Abstract
Improvement of feed efficiency (FE) in pigs is an important milestone in order to reduce the economic and environmental impact of pig production. The goal of finding biomarkers for FE has persisted for decades. However, due to the complexity of the FE trait, these goals have still not been met. Here, we search for quantitative trait loci (QTL), candidate genes, and biological pathways associated with FE using both genotype and RNA-seq data. We obtained genotype and colon epithelium RNA-seq data for 375 and 96 pigs, respectively. In total, a genome-wide association study (GWAS) and differential expression (DE) analysis led to detection of three QTL on SSC9 and 17 DE-genes associated with FE. Possible intersection points between genes located in QTL and DE-genes were found on levels of transcription factor-target interaction. Moreover, cis-eQTL analysis revealed associations between genotype and expression levels of three DE-genes and three genes located in the GWAS QTLs, which may establish the connection between genotype and phenotype through DE. Finally, single nucleotide polymorphism calling using RNA-seq data for genes located in GWAS QTLs revealed 53 polymorphisms of which eleven were missense variants.
Subject terms: Data processing, Gene regulatory networks, Animal breeding, Functional genomics, Gene expression, Genetic association study, Genome
Introduction
As the human population grows, the requirement for effective food production industries intensifies. At the same time, environmental challenges require food production to be sustainable. Extensive agriculture expansion has already led to the usage of one half of habitable land, significant changes in the landscape1 and numerous other challenges, such as greenhouse gas emission, terrestrial acidification, and freshwater eutrophication2. Life cycle assessment studies have shown that feeding accounts for 28–82% of the total impact on climate change from animal production3 and that breeding for feed efficiency (FE) traits can reduce this impact4. This has raised new arguments for improvements in FE in pigs.
FE in pigs is a complex trait with a large number of biological and environmental components. Environmental factors such as feed composition are constantly studied, leading to new understanding and advances in the optimization of feed composition5–7. Furthermore, advancements in molecular biology techniques have led to several studies directed at identifying the biological components involved in FE in pigs. Most of these studies8–12 have been aimed at finding biological markers for FE in pigs using different techniques such as genetic mapping, RNA-sequencing, metabolomics, and metagenomics.
Among these studies, genome-wide association studies (GWAS) are especially interesting, since GWAS results can aid in the understanding of genetic mechanisms for high FE. However, a main drawback is that the heritability of FE is quite low, thus a cohort for a GWAS on this trait must be big to gain enough statistical power for detection of quantitative trait loci (QTL)13. Another issue of GWAS is that in most cases it is not possible to infer the location of causative mutations precisely, thus a list of candidate genes potentially associated with a trait prevails after GWAS. A potential way to overcome this obstacle is to include another omics analysis since consistent results of GWAS and other omics analysis can be regarded as mutual verification.
Omics techniques have found widespread use in porcine FE research, in combination with GWAS or on their own. A number of associations with different types of biological markers have been found in these studies, which overall provide a better understanding of the FE trait complexity. For instance, metagenomics research has shown an association of dozens of bacterial taxonomic units in gut microbiota with FE8,11, and metabolomics studies in combination with transcriptomic9 and GWA studies10 have revealed significant gene-metabolite pairs. Transcriptomic studies have found FE associated differentially expressed genes (DE-genes) in various tissue, including blood14, liver12, brain15 and intestine16.
The colon is a part of the gastrointestinal tract that absorbs water, electrolytes and vitamins from the digesta as it arrives from the small intestine. Furthermore, the colon provides space for a dense and diverse community of mainly anaerobe bacteria with the ability to ferment complex carbohydrates, which otherwise cannot be digested by the host. The bacterial fermentation results in synthesis of short chain fatty acids, which are a rich source of energy for the host17,18. The healthy symbiotic relationship between host and microbiota is tightly regulated by multiple factors and hence, gene expression studies focusing on colon epithelium cells may disclose new aspects of the host-microbiota symbiosis.
Residual feed intake (RFI) can be used to describe FE in pigs. It is calculated as a difference between observed feed intake and expected feed intake as predicted by multiple regression of feed intake on production traits such as body weight gain, and tissue composition to account for production requirements, and average metabolic body weight to account for maintenance requirements19.
In the present study, we combine transcriptome analysis and GWAS to identify candidate genes and pathways linked with FE measured as RFI. Both types of analysis reveal biological markers for FE.
Results
Genome-wide association studies
GWAS was performed using 370 pigs after filtering outlier pigs as described in the methods section. The test model included RFI values (MJ NE/d) as response variable and sex as a fixed effect. Mean and standard deviation for RFI were 0 and 1.111, respectively, and RFI was strictly normally distributed according to the Shapiro–Wilk test. The heritability for RFI was 0.17.
