Abstract
Tomato leaf mold caused by Cladosporium fulvum (C. fulvum) is a serious fungal disease which results in huge yield losses in tomato cultivation worldwide. In our study, we discovered that ROS (reactive oxygen species) burst was triggered by C. fulvum treatment in tomato leaves. RNA-sequencing was used to identify differentially expressed genes (DEGs) induced by C. fulvum inoculation at the early stage of invasion in susceptible tomato plants. Gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used to annotate functions of DEGs in tomato plants. Based on our comparative analysis, DEGs related to plant-pathogen interaction pathway, plant hormone signal transduction pathway and the plant phenylpropanoid pathway were further analyzed. Our results discovered that a number of core defense genes against fungal invasion were induced and plant hormone signal transduction pathways were impacted by C. fulvum inoculation. Further, our results showed that SA (salicylic acid) and ABA (abscisic acid) contents were accumulated while JA (jasmonic acid) content decreased after C. fulvum inoculation in comparison with control, and quantitative real-time PCR to detect the relative expression of genes involved in SA, ABA and JA signaling pathway further confirmed our results. Together, results will contribute to understanding the mechanisms of C. fulvum and tomato interaction in future.
Keywords: Cladosporium fulvum, tomato, RNA-seq, plant hormones, defense responses, differentially expressed genes (DEGs)
Introduction
Tomato (Solanum lycopersicum) is one of the most economically valuable vegetable crops in the world (Krishna et al., 2016). Tomato leaf mold caused by the biotrophic pathogen Cladosporium fulvum (C. fulvum) is a fungal disease which affects the quality and yield of tomato production severely (Griffiths et al., 2018). In China, yield losses due to tomato leaf mold are typically in the range of 10-25% and may exceed 50% when the disease is severe (Wang et al., 2018). Symptoms of tomato leaf mold usually appear in the leaf adaxial surface with irregular yellowish spots and the leaf abaxial surface with white mold layer at the early onset stage. As the disease develops, the spots become yellow-brown, the leaves curled and withered (Iida et al., 2010). As a model system to investigate the mechanism of tomato and C. fulvum interaction, a total of ten C. fulvum effector proteins have been identified, including four avriulence effector proteins (Avr2, Avr4, Avr3E and Avr9) and six extracellular proteins (Ecp1, Ecp2, Ecp4, Ecp5, Ecp6, and Ecp7) (Van Kan et al., 1991; Van Den Ackerveken et al., 1993; Joosten et al., 1994; Lauge et al., 2000; Luderer et al., 2002; Westerink et al., 2004; Bolton et al., 2008). Race-specific resistance responses against C. fulvum follow the typical gene-for-gene hypothesis. Once the avriulence effector was recognized by the dominant C. fulvum (Cf) resistance genes, the host activates the immune response against the pathogen (Rivas, 2005). To date, at least 24 Cf resistance genes have been discovered and applied in resistance breeding of tomato (Jiang et al., 2022b).
Plant hormones play key regulation roles not only in plant growth and development processes but also in plant responses to a wide range of biotic and abiotic stresses (Benoit and Patrick, 2016). More and more evidence has demonstrated that multiple hormones have been involved in plant and pathogen interactions (Denance et al., 2013; Li et al., 2019a; Liu et al., 2021). In general, salicylic acid (SA) signaling regulates plant defense against biotrophic pathogens, while the jasmonic acid (JA)/ethylene (ET) pathway normally defends against necrotrophic pathogens and herbivorous insects (Jane, 2005; Bari and Jones Jonathan, 2009). It has been documented that SA- and JA/ET-mediated defense signaling pathways synergistically or antagonistically depending on different pathogens and plants interaction (Jia et al., 2013; Yang et al., 2015; Li et al., 2019a). Plants were treated with low concentrations of SA and JA resulting in synergistic expression of both the SA target gene PR1 and the JA marker gene PDF1.2, but the antagonism effect was observed when plants were treated with higher concentrations of SA and JA resulting to the antagonistic expression of these genes (Mur et al., 2006). When tomato plants were infected by the pathogen Alternaria alternata f. sp. Lycopersici, SA, JA and ET-dependent pathways synergistically activated the defense pathway (Jia et al., 2013). And the antagonistic mechanism between SA and JA has been demonstrated in many gymnosperm and angiosperm species (Yang et al., 2015). Recently, the synergistic action of SA and JA pathways was found to enhance plant resistance to herbivores in tea plants (Jiao et al., 2022). Abscisic acid (ABA) plays the multifaceted role in disease resistance, a general pattern is that ABA plays a stimulatory role in plant defense during early stages of pathogen invasion (Ton et al., 2009).
Sequencing of RNAs (RNA-seq) has been widely applied in many biological analyses including host and pathogen interaction (Gao and Chen, 2018; Li et al., 2018; Gao and Chen, 2021; Zhang et al., 2022). For examples, it has been conducted RNA-seq to study the mechanism of the interaction between gray leaf spot fungi and tomato (Zhang et al., 2022); it discovered the potential defense pathway of cucumber against downy mildew through RNA-seq (Gao et al., 2021). Previously, the transcriptome profiling analysis has been performed to study the interaction mechanism of C. fulvum and tomato carrying the resistance gene Cf16 (Zhang et al., 2020). In our study, we used a universally susceptible cultivar moneymaker model plant which lacks any Cf resistance gene. We wondering the plant early defenses responses with fungal invasion in order to provide some theoretical basis for biocontrol of leaf mold disease at the early stage in future. Therefore, we analyze the transcriptome change during C. fulvum and tomato interaction at the time point of 24 hours which can be defined the early interaction period that a pathogen completes the invasion of a host plant (Shen et al., 2017). Gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases have been used to annotate functions of differentially expressed genes (DEGs) in tomato after C. fulvum inoculation. We first focused on analysis of DEGs involved in plant-pathogen interaction pathways. As we expected, a set of genes related to plant defense have been induced. Next, we sought to identify DEGs involved in plant hormone signal transduction pathways and DEGs related with secondary metabolism. Further, our results showed that SA and ABA contents increased while JA contents decreased after C. fulvum invasion, which are consistent results with the relative expression level of genes involved in SA, JA and ABA dependent signaling pathways. Together, our results will broaden our understanding for investigating the mechanism of C. fulvum and tomato interaction.
Materials and methods
Plant materials and Cladosporium fulvum infection
The tomato (Solanum lycopersicum) cultivar Moneymaker which is susceptible to C. fulvum was used in this work. All tomato plants were grown in pots containing nutrient soil in a plant growth incubator with photoperiod conditions of 16h/8h, control temperatures of 26°C/18°C, and humidity set at 75%. Cladosporium fulvum was kindly provided by the Institute of Plant Protection, Chinese Academy of Agricultural Sciences. Conidia of C. fulvum were harvested from one-week-old PDA plates with distilled water, and adjust the spore suspensions concentration to 1× 106 conidia/ml (de Wit et al., 2012). Tomato plants grown in soil at 3 weeks old were sprayed with a conidial suspension (1× 106 conidia/ml) on both adaxial and abaxial sides of the leaves (Van Esse et al., 2007). After 24 h of inoculation, the third leaves below the growing point were collected from treated and control plants separately, rapidly frozen in liquid nitrogen, and stored at -80°C (Yu et al., 2015). Remaining plants were monitored for disease progression for up to 20 days. Three biological replicates were performed.
RNA extraction, library preparation, and sequencing
Leaf surface sprayed with C. fulvum (1× 106 conidia/ml) or sterile water. At 24 h after inoculation (hai), leaf samples were harvested and total RNA was isolated using the Trizol method (Meng and Feldman, 2010; Gao et al., 2014a). The RNA concentration was quantified by a NanoDrop-2000 nucleic acid spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). After detecting the RNA integrity in 1% agarose gel, mRNA was enriched from a pool of RNA by Oligo-dT magnetic beads. Based on the manufacturer’s instructions, RNA was sheared into small pieces by using RNA fragmentation kit (Illumina, San Diego, CA, USA). cDNA was synthesized by reverse transcription with N6 primers. The cDNA fragments were subjected to end repair and adapter ligation. Following the PCR amplification, the PCR products were used to generate cDNA libraries. Quality control and library construction were entrusted to Huada Gene Technology Company (Wuhan, Hubei, China). The cDNA sequencing was carried out with BGISEQ-500 platform for generating raw reads. The RNA-seq data for C. fulvum inoculated samples (accession no: SRR21437047、SRR21437048、SRR21437049) and control-inoculated samples (accession no: SRR21437044、SRR21437045、SRR21437046) are available at the NCBI gene expression omnibus server (https://www.ncbi.nlm.nih.gov/geo/).
Processing of sequencing data
Raw data obtained from BGISEQ-500 platform are subsequently subjected to quality control (QC) to determine whether the sequencing data are suitable for further analysis. The filtering is performed by SOAPnuke software as follows: first, remove the reads containing junction (junction contamination); second, remove the reads with unknown base N content greater than 5%; finally, reads with over half of the component bases with a quality score below 15 were defined as low-quality reads and subsequently removed. After quality control, the filtered clean reads were mapped to the reference genome sequence (GCF_000188115.3_SL2.50, https://www.ncbi.nlm.nih.gov/assembly/GCF_000188115.3/) using HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts). After the alignment, the comparison result is judged by the statistics of the mapping rate and the distribution of reads on the reference sequence to pass the second QC of alignment. The dataset analysis was performed after meeting the requirement.
Functional annotation and enrichment pathway analyses of DEGs
DEGs with |Log2 Fold Change| ≥ 1 and Q-value ≤ 0.05 were functionally classified and enriched using the phyper function in R software to calculate P- value, False discovery rate (FDR) correction was then performed to obtain a normalized P-value, also known as Q-value. Gene Ontology (GO, http://geneontology.org/) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) pathway enrichment were used to analysis DEGs. KEGG pathways with an adjusted Q-value of ≤0.05 were regarded as significantly enriched (Gao et al., 2014b).
Quantitative real-time PCR
A total of randomly selected 10 genes were used for the measurement of transcript abundance by quantitative real-time PCR to verify the reliability of the transcriptome data. Total RNA was extracted from frozen leaf tissue using Trizol reagent according to the manufacturer’s instructions. The integrity of RNA was analyzed by 1% (w/v) agarose gel electrophoresis (Gao et al., 2017). The reverse transcription was performed using the All-in-One First-Strand Synthesis MasterMix Kit (with dsDNase) (Lablead, Beijing,China), while qRT-PCR analysis was conducted using the TB GreenTM Premix Ex TaqTM kit (TaKaRa, Shanghai, China) with a RealTime PCR System (Bio-Rad, Hercules, CA, USA). The PCR running procedure was as follows: a pre-denaturation at 95°C for 30 seconds, 40 cycles with each cycle employing a denaturation temperature at 95°C for 5 seconds and an annealing/extension temperature at 60°C for 30 seconds, followed by melt curve analysis. For each biological sample, three technical replicates were used and there was a total of three biological replicates. Relative quantification of genes was carried out using the 2-ΔΔCT method. The primer sequences for the ten genes are listed in Table 1 . SlACTIN (LOC101260631) served as an internal control. The same method was used to detect the relative expression of six genes related to plant hormones signaling pathway with specific primers listed in Table 2 . This experiment was done in three independent biological replicates.
Table 1.
