Summary
Background
Identification of tumor dependencies is important for developing therapeutic strategies for liver cancer.
Methods
A genome-wide CRISPR screen was performed for finding critical vulnerabilities in liver cancer cells. Compounds screen, RNA sequencing, and human phospho-receptor tyrosine kinase arrays were applied to explore mechanisms and search for synergistic drugs.
Findings
We identified mitochondrial translation-related genes associated with proliferation for liver cancer cells. Tigecycline induced deficiency of respiratory chain by disturbing mitochondrial translation process and showed therapeutic potential in liver cancer. For liver cancer cells extremely insensitive to tigecycline, a compounds screen was applied to identify MEK inhibitors as synergistic drugs to tigecycline-insensitive liver cancer cells. Mechanistically, sustained activation of EGFR-ERK1/2-MYC cascade conferred the insensitivity to tigecycline, which was mediated by enhanced secretion of EREG and AREG. Moreover, glycolytic enzymes, such as HK2 and PKM2 were upregulated to stimulate glycolysisin a MYC-dependent manner. Tigecycline induced respiratory chain deficiency in combination with cutting off EGFR-ERK1/2-MYC cascade by MEK inhibitors or EGFR inhibitors, resulting in decrease of both oxidative phosphorylation and glycolysis in liver cancer cells.
Interpretation
Our study proved that blocking EGFR-ERK1/2-MYC cascade combined with tigecycline could be a potential therapeutic strategy for liver cancer.
Funding
This work was funded by grants from the National Natural Science Foundation of China (82073039,82222047, 81920108025), Program of Shanghai Academic/Technology Research Leader (22XD1423100), Shanghai Municipal Science and Technology Project (20JC1411100), 111 Project (B21024), Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZDCX20212700, SHSMU-ZDCX20210802) and Shanghai Jiao Tong University School of Medicine (YG2019GD01).
Keywords: CRISPR screen, Tigecycline, MEK, Glycolysis, Oxidative phosphorylation
Abbreviations: AREG, amphiregulin; BP, biological process; CC, cellular component; CCCP, carbonyl cyanide m-chlorophenylhydrazone; CCK8, cell counting kit-8; CRISPR, clustered regularly interspaced short palindromic repeats; DEGs, differentially expressed genes; 2-DG, 2-Deoxy-D-glucose; ECAR, extracellular acidification rate; EGFR, epidermal growth factor receptor; ELISA, enzyme-linked immunosorbent assay; EREG, epiregulin; GO, gene ontology; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazonec; GSVA, gene set variation analysis; HCC, hepatocellular carcinoma; HGFR, hepatocyte growth factor receptor; HK2, hexokinase 2; IC50, half maximal inhibitory concentration; IHC, Immunohistochemistry; LDHA, lactate dehydrogenase A; LIHC, liver hepatocellular carcinoma; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase kinase; NSCLC, non-small-cell lung cancer; OCR, oxygen consumption rate; OS, overall survival; OXPHOS, oxidative phosphorylation; ROS, reactive oxygen species; RTK, receptor tyrosine kinase; TCGA, The Cancer Genome Atlas; TTR, time to relapse
Research in context.
Evidence before this study
The emergence of functional genetic screens based on CRISPR technology provides a solution to identify genetic vulnerabilities that may represent therapeutic opportunities in cancer and other diseases. However, there are few studies that comprehensively identify potential therapeutic vulnerabilities of liver cancer by genome-wide CRISPR screen. Tigecycline is known to have an inhibitory effect on mitochondrial translation in eukaryotic organisms, resulting in defective oxidative phosphorylation. But in cancer cells, inhibition of mitochondrial translation by tigecycline may cause the metabolic switch to glycolysis due to metabolic reprogramming.
Added value of this study
We identified mitochondrial translation-related genes associated with proliferation for liver cancer cells. Tigecycline induced deficiency of respiratory chain by disturbing mitochondrial translation process and showed therapeutic potential in liver cancer. Mechanistically, the increase of autocrine EREG and AREG activated EGFR-ERK1/2 cascade, thereby enhanced the transcription of MYC downstream genes, which increased glycolysis and conferred the insensitivity to tigecycline.
Implications of all the available evidence
We demonstrated that sustained activation of EGFR-ERK1/2 signaling increased glycolysis and conferred the insensitivity to tigecycline in liver cancer cells. Drugs involved in this study have been approved for clinical use or are part of ongoing clinical trials, therefore, clinical testing of the combination of tigecycline and MEK inhibitors or EGFR inhibitors for the treatment of liver cancer should be feasible.
Introduction
Conventional therapies for liver cancer, such as liver transplantation, tumour resection or ablation, are limited to early-stage tumours.1 For patients with advanced stages of liver cancer, first-line systemic therapies are sorafenib, lenvatinib, oratezolizumab plus bevacizumab. The combination of atezolizumab (anti-PD-L1 antibody) and bevacizumab (anti-VEGF antibody) had better overall and progression-free survival outcomes than single-drug treatment with sorafenib. However, the response rate of the combination therapy was only 27.3%.2 Hence, it is urgent to find further therapeutic approaches and strategies for liver cancer.
The complex regulatory networks that control tumor function make identifying therapeutic targets a major challenge. The use of CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein 9) screens has enabled the exploration of cancer driver genes and molecular mechanisms underlying drug resistance on a large scale.3 For example, using a kinome-wide CRISPR-Cas9 genetic functional screen, we found that activation of epidermal growth factor receptor (EGFR) limits the response of liver cancer to lenvatinib. The combined regimen of the EGFR inhibitor gefitinib and lenvatinib provided a strategy for liver cancer patients with high expression of EGFR.4 Recently, identifying cancer type-specific dependencies is becoming another critical strategy for cancer therapy. The CRISPR-Cas9 screens can provide new insights into therapeutic vulnerabilities of liver cancer. Using a kinome-focused genetic screen, we showed that pharmacological inhibition of the DNA-replication kinase CDC7 induces senescence selectively in liver cancer cells with mutations in TP53.5 In addition, Liang and colleagues performed a genome-wide CRISPR-Cas9 screen and identified WEE1 kinase as a therapeutic vulnerability in liver cancer cells depleted of the ATRX chromatin remodeler gene.6 However, potential therapeutic vulnerabilities of liver cancer remain to be investigated.
Here, we aim to further explore the dependencies of liver cancer through performing genome-wide CRISPR-Cas9 screen. Thus, we performed a genome-wide CRISPR screen in two liver cancer cell lines (SNU398 and HepG2) and identified the vital roles of mitochondrial translation-related genes for liver cancer cells. Emerging evidence suggests high level of mitochondrial translation may be required to support the bioenergetic requirements of cancer cells.7 Besides, some studies showed that abnormal mitochondrial translation was involved in the process of epithelial-to-mesenchymal transition, cell cycle progression and apoptosis signaling.8,9 Therefore, therapeutic approaches that disrupt mitochondrial translation are showing great potentials in combating tumours.
Tigecycline, a FDA-approved broad spectrum antibiotic, can selectively inhibit mitochondrial translation.10,11 Previous studies showed that tigecycline could be used to treat hematological tumors by inhibition of mitochondrial translation.11,12 Limited studies only showed that tigecycline could enhance the activity of chemotherapy in liver cancer, and prevent the tumor relapse after resistance to sorafenib.13,14 However, the roles of mitochondrial translation-related genes and the rationale of tigecycline as a strategy for the treatment of liver cancer are still unclear.
In this study, we report that the sustained activation of EGFR-ERK1/2 signaling pathway confers insensitivity to tigecycline therapy in liver cancer. Tigecycline in combination with MEK inhibitors or EGFR inhibitors achieves synergistic effects both in vitro and in vivo. Therefore, our findings suggest a therapeutic approach for liver cancer, which is a combination of tigecycline and MEK inhibitors or EGFR inhibitors.
Methods
Ethics statement
Ethical approval was gained from the Eastern Hepatobiliary Hospital Research Ethics Committee (EHBHKY2014-03-006) and conducted in accordance with the Helsinki Declaration. All patients provided their written informed consent. All animal experiments complied with the ARRIVE guidelines and were operated according to protocols approved by the Ren Ji Hospital Institutional Animal Care Committee and Shanghai Cancer Institute (DWLB102).
Cells and cell culture conditions
SNU398, Huh6, HepG2, SNU449, SNU423, SNU387, Huh7, Hep3B, PLC/PRF/5, Li7 and MHCC97H cells were cultured in DMEM with 10% FBS, glutamine and penicillin-streptomycin (Gibco) at 37 °C and 5% CO2. All cells were authenticated by Short Tandem Repeat (STR) profiling. Mycoplasma contamination was excluded via a PCR-based method.
