Although the SARS-CoV-2 omicron (BA.1 or B.1.1.529) subvariant BA.5 is dominant worldwide, several new subvariants, including BQ.1, BQ.1.1, BF.7, and BA.4.6, are appearing more frequently in sequenced SARS-CoV-2 infections,1, 2 raising the concern of additional escape neutralisation by antibodies elicited by vaccination or infection. We examined the degree of neutralising antibody escape by omicron subvariants BQ.1, BQ.1.1, BF.7, BA.1, BA.2, BA.2.75, and BA.4 and BA.5 (hereafter referred to as BA.4/5), using 50% neutralisation titres of six serum panels from individuals who had previously had delta BA.1 and BA.2.2 breakthrough infections and more recently had BA.5.1.2, BA.2.76, and BF.7 breakthrough infections (appendix p 2–4, 6).
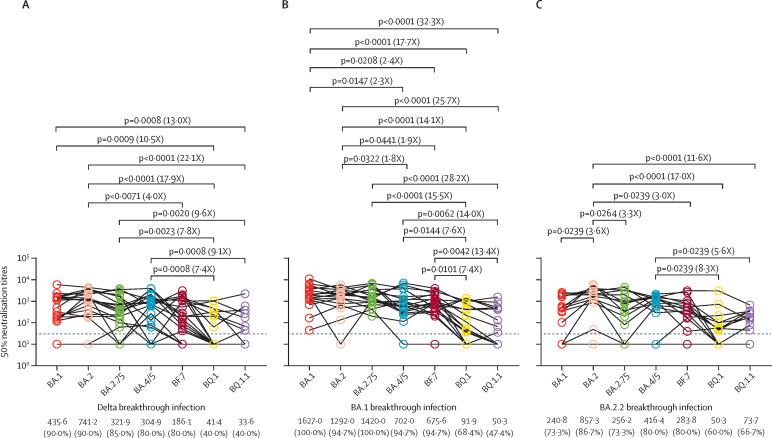
We first examined the resistance of these omicron subvariants to serum samples from 20 individuals with delta breakthrough infections (appendix p 6). We observed a similar neutralisation activity between BQ.1 and BQ.1.1 but a significantly higher neutralisation resistance compared with BA.1, BA.2, BA.2.75, and BA.4/5; and only 40% of serum samples neutralised BQ.1 and BQ.1.1 (figure A ). Specifically, BQ.1 showed a substantially lower neutralisation sensitivity compared with BA.1 (10·5 fold), BA.2 (17·9 fold), BA.2.75 (7·8 fold), and BA.4/5 (7·4 fold); and BQ1.1 showed a lower neutralisation sensitivity compared with BA.1 (13·0 fold), BA.2 (22·1 fold), BA.2.75 (9·6 fold), and BA.4/5 (9·1 fold) (figure A). The serum neutralisation activity was similar against BA.1, BA.2, BA.2.75, BA.4/5, and BF.7, and more than 80% of serum samples neutralised these subvariants (figure A). In addition, BF.7 showed a neutralisation sensitivity 4·0 fold lower than BA.2 (figure A).
Figure.
Neutralisation of omicron subvariants by serum samples from individuals with delta and omicron subvariant breakthrough infections
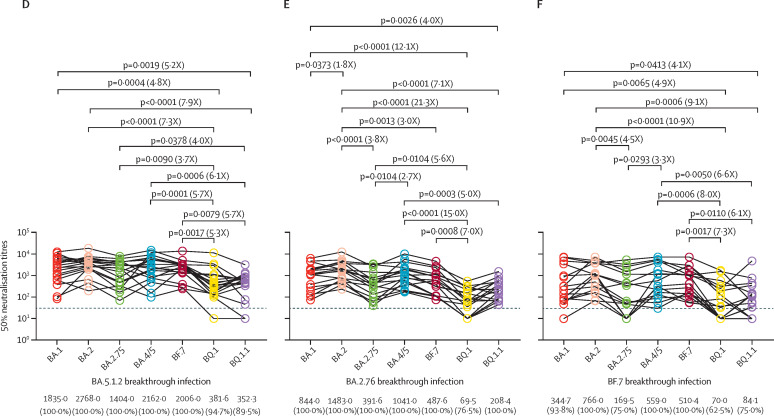
Neutralisation of omicron subvariants, determined by 50% neutralisation titres, by 20 serum samples collected from individuals with delta breakthrough infections (A), 19 serum samples collected from individuals with BA.1 breakthrough infections (B), 15 serum samples collected from individuals with BA.2.2 breakthrough infections (C), 19 serum samples collected from individuals with BA.5.12 breakthrough infections (D), 17 serum samples collected from individuals with BA.2.76 breakthrough infections (E), and 16 serum samples collected from individuals with BF.7 breakthrough infections (F). The horizontal dotted line in all graphs represents a limit of detection of 30, and serum samples with a neutralisation of less than 30 are plotted as 10. The geomatic mean titres and the percentage of individuals with 50% neutralisation titres values above the limit of detection are shown at the bottom of the graph. The fold-change of the geometric mean titre is denoted in brackets. A two-tailed Friedman test with a false discovery rate for multiple comparisons was performed.
