ABSTRACT
Meiosis is essential for evolution and genetic diversity in almost all sexual eukaryotic organisms. The mechanisms of meiotic recombination, such as synapsis, have been extensively investigated. However, it is still unclear whether signals from the cytoplasm or even from outside of the cell can regulate the meiosis process. Cilia are microtubule-based structures that protrude from the cell surface and function as signaling hubs to sense extracellular signals. Here, we reported an unexpected and critical role of cilia during meiotic recombination. During gametogenesis of zebrafish, cilia were specifically present in the prophase stages of both primary spermatocytes and primary oocytes. By developing a germ cell-specific CRISPR/Cas9 system, we demonstrated that germ cell-specific depletion of ciliary genes resulted in compromised double-strand break repair, reduced crossover formation, and increased germ cell apoptosis. Our study reveals a previously undiscovered role for cilia during meiosis and suggests that extracellular signals may regulate meiotic recombination via this particular organelle.
Keywords: cilia, meiosis, homologous recombination, kif3a, zebrafish
Introduction
Meiosis is probably the most key event in the life cycle of sexually reproducing eukaryotes. During the prophase of the first meiotic division, homologous chromosomes (homologs) of the maternal and paternal origins pair and synapse, which creates a context for the exchange of genetic information between homologs through an elaborate process, homologous recombination (HR). Meiotic recombination has been extensively studied in different model systems ranging from yeast, protists, to mammals (Loidl, 2016; Zelkowski et al., 2019). In a classic HR model, DNA double-strand breaks (DSBs) are induced by the meiotic topoisomerase-like protein SPO11, and then the phosphorylation of histone variant H2A.X is induced around the DSB sites (Keeney et al., 1997; Neale et al., 2005; Borde and de Massy, 2013). Meanwhile, DSB ends are processed and the resulting single-strand overhangs are bound by recombinases including Dmc1 and Rad51 (Bishop et al., 1992; Shinohara et al., 1992; Bishop, 1994). The broken DNA ends and these proteins form nucleofilaments and further invade the homologous template for homology searching and HR (Sun et al., 1991). Dmc1-mediated homology searching also promotes the formation of the synaptonemal complex (SC), an evolutionarily conserved tripartite protein structure formed between paired homologs. The SC is indispensable for multiple meiosis processes, including enhancing the interhomolog recombination, ensuring obligate crossover formation, and thus, safeguarding the faithful segmentation of chromosomes (Page and Hawley, 2004; Zickler and Kleckner, 2015). Depending on the resolution of the double holiday junction, repairing of DSBs will be channeled to MutL homolog 1 (Mlh1)-dependent pro-crossover pathway or non-crossover pathway. The crossover recombination event leads to the reciprocal exchange of alleles between two homologs, and thus ensures the genetic diversity of the progeny (Handel and Schimenti, 2010; Hunter, 2015; Dai et al., 2021).
One of the key aspects of the meiotic recombination is the reorganization of the chromosome during prophase to ensure homologous pairing. The formation of the telomere bouquet is an evolutionarily conserved mechanism for promoting homologous pairing (Scherthan, 2001; Loidl, 2016; Blokhina et al., 2019). Telomeres are attached to the nuclear envelope (NE) through the TRF1 and TERB1/2–MAJIN tetrameric complex (Shibuya et al., 2014; Shibuya et al., 2015). During bouquet formation, telomeres are clustered to one side of the nucleus through the linker of nucleoskeleton and cytoskeleton complex, which bridges the movement of chromosome to the dynein motor moving on the perinuclear microtubules (King et al., 2008; Sato et al., 2009; Burke, 2018). Intriguingly, telomeres are usually clustered at the NE near the microtubule organizing center (MTOC), suggesting a potential role of the microtubule network in promoting the homologous pairing (Sawin, 2005; Sato et al., 2009; Elkouby et al., 2016). Although the mechanisms of HR have been extensively studied, it is still an enigma whether the meiotic recombination is modulated by signals from the cytoplasm or extracellular environment.
Cilia are microtubule-based structures that are nucleated by the basal body, a highly conserved structure acting as the MTOC (Song et al., 2016; Zhao et al., 2021). Protruding from the cell surface, cilia can function as a cellular antenna to sense extracellular signals and transfer this information into the cell (Singla and Reiter, 2006; Ishikawa and Marshall, 2011). Dysfunction in cilia will cause a large number of devastating genetic disorders, termed ciliopathies (Hildebrandt et al., 2011; Reiter and Leroux, 2017). In this study, we showed that cilia were present in the prophase stages of both primary spermatocytes and primary oocytes in zebrafish. By generating a germ cell-specific conditional knockout strategy, we demonstrated that cilia play a role during meiotic HR through regulating DSB repair and crossover formation. These results will help add new information for our understanding on the regulatory mechanisms of meiotic recombination.
