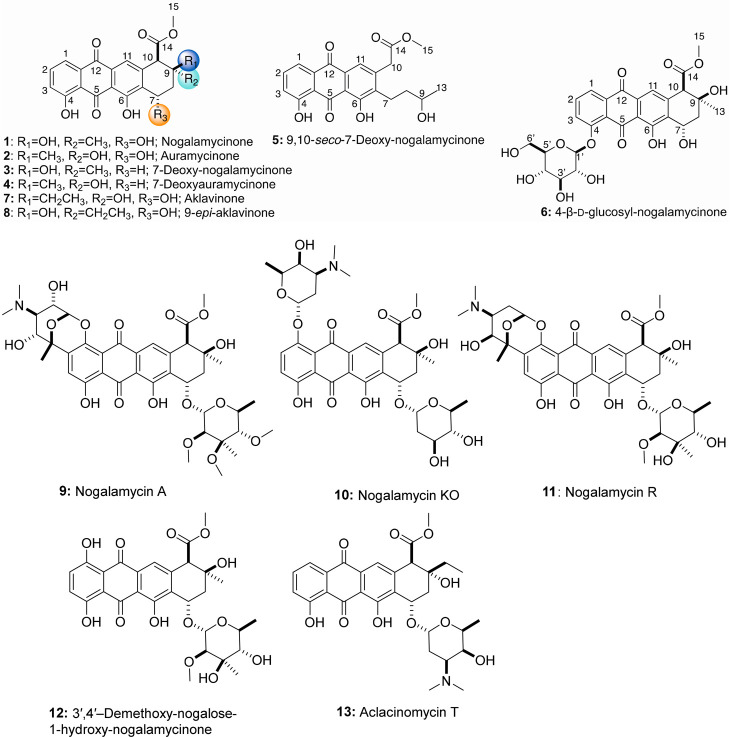
Abstract

Actinomycetes produce a variety of clinically indispensable molecules, such as antineoplastic anthracyclines. However, the actinomycetes are hindered in their further development as genetically engineered hosts for the synthesis of new anthracycline analogues due to their slow growth kinetics associated with their mycelial life cycle and the lack of a comprehensive genetic toolbox for combinatorial biosynthesis. In this report, we tackled both issues via the development of the BIOPOLYMER (BIOBricks POLYketide Metabolic EngineeRing) toolbox: a comprehensive synthetic biology toolbox consisting of engineered strains, promoters, vectors, and biosynthetic genes for the synthesis of anthracyclinones. An improved derivative of the production host Streptomyces coelicolor M1152 was created by deleting the matAB gene cluster that specifies extracellular poly-β-1,6-N-acetylglucosamine (PNAG). This resulted in a loss of mycelial aggregation, with improved biomass accumulation and anthracyclinone production. We then leveraged BIOPOLYMER to engineer four distinct anthracyclinone pathways, identifying optimal combinations of promoters, genes, and vectors to produce aklavinone, 9-epi-aklavinone, auramycinone, and nogalamycinone at titers between 15–20 mg/L. Optimization of nogalamycinone production strains resulted in titers of 103 mg/L. We structurally characterized six anthracyclinone products from fermentations, including new compounds 9,10-seco-7-deoxy-nogalamycinone and 4-O-β-d-glucosyl-nogalamycinone. Lastly, we tested the antiproliferative activity of the anthracyclinones in a mammalian cancer cell viability assay, in which nogalamycinone, auramycinone, and aklavinone exhibited moderate cytotoxicity against several cancer cell lines. We envision that BIOPOLYMER will serve as a foundational platform technology for the synthesis of designer anthracycline analogues.
