Highlights
-
•
DEP is an electrokinetic approach for discriminating and separating floating cells.
-
•
Buffer composition differently influence the viability of Caco-2 and K562 cells.
-
•
NaCl concentration influences both Flow Cytometry outcomes and cell size.
-
•
Buffer composition differently influences cell parameters in Caco-2 and K562 cells.
-
•
Buffer composition differently influences gene expression in Caco-2 and K562 cells.
Keywords: Dielectrophoresis, Cell status, Biochemical parameters, Circulating tumor cells, Cell isolation and separation, Microfluidics
Abstract
Dielectrophoresis (DEP) represents an electrokinetic approach for discriminating and separating suspended cells based on their intrinsic dielectric characteristics without the need for labeling procedure. A good practice, beyond the physical and engineering components, is the selection of a buffer that does not hinder cellular and biochemical parameters as well as cell recovery. In the present work the impact of four buffers on biochemical, morphological, and mechanical parameters was evaluated in two different cancer cell lines (Caco-2 and K562). Specifically, MTT ([3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]) assay along with flow cytometry analysis were used to evaluate the occurring changes in terms of cell viability, morphology, and granulocyte stress formation, all factors directly influencing DEP sorting capability. Quantitative real-time PCR (qRT-PCR) was instead employed to evaluate the gene expression levels of interleukin-6 (IL-6) and inducible nitric oxide synthase (iNOS), two well-known markers of inflammation and oxidative stress, respectively. An additional marker representing an index of cellular metabolic status, i.e. the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, was also evaluated. Among the four buffers considered, two resulted satisfactory in terms of cell viability and growth recovery (24 h), with no significant changes in cell morphology for up to 1 h in suspension. Of note, gene expression analysis showed that in both cell lines the apparently non-cytotoxic buffers significantly modulated IL-6, iNOS, and GAPDH markers, underlining the importance to deeply investigate the molecular and biochemical changes occurring during the analysis, even at apparently non-toxic conditions. The selection of a useful buffer for the separation and analysis of cells without labeling procedures, preserving cell status, represents a key factor for DEP analysis, giving the opportunity to further use cells for additional analysis.
Grpahical abstract
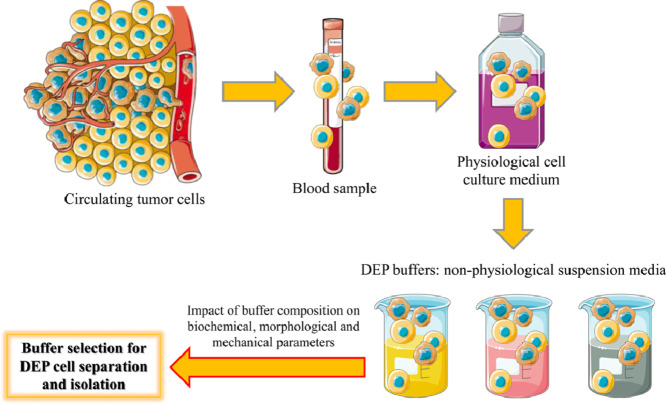
Introduction
The term dielectrophoresis (DEP), coined by Pohl [1], refers to an effect in which a particle is carried based on its dielectric properties. Pohl's early interest in this subject was sparked by an industrial need, specifically the challenge of removing carbon-black filler from polyvinyl chloride samples. Later, as reported in his seminal book [2], he focused his attention on the development of methodologies allowing the dielectrophoretic characterization and separation of bacteria as well as living cells. Since then the number of publications related to DEP has increased considerably [3], [4], [5], [6]. DEP is thus well-established as a label-free process allowing the identification and manipulation of a wide range of particles, from inert particles to proteins and cells. DEP has been used in different applications such as biosensors, cell therapies, drug discovery [7], medical diagnostics [8], microfluidics [9], nano assembly, and particle filtration, just to name a few. Circulating tumor cells (CTCs) isolation is still challenging, mainly due to their heterogeneity and rarity [10]. DEP has been considered as a molecular label-independent method for the isolation of CTCs from the whole blood. During the last decades numerous research studies have been focused on the development of different methods for enriching and isolating CTCs through the use of biomarkers, such as the epithelial cell adhesion molecule (EpCAM) [11], as well as on the development and application of the first automated system (CellSearch) currently approved for clinical use by the US Food and Drug Administration (FDA) for CTC detection, now considered as the gold standard among CTC detection technologies [12]. Despite that, numerous studies have recently shown that a universal marker for CTC detection does not exist [13]. In this context, EpCAM-positive circulating epithelial cells have been seen in patients with benign colon illnesses [14], suggesting that they could represent a source of misleading “positive” results. It is also worth of mention that cancer cells can go through a process known as epithelial-mesenchymal transition (EMT), which results in the loss of epithelial markers including EpCAM and cytokeratin and the emergence of mesenchymal markers [15], potentially leading to false “negative” results, with the additional complications that cells undergoing EMT are known to be highly aggressive and contribute to the development of metastases [16], [17], [18]. Based on the above, EpCAM cannot be considered an universal CTC detection marker. In this context DEP could be an innovative and useful tool, allowing cells’ separation based on both phenotype and membrane capacitance, without the need for labeling and/or modifications [19]. ApoStream™ currently represents the most established commercial DEP-platform [20] allowing for downstream enumeration and characterization of all CTCs from the whole blood, independently of EpCAM-based enrichments. This technology uses a DEP separation scheme known as field-flow-fractionation (DEP-FFF) to isolate non-hematopoietic cells from the peripheral blood mononuclear fraction. To selectively separate a target cell type from a cell mixture by DEP, the field frequency is adjusted to where the target cells exhibit a DEP polarity of the opposite sense to that of the other cells in the mixture. For this so-called “DEP cross-over frequency” to exist, the electrical conductivity of the buffer solution must be much less than the effective conductivity (∼1.2 S/m) of a cell's interior [6]. For the ApoStream™ system the cells are suspended in an optimized isotonic buffer of conductivity 30 mS/m, and subjected to an alternating electric field gradient for a period of around 60 min [21]. Transfer of the cells from a physiological fluid (conductivity ∼1.5 S/m) to the DEP buffer is accomplished without centrifugation using a continuous flow, ion-diffusion, system [21,22]. A similar method in principle to this employs tangential flows of a physiological fluid and a 9.4 mS/m DEP buffer [23]. Such procedures avoid the perceived problem [24] where, as a result of ion leakage from the cells, the buffer conductivity increases to the extent that adjustment of the field frequency is required. The strength of the applied field strength and its gradient are well below the limit to introduce permanent pores in a cell plasma membrane [25] and the rate of fluid flow is such that the Stokes drag force and shear stresses acting on the cells is negligible [21]. For DEP procedures that employ a typical buffer and last for no longer than 1∼2 h, it is generally observed that minimal time-dependent changes occur in a cell's DEP response. Based on the capability of colony formation [26], fluorescent staining, and adherence morphology tests (e.g., [27,28]), exposure of the cells to the imposed electric field, fluid flow stresses, and a typical DEP buffer do not appear to reduce cell viability. However, in our view, the potential risk of a transient, or at worst an irreversible, activation/modification of the cells resulting from their suspension in a typical DEP buffer, has not been thoroughly investigated. In the present study, we focused our attention on the possible stress induced by different non-physiological suspension media on cancer cells of different origin, namely Caco-2 and K562 cells. Our study included four buffers characterized by different compositions and conductivities. In particular, Buffer 4, that has been already employed in a previous study [19], was tested in the presence or absence (Buffer 3) of a conductivity adjustment. The impact of buffers’ composition on cell viability after 1 h or 24 h of incubation was investigated by employing the well-known MTT ([3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]) assay, while the changes related to cell morphology at the same time points were determined by cytofluorimetric analysis. Quantitative real-time PCR (qRT-PCR) was carried out to determine the modulation of the gene expression levels of specific biomarkers representative of cells metabolism (glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) [29], oxidative stress (inducible nitric oxide synthase (iNOS)) [30] and inflammation (interleukin-6 (IL-6)) [31], with the aim to give a better overview of the impact of the different buffers on cell status.
Materials and methods
Materials and reagents
All chemicals were supplied by Merck (Merck KGaA, Darmstadt, Germany) unless specified otherwise. All the reagents and chemicals used in the present study were of analytical grade.
Buffer compositions and electric conductivity measurements
The composition of the four buffers employed for the experiments along with the tested conductivity are reported in Table 1.
Table 1.
