Abstract
Objective
A comprehensive immune landscape for HBV infection is pivotal to achieve HBV cure.
Design
We performed single-cell RNA sequencing of 2 43 000 cells from 46 paired liver and blood samples of 23 individuals, including six immune tolerant, 5 immune active (IA), 3 acute recovery (AR), 3 chronic resolved and 6 HBV-free healthy controls (HCs). Flow cytometry and histological assays were applied in a second HBV cohort for validation.
Results
Both IA and AR were characterised by high levels of intrahepatic exhausted CD8+ T (Tex) cells. In IA, Tex cells were mainly derived from liver-resident GZMK+ effector memory T cells and self-expansion. By contrast, peripheral CX3CR1+ effector T cells and GZMK+ effector memory T cells were the main source of Tex cells in AR. In IA but not AR, significant cell–cell interactions were observed between Tex cells and regulatory CD4+ T cells, as well as between Tex and FCGR3A+ macrophages. Such interactions were potentially mediated through human leukocyte antigen class I molecules together with their receptors CANX and LILRBs, respectively, contributing to the dysfunction of antiviral immune responses. By contrast, CX3CR1+GNLY+ central memory CD8+ T cells were concurrently expanded in both liver and blood of AR, providing a potential surrogate marker for viral resolution. In clinic, intrahepatic Tex cells were positively correlated with serum alanine aminotransferase levels and histological grading scores.
Conclusion
Our study dissects the coordinated immune responses for different HBV infection phases and provides a rich resource for fully understanding immunopathogenesis and developing effective therapeutic strategies.
Keywords: HEPATITIS B, IMMUNE RESPONSE, T LYMPHOCYTES, TOLERANCE, MACROPHAGES
Significance of this study.
What is already known on this subject?
More than 280 million people worldwide are chronically infected with HBV, resulting in >700 000 deaths a year from cirrhosis and hepatocellular carcinoma. Only a minority of patients with chronic HBV (CHB) can achieve functional cure by current treatment regimens.
A comprehensive view of the intrahepatic immune responses to HBV infection at single-cell resolution is still lacking.
CD8+ T cells are widely considered the crucial effectors of viral clearance during acute-resolved HBV infection but are functionally impaired during CHB.
What are the new findings?
We delineated a comprehensive landscape of immune cells across different hepatitis B stages at single-cell resolution.
Exhausted CD8+ T cells were preferentially expanded in immune active (IA) and acute recovery patients with different dynamics.
Network analysis of cell–cell interactions and histological validations revealed that exhausted CD8+ T cells exhibited significant interactions with regulatory CD4+ T cells and FCGR3A+ macrophages in immune tolerant (IT) and IA patients, potentially mediating by HLA class I molecules together with their receptors CANX and leucocyte immunoglobulin-like receptor, respectively.
Significance of this study.
How might it impact on clinical practice in the foreseeable future?
The comprehensive transcriptome atlas is a valuable resource for the understanding of immunology and pathogenesis of hepatitis B.
Our results highlighted the coordinated dysregulation of MAIT, regulatory T-cell, exhausted CD8+ T and myeloid subsets in the liver of IT patients, implicating potential need for early treatment in these patients.
Circulating CX3CR1+GNLY+ CD8+ central memory T cells might serve as a surrogate marker for predicting HBV clearance.
Cellular spatial organisation mapper analysis provided potential cellular and molecular targets for CHB therapy.
Introduction
More than 280 million people are living with chronic HBV (CHB) infection and are at a high risk of developing liver cirrhosis, liver failure or hepatocellular carcinoma (HCC), which results in more than 700 000 deaths a year.1 The availability of effective vaccine and blockade of mother-to-infant transmission has successfully reduced the incidence, but current first-line antiviral treatment rarely attains a functional cure, which is defined as sustained loss of serum HBV surface antigen (HBsAg) despite treatment cessation.2
The vast majority of people encountering HBV in adulthood developed an acute resolved hepatitis, whereas most of the infections acquired in infancy or early childhood become CHB. CD8+ T cells are widely considered as the crucial effectors of viral clearance during acute recovery (AR) but are impaired due to exhaustion sculpted by continuous exposure to high viral antigen and immunosuppressive hepatic microenvironments in CHB.3 The natural history of CHB follows several clinical phases with distinct levels of viral replication and dynamics in liver disease progression.4 The immune tolerant (IT) phase is characterised by high-serum HBV DNA but normal serum alanine aminotransferase (ALT) and mostly normal liver histology. It is still unknown how the host precisely establishes and maintains immunological ignorance to viral replication in this phase. The immune active (IA) phase is featured by liver necroinflammation, fibrosis and fluctuating serum ALT level. During the IA phase, the host immune response is thought to be responsible for liver pathogenesis.5–8 Only a minority of patients with CHB can achieve HBsAg seroclearance (defined as chronic resolved (CR)), either spontaneously or by antiviral treatment. However, the underlying immune mechanisms related to the establishment, maintenance and transition of distinct HBV phases remain elusive.
Although many immunological insights into HBV pathogenesis have been made by studying peripheral blood, it has come to light that tissue-resident immune subsets play vital roles in front-line immunosurveillance in the liver.9 10 For example, the liver is enriched with mucosal-associated invariant T (MAIT) cells, natural killer (NK) cells, and unique tissue-resident CD8+ T-cell subsets.11 12 Due to the infeasibility of liver sampling in most HBV-infected patients, limited information can be accessed. However, single-cell RNA sequencing (scRNA-seq) technologies have greatly advanced our understanding of liver biology and diseases. By dissecting the cellular heterogeneity, novel insights have been made into liver zonation and regeneration.13–15 Multiple novel cell subtypes related to the pathological processes of HCC,16–18 liver fibrosis19 20 and metabolic-associated fatty liver disease21 have also been identified by scRNA-seq. A comprehensive view of the intrahepatic immune responses to HBV infection by using scRNA-seq is still lacking.
To get an unbiased and comprehensive understanding of the intrahepatic immunological features and connections with disease status in HBV-infected patients, we applied scRNA-seq and constructed a comprehensive landscape of immune cells in liver and blood across different hepatitis B stages (IT, IA, AR, CR and healthy controls (HCs)). The expansion of exhausted CD8+ T (Tex) cells was observed in IA and AR phases, with different sources and cellular interactions. Our data highlight the interaction network of T cells and macrophages that contribute to immune failure during HBV persistence, which may be used to guide the development of immunotherapy.
Results
Single-cell immune landscape of HBV-infected individuals
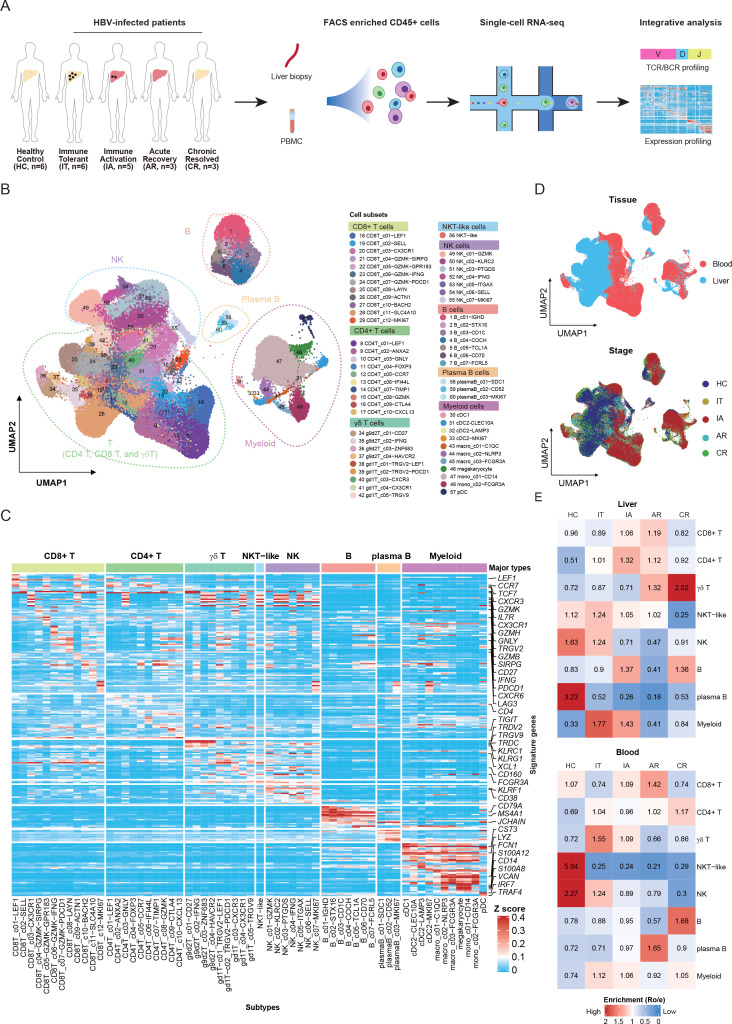
To better understand the heterogeneity within and across individuals infected with HBV, we applied scRNA-seq to characterise the transcriptomic status of intrahepatic immune cells as well as the peripheral blood mononuclear cells (PBMCs) from individuals in IT, IA, AR and CR phases (figure 1A and online supplemental figure 1A). As a control, we profiled both peripheral blood and normal liver tissues obtained from liver transplant donors (HCs). Detailed clinical and pathological information, including time since HBV diagnosis, HBsAg level, HBV DNA level, ALT and AST, is provided in table 1 and online supplemental table 1).
Figure 1.
Single-cell transcriptome map of immune cells in liver and blood. (A) An experimental scheme diagram of the overall study design. Hepatic immune cells and paired PBMCs were collected and subjected for cell barcoding. The cDNA libraries of 5′-mRNA expression, TCR and BCR were constructed independently, followed by high-throughput sequencing and downstream analyses. (B) UMAP plots of the 243 000 single cells from 23 individuals, including 8 major clusters and 60 subclusters. (C) Gene expression heatmap in each cell cluster. EXP, z-score normalised mean expression. (D) UMAP plots of major cell clusters showing tissue distribution and stage distribution. (E) Tissue prevalence of major cell clusters estimated by Ro/e score. AR, acute recovery; BCR, B-cell receptor; cDNA, complementary DNA; CR, chronic resolved; HC, healthy control; IA, immune active; IT, immune tolerant; NK, natural killer; PBMC, peripheral blood mononuclear cell; TCR, T-cell receptor; UMAP, uniform manifold approximation and projection.
Table 1.