Keywords: BioBricks, synthetic biology, natural product biosynthesis, anthracyclinones, Streptomyces coelicolor, anticancer
Introduction
Anthracyclines are clinically important natural products for the treatment of human cancers.1 Anthracyclines influence DNA and chromatin structure via two distinct mechanisms: histone eviction from open chromosomal regions and the inhibition of topoisomerase II, resulting in the formation of double-stranded DNA breaks.2 Recently, several anthracycline biosynthetic pathways have been characterized, which provide genetic tools for the metabolic engineering of new anthracycline derivatives. The nogalamycin, aclacinomycin, and daunorubicin polyketide pathways are canonical examples of anthracycline biosynthesis and have been studied in detail over the past few decades.3−7 For example, anthracyclinones are C20 (e.g., nogalamycin) or C21 (e.g., daunorubicin or aclacinomycin) aromatic polyketides that are synthesized by minimal polyketide synthase (minPKS) composed of a ketoacyl synthase (KSa, AknB/DpsA/Snoa1), chain length factor (KSb, AknC/DpsB/Snoa2), and an acyl carrier protein (ACP, AknD/DpsG/Snoa3) to form an enzyme-linked poly-β-ketothioester intermediate (Figure 1). The AknBCDE2F or DpsABCDG minPKS complex condenses one propionyl-CoA starter unit to nine malonyl-CoA extender units to form a C-21 intermediate,8 whereas the Snoa123 minPKS condenses one acetyl-CoA and nine malonyl-CoA extender units to form a C-20 polyketide. The poly-β-ketothioester is reduced at 9-position by a PKS-associated ketoreductase (KR, AknA/DpsE/SnoaD), and then is aromatized via C7–C12 cyclization (ARO, AknE/DpsF/SnoaE), C5–C14 and C3–C16 cyclization via second-third ring cyclase (CYC, AknW/DpsY/SnoaM),9 and C-12 oxidation via an anthraquinol oxygenase (OXY, AknX/DpsG/SnoaB)10 to afford a tricyclic anthraquinone intermediate (Figure 1). The tricyclic anthracyclinone is O-methylated to form nogalonic acid methyl ester (NAME, SnoaC)11 or aklanonic acid methyl ester (AAME, AknG/DnrC),12 respectively, followed by fourth-ring cyclization to a 9(S)-configured anthracyclinone (Kyc34/SnoaL) and subsequent C7-ketoreduction (SnoaF) to afford nogalamycinone or 9-epi-aklavinone13,14 (Figure 1). Alternatively, the tricyclic intermediate can undergo fourth-ring cyclization to a 9(R)-configured anthracyclinone (AknH/DnrD) and 7-ketoreduction (AknU/DnrE) to yield aklavinone or auramycinone (Figure 1).15 Inspired by the biosynthetic logic of the anthracycline PKS system, the motivation for the present work was to develop an expanded metabolic engineering toolset to improve access to these important natural products and to generate new anthracycline analogues via microbial synthesis.
Figure 1.
Biosynthesis of anthracyclinones. One acetyl-CoA or propionyl-CoA starter unit is condensed with nine malonyl-CoA extender units via iterative Claisen condensation reactions via the minimal PKS to form an enzyme-tethered decaketide intermediate. The polyketide ketoreductase reduces the 9-ketone to a hydroxyl group followed by C7–C12 first-ring aromatization, C5–C14 s-ring cyclization, and C3–C16 third-ring cyclization by the second-third ring cyclase. C-12 oxidation is catalyzed by an anthraquinol monooxygenase, followed by O-methylation to afford a tricyclic anthraquinone methyl ester. The key tricyclic intermediate undergoes fourth-ring cyclization via two distinct routes: SnoaL/Kyc34 cyclizes the fourth ring to afford a 9(S)-anthracyclinone and AknH/DnrD cyclizes the fourth ring to afford a 9(R)-anthracyclinone. The final step is C-7 ketoreduction by ketoreductases to afford C-21 anthracyclinones 9-epi-aklavinone or aklavinone and C-20 anthracyclinones nogalamycinone and auramycinone.
Synthetic biology is a discipline that has been defined as the “engineering-driven building of increasingly complex biological entities for novel applications”.16 Indeed, the field of synthetic biology has resulted in new genetic systems and organisms useful for filling in critical gaps in the pharmaceutical, biofuels, and cosmetics industries.17 Synthetic biology boasts the development of a variety of genetic tools, including chassis hosts, promoters, terminators, genes, and vectors useful for reprogramming model organisms. Synthetic biology is also a promising discipline for increasing access to indispensable natural products, such as polyketides.18 Synthetic biology has also contributed new tools for the refinement of idiosyncratic model organisms, such as Streptomyces spp., that are difficult to transform or feature morphological limitations, such as the formation of mycelial clumps during submerged liquid fermentation.19,20Streptomyces produce valuable bioactive molecules, such as anthracyclines and tetracyclines. Streptomyces spp. exhibit a highly complex life cycle, including sporulation, branching, fragmentation, and adhesion that is regulated and correlated with specialized metabolism, which limits their use as industrial hosts. Recently, the matAB gene cluster was discovered to encode novel glycosyltransferases responsible for the synthesis of extracellular poly-β-1,6-N-acetylglucosamine (PNAG) that leads to cellular clumping.21 Van Dissel et al. inactivated the matAB gene cluster in Streptomyces coelicolor M145 and characterized strains that lost the mycelial aggregation phenotype and exhibited improved biomass accumulation. Here, we developed the derivative strain Streptomyces coelicolor M1152ΔmatAB as an improved cell factory for the metabolic engineering of minimal aromatic polyketides from type II PKS pathways.
