Abstract
Central nervous system (CNS) tumors are the leading cause of cancer death in pediatric patients. Though these tumors typically require invasive surgical procedures to diagnose, cerebrospinal fluid (CSF) liquid biopsy presents a potential method for rapid and noninvasive detection of markers of CNS malignancy. To characterize molecular biomarkers that can be used in the diagnosis, prognosis, and monitoring of pediatric cancer patients, a literature review was conducted in accordance with PRISMA guidelines. PubMed and EMBASE were searched for the terms biomarkers, liquid biopsy, cerebrospinal fluid, pediatric central nervous system tumor, and their synonyms. Studies including pediatric patients with CSF sampling for tumor evaluation were included. Studies were excluded if they did not have full text or if they were case studies, methodology reports, in languages other than English, or animal studies. Our search revealed 163 articles of which 42 were included. Proteomic, genomic, and small molecule markers associated with CNS tumors were identified for further analysis and development of detection tools.
Keywords: Pediatric brain tumors, biomarkers, cerebrospinal fluid, liquid biopsy
Introduction
Tumors of the central nervous system (CNS) are the leading cause of cancer death among pediatric patients, the second most common oncologic condition in children less than 14-years-old, and the most common condition in adolescents age 15-19 years-old [1]. Pathological diagnosis typically requires invasive approaches such as open surgery or stereotactic biopsy which carry the potential for significant morbidity and are subject to sampling error. Monitoring treatment response with imaging remains problematic since magnetic resonance imaging (MRI) is imperfect, often failing to differentiate tumor progression from treatment response [2]. Furthermore, information regarding longitudinal tumoral molecular changes throughout treatment is limited to repeated biopsies.
The ability to rapidly assess biologic specimens for tumor molecular signatures has been aided by the growth of gene sequencing and other high-throughput biochemical testing. Cerebrospinal fluid (CSF) provides an excellent candidate for “liquid biopsies” as CSF is typically in contact with a CNS tumor and provides access to molecules which are sequestered from the rest of the body by the blood brain barrier [3,4]. Liquid biopsies have potential to to diagnose CNS tumors—perhaps obviating the need for surgery in some cases, identify tumor recurrence, monitor response to treatment, and tailor therapy to genetic changes in throughout the patient's treatment.
Here we provide a systematic review of the use of CSF sampling for biomarkers of pediatric CNS tumors. Broadly, these biomarkers can be split into three groups—proteomic markers, genomic markers, and small molecule markers which provide insight into the metabolome and lipidome of the tumor in question.
Methods
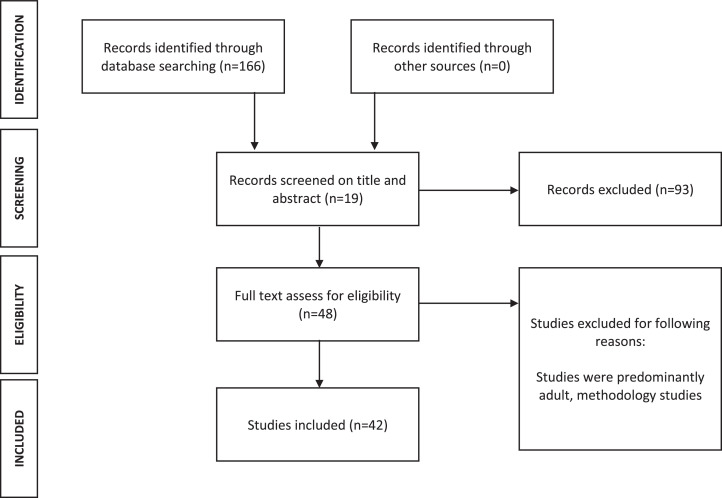
A literature review was conducted according to Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guidelines (Fig. 1). Two independent researchers performed search, selection, data collection, and assessments. PubMed and EMBASE searches were conducted for (“biomarkers” OR “liquid biopsy”) AND (“CSF” OR “cerebrospinal fluid”) AND (“pediatric brain tumor” OR “pediatric CNS tumor” OR “pediatric central nervous system tumor” OR “pediatric glioma” OR “childhood brain tumor” OR “childhood CNS tumor” OR “childhood central nervous system tumor”) in all combinations. Reviews identified in the literature search were checked for references to ensure no studies were missed.
Fig. 1.
Flow diagram demonstrating PRISMA identification, screening, eligibility, and inclusion. 42 studies were included in the final analysis.
Studies including predominantly pediatric patients with CNS tumors who had CSF sampling for biomarkers to diagnose, prognosticate, monitor treatment, and assess for recurrence or tumor evolution over time were included. Studies were excluded if they did not include full text, were methodology reports, were predominantly adult studies, were in languages other than English, were about non-primary CNS malignancies such as leukemia, or were animal studies. Methodology reports were defined as reports that were specific to assay optimization and lacked novel findings. Of note, a single methodology study was identified (Li, et al) and is discussed in the discussion.
Results and review
Our literature search revealed 166 articles. 75 were excluded as reviews, case studies, or non-English. 46 excluded as non-pediatric brain tumor CSF biomarker studies. 42 studies were ultimately included in the review, 26 pertaining to protein biomarkers and the remaining 19 studies evaluating genomic or small molecule markers.
The mean number of patients in each study within the proteomic vs. genomic group was 55 (range: 10–159) vs 34 (range: 4–123), respectively. Since 2020, there has been a trend towards publishing more genomic studies. The majority of studies in the protein category focused on germ cell tumors (GCTs) (36%). The majority of genomic studies focused on midline tumors (35%) including diffuse midline gliomas (DMGs) and diffuse intrinsic pontine gliomas (DIPGs), followed shortly by studies on medulloblastoma (MB) (29%). The majority of proteomic studies focused on one or two proteins (92%). Genomic studies, in contrast, had a wide range of targets and methodology frequently utilizing whole genome sequencing (WGS) (35%), whole exome sequencing (WES), or deep sequencing. The specifics of the review are below.
Genomic biomarkers (Table 1)
Table 1.
Genomic studies.