Patient information
| Group | Immune tolerant | Immune activation | AR | CR | HC |
| Cases | 6 | 5 | 3 | 3 | 6 |
| Age (years) | 34 (21–55) | 29 (25–45) | 37 (28–45) | 47 (45–49) | 52 (33–66) |
| Sex (M/F) | 4/2 | 4/1 | 2/1 | 3/0 | 4/2 |
| Time since HBV diagnosis (years) | 14.7 (1.5–27.0) | 20 (10–20) | <0.5 | 10 (1–20) | NA |
| HBsAg level (IU/mL) | 40 636 (5652–69 050) | 7881 (833.2–32 970.0) | 783 (0–1293) | <0.05 | NA |
| HBV DNA level (log10 IU/mL) | 8.328 (8–8.88) | 6.761 (5.29–8) | 0.9783 (0–2.935) | LDL | NA |
| ALT (U/L) | 36.1 (20–45.7) | 232.2 (60–424) | 108 (25–192) | 18.67 (13–22) | 20.8 (11–31) |
| AST (U/L) | 28.7 (19–42) | 91.2 (34–129) | 44 (21–58) | 19 (16–24) | 19 (15–27) |
ALT, alanine aminotransferase; AR, acute recovery; AST, aspartate transaminase; CR, chronic resolved; F, female; HBsAg, HBV surface antigen; HC, healthy control; LDL, lower detection limit; M, male; NA, not applicable.
gutjnl-2021-325915supp001.pdf (9.8MB, pdf)
gutjnl-2021-325915supp002.xlsx (16.4KB, xlsx)
In addition, the longitudinal information for AR and CR patients is shown in online supplemental figure 1B, C. The sampling timepoints for AR patients were at the convalescent stage, that is, less than 6 months after initial diagnosis with rapid decrease of HBsAg (online supplemental figure 1B, table 1 and online supplemental table 1). The CR patients were diagnosed with a history of CHB and HBsAg loss, either spontaneously or by antiviral treatment (online supplemental figure 1C, table 1 and online supplemental table 1).
After quality control and filtering (online supplemental figure 2A, B, and online supplemental table 2), we obtained 243 000 transcriptomes of single cells from 23 individuals. Eight major cell clusters were identified, including CD4+ T cells (CD3D+CD4+), CD8+ T cells (CD3D+CD8A+), γδ T cells (CD3D+TRDC+), NKT-like cells (CD3E+KLRF1+), NK cells (GNLY+NKG7+), B cells (MZB1+CD79A+), plasma B cells (MZB1+CD38+) and myeloid cells (LYZ+CD14+) (figure 1B). In total, we identified 10 subclusters for CD4+ T cells, 12 for CD8+ T cells, 9 of γδ T cells, 1 of NKT-like cells, 7 of NK cells, 7 of B cells, 3 of plasma B cells and 11 of myeloid cells (figure 1B). Based on top differentially expressed genes (DEGs), we identified unique markers for each of the 60 clusters (figure 1C and online supplemental table 3).
gutjnl-2021-325915supp003.xlsx (23KB, xlsx)
gutjnl-2021-325915supp004.xlsx (1.6MB, xlsx)
The cell clusters showed different tissue origins and enrichment in disease stages (figure 1D, online supplemental figure 3A, B, and online supplemental table 2). To investigate the distinctive immune profiles of different disease phases, we analysed the immune cell composition in each of the individuals. We found that both CD4+ and CD8+ T cells were enriched in the liver of IA and AR phases, both of which are characterised by elevated levels of liver inflammation. Myeloid cells showed preferential enrichment in the liver of IT and IA phases, indicating altered myeloid compartments during chronic HBV infection. γδ T cells and B cells were increased in the liver of CR phase, implicating potential long-term effects of chronic infection on these cells similar to resolved HCV infection.22 23 NK and NKT-like cells showed enrichment in HC and IT (figure 1E and online supplemental figure 3C). Notably, the trends for CD8+ T and myeloid cells among disease stages were consistent between liver and blood (figure 1E and online supplemental figure 3C). These results indicated that each disease stage in HBV-infected patients might be associated with a unique immune signature.
Expansion of Tex and loss of MAIT cells in HBV-infected patients
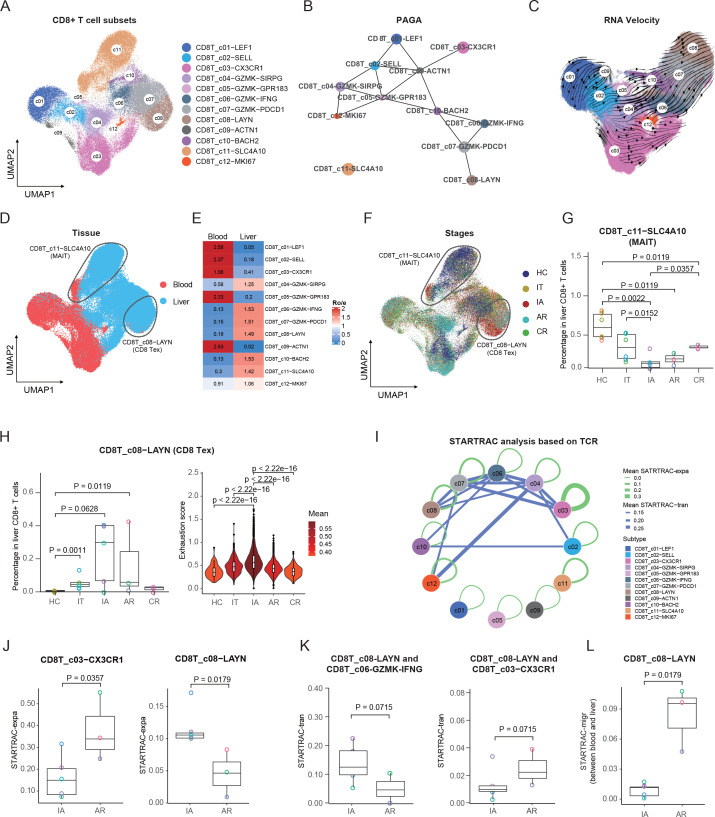
Further clustering of CD8+ T cells yielded 12 subclusters, including naïve (CD8T_c01-LEF1); central memory T (Tcm) (CD8T_c02-SELL, CD8T_c09-ACTN1 and CD8T_c10-BACH2); recently activated effector memory or effector T (Temra) (CD8T_c03-CX3CR1); effector memory (CD8T_c04-GZMK-SIRPG, CD8T_c05-GZMK-GPR183, CD8T_c06-GZMK-IFNG and CD8T_c07-GZMK-PDCD1); exhausted (CD8T_c08-LAYN); MAIT (CD8T_c11-SLC4A10); and a proliferating subset (CD8T_c12-MKI67) (figure 2A, online supplemental figure 4A and online supplemental table 3).
Figure 2.
Immunological features of CD8+ T-cell subsets. (A) UMAP plots of CD8+ T-cell subsets. (B) PAGA analysis showing the potential developmental connectivity between different CD8+ T-cell subsets. (C) RNA velocity analysis showing the transition potential among CD8+ T-cell subsets. (D) UMAP plots of CD8+ T-cell subsets showing tissue distribution. (E) Tissue prevalence of CD8+ T-cell subsets estimated by Ro/e score. (F) UMAP plots of CD8+ T-cell subsets showing stage distribution. (G) Box plots showing the proportions of MAIT cells in liver CD8+ T cells across stages. One-sided unpaired Wilcoxon test. (H) Box plots showing the proportions and violin plots showing the exhaustion scores of Tex cells across stages. One-sided unpaired Wilcoxon test. (I) Clonal expansion and state transition of CD8+ T-cell subsets estimated by STARTRAC analysis based on all patients. (J) Box plots showing the expansion levels (STARTRAC-expa) of Temra and Tex subsets in IA and AR stages. One-sided unpaired Wilcoxon test. (K) Box plots showing the transition scores (STARTRAC-tran) between Tex and CD8T_c06-GZMK-IFNG and CD8T_c03−CX3CR1 in IA and AR stages. One-sided unpaired Wilcoxon test. (L) Box plots showing the migration scores (STARTRAC-migr) of the Tex subset between blood and liver in IA and AR stages. One-sided unpaired Wilcoxon test. AR, acute recovery; CR, chronic resolved; HC, healthy control; IA, immune active; IT, immune tolerant; MAIT, mucosal-associated invariant T; PAGA, partition-based graph abstraction; STARTRAC, single T-cell analysis by RNA sequencing and TCR tracking; Temra, recently activated effector memory or effector T; Tex, exhausted CD8+ T; UMAP, uniform manifold approximation and projection.
By examining signature genes defined previously,24 we observed distinct functional status for each CD8+ T-cell subset, with the highest exhaustion score for CD8T_c08-LAYN (Tex), high inflammatory scores in effector memory subsets and MAIT, the highest cytotoxic score for CD8_C03-CX3CR1 (Temra), and high naive scores in CD8T_c01-LEF1 and two central memory subsets (CD8T_c02-SELL and CD8T_c09-ACTN1) (online supplemental figure 4B and online supplemental table 4).
gutjnl-2021-325915supp005.xlsx (17.6KB, xlsx)
To understand the potential developmental transitions of CD8+ T-cell clusters, we applied partition-based graph abstraction (PAGA) analysis to construct the developmental trajectories of 12 CD8+ T clusters.25 CD8T_c11-SLC4A10 cells stood out as a distinct population outside of the trajectory, consistent with the annotation of MAIT cells. For the other CD8+ T-cell subsets, the Tex (CD8T_c08-LAYN) and Temra (CD8_C03-CX3CR1) subsets were characterised as two different branches (figure 2B). Such developmental features for Tex and Temra were further supported by RNA velocity analysis (figure 2C).26
Further, each subset showed distinct tissue preference: CD8T_c01-LEF1, CD8T_c02-SELL, CD8T_c03-CX3CR1, CD8T_c05-GZMK-GPR183 and CD8T_c09-ACTN1 clusters were mainly derived from blood, while CD8T_c04-GZMK-SIRPG, CD8T_c06_GZMK-IFNG, CD8T_c07-GZMK-PDCD1, CD8T_c08-LAYN, CD8T_c10-BACH2 and CD8T_c11-SLC4A10 clusters were mainly found in the liver (figure 2D, E). As expected, when compared with blood-enriched subsets, these liver-enriched cell subsets expressed higher levels of liver-resident markers, such as CXCR6, CXCR3, CD69 and CD103 15 20 27 28 and exhibited a GZMB low IL-2 high phenotype29 (online supplemental figure 4C). We further compared the transcriptional features of each cluster between liver and blood. Interestingly, we identified a series of commonly upregulated genes in the liver among subsets, with CXCR4 being the most frequent (8 times among 12 subsets, online supplemental figure 4B). It was reported that CXCR4 was expressed at a high level on intrahepatic HBV-specific CD8+ T cells,30 and the CXCL12/CXCR4 pathway was involved in the recruitment and retention of immune cells in the liver during chronic HBV infection.31 Further studies are needed to clarify the significance of CXCR4 for liver residency.
