Abstract
Suspension and imaging cytometry techniques that simultaneously measure hundreds of cellular features are powering a new era of cell biology and transforming our understanding of human tissues and tumors. However, a central challenge remains in learning the identities of unexpected or novel cell types. Cell identification rubrics that could assist trainees, whether human or machine, are not always rigorously defined, vary greatly by field, and differentially rely on cell intrinsic measurements, cell extrinsic tissue measurements, or external contextual information such as clinical outcomes. This challenge is especially acute in the context of tumors, where cells aberrantly express developmental programs that are normally time, location, or cell-type restricted. Well-established fields have contrasting practices for cell identity that have emerged from convention and convenience as much as design. For example, early immunology focused on identifying minimal sets of protein features that mark individual, functionally distinct cells. In neuroscience, features including morphology, development, and anatomical location were typical starting points for defining cell types. Both immunology and neuroscience now aim to link standardized measurements of protein or RNA to informative cell functions such as electrophysiology, connectivity, lineage potential, phospho-protein signaling, cell suppression, and tumor cell killing ability. The expansion of automated, machine-driven methods for learning cell identity has further created an urgent need for a harmonized framework for distinguishing cell identity across fields and technology platforms. Here, we compare practices in the fields of immunology and neuroscience, highlight concepts from each that might work well in the other, and propose ways to implement these ideas to study neural and immune cell interactions in brain tumors and associated model systems.
Keywords: Cell identity, Immunology, Neuroscience, Single cell, Cytometry, Brain tumors
Introduction
Rigorous definitions of cell identity are a sign of field maturity, since they rely on a detailed understanding of connections between cellular phenotype, function, and developmental origins. Cells are the basic unit for organisms, and ways in which they differentiate, become arranged into complex structures or systems, and maintain or shift their function throughout life are processes that are understood incompletely and to different degrees across organ systems. Historically, cells have been defined by features such as morphology, location, and interactions with other cell types. In immunology, advances in multidimensional analyses during the 1970s launched a revolution in understanding the function and origin of immune cells that revealed a spectrum of distinct, highly specialized cell types [1]. Immunology progress was driven in part by access to single cell suspensions and the observation that morphologically similar lymphocyte populations contained cell subsets with highly contrasting functions that could be distinguished by surface antigens. These facts drove the creation of quantitative, single-cell identification systems, including the clusters of differentiation (CD) marker system [2], and closely linked advances in analytical cytometry and fluorescence activated cell sorting (FACS) to advances in immunology [3]. Today, cell type definitions typically include some combination of phenotype, lineage origin, fate potential, and capacity to perform a key function in the future. In immunology, the term “polarization” is used to describe stable, reversible sets of cell states that, with the appropriate signal in the corresponding context, can be switched between contrasting functions [4]. This is similar to the term “plasticity” in neuroscience [5] and cancer biology [6], where the boundary between cell state and cell function is also not well defined. There is now also strong evidence such plasticity exists for both blood and tissue macrophages, which further supports the idea that dysfunctional cells might be reconditioned in settings of an injury or a disease [7]. Other examples of cell states could include signaling ability [8], proliferation or quiescence status, and being memory or naïve, all of which are key refinements to the concept of an identity that can distinguish dramatically different subtypes of cells within an otherwise shared lineage. Critically, the majority of cell states are thought to be encoded in proteins and their posttranslational modifications and are not directly detectable in DNA or RNA sequence reads [9]. A cell’s gene expression program is thus a window into the cell’s potential [10]; however, disconnects between RNA and protein raise the concern that a snapshot of the transcriptome may not reflect a cell’s current functional identity.
Individually, techniques such as histological and morphological assessments, genome-wide profiling, and epigenomic, transcriptomic, proteomic, and metabolomic analyses have been vital tools for defining cell identity [11]. While these techniques have progressed from bulk analysis of sorted or enriched cell populations to true analysis of individual cells, major differences remain in their practicality for single-cell analysis of primary tumors [9]. Furthermore, no one method is widely accepted across fields as sufficient to define a given single cell’s identity. The jargon of field-specific cell definitions also presents a barrier to harmonizing cell classifications across research areas, diseases, and tissue types. This is especially true in tumor microenvironments where complex mixtures of cell types with abnormal functional identities are observed. Additionally, in cancer biology, the goal is frequently to prove a functional identity (viz., “stem,” “malignant,” “suppressor”) that requires study and testing of living cells. The plasticity of mature cells in response to environmental cues and stimuli can lead to changes in cell state and even identity. While studying these processes is especially relevant for understanding cancer [12], it is also challenging to maintain cells in research systems without disrupting the exact processes to be measured. Live cells, whether in a tumor or in a research lab, change over time, especially when the environmental context around them is altered. In addition to epigenetic changes, selective pressures can enrich for mutations in regulatory genes that lead to a shift in cell identity and contribute to cellular reprogramming, as commonly observed in cancer. Thus, oncogenesis might be thought of as a process whereby cells gain the ability to change their identity. For a malignant cell, the advantages of such plasticity include fate flexibility, which creates a diverse pool of cells that can withstand a range of treatments or immune responses, and useful new functions such as stem/progenitor self-renewal abilities [13, 14].
Addressing challenges in stem and cancer cell biology
Links between plasticity of cell identity and malignant transformation have led to widely used terms like “cancer stem cell” (CSC), a term that means very different things in different research contexts. For example, CSCs were originally proposed as a concept to explain clinical observations like therapy evasion [15], but CSCs can also be an allusion to a proposed origin of the cancer from a stem or progenitor cell [16, 17], and CSCs can be used as a name for a cell subtype that is capable of transferring malignancy or repopulating multiple tumor cell types [18, 19]. In brain tumors, multiple proteins have been proposed as defining markers of glioma stem cells, including CD133 [18], SOX2 [20, 21], NESTIN [22], EGFR [23], and CD15 [24] A simultaneous analysis of these proteins reveals that there are multiple subpopulations of glioma stem cells present within individual tumors, and helps to resolve which of these cell subsets are associated with clinical outcomes [25]. Field-specific differences in cell identity definitions will be discussed more below, but as a starting point the authors suggest, as a minimum best practice, that all fields use the cytometry hallmark of formally defining the cell identification system the first time a cell label is used (for the immunologists: show your gates). For example, for the term “helper T cell,” there are multiple ways to identify and even isolate this functional cell group, and immunologists generally are required to “show their work” the first time they mention a cell type by displaying the exact order and set of proteins used to include or exclude cells from that definition label. To go beyond this, we also recommend a table, label, or plot of summary statistics (e.g., heatmap) for all measured features to be reported. As an example, marker enrichment modeling (MEM) labels provide human- and machine-readable reports of enrichment on a 10-point scale [26]. The original goal of the MEM label was to develop an automated, quantitative version of the immunology practice whereby an isolated cell subpopulation is described with a string of observed protein expression values (e.g., CD34hi CD38lo/− as a defining label for blood stem cells or CD3 + CD4 + CD8- FOXP3 + as defining label for regulatory T cells). The MEM algorithm calculates a label based on data and scales values to a 10-point scale, generating a label like “CD44+8 CD38+8 CD8+7 ICOS+7 CD45+6 CD45R0+6 CD26 + 6 PD-1+6…”. This machine-generated label can be used to identify the population as CD38hiICOShi memory CD8 T cells; this example label described the main population of SARS-CoV-2-specific T cells induced by RNA vaccination [27]. Quantitative labels like MEM can be used to compare phenotypes across analysis runs, experiments, or instrument platforms and have been applied in brain tumors to define cells both by total protein and phospho-protein state [25].
