Highlights
-
•
There is a large amount of uncharacterized gene products of prokaryotic origin.
-
•
Evidence suggests that these elements might provide novel biotechnological solutions.
-
•
This review outlines the challenges and potential of these prokaryotic gene products.
Keywords: Functional Genomics, Metagenomics, Microbial Dark Matter, Biotechnological Potential
Graphical abstract

Abstract
Eight-hundred thousand to one trillion prokaryotic species may inhabit our planet. Yet, fewer than two-hundred thousand prokaryotic species have been described. This uncharted fraction of microbial diversity, and its undisclosed coding potential, is known as the “microbial dark matter” (MDM). Next-generation sequencing has allowed to collect a massive amount of genome sequence data, leading to unprecedented advances in the field of genomics. Still, harnessing new functional information from the genomes of uncultured prokaryotes is often limited by standard classification methods. These methods often rely on sequence similarity searches against reference genomes from cultured species. This hinders the discovery of unique genetic elements that are missing from the cultivated realm. It also contributes to the accumulation of prokaryotic gene products of unknown function among public sequence data repositories, highlighting the need for new approaches for sequencing data analysis and classification. Increasing evidence indicates that these proteins of unknown function might be a treasure trove of biotechnological potential. Here, we outline the challenges, opportunities, and the potential hidden within the functional dark matter (FDM) of prokaryotes. We also discuss the pitfalls surrounding molecular and computational approaches currently used to probe these uncharted waters, and discuss future opportunities for research and applications.
1. Introduction
Ever since the dawn of life 3.5 billion years ago (Allwood, Walter, Kamber, Marshall, Burch, 2006, Blaser, Cardon, Cho, Dangl, Donohue, Green, Knight, Maxon, Northen, Pollard, Brodie, 2016), the Earth’s environmental, geochemical, and biological systems of all levels of complexity have relied on microorganisms (McFall-Ngai et al., 2013). Microorganisms are the most abundant, ubiquitous, functionally and metabolically diverse forms of life (Locey and Lennon, 2016); they are responsible for a substantial part of our planet’s biomass (Solden et al., 2016), and an overwhelming fraction of its biodiversity (Locey, Lennon, 2016, Solden, Lloyd, Wrighton, 2016). Previous studies estimated that approximately four-hundred thousand bacterial and archaeal species exist (Yarza et al., 2014). More recent estimates range from eight-hundred thousand (Louca et al., 2019), to over one trillion () (Locey and Lennon, 2016). The actual number, however, is still a subject of debate (Louca, Mazel, Doebeli, Parfrey, 2019, Pedrós-Alió, Manrubia, 2016, Willis, 2016, Zhang, Wang, Wang, Wang, Li, 2020). Nonetheless, only a very small fraction of all prokaryotic species has been described to this day (approximately two-hundred thousand complete and draft genomes (Zhang et al., 2020)), thus exposing our ignorance regarding their diversity. Roughly 85-99% of prokaryotic taxa are unamenable to axenic culture (Lok, 2015), which precludes the in vitro characterization of such species. Yet, it has been known for decades that most microorganisms cannot be cultured (Jannasch, Jones, 1959, Jones, 1970, Rappé, Giovannoni, 2003). In 1985, Norman Pace’s group showed that the diversity of uncultured prokaryotes could be probed by molecular biology methods (Olsen et al., 1986), namely those relying on the 16S ribosomal RNA (16S rRNA) as a marker (Woese, 1987). Microbiology has benefited tremendously from further advances in this field, which became collectively known as “molecular microbial ecology” (Pace, 1995).
Next-generation sequencing (NGS) brought a breadth of new insights into microbial genomics. Through the lens of culture-independent methods like metagenomics, NGS has allowed to investigate the diversity and coding potential of microbial communities in the context of their environment (New and Brito, 2020). Concurrent advances in bioinformatics gave birth to metagenome-assembled genomes (MAGs) (Kayani et al., 2021), which are crucial for broadening our knowledge of the ecology, metabolism, and coding potential of uncultured prokaryotes (Sangwan et al., 2016). Third-generation sequencing (TGS) technologies (i.e., Pacific Biosciences and Oxford Nanopore Technologies) currently allow to gather massive amounts of data with unprecedented detail (van Dijk et al., 2018), and at low cost (Karlsson et al., 2015). The long reads produced by TGS, together with ever-improving de novo assembly algorithms (Dida and Yi, 2021), have streamlined (meta)genome sequencing and assembly endeavors (Athanasopoulou et al., 2021). These developments already allow to attain complete, closed, de novo-assembled prokaryotic genomes and MAGs (Loman, Quick, Simpson, 2015, Moss, Maghini, Bhatt, 2020, Somerville, Lutz, Schmid, Frei, Moser, Irmler, Frey, Ahrens, 2019). Another approach, named single-cell genomics (SCG), relies on amplifying and sequencing the genome of individual cells isolated from environmental samples, instead of bulk sequencing the entire community (Solden et al., 2016), and it has provided numerous insights into the metabolism and evolutionary context of many uncultured groups of Archaea and Bacteria (Santoro et al., 2019). A different method that combines high-throughput culturing with MALDI-TOF mass spectrometry and 16S rRNA sequencing (i.e., culturomics), has allowed to isolate hundreds of new prokaryotic species (Lagier et al., 2018), and to decode their complete genome sequences thereafter. As a result of these advances, a modest part of the microbial “black box” was unveiled over the past decades. Together with continuous improvements in bioinformatics, it has been possible to discover new functions and metabolic features that have: (i) bolstered natural product discovery (Bull, Goodfellow, 2019, Chen, Wong, Burns, 2019, Goodfellow, Nouioui, Sanderson, Xie, Bull, 2018, Lackner, Peters, Helfrich, Piel, 2017, Ling, Schneider, Peoples, Spoering, Engels, Conlon, Mueller, Schäberle, Hughes, Epstein, Jones, Lazarides, Steadman, Cohen, Felix, Fetterman, Millett, Nitti, Zullo, Chen, Lewis, 2015, Owen, Charlop-Powers, Smith, Ternei, Calle, Reddy, Montiel, Brady, 2015, Rust, Helfrich, Freeman, Nanudorn, Field, Rückert, Kündig, Page, Webb, Kalinowski, Sunagawa, Piel, 2020, Ziemert, Alanjary, Weber, 2016), (ii) challenged preconceived boundaries among the three domains of life (Hug, Baker, Anantharaman, Brown, Probst, Castelle, Butterfield, Hernsdorf, Amano, Ise, Suzuki, Dudek, Relman, Finstad, Amundson, Thomas, Banfield, 2016, Parks, Rinke, Chuvochina, Chaumeil, Woodcroft, Evans, Hugenholtz, Tyson, 2017, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013), and (iii) reshaped our understanding of microbial life forms (Brown, Hug, Thomas, Sharon, Castelle, Singh, Wilkins, Wrighton, Williams, Banfield, 2015, Nasir, Kim, Caetano-Anollés, 2015, Wiegand, Jogler, Boedeker, Pinto, Vollmers, Rivas-Marín, Kohn, Peeters, Heuer, Rast, Oberbeckmann, Bunk, Jeske, Meyerdierks, Storesund, Kallscheuer, Lücker, Lage, Pohl, Merkel, Hornburger, Müller, Brümmer, Labrenz, Spormann, Op den Camp, Overmann, Amann, Jetten, Mascher, Medema, Devos, Kaster, Øvreås, Rohde, Galperin, Jogler, 2020).
The unexploited fraction of microbial diversity, along with its functional and metabolic potential, is commonly referred to as the microbial dark matter (MDM). This term was coined by Marcy and colleagues in 2007 (Marcy et al., 2007), alluding to the large amount of unknown microbial taxa and respective genomes inferred by culture-independent approaches. Although most authors use the term interchangeably to refer to the taxonomic tapestry and the coding potential of MDM, we reckon that either concept should be employed separately. There are four categories of sequence novelty based on annotation (Bernard et al., 2018)Fig. 1(A): (i) sequences with known taxonomic provenance and molecular function (e.g., a hydrolase gene from Escherichia coli); (ii) sequences with known taxonomic provenance but unknown molecular function (e.g., an E. coli gene without ascribed function); (iii) sequences with unknown taxonomic provenance but known molecular function (e.g., a hydrolase gene from an unknown prokaryote); and (iv) sequences with unknown taxonomic provenance and unknown molecular function (e.g., a gene with no ascribed function from an unknown prokaryote, i.e., the true dark matter). Abiding by the rationale set forth by Bernard et al. (Bernard et al., 2018), we propose the classification of microbial genomic sequences with unknown taxonomic provenance as “taxonomic dark matter” (TDM), regardless of their functional annotation; and the classification of sequences with unknown molecular function as “functional dark matter” (FDM), regardless of their taxonomic context. Throughout this review we use the terms “microbial” and “microorganisms” to refer to prokaryotes only, comprising the Archaea and the Bacteria domains.
Fig. 1.
(A) Four possible combinations when addressing a protein regarding its taxonomic provenance and molecular function. Both rows and columns depict a binary range: known and unknown. The rows refer to the molecular function of the protein, and the columns refer to its taxonomic provenance. Each quadrant represents one combination that results from the intersection of the rows and columns. The bean-shape represents a prokaryotic cell and the coil-shape represents a protein. A green foreground with a green checkmark represents “known”, and a grey foreground with a red question mark represents “unknown”. Adapted from Bernard et al. (2018). (B) Proportion of CDS annotations of unknown function per genome, for “Known Taxa” (top boxplot, green data-points) and “Candidate Taxa” (bottom boxplot, red data-points). Each data-point corresponds to a genome. To calculate these percentages, we proceeded as follows. We gathered NCBI’s assembly accessions for all complete genomes of Archaea and Bacteria that were either of reference or representatives, from ftp.ncbi.nlm.nih.gov/genomes/refseq/archaea/assembly_summary.txt and ftp.ncbi.nlm.nih.gov/genomes/refseq/bacteria/assembly_summary.txt, on the 8th of June 2022. This amounted to 239 Archaeal and 3,833 Bacterial genomes (n = 4,072). We subset these 4,072 genomes into the “Known Taxa” and “Candidate Taxa” categories according to their taxonomic description. A genome was placed in the “Candidate Taxa” category if its taxonomic description matched any of the following words: ‘uncharacterized’, ‘unclassified’, ‘unidentified’, ‘endosymbiont’, ‘uncultured’, ‘metagenome’, ‘candidatus’, ‘candidate’, or ‘unnamed’. We downloaded the annotation file (i.e., *_feature_table.txt.gz) for each assembly accession, from the corresponding FTP server directory listed in the assembly_summary.txt files. We gathered the total number of CDS annotations for each genome from these annotation files. Each CDS annotation was matched against any of the following words: ‘hypothetical’, ‘predicted’, ‘putative’, ‘uncharacterized’, ‘unknown function’, or ‘unnamed protein’. For each genome, the percentage of CDS’ of unknown function corresponds to the number of CDS annotations that matched these words, divided by the total number of CDS annotations. For each of the “Candidate Taxa” and “Known Taxa” categories, we annotated the data-point whose corresponding genome had the highest percentage of FDM (and therefore, the lowest percentage of known function); and the data-point whose corresponding genome had the lowest percentage of FDM (and therefore, the highest percentage of known function). The icons used in this figure were retrieved from flaticon.com.
Researchers frequently use phylogenetic-driven techniques and canonical molecular markers (e.g., 16S rRNA (Yarza et al., 2014)) to study MDM through targeted community profiling. Still, the coding potential of these bacterial communities often remains inaccessible. Harnessing genomic information from these microorganisms is often limited by gene annotation methods that rely on sequence similarity to proteins characterized from microbial cultures (Michalska et al., 2015). This approach makes it difficult to study the FDM and identify unique functions particular to uncultured organisms (Michalska et al., 2015). Adding to these shortcomings, the genomes of these elusive microorganisms typically diverge from those of well-known species (Grötzinger, Karan, Strillinger, Bader, Frank, Al Rowaihi, Akal, Wackerow, Archer, Rueping, Weuster-Botz, Groll, Eppinger, Arold, 2018, Miller, Weyna, Fong, Lim-Fong, Kwan, 2016, Sysoev, Grötzinger, Renn, Eppinger, Rueping, Karan, 2021). Notwithstanding, several remarkable studies have explored the phylogenetic novelty hidden in the TDM, offering insights into the putative functions it encodes (Gies, Konwar, Beatty, Hallam, 2014, Lackner, Peters, Helfrich, Piel, 2017, Makarova, Wolf, Forterre, Prangishvili, Krupovic, Koonin, 2014, McLean, Lombardo, Badger, Edlund, Novotny, Yee-Greenbaum, Vyahhi, Hall, Yang, Dupont, Ziegler, Chitsaz, Allen, Yooseph, Tesler, Pevzner, Friedman, Nealson, Venter, Lasken, 2013, Mehrshad, Rodriguez-Valera, Amoozegar, López-García, Ghai, 2017, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013, Wegner, Liesack, 2017). Some reports even predict the existence of community-wide metabolic profiles (Anantharaman, Brown, Hug, Sharon, Castelle, Probst, Thomas, Singh, Wilkins, Karaoz, Brodie, Williams, Hubbard, Banfield, 2016, Hawley, Nobu, Wright, Durno, Morgan-Lang, Sage, Schwientek, Swan, Rinke, Torres-Beltrán, Mewis, Liu, Stepanauskas, Woyke, Hallam, 2017, Momper, Jungbluth, Lee, Amend, 2017, Nobu, Narihiro, Rinke, Kamagata, Tringe, Woyke, Liu, 2015, Thrash, Cameron Thrash, Seitz, Baker, Temperton, Gillies, Rabalais, Henrissat, Mason, 2017).
Equally impressive is the diversity of secondary metabolites and enzymes produced by microorganisms, well-known mediators of fundamental biogeochemical cycles (Blaser, Cardon, Cho, Dangl, Donohue, Green, Knight, Maxon, Northen, Pollard, Brodie, 2016, Rust, Helfrich, Freeman, Nanudorn, Field, Rückert, Kündig, Page, Webb, Kalinowski, Sunagawa, Piel, 2020). Insights gained from microbiome research and innovation (Małyska et al., 2019) allow not only to discover new compounds and metabolic pathways with potential applications in agronomic, biotechnological, environmental, and pharmaceutical industries (Blaser, Cardon, Cho, Dangl, Donohue, Green, Knight, Maxon, Northen, Pollard, Brodie, 2016, Bull, Goodfellow, 2019, Chen, Wong, Burns, 2019, Goodfellow, Nouioui, Sanderson, Xie, Bull, 2018, Rust, Helfrich, Freeman, Nanudorn, Field, Rückert, Kündig, Page, Webb, Kalinowski, Sunagawa, Piel, 2020); as they may also promote new biotic solutions to overcome pressing societal challenges (United Nations, Department of Economic and Social Affairs, 2015). Bringing this knowledge to light is critical, as the key to solve some of the current concerns may lie on novel metabolite-producing gene products and enzymes of biotechnological value, such as those hidden in the FDM (Danso, Schmeisser, Chow, Zimmermann, Wei, Leggewie, Li, Hazen, Streit, 2018, Ling, Schneider, Peoples, Spoering, Engels, Conlon, Mueller, Schäberle, Hughes, Epstein, Jones, Lazarides, Steadman, Cohen, Felix, Fetterman, Millett, Nitti, Zullo, Chen, Lewis, 2015, Rashid, Stingl, 2015, Sysoev, Grötzinger, Renn, Eppinger, Rueping, Karan, 2021, Yoshida, Hiraga, Takehana, Taniguchi, Yamaji, Maeda, Toyohara, Miyamoto, Kimura, Oda, 2016, Zrimec, Kokina, Jonasson, Zorrilla, Zelezniak, 2021).
Here, we outline the challenges, opportunities, and unique potential of the hidden world of FDM. We also discuss some limitations of molecular and computational approaches that currently preclude the comprehensive characterization of the MDM. Finally, we offer a perspective on the potential opportunities and directions for future research.
2. Revealing the hidden potential of functional dark matter
Until the turn of the century, genome sequencing of microorganisms depended on their culturability. Researchers often had to isolate the microorganisms from an environmental sample and cultivate them in order to sequence their genome. The progress made in culture-independent DNA sequencing approaches, coupled to NGS technologies, and that of annotation pipelines, has improved our awareness of elusive microorganisms, and our understanding of the genomic content they encode. However, the functions this genomic content codes for are frequently unknown. In this section we will overview studies that aimed to quantify and characterize the FDM, and those that tried to exploit the functional richness it encloses, in order to unearth new knowledge with biotechnological implications.
2.1. Current estimates
The fraction of FDM genes ranges substantially among different prokaryotic genomes. Some authors estimate that the functions of 35% of genes from a given genome remain a mystery (Piao et al., 2014). In other cases, this fraction is thought to amount to as much as 50%, like in newly sequenced genomes (Al-Shahib et al., 2007). Other authors report that, in genomes from uncultured candidate taxa, these proportions range from 46% to 60% (Becraft, Dodsworth, Murugapiran, Ohlsson, Briggs, Kanbar, De Vlaminck, Quake, Dong, Hedlund, Swingley, 2015, Garza, Dutilh, 2015, Marcy, Ouverney, Bik, Lösekann, Ivanova, Martin, Szeto, Platt, Hugenholtz, Relman, Quake, 2007, McLean, Lombardo, Badger, Edlund, Novotny, Yee-Greenbaum, Vyahhi, Hall, Yang, Dupont, Ziegler, Chitsaz, Allen, Yooseph, Tesler, Pevzner, Friedman, Nealson, Venter, Lasken, 2013). Makarova et al. disclosed that archaeal genomes encode from 30% to 80% of FDM (Makarova et al., 2019). In comparison to Bacteria, this represents a greater content of FDM harbored by archaeal genomes (Makarova et al., 2019). These occurrences were attributed to the difficulty in isolating and cultivating most Archaea, which in turn hinders the experimental characterization of their genes (Makarova et al., 2019). The same authors also accounted that in most of the Archaea, the amount of FDM scales linearly with the genome size (Makarova et al., 2019).
More recently, Lobb et al. reported an extreme variation in the proportion of genome annotation incompleteness across distinct bacterial species, according to different annotation tools (Lobb et al., 2020). These authors highlight that certain lineages issuing from the TDM (e.g., Patescibacteria), possessed greater content of FDM genes (Lobb et al., 2020). Moreover, they disclose that the proportion of FDM genes per genome can range from as little as 2.3%, to as high as 87.9% (Lobb et al., 2020). Akin to the results reported by Lobb et al. (2020), we also observe a wide range in the proportion of coding sequences (CDS’) with unknown function for complete prokaryotic genomes from NCBI (Fig. 1B). Interestingly, Mycoplasma haemofelis, a known feline pathogen (Barker et al., 2011), lacks functional annotation for 80.05% of its CDS’; whereas this percentage is just 0.37% for Candidatus ‘Mikella endobia’ (Fig. 1B).
Nevertheless, in order to find FDM genes one does not need to venture into the uncultivated myriad of microorganisms, nor that of candidate taxa. Indeed, in 2016, a minimal synthetic bacterial genome based on that from Mycoplasma mycoides, a well-studied mammalian parasite, was generated (Hutchison et al., 2016). This synthetic genome contained only 473 genes, of which 149 had unknown functions. Yet, each of those genes was considered essential, as deletion of any of them was lethal (Hutchison et al., 2016). Therefore, even in a controlled laboratory environment, the function of nearly one-third of essential genes is unknown.
In more extreme cases, as indicated by metagenomic studies, these percentages can span from 85% (Lobb et al., 2015) to 99% of total gene content (Dutilh, 2014, Mokili, Rohwer, Dutilh, 2012). The reference gene catalog that stemmed from the Tara Oceans initiative, comprised more than 40 million non-redundant sequences from ocean metagenomes (Sunagawa et al., 2015). After mapping these sequences to clusters of orthologous groups, the authors of this study outlined that 40% of these groups were of unknown function (Sunagawa et al., 2015). Focusing on the human gut microbiome, Almeida et al. created the “Unified Human Gastrointestinal Genome” (UHGG), and the “Unified Human Gastrointestinal Protein” (UHGP) catalogs (Almeida et al., 2021). The UHGP contains more than 170 million protein sequences, however 40% of these could not be functionally annotated (Almeida et al., 2021). After exploring the core and accessory gene repertoires for each of the UHGG species, they report that the accessory cohort showed a greater proportion of genes of unknown function, and that 21% of these genes failed to attain a match to any of the reference databases used in their study (Almeida et al., 2021). Other initiatives that aimed to broaden the phylogenetic coverage of prokaryotic genomes also report high percentages of FDM within their genomic catalogs (Mukherjee, Seshadri, Varghese, Eloe-Fadrosh, Meier-Kolthoff, Göker, Coates, Hadjithomas, Pavlopoulos, Paez-Espino, Yoshikuni, Visel, Whitman, Garrity, Eisen, Hugenholtz, Pati, Ivanova, Woyke, Klenk, Kyrpides, 2017, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Wu, Hugenholtz, Mavromatis, Pukall, Dalin, Ivanova, Kunin, Goodwin, Wu, Tindall, Hooper, Pati, Lykidis, Spring, Anderson, D’haeseleer, Zemla, Singer, Lapidus, Nolan, Copeland, Han, Chen, Cheng, Lucas, Kerfeld, Lang, Gronow, Chain, Bruce, Rubin, Kyrpides, Klenk, Eisen, 2009) (see subsection 3.4).
Nonetheless, researchers have striven to push the percentage of annotations for predicted proteins for a long time (Marcotte, Pellegrini, Ng, Rice, Yeates, Eisenberg, 1999, Marcotte, Pellegrini, Thompson, Yeates, Eisenberg, 1999). Recently, Bileschi et al. developed a tool that, by learning known patterns of evolutionary substitutions among protein families, has allowed to extend the coverage of Pfam (Mistry et al., 2021) by more than 9.5% (Bileschi et al., 2022). Another study uncovered and characterized more than four-hundred thousand new protein families whose function was previously unknown from a global genomic dataset (Rodríguez del Río et al., 2022). The authors from this study report that these new protein families increase the total number of current prokaryotic orthologous groups by three-fold (Rodríguez del Río et al., 2022).
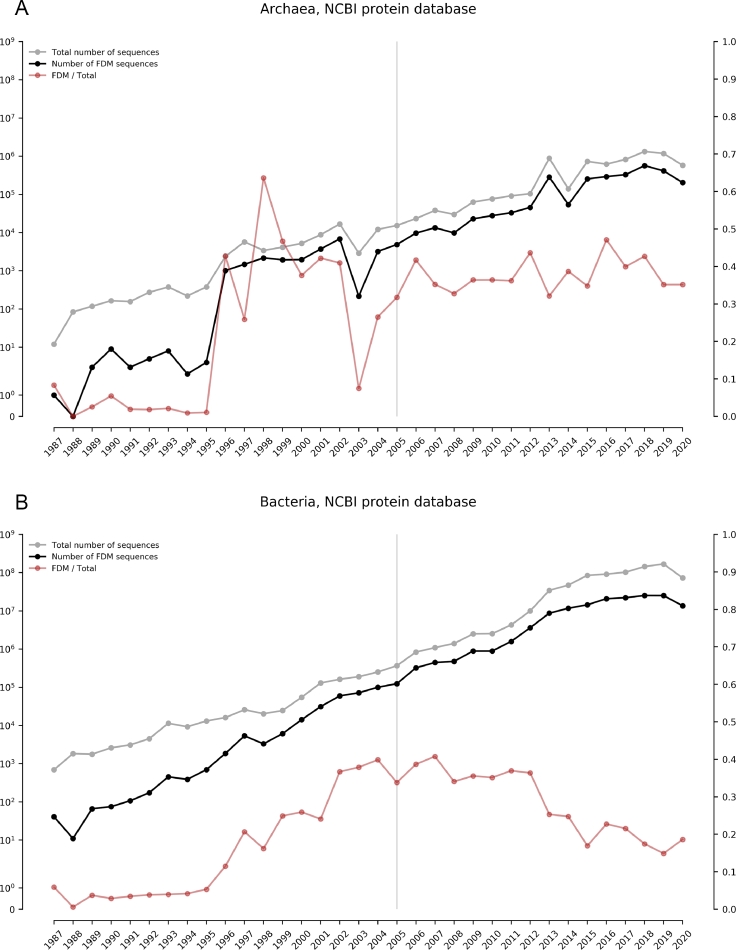
Fig. 2 shows the evolution over time of the number of protein sequences from NCBI’s Protein database that belong to the Archaea and Bacteria FDM. Note that both the number of FDM sequences and the total number of sequences tend to increase over time, regardless of domain. As of 2020, the percentage of FDM sequences in NCBI’s protein database is greater than 30% for Archaea, and nearly 20% for Bacteria. Since the commercialization of next-generation sequencing (NGS; i.e., 2005, see (van Dijk et al., 2014)), these percentages have been steady for Archaea, but decreased in the long run for Bacteria. We speculate that this decrease is a product of the representativeness of the total number of sequences from Bacteria in public databases; and consequently, the improvement and fine-tuning of annotation pipelines aimed particularly at this domain.
