Abstract
Accumulating evidence indicates that gut transit time is a key factor in shaping the gut microbiota composition and activity, which are linked to human health. Both population-wide and small-scale studies have identified transit time as a top covariate contributing to the large interindividual variation in the faecal microbiota composition. Despite this, transit time is still rarely being considered in the field of the human gut microbiome. Here, we review the latest research describing how and why whole gut and segmental transit times vary substantially between and within individuals, and how variations in gut transit time impact the gut microbiota composition, diversity and metabolism. Furthermore, we discuss the mechanisms by which the gut microbiota may causally affect gut motility. We argue that by taking into account the interindividual and intraindividual differences in gut transit time, we can advance our understanding of diet–microbiota interactions and disease-related microbiome signatures, since these may often be confounded by transient or persistent alterations in transit time. Altogether, a better understanding of the complex, bidirectional interactions between the gut microbiota and transit time is required to better understand gut microbiome variations in health and disease.
Keywords: intestinal microbiology, diet, gastrointestinal transit, gastrointestinal physiology
Key messages.
Gut transit time varies considerably between and within individuals, and explains large proportions of the gut microbiota compositional variation between people.
Gut transit time affects substrate availability in the colon affecting the trade-off between saccharolytic and proteolytic fermentation.
The gut microbiota can via production of metabolites stimulate gut motility thus affecting the transit time
Many disease-related microbiome signatures may be confounded by alterations in gut transit time.
By considering interindividual and intraindividual differences in transit time in human studies, diet–microbiota interactions and disease-related microbiome signatures may be better elucidated.
Introduction
The human gastrointestinal tract (GIT) is densely populated by microbes, which play an important role in a broad range of physiological processes from the digestion of complex polysaccharides to the regulation of neural signalling.1 The composition and metabolism of the adult gut microbial communities are affected by a combination of factors including diet,2 3 demographics,4 5 use of medication,6 health status7 and environmental components shaping the gut environment.8 Among these environmental components, gut transit time, that is, the time it takes foods to travel through the GIT, appears to be a major driver of gut microbiome variation.9–12 Gut transit time varies markedly between and within individuals13–15 and has been associated with gut microbial diversity, composition, and metabolism.9–12 16–18 The anatomical segments of the GIT (ie, stomach, small intestine and colon) have segment-specific transit time, affecting the composition of the residing gut microbes.12 Although this knowledge is well established, differences in transit time and pH within and between individuals have largely been neglected when investigating person-specific gut microbiota signatures. Here, we review and discuss the role of gut transit time as a key determinant of the gut microbial composition and metabolism as well as of many diet–microbiota interactions relevant to human health (figure 1). We discuss the implications of altered gut transit time in health and disease, and provide an overview of the currently available methods for assessment of gut transit time in humans.
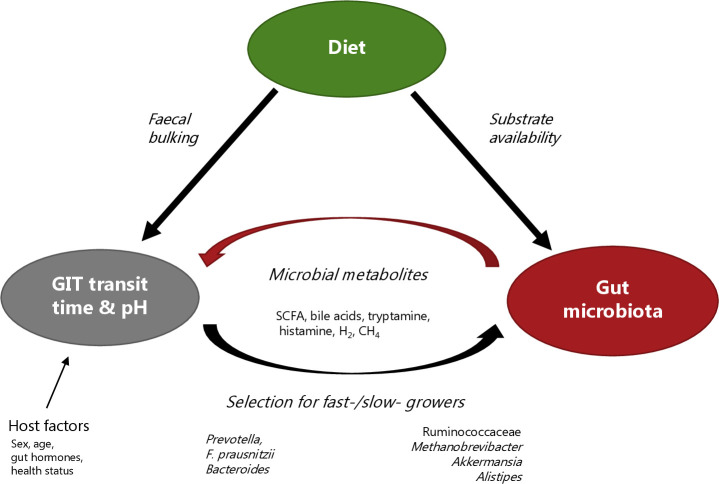
Figure 1.
Illustration of the complex interplay between diet, gut microbiota and gut transit time. Diet can directly affect gastrointestinal motility, especially dietary fibre and osmotically active foods can increase the faecal bulk and thereby accelerate gut transit time. Diet can also affect gut transit time by dictating the substrate availability to the gut microbiota. As a result, the gut microbiota produces metabolites such as short-chain fatty acids (SCFA), secondary bile acids, tryptamine, histamine, H2 or CH4. These microbial-derived metabolites can stimulate gastrointestinal motility and thereby impact gut transit time. In addition, gut transit time affects the gut microbial composition and metabolism and consequently the gut environment (eg, pH). The relation between the gut microbiota and the gut transit time is therefore bidirectional. In addition, host factors including gut hormones, gender, age, health status and physical activity also affect the gut transit time. GIT, gastrointestinal tract.
Transit time throughout the GIT
Interindividual and intraindividual variation in transit time
In healthy populations, whole gut transit time (WGTT) varies substantially between individuals13 15 with a median WGTT of approximately 28 hours.11 19 Segment-specific transit times are commonly referred to as gastric emptying time (GET), small intestinal transit time (SITT) and colonic transit time (CTT). GET is the time it takes for food to empty from the stomach and enter the small intestine in a form of semiliquid chyme.20 SITT is the duration time of the passage of the chyme from the duodenum (i.e. the proximal small intestine) until the ileocaecal region, and similarly, CTT corresponds to the duration time of the chyme’s passage from the caecum until the egestion in a form of stool.21 For GET of solids, a transit time coefficient of variation of 24.5% has been reported between individuals.22 Several human studies have identified large interindividual variation in SITT with a median of approximately 5 hours (range of 2–7.5 hours).21 23–25 Compared with the small intestine, transit through the colon is much slower with a median of 21 hours.19 Consequently, large interindividual variations are often seen in CTT with the minimum and the maximum reported transit times of 0.1–46 hours for the proximal colon, 0.3–80 hours for the distal colon and 1–134 hours for the rectosigmoid colon19 26–29 (figure 2).
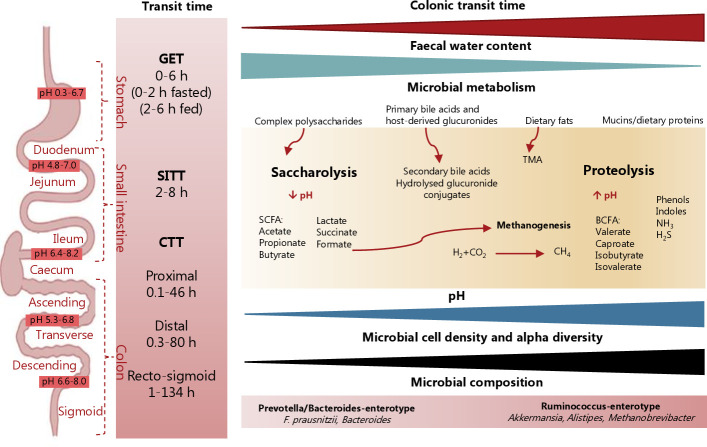
Figure 2.
Segmental transit time and pH throughout the gastrointestinal tract and its association with gut environment and gut microbial metabolism. The transit time varies throughout the gastrointestinal tract with substantial interindividual differences in gastric emptying time (GET), small intestinal transit time (SITT) and colonic transit time (CTT), which account for most of the whole gut transit time. The segmental transit time ranges show the minimum and maximum transit times reported for each segment. Long gut transit time has been associated with higher faecal pH, reduced faecal water content, higher microbial cell density and diversity, and a shift in microbial metabolism from saccharolysis towards proteolysis as reflected by reduced levels of short-chain fatty acids (SCFA) and increased levels of branched-chain fatty acids (BCFA). It is likely that once easy accessible carbohydrate sources become scarce in the colon, the gut microbes switch to ferment dietary and mucin-derived proteins. While saccharolysis by the gut microbiota gives rise to SCFA that are beneficial for the host and a source of energy for the colonocytes, proteolysis can lead to the accumulation of compounds such as BCFA, phenols, indoles, ammonium (NH3) and hydrogen sulphide (H2S) that are generally considered detrimental for health. Moreover, hydrogen (H2) with carbon dioxide (CO2) or formate can be converted into methane (CH4) by methanogenic archaea, which are also linked to slower transit time. In addition, the production and circulation of secondary bile acids and hydrolysis of host-derived glucuronides excreted via bile can also be affected by alterations in gut transit time. Whether microbiota-derived trimethylamine (TMA), produced from mainly choline and carnitine, is linked to transit time remains unknown. Created with Biorender.com.
Gut transit time also varies within individuals over time.13 14 16 For example, repeated measurements of CTT using radio-opaque markers within eight healthy subjects over a period of several months showed that each subject exhibited a wide range of CTT with a mean coefficient of variation of 25%.30 Furthermore, a recent study showed that the percentage of the faecal water content, a proxy of transit time, varied from day to day in both healthy subjects and patients suffering from irritable bowel syndrome (IBS).31 Similarly, intrasubject differences in SITT and CTT have been observed with tandem measurements in 10 healthy adults using the SmartPill capsule13 that can directly assess WGTT and segmental transit times.32
The currently available methods for WGTT and segment-specific transit time assessment include direct methods such as the SmartPill capsule and indirect methods such as stool consistency, stool frequency and faecal water content (table 1). It is important to note that the methods provide a range of outcomes, some of which provide similar results while others may not be comparable. For example, while scintigraphy relies on calculating the geometric centre based on recorded radioactivity in the different GIT regions at certain time points,33 the SmartPill capsule uses landmarks in the gastrointestinal pH to calculate the segmental transit times.34
Table 1.
