Abstract
The worldwide coronavirus disease 2019 pandemic was sparked by the severe acute respiratory syndrome caused by coronavirus 2 (SARS-CoV-2) that first surfaced in December 2019 (COVID-19). The effects of COVID-19 differ substantially not just between patients individually but also between populations with different ancestries. In humans, the human leukocyte antigen (HLA) system coordinates immune regulation. Since HLA molecules are a major component of antigen-presenting pathway, they play an important role in determining susceptibility to infectious disease. It is likely that differential susceptibility to SARS-CoV-2 infection and/or disease course in COVID-19 in different individuals could be influenced by the variations in the HLA genes which are associated with various immune responses to SARS-CoV-2. A growing number of studies have identified a connection between HLA variation and diverse COVID-19 outcomes. Here, we review research investigating the impact of HLA on individual responses to SARS-CoV-2 infection and/or progression, also discussing the significance of MHC-related immunological patterns and its use in vaccine design.
Keywords: Human leukocyte antigens, Polymorphism, COVID-19, Disease susceptibility
SARS-CoV-2 and COVID-19 epidemiology and immunopathology
Three highly pathogenic strains of beta coronaviruses (CoVs) with high mortality have emerged during the past two decades as a result of zoonotic transmission. The first two, severe acute respiratory syndrome virus (SARS-CoV-1) and the Middle Eastern respiratory syndrome virus (MERS-CoV), emerged in 2002 and 2012 respectively (Zhu et al. 2020). SARS-CoV-2 is a new beta coranavirus with positive single-stranded RNA that emerged at the end of 2019 in the Hubei province of China and causes coronavirus disease 2019 (COVID-19) (Zhu et al. 2020; Mackenzie et al. 2020). The incidence of COVID-19 continues to increase and as of August 21, 2022, 600,761,268 infected cases, including 6,471,618 deaths, have been reported globally (COVID Live - Coronavirus Statistics - Worldometer 2022).
The genome organization of SARS-CoV-2 is similar to CoV-1 and other coronaviruses (Chen et al. 2020). Open Reading Frames (ORFs) are found in all beta-coronavirus, including SARS-CoV-2. This comprises ORF1ab, which encodes the vast majority of enzymatic proteins, surface spike glycoproteins (S), envelope proteins (E), membrane proteins (M), and nucleocapsid proteins (N). Other proteins consist of nonstructural proteins encoded by ORF3a, ORF6a, ORF7, and ORF8 and ORF10a. CoVs have a far lower rate of nucleotide alterations than other RNA viruses do because of an enzyme that repairs replication faults (Denison et al. 2011). Even so, there are now reports of multiple variants emerging around the world as the SARS-CoV-2 pandemic continues (Makoni 2021; Korber et al. 2020; Hodcroft et al. 2020; Tang et al. 2021a, b; Quarleri et al. 2021), which are classified by the World Health Organization (WHO) into two types: variants of concern (VOC) and variants of interest (VOI).
SARS-CoV-2 may be transmitted from person to person or sometimes indirectly through contaminated surfaces (Liu et al. 2020). However, SARS-CoV-2 is mostly transmitted via respiratory droplets distributed by coughs, sneezes, or even while conversing. In addition to nonspecific symptoms including headache, fatigue, and muscle pain, as well as digestive problems such as diarrhea and vomiting, fever, cough, shortness of breath, and other breathing difficulties are the most common clinical symptoms of COVID-19 patients (Alimohamadi et al. 2020).
Age (> 60 years), gender, smoking history, prior pneumonia, and major concomitant diseases are linked with COVID-19 mortality (such as immunocompromised states, chronic cardiovascular, cerebrovascular, pulmonary, kidney disease, diabetes mellitus, fulminant inflammation, lactic acid accumulation, and thrombotic events) (Goodman et al. 2019; Henkens et al. 2022; Lacedonia et al. 2021). Despite the global effort to discover various aspects of SARS-CoV-2, such as clinical manifestations, epidemiology, mortality and morbidity, and diagnostics, there are still numerous gaps in our understanding of this disease, and many perspectives with respect to host immune response towards COVID-19 remain unknown.
The pathophysiology of SARS-CoV-2 caused pulmonary illness is substantially similar to that of SARS-CoV-1 and MERS-coV (Zhu et al. 2020; Zhou et al. 2021). Damage to infected lung cells leads to hypoxemia and plasma exudate in alveolar spaces (Bösmüller et al. 2021). Histopathological examinations have revealed hyaline membranes, mononuclear and macrophage infiltration of air gaps, and thickening of the alveoluminal wall (Bösmüller et al. 2021; Mason 2020; Pandey and Agarwal 2020). SARS-CoV-2 also disrupts normal immune responses, leading to an impaired immune system and uncontrolled inflammatory responses in severe and critical patients with COVID-19 (Tay et al. 2020; Rubio-Casillas et al. 2022). These individuals show lymphopenia, lymphocyte activation and dysfunction, granulocyte and monocyte abnormalities, elevated cytokine levels, and a rise in immunoglobulin G (IgG) and total antibodies (Gibson et al. 2020; Lippi and Plebani 2020; Yang et al. 2020a). (Merad et al. 2022) discussed two broad hypotheses: inability to mount a timely antiviral response and to control SARS-CoV-2–driven inflammatory responses to explain severe COVID-19 pathophysiology. Hypercoagulation, endothelial damage, and arterial and venous embolism are very commonly seen in severe COVID-19 (Livanos et al. 2021).
To identify viral infection, the innate immune system employs a variety of pattern recognition receptors (PRRs) while alveolar macrophages patrol the respiratory tract’s lumen and serve as the first line of defense (Madden and Diamond 2022). Coronaviruses are detected by cell types that express endosomal TLR3 and TLR8 and also by specialized immune cells such as plasmacytoid dendritic cells (pDCs) through Toll-like receptor 7 (TLR7) (Merad et al. 2022). Cytosolic RNA sensors such as RIG-I and MDA5, or RIG-I–like receptors (RLRs) within infected cells, recognize dsRNA intermediates during viral replication. Signaling downstream of TLRs and RLRs promotes IRF3/IRF7-dependent transcription of type I and type III interferons (IFNs), as well as NF-B-dependent pro-inflammatory cytokines and chemokines (Merad et al. 2022). A small number of severe patients have been found to have “loss of function” variants in loci that control TLR3- and IRF7-dependent type I IFN immunity (Zhang et al. 2020). Through the expression of a number of viral proteins that block these pathways, SARS-CoV-2 is capable of evading innate recognition, signaling, IFN induction, and IFN-stimulated genes (ISGs) (Kasuga et al. 2021). Patients with genetic mutations or autoantibodies that interfere with IFN pathways suffer from life-threatening COVID-19 disease. pDCs, effector cells like NK cells and alveolar macrophages. and adaptive effector T cells that mediate viral clearance get severely depleted in patients with severe COVID-19 (Sanchez-Cerrillo et al. 2020; Lucas et al. 2020; Wilk et al. 2020, 2021; Zhou et al. 2020; Mathew et al. 2020). Contrary to IFN-I- and IFN-III-mediated early antiviral defense that is hindered, pro-inflammatory cytokines and chemokines also get dramatically elevated (Israelow et al. 2020).
To better understand mechanisms of protection against pathogen, to create immune-based therapeutics, and to design and administer vaccines, it is critical to understand the impact of the acquired immune response in the form of antibody (Ab) and cell-mediated immunity. Antibodies block binding of distinct epitopes mainly in the receptor-binding domain (RBD) of the S protein (primarily to sites 1a and 1b) to the ACE2 receptor and can promote effector function by binding to the complement and Fc receptors (Zohar and Alter 2020). Ab responses of varying magnitudes are observed in infected, preimmune, and vaccinated people, most likely reflecting differences in antigenic load and exposure (Piccoli et al. 2020; Lucas et al. 2021). Cell-mediated immunity protects an organism through the activation of antigen-specific cytotoxic T cells that induce apoptosis in cells displaying foreign antigens, such as virus-infected cells, cells with intracellular bacteria, and cancer cells displaying tumor antigens, and promoting the production of antibodies through their effect on B cells. T cell responses to SARS-CoV-2 are crucial factors for recognizing and killing infected cells (Weiskopf et al. 2020). One of the constraints imposed by severe COVID-19 is extensive T cell lymphopenia, which impairs the host’s ability to develop a powerful immunological response. T cells from patients with severe COVID-19 have been shown to have antigen-dependent phenotypic differences associated with differential cytokine secretion (Altmann and Boyton 2020).
