Abstract
The Major Histocompatibility Complex class I–related protein 1 (MR1) presents small molecule metabolites, drugs, and drug-like molecules that are recognized by MR1-reactive T cells. While we have an understanding of how antigens bind to MR1 and upregulate MR1 cell surface expression, a quantitative, cell-free, assessment of MR1 ligand-binding affinity was lacking. Here, we developed a fluorescence polarization–based assay in which fluorescent MR1 ligand was loaded into MR1 protein in vitro and competitively displaced by candidate ligands over a range of concentrations. Using this assay, ligand affinity for MR1 could be differentiated as strong (IC50 < 1 μM), moderate (1 μM < IC50 < 100 μM), and weak (IC50 > 100 μM). We demonstrated a clear correlation between ligand-binding affinity for MR1, the presence of a covalent bond between MR1 and ligand, and the number of salt bridge and hydrogen bonds formed between MR1 and ligand. Using this newly developed fluorescence polarization–based assay to screen for candidate ligands, we identified the dietary molecules vanillin and ethylvanillin as weak bona fide MR1 ligands. Both upregulated MR1 on the surface of C1R.MR1 cells and the crystal structure of a MAIT cell T cell receptor–MR1–ethylvanillin complex revealed that ethylvanillin formed a Schiff base with K43 of MR1 and was buried within the Aʹ-pocket. Collectively, we developed and validated a method to quantitate the binding affinities of ligands for MR1 that will enable an efficient and rapid screening of candidate MR1 ligands.
Keywords: Immunology, structural biology, fluorescence, antigen presentation
Abbreviations: 5-OP-RU, (5-(2-oxopropylideneamino)-6-D-ribitylaminouracil; 6-FP, 6-formylpterin; Ac-6-FP, acetyl-6-formylpterin; Ags, antigens; BSA, buried surface area; DCF, diclofenac; FP, fluorescence polarization; HMB, 2-hydroxy-5-methoxybenzaldehyde; MAIT, mucosal-associated invariant T; MHC, major histocompatibility complex; MR1, MHC-I related protein 1; TCR, T cell receptor; VDW, van der Waals; 3-F-SA, 3-formylsalicylic acid; RL-6-Me-7-OH, 7-hydroxy-6-methyl-8-D-ribityllumazine; RL-6-Me, 6-methyl-8-D-ribityllumazine
In vertebrates, protective immunity against pathogens relies heavily on a specialized class of molecules, encoded by the Major Histocompatibility Complex (MHC), which present pathogen-derived peptides to T cells for recognition and elimination. In jawed vertebrates, there exist three T cell lineages, namely, αβ, γδ, γμ T cells, which are defined by the cell surface expression of T cell receptor (TCR) heterodimers composed of α- and β- or γ- and δ- or γ- and μ-polypeptide chains, respectively (1, 2, 3, 4). The fundamental principles underscoring the mechanism of recognition of the MHC-peptide complex by αβ TCRs have been extensively studied over the past 25 years (5).
In addition to classical MHC-I and MHC-II molecules, MHC-I–like molecules exist that include the MHC-I related protein 1 (MR1) and the CD1 family of glycoproteins that present small molecules and lipid-derived antigens (Ags) to T cells, respectively (5). Such MHC-I–like molecules can be recognized by specialized immune cell subsets that include αβ and γδ T cells (5, 6, 7, 8).
The monomorphic MR1 molecule is ubiquitously expressed in all human cells and shares structural similarities with the classical MHC-I molecules, although it has evolved to accommodate a chemically distinct class of Ags, namely, vitamin B2- and B9-derived metabolites (9, 10) that are recognized by unconventional subset of T cells, featuring a semi-invariant TCR, called mucosal-associated invariant T (MAIT) cells (11) and more diverse MR1-restricted reactive αβ and γδ T cells (6, 7, 12, 13, 14, 15). The structure and chemical properties of the MR1 Ag-binding cleft are ideally suited to bind small molecules, whereby the ligands are closely sequestered by a cradle of aromatic and polar residues that form the A′-pocket (9). For the majority of known MR1 ligands harboring aldehyde or ketone moieties, a covalent imine (Schiff base) is formed between K43 of MR1 and the reactive aldehyde or ketone functional groups (9). The relative plasticity of the MR1-binding groove is attributable to an A′-pocket capable of accommodating chemically distinct classes of Ags (pterin-, lumazine-, pyrimidine-based ligands). For instance, while both folate- (vitamin B9) and riboflavin-based (vitamin B2) ligands form a Schiff base with K43, their respective conformation within the A′-pocket differed markedly, thus enabling the ribityl moiety of the riboflavin-derived ligands to interact with two arginine residues (R9 and R94) from MR1 via hydrogen bonding.
This evident malleability of the MR1-binding groove has recently led to the exploration of the chemical space for novel MR1-restricted Ags (16, 17, 18, 19), thereby highlighting the chemical diversity of molecules that can be presented by MR1 and activate MAIT cells (20). In-roads have been made in understanding the cellular trafficking of MR1 (21, 22, 23), the molecular basis underpinning antigen presentation by MR1, as well as MAIT TCR activation potency and specificity of MR1-restricted ligands (18). However, our insights into quantitative measurements of ligand affinity for MR1 have not been established. Fluorescence polarization (FP) has previously been used to determine the binding affinity of fluorescently labeled peptides for MHC-I and MHC-II (24, 25). Here, we adapted this approach by using a synthetic fluorescent MR1-restricted ligand (JYM20) (21, 26) to develop a FP-based competitive assay to investigate the binding affinity (IC50) of 16 structurally diverse ligands to MR1 and the molecular determinants that underpinned their affinity hierarchy towards MR1. The FP assay enabled the identification of two diet-derived ligands (vanillin and ethylvanillin) as ‘weak’ MR1 ligands. The crystal structure of a MAIT TCR in complex with MR1-ethylvanillin showed how ethylvanillin was sequestered within the A′-binding pocket of MR1, thereby confirming its ability to bind MR1. This FP-based assay represents a fast and reliable means to determine the affinity of candidate ligands for MR1 and may be a useful screening tool.
Results
An FP-based competitive assay for MR1
While we have an understanding of how Ags bind to MR1 and upregulate MR1 cell surface expression, the affinity of ligands binding to MR1 are yet to be determined. The affinity of ligands was previously correlated to MR1-ligand thermal stability and in vitro–refolding yields (18, 27, 28). However, thermal stability is not a direct measure of ligand binding and is only applicable to ligands that have the ability to be refolded with MR1 in vitro. Based on previous studies whereby FP was used to examine MHC–peptide interactions (24, 26), we sought to develop an FP competitive-binding assay for the quantitative measurement of the affinity of MR1 ligands for MR1 in a cell-free environment. In our assay, we used JYM20, a chemically stable fluorescent analog of the most potent MAIT cell Ags, (5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) (Fig. 1A) (21). JYM20 has its fluorescent tetramethylrhodamine motif sufficiently distant from its MR1-binding motif that it does not interfere with MR1 binding. It has been previously shown that JYM20 competitively inhibits 5-OP-RU activation of MAIT cells and that JYM20 binds to MR1 but not to MHC-II, indicating the MR1-specific binding nature of JYM20 (21).
