Abstract
Histone proteins are highly conserved among all eukaryotes. They have two important functions in the cell: to package the genomic DNA and to regulate gene accessibility. Fundamental to these functions is the ability of histone proteins to interact with DNA and to form the nucleoprotein complex called chromatin. One of the mechanisms the cells use to regulate chromatin and gene expression is through replacing canonical histones with their variants at specific loci to achieve functional consequence. Recent cryo-electron microscope (cryo-EM) studies of chromatin containing histone variants reveal new details that shed light on how variant-specific features influence the structures and functions of chromatin. In this article, we review the current state of knowledge on histone variants biochemistry and discuss the implication of these new structural information on histone variant biology and their functions in transcription.
Keywords: Electron microscopy, Protein structure, Histone variant, Nucleosome, Chromatin
1. Introduction
Proper gene expression is crucial for normal cellular function and organism development. In eukaryotes, the nucleoprotein complex chromatin provides a sophisticated system to control the accessibility of genetic information and, thus the transcription outcome. The nucleosome, the basic building block of chromatin, contains 147 base pairs (bps) of DNA wrapping around an octameric core of histone proteins in 1.7 turns. The incorporation of histone variants (the nonallelic isoforms of canonical histones) is a known epigenetic mechanism to introduce changes into nucleosomes, altering chromatin's chemical and physical properties. Yet, crystal structures of most variant-containing nucleosomes show a highly conserved and relatively stable structural fold that appears insensitive to modification on histones. In recent years there has been a renewed interest in understanding the molecular mechanism of histone variant-dependent chromatin regulation and transcriptional control. It is now well-recognized that histone variants are involved in a plethora of nuclear processes and they are an important part of a cell's epigenetic memory.
Recent advancements in the cryo-EM technique have greatly benefited many areas in biology. Single-particle cryo-EM has been a game changer in our ability to address many long-standing questions in the chromatin field. Compared to X-ray crystallography, cryo-EM can be used to image biological specimens in their native state, which also eliminates the requirement for absolute homogeneity of the sample and allows the sampling of various conformational states of the system [1]. In addition, the advancements in detector technology and image processing algorithm enable single-particle cryo-EM to be used routinely for structural determination of macromolecules at near-atomic resolution. Take histone variant biology for example, recent cryo-EM studies have revealed structural features at both nucleosome and chromatin fiber not observed in previous crystallography studies. In this review, we briefly introduce the hierarchical organization of chromatin and histone variants in transcription. We focus on the most recent cryo-EM discoveries related to histone variants, with particular emphasis on the molecular properties underlying variant-specific changes in chromatin. We end with a discussion on how this information sheds light on our understanding of the mechanism of action of histone variants in vivo and how it shapes our future endeavors in histone variant research.
2. Chromatin organization and dynamics
The histone octamer in a nucleosome contains four dimers defined by H3-H4 and H2A-H2B histone pairs. In the center of the DNA turn along the dyad, two H3-H4 pairs interact through a 4-helix bundle to form the H3-H4 tetramer [2]. Two H2A-H2B heterodimers interact with the H3-H4 tetramer through a similar 4-helix bundle between H2B and H4, forming the histonefold regions that interact with DNA from superhelix location (SHL) −6 to +6 on the nucleosome. The histonefold extension of histone H3 ⍺N helix, with support from the H2A C-terminal docking domain, interacts with DNA at the entry/exit site around the octamer. The H3 aN helix and its preceding N-terminal tail directly contact and stabilize the entry/exit DNA [2]. On the other hand, histones elements on the surface of a nucleosome, such as the L1 loop and the acidic patch (a highly contoured and negatively charged nucleosome surface formed by six H2A residues and two H2B residues) [2], are known to take part in the interactions with nucleosome binding proteins and chromatin regulating enzymes. Linker histone H1, a fifth family of histone and the most abundant chromatin-associated protein, is an integral part of the chromatin higher-order structure, though its precise function remains elusive. Histone H1 stabilizes the nucleosome core particle (NCP) and the chromatin high-order structure [3]. Most eukaryotes have an average of 0.5 to 1.3 H1 per nucleosome [4]. A nucleosome with a bound linker histone H1 is called a chromatosome, where H1 sits at the nucleosome dyad near the DNA entry/exit site.
Oligo-nucleosomes with various linker DNA lengths are often referred to as the 10-nm fiber or nucleosome arrays. This bead-on-a-string nucleosome array represents the transcriptionally active and open form of chromatin, the euchromatin. In vitro, repetitive and highly saturated arrays with strong nucleosome positioning sequences and even spacing can condense into rod-like 30-nm fiber structures in the presence of linker histone H1 or certain divalent cations [5], [6], [7], [8]. Yet, the detailed structure of heterochromatin (the condensed and transcriptionally silent form of chromatin) in vivo is not fully understood [5]. The field, which relies heavily on repetitive nucleosome arrays [9], has evolved over the years regarding the subject. Early studies using X-ray diffraction and Electron Microscopy (EM) supported the existence of 30-nm fiber in diverse nuclei [10], [11], [12]. Several recent microscopy-based studies, however, show that nucleosome arrays in the nuclei generally do not fold into 30-nm fibers [13]. Instead, chromatin in vivo is heterogeneous and amorphous [14], existing as disordered fibers with a range of diameters [15] and at a nanoscale level as local clusters/clutches of short nucleosome stretches [14], [16], [17]. Nevertheless, the in-vitro reconstituted fibers are important as they remain a vital tool for investigating the intrinsic properties of nucleosome array folding and chromatin compaction in the absence of other chromatin constituencies. Lastly, it is worth noting that the same sets of regulatory proteins that influence nucleosome dynamics also alter chromatin higher-order structures and dynamics.
3. Overview of histone variants
Unlike canonical histones expressed from gene clusters in a cell cycle-dependent manner, most histone variants express throughout the cell cycle [18]. Different variants also have distinct genome localization patterns, further underlying their essential and specialized cellular functions. For example, the incorporation of variant H2A.Z at a particular genomic region is shown to facilitate RNA Polymerase II passing and transcription initiation [19]. In addition, histone variant incorporation has many profound implications for the chemical and physical characteristics of the nucleosome [20], as it can erase the existing histone post-translational modifications (PTMs) and affects non-histone proteins binding and their functions. Multiple lines of evidence also show that histone variant incorporations influence chromatin higher-order structures both in vivo and in vitro [21], [22], [23].
Histone variant exchange, a process to deposit or evict the specific variant, is energy-dependent and typically requires the action of histone chaperones or the ATP-dependent chromatin remodelers. Therefore, it is not surprising that the traditional salt-dialysis protocol for in-vitro nucleosome reconstitution is inefficient for some variant nucleosomes [24], [25] in the absence of these enzymes. Chaperons are structurally diverse proteins that bind the histone dimer pair (H3-H4 or H2A-H2B) and facilitate their deposition or eviction from nucleosomes. Though there are variants-specific chaperones, it is important to note that the interactions between chaperons and histones are not always exclusive or binary. For example, chaperone Nap1 is shown to bind to three different dimers, H2A-H2B, H2A.Z-H2B, and H3-H4 [26], [27], [28]. Chaperons Asf1, Daxx, and HIRA facilitate H3.3-H4 dimer deposition at distinct genome localizations [29], [30]. Centromere-specific histone variant CENP-A, on the contrary, requires a specific chaperone for its timely and precise deposit: HJURR in humans [31], [32], Scm3 in yeast [33], [34], [35] and Cal1 in the fly [36]. Compared to the core histones, much less is known about chaperon-mediated linker histone H1 dynamics, despite several proteins (such as Nap1, NASP, and TAF1) being indicated in mediating linker histone H1 deposition [37], [38], [39]. How specific histone is distributed among different chaperones and how functionally overlapping chaperons regulate the dynamics of histone variants in cells remain a topic of future research. Nevertheless, histone chaperones' redundancy and specificity form a complex yet crucial regulatory system that contributes to the spatial and temporal regulation of histone functions in cells. A large amount of literature is available on the roles and regulation of histone chaperons; therefore, we will not discuss this topic further in this article. For those interested in the topic, we would like to point them to a few excellent reviews [40], [41], [42].
At the protein sequence level, a wide range of differences exist among different histone variants. Some variants differ from their canonical counterpart by only a few amino acids, such as variant H3.3. Others have significant variations, such as the existence of an additional domain (in the case of variant macroH2A) or lacking part of a conserved region (in the case of variant H2A.B). Alterations in protein sequence result in changes in nucleosome stability in vitro [43], [44] and are proposed to contribute to distinct chromatin functions in cells. Nevertheless, how such sequence variations result in altered chromatin structures that cause lasting effect on genome functions was not well understood until recently, when multiple high-resolution cryo-EM structures of variant-containing nucleosomes revealed previously undescribed structural changes on the nucleosome. A notable example is the H2A.Z nucleosome, where two cryo-EM studies [25], [45] show structural changes in both DNA and histone surface that were not detected by earlier crystallography studies [22], [46]. The discrepancies are likely the results of stabilizing effect on the complex induced by crystal packing and the advantage of cryo-EM in resolving the conformational dynamics of protein complexes. Table 1 below lists the nucleosome and chromatin fiber structures discussed in this article, representing structures containing variants from two core histone families (H2A and H3) and those of the linker histone H1 family. To our knowledge, no cryo-EM structure is yet published for nucleosomes containing any H4 or H2B variant.
Table 1.
