Abstract
Transforming growth factor β (TGFβ) is a major component of tumor‐derived small extracellular vesicles (TEX) in cancer patients. Mechanisms utilized by TGFβ+ TEX to promote tumor growth and pro‐tumor activities in the tumor microenvironment (TME) are largely unknown. TEX produced by head and neck squamous cell carcinoma (HNSCC) cell lines carried TGFβ and angiogenesis‐promoting proteins. TGFβ+ TEX stimulated macrophage chemotaxis without a notable M1/M2 phenotype shift and reprogrammed primary human macrophages to a pro‐angiogenic phenotype characterized by the upregulation of pro‐angiogenic factors and functions. In a murine basement membrane extract plug model, TGFβ+ TEX promoted macrophage infiltration and vascularization (p < 0.001), which was blocked by using the TGFβ ligand trap mRER (p < 0.001). TGFβ+ TEX injected into mice undergoing the 4‐nitroquinoline‐1‐oxide (4‐NQO)‐driven oral carcinogenesis promoted tumor angiogenesis (p < 0.05), infiltration of M2‐like macrophages in the TME (p < 0.05) and ultimately tumor progression (p < 0.05). Inhibition of TGFβ signaling in TEX with mRER ameliorated these pro‐tumor activities. Silencing of TGFβ emerges as a critical step in suppressing pro‐angiogenic functions of TEX in HNSCC.
Keywords: angiogenesis, exosomes, head and neck squamous cell carcinoma, macrophages, small extracellular vesicles, TGFβ
1. INTRODUCTION
Adequate blood supply is of critical importance for the development, growth, and metastasis of solid tumors. The progression of solid tumors is dependent on accelerated angiogenesis, characterized by an increased functional capillary density and adequate supply of angiogenic growth factors (Carmeliet & Jain, 2011). Tumor angiogenesis is regulated by a complex network of morphogenic and molecular pathways, soluble factors, and extracellular vesicles (EVs) (Carmeliet, 2003; Ludwig & Whiteside, 2018). Recent studies indicate that EVs are present in the tumor microenvironment (TME) and also accumulate in the plasma of most cancer patients, including those with head and neck squamous cell carcinoma (HNSCC) (Hong et al., 2014; Hoshino et al., 2020; Ludwig et al., 2017). In patients with advanced HNSCC, plasma contains large numbers of EVs (in a range from 109 to 1012 particles/ml of plasma). These EVs circulate in all body fluids, including saliva (Xiao & Wong, 2012) and are enriched in tumor‐derived EVs (Sharma et al., 2020). A subset of small (30‐150 nm) EVs (sEVs or exosomes) that tumor cells produce are referred to as TEX (tumor‐derived small extracellular vesicles). Recently, TEX have been of special interest as potential biomarkers in cancer. TEX originate from the endocytic compartment of producer tumor cells and carry biologically‐active factors that define their functional activity (Sharma et al., 2020). Emerging evidence suggests that the promotion of angiogenesis may be a major function of TEX (Lucotti et al., 2022; Ludwig & Whiteside, 2018). Specifically, TEX produced by HNSCC deliver pro‐angiogenic signals to endothelial cells (ECs) and drive the development of blood vessels (Skog et al., 2008), as demonstrated by us (Ludwig et al., 2020; Ludwig, Yerneni et al., 2018) and by others (Sato et al., 2019).
In this study, we provide the first in vitro and in vivo evidence that TGFβ carried by HNSCC‐derived TEX is a major factor in promoting tumor progression by driving angiogenesis in the TME. Interestingly, the pro‐angiogenic effects of TGFβ‐enriched (TGFβ+) TEX are not limited to direct interactions with ECs, which we previously described (Ludwig, Yerneni et al., 2018). TGFβ+ TEX also interact with macrophages and stimulate their chemotaxis and pro‐angiogenic functions in the TME. Further, we show that the pharmacologic blockade of TGFβ+ TEX with the newly developed TGFβ inhibitor – mRER, which acts as a ligand trap, effectively inhibits angiogenesis induced by TGFβ+ TEX as well as tumor growth in vitro and in vivo. In this context, TEX‐associated TGFβ emerges as a target of adjuvant anti‐angiogenic cancer therapies aimed at inhibiting TEX‐mediated effects.
2. MATERIALS AND METHODS
2.1. The cancer genome atlas (TCGA) analysis
The TCGA Head and Neck Cancer database was analyzed using the GEPIA2 analytical tool (Tang et al., 2019). In total, 519 cases of primary HNSCC were included in this study and compared to 44 cases of normal controls. Vesiculation‐related genes were selected based on a published gene set (Fathi et al., 2021). All other gene sets were downloaded from Molecular signatures databases v7.5.1 (https://www.gsea‐msigdb.org/gsea/msigdb/index.jsp). Expression heatmaps of defined gene sets were generated and clustered online, and the data were downloaded for subsequent statistical analysis. TIMER version 2.0 was used to estimate the potential association between candidate genes and macrophage infiltration using the CIBERSORT method (Newman et al., 2015).
2.2. Cell lines
The HPV(‐) cell lines, PCI‐13, and PCI‐30, derived from human HNSCC were established and maintained in our laboratory (Heo et al., 1989). HPV(‐) cell line SCC9 was purchased from ATCC. HPV(+) cell lines UMSCC2, UMSCC47, and UMSCC90 were established by Dr. Thomas Carey (University of Michigan, Ann Arbor, MI). Human UMSCC2, UMSCC47, UMSCC90, and the murine SCCVII head and neck carcinoma cell lines, were obtained from Robert L. Ferris (UPMC Hillman Cancer Center, Pittsburgh, PA). All cell lines were authenticated. Cells were grown in DMEM (Lonza Inc.) supplemented with 1% (v/v) penicillin/streptomycin and 10% (v/v) FBS (Gibco, Thermo Fisher Scientific) at 37°C and in the atmosphere of 5% CO2 in the air. FBS was depleted of EVs by ultracentrifugation at 100,000 × g for 3 h and subsequent filtration using 0.22 μm filters. For TEX isolation, cell culture conditions were optimized as previously described (Ludwig, Razzo et al., 2019) and consistently performed. 2.5 × 106 UMSCC47 cells or 4 × 106 UMSCC2, UMSCC90, SCC9, PCI‐13, PCI‐30, or SCCVII cells were seeded with 25 ml media in 150 cm2 cell culture flasks. Supernatants were collected after 72 h cultures.
SVEC4‐10 lymphoendothelial cells established by O'Connell and Edidin (O'Connell & Edidin, 1990) were purchased from ATCC (Cat. #CRL‐2181) and grown in the same media as cancer cell lines. J774A.1 murine macrophages were purchased from ATCC (Cat. #TIB‐67) and grown in RPMI‐1640 (Gibco), supplemented with 10% (v/v) EV‐depleted and heat‐inactivated FBS (Gibco) at 37°C and in the atmosphere of 5% CO2 in the air. Same conditions were used for HaCaT cells (human immortalized keratinocytes) which were purchased from CLS (#300493).
2.3. TEX isolation and characterization
The detailed step‐by‐step protocol for TEX isolation by size exclusion chromatography (SEC) and subsequent characterization was previously published by us and analogously performed in this study (Ludwig, Hong et al., 2019). Briefly, cell culture supernatants were centrifuged at room temperature (RT) for 10 min at 2000 × g, transferred to new tubes for centrifugation at 10,000 × g at 4°C for 30 min, and filtrated using a 0.22 μm filter. Afterward, aliquots of supernatants were concentrated by using Vivacell 100 concentrators at 2000 × g. Concentrated supernatant (1 ml) was loaded on a 10 cm‐long Sepharose 2‐B column, and individual 1 ml fractions eluted with PBS were collected. Fraction #4 containing non‐aggregated TEX was used in all subsequent assays.
Protein concentrations, transmission electron microscopy (TEM), tuneable resistive pulse sensing (TRPS), and western blotting were used as previously described by us for characterization of TEX in Fraction #4 (Ludwig, Hong et al., 2019). The methodology for TEX isolation and characterization was performed according to current MISEV2018 guidelines (Théry et al., 2019) and EV preparations were routinely tested for purity, profiles, and markers.
2.4. LC‐MS/MS analysis
The analysis of TEX‐associated proteins by LC‐MS/MS was performed as previously described by us (Ludwig, Marczak et al., 2019). A detailed protocol is provided in the Supplementary File #1. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018259.
2.5. On‐bead flow cytometry
The presence of TGFβ1 on TEX was confirmed by on‐bead flow cytometry as previously described (Sharma et al., 2018). A detailed protocol is provided in the Supplementary File #1.
2.6. Dot‐blot analysis
Dot‐blot analysis of TEX was performed as previously described (Sung & Weaver, 2017). Briefly, various concentrations of isolated TEX were absorbed onto nitrocellulose membranes at RT for 30 min. The membranes were blocked with 5% non‐fat milk in PBS in the absence or presence of 0.1% (v/v) Tween‐20 (PBST) at RT for 1 h. The anti‐TGFβ1 Ab (1:1000, ab92486, Abcam) was incubated with the membranes in PBS or PBST blocking buffer at 4°C overnight followed by HRP‐conjugated secondary antibody incubation at RT for 1 h. Blots were developed with ECL detection reagents (GE Healthcare Biosciences).
2.7. TGFβ bioassay
A TGFβ bioassay was used to measure levels of bioactive TGFβ based on MFB‐F11 reporter cells according to the protocols published by Tesseur et al. (Tesseur et al., 2006). Briefly, MFB‐F11 cells were seeded at 4 × 104 cells/well in 96‐well flat‐bottom tissue culture plates (BD Falcon). After overnight incubation, cells were washed twice with PBS and incubated in 50 μl serum‐free DMEM supplemented with penicillin/streptomycin for 2 h before samples were added. For the SEAP assay, a SEAP Reporter Gene Assay Chemiluminescent kit from Roche was used according to the manufacturer's recommendations. For some experiments, MFB‐F11 cells were pre‐incubated at the indicated concentrations with Dynasore, Cytochalasin D or Nystatin (all purchased from Abcam) for 1 h. Cells were washed twice with PBS and the selected tested factors were added. For other experiments, MFB‐F11 cells were pre‐incubated with TGFβ inhibitors added before the start of experiments and used at the indicated concentrations for 2 h.
