Summary
MicroRNAs fine-tune gene regulation and can be targeted for therapeutic purposes. We investigated the physiological roles of miR-146a and miR-200b that are differentially expressed in neurological disorders such as Alzheimer's disease and schizophrenia, particularly in learning and memory mechanisms. Using bioinformatics tools and luciferase assay, we show interaction of these miRNAs with transcripts of N-methyl-D-aspartate receptor (NMDAR) subunits Grin2A and Grin2B. Overexpression of these miRNAs in primary hippocampal neurons caused downregulation of GluN2B and GluN2A proteins. Stereotactic injections of these miRNAs into rat hippocampus caused cognitive deficits in multiple behavioral tests with decreased protein levels of GluN1, GluN2A, GluN2B, AMPAR subunit GluR1, and Neuregulin 1. In pharmacologically treated rat models [MK-801 treated and methylazoxymethanol acetate (MAM) treated], we found upregulated levels of these miRNAs, implying their involvement in downregulating NMDAR subunits in these models. These results suggest the importance of miR-146a-5p and miR-200b-3p in hippocampus-dependent learning and memory.
Subject areas: Biological sciences, Cognitive neuroscience, Cell biology
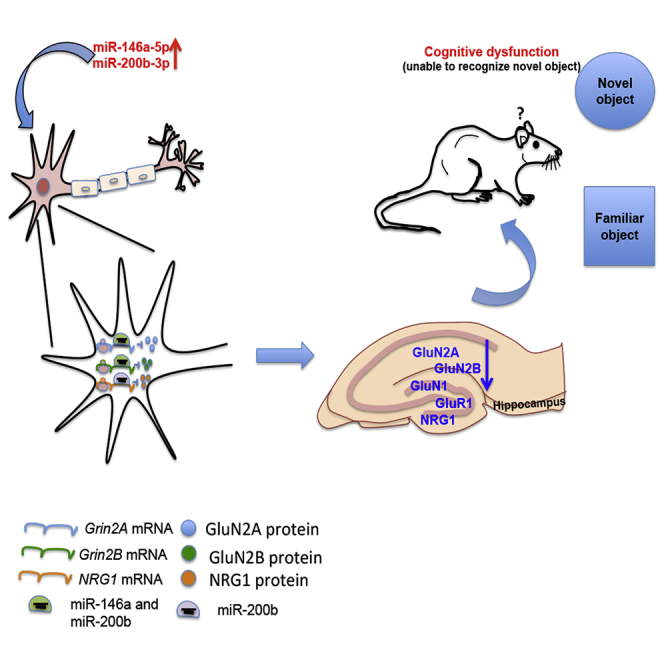
Graphical abstract
Highlights
-
•
miR-146a and miR-200b interact with 3′UTRs of Grin2A and Grin2B subunits of NMDAR
-
•
Overexpressed miRNAs downregulated GluN2A and GluN2B in hippocampal neurons
-
•
Injection of miR-146a or miR-200b in hippocampi led to cognitive deficits
-
•
miRNAs downregulated GluN2A and GluN2B proteins without affecting transcript levels
Biological sciences; Cognitive neuroscience; Cell biology
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that regulate nearly 60% of the mammalian genes by post-transcriptional regulation.1 They influence the stability and/or translation of the target mRNAs by binding to the 3′UTR sequence of the target mRNAs.2,3 Thus, miRNAs fine-tune the expression of various genes and orchestrate different functions of the nervous system.4 They play crucial roles in synaptic plasticity and memory in both vertebrates and invertebrates.5 Differential expression of many miRNAs has been associated with neuropsychiatric and neurodegenerative disorders.6,7 Among the many miRNAs present in the brain, the mechanism of action is known for only a few.8
Glutamatergic signaling through N-methyl-D-aspartate receptor (NMDAR) is essential for several brain functions. NMDARs are excitatory ligand-gated ion channels, which are multiprotein complexes with two obligate GluN1 subunits and two GluN2 (A-D)/one GluN2 and one GluN3 (A and B) subunits. They play a prominent role in synaptic plasticity and synaptic pruning.9 Their regulation and function are impaired in neuropsychiatric diseases such as schizophrenia and Alzheimer's disease.10
There have been reports on miRNAs associated with the NMDAR pathway that act either on NMDAR or on targets upstream or downstream.11,12 miR-223 was shown to directly act on Grin2B using ischemic reperfusion brain injury models in vitro and in vivo. Genetic ablation of miR-223 leads to enhanced expression of GluN2B indicating it to be a potential molecule that protects against neuronal cell death in stroke and other excitotoxic conditions.13 miR-137, a candidate gene in schizophrenia, regulates GluN2A and thus alters synaptic plasticity and imposes an inhibitory effect on post stroke depression.14 miR-19a and miR-539 can also target and regulate NMDAR.15 In our previous study, we found miR-129-2, miR-148a, and miR-296 to be targeting NMDAR in vitro and in animal models in which NMDAR is dysregulated.16
In this study, we have investigated whether miR-146a and miR-200b, which are differentially expressed in diseases such as Alzheimer's disease and schizophrenia,17,18,19,20,21,22,23,24,25,26,27 can regulate NMDAR subunits. We established the interaction of these miRNAs with NMDAR subunits through online bioinformatic analysis followed by experimental approach using luciferase assay. We obtained further validation on these miRNAs by showing their ability to downregulate NMDAR subunits upon overexpressing them in primary hippocampal neurons. We also injected these miRNAs to hippocampi in vivo to understand the physiological role of these miRNAs through behavioral and subsequent biochemical experiments. In addition, we also investigated the levels of these miRNAs in pharmacological rat models in which NMDAR is known to be downregulated.
Results
miR-146a and miR-200b bind to 3′UTRs of NMDAR subunits
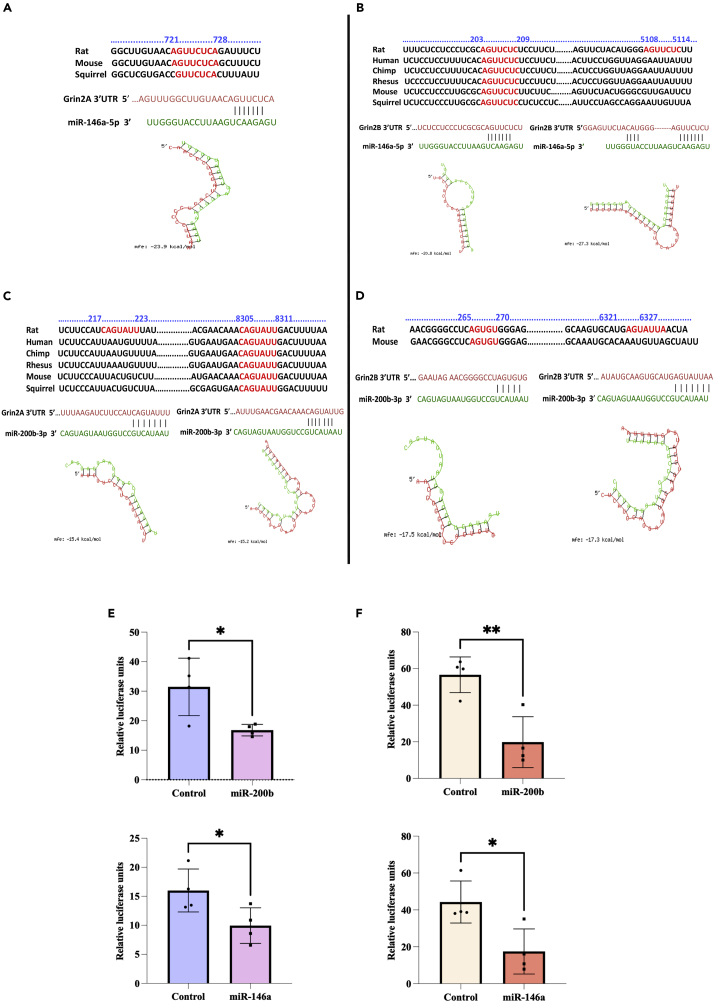
To find the miRNAs that bind to the 3′UTRs of the NMDAR subunits Grin2A and Grin2B, we used 3–5 online prediction tools and found numerous hits. We chose miR-146a and miR-200b for further studies because they were found to be differentially expressed in neurodegenerative disorders such as Alzheimer's disease and also in neuropsychiatric disorders such as schizophrenia and major depressive disorders.17,18,19,20,21,22,23,24,25,26,27,28 Binding sites with conservation across species for miR-146a-5p and miR-200b-3p are found on Grin2B and Grin2A, respectively. Among the binding sites, at least one has strong conservation among other vertebrates including the humans (Figures 1B and 1C). The converse pairing of miR-146a-5p with Grin2A and miR-200b-3p with Grin2B is also possible at different binding sites but the sites are conserved only in mouse, rat, and squirrel, or mouse and rat, respectively (Figures 1A and 1D). The site type was 7mer-m8 or 7mer-A1 or 8mer, implying strong binding of the miRNA seed sequence to the 3′UTR of the target. In case of the complimentary strands of the miRNAs, miR-146a-3p has binding sites on Grin2A and Grin2B and miR-200b-5p has sites on Grin2A, but the sites are not conserved among other species including humans (data not shown). We used RNA hybrid tool that uses minimum free energy (mfe) paradigm to understand the binding stability of the miRNA-mRNA interaction (Figures 1A–1D). The quantitative indicators in terms of scores and other parameters obtained from some of the online tools are presented in Table S1.