In this report, we developed a BioBricks toolkit of promoters, expression vectors, and engineered Streptomyces hosts for the metabolic engineering of anthracyclinones from type II PKS pathways. All genes were assembled according to the BioBricks [RFC10] standard. We designed minimal PKS (minPKS) cassettes based on the snoa123 (C-20 nogalamycin), aknBCDE2F (C-21 aclacinomycin), dpsABGCD (C-21 doxorubicin), and oxyABCD (C-19N oxytetracycline) biosynthetic pathways. First, we compared the production titer resulting from expressing minPKS gene cassettes in different Streptomyces hosts: Streptomyces lividans K4–114, Streptomyces coelicolor M1146, M1152, and M1154, and Streptomyces coelicolor M1152ΔmatAB. These experiments resulted in the production of the expected aromatic minimal polyketides. Second, we performed combinatorial biosynthesis experiments by coexpressing the minPKS gene cassettes with different combinations of KR/ARO/CYC/OXY genes to optimize metabolic flux toward tricyclic anthracyclinones. Third, we coexpressed different orthologs of O-methyltransferases, fourth ring cyclases, and 7-ketoreductases to generate the expected anthracyclinones. Finally, the optimum gene combinations were cloned onto one plasmid to determine the production yields in S. coelicolor M1152ΔmatAB. This work resulted in the production of eight anthracyclinone analogues, including the new compounds 9,10-seco-7-deoxy-nogalamycinone and 4-β-d-glucosyl-nogalamycinone and the unexpected 7-deoxy analogues (Figure 2). Finally, the anticancer activity of the different anthracyclinone derivatives was assessed in a mammalian cell viability assay, which revealed that nogalamycinone, auramycinone, and aklavinone had moderate antiproliferative activity (<30 μM IC50) against several human cancer cell lines.
Figure 2.
Structures of anthracyclin(on)es described in this work.
Results and Discussion
Engineering of the snoa123 MinPKS into Different Streptomyces spp. Hosts
We first synthesized a codon-optimized version of the Streptomyces nogalater minimal polyketide synthase (snoa123) responsible for the synthesis of the nogalamycin C-20 poly-β-ketothioester (Figure 1). The codon optimization was based on the native codon preference for S. coelicolor (Genscript, OptimumGene). Previously, the involvement of snoa123 in the synthesis of nogalamycin has been confirmed via heterologous expression studies and genetic complementation studies in Streptomyces spp. strains.6,22 The snoa123 operon was designed based on principles from the BioBricks [RFC-10] standard: (1) the genes were decoupled from their native translational coupling and synthesized as individual ORFs; (2) the strong BBa_B0034 ribosome binding site was incorporated into the 5′-untranslated region (5′-AAAGAGGAGAAA-3′) to control the rate of translation initiation for each gene in the operon;23,24 (3) the genes were cloned to lack internal EcoRI, PstI, SpeI, and XbaI restriction sites, and if necessary, silent mutations were engineered in these restriction sites to make the genes compatible with the BioBricks [RFC-10] standard; (4) the genes were given BioBricks prefix (5′-GAATTCGCGGCCGCTTCTAGAG-3′) and suffix (5′-TACTAGTAGCGGCCGCTGCAG-3′) sequences to enable isocaudomer cloning25 (Table 1). To facilitate gene expression, we generated a BioBricks compatible version of the ϕC31-based integrating expression vector pSET15226 (e.g., pSET152BB) to allow for cloning into EcoRI/PstI sites and transformation into S. coelicolor via intergeneric conjugation (Table 1).27 The genes were expressed from the strong constitutive promoter kasOp*.28,29 The resulting pSET152BB derivative featured the snoa123 fused to the kasOp* promoter and was transformed into three heterologous hosts: (1) S. lividans K4–114 (lacking the actinorhodin type II polyketide synthase gene cluster);30 (2) S. coelicolor M1146 (lacking the prodiginine, actinorhodin, coelimycin, and cryptic polyketide gene clusters);31 and (3) S. coelicolor M1152 (an RNA polymerase B up-regulated mutant of S. coelicolor M1146) (Table 2).