2.8. Raw‐Blue™ NF‐ĸB reporter assay
The NF‐κB expression assay was performed as previously described (Yerneni et al., 2019). Briefly, RAW‐Blue™ cells (Invivogen) were grown and maintained in high‐glucose DMEM supplemented with 10% (v/v) heat‐inactivated FBS, 1% (v/v) penicillin/streptomycin and 100 μg/ml Normicin™ (Invivogen). RAW‐Blue™ cells (2 × 104 per well) were seeded in wells of a 96‐well plate and treated with TGFβ inhibitors for 2 h at 37°C. Afterwards, 10 μg TEX and as a positive control 100 ng/ml lipopolysaccharide (LPS; Sigma‐Aldrich) were added to the cells in triplicate and incubated for 24 h. After incubation, 20 μl aliquots of conditioned media were collected, incubated with 200 μl QUANTI‐blue™ reagent (Invivogen), and optical density at 655 nm was measured using a TECAN spectrophotometer.
2.9. TEX internalization
SVEC4‐10 or J774A.1 cells (2 × 105 per well) were seeded in a 12‐well plate and incubated overnight at 37°C. TEX were labelled with SYTO®RNASelect™ stain (Invitrogen) according to the manufacturer's recommendations and added to each well at the concentration of 10 μg per well. After incubation of 0, 1, 3, 6, 24, and 48 h, cells were washed twice with PBS and mean MFIs were acquired using Gallios flow cytometer and Kaluza 1.0 software (Beckman Coulter). TEX internalization was validated by confocal microscopy as described by us before (Ludwig, Yerneni et al., 2018).
2.10. Cell viability
SVEC4‐10 cells (5 × 103) were seeded into wells of 96‐well plates with 50 μl of conditioned medium per well and incubated for 48 h. The MTS cell viability assay was performed according to the manufacturer's instructions (Abcam).
2.11. Cell migration
Cell migration by SVEC4–10 or J774A.1 cells was analyzed as previously described (Ludwig, Razzo et al., 2019). Briefly, 5 × 104 cells were starved in serum‐free media overnight and were added to the upper compartment of 24‐well transwell plates with 8 μm pore diameter membranes (Corning). Cells migrated towards the serum‐free medium or the medium supplemented with TEX or conditioned medium in indicated concentrations, which were placed in the lower compartment. After 6 h of incubation at 37°C, non‐migrating cells in the upper chamber were removed with cotton swabs. Migrating cells on the lower surface of the membrane were fixed in methanol and stained with 0.2% crystal violet (Sigma‐Aldrich). The number of migrated cells was counted in a light microscope in six randomly selected regions of interest at 20x magnification using an Olympus BX51 microscope (Olympus America). In addition, cell migration was also analyzed in wound‐healing assays. Confluent cell monolayers in wells of 48‐well plates were wounded mechanically by using pipet tips and were treated with indicated concentrations of conditioned medium. The wound was imaged using an Axiovert 25 CFL inverted microscope at 5× magnification and analyzed using the ImageJ software (http://rsbweb.nih.gov/ij/). Results are expressed as a percent of the recovery.
2.12. Primary macrophage culture
The primary macrophage culture was previously described by us (Ludwig et al., 2020). A detailed protocol is provided in the Supplementary File #1.
2.13. Western blot analysis
For protein analysis, 30 μg of total protein was used per sample. Samples were separated by SDS‐PAGE with a 10% dissolution gel and transferred onto a PVDF membrane. The primary antibody was anti‐MMP‐9 (1:1000, HPA001238, Sigma‐Aldrich). Semiquantitative analysis was performed using ImageJ.
2.14. Angiogenesis antibody arrays
The relative levels of human angiogenesis‐related proteins in cell lysates from treated or untreated macrophages were measured using a Human Angiogenesis Array Kit (R&D Systems Inc.). About 200 μg of cell lysate was added to the array, and the results were analyzed with the ImageJ software.
2.15. TGFβ inhibitors
LY2109761 is a selective TGFβ receptor type I/II (TβRI/II) dual kinase inhibitor and was purchased from Sigma. A TGFβ antibody (1D11) neutralizing all three TGFβ isoforms was purchased from BioXCell. The trivalent TGFβ receptor trap RER was described by us previously and is based on the human TβRII‐rat BG endoglin (orphan) domain and human TβRII (Qin et al., 2016; Zhu et al., 2018). In this study, we used mRER, which is based on an all‐murine sequence, except for linkers, which are non‐natural.
2.16. Basement membrane extract plug assay
C57BL/6 mice aged 6–8 weeks were purchased from Jackson Laboratories. Protocols for animal experiments were approved by the Institutional Animal Care and Use Committee under reference number #18114203. Growth‐factor depleted Cultrex® (500 μl; Trevigen) was mixed with 50 μl PBS (CTRL) or TEX (50 μg of total TEX protein in a volume of 50 μl PBS) and incubated for 24 h on ice. Mice were anesthetized and the skin overlying the area of injection was gently shaved. Each mouse received two subcutaneous injections (CTRL and TEX) in the midventral abdominal region, which were permitted to solidify. Randomly selected mice received intraperitoneal injections of mRER at concentrations of 25 μg/day (mRERlow) or 50 μg/day (mRERhigh) throughout the experiment. Plugs were harvested after 7 d and photographed. Each plug was cut into two halves; one half was used for tissue histology and the other half was used for hemoglobin assessment as an indirect measure of vascularization. For hemoglobin quantification implants were harvested, minced, and digested using the human tumor digestion kit (Miltenyi Biotec) according to the manufacturer's instructions. Hemoglobin content in the supernatants from the digested plugs was quantified using Drabkin's reagent (Sigma Aldrich) as previously described (Ekambaram et al., 2018; Robertson et al., 1991).
2.17. 4‐NQO oral carcinogenesis model
The 4‐NQO oral carcinogenesis model was implemented as described previously (Ludwig, Yerneni et al., 2018; Razzo et al., 2019). A detailed protocol is provided in the Supplementary File #1.
2.18. Tissue histology
Detailed protocols of processing and staining of excised implants/tissues are provided in the Supplementary File #1. The following primary antibodies were used: anti‐CD31 Ab (1:100, #550274, BD Biosciences), anti‐CD68 Ab (1:100, sc‐7084, Santa Cruz), anti‐TGFβ1 Ab (1:100, ab92486, abcam), anti‐phospho‐Smad2 Ab (1:400, #18338, Cell Signaling), anti‐iNOS Ab (1:50, ab178945, abcam) or anti‐Arginase‐1 Ab (1:50, #93668, Cell Signaling).
2.19. Statistical analysis
All quantitative data were analyzed using GraphPad Prism software (v7.0). Values are expressed as mean ± SEM. Differences between groups were assessed by Student t test or by one‐way ANOVA with post hoc analysis when appropriate. Differences were considered significant at p < 0.05.
3. RESULTS
3.1. HNSCC cell‐derived TEX carry TGFβ and angiogenesis‐promoting proteins
Characteristics of TEX in #4 fractions isolated by SEC from supernatants of the HNSCC cell lines, we previously used were described (Ludwig et al., 2020; Ludwig, Razzo et al., 2019; Ludwig, Marczak et al., 2019; Ludwig, Sharma et al., 2018; Razzo et al., 2019). Figures 1A–C and S1A–E show representative data for TEX isolated from the supernatants of HNSCC cell lines used in this study. The nanoparticle analysis showed a size distribution ranging from 60 to 140 nm, and TEX carried TSG101, CD63, and Alix proteins but did not carry Grp94 and Calnexin, negative markers for sEVs (Figure 1A–C). These EV characteristics were in accordance with the criteria specified by MISEV(2018) for EV studies (Théry et al., 2019).
FIGURE 1.

Characterization of HNSCC cell‐derived TEX. (A) Representative TEM image of TEX. (B) Representative TRPS (qNano) size and concentration distribution plot of TEX. (C) Western blots of TEX for small extracellular vesicle markers Alix, CD63, and TSG101 as well as negative markers Calnexin and Grp94 carried by TEX; each lane was loaded with 5 μg protein of TEX lysate. (D) Characterization of TEX proteome by LC‐MS/MS – shown are detected TEX proteins associated with GO term <vasculature development> (GO:0001944); actual and putative functional interactions between these proteins (marked with lines) were found by the STRING database (https://string‐db.org). (E) The relative abundance of the selected vesiculation/angiogenesis‐related proteins in TEX produced by the 5 different HNSCC cell lines; abundances are sorted regarding deciles of all normalized signals and are color‐coded (common sEV markers CD9 and CD63 are shown for reference). (F) Analysis of gene expression of the protein signature shown in E in the TCGA data base for HNSCCs using the GEPIA2 analytical tool. Comparison of normal solid tissue (n = 44) and primary HNSCC tumors (n = 519). (G) Correlation of the gene expression of the protein signature shown in E with vesiculation‐related genes in TCGA database for HNSCCs. (H) Correlation of the gene set HALLMARK_ANGIOGENESIS (GSEA, Molecular Signatures Database M5944) with vesiculation‐related genes in TCGA database for HNSCCs. (I) Correlation of the gene set HALLMARK_TGF_BETA_SIGNALING (GSEA, Molecular Signatures Database, M5896) with vesiculation‐related genes in TCGA database for HNSCCs. (J) Correlation of the TGFB1 gene expression with vesiculation‐related genes in TCGA database for HNSCCs.