Fig. 2.
Number of protein sequences of unknown function in comparison to the total number of sequences in NCBI’s protein database. These counts refer to sequences in the Archaea (A) and Bacteria (B) domains from 1987 to 2020, inclusively. The x-axis represents the date of submission, the y-axis to the left represents the number of sequences in “symlog” scale (i.e., linear from 0 to 1, and logarithmic from 1 upwards), and the y-axis to the right represents the ratio between the number of FDM sequences and the total number of sequences. The light-grey vertical line placed in 2005 marks the commercial availability of the first NGS platform (the pyrosequencing method by 454 Life Sciences, now Roche). In order to obtain these counts we submitted queries to NCBI’s Esearch utility at Entrez (eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?). For each year and for each domain, we retrieved the total number of sequences with the following query: db=protein&term=YEAR[pdat]+AND+DOMAIN[orgn]&rettype=count; whereas as to gather the number of sequences of unknown function we used: db=protein&term=YEAR[pdat]+AND+DOMAIN[orgn]+AND+(hypothetical[title]+OR+predicted[title]+OR+putative[title]+OR+uncharacterized[title]+OR+unknown+function[title])&rettype=count.
2.2. Possible functions
Prokaryotic genes of unknown function were initially considered to be mere “junk” elements, pseudogenes, or misannotations (Andersson, Andersson, 2001, Mira, 2002, Schmid, Aquadro, 2001) owing to the narrow understanding at the time of the functional sequence space (Lobb et al., 2015). But increasing evidence indicates that yet-unclassified elements do encode for specific functions (Hu et al., 2009). For instance, Hanson et al. showed that nearly 15% of E. coli enzymes of unknown function could play roles in metabolite repair (Hanson et al., 2016). Another role thought to be played by these elements is the addition and removal of posttranslational modifications (PTMs) (Ellens et al., 2017). In this respect, it should be emphasized that the enzymes responsible for recently discovered PTMs usually remain unidentified (Choudhary, Weinert, Nishida, Verdin, Mann, 2014, Ellens, Christian, Singh, Satagopam, May, Linster, 2017). Likewise, one can speculate that some uncharacterized proteins may have been enzymes that lost their catalytic properties throughout evolution, and then acquired allosteric regulation functions thereafter (Ellens, Christian, Singh, Satagopam, May, Linster, 2017, Van Schaftingen, Veiga-da Cunha, Linster, 2015).
Other proteins, commonly referred to as “moonlighting”, play multiple biochemical and/or biophysical roles which are not associated with gene fusion events nor proteolytic fragments (Jeffery, 2018). Moonlighting proteins are thought to include a few hundred members and play an extensive range of functions (Jeffery, 2018, Mani, Chen, Amblee, Liu, Mathur, Zwicke, Zabad, Patel, Thakkar, Jeffery, 2015). We posit that the moonlighting phenomenon might explain why some proteins are left uncharacterized in the first place, given the added difficulty in classifying a protein with multiple molecular functions. Proteins of unknown function have also been presumed to encode for ecological or taxon-specific functions (Wilson et al., 2005), including morphological and developmental adaptations (Kaessmann, 2010, Lobb, Kurtz, Moreno-Hagelsieb, Doxey, 2015, Tautz, Domazet-Lošo, 2011), which could explain their lack of homology to annotated genes (Lobb et al., 2015). However, the latter can also be a consequence of the severe undersampling of Earth’s microbiomes, despite the advances in metagenomics in the past decades.
Alternatively, some of these uncharacterized proteins are thought to be gene fragments, or pseudogenes arising naturally through gene degeneration (Makarova et al., 2019). Other studies hypothesized that sequences of unknown function can actually be of viral, integrative, or mobilomic origin (Cortez, Forterre, Gribaldo, 2009, Yin, Fischer, 2006). Dutilh et al. provided additional support for this possibility (Dutilh et al., 2014). This idea makes sense if one considers the fast mutation rates observed in viral DNA (and RNA), and the underrepresentation of sequences from viruses in most public databases (Lobb et al., 2015). Indeed, there is evidence that these unknown elements might be genes that evolve fast, like those associated with anti-parasite defense (Makarova et al., 2014), and those that encode small proteins (Makarova et al., 2019). As an illustration, Sberro et al. reported more than four thousand small ( 50 amino acids in length) protein families, from human-associated metagenomes (Sberro et al., 2019). They describe an abundance of putative functions, namely: housekeeping, mammalian-specific, cell-cell crosstalk, adaptation, anti-parasite defense, secreted or transmembrane proteins, in addition to possible products of horizontal gene transfer (HGT) (Sberro et al., 2019). Despite this breakthrough, they outline that over 90% of these families lack a domain assignment, and nearly half of them are absent from reference genomes (Sberro et al., 2019).
2.3. Biotechnological significance
FDM sequences might encode any potential function waiting to be charted in the microbial sequence space. Some of these sequences may even code for new metabolite-producing proteins, and/or enzymes of biotechnological interest (Bernard, Pathmanathan, Lannes, Lopez, Bapteste, 2018, Chen, Wong, Burns, 2019, Rashid, Stingl, 2015, Sysoev, Grötzinger, Renn, Eppinger, Rueping, Karan, 2021). The FDM (and the MDM in a broader sense) could indeed be an outstanding asset for the discovery of novel biotechnological solutions in a world of ever-increasing societal demands. Pascoal et al. argued that bioremediation and bioprospecting are the two areas holding the most promise from within the MDM, where innovative approaches in biotechnology might arise (Pascoal et al., 2020), like new solutions for the decontamination of environments (Dvořák et al., 2017).
A prominent prospect of the latter is that of bioremediating ecosystems polluted with plastic (Danso et al., 2019). For instance, polyethylene terephthalate (PET) is reported to be the most abundant polyester plastic (Tournier et al., 2020), being mainly used in the textile and packaging industries (Danso, Chow, Streit, 2019, Tournier, Topham, Gilles, David, Folgoas, Moya-Leclair, Kamionka, Desrousseaux, Texier, Gavalda, Cot, Guémard, Dalibey, Nomme, Cioci, Barbe, Chateau, André, Duquesne, Marty, 2020). Nearly 70 million tons of PET are manufactured worldwide per year (Tournier et al., 2020). The common recycling process of PET is through thermomechanical means, resulting in its loss of mechanical properties (Ragaert et al., 2017). As such, PET is preferentially synthesized de novo, and its waste continues to accumulate in ecosystems throughout the globe (Tournier, Topham, Gilles, David, Folgoas, Moya-Leclair, Kamionka, Desrousseaux, Texier, Gavalda, Cot, Guémard, Dalibey, Nomme, Cioci, Barbe, Chateau, André, Duquesne, Marty, 2020, Yoshida, Hiraga, Takehana, Taniguchi, Yamaji, Maeda, Toyohara, Miyamoto, Kimura, Oda, 2016). In 2016, a research team screened natural microbial communities at a PET bottle recycling site and managed to isolate a new bacterium named Ideonella sakaiensis (strain 201-F6) (Yoshida et al., 2016). Characterization of I. sakaiensis revealed that it could use PET as a primary energy and carbon source (Yoshida et al., 2016). After assembling the draft genome sequence of I. sakaiensis, the team identified an ORF that putatively encoded a hydrolase (Yoshida et al., 2016). Upon recombinant expression of this protein, the team observed that it exhibited PET-hydrolytic activity, thus naming it PET hydrolase (PETase) (Yoshida et al., 2016). PETase catalyzes the hydrolysis of PET into its monomeric component mono-2-hydroxyethyl terephthalate (MHET) (Yoshida et al., 2016). The genome of I. sakaiensis also coded for another enzyme that was capable of degrading MHET, which was designated MHET hydrolase (MHETase) (Danso, Chow, Streit, 2019, Yoshida, Hiraga, Takehana, Taniguchi, Yamaji, Maeda, Toyohara, Miyamoto, Kimura, Oda, 2016). MHETase hydrolyzes MHET into its two monomers, terephthalic acid (TPA) and ethylene glycol (EG) (Yoshida et al., 2016), which are used by I. sakaiensis in its metabolism (Danso et al., 2019). Moreover, TPA and EG can potentially serve as novel substrates to be converted into value-added products (Franden et al., 2018). EG, for instance, can be used for numerous applications, such as a coolant in antifreeze (Franden et al., 2018). It is worth noting that even though multiple studies have described enzymes that can degrade PET, the connection of extracellular enzymatic PET degradation to catabolism in a single microbe was hitherto unheard of (Austin et al., 2018). Additionally, the reported crystal structure resolution of MHETase is likely to possess a scaffold that is unprecedented for plastic-degrading enzymes (Palm et al., 2019). This example shows that bioprospection of the MDM offers a promising source for the identification of pollutant-degrading enzymes that could be used for bioremediation (Danso et al., 2019).
Another relevant example of the biotechnological potential of the MDM is that of the isolation chip (Ichip) (Nichols et al., 2010). In 2010, the Ichip was developed for the in situ cultivation of microbes that had eluded previous standard culture efforts (Berdy, Spoering, Ling, Epstein, 2017, Nichols, Cahoon, Trakhtenberg, Pham, Mehta, Belanger, Kanigan, Lewis, Epstein, 2010). The Ichip comprises several hundreds of miniaturized chambers, and each chamber harbors one or few cells from a given environmental sample. Each chip harbors cells from a single environment. Chamber incubation is carried out in the environment from where the cells were taken, allowing growth factors and other molecules to diffuse throughout the semipermeable membranes covering the chambers, thus facilitating growth and increasing the recovery of uncultured microbes (Rashid and Stingl, 2015). The discovery of the antibiotic Teixobactin (Ling et al., 2015b) not only demonstrates the efficacy of the Ichip, but also highlights the hidden potential of the MDM.
2.4. Catalytic prospectives
Enzymes are the backbone of numerous industries (Bruno, Coppola, di Prisco, Giordano, Verde, 2019, Cabrera, Blamey, 2018, Gurung, Ray, Bose, Rai, 2013, Li, Yang, Yang, Zhu, Wang, 2012, Meghwanshi, Kaur, Verma, Dabi, Vashishtha, Charan, Purohit, Bhandari, Bhojak, Kumar, 2020, Ramesh, Harani Devi, Chattopadhyay, Kavitha, 2020, Robinson, 2015, Singh, Kumar, Mittal, Mehta, 2016, Verma, Meghwanshi, Kumar, 2021). Reactions catalyzed by enzymes are thought to follow the rules of green chemistry-they are safer, faster, and generate less waste than traditional methods (Sysoev et al., 2021). The unmatched eco-friendly potential of enzymes is of vital use in the industry to mitigate the rampant overconsumption of our planet’s resources (Sysoev et al., 2021). Presently, there are more than two-hundred types of enzymes of microbial origin that are commercially available (Meghwanshi et al., 2020), of which about 20 types are produced on industrial scales (Li, Yang, Yang, Zhu, Wang, 2012, Verma, Meghwanshi, Kumar, 2021). The discovery of novel enzymes of biotechnological interest is critical for the growth of the industrial enzymes market. This market amounted to 9.9 billion USD in 2019, and it is projected to reach 14.9 billion USD by 2027 (Sysoev et al., 2021).
Most industrial enzymes currently originate from fungi or mesophilic bacteria (Grötzinger, Karan, Strillinger, Bader, Frank, Al Rowaihi, Akal, Wackerow, Archer, Rueping, Weuster-Botz, Groll, Eppinger, Arold, 2018, Sysoev, Grötzinger, Renn, Eppinger, Rueping, Karan, 2021), and the majority of enzymes of industrial relevance are hydrolytic in nature (i.e., hydrolases) (Verma et al., 2021). Examples of industrially-relevant enzymes that were unearthed from the MDM include cellulases (Piao et al., 2014), lipases (Verma et al., 2021), alcohol dehydrogenases (Akal, Karan, Hohl, Alam, Vogler, Grötzinger, Eppinger, Rueping, 2019, Grötzinger, Karan, Strillinger, Bader, Frank, Al Rowaihi, Akal, Wackerow, Archer, Rueping, Weuster-Botz, Groll, Eppinger, Arold, 2018), carbohydrate-active enzymes (Stewart et al., 2018), enzymes that catalyze organophosphorus compounds (Singh, 2009), along with other enzymes displaying enhanced stability under industrial conditions (Sysoev et al., 2021). Table 1 presents a list of enzymes of known biotechnological and/or industrial interest, gathered and manually curated from the literature.
Table 1.
Enzymes of biotechnological and/or industrial interest and their applications sorted by alphabetical order. Adapted and manually curated from (Bruno, Coppola, di Prisco, Giordano, Verde, 2019, Cabrera, Blamey, 2018, Gurung, Ray, Bose, Rai, 2013, Li, Yang, Yang, Zhu, Wang, 2012, Robinson, 2015, Singh, Kumar, Mittal, Mehta, 2016).
| EC number | Enzyme name | Application |
|---|---|---|
| EC:4.1.1.5 | Acetolactate decarboxylase | Converting -acetolactate to acetoin directly. Decreasing fermentation time by avoiding formation of diacetyl. |
| EC:2.7.4.3 | Adenylate kinase | Biological indicator for validation of procedures to inactivate transmissible spongiform encephalopathy agents. |
| EC:1.4.1.1 | Alanine dehydrogenase | Candidate for enantioselective production of optically active amino acids. |
| EC:1.1.1.1 | Alcohol dehydrogenase | Candidate for asymmetric synthesis. Reduction of C-O and C-C bonds. |
| EC:3.1.3.1 | Alkaline phosphatase | Candidate for molecular biology application: dephosphorylation of DNA. |
| EC:3.2.1.212 | Alpha-L-fucosidase | Establishing glycosidic bonds. |
| EC:3.2.1.1 | Alpha-amylase | Additive in food, textile, detergent and bioremediation industries. Waste-water treatment, drainage. Molecular biology applications. Treatment for digestive disorders. |
| EC:3.2.1.22 | Alpha-galactosidase | Additive in soybean foodstuff. |
| EC:3.5.1.4 | Amidase | Acylation, deacylation, enantioseparation. Degradation of nitrile-containing wastes. |
| EC:3.2.1.3 | Amyloglucosidase | Glucose production. Increasing glucose content in beverages. Additive in toothpastes, mouthwashes, and bioremediation. |
| EC:4.3.1.1 | Aspartase | l-aspartic acid production. |
| EC:3.2.1.2 | Beta-amylase | Producing low-molecular weight carbohydrate. Starch hydrolysis. Cleaving -1,4-linkages from non-reducing ends of amylose, amylopectin and glycogen molecules. |
| EC:3.2.1.21 | Beta-glucosidase | Production of ginseng compounds for medical applications. |
| EC:3.5.2.6 | Beta-lactamase | Molecular biology applications by conferring antibiotic resistance to Beta-lactam antibiotics. |
| EC:3.4.22.32 | Bromelain | Additive in the cosmetic industry. |
| EC:4.2.1.1 | Carbonic anhydrase | Candidate for biomedical applications. |
| EC:1.11.1.6 | Catalase | Candidate for textile and cosmetic industries. Antioxidants. Bleach termination. Cheese processing. |
| EC:3.2.1.4 | Cellulase | Additive in food, detergent and textile industries. Deinking. Drainage improvement. Degradation of cellulose in the textile industry. |
| EC:3.4.23.4 | Chymosin | Cheese manufacturing. |
| EC:3.1.1.74 | Cutinase | Triglyceride removal. Degradation of plastics, polycaprolactone. Additive in the textile industry. |
| EC:6.5.1.1 | DNA ligase (ATP) | Candidate for molecular biology applications. |
| EC:2.7.7.7 | DNA-directed DNA polymerase | DNA amplification used in the polymerase chain reaction and recombinant DNA technologies. |
| EC:3.3.2.10 | Epoxide hydratase | Candidate for the production of enantiopure epoxides in the pharmaceutical industry. |
| EC:3.1.11.1 | Exodeoxyribonuclease I | Candidate for molecular biology application: 3’-5’ exonuclease specific for single-stranded DNA. |
| EC:3.1.13.1 | Exoribonuclease II | Antiviral agent. Candidate for molecular biology applications. |
| EC:1.17.1.9 | Formate dehydrogenase | Oxidation of alcohols and oxygenation of C-H and C-C bonds. |
| EC:4.1.2.13 | Fructose-bisphosphate aldolase | Establishes C-C coupling. |
| EC:1.1.3.4 | Glucose oxidase | Dough strengthening. Used in toothpastes and mouthwashes. Oxygen removal from beer. Polymerization of anilines. Detection of glucose in blood. Bleaching agent. |
| EC:3.5.1.2 | Glutaminase | Cancer chemotherapy, particularly for leukemia. |
| EC:1.8.3.3 | Glutathione oxidase | Used in hair waving. |
| EC:1.11.1.9 | Glutathione peroxidase | Antioxidant properties. |
| EC:1.8.1.7 | Glutathione reductase | Candidate as an antioxidant enzyme in heterologous systems. |
| EC:3.2.1.68 | Isoamylase | Hydrolyzing -1,6-linkages in glycogen and amylopectin. |
| EC:3.5.1.1 | L-asparaginase | Cancer chemotherapy, particularly for leukemia. |
| EC:1.10.3.2 | Laccase | Non-chlorine bleaching, delignification. Additive in food, textile, cosmetic, and pesticide industries. Degradation of waste containing olefin unit, polyurethane and phenolic compounds. |
| EC:3.2.1.108 | Lactase | Lactose hydrolysis in dairy products or whey to avoid lactose intolerance. Antitumor agent. |
| EC:1.1.2.4 | Lactic acid dehydrogenase | Reduction of C-O and C-C bonds. |
| EC:1.4.1.9 | Leucine dehydrogenase | Candidate for medical and pharmaceutical industry applications. |
| EC:1.11.1.14 | Lignin peroxidase | Degradation of phenolic compounds. |
| EC:3.1.1.3 | Lipase | Additive in the food, detergent, cosmetic, textile, pharmaceutical, polymer, biodiesel, biosurfactant, pulping, and fossil-fuel industries. |
| EC:1.13.12.8 | Luciferase | Molecular biology applications such as bioluminescent assays involving ATP. |
| EC:3.2.1.17 | Lysozyme | Antibiotic. Disruption of mucopeptide in bacterial cell walls. Cheese manufacturing. |
| EC:1.1.1.37 | Malate dehydrogenase | Candidate for detection and production of malate. |
| EC:3.2.1.20 | Maltase | Additive in detergent and food industries. Production of glucose from maltose. |
| EC:3.2.1.133 | Maltogenic alpha-amylase | Enhances shelf life of bread. |
| EC:1.11.1.13 | Manganese peroxidase | Degradation of phenolic compounds. |
| EC:3.2.1.25 | Mannanase | Additive in food, detergent and textile industries. |
| EC:3.4.24.3 | Microbial collagenase | Treatment for skin ulcers. Wool finishing. |
| EC:3.1.1.102 | Mono(ethylene terephthalate) hydrolase | Conversion of PET monomers into terephthalic acid and ethylene glycol. |
| EC:3.5.1.14 | N-acyl-aliphatic-L-amino acid amidohydrolase | Production of L-amino acids. |
| EC:3.2.1.40 | Naringinase (alpha-L-rhamnosidase) | Acting on compounds that cause bitterness in citrus juices. Debittering. |
| EC:3.2.1.135 | Neopullulanase | Acting on both -1,6- and -1,4-linkages. |
| EC:4.2.1.84 | Nitrile hydratase | Degradation of nitrile-containing wastes. Used in acylation, deacylation, enantioseparation. Synthesis of acrylamide, butyramide, and nicotinamide. |
| EC:3.4.22.2 | Papain | Additive in the cosmetic industry. |
| EC:4.2.2.2 | Pectate lyase | Bioscouring. Candidate for the detergent industry. |
| EC:3.2.1.15 | Pectinase | Destabilizing the outer cell layer to improve fiber extraction via depectinization. Additive in food industries, such as clarification of juice and increasing its overall production, in the process of vinification, and the mashing of fruits. |
| EC:3.5.1.11 | Penicillin acylase | Semi-synthetic penicillin production/broad-spectrum antibiotic production. |
| EC:1.11.1.7 | Peroxidase | Hair dyeing. Quantification of hormones and antibodies. |
| EC:1.11.1.24 | Peroxiredoxin | Candidate for food and pharmaceutical industries. |
| EC:3.1.3.26 | Phytase | Candidate for feed applications, especially in aquaculture. Hydrolysis of phytic acid to release phosphorus, calcium, and magnesium cations. |
| EC:3.1.1.101 | Poly(ethylene terephthalate) hydrolase | Biodegradation of PET polyester plastic into monomers. |
| EC:1.10.3.1 | Polyphenol oxidase | Hair dyeing. |
| EC:5.3.4.1 | Protein disulfide-isomerase | Hair waving. |
| EC:3.2.1.41 | Pullulanase | Additive in food and biofuel industries. Attacking -1,6-linkages, liberating straight-chain oligosaccharides of glucose residues linked by -1,4-bonds. |
| EC:2.8.1.1 | Rhodanese | Cyanide poisoning treatment. |
| EC:1.10.3.6 | Rifamycin-B oxidase | Antibiotic synthesis. |
| EC:3.1.2.12 | S-formylglutathione hydrolase | Candidates for chemical synthesis and industrial pharmaceutics. |
| EC:5.3.1.28 | Sedoheptulose-7-phosphate isomerase | Candidate for biocatalysis under low water conditions. |
| EC:2.1.2.1 | Serine hydroxymethyltransferase | Candidate as a pharmaceutical, agrochemical and food additive. |
| EC:3.4.24.40 | Serralysin | Antiviral and anti-inflammatory properties. |
| EC:3.2.1.18 | Sialidase | Hydrolysis of glycosidic linkages of terminal sialic acid residues in oligosaccharides, glycoproteins, glycolipids, colominic acid and synthetic substrates. |
| EC:3.4.21.62 | Subtilisin | Additive in food, textile, leather, detergent, and cosmetic industries. Degrading protein into its constituent peptides and amino acids to overcome antinutritional factors. |
| EC:1.15.1.1 | Superoxide dismutase | Anti-inflammatory and antioxidant properties. Free radical scavenging. Candidate for applications in agriculture, cosmetics, food, healthcare products and medicines. |
| EC:3.4.24.27 | Thermolysin | Aspartame production. |
| EC:2.3.2.13 | Transglutaminase | Hair waving. Protein cross linking. Laminated dough strengthening. |
| EC:5.3.1.1 | Triosephosphate isomerase | Candidate for biocatalysis under low water conditions. |
| EC:3.4.21.4 | Trypsin | Anti-inflammatory and anti-coagulant properties. Molecular biology applications. Food processing. |
| EC:1.14.18.1 | Tyrosinase | Tumor-associated antigen. Polymerization of lignin and chitosan. |
| EC:3.2.2.27 | Uracil-DNA glycosylase | Candidate for molecular biology application: release of free uracil from uracil-containing single-stranded or double-stranded DNA. |
| EC:3.5.1.5 | Urease | Urea quantification in body fluids. |
| EC:1.7.3.3 | Uricase | Treatment of hyperuricemia. |
| EC:3.4.21.73 | Urokinase | Removal of fibrin clots from bloodstream. Anti-coagulant properties. |
| EC:3.4.24.25 | Vibriolysin | Additive in food, textile, leather, and detergent industries. |
| EC:3.2.1.32 | Xylanase | Additive in food, textile, detergent, pulp and bioremediation industries. Hydrolyzing pentosans of malt, barley and wheat. Enhancing pulp-bleaching efficiency. |
| EC:5.3.1.5 | Xylose isomerase | Production of high-fructose corn syrup. Catalyzing isomerization of glucose to fructose. |
Science has profited tremendously from numerous breakthroughs that relied on enzymes of microbial origin, as substantiated by the work of numerous Nobel Prize laureates (Bernard et al., 2018). Many of these early studies relied on restriction enzymes (Smith and Wilcox, 1970), then on DNA polymerases (Brock and Freeze, 1969) coupled to the advent of the polymerase chain reaction (Saiki et al., 1988), and more recently on the CRISPR-cas9 system (Jinek et al., 2012). Hence, microbial gene discovery can greatly push progress and development of new mechanisms and compounds of pharmaceutical, biotechnological, and biomedical relevance.
