This study provides a roadmap for moving beyond discovery and development of therapeutic strategies based on single driver gene analysis to discovery based on interactions between multiple driver genes.
Abstract
The majority of cancers are driven by multiple genetic alterations, but how these changes collaborate during tumorigenesis remains largely unknown. To gain mechanistic insights into tumor-promoting genetic interactions among tumor suppressor genes (TSG), we conducted combinatorial CRISPR screening coupled with single-cell transcriptomic profiling in human mammary epithelial cells. As expected, different driver gene alterations in mammary epithelial cells influenced the repertoire of tumor suppressor alterations capable of inducing tumor formation. More surprisingly, TSG interaction networks were comprised of numerous cliques—sets of three or four genes such that each TSG within the clique showed oncogenic cooperation with all other genes in the clique. Genetic interaction profiling indicated that the predominant cooperating TSGs shared overlapping functions rather than distinct or complementary functions. Single-cell transcriptomic profiling of CRISPR double knockouts revealed that cooperating TSGs that synergized in promoting tumorigenesis and growth factor independence showed transcriptional epistasis, whereas noncooperating TSGs did not. These epistatic transcriptional changes, both buffering and synergistic, affected expression of oncogenic mediators and therapeutic targets, including CDK4, SRPK1, and DNMT1. Importantly, the epistatic expression alterations caused by dual inactivation of TSGs in this system, such as PTEN and TP53, were also observed in patient tumors, establishing the relevance of these findings to human breast cancer. An estimated 50% of differentially expressed genes in breast cancer are controlled by epistatic interactions. Overall, our study indicates that transcriptional epistasis is a central aspect of multigenic breast cancer progression and outlines methodologies to uncover driver gene epistatic networks in other human cancers.
Significance:
This study provides a roadmap for moving beyond discovery and development of therapeutic strategies based on single driver gene analysis to discovery based on interactions between multiple driver genes.
Introduction
With a few exceptions, such as early-phase chronic myeloid leukemia, human cancer involves the alteration of multiple driver genes (1, 2). For many driver gene alterations, certain features of how they collaborate to promote cancer are known. These features include their relative timing during cancer progression and their functions in different oncogenic signaling pathways (1, 3, 4). However, the degree to which cancer results from genetic interactions (GI; epistasis) between driver genes as opposed to the sum of individual driver gene effects is largely unknown (5, 6). Greater knowledge of epistatic interactions among driver gene alterations is necessary to accurately predict which phenotypes and therapeutic vulnerabilities are to be expected based on a patient's cancer genome (7, 8). However, uncovering these epistatic interactions and understanding their contribution to cancer progression has not previously been possible due to a lack of suitable tools.
Over the past several years, combinatorial CRISPR screening methods have been developed that enable the systematic analysis of GIs in mammalian systems (9–13). However, combinatorial CRISPR on its own cannot address the biology underlying those GIs. Recently, high-content phenotyping approaches that integrate pooled CRISPR screening with single-cell transcriptome readouts have emerged, enabling the growth-based phenotypic effects to be analyzed in parallel with transcriptome-wide changes (14–16). These tools and techniques presented us with an opportunity to systematically analyze how combinations of inactivated tumor suppressor genes (TSG) changed the growth properties and gene expression profiles of human mammary epithelial cells, with the goal of identifying general mechanisms of driver gene cooperation. Human breast cancer genome data guided our study and was also used to benchmark experimental transcriptome results.
Materials and Methods
Cell lines
MCF10A cells were purchased from ATCC (ATCC CRL-10317), and viral packaging cell lines HEK293T and GP2-293 were purchased from Takara, and MCF10A PTEN−/− cells were a generous gift from Michele I. Vitolo. All cells were used within three passages of the original stock. MCF10A derivatives (MCF10A-vector, MCF10A-PIK3CA, and MCF10A-MYC) were generated from MCF10A by retroviral transfection of the plasmids pMSCVneo (Clontech), pLP-LNCX-PIK3CA-H1047R (neo) (Addgene #25635), and MSCV-neo-HA-Myc (deposited with Addgene), respectively. All cells were checked for Mycoplasma contamination on a monthly basis using the LookOut Mycoplasma PCR Detection Kit (Sigma).
Single-guide RNA design
Single-guide RNAs (sgRNA) were designed to target functional protein domains using the online tool developed in Chris Vakoc's laboratory at Cold Spring Harbor Laboratory (17) in conjunction with the optimized sgRNA design tool developed by Doench and colleagues at the Broad Institute (18). Approximately five unique sgRNAs were chosen for each TSG based on the combined ranks using the two design tools. Nontargeting control sgRNAs were selected from a previous publication (19).
In vivo combinatorial CRISPR screening
TSG dual sgRNA cell libraries of MCF10A-vector, MCF10A-PTEN−/−, MCF10A-PIK3CA and MCF10A-MYC were used for in vivo screenings. Three to 5 million cells in 50 μL of Matrigel (Corning, catalog no. 354230)/PBS (1:1) were inoculated orthotopically into the third and fourth mammary glands of athymic nude-Foxn1nu mice. Mice were humanely euthanized at 6 to 8 weeks after injection, such that the maximum volume of the tumors did not exceed 2 cm3 (following Institutional Animal Care and Use Committee procedures and protocols). Dual sgRNA abundance in the tumors was determined by next-generation sequencing.
In vitro combinatorial CRISPR screening
MCF10A-vector TSG dual sgRNA cell libraries were used for in vitro screenings. Two 15-cm dishes containing cells at day 0 were harvested to determine the initial dual sgRNA distribution before screening. Cells were then seeded at different densities in 15 cm dishes for screenings in the three different conditions. In the first condition, 5 million cells were seeded into each of two 15-cm dishes with full medium and passed at a ratio of 1:5 every 3 days. Two plates of cells were sampled at days 3, 6, 9, 12, and 15. In the second condition, 1.5 million cells were seeded into each of 40 15-cm dishes with full medium. After the cells attached overnight, the media were replaced with minimal assay medium. The minimal medium was composed of DMEM/F-12, 1% charcoal-stripped dextran-treated FBS (Hyclone) and 100 units/mL penicillin and 100 μg/mL streptomycin. Eight plates of cells were harvested on days 3, 6, 9, 12, and 15. In the third condition, 5 million cells were seeded into each of two 15-cm dishes with full medium. The media were removed and replaced with MCF10A growth media supplemented with 5 ng/μL TGFβ1 (R&D) after overnight. Cells were cultured and passed at a ratio of 1:5 every 4 days and two plates of cells were sampled on days 4, 8, 12, 16, and 20. Two biological replicates of screens were performed for each condition. Dual sgRNA abundance in the cell samples was determined by next-generation sequencing.
Construction of combinatorial CROP-seq cell library
A cell library expressing dual CROPseq-sgRNAs was constructed by infecting CROPseq-Guide-Puro-sgRNAs viruses and CROPseq-Guide-Blasti-sgRNAs viruses sequentially. MCF10A-vector-Cas9-Venus cells were infected with 15 different CROPseq-Guide-Puro-sgRNA viruses CROPseq-Guide-Puro-(sgNF1_1, sgPTEN_5, sgSMAD4_5, sgCASP8_5, sgCBFB_2, sgCDH1_5, sgRB1_3, sgTP53_3, sgNF2_2, sgTBX3_3, sgUSP9X_3, sgTP53_4, sgTP53_5, sghRosa26_2, sgCTRL0002) individually, and selected in 1.5 μg/mL puromycin for 4 days. Cells were then passed for the next round of infection of CROPseq-Guide-Blasti viruses. After 24 hours of infection, blasticidin (15 ng/μL) was added to the medium for selection. In total, 72 pairwise sgRNA combinations were included in our combinatorial CROP-seq cell library and 68 unique ones were detected by sequencing.
Profiling single-cell transcriptomes with the combinatorial CROP-seq cell library
The combinatorial CROP-seq library of cells was cultured in MCF10A growth medium. When they grew confluent, 2 × 105 cells were seeded into three 10-cm dishes and were further grown in three different conditions: full medium, minimal medium, and medium containing 5 ng/μL TGFβ1 for 6 days. Before seeding, some cells were harvested for the sample for single-cell RNA sequencing (scRNA-seq) on day 0 (S1_full_D0). On day 6, cells in the different conditions were harvested for the other three scRNA-seq. scRNA-seq experiments were performed according to 10× Genomics' protocol, with holding 10 ng of full-length cDNA from the downstream shearing and library prep steps to provide material for barcode-enrichment PCR. Approximately 16,000 cells for each sample were loaded into 10× Chromium chips enabling the profiling of transcriptomes of 5,000 to 9,000 single cells for each sample.
Data availability statement
Raw pair-end amplicon sequencing data in combinatorial screenings are available and can be downloaded from the NCBI Sequence Read Archive (SRA) database with the BioProject ID: PRJNA691742. Raw scRNA-seq, raw enrichment PCR sequencing data, and processed combinatorial CROP-seq data via 10× Cell Ranger count are available and can be downloaded from the NCBI Gene Expression Omnibus (GEO) database with the accession ID: GSE164996. Relevant code and instructions that are used to reproduce the principal results presented in this study, are provided on GitHub: https://github.com/Xiaoyu-Zhao/Oncogenic_GI_screening.git.
Additional information can be found in the Supplementary Materials and Methods.
