Abstract
Objective
To demonstrate that deep learning (DL) methods can produce robust prediction of gene expression profile (GEP) in uveal melanoma (UM) based on digital cytopathology images.
Design
Evaluation of a diagnostic test or technology.
Subjects, Participants, and Controls
Deidentified smeared cytology slides stained with hematoxylin and eosin obtained from a fine needle aspirated from UM.
Methods
Digital whole-slide images were generated by fine-needle aspiration biopsies of UM tumors that underwent GEP testing. A multistage DL system was developed with automatic region-of-interest (ROI) extraction from digital cytopathology images, an attention-based neural network, ROI feature aggregation, and slide-level data augmentation.
Main Outcome Measures
The ability of our DL system in predicting GEP on a slide (patient) level. Data were partitioned at the patient level (73% training; 27% testing).
Results
In total, our study included 89 whole-slide images from 82 patients and 121 388 unique ROIs. The testing set included 24 slides from 24 patients (12 class 1 tumors; 12 class 2 tumors; 1 slide per patient). Our DL system for GEP prediction achieved an area under the receiver operating characteristic curve of 0.944, an accuracy of 91.7%, a sensitivity of 91.7%, and a specificity of 91.7% on a slide-level analysis. The incorporation of slide-level feature aggregation and data augmentation produced a more predictive DL model (P = 0.0031).
Conclusions
Our current work established a complete pipeline for GEP prediction in UM tumors: from automatic ROI extraction from digital cytopathology whole-slide images to slide-level predictions. Our DL system demonstrated robust performance and, if validated prospectively, could serve as an image-based alternative to GEP testing.
Keywords: Artificial intelligence, Cytopathology, Deep learning, Gene expression profile, Uveal melanoma
Abbreviations and Acronyms: ANN, artificial neural network; CAM, class activation map; DA, data augmentation; DL, deep learning; FA, feature aggregation; FNAB, fine-needle aspiration biopsy; GEP, gene expression profile; ML, machine learning; ROI, region-of-interest; UM, uveal melanoma
The recent advent of machine learning (ML) technology has catalyzed the development of novel automatic tools that analyze data to support clinical decision-making. Most ML works focus on clinical tasks routinely performed by human experts, for example, detecting malignant cells and subtyping cancer cells (e.g., lung adenocarcinoma vs. small cell vs. squamous cell carcinoma)1 in pathology whole-slide images. However, tasks that no human experts are capable of are also important in clinical practice, and there exists a high demand for automatic systems for such tasks to further advance the field of medicine. One such “superhuman” task that can provide actionable insights is cancer genetic subtyping from digital cytopathology whole-slide images, which we consider here.
Uveal melanoma (UM), the most common primary intraocular tumor in adults,2 can be divided into 2 classes based on its gene expression profile (GEP) as determined by the commercially available DecisionDx-UM test (Castle Biosciences). There is a stark contrast in long-term survival between the 2 classes. The reported 92-month survival probability of class 1 patients is 95%, in contrast to 31% among class 2 patients.3,4 This GEP test has been validated in numerous prospective clinical trials and has been found to be the most robust prognostication test, independent of other clinicopathology parameters. The information provided by this GEP test could guide clinical management after the local treatment of the tumor because patients at a high risk of death typically undergo more frequent systemic radiologic imaging to monitor for metastasis. However, this GEP test has several drawbacks, including high cost and unavailability to patients outside the United States. Therefore, an ML algorithm that can directly predict the GEP of a tumor from pathology images will be of immense value to patients with UM who do not have access to GEP testing, especially because it is impossible for a human pathologist to directly predict GEP from pathology images for clinical use.
