Integration of ex vivo drug sensitivity testing with genomic and transcriptomic profiling via a functional and molecular tumor board in AML uncovered new opportunities for individualized patient therapy, functional taxonomy, and discovery of biomarkers.
Abstract
We generated ex vivo drug-response and multiomics profiling data for a prospective series of 252 samples from 186 patients with acute myeloid leukemia (AML). A functional precision medicine tumor board (FPMTB) integrated clinical, molecular, and functional data for application in clinical treatment decisions. Actionable drugs were found for 97% of patients with AML, and the recommendations were clinically implemented in 37 relapsed or refractory patients. We report a 59% objective response rate for the individually tailored therapies, including 13 complete responses, as well as bridging five patients with AML to allogeneic hematopoietic stem cell transplantation. Data integration across all cases enabled the identification of drug response biomarkers, such as the association of IL15 overexpression with resistance to FLT3 inhibitors. Integration of molecular profiling and large-scale drug response data across many patients will enable continuous improvement of the FPMTB recommendations, providing a paradigm for individualized implementation of functional precision cancer medicine.
Significance:
Oncogenomics data can guide clinical treatment decisions, but often such data are neither actionable nor predictive. Functional ex vivo drug testing contributes significant additional, clinically actionable therapeutic insights for individual patients with AML. Such data can be generated in four days, enabling rapid translation through FPMTB.
See related commentary by Letai, p. 290.
This article is highlighted in the In This Issue feature, p. 275
INTRODUCTION
High-dose chemotherapy and allogeneic hematopoietic stem cell transplantation (alloHSCT) can cure up to 60% of younger patients with adult acute myeloid leukemia (AML), but many patients relapse and suffer from lifelong toxicities of treatment (1). Patients with refractory or relapsed (R/R) AML, particularly older ones, have limited treatment options, and survival has remained poor (1, 2). Genomic profiling has helped to deconvolute the biological basis, heterogeneity, and clonal evolution of AML and highlighted novel therapeutic targets and subgroups (3–6). The FLT3 inhibitors (FLT3i) midostaurin (7) and gilteritinib (8), the IDH1-mutant inhibitor ivosidenib (9), and the IDH2-mutant inhibitor enasidenib (10) offer new genetically guided treatment options for patients with AML. However, only a fraction of patients harbor these mutations, and even fewer respond to the treatments assigned by genetics (11, 12). Furthermore, often no actionable mutations are seen to guide therapy decisions, and many therapies do not even have any confirmed (genomic) biomarkers (13, 14). For example, the BCL2 inhibitor (BCL2i) venetoclax can provide significant clinical benefits in AML therapy, but we lack effective biomarkers to identify patients likely to benefit (15–17).
We and others have utilized high-throughput ex vivo testing of AML cells to functionally identify drug response patterns (18–24). The Beat AML study reported on the functional testing of 122 small-molecule inhibitors combined with genomic profiling in a cohort of 562 biobanked AML patient samples (23). Snijder and colleagues applied an image-based drug testing assay (pharmacoscopy) to demonstrate that the assay predicted clinical response to chemotherapy (21). Flow cytometry has also been used to quantify responses in distinct cell subpopulations (19, 25–27). Nevertheless, many of these studies are retrospective and lack the integration of functional and molecular data. Prospective implementation of these assays in the clinical decision-making process is needed.
Here, we performed ex vivo drug sensitivity and resistance testing (DSRT) of up to 347 emerging and 168 approved cancer drugs in AML patient cells. Molecular and functional data were interpreted and integrated for individual patients to consider novel therapy options for patients with R/R AML. To implement the results in real time for clinical translation, we designed a multidisciplinary functional precision medicine tumor board (FPMTB). We report here (i) that ex vivo DSRT is informative in highlighting the cancer-specific efficacy of drugs; (ii) that drug responses combined with molecular profiles give actionable data for the treatment of R/R AML; and (iii) that therapies tailored based on FPMTB recommendations are effective and provide clinically meaningful responses. The three-day DSRT assay provided actionable data faster than genomics and transcriptomics profiling, which was advantageous in rapidly recommending treatments for patients for whom standard therapy alternatives had been exhausted and for whom alternative therapy options were urgently needed.
RESULTS
FPMTB Workflow and Criteria
To quickly identify and implement selective treatment options for R/R AML with the help of ex vivo drug testing, we set up a multidisciplinary FPMTB in a large single-center academic hospital setting. The FPMTB prospectively reviewed 61 patients with R/R AML utilizing clinical parameters, ex vivo drug testing from each individual patient, as well as genomic and transcriptomic profiling data when these were available in time. To uncover novel associations of genotypes and drug response phenotypes, we also performed the multiomic profiling of all newly diagnosed patients with AML (Fig. 1).
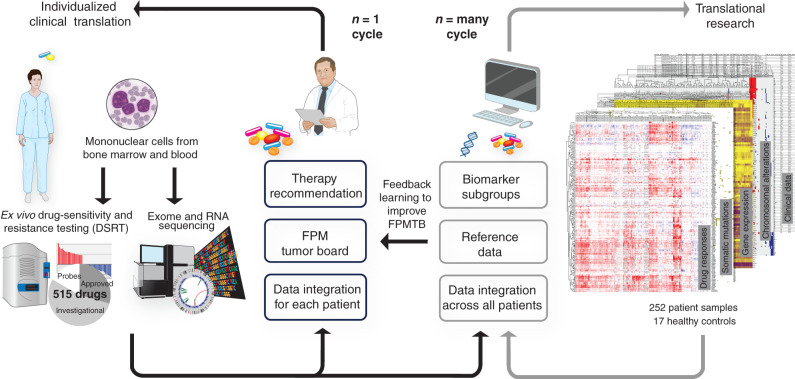
Figure 1.
Schematic of the design for the functional precision systems medicine study. The diagram illustrates how functional molecular precision systems medicine integrates high-throughput drug response assay and molecular profiling, aiming at individualized clinical translation of data for patients with AML. The n = 1 cycle on the left illustrates prospective real-time clinical translation through an FPMTB approach. The drug response and sequencing data are analyzed and integrated within a patient with a goal to tailor therapies in a realistic time frame. The n = many cycle on the right illustrates the data integration across a large sample set. The main goal here is to find possible biomarkers of drug responses, which eventually could also help to refine the rules of the FPMTB.
The FPMTB composition, aims, and treatment selection algorithms are presented in Supplementary Table S1, and the FPMTB patient and sample flow is depicted as a CONSORT diagram in Supplementary Fig. S1 and the meeting scheduling in Supplementary Fig. S2. Data available at the FPMTB meetings included: (i) full clinical patient history, (ii) diagnostic workup (laboratory values, cytogenetics, and clinical mutation data), (iii) ex vivo drug-sensitivity testing with 515 anticancer drugs, (iv) whole-exome sequencing, and (v) transcriptomics sequencing data. Treatments for R/R AML need to be determined quickly, and therefore genomic and transcriptomic profiling data were often not available in time for the clinical decision-making. In contrast, ex vivo drug testing has a turnaround time of four days (three-day assay, one day for data analysis). Thus, the treatment recommendations by the FPMTB were primarily based on drug-sensitivity testing, complemented with clinical disease history, routine molecular diagnostics (e.g., flow cytometry, cytogenetics, FLT3-ITD, NPM1, IDH1/2, and WT1 mutation status), with support from genomics and transcriptomics when such data were available. If the bone marrow blast count was low (<20%), as was observed in five relapsed patients, blast-specific drug sensitivity was assayed using flow cytometry.
FPMTB-Guided Therapy Led to Successful Responses in 59% of Patients with R/R AML
The FPMTB recommendations were implemented for 37 R/R patients, of whom 29 were eligible for objective response evaluation. The criteria for the selection of individual drugs and combinations are shown in Supplementary Table S1. The most frequently used targeted drugs for the clinical implementation of the FPMTB data were venetoclax (BCL2i), dasatinib (inhibitor of ABL1 and other kinases), sunitinib (VEGFR- and FLT3-kinase), and temsirolimus (mTORi). The drugs were administered as a customized combination of two to three drugs based on patient-specific sensitivity to single drugs and molecular data (Supplementary Table S2). Treatment was given to individual consenting patients as prospective n = 1 case studies, not as a formal clinical drug study with case and control groups.