Figure 1.
A fluorescent polarization assay for MR1.A, chemical structure of the fluorescent MR1-restricted ligand JYM20. B, schematic of the process for the generation of ‘empty’ MR1. C, anion-exchange chromatographic profile of MR1-empty (left), SDS-PAGE gel analysis of eluted MR1-empty (middle), and native PAGE of eluted MR1-empty with and without a conformation-specific MR1 antibody (26.5) (44) (right). D, schematic of the FP competitive assay adapted for MR1. BTP, Bis-tris propane; HMB, 2-hydroxy-5-methoxybenzaldehyde; MR1, MHC-I-related protein 1.
To efficiently load JYM20 into MR1 in vitro, we generated an ‘empty’ soluble MR1 molecule (Fig. 1, B and C). This process involved the in vitro refolding of MR1 with the compound 2-hydroxy-5-methoxybenzaldehyde (HMB) and its subsequent displacement through dialysis at pH 6.5 (Fig. 1B) (16) via acidic hydrolysis of the Schiff base between HMB and K43 of MR1 (22), enabling the removal of HMB from the MR1-binding groove. This has been used previously to determine the structure of drug and drug-like molecules binding to MR1 (16). The dialysate was then collected, and the MR1-empty molecule was purified by anion-exchange chromatography (Fig. 1C). The MR1-empty molecule eluted at the expected conductivity for MR1. The stoichiometry and size of MR1 and β2-microglobulin (β2m) were confirmed using SDS-PAGE analysis (Fig. 1C) and its appropriate folding state was confirmed using a conformation-specific MR1 antibody (26.5) (Fig. 1C).
JYM20 can be readily synthesized in high yield and thus, FP was chosen as an ideal and efficient technique for affinity measurements. To determine the optimal conditions for FP, a direct binding assay of JYM20 and MR1 was first performed to identify the minimum concentration of MR1 and JYM20 required to produce a maximal signal, as well as the buffer conditions to maximize stability of the MR1–JYM20 complex. This signal is generated by polarized excitation wavelengths that will be absorbed and emitted as depolarized when JYM20 is free in solution as the molecule tumbles freely, resulting in a low FP value. The light emitted is mostly polarized when JYM20 is bound to MR1, due to the significantly slower tumble speed of JYM20 when bound to the larger MR1 protein (29).
The generation of a refolded ‘empty’ MR1 molecule (Fig. 1B) and the availability of a fluorescent MR1-restricted ligand (JYM20) allowed for the development of an FP-based competitive assay to measure the affinity of a wide range of chemically distinct MR1 ligands (Fig. 1D). Here, a buffer consisting of 25 mM Hepes pH 7.5, 150 mM NaCl, and 5 mM Na-EDTA was used. This was superior to nonphysiological pH buffers such as 25 mM Tris pH 8.5 and 25 mM citric acid pH 6.5 in which MR1 became unstable over time. By adding titrating amounts of MR1-empty to a fixed concentration of JYM20 (Fig. 2A), we determined that a sample volume of 20 μl, a concentration of 100 nM and 10 nM of MR1-empty and JYM20, respectively, and an incubation time of 48 h at 25 °C led to optimal results. Experiments performed at 4 °C did not allow the mixture to reach equilibrium within 96 h, and at 37 °C, the MR1 molecule began to degrade. A 48 h incubation time was chosen to minimize any degradation while still enabling the mixture to reach equilibrium.
Figure 2.
Determination of conditions for the FP competitive assay.A, a dilution series of MR1-empty added to 10 nM of JYM20 allowed for the determination of the binding affinity of JYM20 for MR1 (Kd = 20 nM, 95% CI [18–23 nM]). Refolded MHC-I-NLV was used as a negative control to confirm the specificity of JYM20 for MR1. B, competitive FP-binding assay of Ac-6-FP and a nonbinder peptide NLV. Each data point represents mean values from three independent experiments performed in triplicate. Mean values are plotted with SD represented in error bars. Ac-6-FP, acetyl-6-formylpterin; FP, fluorescence polarization; MHC, major histocompatibility complex; MR1, MHC-I related protein 1.
To determine the specificity of JYM20 towards MR1, an unrelated protein, the MHC-I molecule HLA-A2 (30), was used as a negative control and was shown to not bind to JYM20 (Fig. 2A). Next, we investigated whether the FP competitive-binding assay could differentiate between MR1 binders and nonbinders. We used acetyl-6-formylpterin (Ac-6-FP) as the ligand of choice that was known to bind to MR1 (27), and the native cytomegalovirus MHC-I–restricted peptide (NLV) (30) as a negative control, demonstrating that JYM20 is able to be displaced from the MR1 antigen-binding pocket by known ligand Ac-6-FP but not by nonbinder NLV (Fig. 2B). In summary, this work has identified the optimal experimental conditions for an FP-based competitive assay for MR1 ligand association and demonstrated its specificity for MR1 ligands.
An affinity hierarchy among MR1-binding ligands
Using the FP competitive-binding assay described above, we measured the relative affinity (IC50) of a range of chemically distinct classes of known MR1 ligands (Figs. 3, 4, Table 1 and Table S1). Namely, we investigated 16 ligands (Fig. 3) that included the pterin-based ligands: 6-formylpterin (6-FP) (9) and Ac-6-FP (27); the pyrimidine-based ligands: 5-OP-RU, 2′D-5-OP-RU, 3′D-5-OP-RU, JYM72 (formerly compound 11), the ribityl-less analog of JYM72, 3-[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)formamido]propanoic acid (DB28) and NV18.1 (10, 17, 18, 31); the lumazine-based ligands: 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH), 6-methyl-8-D-ribityllumazine (RL-6-Me), and 2′D-RL-7-Me (9, 32); as well as HMB, 3-formylsalicylic acid (3-F-SA), and the diclofenac (DCF)-based ligands DCF and 5-OH-DCF (16).
Figure 3.
Chemical structures of MR1-restricted ligands. Pterin-based ligands: 6-formylpterin (6-FP) and acetyl-6-formylpterin (Ac-6-FP) (9); pyrimidine-based ligands: (5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), 2′D-5-OP-RU, 3′D-5-OP-RU, JYM72, ribityl-less analog 3-[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)formamido]propanoic acid (DB28), and NV18.1 (17, 18); lumazine-based ligands: 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH), 6-methyl-8-D-ribityllumazine (RL-6-Me), and 2′D-RL-7-Me (9, 32); benzaldehyde-based ligand: 2-hydroxy-5-methoxybenzaldehyde (HMB) (16); salicylate-based ligand: salicylate 3-formylsalicylic acid (3-F-SA) (16); and the diclofenac-derived ligands: diclofenac (DCF) and 5-OH-DCF (16). MR1, MHC-I related protein 1.