HLA overview
The genes encoding the human leukocyte antigen (HLA) molecules have been a primary focus in genetic association studies across a wide range of infectious and immune-mediated diseases due to their critical role in antigen presentation to T cells. The human major histocompatibility complex (MHC) in humans is termed HLA (human leukocyte antigen) because antigens were first found utilizing alloantibodies against leukocytes (Choo 2007). HLA molecules are well-known as transplantation antigens (Crux and Elahi 2017). Human MHC aligns to 6p21 and covers 3600 kilobases of DNA (Tripodis et al. 1998). All the loci of the HLA complex are grouped into four categories of gene status, classified as protein coding, gene candidates, non-coding RNAs, and pseudogenes (Shiina et al. 2009). Human leukocyte antigens are of three main types. Class I HLA antigens include HLA-A, HLA-B, and HLA-C molecules, and are found on the surface of all nucleated cells; class II, which includes HLA-DR, HLA-DQ, and HLA-DP loci, are expressed on the surface of professional antigen-presenting cells; and class III contains genes for proteins that have diverse immune functionality (The MHC sequencing consortium 1999). The HLA-A, HLA-B, and HLA-C class I and HLA-DP, HLA-DQ, and HLA-DR class II molecules exhibit an extraordinary diversity. More than 18,000 class I and 7000 class II alleles have been defined (Choo 2007). Most of their sequence variability is in HLA class I and II antigen-binding sites, indicating diverse and specialized interactions with TCRs and KIR. HLA-E, HLA-F, and HLA-G, the non-classical HLA class I genes, have limited polymorphism and a tissue-restricted expression pattern (Buhler et al. 2016). All the non-classical class I genes encode ligands recognized by NK cell receptors. The HLA class III region is between the class I and II regions and contains genes encoding complement components, heat-shock proteins, and tumor necrosis factor α (TNFα) (Djaoud and Parham 2020). Since HLA genes are closely linked, the entire MHC is inherited as an HLA haplotype in a Mendelian fashion from each parent (Shiina et al. 2009). Possible random combinations of antigens from different HLA loci on an HLA haplotype are enormous, but certain HLA haplotypes are found more frequently in some populations than expected by chance because of linkage disequilibrium (Choo 2007; Meyer et al. 2018). For example, HLA-A1, B8, DR17 is the most common HLA haplotype among individuals with European ancestry, with a frequency of 5% (Abdou et al. 2010). Amino acid variations in several regions change the fine shape of the groove and thus alter the peptide-binding specificity of HLA molecules (Abdou et al. 2010). HLA antigens vary widely by ancestry. This variation of HLA polymorphism may have evolved under distinct geographical pressures. This may be connected to the role of HLA molecules in presenting infectious pathogens in diverse regions (Markov and Pybus 2015).
Almost all nucleated cells have HLA class I molecules on their surface. Only B lymphocytes, antigen-presenting cells (monocytes, macrophages, and dendritic cells), and activated T lymphocytes express class II molecules (Shiina et al. 2009). Class I and class II bind to different natures and sources of peptides. Class I–restricted T cells identify endogenously generated antigens (e.g., cellular, transformed, or virus-induced proteins), while class II–restricted T cells recognize exogenously derived antigens.
The interaction between the HLA complex and the foreign antigen activates T lymphocytes. Upon activation, T cells proliferate and begin to release cytokines, which allows them to initiate an immune response that will recognize and eliminate cells bearing the same foreign antigen/HLA complex when they are next met. The enormous diversity of human leukocyte antigen (HLA) molecules is essential for T cells to receive a variety of antigens. T cell receptors identify the conformational structure of the HLA antigen-binding grove and antigen peptides (Shi et al. 2020; Lee and Koohy 2020; Dutta et al. 2018). The structural determination of disease-relevant peptide-HLA and HLA-peptide-TCR complexes is crucial for the elucidation of the molecular mechanisms responsible for the development of specific protective immunity against pathogens, as well as for the deleterious T cell reactivity that promotes disease. The HLA-peptide-TCR interactions that underpin self and non-self-discrimination are guided by a set of rules that are also implicated in disease. For example, autoreactive T cells are directly generated in autoimmune diseases by mechanisms such as atypical HLA-peptide-TCR binding orientation, low-affinity peptide binding that facilitates thymic escape, TCR-mediated stabilization of weak peptide-HLA interactions, presentation of peptides in a different binding register, post-translational epitope modification, generation of hybrid peptides, and processes that regulate HLA expression and stability (Dendrou et al. 2018). Majorly, HLA can affect three cytotoxic T lymphocyte (CTL) features, which in turn allows for an efficient antiviral response during chronic viral infections. First, targeting a conserved viral sequence or epitope rather than a variable region will increase the effectiveness of CTLs in regulating viral infection (Elahi et al. 2011). Another characteristic that increases the efficiency of viral clearance is polyfunctionality. For instance, CD8 + PD-L1 + CXCR3 + polyfunctional T cell abundances are associated with survival in patients with severe SARS-CoV-2 infection (Adam et al. 2021). Third, the proliferative capability of CTLs is related to their proliferation potential, which may be correlated with their ability to destroy infected targets (Kunwar et al. 2013). Therefore, the immune response may be either beneficial or negative depending on which of these properties an HLA allele can alter.
In humans, the HLA system orchestrates immune regulation. HLA expression during infection may depend on number of other factors, including the presence of specific polymorphic sites in the promoter region that can be targeted by transcription factors; the degree to which post-transcriptional factors may degrade HLA mRNA, particularly microRNAs that can target polymorphic sites at 3′UTR; the structural characteristics of the HLA molecule encoded by that person’s alleles that are available to interact with HLA receptors; and the role of neighboring or distant genes epistatically acting on the HLA gene. Investigating these pathways in depth might help us to comprehend the varying complexity behind immune responses. It is advantageous to have enhanced binding capabilities of HLA molecules for viral peptides such as from SARS-CoV-2, on the cell surface of antigen-presenting cells. For example, Zhang et al. (2022) showed an increased HLA-B18:01/B44:03 allelic fold change in SARS-CoV-2-infected A549 cells (Zhang et al. 2022). New SARS-CoV-2 mutations may modify HLA expression, which may help explain immune response evasion. Benlyamani et al. observed that downregulation of HLA-DR molecules in circulating monocytes creates immunosuppressed circumstances (like lymphopenia) for host response in critically ill individuals (Benlyamani et al. 2020). Thus, HLA haplotypes could be linked to distinct genetic predispositions to disease. Specific HLA genotypes can stimulate the T cell–mediated antiviral response differently and could possibly alter the symptoms and transmission of the disease (COVID-19 Host Genetics Initiative 2020) HLA-B*46:01 possesses the fewest SARS-CoV-2 binding peptides, indicating that persons with this allele could be vulnerable to COVID-19, as found with SARS. HLA-B*15:03 showed the best ability to present highly conserved SARS-CoV-2 peptides shared among human coronaviruses, indicating that this allele may facilitate cross-protective T cell–reliant immunity (Nguyen et al. 2020).
Viruses like hepatitis B virus and SARS-CoV-2 have also been reported to affect cellular splicing machinery (Francies and Dlamini 2021; Wang et al. 2022; Thompson et al. 2020). The virus produces splicing variants that encourage cell division, block signaling pathways, inhibit tumor suppressors, change gene expression through epigenetic alteration, and produce immune-evading mechanisms (Francies and Dlamini 2021). HLA alternate splicing and expression can be impacted by mutation, polymorphism, and gene deletion; for instance, the 14 bp INS/DEL polymorphism is involved in alternative splicing, which results in a better yield of stable HLA-G mRNA transcripts in vitro. Because HLA plays such an important role in the immunological response to infections and the development of infectious illnesses, we hypothesize that population HLA variability could be connected with COVID-19 occurrence. The HLA system affects clinical outcomes in multiple infectious diseases, including HIV and SARS as discussed later in the review (Bardeskar and Mania-Pramanik 2016; Barquera et al. 2020; Kulpa and Collins 2011).
