Abstract
This report describes a single-step extension approach suitable for high-throughput single-nucleotide polymorphism typing applications. The method relies on extension of paired allele-specific primers and we demonstrate that the reaction kinetics were slower for mismatched configurations compared with matched configurations. In our approach we employ apyrase, a nucleotide degrading enzyme, to allow accurate discrimination between matched and mismatched primer-template configurations. This apyrase-mediated allele-specific extension (AMASE) protocol allows incorporation of nucleotides when the reaction kinetics are fast (matched 3′-end primer) but degrades the nucleotides before extension when the reaction kinetics are slow (mismatched 3′-end primer). Thus, AMASE circumvents the major limitation of previous allele-specific extension assays in which slow reaction kinetics will still give rise to extension products from mismatched 3′-end primers, hindering proper discrimination. It thus represents a significant improvement of the allele-extension method. AMASE was evaluated by a bioluminometric assay in which successful incorporation of unmodified nucleotides is monitored in real-time using an enzymatic cascade.
INTRODUCTION
Genome analysis techniques have increasingly been adapted to identify and score single-nucleotide polymorphism (SNP) to elucidate the genetics of individual differences in drug response and disease susceptibility. A number of different techniques have been proposed to scan sequence variations in a high-throughput fashion. Many of these methods are based on hybridization techniques, which discriminate between allelic variants. High-throughput hybridization of allele-specific oligonucleotides can be performed on microarray chips (1), microarray gels (2) or by using allele-specific probes (molecular beacons) in the PCR (3). Other technologies suitable for SNP genotyping are mini-sequencing (4), mass spectrometry (5), dynamic allele-specific hybridization (6) and pyrosequencing (7,8). The use of allele-specific primers with alternating 3′-ends has been employed previously to identify single base variations (9–12); however, it is generally acknowledged that certain mismatches, such as G-T or C-A, are poorly discriminated by the employed DNA polymerase (13). This poor discrimination property of the polymerase has consistently been observed in applications of this technique (12,14–16). In these cases, DNA polymerase extends the mismatched primer-templates in the presence of nucleotides, although as we show here, with slower reaction kinetics in comparison with the extension of the matched primer-template configuration. The kinetic difference is usually not distinguishable in end-point analysis, such as in allele-specific PCR, because extension of a single mismatched substrate in the first cycle will lead to perfectly matched primer-templates in subsequent cycles, yielding comparable amounts of end products for both matched and mismatched configurations.
In this study, SNPs with eight alternative 3′-end primer-template configurations were investigated including the previously reported difficult mismatches G-T and C-A (12). The SNPs were codon 72 of the p53 gene (C or G), wiaf 1764 (G or T), nucleotide position 677 in the MTHFR gene (C or T) and nucleotide position 196 in the GPIIIa gene (G or A). Here we demonstrate that the reaction kinetics were slower for mismatched configurations compared with matched configurations and show that inclusion of apyrase in the extension reaction facilitates accurate SNP genotyping.
MATERIALS AND METHODS
Samples and PCR
Human genomic DNA was extracted from unrelated individuals to analyze four SNPs. The SNPs were wiaf 1764 (G/T) on chromosome 9q (12 samples; four G/G, four G/T and four T/T), codon 72 (C/G) of the p53 gene (13 samples; three C/C, five C/G and five G/G), nucleotide position 677 (C/T) on the MTHFR gene (13 samples; five C/C, five C/T and three T/T) and nucleotide position 196 (A/G) on the GPIIIa gene (13 samples; five A/A, five A/G and three G/G). Two outer PCRs (duplexes) were performed to amplify the SNPs. The outer PCR for co-amplification of wiaf 1764 and p53 gene (94°C 1 min, 50°C 40 s and 72°C 2 min for 35 cycles) was followed by locus-specific inner PCRs (94°C 1 min, 50°C 40 s and 72°C 1 min for 35 cycles) generating ∼80 bp fragments for each SNP according to Ahmadian et al. (7). The outer duplex PCR condition for MTHFR and GPIIIa genes was 95°C 30 s, 60°C 1 min and 72°C 1 min, according to Odeberg et al. (manuscript in preparation). This was followed by individual inner PCRs (95°C 1 min, 66°C 50 s and 72°C 2 min). The outer and inner amplification mixtures comprised of 10 mM Tris–HCl (pH 8.3), 2 mM MgCl2, 50 mM KCl, 0.1% (v/v) Tween-20, 0.2 mM dNTPs, 0.1 µM of each primer and 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, CT, USA) in a total volume of 50 µl. Five microliters of total human DNA (1 ng/µl) were used as an outer PCR template. One of the inner PCR primers in each set was biotinylated at the 5′-end to allow immobilization. Biotinylated inner PCR products (50 µl) were immobilized onto streptavidin-coated super paramagnetic beads (Dynabeads M280; Dynal, Oslo, Norway) and single-stranded DNA was obtained by alkali elution of the non-biotinylated strand.
