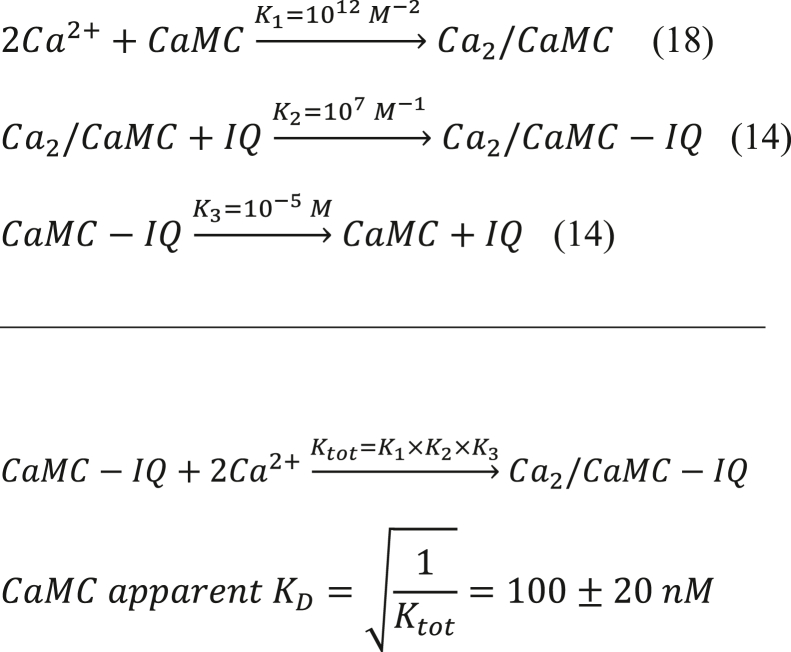Abstract
The L-type Ca2+ channel CaV1.2 controls gene expression, cardiac contraction, and neuronal activity. Calmodulin (CaM) governs CaV1.2 open probability (Po) and Ca2+-dependent inactivation (CDI) but the mechanisms remain unclear. Here, we present electrophysiological data that identify a half Ca2+-saturated CaM species (Ca2/CaM) with Ca2+ bound solely at the third and fourth EF-hands (EF3 and EF4) under resting Ca2+ concentrations (50–100 nM) that constitutively preassociates with CaV1.2 to promote Po and CDI. We also present an NMR structure of a complex between the CaV1.2 IQ motif (residues 1644–1665) and Ca2/CaM12’, a calmodulin mutant in which Ca2+ binding to EF1 and EF2 is completely disabled. We found that the CaM12’ N-lobe does not interact with the IQ motif. The CaM12’ C-lobe bound two Ca2+ ions and formed close contacts with IQ residues I1654 and Y1657. I1654A and Y1657D mutations impaired CaM binding, CDI, and Po, as did disabling Ca2+ binding to EF3 and EF4 in the CaM34 mutant when compared to WT CaM. Accordingly, a previously unappreciated Ca2/CaM species promotes CaV1.2 Po and CDI, identifying Ca2/CaM as an important mediator of Ca signaling.
Keywords: CaV1.2, calmodulin, CDI, NMR, single-channel recordings
Abbreviations: CaM, calmodulin; CDI, Ca2+-dependent inactivation; FP, fluorescence polarization; ITC, isothermal titration calorimetry; PVDF, polyvinylidene difluoride; RDC, residual dipolar coupling
CaV1.2 is the main L-type channel in heart, blood vessels, and brain (1, 2). Ca2+ influx through CaV1.2 triggers cardiac contraction, regulates arterial tone (1), mediates synaptic long-term potentiation (3, 4), controls neuronal excitability (5), and mediates Ca2+-dependent gene expression (6). Defects in inactivation of CaV1.2 cause Timothy syndrome, a rare congenital abnormality leading to lethal arrhythmias, autistic-like behaviors, and immune deficiency (7). Thus, defining mechanisms of CaV1.2 regulation is highly relevant for understanding its physiological and pathological functions. Ca2+ influx through CaV1.2 triggers a rapid negative feedback mechanism by inducing channel inactivation called Ca2+-dependent inactivation (CDI) (8, 9). CDI is mediated by calmodulin (CaM) (8) that is preassociated with CaV1.2 under basal Ca2+ conditions ([Ca2+]i = 100 nM) (10, 11). Ca2+-free apoCaM has been suggested to be preassociated with CaV1.2 (12) and the closely related CaV1.3 (13). However, under physiological conditions, apoCaM binds to the isolated CaV1.2 IQ-motif with a dissociation constant (KD) of ∼10 μM (14, 15) and ∼1 μM for full-length CaV1.2 (11). The concentration of free apoCaM is <100 nM in neurons and cardiomyocytes (15, 16). Accordingly, the fractional binding of CaV1.2 to apoCaM is predicted to be less than 10% and may not be the prevalent CaM species bound to CaV1.2 or the closely related CaV1.3 under basal conditions as proposed previously (12, 13, 17).
To fill a critical gap in our understanding of how CaM governs CaV1.2 function, we used NMR structural analysis, protein biochemistry, and patch-clamp electrophysiology of WT and mutated CaV1.2 bound to CaM. Our studies uncovered a half-calcified form of CaM (with Ca2+ bound solely at EF3 and EF4, called Ca2/CaM) that is functionally preassociated with CaV1.2 under basal conditions. The NMR structure of Ca2/CaM bound to the CaV1.2 IQ-motif (residues 1644–1664) suggests that the Ca2+-bound CaM C-lobe (residues F93, M110, L113, M125) forms intermolecular interactions with the side chain atoms from CaV1.2 residues (Y1649, I1654, Y1657, and F1658), whereas the Ca2+-free CaM N-lobe does not interact with the IQ motif. Electrophysiological data of key mutants of CaV1.2 (I1654A and Y1657E) contrasted with the earlier findings for the K1662E mutant along with the consequences of ectopic expression of CaM34 all suggest that Ca2/CaM, rather than apoCaM, preassociates with CaV1.2 under basal conditions to augment channel open probability (Po) and mediate rapid CDI.
Results
A CaM intermediate with two Ca2+ bound
Isothermal titration calorimetry (ITC) studies have suggested that apoCaM binds to the IQ peptide with submicromolar affinity in the absence of salt (12). However, in the presence of physiological salt levels, apoCaM binds to the CaV1.2 IQ-motif with a dissociation constant (KD) of 10 μM (14, 15). Earlier work suggests that binding of apoCaM to full-length CaV1.2 is ∼10 times stronger than binding to the IQ segment (11). Collectively, these data suggest that apoCaM binds to full-length CaV1.2 with a KD of ∼1 μM, which is outside the physiological concentration range of free CaM (<100 nM) in neurons and cardiomyocytes (15, 16), implying low fractional binding. Furthermore, the recent NMR structure of apoCaM bound to the CaV1.2 IQ-motif revealed an intermolecular salt bridge involving CaV1.2 residue K1662, and the K1662E mutation significantly and selectively weakened apoCaM binding to CaV1.2 (15). At the same time, the K1662E mutation does not affect single-channel Po (15). These previous results suggest that apoCaM may not be the main CaM species to support CaV1.2 activity under basal conditions as proposed previously (12, 13, 17). The current study tested the hypothesis that the CaV1.2 channel may preassociate mostly with a CaM species that is half saturated with Ca2+ under basal Ca2+ conditions ([Ca2+]i = 100 nM).
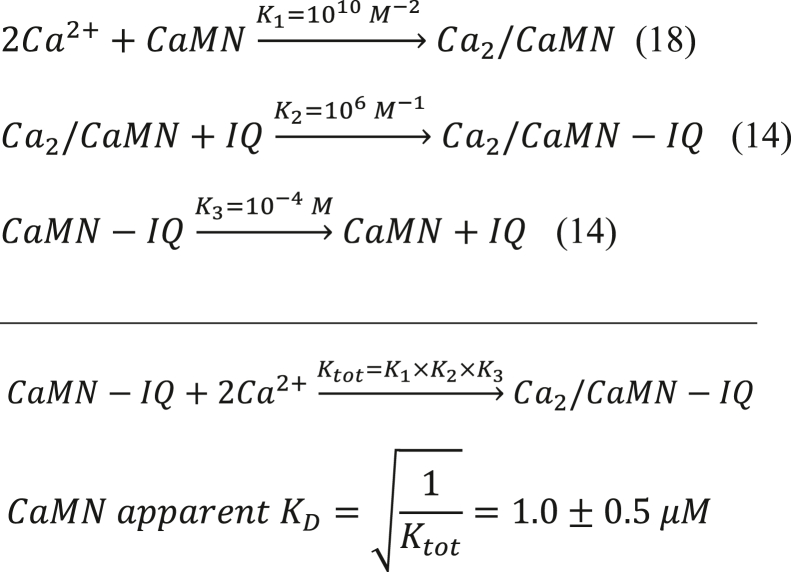
In support of our hypothesis, we find that IQ binding to CaM causes a more than 10-fold increase in the apparent Ca2+ affinity, which allows Ca2+ to bind to the CaM C-lobe under basal conditions (Fig. S1). On the basis of previous binding data (14, 18), the C-lobe under basal conditions is predicted to bind two Ca2+ to form a half-calcified state (called Ca2/CaM) in which the N-lobe is devoid of Ca2+ (19). Indeed, the C-lobe binds Ca2+ as well as the IQ motif with 10-fold higher affinity than the N-lobe (14, 18). Using the binding constants from (14, 18) the relative concentrations of apoCaM, CaM intermediate (Ca2/CaM), and Ca2+-saturated CaM (Ca4/CaM) each bound to the IQ as a function of free Ca2+ concentration are shown in Fig. S1A. The Ca2/CaM intermediate species (red trace in Fig. S1A) has a significant occupancy of ∼50% at 100 nM Ca2+ concentration (basal Ca2+ level). Since the apoCaM N-lobe (CaMN) does not bind to IQ under physiological conditions (14), IQ must instead be bound to the C-lobe (CaMC) of Ca2/CaM. Using binding constants from (14, 18), we calculate that CaMC-IQ (Fig. 1) and CaMN-IQ (Fig. 2) have apparent KD values for Ca2+ binding of 100 nM and 1.0 μM, respectively:
Figure 1.
Apparent Ca2+-binding affinity of the CaM C-lobe bound to IQ (CaMC-IQ).
Figure 2.
Apparent Ca2+-binding affinity of the CaM N-lobe bound to IQ (CaMN-IQ).