Methods available for gut transit measurements and examples of their application in human gut microbiome research
| Method | Region | Subjects | Gut microbiome and/or transit time-related findings | References | |
| Direct transit time measures | Radio-opaque markers | CTT WGTT |
48 healthy subjects | Distal CTT was associated with increased microbial α-diversity, rectosigmoid CTT was negatively associated with faecal SCFA and distal CTT was negatively associated with plasma acetate | 18 |
| 98 subjects | CTT positively associated with microbial community structure, microbial richness and microbial protein catabolism | 10 | |||
| 48 healthy subjects with slow WGTT* | RCT with arabinoxylan-oligosaccharide increased faecal Bifidobacterium and softened stool consistency without changing the WGTT | 46 | |||
| 14 healthy subjects† | WGTT positively correlated with urinary sulphate, faecal methanogens and negatively with total faecal SCFA, sulphate and bile salts | 134 | |||
| Scintigraphy | WGTT GET SITT CTT |
50 healthy and constipated patients | Colonic mucosal microbiota was not associated with CTT and was significantly different between the two groups, faecal microbiota was associated with CTT and breath methane | 88 | |
| 36 healthy and 20 patients with liver cirrhosis | SITT was negatively correlated with B:F ratio and microbial dysbiosis index | 215 | |||
| SmartPill | WGTT GET SITT CTT |
11 obese and 11 normal weight subjects | Shorter SITT was associated with Bact2-enterotype, longer CTT was associated with Rum-enterotype | 12 | |
| 33 healthy and 114 IBS-C patients | Colonic intraluminal pH levels were significantly lower in IBS patients compared with HC, and total faecal SCFA levels correlated negatively with CTT | 216 | |||
| 19 healthy and 9 constipated subjects |
Rectosigmoid pH negatively correlated with Bifidobacterium spp and positively with Coprococcus spp | 108 | |||
| Gas-sensing capsules | WGTT GET SITT CTT |
4 healthy volunteers | SITT was slower with a diet high in fermentable fibre (~34 g/day) compared with a diet low in fermentable fibre (~22 g/day) | 217 | |
| Blue dye | WGTT | 1102 subjects | Gut microbiome composition predicted WGTT, longer WGTT was linked with Akkermansia muciniphila, Bacteroides and Alistipes spp. | 11 | |
| Sweet corn | WGTT | 31 healthy subjects | WGTT positively correlated with faecal BCFA and Coprococcus | 107 | |
| Indirect transit time measures | Stool frequency | WGTT | 69 subjects | B:F ratio and Bacteroides:unclassified_Ruminococcacea positively associated with stool frequency | 218 |
| 60 healthy subjects | B:F ratio was higher in a group with stool frequency of ≤2 times/week compared with one time/day or one time/2 day and ≥2–3 times/day | 219 | |||
| Stool consistency (BSS) | CTT | 53 healthy subjects | Stool consistency was positively correlated with species richness, Akkermansia and Methanobrevibacter abundances, and negatively associated with the B:F ratio, | 17 | |
| 1126 subjects | BSS was associated with the B:F ratio, high BSS score positively correlated with F. prausnitzii | 220 | |||
| Faecal water (stool moisture) | CTT | 31 healthy subjects | Faecal water positively correlated with WGTT | 107 | |
| 40 subjects | Stool moisture accounted for 4.3% of interindividual microbiota variation (absolute abundances) | 79 | |||
| 12 IBS patients and 12 controls | Association between stool consistency and microbial community structure/microbial richness | 31 | |||
| Stool crosslinking | CTT | 170 samples | Faecal acetate and methionine were predictive of stool consistency | 221 | |
| Breath test | Oro-caecal | 14 healthy subjects | Oro-caecal transit time was positively correlated with WGTT | 134 |
*Transit time also measured by BSS, faecal water content and breath test.
†Transit time also measured by the breath test.
BCFA, branched-chain fatty acids; B:F, Bacteroidetes:Firmicutes; BSS, Bristol Stool Scale; CTT, colonic transit time; GET, gastric emptying time; IBS, irritable bowel syndrome; RCT, randomised controlled trial; SCFA, short-chain fatty acids; SITT, small bowel transit time; WGTT, whole gut transit time.
Host and environmental factors influencing transit time
Several factors affect the intraindividual and interindividual variations in gut transit time including sex, ageing, stress, body mass index, colonic anatomy, gut hormones and diet.35 Moreover, the gut microbiota and its metabolites also affect gut transit time, which is discussed in further detail below.
Dietary impact
Dietary patterns, dietary factors such dietary fibres, as well as individual dietary ingredients, can directly affect the gut physiology via stimulation of gastrointestinal motility either independently of the gut microbiota or via gut microbiota dependent pathways, which will be discussed below. Wu et al showed that intestinal transit time was faster on a high-carbohydrate/low-fat diet when compared with a low-carbohydrate/high-fat diet in 10 healthy subjects.3 A randomised control trial with 120 obese participants reported significantly more cases of constipation (68% vs 35%) and diarrhoea (23% vs 7%) in a ketogenic diet group compared with a low-fat diet group.36 However, whether the adverse gastrointestinal outcomes in these studies were caused by the high fat, high protein or low carbohydrate content could not be concluded. Nonetheless, a high intake of fat was associated with constipation and prolonged CTT,35 37 and infusion of fat into the small intestine of healthy subjects has been shown to slow gastric emptying.38 In contrast, a 4-week intervention on a high-fat diet with 12 healthy men resulted in accelerated gastric emptying and orocaecal transit time when compared with a low-fat diet.39 Therefore, the dietary patterns may exert different effects on the GIT depending on their composition and content.
One particularly important component of the dietary patterns is the dietary fibres. Dietary fibres affect the functionality of the GIT including the gut transit.40 Different types of dietary fibres have very different physicochemical characteristics with regards to solubility, fermentability and gel-formation (viscosity). The different characteristics influence their effects on gut transit time, as reviewed in detail elsewhere.41 42 Today, most studies investigating the effects of dietary fibres on the gut transit time have been limited to wheat bran and psyllium. While wheat bran consistently decreases the WGTT,43 the effects of psyllium, which is minimally fermented by the gut microbiota, on WGTT are inconsistent.44 It has been shown that the laxative effects of coarse wheat bran are greater than that of fine wheat bran suggesting that the particle size plays a role for the mechanical stimulation of the intestinal epithelium.45 In line with this, an intervention study with powder arabinoxylan-oligosaccharides (wheat bran-based prebiotics) in 48 subjects resulted in softer stools but did not change the CTT.46 Unlike wheat bran, psyllium contains a soluble type of fibre and has gel-forming properties that increase the water retention in the colon thus increasing the faecal water content and bulk.47 Cellulose, which is also non-fermentable but not gel-forming, has been shown to lower faecal pH, increase the stool outputs and to decrease CTT.48 Fermentable fibres such as inulin seem to alleviate constipation and improve physical discomfort.49 Other human studies have shown that increased intake of cereals and fermentable wheat fibre increased stool output.50 51 Altogether, dietary fibres can accelerate transit time and increase stool outputs through water retention, bulking and via fermentation-mediated effects. However, the effects on transit time depend on the particle size, solubility, fermentability and viscosity of the given fibre. Furthermore, water supplementation has been found to enhance the effect of high-fibre diet on increasing stool frequency in constipated patients,52 suggesting that fluid intake is another important factor to consider with respect to transit time. Indeed, a handful of studies have reported an association between lower fluid intake and constipation.53 However, no human intervention studies have to our knowledge investigated the relationship between water intake, bowel habits and the gut microbiome.
Finally, also individual food ingredients may affect transit time. For instance, carboxymethylcellulose, a widely used emulsifier, has been shown to act as a laxative54 and was recently linked to reduced gut microbiota diversity in humans.55 Another example is sorbitol, a sugar alcohol highly abundant in prunes, which can retain water molecules by osmosis, similar to some types of fibre that increase water content in the gut lumen and may lead to softer stools.56 57 In addition, consumption of turmeric, a spice increasing the bile secretion, has been associated with a significantly longer transit time in gnotobiotic mice.58
Other factors affecting transit time
Another important factor affecting the gut transit time is sex. Gut transit time is shorter in men than in women—even when adjusting for total energy intake, diet, body weight and height.59 The sex difference was shown to be the most pronounced in the distal colon with women having longer transit through the transverse and descending colon,19 possibly as a result of slower gut motility in women.60 Moreover, from epidemiological studies, it has been shown that women are also more prone to developing constipation.61 While there is a strong evidence for gender differences in gut transit time, it remains to be determined whether it is the sex per se or differences in behaviours and habits that drive the differences in transit time. The length of the intestine is also an important factor to consider as it can vary greatly among individuals. Based on a postmortem measurement, men have been found to have a longer intestine than women, which may contribute to longer transit time in women.60 62 The same study reported that the average length of the whole intestine is 7.96±1.3 m with a range of 3.78–13.16 m and found that the intestine’s length positively correlates with body weight but not body height.62 Moreover, ageing is associated with longer gut transit time63 64 and it has been shown that especially transit through the right (ascending) part of the colon is correlated to age,19 perhaps as a consequence of a more refined diet and/or reduced physical activity with older age. Indeed, physical activity may improve bowel habits via increased gut motility.65 Furthermore, social aspects and acute stress are likely to play a role in gut transit time variations, although the contribution of these factors is difficult to assess. In animals, induced stress results in slower transit in the upper gut, while the opposite was observed in the large bowel, where stress increased motility and stool output.65 Also, other exogenous factors such as medication often exert side effects on the gut transit.66 Finally, host genetics has recently been shown to be involved in gut motility as 14 independent loci were found to associate with stool frequency.67
In conclusion, WGTT and segmental transit times depend on a combination of factors, including host genetics, anatomy, physiology, health status and lifestyle as well as external factors such as intake of foods, water and drugs.
Gut transit time: a key determinant of the gut microbiota
Emphasising the importance of transit time, CTT has been linked to gut microbiota diversity and composition in both population-wide analyses and small-scale studies.10 11 17 18 46 68 Moreover, stool consistency assessed by the Bristol Stool Scale (BSS),69 which is a surrogate marker for CTT, has been identified as a top covariate of the faecal microbial composition of a healthy population.9 Considering that CTT varies markedly between individuals and from day to day within individuals,13–16 microbiota-focused investigations should take gut transit time into account. Although faecal microbial species richness has been associated with a diverse diet70 and suggested as an indicator of the host health status,71 72 it has also been associated with long CTT,10 indicating that faecal microbial richness is strongly confounded by CTT.73
Several bacterial groups including Akkermansia, Alistipes, Methanobrevibacter and Ruminococcaceae have consistently been associated with firm stools and a long CTT.9 11 12 17 Moreover, the classification of individuals according to their prevailing microbial community structure (ie, Bacteroides-1, Bacteroides-2, Prevotella or Ruminococcaceae enterotypes) is thought to be driven at least partially by gut transit time.12 74 The Prevotella enterotype, known to prevail on fibre-rich diets,3 has been associated with loose stools while the opposite has been observed for the Ruminococcaceae enterotype,17 75 which has been characterised by increased proteolytic capacity.76 In agreement herewith, a long CTT was associated with an increased prevalence of the Ruminococcaceae enterotype in healthy subjects12 and decreased levels of Prevotella in constipated patients74 and Parkinson’s patients77 that also often suffer from constipation.78 Similarly, the abundance of another saccharolytic bacterium, Eubacterium rectale, was reduced in individuals with a long CTT.11 Furthermore, one study has investigated the associations between the SITT and pH, and the faecal microbial community composition.12 This study found that shorter small intestinal transit is associated with the Bacteroides-2 enterotype,12 which is characterised by a high proportion of Bacteroides and low microbial cell densities in stools.79 Additionally, the abundances of Bacteroides and Flavonifractor were negatively correlated with small intestinal pH, supporting the idea that inter-individual variation in environmental conditions in the small intestine is linked to gut microbial composition and activity.12 Several human studies have established that longer CTT is associated with an increase in distal colonic pH,80–82 suggesting that gut transit time and pH are interrelated. Also in vitro experiments simulating short and long gut transit time by high or low dilution rates, respectively, have shown that dilution rate and pH have a substantial impact on the growth of several bacterial groups. For instance, A. municiphila was present at all pH ranges (6.0–8.0) in low dilution rates, whereas it only grew at high pH (pH>7.0) at a high dilution rate, while M. smithii was only detected at a low dilution rate.83 An emerging body of scientific evidence thus shows that both gut transit time and pH drive the composition of the microbial communities along the GIT (figure 2). However, studies investigating the gut microbiome in relation to gastrointestinal pH or segmental transit time are scarce, and interventional studies to provide further evidence remain to be conducted.