Results
Establishing an in vivo screening platform for testing the tumor-forming ability of combinatorial TSG perturbations
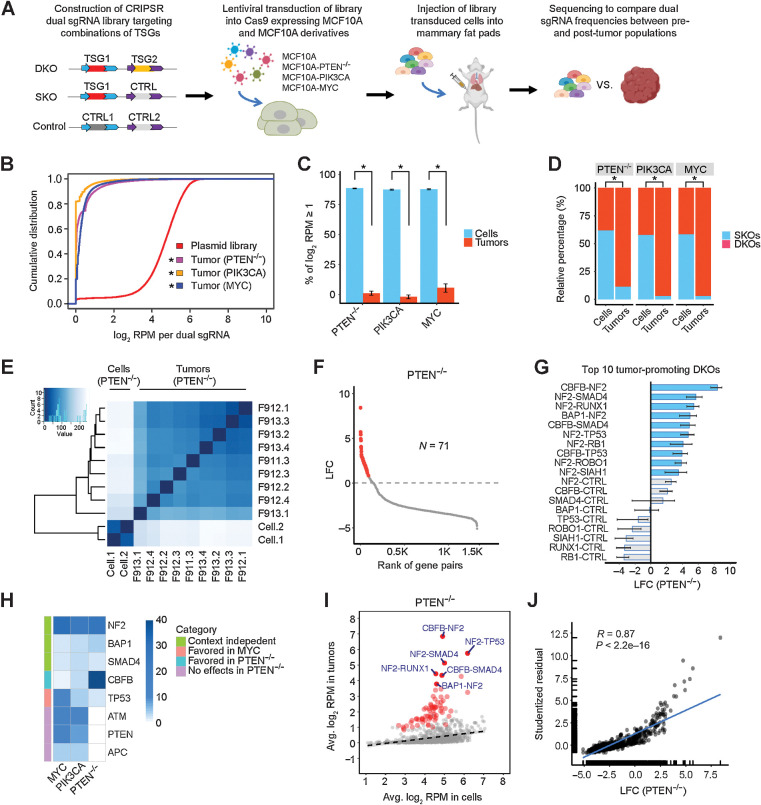
MCF10A, harboring a homozygous deletion of the CDKN2A/B locus encoding p16INK4a, p14ARF, and p15INK4b TSGs (20, 21), is an immortalized, nontumorigenic human breast epithelial cell line (22). Because normal human cells can require five oncogenic alterations to become fully transformed (23), we were concerned that inactivation of two TSGs would not be sufficient to fully transform MCF10A, and therefore constructed three MCF10A derivatives with one additional oncogenic alteration (MCF10A-PTEN−/−, MCF10A-PIK3CA, and MCF10A-MYC). We selected 52 TSGs and/or candidate TSGs based on their mutation and homozygous deletion frequency in breast cancer (Supplementary Table S1). We designed five sgRNAs on average for each TSG and synthesized a pool of dual sgRNA oligonucleotides comprised of 34,937 double knockouts (DKO) targeting all pairwise combinations of the 52 TSGs, 801 single knockouts (SKO) targeting a single TSG, and 88 nontargeting controls (Fig. 1A). We constructed a CRISPR/Cas9 plasmid library from these oligonucleotides and generated lentiviruses that were transduced into MCF10A and its derivatives that stably expressed Cas9 nuclease, followed by selection for stable integration (Fig. 1A). All four library-transduced cell populations successfully integrated over 98% of the dual sgRNAs from the plasmid library, with slight shifts in the cumulative distribution curves of the relative abundance of dual sgRNAs (Supplementary Fig. S1A). Dual sgRNAs containing sgRNA targeting TP53 were noticeably enriched in the library-transduced cell populations compared with the plasmid library, whereas other dual sgRNAs, such as those targeting NF2, PTEN, SMAD4, or RB1, were not enriched (Supplementary Fig. S1B–S1D). However, this enrichment was much lower than that reported for CRISPR screening in the hTERT-immortalized epithelial cell line RPE1, which also retains a wild-type TP53 gene, perhaps because the CDKN2A/B deletion dampened the p53-mediated DNA damage response in MCF10A (24).
Figure 1.
Perturbations in TSG pairs promote tumorigenesis in vivo. A, Schematic of combinatorial CRISPR screening in vivo. B, Cumulative distributions of dual sgRNA abundance in the plasmid library and tumors. *, P < 0.05; Kolmogorov–Smirnov test. C, Percentage of dual sgRNAs with log2 RPM ≥1 in the PTEN−/−, PIK3CA, and MYC cell libraries and tumors. Data are presented as mean ± SEM. Mean values were calculated from nine PTEN−/−, three PIK3CA, and six MYC tumors. *, P < 0.01; one-sided Student t test. D, Relative percentage of DKO/SKO normalized counts in the PTEN−/−, PIK3CA, and MYC cell libraries and tumors. *, P < 0.01; one-sided Student t test. E, Heatmap of Euclidean distances between all the cell and tumor samples (PTEN−/−). F, The effect size (LFC) of DKOs and SKOs in the PTEN−/− context. A total of 71 oncogenic perturbations are highlighted in red. G, The top 10 tumor-promoting DKOs and corresponding SKOs in the PTEN−/− context. The mean is plotted along with the 95% CI. H, Heatmap comparing the single-gene tumorigenic effects in the PTEN−/−, PIK3CA, and MYC contexts. I, Scatter plot of average log2 RPM in the PTEN−/− tumors versus preinjected cell libraries. A total of 71 oncogenic perturbations shown in F are highlighted in red, and the top 6 ones are labeled. The linear regression line is shown (dashed). J, Scatter plot and correlation analysis of the tumorigenic effects measured by quantile analysis versus average based regression (PTEN−/−).
Perturbations in TSG pairs promote tumorigenesis in vivo
Cells from each of the four TSG library-transduced cell populations were orthotopically injected into murine mammary pads (n = 20). At 6 to 8 weeks after injection, none of the library-transduced parental MCF10A cells had formed tumors, while each of the library-transduced MCF10A derivatives had formed tumors. The cumulative frequency curves of the dual sgRNAs in all tumors were remarkably skewed compared with those of the plasmid library or with those of the corresponding transduced cells (Fig. 1B; Supplementary Fig. S1A). Only a small fraction of the dual sgRNAs were detected in the tumors (5%–12%), while over 94% were detected in the cell populations (Fig. 1C). Interestingly, we observed that normalized counts of DKOs relative to SKOs increased substantially in tumors compared with the preinjected cell populations. In the preinjection cells, the ratio of DKO/SKO normalized counts was close to 1:1; in contrast, DKO normalized counts comprised 89% to 97% of the total in tumors (Fig. 1D). These results show that for this platform, perturbing a pair of TSGs promotes tumorigenesis much more strongly than perturbation of single TSGs.
Determining the tumor-promoting strength of TSG perturbation pairs
We used hierarchical clustering to examine the similarities of the dual sgRNAs present in preinjected cells and tumors. In the PTEN−/− group, all nine tumors clustered closely together and separately from the two duplicates of the preinjected cell populations, indicating similar biological selection for the enriched sgRNAs in the tumor replicates (Fig. 1E). Similar results were obtained for tumors obtained from the two other MCF10A derivatives (Supplementary Fig. S2A and S2B). We next employed two different methods to quantify the effect size and statistical significance of perturbations on tumorigenicity at the gene level. The first method used quantiles, specifically the 70th quantile, to minimize the noise contributed by PCR jackpot effects and the preponderance of zero counts, coupled with bootstrap procedures to determine significance. Tumorigenic hits were identified as perturbations that induced a significant positive log2 fold change (LFC) in the relative quantile-based abundance between tumors and cells (Fig. 1F). For all three MCF10A derivatives, between 56 and 71 of the DKOs showed tumor-promoting ability, compared with only zero to three SKOs (Supplementary Table S1). In the PTEN−/− group, the top 10 tumor-promoting effects were all from DKOs, and their effects were notably larger than any SKOs (Fig. 1G). Moreover, the stronger tumorigenic effects of these DKOs were selectively driven by particular TSG partners. For instance, only a few of the 52 NF2-TSG perturbation pairs had much higher tumor-promoting capability than the NF2 SKO, while most of them displayed similar or even less tumorigenicity (Supplementary Fig. S2C). In the other PIK3CA and MYC groups, the top tumor-promoting effects also came from DKOs (Supplementary Fig. S2D and S2E). Notably, some TSG perturbations capable of promoting tumorigenesis were shared by different groups, but others were distinct in the three MCF10A derivatives. Perturbations targeting NF2, BAP1, or SMAD4 were largely independent of the sensitizing oncogenic alterations, whereas perturbations affecting CBFB were found more often in the PTEN−/− tumors, whereas perturbations affecting TP53 were predominant in the MYC tumors, and perturbations affecting APC, ATM, and PTEN were found only in the PIK3CA and MYC tumors (Fig. 1H).
For the second method of quantifying tumorigenic effect size, we employed linear regression modeling of the average relative abundance in tumors and cells. Tumor-promoting perturbations displayed positive residuals in the regression analysis, indicating their overrepresentation in the tumors (Fig. 1I; Supplementary Fig. S2F and S2G). Because there were strong correlations between the LFCs determined by quantile analysis and the residuals computed from the linear regression model, we have confidence in our determination of the tumor-promoting ability of the different dual TSG knockouts (Fig. 1J; Supplementary Fig. S2H and S2I).