Global proteomics profiling of TEX was performed using shot‐gun LC‐MS/MS proteomics. TEX produced by a panel of HPV(+) (UMSCC2, UMSCC47, UMSCC90) and HPV(‐) (PCI‐13 or PCI‐30) HNSCC cell lines were analyzed. This analysis identified 494 proteins encoded by unique genes (see the complete list in Supplementary File #2). Protein content‐based EV characterization revealed a variety of EV markers consistently carried by all five HNSCC cell line‐derived TEX (Figure S2). Among the proteins identified in TEX were 35 proteins associated with the GO term “vasculature development” (GO:0001944); interactions between these proteins are depicted in Figure 1D. Importantly, this subset of proteins consisted of 21 protein species associated with the GO term “angiogenesis” (GO:0001525), and it included TGFβ. Thus, the proteomics analysis confirmed our previous observations of TGFβ presence in TEX (Ludwig, Sharma et al., 2018), and it linked TGFβ to angiogenesis‐related proteins. A vast majority of the detected TEX proteins which associated with the vasculature development and angiogenesis were ubiquitously expressed in all analyzed TEX specimens independently of the HPV status of producer tumor cells (Figure 1E). To test whether this signature of pro‐angiogenic proteins in TEX has clinical relevance in HNSCC, the Cancer Genome Atlas (TCGA) was used to study gene expression of this signature in HNSCC patients (n = 519). The 35 proteins which were detected in TEX were significantly upregulated in tumor tissue compared to neighboring healthy tissue (Figure 1F). To test the correlation between the pro‐angiogenic signature in TEX and the transcriptional signatures for sEV secretion, we used a recently published gene signature consisting of 41 genes known to be involved in sEV secretion (Fathi et al., 2021). The pro‐angiogenic signature detected in TEX significantly and positively correlated with the sEV secretion signature (Figure 1G). Also, the sEV secretion signature correlated with the hallmark angiogenesis (Figure 1H), hallmark TGFβ signaling (Figure 1I), as well as with the expression of the TGFB1 gene (Figure 1J).
These results indicate that TEX released by HNSCC cells are enriched in pro‐angiogenic proteins and these specific proteins correlate with sEV secretion in HNSCC tissues. Also, sEV secretion in HNSCC may be accompanied by upregulation of pro‐angiogenic genes as well as signatures for TGFβ signaling.
3.2. TEX carry TGFβ capable of initiating the TGFβ signaling cascade
To investigate whether TGFβ is present on the surface or in the lumen of TEX, a dot blot assay was used. Prior to the assay, TEX were either permeabilized in 0.1% Tween‐20 v/v (a mild treatment commonly used for western blots) or not permeabilized. As shown in Figure 2A, TGFβ is detectable on the surface of TEX, and it is also present in the vesicle lumen. The detergent used had no adverse effects on the detection of TGFβ, as its intraluminal levels were higher than surface levels in the dot blot assay. Next, on‐bead flow cytometry was used to validate the presence of TGFβ on the EV surface (Figure 2B). Bioactivity of TEX‐associated TGFβ was demonstrated in MFB‐F11 reporter assays, where incubation with TEX produced by UMSCC47 cells resulted in a concentration‐dependent increase in reporter cell activity (Figure 2C). To compare levels of bioactive TGFβ in TEX from all six human HNSCC cell lines (UMSCC2, UMSCC47, UMSCC90, SCC9, PCI‐13, and PCI‐30), and from the murine HNSCC cell line SCCVII, similar co‐incubation experiments of TEX with MFB‐F11 cells were performed. Recombinant TGFβ1 (10 pg) served as a reference. TEX from different HNSCC cell lines contained varying levels of bioactive TGFβ: TEX produced by UMSCC47 had the highest activity levels, while TEX from UMSCC2 or UMSCC90 had very low levels (Figure 2D). The response in the TGFβ bioassay of each cell line correlated with the relative TGFβ abundance obtained by proteomics profiling (Figure 1E). To test whether internalization by responder cells is required for the induction of TGFβ signaling, we pre‐incubated MFB‐F11 cells with the inhibitors of EV uptake Dynasore (blocks clathrin‐ and caveolin‐dependent endocytosis (Mulcahy et al., 2014)), Cytochalasin D (blocks endocytosis (Mulcahy et al., 2014)) or Nystatin (blocks lipid raft‐mediated endocytosis (Tian et al., 2014)) prior to co‐incubation of the cells with TEX. The pre‐treatment with Dynasore and Cytochalasin D significantly increased the response of MFB‐F11 to TEX, whereas no differences were observed after pre‐treatment with Nystatin (Figure 2E). These results suggest that the inhibition of EV uptake leads to greater availability of TEX in the extracellular space, allowing for more extensive TGFβ binding to receptors on recipient cells. These experiments emphasize the importance of surface‐associated TGFβ for effective intercellular signaling mediated by TGFβ+ TEX.
FIGURE 2.

Characterization of TEX‐associated TGFβ. (A) Dot blot analysis shows that TGFβ is present on the surface of TEX derived from UMSCC47 cells and is also enclosed in the vesicle lumen. (B) Representative histograms for TGFβ expression on TEX derived from UMSCC47 cells and analyzed by flow cytometry. (C) Quantification of bioactive TGFβ at the indicated concentrations of TEX derived from UMSCC47 cells using MFB‐F11 reporter cells. (D) Quantification of bioactive TGFβ in 20 μg of total TEX protein isolated from supernatants of the indicated cancer cell lines using MFB‐F11 reporter cells; 10 pg of recombinant TGFβ1 was used as a reference. (E) The response of MFB‐F11 reporter cells to TEX derived from UMSCC47 cells in the presence or absence of the indicated EV uptake inhibitors; no differences were observed between EV uptake inhibitors alone and CTRL (data omitted for simplicity). (F) The response of MFB‐F11 reporter cells to TEX derived from UMSCC47 cells in the presence or absence of the indicated TGFβ inhibitors; no differences were observed between TGFβ inhibitors alone and CTRL (data omitted for simplicity). All experiments were performed at least three times in triplicates. All values represent means ± SEM; **p < 0.01 vs. CTRL;# p < 0.05 vs. TEX; ## p < 0.01 vs. TEX.
To evaluate whether TEX‐induced TGFβ signaling in recipient cells could be inhibited by selective TGFβ inhibitors, we compared the activity of the trivalent TGFβ trap, mRER, the receptor kinase inhibitor, LY2109761, and the neutralizing anti‐TGFβ antibody, 1D11. All the tested inhibitors significantly decreased TEX‐induced responses in MFB‐F11 cells (p < 0.05; Figure 2F). However, mRER and LY2109761 were more effective TGFβ inhibitors compared to 1D11 Abs.
3.3. TEX promote TGFβ‐dependent chemotaxis and activation of macrophages
Since it is well known that TGFβ promotes the differentiation of non‐activated macrophages into a tumor‐associated macrophage‐like (i.e, M2‐like) phenotype, we now focused on the interactions between TEX and macrophages. To study these interactions, a bioinformatics approach utilizing the TCGA head and neck cancer cohort was used to correlate expression of the vesiculation‐related genes with the gene sets ‘positive regulation of macrophage chemotaxis’ (Figure 3A), ‘positive regulation of macrophage migration’ (Figure 3B), as well as the CD68 gene expression (Figure 3C), a macrophage marker. All these correlations were significantly positive, indicating that TEX in HNSCC stimulate the infiltration of macrophages into the tumor tissue. Also, the pro‐angiogenic signature which we detected in TEX (Figure 1D and E) significantly correlated with macrophage infiltration in HNSCC tissues (Figure 3D). To validate these results in functional studies, transwell migration assays were performed. TEX were found to significantly stimulate the migration of SVEC4‐10 ECs as well as J744A.1 macrophages in a concentration‐dependent manner (p < 0.01; Figure 3E and F). This effect was completely blocked by using 50 nM mRER (p < 0.05), indicating that TGFβ is a major factor in TEX‐induced chemotaxis (Figure 3E and F). mRER itself had no effect on cell migration (data not shown). The number of migrating cells was similar for ECs and macrophages. To further highlight the chemotactic effects of TEX‐associated TGFβ, TEX derived from UMSCC47 cells carrying high levels of TGFβ (TGFβhigh) and TEX derived from UMSCC90 cells carrying low levels of TGFβ (TGFβlow) were compared. While TEX from both sources significantly stimulated EC migration (p < 0.01), mRER only partially blocked migration in TGFβlow TEX (p < 0.05). Migration of ECs towards TGFβhigh TEX was significantly blocked by mRER (p < 0.01; Figure 3G and H). sEVs isolated from normal keratinocytes (HaCaT cells), which were shown to have a less angiogenic profile compared to TEX (Głuszko et al., 2021), did not significantly stimulate EC migration (Figure S3A). Flow cytometry was used to study the uptake of TEX by ECs and macrophages. Both the cell types readily internalized TEX; however, J744A.1 macrophages internalized significantly larger quantities of TEX at any measured time point (p < 0.05; Figure 3I). While macrophages internalized TEX at the early time points (1‐6 h), ECs internalized larger quantities of TEX after 24 h of incubation (Figure 3I). The different levels of TGFβ in TEX did not influence their distinct uptake abilities (Figure S3B). In aggregate, these data suggest that substantial differences exist in the optimal time as well as levels of TEX uptake among various cells populating the TME. The Raw‐Blue™ NF‐ĸB reporter assay was used to study macrophage activation after TEX treatment. TEX significantly stimulated NF‐κB activation (p < 0.01) in these macrophages. This effect was inhibited by TGFβ inhibitors (p < 0.05), indicating that NF‐κB activation in macrophages by TEX is partially TGFβ‐dependent. In this assay, mRER had the highest blocking efficiency (p < 0.01; Figure 3J).
FIGURE 3.