Table 1. Plasmids Constructed and/or Used in This Study.
| plasmid | genotype and relevant characteristics | reference |
|---|---|---|
| pSET152 | aac3(IV), oriT, lacZα, ΦC31int, attP, MCS | (26) |
| pSET152BB | BioBricks-compatible vector pSET152; aac3(IV)R, oriT, ϕC31int, attP, MCS | (24) |
| pENSV1 | BioBricks-compatible vector; aadAR, blaR, oriT, ϕSV1int, attP | (24) |
| pENTG1 | BioBricks-compatible vector; vphR, blaR, oriT, ϕTG1int, attP | (24) |
| pOSV808 | BioBricks-compatible vector; hphR, oriT, VWBint, attP, amilCFP | (32) |
| pSnogaori | pKC505-based E. coli–Streptomyces shuttle vector harboring 30.26 kb fragment of nogalamycin biosynthetic gene cluster; aac(3)IV, SCP2* ori, pBR322 ori, λ cos sites | Accession No. OM832358,33 |
| pSET154BB | pSET152BB with strong 5′-tt-sbi-A terminator and 3′-fd phage terminator | This study,8,9 |
| pENSV3 | pENSV1 with strong 5′- ECK120010818-term and 3′- ECK120029600-spy-term terminators | This study,10 |
| pENTG3 | pENTG1 with strong 5′- L3S2P21-term and 3′- L3S3P41-term terminators | This study,10 |
| pSET-S1 | wild-type snoa123 fused to the medium ermE*p promoter; cloned in pSET152BB vector | This study |
| pSET-S2 | wild-type snoa123 fused to the strong kasOp* promoter; cloned in pSET152BB vector | This study |
| pSET-S3 | codon-optimized snoa123 fused to the strong kasOp* promoter; cloned in pSET152BB vector | This study |
| pSET-A1 | wild-type aknBCDE2F fused to the medium ermE*p promoter; cloned in pSET152BB vector | This study |
| pSET-A2 | wild-type aknBCDE2F fused to the strong kasOp* promoter; cloned in pSET152BB vector | This study |
| pSET-D1 | wild-type dpsABCDG fused to the medium ermE*p promoter; cloned in pSET152BB vector | This study |
| pSET-D2 | wild-type dpsABCDG fused to the medium kasOp* promoter; cloned in pSET152BB vector | This study |
| pSET-O2 | wild-type oxyABCD fused to the strong kasOp* promoter; cloned in pSET152BB | This study |
| pSET-S2S5 | kasOp*-snoa123+kasOp*-snoaDEMB cloned in pSET152BB | This study |
| pSET-D1S5 | ermE*p-dpsABCDG+kasOp*-snoaDEMB cloned in pSET152BB | This study |
| pSET-D2S5 | kasOp*-dpsABCDG+kasOp*-snoaDEMB cloned in pSET152BB | This study |
| pSET-A1S5 | ermE*p-aknBCDE2F+kasOp*-snoaDEMB cloned in pSET152BB | This study |
| pSET-A1S5 | kasOp*-aknBCDE2F+kasOp*-snoaDEMB cloned in pSET152BB | This study |
| pSET-D1D5 | ermE*p-dpsABCDG+kasOp*-dpsEFYdnrG cloned in pSET152BB | This study |
| pSET-D2D5 | kasOp*-dpsABCDG+kasOp*-dpsEFYdnrG cloned in pSET152BB | This study |
| pSET-A1D5 | ermE*p-aknBCDE2F+kasOp*-dpsEFYdnrG cloned in pSET152BB | This study |
| pSET-A2D5 | kasOp* -aknBCDE2F+kasOp*-dpsEFYdnrG cloned in pSET152BB | This study |
| pSET-D1A5 | ermE*p-dpsABCDG+kasOp*-aknAE1WX cloned in pSET152BB | This study |
| pSET-D2A5 | kasOp*-dpsABCDG+kasOp*-aknAE1WX cloned in pSET152BB | This study |
| pSET-A1A5 | ermE*p-aknBCDE2F+kasOp*-aknAE1WX cloned in pSET152BB | This study |
| pSET-A2A5 | kasOp*-aknBCDE2F+kasOp*-aknAE1WX cloned in pSET152BB | This study |
| pSV-S4 | kasOp*-snoaDEM cloned in pENSV1 | This study |
| pSV-S5 | kasOp*-snoaDEMB cloned in pENSV1 | This study |
| pSV-A4 | kasOp*-aknAE1W cloned in pENSV1 | This study |
| pSV-A5 | kasOp*-aknAE1WX cloned in pENSV1 | This study |
| pTG-S6 | sp44-snoaLCF cloned in pENTG1 | This study |
| pTG-S7 | sp44-snoaC+kyc34+snoaF cloned in pENTG1 | This study |
| pTG-A6 | sp44-aknGHU cloned in pENTG1 | This study |
| pTG-D6 | sp44-dnrCDE cloned in pENTG1 | This study |
| pEN10001 | kasOp*-aknBCDE2F+kasOp*-aknAE1WX+sp44-aknGHU cloned into pSET154BB | This study |