Information and parameters for cryo-EM structures discussed in this review.
| Sample | EMD | PDB | Resolution | Symmetry |
|---|---|---|---|---|
| canonical NCP | EMD-23632 | 3.8 Å | C2 | |
| canonical NCP | EMD-4297 | 6FQ5 | 3.8 Å | C2 |
| H2A.Z NCP | EMD-23626 | 7M1X | 3.7 Å | C1 |
| H2A.2.2 NCP | EMD-30076 | 6M4D | 4.4 Å | C2 |
| H2A.B NCP | EMD-30078 | 6M4H | 3.9 Å | C2 |
| H2AX-F NCP | EMD-22791 | 7KBE | 3.5 Å | C1 |
| H3.3 NCP | EMD-4692 | 6R0C | 4.2 Å | C1 |
| CENP-A NCP with Widom601 DNA | EMD-10822 | 6SE0 | 3.9 Å | C1 |
| CENP-A NCP with alpha satellite DNA | EMD-0586 | 6O1D | 3.395 Å | C1 |
| H3mm18 NCP | EMD-30631 | 7DBH | 3.6 Å | C1 |
| Cse4 NCP | EMD-20839 | 6UPH | 2.7 Å | C1 |
| Canonical 30 nm fiber | EMD-23631 | 11 Å | C1 | |
| H2A.Z 30 nm fiber | EMD-23630 | 7.5 Å | C1 | |
| CENP-A with CENPN 30 nm fiber | EMD-26333 | 12.5 Å | ||
| H3-CENP-A trinucleosome 22 bp linker | EMD- 0768 | 6L49 | 18.7 Å | C2 |
| H3-CENP-A trinucleosome 30 bp linker | EMD- 0769 | 19.6 Å | C1 | |
| Canonical 12 × 177 30 nm fiber with H1.4 | EMD-2600 | 11 Å | C1 | |
| Canonical 12 × 187 30 nm fiber with H1.4 | EMD-2601 | 11 Å | C1 | |
| Canonical 4 × 177 array with H1.4 | EMD-13356 | 7PET | 9.5 Å | C1 |
| Chromatosome with hH1.0 | EMD-22683 | 7K5X | 2.93 Å | C1 |
| Chromatosome with hH1.4 | EMD-22684 | 7K5Y | 2.76 Å | C1 |
| Chromatosome with hH1.10 | EMD-22685 | 7 K60 | 3.12 Å | C1 |
| Chromatosome with xH1.8 | EMD-22792 | 7KBF | 4.42 Å | C1 |
3.1. Histone H2A family
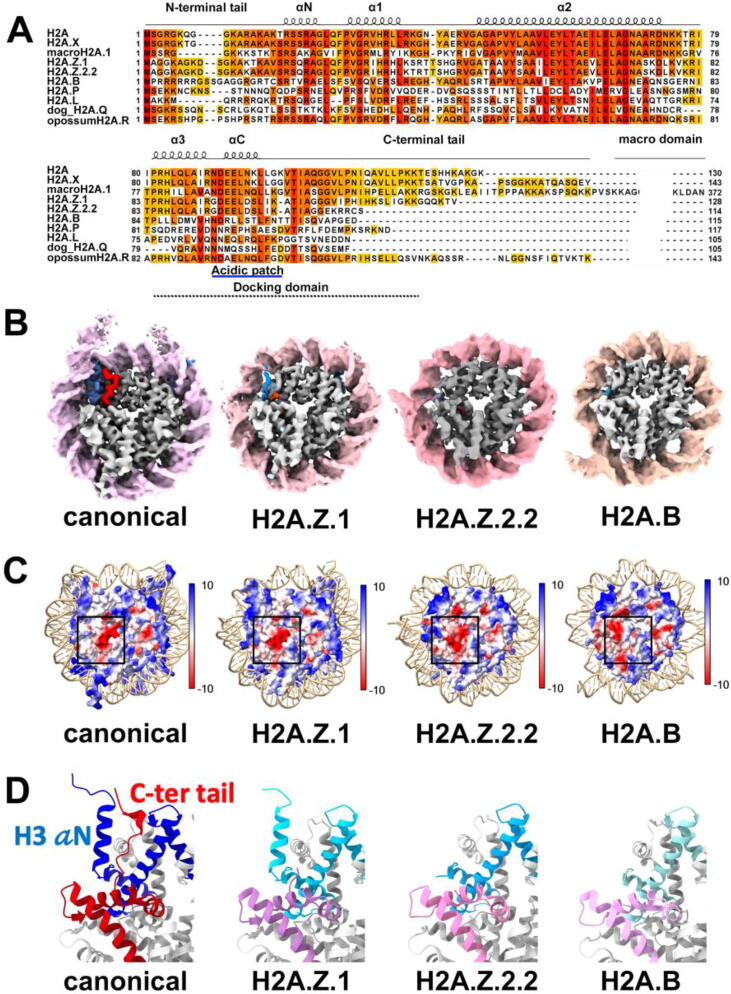
The Histone H2A family has the largest number of variants among core histones. H2A variants are also highly diverse, with only ∼48–60% sequence identity in general compared to canonical H2A (Fig. 1A). Interestingly, H2A variants have divergent C-terminus and acidic patches (Fig. 1A &D). The C-terminus of H2A contains the docking domain crucial for nucleosome stability. It interacts with the aN helix of H3 to stabilize the H2A interactions with the H3-H4 tetramer and the last turn of DNA. On the other hand, the conserved acidic patch on the H2A-H2B dimer has been indicated to play an essential role in mediating nucleosome-nucleosome interactions during chromatin folding [47] and is frequently a site for interactions with many chromatin regulators [48]. In addition, the two H2A molecules interact with each other through the L1 loop region, providing additional stability to two turns of DNA at the back face of the nucleosome. Taken together, H2A plays an important role in nucleosome stability and assembly. Therefore, diversification of the H2A family members confers nucleosomes new characteristics and functions.
Fig. 1.
Structural diversity of histone H2A variants, H2A.Z and H2A.B. A) Sequence alignment of human histone H2A variants prepared using Jalview 1). (B) Cryo-EM density maps of canonical NCP (EMD-23632), H2A.Z.1 (EMD- 23626), H2A.Z.2.2 (EMD-30076) and H2A.B (EMD-30078 ) nucleosomes. Thedensities of H3 αN helix (blue)and H2A C-terminus (red) are highlighted in canonical and H2A.Z.1 nucleosome, but are missing in H2A.Z.2.2 and H2A.B nucleosome. The canonical and H2A.Z.1 nucleosome contain 167 bp 601 Widom sequence, while 147-bp 601 sequence was used for the H2A.Z.2.2 and H2A.B nucleosomes. (C) The electrostatic potential on the nucleosome surface. The acidic patch in each nucleosome is highlighted by a black square for comparison. PDB used: canonical NCP (6FQ5), H2A.Z.1 (7M1X), H2A.2.2 (6M4D), H2A.B (6M4H). (D) Variations in the H3 (blue) αN helix and H2A (red) Cterminal docking domain. H3 in variant nucleosomes are colored in shades of blue. H2A variants are colored in shades of purple. Only one of H3 and one H2A is shown for clarity. (For interpretation of the references to colors in this figure legend, the reader is referred to the web version of this paper.)
3.1.1. H2A.Z
Variant H2A.Z is essential for the survival of mammals [49]. It is also the most extensively studied and best-characterized histone variant, with functions in many nuclear processes, including transcription activation, DNA repair, heterochromatin boundaries, and lineage commitment of embryonic stem (ES) cell [50]. Yet the role of H2A.Z in vivo, especially in transcriptional regulation, remains puzzling, as both facilitating and suppressing functions have been reported [51], [52], [53], [54], [55], [56]. In all eukaryotes, H2A.Z is predominantly found at the distal end of the inducible promoter (the so-called +1 nucleosome adjacent to the nucleosome-free region at the promoter) to poise the gene for rapid activation [54], [55], [56]. Upon induction and transcription activation (through the action of transcription factor and RNA polymerase), H2A.Z is removed and replaced by the canonical histone H2A [57], [58], [59]. Consistent with this view, H2A.Z is found to enrich at unwrapping +1 nucleosomes in mouse ES cells [60]. Notably, H2A.Z is also located at heterochromatin regions such as centromere and pericentromeric heterochromatin [61], [62], [63], [64], [65]. Its accumulation in the gene body is linked to gene suppression in certain plants [51], [52]. In addition, contrasting results from in-vitro biochemical studies exist concerning H2A.Z nucleosome stability [22], [66], [67]. More recent studies suggest that the stability of H2A.Z nucleosome depends on the chromatin context [46], [68]. It is now clear that H2A.Z functions in vivo likely depend on multiple factors such as local enrichment of H2A.Z, the presence of histone post-translational modifications (PTMs), chromatin regulators, and other histone variants.
In mammals, there are three H2A.Z isoforms. H2A.Z.1 and H2A.Z.2 are two nonallelic isoforms [69], while the third isoform, H2A.Z.2.2, is a splice variant of H2A.Z.2 [70]. H2A.Z.2.2 is exclusive to primates and preferentially enriched in the brain [71]. Variant H2A.Z.2.2 contains the shortest C-terminal tail among the three and forms a significantly less stable nucleosome [71]. Nevertheless, all three isoforms display a shorter C-terminal tail than their canonical counterpart. An early study in Drosophila shows that most of the C-terminal region in His2AvD (H2A.Z in Drosophila), especially the M6 cassettes, is essential for His2AvD function [72]. A recent study further confirms this finding by showing the M6 region is responsible for H2A.Z deposition [73]. The acidic patch in all three isoforms is highly conserved, with three residue substitutions and insertion extending this positively charged nucleosome surface (Fig. 1A &C). Zhou et al. recently reported a 4.4 Å cryo-EM structure of H2A.Z.2.2 nucleosome [25]. In the structure, the last 11-bp DNA on each end is missing in the EM density map (Fig. 1B). Accordingly, the atomic model shows only 125 bp of DNA protected by the octamer. In addition, densities for the 14-residue in its C-terminus and the H3 ⍺N helix are absent, indicating their mobile nature. Our cryo-EM structure of variant H2A.Z.1 nucleosome [45] is similar to the H2A.Z.2.2 nucleosome structure but with a better-resolved H3 ⍺N helix and H2A.Z.1 C-terminal tail (Fig. 1B). Since the C-terminus of H2A is known to coordinate the interactions between the H3 ⍺N helix and the major groove of DNA between SHL −6 and SHL −7, these structures indicate that the shorter C-terminus of H2A.Z variants weakens the interaction with H3 ⍺N helix and subsequently the histone-DNA interaction near the entry/exit site (Fig. 1D). Using mutagenesis and restriction enzyme digestion, we demonstrated that the last six residues of H2A.Z.1 are the main structural feature responsible for the enhanced DNA accessibility and nucleosome instability observed in H2A.Z.1 nucleosome [45]. Residue swapping experiments in the docking domains and the L1 loop, on the other hand, show no detectable effects on end-DNA accessibility, indicating that these H2A.Z.1-specific residues do not contribute to the H2A.Z-specific DNA flexibility.