The central premise of our study is that a tumor’s behavior and metastatic potential are dictated by its underlying genetic makeup. There is evidence in the broader oncology literature, suggesting that genetics affect cell morphology that can, in turn, be captured in digital pathology images,5 which can, in turn, be analyzed by ML techniques, such as deep learning (DL). In our pilot study6 that involved only 20 patients, we demonstrated the feasibility of using DL to predict GEP of UM tumors from digital cytopathology whole-slide images. Each whole-slide image contained thousands of smaller unique region-of-interests (ROIs) that were used for model input. However, our pilot study had several major limitations, including low-patient diversity, the use of leave-one-out cross validations, and the labor-intensive process of manual ROI extraction. As an extension, we conducted the current study to address the limitations of our pilot study, for example, by increasing the sample size by more than fourfold and using the more conventional 70/30 train/test data split during the model evaluation. In addition, we incorporated several technical innovations, such as automatic ROI extraction, dual-attention–based neural network, saliency regression, slide-level feature aggregation (FA), and slide-level data augmentation (DA), to address the unique challenges of DL-based analysis of digital cytopathology images (Fig 1).
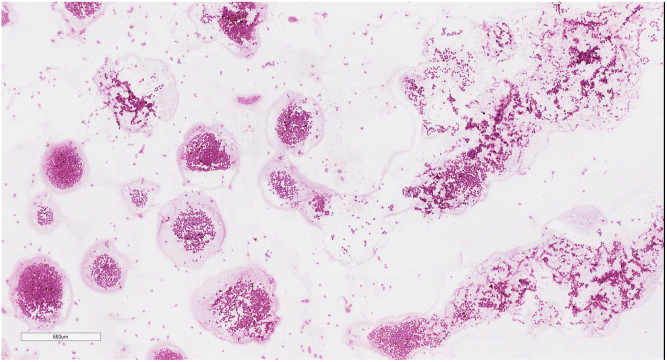
Figure 1.
Sample digital cytopathology whole-slide image at ×3.5 magnification generated by fine-needle aspiration biopsy of a posterior uveal melanoma tumor. Note the presence of variable distribution of smeared tumor cells, empty spaces, and debris.
Methods
We conducted a retrospective study involving deidentified digital pathology whole-slide images from 82 consecutive patients with UM. This study adheres to the Declaration of Helsinki and was approved by the Johns Hopkins University Institutional Review Board as an exempt-from-consent study because the slides and patient information were deidentified.
Data Preparation
Before the treatment of each UM tumor with plaque brachytherapy or enucleation, fine-needle aspiration biopsy (FNAB) of the tumor was performed. Each tumor underwent 2 biopsies: the first biopsy was taken at the tumor apex for GEP testing, and the second biopsy was taken near the tumor apex for cytopathology. Blood specimens were not excluded. These FNABs were performed, using 27-gauge needles for the transvitreal approach and 25-gauge needles for the transscleral approach. The cytology specimen was flushed on a standard pathology glass slide, smeared, and stained with hematoxylin and eosin. The specimen submitted for GEP testing was flushed into a tube containing an extraction buffer and submitted for DecisionDx-UM testing. Whole-slide scanning was performed for each cytology slide at a magnification of ×40, using the Aperio ScanScope AT machine. The data were split into training and testing sets at the patient level. Our training set included 65 slides from 58 patients (28 class 1 tumors; 30 class 2 tumors). Our testing set included 24 slides from 24 patients (12 class 1 tumors; 12 class 2 tumors; 1 slide per patient). Of the class 1 tumors, 22 were class 1A, and 18 were class 1B. Next, we applied a human-interactive computationally assisted tool to automatically extract high-quality ROIs from each slide, although we omitted the boundary refinement step to render the entire process fully automatic. The tool, its use, and adequacy were detailed in a previous report.7 The size of each ROI was 256 × 256 pixels, and in total, 121 388 ROIs were extracted from all slides.
DL System Development
Figure 2 is a schematic representation of the overall DL system development, which involves attention-based feature extraction at the ROI level, FA at the slide level, and GEP prediction at the slide level.
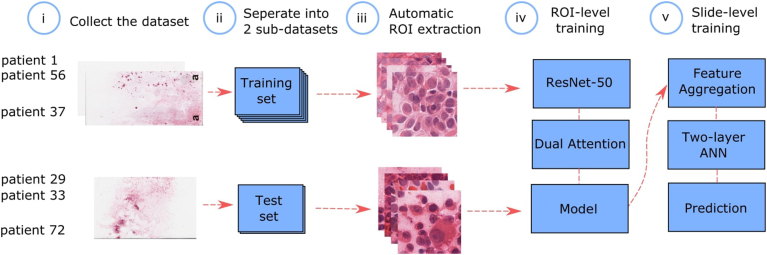
Figure 2.