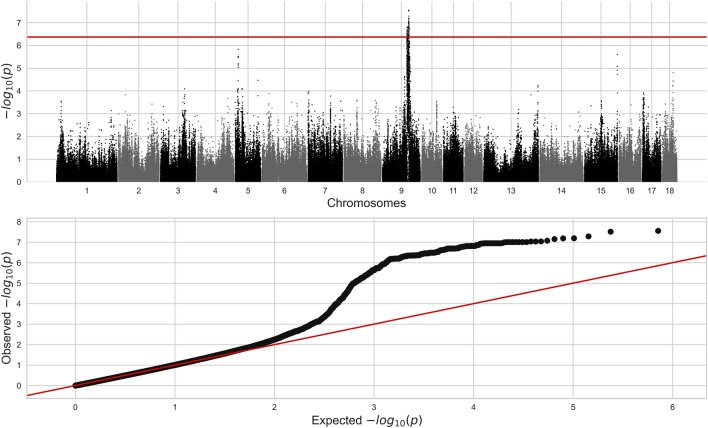
Results of the GWAS are shown in Fig. 1. Genome-wide significance threshold was 4.28e-7 after Bonferroni correction. All genome-wide significantly associated SNPs were located in a region on SSC9 from 83.7 to 95.9 Mb (Table 1). A significant deviation in observed p-values as compared to expected p-values is evident in the qq-plot. This reflects a large number of markers in strong LD with the genome-wide significant SNPs on SSC9.
Figure 1.
Manhattan (top) and Q-Q (bottom) plots of GWAS results.
Table 1.
Description of QTL blocks as defined by linkage disequilibrium.
| Location | Lead SNP | p-value | Number of genes | Number of protein coding genes | Genes flanking the lead SNP | |
|---|---|---|---|---|---|---|
| QTL-1 | SSC9: 83,701,293–86,854,470 | 9:85907846 | 1.54e-7 | 24 | 15 | BZW2 |
| QTL-2 | SSC9: 88,546,817–91,460,506 | 9:90703434 | 2.80e-8 | 19 | 13 | DNAH11 |
| QTL-3 | SSC9: 95,697,312–95,866,333 | 9:95864157 | 1.76e-7 | 0 | 0 |
*ENSSSCG00000042154 *ENSSSCG00000042154 |
*Flanking genes for lead SNP in QTL-3 are located outside the QTL.
All except three SNPs associated with RFI were located in a 10 Mb region on SSC9, which subsequently was studied for LD patterns (Suppl. Figure 1). The LD analysis revealed the majority of SNPs to be located in two LD blocks, SSC9: 83,701,293–86,854,470 (QTL-1) and SSC9: 88,546,817–91,460,506 (QTL-2), whereas one SNP was located outside the two LD blocks in an LD block located at SSC9: 95,697,312–95,866,333(QTL-3). Hence, the LD analysis resulted in identification of three QTLs, two relatively wide (QTL-1 and QTL-2) and one narrow (QTL-3) (Table 1, Suppl. Figure 1).
QTL-1 contained fifteen protein-coding genes and eight non-coding genes, and QTL-2 contained thirteen protein-coding and six non-coding genes (Suppl. Table 1). Inside QTL-3 no genes were found but the nearest flanking genes are given in Table 1.
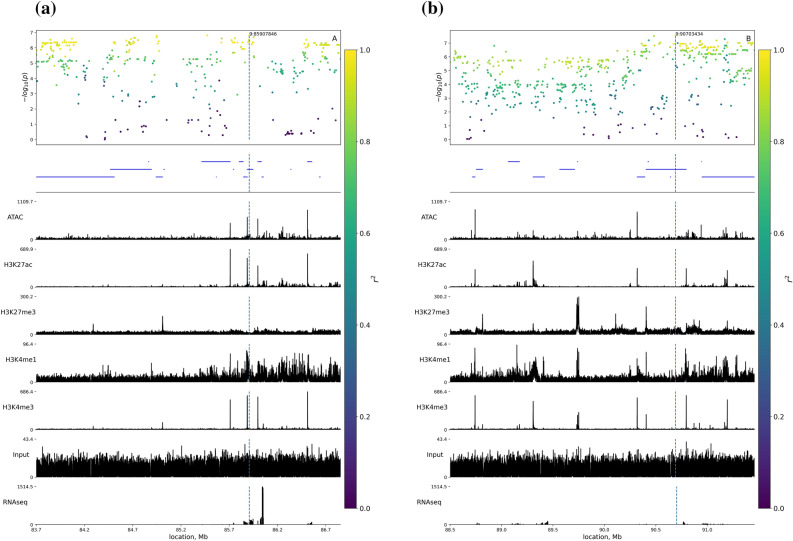
The lead SNP from the QTL-1 block was located in an intron of BZW2 in a region enriched for ATAC-, H3K27ac-, and H3K4me1-signals in colon tissue20 indicating a regulatory role of this region (Fig. 2a).
Figure 2.
Plots of QTL-1 (a) and QTL-2 (b) GWAS results, gene location, and epigenetic markers enrichment. X-axes indicate genome positions, Y-axis in the top panel indicates − log (p values). Linkage disequilibrium between the lead SNP and other SNPs is highlighted by color. Blue lines represent the location of genes, the remaining plots illustrate epigenetic marker enrichment scores. Score values on Y-axes correspond to aligned read counts in the ChIP-seq analysis reported by Pan et al. 202120.
The lead SNP from QTL-2 was located in an intron of the DNAH11 gene. The location of this lead SNP was not enriched for epigenetic markers in colon tissue (Fig. 2b).