Figure 1.
Interactions of miR-146a-5p and miR-200b-3p with Grin2A/Grin2B 3′UTR
Sequence alignment of the binding sites of miR-146a-5p (A and B) and miR-200b-3p (C and D) on Grin2A (A and C) and Grin2B (B and D) indicating conservation among different species is shown. Graphics illustrates the predicted binding of miRNA sequence (green) with the target 3′ UTR sequence (red) obtained using RNA hybrid tool with corresponding minimum free energy (mfe) shown (A–D). E and F show the dual luciferase assay results showing interaction of the miRNAs with the 3′UTR sequences in vitro. The firefly luciferase activity arising from the psiCHECK2 vector was used for normalization and the renilla luciferase activity was used as the reporter of interaction. Statistical analysis was done by unpaired Student’s t test, ∗∗p ≤ 0.01, ∗p ≤ 0.05, n = 4. Data are expressed as mean ±SD.
To experimentally validate the predicted miRNA-mRNA interactions, we performed dual luciferase assay. We constructed plasmids with 3′UTR sequences of Grin2A and Grin2B (Rattus norvegicus) in psiCHECK2 vector. The pre-miRNA sequences for miR-146a and miR-200b were cloned in pRIPM dsRed vector. The primers used for clonning are shown in Table 1. Each pre-miRNA was cotransfected with either Grin2A or Grin2B 3′UTR in HEK-293 cells and after 48 h the cells were subjected to the assay. The control group had co-transfection of empty pRIPM dsRed vector with Grin2A or Grin2B 3′UTR vector. It was observed that both miR-146a and miR-200b showed significant reduction in the renilla luciferase activity against Grin2A 3′UTR implying strong interaction [miR-200b (t = 2.95, df = 6); miR-146a (t = 2.52, df = 6)] (Figure 1E). Similarly, both the miRNAs showed significant interaction with Grin2B 3′UTR also [miR-200b (t = 4.34,df = 6); miR-146a (t = 3.20, df = 6)] (Figure 1F). These results show that the predicted miRNAs can interact with functional binding sites in the 3′UTRs of Grin2A and Grin2B.
Table 1.
Primers used for cloning pre-miRNAs
| miR-146a | Forward – 5′ AAAGGATCCCATGCCCTCTGTGCGTGT-3′ |
|---|---|
| Reverse 5′ AAAAAGCTTCGCAGAGAAACCCCATCTC-3′ | |
| miR-200b | Forward – 5′ AAAGGATCCCACTTAGACATCTGGGCC-3′ |
| Reverse 5′ AAAAAGCTTGGTGAGGTGTGGAACTGG-3′ |
Primers used for cloning the pre-miRNAs into the pRIPM vector were designed using the UCSC genome database.
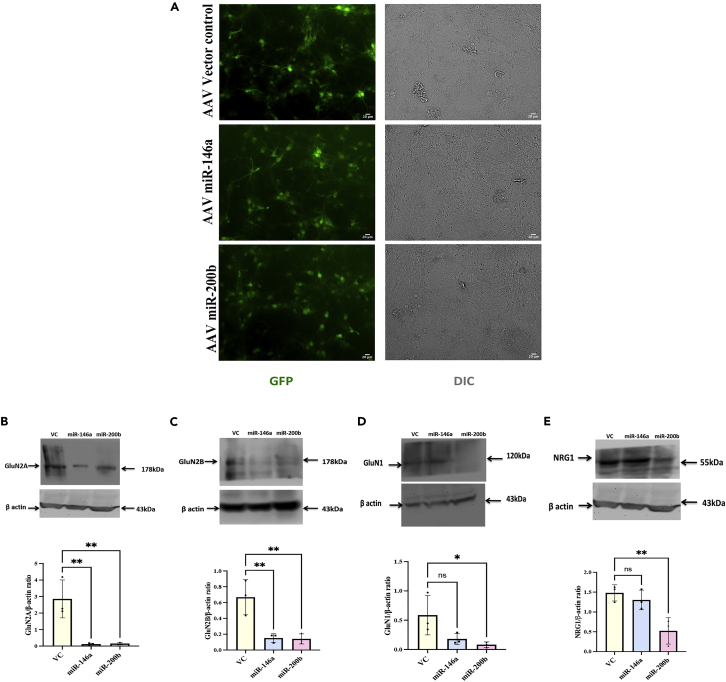
Downregulation of the NMDAR subunits by overexpression of miR-146a or miR-200b in primary hippocampal neurons
We further wanted to investigate whether the miRNA-mRNA binding described above could cause alteration in GluN2A/GluN2B protein levels in a neuronal system. For this, we used primary hippocampal neurons prepared from E−18 rat embryos. The cultures were subjected to overexpression of the miRNAs by treatment with the corresponding AAV constructs, AAV-miR-146a, AAV-miR-200b, or vector control (VC) containing the AAV-EGFP construct on DIV 7 and the cells were harvested on DIV 18–20. The transduction efficiency was more than 70% (Figure 2A) as seen from the expression of the EGFP marker encoded in the viral constructs (Figure S2). We checked for protein expression by Western blotting and found significant reduction of GluN2A and GluN2B in neurons transduced by either miR-146a or miR-200b (Figures 2B and 2C). Analysis using one-way ANOVA showed significant changes among the three groups using the Dunnetts test [F (2,6) = 16.66, p = 0.0036 for GluN2A; F (2,6) = 14.61, p = 0.005 for GluN2B) (Figures 2B and 2C). We also found decrease in GluN1 subunit of the NMDAR implying that the miRNAs caused downregulation of all the major subunits of NMDAR (Figure 2D). The one-way ANOVA showed significant difference between the groups for GluN1 [F (2,6) = 5.13, p = 0.050]. We checked for the other targets of these miRNAs such as neuregulin1 (NRG1) and found significant downregulation by miR-200b [F (2,6) = 10.88, p = 0.01] (Figure 2E). Overall, these results convey that miR-146a and miR-200b can downregulate NMDAR subunits GluN2A and GluN2B in vitro along with downregulating other proteins such as GluN1 and NRG1.
Figure 2.
miR-146a and miR-200b downregulate GluN2B and GluN2A levels in primary hippocampal neurons
The hippocampal neurons were transduced with AAV-miR-146a or AAV-miR-200b or VC on DIV 7, harvested on DIV 18–20 and were processed for western blotting.
(A) Representative images showing the transduction efficiency of the neurons on DIV 20. Scale bar: 20 μm.
(B–E) Representative western blots for GluN2A, GluN2B, GluN1, and NRG1. Since GluN2A and NRG1 were probed in the same blot at different molecular size regions, they share the same actin blot. Quantitation is done using ImageJ software. Data are expressed as mean ± SD. Statistical analysis was done using one-way ANOVA with Dunnett’s test. ∗∗p ≤ 0.01, ∗p ≤ 0.05; n = 3.
To support the western blot data, we also studied expression of GluN2B in primary neurons by immunocytochemical staining after overexpressing the miRNAs. The pRIPM-Ds Red vector construct carrying pre-miR-146a or pre-miR-200b was transfected to cultures at DIV 7. The cells were fixed on DIV 10 and were subjected to immunocytochemical staining. We found decrease in GluN2B expression in miR-146a- and miR-200b-transfected cells (Figure S1) [miR-146a (t = 3.70, df = 38), miR-200b (t = 2.23, df = 38)]. The dsRed present in the constructs marked the transfected neurons. The empty pRIPM-Ds Red vector was used as control for this experiment, which did not show any detectable GluN2B downregulation.16
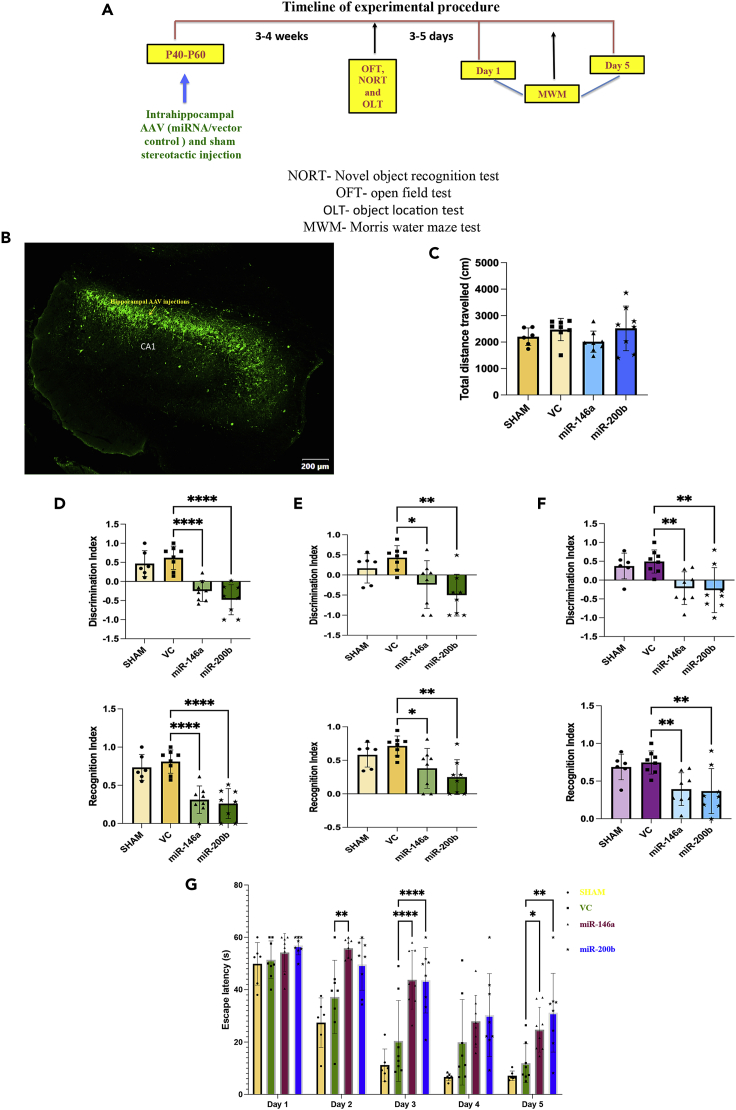
Regulation of NMDAR by miR-146a and miR-200b in vivo
To evaluate the potential role of miR-146a and miR-200b in vivo, we performed stereotactic surgeries to introduce these miRNAs into rat hippocampus. Adult male rats were bilaterally injected with AAV containing the pre-miRNA (miR-146a or miR-200b) at the CA1 region of the hippocampi. Empty vector (VC) and sham injection (using 1X PBS) were used as controls. The animals were allowed to recover for 4–5 weeks followed by behavioral and biochemical experiments. The treatment regime used is shown in Figure 3A. Presence of the virus in the CA1 region was shown by expression of the EGFP marker protein, which is present along with the miRNA sequence in the vector backbone (Figure S2). Figure 3B shows the representative image of AAV-mir-200b-injected hippocampus, 4–5 weeks after injection.