Figure 4.
MR1-restricted ligand affinities (IC50). Titration curves of known ligands binding to MR1. Strong binders (blue), moderate binders (yellow), and weak binders (green). Each data point represents normalized percentage binding from three independent experiments performed in triplicate. Mean values are plotted with SD represented in error bars. Curve fit for ligands are displayed in Table S1. MR1, MHC-I-related protein 1.
Table 1.
IC50 of MR1-restricted ligands
| Ligands | IC50 | 95% CI |
|---|---|---|
| Strong binders | ||
| 5-OP-RU | 4.2 nM | 3.4–5.2 nM |
| JYM72 | 27 nM | 23–31 nM |
| Ac-6-FP | 39 nM | 24–63 nM |
| 2′D-5-OP-RU | 64 nM | 53–77 nM |
| 6-FP | 108 nM | 93–124 nM |
| 3′D-5-OP-RU | 132 nM | 102–172 nM |
| Ribityl-less analog | 491 nM | 415–581 nM |
| 3-F-SA | 563 nM | 497–638 nM |
| Moderate binders | ||
| RL-6-Me-7-OH | 2.6 μM | 2.4–2.9 μM |
| 2′D-RL-7-Me | 11 μM | 9.3–14 μM |
| DB28 | 20 μM | 17–22 μM |
| Weak binders | ||
| RL-6-Me | 113 μM | 88–146 μM |
| Ethylvanillin | 325 μM | 293–361 μM |
| Vanillin | 325 μM | 293–361 μM |
| NV18.1 | 519 μM | 475–567 μM |
| HMB | 775 μM | 690–870 μM |
| 5-OH-DCF | 1.1 mM | 0.8–1.5 mM |
| DCF | 2.6 mM | 1.9–3.7 mM |
Strong binders
All the pterin-, pyrimidine-, and salicylate-based ligands that formed a Schiff base with MR1 exhibited the highest affinity for MR1 with IC50 < 1 μM under the given experimental conditions (Fig. 4 and Table 1). These compounds were considered to be ‘strong’ binders, highlighting the importance of the covalent bond for high affinity MR1 binding. Within this category, the most potent MAIT cell activator 5-OP-RU exhibited by far the highest affinity (IC50 ∼ 4 nM) followed by its analogs (JYM72, 2′D-5-OP-RU, and 3′D-5-OP-RU) with an ∼ 5- to 30-fold decrease in affinity relative to 5-OP-RU.
As the 5-OP-RU–based analogs share the same uracil scaffold, but have slight modifications in the polar ribityl chain, the observed decrease in IC50 further demonstrates the importance of the ribityl chain not only for MAIT TCR activation (18) but also for MR1 binding. Similarly, an ∼200-fold loss in affinity was observed between JYM72 and its ribityl-less analog. This is consistent with the modifications to the ribityl chain flexibility, which in turn, affects the ability of ribityl hydroxyl groups to form interactions with MR1 as observed structurally (33). Other well-characterized strong upregulators of MR1 cell surface expression, 6-FP and Ac-6-FP, also exhibited high affinity (IC50 ∼ 110 and 40 nM respectively) for MR1 that was similar to the 5-OP-RU–based analogs even though 6-FP and Ac-6FP are pterin rather than uracil derivatives and bind differently to MR1. Indeed, while 6-FP and Ac-6-FP do not possess a ribityl chain, they do have a carbonyl group to form a covalent bond with K43 of MR1 as well as a bicyclic ring system that forms additional hydrogen bonds and van der Waals (VDW) interactions to compensate for the absence of hydroxyl substituents on the ribityl chain of 5-OP-RU.
Moderate and weak binders
Within the ‘moderate’ (1 μM < IC50 < 100 μM) to ‘weak’ binders (IC50 >100 μM) (Table 1) are the non-Schiff base ligands that include the lumazines (RL-6-Me-7-OH, RL-6-Me, and 2′D-RL-7-Me), DCF and its analog (5-OH-DCF), and pyrimidine-derived ligands known to downregulate surface MR1 expression (NV18.1 and DB28) (17). Here, while the lumazine-derived ligands contained a ribityl or ribityl-modified moiety and a pterin-based scaffold, the lack of Schiff base formation with K43 clearly impacted their ability to bind tightly to MR1 with IC50 ranging between ∼ 2.5 to 100 μM. Conversely, the benzaldehyde-derived ligand HMB did form a Schiff base with MR1, albeit exhibiting a ‘weak’ binding affinity (IC50 ∼ 750 μM) due to the limited number of hydrogen bonds and VDW interactions (16).
Collectively, the results suggest that the ability of a given ligand to form a covalent bond with MR1 greatly increased its affinity for MR1. However, noncovalent interactions between MR1 and ligand can influence binding affinity for MR1-restricted ligands.
Diet-derived molecules can be presented by MR1
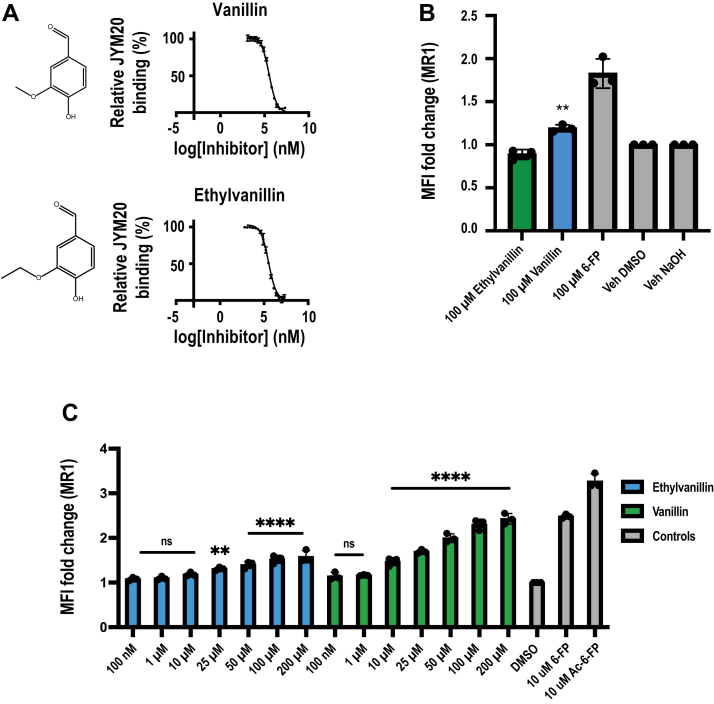
Previously, in silico fragment-based screening identified vanillin and ethylvanillin as aromatic molecules with hydrogen bonding substituents and an aldehyde group that could potentially enable binding to MR1 (16). To confirm this prediction, we used the FP assay to determine their binding affinities for MR1. Vanillin and ethylvanillin were found to have low affinities for MR1 with IC50: 300 to 350 μM categorizing them as weak ligands (Fig. 5A). Next, using flow cytometry to measure the upregulation of surface MR1 on the human lymphoblastoid C1R cell line overexpressing MR1 (C1R.MR1), we first examined MR1 upregulation after a 16 h incubation with ethylvanillin and vanillin (Fig. 5B). Here, under these conditions, we noted that ethylvanillin caused no noticeable upregulation of MR1, whereas vanillin enabled a modest increase in MR1 upregulation. Given that small molecules can be metabolized readily within a cellular environment, we then assayed for MR1 upregulation following a shorter incubation period (3 h). Here, we demonstrated that surface MR1 was upregulated after a 3 h incubation with ethylvanillin and vanillin (Fig. 5C). While both compounds exhibited the same affinity for MR1, vanillin upregulated MR1 significantly at a lower concentration (10 μM) than that of ethylvanillin (25 μM) (Fig. 5C) suggesting that the chemical properties of the compounds (e.g. solubility in aqueous environment) may impact their ability to enter the cells, traffic intracellularly, and to be presented by MR1 on the cell surface.