Primer sequences used for qRT-PCR validation in this study.
| Gene/Gene number | Forward | Reverse | Product length |
|---|---|---|---|
| SlACTIN | GATGGTGGGTATGGGTCAAA | AGGGGCTTCAGTTAGGAGGA | 199 |
| LOC101249624 | ATGCCGATGGATACCGAAACA | CGAGAACTGAACTCCGAAAGA | 131 |
| LOC109119038 | GATGGCTTCACACTTCCCAAG | TACTCTCCATTTTGGTATCCC | 141 |
| LOC101245298 | GTCAAGCCTTTTGGGTTATCG | AGGATCCACTTCGTTCATCAT | 134 |
| LOC101266084 | GAAGCCTTTGAAGGACCATCT | AAGCTTTCCACACTGCTGGTA | 112 |
| LOC101246590 | GGAATGGAATTAGGGTTTGGC | AAATTGAAGGACCTCTTGTGC | 99 |
| LOC101263535 | AAATTGTGGAAGTAATCTCTG | GCATTGACATCATTAAAGTCC | 103 |
| LOC101265854 | GCTCGTGGTCAAGTCGGGGTT | GACCAGAATGAATCAAGTTGC | 112 |
| LOC104648161 | CAAATGCATGTCCCTTGTGTT | CAGATGAAAATCTACCTGACG | 142 |
| LOC101258353 | GTTACTTTGTGCTTCAGCCAA | TCGGAGAGTGAGCTGGTGAGT | 138 |
| LOC101267111 | AGCAGCTTCTTACATGTCAAC | TGGTTATGAGAACACGATCAA | 116 |
Table 2.
Primer sequences for verification of gene expression related to plant hormones signaling pathways used in this study.
| Gene | Forward | Reverse | Product length |
|---|---|---|---|
| SlACTIN | GATGGTGGGTATGGGTCAAA | AGGGGCTTCAGTTAGGAGGA | 199 |
| SlNPR1 | GGTCAGTGTGCTCGCCTAT | TGAAAGGTAAAGGATGCGT | 150 |
| SlPR1 | CACCACTTGATCAAAAAAGTCTAG | TGAATGAATAAGTCTACAATCTTC | 227 |
| SlSRK2C | CGGATATTCGTAGCTGATCCA | TACTAAACTAGCTTCCTCTCC | 120 |
| SlPYR1 | CGTTCATCAGGAAGCAGAAGA | CACTTGATTTGAGCTCATCGG | 98 |
| SlLoxC | AACACCGTTTACTCCGCCCTA | AGTCCTGAAAGATCGACACCC | 131 |
| SlAOC | TCGGAGATCTTGTCCCCTTTA | CGTGCTTGATCAGAATGCAGA | 96 |
Detection of reactive oxygen species burst
Leaves were taken from 3-week-old plant, and a hole punch was used to make a 6 mm diameter round sample from the same part of every leave. The round leave samples were placed in a container with distilled water for 9 h at room temperature to get rid of the mechanical damage for the leaves. Horseradish peroxidase (Shenggong, Shanghai, China) was dissolved in sterile water to make a working solution with a final concentration of 10 mg/ml. C. fulvum were maintained on potato dextrose agar (PDA) at 22°C prior to use, and diluted the spore suspensions in sterile water to 1×106 conidia/ml (de Wit et al., 2012). Mixed solution with 1μl horseradish peroxidase working solution, 1μl conidial suspension (1×106 conidia/ml) as treatment or 1μl sterile water as the control, and 98μl of ECL (enhanced chemiluminescence) solution (Shenggong, Shanghai, China) was as ROS determination solution. For ROS measurement, the cut leaves were removed from water container and put in ROS determination solution immediately. The machine used to measure ROS production is Glomax microplate luminometer (Promega, Wisconsin, USA). One value was detected every 10 seconds, for a total of 160 values per sample. The trend of the values indicates the degree of ROS accumulation (Robineau et al., 2020). The results are presented as means ± SE of three biological and three technical replicates.
Determination of SA, ABA and JA contents
Spore suspensions with C. fulvum (1×106 conidia/ml) were sprayed uniformly on the adaxial and abaxial surfaces of leaves, using sterile water as a control. At 24 h after inoculation, 3-4 leaf samples were taken from three plants for each treatment. The collected samples were immediately frozen in liquid nitrogen and stored at -80°C. The contents of SA, ABA and JA were determined simultaneously by ultra performance liquid chromatography (UPLC) LC-30A coupled with a triple quadrupole mass spectrometer LCMS-8040 (Waters, Massachusetts, USA). The leaf samples prepared in advance were placed on ice and transferred to the rapid nucleic acid extractor. Rapid nucleic acid extraction reagent (900 mg) and 1 ml of ethyl acetate extraction reagent were added and shaken for 45 seconds. The resulting solution was centrifuged at 4°C for 20 min at 16000 g and the supernatant was extracted and evaporated at 30°C under reduced pressure. The dried sample was dissolved in 0.5 ml of 70% methanol solution and centrifuged at 16000 g for 20 min. The supernatant obtained was filtered and put on the machine. The mobile phase consisted of binary gradients of acetonitrile with 0.01% (v/v) formic acid and 0.01% (v/v) aqueous formic acid, with a flowing speed at 0.5 ml/min. A 20 µL portion of each sample was injected into the UPLC-ESI-MS/MS system (Simura et al., 2018; Xin et al., 2020). The results are presented as means ± SE of three biological replicates and every biological repeat includes three technical replicates.
Results
Symptoms induced by Cladosporium fulvum inoculation in tomato plants
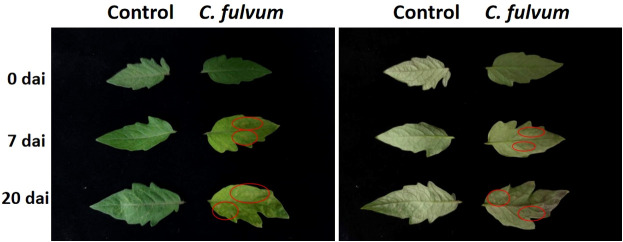
In order to observe the symptoms induced by C. fulvum, a suspension of spores containing the leaf mold pathogen (1× 106 conidia/ml) was sprayed on tomato leaves. Compared to the control group, pathogen-inoculated leaves with yellow spots at 7 days after inoculation. At 20 days after inoculation, yellow spotted area expanded; mold layer can be observed in the abaxial side of leaves and when severe leaves were obviously curled ( Figure 1 ). These symptoms indicated successful infection of C. fulvum and the plant materials we taken at the time point of 24 hours were reliable for experiments.
Figure 1.
Symptoms after tomato leaves infected by Cladosporium fulvum. Tomato leaves were sprayed exogenously with a conidial suspension (1×106 conidia/ml) of C. fulvum on the adaxial and abaxial sides of the leaves, water as a control treatment. Pictures were taken from 0 days, 7 days and 20 days after inoculation. The left half of the figure is the adaxial side of the leaves. The right half of the figure is the abaxial side of the leaves. The disease symptoms are more obvious in the places marked by red circles. dai: days after inoculation.
ROS production triggered by Cladosporium fulvum treatment
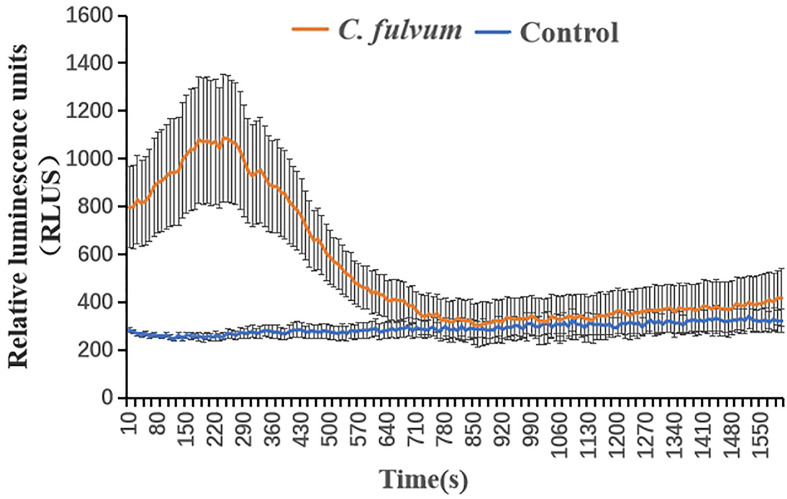
Detection of high levels of ROS, a signal substance, can reflect the ability of the plant to resist pathogen at some extent (Hernandez et al., 2016). The leave samples were firstly put in distilled water up to 9 hours to avoid impact caused by mechanical damage, then we measured the ROS level of tomato leaves treated with C. fulvum. Results showed that the timing and accumulation of ROS production in tomato leaves in response to C. fulvum were different compared with control treatment. Tomato leaves treated with C. fulvum produced a peak of ROS accumulation and the overall value was higher than the control ( Figure 2 ). This indicates that C. fulvum infection activates an early ROS burst in leaves.
Figure 2.
Production of reactive oxygen species (ROS) in tomato leaf disks after treatment with Cladosporium fulvum. Tomato leaf disks were treated with C. fulvum at the concentration of 1×106 conidia/ml and water as control. ROS production was measured using the chemiluminescence of luminol and photon counts were expressed as relative luminescence units (RLUs). The X-axis indicates time, and the instrument measures one value every 10 s, for a total of 160 values, Y-axis indicates the level of ROS accumulation by relative luminescence units. The results are presented as means ± SE of three biological and three technical replicates.
Overview of the Solanum lycopersicum transcriptome
Tomato infected by C. fulvum was used as the treatment group and sterile water treatment was used as the control group for transcriptome sequencing. Three biological repeats were performed for each group. A total of six samples were sequenced using the DNBSEQ platform, and each sample yielded an average of 1.19G of data. We obtained an average of 23.81 M high-quality reads per sample, accounting for > 99% of the raw reads for each sample. The reads with a quality value of 30 accounted for > 95% of the total reads and a quality value of 20 account for > 98% of the total reads, indicating that the sequencing reads are of high quality ( Table 3 ). Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) was used to align the clean reads to the reference genome sequence (http://daehwankimlab.github.io/hisat2/) after obtaining the clean reads. The average alignment rate to the reference genome of Solanum lycopersicum was 97.54%, indicating the high quality of the data, guaranteeing the reliability of subsequent differentially expressed genes (DEGs) analysis.
Table 3.