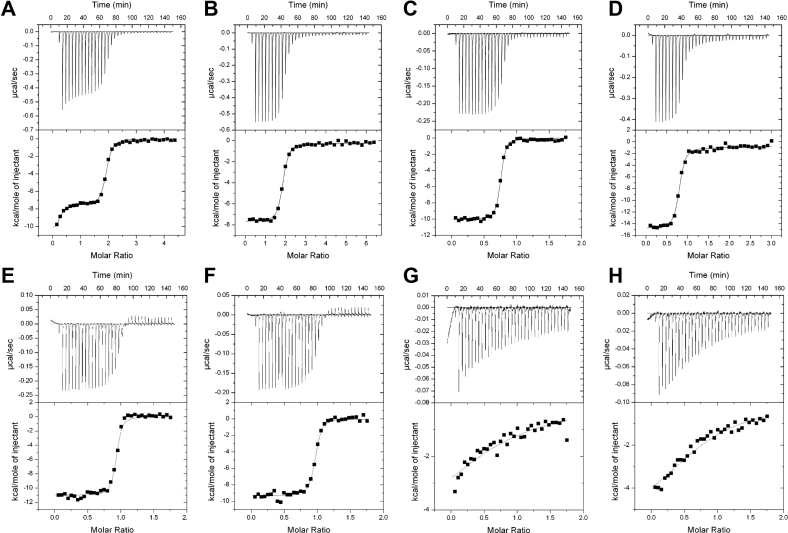
Thus, the CaM C-lobe is calculated to have a 10-fold higher apparent Ca2+ affinity compared to CaM N-lobe. This calculation implies that ∼50% of CaM-IQ complex will have Ca2+ bound to its C-lobe under basal conditions ([Ca2+]i = 100 nM), whereas the N-lobe should be devoid of Ca2+. To test this prediction, we prepared a CaM mutant (D21A/D23A/D25A/E32Q/D57A/D59A/N61A/E68Q, called CaM12’) that completely disabled Ca2+ binding to EF1 and EF2 but retained normal Ca2+ binding to EF3 and EF4. The apparent Ca2+ affinity of CaM12’ in the presence of saturating IQ peptide under physiological conditions (27 °C and 37 °C) was measured by ITC (Fig. 3, A and B). The ITC isotherm at 27 °C is biphasic, suggesting possible sample heterogeneity. The major binding component (N2 = 1.7 ±0.3 Ca2+/protein; Table 1) represents binding of two Ca2+ to CaM12’-IQ as defined by K2, ΔH2, and N2 (Table 1). The other isotherm component is nonstoichiometric (N1 = 0.2 ±0.1 Ca2+/protein) and may be an artifact of IQ partial self-association or other sample heterogeneity. Fitting the ITC isotherm with a two-site model reveals a Ca2+-binding apparent KD () of 60 ± 20 nM (Table 1), which agrees within experimental error with the predicted value in Figure 1 and with previously measured values of obtained by UV fluorescence (20). The Ca2+-binding ITC isotherm became monophasic at 37 °C, which more accurately demonstrates that two Ca2+ bind to CaM12’ with a of 72 ± 20 nM and ΔH = -7.7 ± 1 kcal/mol (Fig. 3B and Table 1). The relatively high apparent Ca2+ affinity ) implies that at least 50% of the CaM/IQ complex will have Ca2+ bound to EF3 and EF4 () at basal Ca2+concentrations (∼100 nM). This analysis predicts that slightly more than half of the CaV1.2 channels should be preassociated with the CaM intermediate, Ca2/CaM, under basal conditions.
Figure 3.
Isothermal titration calorimetry (ITC) binding assays. A and B, ITC measurement of Ca2+ binding to CaM12’-IQ at 27 °C (A) and 37 °C (B). The Ca2+ binding isotherms at 27 °C and 37 °C were fit to a two-site and one-site model, respectively. The apparent Ca2+ affinity () and enthalpy difference (ΔH1 and ΔH2) are given in Table 1. The CaM12’–IQ complex in the sample cell (10 μM at 27 °C or 8.0 μM at 37 °C, 1.5 ml) was titrated with aqueous CaCl2 (0.23 mM at 27 °C or 0.30 mM at 37 °C) using 35 injections of 10 μl each. C–D, ITC measurement of Ca2/CaM12’ binding to IQ at 27 °C (C) and 37 °C (D). The dissociation constant (KD) and enthalpy difference (ΔH) for Ca2/CaM12’ binding to IQ mutants (IQWT, IQY1649A, IQI1654A, IQY1657D, IQF1658D, and IQF1658A) are given in Table 3. The binding of Ca2/CaM12’ to IQK1662E could not be accurately measured by ITC because IQK1662E formed aggregated species under the conditions required for ITC. E–H, ITC measurement at 27 °C of Ca2/CaM12’ binding to IQF1649A (E), IQI1654A (F), IQY1657D (G), and IQF1658D (H). The IQ peptide concentrations for WT, Y1649A, and I1654A were each 10 μM (27 °C) or 7.0 μM (37 °C) in 1.5 ml in the sample cell for titration with 0.1 mM Ca2/CaM12’, and Y1657D and F1658D concentrations were each 50 μM in 1.5 ml for titration with 0.5 mM Ca2/CaM12’ using 35 injections of 10 μl each. CaM, calmodulin.
Table 2.
NMR structural statistics for Ca2/CaM12’-IQ
| NMR restraints | Value (restraint violation) |
|---|---|
| Short-range NOEs | 327 (0.0 ± 0.0) |
| Long-range NOEs | 172 (0.0 ± 0.0) |
| Hydrogen bonds | 81 (not used in water refinement) |
| Dihedral angles | 187 (0.1 ± 0.3) |
| 1DHN RDC | 24 (0.0 ± 0.0) |
| RDC Q-factor | 0.292 |
| Coordinate precision (Å)a | |
| RMSD backbone atoms | 0.83 ± 0.09 |
| RMSD all heavy atoms | 1.56 ± 0.1 |
| Deviation from idealized geometry | |
| Bonds (Å) | 0.007 ± 0.000 |
| Angles (°) | 0.753 ± 0.012 |
| Impropers (°) | 0.927 ± 0.029 |
| Ramachandran Plot (%) | |
| Favored region | 75.0 |
| Allowed region | 19.0 |
| Outlier region | 7.0 |
| Structure qualityb | |
| Clash score | 24 |
| Ramachandran outliers | 6.6% |
| Side chain outliers | 16.3% |
Coordinate precision was calculated for C-lobe residues 85 to 149.
Structure quality metrics assessed by MolProbity (51).
Half-calcified CaM represented by CaM12’
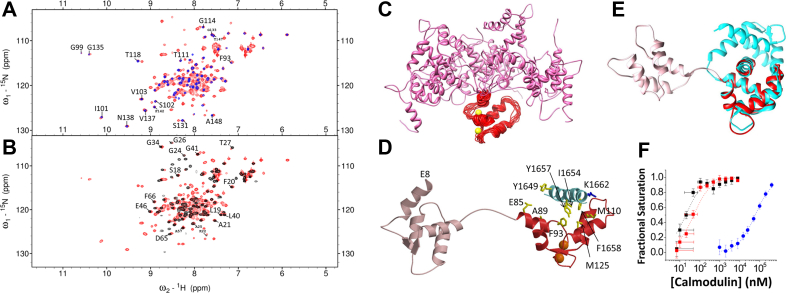
The concentration profiles in Fig. S1A show that half saturated CaM (Ca2/CaM) coexists in an equilibrium mixture with apoCaM and Ca2+-saturated CaM (Ca4/CaM). At a basal Ca2+ concentration of 100 nM, the fractional occupancy of Ca2/CaM is calculated to be 55% compared to 7% occupancy of Ca4/CaM and 38% occupancy of apoCaM. Therefore, under basal conditions, Ca2/CaM cannot be resolved from the other CaM species. To isolate the half Ca2+ saturated species, we performed structural studies on the CaM mutant (D21A/D23A/D25A/E32Q/D57A/D59A/N61A/E68Q, called CaM12’) that completely disables Ca2+ binding to EF1 and EF2 but retains Ca2+ binding to EF3 and EF4. The NMR assignments of Ca2+-bound CaM12’ bound to the IQ peptide (Ca2/CaM12’-IQ) reveal two downfield NMR peaks assigned to G99 (EF3) and G135 (EF4) that indicate Ca2+ is bound to EF3 and EF4 (21). The corresponding Gly residues in EF1 (G26) and EF2 (G62) do not exhibit downfield amide resonances, indicating that EF1 and EF2 in Ca2/CaM12’-IQ are both devoid of Ca2+.
The NMR spectrum of Ca2/CaM12’-IQ is a hybrid of the spectra of Ca2+-bound and Ca2+-free CaM (Fig. 4, A and B). The chemical shifts assigned to the CaM12’ C-lobe (residues 80–149) of Ca2/CaM12’-IQ (peaks labeled red in Fig. 4A) are nearly identical to those of the isolated Ca2+-bound CaM C-lobe bound to IQ (blue peaks in Fig. 4A). NMR peaks assigned to CaM12’ N-lobe (residues 1–79) of Ca2/CaM12’-IQ are similar to those of apoCaM12’ in the absence of IQ (black peaks in Fig. 4B), indicating that the CaM12’ N-lobe is Ca2+ free and does not interact with the IQ peptide. Thus, only the C-lobe, but not N-lobe, residues in Ca2/CaM12’ exhibit IQ-induced spectral shifts.
Figure 4.
NMR-derived structures of Ca2/CaM12’-IQ. A, 15N-1H HSQC NMR spectrum of 15N-labeled Ca2/CaM12’ bound to unlabeled IQ (red) is overlaid with the spectrum of Ca2+-bound CaMWT C-lobe/IQ complex (blue). B, NMR spectrum of 15N-labeled Ca2/CaM12’ bound to unlabeled IQ (black) is overlaid with the spectrum of Ca2+-free CaM12’ (red). C, ensemble of 10 lowest energy NMR structures of Ca2/CaM12’ (PDB ID: 7L8V). Main chain structures are depicted by a ribbon diagram. Structures of the C-lobe (residues 85–149) are overlaid and highlighted in red; N-lobe structures (residues 1–84) are highlighted in pink. Bound Ca2+ ions are yellow. Structural statistics are given in Table 2. D, the lowest energy structure of Ca2/CaM12’-IQ complex is shown as a ribbon diagram of Ca2/CaM12’ bound to the IQ peptide (cyan). The CaM N-lobe and C-lobe are highlighted pink and red, respectively. Side-chain atoms of key residues are depicted by sticks and are colored yellow and blue. E, overlay of the NMR structure of Ca2/CaM12’-IQ (C-lobe in red) with the crystal structure of Ca4/CaM (cyan, 2BE6). The C-lobe structures overlay with an RMSD of 1.8 Å. F, fluorescence polarization assay showing the binding of half Ca2+-saturated CaM mutant (Ca2/CaM12’) with fluorescently labeled IQ peptides (WT: black; K1662E: red; both: KD < 100 nM), and of apoCaM binding to Y1657D (blue, KD = 60 μM). CaM, calmodulin; PDB, Protein Data Bank.