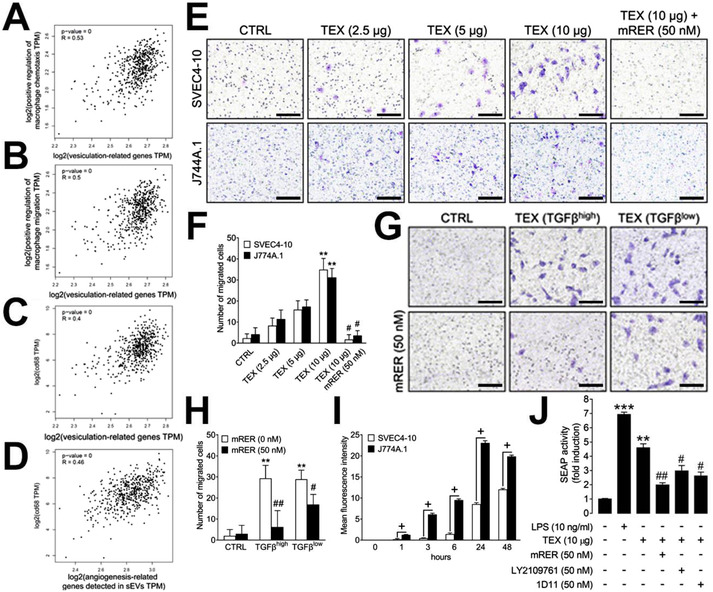
TEX interact with endothelial cells and macrophages and promote their chemotaxis. (A) Correlation of the gene set positive regulation of macrophage chemotaxis (GSEA, Molecular Signatures Database M13699) with vesiculation‐related genes in TCGA database for HNSCCs. (B) Correlation of the gene set positive regulation of macrophage migration (GSEA, Molecular Signatures Database M25365) with vesiculation‐related genes in TCGA database for HNSCCs. (C) Correlation of CD68 gene expression with vesiculation‐related genes in TCGA database for HNSCCs. (D) Correlation of CD68 gene expression with gene expression of the protein signature shown in Figure 1E in TCGA database for HNSCCs. (E) Representative images of migration of SVEC4–10 lymphendothelial cells or J744A.1 macrophages towards serum‐free media (CTRL), 2.5, 5 and 10 μg of TEX protein (derived from SCCVII cells) or a combination of 10 μg of TEX protein and the TGFβ inhibitor mRER (50 nM). Scale bars = 100 μm. (F) Quantification of migrated cells after 6 h of incubation. (G) Representative images of migration of SVEC4–10 cells towards serum‐free media (CTRL), 10 μg of TGFβhigh (UMSCC47) and 10 μg of TGFβlow (UMSCC90) TEX in the presence or absence of mRER (50 nM). Scale bars = 100 μm. (H) Quantification of migrated cells after 6 h of incubation. (I) Internalization of TEX derived from the supernatant of SCCVII cells by SVEC4‐10 or J744A.1 cells; internalization was quantified by flow cytometry at the indicated time points. (H) Raw‐Blue™ NF‐ĸB reporter cells were treated with PBS (Neg. CTRL), LPS (100 ng/ml, Pos. CTRL), TEX from UMSCC47 cells (20 μg/ml TEX protein) and TEX in the presence of indicated TGFβ inhibitors. Experiments were performed two times in triplicates. All values represent means ± SEM; *p < 0.05 vs. CTRL; **p < 0.01 vs. CTRL; ***p < 0.001 vs. CTRL; + p < 0.05 vs. SVEC4‐10; # p < 0.05 vs. TEX; ## p < 0.01 vs. TEX.
3.4. TGFβ+ TEX reprogram macrophages towards a pro‐angiogenic phenotype and pro‐angiogenic functions
Macrophages are known to adopt different functional programs in response to the signals from the TME, a process which involves polarization towards the M1 or M2 phenotype. In our hands, co‐incubation of TEX with human primary macrophages freshly isolated from the blood of healthy donors resulted in a significant upregulation of various surface markers, either those associated with an M1 phenotype (CD86, CD80, and EGFR) or an M2 phenotype (CD206, Arginase‐1, and LAP) (Figure 4A). This suggested that TEX promoted macrophage differentiation resulting in expression of the mixed M1/M2 phenotype in recipient cells. However, no differences were observed for HLA‐DR and IL‐10 expression in these cells (Figure 4A). To evaluate the role of TGFβ in this co‐incubation experiment, we followed the fate of CD86, the M1 marker, and Arginase‐1, the M2 marker, since these markers were most drastically altered after co‐incubation of primary macrophages with TEX. The same experiment was then repeated after pre‐incubating the macrophages with TGFβ inhibitors prior to addition of TEX. Inhibition of TGFβ signaling with mRER and LY2109761 resulted in the further upregulation of CD86 (p < 0.05), whereas inhibition with 1D11 Abs showed no effects on CD86 expression (Figure 4B). The TEX‐induced upregulation of the M2 marker, Arginase‐1, was significantly reduced by all three inhibitors (Figure 4C). In aggregate, these results suggest that while TEX promote expression of the mixed M1/M2 phenotype, TGFβ appears to be the TEX cargo component responsible for mediating polarization of naïve macrophages towards the M2 phenotype. This finding was validated using the TCGA data, showing that expression of vesiculation‐related genes is associated with the infiltration of naïve macrophages (Mϕ) into HNSCC tissue, however, no clear polarization towards M1 or M2 could be observed (Figure 4D).
FIGURE 4.
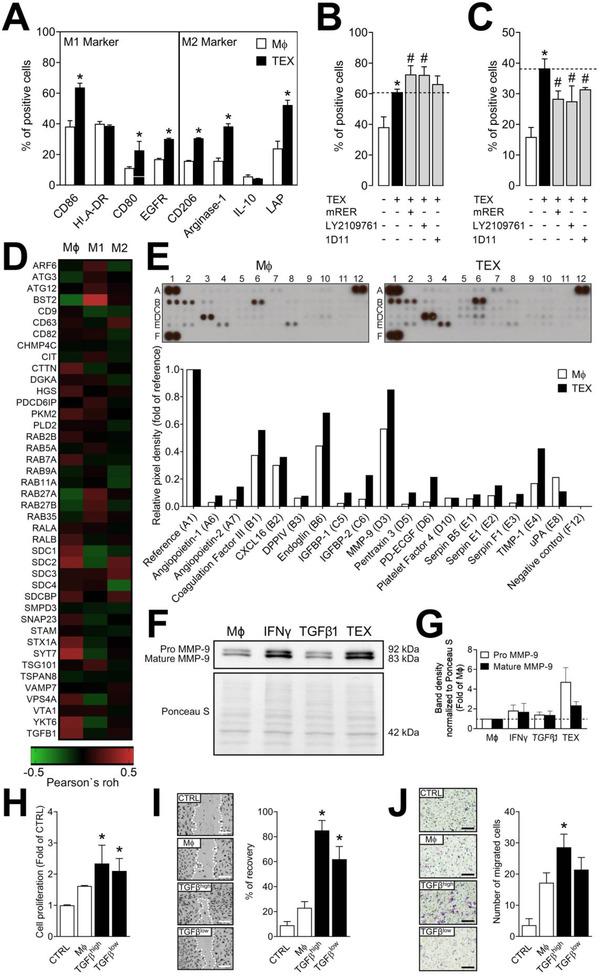
TGFβ+ TEX interact with macrophages and reprogram them to a pro‐angiogenic phenotype. (A) Expression of established M1‐ and M2‐markers was analyzed by flow cytometry in naïve human primary macrophages (Mϕ) and macrophages treated with TEX (UMSCC47‐derived). (B) Expression of CD86 by macrophages treated with UMSCC47‐derived TEX in the presence or absence of the TGFβ inhibitors mRER, LY2109761 or 1D11 (all used at 50 nM). (C) Expression of Arginase‐1 by macrophages treated with UMSCC47‐derived TEX in the presence or absence of the TGFβ inhibitors mRER, LY2109761 or 1D11 (all used in 50 nM). (D) Correlation of vesiculation‐related genes with infiltration of Mϕ, M1, and M2 macrophages in the TCGA head and neck cancer cohort analyzed using TIMER2.0. (E) Cell lysates of naïve macrophages or macrophages treated with UMSCC47‐derived TEX were analyzed by human angiogenesis arrays; the arrays were quantified using ImageJ, values are normalized to reference spots on the membranes (A1, A12, and F1). (F) Immunoblotting of MMP‐9 in macrophages treated with PBS (Mϕ CTRL), IFNγ (M1 CTRL), TGFβ1 (M2 CTRL) and TEX (UMSCC47‐derived). (G) Quantification of band intensities shown in F normalized to Ponceau S staining. (H) Proliferation of SVEC4‐10 cells in response to the treatment with conditioned medium from macrophages treated with PBS (Mϕ), TGFβhigh TEX (UMSCC47‐derived) or TGFβlow TEX (UMSCC90‐derived); proliferation was determined after 48 h, values are presented as fold of CTRL. (I) Wound healing of SVEC4‐10 cells after 12 h of incubation with PBS (CTRL) or with conditioned medium from macrophages treated with PBS (Mϕ), TGFβhigh TEX (UMSCC47‐derived) or TGFβlow TEX (UMSCC90‐derived). The left panel shows representative images 12 h after scratching; dotted lines indicate the border of SVEC4‐10, scale bars = 100 μm. The right panel shows quantified data expressed as a percent of the recovery. (J) The left panel shows representative images of SVEC4–10 cell migration towards serum‐free media (CTRL) or conditioned medium from macrophages treated with PBS (Mϕ), TGFβhigh TEX (UMSCC47‐derived) or TGFβlow TEX (UMSCC90‐derived); scale bars = 100 μm. The right panel shows the quantification of migrated cells after 6 h incubation. Experiments in A, B, C were performed two times in duplicates. Experiments in F and G were performed in duplicates. Experiments in H, I and J were performed three times in triplicates. All values in this figure represent means ± SEM. *p < 0.05 vs. Mϕ; # p < 0.05 vs. TEX.
Macrophages are known as important stimulators of angiogenesis in the TME due to their ability to produce pro‐angiogenic factors (Denardo & Ruffell, 2019; Ludwig et al., 2020). Therefore, angiogenesis arrays were used to analyze the pro‐angiogenic content of macrophages in response to treatments with TEX. TEX increased the quantities of several pro‐angiogenic factors in macrophages, including Angiopoietin‐2, MMP‐9, PD‐ECGF and TIMP‐1 (Figure 4E). These results were validated by immunoblotting in macrophages treated with PBS (Mϕ CTRL), IFNγ (M1 CTRL), TGFβ1 (M2 CTRL) and TEX (Figure 4F and G). Both, IFNγ and TGFβ1 stimulated upregulation of pro‐MMP‐9 and mature MMP‐9 in comparison to naïve macrophages. TEX stimulated upregulation of mature MMP‐9 by ∼2.5‐fold and pro‐MMP‐9 by ∼4.7‐fold (Figure 4F and G). While TEX increased the quantities of pro‐angiogenic factors in macrophages, we previously showed that the macrophages also secreted these factors (Ludwig et al., 2020). To validate this finding in functional studies, conditioned media (CM) were collected from treated and untreated macrophages. CM from macrophages treated with TEX enriched in TGFβ (derived from TGFβhigh UMSCC47 cells) significantly stimulated the proliferation, migration, and chemotaxis of ECs compared to CM from untreated control macrophages (p < 0.05; Figure 4H, I and J). CM from macrophages treated with TGFβlow TEX (UMSCC90‐derived) also significantly stimulated cell proliferation and migration of ECs, however, not as robustly as the TGFβhigh group (p < 0.05; Figure 4H and I). Similar effects were observed for tumor cells since treatment with CM from TEX‐treated macrophages (TGFβhigh and TGFβlow TEX) resulted in an increased proliferation and migration of UMSCC47 cells (p < 0.05; Figure S4). These results indicate that TEX‐treated macrophages may not only induce neovascularization, but also have a direct impact on tumor growth.
