Abstract

Systems that allow researchers to precisely control the expression of genes are fundamental to biological research, biotechnology, and synthetic biology. However, few inducible gene expression systems exist that can enable simultaneous multigene control under common nutritionally favorable conditions in the important model organism and chassis Saccharomyces cerevisiae. Here we repurposed ligand binding domains from mammalian type I nuclear receptors to establish a family of up to five orthogonal synthetic gene expression systems in yeast. Our systems enable tight, independent, multigene control through addition of inert hormones and are capable of driving robust and rapid gene expression outputs, in some cases achieving up to 600-fold induction. As a proof of principle, we placed expression of four enzymes from the violacein biosynthetic pathway under independent expression control to selectively route pathway flux by addition of specific inducer combinations. Our results establish a modular, versatile, and potentially expandable toolkit for multidimensional control of gene expression in yeast that can be used to construct and control naturally occurring and synthetic gene networks.
Keywords: yeast, gene expression control, inducible promoters, transcription factors, MoClo Toolkit, nuclear receptors
Cells activate gene expression programs in response to internal and external signals. Manipulating this fundamental process in order to control cellular behavior is a principal goal of synthetic biology and serves as the basis of widely used tools in the life sciences and biotechnology. In particular, tools that enable a single gene to be turned on/off and its expression to be precisely controlled with external stimuli in the yeast Saccharomyces cerevisiae have played an outsized role in facilitating important discoveries and applications. For example, inducible gene expression systems in yeast have been critical for probing gene function and regulatory network architecture, establishing models of gene regulation, elucidating the origins and consequences of noise in gene expression, and even developing yeast models of human proteinopathies that paved the way for the discovery of potential therapeutic agents.1,2,11−15,3−10 In addition, these “parts” have served as essential building blocks for constructing and controlling synthetic genetic circuits and metabolic pathways.16−24 Overall, these systems have transformed our ability to control and investigate the roles of individual genes within naturally occurring and synthetic cellular networks.
The function of cellular networks arises from the coordinated behavior of individual molecular components. Thus, while single-gene perturbations enabled by existing inducible expression systems have been highly valuable, they are unable to achieve the simultaneous, orthogonal-to-host, multigene control necessary to study and manipulate higher-order networks. Moving forward, a common toolkit is needed to control multiple genes simultaneously and independently using a harmonized and modular architecture that provides organizational simplicity and expandability. Current technologies for controlling gene expression lack systemization and fall short of this goal, a limitation that motivated the development of the “Marionette” strains in Escherichia coli, which can allow for robust and independent inducible expression of many genes within a single bacterial cell.25 This advance opens up the possibility to study and control multigene pathways with precision and ease and to implement elaborated multi-input synthetic circuits; however, to date, no similar system has been developed for yeast or higher-order eukaryotes.26,27
In yeast, historically, the most widely used inducible systems have relied on endogenous metabolite-controlled promoters: the MET25, CUP1, and GAL1 promoters. In particular, the pGAL1 system has been a workhorse for yeast genetics. However, these systems suffer considerable drawbacks that limit their applicability and generalizability. For example, pMET25 is restricted to synthetic minimal-medium conditions; pCUP1 is induced by the addition of toxic copper; and pGAL1 induction requires a major metabolic shift and growth on alternative (non-glucose) carbon sources.28−30 The restrictive nature of the induction and nutritional conditions makes combining these systems for multigene control virtually impossible. Furthermore, and importantly, because all three are native to yeast, induction triggers substantial genome-wide transcriptional changes. These limitations motivated efforts to port heterologous transcription factor (TF) systems, such as LacI- and TetR-based systems, from bacteria into yeast. The mutual orthogonality and heterologous nature of these TFs offer the possibility of imposing two-gene control with minimal effects on the host. However, expanding on these to create a larger set of TFs for higher-order control has proven challenging. Bacterial TFs are highly integrated proteins with intertwined DNA recognition, dimerization, and ligand recognition domains; thus, decoupling these to engineer additional mutually orthogonal, small-molecule-responsive TFs with high activity in yeast requires extensive directed evolution or structure-based design.31,32
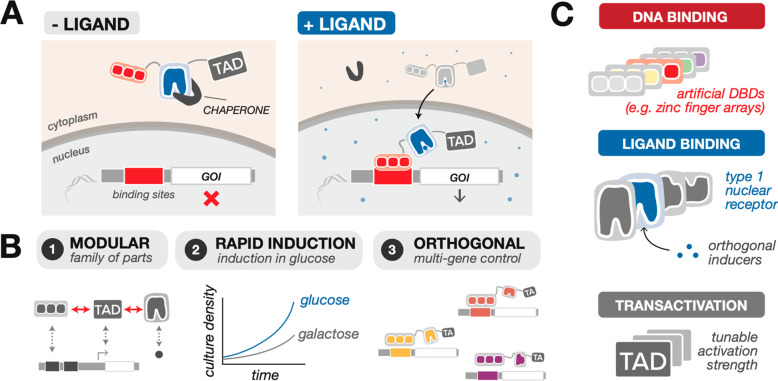
We instead drew inspiration from the nuclear receptor superfamily of ligand-activated transcriptional regulators found in metazoans. In particular, type I nuclear receptors (NRs) are a unique family of transcriptional regulators with a largely modular structure, composed of independent hormone ligand binding (LBD), DNA binding (DBD), and transcriptional activation (TAD) domains.33 In the presence of hormone, NRs translocate from the cytoplasm to the nucleus, bind to a cognate DNA sequence, and recruit transcriptional machinery to activate gene expression (Figure 1A).34−37 Given their modular tripartite architecture, NRs are an ideal framework from which to build a suite of ligand-responsive synthetic TFs (synTFs); moreover, their ligands are not yeast metabolites, making them effectively inert inducers. Important recent advances have shown the potential of this strategy: the development of β-estradiol-inducible gene expression systems (e.g., GEV and ZEV) based on the human estrogen receptor have been shown to enable tight, robust, and rapid induction of gene expression in yeast,21−23,38 and a progesterone-inducible system indicates the potential for expandability.39
Figure 1.