In a seminal report, Grotzinger et al. proposed a workflow targeting protein production based on single amplified genomes (SAGs) from species that we cannot culture yet (Grötzinger et al., 2018). As a proof-of-concept, they used the method described in their paper to unearth an alcohol dehydrogenase (ADH) from an uncharacterized polyextremophilic archaeon sampled from a brine pool at the bottom of the Red Sea (Grötzinger et al., 2018). ADHs are of industrial interest, given their ability to produce chiral compounds for pharmaceuticals and fine chemicals (Akal et al., 2019). ADHs can also be used in biosensor-based diagnostics and fuel-cell technologies (Grötzinger et al., 2018). Characterization of this ADH not only demonstrated its thermostability, halotolerance, and the ability to withstand the presence of different solvents, but also the prospect of it being stored and used as a powder; all of which are features of utmost biotechnological significance (Grötzinger et al., 2018). A more recent paper identified and characterized another ADH of similar polyextremophilic nature and with solvent tolerance, presumed to be a member of a rare enzyme family-that of microbial cinnamyl alcohol dehydrogenases (Akal et al., 2019).
Another notorious example is that of Stewart et al., who assembled 913 near-complete and draft-quality prokaryotic genomes from a rumen metagenome sequencing study (Stewart et al., 2018). These assembled genomes encoded over 69,000 novel enzymes presumed to be carbohydrate-active, and over 90% of which lacked a significant match in public databases (Stewart et al., 2018). These authors further highlight that their rumen metagenomic dataset not only offers a valuable resource for the discovery of biomass-degrading enzymes, but also that these novel enzymes might be potential candidates for application in the biofuels and biotechnology industries (Stewart et al., 2018).
More recently, there were studies that created hidden Markov models (HMMs) based on the protein sequences of enzymes whose plastic-degrading abilities had been experimentally validated (Danso, Schmeisser, Chow, Zimmermann, Wei, Leggewie, Li, Hazen, Streit, 2018, Zrimec, Kokina, Jonasson, Zorrilla, Zelezniak, 2021). By mining ocean and soil metagenomes, Zrimec et al. were able to compile a catalogue with more than 30 thousand non-redundant sequences that potentially coded for enzymes with the ability to degrade 10 different types of plastic (Zrimec et al., 2021). They report a significant correlation between the abundance of these enzymes in the two sampled biomes, and both marine and country-specific plastic pollution measurements (Zrimec et al., 2021). They suggest that these results might indicate signs of adaptation to current global plastic pollution trends by the Earth’s microbiome (Zrimec et al., 2021), thus emphasizing the potential within global microbiomes in providing solutions to contemporary concerns.
2.5. Biosynthetic Gene Clusters
Prokaryotes and other microorganisms (e.g., Fungi) are known to produce many secondary metabolites (SM) (Chen et al., 2019). SMs are natural products that encompass diverse chemical structures (Chen et al., 2019). This chemical diversity allows SMs to perform a plethora of functions (Chen et al., 2019). SMs may have antibiotic, anti-cancer, anti-viral, antifungal, antioxidant, anti-trypanosome, cholesterol-lowering, immunosuppressant, insecticide, and herbicide properties, among many others (Bull, Goodfellow, 2019, Chavali, Rhee, 2018, Chen, Wong, Burns, 2019, Newman, Cragg, 2016). Biosynthetic gene clusters (BGCs) are the physical grouping of the genes in a given genome that encode all enzymes required to produce a SM (Medema et al., 2015). Previous evidence has suggested that microorganisms may harbor up to one million BGCs (Hadjithomas, Chen, Chu, Ratner, Palaniappan, Szeto, Huang, Reddy, Cimermančič, Fischbach, Ivanova, Markowitz, Kyrpides, Pati, 2015, Ziemert, Alanjary, Weber, 2016), few of which have been thoroughly described (Ziemert et al., 2016).
Two major biosynthetic systems are those of polyketide synthases (PKS), and nonribosomal peptide synthases (NRPS) (Chen, Wong, Burns, 2019, Weber, Kim, 2016). PKS and NRPS synthesize the two major classes of SMs: polyketides (PK), and nonribosomal peptides (NRP) (Chen et al., 2019). PKS and NRPS are popular targets for bioprospection, given their reputation as producers of a broad range of SMs with important applications in healthcare and research (Chen et al., 2019). PK and NRP, together with terpenoids and alkaloids, were regarded as the four major groups of SMs throughout the 20th century (Arnison et al., 2013).
At the turn of the 21st century, the NGS revolution unveiled another major class of SMs, that of ribosomally synthesized and post-translationally modified peptides (RiPPs), which have since attracted increasing interest (Arnison, Bibb, Bierbaum, Bowers, Bugni, Bulaj, Camarero, Campopiano, Challis, Clardy, Cotter, Craik, Dawson, Dittmann, Donadio, Dorrestein, Entian, Fischbach, Garavelli, Göransson, Gruber, Haft, Hemscheidt, Hertweck, Hill, Horswill, Jaspars, Kelly, Klinman, Kuipers, Link, Liu, Marahiel, Mitchell, Moll, Moore, Müller, Nair, Nes, Norris, Olivera, Onaka, Patchett, Piel, Reaney, Rebuffat, Ross, Sahl, Schmidt, Selsted, Severinov, Shen, Sivonen, Smith, Stein, Süssmuth, Tagg, Tang, Truman, Vederas, Walsh, Walton, Wenzel, Willey, van der Donk, 2013, Hetrick, van der Donk, 2017). This interest stems from academic and industrial sectors alike, due to the structural variability and functional diversity shown by RiPPs (Zhong et al., 2020). The chemical space of RiPPs is determined by their nucleotide sequence, therefore linking the diversity of these small molecules with that of genes (Zhong et al., 2020). The genetically-encoded nature of RiPPs enables researchers to freely manipulate the scaffolds of the peptides by site-directed mutagenesis, and efficiently screen the targets for those possessing characteristics of interest (Zhong et al., 2020).
Several computational tools are able to accurately identify BGCs (see Chavali and Rhee (2018)), albeit not without limitations. These tools might rely on external databases, and/or rules extracted from previous knowledge, implying that only known biosynthetic pathways whose rules are implemented in the software are detected (Chen et al., 2019). Thus, biosynthetic pathways that make use of enzymes from the FDM will be missed (Blin, Kim, Medema, Weber, 2019, Chen, Wong, Burns, 2019). Bioprospecting BGC data from metagenomes is also challenging, as the computational tools that do so commonly require high-quality genomes, or those resolved from metagenomes as input (Blin, Kim, Medema, Weber, 2019, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Skinnider, Johnston, Edgar, Dejong, Merwin, Rees, Magarvey, 2016, Youngblut, de la Cuesta-Zuluaga, Reischer, Dauser, Schuster, Walzer, Stalder, Farnleitner, Ley, 2020). Another major challenge in natural product discovery is that a substantial portion of BGCs are transcriptionally silent, or expressed at very low levels when in a standard laboratory setting (Chen, Wong, Burns, 2019, Ren, Wang, Zhao, 2017). Strategies designed to activate these silent BGCs are crucial for discovering new chemical scaffolds (Goodfellow et al., 2018).
Nonetheless, there are successes arising from the systematic interrogation of BGCs from within the FDM (Bull, Goodfellow, 2019, Goodfellow, Nouioui, Sanderson, Xie, Bull, 2018, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Skinnider, Johnston, Edgar, Dejong, Merwin, Rees, Magarvey, 2016, Youngblut, de la Cuesta-Zuluaga, Reischer, Dauser, Schuster, Walzer, Stalder, Farnleitner, Ley, 2020). As an example, a research team developed a novel algorithm that catalogs RiPP biosynthetic gene clusters (Skinnider et al., 2016). Upon analyzing 65,000 prokaryotic genomes, they unearthed RiPP BGCs that coded for more than two-thousand novel natural products (Skinnider et al., 2016). Bull and Goodfellow have studied BGCs while focusing their bioprospecting efforts on the phylum Actinobacteria (Bull and Goodfellow, 2019). Their rationale for focusing on this taxon is fivefold: (i) the recurrent and foundational role of Actinobacteria in soil ecosystems; (ii) the size and diversity of the taxon; (iii) the ceaseless discovery of new taxonomic radiations; (iv) the BGC-rich genomes of Actinobacteria; (v) and their unparalleled track record as producers of bioactive compounds of notable ecological and economic value (Bull and Goodfellow, 2019). SMs discovered from Actinobacteria–especially Streptomyces strains–account for two thirds of known antibiotics (Bérdy, 2012), including those in clinical use today (Goodfellow et al., 2018). Actinobacteria are also known to produce roughly tenfold as many specialized metabolites as those known from laboratory experiments (Goodfellow et al., 2018). Consequently, this ability has renewed interest in these prokaryotes as producers of new chemical entities (Goodfellow et al., 2018).
Bull and Goodfellow emphasize that SMs have a significantly greater diversity and quantity of chemical scaffolds than those produced by combinatorial synthetic compounds (Bull and Goodfellow, 2019). Thus providing a compelling reason for prioritizing them in the search of novel drugs (Bull and Goodfellow, 2019). Their research into Actinobacteria recovered from two extreme environments has uncovered a remarkable assortment of new chemical class members, and each of those products is either a new-in-a-class or first-in-a-class chemical entity (Bull and Goodfellow, 2019). The most widely distributed bioactivity of these compounds is that of antibacterial and anticancer activities (Bull and Goodfellow, 2019). They report that these compounds are also putative drug hits that could provide potential therapeutic targets for inflammatory diseases, Alzheimer’s disease, and type II diabetes (Bull and Goodfellow, 2019). The discoveries disclosed above further elevate the foregoing rationale: the exploitation of biological know-how from within the FDM can offer an unprecedented range of biotechnological solutions. These solutions might not only be at the core of new markets and business models (Cornelissen et al., 2021), but also at the bleeding edge of innovation in therapeutics, industrial applications, and bioremediation strategies.
2.6. Additional insights into the functional dark matter
Deep investigation of the FDM has prompted an exciting scientific revolution. Besides presenting evidence for biotechnological exploitability, this revolution has provided invaluable insights into the MDM in a broader sense. For instance, in a study of uncharacterized genomic “islands” from archaeal genomes, Makarova et al. found that besides being abundant and comprising a heterogeneous gene pool of diverse putative functions; these islands also code for defense systems, along with new variants of the CRISPR-Cas genome editing system (Makarova et al., 2014). Other articles described synthrophic networks in anaerobic methanogenic consortia of uncultured microorganisms (Gies, Konwar, Beatty, Hallam, 2014, Nobu, Narihiro, Rinke, Kamagata, Tringe, Woyke, Liu, 2015), as well as in benzene-degrading settings (Luo et al., 2016). In this way, they highlight the potential applications for anaerobic bioreactors aimed at bioremediation and energy generation (Gies et al., 2014). Another study predicted metabolic roles for multifaceted chemoorganoheterotrophic bacterioplankton, that would be involved with degradation of complex carbon compounds and the nitrogen cycle (Thrash et al., 2017). A different study revealed that uncultivated ultra small marine prokaryotes encoded for a wealth of gene homologs associated with diverse metabolic pathways, such as: carbon, methane, nitrogen, and sulfur (Lannes et al., 2021). These authors highlight that these little known prokaryotes presumably contribute to elemental cycling (Lannes et al., 2021).
Wong et al. reconstructed 115 genomes assembled from hypersaline microbial mat metagenomes (Wong et al., 2020). They uncovered novel eukaryotic signature proteins in the Asgard archaeal superphylum, many forms of RuBisCo (ribulose-1,5-bisphosphate carboxylase-oxygenase), high hydrogen production capacity, putative schizorhodopsins, and diversity-generating retroelements, among many other findings (Wong et al., 2020). Around the same time, Wiegand et al. characterized and sequenced the genome of 79 bacterial strains from the enigmatic bacterial phylum Planctomycetes (Wiegand et al., 2020). These authors identified previously unknown modes of bacterial cell division, such as lateral budding and binary fission; as well as new cell signaling and small-molecule production processes (Wiegand et al., 2020). Their study also advanced that the vast majority of putative BGCs encoded by planctomycetes differ from known BGCs, hinting at an untapped potential for small-molecule production (Wiegand et al., 2020). Two articles reported extensive drug discovery potential amidst the microbiome of distinct marine sponges, evidencing a wealth and breadth of untapped resources for novel chemistry (Lackner, Peters, Helfrich, Piel, 2017, Rust, Helfrich, Freeman, Nanudorn, Field, Rückert, Kündig, Page, Webb, Kalinowski, Sunagawa, Piel, 2020).
Also worthy of mention are several other studies that: identified hundreds of metalloproteases with signature catalytic motifs within ORFs of previous unknown function (Lobb et al., 2015); probed uncharacterized groups of Acidobacteria displaying extensive carbon catabolic abilities, including polysaccharide breakdown and metabolism of lignin derivatives (Wegner and Liesack, 2017); characterized new members of the Oceanospirillales order whose genomes code for enzymes capable of metabolizing crude oil (Mason et al., 2012); and described exceptionally high diversity of actinobacteria in the arid Atacama desert, outlining its remarkable significance for future biodiscovery campaigns (Bull, Idris, Sanderson, Asenjo, Andrews, Goodfellow, 2018, Idris, Goodfellow, Sanderson, Asenjo, Bull, 2017).
3. Progress and pitfalls
There are numerous reasons why the function of many proteins has not been characterized yet. This problem is manifold. It starts with DNA extraction protocols, and sequencing technologies, which have limitations. Yet, also significant are most mainstream public databases, and associated computational tools, that in many cases are prone to unsupervised dissemination of (mis)information. In the following section we will briefly overview the progress made by current approaches and technologies, and discuss “how” and “why” they might be fostering an ever-increasing inflation in the number of protein sequences of unknown function.
3.1. Shotgun metagenomics
Shotgun metagenomics is the untargeted DNA sequencing performed from an environmental sample (Quince et al., 2017). This approach consists on extracting the DNA from a sample, which can be made into libraries, and then sequencing that DNA using either a short-read (i.e., NGS) or long-read platform (i.e., TGS) (New and Brito, 2020). A metagenome by itself, represents a bulk of fragmented genomic data, from a multitude of microorganisms, at different abundances. Two of the major assets of shotgun metagenomics are its versatility, as it can be used with different types of samples, and applied when other approaches such as SCG, have failed (Hedlund et al., 2014); and its simplicity in terms of sample preparation and data acquisition, provided that a suitable amount of DNA has been extracted.
Many discoveries were only made possible by the advances in metagenomics. Examples of this are the identification of bacteria that can perform complete oxidation of ammonia to nitrate (Daims, Lebedeva, Pjevac, Han, Herbold, Albertsen, Jehmlich, Palatinszky, Vierheilig, Bulaev, Kirkegaard, von Bergen, Rattei, Bendinger, Nielsen, Wagner, 2015, van Kessel, Speth, Albertsen, Nielsen, Op den Camp, Kartal, Jetten, Lücker, 2015), and BGCs that code for antibiotics in the human gut microbiome (Donia et al., 2014), among many others. Metagenomic studies have disclosed a momentous portion of MDM. One study created a ocean microbiome gene catalog comprising more than 40 million nonredundant and mostly new sequences from prokaryotes, viruses, and picoeukaryotes (Sunagawa et al., 2015). More recently, a different study has created a non-redundant gene catalogue of 303 million genes from 13,174 metagenomes spanning 14 major biomes (Coelho et al., 2022). Another project allowed to gather genomic information on hundreds of bacterial species with no sequenced representatives, enabling their subsequent use in reference-based metagenomic studies (Human Microbiome Project Consortium, 2012). Outstandingly, metagenomics allowed to uncover a novel branch of bacteria: the Patescibacteria, previously known as Candidate Phyla Radiation (CPR) (Brown, Hug, Thomas, Sharon, Castelle, Singh, Wilkins, Wrighton, Williams, Banfield, 2015, Parks, Rinke, Chuvochina, Chaumeil, Woodcroft, Evans, Hugenholtz, Tyson, 2017); and two new archaeal superphyla: the DPANN (i.e., Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaeota) (Castelle, Banfield, 2018, Dombrowski, Lee, Williams, Offre, Spang, 2019, Dombrowski, Williams, Sun, Woodcroft, Lee, Minh, Rinke, Spang, 2020, Parks, Rinke, Chuvochina, Chaumeil, Woodcroft, Evans, Hugenholtz, Tyson, 2017, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013), and the Asgard superphylum (Eme, Spang, Lombard, Stairs, Ettema, 2018, Imachi, Nobu, Nakahara, Morono, Ogawara, Takaki, Takano, Uematsu, Ikuta, Ito, Matsui, Miyazaki, Murata, Saito, Sakai, Song, Tasumi, Yamanaka, Yamaguchi, Kamagata, Tamaki, Takai, 2020, López-García, Moreira, 2020, López-García, Moreira, 2020, Spang, Saw, Jørgensen, Zaremba-Niedzwiedzka, Martijn, Lind, van Eijk, Schleper, Guy, Ettema, 2015, Zaremba-Niedzwiedzka, Caceres, Saw, Bäckström, Juzokaite, Vancaester, Seitz, Anantharaman, Starnawski, Kjeldsen, Stott, Nunoura, Banfield, Schramm, Baker, Spang, Ettema, 2017).
Even though metagenomics has expanded our knowledge on the function and diversity of microbial communities, it has its limitations. Metagenomic data suffers from several confounders that can render it incomplete, biased, and difficult to interpret (New and Brito, 2020). These confounding factors include, but are not limited to, sequencing noise, phenotypic noise, spatial microenvironments, and contaminant DNA (Ackermann, 2013, Doud, Woyke, 2017, Quince, Walker, Simpson, Loman, Segata, 2017). In addition, a metagenome only renders a snapshot of a microbial community at a given time, and it may not depict the active microbial population (i.e., the DNA might issue from dead or dying cells) (Bellali et al., 2021). These factors may consequently: (i) mask population (i.e., operational taxonomic units–OTUs) and (ii) genetic diversity (e.g., SNPs); (iii) hinder taxonomic assignment, (iv) functional profiling, and (v) the recovery of individual genomes; (vi) overlook mobile genetic elements; (vii) neglect rare taxa; (viii) misconceive spatially divergent populations; (ix) ignore ecological relationships,(x) phenotypic traits, and (xi) metabolic output; and (xii) restrain genomic context analysis (D Ainsworth, Krause, Bridge, Torda, Raina, Zakrzewski, Gates, Padilla-Gamiño, Spalding, Smith, Woolsey, Bourne, Bongaerts, Hoegh-Guldberg, Leggat, 2015, Doud, Woyke, 2017, Engel, Stepanauskas, Moran, 2014, New, Brito, 2020, Quince, Walker, Simpson, Loman, Segata, 2017). This makes it so that some information relating to microbial communities, and its constituents, is prone to be lost among these data.
Arguably, the greatest limitation of metagenomes is the difficulty in annotating the function of most of the genomic data (Dutilh, 2014, Lobb, Kurtz, Moreno-Hagelsieb, Doxey, 2015, Mokili, Rohwer, Dutilh, 2012). There are several variables that influence metagenome function annotation besides those enumerated above. These include the complexity of the metagenome (i.e., richness and homogeneity of species), read length and format (i.e., single or paired end), sequencing depth, coverage, the size and quality of the data, taxonomic novelty, environmental context, and the sensitivity and accuracy of the computational methods used for downstream annotation (Lobb et al., 2015). The foremost reason however stems from metagenomic data itself, which consists on short-reads, in case of NGS-sequenced metagenomes. Usually, these short-reads are assembled into longer fragments named contigs, then genes are predicted and mapped to reference sequence databases (Treiber et al., 2020). However, this process oftentimes generates many short contigs, chimeric contigs (i.e., assemblies between different species), and reads that could not be assembled at all. Given that contig function prediction usually relies on homology-estimation algorithms (Doud and Woyke, 2017), the efforts to annotate these short contigs, chimeric contigs, or unassembled reads may prove unsuccessful. The reasons for this include the fact that short query lengths could lead to inaccurate assignments (Wommack et al., 2008), be deprived of significant matches against reference sequence databases (Prakash and Taylor, 2012), or fail to differentiate between functions, because they might align to promiscuous domains shared among distinct proteins (Tamames et al., 2019).
3.2. Metagenome-assembled genomes
Gene neighborhoods allow to deduce the function of FDM genes (Cotroneo et al., 2021). This may be achieved by analyzing the genes of known function with whom the FDM genes are consistently linked (i.e., across multiple genomes) (Koonin et al., 2021). Contiguous genomic data also enhances both functional profiling and taxonomic classification (Kayani, Huang, Feng, Chen, 2021, Somerville, Lutz, Schmid, Frei, Moser, Irmler, Frey, Ahrens, 2019, Tamames, Cobo-Simón, Puente-Sánchez, 2019). Nonetheless, metagenomic reads lack genomic context, because they have yet to be associated with their neighboring genes, and ultimately their genome of origin. To reconstruct the genomic context of metagenomic reads, two main computational procedures are usually required: metagenomic assembly, and “binning”. Metagenomic assembly allows not only to gather contigs from metagenomic data, but also operons, gene arrays, syntenic blocks, or even putative metagenome-assembled genomes (MAGs) (Almeida, Nayfach, Boland, Strozzi, Beracochea, Shi, Pollard, Sakharova, Parks, Hugenholtz, Segata, Kyrpides, Finn, 2021, Olson, Treangen, Hill, Cepeda-Espinoza, Ghurye, Koren, Pop, 2019). Contigs are usually grouped (i.e., “binned”) together after assembly, so taxon-specific gene inventories can be created (Teeling and Glöckner, 2012). Binning is achieved by grouping contigs according to their intrinsic nucleotide composition and/or co-abundance profiles (i.e., unsupervised binning) (Hedlund, Dodsworth, Murugapiran, Rinke, Woyke, 2014, Nielsen, Almeida, Juncker, Rasmussen, Li, Sunagawa, Plichta, Gautier, Pedersen, Le Chatelier, Pelletier, Bonde, Nielsen, Manichanh, Arumugam, Batto, Quintanilha Dos Santos, Blom, Borruel, Burgdorf, Boumezbeur, Casellas, Doré, Dworzynski, Guarner, Hansen, Hildebrand, Kaas, Kennedy, Kristiansen, Kultima, Léonard, Levenez, Lund, Moumen, Le Paslier, Pons, Pedersen, Prifti, Qin, Raes, Sørensen, Tap, Tims, Ussery, Yamada, MetaHIT Consortium, Renault, Sicheritz-Ponten, Bork, Wang, Brunak, Ehrlich, MetaHIT Consortium, 2014, Plaza Oñate, Le Chatelier, Almeida, Cervino, Gauthier, Magoulès, Ehrlich, Pichaud, 2019); or by assigning a taxon to a contig based on its homology to sequences of known taxonomy (i.e., supervised binning) (Teeling and Glöckner, 2012). When combined, metagenomic assembly and binning have allowed to obtain closed MAGs from candidate phyla (Brown, Hug, Thomas, Sharon, Castelle, Singh, Wilkins, Wrighton, Williams, Banfield, 2015, Kantor, Wrighton, Handley, Sharon, Hug, Castelle, Thomas, Banfield, 2013). However, supervised binning often leads to numerous genomic fragments that cannot be mapped to reference genomes (Quince et al., 2017). This is mainly due to the magnitude of prokaryotes from the TDM that lack representative genomes (New and Brito, 2020). Thus, the genomic fragments from these prokaryotes cannot be classified by supervised approaches. Furthermore, the assembly and binning processes often fail to distinguish between related community members, resulting in MAGs that erroneously include contigs from distinct strains and/or species (Parks et al., 2017). This has repercussions on downstream function imputation and taxonomic assignments (Almeida et al., 2021).