Today, most research on the human gut microbiota relies on stool samples. While stool samples are generally considered to be representative of the luminal colonic microbiota,84 85 the faecal microbiota represent an ‘end-product’ of the whole gut microbial community. Thus, the relative microbial community composition in the faeces is more similar to that of the distal colon compared with the proximal colon and small intestine.86 87 However, the question remains whether absolute numbers of saccharolytic bacterial groups are similar in the proximal colon and in faeces (mirroring the distal colon), and only change in terms of relative abundance due to an increase in abundance of slow-growing and proteolytic bacteria in the distal colon. Studies including sampling throughout the human GIT are needed to deduce how transit time associate with the quantitative human gut microbiome composition in the proximal and distal colon, respectively. This may be important as the state of the gut microbial community maturation (ie, life-time of the bolus in the colon) could explain a considerable fraction of intraindividual and interindividual microbiota variation in healthy individuals.73 Finally, the mucosal microbial communities, which occupy the outer mucus layer of the intestinal epithelium, are distinct from the luminal microbial communities and their composition appears to be less affected by the gut transit time.88
Gut transit time and gut microbial metabolism
Transit time not only affects the gut microbiota composition but also affects the gut microbial metabolism since differences in transit time have consequences for substrate availability throughout the GIT (figure 2).
A trade-off between saccharolytic and proteolytic fermentation
Non-digestible polysaccharides reach the caecum and the proximal colon where they undergo fermentation by the residential microbes resulting in the generation of gases (H2 and CO2) and metabolites such as short-chain fatty acids (SCFA), mostly acetate, propionate and butyrate, which are generally considered beneficial for health.89 However, when easy accessible carbohydrates become scarce the microbial activity shifts towards fermentation of dietary or mucosal proteins instead.90 This results in the formation of potentially deleterious compounds such as phenols, indoles, ammonia or hydrogen sulphide (H2S).90 The depletion of carbohydrates ultimately leads to a decrease in SCFA consequently increasing the luminal pH that creates a selective pressure on the microbial community thereby redirecting the microbial metabolism towards proteolysis.83 Long CTT has been associated with reduced faecal SCFA indicating either increased absorption, lower availability of fermentable polysaccharides in the colon, and/or changed activity.18 81 91 92 In a recent publication, faecal SCFA concentrations and microbial diversity clustered according to stool consistency assessed by the BSS with higher levels of SCFA detected in looser stools, reflecting a shorter colonic transit.93
Furthermore, increased CTT has also been associated with increased proteolytic fermentation in the colon in both healthy subjects10 and patients with Parkinson’s disease.94 In Parkinson’s disease, constipation and long CTT are common complications.78 One study observed elevated serum levels of host–microbial coproducts derived from bacterial proteolysis (p-cresol sulphate and phenylacetylglutamine) in a cross-sectional cohort with 197 Parkinson’s patients.94 Furthermore, the authors also showed that bacterial taxa (Oscillospira and Ruminococcus) positively associated with these proteolytic metabolites were also positively associated with firm stools.94 Likewise, elevated levels of p-cresol sulphate have been observed in autistic children95 and in patients with end-stage renal disease,96 patient groups who also often suffer from prolonged CTT.97 98 These findings suggest that proteolytic metabolites including the host–microbial co-metabolite p-cresol sulphate might be markers of constipation and slow transit rather than indicators of disease. In support hereof, urinary levels of p-cresol sulphate, as well as phenylacetylglutamine, were also associated with longer CTT in healthy individuals, clearly indicating a shift towards microbial proteolysis with the prolonged colonic transit.10 Additionally, recent evidence shows that even on a homogenous diet, urinary levels of these metabolites remain highly variable between individuals,99 further emphasising that other factors than diet modulate the concentrations of these metabolites. Altogether, this suggests that intestinal transit time plays a role in the highly individual diet–microbiota responses.
Prolonged transit time has also been linked to increased urinary sulphate excretion,100 elevated levels of urinary phenol and increased excretion of faecal ammonia,101 another microbial by-product of protein degradation. Ammonia has been shown to increase mucosal damage and promote colonic cancer in rats at relatively low concentrations.102 103 Similarly, ammonia, as well as phenol, have been shown to disrupt tight junctions of cultured colon cells (Caco-2).104 However, there is a lack of evidence from human studies to link ammonia to colon cancer. Impaired mucosal integrity and breakdown of the mucosal barrier are also caused by H2S via inhibition of butyrate oxidation by the colonocytes.105 Importantly, we have previously observed a negative correlation between mucus-degradation-associated metabolites in urine and CTT, suggesting that prolonged colonic transit may also lead to enhanced degradation of the mucus layer by the microbiota10 in line with observations from fibre-depleted diets.106
Elevated concentrations of branched-chain fatty acids (BCFA) are positively associated with CTT in healthy adults.107 BCFA such as isobutyrate, isovalerate, or 2-methylbutyrate are products of bacterial fermentation of branched-chain amino acids and has been positively correlated with the relative abundance of Coprococcus and Blautia.107 Interestingly, a study that employed the SmartPill to measure gastrointestinal pH, showed that Coprococcus spp. was positively associated with recto-sigmoid pH, where pH is slightly alkaline (>7) while the inverse was observed for Bifidobacterium spp,108 a saccharolytic species109 thriving at neutral pH.110 This supports the hypothesis that prolonged transit time may lead to a less acidic environment in the colon as a result of SCFA depletion and accumulation of alkaline compounds from the proteolytic processes. In line with this, in vitro studies indicate that higher concentrations of BCFA were produced at high pH and low dilution rate simulating slow luminal washout and thereby slow CTT.83 This increase of intraluminal pH towards the distal colon was not observed in rural Africans111 who have shorter CTT112 113 and a habitual diet rich in dietary fibres, which likely provides a surplus of SCFA.114 In addition, it is important to mention that microbial metabolites of dietary fats remain an underexplored area, where only a few products of microbial origin are known including some sphingolipids, endocannabinoids or trimethylamine (TMA). TMA is produced by gut microbes from methylamine-containing nutrients (eg, choline, lecithin, L-carnitine) and further processed in the liver to trimethylamine N-oxide (TMAO).115 While TMAO levels have been correlated with the risk of cardiovascular events,116 no study has investigated links between TMAO and gut transit time.
Taken together, colonic fermentation is in essence a trade-off between saccharolytic and proteolytic metabolism, which depends on a complex interplay between the composition of the gut microbiome, the substrate availability and colonic pH—all of which are affected by transit time. A slow colonic transit limits the carbohydrate availability in the colon, favouring bacteria that can use other sources of energy such as dietary or host-derived proteins. Moreover, the nature of the microbial products also changes the physicochemical properties of the colonic environment for example, by changing pH and thus altering the microbial composition and metabolism.
Cross-feeding and gas metabolism
While most of the microbes residing in the colon belong to Bacteroidetes or Firmicutes phyla117 including species from the classes Bacteroidia and Clostridia that possess a large variety of carbohydrate-active enzymes,118 less abundant species in the colon include those using secondary products of carbohydrate fermentation (eg, hydrogen, lactate, succinate, formate or ethanol). These include hydrogen-consumers such acetogenic bacteria, which comprise a phylogenetically diverse group of bacteria including Blautia hydrogenotrophica (previously known as R. hydrogenotrophicus 119 ), methanogenic archaea with the predominance of Methanobrevibacter smithii, and sulphate-reducing bacteria (SRB), mostly represented by Desulfovibrio genus.120–122 Although many species can produce lactate, it does not accumulate in the colon under healthy conditions due to the presence of lactate utilisers that use lactate for growth and produce SCFA.123 Lactate can be converted into propionate by Coprococcus catus, while Anaerostipes and Anaerobutyricum spp can convert lactate into butyrate.124 Both lactate and succinate can be converted into propionate by Veillonella spp. Some of the other succinate utilisers include Dialister and species, although some Bacteroides, for example, B. vulgatus can also produce succinate.125 Lactate accumulates in stools of subjects with chronic diarrhoea, especially during severe ulcerative colitis suggesting a perturbation of the balance between lactate-producers and utilisers in those patients.126 127The production of H2 by the gut microbes seems to be coupled with low pH.83 Given that CTT is linked to pH, CTT likely affects the H2 production and competition between hydrogen-using species including acetogens, methanogens and SRB.128 While acetogens can use H2 (and CO2) or formate to generate acetate at low pH,129 methanogens can use H2 (and CO2) or formate to produce methane (CH4) and SRB can use H2 or lactate to produce H2S (in a presence of sulphate) at neutral or slightly alkaline pH.122 H2S and CH4 have both been found in higher concentrations at low dilution rates (simulating long gut transit time) in vitro suggesting that SRB and methanogens are affected by the rate of the luminal washout. In support hereof, high breath levels of CH4 as well as the faecal abundance of M. smithii, a slow-growing methanogen unable to degrade sugars,130 have repeatedly been associated with constipation and slow CTT.9 11 12 17 130 Yet, a recent study reported that breath CH4 was associated with both the faecal and mucosal microbiota even after adjusting for transit time, emphasising that other factors beside transit time affect the presence of methanogens. Indeed, the abundance of hydrogen-using species is dependent on the growth and competition of hydrogen-producers, and intraluminal factors such as substrate availability and pH.83 128
Bile acids and enterohepatic circulation
During meals, bile is released from the gallbladder into the duodenum. Although 95% of the bile acids are reabsorbed in the terminal ileum, the rest escapes to the colon and becomes available to the colonic microbial community131 that forms a large variety of secondary bile acids that upon re-absorption can re-enter the bile acid pool via the enterohepatic circulation.132 Yet, the bile acid composition may be affected by CTT. Increased levels of deoxycholic acid in bile133 and serum82 were observed with longer colonic transit and less bile was egested via stool,134 suggesting that prolonged CTT provides a longer time for the conversion into secondary bile acids and/or reabsorption of the secondary bile acids. Notably, the intestinal transit time is longer in patients with cholesterol gallbladder stones,135 136 who have been shown to have higher deoxycholic acid in their bile and higher intraluminal pH in the proximal and distal colon, compared with healthy controls.82 In addition, the 7α-dehydroxylases, responsible for microbial conversion of primary bile acids into deoxycholic acid and lithocholic acid, are pH-sensitive and only active at pH above 6.5.137 138 Since the intraluminal pH is related to CTT, interindividual differences in pH and transit time could be important for personal diet–microbiome–host interactions also concerning the microbial conversion of bile acids.
Prolonged colonic transit has also been associated with an increased concentration of circulating oestrogens,139 140 which has been associated with an increased breast cancer risk in postmenopausal women.141 Interestingly, a large cohort study reported that higher stool frequency, typically reflecting shorter transit time, was associated with decreased risk of breast cancer.142 Steroids as well as drugs, food additives and some other dietary compounds, for example, heterocyclic amines from protein-rich diets,143 undergo glucuronidation or sulphation in the liver prior to excretion to the bile. However, the glucuronide conjugates can be hydrolysed by bacterial β-glucuronidases in the gut, which in return increases their reabsorption and retention time in the body.144 Long colonic transit may therefore increase the hydrolysis of these glucuronide-conjugates in the colon thus increasing their bioavailability.145 Bacterial β-glucuronidases seem to be inhibited by low pH137 138 and acidification of the colon (eg, by high intake of dietary fibre) may prevent accumulation of glucuronide de-conjugates. Similarly, the sulphate-conjugates excreted via bile provide substrate for the SRB that are inversely associated with transit time.92 Although the relationship between transit time and the enterohepatic circulation is still not well understood, changes in transit time may alter the bile acid pool and affect enterohepatic circulation, which could have implications for both the resident gut microbes and human metabolism.