Chemical composition of the four buffers used in the present study. All buffers were prepared in Ultra-Pure MilliQ® Water.
| Buffer 1 | Buffer 2 | Buffer 3 | Buffer 4 [19] |
|---|---|---|---|
| ü 1X phosphate buffered saline (PBS) pH 7.0, 1%; ü 1X fetal bovine serum (FBS), 2%; ü 0.1% bovine serum albumin (BSA), 2%. § Tested conductivity: 33 mS/m. |
ü 1X FBS, 2%; ü Sucrose 9.5%; ü Glucose 0.1 mg/mL; ü 0.1 BSA, 2%. § Tested conductivity: 58 mS/m. |
ü Sucrose 9.5%; ü Dextrose 0.1 mg/mL; ü 0.1% BSA, 2%. ü 1X PBS pH 7.0, 1%; ü (CH3COO)2Ca 0.1 mM. § Tested conductivity: 296 mS/m. |
ü Sucrose 9.5%; ü Dextrose 0.1 mg/mL; ü 0.1% BSA, 2%. ü 1X PBS pH 7.0, 1%; ü (CH3COO)2Ca 0.1 mM. § Tested conductivity: 33 mS/m. |
Once prepared, all buffers were filtered by using a Millex Millipore syringe with 0.22 μm pores (Biosigma S.p.A., Cona, Italy). Buffer 1 and Buffer 4 conductivities were adjusted with a potassium chloride (KCl) solution (0.1 mM) obtaining a final conductivity of 33 mS/m (representing the most widely used [23]) that was measured with an Electric Conductometer DDS Digital Lab (DDS-307, Changzhou W&J Instrument Co., Ltd., Changzhou, Jiangsu, China).
Cell culture, treatment protocol, and evaluation of cell viability
Caco-2 cells (HTB-37TM; human colorectal adenocarcinoma, American Type Culture Collection, Manassas (ATCC), VA, USA) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with heat-inactivated FBS (10%), L-Glutamine (2 mM), penicillin-streptomycin (50 units-50 µg for mL) and maintained in 25 or 75 cm2 polystyrene cell culture flasks with a vent cap at 37 °C in a humidified atmosphere of 5% CO2/95% air. K562 cells (CCL-243TM; chronic myelogenous leukemia from human blood cells, ATCC) were grown in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with heat-inactivated FBS (10%), L-Glutamine (2 mM), penicillin-streptomycin (50 units-50 µg for mL) and maintained in 25 or 75 cm2 polystyrene cell culture flasks with a vent cap at 37 °C in a humidified atmosphere of 5% CO2/95% air. Despite being both cancer cell lines their origin as well as their characteristics are very different. Caco-2 are epithelial cells isolated from colon tissue derived from a 72-year-old, White, male with colorectal adenocarcinoma, while K-562 are lymphoblast cells isolated from the bone marrow of a 53-year-old chronic myelogenous leukemia patient. Additionally, the selected cell lines are enough distant in terms of crossover frequency [10].
Twenty-four hours prior to treatment, cells were harvested, counted by using a C-Chip disposable hemocytometer, and seeded in 96-well plates at the appropriate density. Cell culture medium was then replaced with 100 µL of one of the four different buffers and cells were incubated for 1 h or 24 h. At the end of the incubation time, to evaluate the effect of the different buffers on the viability of Caco-2 and K562 cells, an MTT assay was performed as previously described [32].
Analysis of discrimination capacity
In order to evaluate the ability of FlowSight® Imaging Flow Cytometer (Amnis® FlowSight® Millipore, Merck KGaA, Darmstadt, Germany) to discriminate cells features in buffers containing increasing concentrations of sodium chloride (NaCl), seven NaCl solutions (0%, 0.3%, 0.6%, 0.9% (NaCl concentration normally used for “saline” [33]), 1.5%, 3%, and 4%) were prepared. Caco-2 cells were incubated for 1 h at 37 °C with each of the above-mentioned solutions at the end of which they were analyzed by using the FlowSight® Imaging Flow Cytometer, which quantitatively detects brightfield, darkfield, and fluorescent images with a high sensitivity. A detailed analysis of the intensity was achieved by using IDEAS® (Image Data Exploration and Analysis Software), a powerful tool allowing the robust statistical analyses of images as well as of hundreds of morphological features (in addition to intensity). The two main parameters selected and analyzed were perimeter and side scatter (SSC). The measurement of the SSC provides information about the internal complexity (i.e. granularity) of a cell.
Sample preparation protocol for flow cytometry
Caco-2 and K562 cells were centrifuged, the supernatant was removed, and cells were resuspended in 300 μL of 1X PBS. One hundred μL of each cell suspension were added into 1.9 mL of Buffer 1, Buffer 4, or Minimum Essential Medium (MEM) without phenol red (control solution). An aliquot of each cell suspension was taken at four different time points indicated as T0 (0 min), T1 (15 min), T2 (30 min), T3 (45 min), and T4 (60 min) and analyzed in terms of Circularity, Area vs. Diameter, and SSC, representing parameters that are normally quickly modulated. The withdrawals made and the related time points are reported in Supplementary Table 1. The cell suspension aliquots needed for qRT-PCR analysis were stored in 350 μL of Buffer RLT, part of the RNA extraction kit (Qiamp RNeasy Mini Kit, Qiagen, Hilden, Germany) and β-Mercaptethanol (1%) until used.
Gene expression analysis by qRT-PCR
Gene expression analysis by qRT-PCR was performed at T4 (120 min) and T5 (240), since significant changes in the expression of our targets, representative of metabolism, oxidative stress, and inflammation, are already detectable at these time points [29,34]. The RNA was extracted from the previously stored samples by using the Qiamp RNeasy Mini Kit following the manufacturer's instructions. Both integrity and quantification of the RNA were assessed by using Agilent RNA 6000 Nano Kit (Agilent, Santa Clara, CA, USA) on a 2100 Bioanalyzer Instrument (Agilent). In order to peform reverse transcription, sample amplification, fluorescence data collection, and sample quantification, the same protocol as previously described was used [35,36]. The following QuantiTect Primer Assays (Qiagen) were used for the expression analysis of β-actin (Cat. No. QT00095431), iNOS (also known as nitric oxide synthase 2: Cat. No. QT00100275), IL-6 (Cat. No. QT00083720), and GAPDH (Cat. No. QT01658692). Additional information regarding QuantiTect Primer Assays employed for the gene expression analysis are reported in Supplementary Table 2. As a negative control, a reaction in the absence of cDNA (no template control, NTC) was performed. The relative RNA expression level for each sample was calculated using the well-known 2−∆∆CT method in which the threshold cycle (CT) value of the gene of interest is compared to the CT value of our selected internal control (β-actin). qRT-PCR amplifications were performed in quadruplicate.
Measurement of osmolarity
The osmolarity of both Buffer 1 and Buffer 4 was determined by analyzing 50 μL of each solution in a Gonotec® Osmomat® Model 030-D freezing point Osmometer (ELITech Group, Logan, Utah, USA).
Statistical analysis
In the case of qRT-PCR data, statistical analysis was performed by using version 8 of GraphPad Prism software. In the case of multiple comparisons, one-way ANOVA followed by Bonferroni's post hoc test was used. Only two-tailed p-values less than 0.05 were considered statistically significant. All experiments were performed at least in triplicate unless specified otherwise.
Results
Buffer composition differently influence the viability of Caco-2 and K562 cells
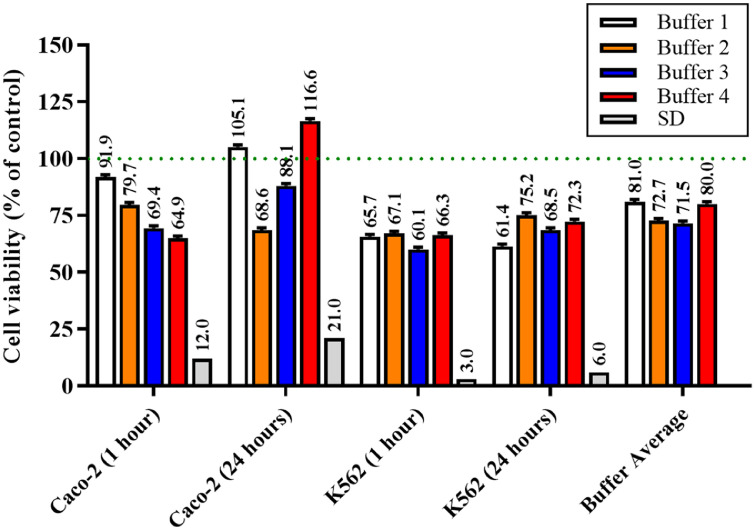
The effects of the four different buffers on the viability of Caco-2 and K562 cells at 1 h and 24 h are reported in Fig. 1.
