Abstract
The relatedness of group A streptococcal (GAS) strains isolated from 35 Canadian patients with invasive disease of different severity was investigated by a variety of molecular methods. All patients were infected with M1T1 strains and, based on clinical criteria, were classified as severe (n = 21) and nonsevere (n = 14) invasive GAS infection cases. All the M1 strains studied had the emm1.0 allele and the same streptococcal pyrogenic exotoxin (Spe) genotype, speA+ speB+ speC speF+ speG+ speH smeZ+ ssa. All isolates had the same speA allotype, speA2. The randomly amplified polymorphic DNA banding pattern with two different primers was identical for all strains, and pulsed field gel electrophoresis analysis showed that 33 and 30 isolates had identical banding patterns after DNA digestion with SfiI or SmaI, respectively; the nonidentical isolates differed from the main pattern by only one band. A relatively high degree of polymorphism in specific regions of the sic gene was observed among isolates; however, this polymorphism was not associated with disease severity. Likewise, although the phenotypic expression of SpeA, SpeB, and SpeF proteins varied among the M1T1 isolates, there was no correlation between the amount of Spe expressed and disease severity. Importantly, mitogenic and cytokine responses induced by partially purified bacterial culture supernatants containing a mixture of expressed superantigens were very similar for isolates from severe and nonsevere cases (P > 0.1). Together, the data indicate that highly related invasive M1T1 isolates, some indistinguishable, can cause disease of varying severity in different individuals. These findings underscore the contribution of host factors to the outcome of invasive GAS infections.
Group A streptococci (GAS) are responsible for a variety of human diseases ranging from simple pharyngitis to highly severe infections, such as necrotizing fasciitis (NF) and streptococcal toxic shock syndrome (STSS) (16). Since the late 1980s, a marked increase in the incidence and severity of invasive infections has been reported in the United States, Canada, Japan, and many regions of Europe (11, 13, 16, 17, 20, 22, 37). Whether this dramatic rise in the incidence of invasive GAS infections resulted from changes in virulence properties of the bacteria and/or alterations of host protective immunity against specific strains or specific virulence factors remains an area of intense investigation.
Among the many virulence factors produced by GAS, the M protein and the streptococcal pyrogenic exotoxins (Spes) are considered important virulence factors in the pathogenesis of invasive GAS infections. The Spes belong to the superantigen (SAg) family and thus can induce massive secretion of inflammatory cytokines, such as gamma interferon (IFN-γ), interleukin-1 (IL-1), and tumor necrosis factor α. Overproduction of these cytokines can lead to tissue damage, organ failure, and shock (reviewed by Kotb [15]).
GAS are classified by their surface M protein type (4, 10). To date, more than 100 different M types have been identified (9); however, epidemiological studies have indicated that the majority of recent invasive GAS infections are caused by strains of the M1 or M3 serotype (6, 8, 13, 14, 17, 22, 37, 39). However, further genotypic characterizations of GAS strains by molecular methods such as multilocus enzyme electrophoresis (MLEE), restriction endonuclease analysis, ribotyping, pulsed-field gel electrophoresis (PFGE), and random amplified polymorphic DNA (RAPD) analysis have demonstrated that strains expressing the same M protein serotype are not necessarily genetically homogeneous (5, 7, 21, 22, 24, 33, 34, 39).
Despite the fact that invasive GAS disease may not be restricted to the spread of one or two virulent clones, recent studies have suggested that the same serotype or the same subtype of a serotype can cause invasive as well as noninvasive infections (6, 14, 20). Furthermore, ongoing surveillance of invasive GAS infections in Ontario, Canada, revealed that a large number of severe and nonsevere invasive cases appear to be caused by related M1T1 strains. The main question was whether M1T1 strains causing invasive disease of varying severity are similar or whether they represent distinct subtypes of the M1T1 serotype. To address this question, we examined the genetic diversity, SAg expression, and mitogenic and cytokine-inducing capacity of 35 M1T1 GAS strains recovered from severe and nonsevere invasive GAS infection cases. The data indicate that highly related strains that are immunologically indistinguishable can cause disease with starkly varying severity in different individuals. The findings of this study underscore the importance of host factors in potentiating the outcome of invasive GAS infections.
MATERIALS AND METHODS
Bacterial strains.
The demographics of the invasive GAS infection patients from whom the M1T1 GAS strains were isolated are described in Table 1. All isolates were obtained between December 1994 and December 1996 through ongoing active surveillance of invasive diseases in Ontario, Canada. Twenty-one strains were obtained from patients with severe GAS invasive infections, and 14 strains were from patients with nonsevere GAS invasive infections. The severe cases were those with STSS and/or NF; nonsevere cases had neither. Additional GAS isolates of serotypes M1, M3, M12, and M28, from the American Navy Health Research Center, were also tested for comparison with the Canadian M1 strains.
TABLE 1.
Demographics of invasive GAS infection patients from whom M1T1 isolates were obtained
| Group and diagnosis | No. of isolates | Patient age, yr (mean ± SD) | Gender (no.)
|
|
|---|---|---|---|---|
| M | F | |||
| Severe invasive | 21 | 52 ± 26 | 11 | 10 |
| NF | 8 | 48 ± 34 | 5 | 3 |
| STSS | 9 | 58 ± 22 | 4 | 5 |
| STSS and NF | 4 | 45 ± 18 | 2 | 2 |
| Nonsevere invasive | 14 | 36 ± 15 | 7 | 7 |
| Soft tissue infection, cellulitis, erysepelas | 10 | 35 ± 18 | 5 | 5 |
| Septicemia ± pneumonia | 3 | 39 ± 5 | 1 | 2 |
| Septicemia/arthritis | 1 | 35 | 1 | 0 |
Serotyping.
M protein and T protein serotyping was performed at the National Reference Center for the Streptococci, Edmonton, Canada. Verification of M1 type was performed by direct sequencing of the emm gene as described below.
DNA preparation.
GAS strains were grown overnight at 37°C on blood agar plates, and pure colonies were picked and inoculated into Todd-Hewitt broth (THB) and grown for 18 h. Genomic DNA was extracted from the bacterial pellet as previously described (30). Briefly, cells were harvested, resuspended in 100 μl of fresh lysozyme at 50 mg/ml in 1× TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 7.5]), and incubated at 37°C for 30 min. Cell lysis was completed in guanidium thiocyanate buffer. DNA was extracted with phenol, precipitated with ethanol, and resuspended in 1× Tris-EDTA buffer.
RAPD analysis.
RAPD was performed with Ready-to-Go RAPD beads from Pharmacia (Uppsala, Sweden). Briefly, 20 pmol (4 μl) of primer P1 (5′-GGTGCGGGAA-3′) or primer P5 (5′-AACGCGCAAC-3′), 10 ng (2 μl) of template DNA, and 20.5 μl of H2O were added to the RAPD analysis bead. The PCR cycling program was 1 cycle at 95°C for 5 min, followed by 45 cycles at 95°C for 1 min, 36°C for 1 min, and 72°C for 2 min. The amplified fragments were separated on a 2% agarose gel and visualized by ethidium bromide staining. The gels were photographed, and the banding patterns were recorded.
PFGE analysis.