Compounds
Tigecycline (S1403), Cobimetinib (S8041), Gefitinib (S1025) and 2-Deoxy-d-glucose (S4701) were purchased from Selleck Chemicals. Trametinib (HY-10999) was purchased from MedChemExpress.
Genome-wide CRISPR screen
The genome-wide CRISPR library, around 75,000 sgRNAs (along with 1000 non-targeting controls) targeting 19,114 human genes, including 50 negative genes (non-essential genes) and 50 positive genes (essential genes), was applied. By lentiviral transduction, the genome-wide CRISPR library was ferried into SNU398 and HepG2 cells (three independent replicates). The coverage of the genome-wide library screening was 800. After puromycin selection (2 μg/ml), cells were treated with trypsin and counted for T0 (day 0) samples and Tu (day 14) samples incubated for another 14 days. Genomic DNA isolation was performed by the ZYMO gDNA isolation kit according to the manufacturer's protocol. After PCR amplification, gRNA barcodes from T0 and Tu were detected by deep sequencing in an Illumina HiSeq-2500. A differential test between T0 and Tu for each sgRNA was conducted by DESeq2. The MAGeCK Robust Rank Algorithm was used to calculate to represent sgRNA prioritization. The detailed method was described in our previous study.4
Short hairpin RNA (shRNA)-mediated knockdown
Lentiviral vectors containing shRNA against GFM1, MRPL4, MRPS23, SHMT2, MYC, and HK2 came from the Human TRC shRNA Library. PLKO is TRC cloning vector. Lentivirus transduction was conducted using polyplus regents according to the instructions and followed by drug selection (2 μg/ml puromycin for 3–5 days). The sequences for shRNA were provided in Supplementary Materials and Methods.
Quantitative reverse-transcription PCR
EZ-press RNA Purification Kit (EZBioscience) was used to extract total RNA from cells. cDNA was acquired using Color Reverse Transcription Kit with gDNA remover (EZBioscience). Quantitative PCR reactions were conducted using 2 × Color SYBR Green qPCR Master Mix (EZBioscience). The sequences of the primers for quantitative reverse-transcription PCR (qRT-PCR) were shown in Supplementary Materials and Methods.
Immunohistochemistry (IHC)
Tissue microarray contained 243 hepatocellular carcinoma (HCC) samples from the Naval Military Medical University Affiliated Eastern Hepatobiliary Hospital in Shanghai, China. Diagnosis of HCC was based on clinical manifestations, laboratory and pathological examinations, combined with endoscopic and imaging examinations by experienced clinicians according to clinical guidelines. Patients did not receive any preoperative anticancer therapy, and clinical and follow-up information were completed. The demographic data were self-reported by study participants. The method of IHC staining and H score evaluation referred to our previous study.4 Due to the differences in gene expression, we divided the patients into high and low expression groups according to the median value of H score.15 The demographic data including gender and age of the 243 HCC samples were shown in Table S1.
Gene set variation analysis (GSVA)
GSVA is a method to evaluate the variation of pathway activity in an unsupervised manner.16 The list of 134 mitochondrial translation-related genes was downloaded from http://www.gsea-msigdb.org/gsea/downloads.jsp. For 50 pairs of tumour and corresponding non-tumour tissues in HCC, GSVA scores of mitochondrial translation process of all mitochondrial translation-related genes were estimated by “GSVA” R package. Besides, GSVA score of mitochondrial translation was also calculated in subclusters stratified by 26 mitochondrial translation-related genes.
Long-term and short-term cell growth assays
Long-term cell growth assays
Cells were seeded into six-well plates at a density of 20,000–50,000 cells per well. After static adherence, drugs were added into wells at the given concentrations for 10–14 days. Culture medium was replaced twice a week and drugs were supplemented accordingly. Cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet.
Short-term cell growth assays
Cells were seeded into 96-well plates at a density of 2000–5000 cells per well. After 24 h, drugs were added at the indicated concentrations for three days. Cell Counting Kit-8 (CCK8) (Dojindo) was used to measure the absorbance at 450 nm according to the instructions.
Western blots
Cells were lysed in RIPA buffer contained with phosphatase and protease inhibitor cocktails (Sigma). Protein quantification was conducted using BCA protein assay Kit. Quantified proteins lysates were separated with 10%–15% SDS-PAGE and transferred to PVDF membrane. After blocking with 5% skim milk, the PVDF membranes were incubated with primary antibody (1:1000 dilution) overnight at 2–8 °C on a rocking platform shaker. Secondary antibodies were used at a 1:5000–5,0000 dilution of anti-rabbit or anti-mouse HRP-conjugated secondary antibodies (Bioworld). The information on antibodies applied in this study referred to Supplementary Materials and Methods.
Compounds screen
A FDA-approved drug library containing 1957 compounds (MedChemExpress) was used to perform compounds screen in MHCC97H cells in the initial screen process. MHCC97H cells were seeded into 96-well plates at a density of 3500 cells per well and divided into two groups: control and tigecycline treatment (25 μM). 2 μM compounds were added into each well of these groups for three days respectively. Compared with control group, top 20 compounds that inhibited cell proliferation in tigecycline group were chosen for the second-round screen in MHCC97H (tigecycline, 25 μM), PLC/PRF/5 (tigecycline, 10 μM), and Li7 (tigecycline, 25 μM) cells. The screen was conducted with two replicates of each cell line. Under treatment of compounds, the cell viability of control group was normalized against control conditions (untreated cells) after subtraction of background signal, and tigecycline group was normalized against single tigecycline-treated cells. The proliferation inhibition was calculated between normalized control cells and tigecycline-treated cells under treatment of compounds.
Incucyte cell proliferation assays
MHCC97H, PLC/PRF/5 and Li7 cells were counted and seeded into 96-well plates at a density of 2000–4000 cells per well. After static adherence, drugs were added into wells at the given concentrations. The confluence of cells was monitored automatically using IncuCyte ZOOM every 4 h (Essen Bioscience).
Seahorse analysis
Cells were seeded into XFe96 microplate at the density of 4000–10,000 cells per well. Tigecyline or/and MEK inhibitors were added to wells for three days before Seahorse glycolysis or mitochondrial stress analyses. The Seahorse glycolysis and mitochondrial stress test kits were used in combination with the Seahorse XFe96 Analyzer (Agilent Technologies, Santa Clara, CA) as described by the manufacturer. Glucose (10 mM), oligomycin (1 μM), and 2-DG (50 mM) were used in the glycolysis stress test. Oligomycin (1.5 μM), Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 1 μM), rotenone and antimycin A (0.5 μM) were applied in mitochondrial stress test. Results were analyzed by the Seahorse Wave 2.6.1 software.
RNA sequencing and data analysis
MHCC97H and PLC/PRF/5 cells were treated with DMSO or 10 μM tigecycline for three days followed by RNA-sequencing. Compared tigecycline to control groups, differentially expressed genes (DEGs) were identified and KEGG analysis of these genes was performed. Top enriched pathways and hub genes were visualized using Sankey diagram.
Human phospho-receptor tyrosine kinase arrays
Human phospho-receptor tyrosine kinase (Phospho-RTK) arrays were applied in the detection of kinase signaling after treatment of tigecycline in MHCC97H cells, according to the instructions (R&D).
Enzyme-linked immunosorbent assay (ELISA)
Cell culture supernatants were collected and analyzed for relative autocrine EREG and AREG by ELISA analyses according to the manufacturer's instructions. Human EREG and AREG ELISA kits were purchased from Mlbio.
In vivo experiments
SNU398 and MHCC97H cells (5 × 106 per mouse) were injected into right shoulder of 6-week-old BALB/c nude mice (male, 6 mice per group), and Hepa1-6 cells were injected into right shoulder of 4-6-week-old C57BL/6 mice (male, 6 mice per group). Based on the measurement of vernier caliper, tumour volume was assessed by the modified ellipsoidal formula: tumour volume = ½ length × width2. When tumours grew to about 100 mm3, nude mice were randomly assigned for 5 days/week to receive either solvent, tigecycline (50 mg/kg, intraperitoneal injection), trametinib (0.25 mg/kg, oral gavage), and combination of tigecycline plus trametinib once daily. Immune-competent mice were assigned for 5 days/week to receive either solvent, tigecycline (50 mg/kg, intraperitoneal injection), trametinib (0.25 mg/kg, oral gavage), gefitinib (80 mg/kg, oral gavage), tigecycline plus trametinib, and tigecycline plus gefitinib once daily. In the combination group, drugs with each compound administered at the same dose and schedule as a single drug. Mice were euthanized when they lost more than 15 percent of their body weights or failed to thrive. All mice were placed on a 12 h light/dark cycle and had unlimited access to food and water. Experiments were carried out with the minimization of animal suffering and pain.
Statistics
In vitro and in vivo data are shown as the mean ± SEM. The Student's t-test was applied for statistical discrepancy between two groups. Kaplan-meier analysis was performed to indicate overall survival of patients and the log-rank (Mantel–Cox) test was conducted for statistical difference comparisons.