3.1.2. H2A.B
Variant H2A.B (also known as H2A.Bbd) was found in mammals but not in invertebrates. It represents one of the lowest similarities (∼48 % identical to H2A) to its canonical counterpart among the H2A family. H2A.B was first identified in a screen for genes involved in female X-chromosome inactivation and was thought to be excluded from the inactive X-chromosome [74]. However, subsequent studies in the mouse testis revealed that it is in fact, present on the inactive X chromosome following meiotic sex chromosome inactivation [75]. Later ChIP-Seq and RNA-Seq experiments revealed that H2A.B is localized at the transcription start site (TSS) of prior activated genes, and such localization correlates with the higher expression level of the genes [75], [76], [77], [78]. More recent studies also revealed H2A.B functions in mRNA processing [79], DNA synthesis, and DNA repairs [80].
H2A.B is one of the four classes (H2A.B, H2A.L, H2A.P, and H2A.Q) of short H2A variants [81], which lacks part of the H2A C-terminal tail and its characteristic docking domain (Fig. 1A). It is not surprising that biochemical studies showed that H2A.B-containing nucleosome is less stable than the major type nucleosome in vitro and only organizes 118 bp of DNA [82]. The flexible and accessible DNA ends were confirmed by Zhou et al. recently with a 2.9 Å cryo-EM structure of NCP containing variant H2A.B [25]. In the study, the DNA-binding domain (DBD) of poly(ADP-ribose) polymerase 1 (PARP1) was used to stabilize the H2A.B nucleosome for structural study. 3D classification revealed a class showing that PARP1-DBD tethers the DNA at SHL 6 – SHL 7 to DNA at SHL −2, likely preventing the DNA from unwrapping. This interaction was speculated to stabilizes H2A.B nucleosome to enable the complex being resolved to a much higher resolution than the complex without bound PARP1 [25]. Nevertheless, the two density maps are very similar in overall architecture, indicating that PARP1 binding does not perturb the H2A.B nucleosome structure. Probably the most notable feature of the structure is that H2A.B compacts significantly less DNA around the octamer than other known nucleosome structures. The structure shows the last ∼5-bp DNA at each end, tilting outward from the octamer [25]. Through Micrococcal Nuclease (MNase) digestion, the authors showed that ∼ 103 bp of DNA was protected in the H2A.B nucleosome. Similar to the two H2A.Z nucleosome structures described in the previous section, the H3 ⍺N-helix and the C-terminal region (residues 109–114) of the H2A.B docking domain are both absent from the density map (Fig. 1B &D). Though H2A.B and H2A.Z have not been studied simultaneously under the same setting, based on these cryo-EM structures and the shorter DNA in the H2A.B nucleosome, it is reasonable to speculate that the H2A.B nucleosome is less compact and more unstable than variant H2A.Z nucleosome. In addition, densities for the H2B N-terminal region (residue 1–32) are absent, and the H2A.B-H2B dimer tilts away from the H3-H4 tetramer. These structural features were speculated to disrupt the interactions between H2B residues 26–32 and DNA, negatively affecting H2A.B-H2A dimers and other core histones [25].
Through multiple mutagenesis and domain-swapping experiments, the authors validated the structural prediction. They confirmed that seven H2A.B-specific residues at the N-terminal domain and the lack of H2A-like 19 residues of its C-terminus are mainly responsible for the highly unstable H2A.B nucleosome. Notably, H2A.B incorporation also altered the nucleosome surface, with its specific residue replacement that renders a more neutral acidic patch (Fig. 1C). Though it did not cause detectable differences in the overall structure of the nucleosome, it likely has far more implications on chromatin folding and higher-order structure maintenance. Future studies on H2A.B chromatin fibers and H2A.B nucleosomes in complex with chromatin-associated factors are required to test these hypotheses.
3.1.3. H2A.X-F
H2A.X, a variant that shares 90 % homology with the canonical H2A, is best known for its function in DNA damage response. H2A.X is subjected to numerous post-translational modifications, including the rapid phosphorylation at its Ser-139 upon DNA double-strand break (DSB). The phosphorylated H2A.X, called γH2A.X, is important for initiating the DSB repairs cascade [83]. The formation of γH2AX was first discovered in yeast where the loss of the H2A C-terminus, which contained Ser-129 (the yeast homolog of mammalian Ser-139), led to impairment of non-homologous end joining [84]. In-vitro functional studies suggested that γH2A.X contributes to the efficient recruitment of downstream repair factors [85] by destabilizing nucleosomes and enhancing DNA accessibility [43], [44]. A recent cryo-EM study of nucleosomes isolated from Xenopus egg extracts showed that H2A.X-F1/F2 (a H2A.X isoform in Xenopus) nucleosome are identical to the canonical nucleosome [86], indicating that H2A.X variant itself does not alter the nucleosome structure. It remains to be seen whether future cryo-EM studies of phosphorylated H2A.X nucleosomes will reveal enhanced DNA flexibility near the nucleosome DNA entry/exit site. Furthermore, how γH2A.X influence array folding and chromatin secondary structure is largely unknown and remains a subject for future studies.
3.2. Histone H3 family
Histone H3 partners with H4 to form a tetramer, which is incorporated into nucleosome immediately after DNA synthesis and before H2A-H2B dimers incorporation [2]. Histone H3 has the largest number of PTMs among the four core histones. Contrary to H2A variants, most histone H3 variants contain minor sequence differences, often with only a few amino acid substitutions within the globular domain of the protein (Fig. 2A). An exception to this rule is the centromeric-specific H3 variant CENP-A, which harbors a large degree of sequence variation (∼50 % identity) compared to canonical H3.1. This likely reflects its special cellular localization and function. Structures of CENP-A nucleosome alone and in complex with specific centromeric proteins are widely available in PDB and EMDB. We will discuss recent development in understanding CENP-A, variant H3.3, and a less-known H3.3 derivative variant H3mm8.
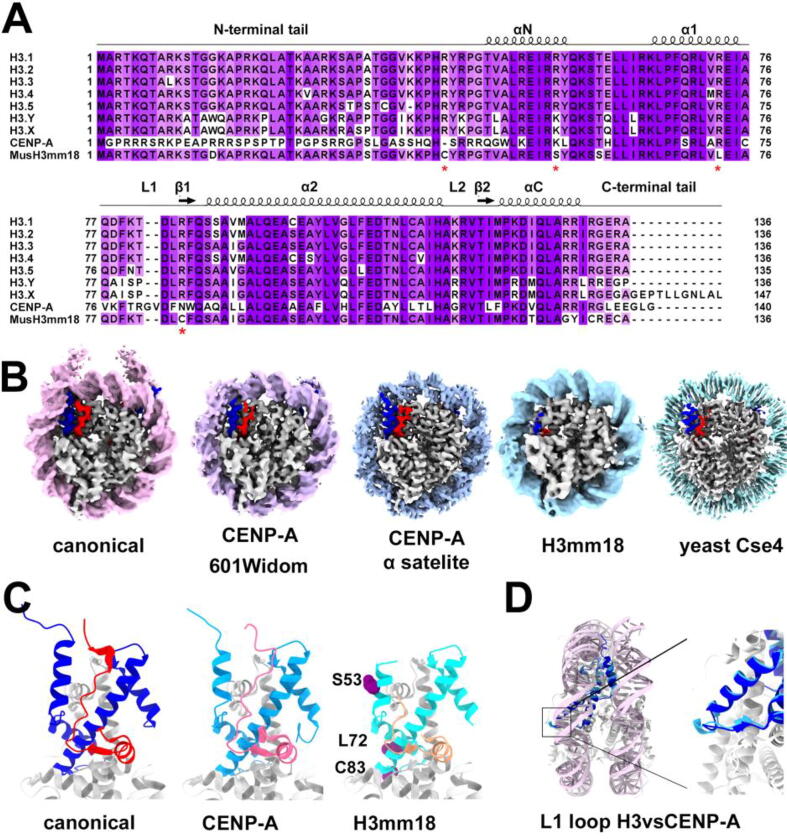
Fig. 2.