PFGE was carried out by the following modification of the method described by McCarthy and Khambaty (18). The cultures were grown in brain-heart infusion broth at 37°C in a blood-gas environment until an optical density at 610 nm (OD610) of 0.8 was reached. Cells from 1 ml of culture were harvested by centrifugation (12,000 × g for 2 min), washed sequentially with TSE buffer (10 mM Tris-Cl, 1.0 M NaCl, 50 mM EDTA [pH 8.0]) and then resuspended in 1 ml of TE buffer. The cells were then embedded in 0.5% PFGE-grade agarose. After solidification, the plugs were treated for 4 h with 2.0 ml of lysis buffer (0.5% sarcosyl, 0.2% sodium deoxycholate, 6 mM Tris-Cl, 100 mM EDTA [pH 8.0]) containing freshly added lysozyme (2 mg/ml) and mutanolysin (100 U/ml). The lysis buffer was replaced with 2.0 ml of ESP buffer (1 mg of proteinase K per ml in 0.1% sarcosyl, 0.5 M EDTA [pH 9.0]), and the plugs were deproteinated by agitating for 4 h at 55°C. The plugs were then washed twice with 2.0 ml of phenylmethylsulfonyl fluoride (PMSF) solution (TSE buffer with freshly added 1.5 mM PMSF) with gentle agitation for 30 min each time to facilitate inactivation and removal of the proteinase K. The plugs were then washed three times with 10 mM Tris-Cl–50 mM EDTA (pH 8.0) for 30 min each and stored at 4°C until used. A slice of the plugs (1 to 2 mm) was used for DNA digestion with restriction enzymes.
The slices, prepared aseptically, were transferred to a microcentrifuge tube and rinsed for 10 min with sterile distilled water, followed by 10 min in the appropriate restriction buffer. The slices were then digested with 200 μl of fresh restriction buffer containing 20 U of the restriction enzyme for 6 to 8 h. SfiI-digested DNA was resolved by PFGE with a Bio-Rad Chef Mapper system (Bio-Rad Laboratories, Richmond, Calif.) employing the following parameters: 6 V/cm, 4 to 40 s linearly ramped switch times, at 10°C for 23 h. SmaI-digested DNA was resolved using the following parameters: 6 V/cm, 2 to 36 s linearly ramped switch times, at 10°C for 23 h. The DNA patterns were visualized and recorded under UV illumination after staining for 30 min in ethidium bromide (1 μg/ml). Patterns were compared visually; the match between isolates appearing to exhibit similar patterns was confirmed by repeated analysis of the same gel.
Strains were considered identical when their PFGE patterns contained the same number and sizes of fragments. Patterns varying by one band were considered closely related (5).
PCR amplification and sequence analysis of the emm gene, encoding the M1 protein.
The emm gene encoding M1 protein was amplified by PCR using genomic DNA of GAS isolates and the primers 5′AllM (5′-ATAAGGAGCATAAAAATGGCT-3′) and EmmR (5′-GCAAGTTCTTCAGCTTGTTT-3′), specific to the conserved region of emm genes, as previously described (31). The PCR products were purified using the QUIAquick PCR purification kit from Quiagen (Valencia, Calif.) and used as the DNA matrix for sequencing of the emm gene. Sequencing of the variable N-terminal domain was performed with a fluorescent-labeled primer (5′Cy5.5-TATTCGCTTAGAAAATTAAAAACAGG-3′) and the automated DNA sequencing system OpenGene from Visible Genetics (Toronto, Canada). The DNA analysis program DNAStar was used to compare the sequences of the isolates with the GenBank database of nucleotide sequences encoding N-terminal variable regions of other M proteins.
Detection of speA, speB, speC, speF, speG, speH, smeZ, and ssa genes by PCR.
GAS strains were tested for the presence of speA, speB, speC, speF, speG, speH, smeZ, and ssa genes by PCR with primer pairs specific to each gene as previously described (19, 27, 32, 38). The primers used for spe genotyping are described in Table 2.
TABLE 2.
Primers used for genotyping of genes encoding different Spes
| Gene | Primer direction | Primer sequencea | Size of PCR product (bp) |
|---|---|---|---|
| speA | Forward | 5′-ACT TAA GAA CCA AGA GAT GG-3′ | 353 |
| Reverse | 5′-CCT TAT TCT TAG GTA TGA AC-3′ | ||
| speB | Forward | 5′-GGA TCC CAA CCA GTT GTT AAA TCT CT-3′ | 762 |
| Reverse | 5′-AAC GTT CTA AGG TTT GAT GCC TAC AA-3′ | ||
| speC | Forward | 5′-AAG TGA CTC TAA GAA AGA CA-3′ | 130 |
| Reverse | 5′-TTG AGT ATC AAT GTT TAA TG-3′ | ||
| speF | Forward | 5′-CGA AAT TAG AAA AGA GGA C-3′ | 1,193 |
| Reverse | 5′-GGC TGA GCA AAA GTG TGT G-3′ | ||
| speG | Forward | 5′-CTG GAT CCG ATG AAA ATT TAA AAG ATT TAA-3′ | 652 |
| Reverse | 5′-AAG AAT TCG GGG GGA GAA TAG-3′ | ||
| speH | Forward | 5′-TTG GAT CCA ATT CTT ATA ATA CAA CC-3′ | 630 |
| Reverse | 5′-AAA AGC TTT TAG CTG ATT GAC AC-3′ | ||
| smeZ/2 | Forward | 5′-TGG GAT CCT TAG AAG TAG ATA ATA-3′ | 645 |
| Reverse | 5′-AAG AAT TCT TAG GAG TCA ATT TC-3′ | ||
| ssa | Forward | 5′-AGT CAG CCT GAC CCT AC-3′ | 691 |
| Reverse | 5′-TAA GGT GAA CCT CTA T-3′ |
DNA sequence analysis of the sic gene encoding a streptococcal inhibitor of complement.
The sic gene was amplified with primers sic-forward (5′-TAAGGAGAGGTCACAAACTA-3′) and sic-reverse (5′-TTACGTTGCTGATGGTGTAT-3′) as previously described (1). The amplified products were purified using the QIAquick PCR purification kit and sequenced with the same primers on an ABI model 373A Stretch DNA sequencer (Perkin-Elmer, Foster City, Calif.). Sequence data analysis was performed with the EDITSEQ, ALIGN, and MEGALIGN programs (DNAstar, Madison, Wis.). The sequences were compared to the sic gene sequence deposited in the GenBank database under accession no. X92968 (1).
Sequencing of speA gene.
Manual sequencing of the speA PCR product was performed with a [32P]ATP-labeled primer (5′-GACATGGATAACAATTTCACACAGG-3′) that extends towards the area differentiating the four alleles of the speA gene as previously described (23).
Preparation of bacterial culture supernatants.
Bacterial isolates were cultured overnight in 12.5 ml of THB supplemented with 1.5% yeast extract (Difco, Detroit, Mich.). The bacteria were then removed by centrifugation, and the culture supernatants were recovered. For detection of SpeA and SpeB, the culture supernatants were concentrated on Amicon ultraconcentrators with a membrane cutoff of 10 kDa (Amicon, Beverly, Mass.). For detection of SpeA and SpeF and for proliferation assay experiments, proteins present in the supernatants were precipitated by addition of ice-cold absolute ethanol (1 part supernatant to 3 parts ethanol). The precipitate was dissolved in 1 ml of distilled H2O and dialyzed extensively against multiple changes of distilled H2O. The dialysates were filter sterilized and stored at −20°C.
Quantification of SpeA, SpeB, and SpeF production by SDS-PAGE and immunoblotting.
Protein samples from culture supernatants of GAS isolates were separated by sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto nitrocellulose membranes (Hybond-C Super; Amersham, Arlington Heights, Ill.). SpeA, SpeB, and SpeF toxins were detected by probing nitrocellulose blots with rabbit polyclonal antibodies (Abs) to SpeA (1:2,000) or to SpeF (1:2,000) and a mouse monoclonal Ab to SpeB (1:25,000). The latter was a generous gift from J. Musser, Rocky Mountain Laboratories, Hamilton, Mont. All Abs were specific to their respective Spe, and none showed any cross-reactivity with other Spes. Proteins were visualized by incubating the blots with a horseradish peroxidase-linked goat anti-mouse immunoglobulin G (IgG) or goat anti-rabbit IgG as the second Ab, followed by treatment with the chemiluminescent Western system (Amersham Life Science Inc.).