An alternative consists in reconstructing MAGs directly from read data (i.e., de novo) (Olson et al., 2019). Prominent discoveries have been made based on de novo reconstruction of MAGs (Almeida, Mitchell, Boland, Forster, Gloor, Tarkowska, Lawley, Finn, 2019, Anantharaman, Brown, Hug, Sharon, Castelle, Probst, Thomas, Singh, Wilkins, Karaoz, Brodie, Williams, Hubbard, Banfield, 2016, Delmont, Quince, Shaiber, Esen, Lee, Rappé, McLellan, Lücker, Eren, 2018, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Parks, Rinke, Chuvochina, Chaumeil, Woodcroft, Evans, Hugenholtz, Tyson, 2017, Pasolli, Asnicar, Manara, Zolfo, Karcher, Armanini, Beghini, Manghi, Tett, Ghensi, Collado, Rice, DuLong, Morgan, Golden, Quince, Huttenhower, Segata, 2019). Despite these developments, many NGS-sequenced MAGs–and prokaryotic genomes in a general sense–consist on draft assemblies spanning hundreds to thousands of contigs (Koren et al., 2013), from which even core genes might be absent (Schmid et al., 2018). The primary reason for these incomplete assemblies is that NGS-generated reads are usually shorter than intragenomic repeats (Kingsford et al., 2010). Assembly complexity has also been directly associated with the ratio between the length of reads and that of repeats (Nagarajan and Pop, 2009). Repeat elements pose a challenge to de novo assembly algorithms (Koren et al., 2013), that typically fail to resolve them (Derakhshani et al., 2020), leading to fragmented assemblies, or misassemblies altogether. This challenge becomes even harder to overcome in MAG de novo assembly. In this case, the assemblers must not only resolve intragenomic repeats, but also intergenomic repeats (i.e., genomic fragments shared across strains) (Olson, Treangen, Hill, Cepeda-Espinoza, Ghurye, Koren, Pop, 2019, Somerville, Lutz, Schmid, Frei, Moser, Irmler, Frey, Ahrens, 2019). Failure to resolve intergenomic repeats may lead to the creation of composite MAGs, which complicate the ensuing interpretation of results (e.g., inaccurate functional profiles and taxonomic diversity, and distorted population abundance and prevalence) (New and Brito, 2020). Other factors that contribute to MAG incompleteness are sequencing depth and coverage. The coverage of a MAG depends on the abundance of the prokaryote of origin within the sampled community, and low-abundance prokaryotes may give rise to fragmented MAGs if the sequencing depth does not allow for genomic contiguity (Olson, Treangen, Hill, Cepeda-Espinoza, Ghurye, Koren, Pop, 2019, Quince, Walker, Simpson, Loman, Segata, 2017). Draft assemblies are also error-prone (Derakhshani, Bernier, Marko, Surette, 2020, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021). This limitation is irrespective of assembly algorithm and/or sequencing method–it is a result of the inherent complexity of the metagenomic assembly process (Olson et al., 2019).
MAG quality is usually assessed by measures, namely completeness and contamination (Bowers et al., 2017). Completeness and contamination have the standard English meanings, and these measures can be estimated in different ways (see Bowers et al. (2017); Nayfach et al. (2019); Orakov et al. (2021); Vollmers et al. (2022)). As an example, completeness might be calculated as the ratio between the number of observed and total single-copy marker genes from a marker gene set; and contamination as the ratio between the number of observed copies and total single-copy marker genes from a marker gene set (Bowers et al., 2017). Nonetheless, these estimates might be more appropriate to assess the quality of MAGs from known prokaryotes, as they might fail to attain the same resolution with MAGs issuing from the TDM (New and Brito, 2020), leading to MAGs whose quality cannot be accurately ascertained. The compilation of MAG-associated problems described hitherto might compromise downstream prediction of coding sequences, and the imputation of their function (Mavromatis et al., 2012).
To circumvent MAG fragmentation and incompleteness, some studies increase the minimum contig length threshold for MAG de novo assembly (Almeida, Mitchell, Boland, Forster, Gloor, Tarkowska, Lawley, Finn, 2019, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Pasolli, Asnicar, Manara, Zolfo, Karcher, Armanini, Beghini, Manghi, Tett, Ghensi, Collado, Rice, DuLong, Morgan, Golden, Quince, Huttenhower, Segata, 2019). Other studies use sequencing data gathered from TGS technologies in the first place (Moss, Maghini, Bhatt, 2020, Somerville, Lutz, Schmid, Frei, Moser, Irmler, Frey, Ahrens, 2019). TGS is characterized by single-molecule, real-time sequencing of nucleotides, without the need for template amplification (Athanasopoulou et al., 2021). These traits allow to overcome amplification, sequencing, and compositional biases inherent to other molecular approaches (Sims et al., 2014). The most attractive feature of TGS is the production of long reads, with average read lengths surpassing 10kb (van Dijk et al., 2018), and reported maxima of 1Mb (Jain et al., 2018). Increasing read length and contiguity improves the quality and accuracy of de novo assemblies (Koren et al., 2013), because fewer contigs are required to close a genome (Moss et al., 2020). The ability of long reads to span past repeat elements and anchor their copies to unique parts of the genome has allowed to overcome the major drawback of NGS-generated assemblies (Loman et al., 2015). The long reads produced by TGS, together with ever-improving de novo assembly algorithms (Dida and Yi, 2021), have already allowed to attain complete, closed, de novo-assembled prokaryotic genomes (Loman et al., 2015), and MAGs (Moss, Maghini, Bhatt, 2020, Somerville, Lutz, Schmid, Frei, Moser, Irmler, Frey, Ahrens, 2019). Fully resolved genomes are particularly valuable (Somerville, Lutz, Schmid, Frei, Moser, Irmler, Frey, Ahrens, 2019, Varadarajan, Allan, Valentin, Castañeda Ocampo, Somerville, Pietsch, Buhmann, West, Skipp, van der Mei, Ren, Schreiber, Webb, Ahrens, 2020). Namely because these offer a complete landscape where the ensuing characterization of functional elements can take place. However, one should mind that even though MAGs may act as genomic placeholders for the TDM, these should eventually, and ideally, be replaced by genomes sequenced from clonal isolates (Nayfach et al., 2021).
3.3. Nucleic acid extraction
In studies of molecular microbial ecology, it is reasonable to favor the enrichment of prokaryotic DNA with a specific functional activity, prior to sequencing (Healy et al., 1995). However, in many cases (e.g., metagenomics) the interest may not be in favoring the DNA extraction of a particular type of prokaryote, but rather to recover the bulk DNA of the whole community. In these cases, the extraction method must allow to recover DNA from prokaryotes with distinct characteristics (e.g., cell wall morphology). Otherwise, sequencing data might be biased towards the prokaryotes that were the most susceptible to lysis (Quince et al., 2017). Thus, the DNA extraction method must suit the goal of the study, without compromising downstream sequence data interpretation.
When extracting DNA from an environmental sample, two main approaches are usually considered. Either the DNA is directly extracted in bulk from the sample (i.e., “direct extraction”); or cells are recovered from the sample, and only then are they lysed and the DNA extracted (i.e., “indirect extraction”) (Kakirde et al., 2010). The “direct extraction” method has significant advantages: it is less time-consuming, and yields greater amounts of DNA (Kakirde et al., 2010). However, this method also isolates a high yield of non-prokaryotic contaminant DNA, and it produces shorter DNA fragments as a result of harsh lysis and purification steps (Kakirde et al., 2010). The “indirect extraction” method can overcome these limitations to a certain extent, since it tends to preserve the genomic DNA in larger fragments that may be further used to build metagenomic libraries and large prokaryotic DNA templates (Kakirde et al., 2010). It has also been shown to yield less non-prokaryotic DNA than the “direct extraction” method, while still collecting DNA from diverse prokaryotic taxa (Gabor et al., 2003). Nonetheless, the “indirect extraction” method is more laborious and yields less DNA, as it is potentially biased towards microorganisms that are not assorted in an environmental matrix, biofilm, or aggregated together with sample material (Kakirde et al., 2010).
Consequently, metagenomics has typically relied on short fragments from “direct extraction” protocols, followed by high-throughput NGS. Despite being a robust approach, it falls short when factors such as sequencing depth are considered (Sims et al., 2014), as its potential to unveil the taxonomic and functional profiles of a given community is restrained by read length (Brown et al., 2017). Although DNA extraction protocols can be optimized for a particular prokaryote of interest, this is more difficult to achieve when dealing with metagenomic mixtures (Olson et al., 2019). Additionally, in order to increase DNA yield, extraction protocols that achieve cell lysis through physical disruption (e.g., bead beating and sonication) have been preferred over those that use chemical lysis (Yuan et al., 2012). Yet, not only do these approaches vary in efficiency (Kennedy et al., 2014), they also shear the DNA, thus contributing to its loss during library preparation methods that select for fragment size (Quince et al., 2017). The efficacy of long-read TGS technologies depends on pure high-molecular-weight (HMW) DNA being provided to the sequencing instrument in the first place (Jones et al., 2021). Therefore, DNA extraction methods that generate small fragments for short-read sequencing are inappropriate for TGS (Mayjonade et al., 2016). To circumvent this issue, protocols to create bacterial artificial chromosome (BAC) libraries might be adapted; but these are laborious, and the commercial kits that are available are expensive (Mayjonade et al., 2016). Extraction methods that make use of agarose gel matrices (i.e., agarose plugs) can yield megabase-sized DNA molecules (Zhang et al., 2012); yet they span several days, and it is difficult to retrieve the DNA from the agarose plug (Mayjonade et al., 2016).
With these constraints in mind, Mayjonade et al. developed a protocol for fast and inexpensive extraction of HMW DNA that is suitable for TGS (Mayjonade et al., 2016). First, cell lysis is achieved through a combination of mechanical and chemical disruption (Mayjonade et al., 2016). The biological material is homogenized with SDS-based buffer, which disrupts cell membranes and inactivates DNA-degrading proteins (e.g., nuclease); and RNase A is added into the mixture, thereby digesting RNA (Mayjonade et al., 2016). Then, potassium acetate is added to the solution, leading to the formation of potassium dodecyl sulfate precipitate (Mayjonade et al., 2016). This compound binds contaminants like proteins and polysaccharides, that are removed via centrifugation (Mayjonade et al., 2016). DNA purification ensues by binding it to carboxylated magnetic beads with polyethylene glycol (Mayjonade et al., 2016). The authors emphasize that this combination is preferred, because carboxylated magnetic beads are inexpensive and cause minimal shearing; and polyethylene glycol precipitates less contaminants than other reactants like isopropanol and ethanol (Mayjonade et al., 2016). After ethanol washing, and resuspension in a buffer solution, highly pure HMW DNA is retrieved (Mayjonade et al., 2016). Many other DNA extraction protocols targeting TGS have been recently developed, adapted, and outlined (Jones, Torkel, Stanley, Nasim, Borevitz, Schwessinger, 2021, Maghini, Moss, Vance, Bhatt, 2021, Moss, Maghini, Bhatt, 2020, Trigodet, Lolans, Fogarty, Shaiber, Morrison, Barreiro, Jabri, Eren, 2022). With TGS technologies at the forefront of current advancements (Jones et al., 2021), one can envision that DNA extraction protocols directed towards long-read sequencing will keep being improved and refined, allowing for increased levels of DNA purity and yield, and thus enhancing downstream sequencing resolution and read contiguity.
3.4. Rare taxa
Culture-independent methods are bounded by protocol heterogeneity (e.g., DNA extraction protocols, primers used for amplification), and are especially susceptible to depth bias (Lagier, Dubourg, Million, Cadoret, Bilen, Fenollar, Levasseur, Rolain, Fournier, Raoult, 2018, Sims, Sudbery, Ilott, Heger, Ponting, 2014). Thus, they make it difficult to detect prokaryotes that exist in low numbers, also known as the microbial “rare biosphere” (Sogin et al., 2006). Although scarcely represented, this minority might perform important functions, like that of a “seed bank” (i.e., dormant or metabolically inactive cells that behave as a genomic reservoir), or as “keystone species” (i.e., disproportional ecological roles in relation to their numbers) (Pascoal et al., 2020). A significant proportion of this observed, yet unculturable, diversity might result from stochastic phenomena, like random dispersal, or transient existence (Pascoal et al., 2020). These prokaryotes might also require long incubation times to form visible colonies, or require specific nutrients and physical conditions for growth (Bellali et al., 2021). Alternatively, they might be composed of dead or dying cells (Pedrós-Alió, 2012).
As an example, Bellali et al. set out to test the hypothesis that some species from the human gut microbiota remain uncultured because their cells are dead before reaching the end of the gastrointestinal tract, and not exclusively because of culture limitations (Bellali et al., 2021). By combining fluorescence-activated cell sorting (FACS) and culturomics, these authors were able to discriminate between live, injured, and dead bacterial groups (Bellali et al., 2021). They outlined that minority species constituted a substantial portion of the live TDM in human fecal samples, and that 28% of bacterial OTUs in the total fecal samples were either dead or injured (Bellali et al., 2021). Moreover, roughly two-thirds of the latter group were members of the TDM, consisting in part of anaerobic bacteria (Bellali et al., 2021). This result might explain why some taxa were missing in culture, given that culturing obligate anaerobes often requires specific laboratory equipment (e.g., anaerobic chamber) so as to provide an anoxic environment for these organisms (Lagier et al., 2018). Techniques like metatranscriptomics, or compounds that bind free DNA (e.g., propidium monazide), coupled to “indirect” DNA extraction protocols, might be useful to prevent potential biases arising from injured or dead cells (Quince et al., 2017).
The unculturability of most prokaryotes is clearly a deterring factor towards a comprehensive characterization of the microbial realm. Yet, one might argue that the underrepresentation of rare, uncultured, or unclassified prokaryotic taxa among reference genome catalogs is a more pressing concern (Zhang et al., 2020). This predicament impairs our ability to classify genomic sequences that were gathered from culture-independent approaches to begin with (Carr, Borenstein, 2014, Quince, Walker, Simpson, Loman, Segata, 2017, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013). Most reference genome catalogs are biased towards pathogens, model organisms, and those amenable to cultivation (Quince et al., 2017). Taken together, these circumstances provide a distorted view of prokaryotic functional and taxonomic diversity (Mukherjee et al., 2017). Accurate functional annotation itself depends on the phylogenetic breadth of reference genomes (Carr and Borenstein, 2014). In addition, the discovery of functional novelty has been correlated with phylogenetic distance (Kunin, Cases, Enright, de Lorenzo, Ouzounis, 2003, Mukherjee, Seshadri, Varghese, Eloe-Fadrosh, Meier-Kolthoff, Göker, Coates, Hadjithomas, Pavlopoulos, Paez-Espino, Yoshikuni, Visel, Whitman, Garrity, Eisen, Hugenholtz, Pati, Ivanova, Woyke, Klenk, Kyrpides, 2017, Wu, Hugenholtz, Mavromatis, Pukall, Dalin, Ivanova, Kunin, Goodwin, Wu, Tindall, Hooper, Pati, Lykidis, Spring, Anderson, D’haeseleer, Zemla, Singer, Lapidus, Nolan, Copeland, Han, Chen, Cheng, Lucas, Kerfeld, Lang, Gronow, Chain, Bruce, Rubin, Kyrpides, Klenk, Eisen, 2009). By broadening our genome repertoire one might not only improve functional annotation and taxonomic assignment of genomic sequences, but also extend the genomic landscape where genes and functions of interest can be mined (Nayfach et al., 2021), or even unravel biologic novelty altogether (Rinke et al., 2013). While doing so, one also provides phylogenetic anchors for the posterior identification of genomic fragments by forthcoming (meta)genomic studies (Mukherjee et al., 2017). One way of rectifying the underrepresentation of rare prokaryotic taxa is to specifically target these organisms for genome sequencing (i.e., if cultivable); or alternatively single-cell sequencing, and metagenomic-assembly (i.e., if unculturable).
Numerous initiatives have been carried out to meet the need for a broader phylogenetic coverage of prokaryotic genomes (Almeida, Nayfach, Boland, Strozzi, Beracochea, Shi, Pollard, Sakharova, Parks, Hugenholtz, Segata, Kyrpides, Finn, 2021, Anantharaman, Brown, Hug, Sharon, Castelle, Probst, Thomas, Singh, Wilkins, Karaoz, Brodie, Williams, Hubbard, Banfield, 2016, Brown, Hug, Thomas, Sharon, Castelle, Singh, Wilkins, Wrighton, Williams, Banfield, 2015, Mukherjee, Seshadri, Varghese, Eloe-Fadrosh, Meier-Kolthoff, Göker, Coates, Hadjithomas, Pavlopoulos, Paez-Espino, Yoshikuni, Visel, Whitman, Garrity, Eisen, Hugenholtz, Pati, Ivanova, Woyke, Klenk, Kyrpides, 2017, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Parks, Rinke, Chuvochina, Chaumeil, Woodcroft, Evans, Hugenholtz, Tyson, 2017, Pasolli, Asnicar, Manara, Zolfo, Karcher, Armanini, Beghini, Manghi, Tett, Ghensi, Collado, Rice, DuLong, Morgan, Golden, Quince, Huttenhower, Segata, 2019, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013, Tully, Graham, Heidelberg, 2018, Wu, Hugenholtz, Mavromatis, Pukall, Dalin, Ivanova, Kunin, Goodwin, Wu, Tindall, Hooper, Pati, Lykidis, Spring, Anderson, D’haeseleer, Zemla, Singer, Lapidus, Nolan, Copeland, Han, Chen, Cheng, Lucas, Kerfeld, Lang, Gronow, Chain, Bruce, Rubin, Kyrpides, Klenk, Eisen, 2009). Wu et al. sequenced the genomes of 56 prokaryotic species, thereby creating the “Genomic Encyclopedia of Bacteria and Archaea” (GEBA) (Wu et al., 2009). By analyzing this genome set, these authors discovered new protein families, and improved function prediction for genes issuing from other organisms (Wu et al., 2009). More than 10% of the protein families identified in this study showed no significant sequence similarity to any known proteins at the time, thus hinting at functional novelty (Wu et al., 2009). Rinke et al. created GEBA-MDM, an extension of GEBA, that targeted candidate phyla (Rinke et al., 2013). They recovered draft SAGs of 201 uncultivated prokaryotic cells, issuing from 29 major branches of the phylogenetic tree (Rinke et al., 2013). The GEBA-MDM dataset enabled a twofold increase in phylogenetic diversity in relation to GEBA, the disclosure of 20 thousand new hypothetical protein families, among many other notable findings that significantly expanded the boundaries of biological knowledge (Rinke et al., 2013). Mukherjee et al. presented a significantly augmented version of the GEBA catalog (GEBA-I) (Mukherjee et al., 2017). GEBA-I includes reference genomes for 974 bacterial and 29 archaeal type strains, which increased the number of type strains at that time by twofold, and the overall phylogenetic diversity of prokaryotes by 25% (Mukherjee et al., 2017). By comparing the GEBA-I genomes with previously available ones, they reported an increase of 10.5% in new protein families as a consequence of increased phylogenetic diversity (Mukherjee et al., 2017). They disclose that the GEBA-I genomes improved the phylogenetic and functional interpretation of 25 million previously unassigned proteins from 4,650 metagenomes (Mukherjee et al., 2017). These authors also predicted more than 23 thousand BGCs from these genomes, most of which were of unknown biosynthetic function, highlighting the biotechnological potential encased in these organisms (Mukherjee et al., 2017). Nayfach et al. created the “Genomes from Earth’s Microbiomes” (GEM) catalog (Nayfach et al., 2021). GEM comprises 52,515 MAGs, representing 12,556 new candidate species, and spanning 135 phyla, as gathered from more than 10 thousand metagenomes, covering a myriad habitats worldwide (Nayfach et al., 2021). They report that the GEM catalog increases the known phylogenetic diversity of prokaryotes by 44% (Nayfach et al., 2021). To ascertain the potential for new functions, these authors gathered more than 5 million protein clusters from the GEM catalog (Nayfach et al., 2021). Upon annotating these clusters, they found that nearly 70% could not have their functions predicted by either TIGRFAM (Haft et al., 2013), KEGG Orthology (Kanehisa et al., 2016), nor the Pfam database (Mistry et al., 2021); and that 47% had no significant similarity to UniRef (Nayfach et al., 2021).
The results reported by these studies reinforce the hypothesis that functional novelty is far from saturated among sequence space (Mukherjee, Seshadri, Varghese, Eloe-Fadrosh, Meier-Kolthoff, Göker, Coates, Hadjithomas, Pavlopoulos, Paez-Espino, Yoshikuni, Visel, Whitman, Garrity, Eisen, Hugenholtz, Pati, Ivanova, Woyke, Klenk, Kyrpides, 2017, Nayfach, Roux, Seshadri, Udwary, Varghese, Schulz, Wu, Paez-Espino, Chen, Huntemann, Palaniappan, Ladau, Mukherjee, Reddy, Nielsen, Kirton, Faria, Edirisinghe, Henry, Jungbluth, Chivian, Dehal, Wood-Charlson, Arkin, Tringe, Visel, IMG/M Data Consortium, Woyke, Mouncey, Ivanova, Kyrpides, Eloe-Fadrosh, 2021, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013, Wu, Hugenholtz, Mavromatis, Pukall, Dalin, Ivanova, Kunin, Goodwin, Wu, Tindall, Hooper, Pati, Lykidis, Spring, Anderson, D’haeseleer, Zemla, Singer, Lapidus, Nolan, Copeland, Han, Chen, Cheng, Lucas, Kerfeld, Lang, Gronow, Chain, Bruce, Rubin, Kyrpides, Klenk, Eisen, 2009). They also strongly suggest that future endeavors aiming to increase the genomic representation of elusive taxa will continue to provide improvements towards the functional and taxonomic assignment of unclassified genomic sequences throughout public databases.
3.5. Culturomics
Metagenomics and SCG do not allow for the effortless differentiation among strains of the same species, nor do they provide biological material for subsequent research (Lagier et al., 2018). Axenic culture remains indispensable in the complete characterization of a given prokaryote and its physiology (Wiegand et al., 2020). An approach that aims to reignite the pure culture of microorganisms, particularly those that have eluded previous cultivation efforts, is that of culturomics (Lagier et al., 2012). Culturomics is a high-throughput culturing method that makes use of multiple culture conditions, together with MALDI-TOF mass spectrometry and 16S rRNA gene amplicon sequencing (Sood et al., 2021). So far it has allowed to identify hundreds of new species (Lagier, Armougom, Million, Hugon, Pagnier, Robert, Bittar, Fournous, Gimenez, Maraninchi, Trape, Koonin, La Scola, Raoult, 2012, Lagier, Dubourg, Million, Cadoret, Bilen, Fenollar, Levasseur, Rolain, Fournier, Raoult, 2018, Lagier, Khelaifia, Alou, Ndongo, Dione, Hugon, Caputo, Cadoret, Traore, Seck, Dubourg, Durand, Mourembou, Guilhot, Togo, Bellali, Bachar, Cassir, Bittar, Delerce, Mailhe, Ricaboni, Bilen, Dangui Nieko, Dia Badiane, Valles, Mouelhi, Diop, Million, Musso, Abrahão, Azhar, Bibi, Yasir, Diallo, Sokhna, Djossou, Vitton, Robert, Rolain, La Scola, Fournier, Levasseur, Raoult, 2016). Besides expanding our knowledge of the repertoire of the prokaryotic biosphere, culturomics provides pure, viable cultures that can be further used for in vitro and in vivo experiments (Bellali, Lagier, Million, Anani, Haddad, Francis, Kuete Yimagou, Khelaifia, Levasseur, Raoult, Bou Khalil, 2021, Lagier, Dubourg, Million, Cadoret, Bilen, Fenollar, Levasseur, Rolain, Fournier, Raoult, 2018).