In general, host genetic variability may help explain the multiplicity of immune responses to a virus within a community. Knowing how variability in HLA can impact the progression of COVID-19, in particular, may help distinguish individuals at higher risk for the disease. In the following section, we detail genetic findings relevant to SARS-CoV-2 infection and COVID-19 disease outcomes, with specific emphasis on HLA. In addition to this, in silico epitope prediction contributes new knowledge to the creation of a COVID-19 vaccine (Grifoni et al. 2020).
HLA associations in viral disease
Numerous studies have established links between variation in HLA and susceptibility to, and disease course in, viral disease. HLAs play a complex role in immunomodulation during HIV infection, and variations at the HLA class I locus have been linked to the efficiency of CD8 + T cell control of viremia (Martin and Carrington 2013). Polymorphisms within HLA class I and II loci have been identified as the host genetic modifier of HIV disease progression in several populations. HLA-B*39 allele frequency was considerably greater in HIV-1-positive participants compared to controls, but the HLA-B*44 allele was not present in the Argentinian population’s HIV-1-positive subjects (Sorrentino et al. 2000). In a Zambian early infection cohort, two HLA class II alleles (DRB1*15) and four HLA class I alleles (B*14:01, B*57, B*58:01, and B*81) were discovered to be linked with protection from rapid CD4 + T cell decline without affecting early plasma viral load (Claiborne et al. 2019). A GWAS showed HLA-B ∗ 57:01 rs2395029 and HLA-C rs9264942 and HLA-B ∗ 27:02 associated with HIV-1 disease progression in Caucasian subjects (Fellay et al. 2009; Teixeira et al. 2014) while the presence of the HLA-B ∗ 52 allele was shown to be favorable to slow AIDS progression in Brazilian subjects (Sorrentino et al. 2000).
A number of HLA alleles are identified as protective (A*03:01, B*15:39, B*27:05, B*39:02, B*57:01/02/03, HLA-B*44, and B*58:01) and risk (A*68:03/05, B*15:30, B*35:02, B*35:12/14, B*39:01/06, B*39:05, and B*40:01) factors for disease progression (Valenzuela-Ponce et al. 2018).
Infection with hepatitis B virus (HBV) increases the risk of cirrhosis and liver cancer (Yuen et al. 2018). HBV-specific CD8 + cytotoxic T cells are crucial for viral clearance and liver damage, and HLA polymorphisms modify their responses. For example, HLA-DRB1*13:02 was associated with protection against persistent HBV infection among children and adults in Gambia (Thursz et al. 1995) while HLA-DQ rs9275319C, HLA-DQB1*06:03, HLA-A*03:01, and HLA-DQB1*06:03 allele were found to decrease HBV infection risk and an increased HBV clearance (Ou et al. 2018; Fan et al. 2016). HLA-DPB1 rs9277535A and HLA-B*08 were associated with the risk of persistent HBV infection (Akgöllü et al. 2017; Thio et al. 2003) while HLA-DQB1*06:01 was associated with chronic HBV infection in Japanese patients (Nishida et al. 2016).
Likewise, infection with the hepatitis C virus (HCV) is a substantial contributor to both acute and chronic hepatitis. Hepatocellular carcinoma and liver cirrhosis are brought on by chronic hepatitis (HCC) (Saito et al. 2018). HLA alleles (B*57:01, B*57:03, Cw*01:02, and DRB1*01:01) were associated with the absence of HCV RNA in a large multiracial cohort of US women (Kuniholm et al. 2010). HLA-A*02:01 and HLA-DRB1*11:01 were associated with HCV spontaneous clearance in the Chinese population (Huang et al. 2016) and HLA-DQB1*02:02 and HLA-DRB1 allele associations with HCV were reported (DRB1*03:01:01 and DRB1*13:01:01 alleles as the risk of progression to chronic hepatitis C infection and DRB1*04:01:01, DRB1*04:05:01, DRB1*07:01:01, and DRB1*11:01:01 and protection against HCV infection) (El-Bendary et al. 2019; Vejbaesya et al. 2000).
HLA-A (HLA-A*02:01), the amino acid polymorphism at position 107 of HLA-A protein (HLA-A Gly107) and HLA-B (rs9266089), has been accounted for association with chickenpox. Disease associations have also been seen in the class III region for variant rs41316748), and IFNA21 (rs7047299) in a GWS association study with shingles (Tian et al. 2017). IFNA21 encodes type I interferon and is mainly involved in innate immune response against viral infection. It has been shown to be involved in the pathogenesis of rubella (Mo et al. 2007) and may also influence susceptibility to asthma and atopy (Chan et al. 2006). The amino acid polymorphisms HLA-A Gln43 (rs114193679) and HLA-A*02:05 have been found to be significantly associated with mumps. A variant in the HLA class I region (rs3131623) HLA-DRB1 LS11 in class II region, rs3131623, HLA-DRB1 LS11 has showed GWAS association with pneumonia (Tian et al. 2017).
HLA associations in SARS-CoV-2 infection and COVID-19 disease outcome
Particularly early in the pandemic, many investigations of the role of HLA in disease focused on bioinformatic prediction of HLA binding affinity to SARS-CoV-2 peptides. These studies have shown that mutations in numerous viral strains, mostly in the Spike protein, alter the affinity between these mutant peptides and HLA molecules (Augusto and Hollenbach 2022). For example, The HLA-B*46:01 allele has a low binding affinity, suggesting that subjects with this allele may have a higher risk of developing the more severe forms of COVID-19 (Migliorini et al. 2021). In contrary, the HLA-B*15:03 allele is reported to have the highest binding affinity for viral peptides (Nguyen et al. 2020; Lin et al. 2003). A recent study examining HLA binders reported that five B22 alleles (B*54:01, B*55:01, B*55:07, B*55:12, and B*56:01) were among the 94 weakest HLA-B binders to SARS-CoV-2, further suggesting B22 as a susceptibility marker (Barquera et al. 2020). An in silico analysis of viral peptide-major histocompatibility complex (MHC) class I binding affinity was conducted by Nguyen et al., which revealed that HLA-A*02:02, HLA-B*15:03, and HLA-C*12:03 effectively presented a larger amount of peptides whereas A*25:01, B*46:01, and C*01:02 were the least efficient for of SARS-CoV-2 peptide presentation (Nguyen et al. 2020). Finally, another bioinformatic prediction revealed high binding affinity between SARS-CoV-2 epitopes and the HLA-A*02:06, HLA-B*52:01, HLA-A*24:02, HLA-A*02:01, and HLA-C*12:02 alleles (Kiyotani et al. 2020).
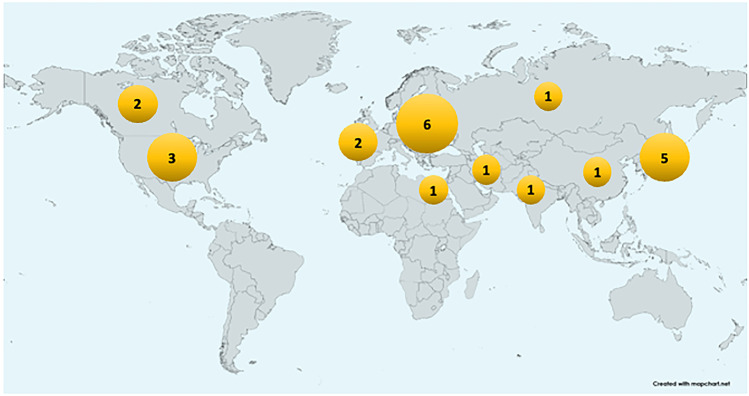
More recent investigations have suggested an association between HLA variation and COVID-19 outcomes, and between HLA genotypes and varying immune responses to SARS-CoV-2. While the number of studies continues to increase, the need for studies in more diverse global regions is evident. HLA Association studies for COVID-19 reported as of this writing by geographic area are shown in Fig. 1.
Fig. 1.