NMR structure of Ca2/CaM12’-IQ
NMR spectral assignments for Ca2/CaM12’-IQ were reported previously (BMRB accession number 27692) (21). These previous NMR assignments were used in the current study to obtain NMR-derived structural restraints from NOESY and residual dipolar coupling (RDC) data (Fig. S4). NMR structures of Ca2/CaM12’-IQ were then calculated on the basis of distance restraints derived from analysis of NOESY (22) and long-range orientational restraints derived from RDC data (23) as described in the Experimental procedures. The final NMR-derived structures of Ca2/CaM12’ are overlaid in Figure 4C and structural statistics summarized in Table 2. The two domains of Ca2/CaM12’ (N-lobe in pink and C-lobe in red, Fig. 4C) are separately folded and noninteracting, as was seen previously for the NMR structures of apoCaM (24, 25, 26). The overall precision of the NMR ensemble is expressed by a RMSD of 0.83 ± 0.09 Å calculated from the coordinates of the main chain atoms in the C-lobe (Fig. 4C) and 0.9 ± 0.1 Å from the main chain atoms in the N-lobe. The lowest energy NMR structure of Ca2/CaM12’ bound to the IQ peptide is shown in Figure 4D. The quality of the NMR structures of Ca2/CaM12’-IQ was assessed using PROCHECK-NMR (27), which shows that 93% of the residues occur in the allowed or favorable regions from the Ramachandran plot. The NMR structure of the Ca2+-bound CaM C-lobe (residues 80–149) of Ca2/CaM12’-IQ (dark red in Fig. 4, D and E) looks similar to that observed in the crystal structure of Ca2+-saturated CaM bound to the IQ (cyan in Fig. 4E) (28). The structure of the Ca2+-free CaM N-lobe (residues 1–78) of Ca2/CaM12’-IQ (light red in Fig. 4D) adopts a closed conformation and looks similar to that of apoCaM (26). The IQ peptide was verified by NMR to have a helical conformation (cyan in Fig. 4D). In the Ca2/CaM12’-IQ structure (Fig. 4D), the IQ residues (Y1649, I1654, Y1657, and F1658) point toward CaM and make extensive contacts with CaM C-lobe residues (E85, A89, F93, M110, L113, M125). The IQ peptide in the Ca2/CaM12’-IQ structure does not make any contacts with the Ca2+-free N-lobe, in contrast to the crystal structure of Ca4/CaM12’-IQ (28, 29, 30) where IQ aromatic residues (F1648, Y1649, and F1652) make extensive contacts with N-lobe residues (F13, F69, M73).
Table 1.
ITC thermodynamic parameters for Ca2+ binding to CaM12’-IQ
| Temp (° C) | N1 | K1 (x108 M-1) | ΔH1 (kcal/mol) | N2 | K2 (x107 M-1) | ΔH2 (kcal/mol) | (nM) |
|---|---|---|---|---|---|---|---|
| 27 | 0.2 ± 0.1 | 6 ± 4 | −10 ± 1 | 1.7 ± 0.3 | 1.7 ± 0.4 | −7.5 ± 1 | 60 ± 20 |
| 37 | − | − | − | 1.8 ± 0.3 | 1.4 ± 0.3 | −7.7 ± 1 | 72 ± 20 |
IQ residue K1662 interacts with apoCaM more strongly than Ca2/CaM12’
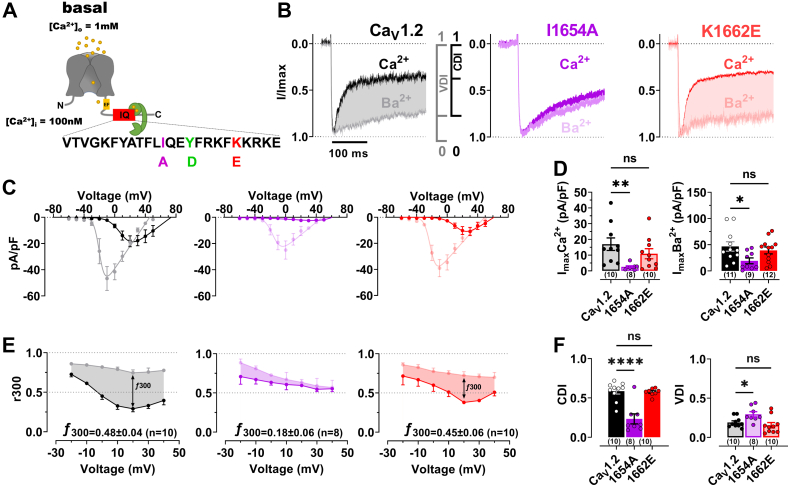
The NMR structure of Ca2/CaM12’-IQ (Fig. 4D) looks quite different from the recent NMR structure of apoCaM bound to IQ (15). In the apoCaM-IQ structure, K1662 forms intermolecular salt bridges with CaM residues, E85 and E88. By contrast, K1662 is mostly solvent exposed in the Ca2/CaM12’-IQ structure and does not contact either E85 or E88 (Fig. 4D). This analysis predicts that the CaV1.2 mutation K1662E weakens binding to apoCaM more than it does to Ca2/CaM12’. Because the K1662E peptide (IQK1662E) was not soluble enough for ITC with Ca2/CaM12’, we used fluorescence polarization (FP) to measure binding affinity in the nanomolar range. As predicted, titration of the IQ peptides with Ca2/CaM12’ reached full saturation at 100 nM Ca2/CaM12’, indicating a KD < 100 nM for both, IQWT and IQK1662E (Fig. 4F). It was not possible to more accurately determine the actual KD because the IQ peptide concentration in Figure 4F had to be 100 nM due to limited detection sensitivity. This concentration is much larger than the KD for IQWT (16 nM in Table 3) and apparently also for IQK1662E, as binding was clearly saturated at 100 nM for both peptides. The free concentrations of Ca2/CaM12’ are within the sample noise level during the first half of the titration when (see SD bars in Fig. 4F). During the second half of the titration, was above the noise level and the titration curves show clear saturation at 100 nM providing an upper limit of 100 nM for the KD of both, IQWT and IQK1662E, consistent with the 16 nM KD for IQWT as seen by ITC (Table 3). As a result, Ca2/CaM12’ can bind to IQK1662E in the nanomolar range in contrast to apoCaM, which binds to IQK1662E with a KD in the high micromolar range (60 μM) that is 6-fold higher than that of IQWT (15). Thus, the K1662E mutation weakens IQ binding to apoCaM to a degree that is outside the physiological range of its concentration (16) (<100 nM), in contrast to the nanomolar binding of IQK1662E with Ca2/CaM12’ (Fig. 4F). Accordingly, the K1662E mutation can be used to selectively disable apoCaM binding to CaV1.2, while retaining CaV1.2 binding to Ca2/CaM.
Table 3.
Dissociation constants (KD), enthalpy differences (ΔH), and stoichiometry (n) for Ca2/CaM12’ binding to IQ variants as measured by ITC
| Temp (° C) | IQ peptide | KD (nM) | ΔH (kcal/mol) | n-value |
|---|---|---|---|---|
| 37 | WT | 37 ± 10 | −15 ± 0.2 | 0.76 ± 0.25 |
| 27 | WT | 16 ± 5 | −10 ± 0.2 | 0.77 ± 0.25 |
| 27 | Y1649A | 26 ± 5 | −9.7 ± 0.2 | 0.88 ± 0.25 |
| 27 | I1654A | 60 ± 10 | −9.2 ± 0.2 | 0.89 ± 0.25 |
| 27 | Y1657D | 8000 ± 900 | −5.6 ± 0.7 | 0.72 ± 0.5 |
| 27 | F1658A | 32 ± 5 | −9.5 ± 0.2 | 1.0 ± 0.25 |
| 27 | F1658D | 4000 ± 700 | −5.9 ± 0.7 | 0.8 ± 0.5 |
The errors are the SD calculated from three independent trials.
IQ residues Y1649, I1654, Y1657, and F1658 interact with Ca2/CaM12’
The NMR structure of Ca2/CaM12’-IQ reveals intermolecular contacts with IQ residues, Y1649, I1654, Y1657, and F1658, that are each located on the same side of the IQ helix pointing toward the Ca2+-occupied C-lobe of Ca2/CaM12’ (Fig. 4D). As predicted by this analysis, the IQ peptide mutants IQY1649A, IQF1654A, IQY1657D, and IQF1658D each exhibited weaker binding to Ca2/CaM12’ compared to IQWT. The KD was 16 ± 5 nM for IQWT, 26 ± 5 nM for IQY1649A, 60 ± 10 nM for IQI1654A, 8000 ± 10 nM for IQY1657D, 4000 ± 10 nM for IQF1658D, and 32 ± 5 nM for IQF1658A (Fig. 3, C and E–H and Table 3). These findings validate our structural analysis and verify that Y1657 makes the strongest contact with CaM.
The highly exothermic binding of the IQ peptide to Ca2/CaM12’ (ΔH° = -15 kcal/mol in Figure 3D and Table 3) predicts the KD to increase by 2.3-fold when the temperature is increased from 27 °C to 37 °C. As predicted, the KD for IQ binding to Ca2/CaM12’ increased from 16 ± 5 nM (at 27 °C) to 37 ± 10 nM at 37 °C. Also, the temperature dependence of ΔH (-10 kcal/mol at 27 °C versus −15 kcal/mol at 37 °C) indicates a negative ΔCp value, which is consistent with the relatively large change in solvent accessible hydrophobic surface area that occurs when Ca2/CaM12’ binds to the IQ peptide.
The K1662E mutation affects binding of apoCaM but not CDI of CaV1.2
The aforementioned analysis suggests that K1662E retains binding to Ca2/CaM12’ but not apoCaM under physiological conditions (i.e., with free CaM < 100 nM (16)) (Fig. 4F). This differential effect informs interpretation of recently published data that showed that the K1662E mutation has no effect on Po (15), while the I1654A mutation, which affects binding of both apoCaM and Ca/CaM, decreased Po by 6-fold (15). A similar effect has been seen for an analogous Ile to Ala mutation in the closely related CaV1.3 (17). Collectively, these findings suggest that CaM promotes Po when it forms a complex with CaV1.2 with Ca2+ bound to EF3 and EF4 to give rise to a half-saturated Ca2/CaM state in this complex. To further test the idea of preassociation of half Ca2+-saturated Ca2/CaM with CaV1.2 at basal Ca2+ concentrations, we wanted to compare CDI of CaV1.2K1662E with WT and also CaV1.2I1654A, which served as a well-established reference point for loss of CDI (8, 13, 17). For that purpose, we measured whole-cell current density for IBa and ICa. Consistent with the earlier Po analysis, IBa and ICa were reduced by the I1654A but not K1662E mutation (Fig. 5, A–D and Table S1A). Strikingly, the K1662E mutation had no significant effect on CDI (nor on voltage-dependent inactivation), in contrast to the I1654A mutation, which reduced CDI by ∼75% (Fig. 5, B, E, and F and Table S1B). The small, remaining CDI seen for the I1654A mutant channel may be due to N-lobe effects such as its binding to the N terminus of the CaV1.2 α1 subunit (31). The differential effect on IBa, ICa, and CDI by the K1662E versus I1654A mutation is consistent with the differential effect of the K1662E versus I1654A mutation on Po (15) and suggests that formation of a complex of CaV1.2 with half Ca2+-saturated Ca2/CaM is important for Po and for predisposing CaV1.2 to CDI.