Recombinant SpeA (rSpeA) and rSpeF were expressed and purified as histidine-tagged fusion proteins as detailed elsewhere (3). The clone used for SpeA production was kindly provided by C. Collins, University of Miami, Fla. Pure rSpeA and rSpeF were used for the generation of polyclonal Abs as previously described (3). Purified SpeB was kindly provided by J. Musser. Serial dilutions of known concentrations of purified rSpeA, SpeB, and rSpeF were included in each gel to generate a standard curve for quantification purposes. In addition, one selected GAS culture supernatant was included in each gel and used as an internal control to correct for intergel variation. The blots were exposed to X-OMAT films (Eastman Kodak, Rochester, N.Y.), and the autoradiograms were scanned on a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Densitometry was performed using ImageQuant software (Molecular Dynamics), and bands corresponding to SpeA, SpeB, and SpeF were converted from pixel values to concentrations (in nanograms or micrograms per milliliter) based on the pixel values obtained with the Spe standards.
Proliferation assays.
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy individuals by Ficoll-Hypaque gradient centrifugation. The PBMCs from three different individuals were used to assess differences in mitogenic and cytokine responses of culture supernatants of isolates. Pilot experiments were performed to determine the optimal concentrations of culture supernatant to be used in mitogenic and cytokine assays. PBMCs (106 cells/ml) were stimulated with different dilutions of sterile culture supernatants (1:20, 1:50, 1:100, and 1:500 [final dilution] per well) or with phytohemagglutinin A (PHA) at 1 μg/ml (final concentration). After 72 h of culture, the cells were pulsed for 6 h with 1 μCi of [3H]thymidine (specific activity, 5.0 Ci/mmol; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), harvested onto glass fiber filters, and counted in a beta-scintillation counter. All samples were assayed in triplicate, and the experiments were repeated three or four times using PBMCs from three different individuals. The results of these dose-response experiments revealed that a 1:100 dilution was optimal for all supernatants, and this concentration was thus used for further mitogenic and cytokine analyses. All proliferation data were normalized to the PHA response and are presented as percent relative response, calculated by the following equation: [(cpmsupernatant − cpmunstimulated)/(cpmPHA − cpmunstimulated)] × 100.
Analyses of cytokine production at a single-cell level.
PBMCs (106 cells/ml) from healthy individuals (same as those tested for mitogenic responses) were stimulated with the predetermined optimal concentration of sterile GAS supernatants (1:100 dilution) and harvested after 24 and 72 h of culture. Harvested cells were washed three times with balanced salt solution and allowed to adhere to adhesion glass slides (Erie Scientific, Portsmouth, N.H.) for 10 min at room temperature. The cells were fixed with 2% formaldehyde (Fisher Scientific Co., Fairlawn, N.J.) in phosphate-buffered saline for 20 min and stained for specific cytokines as previously described (25). Cells harvested after 24 h were used for studies of IL-2 production, whereas the 72-h cultures were used to assess IFN-γ production. For detection of IL-2-producing cells, a mixture of 2 μg each of A6493-1, A7050-1, and A7050-2 (Pharmingen, San Diego, Calif.) per ml was used, and for IFN-γ a mixture of 2 μg/ml each of mab1-DIK and mab7-B6-1 (MabTek, Stockholm, Sweden) was used. Biotinylated goat anti-mouse IgG1 (Caltag Laboratories, Burlingame, Calif.) diluted 1:300 and biotinylated goat anti-rat IgG Ab (Vector Laboratories, Burlingame, Calif.) diluted 1:500 were used as secondary Abs. Immunohistochemical staining was achieved by Vectastain-Elite (Vector Laboratories) in combination with the substrate 3,3-diaminobenzidine (Vector Laboratories). Identification of cytokine-producing cells was facilitated by a local dense juxtanuclear staining pattern, which identifies cytokine producer cells (2). The cytokine-inducing capacity of each supernatant was evaluated using cells from three different individuals, each test performed in duplicate. The data are presented as mean percent cytokine-producing cells ± standard deviation (SD).
Statistical analyses of data.
Evaluation of statistical differences was performed with the Student t test.
RESULTS
Selection of a cohort of patients infected by apparently identical M1 strains.
A population-based active surveillance for all invasive GAS infections has been ongoing since 1992 in Ontario, Canada. Since the beginning of the study period, serotype M1 has been the most prevalent serotype isolated from invasive severe and nonsevere cases (16). To determine if the M1 strains responsible for severe invasive infections were different from those isolated from nonsevere cases, we analyzed 35 isolates (21 from severe and 14 from nonsevere cases) for genotypic, phenotypic, and immunologic characterization. The demographics of the patients from whom the isolates were obtained are described in Table 1. There was no significant difference between severe and nonsevere cases with respect to age or gender. Likewise, underlying diseases were present in some cases, but were similarly distributed among the severe and nonsevere groups (data not shown).
Isolates from severe and nonsevere M1T1 invasive cases have identical emm genes and Spe genotypes.
Since several alleles have been described for the emm1 gene encoding the M1 protein (21, 23), it was important to determine the emm1 allotype of the 35 M1 strains studied here. The sequence of the 5′ end of the emm gene of all isolates was identical and revealed that all isolates possessed the emm1.0 allele.
The 35 GAS strains were further characterized for Spe genotype by sequence-specific PCR. All isolates had the same spe genotype, speA+ speB+ speC speF+ speG+ speH smeZ+ ssa. Furthermore, the speA gene was sequenced in all 35 isolates, and the data revealed that all harbored the speA2 allele.
RAPD fingerprint patterns of M1 isolates from severe and nonsevere cases.
To identify primers that generate informative and discriminatory arrays of PCR products for GAS isolates, purified DNAs from four M1T1 strains, three M3 strains, one M12 strain, and one M28 strain were tested with seven primers. Strains of different serotypes were characterized by distinct RAPD patterns. The four M1T1 strains tested in the pilot experiments all exhibited the same pattern independent of the primer used. However, the most discriminant patterns, with a limited number of low-intensity bands, were obtained using primers P1 (5′-GGTGCGGGAA-3′) and P5 (5′-AACGCGCAAC-3′); therefore, these primers were selected to test all M1T1 isolates as well as the M3, M12, and M28 strains.
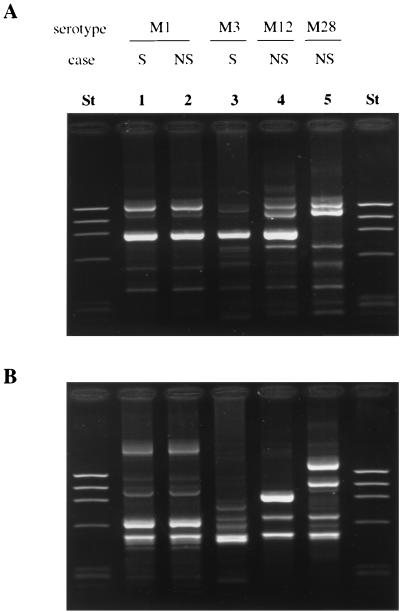
A distinct RAPD pattern generated by primers P1 and P5 was seen for the different M serotypes, including M1, M3, M28, and M12 (Fig. 1). However, all 35 M1T1 isolates included in this study showed an identical pattern, 1A or 5A, following DNA amplification with primer P1 or P5, respectively.
FIG. 1.
RAPD analysis of GAS isolates. RAPD was performed with primers P1 and P5. (A) Patterns obtained with primer P1; (B) patterns obtained with primer P5. Lanes St, lambda ladder used as molecular size markers; lanes 1 and 2, isolates of serotype M1T1; lane 3, M3 isolate; lane 4, M12 isolate; lane 5, M28 isolate. S, strain from severe invasive case; NS, strain from nonsevere invasive case.