Fig. 1.
Changes in cell viability of Caco-2 and K562 cells in the presence of the four different buffers after 1 h and 24 h. Data are the mean of four values and are expressed as the percent variation with respect to the absorbance at 569 nm detected in cells grown in the presence of MEM (physiological cell culture medium; dotted line). SD = standard deviations among the four buffers for a specific condition; Buffer Average = mean of the four cell viability (%) values (Caco-2 at 1 h and 24 h; K562 at 1 h and 24 h) measured for each buffer.
Buffer 1 slightly effects Caco-2 cell viability at both time points, giving values comparable to that observed for controls (cells grown in the presence of MEM) (91.9% at 1 h; 105.1% at 24 h). Buffer 2, normally employed for flow cytometry experiments, decreased Caco-2 cell viability in a time-dependent manner (79.7% at 1 h; 68.6% at 24 h), suggesting that cells are suffering as a consequence of the incubation with this buffer. A different effect was observed when employing Buffer 3; in fact, despite the decrease in cell viability observed at 1 h (69.4%), cells were able to recover at 24 h, giving a value of cell viability equal to 88.1% compared to control conditions. Interestingly, Buffer 4, showing the highest decrease among all the buffers used at 1 h (64.9%), did not affect the cell viability at 24 h, giving a value even higher (116.6%) than that observed in control cells, sign of a complete recovery within 24 h. When considering the effects of the buffer composition on the viability of K562 cells at 1 h, a similar decrease in cell viability (∼25%) was observed for all the buffers tested. Differently from what observed for Caco-2 cells, K562 cells were unable to completely recover under the four experimental conditions at 24 h. The lowest cell viability decrease was observed for Buffer 2 (75.2% vs. control) and, once again, for Buffer 4 (72.3% vs. control). From this set of data, we were able to select the buffers (1 and 4) that have the least influence on cell viability, hence being the most suitable for subsequent experiments.
NaCl concentration influences both FlowSight® Imaging Flow Cytometer outcomes and cell size
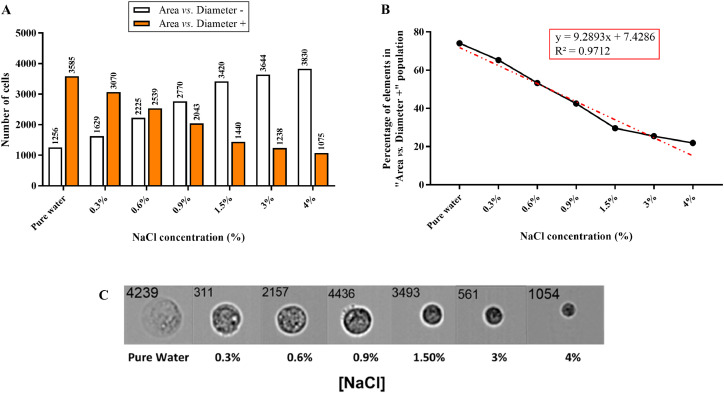
As previously mentioned, to evaluate the ability of FlowSight® Imaging Flow Cytometer to discriminate cells features in solutions containing increasing concentrations of NaCl, seven NaCl solutions (0%, 0.3%, 0.6%, 0.9%, 1.5%, 3%, and 4%) were prepared and their effects tested on Caco-2 cells.
For each of the analyzed solutions, a total of 5000 events were recorded. The tool defines a cut-off by combining “Area” and “Diameter” parameters, giving two populations as an output: a population in which the aforementioned geometric parameters have a value that is lower than that of the cut-off (Area vs. Diameter –) and a population in which they have a value higher than that of the cut-off (Area vs. Diameter +). With regard to Area vs. Diameter – population, the number of cells included in this population increases by increasing the concentration of NaCl (Fig. 2A).
Fig. 2.
Effects of increasing concentrations of NaCl on the ability of FlowSight® Imaging Flow Cytometer to discriminate cells features. (A) Variation of cells’ number in “Area vs. Diameter –” and “Area vs. Diameter +” populations as a function of NaCl concentration. (B) Variation of the percentage of elements (cells) in “Area vs. Diameter +” (5000 acquisitions total) as a function of NaCl concentration. (C) Effect of NaCl concentration salt on cell size.
This is most probably the consequence of the osmotic effect, causing a flow of water from the weak solution (inside the cells) to the strong solution (outside the cells) and leading to a reduction of cells’ size (Fig. 2C). This phenomenon is strengthen by the results plotted in Fig. 2B, showing a linear correlation between the independent variable “NaCl concentration” and the dependent variable “Percentage of elements in “Area vs. Diameter +” population”. The equation of the trend line is: y = 9.2893x + 7.4286 (R2 = 0.9712).
Buffer composition differently influences circularity, SSC, and Area vs. Diameter parameters in Caco-2 and K562 cells
Table 2 reports the measurements of circularity, SSC, and Area vs. Diameter in both populations (“Area vs. Diameter –“ and “Area vs. Diameter +”) of Caco-2 and K562 cells at different time points (from T1 to T4).
Table 2.
Measurements of Circularity, SSC, and Area vs. Diameter parameters at four different time points (T1 = 15 min, T2 = 30 min, T3 = 45 min, T4 = 60 min) in Caco-2 and K562 cells. Percentages were calculated on a total of 5000 acquisitions (cells).
| Caco-2 | K562 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | |||||||||||
| T1 | T2 | T3 | T4 | T1 | T2 | T3 | T4 | ||||
| Circularity | - | 13.9 | 15.7 | 13.6 | 16.8 | Circularity | - | 15.1 | 14.6 | 15.8 | 13.3 |
| + | 85.9 | 84.3 | 86.3 | 83.1 | + | 84.9 | 85.4 | 84.2 | 86.7 | ||
| SSC | - | 45.9 | 85.5 | 86.1 | 87.7 | SSC | - | 97.6 | 97.8 | 97.5 | 98.1 |
| + | 53.5 | 13.1 | 12.7 | 11.6 | + | 2.27 | 2.07 | 2.23 | 1.47 | ||
| Area vs. Diameter | - | 17.7 | 44.4 | 48.8 | 51.8 | Area vs. Diameter | - | 34.9 | 43.7 | 39.8 | 40.6 |
| + | 78.4 | 50.6 | 46.1 | 44.2 | + | 60.9 | 52.9 | 56 | 55.2 | ||
| Buffer 1 | |||||||||||
| T1 | T2 | T3 | T4 | T1 | T2 | T3 | T4 | ||||
| Circularity | - | 11.2 | 9.4 | 6.53 | 7.2 | Circularity | - | 19.8 | 26.6 | 25 | 19.6 |
| + | 88.7 | 90.5 | 93.4 | 92.5 | + | 80.2 | 73.3 | 74.9 | 80.2 | ||
| SSC | - | 89 | 89.4 | 95.8 | 69 | SSC | - | 94 | 92.6 | 90.3 | 92.5 |
| + | 9.73 | 9.3 | 3.57 | 30.2 | + | 1.47 | 1.03 | 2.25 | 1.46 | ||
| Area vs. Diameter | - | 17.5 | 19.6 | 8.8 | 13.7 | Area vs. Diameter | - | 55.1 | 66.3 | 66.5 | 58.8 |
| + | 70.9 | 68.8 | 69.6 | 69.7 | + | 38.3 | 28.1 | 27.3 | 34.7 | ||
| Buffer 4 | |||||||||||
| T1 | T2 | T3 | T4 | T1 | T2 | T3 | T4 | ||||
| Circularity | - | 28.1 | 29.8 | 34.6 | 30 | Circularity | - | 24.8 | 19 | 26.5 | 27 |
| + | 71.9 | 70.2 | 65.4 | 70 | + | 75.2 | 81 | 73.5 | 73 | ||
| SSC | - | 72.9 | 70.5 | 62.8 | 64.9 | SSC | - | 98 | 97.7 | 97.9 | 98.1 |
| + | 23.8 | 26.1 | 34.4 | 31.4 | + | 1.73 | 2.13 | 1.9 | 1.63 | ||
| Area vs. Diameter | - | 64.3 | 67 | 70.8 | 68.2 | Area vs. Diameter | - | 57.8 | 54.2 | 64.5 | 65.8 |
| + | 30.9 | 28.3 | 25.4 | 27.7 | + | 38.4 | 41.4 | 32.1 | 30.5 | ||
The same data has been plotted in the Supplementary Fig. 1.
