine dimers in DNA. In Friedberg,E.C. and Hanawalt,P.R. (eds), DNA Repair, a Laboratory Manual of Research Procedures. Marcel Dekker, New York, NY, Vol. 2, pp. 75–85.
- 20.Patrick M.H. and Snow,J.M. (1977) Studies on thymine-derived UV photoproducts in DNA - II. A comparative analysis of damage caused by 254 nm irradiation and triplet-state photosensitization. Photochem. Photobiol., 25, 373–384. [DOI] [PubMed] [Google Scholar]
- 21.Lamola A.A., Guéron,M., Yamane,T., Eisinger,J. and Shulman,R.G. (1967) Triplet state of DNA. J. Chem. Phys., 47, 2210–2217. [DOI] [PubMed] [Google Scholar]
- 22.Gut I.G., Wood,P.D. and Redmond,R.W. (1996) Interaction of triplet photosensitizers with nucleosides and DNA in aqueous solution at room temperature. J. Am. Chem. Soc., 118, 2366–2373. [Google Scholar]
- 23.Umlas M.E., Franklin,W.A., Chan,G.L. and Haseltine,W.A. (1985) Ultraviolet light irradiation of defined-sequence DNA under conditions of chemical photosensitization. Photochem. Photobiol., 42, 265–273. [DOI] [PubMed] [Google Scholar]
- 24.Epe B., Henzl,H., Adam,W. and Saha-Möller,C.R. (1993) Endonuclease-sensitive DNA modifications induced by acetone and acetophenone as photosensitizers. Nucleic Acids Res., 21, 863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douki T. and Cadet,J. (1999) Modification of DNA bases by photosensitized one-electron oxidation. Int. J. Radiat. Biol., 75, 571–581. [DOI] [PubMed] [Google Scholar]
- 26.Adam W., Andler,S., Nau,W.M. and Saha-Möller,C.R. (1998) Oxidative DNA damage by radicals generated in the thermolysis of hydroxymethyl-substituted 1,2-dioxetanes through the α cleavage of chemiexcited ketones. J. Am. Chem. Soc., 120, 3549–3559. [Google Scholar]
- 27.Adam W., Saha-Möller,C.R., Schönberger,A., Berger,M. and Cadet,J. (1995) Formation of 7,8-dihydro-8-oxoguanine in the 1,2-dioxetane-induced oxidation of calf-thymus DNA: evidence for photosensitized DNA damage by thermally generated triplet ketones in the dark. Photochem. Photobiol., 62, 231–238. [DOI] [PubMed] [Google Scholar]
- 28.Adam W., Saha-Möller,C.R. and Schönberger,A. (1996) Photooxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine by thermally generated triplet-excited ketones from 3-(hydroxymethyl)-3,4,4-trimethyl-1,2-dioxetane and comparison with type I and type II photosensitizers. J. Am. Chem. Soc., 118, 9233–9238. [Google Scholar]
- 29.Adam W., Saha-Möller,C.R. and Schönberger,A. (1997) Type I and type II photosensitized oxidative modification of 2′-deoxyguanosine (dGuo) by triplet-excited ketones generated thermally from the 1,2-dioxetane HTMD. J. Am. Chem. Soc., 119, 719–723. [Google Scholar]
- 30.Andler S. (1997) Untersuchungen zur DNA-Schädigung durch Freie Radikale aus Lipidhydroperoxiden und Hydroxymethyl-substituierten 1,2-Dioxetanen. PhD Thesis, Universität Würzburg.