Tumor-promoting TSGs form GI networks
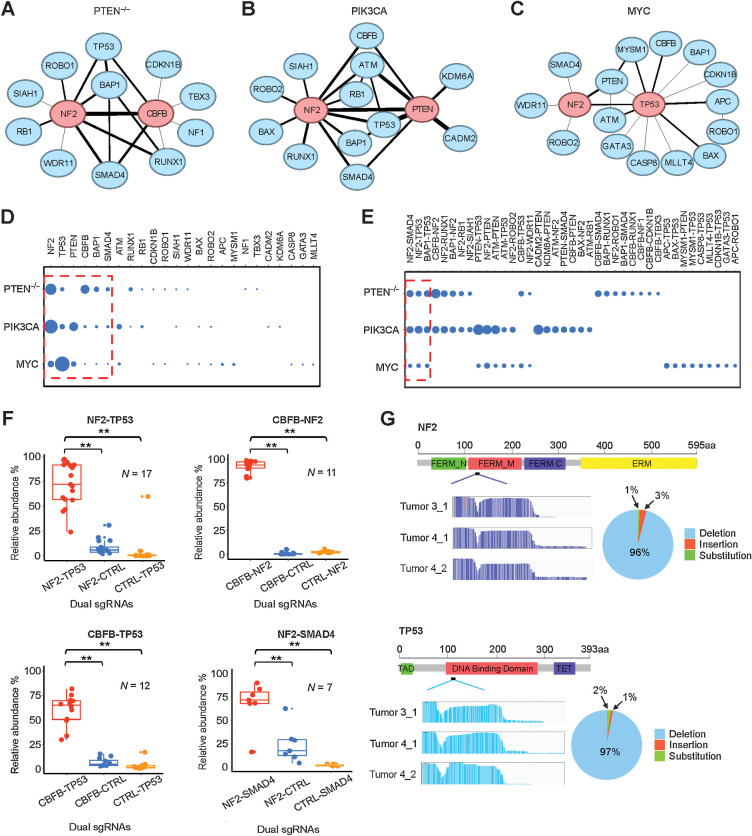
We next determined which of the tumor-promoting dual TSG alterations showed evidence of positive epistasis, such that the DKO effect was significantly greater than the individual SKO effects (Supplementary Fig. S3A–S3C). We computed the GI scores π of 1,325 TSG pairs and graphed the tumor-promoting epistatic networks for all three MCF10A derivatives (Fig. 2A–C). One of the most striking features of these networks is the absence of any bipartite subgraphs (25), which often correspond to between-pathway interactions—a common feature of the global synthetic lethal GI network in yeast (26) and the type of interaction proposed to underlie PARP and BRCA synthetic lethality (27). Instead, we observed numerous three-gene cliques, such that all three genes within the clique show oncogenic cooperation (positive epistasis) with each other. In the PIK3CA network, there was a single four-gene clique, where ATM, PTEN, TP53, and NF2 each displayed oncogenic cooperation with each other (Fig. 2B). This latter type of network subgraph has been termed within-pathway interactions and in the yeast global interaction network has been observed for genes encoding members of the same complex, such as the spliceosome (26). For all three networks, the genes with the most interactions or hubs interacted with each other (Fig. 2A–C), a network property that is termed assortative and that is a property of social networks but not a property of most described biological networks (25).
Figure 2.
Tumor-promoting TSGs form GI networks. A–C, Oncogenic GI networks in the PTEN−/−, PIK3CA, and MYC contexts, respectively. The top two central nodes are highlighted in red. D, Comparison of degree centrality scores of TSG nodes in the three GI networks. Six nodes commonly shared in the three networks are enclosed in the red dashed frame. E, Comparison of edges (interactions) in the three networks. Three common interactions are enclosed in the red dashed frame. F, Validation of four oncogenic GIs in vivo. **, P < 0.05; one-sided Student t test. G, CRISPR mutational profiles induced by sgNF2_2 and sgTP53_4 in the PTEN−/− tumors.
To further compare the three networks, we determined the degree centrality score of each TSG node, which reflects the number of its connections. The two most central nodes were different for each of the three networks: NF2 and CBFB were most central for the PTEN−/− network; NF2 and PTEN were most central for the PIK3CA network; and TP53 was by far the most central node in the MYC network, with PTEN and NF2 approximately tied for the second most central node (Fig. 2D). The absence of PTEN in the PTEN−/− network was because its sgRNAs had no or little tumorigenicity in PTEN−/− MCF10A cells. Moreover, we found that the edges or epistatic relationships among TSGs also varied considerably. Only three of the total 39 interactions—NF2-TP53, NF2-SMAD4, and BAP1-TP53—commonly occurred in all three networks; 12 interactions were shared by only two of the networks and 24 were unique to specific networks (Fig. 2E). The most common reason for the absence of TSG pairs in a particular network was that they were nontumorigenic in that particular background (∼73% of the cases; Supplementary Table S1). For example, the two pairs APC-ROBO1 and APC-TP53 were only tumorigenic in the MYC background, possibly because of the tight interaction of MYC and Wnt signaling in breast cancer (28). In addition, it is likely that the reason why TSG pairs containing ATM did not appear in the PTEN−/− network is the synthetic lethality between ATM inhibition and PTEN loss (29). This synthetic lethality is related to PTEN's function in DNA repair that is independent of Akt, which explains why ATM pairs are present in the PIK3CA network. Another relatively common reason for missing TSG pairs (∼22% of the cases) was that although the TSG pair was tumorigenic, there was not any positive epistasis between the pair. The tumorigenic effects were either additive or suppressive when one TSG perturbation significantly inhibited the tumorigenicity of the other—for example, BAX significantly inhibited NF2 in the PTEN−/− background (Supplementary Table S1). A minor technical point is that in 5% of the cases, to have the PIK3CA network be of the same size as the other two, we only included interactions with strong epistasis (the full PIK3CA network including the weaker epistatic interactions is included in Supplementary Table S1). Interestingly, the interactive degree of the TSG nodes did not always fall in line with the size of their tumor-promoting effects. For example, CBFB not only showed strong tumor-promoting function but also cooperated with many other TSGs, particularly in the PTEN−/− network, whereas TP53 acted as the master central node in the MYC network, though its perturbation-induced tumorigenicity was not as robust as that seen with NF2.
Functional validation of cooperative tumor-promoting dual TSG perturbations
We next performed validation experiments to ensure that our CRISPR/Cas9 GI screening platform was pinpointing bona fide cooperative TSGs. We chose four dual TSG perturbations (NF2-TP53, CBFB-NF2, CBFB-TP53, and NF2-SMAD4) that were detected in the MCF10A PTEN−/− derivative and at least one of the other two derivatives. For each TSG pair, equal mixtures of DKO, SKO1, SKO2 and control MCF10A PTEN−/− cells were orthotopically injected into mammary fat pads, and tumors formed 6 to 8 weeks after injection were harvested to determine by deep sequencing their relative abundance. All tumors displayed the similar histopathologic features of invasive carcinoma (Supplementary Fig. S3D). For all four groups, the abundance of DKOs in the resultant tumors were significantly greater than their respective SKO counterparts and were in fact greater than the additive effect of two SKOs, validating both their tumor-promoting ability and the epistatic interactions (Fig. 2F). We also performed validation experiments to ensure that our CRISPR/Cas9 GI screening platform was creating the expected Cas9-mediated deletions. We determined the sequence of the target regions in tumors formed in the NF2-TP53 group. We found that the NF2 sgRNA (sgNF2_2) induced mutations within the FERM-M domain of the NF2 gene in approximately 80% of the tumor cells, consisting of 96% deletions, 1% insertions, and 3% substitutions. We found that the TP53 sgRNA (sgTP53_4) induced mutations within the DNA-binding domain of TP53 in approximately 78% of the tumor cells, consisting of 97% deletions, 2% insertions, and 1% substitutions (Fig. 2G).
Ascertaining in vitro growth-promoting GI networks
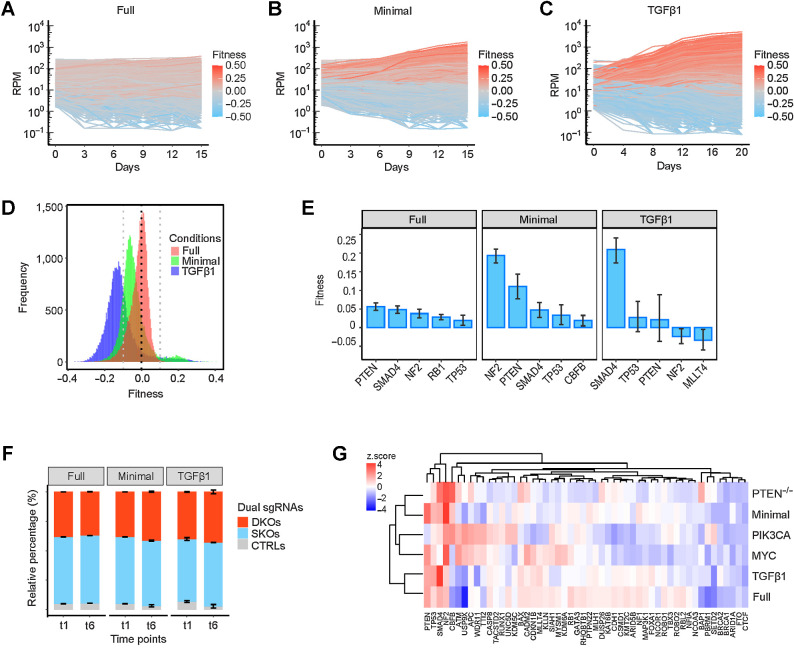
As stated in the Introduction, we wanted to address the biology underlying TSG GIs by high-content phenotypic screening using single-cell CRISPR-Cas9 knockouts integrated with scRNA-seq, so that growth-based phenotypic effects could be analyzed in parallel with transcriptome-wide effects. For this purpose, in vivo screening was problematic, since only 5% to 12% of the TSG perturbation pairs were detected in tumors (Fig. 1B) and we would not be able to profile transcriptomes of the underrepresented nontumorigenic cells, which is the cornerstone of our approach. Therefore, we tested different in vitro culture growth conditions for their ability to represent the tumor-promoting GIs we observed in vivo. To generate results that would not be biased to any specific MCF10A derivative we used in vivo, we employed the parental MCF10A cell line. Two important hallmarks of cancer are sustaining proliferative signaling and evading anti-growth signaling (30). To test for the ability to sustain proliferative signaling, we assayed growth in medium deprived of multiple growth factors (minimal medium). To test for the ability to evade antigrowth signaling, we assayed growth in the presence of TGFβ1 (5 ng/μL). These two restrictive growth conditions were compared with growth in standard full medium. Dual sgRNA representation was tracked at six different time points, and two independent screenings were performed for each condition. The cell trajectories over time revealed that the growth dynamics of the approximately 36K cell lineages expressing unique dual sgRNAs were noticeably different in the two restrictive growth conditions: some cell lineages appeared to have acquired strong growth advantages in contrast to the more even trajectories in the full medium (Fig. 3A–C).
Figure 3.
Growth-promoting effects of TSG perturbations in vitro. A–C, Representative trajectories of cell lineages expressing unique dual sgRNAs grown in full, minimal, and TGFβ1-supplemented media, respectively. D, Distribution of gene-level fitness of 1,378 perturbations in the three conditions. E, The top five single-gene effects on fitness in the three conditions. Data are presented as mean ± SEM. F, Relative percentage of DKO/SKO normalized counts in the three conditions. Data are presented as mean ± SEM. G, A heatmap that shows the comparison of growth-promoting effects in vitro and tumorigenic effects in vivo of SKOs.