Schematic representation of our DL system development. (i) Cytopathology slides obtained from fine-needle aspiration biopsies (FNABs) of UM tumors were digitized by whole-slide scanning at ×40 magnification. (ii) Slides were partitioned on a patient level for training and testing. (iii) Automatic high-quality ROI extraction. (iv) ResNet-50 architecture and dual-attention mechanism were used for training at an ROI level. (v) Downsampled, pixelwise features were extracted from every ROI and aggregated as slide-level features that were used as input to 2, 2-layer AANs to directly produce slide-level GEP prediction. ANN = artificial neural network; DL = deep learning; GEP = gene expression profile; ROI = region-of-interest; UM = uveal melanoma.
Attention-Based Neural Networks
After the ROIs were extracted from the whole-slide images, we annotated the ROIs using slide-level labels. That is, the GEP classification label for a particular slide (tumor) was propagated to all ROIs generated from that slide. We trained a ResNet-508 network for the binary classification task of differentiating between GEP classes 1 and 2 for every ROI with the slide-level annotations. A dual-attention mechanism9 that contained both location attention and channel attention was applied directly to the output of the final residual block of ResNet-50. This was done to encourage the network to focus more on salient ROIs, for example, high-quality ROIs that contain multiple in-focus UM cells. Finally Log–Sum–Exp pooling was applied to obtain ROI-level predictions, which were supervised by the corresponding slide-level labels.
Attention-Based FA at the Slide Level
Instead of generating slide-level predictions from ROI-level predictions directly, we aimed to take into account richer information, compared with only ROI-level predictions, to make slide-level decisions. First, we aggregated ROI feature maps into ROI-level features. The output features of the dual-attention mechanism, Fdual, were used to generate the class activation maps (CAMs) {Mk}{k = 1,2}, which were treated as the saliency of each pixel to a specific class. For each slide, 2 CAMs were generated, 1 for each GEP class. Because some ROIs within each slide were less informative, the 2 CAMs were not complementary. Finally, ROI-level features were aggregated by weighted averaging of Fdual using {Mk}{k = 1,2}.
in which i, j was the spatial index and k = 1, 2 was the GEP class index. ∗ was the spatial elementwise multiplication.
Then, we further aggregated ROI-level features into slide-level features by weighted averaging of using .
in which n = 1, N was the ROI index for every slide.
Slide-Level GEP Prediction
The last stage of our DL system involved 2, 2-layer artificial neural networks (ANNs), whose predictions were combined to directly output slide-level GEP classification. Given that there were only 65 slides in our training set, we performed DA to increase the data diversity for this stage of the training. Synthetic whole-slide images were generated from real whole-slide images (in the training set). We randomly chose Nrand ROIs from a particular real slide and created aggregated slide-level features in a similar way as mentioned above for each corresponding synthetic slide, in which 100 <= Nrand <= N, and N was the actual total number of extracted ROIs from that particular real slide.
After slide-level features Fslide were generated, they were used to train a 2-level ANN for the binary classification task of predicting GEP status on a slide level. Because the features were generated from CAMs for each GEP class, 2 set slide-level features for each whole-slide image. Furthermore, because the 2 GEP classes had distinct cell representation, features were encoded and decoded separately for each GEP class. As a result, we trained 2, 2-layer ANNs, each with the same structure but different parameters, for each class. Each ANN was a 2-layer neural network, in which the first fully connected layer was followed by batch normalization and rectified linear unit activation function. A dropout layer was also inserted right before the final fully connected layer with a dropout rate of 0.5. The output of each ANN was yi, in which i = 1,2 was the GEP class index. Cross-entropy loss was used in training, and the softmax function was applied to {yi}{i=1,2} to predict the GEP status for each slide. The output from each of the 2 ANNs was concatenated to provide a single prediction at the slide level.