Figure 5.
Initial and functional characterization of the presentation of ethylvanillin and vanillin by MR1.A, titration curves of vanillin (left) and ethylvanillin (right). Each data point represents normalized percentage binding from three independent experiments. Mean values are plotted with SD represented by error bars. B, bar graphs depicting upregulation of surface MR1 on C1R.MR1 cells incubated for 16 h with titrated quantities of ligand. C, bar graphs depicting upregulation of surface MR1 on C1R.MR1 cells incubated for 3 h with titrated quantities of ligand. Upregulation is expressed as a fold change from basal surface MR1 upregulation upon incubation with the vehicle (DMSO). Each column represents the mean from three independent experiments with SD represented by error bars. Each independent experiment was performed in triplicate. One-way ANOVA statistical analysis was performed for all samples with multiple comparisons performed using DMSO as a control (ns: not significant, ∗∗p = 0.0013, ∗∗∗∗p < 0.0001). MR1, MHC-I related protein 1.
To gain insight into the molecular presentation of ethylvanillin by MR1, a ligand-exchange approach was used to load ethylvanillin in vitro into MR1-empty (16). Using high performance liquid chromatography tandem mass spectrometry analysis of the MAIT TCR–MR1–ethylvanillin complex crystals, we detected a major MS signal (167.1 m/z, [M + H]+) (Fig. S1A) that corresponded to intact ethylvanillin. This was an equivalent fragmentation to the ethylvanillin stock (Fig. S1B), confirming the presence of ethylvanillin within the MAIT TCR–MR1 complex.
Crystal structure of MAIT TCR–MR1–ethylvanillin complex
Next, we determined the crystal structure of the A-F7 MAIT TCR–MR1–ethylvanillin ternary complex to 2.5 Å resolution (Fig. 6A and Table S2). The electron density at the MR1/TCR interface and for the bound ethylvanillin was unambiguous (Fig. 6, B and C). The docking mode adopted by the A-F7 MAIT TCR to recognize the MR1–ethylvanillin binary complex was identical to previously published A-F7 MAIT TCR–MR1–Ag ternary complexes (16, 17, 18, 27, 32) (Table S3). Here, the A-F7 TCR adopted a central and orthogonal docking mode atop the MR1-binding groove formed by the α1- and α2-helices (Fig. 6A). The CDR3α and CDR3β loops dominated the interactions at the TCR/MR1 interface with a contribution of 19.6 % and 27 % to the buried surface area (BSA), respectively. The majority of the molecular interactions found at the TCR/MR1-Ag interface (Fig. 6, D–H) were conserved with those in all other A-F7 MAIT TCR–MR1–Ag ternary complexes.
Figure 6.
Molecular insights into the presentation of ethylvanillin by MR1. A, cartoon representation of the crystal structure of the A-F7 TCR–MR1–ethylvanillin ternary complex. MR1, gray; β2m, light green; TCRα, pink; CDR1α, olive; CDR2α, orange; CDR3α, yellow; TCRβ, cyan; CDR1β, blue; CDR2β, magenta; CDR3β, brown. Ethylvanillin is shown as green spheres. Electron density of the bound ethylvanillin are 2Fo-Fc (0.8σ level) (B) and Fo-Fc (2.2σ level) (C). Electron density maps are shown in blue and green, respectively. D and E, molecular interactions of CDR2β and CDR3β loops with MR1. CDR2β, magenta; CDR3β, brown; Framework (FW) residues, cyan; MR1, gray. F–H, molecular interactions of CDR1α, CDR2α, and CDR3α loops with MR1. CDR1α, olive; CDR2α, orange; CDR3α, yellow; Framework (FW) residues, pink; MR1, gray. I, molecular interactions of the ethylvanillin and MR1 residues. MR1, gray; ethylvanillin, green sticks. J, superposition of the bound ethylvanillin (EV) (green), HMB (orange), 6-FP (blue), and RL-6-Me-7-OH (pink). Superposition was based on the α1- and α2-domains of MR1. FP, formylpterin; HMB, 2-hydroxy-5-methoxybenzaldehyde; MR1, MHC-I-related protein 1.
The bound ethylvanillin was fully buried (BSA ∼290 Å2) within the A′-pocket of the MR1-binding cleft and was therefore inaccessible for direct MAIT TCR contacts. Its aldehyde moiety formed a Schiff base with the K43 residue of MR1. Similar to other MR1-bound ligands, ethylvanillin was sequestered within an aromatic cradle formed by Y7, W69, and W156 residues (Fig. 6I and Table S4). While the hydrophobic aromatic scaffold of ethylvanillin mainly interacted via VDW contacts with Y7, R9, S24, L66, W69, R94, and W156 residues, the hydroxyl group of the benzaldehyde scaffold of ethylvanillin also formed direct and water-mediated hydrogen bonds with R9 and R94, respectively (Fig. 6I). Upon binding of ethylvanillin, there was no significant structural rearrangement of the residues forming the A′-binding pocket of MR1 relative to the other available crystal structures of A-F7 TCR-MR1-Ag (16, 17, 18, 27, 32). However, ethylvanillin adopted an overall position that was distinct from other Schiff base–forming ligands such as HMB, 6-FP, and RL-6-Me-7-OH (Fig. 6J), being that ethylvanillin is positioned more towards the F′-pocket of MR1 in comparison to other ligands. This further highlights the capacity of the A′-pocket to accommodate a diverse range of ligands without disrupting the overall architecture of the binding pocket.