Structural diversity of histone H3 variants, CENP-A and H3mm18. (A) Sequence alignment of human histone H3 variants. (B) Cryo-EM density maps of canonical NCP with 167-bp 601 sequence, CENP-A nucleosomes with 145-bp 601 DNA (EMD-10822), CENP-A nucleosome with 145-bp α satellite DNA (EMD-0586), H3mm18 nucleosome with 145-bp 601 DNA (EMD- 30631), and yeast Cse4 nucleosome with 147-bp 601 DNA (EMD-20839) respectively. Densities of H3 αN helix (blue) and H2A Cterminus (red) are highlighted. (C) Comparison of the H3 αN helix and H2A (red) Cterminal tail showing the αN helix is shorter in CENP-A (light blue) nucleosome and mobile in H3mm18 (cyan) nucleosome. Note the H2A C-terminal tail is not modeled in the H3mm18 nucleosome. (D) Comparison of the canonical (light blue) and CENP-A (blue) nucleosomes showing the protruding RG loop (red arrow) in CENP-A nucleosome. (For interpretation of the references to colors in this figure legend, the reader is referred to the web version of this paper.)
3.2.1. CENP-A (also known as cenH3)
The centromere-specific histone H3 variant CENP-A (CENP-A in metazoan and Cse4 in yeast) is an essential component of the eukaryotic centromere, the specialized chromosome regions that connect with mitotic spindles [87], [88]. CENP-A is only found in centromeric nucleosomes and serves as the epigenetic mark of centromere. The primary function of the CENP-A nucleosome is to recruit centromere-specific proteins to form a complete and functional kinetochore complex to ensure accurate chromosome segregation during cell division. It is found that a decrease in CENP-A level in cells impairs kinetochore assembly [89], [90], [91]. On the other hand, overexpression of CENP-A has been linked to multiple aggressive cancers, and mislocalization of CENP-A leads to chromosome instability, a hallmark of cancers [92], [93]. CENP-A is the most extensively studied H3 variant for its specialized and essential function in cells and has garnered much interest among chromatin structural biologists. Early crystallographic and biochemical studies showed that the CENP-A nucleosome contains several unique features, including a truncated ⍺N helix and substitution of the R49 residue essential for DNA binding [94]. These features confer an unstable CENP-A nucleosome in vitro. For a comprehensive overview on centromere chromatin and kinetochore assemble, we recommend readers to other excellent reviews [95], [96].
CENP-N and CENP-C are both CENP-A interaction proteins and components of the inner kinetochore CCAN (constitutive centromere associated network) complex. CENP-N bound CENP-A nucleosome forms a critical part of the CENP-A targeting domain (CATD) responsible for directing CENP-A to centromeres. CENP-C has also been shown to promote CENP-A nucleosome targeting in vivo. High-resolution cryo-EM structures of CENP-A nucleosome alone [97], [98], in complex with human CENP-C [99], or complex with human CENP-N, have been reported [100], [101], [102]. These cryo-EM structures reveal a similar structural fold, with poorly ordered DNA densities from each end of the CENPA nucleosome (Fig. 2B). Flexible DNA ends are essential for the proper function of CENP-A, as mutants that abolish the terminal DNA flexibility also cause a severe defect in the centromere in cells [103]. Another feature that differentiates metazoan CENP-A from the canonical H3 nucleosomes is the ⍺N helix in CENP-A, which comprises only two helical turns compared to three in H3.1 (Fig. 2C). This shorter aN helix and its specific N-terminal tail are shown to be responsible for the terminal DNA flexibility in the CENP-A nucleosome [99]. In addition, CENP-A contains a unique insertion of three CENP-A specific residues (R80, G81, V82) in the L1 loop (also known as RG loop) (Fig. 2D). The RG loop is seen protruding from the CENP-A nucleosomal DNA to allow direct contact with CENP-N in the cryo-EM structures of the CENP-N-CENP-A nucleosome complex [99], [102]. The RG loop, together with ⍺2, forms the so-called CENP-A targeting domain (or CATD), which binds CENP-N and its specific chaperone HJURP [31], [32]. It is worth noting that in the cryo-EM structure of yeast CCAN-CENP-A-nucleosome complex, CENP-N binding is also through an additional feature of CENP-A nucleosome, the entry/exit DNA [104]. Interestingly, this interaction was not observed in the cryo-EM structure of the human CCAN-CENP-A-nucleosome complex [105].
3.2.2. H3.3
H3.3 is nearly identical (96 % sequence identity) to canonical histone H3 (H3.1 and H3.2), with only five amino acids difference (Fig. 2A). These amino acids are the binding site for histone chaperone Daxx, responsible for H3.3 deposition [106] and the proper genome localization of H3.3 in vivo. Variant H3.3 is mainly found in the promoter region of actively transcribed genes and is best known for its role in transcription activation [29], [107]. Such nuclear localization is not exclusive, as H3.3 is also found in heterochromatin regions [106], [108]. Enrichment of H3.3 at the promoters and gene enhancers is proposed to keep the sites open and accessible for transcription factor binding [109]. However, H3.3 alone does not seem to affect nucleosome structure [110] and stability in vitro [111]. It is also worth noting that nucleosomes containing both variants H2A.Z and H3.3 have recently been identified and found to be enriched at the TSS in human cells [112], [113], suggesting the potential interplay and positive reinforcement between the two variants in transcription regulation.
Cryo-EM structure of H3.3 containing nucleosome has been reported recently [53]. The structure is identical to that of the canonical NCP and the crystal structure of H3.3 NCP [110], an observation not surprising given the minor sequence alteration in H3.3 and the fact that H3.3 alone does not impart detectible structural alteration to the nucleosome [110]. It remains to be seen whether the presence of both variants H3.3 and H2A.Z would induce structural changes on the nucleosome beyond those observed in the H2A.Z nucleosome. In addition, further cryo-EM studies are needed to elucidate the structural mechanism of how variant H3.3 impairs chromatin compaction and counteracts H2A.Z-mediated array folding [111].
3.2.3. H3.3 H3mm18
During an in-silico hybridization screening in mice, Maehara et al. discovered 14 mouse-specific H3 variants. Except for one, all these variants are highly similar to H3.3 [114]. One of these mouse H3.3-derived variants, H3mm18, is expressed in skeletal muscle and brain [114] and is suggested to function in regulating gene expression during muscle differentiation [24]. However, its precise functions remain to be determined. Compared to H3.3, it contains 12 amino acid differences (Fig. 2A), many of which replace basic Arginine in H3.3 with neutral residues in H3mm18. A recent cryo-EM study by Hirai et al. reveals that the incorporation of H3mm18 also leads to a drastically disordered DNA ends on nucleosome [24] where the histone octamer protects ∼ 125–130 bp of DNA (Fig. 2B). Similar to the CENP-A nucleosome, the ⍺N helix of H3mm18 was poorly resolved in the cryo-EM density map (Fig. 2B). The authors also showed the substitution of four Arginine residues at helix ⍺N, and ⍺1 on canonical H3.1 with the H3mm18-specific neutral residues (R40C, R53C, R72L, and R83C) (Fig. 2C) enhances the end-DNA flexibility, further validating the cryo-EM structure. On the other hand, mutations on the H3.3 C-terminus and substitution of I124T did not substantially affect the MNase digestion pattern of the DNA ends. Overall, results from the studies of the two H3 variant members underlie the importance of the histone H3 ⍺N helix in mediating interactions with entry/exit DNAs and the structural and stability of the nucleosome.
3.3. Linker histone H1 family
Linker histones are structural components of chromatin, whose primary function is to control chromatin compaction and gene accessibility through the formation of higher-order structures. They have also been implicated in DNA replication, recombination, and repairs [3]. The existence of multiple copies of linker histone H1 inside the cells presented a challenge to pinpoint their precise function in cells. Early cell biology studies led to the belief that linker histone H1 is nonessential for an organism's survival until a study demonstrated that triple knockout of all three H1 subtypes causes embryonic lethality in mice [115]. The study further shows that reduced H1 in embryos leads to a global reduction in nucleosome repeat length and local decompaction of chromatin. Together with several subsequent studies, these results established the crucial role of linker histone H1 in maintaining chromatin structure. Regarding transcription, studies in organisms from Tetrahymena to vertebrates indicate that H1 depletion does not alter global gene expression but affects the expression of a subset of genes [116], [117], [118], [119], [120], [121].