Figure 3.
Animals injected with miRNAs show impaired learning and memory
(A) Timeline of the experimental procedures.
(B) Representative fluorescence microscopy image of a coronal section of the brain of rat 4 weeks after stereotactic injection in the CA1 area of hippocampus with AAV-mir-200b particles marked by EGFP expression. The presented image was obtained by automatically aligning and stitching 50 tiled images into a mosaic (using Olympus confocal microscope). Scale bar: 200 μm.
(C–G) Behavioral analysis of the animals, four weeks after stereotactic surgeries for AAV-miR-146a, AAV-miR-200b, VC construct, and Sham (1X PBS) injections. (C): OFT for 10–11 min (D and E): Results of NORT showing DI and RI, 1 (D) and 24 h (E) after training. (F): Results of OLT conducted 1 h after training. Quantitation is shown as mean ± SD. Statistical analysis was done by one-way ANOVA followed by Dunnett’s post hoc multiple comparison test with p values indicated: ∗∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.01, ∗p ≤ 0.05, n = 6 per group for sham, n = 8 per group for VC, AAV-miR-146a, and AAV-miR-200b. (G): MWM test showing the latency to reach the platform for 5 days. Quantitation is shown as mean ± SD. Statistical analysis was done by repeated measures two-way ANOVA with Dunnett’s multiple comparison test with p values indicated: ∗∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.01, ∗p ≤ 0.05; n = 6 per group for sham, n = 8 per group for VC, AAV-miR-146a, and AAV-miR-200b.
miR-146a- and miR-200b-injected animals show cognitive disability
The animals were first subjected to open field test to assess the anxiety levels and locomotor ability. It was observed that the animals injected with AAV-miR-146a or AAV-miR-200b performed similar to sham and VC, implying no major impairment in locomotor activity (Figure 3C). The miRNA-injected animals did not seem to have increased anxiety as observed by the percentage of time spent in the peripheral and central zone, which showed no significant differences among the groups (Figure S3). The animals then underwent tests for learning and memory—NORT and OLT. It was observed that in the NORT tests done 1 h and 24 h after training, both the miRNA-injected groups had significant reduction in the discrimination index (DI) and recognition index (RI) when compared to VC (Figures 3D and 3E) indicating that the miRNA-injected animals did not spend more time in exploring the novel object, implying memory deficits. The one-way ANOVA showed significant differences between the groups for tests done 1 h [DI (F (3,26) = 20.31; p < 0.0001) and RI (F(3,26) = 19.9; p < 0.0001)] and 24 h [DI (F(3,26) = 6.18; p = 0.0026) and RI (F(3,26) = 6.18; p = 0.0026)] after training.
In OLT, the miRNA-injected animals did not spend more time in exploring the displaced object. RI and DI were significantly reduced in the miRNA-injected animals than VC indicating memory impairment (Figure 3F). The one-way ANOVA analysis showed significant difference among the groups [DI (F (3,26) = 6.07; p = 0.0028) and RI (F (3,26) = 6.07; p = 0.0028)].
Lastly, Morris water maze experiments were conducted to check the hippocampal dependent spatial learning and memory.29 The miRNA-treated animals showed significant learning impairment on day 3 and day 5 (Figure 3G). Although performance appears impaired on day 4, the data were not statistically significant. Animals injected with AAV-miR-146a showed significant learning impairment even on day 2 of the experiment. The repeated measures two-way ANOVA indicated significant differences between the time and groups (F (12,130) = 1.97; p = 0.031).
These results showed that the animals injected with AAV-miR-146a or AAV-miR-200b had severe cognitive impairment with learning and memory deficits. Because there was no significant difference between sham and VC animals in the behavior experiments, only VC animals were used as controls for subsequent miRNA, mRNA, and protein analysis.
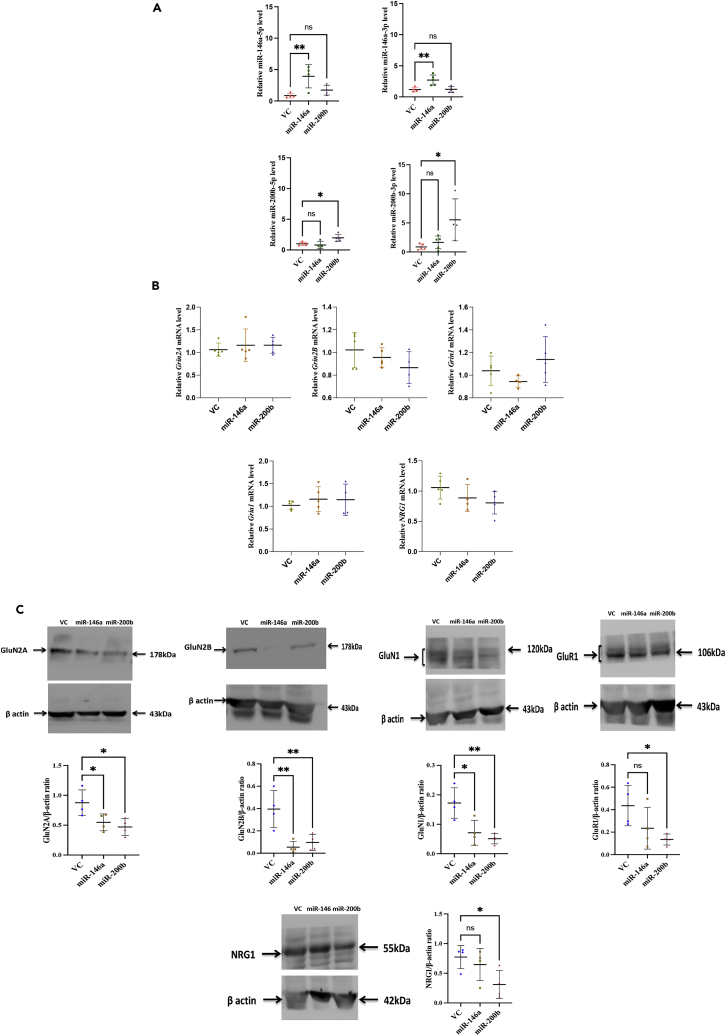
Biochemical analyses indicate miR-146a and miR-200b downregulate NMDAR in hippocampus
The 5p and 3p forms of both miR-146a and miR-200b were significantly high in the animals injected with the AAV constructs of the respective miRNAs (Figure 4A). The one-way ANOVA analysis showed significant differences among the groups injected with the AAV constructs [F (2,11) = 9.96; p = 0.003 for miR-146a-3p; F (2,11) = 7.53; p = 0.008 for miR-146a-5p; F (2,11) = 6.55; p = 0.013 for miR-200b-3p; F (2,11) = 7.81; p = 0.007 for miR-200b-5p]. As expected, the level of only the injected miRNA increased whereas the other miRNA was not significantly altered in the respective experimental group (Figure 4A). The transcript levels for the proteins that were downregulated by the miRNAs in hippocampal neurons in culture (Figure 2) were checked in the miRNA-injected animals. The NMDAR subunit transcripts, Grin2A and Grin2B, that are targets of miR-146a and miR-200b, were not significantly altered when compared to the control groups (Figure 4B). There was no significant change in the levels of Grin1 transcript (Figure 4B). The levels of NRG1 were also largely unchanged, implying that the injected miRNAs are not altering the transcript levels of the target proteins. We also checked the expression of AMPA receptor Gria1 due to impaired memory in the miRNA-overexpressed animals and found no significant change in the transcript level. The primers used for performing miRNA and transcript analysis are shown in Table 2.
