Abstract
Nicotinamide adenine dinucleotide (NAD+) kinase (NADK) phosphorylates NAD+, thereby producing nicotinamide adenine dinucleotide phosphate (NADP). Both NADK genes and the NADP(H)-producing mechanism are evolutionarily conserved among archaea, bacteria, plants and mammals. In mammals, NADK is activated by phosphorylation and protein-protein interaction. Recent studies conducted using genetically altered models validate the essential role of NADK in cellular redox homeostasis and metabolism in multicellular organisms. Here, we describe the evolutionary conservation, molecular properties, and signaling mechanisms and discuss the pathophysiological significance of NADK.
Keywords: NADK, NADK2, NAD+, NADPH, Thioredoxin, Nampt
Highlights
-
•
NADK phosphorylates NAD+, thereby producing NADP(H).
-
•
NADK plays a crucial role in anabolic process and redox homeostasis through production of NADP(H).
-
•
NADK is regulated by posttranslational modifications such as phosphorylation.
-
•
NADK could be a therapeutic target in human disease, such as cancer.
List of abbreviations
- AGC
(cAMP- and cGMP-dependent protein kinases and protein kinase C (PKC))
- BAD
benzamide adenine nucleoside
- CaM kinase
Ca2+/calmodulin-dependent protein kinase
- DHFR
dihydrofolate reductase
- EMKA
Element conserved in mitochondrial kinases of animals
- G6PD
glucose-6-phosphate dehydrogenase
- GLUD
glutamate dehydrogenase
- IDH
isocitrate dehydrogenase
- IR
ischemia reperfusion
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- ME
malic enzyme
- NA
nicotinic acid
- NAD+
NADH: nicotinamide adenine dinucleotide, oxidized and reduced forms
- NADK
NAD kinase
- NADP
NADPH: nicotinamide adenine dinucleotide phosphate, oxidized and reduced forms
- NADPS
thionicotinamide adenine dinucleotide phosphate
- Nampt
nicotinamide phosphoribosyltransferase
- NCBI
National Center for Biological Information
- NMN
nicotinamide mononucleotide
- NNT
nicotinamide nucleotide transhydrogenase
- Nox
NADPH oxidase
- NR
nicotinamide ribose
- PARG
poly-ADP-ribose glycohydrolase
- PARP
poly (ADP-ribose) polymerase
- PDAC
pancreatic ductal adenocarcinoma
- PKC
protein kinase C
- ROS
reactive oxygen species
- TN
Thionicotinamide
- Trx
Thioredoxin
- 6PGD
6-phosphogluconate dehydrogenase
1. Introduction
Nicotinamide adenine dinucleotide (NAD+) phosphate (NADP(H)) plays a crucial role in redox homeostasis and metabolism [1]. Phosphorylation of NAD+ by NAD kinase (NADK) is the only known mechanism by which NADP is produced de novo [2]. NADK plays an essential role in NADP(H) production and subsequent biological processes. Importantly, however, the pathophysiological significance and regulation of NADK remain underinvestigated, particularly in mammals and in the context of human disease. In this review, we describe the evolutionary conservation, molecular properties, and posttranslational regulation of NADK. We also discuss recent studies that demonstrate the pathophysiological significance of NADK in human diseases.
2. NADP(H) may mediate the beneficial effects of NAD+
NAD+ plays a crucial role in a wide range of metabolic processes, including energy production in mitochondria. NAD+ is synthesized de novo or through the Preuss-Hander or salvage pathway (Fig. 1A). Dietary tryptophan is converted to NAD+ in the de novo pathway, whereas nicotinic acid is used in the Preuss-Hander pathway [3]. NAD+ is degraded to nicotinamide and ADP-ribose derivatives, such as O-acetyl-ADP-ribose and poly-ADP-ribose, by the NAD+ consuming enzymes, sirtuins and poly-ADP-ribose polymerases (PARPs), respectively [4]. In addition, CD38, also known as cyclic ADP ribose hydrolase, a glycoprotein found on the surface of immune cells, degrades NAD+ to produce nicotinamide and ADP-ribose [5]. In the salvage pathway, NAD+ is synthesized from nicotinamide and ADP-ribose, which is produced either from poly-ADP-ribose by poly-ADP-ribose glycohydrolases (PARG) [6] or by conversion of O-acetyl-ADP-ribose [7]. Nicotinamide phosphoribosyltransferase (Nampt) is the rate limiting enzyme of the salvage pathway of NAD+ production. NAD+ can accept two electrons to form NADH. The levels of NAD+ decline in multiple tissues, including muscle, adipose, brain, skin, liver and pancreas, in rodents during aging [8]. This aging-dependent decrease in NAD+ is also observed in the brain, liver and blood plasma in humans [9,10]. Depletion of NAD+ promotes diabetes [11], fatty liver diseases [12], neurodegenerative disorders [[13], [14], [15]] and heart disease [16,17]. Thus, the decline in NAD+ may induce functional impairment in cells and tissues during aging. Indeed, replenishment of NAD+ prolongs life span in worms and mice [18,19]. In contrast, recent studies demonstrated that the replenishment of NAD+ prolongs heath span, but not life span, in mice [20,21]. Replenishment of NAD+ also ameliorates diabetes, obesity, cardiac disease, a neurodegenerative disorder and renal degeneration [16,22,23]. Thus, NAD+ may contribute to longevity and prolonged health span.
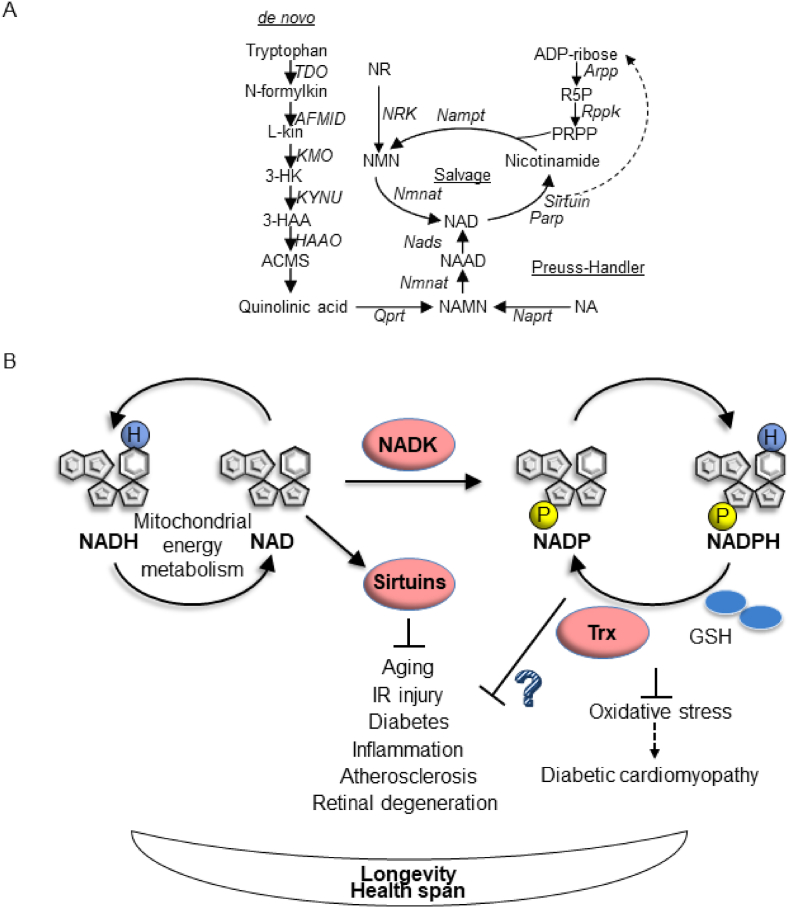
Fig. 1.