Schiff base formation and the number of noncovalent interactions correlate with ligand affinity
Next, we investigated the structural determinants that underpinned the observed affinity hierarchy for the 16 MR1-restricted ligands. We used the available crystal structures of the A-F7 MAIT TCR in complex with MR1 presenting each of those 16 ligands (PDB codes: 4L4T, 4L4V, 4PJ5, 5U1R, 5U2V, 5U6Q, 5U72, 6PUC, 6PUH, 6PUK, 6PUL, 6PUM, 6PVC, and 6PVD) (16, 17, 18, 27, 32) to analyze the number of molecular contacts that included hydrogen bonds (3.5 Å cut-off), salt bridges (4 Å cut-off) (Fig. 7A), and VDW interactions (4 Å cut-off) (Fig. 7B) and BSA (Fig. 7B) between MR1 and the bound ligand. These data were plotted against the binding affinity (log IC50) of each ligand to produce two linear regression models for the Schiff base- and non-Schiff base–forming ligands (Fig. 7, A–C). In all three plots, intercept analysis clearly indicated that the regression lines for Schiff base– and non-Schiff base–forming ligands were significantly different with p = 0.0003 (Fig. 7A), p = 0.0012 (Fig. 7B), and p = 0.0019 (Fig. 7C), demonstrating the significant contribution of Schiff base formation to ligand affinity. Furthermore, a correlation was observed between the high number of MR1/ligand polar interactions and the high affinity of a given ligand (Fig. 7A) with correlation of determination of r2 = 0.57 (Schiff base ligands) and r2 = 0.84 (Non-Schiff base ligands). The Schiff base– and non-Schiff base–forming ligands also showed a significant slope deviation with p = 0.011 and p = 0.028, respectively. For ligand affinity and its correlation with both the VDW interactions and the BSA of the ligand (Fig. 7, B and C), only the covalently bound ligands showed significant correlation, showing a slope deviating from zero with p = 0.0093 (Fig. 7B) and p = 0.043 (Fig. 7C) and associated with a moderate correlation strength with r2 = 0.59 and r2 = 0.42, respectively. This shows that the hierarchy of ligand affinities derived from the FP competitive assay corroborate with previously published structural data and establish that ligand affinity is based on the number and strength of intermolecular interactions between ligand and MR1.
Figure 7.
MR1-restricted ligands’ affinity hierarchy and structural correlates. Correlative plots of the log IC50 versus the number of hydrogen bonds and salt bridges between MR1 and ligand (A), the number of van der Waals contacts between MR1 and ligand (B) and the ligand BSA (C). The linear regression analysis was divided into Schiff (orange squares) and non-Schiff base–bound ligands (blue circles). BSA, buried surface area; MR1, MHC-I-related protein 1.
Discussion
The A′-binding pocket of MR1 is known to accommodate a wide range of chemical classes of ligands recognized by MAIT cells and other subsets of MR1-reactive T cells (6, 7, 12, 13, 14, 15). Yet, there is currently no established method for directly quantifying ligand affinity for MR1. However, for other MHC molecules, MHC-I and MHC-II (24), FP assays have been extremely informative, for instance, in MHC-II-DM–mediated peptide exchange studies (24), real-time peptide-binding kinetics (34), and peptide dissociation rate studies (35). Together with peptide elution studies, a robust characterization of both peptide motifs that MHC molecules bind and the relative affinities with which the peptides bind to MHC molecules can be performed (26).
Here, we have developed an FP assay for quantitating the relative binding affinities of putative ligands for MR1, including both activators and inhibitors of MAIT cells. In addition, while efforts have been made to identify MR1-restricted Ags, the process has involved cellular assays, crystal structure determination, and functional validation, which is highly time- and resource-intensive. Our developed cell-free MR1-binding assay using FP has enabled the measurement and quantification of the affinity of ligands for the MR1 protein. This approach provided an efficient and rapid screening method for identifying novel MR1-restricted ligands that can be prioritized for subsequent evaluation of MAIT cell activation or inhibition. We demonstrated that the competitive ligand-binding FP assay is suitable for quantifying the potency of MR1-restricted ligands with IC50 in the nM to mM concentration range. This assay reflects the clear correlation between Schiff base formation, the number of MR1/ligand non covalent interactions, and the increase in binding affinity of a given ligand.
The affinity of a peptide for MHC typically falls within the nM to the low μM concentration range (36). While some peptides exhibited weak to moderate affinity for MHC, they are still immunologically relevant and exert an immune response in the context of MHC-peptide–mediated T cell immunity. For instance, the citrullinated fibrinogen peptide (α-84Cit79–81) binds to the MHC-II RB1∗0401 with a weak affinity (∼90 μM) (26) and can still induce an IFN-γ release in MHC-II DRB1∗0401 transgenic mice (37). Similarly, low affinity MR1 ligand such as DCF (IC50 of 2.6 mM) have been shown to be functionally relevant via their ability to activate a transduced MAIT TCR cell line while being presented by a C1R cell line overexpressing MR1 (16).
Further, the FP assay enabled the identification of novel MR1 ligands such as the common dietary molecules vanillin and ethylvanillin. Both, vanillin and ethylvanillin, are present in vanilla extract, a common additive in food, medicine, and perfume (38). Ethylvanillin is synthetic and while vanillin can be synthetic too, it can also be extracted from the seed pods of Vanilla planifolia and is furthermore found in plants such as buckwheat, green pea, fava bean, hemp, lupin, and wheat (39). Our structural investigations confirm the ability of ethylvanillin to bind within the A′-pocket of the MR1-binding groove without contacting the MAIT TCR. Yet, while both compounds exhibited similar affinities for MR1, their upregulation profile did differ suggesting that a direct correlation between the affinity of a given ligand for MR1 and its ability to upregulate MR1 cannot be always fully drawn. Thus, the ability of a ligand to enter and traffic within, the cell may explain the differences in surface MR1 upregulation observed between ligands.
Considering that MAIT cells exhibit tissue homing towards the intestine and comprise 4 to 10% of intestinal T cells (40), this FP approach as an initial screening tool for the discovery of MR1 ligands may form the basis of further investigations into the discovery to novel Ags present within the gastrointestinal tract that may activate MR1-reactive T cells, particularly, those of endogenous, bacterial, or dietary origin. Collectively, this study provides a new, simple, cost-effective, and rapid method for discovering MR1-restricted ligands to advance our knowledge of the biology of the TCR–MR1 axis.
Experimental procedures
MR1-restricted ligands
Vanillin (Cat. No. V1104), ethylvanillin (Cat. No. PHR1268), DCF (Cat. No. D6899), and HMB (Cat. No. 146862) were supplied by Sigma-Aldrich; 6-FP (Cat. No. 11.415) and Ac-6-FP (Cat. No. 11.418) were synthesized by Schircks Laboratories. NLV (Cat. No. 181329) was synthesized by GL Biochem. 5-OP-RU (20), JYM20 (21, 31), 2′D-5-OP-RU, 3′D-5-OP-RU (33), JYM72 (previously reported as compound 11) (18, 31), the ribityl-less analog of JYM72 (18), 3-F-SA (16), RL-6-Me-7-OH (9), 2′D-RL-7-Me (33), and RL-6-Me (31) were synthesized as previously described. The NV18.1 and DB28 ligands (17) were provided by D. Besra (University of Birmingham). JYM72 was dissolved in H2O and diluted when required. Each other ligand was dissolved in dimethyl sulfoxide and diluted when required. Alternatively, 6-FP was dissolved at 5 mM in water, supplemented with 17 mM NaOH.