- 31.Adam W., Arnold,M.A. and Saha-Möller,C.R. (2001) Photooxidative damage of guanine in dG and DNA by the radicals derived from the α cleavage of the electronically excited carbonyl products generated in the thermolysis of alkoxymethyl-substituted dioxetanes and the photolysis of alkoxyacetones. J. Org. Chem., 66, 597–604. [DOI] [PubMed] [Google Scholar]
- 32.Schuster G. and Turro,N.J. (1975) Energy migration. Energy hopping and self-quenching reaction involving carbonyl chromophores. Tetrahedron Lett., 27, 2261–2264. [Google Scholar]
- 33.Barwise A.J.G., Gorman,A.A. and Rodgers,M.A.J. (1978) Evidence against reversible triplet energy transfer between acetophenone and norbornene. J. Photochem., 8, 11–16. [Google Scholar]
- 34.Schuster D.I. and Weil,T.M. (1973) Photochemistry of ketones in solution. Mechanism for photoreduction of benzophenone in benzene. Evidence for self-quenching of benzophenone triplets in solution and for hydrogen abstraction from benzophenone ground state. J. Am. Chem. Soc., 95, 4091–4092. [Google Scholar]
- 35.Xu Y. and Langford,C.H. (1997) A comparison of acetophenone photooxidation in aqueous media via direct photolysis and TiO2 photocatalysis. J. Adv. Oxid. Technol., 2, 408–414. [Google Scholar]
- 36.Zhang X., Erb,C., Flammer,J. and Nau,W.M. (2000) Absolute rate constants for the quenching of reactive excited states by melanin and related 5,6-dihydroxyindole metabolites: implications for their antioxidant activity. Photochem. Photobiol., 71, 524–533. [DOI] [PubMed] [Google Scholar]
- 37.Floyd R.A., West,M.S., Eneff,K.L., Schneider,J.E., Wong,P.K., Tingley,D.T. and Hogsett,W.E. (1990) Conditions influencing yield and analysis of 8-hydroxy-2′-deoxyguanosine in oxidatively damaged DNA. Anal. Biochem., 188, 155–158. [DOI] [PubMed] [Google Scholar]
- 38.Ravanat J.-L., Berger,M., Benard,F., Langlois,R., Ouellet,R., van Lier,J.E. and Cadet,J. (1992) Phthalocyanine and naphthalocyanine photosensitized oxidation of 2′-deoxyguanosine: distinct type I and type II products. Photochem. Photobiol., 55, 809–814. [Google Scholar]
- 39.Buchko G.W., Cadet,J., Ravanat,J.-L. and Labataille,P. (1993) Isolation and characterization of a new product produced by ionizing irradiation and type I photosensitization of 2′-deoxyguanosine in oxygen-saturated aqueous solution: (2s)-2,5′-anhydro-1-(2′-deoxy-β-d-erythro-pentofuranosyl)-5-guanidinylidene-2-hydroxy-4-oxoimidazolidine. Int. J. Radiat. Biol., 63, 669–676. [DOI] [PubMed] [Google Scholar]
- 40.Cadet J., Berger,M., Buchko,G.W., Joshi,P.C., Raoul,S. and Ravanat,J.-L. (1994) 2,2-Diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-D-erythro-pentofuranosyl)amino]-5-(2H)-oxazolone: a novel and predominant radical oxidation product of 3′,5′-Di-O-acetyl-2′-deoxyguanosine. J. Am. Chem. Soc., 116, 7403–7404. [Google Scholar]
- 41.Wang T.P., Kagan,J., Tuveson,R.W. and Wang,G.R. (1991) α-Terthienyl photosensitizes damage to pBR322 DNA. Photochem. Photobiol., 53, 463–467. [DOI] [PubMed] [Google Scholar]
- 42.Shubsda M.F., Goodisman,J. and Dabrowiak,J.C. (1997) Quantitation of ethidium-stained closed circular DNA in agarose gels. J. Biochem. Biophys. Methods, 34, 73–79. [DOI] [PubMed] [Google Scholar]
- 43.Shizuka H. and Kimura,E. (1984) Acid-base properties in the triplet state of aromatic ketones studied by nanosecond-laser-flash photolysis. Can. J. Chem., 62, 2041–2046. [Google Scholar]
- 44.Schönberger A. (1995) DNA-Photooxidation ohne Licht durch thermisch erzeugte triplettangeregte Ketone aus 1,2-Dioxetanen. PhD Thesis, Universität Würzburg.
- 45.Cullis P.M., Malone,M.E. and Merson-Davies,L.A. (1996) Guanine radical cations are precursors of 7,8-dihydro-8-oxo-2′-deoxyguanosine but are not precursors of immediate strand breaks in DNA. J. Am. Chem. Soc., 118, 2775–2781. [Google Scholar]
- 46.Hickerson R.P., Prat,F., Muller,J.G., Foote,C.S. and Burrows,C.J. (1999) Sequence and stacking dependence of 8-oxoguanine oxidation: comparison of one-electron vs. singlet oxygen mechanisms. J. Am. Chem. Soc., 121, 9423–9428. [Google Scholar]
- 47.Melvin T., Botchway,S.W., Parker,A.W. and O′Neill,P. (1996) Induction of strand breaks in single-stranded polyribonucleotides and DNA by photoionization: one electron oxidized nucleobase radicals as precursors. J. Am. Chem. Soc., 118, 10031–10036. [Google Scholar]
- 48.Meggers E., Dussy,A., Schäfer,T. and Giese,B. (2000) Electron transfer in DNA from guanine and 8-oxoguanine to a radical cation of the carbohydrate backbone. Chemistry, 6, 485–492. [DOI] [PubMed] [Google Scholar]