Quantitative analysis and comparison ratio of raw RNA-seq data.
| Sample | Total Raw Reads (M) | Total Clean Reads (M) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Clean Reads Ratio (%) | Genome Total Mapping (%) |
|---|---|---|---|---|---|---|
| Control_1 | 23.92 | 23.79 | 98.43 | 95.28 | 99.44 | 97.71 |
| Control_2 | 23.92 | 23.71 | 98.43 | 95.23 | 99.12 | 97.02 |
| Control_3 | 23.92 | 23.86 | 98.5 | 95.41 | 99.75 | 97.7 |
| C. fulvum_1 | 23.92 | 23.76 | 98.42 | 95.25 | 99.31 | 97.53 |
| C. fulvum_2 | 23.92 | 23.85 | 98.54 | 95.54 | 99.69 | 97.64 |
| C. fulvum_3 | 23.92 | 23.87 | 98.49 | 95.43 | 99.76 | 97.66 |
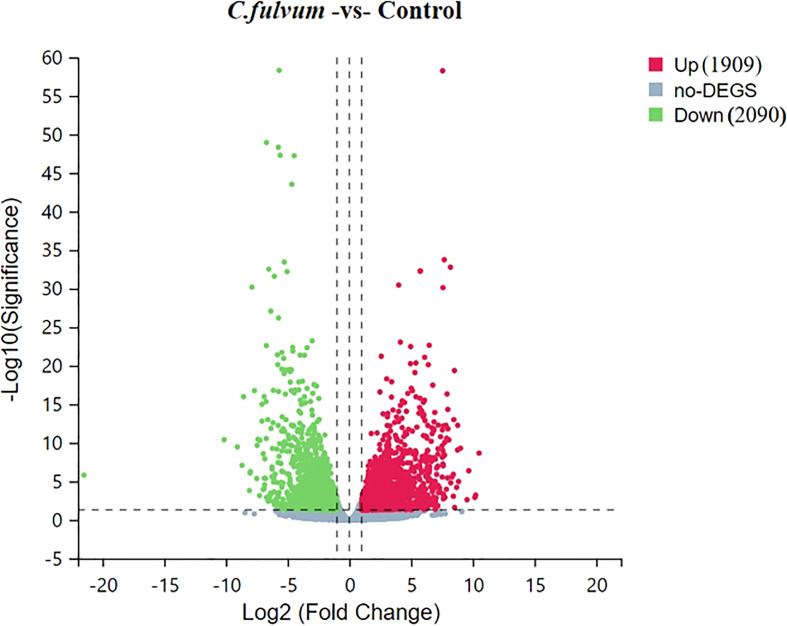
Clean reads were compared to reference gene sequences by Bowtie2 software, and then gene expression levels of individual samples were calculated using RNA-Seq by Expectation-Maximization (RSEM) (deweylab.github.io), followed by DEGs screening according to the method described by Michael (Love et al., 2014). The significant DEGs were determined based on the criteria of |Log2 Fold Change| ≥ 1 and Q-value ≤0.05, the distribution of DEGs based on degree of difference and the significance of the difference were visualized with a volcanic plot ( Figure 3 ). Compared to the control, C. fulvum inoculation plants induced 1909 upregulated DEGs and 2090 downregulated DEGs ( Tables S1 , S2 ).
Figure 3.
Volcano plot of upregulated and downregulated differentially expressed genes (DEGs) after Cladosporium fulvum inoculation. X-axis represents log2 transformed difference multiplier values and Y-axis represents -log10 transformed significance values. Red dots represent upregulated DEGs, green dots represent downregulated DEGs, and gray dots represent that were not significantly different between C.fulvum and control treatment.
Gene ontology enrichment analysis of differentially expressed genes
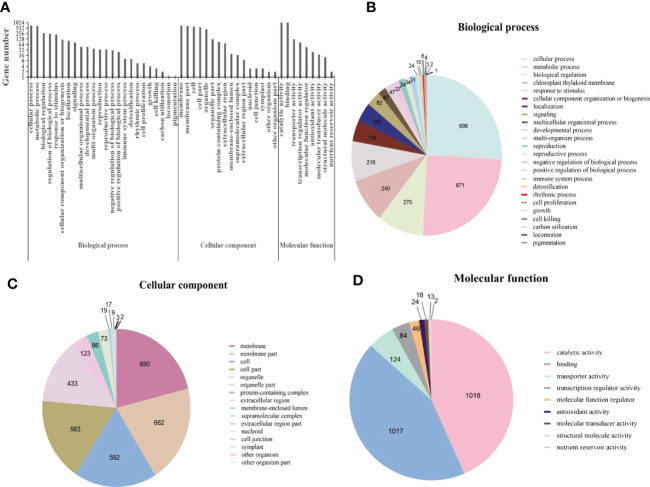
Gene Ontology (GO) is an international standardized gene function classification system. Based on GO terms, DEGs were classified into three major categories: biological process, cellular component, and molecular function. Within the broad GO category of biological process, a total of 2693 DEGs were involved in 24 GO terms ( Figure 4 ). According to previous reports, terms with “cellular process”, “metabolic process”, “biological regulation”, “regulation of biological process”, “response to stimulus” were associated with plant disease resistance (Zhang et al., 2020). In the classification of cellular component, there were 3280 DEGs involving 16 GO terms ( Figure 4 ), DEGs mainly concentrated in the integral component of membrane. In the molecular function category, more than 85% of the 2346 DEGs were mainly enriched in the “catalytic activity” and “binding” ( Figure 4 ). The enrichment of DEGs in these two terms has been found to associate with the induction of many genes involved in plant hormone signal transduction pathway (Zhang et al., 2020; Singh et al., 2021).
Figure 4.
Gene ontology (GO) annotations for differentially expressed genes (DEGs) in Cladosporium fulvum inoculated tomato leaves vs control treatment. (A) GO classifications of differential genes in C fulvum vs control treatment. Pie charts showing the breakdown and prevalence of enriched GO terms in the following categories: biological process (B), cellular component (C) and molecular function (D).
Kyoto Encyclopedia of Genes and Genomes enrichment analysis of differentially expressed genes
The KEGG (Kyoto Encyclopedia of Genes and Genomes) database is a database that systematically analyzes gene functions, links genomic information and functional information. Through KEGG enrichment analysis, C. fulvum infection induced 830 DEGs involved in 125 KEGG pathways. Fifteen pathways with P- values ≤0.05 were selected based on the number of involved DEGs of corresponding pathways arranged in a descending order ( Table 4 ). The KEGG pathway with the highest number of enriched DEGs was Plant hormone signal transduction pathway (ko04075) involving in 61 DEGs, which includes 40 upregulated DEGs and 21 downregulated DEGs ( Table S3 ). The results above suggest that plant hormone pathways were significantly affected by C. fulvum infection. More importantly, many pathways associated with plant disease resistance were activated, such as Plant-pathogen interaction (ko04626), MAPK signaling pathway – plant (ko04016), and Peroxisome (ko04146) (Nyathi and Baker, 2006; Jiang et al., 2022a). In addition, many metabolic pathways such as Starch and sucrose metabolism (ko00500) and Circadian rhythm – plant (ko04712) were also altered, indicating that pathogenic fungal infection can also affect normal plant growth and development.
Table 4.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (top 15) of differentially expressed genes (DEGs) enrichment by C. fulvum infection.
| PathwayID | KEGG Pathway | Gene Num | upregulatedDEGs | downregulatedDEGs | P- value |
|---|---|---|---|---|---|
| 04075 | Plant hormone signal transduction | 61 | 40 | 21 | 0.02 |
| 00940 | Phenylpropanoid biosynthesis | 47 | 26 | 21 | 0.03 |
| 04626 | Plant-pathogen interaction | 45 | 33 | 12 | 0.05 |
| 04016 | MAPK signaling pathway - plant | 45 | 34 | 11 | 2.56E-3 |
| 00500 | Starch and sucrose metabolism | 42 | 12 | 30 | 8.12E-5 |
| 00480 | Glutathione metabolism | 31 | 19 | 12 | 2.39E-3 |
| 00230 | Purine metabolism | 25 | 9 | 16 | 9.00E-3 |
| 04146 | Peroxisome | 23 | 13 | 10 | 0.02 |
| 00561 | Glycerolipid metabolism | 22 | 17 | 5 | 0.01 |
| 00562 | Inositol phosphate metabolism | 20 | 10 | 10 | 0.02 |
| 01212 | Fatty acid metabolism | 20 | 10 | 10 | 0.03 |
| 00941 | Flavonoid biosynthesis | 19 | 9 | 10 | 2.09E-3 |
| 04712 | Circadian rhythm - plant | 17 | 8 | 9 | 4.04E-4 |
| 00908 | Zeatin biosynthesis | 17 | 9 | 8 | 0.04 |
| 00592 | Alpha-Linolenic acid metabolism | 14 | 7 | 7 | 0.03 |
A set of core genes involved in plant defense were upregulated after Cladosporium fulvum inoculation
In our study, a total of 45 DEGs related with the plant defense responses were identified after C. fulvum inoculation, in which 33 genes were upregulated and 12 genes were downregulated ( Table S4 ). These genes can be considered indicators of plant defense in response to C. fulvum invasion. A total of six serine/threonine-protein kinase receptors or receptor like kinases were identified ( Table 5 ), all of which were upregulated after C. fulvum inoculation. In particular, two genes encoding of LRR receptor-like serine/threonine-protein kinase FLS2 (flagellin sensing 2) and FLS3 (flagellin sensing 3) that binds to flg22 and flgII-28 respectively (Hind et al., 2016; Chi et al., 2021) were significantly induced to 6-fold and 27-fold compared to the control treatment. And gene encoding SERK3B (somatic embryogenesis receptor kinase 3B), which interacts with FLS2 for activating downstream signaling (Ma et al., 2022), was also slightly upregulated. This result indicated that tomato activated the intracellular immune signaling responses after C. fulvum invasion.
Table 5.
Differentially expressed genes related with the plant defense responses after Cladosporium fulvum treatment.
| Gene ID | Gene Symbol | Seq Description | Log2 (C.fulvum/Control) | P-value |
|---|---|---|---|---|
| 101246110 | LOC101246110 | LRR receptor-like serine/threonine-protein kinase EFR | 2.20 | 1.97E-04 |
| 101248095 | FLS3 | FLAGELLIN-SENSING 3 protein | 4.76 | 1.80E-08 |
| 101256183 | LOC101256183 | Serine/threonine-protein kinase PBS1 | 0.81 | 0.003420758 |
| 101257866 | LOC101257866 | Probable serine/threonine-protein kinase PBL7 | 4.75 | 1.48E-08 |
| 101260980 | LOC101260980 | Probable LRR receptor-like serine/threonine-protein kinase At3g47570 | 3.41 | 0.003189953 |
| 101263667 | FLS2 | LRR receptor-like serine/threonine-protein kinase FLS2 | 2.62 | 1.82E-04 |
| 100736531 | SERK3B | Somatic embryogenesis receptor kinase 3B | 0.87 | 0.002665399 |
| 101055527 | LOC101055527 | Hop-interacting protein THI080 | 1.45 | 9.70E-04 |
| 101260391 | LOC101260391 | Calcium-dependent protein kinase 18-like | 2.10 | 0.008481838 |
| 101244290 | LOC101244290 | Calmodulin-like protein 3 | 3.15 | 1.68E-09 |
| 101245298 | LOC101245298 | Calmodulin | 4.78 | 4.67E-06 |
| 101245539 | LOC101245539 | Probable calcium-binding protein CML44 | 1.24 | 0.00819727 |
| 101257476 | LOC101257476 | Calmodulin-like protein 8 | 4.16 | 2.38E-04 |
| 101264550 | LOC101264550 | Calcium-binding protein CP1 | 5.09 | 2.52E-09 |
| 101265816 | LOC101265816 | Caltractin | 1.06 | 0.007541617 |
| 112940015 | LOC112940015 | Calmodulin-like protein 1 | 1.81 | 4.66E-04 |
| 101245220 | CER6 | 3-ketoacyl-CoA synthase 6 | 0.74 | 0.007012538 |
| 101262858 | LOC101262858 | 3-ketoacyl-CoA synthase 20-like | 1.67 | 1.40E-05 |
| 101268230 | LOC101268230 | 3-ketoacyl-CoA synthase 1 | 2.67 | 1.06E-09 |
| 101268257 | LOC101268257 | 3-ketoacyl-CoA synthase 11-like | 4.88 | 0.004680944 |
| 101252097 | LOC101252097 | WRKY transcription factor 1 | 2.12 | 5.29E-05 |
| 101260537 | LOC101260537 | Probable WRKY transcription factor 26 | 3.09 | 3.29E-10 |
| 100191111 | LOC100191111 | PR1 protein | 6.51 | 2.22E-09 |
| 544123 | PR1b1 | Pathogenesis-related leaf protein 6 | 7.54 | 1.38E-14 |
| 544185 | P4 | Pathogenesis-related protein P4 | 8.69 | 2.47E-06 |
| 101246133 | LOC101246133 | Calcium-dependent protein kinase 29 | -2.37 | 1.23E-08 |
| 101250418 | LOC101250418 | Calcium-dependent protein kinase 24 | -1.81 | 0.006829089 |
| 101255379 | LOC101255379 | Calcium-dependent protein kinase 17-like | -3.60 | 4.70E-08 |
| 101256200 | LOC101256200 | Calcium-dependent protein kinase 1 | -2.30 | 2.84E-12 |
| 101244728 | LOC101244728 | Caltractin | -3.98 | 4.44E-06 |
| 543984 | CaM6 | Calmodulin 6 | -1.13 | 0.005832385 |
In response to various stimuli in plants, Ca2+ acts as a second messenger to modulate different target protein activities through CAM/CML receptors, thereby regulating a variety of cellular functions (Zhang et al., 2014). A total of 15 DEGs related with Ca2+ signaling were identified upon C. fulvum infection with 9 upregulated and 6 downregulated genes ( Table 5 ). Genes encoding a few of calmodulin (CML) or calmodulin like proteins such as calmodulin (LOC101245298), CML44 (LOC101245539), calcium-binding protein (LOC101264550, CP1), and one potential CDPK protein (LOC101260391, calcium-dependent protein kinase 18-like) were upregulated. Four genes encoding CDPK were downregulated after C. fulvum infection, including CDPK1 (calcium-dependent protein kinase 1), CDPK17-like, CDPK24 and CDPK29, which is consistent with previous report that CDPK1 expression was decreased after 1.5 h treatment with C. fulvum (Chico et al., 2002). This result indicated that differently expression of CDPK genes are involved in the complex signaling network of tomato in response to C. fulvum invasion.