As reported in Table 2 and Supplementary Fig. 1, both Circularity and Area vs. Diameter parameters in Caco-2 cells were not affected by the incubation with the two buffers (1 and 4), while a different outcome was observed in the case of MEM giving a decrease from 78.4 (T1) to 50.6 (T2), 46.1 (T3), and finally 44.2 (T4), probably as a consequence of the trypsinization step required for cell detachment before the resuspension in one of the three solutions. With regard to SSC, this parameter was not affected by Buffer 4 that did not particularly affect cellular granularity, while this parameter increased in the presence of Buffer 1 (T4) and decreased when the cells were resuspended in MEM (T2, T3, and T4). Interestingly, Buffer 1 and Buffer 4 led to the same value of SSC at T4, hence leading to the same level of cell granularity. As expected, MEM, the medium normally used for cell culture, gave SSC values lower compared to that observed for Buffer 1 and Buffer 4, indicating a lower or absent stress.
Table 2 and Supplementary Fig. 1 also report the measurements of the same three parameters in K562 cells. In general, K562 cells were less susceptible to the solutions tested. Buffer 4 did not affect Circularity and SSC parameters, while a decrease of Area vs. Diameter was observed starting from T3. Some variations of the three parameters was measured at T2 and T3 in K562 cells resuspended in Buffer 1, but at T4 the measured values were comparable to that observed at T1, suggesting a transient modulation of them followed by a complete recover of the initial conditions. None of the parameters were affected by the resuspension of K562 cells in MEM. Illustrations of the three parameters evaluated for a random sample, obtained by using FlowSight® Imaging Flow Cytometer, are shown in Supplementary Fig. 2.
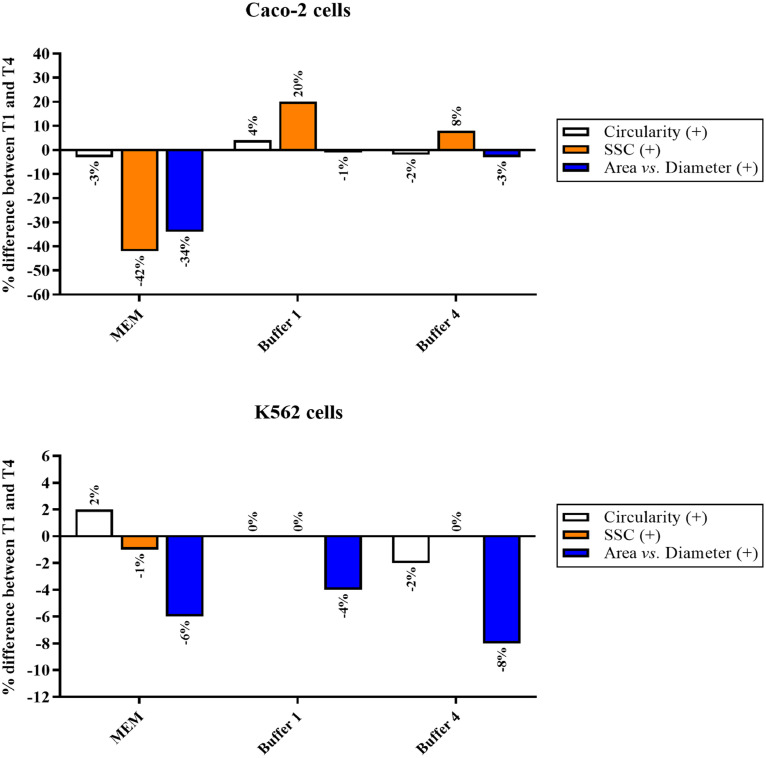
With regard to the three parameters evaluated (Circularity, SSC, and Area vs. Diameter), in order to better show the differences occurring between the start time point (T1) and the endpoint (T4), depending on the solution considered (MEM, Buffer 1, and Buffer 4), the above described results were expressed as a percentage difference (Fig. 3).
Fig. 3.
Percentage (%) difference of the three parameters between the start time point (T1) and the endpoint (T4) in Caco-2 and K562 cells. Each % difference was calculated as follows: (Parameter value at T4 – Parameter value at T1)/100.
Fig. 3 clearly shows how Buffer 4 represents the most suitable for Caco-2 cells, while Buffer 1 should be selected in the case of K562 cells. It is worth noting that in the case of Caco-2 cells the highest percentage difference for both Circularity and SSC was observed when resuspending the cells in MEM.
Buffer 1 and Buffer 4 differently effect the gene expression levels of GAPDH, iNOS, and IL-6 in Caco-2 and K562 cells
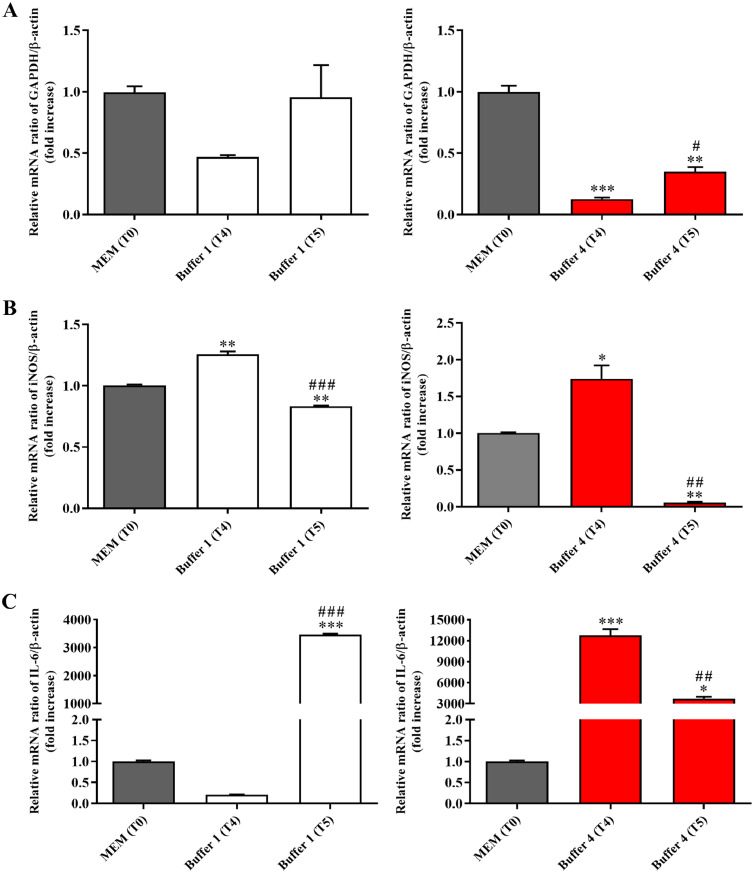
The quantification of each RNA extracted along with the related integrity value (RIN) are reported in Supplementary Table 3. In order to better understand the impact of the different buffers on cell status, the modulation of the gene expression levels of GAPDH (metabolism) [29], iNOS (oxidative stress) [30], and IL-6 (inflammation) [31] was investigated by qRT-PCR. In particular, the mRNA ratio of target gene/β-actin (fold increase) at T4 and T5 was compared to that observed in control conditions, identified as the same ratio in cells resuspended in MEM at T0. As shown in Fig. 4A, Buffer 1 did not alter GAPDH gene expression levels in Caco-2 cells at both time points, even though a trend towards the reduction of metabolism was observed in the case of T4.
Fig. 4.
Effects of the exposure to Buffer 1 or Buffer 4 for 60 min (T4) or 120 min (T5) on expression levels of targets related to (A) metabolism (GAPDH), (B) oxidative stress (iNOS), and (C) inflammation (IL-6) in Caco-2 cells. The abundance of each mRNA of interest was expressed relatively to the abundance of β-actin mRNA, as an internal control. The relative mRNA ratio of target gene/β-actin (fold increase) measured for Buffer 1 or Buffer 4 at both time points was compared to that of control conditions identified as cells resuspended in MEM at T0. Data are represented as means ± standard deviation (SD). *significantly different from T0, p < 0.05; **significantly different from T0, p < 0.01, ***significantly different from T0, p < 0.001; #significantly different from T4, p < 0.05; ##significantly different from T4, p < 0.01, ###significantly different from T4, p < 0.001.