Cancer research has focused on the cellular and molecular characterizations of bulk tumor masses in the past. However, there is strong evidence that many tumors, including glioblastoma, are composed of heterogeneous cell types. It remains unclear whether this heterogeneity is strictly genetic or if it arises from a cellular hierarchy of growth and differentiation within the tumor that allows for cells to employ new and diverse cell identity programs [28, 29]. For example, the disruption of pathways regulating self-renewal and differentiation through the acquisition of transforming mutations in leukemia generates leukemic stem cells that possess an altered differentiation program. This was demonstrated by aberrant expression of surface markers and ability to give rise to an altered developmental hierarchy that retained aspects of its normal counterpart [30]. To further dissect if this mechanism also holds true across many tumors, it will be valuable to conduct functional experiments with subpopulations of viable cells isolated from tumors that have been rigorously identified by quantitative cytometry.
Historical and modern best practices
Both immune and neural cells are well characterized based on phenotype, structure, and function. Tumor cells, however, are often found in intermediate differentiation steps that coopt different phenotypes and functions, enabling them to survive and proliferate even in unfavorable conditions. Tumors can achieve complexity at the level of organs or tissues with dynamic regulation, organization of immune structures, and support of other nonmalignant cellular populations and structures during tumor initiation, maintenance, and progression. Here, we propose a combination of approaches from the fields of immunology and neuroscience that may be helpful for characterizing and aligning glioblastoma cell subsets to their nearest neural cell cognate (Fig. 1).
Fig. 1.
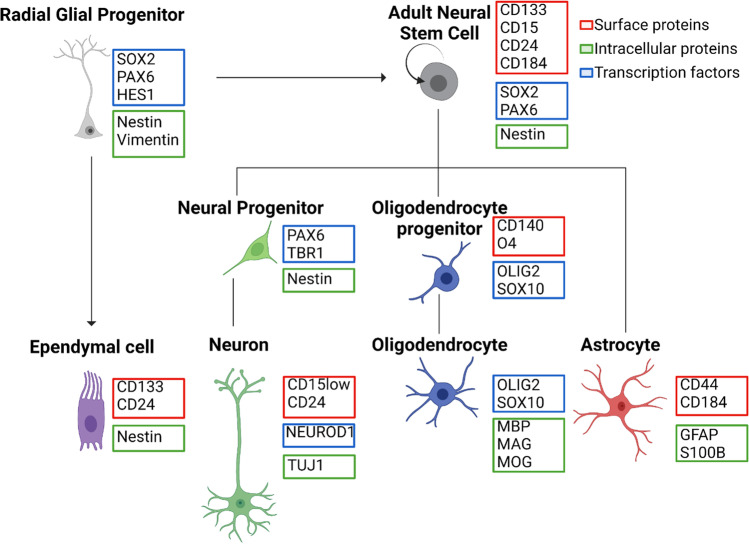
Phenotypic identifiers of cells in the neural lineage. A diagram of human neural cell identities is shown in the style of hematopoietic immune cell identity maps. Bold labels indicate neural cell types. Protein name labels highlight markers that are proposed to define neural cell identities, including cell surface proteins (red), transcription factors (blue), and other intracellular proteins (green). Lines connect multipotent stem and progenitor cells (top) with the neural lineage cell types they can produce (middle and bottom). This diagram attempts to be cell-intrinsic and to highlight surface marker sets that distinguish each major cell type, as in traditional immunology views of cell identity where FACS separation of live cells can be used for functional testing. Thus, the diagram does not explicitly consider other features that are classically important to understanding neural cell identity, such as morphology, location, and structure [31–33]. Notably, radial glial progenitors, a type of neural stem cell, are conceptually separated from adult neural stem cells both by a key difference in potency (only radial glial progenitors are normally thought to produce ependymal cells) and by developmental time (radial glial progenitors are only observed prenatally in humans)
Historically, the hematopoietic system has been used as a paradigm to illustrate a developmental hierarchy of cells that is sustained by a population of long-lived, quiescent, pluripotent stem cells capable of self-renewal and contributing to the replenishment of a spectrum of mature immune cell types [34]. The discovery and characterization of hematopoietic stem cells (HSCs) relied heavily on the ability to identify and purify such cells [35], as these cells were first defined by their functional ability to regenerate all hematopoietic lineages in vivo [36]. Repopulation assays using hematopoietic stem cells have since been performed to help investigate heterogeneity in HSCs [37]. This highlights how functional assays played a major role in the identification of different immune cells [38, 39]. Table 1 summarizes and highlights different ways in which the field of immunology established cell identity definitions across cells of a hematopoietic lineage. The establishment of the CD marker system was central in reshaping the way immune cells are identified, in this case based on the molecules expressed on their surface [40]. CD molecules are now routinely used as definitional cell markers, allowing for the scoring of the presence and proportions of specific leukocyte cell subsets. For example, CD45, also known as the leukocyte common antigen, is a receptor-linked protein tyrosine phosphatase that is expressed on nucleated cells of a hematopoietic lineage and can be used to distinguish immune cells from other cell types [41]. As with cell surface signaling proteins, transcription factor proteins can greatly influence the molecular content and function of cells. Measuring the expression of a specific transcription factor can provide information about the state of a cell and how it is likely to respond to signaling cues or regulate the expression of important functional proteins. For example, T cells can functionally be defined by the expression of CD3, a signaling subunit of the T cell antigen receptor [42]; the expression of transcription factors such as FOXP3 can give additional context and suggest a functional identity as a regulatory T cell [43]. Distinct patterns of transcription factor expression are also illustrated across B cell maturation. While all B cells express CD19, a defining B lymphocyte antigen, B cells express different transcription factors such as PAX5, BCL6, and BLIMP throughout development and maturity. These transcription factors mark key B cell maturation events including commitment to the B lineage in early B cells (PAX5) [44, 45], encountering antigen, receiving T cell help, becoming a germinal center B cell (BCL6) [46], and specializing into a plasmablast (BLIMP1) [47] that will ultimately turn off much of the B cell program and generate an antibody-producing plasma cell. It has been shown that the activation of transcription factors not only marks cell maturation/differentiation, but activation of certain transcription factors can also lead to the dedifferentiation of hematopoietic cells [48]. Lineage inference techniques including single cell RNA sequencing have also suggested new models of cellular development or markers identifying transitional states [49]. Critically, inferred lineages based on protein expression pattern must be tested experimentally, as these approaches can misclassify cells when expression programs follow an off–on-off type of pattern where many genes are coordinated together, as with lymphocytes.
Table 1.