Figure 5.
Effects of IQ mutants I1654A and K1662E on CaV1.2 activity and inactivation. A, topology of the hypothetical CaV1.2 Ca2+ channel pore and localization of the IQ domain and its mutations in the α11.2 subunit. At rest with [Ca2+]i ≤100 nM the C-lobe (green) of half-calcified Ca2+/CaM is predicted to bind to the C-terminal portion of the IQ motif, making hydrophobic contacts with I1654 and Y1657 but not with K1662. B–F,) HEK 293T/17 cells were transfected with α11.2, α2δ1, and β2A. Shown are representative whole-cell current traces (B), population data of current-voltage relationships (I/V curves) (C), their respective peak current density plots (D), and currents remaining after 300 ms of depolarization (r300; bottom) of IBa (10 mM Ba2+; gray or light colors) and ICa (10 mM Ca2+; black or dark colors), for WT (black), I1654A (purple), and K1662E (red). Statistical significance was determined by a one-way ANOVA with Bonferroni correction, (∗p < 0.05, F(DFn, DFd), F(2,29), F3.3and p∗∗<0.01, F(DFn, DFd), F(2,25) = 4.9. E, peak currents in (B) were normalized to the respective current maxima (Imax). Shaded areas indicate differences between IBa and ICa as read out for CDI (f300: difference between IBa and ICa remaining after 300 ms). Quantification of peak current densities of IBa and ICa at potential of respective Imax reveals a strong decrease in current density for I1654A but not K1662E versus WT (D). F, quantification of CDI and VDI reveals a strong decrease in CDI for I1654A but not K1662E versus WT. Additionally, I1654A showed a robust and significant increase in VDI versus WT, whereas K1662E remained unaffected. Numbers in parenthesis under bars reflect n independent recordings and error bars SEM (∗p < 0.05, F(DFn, DFd), F(2,25) = 5.5, and ∗∗∗∗p < 0.0001, F(DFn, DFd), F(2,23) = 20.9, One-way ANOVA with Bonferroni correction). CaM, calmodulin; CDI, Ca2+ dependant inactivation; VDI, voltage dependent inactivation.
The Y1657D mutation strongly affects binding of half-saturated Ca2/CaM as well as IBa, ICa, Po, and CDI of CaV1.2
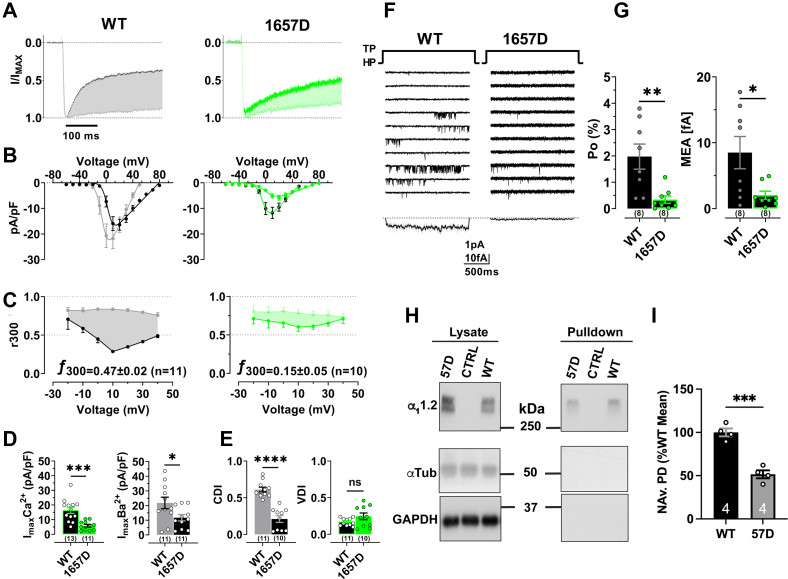
Our new Ca2/CaM12’-IQ structure indicates that Y1657 makes the most and closest contacts among all IQ residues with Ca2/CaM12’ (Fig. 4). In support of its central role in mediating this interaction, binding studies indicate that the Y1657D mutation has the strongest negative effect on the affinity of the Ca2/CaM12’-IQ interaction of all tested IQ peptides (KD for IQWT is 16 nM and for IQY1657D 8 μM; Table 3). The Y1657D mutation decreased whole-cell currents, IBa and ICa, as well as CDI with no apparent effect on voltage dependent inactivation (Fig. 6, A–E). Single-channel recordings show a remarkably strong decrease in Po for Y1657D versus WT CaV1.2 (Fig. 6, F and G). This loss in Po and CDI is comparable to similarly strong effects for the I1654A mutation on Po (15) and CDI (9) but the K1662E mutation, which specifically affects apoCaM but not Ca/CaM binding, did not affect Po (15) or CDI (Fig. 5). The decrease in Po is also well reflected when calculating the ensemble averages of unitary single-channel currents (Fig. 6F and Table S2). To test whether there is also a change in channel surface expression in addition to a decrease in Po of individual channels, we conducted surface biotinylation experiments. We determined that CaV1.2 surface expression was reduced by almost 50% (Fig. 6, H and I), which can explain some, but not all, of the 80% loss in Po.
Figure 6.
Effects of IQ mutant Y1657D on CaV1.2 activity, inactivation, and surface expression. HEK 293T/17 cells were transfected with α11.2, α2δ1, and β2A. Shown are representative whole-cell current traces (A), population data of I/V curves (B) currents remaining after 300 ms of depolarization (r300; bottom) of IBa (gray or light green), and ICa (black or dark green), for WT (black) and Y1657D (green) (C), and peak current density plots (D). Peak currents in (A) were normalized to the respective current maxima (Imax). Shaded areas indicate differences between IBa and ICa as read out for CDI (f300: difference between IBa and ICa remaining after 300 ms). D, quantification of peak IBa and ICa at potential of respective Imax reveals a strong decrease in current density for Y1657D versus WT. E, quantification of CDI and VDI reveals a strong decrease in CDI but not VDI for Y1657D versus WT. F, 10 consecutive representative single-channel traces of WT and Y1657D. Below: mean ensemble average currents (MEA) calculated from a total of 857 superimposed traces for WT (n = 8 cells) and 1366 traces for Y1657D (n= 8 cells). G, quantification of single-channel open probability Po (left) and MEA (right) reveals a strong decrease in channel activity for Y1657D versus WT. Statistical difference was determined by an unpaired, two-tailed Student’s t test, p∗<0.05, p∗∗<0.01, p∗∗∗<0.001 and p∗∗∗∗<0.0001. H, surface biotinylation of CaV1.2 was followed by Neutravidin pull downs and immunoblotting (right panels) with antibodies against the proteins indicated at the left. Left panels show immunoblots of total lysate. Tubulin (α-Tub) and GAPDH were used as loading controls for lysate samples (left) and assessment of membrane integrity (right; all left and right panels were from same gels and exposures). Absence of tubulin and GAPDH immunoreactivity indicates that the biotin reagent did not leak into cells ruling out biotinylation of intracellular proteins. I, quantification of α11.2 immunosignals in Neutravidin pull downs (NAv.PD) normalized to WT α11.2 (set to 100%). The 1657D α11.2 mutant exhibits a decrease in surface biotinylation relative to the WT subunit (n = 4; p = 0.0003, two-tailed unpaired t test). Numbers in parenthesis under bars or inside bars reflect “n” independent recordings or pull downs and error bars SEM (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, t test). VDI, voltage dependent inactivation.
CaM intermediate (Ca2/CaM) increases Po of CaV1.2
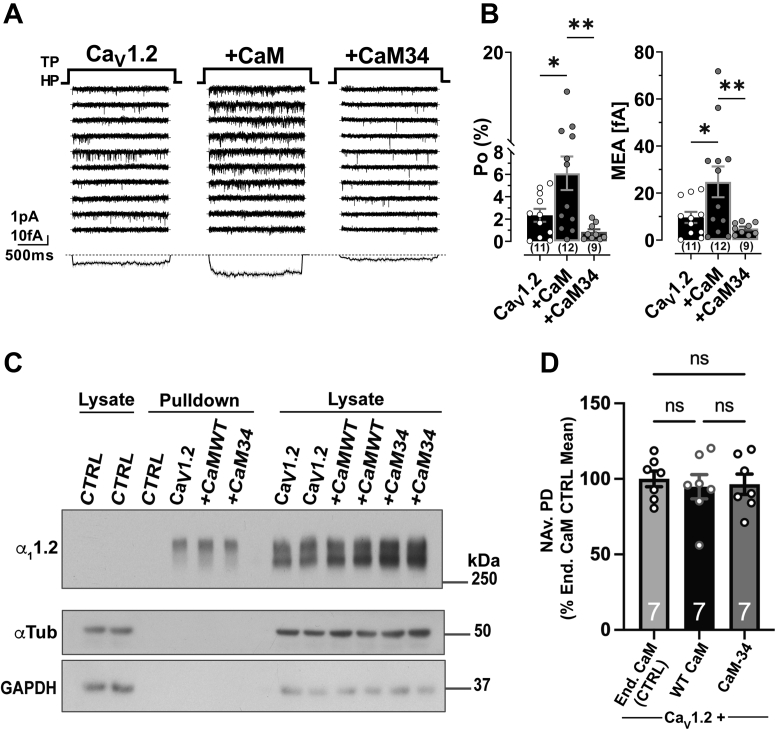
To further analyze the role of CaM in Po, we ectopically expressed CaM in HEK 293T cells. Although this approach has been used before to define the role of CaM in CDI, the level to which exogenous CaM was expressed in these CDI studies had not been thoroughly assessed (32). Thus, we investigated whether the expression of CaM34 (described by (8)) was sufficient to allow detection of an effect (i.e., many fold greater than endogenous CaM) by immunoblotting extracts of 293T cells transfected with CaV1.2 expression constructs ± WT CaM or CaM34 plasmids (Fig. S2). We found that overexpression of WT compared to endogenous CaM is about ∼10 fold, while CaM34 is ∼20 fold (Fig. S2, A–D). To test whether ectopic expression of CaM affects levels of endogenous CaM, we expressed YFP-tagged WT CaM or CaM34, which migrate at an MR of ∼ 45 kDa (verified by anti-YFP immunoblotting; Fig. S2E). Probing immunoblots with anti-CaM identifies a prominent 45 kDa band and a weaker signal for the endogenous 17 kDa band. Comparison of the 17 kDa band in mock-transfected (no CaM vectors) cell lysate to the same MR immunoreactive band in the CaM plasmid-transfected samples did not indicate a significant effect of ectopic CaM on endogenous CaM levels (Fig. S2, E and F).