Analysis of differential expression
For analysis of differential gene expression (DE-analysis) RNA sequencing data of colon epithelium was obtained from 96 preselected pigs with high or low RFI as described in the methods section. Four pigs were excluded from the analysis because they were found to be outliers with regard to their RFI based on statistics described in the methods section. Furthermore, one pig was found to be an outlier in respect to gene expression and was excluded from further analysis. Hence, after filtering, 47 (24 females and 23 males) low RFI pigs and 44 (22 females and 22 males) high RFI pigs were left for DE-analysis. For these pigs, mean values of RFI for the low and high RFI groups were − 1.56 and 1.56, respectively, standard deviation within groups were 0.57 and 0.88, respectively. The distribution of p-values for the DE model was anti-conservative (Suppl. Figure 2).
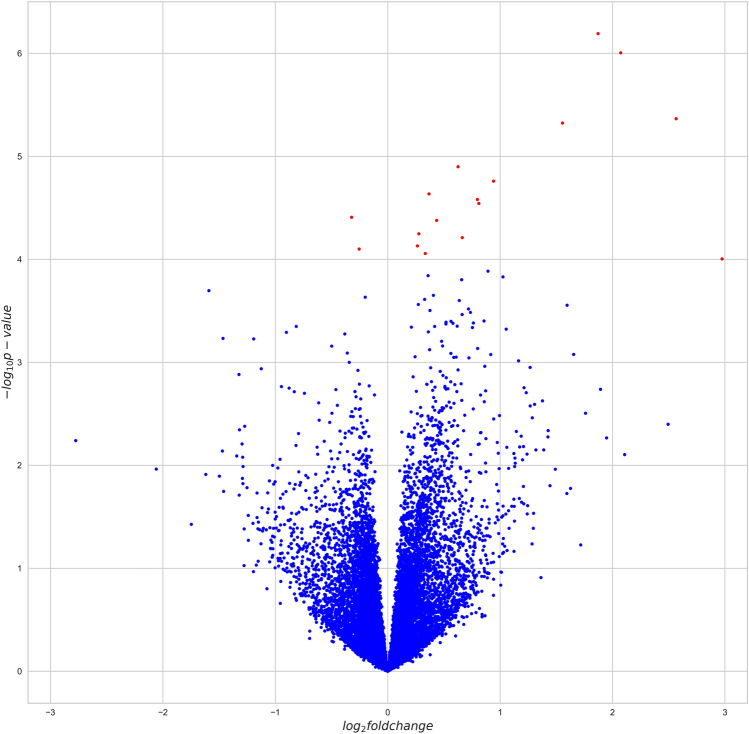
A False Discovery Rate of 0.1 was used as a threshold for significance in the DE-analysis (Fig. 3). This resulted in identification of 17 DE-genes with 15 genes upregulated in the high RFI group and two genes downregulated in the high RFI group (Suppl. Table 2). Immune system associated terms were found in the rank-based enrichment test for p-values of all genes tested in the DE-analysis. These functions include regulation of T-cell activation, regulation of leukocyte cell–cell adhesion, and antigen processing and presentation (Suppl. Table 3). The same type of enrichment test but for log fold change values of genes showed that immune system associated terms were characteristic for upregulated genes in the high RFI group (Suppl. Table 4). Downregulated genes were characterized by a diverse group of terms, including terms related to mitosis, protein synthesis, etc.
Figure 3.
Differentially expressed genes between the high RFI and the low RFI. Red dots represent significantly up- or downregulated genes.
eQTL analysis
For the eQTL analysis, 94 pigs with both RNA-seq and genotype data available were used. Only cis-eQTL mapping was performed and SNPs located no more than 1 Mb from a gene were considered. Significance threshold was set based on a three step correction procedure as described by Huand et al.21. Genes and SNPs passing the significance threshold are called eGenes and eQTLs, respectively in the following.
In total, 2016 eGenes were identified and their expression was associated with 5263 different cis-eQTLs. The 2016 eGenes represent: 1701 protein-coding genes, 269 lncRNA genes, 35 pseudogenes, 3 snRNA genes, 3 processed pseudogenes, 2 IG V genes, 1 miRNA gene, 1 snoRNA gene, and 1 TR V gene.
eQTL were found for two out of the 17 DE-genes, ENSSSCG00000025271, and ENSSSCG00000034614. None of these eGenes had a paired eSNP with a p-value in GWAS lower than 0.05.
Three eGenes, AGMO, SOSTDC1, and CRPPA, were found in QTL-1 (Table 2). Only AGMO had a relatively low p-value in the DE-analysis.