Design of a toolkit for multigene inducible gene expression control in yeast based on hormone-responsive type I nuclear receptors (NRs). (A) Schematic of the mechanism of action of NR-based gene regulation. Upon ligand binding, the transcriptional regulator dissociates from a chaperone, enters the nucleus, and initiates transcription of a target gene downstream of response elements. (B) Features of the NR-based synthetic transcription factor (synTF) toolkit: modularity, rapid induction in a glucose-based medium, and mutual orthogonality for simultaneous multigene control. (C) Modular synTF design enables mix-and-match construction to obtain desired properties. The synTFs are composed of interchangeable artificial DNA binding domains (DBDs), type I NR ligand binding domains (LBDs), and transactivation domains (TADs).
In this study, we asked whether this concept could be expanded to construct a toolkit of synthetic transcriptional systems for simultaneous and independent control of multiple genes based on orthogonal hormone inducers. We screened combinations of established artificial DBDs (derived from artificial zinc fingers (ZFs)), TADs, and LBDs for synTFs that, when paired with synthetic responsive promoters, were capable of driving robust and rapid gene expression outputs.40 We created a set of five orthogonal inducible systems in yeast that deliver robust, titratable gene induction and offer the ability to simultaneously control expression of several genes in a single cell. To illustrate the versatility of our toolkit, we engineered independent expression control over four enzymes from the violacein biosynthetic pathway and selectively routed pathway flux by inducing with specific inducer combinations. To facilitate adoption and widespread use, we domesticated all of the components into the common Yeast MoClo Toolkit.41
We first created a panel of candidate synTFs by mining modular DBDs, LBDs, and TADs from the literature and assembling them combinatorially (Figure 1). We chose five well-characterized, mutually orthogonal synthetic ZFs for DBDs.40 For LBDs, we surveyed natural and engineered NR LBDs predicted to have low crosstalk, nominating the estradiol-responsive (ER), aldosterone-responsive (MR), 1,2-bis(4-hydroxyphenyl)ethane-1,2-dione (DHB)-responsive (DHBR), testosterone-responsive (AR), and dexamethasone-responsive (GR) domains as potential candidates.21−23,39,42,43 Finally, we selected three commonly used TADs—VP16, Rta, and Msn2—to stimulate transcription activation. We then constructed a full set of candidate synTFs by fusing each LBD to a unique DBD and to one of each of the three TADs, in total producing 15 variants for initial characterization.
We cloned the synTFs alongside fluorescent protein reporter cassettes in yeast expression vectors, incorporating a number of design elements for optimal performance. First, we drove synTF expression with the strong constitutive promoter pTEF1 to maximize sensitivity. Second, we constructed corresponding responsive promoters for each hormone-inducible synTF by encoding six to eight cognate ZF binding sites upstream of a minimal CYC1 promoter sequence. We generated reporters by encoding an mKate2 gene downstream of the synthetic promoters. Finally, to avoid fluctuations in gene expression level due to copy number variability, we chromosomally cointegrated the synTF and reporter cassettes into yeast.
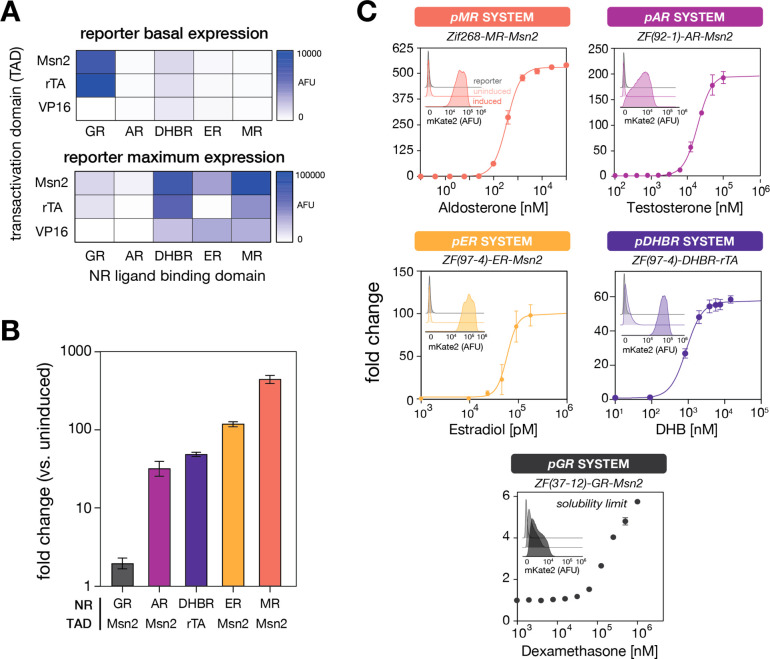
We screened the candidate synTF variants by measuring fluorescent reporter expression using flow cytometry following induction with the respective inducer hormones (Figures 2A and S1). Maximum reporter expression varied across 3 orders of magnitude, with the top-performing GR-, AR-, DHBR-, ER-, and MR-based synTFs delivering 2-, 33-, 51-, 124-, and 465-fold reporter changes, respectively, at inducer concentrations of 1 mM dexamethasone, 20 μM testosterone, 10 μM DHB, 100 nM estradiol, and 10 μM aldosterone, respectively. (Figure 2B). Dose–response curves for these five selected systems showed smooth and titratable reporter outputs, capable of accessing a large range of expression levels between basal and maximum (Figure 2C). In every case, populations shifted unimodally from uninduced to induced states. Encouraged by the results, we advanced these five synTF systems for further study.