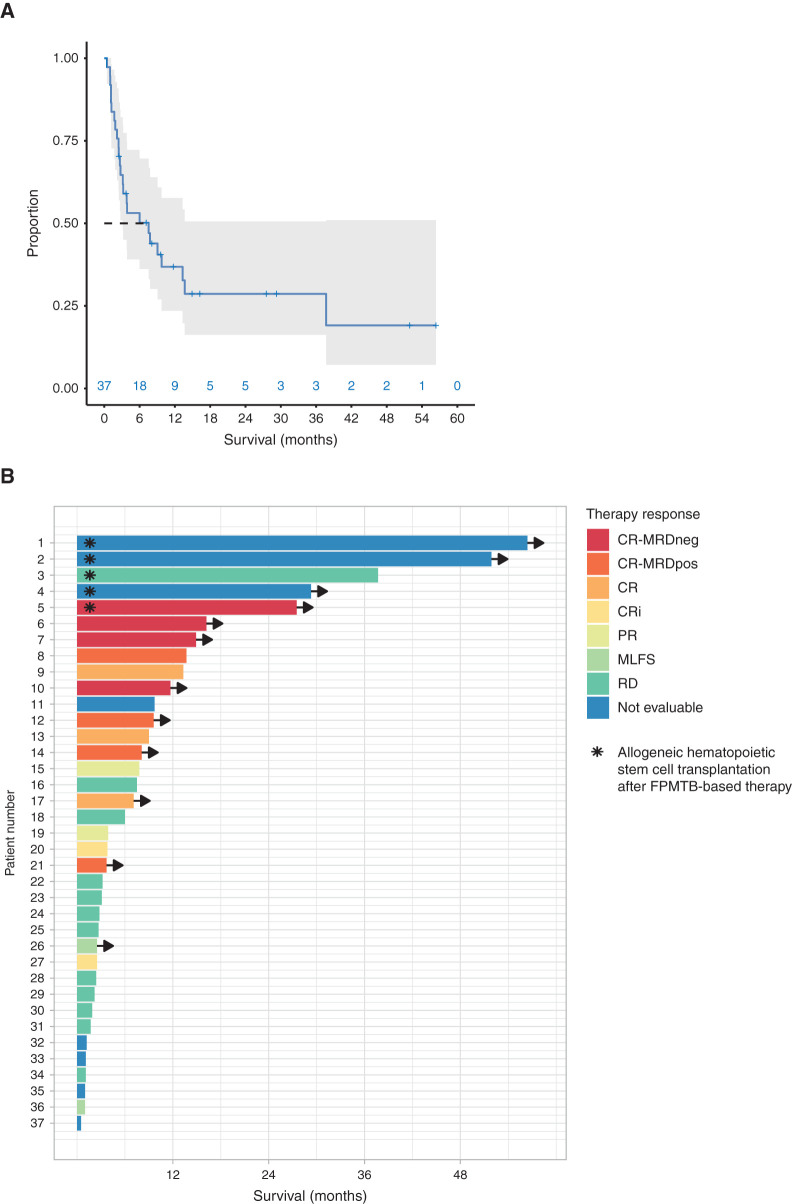
Implementing the FPMTB-recommended therapy resulted in clinically meaningful responses (complete and partial responses, morphologic leukemia-free state, evaluated per ELN2017 criteria; ref. 28) in 17 of 29 patients (59% objective response rate), including 13 (45%) patients achieving complete remission/complete remission with incomplete hematologic recovery (CR/CRi) after treatment (Fig. 2A). The remaining 12 patients progressed with resistant disease. The median time from the beginning of therapy to CR was 36 days (range, 23–110 days). In five R/R patients with no other conventional treatment options, the targeted treatments given enabled the patient to be bridged to alloHSCT, which then resulted in a long-term remission and survival. The time from the beginning of therapy to transplantation ranged from 1.3 to 37.7 months (mean, 10.7). The median overall survival for all 37 patients was 7.5 months (range, 14 days–4.7+ years), with a median follow-up for surviving patients of 23 months (range, 13–66 months; Fig. 2B). Comparing the drugs applied in clinical translation, the best predictive value of the DSRT was observed for venetoclax: 11 of 15 patients who showed ex vivo sensitivity had a CR/CRi response to venetoclax therapy (positive predictive value 73%; Supplementary Fig. S3A). We further explored expression of BCL2 family members and found that the expression of BFL1 may be associated with venetoclax resistance whereas expression of MCL1 does not associate with ex vivo response to venetoclax (Supplementary Fig. S3B–S3E).
Figure 2.
The outcome of patients treated with FPMTB-guided personalized therapies. A, The overall survival estimated by the Kaplan–Meier method of all patients (gray area denotes 95% confidence interval). B, Swimmer plot illustrates survival and therapy responses in 37 patients with R/R AML upon FPMTB-guided therapies, where the asterisk represents patients who received allogeneic hematopoietic stem cell transplantation after the treatment, and arrows represent the patients who are alive. The zero month represents the starting time point of the FPMTB-recommended therapy. The therapy responses—CR-MRDneg, complete response with minimal residual disease negative; CR-MRDpos, complete response with minimal residual disease positive; CR, complete remission; CRi, complete remission with incomplete hematologic recovery; PR, progressive disease; MLFS, bone marrow blasts <5%; RD, resistant disease—were defined by ELN-2017 criteria.
Novel drug combinations, such as those used in this study, may result in novel, unexpected toxicities. However, as most of the drugs used in clinical translation were molecularly targeted agents, with limited toxicity profiles as single-agent drugs, we did not observe any grade 3 to 4 adverse effects attributable to the drug combinations used.
Overview of Drug Response and Molecular Profiling Data
In order to reveal molecular patterns underlying drug responses, we generated comprehensive functional, genomic, and transcriptomic data from 252 consecutive samples from 186 individual patients with AML. By assembling a large database where molecular and functional data can be mined, we think we can eventually improve the FPMTB rules and clinical implementation. The data set includes (i) disease status information for 252 samples; (ii) DSRT assay details for 164 patient and 17 healthy control samples; (iii) drug-sensitivity data for each sample, along with full annotated data from the 515 chemical compounds; (iv) exome-sequencing genomic data for 226 samples; (v) RNA-sequencing (RNA-seq) gene-expression and fusion gene data for 163 patient samples; and (vi) DSRT and RNA-seq data for four healthy control samples. The AML patient cohort and sample information are given in Supplementary Tables S3 and S4. The overview of mutation and gene-expression patterns across all samples is given in Supplementary Figs. S4 and S5A–S5C.
Ex Vivo Drug Responses Providing Functional Insights in AML
Ex vivo drug-sensitivity and resistance profiles were determined for 164 consecutive AML patient samples by high-throughput testing of a library of 515 chemical compounds (Supplementary Tables S5 and S6). This analysis revealed selective efficacy profiles for individual drugs in individual patients, hence enabling us to determine the proportion of samples sensitive to each of the drugs in the library (Supplementary Tables S7 and S8). The DSRT was performed in mononuclear cell medium (MCM) or conditioned medium (CM), which provide different response levels for the FLT3, BET, and JAK inhibitors, as shown previously (29). The data for the two media types were therefore analyzed separately. The drug responses were quantified as an area under the curve (calculated as described previously; ref. 30) and then normalized using data from healthy control and expressed as selective drug-sensitivity scores (sDSS). The distribution of blast cell percentage and cell viability without drug treatment is given in Supplementary Fig. S6A. These two variables were not significantly associated with drug responses in the 164 samples (Supplementary Fig. S6B). The cutoff for a significant drug response was defined by an sDSS value of 8.7 that represents the 95th percentile of the sDSS distribution of all drugs in all cases (Supplementary Fig. S7A). The most often effective 50 drugs that were most frequently effective were further categorized into subclasses based on their known mechanisms of action, such as BCL2i, PI3Ki, HSP90i, JAKi, MEKi, CDKi, and BETi (Supplementary Fig. S7B). We observed that about half of all the chemical compounds (n = 272) in the drug library were effective in three or more samples (Supplementary Fig. S7C). Of these ex vivo effective drugs, 15% (n = 77) were drugs that were already approved for some oncology indications and hence could readily be repurposed for AML. The effective drugs included chemotherapeutics such as topoisomerase inhibitors but also targeted drugs, e.g., tyrosine kinase inhibitors, immunomodulators, mTORi, JAKi, MEKi, BCL2i, CDKi, and PI3Ki (Supplementary Fig. S7D).