Map showing number of significant association studies for HLA loci in different geographic regions
Studies with candidate gene approaches suggested roles for several alleles in the HLA region, C*07:29, B*15:27, B*27:07, DRB1*15:01, DQB1*06:02, C*06:02, and DRB1*07:01 in Novelli et al. (Novelli et al. 2020); HLA-C*07:29 and HLA-B*15:27 in (Wang et al. 2020a); and HLA-C*04:01 in (Littera et al. 2020) and in genes associated with the viral cell. HLA-DRB1*08 conferred risk for death due to COVID-19 (Amoroso et al. 2021). Peptide-binding prediction analyses showed that these DRB1*08 alleles were unable to bind any of the viral peptides with high affinity. The extended haplotypes HLA-A*02:05, B*58:01, C*07:01, and DRB1*03:01 were also shown to have a protective effect against SARS-CoV-2 infection in the Sardinian population (Littera et al. 2020).
Several HLA associations in COVID-19 outcomes have also been inferred from genome-wide studies. HLA-A*11:01 (p value = 0.009, OR 2.3), HLA-B*51:01 (p value = 0.007, OR 3.3), and HLA-C*14:02 (p value = 0.003, OR 4.7) were identified as top signals in the HLA class I region in the Chinese population by deeply sequencing and analyzing 332 COVID-19 patients categorized by varying levels of severity (Wang et al. 2020b). HLA-A*11:01:01:01 was also identified as a risk factor for COVID-19 severity (p value = 0.003, OR 3.4), in a study involving 190 Japanese patients and 423 controls, after controlling for comorbidities and other confounding factors (Khor et al. 2021). Three different groups conducted association analyses against a healthy control group to identify susceptibility of infection to SARS-CoV-2 compared 82 COVID-19 vs 3548 controls from China and found HLA-B*15:27 as associated (p value = 0.001, OR 3.6) (Wang et al. 2020a), compared 99 COVID-19 vs 1017 controls from Italy and found 3 significant association (HLA-B*27:07, p value = 0.00001; HLA-DRB1*15:01, p value = 0.002; HLA-DQB1*06:02, p value = 0.0001) (Novelli et al. 2020).
Examination of HLA associations was also performed by the meta-analysis on COVID-19 severity performed by the COVID-19 Human Genetic Initiative (HGI), where a variant in HLA-G was found to be significantly associated but not replicated in a genome-wide association study of 2244 critically ill patients with COVID-19 from 208 UK intensive care units (Douillard 2021; Pairo-Castineira et al. 2021). Five variants reaching statistical significance of association within the CCHCR1 gene, situated 110 kb downstream of HLA-C (top SNP: rs111837807, p value = 2.2 × 10 − 11, OR meta 1.23) as well as a variant within HLA-DPB1 3′UTR (rs9501257, p value = 4.1 × 10 − 8, OR meta 1.19), when comparing the general population to patients with critical COVID-19 (n cases = 8779, n control = 1,001,875, from 25 studies of various ancestries) (Douillard 2021) were identified.
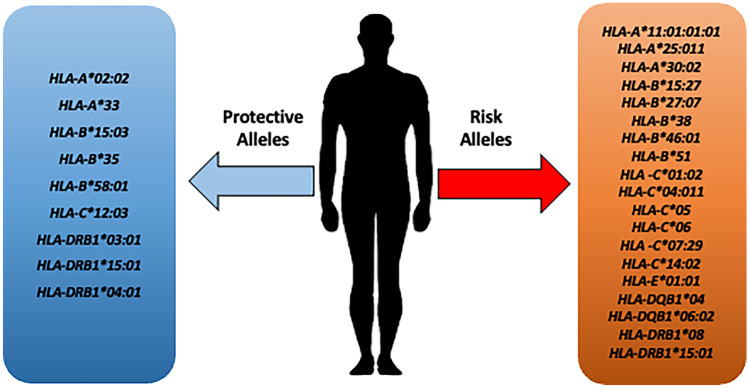
A recent extended GWAS meta-analysis of a well-characterized cohort of 3255 COVID-19 patients with respiratory failure and 12,488 population controls from Italy, Spain, Norway, and Germany/Austria, including stratified analyses based on age, sex, and disease severity, yielded no association for HLA loci at the genome-wide or nominal (p < 10–5) significance threshold, indicating no major role for HLA variability in mediating the severity of COVID-19 in our cohorts (Degenhardt, et al. 2022). In total, 29 HLA alleles—eight protective and 21 risk alleles—have been identified as being linked with COVID-19 susceptibility (Robinson et al. 2020). Figure 2 shows a summary of risk and protective HLA alleles identified across studies as of this writing.
Fig. 2.
Risk and protective alleles for HLA loci identified to date
In addition to classical HLA loci, several non-classical HLA associations with COVID-19 have been identified. Human leukocyte antigen-G (HLA-G) is a ligand for multiple immune inhibitory receptors, whose expression can be upregulated by viral infections (Jasinski-Bergner et al. 2022). Synergistic suppression effects induced by HLA-G/receptor signaling include the inhibition of cell proliferation and differentiation and the induction of cell apoptosis and senescence, resulting in significant decrease or even exhaustion of immune-competent cells such as T cells, NK cells, B cells, and macrophages in patients with COVID-19 (Lin and Yan 2021). Critical COVID-19 patients had a significantly lower frequency of CD4 + HLA-G + T lymphocytes compared with moderate/severe COVID-19 patients. The increased amount of immunomodulatory HLA-G + cells may reduce the severity of the disease in moderate/severe COVID-19 patients compared with critical COVID-19 patients (Ramzannezhad et al. 2022). Furthermore, it is suggested that HLA-G 14-bp Ins/Del polymorphism is associated with COVID-19 risk (Ad’hiah and Al-Bayatee 2019).
SARS-CoV-2 Nsp13232–240 presented by HLA-E abrogates inhibition of NKG2A + NK cells. HLA-E/Nsp13232–240 complexes are formed efficiently but fail to engage NKG2A, thereby enabling unleashed NKG2A + NK cell effector functions by missing self-recognition (Hammer 2022). NP105-113-B*07:02-specific T cell responses associate with mild disease and high antiviral efficacy, pointing to inclusion for future vaccine design (Peng et al. 2022).
While HLA-C presents various antigens to cytotoxic T lymphocytes (CTLs), it also plays a key role in modulation of natural killer (NK) cell activity, which are primary cells in viral and inflammatory diseases (Kulpa and Collins 2011). A study reported the higher rate of HLA-C methylation in severe patients compared to moderate, critical, and total patients (p < 0.05). Both male and female patients had lower methylation levels than healthy volunteers. (Sharif-zak 2022) Antiviral defense relies heavily on natural killer (NK) cells. Killer cell immunoglobulin–like receptors (KIR) and associated HLA class I ligands are critical regulators of NK cell development and function. Inadequate NK cell maturation and reduced anti-SARS-CoV-2 defense are likely because of a lack of KIR3DL1 + HLA-Bw4 + and KIR3DL2 + HLA-A3/11 + combinations, which cause COVID-19 (Maruthamuthu et al. 2022). The KLRC2 gene encodes NKG2C, an activating NK cell receptor that interacts to HLA-E on infected cells to activate NK cells. Naturally occurring heterozygous or homozygous KLRC2 deletion (KLRC2del) is connected to a considerably reduced or nonexistent amount of NKG2C expression. When compared to patients with minor symptoms, hospitalized patients were markedly overrepresented in both the KLRC2del allele and, to a lesser extent, the HLA-E*0101 allele (p = 0.0006 and p = 0.01, respectively). This was especially true for patients needing intensive care (p 0.0001 and p = 0.01). The two genetic variants each served as a separate risk factor for severe COVID-19 (Vietzen et al. 2021).
Non-HLA genetic findings in COVID-19
Variation in COVID-19 infection and mortality have been documented across different groups as the pandemic has progressed. For example, total cumulative data shows that Black, Hispanic, American Indian, and Native Hawaiian people have experienced higher rates of COVID-19 cases and deaths than White and Asian people when data was adjusted to account for differences in age by race and ethnicity (COVID-19 cases and deaths by race, ethnicity 2022). COVID-19 is associated with a number of risk factors, including human genetic polymorphisms. Polymorphisms in ACE2, TMPRSS2, CCR5, ACE1, ABO locus, CCR5, APOE, IL-6, IL-10, MBL2, TLR4, NLRP3, TNFA, SIR, IL1RN, IL1B, CX3CR1, and INK4A/ARF have been reported to be potential risk predictors of COVID-19 (Feng et al. 2022; Dieter et al. 2022).