Gut microbial metabolism impacting gut transit time
Evidently, the gut transit time is a major driver of heterogeneity of the intestinal microbial community and either directly or indirectly impacts host–microbial cometabolism.124 Adding to the complexity, there is evidence that the interaction between the microbiota and the intestinal transit time is bidirectional, as the presence of microbes and their excreted compounds may affect gut motility (figure 3).146 147 Germ-free mice exhibit impaired peristalsis in the GIT, which is restored by colonisation of the gut.148 It has also been shown that intestinal peristalsis and colonic serotonin levels were decreased in mice that received faecal microbiota from patients with constipation.149 Moreover, administration of some probiotics has been shown to improve constipation symptoms, suggesting that presence of certain species might change gut transit.150 151 One of the possible mechanisms by which microbes impact gut transit is through the host cells’ recognition of bacterial molecular components by the toll-like receptors, which mediate interactions between the microbiota and the enteric neuromuscular apparatus. For example, lipopolysaccharides from the outer membrane of gram-negative bacteria impair intestinal contractility by activating oxidative stress in the mucosa.152 Other mechanisms may involve the microbial-derived metabolites such as SCFA, neurotransmitter homologs and gases, which can act on the enteric neuromuscular apparatus.153 SCFA can bind to G-protein coupled receptors for free fatty acids (GPR41, GPR43, OLFR78, GPR109A)154 and consequently stimulate the release of serotonin (5-hydroxytryptamine, 5-HT) from endocrine cells present in the colonic epithelium, a process which promotes the peristalsis via the enteric nervous system.155 However, the presence of SCFA in the gut lumen also stimulates the release of gut hormones, for example, PYY156 that may slow down gastrointestinal transit.157 Butyrate and acetate may also affect the GIT motility through smooth muscle and myenteric neuron activation.158 159 Moreover, absorption of SCFA in the colon is linked to fluid and electrolyte uptake,160 which, if disrupted, can lead to altered CTT.124 Recent evidence shows that secondary bile acids regulate CTT in mice with lithocholic acid inducing faster transit.161 One of the mechanisms by which bile acids affect the gut transit time is via the activation of the G protein-coupled bile acid receptor 1 (TGR5) leading to increased colonic motility.162 Furthermore, bile acids, both the host-derived and the microbiome-modified, can act as signalling molecules on the Farnesoid X receptor (FXR) and TGR5 receptor, which are expressed not only on epithelial cells throughout the GIT, but also outside of the GIT.163 164 Through these receptors, bile acids also act as regulators of lipid, glucose and energy metabolism.165–167 Furthermore, tryptophan catabolites (eg, tryptamine, indoleacetic acid, indolelactic acid or indolealdehyde)168–171 may affect intestinal motility via activation of the aryl hydrocarbon receptor.172 Tryptamine can activate the serotonin receptor-4 (5-HT4R)173 thereby accelerating the gut transit. Histamine, produced by M. morganii and L. reuteri from histidine, has also been observed to increase colonic motility in monocolonised mice.174 Finally, the gases H2 and CH4 are known to exert effects on the intestinal muscle contractile activity thereby affecting the gut transit time in animal models.175 While the infusion of H2 into the colon of guinea pigs shortened CTT, the inverse has been shown for CH4.176 Altogether, the gut microbiota can modulate gastrointestinal motility via production of small molecules interacting with the host-receptors on enteroendocrine cells and other cell types such as enteric neurons.172
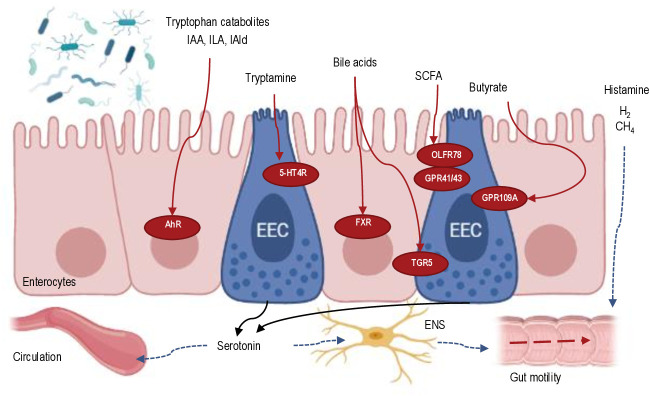
Figure 3.
Schematic overview of microbial-derived signalling metabolites in the intestinal epithelium and their effect on gut motility. Microbial-derived metabolites interact with various metabolite receptors expressed on enterocytes or enteroendocrine cells (EEC) and stimulate serotonin secretion from the EEC cells. The released serotonin activates the enteric neurons that promote gut motility. Other metabolites (eg, histamine) can modulate gut motility via other mechanisms. 5-HT4R, serotonin receptor-4; AhR, aryl hydrocarbon receptor; FXR, Farnesoid X receptor; IAA, indoleacetic acid; IAld, indolealdehyde; ILA, indolelactic acid; SCFA, short-chain fatty acids.Created with Biorender.com.
The role of gut transit time in health and disease
Sustained prolonged or shortened transit time could have consequences for host health due to the effects of gut microbial composition and metabolism.177–179 Here, we discuss several areas in which transit time may play a vital role.
Gastrointestinal diseases
Gastrointestinal diseases such as constipation or IBS are highly prevalent worldwide. According to a recent large-scale study, more than 40% of persons worldwide suffer from at least one functional gastrointestinal disorder,180 which are disorders related to any combination of motility disturbance, visceral hypersensitivity, altered mucosal, and immune function, altered gut microbiota and altered central nervous system processing.181 Here, we focus on gastrointestinal diseases that exhibit altered transit time.
Slow transit through the small bowel may result in the overgrowth of bacteria in the small intestine, a condition known as small intestinal bacterial overgrowth (SIBO). Patients with SIBO typically have high bacterial densities in the small intestine (>105 colony forming unit (CFU)/mL) due to impaired peristalsis and insufficient washout of the bacterial mass into the colon.182 Moreover, slow transit through the small intestine provides a longer time for absorption of chyme resulting in reduced flow of the chyme into the colon, which in turn also slows down the transit rate through the colon.183 SIBO is prevalent among patients with IBS,184 a condition characterised by abdominal pain or discomfort and associated with changes in bowel habits affecting more than a tenth of the general population.185 Patients with constipation-predominant IBS (IBS-C) exhibit prolonged CTT when compared with healthy controls in all regions of the colon.186 A recent study showed that IBS-C and diarrhoea-predominant IBS (IBS-D) patients have distinct faecal microbiome compositions and metabolomes. The microbiome of the patients was found to be classified according to predominant bowel habits, but not the severity of IBS symptoms.187 The microbiome differences observed between the two phenotypes of IBS are thus likely to be explained by differences in transit times,188 as similar compositional differences have been observed when comparing healthy individuals with long and short gut transit, respectively.11
Diarrhoea and/or constipation are also often experienced by patients with inflammatory bowel disease (IBD) including Crohn’s disease and ulcerative colitis, both characterised by inflammation in the gut.189 It has been observed that faecal levels of secondary bile acids in IBD patients were decreased during the flare episodes but not in the remission state.190 This may be explained by changes in CTT between the two states since flares are often accompanied by diarrhoea, which could limit the gut microbiota’s conversion of bile acids into secondary bile acids. Nonetheless, both faecal water content and inflammatory markers were needed to predict the microbial enterotypes in IBD patients.191
Both constipation192 and IBD189 are risk factors for the development of colon cancer, which is the third most common cause of cancer worldwide.193 Colon cancer is, among other lifestyle factors, associated with Western-type diets and prolonged gut transit time, both of which can lead to an altered bile acid pool.82 194 Secondary bile acids at high physiological concentrations, especially deoxycholic acid and lithocholic acid are toxic to colonic cells as they induce apoptosis and cause DNA damage.195–198 Notably, tumours often occur in the distal part of the colon,199 where fermentation of complex carbohydrates is less active and the microbiota is switching to proteolysis.123 200 Long CTT192 201 as well as lack of fermentable dietary fibres202 may lead to enhanced proteolysis in the colon, which potentially could play a role in the pathophysiology of colon cancer.203 Conclusive results for the latter are scarce, but the interplay between CTT, diet and gut microbiome could be key in the prevention and management of gastrointestinal diseases.
Diseases beyond the gut
Constipation and altered bowel habits have also been associated with neurological and metabolic diseases, and the use of several medications.61 In Parkinson’s disease, constipation affects up to 80% of the patients and often precedes the onset of motor symptoms by years.204 A recent meta-analysis on gut microbiota in Parkinson’s patients has shown higher species richness, an increase in relative abundances of the genera Akkermansia and Methanobrevibacter as well as the family Christensenellaceae, depletion of butyrate producers, and low faecal SCFA when compared with healthy controls.205 These changes are very similar to the associations seen between transit time and gut microbiota composition and metabolism in healthy individuals.9 11 12 17 Therefore, the observed microbiome differences between Parkinson’s patients and healthy controls are likely to be confounded by differences in transit time. Similar to Parkinson’s disease, constipation is a common complication among patients with Alzheimer’s disease78 and multiple sclerosis206. Investigations of microbial compositional changes in these patients207 208 could therefore be confounded by an altered gut transit time as well.
Delayed gastric emptying, as well as episodes of constipation and diarrhoea, have been reported for patients with both type 1 and type 2 diabetes mellitus.209–211 Although the changes in the gut motility of these patients may be a consequence of their treatment (e.g. metformin).66 In obesity, accelerated gastric emptying and changes in the transit in the small intestine, as well as both constipation and diarrhoea, have been reported.212 213 Altered gut motility affects the time for nutrient absorption and may contribute to changes in hormonal responses and glucose homeostasis.107 213 A recent cohort study has shown associations between stool frequency and vascular and non-vascular diseases in a Chinese population.214 The authors found that ‘less than three stools per week’ were associated with a higher risk of ischaemic heart disease and chronic kidney disease, further suggesting a link between bowel habits and health.214 These findings together suggest that gut transit time can confound investigations of microbiota composition when comparing patient groups. Whether altered transit time and bowel habits play a role in the early onset and development of diseases remains unknown.
Concluding remarks and future perspectives
Taken together, there is convincing evidence that gut transit time varies not only between healthy individuals but also within subjects from day-to-day and that many diseases are associated with altered gut transit time. Changes in gut transit time have been associated with changes in faecal pH, faecal microbial load and composition but most importantly with diet–microbe interactions and microbial metabolism including shifts from saccharolytic to proteolytic fermentation. Since microbial-derived metabolites are important regulators of host physiology, gut transit time is likely to play a key role in host health. Although gut transit time remains largely overlooked in many gut microbiome studies, an increasing number of human studies have included WGTT or segmental transit time (SITT or CTT) and evaluated its impact on microbial composition and other target outcomes confirming the importance of this factor. By including gut transit time measurements in gut microbiome-related studies, we can advance our understanding of the links between the gut microbiome, diet and disease. Such insights may be key for the prevention, diagnosis and treatment of several diseases in the gut and beyond throughout the lifespan.
Footnotes
Twitter: @nicolaproch, @hroager
Contributors: NP wrote the manuscript. NP and HMR conceptualised the manuscript. All authors critically revised the manuscript. All authors approved the final version of the manuscript.