Table 2.
eGenes among Differentially Expressed RFI QTL genes.
| eGene | eSNPs | baseMean | log2FoldChange | PDE | FDRDE |
|---|---|---|---|---|---|
| AGMO | 9:84047949 | 3.41 | −1.27211 | 0.00418 | 0.41600 |
| CRPPA | 9:85565144 | 32.77 | 0.26116 | 0.15780 | 0.81436 |
| SOSTDC1 | 9:84672568 | 8.15 | −0.67361 | 0.19125 | 0.84661 |
SNP calling using RNA-seq data
SNP calling revealed 53 polymorphisms in genes located in QTL-1 and QTL-2. Using the Ensembl Variant Effect Predictor tool22 it was found that eleven polymorphisms were missense variants (Suppl. Table 5). Among those, two variants were deleterious based on SIFT score, both of them were located in a conserved domain of the DNAH11 gene. Moreover, four missense mutations were located inside the AHR gene.
Transcription factor regulation
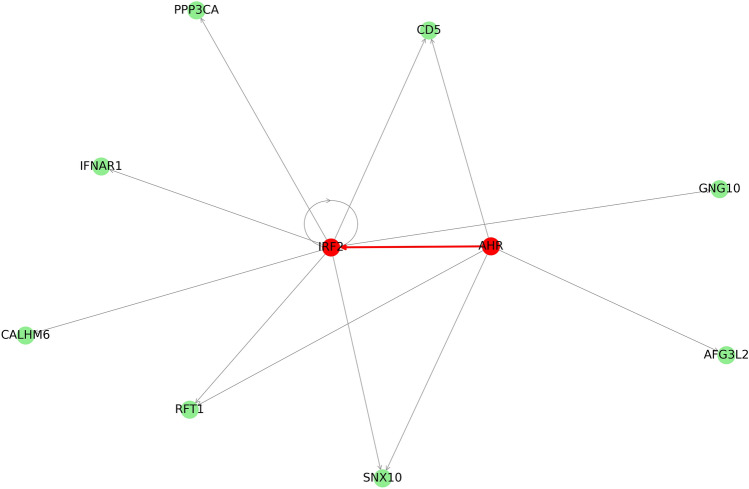
A survey of the GeneHancer database revealed that the AHR transcription factor encoded by a gene located in QTL-1 has five targets among proteins encoded by DE-genes (Fig. 4). Among target DE-genes for AHR, one transcription factor was found. This transcription factor, IRF2, in turn, regulates the expression of four additional DE-genes.
Figure 4.
Graph of transcription transcription factor regulation pathways among DE-genes (green) and of RFI QTL gene (red). Interactions between transcription factors highlighted by red color.
Discussion
The combination of GWAS and RNA-seq data analysis provided an opportunity for making a multi-faceted analysis of biological markers for RFI. Using the two methods in tandem, we obtained mutual support for results from multiple analyses.
The priority in our study was the identification of QTLs for RFI and the biological pathways on which they have an impact, thus RNA-seq data was mostly used for verification and interpretation of GWAS results. We have achieved this in three different ways: identification of transcription factors in QTLs with targets in DE-genes, finding cis-eQTLs in QTL regions for RFI, and SNP calling based on RNA-seq data.
The present GWAS analysis revealed three QTL regions on SSC9, which overlap with a number of QTL found in previous studies according to the Animal QTL Database23. In total, QTLs from 26 previous studies are overlapping with the QTLs identified in the present study. Two of these overlapping QTLs are associated with production traits such as body weight at day 21 (QTL-1,2,3)24, lipid (QTL-2) and protein (QTL-1,2,3) accretion rate, and average daily gain (QTL-1,2,3)25. However, since FE is a genetically highly complex trait, the QTLs identified in the present study are at best small pieces in the genetic puzzle underlying FE. A more complete picture of the genetic mechanisms underlying this trait must include the many other QTLs found in previous studies on this subject [e.g.13,26,27].
In our GWAS analysis the lead SNP for QTL-1 was located in a region enriched for ATAC-, H3K27ac-, and H3K4me1-signals in colon tissue. The lead SNP for QTL-2 was located in an intron region of the DNAH11 gene. A potential functional importance of this gene is supported by SNP calling based on RNA-seq data which revealed two deleterious variants in conserved domains of DNAH11. SNPs located in QTLs were associated with the expression of three genes, however, only one gene, AGMO, shows a p-value in the DE-analysis lower than 0.05.
Analysis of DE in the RNA-seq data revealed 17 DE-genes for RFI. Fifteen DE-genes were upregulated in the high RFI group of pigs. Rank-based enrichment test for log fold change values showed that expression upregulation in the high RFI group was enriched for immune system associated annotations in agreement with previous studies14,28. The identified immune system response in colon epithelium might indicate a link between gut immune function, the gut microbiome and FE. The causation of this link can be explained in two different ways. On one hand, the microbiome might modulate immune system response. For example, it has been shown that short-chain fatty acids (SCFAs) are involved in immune response inhibition29,30. Energy rich SCFAs are also product of fiber fermentation by the gut microbiome and therefore increase of their production might lead to increased FE. On the other hand, a modified immune response might be determined genetically and subsequently modulate the microbiome. The potential role of genetics in this link is supported by interactions found between GWAS and DE results using transcription factor–target networks. In this way, a transcription factor encoding gene located in QTL-1, AHR, may explain the differential expression of nine out of 17 DE-genes. The AHR gene has to our knowledge not been linked to FE before, but it has been considered a candidate gene for reproductive traits in previous studies31–33. SNP calling based on RNA-seq data revealed four missense mutations in the protein-coding part of the AHR gene. Thus, we conclude that AHR is a promising candidate gene for a RFI genetic determinant.