Drugs with the same mode of action or with the same molecular targets often clustered together as expected (Supplementary Fig. S8A). Interestingly, drugs representing different molecular mechanisms also sometimes showed clustering. For example, we found a strong association of responses to the BCL2i venetoclax and the MDM2 antagonist idasanutlin. These two drugs clustered in their own branch of the dendrogram with a significant correlation (Supplementary Fig. S8B). Another example of drugs with different molecular mechanisms showing similar response patterns was the coclustering of BETi, MEKi, and HDACi (Supplementary Fig. S8C), with correlation coefficients between the individual drugs ranging from 0.64 to 0.71 (P < 0.001; Supplementary Fig. S8D).
We then analyzed drug responses in paired diagnostic and relapsed samples from the same patients, tested in identical conditions. We found that the average responses to BCL2 and PI3K/mTOR inhibitors were higher at the time of diagnosis. In contrast, responses to MEK inhibitors and dexamethasone responses were stronger in the relapsed samples (Supplementary Fig. S9).
Drug Sensitivities Associated with Mutations
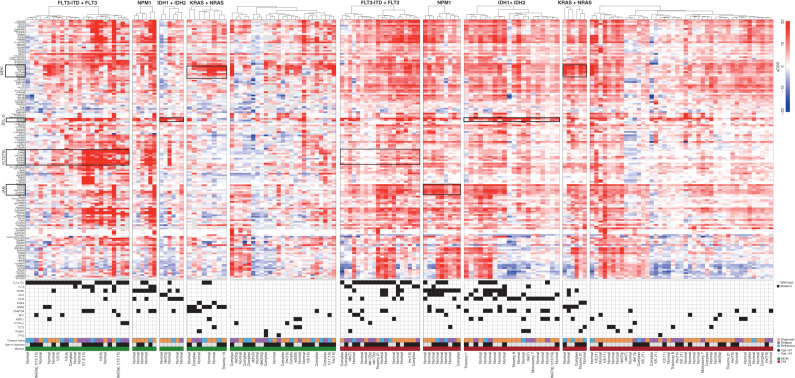
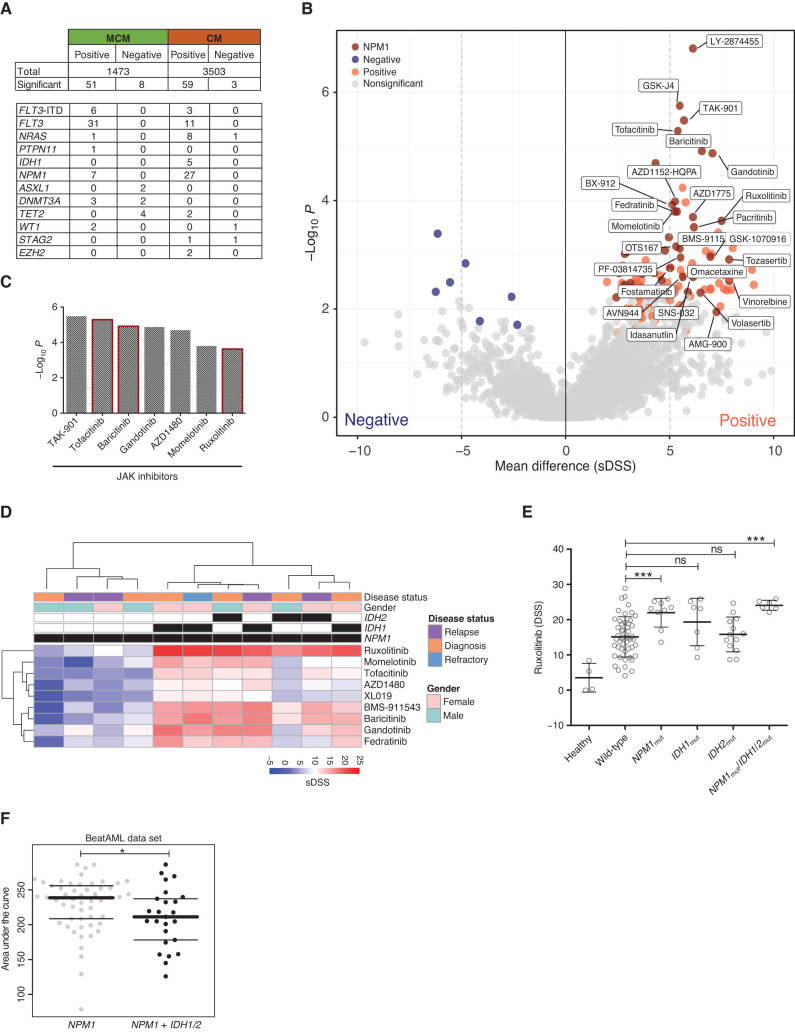
We first analyzed ex vivo drug-response profiles of all AML patient samples in distinct molecular groups defined by the common AML mutations (FLT3, NPM1, IDH1 or IDH2, and NRAS or KRAS) to obtain an overview of genomic subset-specific drug response (Fig. 3). For this analysis, 146 drugs were selected based on sample-wise average sDSS values (>5) and variance (>10; Supplementary Fig. S10A). A total of 121 significant associations were observed between mutations (Supplementary Fig. S10B) and responses of individual drugs. Many of these associations were between a mutation and several drugs of the same class (Fig. 4A; Supplementary Table S9). Previously known findings included the association of FLT3 mutations (point mutations and ITD) with response to several FLT3 and tyrosine kinase inhibitors (23), or between RAS mutations with different MEKi (ref. 31; Supplementary Fig. S11A and S11B). The FLT3-mutated samples, including ITD and point mutations, clustered functionally in two distinct subgroups based on the response patterns to FLT3i and other multikinase inhibitors. When we compared the average sDSS values for all drugs in FLT3-mutant and RAS-mutant samples, a clear pattern emerged where all MEKi were more effective in RAS-mutant and all FLT3i in FLT3-mutant AML samples (Supplementary Fig. S11C). Similar segregation between key genetic lesions causing constitutive activation of cytokine receptor STAT5 and RAS–MEK signaling pathways was found in B-cell acute lymphoblastic leukemia (B-ALL; ref. 32). NPM1-mutated samples showed ex vivo sensitivity to JAKi. This was observed for drugs targeting JAK1i, JAK2i, and pan-JAKi, particularly in the CM (Fig. 4B and C). NPM1-mutated cases that also harbored IDH1 or IDH2 mutations were even more strongly sensitive to all JAKi (Fig. 4D), including the clinically approved JAK1/2i ruxolitinib (Fig. 4E). This observation was also validated in the Beat AML data set (Fig. 4F). We then analyzed combinations of two mutations predicting stronger sensitivity to individual drugs using the LOBICO method (33). For example, navitoclax sensitivity was strongly associated with IDH1 and IDH2 mutations (Supplementary Fig. S12).
Figure 3.
Drug-response patterns in molecular subsets of AML. AML patient samples were categorized into molecular subclasses according to mutation status in common AML driver genes. Hierarchical clustering of samples using Euclidean distance and ward linkage for sDSS of 146 drugs in individual molecular subsets. The drugs were selected considering variance >10 and data points available in at least 20% of the samples. Gray bars in the drug-response heatmap indicate missing data. Fourteen recurrent AML driver genes, with at least three samples recurrently mutated and VAF > 25%, were displayed to indicate the mutation patterns in the molecular subsets. The disease status, age, medium used for drug testing, and cytogenetics information for each patient are displayed in the lower panel.
Figure 4.
Somatic mutations as a molecular denominator of drug sensitivities. A, An overview of the mutation–drug response association analysis results. The top part of the table lists the total number of positive (drug sensitivity) and negative (drug resistance) gene–drug associations identified separately in the MCM and CM sample sets. The associations at FDR < 0.1 were considered significant. The lower part shows the number of drugs significantly associated with each selected gene mutation. For example, the FLT3-ITD mutation is positively associated with six drug responses in the MCM sample set and with three drugs in the CM sample set. B, The volcano plot illustrates the mean difference of sDSS values on the x-axis and adjusted P values on y-axis for each drug–gene pair in the CM sample set. The significant (FDR < 0.1, mean difference >5 or ≤5) drug–gene pairs were highlighted in blue (negative associations) or red (positive associations) where dark red dots show significant positive associations with the NPM1 gene. C,P values for NPM1 mutation–associated JAKi including the approved drugs ruxolitinib, baricitinib, and tofacitinib in the CM sample set. D, Hierarchical clustering of NPM1-mutant samples and sDSS of nine JAKi divide samples into two distinct subsets based on the presence of IDH1 or IDH2. E, The coexisting IDH1 or IDH2 in NPM1-mutated samples were significantly associated with strong JAKi sensitivity. F, The same association was significantly observed for ruxolitinib in the Beat AML data set (23).