Figure 2.
Development and characterization of a collection of hormone-inducible synthetic gene expression systems. (A) Heat maps of (top) basal and (bottom) maximum reporter expression levels for synTFs featuring different ligand binding and transactivation domains. Values are normalized to the maximum recorded fluorescence. Data were obtained by flow cytometry following induction for 16 h (see Methods) (B) Fold-change reporter induction for the five top-performing hormone-inducible synTF systems. Bars represent mean ± SEM for N = 4 biological replicates. (C) Dose–response curves and (insets) flow cytometry histograms for the five top-performing inducible systems. Data were obtained following induction for 16 h (see Methods). Points represent mean ± SEM for N = 4 biological replicates.
To benchmark our systems, we compared them against the workhorse yeast inducible promoter pGAL1. Although pGAL1 is celebrated for its strong transcriptional output and low basal activity, it is induced by shifting cells from glucose- to galactose-based growth, a nutritional change that alters the expression of hundreds of native genes for galactose utilization, resulting in physiological changes in the cell.44 Furthermore, on a practical level, pGAL1-based induction can be slow, taking several hours before a gene of interest is fully induced. We compared our ER-based system (pER promoter) to pGAL1 in a time-course experiment and observed that estradiol induction was as strong as galactose induction, has lower basal expression, is more uniform, and has faster induction kinetics regardless of the carbon source (Figure S2). The rapid onset likely stems from estradiol’s ability to passively diffuse into yeast, whereas galactose relies on metabolically controlled transporters, such as GAL2, to enter the cell.45 This highlights the benefits of transitioning away from legacy inducible systems controlled by endogenous metabolites toward synthetic systems regulated by non-native small-molecule inducers.
Given the modularity of our promoter design, additional TF binding sites can be incorporated into the promoter sequences to create hybrid promoters capable of responding to two inducers either separately or in combination. We demonstrated this by adding GAL4 binding sites to the aldosterone-inducible MR promoter (pMR) (Figure S3A). Without changing the aldosterone responsiveness of the promoter, the additional binding sites endowed the promoter with galactose inducibility. When induced simultaneously with aldosterone and galactose, the reporter output rose by 1.5-fold over aldosterone induction in glucose-containing media to reach a new maximum expression level of ∼600-fold over basal (Figure S3B,C). These results demonstrate how one can leverage the modularity of the system to unlock new and desired gene expression properties.
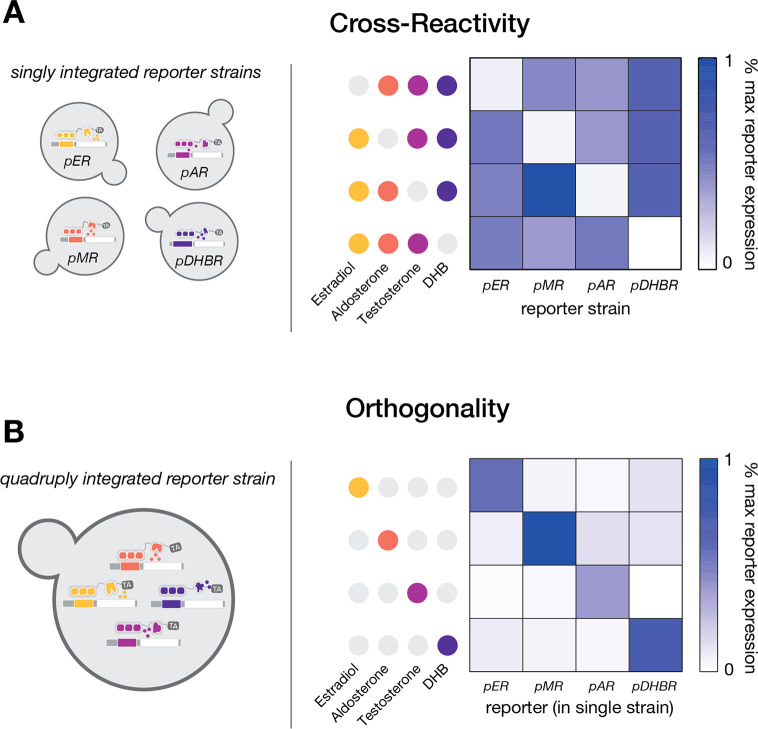
In a final set of characterization experiments, we tested the mutual orthogonality of our four top-performing inducible expression systems (ER, MR, AR, and DHBR). First, we subjected strains singly integrated with synTF–reporter constructs for each system to four induction cocktails, in each case omitting one of the four inducers (Figure 3A). As expected, each synTF drives gene expression only when its cognate inducer is present, regardless of additional hormones present in the medium, demonstrating a lack of inducer cross-reactivity. In a complementary experiment, we created a single strain encoding all four synTF systems, each reporting through a spectrally distinct fluorescent protein. We found that when induced with the respective inducers, the synTFs exclusively drove transcription of their cognate promoters, demonstrating mutual orthogonality (Figure 3B; see Methods). Additionally, we verified that none of the synTFs or inducers trigger significant cell growth defects when expressed or applied at concentrations used in this study (Figure S4). Taken together, these results establish a collection of inducible synTF systems for simultaneous, selective, and orthogonal expression control of up to four genes in yeast.