Image Appearance Distribution
To visualize the relationship between cytology characteristics and GEP prediction by our DL system, we used ROIs extracted from the testing slides and t-distributed stochastic neighbor embedding (t-SNE)10 (a nonlinear dimensionality reduction technique well suited for embedding high-dimensional data for visualization in a low-dimensional space), downsampled pixel features, and generated a distribution map based on image appearance. This experiment was inspired by the historical belief that low- and high-risk tumors differ cytologically. After training a ResNet-50 (parameters initialized by ImageNet pretrained weights) as the first stage attention model backbone, we clustered subareas within ROIs by corresponding ResNet-50 output features (Fdual) to visualize model behavior using t-SNE. By treating the 2-channel output of the attention module as the CAMs for each GEP class, all subareas were split into class-1 informative, class-2 informative, and noninformative subareas. Only class-1 informative and class-2 informative subareas were clustered and colored differently in the visualization (Fig 3). All other areas were treated as noninformative areas, where no strong signals for GEP class 1 or 2 were present.
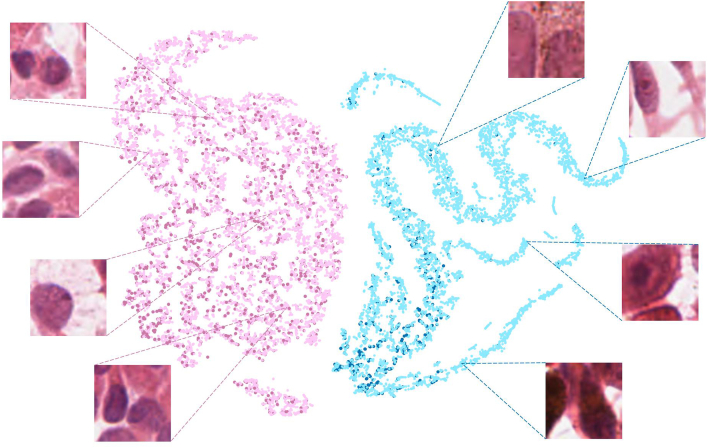
Figure 3.
t-distributed stochastic neighbor embedding (t-SNE)10 with downsampled pixel features. This is a composite image with 4 colors representing 4 populations of cells: light pink, maroon, teal, and blue. Light pink represents GEP class 1 samples that are correctly predicted as GEP class 1. Maroon represents GEP class 2 samples that are incorrectly predicted as GEP class 1. Teal represents GEP class 2 samples that are correctly predicted as GEP class 2. Blue represents GEP class 1 samples that are incorrectly predicted as GEP class 2. Note that the samples predicted to be GEP class 1 (light pink/maroon) typically contain less-aggressive–appearing UM cells, whereas samples predicted to be GEP class 2 (teal/blue) typically contain more-aggressive–appearing UM cells. GEP = gene expression profile; UM = uveal melanoma.
Results
In total, our study included 89 whole-slide images from 82 patients and 121 388 unique ROIs (image tiles). Of the 82 patients, 32 were women, and the mean age was 62.2 years at the time of diagnosis. Our training set included 65 slides from 58 patients. Of the 58 tumors, there were 28 class 1 tumors, the mean thickness was 6.3 mm (range, 0.7–16.2 mm), and the mean largest basal diameter was 12.1 mm (range, 2.2–20 mm). Of the 58 tumors, 8, 38, and 12 were small, medium, and large tumors by the COMS standard, respectively. Our testing set included 24 slides from 24 patients (12 class 1 tumors; 12 class 2 tumors; 1 slide per patient). Of the 24 tumors, the mean thickness was 6.0 mm (range, 1.0–11.5 mm), and the mean largest basal diameter was 12.6 mm (range, 3.5–18 mm). Of the 24 tumors, 2, 15, and 7 were small, medium, and large tumors by the COMS standard, respectively. Our DL system for GEP status prediction achieved an area under the receiver operating characteristic curve of 0.944, an accuracy of 91.7%, a sensitivity of 91.7%, and a specificity of 91.7% on a patient-level analysis. The accuracy, sensitivity, and specificity numbers were identical by chance because there was 1 false-positive and 1 false-negative during testing.