Actionability of Mutation Data in AML
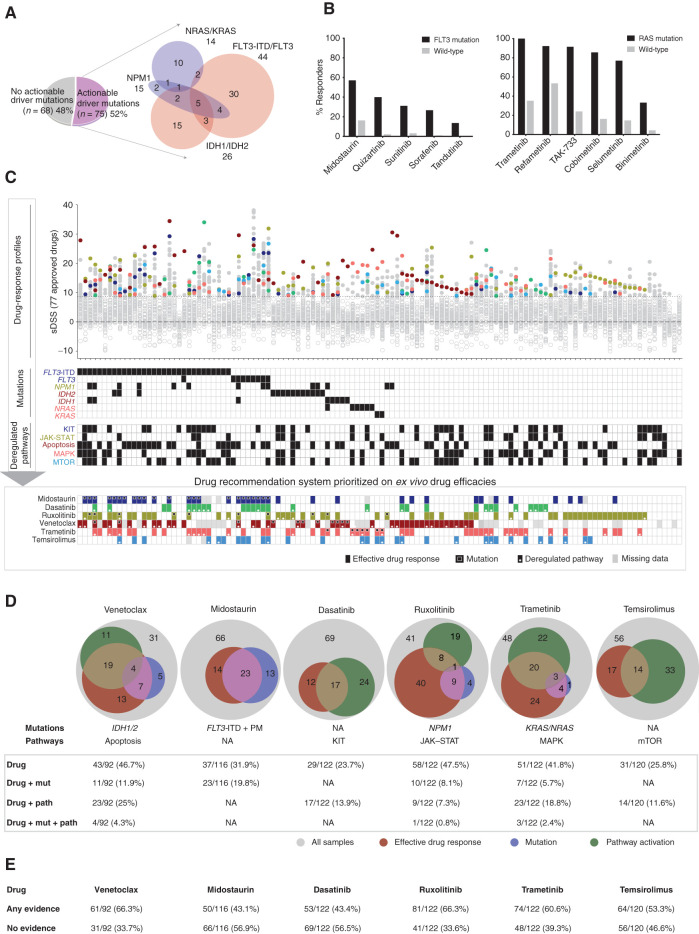
Samples were divided into those with an actionable driver mutation (52%) and those without (48%; Fig. 5A). The mutations considered “potentially actionable” included FLT3-ITD and FLT3 (point mutation), IDH1/2, NPM1, and KRAS/NRAS. The efficacy of FLT3i in FLT3-mutant AML and venetoclax in patients with IDH1/2-mutant AML, as well as increased efficacy of JAKi in NPM1-mutated samples and of MEKi in KRAS/NRAS-mutated samples represent significant associations of mutations with ex vivo drug efficacies in our study. However, not all samples carrying these mutations were sensitive to the corresponding drugs (Fig. 5B). Furthermore, efficacies of these drugs were also observed in cases where no mutations in the corresponding genes were found. For example, 16% of the FLT3 wild-type cases responded to the FLT3i midostaurin, and 35% of the RAS wild-type cases also responded to the MEKi trametinib.
Figure 5.
Genomic and transcriptomics-based prediction of ex vivo drug efficacies. A, The division of 143 AML patient samples in actionable and nonactionable subsets. B,Ex vivo drug sensitivity of FLT3i in FLT3-ITD and point mutation–positive samples and of MEKi in KRAS/NRAS mutation–positive samples. C, Samples with complete molecular profiling and drug-response data ordered as per actionable driver mutations and subsequently nonactionable mutations. Selective drug responses for FDA/EMA-approved 77 drugs are depicted on the Y-axis and individual patient samples on the X-axis, where ineffective drugs below sDSS 8.7 were marked with gray rings. The common effective drugs were highlighted for integration with mutation and pathway activation. The bottom panel illustrates integrated ex vivo efficacy and the presence of respective mutations and pathways for each sample. D, Statistics of patient samples showing evidence of drug sensitivity, the presence of mutation, and pathway activation for key targeted drugs in AML including the BCL2i venetoclax, FLT3i midostaurin, TKi dasatinib, JAKi ruxolitinib, MEKi trametinib, and JAKi ruxolitinib. E, The drug-wise percentage of samples showing any evidence and no evidence from effective drug response, mutation, and/or pathway upregulation.
Integration of Drug Response with Mutations and Pathway Data: A Basis for Continuous Development and Refinement of FPMTB Rules
In 97% of the samples (119/122), drug response data alone provided potential clinically applicable information for approved drugs (Fig. 5C, drug response panel). We found that activity of key pathways measured by gene expression (34) was associated with drug sensitivities in cases where mutations were not informative. Thus, by incorporation of ex vivo DSRT, mutations, and gene-expression data, we were able to define groups of patients responding to drugs based on three lines of evidence. For example, such associations were defined for midostaurin with FLT3mut, ruxolitinib with NPM1mut–JAK–STAT pathway, venetoclax with IDH1/2mut apoptosis pathway, trametinib with RASmut–MAPK pathway, dasatinib with KIT pathway (as was previously observed in a smaller sample set; ref. 35), and temsirolimus with mTOR pathway activation (Fig. 5C; Supplementary Fig. S13A–S13C). We believe that the combination of DSRT, genomic, and transcriptomic data will provide a means to further improve the reliability of the FPMTB recommendations for specific therapeutic alternatives. To demonstrate this, we quantified the overlap among drug efficacies and corresponding mutation and pathway activities. For example, expression of the BCL2 pathway genes (n = 23) was more concordant with ex vivo efficacy of venetoclax as compared with IDH1/IDH2 mutation (n = 4). The combination of mutation and deregulated pathways together gave complementary data in support of ex vivo drug responses (Fig. 5D and E). The results suggested that a systematic data-driven strategy combining all the profiling data will enable further refinement of drug response predictions.