Consistent with earlier work on CaV1.3 by Adams et al. (17), we find that overexpression of WT CaM strongly increases Po by ∼300% as compared to expression of CaV1.2 alone (Fig. 7, A and B and Table S3). This effect could be due to increased binding of apoCaM, half Ca2+-saturated Ca2/CaM, or both. Because earlier work did not differentiate between these possibilities (17), we tested the effect of ectopic expression of CaM34 and found no increase at all in Po as compared to expression of CaV1.2 alone. This result demonstrates that Ca2+ binding to EF3 and EF4 in CaM is essential for promoting the increased Po. There was no detectable effect on surface expression of CaV1.2 by either WT CaM or CaM34 (Fig. 7, C and D and Table S3). Given the ∼20-fold higher expression levels of CaM34 versus endogenous CaM, it seems especially remarkable that this overexpression had no effect at all on Po when a lesser degree of overexpression of WT CaM induced a ∼3-fold increase in Po (Fig. 7). Collectively, these data indicate that binding of Ca2/CaM and not apoCaM to CaV1.2 at basal Ca2+ concentrations mediates the observed increase in Po.
Figure 7.
Effects of ectopic expression of WT CaM and CaM34on CaV1.2 activity, inactivation, and surface expression. HEK 293T/17 cells were transfected with α11.2, α2δ1, and β2A plus, if indicated, WT CaM or CaM34. A, 10 consecutive representative single-channel traces of WT CaV1.2 expressed alone (left) or together with WT CaM (middle) or CaM34 (right). Bottom: MEA calculated from a total of 2009 superimposed traces for CaV1.2 expressed without CaM (n = 11 cells), 2327 traces for CaV1.2 expressed with WT CaM (n = 12 cells), and 1655 traces for CaV1.2 expressed with CaM34 (n = 9 cells). B, quantification of Po (left) and MEA (right) reveals a strong increase in channel activity for ectopic expression of WT CaM but not CaM34 (numbers in parenthesis under bars reflect n independent recordings and error bars SEM; ∗p < 0.05, and ∗∗p < 0.01, F(DFn, DFd), F(2,29) = 6.8 and ∗p < 0.05 and ∗∗p < 0.01, F(DFn, DFd), F(2,29) = 5.4, one-way ANOVA with Bonferroni correction). C, surface biotinylation of CaV1.2 was followed by Neutravidin pull downs (middle of blot) and immunoblotting with antibodies against the proteins indicated at the left. Right side shows respective total lysate samples in duplicate and left side total lysate samples from mock-transfected cells. Cells expressing CaV1.2 only or CFP-tag empty vector only were used as controls (CTRL). Tubulin (α-Tub) and GAPDH were used as loading controls for lysate samples and assessment of membrane integrity for pull-down samples. Absence of tubulin and GAPDH immunoreactivity ruled out biotinylation of intracellular proteins. D, quantification of α11.2 immunosignals in Neutravidin pull downs (NAv.PD) normalized to mean (set to 100%) of the signal in CaV1.2 only samples (control, only endogenous CaM); n = 7; one-way ANOVA (F = 0.1547, p = 0.8578), followed by Tukey’s post-hoc test, ns = p > 0.05). CaM, calmodulin; MEA, mean ensemble average.
Discussion
Preassociation of CaM with CaV1.2 and the highly homologous CaV1.3 under basal conditions has been suggested to both augment channel activity at low Ca2+ levels (17) and facilitate rapid CDI (8, 9). We provide multiple lines of evidence that CaV1.2 preassociates with half-calcified Ca2/CaM that contains two Ca2+ bound to the CaM C-lobe. The fact that the CaM34 mutant abolished the 300% increase in channel open probability of CaV1.2 caused by WT CaM (Fig. 7, A and B) implies that Ca2+ binding to EF3 and EF4 (hence half-calcified CaM) is essential for CaV1.2 channel function. Also, our binding analysis reveals that IQ binding to CaM increases the apparent Ca2+ affinity by at least 10-fold (see Fig. 1 and Table 3), consistent with observations from previous binding studies (14, 20). Hence, the IQ-bound CaM C-lobe is more than 50% saturated with Ca2+ at basal Ca2+ concentrations when CaM is saturated with the IQ peptide (Fig. S1). The concentration of free endogenous CaM inside a cell is estimated to be between 50 to 100 nM (16). As Ca2/CaM binds to the IQ motif with a KD of 16 nM, we estimate that ∼50% of CaV1.2 is bound to Ca2/CaM under basal conditions, which would put the channel regulation by CaM in the middle of its dynamic range.
The NMR structure of Ca2/CaM12’-IQ reveals that half Ca2+-saturated CaM (Ca2/CaM) has a closed conformation (26) in the Ca2+-free N-lobe and a Ca2+-bound open conformation (28) in the C-lobe (Fig. 4). The N- and C-lobe structures of Ca2/CaM12’-IQ are separately folded and do not exhibit interdomain contacts (Fig. 4C). The two separate lobes in Ca2/CaM12’-IQ are dynamically independent, similar to apoCaM (26, 33, 34). The Ca2+-free N-lobe structure in Ca2/CaM12’-IQ does not interact with the IQ peptide, in contrast to the IQ contacts with the N-lobe observed in the crystal structure of Ca2+-saturated CaM (28, 29, 30). The IQ peptide binds exclusively to the Ca2+-bound C-lobe of Ca2/CaM (Fig. 4D), whose structure is similar to the C-lobe of Ca4/CaM bound to the IQ (Fig. 4E) (28, 29, 30). The IQ peptide bound to Ca2/CaM12’ is rotated 180° compared to the orientation of the IQ bound to apoCaM (15). The opposite binding orientation may explain in part why the IQ binds to Ca2/CaM with at least 100-fold higher affinity (Fig. 4F) compared to that of apoCaM (14, 15). The contrasting binding orientation also suggests why the preassociation of CaV1.2 with Ca2/CaM (rather than with apoCaM) predisposes CaV1.2 for CDI. Since Ca2/CaM and Ca4/CaM both bind to CaV1.2 with the same orientation, CaM can remain bound to CaV1.2 upon Ca2+ influx to facilitate rapid CDI. By contrast, preassociated apoCaM would first need to dissociate from CaV1.2 upon Ca2+ influx and then subsequently rebind in the conformation adopted by Ca2+-saturated Ca4/CaM to engage CDI (28, 29, 30). This unbinding of apoCaM and rebinding of Ca4/CaM would likely prevent rapid CDI and defeat the purpose of the CaM preassociation.
Our functional analysis fully supports the relevance of prebinding of Ca2/CaM to the CaV1.2 IQ motif. The K1662E mutation, which impaired binding of apoCaM (15) but retained binding to Ca2/CaM at physiological CaM concentrations of ∼100 nM (16) (Fig. 4F), did not affect Po (15), CDI, IBa, or ICa (Fig. 5). Furthermore, the Y1657D mutation impaired binding of apoCaM (KD = 60 μM, Fig. 4F), as well as Ca2/CaM (KD = 8 μM, Table 3), and reduced Po, CDI, IBa, and ICa (Fig. 6). We also tested the effect of ectopic expression of CaM34 and CaM1234. Consistent with the earlier work on the closely related CaV1.3 (17), overexpression of WT CaM strongly augmented Po (Fig. 7 and Table S3). The main finding of these authors (17) was that substitution of the eponymous Ile in the IQ motif by Met reduced Po and overexpression of WT CaM rescued this loss. Because mutating this Ile reduces binding of apoCaM, these authors concluded that it is apoCaM that binds to the IQ motif under resting Ca2+ concentrations to augment Po. However, they did not test the effect of overexpression of CaM1234 or CaM34 on single channel activity as is required for measuring Po and thus did not rule out that Po is driven by the binding of Ca2/CaM, whose binding to the IQ motif is also strongly impaired by mutating this Ile. Importantly, we found that neither CaM34 (Fig. 7 and Table S3) nor CaM1234 (Fig. S3 and Table S4) increased Po, despite the fact that the exogenous CaM levels were much higher (by 20-fold) than that of endogenous CaM. In addition, the differential effects of (1) the K1662E mutation on CaV1.2 binding to apoCaM versus Ca2/CaM; (2) K1662E versus Y1657D on Po and CDI; and (3) WT CaM versus CaM34 or CaM1234 on Po collectively indicate that preassociated Ca2/CaM is an important factor in determining channel Po.
As discussed previously, we estimate that ∼50% of CaV1.2 is occupied by Ca2/CaM with little occupancy by apoCaM due to its low concentration in the cytosol (50–100 nM (16)) and low affinity binding to the IQ (KD = 10 μM (15)) and full-length CaV1.2 (KD = 1 μM (11)). How then can the remainder of the CaV1.2 population possess a reasonable level of activity? We previously found that binding of α-actinin to the IQ motif also strongly augments Po (15). Thus, we propose a model in which CaV1.2 is either occupied by α-actinin, which at the same time anchors CaV1.2 at the cell surface and especially in dendritic spines where α-actinin is concentrated (35) or by Ca2/CaM. Accordingly, in addition to strongly promoting Po, α-actinin also augments the CaV1.2 surface expression (15), perhaps by connecting to F-actin (36). On the other hand, Ca2/CaM augments Po with apparently little if any effect on surface expression. Channel occupancy by Ca2/CaM could be increased upon modest increases of basal Ca2+ influx potentially in a positive feedback loop at low Ca2+ levels and low channel activity. However, prolonged displacement of α-actinin by Ca4/CaM also triggers endocytosis of CaV1.2 as a negative feedback mechanism (35). At this point, we cannot be certain about how α-actinin and CaM intersect at the IQ motif to govern CaV1.2 activity, and much needs to be learned with respect to the exact function of these interactions.
In conclusion, our analysis provides novel mechanistic insight into preassociation of CaM with CaV1.2 and its role in controlling channel activity and CDI. These findings are not only of functional relevance for understanding the physiological effects of CaV1.2 but also inform the current understanding of pathological events such as arrhythmias due to impaired CDI (37, 38).