Buffer 1 significantly increased the expression of iNOS at T4 but, as observed at T5, the modulation of this gene was transient, being the gene expression levels at this time point comparable to that observed at T0 (Fig. 4B). A very interesting result was observed when measuring the effects of Buffer 1 on IL-6 expression; in fact, as reported in Fig. 4C, differently from T4, Buffer 1 at T5 strongly enhanced the expression levels of IL-6 compared to T0. Buffer 4 significantly decreased GAPDH expression levels at both time points in Caco-2 cells compared to control conditions (Fig. 4A). In line with what observed for buffer 1 at T4, Buffer 4 significantly increased the expression of iNOS at the same time point (stronger induction compared to Buffer 1), but an opposite effect was observed at T5, where the expression levels were even lower than that observed at T0 (Fig. 4B). A very signifcant induction of IL-6 gene expression was measured in Caco-2 cells resuspended in Buffer 4, at both time points (Fig. 4C).
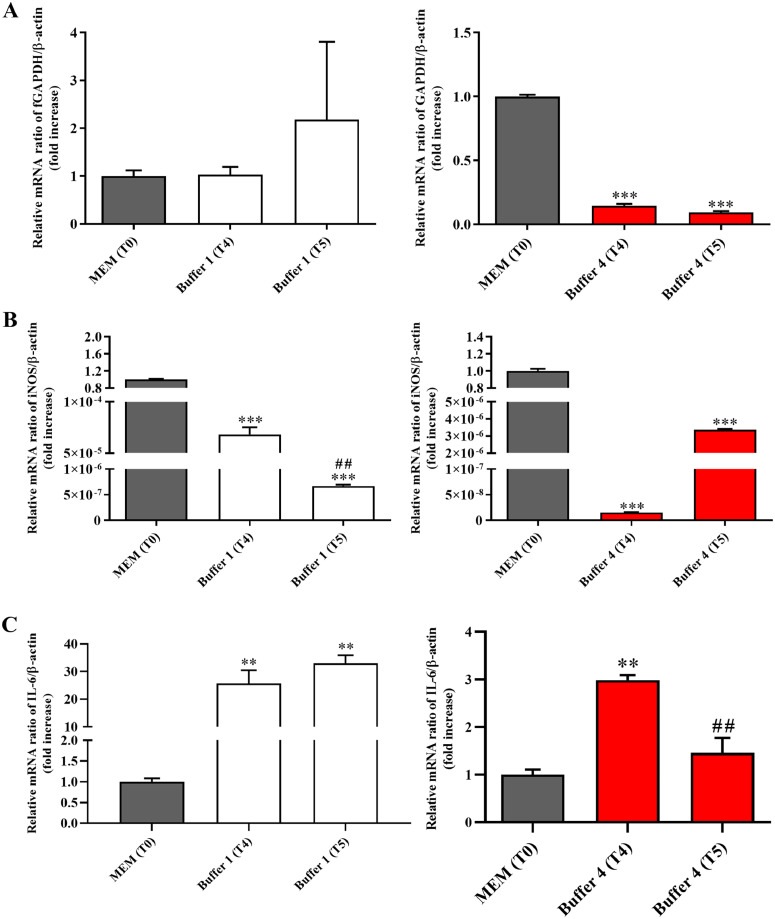
As observed in the case of Caco-2 cells, Buffer 1 did not alter GAPDH gene expression levels in K562 cells at both time points (Fig. 5A).
Fig. 5.
Effects of the exposure to Buffer 1 or Buffer 4 for 60 min (T4) or 120 min (T5) on expression levels of targets related to (A) metabolism (GAPDH), (B) oxidative stress (iNOS), and C) inflammation (IL-6) in K562 cells. The abundance of each mRNA of interest was expressed relatively to the abundance of β-actin mRNA, as an internal control. The relative mRNA ratio of target gene/β-actin (fold increase) measured for Buffer 1 or Buffer 4 at both time points was compared to that of control conditions identified as cells resuspended in MEM at T0. Data are represented as means ± standard deviation (SD). **significantly different from T0, p < 0.01, ***significantly different from T0, p < 0.001; ##significantly different from T4, p < 0.01.
Opposite results were observed when comparing the effects of Buffer 1 on the expression of iNOS and IL-6; in fact, from one hand, Buffer 1 significantly decreased iNOS expression levels at both time points in K562 cells (Fig. 5B), from another hand the same buffer strongly increased the expression of IL-6 at T4 and T5 (Fig. 5C). When considering the effect of Buffer 4 on K562 cells, a significant decrease in terms of GAPDH (Fig. 5A) and iNOS (Fig. 5B) expression levels was observed at both time points when compared to control conditions. An opposite effect, i.e. an up-regulation, was detected in the case of IL-6 at T4, while the longest incubation time (T5) gave results comparable to that observed at T0.
Caco-2 cells are more susceptible to osmolarity changes than K562 cells
The osmolarity of Buffer 1, determined by freezing point, was equal to 0.012 osmol/Kg, while that measured for Buffer 4 was 0.320 osmol/Kg. As already described in literature [37,38], osmolarity represents an important parameter able to modulate cells features and morphological parameters. The lower value of osmolarity of Buffer 1 is most probably connected to the higher percentage of Caco-2 cells part of the “Area vs. Diameter +” population (Table 2). An opposite situation, according to the higher osmolarity, is observed in the case of Buffer 4, with a higher percentage of Caco-2 cells part of the “Area vs. Diameter -” population. Interestingly, K562 cells are less susceptible to the changes in osmolarity, giving almost superimposable percentage values of “Area vs. Diameter +” and “Area vs. Diameter -” populations for both buffers (1 and 4).
Discussion
CTCs represent biomarkers allowing the non-invasive measure of the evolution of tumor genotypes during treatment and disease progression, thus offering the possibility to obtain key biological information extremely important in the context of personalized medicine [39]. As previously mentioned, numerous current methods are based on EpCAM detection, even though it has been shown that it does not represent an universal marker for CTCs detection, failing, for example, in the detection of carcinoma cells undergoing EMT as well as in the case of CTCs of mesenchymal origin [39,40]. These issues along with the fact that EpCAM expression has also been found in patients with benign diseases emphasizes the need for non-EpCAM (and, more broadly, label-free) approaches for the detection and isolation of CTCs.
During the last decade, DEP, a label-free method taking advantage of the intrinsic dielectric properties of suspended cells, has emerged as a promising tool for the isolation of CTCs from the whole blood [10,41]. Moreover, it is important to mention that, once isolated, CTCs are viable and can be maintained in culture, suggesting that DEP-based methods could be more generally applicable than the well-known antibody-based affinity methods [42]. One challenge in employing any DEP device is the sample being separated must be transferred into an ultralow conductivity medium, which can be detrimental in retaining cells’ native phenotypes (e.g., alterations of biochemical, morphological, and mechanical parameters) for separation [43]. In fact, since during DEP analysis cells are suspended in a buffer, it is important to examine whether the selected buffer in any way damages cells or affects their characteristics in subsequent analytical procedures. Several research studies have been devoted to this matter. For example, in a study carried out by Becker et al., the azo dye trypan blue was used to monitor the survival of the erythrocytes’ population separated from leukemia cells [44]. In a different study, the CD34+ cells obtained by DEP enrichment from bone marrow and peripheral stem cell harvests were successfully grown [33]. It was also demonstrated that even though fibroblasts are exposed to DEP fields for up to three days, cells can still be grown without significant changes in terms of viability, motility, anchoring, or cell-cycle time [34]. On the other hand, it has been shown that a 1 h DEP treatment/manipulation can increase cell immuno-reactivity, while a longer time (up to 4 h) can reduce the number of viable cells [45].
In order to shed more light on the above described conflicting results, in the present work the impact of four different buffers, including the one normally used for flow cytometry analysis (Buffer 2) on biochemical, morphological, and mechanical parameters was evaluated in two different cancer cell lines, Caco-2 and K562. Caco-2 were selected as a model of solid tumor cells [46], while K652 were selected as a model of liquid (blood) tumor cells [47].