| Author and Year | Neoplasms | Targets | Techniques | N | Findings |
|---|---|---|---|---|---|
| Vu Han et al, 2014 [27] | AT/RT | SMARCB1 | rtPCR | 7 | ctDNA present in 2/7 patients |
| Wang et al, 2015 [3] | Mixed CNS tumors | TP53, IDH1, TERT, NF2, PIK3R1, PTCH1, PTEN | Targeted sequencing, WES | 35 tumors, 14 pediatric | ctDNA was common in high grade neoplasms |
| Murray et al, 2016 [64] | GCT | miR–371a–3p, miR–372–3p, miR–373–3p, miR–367–3p | rtPCR | 45 samples from 25 patients | miRNA used for GCT diagnosis |
| Huang et al, 2017 [10] | Midline pediatric tumors | H3K27M | Sanger sequencing | 11 | H3K27M and H3G34V were present in tumor and CSF |
| Stallard et al, 2018 [12] | DIPG | H3K27M | ddPCR | 2 DIPG, 2 HGG | H3K27M levels correlated with tumor size, location, treatment |
| Panditharatna et al, 2018 [14] | DMG | H3K27M, ACVR1, PIK3R1, BRAF | ddPCR | 110 samples from 48 DMGs | H3K27M correlated with tumor size |
| Pan et al, 2019 [17] | DIPG, HGG | H3F3, TP53, ATRX, PDGFRA, FAT1, HIST1H3B, PPM1D, IDH1, NF1, PIK3CA, ACVR1 | Deep sequencing | 57 brainstem tumors | Mutations detected by liquid biopsy not present in tissue sample |
| Garcia-Romero et al, 2019 [25] | Mixed CNS tumors | BRAF V600E | ddPCR | 14 samples from 29 patients, 2 with confirmed BRAF V600E | 2 samples with BRAF V600E present |
| Escudero et al, 2020 [18] | MB | Tumor-specific mutations | WES, ddPCR | 13 | Somatic mutations in ctDNA correlated with disease progression |
| Li et al, 2020 [28] | MB | Epigenome | Whole genome bisulfate screening | 4 | Epigenomes determined from ctDNA, CpG islands used monitor treatment |
| Bruzek et al, 2020 [11] | HGG | Histone mutations | Nanopore, ddPCR, NGS | 12 tumor, 6 control | Nanopore sequencing had comparable results to NGS for detection of H3F3A and H3C2 mutations |
| Izquierdo et al, 2021 [26] | DMG, HGG | H3K27M, BRAF V600E | ddPCR | 41 non-brainstem DMG or HGG | ctDNA correlated with tumor progression |
| Liu et al, 2021 [19] | MB | Copy number varients | WGS | 476 samples from 123 patients | CNVs correlated with measurable residual disease |
| Sun et al, 2021 [20] | MB | Tumor-specific mutations | NGS | 58 | ctDNA in CSF correlated with progression |
| Pagès et al, 2022 [21] | Mixed CNS tumors | 46 commonly mutated genes | WGS, deep sequencing | 46 CSF specimens compared to urine and serum (564 specimens total) | Low levels of ctDNA identified in CSF, serum, and urine |
| Miller et al, 2022 [22] | Mixed CNS tumors | 468 cancer-associated genes | NGS | 45 | ctDNA used for diagnosis, treatment monitoring, and tumor monitoring |
| Cantor et al, 2022 [15] | DMG | H3K27M | ddPCR | 24 | H3K27M used for longitudinal tumor monitoring, differentiated pseudoprogression |
| Lee et al, 2022 [31] | MB | Metabolomes, transcriptomes | RNAseq, MS | 40 | Combination of metabolomic and trancriptomic information differentiates MB CSF |
| Majzner et al, 2022 [16] | DMG | H3K27M | Not discussed | 4 | GD2 expressed in H3K27M can be targeted with CAR-T cell therapy for clinical benefit |
Circulating tumor DNA (ctDNA) are 150-200 nucleotide fragments which exist as a small proportion of cell free DNA (cfDNA) that is primarily released when circulating cells undergo apoptosis [5,6]. ctDNA in the blood is frequently overwhelmed by cell-free DNA from other cells which compromise the majority of the circulating DNA mass. The use of CSF to sample ctDNA circumvents this problem, especially in the case of CNS malignancy where it is relatively excluded from the circulation and the blood-brain barrier prevents the circulation of cells which contribute to cfDNA [7]. Next-generation sequencing (NGS) and polymerase chain reaction (PCR) technologies are invaluable tools to detect the presence of genetic targets which exist in low quantities and sequence tumor targets enriched in the CSF [3,8]. Applications of these techniques specific to pediatric CNS malignancy are reviewed below. As many of these studies are small, the details are outlined to a greater extent.
Histone gene mutations
Diffuse midline gliomas (DMGs) occur in the thalamus or brainstem in young children. Their location makes surgical access difficult, and biopsy has potential for high morbidity. Most of these tumors carry a mutation in the histone isoform H3 (H3F3A or HIST1H3B) resulting in a lysine to methionine substitution (H3K27M) which alters the N-terminal tail of the H3 protein and post-translational modification. The presence of this mutation is associated with resistance to treatment and a poor prognosis, and makes it an attractive target for CSF liquid biopsy examining ctDNA [9].
The detection of H3 mutations in the CSF was first described in a study of 11 children with brain tumors, six of which had DMGs. ctDNA was isolated from five patients, four of which were found to have the H3K27M mutation by Sanger sequencing, all of whom had DMGs. Another H3 mutation, H3G34V, was detected in one patient with supratentorial HGG [10]. Mutations in histone genes have also been detected using Nanopore technology, a fast, less expensive alternative to next-generation sequencing, in patients with high grade gliomas with H3F3A and H3C2 mutations [11].
This finding was reproduced by another group who described their experience utilizing CSF liquid biopsy to detect H3K27M in two patients with diffuse intrinsic pontine glioma (DIPG) and two patients with HGG utilizing droplet digital PCR (ddPCR) and probes specific to the K27M mutation. They found that tumors with a large cross-sectional area of enhancement on MRI were associated with the highest number of copies of the K27M mutation. Higher levels of K27M were also detected in the lateral ventricles compared to lumbar puncture in one patient, indicating that the amount of ctDNA obtained may be dependent on the location of the tumor. Cells collected from one patient were cultured and radiated; copies of K27M were found to increase with tumor growth and decrease with treatment, indicating the potential utility in this marker for monitoring treatment [12]. The ability to detect K27M was again verified in another study of 2 pediatric patients with DMG [13].
The largest study examining the role of CSF liquid biopsy to detect H3K27M as a biomarker for DMG in pediatric patients utilized 110 CSF and plasma specimens from 48 subjects. ctDNA was quantified using ddPCR with H3K27M detected in 88% of patients, and the highest levels were found in CSF. Longitudinal samples with MRI comparison were available in 12 patients, and a significant decrease in H3K27M ctDNA was observed in 10/12 patients undergoing radiotherapy, providing the first in vivo evidence of clinical utility [14].
The clinical utility of CSF liquid biopsy was demonstrated in a longitudinal assessment of CSF for the H3K27M mutation in 24 patients with DMG. Patients without recurrence of their DMG following treatment were associated with a decrease in concentrations of H3K27M, and sudden increases in H3K27M preceded tumor progression in 5/11 cases. Early decreases in H3K27M were associated with longer progression-free survival, and levels of H3K27M did not increase in patients found to have pseudo-progression [15]. This biomarker has also been used for targeting of GD2-CAR T-cell therapy to H3K27M mutant diffuse midline gliomas in a study of four patients, three of which exhibited clinical and radiographic improvement in their tumors [16].
Untargeted sequencing
Tumor-specific genes provide an attractive target for CSF liquid biopsies. In a study of 35 patients with primary CNS tumors, 14 of which were pediatric patients, tumors underwent WES or sequencing for a range of known mutations to identify somatic mutations in tumor tissue obtained at biopsy. CSF collected from patients was then assessed using a sensitive sequencing-based method to identify the presence of ctDNA. MBs, ependymomas, or HGGs abutting CSF spaces had detectable ctDNA, however, tumors isolated from CSF did not have detectable levels of ctDNA. ctDNA mutations identified included mutations in TP53, IDH1, TERT, NF2, PIK3R1, PTCH1, PTEN [3].