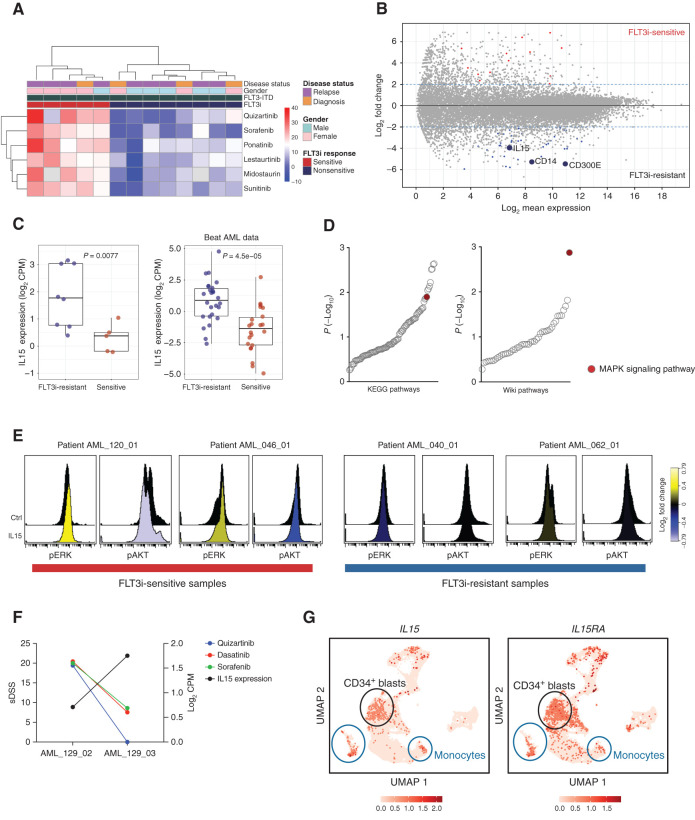
IL15 Overexpression as a Functional Biomarker for Resistance to FLT3 Inhibitors
This integrated data resource on AML provides insights on biomarkers of drug response and potential mechanistic insights that could help us to understand the sensitivity, resistance, and development of combinatorial therapies. We analyzed here the value of transcriptomic data in predicting the response to FLT3i in the subgroup of FLT3-mutant AML cases. Analogous to the clinical situation, FLT3-ITD–mutant AML samples clustered in two distinct groups based on DSRT data, indicating that about half of these patients are responsive (Fig. 6A). Analysis of differential gene expression between FLT3i-sensitive versus FLT3i-resistant AML patient samples resulted in 57 genes with FDR < 0.1 and a log fold change >2 (Supplementary Table S10). We discovered IL15 as one of the most significantly overexpressed (average 4-fold upregulated) genes in FLT3i-resistant samples (Fig. 6B). This was validated in FLT3i-resistant samples by RT-qPCR (Supplementary Fig. S14A). Furthermore, data mining of the Beat AML data set confirmed this association (Fig. 6C; Supplementary Fig. S14B). In line with Mathew and colleagues (36), we observed that the addition of recombinant IL15 protein reduced the sensitivity of FLT3-ITD–mutated AML cell lines to FLT3i, sorafenib in particular (Supplementary Fig. S14C). The MAPK pathway was the most upregulated [false discovery rate (FDR) < 0.001] pathway in the FLT3i-resistant cases (Fig. 6D). We then tested ERK phosphorylation after IL15 stimulation in AML patient samples using phospho-flow cytometry analysis. The treatment of human recombinant IL15 markedly increased phosphorylation of ERK and reduced phosphorylation of AKT in FLT3i-sensitive samples compared with FLT3i-resistant samples (Fig. 6E; Supplementary Fig. S14D). The results suggest not only that IL15 may be a biomarker but that the overexpression and production of IL15 protein may activate the ERK–MAPK pathway and hence contribute to FLT3i resistance. Finally, our ex vivo DSRT data from the same samples indicated a higher efficacy of MEKi in the FLT3i-resistant samples compared with the sensitive samples, suggesting activation of this pathway leads to subsequent MEKi sensitivity, which could provide novel treatment options to be tested in clinical trials (Supplementary Fig. S14E and S14F). We retrospectively assessed whether IL15 expression could predict the clinical response to FLT3i. We validated the high expression of IL15 in the patients who did not respond to the FLT3i-based treatments (Supplementary Table S11). Also, higher expression of IL15 along with the development of resistance to FLT3i was seen in serial samples from patient AML_129, thus further validating a potential functional link between IL15 and FLT3i resistance (Fig. 6F).
Figure 6.
IL15 overexpression is associated with resistance to FLT3i in FLT3-ITD+ cells. A, Hierarchical clustering of FLT3-ITD–positive AML patient samples and six FLT3i resulted in two groups of the samples with high (sensitive group) and low (resistant group) efficacy to FLT3 inhibitors (FLT3i). B, Differential gene expression of FLT3i-sensitive versus FLT3i-resistant samples depicts upregulation of the IL15, CD14, and CD300E genes in the FLT3i-resistant group. C, The overexpression of IL15 was significant in FLT3-ITD–mutant FLT3i-resistant samples in our data and in the Beat AML data set. D, Gene set enrichment analysis of the genes upregulated in FLT3i-resistant samples depicts the MAPK pathway as the top enriched pathway. E, AML patient cells stimulated with human recombinant IL15 had increased phosphorylation of ERK compared with unstimulated control cells using phospho-flow cytometry. The color bar displays phosphorylation ratio to the control cells. F, FLT3i sensitivity and expression of IL15 in serial samples from the patient AML_129. G, The UMAP plots demonstrate expression of IL15 and IL15 receptor (IL15RA) in eight AML patient samples from published study (Dufva et al.; ref. 37).
To explore the origin of the IL15 and the nature of the signaling in patient samples in vivo, we explored both the original RNA-seq data and a new data set of single-cell RNA-seq (scRNA-seq) data from eight AML patient samples in a previously published study (37). scRNA-seq data indicated that both IL15 and IL15 receptor are expressed not only in many cell types, including the monocytic lineage as expected, but also in the CD34-positive AML blast cells (Fig. 6G; Supplementary Fig. S15A–S15C). The bulk RNA-seq data indicated that the FLT3i-resistant FLT3-ITD–mutant AML cells have a higher expression of not just IL15, but also the monocyte markers CD14 and CD300E (Fig. 6B; Supplementary Table S10). Taken together, these findings suggest that in the FLT3i-resistant cases, production of IL15 takes place in the blast cells and/or within the monocytic lineage and that these cells also harbor the IL15 receptor. These findings are compatible with an autocrine signaling hypothesis, although paracrine signaling from other cell types or the stroma cannot be excluded.
DISCUSSION
Multidisciplinary molecular tumor boards increasingly interpret cancer genomic data in order to match patients with cancer to clinical trials with targeted agents or to allocate novel clinical treatments to patients with cancer (38, 39). However, for many patients, genomic analyses often fail to provide clues on clinically actionable therapies (13, 14). Furthermore, across all cancer types, fewer than half of the patients receiving genetically assigned approved therapies successfully respond as predicted (40, 41).
Here, we developed and implemented an FPMTB to integrate functional drug testing with genomics, transcriptomics, and clinical laboratory data to define patient treatments. The FPMTB processed consecutive patients with AML during 2011 to 2019 and recommended therapeutic options for individual R/R patients. The outcome of FPMTB-recommended individualized treatments in 37 patients with multirefractory, often end-stage AML was encouraging, with an overall response rate of 59%. Five patients could be bridged to curative hematopoietic stem cell transplantation therapy. In many of these patients, the FPMTB-guided therapy was started at a low disease burden [minimal residual disease (MRD)]. Persistent MRD is a major cause for treatment failure in AML and may be the ideal setting for implementing personalized targeted therapies.
The response and survival rates of these patients warrant a randomized, controlled clinical trial to be launched to formally validate the benefit of FPMTB-based therapeutic recommendations. The approach has become more and more informative over the years, as the number of clinically approved, better-tolerated drugs has increased (13, 42). We found that the clinical efficacy of venetoclax could be predicted by ex vivo testing in AML. This is of particular importance as this drug is a major advance in AML, and also had a significant impact on the positive clinical responses in the present study. Particularly in the R/R AML setting, where many patients do not respond well to venetoclax (or experience short responses), an ex vivo drug-sensitivity assay may prove valuable for selecting patients most likely to respond and directing nonresponding patients to alternative therapies.
Compared with genomics-based precision medicine, ex vivo testing provides informative results in a substantially higher fraction of patients as well as for more drugs. One or more clinically applicable drugs were considered selectively effective in 97% of the evaluated AML cases. Furthermore, ex vivo drug testing assay provided results in a clinically applicable time frame (median of 4 days), and with comparative efficacy estimates across all the 515 tested drugs in each sample. This timescale is particularly relevant for aggressive, rapidly progressing cancers such as AML.
There is a need to further standardize the approaches used for functional laboratory testing and the molecular analyses used to characterize each patient sample. For example, we have documented the effect of different media types (regular and stromal cell conditioned media; ref. 29) and readouts (cell viability and flow-cytometric assays) on drug testing results (26). Further refinement and standardization will improve our ability to predict drug responses in the clinic as well as to understand the driver signals and vulnerabilities of each patient with AML.
The large integrated data set described here enables continuous improvement of the FPMTB rules as well as exploration of the data to identify potential biological insights and biomarkers of drug efficacy in subsets of patients. The continuous improvement of FPMTB could, in the future, include machine learning–based decision trees as key components of a learning health care infrastructure. Our analysis of these data has already revealed insights on mechanisms of action that could be clinically applied. For example, our data suggested that IL15 may both act as a biomarker and functionally contribute to FLT3i resistance in FLT3-ITD–mutant AML. This observation was validated in independent gene-expression data from the Beat AML data set (23). The administration of recombinant IL15 protein reduced efficacy of FLT3i in FLT3-ITD–positive AML cells in vitro. The mechanistic link between IL15 and FLT3 was previously proposed by Mathew and colleagues (36). We showed how IL15 increased phosphorylation of ERK in FLT3i-resistant but not in FLT3i-sensitive samples, pointing to the ERK–MAPK pathway as a possible escape route for FLT3 inhibition (43, 44). FLT3i-resistant FLT3-ITD–mutant AML cells showed ex vivo sensitivity to MEK inhibition, suggesting potential combinatorial strategies for future clinical studies. Data integration also revealed insights into the role of AML blasts and monocytes in producing IL15, including a potential signaling loop involving the IL15 receptor.