| pEN10002 | kasOp*-aknBCDE2F+kasOp*-aknAE1WX+sp44-snoaC+kyc34+snoaF cloned into pSET154BB | This study |
| pEN10003 | kasOp*-snoa123+kasOp*-snoaDEMB+sp44-aknGHU cloned into pSET154BB | This study |
| pEN10004 | kasOp*-snoa123+kasOp*-snoaDEMB+sp44-snoaC+kyc34+snoaF | This study |
| pRW10000 | codon-optimized kasOp*-snoa123+kasOp*-aknAE1WX+sp44-snoaLCF | This study |
Table 2. Bacterial Strains Used in This Study.
| strain | genotype or comments | source or reference |
|---|---|---|
| Escherichia coli JM109 | F′ traD36 proA+B+lacIq Δ(lacZ)M15/ Δ(lac-proAB) gln V44 e14- gyrA96 recA1 relA1 endA1 thi hsdR17 | Promega |
| Escherichia coli ET12567/(pUZ8002) | dam- dcm- hsdM hsdS hsdR cat tet; carrying plasmid pUZ8002 | (34) |
| Streptomyces lividans K4–114 | pro-2 str-6 SLP2- SLP3- act::ermE | (30) |
| Streptomyces coelicolor M1146 | SCP1- SCP2- Δact Δred Δcpk Δcda | (31) |
| Streptomyces coelicolor M1152 | SCP1- SCP2- Δact Δred Δcpk Δcda rpoB(C1298T) | (31) |
| Streptomyces coelicolor M1152ΔmatAB | SCP1- SCP2- Δact Δred Δcpk Δcda rpoB(C1298T) Δsco2961–2962 | This study,19 |
The heterologous expression of the codon-optimized snoa123 operon resulted in significant production of yellow-orange pigments on SFM agar plates. Each of the recombinant strains was plated in triplicate on R5 agar plates for 5 days and then extracted to analyze the production of polyketides (Figure 3). Each strain produced copious amounts of known polyketides SEK15 (e.g., C20H16O8, calc. m/z = 385.0923 [M + H]+; found m/z = 385.0910 [M + H]+) and SEK15b (e.g., C20H12O8, calc. m/z = 381.0610 [M + H]+; found m/z = 381.0595 [M + H]+) as determined by HRESI-MS analysis (Supporting Information Figures S11 and S12). S. lividans K4–114 also produced large quantities of undesired prodiginines, based on HRESI-MS total ion counts, and therefore this strain was excluded from further experimentation. The production titer of SEK15 was strain-dependent and media-dependent. On R5 agar plates, the strains transformed with empty pSET152BB vector produced no detectable SEK15; however, S. lividans K4–114 expressing the construct produced 39.6 mg/L SEK15, S. coelicolor M1146 produced 39.7 mg/L SEK15, and S. coelicolor M1152 produced 34.8 mg/L SEK15 (Figure 3). In SG liquid media, S. coelicolor M1146 and M1152 strains transformed with empty pSET152BB produced no SEK15, whereas S. coelicolor M1152 expressing the snoa123 operon produced 55 mg/L SEK15 and S. coelicolor M1146 produced the highest SEK15 titer at 79 mg/L (Figure 3).
Figure 3.
Metabolic engineering of SEK15. (A) Scheme for the biosynthesis of SEK15 via the Snoa123 minPKS. (B) Production titers of SEK15 on R5 solid agar plates from the heterologous expression of codon-optimized snoa123 in S. lividans K4–114, S. coelicolor M1146, and S. coelicolor M1152. (C) Production titers of SEK15 in SG liquid media shake flask experiments from the heterologous expression of codon-optimized snoa123 in S. coelicolor M1146 and S. coelicolor M1152. (D) Production titers of SEK15 in SG liquid media shake flask experiments from the heterologous expression of wild-type snoa123 in S. coelicolor M1152 and S. coelicolor M1152ΔmatAB. Experiments were carried out in double triplicate and the error bars reflect the standard deviation. ANOVA was carried out to determine the statistical significance between strains. The statistically significant comparisons are reflected with asterisks. The statistical significance of observed results was established with a p < 0.05. * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001.