Culturomics itself is a workflow composed of several steps. The first step consists on dividing and diversifying the environmental sample of interest into multiple culture conditions (e.g., complex media, broad range of temperatures and incubation times, addition of growth factors, supplements or metals, aerobic/anaerobic setting, etc) (Lagier et al., 2018). These conditions are designed to suppress the growth of the most represented populations, and to promote the growth of fastidious or rare prokaryotes that exist at lower concentrations (Lagier et al., 2018). Targeted culture conditions can also be employed to promote the growth of specific taxa (Lagier et al., 2018). Secondly, taxa identification is achieved by MALDI-TOF mass spectrometry, i.e., comparing the protein mass spectra of the target isolate with that of a preexistent database (Lagier et al., 2018). If MALDI-TOF fails to identify the isolate, the latter undergoes 16S rRNA gene amplicon sequencing (Lagier et al., 2018). Finally, the discovery of novelty is confirmed by sequencing the isolate’s genome (Lagier et al., 2018).
However, no technique is without drawbacks. In the case of culturomics these include: (i) the sheer workload posed by the multiplex culturing conditions; (ii) the inability to test a multitude of samples like in metagenomics; and (iii) its incapability to provide data on gene expression and/or the functional potential enclosed within prokaryotic species by itself, as this always requires genome sequencing of the newly isolated organisms (Lagier et al., 2018).
3.6. Heterologous expression
Another problem associated with FDM proteins is that these might be spurious predictions–as discussed later on subsection 3.7. The (supposed) CDS’ predicted for these sequences might instead encode genomic information that is not transcribed. One way of overcoming this problem is through large scale experimental functional genomics initiatives, like the Keio collection of E. coli single-gene knockout mutants (Yamamoto et al., 2009), and the transposon mutant library of Pseudomonas aeruginosa (Jacobs et al., 2003). These endeavors allow for an accurate characterization of proteins whose function is yet unknown. Nonetheless, these initiatives are costly and resource-demanding.
Likewise, heterologous expression of gene products is a promising approach for the study of particular gene products from uncultivated taxa (Lloyd et al., 2013). Together with increasing developments in synthetic biology, heterologous expression offers great promise as a strategy for re-coding entire operons for expression in model organisms (Hedlund, Dodsworth, Murugapiran, Rinke, Woyke, 2014, Temme, Zhao, Voigt, 2012). In silico analyses have predicted that E. coli can transcribe roughly 40% of genes from well-studied cultivated taxa if cloned in expression libraries (Doud, Woyke, 2017, Gabor, Alkema, Janssen, 2004).
However, there are many obstacles inherent to this approach. Examples of these obstacles are: the presence of accessory proteins, ambiguous decoding or recoding, apoprotein activation, codon bias, codon reassignment, different promoter structures, inaccurate protein folding, low expression rates, lack of essential post-translational modifications, lack of essential protein secretion, rare codon utilization, shared metabolic pools, or even unforeseen genetic codes (Ling, O’Donoghue, Söll, 2015, Sysoev, Grötzinger, Renn, Eppinger, Rueping, Karan, 2021). These limitations indicate that only a minute percentage of the functional sequence space can be easily identified with this approach (Doud and Woyke, 2017). These problems become even more acute when taking into account uncultured taxa, which may harbor extremely divergent and mostly uncharted DNA sequences and physiologies. This is particularly true for extremophiles, because the experimental characterization of their gene products is further restrained by the lack of expression systems that allow for the production of recombinant proteins under the same extreme conditions required by these organisms to begin with (Grötzinger et al., 2018). These issues reduce the success rate of current methods in accessing the functional novelty that may lie within the FDM, leaving most of unknown functions from uncultivated microorganisms concealed within their native expression hosts (Doud and Woyke, 2017).
3.7. Annotation, public databases, and canonical computational approaches
Over the past decade, the growth in (meta)genomic data acquisition has been staggering. Previous estimates indicated that the total amount of sequencing data doubles approximately every seven months, spanning from small groups producing a couple of terabases per year, to large, dedicated institutes generating several petabases a year (Stephens et al., 2015). Yet, this enormous amount of sequence data is useless, unless it is accompanied by information regarding where the CDS’ are, what ORFs do they belong to, and what products might they code for (Koonin, Makarova, Wolf, 2021, Salzberg, 2019). Besides allowing to understand the biology of genomes (Salzberg, 2019), functional annotation is critical for the growth of biological knowledge itself (Danchin et al., 2018). Unfortunately, it has not kept up with the progress witnessed for sequencing technologies (Salzberg, 2019), arguably due to two interdependent factors. First, the rampant accumulation of (meta)genomic data as described above. Second, a shift from automatic annotation, complemented by manual curation; to complete, unsupervised, automation of the entire annotation process (Koonin et al., 2021)–as currently witnessed for sequence submission portals and public databases.
Sequence submission portals often redirect sequence data to integrated annotation pipelines (Clum, Huntemann, Bushnell, Foster, Foster, Roux, Hajek, Varghese, Mukherjee, Reddy, Daum, Yoshinaga, O’Malley, Seshadri, Kyrpides, Eloe-Fadrosh, Chen, Copeland, Ivanova, 2021, Li, O’Neill, Haft, DiCuccio, Chetvernin, Badretdin, Coulouris, Chitsaz, Derbyshire, Durkin, Gonzales, Gwadz, Lanczycki, Song, Thanki, Wang, Yamashita, Yang, Zheng, Marchler-Bauer, Thibaud-Nissen, 2021, Overbeek, Olson, Pusch, Olsen, Davis, Disz, Edwards, Gerdes, Parrello, Shukla, Vonstein, Wattam, Xia, Stevens, 2014). Sometimes, this data is manually curated thereafter (Famiglietti et al., 2014). After annotation, the information is added to, or updated, in different databases harboring distinct biological sequence data (e.g., NCBI’s Nucleotide database (NCBI Resource Coordinators, 2018), and the Translated EMBL Nucleotide Sequence Data Library (Boeckmann, 2003)). A previous report on the Reference sequence database (RefSeq) at NCBI (O’Leary et al., 2016), established that prokaryotic genomes and proteins form most of this dataset, even though only a fraction of their reference bacterial genomes were manually curated. Even though small-scale community-based manual curation was reported to drastically improve gene annotations (Rödelsperger et al., 2019), it is still not a viable option to deal with large amounts of data, for two main reasons: it does not scale, nor does it always offer a solution (Salzberg, 2019). To characterize and validate the function of each individual protein in an experimental setting (e.g., gene-specific screening assays), is not a suitable alternative either (Ellens, Christian, Singh, Satagopam, May, Linster, 2017, Quince, Walker, Simpson, Loman, Segata, 2017). These constraints leave no choice but to rely on computer-automated and unreviewed annotation methods as the sole alternative to predict the function of these sequences. However, the crux of the problem lies when annotation tools cannot assign a molecular function to a given sequence. In these cases, the imputed gene products are often classified as “uncharacterized”, “hypothetical”, or simply “unknown” (i.e., FDM) (Erdin, Lisewski, Lichtarge, 2011, Makarova, Wolf, Koonin, 2019). There are a multitude of factors that influence our current inability to infer the function of these sequences. These factors include, but are not restricted to, the annotation methods and tools that are used, the taxonomic classification of the organism of origin, the size of the genome, genome completeness, research bias towards model organisms and pathogens, HGT events, mobile elements (e.g., transposons), location in core- versus accessory-genome, contamination of the sequencing data, sequencing errors, data and metadata collection, pre-processing, quality, and structure, among countless others (Arkhipova, 2020, Danchin, Ouzounis, Tokuyasu, Zucker, 2018, Dimonaco, Aubrey, Kenobi, Clare, Creevey, 2021, Lobb, Tremblay, Moreno-Hagelsieb, Doxey, 2020). Below we will discuss the most blatant ones, with a focus on standard annotation workflows.
Given a genomic fragment, an annotation workflow will often start by identifying CDS genes, and then translating them (Lobb et al., 2020). Several problems may arise just in these two steps alone, such as: incorrect gene calling (e.g., prediction of spurious ORFs), prediction of genes in the wrong DNA strand, incorrect prediction of ORFs in non-coding sequences (e.g., CRISPR arrays), erroneous start codon assignment, under-prediction of short ORFs (sORFs) and alternative ORFs, and the failure to identify non-standard codon usage, as well as overlapping genes (Dimonaco, Aubrey, Kenobi, Clare, Creevey, 2021, Makarova, Wolf, Koonin, 2019, Omasits, Varadarajan, Schmid, Goetze, Melidis, Bourqui, Nikolayeva, Québatte, Patrignani, Dehio, Frey, Robinson, Wollscheid, Ahrens, 2017, Orr, Mao, Storz, Qian, 2020). Besides severely compromising downstream functional annotation, these issues might also lead to inconsistent numbers of reported CDS genes among distinct annotation centers, over-prediction of non-coding genes, and the absence or under-representation of many gene types from public databases (Dimonaco, Aubrey, Kenobi, Clare, Creevey, 2021, Omasits, Varadarajan, Schmid, Goetze, Melidis, Bourqui, Nikolayeva, Québatte, Patrignani, Dehio, Frey, Robinson, Wollscheid, Ahrens, 2017). As an illustration, increasing evidence shows that sORFs code for a broad range of important functions (see Duval and Cossart (2017); Khitun et al. (2019); Plaza et al. (2017); Saghatelian and Couso (2015) and references therein). Moreover, there are studies providing evidence for the existence of these sORFs using proteomics data (Omasits, Varadarajan, Schmid, Goetze, Melidis, Bourqui, Nikolayeva, Québatte, Patrignani, Dehio, Frey, Robinson, Wollscheid, Ahrens, 2017, Sberro, Fremin, Zlitni, Edfors, Greenfield, Snyder, Pavlopoulos, Kyrpides, Bhatt, 2019). Yet, true protein-coding sORFs are poorly covered in public databases to begin with (Dimonaco et al., 2021). In addition to–and perhaps because of–their poor coverage, it is often difficult to discriminate between true sORFs, random in-frame genomic fragments, spurious sORFs, and pseudogenes (Lobb, Tremblay, Moreno-Hagelsieb, Doxey, 2020, Omasits, Varadarajan, Schmid, Goetze, Melidis, Bourqui, Nikolayeva, Québatte, Patrignani, Dehio, Frey, Robinson, Wollscheid, Ahrens, 2017, Orr, Mao, Storz, Qian, 2020, Sberro, Fremin, Zlitni, Edfors, Greenfield, Snyder, Pavlopoulos, Kyrpides, Bhatt, 2019). Therefore, gene calling and annotation tools frequently impose a minimum sequence length cutoff to prevent spurious ORF annotation (Orr et al., 2020). In turn, this keeps perpetuating the under-representation of true sORFs in sequence databases (Sberro et al., 2019).
Most often, after the gene-calling and translation steps, a function is predicted for a given putative protein based on the similarity of its sequence to those of other proteins whose function is already known (Zallot et al., 2016). This is usually achieved by homology-based methods, also known as sequence similarity-based methods (e.g., BLAST (Altschul et al., 1990)). Annotation pipelines frequently rely on homology-based methods because these are straightforward to use and well-established. These methods also allow for huge numbers of query sequences to inherit the function annotations of homologous proteins from a given database (i.e., inheritance through homology) (Lee et al., 2007), provided certain conditions are met (e.g., percent identity and e-value thresholds). Yet, in many cases, the function of a given gene-product cannot be inferred by homology-based methods, due to a lack of recognizable homologs in sequence databases (Lobb et al., 2015). This may happen in the cases of highly divergent genes, or truly novel ones (Lobb et al., 2020). As such, annotation tools that rely solely on inheritance through homology are not applicable if homologous regions among targets of interest are undetectable (Coutinho et al., 2015). An alternative to homology-based methods is to compare the query sequence against position-specific scoring matrices (PSSMs), or HMMs. PSSMs and HMMs are gathered from curated collections of protein families (e.g., Clusters of Orthologous Genes (Galperin et al., 2021)), and protein domains (e.g., Conserved Domain Database (Lu et al., 2020)). These methods are more sensitive, because they are able to detect remote matches to protein or domain families that were conserved throughout long evolutionary distances (Koonin, Makarova, Wolf, 2021, Lobb, Tremblay, Moreno-Hagelsieb, Doxey, 2020). Nevertheless, numerous protein and domain families lack functional annotation themselves–i.e., the so-called domains of unknown function (DUFs), and uncharacterized protein families (UPFs) (Mistry et al., 2021). This entails that, even if a FDM protein manages to attain a match to a distantly related protein family or domain, there is a chance that the function for that protein family or domain is also unknown–e.g., as of 2021, 23% of all Pfam families were either DUF or UPF families (Mistry et al., 2021).
In many cases, when function is inferred solely via inheritance through homology, erroneous predictions arise leading to incorrect annotations (Promponas et al., 2015). The same can happen with more sensitive methods that rely on protein domains instead–in these cases, annotation inheritance might even occur in the absence of a full protein match (Lobb et al., 2020). Misannotations are specially pernicious for FDM sequences, as they might contribute to false-positive predictions, and therefore inaccurate annotation coverage reports. The quality of an annotation depends not only on the tools that generated it, but also on the fast and comprehensive inclusion of new data on the preexistent database annotations that are used by these tools in the first place (Makarova et al., 2019). This implies that when a misannotation is identified, not only should this error be corrected, but also that every other (mis)annotation that relied upon the latter as a source should be corrected as well (Salzberg, 2019). One can thus make the case that homology-based methods are only as good as the databases they rely upon (Dimonaco et al., 2021). Unfortunately, misannotation has not only become commonplace in public databases, as it is also an ongoing problem (Arkhipova, 2020, Girardi, Thoden, Holden, 2020, Impey, Lee, Hawkins, Sutton, Panjikar, Perugini, Soares da Costa, 2020, Meyer, Scalzitti, Jeannin-Girardon, Collet, Poch, Thompson, 2020, Nobre, Campos, Lucic-Mercy, Arnholdt-Schmitt, 2016, Rembeza, Engqvist, 2021, Schnoes, Brown, Dodevski, Babbitt, 2009). To make matters worse, misannotations are seldomly confined to the database where the error first occurred, they often percolate throughout several databases as well (Promponas et al., 2015). Previous reports showed that the percentage of incorrect functional assignments in public databases soared from less than 5% in 1998 to as high as 40% in 2005 (Schnoes et al., 2009). More recently, it was reported that up to 50% of protein sequences from public databases contain at least one error (Meyer et al., 2020). As a case in point, some authors express particular concern towards error propagation issuing from draft genome assemblies (e.g., MAGs) (Arkhipova, 2020, Koonin, Makarova, Wolf, 2021, Salzberg, 2019). However, this issue is not exclusive to predicted genes issuing from draft genomes nor newly-discovered proteins. Well-known enzyme classes of industrial relevance (Rembeza and Engqvist, 2021), and genes encoding antibiotic targets from clinically important pathogens (Impey et al., 2020), also suffer from annotation error. Therefore, the degree to which one may rely on annotations from public databases is, for the most part, currently unknown (Rembeza and Engqvist, 2021). One may conclude that, in addition to the predicaments discussed throughout this section, the accumulation of misannotations in public databases may also restrain our ability to uncover biological novelty. Hence, truly innovative frameworks, spanning the entire process from functional (re)annotation to public data delivery, are desperately needed.
3.8. Logistical challenges
Besides the foregoing reasons, there is a more logistical factor for this large quantity of (meta)genomic data to be left unannotated: only a few research projects target the FDM in public datasets (Dutilh, 2014). This low number of projects is understandable. Searching for patterns amidst the data, or aiming to discover new phenomena, is essentially an exploratory science venture, following a “bottom-up” approach that goes from the data to the hypotheses, and that not always produces publishable results (Bernard, Pathmanathan, Lannes, Lopez, Bapteste, 2018, Burian, 2013). Since exploratory science is not hypothesis-driven, it can either reveal new knowledge or simply fail in revealing any knowledge at all.
In addition to this, researchers commonly find themselves under pressure to publish their results and move to the next project, leaving numerous uncharacterized sequences drowned amidst the contigs that were able to be annotated via inheritance through homology. This happens since using previously annotated sequences leads to straightforward conclusions (Dutilh, 2014). Thus, scientists contemplating the investment in purely exploratory studies take great risk and face a precarious situation (Bernard et al., 2018), that in turn perpetuates the FDM problem, as explained in this review.
4. Outlook
The rate at which proteins from the FDM keep accumulating in public sequence data repositories is nothing short of alarming. To experimentally validate the function of each of these proteins would take so long, that it would be impossible to keep abreast with the pace at which they are being discovered. There are numerous ways to tackle the FDM without resorting to in vitro/in vivo experimental characterization (see Danchin et al. (2018)). Moreover, these measures can be undertaken either during the upstream stage of a given (meta)genomic workflow (e.g., by improving the quality and interpretability of the data being generated); or downstream from that (e.g., by refining function prediction methods).
An example of a technique that can be applied upstream, and although not properly addressed in this review, is SCG. SCG is a powerful tool that holds great promise, not only towards MDM disclosure (Marcy, Ouverney, Bik, Lösekann, Ivanova, Martin, Szeto, Platt, Hugenholtz, Relman, Quake, 2007, Martijn, Schulz, Zaremba-Niedzwiedzka, Viklund, Stepanauskas, Andersson, Horn, Guy, Ettema, 2015, Rinke, Schwientek, Sczyrba, Ivanova, Anderson, Cheng, Darling, Malfatti, Swan, Gies, Dodsworth, Hedlund, Tsiamis, Sievert, Liu, Eisen, Hallam, Kyrpides, Stepanauskas, Rubin, Hugenholtz, Woyke, 2013, Woyke, Xie, Copeland, González, Han, Kiss, Saw, Senin, Yang, Chatterji, Cheng, Eisen, Sieracki, Stepanauskas, 2009), but also in bioprospecting, natural product discovery, and in the search for heterologous expression systems, among many other applications (Kaster, Sobol, 2020, Mauger, Monard, Thion, Vandenkoornhuyse, 2022). When SCG and metagenomics are combined in the same study, they provide a highly complementary and synergistic approach, allowing for a fine-scale analysis and high throughput (Alneberg, Karlsson, Divne, Bergin, Homa, Lindh, Hugerth, Ettema, Bertilsson, Andersson, Pinhassi, 2018, Mauger, Monard, Thion, Vandenkoornhuyse, 2022, Probst, Ladd, Jarett, Geller-McGrath, Sieber, Emerson, Anantharaman, Thomas, Malmstrom, Stieglmeier, Klingl, Woyke, Ryan, Banfield, 2018). SAGs can improve metagenomic bins (Pachiadaki et al., 2019), as the former are able to capture HGT events, and allow to cast light on rare prokaryotes that might have been otherwise overlooked during the binning process, in addition to cell-cell associations missed in the course of MAG assembly (Dam, Vollmers, Sobol, Cabezas, Kaster, 2020, Wiegand, Dam, Riba, Vollmers, Kaster, 2021, Woyke, Doud, Schulz, 2017). In the same way, metagenomic data enables the recovery of genomic fragments that may be missing from SAGs, thus improving subsequent assemblies (Becraft, Dodsworth, Murugapiran, Ohlsson, Briggs, Kanbar, De Vlaminck, Quake, Dong, Hedlund, Swingley, 2015, Dodsworth, Blainey, Murugapiran, Swingley, Ross, Tringe, Chain, Scholz, Lo, Raymond, Quake, Hedlund, 2013, Kaster, Sobol, 2020, Nurk, Bankevich, Antipov, Gurevich, Korobeynikov, Lapidus, Prjibelski, Pyshkin, Sirotkin, Sirotkin, Stepanauskas, Clingenpeel, Woyke, McLean, Lasken, Tesler, Alekseyev, Pevzner, 2013, Xu, Zhao, 2018). Functional-driven SCG allows to isolate and characterize individual cells that exhibit a functional trait of interest, before and/or during genome sequencing (Couradeau, Sasse, Goudeau, Nath, Hazen, Bowen, Chakraborty, Malmstrom, Northen, 2019, Doud, Woyke, 2017, Hatzenpichler, Krukenberg, Spietz, Jay, 2020, Kaster, Sobol, 2020, Lee, Men, Wang, He, Alvarez-Cohen, 2015, Woyke, Jarett, 2015). In this way, it has enabled to uncover gene functions that were previously encased in the FDM (Woyke et al., 2019). The development of new SCG-based technologies like the “mini-metagenomic” approach (Alteio, Schulz, Seshadri, Varghese, Rodriguez-Reillo, Ryan, Goudeau, Eichorst, Malmstrom, Bowers, Katz, Blanchard, Woyke, 2020, Geesink, Wegner, Probst, Herrmann, Dam, Kaster, Küsel, 2020, Grieb, Bowers, Oggerin, Goudeau, Lee, Malmstrom, Woyke, Fuchs, 2020); and the use of a single-cell printers (Gross et al., 2013), to investigate elusive taxa from complex environmental samples (Wiegand et al., 2021), emphasize on the exciting opportunities stemming from SCG research.
A multitude of in silico approaches have been designed to predict the function of the gene products inferred downstream from a given workflow, without the need to rely on inheritance through homology. Some examples include, but are not restricted to, predicting protein function by (i) inferring known patterns of evolutionary substitutions (Bileschi et al., 2022); (ii) integrative mapping of metabolic pathways (Calhoun et al., 2018); (iii) using domain-function associations (Rojano et al., 2022); (iv) leveraging template-based protein structure prediction (Zheng et al., 2022); (v) integration of multiple data sources (Zohra Smaili et al., 2021); and (vi) genomic context analysis (Cotroneo et al., 2021), among many others. Most current protein function prediction tools are based upon machine-learning (ML) (Bernardes, Pedreira, 2013, Bonetta, Valentino, 2020, Libbrecht, Noble, 2015), to some extent. ML approaches are built upon substantial knowledge of the subject at hand, and possess the ability to identify unforeseen patterns within the data itself (LeCun et al., 2015). These approaches often use features gathered from metadata associated with the primary protein sequences in order to train their models, and to predict the features of the protein sequence used as input, including its domains, protein-protein interactions, subcellular location, physico-chemical properties, secondary structure, and so forth (Cao et al., 2017). In Fig. 3 we show an overview of some of the metadata features that can be gathered for protein sequences.
Fig. 3.
Mind-map of in silico concepts relating to FDM disclosure. The three main hubs depict the general categories each concept belongs to. A non-exhaustive overview of some of the major concepts is shown for each hub, as well as how/whether these concepts hierarchically relate to one another. The first concept (i.e., node) that has “offspring” concepts relating to it has its respective node and text shown with a darker color. A dashed line represents a relation between concepts that are shared by more than one “parent”. CDS: coding sequence; COGs: clusters of orthologous genes; eggNOG: evolutionary genealogy of genes, non-supervised orthologous groups; FDM: functional dark matter; HMMs: hidden Markov models; KEGG: Kyoto encyclopedia of genes and genomes; KNN: k-nearest neighbors algorithm; MAGs: metagenome-assembled genomes; ML: machine-learning; PPI: protein-protein interaction; PSSMs: position-specific scoring matrices; SVM: support-vector machine.
The state-of-the-art of protein function prediction makes use of a broad family of ML methods that rely on artificial neural networks, called Deep Learning (DL) (LeCun et al., 2015). DL has already revolutionized many scientific fields, with particular emphasis on the biological sciences (Ching et al., 2018), especially computational biology (Sapoval et al., 2022). Recently, it has allowed for a groundbreaking advance towards the solution of the protein folding prediction problem (Jumper et al., 2021). Furthermore, DL model-based methods are substantially faster than non-DL methods, and can be leveraged for large-scale (meta)genomic data mining towards accurate and efficient MDM disclosure (Zha et al., 2022).
It is critical to understand however, that DL suffers from restraints just as other computational approaches. For instance, the classification outputs are inherently intertwined with the methodological design, and the duality of correlation and causality are still present (Ching et al., 2018). In the same way, DL models are only as good as the quantity and quality of the data they are trained upon (Robinson et al., 2021). DL algorithms also require huge amounts of data to train their models, which might be a deterring factor to its success in some occasions (Bonetta and Valentino, 2020). Another setback is their inability to predict true biological novelty, as these models are built upon preexisting knowledge (Robinson, 2022, Robinson, Piel, Sunagawa, 2021).