Results
Cilia are present in the prophase stages of primary spermatocytes and primary oocytes
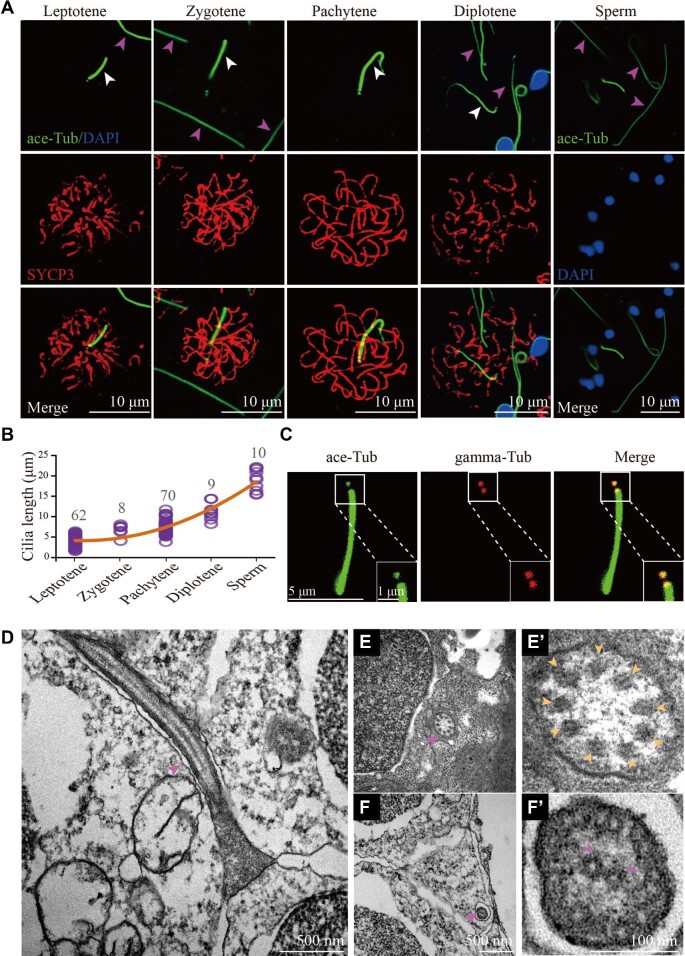
When examining the spermatogenesis process in zebrafish, we observed a special cilia-like structure that was present in the spermatocytes (Figure 1A). This cilium is shorter than the sperm flagellum, a special type of motile cilia that is essential for sperm motility (Figure 1A). Double immunostaining with anti-acetylated tubulin (a cilia marker) and SYCP3 (a lateral element of SC) antibodies showed that these ciliary structures were initially formed at leptotene stage, and gradually elongated until diplotene stage (Figure 1A). The diplotene cilia were about half-length of sperm flagella (Figure 1A and B). Interestingly, these cilia disappeared at diakinesis stage, implying that they are not the rudiments of sperm flagella. Further immunostaining with the antibody against gamma tubulin, the centrosomal marker, indicated that these microtubules were nucleated by one of the centrioles (Figure 1C; Supplementary Figure S1). Moreover, these cilia could also be labelled with anti-glutamylated tubulin antibody (Supplementary Figure S2A). In contrast, tubulin glycylation was absent in the spermatocyte cilia, compared with the strong signals in sperm flagella (Supplementary Figure S2B). Furthermore, ultrastructural analysis showed that these cilia had a ‘9 + 0’ axonemal configuration, in contrast to the ‘9 + 2’ arrangement of sperm flagella (Figure 1D–F’). Finally, cilia were also present in the leptotene and zygotene stages of primary oocytes (Supplementary Figure S3).
Figure 1.
Cilia of primary spermatocytes in meiosis. (A) Confocal images showing cilia (white arrowhead) and sperm flagella (pink arrowhead) labelled with anti-acetylated tubulin (ace-Tub, green) antibody. The synaptonemal complexes were labelled with SYCP3 (red) and nuclei were counter-stained with DAPI in blue. (B) Statistical results showing the length of cilia in different stages of primary spermatocytes and sperms. (C) Confocal images showing cilia and basal bodies labelled with anti-acetylated tubulin (ace-Tub, green) and anti-gamma tubulin (gamma-Tub, red) antibodies on primary spermatocyte. (D–F’) Transmission electron microscopy results showing the ultrastructure of primary spermatocyte cilia (D–E’) and sperm flagella (F and F’). Cross section showing the ‘9 + 0’ configuration of spermatocyte cilia (E’).
A high-efficiency approach for generating germ cell-specific conditional knockout mutants
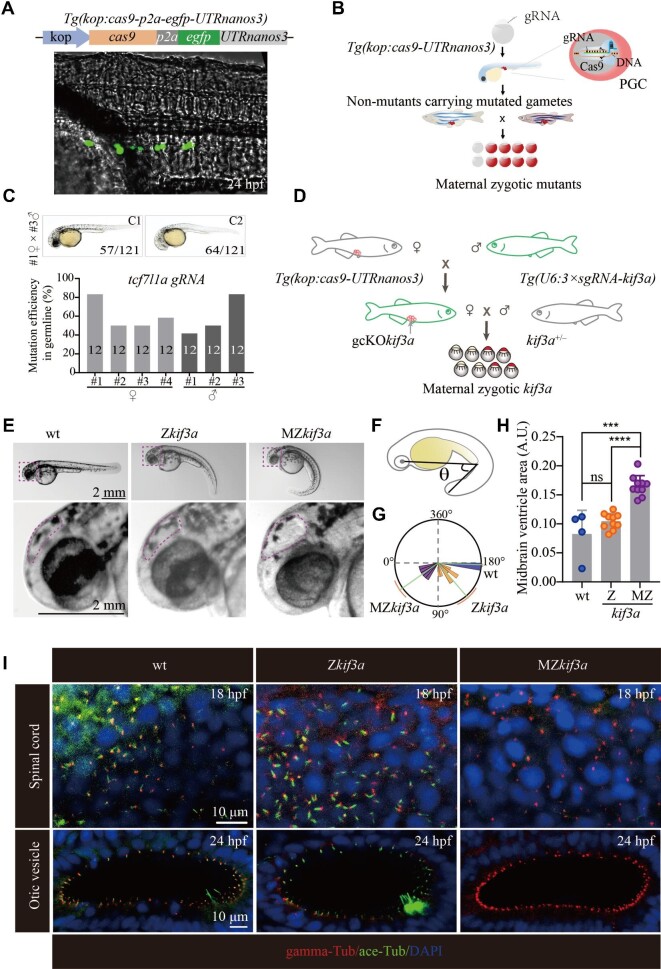
The presence of cilia during prophase I of the first meiotic division implies that cilia may participate in the regulation of HR. As cilia are essential for early zebrafish embryo development (Song et al., 2016), we seek to design a strategy to remove these cilia specially in the spermatocytes. To reach this goal, we developed a germ cell-specific CRISPR/Cas9 system by generating a primordial germ cell (PGC)-specific Cas9-transgenic line, Tg(kop:cas9-p2a-egfp-UTRnanos3), abbreviated here as Tg(kop:cas9-UTRnanos3) (Figure 2A), in which a Cas9-2A-EGFP expression cassette was placed under the control of the PGC-specific askopos (kop) promoter and nanos3 3′-UTR (Xiong et al., 2013). Since the Cas9 protein was expressed only in the PGCs of Tg(kop:cas9-UTRnanos3) embryos (Figure 2A), injection of single guide RNA (sgRNA) into the embryos will result in PGCs-specific gene knockout. Maternal derived RNAs and proteins are synthesized and accumulated during oogenesis, which play vital roles during early embryonic development. The presence of these maternal products often partially compensates for the loss of zygotic mutations in many organisms including zebrafish and xenopus. Generating maternal zygotic mutants will help clarify the role of maternal genes in regulating early development. Since the candidate genes were mutated in the PGCs of Tg(kop:cas9-UTRnanos3) fish, we hypothesized that crossbreeding of the sgRNA-injected F0 female and male fish could generate maternal zygotic mutants in the F1 generation, even if the zygotic mutants were lethal (Figure 2B).