FP-based assay: saturation curve and competitive-binding assay
To determine the optimal concentration of MR1 for use in the competitive assay, JYM20 (10 nM) was incubated with a range of MR1 concentrations (0 μM–2 μM) in assay buffer (25 mM Hepes pH 7.5, 150 mM NaCl, 5 mM Na-EDTA). FP was measured (Excitation λ: 520–560 nm, emission λ: 570–610 nm) after 24 h and 48 h incubation at 25 °C using the PHERAstar microplate reader (BMG LabTech). The ligand-binding curves were plotted using Prism (Version 9.0.2, GraphPad Software Inc.) and graphed as a one-site saturation curve. Kd values were calculated at the concentration of JYM20 required for 50% relative binding to MR1. The MR1 ligands were tested at concentrations ranging from 0 to 40 mM and were incubated with JYM20 (10 nM) and MR1 (100 nM) in assay buffer. FP was measured after 24 h and 48 h incubation at 25 °C using the PHERAstar microplate reader. Ligand-binding curves were graphed as a sigmoidal concentration-response curve using Prism. IC50 values for binding affinity were calculated at the ligand concentration required for 50% of inhibition for JYM20 binding to MR1.
Correlation between the structural determinants and the binding affinities of the ligands
The coordinates of the crystal structure of the A-F7 TCR–MR1–Ags ternary complexes were obtained from the Protein Data Bank (PDB codes: 4L4T, 4L4V, 4PJ5, 5U1R, 5U2V, 5U6Q, 5U72, 6PUC, 6PUH, 6PUK, 6PUL, 6PUM, 6PVC, and 6PVD) (16, 17, 18, 27, 32) and were used for the structural analysis. The number of direct hydrogen bonds, salt bridges, and VDW contacts were calculated using CONTACT from the CCP4 suite of programs (41). A cut-off at 4 Å for VDW interactions, 4 Å for salt bridges, and 3.5 Å for hydrogen bonds was applied. BSA was calculated using Areaimol from CCP4. The data was then plotted against the IC50 of each ligand using Prism software to produce a linear regression model.
Recombinant expression and purification of MR1, HLA-A2, and the A-F7 MAIT TCR
DNA encoding for the extracellular domains of MR1, β2m, HLA-A2, and the A-F7 TCRα and TCRβ chains (9, 32, 42) were transformed into Escherichia coli BL21(DE3) cells. The proteins were then overexpressed and purified as inclusion bodies. For MR1 and A-F7, the inclusion bodies were refolded through rapid dilution and purified using size-exclusion (Superdex 200, GE Healthcare) and anion-exchange (HiTrapQ, GE Healthcare) chromatography techniques as previously described (16). For HLA-A2-NLV, 4 mg of NLV peptide were dissolved in 200 μl of dimethyl sulfoxide, 30 mg of HLA-A2, and 10 mg of β2m and placed in a 500 ml of refold buffer consisting of 0.1 M Tris pH 8.0, 3 M of urea, 0.4 M of L-arginine, 2 mM EDTA, 2.5 mM oxidized glutathione, and 20 mM reduced glutathione for 3 h at 4 °C. The refolding solution was dialyzed against 10 mM Tris pH 8.0, and the HLA-A2–NLV complex was purified using anion-exchange (HiTrapQ) chromatography.
Removal of HMB from MR1
MR1-HMB was diluted to 0.1 mg/ml with 10 mM Tris pH 8.5, 150 mM NaCl and dialyzed against 10 mM Bis-Tris propane pH 6.5, 150 mM NaCl at 4 °C for 16 h, changing dialysis buffer after 8 h. The resultant MR1-empty was buffer exchanged to 10 mM Tris pH 8.5 and purified using anion-exchange (HiTrapQ) chromatography as previously described (16).
Upregulation of surface MR1
105 C1R cells overexpressing MR1 (C1R.MR1), described previously (43), were incubated with ligand for 3 or 16 h at 37 °C and 5% CO2 in 100 μl or 200 μl RPMI 1640 medium from Gibco (Cat. No. 11875-093) supplemented with 10% fetal calf serum, 2% penicillin (100 U/ml), streptomycin (100 mg/ml), glutamax (2 mmol/L), sodium pyruvate (1 mmol/L), nonessential amino acids (0.1 mmol/L), Hepes buffer (15 mmol/L), pH 7.2 to 7.5 (all from ThermoFisher, Life Technologies), and 2-mercaptoethanol (50 mmol/L, Sigma) (RF-10). The cells were optionally stained with either Zombie Aqua Fixable Viability Kit from BioLegend (Cat. No. 423102) or Fixable Viability Dye eFluor 780 from ThermoFisher Scientific (Cat. No. 65-0865-14) and then incubated with biotinylated αMR1 antibody 26.5 (44). Unbound antibody was washed off with 2% fetal calf serum/PBS and the cells were incubated with PE- or APC-conjugated streptavidin (BioLegend, 30 μg/ml), washed again and fixed in 1% PFA/PBS. Data was acquired with an LSRFortessa X-20 (BD Biosciences) and Diva software (BD Biosciences) and analyzed using FlowJo software (BD Biosciences).
Crystallization, structure determination, and refinement
Purified MR1-empty and A-F7 TCR were concentrated and mixed at a final concentration of 3 mg/ml in a 1:1 M ratio and 300 μM of ethylvanillin was added. The ternary complex crystallized in 100 mM Bis-Tris propane (pH 6.1–6.5), 10 to 16% w/v PEG3350, and 200 mM sodium acetate. The crystals were washed in mother liquor supplemented with 15% (v/v) glycerol and flash-cooled in liquid nitrogen. A data set was collected on the MX2 beamline (Australian Synchrotron) (45) to 2.5 Å resolution. Data were integrated using XDS (46) and scaled using Aimless from the CCP4 suite of programs (41). The ternary complex structure was determined by molecular replacement using PHASER (47) and the A-F7 TCR-MR1-HMB structure without the ligand (PDB code: 5U2V) (16) as search model. After iterative model improvement with COOT (48) and refinement with Phenix.refine (49), the ternary complex crystal structure refinement led to an R/R-free (%) of 17.7/23.6 (Table S2). The quality of the structure was confirmed at the Research Collaboratory for Structural Bioinformatics Protein Data Bank Data Validation and Deposition Services website. All presentations of molecular graphics were created with the PyMOL molecular visualization system.