3.5. TGFβ+ TEX promote TGFβ‐dependent vascularization in vivo
To assess the pro‐angiogenic potential of TEX in vivo, TEX mixed with growth‐factor‐depleted Cultrex were injected subcutaneously into C57BL/6 mice. To address potential species‐dependent differences, TEX from a murine HNSCC cell line (SCCVII) and a human HNSCC cell line (UMSCC47) were injected into mice. The plugs with TEX from both sources induced a grossly obvious, enhanced vascularization, which was validated by significantly enhanced hemoglobin levels in the plugs (p < 0.001) and the presence of CD31+ signals (Figures 5A, C and S5). Furthermore, plugs with TEX showed a massive infiltration of macrophages as demonstrated by immunostaining for CD68 (Figure 5A). Control plugs (CTRL) injected with a mixture of Cultrex and PBS were free of any signs of vascularization or macrophage infiltration. To show the relevance of TGFβ signaling in this process, double staining for phosphorylated Smad2 (pSmad2) was performed either with CD31 for ECs or CD68 for macrophages. As presented in Figure 5B, ECs and macrophages showed pSmad2‐positive signals in the nuclear region, indicating that TGFβ signaling was ongoing in these cell types.
FIGURE 5.

TEX promote TGFβ‐dependent angiogenesis in vivo. Mice received subcutaneous injections of growth‐factor depleted Cultrex®. Each animal received 2 plugs (CTRL: 500 μl Cultrex® + 100 μl PBS; TEX: 500 μl Cultrex® + 50 μg TEX protein in 50 μl PBS). TEX derived either from SCCVII cells (TEX murine) or from UMSCC47 cells (TEX). Plugs were harvested after 7 days. (A) Representative photographs of harvested plugs and representative images of immunofluorescence staining for CD31 for the detection of vascular structures and CD68 for the detection of macrophages (green fluorescence); nuclei were counterstained with DAPI (blue fluorescence). (B) Representative images of immunofluorescence staining for CD31 or CD68 (green fluorescence) in combination with staining for pSmad2 (red fluorescence); nuclei were counterstained with DAPI (blue fluorescence). (C) Hemoglobin content in plugs. (D) Representative images of immunofluorescence staining for CD68 (green fluorescence) in combination with iNOS or ARG1 (red fluorescence) and DAPI (blue fluorescence). (E) Histology‐based quantification of macrophage infiltration into plugs. The following phenotypes were quantified: CD68 positive cells, CD68/iNOS double‐positive cells, and CD68/ARG1 double‐positive cells. Black scale bars = 1 cm. White scale bars = 100 μm. Data are presented as means ± SEM. ***p < 0.001 vs. CTRL; ## p < 0.01 vs. TEX; + p < 0.05 vs. TEX; & p < 0.01 vs. TEX.
To further evaluate the role of TGFβ in TEX‐mediated signaling, mice received daily injections of 25 or 50 μg of the mRER inhibitor (mRERlow and mRERhigh, respectively). The treatment of mice harboring CTRL plugs with mRER (at either concentration) did not show any differences compared to CTRL alone (Figure 5A and C). The mRER treatment of mice harboring plugs with TEX resulted in a concentration‐dependent decrease of the gross vascularization, lower CD31 signals inside of plugs and a significantly reduced hemoglobin content compared to mice which received no mRER treatment but harbored plugs with TEX (p < 0.01; Figure 5A and C). The infiltration of macrophages was reduced by mRER treatment in a concentration‐dependent manner and was especially reduced in mice treated with mRERhigh (p < 0.05; Figure 5A and E). To further evaluate the phenotype of infiltrating macrophages, double staining for CD68/iNOS was performed to quantify M1 macrophages, while double staining for CD68/ARG1 was performed to quantify M2 macrophages. The data show that M1 macrophages were significantly elevated in TEX and mRERlow‐treated mice but were significantly reduced in TEX and mRERhigh‐treated mice (p < 0.05; Figure 5D and E). Infiltration of M2 macrophages was significantly reduced depending on the concentration of mRER used (p < 0.05; Figure 5D and E).
Our results demonstrate that TEX promoted angiogenesis in vivo and that mRER effectively inhibited these effects. The data explicitly link the pro‐angiogenic functions of TEX to their TGFβ content and TGFβ signaling. Additionally, TGFβ‐rich TEX promoted the infiltration of pro‐angiogenic M2 macrophages which was blocked by mRER.
3.6. TGFβ+ TEX promote tumor progression in a murine oral carcinogenesis model
The in vivo effects of TEX were also tested in the immunocompetent carcinogenesis model, 4‐NQO, previously used by us (Ludwig, Yerneni et al., 2018; Ludwig et al., 2021; Razzo et al., 2019). Mice spontaneously developing orthotopic tumors were sacrificed at week 25 of 4‐NQO treatment, as indicated in Figure 6A. Tumors developed on the tongue of the mice, starting with dysplastic lesions at week 16, early carcinomas at week 18, and advanced carcinomas at week 20 (Figure 6A). TEX isolated from supernatants of HNSCC cell lines were injected intravenously at week 18 (i.e., time of the first carcinomas appearance; Figure 6A), TEX with high levels of bioactive TGFβ (PCI‐13‐derived), low levels of bioactive TGFβ (UMSCC90‐derived) and murine TEX (SCCVII‐derived) were used (see Figure 2D). TEX from all three sources increased number of tumors and the total tumor volume per mouse compared to control mice not injected with TEX, however, only SCCVII‐derived TEX induced statistically significant differences (p < 0.05; Figure 6B and C). In contrast, mRER which was intraperitoneally injected daily starting on week 18, significantly reduced the number of tumors and total tumor volumes (p < 0.05; Figure 6B and C). The levels of TGFβ expression in tumors were unaffected by treatment with UMSCC90‐ or SCCVII‐derived TEX and were slightly lower after treatment with mRER but were significantly enhanced by treatment with PCI‐13‐derived TEX relative to control mice (p < 0.05; Figure 6D and E). TEX from all three sources significantly increased levels of vascularization in 4‐NQO tumors compared to control mice not injected with TEX (Figure 6F and S5; p < 0.05) as also previously described by us (Ludwig, Yerneni et al., 2018). The greatest increase was observed in mice treated with PCI‐13‐derived TEX and the lowest increase after treatment with UMSCC90‐derived TEX, and these results correlated with TGFβ levels carried by the respective TEX (Figures 2D and 6D). The examination of macrophage infiltration indicated that tumors treated with TEX carrying high TGFβ levels (PCI‐13‐ and SCCVII‐derived) were infiltrated with significantly larger numbers of M2‐like (CD68+/ARG1+) macrophages than control mice or mice treated with TEX carrying low TGFβ levels (i.e., UMSCC90‐derived TEX; Figure 6D and G). Levels of infiltrating M2‐like macrophages were reduced in mice treated with mRER (Figure 6D and G). These data show that TGFβ levels in TEX delivered intravenously to tumor‐bearing mice determine the ability of tumors to progress, accelerate angiogenesis and also induce infiltration of macrophages with the tumor‐invasive M2 phenotype.
FIGURE 6.

TGFβ+ TEX promote reprogramming of macrophages of tumors in 4‐NQO‐treated mice. (A) A schematic is provided for 4‐NQO oral administration in water for the initiation of oral carcinomas. Green arrow indicates time‐point for intravenous injection of TEX or daily intraperitoneal injections of mRER into mice. (B) Number of tumors per mouse at the experimental end‐point. (C) Aggregate volumes of tumors in mm3 per mouse at the experimental end‐point. (D) Representative immunofluorescence staining of tumor sections at week 27/28 from the various indicated treatment cohorts for CD31 and CD68 (green fluorescence), TGFβ, iNOS, and ARG1 (red fluorescence) and counterstaining of nuclei with DAPI (blue fluorescence). Scale bars = 100 μm. (E) Quantitative analysis of immunofluorescence staining for TGFβ. All data are expressed as the percentage of the area positively stained from the region of interest (% ROI). (F) Quantitative analysis of immunofluorescence staining for CD31. All data are expressed as the area positively stained from the region of interest (% ROI). (G) Histology‐based quantification of macrophages at the border zones of the tumor. The following phenotypes were quantified: CD68 positive cells, CD68/iNOS double‐positive cells, and CD68/ARG1 double‐positive cells. Values represent means ± SEM. *p < 0.05 vs. CTRL; # p < 0.05 vs. number of CD68+/ARG1+ cells in CTRL.
4. DISCUSSION
The current literature amply illustrates the significant and varied roles of TGFβ in cancer progression (Batlle & Massagué, 2019). The Yin/Yang effects of TGFβ as the tumor suppressor or the tumor promoter have been extensively documented and may be dependent on the tumor type, stage, aggressiveness, or various host factors (Syed, 2016). Like many other solid tumors, HNSCCs produce, express, and release TGFβ into body fluids. Previous reports have shown that TGFβ carried by TEX induces Smad‐dependent signaling and pro‐tumor reprogramming in the TME (Webber et al., 2010). Specifically, TEX‐associated TGFβ was shown to trigger the formation of tumor‐associated fibroblasts, alter immune cell functions, and initiate pre‐metastatic niche formation (Costa‐Silva et al., 2015; Hong et al., 2014; Webber et al., 2010). In this study, we focused on the pro‐angiogenic role of TGFβ+ TEX. These small, virus‐size EVs were isolated from supernatants of HNSCC cell lines (Ludwig, Yerneni et al., 2018). Unlike EVs in cancer patients’ plasma, vesicles produced by cultured tumor cells are “pure” TEX, and they serve in this study as an experimental model for TGFβ+ TEX accumulating in cancer patients’ plasma or the TME.