Figure 2.
Generation of MZkif3a mutants via germ cell-specific expression of Cas9. (A) Bright field image showing a 24 hpf Tg(kop:cas9-p2a-egfp-UTRnanos3) zebrafish embryo with EGFP fluorescence in the PGCs. Top: diagram of the construct for making this transgene. (B) Schematic workflow showing process of generating MZ mutants using PGC-specific Cas9-expressing Tg(kop:cas9-UTRnanos3) embryos. (C) Mutation efficiencies of gametes and the phenotypes of offspring of tcf7l1a sgRNA-injected Tg(kop:cas9-UTRnanos3) fish. C1 shows the wild type-like phenotype, and C2 shows complete loss of eyes. The mutation efficiencies were calculated by the number of mutated heterozygotic embryos from crossing between F0 and wild type fish. A total of 12 embryos were genotyped from each cross and the mutation efficiency was calculated. (D) Schematic diagram showing the strategy for generating germ cell-specific knockout mutants of kif3a (gcKOkif3a). (E) External phenotypes of 48 hpf Zkif3a and MZkif3a mutants. The enlarged boxes are shown in the bottom, and dots outline the midbrain ventricles. (F and G) Statistical analysis showing the increased body curvature severity as demonstrated by the reduced angles of MZkif3a mutants. (H) Bar graph showing relative size of midbrain ventricles in different mutants as indicated. A.U., arbitrary unit. (I) Confocal images showing cilia in the spinal cord and otic vesicle of wild type, Zkif3a and MZkif3a mutants as indicated. Cilia were labelled with acetylated tubulin in green and the basal bodies were stained with gamma tubulin in red. Nuclei were counterstained with DAPI. ***P < 0.001; ****P < 0.0001. ns, not significant; hpf, hours post-fertilization; wt, wild type.
To test the efficacy and penetrance of PGCs-specifically expressed Cas9 in the Tg(kop:cas9-UTRnanos3) embryos, we first injected a validated sgRNA targeting tcf7l1a (Zhang et al., 2020), whose maternal zygotic mutants (MZtcf7l1a), but not maternal (Mtcf7l1a) or zygotic mutants (Ztcf7l1a), showed a headless phenotype (Kim et al., 2000). Crossing two germline-mutated females and males resulted in a typical headless phenotype in >50% of the F1 embryos (Figure 2C; Supplementary Figure S4A), suggesting the high efficiency of PGC-expressed Cas9. Similarly, we were also able to generate MZpou5f3 mutants, which exhibited severe developmental defects (Reim and Brand, 2006; Schulz and Harrison, 2019) (Supplementary Figure S4B and C). These data demonstrate that the Tg(kop:cas9-UTRnanos3) line expresses a highly efficient Cas9 protein, which enables germ cell-specific gene knockout. Noticeably, albeit with high efficiency, the penetrance of mutations in the gametes was not 100% (Figure 2C; Supplementary Figure S4B), which may be due to the temporal presence of sgRNAs.
Generation of MZkif3a mutants
To further improve the efficiency of gene editing in the PGCs, we generated a stable Tg(U6:3×sgRNA-kif3a) transgenic line, in which three sgRNAs targeting zebrafish kif3a were each driven by an individual U6 promoter in tandem. Kif3a is a plus-end motor protein crucial for cilia assembly (Zhao et al., 2012). We further crossed the Tg(kop:cas9-UTRnanos3) line with the Tg(U6:3×sgRNA-kif3a) line (Figure 2D) to generate the Tg(kop:cas9-UTRnanos3;U6:3×sgRNA-kif3a) double transgenic fish. By crossing the double transgenic female with kif3a heterozygotic male, we have successfully generated maternal-zygotic kif3a mutants (MZkif3a). Around 50% of embryos (340 of 707) displayed body curvature defects, strongly implying that Kif3a was deficient in almost all of the eggs produced from the double transgenics. Compared with Zkif3a mutants, MZkif3a mutants displayed strong body curvature defects, together with the increased size of the brain ventricles, a typical feature of hydrocephalous (Figure 2E–H; Feng et al., 2017). Further whole-mount staining results confirmed that cilia were completely absent in the MZkif3a mutants, while cilia were still formed in the spinal cord and otic vesicle of Zkif3a mutants at early stages due to the maternal effects of Kif3a (Figure 2I).
Ciliogenesis defects in both sperms and spermatocytes of germ cell-specific kif3a knockout mutants
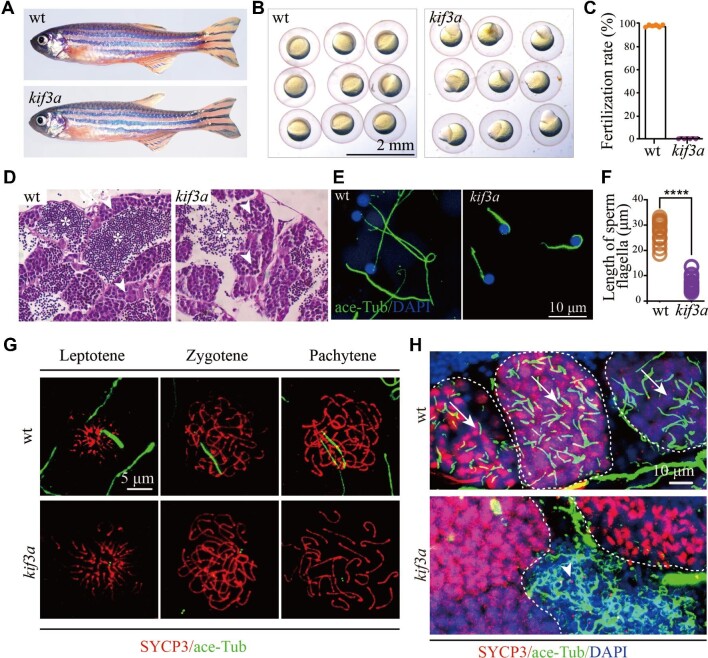
Next, we further analyzed the phenotypes of mutant males. Although the double transgenic males can develop to adulthood normally as the wild type males, their sperms failed to fertilize wild type eggs due to loss of Kif3a (Figure 3A–C). The spermatozoa were able to be produced in the testis, while all the flagella were significantly shorter than those of control siblings (Figure 3D–F). These data also suggest that sperm flagellum can still be partially assembled in the absence of Kif3a.