Funding: The work was supported by the Novo Nordisk Foundation (PRIMA; NNF19OC0056246). In addition, HMR was supported by the Sapere Aude: DFF-Starting Grant (MOTILITY; 0171-00006B) from the Independent Research Fund Denmark.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Sekirov I, Russell SL, Antunes LCM, et al. Gut microbiota in health and disease. Physiol Rev 2010;90:859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 2. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mueller S, Saunier K, Hanisch C, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 2006;72:1027–33. 10.1128/AEM.72.2.1027-1033.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–6. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manor O, Dai CL, Kornilov SA, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun 2020;11:5206. 10.1038/s41467-020-18871-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tropini C. How the physical environment shapes the microbiota. mSystems 2021;6:e0067521. 10.1128/mSystems.00675-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falony G, Joossens M, Vieira-Silva S, et al. Population-Level analysis of gut microbiome variation. Science 2016;352:560–4. 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- 10. Roager HM, Hansen LBS, Bahl MI, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol 2016;1:16093. 10.1038/nmicrobiol.2016.93 [DOI] [PubMed] [Google Scholar]
- 11. Asnicar F, Leeming ER, Dimidi E, et al. Blue poo: impact of gut transit time on the gut microbiome using a novel marker. Gut 2021;70:1665–74. 10.1136/gutjnl-2020-323877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steenackers N, Falony G, Augustijns P, et al. Specific contributions of segmental transit times to gut microbiota composition. Gut 2022;71:1443–4. 10.1136/gutjnl-2021-325916 [DOI] [PubMed] [Google Scholar]
- 13. Mikolajczyk AE, Watson S, Surma BL, et al. Assessment of tandem measurements of pH and total gut transit time in healthy volunteers. Clin Transl Gastroenterol 2015;6:e100. 10.1038/ctg.2015.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mättö J, Maunuksela L, Kajander K, et al. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol 2005;43:213–22. 10.1016/j.femsim.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 15. Stephen AM, Wiggins HS, Cummings JH. Effect of changing transit time on colonic microbial metabolism in man. Gut 1987;28:601–9. 10.1136/gut.28.5.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vandeputte D, De Commer L, Tito RY, et al. Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat Commun 2021;12:6740. 10.1038/s41467-021-27098-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016;65:57–62. 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Müller M, Hermes GDA, Canfora EE, et al. Distal colonic transit is linked to gut microbiota diversity and microbial fermentation in humans with slow colonic transit. Am J Physiol Gastrointest Liver Physiol 2020;318:G361–9. 10.1152/ajpgi.00283.2019 [DOI] [PubMed] [Google Scholar]
- 19. Nandhra GK, Mark EB, Di Tanna GL, et al. Normative values for region-specific colonic and gastrointestinal transit times in 111 healthy volunteers using the 3D-Transit electromagnet tracking system: influence of age, gender, and body mass index. Neurogastroenterol Motil 2020;32:e13734. 10.1111/nmo.13734 [DOI] [PubMed] [Google Scholar]
- 20. Hellström PM, Grybäck P, Jacobsson H. The physiology of gastric emptying. Best Pract Res Clin Anaesthesiol 2006;20:397–407. 10.1016/j.bpa.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 21. Maurer AH. Gastrointestinal motility, part 2: small-bowel and colon transit. J Nucl Med Technol 2016;44:12–18. 10.2967/jnumed.113.134551 [DOI] [PubMed] [Google Scholar]
- 22. Camilleri M, Iturrino J, Bharucha AE, et al. Performance characteristics of scintigraphic measurement of gastric emptying of solids in healthy participants. Neurogastroenterol Motil 2012;24:1076–562. 10.1111/j.1365-2982.2012.01972.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang YT, Mohammed SD, Farmer AD, et al. Regional gastrointestinal transit and pH studied in 215 healthy volunteers using the wireless motility capsule: influence of age, gender, study country and testing protocol. Aliment Pharmacol Ther 2015;42:761–72. 10.1111/apt.13329 [DOI] [PubMed] [Google Scholar]
- 24. Miller MA, Parkman HP, Urbain JL, et al. Comparison of scintigraphy and lactulose breath hydrogen test for assessment of orocecal transit: lactulose accelerates small bowel transit. Dig Dis Sci 1997;42:10–18. 10.1023/a:1018864400566 [DOI] [PubMed] [Google Scholar]
- 25. Bouras EP, Burton DD, Camilleri M, et al. Effect of cyclooxygenase-2 inhibitors on gastric emptying and small intestinal transit in humans. Neurogastroenterol Motil 2004;16:729–35. 10.1111/j.1365-2982.2004.00547.x [DOI] [PubMed] [Google Scholar]
- 26. Martelli H, Devroede G, Arhan P, et al. Some parameters of large bowel motility in normal man. Gastroenterology 1978;75:612–8. [PubMed] [Google Scholar]
- 27. Haase AM, Gregersen T, Christensen LA, et al. Regional gastrointestinal transit times in severe ulcerative colitis. Neurogastroenterol Motil 2016;28:217–24. 10.1111/nmo.12713 [DOI] [PubMed] [Google Scholar]
- 28. Gregersen T, Haase A-M, Schlageter V, et al. Regional gastrointestinal transit times in patients with carcinoid diarrhea: assessment with the novel 3D-transit system. J Neurogastroenterol Motil 2015;21:423–32. 10.5056/jnm15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abrahamsson H, Antov S, Bosaeus I. Gastrointestinal and colonic segmental transit time evaluated by a single abdominal X-ray in healthy subjects and constipated patients. Scand J Gastroenterol Suppl 1988;152:72–80. 10.3109/00365528809095938 [DOI] [PubMed] [Google Scholar]
- 30. Cummings JH. Diet and transit through the gut. Journal of Plant Foods 1978;3:83–95. 10.1080/0142968X.1978.11904206 [DOI] [Google Scholar]
- 31. Vork L, Penders J, Jalanka J, et al. Does day-to-day variability in stool consistency link to the fecal microbiota composition? Front Cell Infect Microbiol 2021;11:639667. 10.3389/fcimb.2021.639667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diaz Tartera HO, Webb D-L, Al-Saffar AK, et al. Validation of SmartPill® wireless motility capsule for gastrointestinal transit time: Intra-subject variability, software accuracy and comparison with video capsule endoscopy. Neurogastroenterol Motil 2017;29:1–9. 10.1111/nmo.13107 [DOI] [PubMed] [Google Scholar]
- 33. Mariani G, Pauwels EKJ, AlSharif A, et al. Radionuclide evaluation of the lower gastrointestinal tract. J Nucl Med 2008;49:776–87. 10.2967/jnumed.107.040113 [DOI] [PubMed] [Google Scholar]
- 34. Saad RJ, Hasler WL. A technical review and clinical assessment of the wireless motility capsule. Gastroenterol Hepatol 2011;7:795–804. [PMC free article] [PubMed] [Google Scholar]
- 35. Taba Taba Vakili S, Nezami BG, Shetty A, et al. Association of high dietary saturated fat intake and uncontrolled diabetes with constipation: evidence from the National health and nutrition examination survey. Neurogastroenterol Motil 2015;27:1389–97. 10.1111/nmo.12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yancy WS, Olsen MK, Guyton JR, et al. A low-carbohydrate, ketogenic diet versus a low-fat diet to treat obesity and hyperlipidemia: a randomized, controlled trial. Ann Intern Med 2004;140:769–79. 10.7326/0003-4819-140-10-200405180-00006 [DOI] [PubMed] [Google Scholar]
- 37. vd Baan-Slootweg OH, Liem O, Bekkali N, et al. Constipation and colonic transit times in children with morbid obesity. J Pediatr Gastroenterol Nutr 2011;52:442–5. 10.1097/MPG.0b013e3181ef8e3c [DOI] [PubMed] [Google Scholar]
- 38. Heddle R, Collins PJ, Dent J, et al. Motor mechanisms associated with slowing of the gastric emptying of a solid meal by an intraduodenal lipid infusion. J Gastroenterol Hepatol 1989;4:437–47. 10.1111/j.1440-1746.1989.tb01741.x [DOI] [PubMed] [Google Scholar]
- 39. Boyd KA, O'Donovan DG, Doran S, et al. High-Fat diet effects on gut motility, hormone, and appetite responses to duodenal lipid in healthy men. Am J Physiol Gastrointest Liver Physiol 2003;284:G188–96. 10.1152/ajpgi.00375.2002 [DOI] [PubMed] [Google Scholar]
- 40. Stephen AM, Cummings JH. Mechanism of action of dietary fibre in the human colon. Nature 1980;284:283–4. 10.1038/284283a0 [DOI] [PubMed] [Google Scholar]
- 41. So D, Gibson PR, Muir JG, et al. Dietary fibres and IBS: translating functional characteristics to clinical value in the era of personalised medicine. Gut 2021;70:2383–94. 10.1136/gutjnl-2021-324891 [DOI] [PubMed] [Google Scholar]
- 42. Gill SK, Rossi M, Bajka B, et al. Dietary fibre in gastrointestinal health and disease. Nat Rev Gastroenterol Hepatol 2021;18:101–16. 10.1038/s41575-020-00375-4 [DOI] [PubMed] [Google Scholar]
- 43. Stevens J, Vansoest PJ, Robertson JB, et al. Comparison of the effects of psyllium and wheat bran on gastrointestinal transit time and stool characteristics. J Am Diet Assoc 1988;88:323–6. 10.1016/S0002-8223(21)01971-4 [DOI] [PubMed] [Google Scholar]
- 44. Major G, Murray K, Singh G, et al. Demonstration of differences in colonic volumes, transit, chyme consistency, and response to psyllium between healthy and constipated subjects using magnetic resonance imaging. Neurogastroenterol Motil 2018;30:e13400. 10.1111/nmo.13400 [DOI] [PubMed] [Google Scholar]
- 45. Tomlin J, Read NW. Laxative properties of indigestible plastic particles. BMJ 1988;297:1175–6. 10.1136/bmj.297.6657.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Müller M, Hermes GDA, Emanuel E C, et al. Effect of wheat bran derived prebiotic supplementation on gastrointestinal transit, gut microbiota, and metabolic health: a randomized controlled trial in healthy adults with a slow gut transit. Gut Microbes 2020;12:1704141. 10.1080/19490976.2019.1704141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stephen AM, Cummings JH. Water-Holding by dietary fibre in vitro and its relationship to faecal output in man. Gut 1979;20:722–9. 10.1136/gut.20.8.722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hillman L, Peters S, Fisher A, et al. Differing effects of pectin, cellulose and lignin on stool pH, transit time and weight. Br J Nutr 1983;50:189–95. 10.1079/bjn19830088 [DOI] [PubMed] [Google Scholar]
- 49. Vandeputte D, Falony G, Vieira-Silva S, et al. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 2017;66:1968–74. 10.1136/gutjnl-2016-313271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Vries J, Birkett A, Hulshof T, et al. Effects of cereal, fruit and vegetable fibers on human fecal weight and transit time: a comprehensive review of intervention trials. Nutrients 2016;8:130. 10.3390/nu8030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Vries J, Miller PE, Verbeke K. Effects of cereal fiber on bowel function: a systematic review of intervention trials. World J Gastroenterol 2015;21:8952–63. 10.3748/wjg.v21.i29.8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anti M, Pignataro G, Armuzzi A, et al. Water supplementation enhances the effect of high-fiber diet on stool frequency and laxative consumption in adult patients with functional constipation. Hepatogastroenterology 1998;45:727–32. [PubMed] [Google Scholar]