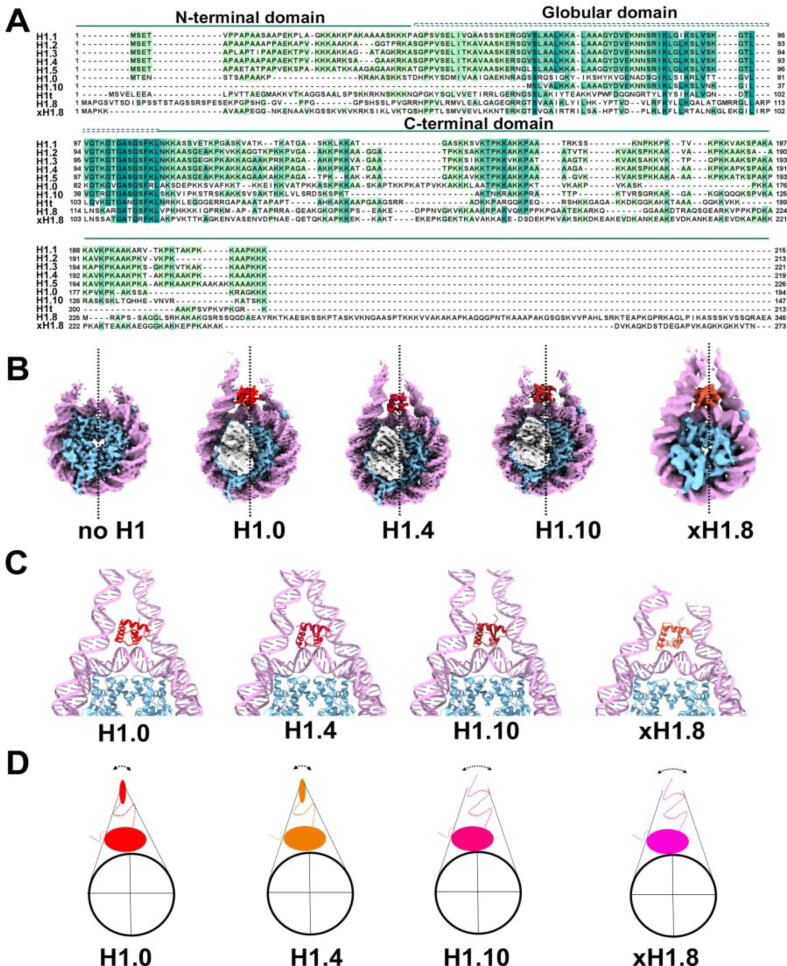
Members of the linker histone H1 family are highly divergent in sequence (Fig. 3A). In mammals, there are 11 isoforms of H1, seven expressed in somatic cells and four in germ cells. The protein level of the somatic linker histones appears to be tissue- and cell-type specific, suggesting the existence of H1 isoform-specific functions in vivo [122]. These seven somatic linker histones all share a conserved tripartite structure: a basic N-terminal tail (∼20–35 aa), a central globular domain (∼70 aa), followed by a long and disordered C-terminal tail (∼100 aa). On the other hand, the four linker histones in germ cells do not have the corresponding globular domain. The globular domain of H1 is sufficient for binding and protecting the nucleosome's entry/exit DNA from nuclease digestion, while the N-terminal tail contributes little to nucleosome binding [123], [124]. On the other hand, the C-terminal tail enhances the interactions of H1 to nucleosome further and stabilizes chromatin secondary structure [123], [125]. Recently, Zhou et al. reported the cryo-EM structures of chromatosomes containing 197-bp DNA and three full-length human H1 variants, H1.0, H1.4, and H1.10 [126]. These variants have diverse C-terminal domain sequences but a conserved globular domain. Another study using endogenous nucleosomes isolated from Xenopus egg extracts revealed a 4.4 Å cryo-EM structure of chromatosome with linker histone H1.8 [86]. We will discuss the revelation from these studies regarding domain-specific functions and how variations in linker histone H1 domains influence chromatosome structure and functions.
Fig. 3.
Structural diversity of linker histone H1 and chromatosome. (A) Sequence alignment of human H1 histone variants and Xenopus H1.8. (B) cryo-EM density maps of nucleosomes (EMD- 23632), and H1.0 (EMD- 22683), H1.4 (EMD-22684), H.10 (EMD-22685) and xH1.8 (EMD-22792) chromatosome. Nucleosome dyad is indicated with a dotted line. The Globular domain of H1 in each structure is in red, with the octamer in blue and DNA in purple. The antibody scFv used stabilize chromatosome is colored grey. (C) Closeup view of the atomic coordinates of nucleosomes, chromatosome with H1.0, H1.4, and H1.10 linker histone respectively, showing the interactions between the H1 globular domain and the linker DNA. (D) Linker DNAs adopt different angles (relative to the dyad) and conformations in different chromatosomes, Figure adapted from [127].
3.3.1. Globular domains
Earlier crystallographic studies were done on chromatosomes containing only the H1 globular domain. These studies show that all human somatic linker histone adopts the on-dyad binding mode [126], [127], where H1 binds at the center of the nucleosome DNA. The cryo-EM structure of frog H1.8 chromatosome reveals a similar on-dyad binding of linker histone H1.8 [86]. Drosophila H1, on the other hand, binds off-centered of the nucleosome DNA, adopting the so-called off-dyad model [128]. Yet, how the C-terminal domains of linker histone H1 contribute to nucleosome binding and linker DNA conformation remain poorly understood. The cryo-EM structures of chromatosome show that all three somatic linker histone H1 isoforms (H1.0, H1.4, and H1.10) bind on the dyad through the globular domains (Fig. 3B), make close interactions with the nucleosomal DNA via their L1 loop and with linker DNA through their ⍺3 helix. Comparison of the structures also reveals small differences in the orientation of the globular domains between H1.10 and H1.0 chromatosome, which supports the fluorescent recovery results after photobleaching (FRAP) experiment showing a highly dynamic globular domain in chromatosome [129]. Overall, this binding mode is consistent with the crystal structures of chromatosome containing the globular domain of chicken H5 (cH1.0) [130] and that of chromatosome containing full-length Xenopus H1.0 (xH1.0) [131].
Interestingly, when the density maps were low-pass filtered to ∼ 6 Å, additional densities were observed between the DNA gyres of SHL 1 to 2 and −6 to −7. Based on their location and with support from the NMR experiments, the authors attributed the extra densities to part of the histone H3 N-terminal tail and the C-terminal tail of H2A [126].
3.3.2. C-terminal tail
The most notable difference among the three chromatosome cryo-EM structures is the different conformations adopted by the linker DNAs. The two DNA linkers in the H1.10 chromatosome have a more open conformation than those in the H1.0 and H1.4 chromatosome, though it is still closer to each other than those in the free nucleosomes (Fig. 4D). The authors performed tail-swap experiments and showed that the C-terminal tails control the exit angle and openness of the two DNA linkers. The study further indicated that the T/SPKK motifs in H1.0 and H1.4 chromatosomes are responsible for controlling the linker DNA angle. They suggested that the closer linker DNA conformation implies a tighter association of H1 to the nucleosome, consistent with the observation that chromatin with H1.0 and H1.4 have a much longer residence time measured by FRAP [129], [132], [133], [134]. Notably, a frameshift mutation in the H1.4 C-terminal tail disrupted chromatin structure and nuclear lamina organization [135]. Furthermore, clinical studies of a large cohort of individuals sharing these mutations suggest a strong link between mutations in H1.4 C-terminus to autism and premature aging [135], [136]. Taken together, these studies underlie the importance of linker histone H1 C-terminal tails in mediating linker DNA conformations in chromatin. Since linker DNA conformation is an important player in chromatin compaction, linker histone H1 variants with diverse C-terminal tails likely evolve to meet the need of the cell to fine-tune the chromatin structures and functions.
Fig. 4.
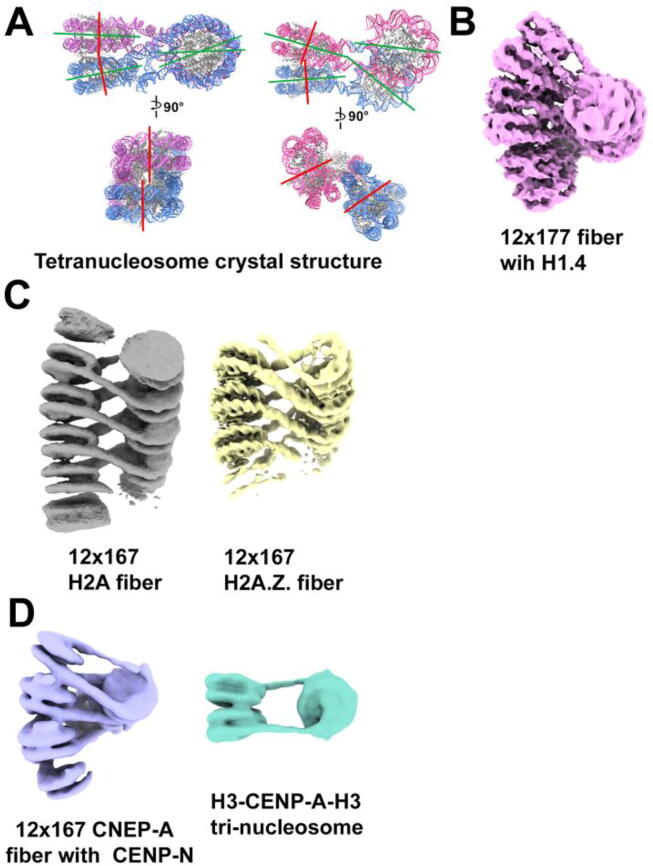
Reconstituted poly-nucleosome fiber. (A) Crystal structures of tetranucleosome with 167-bp and 157-bp 601 sequence (PDB 1ZBB and 5OY7) respectively in two different views. The dyad is green and the nucleosome superhelical axis is red. (B) Cryo-EM density map of dodeca-nucleosome fiber with 177-bp NRL (EMD-2600). (C) Cryo-EM density map of canonical (left) and H2A.Z (right) dodeca-nucleosome fibers (EMD- 23631 and EMD-23630). (D) Cryo-EM density map of CENP-A-CENP-N dodeca-nucleosome fiber (EMD-26333) is shown on the left. The Cryo-EM density map of compact H3-CENP-A-H3 tri-nucleosome (containing 22-bp linker DNA) is shown on the right (EMD-0768).
3.4. Histone variants and chromatin higher-order structures.
3.4.1. Reconstituted chromatin fibers
As mentioned earlier, nucleosome arrays reconstituted with tandem-repeat nucleosome positioning sequence can condense and form 30-nm fibers in vitro under carefully tuned conditions in the presence of divalent cations [137] or stochiometric amount of linker histone H1 [138]. Factors such as linker DNA length and the incorporation of histone variants are known to influence the architecture of 30-nm fibers and chromatin higher-order structures in vivo.