In addition to the main outcome reported above, we analyzed the contributions of 2 of the core modules: slide-level FA and slide-level DA. In the FA module, we aggregated ROI features based on CAMs11 to generate slide-level features. We then trained a 2-layer ANN to predict GEP class on the slide level based on slide-level features. In the DA module, we generated 100 sets of synthetic slide-level features for each class to enrich the training set diversity for the 2-layer ANN. Our “basic” model did not include the FA or DA module and generated slide-level predictions by simple averaging of ROI predictions. Our “full” model included both the FA and DA modules. We performed 10-fold cross-validation for our basic and full models and compared the performance using a nonparametric test for the area under the curve (AUC)12 and the StAR software implementation.13 The basic model achieved an average AUC of 0.800, the full model achieved an average AUC of 0.873, and the difference was statistically significant (P = 0.0031).
In our image appearance distribution experiment (Fig 3) that was performed to visualize the relationship between cytology characteristics and GEP prediction by our DL system, 94.6% of the subareas generated from GEP class 1 slides were correctly classified, and 82.3% of the subareas generated from GEP class 2 slides were correctly classified.
Discussion
We have developed a DL system that is capable of robust GEP prediction based on digital cytopathology whole-slide images from UM tumor cell aspirates. Our DL system achieved an AUC of 0.944 and an accuracy of 91.7% on a slide-level analysis. Gene expression profile testing is the most robust survival predictor in UM, independent of other clinicopathologic parameters; however, the commercially available GEP test is only available in the United States, requires specialized storage and transport, and has a turnover time of days. Our proposed methods, if validated prospectively, could serve as an image-based alternative to GEP testing. In addition, our technical pipeline is able to analyze digital cytopathology images sent remotely and can produce results within hours.
Similar to the underlying scientific premise of our current study, several studies have attempted to predict genetic or molecular information directly from digital histopathology images, which, in contrast to cytopathology images, are more amenable to DL analyses because of the preservation of tissue architecture. For example, Chen et al14 predicted 4 common prognostic gene mutations in hepatocellular carcinoma, with an AUC range of 0.71 to 0.89. Coudray et al15 predicted 6 commonly mutated genes in lung adenocarcinoma, with an AUC range of 0.73 to 0.86. Kather et al16 predicted microsatellite instability in gastrointestinal cancer, with an AUC of 0.84. Although it is difficult to compare results across studies given the difference in diseases and datasets, our DL system achieved an AUC of 0.944, suggesting a robust performance when compared with other published studies with similar methodologies.
Currently, most UM tumors do not require enucleation for the treatment. Instead, FNABs are frequently performed to either confirm the diagnosis or obtain prognostication. Hence, cytopathology images, instead of histopathology images, are the predominant form of pathology images generated in the management of UM. Unlike histopathology images, the tissue architecture is not preserved in cytopathology images, and the unpredictable nature of cell smearing in cytopathology slides creates a myriad of qualitative issues, such as uneven cell distribution, cell stacking, and the presence of cell aspirate medium and debris. These qualitative issues, in turn, render DL analysis difficult. To address these challenges, we applied several technical innovations that were not present in our pilot study.
First, we developed a fully automatic method to perform large-scale, efficient extraction of high-quality ROIs from digital whole-slide images that rendered the current study feasible in the first place. Extraction of high-quality ROIs was performed manually in our pilot study to ensure the exclusion of unusable image tiles, for example, image tiles with only empty space, out-of-focus cells, and significant artifacts. For comparison, 26 351 unique ROIs were extracted over weeks in our pilot study, whereas 121 388 unique ROIs were extracted over hours in our current study. Second, to account for the variable nature of cell smearing and the fact that some regions in a cytopathology whole-slide image were more informative than the others, we trained our system to regress the saliency, or relevance, of specific image regions for GEP prediction. That is, not only did we trained our system to learn from the “trees” (individual ROI), but we also trained our system to learn from the “forest” (clusters of salient ROIs). Third, we demonstrated that the incorporation of more sophisticated techniques—slide-level FA and slide-level DA—further improved the accuracy of our system on a patient (slide) level.