In a similar study, CSF was collected from 57 patients with brainstem gliomas prior to biopsy in 47 of those patients. Genomic DNA libraries including 68 commonly mutated brain tumor genes were generated from tumor DNA and compared with ctDNA from the CSF using NGS. H3F3A, TP53, ATRX, PDGFRA, FAT1, HIST1H3B, PPM1D, IDH1, NF1, PIK3CA, and ACVR1 were the most commonly identified gene mutations in the CSF samples. In 97% of cases with detectable primary tumor mutations in tissue, a single detectable tumor-specific mutation was identified in the CSF; all primary tumor alterations in CSF were identifiable in 84% of cases. At least half of the tumor-specific alterations in the CSF were identified in 92% of cases, demonstrating the power of this tool for molecular diagnosis [17]. Furthermore, a study of 13 MB patients using WES of tumor tissue and ctDNA isolated from CSF samples successfully risk stratified patients based on the molecular profile of ctDNA in CSF [18].
The use of CSF liquid biopsy in MB patients was further explored in a large study with 123 patients and 476 CSF samples collected longitudinally during treatment. MB has relatively fewer driver mutations compared to other CNS tumors, however, the MB genome is characterized by large numbers of chromosomal copy number variations (CNVs). WGS was used to detect tumor-associated CNVs in CSF, which was then used as a marker for measurable residual disease (MRD). MRD was detected in 64% of baseline samples, 39% of post-radiation therapy, 28% mid-chemotherapy, and 34% at the end of therapy. 32 patients achieved complete radiographic response, and 50% of these patients had MRD detected via CNVs at least 3 months prior to radiographic appearance of recurrence [19]. Similar use of liquid biopsy was demonstrated in another study of 58 patients utilizing deep sequencing based on an NGS platform. Variations were detected in the following genes: KMT2D, KMT2C, SMARCA4, BCOR, TP53, PTCH1, EP300, NF1, SETD2, MED12, and SPEN. Interestingly, only 15/58 patients had detectable ctDNA in CSF despite tumors being located in the fourth ventricle in 48/58 patients. Notably, this was low-coverage whole genome sequencing [20].
It is unclear why there is such a difference in detection of ctDNA between the two above studies. This could be secondary to lack of standardization of CSF collection or storage methods, methods used to detect the ctDNA, or the difference in searching for CNVs versus specific MB-associated mutations. Pagès et al. detected similarly low levels of ctDNA in 67 CSF specimens collected from pediatric brain tumor patients with a range of tumor types. They utilized ultra-low pass WGS and were able to detect ctDNA via CNVs in 9/46 CSF samples and a deep sequencing of specific mutations associated with pediatric CNS tumors were able to detect ctDNA in 3/10 CSF samples. Only patients with high grade tumors had positive results [21].
The multiple uses of CSF liquid biopsy were recently described by Miller et al. in a study which included 64 CSF samples from 45 patients, 20 of whom were pediatric, using NGS and the Memorial Sloan Kettering-Integrated Molecular Profiling of Actionable Cancer Targets (MSK-IMPACT) assay which captures all protein-coding exons of 468 cancer-associated genes and select introns. 30/64 samples were found to be positive for at least one somatic mutation described in the MSK-IMPACT panel, and 20/45 patients were found to have a positive CSF sample. Specifically, 5/5 pineoblastomas, 4/4 diffuse leptomeningeal glioneuronal tumors, 7/10 HGG, 3/10 MB, 1/4 retinoblastoma, and 0/4 low grade glioma CSF samples were positive. They further described three categories where liquid biopsy altered the patients care. In the first, ctDNA was used to establish a diagnosis or identify actionable mutations in patients with nondiagnostic biopsies or surgically inaccessible disease. In the second, cfDNA was used to monitor the tumor's response to therapy. In the third, cfDNA was used to monitor the evolution of the tumor over time [22].
Targeted sequencing
BRAF V600E
The BRAF V600E mutation leads to constitutive activation of the MAPK/ERK pathway and is found in up to 33% of pediatric midline tumors with the highest incidence in pleomorphic xanthoastrocytomas (PXAs) [23]. Tumors harboring this mutation have BRAF-targeted therapies available which may increase survival and increases the utility of detecting this mutation [24]. This mutation was detected in 2 CSF samples using ddPCR in a study consisting of 14 CSF samples in a cohort of 29 pediatric patients, four of which were confirmed to harbor the BRAF V600E mutation [25]. A similar study included 32 patients with pediatric HGGs or DMGs. 9 CSF samples were analyzed, 6 of which demonstrated molecular alterations in ctDNA. The presence of TP53, H3K27M, BRAF V600E, H3G34R, and a known ETV6:NTRK3 fusion gene was detected. Longitudinal CSF samples were not available to assess the response to treatment, however, plasma samples demonstrated a decrease in levels of BRAF V600E in response to BRAF-targeted therapy, indicating a potential role for CSF liquid biopsy [26].
SMARC-B1
AT/RTs are characterized by loss of function mutations in the SMARCB1 tumor suppressor gene which forms the SMARCB1/INI1 subunit of the SWI/SNF chromatin remodeling complex. A single study analyzed 5 CSF samples from patients with AT/RTs utilizing real time-PCR (rtPCR) and mutation-specific primers based on DNA extracted from tumor tissue collected at time of surgery. Tumor-specific DNA was identified in the CSF of 2 patients [27].
Epigenetic studies
A single study examining the epigenetic profile of patients with pediatric CNS tumors was identified. CSF samples from four patients were collected via lumbar puncture. DNA methylomes and hydroxymethylomes were obtained with whole genome bisulfite screening and anti-cytosine-5-methylenesulfonate immunoprecipitation sequencing resulting in an average of 600 million read over 13 million CpG sites. There was a high correlation observed with epigenetic modification of transcription start sites, exons, CpG islands, and promoters between tumor tissue and cfDNA from CSF, and the DNA methylation levels within the CpG islands were highly consistent within individuals, suggesting their potential utility as a biomarker. Compared to normal cerebellum, cfDNA from CSF showed lower overall levels of methylation but higher levels of methylation at CpG islands, consistent with data regarding MB epigenetic patterns. Two patients had CSF samples collected at diagnosis and treatment, and the methylation patterns of CpG islands were seen to change, suggesting a use to monitor response to treatment. Differences in the epigenome were also observed between MB subtypes [28].
RNA and miRNAs
miRNAs are small, non-coding RNAs identifiable in CSF which modulate post-transcriptional gene expression and act as both tumor suppressors and oncogenes [29,30]. There was a single study utilizing miRNA as a biomarker for pediatric CNS tumors. As discussed above, while AFP and bHCG are useful biomarkers for malignant GCTs, they have a limited sensitivity and specificity for diagnosis. Malignant GCTs overexpress miRNAs from the miR–371–373 and miR–302/367 clusters. Murray et al. quantified these miRNAs in serum and CSF of 25 pediatric patients with GCTs [62]. Utilizing quantitative real time-PCR (qRT-PCR) and a miRNA panel with miR–371a–3p, miR–372–3p, miR–373–3p and miR–367–3p, they were able to distinguish intracranial malignant GCTs from intracranial non-GCTs at diagnosis, indicating potential for diagnosing and monitoring disease treatment.