Figure 3.
Phenotypes of germ cell-specific kif3a conditional knockout mutants. (A) External phenotypes of wild type and Tg(kop:cas9-UTRnanos3;U6:3×sgRNA-kif3a) double transgenic male fish. (B) The embryos produced from wild type female crossed with control or double transgenic male at 6 hpf. (C) Statistical results showing the percentages of fertilization rates of wild type and double transgenic males as indicated. (D) Histological analysis of testis of wild type and Tg(kop:cas9-UTRnanos;U6:3×sgRNA-kif3a) double transgenic fish. Arrowheads point to spermatocytes and asterisks indicate spermatozoa. (E) Confocal images showing sperm flagella in wild type and double transgenic fish labelled with anti-acetylated tubulin antibody (green). (F) Statistical analysis of the length of sperm flagella in wild type and double transgenic fish. (G) Confocal images showing cilia in primary spermatocytes of wild type and double transgenic fish. Spermatocyte cilia (green) were absent in the double transgenic fish. The stages of spermatocytes were distinguished by the staining of SYCP3 (red). (H) Staining of SYCP3 (red) and acetylated-tubulin (green) in the testis of wild type and double transgenic fish. Arrows indicate spermatocyte cilia, while arrowheads indicate the staining of abnormal sperm flagella in the mutant testis. ****P < 0.0001. wt, wild type.
Since Cas9 protein was expressed in PGCs, it is conceived that kif3a gene should be disrupted at earlier stages. Indeed, cilia failed to form and the staining of acetylated tubulin was only detected in the centrosomes of the mutant spermatocytes (Figure 3G). Furthermore, staining with SYCP3 on the sections of testis showed that cilia were completely absent in all the earlier stage primary spermatocytes (Figure 3H), which further confirmed the nearly 100% efficiency in inducing germ cell-specific knockout of kif3a (gcKOkif3a) by the double transgenic approach (Figure 2D). Noticeably, strong signals were still present in those regions that were rich for spermatozoa, which were related to the remaining shorter sperm flagella (Figure 3H). Moreover, cilia in the primary oocytes were also absent in the double transgenic females (Supplementary Figure S3).
The repair of DNA DSBs is compromised in the absence of cilia
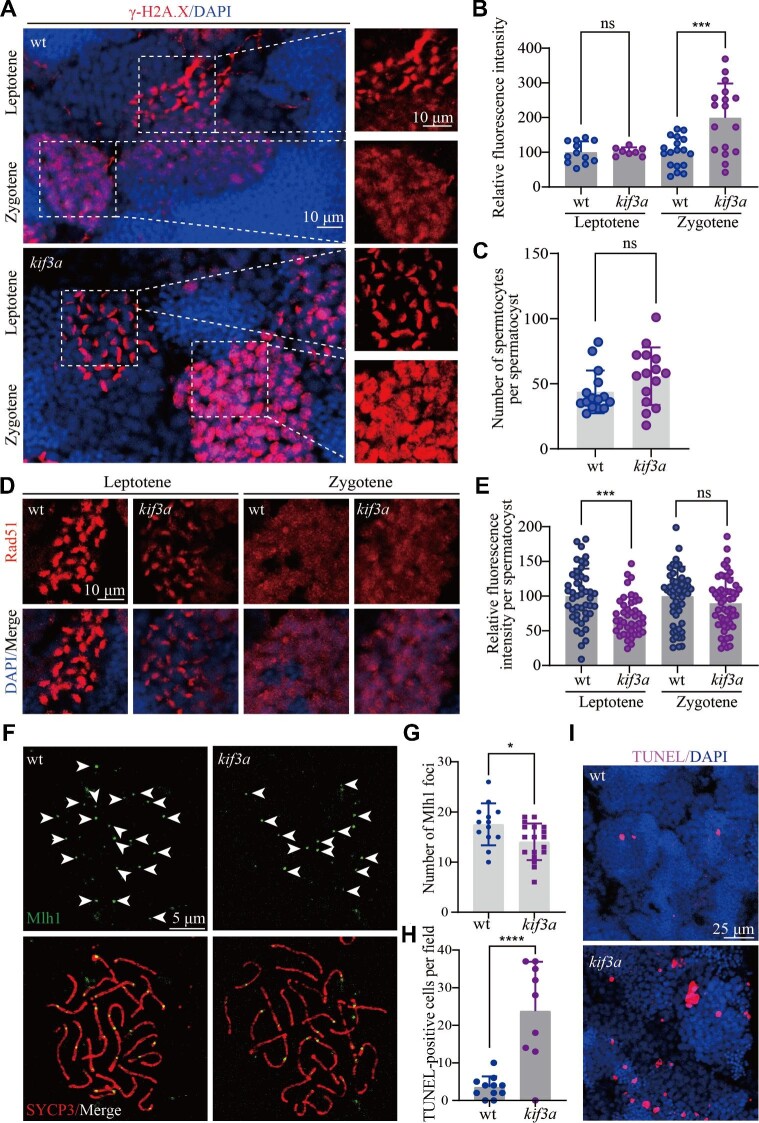
During meiotic recombination, the formation of DSB provokes a rapid DNA damage response, leading to the phosphorylation of a histone H2A isoform at sites of DSBs. By using the anti-γ-H2A.X antibody, we can distinguish spermatocytes at both leptotene and zygotene stages. The staining of γ-H2A.X disappeared from mid-zygotene stage (Xie et al., 2020). The staining signals were similar between gcKOkif3a spermatocytes and control siblings at leptotene stage, while unexpectedly, the spermatocytes had strong γ-H2A.X staining throughout their nuclei at zygotene stage in gcKOkif3a mutants (Figure 4A and B). By counting the number of γ-H2A.X-positive germ cells, we further found that the number of spermatocytes within each spermatocyst was comparable between the gcKOkif3a and control fish (Figure 4C), suggesting that loss of spermatocyte cilia did not affect the mitotic division of spermatogonium.
Figure 4.