The 12 CD8+ T-cell subsets showed stage heterogeneities (figure 2F and online supplemental figure 5A, B). Among CD8+ T subsets, the MAIT population decreased significantly in the liver of all four groups of HBV-infected patients, with that in the IA group being the lowest (figure 2G). The results are consistent with a prior study reporting the loss of hepatic MAIT cells in patients with HBV-related HCC.11 Strikingly, the frequency of MAIT cells in CR failed to restore to the level of HC (figure 2G), although these patients had achieved HBsAg loss for more than 8 months, suggesting that the loss of intrahepatic MAIT induced by HBV infection is long-lasting. Notably, the frequencies of CD8T_c08-LAYN cells increased in IT, IA and AR patients (figure 2H), of which CD8T_c08-LAYN cells from IA patients also displayed the highest exhaustion score (figure 2H). Notably, CD8T_c08-LAYN cells from IA patients showed higher expression of exhaustion genes (such as LAG3 and LAYN), while AR patients had higher expression of cytotoxic genes (such as GZMH, GZMA, GZMB and GZMK) (online supplemental figure 6A), suggesting a cytotoxic CD8+ T-cell response might contribute to control of HBV infection.
Since exhaustion markers are also upregulated in CD8+ T cells by T-cell receptor (TCR)-induced activation, it is also possible that CD8T_c08-LAYN cells from AR are activated rather than exhausted, reminiscent of that observed in patients with COVID-19.32 As for transcriptional level, CD8T_c08-LAYN cells from AR displayed a typical exhausted state, which is featured by high expression of inhibitory markers (TOX, HAVCR2, LAG3, LAYN, PDCD1 and TIGIT) and low expression of effector markers (TBX21 and IFNG) (online supplemental figure 6B). Moreover, the phenotypes of exhausted CD8+ T cells in AR were also validated by flow cytometry (online supplemental figure 6C, D). Therefore, CD8T_c08-LAYN cells represent the exhausted cluster in AR, as well as in other groups.
To further understand the changes of CD8+ T-cell subsets among liver immune cells, we also compared the abundance of each CD8+ T-cell cluster among liver CD45+ cells across stages (online supplemental figure 5C), which confirmed the dramatic loss of MAIT cells and significant expansion of CD8+ Tex cells in IA and AR patients (online supplemental figure 5A). Taken together, these results indicated that exhausted CD8+ T cells in the liver were expanded in both chronic (IT and IA) and acute (AR) HBV-infected patients.
Dynamics differences of Tex cells between IA and AR patients
Since we also captured TCR sequences from scRNA-seq (online supplemental table 5), this enabled us to trace the dynamics of T cells, such as clonal expansion (expa), migration (migr) and developmental transition (tran), by previously developed bioinformatics metrics, that is, single T-cell analysis by RNA sequencing and TCR tracking (STARTRAC) indices (figure 2I).33
gutjnl-2021-325915supp006.xlsx (10.7MB, xlsx)
The STARTRAC-expa index revealed that CD8T_c03-CX3CR1 and CD8T_c08-LAYN displayed a high degree of clonal expansion, mainly in the AR and IA stages (online supplemental figure 7A). Of note, CD8T_c03-CX3CR1 was more expanded in AR patients, whereas CD8T_c08-LAYN was more expanded in IA patients (figure 2J).
STARTRAC-tran analysis indicated that CD8T_c08-LAYN was highly associated with GZMK+ subsets, including CD8T_c07-GZMK-PDCD1, CD8T_c04-GZMK-SIRPG and CD8T_c06_GZMK-IFNG (online supplemental figure 7B, C). Notably, CD8T_c08-LAYN in IA was more closely linked to CD8T_c06_GZMK-IFNG but less closely to CD8T_c03-CX3CR1 compared with that in AR (figure 2K), suggesting different sources of exhausted CD8+ T cells in IA and AR patients.
Overall, CD8T_c08-LAYN displayed modest mobility as revealed by the STARTRAC-migr analysis (online supplemental figure 7D), similar to the observation in tumours.29 However, exhausted T cells in AR patients exhibited higher STARTRAC-migr scores when compared with that in IA patients (figure 2L). Moreover, we found that the cells that shared the same TCR clonal type with liver Tex cells were mainly CD8T_c03-CX3CR1 and CD8T_c04-GZMK-SIRPG subsets (online supplemental figure 7E).
Furthermore, RNA velocity analysis of CD8+ T-cell clusters also corroborated the strong self-expansion of CD8+ Tex in IA patients and the transition of liver Tem to CD8+ Tex in IA together with the transition of blood Tem to CD8+ Tex in AR patients (online supplemental figure 8A, B). The different dynamics of Tex in IA and AR patients suggest that the transition of CD8+ T cells between liver and blood might facilitate viral clearance, implicating vital roles of factors that affect the function and differentiation of CD8+ T cells on HBV infection.
Increased intrahepatic regulatory CD4+ T cells in IT and IA patients
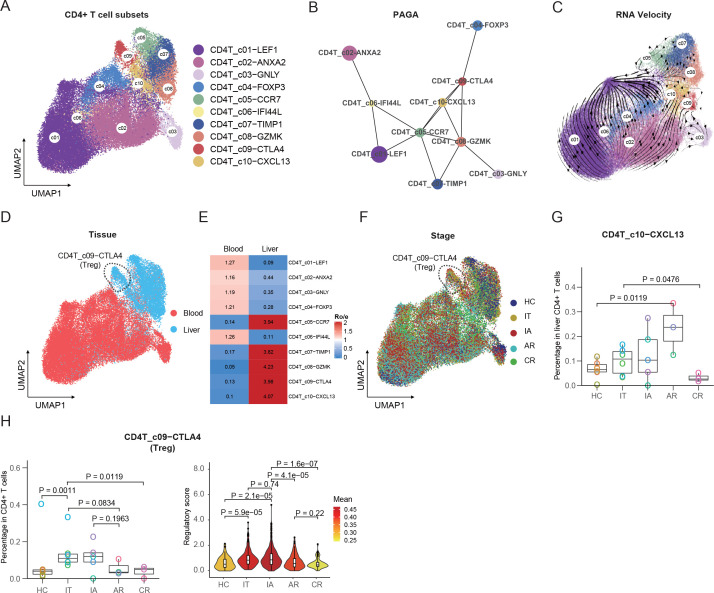
A total of 10 CD4+ T-cell clusters were identified, including 3 naive (CD4T_c01-LEF1, CD4T_c05-CCR7 and CD4T_c06-IFI44L), central memory (CD4T_c02-ANXA2), effector (CD4T_c03-GNLY), 2 regulatory (CD4T_c04-FOXP3 and CD4T_c09-CTLA4), transient memory (CD4T_c07-TIMP1), effector memory (CD4T_c08-GZMK) and Th1-like (CD4T_c10-CXCL13) T cells (figure 3A, online supplemental figure 9A and online supplemental table 3). PAGA trajectory analysis revealed four different fates of CD4+ T cells, with CD4T_c02-ANXA2, CD4T_c03-GNLY, CD4T_c04-FOXP3 and CD4T_c06-TIMP1 as different ends (figure 3B). The developmental direction towards the effector and regulatory subset was also informed by RNA velocity analysis (figure 3C). The developmental trajectory was correlated with the functional scores in various cell clusters, with CD4T_c08-CTLA4 cluster being the most exhausted and CD4T_c03-GNLY cluster being the most cytotoxic (online supplemental figure 9B). Tissue preference analysis revealed that CD4T_c05-CCR7, CD4T_c07-TIMP1, CD4T_c08-GZMK, CD4T_c09-CTLA4 and CD4T_c10-CXCL13 accumulated in the liver, and the rest of the five subsets were mainly found in blood (figure 3D, E).
Figure 3.
Immunological features of CD4+ T-cell subsets and interaction with CD8+ T cells. (A) The UMAP plots of CD4+ T-cell subsets. (B) PAGA analysis showing the potential developmental connectivity between different CD4+ T-cell subsets. (C) RNA velocity analysis showing the transition potential among CD4+ T-cell subsets. (D) The UMAP plots of CD4+ T-cell subsets showing tissue distribution. (E) Tissue prevalence of CD4+ T-cell subsets estimated by Ro/e score. (F) The UMAP plots of CD4+ T-cell subsets showing stage distribution. (G) Box plots showing the proportions of CXCL13+ CD4+ T cells in liver CD4+ T cells across stages. One-sided unpaired Wilcoxon test. (H) Box plots showing the proportions and violin plots showing the regulatory scores of liver-resident Treg cells across stages. One-sided unpaired Wilcoxon test. AR, acute recovery; CR, chronic resolved; HC, healthy control; IA, immune active; IT, immune tolerant; PAGA, partition-based graph abstraction; Treg, regulatory T-cell; UMAP, uniform manifold approximation and projection.
We then compared the fractions of CD4+ T-cell subsets among CD4+ T cells (figure 3F and online supplemental figure 10A, B) or liver CD45+ cells (online supplemental figure 10C) across different groups. Among these CD4+ T subsets, the CD4T_c10-CXCL13 subset was enriched in AR patients in both liver CD4+ T cells (figure 3G) and liver CD45+ immune cells (online supplemental figure 10C). CD4T_c10-CXCL13 cells exhibited transcriptional features of T-follicular helper cells (PDCD1, ICOS, TIGIT, TNFRSF4 and CXCL13, MAF) (online supplemental figure 9A and online supplemental table 3), implicating the potential role of this subset in enabling protective B-cell immune responses. Moreover, CD4T_c09-CTLA4, referred to as the liver-resident regulatory T-cell (Treg) subset, was increased in both IT and IA patients among liver CD4+ T cells (figure 3H) or liver CD45+ immune cells (online supplemental figure 10C). In addition, regulatory effector score analysis of CD4T_c09-CTLA4 cells among stages revealed high levels of regulatory effector function in IT and IA patients (figure 3H). These results suggested that immune regulation by Treg cells might be associated with immune tolerance and viral persistence in CHB.