PFGE banding patterns of M1 isolates from severe and nonsevere cases.
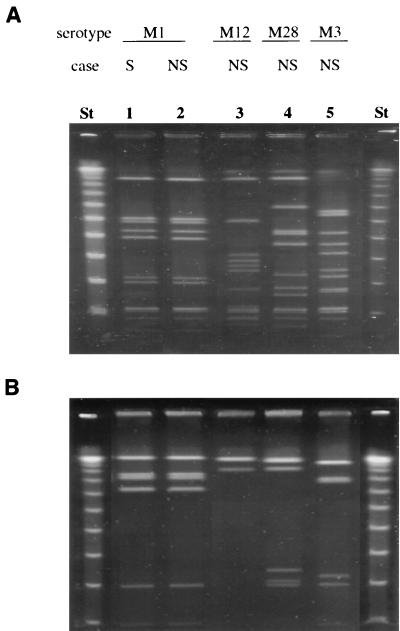
The endonucleases SmaI and SfiI were chosen for this analysis because the restriction sites for these enzymes are GC rich and they have been shown to generate discriminatory PFGE patterns for GAS, a low-GC-content bacterial species. As shown in Fig. 2, both enzymes generated distinct banding patterns for the different M serotypes. However, 30 of the 35 M1 isolates showed an identical pattern (S1) after SmaI digestion, and all but two of the M1 isolates digested with SfiI showed identical banding patterns (Sf1). The five divergent strains were assigned to two SmaI patterns (S2 and S3). One of the isolates from the severe cases showed pattern S2, which differed from the main pattern S1 by the absence of only one band. Four of 14 nonsevere isolates showed pattern S3, which differed from the main pattern S1 by one band. Taken together, the data indicate that the 35 M1T1 Canadian isolates show a high degree of genetic relatedness.
FIG. 2.
Representative PFGE patterns of different GAS isolates. Chromosomal DNA was digested with the restriction endonuclease SmaI (A) or SfiI (B). Lanes St, lambda ladder used as molecular size markers. Lanes 1 and 2, isolates of serotype M1T1; lane 3, M12 isolate; lane 4, M28 isolate; lane 5, M3 isolate. S, strain from severe invasive case; NS, strain isolated from nonsevere invasive case.
Similar sequence variations in sic gene and Sic protein among M1T1 isolates from severe and nonsevere cases.
Inasmuch as previous studies have indicated that M1 strains that could not be differentiated on the basis of M serotyping, Spe genotyping, MLEE, and PFGE can still display a high level of polymorphism for the streptococcal complement-inhibiting protein Sic (29, 36), we investigated whether variations in the sic gene or the Sic protein correlated with disease severity. The sic gene was completely sequenced in 20 selected isolates (10 from severe cases and 10 from nonsevere cases). Ten variants of the sic gene were identified, and none of them was identical to the sic gene sequence deposited in GenBank under accession number X92968, but the variations were found in positions similar to those previously reported to be highly polymorphic (36). Four strains from severe cases and six strains from nonsevere cases had the identical sic gene sequence, and the remaining 10 strains had different sequences. Variations were mainly introduced by either deletions or insertions of nucleotides at different locations throughout the entire gene sequence (Fig. 3). The sequences obtained in the present study were compared to the sic gene sequence X92968, and the major differences are summarized in Fig. 3 and Table 3. All DNA polymorphisms detected in the sic genes sequenced resulted in amino acid substitutions, and as shown in Table 3, these substitutions were similar in isolates from both groups. Therefore, sequence variations or specific patterns of polymorphism in the sic gene or Sic protein were not correlated with the severity of invasive GAS infections.
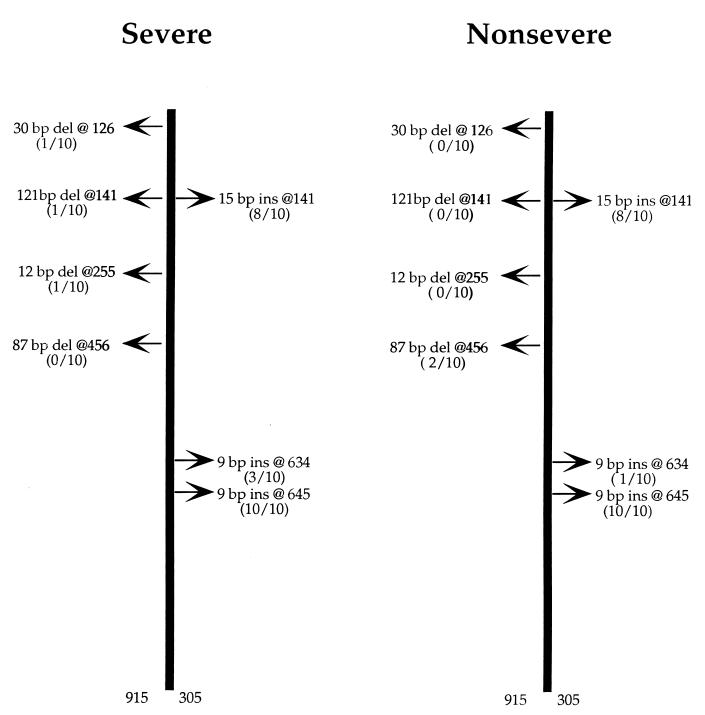
FIG. 3.
Nucleotide insertions and deletions identified in the sic gene of selected M1T1 isolates. The sic gene was completely sequenced in 20 selected isolates (10 from severe cases and 10 from nonsevere cases). Deletions (del) and insertions (ins) are with reference to the sequence of the sic allele deposited in the GenBank database (accession no. X92968). Among the severe isolates, one had a 30-bp deletion at 126, and another isolate had two deletions at 141 (121 bp) and 255 (12 bp). Among the nonsevere isolates, two isolates showed an 87-bp deletion at 456. All isolates showed an insertion of 9 bp at 645, and with the exception of two isolates from each group, all isolates showed an insertion of 15 bp at 141. A third insertion of 9 bp at 634 was seen in two isolates from the severe group and one from the nonsevere group.
TABLE 3.
Amino acid substitutions in the Sic protein of selected M1T1 isolatesa
| Isolate no. | Amino acid substitution
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| K74Q | T160I | G169E | R209W | G217E | S220F | F228S | T231A | T241P | |
| Severe cases | |||||||||
| 5447 | + | + | + | − | + | + | − | + | + |
| 5449 | + | + | + | − | + | + | − | + | + |
| 5442 | + | + | + | − | + | + | − | + | + |
| 5443 | + | + | + | − | + | + | − | + | + |
| 5444 | + | + | + | − | − | + | − | + | + |
| 5448 | + | + | + | − | + | + | − | + | + |
| 5628 | − | + | + | − | + | + | − | + | + |
| 6021 | + | + | + | − | − | + | − | + | + |
| 6043 | + | + | + | − | + | + | − | + | + |
| 8004 | + | + | + | − | + | + | − | + | + |
| Nonsevere cases | |||||||||
| 8143 | + | − | − | − | + | + | − | + | + |
| 5438 | + | + | + | − | + | + | − | + | + |
| 5441 | + | + | + | − | + | + | − | + | + |
| 5622 | + | + | + | − | + | + | − | + | + |
| 5632 | + | + | + | − | + | + | − | + | + |
| 5649 | + | + | + | − | + | + | − | + | + |
| 5650 | + | − | − | − | + | + | + | + | + |
| 5873 | + | + | + | − | + | + | − | + | + |
| 6054 | + | + | + | + | − | + | − | + | + |
| 6072 | + | + | + | − | + | + | − | + | + |
Presence (+) or absence (−) of amino acid substitutions is with reference to the sequence deposited in the GenBank database under accession no. X92968.