We determined the fitness “f” of each dual sgRNA lineage by fitting a linear model to its growth curve, and then used the average sgRNA fitness to determine gene-level fitness. The gene-level fitness values of the 1,378 perturbations, including SKOs, DKOs, and nontargeting controls, ranged from −0.09 to 0.07 in full medium; however, a much broader fitness range was detected in both minimal medium (−0.18 to 0.22) and TGFβ1 medium (−0.19 to 0.16; Fig. 3D). The gene-level fitnesses in replicate library screens were highly correlated (Supplementary Fig. S4A). The top five single-gene effects on growth promotion in full medium came from deletions of PTEN, SMAD4, NF2, RB1, or TP53 (Fig. 3E; Supplementary Fig. S4B). In minimal medium, the top five genes were nearly identical to the full-medium genes, but their effects on growth were more pronounced (Fig. 3E; Supplementary Fig. S4B). CRISPR-mediated inactivation of SMAD4 was the only single-gene effect that significantly increased fitness in the presence of TGFβ1 (Fig. 3E; Supplementary Fig. S4B). In contrast to the strong enrichment for DKOs seen in tumor formation, we did not observe this for the in vitro conditions (Fig. 3F). We used unsupervised clustering to examine the relationship between the 52 SKO effects in the three in vitro conditions and three in vivo conditions. Notably, we found that the single-gene effects in minimal medium resembled the in vivo effects more than those of the other two in vitro conditions (Fig. 3G).
Determining which in vitro growth-promoting networks resemble in vivo tumor-promoting networks
We computed the GI scores π of 1,325 TSG pairs under the three growth conditions. As was the case for gene-level fitness, the GI scores in the two biological replicate library screens were highly correlated demonstrating the reproducibility of our experimental and analytic procedures (Supplementary Fig. S4C). We observed in total nine growth-promoting GIs in full medium, in which TP53 acted as a central node in the network, and nine GIs that cooperatively promoted growth factor independence in minimal medium (Fig. 4A and B). There were no significant epistatic interactions detected in the presence of TGFβ1, likely because the single deletion of SMAD4 had already conferred upon MCF10A cells the maximal ability to evade TGFβ1 growth suppression.
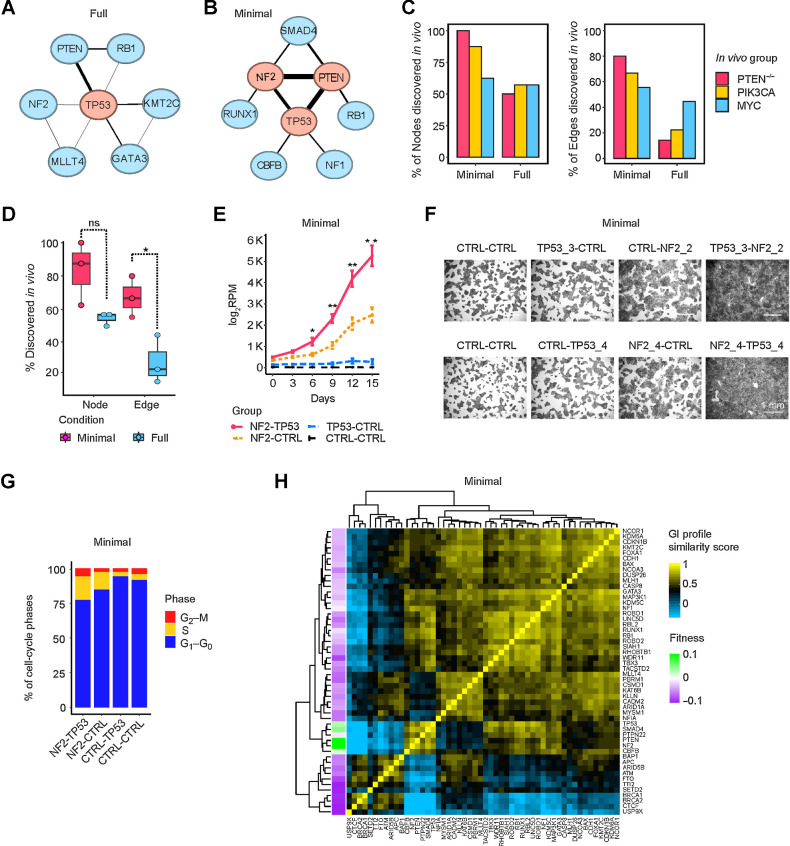
Figure 4.
Growth-promoting networks in minimal medium resemble in vivo tumor-promoting networks. A and B, Growth-promoting GI network in full and minimal media. The top three central nodes are highlighted in red. C, Comparison of growth/tumor-promoting GI networks in vitro and in vivo, with respect to nodes (left) and edges (right). D, Percentage of nodes and edges in vitro that were discovered in GI networks in vivo. Data are presented as mean ± SEM. *, P < 0.05; one-sided Student t test. ns, not significant. E–G, Growth curves, crystal-violet staining, and cell-cycle cytometric analysis of NF2-TP53 DKO, NF2-CTRL, or TP53-CTRL SKO and control cells in minimal medium. Data are presented as mean ± SEM in E. *, P < 0.1; **, P < 0.05; one-sided Student t test. H, GI profiles of 52 TSGs in minimal medium.
We compared the full and minimal medium in vitro growth-promoting GI networks to the in vivo tumor-promoting GI networks with respect to both nodes (single TSGs) and edges (interactions). Most nodes in the growth factor–independent in vitro network were also found in the tumor-promoting GI networks, ranging from 5 of 8 to 7 of 7; in contrast, fewer nodes of the full medium network were found in the in vivo networks (between 3 of 6 to 4 of 7; Fig. 4C). The difference between full and minimal medium was even more pronounced when comparing edges. Between 4 to 6 of the nine growth factor–independent edges were also found in the in vivo networks, compared with only one to four edges of nine edges in the full medium network (Fig. 4C). The MYC tumor-promoting network showed less of a difference than the other two networks in matching nodes and edges between the control and minimal growth factor conditions, perhaps because both PIK3CA and PTEN are more directly involved in growth factor signaling. Overall, 83% of the TSG nodes and 67% of the edges of the network in minimal medium were found in the in vivo networks, while 55% of the TSG nodes and 27% of the edges in the standard culture condition were detected in vivo (Fig. 4D). This GI analysis further supports the conclusion that growth in minimal medium is the best match for in vivo growth. The perturbation pairs of NF2-TP53, PTEN-TP53, and NF2-PTEN were particularly effective at promoting both growth factor independence and tumorigenicity. We validated the synergy between dual inactivation of NF2 and TP53 in conferring growth factor independence by comparing the growth curves and the degree to which cells could reach confluence in minimal medium (Fig. 4E and F). Dual deletion of NF2 and TP53 cooperatively reduced the percentage of cells in the G1-phase of the cell cycle (Fig. 4G).
GI profiles segregate TSGs according to function
One of the most useful insights gained from genome-scale GI analyses in yeast is that genes that had similar GI profiles (for a given gene, the set of GI scores for every other gene) shared common cellular functions, and that GI profile similarities provided a means to construct a functional map of the cell (31). To explore the utility of GI profiling of TSGs, we constructed profiles of the GIs of each TSG with all other TSGs, and by correlation and clustering analysis, generated heatmaps displaying relationships between the TSGs for each of the three in vitro conditions (Fig. 4H; Supplementary Fig. S4D and S4E). Notably, all three paralog pairs that are functionally similar (BRCA1 and BRCA2; RB1 and RBL2; ROBO1 and ROBO2) clustered together in all three conditions with one informative exception. In two cases, the paralogs formed a two-gene clade, indicating the highest degree of similarity (such as BRCA1 and BRCA2 in Fig. 4H), in two cases a three-gene clade, and another two cases a five-gene or eight-gene clade (RB1 and RBL2 and ROBO1 and ROBO2 in Fig. 4H). The lone exception was found with TGFβ1 medium, where BRCA1 and BRCA2 were completely separated (Supplementary Fig. S4E). Unlike BRCA1, BRCA2 binds to and enhances the transcriptional function of SMAD3, so its loss of function would be expected to decrease the growth-inhibitory signaling of TGFβ1 (32). In TGFβ1 medium, BRCA2 co-clusters with NCOA3, a transcriptional coactivator that binds directly to SMAD3 (33). These results demonstrate that GI profiling can pinpoint functionally similar TSGs and that these relationships change depending upon the physiologic context.
Interestingly, five of the six top nodes of the in vivo networks, PTEN, NF2, TP53, SMAD4, and CBFB are tightly clustered within a six-gene clade in minimal medium (Fig. 4H). With few exceptions, all five of these TSGs cooperate with each other, indicating that these predominant cooperating TSGs share overlapping function, rather than distinct or complementary function.