Mass spectrometry analysis of the A-F7 TCR-MR1-ethylvanillin crystals
A-F7 TCR-MR1-ethylvanillin crystals were solubilized in 10 mM Tris pH 8.5. To extract bound ligands, 7 μg of total protein was precipitated with acidified acetonitrile (99.9% acetonitrile, 0.1% formic acid) at a 4:1 acetonitrile:sample ratio and vortexed. After incubation for 10 min (room temperature, dark), protein was removed by centrifugation at 21,000g for 10 min. The supernatant was collected, and the acetonitrile was removed by vacuum concentration, prior to the sample being reconstituted in 2% acetonitrile, 0.1% formic acid. The extracted small molecules were analyzed by liquid chromatography tandem mass spectrometry using an Eksigent ekspert nanoLC 400 paired with a SCIEX TripleTOF 6600+ mass spectrometer equipped with a DuoSpray ion source. The sample was loaded onto a ProteCol Trap Column (C18G 200 Å 3 μm 10 mm × 0.3 mm) in 2% acetonitrile, 0.1% formic acid at 10 μl/min for 5 min. The sample was eluted from a ProteCol 250 mm × 0.3 mm C18 analytical column at 5 μl/min using an increasing gradient of acetonitrile in 0.1% formic acid over 18 min. The mass spectrometer was operated in positive mode with the following parameters: Curtain gas (CUR) 30 L/min, ion source gas 1 (GS1) 10 L/min, ion source gas 2 (GS2) 25 L/min, ion spray voltage (ISVF) 5500 V, temperature (TEM) 400 °C. The MS1 m/z range was set to 100 to 900, with a selection of 15 ions per cycle meeting the following criteria for fragmentation at a set collision energy of 30 eV: charge state +1 to +5, m/z >150 Da, intensity > 500 cps. Ions were excluded from fragmentation for 10 s after two occurrences of fragmentation. For comparison, an ethylvanillin standard was analyzed by the same method.
Data availability
Atomic coordinate and structure factors of the A-F7 MAIT TCR-MR1-ethylvanillin ternary complex were deposited in the Protein Data Bank (PDB) under the ID 7UFJ.
Supporting information
This article contains supporting information.
Conflicts of interest
J. R., L. L., D. P. F., S. B. G. E., J. M., and J. Y. W. M. are inventors on patents describing MR1 tetramers and MR1-ligand complexes. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the staff at the National Synchrotron for assistance with data collection and at the Monash Macromolecular Crystallization Facility (MMCF). We thank Ms Daphne Ju for technical assistance with the MR1 upregulation assay. We thank Dr Jia Jia Lim and Ms Keshia Kim for their assistance in the development of the MR1 FP competitive binding assay. The crystallography research was undertaken on the MX2 beamline at the Australian Synchrotron, part of ANSTO (45). We thank the Australian Research Council (ARC) (DP220102401 and Centre of Excellence in Advanced Molecular Imaging Grant CE140100011), the NHMRC (SPRF1027369, SPRF1117017, and APP1125493), the Allergy and Immunology Foundation of Australasia (AIFA), and the U.S. National Institute of Health (RO1 Grant AI148407-01A1) for funding.
Author contributions
C. J. H. W. methodology; C. J. H. W., W. A., L. L., J. Y. W. M., P. T. I., A. W. P., S. B. G. E., J. M., G. S. B., and D. P. F. investigation; L. L., J. Y. W. M., S. B. G. E., J. M., G. S. B., and D. P. F. resources; C. J. H. W., N. V., J. R., and J. L. N. data curation; C. J. H. W., J. R., and J. L. N. writing–original draft.
Funding and additional information
C. J. H. W. is supported by an Australian Government Research Training Program (RTP) Scholarship. W. A. is supported by an ARC Discovery Early Career Researcher Award (DE220101491). L. L., J. Y. W. M, and D. P. F. were supported by NHMRC Fellowship (1117017) and Investigator Grant (2009551). A. W. P. is supported by an NHMRC Principal Research Fellowship (1137739). J. M. and J. R. (2008616) are supported by NMHRC Leadership Investigator Grants. S. B. G. E. is supported by an NHMRC Emerging Leadership Investigator Award (1196881). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Peter Cresswell
Contributor Information
Jamie Rossjohn, Email: Jamie.rossjohn@monash.edu.
Jérôme Le Nours, Email: jerome.lenours@monash.edu.
Supporting information
References
- 1.Morrissey K.A., Wegrecki M., Praveena T., Hansen V.,L., Bu L., Sivaraman K.,K., et al. The molecular assembly of the marsupial γμ T cell receptor defines a third T cell lineage. Science. 2021;371:1383–1388. doi: 10.1126/science.abe7070. [DOI] [PubMed] [Google Scholar]
- 2.Chien Y., Becker D.M., Lindsten T., Okamura M., Cohen D.I., Davis M.M. A third type of murine T-cell receptor gene. Nature. 1984;312:31–35. doi: 10.1038/312031a0. [DOI] [PubMed] [Google Scholar]
- 3.Yanagi Y., Yoshikai Y., Leggett K., Clark S.P., Aleksander I., Mak T.W. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984;308:145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- 4.Brenner M.B., McLean J., Dialynas D.P., Strominger J.L., Smith J.A., Owen F.L., et al. Identification of a putative second T-cell receptor. Nature. 1986;322:145–149. doi: 10.1038/322145a0. [DOI] [PubMed] [Google Scholar]
- 5.Rossjohn J., Gras S., Miles J.J., Turner S.J., Godfrey D.I., McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 6.Rice M.T., von Borstel A., Chevour P., Awad W., Howson L.J., Littler D.R., et al. Recognition of the antigen-presenting molecule MR1 by a Vδ3(+) γδ T cell receptor. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2110288118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Nours J., Gherardin N.A., Ramarathinam S.H., Awad W., Wiede F., Gully B.S., et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science. 2019;366:1522–1527. doi: 10.1126/science.aav3900. [DOI] [PubMed] [Google Scholar]
- 8.Willcox B.E., Willcox C.R. Γδ TCR ligands: the quest to solve a 500-million-year-old mystery. Nat. Immunol. 2019;20:121–128. doi: 10.1038/s41590-018-0304-y. [DOI] [PubMed] [Google Scholar]
- 9.Kjer-Nielsen L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 10.Corbett A.J., Eckle S.B.G., Birkinshaw R.W., Liu L., Patel O., Mahony J., et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 11.Eckle S.B.G., Corbett A.J., Keller A.N., Chen Z., Godfrey D.I., Liu L., et al. Recognition of vitamin B precursors and byproducts by mucosal associated invariant T cells. J. Biol. Chem. 2015;290:30204–30211. doi: 10.1074/jbc.R115.685990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gherardin N.A., Keller A.N., Woolley R.E., Le Nours J., Ritchie D.S., Neeson P.J., et al. Diversity of T Cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. 2016;44:32–45. doi: 10.1016/j.immuni.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Lepore M., Kalinichenko A., Calogero S., Kumar P., Paleja B., Schmaler M., et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. eLife. 2017;18 doi: 10.7554/eLife.24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowther M.D., Dolton G., Legut M., Caillaud M.E., Lloyd A., Attaf M., et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 2020;21:178–185. doi: 10.1038/s41590-019-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koay H.F., Gherardin N.A., Xu C., Seneviratna R., Zhao Z., Chen Z., et al. Diverse MR1-restricted T cells in mice and humans. Nat. Commun. 2019;10:2243. doi: 10.1038/s41467-019-10198-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller A.N., Eckle S.B.G., Xu W., Liu L., Hughes V.A., Mak J.Y.W., et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol. 2017;18:402. doi: 10.1038/ni.3679. [DOI] [PubMed] [Google Scholar]