Apyrase-mediated allele-specific extension using a bioluminometric assay
The immobilized single-stranded DNA was resuspended in H2O and 10× annealing buffer (100 mM Tris–acetate pH 7.75, 20 mM Mg–acetate) and was divided into two parallel reactions. Allele-specific primers were added to the single-stranded templates (final concentration 0.1 µM). The allele-specific primers were 5′-GCTGCTGGTGCAGGGGCCACGC-3′ (extension 1) and 5′-GCTGCTGGTGCAGGGGCCACGG-3′ (extension 2) for p53, 5′-ACTCCCTTCAGATCA-3′ (extension 1) and 5′-ACTCCCTTCAGATCC-3′ (extension 2) for wiaf 1764, 5′-GCTGCGTGATGATGAAATCGA-3′ (extension 1) and 5′-GCTGCGTGATGATGAAATCGG-3′ (extension 2) for MTHFR and 5′-TCTTACAGGCCCTGCCTCC-3′ (extension 1) and 5′-TCTTACAGGCCCTGCCTCT-3′ (extension 2) for GPIIIa. Allele discrimination between the allelic variants was investigated by the use of Klenow DNA polymerase using the two separate SNP primers that differed in the 3′-end position. Hybridization of the template and primers was performed (by incubation at 72°C for 5 min and then cooling to room temperature) and the content of each well was then further divided into two separate reactions for comparison of extension analysis with and without apyrase. Extension and real-time luminometric monitoring was performed at 25°C in a Luc96 pyrosequencer instrument (Pyrosequencing, Uppsala, Sweden). A luminometric reaction mixture was added to the single-stranded DNA with annealed primer (the substrate). The extension reaction mixture (40 µl) contained: 10 U exonuclease-deficient (exo–) Klenow DNA polymerase (Amersham Pharmacia Biotech, Uppsala, Sweden), 0.4 µg luciferase (BioThema, Dalarö, Sweden), 15 mU recombinant ATP sulfurylase, 0.1 M Tris–acetate (pH 7.75), 0.5 mM EDTA, 5 mM Mg–acetate, 0.1% (w/v) bovine serum albumin (BioThema), 1 mM dithiothreitol, 10 µM adenosine 5′-phosphosulfate (APS), 0.4 mg polyvinylpyrrolidone/ml (molecular weight 360 000), 100 µg d-luciferin/ml (BioThema) and 8 mU apyrase (Sigma Chemical Co., St Louis, MO, USA) when applicable. Prior to nucleotide addition and measuring of emitted light, pyrophosphate was added to the reaction mixture (0.02 µM), which served as a positive control for the reaction mixture and was also used for peak calibration. All 4 nucleotides were mixed and dispensed to the extension mixture (1.4 µM, final concentration) and the emitted light was detected in real-time and measured after 3 min.
Pyrosequencing
Prior to allele-specific extension analysis, the samples were genotyped by pyrosequencing. Single-stranded DNA with annealed sequence primer was used for pyrosequencing. The pyrosequencing primers were 5′-GCTGCTGGTGCAGGGGCCA-3′ for p53, 5′-CATTTGTTAAGCTTTT-3′ for wiaf 1764, 5′-AAGCTGCGTGATGATGAAA-3′ for MTHFR and 5′-CCTGTCTTACAGGCCCTGCC-3′ for GPIIIa. Real-time pyrosequencing was performed at 28°C in a total volume of 50 µl in an automated 96-well PyroSequencer using PSQ™ SNP 96 enzymes and substrates (Pyrosequencing AB, Uppsala, Sweden).