Some genes with top associations in GWAS studies have also been reported: ACE2 (p value = 3.63 10), LZTFL1 (p value = 1.05 × 10−80), SLC6A20 (p value = 1.82 × 10−47), CCR3 (p value = 1.86 × 10−19), ABO (p value = 1.15 × 10−9), TYK2 (p value = 3.7 × 10−9), OAS1 (p value = 2.73 × 10−10), FOXP4 (p value = 3.41 × 10−11), KANSL1 (p value = 1.7 × 10−11), TAC4 (p value = 3.88 × 10−9), TMPRSS2 (p value = 1.6 × 10−6), DPP9 (p value = 6.19 × 10−22), CCHCR1 (p value = 9.46 × 10−10) (https://www.covid19hg.org/).
A recent extensive review provides all association studies on COVID-19 (Dieter et al. 2022), some key findings which we will summarize here.
The gene encoding the key receptor ACE2 for SARS-CoV-2 binding is located on the X chromosome. Because males are hemizygous, they have a higher risk of overexpressing ACE2 mutations, resulting in increased susceptibility or disease severity (Feng et al. 2022). Two genome-wide association studies (GWAS) have shown that 3p21.31 and the 9q34 region containing ABO blood group sites are significantly associated with severe COVID-19 (Wu et al. 2020; Ellinghaus et al. 2020). TMPRSS2 gene polymorphisms (rs12329760, rs61735792, rs61735794) were significantly associated with COVID-19 infection (Monticelli et al. 2021; Torre-Fuentes et al. 2021). Furthermore, another study of an Italian population has shown that the furin encoded by the PCSK3 variant rs769208985 is associated with COVID-19 infection (Latini et al. 2020).
Independent of gender, age, or the severity of the illness, the IFNL4 gene (that encodes the interferon (IFN) lambda 4 protein) mutation rs12979860 is linked to the presence of COVID-19 in the Spanish population, which may be due to a decreased virus clearance (Saponi-Cortes et al. 2021). Variant rs738409 (GG genotype) in PNPLA3 gene (Patatin-like phospholipase domain-containing protein 3 PNPLA3) involved in triacylglycerol hydrolysis in adipocytes and the variant rs17047200 (TT genotype) of TLL-1 (Tolloid like-1) which is involved in activating complement system and spike protein cleavage have been correlated with SARS-COV-2-induced infection (Grimaudo et al. 2021). Subjects with PNPLA3 GG genotype have constitutive NLRP3 inflammasome upregulation and be more prone to SARS-CoV-2 tissue damage. TT genotype in TLL-1 gene may influence protease activity on SARS-CoV-2 Spike protein, increasing potential to infect or re-infect host cells (Grimaudo et al. 2021). TLE1 that encodes for TLE1 encodes for the transducin-like enhancer involved in negative regulation of negatively regulate NF-κB has been recently reported to be associated` with critical COVID-19 illness in females (Cruz et al. 2022; Kousathanas et al. 2022). Also, presence of X-linked deleterious variants in the TLR7 gene is reported to be causal for life-threatening COVID-19 only affecting males (Asano et al. 2021; Made et al. 2020; Fallerini et al. 2021). Fine-mapping shows a significant association with an independent missense variant in IL10RB, a receptor for type III (lambda) interferons (rs8178521). A lead risk variant in phospholipid scramblase 1 (chr3:146,517,122:G:A, rs343320; PLSCR1) which disrupts a nuclear localization signal that is important for the antiviral effect of interferons was also reported to be associated with critical COVID-19 outcomes (Kousathanas et al. 2022). Genes like BCL11A and TAC4 involved in lymphopoesis and differentiation of myeloid cells were also to be significantly associated in the same study (Kousathanas et al. 2022). Granulocyte macrophage colony-stimulating factor (GM-CSF) is strongly upregulated in critical COVID-19, and proposed to be target for therapy (Bonaventura et al. 2020). Mendelian randomization supported a causal involvement for coagulation factors (F8) and platelet activation (PDGFRL) in critical COVID-19 (Kousathanas et al. 2022).
A study by the Severe COVID-19 Consortium conducted a genome-wide association study of 1980 patients of European ancestry and found chromosome 3 (SLC6A20, LZTFL1, CCR9, FYC O 1, CXCR6, and XCR1) as well as in the ABO locus (with A as risk and O protective) as the only significantly associated loci (Severe Covid-19 GWAS Group et al. 2020). ELF5 has been recently reported as a new locus associated with critical illness in Europeans (Kousathanas et al. 2022). The DNA methylation status of 44 CpG sites was associated with the clinical severity of COVID-19. The gain-of-function risk A allele of a single-nucleotide polymorphism (SNP), rs17713054G > A, is identified as a probable causative variant and LZTFL1 as a candidate effector gene in pulmonary epithelial cells as contributing to the strong COVID-19 association at the 3p21.31 locus (Downes et al. 2021). Several studies imply that the androgen receptor (AR) pathway is implicated in the severity of SARS-CoV-2 infection (Cruz et al. 2022; Lamy et al. 2021). Finally, fine-mapping, colocalization, and TWAS studies show enhanced MUC1 expression due to rs41264915. This shows that mucins could play a therapeutically essential function in COVID-19 critical disease (Kousathanas et al. 2022).
Importantly, most of these association studies were conducted in European and East Asian cohorts, with a very few studies in South Asian cohorts (Srivastava et al. 2020b; Pandey et al. 2022; Srivastava 2020a; Ahmad et al. 2021). This highlights the necessity of conducting more association studies in increasingly diverse populations.
Future directions
It is abundantly clear that the HLA and COVID-19 link needs to be studied using more diverse patient populations. Due to a lack of diversity, relatively few HLA allele sets have actually been examined in relation to disease. The analysis of allelic and haplotypic variation in populations with varying patterns of linkage disequilibrium ought to aid in identifying those loci and alleles that are responsible for the observed HLA disease associations as more immunological data becomes available and DNA sequences are interpreted in the area of disease susceptibility. Moreover, the issue of genotyping ambiguity will become much more prevalent when more alleles (as well as extra exonic and intronic sequences for currently listed alleles) are discovered in more and more populations (Erlich 2012). Another potential drawback of the studies performed to date is difficulty in evaluating the significance of HLA type in connection to well-established risk variables for disease modification, such as age and clinical comorbidities (Migliorini et al. 2021; Li et al. 2020; Jain and Yuan 2020; Yang et al. 2020b; Guan et al. 2020).
Based on evidence for multigenic control of immune responses to infectious pathogens, there have been some studies where not just single HLA polymorphism but an whole extended haplotype has been reported to influence humoral responses even in vaccinated subjects (Pajewski et al. 2011; Ovsyannikova et al. 2010). Thus, in addition to specific genotypes, the entire HLA region and its associated haplotypes will determine susceptibility, making knowledge of these haplotypes crucial for the creation of future vaccines. Identification of high-risk subjects susceptible to SARS-CoV-2 infection could help to design more effective vaccines and treatments, reducing public health burden and prioritizing preventive medicine. In light of the available data, it seems prudent to include HLA testing in clinical trials, and to combine HLA typing with COVID-19 testing in order to more rapidly identify a predictor of viral severity in the population and to possibly adapt vaccination strategies to genotypically at risk populations.
Platforms for storing, processing, displaying, aggregating, and accessing data across numerous studies can be used for studying the etiology of human diseases, building predictive risk models and finding potential biomarkers and therapeutic interventions. Other GWAS portals like GRASP: Genome-Wide Repository of Associations Between SNPs and Phenotypes (Leslie et al. 2014), NHGRI GWAS catalog (Welter et al. 2014), HVGbaseG2P (Retter et al. 2005), and GWASdb (Li et al. 2016) have proven invaluable for genome-wide association studies. Due to the highly variable nature of the HLA region, and its complex nomenclature, a dedicated platform to aggregate HLA studies in SARS-CoV-2 infection and COVID-19 disease outcomes has been developed by our group and others (http://www.hlacovid19.org/database/). As we continue to unravel the fine details of the role of HLA variation in the ongoing pandemic, a resource dedicated to these studies should improve our ability to better understand the important role that these critical immune genes play in disease.