As reported in Fig. 1, the selection of the buffer represents a key point for the subsequent analysis. In fact, Buffer 2, normally employed for flow cytometry experiments, and Buffer 3, characterized by a very high conductivity (296 mS/m) strongly affected cell viability, especially in the case of Caco-2 cells. These results are important in the light of the fact that: (1) CTCs are rare in the blood (1 CTC/109 blood cells) [48], therefore even the loss of a small number of cells can be crucial; (2) even if still alive, cells could undergo changes that modify the state they had at the time of collection from the patient. It is also worth mentioning that Buffer 4, the one giving the highest decrease in cell viability among all the buffers used at 1 h, did not affect the cell viability at 24 h, giving a value even higher than that observed in control cells, sign of a complete recovery within 24 h. A cell viability value (%) higher than that observed for control cells can be considered an increase in metabolic rate and/or proliferation rate [49], in line with cells that are trying to recovery from transient adverse conditions. Different factors could be responsible for the observed transient decrease of cell viability. The first thing to take into account is the change of environment, i.e. from the complete growing medium to one of the non-physiological suspension media (satisfactory even though not corresponding to the ideal conditions). It can lead to transient alterations regarding the cell metabolism as well as the cellular structure. One of the reason why cells can transiently suffer after switching the medium can be the modulation of the intracellular calcium (Ca2+) concentrations; in fact, it has been already shown that the presence of exogenous buffer is able to modify the Ca2+ transients to a variable extent depending on its proportion relative to the natural, intrinsic buffers [50]. Additionally, most cell culture media are formulated with a CO2-carbonate buffering system that is optimized for tissue culture CO2 partial pressure, which is higher than the typical ambient CO2 partial pressure. When these types of buffers are left in typical atmospheric conditions (e.g., “room temperature”), CO2 will evaporate from the medium and cause the pH of the medium to rise into the alkaline range, which expectedly can significantly affect cell viability. A third noteworthy variable relates to cooling and freezing; in fact, as recently described by Wolkers et al. [51], cellular membranes can undergo thermotropic and lyotropic phase transitions, which significantly alter membrane permeability and barrier function. Membrane permeability to water and solutes could be affected by temperature (e.g., from 37 °C to RT), medium osmolality, solute type, and cell hydration level. If the above-mentioned factors along with other cell-specific susceptibility factors do not particularly alter cellular metabolism, cell growth and replication can resume even if the cells are still exposed to disturbing factors [52].
Another factor to be considered during DEP analysis is the concentration of salt ions [53]. In fact, as shown in Fig. 2, the increasing concentrations of NaCl strongly influenced both the ability of FlowSight® Imaging Flow Cytometer to discriminate cells features (e.g., number of cells within a specific cell population) and cell size, with the 0.9% NaCl (normally used for “saline”) which appears to be a good compromise; despite that, it has been demonstrated as large-volume saline infusion in healthy people is able to induce hyperchloremia which in turn is associated with metabolic acidosis, hyperkalemia, and negative protein balance [33], reason why the use of this highly acidic solution should be carefully evaluated in both preclinical and clinical settings. It is well-know that cancer disease represents a heterogenous pathology, the development of which often depends on the synergic interaction between molecular features and phenotypic context. These complex and altered molecular pathways are potentially different between patients, and in some case even in the same patient at a different stage of disease development [54]. One key characteristic of a tumor is the intra-tumor heterogeneity; the differences identified between the same tumors coming from different patients are referred to as “inter-tumor heterogeneity”, while “intra-tumor heterogeneity” refers to different tumor cell populations, often characterized by very different genetic and phenotypic profiles, within the same tumor specimen [55]. Taking into consideration this information, it becomes clear how the maintenance of the cell characteristics immediately before, during, and, in the best scenario, at the end of the analysis is of utmost importance. As previously mentioned, CTCs preserve primary tumor heterogeneity and mimic tumor properties, and may be considered a therapeutic target, being a component of liquid biopsy [56]. We investigated the effects of the buffer composition on our cell models at two levels: (1) evaluating Circularity, SSC, and Area vs. Diameter through flow cytometry; (2) measuring the expression levels of GAPDH (metabolism marker), iNOS (oxidative stress marker), and IL-6 (inflammation marker) by using qRT-PCR. The results reported in Table 2 and Supplementary Figs. 1 and 2 highlight the importance of the selection of buffer as a consequence of the cell model selected. In fact, as previously described in the pertinent section, Buffer 4 seems to be the most suitable in the case of Caco-2 cells, while Buffer 1 should be selected for studies employing K562 cells. These results along with that reported in Figs. 4 and 5, showing that Buffer 1 and Buffer 4 differently, and very often significantly, affect the gene expression levels of GAPDH, iNOS, and IL-6 in Caco-2 and K562 cells, underline, once again, the importance of monitoring analysis’ conditions on cell status. This point is crucial, since once CTCs are extracted from the blood of cancer patients, reculturing them give the opportunity to perform studies regarding phenotypic heterogeneity, along with the measurements of parameters related to metabolism and cell replication [57]. Culturing CTCs also represents a promising approach for identfying a pharmacological target and individualize drug susceptibility tests in cancer therapy [58].
The coupling of microfluidic-based systems to DEP has already shown the potential to enhance cancer cell discrimination [59,60]. The rapid development of fluorescent analysis techniques combined with microfluidic chips have offered a widely applicable solution, including the possibility to monitor the production or uptake of bioactive molecules, thus allowing a better understanding of cell biology, exploring cell heterogeneity, and enhancing the ability to detect a disease at an early stage [61], [62], [63]. Therefore it appears clear how microfluidics coupled to fluorescence can increase not only the ability to monitor cell status (viability, morphology, etc.) during the analysis, but also cancer cell discrimination, independently from the differences of membrane permeability and mitochondrial membrane potential between cancer (higher) and normal (healthy) cells [64]. In this context, Yan et al. developed a microfluidic DEP-active hydrophoretic sorter for particle and cells sorting based on size and on dielectric properties of the particles or cells of interest without any labeling [65]. By using this hybrid device and according to the different Re[fCM] [66] of live and dead Chinese Hamster Ovary cells at the medium conductivity of 0.03 S/m, it was possible to separate and collect the cells of interest with high separation efficiency (99.6 ± 0.2%). Still in the context of microfluidics, a very innovative approach able to ensure that sensitive cells are not subject to centrifugation for resuspension and spend minimal time outside of their culture media, reducing cellular stress has been recently published by Huang et al. [23]. The integration of this on-chip sample preparation platform prior to or post-DEP, in-line with on-chip monitoring of several factors such as the outlet sample for metrics of media conductivity as well as cell velocity and viability, has been proposed. A continuous flow microfluidic processing chamber into which the peripheral blood mononuclear cell fraction of a clinical specimen is slowly injected, deionized by diffusion, and then subjected to a balance of DEP, sedimentation and hydrodynamic lift forces has also been described [22]. Several additional microfluidic-based technologies taking advantage of using impedance cytometry for label-free single-cell monitoring based on plasma membrane capacitance and apoptotic states have been proposed, allowing label-free separation and monitoring of live CTC sub-populations [67,68], strengthening the usefulness of microfluidics in the context of DEP.
An additional help to improve cell sorting and monitoring cells’ conditions is represented by next-generation sequencing (NGS). In fact, the most powerful genetic tool available for mutational analysis of single CTCs is currently represented by the combination of NGS to whole genome amplification (WGA) [69]. The rapid advancement of NGS has resulted in a significant improvement in the molecular profiling accuracy, allowing non-invasive and real-time detection of novel biomarkers for cancer screening and dynamic disease monitoring [70].
Conclusions
Cell sorting and separation have become key diagnostic, research, and treatment tools for personalized medicine. DEP provides a biophysical separation technique able to target different cell populations based on their phenotypes without labels, returning native cells for downstream analysis. One of the hardest challenges when using DEP is represented by the fact that the cells being separated need to be transferred form a “normal” culture medium into an ultralow conductivity medium, which can be detrimental for the retention of the “original” cell phenotypes. In the present work the impact of four buffers on biochemical, morphological, and mechanical parameters was evaluated in Caco-2 (solid tumor) and K562 (liquid tumor) cells. Among the four buffers considered, two (Buffer 1 and 4) were considered satisfactory in terms of cell viability and growth recovery (24 h), with no significant changes of morphology for up to 1 h in suspension. Despite that, the analysis of IL-6, iNOS, and GAPDH markers at gene level showed significant variations as a consequence of the transfer of the cells from MEM (normal culture medium) to the DEP buffers, strengthening the importance to select the appropriate experimental conditions (including buffer selection), allowing not only the discrimination between different cell populations (e.g., CTCs vs. healthy cells), but also giving the opportunity to analyze the cells without stressing them as well as to further use cells for additional analysis.
Funding
G.C. is a researcher at the University of Catania within the EU-funded PON REACT project (Azione IV.4—“Dottorati e contratti di ricerca su tematiche dell'innovazione”, nuovo Asse IV del PON Ricerca e Innovazione 2014–2020 “Istruzione e ricerca per il recupero—REACT—EU”; Progetto “Identificazione e validazione di nuovi target farmacologici nella malattia di Alzheimer attraverso l'utilizzo della microfluidica”, CUP E65F21002640005). This work has been partially funded by European Union (NextGeneration EU), through the MUR-PNRR project SAMOTHRACE (ECS00000022) and through the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT), and by Italian Ministry of Health Research Program (grant number RC2022-N4).