Cellular functions of NAD+. (A) A schematic representation of the NAD+ synthetic pathways. NAD+ is synthesized from tryptophan via the de novo pathway, nicotinic acid via the Preuss-Handler pathway, or nicotinamide and ADP-ribose via the salvage pathway. Abbreviated metabolites and enzymes are as follows. L-Kyn: l-kynurenine, 3-HK: 3-hydroxykynurenine, 3-HAA: 3-hydroxyanthranilic acid, ACMS: 2-amino-3-carboxymuconic semialdehyde, NAMN: nicotinic acid mononucleotide, NAAD: nicotinic acid adenine dinucleotide, NMN: nicotinamide mononucleotide, NR: nicotinamide ribose, NA: nicotinic acid, R5P: ribose 5 phosphate, PRPP: 5-phosphoribosyl-1-pyrophosphate, TDO: tryptophan 2,3-dioxygenase, AFMID: arylformamidase, KMO: Kynurenine 3-monooxygenase, KYNU: Kynureinase, HAAO: 3-hydroxyanthranilate 3,4-dioxygenase, Qprt: quinolinate phosphoribosyl transferase, Naprt: nicotinate phosphoribosyltransferase, Nmnat: nicotinamide nucleotide adenylyltransferase, NADS: NAD+ synthase, Nampt: nicotinamide phosphoribosyltransferase, Parp: poly ADP ribose polymerase, Arpp: ADP-ribose pyrophosphatase, Rppk: ribose phosphate pyrophosphokinase and NRK: nicotinamide ribose kinase. (B) Cell protective and adaptive effects of NAD+. NAD(H) functions as an electron carrier that contributes to metabolic processes, including mitochondrial energy metabolism. NAD+ also functions as a co-substrate of NAD+-consuming enzymes, including sirtuins and Parps. Sirtuins mediate beneficial effects of NAD+ in aging and disease conditions. Although NAD+ is a precursor of NADP, which plays an essential role in cellular redox homeostasis and metabolic processes, the extent to which NADK-induced NADP production contributes to the salutary effects of NAD+ is not fully understood.
Interventions to increase NAD+ levels include 1) supplementation of NAD+ precursors, such as nicotinamide mononucleotide (NMN), nicotinamide ribose (NR), nicotinic acid (NA) and nicotinamide, 2) reduction of NAD+ consumption through inhibition of NAD+ consuming enzymes, 3) manipulation of the NAD+ biosynthesis pathways, including activation of Nampt and 4) interventions that improve the bioavailability of NAD+, including calorie restriction and exercise [24]. Among the NAD+ consuming enzymes, sirtuins play a major role in the beneficial effects of NAD+ [25]. For instance, Sirt1 mediates the salutary effect of NAD+ in axonal degeneration, and cardiac ischemia reperfusion and acute kidney injuries [[26], [27], [28]]. However, given that NAD+ is the sole precursor of NADP(H) and that the NADP(H)-dependent antioxidant system exerts protective effects, including prevention of aging and disease, NADK-induced NADP(H) production may also account for the salutary actions of NAD+. Indeed, we have shown recently that the salutary effect of NAD+ in diabetic cardiomyopathy is partly mediated by NADK-dependent NADP(H) production [29] (Fig. 1B).
3. Evolutionary conservation of NADK
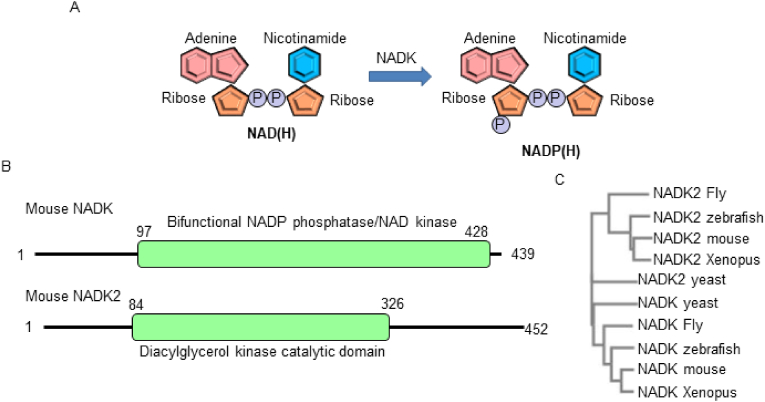
NADK genes are evolutionarily conserved from archaea to mammals (Fig. 2). NADK phosphorylates the 2’ position of the ribose ring connected to the adenine moiety in NAD(H) (Fig. 3A). Because NADP and NADPH are membrane impermeable, the presence of organelle-specific NADP(H) synthesis systems is critical [30]. Plants possess three NADK genes, localized in the cytoplasm (NADK1), chloroplasts (NADK2) and peroxisomes (NADK3) [31]. In mammals, NADK is predominantly localized in the cytoplasm, whereas NADK2 is found in mitochondria [31,32]. The NCBI conserved domain database indicates that NADK contains a bifunctional NADP phosphatase/NAD+ kinase domain, while NADK2 contains a diacylglycerol kinase catalytic domain (Fig. 3B). In addition, phylogenetic trees indicate that NADK and NADK2 developed independently during evolution (Fig. 3C). Whether or not eukaryotes possess other organelle specific NADKs, including in peroxisomes and the endoplasmic reticulum, remains unknown.
Fig. 2.
Conservation of NADK in eukaryotes. Alignment of NADK proteins derived from yeast, fly, zebrafish, xenopus and mouse was conducted with the Clustal Omega program.
Fig. 3.
Structure of NADK and NADK2. (A) NADK phosphorylates the 2′ position of the ribose ring connected to the adenine moiety in NAD(H). (B) The catalytic domains of mouse NADK and NADK2, as assigned by the NCBI conserved domain database. (C) Phylogenetic trees of NADK and NADK2 provided by the Clustal Omega program.
Some NADKs found in lower organisms such as bacteria and archaea use not only ATP but also polyphosphate to generate NADP. Polyphosphate is a negatively charged linear or cyclic homopolymer of orthophosphate (PO4−3), ranging in size from a few to several hundred subunits. It may have served as an energy carrier in ancient primitive organisms and is referred to as a fossil molecule or bioenergy fossil [33]. In contrast, in eukaryotes such as humans, NADK specifically uses ATP but not polyphosphate [31]. Interestingly, however, human mitochondrial NADK2 retains the ability to use polyphosphate to generate NADP [34]. This might be because mitochondria are derived from ancient organisms, even though the NADK2 gene is encoded in the nuclear genome. In addition, some NADKs in lower organisms phosphorylate not only NAD+ but also NADH, whereas those in higher organisms, such as human NADK, phosphorylate only NAD+ but not NADH. On the other hand, human NADK2 phosphorylates both NAD+ and NADH, although NAD+ is the preferred substrate [31,32]. Thus, NADK2 might be a more ancient type of enzyme than NADK in multicellular organisms.
4. Molecular properties of NADK
The crystal structure of NADK in Listeria monocytogenes and Mycobacterium tuberculosis has been determined, and the critical residues and domains for NADK function have been proposed [35,36]. These are evolutionarily conserved from bacteria to mammals. Thus, the molecular properties and catalytic mechanisms are likely also preserved in mammalian NADK. Based on this assumption, we describe the residues and domains critical for NADK function in mouse NADK.
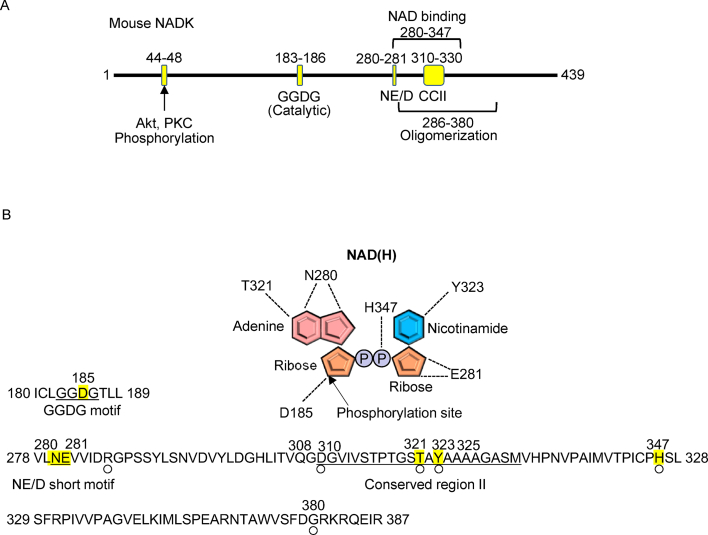
There are three conserved domains: the GGDG motif, NE/D short motif and conserved region II (Fig. 4A). In addition, mammalian NADK possesses serine residues at 44, 46 and 48 that are phosphorylated by Akt and PKC [37,38]. The NAD+ binding and oligomerization domains are located at 280–347 and 286–380, respectively (Fig. 4A). As shown in Fig. 4B, D185 in the GGDG motif serves as a catalytic center that interacts with the phosphorylation site of NAD+, namely the 2’ position of ribose. N280 and E281 in the NE/D short motif recognize adenine and ribose connected to nicotinamide, respectively. T321 and Y323 in the conserved region II recognize adenine and nicotinamide, respectively. H347 recognizes the phosphate group. Site directed mutagenesis of NADK in Mycobacterium tuberculosis demonstrated that D185, N280, E281, T321 and Y323 are critical for NADK function [35,39]. In addition, several residues have been demonstrated to play a role in the substrate specificity of NADK. NADKs that possess arginine corresponding to Q308, serine corresponding to A325 and arginine corresponding to H347 tend to use NADH as a substrate [35,40]. Thus, the region from 280 to 347 is involved in substrate binding and selectivity.