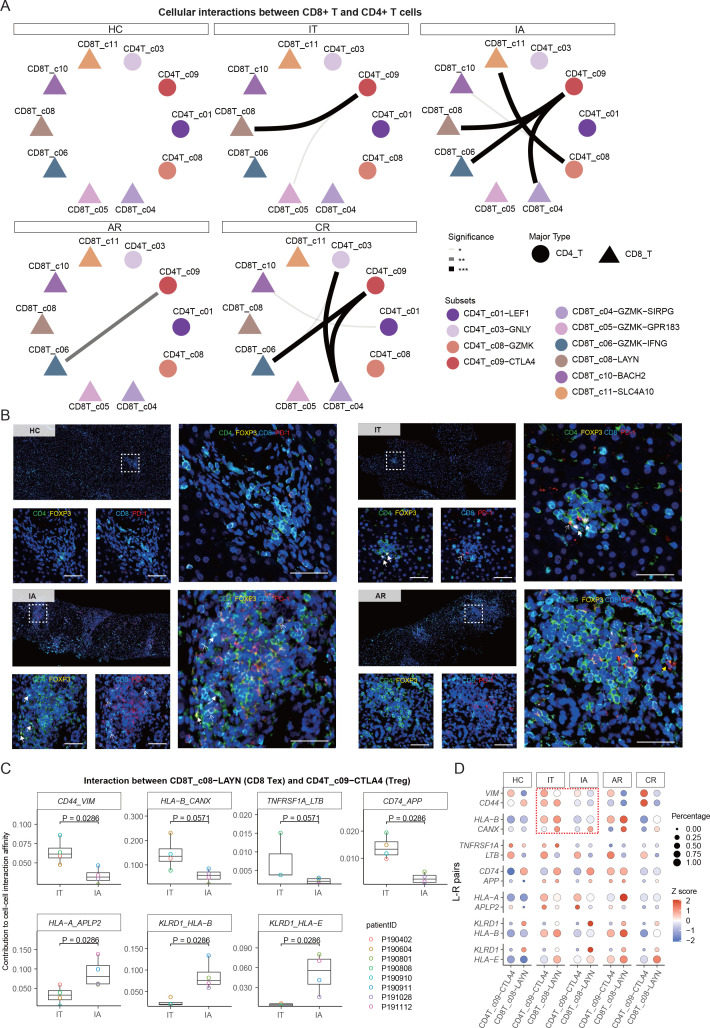
Potential cell–cell interactions between Treg and Tex cells
Using cellular spatial organisation mapper (CSOmap),34 a bioinformatics tool to infer spatial organisation of tissues and molecular determinants of cellular interaction, we estimated the cellular interaction potentials between CD4+ T-cell and CD8+ T-cell subsets in a computationally constructed pseudo-space. Significant interactions were found between Treg (CD4T_c09-CTLA4) and Tex (CD8T_c08-LAYN) in IT and IA stages (figure 4A and online supplemental table 6). Notably, Tex and Treg were expanded coordinately in the liver of IT and IA patients (figure 2H). We further confirmed the adjacent spatial relationship between Tex (CD8+PD-1+) and Treg (CD4+FOXP3+) cells in IT and IA patients by multicolour immunohistochemistry (IHC) staining (figure 4B and online supplemental figure 11).
Figure 4.
Multicolour IHC staining of Treg and Tex cells in the liver. (A) CSOmap analysis showing the interaction between CD8+ T-cell and CD4+ T-cell subsets. (B) Multicolour IHC staining in the liver across HC, IT, IA and AR groups. One representative image for each group is shown. White dotted arrows indicate Tex cells, white solid arrows indicated Tregs, yellow solid arrows indicated CD4 +PD1+cells. scale bar, 50 µm. (C) Box plots showing the contributions of indicated L-R pairs to interaction affinity between Tex and Treg in IT and IA stages. one-sided unpaired Wilcoxon test. (D) Bubble heatmap showing the selected L-R pairs between Treg and Tex cells. AR, acute recovery; CR, chronic resolved; CSOmap, cellular spatial organisation mapper; HC, healthy control; IA, immune active; IHC, immunohistochemistry; IT, immune tolerant; L-R, ligand–receptor; Tex, exhausted CD8+ T; Treg, regulatory T-cell.
gutjnl-2021-325915supp007.xlsx (37.3KB, xlsx)
We then compared the strength of potential ligand–receptor (L-R) pairs contributing to Treg-CD8 Tex interaction in IT and IA patients. We found that several L-R pairs, CD44_VIM, HLA-B_CANX, TNFRSF1A_LTB and CD74_APP, showed higher interaction potentials in the IT phase, while HLA-A_APLP2, KLRD1_HLA and KLRD1_HLA-E showed higher interaction potentials in the IA phase (figure 4C, D). It was recently reported that the HLA-B_CANX axis is able to restrain antitumor immunity of T cells.35 Vimentin, encoded by the VIM gene, is a key metabolic and functional controller of Treg activity,36 and CD44 immune checkpoint controls CD8+ T-cell activation.37 Moreover, the HLA-E_KLRD1 signal is involved in mediating T-cell dysfunction and viral persistence in chronic HBV and HCV infections.38 39 Together, these data provide valuable information for functional verification in future studies regarding immune pathogenesis as well as therapeutic attempts for CHB.
Intrahepatic myeloid cells and their interactions with T cells in HBV-infected patients
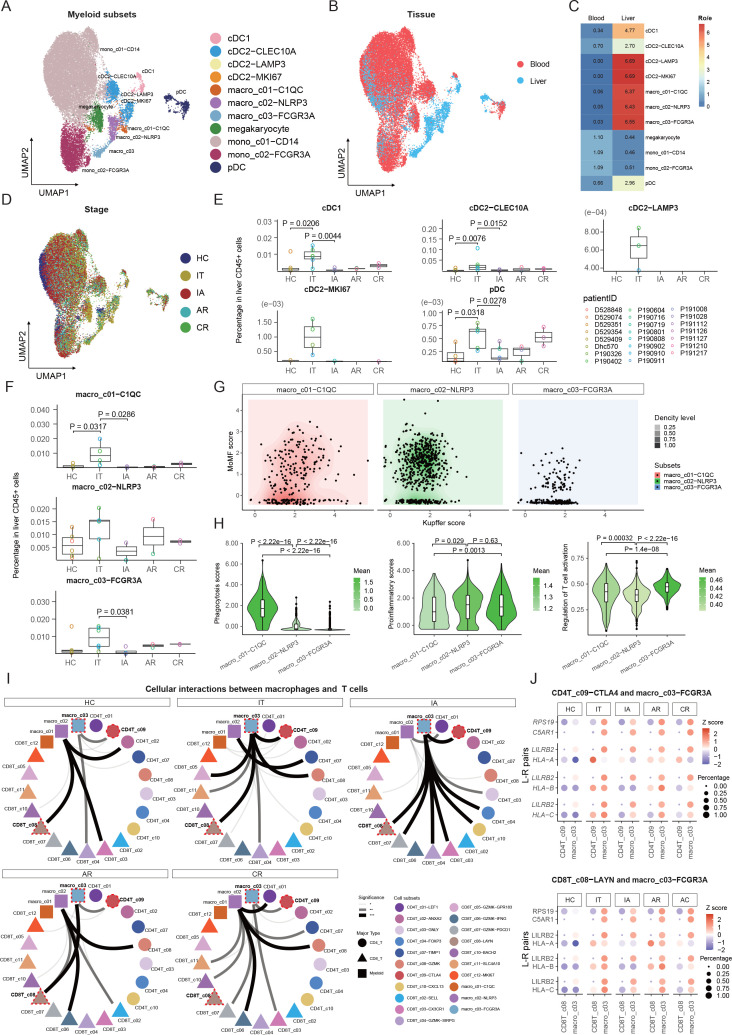
Myeloid cells were clustered into 11 cell subsets belonging to four common lineages (dendritic cells (DCs), macrophages, megakaryocytes and monocytes) based on canonical cell markers (figure 5A, online supplemental figure 12A, B, and online supplemental table 3). DCs were further divided into two classical cDC subsets (CLEC9A+cDC1 and CLEC10A+cDC2), a mature cDC subset (cDC2-LAMP3), a proliferating subset (cDC2-MKI67) and pDC. All the five DC subsets were mainly found in the liver (figure 5B, C) and were enriched in IT patients (figure 5D,E), suggesting these cells might be driven by the high level of viral replication in IT patients. Macrophages were further divided into macro_c01-C1QC, macro_c02-NLRP3 and macro_c03-FCGR3A. Similar to DCs, macrophage subsets were also mainly distributed in the liver (figure 5B, C). Among the five patient groups, macro_c01-C1QC and macro_c03-FCGR3A showed significant expansion among liver CD45+ cells of IT patients (figure 5F).
Figure 5.
Immunological features of myeloid cell subsets and interaction with T cells. (A) UMAP plots of myeloid cell subsets. (B) UMAP plots of myeloid cell subsets showing tissue distribution. (C) Tissue prevalence of myeloid cell subsets estimated by the Ro/e score. (D) UMAP plots of myeloid cell subsets showing stage distribution. (E) Box plots showing the proportions of DC subsets in liver CD45+ cells across stages. One-sided unpaired Wilcoxon test. (F) Box plots showing the proportions of macrophage subsets in liver CD45+ cells across stages. One-sided unpaired Wilcoxon test. (G) Dot plots of macrophages defined by Kupffer score and MoMF score. (H) Violin plots showing the functional scores (phagocytosis, proinflammatory, regulation of T-cell activation) of macrophage subsets across stages. One-sided unpaired Wilcoxon test. (I) CSOmap analysis showing the interaction between macrophage and T-cell subsets. (J) Bubble heatmap showing the selected ligand-receptor pairs between FCGR3A+ macrophage and Treg or Tex cells. AR, acute recovery; CR, chronic resolved; CSOmap, cellular spatial organisation mapper; DC, dendritic cell; HC, healthy control; IA, immune active; IT, immune tolerant; Tex, exhausted CD8+ T; Treg, regulatory T-cell; UMAP, uniform manifold approximation and projection.
To understand the origins of the macrophage subsets, we defined scores regarding Kupffer-derived source (Kupffer score) or monocyte-derived macrophages (MoMF score) based on reported markers (online supplemental table 4). Expression analyses based on genes associated with Kupffer/monocyte origin did not fully explain the heterogeneity of macrophages (figure 5G, online supplemental figure 12C, D, and online supplemental table 4). Further comparison of the DEGs between macrophage populations revealed that macro_c01-C1QC expressed high levels complement C1Q genes and class II HLA molecules, suggesting capacities of phagocytosis (figure 5H) and antigen presentation in this cluster (online supplemental figure 12E and online supplemental table 4). Macro_c02-NLRP3 expressed high levels of proinflammatory genes, such as S100 genes, IL1B, VCAN and LYZ (online supplemental figure 12E and online supplemental table 3) and was featured by a high proinflammatory score (figure 5H). Macro_c03-FCGR3A expressed high levels of antiviral ISGs (IFITM2, IFITM3, SPN, WARS and CTSC) and immunoregulatory genes (SOD1, MT2A, LST1, LYN, MTSS1, PECAM1, LILRB2 and LILRA5) (online supplemental figure 12E and online supplemental table 3), exhibiting a high proinflammatory score and high ability to inhibit T-cell activation (figure 5H).