A major hurdle in implementing precision medicine is the limited access to potentially effective drugs for patients. Many drugs showing ex vivo efficacy are neither available for off-label indications nor approved at all, or not even in clinical trials. There are also financial, legislative, and policy-related implications that make the design of clinical drug studies for individually tailored (combinatorial) therapies challenging. However, n = 1 proof-of-concept studies as described here should be highly encouraging and informative for the design of formal clinical studies. The FPMTB approach and the specific findings on ex vivo drug response described here should be explored to set up international multicenter collaborations between private and public stakeholders to solve issues that currently hinder the application of individually tailored functional precision medicine.
In conclusion, ex vivo drug testing is a powerful approach for understanding AML biology and drug sensitivity as well as for facilitating repositioning known and emerging drugs for AML therapy. Systematic data integration prioritizes the most promising drugs and biomarkers for drug development and clinical trials. Although further research is warranted, the combination of molecular and functional assays is warranted in a clinical cancer drug trial setting.
METHODS
AML Patient Cohort and Samples
Samples (n = 252) from 186 adult patients with AML and 17 healthy donors were collected with signed informed consent with protocols in accordance with the Declaration of Helsinki [study acronym HRUHLAB2, HUS Ethical Committee Statement 303/13/03/01/2011 (original), latest amendment 7 dated June 15, 2016. Latest HUS study permit HUS/395/2018 dated February 13, 2018]. Mononuclear cells (MNC) were isolated by Ficoll-Paque centrifugation from freshly collected bone marrow and peripheral blood specimens of 133 diagnosed, 78 relapsed, 41 chemorefractory-stage patients. For 42 patients we profiled two or more consecutive samples. Skin biopsies were collected from all patients for germline DNA analysis. The median age at diagnosis of the patient cohort was 62 years. Other clinical details of the patients with AML are given in Supplementary Table S1. A summary of the cohort of patients including demographic information and clinical and treatment data is given in Supplementary Table S1.
FPMTB
The FPMTB consisted of the AML tumor group chair and clinicians managing the patients, clinical laboratory specialists, translational scientists familiar with the functional assays and multiomics data, bioinformaticians, study nurses, and by referral a genetic counselor for actionable germline variants (Supplementary Table S1). The meetings were scheduled every week (Wednesdays) and also ad hoc if necessary, when a patient case was submitted (meeting within 1 week of sampling). The task of the FPMTB was to overview and analyze clinical, molecular, and functional characterization of all consecutive patients with newly diagnosed or R/R AML, assign risk groups, evaluate standard-of-care options, and open clinical trials. In addition, in the case of R/R AML, candidate drugs were evaluated for on- or off-label treatment to make rational therapy recommendations based on DSRT and other profiling data. The board also analyzed treatment follow-up and responses for eligible patients, and recommended bridging to alloHSCT (Supplementary Fig. S1). More detailed criteria for patient and treatment selection are shown in Supplementary Table S1.
DSRT
A library of 515 commercially available chemotherapeutic and targeted oncology compounds consisted of 168 approved drugs, 261 investigational compounds, and 86 probes (Supplementary Table S2). The chemical compounds DMSO (negative control) and benzethonium chloride (positive control) were added to 384-well plates using an acoustic liquid dispensing system Echo 500/550 (Labcyte). Freshly isolated MNCs were counted and resuspended in MCM (PromoCell) with 0.5 μg/mL gentamicin and 2.5 μg/mL amphotericin or in CM constituted of 77.5% RPMI 1640, 10% FCS, 12.5% human HS-5 bone marrow stromal cell line–derived CM, and 1% penicillin and streptomycin. A 5-μL cell-free medium was added to dissolve compounds followed by 20 μL cell suspension containing 5,000 to 10,000 cells to each well using multidrop (Thermo Fisher). The plates were incubated at 37°C in 5% CO2 for 72 hours. Subsequently, CellTiter-Glo (Promega) reagent was added to all wells, and cell viability as luminescence generated by total cellular ATP was measured using a PHERAstar (BMG Labtech).
The drug responses passing the data quality assessment were included in further analysis (45). DSS were calculated as shown previously (30) and sDSS were calculated by normalizing drug responses against 17 healthy controls.
Exome and RNA Sequencing
Exome and RNA sequencing analyses were performed in real time under individualized systems medicine program using DNA and RNA materials isolated from MNCs as described previously (20, 46). The skin biopsies from the same patient were used as germline control for exome-sequencing data analysis. Detailed methods are given in the Supplementary text.
Mutation and Drug Response Association Analysis
Cancer and AML-specific genes were selected for the drug–mutation association analysis. The AML genes were collected from published studies, TCGA (ref. 5; n = 23), InToGen (n = 32), and Papeammanuil and colleagues (ref. 6; n = 111); and other cancer-associated genes (n = 616) were obtained from the Census database (47). Out of altogether 667 genes, 340 genes were found mutated in our data set. Furthermore, the genes with mutation in at least two samples and tumor variant allele frequency (VAF) >25% were selected for the analysis. The VAF below 25% was disregarded for the analysis considering no significant impact on drug responses. The drugs were selected based on effectiveness across all samples. The analysis was done independently on MCM (n = 61) and CM (n = 82) subsets to avoid the impact of media on the efficacy of key AML drugs and misleading the biological signal. To test significant differences in drug responses, the Wilcoxon signed-rank test was applied using R package “exactRankTests” (version 0.8-29) between mutated and wild-type samples for each gene. For adjusting drug-wise multiple comparisons, FDR was calculated using the Benjamini and Hochberg (BH) method. FDR < 0.1 and mean sDSS difference ≤5 or ≥5 between wild-type and mutated samples was considered significant.
Gene Set Variance Analysis
Gene set variance analysis (GSVA; ref. 48) was used to calculate pathway activation scores (R package version 1.22.4). As an input, log2 CPM of protein-coding genes from all the AML patient samples and four healthy controls (CD34+ sorted cells) was used. GSVA calculates relative enrichment of a gene set for each sample across the sample space, allowing for sample-wise comparison of gene set enrichment within a data set. A positive enrichment value for a sample indicates overall higher expression of the genes in the pathway in the sample, compared with the other samples analyzed. Pathway definitions were taken for canonical pathways (CP) that had 1,329 gene sets (MSigDB database v6.2). The gene sets used were (i) CP:BIOCARTA (n = 217), (ii) CP:KEGG (n = 186), and (iii) CP:REACTOME (n = 674). To consider a pathway to be active, we used a robust, four-step methodology. First, to get the highly significant active pathways (P values) in a sample, we applied 1,000 bootstrap iterations on GSVA scores. Pathway-wise P values were corrected by applying the BH method, and FDR < 0.01 was considered significant. Second, we chose highly variable pathways that had a GSVA score > 0.2 (cutoff based on overall distribution). Third, these significant pathways were further normalized to four healthy controls (CD34+). A given pathway was considered active only when it had a GSVA score more than the average GSVA score of healthy controls. The final step involved choosing only those active pathways that passed the above three criteria and were also active in at least two databases. For example, apoptosis pathway was considered active if found to be deregulated in at least two of the databases: BIOCARTA_TCAPOPTOSIS_PATHWAY, KEGG_APOPTOSIS, and REACTOME_APOPTOSIS.
Differential Gene-Expression and Pathway Enrichment Analysis
Differential gene-expression analysis was performed using the R package DESeq2 (49). The analysis was performed using raw read counts from FLT3i-sensitive and FLT3i-resistant samples. To remove any batch effects in the data, we corrected for RNA-seq library preparation protocols and gender by modifying the design formula (∼batch + condition) and then applied a likelihood ratio test to get the differentially expressed genes. The BH method was used to control the FDR. A cutoff value of absolute log2 fold change of greater than or equal to 2 and FDR < 0.1 were used as additional filters to select differentially expressed genes for the downstream analysis. Pathway analysis was performed using the genes upregulated in FLT3i-resistant samples. Enrichr web-tool was used for pathway enrichment analysis. Outputs from KEGG 2016 and Wiki pathways were considered for further analysis.