Moreover, one gene located in QTL-1, AGMO, showed an expression level significantly associated with the genotype and a relatively low p-value in DE-analysis for RFI. This gene encodes alkylglycerol monooxygenase which appears to play a role in immunity, energy homeostasis, and development34. In our study AGMO was down-regulated in the high RFI group of pigs.
In conclusion, we have identified a number of genes of potential importance for RFI. Of particular interest is the gene encoding the AHR transcription factor, which is located in QTL-1 and targeting some of the identified DE-genes. Another gene of interest is DNAH11, in which the QTL-2 lead SNP was located in one of the introns. Missense mutations were found in both AHR and DNAH11 genes. Further studies in independent cohorts should be performed to confirm the relevance of the candidate genes for feed efficiency in pigs. Furthermore, the hypotheses concerning interactions between immune system response in colon and microbiome could be studied by integration of metagenomics data. Such data will soon be available for the pigs in the present study and an analysis of these data will be presented in a separate paper.
Materials and methods
Animals
In the present study, 409 cross-bred animals were used. Study animals were produced by insemination of (Landrace x Yorkshire) crossbred sows by mixed semen from Duroc boars. DanBred (Herlev, Denmark) provided all animals. Conditions from the Danish “Animal Maintenance Act” (Act 432 dated 09/06/2004) and the “Order regarding animal experimentation” (BEK nr 12 af 07/01/2016) were met and approved by the Danish Veterinary and Food Administration. Individual IDs were assigned to each pig at weaning and each pig was equipped with an electronic ear-tag. An automatic feeding station was used to record the time, duration, and feed consumption at every visit of the pig using ear-tag screening. The trial period started when pigs reached the approximate body weight of 30 kg and lasted for 33 days. The same diet was given to all pigs. None of the pigs were treated during the trial period. The diet was composed of wheat (49%) barley (25%) and soybean meal (17%). The crude protein (CP) content was 16% and the content of standardized ileal digestible protein was 132 g per kg feed. The energy content was 9.6 MJ NE per kg feed. Individual feed intake was measured in MJ NE. All pigs were slaughtered after overnight fasting at an age of approximately 6 months and a bodyweight of approximately 100 kg in a commercial abattoir. Blood from each pig was sampled in Thermo Fischer BD K2E (EDTA) tubes immediately after exsanguination.
DNA isolation, genotyping and imputation to whole genome sequence variants
High quality DNA was isolated from EDTA stabilized blood using a classic salting out procedure35. SNP genotyping was performed by Edinburgh Genomics, Ashworth Laboratories (Edinburgh) using the 700 K Affymetrix Axiom PigHD chip. Sequences of the probes for Affymetrix Axiom PigHD SNPs were mapped to the new assembly using BWA36 to retrieve marker positions in the newest version of the pig genome assembly (Sscrofa11.181). Markers that were assigned to multiple positions across the genome were excluded from the analyses. Only autosomal markers were retained. The haplotype phasing for the HD marker set was performed using Eagle37 with default parameters. Finally, Minimac338 was used to impute missing values of the HD marker set. Imputation of missing data was aided by exploitation of DNA sequence information from 217 purebred pigs of the three involved breeds.
Phenotype calculation
Residual feed intake (RFI) was defined as the difference between an observed average daily feed intake (ADFI) and expected ADFI. Expected ADFI was estimated using multiple linear regression of ADFI on average daily gain and metabolic body weight (MBW). MBW was calculated under an assumption of a linear growth rate over the experiment period using formula from39:
where W1 and W0 are weights at start and end of the test, respectively.
The linear model was built using the LinearRegression function in the Sklearn package V.1.1.240. At first, data for 409 pigs were used, however nine pigs were excluded from calculation of expected ADFI and further analysis as outliers. Outlier pigs were identified using Cook’s distance values in the abovementioned multiple linear regression model. Cook’s distance estimates the impact of separate data points in a least squares regression and identify extreme observations for elimination41. Cook’s distance threshold was set on the level of the 98th percentile (Suppl. Figure 3). Outliers were excluded prior to building the final linear model used for RFI calculation.
The 96 pigs selected for RNA sequencing (see below) contained four outliers, thus 92 pigs were left after filtering.
Genome-wide association study
375 pigs were genotyped, but 5 pigs were excluded from GWAS as phenotype outliers as described above. Quality control for the genotypes was conducted based on the criteria of Hardy–Weinberg equilibrium (HWE > 10–8) and minor allele frequency (MAF > 0.05) by PLINK software42. After quality control, 357,735 SNPs remained for further analysis. We estimated the genomic relationship matrix (GRM) using GCTA43 with the filtered autosomal marker set. GWAS analyses were performed using the “leave-one-chromosome-out” procedure in GCTA. Number of independent tests (Nindep) was estimated using SimpleM44 and a Bonferroni corrected genome wide significance level was defined as 0.05/Nindep. Genome-wide significance threshold was 4.28e-7 after Bonferroni correction.