Immunology and neurobiology examples of class and modern cell identity definitions
| Identifier category | Immune cells | Neural cells | Notes on identifiers** | |||||
|---|---|---|---|---|---|---|---|---|
| Feature(s) | Cell | Proof type | Feature(s) | Cell | Proof type | Pros | Cons | |
| Morphology and location | Star shaped | Dendritic cells | Functional | Star shaped | Astrocytes | Functional [50] | Easily visualized | Not connected to specific molecules or functions |
| - | - | - | Large, extensive dendritic tree; located in cerebellum | Purkinje neurons | Functional [51] | |||
| - | - | - | Elongated radial shape, apical membrane contacting lateral ventricle SVZ | Adult neural stem cells | Functional [52, 53] | |||
| - | - | - | Columnar shaped, multiciliated cells that line the ventricles | Ependymal cells | Functional [54] | |||
| Surface protein, extracellular epitope | CD3 + | T lymphocytes | Definitional, functional [42] | E-cadherin + , CD133, CD24 | Ependymal cells | Strong evidence of epithelial barrier function [55–57] | Can be used to physically isolate live cells | Many surface proteins are not cell type restricted or have unknown functions |
| CD19 + | B lymphocytes | Definitional, functional [58] | CD133, CD15, CD24, CD184 | Adult neural stem cell | Strong evidence, not definitional, requires validation [59, 60] | |||
| CD45 + | Mature, hematopoietic | Good evidence [41] | CD24, CD44 | Astrocytes | Strong evidence, not definitional, requires validation [61] | |||
| CD33 + CD16 + CD14- | Non-classical monocyte | Definitional [56] | CD140a (PDGFRa), NG2 | Oligodendrocytes | Strong evidence, not definitional, requires validation [57, 62] | |||
| Transcription factor protein | FOXP3 + | Regulatory T cell (Treg) | Strong evidence, functional[43] | SOX2, PAX6 | Adult neural stem cells | Strong evidence, but functional assays required [63, 64] | Transcription factors can directly change cell identity | Low expression levels, intracellular |
| PAX5 + | Early B lymphocytes | Definitional, functional[44, 45, 65] | OLIG2, SOX10 | Oligodendrocytes | Strong evidence, but functional assays required [62] | |||
| BCL6 + | Germinal center B cell | Definitional, functional (in tissue context) [46] | NEUROD1 | Neurons | Strong evidence, but functional assays required [66] | |||
| Other intracellular protein | CD20 | B lymphocyte | Strong evidence [67] | TUJ1 (β-III-tubulin) | Neurons | Strong evidence (in tissue context) [68] | Commonly available | Low expression levels, intracellular |
| LYN | B lymphocyte | Good evidence, functional [69] | MBP | Oligodendrocytes | Strong evidence (in tissue context) [70] | |||
| TdT | (pre-B cells) | Definitional, functional [71] | GFAP, S100B | Astrocytes | Strong evidence (in tissue context) [72] | |||
| Protein production | Secretes IL-4 | Th2 Helper T cell |
Functional [73] |
Produces myelin | Oligodendrocytes | Definitional, functional [70] | Functional test | Requires live cells, difficult assay, can require Golgi blocking agents |
| Secretes immunoglobulin | Plasma cell |
Definitional, functional [74] |
Secretes neurotransmitters | Neurons | Definitional, functional [75] | |||
| Secretes iNOS | Myeloid derived suppressor cell |
Functional [76] |
- | - | - | |||
| Intracellular feature | Engulfs bacteria, beads, or other cells | Phagocyte |
Definitional, functional [39] |
Electrical activity | Neurons | Definitional, functional [54] | Widely used | Requires live cells or tissue |
| Cell mixing assay | Suppresses T cell proliferation | Myeloid derived suppressor cell |
Definitional, functional [38] |
Oligodendrocyte and neuron co-culture to assess myelination | Oligodendrocytes | Definitional, functional [77, 78] | Commonly available | Requires cell culture and expertise |
| Lineage tracing assay | Produces all lymphocytes, no myeloid cells | Common lymphocyte progenitor |
Definitional, functional [79] |
Generates astrocytes, neurons and oligodendrocytes | Adult neural stem cells | Definitional, functional [80] | Commonly available | Requires animal model and/or live cells |
| DNA mutations | Somatic hypermutation in IgH gene | Antigen-experienced, mature B cells | Definitional, functional [81] | - | - | - | Commonly available | Low expression levels; not functional |
| Transcription factor mRNA | Surrogate, requires validation | Surrogate, requires validation | Commonly available | Low expression levels; not functional | ||||
* Definitions may have exceptions in less commonly studied tissues or in the context of diseases. ** Other terms that do not mark cell identities (and explanation): stromal (in epithelial biology it marks a layer containing multiple cell types, in cancer it means "all other cell types'), mesenchymal (adjective applied to distinct types of cells, including adult cells of epithelial or hematopoietic origin), glial (typically denotes cells derived from a neural stem cell that are not neuronal, such as astrocytes and oligodendrocytes; historically denotes cells in nervous tissue that are not neurons), immune (marks a function seen on epithelial and hematopoietic cells), antigen presenting cell (marks a function, not a cell type)
Use of brain anatomy in neural cell identity
Neural cell identity characterization is based on both classic features such as cell shape/morphology, physiological location, and structure, as well as per-cell measurement of RNA or protein [54]. Table 1 reviews some of the classic ways of identifying cells and how they have been applied to neural cell identity. Given the complex architecture of the human brain, the effort to categorize neural stem cells and their progeny has focused extensively on their location across developmental time and space [31]. There are two main germinal structures where a series of distinct stages of neural progeny maturation have been well characterized: the ventricular-subventricular zone (V-SVZ) lining the lateral ventricles and the subgranular zone (SGZ) in the dentate gyrus of the hippocampus [82, 83]. Here, we have chosen to focus on the larger of the two niches, the V-SVZ, and its developmental antecedents, radial glia, as an example case of neural stem cells [84]. Radial glia are essential neural and glial progenitor cells in the prenatal brain. Their hallmark radial process serves as a physical guide for migrating neurons during the structural development of the brain [85]. The generation of radial glial cells is marked by the expression of several intermediate filament proteins including nestin and vimentin, which are known stem cell markers [86]. However, neural stem cells and their progeny highlight one of the major challenges of characterizing neural cell identity using protein expression markers as the hierarchy of stem, progenitor, and differentiated neural cells contains many areas of exception and overlap. For example, the simple category of “radial glia” in the prenatal brain has, of late, been expanded and subdivided as different subclasses have been found across developmental stages and species [87, 88]. Adult neural stem cells in the SVZ express glial fibrillary acidic protein (GFAP), which is also a historically well-established marker of most astrocytes [29, 72], and a subset of quiescent neural stem cells lacks nestin [59]. Similarly, ependymal cells express other markers, such as CD133/prominin-1, which are also seen on neural stem cells. However, ependymal cells have historically been considered separately from other neural original cells, since ependymal cells are multiciliated, contiguous with choroid plexus epithelial cells, and form a monolayered barrier between the V-SVZ and the ventricle lumen [59, 89]. Thus, ependymal cells arise from a neural stem cell and provide epithelial functions. Another example of this apparent disconnect between function and lineage is seen in pulmonary neuroendocrine cells, which arise from an epithelial cell and provide neuroendocrine functions [90]. Thus, terms like “epithelial” should be clearly defined as referring to a current functional identity or a prior lineage or tissue origin.
Most neural cell types are functionally characterized and have a specific assay that is considered definitional. For example, neural stem cells are defined in vivo by their ability to self-renew and give rise to neural and glial progeny [91]. Stem-ness features are commonly tested in vitro using neurosphere assays, although extensive evidence has shown that the culture conditions for such assays can change the assay’s outcome [92–94]. For example, detection of long-term stem cells that are quiescent in vivo is especially challenging [59]. Astrocytes are the most abundant cell type in the brain and help regulate axons and blood flow and maintain homeostasis [95, 96]. Oligodendrocytes myelinate cells and regulate neuronal activities [97]. Neurons are electrically excitable, and their primary function is to relay electrochemical signals to, from, and within the brain [75]. Much effort has been spent to identify definitive markers of neural identity that will help distinguish them from other neural cells and further study them in the context of cancer. An extensive transcriptome analysis of neural cells has brought greater understanding of each cell type and their gene expression programs [98]. Transiently expressed transcription factors have been identified as definitional markers of cell identity, but transcription factors may be expressed in different cell subtypes across different stages of cell development or be expressed in an oscillatory fashion, complicating the interpretation of a single transcriptional snapshot [99]. For example, SOX2 is a well-established but not exclusive functional marker of neural stem cells, while PAX6 can be expressed both in neural stem cells as well as intermediate cell types like neural progenitors during neuronal differentiation [63, 66]. Lastly, immunophenotyping screens have aided in the identification of potential cell surface signatures of neural cells [32, 59, 72]; however, a better understanding of CD marker expression would help bridge the gap between descriptive and functional single cell analyses for neural origin cells.