Our technological innovations also have implications beyond UM. Published DL studies using digital pathology images typically made slide-level (patient-level) predictions by simple averaging of14,15,17 or majority voting of15,16 ROI-level predictions and implying that all ROIs carry the same amount of information. This is not ideal for cytopathology because the high variability in quality within each cytopathology slide image inevitably renders some ROIs less informative or important for classification purposes. Our DL system took into account both intra-ROI and inter-ROI variations by devising an attention mechanism to score the saliency of each pixel within every ROI and the relevance of each ROI (considered in its entirety). This attention-scoring technique, combined with the aggregation of ROI-level features into slide-level features, is a solution specifically for the high variability seen in digital cytopathology images, enables slide-level predictions to focus more on highly informative areas, and can be applied to other diseases beyond UM.
The image appearance distribution map (Fig 3) allowed us to visualize the relationship between cytology appearance and GEP prediction and to gain insights into the biologic rationale behind the predictions made by our DL system. We noticed that some “GEP class 1” samples that were incorrectly predicted as “GEP class 2” contained more atypical epithelioid cells that had an aggressive phenotype. Similarly, some “GEP class 2” samples that were incorrectly predicted as “GEP class 1” contained more uniform-appearing cells with small nuclei and a less-aggressive phenotype. This observation is consistent with the fact that both GEP class 1 and 2 tumors can be cytologically heterogeneous. In fact, GEP class 2 tumors are more heterogeneous cytologically, which could explain the lower accuracy on a subarea/ROI level in our class 2 samples. Given that GEP class 1 tumors typically contain a majority of less-aggressive tumor cells and vice versa for class 2 tumors, we hypothesize that our algorithm learned to equate “GEP class 1” with cytologically less-aggressive tumors cells (e.g., spindle-shaped cells) and equate “GEP class 2” with cytologically more aggressive tumors cells (e.g., epithelioid cells).
In conclusion, our current work established a complete pipeline for GEP prediction in UM tumors: from ROI extraction to slide-level predictions. Briefly, we applied an automatic technique to extract high-quality ROIs in each digital cytopathology whole-slide image. Because of the sparsity and unpredictability of high-quality information in cytopathology images, we introduced an attention mechanism to help the network focus on informative regions and proposed a novel method for slide-level prediction by aggregating ROI features to preserve as much information as possible for the final decision inference. The significance of our study lies in our groundbreaking effort to successfully address the unique challenges of automatic cytopathology ML analysis. The strength of our study lies in the genetic ground truth that we used to train and test our DL algorithm, given that GEP status is the most robust survival predictor in UM independent of all other clinicopathologic parameters. Very few malignancies have such a strong correlation between specific genetic signatures and actual patient survival, making our algorithm a prime candidate for clinical impact. Our study has several limitations. First, small or thin UM tumors may not be amenable to FNABs that are required for the generation of cytopathology slides in the first place. Also, FNABs could be technically challenging to obtain and, at times, only yield a minuscule amount of cell aspirates, which may be sufficient for GEP testing but not adequate for cell smearing and cytopathology slide generation. In our experience, a minimal yield of ∼ 5 tumor cells is necessary for a diagnostic GEP testing, whereas a minimal yield of ∼ 20 tumors cells is necessary to create a cytopathology slide. In paucicellular specimens, it is possible that our automatic ROI extraction method may fail to extract any ROI, thus necessitating manual extraction of usable ROIs as was performed in our pilot study. Second, our DL algorithm was trained and tested with cellular aspirates prepared with hematoxylin and eosin staining. Cellular aspirations from UM tumors could be prepared with other methods, such as Pap stain. Our algorithm will need to be updated before it can be used to analyze specimens prepared with Pap stain. Third, our study is limited by the relatively low patient diversity; however, UM is inherently a rare disease, and large datasets that contain both genetic and pathology information are not readily available. For example, the frequently cited and used UM dataset, which was collected by the National Cancer Institute as part of the Cancer Genome Atlas, only contains 80 UM tumors.18 By comparison, our study contained tumor samples from 82 patients. Fourth, although DL-based prediction of GEP class 1 versus class 2 could help identify candidate patients for adjuvant systemic therapy or adjusted systemic surveillance protocols, our dataset was not large enough to train a model capable of more fine-grained predictions of class 1A versus class 1B versus class 2.