Role of the funding source
The funding sources were not involved in study design, analysis, interpretation, tissues collection, and submission decision.
Results
A genome-wide CRISPR-Cas9 screen identifies requirement of mitochondrial translation-related genes for liver cancer cells
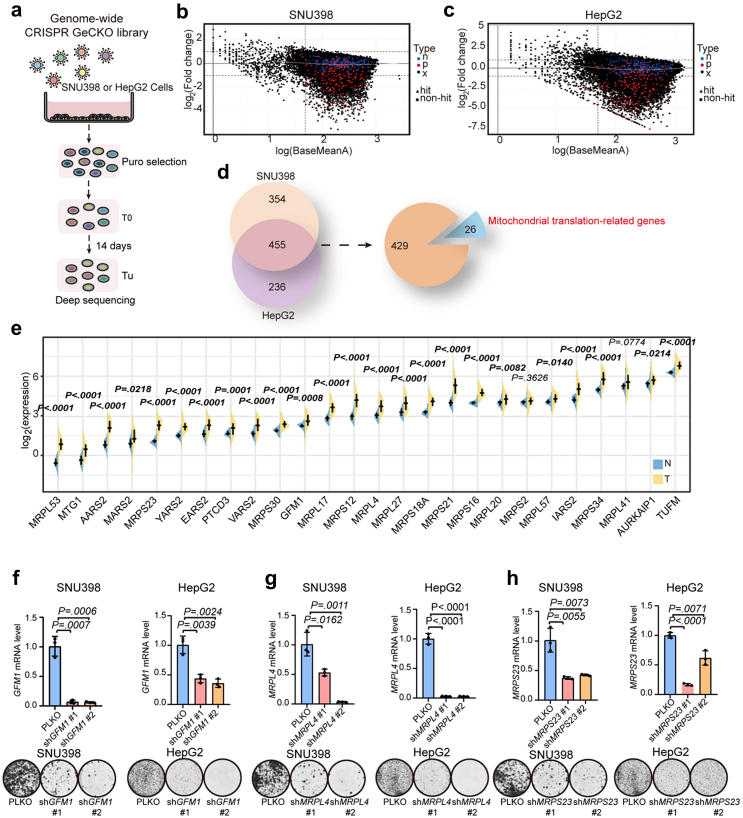
To identify new vulnerabilities of liver cancer cells, we performed a genome-wide CRISPR-Cas9 genetic screen in SNU398 and HepG2 cells (Fig. 1a–c, Fig. S1, Table S2) and identified 455 genes commonly required in the two cells (Fig. 1d). According to the Gene Ontology (GO) gene set enrichment analyses of cellular component (CC) and biological process (BP), mitochondrial ribosome cellular component and mitochondrial translation biological process were highly enriched (Fig. S2a and b). MitoCarta3.0 is an updated inventory of mammalian mitochondrial proteins using multiple experimental and computational approaches.17 By comparing the common hits with the MitoCarta3.0 inventory, we found that 26 of these genes were implicated in mitochondrial translation process (Fig. 1d, Table S3). The process of mitochondrial translation mainly depends on mitochondrial translation factors and mitochondrial ribosomes, which are encoded by nuclear genome and they are called as mitochondrial translation-related genes.18 The differential expression of mitochondrial translation-related genes in The Cancer Genome Atlas (TCGA) liver hepatocellular carcinoma (LIHC) cohort was investigated. 24 mitochondrial translation-related genes were significantly upregulated in 50 tumour (T) tissues compared with corresponding non-tumour (N) tissues (Fig. 1e). Using GSVA to analyse the enrichment scores of the 26 mitochondrial translation-related genes between tumour and non-tumour tissues of HCC in TCGA, we found that these genes were highly enriched in tumour tissues, indicating that mitochondrial translation may be an important pathway for liver cancer cells (Fig. S2c).
Fig. 1.
A genome-wide CRISPR-Cas9 screen identifies requirement of mitochondrial translation-related genes for liver cancer cells. (a) Schematic diagram of the genome-wide screen. SNU398 and HepG2 cells were transduced with a lentiviral genome gRNA library (three independent replicates). gRNA barcodes from T0 (day 0) and Tu (day 14) were detected by deep sequencing. (b–c) Graphical representation for the abundance of the gRNA sequences in the screen of SNU398 (b) and HepG2 (c) cells. The x-axis shows log (Base Mean). The y-axis shows log2(Fold change) (gRNA abundance of day-14 sample/day-0 sample). For one sgRNA, when |log2 (fold change)|>1, false discovery rate (FDR) ≤ 0.1, and log (Base Mean) ≥ 1.7, it was considered as a “hit”. Otherwise, it was a “non-hit”. ‘n’, sgRNAs targeting 50 non-essential genes (blue); ‘p’, sgRNAs targeting 50 essential genes (red); ‘x’, sgRNAs targeting studied human genes (grey). (d) Identification of 26 genes related to mitochondrial translation process in the screen in SNU398 and HepG2 cells. FDR≤ 0.1. (e) Expression analysis of 26 mitochondrial translation-related genes in 50 paired samples of tumour (T) and corresponding non-tumour (N) tissues in liver hepatocellular carcinoma (LIHC) cohort of The Cancer Genome Atlas (TCGA) (P values are calculated by two-tailed paired t test). Values in bold are statistically significant. (f–h) Gene knockdown efficiency and long-term cell growth assays of GFM1 (f), MRPL4 (g), and MRPS23 (h) in SNU398 (left) and HepG2 (right) cells. Data are mean ± SEM (P values are calculated by two-tailed unpaired t test).
Mitochondrial translation products encoded by mitochondrial DNA are only 13 hydrophobic subunits of oxidative phosphorylation system, as well as several tRNAs and rRNAs.19 Thus, detection of protein levels of mitochondrial translation products is a common method to reflect the function of mitochondrial translation.11,20,21 To validate the results of the CRISPR screen, we selected three representative mitochondrial translation-related genes, GFM1 (a mitochondrial translation factor), MRPL4 (a protein of large mitochondrial ribosomal subunit), and MRPS23 (a protein of small mitochondrial ribosomal subunit), by interfering with their expression through shRNA-mediated knockdown. Knockdown of GFM1, MRPL4, or MRPS23 in SNU398 and HepG2 cells resulted in not only short-term and long-term cell proliferation inhibition (Fig. 1f–h, Fig. S3a and b), but also the downregulation of protein levels of translation products, such as CYTB (complex III), MTCO2 (complex IV), and ATP6 (complex V), which are components of the respiratory chain complex (Fig. S3c and d), whereas mRNA levels of these genes of respiratory chain complex were unchanged (Fig. S3e–g), indicating that knockdown of mitochondrial translation-related genes leads to the reduced expression of mitochondrial proteins such as CYTB, MTCO2, and ATP6.
Clinical significance of mitochondrial translation-related genes
To investigate the clinical value of mitochondrial translation-related genes, we collected tumour material and clinical data from 243 HCC patients from the Naval Military Medical University Affiliated Eastern Hepatobiliary Hospital. IHC analyses indicated that the protein levels of GFM1 (P = 0.0005, two-tailed paired t test), MRPL4 (P < 0.0001, two-tailed paired t test), and MRPS23 (P < 0.0001, two-tailed paired t test) were significantly high in the tumour tissues compared to corresponding non-tumour tissues in liver cancer (Fig. 2a–c). Univariate and multivariate analyses for protein levels (as determined by IHC on tumour material) and clinicopathological features showed that GFM1, MRPL4, and MRPS23 were independent prognostic markers of overall survival (OS) and time to relapse (TTR) (Tables S4–S6). Kaplan–Meier survival analyses showed that HCC patients with high expression of GFM1, MRPL4, or MRPS23, had worse OS and shorter TTR (Fig. 2d–i). Using consensus clustering analysis for the 26 genes, the HCC cohort from TCGA could be divided into three subclusters. Patients in the cluster 2 represented generally low levels of 26 mitochondrial translation-related genes and low GSVA score of mitochondrial translation, but high survival rate (Fig. 2j–l). These data support the notion that high expression of mitochondrial translation-related genes is associated with poor outcome in HCC.
Fig. 2.