Fig. 4.
Structure and function of NADK. (A) A schematic representation of the NADK domains, including phosphorylation sites, GGDG motif, NE/D short motif, conserved region II (CCII), NAD+ binding region and oligomerization region. (B) Critical amino acid residues for catalysis, NAD+ recognition and oligomerization. Underlines indicate the GGDG motif, NE/D short motif, and conserved region II. Highlighting indicates residues involved in the kinase reaction and binding to NAD+. Open circles indicate residues involved in oligomerization.
Mammalian NADK forms a homotetramer, whereas NADK2 forms a homodimer [31,34]. The region critical for the oligomerization partly overlaps with the NAD+ binding region, from 286 to 380. Residues that may be responsible for oligomerization include R286, D310, T321, Y323, H347 and G380 [35]. Several of the residues necessary for oligomerization in bacteria are not conserved in mammalian NADK [35]. Given the high homology among species, the molecular properties are likely evolutionarily conserved but the significance of these residues and domains should be validated in mammalian NADKs.
5. Validation of the NADP(H) producing mechanism in multicellular organisms
Phosphorylation of NAD+ by NADK is the only known mechanism of de novo NADP production. Yeast possesses three NADK genes. An NADK triple mutant was shown to be lethal, suggesting that NADK-induced NAD(H) phosphorylation is the primary mechanism of NADP(H) production [41]. Similarly, systemic NADK knockout mice exhibit embryonic and preweaning lethality (http://www.informatics.jax.org/marker/phenotypes/MGI:2183149). Phenotypes of organisms where NADK is genetically-modified are summarized in Table 1. Recent studies have validated NADK-induced NAD+ phosphorylation as a major pathway and mechanism responsible for NADP production in multicellular organisms. Knockdown of NADK significantly downregulates NADP in HEK293 cells [37,42], the fat body in flies [43], and cancer cells [38]. Inhibition of Nampt in HEK293 cells with FK866 significantly downregulates not only NAD+ but also NADP [37]. Similarly, systemic Nampt heterozygous knockout downregulates not only NAD+ but also NADP in the heart [29]. Conversely, cardiac specific Nampt overexpression upregulates not only NAD+ but also NADP [29]. However, Nampt-induced NADP(H), but not NAD+, upregulation is inhibited by knockdown of NADK in cardiomyocytes [29]. Taken together, these studies show that NADK-induced NAD+ phosphorylation is a major mechanism responsible for NADP production in multicellular organisms. Furthermore, NAD+ production through Nampt appears to be coupled to NADK-induced NADP production in some mammalian cells.
Table 1.
Genetic models of NADK and NADK2 and their phenotype.
| Organism | Genotype | Phenotype | References |
|---|---|---|---|
| Saccharomyces cerevisiae | POS5, UTR1 and YEF1 |
|
[41,82] |
| Drosophila melanogaster | CG6145/NADK RNAi | Lower lipid storage in fat body, reduced mitochondrial mass and ROS (mitochondrial) level, mitochondrial cristae disruption | [43] |
| Mus musculus | C57BL/6N-Nadkem2(IMPC)Bay (Endonuclease-mediated mutation 2, Baylor College of Medicine) |
|
Nadk Mouse Gene Details | NAD kinase | International Mouse Phenotyping Consortium (mousephenotype.org) and Nadk < em2(IMPC)Bay > Endonuclease-mediated Allele Detail MGI Mouse (MGI:6257722) (jax.org) |
| Chemically-induced NADK2 point mutations S326L and S330P |
Severe neuromuscular disease Short lifespan |
[83] | |
| NADK2-KO (C57BL/6NTac background) |
|
[65] | |
| Homo sapiens (Clinical studies) | a homozygous nonsense mutation, c.1018C > T (NM_001085411.1), in exon 10 of NADK2, which leads to a premature stop codon at position 340 (p.R340X) and start loss mutation in the NADK2 gene |
Developmental defects, hyperlysinemia, severe mitochondrial dysfunction, metabolic defects, metabolic acidosis, CNS anomalies, ataxia, and incoordination | [66,75] |
| Gain-of-function mutation: NADK(I90F) | Pro-tumor effects due to increased kinase activities and induction of NADP(H) level-dependent enhanced antioxidant defense response | [62] |
6. Redox cycle of NADP(H)
6.1. NADPH dependent cellular mechanisms
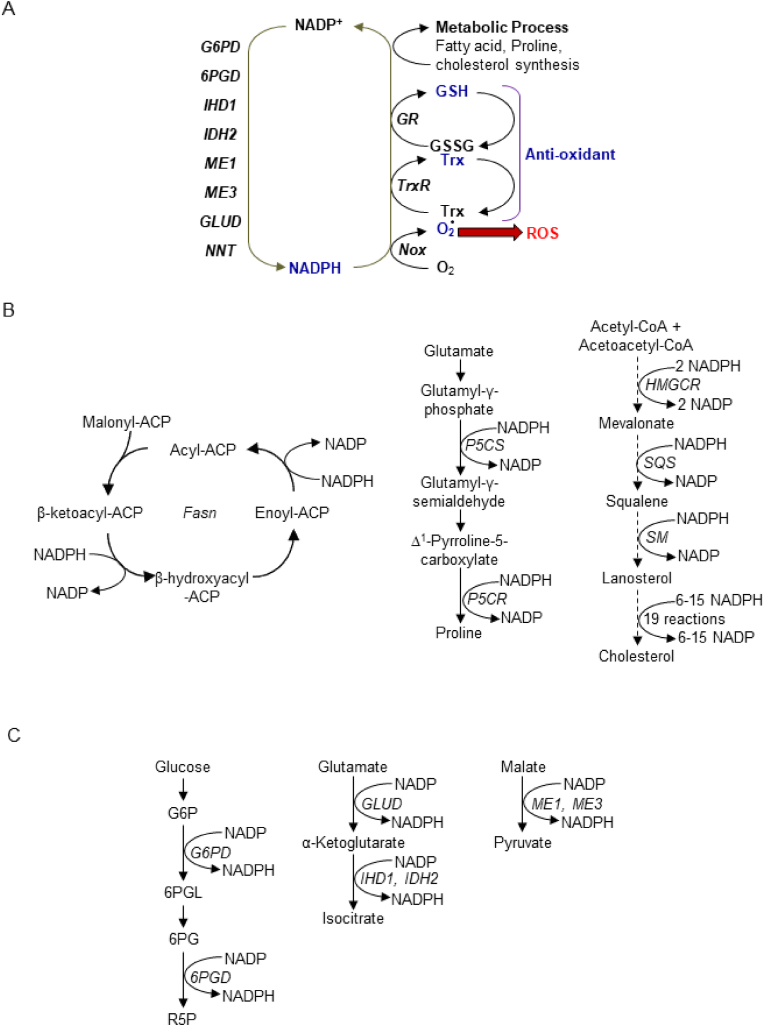
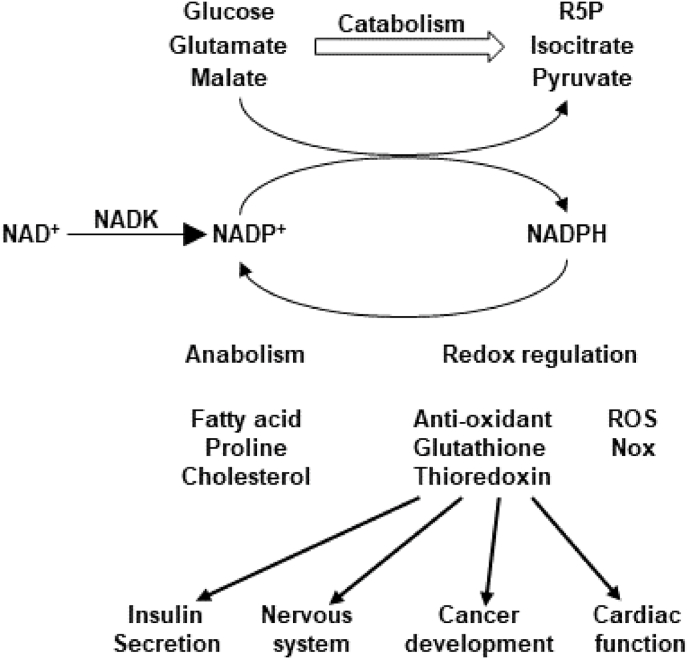
NADP is subject to two-electron reduction at the nicotinamide ring, thereby forming NADPH. NADPH functions as an electron donor for redox regulating enzymes, including NADPH oxidase (Nox) and the thioredoxin and glutathione systems [1], and metabolic enzymes, including those involved in fatty acid, proline and cholesterol synthesis [43,44] (Fig. 5A and B). In addition, NADPH donates electrons to aspartate semialdehyde dehydrogenase in the aspartate catabolism pathway and to dihydrouracil dehydrogenase in the nucleotide catabolism pathway [45,46].