Figure 4.
Alterations in miRNA, mRNA, and protein levels in the hippocampal region of the animals injected with AAV particles
(A) Expression levels of miR-146a-5p, miR-146a-3p, miR-200b-5p, and miR-200b-3p in animals injected with AAV as estimated by real-time PCR.
(B) Quantitation of transcript levels done by real-time PCR for Grin2A, Grin2B, Grin1, Gria1, and NRG1.
(C) Protein expression levels of GluN2A, GluN2B, GluN1, GluR1, and NRG1. Representative Western blots along with quantitation are shown. Quantitation is done using ImageJ software. Statistical analysis was done by one-way ANOVA with Dunnett’s post hoc test. Data is represented as mean ± SD. ∗∗p ≤ 0.01, ∗p ≤ 0.05; n = 4–5.
Table 2.
Primers used for real-time PCR of miRNAs and genes
| A: Primers used for amplifying the miRNAs | |
| Rno-miR-146a-5p | 5′GGGTGAGAACTGAATTCCATGG-3′ |
| Rno-miR-146a -3p | 5′GACCTGTGAAGTTCAGTTCTT-3′ |
| Rno-miR-200b-5p | 5′GCATCTTACTGGGCAGCAT-3′ |
| Rno-miR-200b -3p | 5′GTAATACTGCCTGGTAATGATGAC-3′ |
| U6 | Forward 5′- ACAGAGAAGATTAGCATGGCC-3' Reverse 5′-GACCAATTCTCGATTTGTGCG-3' |
| B: Primers used for amplifying the transcripts of the genes | |
| GRIN2A | Forward – 5′ GATCAACAATTCAACCAACG-3′ |
| Reverse 5′ AGACCACTTCACCTATCATTC-3′ | |
| GRIN2B | Forward – 5′ GTTTAACAACTCCGTACCTG-3′ |
| Reverse 5′ TCTGGAACTTGTCACTC-3′ | |
| NRG1 | Forward – 5′ CCTACTGCAAAACCAAGAAG-3′ |
| Reverse 5′ CAACAATATGCTCACTGGAG-3′ | |
| GRIN1 | Forward – 5′ AAGGAGAATATCACTGACCC-3′ |
| Reverse 5′TACTTAGAAGACATCAGAACC-3′ | |
| GRIA1 | Forward – 5′ AAAAGGAATATGCCGTACATC-3′ |
| Reverse 5′ACAATATCGATCTGGGGAAG-3′ | |
| BETA ACTIN | Forward – 5′ CCGCGAGTACAACCTTCTTG-3′ |
| Reverse 5′ GCAGCGATATCGTCATCCAT-3′ | |
The protein levels of these transcripts were analyzed and it was found that GluN2A and GluN2B levels for both the miRNA-injected groups were significantly reduced (Figure 4C). The one-way ANOVA showed significant differences among the groups (F (2,9) = 6.43; p = 0.018 for GluN2A; F (2,9) = 11.72; p = 0.003 for GluN2B). NRG1 protein was found to be significantly downregulated in the miR-200b-injected animals. GluN1 was also found to be significantly reduced in both the miRNA-injected groups with one-way ANOVA showing significant difference between the groups (F (2,9) = 10.58; p = 0.004). GluR1 was significantly decreased in AAV-miR-200b-injected animals.
These results indicate that NMDAR subunits GluN2A and GluN2B are downregulated by miR-146a-5p and miR-200b-3p, without altering the levels of their transcripts.
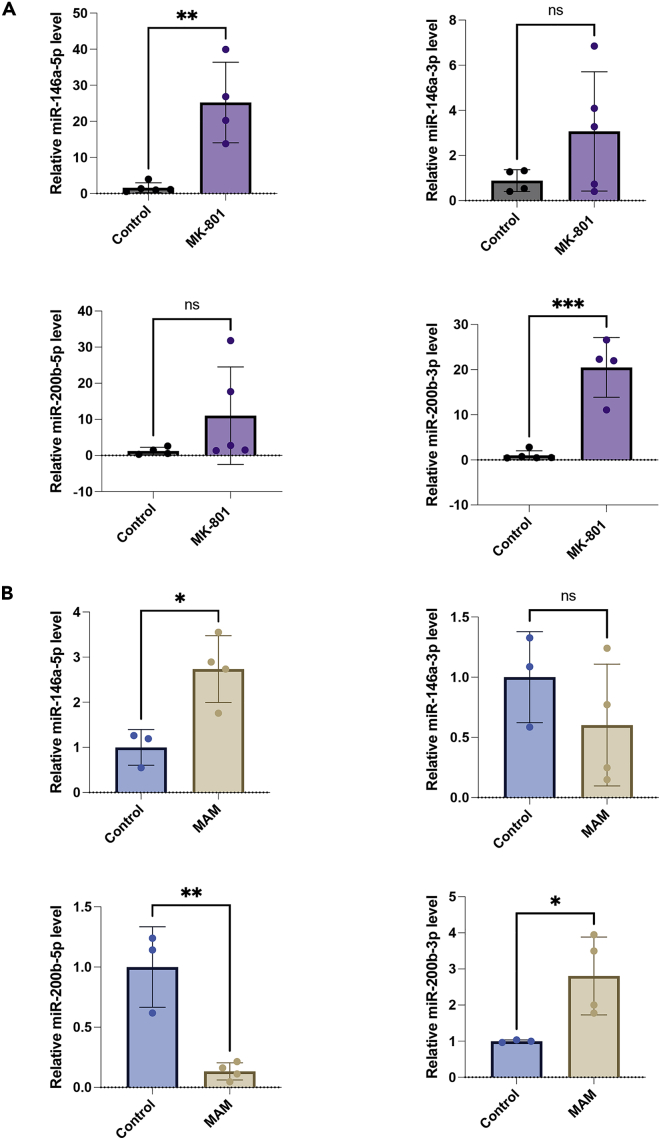
Altered expression levels of miR-146a and miR-200b in pharmacological models with NMDAR downregulation
The data so far show that miR-146a and miR-200b can downregulate NMDAR subunits in vitro and in vivo, upon overexpression. We then checked if these miRNAs play a role in downregulating NMDARs in other physiological conditions of NMDAR hypofunction. For this, we chose two pharmacological rat models in which NMDAR is downregulated by physiological signals that are indirectly induced by the experimental treatment.30 We used the MK-801 model and MAM model created as reported before.16 MK-801 is an NMDAR antagonist, and is used for creating a pharmacological model by a short course of administration and MAM, a neurotoxin, is used for generating a neurodevelopmental model by administering the compound at the embryonic stage.
miR-146a-5p is significantly upregulated in MK-801 model whereas expression of miR-146a-3p exhibited a lesser extent of change and was not statistically significant (Figure 5A) (t = 4.15, df = 6 for miR-146a-5p; t = 2.22, df = 6 for miR-146a-3p). miR-200b-3p was significantly upregulated several folds in MK-801 model but the change in miR-200b-5p was not significant (Figure 5A) (t = 6.6, df = 7 for miR-200b-3p; t = 1.42, df = 7 for miR-200b-5p).
Figure 5.
Upregulation of miR146a-5p and miR-200b-3p in MK-801-injected and MAM-injected animals
The MK-801 and MAM injected animal models were created as described before16 and is mentioned in brief in the methods section. Real-time PCR analysis was done for quantitating the levels of miR-146a-5p, miR-146a-3p, miR-200b-5p, and miR-200b-3p in the MK-801 model (A) and in the MAM model (B), in the hippocampal region. Data are expressed as mean ± SD. Statistical analysis was done using Student’s t test, ∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05, n = 3–5.
With MAM model, we observed significant upregulation of miR-146a-5p and miR-200b-3p (Figure 5B) (t = 3.63, df = 5 for miR-146a-5p; t = 2.89, df = 5 for miR-200b-3p). Interestingly, we found significant downregulation in one of the complimentary forms, miR-200b-5p, but no significant change was observed in miR-146a-3p expression (Figure 5B) (t = 5.19, df = 5 for miR-200b-5p; t = 1.21, df = 5 for miR-146a-3p). These results showed that miR-146a-5p and miR-200b-3p were upregulated in both the models in which NMDAR subunits GluN2A and GluN2B were downregulated,16 consistent with a role of these miRNAs in causing decrease in the protein expression of GluN2A and GluN2B in these models.
Discussion
Numerous studies have reported differential expression of miRNAs in the brain under various conditions, but very few have addressed their physiological role in brain development and cognitive functions. Elucidating the specific roles of brain-enriched miRNAs would help in understanding neural mechanisms underlying brain functions and their importance in neurological disorders. This study examines the roles of miR-146a and miR-200b in regulating NMDAR subunits. These miRNAs were identified as candidates that can target Grin2A and Grin2B, by bioinformatics-assisted screening. Another major reason for choosing these miRNAs was their altered expression levels in neurological disorders.17,18,19,20,21,22,23,24,25,26,27 Using both in vitro and in vivo models, we showed interaction of the miRNAs with Grin2A and Grin2B and attenuation of their translation. We also found that in the two animal models where GluN2A and GluN2B proteins were downregulated in the hippocampus, miR-146a-5p and miR-200b-3p were upregulated (Figure 5) consistent with their involvement in downregulating NMDAR subunits. Complementary strands, miR-146a-3p and miR-200b-5p which do not target Grin2A and Grin2B, did not show a major change in expression, thereby further supporting the mechanism (Figure 5). We show severe learning and memory impairment upon overexpression of both the miRNAs in vivo (Figure 3) along with decreased levels of GluN2A and GluN2B protein expression (Figure 4C) indicating possible synaptic dysfunction.