- 53. Boilesen SN, Tahan S, Dias FC, et al. Water and fluid intake in the prevention and treatment of functional constipation in children and adolescents: is there evidence? J Pediatr 2017;93:320–7. 10.1016/j.jped.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 54. Schultz J. Carboxymethylcellulose as a colloid laxative. Am J Dig Dis 1949;16:319–22. 10.1007/BF03002733 [DOI] [PubMed] [Google Scholar]
- 55. Chassaing B, Compher C, Bonhomme B, et al. Randomized Controlled-Feeding study of dietary emulsifier carboxymethylcellulose reveals detrimental impacts on the gut microbiota and metabolome. Gastroenterology 2022;162:743–56. 10.1053/j.gastro.2021.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Katsirma Z, Dimidi E, Rodriguez-Mateos A, et al. Fruits and their impact on the gut microbiota, gut motility and constipation. Food Funct 2021;12:8850–66. 10.1039/d1fo01125a [DOI] [PubMed] [Google Scholar]
- 57. Lu Y. Humectancies of D-tagatose and D-sorbitol. Int J Cosmet Sci 2001;23:175–81. 10.1046/j.1467-2494.2001.00084.x [DOI] [PubMed] [Google Scholar]
- 58. Dey N, Wagner VE, Blanton LV, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell 2015;163:95–107. 10.1016/j.cell.2015.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Degen LP, Phillips SF. Variability of gastrointestinal transit in healthy women and men. Gut 1996;39:299–305. 10.1136/gut.39.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lampe JW, Fredstrom SB, Slavin JL, et al. Sex differences in colonic function: a randomised trial. Gut 1993;34:531–6. 10.1136/gut.34.4.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mugie SM, Benninga MA, Di Lorenzo C. Epidemiology of constipation in children and adults: a systematic review. Best Pract Res Clin Gastroenterol 2011;25:3–18. 10.1016/j.bpg.2010.12.010 [DOI] [PubMed] [Google Scholar]
- 62. Hounnou G, Destrieux C, Desmé J, et al. Anatomical study of the length of the human intestine. Surg Radiol Anat 2002;24:290–4. 10.1007/s00276-002-0057-y [DOI] [PubMed] [Google Scholar]
- 63. Graff J, Brinch K, Madsen JL. Gastrointestinal mean transit times in young and middle-aged healthy subjects. Clin Physiol 2001;21:253–9. 10.1046/j.1365-2281.2001.00308.x [DOI] [PubMed] [Google Scholar]
- 64. Madsen JL, Graff J. Effects of ageing on gastrointestinal motor function. Age Ageing 2004;33:154–9. 10.1093/ageing/afh040 [DOI] [PubMed] [Google Scholar]
- 65. Bingham SA, Cummings JH. Effect of exercise and physical fitness on large intestinal function. Gastroenterology 1989;97:1389–99. 10.1016/0016-5085(89)90381-8 [DOI] [PubMed] [Google Scholar]
- 66. Fosnes GS, Lydersen S, Farup PG. Constipation and diarrhoea - common adverse drug reactions? A cross sectional study in the general population. BMC Clin Pharmacol 2011;11:2. 10.1186/1472-6904-11-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bonfiglio F, Liu X, Smillie C, et al. Gwas of stool frequency provides insights into gastrointestinal motility and irritable bowel syndrome. Cell Genom 2021;1:100069. 10.1016/j.xgen.2021.100069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vujkovic-Cvijin I, Sklar J, Jiang L, et al. Host variables confound gut microbiota studies of human disease. Nature 2020;587:448–54. 10.1038/s41586-020-2881-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 1997;32:920–4. 10.3109/00365529709011203 [DOI] [PubMed] [Google Scholar]
- 70. McDonald D, Hyde E, Debelius JW, et al. American gut: an open platform for citizen science microbiome research. mSystems 2018;3:e00031–18. 10.1128/mSystems.00031-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 72. Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–30. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Falony G, Vieira-Silva S, Raes J. Richness and ecosystem development across faecal snapshots of the gut microbiota. Nat Microbiol 2018;3:526–8. 10.1038/s41564-018-0143-5 [DOI] [PubMed] [Google Scholar]
- 74. Zhu L, Liu W, Alkhouri R, et al. Structural changes in the gut microbiome of constipated patients. Physiol Genomics 2014;46:679–86. 10.1152/physiolgenomics.00082.2014 [DOI] [PubMed] [Google Scholar]
- 75. Adamberg K, Jaagura M, Aaspõllu A, et al. The composition of faecal microbiota is related to the amount and variety of dietary fibres. Int J Food Sci Nutr 2020;71:845–55. 10.1080/09637486.2020.1727864 [DOI] [PubMed] [Google Scholar]
- 76. Vieira-Silva S, Falony G, Darzi Y, et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat Microbiol 2016;1:16088. 10.1038/nmicrobiol.2016.88 [DOI] [PubMed] [Google Scholar]
- 77. Scheperjans F, Aho V, Pereira PAB, et al. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord 2015;30:350–8. 10.1002/mds.26069 [DOI] [PubMed] [Google Scholar]
- 78. Fu P, Gao M, Yung KKL. Association of intestinal disorders with Parkinson's disease and Alzheimer's disease: a systematic review and meta-analysis. ACS Chem Neurosci 2020;11:395–405. 10.1021/acschemneuro.9b00607 [DOI] [PubMed] [Google Scholar]
- 79. Vandeputte D, Kathagen G, D'hoe K, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017;551:507–11. 10.1038/nature24460 [DOI] [PubMed] [Google Scholar]
- 80. Abbas A, Wilding G, Semler J, et al. Does colonic transit time affect colonic pH? Am J Gastroenterol 2010;105:S123. 10.14309/00000434-201010001-00333 [DOI] [Google Scholar]
- 81. Lewis SJ, Heaton KW. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut 1997;41:245–51. 10.1136/gut.41.2.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Thomas LA, Veysey MJ, Bathgate T, et al. Mechanism for the transit-induced increase in colonic deoxycholic acid formation in cholesterol cholelithiasis. Gastroenterology 2000;119:806–15. 10.1053/gast.2000.16495 [DOI] [PubMed] [Google Scholar]
- 83. Raba G, Adamberg S, Adamberg K. Acidic pH enhances butyrate production from pectin by faecal microbiota. FEMS Microbiol Lett 2021;368:1–8. 10.1093/femsle/fnab042 [DOI] [PubMed] [Google Scholar]
- 84. Yasuda K, Oh K, Ren B, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 2015;17:385–91. 10.1016/j.chom.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gu S, Chen D, Zhang J-N, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 2013;8:e74957. 10.1371/journal.pone.0074957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sommer F, Bäckhed F. Know your neighbor: microbiota and host epithelial cells interact locally to control intestinal function and physiology. Bioessays 2016;38:455–64. 10.1002/bies.201500151 [DOI] [PubMed] [Google Scholar]
- 87. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016;14:20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parthasarathy G, Chen J, Chen X, et al. Relationship between microbiota of the colonic mucosa vs feces and symptoms, colonic transit, and methane production in female patients with chronic constipation. Gastroenterology 2016;150:367–79. 10.1053/j.gastro.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Koh A, De Vadder F, Kovatcheva-Datchary P, et al. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 90. Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 1991;70:443–59. 10.1111/j.1365-2672.1991.tb02739.x [DOI] [PubMed] [Google Scholar]
- 91. Tian H, Chen Q, Yang B, et al. Analysis of gut microbiome and metabolite characteristics in patients with slow transit constipation. Dig Dis Sci 2021;66:3026–35. 10.1007/s10620-020-06500-2 [DOI] [PubMed] [Google Scholar]
- 92. El Oufir L, Flourié B, Bruley des Varannes S, et al. Relations between transit time, fermentation products, and hydrogen consuming flora in healthy humans. Gut 1996;38:870–7. 10.1136/gut.38.6.870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jones J, Reinke SN, Ali A, et al. Fecal sample collection methods and time of day impact microbiome composition and short chain fatty acid concentrations. Sci Rep 2021;11:1–16. 10.1038/s41598-021-93031-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cirstea MS, Yu AC, Golz E, et al. Microbiota composition and metabolism are associated with gut function in Parkinson's disease. Mov Disord 2020;35:1208–17. 10.1002/mds.28052 [DOI] [PubMed] [Google Scholar]
- 95. Gabriele S, Sacco R, Altieri L. Slow intestinal transit contributes to elevate urinary p-cresol level in Italian autistic children. John Wiley & Sons, Ltd, 2016. [DOI] [PubMed] [Google Scholar]
- 96. Nakabayashi I, Nakamura M, Kawakami K, et al. Effects of synbiotic treatment on serum level of p-cresol in haemodialysis patients: a preliminary study. Nephrol Dial Transplant 2011;26:1094–8. 10.1093/ndt/gfq624 [DOI] [PubMed] [Google Scholar]
- 97. McElhanon BO, McCracken C, Karpen S, et al. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics 2014;133:872–83. 10.1542/peds.2013-3995 [DOI] [PubMed] [Google Scholar]
- 98. Cano AE, Neil AK, Kang J-Y, et al. Gastrointestinal symptoms in patients with end-stage renal disease undergoing treatment by hemodialysis or peritoneal dialysis. Am J Gastroenterol 2007;102:1990–7. 10.1111/j.1572-0241.2007.01321.x [DOI] [PubMed] [Google Scholar]
- 99. Guthrie L, Spencer SP, Perelman D. Clinical and translational report impact of a 7-day homogeneous diet on interper- sonal variation in human gut microbiomes and Me- tabolomes clinical and translational report impact of a 7-day homogeneous diet on interpersonal variation in human gut micro. Cell Host Microbe 2022:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Macy IG. Nutrition and chemical growth in Childhood—Vol. 1. evaluation. Am J Public Health Nations Health 1943;33:88. [Google Scholar]
- 101. Cummings JH, Hill MJ, Bone ES, et al. The effect of meat protein and dietary fiber on colonic function and metabolism. II. bacterial metabolites in feces and urine. Am J Clin Nutr 1979;32:2094–101. 10.1093/ajcn/32.10.2094 [DOI] [PubMed] [Google Scholar]
- 102. Lin HC, Visek WJ. Colon mucosal cell damage by ammonia in rats. J Nutr 1991;121:887–93. 10.1093/jn/121.6.887 [DOI] [PubMed] [Google Scholar]
- 103. Topping DC, Visek WJ. Nitrogen intake and tumorigenesis in rats injected with 1,2-dimethylhydrazine. J Nutr 1976;106:1583–90. 10.1093/jn/106.11.1583 [DOI] [PubMed] [Google Scholar]
- 104. Hughes R, Kurth MJ, McGilligan V, et al. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer 2008;60:259–66. 10.1080/01635580701649644 [DOI] [PubMed] [Google Scholar]
- 105. Roediger WE, Duncan A, Kapaniris O, et al. Reducing sulfur compounds of the colon impair colonocyte nutrition: implications for ulcerative colitis. Gastroenterology 1993;104:802–9. 10.1016/0016-5085(93)91016-b [DOI] [PubMed] [Google Scholar]
- 106. Desai MS, Seekatz AM, Koropatkin NM, et al. A dietary Fiber-Deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016;167:1339–53. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nestel N, Hvass JD, Bahl MI, et al. The gut microbiome and abiotic factors as potential determinants of postprandial glucose responses: a single-arm meal study. Front Nutr 2020;7:1–9. 10.3389/fnut.2020.594850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Blatchford P, Stoklosinski H, Eady S, et al. Consumption of kiwifruit capsules increases Faecalibacterium prausnitzii abundance in functionally constipated individuals: a randomised controlled human trial. J Nutr Sci 2017;6:1–10. 10.1017/jns.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bottacini F, Ventura M, van Sinderen D, et al. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact 2014;13 Suppl 1:S4. 10.1186/1475-2859-13-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bacteria SNP. Beneficial: Bifidobacterium spp.: Morphology and Physiology. In: Encyclopedia of dairy sciences. Second Edition, 2011: 381–7. [Google Scholar]