Figure 3.
Hormone-inducible synthetic gene expression systems exhibit minimal cross-reactivity and mutual orthogonality. (A) Reporter expression for strains harboring each of the inducible systems (ER, MR, AR, and DHBR) following induction for 16 h with hormone cocktails featuring three out of the four inducers (N = 4 biological replicates per condition). The heat map represents reporter expression averages collected from three independent experiments, and the reporter outputs for each given condition were not significantly different between experiments (p > 0.05). (B) Reporter expression for a single strain quadruply integrated with all four inducible systems (ER, MR, AR, and DHBR) following induction for 16 h with the indicated hormone inducers (N = 4 biological replicates per condition). The heat map represents reporter expression averages collected from three independent experiments, and the reporter outputs for each given condition were not significantly different between experiments (p > 0.05).
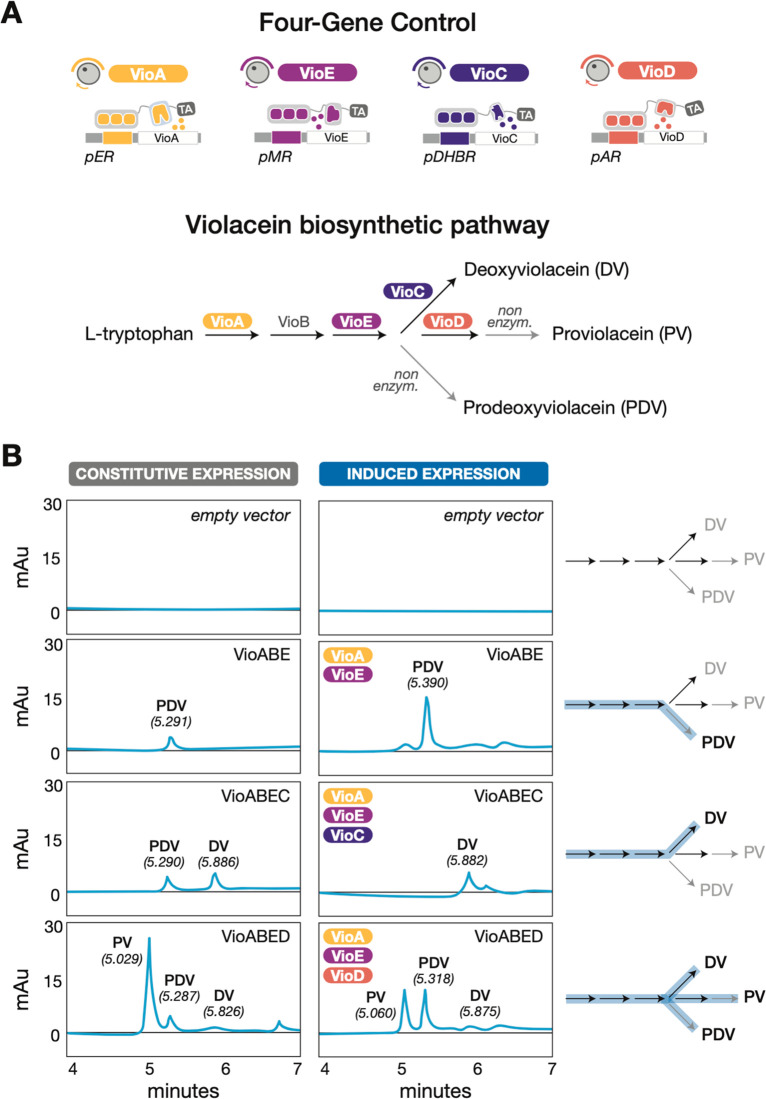
Our toolkit should be of immediate use in metabolic engineering studies that explore the full range of intermediate and end-point products generated from linear or branched pathways. Traditionally, researchers have studied and perturbed metabolic flux by building combinatorial strain libraries in which each enzyme of interest is expressed by a panel of constitutive promoters. Instead, we can now create a single strain with multiplexed, inducible control of pathway enzymes. In effect, our synTFs can act as levers that precisely shunt flux down various metabolic branches. To demonstrate this concept, we turned to the model violacein biosynthetic pathway, which is highly branched and whose outputs are governed by five enzymes: VioA, VioB, VioC, VioD, and VioE (Figure 4A).46 We predicted that by inducing subsets of these enzymes, we could selectively route flux along different pathway branches to yield distinct products that would be detectable via high-performance liquid chromatography (HPLC).47 To do this, we placed expression of VioA, VioB, VioC, VioD, and VioE under estradiol, constitutive, DHB, aldosterone, and testosterone control, respectively (Figure 4A). We then induced different enzyme combinations by subjecting the strain to inducer cocktails and measured the resulting metabolite production via HPLC (Figure 4B). In the absence of induction, we observed no metabolite production. By inducing the VioABE combination, we were able to route flux to produce prodeoxyviolacein (PDV). By adding DHB to the inducer cocktail, we rerouted flux away from PDV toward deoxyviolacein (DV). Finally, after adding aldosterone to allow expression of the full VioABECD set, we observed the production of PDV, DV, and proviolacein (PV). These results demonstrate that our toolkit can be used to control multiple genes in a single pathway in a user-defined manner to produce distinct outputs in a single strain. In future work, exercising the promoter titratability demonstrated here would allow a user to optimize flux through cascading metabolic pathways of interest.