Fig. 5.
The redox cycle of NADP(H) couples with cellular redox homeostasis and metabolism. (A) NADP is reduced by multiple enzymes, whereas
NADPH serves as an electron donor for Nox enzymes, the Trx and Glutathione systems, and metabolic processes. (B) Metabolic reactions in which NADPH donates electrons, including fatty acid (left), proline (middle) and cholesterol (right) syntheses. In the last 19 reactions of cholesterol synthesis, 6 reactions require NADPH and 9 reactions redundantly use NADPH and NADH. As a result, 6–15 NADPH can be used for cholesterol synthesis from lanosterol. (C) Metabolic reactions in which NADP accepts electrons, including catabolic processes of glucose, glutamate, and malate. Abbreviated metabolites and enzymes are as follows. ACP: acyl carrier protein, G6P: Glucose 6 phosphate, 6PGL: 6-phosphogluconolaconase, 6 PG: 6-phosphogluconate, R5P: ribose 5 phosphate, G6PD: glucose 6 phosphate dehydrogenase, 6PGD: 6 phosphogluconate dehydrogenase, IDH: isocitrate dehydrogenase, ME: malic enzyme, GLUD: glutamate dehydrogenase, NNT: nicotinamide nucleotide transhydrogenase, Fasn: fatty acid synthase, P5CS: pyrroline 5 carboxylate synthase, P5CR: pyrroline 5 carboxylate reductase. HMGCR: HMG-CoA reductase, SQS: squalene synthase and SM: squalene monooxygenase.
6.2. Production of NADPH from NADP
Oxidized NADP is reduced by multiple enzymes (Fig. 5C). In the cytosol, NADP is reduced by glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD) in the pentose phosphate pathway, cytosolic isocitrate dehydrogenase (IDH1) and cytosolic malic enzyme (ME1). Of these, G6PD is the major mechanism responsible for the production of NADPH [47]. In mitochondria, mitochondrial isocitrate dehydrogenase (IDH2), glutamate dehydrogenase (GLUD), mitochondrial malic enzyme (ME3) and nicotinamide nucleotide transhydrogenase (NNT) reduce NADP to NADPH [47]. Of these, IDH2 plays a major role in NADPH production [48]. Among the NADPH producing enzymes, only NADK has been demonstrated to have an indispensable role in fatty acid and proline syntheses in vivo [43,44]. Although the involvement of NADK is highly likely, whether NADK-mediated production of NADP(H) is also essential for other enzymatic reactions, including those mediated by IDHs and MEs, or in the pentose phosphate pathway and cholesterol synthesis has yet to be clearly demonstrated in vivo. Further investigation with targeted metabolomic analyses is needed to address these issues.
6.3. NADK-dependent upregulation of NADPH
Overexpression of NADK upregulates NADPH, but not NADP, in HEK293 cells [42]. Similarly, overexpression of Nampt upregulates NADPH, but not NADP, in primary cultured cardiomyocytes [29]. NADP produced by NADK can be immediately converted to NADPH by the NADP-dependent dehydrogenases. Feedback regulation may prevent NADK from functioning except when NADP-dependent dehydrogenases are functional, thereby limiting accumulation of NADP when dehydrogenase activity is limited. Indeed, G6PD binds to and stimulates NADK in pancreatic cancer cells [49]. Alternatively, NADK may directly phosphorylate NADH in cells. NADK specifically phosphorylates NADH when NAD+ and NADH concentrations are low and the NAD+/NADH ratio is high in vitro [42]. Further investigation is required to clarify the mechanism through which NADK produces NADPH without NADP upregulation.
7. Redox regulation via NADK
NADPH plays a crucial role in redox homeostasis as an electron donor. Since NADPH donates electrons not only to antioxidant systems, including thioredoxin and glutathione, but also to Noxs to produce superoxide, in theory, NADK can potentiate both the antioxidant defense and oxidative stress. However, recent studies have demonstrated the significance of NADK in the antioxidant defense rather than in oxidative stress. Overexpression of NADK attenuates, whereas knockdown of NADK enhances, H2O2-induced oxidative stress in HEK293 cells [42], and increased oxidative stress is observed in HEK293 cells with NADK2 knockdown [34]. NADK knockdown also promotes high sugar diet-induced oxidative stress in fly fat body [43] and promotes glutathione oxidation in cancer cell lines [38]. In addition, we have shown that cardiac-specific Nampt overexpression potentiates the antioxidant defense partly through NADK-mediated NADP(H) production [29]. These observations suggest that NADK plays an essential role in the antioxidant defense via NADPH production.
In contrast, whether NADK is essential for Nox-induced ROS production has yet to be fully investigated. Although NADPH is the preferred electron donor, Nox enzymes also utilize NADH [50]. Interferon-γ (INFγ) induces NADP(H) and oxidative stress in a Nampt-dependent manner, but the extent to which NADP(H) and NADK are required for oxidative stress has not been demonstrated [51]. Thus, NADK and NADPH may not be essential for Nox function. In addition, Nox-induced ROS production is tightly regulated by cellular signal transduction [52]. For instance, Nox4 is negatively regulated by Fyn kinase via direct phosphorylation [53]. Thus, an NADK-induced increase in NADPH may not be sufficient to activate Nox enzymes.
If NADPH originating from the NAD+-NADK pathway is utilized by both the antioxidant system and Nox, the cellular mechanism regulating the coupling remains to be elucidated. Since the antioxidant system can be located in the cytosol and Noxs are membrane-associated enzymes, it is possible that the NAD+ synthesis system, NADP-dependent dehydrogenases and electron acceptors are compartmentalized, thereby forming a signaling complex. Alternatively, their coupling may be regulated through unknown mechanisms. Further investigation is required to address this issue.
8. Cellular signaling regulating NADK
NADKs are highly important in the regulation of anabolic processes and the antioxidant defense in cells. NADK affects the NAD+/NADH pool for both catabolic processes leading to ATP production and anabolic processes, including maintenance of the antioxidant system. However, the mechanisms by which NADK maintains or changes the balance between the catabolic and anabolic processes are not well understood.
8.1. Upregulation of endogenous NADK
In the liver and white adipose tissue (WAT), fasting upregulates NADK2, and the opposite occurs in response to high fat diet consumption [32]. In WAT, downregulation of CD38, cyclic ADP ribose hydrolase, upregulates NADK and NADP(H) levels [54].