Lee et al. examined CSF samples from 40 patients with MB with RNAseq and high-resolution MS to identify RNA profiles associated with the tumor. A multivariate analysis utilizing the Data Integration Analysis for Biomarker discovery using a Latent component method for Omics studies (DIABLO) package identified RNA changes within CSF that accurately characterized tumor CSF compared to non-tumor using the 48 most differentially expressed genes. A large scale approach such as this was able to determine transcripts which clustered together, and a novel circular RNA circ_463 was identified as a potential future MB biomarker [31].
Proteomic markers (Table 2)
Table 2.
Proteomic studies.
| Author and Year | Neoplasm | Protein identified | Technique | N | Findings |
|---|---|---|---|---|---|
| Müller et al, 1993 [46] | CNS tumors | IGF family proteins | Radioimmunoassay | 23 CNS tumors, 18 leukemia, 13 meningitis, 38 control | Elevated IGFBP-3 in CNS tumors |
| Müller et al, 1994 [65] | CNS tumors | IGFBP-2 | Western blot | 21 CNS tumors, 25 leukemia, 4 peripheral solid tumors | Elevated IGFBP-2 in CNS tumors |
| Nishizaki et al, 2001 [36] | GCT | AFP/bHCG | ELISA | 19 GCTs | Disseminated disease can be prevented with surgery in GCTs with elevated bHCG and AFP |
| Seregni et al, 2002 [32] | GCT | AFP/bHCG | Radioimmunoassay | 30 | bHCG and AFP can be used to monitor response to therapy |
| Miyanohara et al, 2002 [40] | GCT | s-kit | ELISA | 47 samples from 32 pts | Elevated s-kit in germinanomatous GCT's |
| Kao 2005 [52] | CNS tumors | Osteopontin | ELISA | 16 MB, 8 AT/RT, 15 control | Osteopontin elevated in AT/RT |
| de Bont 2006 | CNS tumors | Apolipoprotein A2 | SELDI-TOF, Q10 ProteinChip arrays | 32 CNS tumor, 70 control | Apolipoprotein A2 elevated in CNS tumors |
| Figarella-Branger et al, 2006 [45] | MB | PSA-NCAM | ELISA | 145 samples from 29 MB, 14 control | PSA-NCAM correlated with tumor and response to treatment |
| de Bont 2008 [54] | CNS tumors | t-Tau | ELISA | 37 CNS tumor, 51 control | Elevated t-Tau in CNS tumors, especially MB |
| de Bont et al, 2008 [47] | MB, ependymoma | IGF | Radioimmunoassay | 16 MB, 4 Ependymoma, 23 control | IGF system is a source of biomarkers |
| Rajagopal et al, 2011 [49] | MB | PGD2-S | ELISA, MALDI-TOF | 33 MB, 25 control | PGD2-S levels negatively correlated with presence of tumor |
| Desiderio et al, 2012 [50] | MB, ependymoma, PA | LVV-hemorphin-7, VV-hemorphin-7 | LC-MS | 14 PA, 1 MB, 5 ependymoma, 5 control | Hemorphins were present post-resection if residual or with spinal dissemination |
| Watanabe et al, 2012 [41] | GCT | PLAP | Chemi-luminescence | 36 GCTs, 3 nongerminomatous GCTS, 21 glioma, 12 other, 37 control | Elevated PLAP in GCT's |
| Qaddoumi et al, 2012 [35] | GCT | AFP/bHCG | 67 GCTs | CSF and serum bHCG and AFP correspond | |
| Saratsis 2012 [57] | DIPG | CypA, DDAH1 | MS | 10 DIPG, 1 GBM, 4 control | CypA and DDAH1 marker of DIPG |
| Legault et al, 2013 [34] | GCT | AFP/bHCG | 86 | Lumbar CSF has better yield of AFP and bHCG relative to ventricular CSF | |
| Cengiz 2015 [53] | CNS tumors | C-tau levels | ELISA | 26 | c-Tau elevated with tumors, correlated axonal damage, not diseasde extent |
| Fukuoka et al, 2016 [33] | GCT | bHCG | ELISA | 35 | High levels of bHCG are associated with GCT recurrence |
| Hu et al, 2016 [37] | GCT | AFP/bHCG | ELISA (bHCG), Chemi-luminescence (PLAP) | 58 GCTs, 17 suspected GCTs, 17 controls | Establishes cut-off alues of bHCG and AFP for diagnosis |
| Spreafico 2017 [48] | CNS tumors | CSF Proteome | Core-shell hydrogel nanoparticles, LC-MS, ELISA, Westernblot | 27 brain tumors, 13 control | Type 1 collagen, IGFBP-4, procollagen C-endopeptidase enhancer 1, GDNF α2, ITIH4, NPDC1 elevated with metastatis |
| Low 2020 [55] | Metastatic MB | Cytokines | Proteome array for 43 proteins | 10 metastatic MB | CCL2, CXCL1, IL6, IL8 elevated in metastatic MB |
| Okamoto et al, 2021 [42] | GCT | PLAP | 37 intra- or periventricular tumors | PLAP can be used to monitor response to treatment and recurrence | |
| Bruschi et al, 2021 [59] | All CNS tumors requiring ventriculostomy | CSF Proteome | ELISA | 31 brain tumors, 37 control | TAF15 and S100B differentiate tumor and nontumor |
Germ cell tumors (GCTs)
Intracranial GCTs are primary malignant CNS tumors in the pineal or suprasellar regions with variable prognoses depending on their classification as germinomas or nongerminomatous (NGGCTs). Germinomas are the most common form of GCT and are highly responsive to chemoradiotherapy with >90% remission rates. NGGCTs are relatively less responsive to treatment with remission rates of 65-85% and poorer survival. Diagnostic tool development has focused on improving early detection of GCTs, distinguishing germinoma and NGGCTs, and monitoring treatment response.
Beta-human chorionic gonadotropin (bHCG) and alpha fetoprotein (AFP)
Patients with intracranial GCTs have elevated levels of bHCG in CSF which decrease after treatment and predict treatment success better than MRI which cannot distinguish necrotic masses from residual tumor [32]. In germinomas, higher levels of CSF bHCG are also correlated with more frequent recurrence [33].
There is some disagreement regarding the diagnostic sensitivity of biomarker detection and CSF biomarker yield depending on the location of CSF access (e.g. lumbar vs ventricular). In a study of 86 patients with GCTs, lumbar bHCG values were equal to or greater than values in ventricular CSF or serum, suggesting that lumbar CSF may be a more reliable source of bHCG [34]. Another study on 67 patients found 96.2% agreement in serum and CSF, but did not specify the location of CSF access [35].
AFP is used alongside bHCG for diagnosis, is detected in both serum and CSF, and correlates with treatment response. The diagnostic sensitivity of AFP across different sources of CSF is also debated. For patients with mixed malignant GCTs, lumbar AFP levels are greater than ventricular values, but serum AFP values remain the highest—a rare example where serum values are higher than CSF values [32]. Nevertheless, aggressive removal of tumor is effective as a first-line therapy in preventing craniospinal dissemination in patients with GCTs who have a relative increase in levels of bHCG or AFP [36].