Finally, incorporation of data regarding HLA associations may aid in vaccine design and development. Binding of promiscuous epitopes to a range of human leukocyte antigen (HLA) alleles is crucial for immune control of COVID-19. When the pathogen epitope(s) triggering protective responses is unknown, genetic variation associated to resistance to infection or with less severe clinical manifestations are potentially powerful tools for the design of vaccines (Blackwell et al. 2009). For vaccine design, predictions can be made using immunoinformatic methods, which combine predictors of proteasomal processing, TAP transport, and MHC binding to produce an overall score indicating the intrinsic potential of each peptide as a T cell epitope (Cun et al. 2021). For example, an in silico analysis focused on vaccine design predicted the binding affinity of epitopes of the spike-glycoprotein to 66 common HLA class II alleles (frequency ≥ 0.01) and found that DPB1 had highest binding affinities, followed by DRB1 and DQB1. A cohort-based study, 17,440 participants of European-based ancestory, identifies HLA-A*03:01, with a strong association with poorer clinical reactions to Pfizer-BioNTech vaccines (Bolze et al. 2022) thus shows the importance of understanding the role of HLA variation in vaccine design.
Conclusion
It is becoming increasingly clear that HLA genotypes affect COVID-19 morbidity and mortality between individuals. Harnessing HLA function clinically is a challenging mission (Nguyen et al. 2020; Dendrou et al. 2018; Lin et al. 2003). Our knowledge of HLA function for clinical benefit has improved as a result of the advancements in characterizing HLA variation and HLA correlations with human disease (Tomita et al. 2020). Identifying the HLA genotype associated with the severity of COVID‐19 or susceptibility to SARS‐CoV‐2 may support future vaccination strategies to genotypically at‐risk populations. Individuals with high‐risk HLA types could be prioritized for vaccines against SARS‐CoV‐2, which may lead to effectively reduce the COVID‐19 morbidity and mortality (Tomita et al. 2020). Increased future cohort size or HLA-centric explorations may yield more meaningful signals. We also propose that more studies examining population-specific binding affinity predictions across different HLA alleles can help to assess the potential for cross‐protective immunity conferred by prior exposure. We hope that the studies detailed here may provide new insights into the COVID‐19 pandemic and improve our understanding of the relationship between HLA and COVID-19.
Footnotes
The original online version of this article was revised due to change of authorship orders of authors.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/23/2023
A Correction to this paper has been published: 10.1007/s00251-023-01300-0
References
- Abdou A et al (2010) Human leukocyte antigen (HLA) A1-B8-DR3 (8.1) haplotype, tumor necrosis factor (TNF) G-308A, and risk of non-Hodgkin lymphoma. Leukemia 24:1055–1058 [DOI] [PMC free article] [PubMed]
- Adam L et al (2021) CD8+PD-L1+CXCR3+ polyfunctional T cell abundances are associated with survival in critical SARS-CoV-2–infected patients. JCI Insight 6:e151571 [DOI] [PMC free article] [PubMed]
- Ad’hiah AH, Al-Bayatee NT (2022) HLA-G 14-bp insertion/deletion polymorphism and risk of coronavirus disease 2019 (COVID-19) among Iraqi patients. Hum Immunol 83:521–527 [DOI] [PMC free article] [PubMed]
- Ahmad SU, et al. Genome wide association study and phylogenetic analysis of novel SARS-COV-2 virus among different countries. Pak J Pharm Sci. 2021;34:1305–1313. [PubMed] [Google Scholar]
- Akgöllü E, et al. Association between chronic hepatitis B virus infection and HLA-DP gene polymorphisms in the Turkish population. Virus Res. 2017;232:6–12. doi: 10.1016/j.virusres.2017.01.017. [DOI] [PubMed] [Google Scholar]
- Alimohamadi Y, Sepandi M, Taghdir M, Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J Prev Med Hyg. 2020;61:E304–E312. doi: 10.15167/2421-4248/jpmh2020.61.3.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann DM, Boyton RJ (2020) SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci Immunol 5:eabd6160 [DOI] [PubMed]
- Amoroso A, et al. HLA and AB0 polymorphisms may influence SARS-CoV-2 infection and COVID-19 severity. Transplantation. 2021;105:193–200. doi: 10.1097/TP.0000000000003507. [DOI] [PubMed] [Google Scholar]
- Asano T et al (2021) X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci Immunol 6:eabl4348 [DOI] [PMC free article] [PubMed]
- Augusto DG, Hollenbach JA. HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection. Curr Opin Immunol. 2022;76:102178. doi: 10.1016/j.coi.2022.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeskar NS, Mania-Pramanik J. HIV and host immunogenetics: unraveling the role of HLA-C. HLA. 2016;88:221–231. doi: 10.1111/tan.12882. [DOI] [PubMed] [Google Scholar]
- Barquera R, et al. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020;96:277–298. doi: 10.1111/tan.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlyamani I, Venet F, Coudereau R, Gossez M, Monneret G (2020) Monocyte HLA-DR measurement by flow cytometry in COVID-19 patients: an interim review. Cytom Part J Int Soc Anal Cytol 97:1217–1221 [DOI] [PubMed]
- Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolze A, et al. HLA-A∗03:01 is associated with increased risk of fever, chills, and stronger side effects from Pfizer-BioNTech COVID-19 vaccination. Hum Genet Genomics Adv. 2022;3:100084. doi: 10.1016/j.xhgg.2021.100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura A, et al. Targeting GM-CSF in COVID-19 pneumonia: rationale and strategies. Front Immunol. 2020;11:1625. doi: 10.3389/fimmu.2020.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösmüller H, Matter M, Fend F, Tzankov A. The pulmonary pathology of COVID-19. Virchows Arch. 2021;478:137–150. doi: 10.1007/s00428-021-03053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler S, Nunes JM, Sanchez-Mazas A. HLA class I molecular variation and peptide-binding properties suggest a model of joint divergent asymmetric selection. Immunogenetics. 2016;68:401–416. doi: 10.1007/s00251-016-0918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A, et al. Variation in the type I interferon gene cluster on 9p21 influences susceptibility to asthma and atopy. Genes Immun. 2006;7:169–178. doi: 10.1038/sj.gene.6364287. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo SY. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med J. 2007;48:11–23. doi: 10.3349/ymj.2007.48.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborne DT, et al. Protective HLA alleles are associated with reduced LPS levels in acute HIV infection with implications for immune activation and pathogenesis. PLOS Pathog. 2019;15:e1007981. doi: 10.1371/journal.ppat.1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative (2020) The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet EJHG 28:715–718 [DOI] [PMC free article] [PubMed]
- COVID Live - Coronavirus Statistics (2022) - Worldometer. https://www.worldometers.info/coronavirus/
- COVID-19 cases and deaths by race/ethnicity: current data and changes over time | KFF (2022). https://www.kff.org/coronavirus-covid-19/issue-brief/covid-19-cases-and-deaths-by-race-ethnicity-current-data-and-changes-over-time/
- Crux NB, Elahi S (2017) Human leukocyte antigen (HLA) and immune regulation: how do classical and non-classical HLA alleles modulate immune response to human immunodeficiency virus and hepatitis C virus infections? Front Immunol 8 [DOI] [PMC free article] [PubMed]
- Cruz R et al (2022) Novel genes and sex differences in COVID-19 severity. Hum Mol Genet ddac132. 10.1093/hmg/ddac132 [DOI] [PMC free article] [PubMed]
- Cun Y, et al. COVID-19 coronavirus vaccine T cell epitope prediction analysis based on distributions of HLA class I loci (HLA-A, -B, -C) across global populations. Hum Vaccines Immunother. 2021;17:1097–1108. doi: 10.1080/21645515.2020.1823777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sorrentino AH, et al. HLA class I alleles associated with susceptibility or resistance to human immunodeficiency virus type 1 infection among a population in Chaco Province. Argentina J Infect Dis. 2000;182:1523–1526. doi: 10.1086/315854. [DOI] [PubMed] [Google Scholar]
- Degenhardt F et al (2022) Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Hum Mol Genet ddac158. 10.1093/hmg/ddac158 [DOI] [PMC free article] [PubMed]
- Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol. 2018;18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter C, et al. Genetic polymorphisms associated with susceptibility to COVID-19 disease and severity: a systematic review and meta-analysis. PLoS ONE. 2022;17:e0270627. doi: 10.1371/journal.pone.0270627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djaoud Z, Parham P. HLAs, TCRs, and KIRs, a triumvirate of human cell-mediated immunity. Annu Rev Biochem. 2020;89:717–739. doi: 10.1146/annurev-biochem-011520-102754. [DOI] [PubMed] [Google Scholar]