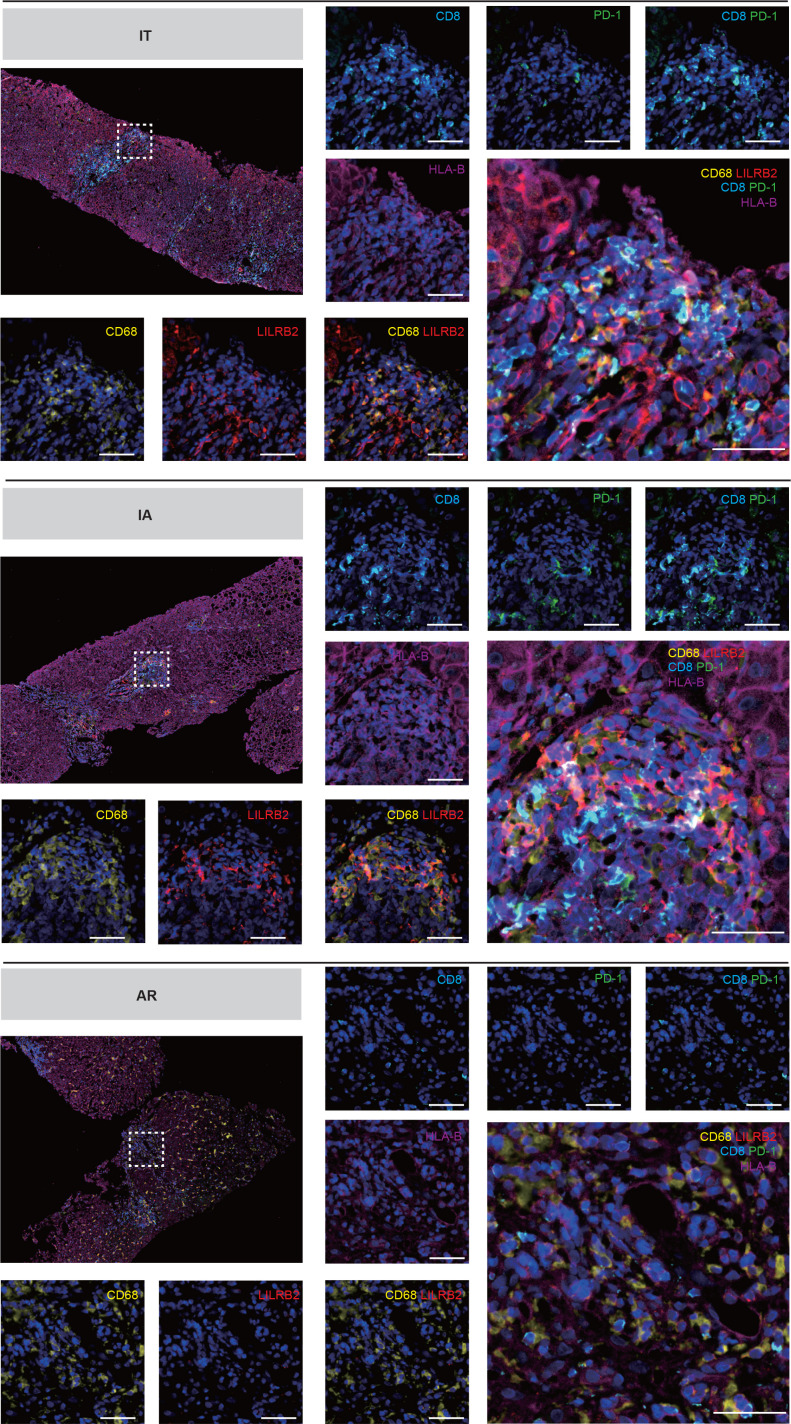
To dissect the potential crosstalk of macrophages and T cells, we used CSOmap to infer cell–cell interaction between macrophage subsets and T cells. Strikingly, CSOmap analysis revealed that macro_c03-FCGR3A dominated the interaction with T cells in two chronic phases (IT and IA), and the intensity of such interaction decreased in two resolved phases (AR and CR), while macro_c02-NLRP3 dominated the interaction with T cells in resolved phases (figure 5I). By examining L-R pair interactions between macro_03-FCGR3A and CD8T_c08-LAYN, several L-R pairs between the HLA class I molecules (HLA-I) and leucocyte immunoglobulin-like receptor (LILR) family exhibited strong potential interaction (figure 5J). Consistently, we observed the enrichment of LILRB2-positive macrophages with adjacent locations to Tex cells in the liver of IT and IA patients (figure 6). These potential interactions might be functional, considering previous findings that LILRB1 and LILRB2 can facilitate immune evasion in the tumour microenvironment by competing with CD8+ T cells for HLA-I binding as well as by recruiting inhibitory molecules through their immunoreceptor tyrosine-based inhibitory receptor motif.40 Taken together, these results indicate a central role of FCGR3A+ macrophages in modulating T-cell responses in CHB, potentially through HLA-I_LILRB signalling.
Figure 6.
Multicolour IHC staining of Tex cells and macrophages in the liver across IT, IA and AR groups. Multicolour IHC staining in the liver of HBV-infected patients. One representative image for each group is shown. Scale bar, 50 µm. AR, acute recovery; IA, immune active; IHC, immunohistochemistry; IT, immune tolerant; Tex, exhausted CD8+ T.
Correlations of liver immune subsets with clinical parameters
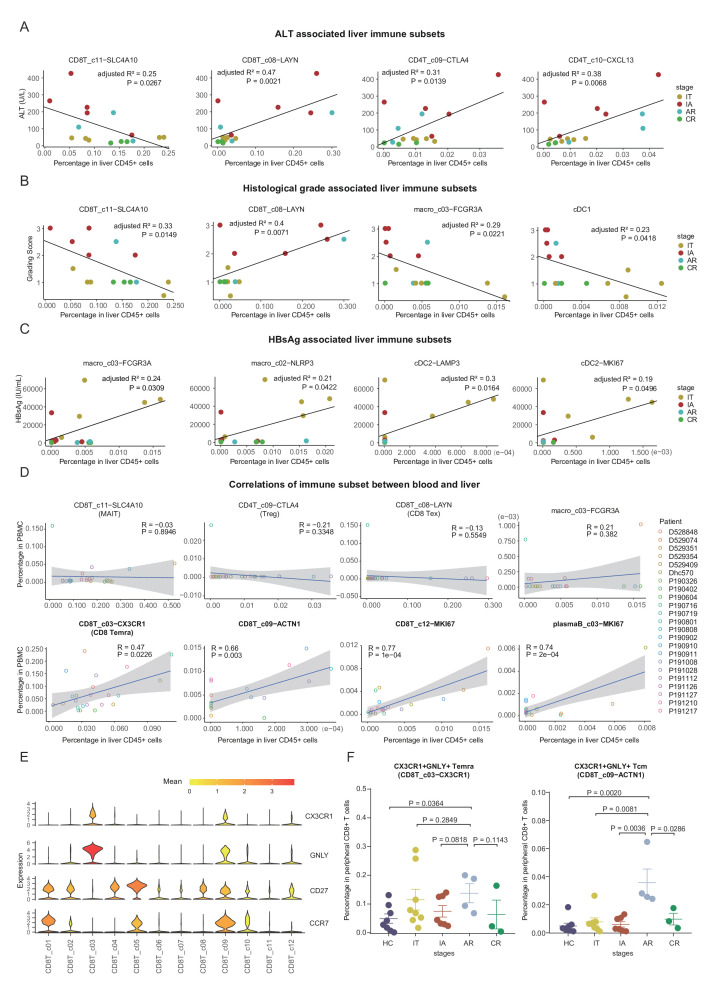
To understand the relationships of hepatic immune subsets with clinical parameters in HBV-infected patients, we applied correlation analysis of cell frequencies with the levels of HBsAg, HBV DNA, ALT, AST, and histological grade and stage among all the 17 HBV-infected patients. Age and time since diagnosis were also included in the analysis to set aside factors that are affected by these two parameters (online supplemental figure 13).
Interestingly, the levels of ALT showed negative correlation with the frequencies of MAIT cell and positive correlations with the frequencies of CD8+ Tex, Treg and CD4T_c10-CXCL13 in the liver (figure 7A and online supplemental figure 13), suggesting these cell subsets could be the key executors or responders of liver injury. Moreover, the frequencies of NK_c03-PTGDS and pDC in liver CD45+ cells also negatively correlated with ALT levels (online supplemental figure 13). Consistently, we observed a similar pattern for the correlations of AST levels with hepatic immune subsets compared with that of ALT (online supplemental figure 13).
Figure 7.
Associations between intrahepatic immune features and clinical parameters in HBV-infected patients. (A) Correlations of the compositions of cell subsets in liver CD45+ cells with ALT levels in HBV-infected patients. (B) Correlations of the compositions of cell subsets in liver CD45+ cells with histological grading of liver biopsies in HBV-infected patients. (C) Correlations of the compositions of cell subsets in liver CD45+ cells with serum HBsAg levels in HBV-infected patients. (D) Correlations of immune subsets between blood and liver. (E) Violin plots showing the expression levels of indicated genes in each CD8+ T-cell subset. (F) Dot plots showing the frequencies of CX3CR1+GNLY+ Temra or Tcm cells in CD8+ T cells from blood across stages. One-sided unpaired Wilcoxon test. ALT, alanine aminotransferase; AR, acute recovery; CR, chronic resolved; HbsAg, HBV surface antigen; HC, healthy control; IA, immune active; IT, immune tolerant; PBMC, peripheral blood mononuclear cell; Tcm, central memory T; Temra, recently activated effector memory or effector T.
Similar to ALT, a negative correlation for MAIT cell and a positive correlation for CD8+ Tex were observed with liver histological grading scores of liver biopsies (figure 7B). In addition, histological grade positively correlated with the frequencies of CD8T_c05-GZMK-GPR183 and negatively with plasmaB_c01-SDC1 and two myeloid subsets (cDC1 and macro_c03-FCGR3A) (figure 7B and online supplemental figure 13). In contrast, the histological stage only showed positive correlation with the frequencies of CD8T_c05-GZMK-GPR183 in liver CD45+ cells (online supplemental figure 13).