Relation between expression of SpeA, SpeB, and SpeF and severity of invasive infections.
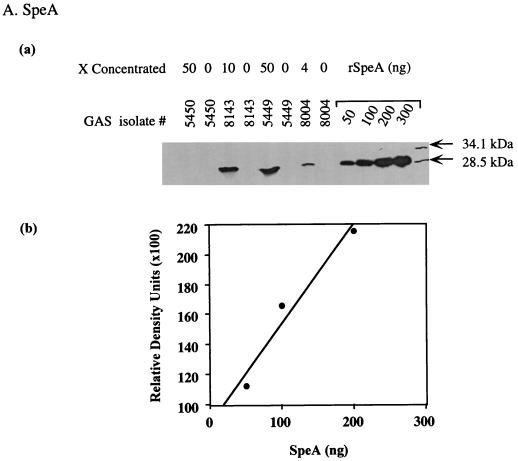
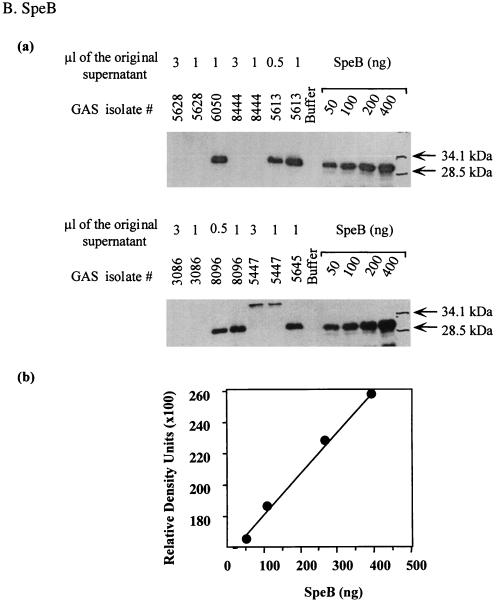
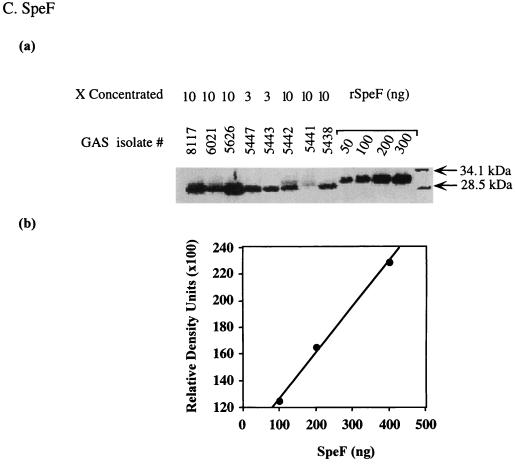
To determine whether isolates from severe cases produce higher levels of streptococcal SAgs than isolates from nonsevere cases, we compared the production of SpeA, SpeF, and the zymogen and active forms of SpeB produced by the M1T1 GAS isolates. The amount of Spe produced in the culture supernatants of isolates was quantified by Western blots, with known amounts of standard purified rSpeA, SpeB, and rSpeF included in each gel to generate a standard curve (Fig. 4).
FIG. 4.
Method for measurement of Spe production in the culture supernatant of GAS isolates. Bacterial culture supernatants were prepared as described in Materials and Methods and subjected to Western blot analysis. To allow quantification of Spe production, each gel contained serial dilutions of standard amounts of rSpeA (50 to 300 ng) or purified SpeB (50 to 400 ng) or rSpeF (50 to 300 ng) along with the unknown test samples. The blots were sequentially probed with specific Ab to each Spe tested and subjected to autoradiography, and the bands corresponding to the specific Spe being analyzed were visualized. The amount of Spe produced in each supernatant was quantified from a standard curve generated by scanning values of known concentrations of standards that were included in the same gel.
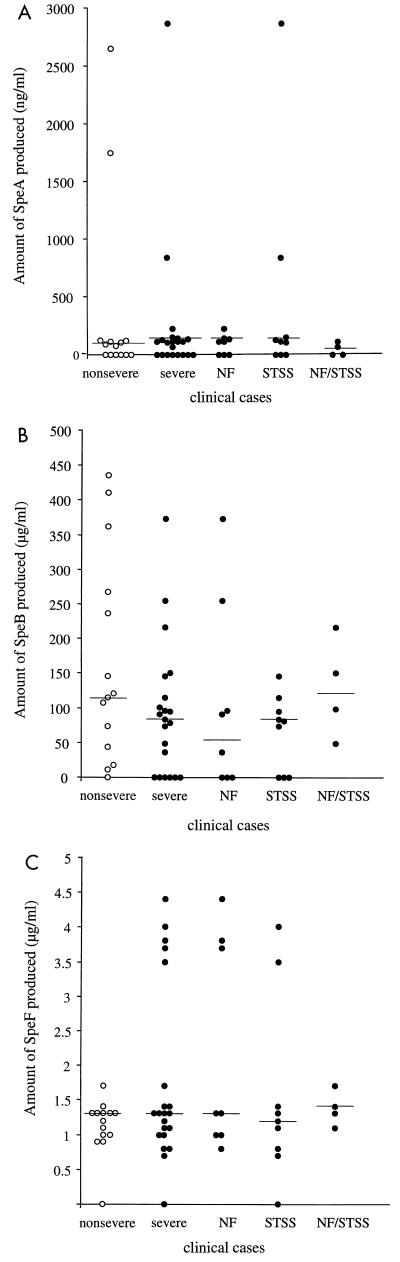
The amount of SpeA produced varied considerably among isolates (Fig. 5A and Table 4). Expression of SpeA was either very low (<20 ng/ml) or undetectable in 38% (8 of 21) of severe and 43% (6 of 14) of nonsevere invasive isolates (not significant). Moderate SpeA production (70 to 225 ng/ml) was found in 53% (11 of 21) of severe and 43% (6 of 14) of nonsevere isolates (not significant). Two isolates from each group produced very high levels of SpeA (≥800 ng/ml). The two isolates from severe cases came from patients with STSS, and the two isolates from nonsevere cases came from a patient with septicemia/pneumonia and a patient with erysipelas.
FIG. 5.
Comparison of amounts of SpeA, SpeB, and SpeF in culture supernatant of GAS isolates from severe and nonsevere invasive cases. SpeA, SpeB, and SpeF toxins were detected by Western blot and quantified after scanning the autoradiograms as detailed in the legend to Fig. 3 and in Materials and Methods. The amount of each Spe was calculated from a standard curve generated by scanning values of known amounts of standards included in each run, as detailed in the legend to Fig. 3. Horizontal lines denote median values of Spe production from severe and nonsevere cases.
TABLE 4.
Amount of Spes detected in culture supernatant of M1T1 isolates from severe and nonsevere invasive casesa
| Group and toxin | % of isolates producing the toxin | Amount of Spe produced (ng or μg/ml)
|
|||
|---|---|---|---|---|---|
| All isolates
|
Positive isolates only (mean ± SD) | ||||
| Range | Median | Mean ± SD | |||
| Severe invasive cases | |||||
| SpeA (ng/ml) | 57 | 0–2,863 | 109 | 242.3 ± 626.6 | 391.4 ± 768.8 |
| SpeB (μg/ml) | 71 | 0–373 | 83 | 93.1 ± 96 | 130.4 ± 89.2 |
| SpeF (μg/ml) | 95 | 0–4.4 | 1.3 | 1.8 ± 1.3 | 1.8 ± 1.2 |
| Nonsevere invasive cases | |||||
| SpeA (ng/ml) | 64 | 0–2,646 | 84.5 | 357.5 ± 799.7 | 625.6 ± 998.2 |
| SpeB (μg/ml) | 93 | 0–435 | 118 | 167.8 ± 149.3 | 180.7 ± 147.1 |
| SpeF (μg/ml) | 93 | 0–1.7 | 1.3 | 1.1 ± 0.4 | 1.2 ± 0.2 |
Differences in Spe production between isolates from severe and nonsevere were evaluated by a Student t test and were not statistically significant (P value equal to 0.6, 0.08, and 0.08 for SpeA, SpeB, and SpeF, respectively).