Ultimately, while we note the immense amount of work that has been put toward characterizing neural cell identity, we think it is important that we continue to link more routine measurements of protein and RNA transcripts to critical functional determinants of neural cell identity, a gap noted by others in the field [100]. However, to achieve this, several challenges must be overcome. Currently, the most common source of healthy human brain tissue available for research is limited to formalin fixed paraffin embedded (FFPE) or fixed frozen tissues. Thus, it is especially hard to study and assess changes in signaling, metabolism, function, and overall state across developmental times in human due to the lack of living cells available for experimentation. One way in which neuroscientists have attempted to overcome this challenge is by using animal models including rodents, ferrets, pigs, and, in a few cases, primates. However, the use of non-murine organisms significantly raises the monetary and temporal cost of the research. The rise of organoid-based model systems in neuroscience has also been rapid. However, the field of human organoids has challenges, including 1) known ground truth in vivo in human tissue has not been well established enough to validate the organoid models, 2) most organoids lack tissue resident immune cells (e.g., microglia in the brain) that are increasingly understood to be critical to normal function of non-immune organ cells, and 3) organoids can take months to generate and the very heterogeneity that makes them outstanding models means that many more examples must be studied than in genetically homogeneous animal models or cell lines. However, organoid research is likely essential given that the rodent brain is a suboptimal comparator to the human brain, both at a molecular cell biology level and an anatomic level. Immunology, by contrast, had a rapid start as healthy human blood was more widely available and ethically reasonable to collect across most stages of human development. However, immunology is now encountering the same challenge as the field seeks to understand the role of immune cells in tissues, including tissue resident immune cells of non-hematopoietic origin.
Is “cell type” different from “cell state”?
It has been proposed that there are three pillars central to the concept of cell identity: lineage, phenotype (which here includes function), and cell state [101]. For example, a regulatory helper T cell might be of the hematopoietic stem cell and T lymphocyte lineages, might currently express proteins like CD3, CD4, and FOXP3, and could be in the states of actively signaling via its T cell antigen receptor and in the G1 phase of the cell cycle. We propose here that the borders between these concepts of lineage, phenotype, and state are not well defined at a chemical or temporal level and the concepts may thus have overlapping domains. In particular, the boundary for when a feature is considered to mark a distinct cell type (an identity) versus a state, which exists within an identity and a lineage, could be much better defined. Furthermore, while a given cell type as a population might be expected to express a set of genes, individual cells vary from their population’s statistical norm. If the RNA transcripts of two cells have detectably different levels of different transcripts, does this indicate that they are of different cell types or could they be of the same cell type and in a different state? Cells exist in flux across a spectrum of states, including the cell cycle, reversible transitions like metabolic programming and mTOR pathway activity, flux of ions like calcium, redox states including production of species like H2O2, and activity of phospho-protein-driven signaling networks that control the function of identity-defining transcription factors and other proteins. To what extent can a cell deviate from its population’s norm and still be considered a member of that group?
Understanding how to define a normal cell’s states is particularly important for identifying when a cell travels out of normal physiological bounds into a pathological state. A prime example is seen in oncogenesis, and much attention has been paid to considering the boundary between healthy cells and malignant cells. Cells that exist in liminal spaces between cell identity groups or with the potential to shift into multiple identities are especially important to understand fields where we seek to trigger or prevent specific cell identity changes, such as regenerative medicine and cancer biology. The degree to which a cell is biochemically constrained or encouraged to explore different identities, i.e., its intrinsic plasticity in identity, maybe a critical piece of information for understanding stem cells, immune evolution, and cancer. In thinking about factors that influence a cell’s state, the tissue context is critical. The tissue environment includes inputs that can alter signaling and metabolism, and broadly dictates a cell’s functional capacities. Thus, mapping specific cell states to their corresponding cell identities might help define the changes seen in cancer or other disease contexts (explored for glioblastoma in Table 2). A leading example of this is seen in measurements of phospho-proteins, which are now well established as superior markers of clinically relevant blood cancer cells [8, 102–105], a finding recently extended to identify risk stratifying cells by signaling in glioblastoma [25], surgical recovery [106], and pregnancy [107]. It will likely also be critical to understand cell state identities to ensure the reliable performance of cell-based therapies in which functions such as cytokine production, proliferation, and cell killing are critical to their function and likely defined by cell state identity.
Table 2.
Cell identity approaches used in glioblastoma brain tumors
| Identifying category | Feature(s) | Implications | Proof type |
|---|---|---|---|
| Morphology and location* | SVZ contact | Poor prognosis | Correlative, not functional [108] |
| Surface epitopes |
CD44, CD133, EGF |
GBM stem cells | Good evidence, not definitional [109, 110] |
| Transcription factors | POU3F2, SOX2, SALL2, and OLIG2 | GBM stem-like, tumor propagating cells | Good evidence, functional, not definitional [21] |
| Functional test or assay | Self-renewal, neurosphere formation | GBM stem cells | Functional, not definitional [111] |
| Lineage tracing assay | Serial xenotransplantation | GSCs as tumor initiators | Functional [18, 111] |
| Phospho-protein signaling response |
▲ Basal p-STAT5 ▲ Basal p-S6 |
Negative prognostic GBM cells | Definitional, functional [25] |
| DNA mutations | IDH1 or IDH2 mutation | Metabolically reprogrammed glioma cells | Correlative, not functional [112] |
| Transcription factor mRNA and genetics | Signatures including PDGFRA, IDH1, EGFR, and NF1 | Classical, mesenchymal, and proneural subtypes ** | Correlative, not functional [113] |
*Additional features to consider here could include location in tumor core or periphery, association with vasculature, degree of pathology-defined necrosis, and ability to form gap junctions with tumor cells or synapses with neurons. **Classification system based originally on a set of 200 + transcripts, DNA mutation, and copy number status
One persistent challenge forecast by studies discussed above is defining the differences between a stem cell, a cancer cell, and a cancer stem cell, and inferring a possible lineage, when these cell types are detected. Broadly speaking, it is difficult to determine whether a CSC is a cancer cell that has shifted its identity into a stem-like cell as a mechanism for more favorable survival, or rather a cell that arises from a population of healthy stem cells that have transformed into a malignant state [28]. When evaluating the existence of CSCs, it is important to keep in mind their potential for differentiation or plasticity, including the reemergence of states that resemble cells normally seen in earlier development [114]. Subsets of cells with different phenotypes are observed within and between tumors from different patients, and only some of these cell subsets will behave as CSCs in functional assays [17, 115–117]. Such cells are often able to undergo genetic changes that make it difficult to establish a standardized set of markers that would canonically define them. CSCs were first described in acute myeloid leukemia [118] and were later shown to be present in solid tumors including glioblastoma [18], where they are thought to contribute to therapy resistance and drive tumor growth [119]. CSCs have been functionally defined by assays such as xenotransplantation to assess their ability to self-renew and differentiate like stem cells [18, 103, 120]. However, relying solely on such functional assays to evaluate these cells makes it quite difficult to study them further considering the large number of resources required to validate one subset of cells. Although currently there are no individual markers that exclusively or definitively mark CSCs for all patients, markers including CD133, CD44, and CD15 have been useful in prospectively isolating or enriching some subsets of glioblastoma cancer stem cells (GSCs) [109]. Unfortunately, these markers are not exclusively expressed in GSCs. Using single cell technologies including mass cytometry, researchers can continue to characterize and further define markers that identify these cells and different transient states that are associated with them. By assessing a variety of surface markers, transcription factors, signaling molecules, and metabolic markers, profiles that will identify such cells from within heterogenous populations can be established. One intriguing recent example used functional features (uptake and cross-cell transfer of specific dyes) to distinguish glioblastoma cells that are or are not enmeshed in a gap junction-coupled network, and then determined the transcriptional and invasive features of each subgroup [121]. Such profiles will enable the targeting and directing of cells into more favorable states or identities in the context of cancer.