Single-cell transcriptomic analysis provides insights into the growth-promoting mechanisms underlying TSG perturbations
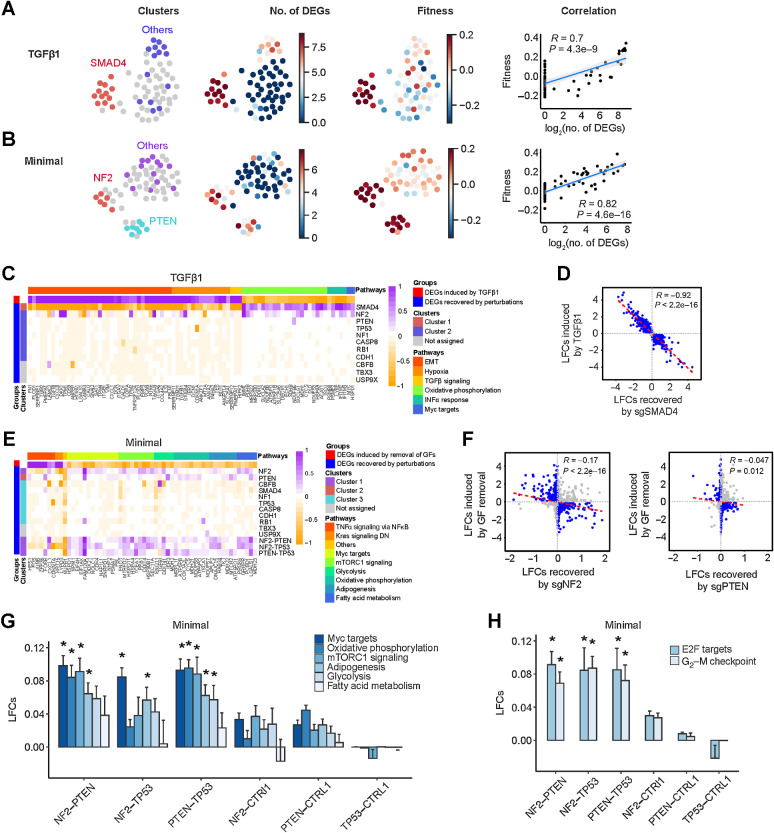
To address the biology underlying TSG GIs, we performed combinatorial CROP-seq, such that the single-cell transcriptomes of thousands of cells with different dual and single TSG perturbations could be assayed. We focused on 11 genes and their pairwise combinations (55 DKOs), including six genes that were present in all or nearly all of the growth-promoting and tumor-promoting networks (TP53, NF2, SMAD4, PTEN, RB1, CBFB), three genes in only one or two of the networks (CASP8, NF1, TBX3) and two negative controls (CDH1, USP9X). We screened in the three different in vitro conditions: full medium, TGFβ1-supplemented medium, and minimal medium (Supplementary Fig. S5A). We used Uniform Manifold Approximation and Projection (UMAP) to project mean expression profile and identified stable clusters of transcriptionally similar perturbations using the Hierarchical Density-Based Spatial Clustering of Applications with Noise (HDBSCAN) algorithm (Fig. 5A and B; Supplementary Fig. S5B). Perturbations affecting SMAD4 formed a distinct cluster in the presence of TGFβ1 and showed an increased ability to grow in the presence of TGFβ1 and an increased number of differentially expressed genes (DEG; Fig. 5A). Similarly, when cultured in minimal medium, the transcriptomes of perturbations affecting NF2 and PTEN formed two separate clusters that both showed increases in transcriptional alterations and fitness (Fig. 5B). We observed that fitness measurements were strongly correlated with the extent of transcriptional changes in both restrictive conditions, but less so in full medium (Fig. 5A and B; Supplementary Fig. S5B). Collectively, the close relationship between fitness and transcriptional alterations suggests that transcriptional changes can shed light on the mechanisms by which single and dual TSG knockouts overcome growth restrictions.
Figure 5.
Single-cell transcriptional analysis reveals growth-promoting mechanisms underlying TSG perturbations. A and B, Relationship between transcriptional states and fitness in TGFβ1 and minimal medium, respectively. First column, UMAP projection of mean expression profiles for all the perturbations where transcriptionally similar clusters are highlighted in an identical color; second column, number of significant DEGs induced by each perturbation; third column, fitness measurements of each unique perturbation; and fourth column, correlation between fitness and number of significant DEGs. C, Representative DEGs in different biological pathways induced by TGFβ1 and recovered by the 11 SKO dual sgRNAs. EMT, epithelial–mesenchymal transition. D, Scatter plot of TGFβ1-induced LFCs and LFCs recovered by sgSMAD4. Genes with LFCs > 0.1 are highlighted in blue. E, DEGs in different biological pathways induced by growth factor (GF) deprivation and recovered by the 11 SKO and 3 DKO dual sgRNAs. DN, down. F, Scatter plots of growth factor withdrawal–induced LFCs and LFCs recovered by sgNF2 and sgPTEN. Genes with LFCs > 0.1 are highlighted in blue. G and H, Comparison of gene expression pathways involved in growth factor independence and cell cycle induced by sgNF2, sgPTEN, and sgTP53 and their dual combinations. Data are presented as mean ± SEM. *, P < 0.05; one-sided Student t test.
Pathway analysis indicated that the major transcriptional alterations induced by TGFβ1 included the induction of epithelial–mesenchymal transition and hypoxic response genes and the suppression of genes involved in oxidative phosphorylation (Fig. 5C; Supplementary Fig. S5C and S5D). These alterations appeared to be either entirely or largely due to canonical TGFβ1 pathway signaling, as CRISPR-mediated SKO of SMAD4 counteracted all of these changes (Fig. 5C). CRISPR-mediated SKO of NF2 reversed some of the suppression of genes involved in oxidative phosphorylation, but was otherwise ineffective, as were other SKOs (Fig. 5C). By examining all expression changes induced by TGFβ1, and the corresponding effects of SMAD4 inactivation, we observed a remarkably complete reversal of TGFβ1-induced expression changes (Fig. 5D). These results substantiated the sensitivity and reliability of our combinatorial CROP-seq platform and analytical pipeline.
Pathway analysis showed that growth factor withdrawal resulted in downregulation of genes involved in mTOR signaling, glycolysis, oxidative phosphorylation, adipogenesis, fatty acid metabolism, and Myc signaling, as well as upregulation of inflammatory genes and the cell-cycle inhibitor CDKN1A (Fig. 5E; Supplementary Fig. S5E and S5F). The SKOs that conferred significant growth factor independence displayed variable ability to reverse the gene expression changes caused by growth factor withdrawal (Fig. 5E). The NF2 SKO showed the greatest increase in fitness in minimal medium and also had the biggest effect in reversing expression changes caused by growth factor withdrawal, followed by PTEN SKO, which was especially effective at reversing changes affecting oxidative phosphorylation (Fig. 5E and F). Interestingly, the endoplasmic reticulum stress response gene NUPR1 was upregulated by growth factor removal, but downregulated by all five SKOs that increased fitness, suggesting that relieving growth factor deprivation-induced stress is a prerequisite to increasing fitness (Fig. 5E). Nevertheless, the ability of SKOs to reverse the expression changes caused by growth factor deprivation was not as comprehensive as that seen in TGFβ1 medium with the SMAD4 SKO.
Notably, the double deletions of NF2-PTEN, NF2-TP53, and PTEN-TP53 induced stronger reversal of the downregulation of genes caused by growth factor withdrawal when compared with SKOs (Fig. 5G; Supplementary Table S1). These three DKOs also induced significantly higher expression of E2F targets and G2–M checkpoints than their SKO counterparts (Fig. 5H). Compared with the simpler growth barrier induced by a single factor (TGFβ1), the more complex barrier created by deprivation of multiple growth factors was better counteracted by two driver gene alterations.
Transcriptional GIs correlate with fitness GIs
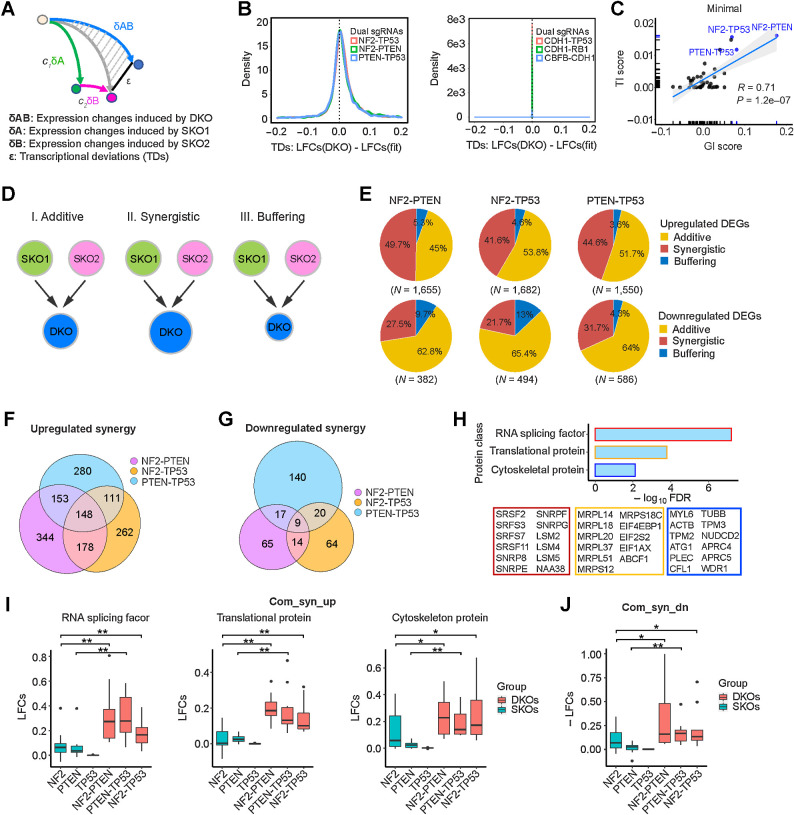
Because several tumor-promoting DKOs showed positive epistatic effects on tumor growth as well as growth in minimal medium, we wanted to investigate whether DKOs also had epistatic effects on transcription. Toward this end, we determined for each expressed gene the transcriptional deviations (TD) of the DKOs from their corresponding SKOs using a linear regression model developed by the Weissman group (Fig. 6A; ref. 16). For some DKOs like CDH1-TP53, there was little divergence of the observed expression changes from the changes predicted by a linear fit of the two SKOs, whereas for other DKOs, such as NF2-TP53, there was noticeable divergence (Fig. 6B). We calculated a summary statistic of the TDs for each DKO, which we termed the transcriptional interaction (TI) score. We observed that TI magnitudes were significantly correlated with fitness GI scores for the DKOs cultured in minimal medium (Fig. 6C) and to some extent for TGFβ1-containing medium (Supplementary Fig. S6A). In contrast, there was not a significant correlation between the GI and TI scores of DKOs cultured in full medium (Supplementary Fig. S6A).
Figure 6.