In addition, four genes encoding 3-Ketoacyl-CoA synthase (KCS), which catalyzes a condensation reaction to form 3-ketoacyl-CoA during very long chain fatty acid synthesis, including KCS1, KCS6, 3-ketoacyl-CoA synthase 20-like (LOC101262858) and 3-ketoacyl-CoA synthase 11-like (LOC101268257) were all upregulated in our study ( Table 5 ). These gene expression might contribute to plant cuticular wax and suberin biosynthesis after pathogen invasion (Edqvist et al., 2018). Moreover, two transcriptional factors WRKY26 and WRKY1, which have been reported as key components of resistance in tomato against Alternaria solani (Shinde et al., 2018), were induced in our study. Further, several defense responses related genes such as those encoding pathogenesis-related (PR)1 protein (LOC100191111) (Lavrova et al., 2017), PR1b (Hoegen et al., 2002), and P4 (Pieterse and van Loon, 1999) were significantly induced. Together, our data analysis suggested that plant defense pathways are activated after C. fulvum inoculation in tomato plant.
Expression of genes related to multiple hormones affected by Cladosporium fulvum inoculation
Next, we chose to analyze DEGs related to plant hormone signal transduction pathway which play key roles in regulating plant defense response against pathogen ( Table S3 ). In the SA signaling pathway, a total of 9 DEGs were identified; 6 genes were upregulated and 3 genes were downregulated. The downregulated genes include encoding the transcription factor TGA1 (TGACG MOTIF-BINDING FACTOR 1), pathogenesis-related leaf protein 4 (LOC101265854) and BOP2 (BLADE-ON-PETIOLE protein), which is associated with leaf and flower development in Arabidopsis (Zhang et al., 2017). The upregulated genes include a SA receptor NPR1 (Nonexpressor of PR1, also known as NIM1) (Li et al., 2019b), NML2 (NPR1/NIM1-like protein), TGA2.2 (Hou et al., 2019) and PR genes (PR-1b1 and P4) ( Table 6 ). This result suggested that SA signaling pathway are activated after C. fulvum invasion given that PR1 is an indicator for activation of the SA signaling pathway in plants.
Table 6.
Differentially expressed genes related with plant hormones signaling pathway affected by Cladosporium fulvum treatment.
| Pathway | Gene ID | Gene Symbol | Seq Description | Log2 (C.fulvum/Control) | P-value |
|---|---|---|---|---|---|
| SA | 543939 | NPR1 | Regulatory protein NPR1 | 1.39 | 2.07E-04 |
| 544270 | NML2 | NIM1-like protein 2 | 1.94 | 1.83E-07 | |
| 543600 | TGA2.2 | Transcription factor TGA2.2 | 1.15 | 0.001692675 | |
| 544123 | PR1b1 | Pathogenesis-related leaf protein 6 | 7.54 | 1.38E-14 | |
| 544185 | P4 | Pathogenesis-related protein P4 | 8.69 | 2.47E-06 | |
| 100191111 | LOC100191111 | PR1 protein | 6.51 | 2.22E-09 | |
| 101246001 | BOP2 | BLADE-ON-PETIOLE protein BOP2 | -3.21 | 0.006220984 | |
| 101265431 | LOC101265431 | Transcription factor TGA1 | -2.12 | 5.31E-04 | |
| 101265854 | LOC101265854 | Pathogenesis-related leaf protein 4 | -2.38 | 1.94E-06 | |
| ABA | 101246807 | LOC101246807 | Abscisic acid receptor PYL9 | 1.06 | 0.008230946 |
| 101258886 | LOC101258886 | Abscisic acid receptor PYL3 | 1.07 | 0.003742465 | |
| 101267127 | LOC101267127 | Abscisic acid receptor PYR1 | 0.97 | 0.005559441 | |
| 101265524 | LOC101265524 | Protein phosphatase 2C 53 | 2.68 | 1.60E-09 | |
| 100037510 | SRK2C | SNF1-related kinase | 1.21 | 0.003101157 | |
| 101249794 | LOC101249794 | Protein phosphatase 2C 51-like | -2.19 | 0.004197722 | |
| 101251432 | LOC101251432 | Serine/threonine-protein kinase SRK2I | -2.84 | 3.23E-12 | |
| 100820704 | ABF4 | ABA responsive transcription factor | -1.10 | 0.006243886 | |
| JA | 101266902 | AOS2 | Allene oxide synthase 2 | -2.05 | 2.30E-3 |
| 544008 | LOXC | Lipoxygenase | -3.31 | 3E-8 | |
| 544306 | AOC | Allene oxide cyclase | -1.40 | 8.09E-3 | |
| ET | 101249950 | LOC101249950 | ETHYLENE INSENSITIVE 3-like 3 protein | 2.11 | 0.002604271 |
| 101246590 | LOC101246590 | Ethylene-responsive transcription factor 1B | 4.73 | 1.18E-19 | |
| 543712 | EREB | Ethylene responsive element binding protein | 5.26 | 5.03E-05 | |
| 606712 | LOC606712 | Ethylene-responsive transcription factor 1 | 3.19 | 3.68E-05 |
We further analyzed genes involved in the ABA signaling pathway affected by C. fulvum invasion. The complex consisting of the ABA receptor PYR/PYL/RCAR (Pyrabactin resistance/PYR-like/regulatory components of ABA receptor), PP2C (type 2C protein phosphatase), and SnRK2s (Sucrose Non-fermentation Kinase Subfamily 2) has a key role in ABA signaling (Guo et al., 2011). In our study, genes encoding abscisic acid receptor PYL9 (LOC101246807), PYL3 (LOC101258886), PYR1(LOC101267127), PP2C (protein phosphatase 2C 53)and SRK2C (SNF1-related kinase), analogous to subclass III SnRK2s (Mizoguchi et al., 2010), were all upregulated after C. fulvum invasion. In addition, LOC101249794 encoding protein phosphatase 2C 51-like, SRK21 (LOC101251432) encoding serine/threonine-protein kinase SnRK21 and ABF4 (ABA responsive transcription factor) (Orellana et al., 2010) were downregulated ( Table 6 ). Previous study suggested that once PYR/PYLs binds to ABA, it represses PP2C activity and releases SnRK2 suppression, triggering downstream ABA responses (Yang et al., 2017). Our data analysis indicated that ABA signaling might be activated after C. fulvum invasion.
In the JA synthesis pathway, the genes LoxC (lipoxygenase), AOS2 (allene oxide synthase 2), and AOC (allene oxide cyclase) related to JA synthesis (Liu et al., 2012) were downregulated 2-8 fold ( Table 6 ). And four genes involved in the ethylene signaling pathway were upregulated, including ERF1 (ethylene response transcription factor 1), LOC101246590 (ethylene-responsive transcription factor 1B), EREB (ethylene responsive element binding protein) (Mingchun et al., 2016) and LOC101249950(ETHYLENE INSENSITIVE 3-like 3 protein) ( Table 6 ). These results suggested that C. fulvum inoculation impacted the JA and ET signaling pathway differently in plants.
A total of 24 DEGs related to auxin signaling pathway were identified after C. fulvum invasion, of which 13 were upregulated and 11 were downregulated ( Table 7 ). The SAUR (small auxin-up RNA) genes can respond not only to auxin but also to internal and environmental stress with mounting dynamic spatial-temporal responses (Wang et al., 2020). In particular, 11 SAUR genes were identified to be affected after C. fulvum treatment. Among these 11 DEGs, genes such as SAUR32, SAUR58, an SAUR71 were upregulated and those SAUR71-like (LOC101248065, LOC101257321) genes were downregulated. The other class of DEGs affected by C. fulvum inoculation was IAAs; upregulated genes include IAA4, IAA14, IAA35, IAA7, and downregulated genes include IAA15, IAA17, and IAA19 (Audran-Delalande et al., 2012). One gene ARF5 (auxin response factor 5) was inhibited while the gene ARF1 (auxin response factor 1) was slightly induced (Zouine et al., 2014). Two genes encoding Auxin transporter-like protein LAX2 and LAX5 were downregulated (Pattison and Catala, 2012). The above results indicate that C. fulvum infection does affect the auxin-related pathways in tomato plants in a complicated manner.
Table 7.