8.2. Posttranslational modification of NADK
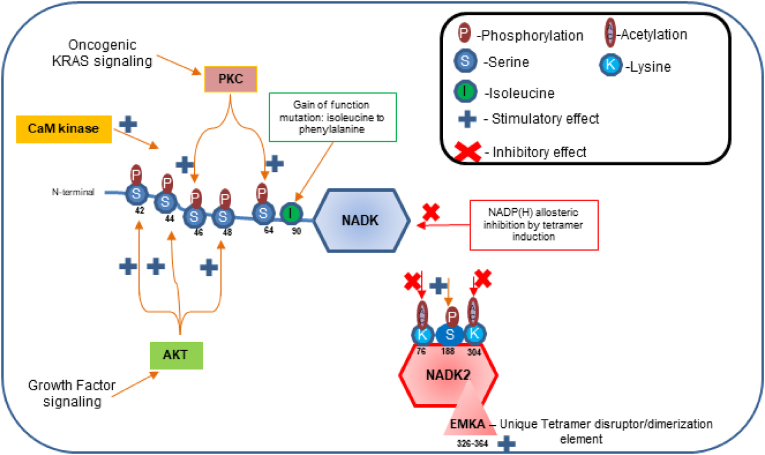
At the post-translational level, NADK is activated via phosphorylation of its N-terminal regulatory domain by various AGC kinases (cAMP- and cGMP-dependent protein kinases and the protein kinase C (PKC) family of protein kinases) [43] and Ca2+/calmodulin-dependent protein kinase (CaM kinase) [3]. In human neutrophils, calmodulin and PKC induce NADK phosphorylation and activation, triggering robust host defense mechanisms [55,56]. In pancreatic ductal adenocarcinoma (PDAC), oncogenic Kirsten rat sarcoma viral oncogene homolog (KRAS) signaling through PKC induces NADK phosphorylation at serines 46 and 48, leading to hyperactivation of NADK [38]. Growth factor-mediated Akt signaling similarly relieves the auto-inhibitory action of the regulatory N-terminal domain of NADK through phosphorylation of serines 44, 46 and 48 and activates NADK [37]. In humans, phosphorylation at serine 188 activates NADK2, but the responsible kinase and phosphatase have yet to be identified [57]. In addition, NADK2 may be regulated by acetylation at lysines 76 and 304 [44].
Ca2+/calmodulin binds to the N-terminal region to activate NADK [58]. Furthermore, NADPH allosterically inhibits NADK by enhancing the tetramer formation [59]. NADK2 possesses a unique tetramer disruptor/dimerization element conserved in mitochondrial kinases of animals (EMKA), which prevents NADK2 oligomerization, promotes dimerization and maintains NADK2 in a constitutively active conformation [60]. Thus, mammalian NADKs are regulated by phosphorylation, acetylation and protein-protein interaction (Fig. 6). Further investigation is required to clarify the upstream regulatory mechanisms and the functional significance of post-translational modifications of NADKs.
Fig. 6.
Cellular signaling mechanisms regulating NADKs. KRAS signaling, growth factor signaling, and CaM kinase stimulate NADK. High NADP(H) levels exert allosteric feedback inhibition of NADK by inducing tetramerization. NADK2 has a unique tetramer disruptor/dimerization Element conserved in Mitochondrial Kinases of Animals (EMKA) that restricts its oligomerization and instead promotes dimerization to maintain a constitutively active conformation. Mutational studies have shown that acetylation of lysines 76 and 304 and phosphorylation of serine 188 regulate NADK2 activity. Amino acid positions are indicated with numbers at their sites.
9. Pathophysiological role of NADK
9.1. Cancer development
NADK contributes to cancer development in PDACs and colon cancers [38,61]. Through screening of patients with PDAC, NADK(I90F) has been identified as a gain-of-function mutation: NADK(I90F) possesses increased kinase activity and induces a greater antioxidant defense response through increased NADP(H) levels [62] compared to wild type NADK.
9.2. Redox homeostasis and metabolism
NADK plays an important role in regulating the intracellular redox homeostasis, thereby regulating insulin secretion in the pancreatic β-cell [63]. NADK also plays an essential role in mediating the antioxidant defense through NADP(H) production in cardiomyocytes, thereby ameliorating cardiomyopathy associated with obesity [29]. Loss of NADK in fly fat body impairs lipid storage, as well as mitochondrial function and mass [43]. Interestingly, however, inhibition of NADK protects against acetaminophen-induced acute liver injury by elevating NAD+ levels, which improves the antioxidant capacity through unknown mechanisms [64]. On the other hand, loss of NADK2 promotes stress-induced liver steatosis in mice [65].
9.3. Nervous system and aging
Loss of NADK2 due to mutation of the start codon is responsible for early-stage lethality and central nervous system anomalies, ataxia, and loss of coordination in humans [66]. Furthermore, both NADK and Akt levels were upregulated in aging mice [67,68]. Thus, Akt-induced NADK activation may account for the decrease in NAD+ during aging, although further investigation is necessary to confirm this hypothesis. In summary, although only a limited number of studies have been conducted, increasing lines of evidence suggest that endogenous NADKs play a significant role in regulating cancer, insulin secretion, heart disease, anabolic and catabolic processes, neuronal development, and aging (Fig. 7).
Fig. 7.
Cellular effects and pathophysiological roles of NADKs. NADK is essential for NADP production. NADP is reduced to NADPH, which couples with catabolic processes, including the pentose phosphate pathway. NADPH donates electrons to anabolic pathways and enzymes for redox homeostasis. Anti-oxidant pathways play a role in insulin secretion, cancer development and the maintenance of nervous and cardiac function.
10. NADKs as therapeutic targets in cancer
Because of the differences in the active structure and substrates between prokaryotic and mammalian NADK, researchers have focused on the development of pharmacological inhibitors as a treatment strategy for pathogenic bacterial infections, particularly against multidrug-resistant bacteria [[69], [70], [71]]. The idea of targeting human NADK as a treatment strategy for inflammation and cancer has only recently gained traction [72].
The role of NADK in modulating cellular ROS through NADPH, knock-down or over-expression of cytosolic NADK has only a modest effect on levels of ROS [42,73]. However, mitochondrial NADK2 might also play a role in NADPH generation and protect mitochondria from ROS [34,74]. On the other hand, although NADK2 is possibly a better target in cancers, because of its critical role in protection against ROS in normal tissues like heart and brain, inhibition of NADK2 might have detrimental effects. Indeed, NADK2 deficiency due to a homozygous nonsense mutation was identified in a pediatric patient with developmental defects and early death [75]. Thus, NADK might be a better target for the development of pharmacological inhibitors. To this end, various mutants of NADK, including NADK(I90F), have been isolated from PDAC that render pro-tumor effects by enhancing NADPH levels and ROS protection [62].
11. NAD analog as potential NADK inhibitors
Approximately 30 clinically approved NADK inhibitors targeting the ATP binding site have been developed thus far. However, as with many chemical inhibitors targeting ATP binding domains, their selectivity is not optimal [70,72,76]. Targeting the NAD+ binding site instead could increase selectivity, since human NADK possesses a unique NAD+ binding domain that differs from those of other NAD+-binding enzymes [72]. Benzamide riboside is metabolized in cells to benzamide adenine nucleoside (BAD), an NAD+ analog, thereby acting as a prodrug. BAD inhibits NADKs, thereby lowering NADPH levels and inhibiting dihydrofolate reductase (DHFR), an enzyme that uses NADPH as an electron donor [77]. Due to skeletal muscle loss and liver toxicity, however, benzamide riboside was not advanced in the drug development process. Screenings with small compound libraries for NAD+ analogs showed that thionicotinamide adenine dinucleotide and thionicotinamide adenine dinucleotide phosphate (NADPS) exhibit cytotoxicity due to NADK inhibition [78]. Thionicotinamide (TN), another NAD+ analog, is converted to NADPS upon cell entry and also exhibits cytotoxicity in various cancer cell lines due to NADK inhibition [79]. TN upregulates ROS in C85 cells through downregulation of NADPH [79]. TN also inhibits fatty acid synthesis and nucleotide synthesis pathways, both of which are NADPH-dependent [73]. TN effectively sensitizes cells to chemotherapeutic agents, including methotrexate, gemcitabine, docetaxel, and irinotecan, due to its ability to increase ROS levels and inhibit NADPH-dependent synthetic pathways [73]. TN administration as a single dose in human xenograft mouse models produced a promising result comparable to that of NADK downregulation (50%) with shRNA [73]. Another NAD+ analog, 6-amino nicotinamide, showed promising pre-clinical anti-cancer activity but was associated with neurological toxicity [80]. The neurotoxicity of 6-amino nicotinamide was ascribed to its ability to block the pentose phosphate pathway via phosphogluconate dehydrogenase inhibition [81] and inhibit NADK2 phosphorylation [79]. Nicotinamide administration was found to reverse the cytotoxic effects of NAD+ analogs [79]. On the other hand, TN does not show neurotoxic effects, suggesting that not all NAD+ analogs are neurotoxic [79]. Aside from the neurotoxicity of 6-amino nicotinamide, interventions that inhibit NADPH synthesis via NADK inhibition demonstrate safe therapeutic anti-cancer activity by lowering NADPH levels, which hinders the protection against ROS and negatively affects NADP(H)-dependent metabolic pathways. However, the development of more selective inhibitors that can differentiate between cytoplasmic and mitochondrial forms of NADKs is important for the development of more effective cancer treatments.