As the next step, we plan to prospectively validate our algorithm with an external independent dataset and to compare the prognosticative ability of our algorithm with nuclear BAP1 staining. In addition, our current technical pipeline is set up to allow for straightforward incorporation of additional inputs, such as patient age, tumor basal diameter, tumor maximum height, and tumor location within the eye. These variables are known to carry prognostic significance. We plan to use the current technical pipeline to develop an ensemble ML model that incorporates multimodal data and uses actual patient survival data for training and testing.
Manuscript no. XOPS-D-22-00167R1.
Footnotes
Disclosures:
All authors have completed and submitted the ICMJE disclosures form.
The authors have made the following disclosures:
T.Y.A.L.: Grants – Emerson Collective Cancer Research Fund, Research to Prevent Blindness Unrestricted Grant to Wilmer Eye Institute; Patents – Pending patent: MH2# 0184.0097-PRO.
Z.C.: Grants – Emerson Collective Cancer Research Fund; Patents – Pending patent: MH2# 0184.0097-PRO.
The other authors have no proprietary or commercial interest in any materials discussed in this article.
Supported by Emerson Collective Cancer Research Fund #642653 and Research to Prevent Blindness Unrestricted Grant to Wilmer Eye Institute.
HUMAN SUBJECTS: Human subjects were included in this study. This study adheres to the Declaration of Helsinki and is approved by the Johns Hopkins University Institutional Review Board as an exempt-from-consent study because the slides and patient information were deidentified.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Lui, Chen, Correa, Unberath.
Data collection: Lui, Correa.
Analysis and interpretation: Lui, Chen, Gomez, Correa, Unberath.
Obtained funding: N/A.
Overall responsibility: Lui, Chen, Gomez, Correa, Unberath.
References
- 1.Teramoto A., Tsukamoto T., Kiriyama Y., Fujita H. Automated classification of lung cancer types from cytological images using deep convolutional neural networks. Biomed Res Int. 2017;2017 doi: 10.1155/2017/4067832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh A.D., Turell M.E., Topham A.K. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Onken M.D., Worley L.A., Ehlers J.P., Harbour J.W. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onken M.D., Worley L.A., Char D.H., et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ash J.T., Darnell G., Munro D., Engelhardt B.E. Joint analysis of expression levels and histological images identifies genes associated with tissue morphology. Nat Commun. 2021;12:1609. doi: 10.1038/s41467-021-21727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu T.Y.A., Zhu H., Chen H., et al. Gene expression profile prediction in uveal melanoma using deep learning: a pilot study for the development of an alternative survival prediction tool. Ophthalmol Retina. 2020;4:1213–1215. doi: 10.1016/j.oret.2020.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Liu T.Y.A., Correa Z., Unberath M. Ophthalmic Medical Image Analysis. Springer International Publishing; 2020. An interactive approach to region of interest selection in cytologic analysis of uveal melanoma based on unsupervised clustering; pp. 114–124. [Google Scholar]
- 8.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 9.Fu J., Liu J., Tian H., et al. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. Dual attention network for scene segmentation; pp. 3146–3154. [Google Scholar]
- 10.Van der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 11.Selvaraju R.R., Cogswell M., Das A., et al. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 12.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 13.Vergara I.A., Norambuena T., Ferrada E., et al. StAR: a simple tool for the statistical comparison of ROC curves. BMC Bioinformatics. 2008;9:265. doi: 10.1186/1471-2105-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M., Zhang B., Topatana W., et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. doi: 10.1038/s41698-020-0120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coudray N., Ocampo P.S., Sakellaropoulos T., et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kather J.N., Pearson A.T., Halama N., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skrede O.J., De Raedt S., Kleppe A., et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 18.Bakhoum M.F., Esmaeli B. Molecular characteristics of uveal melanoma: insights from the Cancer Genome Atlascancer genome atlas (TCGA) project. Cancers. 2019;11:1061. doi: 10.3390/cancers11081061. [DOI] [PMC free article] [PubMed] [Google Scholar]