- Douillard V et al (2021) Current HLA investigations on SARS-CoV-2 and perspectives. Front Genet 12 [DOI] [PMC free article] [PubMed]
- Downes DJ, et al. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat Genet. 2021;53:1606–1615. doi: 10.1038/s41588-021-00955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta M, Dutta P, Medhi S, Borkakoty B, Biswas D. Polymorphism of HLA class I and class II alleles in influenza A(H1N1)pdm09 virus infected population of Assam. Northeast India J Med Virol. 2018;90:854–860. doi: 10.1002/jmv.25018. [DOI] [PubMed] [Google Scholar]
- Elahi S, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bendary M, et al. HLA class II-DRB1 alleles with hepatitis C virus infection outcome in Egypt: a multicentre family-based study. Ann Hepatol. 2019;18:68–77. doi: 10.5604/01.3001.0012.7864. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D et al (2020) Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med NEJMoa2020283. 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed]
- Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80:1–11. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- Fan J-H, et al. Association of HLA-DQ and IFNL4 polymorphisms with susceptibility to hepatitis B virus infection and clearance. Ann Hepatol. 2016;15:532–539. [PubMed] [Google Scholar]
- Fallerini C et al (2021) Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. eLife 10:e67569 [DOI] [PMC free article] [PubMed]
- Fellay J, et al. Common genetic variation and the control of HIV-1 in humans. PLOS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, et al. Potential genes associated with COVID-19 and comorbidity. Int J Med Sci. 2022;19:402–415. doi: 10.7150/ijms.67815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francies FZ, Dlamini Z. Aberrant splicing events and epigenetics in viral oncogenomics: current therapeutic strategies. Cells. 2021;10:239. doi: 10.3390/cells10020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. 2020;213:54–56.e1. doi: 10.5694/mja2.50674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman KE et al (2021) Impact of sex and metabolic comorbidities on coronavirus disease 2019 (COVID-19) mortality risk across age groups: 66 646 inpatients across 613 U.S. hospitals. Clin Infect Dis Off Publ Infect Dis Soc Am 73:e4113–e4123 [DOI] [PMC free article] [PubMed]
- Grifoni A, et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaudo S, et al. PNPLA3 and TLL-1 polymorphisms as potential predictors of disease severity in patients with COVID-19. Front Cell Dev Biol. 2021;9:627914. doi: 10.3389/fcell.2021.627914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W-J, et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55:2000547. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Q et al (2022) SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide that abrogates inhibition of NKG2A-expressing NK cells. Cell Rep 38 [DOI] [PMC free article] [PubMed]
- Henkens MTHM, et al. Age is the main determinant of COVID-19 related in-hospital mortality with minimal impact of pre-existing comorbidities, a retrospective cohort study. BMC Geriatr. 2022;22:184. doi: 10.1186/s12877-021-02673-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft EB et al (2020) Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. MedRxiv Prepr Serv Health Sci 2020.10.25.20219063 (2021). 10.1101/2020.10.25.20219063
- Huang J, et al. The associations of HLA-A*02:01 and DRB1*11:01 with hepatitis C virus spontaneous clearance are independent of IL28B in the Chinese population. Sci Rep. 2016;6:31485. doi: 10.1038/srep31485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Yuan J-M. Predictive symptoms and comorbidities for severe COVID-19 and intensive care unit admission: a systematic review and meta-analysis. Int J Public Health. 2020;65:533–546. doi: 10.1007/s00038-020-01390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski-Bergner S, Schmiedel D, Mandelboim O, Seliger B (2022) Role of HLA-G in viral infections. Front Immunol 13 [DOI] [PMC free article] [PubMed]
- Kasuga Y, Zhu B, Jang K-J, Yoo J-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp Mol Med. 2021;53:723–736. doi: 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor S-S, et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, age and sex are associated with severity of Japanese COVID-19 with respiratory failure. Front Immunol. 2021;12:658570. doi: 10.3389/fimmu.2021.658570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607:97–103. doi: 10.1038/s41586-022-04576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa DA, Collins KL. The emerging role of HLA-C in HIV-1 infection. Immunology. 2011;134:116–122. doi: 10.1111/j.1365-2567.2011.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuniholm MH, et al. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology. 2010;51:1514–1522. doi: 10.1002/hep.23515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar P, et al. Superior control of HIV-1 replication by CD8+ T cells targeting conserved epitopes: implications for HIV vaccine design. PLoS ONE. 2013;8:e64405. doi: 10.1371/journal.pone.0064405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacedonia D, et al. Impact of smoking, COPD and comorbidities on the mortality of COVID-19 patients. Sci Rep. 2021;11:19251. doi: 10.1038/s41598-021-98749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy P-J, Rébillard X, Vacherot F, de la Taille A. Androgenic hormones and the excess male mortality observed in COVID-19 patients: new convergent data. World J Urol. 2021;39:3121–3123. doi: 10.1007/s00345-020-03284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini A, et al. COVID-19 and genetic variants of protein involved in the SARS-CoV-2 entry into the host cells. Genes. 2020;11:E1010. doi: 10.3390/genes11091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Koohy H (2020) In silico identification of vaccine targets for 2019-nCoV. F1000Research 9:145 [DOI] [PMC free article] [PubMed]
- Leslie R, O’Donnell CJ, Johnson AD. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinforma Oxf Engl. 2014;30:i185–194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016;44:D869–876. doi: 10.1093/nar/gkv1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146:110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Yan W-H (2021) Perspective of HLA-G induced immunosuppression in SARS-CoV-2 infection. Front Immunol 12 [DOI] [PMC free article] [PubMed]
- Lin M, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58:1131–1134. doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- Littera R, et al. Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. The Sardinian Experience Front Immunol. 2020;11:605688. doi: 10.3389/fimmu.2020.605688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Livanos AE, et al. Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 2021;160:2435–2450.e34. doi: 10.1053/j.gastro.2021.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C, et al. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature. 2021;600:523–529. doi: 10.1038/s41586-021-04085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Smith DW, Mackenzie JS, Smith DW. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol Aust. 2020;41:45–50. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden EA, Diamond MS. Host cell-intrinsic innate immune recognition of SARS-CoV-2. Curr Opin Virol. 2022;52:30–38. doi: 10.1016/j.coviro.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoni M. South Africa responds to new SARS-CoV-2 variant. Lancet Lond Engl. 2021;397:267. doi: 10.1016/S0140-6736(21)00144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov PV, Pybus OG. Evolution and diversity of the human leukocyte antigen(HLA) Evol Med Public Health. 2015;2015:1. doi: 10.1093/emph/eou033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol Rev. 2013;254:245–264. doi: 10.1111/imr.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu S, et al. Individualized constellation of killer cell immunoglobulin-like receptors and cognate HLA class I ligands that controls natural killer cell antiviral immunity predisposes COVID-19. Front Genet. 2022;13:845474. doi: 10.3389/fgene.2022.845474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RJ (2020) Thoughts on the alveolar phase of COVID-19. Am J Physiol - Lung Cell Mol Physiol 319:L115–L120 [DOI] [PMC free article] [PubMed]
- Mathew D et al (2020) Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369:eabc8511 [DOI] [PMC free article] [PubMed]
- Merad M, Blish CA, Sallusto F, Iwasaki A. The immunology and immunopathology of COVID-19. Science. 2022;375:1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- Meyer DC, Aguiar VR, Bitarello BDC, Brandt DY, Nunes KA (2018) genomic perspective on HLA evolution. Immunogenetics 70:5–27 [DOI] [PMC free article] [PubMed]
- Migliorini F, et al. Association between HLA genotypes and COVID-19 susceptibility, severity and progression: a comprehensive review of the literature. Eur J Med Res. 2021;26:84. doi: 10.1186/s40001-021-00563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, et al. Microarray analyses of differentially expressed human genes and biological processes in ECV304 cells infected with rubella virus. J Med Virol. 2007;79:1783–1791. doi: 10.1002/jmv.20942. [DOI] [PubMed] [Google Scholar]