Phospho-Flow Cytometry and Data Analysis
Viably frozen MNCs from patients with AML were thawed and resuspended in RPMI 1640 medium supplemented with 10% FBS and penicillin and streptomycin. Cells were treated with 50 μL of DNase (Promega) for two hours at 37°C to dissolve dead cell clumps. Cells were centrifuged and resuspended in RPMI 1640 medium with penicillin and streptomycin without serum. Cells were stained with Zombie violet cell viability dye (423113, BioLegend) stimulated with 100 ng/mL human recombinant IL15 (PeproTech) for 20 minutes at 37°C. Subsequently, cells were washed with ice-cold PBS, centrifuged at 1,000 × g, fixed with 500 μL of 4% formaldehyde, and incubated at 37°C for 10 minutes. PBS was directly added to the fixed cells and centrifuged at 1,000 × g, and the supernatant was discarded. Ice-cold methanol was added dropwise to the cell pellets and incubated on ice for 30 minutes to permeabilize the cell membrane. Cells were washed with PBS, counted, and added to 96-well V bottom plates. The surface IgG was blocked using human IgG Fc receptor inhibitor (Invitrogen) in staining buffer (0.5% bovine serum albumin in PBS) for 15 minutes on shaker at room temperature and washed with PBS. The antibodies for CD45 (563716, BD Biosciences), pERK (612566, BD Biosciences), pAKT S473 (4075S, Cell Signaling Technologies), and isotype controls were added to the respective wells and incubated for 30 minutes on a plate shaker. The cells were washed with staining buffer and PBS before flow cytometry analysis using iQue Screener Plus (Intellicyte). Antibody-stained UltraComp beads (01-2222-41, Invitrogen) and cells without viability staining were used for compensation. The data were analyzed using Cytobank cellmass software.
Testing of FLT3 Inhibitors in FLT3-ITD–Mutated AML Cell Lines
FLT3-ITD–mutated AML cell lines MOLM-13 and MV4-11 were purchased from DSMZ and were cultured in recommended media. Sorafenib was dispensed in nine different doses in 384-well plates. MOLM-13 and MV4-11 cells were stimulated with 100 ng/mL human recombinant IL15 (PeproTech) at 37°C for one hour. AML cell lines were resuspended in their respective medium with CellTox Green reagent (Promega). Cells (3,000 per well) were dispensed in predrugged plates and incubated at 37°C for 72 hours. Fluorescence was detected using a Phearastar plate reader (BMG LabTech), and dose response was generated using the four-parameter logistic regression.
Data Access
Basic demographics, clinical laboratory values, drug therapies, treatment responses, ex vivo drug testing, and sequencing (exome and RNA sequencing) data will be available at the publication-specific analysis environment at the Helsinki University Hospital datalake. This is an EU GDPR-compliant (General Data Protection Regulation, https://gdpr.eu/), secure, cloud-based data environment accessible by a virtual machine (IP-restricted, 2-level authentication), including all key analytical tooling. Datalake onboarding commences by sending an e-mail request to tietopalvelu@hus.fi.
Authors' Disclosures
O. Brück reports personal fees from Novartis, Amgen, and Sanofi outside the submitted work. M. Kontro reports grants from Finnish Medical Foundation, Finnish Cancer Institute, Cancer Foundation Finland, and FiCAN South and iCAN during the conduct of the study. H. Kuusanmäki reports grants from AbbVie Investigator Initiated Study grant outside the submitted work. I. Västrik reports grants from Business Finland and Academy of Finland during the conduct of the study; grants from Business Finland and Academy of Finland outside the submitted work. Janna Saarela reports grants from Sanofi-Genzyme and personal fees from Sanofi-Genzyme outside the submitted work. M. Wolf reports other support from Novartis Finland Oy outside the submitted work; and M. Wolf's contribution to the work presented in this manuscript is based on work conducted during her employment at FIMM and is not conflicted by her current position at Novartis. B.T. Gjertsen reports other support from KinN Therapeutics AS, Alden Cancer Therapy II, personal fees from AbbVie, Jazz Pharmaceutical, Incyte, Curamir, and other support from BMS outside the submitted work; in addition, B.T. Gjertsen has a patent for repurposing in leukemia therapy pending. S. Mustjoki reports grants from Cancer Foundation Finland during the conduct of the study; grants from Novartis, Pfizer, and BMS outside the submitted work. K. Wennerberg reports grants from Novo Nordisk Foundation during the conduct of the study. C.A. Heckman reports grants from Sigrid Jusélius Foundation, Cancer Society of Finland, and Academy of Finland during the conduct of the study; grants from BMS/Celgene, Kronos Bio, Novartis, Oncopeptides, Orion Pharma, Innovative Medicines Initiative Joint Undertaking project HARMONY, grants from Innovative Medicines Initiative Joint Undertaking project HARMONYPLUS, and grants from Erasmus+ Knowledge Alliance NEMHESYS outside the submitted work. O. Kallioniemi reports other support from Vinnova, AstraZeneca, Pelago, Lucerobio, MediSapiens, Sartar Therapeutics, and Abbott, Vysis during the conduct of the study. K. Porkka reports grants from Novartis and BMS/Celgene during the conduct of the study; grants from Incyte and personal fees from Astellas outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
D. Malani: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. A. Kumar: Data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. O. Brück: Data curation, software. M. Kontro: Resources, data curation. B. Yadav: Formal analysis, visualization, methodology. M. Hellesøy: Validation. H. Kuusanmäki: Validation. O. Dufva: Visualization. M. Kankainen: Data curation. S. Eldfors: Data curation, software, methodology. S. Potdar: Formal analysis. J. Saarela: Methodology. L. Turunen: Methodology. A. Parsons: Validation. I. Västrik: Software. K. Kivinen: Methodology. J. Saarela: Methodology. R. Räty: Resources. M. Lehto: Resources. M. Wolf: Writing–review and editing. B.T. Gjertsen: Resources. S. Mustjoki: Supervision, funding acquisition, writing–review and editing. T. Aittokallio: Conceptualization, writing–review and editing. K. Wennerberg: Conceptualization, writing–review and editing. C.A. Heckman: Conceptualization, resources, investigation, methodology, project administration, supervision, funding acquisition, writing–review and editing. O. Kallioniemi: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing. K. Porkka: Conceptualization, resources, formal analysis, supervision, funding acquisition, visualization, writing–review and editing.