Figure 4.
Engineering hormone-inducible, multigene control over the violacein biosynthetic pathway to selectively route metabolic flux. (A) Schematic of the violacein pathway, wherein four enzymes (VioA, VioE, VioC, VioD) are placed under orthogonal, hormone-inducible gene expression control in a single strain. (B) HPLC chromatograms for strains in which designated enzyme combinations are either expressed constitutively (left) or induced with the corresponding hormone inducer cocktails (right), leading to the selective production of compounds in the violacein pathway.
In this work, we developed and demonstrated a set of mutually orthogonal, hormone-inducible synTFs and responsive promoters that enable multigene control under common nutritionally favorable conditions in the yeast S. cerevisiae. The toolkit is inspired by and leverages regulatory mechanisms from type I NRs, a subtype of the nuclear receptor superfamily of transcriptional regulators that are specific to metazoans. Our synTF systems feature a modular design consisting of artificial DBDs, hormone-responsive LBDs from type I NRs, and TADs. Using fluorescent reporters, we characterized the maximum transcriptional output and dose-dependent behavior of our systems. We further demonstrated that our inducible systems all commonly function in cells grown in glucose media and provided evidence that they can drive more rapid and complete induction than the widely used pGAL1 system in galactose media. Critically, these hormone-inducible synTF systems are mutually orthogonal and multiplexable, which we leveraged in a proof-of-principle experiment to selectively route metabolic flux through distinct branches of the violacein biosynthetic pathway. To our knowledge, this is one of the largest collections of genes placed under simultaneous orthogonal inducer control in S. cerevisiae.
While our toolkit undoubtedly provides new capabilities for gene expression control in yeast and demonstrates comparable and in many cases superior performance relative to existing systems, it is also not without some drawbacks. For example, the solubility limit of dexamethasone in yeast media may limit that system’s maximum achievable induction. Additionally, while the AR- and MR-based systems do not cross-activate one another and therefore can be used simultaneously, testosterone application appears to have an antagonistic effect on the MR-based system (Figure S5). Finally, for U.S. Drug Enforcement Agency compliance purposes, we used testosterone stocks dissolved in acetonitrile, which becomes toxic to cells at high concentrations and limits the maximum practical induction of the AR-based system. Despite these drawbacks, our system provides a flexible, easy-to-use set of tools that will provide researchers additional levers of control to build increasingly complex genetic programs in yeast with potential applications to synthetic circuit design, gene perturbation studies, dynamic control in metabolic engineering, and yeast-surface display.48−50 Important future work will domesticate additional components into the platform, such as DBDs beyond ZFs and additional type I NR LBDs, and leverage directed evolution techniques to improve synTF activity while maintaining specificity, expanding the toolkit’s ability to control large genetic networks.
Methods
Strains and Growth Media
The S. cerevisiae strain used in all experiments was W303 (MATa, leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15).
All of the hormone experiments were performed in synthetic media with 2% w/v Dextrose (Chem-Impex), 0.67% w/v Yeast Nitrogen Base without Amino Acids (Sunrise Science Products), 0.2% w/v Dropout Mix without Yeast Nitrogen Base Minus Appropriate Amino Acids (Sunrise Science Products). Hormone inductions were performed with β-estradiol (Sigma), Aldosterone (Sigma), Testosterone (Sigma), DHB (Ambeed), and Dexamethasone (Sigma).
Galactose inductions were performed in synthetic media with 2% w/v galactose (Chem-Impex), 0.67% w/v Yeast Nitrogen Base without Amino Acids (Sunrise Science Products), and 0.2% w/v Dropout Mix without Yeast Nitrogen Base Minus Appropriate Amino Acids (Sunrise Science Products).
2XYPAD was used to prepare cells for integration and recovery after heat shock: 2% w/v yeast extract (Fisher Scientific), 4% w/v Bacto peptone, and 4% w/v dextrose.
All of the cloning experiments were performed with TG1 chemically competent E. coli. Transformed cells were plated on Lysogeny Broth (LB) plates containing the appropriate antibiotic (chloramphenicol, carbenicillin, or kanamycin).