While the detailed architectures of chromatin in the nucleus remain somewhat controversial, it is clear that chromatin is highly dynamic at all hierarchical levels. The mechanisms that regulate mono-nucleosome structure–function also influence chromatin higher-order structures. Over the years, many attempts have been made to obtain high-resolution structures of various forms of reconstituted 30-nm fibers to understand the intrinsic properties of array folding and to infer chromatin condensation in the nucleus. An early crystal structure of tetra-nucleosomes compacted by using a high concentration of divalent cation was resolved to 9 Å resolution by Richmond et al. [139]. The structure revealed two-stack di-nucleosomes connected by a zigzagging linker DNA, one straight and one bent [140] (Fig. 4A left). It is unclear if the difference in linker DNA conformations results from the high-concentration (25 mM) of Mg2 + used or due to crystal packing. Dodeca-nucleosome fiber with the same 167-bp nucleosome-repeat-length (NRL) but compacted under lower divalent cation concentration (2 mM Mg2 + ) shows only straight DNA linkers in our cryo-EM study [45] (Fig. 4C). The two-start helical tetra-nucleosome structure seems to be a common feature for reconstituted 30-nm fibers, as a more recent crystallographic study (Fig. 4A right) also reported similar two-start models of compact tetra-nucleosomes with a range of linker DNA lengths [141]. This study, however, further revealed structural heterogeneity in compact tetra-nucleosomes. These studies, combined with our cryo-EM analysis of the canonical fiber that displays a range of conformations (from a flat array to a twisted compact form) [45], underlie the structural plasticity of reconstituted chromatin fibers.
Using cryo-EM, Song et al. resolved the structure of dodeca-nucleosome 30-nm fiber containing linker histone H1 to 11 Å resolution. [142] (Fig. 4B). The study shows that despite different NRL used (177-bp and 187-bp), fibers form a left-handed zigzag two-start helix, similar to the tetra-nucleosome crystal structure. The overall structure of the dodeca-nucleosome H1 fibers comprises three tetra-nucleosome units that are twisted against each other with straight linker DNA connecting the adjacent nucleosomes. Compared to the tetra-nucleosome crystal structures, the diameter of the H1 fiber increases with longer NRL, while the rotation angle between two nucleosome stacks is smaller. Notably, a relatively strong density was detected at the inter-nucleosome interface where the H4 N-terminus meets the adjacent H2A-H2B dimer. The author attributed the density to the H4 tail-acidic patch interaction, which is also known to play a role in chromatin folding. A recent structural study on hexa-nucleosome fiber containing linker histone H1 further revealed the existence of conformational heterogeneity in the H1 fiber [143], similar to the canonical fiber without H1 observed in our study [45]. Taken together, these findings confirm the polymorphic nature of the reconstituted 30-nm fibers and underlie the importance of linker DNA angles in chromatin compaction. A major limitation of these studies is the low resolution of the EM maps due to the structural mobility within the fiber, which is insufficient to resolve features (such as the H4 tail-acidic patch interaction) deemed critical for chromatin compaction. Future studies that can resolve sample homogeneity are required to push the resolution and obtain atomic structural information at the inter-nucleosome interface. Compared to the structural information available on 30-nm fibers from the major-type nucleosome, much less is known about how histone variants alter chromatin higher-order structures. We will summarize the two available studies below on variants H2A.Z and CENP-A.
3.4.2. H2A.Z
We have recently determined a 10.8 Å resolution structure of H2A.Z dodeca-nucleosome fiber using single-particle cryo-EM [1] (Fig. 4C). Compared to the H2A dodeca-nucleosome fiber in the same study, the H2A.Z fiber is more homogeneous in conformation where all 3D classes adopting a twisted structure. This suggests that H2A.Z incorporation stabilizes inter-nucleosome interactions to facilitate a regular fiber formation. The H2A.Z fiber also follow a zigzag two-start helical organization similar to the tetra-nucleosome crystal structure [2] and the cryo-EM structure of the H1 fiber [3]. In both the H2A and H2A.Z fiber structures, di-nucleosome is the structural unit. Detailed analyses on the rotation and shift between nucleosome stacks reveal that H2A.Z fiber is more twisted and thus more compact than the H2A fiber, though it is less compact than the tetra-nucleosome crystal structure [1]. Our study suggests that variant H2A.Z alone can significantly alter the structure of chromatin fibers, where it enables nucleosome arrays to fold into a regular and condensed structure. This explains why H2A.Z is also found to enrich in heterochromatin regions and is linked to transcription repression.
Notably, densities in interface between nucleosome-stacks where the H4 N-terminal tail meets the H2A-H2B acidic patch of adjacent nucleosome are observed in our H2A.Z fiber map, despite the limited resolution [3]. We speculate that the distinct H2A.Z features of flexible DNA ends and the extended acidic patch are responsible for its role in mediating chromatin compaction. It remains to be seen how the H4 tail-acidic patch interaction contributes to chromatin compaction and how modification of the acidic patch found in some histone variants could fine-tune the structure-dynamics of chromatin fiber.
3.4.3. CENP-A
CENP-A nucleosomes are found to be interspersed among H3 nucleosomes along the centromeric DNA [144]. In three-dimension, CENP-A nucleosomes likely form clusters in the centromere chromosome [145]. Despite extensive biochemical and structural studies on this centromeric-specific H3 variant, the molecular mechanism of how CENP-A facilitates the assembly of centromeric chromatin remains elusive. Two recent cryo-EM studies shed light on this question by investigating the effects of CENP-A on chromatin higher-order structures. One study presented a mononucleosome stack formed by two CENP-A nucleosomes connected by two copies of CENP-N. It showed that CENP-N promotes the stacking of CENP-A nucleosomes through a previously undefined interaction between its ⍺6 helix and the DNA of the neighboring nucleosome at SHL 4 &5 [146]. Using Analytical Ultracentrifugation (AUC), the authors showed that single mutations in the ⍺6 helix affect nucleosome-nucleosome interactions. The structure indicates the importance of the histone H4 tail in mediating nucleosome compaction by promoting CENP-A nucleosome stacks. This study also presented a cryo-EM structure of CENP-A dodeca-nucleosome fiber compacted by CENP-N, showing a two-start helix-like structure (Fig. 4D). Notably, the interaction of ⍺6 helix of CENP-N with the DNA from adjacent nucleosomes in the fiber is distinct from those in the CENP-A mononucleosome stack. Compared with the H1 canonical fiber, it was also found that the relative orientation of n and n + 2 nucleosomes are conserved between the two fibers. Yet, the inter-nucleosome distance between the di-nucleosome stacks is larger in the CENP-A fiber. This likely reflects the need to accommodate the extra CENP-N proteins between the nucleosomes. The study also shows that the CENP-A nucleosome array without CENP-N forms a more relaxed parallel structure, confirming that CENP-N promotes the stacking of CENP-A mononucleosome and the compaction of CENP-A arrays. The former may reflect the interactions between distanced CENP-A nucleosomes clustered in the centromere. In contrast, the latter may reflect how CENP-A interacts with neighboring nucleosomes at the centromere to promote centromeric-specific chromatin structure [146].
On the other hand, the cryo-EM study by Takizawa et al. used reconstituted tri-nucleosomes containing CENP-A nucleosome flanking by two H3 nucleosomes to mimic the interspersed CENP-A nucleosome arrangement in vivo [147]. The subsequent cryo-EM structure shows that the H3-CENP-A-H3 tri-nucleosome array is flexible and similar to H3-H3-H3 arrays in vitro without divalent cations. When the physiological concentration of Mg2+ was added, the H3-CENP-A-H3 tri-nucleosome array adopted a more compact structure where the CENP-A nucleosome exhibits an untwisted conformation with an outward-facing linker DNA path (Fig. 4D). The H3-H3-H3 tri-nucleosome arrays, on the other hand, use an inward-facing DNA path and adopt a twisted conformation under the same condition. The authors propose that the untwisted organization allows the CENP-A nucleosome to be exposed to solvent and thus be accessible to interacting partners in a condensed centromere chromatin environment.
4. Concluding remarks and future perspectives
Histone variant exchange is a major mechanism the cell uses to diversify the chromatin building block and to fine-tune the structure and dynamics of chromatin. Some variants serve as a specific binding site for chromatin regulators and chromatin-modifying enzymes. Other variants play a more active role in transcription, replication, and DNA repair by modulating genome accessibility through differential recruitment of trans-associated factors. Despite their well-established functions, much remains unanswered regarding the molecular mechanism of histone variants' action and their regulation. In this review, we highlighted recent work on histone variants involving cryo-EM to dissect their versatility in structures and functions. The cryo-EM studies provide direct structural evidence to support the long-standing speculation that variant-specific sequence alterations lead to structural changes in chromatin. These structural changes, primarily in regions that directly influence the dynamics of entry/exit DNAs, are more substantial than those reported by previous crystallographic studies. These regions, including the histone H2A C-terminus and the histone H3 ⍺N helix, are highly divergent among variants from the H2A and the H3 families. They are also hotspots for histone PTMs. Interestingly, the recent cryo-EM study on endogenous nucleosomes isolated from Xenopus egg extract indicated that linker DNA and H2A C-terminal tail are among the structural variations observed across different nucleosome samples [86]. Diversifying and modulating these structural elements appears to be a common principle shared by various epigenetic mechanisms to control gene accessibility and functions.
Recent advances in single-particle cryo-EM are instrumental to the remarkable progress we have experienced in histone variant research. With more advancements and the rise of cryo-electron tomography, the technique will continue to play an important role in the histone variant field, especially for mechanistic questions that have proven challenging to tackle by other structural methods. For example, nucleosomes containing full-length (FL) variant macroH2A have proven challenging for structural studies due to their flexible linker region and the unique macro domain outside its histone fold. Single-particle cryo-EM may be able to capture intermediate structural states of nucleosome and chromatin fiber containing FL-macroH2A, which is expected to provide valuable insights into the mechanism-of-action of macro-H2A related to X-chromosome inactivation. In addition, cryo-EM is an ideal tool to investigate how different PTMs and oncogenic mutations on histone variants modulate chromatin structure and how the changes infer their functions. Finally, it remains to be seen how interplays of histone variants with each other and with non-histone proteins affect the structure and dynamics of chromatin at the molecular level.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the members of the Tan lab for fruitful discussions and support. Research in the Tan lab is supported by funding from the National Institute of General Medical Sciences of NIH under award number 1R35GM133611 and by the National Science Foundation under award number 194204.