The repair of DSBs is compromised in the absence of spermatocyte cilia. (A) Confocal images showing the staining of γ-H2A.X (red) in the testis of wild type and Tg(kop:cas9-UTRnanos3;U6:3×sgRNA-kif3a) double transgenic fish. (B) Statistical analysis showing relative fluorescence intensity of γ-H2A.X staining on leptotene and zygotene spermatocytes of wild type and double transgenic fish. (C) Statistical analysis of the number of spermatocytes per spermatocyst in wild type and double transgenic fish. (D) Staining of Rad51 in the testis of wild type and double transgenic fish. (E) Statistical analysis of relative fluorescence intensity of Rad51 staining on leptotene and zygotene spermatocytes. (F) Staining of Mlh1 in primary spermatocytes of wild type and double transgenic fish. (G) Statistical analysis of the Mlh1 foci number per spermatocyte. (H and I) Apoptotic cells stained by TUNEL assay in wild type and double transgenic fish. *P < 0.05; ***P < 0.001; ****P < 0.0001. ns, not significant; wt, wild type.
The strong signals of γ-H2A.X staining suggest that DSB repair may be affected in the absence of spermatocyte cilia. Next, we checked the distribution of Rad51, one of the key recombinases recruited to the break sites for DSB repair (Shinohara et al., 1992). Compared with those of control cells, Rad51 signals decreased substantially in leptotene-stage spermatocytes of gcKOkif3a males (Figure 4D and E). Rad51 signals became weaker at zygotene stage and there was no difference in the staining of Rad51 between mutant and control siblings at later zygotene stage (Figure 4D). Rad51 is required for the formation of crossovers. Next, we evaluated the crossover events by investigating the number of Mlh1 foci on surface spread spermatocytes. The number of Mlh1 foci decreased significantly in the mutant spermatocytes (Figure 4F and G). Finally, TUNEL assay results indicated that mutant testis contained a higher number of apoptotic spermatocytes than those of control siblings (Figure 4H and I). Altogether, these data suggested that loss of spermatocyte cilia affected the DSB repair process, leading to decreased crossover formation and cell apoptosis.
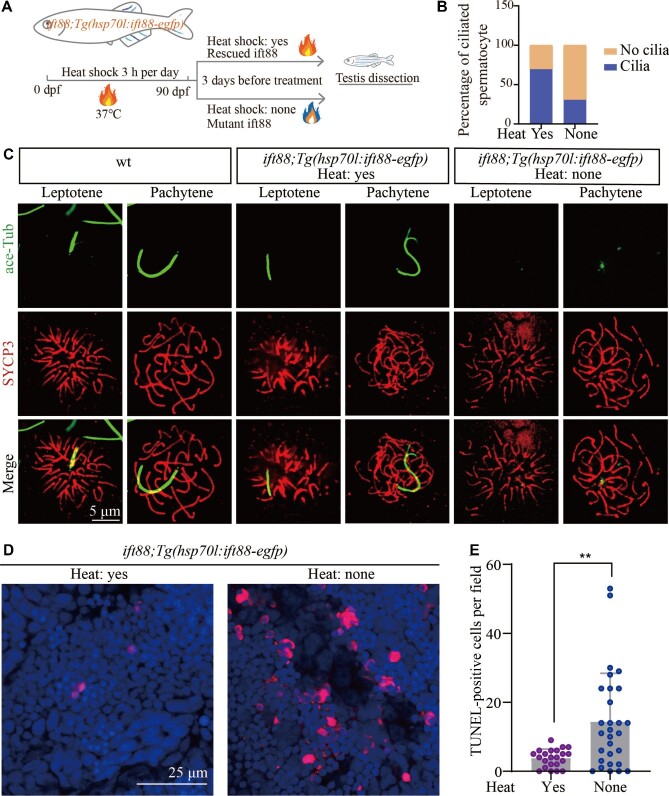
Loss of ift88 inhibits spermatocyte cilia formation and leads to cell apoptosis
To exclude the non-ciliary role of Kif3a during meiotic progression, we further generated Tg(hsp70l:ift88-egfp) transgenic line in which ift88, another essential gene for ciliogenesis (Tsujikawa and Malicki, 2004), was driven by a heat shock promoter. By performing routine heat shock, we were able to rescue ovl(ift88) mutants with this transgene (Figure 5A). Interestingly, the adult mutants can still survive for a short period even without heat inducement. By comparing mutants between heat-induced and untreated fish, we found that the spermatocyte cilia were largely lost in the testis of non-heat-shocked controls (Figure 5B and C). Moreover, mutants generated from these non-heat-shocked adult females and heterozygotic males showed stronger body curvature and ciliogenesis defects, resembling those of MZovl embryos (Supplementary Figure S5; Huang and Schier, 2009). Similar to those of kif3a mutants, loss of ift88 also caused abnormal cell apoptosis of the spermatocytes (Figure 5D and E), further confirming the role of cilia during meiosis processing.
Figure 5.
Ift88 deficiency results in ciliogenesis defects and apoptosis of spermatocytes. (A) Schematic workflow showing the strategy of generating Tg(hsp70l:ift88-egfp) transgene to rescue ovl(ift88) mutants. (B) Statistical analysis of the percentage of ciliated spermatocytes in heat-shocked and non-heat-shocked ift88;Tg(hsp70l:ift88-egfp) transgenic fish. (C) Confocal images showing cilia and synaptonemal complexes labelled with anti-acetylated tubulin (green) and anti-SYCP3 (red) antibodies on primary spermatocytes from different fish as indicated. (D and E) Confocal images showing apoptotic cells stained by TUNEL assay (D) and the statistical results (E) of TUNEL-positive cells in heat-shocked and non-heat-shocked fish as indicated. **P < 0.01. wt, wild type.