Epistatic transcriptional interactions correlate with fitness GIs. A, Schemata of the quantitative model used to measure TDs. B, TDs of DKO examples—NF2-PTEN, NF2-TP53, PTEN-TP53, CDH1-TP53, CDH1-RB1, and CBFB-CDH1—in minimal medium. C, Correlation of TI scores and fitness GI scores in minimal medium. D, Categorization of DEGs based on their TDs. E, Percentages of synergistic, additive, and buffering genes of upregulated DEGs (top) and downregulated DEGs (bottom) for NF2-PTEN, NF2-TP53, and PTEN-TP53 DKOs. F, Number of upregulated synergistic DEGs in NF2-PTEN, NF2-TP53, and PTEN-TP53 DKOs. G, Number of downregulated synergistic DEGs in NF2-PTEN, NF2-TP53, and PTEN-TP53 DKOs. H, Gene ontology term enrichment analysis of the 148 common upregulated synergistic genes. I, Expression of the enriched protein classes in NF2-PTEN, NF2-TP53, and PTEN-TP53 DKOs and their SKO counterparts. *, P < 0.1; **, P < 0.05; one-sided Student t test. J, Expression of the nine common downregulated synergistic genes in NF2-PTEN, NF2-TP53, and PTEN-TP53 DKOs and their SKO counterparts. *, P < 0.1; **, P < 0.05; one-sided Student t test.
To probe deeper into the biology underlying these epistatic interactions, we further examined three DKO perturbations (NF2-PTEN, NF2-TP53, and PTEN-TP53), which showed significant growth-promoting genetic and transcriptional interactions in minimal medium (Fig. 6C). We classified upregulated genes (LFC > 0.03) into three categories: additive, showing little or no transcriptional deviations from the linear fit (−0.05 <TDs <0.05); synergistic, with positive TD scores (TDs ≥0.05); and buffering, with negative TD scores (TDs ≤ −0.05; Fig. 6D). The percentage of upregulated genes that fell into these three categories were largely similar for the three DKOs: 41% to 50% for synergistic, 3% to 5% for buffering, and 45% to 54% for additive (Fig. 6E). Although most of these upregulated genes were unique to each of the DKOs, there were a number of shared upregulated genes, including 148 synergistic and 74 additive genes commonly shared between all three DKOs (Fig. 6F; Supplementary Fig. S6B). Overall, we observed 3- to 4-fold less downregulated genes (LFC < 0.03) than upregulated genes, with the majority (62%–66%) being additive (−0.05 <TDs <0.05), 21% to 32% synergistic (TDs ≤−0.05), and 4% to 13% buffering (TDs ≥0.05; Fig. 6E). Most of the downregulated genes were unique to each of the DKOs; however, there were nine commonly shared synergistic and 28 commonly shared additive genes (Fig. 6G; Supplementary Fig. S6B).
The result that oncogenic GIs were associated with epistatic transcriptional alterations led us to hypothesize that epistatic transcriptional alterations induced stronger oncogenic as well as weaker tumor-suppressive functions, underpinning the phenotypic cooperation of two TSGs in promoting both growth in minimal growth factor medium and tumor progression. To test this hypothesis, we performed gene set enrichment analysis. We found that the 148 commonly upregulated synergistic genes were enriched in multiple protein classes including RNA splicing factors, translational and cytoskeletal proteins (Fig. 6H). Their comparative expression in the three SKOs versus three DKOs clearly shows their synergistic upregulation (Fig. 6I). Splicing factors play a key role in mammary cell transformation and breast cancer metastasis (34), as do proteins in the other enriched categories (35–37). The 74 common additive genes showed enrichment only for translational proteins but to a much lesser degree of significance (FDR = 3.6e-02). The nine commonly shared downregulated synergistic genes included TP53TG1 and MALAT1, which have been reported to suppress breast cancer progression and metastasis (38, 39) and were synergistically downregulated in all three DKOs (Fig. 6J).
Transcriptional synergies caused by dual inactivation of cooperating TSGs in our model system are also observed in patient tumors
To determine whether the synergistic expression alterations we observed for the three DKOs in our experimental system could be observed in human breast cancer, we examined the METABRIC human breast cancer genomic dataset that integrates RNA expression, mutational status, and copy-number status (40, 41). Tumors were categorized as being altered for TP53 and PTEN based on their mutational and copy-number status. Because NF2 mutations or homozygous deletions are rare in breast cancer, and because NF2 belongs to the Hippo pathway, we instead used the much more common genetic alterations of the Hippo pathway in human breast cancer—amplification status of the interacting transcriptional activators YAP1 and TAZ (WWTR1) for the categorization of NF2 alteration (42). To examine whether the oncogenic alterations of PTEN, TP53, and YAP1/TAZ show signs of cooperation in human cancer data, we performed co-occurrence/mutual exclusivity analysis using cBioPortal (43). This analysis indicated that these oncogenic alterations had a strong tendency to co-occur (Supplementary Table S1).
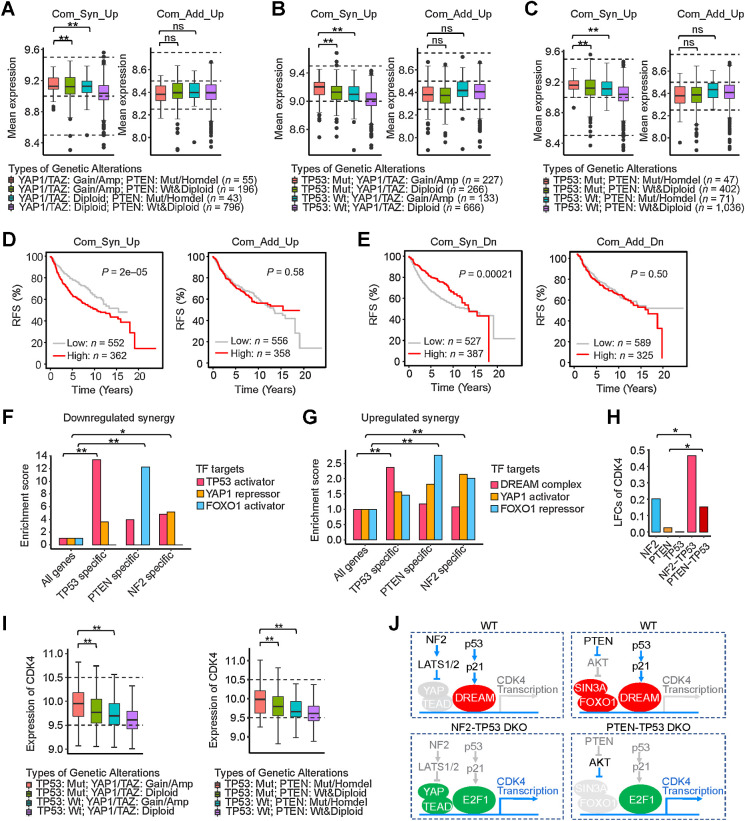
The average expression level of the 148 common synergistically upregulated genes was significantly higher in tumors with dual alterations than in tumors with single alterations (Fig. 7A–C). As further evidence of relevance to human breast cancer, the expression of the 148 common synergistically upregulated genes, but not the 74 common additively upregulated genes, was associated with poorer relapse-free survival in patients with breast cancer (Fig. 7D). In parallel, the average expression of the nine commonly shared synergistically downregulated DEGs was lower in tumors with dual alterations than in tumors with single alterations (Supplementary Fig. S6C) and was also associated with poorer relapse-free survival in patients with breast cancer, and as before, this association was not seen with the 28 common additive downregulated genes (Fig. 7E). These results demonstrate that the epistatic expression effects observed in our experimental system reflect the transcriptional synergies induced by epistatic interactions of driver gene alterations in human breast cancer.
Figure 7.
Transcriptional synergies in patients with breast cancer. A–C, Average expression of the 148 common upregulated synergistic DEGs and 74 common upregulated additive DEGs in human tumors with the indicated dual and single genetic alterations. **, P < 0.05; one-sided Student t test. ns, not significant. D, Relapse-free survival (RFS) of patients with breast cancer with differential expression of 148 common upregulated synergistic and 74 common upregulated additive genes. E, RFS of patients with breast cancer with differential expression of 9 common downregulated synergistic and 28 common downregulated additive genes. F, Enrichment of targets of TFs (p53 activator, FOXO1 activator, and YAP repressor) in the TP53-specific, PTEN-specific, and NF2-specific downregulated synergies. *, P < 0.1; **, P < 0.01; χ2 test. G, Enrichment of targets of TFs (DREAM complex, FOXO1 repressor, and YAP activator) in the TP53-specific, PTEN-specific, and NF2-specific upregulated synergies. **, P < 0.01; χ2 test. H and I, Synergistic expression of CDK4 in TP53-deleted DKO cells and TP53-mutated patients with double-mutant breast cancer. *, P < 0.1; **, P < 0.05; one-sided Student t test. J, Proposed mechanistic models for transcriptional synergy of CDK4 induced in TP53-deleted DKOs—NF2-TP53 and PTEN-TP53.
Transcriptional epistasis is associated with specific transcription factors
We hypothesized that the established primary transcription factors (TF) of each of the three pathways—p53, FOXO1, and YAP—would play a role in mediating the epistatic expression alterations we observed. To test this, we counted how many epistatic expressed genes were direct transcriptional targets of these major individual TFs by querying the ChIP-Atlas database (44). For the downregulated genes, there was selective enrichment of p53 targets among the 20 synergistically downregulated genes shared between PTEN-TP53 and NF2-TP53 DKOs (TP53-specific DKOs) and also selective enrichment of FOXO1 targets among the 17 synergistically downregulated genes in the PTEN-specific DKOs (Fig. 7F; Supplementary Fig. S6D and S6E). Although YAP is generally considered to be a transcriptional activator, it has been shown to also be a transcriptional repressor at roughly equal frequency (45), and we found over 4-fold enrichment of YAP targets for the synergistically downregulated genes in the NF2-specific DKOs (Fig. 7F; Supplementary Fig. S6F).