GWAS was performed for each chromosome using RFI as a response phenotype and including sex as a fixed effect. Genome-wide significantly associated SNPs were further used for QTL region identification.
QTL regions were identified based on linkage disequilibrium (LD) between lead SNPs in a region and surrounding SNPs. Pairwise LD (r2) was calculated using the –r2 function in PLINK42. Borders of QTLs were defined based on position of the most distant SNP from the lead SNP with an r2 score above 0.85.
Genomic heritability was estimated for RFI using sex as a fixed effect. For estimation, the GCTA –reml function and the GRM established above were used.
Identified QTL regions were screened for epigenetic marker tracks in colon tissue based on published pig genome functional annotations20. Epigenetic marker tracks were obtained from the UCSC database45 and visualized based on scores. Scores were derived from ChIP-seq data and represent processed counts of reads aligned to the given location in the genome.
RNA extraction and RNA sequencing
At a commercial slaughterhouse, the gastrointestinal organs were removed from the pigs within 20 min after bleeding. Approximately 5 cm of the middle part of the colon was cut out and rinsed in 0.9% saline. The mucosal lining was scraped off with a scalpel, transferred to cryo-tubes, and snap-frozen in liquid nitrogen. Samples were collected from 325 pigs. Among those pigs, 48 animals with high FE and 48 animals with low FE were selected for RNA sequencing. An equal number of females and males were selected for both high and low FE. The animals were selected based on feed conversion ratio (FCR) adjusted to the weight of pigs at the start of the trial. Mean values of adjusted FCR in high and low FCR groups were 2.291 and − 1.799 MJ NE/kg, respectively. The mean and standard deviation value for adjusted FCR in the entire population were 0 and 1.593, respectively. Thus, a ratio of sample difference between groups to standard deviation in the entire population was 2.567.
For RNA isolation, 50 mg of tissue per sample were homogenized in a GentleMACS™ Octo Dissociator machine (Miltenyi Biotec) using the RNeasy® Mini Kit (Qiagen) with DNase digestion, following manufacturer’s instructions. The concentration and purity of the RNA samples were measured on a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). RNA integrity was assessed on a BIO-RAD Experion machine using the RNA stdSens kit. All samples had an RNA-quality index (RQI) above 8.
RNA-seq library construction was performed by purifying the mRNA by oligo(dT)beads, the resulting library was sequenced on an Illumina HiSeq 2000 sequencing platform with TruSeqV3 sequencing reagents at the Beijing Genomics Institute, Shenzhen, PR China as previously described46.
Differential expression analysis
Quality control for total RNA-Seq reads was performed using FastQC software. Reads were aligned to the pig reference genome (Sscrofa 11.1) using the Bioconductor Rsubread package47 with default parameters. Genes were annotated using the Ensembl database48 and filtered excluding genes which were expressed in less than twenty percent of the animals. The same phenotype as for the GWAS (RFI) was used as a response variable in the DE-analysis, i.e., the animals were divided into high and low RFI groups using 0 MJ NE per day as a threshold. As mentioned above, pigs for RNA sequencing were initially selected based on FCR values adjusted to the body weight at the start of the trial. However, high and low FE grouping based on FCR and RFI matched for all pigs. Mean values of RFI for the low and high RFI groups were -1.56 and 1.56, respectively, standard deviation were 0.57 and 0.88, respectively. Analysis of differential expression was carried out using DESeq2 platform V.1.3649. After the first fit of the model, Cook’s distances for gene expression were retrieved using the assays () function in DESeq2 in order to detect outliers. In this way one animal was considered as an outlier based on average Cook’s distance value (Suppl. Figure 4). This pig was excluded from further analysis and the final DE model was fitted using the remaining dataset. Features with a False Discovery Rate (FDR) ≤ 0.1 were considered to be differentially expressed genes (DE-genes).
Functional enrichment analysis for the full result table of DE association test was carried out using the STRING V. 11.5 web-tool (www.string-db.org)50 based on Kolmogorov–Smirnov test statistics. Functional enrichment was run on two estimates of DE analysis, namely nominal p-values and log fold changes for genes.
Cis-eQTL analysis
For cis-eQTL analysis, transcriptome and genotype data for 94 out of the 96 pigs with transcriptome data were used. One pig was excluded after DE-analysis as described above and another pig lacked genotype data. The RNA-seq data were mapped and filtered as for the DE-analysis and the data was normalized by size factor using the DESeq2 platform V.1.3649. Quality control of SNPs was conducted as for GWAS. Cis-QTL were mapped using the BootstrapQTL method V 1.0.521. BootstrapQTL uses three steps of multiple testing correction. At first Bonferroni correction is used to adjust nominal p-values from MatrixEQTL cis-SNPs separately for each gene, after that FDR correction is applied to the lowest p-values for all genes, and in the end SNPs with locally adjusted p-values corresponding to globally corrected p-value threshold of 0.05 are considered as eSNPs. The bootstrap procedure was used to correct for the overestimation of effect sizes. Cis-eQTL associations were searched within a 2 Mb region around each gene.