Discussion
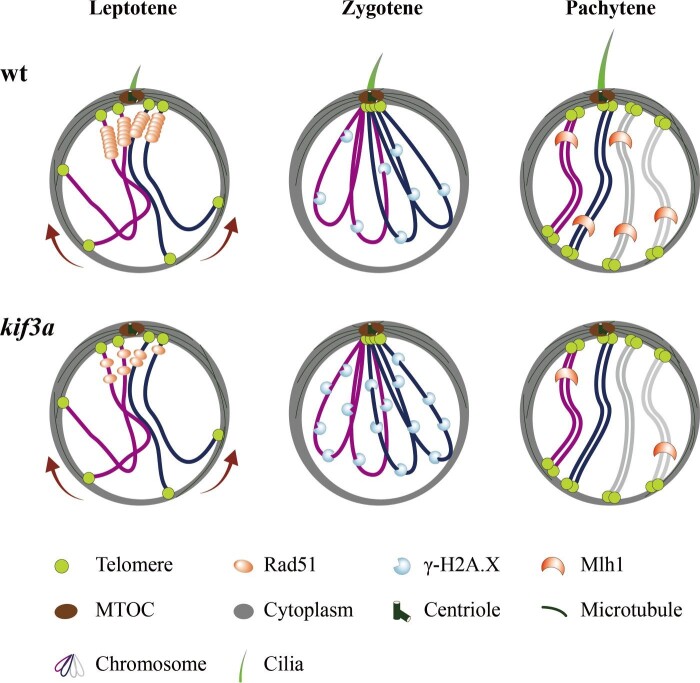
Cilia are tiny organelles that protrude from the cell surface to sense extracellular signals. Motile cilia can drive fluid flow or propel cells (sperms), while primary cilia play diverse roles in many aspects such as cell proliferation, migration, differentiation, and neural axon guidance (Bisgrove and Yost, 2006; Zhao et al., 2021). Here, we report that cilia may also regulate the repair of DSBs and crossover formation during meiosis. Our data demonstrate that cilia are present in the primary spermatocytes and oocytes during meiotic recombination (Figure 6). By developing a germ cell-specific gene knockout approach, we show that depletion of these early germ cell-specific cilia results in a compromised processing of DSB repair, impaired crossover formation, and increased cell apoptosis as well (Figure 6). It is likely that this cilium may regulate DSB repair through recruiting recombinases such as Rad51 to the DSB sites to ensure crossover formation, whereas it is not essential for the processing of meiosis (Figure 6). Actually, both mature eggs and spermatozoa could be still produced in the absence of germ cell-specific cilia, albeit with cell apoptosis and flagellar defects in the spermatozoa.
Figure 6.
Model of spermatocyte cilia during meiosis. During the prophase of meiosis I, cilia are present in the spermatocyte from leptotene to pachytene stages. This cilium participates in the regulation of HR. In the absence of cilia, less Rad51 proteins are recruited to the DSBs, which compromises the efficiency of DSB repair, causing accumulation of γ-H2A.X signals and decreased level of crossover formation. wt, wild type.
Although we do not know yet the detailed mechanisms of these cilia in the regulation of meiotic recombination, our data provide an important message that meiotic recombination can be regulated by signals outside the nucleus. Cilia can act as a cellular antenna to sense extracellular signals including Hedgehog, Notch/Delta, and canonical and non-canonical Wnt signals (Ishikawa and Marshall, 2011; Anvarian et al., 2019). Thus, the spermatocyte cilia may function as signal hubs to sense these signals outside the cell to help recruit DSB repair components, such as Rad51 and Dmc1, to the DSB sites and ensure the HR processing. On the other hand, these cilia are nucleated by the basal body, from which the bouquet microtubule originates. It is possible that the cilia may supply forces for dynein/kinesin motors to support telomere movement and homolog pairing.
During the preparation of this paper, we noticed a similar paper was published recently (Mytlis et al., 2022). This study reported the presence of zygotene cilium in the oocyte, which they think may regulate chromosomal paring, germ cell morphogenesis, and ovarian development and fertility. It is most likely that the zygotene cilium is similar to the spermatocyte cilium discussed here. Of note, all the mutants generated from their study contain mutations of genes related to centrosome (basal body) (cep290, cc2d2a) or Hedgehog signaling (kif7) (Mytlis et al., 2022). These genes have been shown earlier to participate in the regulation of cell cycle, which may also affect meiosis (Tsang et al., 2008; He et al., 2014; Veleri et al., 2014; Prosser et al., 2022). Moreover, all the mutants are global knockout in their study, which may lead to defects in other cells such as granulosa cells and defects in these cells may also contribute to the morphogenesis defects of germ cells. In contrast, we generated PGC-specific conditional knockout mutants and eliminated the possibility of cilia defects in other tissues. Our data suggest that the primary spermatocyte cilia participate in the regulation of meiotic recombination and its defects can delay, but not inhibit, the meiosis process. Indeed, mature eggs can still be produced in our mutants, in sharp contrast to the oogenesis defects observed in either cep290 or cc2d2a mutants. Similarly, zebrafish eggs can also be produced normally in the absence of ift88 in the PGCs (Huang and Schier, 2009; Borovina and Ciruna, 2013).
In summary, we reported here the presence of cilia in both primary spermatocytes and oocytes, whose defects may affect HR during meiosis. Further experiments are needed to dissect the precise signaling molecules underlying this intriguing organelle.
Materials and methods
Ethics statement
All zebrafish studies were conducted according to standard animal guidelines and approved by the Animal Care Committee of Ocean University of China and the Animal Care Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
Zebrafish strains
All zebrafish strains were maintained at 14-h light/10-h dark cycle at 28.5̊C. The plasmid for generating Tg(kop:cas9-p2a-egfp-UTRnanos3) transgenic fish was generated by replacing the egfp sequence in the pTol2(kop:egfp-UTRnanos3) construct (Xiong et al., 2013), with an in frame sequence, cas9-p2A-egfp, which encodes zebrafish codon optimized Cas9, p2A peptide, and EGFP. The plasmid for Tg(U6:3×sgRNA-kif3a) transgene was generated by Golden Gate cloning as previously described (Yin et al., 2016). Three sgRNAs targeting kif3a (sgRNA1, GGGAAAACGTTCACTATGGA; sgRNA2, GGCCAAACTCGACATGGAGG; sgRNA3, GAGGAGGTGAGAGATCTGTT) were each driven by a distinct zebrafish U6 promoter. The Tg(U6:3×sgRNA-kif3a) transgenic fish were further crossed to the Tg(kop:cas9-p2a-egfp-UTRnanos3) transgenic line to get the double transgenic fish. The pTol2-hsp70l:ift88-egfp plasmid was constructed through Multisite Gateway cloning and injected into the homozygous ovl(ift88) mutants at 1-cell stage. The injected embryos were heat-induced for 3 h every day until adulthood for further analysis.