Prognostic significance of mitochondrial translation-related genes for patients with HCC. (a–c) Differential protein levels of GFM1 (a), MRPL4 (b), or MRPS23 (c) between tumour (T) and corresponding non-tumour (N) tissues in liver cancer from 243 HCC patients (P values are calculated by two-tailed paired t test). (d–i) IHC analyses for GFM1, MRPL4, and MRPS23 in patients with HCC. Representative images for high or low expression of GFM1, MRPL4, and MRPS23 are shown in (d), (f), and (h), respectively. Kaplan–Meier analyses of overall survival (left) and recurrence (right) for patients with HCC were performed based on their protein levels of GFM1 (e), MRPL4 (g), and MRPS23 (i) High (n = 121) or low (n = 122) expression of genes was defined according to their median values of H score. (P values are calculated by log-rank (Mantel–Cox) test.) (j) Identification of HCC subclusters by 26 mitochondrial translation-related genes from the genome-wide CRISPR-Cas9 screen. 193 HCC patients with relatively complete clinical information were from TCGA. Expression level of genes was transformed by log2 with the scale into −4 to 4. (k) Gene set variation analysis (GSVA) score of mitochondrial translation process for the subclusters (Unpaired two-sided t-test). (l) Kaplan–Meier analysis of overall survival according to the subclusters (P values are calculated by log-rank (Mantel–Cox) test).
Tigecycline reduces mitochondrial translation products in liver cancer cells
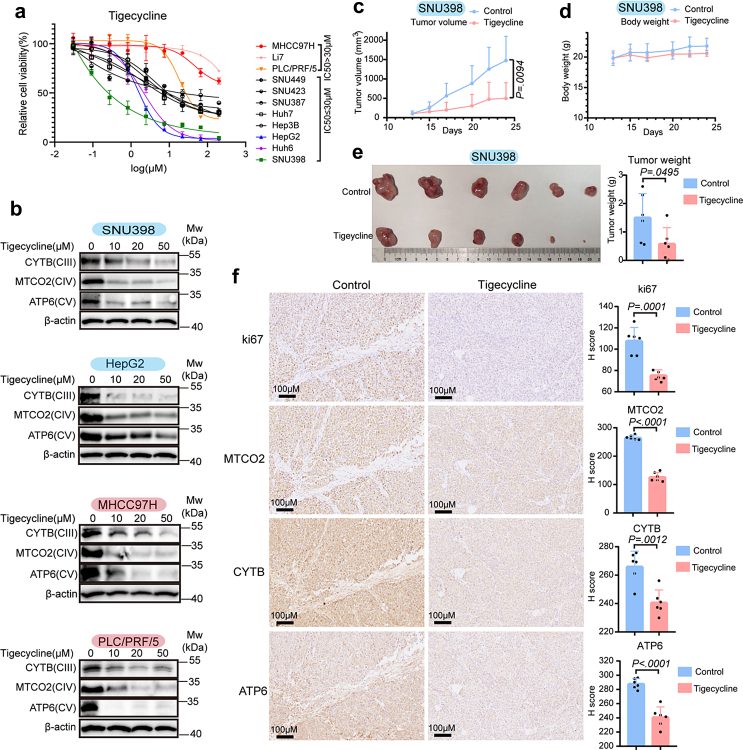
Tigecycline, the only FDA-approved antibiotic of the glycylcyclines, has been shown to specifically inhibit mitochondrial translation.11,22 According to cell viability assays, we found tigecycline had significant activity against the majority of liver cancer cells tested (Fig. 3a). Most liver cancer cells were sensitive to tigecycline, especially SNU398, Huh6, and HepG2 cells. In contrast, PLC/PRF/5, Li7, and MHCC97H cells were considered to be relatively insensitive because their half maximal inhibitory concentration (IC50) exceeded 30 μM (Fig. 3a). Moreover, long-term cell proliferation assays using tigecycline also validated that SNU398, Huh6, and HepG2 cells were sensitive to tigecycline, while PLC/PRF/5, Li7, and MHCC97H cells were not (Fig. S4).
Fig. 3.
Tigecycline inhibits cell proliferation in liver cancer cells. (a) Short-term cell growth assays of tigecycline in liver cancer cell lines. Cells with half maximal inhibitory concentration (IC50) > 30 μM were considered as relatively insensitive cells, while those with IC50 ≤ 30 μM were regarded as sensitive cells. (b) Western blot analyses for subunits of respiratory complex (CIII-CV) from sensitive (SNU398 and HepG2) and insensitive (MHCC97H and PLC/PRF/5) cells treated with increasing concentration of tigecycline. (c) Tumour volumes of subcutaneous xenografts for control (n = 6) and tigecycline-treated groups (50 mg/kg, intraperitoneal injection, n = 6) of SNU398 cells. (d) Body weight for mice of control and tigecycline-treated groups. (e) Tumour image and tumour weight. (f) Representative IHC staining for ki67, MTCO2, CYTB, and ATP6 on sequential slides in xenografts tumour tissues and H score analyses. Scale bar = 100 μM. Data are mean ± SEM (Unpaired two-sided t-test).
Tigecycline could reduce mitochondrial translation products of both sensitive and insensitive cells, as evidenced by the notion that this drug can downregulate protein levels of CYTB, MTCO2, and ATP6 in both sensitive and insensitive cells (Fig. 3b). In vivo experiments showed that tigecycline could inhibit the growth of liver cancer cells without affecting the body weight of mice (Fig. 3c–e). IHC staining analyses confirmed that protein levels of mitochondrially-translated MTCO2, CYTB, and ATP6 were decreased in tumour tissues removed from mice treated with tigecycline (Fig. 3f). Therefore, we conclude that tigecycline reduced the mitochondrial translation products of all liver cancer cells but has major anti-proliferative effects in a subset of liver cancer cells.
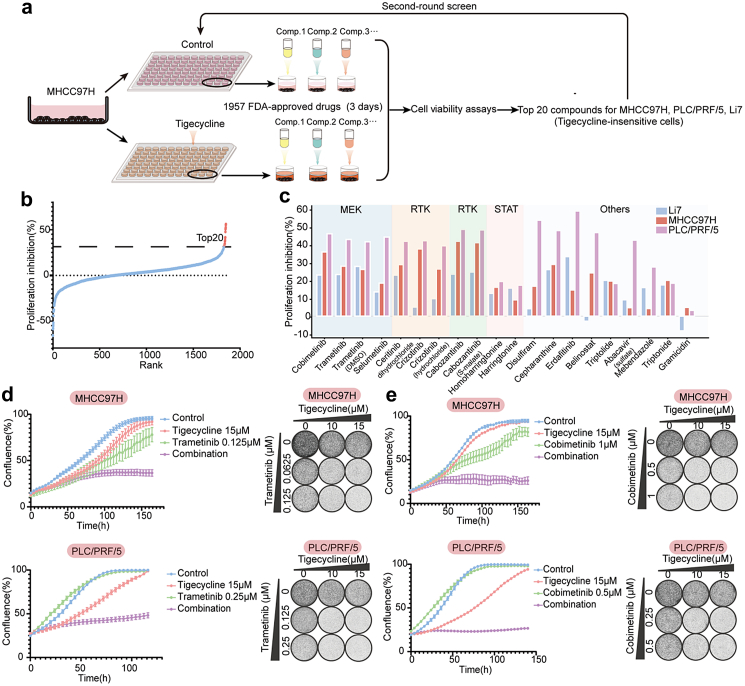
Compound screen identifies MEK inhibitors as synergistic with tigecycline
To find drugs that have synergistic effects of tigecycline in liver cancer cells, we performed a compound screen in tigecycline-insensitive cells (Fig. 4a). The top 20 compounds with synergistic effects in MHCC97H cells were then selected for the second-round screen in three insensitive cell lines: MHCC97H, PLC/PRF/5, and Li7 (Fig. 4b). The second-round screen showed mitogen-activated protein kinase kinase (MEK) inhibitors and receptor tyrosine kinase (RTK) inhibitors such as trametinib, cobimetinib, selumetinib, ceritinib, crizotinib, and cabozantinib had favorable synergies of tigecycline (Fig. 4c). Incucyte cell proliferation, short-term and long-term cell proliferation assays showed that the MEK inhibitors trametinib and cobimetinib increased the sensitivity of MHCC97H, PLC/PRF/5, and Li7 cells to tigecycline (Fig. 4d–e, Fig. S5a–d). However, ceritinib, crizotinib, and cabozantinib had no synergistic effects in long-term cell growth assays (data not shown). Thus, we identify MEK inhibitors as drugs that display synergy with tigecycline.
Fig. 4.
Compound screen identifies MEK inhibitors as synergistic drugs for tigecycline-insensitive cells. (a) Schematic diagram of two-rounds compounds screen. (b) Proliferation inhibition of the first-round screen in MHCC97H cells. (c) Proliferation inhibition between normalized control cells and tigecycline-treated cells with application of top 20 compounds in MHCC97H, PLC/PRF/5, and Li7 cells. Main targets of top 20 compounds are shown on the bar chart. (d) Incucyte cell proliferation assays (left) and long-term cell growth assays (right) showed the synergistic effect of a combined regimen of tigecycline and trametinib in MHCC97H and PLC/PRF/5 cells (n = 3). (e) Incucyte cell proliferation assays (left) and long-term cell growth assays (right) showed the synergistic effect of drug combination of tigecycline and cobimetinib in MHCC97H and PLC/PRF/5 cells (n = 3).