- Monticelli M, et al. Protective role of a TMPRSS2 variant on severe COVID-19 outcome in young males and elderly women. Genes. 2021;12:596. doi: 10.3390/genes12040596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94:e00510–e520. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N, et al. Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci Rep. 2016;6:24767. doi: 10.1038/srep24767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A, et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA. 2020;96:610–614. doi: 10.1111/tan.14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, et al. The roles of HLA-DQB1 gene polymorphisms in hepatitis B virus infection. J Transl Med. 2018;16:362. doi: 10.1186/s12967-018-1716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Extended LTA, TNF, LST1 and HLA gene haplotypes and their association with Rubella vaccine-induced immunity. PLoS ONE. 2010;5:e11806. doi: 10.1371/journal.pone.0011806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E, et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pajewski NM, et al. The role of HLA DR-DQ haplotypes in variable antibody responses to Anthrax Vaccine Adsorbed. Genes Immun. 2011;12:457–465. doi: 10.1038/gene.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P, Agarwal S, Rajkumar (2020) Lung pathology in COVID-19: a systematic review. Int J Appl Basic Med Res 10:226–233 [DOI] [PMC free article] [PubMed]
- Pandey RK, Srivastava A, Singh PP, Chaubey G. Genetic association of TMPRSS2 rs2070788 polymorphism with COVID-19 case fatality rate among Indian populations. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2022;98:105206. doi: 10.1016/j.meegid.2022.105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, et al. An immunodominant NP105-113-B*07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID-19 disease. Nat Immunol. 2022;23:50–61. doi: 10.1038/s41590-021-01084-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarleri J, Galvan V, Delpino MV. Omicron variant of the SARS-CoV-2: a quest to define the consequences of its high mutational load. GeroScience. 2021;44:53–56. doi: 10.1007/s11357-021-00500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramzannezhad S, et al. The association of decreased HLA-G+ immune cell frequencies with critical COVID-19 patients. Microb Pathog. 2022;167:105550. doi: 10.1016/j.micpath.2022.105550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retter I, Althaus HH, Münch R, Müller W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33:D671–674. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, et al. IPD-IMGT/HLA database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Casillas A, Redwan EM, Uversky VN. SARS-CoV-2: a master of immune evasion. Biomedicines. 2022;10:1339. doi: 10.3390/biomedicines10061339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, et al. KIR2DL2 combined with HLA-C1 confers risk of hepatitis C virus-related hepatocellular carcinoma in younger patients. Oncotarget. 2018;9:19650–19661. doi: 10.18632/oncotarget.24752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cerrillo I et al (2020) Differential redistribution of activated monocyte and dendritic cell subsets to the lung associates with severity of COVID-19. medRxiv 2020.05.13.20100925. 10.1101/2020.05.13.20100925
- Saponi-Cortes JMR, et al. IFNL4 genetic variant can predispose to COVID-19. Sci Rep. 2021;11:21185. doi: 10.1038/s41598-021-00747-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe Covid-19 GWAS Group et al (2020) Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med 383:1522–1534 [DOI] [PMC free article] [PubMed]
- Sharif-zak, M et al (2022) Influence of disease severity and gender on HLA-C methylation in COVID-19 patients. Iran J Sci Technol Trans Sci 1–8. 10.1007/s40995-022-01334-1 [DOI] [PMC free article] [PubMed]
- Shi Y, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- Srivastava A et al (2020a) Genetic association of ACE2 rs2285666 polymorphism with COVID-19 spatial distribution in India. Front Genet 11 [DOI] [PMC free article] [PubMed]
- Srivastava A, et al. Most frequent South Asian haplotypes of ACE2 share identity by descent with East Eurasian populations. PLoS ONE. 2020;15:e0238255. doi: 10.1371/journal.pone.0238255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JW, Tambyah PA, Hui DS. Emergence of a new SARS-CoV-2 variant in the UK. J Infect. 2021;82:e27–e28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JW, Toovey OTR, Harvey KN, Hui DSC (2021b) Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect 82:e8–e10 [DOI] [PMC free article] [PubMed]
- Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira SLM, et al. Association of the HLA-B*52 allele with non-progression to AIDS in Brazilian HIV-1-infected individuals. Genes Immun. 2014;15:256–262. doi: 10.1038/gene.2014.14. [DOI] [PubMed] [Google Scholar]
- The MHC sequencing consortium Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Thio CL, et al. Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol. 2003;77:12083–12087. doi: 10.1128/JVI.77.22.12083-12087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MG et al (2020) Viral-induced alternative splicing of host genes promotes influenza replication. eLife 9:e55500 [DOI] [PMC free article] [PubMed]
- Thursz MR, et al. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–1069. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- Tian C, et al. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun. 2017;8:599. doi: 10.1038/s41467-017-00257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Ikeda T, Sato R, Sakagami T. Association between HLA gene polymorphisms and mortality of COVID-19: an in silico analysis. Immun Inflamm Dis. 2020;8:684–694. doi: 10.1002/iid3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre-Fuentes L, et al. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid. Spain J Med Virol. 2021;93:863–869. doi: 10.1002/jmv.26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodis N et al (1998) Physical map of human 6p21.2–6p21.3: region flanking the centromeric end of the major histocompatibility complex. Genome Res 8:631–643 [DOI] [PMC free article] [PubMed]
- Valenzuela-Ponce H, et al. Novel HLA class I associations with HIV-1 control in a unique genetically admixed population. Sci Rep. 2018;8:6111. doi: 10.1038/s41598-018-23849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made CI, et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejbaesya S, et al. HLA association with hepatitis C virus infection. Hum Immunol. 2000;61:348–353. doi: 10.1016/S0198-8859(99)00131-7. [DOI] [PubMed] [Google Scholar]
- Vietzen H, et al. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA-E variants are risk factors for severe COVID-19. Genet Med off J Am Coll Med Genet. 2021;23:963–967. doi: 10.1038/s41436-020-01077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, et al. Abnormal global alternative RNA splicing in COVID-19 patients. PLOS Genet. 2022;18:e1010137. doi: 10.1371/journal.pgen.1010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6:1–16. doi: 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang W, Zhang J, He J, Zhu F. Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19) HLA. 2020;96:194–196. doi: 10.1111/tan.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D et al (2020) Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 5:eabd2071 [DOI] [PMC free article] [PubMed]
- Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk AJ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk AJ, et al. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J Exp Med. 2021;218:e20210582. doi: 10.1084/jem.20210582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B-B, Gu D-Z, Yu J-N, Yang J, Shen W-Q. Association between ABO blood groups and COVID-19 infection, severity and demise: a systematic review and meta-analysis. Infect Genet Evol. 2020;84:104485. doi: 10.1016/j.meegid.2020.104485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, et al. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct Target Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen M-F, et al. Hepatitis B virus infection. Nat Rev Dis Primer. 2018;4:1–20. doi: 10.1038/nrdp.2018.35. [DOI] [PubMed] [Google Scholar]
- Zhang Q et al (2020) Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570 [DOI] [PMC free article] [PubMed]
- Zhang Y, et al. Allelic imbalance of HLA-B expression in human lung cells infected with coronavirus and other respiratory viruses. Eur J Hum Genet. 2022;30:922–929. doi: 10.1038/s41431-022-01070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. A review of SARS-CoV2: compared with SARS-CoV and MERS-CoV. Front Med. 2021;8:628370. doi: 10.3389/fmed.2021.628370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, et al. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T, Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat Rev Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]