Our proteomics analysis of TEX showed that TGFβ is among the proteins participating in the promotion of angiogenesis in HNSCC. As angiogenesis is the sine qua non of the tumor development and progression, TEX‐associated TGFβ emerges as a significant component of the tumorigenesis. Interestingly, the angiogenesis‐promoting TGFβ pathway was induced by TEX not only in ECs but simultaneously in other cell types known to reside in the TME, such as macrophages. Thus, TEX‐associated TGFβ can induce a direct pro‐angiogenic pathway in ECs and/or reprogram macrophages to a pro‐angiogenic phenotype and indirectly promote angiogenesis. Such reprogramming of macrophages in the TME by TGFβ+ TEX was also reported by Costa‐Silva et al. (Costa‐Silva et al., 2015). However, while these data suggest that macrophages stimulated by TEX acquire an M2 phenotype, the TEX‐induced reprogramming from M1 to M2 might not always be complete, resulting in a mixed M1/M2 phenotype. Specific molecular mechanisms involved in the reprogramming of macrophages by TGFβ+ TEX require further analysis. Nevertheless, by ready access to all cells in the TME, TEX carrying TGFβ are likely to be highly effective in reprogramming cellular functions, especially when these cells express TGFβ receptors. For instance, CD4(+) and CD8(+) T cells might be affected by TEX‐associated TGFβ signalling since the injection of TGFβ+ TEX into tumor‐bearing 4‐NQO mice decreased their infiltration abundance in the tumor tissue (Razzo et al., 2019).
TEX released by solid tumors, including HNSCC, are known to carry multiple proteins or miRNAs that promote tumor progression (Ludwig & Whiteside, 2018). Recent literature suggests several TEX‐associated factors which are involved in the reprogramming of macrophages in HNSCC including miR‐9 (Tong et al., 2020), CMTM6 (Pang et al., 2021), CDC37/HSP90α/HSP90β (Ono et al., 2020), PD‐L1 (Yuan et al., 2022) or THBS1 (Chen et al., 2018). While these reports are not focused on the pro‐angiogenic functions of macrophages, we were able to show that TEX‐associated adenosine is also involved in the generation of pro‐angiogenic macrophages in HNSCC (Ludwig et al., 2020). Thus, TEX carry signals able to activate various molecular pathways which contribute to macrophage reprogramming. The involvement of additional factors besides TGFβ was also emphasized in our experiments since TGFβlow TEX still possessed considerable biological activity that was only partially inhibited by mRER. It will be important to study potential synergies between the TGFβ pathway and pathways of the other discovered factors to potently block the TEX‐induced effects in HNSCC.
The in vitro mechanistic and in vivo studies of TGFβ+ TEX we performed illustrate angiogenesis promoting and tumor‐growth enhancing activities of TEX in HNSCC. We showed that TGFβ is not only present on the surface of TEX but is also detected in the vesicle lumen. While the surface‐associated TGFβ is signaling‐competent and may be mostly responsible for inducing recipient cell reprogramming, intra‐vesicular TGFβ is carried and delivered to near and distant sites in a protected, stable form (Whiteside, 2016). It has been suggested that vesicular membrane‐associated proteins may be more stable than their soluble counterparts (Ludwig, Marczak et al., 2019) and thus may have stronger bioactivity. However, future studies need to carefully address the possibility that inhibition of endocytosis, as shown in Figure 2E, may impact the recycling of TGFβ receptors from the cell surface. Increased levels of TGFβ receptors present at the cell surface might also increase cellular responses to TEX‐associated TGFβ.
In performing the described experiments, we strived to avoid potential effects of species‐dependent differences between human and murine TGFβ+ TEX. Therefore, in all in vitro assays, TEX and recipient cells were species matched, and for in vivo assays, murine TEX served as controls. Mature human TGFβ1 shares 99% amino acid identity with mouse TGFβ1 and demonstrates cross‐species activity as previously observed (Derynck & Miyazono, 2017). Also, TGFβ stimulation in human and murine cells activates common biological processes and pathways at the transcriptional level (Abnaof et al., 2014). Active TGFβ1 can be detected in human and murine TEX; however, to the best of our knowledge, no distinct species‐dependent characteristics of TGFβ+ TEX were reported in the literature.
While clustering of the tumor cell lines based on the levels of active TGFβ in TEX was functionally relevant, we were unable to detect any clear differences between the pro‐angiogenic functions of TEX isolated from HPV(+) and HPV(‐) HNSCC cells lines. The proteomic cargo components as well as the results of the functional in vitro and in vivo assays did not reveal significant differences in TEX produced by these cell lines. The results are counterintuitive in the light of recent studies reporting major differences in the TME, the immune landscape, and the therapeutic aspects of HPV(+) versus HPV(‐) HNSCC (Cillo et al., 2020; Eberhardt et al., 2021; Ferris et al., 2021). Our previous studies showed that the TEX protein profiles resemble those of respective parent HPV(+) or HPV(‐) tumor cells, and that this distinct protein profile might reflect distinct immunoregulatory functions (Ludwig, Sharma et al., 2018; Ludwig, Marczak et al., 2019). Therefore, future studies are necessary to compare TEX produced by tumor cell lines and circulating total EVs in HPV(+) and HPV(‐) samples, derived either from tumor biopsies or isolated from cancer patients’ blood.
A large body of evidence suggested that targeting angiogenesis could provide a clinical benefit in the treatment of solid tumors and, therefore, two major categories of agents have been developed to target this pathway: antibody‐based agents and VEGF receptor tyrosine kinase inhibitors (TKIs) (Hyytiäinen et al., 2021; Munn & Jain, 2019). While the US Food and Drug Administration (FDA) approved several anti‐angiogenic agents to treat solid tumours, there are no FDA‐approved anti‐angiogenic drugs for HNSCC, although these cancers arise in highly vascularized tissues (Salem et al., 2021). Most clinical studies did not show benefit of angiogenesis inhibitors in HNSCC treatment and additionally, angiogenesis inhibitors were associated with considerable toxicity (Hyytiäinen et al., 2021). Nevertheless, newer results are more encouraging and, therefore, novel anti‐angiogenic agents and combinatorial treatment strategies are under intense investigation. Specifically, the combinations of immune checkpoint inhibitors with anti‐angiogenic agents are actively being pursued, including ongoing phase II trials of ramucirumab plus pembrolizumab and bevacizumab plus atezolizumab in recurrent/metastatic HNSCC (NCT03650764, NCT03818061) (Micaily et al., 2020). In view of these developments, perhaps the most important results that emerge from our preclinical experiments relate to in vivo blocking of TEX‐induced angiogenesis and macrophage infiltration into the tumors by a novel TGFβ trap, mRER (Qin et al., 2016; Zhu et al., 2018). mRER, which binds to TGFβ1, β2, and β3 isoforms and blocks their respective interactions with the cognate receptors, inhibited signaling of TGFβ+ TEX in vitro and in vivo. This mRER‐mediated silencing of TGFβ signaling not only confirms that TEX binding to cells is the receptor‐ligand interaction, but it also introduces mRER as a promising inhibitor able to reverse the adverse pro‐angiogenic stimuli delivered by TGFβ+ TEX. Our data showed that among the known TGFβ inhibitors, mRER had the best potential for inhibiting pro‐angiogenic and tumor growth‐promoting effects of TGFβ+ TEX. Given the multifaceted role of TGFβ in driving malignant progression, for example by increasing tumor invasiveness and metastasis, the blockade of TGFβ in HNSCC appears promising also indicated by the significant reduction of tumor burden below the control group in 4‐NQO mice. Analogous to our findings, previous studies reported that RER inhibited early‐stage tumorigenesis and tumor cell invasion in murine PTEN‐deficient prostate glands (Qin et al., 2016). Also, RER was shown to block chemotherapeutics‐induced TGFβ1 signaling and enhance their anti‐cancer activity in gynecologic cancers (Zhu et al., 2018). The therapeutic assessments of TGFβ inhibitors, such as mRER, that target not only soluble but also membrane‐tethered isoforms of TGFβ carried by TEX are in progress (Kim et al., 2017; Qin et al., 2016; Zhu et al., 2018). The future clinical use of TGFβ inhibitors will require the selection of agents that are safe and inhibit functions of most forms of TGFβ, including those carried by TEX accumulating in body fluids of patients with cancer. The expectation is that such inhibitors in combination with current therapeutic approaches, including the PD‐L1 blockade (Ludwig et al., 2021; Mariathasan et al., 2018), will be highly effective in improving response to therapy and may represent a new category of anti‐angiogenic agents.
The role of TEX in the promotion of tumor progression is of great current interest, as is the development of strategies for silencing metastasis‐promoting TEX (Bhatta et al., 2021). Our results emphasize the key pro‐angiogenic role of TGFβ+ TEX and the importance of suppressing TEX‐mediated pro‐tumor/pro‐angiogenic activities for anti‐tumor strategies. TEX have been reported to carry biologically‐active VEGF (Ko et al., 2019; Skog et al., 2008) in addition to TGFβ, PD‐L1 (Sharma et al., 2020), and other proteins or miRNAs facilitating tumor progression (Ludwig & Whiteside, 2018). This emphasizes the critically important pro‐tumor role TEX play in cancer and the need for therapeutic silencing of circulating TEX. Inadequate therapeutic benefits of anti‐angiogenic agents in patients with HNSCC suggest that more effective inhibitors of angiogenesis or novel combinatorial therapies are needed (Hyytiäinen et al., 2021).
AUTHOR CONTRIBUTIONS
Nils Ludwig: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Writing – original draft; Writing – review & editing. Saigopalakrishna S. Yerneni: Data curation; Formal analysis; Investigation; Methodology; Writing – review & editing. Juliana H. Azambuja: Data curation; Formal analysis; Investigation; Writing – review & editing; Monika Pietrowska: Data curation; Formal analysis; Funding acquisition; Investigation; Methodology. Piotr Widłak: Supervision; Writing – review & editing. Cynthia S. Hinck: Data curation; Methodology. Alicja Głuszko: Data curation; Investigation; Writing – review & editing. Mirosław J. Szczepański: Investigation; Resources; Supervision; Writing – review & editing. Teresa Kärmer: Data curation; Investigation. Isabella Kallinger: Data curation; Investigation. Daniela Schulz: Formal analysis; Supervision. Richard J. Bauer: Resources; supervision; Writing – review & editing Gerrit Spanier: supervision; Writing – review & editing. Steffen Spoerl: Supervision; Writing – review & editing. Johannes K. Meier: Supervision; Writing – review & editing. Tobias Ettl: supervision; Writing – review & editing. Beatrice M. Razzo: Data curation; formal analysis; Investigation. Torsten E. Reichert: Resources; Supervision; Writing – review & editing. Andrew P. Hinck: Methodology; Resources; Supervision; Writing – review & editing. Theresa L. Whiteside: Conceptualization; Project administration; Funding acquisition; Resources; Supervision; Writing – review & editing.