Protein function prediction is indubitably a grand challenge of the post-genomic era. The progress made so far has been substantial, and despite the fact that year upon year new protein function prediction tools keep surfacing, with ever-improving performance and accuracy, this biological conundrum is far from being solved (Makrodimitris et al., 2020). There is tremendous potential for future innovation, and plenty of room for improvement, in order to enhance protein function prediction and genome annotation in the years to come (Bonetta, Valentino, 2020, Makrodimitris, van Ham, Reinders, 2020). We envision that new developments in DL will continue to expedite opportunities for (meta)genomic data mining, thereby allowing to characterize the massive breadth of “known unknowns” that lie overlooked amidst sequence data repositories. An optimistic outlook from this endeavor is that, a breakthrough of new molecular functions and metabolites crucial for the biotechnology industry will be made, helping to achieve ahead of time the sustainable development goals for 2030 adopted by the United Nations. Undoubtedly, the horizon goes far and wide, while the potential treasures that can be unearthed by prospecting this information are hidden in plain sight.
CRediT authorship contribution statement
Pedro Escudeiro: Conceptualization, Writing – original draft, Writing – review & editing, Visualization, Data curation. Christopher S. Henry: Supervision, Funding acquisition. Ricardo P.M. Dias: Conceptualization, Writing – review & editing, Supervision, Funding acquisition, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Professor Rogério Tenreiro for proof-reading an early version of the manuscript, and for his comments and suggestions. We would also like to thank the anonymous reviewers for their helpful and constructive comments and suggestions. Funding: This work was supported by UIDB/04046/2020 and UIDP/04046/2020 grant from FCT, Portugal (to BioISI). P.E. is recipient of a fellowship from BioSys PhD programme PD65-2012 (Ref SFRH/BD/131416/2017) from FCT (Portugal).
References
- Ackermann M. Microbial individuality in the natural environment. ISME J. 2013;7(3):465–467. doi: 10.1038/ismej.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akal A.L., Karan R., Hohl A., Alam I., Vogler M., Grötzinger S.W., Eppinger J., Rueping M. A polyextremophilic alcohol dehydrogenase from the atlantis II deep red sea brine pool. FEBS Open Bio. 2019;9(2):194–205. doi: 10.1002/2211-5463.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahib A., Breitling R., Gilbert D.R. Predicting protein function by machine learning on amino acid sequences – a critical evaluation. BMC Genomics. 2007;8(1):78. doi: 10.1186/1471-2164-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwood A.C., Walter M.R., Kamber B.S., Marshall C.P., Burch I.W. Stromatolite reef from the early archaean era of australia. Nature. 2006;441(7094):714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A., Nayfach S., Boland M., Strozzi F., Beracochea M., Shi Z.J., Pollard K.S., Sakharova E., Parks D.H., Hugenholtz P., Segata N., Kyrpides N.C., Finn R.D. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021;39(1):105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alneberg J., Karlsson C.M.G., Divne A.-M., Bergin C., Homa F., Lindh M.V., Hugerth L.W., Ettema T.J.G., Bertilsson S., Andersson A.F., Pinhassi J. Genomes from uncultivated prokaryotes: a comparison of metagenome-assembled and single-amplified genomes. Microbiome. 2018;6(1):173. doi: 10.1186/s40168-018-0550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteio L.V., Schulz F., Seshadri R., Varghese N., Rodriguez-Reillo W., Ryan E., Goudeau D., Eichorst S.A., Malmstrom R.R., Bowers R.M., Katz L.A., Blanchard J.L., Woyke T. Complementary metagenomic approaches improve reconstruction of microbial diversity in a forest soil. mSystems. 2020;5(2) doi: 10.1128/mSystems.00768-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anantharaman K., Brown C.T., Hug L.A., Sharon I., Castelle C.J., Probst A.J., Thomas B.C., Singh A., Wilkins M.J., Karaoz U., Brodie E.L., Williams K.H., Hubbard S.S., Banfield J.F. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.O., Andersson S.G. Pseudogenes, junk DNA, and the dynamics of rickettsia genomes. Mol. Biol. Evol. 2001;18(5):829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- Arkhipova I.R. Metagenome proteins and database contamination. mSphere. 2020;5(6) doi: 10.1128/mSphere.00854-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., Camarero J.A., Campopiano D.J., Challis G.L., Clardy J., Cotter P.D., Craik D.J., Dawson M., Dittmann E., Donadio S., Dorrestein P.C., Entian K.-D., Fischbach M.A., Garavelli J.S., Göransson U., Gruber C.W., Haft D.H., Hemscheidt T.K., Hertweck C., Hill C., Horswill A.R., Jaspars M., Kelly W.L., Klinman J.P., Kuipers O.P., Link A.J., Liu W., Marahiel M.A., Mitchell D.A., Moll G.N., Moore B.S., Müller R., Nair S.K., Nes I.F., Norris G.E., Olivera B.M., Onaka H., Patchett M.L., Piel J., Reaney M.J.T., Rebuffat S., Ross R.P., Sahl H.-G., Schmidt E.W., Selsted M.E., Severinov K., Shen B., Sivonen K., Smith L., Stein T., Süssmuth R.D., Tagg J.R., Tang G.-L., Truman A.W., Vederas J.C., Walsh C.T., Walton J.D., Wenzel S.C., Willey J.M., van der Donk W.A. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasopoulou K., Boti M.A., Adamopoulos P.G., Skourou P.C., Scorilas A. Third-generation sequencing: The spearhead towards the radical transformation of modern genomics. Life (Basel) 2021;12(1):30. doi: 10.3390/life12010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin H.P., Allen M.D., Donohoe B.S., Rorrer N.A., Kearns F.L., Silveira R.L., Pollard B.C., Dominick G., Duman R., El Omari K., Mykhaylyk V., Wagner A., Michener W.E., Amore A., Skaf M.S., Crowley M.F., Thorne A.W., Johnson C.W., Woodcock H.L., McGeehan J.E., Beckham G.T. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. U. S. A. 2018;115(19):E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker E.N., Helps C.R., Peters I.R., Darby A.C., Radford A.D., Tasker S. Complete genome sequence of mycoplasma haemofelis, a hemotropic mycoplasma. J. Bacteriol. 2011;193(8):2060–2061. doi: 10.1128/JB.00076-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft E.D., Dodsworth J.A., Murugapiran S.K., Ohlsson J.I., Briggs B.R., Kanbar J., De Vlaminck I., Quake S.R., Dong H., Hedlund B.P., Swingley W.D. Single-Cell-Genomics-Facilitated read binning of candidate phylum EM19 genomes from geothermal spring metagenomes. Appl. Environ. Microbiol. 2015;82(4):992–1003. doi: 10.1128/AEM.03140-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellali S., Lagier J.-C., Million M., Anani H., Haddad G., Francis R., Kuete Yimagou E., Khelaifia S., Levasseur A., Raoult D., Bou Khalil J. Running after ghosts: are dead bacteria the dark matter of the human gut microbiota? Gut Microbes. 2021;13(1):1–12. doi: 10.1080/19490976.2021.1897208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdy B., Spoering A.L., Ling L.L., Epstein S.S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 2017;12(10):2232–2242. doi: 10.1038/nprot.2017.074. [DOI] [PubMed] [Google Scholar]
- Bérdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 2012;65(8):385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- Bernard G., Pathmanathan J.S., Lannes R., Lopez P., Bapteste E. Microbial dark matter investigations: how microbial studies transform biological knowledge and empirically sketch a logic of scientific discovery. Genome Biol. Evol. 2018 doi: 10.1093/gbe/evy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes J.S., Pedreira C.E. A review of protein function prediction under machine learning perspective. Recent Pat. Biotechnol. 2013;7(2):122–141. doi: 10.2174/18722083113079990006. [DOI] [PubMed] [Google Scholar]
- Bileschi M.L., Belanger D., Bryant D.H., Sanderson T., Carter B., Sculley D., Bateman A., DePristo M.A., Colwell L.J. Using deep learning to annotate the protein universe. Nat. Biotechnol. 2022 doi: 10.1038/s41587-021-01179-w. [DOI] [PubMed] [Google Scholar]
- Blaser M.J., Cardon Z.G., Cho M.K., Dangl J.L., Donohue T.J., Green J.L., Knight R., Maxon M.E., Northen T.R., Pollard K.S., Brodie E.L. Toward a predictive understanding of earth’s microbiomes to address 21st century challenges. MBio. 2016;7(3) doi: 10.1128/mBio.00714-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Kim H.U., Medema M.H., Weber T. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 2019;20(4):1103–1113. doi: 10.1093/bib/bbx146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31(1):365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetta R., Valentino G. Machine learning techniques for protein function prediction. Proteins. 2020;88(3):397–413. doi: 10.1002/prot.25832. [DOI] [PubMed] [Google Scholar]
- Bowers R.M., The Genome Standards Consortium, Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K., Schulz F., Jarett J., Rivers A.R., Eloe-Fadrosh E.A., Tringe S.G., Ivanova N.N., Copeland A., Clum A., Becraft E.D., Malmstrom R.R., Birren B., Podar M., Bork P., Weinstock G.M., Garrity G.M., Dodsworth J.A., Yooseph S., Sutton G., Glöckner F.O., Gilbert J.A., Nelson W.C., Hallam S.J., Jungbluth S.P., Ettema T.J.G., Tighe S., Konstantinidis K.T., Liu W.-T., Baker B.J., Rattei T., Eisen J.A., Hedlund B., McMahon K.D., Fierer N., Knight R., Finn R., Cochrane G., Karsch-Mizrachi I., Tyson G.W., Rinke C., Lapidus A., Meyer F., Yilmaz P., Parks D.H., Murat Eren A., Schriml L., Banfield J.F., Hugenholtz P., Woyke T. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017;35(8):725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock T.D., Freeze H. Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J. Bacteriol. 1969;98(1):289–297. doi: 10.1128/jb.98.1.289-297.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.L., Watson M., Minot S.S., Rivera M.C., Franklin R.B. MinION™nanopore sequencing of environmental metagenomes: a synthetic approach. Gigascience. 2017;6(3):1–10. doi: 10.1093/gigascience/gix007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.T., Hug L.A., Thomas B.C., Sharon I., Castelle C.J., Singh A., Wilkins M.J., Wrighton K.C., Williams K.H., Banfield J.F. Unusual biology across a group comprising more than 15% of domain bacteria. Nature. 2015;523(7559):208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- Bruno S., Coppola D., di Prisco G., Giordano D., Verde C. Enzymes from marine polar regions and their biotechnological applications. Mar. Drugs. 2019;17(10) doi: 10.3390/md17100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull A.T., Goodfellow M. Dark, rare and inspirational microbial matter in the extremobiosphere: 16 000 m of bioprospecting campaigns. Microbiology. 2019;165(12):1252–1264. doi: 10.1099/mic.0.000822. [DOI] [PubMed] [Google Scholar]
- Bull A.T., Idris H., Sanderson R., Asenjo J., Andrews B., Goodfellow M. High altitude, hyper-arid soils of the Central-Andes harbor mega-diverse communities of actinobacteria. Extremophiles. 2018;22(1):47–57. doi: 10.1007/s00792-017-0976-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian R.M. Encyclopedia of Systems Biology. 2013. Exploratory experimentation; pp. 720–723. [Google Scholar]
- Cabrera M.Á., Blamey J.M. Biotechnological applications of archaeal enzymes from extreme environments. Biol. Res. 2018;51(1):37. doi: 10.1186/s40659-018-0186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun S., Korczynska M., Wichelecki D.J., San Francisco B., Zhao S., Rodionov D.A., Vetting M.W., Al-Obaidi N.F., Lin H., O’Meara M.J., Scott D.A., Morris J.H., Russel D., Almo S.C., Osterman A.L., Gerlt J.A., Jacobson M.P., Shoichet B.K., Sali A. Prediction of enzymatic pathways by integrative pathway mapping. Elife. 2018;7 doi: 10.7554/eLife.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Freitas C., Chan L., Sun M., Jiang H., Chen Z. ProLanGO: Protein function prediction using neural machine translation based on a recurrent neural network. Molecules. 2017;22(10) doi: 10.3390/molecules22101732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr R., Borenstein E. Comparative analysis of functional metagenomic annotation and the mappability of short reads. PLoS One. 2014;9(8):e105776. doi: 10.1371/journal.pone.0105776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle C.J., Banfield J.F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 2018;172(6):1181–1197. doi: 10.1016/j.cell.2018.02.016. [DOI] [PubMed] [Google Scholar]
- Chavali A.K., Rhee S.Y. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief. Bioinform. 2018;19(5):1022–1034. doi: 10.1093/bib/bbx020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wong H.L., Burns B.P. New approaches to detect biosynthetic gene clusters in the environment. Medicines (Basel) 2019;6(1) doi: 10.3390/medicines6010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching T., Himmelstein D.S., Beaulieu-Jones B.K., Kalinin A.A., Do B.T., Way G.P., Ferrero E., Agapow P.-M., Zietz M., Hoffman M.M., Xie W., Rosen G.L., Lengerich B.J., Israeli J., Lanchantin J., Woloszynek S., Carpenter A.E., Shrikumar A., Xu J., Cofer E.M., Lavender C.A., Turaga S.C., Alexandari A.M., Lu Z., Harris D.J., DeCaprio D., Qi Y., Kundaje A., Peng Y., Wiley L.K., Segler M.H.S., Boca S.M., Swamidass S.J., Huang A., Gitter A., Greene C.S. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface. 2018;15(141) doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Clum A., Huntemann M., Bushnell B., Foster B., Foster B., Roux S., Hajek P.P., Varghese N., Mukherjee S., Reddy T.B.K., Daum C., Yoshinaga Y., O’Malley R., Seshadri R., Kyrpides N.C., Eloe-Fadrosh E.A., Chen I.-M.A., Copeland A., Ivanova N.N. DOE JGI metagenome workflow. mSystems. 2021;6(3) doi: 10.1128/mSystems.00804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho L.P., Alves R., Del Río Á.R., Myers P.N., Cantalapiedra C.P., Giner-Lamia J., Schmidt T.S., Mende D.R., Orakov A., Letunic I., Hildebrand F., Van Rossum T., Forslund S.K., Khedkar S., Maistrenko O.M., Pan S., Jia L., Ferretti P., Sunagawa S., Zhao X.-M., Nielsen H.B., Huerta-Cepas J., Bork P. Towards the biogeography of prokaryotic genes. Nature. 2022;601(7892):252–256. doi: 10.1038/s41586-021-04233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen M., Małyska A., Nanda A.K., Lankhorst R.K., Parry M.A.J., Saltenis V.R., Pribil M., Nacry P., Inzé D., Baekelandt A. Biotechnology for tomorrow’s world: Scenarios to guide directions for future innovation. Trends Biotechnol. 2021;39(5):438–444. doi: 10.1016/j.tibtech.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Forterre P., Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10(6):R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotroneo C.E., Gormley I.C., Shields D.C., Salter-Townshend M. Computational modelling of chromosomally clustering protein domains in bacteria. BMC Bioinformatics. 2021;22(1):593. doi: 10.1186/s12859-021-04512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couradeau E., Sasse J., Goudeau D., Nath N., Hazen T.C., Bowen B.P., Chakraborty R., Malmstrom R.R., Northen T.R. Probing the active fraction of soil microbiomes using BONCAT-FACS. Nat. Commun. 2019;10(1):2770. doi: 10.1038/s41467-019-10542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho T.J.D., Franco G.R., Lobo F.P. Homology-independent metrics for comparative genomics. Comput. Struct. Biotechnol. J. 2015;13:352–357. doi: 10.1016/j.csbj.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D Ainsworth T., Krause L., Bridge T., Torda G., Raina J.-B., Zakrzewski M., Gates R.D., Padilla-Gamiño J.L., Spalding H.L., Smith C., Woolsey E.S., Bourne D.G., Bongaerts P., Hoegh-Guldberg O., Leggat W. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9(10):2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Lebedeva E.V., Pjevac P., Han P., Herbold C., Albertsen M., Jehmlich N., Palatinszky M., Vierheilig J., Bulaev A., Kirkegaard R.H., von Bergen M., Rattei T., Bendinger B., Nielsen P.H., Wagner M. Complete nitrification by nitrospira bacteria. Nature. 2015;528(7583):504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam H.T., Vollmers J., Sobol M.S., Cabezas A., Kaster A.-K. Targeted cell sorting combined with single cell genomics captures low abundant microbial dark matter with higher sensitivity than metagenomics. Front. Microbiol. 2020;11:1377. doi: 10.3389/fmicb.2020.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin A., Ouzounis C., Tokuyasu T., Zucker J.-D. No wisdom in the crowd: genome annotation in the era of big data - current status and future prospects. Microb. Biotechnol. 2018;11(4):588–605. doi: 10.1111/1751-7915.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danso D., Chow J., Streit W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019;85(19) doi: 10.1128/AEM.01095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., Leggewie C., Li X., Hazen T., Streit W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018;84(8) doi: 10.1128/AEM.02773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont T.O., Quince C., Shaiber A., Esen Ö.C., Lee S.T., Rappé M.S., McLellan S.L., Lücker S., Eren A.M. Nitrogen-fixing populations of planctomycetes and proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 2018;3(7):804–813. doi: 10.1038/s41564-018-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshani H., Bernier S.P., Marko V.A., Surette M.G. Completion of draft bacterial genomes by long-read sequencing of synthetic genomic pools. BMC Genomics. 2020;21(1):519. doi: 10.1186/s12864-020-06910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dida F., Yi G. Empirical evaluation of methods for de novo genome assembly. PeerJ Comput. Sci. 2021;7(e636):e636. doi: 10.7717/peerj-cs.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- van Dijk E.L., Jaszczyszyn Y., Naquin D., Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34(9):666–681. doi: 10.1016/j.tig.2018.05.008. [DOI] [PubMed] [Google Scholar]
- Dimonaco N.J., Aubrey W., Kenobi K., Clare A., Creevey C.J. No one tool to rule them all: Prokaryotic gene prediction tool annotations are highly dependent on the organism of study. Bioinformatics. 2021;38(5):1198–1207. doi: 10.1093/bioinformatics/btab827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth J.A., Blainey P.C., Murugapiran S.K., Swingley W.D., Ross C.A., Tringe S.G., Chain P.S.G., Scholz M.B., Lo C.-C., Raymond J., Quake S.R., Hedlund B.P. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat. Commun. 2013;4(1):1854. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N., Lee J.-H., Williams T.A., Offre P., Spang A. Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol. Lett. 2019;366(2) doi: 10.1093/femsle/fnz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N., Williams T.A., Sun J., Woodcroft B.J., Lee J.-H., Minh B.Q., Rinke C., Spang A. Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution. Nat. Commun. 2020;11(1):3939. doi: 10.1038/s41467-020-17408-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M.S., Cimermancic P., Schulze C.J., Wieland Brown L.C., Martin J., Mitreva M., Clardy J., Linington R.G., Fischbach M.A. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158(6):1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud D.F.R., Woyke T. Novel approaches in function-driven single-cell genomics. FEMS Microbiol. Rev. 2017;41(4):538–548. doi: 10.1093/femsre/fux009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh B.E. Metagenomic ventures into outer sequence space. Bacteriophage. 2014;4(4):e979664. doi: 10.4161/21597081.2014.979664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G.Z., Boling L., Barr J.J., Speth D.R., Seguritan V., Aziz R.K., Felts B., Dinsdale E.A., Mokili J.L., Edwards R.A. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M., Cossart P. Small bacterial and phagic proteins: an updated view on a rapidly moving field. Curr. Opin. Microbiol. 2017;39:81–88. doi: 10.1016/j.mib.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Dvořák P., Nikel P.I., Damborský J., de Lorenzo V. Bioremediation 3. 0 : Engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 2017;35(7):845–866. doi: 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Ellens K.W., Christian N., Singh C., Satagopam V.P., May P., Linster C.L. Confronting the catalytic dark matter encoded by sequenced genomes. Nucleic Acids Res. 2017;45(20):11495–11514. doi: 10.1093/nar/gkx937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L., Spang A., Lombard J., Stairs C.W., Ettema T.J.G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2018;16(2):120. doi: 10.1038/nrmicro.2017.154. [DOI] [PubMed] [Google Scholar]
- Engel P., Stepanauskas R., Moran N.A. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 2014;10(9):e1004596. doi: 10.1371/journal.pgen.1004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdin S., Lisewski A.M., Lichtarge O. Protein function prediction: towards integration of similarity metrics. Curr. Opin. Struct. Biol. 2011;21(2):180–188. doi: 10.1016/j.sbi.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiglietti M.L., Estreicher A., Gos A., Bolleman J., Géhant S., Breuza L., Bridge A., Poux S., Redaschi N., Bougueleret L., Xenarios I., UniProt Consortium Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation. Hum. Mutat. 2014;35(8):927–935. doi: 10.1002/humu.22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franden M.A., Jayakody L.N., Li W.-J., Wagner N.J., Cleveland N.S., Michener W.E., Hauer B., Blank L.M., Wierckx N., Klebensberger J., Beckham G.T. Engineering pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018;48:197–207. doi: 10.1016/j.ymben.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Gabor E.M., Alkema W.B.L., Janssen D.B. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 2004;6(9):879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- Gabor E.M., de Vries E.J., Janssen D.B. Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol. Ecol. 2003;44(2):153–163. doi: 10.1016/S0168-6496(02)00462-2. [DOI] [PubMed] [Google Scholar]
- Galperin M.Y., Wolf Y.I., Makarova K.S., Vera Alvarez R., Landsman D., Koonin E.V. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021;49(D1):D274–D281. doi: 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza D.R., Dutilh B.E. From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems. Cell. Mol. Life Sci. 2015;72(22):4287–4308. doi: 10.1007/s00018-015-2004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geesink P., Wegner C.-E., Probst A.J., Herrmann M., Dam H.T., Kaster A.-K., Küsel K. Genome-inferred spatio-temporal resolution of an uncultivated roizmanbacterium reveals its ecological preferences in groundwater. Environ. Microbiol. 2020;22(2):726–737. doi: 10.1111/1462-2920.14865. [DOI] [PubMed] [Google Scholar]
- Gies E.A., Konwar K.M., Beatty J.T., Hallam S.J. Illuminating microbial dark matter in meromictic sakinaw lake. Appl. Environ. Microbiol. 2014;80(21):6807–6818. doi: 10.1128/AEM.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi N.M., Thoden J.B., Holden H.M. Misannotations of the genes encoding sugar n-formyltransferases. Protein Sci. 2020;29(4):930–940. doi: 10.1002/pro.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M., Nouioui I., Sanderson R., Xie F., Bull A.T. Rare taxa and dark microbial matter: novel bioactive actinobacteria abound in atacama desert soils. Antonie Van Leeuwenhoek. 2018;111(8):1315–1332. doi: 10.1007/s10482-018-1088-7. [DOI] [PubMed] [Google Scholar]
- Grieb A., Bowers R.M., Oggerin M., Goudeau D., Lee J., Malmstrom R.R., Woyke T., Fuchs B.M. A pipeline for targeted metagenomics of environmental bacteria. Microbiome. 2020;8(1):21. doi: 10.1186/s40168-020-0790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A., Schöndube J., Niekrawitz S., Streule W., Riegger L., Zengerle R., Koltay P. Single-cell printer: automated, on demand, and label free. J. Lab. Autom. 2013;18(6):504–518. doi: 10.1177/2211068213497204. [DOI] [PubMed] [Google Scholar]
- Grötzinger S.W., Karan R., Strillinger E., Bader S., Frank A., Al Rowaihi I.S., Akal A., Wackerow W., Archer J.A., Rueping M., Weuster-Botz D., Groll M., Eppinger J., Arold S.T. Identification and experimental characterization of an extremophilic brine pool alcohol dehydrogenase from single amplified genomes. ACS Chem. Biol. 2018;13(1):161–170. doi: 10.1021/acschembio.7b00792. [DOI] [PubMed] [Google Scholar]
- Gurung N., Ray S., Bose S., Rai V. A broader view: microbial enzymes and their relevance in industries, medicine, and beyond. Biomed Res. Int. 2013;2013:329121. doi: 10.1155/2013/329121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjithomas M., Chen I.-M.A., Chu K., Ratner A., Palaniappan K., Szeto E., Huang J., Reddy T.B.K., Cimermančič P., Fischbach M.A., Ivanova N.N., Markowitz V.M., Kyrpides N.C., Pati A. IMG-ABC: A knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio. 2015;6(4):e00932. doi: 10.1128/mBio.00932-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D.H., Selengut J.D., Richter R.A., Harkins D., Basu M.K., Beck E. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41(Database issue):D387–95. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson A.D., Henry C.S., Fiehn O., de Crécy-Lagard V. Metabolite damage and metabolite damage control in plants. Annu. Rev. Plant Biol. 2016;67(1):131–152. doi: 10.1146/annurev-arplant-043015-111648. [DOI] [PubMed] [Google Scholar]