Role of technology
Immunology largely owes its present status to multidimensional single cell analysis using cytometers [1], and this technology has long driven refinements in concepts of cell identity [3], beginning with the ability to prospectively sort marker-defined populations of cells for bulk analysis of transcripts and/or genomes. These initial approaches, while offering improved resolution of abundant or antigenically and functionally distinct cell populations, may not have been powered to identify fine differences within subsets cells, such as cell state identities. Increases in dimensionality, throughput, and intracellular detection abilities in single cell technologies have driven a tremendous expansion in the mapping of cell lineages and trajectories that span a continuum between stem/progenitor cells and fully differentiated progeny.
While a comprehensive review of technologies is beyond the scope of this review, it is worth noting the current state of the art in some key single cell and cytometry tools that have driven recent discoveries. Each technology presents strengths and weaknesses that should be considered in experimental design. Single-cell RNA sequencing and its relatives have the ability to examine thousands of parameters (transcripts) but currently has a much higher cost and lower per-feature dynamic range than antibody-based imaging or suspension cytometry [9], and sequencing-based detection especially suffers from signal dropouts where present molecules are not measured in cells, leading to the artificial appearance of heterogeneity even for commonly expressed molecules [52]. Flow cytometry has enabled the analysis of small panels of proteins/markers in individual single cells, and FACS-based isolation has long been employed for the functional and molecular profiling of heterogeneous cell populations [11]. Suspension cytometry, including mass cytometry, spectral flow cytometry, and fluorescence flow cytometry, lack spatial and cytoarchitectural information but are able to quantify protein expression and posttranslational modifications in a sufficient number of cells to enable the detection of rare populations of cells, such as quiescent stem cells, while also including additional parameters that identify cell function and phenotype. Single cell approaches including spectral flow cytometry and mass cytometry offer greater resolution and more information per cell than conventional fluorescence cytometry [9, 50, 53, 122]. Multiplex imaging allows for morphology assessment, tissue structure, provides spatial resolution and subcellular resolution like the location of protein expression within a cell, and can be multiplexed to measure multiple proteins [123–126], whereas, subcellular imaging techniques, such as super-resolution imaging, lack the throughput and higher dimensionality that existing multiplex fluorescence or mass imaging can provide. While multiplex imaging allows for sub-cellular comparison of co-expression, it generally relies on fixed tissues. Thus, the vast majority of tissue collection does not include live cells that can be used in functional experiments. Furthermore, the analysis for high dimensional imaging can be a computationally intensive process [127].
Historically, genome-wide profiling of single cells has enabled the unbiased exploration of cell identity, allowing for the discovery of possible known and unknown cell types at single-cell resolution. Yet inferring the identity of cells has become a renewed challenge as the expanding breadth and depth of single-cell data can now provide an unprecedented lens into the complexities and nuances of cell identities [128]. As an example, mass cytometry has recently been developed for studies on the nervous system, and a central finding of this work is that RNA transcript and protein expression do not align well in all cases [60]. Single cell technologies will continue to improve and allow for us to better understand the cellular and molecular processes that contribute to tumorigenesis and the tumor microenvironment and develop novel therapies and delivery mechanisms that will help treat refractory tumors.
Concluding thoughts
Dissecting the functional identity of cells in tissues is a central goal of cell biology, and a central goal of modern cytometry is to enable automated cell identification. However, distinct fields have different rules and conventions surrounding what distinguishes key cell subtypes, whose features are definitional and whose functional tests are the ne plus ultra for each cell type. On one end of the spectrum is a cell like the T lymphocyte, which is distinct in DNA sequence, transcript, protein expression, and function, although it is largely lacking in distinguishing morphology. On the other end might be subtypes of neurons that were described largely by position and function, such as neurotransmitter responsiveness or calcium signaling patterns, but which lack a strict, known protein or DNA sequence identity.
Perhaps a helpful thought experiment would be to imagine we are a computer algorithm whose job is to correctly identify cells. What information would this algorithm need to be satisfied, and what is the level of confidence it needs? This quickly leads to a challenge: biologists have invented diverse systems to say when a given cell has shifted from one identity to another. In which cases might the algorithm be confident of a cell’s function without measuring that function? In this review, the goal was to highlight useful aspects of measuring diverse cellular features, from easily detectable surface markers to functions that must be measured over time in living cells. We hope this has especially brought out the usefulness of measuring surrogates of identity and developing realistic model systems, as well as the value of working with living human cells from primary tissues. This system of defining cell identity is urgently needed in the study of cellular diseases, especially cancers and neurodegenerative diseases, as it is crucial to distinguishing abnormal cell functions from healthy ones misplaced in space or developmental time.
Abbreviations
- CD
Clusters of differentiation
- CSC
Cancer stem cell
- FACS
Fluorescence activated cell sorting
- FFPE
Formalin-fixed, paraffin-embedded
- GFAP
Glial fibrillary acidic protein
- GSC
Glioblastoma stem cell
- HSC
Hematopoietic stem cell
- MEM
Marker enrichment modeling
- SGZ
Subgranular zone
- V-SVZ
Ventricular subventricular zone
Author contribution
S.M. and J.M.I. conceived the work and developed the initial figures. All the authors contributed to the writing, reviewing, and editing of the manuscript.
Funding
This work was supported by the following funding resources: R01 NS096238 (R.A.I., J.M.I.), R01 NS118580 (R.A.I.), R01 CA226833 (J.M.I.), U01 AI125056 (J.M.I.), U54 CA217450 (J.M.I.), the Michael David Greene Brain Cancer Fund (R.A.I., J.M.I.), the Ben & Catherine Ivy Foundation (R.A.I.), and the Vanderbilt-Ingram Cancer Center (VICC, P30 CA68485).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
This article is a contribution to the special issue on: Single-cell and spatial multi-omics in clinical outcomes studies - Guest Editor: Brice Gaudillière
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Van Epps HL. Bringing order to early B cell chaos. J Exp Med. 2006;203(6):1389–1389. doi: 10.1084/jem.2036fta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(1984) Nomenclature for clusters of differentiation (CD) of antigens defined on human leukocyte populations. IUIS-WHO Nomenclature Subcommittee. Bull World Health Organ 62(5):809–15 [PMC free article] [PubMed]
- 3.Roussel M, Greenplate AR, Irish JM (2016) Dissecting complex cellular systems with high dimensional single cell mass cytometry. In: Montgomery RR, Bucala R (eds) Experimental Approaches for the Investigation of Innate Immunity. World Sci pp, 15–26. 10.1142/9789814678735_0002
- 4.DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat Rev Immunol. 2016;16(3):149–163. doi: 10.1038/nri.2015.18. [DOI] [PubMed] [Google Scholar]