Yeast Transformations
Yeast colonies were grown to saturation overnight in YPD and then diluted to OD600 = 0.1 in 10 mL of fresh medium. Cultures were grown for approximately 6 h to OD600 = 0.6–0.8. Cultures were pelleted and washed twice with sterile DI water. Washed cultures were resuspended in 1 mL of sterile DI water and then divided into 100 μL aliquots and spun down. Pellets were resuspended with 34 μL of DNA digestion mixture, 36 μL of 1 M lithium acetate (Sigma), 50 μL of salmon sperm DNA (Invitrogen), and 240 μL of 50% w/v PEG 3350 (Fisher). The transformation mixture was then incubated at 42 °C for 35 min. When selecting for prototrophy, the transformation mixture was spun down, resuspended in synthetic medium with appropriate amino acid dropouts, and plated directly on solid agar plates. When selecting for drug resistance, the transformation mixture was spun down, resuspended in YPD, rescued at 30 °C for 1 h with shaking, spun down, resuspended with 100 μL of YPD, and plated directly onto solid agar plates.
All of the plasmids were designed for chromosomal integration (i.e., containing 5′ and 3′ genomic homology regions and lacking a yeast origin of replication). For integration, they were linearized with NotI for 1 h prior to transformation to stimulate homologous recombination. A 34 μL aliquot of this reaction mixture, without DNA cleanup, was used in the transformations.
Plasmid Assembly Protocol
All of the plasmids used in this study were assembled with the Golden Gate assembly protocol developed for the Yeast Toolkit.41
Yeast Induction Protocol
In all of the hormone induction experiments, yeast colonies were picked from solid agar plates, placed into individual wells in a 96-deepwell plate containing 500 mL of the appropriate synthetic selective media, and allowed to grow to saturation overnight. Cultures were then diluted 1:100 into 500 mL of nonselective synthetic medium and allowed to grow for 8 h. While the yeast cultures grew, induction media were prepared by diluting concentrated inducers into nonselective synthetic medium. The outgrowth cultures were then diluted 1:100 into 500 mL of induction media and allowed to grow for 16 h.
Promoter Characterization
Following overnight induction, yeast cultures were fixed in PBS + cycloheximide for 1 h and then run on an Attune Nxt flow cytometer (Invitrogen). All of the samples were run at a flow rate of 200 μL/min. For promoter characterization, mKate2 was read on YL2 at 460 V. Expression fold-change values were determined by calculating the geometric means of the fluorescence of induced and uninduced cultures and dividing the induced mean by the uninduced mean.
Orthogonality Characterization
For orthogonality characterization, we placed mTagBFP under pMR control, mNeonGreen under pAR control, mRuby under pER control, and TagRFP657 under pDHBR control. Following overnight induction with inducer concentrations held at 100 nM estradiol, 10 μM aldosterone, 20 μM testosterone, and 10 μM DHB, yeast cultures were fixed in PBS + cycloheximide for 1 h and then run on an Attune Nxt flow cytometer (Invitrogen). All of the samples were run at a flow rate of 200 μL/min. mTagBFP was read on VL1 at 420 V, mNeonGreen on BL1 at 380 V, mRuby2 on YL1 at 420 V, and TagRFP657 on RL1 at 640 V. Raw fluorescence values were normalized to the background fluorescence of cells not expressing any fluorescent protein. Expression fold-change values were determined by calculating the geometric means of fluorescence of normalized induced and uninduced cultures and dividing the induced mean by the uninduced mean.
Growth Rate Measurement
All of the growth rate measurements were performed using the eVOLVER continuous culture platform.51 Glycerol stocks were used to inoculate synthetic selective medium overnight cultures of the chassis strain or singly integrated mKate2 reporter strains. Overnight cultures were then used to inoculate eVOLVER vials containing appropriate induction media. The inducer concentrations were held at 100 nM estradiol, 10 μM aldosterone, 20 μM testosterone, 1 mM dexamethasone, and 10 μM DHB. Cultures were allowed to grow to steady state before growth rate measurements were begun. The eVOLVER calculates the growth rate by tracking culture OD changes to find the culture doubling time.
Violacein Flux Routing
Yeast colonies were picked from solid agar plates, placed into individual wells in a 96-deepwell plate containing 500 mL of appropriate synthetic selective media, and allowed to grow to saturation overnight. Cultures were then diluted 1:100 into 2 mL of nonselective synthetic medium and allowed to induce for 48 h with inducer concentrations held at 100 nM estradiol, 10 μM aldosterone, 20 μM testosterone, and 10 μM DHB, as appropriate to the pathway being induced. Following growth, cultures were spun down and resuspended in 500 μL of methanol. The resuspended cultures were boiled at 95 °C for 15 min with a vortex halfway through the boil. The boiled cultures were then spun down, had the supernatant removed, spun down, and packaged into HPLC vials for analysis.
HPLC analysis took place on an HP 1090 chromatograph with fluorescence detection (Agilent) and a Rapid Resolution SB-C18 column (30 mm × 2.1 mm, 3.5 μm particle size; Agilent). The column temperature was held at 30 °C. Mobile phase A was 0.1% formic acid in water (Fisher), and mobile phase B was 0.1% formic acid in acetonitrile (Fisher). The HPLC method was started at 5% B, held for 1.5 min, ramped at 16.9% min–1 to 98% B, held for 2 min, ramped at 3.1% s–1 to 5% B, and held for 2.5 min. The method maintained a flow rate of 0.5 mL/min and injected 20 μL of sample. ChemStation (Agilent) was used to run the HPLC and analyze the chromatograms.