Experimental procedures
CaM12’ mutagenesis and purification and IQ peptide for NMR
The CaM12’ mutation ((D21A/D23A/D25A/E32Q/D57A/D59A/N61A/E68Q) was introduced into Xenopus CaM complementary DNA by PCR QuickChange procedure (39). The mutated complementary DNA was inserted into the NcoI/BamHI sites of a pET11d vector and verified by automated Sanger sequencing. The recombinant CaM12’ protein was expressed from a pET11d vector in a BL21(DE3) Codon Plus Escherichia coli strain (Stratagene) and purified as described previously (40). The CaV1.2 IQ peptide (residues 1644–1664) was purchased from ChinaPeptides. The peptide was dissolved in d6-dimethyl sulfoxide to give a peptide concentration of 7.8 mM. The peptide concentration was determined by measuring absorbance at 280 nm with ε280 = 2980 M−1 cm−1. An aliquot of peptide (1.5 equivalents) was added to a dilute solution of CaM12’ (50 μM protein dissolved in 20 mM 2-amino−2−hydroxymethyl-propane-1,3-diol-d11 (Tris-d11) with 95% H2O/5% D2O). The complex was then concentrated to a final concentration of 500 μM in a final volume of 500 μl for NMR experiments. The 1.5-fold excess of IQ peptide in the NMR sample of Ca2/CaM12’-IQ was necessary to minimize the occupancy of a 2:1 complex, in which two molecules of CaM12’ were bound to one IQ. The HSQC spectrum of a sample that contained an equal concentration of CaM12’ and IQ revealed two distinct peaks for each C-lobe residue of CaM12’ (Fig. S4D). The most intense peak represented a 1:1 complex (∼90% occupancy) and a weaker second peak (marked by arrows in Fig. S4D) represented a second CaM12’ molecule bound to IQ in a 2:1 complex (∼10% occupancy). The relative occupancy of the 2:1 complex could approach nearly 100% when the CaM12 concentration is more than 10-fold higher than that of CaV1.2, like what exists inside HEK293 cells used in the CaV1.2 electrophysiological experiments (Fig. S2). The 2:1 complex likely consists of a single IQ peptide that binds tightly to a Ca2+-bound C-lobe on one side of the IQ helix (CaM12’ C-lobe contacting I1654 and Y1657) as well as a second CaM12’ C-lobe that binds with lower affinity to the opposite side of the IQ helix (CaM12’ C-lobe contacting F1648 and F1652). The binding of a second C-lobe from CaM12’ mimics the binding of the Ca2+-bound N-lobe from WT CaM. Therefore, we suggest that the CDI observed for CaV1.2 in the presence of CaM12’ (13) is likely an artifact of the formation of a 2:1 complex in HEK293 cells involving two of the overexpressed CaM12’ molecules bound to a single CaV1.2.
ITC
ITC experiments were performed using a VP-ITC calorimeter (Micro-Cal) at 27 °C and 37 °C. The data were acquired and processed with MicroCal software (https://www.originlab.com) as described previously (41). The first data point from each ITC isotherm was deleted because the amount of injectant delivered during the first injection has significant error caused by a dead volume void in the injection syringe. For ITC experiments in Figure 3, A and B, samples of Ca2+ (injectant) and CaM12’–IQ complex (titrant) were prepared by exchanging each into buffer containing 20 mM Tris, pH 7.4, and 100 mM KCl. The CaM12’–IQ complex in the sample cell (10 μM at 27 °C or 8.0 μM at 37 °C in 1.5 ml) was titrated with aqueous CaCl2 (0.23 mM at 27 °C or 0.3 mM at 37 °C) using 35 injections of 10 μl each. For the ITC experiments in Fig. 3, C, E–H, samples of Ca2/CaM12’ (injectant) and IQ peptide (titrant) were prepared by exchanging each into buffer containing 20 mM Tris, pH 7.4, 100 mM KCl, and 1 mM CaCl2. The concentrations of the IQ peptides (WT, Y1649A, I1654A, or F1658A) were each 10 μM in 1.5 ml in the sample cell for titration with 0.1 mM Ca2/CaM12’ and the concentrations of Y1657D and F1658D were each 50 μM in 1.5 ml for titration with 0.5 mM Ca2/CaM12’ using 35 injections of 10 μl each.
NMR spectroscopy
All NMR measurements were performed at 303 K using a Bruker Avance III 600 MHz spectrometer equipped with a four-channel interface and triple-resonance cryoprobe. NMR sample preparation of Ca2/CaM12’-IQ was described previously (21). Two-dimensional NMR experiments (heteronuclear single quantum coherence [HSQC] and HSQC-IPAP) were recorded on samples of 15N-labeled Ca2/CaM12’ (0.5 mM) bound to unlabeled IQ (0.75 mM). Each sample was dissolved in 20 mM 2-Amino−2−hydroxymethyl-propane-1,3-diol-d11 (Tris-d11 at pH 7.5), 1.0 mM CaCl2, and 95% H2O/5% D2O. Three-dimensional NMR experiments for assigning backbone and side-chain resonances, and NOESY distance restraints were analyzed as described previously (42). NMR data were processed using NMRPipe (43) and analyzed with SPARKY (Goddard T.D. and Kneller D.G., University of California at San Francisco). To measure RDCs (23) of Ca2/CaM12’ bound to the IQ peptide, the filamentous bacteriophage Pf1 (Asla Biotech Ltd) was used as an orienting medium. Pf1 (12′ mg/ml) was added to an NMR sample that contained either 15N-labeled Ca2/CaM12’ bound to unlabeled IQ. 1H-15N residual dipolar coupling constants (DNH) were measured using a 2D IPAP (inphase/antiphase) 1H-15N HSQC experiment as described by (44). Representative IPAP-HSQC spectra of 15N-labeled Ca2/CaM12’ bound to the IQ peptide are shown in Fig. S4A. Briefly, the backbone N-H RDCs were calculated by measuring the difference in 15N splitting for each amide resonance, both in the presence and absence of the orienting medium. The RDC Q-factor and analysis of RDC data were calculated by PALES (45). The Q-factor is calculated as Q = RMS(Dmeas-Dcalc)/RMS(Dmeas), where Dmeas is the measured RDC, Dcalc is the calculated RDC, and RMS is the root mean square difference. A Q-factor of 30% corresponds to 2 Å resolution.
NMR structure calculation
NMR-derived structures of Ca2/CaM12’ bound to the IQ peptide were calculated using restrained molecular dynamics simulations within Xplor-NIH (46). RDCs, NOE distances, dihedral angles from TALOS+ (47), and backbone hydrogen bonds were used as structural restraints. NOEs were obtained from 15N-edited NOESY-HSQC, 13C-edited NOESY-HSQC (aliphatic), and 13C-filtered NOESY-HSQC as described by (48). Representative 13C-edited NOESY-HSQC and 13C-filtered NOESY-HSQC spectra of 13C-labeled Ca2/CaM12’ bound to unlabeled IQ peptide are shown in Fig. S4, B and C, respectively. Backbone dihedral angles were calculated by TALOS+ (47) using backbone chemical shifts (Hα, Cα, Cβ, CO, 15N, and HN) as input. Hydrogen bond restraints in helices and β-sheets were verified by measuring amide hydrogen-deuterium exchange rates as described by (49). The Xplor-NIH structure calculation was performed in three stages: annealing, refinement, and water refinement (50). Annealing started from an extended random structure. A total of 200 structures were calculated and the one with lowest energy was used as a starting structure during the refinement. The lowest energy structure was refined in an explicit water environment. A Ramachandran plot was generated by PROCHECK-NMR (27) and structure quality was assessed by MolProbity (51).
FP assays
Fluorescein-labeled peptides (100 nM; ChinaPeptides) were titrated with increasing concentrations of purified Ca2/CaM12’ in FP buffer (20 mM Tris, pH 7.4, 100 mM KCl, 1 mM MgCl2, 1.0 mM CaCl2) or apoCaM in Ca2+-free buffer (20 mM Tris, pH 7.4, 100 mM KCl, 1 mM MgCl2, 2.0 mM EGTA). FP was measured with a Synergy 2 plate reader (BioTek) as described (52). FP was calculated as P = (Iv - g∗Ih)/(Iv + g∗Ih); Iv and Ih are vertical and horizontal fluorescence intensity, respectively, and g is the correction factor for fluorescein. To obtain binding curves and KD values, data were fitted in GraphPad Prism 5 (GraphPad Software Inc) to the equation Y = B∗X/(Kd + X); B is maximal FP value that would be reached at saturation as determined by extrapolation of the fitted curve.
Concentration profiles of CaM species versus [Ca2+]
The concentration profiles of apoCaM-IQ, Ca2/CaM-IQ, and Ca4/CaM-IQ as a function of the free Ca2+ concentration were calculated according to the following scheme in Figure 8.
Figure 8.
Kinetic scheme for the sequential binding of Ca2+to the CaM C-lobe (K1) and CaM N-lobe (K2).
Expression of CaV1.2 IQ domain mutants and CaM species in HEK 293T/17 cells
HEK 293T/17 cells (ATCC) were maintained as previously described (15, 53). For electrophysiology, Lipofectamine 2000 (Invitrogene) or JetPrime (Polyplus Transfection) was used to transiently transfect cells with indicated plasmid DNAs in 35 mm dishes. For biochemistry experiments, transient transfection of HEK 293T/17 cells in 100 mM dishes was achieved using either JetPrime or, as previously described (15, 53), the calcium phosphate method. Cells were cotransfected with plasmids encoding the pore-forming α11.2 subunit N-terminally tagged with eCFP (15, 53) or mCherry (54) plus pGWIH-based plasmids encoding the auxiliary subunits rat β2A (55) and rabbit α2δ-1 (56) as previously described (15, 53). For all transfections, equimolar ratio of 1:1:1 was used for CaV1.2 channel subunits and later further optimized (JetPrime) for CaM (at ratio of 1:1:1:0.5 for α11.2:β2A:α2δ−1:CaM). Rat brain α11.2 (GenBank ID: M67515.1) N-terminally fused to eCFP was utilized as previously described (15). The point mutations in plasmids encoding single-residue I1654A, Y1657D (this report), and K1662E exchanges in α11.2 were generated via QuikChange II as previously described (15, 53) using N-terminally eCFP (15, 53) or mCherry tagged (54) rat brain α11.2 plasmid template DNAs. We studied CDI using mCherry-tagged α11.2 subunit coexpressed with the other, untagged CaV1.2 subunits and WT CaM or the calmodulin 34 mutant CaM34 (kindly provided by JP Adelman, (8)). For some biochemical experiments shown in Fig. S2, YFP-tagged CaM was used (32).
Whole-cell patch clamp recording
Macroscopic Ba2+- (IBa) and Ca2+ currents (ICa) of CaV1.2 L-type Ca2+ channels were obtained in the whole-cell configuration using external bath solution containing (in mM) 134 N-methyl-D-glucamine, 10 BaCl2 (for CDI, 10 CaCl2), 1 MgCl2, 10 Hepes, and 10 glucose with an adjusted pH of 7.4 (Cs-OH) and an osmolarity of 300 to 310 mOsm (sucrose). Intracellular pipette solution contained (in mM) 125 Cs-MeSO3, 5 CsCl, 10 EGTA, 10 Hepes, 1 MgCl2, 4 Mg-ATP, and pH 7.3 (CsOH), mOsm 290 to 300 (sucrose). Cells were clamped at a holding potential of -80 mV and depolarized for 900 ms to a series of activating potentials, from −60 mV to +50 mV (or +80 mV for Ca2+ currents), in increments of 10 mV at an interval of 0.033 Hz. The series resistance and the cell capacitance were directly taken from the Amplifier (Axopatch 200B, Molecular Device) and compensated to ∼40%. Data were sampled at 10 kHz and lowpass filtered at 2 kHz. Leak subtracted raw data were analyzed with Pclamp10 and GraphPad Prism IX software. All recordings were performed at room temperature (RT).