Although overexpression of miR-146a and miR-200b in vivo caused cognitive impairment, it did not lead to anxiety and depression-related phenotype (Figures 3C and S3) indicating that upregulation of these miRNAs may only contribute to the cognitive component in disease conditions. Our study suggests that the association of miR-146a and miR-200b to neurological disorders could be, at least partly, due to their ability to downregulate NMDARs,20,21,27 thus offering a mechanistic explanation for diseases such as Alzheimer's disease and schizophrenia that have an NMDAR component in their pathophysiology.31
miR-146a and miR-200b are known to affect and regulate other pathways. miR-146a is reported to also be associated with cerebrovascular diseases, neuroinflammation, CNS trauma, neuroautoimmune diseases, neuroviral infections, peripheral neuropathy, neurological tumor, ischemic stroke, epilepsy, and multiple sclerosis.32,33,34 It has roles in neuronal survival and axonal growth35,36 and in development by targeting NF-κB, NOTCH, and WNT/β-catenin.37,38,39,40 Numerous studies showed miR-200 family to be associated with the pathogenesis of neurodegenerative diseases such as Parkinson's disease, ALS, and Huntington's disease.27 A recent report suggests a role for miR-200b-3p in hypoxia-ischemia brain damage by upregulating Slit2 expression. Inhibiting miR-200b-3p alleviates brain injury in neonatal rat model of hypoxia-ischemic brain damage.41 miR-200 family is found to play key roles in the control of epithelial-mesenchymal transition in neurogenesis42 and in processes such as initiation, progression, and metastasis in gliomas.43 We also found other putative targets of these miRNAs that are involved in synaptic plasticity-related pathways using computational tools (Table S2).
Interestingly, we obtained data on inverse correlation of miR-200b-5p and miR-200b-3p levels in the MAM treatment model (Figure 5B). This could be due to two mechanisms which are recently gaining attention—target-mediated miRNA protection and/or target RNA-directed miRNA degradation.44,45,46
MiRNAs are post-transcriptional regulators which bind to their target mRNAs resulting in mRNA destabilization and translational repression.47 But there are contradictory reports on decreased mRNA levels with elevated miRNA expression. Hill et al. proposed a regulatory network model in which miRNAs might not profoundly change the target mRNA levels. It states that the downregulation of the target mRNA might be masked because of increased mRNA transcription or by downregulation of its repressor genes that are targeted by the same miRNAs.48 Our data show that the mRNA levels were not affected in the animals in which miRNAs were overexpressed in the hippocampal region (Figure 4B). The protein levels were significantly reduced for these transcripts leading to downregulation of NMDARs (Figures 4B and 4C).
We also checked for GluN1 protein levels as it is one of the major subunits of NMDAR. There was reduction in GluN1 protein which might be a secondary effect of reduced GluN2A/GluN2B protein levels (Figures 2D and 4C) as GluN1 is not a direct target of these miRNAs based on bioinformatic analysis. Because of the persistent decrease in the levels of GluN2A/GluN2B proteins, the functional NMDAR heteromer, GluN1/GluN2, would be low leading to impairment in learning and memory tasks. The majority of native NMDA receptors are tetrameric assemblies of two GluN1 subunits and two GluN2 subunits,49 in particular GluN1/GluN2A/GluN2B triheteromers. This combination accounts for 50% of the total NMDAR in hippocampus and cortex of an adult rodent brain.50 GluN1/GluN2/GluN3 heteromers are also present but their relative abundance is unknown. From our data, we can extrapolate that miR-146a and miR-200b can independently downregulate GluN1/GluN2A/GluN2B triheteromers which form a major proportion of NMDARs leading to significant functional alterations in vivo. It will be relevant to investigate the role of these miRNAs in synaptic plasticity mechanisms that are likely to be hampered in our miRNA overexpression models.
NRG1 was downregulated by miR-200b-3p as predicted by bioinformatic analysis (Figure S4). This was observed in primary hippocampal neurons and in AAV-miR-200b-injected animals (Figures 2E and 4C). There are two sites of interaction in NRG1 3′UTR and both are highly conserved among vertebrates (Figure S4) with site type 7mer-m8. miR-146a-5p was also predicted to show interaction with NRG1 at sites conserved only in rat and mouse, but there was no significant effect on the protein level in our experiments in vitro and in vivo in which miR-146a was overexpressed. We also observed decrease in the GluR1 levels in miRNA-injected animals (data statistically significant for only AAV-miR-200b-injected animals) (Figure 4C). GluR1 is an important AMPA receptor subunit playing crucial roles in synaptic plasticity.51 Decrease in GluR1 could be a secondary effect of either NMDAR downregulation or overexpression of these miRNAs, as there are no direct binding sites for miR-200b in Gria1.
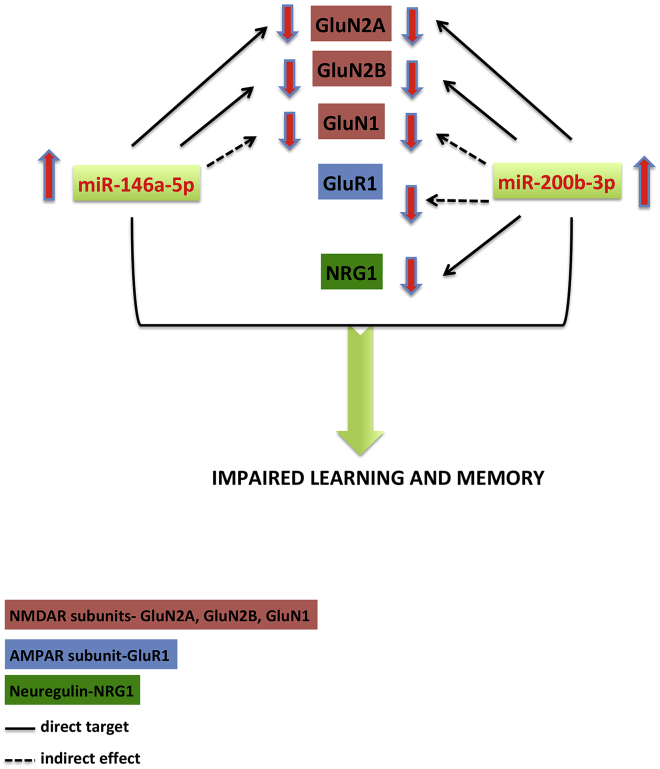
Understanding the role of miRNAs will provide insights on how to manipulate the levels of certain proteins which are altered in neurological diseases. Manipulating NMDAR activity as such leads to severe side effects in patients.52 Hence alterative therapeutic strategies such as using miRNAs could play a significant role in modulating these proteins without much side effects. Our study provides insights on how upregulated miR-146a-5p and miR-200b-3p can contribute to severe cognitive impairment in neurological disorders (Figure 6).
Figure 6.
Schematic representation of the downregulation of proteins by overexpressed miR-146a-5p and miR-200b-3p
Overexpression of miR-146a-5p or miR-200b-3p in rats leads to downregulation of GluN1, GluN2A, and GluN2B subunits of NMDAR along with downregulation of GluR1 and NRG1 only in miR-200b-3p-injected animals thus affecting cognition.
Conclusions
Our objective was to identify miRNAs that target the NMDA receptors and study their role in learning and memory. For this, we selected miR-146a and miR-200b, which were differentially expressed in neuropsychiatric and neurodegenerative disorders. By bioinformatics analysis, it was found that the two miRNAs have target sites on the transcripts of the NMDAR subunits GluN2A and GluN2B. Interaction of these miRNAs with Grin2A and Grin2B was shown by luciferase assay. Western blot analysis of primary hippocampal neurons overexpressed with these miRNAs showed downregulation of GluN2A and GluN2B. Overexpression of these miRNAs in vivo in the hippocampus of rats through stereotactic injection of AAV particles caused downregulation of GluN2A, GluN2B, and GluN1 proteins without significant alterations in their transcript levels. Additionally, protein levels of NRG1 and GluR1 were also significantly downregulated in AAV-miR-200b-injected animals. Both the miRNAs caused learning and memory defects upon overexpression in animals as expected from the molecular level changes. The levels of miR-146a-5p and miR-200b-3p were also upregulated in two rat models in which the NMDAR subunits were downregulated. These results highlight the key roles of small non-coding RNAs in modulating NMDAR and learning and memory processes thus bringing out the possibility of using them in therapeutic interventions.