CONFLICT OF INTERESTS
Dr. Andrew Hinck is the Co‐Inventor of RER, which is covered by U.S. patent 9,611,306, and holds royalty rights for the clinical deployment of RER, which is currently being pursued.
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants R01‐CA 168628 and U01‐DE‐029759 to TLW. NL was supported by the Leopoldina Fellowships LPDS 2017‐12 and LPDR 2019‐02 from German National Academy of Sciences Leopoldina and by the Walter Schulz Foundation. AG was supported by Medical University of Warsaw MB/M/48(79)#. MJS was supported by National Science Centre, Poland UMO‐2017/26/M/NZ5/00877#. MP who performed LC‐MS/MS analyses was supported by the National Science Centre, Poland, grant no. 2016/22/M/NZ5/00667. TK and IK were supported by the Verein zur Förderung der wissenschaftlichen Zahnheilkunde (VFwZ).
Ludwig, N. , Yerneni, S. S. , Azambuja, J. H. , Pietrowska, M. , Widłak, P. , Hinck, C. S. , Głuszko, A. , Szczepański, M. J. , Kärmer, T. , Kallinger, I. , Schulz, D. , Bauer, R. J. , Spanier, G. , Spoerl, S. , Meier, J. K. , Ettl, T. , Razzo, B. M. , Reichert, T. E. , Hinck, A. P. , & Whiteside, T. L. (2022). TGFβ+ small extracellular vesicles from head and neck squamous cell carcinoma cells reprogram macrophages towards a pro‐angiogenic phenotype. Journal of Extracellular Vesicles, 11, e12294. 10.1002/jev2.12294
REFERENCES
- Abnaof, K. , Mallela, N. , Walenda, G. , Meurer, S. K. , Seré, K. , Lin, Q. , Smeets, B. , Hoffmann, K. , Wagner, W. , Zenke, M. , Weiskirchen, R. , & Fröhlich, H. (2014). TGF‐β stimulation in human and murine cells reveals commonly affected biological processes and pathways at transcription level. BMC Systems Biology, 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle, E. , & Massagué, J. (2019). Transforming Growth Factor‐β Signaling in Immunity and Cancer. Immunity, 50, 924–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatta, M. , Shenoy, G. N. , Loyall, J. L. , Gray, B. D. , Bapardekar, M. , Conway, A. , Minderman, H. , Kelleher Jr, R. J. , Carreno, B. M. , Linette, G. , Shultz, L. D. , Odunsi, K. , Balu‐Iyer, S. V. , Pak, K. Y. , & Bankert, R. B. (2021). Novel phosphatidylserine‐binding molecule enhances antitumor T‐cell responses by targeting immunosuppressive exosomes in human tumor microenvironments. Journal for ImmunoTherapy of Cancer, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet, P. (2003). Angiogenesis in health and disease. Nature Medicine, 9, 653–660. [DOI] [PubMed] [Google Scholar]
- Carmeliet, P. , & Jain, R. K. (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature, 473, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Xiao, M. , Zhang, J. , & Chen, W. (2018). M1‐like tumor‐associated macrophages activated by exosome‐transferred THBS1 promote malignant migration in oral squamous cell carcinoma. Journal of Experimental & Clinical Cancer Research, 37, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo, A. R. , Kürten, C. H. L. , Tabib, T. , Qi, Z. , Onkar, S. , Wang, T. , Liu, A. , Duvvuri, U. , Kim, S. , Soose, R. J. , Oesterreich, S. , Chen, W. , Lafyatis, R. , Bruno, T. C. , Ferris, R. L. , & Vignali, D. A. A. (2020). Immune Landscape of Viral‐ and Carcinogen‐Driven Head and Neck Cancer. Immunity, 52, 183–199.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa‐Silva, B. , Aiello, N. M. , Ocean, A. J. , Singh, S. , Zhang, H. , Thakur, B. K. , Becker, A. , Hoshino, A. , Mark, M. T. , Molina, H. , Xiang, J. , Zhang, T. , Theilen, T.‐. M. , García‐Santos, G. , Williams, C. , Ararso, Y. , Huang, Y. , Rodrigues, G. , Shen, T.‐. L. , … Lyden, D. (2015). Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nature Cell Biology, 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denardo, D. G. , & Ruffell, B. (2019). Macrophages as regulators of tumour immunity and immunotherapy. Nature Reviews Immunology, 19, 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck, R. , & Miyazono, K. (2017). The Biology of the TGF‐β Family. Cold Spring Harbor Perspectives in Biology. [DOI] [PMC free article] [PubMed]
- Eberhardt, C. S. , Kissick, H. T. , Patel, M. R. , Cardenas, M. A. , Prokhnevska, N. , Obeng, R. C. , Nasti, T. H. , Griffith, C. C. , Im, S. J. , Wang, X. , Shin, D. M. , Carrington, M. , Chen, Z. G. , Sidney, J. , Sette, A. , Saba, N. F. , Wieland, A. , & Ahmed, R. (2021). Functional HPV‐specific PD‐1+ stem‐like CD8 T cells in head and neck cancer. Nature, 597, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekambaram, P. , Lee, J. L. , Hubel, N. E. , Hu, D. , Yerneni, S. , Campbell, P. G. , Pollock, N. , Klei, L. R. , Concel, V. J. , Delekta, P. C. , Chinnaiyan, A. M. , Tomlins, S. A. , Rhodes, D. R. , Priedigkeit, N. , Lee, A. V. , Oesterreich, S. , Mcallister‐Lucas, L. M. , & Lucas, P. C. (2018). The CARMA3‐Bcl10‐MALT1 Signalosome Drives NF‐κB Activation and Promotes Aggressiveness in Angiotensin II Receptor‐positive Breast Cancer. Cancer Research, 78, 1225–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi, M. , Joseph, R. , Adolacion, J. R. T. , Martinez‐Paniagua, M. , An, X. , Gabrusiewicz, K. , Mani, S. A. , & Varadarajan, N. (2021). Single‐cell cloning of breast cancer cells secreting specific subsets of extracellular vesicles. Cancers (Basel), 13, 4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, R. L. , Spanos, W. C. , Leidner, R. , Gonã§Alves, A. , Martens, U. M. , Kyi, C. , Sharfman, W. , Chung, C. H. , Devriese, L. A. , Gauthier, H. , Chiosea, S. I. , Vujanovic, L. , Taube, J. M. , Stein, J. E. , Li, J. , Li, B. , Chen, T. , Barrows, A. , & Topalian, S. L. (2021). Neoadjuvant nivolumab for patients with resectable HPV‐positive and HPV‐negative squamous cell carcinomas of the head and neck in the CheckMate 358 trial. Journal for ImmunoTherapy of Cancer, 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głuszko, A. , Szczepański, M. J. , Whiteside, T. L. , Reichert, T. E. , Siewiera, J. , & Ludwig, N. (2021). Small extracellular vesicles from head and neck squamous cell carcinoma cells carry a proteomic signature for tumor hypoxia. Cancers (Basel), 13, 4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, D. S. , Snyderman, C. , Gollin, S. M. , Pan, S. , Walker, E. , Deka, R. , Barnes, E. L. , Johnson, J. T. , Herberman, R. B. , & Whiteside, T. L. (1989). Biology, Cytogenetics, and Sensitivity to Immunological Effector Cells of New Head and Neck Squamous Cell Carcinoma Lines. Cancer Research, 49, 5167–5175. [PubMed] [Google Scholar]
- Hong, C.‐. S. , Muller, L. , Whiteside, T. L. , & Boyiadzis, M. (2014). Boyiadzis M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Frontiers in immunology, 5, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , Mark, M. T. , Steiner, L.‐. C. , Benito‐Martin, A. , Lucotti, S. , Di Giannatale, A. , Offer, K. , Nakajima, M. , Williams, C. , Nogués, L. , … Lyden, D. (2020). Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell, 182, 1044–1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytiäinen, A. , Wahbi, W. , Väyrynen, O. , Saarilahti, K. , Karihtala, P. , Salo, T. , & Al‐Samadi, A. (2021). Angiogenesis Inhibitors for Head and Neck Squamous Cell Carcinoma Treatment: Is There Still Hope? Frontiers in oncology, 11, 683570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. K. , Barron, L. , Hinck, C. S. , Petrunak, E. M. , Cano, K. E. , Thangirala, A. , Iskra, B. , Brothers, M. , Vonberg, M. , Leal, B. , Richter, B. , Kodali, R. , Taylor, A. B. , Du, S. , Barnes, C. O. , Sulea, T. , Calero, G. , Hart, P. J. , Hart, M. J. , … Hinck, A. P. (2017). An engineered transforming growth factor β (TGF‐β) monomer that functions as a dominant negative to block TGF‐β signaling. Journal of Biological Chemistry, 292, 7173–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, S. Y. , Lee, W. , Kenny, H. A. , Dang, L. H. , Ellis, L. M. , Jonasch, E. , Lengyel, E. , & Naora, H. (2019). Cancer‐derived small extracellular vesicles promote angiogenesis by heparin‐bound, bevacizumab‐insensitive VEGF, independent of vesicle uptake. Communications Biology, 2, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucotti, S. , Kenific, C. M. , Zhang, H. , & Lyden, D. C. (2022). Extracellular vesicles and particles impact the systemic landscape of cancer. EMBO Journal, 41, e109288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N. , Hong, C. S. , Ludwig, S. , Azambuja, J. H. , Sharma, P. , Theodoraki, M. N. , & Whiteside, T. L. (2019). Isolation and Analysis of Tumor‐Derived Exosomes. Current protocols in immunology, 127, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N. , Razzo, B. M. , Yerneni, S. S. , & Whiteside, T. L. (2019). Optimization of cell culture conditions for exosome isolation using mini‐size exclusion chromatography (mini‐SEC). Experimental Cell Research, 378, 149–157. [DOI] [PubMed] [Google Scholar]