12. Conclusions
NADKs are evolutionarily conserved enzymes that phosphorylate NAD+ and produce NADP(H). NADK is the only known mechanism by which NADP(H) is produced de novo. In eukaryotes, both cytosolic and mitochondrial NADKs exist, but other organelle specific NADKs have yet to be identified. Recent studies using genetically modified mouse models validate NADKs as the major enzymes that produce NADP(H) across species. Moreover, these studies validate the significance of NADP(H) in cellular redox homeostasis. Although NADP(H) plays an essential role in many metabolic processes, its functional significance requires further validation in vivo with NADK loss-of-function models. In addition, the mechanisms by which NADK balances the NAD(H) and NADP(H) pools remain to be clarified, and how cells sense the status of the NAD(H) vs. NADP(H) pools and regulate NADK function is not currently well understood. The molecular mechanisms that regulate NADK include protein-protein interaction and post-translational modifications, such as phosphorylation and acetylation. However, other than Akt, CaMK and PKC, the phosphatases, acetylases and deacetylases involved in post-translational modification of NADKs remain to be identified. How NADKs are differentially regulated in tissues, biological processes and disease conditions also remains unclear. Considering the wealth of knowledge about NAD+ biology, investigation of NADKs would dramatically advance our understanding of how NAD+ is utilized during disease and aging in humans.
Author contributions
Author contributions: S.O., A.S.T., D.Z. and J.S. wrote the manuscript, and the authors reviewed and approved the manuscript for publication.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This work was supported in part by U.S. Public Health Service Grants HL67724, HL91469, HL102738, HL112330, HL138720, HL144626, HL150881, and AG23039 (J.S.), the American Heart Association (AHA) Grant in Aid 17GRNT33440031 (S.O.), Transformational Project Award 19TPA34850170 (S.O.), Merit Award 20 Merit 35120374 (J.S.), and Foundation Leducq Transatlantic Network of Excellence 15CVD04 (J.S.).
Data availability
No data was used for the research described in the article.
References
- 1.Oka S., Hsu C.P., Sadoshima J. Regulation of cell survival and death by pyridine nucleotides. Circ. Res. 2012;111:611–627. doi: 10.1161/CIRCRESAHA.111.247932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burch H.B., Bradley M.E., Lowry O.H. The measurement of triphosphopyridine nucleotide and reduced triphosphopyridine nucleotide and the role of hemoglobin in producing erroneous triphosphopyridine nucleotide values. J. Biol. Chem. 1967;242:4546–4554. [PubMed] [Google Scholar]
- 3.Okabe K., Yaku K., Tobe K., Nakagawa T. Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 2019;26:34. doi: 10.1186/s12929-019-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai S.I. The NAD World 2.0: the importance of the inter-tissue communication mediated by NAMPT/NAD(+)/SIRT1 in mammalian aging and longevity control. NPJ Syst Biol Appl. 2016;2 doi: 10.1038/npjsba.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camacho-Pereira J., Tarrago M.G., Chini C.C.S., Nin V., Escande C., Warner G.M., Puranik A.S., Schoon R.A., Reid J.M., Galina A., et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metabol. 2016;23:1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassa P.O., Haenni S.S., Elser M., Hottiger M.O. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasamatsu A., Nakao M., Smith B.C., Comstock L.R., Ono T., Kato J., Denu J.M., Moss J. Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J. Biol. Chem. 2011;286:21110–21117. doi: 10.1074/jbc.M111.237636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braidy N., Guillemin G.J., Mansour H., Chan-Ling T., Poljak A., Grant R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clement J., Wong M., Poljak A., Sachdev P., Braidy N. The plasma NAD(+) metabolome is dysregulated in "normal" aging. Rejuvenation Res. 2019;22:121–130. doi: 10.1089/rej.2018.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C.C., Yang X., Hua X., Liu J., Fan M.B., Li G.Q., Song J., Xu T.Y., Li Z.Y., Guan Y.F., et al. Hepatic NAD(+) deficiency as a therapeutic target for non-alcoholic fatty liver disease in ageing. Br. J. Pharmacol. 2016;173:2352–2368. doi: 10.1111/bph.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshino J., Mills K.F., Yoon M.J., Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metabol. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarino M., Dufour J.F. Nicotinamide, NAFLD Is there nothing new under the sun? Metabolites. 2019;9 doi: 10.3390/metabo9090180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conforti L., Gilley J., Coleman M.P. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 14.Hikosaka K., Yaku K., Okabe K., Nakagawa T. Implications of NAD metabolism in pathophysiology and therapeutics for neurodegenerative diseases. Nutr. Neurosci. 2021;24:371–383. doi: 10.1080/1028415X.2019.1637504. [DOI] [PubMed] [Google Scholar]
- 15.Liu D., Pitta M., Jiang H., Lee J.H., Zhang G., Chen X., Kawamoto E.M., Mattson M.P. Nicotinamide forestalls pathology and cognitive decline in Alzheimer mice: evidence for improved neuronal bioenergetics and autophagy procession. Neurobiol. Aging. 2013;34:1564–1580. doi: 10.1016/j.neurobiolaging.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdellatif M., Trummer-Herbst V., Koser F., Durand S., Adao R., Vasques-Novoa F., Freundt J.K., Voglhuber J., Pricolo M.R., Kasa M., et al. Nicotinamide for the treatment of heart failure with preserved ejection fraction. Sci. Transl. Med. 2021:13. doi: 10.1126/scitranslmed.abd7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byun J., Oka S.I., Imai N., Huang C.Y., Ralda G., Zhai P., Ikeda Y., Ikeda S., Sadoshima J. Both gain and loss of Nampt function promote pressure overload-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 2019;317:H711–H725. doi: 10.1152/ajpheart.00222.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Canto C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H., Ryu D., Wu Y., Gariani K., Wang X., Luan P., D'Amico D., Ropelle E.R., Lutolf M.P., Aebersold R., et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 20.Harrison D.E., Strong R., Reifsnyder P., Kumar N., Fernandez E., Flurkey K., Javors M.A., Lopez-Cruzan M., Macchiarini F., Nelson J.F., et al. 17-a-estradiol late in life extends lifespan in aging UM-HET3 male mice; nicotinamide riboside and three other drugs do not affect lifespan in either sex. Aging Cell. 2021;20 doi: 10.1111/acel.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell S.J., Bernier M., Aon M.A., Cortassa S., Kim E.Y., Fang E.F., Palacios H.H., Ali A., Navas-Enamorado I., Di Francesco A., et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metabol. 2018;27:667–676 e664. doi: 10.1016/j.cmet.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdellatif M., Sedej S., Kroemer G. NAD(+) metabolism in cardiac health, aging, and disease. Circulation. 2021;144:1795–1817. doi: 10.1161/CIRCULATIONAHA.121.056589. [DOI] [PubMed] [Google Scholar]