Acknowledgments
This work was supported by the Department of Defense (Vannevar Bush Faculty Fellowship N00014-20-1-2825), the National Science Foundation (Grants CCF-2027045 and EF-1921677), and the National Institutes of Health (NIH) (Grants R01EB029483 and R01EB027793). We thank the Boston University Chemical Instrumentation Center for assistance with HPLC method development and measurements. We are grateful to Arjun Ravikumar and Nikit Patel for helpful discussions and feedback on the manuscript and to Brandon Wong for helpful consultations with figure preparation. A.S. was supported by an NIH-funded predoctoral training fellowship (T32GM130546).
Data Availability Statement
The toolkit of hormone-inducible gene expression systems described in this paper are available from Addgene (http://www.addgene.org/).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00423.
Supporting tables that list sequencing primer sequences and the plasmids available from this toolkit, sequence files in GenBank format for the plasmids included in this toolkit, and supporting figures that provide additional data from experiments described in the text (PDF)
Author Contributions
S.K. and A.S.K. conceived the project. A.S. and S.K. designed and generated all constructs. A.S. performed all experiments and data analysis. A.S. and A.S.K. drafted the manuscript with input from S.K.
The authors declare the following competing financial interest(s): A.S.K. is a scientific advisor for and holds equity in Senti Biosciences and Chroma Medicine and is a cofounder of Fynch Biosciences and K2 Biotechnologies.
Supplementary Material
References
- Hickman M. J.; Petti A. A.; Ho-Shing O.; et al. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol. Biol. Cell 2011, 22, 4192–4204. 10.1091/mbc.e11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R.; Huang D.; Preston N.; et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 2006, 21, 319–330. 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Douglas A. C.; Smith A. M.; Sharifpoor S.; et al. Functional analysis with a barcoder yeast gene overexpression system. G3: Genes, Genomes, Genet. 2012, 2, 1279–1289. 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita Y.; Kim G.; Li Z. A genome-scale yeast library with inducible expression of individual genes. Mol. Syst. Biol. 2021, 17, e10207. 10.15252/msb.202110207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. D.; O’Shea E. K. A quantitative model of transcription factor-activated gene expression. Nat. Struct. Mol. Biol. 2008, 15, 1192–1198. 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake W. J.; Kærn M.; Cantor C. R.; Collins J. J. Noise in eukaryotic gene expression. Nature 2003, 422, 633–637. 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Stewart-Ornstein J.Dimensionality and the Stress/Growth Axis of Gene Expression in S. cerevisiae. Ph.D. Dissertation, University of California, San Francisco, 2012. [Google Scholar]
- To T.-L.; Maheshri N. Noise Can Induce Bimodality in Positive Transcriptional Feedback Loops without Bistability. Science 2010, 327, 1142–1145. 10.1126/science.1178962. [DOI] [PubMed] [Google Scholar]
- Lee T. H.; Maheshri N. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol. Syst. Biol. 2012, 8, 576. 10.1038/msb.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro T. F.; Lindquist S. Yeast Cells Provide Insight into Alpha-Synuclein Biology and Pathobiology. Science 2003, 302, 1772–1775. 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham S.; Outeiro T. F.; DeVit M. J.; Lindquist S. L.; Muchowski P. J. Yeast Genes that Enhance the Toxicity of a Mutant Huntingtin Fragment or α-Synuclein. Science 2003, 302, 1769–1772. 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- Johnson B. S.; McCaffery J. M.; Lindquist S.; Gitler A. D. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 6439–6444. 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff D. F.; Lindquist S. Phenotypic screens for compounds that target the cellular pathologies underlying Parkinson’s disease. Drug Discovery Today: Technol. 2013, 10, e121. 10.1016/j.ddtec.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B.; Neuert G.; Van Oudenaarden A. Using gene expression noise to understand gene regulation. Science 2012, 336, 183–187. 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasi E. A.; Eisen S. L.; Wang H.; et al. Rapid Deorphanization of Human Olfactory Receptors in Yeast. Biochemistry 2019, 58, 2160–2166. 10.1021/acs.biochem.8b01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E. M.; Lalwani M. A.; Lovelett R. J.; et al. Design and Characterization of Rapid Optogenetic Circuits for Dynamic Control in Yeast Metabolic Engineering. ACS Synth. Biol. 2020, 9, 3254–3266. 10.1021/acssynbio.0c00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endalur Gopinarayanan V.; Nair N. U. A semi-synthetic regulon enables rapid growth of yeast on xylose. Nat. Commun. 2018, 9, 1233. 10.1038/s41467-018-03645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam F. H.; Turanli-Yildiz B.; Liu D.; et al. Engineered yeast tolerance enables efficient production from toxified lignocellulosic feedstocks. Sci. Adv. 2021, 7, eabf7613. 10.1126/sciadv.abf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H.; Fischer C.; Nevoigt E.; et al. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 12678–12683. 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby C. M.; Li R. A.; Vu V. T.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965. 10.1038/s41467-018-03293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R. S.; Oakes B. L.; Wang X.; et al. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013, 41, e57. 10.1093/nar/gks1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R. S.; Gibney P. A.; Chandran S. S.; Benjamin K. R.; Botstein D. Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 2014, 42, e48. 10.1093/nar/gkt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R. S.; Silverman S. J.; McClean M. N.; et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 2011, 22, 4447–4459. 10.1091/mbc.e11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasi E. A.; Kruyer N. S.; Peralta-Yahya P. Advances in G protein-coupled receptor high-throughput screening. Curr. Opin. Biotechnol. 2020, 64, 210–217. 10.1016/j.copbio.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. J.; Segall-Shapiro T. H.; Glassey E.; Zhang J.; Voigt C. A. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 2019, 15, 196–204. 10.1038/s41589-018-0168-3. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Zhang Y.; Nielsen J. Synthetic Biology of Yeast. Biochemistry 2019, 58, 1511–1520. 10.1021/acs.biochem.8b01236. [DOI] [PubMed] [Google Scholar]