- 111. Evans D, Pye G, Opare-Sem P. Is colorectal cancer risk increasing in urban Ghanaians? Ghana Med J 1998;22:8–16. [Google Scholar]
- 112. Walker AR, Diet WAR. Diet, bowel motility, faeces composition and colonic cancer. S Afr Med J 1971;45:377–9. [PubMed] [Google Scholar]
- 113. Schäpe SS, Krause JL, Engelmann B, et al. The Simplified Human Intestinal Microbiota (SIHUMIx) Shows High Structural and Functional Resistance against Changing Transit Times in In Vitro Bioreactors. Microorganisms 2019;7:1–19. 10.3390/microorganisms7120641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr 2013;98:111–20. 10.3945/ajcn.112.056689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lamichhane S, Sen P, Alves MA, et al. Linking gut microbiome and lipid metabolism: moving beyond associations. Metabolites 2021;11:1–15. 10.3390/metabo11010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472:57–65. 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tropini C, Earle KA, Huang KC, et al. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 2017;21:433–42. 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. El Kaoutari A, Armougom F, Gordon JI, et al. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 2013;11:497–504. 10.1038/nrmicro3050 [DOI] [PubMed] [Google Scholar]
- 119. Liu C, Finegold SM, Song Y, et al. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2008;58:1896–902. 10.1099/ijs.0.65208-0 [DOI] [PubMed] [Google Scholar]
- 120. Bernalier A, Willems A, Leclerc M. A new H 2 / CO 2 -utilizing acetogenic bacterium isolated from human feces. New York, 1996: 176–83. [DOI] [PubMed] [Google Scholar]
- 121. Nava GM, Carbonero F, Croix JA, et al. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. Isme J 2012;6:57–70. 10.1038/ismej.2011.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gibson GR, Cummings JH, Macfarlane GT, et al. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 1990;31:679–83. 10.1136/gut.31.6.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Macfarlane GT, Gibson GR, Cummings JH. Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 1992;72:57–64. 10.1111/j.1365-2672.1992.tb04882.x [DOI] [PubMed] [Google Scholar]
- 124. Krautkramer KA, Fan J, Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol 2021;19:77–94. 10.1038/s41579-020-0438-4 [DOI] [PubMed] [Google Scholar]
- 125. Fernández-Veledo S, Vendrell J. Gut microbiota-derived succinate: friend or foe in human metabolic diseases? Rev Endocr Metab Disord 2019;20:439–47. 10.1007/s11154-019-09513-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bjerrum JT, Wang Y, Hao F, et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics 2015;11:122–33. 10.1007/s11306-014-0677-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Vernia P, Caprilli R, Latella G, et al. Fecal lactate and ulcerative colitis. Gastroenterology 1988;95:1564–8. 10.1016/s0016-5085(88)80078-7 [DOI] [PubMed] [Google Scholar]
- 128. Nakamura N, Lin HC, McSweeney CS, et al. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Annu Rev Food Sci Technol 2010;1:363–95. 10.1146/annurev.food.102308.124101 [DOI] [PubMed] [Google Scholar]
- 129. Laverde Gomez JA, Mukhopadhya I, Duncan SH, et al. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol 2019;21:259–71. 10.1111/1462-2920.14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Samuel BS, Hansen EE, Manchester JK, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A 2007;104:10643–8. 10.1073/pnas.0704189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ridlon JM, Harris SC, Bhowmik S, et al. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016;7:22–39. 10.1080/19490976.2015.1127483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J Lipid Res 2015;56:1085–99. 10.1194/jlr.R054114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Marcus SN, Heaton KW. Effects of a new, concentrated wheat fibre preparation on intestinal transit, deoxycholic acid metabolism and the composition of bile. Gut 1986;27:893–900. 10.1136/gut.27.8.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lewis S, Cochrane S. Alteration of sulfate and hydrogen metabolism in the human colon by changing intestinal transit rate. Am J Gastroenterol 2007;102:624–33. 10.1111/j.1572-0241.2006.01020.x [DOI] [PubMed] [Google Scholar]
- 135. Van Erpecum KJ, Van Berge-Henegouwen GP. Gallstones: an intestinal disease? Gut 1999;44:435–8. 10.1136/gut.44.3.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Heaton KW, Emmett PM, Symes CL, et al. An explanation for gallstones in normal-weight women: slow intestinal transit. Lancet 1993;341:8–10. 10.1016/0140-6736(93)92479-d [DOI] [PubMed] [Google Scholar]
- 137. Stellwag EJ, Hylemon PB. 7alpha-Dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J Lipid Res 1979;20:325–33. 10.1016/S0022-2275(20)40615-7 [DOI] [PubMed] [Google Scholar]
- 138. Masuda N, Oda H, Hirano S, et al. 7 alpha-dehydroxylation of bile acids by resting cells of a Eubacterium lentum-like intestinal anaerobe, strain C-25. Appl Environ Microbiol 1984;47:735–9. 10.1128/aem.47.4.735-739.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lewis SJ, Heaton KW, Oakey RE, et al. Lower serum oestrogen concentrations associated with faster intestinal transit. Br J Cancer 1997;76:395–400. 10.1038/bjc.1997.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lewis SJ, Heaton KW. The metabolic consequences of slow colonic transit. Am J Gastroenterol 1999;94:2010–6. 10.1111/j.1572-0241.1999.01271.x [DOI] [PubMed] [Google Scholar]
- 141. Kaaks R, Rinaldi S, Key TJ, et al. Postmenopausal serum androgens, oestrogens and breast cancer risk: the European prospective investigation into cancer and nutrition. Endocr Relat Cancer 2005;12:1071–82. 10.1677/erc.1.01038 [DOI] [PubMed] [Google Scholar]
- 142. Maruti SS, Lampe JW, Potter JD, et al. A prospective study of bowel motility and related factors on breast cancer risk. Cancer Epidemiol Biomarkers Prev 2008;17:1746–50. 10.1158/1055-9965.EPI-07-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Humblot C, Murkovic M, Rigottier-Gois L, et al. Beta-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Carcinogenesis 2007;28:2419–25. 10.1093/carcin/bgm170 [DOI] [PubMed] [Google Scholar]
- 144. Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006;47:241–59. 10.1194/jlr.R500013-JLR200 [DOI] [PubMed] [Google Scholar]
- 145. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014;12:661–72. 10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- 146. Quigley EMM. Microflora modulation of motility. J Neurogastroenterol Motil 2011;17:140–7. 10.5056/jnm.2011.17.2.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Barbara G, Stanghellini V, Brandi G, et al. Interactions between commensal bacteria and gut sensorimotor function in health and disease. Am J Gastroenterol 2005;100:2560–8. 10.1111/j.1572-0241.2005.00230.x [DOI] [PubMed] [Google Scholar]
- 148. Husebye E, Hellström PM, Midtvedt T. Intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of migrating myoelectric complex. Dig Dis Sci 1994;39:946–56. 10.1007/BF02087542 [DOI] [PubMed] [Google Scholar]
- 149. Cao H, Liu X, An Y, et al. Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci Rep 2017;7:10322. 10.1038/s41598-017-10835-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Dimidi E, Christodoulides S, Scott SM, et al. Mechanisms of action of probiotics and the gastrointestinal microbiota on gut motility and constipation. Adv Nutr 2017;8:484–94. 10.3945/an.116.014407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhang C, Jiang J, Tian F, et al. Meta-Analysis of randomized controlled trials of the effects of probiotics on functional constipation in adults. Clin Nutr 2020;39:2960–9. 10.1016/j.clnu.2020.01.005 [DOI] [PubMed] [Google Scholar]
- 152. Matarrese P, Petitta C, Scirocco A, et al. Antioxidants counteract lipopolysaccharide-triggered alterations of human colonic smooth muscle cells. Free Radic Biol Med 2012;53:2102–11. 10.1016/j.freeradbiomed.2012.09.022 [DOI] [PubMed] [Google Scholar]
- 153. Mayer EA, Tillisch K, Gupta A. Gut/Brain axis and the microbiota. J Clin Invest 2015;125:926–38. 10.1172/JCI76304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Pryde SE, Duncan SH, Hold GL, et al. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 2002;217:133–9. 10.1111/j.1574-6968.2002.tb11467.x [DOI] [PubMed] [Google Scholar]
- 155. Soret R, Chevalier J, De Coppet P, et al. Short-Chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 2010;138:1772–82. 10.1053/j.gastro.2010.01.053 [DOI] [PubMed] [Google Scholar]
- 156. Larraufie P, Martin-Gallausiaux C, Lapaque N, et al. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep 2018;8:74. 10.1038/s41598-017-18259-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. El-Salhy M, Hatlebakk JG, Hausken T. Possible role of peptide YY (PYY) in the pathophysiology of irritable bowel syndrome (IBS). Neuropeptides 2020;79:101973. 10.1016/j.npep.2019.101973 [DOI] [PubMed] [Google Scholar]
- 158. Haschke G, Schäfer H, Diener M. Effect of butyrate on membrane potential, ionic currents and intracellular Ca2+ concentration in cultured rat myenteric neurones. Neurogastroenterol Motil 2002;14:133–42. 10.1046/j.1365-2982.2002.00312.x [DOI] [PubMed] [Google Scholar]
- 159. Guarino MPL, Cicala M, Putignani L, et al. Gastrointestinal neuromuscular apparatus: an underestimated target of gut microbiota. World J Gastroenterol 2016;22:9871–9. 10.3748/wjg.v22.i45.9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tazoe H, Otomo Y, Kaji I, et al. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol 2008;59 Suppl 2:251–62. [PubMed] [Google Scholar]
- 161. Li N, Koester ST, Lachance DM, et al. Microbiome-encoded bile acid metabolism modulates colonic transit times. iScience 2021;24:102508. 10.1016/j.isci.2021.102508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Alemi F, Poole DP, Chiu J, et al. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 2013;144:145–54. 10.1053/j.gastro.2012.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kast HR, Nguyen CM, Sinal CJ, et al. Farnesoid X-activated receptor induces apolipoprotein C-II transcription: a molecular mechanism linking plasma triglyceride levels to bile acids. Mol Endocrinol 2001;15:1720–8. 10.1210/mend.15.10.0712 [DOI] [PubMed] [Google Scholar]
- 164. Cariou B, van Harmelen K, Duran-Sandoval D, et al. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem 2006;281:11039–49. 10.1074/jbc.M510258200 [DOI] [PubMed] [Google Scholar]
- 165. Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun 2005;329:386–90. 10.1016/j.bbrc.2005.01.139 [DOI] [PubMed] [Google Scholar]
- 166. Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006;439:484–9. 10.1038/nature04330 [DOI] [PubMed] [Google Scholar]