We then assessed the relationship of intrahepatic immune subsets with levels of serum HBsAg and serum HBV DNA. Serum HBsAg levels positively correlated with the frequencies of macro_c03-FCGR3A, macro_c02-NLRP3, cDC-LAMP3 and cDC-MKI67 (figure 7C). Moreover, HBV DNA levels showed a positive correlation with the frequencies of NK_c07-MKI67, CD4T_c08-GZMK and pDC in liver CD45+ cells (online supplemental figure 13).
Taken together, these results suggest that the massive HBV DNA and HBsAg in IT patients might be related to the alterations in the myeloid compartment, whereas liver injury in IA and AR patients is likely closely associated with T-cell dysfunction. This notion is consistent with recent findings that HBsAg has minimal impact on virus-specific or global T cells in humans41 42 or animal models.43
To dissect if there were coordinated immune changes between blood and liver, we applied correlation analysis for all identified immune subsets between blood and liver in 17 HBV-infected patients. Among 60 subsets, four subsets (CD8T_c03-CX3CR1, CD8T_c09-ACTN1, CD8T_c12-MKI67 and plasmaB_c03-MKI67) showed significant correlations between blood and liver (figure 7D and online supplemental table 7). Intrahepatic immune cell subsets with significant alterations across stages, such as MAIT, Treg and CD8+ Tex, could barely be found in the blood (figure 7D). Notably, the frequencies of both CD8T_c03-CX3CR1 and CD8T_c09-ACTN1 showed positive correlations between blood and liver (figure 7D). Both CD8T_c03-CX3CR1 (resembles of Temra) and CD8T_c09-ACTN1 (a Tcm subset) cells showed similar functional features, including low exhaustion and high cytotoxicity levels (figure 7E and online supplemental figure 7A, B). Circulating CX3CR1+CD8+ T cells were recently identified as a predictor for the response of checkpoint therapy in cancer.44 By using an additional validation cohort (online supplemental table 1), we confirmed that CX3CR1+GNLY+ Tcm cells (potential surrogates for CD8T_c09-ACTN1 cells) preferentially expanded in the blood of AR patients (figure 7F and online supplemental figure 14). The possible roles of the CX3CR1+GNLY+ CD8+Tcm cells in the cure of HBV should be investigated.
gutjnl-2021-325915supp008.xlsx (12.2KB, xlsx)
Discussion
In this study, we employed scRNA-seq to generate a comprehensive immune landscape of both hepatic and peripheral immune microenvironment at single-cell resolution in patients with acute or chronic HBV infection. The findings will not only get more insights into further understanding of HBV pathogenesis at different phases of HBV infection in humans but also help the discovery of novel immune targets or the development of novel therapeutic strategies for the clinical cure of CHB.
Our data provide an unbiased illustration of the immunological hallmarks for HBV-infected patients (online supplemental figure 15). Specifically, IT patients were characterised by a decrease of MAIT cells and increases of Treg and Tex in the liver. These changes became more pronounced in IA patients and were associated with liver inflammation. Notably, Tex, Treg and FCGR3A+ macrophage subsets showed strong interactions in the IT and IA phases. These alterations might collectively contribute to immune tolerance and viral persistence. As for acute HBV infection, AR patients also showed expansion of Tex but with different sources compared with IA patients. Importantly, we found that such immune alterations in the liver can hardly be simultaneously observed in the blood, highlighting the requirement of evaluating immunological changes compartmentalised at the site of infection. Several recent studies have suggested the use of fine-needle aspirates and flow cytometry to analyse the local immune cells in the liver,45 46 which showed good clinical manoeuvrability. Our data provide an in-depth insight into liver immunology and might be an essential resource for drug discovery in the future.
Current guidelines recommend observation rather than active treatment for IT patients.47–49 However, it is becoming realised that there might be profound immune alterations at least in some IT patients, which can subsequently link to disease progression. For example, both NK cells and T cells are functionally impaired in IT patients.50 Our study incisively strengthened this notion and highlighted the coordinated dysregulation of MAIT, Treg, Tex and myeloid subsets in the liver of IT patients, which preceded the elevation of aminotransferases and histological alterations. For this reason, we recommend taking the immunological parameters into account for the decision to start antiviral therapy in the IT phase.
Clear differences in immune features between IA and AR phases were unveiled in this study, representing ‘inadequate response’ and ‘successful response’, respectively. First, the sources of expanded liver Tex cells in IA and AR phases were different. Specifically, intrahepatic Tex cells in AR but not IA were observed to be derived from peripheral CX3CR1+ Temra cells. Moreover, AR exhibited stronger expansion of CX3CR1+GNLY+ Tcm cells than IA, which were consistently observed in both liver and blood, suggesting that CX3CR1+GNLY+ Tcm cells may serve as a candidate marker for prediction of HBV clearance. Second, Tex cells in IA and AR displayed different patterns of cellular interaction. In particular, Tex showed more significant interaction with Treg and FCGR3A+ macrophage in IA than that in AR, suggesting potential roles of these cell–cell communications in shaping CD8+ T-cell dysfunction and viral persistence in CHB. Furthermore, several L-R pairs, such as HLA-B_CANX and RPS19_C5aR1, were potentially involved in mediating the interactions of Tex cells with Treg and/or FCGR3A+ macrophages, respectively. Third, the CD4T_c09-CXCL13 subset was preferentially expanded in AR. CD4T_c09-CXCL13 is referred to a CXCL13-producing PD-1hiCXCR5− (Tfh-like or peripheral helper) CD4+ T-cell cluster, which was first described in synovial tissues of rheumatoid arthritis,51 promoting the formation of tertiary lymphoid-like structures and B-cell responses.52 53 Taken together, the different immune features underlying the active hepatitis of IA and AR might be leveraged to the understanding of active hepatitis in various clinical settings, including on-treatment flares, treatment withdrawal flares, reactivation flares with immunosuppressive therapy, etc, and further to optimal management.
In summary, our study revealed the intrahepatic landscape of immune cells across different phases of HBV infection. Despite the limitations from the cross-sectional design with small sample size and lack of hepatocytes and CD45 negative non-parenchymal cells, our data will serve as a rich resource to gain a deeper understanding of immune responses in HBV infection and might provide valuable insights for the development of novel immunotherapeutic strategies for HBV cure.
Methods
Study subjects
The present study recruited patients with CHB and patients with resolved HBV infection from the Fifth Medical Centre of Chinese PLA General Hospital. Patients were classified into different clinical phases of HBV infection according to the European Association for the Study of the Liver guideline of 2017,49 which considers the presence of HBeAg, HBV DNA levels, transaminase levels (ALT and AST) and the presence or absence of liver inflammation. The time window to enrol acute HBV patients was ‘just achieved or about to achieve HBsAg negativity’ (online supplemental figure 1B), which enables us to observe the ongoing immune responses against HBV, with good safety for liver biopsy. Patients would not be enrolled for the study if they were pregnant or had concomitant hepatitis C or D virus, HIV infection, Wilson’s disease, autoimmune hepatitis, primary biliary cholangitis, non-alcoholic steatohepatitis, significant alcohol intake or history of HCC. Control liver tissue was obtained from livers procured from deceased donors deemed acceptable for liver transplantation from the Department of Organ Transplantation, Tianjin First Central Hospital. The basic characteristics of the enrolled subjects are listed in table 1 and online supplemental table 1.
Sample collection and cell isolation
PBMCs were isolated using the HISTOPAQUE-1077 (Sigma-Aldrich) solution according to the manufacturer’s instructions. Briefly, 2 mL fresh peripheral blood was collected prior to surgery in EDTA anticoagulant tubes and subsequently layered onto HISTOPAQUE-1077. After centrifugation, immune cells remained at the plasma-HISTOPAQUE-1077 interface and were transferred to a new tube, with two times washing using phosphate-buffered saline (PBS) (Gibco). Cells were treated with a red blood cell lysis buffer (Miltenyi Biotec) on ice for 1~2 min. After being washed twice, the cell pellets were resuspended in the fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 2% fetal bovine serum (FBS)) at the approximate concentration of 106 cells/mL.
Liver tissue samples were obtained through ultrasound-guided percutaneous liver biopsy with 21G needle and treated as previously described.16 Briefly, tissues were cut into small pieces in the RPMI-1640 medium (Invitrogen) with 10% FBS (ScienCell), and enzymatically digested with MACS tumour dissociation kit (Miltenyi Biotec) for 60 min on a rotor at 37°C, according to the manufacturer’s instructions. Cells were then stained with APC anti-human CD45 antibody (clone HI30, BioLegend) and 7-AAD (ThermoFisher). Live CD45+ cells were sorted into 0.2 mL low binding tubes (Eppendorf) with a BD FACSAria III instrument (BD Biosciences). Representative images of flow cytometry sorting are shown in online supplemental figure 1A). The availability of cells was generally above 90% and the percentages for CD45+ fraction were about 60%–80%.
Library preparation and sequencing (10× Genomics)
Library construction was performed using the 10× Chromium Single-cell 5′ and VDJ Library Construction Kits V.1.1 (10× Genomics, Pleasanton) as per the manufacturer’s protocol. Purified libraries were analysed by an Illumina Hiseq 4000 sequencer with 150 bp paired-end reads.
Single-cell RNA-seq data processing
The Cell Ranger toolkit V.3.1.0 provided by 10× Genomics was applied to align reads and generate the gene-cell unique molecular identifier (UMI) matrix against the reference genome GRCh38 downloaded from 10× Genomics official website. For each cell, we quantified the number of genes and UMIs and kept high-quality cells with 600–10 000 genes detected and no more than 10% of mitochondrial gene counts.54
Unsupervised clustering analysis and identification of signature genes
To comprehensively assign cell clusters, we performed three rounds of clustering for most of the cell types. Each round of clustering starts from a raw count matrix. Using Scanpy V.1.4.3, we normalised raw counts and transformed these into logarithmic scales. Highly variable genes were selected using the function sc.pp.highly_variable_genes with the parameter ‘batch_key’ set as patient ID. Principal component analysis (PCA) matrix was then calculated and the top 50 PCA components were used to build a batch balanced k nearest neighbour graph using the BBKNN algorithm.55 With the batch effects adjusted, cell clusters were identified by Leiden algorithm.56 DEGs were identified using the Scanpy function sc.tl.rank_genes_group with the parameter ‘use_raw=True’ based on clusters.