The amount of 28-kDa SpeB produced by isolates from severe cases varied between 0 and 300 μg/ml, with a median value of 170 μg/ml (Fig. 5B and Table 4). Only two isolates from the severe group produced the 40-kDa zymogen, and five isolates failed to produce detectable amounts of either zymogen or active SpeB. All but one isolate from nonsevere cases (isolate 8143) produced active 28-kDa SpeB; this isolate produced only the 40-kDa zymogen form of SpeB. Isolates from nonsevere cases produced the 28-kDa SpeB (0 to 354 μg/ml, with a median value of 218 μg/ml). Although there was a trend towards higher production of SpeB by isolates from nonsevere invasive cases compared to isolates from severe cases, the difference did not reach statistical significance (P = 0.08) possibly due to sample size.
With the exception of one isolate from the severe and nonsevere groups, all M1T1 isolates expressed SpeF (Fig. 5C and Table 4). The amount of SpeF produced ranged from 0.7 to 4.4 μg/ml and 0.9 to 1.4 μg/ml for isolates from severe and nonsevere cases, respectively, and both groups had the same median value of 1.3 μg/ml. There was a trend towards higher SpeF production by isolates from severe cases compared to those from nonsevere cases, but again statistical significance was not reached (P = 0.08).
Analyses of mitogenic and cytokine-inducing activity in culture supernatants of M1T1 GAS isolates.
Our studies have shown no direct correlation between the amount of a particular known Spe produced and the mitogenic or cytokine-inducing capacity of isolate supernatants (data not shown). Indeed, recent studies revealed that in addition to the well-known Spes, M1 strains may also produce a large number of novel SAgs, some not fully characterized (32). To assess the activity of mixtures of SAgs produced by M1T1 isolates, we evaluated the mitogenic and cytokine-inducing capacity of the culture supernatants containing the mixture of SAgs. To focus on whether isolates from severe and nonsevere cases differ in their mitogenic capacity, we selected 20 isolates (10 from each group) that had an identical PFGE pattern, Spe genotype, and comparable Spe expression profile (Table 4). These isolates were tested for mitogenic and cytokine-inducing activity using PBMCs from healthy individuals as responders. Since it has been reported that there is a high degree of interindividual variation in responses to SAgs (26, 27), the analyses were performed using cells from three different individuals to confirm the results.
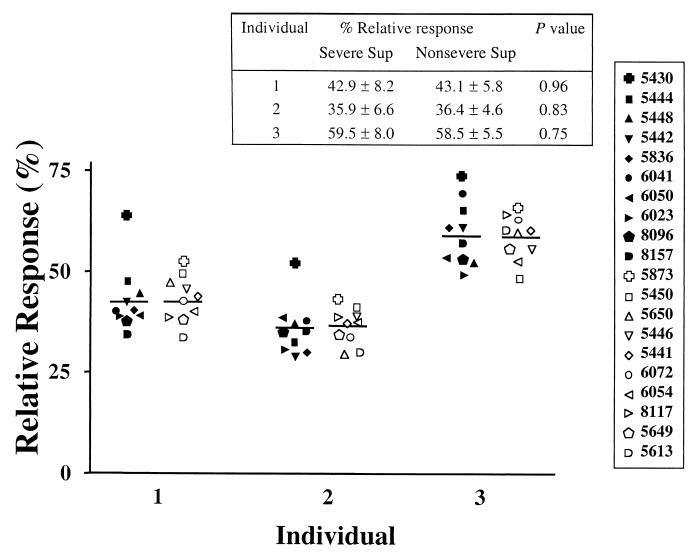
Proliferation experiments revealed that all 20 GAS culture supernatants tested induced strong proliferative responses in PBMCs from the three individuals tested. Importantly, there was no significant difference in mitogenic activity between culture supernatants prepared from severe and nonsevere invasive isolates (Fig. 6). For example, in the mitogenic assay, isolate 5430 (severe, SpeA protein negative) and isolate 5873 (nonsevere, SpeA protein positive) elicited the strongest response in PBMCs from all three individuals tested. PBMCs from individual 3 mounted a higher response than those from individuals 1 and 2 to all isolates tested; however, the response of PBMCs from individual 3 to isolates from severe and nonsevere cases was very similar. Therefore, despite the expected interindividual variation in response to the culture supernatants, each individual responded similarly to supernatants of isolates from severe and nonsevere cases (Fig. 6).
FIG. 6.
Mitogenic activity of culture supernatants of M1T1 isolates. PBMCs from three healthy individuals were stimulated with sterile culture supernatants (1:100 dilution) prepared from 20 different M1T1 GAS strains (10 each from severe and nonsevere cases). Different symbols are used for each isolate; solid symbols are for severe and open symbols are for nonsevere isolates. Proliferative responses were assessed after 72 h of culture, after a 6-h [3H]thymidine pulse. The data are presented as percent relative response as defined in the text. Horizontal lines denote mean values for culture supernatants from severe and nonsevere cases.
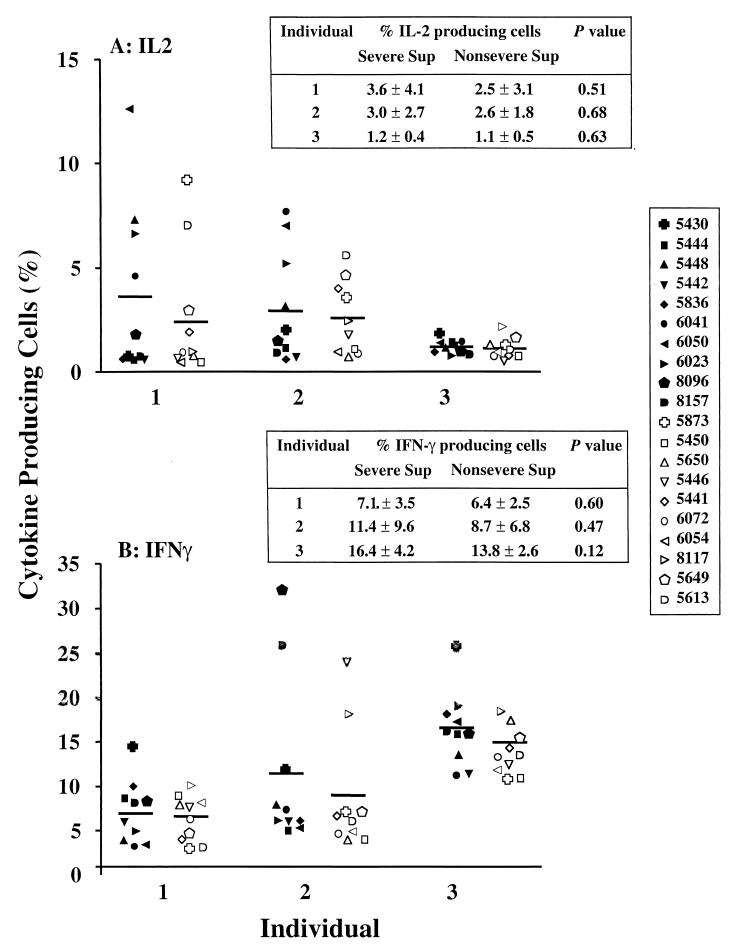
Differences in cytokine-inducing capacity among the M1T1 isolates were also investigated. The capacity of the GAS supernatants to induce IL-2 and IFN-γ production in PBMCs from the three responders was assessed by counting the percentage of cytokine-producing cells (Fig. 7). Consistent with the proliferation data, the supernatants were found to induce high levels of IL-2 and IFN-γ production. Despite the high degree of interindividual variation in the response, PBMCs from each individual responded similarly to culture supernatants of isolates from severe and nonsevere cases (Fig. 7). For example, the percentage of mean IFN-γ-producing cells from individual 1 was 7.1 ± 3.5% and 6.4 ± 2.5% in response to isolates from severe and nonsevere cases, respectively (inset table in Fig. 7).