SNP calling using RNA-seq data
For SNP calling, increased requirements for reads were applied. Read head cutting (12 bases) and sliding window trimming (SLIDINGWINDOW:4:20) were applied for all reads using Trimmomatic51. Then reads were aligned to the pig reference genome (Sscrofa 11.1) using the STAR package V.2.7.1052 with default parameters. SNP calling was performed using the Rsubread package V.2.0.347. Only polymorphisms with a score higher than 0.2 were kept. The identified polymorphisms were annotated using Ensembl Variant Effect Predictor tool22.
Transcription factor regulation
The GeneHancer database V.5.953 was used to build transcription factor regulatory networks. The database was filtered to include only promoters and promoter genes with “double elite” confidence scores53. Genes located in QTL regions were tested as transcription factors regulating the expression of DE-genes. If the target DE gene was a transcription factor, its potential role in regulating expression of other DE-genes was also tested. In this way, a network was built starting with the QTL transcription factor coding gene and ending with all targets found.
Supplementary Information
Acknowledgements
This research was funded by Innovation Fund Denmark, Grant ID: 4105-00011B, and the Danish Veterinary and Food Administration (FVST) as part of the agreement of commissioned work between the Ministry of Food and Agriculture and Fisheries of Denmark and the University of Copenhagen and Statens Serum Institute. Tina N. Mahler, Minna Jakobsen and Charlotte B. Larsen are acknowledged for the great contributions to the laboratorial work.
Author contributions
M.F. and P.K.M. conceived and designed the study. A.P. was in charge of the pig cohort and sampling. L.X. generated and processed the sequencing data. S.C. was in charge of RNA isolation and gave conceptual advice. E.I. performed the computational analysis. All authors edited and contributed to the final version of manuscript.
Data availability
The datasets generated and analyzed during the current study are available from the Genome Sequence Archive (GSA) repository with the primary accession code CRA007239.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-26496-1.
References
- 1.Ellis EC, et al. Anthropogenic transformation of the biomes, 1700 to 2000. Glob. Ecol. Biogeogr. 2010;19(5):589–606. [Google Scholar]
- 2.Soleimani T, Hermesch S, Gilbert H. Economic and environmental assessments of combined genetics and nutrition optimization strategies to improve the efficiency of sustainable pork production. J. Anim. Sci. 2021;99(3):skab051. doi: 10.1093/jas/skab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andretta I, et al. Environmental impacts of pig and poultry production: Insights From a systematic review. Front. Vet. Sci. 2021;8:750733. doi: 10.3389/fvets.2021.750733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monteiro A, et al. Environmental impacts and their association with performance and excretion traits in growing pigs. Front. Vet. Sci. 2021;8:677857. doi: 10.3389/fvets.2021.677857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiarie EG, et al. Significance of single beta-mannanase supplementation on performance and energy utilization in broiler chickens, laying hens, turkeys, sows, and nursery-finish pigs: A meta-analysis and systematic review. Trans. Anim. Sci. 2021;5(4):txab160. doi: 10.1093/tas/txab160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veira AM, et al. Effects of sequential feeding with adjustments to dietary amino acid concentration according to the circadian rhythm on the performance, body composition, and nutrient balance of growing-finishing pigs. PLoS ONE. 2021;16(12):e0261314. doi: 10.1371/journal.pone.0261314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andretta I, et al. Precision feeding can significantly reduce lysine intake and nitrogen excretion without compromising the performance of growing pigs. Animal. 2016;10(7):1137–1147. doi: 10.1017/S1751731115003067. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, et al. Identification of the relationship between the gut microbiome and feed efficiency in a commercial pig cohort. J. Anim. Sci. 2021;99(3):skab045. doi: 10.1093/jas/skab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee P, Carmelo VAO, Kadarmideen HN. Integrative analysis of metabolomic and transcriptomic profiles uncovers biological pathways of feed efficiency in pigs. Metabolites. 2020;10(7):275. doi: 10.3390/metabo10070275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Kadarmideen HN. Metabolite genome-wide association study (mGWAS) and gene-metabolite interaction network analysis reveal potential biomarkers for feed efficiency in pigs. Metabolites. 2020;10(5):201. doi: 10.3390/metabo10050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormack UM, et al. porcine feed efficiency-associated intestinal microbiota and physiological traits: Finding consistent cross-locational biomarkers for residual feed intake. mSystems. 2019;4(4):e00324-18. doi: 10.1128/mSystems.00324-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C, et al. A transcriptome analysis reveals that hepatic glycolysis and lipid synthesis are negatively associated with feed efficiency in DLY pigs. Sci. Rep. 2020;10(1):9874. doi: 10.1038/s41598-020-66988-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delpuech E, et al. Identification of genomic regions affecting production traits in pigs divergently selected for feed efficiency. Genet. Sel. Evol. 2021;53(1):49. doi: 10.1186/s12711-021-00642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messad F, et al. Analysis of merged whole blood transcriptomic datasets to identify circulating molecular biomarkers of feed efficiency in growing pigs. BMC Genomics. 2021;22(1):501. doi: 10.1186/s12864-021-07843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, et al. Brain transcriptome analysis reveals potential transcription factors and biological pathways associated with feed efficiency in commercial DLY pigs. DNA Cell Biol. 2021;40(2):272–282. doi: 10.1089/dna.2020.6071. [DOI] [PubMed] [Google Scholar]