Combined regimen of tigecycline and trametinib inhibits both oxidative phosphorylation and glycolysis
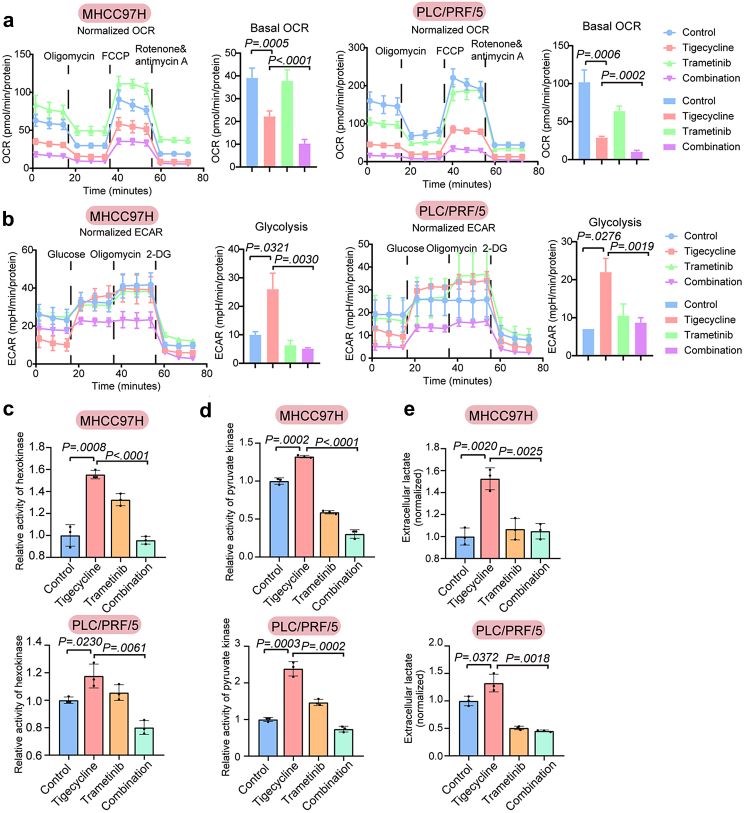
Given that tigecycline inhibited mitochondrial translation, thereby potentially affecting oxidative phosphorylation (OXPHOS), we measured the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of sensitive and insensitive cells treated with tigecycline. These results showed that tigecycline could reduce the basal OCR, maximal respiration and ATP production of OXPHOS of both tigecycline-sensitive and -insensitive cells (Fig. S6a and b). However, the effects on glycolysis were quite different. Compensatory enhancement of glycolysis and glycolytic capacity was only observed in tigecycline-insensitive cells, which might explain the differences in sensitivity of liver cancer cells to tigecycline (Fig. S6c and d). Compared to tigecycline treatment alone, the combination of trametinib and tigecycline could further reduce the basal OCR, maximal respiration, and ATP production of OXPHOS in tigecycline-insensitive cells. Additionally, compensatory enhancement of glycolysis and glycolytic capacity was inhibited (Fig. 5a and b, Fig. S7a–e). Besides, tigecycline caused changes in the activities of key enzymes in glycolysis pathway. After treatment with tigecycline, the activities of hexokinase, pyruvate kinase, and the content of extracellular lactate increased in MHCC97H and PLC/PRF/5 cells, but decreased after addition of trametinib (Fig. 5c–e).
Fig. 5.
Combined regimen of tigecycline and trametinib inhibits both oxidative phosphorylation and glycolysis. (a–b) Oxygen consumption rate (OCR) (a) and Extracellular acidification rate (ECAR) (b) with the regimen of tigecycline and trametinib in MHCC97H and PLC/PRF/5 cells. (c–e) Activity of hexokinase (c) and pyruvate kinase (d), and concentration of extracellular lactate (e) in MHCC97H and PLC/PRF/5 cells with the combined treatment of tigecycline and trametinib. (n = 3). Data are mean ± SEM (P values are calculated by two-tailed unpaired t test). 2-DG, 2-Deoxy-d-glucose; FCCP, Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone.
Tigecycline activates the EGFR-ERK1/2-MYC cascade
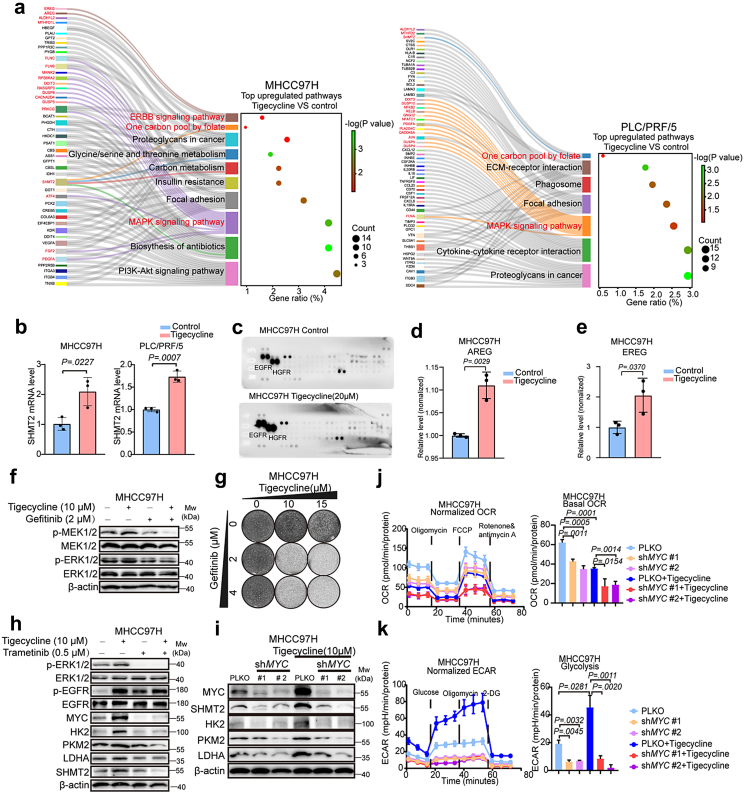
To explore the molecular basis of liver cancer cells insensitivity to tigecycline and how trametinib might enhance this sensitivity, we conducted RNA sequencing of MHCC97H and PLC/PRF/5 cells following treatment with tigecycline. Compared tigecycline to control groups, we found 357 upregulated and 261 downregulated genes in MHCC97H cells, and 603 upregulated and 574 downregulated genes in PLC/PRF/5 cells (Fig. S8a, Tables S7 and S8). KEGG pathways analyses showed shared upregulated pathways and no shared downregulated pathway of MHCC97H and PLC/PRF/5 cells (Fig. 6a, Fig. S8b). Sankey diagrams of upregulated genes showed that tigecycline treatment significantly enriched the mitogen-activated protein kinase (MAPK) signaling pathway and one carbon pool by folate pathway compared to control treated cells (Fig. 6a). qRT-PCR was then performed to investigate the expression of genes involved in one carbon pool by folate pathway, including SHMT2, ALDH1L2, MTHFD1L and MTHFD2. Results showed that mRNA level of SHMT2 was markedly elevated with treatment of tigecycline in MHCC97H and PLC/PRF/5 cells (Fig. 6b, Fig. S8c). These results suggest that the MAPK cascade is enhanced in tigecycline-insensitive cells with treatment of tigecycline and SHMT2 may be involved in the low response of liver cancer cells to tigecycline.
Fig. 6.
Feedback activation of the EGFR-ERK1/2-MYC cascade limits response to tigecycline of liver cancer cells. (a) Sankey Diagram of top upregulated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways on the tigecycline group versus control group for MHCC97H and PLC/PRF/5 cells. (b) mRNA levels of SHMT2 in MHCC97H and PLC/PRF/5 cells with treatment of tigecycline. (c) Relative levels of tyrosine phosphorylation of human receptor tyrosine kinases with treatment of tigecycline. MHCC97H cells were treated with tigecycline (20 μM) for three days and then human Phospho-RTK arrays analyses were performed. (d–e) ELISA measured secretion levels of AREG (d) and EREG (e) in supernatants of MHCC97H cells that were treated for three days with DMSO or 10 μM tigecycline. (f) Western blot analyses of MAPK cascade in MHCC97H cells treated with tigecycline (10 μM), gefitinib (2 μM) or their combination for three days. (g) Long-term cell growth assays for the combined regimen of tigecycline and gefitinib. (h) Biochemical responses of MHCC97H cells with treatment of tigecycline (10 μM), trametinib (0.5 μM) or their combination were recorded by Western blot analysis. (i) Western blot analyses of MYC, SHMT2, HK2, PKM2, and LDHA for MHCC97H cells after knockdown of MYC. (j–k) Oxygen consumption rate (OCR) (j) and extracellular acidification rate (ECAR) (k) of MHCC97H cells that were transfected with MYC shRNAs and treated with 10 μM tigecycline. Data are shown as mean ± SEM (P values are calculated by two-tailed unpaired t test).