Differentially expressed genes related with auxin signaling pathway affected by Cladosporium fulvum treatment.
| Gene ID | Gene Symbol | Seq Description | Log2 (C.fulvum/Control) | P-value |
|---|---|---|---|---|
| 101246226 | LOC101246226 | Auxin-responsive protein SAUR32 | 2.02 | 2.33E-04 |
| 101055583 | LOC101055583 | Small auxin-up protein 58 | 3.18 | 2.27E-12 |
| 101253234 | LOC101253234 | Auxin-responsive protein SAUR50-like | 2.23 | 0.00387351 |
| 101250847 | LOC101250847 | Auxin-responsive protein SAUR50-like | 6.60 | 2.18E-07 |
| 101265243 | LOC101265243 | Auxin-responsive protein SAUR71 | 3.24 | 8.21E-04 |
| 104645436 | LOC104645436 | Auxin-responsive protein SAUR21-like | 1.81 | 0.001693518 |
| 101255303 | IAA4 | Auxin-responsive protein IAA4 | 3.22 | 1.28E-10 |
| 543542 | IAA7 | IAA7 protein | 1.88 | 3.94E-05 |
| 101055547 | LOC101055547 | IAA14 | 0.84 | 0.003315808 |
| 101055555 | LOC101055555 | IAA35 | 1.80 | 0.009160947 |
| 104645435 | LOC104645435 | Auxin-induced protein 15A-like | 1.98 | 5.94E-04 |
| 109118704 | LOC109118704 | Auxin-induced protein 15A-like | 2.31 | 4.33E-04 |
| 100736509 | ARF1 | Auxin response factor 1 | 1.17 | 0.00645183 |
| 101246270 | LOC101246270 | Auxin-responsive protein SAUR36-like | -2.09 | 6.36E-04 |
| 101248065 | LOC101248065 | Auxin-responsive protein SAUR71-like | -5.45 | 0.002862421 |
| 101257321 | LOC101257321 | Auxin-responsive protein SAUR71-like | -3.85 | 4.31E-05 |
| 101248844 | LOC101248844 | Auxin-responsive protein SAUR50 | -5.53 | 8.48E-04 |
| 101255313 | LOC101255313 | Auxin-responsive protein SAUR71 | -1.74 | 0.006481308 |
| 101055548 | IAA15 | Auxin-regulated IAA15 | -1.83 | 0.002256359 |
| 543544 | IAA17 | Auxin-responsive protein IAA17 | -3.02 | 6.02E-05 |
| 101055549 | IAA19 | Auxin-responsive protein IAA19 | -1.92 | 0.004643909 |
| 100736448 | ARF5 | Auxin response factor 5 | -3.50 | 3.37E-11 |
| 100736477 | LAX2 | Auxin transporter-like protein 2 | -3.38 | 4.80E-14 |
| 100736541 | LAX5 | Auxin transporter-like protein 5 | -4.55 | 6.05E-04 |
Genes involved in the plant phenylpropanoid pathway are affected by Cladosporium fulvum inoculation
Plant phenylpropanoid pathway is an important pathway for the synthesis of plant secondary metabolites, some of which have been proposed as important components of plant defense responses (Naoumkina et al., 2010; Piasecka et al., 2015). Its downstream metabolites mainly include coumarins, flavonoids, terpenoids, anthocyanins, lignin and other phenylpropanoids (Deng and Lu, 2017; Pratyusha and Sarada, 2022). In this study, many tomato DEGs were related with plant phenylpropanoid pathway after C. fulvum invasion. A total of 47 genes was enriched in the phenylpropanoid biosynthesis pathway, with 26 DEGs upregulated and 21 DEGs downregulated ( Table S5 ). Phenylalanine ammonia lyase (PAL) is the first rate-limiting enzyme in the phenylpropane metabolic pathway (Ruili et al., 2016). PAL2 (LOC101249824) was found to be slightly induced after invasion. The other two genes (LOC101244496, LOC101248210) encoding trans-cinnamate 4-monooxygenase and 4-coumarate-CoA ligase, two major players in this pathway, were also upregulated ( Table 8 ). The gene encoding cinnamoyl-CoA reductase (CCR, LOC778359), a key enzyme in the formation of lignin monomers (Fan et al., 2015), was upregulated. Peroxidases (PRXs) are often found in lignifying tissues and they have the capability to oxidize a wide variety of small phenolic compounds, including monolignols (Marjamaa et al., 2009). In our study, there are 9 related genes upregulated such as TAP2 (tomato anionic peroxidase 2, LOC101245316) (Melillo et al., 2014) and CEVI-1 (citrus exocortis viroid) (Vera et al., 1993) encoding an anionic peroxidase in tomato, which could be induced by compatible viral infection (Mayda et al., 2000), while peroxidase 12, peroxidase 18, peroxidase 3, peroxidase 44 like and 45 like genes were downregulated ( Table 8 ). Four genes related with flavonoid biosynthesis, CHS1 (chalcone synthase 1), CHS2 (Heredia et al., 2015), CHI1 (chalcone-flavonoid isomerase 1) (Kang et al., 2014) and DFR (dihydroflavonol 4-reductase) (Bongue-Bartelsman et al., 1994) were significantly downregulated ( Table 8 ). Both ANS (anthocyanidin synthase) and F3H (flavanone 3-dioxygenase) (Li et al., 2019c) involved in anthocyanin synthesis were also downregulated ( Table 8 ). In addition, a few genes encoding beta-glucosidase, catalyzing the hydrolysis of cellobiose and cello-oligosaccharides containing (1 → 4)-beta-glycosidic bonds to glucose, which is crucial in cellulosic ethanol production were downregulated, such as LOC101254239, LOC101263519, LOC101256554, whereas only 1 beta-glucosidase 18-like (LOC101248595) was upregulated ( Table 8 ), suggesting cellulosic ethanol production pathway was inhibited after C. fulvum invasion. Our results indicated that a variety of secondary metabolites related to plant disease resistance was affected by fungal infection.
Table 8.
Differentially expressed genes induced by C. fulvum infection are involved in the production of secondary metabolites.
| Pathway | Gene ID | Gene symbol | Seq Description | Log2(C.fulvum/Control) | P-value |
|---|---|---|---|---|---|
| Phenylpropanoid | 101249824 | PAL2 | Phenylalanine ammonia-lyase 2 | 0.96 | 0.17 |
| 101244496 | LOC101244496 | Trans-cinnamate 4-monooxygenase | 1.38 | 1.32E-3 | |
| 101248210 | LOC101248210 | 4-coumarate–CoA ligase | 2.29 | 1.37E-6 | |
| Lignin | 778359 | CCR2 | Cinnamoyl-CoA reductase | 2.03 | 2.74E-4 |
| 101267754 | LOC101267754 | Peroxidase 51 | 6.23 | 1.46E-4 | |
| 101265511 | LOC101265511 | Suberization-associated anionic peroxidase 2-like | 6.22 | 5.3E-10 | |
| 101251503 | LOC101251503 | Peroxidase 21 | 5.82 | 1.94E-6 | |
| 101258529 | LOC101258529 | Peroxidase P7-like | 5.28 | 4E-9 | |
| 101264425 | LOC101264425 | Peroxidase P7 | 5.09 | 9E-9 | |
| 101243845 | LOC101243845 | Peroxidase 72 | 4.83 | 2.86E-3 | |
| 101245316 | TAP2 | Suberization-associated anionic peroxidase 2 | 3.70 | 5.34E-4 | |
| 101261825 | LOC101261825 | Peroxidase P7 | 2.95 | 1.39E-3 | |
| 544084 | CEVI-1 | Peroxidase | 2.58 | 3E-8 | |
| 101253377 | LOC101253377 | Peroxidase 12 | -1.45 | 8.97E-4 | |
| 101253684 | LOC101253684 | Peroxidase 12 | -1.77 | 2.55E-3 | |
| 101247048 | LOC101247048 | Peroxidase 45-like | -3.28 | 4.83E-4 | |
| 101254073 | LOC101254073 | Peroxidase 18 | -3.57 | 7.70E-3 | |
| 101250356 | LOC101250356 | Peroxidase 44-like | -5.44 | 3.25E-3 | |
| 101250523 | LOC101250523 | Peroxidase 3 | -6.39 | 1.84E-5 | |
| Flavonoid | 778294 | CHS1 | Chalcone synthase | -5.13 | 1.4E-12 |
| 778295 | CHS2 | Chalcone synthase | -4.11 | 9.1E-13 | |
| 101249265 | CHI1 | Chalcone–flavonone isomerase 1 | -3.06 | 2.63E-5 | |
| 544150 | DFR | Dihydroflavonol 4-reductase | -5.69 | 7.5E-11 | |
| Anthocyanin | 101251607 | ANS | Anthocyanidin synthase | -3.11 | 1E-7 |
| 100736482 | F3H | Flavanone 3-dioxygenase | -2.40 | 5.11E-6 | |
| Cellulosic ethanol | 101248595 | LOC101248595 | Beta-glucosidase 18-like | 2.08 | 1.01E-08 |
| 101254239 | LOC101254239 | Beta-glucosidase BoGH3B | -5.13 | 5.09E-16 | |
| 101263519 | LOC101263519 | Beta-glucosidase BoGH3B | -2.17 | 0.005989607 | |
| 101256554 | LOC101256554 | Beta-glucosidase BoGH3B | -1.67 | 7.03E-05 |
Quantitative real-time PCR validation of transcriptome results
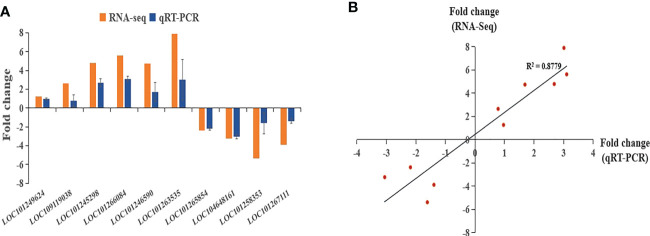
To validate the reliability of transcriptome data, we randomly picked some upregulated and downregulated genes with one gene related to plant resistance. A total of 10 genes was selected including LOC101249624, LOC109119038, LOC101245298, LOC101266084, LOC101246590, LOC101263535, LOC101265854, LOC104648161, LOC101258353, LOC101267111 ( Figure 5A ). The expression data obtained from qRT-PCR verification had consistent trend with RNA-Seq, with a correlation coefficient of 0.8779 ( Figure 5B ), indicating the RNA-seq data were reliable.
Figure 5.
qRT-PCR validation of RNA-seq sequencing results. (A) qRT-PCR and RNA-seq comparison between ten randomly selected differentially expressed genes. The qRT-PCR values represent mean ± SE from three biological replicates. (B) Regression analysis comparing gene expression ratios by qRT-PCR and RNA-seq analysis.
SA, ABA and JA contents in plants are affected by Cladosporium fulvum inoculation
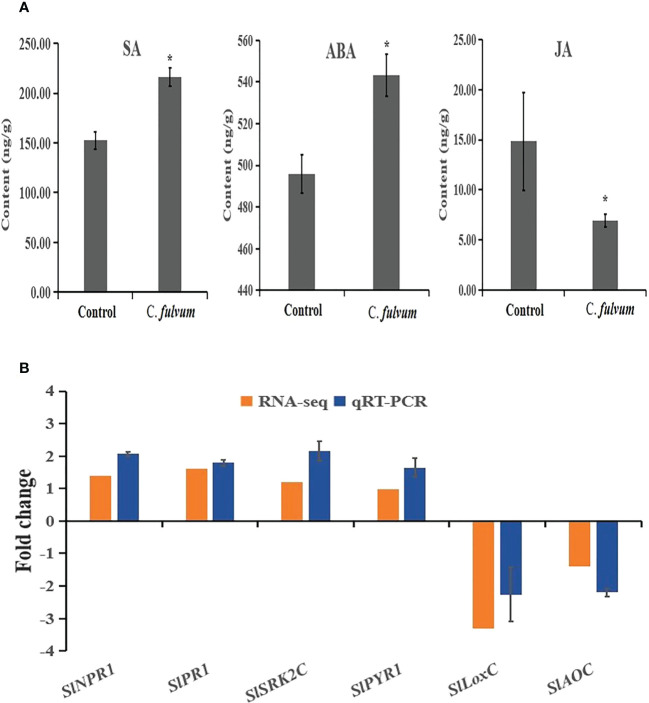
According to our RNA-seq data analysis, some key genes involved in SA and ABA signaling were upregulated after C. fulvum invasion. Thus, we decided to measure the content of SA and ABA. Our results showed that both SA and ABA contents increased after fungal invasion, indicating SA and ABA singling pathways were activated after C. fulvum treatment. We further tested the JA content and showed that JA level decreased after fungal invasion in comparison to the control treatment ( Figure 6A ). This is in consistence with the RNA-seq data in which a few genes related with JA synthesis was inhibited after C. fulvum inoculation.