- Ludwig, N. , & Whiteside, T. L. (2018). Potential Roles of Tumor‐derived Exosomes in Angiogenesis. Expert Opinion on Therapeutic Targets, 22, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N. , Wieteska, Ł. , Hinck, C. S. , Yerneni, S. S. , Azambuja, J. H. , Bauer, R. J. , Reichert, T. E. , Hinck, A. P. , & Whiteside, T. L. (2021). Novel TGFβ Inhibitors Ameliorate Oral Squamous Cell Carcinoma Progression and Improve the Antitumor Immune Response of Anti–PD‐L1 Immunotherapy. Molecular Cancer Therapeutics, 20, 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N. , Yerneni, S. S. , Azambuja, J. H. , Gillespie, D. G. , Menshikova, E. V. , Jackson, E. K. , & Whiteside, T. L. (2020). Tumor‐derived exosomes promote angiogenesis via adenosine A2B receptor signaling. Angiogenesis, 23, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N. , Yerneni, S. S. , Razzo, B. M. , & Whiteside, T. L. (2018). Exosomes from HNSCC Promote Angiogenesis through Reprogramming of Endothelial Cells. Molecular Cancer Research, 16, 1798–1808. [DOI] [PubMed] [Google Scholar]
- Ludwig, S. , Floros, T. , Theodoraki, M. ‐N. , Hong, C. ‐S. , Jackson, E. K. , Lang, S. , & Whiteside, T. L. (2017). Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clinical Cancer Research, 23, 4843–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S. , Marczak, L. , Sharma, P. , Abramowicz, A. , Gawin, M. , Widlak, P. , Whiteside, T. L. , & Pietrowska, M. (2019). Proteomes of exosomes from HPV(+) or HPV(‐) head and neck cancer cells: Differential enrichment in immunoregulatory proteins. Oncoimmunology, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S. , Sharma, P. , Theodoraki, M.‐. N. , Pietrowska, M. , Yerneni, S. S. , Lang, S. , Ferrone, S. , & Whiteside, T. L. (2018). Molecular and Functional Profiles of Exosomes From HPV(+) and HPV(−) Head and Neck Cancer Cell Lines. Frontiers in oncology, 8, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan, S. , Turley, S. J. , Nickles, D. , Castiglioni, A. , Yuen, K. , Wang, Y. , Kadel Iii, E. E. , Koeppen, H. , Astarita, J. L. , Cubas, R. , Jhunjhunwala, S. , Banchereau, R. , Yang, Y. , Guan, Y. , Chalouni, C. , Ziai, J. , Şenbabaoğlu, Y. , Santoro, S. , Sheinson, D. , … Powles, T. (2018). TGF‐β attenuates tumour response to PD‐L1 blockade by contributing to exclusion of T cells. Nature, 554, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micaily, I. , Johnson, J. , & Argiris, A. (2020). An update on angiogenesis targeting in head and neck squamous cell carcinoma. Cancers Head Neck, 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, L. A. , Pink, R. C. , & Carter, D. R. F. (2014). Routes and mechanisms of extracellular vesicle uptake. Journal of extracellular vesicles, 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn, L. L. , & Jain, R. K. (2019). Vascular Regulation of Anti‐Tumor Immunity. Science (80‐), 365, 544–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A. M. , Liu, C. L. , Green, M. R. , Gentles, A. J. , Feng, W. , Xu, Y. , Hoang, C. D. , Diehn, M. , & Alizadeh, A. A. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, K. , & Edidin, M. (1990). A mouse lymphoid endothelial cell line immortalized by simian virus 40 binds lymphocytes and retains functional characteristics of normal endothelial cells. Journal of Immunology, 144, 521–525. [PubMed] [Google Scholar]
- Ono, K. , Sogawa, C. , Kawai, H. , Tran, M. T. , Taha, E. A. , Lu, Y. , Oo, M. W. , Okusha, Y. , Okamura, H. , Ibaragi, S. , Takigawa, M. , Kozaki, K. I. , Nagatsuka, H. , Sasaki, A. , Okamoto, K. , Calderwood, S. K. , & Eguchi, T. (2020). Triple knockdown of CDC37, HSP90‐alpha and HSP90‐beta diminishes extracellular vesicles‐driven malignancy events and macrophage M2 polarization in oral cancer. Journal of extracellular vesicles, 9, 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, X. , Wang, S. ‐S. , Zhang, M. , Jiang, J. , Fan, H. ‐Y. , Wu, J. ‐S. , Wang, H. ‐F. , Liang, X. ‐H. , & Tang, Y. ‐L. (2021). OSCC cell‐secreted exosomal CMTM6 induced M2‐like macrophages polarization via ERK1/2 signaling pathway. Cancer Immunology, Immunotherapy, 70, 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, T. , Barron, L. , Xia, L. , Huang, H. , Villarreal, M. M. , Zwaagstra, J. , Collins, C. , Yang, J. , Zwieb, C. , Kodali, R. , Hinck, C. S. , Kim, S. K. , Reddick, R. L. , Shu, C. , O'Connor‐McCourt, M. D. , Hinck, A. P. , & Sun, L. ‐Z. (2016). A novel highly potent trivalent TGF‐beta receptor trap inhibits early‐stage tumorigenesis and tumor cell invasion in murine Pten‐deficient prostate glands. Oncotarget, 7, 86087–86102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzo, B. M. , Ludwig, N. , Hong, C. , Sharma, P. , Fabian, K. P. , Fecek, R. J. , Storkus, W. J. , & Whiteside, T. L. (2019). Tumor‐derived exosomes promote carcinogenesis of murine oral squamous cell carcinoma. Carcinogenesis, 41(5), 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, N. E. , Discafani, C. M. , Downs, E. C. , Hailey, J. A. , Sarre, O. , Runkle, R. L. , Popper, T. L. , & Plunkett, M. L. (1991). A Quantitative in Vivo Mouse Model Used to Assay Inhibitors of Tumor‐induced Angiogenesis. Cancer Research, 51, 1339–1344. [PubMed] [Google Scholar]
- Salem, A. , Hadler‐Olsen, E. , & Al‐Samadi, A. (2021). Editorial: Angiogenesis and Angiogenesis Inhibitors in Oral Cancer. Frontiers in Oral Health, 2, 816963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S. , Vasaikar, S. , Eskaros, A. , Kim, Y. , Lewis, J. S. , Zhang, B. , Zijlstra, A. , & Weaver, A. M. (2019). EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI insight, 4, e132447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P. , Diergaarde, B. , Ferrone, S. , Kirkwood, J. M. , & Whiteside, T. L. (2020). Melanoma cell‐derived exosomes in plasma of melanoma patients suppress functions of immune effector cells. Scientific Reports, 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P. , Ludwig, S. , Muller, L. , Hong, C. S. , Kirkwood, J. M. , Ferrone, S. , & Whiteside, T. L. (2018). Immunoaffinity‐based isolation of melanoma cell‐derived exosomes from plasma of patients with melanoma. Journal of extracellular vesicles, 7, 1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog, J. , Würdinger, T. , Van Rijn, S. , Meijer, D. H. , Gainche, L. , Curry, W. T. , Carter, B. S. , Krichevsky, A. M. , & Breakefield, X. O. (2008). Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nature Cell Biology, 10, 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, B. H. , & Weaver, A. M. (2017). Exosome secretion promotes chemotaxis of cancer cells. Cell Adhesion Migration, 11, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed, V. (2016). TGF‐β Signaling in Cancer. Journal of Cellular Biochemistry, 117, 1279–1287. [DOI] [PubMed] [Google Scholar]
- Tang, Z. , Kang, B. , Li, C. , Chen, T. , & Zhang, Z. (2019). GEPIA2: An enhanced web server for large‐scale expression profiling and interactive analysis. Nucleic acids research, 47, W556–W560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesseur, I. , Zou, K. , Berber, E. , Zhang, H. , & Wyss‐Coray, T. (2006). Highly sensitive and specific bioassay for measuring bioactive TGF‐β. BMC Cell Biology [Electronic Resource], 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. ‐. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2019). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles, 8, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, T. , Zhu, Y. ‐L. , Zhou, Y. ‐Y. , Liang, G. ‐F. , Wang, Y. ‐Y. , Hu, F. ‐H. , & Xiao, Z. ‐D. (2014). Exosome uptake through clathrin‐mediated endocytosis and macropinocytosis and mediating miR‐21 delivery. Journal of Biological Chemistry, 289, 22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, F. , Mao, X. , Zhang, S. , Xie, H. , Yan, B. , Wang, B. , Sun, J. , & Wei, L. (2020). HPV+ HNSCC‐derived exosomal miR‐9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Letters, 478, 34–44. [DOI] [PubMed] [Google Scholar]
- Webber, J. , Steadman, R. , Mason, M. D. , Tabi, Z. , & Clayton, A. (2010). Cancer Exosomes Trigger Fibroblast to Myofibroblast Differentiation. Cancer Research, 70, 9621–9630. [DOI] [PubMed] [Google Scholar]
- Webber, J. , Steadman, R. , Mason, M. D. , Tabi, Z. , & Clayton, A. (2010). Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Research, 70, 9621–9630. [DOI] [PubMed] [Google Scholar]
- Whiteside, T. L. (2016). Tumor‐Derived Exosomes and Their Role in Cancer Progression. Advances in Clinical Chemistry, 74, 103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H. , & Wong, D. T. W. (2012). Proteomic analysis of microvesicles in human saliva by gel electrophoresis with liquid chromatography‐mass spectrometry. Analytica Chimica Acta, 723, 61–67. [DOI] [PubMed] [Google Scholar]
- Yerneni, S. S. , Whiteside, T. L. , Weiss, L. E. , & Campbell, P. G. (2019). Bioprinting exosome‐like extracellular vesicle microenvironments. Bioprinting, 13, 1–12. [Google Scholar]
- Yuan, Y. , Jiao, P. , Wang, Z. , Chen, M. , Du, H. , Xu, L. , Xu, J. , Dai, Y. , Wu, F.‐. G. , Zhang, Y. , & Wu, H. (2022). Endoplasmic reticulum stress promotes the release of exosomal PD‐L1 from head and neck cancer cells and facilitates M2 macrophage polarization. Cell communication and signaling : CCS, 20, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Gu, X. , Xia, L. , Zhou, Y. , Bouamar, H. , Yang, J. , Ding, X. , Zwieb, C. , Zhang, J. , Hinck, A. P. , Sun, L.‐Z. , & Zhu, X. (2018). A novel TGFβ trap blocks chemotherapeutics‐induced TGFβ1 signaling and enhances their anticancer activity in gynecologic cancers. Clinical Cancer Research, 24, 2780–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information