GCTs are traditionally diagnosed by CSF bHCG >= 50 IU/L or AFP >= 10 ng/mL. However, this diagnostic cutoff has a very poor sensitivity of 34.6%. A recent study of predominantly pediatric patients found that reducing the diagnostic threshold to bHCG >= 8.2 IU/L increased sensitivity to 47% while maintaining specificity at 100%. This bHCG threshold, taken together with a cutoff of AFP >= 3.8 ng/mL, creates a total diagnostic sensitivity of 65.4% [37].
s-kit/PLAP
Our review found that bHCG and AFP are the most commonly used biomarkers for GCTs, however, recent studies have identified new markers. C-kit is a protooncogene involved in stem cell growth factor signaling and has been implicated in a variety of extra-CNS cancers such as leukemia and breast cancer [38,39]. The soluble form of c-kit (s-kit) is found on germinoma cells and is elevated in CSF of germinomas and syncytiotrophoblastic giant cells, but not teratomas, non-germ cell brain tumors, or controls. Furthermore, the level of s-kit is correlated with response or non-response to chemoradiation [40].
Placental alkaline phosphatase (PLAP) has also been proposed as a tumor marker for intracranial germinomas. Watanabe et al. reported high CSF PLAP in germinoma patients which decreased after remission following radiochemotherapy. The assay achieved a sensitivity of 94% and a specificity of 97% [41]. A similar assay detected higher CSF PLAP levels in patients with germinomas compared to NGGCTs which remained elevated in patients with recurrence [42].
Posterior fossa tumors
Posterior fossa tumors comprise up to 70% of all childhood brain tumors [43]. There are a few studies examining specific protein markers of posterior fossa tumors, the most common of which are MB, ependymomas, and cerebellar astrocytomas [44].
polysialic-neural cell adhesion molecule (PSA-NCAM)
PSA-NCAM is a marker of developing neurons and plays a role in neural plasticity. PSA-NCAM levels are increased in the CSF of patients with MB and levels correspond to the extent of disease. Following treatment, elevated levels of PSA-NCAM remain present in patients refractory to treatment. The PSA-NCAM test correlates with cytology results and has a higher sensitivity than cytology [45].
Insulin-like growth factors (IGFs) and insulin-like growth factor binding proteins (IGFBPs)
Insulin-like growth factor (IGF) and insulin-like growth factor binding proteins (IGFBP) play an important role in CNS development and may have a role in brain tumor development, but can become imbalanced in pathologic states. In normal CSF, IGF-1 is detectable only in trace amounts and IGF-2 is abundant. Likewise, IGFBP-2 and IGFBP-4 are abundant in normal CSF and IGFBP-3 is typically only a minor component. In MBs and ependymomas, IGFBP-2 and IGFBP-3 are overexpressed. IGFBP-3 is elevated in children with highly malignant CNS tumors and returns to normal following chemotherapy. Elevated IGFBP-3 levels correlate with microscopically detectable malignant cells in CSF in MBs and ependymomas [46]. Though studies are limited, IGFBP-4 is also reported to be significantly elevated in patients with CNS metastasis compared to controls [47,48].
Prostaglandin D2 Synthase and hemorphins
While most biomarkers are detected in elevated levels, prostaglandin D2 synthase (PGD2S is six-fold decreased in MB samples [49]. PGD2S is one of the most abundant glycoproteins in healthy CSF and its reduction in cancer patients is speculated to be a host response to the tumor. Similarly, LVV-hemorphin-7 (LVV) and VV-hemorphin-7 (VV-h7), derived from the hemoglobin family, act as negative biomarkers. These peptides are absent in posterior fossa tumor patients prior to treatment and in patients with residual tumor or metastatic disease, yet are elevated immediately following surgery [50]. Though studies have been limited, preliminary data on the reduction of PGD2S, LVV, and VV-h7 in posterior fossa tumor patients suggest that the absence of these CSF protein markers may serve clinical utility in the monitoring of response to treatment and early detection of recurrence [49,50].
Other protein biomarkers
Osteopontin
Osteopontin (OPN) is a bone-related extracellular protein which mediates immune responses and cell migration critical to cancer progression. OPN has been evaluated in atypical teratoid/rhabdoid tumors (AT/RTs) which are WHO grade IV tumors comprising approximately 50% of malignant brain tumors in children less than 1-year-old [51]. OPN in CSF is significantly higher in AT/RTs compared with MB, and correlates with treatment response [52].
Tau
Tau is an internal skeleton protein found in neurons commonly associated with neurodegeneration. Total tau is a general marker of neurodegeneration, elevated in lumbar but not ventricular CSF in children with brain tumors. Cleaved tau is associated with neuronal damage and is found to be elevated by 400-fold in children with newly diagnosed brain tumors. Of note, elevated tau in CSF is a non-specific indicator of malignancy, as it is also an indicator of other CNS insults like hydrocephalus, infection, and traumatic brain injury [53,54].
Cytokines
Cytokines are signaling proteins released in response to inflammation and immune function. These proteins are increasingly studied with regard to pediatric brain tumors. CCL2 is involved in regulating macrophage recruitment during inflammation and is increased in the CSF of patients with metastatic MB compared to non-metastatic. In group 3 MB, MYC-amplified patients express higher levels of CXCL1, IL6, and IL8 in CSF [55].
Increasing interest in molecular signatures of pediatric brain tumors has uncovered novel markers for potential use in liquid biopsy. In diffuse intrinsic pontine glioma (DIPG), the overexpression of tenascin-C (TNC) gene and protein, an extracellular matrix glycoprotein involved in neuronal development, has been linked to higher tumor grade and poorer survival [56]. DIPG has also been associated with an increase in secreted cyclophillin A and dimethylarginase 1 in CSF [57].
Proteomic analyses
Large-scale proteomic analysis has also been utilized. SELDI-TOF mass spectrometry (MS) was used on CSF from 32 pediatric brain tumor patients to identify protein expression profiles using Q10 ProteinChip arrays. 247 proteins were identified with MS and 123 were differentially expressed in tumors. The authors selected one of the proteins overexpressed in tumors and identified it as apolipoprotein A2a. The levels were correlated with CSF albumin levels, and it was thought that the elevated values were secondary to blood-brain barrier disruption [58]. An additional study examining 29 patients with tumors requiring CSF diversion utilized liquid chromatography-MS to evaluate the proteome of CSF obtained. TATA-binding protein assoicated factor 15 (TAF15) and S100B were identified as discriminating tumor from non-tumor among 1598 proteins. Additional proteins which could discriminate specific types of tumors included thymosin-beta 4, CD109, 14.3.3, and HSP90 alpha [59].
In a study on 27 patients with brain tumors, Spreafico et al. processed CSF samples using nanoparticles to identify low abundance and low molecular weight proteins associated with the tumors. Of 558 proteins identified using reverse phase protein array, 14 were found to discriminate metastatic CNS tumors from extra-CNS cancer controls. Proteins identified included type 1 collagen, IGFBP4, procollagen C-endopeptidase enhancer 1, glial cell-line derived neurotrophic factor receptor α2, inter-alpha-trypsin inhibitor heavy chain 4, and neural proliferation and differentiation control protein-1 [48]. Thus, novel biomarkers are rapidly being identified with high throughput methods of protein analysis.
Small molecule and metabolomic biomarkers
The ability to detect and profile large numbers of small molecule metabolites and lipids has been greatly facilitated by high resolution MS, and these molecules can now be profiled to develop a picture of a tumor's microenvironment [60]. One study examining the metabolome of CSF in pediatric CNS tumors was identified in our review. Lee et al. examined CSF samples from 40 patients with MB with high-resolution MS (in addition to RNAseq, as mentioned above) to identify metabolite and lipid profiles associated with the tumor. Metabolomic analysis demonstrated alterations in metabolites of the tricarboxylic acid cycle and amino acid metabolism in MB CSF, and total triacylglycerols were also found to be upregulated. These profiles provided evidence of a hypoxic environment within the tumor and allowed accurate categorization of MB CSF [31].