Cell-attached patch clamp recording
Single-channel recordings were performed as described previously (15, 31). In brief, low noise raw data were recorded with an Axopatch 200B amplifier and data were sampled at 10 kHz with a low-pass filter at 2 kHz (3 dB, four pole Bessel) and digitalized with a Digidata 1440 digitizer. Recording electrodes were pulled from borosilicate capillary glass (0.86 OD/1.25 ID) with a Flaming/Brown micropipette puller (Model P-97, Sutter Instruments), heat polished, and coated with Sylgard (Sylgard 289) until close to the electrode tip. Electrode resistance in solution was usually 5 to 10 MΩ. To keep the membrane potential close to 0 mV the extracellular bath solution contained (in mM) 120 K-Glutamate, 25 KCl, 2 MgCl2, 1 CaCl2, 10 EGTA, 10 Hepes, and 2 Na2-ATP pH 7.4 (KOH). The intracellular pipette solution contained (in mM) 110 BaCl2 and 10 Hepes, adjusted to pH 7.4 (TEA-OH). Cells were depolarized for 2 s from a holding potential of -80 mV to 0 mV every 7 s. Event lists were created from raw Ba2+ currents after leak and capacity transients were digitally subtracted by pClamp 10. Unitary current events were then analyzed based on the half-height criterium (57) using the single-channel software provided by pClamp 10.
For statistical analysis, single-channel parameters were corrected by the channel number (k), respectively, the maximum of simultaneously open channels (PMAX). The number of channels in the patch was estimated based on the observed simultaneous openings and is a precise parameter for k < 4, as included in this article and originally described by R. Horn (58). On average, 100 to 200 Ba2+ current traces were recorded for each cell for each experimental condition for an appropriate statistical analysis.
Surface biotinylation, NeutrAvidin pull downs, and immunoblotting
Surface biotinylation and analysis of CaV1.2 surface expression was carried out essentially as described (15, 53) with the following modifications. Twenty-two to twenty-four hours post transfection, HEK 293T/17 cells plated in 100 mm diameter dishes were rinsed with RT PBS-CM (PBS supplemented with 1 mM Ca2+ and 0.5 mM Mg2+) and placed on ice. Cell were incubated with freshly prepared 0.4 mg/ml of EZ-Link-Sulfo-NHS-LC-biotin (Thermo Fisher Scientific) in PBS-CM for 30 min, followed by quenching of remaining NHS reactive groups with ice-cold 100 mM glycine in PBS-CM, four separate washes with quenching buffer, and a final rinse with PBS alone. Labeled and quenched cells were dislodged by scrapping and directly lysed into ice-cold radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EGTA, 10 mM EDTA, 1% NP−40, 0.05% SDS, 0.4% DOC, and 10% glycerol) supplemented with protease inhibitors: 1 μg/ml leupeptin (Merck Millipore), 2 μg/ml aprotinin (Merck Millipore), 1 μg/ml pepstatin A (Merck Millipore), and 34 μg/ml PMSF (Sigma). Lysates were cleared of insoluble material via centrifugation at 200,000g for 30 min at 4 °C. The protein concentration of the solubilized material in the cleared lysate was determined by a standard bicinchoninic acid assay (Thermo Fisher Scientific). Biotinylated constituents in equal amount protein lysates (e.g., 400 μg/sample) were affinity purified by incubation with 30 μl of NeutrAvidin-conjugated Sepharose beads (Thermo Fisher Scientific) for 2 h at 4 °C. Bead-bound material was sedimented by centrifugation, washed several times with ice-cold buffer, and bound proteins extracted in SDS sample buffer (with shaking at 65 °C for 15 min). Proteins from pull downs as well as directly loaded lysates were fractionated by 7.5% acrylamide SDS–PAGE and transferred onto polyvinylidene difluoride (PVDF; Bio-Rad) membranes. For experiments used for analysis of CaM expression levels in directly loaded lysates (Fig. 6), 12% acrylamide gels were used. PVDF membranes were stained with Ponceau S, imaged, washed, and then incubated in blocking buffer (150 mM NaCl, 10 mM Tris–HCl, pH 7.4 (TBS) with 0.1% Tween (TBST) and 2% bovine serum albumin (RPI Corp.)) for 1 h at RT and then incubated with primary antibodies in blocking buffer for 3 h at RT. For analysis of surface expressed CaV1.2, α11.2 was detected using rabbit antibodies against epitopes in the intracellular loop II/III (FP1 or CNC1) and the CNC2 epitope near the C terminus of α11.2 (59). When CaM expression in directly loaded lysates was assessed, the membranes were probed with a mouse anti-CaM monoclonal primary antibody (made against a synthetic peptide corresponding to the 21 carboxy terminal amino acids (128–148) of bovine calmodulin) obtained from from Sigma Millipore (catalog no.: # 05-173, Lot # 2717626). YFP-tagged CaM signals were further verified by the NeuroMab mouse anti-GFP monoclonal antibody N86/8 (UC Davis). Signals obtained from probing with antibodies against the cytosolic proteins GAPDH (mouse monoclonal, Sigma/Millipore 214592) and α-tubulin (DM1A mouse monoclonal, Santa Cruz Biotechnology SC32293) were used (along with Ponceau S-stained bands) as loading controls for correction of variation in protein content between lysate samples. The absence of GAPDH and α-tubulin antibody signals in NeutrAvidin-pull down samples also served as intracellular protein controls for assurance of plasma membrane integrity during the biotinylation of plated cells. PVDF membranes were washed for 40 min with at least five exchanges of TBST, incubated with horseradish peroxidase–conjugated secondary goat antimouse antibodies (Jackson) or mouse anti-rabbit antibodies (Jackson) for 1 h at RT, and washed again with TBST with at least five exchanges for 1.5 h. Immunosignals were detected using the horseradish peroxidase substrates Luminata Classico or Crescendo (Merck Millipore) or Femto (Thermo Fisher Scientific) by X-ray film (Denville Scientific Inc). Multiple exposures over increasing time periods were taken to ensure that all signals were in the linear range (60, 61).
Analysis of immunoblots
Signal intensity for each band in scanned film images of immunoblots were assessed using ImageJ (https://imagej.nih.gov). Background signals in individual lanes were subtracted from the band signal prior to quantitative analysis. Differences in immunosignal strengths were corrected for potential immunoblotting and film exposures differences between experiments, as described (15, 53). Loading control (e.g., GAPDH, α-tubulin) lysate immunosignals were used to correct for minor differences in protein amounts loaded in individual sample lanes. To correct for variation in test immunosignals (e.g., α11.2, CaM) between experimental replicates, normalization was done according to the ‘sum of the replicates’ method as described (62). Each immunosignal for a protein (e.g., α11.2, CaM) on one blot was divided by the sum of all immunosignals from the same immunoblot exposure for that experimental run to obtain the relative signal fraction for each band (62). The means of these signal intensity fractions were calculated for each condition (e.g., α11.2 WT, Y1657D) from all experiments (e.g., α11.2 WT, Y1657D) and these means then divided by the mean value of the test control (e.g., α11.2 WT, which is now equal to 1% or 100%). All data were statistically analyzed (GraphPad Prism IX software) applying either a Student's t test (two-sample comparison) or ANOVA with Tukey post hoc test.
Data availability
Atomic coordinates were deposited in the Protein Databank (accession no. 7L8V), and all other data are contained within the article.
Supporting information
This article contains supporting information (13, 19, 28, 63).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Derrick Kaseman and Ping Yu from the UC Davis NMR Facility for help with NMR experiments.
Author contributions
M. C. H, J. W. H., and J. B. A. methodology; P. B., I. S., A. M. C., D. E. A., Q. Y., E. K., M. N.-C., M. F. N., M. C. H, J. W. H., and J. B. A. formal analysis; P. B., I. S., A. M. C., D. E. A., G. J., Z. M. E.-T., K. N. M. M., M. N.-C., M. F. N., M. C. H., and J. B. A. investigation; M. C. H, J. W. H., and J. B. A. writing–original draft.
Funding and additional information
This work was supported by NIH grants R01 HL121059 (M. F. N.), R01 EY012347 and R01 GM130925 (J. B. A.), and RF1 AG055357 and R01 NS123050 (J. W. H.). A. M. C. was supported by R25 GM056765 and T32 GM113770 and ZME-T by T32 GM 007377. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Roger Colbran
Contributor Information
Mary C. Horne, Email: mhorne@ucdavis.edu.
Johannes W. Hell, Email: jwhell@ucdavis.edu.
James B. Ames, Email: jbames@ucdavis.edu.