- 5.Ottoboni L, Merlini A, Martino G. Neural stem cell plasticity: advantages in therapy for the injured central nervous system. Front Cell Dev Biol. 2017;5:52. doi: 10.3389/fcell.2017.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen S, Clairambault J. Cell plasticity in cancer cell populations. F1000Res. 2020;9:635. doi: 10.12688/f1000research.24803.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233(9):6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 8.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6(2):146–155. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 9.Mistry AM, Greenplate AR, Ihrie RA, Irish JM. Beyond the message: advantages of snapshot proteomics with single-cell mass cytometry in solid tumors. FEBS J. 2019;286(8):1523–1539. doi: 10.1111/febs.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, Sabeti PC (2019) Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. 8:e43803. 10.7554/eLife.43803 [DOI] [PMC free article] [PubMed]
- 11.Mincarelli L, Lister A, Lipscombe J, Macaulay IC. Defining cell identity with single-cell omics. Proteomics. 2018;18(18):1700312. doi: 10.1002/pmic.201700312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baslan T, Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer. 2017;17(9):557–569. doi: 10.1038/nrc.2017.58. [DOI] [PubMed] [Google Scholar]
- 13.Flitsch LJ, Laupman KE, Brustle O. Transcription factor-based fate specification and forward programming for neural regeneration. Front Cell Neurosci. 2020;14:121. doi: 10.3389/fncel.2020.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto T, Akashi K. Lineage promiscuous expression of transcription factors in normal hematopoiesis. Int J Hematol. 2005;81(5):361–367. doi: 10.1532/IJH97.05003. [DOI] [PubMed] [Google Scholar]
- 15.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 16.Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, Nolan GP, Shannon K, Braun BS. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109(9):3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhaduri A, Di Lullo E, Jung D, Müller S, Crouch EE, Espinosa CS, Ozawa T, Alvarado B, Spatazza J, Cadwell CR, Wilkins G, Velmeshev D, Liu SJ, Malatesta M, Andrews MG, Mostajo-Radji MA, Huang EJ, Nowakowski TJ, Lim DA, Diaz A, Raleigh DR, Kriegstein AR. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26(1):48–63.e6. doi: 10.1016/j.stem.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 19.Torre-Healy LA, Berezovsky A, Lathia JD. Isolation, characterization, and expansion of cancer stem cells. Methods Mol Biol. 2017;1553:133–143. doi: 10.1007/978-1-4939-6756-8_10. [DOI] [PubMed] [Google Scholar]
- 20.Berezovsky AD, Poisson LM, Cherba D, Webb CP, Transou AD, Lemke NW, Hong X, Hasselbach LA, Irtenkauf SM, Mikkelsen T, deCarvalho AC (2014) Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia 16(3):193–206, 206.e19–25 [DOI] [PMC free article] [PubMed]
- 21.Stevanovic M, Kovacevic-Grujicic N, Mojsin M, Milivojevic M, Drakulic D. SOX transcription factors and glioma stem cells: choosing between stemness and differentiation. World J Stem Cells. 2021;13(10):1417–1445. doi: 10.4252/wjsc.v13.i10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Song T, Yang L, Chen R, Wu L, Yang Z, Fang J. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 2008;27(1):85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard BM, Gursel DB, Bleau AM, Beyene RT, Holland EC, Boockvar JA. EGFR signaling is differentially activated in patient-derived glioblastoma stem cells. J Exp Ther Oncol. 2010;8(3):247–260. [PubMed] [Google Scholar]
- 24.Yamamuro S, Okamoto Y, Sano E, Ochiai Y, Ogino A, Ohta T, Hara H, Ueda T, Nakayama T, Yoshino A, Katayama Y. Characterization of glioma stem-like cells from human glioblastomas. Int J Oncol. 2015;47(1):91–96. doi: 10.3892/ijo.2015.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leelatian N, Sinnaeve J, Mistry AM, Barone SM, Brockman AA, Diggins KE, Greenplate AR, Weaver KD, Thompson RC, Chambless LB, Mobley BC, Ihrie RA, Irish JM (2020) Unsupervised machine learning reveals risk stratifying glioblastoma tumor cells. Elife 9:e56879. 10.7554/eLife.56879 [DOI] [PMC free article] [PubMed]
- 26.Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets using marker enrichment modeling. Nat Methods. 2017;14(3):275–278. doi: 10.1038/nmeth.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer KJ, Wilfong EM, Voss K, Barone SM, Shiakolas AR, Raju N, Roe CE, Suryadevara N, Walker LM, Wall SC, Paulo A, Schaefer S, Dahunsi D, Westlake CS, Crowe JE, Jr, Carnahan RH, Rathmell JC, Bonami RH, Georgiev IS, Irish JM. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat Commun. 2022;13(1):3466. doi: 10.1038/s41467-022-31142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Dirks PB. Brain tumour stem cells: the undercurrents of human brain cancer and their relationship to neural stem cells. Philos Trans R Soc Lond B Biol Sci. 2008;363(1489):139–152. doi: 10.1098/rstb.2006.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JCY, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15(9):494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Rushing G, Ihrie RA. Neural stem cell heterogeneity through time and space in the ventricular-subventricular zone. Front Biol. 2016;11(4):261–284. doi: 10.1007/s11515-016-1407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, Vidal JG, Mu Y, Killian RL, Israel MA, Emre N, Marsala S, Marsala M, Gage FH, Goldstein LSB, Carson CT. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS ONE. 2011;6(3):e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Y, Yu P, Cheng L. Current progress in the derivation and therapeutic application of neural stem cells. Cell Death Dis. 2017;8(10):e3108–e3108. doi: 10.1038/cddis.2017.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glauche I, Marr C. Mechanistic models of blood cell fate decisions in the era of single-cell data. Curr Opin Syst Biol. 2021;28:100355. doi: 10.1016/j.coisb.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wognum AW, Eaves AC, Thomas TE. Identification and isolation of hematopoietic stem cells. Arch Med Res. 2003;34(6):461–475. doi: 10.1016/j.arcmed.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Zanjani ED, Almeida-Porada G, Ascensao JL, Mackintosh FR, Flake AW. Transplantation of hematopoietic stem cells in utero. Stem Cells. 2009;15(S2):79–93. doi: 10.1002/stem.5530150812. [DOI] [PubMed] [Google Scholar]
- 37.Sieburg HB, Cho RH, Dykstra B, Uchida N, Eaves CJ, Muller-Sieburg CE. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107(6):2311–2316. doi: 10.1182/blood-2005-07-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010;70(1):68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Pan M, Jin T. How Phagocytes acquired the capability of hunting and removing pathogens from a human body: lessons learned from chemotaxis and phagocytosis of Dictyostelium discoideum (review) Front Cell Dev Biol. 2021;9:724940. doi: 10.3389/fcell.2021.724940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalina T, Fišer K, Pérez-Andrés M, Kuzílková D, Cuenca M, Bartol SJW, Blanco E, Engel P, van Zelm MC. CD maps-dynamic profiling of CD1-CD100 surface expression on human leukocyte and lymphocyte subsets. Front Immunol. 2019;10:2434. doi: 10.3389/fimmu.2019.02434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altin JG, Sloan EK. The role of CD45 and CD45-associated molecules in T cell activation. Immunol Cell Biol. 1997;75(5):430–445. doi: 10.1038/icb.1997.68. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, Parkhouse RME, Wileman T. Monoclonal antibodies that identify the CD3 molecules expressed specifically at the surface of porcine gammadelta-T cells. Immunology. 2005;115(2):189–196. doi: 10.1111/j.1365-2567.2005.02137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudensky AY. Regulatory T cells and Foxp 3. Immunol Rev. 2011;241(1):260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desouki MM, Post GR, Cherry D, Lazarchick J. PAX-5: a valuable immunohistochemical marker in the differential diagnosis of lymphoid neoplasms. Clin Med Res. 2010;8(2):84–88. doi: 10.3121/cmr.2010.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14(6):779–790. doi: 10.1016/S1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 46.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247(1):172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 47.Nutt SL, Fairfax KA, Kallies A. BLIMP1 guides the fate of effector B and T cells. Nat Rev Immunol. 2007;7(12):923–927. doi: 10.1038/nri2204. [DOI] [PubMed] [Google Scholar]