- 23.Yoshino J., Baur J.A., Imai S.I. NAD(+) intermediates: the biology and therapeutic potential of NMN and NR. Cell Metabol. 2018;27:513–528. doi: 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie N., Zhang L., Gao W., Huang C., Huber P.E., Zhou X., Li C., Shen G., Zou B. NAD(+) metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Targeted Ther. 2020;5:227. doi: 10.1038/s41392-020-00311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson S., Imai S.I. NAD (+) biosynthesis, aging, and disease. F1000Res. 2018;7:132. doi: 10.12688/f1000research.12120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araki T., Sasaki Y., Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 27.He S., Gao Q., Wu X., Shi J., Zhang Y., Yang J., Li X., Du S., Zhang Y., Yu J. NAD(+) ameliorates endotoxin-induced acute kidney injury in a sirtuin1-dependent manner via GSK-3beta/Nrf2 signalling pathway. J. Cell Mol. Med. 2022;26:1979–1993. doi: 10.1111/jcmm.17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto T., Byun J., Zhai P., Ikeda Y., Oka S., Sadoshima J. Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka S.I., Byun J., Huang C.Y., Imai N., Ralda G., Zhai P., Xu X., Kashyap S., Warren J.S., Alan Maschek J., et al. Nampt potentiates antioxidant defense in diabetic cardiomyopathy. Circ. Res. 2021;129:114–130. doi: 10.1161/CIRCRESAHA.120.317943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R.Mnadk. A long-awaited human mitochondrion-localized NAD kinase. J. Cell. Physiol. 2015;230:1697–1701. doi: 10.1002/jcp.24926. [DOI] [PubMed] [Google Scholar]
- 31.Murata K. Polyphosphate-dependent nicotinamide adenine dinucleotide (NAD) kinase: a novel missing link in human mitochondria. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021;97:479–498. doi: 10.2183/pjab.97.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R.Mnadk. A novel liver-enriched mitochondrion-localized NAD kinase. Biol Open. 2013;2:432–438. doi: 10.1242/bio.20134259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Achbergerova L., Nahalka J. Polyphosphate--an ancient energy source and active metabolic regulator. Microb. Cell Factories. 2011;10:63. doi: 10.1186/1475-2859-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohashi K., Kawai S., Murata K. Identification and characterization of a human mitochondrial NAD kinase. Nat. Commun. 2012;3:1248. doi: 10.1038/ncomms2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori S., Yamasaki M., Maruyama Y., Momma K., Kawai S., Hashimoto W., Mikami B., Murata K. NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex. Biochem. Biophys. Res. Commun. 2005;327:500–508. doi: 10.1016/j.bbrc.2004.11.163. [DOI] [PubMed] [Google Scholar]
- 36.Poncet-Montange G., Assairi L., Arold S., Pochet S., Labesse G. NAD kinases use substrate-assisted catalysis for specific recognition of NAD. J. Biol. Chem. 2007;282:33925–33934. doi: 10.1074/jbc.M701394200. [DOI] [PubMed] [Google Scholar]
- 37.Hoxhaj G., Ben-Sahra I., Lockwood S.E., Timson R.C., Byles V., Henning G.T., Gao P., Selfors L.M., Asara J.M., Manning B.D. Direct stimulation of NADP(+) synthesis through Akt-mediated phosphorylation of NAD kinase. Science. 2019;363:1088–1092. doi: 10.1126/science.aau3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schild T., McReynolds M.R., Shea C., Low V., Schaffer B.E., Asara J.M., Piskounova E., Dephoure N., Rabinowitz J.D., Gomes A.P., et al. NADK is activated by oncogenic signaling to sustain pancreatic ductal adenocarcinoma. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109238. [DOI] [PubMed] [Google Scholar]
- 39.Raffaelli N., Finaurini L., Mazzola F., Pucci L., Sorci L., Amici A., Magni G. Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by site-directed mutagenesis. Biochemistry. 2004;43:7610–7617. doi: 10.1021/bi049650w. [DOI] [PubMed] [Google Scholar]
- 40.Ando T., Ohashi K., Ochiai A., Mikami B., Kawai S., Murata K. Structural determinants of discrimination of NAD+ from NADH in yeast mitochondrial NADH kinase Pos5. J. Biol. Chem. 2011;286:29984–29992. doi: 10.1074/jbc.M111.249011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bieganowski P., Seidle H.F., Wojcik M., Brenner C. Synthetic lethal and biochemical analyses of NAD and NADH kinases in Saccharomyces cerevisiae establish separation of cellular functions. J. Biol. Chem. 2006;281:22439–22445. doi: 10.1074/jbc.M513919200. [DOI] [PubMed] [Google Scholar]
- 42.Pollak N., Niere M., Ziegler M. NAD kinase levels control the NADPH concentration in human cells. J. Biol. Chem. 2007;282:33562–33571. doi: 10.1074/jbc.M704442200. [DOI] [PubMed] [Google Scholar]
- 43.Xu M., Ding L., Liang J., Yang X., Liu Y., Wang Y., Ding M., Huang X. NAD kinase sustains lipogenesis and mitochondrial metabolism through fatty acid synthesis. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.110157. [DOI] [PubMed] [Google Scholar]
- 44.Mary C., Soflaee M.H., Kesavan R., Gelin M., Brown H., Zacharias G., Mathews T.P., Lemoff A., Lionne C., Labesse G., et al. Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation. Mol Cell. 2022 doi: 10.1016/j.molcel.2022.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco J., Moore R.A., Kabaleeswaran V., Viola R.E. A structural basis for the mechanism of aspartate-beta-semialdehyde dehydrogenase from Vibrio cholerae. Protein Sci. 2003;12:27–33. doi: 10.1110/ps.0230803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith A.E., Yamada E.W. Dihydrouracil dehydrogenase of rat liver. Separation of hydrogenase and dehydrogenase activities. J. Biol. Chem. 1971;246:3610–3617. [PubMed] [Google Scholar]
- 47.Xiao W., Wang R.S., Handy D.E., Loscalzo J. NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxidants Redox Signal. 2018;28:251–272. doi: 10.1089/ars.2017.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rydstrom J. Mitochondrial NADPH, transhydrogenase and disease. Biochim. Biophys. Acta. 2006;1757:721–726. doi: 10.1016/j.bbabio.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Xu Y., Lu W., Li J., Yu S., Brown E.J., Stanger B.Z., Rabinowitz J.D., Yang X. G6PD-mediated increase in de novo NADP(+) biosynthesis promotes antioxidant defense and tumor metastasis. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 51.McCann K.J., Christensen S.M., Colby D.H., McGuire P.J., Myles I.A., Zerbe C.S., Dalgard C.L., Sukumar G., Leonard W.J., McCormick B.A., et al. IFNgamma regulates NAD+ metabolism to promote the respiratory burst in human monocytes. Blood Adv. 2022;6:3821–3834. doi: 10.1182/bloodadvances.2021005776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermot A., Petit-Hartlein I., Smith S.M.E., Fieschi F. NADPH oxidases (NOX): an overview from discovery, molecular mechanisms to physiology and pathology. Antioxidants. 2021;10 doi: 10.3390/antiox10060890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsushima S., Kuroda J., Zhai P., Liu T., Ikeda S., Nagarajan N., Oka S., Yokota T., Kinugawa S., Hsu C.P., et al. Tyrosine kinase FYN negatively regulates NOX4 in cardiac remodeling. J. Clin. Invest. 2016;126:3403–3416. doi: 10.1172/JCI85624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benzi A., Sturla L., Heine M., Fischer A.W., Spinelli S., Magnone M., Sociali G., Parodi A., Fenoglio D., Emionite L., et al. CD38 downregulation modulates NAD(+) and NADP(H) levels in thermogenic adipose tissues. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1866 doi: 10.1016/j.bbalip.2020.158819. [DOI] [PubMed] [Google Scholar]
- 55.Rabani R., Cossette C., Graham F., Powell W.S. Protein kinase C activates NAD kinase in human neutrophils. Free Radic. Biol. Med. 2020;161:50–59. doi: 10.1016/j.freeradbiomed.2020.09.022. [DOI] [PubMed] [Google Scholar]
- 56.Williams M.B., Jones H.P. Calmodulin-dependent NAD kinase of human neutrophils. Arch. Biochem. Biophys. 1985;237:80–87. doi: 10.1016/0003-9861(85)90256-5. [DOI] [PubMed] [Google Scholar]