Based on the DEGs for each cluster, the first round of clustering identified major cell types, such as T cells, NK cells, B cells, myeloid cells and non-immune cells. An additional round of clustering was performed within each major cell type. The cells of each major cell type were extracted and clustered at a relatively large resolution (2–10). The resulting clusters with more than 10% of cells with doublet scores larger than 0.5 (defined by Scrublet57) were flagged and further examined by the expression of canonical markers. The cells from the flagged cluster would be identified as doublet if they expressed more than one set of canonical markers. Doublets and non-immune cells were then excluded, resulting in 243 000 cells for further analysis. A third round of clustering was then performed to identify the subtypes of cell, following a similar process as the first round within each major type of cells. For cDC2 cells, a fourth round of clustering was also performed to investigate the detailed status.
Dimensionality reduction
For visualisation, the dimensionality of each dataset was further reduced using either the Barnes-Hut t-distributed stochastic neighbour embedding or uniform manifold approximation and projection (UMAP) with Seurat 58 functions RunTSNE and RunUMAP on R and scanpy 59 tool kits with functions sc.pl.umap and sc.pl.tsne on Python. The principle components used to calculate the embedding were the same as those used for clustering.
TCR/B-cell receptor (BCR) analysis
The TCR/BCR-seq sequencing data were processed using the Cell Ranger vdj pipeline with GRCh38 as reference. In all TCR/BCR contigs assembled, we first discarded the low-confidence, non-productive or those UMIs<2. For T cells with two or more α or β chains assembled, the α–β pair showing the highest expression level (UMI) was defined as the dominant α–β pair in the corresponding T cell. If two or more cells had identical dominant α–β pairs, the dominant α–β pairs were identified as clonal TCRs, and these T cells were identified as clonal T cells. To integrate TCR results with the gene expression data, the TCR-based analysis was performed only for cells that were identified as T cells.
A similar protocol was applied to identify BCR clones. For B cells with two or more heavy or light (kappa or lambda) chains assembled, the heavy-light chain with the highest expression level (UMI) was defined as the dominant heavy–light pair.
TCR clonal information and the cell cluster annotations were then used to perform the STARTRAC analysis as previously described.33 In brief, the degree of clonal expansion, migration and transition of T-cell clusters were determined using three STARTRAC indices, STARTRAC-expa, STARTRAC-migr and STARTRAC-tran, respectively. For a detailed pipeline, please refer to the website (https://github.com/Japrin/STARTRAC).
Cell–cell interaction analysis by CSOmap
CSOmap34 was used to construct a three-dimensional (3D) pseudo space and calculate the cell–cell interaction based on our scRNA-seq data. For each sample, CSOmap was applied individually to reconstruct the cell spatial organisation based on the corresponding scRNA-seq data. Then, a distance matrix among cells was calculated based on the 3D coordinates generated by CSOmap. The shorter the distance of a pair of cells was, the closer the cells were located in the 3D space. A connection matrix was further constructed for cell clusters, of which the connections of a specific pair of cell clusters was defined as the number of cell pairs that belonged to the specific cell clusters with their distance smaller than the cut-off estimated by CSOmap according to the median number of cell neighbours. For a cluster pair, normalised connection was further calculated by dividing its corresponding connection value by the product of their respective cell numbers and then multiplying 10 000 to eradicate the potential biases resulting from cell numbers of different cell clusters. Meanwhile, to highlight the key L-R pairs underlying the cellular interactions, we also examined the contribution of each L-R pair to the cell–cell affinity estimation with the function provided by CSOmap. During the analysis, both MATLAB and R version (https://github.com/lijxug/CSOmapR) of CSOmap were used.
Definition of functional scores
To illustrate the functional properties of each cell type, we collected sets of genes from the literature and calculated functional scores for each cell. For T cells, exhaustion-related and naiveness-related genes were defined as in Guo et al 24; effector genes were extracted from Herbst et al 60; inflammatory genes were previously defined in Spranger et al 61; and regulatory genes were from Zemmour et al.62 As for macrophages, the genes for Kupffer score and MoMF score were adapted from Guillot, et al,63 and genes for phagocytosis and proinflammatory were extracted from.14 14 Each score was defined as the mean expression of function-related genes (normalised by z-score). The genes for each score are listed in online supplemental table 4.
Tissue enrichment analysis
To quantify the enrichment of immune cell types across different tissues, we compared the observed and expected cell numbers for each cluster in each tissue according to the following formula as we previously described,24 33 Ro/e=(observed/expected), where the expected cell numbers of immune cell clusters in a given tissue were calculated from the Chi-square test. We assumed that one cluster was enriched in a specific tissue if Ro/e>1. Fisher exact tests were applied and p-values were adjusted using Bonferroni method to acquire the significance levels in online supplemental figure 3C.
Correlations between proportion of cell types and clinical features
We investigate the relationship between proportions of cell types and clinical features by fitting linear models. For each cell type and each clinical feature, a linear model was fitted with the proportion of cell type as the predictor and the values of the clinical feature as the response variable using R function lm. Cell types with the output coefficient ‘Pr(>t)’ <0.05 were considered significant.
Flow cytometric analysis
For phenotypicl staining in figure 6E, PBMCs were stained extracellularly using primary antibodies for specific detection of surface markers for 30 min at 4°C. The cells were then washed with FACS buffer. For subsequent intracellular staining, the cells were permeabilised, fixed and stained using the eBioscience Permeabilization/Fixation Kit according to the manufacturer’s instructions. The cells were then incubated for 30 min at 4°C with antibodies for specific detection of intracellular antigens. Information for the antibodies and reagents used is listed in online supplemental table 8. Samples were analysed by flow cytometry using a BD Canto II flow cytometer (BD Biosciences) with the FlowJo software (BD Biosciences).
gutjnl-2021-325915supp009.xlsx (12.1KB, xlsx)
For spectrum flow cytometric analysis in online supplemental figure 6B, C, cryopreserved PBMCs were thawed and incubated in RPMI 1640 media containing 10% FBS at 37°C for 3 hours resting. PBMCs were then washed twice in PBS and incubated in Fc block and True Stain Monocyte Blocker for 5 min at room temperature. Cells were stained with surface antibodies for 15 min at 4°C and intracellular antibodies for 30 min at 4°C with Brilliant Buffer Plus (eBioscience) after fixation and permeabilisation for 30 min at room temperature. The information for antibodies is listed in online supplemental table 8). Data were acquired on a five-laser Aurora spectral flow cytometer (Cytek) then analysed using FlowJo software V.10.8.1 (BD Biosciences). For UMAP generation and visualisation as in online supplemental figure 6B, FCS files from one IA, one AR and one CR patients were used for downstream analysis. First, the FlowAI plugin was used to remove aberrant events. CD8+ T cells in each sample were randomly down-sampled to 100 000 cells using the DownSample plugin, and all the downsampled CD8+ T cells were concatenated into a single FCS file. The UMAP plugin was used to calculate the UMAP coordinates (with 15 neighbours, metric=euclidian and minimum distance=0.5 as default parameters). Then the FlowSOM plugin was used in parallel on the same dataset to create a self-organising map (using n=12 clusters as default parameter). Cell populations were annotated according to conventional gating strategy and colour coded as indicated.
Multicolour IHC
Fresh tissues were fixed in 4% neutral buffered formalin and embedded in paraffin. Multiplex immunofluorescence staining was performed using PANO 7-plex IHC Kit (cat#0004100100, Panovue) according to the manufacturer’s protocol. In brief, tissue sections (4 µm) were melted at 60 °C for 1 hour followed by deparaffinisation and rehydration. Heat-mediated antigen retrieval was performed in citrate acid buffer (pH 6.0) using microwave incubation. After incubation with blocking buffer at room temperature for 10 min, the sections were incubated with different primary antibodies, including CD4 (ZM0418, Zsbio), CD8 (CST70306, CST), FOXP3 (BLG320202, BioLegend), PD-1 (CST43248, CST), CD68 (CST76437, CST), LILRB2 (AF2078, R&D) and HLA-B (17 260–1-AP, ProteinTech). Sections were incubated for 10 min at room temperature with an anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibody. The stained signals were further amplified using tyramide signal amplification reagents (0004100100, Panovue). Finally, nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). The stained slides were scanned with the Mantra System (PerkinElmer).
Acknowledgments
We thank the contribution by the BIOPIC high-throughput sequencing facility (Peking University), and the Computing Platform of the Center for Life Science (Peking University). We thank Junqing Luan (Chinese PLA General Hospital) and Xin Wang (Tianjin First Central Hospital) for the management of patients. We thank Chunbao Zhou (Chinese PLA General Hospital) for his technical assistance with FACS.
Footnotes
Correction notice: This article has been corrected since it published Online First. The supplementary figure file has been replaced.
Contributors: F-SW is responsible for the overall content as the guarantor. F-SW, ZZ and XR conceived and supervised this study. CZ, YC, FM, J-WS, XF, HF, JL, S-YW, Y-JF and XH performed sample preparation, library construction and data generation. JL performed bioinformatics analyses under the supervision of XR and ZZ. CZ, JL and XR wrote the manuscript. All authors read and approved the final manuscript.
Funding: This work was supported by National Natural Science Foundation of China (grant numbers 81721002, 32 022 016 and 82130019), Outstanding Youth Training Fund of the Chinese PLA General Hospital (grant number 2019-JQPY-009) and the National Key Research and Development Program of China (grant number 2019YFC0840704).
Competing interests: ZZ is a founder of Analytical Bioscience and an advisor for InnoCare. All financial interests are unrelated to this study. The remaining authors declare no competing interests.
Patient and public involvement: Patients and/or the public were not involved in the design, conduct, reporting or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information, except for the raw data for single-cell RNA sequencing reported in this publication can be accessed under the Gene Expression Omnibus (GSE182159) and the Genome Sequence Archive (https://ngdc.cncb.ac.cn, accession number HRA001730) on request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the ethical review community of the Fifth Medical Center of Chinese PLA General Hospital (number 2018-006-D). The participants or their guardians gave written informed consent to participate in the study before taking part.