Figure 6.
Hormone contents and relative gene expression levels impacted by Cladosporium fulvum treatment at the time points of 24 hours. (A) The contents of SA, ABA and JA in tomato leaves were detected 24 hours after sprayed exogenously with a conidial suspension (1X106 conidia/ml) of C fulvum, water treatment as a control. Shown are the mean and SE of three biological replicates. Statistical differences were determined Student’s t-test (p<0.05). (B) Six genes related to plant hormone signal transduction pathway were tested expression level by C fulvum inoculation after 24 hours. SlNPR1 and SlPR1 are SA-related pathway genes, SlSRK2C and SlPYR1 are ABA-related pathway genes, SlLoxC and SlAOC are JA-related pathway genes. The qRT-PCR values represent mean ± SE from three biological replicates. * indicate signifificant differences between control and C.fulvum with p < 0.05, as determined by t-test.
The related expression of genes involved in SA, ABA and JA signaling pathways after Cladosporium fulvum inoculation
To further confirm the impact to SA, ABA and JA hormone pathways after C. fulvum invasion, two genes related to each plant hormone signal transduction pathway were selected for qRT-PCR gene expression analysis. Results showed that the relative expression of genes with RNA-seq and qRT-PCR are the similar trend ( Figure 6B ). The qRT-PCR results showed that expression of ABA-related SlSRK2C and SlPYR1) and SA-related genes (SlNPR1 and SlPR1) were upregulated 2-4 folds. In contrast, expression of synthetic genes of JA (SlLoxC and SlAOC) were significantly downregulated by more than 3-fold. Our results further confirmed that these three hormone pathways are affected after C. fulvum invasion.
Discussion
Tomato leaf mold caused by C. fulvum results in seriously yield losses in tomato cultivation worldwide (Jia, 2010). Considering that the ability of resistance of some tomato cultivar has decreased because of many physiological races of C. fulvum rapidly mutated, we focused on investigating the transcriptome profiles during early stage of interaction with C. fulvum in tomato leaves by using susceptible tomato plants (Jiang et al., 2022b). As we expected, a variety of defense related genes are identified after C. fulvum inoculation, such as PR1, HSP90, and WRKY1, which is consistence with the previous findings (Zhao et al., 2019; Wang et al., 2022). Based on previous reported that flg22 (flagellin 22) binding induces FLS2-BAK1 (BRI1-associated kinase1) heteromerization to activate immune responses in Arabidopsis thaliana (Sun et al., 2013). In our study, we found that genes FLS2, FLS3 and EFR were all upregulated after C. fulvum inoculation, indicating that tomato plants might share the common PAMP pathways in response to biotroph fungi and bacterial pathogens at this early stage of invasion, which needs to be further study.
Facing pathogen invasion, plant have evolved a complex signal transduction network to activate plant resistance. Ca2+ is a conserved second messenger involved in nearly all aspects of cellular signaling programs including regulation of plant development as well as stress resistance (Zhang et al., 2014). CDPK senses Ca2+ signals and translates them into protein phosphorylation (Liese and Romeis, 2013). In our study, several of genes encoding CDPKs were identified to differently express after C. fulvum invasion, either upregulated or downregulated, which are in consistence with many previous reports that CDPKs are widely involved in the regulation of different kinds of disease resistance (Boudsocq et al., 2010; Boudsocq and Sheen, 2013; Romeis and Herde, 2014). For example, the AtCDPK4/5/6/11 phosphorylate specific WRKY transcription factor regulates immune response by restricting pathogen growth (Gao et al., 2013). The StCDPK4/5 mediated ROS production by phosphorylating NADPH oxidases (RBOH, Respiratory Burst Oxidase Homolog) in potato (Kobayashi et al., 2007). In our study, RBOH1 was also upregulated and ROS was produced after C. fulvum invasion, suggesting that CDPK may have activated ROS production and that Ca2+ signal pathway could play an essential role in plant early defense against C. fulvum.
More and more evidence demonstrate that multiple hormones are involved in plant and pathogen interactions (Denance et al., 2013; Li et al., 2019a; Liu et al., 2021). In our study, results showed that genes involved in SA signaling pathway, such as SA receptor NPR1, transcription factor TGA and PR1, were all upregulated. Previous reports showed that SA signaling pathway may be a unique Cf12 dependent resistance pathway (Xue et al., 2017). When the biotrophic pathogen P. syringae infects Arabidopsis, plants also activate SA dependent defenses against invasion (DebRoy et al., 2004; Geng et al., 2012). Our results provide further evidence for the important role of SA signaling pathway in plant defense against biotroph pathogens. A few genes involved in SA signaling pathway were downregulated expression such as BOP2, a NPR1 like gene, and the function for BOP2 in plant is to control growth asymmetry (Hepworth et al., 2005). Therefore, we also speculate that SA may regulate the balance between cell growth and cell death, but the significance on how to regulate plant development is unclear (Hepworth et al., 2005). It is well known that SA and JA-mediate defense singling pathways have antagonistic effect during pathogen invasion (Thaler et al., 2012). In our study, we also found that genes involved in JA synthesis were downregulated after C. fulvum invasion. With the measurement of SA and JA contents after fungal invasion, our results revealed that SA accumulated significantly and the JA level decreased significantly upon C. fulvum invasion, indicating the existence of crosstalk between SA and JA signaling pathway at the early stage of invasion. In addition, our results revealed that C. fulvum invasion also activated the ABA signaling pathway, which may be induced to trigger the ABA-dependent stomata immune response against pathogen entry (Ou et al., 2022). There are also many DEGs involved in the auxin signaling pathway. Given the tradeoff between development and plant defense, it is not surprised that DEGs in so many pathways were identified after C. fulvum invasion.
Plant metabolism can be divided into primary and secondary metabolism, and among the secondary metabolism, the plant phenylpropanoid pathway is one of the most important secondary metabolic pathways (Li et al., 2007). Previous studies have found that the expression of genes related to the phenylpropanoid pathway is also stimulated by Asian soybean rust in Arabidopsis (Beyer et al., 2019), suggesting that the phenylpropanoid pathway plays an important role in plant response to pathogen infection (Dong and Lin, 2021). It has been reported that the expression of PAL and 4CL, key genes in the phenylpropanoid pathway, were upregulated both in susceptible and resistant varieties upon C. fulvum invasion (Xue et al., 2017), which is in consistence with our results. In this study, ANS as the key gene for anthocyanin synthesis (Mattus-Araya et al., 2022), was downregulated, indicating the inhibition of anthocyanin synthesis in plants in response to C. fulvum. Interestingly, CCR2 and some genes encoding peroxidases involved in lignin synthesis pathway were upregulated ( Table 8 ); we hypothesized that tomato plants might activate the lignin synthesis pathway to synthesize lignin as a physical barrier to prevent pathogen invasion.
Conclusion
The plant pathogen C. fulvum causes a large amount of yield loss in global tomato production, which can cause more than 50% reduction in tomato production in severe cases. In the present study, we carried out RNA-seq experiments to identify DEGs induced in C. fulvum inoculated tomato leaves. We analyzed DEGs related to plant and pathogen interaction pathway, plant hormones signaling pathway and plant phenylpropanoid pathway. Our results discovered that a number of core defense genes against fungal invasion were induced. Moreover, we found SA and ABA accumulation in tomato leaves after C. fulvum invasion, while JA content decreased after C. fulvum invasion. Together, our results will broaden our understanding for investigating the mechanism of C. fulvum and tomato interaction in future.
Data availability statement
The data presented in the study are deposited in the NCBI gene expression omnibus server (https://www.ncbi.nlm.nih.gov/geo/), accession number SRR21437044, SRR21437045, SRR21437046, SRR21437047, SRR21437048, SRR21437049.
Author contributions
Conceptualization, RP and SS; Formal analysis, RP, SS and LK; Funding acquisition, SS, ZC and XG; Methodology, PW, ZC, NL, LX and XG; Supervision, SS and XG; Writing – original draft, SS, XG and RP; Writing review & editing, HW and XG. All authors contributed to the article and approved the submitted version.
Acknowledgments
We would like to thank Huada gene institution in China for providing technical assistance with bioinformatics analysis.