- 48.Heyworth C. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 2002;21(14):3770–3781. doi: 10.1093/emboj/cdf368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinreb C, Rodriguez-Fraticelli A, Camargo FD, Klein AM. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. 2020;367(6479):eaaw3381. doi: 10.1126/science.aaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33(7):323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirano T. Purkinje Neurons: development, morphology, and function. Cerebellum. 2018;17(6):699–700. doi: 10.1007/s12311-018-0985-7. [DOI] [PubMed] [Google Scholar]
- 52.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matthew G. Mass cytometry: single cells, many features. Cell. 2016;165(4):780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fishell G, Heintz N. The neuron identity problem: form meets function. Neuron. 2013;80(3):602–612. doi: 10.1016/j.neuron.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 55.Wells A, Yates C, Shepard CR. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin Exp Metastasis. 2008;25(6):621–628. doi: 10.1007/s10585-008-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gustafson MP, Lin Y, Maas ML, Van Keulen VP, Johnston PB, Peikert T, Gastineau DA, Dietz AB. A method for identification and analysis of non-overlapping myeloid immunophenotypes in humans. PLoS ONE. 2015;10(3):e0121546. doi: 10.1371/journal.pone.0121546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergles DE, Richardson WD. Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol. 2016;8(2):a020453. doi: 10.1101/cshperspect.a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang K, Wei G, Liu D. CD19: a biomarker for B cell development, lymphoma diagnosis and therapy. Exp Hematol Oncol. 2012;1(1):36. doi: 10.1186/2162-3619-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mich JK, Signer RA, Nakada D, Pineda A, Burgess RJ, Vue TY, Johnson JE, Morrison SJ (2014) Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife 3:e02669. 10.7554/eLife.02669 [DOI] [PMC free article] [PubMed]
- 60.Keeler AB, Van Deusen AL, Cheng I, Williams CM, Goggin SM, Hirt AK, Vradenburgh SA, Fread KI, Puleo EA, Jin L, Deppmann CD, Zunder ER (2022) A developmental atlas of somatosensory diversification and maturation in the dorsal root ganglia by single-cell mass cytometry, bioRxiv 2022.06.01.494445 [DOI] [PMC free article] [PubMed]
- 61.Wei J, Wu A, Kong L-Y, Wang Y, Fuller G, Fokt I, Melillo G, Priebe W, Heimberger AB. Hypoxia potentiates glioma-mediated immunosuppression. PLoS ONE. 2011;6(1):e16195. doi: 10.1371/journal.pone.0016195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y-I. The basic helix-loop-helix factor Olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12(13):1157–1163. doi: 10.1016/S0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 63.Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26(2–4):148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 64.Miyagi S, Saito T, Mizutani K-I, Masuyama N, Gotoh Y, Iwama A, Nakauchi H, Masui S, Niwa H, Nishimoto M, Muramatsu M, Okuda A. The Sox-2 regulatory regions display their activities in two distinct types of multipotent stem cells. Mol Cell Biol. 2004;24(10):4207–4220. doi: 10.1128/MCB.24.10.4207-4220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of <i>Pax5</i> expression. Science. 2002;297(5578):110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- 66.Englund C. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25(1):247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kläsener K, Jellusova J, Andrieux G, Salzer U, Böhler C, Steiner SN, Albinus JB, Cavallari M, Süß B, Voll RE, Boerries M, Wollscheid B, Reth M. CD20 as a gatekeeper of the resting state of human B cells. Proc Natl Acad Sci. 2021;118(7):e2021342118. doi: 10.1073/pnas.2021342118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hodge RD, Nelson BR, Kahoud RJ, Yang R, Mussar KE, Reiner SL, Hevner RF. Tbr2 is essential for hippocampal lineage progression from neural stem cells to intermediate progenitors and neurons. J Neurosci. 2012;32(18):6275–6287. doi: 10.1523/JNEUROSCI.0532-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shahaf G, Gross AJ, Sternberg-Simon M, Kaplan D, DeFranco AL, Mehr R. Lyn deficiency affects B-cell maturation as well as survival. Eur J Immunol. 2012;42(2):511–521. doi: 10.1002/eji.201141940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michalski J-P, Kothary R (2015) Oligodendrocytes in a nutshell. Front Cell Neurosci 9:340. 10.3389/fncel.2015.00340 [DOI] [PMC free article] [PubMed]
- 71.Gholami S, Mohammadi SM, MovasaghpourAkbari A, Abedelahi A, Alihemmati A, Fallahi S, NozadCharoudeh H. Terminal deoxynucleotidyl transferase (TdT) inhibition of cord blood derived B and T cells expansion. Adv Pharm Bull. 2017;7(2):215–220. doi: 10.15171/apb.2017.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.John Lin C-C, Yu K, Hatcher A, Huang T-W, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, Patel AJ, Arenkiel BR, Noebels JL, Creighton CJ, Deneen B. Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci. 2017;20(3):396–405. doi: 10.1038/nn.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi P, Reiser H. IL-4: role in disease and regulation of production. Clin Exp Immunol. 1998;113(3):317–319. doi: 10.1046/j.1365-2249.1998.00690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8(3):363–372. doi: 10.1016/S1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 75.Peterka DS, Takahashi H, Yuste R. Imaging voltage in neurons. Neuron. 2011;69(1):9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xue Q, Yan Y, Zhang R, Xiong H. Regulation of iNOS on immune cells and its role in diseases. Int J Mol Sci. 2018;19(12):3805. doi: 10.3390/ijms19123805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Z, Ma Z, Wang Y, Li Y, Lü H, Fu S, Hang Q, Lu PH. Oligodendrocyte-spinal cord explant co-culture: an in vitro model for the study of myelination. Brain Res. 2010;1309:9–18. doi: 10.1016/j.brainres.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 78.Kerman BE, Kim HJ, Padmanabhan K, Mei A, Georges S, Joens MS, Fitzpatrick JAJ, Jappelli R, Chandross KJ, August P, Gage FH. In vitro myelin formation using embryonic stem cells. Development. 2015;142(12):2213–2225. doi: 10.1242/dev.116517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlenner SM, Madan V, Busch K, Tietz A, Läufle C, Costa C, Blum C, Fehling HJ, Rodewald H-R. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32(3):426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 80.Liu X, Bolteus AJ, Balkin DM, Henschel O, Bordey A. GFAP-expressing cells in the postnatal subventricular zone display a unique glial phenotype intermediate between radial glia and astrocytes. Glia. 2006;54(5):394–410. doi: 10.1002/glia.20392. [DOI] [PubMed] [Google Scholar]
- 81.Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogné M, Denizot Y. The IgH 3’ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210(8):1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quiñones-Hinojosa A, Sanai N, Soriano-Navarro M, Gonzalez-Perez O, Mirzadeh Z, Gil-Perotin S, Romero-Rodriguez R, Berger MS, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and cytoarchitecture of the adult human subventricular zone: a niche of neural stem cells. J Comp Neurol. 2006;494(3):415–434. doi: 10.1002/cne.20798. [DOI] [PubMed] [Google Scholar]
- 83.Seri B, García-Verdugo JM, Collado-Morente L, McEwen BS, Alvarez-Buylla A. Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J Comp Neurol. 2004;478(4):359–378. doi: 10.1002/cne.20288. [DOI] [PubMed] [Google Scholar]
- 84.Fiorelli R, Azim K, Fischer B, Raineteau O. Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development. 2015;142(12):2109–2120. doi: 10.1242/dev.119966. [DOI] [PubMed] [Google Scholar]
- 85.Malatesta P, Appolloni I, Calzolari F. Radial glia and neural stem cells. Cell Tissue Res. 2008;331(1):165–178. doi: 10.1007/s00441-007-0481-8. [DOI] [PubMed] [Google Scholar]
- 86.Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Götz M. Neuronal or glial progeny. Neuron. 2003;37(5):751–764. doi: 10.1016/S0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 87.Cárdenas A, Borrell V. Molecular and cellular evolution of corticogenesis in amniotes. Cell Mol Life Sci. 2020;77(8):1435–1460. doi: 10.1007/s00018-019-03315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron. 2016;91(6):1219–1227. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coskun V, Wu H, Blanchi B, Tsao S, Kim K, Zhao J, Biancotti JC, Hutnick L, Krueger RC, Jr, Fan G, de Vellis J, Sun YE. CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc Natl Acad Sci U S A. 2008;105(3):1026–1031. doi: 10.1073/pnas.0710000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noguchi M, Furukawa KT, Morimoto M (2020) Pulmonary neuroendocrine cells: physiology, tissue homeostasis and disease. Dis Model Mech 13(12):dmm.046920. 10.1242/dmm.046920 [DOI] [PMC free article] [PubMed]