References
- 1.Cheng Y. Single-particle Cryo-EM at crystallographic resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Hergeth S.P., Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439–1453. doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 5.Finch J.T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huynh V.A., Robinson P.J., Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 7.Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widom J., Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 9.Simpson R.T., Thoma F., Brubaker J.M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 10.Andersson K., Mahr R., Bjorkroth B., Daneholt B. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma. 1982;87:33–48. doi: 10.1007/BF00333508. [DOI] [PubMed] [Google Scholar]
- 11.Langmore J.P., Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 12.Marsden M.P., Laemmli U.K. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 13.Maeshima K., Ide S., Babokhov M. Dynamic chromatin organization without the 30-nm fiber. Curr Opin Cell Biol. 2019;58:95–104. doi: 10.1016/j.ceb.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Ricci M.A., Manzo C., Garcia-Parajo M.F., Lakadamyali M., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 15.Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., O'Shea C.C. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357 doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohno M., Ando T., Priest D.G., Kumar V., Yoshida Y., Taniguchi Y. Sub-nucleosomal Genome Structure Reveals Distinct Nucleosome Folding Motifs. Cell. 2019;176(520–534):e525. doi: 10.1016/j.cell.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Risca V.I., Denny S.K., Straight A.F., Greenleaf W.J. Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature. 2017;541:237–241. doi: 10.1038/nature20781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzluff W.F., Gongidi P., Woods K.R., Jin J., Maltais L.J. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 19.Weber C.M., Ramachandran S., Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Talbert P.B., Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 21.Fan J.Y., Gordon F., Luger K., Hansen J.C., Tremethick D.J. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- 22.Suto R.K., Clarkson M.J., Tremethick D.J., Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarthy S., Luger K. The histone variant macro-H2A preferentially forms “hybrid nucleosomes”. J Biol Chem. 2006;281:25522–25531. doi: 10.1074/jbc.M602258200. [DOI] [PubMed] [Google Scholar]
- 24.Hirai S., Tomimatsu K., Miyawaki-Kuwakado A., Takizawa Y., Komatsu T., Tachibana T., et al. Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant. Nucleic Acids Res. 2022;50:72–91. doi: 10.1093/nar/gkab1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou M., Dai L., Li C., Shi L., Huang Y., Guo Z., et al. Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2. EMBO J. 2021;40:e105907. doi: 10.15252/embj.2020105907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews A.J., Downing G., Brown K., Park Y.J., Luger K. A thermodynamic model for Nap1-histone interactions. J Biol Chem. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosammaparast N., Ewart C.S., Pemberton L.F. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selth L., Svejstrup J.Q. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green E.M., Antczak A.J., Bailey A.O., Franco A.A., Wu K.J., Yates J.R., 3rd, et al. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 32.Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., 3rd, Bassett E.A., Wood S., et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R.E. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C.C., Dechassa M.L., Bettini E., Ledoux M.B., Belisario C., Heun P., et al. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson R.T., Alekseev O.M., Grossman G., Widgren E.E., Thresher R., Wagner E.J., et al. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281:21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- 38.Shintomi K., Iwabuchi M., Saeki H., Ura K., Kishimoto T., Ohsumi K. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc Natl Acad Sci U S A. 2005;102:8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato K., Okuwaki M., Nagata K. Role of Template Activating Factor-I as a chaperone in linker histone dynamics. J Cell Sci. 2011;124:3254–3265. doi: 10.1242/jcs.083139. [DOI] [PubMed] [Google Scholar]
- 40.Mattiroli F., D'Arcy S., Luger K. The right place at the right time: chaperoning core histone variants. EMBO Rep. 2015;16:1454–1466. doi: 10.15252/embr.201540840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond C.M., Stromme C.B., Huang H., Patel D.J., Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18:141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess R.J., Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heo K., Kim H., Choi S.H., Choi J., Kim K., Gu J., et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 44.Li A., Yu Y., Lee S.C., Ishibashi T., Lees-Miller S.P., Ausio J. Phosphorylation of histone H2A.X by DNA-dependent protein kinase is not affected by core histone acetylation, but it alters nucleosome stability and histone H1 binding. J Biol Chem. 2010;285:17778–17788. doi: 10.1074/jbc.M110.116426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis T.S., Sokolova V., Jung H., Ng H., Tan D. Structural basis of chromatin regulation by histone variant H2A.Z. Nucleic Acids Res. 2021;49:11379–11391. doi: 10.1093/nar/gkab907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horikoshi N., Sato K., Shimada K., Arimura Y., Osakabe A., Tachiwana H., et al. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Crystallogr D Biol Crystallogr. 2013;69:2431–2439. doi: 10.1107/S090744491302252X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J., Fan J.Y., Rangasamy D., Tremethick D.J. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 48.McGinty R.K., Tan S. Recognition of the nucleosome by chromatin factors and enzymes. Curr Opin Struct Biol. 2016;37:54–61. doi: 10.1016/j.sbi.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faast R., Thonglairoam V., Schulz T.C., Beall J., Wells J.R., Taylor H., et al. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 50.Colino-Sanguino Y., Clark S.J., Valdes-Mora F. The H2A.Z-nuclesome code in mammals: emerging functions. Trends Genet. 2022;38:273–289. doi: 10.1016/j.tig.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Lashgari A., Millau J.F., Jacques P.E., Gaudreau L. Global inhibition of transcription causes an increase in histone H2A.Z incorporation within gene bodies. Nucleic Acids Res. 2017;45:12715–12722. doi: 10.1093/nar/gkx879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Latorre I., Chesney M.A., Garrigues J.M., Stempor P., Appert A., Francesconi M., et al. The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev. 2015;29:495–500. doi: 10.1101/gad.255810.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marques M., Laflamme L., Gervais A.L., Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010;5:267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- 54.Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., et al. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H., Roberts D.N., Cairns B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.John S., Sabo P.J., Johnson T.A., Sung M.H., Biddie S.C., Lightman S.L., et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Sutcliffe E.L., Parish I.A., He Y.Q., Juelich T., Tierney M.L., Rangasamy D., et al. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol. 2009;29:1972–1986. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong M.M., Cox L.K., Chrivia J.C. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J Biol Chem. 2007;282:26132–26139. doi: 10.1074/jbc.M703418200. [DOI] [PubMed] [Google Scholar]
- 60.Wen Z., Zhang L., Ruan H., Li G. Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells. Nucleic Acids Res. 2020;48:5939–5952. doi: 10.1093/nar/gkaa360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greaves I.K., Rangasamy D., Ridgway P., Tremethick D.J. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci U S A. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rangasamy D., Berven L., Ridgway P., Tremethick D.J. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farris S.D., Rubio E.D., Moon J.J., Gombert W.M., Nelson B.H., Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J Biol Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 64.Gevry N., Chan H.M., Laflamme L., Livingston D.M., Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotekar A.S., Weissman J.D., Gegonne A., Cohen H., Singer D.S. Histone modifications, but not nucleosomal positioning, correlate with major histocompatibility complex class I promoter activity in different tissues in vivo. Mol Cell Biol. 2008;28:7323–7336. doi: 10.1128/MCB.00889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horikoshi N., Arimura Y., Taguchi H., Kurumizaka H. Crystal structures of heterotypic nucleosomes containing histones H2A.Z and H2A. Open Biol. 2016;6 doi: 10.1098/rsob.160127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park Y.J., Dyer P.N., Tremethick D.J., Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 68.Horikoshi N., Kujirai T., Sato K., Kimura H., Kurumizaka H. Structure-based design of an H2A.Z.1 mutant stabilizing a nucleosome in vitro and in vivo. Biochem Biophys Res Commun. 2019;515:719–724. doi: 10.1016/j.bbrc.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 69.Dryhurst D., Ishibashi T., Rose K.L., Eirin-Lopez J.M., McDonald D., Silva-Moreno B., et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wratting D., Thistlethwaite A., Harris M., Zeef L.A., Millar C.B. A conserved function for the H2A.Z C terminus. J Biol Chem. 2012;287:19148–19157. doi: 10.1074/jbc.M111.317990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonisch C., Schneider K., Punzeler S., Wiedemann S.M., Bielmeier C., Bocola M., et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 2012;40:5951–5964. doi: 10.1093/nar/gks267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clarkson M.J., Wells J.R., Gibson F., Saint R., Tremethick D.J. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. doi: 10.1038/21436. [DOI] [PubMed] [Google Scholar]
- 73.Tachiwana H., Dacher M., Maehara K., Harada A., Seto Y., Katayama R., et al. Chromatin structure-dependent histone incorporation revealed by a genome-wide deposition assay. Elife. 2021;10 doi: 10.7554/eLife.66290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chadwick B.P., Willard H.F. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soboleva T.A., Nekrasov M., Pahwa A., Williams R., Huttley G.A., Tremethick D.J. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol. 2011;19:25–30. doi: 10.1038/nsmb.2161. [DOI] [PubMed] [Google Scholar]