Future directions
As described above, techniques for profiling the proteomic, genomic, and metabolomic profile of pediatric CNS malignancies are becoming increasingly used and are enabling the rapid identification of large numbers of molecules to facilitate diagnosis, monitor treatment, and evaluate recurrence of disease. We briefly review a selection of these methods in Table 3. Despite this vast toolbox, the research has yet to be implemented in neuro-oncology clinics as standard of care. Disparate methods have been used to build this body of literature. This is necessary given the variety of genetic changes that have been identified, and clinicians and researchers will need to be familiar with the basics of these methods and their benefits and limitations to facilitate their use and contextualize comparison of studies. It is also possible that the current data is biased since most studies occurred at tertiary care centers where patients likely have more severe disease. Larger prospective studies are needed to validate the utility of the above studies.
Table 3.
Summary of selection of techniques discussed.
| Technique | Description | Benefits | Limitations |
|---|---|---|---|
| Genetic | |||
| Sanger sequencing [66] | Sequences DNA with primers using fluorescent nucleotides and electrophoresis to generate sequence | Highly accurate, long read length (400–900 bp) | Low throughput, high cost |
| Droplet digital PCR (ddPCR) [67] | Allows for detection of known variants based on primer, best for point mutations | Small amount of CSF necessary, quantitative assay | Assay is expensive, time-consuming set-up |
| Next generation sequencing (NGS) [66,68] | Simultaneously sequences multiple DNA fragments for larger genome coverage | Multiple platforms, can provide whole-genome/exome, allows discovery of new genes, low error rate | Lower sensitivity, may require larger volumes of CSF, expensive, complex data analysis |
| Nanopore sequencing [11,69] | Direct electronic analysis of nucleic acids fed through biologic pores | Inexpensive, handheld; high sensitivity, requires small amounts of CSF | Potentially high error rate |
| Proteins | |||
| Enzyme linked immunosorbent assay (ELISA) [70] | Immuno assay to detect specific proteins of interest | Rapid, standardized, high sensitivity, can be multiplexed | Requires well-defined antibody to protein(s) of interest, no discovery ability |
| Liquid Chromatography-Mass Spectrometery (LC-MS) [71,72] | Separates proteins in sample with chromatography and analyzes with MS | High throughput, quantitative | Extensive data processing, difficult to identify specific proteins, expensive equipment |
To facilitate larger studies, standardization of CSF collection and of assays will be necessary to ensure results are translatable between institutions. Indeed, only a single paper describing a standardized method for detection of the H3K27 mutation was identified in our review, indicating the need for further research to standardize and validate findings [61]. Additionally, the tools to perform high-throughput molecular analysis are frequently expensive and not readily available, so less expensive technology such as the Nanopore sequencing mentioned above is critical for dissemination of these methods. Newer high-throughput methods are able to sequence genomes, transcriptomes, proteomes, and metabolomes, as opposed to prior methods which could only examine a few biomarkers of interest. With increasing data, computational biology approaches are becoming increasingly useful to provide a deeper understanding of tumor biology and cancer treatment [62,63,16]. In the future, approaches that combine insights from DNA, RNA, protein, and small molecule diagnostic tests may allow for excellent monitoring of tumor progression and treatment and provide further targets for management of childhood brain tumors.
CRediT authorship contribution statement
Kurt R. Lehner: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. Kelly Jiang: Investigation, Writing – original draft, Writing – review & editing. Jordina Rincon-Torroella: Writing – review & editing. Ranjan Perera: Writing – review & editing. Chetan Bettegowda: Conceptualization, Writing – review & editing.
Footnotes
Disclosures: The authors did not receive any financial support for the review and have no competing interests to report. All data is publicly available.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Warmuth-Metz M., Bison B., Leykamm S. Neuroradiologic review in pediatric brain tumor studies. Clin. Neuroradiol. 2009;19(4):263–273. doi: 10.1007/s00062-009-9029-5. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Springer S., Zhang M., et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl Acad. Sci. 2015;112(31):9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattox A.K., Yan H., Bettegowda C. The potential of cerebrospinal fluid–based liquid biopsy approaches in CNS tumors. Neuro. Oncol. 2019;21(12):1509–1518. doi: 10.1093/neuonc/noz156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronkhorst A.J., Ungerer V., Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019;17 doi: 10.1016/j.bdq.2019.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corcoran R.B., Chabner B.A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 2018;379(18):1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 7.Pentsova E.I., Shah R.H., Tang J., et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J. Clin. Oncol. 2016;34(20):2404–2415. doi: 10.1200/JCO.2016.66.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunda S., Zuccato J.A., Voisin M.R., et al. Liquid biomarkers for improved diagnosis and classification of CNS tumors. Int. J. Mol. Sci. 2021;22(9):4548. doi: 10.3390/ijms22094548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azad T.D., Jin M.C., Bernhardt L.J., Bettegowda C. Liquid biopsy for pediatric diffuse midline glioma: a review of circulating tumor DNA and cerebrospinal fluid tumor DNA. Neurosurg. Focus. 2020;48(1):E9. doi: 10.3171/2019.9.FOCUS19699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T.Y., Piunti A., Lulla R.R., et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol. Commun. 2017;5(1):28. doi: 10.1186/s40478-017-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruzek A.K., Ravi K., Muruganand A., et al. Electronic DNA analysis of CSF cell-free tumor DNA to quantify multi-gene molecular response in pediatric high-grade glioma. Clin. Cancer Res. 2020;26(23):6266–6276. doi: 10.1158/1078-0432.CCR-20-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stallard S., Savelieff M.G., Wierzbicki K., et al. CSF H3F3A K27M circulating tumor DNA copy number quantifies tumor growth and in vitro treatment response. Acta Neuropathol. Commun. 2018;6(1):80. doi: 10.1186/s40478-018-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Ricarte F., Mayor R., Martínez-Sáez E., et al. Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumor DNA from cerebrospinal fluid. Clin. Cancer Res. 2018;24(12):2812–2819. doi: 10.1158/1078-0432.CCR-17-3800. [DOI] [PubMed] [Google Scholar]