Microinjection of sgRNA
For microinjection-based PGC-targeted gene knockout experiment, sgRNAs against tcf7l1a and pou5f3 were GGAGGAGGAGGTGATGACCT and GGGTGAACTACTACACGCCA, respectively, as previously described (Zhang et al., 2020). The sgRNAs were injected with a dosage of 80 pg per embryo into the Tg(kop:cas9-p2a-egfp-UTRnanos3) transgenic embryo at 1-cell stage.
Chromosome spreading and immunostaining
Chromosome spreading steps were similar to previously published (Xie et al., 2020). Briefly, the dissected testis was first incubated in 50 μl 1× phosphate-buffered saline (PBS) for 5 min, and then transferred into 300 μl hypotonic solution (100 μl 1× PBS plus 200 μl double distilled water) for 25 min on an adhesive slide. After fixation with 4% paraformaldehyde (PFA) for 5 min, the cells were washed three times with PBS, blocked with blocking solution (10% goat serum, 3% bovine serum albumin, and 0.05% Triton X-100) for 30 min, then followed by regular antibody staining.
Immunofluorescence and TUNEL assay
Zebrafish testes were first dissected and fixed in 4% PFA overnight at 4̊C. After three times wash with PBS for 10 min each, the testes were infiltrated in 30% sucrose overnight at 4̊C, and then embedded with tissue freezing medium (OCT). Cryosectioning was performed using LEICA cryostat (CM1860). TUNEL assay was performed using the in situ cell death detection kit (Roche) according to standard protocols from the manufacturer. Images were collected with a Leica Sp8 confocal microscope.
To visualize cilia in the primary ovaries, zebrafish females ∼1.5-month-old were euthanized with ice-cold water, and ovaries were dissected and transferred into 4% PFA overnight at 4̊C. After three times wash with PBST (1× PBS + 0.1% Tween20), the ovaries were further blocked with blocking solution (0.3% Triton X-100, 10% fetal bovine serum, and 1× PBS) for 1.5 h at room temperature. Antibody staining steps were similar to those of testes.
For immunofluorescence, the following antibodies were used: mouse anti-acetylated tubulin (Sigma, T6793), rabbit anti-SYCP3 (Abcam, ab150292), rabbit anti-γ-H2A.X (Genetex, GTX127342), rabbit anti-RAD51 (THERMO, PA5-27195), mouse anti-MLH1 (Abcam, ab14206), mouse anti-gamma tubulin (Sigma, T5326), rabbit anti-acetylated tubulin (Cell Signaling Technology, 5335S), mouse anti-glutaminated tubulin (AdipoGen, GT335), mouse anti-monoglycylated tubulin (Sigma, MABS277), and mouse anti-polyglycylated tubulin (Sigma, MABS276).
Immunohistochemical staining
Zebrafish testes were dissected and fixed in 4% PFA overnight at 4̊C. After gradual dehydration through 30%, 50%, 75%, 95%, and 100% ethanol, the testes were embedded in JB4 embedding medium (Polysciences Inc.). Transverse sections through the testes were collected using Leica RM2235 microtome and stained with hematoxylin and eosin. Images were taken using a Leica DM2500 microscope.
Ultrastructural analysis
Zebrafish testis were dissected and fixed overnight in 2.5% glutaraldehyde. The testis was washed three times in PBS, for 10 min each, and fixed again in 1% osmium tetroxide. After that, the testis was washed, gradually dehydrated to 100% acetone, and then embedded in Epon812 resin. Ultrathin sections were collected and stained with uranyl acetate and lead citrate. The sections were observed on a transmission electron microscope (JEM-1200EX) and imaged with Olympus Soft Imaging Solutions.
Statistical analysis
All the confocal images were captured using Leica Sp8 confocal microscope. To compare the fluorescence signals between control and mutants, all the images were collected at the same parameters within each experiment. Image J software was used to measure the mean fluorescence intensity per spermatocyst in staining of Rad51 and γ-H2A.X. Spermatocyst refers to a cluster of primary spermatocytes that have similar protein localization in the immunofluorescence of Rad51 and γ-H2A.X. Then, the average fluorescence intensity of wild type was set as 100%, the relative fluorescence intensity of kif3a mutant was calculated by Excel software, and the graph was made by GraphPad Prism 8.0 software. All experiments were repeated for at least three times. Statistical analysis was performed using ImageJ, Microsoft Excel, or GraphPad Prism 8.0 software. Statistical significance was evaluated by means of the two-tailed Student's t-test for unpaired data. All data were presented as mean ± SD as indicated in the figure legends. A value of P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank Dr Feng Xiong at the China Zebrafish Resource Center for generating the pTol2(kop:cas9-p2a-egfp-UTRnanos3) construct. We thank Drs Liangran Zhang, Wei Li, and Ming Shao for providing DNA constructs and reagents. We also thank Drs Miao Tian and Tao Qiu for their help during the preparation of this manuscript.
Contributor Information
Haibo Xie, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China; Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266003, China; Sars-Fang Centre, Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China.
Xiaosi Wang, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Innovation Academy for Seed Design, Hubei Hongshan Laboratory, Chinese Academy of Sciences, Wuhan 430072, China.
Minjun Jin, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China; Sars-Fang Centre, Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China.
Lanqin Li, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China; Sars-Fang Centre, Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China.
Junwen Zhu, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Innovation Academy for Seed Design, Hubei Hongshan Laboratory, Chinese Academy of Sciences, Wuhan 430072, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yunsi Kang, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China; Sars-Fang Centre, Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China.
Zhe Chen, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China.
Yonghua Sun, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Innovation Academy for Seed Design, Hubei Hongshan Laboratory, Chinese Academy of Sciences, Wuhan 430072, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Chengtian Zhao, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China; Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266003, China; Sars-Fang Centre, Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31991194, 32025037, and 32125015), the Fundamental Research Funds for Central Universities (201941004), and the State Key Laboratory of Freshwater Ecology and Biotechnology (2019FBZ05).
Conflict of interest: none declared.
Author contributions: C.Z., Y.S., and H.X. designed the project; H.X. discovered the spermatocyte cilia and performed most of the initial analysis; H.X., L.L., and Y.K. performed data analysis on kif3a mutants; X.W. and J.Z. performed the screening and efficiency test of the Tg(kop:cas9-UTRnanos3) transgene; M.J. and Z.C. performed analysis on the ift88 mutants; C.Z., Y.S., and H.X. analyzed the data and wrote the manuscript.