RESULTS
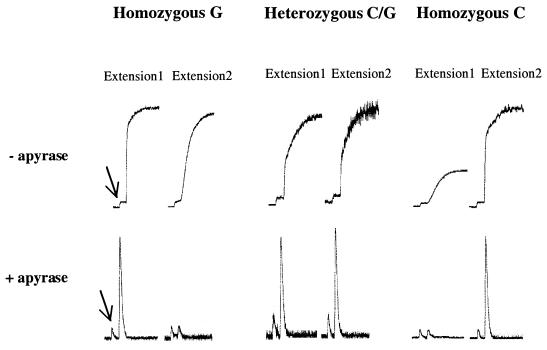
Single-stranded templates were obtained using biotinylated PCR products and streptavidin-coated super paramagnetic beads as described previously (7). The templates were separately hybridized with two alternative allele-specific primers (Table 1) and used in extension experiments. Figure 1 illustrates the real-time data obtained with the bioluminometric assay, with and without apyrase, for codon 72 of the p53 gene. When codon 72 of the p53 gene is analyzed in a homozygous G sample (without apyrase) (see Table 1), the mismatch signal (G-G) is as high as the match signal but the slope of the curve indicates slower reaction kinetics (Fig. 1, upper panel, extension 2). Similar mismatch signals were observed for wiaf 1764 (C-T mismatch) and MTHFR (G-T mismatch). However, when apyrase was included in the allele-specific extension of the matched and mismatched samples (Fig. 1, lower panel) a significant difference was observed. The previously high signals for mismatched configurations did not appear with the addition of apyrase. In these mismatched primer-template configurations, the reaction kinetics are slow leading to the degradation of nucleotides by apyrase before any incorporation by DNA polymerase can occur. However, in the case of matched primer-template configurations, the reaction kinetics were fast and incorporation of nucleotides by DNA polymerase takes place before degradation by apyrase.
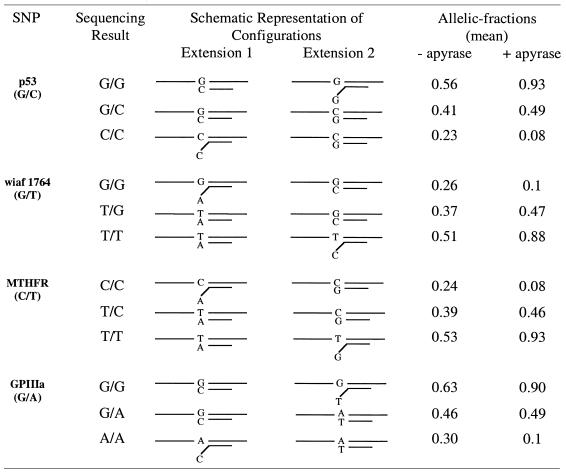
Table 1. Allele-specific extension configurations and corresponding allelic fractions with and without apyrase.
Figure 1.
Allele-specific extension of the three variants of the SNP at codon 72 of the p53 gene. The SNP status was determined separately for each sample and extension primers (1 and 2) (as listed in Table 1) are shown at the top of the figure. The top panel shows the raw-data obtained using the bioluminometric assay without apyrase and the lower panel shows the raw-data with the use of apyrase in the allele-specific extension reaction. The arrows point out the signal of pyrophosphate, which was added to the reaction mixture in all samples prior to nucleotide addition to serve as a positive control as well as for peak calibration.