- 14.Panditharatna E., Kilburn L.B., Aboian M.S., et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin. Cancer Res. 2018;24(23):5850–5859. doi: 10.1158/1078-0432.CCR-18-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantor E., Wierzbicki K., Tarapore R.S., et al. Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro. Oncol. 2022 doi: 10.1093/neuonc/noac030. Published online February 6noac030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majzner R.G., Ramakrishna S., Yeom K.W., et al. GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature. 2022;603(7903):934–941. doi: 10.1038/s41586-022-04489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan C., Diplas B.H., Chen X., et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol. 2019;137(2):297–306. doi: 10.1007/s00401-018-1936-6. (Berl) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escudero L., Llort A., Arias A., et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat. Commun. 2020;11(1):5376. doi: 10.1038/s41467-020-19175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu A.P.Y., Smith K.S., Kumar R., et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39(11):1519–1530. doi: 10.1016/j.ccell.2021.09.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y., Li M., Ren S., et al. Exploring genetic alterations in circulating tumor DNA from cerebrospinal fluid of pediatric medulloblastoma. Sci. Rep. 2021;11(1):5638. doi: 10.1038/s41598-021-85178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagès M., Rotem D., Gydush G., et al. Liquid biopsy detection of genomic alterations in pediatric brain tumors from cell-free DNA in peripheral blood, CSF, and urine. Neuro. Oncol. 2022:noab299. doi: 10.1093/neuonc/noab299. Published online January 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller A.M., Szalontay L., Bouvier N., et al. Next-generation sequencing of cerebrospinal fluid for clinical molecular diagnostics in pediatric, adolescent and young adult brain tumor patients. Neuro. Oncol. 2022:noac035. doi: 10.1093/neuonc/noac035. Published online February 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindler G., Capper D., Meyer J., et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6. (Berl) [DOI] [PubMed] [Google Scholar]
- 24.Sanchez J.N., Wang T., Cohen M.S., BRAF and MEK Inhibitors Use and resistance in BRAF-mutated cancers. Drugs. 2018;78(5):549–566. doi: 10.1007/s40265-018-0884-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Romero N., Carrión-Navarro J., Areal-Hidalgo P., et al. BRAF V600E detection in liquid biopsies from pediatric central nervous system tumors. Cancers. 2019;12(1):66. doi: 10.3390/cancers12010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izquierdo E., Proszek P., Pericoli G., et al. Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neuro-Oncol. Adv. 2021;3(1):vdab013. doi: 10.1093/noajnl/vdab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vu-Han T.L., Frühwald M.C., Hasselblatt M., et al. Identifying molecular markers for the sensitive detection of residual atypical teratoid rhabdoid tumor cells. Cancer Genet. 2014;207(9):390–397. doi: 10.1016/j.cancergen.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Zhao S., Lee M., et al. Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma. Sci. Adv. 2020;6(42):eabb5427. doi: 10.1126/sciadv.abb5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16(7):421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q., Li P., Li A., et al. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J. Exp. Clin. Cancer Res. 2012;31(1):97. doi: 10.1186/1756-9966-31-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee B., Mohamad I., Pokhrel R., et al. Medulloblastoma cerebrospinal fluid reveals metabolites and lipids indicative of hypoxia and cancer-specific RNAs. Acta Neuropathol. Commun. 2022;10(1):25. doi: 10.1186/s40478-022-01326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seregni E., Massimino M., Nerini Molteni S., et al. Serum and cerebrospinal fluid human chorionic gonadotropin (hCG) and alpha-fetoprotein (AFP) in intracranial germ cell tumors. Int. J. Biol. Markers. 2002;17(2):112–118. doi: 10.5301/jbm.2008.3768. [DOI] [PubMed] [Google Scholar]
- 33.Fukuoka K., Yanagisawa T., Suzuki T., et al. Human chorionic gonadotropin detection in cerebrospinal fluid of patients with a germinoma and its prognostic significance: assessment by using a highly sensitive enzyme immunoassay. J. Neurosurg. Pediatr. 2016;18(5):573–577. doi: 10.3171/2016.4.PEDS1658. [DOI] [PubMed] [Google Scholar]
- 34.Legault G., Allen J.C. Potential role of ventricular tumor markers in CNS germ cell tumors: ventricular tumor markers in CNS GCT. Pediatr. Blood Cancer. 2013;60(10):1647–1650. doi: 10.1002/pbc.24620. [DOI] [PubMed] [Google Scholar]
- 35.Qaddoumi I., Sane M., Li S., et al. Diagnostic utility and correlation of tumor markers in the serum and cerebrospinal fluid of children with intracranial germ cell tumors. Childs Nerv. Syst. 2012;28(7):1017–1024. doi: 10.1007/s00381-012-1762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishizaki T., Kajiwara K., Adachi N., et al. Detection of craniospinal dissemination of intracranial germ cell tumours based on serum and cerebrospinal fluid levels of tumour markers. J. Clin. Neurosci. 2001;8(1):27–30. doi: 10.1054/jocn.2000.0750. [DOI] [PubMed] [Google Scholar]
- 37.Hu M., Guan H., Lau C.C., et al. An update on the clinical diagnostic value of β-hCG and αFP for intracranial germ cell tumors. Eur. J. Med. Res. 2016;21(1):10. doi: 10.1186/s40001-016-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda H., Kanakura Y., Tamaki T., et al. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 1991;78(11):2962–2968. doi: 10.1182/blood.V78.11.2962.2962. [DOI] [PubMed] [Google Scholar]
- 39.Natali P.G., Nicotra M.R., Sures I., Mottolese M., Botti C., Ullrich A. Breast cancer is associated with loss of the c-kit oncogene product. Int. J. Cancer. 1992;52(5):713–717. doi: 10.1002/ijc.2910520508. [DOI] [PubMed] [Google Scholar]
- 40.Miyanohara O., Takeshima H., Kaji M., et al. Diagnostic significance of soluble c-kit in the cerebrospinal fluid of patients with germ cell tumors. J. Neurosurg. 2002;97(1):177–183. doi: 10.3171/jns.2002.97.1.0177. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe S., Aihara Y., Kikuno A., et al. A highly Sensitive and specific chemiluminescent enzyme immunoassay for placental alkaline phosphatase in the cerebrospinal fluid of patients with intracranial germinomas. Pediatr. Neurosurg. 2012;48(3):141–145. doi: 10.1159/000345632. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto M., Yamaguchi S., Ishi Y., et al. Diagnostic capability of cerebrospinal fluid-placental alkaline phosphatase value in intracranial germ cell tumor. Oncology. 2021;99(1):23–31. doi: 10.1159/000509395. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien D.F., Caird J., Kennedy M., Roberts G.A., Marks J.C., Allcutt D.A. Posterior fossa tumours in childhood: evaluation of presenting clinical features. Ir. Med. J. 2001;94(2):52–53. [PubMed] [Google Scholar]
- 44.Vara Prasad K., Ravi D., Pallikonda V., Raman B.S. Clinicopathological study of pediatric posterior fossa tumors. J. Pediatr. Neurosci. 2017;12(3):245. doi: 10.4103/jpn.JPN_113_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figarella-Branger D., Dubois C., Chauvin P., De Victor B., Gentet J.C., Rougon G. Correlation between polysialic-neural cell adhesion molecule levels in CSF and medulloblastoma outcomes. J. Clin. Oncol. 1996;14(7):2066–2072. doi: 10.1200/JCO.1996.14.7.2066. [DOI] [PubMed] [Google Scholar]