- Hatzenpichler R., Krukenberg V., Spietz R.L., Jay Z.J. Next-generation physiology approaches to study microbiome function at single cell level. Nat. Rev. Microbiol. 2020;18(4):241–256. doi: 10.1038/s41579-020-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley A.K., Nobu M.K., Wright J.J., Durno W.E., Morgan-Lang C., Sage B., Schwientek P., Swan B.K., Rinke C., Torres-Beltrán M., Mewis K., Liu W.-T., Stepanauskas R., Woyke T., Hallam S.J. Diverse marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 2017;8(1):1507. doi: 10.1038/s41467-017-01376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy F.G., Ray R.M., Aldrich H.C., Wilkie A.C., Ingram L.O., Shanmugam K.T. Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose. Applied Microbiology and Biotechnology. 1995;43(4):667–674. doi: 10.1007/bf00164771. [DOI] [PubMed] [Google Scholar]
- Hedlund B.P., Dodsworth J.A., Murugapiran S.K., Rinke C., Woyke T. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter”. Extremophiles. 2014;18(5):865–875. doi: 10.1007/s00792-014-0664-7. [DOI] [PubMed] [Google Scholar]
- Hetrick K.J., van der Donk W.A. Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol. 2017;38:36–44. doi: 10.1016/j.cbpa.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Janga S.C., Babu M., Díaz-Mejía J.J., Butland G., Yang W., Pogoutse O., Guo X., Phanse S., Wong P., Chandran S., Christopoulos C., Nazarians-Armavil A., Nasseri N.K., Musso G., Ali M., Nazemof N., Eroukova V., Golshani A., Paccanaro A., Greenblatt J.F., Moreno-Hagelsieb G., Emili A. Global functional atlas of escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7(4):e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug L.A., Baker B.J., Anantharaman K., Brown C.T., Probst A.J., Castelle C.J., Butterfield C.N., Hernsdorf A.W., Amano Y., Ise K., Suzuki Y., Dudek N., Relman D.A., Finstad K.M., Amundson R., Thomas B.C., Banfield J.F. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison 3rd, C. A., Chuang R.-Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L., Pelletier J.F., Qi Z.-Q., Richter R.A., Strychalski E.A., Sun L., Suzuki Y., Tsvetanova B., Wise K.S., Smith H.O., Glass J.I., Merryman C., Gibson D.G., Venter J.C. Design and synthesis of a minimal bacterial genome. Science. 2016;351(6280):aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- Idris H., Goodfellow M., Sanderson R., Asenjo J.A., Bull A.T. Actinobacterial rare biospheres and dark matter revealed in habitats of the chilean atacama desert. Sci. Rep. 2017;7(1):8373. doi: 10.1038/s41598-017-08937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H., Nobu M.K., Nakahara N., Morono Y., Ogawara M., Takaki Y., Takano Y., Uematsu K., Ikuta T., Ito M., Matsui Y., Miyazaki M., Murata K., Saito Y., Sakai S., Song C., Tasumi E., Yamanaka Y., Yamaguchi T., Kamagata Y., Tamaki H., Takai K. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature. 2020;577(7791):519–525. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey R.E., Lee M., Hawkins D.A., Sutton J.M., Panjikar S., Perugini M.A., Soares da Costa T.P. Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens. FEBS Lett. 2020;594(9):1453–1463. doi: 10.1002/1873-3468.13733. [DOI] [PubMed] [Google Scholar]
- Jacobs M.A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M.V., Manoil C. Comprehensive transposon mutant library of pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T., Malla S., Marriott H., Nieto T., O’Grady J., Olsen H.E., Pedersen B.S., Rhie A., Richardson H., Quinlan A.R., Snutch T.P., Tee L., Paten B., Phillippy A.M., Simpson J.T., Loman N.J., Loose M. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018;36(4):338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannasch H.W., Jones G.E. Bacterial populations in sea water as determined by different methods of enumeration1. Limnol. Oceanogr. 1959;4(2):128–139. [Google Scholar]
- Jeffery C.J. Protein moonlighting: what is it, and why is it important? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373(1738) doi: 10.1098/rstb.2016.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A., Torkel C., Stanley D., Nasim J., Borevitz J., Schwessinger B. High-molecular weight DNA extraction, clean-up and size selection for long-read sequencing. PLoS One. 2021;16(7):e0253830. doi: 10.1371/journal.pone.0253830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.G. Studies on freshwater bacteria: Effect of medium composition and method on estimates of bacterial population. J. Appl. Bacteriol. 1970;33(4):679–686. doi: 10.1111/j.1365-2672.1970.tb02250.x. [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20(10):1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakirde K.S., Parsley L.C., Liles M.R. Size does matter: Application-driven approaches for soil metagenomics. Soil Biol. Biochem. 2010;42(11):1911–1923. doi: 10.1016/j.soilbio.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Wrighton K.C., Handley K.M., Sharon I., Hug L.A., Castelle C.J., Thomas B.C., Banfield J.F. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 2013;4(5):e00708–13. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson E., Lärkeryd A., Sjödin A., Forsman M., Stenberg P. Scaffolding of a bacterial genome using MinION nanopore sequencing. Sci. Rep. 2015;5:11996. doi: 10.1038/srep11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaster A.-K., Sobol M.S. Microbial single-cell omics: the crux of the matter. Appl. Microbiol. Biotechnol. 2020;104(19):8209–8220. doi: 10.1007/s00253-020-10844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayani M.U.R., Huang W., Feng R., Chen L. Genome-resolved metagenomics using environmental and clinical samples. Brief. Bioinform. 2021;22(5) doi: 10.1093/bib/bbab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N.A., Walker A.W., Berry S.H., Duncan S.H., Farquarson F.M., Louis P., Thomson J.M., Satsangi J., Flint H.J., Parkhill J., Lees C.W., Hold G.L., Other members not named within the manuscript author list (alphabetical by surname): The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9(2):e88982. doi: 10.1371/journal.pone.0088982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel M.A.H.J., Speth D.R., Albertsen M., Nielsen P.H., Op den Camp H.J.M., Kartal B., Jetten M.S.M., Lücker S. Complete nitrification by a single microorganism. Nature. 2015;528(7583):555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khitun A., Ness T.J., Slavoff S.A. Small open reading frames and cellular stress responses. Mol. Omics. 2019;15(2):108–116. doi: 10.1039/c8mo00283e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford C., Schatz M.C., Pop M. Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics. 2010;11(1):21. doi: 10.1186/1471-2105-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Wolf Y.I. Evolution of microbial genomics: Conceptual shifts over a quarter century. Trends Microbiol. 2021;29(7):582–592. doi: 10.1016/j.tim.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Harhay G.P., Smith T.P.L., Bono J.L., Harhay D.M., Mcvey S.D., Radune D., Bergman N.H., Phillippy A.M. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14(9):R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., Cases I., Enright A.J., de Lorenzo V., Ouzounis C.A. Genome Biol. 2003;4(2):401. doi: 10.1186/gb-2003-4-2-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G., Peters E.E., Helfrich E.J.N., Piel J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. U. S. A. 2017;114(3):E347–E356. doi: 10.1073/pnas.1616234114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C., Bittar F., Fournous G., Gimenez G., Maraninchi M., Trape J.-F., Koonin E.V., La Scola B., Raoult D. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012;18(12):1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- Lagier J.-C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F., Levasseur A., Rolain J.-M., Fournier P.-E., Raoult D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P., Caputo A., Cadoret F., Traore S.I., Seck E.H., Dubourg G., Durand G., Mourembou G., Guilhot E., Togo A., Bellali S., Bachar D., Cassir N., Bittar F., Delerce J., Mailhe M., Ricaboni D., Bilen M., Dangui Nieko N.P.M., Dia Badiane N.M., Valles C., Mouelhi D., Diop K., Million M., Musso D., Abrahão J., Azhar E.I., Bibi F., Yasir M., Diallo A., Sokhna C., Djossou F., Vitton V., Robert C., Rolain J.M., La Scola B., Fournier P.-E., Levasseur A., Raoult D. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016;1(12) doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lannes R., Cavaud L., Lopez P., Bapteste E. Marine ultrasmall prokaryotes likely affect the cycling of carbon, methane, nitrogen, and sulfur. Genome Biol. Evol. 2021;13(1) doi: 10.1093/gbe/evaa261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lee D., Redfern O., Orengo C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8(12):995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- Lee P.K.H., Men Y., Wang S., He J., Alvarez-Cohen L. Development of a fluorescence-activated cell sorting method coupled with whole genome amplification to analyze minority and trace dehalococcoides genomes in microbial communities. Environ. Sci. Technol. 2015;49(3):1585–1593. doi: 10.1021/es503888y. [DOI] [PubMed] [Google Scholar]
- Li S., Yang X., Yang S., Zhu M., Wang X. Technology prospecting on enzymes: application, marketing and engineering. Comput. Struct. Biotechnol. J. 2012;2:e201209017. doi: 10.5936/csbj.201209017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., O’Neill K.R., Haft D.H., DiCuccio M., Chetvernin V., Badretdin A., Coulouris G., Chitsaz F., Derbyshire M.K., Durkin A.S., Gonzales N.R., Gwadz M., Lanczycki C.J., Song J.S., Thanki N., Wang J., Yamashita R.A., Yang M., Zheng C., Marchler-Bauer A., Thibaud-Nissen F. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49(D1):D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht M.W., Noble W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16(6):321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., O’Donoghue P., Söll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 2015;13(11):707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., Mueller A., Schäberle T.F., Hughes D.E., Epstein S., Jones M., Lazarides L., Steadman V.A., Cohen D.R., Felix C.R., Fetterman K.A., Millett W.P., Nitti A.G., Zullo A.M., Chen C., Lewis K. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517(7535):455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd K.G., Schreiber L., Petersen D.G., Kjeldsen K.U., Lever M.A., Steen A.D., Stepanauskas R., Richter M., Kleindienst S., Lenk S., Schramm A., Jørgensen B.B. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496(7444):215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- Lobb B., Kurtz D.A., Moreno-Hagelsieb G., Doxey A.C. Remote homology and the functions of metagenomic dark matter. Front. Genet. 2015;6:234. doi: 10.3389/fgene.2015.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb B., Tremblay B.J.-M., Moreno-Hagelsieb G., Doxey A.C. An assessment of genome annotation coverage across the bacterial tree of life. Microb. Genom. 2020;6(3) doi: 10.1099/mgen.0.000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locey K.J., Lennon J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 2016;113(21):5970–5975. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok C. Mining the microbial dark matter. Nature. 2015;522(7556):270–273. doi: 10.1038/522270a. [DOI] [PubMed] [Google Scholar]
- Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12(8):733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- López-García P., Moreira D. Cultured asgard archaea shed light on eukaryogenesis. Cell. 2020;181(2):232–235. doi: 10.1016/j.cell.2020.03.058. [DOI] [PubMed] [Google Scholar]
- López-García P., Moreira D. The syntrophy hypothesis for the origin of eukaryotes revisited. Nat. Microbiol. 2020;5(5):655–667. doi: 10.1038/s41564-020-0710-4. [DOI] [PubMed] [Google Scholar]
- Louca S., Mazel F., Doebeli M., Parfrey L.W. A census-based estimate of earth’s bacterial and archaeal diversity. PLoS Biol. 2019;17(2):e3000106. doi: 10.1371/journal.pbio.3000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., Thanki N., Yamashita R.A., Yang M., Zhang D., Zheng C., Lanczycki C.J., Marchler-Bauer A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Devine C.E., Edwards E.A. Cultivating microbial dark matter in benzene-degrading methanogenic consortia. Environ. Microbiol. 2016;18(9):2923–2936. doi: 10.1111/1462-2920.13121. [DOI] [PubMed] [Google Scholar]
- Maghini D.G., Moss E.L., Vance S.E., Bhatt A.S. Improved high-molecular-weight DNA extraction, nanopore sequencing and metagenomic assembly from the human gut microbiome. Nat. Protoc. 2021;16(1):458–471. doi: 10.1038/s41596-020-00424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Wolf Y.I., Forterre P., Prangishvili D., Krupovic M., Koonin E.V. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles. 2014;18(5):877–893. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Wolf Y.I., Koonin E.V. Towards functional characterization of archaeal genomic dark matter. Biochem. Soc. Trans. 2019;47(1):389–398. doi: 10.1042/BST20180560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrodimitris S., van Ham R.C.H.J., Reinders M.J.T. Automatic gene function prediction in the 2020’s. Genes. 2020;11(11) doi: 10.3390/genes11111264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małyska A., Markakis M.N., Pereira C.F., Cornelissen M. The microbiome: A life science opportunity for our society and our planet. Trends Biotechnol. 2019;37(12):1269–1272. doi: 10.1016/j.tibtech.2019.06.008. [DOI] [PubMed] [Google Scholar]
- Mani M., Chen C., Amblee V., Liu H., Mathur T., Zwicke G., Zabad S., Patel B., Thakkar J., Jeffery C.J. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 2015;43(Database issue):D277–82. doi: 10.1093/nar/gku954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285(5428):751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Marcotte E.M., Pellegrini M., Thompson M.J., Yeates T.O., Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402(6757):83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- Marcy Y., Ouverney C., Bik E.M., Lösekann T., Ivanova N., Martin H.G., Szeto E., Platt D., Hugenholtz P., Relman D.A., Quake S.R. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U. S. A. 2007;104(29):11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J., Schulz F., Zaremba-Niedzwiedzka K., Viklund J., Stepanauskas R., Andersson S.G.E., Horn M., Guy L., Ettema T.J.G. Single-cell genomics of a rare environmental alphaproteobacterium provides unique insights into rickettsiaceae evolution. ISME J. 2015;9(11):2373–2385. doi: 10.1038/ismej.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason O.U., Hazen T.C., Borglin S., Chain P.S.G., Dubinsky E.A., Fortney J.L., Han J., Holman H.-Y.N., Hultman J., Lamendella R., Mackelprang R., Malfatti S., Tom L.M., Tringe S.G., Woyke T., Zhou J., Rubin E.M., Jansson J.K. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to deepwater horizon oil spill. ISME J. 2012;6(9):1715–1727. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger S., Monard C., Thion C., Vandenkoornhuyse P. Contribution of single-cell omics to microbial ecology. Trends Ecol. Evol. 2022;37(1):67–78. doi: 10.1016/j.tree.2021.09.002. [DOI] [PubMed] [Google Scholar]
- Mavromatis K., Land M.L., Brettin T.S., Quest D.J., Copeland A., Clum A., Goodwin L., Woyke T., Lapidus A., Klenk H.P., Cottingham R.W., Kyrpides N.C. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS One. 2012;7(12):e48837. doi: 10.1371/journal.pone.0048837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayjonade B., Gouzy J., Donnadieu C., Pouilly N., Marande W., Callot C., Langlade N., Muños S. Extraction of high-molecular-weight genomic DNA for long-read sequencing of single molecules. BioTechniques. 2016;61(4):203–205. doi: 10.2144/000114460. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F., Hentschel U., King N., Kjelleberg S., Knoll A.H., Kremer N., Mazmanian S.K., Metcalf J.L., Nealson K., Pierce N.E., Rawls J.F., Reid A., Ruby E.G., Rumpho M., Sanders J.G., Tautz D., Wernegreen J.J. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean J.S., Lombardo M.-J., Badger J.H., Edlund A., Novotny M., Yee-Greenbaum J., Vyahhi N., Hall A.P., Yang Y., Dupont C.L., Ziegler M.G., Chitsaz H., Allen A.E., Yooseph S., Tesler G., Pevzner P.A., Friedman R.M., Nealson K.H., Venter J.C., Lasken R.S. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc. Natl. Acad. Sci. U. S. A. 2013;110(26):E2390–9. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C., Cruz-Morales P., Duddela S., Düsterhus S., Edwards D.J., Fewer D.P., Garg N., Geiger C., Gomez-Escribano J.P., Greule A., Hadjithomas M., Haines A.S., Helfrich E.J.N., Hillwig M.L., Ishida K., Jones A.C., Jones C.S., Jungmann K., Kegler C., Kim H.U., Kötter P., Krug D., Masschelein J., Melnik A.V., Mantovani S.M., Monroe E.A., Moore M., Moss N., Nützmann H.-W., Pan G., Pati A., Petras D., Reen F.J., Rosconi F., Rui Z., Tian Z., Tobias N.J., Tsunematsu Y., Wiemann P., Wyckoff E., Yan X., Yim G., Yu F., Xie Y., Aigle B., Apel A.K., Balibar C.J., Balskus E.P., Barona-Gómez F., Bechthold A., Bode H.B., Borriss R., Brady S.F., Brakhage A.A., Caffrey P., Cheng Y.-Q., Clardy J., Cox R.J., De Mot R., Donadio S., Donia M.S., van der Donk W.A., Dorrestein P.C., Doyle S., Driessen A.J.M., Ehling-Schulz M., Entian K.-D., Fischbach M.A., Gerwick L., Gerwick W.H., Gross H., Gust B., Hertweck C., Höfte M., Jensen S.E., Ju J., Katz L., Kaysser L., Klassen J.L., Keller N.P., Kormanec J., Kuipers O.P., Kuzuyama T., Kyrpides N.C., Kwon H.-J., Lautru S., Lavigne R., Lee C.Y., Linquan B., Liu X., Liu W., Luzhetskyy A., Mahmud T., Mast Y., Méndez C., Metsä-Ketelä M., Micklefield J., Mitchell D.A., Moore B.S., Moreira L.M., Müller R., Neilan B.A., Nett M., Nielsen J., O’Gara F., Oikawa H., Osbourn A., Osburne M.S., Ostash B., Payne S.M., Pernodet J.-L., Petricek M., Piel J., Ploux O., Raaijmakers J.M., Salas J.A., Schmitt E.K., Scott B., Seipke R.F., Shen B., Sherman D.H., Sivonen K., Smanski M.J., Sosio M., Stegmann E., Süssmuth R.D., Tahlan K., Thomas C.M., Tang Y., Truman A.W., Viaud M., Walton J.D., Walsh C.T., Weber T., van Wezel G.P., Wilkinson B., Willey J.M., Wohlleben W., Wright G.D., Ziemert N., Zhang C., Zotchev S.B., Breitling R., Takano E., Glöckner F.O. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015;11(9):625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meghwanshi G.K., Kaur N., Verma S., Dabi N.K., Vashishtha A., Charan P.D., Purohit P., Bhandari H.S., Bhojak N., Kumar R. Enzymes for pharmaceutical and therapeutic applications. Biotechnol. Appl. Biochem. 2020;67(4):586–601. doi: 10.1002/bab.1919. [DOI] [PubMed] [Google Scholar]
- Mehrshad M., Rodriguez-Valera F., Amoozegar M.A., López-García P., Ghai R. The enigmatic SAR202 cluster up close: shedding light on a globally distributed dark ocean lineage involved in sulfur cycling. ISME J. 2017 doi: 10.1038/s41396-017-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Scalzitti N., Jeannin-Girardon A., Collet P., Poch O., Thompson J.D. Understanding the causes of errors in eukaryotic protein-coding gene prediction: a case study of primate proteomes. BMC Bioinformatics. 2020;21(1):513. doi: 10.1186/s12859-020-03855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K., Steen A.D., Chhor G., Endres M., Webber A.T., Bird J., Lloyd K.G., Joachimiak A. New aminopeptidase from “microbial dark matter” archaeon. FASEB J. 2015;29(9):4071–4079. doi: 10.1096/fj.15-272906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller I.J., Weyna T.R., Fong S.S., Lim-Fong G.E., Kwan J.C. Single sample resolution of rare microbial dark matter in a marine invertebrate metagenome. Sci. Rep. 2016;6(1) doi: 10.1038/srep34362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A. Microbial genome evolution: sources of variability. Curr. Opin. Microbiol. 2002;5(5):506–512. doi: 10.1016/s1369-5274(02)00358-2. [DOI] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2(1):63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momper L., Jungbluth S.P., Lee M.D., Amend J.P. Energy and carbon metabolisms in a deep terrestrial subsurface fluid microbial community. ISME J. 2017;11(10):2319–2333. doi: 10.1038/ismej.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E.L., Maghini D.G., Bhatt A.S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 2020;38(6):701–707. doi: 10.1038/s41587-020-0422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Seshadri R., Varghese N.J., Eloe-Fadrosh E.A., Meier-Kolthoff J.P., Göker M., Coates R.C., Hadjithomas M., Pavlopoulos G.A., Paez-Espino D., Yoshikuni Y., Visel A., Whitman W.B., Garrity G.M., Eisen J.A., Hugenholtz P., Pati A., Ivanova N.N., Woyke T., Klenk H.-P., Kyrpides N.C. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat. Biotechnol. 2017;35(7):676–683. doi: 10.1038/nbt.3886. [DOI] [PubMed] [Google Scholar]
- Nagarajan N., Pop M. Parametric complexity of sequence assembly: theory and applications to next generation sequencing. J. Comput. Biol. 2009;16(7):897–908. doi: 10.1089/cmb.2009.0005. [DOI] [PubMed] [Google Scholar]
- Nasir A., Kim K.M., Caetano-Anollés G. Lokiarchaeota: eukaryote-like missing links from microbial dark matter? Trends Microbiol. 2015;23(8):448–450. doi: 10.1016/j.tim.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., Wu D., Paez-Espino D., Chen I.-M., Huntemann M., Palaniappan K., Ladau J., Mukherjee S., Reddy T.B.K., Nielsen T., Kirton E., Faria J.P., Edirisinghe J.N., Henry C.S., Jungbluth S.P., Chivian D., Dehal P., Wood-Charlson E.M., Arkin A.P., Tringe S.G., Visel A., IMG/M Data Consortium, Woyke T., Mouncey N.J., Ivanova N.N., Kyrpides N.C., Eloe-Fadrosh E.A. A genomic catalog of earth’s microbiomes. Nat. Biotechnol. 2021;39(4):499–509. doi: 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S., Shi Z.J., Seshadri R., Pollard K.S., Kyrpides N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568(7753):505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018;46(D1):D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New F.N., Brito I.L. What is metagenomics teaching us, and what is missed? Annu. Rev. Microbiol. 2020;74(1):117–135. doi: 10.1146/annurev-micro-012520-072314. [DOI] [PubMed] [Google Scholar]
- Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Nichols D., Cahoon N., Trakhtenberg E.M., Pham L., Mehta A., Belanger A., Kanigan T., Lewis K., Epstein S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010;76(8):2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H.B., Almeida M., Juncker A.S., Rasmussen S., Li J., Sunagawa S., Plichta D.R., Gautier L., Pedersen A.G., Le Chatelier E., Pelletier E., Bonde I., Nielsen T., Manichanh C., Arumugam M., Batto J.-M., Quintanilha Dos Santos M.B., Blom N., Borruel N., Burgdorf K.S., Boumezbeur F., Casellas F., Doré J., Dworzynski P., Guarner F., Hansen T., Hildebrand F., Kaas R.S., Kennedy S., Kristiansen K., Kultima J.R., Léonard P., Levenez F., Lund O., Moumen B., Le Paslier D., Pons N., Pedersen O., Prifti E., Qin J., Raes J., Sørensen S., Tap J., Tims S., Ussery D.W., Yamada T., MetaHIT Consortium, Renault P., Sicheritz-Ponten T., Bork P., Wang J., Brunak S., Ehrlich S.D., MetaHIT Consortium Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014;32(8):822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- Nobre T., Campos M.D., Lucic-Mercy E., Arnholdt-Schmitt B. Misannotation awareness: A tale of two gene-groups. Front. Plant Sci. 2016;7:868. doi: 10.3389/fpls.2016.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobu M.K., Narihiro T., Rinke C., Kamagata Y., Tringe S.G., Woyke T., Liu W.-T. Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J. 2015;9(8):1710–1722. doi: 10.1038/ismej.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A.A., Korobeynikov A., Lapidus A., Prjibelski A.D., Pyshkin A., Sirotkin A., Sirotkin Y., Stepanauskas R., Clingenpeel S.R., Woyke T., McLean J.S., Lasken R., Tesler G., Alekseyev M.A., Pevzner P.A. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013;20(10):714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C.M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V.S., Kodali V.K., Li W., Maglott D., Masterson P., McGarvey K.M., Murphy M.R., O’Neill K., Pujar S., Rangwala S.H., Rausch D., Riddick L.D., Schoch C., Shkeda A., Storz S.S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R.E., Vatsan A.R., Wallin C., Webb D., Wu W., Landrum M.J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T.D., Pruitt K.D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G.J., Lane D.J., Giovannoni S.J., Pace N.R., Stahl D.A. Microbial ecology and evolution: A ribosomal RNA approach. Annu. Rev. Microbiol. 1986;40(1):337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Olson N.D., Treangen T.J., Hill C.M., Cepeda-Espinoza V., Ghurye J., Koren S., Pop M. Metagenomic assembly through the lens of validation: recent advances in assessing and improving the quality of genomes assembled from metagenomes. Brief. Bioinform. 2019;20(4):1140–1150. doi: 10.1093/bib/bbx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U., Varadarajan A.R., Schmid M., Goetze S., Melidis D., Bourqui M., Nikolayeva O., Québatte M., Patrignani A., Dehio C., Frey J.E., Robinson M.D., Wollscheid B., Ahrens C.H. An integrative strategy to identify the entire protein coding potential of prokaryotic genomes by proteogenomics. Genome Res. 2017;27(12):2083–2095. doi: 10.1101/gr.218255.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orakov A., Fullam A., Coelho L.P., Khedkar S., Szklarczyk D., Mende D.R., Schmidt T.S.B., Bork P. GUNC: detection of chimerism and contamination in prokaryotic genomes. Genome Biol. 2021;22(1):178. doi: 10.1186/s13059-021-02393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr M.W., Mao Y., Storz G., Qian S.-B. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res. 2020;48(3):1029–1042. doi: 10.1093/nar/gkz734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R., Xia F., Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen J.G., Charlop-Powers Z., Smith A.G., Ternei M.A., Calle P.Y., Reddy B.V.B., Montiel D., Brady S.F. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2015;112(14):4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.R. Opening the door onto the natural microbial world: molecular microbial ecology. Harvey Lect. 1995;91:59–78. [PubMed] [Google Scholar]