Next, we examined the resistance of omicron subvariants to neutralisation by serum samples from individuals with BA.1 (n=19) or BA.2.2 (n=15) breakthrough infections (appendix p 6). We found that BA.1 serum samples more efficiently neutralised BA.2, BA.2.75, BA.4/5, and BF.7 compared with a delta breakthrough infection, and more than 90% of serum samples neutralised these subvariants (figure B). However, neutralisation activity against BQ.1 was substantially decreased compared with BA.1 (17·7 fold), BA.2 (14·1 fold), BA.2.75 (15·5 fold), BA.45 (7·6 fold), and BF.7 (7·4 fold); and was also substantially decreased against BQ.1.1 compared with BA.1 (32·3 fold), BA.2 (25·7 fold), BA.2.75 (28·2 fold), BA.45 (14·0 fold), and BF.7 (13·4 fold; figure B). In addition, neutralisation activity against BA.4/5 was substancially reduced compared with the neutralisation activity against BA.1 (2·3 fold) and BA.2 (1·8 fold); and the neutralisation activity against BF.7 was substantially reduced compared with the neutralisation activity against BA.1 (2·4 fold) and BA.2 (1·9 fold). In contrast, BA.2.2 serum samples less efficiently neutralised BA.1, BA.2.75, BA.4/5, and BF.7, and approximately 80% of all serum samples neutralised these variants. Similarly, BQ.1 and BQ.1.1 were the most resistant subvariants, and only approximately 60% of serum samples were susceptible to them, with a 17-fold reduction in geometric mean titres for BQ.1 and 11·6-fold reduction for BQ.1.1 compared with BA.2 (figure C).
We next examined serum samples from individuals infected with BA.5.1.2 (n=19), BA.2.76 (n=17), or BF.7 (n=16) (appendix p 6). We observed an overall improvement in the neutralising antibody titre, and all serum samples of the three panels neutralised BA.1, BA.2, BA.2.75, BA.4/5, and BF.7, except for serum samples one and four from individuals infected with BF.7, which showed complete loss of neutralising ability against BA.1 and BA.2.75 (figure D–F). Similarly, BQ.1 and BQ.1.1 were significantly resistant to neutralisation, although most serum samples could neutralise these subvariants (figure D–F). We found that the serum samples of a BA.5.1.2 breakthrough infection could not only efficiently neutralise BA.1, BA.2, BA.2.75, and BF.7, but also the majority of these serum samples could neutralise BQ.1 (94·7%) and BQ.1.1 (89·5%), although the neutralisation sensitivity against BQ.1 and BQ.1.1 was significantly lower than other tested variants (figure D). Additionally, BA.1, BA.2, BA.4/5, and BF.7 exhibited susceptibility to BA.2.76 breakthrough infection serum samples; however, BA.2.75 showed more resistance than BA.2 and BA.4/5 (figure E). Moreover, BA.2.75 is more resistant to breakthrough BF.7 infection neutralisation than BA.2 and BA.4/5. Further comparisons showed that BA.5.1.2 breakthrough infections induced a broader antibody response against the tested subvariants and induced significantly higher geometric mean titres against BQ.1 and BQ.1.1 compared with delta, BA.1, BA.2.2, BA.2.76, or BF.7 breakthrough infections (figure; appendix p 7).
Omicron subvariants BQ.1 and BQ.1.1 with increased resistance to neutralising antibodies can pose a challenge to immunity induced by vaccination or infection and render therapeutic monoclonal antibodies ineffective.3, 4, 5, 6 Our results suggest that BQ.1 and BQ.1.1 extensively, but incompletely, escape omicron subvariant breakthrough infection neutralisation, including the most recent BA.5.1.2, BA.2.76, and BF.7 infections. However, serum samples of BA.5.1.2 breakthrough infection were effectively neutralised by BQ.1 and BQ.1.1, suggesting that previous BA.5 breakthrough infection might prevent BQ.1 and BQ.1.1, and BQ.1 and BQ.1.1 might not completely replace BA.5.
X-LJ, K-LZ, and X-JW contributed equally as joint first authors. E-HD and M-JM contributed equally as joint last authors. We declare no competing interests. We thank all study subjects for their participation in our study. This work was supported by grants from the National Natural Science Foundation of China (82273692, 92169207, 81621005, and 81830101), the Beijing Natural Science Foundation (L202038), and the Key Research and Development Plan of Shandong Province (2021RZA01021).
Supplementary Material
References
- 1.US Department of Health and Human Services. CDC Prevention CfDCa. COVID data tracker. 2022. https://covid.cdc.gov/covid-data-tracker/#datatracker-home
- 2.GISAID Genomic epidemiology of SARS-CoV-2 with subsampling focused globally over the past 6 months. 2022. https://gisaid.org/phylodynamics/global/nextstrain/
- 3.Qu P, Evans JP, Faraone J, et al. Distinct neutralizing antibody escape of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. bioRxiv. 2022 doi: 10.1101/2022.10.19.512891. published online Oct 20. (preprint). [DOI] [Google Scholar]
- 4.Cao Y, Jian F, Wang J, et al. Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. published online Oct 4. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurhade C, Zou J, Xia H, et al. Low neutralization of SARS-CoV-2 omicron BA.2.75.2, BQ.1.1, and XBB.1 by 4 doses of parental mRNA vaccine or a BA.5-bivalent booster. bioRxiv. 2022 doi: 10.1101/2022.10.31.514580. published online Nov 2. (preprint). [DOI] [PubMed] [Google Scholar]
- 6.Miller J, Hachmann NP, Collier A-rY, et al. Substantial neutralization escape by the SARS-CoV-2 omicron variant BQ.1.1. bioRxiv. 2022 doi: 10.1101/2022.11.01.514722. published online Nov 2. (preprint). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.