- 76.Nekrasov M., Soboleva T.A., Jack C., Tremethick D.J. Histone variant selectivity at the transcription start site: H2A.Z or H2A.Lap1. Nucleus. 2013;4:431–438. doi: 10.4161/nucl.26862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soboleva T.A., Nekrasov M., Ryan D.P., Tremethick D.J. Histone variants at the transcription start-site. Trends Genet. 2014;30:199–209. doi: 10.1016/j.tig.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Soboleva T.A., Parker B.J., Nekrasov M., Hart-Smith G., Tay Y.J., Tng W.Q., et al. A new link between transcriptional initiation and pre-mRNA splicing: The RNA binding histone variant H2A.B. PLoS Genet. 2017;13:e1006633. doi: 10.1371/journal.pgen.1006633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tolstorukov M.Y., Goldman J.A., Gilbert C., Ogryzko V., Kingston R.E., Park P.J. Histone variant H2A.Bbd is associated with active transcription and mRNA processing in human cells. Mol Cell. 2012;47:596–607. doi: 10.1016/j.molcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sansoni V., Casas-Delucchi C.S., Rajan M., Schmidt A., Bonisch C., Thomae A.W., et al. The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 2014;42:6405–6420. doi: 10.1093/nar/gku303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Molaro A., Young J.M., Malik H.S. Evolutionary origins and diversification of testis-specific short histone H2A variants in mammals. Genome Res. 2018;28:460–473. doi: 10.1101/gr.229799.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bao Y., Konesky K., Park Y.J., Rosu S., Dyer P.N., Rangasamy D., et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 84.Downs J.A., Lowndes N.F., Jackson S.P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 85.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 86.Arimura Y., Shih R.M., Froom R., Funabiki H. Structural features of nucleosomes in interphase and metaphase chromosomes. Mol Cell. 2021;81(4377–4397):e4312. doi: 10.1016/j.molcel.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palmer D.K., O'Day K., Margolis R.L. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma. 1990;100:32–36. doi: 10.1007/BF00337600. [DOI] [PubMed] [Google Scholar]
- 88.Palmer D.K., O'Day K., Trong H.L., Charbonneau H., Margolis R.L. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fachinetti D., Han J.S., McMahon M.A., Ly P., Abdullah A., Wong A.J., et al. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Regnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., et al. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 93.Shrestha R.L., Ahn G.S., Staples M.I., Sathyan K.M., Karpova T.S., Foltz D.R., et al. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget. 2017;8:46781–46800. doi: 10.18632/oncotarget.18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 95.McKinley K.L., Cheeseman I.M. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 2016;17:16–29. doi: 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Westhorpe F.G., Fuller C.J., Straight A.F. A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J Cell Biol. 2015;209:789–801. doi: 10.1083/jcb.201503132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boopathi R., Danev R., Khoshouei M., Kale S., Nahata S., Ramos L., et al. Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends. Nucleic Acids Res. 2020;48:5735–5748. doi: 10.1093/nar/gkaa246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Migl D., Kschonsak M., Arthur C.P., Khin Y., Harrison S.C., Ciferri C., et al. Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 A Resolution. Structure. 2020;28:363–370 e363. doi: 10.1016/j.str.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ali-Ahmad A., Bilokapic S., Schafer I.B., Halic M., Sekulic N. CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail. EMBO Rep. 2019;20:e48913. doi: 10.15252/embr.201948913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A.E., et al. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 2018;359:339–343. doi: 10.1126/science.aar2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G.P., et al. Decoding the centromeric nucleosome through CENP-N. Elife. 2017;6 doi: 10.7554/eLife.33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tian T., Li X., Liu Y., Wang C., Liu X., Bi G., et al. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 2018;28:374–378. doi: 10.1038/cr.2018.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roulland Y., Ouararhni K., Naidenov M., Ramos L., Shuaib M., Syed S.H., et al. The Flexible Ends of CENP-A Nucleosome Are Required for Mitotic Fidelity. Mol Cell. 2016;63:674–685. doi: 10.1016/j.molcel.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 104.Yan K., Yang J., Zhang Z., McLaughlin S.H., Chang L., Fasci D., et al. Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature. 2019;574:278–282. doi: 10.1038/s41586-019-1609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yatskevich S., Muir K.W., Bellini D., Zhang Z., Yang J., Tischer T., et al. Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome. Science. 2022;376:844–852. doi: 10.1126/science.abn3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Heijden G.W., Derijck A.A., Posfai E., Giele M., Pelczar P., Ramos L., et al. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 107.Schwartz B.E., Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams J.S., Hayashi T., Yanagida M., Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 110.Tachiwana H., Osakabe A., Shiga T., Miya Y., Kimura H., Kagawa W., et al. Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr D Biol Crystallogr. 2011;67:578–583. doi: 10.1107/S0907444911014818. [DOI] [PubMed] [Google Scholar]
- 111.Chen P., Zhao J., Wang Y., Wang M., Long H., Liang D., et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maehara K., Harada A., Sato Y., Matsumoto M., Nakayama K.I., Kimura H., et al. Tissue-specific expression of histone H3 variants diversified after species separation. Epigenetics Chromatin. 2015;8:35. doi: 10.1186/s13072-015-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fan Y., Nikitina T., Morin-Kensicki E.M., Zhao J., Magnuson T.R., Woodcock C.L., et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 117.Hashimoto H., Takami Y., Sonoda E., Iwasaki T., Iwano H., Tachibana M., et al. Histone H1 null vertebrate cells exhibit altered nucleosome architecture. Nucleic Acids Res. 2010;38:3533–3545. doi: 10.1093/nar/gkq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hellauer K., Sirard E., Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- 119.Sancho M., Diani E., Beato M., Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4:e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shen X., Gorovsky M.A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 121.Vujatovic O., Zaragoza K., Vaquero A., Reina O., Bernues J., Azorin F. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012;40:5402–5414. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pan C., Fan Y. Role of H1 linker histones in mammalian development and stem cell differentiation. Biochim Biophys Acta. 2016;1859:496–509. doi: 10.1016/j.bbagrm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Allan J., Hartman P.G., Crane-Robinson C., Aviles F.X. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 124.Simpson R.T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 125.Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 126.Zhou B.R., Feng H., Kale S., Fox T., Khant H., de Val N., et al. Distinct structures and dynamics of chromatosomes with different human linker histone isoforms. Mol Cell. 2021;81(166–182):e166. doi: 10.1016/j.molcel.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou B.R., Feng H., Ghirlando R., Li S., Schwieters C.D., Bai Y. A small number of residues can determine if linker histones are bound on or off dyad in the chromatosome. J Mol Biol. 2016;428:3948–3959. doi: 10.1016/j.jmb.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou B.R., Feng H., Kato H., Dai L., Yang Y., Zhou Y., et al. Structural insights into the histone H1-nucleosome complex. Proc Natl Acad Sci U S A. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 130.Zhou B.R., Jiang J., Feng H., Ghirlando R., Xiao T.S., Bai Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Mol Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bednar J., Garcia-Saez I., Boopathi R., Cutter A.R., Papai G., Reymer A., et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol Cell. 2017;66(384–397):e388. doi: 10.1016/j.molcel.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brown D.T., Izard T., Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Takata H., Matsunaga S., Morimoto A., Ono-Maniwa R., Uchiyama S., Fukui K. H1.X with different properties from other linker histones is required for mitotic progression. FEBS Lett. 2007;581:3783–3788. doi: 10.1016/j.febslet.2007.06.076. [DOI] [PubMed] [Google Scholar]
- 134.Hergeth S.P., Dundr M., Tropberger P., Zee B.M., Garcia B.A., Daujat S., et al. Isoform-specific phosphorylation of human linker histone H1.4 in mitosis by the kinase Aurora B. J Cell Sci. 2011;124:1623–1628. doi: 10.1242/jcs.084947. [DOI] [PubMed] [Google Scholar]
- 135.Flex E., Martinelli S., Van Dijck A., Ciolfi A., Cecchetti S., Coluzzi E., et al. Aberrant Function of the C-Terminal Tail of HIST1H1E Accelerates Cellular Senescence and Causes Premature Aging. Am J Hum Genet. 2019;105:493–508. doi: 10.1016/j.ajhg.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Duffney L.J., Valdez P., Tremblay M.W., Cao X., Montgomery S., McConkie-Rosell A., et al. Epigenetics and autism spectrum disorder: A report of an autism case with mutation in H1 linker histone HIST1H1E and literature review. Am J Med Genet B Neuropsychiatr Genet. 2018;177:426–433. doi: 10.1002/ajmg.b.32631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dorigo B., Schalch T., Bystricky K., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 138.Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977;12:101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- 139.Schalch T., Duda S., Sargent D.F., Richmond T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 140.Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R.R., Richmond T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 141.Ekundayo B., Richmond T.J., Schalch T. Capturing Structural Heterogeneity in Chromatin Fibers. J Mol Biol. 2017;429:3031–3042. doi: 10.1016/j.jmb.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 142.Song F., Chen P., Sun D., Wang M., Dong L., Liang D., et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 143.Garcia-Saez I., Menoni H., Boopathi R., Shukla M.S., Soueidan L., Noirclerc-Savoye M., et al. Structure of an H1-Bound 6-nucleosome array reveals an untwisted two-start chromatin fiber conformation. Mol Cell. 2018;72(902–915):e907. doi: 10.1016/j.molcel.2018.09.027. [DOI] [PubMed] [Google Scholar]
- 144.Nishimura K., Komiya M., Hori T., Itoh T., Fukagawa T. 3D genomic architecture reveals that neocentromeres associate with heterochromatin regions. J Cell Biol. 2019;218:134–149. doi: 10.1083/jcb.201805003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Allshire R.C., Karpen G.H. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou K., Gebala M., Woods D., Sundararajan K., Edwards G., Krzizike D., et al. CENP-N promotes the compaction of centromeric chromatin. Nat Struct Mol Biol. 2022;29:403–413. doi: 10.1038/s41594-022-00758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Takizawa Y., Ho C.H., Tachiwana H., Matsunami H., Kobayashi W., Suzuki M., et al. Cryo-EM structures of centromeric tri-nucleosomes containing a central CENP-A nucleosome. Structure. 2020;28(44–53):e44. doi: 10.1016/j.str.2019.10.016. [DOI] [PubMed] [Google Scholar]