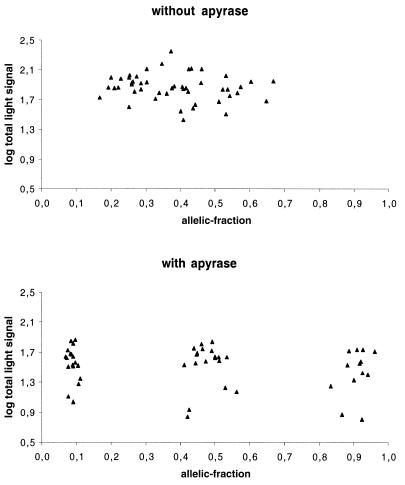
To represent the extension signals from matched and mismatched primer pairs, allelic-fractions were calculated as wt/(wt + mut), where wild type (wt) and mutant (mut) correspond to the light signals from extension primers 1 and 2, respectively (see Table 1). Using this formula, heterozygous samples are expected to have allelic-fractions near 0.5 (ratio of 1:1) and homozygous wt and mut are expected to present allelic-fractions >0.8 and <0.2, respectively (as a signal to noise ratio of 4:1 is considered significant). In Table 1, the mean allelic-fraction values for extensions with and without apyrase for each SNP variant (12–13 samples for each SNP) are listed. However, as expected, some deviation from the predicted allelic-fractions was observed. Table 2 gives detailed information of the range of allelic-fractions obtained for the SNP variants with and without the use of apyrase. The mean allelic-fractions and the standard deviation (SD) for all homozygous (wt and mut) and heterozygous samples are also given. The mean values and SD were then used to set scoring boundaries for the method. These boundaries determine the range of allelic-fraction that a sample should fall between to be considered homozygous or heterozygous (Table 2). For homozygous mut the SD has been added to the mean allelic-fraction while in the case of homozygous wt the SD has been subtracted from the mean allelic-fraction. For heterozygous variants the SD has been added to or subtracted from mean allelic-fraction, defining the upper and lower boundaries, respectively. The overall mean allelic-fraction with apyrase for homozygous wt samples was 0.907 while the allelic-fraction for the same samples without apyrase was 0.562 (Table 2). The mean allelic-fraction for homozygous mut samples with and without apyrase was 0.088 and 0.254, respectively (Table 2). Heterozygous samples with apyrase showed a mean allelic-fraction of 0.477, while without apyrase the mean allelic-fraction was 0.412 (Table 2). As a result of these deviations, scoring boundaries were calculated by taking 1–5-fold SD. When apyrase was included in the allele-specific extensions all the samples fell inside the boundaries even when up to 5-fold SD was taken into account. In these experiments, to stringently include all allelic-fractions, we chose to set scoring boundaries that correspond to 3-fold SD. Thus, the allelic-fraction for a heterozygous sample has to fall between 0.35 and 0.60, while the allelic-fraction for a wt and mut homozygous should be ≥0.81 and ≤0.12, respectively. However, if the allelic-fraction for a sample falls outside these boundaries (0.13–0.34 and 0.61–0.80) the sample will be scored as ambiguous. Note that when apyrase was excluded from the allele-specific extensions, many of the samples fell outside the scoring boundaries giving both ambiguous and false genotyping results. For example, the mean allelic-fraction for homozygous G/G in codon 72 of the p53 gene was 0.56 without apyrase (incorrect genotype) and 0.93 with apyrase (correct genotype) and for homozygous C/C the mean allelic-fraction was 0.23 without apyrase (ambiguous genotype) and 0.08 with apyrase (correct genotype) (see Table 1). This clearly shows that the addition of apyrase minimizes the extension of mismatched primer configurations by removal of nucleotides before incorporation. Figure 2 shows cluster analysis of all samples with and without apyrase. As shown in Figure 2, when apyrase is included, three distinct clusters are observed for the three possible variants of the SNPs, while no distinguishable clusters are detected when apyrase is omitted. A summary of the investigated SNPs presented here shows that all extension signals and allelic-fractions with apyrase results in genotypes that are in agreement with the pyrosequencing data. In fact, the allelic-fractions for all heterozygous SNPs were between 0.41 and 0.56, while the lowest allelic-fraction for a wt homozygous sample was 0.83 and the highest allelic-fraction for a mut homozygous was 0.11. In contrast, when apyrase was excluded, three mismatches (G-G, C-T and G-T) contributed such high extension signals that these three variants were incorrectly scored (according to scoring boundaries of 3-fold SD). The remaining five primer-template mismatches contributed somewhat lower signals, scoring these variants as ambiguous (Table 1).