- Pachiadaki M.G., Brown J.M., Brown J., Bezuidt O., Berube P.M., Biller S.J., Poulton N.J., Burkart M.D., La Clair J.J., Chisholm S.W., Stepanauskas R. Charting the complexity of the marine microbiome through single-cell genomics. Cell. 2019;179(7):1623–1635.e11. doi: 10.1016/j.cell.2019.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm G.J., Reisky L., Böttcher D., Müller H., Michels E.A.P., Walczak M.C., Berndt L., Weiss M.S., Bornscheuer U.T., Weber G. Structure of the plastic-degrading ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019;10(1):1717. doi: 10.1038/s41467-019-09326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2(11):1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- Pascoal F., Magalhães C., Costa R. The link between the ecology of the prokaryotic rare biosphere and its biotechnological potential. Front. Microbiol. 2020;11:231. doi: 10.3389/fmicb.2020.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P., Collado M.C., Rice B.L., DuLong C., Morgan X.C., Golden C.D., Quince C., Huttenhower C., Segata N. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649–662.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrós-Alió, C., 2012. The rare bacterial biosphere. [DOI] [PubMed]
- Pedrós-Alió C., Manrubia S. The vast unknown microbial biosphere. Proc. Natl. Acad. Sci. U. S. A. 2016;113(24):6585–6587. doi: 10.1073/pnas.1606105113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H., Froula J., Du C., Kim T.-W., Hawley E.R., Bauer S., Wang Z., Ivanova N., Clark D.S., Klenk H.-P., Hess M. Identification of novel biomass-degrading enzymes from genomic dark matter: Populating genomic sequence space with functional annotation. Biotechnol. Bioeng. 2014;111(8):1550–1565. doi: 10.1002/bit.25250. [DOI] [PubMed] [Google Scholar]
- Plaza S., Menschaert G., Payre F. In search of lost small peptides. Annu. Rev. Cell Dev. Biol. 2017;33(1):391–416. doi: 10.1146/annurev-cellbio-100616-060516. [DOI] [PubMed] [Google Scholar]
- Plaza Oñate F., Le Chatelier E., Almeida M., Cervino A.C.L., Gauthier F., Magoulès F., Ehrlich S.D., Pichaud M. MSPminer: abundance-based reconstitution of microbial pan-genomes from shotgun metagenomic data. Bioinformatics. 2019;35(9):1544–1552. doi: 10.1093/bioinformatics/bty830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash T., Taylor T.D. Functional assignment of metagenomic data: challenges and applications. Brief. Bioinform. 2012;13(6):711–727. doi: 10.1093/bib/bbs033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.J., Ladd B., Jarett J.K., Geller-McGrath D.E., Sieber C.M.K., Emerson J.B., Anantharaman K., Thomas B.C., Malmstrom R.R., Stieglmeier M., Klingl A., Woyke T., Ryan M.C., Banfield J.F. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat. Microbiol. 2018;3(3):328–336. doi: 10.1038/s41564-017-0098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promponas V.J., Iliopoulos I., Ouzounis C.A. Annotation inconsistencies beyond sequence similarity-based function prediction - phylogeny and genome structure. Stand. Genomic Sci. 2015;10(1):108. doi: 10.1186/s40793-015-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017;35(9):833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- Ragaert K., Delva L., Van Geem K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017;69:24–58. doi: 10.1016/j.wasman.2017.07.044. [DOI] [PubMed] [Google Scholar]
- Ramesh A., Harani Devi P., Chattopadhyay S., Kavitha M. Microorganisms for Sustainability. Springer Singapore; Singapore: 2020. Commercial applications of microbial enzymes; pp. 137–184. [Google Scholar]
- Rappé M.S., Giovannoni S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57(1):369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Rashid M., Stingl U. Contemporary molecular tools in microbial ecology and their application to advancing biotechnology. Biotechnol. Adv. 2015;33(8):1755–1773. doi: 10.1016/j.biotechadv.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Rembeza E., Engqvist M.K.M. Experimental and computational investigation of enzyme functional annotations uncovers misannotation in the EC 1.1.3.15 enzyme class. PLoS Comput. Biol. 2021;17(9):e1009446. doi: 10.1371/journal.pcbi.1009446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Wang B., Zhao H. Breaking the silence: new strategies for discovering novel natural products. Curr. Opin. Biotechnol. 2017;48:21–27. doi: 10.1016/j.copbio.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.-F., Darling A., Malfatti S., Swan B.K., Gies E.A., Dodsworth J.A., Hedlund B.P., Tsiamis G., Sievert S.M., Liu W.-T., Eisen J.A., Hallam S.J., Kyrpides N.C., Stepanauskas R., Rubin E.M., Hugenholtz P., Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Rodríguez del Río, Á., Giner-Lamia, J., Cantalapiedra, C. P., Botas, J., Deng, Z., Hernández-Plaza, A., Paoli, L., Schmidt, T. S. B., Sunagawa, S., Bork, P., Coelho, L. P., Huerta-Cepas, J., 2022. Functional and evolutionary significance of unknown genes from uncultivated taxa. [DOI] [PMC free article] [PubMed]
- Robinson P.K. Enzymes: principles and biotechnological applications. Essays Biochem. 2015;59:1–41. doi: 10.1042/bse0590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.L. Artificial intelligence for microbial biotechnology: beyond the hype. Microb. Biotechnol. 2022;15(1):65–69. doi: 10.1111/1751-7915.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.L., Piel J., Sunagawa S. A roadmap for metagenomic enzyme discovery. Nat. Prod. Rep. 2021;38(11):1994–2023. doi: 10.1039/d1np00006c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C., Athanasouli M., Lenuzzi M., Theska T., Sun S., Dardiry M., Wighard S., Hu W., Sharma D.R., Han Z. Crowdsourcing and the feasibility of manual gene annotation: A pilot study in the nematode pristionchus pacificus. Sci. Rep. 2019;9(1):18789. doi: 10.1038/s41598-019-55359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojano E., Jabato F.M., Perkins J.R., Córdoba-Caballero J., García-Criado F., Sillitoe I., Orengo C., Ranea J.A.G., Seoane-Zonjic P. Assigning protein function from domain-function associations using DomFun. BMC Bioinformatics. 2022;23(1):43. doi: 10.1186/s12859-022-04565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M., Helfrich E.J.N., Freeman M.F., Nanudorn P., Field C.M., Rückert C., Kündig T., Page M.J., Webb V.L., Kalinowski J., Sunagawa S., Piel J. A multiproducer microbiome generates chemical diversity in the marine sponge. Proc. Natl. Acad. Sci. U. S. A. 2020;117(17):9508–9518. doi: 10.1073/pnas.1919245117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A., Couso J.P. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 2015;11(12):909–916. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R.K., Gelfand D.H., Stoffel S., Scharf S.J., Higuchi R., Horn G.T., Mullis K.B., Erlich H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Salzberg S.L. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 2019;20(1):92. doi: 10.1186/s13059-019-1715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan N., Xia F., Gilbert J.A. Recovering complete and draft population genomes from metagenome datasets. Microbiome. 2016;4(1):8. doi: 10.1186/s40168-016-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A.E., Kellom M., Laperriere S.M. Contributions of single-cell genomics to our understanding of planktonic marine archaea. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374(1786):20190096. doi: 10.1098/rstb.2019.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapoval N., Aghazadeh A., Nute M.G., Antunes D.A., Balaji A., Baraniuk R., Barberan C.J., Dannenfelser R., Dun C., Edrisi M., Elworth R.A.L., Kille B., Kyrillidis A., Nakhleh L., Wolfe C.R., Yan Z., Yao V., Treangen T.J. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 2022;13(1):1728. doi: 10.1038/s41467-022-29268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sberro H., Fremin B.J., Zlitni S., Edfors F., Greenfield N., Snyder M.P., Pavlopoulos G.A., Kyrpides N.C., Bhatt A.S. Large-Scale analyses of human microbiomes reveal thousands of small, novel genes. Cell. 2019;178(5):1245–1259.e14. doi: 10.1016/j.cell.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K.J., Aquadro C.F. The evolutionary analysis of “orphans” from the drosophila genome identifies rapidly diverging and incorrectly annotated genes. Genetics. 2001;159(2):589–598. doi: 10.1093/genetics/159.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Muri J., Melidis D., Varadarajan A.R., Somerville V., Wicki A., Moser A., Bourqui M., Wenzel C., Eugster-Meier E., Frey J.E., Irmler S., Ahrens C.H. Comparative genomics of completely sequenced lactobacillus helveticus genomes provides insights into strain-specific genes and resolves metagenomics data down to the strain level. Front. Microbiol. 2018;9:63. doi: 10.3389/fmicb.2018.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes A.M., Brown S.D., Dodevski I., Babbitt P.C. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009;5(12):e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15(2):121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- Singh B.K. Organophosphorus-degrading bacteria: ecology and industrial applications. Nat. Rev. Microbiol. 2009;7(2):156–164. doi: 10.1038/nrmicro2050. [DOI] [PubMed] [Google Scholar]
- Singh R., Kumar M., Mittal A., Mehta P.K. Microbial enzymes: industrial progress in 21st century. 3 Biotech. 2016;6(2):174. doi: 10.1007/s13205-016-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider M.A., Johnston C.W., Edgar R.E., Dejong C.A., Merwin N.J., Rees P.N., Magarvey N.A. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc. Natl. Acad. Sci. U. S. A. 2016;113(42):E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.O., Wilcox K.W. A restriction enzyme from hemophilus influenzae. i. purification and general properties. J. Mol. Biol. 1970;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Sogin M.L., Morrison H.G., Huber J.A., Mark Welch D., Huse S.M., Neal P.R., Arrieta J.M., Herndl G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U. S. A. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solden L., Lloyd K., Wrighton K. The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 2016;31:217–226. doi: 10.1016/j.mib.2016.04.020. [DOI] [PubMed] [Google Scholar]
- Somerville V., Lutz S., Schmid M., Frei D., Moser A., Irmler S., Frey J.E., Ahrens C.H. Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol. 2019;19(1):143. doi: 10.1186/s12866-019-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood U., Kumar R., Hira P. Expanding culturomics from gut to extreme environmental settings. mSystems. 2021;6(4):e0084821. doi: 10.1128/msystems.00848-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Saw J.H., Jørgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E., van Eijk R., Schleper C., Guy L., Ettema T.J.G. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521(7551):173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens Z.D., Lee S.Y., Faghri F., Campbell R.H., Zhai C., Efron M.J., Iyer R., Schatz M.C., Sinha S., Robinson G.E. Big data: Astronomical or genomical? PLoS Biol. 2015;13(7):e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R.D., Auffret M.D., Warr A., Wiser A.H., Maximilian O. Press, Langford K.W., Liachko I., Snelling T.J., Dewhurst R.J., Walker A.W., Roehe R., Watson M. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 2018;9(1) doi: 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D.R., Alberti A., Cornejo-Castillo F.M., Costea P.I., Cruaud C., d’Ovidio F., Engelen S., Ferrera I., Gasol J.M., Guidi L., Hildebrand F., Kokoszka F., Lepoivre C., Lima-Mendez G., Poulain J., Poulos B.T., Royo-Llonch M., Sarmento H., Vieira-Silva S., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Tara Oceans coordinators, Bowler C., de Vargas C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Jaillon O., Not F., Ogata H., Pesant S., Speich S., Stemmann L., Sullivan M.B., Weissenbach J., Wincker P., Karsenti E., Raes J., Acinas S.G., Bork P. Ocean plankton. structure and function of the global ocean microbiome. Science. 2015;348(6237):1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Sysoev M., Grötzinger S.W., Renn D., Eppinger J., Rueping M., Karan R. Bioprospecting of novel extremozymes from Prokaryotes-The advent of Culture-Independent methods. Front. Microbiol. 2021;12:630013. doi: 10.3389/fmicb.2021.630013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J., Cobo-Simón M., Puente-Sánchez F. Assessing the performance of different approaches for functional and taxonomic annotation of metagenomes. BMC Genomics. 2019;20(1):960. doi: 10.1186/s12864-019-6289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Domazet-Lošo T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011;12(10):692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Teeling H., Glöckner F.O. Current opportunities and challenges in microbial metagenome analysis–a bioinformatic perspective. Brief. Bioinform. 2012;13(6):728–742. doi: 10.1093/bib/bbs039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K., Zhao D., Voigt C.A. Refactoring the nitrogen fixation gene cluster from klebsiella oxytoca. Proc. Natl. Acad. Sci. U. S. A. 2012;109(18):7085–7090. doi: 10.1073/pnas.1120788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash J.C., Cameron Thrash J., Seitz K.W., Baker B.J., Temperton B., Gillies L.E., Rabalais N.N., Henrissat B., Mason O.U. Metabolic roles of uncultivated bacterioplankton lineages in the northern gulf of mexico “dead zone”. MBio. 2017;8(5):e01017–17. doi: 10.1128/mBio.01017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E., Kamionka E., Desrousseaux M.-L., Texier H., Gavalda S., Cot M., Guémard E., Dalibey M., Nomme J., Cioci G., Barbe S., Chateau M., André I., Duquesne S., Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580(7802):216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Treiber M.L., Taft D.H., Korf I., Mills D.A., Lemay D.G. Pre- and post-sequencing recommendations for functional annotation of human fecal metagenomes. BMC Bioinformatics. 2020;21(1):74. doi: 10.1186/s12859-020-3416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigodet F., Lolans K., Fogarty E., Shaiber A., Morrison H.G., Barreiro L., Jabri B., Eren A.M. High molecular weight DNA extraction strategies for long-read sequencing of complex metagenomes. Mol. Ecol. Resour. 2022 doi: 10.1111/1755-0998.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully B.J., Graham E.D., Heidelberg J.F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data. 2018;5:170203. doi: 10.1038/sdata.2017.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations, Department of Economic and Social Affairs, 2015. Transforming our world: the 2030 agenda for sustainable development. sustainable development knowledge platform. https://sustainabledevelopment.un.org/post2015/transformingourworldAccessed: 2018-6-26.
- Van Schaftingen E., Veiga-da Cunha M., Linster C.L. Enzyme complexity in intermediary metabolism. J. Inherit. Metab. Dis. 2015;38(4):721–727. doi: 10.1007/s10545-015-9821-0. [DOI] [PubMed] [Google Scholar]
- Varadarajan A.R., Allan R.N., Valentin J.D.P., Castañeda Ocampo O.E., Somerville V., Pietsch F., Buhmann M.T., West J., Skipp P.J., van der Mei H.C., Ren Q., Schreiber F., Webb J.S., Ahrens C.H. An integrated model system to gain mechanistic insights into biofilm-associated antimicrobial resistance in pseudomonas aeruginosa MPAO1. NPJ Biofilms Microbiomes. 2020;6(1):46. doi: 10.1038/s41522-020-00154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Meghwanshi G.K., Kumar R. Current perspectives for microbial lipases from extremophiles and metagenomics. Biochimie. 2021;182:23–36. doi: 10.1016/j.biochi.2020.12.027. [DOI] [PubMed] [Google Scholar]
- Vollmers J., Wiegand S., Lenk F., Kaster A.-K. How clear is our current view on microbial dark matter? (re-)assessing public MAG & SAG datasets with MDMcleaner. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Kim H.U. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol. 2016;1(2):69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner C.-E., Liesack W. Unexpected dominance of elusive acidobacteria in early industrial soft coal slags. Front. Microbiol. 2017;8:1023. doi: 10.3389/fmicb.2017.01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S., Dam H.T., Riba J., Vollmers J., Kaster A.-K. Printing microbial dark matter: Using single cell dispensing and genomics to investigate the Patescibacteria/Candidate phyla radiation. Front. Microbiol. 2021;12:635506. doi: 10.3389/fmicb.2021.635506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S., Jogler M., Boedeker C., Pinto D., Vollmers J., Rivas-Marín E., Kohn T., Peeters S.H., Heuer A., Rast P., Oberbeckmann S., Bunk B., Jeske O., Meyerdierks A., Storesund J.E., Kallscheuer N., Lücker S., Lage O.M., Pohl T., Merkel B.J., Hornburger P., Müller R.-W., Brümmer F., Labrenz M., Spormann A.M., Op den Camp H.J.M., Overmann J., Amann R., Jetten M.S.M., Mascher T., Medema M.H., Devos D.P., Kaster A.-K., Øvreås L., Rohde M., Galperin M.Y., Jogler C. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. 2020;5(1):126–140. doi: 10.1038/s41564-019-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A. Extrapolating abundance curves has no predictive power for estimating microbial biodiversity. Proc. Natl. Acad. Sci. U. S. A. 2016;113(35):E5096. doi: 10.1073/pnas.1608281113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.A., Bertrand N., Patel Y., Hughes J.B., Feil E.J., Field D. Orphans as taxonomically restricted and ecologically important genes. Microbiology. 2005;151(Pt 8):2499–2501. doi: 10.1099/mic.0.28146-0. [DOI] [PubMed] [Google Scholar]
- Woese C.R. Bacterial evolution. Microbiol. Rev. 1987;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack K.E., Bhavsar J., Ravel J. Metagenomics: Read length matters. Appl. Environ. Microbiol. 2008;74(5):1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.L., MacLeod F.I., White 3rd, R. A., Visscher P.T., Burns B.P. Microbial dark matter filling the niche in hypersaline microbial mats. Microbiome. 2020;8(1):135. doi: 10.1186/s40168-020-00910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T., Doud D.F.R., Eloe-Fadrosh E.A. Reference Module in Life Sciences. Elsevier; 2019. Genomes from uncultivated microorganisms. [Google Scholar]
- Woyke T., Doud D.F.R., Schulz F. The trajectory of microbial single-cell sequencing. Nat. Methods. 2017;14(11):1045–1054. doi: 10.1038/nmeth.4469. [DOI] [PubMed] [Google Scholar]
- Woyke T., Jarett J. Function-driven single-cell genomics. Microb. Biotechnol. 2015;8(1):38–39. doi: 10.1111/1751-7915.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T., Xie G., Copeland A., González J.M., Han C., Kiss H., Saw J.H., Senin P., Yang C., Chatterji S., Cheng J.-F., Eisen J.A., Sieracki M.E., Stepanauskas R. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4(4):e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Hugenholtz P., Mavromatis K., Pukall R., Dalin E., Ivanova N.N., Kunin V., Goodwin L., Wu M., Tindall B.J., Hooper S.D., Pati A., Lykidis A., Spring S., Anderson I.J., D’haeseleer P., Zemla A., Singer M., Lapidus A., Nolan M., Copeland A., Han C., Chen F., Cheng J.-F., Lucas S., Kerfeld C., Lang E., Gronow S., Chain P., Bruce D., Rubin E.M., Kyrpides N.C., Klenk H.-P., Eisen J.A. A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature. 2009;462(7276):1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhao F. Single-cell metagenomics: challenges and applications. Protein Cell. 2018;9(5):501–510. doi: 10.1007/s13238-018-0544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Nakahigashi K., Nakamichi T., Yoshino M., Takai Y., Touda Y., Furubayashi A., Kinjyo S., Dose H., Hasegawa M., Datsenko K.A., Nakayashiki T., Tomita M., Wanner B.L., Mori H. Update on the keio collection of escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009;5(1):335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E., Glöckner F.O., Ludwig W., Schleifer K.-H., Whitman W.B., Euzéby J., Amann R., Rosselló-Móra R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014;12(9):635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- Yin Y., Fischer D. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol. Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y., Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351(6278):1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- Youngblut N.D., de la Cuesta-Zuluaga J., Reischer G.H., Dauser S., Schuster N., Walzer C., Stalder G., Farnleitner A.H., Ley R.E. Large-scale metagenome assembly reveals novel animal-associated microbial genomes, biosynthetic gene clusters, and other genetic diversity. mSystems. 2020;5(6) doi: 10.1128/mSystems.01045-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Cohen D.B., Ravel J., Abdo Z., Forney L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7(3):e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R., Harrison K., Kolaczkowski B., de Crécy-Lagard V. Functional annotations of paralogs: A blessing and a curse. Life. 2016;6(4):39. doi: 10.3390/life6030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K., Caceres E.F., Saw J.H., Bäckström D., Juzokaite L., Vancaester E., Seitz K.W., Anantharaman K., Starnawski P., Kjeldsen K.U., Stott M.B., Nunoura T., Banfield J.F., Schramm A., Baker B.J., Spang A., Ettema T.J.G. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541(7637):353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- Zha Y., Chong H., Yang P., Ning K. Microbial dark matter: from discovery to applications. Genomics Proteomics Bioinformatics. 2022 doi: 10.1016/j.gpb.2022.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang Y., Scheuring C.F., Wu C.-C., Dong J.J., Zhang H.-B. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat. Protoc. 2012;7(3):467–478. doi: 10.1038/nprot.2011.455. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang J., Wang J., Wang J., Li Y. Estimate of the sequenced proportion of the global prokaryotic genome. Microbiome. 2020;8(1):134. doi: 10.1186/s40168-020-00903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Wuyun Q., Zhou X., Li Y., Freddolino P.L., Zhang Y. LOMETS3: integrating deep learning and profile alignment for advanced protein template recognition and function annotation. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., He B., Li J., Li Y.-X. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs) Synth Syst Biotechnol. 2020;5(3):155–172. doi: 10.1016/j.synbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N., Alanjary M., Weber T. The evolution of genome mining in microbes – a review. Nat. Prod. Rep. 2016;33(8):988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- Zohra Smaili F., Tian S., Roy A., Alazmi M., Arold S.T., Mukherjee S., Scott Hefty P., Chen W., Gao X. QAUST: Protein function prediction using structure similarity, protein interaction, and functional motifs. Genomics Proteomics Bioinformatics. 2021 doi: 10.1016/j.gpb.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrimec J., Kokina M., Jonasson S., Zorrilla F., Zelezniak A. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. MBio. 2021;12(5):e0215521. doi: 10.1128/mBio.02155-21. [DOI] [PMC free article] [PubMed] [Google Scholar]