- 91.Gonzalez-Perez O. Neural stem cells in the adult human brain. Biol Biomed Rep. 2012;2(1):59–69. [PMC free article] [PubMed] [Google Scholar]
- 92.Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8(5):486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P, Doetsch F. Age-dependent niche signals from the choroid plexus regulate adult neural stem cells. Cell Stem Cell. 2016;19(5):643–652. doi: 10.1016/j.stem.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 94.Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82(3):545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allen NJ. Astrocyte regulation of synaptic behavior. Annu Rev Cell Dev Biol. 2014;30(1):439–463. doi: 10.1146/annurev-cellbio-100913-013053. [DOI] [PubMed] [Google Scholar]
- 96.Khakh BS, McCarthy KD. Astrocyte calcium signaling: from observations to functions and the challenges therein. Cold Spring Harb Perspect Biol. 2015;7(4):a020404. doi: 10.1101/cshperspect.a020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jang M, Gould E, Xu J, Kim EJ, Kim JH (2019) Oligodendrocytes regulate presynaptic properties and neurotransmission through BDNF signaling in the mouse brainstem. eLife 8:e42156. 10.7554/eLife.42156 [DOI] [PMC free article] [PubMed]
- 98.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A Transcriptome Database for Astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342(6163):1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- 100.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505(7483):318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morris SA (2019) The evolving concept of cell identity in the single cell era. Development 146(12):dev169748. 10.1242/dev.169748 [DOI] [PubMed]
- 102.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 103.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe’er D, Nolan GP. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–97. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK, Nolan GP, Levy R. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci USA. 2010;107(29):12747–12754. doi: 10.1073/pnas.1002057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Myklebust JH, Brody J, Kohrt HE, Kolstad A, Czerwinski DK, Walchli S, Green MR, Troen G, Liestol K, Beiske K, Houot R, Delabie J, Alizadeh AA, Irish JM, Levy R. Distinct patterns of B-cell receptor signaling in non-Hodgkin lymphomas identified by single-cell profiling. Blood. 2017;129(6):759–770. doi: 10.1182/blood-2016-05-718494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gaudilliere B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, Silva J, Ganio EA, Yeh CG, Maloney WJ, Huddleston JI, Goodman SB, Davis MM, Bendall SC, Fantl WJ, Angst MS, Nolan GP. Clinical recovery from surgery correlates with single-cell immune signatures. Sci Transl Med. 2014;6(255):255ra131. doi: 10.1126/scitranslmed.3009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aghaeepour N, Ganio EA, Mcilwain D, Tsai AS, Tingle M, Van Gassen S, Gaudilliere DK, Baca Q, McNeil L, Okada R. An immune clock of human pregnancy. Sci Immunol. 2017;2(15):eaan2946. doi: 10.1126/sciimmunol.aan2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mistry AM, Dewan MC, White-Dzuro GA, Brinson PR, Weaver KD, Thompson RC, Ihrie RA, Chambless LB. Decreased survival in glioblastomas is specific to contact with the ventricular-subventricular zone, not subgranular zone or corpus callosum. J Neurooncol. 2017;132(2):341–349. doi: 10.1007/s11060-017-2374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brown DV, Filiz G, Daniel PM, Hollande F, Dworkin S, Amiridis S, Kountouri N, Ng W, Morokoff AP, Mantamadiotis T. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS ONE. 2017;12(2):e0172791. doi: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tome-Garcia J, Doetsch F, Tsankova N (2017) FACS-based isolation of neural and glioma stem cell populations from fresh human tissues utilizing EGF ligand. Bio-Protocol 7(24):2659. 10.21769/BioProtoc.2659 [DOI] [PMC free article] [PubMed]
- 111.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, Modrusan Z, Nacu S, Guerrero S, Edgar KA, Wallin JJ, Lamszus K, Westphal M, Heim S, James CD, Vandenberg SR, Costello JF, Moorefield S, Cowdrey CJ, Prados M, Phillips HS. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17(4):362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 112.Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, Gilbert MR, Yang C. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer. 2020;122(11):1580–1589. doi: 10.1038/s41416-020-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hamed AA, Kunz DJ, El-Hamamy I, Trinh QM, Subedar OD, Richards LM, Foltz W, Bullivant G, Ware M, Vladoiu MC, Zhang J, Raj AM, Pugh TJ, Taylor MD, Teichmann SA, Stein LD, Simons BD, Dirks PB. A brain precursor atlas reveals the acquisition of developmental-like states in adult cerebral tumours. Nat Commun. 2022;13(1):4178. doi: 10.1038/s41467-022-31408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Piccirillo SG, Dietz S, Madhu B, Griffiths J, Price SJ, Collins VP, Watts C. Fluorescence-guided surgical sampling of glioblastoma identifies phenotypically distinct tumour-initiating cell populations in the tumour mass and margin. Br J Cancer. 2012;107(3):462–468. doi: 10.1038/bjc.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Piccirillo SG, Combi R, Cajola L, Patrizi A, Redaelli S, Bentivegna A, Baronchelli S, Maira G, Pollo B, Mangiola A, DiMeco F, Dalprà L, Vescovi AL. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28(15):1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 117.Wang L, Babikir H, Müller S, Yagnik G, Shamardani K, Catalan F, Kohanbash G, Alvarado B, Di Lullo E, Kriegstein A, Shah S, Wadhwa H, Chang SM, Phillips JJ, Aghi MK, Diaz AA. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 2019;9(12):1708–1719. doi: 10.1158/2159-8290.CD-19-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 119.Lan X, Jörg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, Park NI, Coutinho FJ, Whetstone H, Selvadurai HJ, Che C, Luu B, Carles A, Moksa M, Rastegar N, Head R, Dolma S, Prinos P, Cusimano MD, Das S, Bernstein M, Arrowsmith CH, Mungall AJ, Moore RA, Ma Y, Gallo M, Lupien M, Pugh TJ, Taylor MD, Hirst M, Eaves CJ, Simons BD, Dirks PB. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549(7671):227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 121.Venkataramani V, Yang Y, Schubert MC, Reyhan E, Tetzlaff SK, Wißmann N, Botz M, Soyka SJ, Beretta CA, Pramatarov RL, Fankhauser L, Garofano L, Freudenberg A, Wagner J, Tanev DI, Ratliff M, Xie R, Kessler T, Hoffmann DC, Hai L, Dörflinger Y, Hoppe S, Yabo YA, Golebiewska A, Niclou SP, Sahm F, Lasorella A, Slowik M, Döring L, Iavarone A, Wick W, Kuner T, Winkler F. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185(16):2899–2917.e31. doi: 10.1016/j.cell.2022.06.054. [DOI] [PubMed] [Google Scholar]
- 122.Park LM, Lannigan J, Jaimes MC (2020) OMIP-069: forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry A 97(10):1044–1051. 10.1002/cyto.a.24213 [DOI] [PMC free article] [PubMed]
- 123.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci USA. 2013;110(29):11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 125.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Phillips D, Schurch CM, Khodadoust MS, Kim YH, Nolan GP, Jiang S. Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX tissue imaging. Front Immunol. 2021;12:687673. doi: 10.3389/fimmu.2021.687673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ye Z, Sarkar CA. Towards a quantitative understanding of cell identity. Trends Cell Biol. 2018;28(12):1030–1048. doi: 10.1016/j.tcb.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim HJ, Tam PPL, Yang P (2021) Defining cell identity beyond the premise of differential gene expression. Cell Regen 10(1):20. 10.1186/s13619-021-00083-7 [DOI] [PMC free article] [PubMed]