Limitations of the study
In this study, only male Wistar rats were used for manipulating the levels of miRNAs in hippocampus to study their cognitive behavior. It would be interesting to understand the role of miR-146a and miR-200b in female Wistar rats as there are reports suggesting sexual dimorphism of miRNA expression.53 In addition, incorporating more tests for emotional and cognitive behavior such as elevated plus maze test, fear conditioning, or forced swim test will uncover the involvement of these miRNAs in a wider range of behavioral phenotypes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GluR1 | Santa Cruz Biotechnology | Cat# sc-55509; RRID:AB_629532 |
| GluN2B | Abcam | Cat# ab65783; RRID:AB_1658870 |
| GluN2B | Santa Cruz Biotechnology | Cat# sc-9057; RRID:AB_670232 |
| GluN2A | Millipore | Cat# MAB5216; RRID:AB_95169 |
| GluN2A | Cell Signaling Technologies | Cat# 4205-1; RRID:AB_764455 |
| GluN1 | Abcam | Cat# ab17345; RRID:AB_776808 |
| β-actin | Sigma | Cat# A5316; RRID:AB_476743 |
| NRG1 | Santacruz | Cat# sc-28916; RRID:AB_2154793 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150077; RRID:AB_2630356 |
| Anti-Rabbit IgG (whole molecule)–Peroxidase antibody produced in goat | Sigma | Cat# A0545; RRID:AB_257896 |
| Anti-Mouse IgG (Fab specific)–Peroxidase antibody produced in goat | Sigma | Cat# A3682; RRID:AB_258100 |
| Bacterial and virus strains | ||
| AAV-mir-GFP-Blank Control Virus (Serotype 8) | Applied Biological Materials | Cat# Am00108 |
| GFP rno-mir-200b AAV miRNA Virus (Serotype 8) | Applied Biological Materials | Cat# Amr1010308 |
| GFP rno-mir-146a AAV miRNA Virus (Serotype 8) | Applied Biological Materials | Cat# Amr1005808 |
| Chemicals, peptides, and recombinant proteins | ||
| Bovine serum albumin | SRL | Cat# 97350 |
| B27 | Thermo Fisher Scientific | Cat# 17504044 |
| Neurobasal medium | Thermo Fisher Scientific | Cat# 21103049 |
| Hank’s Balanced Salt Solution | Thermo Fisher Scientific | Cat# 14170112 |
| Antibiotic-Antimycotic solution | Thermo Fisher Scientific | Cat# 15240062 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat# 10270106 |
| Lipofectamine™ Transfection Reagent | Thermo Fisher Scientific | Cat# 18324012 |
| Lipofectamine™ 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668019 |
| TB Green® Advantage® qPCR Premix | Takara | Cat# 639676 |
| Trizma base | Merck | Cat# T1503-1KG |
| Poly-D-lysine hydrobromide | Merck | Cat# P0899-50MG |
| BioTrace™ NT Nitrocellulose Transfer Membrane | Pall Life Sciences | Cat# 66485 |
| Clarity™ Western ECL Substrate | BioRad | Cat# 1705060 |
| Methylazoxymethanol acetate (MAM) | FUJIFILM Wako Pure Chemical Corporation | Cat# 136-16303 |
| MK-801 (dizocilpine) | Sigma | Cat# M107-25MG |
| RIPA Lysis Buffer | Sigma | Cat# 20-188 |
| Critical commercial assays | ||
| Mir-X™ miRNA First Strand Synthesis Kit | Takara | Cat# 638315 |
| miRVana Isolation kit | Thermo Fisher Scientific | Cat# AM1560 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems™ | Cat# 4368814 |
| PureLink™ Quick Plasmid Miniprep Kit | Thermo Fisher Scientific | Cat# K210010 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat# CRL-3216 |
| Experimental models: Organisms/strains | ||
| Wistar rats | RGCB animal facility | N/A |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad Software Inc | https://www.graphpad.com/scientificsoftware/prism/ |
| Noldus EthoVision XT | Noldus Information Technology, Wageningen, The Netherlands | https://www.noldus.com/ethovision-xt |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr.R.V.Omkumar (omkumar@rgcb.res.in; omkumar18@hotmail.com).
Materials availability
Plasmid constructs generated in this study are available from the lead contact, Dr.R.V.Omkumar (omkumar@rgcb.res.in; omkumar18@hotmail.com).
Data and code availability
-
•
All the data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
We used Wistar strain of rats for our study. For primary cultures of hippocampal neurons, rat embryos of embryonic day 18–19 (E18-19) were used. AAVs carrying the pre-miRNAs were streotactically injected into male Wistar rats of adult stage (60–70-day-old, 200–300g). For the MK-801 model, male rats of adolescent stage (40–45-day-old, 100–150g) were used. For generating the MAM model, female dams were subjected to the treatment and both sexes of the pups delivered were used. All the animals were housed with 12 h light-dark cycle. Access to chow and water were ad libitum to the animals. The procedures followed the rules and regulations of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India. The protocols were approved by the Institutional Animal Ethics Committee (IAEC) of Rajiv Gandhi Center for Biotechnology.
Method details
Bioinformatic analysis
The sequences of the miRNAs were obtained using miRBase database (http://www.mirbase.org/) and the sequences of the 3′UTR of Grin2A/Grin2B were obtained from UCSC Genome browser (https://genome.ucsc.edu/).
The tools used for mining the miRNAs that target the genes Grin2A and Grin2B were – TargetScan54 (http://www.targetscan.org/vert_72/), miRanda55 (http://www.microrna.org/), Micro-Cosm56 (https://tools4mirs.org/software/mirna_databases/microcosm-targets/), miRDB57 (http://mirdb.org/), Segal/PITA58 (https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html) and RNAhybrid tool59 (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/).
The foundations on which the computational tools rely are miRNA sequence datasets, the 3′UTR sequences of the target sites, the seed region at the 5′ end of the miRNA sequence which has perfect Watson-Crick complementarity with the 3′UTR of the target sequence, the minimum free energy measurement which reflects the strength of the miRNA-mRNA binding, the 3′ region of the miRNA sequence that also has the Watson-Crick pairing with the target mRNA, the conservation of the miRNA-mRNA interaction within the species, the miRNA sequence interaction with other regions of the mRNA such as 5′UTR and coding sequences and the factors that affect the miRNA-mRNA interaction other than the Watson-Crick base pairing.60 The prediction of miRNA-mRNA interactions by these online tools involve different algorithms, which incorporates some of the fundamentals mentioned above. The tools have picked up the sequences from databases such as Rfam for non-coding sequences and for obtaining the mRNA sequences (containing the 3′UTR), databases such as Ensembl, RefSeq database or UCSC genome database were used. All these tools have strengths and weakness and hence using more tools will help in decreasing false positives. Therefore we used multiple tools for the mining of miRNAs against NMDAR subunits.
DNA constructs
UCSC database (https://genome.ucsc.edu/) was used for obtaining precursor miRNA (pre-miRNA) sequences for miR-146a and miR-200b. Suitable primers were designed (Table 1) and were used for cloning these pre-miRNAs from rat genomic DNA. The amplified pre-miRNAs were cloned into the expression vector, pRIPM that has dsRed as marker. The 3′UTRs of Grin2A and Grin2B in the vector backbone psiCHECK2 described before16 were also used for performing the dual luciferase assay. All the clones were confirmed by restriction digestion and sequencing.
Luciferase assay
Dual luciferase assay was performed using human embryonic kidney-293 (HEK-293) cells as described before.16 In brief, psiCHECK2 and pRIPM vector constructs carrying the 3′UTRs and the pre-miRNAs respectively were used for the assay. These vectors were cotransfected and after 48hrs, reporter assay was performed. The empty pRIPM vector along with Grin2A/Grin2B 3′UTR psiCHECK2 was used as control for this experiment.
Primary neuronal culture and transfection
Pregnant female dams of E18-19 were sacrificed and the hippocampi from the embryos were dissected, trypsinized and the dissociated cells were cultured in coated dishes. The primary neurons were maintained up to 20 days in vitro (DIV 20).16,61,62
Neurons were transduced using AAVs containing the pre-miRNAs, miR-146a (AAV-miR-146a), miR-200b (AAV-miR-200b) or vector control (VC) on DIV 7. The viral titer for all the AAVs were 108 GC/mL. The vector maps for these constructs are shown in Figure S2. The cells were harvested on DIV 18–20 and were further processed for conducting western blotting experiments.
Treatment regimes for the animal models
Stereotactic injections of AAV to male Wistar rats
The rats were deeply anesthetized by isoflurane (2–4%) inhalation. The head was mounted on a stereotaxic frame (Company name: BenchMarkTM, Coretech Holding Scientific, USA currently merged to Leica Microsystems, Germany). The coordinates used for bilateral intrahippocampal injections at the CA1 site were: AP = −3.8 mm, ML = +1.4 mm and DV = −2.7 mm.63 All AAV constructs carried EGFP as marker. The viral titer used was 1 × 108 GC/mL and around 5 μL was injected per side using a 25 μL Hamilton syringe. The solution was injected very slowly and the needle was kept undisturbed for another 2 min to avoid backflow of the fluid. Sham control was used with PBS injection. The animals were kept for 4–5 weeks for recovery followed by behavioral tests. These animals were then sacrificed by cervical dislocation and the hippocampal region was dissected to measure the protein and mRNA levels of different targets as well as miRNA levels using western blotting and qPCR.