- 16.Ramayo-Caldas Y, et al. Integrative approach using liver and duodenum RNA-Seq data identifies candidate genes and pathways associated with feed efficiency in pigs. Sci. Rep. 2018;8(1):558. doi: 10.1038/s41598-017-19072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu Q, et al. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015;5:9938. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jandhyala SM, et al. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21(29):8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy BW, van der Werf JH, Meuwissen TH. Genetic and statistical properties of residual feed intake. J. Anim. Sci. 1993;71(12):3239–3250. doi: 10.2527/1993.71123239x. [DOI] [PubMed] [Google Scholar]
- 20.Pan Z, et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat. Commun. 2021;12(1):5848. doi: 10.1038/s41467-021-26153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang QQ, et al. Power, false discovery rate and winner's curse in eQTL studies. Nucleic Acids Res. 2018;46(22):e133. doi: 10.1093/nar/gky780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu ZL, Park CA, Reecy JM. Bringing the animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022;50(D1):D956–D961. doi: 10.1093/nar/gkab1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ai H, et al. Detection of quantitative trait loci for growth- and fatness-related traits in a large-scale White Duroc x Erhualian intercross pig population. Anim. Genet. 2012;43(4):383–391. doi: 10.1111/j.1365-2052.2011.02282.x. [DOI] [PubMed] [Google Scholar]
- 25.Duthie C, et al. Quantitative trait loci for chemical body composition traits in pigs and their positional associations with body tissues, growth and feed intake. Anim. Genet. 2008;39(2):130–140. doi: 10.1111/j.1365-2052.2007.01689.x. [DOI] [PubMed] [Google Scholar]
- 26.Onteru SK, et al. Whole genome association studies of residual feed intake and related traits in the pig. PLoS ONE. 2013;8(6):e61756. doi: 10.1371/journal.pone.0061756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do, D.N., et al., Genome-wide association and pathway analysis of feed efficiency in pigs reveal candidate genes and pathways for residual feed intake. Front. Genet.5 (2014). [DOI] [PMC free article] [PubMed]
- 28.Vigors S, et al. Analysis of the basal colonic innate immune response of pigs divergent in feed efficiency and following an ex vivo lipopolysaccharide challenge. Physio.l Genomics. 2019;51(9):443–448. doi: 10.1152/physiolgenomics.00013.2019. [DOI] [PubMed] [Google Scholar]
- 29.Cait A, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018;11(3):785–795. doi: 10.1038/mi.2017.75. [DOI] [PubMed] [Google Scholar]
- 30.Santos AFP, et al. Proof-of-principle study suggesting potential anti-inflammatory activity of butyrate and propionate in periodontal Cells. Int. J. Mol. Sci. 2022;23(19):785. doi: 10.3390/ijms231911006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, et al. Association of Rs339939442 in the AHR Gene with litter size are inconsistent among chinese indigenous pigs and western commercial pigs. Animals (Basel) 2019;10(1):11006. doi: 10.3390/ani10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X, et al. CNV analysis of Meishan pig by next-generation sequencing and effects of AHR gene CNV on pig reproductive traits. J. Anim. Sci. Biotechnol. 2020;11:42. doi: 10.1186/s40104-020-00442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonneman D, et al. Potential functional variants in AHR signaling pathways are associated with age at puberty in swine. Anim. Genet. 2021;52(3):284–291. doi: 10.1111/age.13051. [DOI] [PubMed] [Google Scholar]
- 34.Sailer, S., et al., The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification. Life (Basel) 11(2) (2021). [DOI] [PMC free article] [PubMed]
- 35.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh PR, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 2016;48(7):811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howie B, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen NH, McPhee CP, Wade CM. Responses in residual feed intake in lines of large white pigs selected for growth rate on restricted feeding (measured on ad libitum individual feeding) J. Anim. Breed. Genet. 2005;122(4):264–270. doi: 10.1111/j.1439-0388.2005.00531.x. [DOI] [PubMed] [Google Scholar]
- 40.Pedregosa F, et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 41.Cook RD. Influential observations in linear-regression. J. Am. Stat. Assoc. 1979;74(365):169–174. doi: 10.1080/01621459.1979.10481634. [DOI] [Google Scholar]
- 42.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, et al. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao X. Multiple testing corrections for imputed SNPs. Genet. Epidemiol. 2011;35(3):154–158. doi: 10.1002/gepi.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee BT, et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50(D1):D1115–D1122. doi: 10.1093/nar/gkab959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Z, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol. Psychiatry. 2015;20(5):563–572. doi: 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8):e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howe KL, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D, et al. Correction to 'The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets'. Nucleic Acids Res. 2021;49(18):10800. doi: 10.1093/nar/gkab835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fishilevich, S., et al., GeneHancer: genome-wide integration of enhancers and target genes in GeneCards.Database (Oxford), 2017 (2017). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the Genome Sequence Archive (GSA) repository with the primary accession code CRA007239.