- 57.Kawabata Y., Murata K., Kawai S. Significance of Ser-188 in human mitochondrial NAD kinase as determined by phosphomimetic and phosphoresistant amino-acid substitutions. Biochem. Biophys. Res. Commun. 2015;468:691–695. doi: 10.1016/j.bbrc.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 58.Love N.R., Pollak N., Dolle C., Niere M., Chen Y., Oliveri P., Amaya E., Patel S., Ziegler M. NAD kinase controls animal NADP biosynthesis and is modulated via evolutionarily divergent calmodulin-dependent mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1386–1391. doi: 10.1073/pnas.1417290112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grose J.H., Joss L., Velick S.F., Roth J.R. Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7601–7606. doi: 10.1073/pnas.0602494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du J., Estrella M., Solorio-Kirpichyan K., Jeffrey P.D., Korennykh A. Structure of human NADK2 reveals atypical assembly and regulation of NAD kinases from animal mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2200923119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yau E.H., Kummetha I.R., Lichinchi G., Tang R., Zhang Y., Rana T.M. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res. 2017;77:6330–6339. doi: 10.1158/0008-5472.CAN-17-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsang Y.H., Dogruluk T., Tedeschi P.M., Wardwell-Ozgo J., Lu H., Espitia M., Nair N., Minelli R., Chong Z., Chen F., et al. Functional annotation of rare gene aberration drivers of pancreatic cancer. Nat. Commun. 2016;7 doi: 10.1038/ncomms10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray J.P., Alavian K.N., Jonas E.A., Heart E.A. NAD kinase regulates the size of the NADPH pool and insulin secretion in pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 2012;303:E191–E199. doi: 10.1152/ajpendo.00465.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao C., Zhang L., Jiang R., Xu J., Tang J., Hu K., Jiang S., Li L., Yang Y., Huang J., et al. Inhibition of NAD kinase elevates the hepatic NAD(+) pool and alleviates acetaminophen-induced acute liver injury in mice. Biochem. Biophys. Res. Commun. 2022;612:70–76. doi: 10.1016/j.bbrc.2022.04.079. [DOI] [PubMed] [Google Scholar]
- 65.Zhang K., Kim H., Fu Z., Qiu Y., Yang Z., Wang J., Zhang D., Tong X., Yin L., Li J., et al. Deficiency of the mitochondrial NAD kinase causes stress-induced hepatic steatosis in mice. Gastroenterology. 2018;154:224–237. doi: 10.1053/j.gastro.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pomerantz D.J., Ferdinandusse S., Cogan J., Cooper D.N., Reimschisel T., Robertson A., Bican A., McGregor T., Gauthier J., Millington D.S., et al. Clinical heterogeneity of mitochondrial NAD kinase deficiency caused by a NADK2 start loss variant. Am. J. Med. Genet. 2018;176:692–698. doi: 10.1002/ajmg.a.38602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dall M., Penke M., Sulek K., Matz-Soja M., Holst B., Garten A., Kiess W., Treebak J.T. Hepatic NAD(+) levels and NAMPT abundance are unaffected during prolonged high-fat diet consumption in C57BL/6JBomTac mice. Mol. Cell. Endocrinol. 2018;473:245–256. doi: 10.1016/j.mce.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 68.Baar E.L., Carbajal K.A., Ong I.M., Lamming D.W. Sex- and tissue-specific changes in mTOR signaling with age in C57BL/6J mice. Aging Cell. 2016;15:155–166. doi: 10.1111/acel.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bi J., Wang H., Xie J. Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets. J. Cell. Physiol. 2011;226:331–340. doi: 10.1002/jcp.22419. [DOI] [PubMed] [Google Scholar]
- 70.Petrelli R., Sham Y.Y., Chen L., Felczak K., Bennett E., Wilson D., Aldrich C., Yu J.S., Cappellacci L., Franchetti P., et al. Selective inhibition of nicotinamide adenine dinucleotide kinases by dinucleoside disulfide mimics of nicotinamide adenine dinucleotide analogues. Bioorg. Med. Chem. 2009;17:5656–5664. doi: 10.1016/j.bmc.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 71.Shi F., Li Y., Li Y., Wang X. Molecular properties, functions, and potential applications of NAD kinases. Acta Biochim. Biophys. Sin. 2009;41:352–361. doi: 10.1093/abbs/gmp029. [DOI] [PubMed] [Google Scholar]
- 72.Petrelli R., Felczak K., Cappellacci L. NMN/NaMN adenylyltransferase (NMNAT) and NAD kinase (NADK) inhibitors: chemistry and potential therapeutic applications. Curr. Med. Chem. 2011;18:1973–1992. doi: 10.2174/092986711795590048. [DOI] [PubMed] [Google Scholar]
- 73.Tedeschi P.M., Lin H., Gounder M., Kerrigan J.E., Abali E.E., Scotto K., Bertino J.R. Suppression of cytosolic NADPH pool by thionicotinamide increases oxidative stress and synergizes with chemotherapy. Mol. Pharmacol. 2015;88:720–727. doi: 10.1124/mol.114.096727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lerner F., Niere M., Ludwig A., Ziegler M. Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 2001;288:69–74. doi: 10.1006/bbrc.2001.5735. [DOI] [PubMed] [Google Scholar]
- 75.Houten S.M., Denis S., Te Brinke H., Jongejan A., van Kampen A.H., Bradley E.J., Baas F., Hennekam R.C., Millington D.S., Young S.P., et al. Mitochondrial NADP(H) deficiency due to a mutation in NADK2 causes dienoyl-CoA reductase deficiency with hyperlysinemia. Hum. Mol. Genet. 2014;23:5009–5016. doi: 10.1093/hmg/ddu218. [DOI] [PubMed] [Google Scholar]
- 76.Pankiewicz K.W., Watanabe K.A., Lesiak-Watanabe K., Goldstein B.M., Jayaram H.N. The chemistry of nicotinamide adenine dinucleotide (NAD) analogues containing C-nucleosides related to nicotinamide riboside. Curr. Med. Chem. 2002;9:733–741. doi: 10.2174/0929867024606920. [DOI] [PubMed] [Google Scholar]
- 77.Roussel B., Johnson-Farley N., Kerrigan J.E., Scotto K.W., Banerjee D., Felczak K., Pankiewicz K.W., Gounder M., Lin H., Abali E.E., et al. A second target of benzamide riboside: dihydrofolate reductase. Cancer Biol. Ther. 2012;13:1290–1298. doi: 10.4161/cbt.21786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsieh Y.C., Tedeschi P., Adebisi Lawal R., Banerjee D., Scotto K., Kerrigan J.E., Lee K.C., Johnson-Farley N., Bertino J.R., Abali E.E. Enhanced degradation of dihydrofolate reductase through inhibition of NAD kinase by nicotinamide analogs. Mol. Pharmacol. 2013;83:339–353. doi: 10.1124/mol.112.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tedeschi P.M., Bansal N., Kerrigan J.E., Abali E.E., Scotto K.W., Bertino J.R. NAD+ kinase as a therapeutic target in cancer. Clin. Cancer Res. 2016;22:5189–5195. doi: 10.1158/1078-0432.CCR-16-1129. [DOI] [PubMed] [Google Scholar]
- 80.Herken H., Meyer-Estorf G., Halbhubner K., Loos D. Spastic paresis after 6-aminonicotinamide: metabolic disorders in the spinal cord and electromyographically recorded changes in the hind limbs of rats. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1976;293:245–255. doi: 10.1007/BF00507347. [DOI] [PubMed] [Google Scholar]
- 81.Koutcher J.A., Alfieri A.A., Matei C., Meyer K.L., Street J.C., Martin D.S. Effect of 6-aminonicotinamide on the pentose phosphate pathway: 31P NMR and tumor growth delay studies. Magn. Reson. Med. 1996;36:887–892. doi: 10.1002/mrm.1910360611. [DOI] [PubMed] [Google Scholar]
- 82.Outten C.E., Culotta V.C. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2003;22:2015–2024. doi: 10.1093/emboj/cdg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murray G., Bais P., Hatton C., Tadenev A.L.D., Hoffmann B.R., Stodola T.J., Morelli K.H., Pratt S.L., Schroeder D., Doty R., et al. Mouse models of NADK2 deficiency analyzed for metabolic and gene expression changes to elucidate pathophysiology. Hum. Mol. Genet. 2022 doi: 10.1093/hmg/ddac151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.