Funding
This research was funded by Shanghai agricultural and rural committee foundation (No. 2021-02-08-00-12-F00799), Key research and Development Project of Shanxi Province (No. 202102140601015), the Specific Funding for Modern Agricultural (Vegetable) Industrial System Construction of Shanxi Province, Jinzhong National Agricultural Advanced Area Tomato Wisdom Standardization Technology Research Professor and Doctor Workstation (JZNGQBSGZZ004), Guizhou Province Science and Technology Plan Project (QKHZC (2021) No.207), Guizhou Province Youth Science and Technology Top Talent Project (QJJ (2022) No.89).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1085395/full#supplementary-material
References
- Audran-Delalande C., Bassa C., Mila I., Regad F., Zouine M., Bouzayen M. (2012). Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 53 (4), 659–672. doi: 10.1093/pcp/pcs022 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones Jonathan D. G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 (4), 473–488. doi: 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Benoit L., Patrick A. (2016). Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 34 (1369-5266), 1–8. doi: 10.1016/j.pbi.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Beyer S. F., Beesley A., Rohmann P. F. W., Schultheiss H., Conrath U., Langenbach C. J. G. (2019). The arabidopsis non-host defence-associated coumarin scopoletin protects soybean from Asian soybean rust. Plant J. 99 (3), 397–413. doi: 10.1111/tpj.14426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M. D., Van Esse H. P., Vossen J. H., De Jonge R., Stergiopoulos I., Stulemeijer I. J. E., et al. (2008). The novel cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69 (1), 119–136. doi: 10.1111/j.1365-2958.2008.06270.x [DOI] [PubMed] [Google Scholar]
- Bongue-Bartelsman M., O'Neill S. D., Tong Y., Yoder J. I. (1994). Characterization of the gene encoding dihydroflavonol 4-reductase in tomato. Gene (Amsterdam) 138 (1-2), 153–157. doi: 10.1016/0378-1119(94)90799-4 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18 (1), 30–40. doi: 10.1016/j.tplants.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M. R., McCormack M., Lee H., Shan L., He P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464 (7287), 418–U116. doi: 10.1038/nature08794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J. M., Raices M., Tellez-Inon M. T., Ulloa R. M. (2002). A calcium-dependent protein kinase is systemically induced upon wounding in tomato plants. Plant Physiol. 128 (1), 256–270. doi: 10.1104/pp.010649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Wang C., Wang M., Wan D., Huang F., Jiang Z., et al. (2021). Flg22-induced Ca2+ increases undergo desensitization and resensitization. Plant Cell Environ. 44 (12), 3563–3575. doi: 10.1111/pce.14186 [DOI] [PubMed] [Google Scholar]
- DebRoy S., Thilmony R., Kwack Y. B., Nomura K., He S. Y. (2004). A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. United States America 101 (26), 9927–9932. doi: 10.1073/pnas.0401601101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denance N., Sanchez-Vallet A., Goffner D., Molina A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Lu S. (2017). Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 36 (4), 257–290. doi: 10.1080/07352689.2017.1402852 [DOI] [Google Scholar]
- de Wit P., van der Burgt A., Okmen B., Stergiopoulos I., Abd-Elsalam K. A., Aerts A. L., et al. (2012). The genomes of the fungal plant pathogens cladosporium fulvum and dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PloS Genet. 8 (11). doi: 10.1371/journal.pgen.1003088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N.-Q., Lin H.-X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63 (1), 180–209. doi: 10.1111/jipb.13054 [DOI] [PubMed] [Google Scholar]
- Edqvist J., Blomqvist K., Nieuwland J., Salminen T. A. (2018). Plant lipid transfer proteins: are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 59 (8), 1374–1382. doi: 10.1194/jlr.R083139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F., Li J., Zhan Q., Wang L., Liu Y. (2015). Research progress of cinnamoyl-CoA reductase (CCR) gene in plants. Chin. Biotechnol. 35 (12), 96–102. doi: 10.13523/j.cb.20151215 [DOI] [Google Scholar]
- Gao J.-X., Chen J. (2018). Transcriptome analysis identifies candidate genes associated with melanin and toxin biosynthesis and pathogenicity of the maize pathogen, curvularia lunata. J. Phytopathol. 166 (4), 233–241. doi: 10.1111/jph.12680 [DOI] [Google Scholar]
- Gao J., Chen J. (2021). The role of Clt1-regulated xylan metabolism in melanin and toxin formation for the pathogenicity of curvularia lunata in maize. Mol. Plant-Microbe Interact. 34 (6), 617–630. doi: 10.1094/mpmi-08-20-0235-r [DOI] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., et al. (2013). Bifurcation of arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PloS Pathog. 9 (1), e1003127. doi: 10.1371/journal.ppat.1003127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Guo P., Wang Z., Chen C., Ren Z. (2021). Transcriptome profiling reveals response genes for downy mildew resistance in cucumber. Planta 253 (5), 112. doi: 10.1007/s00425-021-03603-6 [DOI] [PubMed] [Google Scholar]
- Gao S., Li Y., Gao J., Suo Y., Fu K., Li Y., et al. (2014. b). Genome sequence and virulence variation-related transcriptome profiles of curvularia lunata, an important maize pathogenic fungus. BMC Genomics 15:627. doi: 10.1186/1471-2164-15-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.-X., Liu T., Chen J. (2014. a). Insertional mutagenesis and cloning of the gene required for the biosynthesis of the non-Host-Specific toxin in cochliobolus lunatus that causes maize leaf spot. Phytopathology 104 (4), 332–339. doi: 10.1094/phyto-07-13-0190-r [DOI] [PubMed] [Google Scholar]
- Gao J.-X., Yu C.-J., Wang M., Sun J.-N., Li Y.-Q., Chen J. (2017). Involvement of a velvet protein ClVelB in the regulation of vegetative differentiation, oxidative stress response, secondary metabolism, and virulence in curvularia lunata. Sci. Rep. 7:46054. doi: 10.1038/srep46054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X. Q., Cheng J. Y., Gangadharan A., Mackey D. (2012). The coronatine toxin of pseudomonas syringae is a multifunctional suppressor of arabidopsis defense. Plant Cell 24 (11), 4763–4774. doi: 10.1105/tpc.112.105312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S., Mesarich C. H., Overdijk E. J. R., Saccomanno B., de Wit P. J. G. M., Collemare J. (2018). Down-regulation of cladofulvin biosynthesis is required for biotrophic growth of cladosporium fulvum on tomato. Mol. Plant Pathol. 19 (1464-6722), 369–380. doi: 10.1111/mpp.12527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Yang X., Weston D. J., Chen J.-G. (2011). Abscisic acid receptors: Past, present and future. J. Integr. Plant Biol. 53 (6), 469–479. doi: 10.1111/j.1744-7909.2011.01044.x [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Zhang Y. L., McKim S., Li X., Haughn G. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in arabidopsis. Plant Cell 17 (5), 1434–1448. doi: 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia A., Heredia-Guerrero J. A., Dominguez E. (2015). CHS silencing suggests a negative cross-talk between wax and flavonoid pathways in tomato fruit cuticle. Plant Signaling Behav. 10 (5), e1019979. doi: 10.1080/15592324.2015.1019979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. A., Gullner G., Clemente-Moreno M. J., Kunstler A., Juhasz C., Diaz-Vivancos P., et al. (2016). Oxidative stress and antioxidative responses in plant-virus interactions. Physiol. Mol. Plant Pathol. 94, 134–148. doi: 10.1016/j.pmpp.2015.09.001 [DOI] [Google Scholar]
- Hind S. R., Strickler S. R., Boyle P. C., Dunham D. M., Bao Z., O'Doherty I. M., et al. (2016). Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2 (9), 16128. doi: 10.1038/nplants.2016.128 [DOI] [PubMed] [Google Scholar]
- Hoegen E., Stromberg A., Pihlgren U., Kombrink E. (2002). Primary structure and tissue-specific expression of the pathogenesis-related protein PR-1b in potato. Mol. Plant Pathol. 3 (5), 329–345. doi: 10.1046/j.1364-3703.2002.00126.x [DOI] [PubMed] [Google Scholar]
- Hou J., Sun Q., Li J., Ahammed G. J., Yu J., Fang H., et al. (2019). Glutaredoxin S25 and its interacting TGACG motif-binding factor TGA2 mediate brassinosteroid-induced chlorothalonil metabolism in tomato plants. Environ. pollut. 255:113256. doi: 10.1016/j.envpol.2019.113256 [DOI] [PubMed] [Google Scholar]
- Iida Y., Iwadate Y., Kubota M., Terami F. (2010). Occurrence of a new race 2.9 of leaf mold of tomato in Japan. J. Gen. Plant Pathol. 76 (1), 84–86. doi: 10.1007/s10327-009-0207-8 [DOI] [Google Scholar]
- Jane G. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Jia Y. G. (2010). Technical study on controlling leaf mold of tomato in the protected area. Heilongjiang Agric. Sci. 2), 44–45. doi: 10.3969/j.issn.1002-2767.2010.02.017 [DOI] [Google Scholar]
- Jiang X., Li Y., Li R., Gao Y., Liu Z., Yang H., et al. (2022. b). Transcriptome analysis of the cf-13-Mediated hypersensitive response of tomato to cladosporium fulvum infection. Int. J. Mol. Sci. 23 (9), 4844. doi: 10.3390/IJMS23094844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Zhang Y. Z., Li P., Jian J. J., Zhao C. L., Wen G. S. (2022. a). Mitogen-activated protein kinase and substrate identification in plant growth and development. Int. J. Mol. Sci. 23 (5), 2744. doi: 10.3390/ijms23052744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L., Bian L., Luo Z., Li Z., Xiu C., Fu N., et al. (2022). Enhanced volatile emissions and anti-herbivore functions mediated by the synergism between jasmonic acid and salicylic acid pathways in tea plants. Horticult. Res. 9, uhac144–uhac144. doi: 10.1093/hr/uhac144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C., Zhang L., Liu L., Wang J., Li C., Wang Q. (2013). Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to alternaria alternata f. sp lycopersici. J. Exp. Bot. 64 (2), 637–650. doi: 10.1093/jxb/ers360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M. H. A. J., Cozijnsen T. J., De Wit P. J. G. M. (1994). Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nat. (London) 367 (6461), 384–386. doi: 10.1038/367384a0 [DOI] [PubMed] [Google Scholar]
- Kang J.-H., McRoberts J., Shi F., Moreno J. E., Jones A. D., Howe G. A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164 (3), 1161–1174. doi: 10.1104/pp.113.233395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Ohura I., Kawakita K., Yokota N., Fujiwara M., Shimamoto K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19 (3), 1065–1080. doi: 10.1105/tpc.106.048884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna B., J L. F., D W. J., R P. D. (2016). Diversity analysis of tomato genotypes based on morphological traits with commercial breeding significance for fresh market production in eastern USA. Aust. J. Crop Sci. 10 (8), 1098–1103. doi: 10.21475/ajcs.2016.10.08.p7391 [DOI] [Google Scholar]
- Lauge R., Goodwin P. H., De Wit P., Joosten M. (2000). Specific HR-associated recognition of secreted proteins from cladosporium fulvum occurs in both host and non-host plants. Plant J. 23 (6), 735–745. doi: 10.1046/j.1365-313x.2000.00843.x [DOI] [PubMed] [Google Scholar]
- Lavrova V. V., Zinovieva S. V., Udalova Z. V., Matveeva E. M. (2017). Expression of PR genes in tomato tissues infected by nematode meloidogyne incognita (Kofoid et white 1919) chitwood 1949. Doklady Biochem. Biophys. 476 (1), 306–309. doi: 10.1134/s1607672917050064 [DOI] [PubMed] [Google Scholar]
- Liese A., Romeis T. (2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Et Biophys. Acta-Mol. Cell Res. 1833 (7), 1582–1589. doi: 10.1016/j.bbamcr.2012.10.024 [DOI] [PubMed] [Google Scholar]
- Li N., Han X., Feng D., Yuan D., Huang L.-J. (2019. a). Signaling crosstalk between salicylic acid and Ethylene/Jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 20 (3), 671. doi: 10.3390/ijms20030671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-R., Liu C.-C., Sun C.-H., Chen Y.-T. (2018). Plant stress RNA-seq nexus: a stress-specific transcriptome database in plant cells. BMC Genomics 19:966. doi: 10.1186/s12864-018-5367-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liu C., Zhao R., Wang L., Chen L., Yu W., et al. (2019. b). CRISPR/Cas9-mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19:38. doi: 10.1186/s12870-018-1627-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Atta U. R., Wang R., Liu K., Ma X., Weng Q. (2021). Defense-related hormone signaling coordinately controls the role of melatonin during arabidopsis thaliana-pseudomonas syringae interaction. Eur. J. Plant Pathol. 160 (3), 707–716. doi: 10.13592/j.cnki.ppj.2012.09.013 [DOI] [Google Scholar]