Next, we performed an assay to measure activation (tyrosine phosphorylation) of human receptor tyrosine kinases. Results showed that phosphorylation of EGFR and hepatocyte growth factor receptor (HGFR) are further activated following treatment with tigecycline (Fig. 6c). Combined data analysis for RNA sequencing, mRNA levels of amphiregulin (AREG) and epiregulin (EREG), ligands of EGFR in ERBB signaling pathway, were upregulated in tigecycline-treated cells. Therefore, we focused on EGFR signaling in subsequent studies (Fig. 6a). qRT-PCR experiments confirmed that mRNA levels of AREG and EREG were increased following treatment with tigecycline (Fig. S9a and b). It has been reported that oxidative phosphorylation inhibitors-mediated mitochondrial stress (such as the inhibitors of oxidative phosphorylation: Roteone, Carbonyl cyanide m-chlorophenylhydrazone (CCCP), and Antimycin A, etc.) might increase intracellular levels of reactive oxygen species (ROS) and calcium, which promotes gene transcription, translation and secretion of AREG by mitonuclear retrograde signaling.23 We detected the effects of BAPTA-AM (a cell membrane-permeable calcium chelator) and NAC (N-Acetylcysteine, a ROS inhibitor) on mRNA levels of EREG and AREG under treatment of tigecycline. Results showed that BAPTA-AM and NAC could decrease the expression of EREG and AREG which were stimulated by the treatment of tigecycline (Fig. S9a and b). The results suggest ROS and calcium may be involved in the tigecycline-induced expression of EREG and AREG.
Since AREG and EREG are growth factors that can interact with EGFR in an autocrine fashion, ELISA was then performed to measure the levels of AREG and EREG in the supernatants. Results showed that AREG and EREG were significantly increased in the cell culture supernatants with treatment of tigecyline (Fig. 6d–e, Fig. S9c and d). Moreover, Western blot experiments indicated that EGFR pathway was activated, which subsequently the levels of p-MEK1/2 and p-ERK1/2 were increased in tigecycline-treated cells. Inhibition of EGFR by gefitinib could also reduce the levels of p-MEK1/2 and p-ERK1/2 and had a synergistic effect of inhibition on liver cancer cells with tigecycline (Fig. 6f–g, Fig. S9e and f). It has been described that MAPK cascade can transcriptionally activate MYC expression, thereby increasing the transcription of MYC downstream genes.24,25 Considering the increase of glycolysis after treatment with tigecycline, protein levels of hexokinase 2 (HK2), pyruvate kinase M2 (PKM2), and lactate dehydrogenase A (LDHA) were detected by Western blot. Results showed the increase in their protein levels after treatment with tigecycline and a decrease after addition of trametinib (Fig. 6h, Fig. S9g). Our data suggest that a feedback activation loop involving in the EGFR-ERK1/2-MYC cascade may mediate insensitivity of liver cancer cells to tigecycline. Now that AREG and EREG resulted in activation of EGFR and downstream signaling, we detected baseline mRNA levels of AREG and EREG in liver cancer cells to see whether these genes could predict response to tigecycline. Results showed that mRNA levels of AREG and EREG were generally high in tigecycline-insensitive cells PLC/PRF/5, MHCC97H, and Li7 (Fig. S10a and b), which suggests these two genes may be related to the sensitivity of tigecycline. Whereas, further studies are needed to test for association between sensitivity of tigecycline and expressions of AREG and EREG.
To explore whether responses of liver cancer cells to tigecycline could be identified by the level of activation of the MAPK cascade, we performed Western blot analyses and found sustained activation of ERK1/2 signaling by tigecycline in tigecycline-insensitive cells, while this signaling was only transiently activated in tigecycline-sensitive cells, indicating that sustained feedback activation of ERK1/2 is crucial for liver cancer cells insensitivity to tigecycline (Fig. S10c and d).
To understand how MYC, an important downstream target of the MAPK cascade, influenced OXPHOS and glycolysis after treatment with tigecycline, we conducted shRNA-mediated knockdown of MYC in MHCC97H and PLC/PRF/5 cells. After addition of tigecycline, knockdown of MYC not only reduced the expression of SHMT2 and glycolytic enzymes-encoding genes such as HK2, PKM2, and LDHA (Fig. 6i, Fig. S11a), but also decreased the levels of OCR and ECAR in MHCC97H and PLC/PRF/5 cells (Fig. 6j–k, Fig. S11b and d).
To confirm feedback activation of glycolysis when treated with tigecycline, we knocked down HK2 in MHCC97H and PLC/PRF/5 cells with shRNA (Fig. S12a). Results exhibited that the levels of OCR were enhanced and ECAR were declined after knockdown of HK2, while the levels of OCR and ECAR were decreased in the presence of tigecycline (Fig. S12b and c). Our findings indicate that inhibition of feedback activation of glycolysis might result in energy depletion when treated with tigecycline, which was further supported by the synergistic effect of glycolysis inhibitor 2-Deoxy-d-glucose (2-DG) and tigecycline in liver cancer cells (Fig. S12d).
To explore the role of SHMT2 in the treatment of tigecycline, we stably knocked down SHMT2 in liver cancer cells. In the presence of tigecycline, the expression of SHMT2 increased to partially maintain the suppressed mitochondrial function, while knockdown of SHMT2 in MHCC97H and PLC/PRF/5 cells led to downregulation of mitochondrial translation products, such as CYTB, MTCO2 (Fig. S13a). The usage of tigecycline in SHMT2-knockdown cells further reduced the OCR levels compared with only treated with tigecycline (Fig. S13b). Thus, knockdown of mitochondrial folate enzyme SHMT2 in treatment of tigecycline resulted in further defective OXPHOS.
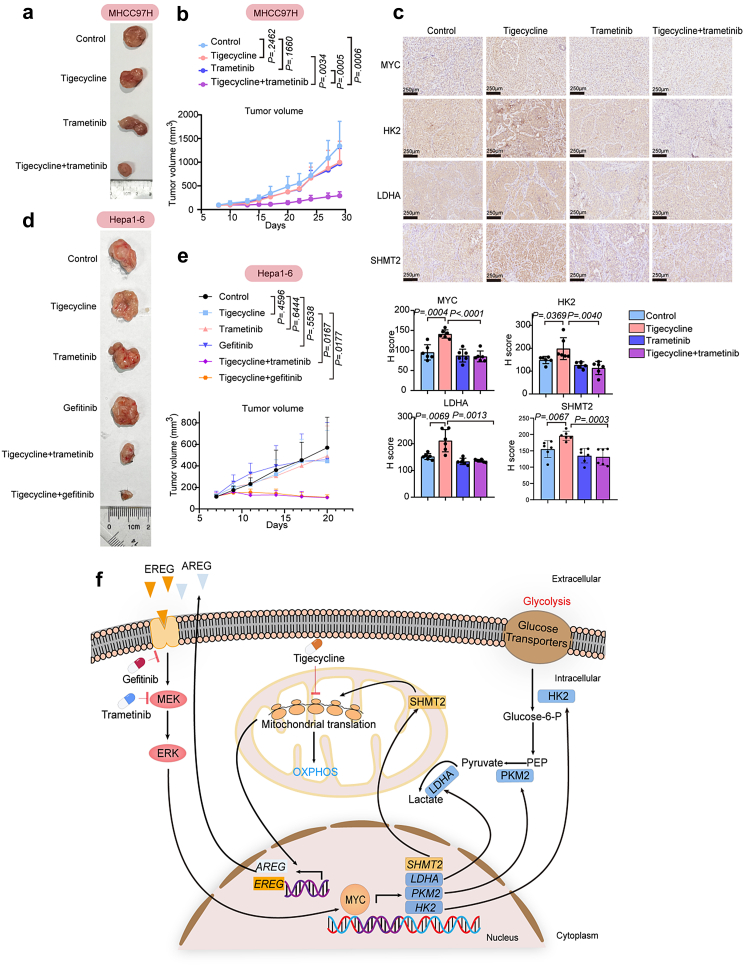
Blocking EGFR-ERK1/2-MYC cascade sensitizes to tigecycline in vivo
To validate the tigecycline drug combination in vivo, we used the tigecycline-insensitive MHCC97H subcutaneous tumour model. Compared to the control group, treatment of tigecycline or trametinib alone represented no obvious tumour suppressive effect, while the combination of tigecycline and trametinib showed a more effective inhibition of tumour growth without affecting the weight of mice (Fig. 7a and b, Fig. S14a and b). Protein levels of MYC, glycolytic enzymes such as HK2 and LDHA, SHMT2, and p-ERK1/2 in tumour tissues from tigecycline group were increased compared to control. The combination of tigecycline and trametinib reduced the protein levels of these genes induced by feedback activation (Fig. 7c, Fig. S14c and d). On the other hand, the combination treatment further inhibited the expression of mitochondrial translation products such as MTCO2, ATP6, and CYTB compared to tigecycline monotherapy, indicating the involvement of defective OXPHOS in the xenografts treated with the drug combination (Fig. S14c and d). Our data suggest that combined regimen of tigecycline and trametinib can significantly suppress tumour growth in vivo by reducing glycolysis and OXPHOS.