FIG. 7.
Cytokine-inducing activity of culture supernatants of M1T1 isolates from severe versus nonsevere cases. PBMCs from three healthy individuals were stimulated with culture supernatants (1:100) prepared from 20 different M1T1 GAS strains (10 each from severe and nonsevere cases). Different symbols are used for each isolate; solid symbols are for severe and open symbols are for nonsevere isolates. Cells were harvested after 24 h for analyses of IL-2 production (A) and after 72 h for IFN-γ production (B). Horizontal lines denote mean values of culture supernatants from severe and nonsevere cases.
DISCUSSION
In this study, we have demonstrated that M1T1 GAS isolates that are highly related can cause disease of varying severity. It is clear that the isolates studied here are derived from the same parental strain, as they have identical emm1.0 gene sequences and the same spe genotype. In addition, the isolates had the same RAPD profile, and most had an identical PFGE banding pattern after digestion with two enzymes.
A high degree of sic gene variation was found among the M1 isolates studied here. The observed variation in the sic gene is to be expected, as this gene shows a high degree of polymorphism (29, 36). Despite this variation, there was no correlation between certain mutations, deletions, or insertions and the severity of invasive disease.
Significant variations in Spe expression were also found among the M1T1 isolates, but no particular pattern was significantly associated with severe disease. The variation in SpeA production was comparable among isolates from severe and nonsevere cases, and while SpeB production was higher for isolates from nonsevere cases and SpeF production was lower for the same group of isolates compared to isolates from severe cases, the differences were not significant (P = 0.08). Additional studies with a larger sample size may reveal whether expression of SpeB by isolates from severe and nonsevere cases are significantly different. The variation in Spe expression among highly related isolates may be immunologically important, and it is tempting to propose that host factors may be involved in the differential regulation of spe gene expression. This hypothesis will be tested directly in future studies.
Inasmuch as novel SAgs continue to be discovered (32), Western blot detection of known SAgs does not account for all SAgs present in the culture supernatant of GAS isolates. In fact, there was often no correlation between the amount of a particular known Spe produced and the mitogenic or cytokine-inducing capacity of the isolates studied here. For example, several isolates elicited very comparable responses despite having very different known SpeA production profiles. This underscored the importance of novel SAgs in GAS pathogenesis. Therefore, it was important to compare the mitogenic and cytokine-inducing responses of the partially purified M1T1 isolate supernatants. In agreement with our previous studies, PBMCs from different individuals responded differently to the GAS supernatants in both proliferation and cytokine production assays. However, despite this interindividual variation, PBMCs from each individual responded similarly to isolates from severe and nonsevere cases. In other words, the mitogenic, IL-2, and IFN-γ responses were not significantly different between isolates from severe and nonsevere invasive cases.
Together, the data presented here show that despite variations in the sic gene and Spe expression among highly related isolates, there is no indication that any particular pattern correlates with the severity of invasive disease. The data strongly support the hypothesis that, faced with the same strain of GAS, different individuals will respond differently to the streptococcal virulence factors, and this will result in starkly variable outcomes of the infection.
Several host factors can contribute to the immunologic response to streptococcal virulence factors. Differences in the levels of protective anti-M1 protein and neutralizing anti-Spe antibodies have been considered as possible factors in determining disease severity (8, 12, 28, 35). However, a recent study which investigated levels of protective antibodies to the infecting isolates showed that low levels of protective Abs increase the risk for invasive disease severity but are not a factor in determining the severity of invasive infection (3).
Age and underlying diseases are also important factors that can influence infection outcome. However, the severe and nonsevere cases included in this study were matched for age, gender, and underlying illness. We believe that allelic variation in certain immunogenetic factors involved in regulating the inflammatory response to streptococcal virulence factors may play a major role in determining disease outcome. In particular, variation in HLA class II and differences in T-cell receptor Vβ frequency among individuals may contribute to the risk of or protection from severe disease, as these cell receptors can directly influence the SAg response. Indeed, recent studies from our laboratory suggest that allelic and haplotype variations of major histocompatibility complex class II antigens, which serve as receptors for SAgs, may contribute to risk of or protection from severe disease (Y. Guedez, A. Norrby-Teglund, D. E. Low, A. McGeer, and M. Kotb, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. G050, 1997). Ongoing studies in our laboratory suggest that the molecular basis for this association is related to a significant difference in the magnitude of inflammatory cytokine response elicited to the same SAg when presented by different HLA class II alleles. Therefore, the data indicate that highly related strains of GAS can cause disease with varying severity in different individuals. This underscores the role of host factors in potentiating the outcome of GAS severe invasive disease.
ACKNOWLEDGMENTS
This work was supported by an award from the American Heart Association, southeast affiliate (to S.C.), grant AI40198 from the National Institutes of Health, NIAID (to M.K.), the Office of Research and Development, Medical Research Service Department of Veterans Affairs' Merit Award (to M.K.), a grant from VA/DOD joint research funds (Emerging Pathogen Research Initiative, to M.K.), and funds from the Swedish Medical Research Council and Swedish Society for Medicine (to A.N.T.).