Institutional review board statement
N/A.
Informed consent statement
N/A.
CRediT authorship contribution statement
Paolo G. Bonacci: Investigation, Formal analysis, Visualization, Methodology, Writing – original draft, Writing – review & editing. Giuseppe Caruso: Conceptualization, Visualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Grazia Scandura: Investigation, Writing – review & editing. Clarissa Pandino: Investigation, Writing – review & editing. Alessandra Romano: Visualization, Methodology, Writing – review & editing. Giorgio I. Russo: Visualization, Methodology, Writing – review & editing. Ronald Pethig: Visualization, Methodology, Writing – review & editing. Massimo Camarda: Conceptualization, Visualization, Methodology, Writing – original draft, Writing – review & editing. Nicolò Musso: Conceptualization, Visualization, Investigation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
The authors would like to acknowledge the helpful discussion with Prof. Susan M. Lunte (University of Kansas, Lawrence, Kansas, USA). The authors would also like to thank the BRIT laboratory at the University of Catania (Italy) for the valuable technical assistance and use of their laboratories.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101599.
Appendix. Supplementary materials
Supplementary Materials: Supplementary Fig. 1: Measurements of Circularity, SSC, and Area vs. Diameter parameters at four different time points (T1 = 15 min, T2 = 30 min, T3 = 45 min, T4 = 60 min) in Caco-2 and K562 cells. Supplementary Fig. 2: Illustration of the three parameters evaluated for a random sample, obtained by using FlowSight® Imaging Flow Cytometer. Supplementary Table 1: Sampling scheme for single cell line. Supplementary Table 2: List of primers used for quantitative real-time PCR (qRT-PCR). Supplementary Table 3: Quantification of each RNA extracted and related integrity value (RIN). Row SSC Datasets: Row SSC data for Caco-2 cells and Row SSC data for Caco-2 K562 cells.
References
- 1.Pohl H.A. The motion and precipitation of suspensoids in divergent electric fields. J. Appl. Phys. 1951;22(7):869–871. [Google Scholar]
- 2.Pohl, H.A., Dielectrophoresis. The behavior of neutral matter in nonuniform electric fields, 1978.
- 3.Yao J., et al. Microfluidic device embedding electrodes for dielectrophoretic manipulation of cells-A review. Electrophoresis. 2018 doi: 10.1002/elps.201800440. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Duncan J.L., Davalos R.V. A review: dielectrophoresis for characterizing and separating similar cell subpopulations based on bioelectric property changes due to disease progression and therapy assessment. Electrophoresis. 2021;42(23):2423–2444. doi: 10.1002/elps.202100135. [DOI] [PubMed] [Google Scholar]
- 5.Velmanickam L., et al. Recent advances in dielectrophoresis toward biomarker detection: a summary of studies published between 2014 and 2021. Electrophoresis. 2022;43(1-2):212–231. doi: 10.1002/elps.202100194. [DOI] [PubMed] [Google Scholar]
- 6.Pethig R.R. John Wiley & Sons; 2017. Dielectrophoresis: Theory, methodology and Biological Applications. [Google Scholar]
- 7.Pethig R. Dielectrophoresis: an assessment of its potential to aid the research and practice of drug discovery and delivery. Adv. Drug. Deliv. Rev. 2013;65(11-12):1589–1599. doi: 10.1016/j.addr.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Yang L., et al. Effects of dielectrophoresis on growth, viability and immuno-reactivity of listeria monocytogenes. J. Biol. Eng. 2008;2(1):1–14. doi: 10.1186/1754-1611-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kung Y.C., et al. Tunnel dielectrophoresis for tunable, single-stream cell focusing in physiological buffers in high-speed microfluidic flows. Small. 2016;12(32):4343–4348. doi: 10.1002/smll.201600996. [DOI] [PubMed] [Google Scholar]
- 10.Russo G.I., et al. The role of dielectrophoresis for cancer diagnosis and prognosis. Cancers. 2021;14(1) doi: 10.3390/cancers14010198. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liljefors M., et al. Clinical effects of a chimeric anti-EpCAM monoclonal antibody in combination with granulocyte-macrophage colony-stimulating factor in patients with metastatic colorectal carcinoma. Int. J. Oncol. 2005;26(6):1581–1589. [PubMed] [Google Scholar]
- 12.Hofman V., et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int. J. Cancer. 2011;129(7):1651–1660. doi: 10.1002/ijc.25819. [DOI] [PubMed] [Google Scholar]
- 13.Gabriel M.T., et al. Circulating tumor cells: a review of non–EpCAM-based approaches for cell enrichment and isolation. Clin. Chem. 2016;62(4):571–581. doi: 10.1373/clinchem.2015.249706. [DOI] [PubMed] [Google Scholar]
- 14.Pantel K., et al. Circulating epithelial cells in patients with benign colon diseases. Clin. Chem. 2012;58(5):936–940. doi: 10.1373/clinchem.2011.175570. [DOI] [PubMed] [Google Scholar]
- 15.Mikolajczyk S.D., et al. Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J. Oncol. 2011 doi: 10.1155/2011/252361. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onder T.T., et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 17.Tam W.L., Weinberg R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013;19(11):1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 19.Shim S., et al. Dielectrophoresis has broad applicability to marker-free isolation of tumor cells from blood by microfluidic systems. Biomicrofluidics. 2013;7(1) doi: 10.1063/1.4774307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Du F., et al. EpCAM-independent isolation of circulating tumor cells with epithelial-to-mesenchymal transition and cancer stem cell phenotypes using ApoStream® in patients with breast cancer treated with primary systemic therapy. PLoS One. 2020;15(3) doi: 10.1371/journal.pone.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta V., et al. ApoStream™, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6(2) doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shim S., et al. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics. 2013;7(1):11807. doi: 10.1063/1.4774304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., et al. On-chip microfluidic buffer swap of biological samples in-line with downstream dielectrophoresis. Electrophoresis. 2022;43(12):1275–1282. doi: 10.1002/elps.202100304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabuncu A.C., et al. Differential dielectric responses of chondrocyte and Jurkat cells in electromanipulation buffers. Electrophoresis. 2015;36(13):1499–1506. doi: 10.1002/elps.201500119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menachery A., Pethig R. Controlling cell destruction using dielectrophoretic forces. IEE Proc. Nanobiotechnol. 2005;152(4):145–149. doi: 10.1049/ip-nbt:20050010. [DOI] [PubMed] [Google Scholar]
- 26.Stephens M., et al. The dielectrophoresis enrichment of CD34+ cells from peripheral blood stem cell harvests. Bone Marrow Transplant. 1996;18(4):777–782. [PubMed] [Google Scholar]
- 27.Guo X., Zhu R. A biocompatible microchip and methodology for efficiently trapping and positioning living cells into array based on negative dielectrophoresis. J. Appl. Phys. 2015;117(21):214702. [Google Scholar]
- 28.Puttaswamy S.V., et al. Enhanced cell viability and cell adhesion using low conductivity medium for negative dielectrophoretic cell patterning. Biotechnol. J. 2010;5(10):1005–1015. doi: 10.1002/biot.201000194. [DOI] [PubMed] [Google Scholar]
- 29.Musso N., et al. Different modulatory effects of four methicillin-resistant staphylococcus aureus clones on MG-63 osteoblast-like cells. Biomolecules. 2021;11(1):72. doi: 10.3390/biom11010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fresta C.G., et al. Modulation of pro-oxidant and pro-inflammatory activities of M1 macrophages by the natural dipeptide carnosine. Int. J. Mol. Sci. 2020;21(3):776. doi: 10.3390/ijms21030776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fresta C.G., et al. A new human blood-retinal barrier model based on endothelial cells, pericytes, and astrocytes. Int. J. Mol. Sci. 2020;21(5):1636. doi: 10.3390/ijms21051636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caruso G., et al. Receptor-mediated toxicity of human amylin fragment aggregated by short- and long-term incubations with copper ions. Mol. Cell. Biochem. 2017;425(1-2):85–93. doi: 10.1007/s11010-016-2864-1. [DOI] [PubMed] [Google Scholar]