Fig. 7.
Blocking EGFR-ERK1/2-MYC cascade sensitizes liver cancer cells to tigecycline in vivo. (a) Representative tumour images of each group in MHCC97H subcutaneous tumour model. (b) Tumour volumes of subcutaneous xenografts of control, tigecycline group, trametinib group, and their combined regimen group (n = 6). (c) Representative IHC images of MYC, HK2, LDHA, SHMT2 in xenografts tissues and H score analyses. (d) Representative tumour images of each group in immune-competent mice with mouse liver cancer cells (Hepa1-6) transplanted subcutaneously. (e) Tumour volumes of subcutaneous Hepa1-6 cells of control, tigecycline group, trametinib group, gefitinib group, tigecycline plus trametinib group, and tigecycline plus gefitinib group (n = 6). (f) Schematic outline of this study. Values are shown as mean ± SEM (Unpaired two-sided t-test).
We then investigated the combination therapies in immune-competent mice with mouse liver cancer cells (Hepa1-6) transplanted subcutaneously. Trametinib or gefitinib had markedly synergistic effects with tigecycline in immune-competent mice, without affecting the weight of mice (Fig. 7d and e, Fig. S14e). IHC analyses of markers of mouse immune cells showed the increased CD4+ cells, CD8+ cells, and NKG2D + cells (a marker of NK cells) in the combination groups (tigecycline combined with trametinib or tigecycline combined with gefitinib) compared to control or tigecycline group (Fig. S14f).
Discussion
Tumours exhibit reprogrammed metabolic activities that promote cancer progression.26 Research on cancer metabolism has mainly focused on glycolysis in the past. The rapid rise in studies on the role of mitochondria in tumour reprograming has uncovered new insights and therapeutic opportunities.27 Recent studies have highlighted the heterogeneity and flexibility of tumour metabolism between tumours and even within different regions of solid tumours. A rigid or static focus on cellular autonomic pathways, such as glycolysis, would not be conducive to the precise treatment of tumours.28 However, identifying metabolic vulnerabilities susceptible to therapeutic targeting remains a challenge.3 The emergence of functional genetic screens provides a possible solution to this conundrum.29 Through a genome-wide CRISPR screen, we found that mitochondrial translation-related genes are essential for proliferation in liver cancer cells. Our data indicate that liver cancer cells can have different dependencies on mitochondrial translation-related genes, indicating the heterogeneity in tumour cell metabolism and the need for different strategies targeting tumour vulnerabilities.
Based on a high-resolution CRISPR screen, it was found that colorectal carcinoma cell HCT116 was specifically dependent on mitochondrial activity. However, the specific mechanism and its clinical significance were not described.30 In this study, we found through a genome-wide CRISPR screen that mitochondrial translation-related genes were closely involved in cell proliferation and may become a therapeutic strategy for liver cancer. This finding has clinical significance because FDA-approved tigecycline has inhibitory properties on mitochondrial translation. For tigecycline-sensitive liver cancer cells, tigecycline serves as a potential therapeutic agent to inhibit OXPHOS. As for tigecycline-insensitive cells, tigecycline inhibited OXPHOS as well. However, due to the increase of compensatory glycolysis and the decrease in dependence on OXPHOS, cells still survived under tigecycline treatment. Through a compound library screen, we identified MEK inhibitors as a class of drugs that can sensitize tigecycline-insensitive cells to this drug. From a perspective of energy metabolism, the combination regimen of tigecycline and MEK inhibitors reduced the increase of compensatory glycolysis and avoided the energy shift between glycolysis and OXPHOS. For such metabolic plasticity or hybrid metabolism of cancer cells, our strategy is to inhibit the main energy metabolism of the tumour to achieve tumour starvation.
In this study, we observed that secretion of EREG and AREG increased following treatment with tigecycline, resulting in activation of EGFR and downstream signaling. We found that EREG and AREG induced glycolysis by activating the EGFR-ERK1/2 cascade and the subsequent transcription of its downstream gene MYC, which represents a highlight of mechanism of our results. MYC is involved in the development and maintenance of most human tumors, including HCC.31 MYC regulates the expression of glycolytic enzymes, such as HK2 and PKM2, to stimulate glycolysis. Besides, we found that the expression of SHMT2 was elevated by MYC after treatment with tigecycline. Ye et al. reported that the mitochondrial enzyme SHMT2 was induced in MYC-transformed cells under hypoxia.32 However, their studies did not report that SHMT2 can be regulated by MYC through mitochondrial translation inhibitors or inhibition of OXPHOS.
Loss of the mitochondrial folate enzyme SHMT2 (rather than other folate enzymes) led to the inhibition of mitochondrial translation and defective OXPHOS.33 Further research showed that methyltetrahydrofolate catalyzed by SHMT2 could lead to methyl modification to form the taurinomethyluridine base, which is important for mitochondrial tRNAs. Lacking of this modified base caused defective translation, which may block the elongation of polypeptide chains.33 In addition, SHMT2 participated in providing formylmethionyl-tRNAs, which is utilized to initiate translation in prokaryotes and eukaryotic mitochondria. Specifically, from serine via the SHMT2 reaction, Met-tRNAMet is modified by a formyl group to form fMet-tRNAMet. Under the action of initiation factors, the small ribosomal subunit of mitochondria, mRNA, fMet-tRNAMet, and large ribosomal subunit combine to form initiation complex to initiate the synthesis of polypeptide chains.34 Tigecycline inhibits mitochondrial translation by competing with charged aminoacyl-tRNAs to bind the small ribosomal subunit, resulting in a blockage of the entry of (EF-Tu)·GTP aminoacyl (AA)-tRNA complex into the A site of the ribosome, thereby blocking the initial codon recognition step of tRNA accommodation and elongation process.10,35 In this study, under treatment of tigecycline the expression of SHMT2 was reactively elevated to compensate for the assembly of initiation complex by providing fMet-tRNAMet and guarantee the elongation of polypeptide chains by providing methyl donors for mitochondrial tRNAs as possible. Thus, the increased secretion of EREG and AREG activated EGFR-ERK1/2-MYC cascade to upregulate SHMT2 that may partially maintain the suppressed mitochondrial function in the tigecycline-insensitive cells under treatment of tigecycline.
This study has potential clinical and translational significance in liver cancer. Tigecycline is a broad-spectrum antibiotic used against many Gram-positive and Gram-negative pathogens. Its safety should be similar to standard antimicrobial therapy for complicated skin and soft tissue infections and complicated intra-abdominal infections in adults.36 A multicenter, controlled, randomized clinical trial uncovered that the combination of piperacillin/tazobactam and tigecycline (50 mg/12 h intravenously) was safe, effective and well tolerated in high-risk hematologic patients with cancer with febrile neutropenia.37 Though tumour suppression was not disclosed in the trial, it appears that tigecycline (50 mg/12 h intravenously) is tolerated in cancer patients. Recent studies have revealed the anti-tumour activity of tigecycline and an in vitro study of tigecycline in the treatment of chronic myeloid leukemia is under way (NCT02883036). More efforts are still needed to bring tigecycline to the clinical treatment of cancer. In this study, we identified the feedback activation of EGFR-ERK1/2-MYC cascade as the primary mechanism of the low response of some liver cancer cells to mitoribosome-targeting antibiotic tigecycline. MEK inhibitors or EGFR inhibitors conferred tigecycline sensitivity by blocking this feedback activation. Clinical trials of dabrafenib and MEK inhibitor trametinib in patients with advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer, melanoma, and non-small-cell lung cancer (NSCLC) revealed that this therapeutic regimen had strong clinical activity and good tolerability.38, 39, 40 EGFR inhibitor gefitinib has been approved by FDA for the treatment of NSCLC.41 Our previous study showed that the combined treatment of lenvatinib plus gefitinib had favorable clinical responses in 12 patients with advanced HCC (NCT04642547).4 Since these drugs have been approved for clinical use or are part of ongoing clinical trials, clinical testing of the combination of tigecycline and MEK inhibitors or EGFR inhibitors for the treatment of liver cancer should be feasible in the near future.