Supplementary Material
Supplement text file including supplement figures, supplementary table legends and supplement methods
15 Supplement figures and legends
11 supplement tables
Acknowledgments
We are most grateful to the patients and their families for participating in the study. We thank the Academy of Finland grant, Academy of Finland Center of Excellence on Translational Cancer Biology, the iCAN Digital Precision Cancer Medicine Flagship, Sigrid Jusélius Foundation, Cancer Society of Finland, Tekes/Business Finland Salwe program, HUS-EVO, and HUCS Institute for support to this study. We utilized FIMM/HiLife/Biocenter Finland local and national infrastructures (Genome-wide, Bioinformatics, and DDCB National infrastructures) as well as national networks for EU infrastructures BBMRI, ELIXIR, EATRIS, and EU-OPENSCREEN. We thank the excellent staff and resources provided by the FIMM Technology Centre and the FIMM High-Throughput Biomedicine Unit. We thank Minna Suvela, Alun Parsons, and Siv Knaappila for processing patient samples. The authors acknowledge the assistance and support of the personnel at the FIMM Sequencing Unit, especially Pekka Ellonen and Pirkko Mattila. The authors acknowledge the support of personnel at the FIMM High-Throughput Biomedicine Unit: Elina Huovari, Aleksandr Ianevski, Meri Kokkonen, Evgeny Kulesskiy, Sergey Kuznetsov, Karoliina Laamanen, Elina Lehtinen, Piia Mikkonen, Maria Nurmi, Katja Närhi, Katja Suomi, and Sanna Timonen. D. Malani received personal funding support from the Finnish Cultural Foundation, Blood Disease Research Foundation, Finnish Hematology Association, K. Albin Johansson Foundation, Paivikki ja Sakari Sohlberg Foundation, Maud Kuistila Memorial Foundation, and Helsinki Biomedicum foundation to support this research. M. Hellesøy and B.T. Gjertsen received funding from The Norwegian Cancer Society and Helse Vest Health Trust. A. Kumar received an EMBO short-term lab visit fellowship for this study. S. Mustjoki received funding from European Research Council (M-IMM project), Academy of Finland, Cancer Society of Finland, and Sigrid Jusélius Foundation. T. Aittokallio received funding from Academy of Finland (grants 292611, 310507 and 313267), Cancer Society of Finland, and Sigrid Jusélius Foundation. K. Wennerberg received funding from Novo Nordisk Foundation (Grant Number NNF17CC0027852). M. Kontro received a University of Helsinki Early Career Grant. C.A. Heckman received funding from the Sigrid Jusélius Foundation, Cancer Society of Finland, and Academy of Finland (grants 1320185 and 334781). O. Kallioniemi received support from Academy of Finland (259777, 278741, 271845, and 333050), Cancer Society of Finland (120089, 140114, 160080, 170112, and 190116) and Sigrid Jusélius Foundation, Knut and Alice Wallenberg Foundation (KAW; 2015.0291), Swedish Foundation for Strategic Research (SSF; SB16-0058), VR environment grant (2017-06095), and Vinnova (2018-03338).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
References
- 1. Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 2. Estey E. Acute myeloid leukemia: 2016 update on risk-stratification and management. Am J Hematol 2016;91:824–46. [DOI] [PubMed] [Google Scholar]
- 3. Assi SA, Imperato MR, Coleman DJL, Pickin A, Potluri S, Ptasinska Aet al. Subtype-specific regulatory network rewiring in acute myeloid leukemia. Nat Genet 2019;51:151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gerstung M, Papaemmanuil E, Martincorena I, Bullinger L, Gaidzik VI, Paschka Pet al. Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat Genet 2017;49:332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson Aet al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts NDet al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CDet al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med 2017;377:454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini Set al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med 2019;381:1728–40. [DOI] [PubMed] [Google Scholar]
- 9. DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims ASet al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med 2018;378:2386–98. [DOI] [PubMed] [Google Scholar]
- 10. Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JKet al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amatangelo MD, Quek L, Shih A, Stein EM, Roshal M, David MDet al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 2017;130:732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cortes JE, Kantarjian H, Foran JM, Ghirdaladze D, Zodelava M, Borthakur Get al. Phase I study of quizartinib administered daily to patients with relapsed or refractory acute myeloid leukemia irrespective of FMS-like tyrosine kinase 3-internal tandem duplication status. J Clin Oncol 2013;31:3681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Letai A. Functional precision cancer medicine-moving beyond pure genomics. Nat Med 2017;23:1028–35. [DOI] [PubMed] [Google Scholar]
- 14. Prasad V. Perspective: the precision-oncology illusion. Nature 2016;537:S63. [DOI] [PubMed] [Google Scholar]
- 15. DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AHet al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med 2020;383:617–29. [DOI] [PubMed] [Google Scholar]
- 16. Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman Tet al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov 2016;6:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang H, Nakauchi Y, Köhnke T, Stafford M, Bottomly D, Thomas Ret al. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat Cancer 2020;1:826–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collignon A, Hospital MA, Montersino C, Courtier F, Charbonnier A, Saillard Cet al. A chemogenomic approach to identify personalized therapy for patients with relapse or refractory acute myeloid leukemia: results of a prospective feasibility study. Blood Cancer J 2020;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin L, Tong Y, Straube J, Zhao J, Gao Y, Bai Pet al. Ex-vivo drug testing predicts chemosensitivity in acute myeloid leukemia. J Leukoc Biol 2020;107:859–70. [DOI] [PubMed] [Google Scholar]
- 20. Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda Aet al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov 2013;3:1416. [DOI] [PubMed] [Google Scholar]
- 21. Snijder B, Vladimer GI, Krall N, Miura K, Schmolke A-S, Kornauth Cet al. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. Lancet Haematol 2017;4:e595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malani D, Murumägi A, Yadav B, Kontro M, Eldfors S, Kumar Aet al. Enhanced sensitivity to glucocorticoids in cytarabine-resistant AML. Leukemia 2016;31:1187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SLet al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tyner JW, Yang WF, Bankhead A, Fan G, Fletcher LB, Bryant Jet al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res 2013;73:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hernández P, Gorrochategui J, Primo D, Robles A, Rojas JL, Espinosa ABet al. Drug discovery testing compounds in patient samples by automated flow cytometry. SLAS Technol 2017;22:325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuusanmäki H, Leppä A-M, Pölönen P, Kontro M, Dufva O, Deb Det al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020;105:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spinner MA, Aleshin A, Santaguida MT, Schaffert SA, Zehnder JL, Patterson ASet al. Ex vivo drug screening defines novel drug sensitivity patterns for informing personalized therapy in myeloid neoplasms. Blood Adv 2020;4:2768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner Tet al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karjalainen R, Pemovska T, Popa M, Liu M, Javarappa KK, Majumder MMet al. JAK1/2 and BCL2 inhibitors synergize to counteract bone marrow stromal cell-induced protection of AML. Blood 2017;130:789–802. [DOI] [PubMed] [Google Scholar]
- 30. Yadav B, Pemovska T, Szwajda A, Kulesskiy E, Kontro M, Karjalainen Ret al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep 2014;4:5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burgess MR, Hwang E, Firestone AJ, Huang T, Xu J, Zuber Jet al. Preclinical efficacy of MEK inhibition in Nras-mutant AML. Blood 2014;124:3947–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan LN, Murakami MA, Robinson ME, Caeser R, Sadras T, Lee Jet al. Signalling input from divergent pathways subverts B cell transformation. Nature 2020;583:845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Knijnenburg TA, Klau GW, Iorio F, Garnett MJ, McDermott U, Shmulevich Iet al. Logic models to predict continuous outputs based on binary inputs with an application to personalized cancer therapy. Sci Rep 2016;6:36812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laganà A, Beno I, Melnekoff D, Leshchenko V, Madduri D, Ramdas Det al. Precision medicine for relapsed multiple myeloma on the basis of an integrative multiomics approach. JCO Precis Oncol 2018;2:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malani D, Yadav B, Kumar A, Potdar S, Kontro M, Kankainen Met al. KIT pathway upregulation predicts dasatinib efficacy in acute myeloid leukemia. Leukemia 2020;34:2780–4. [DOI] [PubMed] [Google Scholar]
- 36. Mathew NR, Baumgartner F, Braun L, O'Sullivan D, Thomas S, Waterhouse Met al. Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells. Nat Med 2018;24:282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dufva O, Pölönen P, Brück O, Keränen MAI, Klievink J, Mehtonen Jet al. Immunogenomic landscape of hematological malignancies. Cancer Cell 2020;38:380–99. [DOI] [PubMed] [Google Scholar]
- 38. Bryce AH, Egan JB, Borad MJ, Stewart AK, Nowakowski GS, Chanan-Khan Aet al. Experience with precision genomics and tumor board, indicates frequent target identification, but barriers to delivery. Oncotarget 2017;8:27145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harada S, Arend R, Dai Q, Levesque JA, Winokur TS, Guo Ret al. Implementation and utilization of the molecular tumor board to guide precision medicine. Oncotarget 2017;8:57845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell 2017;168:584–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marquart J, Chen EY, Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol 2018;4:1093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Short NJ, Kantarjian H, Ravandi F, Daver N. Emerging treatment paradigms with FLT3 inhibitors in acute myeloid leukemia. Ther Adv Hematol 2019;10:2040620719827310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McMahon CM, Canaani J, Rea B, Sargent RL, Morrissette JJD, Lieberman DBet al. Mechanisms of acquired resistance to gilteritinib therapy in relapsed and refractory FLT3-mutated acute myeloid leukemia. Blood 2017;130:295. [Google Scholar]
- 44. Traer E, Martinez J, Javidi-Sharifi N, Agarwal A, Dunlap J, English Iet al. FGF2 from marrow microenvironment promotes resistance to FLT3 inhibitors in acute myeloid leukemia. Cancer Res 2016;76:6471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Potdar S, Ianevski A, Mpindi JP, Bychkov D, Fiere C, Ianevski Pet al. Breeze: an integrated quality control and data analysis application for high-throughput drug screening. Bioinformatics 2020;36:3602–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar A, Kankainen M, Parsons A, Kallioniemi O, Mattila P, Heckman CA. The impact of RNA sequence library construction protocols on transcriptomic profiling of leukemia. BMC Genomics 2017;18:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal Net al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement text file including supplement figures, supplementary table legends and supplement methods
15 Supplement figures and legends
11 supplement tables