- Liu Q., Li M., Guo J. (2012). Regulation of jasmonic acid biosynthesis and jasmonic acid signaling pathway. Plant Physiol. J. 48 (9), 837–844. doi: 10.3969/j.issn.2095-1736.2014.02.069 [DOI] [Google Scholar]
- Li Z., Vickrey T. L., McNally M. G., Sato S. J., Clemente T. E., Mower J. P. (2019. c). Assessing anthocyanin biosynthesis in solanaceae as a model pathway for secondary metabolism. Genes 10 (8), 559. doi: 10.3390/genes10080559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., Ma J.L. (2007). Recent progress on key enzymes: PAL, C4H, 4CL of phenylalanine metabolism pathway. China J. Bioinf. 5 (4), 187–189. doi: 10.3969/j.issn.1672-5565.2007.04.015 [DOI] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer R., Takken F. L. W., De Wit P., Joosten M. (2002). Cladosporium fulvum overcomes cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45 (3), 875–884. doi: 10.1046/j.1365-2958.2002.03060.x [DOI] [PubMed] [Google Scholar]
- Ma Q., Hu Z., Mao Z., Mei Y., Feng S., Shi K. (2022). The novel leucine-rich repeat receptor-like kinase MRK1 regulates resistance to multiple stresses in tomato. Horticult. Res. 9:uhab088. doi: 10.1093/hr/uhab088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjamaa K., Kukkola E. M., Fagerstedt K. V. (2009). The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60 (2), 367–376. doi: 10.1093/jxb/ern278 [DOI] [PubMed] [Google Scholar]
- Mattus-Araya E., Guajardo J., Herrera R., Moya-Leon M. A. (2022). ABA speeds up the progress of color in developing f. chiloensis fruit through the activation of PAL, CHS and ANS, key genes of the Phenylpropanoid/Flavonoid and anthocyanin pathways. Int. J. Mol. Sci. 23 (7), 3854. doi: 10.3390/ijms23073854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayda E., Marques C., Conejero V., Vera P. (2000). Expression of a pathogen-induced gene can be mimicked by auxin insensitivity. Mol. Plant-Microbe Interact. 13 (1), 23–31. doi: 10.1094/mpmi.2000.13.1.23 [DOI] [PubMed] [Google Scholar]
- Melillo M. T., Leonetti P., Veronico P. (2014). Benzothiadiazole effect in the compatible tomato-meloidogyne incognita interaction: changes in giant cell development and priming of two root anionic peroxidases. Planta 240 (4), 841–854. doi: 10.1007/s00425-014-2138-7 [DOI] [PubMed] [Google Scholar]
- Meng L., Feldman L. (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from arabidopsis siliques and dry seeds. Biotechnol. J. 5 (2), 183–186. doi: 10.1002/biot.200900211 [DOI] [PubMed] [Google Scholar]
- Mingchun L., Gomes B. L., Mila I., Purgatto E., Peres L. E. P., Frasse P., et al. (2016). Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 170 (3), 1732–1744. doi: 10.1104/pp.15.01859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi M., Umezawa T., Nakashima K., Kidokoro S., Takasaki H., Fujita Y., et al. (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 51 (5), 842–847. doi: 10.1093/pcp/pcq041 [DOI] [PubMed] [Google Scholar]
- Naoumkina M. A., Zhao Q. A., Gallego-Giraldo L., Dai X. B., Zhao P. X., Dixon R. A. (2010). Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11 (6), 829–846. doi: 10.1111/j.1364-3703.2010.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y., Baker A. (2006). Plant peroxisomes as a source of signalling molecules. Biochim. Et Biophys. Acta-Mol. Cell Res. 1763 (12), 1478–1495. doi: 10.1016/j.bbamcr.2006.08.031 [DOI] [PubMed] [Google Scholar]
- Orellana S., Yanez M., Espinoza A., Verdugo I., Gonzalez E., Ruiz-Lara S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33 (12), 2191–2208. doi: 10.1111/j.1365-3040.2010.02220.x [DOI] [PubMed] [Google Scholar]
- Ou X., Li T., Zhao Y., Chang Y., Wu L., Chen G., et al. (2022). Calcium-dependent ABA signaling functions in stomatal immunity by regulating rapid SA responses in guard cells. J. Plant Physiol. 268:153585. doi: 10.1016/j.jplph.2021.153585 [DOI] [PubMed] [Google Scholar]
- Pattison R. J., Catala C. (2012). Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 70 (4), 585–598. doi: 10.1111/j.1365-313X.2011.04895.x [DOI] [PubMed] [Google Scholar]
- Piasecka A., Jedrzejczak-Rey N., Bednarek P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206 (3), 948–964. doi: 10.1111/nph.13325 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., van Loon L. C. (1999). Salicylic acid-independent plant defence pathways. Trends Plant Sci. 4 (2), 52–58. doi: 10.1016/s1360-1385(98)01364-8 [DOI] [PubMed] [Google Scholar]
- Pratyusha D. S., Sarada D. V. L. (2022). MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 41, 2245–2260 doi: 10.1007/s00299-022-02927-1 [DOI] [PubMed] [Google Scholar]
- Rivas S. (2005). Molecular interactions between tomato and the leaf mold pathogen cladosporium fulvum. Annu. Rev. Phytopathol. 43 (0066-4286), 395–436. doi: 10.1146/annurev.phyto.43.040204.140224 [DOI] [PubMed] [Google Scholar]
- Robineau M., Le Guenic S., Sanchez L., Chaveriat L., Lequart V., Joly N., et al. (2020). Synthetic mono-rhamnolipids display direct antifungal effects and trigger an innate immune response in tomato againstBotrytis cinerea. Molecules 25 (14), 3108. doi: 10.3390/molecules25143108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Herde M. (2014). From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 20, 1–10. doi: 10.1016/j.pbi.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Ruili Z., Wenhua W., Shaodong R., Qunxia W., Huaying L. (2016). Study on separation, purification and enzymatic properties of phenylalanine ammonia-lyase (PAL) from kuqa apricot. J. Henan Univ. Technol. Natural Sci. Edition 37 (4), 74–78. doi: 10.16433/j.cnki.issn1673-2383.2016.04.014 [DOI] [Google Scholar]
- Shen Y., Liu N., Li C., Wang X., Xu X., Chen W., et al. (2017). The early response during the interaction of fungal phytopathogen and host plant. Open Biol. 7 (5), 170057. doi: 10.1098/rsob.170057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde B. A., Dholakia B. B., Hussain K., Aharoni A., Giri A. P., Kamble A. C. (2018). WRKY1 acts as a key component improving resistance against alternaria solani in wild tomato, solanum arcanum peralta. Plant Biotechnol. J. 16 (8), 1502–1513. doi: 10.1111/pbi.12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simura J., Antoniadi I., Siroka J., Tarkowska D., Strnad M., Ljung K., et al. (2018). Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 177 (2), 476–489. doi: 10.1104/pp.18.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Aggarwal R., Bashyal B. M., Darshan K., Parmar P., Saharan M. S., et al. (2021). Transcriptome reprogramming of tomato orchestrate the hormone signaling network of systemic resistance induced by chaetomium globosum. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.721193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A. P., Han Z., Hu Z., Zipfel C., et al. (2013). Structural basis for flg22-induced activation of the arabidopsis FLS2-BAK1 immune complex. Science 342 (6158), 624–628. doi: 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- Thaler J. S., Humphrey P. T., Whiteman N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17 (5), 260–270. doi: 10.1016/j.tplants.2012.02.010 [DOI] [PubMed] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14 (6), 310–317. doi: 10.1016/j.tplants.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Van Den Ackerveken G. F., Van Kan J. A., Joosten M. H., Muisers J. M., Verbakel H. M., De Wit P. J. (1993). Characterization of two putative pathogenicity genes of the fungal tomato pathogen cladosporium fulvum. Mol. Plant-Microbe Interact. 6 (2), 210–215. doi: 10.1094/MPMI-6-210 [DOI] [PubMed] [Google Scholar]
- Van Esse H. P., Bolton M. D., Stergiopoulos I., De Wit P. J., Thomma B. P. (2007). The chitin-binding cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20 (9), 1092–1101. doi: 10.1094/mpmi-20-9-1092 [DOI] [PubMed] [Google Scholar]
- Van Kan J. A., Van Den Ackerveken G. F., De Wit P. J. (1991). Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant-Microbe Interact. 4 (1), 52–59. doi: 10.1094/mpmi-4-052 [DOI] [PubMed] [Google Scholar]
- Vera P., Tornero P., Conejero V. (1993). Cloning and expression analysis of a viroid-induced peroxidase from tomato plants. Mol. Plant-Microbe Interact. 6 (6), 790–794. doi: 10.1094/MPMI-6-786 [DOI] [PubMed] [Google Scholar]
- Wang P., Sun S., Liu K., Peng R., Li N., Hu B., et al. (2022). Physiological and transcriptomic analyses revealed gene networks involved in heightened resistance against tomato yellow leaf curl virus infection in salicylic acid and jasmonic acid treated tomato plants. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.970139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun N., Zhang F., Yu R., Chen H., Deng X. W., et al. (2020). SAUR17 and SAUR50 differentially regulate PP2C-D1 during apical hook development and cotyledon opening in arabidopsis. Plant Cell 32 (12), 3792–3811. doi: 10.1105/tpc.20.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Y., Yin Q.-S., Qu Y., Li G.-Z., Hao L. (2018). Arbuscular mycorrhiza-mediated resistance in tomato against cladosporium fulvum-induced mould disease. J. Phytopathol. 166 (1), 67–74. doi: 10.1111/jph.12662 [DOI] [Google Scholar]
- Westerink N., Brandwagt B. F., de Wit P., Joosten M. (2004). Cladosporium fulvum circumvents the second functional resistance gene homologue at the cf-4 locus (Hcr9-4E) by secretion of a stable avr4E isoform. Mol. Microbiol. 54 (2), 533–545. doi: 10.1111/j.1365-2958.2004.04288.x [DOI] [PubMed] [Google Scholar]
- Xin P., Guo Q., Li B., Cheng S., Yan J., Chu J. (2020). A tailored high-efficiency sample pretreatment method for simultaneous quantification of 10 classes of known endogenous phytohormones. Plant Commun. 1 (3), 100047. doi: 10.1016/j.xplc.2020.100047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D.-Q., Chen X.-L., Zhang H., Chai X.-F., Jiang J.-B., Xu X.-Y., et al. (2017). Transcriptome analysis of the cf-12-Mediated resistance response to cladosporium fulvum in tomato. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-X., Ahammed G. J., Wu C., Fan S.-y., Zhou Y.-H. (2015). Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 16 (1389-2037), 450–461. doi: 10.2174/1389203716666150330141638 [DOI] [PubMed] [Google Scholar]
- Yang W. Q., Zhang W., Wang X. X. (2017). Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 15 (1), 4–14. doi: 10.1111/pbi.12652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. J., Fan L. L., Gao J. X., Wang M., Wu Q., Tang J., et al. (2015). The platelet-activating factor acetylhydrolase gene derived from trichoderma harzianum induces maize resistance to curvularia lunata through the jasmonic acid signaling pathway. J. Environ. Sci. Health Part B-Pestic. Food Contam. Agric. Waste. 50 (10), 708–717. doi: 10.1080/03601234.2015.1048104 [DOI] [PubMed] [Google Scholar]
- Zhang D., Bao Y., Sun Y., Yang H., Zhao T., Li H., et al. (2020). Comparative transcriptome analysis reveals the response mechanism of cf-16-mediated resistance to cladosporium fulvum infection in tomato. BMC Plant Biol. 20 (1), 33. doi: 10.1186/s12870-020-2245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Chi W., Wang C., Dai H., Li J., Li C., et al. (2022). Pathogenic process-associated transcriptome analysis of stemphylium lycopersici from tomato. Int. J. Genomics 2022:4522132. doi: 10.1155/2022/4522132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Holmlund M., Lorrain S., Norberg M., Bako L., Fankhauser C., et al. (2017). BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. Elife 6:e26759. doi: 10.7554/eLife.26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhao Q., Zhang Z., Song S. (2014). The function of Ca~(2 +) and CaM/CML in plant immunity. J. Biol. 31 (2), 69–72. doi: 10.3969/j.issn.2095-1736.2014.02.069 [DOI] [Google Scholar]
- Zhao T., Liu W., Zhao Z., Yang H., Bao Y., Zhang D., et al. (2019). Transcriptome profiling reveals the response process of tomato carrying cf-19 and cladosporium fulvum interaction. BMC Plant Biol. 19 (1), 572. doi: 10.1186/s12870-019-2150-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouine M., Fu Y., Chateigner-Boutin A.-L., Mila I., Frasse P., Wang H., et al. (2014). Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PloS One 9 (1), e84203. doi: 10.1371/journal.pone.0084203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the NCBI gene expression omnibus server (https://www.ncbi.nlm.nih.gov/geo/), accession number SRR21437044, SRR21437045, SRR21437046, SRR21437047, SRR21437048, SRR21437049.