In conclusion, we characterized the crucial role of mitochondrial translation-related genes in liver cancer cells. Tigecycline is a potential therapeutic agent for liver cancer by impairing mitochondrial respiratory. Sustained activation of EGFR-ERK1/2-MYC cascade limits the response of liver cancer cells to tigecycline. Blocking the EGFR-ERK1/2-MYC cascade by EGFR inhibitor gefitinib or MEK inhibitor trametinib sensitizes the effects of liver cancer cells to tigecycline both in vitro and in vivo, which inhibits the energy metabolism of glycolysis and OXPHOS (Fig. 7f).
Contributors
W. Q., H. J., and R. B. conceptualized and supervised the study. Y. Z., S. W., and W. W. performed all the experimental assays and wrote the original draft. They have verified the underlying data. J. L., H. L., Q. J., J. Z., X. Z., R. Y., Q. W., and Y. S. provided constructive comments and discussion. C. L., and R. L. B. performed bioinformatics analyses. S. Y. collected the HCC tissues and supervised the study. All authors read and approved the final version of the manuscript.
Data sharing statement
All data associated with this study are present in the paper or the Supplementary Materials. RNA-seq data are accessible at the GEO under accession number: GSE199842. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
This work was funded by grants from the National Natural Science Foundation of China (82073039, 82222047, 81920108025), Program of Shanghai Academic/Technology Research Leader (22XD1423100), Shanghai Municipal Science and Technology Project (20JC1411100), 111 Project (B21024), Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZDCX20212700, SHSMU-ZDCX20210802) and Shanghai Jiao Tong University School of Medicine (YG2019GD01).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104397.
Contributor Information
Shengxian Yuan, Email: yuanshengx@126.com.
René Bernards, Email: r.bernards@nki.nl.
Haojie Jin, Email: hjjin1986@126.com.
Wenxin Qin, Email: wxqin@sjtu.edu.cn.
Appendix A. Supplementary data
References
- 1.Galle P.R., Forner A., Llovet J.M., et al. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Finn R.S., Qin S., Ikeda M., et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382(20):1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 3.Wang C., Cao Y., Yang C., Bernards R., Qin W. Exploring liver cancer biology through functional genetic screens. Nat Rev Gastroenterol Hepatol. 2021;18(10):690–704. doi: 10.1038/s41575-021-00465-x. [DOI] [PubMed] [Google Scholar]
- 4.Jin H., Shi Y., Lv Y., et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature. 2021;595(7869):730–734. doi: 10.1038/s41586-021-03741-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang C., Vegna S., Jin H., et al. Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature. 2019;574(7777):268–272. doi: 10.1038/s41586-019-1607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J., Zhao H., Diplas B.H., et al. Genome-wide CRISPR-cas9 screen reveals selective vulnerability of ATRX-mutant cancers to WEE1 inhibition. Cancer Res. 2020;80(3):510–523. doi: 10.1158/0008-5472.CAN-18-3374. [DOI] [PubMed] [Google Scholar]
- 7.Criscuolo D., Avolio R., Matassa D.S., Esposito F. Targeting mitochondrial protein expression as a future approach for cancer therapy. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.797265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F., Zhang D., Zhang D., Li P., Gao Y. Mitochondrial protein translation: emerging roles and clinical significance in disease. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.675465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pecoraro A., Pagano M., Russo G., Russo A. Ribosome biogenesis and cancer: overview on ribosomal proteins. Int J Mol Sci. 2021;22(11):5496. doi: 10.3390/ijms22115496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z., Abbas M.N., Kausar S., et al. Biological functions and molecular mechanisms of antibiotic tigecycline in the treatment of cancers. Int J Mol Sci. 2019;20:3577. doi: 10.3390/ijms20143577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Škrtić M., Sriskanthadevan S., Jhas B., et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20(5):674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuntz E.M., Baquero P., Michie A.M., et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med. 2017;23(10):1234–1240. doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan J., Song M., Zhou M., Hu Y. Antibiotic tigecycline enhances cisplatin activity against human hepatocellular carcinoma through inducing mitochondrial dysfunction and oxidative damage. Biochem Biophys Res Commun. 2017;483(1):17–23. doi: 10.1016/j.bbrc.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Meßner M., Schmitt S., Ardelt M.A., et al. Metabolic implication of tigecycline as an efficacious second-line treatment for sorafenib-resistant hepatocellular carcinoma. FASEB J. 2020;34(9):11860–11882. doi: 10.1096/fj.202001128R. [DOI] [PubMed] [Google Scholar]
- 15.Mazières J., Brugger W., Cappuzzo F., et al. Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: Re-analysis of the SATURN study. Lung Cancer. 2013;82(2):231–237. doi: 10.1016/j.lungcan.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rath S., Sharma R., Gupta R., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49(D1):D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza A.R., Minczuk M. Mitochondrial transcription and translation: overview. Essays Biochem. 2018;62(3):309–320. doi: 10.1042/EBC20170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Malley J., Kumar R., Inigo J., Yadava N., Chandra D. Mitochondrial stress response and cancer. Trends Cancer. 2020;6(8):688–701. doi: 10.1016/j.trecan.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arroyo J.D., Jourdain A.A., Calvo S.E., et al. A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 2016;24(6):875–885. doi: 10.1016/j.cmet.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yousefi R., Fornasiero E.F., Cyganek L., et al. Monitoring mitochondrial translation in living cells. EMBO Rep. 2021;22(4) doi: 10.15252/embr.202051635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H.J., Maiti P., Barrientos A. Mitochondrial ribosomes in cancer. Semin Cancer Biol. 2017;47:67–81. doi: 10.1016/j.semcancer.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S.F., Chang Y.L., Tzeng Y.D., et al. Mitochondrial stress adaptation promotes resistance to aromatase inhibitor in human breast cancer cells via ROS/calcium up-regulated amphiregulin-estrogen receptor loop signaling. Cancer Lett. 2021;523:82–99. doi: 10.1016/j.canlet.2021.09.043. [DOI] [PubMed] [Google Scholar]
- 24.Yang W., Zheng Y., Xia Y., et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Z., Liu C., Zhi Y., et al. LIMK1 nuclear translocation promotes hepatocellular carcinoma progression by increasing p-ERK nuclear shuttling and by activating c-Myc signalling upon EGF stimulation. Oncogene. 2021;40(14):2581–2595. doi: 10.1038/s41388-021-01736-2. [DOI] [PubMed] [Google Scholar]
- 26.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira G.L., Coelho A.R., Marques R., Oliveira P.J. Cancer cell metabolism: rewiring the mitochondrial hub. Biochim Biophys Acta Mol Basis Dis. 2021;1867(2) doi: 10.1016/j.bbadis.2020.166016. [DOI] [PubMed] [Google Scholar]
- 28.Kim J., DeBerardinis R.J. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 2019;30(3):434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei L., Lee D., Law C.T., et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. 2019;10(1):4681. doi: 10.1038/s41467-019-12606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart T., Chandrashekhar M., Aregger M., et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163(6):1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Dauch D., Rudalska R., Cossa G., et al. A MYC–aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med. 2016;22(7):744–753. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- 32.Ye J., Fan J., Venneti S., et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4(12):1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morscher R.J., Ducker G.S., Li S.H., et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554(7690):128–132. doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minton D.R., Nam M., McLaughlin D.J., et al. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol Cell. 2018;69(4):610–621.e5. doi: 10.1016/j.molcel.2018.01.024. https://doi:10.1016/j.molcel.2018.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenner L., Starosta A.L., Terry D.S., et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci USA. 2013;110(10):3812–3816. doi: 10.1073/pnas.1216691110. https://doi:10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein G.E., Babinchak T. Tigecycline: an update. Diagn Microbiol Infect Dis. 2013;75(4):331–336. doi: 10.1016/j.diagmicrobio.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Bucaneve G., Micozzi A., Picardi M., et al. Results of a multicenter, controlled, randomized clinical trial evaluating the combination of piperacillin/tazobactam and tigecycline in high-risk hematologic patients with cancer with febrile neutropenia. J Clin Oncol. 2014;32(14):1463–1471. doi: 10.1200/JCO.2013.51.6963. [DOI] [PubMed] [Google Scholar]
- 38.Subbiah V., Kreitman R.J., Wainberg Z.A., et al. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600–mutant anaplastic thyroid cancer. J Clin Oncol. 2018;36(1):7–13. doi: 10.1200/JCO.2017.73.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robert C., Karaszewska B., Schachter J., et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 40.Planchard D., Smit E.F., Groen H.J.M., et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 2017;18(10):1307–1316. doi: 10.1016/S1470-2045(17)30679-4. [DOI] [PubMed] [Google Scholar]
- 41.Du X., Yang B., An Q., Assaraf Y.G., Cao X., Xia J. Acquired resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors. Innovation (Camb) 2021;2(2) doi: 10.1016/j.xinn.2021.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.