Supporting information
References
- 1.Ghosh D., Syed A.U., Prada M.P., Nystoriak M.A., Santana L.F., Nieves-Cintron M., et al. Calcium channels in vascular smooth muscle. Adv. Pharmacol. 2017;78:49–87. doi: 10.1016/bs.apha.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hell J.W., Westenbroek R.E., Warner C., Ahlijanian M.K., Prystay W., Gilbert M.M., et al. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J. Cell. Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moosmang S., Haider N., Klugbauer N., Adelsberger H., Langwieser N., Muller J., et al. Role of hippocampal Cav1.2 Ca2+ channels in NMDA receptor-independent synaptic plasticity and spatial memory. J. Neurosci. 2005;25:9883–9892. doi: 10.1523/JNEUROSCI.1531-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian H., Patriarchi T., Price J.L., Matt L., Lee B., Nieves-Cintron M., et al. Phosphorylation of Ser1928 mediates the enhanced activity of the L-type Ca2+ channel Cav1.2 by the beta2-adrenergic receptor in neurons. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aaf9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkefeld H., Sailer C., Bildl W., Rohde V., Thumfart J., Eble S., et al. BKCa-Cav channel complexes mediate rapid and localized Ca2+-activated K+ signaling. Science. 2006;314:615–620. doi: 10.1126/science.1132915. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S.M., Suutari B., He X., Wang Y., Sanchez S., Tirko N.N., et al. Calmodulin shuttling mediates cytonuclear signaling to trigger experience-dependent transcription and memory. Nat. Commun. 2018;9:2451. doi: 10.1038/s41467-018-04705-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Splawski I., Timothy K.W., Sharpe L.M., Decher N., Kumar P., Bloise R., et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Peterson B., DeMaria C., Adelman J., Yue D. Calmodulin is the Ca2+ sensor for Ca2+ -dependent inactivation of L-type calcium channels. Nature. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 9.Zuhlke R.D., Pitt G.S., Deisseroth K., Tsien R.W., Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 10.Erickson M., Alseikhan B., Peterson B., Yue D. Preassociation of calmodulin with voltage-gated Ca(2+) channels revealed by FRET in single living cells. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 11.Erickson M.G., Liang H., Mori M.X., Yue D.T. FRET two-hybrid mapping reveals function and location of L-type Ca2+ channel CaM preassociation. Neuron. 2003;39:97–107. doi: 10.1016/s0896-6273(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 12.Findeisen F., Rumpf C.H., Minor D.L., Jr. Apo states of calmodulin and CaBP1 control CaV1 voltage-gated calcium channel function through direct competition for the IQ domain. J. Mol. Biol. 2013;425:3217–3234. doi: 10.1016/j.jmb.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben Johny M., Yang P.S., Bazzazi H., Yue D.T. Dynamic switching of calmodulin interactions underlies Ca2+ regulation of CaV1.3 channels. Nat. Commun. 2013;4:1717. doi: 10.1038/ncomms2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans T.I., Hell J.W., Shea M.A. Thermodynamic linkage between calmodulin domains binding calcium and contiguous sites in the C-terminal tail of Ca(V)1.2. Biophys. Chem. 2011;159:172–187. doi: 10.1016/j.bpc.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner M., Anderson D.E., Nieves-Cintron M., Bartels P., Coleman A.M., Yarov V., et al. a-Actinin-1 promotes gating of the L-type Ca2+ Channel CaV1.2. EMBO J. 2020;39 doi: 10.15252/embj.2020106171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X., Bers D.M. Free and bound intracellular calmodulin measurements in cardiac myocytes. Cell Calcium. 2007;41:353–364. doi: 10.1016/j.ceca.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams P.J., Ben-Johny M., Dick I.E., Inoue T., Yue D.T. Apocalmodulin itself promotes ion channel opening and Ca(2+) regulation. Cell. 2014;159:608–622. doi: 10.1016/j.cell.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilli R., Lafitte D., Lopez C., Kilhoffer M., Makarov A., Briand C., et al. Thermodynamic analysis of calcium and magnesium binding to calmodulin. Biochemistry. 1998;37:5450–5456. doi: 10.1021/bi972083a. [DOI] [PubMed] [Google Scholar]
- 19.Ames J.B. L-type Ca(2+) channel regulation by calmodulin and CaBP1. Biomolecules. 2021;11:1811. doi: 10.3390/biom11121811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halling D.B., Georgiou D.K., Black D.J., Yang G., Fallon J.L., Quiocho F.A., et al. Determinants in CaV1 channels that regulate the Ca2+ sensitivity of bound calmodulin. J. Biol. Chem. 2009;284:20041–20051. doi: 10.1074/jbc.M109.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salveson I., Anderson D.E., Hell J.W., Ames J.B. Chemical shift assignments of a calmodulin intermediate with two Ca2+ bound in complex with the IQ-motif of voltage-gated Ca2+ channels (CaV1.2) Biomol. NMR Assign. 2019;13:233–237. doi: 10.1007/s12104-019-09883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clore G.M., Gronenborn A.M. Determining the structures of large proteins and protein complexes by NMR. Curr. Opin. Chem. Biol. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 23.Tjandra N., Bax A. Direct measurement of disances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 24.Finn B.E., Evenas J., Drakenberg T., Waltho J.P., Thulin E., Forsen S. Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 1995;2:777–783. doi: 10.1038/nsb0995-777. [DOI] [PubMed] [Google Scholar]
- 25.Kuboniwa H., Tjandra N., Grzesiek S., Ren H., Klee C.B., Bax A. Structure of calcium-free calmodulin. Nat. Struct. Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M., Tanaka T., Ikura M. Calcium-induced conformational transition revealed by the solution structures of apo calmodulin. Nat. Struct. Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R.A., Rullmann J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 28.Van Petegem F., Chatelain F.C., Minor D.L., Jr. Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Nat. Struct. Mol. Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fallon J.L., Baker M.R., Xiong L., Loy R.E., Yang G., Dirksen R.T., et al. Crystal structure of dimeric cardiac L-type calcium channel regulatory domains bridged by Ca2+∗ calmodulins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5135–5140. doi: 10.1073/pnas.0807487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fallon J.L., Halling D.B., Hamilton S.L., Quiocho F.A. Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure. 2005;13:1881–1886. doi: 10.1016/j.str.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Bartels P., Yu D., Huang H., Hu Z., Herzig S., Soong T.W. Alternative splicing at N terminus and domain I modulates CaV1.2 inactivation and surface expression. Biophys. J. 2018;114:2095–2106. doi: 10.1016/j.bpj.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iacobucci G.J., Popescu G.K. Spatial coupling tunes NMDA receptor responses via Ca(2+) diffusion. J. Neurosci. 2019;39:8831–8844. doi: 10.1523/JNEUROSCI.0901-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baber J.L., Szabo A., Tjandra N. Analysis of slow interdomain motion of macromolecules using NMR relaxation data. J. Am. Chem. Soc. 2001;123:3953–3959. doi: 10.1021/ja0041876. [DOI] [PubMed] [Google Scholar]
- 34.Tjandra N., Kuboniwa H., Ren H., Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur. J. Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 35.Hall D.D., Dai S., Tseng P.Y., Malik Z., Nguyen M., Matt L., et al. Competition between a-actinin and Ca2+-calmodulin controls surface retention of the L-type Ca2+ channel Ca(V)1.2. Neuron. 2013;78:483–497. doi: 10.1016/j.neuron.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson B.D., Byerly L. A cytoskeletal mechanism for Ca2+ channel metabolic dependence and inactivation by intracellular Ca2+ Neuron. 1993;10:797–804. doi: 10.1016/0896-6273(93)90196-x. [DOI] [PubMed] [Google Scholar]
- 37.Jensen H.H., Brohus M., Nyegaard M., Overgaard M.T. Human calmodulin mutations. Front. Mol. Neurosci. 2018;11:396. doi: 10.3389/fnmol.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K., Holt C., Lu J., Brohus M., Larsen K., Overgaard M., et al. Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E10556–E10565. doi: 10.1073/pnas.1808733115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Li Z., Sacks D.B., Ames J.B. Structural basis for Ca2+-induced activation and dimerization of estrogen receptor a by calmodulin. J. Biol. Chem. 2012;287:9336–9344. doi: 10.1074/jbc.M111.334797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingard J.N., Chan J., Bosanac I., Haeseleer F., Palczewski K., Ikura M., et al. Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J. Biol. Chem. 2005;280:37461–37470. doi: 10.1074/jbc.M508541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim S., Cudia D., Yu Q., Peshenko I., Dizhoor A., Ames J. Chemical shift assignments of retinal degeneration 3 protein (RD3) Biomol. NMR Assign. 2018;12:167–170. doi: 10.1007/s12104-018-9802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeiffer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Ottiger M., Delaglio F., Marquardt J.L., Tjandra N., Bax A. Measurement of dipolar couplings for methylene and methyl sites in weakly oriented macromolecules and their use in structure determination. J. Magn. Reson. 1998;134:365–369. doi: 10.1006/jmre.1998.1546. [DOI] [PubMed] [Google Scholar]
- 45.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 46.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 47.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T., Ames J.B., Kainosho M., Stryer L., Ikura M. Differential isotype labeling strategy for determining the structure of myristoylated recoverin by NMR spectroscopy. J. Biomol. NMR. 1998;11:135–152. doi: 10.1023/a:1008212316986. [DOI] [PubMed] [Google Scholar]
- 49.Ames J.B., Tanaka T., Stryer L., Ikura M. Secondary structure of myristoylated recoverin determined by three-dimensional heteronuclear NMR: implications for the calcium-myristoyl switch. Biochemistry. 1994;33:10743–10753. doi: 10.1021/bi00201a023. [DOI] [PubMed] [Google Scholar]
- 50.Nilges M., Gronenborn A.M., Brunger A.T., Clore G.M. Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints. Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 1988;2:27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- 51.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Matt L., Patriarchi T., Malik Z.A., Chowdhury D., Park D.K., et al. Capping of the N-terminus of PSD-95 by calmodulin triggers its postsynaptic release. EMBO J. 2014;33:1341–1353. doi: 10.1002/embj.201488126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tseng P.Y., Henderson P.B., Hergarden A.C., Patriarchi T., Coleman A.M., Lillya M.W., et al. Alpha-actinin promotes surface localization and current density of the Ca(2+) channel CaV1.2 by binding to the IQ region of the alpha1 subunit. Biochemistry. 2017;56:3669–3681. doi: 10.1021/acs.biochem.7b00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen A., Nieves-Cintron M., Deng Y., Shi Q., Chowdhury D., Qi J., et al. Functionally distinct and selectively phosphorylated GPCR subpopulations co-exist in a single cell. Nat. Commun. 2018;9:1050. doi: 10.1038/s41467-018-03459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Reyes E., Castellano A., Kim H.S., Bertrand P., Baggstrom E., Lacerda A.E., et al. Cloning and expression of a cardiac/brain beta subunit of the L-type calcium channel. J. Biol. Chem. 1992;267:1792–1797. [PubMed] [Google Scholar]
- 56.Ellis S.B., Williams N.R., Ways N.R., Brenner R., Sharp A.H., Leung A.T., et al. Sequence and expression of mRNAs encoding the alpha 1 and alpha 2 subunits of a DHP-sensitive calcium channel. Science. 1988;241:1661–1664. doi: 10.1126/science.2458626. [DOI] [PubMed] [Google Scholar]
- 57.Sachs F., Neil J., Barkakati N. The automated analysis of data from single ionic channels. Pflugers Archiv. 1982;395:331–340. doi: 10.1007/BF00580798. [DOI] [PubMed] [Google Scholar]
- 58.Horn R. Estimating the number of channels in patch recordings. Biophys. J. 1991;60:433–439. doi: 10.1016/S0006-3495(91)82069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buonarati O.R., Henderson P.B., Murphy G.G., Horne M.C., Hell J.W. Proteolytic processing of the L-type Ca 2+ channel alpha 11.2 subunit in neurons. F1000 Res. 2017;6:1166. doi: 10.12688/f1000research.11808.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davare M.A., Hell J.W. Increased phosphorylation of the neuronal L-type Ca(2+) channel Ca(v)1.2 during aging. Proc. Natl. Acad. Sci. U. S. A. 2003;100:16018–16023. doi: 10.1073/pnas.2236970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall D.D., Feekes J.A., Arachchige A.S., Shi M., Hamid J., Chen L., et al. Binding of protein phosphatase 2A to the L-type calcium channel Cav1.2 next to Ser1928, its main PKA site, is critical for Ser1928 dephosphorylation. Biochemistry. 2006;45:3448–3459. doi: 10.1021/bi051593z. [DOI] [PubMed] [Google Scholar]
- 62.Degasperi A., Birtwistle M.R., Volinsky N., Rauch J., Kolch W., Kholodenko B.N. Evaluating strategies to normalise biological replicates of Western blot data. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J., Yan Z., Li Z., Qian X., Lu S., Dong M., et al. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 Å resolution. Nature. 2016;537:191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates were deposited in the Protein Databank (accession no. 7L8V), and all other data are contained within the article.