Table 2. Range of allelic fractions and scoring boundries obtained with and without apyrase.
| Without apyrase | With apyrase | |||||
|---|---|---|---|---|---|---|
| Homozygous mut | Heterozygous | Homozygous wt | Homozygous mut | Heterozygous | Homozygous wt | |
| Range of allelic-fractions |
0.167–0.339 |
0.302–0.521 |
0.461–0.646 |
0.068–0.111 |
0.412–0.562 |
0.833–0.961 |
| Mean allelic-fraction |
0.254 |
0.412 |
0.562 |
0.088 |
0.477 |
0.907 |
| SD |
0.0446 |
0.0522 |
0.0581 |
0.0116 |
0.0423 |
0.0336 |
| Mean ± 1 × SD |
0.298 |
0.359–0.464 |
0.504 |
0.099 |
0.435–0.519 |
0.874 |
| Mean ± 2 × SD |
0.343 |
0.307–0.516 |
0.446 |
0.111 |
0.393–0.562 |
0.840 |
| Mean ± 3 × SD |
0.388 |
0.255–0.568 |
0.388 |
0.123 |
0.351–0.604 |
0.806 |
| Mean ± 4 × SD |
0.432 |
0.203–0.621 |
0.323 |
0.134 |
0.308–0.646 |
0.773 |
| Mean ± 5 × SD | 0.477 | 0.150–0.673 | 0.272 | 0.146 | 0.266–0.689 | 0.739 |
Figure 2.
Cluster analysis of all samples without (upper chart) and with (lower chart) the use of apyrase in the allele-specific extension reactions. Allelic-fractions on the x-axis are calculated as wt/(wt + mut), where wt and mut correspond to the light signals from extension primers 1 and 2 respectively. The y-axis is a logarithmic scale of log(wt + mut).
DISCUSSION
One of the great challenges in the post-genome era is to develop and apply large-scale techniques to score SNPs. A number of issues are important in this respect, such as multiplexing and assay format. The latter should be simple, accurate and preferably performed in a microarray format. The report here presents a solution to one of the major obstacles in allele-specific primer extensions. Certain mismatch configurations are known to yield extension products and hinder proper discrimination between genotypes, and consequently limit the use of this very convenient approach. In this report, using a real-time bioluminometric approach, we have first demonstrated that the reaction kinetics are slower for mismatched configurations compared with matched configurations (Fig. 1) and this feature has been exploited by the introduction of apyrase, a nucleotide-degrading enzyme, to perform accurate SNP genotyping. In apyrase-mediated allele-specific extension (AMASE), when the reaction kinetics are fast the primer-template is extended by DNA polymerase before apyrase degrades the nucleotides. However, when the reaction kinetics are slow, due to mismatched 3′-end primer, apyrase degrades the nucleotides and prevents the extension of mismatched primer-template. Based on these findings, four different SNPs with eight possible mismatched 3′-end primer-templates were investigated. Some of these mismatched primer-template types are documented as difficult mismatches (13–15). The analyses were carried out both with and without the use of apyrase in the allele-specific extensions, and despite the relative short extension time (3 min), we could not discriminate between these difficult mismatches and the corresponding matched primer-templates unless apyrase was included. Cluster analysis of samples that were typed by AMASE showed three distinct clusters for each set of SNP variants while no distinguishable clusters were obtained when apyrase was omitted from the reactions (Fig. 2). In the cluster analysis, one of the homozygous variants was set to correspond to wt sequence (signal obtained from extension primer 1 in Table 1) and the other homozygous variant was set to correspond to the mut sequence (signal obtained from extension primer 2 in Table 1). Thus, allelic-fractions were calculated as wt/(wt + mut). By taking a 3-fold SD, the allelic-fraction boundaries were defined as 0.01–0.12 for homozygous mut samples, 0.35–0.60 for heterozygous samples and 0.81–0.99 for homozygous wt samples (see Table 2).
While apyrase is not a thermostable enzyme, it should be mentioned that AMASE is not a solution for allele-specific PCR methods until a thermostable nucleotide-degrading enzyme is engineered. However, high-throughput analysis of SNPs in microarray format may be performed with non-thermostable DNA polymerases and therefore AMASE can be applicable to typing on oligonucleotide arrays using fluorescently labeled nucleotides. Thus, to investigate the ability of apyrase to degrade dye-labeled nucleotides, fluorescent-based detection of extension products (in solution), was performed on glass slides. In this fluorescent-based detection, three samples of each SNP were selected to represent each variant. The results were fully comparable with the bioluminometric assay as the same primer-template mismatches gave high fluorescent signals in the absence of apyrase. However, in the presence of apyrase, no ambiguities or discordant results were observed (data not shown).