Unlike FOXO1 or YAP, which can act as either activators or repressors, p53 does not act as a direct transcriptional repressor. Instead, the genes that are repressed by p53 function, including many cell-cycle genes, are mediated by promoter binding of the transcriptional repressor complex DREAM (DP, RB-like, E2F4, and MuvB; refs. 46, 47). Accordingly, we tested whether the genes synergistically upregulated in the TP53-specific pair of DKOs were selectively enriched for genes that are targets of the DREAM complex and found that they were more than 2-fold enriched (Fig. 7G; Supplementary Fig. S6D). Although FOXO1 is generally considered to be a transcriptional activator, it has been shown to also be a transcriptional repressor when bound to the SIN3A corepressor factor (48), and FOXO1 targets were enriched in the genes synergistically upregulated in the PTEN-specific pair of DKOs (Fig. 7G; Supplementary Fig. S6E). In addition, YAP targets were enriched in the genes synergistically upregulated in the NF2-specific pair of DKOs (Fig. 7G; Supplementary Fig. S6F).
TP53 cooperated with NF2 and/or PTEN to synergistically regulate expression of therapeutic drug target genes
Intriguingly, expression of genes encoding multiple drug targets were synergistically regulated by DKOs. For instance, CDK4, a potent drug target in estrogen receptor–positive, Her2-negative (ER+/HER2−) breast cancer and DNMT1, a drug target in triple-negative breast cancer were synergistically increased in the two TP53-specific DKOs (Fig. 7H; Supplementary Fig. S6G). Expression of CDK4 or DNMT1 was also higher in patients with breast cancer with mutant TP53 in combination with alterations affecting the PTEN or Hippo pathways, such that patients with both alterations had higher expression than patients with single alterations (Fig. 7I; Supplementary Fig. S6H; Supplementary Table S1). Expression of SRPK1, another potential breast cancer drug target, was synergistically upregulated in NF2-TP53 DKO (Supplementary Fig. S6I) as well as patients with breast cancer with dual alterations in TP53 and Hippo pathways (Supplementary Fig. S6J; Supplementary Table S1). In addition, according to previous studies and the ChIP-Atlas database (44), the CDK4 promoter contains binding sites for the DREAM complex, E2F1, FOXO1, and YAP in human cells (46, 49). Thus, we propose mechanistic models for the transcriptional synergy of CDK4 expression induced by NF2-TP53 and PTEN-TP53 DKOs (Fig. 7J). Dual deletion of TP53 and NF2 induces dissociation of the DREAM repressor from E2F-binding sites and recruits free activating E2F1–3 and YAP to the CDK4 promoter, cooperatively activating its transcription. Similarly, simultaneous loss of PTEN and TP53 causes the repressive TFs FOXO1/SIN3A and DREAM complex to be replaced with the transcriptional activator E2F1 on the CDK4 promoter, synergistically boosting its expression. These results suggest that oncogenic cooperation between TSGs can be mediated by corresponding TF interactions.
Discussion
The goal of our study was to identify general mechanisms of driver gene cooperation by systematically analyzing how combinations of inactivated TSGs altered the tumorigenic growth characteristics and transcriptomes of breast epithelial cells. Our experimental approach combined combinatorial CRISPR screening for in vivo and in vitro growth phenotypes with parallel profiling for transcriptional alterations using scRNA-seq. We found that the driver gene combinations that cooperatively promoted tumorigenicity were those that showed transcriptional epistasis—that is, hundreds of genes that were differentially expressed in a way that could not be predicted by the additive combination of the effects of the individual driver genes. The same patterns of epistatic expression changes were observed in patients with breast cancer, establishing that these cooperative transcriptional interactions do occur in patients and were not just an anomaly of our experimental system. Our results indicate that approximately 50% of DEGs in cancer cells are influenced by transcriptional epistasis. On the basis of gene set enrichment analysis and association with poor prognosis, transcriptional epistasis is a functionally important mediator of oncogenesis. Together, these results argue that transcriptional epistasis is a central aspect of multigenic cancer progression and a general mechanism for how driver genes cooperate to promote tumorigenicity. In addition, we found shared epistatic transcriptional changes that are independent of specific driver genes, affecting alternative splicing, cytoskeletal assembly, and protein translation—processes that are commonly altered in cancer (37, 50, 51). This result suggests that shared epistatic transcriptional alterations may underlie the ability of diverse configurations of driver gene alterations to converge upon common cancer phenotypes.
Our results also suggest that cooperation between driver genes involves some degree of functional relatedness. GI profiling indicated shared function(s) between the five genes with the most tumor-promoting interactions (PTEN, NF2, TP53, SMAD4, and CBFB). In addition, tumor-promoting epistatic networks contained numerous three-gene cliques, such that all three genes showed oncogenic cooperation with each other, as in the “within-pathway” groups of genes found in the yeast global GI network (26, 52). These results appear to conflict with earlier proposed mechanisms of oncogenic cooperation that posited separate functions rather than functional relatedness. The mechanism of distinct complementary function was based on the observation that oncogenes that cooperated to transform primary rodent fibroblasts either complemented mutant HRAS or complemented overexpressed MYC, but never both (53). However, this functional distinction was later blurred by the experiments showing that overexpression of BMI1 or homozygous deletion of CDKN2A could cooperate with both mutant HRAS and MYC in transforming primary rodent fibroblasts (54, 55). Similarly, the oncogene complementation groups initially identified by retroviral tagging were called into question by the discovery that overexpression of RUNX2 cooperated with all of the groups (56). Another earlier idea at odds with our results was the notion that alterations in driver genes within the same functional pathway rarely, if ever, occurred; however, as more cancer genomes have been sequenced, co-occurrences of driver gene alterations within the same functional pathway have become more the rule rather than the exception. For example, PIK3CA or PIK3R1 aberrations frequently co-occur with PTEN mutations in breast cancer, consistent with our finding that inactivation of PTEN is a powerful driver in MCF10A cells overexpressing mutant PIK3CA (41). Apart from RB1 mutations, other alterations in the RB1 pathway, such as amplification of CCND1 or CDK4 amplification or CDKN2A deletion can frequently co-occur (57). Finally, driver genes that have been studied extensively, such as TP53 and KRAS, are highly pleiotropic and are involved in most of the hallmarks of cancer, including metabolism, apoptosis, invasion, and the tumor microenvironment (58–60). Given such extensive pleiotropy, it is not surprising that there is some degree of functional overlap between many driver genes.
We found that tumor suppressor GI networks have properties that are different than those observed in most molecular interaction networks (e.g., protein–protein interaction or synthetic lethal interaction networks). In these latter networks, highly connected nodes (hubs) tend to link to nodes with fewer interaction partners rather than to other hubs, and this has been proposed to enhance the robustness of the network by localizing the effects of deleterious perturbations (61). In contrast, the hubs of TSG networks tend to link to other hubs, a network property termed assortative, and assortative networks (which include social networks) show superior resilience to the removal of nodes (62). We propose that during the evolution of TSG networks that an assortative network structure provided optimal protection from mutations of one, two, or even multiple TSGs.
There was a striking difference between the dramatic enrichment for double TSG knockouts relative to single knockouts during tumor formation, compared with the lack of any corresponding enrichment in the in vitro conditions. We believe that in vivo tumor formation is a more complex phenotype than restrictive growth in culture. Others have found that by imaging with luciferin that when luciferase-tagged, weakly oncogenic derivatives of MCF10A cells are injected into mice that there is a loss in the luciferin signal over the first week, which likely results from cell death (63). Unlike cells in tissue culture, the cells in vivo are in hypoxic conditions, have less nutrients (e.g., glucose), and lack the cell–cell and substrate contact found in culture. Our data indicate that double TSG knockouts are superior to single TSG knockouts for inducing this complex phenotype. While minimal growth factor medium reflects the in vivo effects better than any other in vitro condition tested, it does not provide a complete explanation.
One of the limitations of our current study is that injection of a single-cell suspension into the mammary gland is not how breast cancer normally develops. Future work using different models that select for tumor progression in a more physiological relevant tissue context will be needed. However, it is clear from our results that GI fitness networks determined using standard cell culture conditions are not representative of GIs in vivo, and that using the restrictive condition of growth factor deprivation generates results that are significantly more representative of in vivo networks. An additional limitation is that we compared networks from different cell culture conditions using the original MCF10A cell line, whereas the in vivo networks were generated out of necessity with MCF10A derivatives containing an additional oncogenic alteration. How these additional oncogenic alterations would have influenced the in vitro results will need to be addressed in the future. Another limitation of our study is that we did not determine a detailed molecular mechanism of how specific transcriptional epistasis occurs. We showed that for some genes, transcriptional epistasis can be linked to the promoters binding the primary transcription factors that mediate driver gene function, but more detailed work needs to be done regarding molecular mechanism(s).
One of the important outcomes of our study is that it demonstrates that driver gene interactions in any cancer can be systematically mapped using combinatorial CRISPR screening of tumor growth or other physiologically relevant growth effects in parallel with transcriptome-wide changes (14–16). While it is possible to find epistatic expression changes without screening, we believe it is important that future work be systematic. Three groups have previously published on genes differentially expressed as a result of alterations in both TP53 and KRAS, and although these reports describe interesting cancer biology associated with these expression changes, they were not systematic analyses comparing several paired alterations, and their relevance to understanding driver gene cooperation and multigenic tumor progression was limited (64–66). It should be noted that one of these groups went on to show synergistic transcriptional alterations induced by the cooperating genetic alterations BCR-ABL and NUP98-HOXA9 in leukemia, suggesting that the synergy they observed with TP53 and KRAS may be a more general phenomena of oncogenic cooperation (67). Their work and our work here suggest that new cancer drug targets might be identified from systematic analyses of transcriptional epistasis caused by driver gene interactions. One of the genes that we found synergistically upregulated in breast cancer cells by the combination of alterations in TP53 and other TSGs was CDK4, which encodes the target of FDA-approved drugs used to treat metastatic ER+/HER2− breast cancer. However, we have no data at this time to indicate whether TP53 status would serve as useful biomarker for CDK4 inhibitors. We acknowledge that our work is only the first step in opening this field and understand that clinical applications are a long way off. In the near term, it will be important to rigorously test the functional impact and druggability of synergistically altered genes to determine whether they could be useful targets for inhibiting tumorigenicity.
Authors' Disclosures
No disclosures were reported.