- 167. Sepe V, Festa C, Renga B, et al. Insights on FXR selective modulation. speculation on bile acid chemical space in the discovery of potent and selective agonists. Sci Rep 2016;6:19008. 10.1038/srep19008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hubbard TD, Murray IA, Bisson WH, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep 2015;5:1–13. 10.1038/srep12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Cheng Y, Jin U-H, Allred CD, et al. Aryl hydrocarbon receptor activity of tryptophan metabolites in young adult mouse colonocytes. Drug Metab Dispos 2015;43:1536–43. 10.1124/dmd.115.063677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Cervantes-Barragan L, Chai JN, Tianero MD. Lactobacillus reuteri induces gut intraepithelial CD4 + CD8αα + T cells. Science 2017;357:806–10. 10.1126/science.aah5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013;39:372–85. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 172. Obata Y, Castaño Álvaro, Boeing S, et al. Neuronal programming by microbiota regulates intestinal physiology. Nature 2020;578:284–9. 10.1038/s41586-020-1975-8 [DOI] [PubMed] [Google Scholar]
- 173. Bhattarai Y, Schmidt BA, Linden DR, et al. Human-derived gut microbiota modulates colonic secretion in mice by regulating 5-HT3 receptor expression via acetate production. Am J Physiol Gastrointest Liver Physiol 2017;313:G80–7. 10.1152/ajpgi.00448.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Chen H, Nwe P-K, Yang Y, et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 2019;177:1217–31. 10.1016/j.cell.2019.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Pimentel M, Lin HC, Enayati P, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol 2006;290:1089–95. 10.1152/ajpgi.00574.2004 [DOI] [PubMed] [Google Scholar]
- 176. Jahng J, Jung IS, Choi EJ, et al. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol Motil 2012;24:185–91. 10.1111/j.1365-2982.2011.01819.x [DOI] [PubMed] [Google Scholar]
- 177. Byndloss MX, Pernitzsch SR, Bäumler AJ. Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol 2018;11:1299–305. 10.1038/s41385-018-0010-y [DOI] [PubMed] [Google Scholar]
- 178. Litvak Y, Byndloss MX, Tsolis RM, et al. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol 2017;39:1–6. 10.1016/j.mib.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 179. Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science 2018;362:362. 10.1126/science.aat9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Sperber AD, Bangdiwala SI, Drossman DA, et al. Worldwide prevalence and burden of functional gastrointestinal disorders, results of Rome Foundation global study. Gastroenterology 2021;160:99–114. 10.1053/j.gastro.2020.04.014 [DOI] [PubMed] [Google Scholar]
- 181. Drossman DA. Functional gastrointestinal disorders: history, pathophysiology, clinical features and Rome IV. Gastroenterology 2016;150:1262–79. 10.1053/j.gastro.2016.02.032 [DOI] [PubMed] [Google Scholar]
- 182. Maneerattanaporn M, Chey WD. Small intestinal bacterial overgrowth. In: Practical gastroenterology and hepatology: small and large intestine and pancreas. 3, 2010: 244–51. [Google Scholar]
- 183. Roland BC, Ciarleglio MM, Clarke JO, et al. Small intestinal transit time is delayed in small intestinal bacterial overgrowth. J Clin Gastroenterol 2015;49:571–6. 10.1097/MCG.0000000000000257 [DOI] [PubMed] [Google Scholar]
- 184. Chen B, Kim JJ-W, Zhang Y, et al. Prevalence and predictors of small intestinal bacterial overgrowth in irritable bowel syndrome: a systematic review and meta-analysis. J Gastroenterol 2018;53:807–18. 10.1007/s00535-018-1476-9 [DOI] [PubMed] [Google Scholar]
- 185. Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol 2012;10:712–21. 10.1016/j.cgh.2012.02.029 [DOI] [PubMed] [Google Scholar]
- 186. Raahave D, Jensen AK. Increased colon transit time and faecal load in irritable bowel syndrome. World J Gastrointest Pharmacol Ther 2021;12:13–20. 10.4292/wjgpt.v12.i1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Mars RAT, Yang Y, Ward T, et al. Longitudinal multi-omics reveals Subset-Specific mechanisms underlying irritable bowel syndrome. Cell 2020;182:1460–73. 10.1016/j.cell.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Ahluwalia B, Iribarren C, Magnusson MK, et al. A distinct faecal microbiota and metabolite profile linked to bowel habits in patients with irritable bowel syndrome. Cells 2021;10:1459–16. 10.3390/cells10061459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Fakhoury M, Negrulj R, Mooranian A, et al. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res 2014;7:113–20. 10.2147/JIR.S65979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013;62:531–9. 10.1136/gutjnl-2012-302578 [DOI] [PubMed] [Google Scholar]
- 191. Vieira-Silva S, Sabino J, Valles-Colomer M, et al. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol 2019;4:1826–31. 10.1038/s41564-019-0483-9 [DOI] [PubMed] [Google Scholar]
- 192. Guérin A, Mody R, Fok B, et al. Risk of developing colorectal cancer and benign colorectal neoplasm in patients with chronic constipation. Aliment Pharmacol Ther 2014;40:83–92. 10.1111/apt.12789 [DOI] [PubMed] [Google Scholar]
- 193. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 194. Yokota A, Fukiya S, Islam KBMS, et al. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 2012;3:455–9. 10.4161/gmic.21216 [DOI] [PubMed] [Google Scholar]
- 195. Farhana L, Nangia-Makker P, Arbit E, et al. Bile acid: a potential inducer of colon cancer stem cells. Stem Cell Res Ther 2016;7:1–10. 10.1186/s13287-016-0439-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol 2014;12:1–5. 10.1186/1477-7819-12-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Diether N, Willing B. Microbial fermentation of dietary protein: an important factor in diet–microbe–host interaction. Microorganisms 2019;7:19. 10.3390/microorganisms7010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Hughes R, Magee EA, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr Issues Intest Microbiol 2000;1:51–8. [PubMed] [Google Scholar]
- 199. Rabeneck L, Davila JA, El-Serag HB. Is there a true "shift" to the right colon in the incidence of colorectal cancer? Am J Gastroenterol 2003;98:1400–9. 10.1111/j.1572-0241.2003.07453.x [DOI] [PubMed] [Google Scholar]
- 200. Macfarlane GT, Gibson GR, Beatty E, et al. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol Lett 1992;101:81–8. 10.1111/j.1574-6968.1992.tb05764.x [DOI] [Google Scholar]
- 201. Kojima M, Wakai K, Tokudome S, et al. Bowel movement frequency and risk of colorectal cancer in a large cohort study of Japanese men and women. Br J Cancer 2004;90:1397–401. 10.1038/sj.bjc.6601735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Burkitt DP, Walker AR, Painter NS. Effect of dietary fibre on stools and the transit-times, and its role in the causation of disease. Lancet 1972;2:1408–11. 10.1016/S0140-6736(72)92974-1 [DOI] [PubMed] [Google Scholar]
- 203. Zhang W, An Y, Qin X, et al. Gut Microbiota-Derived metabolites in colorectal cancer: the bad and the challenges. Front Oncol 2021;11:4287. 10.3389/fonc.2021.739648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Fasano A, Visanji NP, Liu LWC, et al. Gastrointestinal dysfunction in Parkinson's disease. Lancet Neurol 2015;14:625–39. 10.1016/S1474-4422(15)00007-1 [DOI] [PubMed] [Google Scholar]
- 205. Romano S, Savva GM, Bedarf JR, et al. Meta-Analysis of the Parkinson's disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis 2021;7:27. 10.1038/s41531-021-00156-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Hinds JP, Eidelman BH, Wald A. Prevalence of bowel dysfunction in multiple sclerosis. A population survey. Gastroenterology 1990;98:1538–42. 10.1016/0016-5085(90)91087-m [DOI] [PubMed] [Google Scholar]
- 207. Vogt NM, Kerby RL, Dill-McFarland KA, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep 2017;7:1–11. 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 2016;6:1–10. 10.1038/srep28484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Horowitz M, Harding PE, Maddox AF, et al. Gastric and oesophageal emptying in patients with type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia 1989;32:151–9. 10.1007/BF00265086 [DOI] [PubMed] [Google Scholar]
- 210. Coleski R, Wilding GE, Semler JR, et al. Blunting of colon contractions in diabetics with gastroparesis quantified by wireless motility capsule methods. PLoS One 2015;10:1–15. 10.1371/journal.pone.0141183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Farmer AD, Pedersen AG, Brock B, et al. Type 1 diabetic patients with peripheral neuropathy have pan-enteric prolongation of gastrointestinal transit times and an altered caecal pH profile. Diabetologia 2017;60:709–18. 10.1007/s00125-016-4199-6 [DOI] [PubMed] [Google Scholar]
- 212. Steenackers N, Wauters L, Van der Schueren B, et al. Effect of obesity on gastrointestinal transit, pressure and pH using a wireless motility capsule. Eur J Pharm Biopharm 2021;167:1–8. 10.1016/j.ejpb.2021.07.002 [DOI] [PubMed] [Google Scholar]
- 213. Mushref MA, Srinivasan S. Effect of high fat-diet and obesity on gastrointestinal motility. Ann Transl Med 2013;1:14. 10.3978/j.issn.2305-5839.2012.11.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Yang S, Yu C, Guo Y, et al. Bowel movement frequency and risks of major vascular and non-vascular diseases: a population-based cohort study among Chinese adults. BMJ Open 2020;10:e031028. 10.1136/bmjopen-2019-031028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Liu Y, Jin Y, Li J, et al. Small bowel transit and altered gut microbiota in patients with liver cirrhosis. Front Physiol 2018;9:470. 10.3389/fphys.2018.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Ringel-Kulka T, Choi CH, Temas D, et al. Altered colonic bacterial fermentation as a potential pathophysiological factor in irritable bowel syndrome. Am J Gastroenterol 2015;110:1339–46. 10.1038/ajg.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Kalantar-Zadeh K, Berean KJ, Ha N, et al. A human pilot trial of ingestible electronic capsules capable of sensing different gases in the gut. Nat Electron 2018;1:79–87. 10.1038/s41928-017-0004-x [DOI] [Google Scholar]
- 218. Hadizadeh F, Walter S, Belheouane M, et al. Stool frequency is associated with gut microbiota composition. Gut 2017;66:559–60. 10.1136/gutjnl-2016-311935 [DOI] [PubMed] [Google Scholar]
- 219. Kwon HJ, Lim JH, Kang D, et al. Is stool frequency associated with the richness and community composition of gut microbiota? Intest Res 2019;17:419–26. 10.5217/ir.2018.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Tigchelaar EF, Bonder MJ, Jankipersadsing SA, et al. Gut microbiota composition associated with stool consistency. Gut 2016;65:540–2. 10.1136/gutjnl-2015-310328 [DOI] [PubMed] [Google Scholar]
- 221. Deutsch L, Stres B. The importance of objective stool classification in fecal 1H-NMR metabolomics: exponential increase in stool crosslinking is mirrored in systemic inflammation and associated to fecal acetate and methionine. Metabolites 2021;11:1–16. 10.3390/metabo11030172 [DOI] [PMC free article] [PubMed] [Google Scholar]