References
- Anvarian Z., Mykytyn K., Mukhopadhyay S.et al. (2019). Cellular signaling by primary cilia in development, organ function, and disease. Nat. Rev. Nephrol. 15, 199–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove B.W., Yost H.J. (2006). The roles of cilia in developmental disorders and disease. Development 133, 4131–4143. [DOI] [PubMed] [Google Scholar]
- Bishop D.K. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L.et al. (1992). DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Blokhina Y.P., Nguyen A.D., Draper B.W.et al. (2019). The telomere bouquet is a hub where meiotic double-strand breaks, synapsis, and stable homolog juxtaposition are coordinated in the zebrafish, Danio rerio. PLoS Genet. 15, e1007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., de Massy B. (2013). Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 23, 147–155. [DOI] [PubMed] [Google Scholar]
- Borovina A., Ciruna B. (2013). IFT88 plays a cilia- and PCP-independent role in controlling oriented cell divisions during vertebrate embryonic development. Cell Rep. 5, 37–43. [DOI] [PubMed] [Google Scholar]
- Burke B. (2018). LINC complexes as regulators of meiosis. Curr. Opin. Cell Biol. 52, 22–29. [DOI] [PubMed] [Google Scholar]
- Dai J., Sanchez A., Adam C.et al. (2021). Molecular basis of the dual role of the Mlh1–Mlh3 endonuclease in MMR and in meiotic crossover formation. Proc. Natl Acad. Sci. USA 118, e2022704118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkouby Y.M., Jamieson-Lucy A., Mullins M.C. (2016). Oocyte polarization is coupled to the chromosomal bouquet, a conserved polarized nuclear configuration in meiosis. PLoS Biol. 14, e1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Chen Z., Yang K.et al. (2017). The cytoplasmic tail of rhodopsin triggers rapid rod degeneration in kinesin-2 mutants. J. Biol. Chem. 292, 17375–17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel M.A., Schimenti J.C. (2010). Genetics of mammalian meiosis: regulation, dynamics, and impact on fertility. Nat. Rev. Genet. 11, 124–136. [DOI] [PubMed] [Google Scholar]
- He M., Subramanian R., Bangs F.et al. (2014). The kinesin-4 protein Kif7 regulates mammalian Hedgehog signaling by organizing the cilium tip compartment. Nat. Cell Biol. 16, 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Benzing T., Katsanis N. (2011). Ciliopathies. N. Engl. J. Med. 364, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Schier A.F. (2009). Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development 136, 3089–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. (2015). Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W.F. (2011). Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Kim C.-H., Oda T., Itoh M.et al. (2000). Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407, 913–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Drivas T.G., Blobel G. (2008). A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. (2016). Conservation and variability of meiosis across the eukaryotes. Annu. Rev. Genet. 50, 293–316. [DOI] [PubMed] [Google Scholar]
- Mytlis A., Kumar V., Qiu T.et al. (2022). Control of meiotic chromosomal bouquet and germ cell morphogenesis by the zygotene cilium. Science 376, eabh3104. [DOI] [PubMed] [Google Scholar]
- Neale M.J., Pan J., Keeney S. (2005). Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558. [DOI] [PubMed] [Google Scholar]
- Prosser S.L., Tkach J., Gheiratmand L.et al. (2022). Aggresome assembly at the centrosome is driven by CP110–CEP97–CEP290 and centriolar satellites. Nat. Cell Biol. 24, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim G., Brand M. (2006). Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development 133, 2757–2770. [DOI] [PubMed] [Google Scholar]
- Reiter J.F., Leroux M.R. (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M.et al. (2009). Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E. (2005). Meiosis: organizing microtubule organizers. Curr. Biol. 15, R633–R635. [DOI] [PubMed] [Google Scholar]
- Scherthan H. (2001). A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2, 621–627. [DOI] [PubMed] [Google Scholar]
- Schulz K.N., Harrison M.M. (2019). Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H., Hernandez-Hernandez A., Morimoto A.et al. (2015). MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163, 1252–1266. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Ishiguro K., Watanabe Y. (2014). The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 16, 145–156. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. (1992). Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J.F. (2006). The primary cilium as the cell's antenna: signaling at a sensory organelle. Science 313, 629–633. [DOI] [PubMed] [Google Scholar]
- Song Z., Zhang X., Jia S.et al. (2016). Zebrafish as a model for human ciliopathies. J. Genet. Genomics 43, 107–120. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J.W. (1991). Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Tsang W.Y., Bossard C., Khanna H.et al. (2008). CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell 15, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa M., Malicki J. (2004). Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 42, 703–716. [DOI] [PubMed] [Google Scholar]
- Veleri S., Manjunath S.H., Fariss R.N.et al. (2014). Ciliopathy-associated gene Cc2d2a promotes assembly of subdistal appendages on the mother centriole during cilia biogenesis. Nat. Commun. 5, 4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Kang Y., Wang S.et al. (2020). E2f5 is a versatile transcriptional activator required for spermatogenesis and multiciliated cell differentiation in zebrafish. PLoS Genet. 16, e1008655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Wei Z.Q., Zhu Z.Y.et al. (2013). Targeted expression in zebrafish primordial germ cells by Cre/loxP and Gal4/UAS systems. Mar. Biotechnol. 15, 526–539. [DOI] [PubMed] [Google Scholar]
- Yin L., Maddison L.A., Chen W. (2016). Multiplex conditional mutagenesis in zebrafish using the CRISPR/Cas system. Methods Cell Biol. 135, 3–17. [DOI] [PubMed] [Google Scholar]
- Zelkowski M., Olson M.A., Wang M.et al. (2019). Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35, 359–370. [DOI] [PubMed] [Google Scholar]
- Zhang F., Li X., He M.et al. (2020). Efficient generation of zebrafish maternal-zygotic mutants through transplantation of ectopically induced and Cas9/gRNA targeted primordial germ cells. J. Genet. Genomics 47, 37–47. [DOI] [PubMed] [Google Scholar]
- Zhao C., Omori Y., Brodowska K.et al. (2012). Kinesin-2 family in vertebrate ciliogenesis. Proc. Natl Acad. Sci. USA 109, 2388–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Gao F., Gao S.et al. (2021). Biodiversity-based development and evolution: the emerging research systems in model and non-model organisms. Sci. China Life Sci. 64, 1236–1280. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (2015). Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.