MK-801 model and MAM model
MK-801 (0.5 mg/kg) was dissolved in saline and was injected intraperitoneally for 5 consecutive days (one injection a day) at consistent time period of 2–5 pm to male adolescent rats. MAM (20 mg/kg) was dissolved in saline and was injected intraperitoneally to pregnant dams on gestational day 17 (GD 17). Weaning of the pups was performed on postnatal day 30 (P30). Detailed procedure was described before.16 The experimental rats were subjected to behavioral testing to confirm impairments, sacrificed by cervical dislocation and the hippocampi were dissected for performing biochemical analysis.
Behavioral analysis
Open field test (OFT), novel object recognition (NORT), object location tests (OLT) and Morris water maze (MWM) test were used for behavioral analysis as described before.16 The animals were made to acclimatize to the behavior chamber 1 h prior to the start of each experiment. In brief, OFT was carried out in an open square box. The total distance traveled and the time spent in central/peripheral zone by the animal in the box for 10–11 min was recorded. NORT was also conducted in the open square box with 2–3 non-toxic objects. The initial habituations for the animals were carried out for 10 min in the empty arena. About 24 h after habituation, the animal was given training in the familiar arena where they were exposed to two identical non-toxic objects for 5 min. During the test period, which is 1 h and 24 h after the training period, the animals were exposed to one novel object and one familiar object for 5 min in the same arena. The time taken for exploring the novel and familiar objects were noted and accordingly discrimination index (DI) and recognition index (RI) were calculated.
In OLT, the animal was subjected to training wherein it was made to explore two familiar objects for 5 min in the open square box. After 1 h, the animal was put in the same arena with one of the objects displaced to a new location. This is the test period, which was 5 min in duration and the time taken to explore the displaced object and the non-displaced object by the animal were noted. DI and RI were calculated.
The water maze test was performed in a white tank having a small circular platform inside. The whole tank was filled with water, which was translucent enough to make the platform invisible. The animals were trained for 5 days to locate the hidden platform using spatial cues. Each day, the animals underwent five trials of 1 min each with intertrial intervals of 20–30 s. The time taken to reach the platform was noted and the average escape latency was calculated for each animal on each day.
Noldus EthoVision XT software (Noldus Information Technology, Wageningen, The Netherlands) was used for analyses of the video files. After the MWM test, animals were sacrificed and the brains were immediately removed, dissected, snap frozen and stored in −80°C.
Western blotting
The hippocampal neurons were scrapped and were washed with 1X PBS. The cell pellet was lysed using RIPA lysis buffer with 50 mM β-glycerophosphate, 50 mM sodium fluoride, 200 μM sodium orthovanadate and 5 mM EDTA. The hippocampal tissues from animals were homogenized using liquid nitrogen followed by solubilizing them in lysis buffer [150 mM NaCl, 1 mM dithiothreitol (DTT), 1% NP-40, 50 mM Tris-HCl, pH 7.4, 5 mM EDTA, 10 mM staurosporine, 50 mM β-glycerophosphate, 5 mM sodium orthovanadate, 1% SDS, 0.2 mM phenylmethylsulphonylfluoride (PMSF), and 1X complete protease inhibitor cocktail]. Bicinchoninic acid (BCA) method was used to quantitate the protein concentration in the sample and 60–100 μg of protein was subjected to SDS-PAGE for each sample. The whole procedure was carried out as described before.16 After the transfer procedure, the membrane has been cut horizontally. Different molecular size regions were processed separately. The blots were incubated overnight with the corresponding primary antibodies. The primary antibodies used were: rabbit anti-GluN1 (1:1000, Abcam), rabbit anti-GluN2B (1:1000, Abcam), mouse anti-GluN2A (1:1000, Millipore), mouse anti-GluR1 (1:750, Santacruz), mouse anti-beta actin (1:3000, Sigma) and rabbit anti-NRG1 (1:1000, Santacruz). Subsequently, the blots were incubated for 2 h at room temperature with secondary antibodies, which are horseradish peroxidase (HRP)-conjugated (1:3000 or 1:10,00, Sigma). The blots were developed using Clarity ECL reagent in Chemidoc gel apparatus (Biorad, USA). The band intensity was quantitated using ImageJ software and the band intensity of β-actin was used for normalization.
Real-time PCR analysis of hippocampal tissues
Extraction and measurement of transcripts (mRNAs and miRNAs) in the brain tissues were carried out by Quantitative real-time PCR (qPCR) as described before16 according to manufacturer’s instructions.
KiCqStart SYBR green primers (Sigma) were used for quantitating the mRNA levels. Table 2 shows the primers used for quantitating miRNAs and mRNAs. From the 2ˆΔCT values, relative expression levels were calculated in each sample. Data presented is the average value from minimum three independent experiments and β-actin was used for normalization.
Quantification and statistical analysis
The data is presented as mean ± standard deviation (SD) for independent experiments/replicates. Luciferase assay data (n = 4) was analyzed using parametric unpaired Student’s t test between each miRNA group and the corresponding control group. Behavioral data analysis (n = 6–8 per group) for stereotactic injection experiments was done using one-way ANOVA followed by Dunnett’s post hoc test between the control and the treatment groups. Data from the MWM tests were analyzed using repeated measures two-way ANOVA followed by Dunnett’s post hoc test. The data from the biochemical analysis (n = 4–5 for qPCR and western blotting) was subjected to one-way ANOVA followed by Dunnett’s post hoc test. The statistical analysis for the behavior experiments was completely blinded. Data from the qPCR (n = 3–5 per group) for MK-801 and MAM model were statistically analyzed using unpaired parametric Student’s t test. The data was analyzed using the GraphPad prism 8 software (8.4.2). Statistical significance was set at p < 0.05.
Acknowledgments
We are grateful to Dr. Ani V Das and Dr. Jackson James for providing luciferase plasmid psiCHECK2 (Promega) and pRIPM plasmid and for help. We thank Ms. Reena Sarah Jacob for performing the statistical analysis. We thank Mr. K.C. Sivakumar for technical advice on bioinformatics. We thank Dr. Mayadevi, Dr. Mantosh Kumar, and Dr. Lakshmi K for their support and advice. We also thank the funding agencies- RGCB and Department of Science and Technology (DST), India.
Author contributions
S.G. and R.V.O. contributed to conception and design of the study. S.G. and R.V.O. developed the methodology. S.G. performed the experiments. R.V.O. acquired and provided resources and was responsible for overall supervision for the study. S.G. wrote the first draft of the manuscript. Both the authors contributed to manuscript revision and approved the submitted version.
Declaration of interests
The authors declare no competing interests.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105515.