- 46.Müller H.L., Oh Y., Gargosky S.E., Lehrnbecher T., Hintz R.L., Rosenfeld R.G. Concentrations of insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3), IGF, and IGFBP-3 protease activity in cerebrospinal fluid of children with leukemia, central nervous system tumor, or meningitis. J. Clin. Endocrinol. Metab. 1993;77(5):1113–1119. doi: 10.1210/jcem.77.5.7521338. [DOI] [PubMed] [Google Scholar]
- 47.de Bont J.M., van Doorn J., Reddingius R.E., et al. Various components of the insulin-like growth factor system in tumor tissue, cerebrospinal fluid and peripheral blood of pediatric medulloblastoma and ependymoma patients: the IGF system in pediatric medulloblastoma and ependymoma. Int. J. Cancer. 2008;123(3):594–600. doi: 10.1002/ijc.23558. [DOI] [PubMed] [Google Scholar]
- 48.Spreafico F., Bongarzone I., Pizzamiglio S., et al. Proteomic analysis of cerebrospinal fluid from children with central nervous system tumors identifies candidate proteins relating to tumor metastatic spread. Oncotarget. 2017;8(28):46177–46190. doi: 10.18632/oncotarget.17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajagopal M.U., Hathout Y., MacDonald T.J., et al. Proteomic profiling of cerebrospinal fluid identifies prostaglandin D2 synthase as a putative biomarker for pediatric medulloblastoma: a pediatric brain tumor consortium study. Proteomics. 2011;11(5):935–943. doi: 10.1002/pmic.201000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desiderio C., D'Angelo L., Rossetti D.V., et al. Cerebrospinal fluid top-down proteomics evidenced the potential biomarker role of LVV- and VV-hemorphin-7 in posterior cranial fossa pediatric brain tumors. Proteomics. 2012;12(13):2158–2166. doi: 10.1002/pmic.201100499. [DOI] [PubMed] [Google Scholar]
- 51.Packer R.J., Biegel J.A., Blaney S., et al. Atypical teratoid/rhabdoid tumor of the central nervous system: report on workshop. J. Pediatr. Hematol. Oncol. 2002;24(5) doi: 10.1097/00043426-200206000-00004. https://journals.lww.com/jpho-online/Fulltext/2002/06000/Atypical_Teratoid_Rhabdoid_Tumor_of_the_Central.4.aspx [DOI] [PubMed] [Google Scholar]
- 52.Kao C.L., Chiou S.H., Ho D.M.T., et al. Elevation of plasma and cerebrospinal fluid osteopontin levels in patients with atypical teratoid/rhabdoid tumor. Am. J. Clin. Pathol. 2005;123(2):297–304. doi: 10.1309/0FTKBKVNK4T5P1L1. [DOI] [PubMed] [Google Scholar]
- 53.Cengiz P., Zemlan F., Eickhoff J.C., Ellenbogen R., Zimmerman J.J. Increased cerebrospinal fluid cleaved tau protein (C-tau) levels suggest axonal damage in pediatric patients with brain tumors. Childs Nerv. Syst. 2015;31(8):1313–1319. doi: 10.1007/s00381-015-2705-7. [DOI] [PubMed] [Google Scholar]
- 54.de Bont J.M., Vanderstichele H., Reddingius R.E., Pieters R., van Gool S.W. Increased total-Tau levels in cerebrospinal fluid of pediatric hydrocephalus and brain tumor patients. Eur. J. Paediatr. Neurol. 2008;12(4):334–341. doi: 10.1016/j.ejpn.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 55.Low S.Y.Y., Bte Syed Sulaiman N., Tan E.E.K., et al. Cerebrospinal fluid cytokines in metastatic group 3 and 4 medulloblastoma. BMC Cancer. 2020;20(1):554. doi: 10.1186/s12885-020-07048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi J., Esfahani D.R., Huang T., et al. Tenascin-C expression contributes to pediatric brainstem glioma tumor phenotype and represents a novel biomarker of disease. Acta Neuropathol. Commun. 2019;7(1):75. doi: 10.1186/s40478-019-0727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saratsis A.M., Yadavilli S., Magge S., et al. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol. 2012;14(5):547–560. doi: 10.1093/neuonc/nos067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Bont J.M., den Boer M.L., Reddingius R.E., et al. Identification of apolipoprotein A-II in cerebrospinal fluid of pediatric brain tumor patients by protein expression profiling. Clin. Chem. 2006;52(8):1501–1509. doi: 10.1373/clinchem.2006.069294. [DOI] [PubMed] [Google Scholar]
- 59.Bruschi M., Petretto A., Cama A., et al. Potential biomarkers of childhood brain tumor identified by proteomics of cerebrospinal fluid from extraventricular drainage (EVD) Sci. Rep. 2021;11(1):1818. doi: 10.1038/s41598-020-80647-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh A., Nishtala K. Biofluid lipidome: a source for potential diagnostic biomarkers. Clin. Transl. Med. 2017;6(1) doi: 10.1186/s40169-017-0152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li D., Bonner E.R., Wierzbicki K., et al. Standardization of the liquid biopsy for pediatric diffuse midline glioma using ddPCR. Sci. Rep. 2021;11(1):5098. doi: 10.1038/s41598-021-84513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohart F., Gautier B., Singh A., Lê Cao K.A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13(11) doi: 10.1371/journal.pcbi.1005752. Schneidman D, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang M., Li Z., Chen T., Zeng J. Integrative data analysis of multi-platform cancer data with a multimodal deep learning approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;12(4):928–937. doi: 10.1109/TCBB.2014.2377729. [DOI] [PubMed] [Google Scholar]
- 64.Murray M.J., Bell E., Raby K.L., et al. A pipeline to quantify serum and cerebrospinal fluid microRNAs for diagnosis and detection of relapse in paediatric malignant germ-cell tumours. Br. J. Cancer. 2016;114(2):151–162. doi: 10.1038/bjc.2015.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh Y., Müller H.L., Zhang H., Ling N., Rosenfeld R.G., Le Roith D., Raizada M.K. Current Directions in Insulin-Like Growth Factor Research. Vol 343. Advances in Experimental Medicine and Biology. Springer US; 1994. Synthesis and characterization of IGF-II analogs: applications in the evaluation of IGF receptor function and IGF-independent actions of IGFBPS; pp. 41–54. In: eds. [DOI] [PubMed] [Google Scholar]
- 66.Moorcraft S.Y., Gonzalez D., Walker B.A. Understanding next generation sequencing in oncology: a guide for oncologists. Crit. Rev. Oncol. Hematol. 2015;96(3):463–474. doi: 10.1016/j.critrevonc.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Olmedillas-López S., García-Arranz M., García-Olmo D. Current and emerging applications of droplet digital PCR in oncology. Mol. Diagn. Ther. 2017;21(5):493–510. doi: 10.1007/s40291-017-0278-8. [DOI] [PubMed] [Google Scholar]
- 68.Mardis E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017;12(2):213–218. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- 69.Euskirchen P., Bielle F., Labreche K., et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017;134(5):691–703. doi: 10.1007/s00401-017-1743-5. (Berl) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wulfkuhle J.D., Liotta L.A., Petricoin E.F. Proteomic applications for the early detection of cancer. Nat. Rev. Cancer. 2003;3(4):267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 71.Boys E.L., Liu J., Robinson P.J., Reddel R.R. Clinical applications of mass spectrometry-based proteomics in cancer: where are we? Proteomics. 2022 doi: 10.1002/pmic.202200238. Published online August 26. [DOI] [PubMed] [Google Scholar]
- 72.Samuel N., Remke M., Rutka J.T., Raught B., Malkin D. Proteomic analyses of CSF aimed at biomarker development for pediatric brain tumors. J. Neurooncol. 2014;118(2):225–238. doi: 10.1007/s11060-014-1432-3. [DOI] [PubMed] [Google Scholar]