In conclusion, our study shows that SNPs can rapidly be scored with allele-specific primers in simple extension reactions, which is a convenient alternative for high-throughput applications. Here we have shown that the reaction kinetics are slower for mismatched configurations compared with matched configurations and, consequently, have introduced apyrase to accomplish correct SNP genotyping. Furthermore, apyrase is able to degrade fluorescent-labeled nucleotides opening up the possibility to perform AMASE in a microarray format.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by The Swedish Research Council for Engineering Science (TFR) and the Knut and Alice Wallenberg Foundation.
REFERENCES
- 1.Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 2.Yershov G., Barsky,V., Belgovskiy,A., Kirillov,E., Kreindlin,E., Ivanov,I., Parinov,S., Guschin,D., Drobishev,A., Dubiley,S. et al. (1996) DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl Acad. Sci. USA, 93, 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nature Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 4.Pastinen T., Kurg,A., Metspalu,A., Peltonen,L. and Syvanen,A.C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res., 7, 606–614. [DOI] [PubMed] [Google Scholar]
- 5.Laken S.J., Jackson,P.E., Kinzler,K.W., Vogelstein,B., Strickland,P.T., Groopman,J.D. and Friesen,M.D. (1998) Genotyping by mass spectrometric analysis of short DNA fragments [see comments]. Nature Biotechnol., 16, 1352–1356. [DOI] [PubMed] [Google Scholar]
- 6.Howell W.M., Jobs,M., Gyllensten,U. and Brookes,A.J. (1999) Dynamic allele-specific hybridization. A new method for scoring single nucleotide polymorphisms. Nature Biotechnol., 17, 87–88. [DOI] [PubMed] [Google Scholar]
- 7.Ahmadian A., Gharizadeh,B., Gustafsson,A.C., Sterky,F., Nyren,P., Uhlen,M. and Lundeberg,J. (2000) Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem., 280, 103–110. [DOI] [PubMed] [Google Scholar]
- 8.Ronaghi M., Uhlen,M. and Nyren,P. (1998) A sequencing method based on real-time pyrophosphate. Science, 281, 363–365. [DOI] [PubMed] [Google Scholar]
- 9.Higgins G.S., Little,D.P. and Koster,H. (1997) Competitive oligonucleotide single-base extension combined with mass spectrometric detection for mutation screening. Biotechniques, 23, 710–714. [DOI] [PubMed] [Google Scholar]
- 10.Newton C.R., Heptinstall,L.E., Summers,C., Super,M., Schwarz,M., Anwar,R., Graham,A., Smith,J.C. and Markham,A.F. (1989) Amplification refractory mutation system for prenatal diagnosis and carrier assessment in cystic fibrosis. Lancet, 2, 1481–1483. [DOI] [PubMed] [Google Scholar]
- 11.Goergen B., Meyer zum Buschenfeld,K.H. and Gerken,G. (1994) Mutation specific PCR and direct solid phase sequencing assay for the detection of hepatitis B virus pre-C/C mutants in anti-HBe-positive, chronic hepatitis B. J. Med. Virol., 43, 97–102. [DOI] [PubMed] [Google Scholar]
- 12.Newton C.R., Graham,A., Heptinstall,L.E., Powell,S.J., Summers,C., Kalsheker,N., Smith,J.C. and Markham,A.F. (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res., 17, 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day J.P., Bergstrom,D., Hammer,R.P. and Barany,F. (1999) Nucleotide analogs facilitate base conversion with 3′ mismatch primers. Nucleic Acids Res., 27, 1810–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok S., Kellogg,D.E., McKinney,N., Spasic,D., Goda,L., Levenson,C. and Sninsky,J.J. (1990) Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res., 18, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayyadevara S., Thaden,J.J. and Shmookler Reis,R.J. (2000) Discrimination of primer 3′-nucleotide mismatch by taq DNA polymerase during polymerase chain reaction. Anal. Biochem., 284, 11–18. [DOI] [PubMed] [Google Scholar]
- 16.Nyren P., Karamohamed,S. and Ronaghi,M. (1997) Detection of single-base changes using a bioluminometric primer extension assay. Anal. Biochem., 244, 367–373. [DOI] [PubMed] [Google Scholar]