- 17.Salio M., Awad W., Veerapen N., Gonzalez-Lopez C., Kulicke C., Waithe D., et al. Ligand-dependent downregulation of MR1 cell surface expression. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10465–10475. doi: 10.1073/pnas.2003136117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awad W., Ler G.J.M., Xu W., Keller A.N., Mak J.Y.W., Lim X.Y., et al. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat. Immunol. 2020;21:400–411. doi: 10.1038/s41590-020-0616-6. [DOI] [PubMed] [Google Scholar]
- 19.Harriff M.J., McMurtrey C., Froyd C.A., Jin H., Cansler M., Null M., et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aao2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mak J.Y.W., Liu L., Fairlie D.P. Chemical modulators of mucosal associated invariant T cells. Acc. Chem. Res. 2021;54:3462–3475. doi: 10.1021/acs.accounts.1c00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWilliam H.E.G., Mak J.Y.W., Awad W., Zorkau M., Cruz-Gomez S., Lim H.J., et al. Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl. Acad. Sci. U. S. A. 2020;117 doi: 10.1073/pnas.2011260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McWilliam H.E.G., Eckle S.B.G., Theodossis A., Liu L., Chen Z., Wubben J.M., et al. The intracellular pathway for the presentation of vitamin B–related antigens by the antigen-presenting molecule MR1. Nat. Immunol. 2016;17:531. doi: 10.1038/ni.3416. [DOI] [PubMed] [Google Scholar]
- 23.Harriff M.J., Karamooz E., Burr A., Grant W.F., Canfield E.T., Sorensen M.L., et al. Endosomal MR1 trafficking plays a key role in presentation of Mycobacterium tuberculosis ligands to MAIT cells. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin L., Stern L.J. Measurement of peptide binding to MHC class II molecules by fluorescence polarization. Curr. Protoc. Immunol. 2014;106:5. doi: 10.1002/0471142735.im0510s106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dédier S., Reinelt S., Rion S., Folkers G., Rognan D. Use of fluorescence polarization to monitor MHC–peptide interactions in solution. J. Immunol. Met. 2001;255:57–66. doi: 10.1016/s0022-1759(01)00423-9. [DOI] [PubMed] [Google Scholar]
- 26.Ting Y.T., Petersen J., Ramarathinam S.H., Scally S.W., Loh K.L., Thomas R., et al. The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis. J. Biol. Chem. 2018;293:3236–3251. doi: 10.1074/jbc.RA117.001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckle S.B.G., Birkinshaw R.W., Kostenko L., Corbett A.J., McWilliam H.E.G., Reantragoon R., et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 2014;211:1585–1600. doi: 10.1084/jem.20140484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McShan A.C., Devlin C.A., Papadaki G.F., Sun Y., Green A.I., Morozov G.I., et al. TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules. Nat. Chem. Biol. 2022;18:859–868. doi: 10.1038/s41589-022-01049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jameson D.M., Ross J.A. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gras S., Saulquin X., Reiser J.B., Debeaupuis E., Echasserieau K., Kissenpfennig A., et al. Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope. J. Immunol. 2009;183:430–437. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- 31.Mak J.Y., Xu W., Reid R.C., Corbett A.J., Meehan B.S., Wang H., et al. Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun. 2017;8 doi: 10.1038/ncomms14599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel O., Kjer-Nielsen L., Le Nours J., Eckle S.B.G., Birkinshaw R., Beddoe T., et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 2013;4:2142. doi: 10.1038/ncomms3142. [DOI] [PubMed] [Google Scholar]
- 33.Ler G.J.M., Xu W., Mak J.Y.W., Liu L., Bernhardt P.V., Fairlie D.P. Computer modelling and synthesis of deoxy and monohydroxy analogues of a ribitylaminouracil bacterial metabolite that potently activates human T cells. Eur. J. Chem. 2019;25:15594–15608. doi: 10.1002/chem.201903732. [DOI] [PubMed] [Google Scholar]
- 34.Pos W., Sethi D.K., Call M.J., Schulze M.-S.E.D., Anders A.-K., Pyrdol J., et al. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 2012;151:1557–1568. doi: 10.1016/j.cell.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z., Callaway K.A., Weber D.A., Jensen P.E. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J. Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchli R., VanGundy R.S., Hickman-Miller H.D., Giberson C.F., Bardet W., Hildebrand W.H. Development and validation of a fluorescence polarization-based competitive peptide-binding assay for HLA-A∗0201--a new tool for epitope discovery. Biochemistry. 2005;44:12491–12507. doi: 10.1021/bi050255v. [DOI] [PubMed] [Google Scholar]
- 37.Hill J.A., Bell D.A., Brintnell W., Yue D., Wehrli B., Jevnikar A.M., et al. Arthritis induced by posttranslationally modified (citrullinated) fibrinogen in DR4-IE transgenic mice. J. Exp. Med. 2008;205:967–979. doi: 10.1084/jem.20072051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali L., Perfetti G., Diachenko G. Rapid method for the determination of coumarin, vanillin, and ethyl vanillin in vanilla extract by reversed-phase liquid chromatography with ultraviolet detection. J. AOAC Int. 2008;91:383–386. [PubMed] [Google Scholar]
- 39.Multari S., Neacsu M., Scobbie L., Cantlay L., Duncan G., Vaughan N., et al. Nutritional and phytochemical content of high-protein crops. J. Agric. Food Chem. 2016;64:7800–7811. doi: 10.1021/acs.jafc.6b00926. [DOI] [PubMed] [Google Scholar]
- 40.Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood. 2011;117:1250. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 41.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatzileontiadou D.S.M., Szeto C., Jayasinghe D., Gras S. Protein purification and crystallization of HLA-A∗02:01 in complex with SARS-CoV-2 peptides. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reantragoon R., Kjer-Nielsen L., Patel O., Chen Z., Illing P.T., Bhati M., et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 2012;209:761. doi: 10.1084/jem.20112095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S., Gilfillan S., Cella M., Miley M.J., Lantz O., Lybarger L., et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 2005;280:21183–21193. doi: 10.1074/jbc.M501087200. [DOI] [PubMed] [Google Scholar]
- 45.Aragao D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N.P., et al. MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2018;25:885–891. doi: 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinate and structure factors of the A-F7 MAIT TCR-MR1-ethylvanillin ternary complex were deposited in the Protein Data Bank (PDB) under the ID 7UFJ.