Supplementary Material
Supplementary Methods, Description of supplementary table and Supplementary Figures/legends
This table lists 52 TSG names, TSG sgRNA sequences, all the oncogenic interactions in vivo, full PIK3CA network, co-occurrence analysis and the epistatic expression of CDK4, DNMT1 and SPRK1in breast cancer patients
Acknowledgments
This project was supported by funding from the NCI (R01 CA217206 to S. Powers) and the National Human Genome Research Institute (R21HG009255 to S. Powers). The authors thank their colleagues at Stony Brook University and Cold Spring Harbor, Gabor Balazsi, Amy Caudy, Pei-Fen Kuan, Adam Rosebrock, and Chris Vakoc for useful discussions and advice. They thank Dongyan Song at Cold Spring Harbor laboratory for help with sgRNA design. The authors thank Xueyan He in the Egeblad laboratory for teaching mammary pad injections. They thank Michele I. Vitolo for providing MCF10A-PTEN−/− cells. They thank lab members Abhay Kanodia and Shaobo Qin for useful comments on the article. The authors thank the staff in the research histology core, the flow cytometry facility, and the Division of Laboratory Animal Resources (DLAR) of the school of Stony Brook Medicine for their technical support, as well as the staff in the Tissue Analytics and Biostatistics & Data Science shared resources of the Stony Brook Cancer Center.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
X. Zhao: Conceptualization, data curation, formal analysis, validation, visualization, methodology, writing–original draft, writing–review and editing. J. Li: Formal analysis. Z. Liu: Formal analysis. S. Powers: Conceptualization, resources, formal analysis, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer 2007;7:441–53. [DOI] [PubMed] [Google Scholar]
- 3. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogelstein B, Kinzler KW. The path to cancer — three strikes and you're out. N Engl J Med 2015;373:1895–8. [DOI] [PubMed] [Google Scholar]
- 5. Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell 2011;145:30–8. [DOI] [PubMed] [Google Scholar]
- 6. Persi E, Wolf YI, Horn D, Ruppin E, Demichelis F, Gatenby RA, et al. Mutation–selection balance and compensatory mechanisms in tumour evolution. Nat Rev Genet 2021;22:251–62. [DOI] [PubMed] [Google Scholar]
- 7. van de Haar J, Canisius S, Yu MK, Voest EE, Wessels LFA, Ideker T. Identifying epistasis in cancer genomes: a delicate affair. Cell 2019;177:1375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rauscher B, Heigwer F, Henkel L, Hielscher T, Voloshanenko O, Boutros M. Toward an integrated map of genetic interactions in cancer cells. Mol Syst Biol 2018;14:e7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boettcher M, Tian R, Blau JA, Markegard E, Wagner RT, Wu D, et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat Biotechnol 2018;36:170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chow RD, Wang G, Ye L, Codina A, Kim HR, Shen L, et al. In vivo profiling of metastatic double knockouts through CRISPR–Cpf1 screens. Nat Methods 2019;16:405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 2017;35:463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, et al. Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat Methods 2017;14:573–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wong ASL, Choi GCG, Cui CH, Pregernig G, Milani P, Adam M, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci U S A 2016;113:2544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 2016;167:1867–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 2017;14:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Norman TM, Horlbeck MA, Replogle JM, Ge AY, Xu A, Jost M, et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 2019;365:786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 2015;33:661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science 2015;350:1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cowell JK, LaDuca J, Rossi MR, Burkhardt T, Nowak NJ, Matsui S. Molecular characterization of the t(3;9) associated with immortalization in the MCF10A cell line. Cancer Genet Cytogenet 2005;163:23–9. [DOI] [PubMed] [Google Scholar]
- 21. Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003;30:256–68. [DOI] [PubMed] [Google Scholar]
- 22. Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 1990;50:6075–86. [PubMed] [Google Scholar]
- 23. Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol 2005;25:6464–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 2018;24:927–30. [DOI] [PubMed] [Google Scholar]
- 25. Pavlopoulos GA, Secrier M, Moschopoulos CN, Soldatos TG, Kossida S, Aerts J, et al. Using graph theory to analyze biological networks. BioData Min 2011;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol 2005;23:561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 2011;5:387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu J, Chen Y, Olopade OI. MYC and breast cancer. Genes Cancer 2010;1:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCabe N, Hanna C, Walker SM, Gonda D, Li J, Wikstrom K, et al. Mechanistic rationale to target PTEN-deficient tumor cells with inhibitors of the DNA damage response kinase ATM. Cancer Res 2015;75:2159–65. [DOI] [PubMed] [Google Scholar]
- 30. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 31. Costanzo M, Baryshnikova A, Myers CL, Andrews B, Boone C. Charting the genetic interaction map of a cell. Curr Opin Biotechnol 2011;22:66–74. [DOI] [PubMed] [Google Scholar]
- 32. Preobrazhenska O, Yakymovych M, Kanamoto T, Yakymovych I, Stoika R, Heldin CH, et al. BRCA2 and Smad3 synergize in regulation of gene transcription. Oncogene 2002;21:5660–4. [DOI] [PubMed] [Google Scholar]
- 33. Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell 2004;119:157–67. [DOI] [PubMed] [Google Scholar]
- 34. Park S, Brugiolo M, Akerman M, Das S, Urbanski L, Geier A, et al. Differential functions of splicing factors in mammary transformation and breast cancer metastasis. Cell Rep 2019;29:2672–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia A, et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 2020;367:1468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol 2014;171:5507–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev 2009;28:5–14. [DOI] [PubMed] [Google Scholar]
- 38. Diaz-Lagares A, Crujeiras AB, Lopez-Serra P, Soler M, Setien F, Goyal A, et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc Natl Acad Sci U S A 2016;113:E7535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 2018;50:1705–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pereira B, Chin SF, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 2016;7:11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maugeri-Saccà M, Barba M, Pizzuti L, Vici P, Lauro LD, Dattilo R, et al. The Hippo transducers TAZ and YAP in breast cancer: oncogenic activities and clinical implications. Expert Rev Mol Med 2015;17:e14. [DOI] [PubMed] [Google Scholar]
- 43. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oki S, Ohta T, Shioi G, Hatanaka H, Ogasawara O, Okuda Y, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep 2018;19:e46255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim M, Kim T, Johnson RL, Lim D-S. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep 2015;11:270–82. [DOI] [PubMed] [Google Scholar]
- 46. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ 2018;25:114–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 2016;44:6070–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Langlet F, Haeusler RA, Lindén D, Ericson E, Norris T, Johansson A, et al. Selective inhibition of FOXO1 activator/repressor balance modulates hepatic glucose handling. Cell 2017;171:824–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jang W, Kim T, Koo JS, Kim S, Lim DS. Mechanical cue-induced YAP instructs Skp2-dependent cell cycle exit and oncogenic signaling. EMBO J 2017;36:2510–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov 2015;14:261–78. [DOI] [PubMed] [Google Scholar]
- 51. Urbanski LM, Leclair N, Anczuków O. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 2018;9:e1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boucher B, Jenna S. Genetic interaction networks: better understand to better predict. Front Genet 2013;4:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinberg RA. Oncogenes, antioncogenes, and the molecular bases of multistep carcinogenesis. Cancer Res 1989;49:3713–21. [PubMed] [Google Scholar]
- 54. Jacobs JJL, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 1999;13:2678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999;397:164–8. [DOI] [PubMed] [Google Scholar]
- 56. Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER, et al. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 2001;20:295–302. [DOI] [PubMed] [Google Scholar]
- 57. Knudsen ES, Nambiar R, Rosario SR, Smiraglia DJ, Goodrich DW, Witkiewicz AK. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun Biol 2020;3:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boutelle AM, Attardi LD. p53 and tumor suppression: it takes a network. Trends Cell Biol 2021;31:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ischenko I, D'Amico S, Rao M, Li J, Hayman MJ, Powers S, et al. KRAS drives immune evasion in a genetic model of pancreatic cancer. Nat Commun 2021;12:1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mukhopadhyay S, Vander Heiden MG, McCormick F. The metabolic landscape of RAS-driven cancers from biology to therapy. Nat Cancer 2021;2:271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science 2002;296:910–3. [DOI] [PubMed] [Google Scholar]
- 62. Newman MEJ. Mixing patterns in networks. Phys Rev E Stat Nonlin Soft Matter Phys 2003;67:026126. [DOI] [PubMed] [Google Scholar]
- 63. Zhou Z, Li M, Zhang L, Zhao H, Şahin Ö, Chen J, et al. Oncogenic kinase–induced PKM2 tyrosine 105 phosphorylation converts nononcogenic PKM2 to a tumor promoter and induces cancer stem–like cells. Cancer Res 2018;78:2248–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Buganim Y, Solomon H, Rais Y, Kistner D, Nachmany I, Brait M, et al. p53 regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res 2010;70:2274–84. [DOI] [PubMed] [Google Scholar]
- 65. Coppé J-P, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 2008;6:2853–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature 2008;453:1112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ashton JM, Balys M, Neering SJ, Hassane DC, Cowley G, Root DE, et al. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell Stem Cell 2012;11:359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Description of supplementary table and Supplementary Figures/legends
This table lists 52 TSG names, TSG sgRNA sequences, all the oncogenic interactions in vivo, full PIK3CA network, co-occurrence analysis and the epistatic expression of CDK4, DNMT1 and SPRK1in breast cancer patients
Data Availability Statement
Raw pair-end amplicon sequencing data in combinatorial screenings are available and can be downloaded from the NCBI Sequence Read Archive (SRA) database with the BioProject ID: PRJNA691742. Raw scRNA-seq, raw enrichment PCR sequencing data, and processed combinatorial CROP-seq data via 10× Cell Ranger count are available and can be downloaded from the NCBI Gene Expression Omnibus (GEO) database with the accession ID: GSE164996. Relevant code and instructions that are used to reproduce the principal results presented in this study, are provided on GitHub: https://github.com/Xiaoyu-Zhao/Oncogenic_GI_screening.git.
Additional information can be found in the Supplementary Materials and Methods.