- 33.Li H., et al. 0.9% saline is neither normal nor physiological. J. Zhejiang Univ. Sci. B. 2016;17(3):181–187. doi: 10.1631/jzus.B1500201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigillo G., et al. LPS-induced histone H3 phospho(Ser10)-acetylation(Lys14) regulates neuronal and microglial neuroinflammatory response. Brain Behav. Immun. 2018;74:277–290. doi: 10.1016/j.bbi.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Caruso G., et al. Carnosine decreases PMA-induced oxidative stress and inflammation in murine macrophages. Antioxidants (Basel) 2019;8(8):281. doi: 10.3390/antiox8080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torrisi S.A., et al. Fluoxetine and vortioxetine reverse depressive-like phenotype and memory deficits induced by abeta1-42 oligomers in mice: a key role of transforming growth factor-beta1. Front. Pharmacol. 2019;10:693. doi: 10.3389/fphar.2019.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bush P.G., et al. Viability and volume of in situ bovine articular chondrocytes—changes following a single impact and effects of medium osmolarity. Osteoarthr. Cartil. 2005;13(1):54–65. doi: 10.1016/j.joca.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Amin A.K., et al. Osmolarity influences chondrocyte death in wounded articular cartilage. JBJS. 2008;90(7):1531–1542. doi: 10.2106/JBJS.G.00857. [DOI] [PubMed] [Google Scholar]
- 39.Gabriel M.T., et al. Circulating tumor cells: a review of Non-EpCAM-based approaches for cell enrichment and isolation. Clin. Chem. 2016;62(4):571–581. doi: 10.1373/clinchem.2015.249706. [DOI] [PubMed] [Google Scholar]
- 40.Gorges T.M., et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gascoyne P.R., Shim S. Isolation of circulating tumor cells by dielectrophoresis. Cancers. 2014;6(1):545–579. doi: 10.3390/cancers6010545. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahneh F.Z. Sensitive antibody-based CTCs detection from peripheral blood. Hum. Antibodies. 2013;22(1-2):51–54. doi: 10.3233/HAB-130270. [DOI] [PubMed] [Google Scholar]
- 43.Hyler A.R., et al. A novel ultralow conductivity electromanipulation buffer improves cell viability and enhances dielectrophoretic consistency. Electrophoresis. 2021;42(12-13):1366–1377. doi: 10.1002/elps.202000324. [DOI] [PubMed] [Google Scholar]
- 44.Becker F., et al. The removal of human leukaemia cells from blood using interdigitated microelectrodes. J. Phys. D Appl. Phys. 1994;27(12):2659. [Google Scholar]
- 45.Yang L., et al. Effects of dielectrophoresis on growth, viability and immuno-reactivity of listeria monocytogenes. J. Biol. Eng. 2008;(2):6. doi: 10.1186/1754-1611-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engür S., Dikmen M. The evaluation of the anti-cancer activity of ixazomib on Caco2 colon solid tumor cells, comparison with bortezomib. Acta Clin. Belg. 2017;72(6):391–398. doi: 10.1080/17843286.2017.1302623. [DOI] [PubMed] [Google Scholar]
- 47.Richter F., et al. Improved gene delivery to K-562 leukemia cells by lipoic acid modified block copolymer micelles. J. Nanobiotechnol. 2021;19(1):70. doi: 10.1186/s12951-021-00801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw Bagnall J., et al. Deformability of tumor cells versus blood cells. Sci. Rep. 2015;5:18542. doi: 10.1038/srep18542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caruso G., et al. Sub-toxic human amylin fragment concentrations promote the survival and proliferation of SH-SY5Y cells via the release of VEGF and HspB5 from Endothelial RBE4 Cells. Int. J. Mol. Sci. 2018;(11):19. doi: 10.3390/ijms19113659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sala F., Hernández-Cruz A. Calcium diffusion modeling in a spherical neuron. Relevance of buffering properties. Biophys. J. 1990;57(2):313–324. doi: 10.1016/S0006-3495(90)82533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolkers W.F., et al. Factors affecting the membrane permeability barrier function of cells during preservation technologies. Langmuir. 2019;35(23):7520–7528. doi: 10.1021/acs.langmuir.8b02852. [DOI] [PubMed] [Google Scholar]
- 52.Cherubin P., Quiñones B., Teter K. Cellular recovery from exposure to sub-optimal concentrations of AB toxins that inhibit protein synthesis. Sci. Rep. 2018;8(1):2494. doi: 10.1038/s41598-018-20861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Q., et al. The impact of the ionic concentration on electrocoalescence of the nanodroplet driven by dielectrophoresis. J. Mol. Liq. 2019;290 [Google Scholar]
- 54.Zuo S., Dai G., Ren X. Identification of a 6-gene signature predicting prognosis for colorectal cancer. Cancer Cell Int. 2019;19(1):1–15. doi: 10.1186/s12935-018-0724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamal-Hanjani M., et al. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015;21(6):1258–1266. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C., et al. Circulating tumor cells in precision oncology: clinical applications in liquid biopsy and 3D organoid model. Cancer Cell Int. 2019;19:341. doi: 10.1186/s12935-019-1067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saxena K., Subbalakshmi A.R., Jolly M.K. Phenotypic heterogeneity in circulating tumor cells and its prognostic value in metastasis and overall survival. EBioMedicine. 2019;46:4–5. doi: 10.1016/j.ebiom.2019.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smit D.J., Pantel K., Jücker M. Circulating tumor cells as a promising target for individualized drug susceptibility tests in cancer therapy. Biochem. Pharmacol. 2021;188 doi: 10.1016/j.bcp.2021.114589. [DOI] [PubMed] [Google Scholar]
- 59.Alshareef M., et al. Separation of tumor cells with dielectrophoresis-based microfluidic chip. Biomicrofluidics. 2013;7(1) doi: 10.1063/1.4774312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiriac E., Avram M., Bălan C. Proceedings of the International Semiconductor Conference (CAS) IEEE; 2020. Dielectrophoretic separation of circulating tumor cells and red blood cells in a microfluidic device. [Google Scholar]
- 61.Fan Y., et al. Fluorescent analysis of bioactive molecules in single cells based on microfluidic chips. Lab Chip. 2018;18(8):1151–1173. doi: 10.1039/c7lc01333g. [DOI] [PubMed] [Google Scholar]
- 62.de Campos R.P., et al. Indirect detection of superoxide in RAW 264.7 macrophage cells using microchip electrophoresis coupled to laser-induced fluorescence. Anal. Bioanal. Chem. 2015;407(23):7003–7012. doi: 10.1007/s00216-015-8865-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fresta C.G., et al. Monitoring carnosine uptake by RAW 264.7 macrophage cells using microchip electrophoresis with fluorescence detection. Anal. Methods. 2017;9(3):402–408. doi: 10.1039/C6AY03009B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang R., et al. Cancer cell discrimination and dynamic viability monitoring through wash-free bioimaging using AIEgens. Chem. Sci. 2020;11(29):7676–7684. doi: 10.1039/d0sc01213k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan S., et al. A hybrid dielectrophoretic and hydrophoretic microchip for particle sorting using integrated prefocusing and sorting steps. Electrophoresis. 2015;36(2):284–291. doi: 10.1002/elps.201400397. [DOI] [PubMed] [Google Scholar]
- 66.Hu X., et al. Marker-specific sorting of rare cells using dielectrophoresis. Proc. Natl. Acad. Sci. U. S. A. 2005;102(44):15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang X., et al. Self-aligned sequential lateral field non-uniformities over channel depth for high throughput dielectrophoretic cell deflection. Lab Chip. 2021;21(5):835–843. doi: 10.1039/d0lc01211d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Honrado C., et al. Automated biophysical classification of apoptotic pancreatic cancer cell subpopulations by using machine learning approaches with impedance cytometry. Lab Chip. 2022;22(19):3708–3720. doi: 10.1039/d2lc00304j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palmirotta R., et al. Next-generation sequencing (NGS) analysis on single circulating tumor cells (CTCs) with no need of whole-genome amplification (WGA) Cancer Genom. Proteom. 2017;14(3):173–179. doi: 10.21873/cgp.20029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonzalez-Kozlova E.E. Computational Methods for Precision Oncology. Springer; 2022. Molecular profiling of liquid biopsies for precision oncology; pp. 235–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials: Supplementary Fig. 1: Measurements of Circularity, SSC, and Area vs. Diameter parameters at four different time points (T1 = 15 min, T2 = 30 min, T3 = 45 min, T4 = 60 min) in Caco-2 and K562 cells. Supplementary Fig. 2: Illustration of the three parameters evaluated for a random sample, obtained by using FlowSight® Imaging Flow Cytometer. Supplementary Table 1: Sampling scheme for single cell line. Supplementary Table 2: List of primers used for quantitative real-time PCR (qRT-PCR). Supplementary Table 3: Quantification of each RNA extracted and related integrity value (RIN). Row SSC Datasets: Row SSC data for Caco-2 cells and Row SSC data for Caco-2 K562 cells.