REFERENCES
- 1.Akesson P, Sjoholm A G, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Andersson J, Nagy S, Bjork L, Abrams J, Holm S E, Andersson U. Bacterial toxin-induced cytokine production studied at a single cell level. Immunol Rev. 1992;127:69–96. doi: 10.1111/j.1600-065x.1992.tb01409.x. [DOI] [PubMed] [Google Scholar]
- 3.Basma H, Norrby-Teglund A, Guedez Y, McGeer A, Low D E, El-Ahmedy O, Schwartz B, Kotb M. Risk factors in the pathogenesis of invasive group A streptococcal infections: role of protective humoral immunity. Infect Immun. 1999;67:1871–1877. doi: 10.1128/iai.67.4.1871-1877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beachey E H, Seyer J M. Primary structure and immunochemistry of group A streptococcal M proteins. Semin Infect Dis. 1982;4:401–410. [Google Scholar]
- 5.Chaussee M S, Liu J, Stevens D L, Ferretti J J. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J Infect Dis. 1996;173:901–908. doi: 10.1093/infdis/173.4.901. [DOI] [PubMed] [Google Scholar]
- 6.Cockerill F R, 3rd, MacDonald K L, Thompson R L, Roberson F, Kohner P C, Besser-Wiek J, Manahan J M, Musser J M, Schlievert P M, Talbot J, Frankfort B, Steckelberg J M, Wilson W R, Osterholm M T. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA. 1997;277:38–43. [PubMed] [Google Scholar]
- 7.Desai M, Tanna A, Efstratiou A, George R, Clewley J, Stanley J. Extensive genetic diversity among clinical isolates of Streptococcus pyogenes serotype M5. Microbiology. 1998;144:629–637. doi: 10.1099/00221287-144-3-629. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson B K, Andersson J, Holm S E, Norgren M. Invasive group A streptococcal infections: T1M1 isolates expressing pyrogenic exotoxins A and B in combination with selective lack of toxin-neutralizing antibodies are associated with increased risk of streptococcal toxic shock syndrome. J Infect Dis. 1999;180:410–418. doi: 10.1086/314872. [DOI] [PubMed] [Google Scholar]
- 9.Facklam R, Beall B, Efstratiou A, Fischetti V, Johnson D, Kaplan E, Kriz P, Lovgren M, Martin D, Schwartz B, Totolian A, Bessen D, Hollingshead S, Rubin F, Scott J, Tyrrell G. emm typing and validation of provisional M types for group A streptococci. Emerg Infect Dis. 1999;5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoge C W, Schwartz B, Talkington D F, Breiman R F, MacNeill E M, Englender S J. The changing epidemiology of invasive group A streptococcal infections and the emergence of streptococcal toxic shock-like syndrome. A retrospective population-based study. JAMA. 1993;269:384–389. [PubMed] [Google Scholar]
- 12.Holm S E, Norrby A, Bergholm A M, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- 13.Kaul R, McGeer A, Low D E, Green K, Schwartz B. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Ontario Group A Streptococcal Study. Am J Med. 1997;103:18–24. doi: 10.1016/s0002-9343(97)00160-5. [DOI] [PubMed] [Google Scholar]
- 14.Kiska D L, Thiede B, Caracciolo J, Jordan M, Johnson D, Kaplan E L, Gruninger R P, Lohr J A, Gilligan P H, Denny F W., Jr Invasive group A streptococcal infections in North Carolina: epidemiology, clinical features, and genetic and serotype analysis of causative organisms. J Infect Dis. 1997;176:992–1000. doi: 10.1086/516540. [DOI] [PubMed] [Google Scholar]
- 15.Kotb M. Bacterial pyrogenic exotoxins as superantigens. Clin Microbiol Rev. 1995;8:411–426. doi: 10.1128/cmr.8.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Low D E, Schwartz B, McGeer A, editors. The reemergence of severe group A streptococcal disease: an evolutionary perspective. Vol. 7. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 17.Martin P R, Høiby E A. Streptococcal serogroup A epidemic in Norway 1987–1988. Scand J Infect Dis. 1990;22:421–429. doi: 10.3109/00365549009027073. [DOI] [PubMed] [Google Scholar]
- 18.McCarthy S A, Khambaty F M. International dissemination of epidemic Vibrio cholerae by cargo ship ballast and other nonpotable waters. Appl Environ Microbiol. 1994;60:2597–2601. doi: 10.1128/aem.60.7.2597-2601.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mollick J, Miller G, Musser J, Cook R, Grossman D, Rich R. A novel superantigen isolated from pathogenic strains of Streptococcus pyogenes with aminoterminal homology to staphylococcal enterotoxins B and C. J Clin Investig. 1993;92:710–719. doi: 10.1172/JCI116641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muotiala A, Seppala H, Huovinen P, Vuopio-Varkila J. Molecular comparison of group A streptococci of T1M1 serotype from invasive and noninvasive infections in Finland. J Infect Dis. 1997;175:392–399. doi: 10.1093/infdis/175.2.392. [DOI] [PubMed] [Google Scholar]
- 21.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Genetic diversity and relationship among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima K, Ichiyama S, Iinuma Y, Hasegawa Y, Ohta M, Ooe K, Shimizu Y, Igarashi H, Murai T, Shimokata K. A clinical and bacteriologic investigation of invasive streptococcal infections in Japan on the basis of serotypes, toxin production, and genomic DNA fingerprints. Clin Infect Dis. 1997;25:260–266. doi: 10.1086/514543. [DOI] [PubMed] [Google Scholar]
- 23.Nelson K, Schlievert P M, Selander R K, Musser J M. Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J Exp Med. 1991;174:1271–1274. doi: 10.1084/jem.174.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norgren M, Norrby A, Holm S E. Genetic diversity in T1M1 group A streptococci in relation to clinical outcome of infection. J Infect Dis. 1992;166:1014–1020. doi: 10.1093/infdis/166.5.1014. [DOI] [PubMed] [Google Scholar]
- 25.Norrby-Teglund A, Kaul R, Low D E, McGeer A, Andersson J P, Andersson U G, Kotb M. Plasma from patients with severe invasive group A streptococcal infections treated with polyspecific IgG (IVIG) inhibits streptococcal superantigen-induced T cell proliferation and cytokine production. J Immunol. 1996;156:3057–3064. [PubMed] [Google Scholar]
- 26.Norrby-Teglund A, Lustig R, Kotb M. Differential induction of Th1 versus Th2 cytokines by group A streptococcal toxic shock syndrome isolates. Infect Immun. 1997;65:5209–5215. doi: 10.1128/iai.65.12.5209-5215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norrby-Teglund A, Newton D, Kotb M, Holms S, Norgren M. Superantigenic properties of the group A streptococcal exotoxin MF (SPeF) Infect Immun. 1994;62:5227–5233. doi: 10.1128/iai.62.12.5227-5233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norrby-Teglund A, Pauskens K, Holm S E, Norgren M. Relation between low capacity of human sera to inhibit streptococcal mitogens and serious manifestation of disease. J Infect Dis. 1994;170:585–591. doi: 10.1093/infdis/170.3.585. [DOI] [PubMed] [Google Scholar]
- 29.Perea Mejia L M, Stockbauer K E, Pan X, Cravioto A, Musser J M. Characterization of group A Streptococcus strains recovered from Mexican children with pharyngitis by automated DNA sequencing of virulence-related genes: unexpectedly large variation in the gene encoding a complement-inhibiting protein. J Clin Microbiol. 1997;35:3220–3224. doi: 10.1128/jcm.35.12.3220-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 31.Podbielski A, Melzer B, Lutticken R. Application of the polymerase chain reaction to study the M protein(-like) gene family in beta-hemolytic streptococci. Med Microbiol Immunol (Berl) 1991;180:213–227. doi: 10.1007/BF00215250. [DOI] [PubMed] [Google Scholar]
- 32.Proft T, Moffatt S L, Berkahn C J, Fraser J D. Identification and characterization of novel superantigens from Streptococcus pyogenes. J Exp Med. 1999;189:89–102. doi: 10.1084/jem.189.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seppala H, He Q, Osterblad M, Huovinen P. Typing of group A streptococci by random amplified polymorphic DNA analysis. J Clin Microbiol. 1994;32:1945–1948. doi: 10.1128/jcm.32.8.1945-1948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley J, Linton D, Desai M, Efstratiou A, George R. Molecular subtyping of prevalent M serotypes of Streptococcus pyogenes causing invasive disease. J Clin Microbiol. 1995;33:2850–2855. doi: 10.1128/jcm.33.11.2850-2855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens D L. Invasive group A streptococcus infections. Clin Infect Dis. 1992;14:2–13. doi: 10.1093/clinids/14.1.2. [DOI] [PubMed] [Google Scholar]
- 36.Stockbauer K E, Grigsby D, Pan X, Fu Y X, Mejia L M, Cravioto A, Musser J M. Hypervariability generated by natural selection in an extracellular complement-inhibiting protein of serotype M1 strains of group A Streptococcus. Proc Natl Acad Sci USA. 1998;95:3128–3133. doi: 10.1073/pnas.95.6.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stromberg A, Romanus V, Burman L G. Outbreak of group A streptococcal bacteremia in Sweden: an epidemiologic and clinical study. J Infect Dis. 1991;164:959–968. doi: 10.1093/infdis/164.3.595. [DOI] [PubMed] [Google Scholar]
- 38.Tyler S D, Johnson W M, Huang J C, Ashton F E, Wang G, Low D E, Rozee K R. Streptococcal erythrogenic toxin genes: detection by polymerase chain reaction and association with disease in strains isolated in Canada from 1940 to 1991. J Clin Microbiol. 1992;30:3127–3131. doi: 10.1128/jcm.30.12.3127-3131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Upton M, Carter P E, Orange G, Pennington T H. Genetic heterogeneity of M type 3 group A streptococci causing severe infections in Tayside, Scotland. J Clin Microbiol. 1996;34:196–198. doi: 10.1128/jcm.34.1.196-198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]