References
- 1. Seto W-K, Lo Y-R, Pawlotsky J-M, et al. Chronic hepatitis B virus infection. The Lancet 2018;392:2313–24. 10.1016/S0140-6736(18)31865-8 [DOI] [PubMed] [Google Scholar]
- 2. Thomas DL. Global elimination of chronic hepatitis. N Engl J Med 2019;380:2041–50. 10.1056/NEJMra1810477 [DOI] [PubMed] [Google Scholar]
- 3. Gehring AJ, Protzer U. Targeting innate and adaptive immune responses to cure chronic HBV infection. Gastroenterology 2019;156:325–37. 10.1053/j.gastro.2018.10.032 [DOI] [PubMed] [Google Scholar]
- 4. Wang Fu‐Sheng, Fan Jian‐Gao, Zhang Z, Wang FS, Fan JG, et al. The global burden of liver disease: the major impact of China. Hepatology 2014;60:2099–108. 10.1002/hep.27406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Z, Zhang J-Y, Wang L-F, et al. Immunopathogenesis and prognostic immune markers of chronic hepatitis B virus infection. J Gastroenterol Hepatol 2012;27:223–30. 10.1111/j.1440-1746.2011.06940.x [DOI] [PubMed] [Google Scholar]
- 6. Ferrari C. Hbv and the immune response. Liver Int 2015;35:121–8. 10.1111/liv.12749 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Z, Zhang S, Zou Z, et al. Hypercytolytic activity of hepatic natural killer cells correlates with liver injury in chronic hepatitis B patients. Hepatology 2011;53:73–85. 10.1002/hep.23977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tseng T-C, Huang L-R. Immunopathogenesis of hepatitis B virus. J Infect Dis 2017;216:S765–70. 10.1093/infdis/jix356 [DOI] [PubMed] [Google Scholar]
- 9. Pallett LJ, Burton AR, Amin OE, et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J Exp Med 2020;217. 10.1084/jem.20200050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartsch LM, Damasio MPS, Subudhi S, et al. Tissue-Resident memory T cells in the Liver—Unique characteristics of local specialists. Cells 2020;9:2457. 10.3390/cells9112457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Zhu P, Wang W, et al. Mucosal‐Associated invariant T cell dysregulation correlates with conjugated bilirubin level in chronic HBV infection. Hepatology 2021;73:1671–87. 10.1002/hep.31602 [DOI] [PubMed] [Google Scholar]
- 12. Peng H, Sun R. Liver-resident NK cells and their potential functions. Cell Mol Immunol 2017;14 10.1038/cmi.2017.72. [Epub ahead of print: 18 Sep 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacParland SA, Liu JC, Ma X-Z, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aizarani N, Saviano A, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199–204. 10.1038/s41586-019-1373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gola A, Dorrington MG, Speranza E, et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 2021;589:131–6. 10.1038/s41586-020-2977-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng C, Zheng L, Yoo J-K, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:e16:1342–56. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Q, He Y, Luo N, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 2019;179:e20:829–45. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 18. Xue R, Chen L, Zhang C, et al. Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell 2019;35:932–47. 10.1016/j.ccell.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobie R, Wilson-Kanamori JR, Henderson BEP, et al. Single-Cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep 2019;29:1832–47. 10.1016/j.celrep.2019.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575:512–8. 10.1038/s41586-019-1631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krenkel O, Hundertmark J, Abdallah AT, et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 2020;69:551–63. 10.1136/gutjnl-2019-318382 [DOI] [PubMed] [Google Scholar]
- 22. Ghosh A, Mondal RK, Romani S, et al. Persistent gamma delta T-cell dysfunction in chronic HCV infection despite direct-acting antiviral therapy induced cure. J Viral Hepat 2019;26:1105–16. 10.1111/jvh.13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Visentini M, Del Padre M, Colantuono S, et al. Long‐lasting persistence of large B‐cell clones in hepatitis C virus‐cured patients with complete response of mixed cryoglobulinaemia vasculitis. Liver International 2019;39:628–32. 10.1111/liv.14053 [DOI] [PubMed] [Google Scholar]
- 24. Guo X, Zhang Y, Zheng L, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018;24:978–85. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- 25. Wolf FA, Hamey FK, Plass M, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 2019;20:59. 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. La Manno G, Soldatov R, Zeisel A, et al. Rna velocity of single cells. Nature 2018;560:494–8. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stelma F, de Niet A, Sinnige MJ, et al. Human intrahepatic CD69 + CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity. Sci Rep 2017;7:1–10. 10.1038/s41598-017-06352-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim JH, Han JW, Choi YJ, et al. Functions of human liver CD69+CD103-CD8+ T cells depend on HIF-2α activity in healthy and pathologic livers. J Hepatol 2020;72:1170–81. 10.1016/j.jhep.2020.01.010 [DOI] [PubMed] [Google Scholar]
- 29. Pallett LJ, Davies J, Colbeck EJ, et al. IL-2high tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J Exp Med 2017;214:1567–80. 10.1084/jem.20162115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheng Y, Gunasegaran B, Singh HD, et al. Non-terminally exhausted tumor-resident memory HBV-specific T cell responses correlate with relapse-free survival in hepatocellular carcinoma. Immunity 2021;54:e7:1825–40. 10.1016/j.immuni.2021.06.013 [DOI] [PubMed] [Google Scholar]
- 31. Wald O, Pappo O, Safadi R, et al. Involvement of the CXCL12/CXCR4 pathway in the advanced liver disease that is associated with hepatitis C virus or hepatitis B virus. Eur J Immunol 2004;34:1164–74. 10.1002/eji.200324441 [DOI] [PubMed] [Google Scholar]
- 32. Rha M-S, Jeong HW, Ko J-H, et al. PD-1-Expressing SARS-CoV-2-Specific CD8+ T Cells Are Not Exhausted, but Functional in Patients with COVID-19. Immunity 2021;54:e3:44–52. 10.1016/j.immuni.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- 34. Ren X, Zhong G, Zhang Q, et al. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res 2020;30:763–78. 10.1038/s41422-020-0353-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y, Ma D, Wang X. Calnexin Impairs the Antitumor Immunity of CD4(+) and CD8(+) T Cells. Cancer Immunol Res 2019;7:123–35. 10.1158/2326-6066.CIR-18-0124 [DOI] [PubMed] [Google Scholar]
- 36. McDonald-Hyman C, Muller JT, Loschi M, et al. The vimentin intermediate filament network restrains regulatory T cell suppression of graft-versus-host disease. J Clin Invest 2018;128:4604–21. 10.1172/JCI95713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klement JD, Paschall AV, Redd PS, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest 2018;128:5549–60. 10.1172/JCI123360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang C, Wang X-M, Li S-R, et al. Nkg2A is a NK cell exhaustion checkpoint for HCV persistence. Nat Commun 2019;10:1507. 10.1038/s41467-019-09212-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li F, Wei H, Wei H, et al. Blocking the natural killer cell inhibitory receptor NKG2A increases activity of human natural killer cells and clears hepatitis B virus infection in mice. Gastroenterology 2013;144:392–401. 10.1053/j.gastro.2012.10.039 [DOI] [PubMed] [Google Scholar]
- 40. Shiroishi M, Tsumoto K, Amano K, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A 2003;100:8856–61. 10.1073/pnas.1431057100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le Bert N, Gill US, Hong M, et al. Effects of hepatitis B surface antigen on virus-specific and global T cells in patients with chronic hepatitis B virus infection. Gastroenterology 2020;159:652–64. 10.1053/j.gastro.2020.04.019 [DOI] [PubMed] [Google Scholar]
- 42. Aliabadi E, Urbanek-Quaing M, Maasoumy B, et al. Impact of HBsAg and HBcrAg levels on phenotype and function of HBV-specific T cells in patients with chronic hepatitis B virus infection. Gut 2022;71:2300–12. 10.1136/gutjnl-2021-324646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fumagalli V, Di Lucia P, Venzin V, et al. Serum HBsAg clearance has minimal impact on CD8+ T cell responses in mouse models of HBV infection. J Exp Med 2020;217. 10.1084/jem.20200298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamauchi T, Hoki T, Oba T, et al. T-Cell CX3CR1 expression as a dynamic blood-based biomarker of response to immune checkpoint inhibitors. Nat Commun 2021;12:1402. 10.1038/s41467-021-21619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gill US, Pallett LJ, Thomas N, et al. Fine needle aspirates comprehensively sample intrahepatic immunity. Gut 2019;68:1493–503. 10.1136/gutjnl-2018-317071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gill US, Pallett LJ, Kennedy PTF, et al. Liver sampling: a vital window into HBV pathogenesis on the path to functional cure. Gut 2018;67:767–75. 10.1136/gutjnl-2017-314873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 2016;10:1–98. 10.1007/s12072-015-9675-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560–99. 10.1002/hep.29800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49., European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu, European Association for the Study of the Liver . EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370–98. 10.1016/j.jhep.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 50. Wang W-T, Zhao X-Q, Li G-P, et al. Immune response pattern varies with the natural history of chronic hepatitis B. World J Gastroenterol 2019;25:1950–63. 10.3748/wjg.v25.i16.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manzo A, Vitolo B, Humby F, et al. Mature antigen-experienced T helper cells synthesize and secrete the B cell chemoattractant CXCL13 in the inflammatory environment of the rheumatoid joint. Arthritis Rheum 2008;58:3377–87. 10.1002/art.23966 [DOI] [PubMed] [Google Scholar]
- 52. Rao DA, Gurish MF, Marshall JL, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 2017;542:110–4. 10.1038/nature20810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pitzalis C, Jones GW, Bombardieri M, et al. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat Rev Immunol 2014;14:447–62. 10.1038/nri3700 [DOI] [PubMed] [Google Scholar]
- 54. Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:281–91. 10.1016/j.cels.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Polański K, Young MD, Miao Z, et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 2020;36:964–5. 10.1093/bioinformatics/btz625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 2019;9:1–12. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:e9:281–91. 10.1016/j.cels.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Herbst RS, Soria J-C, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231–5. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 62. Zemmour D, Zilionis R, Kiner E, et al. Single-Cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol 2018;19:291–301. 10.1038/s41590-018-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guillot A, Tacke F. Liver macrophages: old dogmas and new insights. Hepatol Commun 2019;3:730–43. 10.1002/hep4.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-325915supp001.pdf (9.8MB, pdf)
gutjnl-2021-325915supp002.xlsx (16.4KB, xlsx)
gutjnl-2021-325915supp003.xlsx (23KB, xlsx)
gutjnl-2021-325915supp004.xlsx (1.6MB, xlsx)
gutjnl-2021-325915supp005.xlsx (17.6KB, xlsx)
gutjnl-2021-325915supp006.xlsx (10.7MB, xlsx)
gutjnl-2021-325915supp007.xlsx (37.3KB, xlsx)
gutjnl-2021-325915supp008.xlsx (12.2KB, xlsx)
gutjnl-2021-325915supp009.xlsx (12.1KB, xlsx)
Data Availability Statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information, except for the raw data for single-cell RNA sequencing reported in this publication can be accessed under the Gene Expression Omnibus (GSE182159) and the Genome Sequence Archive (https://ngdc.cncb.ac.cn, accession number HRA001730) on request.