- Bashor C. J.; Horwitz A. A.; Peisajovich S. G.; Lim W. A. Rewiring cells: Synthetic biology as a tool to interrogate the organizational principles of living systems. Annu. Rev. Biophys. 2010, 39, 515–537. 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solow S. P.; Sengbusch J.; Laird M. W. Heterologous protein production from the inducible MET25 promoter in Saccharomyces cerevisiae. Biotechnol. Prog. 2005, 21, 617–620. 10.1021/bp049916q. [DOI] [PubMed] [Google Scholar]
- Etcheverry T. Induced Expression Using Yeast Copper Metallothionein Promoter. Methods Enzymol. 1990, 185, 319–329. 10.1016/0076-6879(90)85028-M. [DOI] [PubMed] [Google Scholar]
- Lohr D.; Venkov P.; Zlatanova J.; et al. Transcriptional Regulation in the Yeast GAL Gene Family: A Complex Genetic Network. FASEB J. 1995, 9, 777–787. 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- Cuperus J. T.; Lo R. S.; Shumaker L.; Proctor J.; Fields S. A tetO Toolkit to Alter Expression of Genes in Saccharomyces cerevisiae. ACS Synth. Biol. 2015, 4, 842–852. 10.1021/sb500363y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T.; Wang X.; Collins J. J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009, 27, 465–471. 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander S. P. H.; Cidlowski J. A.; Kelly E.; et al. The Concise Guide To Pharmacology 2021/22: Nuclear hormone receptors. Br. J. Pharmacol. 2021, 178, S246–S263. 10.1111/bph.15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W. B. The hsp90-based chaperone system: Involvement in signal transduction from a variety of hormone and growth factor receptors. Proc. Soc. Exp. Biol. Med. 1998, 217, 420–434. 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- Pratt W. B.; Toft D. O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997, 18, 306–360. 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Smith D. F.; Toft D. O. The intersection of steroid receptors with molecular chaperones: Observations and questions. Mol. Endocrinol. 2008, 22, 2229–2240. 10.1210/me.2008-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss A. E.; Benzeno S.; Rao J.; Caplan A. J. Control of estrogen receptor ligand binding by Hsp90. J. Steroid Biochem. Mol. Biol. 2000, 72, 223–230. 10.1016/S0960-0760(00)00037-6. [DOI] [PubMed] [Google Scholar]
- Louvion J. F.; Havaux-Copf B.; Picard D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 1993, 131, 129–134. 10.1016/0378-1119(93)90681-R. [DOI] [PubMed] [Google Scholar]
- Aranda-Díaz A.; Mace K.; Zuleta I.; Harrigan P.; El-Samad H. Robust Synthetic Circuits for Two-Dimensional Control of Gene Expression in Yeast. ACS Synth. Biol. 2017, 6, 545–554. 10.1021/acssynbio.6b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. S.; Lu T. K.; Bashor C. J.; et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell 2012, 150, 647–658. 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. E.; DeLoache W. C.; Cervantes B.; Dueber J. E. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 2015, 4, 975–986. 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- Mazumder M.; McMillen D. R. Design and characterization of a dual-mode promoter with activation and repression capability for tuning gene expression in yeast. Nucleic Acids Res. 2014, 42, 9514–9522. 10.1093/nar/gku651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chockalingam K.; Chen Z.; Katzenellenbogen J. A.; Zhao H. Directed evolution of specific receptor–ligand pairs for use in the creation of gene switches. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 5691–5696. 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Chong R.; Savir Y.; Carroll S. M.; et al. Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 1636–1641. 10.1073/pnas.1418058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J. F.; Emr S. D.; Field C.; Schekman R. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J. Bacteriol. 1986, 166, 313–318. 10.1128/jb.166.1.313-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. E.; Aswani A.; Han A. S.; Tomlin C. J.; Dueber J. E. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013, 41, 10668–10678. 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan J. G.; Lee M. E.; Almeida R.; et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015, 160, 339–350. 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor C. J.; Patel N.; Choubey S.; et al. Complex Signal Processing in Synthetic Gene Circuits Using Cooperative Regulatory Assemblies. Science 2019, 364, 593–597. 10.1126/science.aau8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby G. A.; Kiriakov S.; Hallacli E.; et al. A Genetic Tool to Track Protein Aggregates and Control Prion Inheritance. Cell 2017, 171, 966–979. 10.1016/j.cell.2017.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartline C. J.; Schmitz A. C.; Han Y.; Zhang F. Dynamic control in metabolic engineering: Theories, tools, and applications. Metab. Eng. 2021, 63, 126–140. 10.1016/j.ymben.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong B. G.; Mancuso C. P.; Kiriakov S.; Bashor C. J.; Khalil A. S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018, 36, 614–623. 10.1038/nbt.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The toolkit of hormone-inducible gene expression systems described in this paper are available from Addgene (http://www.addgene.org/).