Supplemental information
References
- 1.Friedman R.C., Farh K.K.H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 4.Rajman M., Schratt G. MicroRNAs in neural development: from master regulators to fine-tuners. Development. 2017;144:2310–2322. doi: 10.1242/dev.144337. [DOI] [PubMed] [Google Scholar]
- 5.Schratt G. microRNAs at the synapse. Nat. Rev. Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 6.O’Connor R.M., Gururajan A., Dinan T.G., Kenny P.J., Cryan J.F. All roads lead to the miRNome: miRNAs have a central role in the molecular pathophysiology of psychiatric disorders. Trends Pharmacol. Sci. 2016;37:1029–1044. doi: 10.1016/j.tips.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Maciotta S., Meregalli M., Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front. Cell. Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. Expression profiling of mammalian microRNAs uncovers a sub- set of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monyer H., Sprengel R., Schoepfer R., Herb A., Higuchi M., Lomeli H., Burnashev N., Sakmann B., Seeburg P.H. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 10.Javitt D.C. Glutamate as a therapeutic target in psychiatric disorders. Mol. Psychiatry. 2004;9 doi: 10.1038/sj.mp.4001551. 984-97, 979. [DOI] [PubMed] [Google Scholar]
- 11.Kocerha J., Faghihi M.A., Lopez-Toledano M.A., Huang J., Ramsey A.J., Caron M.G., Sales N., Willoughby D., Elmen J., Hansen H.F., et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. USA. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller B.H., Zeier Z., Xi L., Lanz T.A., Deng S., Strathmann J., Willoughby D., Kenny P.J., Elsworth J.D., Lawrence M.S., et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. USA. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harraz M.M., Eacker S.M., Wang X., Dawson T.M., Dawson V.L. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc. Natl. Acad. Sci. USA. 2012;109:18962–18967. doi: 10.1073/pnas.1121288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L., Li H., Guo R., Ma T., Hou R., Ma X., Du Y. miR-137, a new target for post-stroke depression. Neural Regen. Res. 2013;8:2441–2448. doi: 10.3969/j.issn.1673-5374.2013.26.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbel C., Hernandez I., Wu B., Kosik K.S. Developmental attenuation of N-methyl-D-aspartate receptor subunit expression by microRNAs. Neural Dev. 2015;10:20–29. doi: 10.1186/s13064-015-0047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunasekaran S., Jacob R.S., Omkumar R.V. Differential expression of miR-148b, miR-129-2 and miR-296 in animal models of schizophrenia-Relevance to NMDA receptor hypofunction. Neuropharmacology. 2022;210 doi: 10.1016/j.neuropharm.2022.109024. [DOI] [PubMed] [Google Scholar]
- 17.Beveridge N.J., Gardiner E., Carroll A.P., Tooney P.A., Cairns M.J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins D.O., Jeffries C.D., Jarskog L.F., Thomson J.M., Woods K., Newman M.A., Parker J.S., Jin J., Hammond S.M. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angelucci F., Cechova K., Valis M., Kuca K., Zhang B., Hort J. MicroRNAs in Alzheimer’s disease: diagnostic markers or therapeutic agents? Front. Pharmacol. 2019;10:665. doi: 10.3389/fphar.2019.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghazaryan H., Zakharyan R., Petrek M., Navratilova Z., Chavushyan A., Novosadova E., Arakelyan A. Expression of micro-RNAs miR-31, miR-146a, miR-181c and miR-155 and their target gene IL-2 are altered in schizophrenia: a case-control study. F1000Res. 2019;8:2077. [Google Scholar]
- 21.Ibrahim R.R., Amer R.A., Abozeid A.A., Elsharaby R.M., Shafik N.M. Micro RNA 146a gene variant/TNF-α/IL-6/IL-1 β; A cross-link axis inbetween oxidative stress, endothelial dysfunction and neuroinflammation in acute ischemic stroke and chronic schizophrenic patients. Arch. Biochem. Biophys. 2020;679 doi: 10.1016/j.abb.2019.108193. [DOI] [PubMed] [Google Scholar]
- 22.Moreau M.P., Bruse S.E., David-Rus R., Buyske S., Brzustowicz L.M. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santarelli D.M., Beveridge N.J., Tooney P.A., Cairns M.J. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 24.Wang G., Huang Y., Wang L.L., Zhang Y.F., Xu J., Zhou Y., Lourenco G.F., Zhang B., Wang Y., Ren R.J., et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci. Rep. 2016;6:26697. doi: 10.1038/srep26697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mai H., Fan W., Wang Y., Cai Y., Li X., Chen F., Chen X., Yang J., Tang P., Chen H., et al. Intranasal administration of miR-146a agomir rescued the pathological process and cognitive impairment in an AD mouse model. Mol. Ther. Nucleic Acids. 2019;18:681–695. doi: 10.1016/j.omtn.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C.G., Wang J.L., Li L., Xue L.X., Zhang Y.Q., Wang P.C. MicroRNA-135a and-200b, potential Biomarkers for Alzheimer׳ s disease, regulate β secretase and amyloid precursor protein. Brain Res. 2014;1583:55–64. doi: 10.1016/j.brainres.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Fu J., Peng L., Tao T., Chen Y., Li Z., Li J. Regulatory roles of the miR-200 family in neurodegenerative diseases. Biomed. Pharmacother. 2019;119 doi: 10.1016/j.biopha.2019.109409. [DOI] [PubMed] [Google Scholar]
- 28.Geaghan M., Cairns M.J. MicroRNA and posttranscriptional dysregulation in psychiatry. Biol. Psychiatry. 2015;78:231–239. doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Garthe A., Kempermann G. An old test for new neurons: refining the Morris water maze to study the functional relevance of adult hippocampal neurogenesis. Front. Neurosci. 2013;7:63. doi: 10.3389/fnins.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyle J.T. NMDA receptor and schizophrenia: a brief history. Schizophr. Bull. 2012;38:920–926. doi: 10.1093/schbul/sbs076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Chang L., Song Y., Li H., Wu Y. The role of NMDA receptors in Alzheimer’s disease. Front. Neurosci. 2019;13:43. doi: 10.3389/fnins.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan W., Liang C., Ou M., Zou T., Sun F., Zhou H., Cui L. MicroRNA-146a is a wide-reaching neuroinflammatory regulator and potential treatment target in neurological diseases. Front. Mol. Neurosci. 2020;13:90. doi: 10.3389/fnmol.2020.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shomali N., Mansoori B., Mohammadi A., Shirafkan N., Ghasabi M., Baradaran B. MiR-146a functions as a small silent player in gastric cancer. Biomed. Pharmacother. 2017;96:238–245. doi: 10.1016/j.biopha.2017.09.138. [DOI] [PubMed] [Google Scholar]
- 34.Juźwik C.A., S Drake S., Zhang Y., Paradis-Isler N., Sylvester A., Amar-Zifkin A., Douglas C., Morquette B., Moore C.S., Fournier A.E. microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog. Neurobiol. 2019;182 doi: 10.1016/j.pneurobio.2019.101664. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X., Su S., Li S., Pang X., Chen C., Li J., Liu J. MicroRNA-146a down-regulation correlates with neuroprotection and targets pro-apoptotic genes in cerebral ischemic injury in vitro. Brain Res. 2016;1648:136–143. doi: 10.1016/j.brainres.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Jia L., Wang L., Chopp M., Zhang Y., Szalad A., Zhang Z.G. MicroRNA 146a locally mediates distal axonal growth of dorsal root ganglia neurons under high glucose and sildenafil conditions. Neuroscience. 2016;329:43–53. doi: 10.1016/j.neuroscience.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C., Liu X.J., Xie J., Xie J., Ma T.T., Meng X.M., Li J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264. 7 macrophages. Int. Immunopharmacol. 2016;32:46–54. doi: 10.1016/j.intimp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Ghahhari N.M., Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial–mesenchymal transition in cancer. Eur. J. Cancer. 2015;51:1638–1649. doi: 10.1016/j.ejca.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Hwang S.J., Seol H.J., Park Y.M., Kim K.H., Gorospe M., Nam D.H., Kim H.H. MicroRNA-146a suppresses metastatic activity in brain metastasis. Mol. Cells. 2012;34:329–334. doi: 10.1007/s10059-012-0171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N., Yang L., Meng L., Cui H. Inhibition of miR-200b-3p alleviates hypoxia-ischemic brain damage via targeting Slit2 in neonatal rats. Biochem. Biophys. Res. Commun. 2020;523:931–938. doi: 10.1016/j.bbrc.2020.01.029. [DOI] [PubMed] [Google Scholar]
- 42.Beclin C., Follert P., Stappers E., Barral S., Coré N., De Chevigny A., Magnone V., Lebrigand K., Bissels U., Huylebroeck D., et al. miR-200 family controls late steps of postnatal forebrain neurogenesis via Zeb2 inhibition. Sci. Rep. 2016;6:35729. doi: 10.1038/srep35729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng L., Fu J., Ming Y. The miR-200 family: multiple effects on gliomas. Cancer Manag. Res. 2018;10:1987–1992. doi: 10.2147/CMAR.S160945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D. Target RNA–directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hass J., Walton E., Wright C., Beyer A., Scholz M., Turner J., Liu J., Smolka M.N., Roessner V., Sponheim S.R., et al. Associations between DNA methylation and schizophrenia-related intermediate phenotypes—a gene set enrichment analysis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2015;59:31–39. doi: 10.1016/j.pnpbp.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de la Mata M., Gaidatzis D., Vitanescu M., Stadler M.B., Wentzel C., Scheiffele P., Filipowicz W., Großhans H. Potent degradation of neuronal mi RNA s induced by highly complementary targets. EMBO Rep. 2015;16:500–511. doi: 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill C.G., Matyunina L.V., Walker D., Benigno B.B., McDonald J.F. Transcriptional override: a regulatory network model of indirect responses to modulations in microRNA expression. BMC Syst. Biol. 2014;8:36. doi: 10.1186/1752-0509-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen K.B., Ogden K.K., Yuan H., Traynelis S.F. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chater T.E., Goda Y. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Front. Cell. Neurosci. 2014;8:401. doi: 10.3389/fncel.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipton S.A. Failures and successes of NMDA receptor antagonists: molecular basis for the use of open-channel blockers like memantine in the treatment of acute and chronic neurologic insults. NeuroRx. 2004;1:101–110. doi: 10.1602/neurorx.1.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mavrikaki M., Pantano L., Potter D., Rogers-Grazado M.A., Anastasiadou E., Slack F.J., Amr S.S., Ressler K.J., Daskalakis N.P., Chartoff E. Sex-dependent changes in miRNA expression in the bed nucleus of the stria terminalis following stress. Front. Mol. Neurosci. 2019;12:236. doi: 10.3389/fnmol.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 55.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Griffiths-Jones S., Grocock R.J., Van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 59.Rehmsmeier M., Steffen P., Höchsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riffo-Campos Á.L., Riquelme I., Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? Int. J. Mol. Sci. 2016;17:1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Remya C., Dileep K.V., Variyar E.J., Zhang K.Y.J., Omkumar R.V., Sadasivan C. Chemical similarity assisted search for acetylcholinesterase inhibitors: molecular modeling and evaluation of their neuroprotective properties. Int. J. Biol. Macromol. 2021;174:466–476. doi: 10.1016/j.ijbiomac.2021.01.148. [DOI] [PubMed] [Google Scholar]
- 62.Salazar I.L., Mele M., Caldeira M.V., Costa R.O., Correia B., Frisari S., Duarte C.B. Preparation of primary cultures of embryonic rat hippocampal and cerebrocortical neurons. Bio. Protoc. 2017;7:e2551. doi: 10.21769/BioProtoc.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paxinos G., Watson C.R., Emson P.C. AChE-stained horizontal sections of the rat brain in stereotaxic coordinates. J. Neurosci. Methods. 1980;3:129–149. doi: 10.1016/0165-0270(80)90021-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All the data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.