Engineering of aknBCDE2F, dpsABCDG, and oxyABCD MinPKS Operons into S. coelicolor
To compare the biosynthetic capacity of S. coelicolor M1152 and the S. coelicolor M1152ΔmatAB mutant, heterologous expression experiments were conducted to produce minimally cyclized aromatic polyketides using propionyl-CoA, acetyl-CoA, or malonamyl-CoA starter units. We anticipated that the strains resulting from these experiments would serve as a proof-of-concept for the use of S. coelicolor M1152ΔmatAB as a production host for pathway engineering of anthracyclines. The original aknBCDE2F gene annotations from Streptomyces galilaeus ATCC 31615 (Accession No. AF257324.2) were revised using next-generation sequencing (Supporting Information, Table S1). Next-generation sequencing of pSnogaori, which encodes the majority of the nogalamycin biosynthetic pathway, identified several mutations in the original annotations of snoaD, snoaE, snoaM, and snoaB genes. The revised sequence was deposited in the NCBI database (Accession No. OM832358). Despite several attempts to express codon-optimized versions of the aknBCDE2F, dpsABCDG, and oxyABCD minPKS operons, attempts to produce the expected minimal aromatic polyketides were unsuccessful. These attempts also included the preservation of the native translational coupling and ribosome binding site of the KSα-KSβ subunits, which Liu et al. previously demonstrated to be indispensable for the expression of the alpABC and whiE–III–IV–V type II PKS subunits in E. coli.35 As a result, we focused further construct development on the expression of wild-type versions of the snoa123, aknBCDE2F, dpsABCDG, and oxyABCD minPKS operons.36 The expression of aknBCDE2F and dpsABCDG operons was expected to produce the C-21 polyketide UWM7 and the oxyABCD operon was expected to produce the amidated polyketide WJ85.37,38 We cloned the aknBCDE2F, snoa123, and oxyABCD genes under the control of the intermediate strength ermE*p promoter or the strong kasOp* promoter and spliced these cassettes into pSET152BB.28,39 The resulting plasmids were transformed into S. coelicolor M1152 and S. coelicolor M1152ΔmatAB via intergeneric conjugation. S. coelicolor M1152/pSET152BB and S. coelicolor M1152ΔmatAB/pSET152BB were included in the analysis as negative controls.
Expression of the wild-type snoa123 minPKS in S. coelicolor M1152 resulted in the production of 9.2 mg/L SEK15, S. coelicolor M1152ΔmatAB harboring the same construct produced 3-fold more SEK15 at 41.8 mg/L (p < 0.0001) (Figure 3). The codon-optimized version of snoa123 was more productive than the wild-type version of snoa123 (e.g., 79 mg/L), which indicates that codon-optimization likely enhances translation of the Snoa123 minPKS complex and results in greater metabolic flux toward SEK15 via mass action. We previously observed a similar result when comparing wild-type and codon-optimized versions of the valerena-1,10-diene synthase, VoTPS1, in E. coli.40 Expression of the codon-optimized version resulted in a 3-fold greater production titer of valerena-1,10-diene than the wild-type version. Additionally, this result demonstrates that the dispersed growth phenotype of S. coelicolor M1152ΔmatAB greatly enhanced polyketide production.
We are only beginning to unravel the mechanisms that control morphogenesis of streptomycetes in submerged cultures. Productivity of streptomycetes in industrial fermentation depends on the mycelial morphology in a product-dependent manner; in other words, less favorable growth conditions may have to be accepted to obtain good productivity.41−42 Considering that it is hard to predict how production responds to changes in growth and morphology, we need to have more tools at our disposal to change the growth characteristics and thus optimize the chance of success. Overexpression of the cell division activator SsgA increases fragmentation of streptomycetes, which leads to faster growth. A strain of S. coelicolor overexpressing SsgA produced large amounts of prodigionines, but hardly any actinorhodin.43 However, SsgA affects the intracellular architecture, making the hyphae less robust. Deletion of matAB prevents the production of poly-N-acetylglucosamine (PNAG), an EPS that “glues” the hyphae together, promoting pellet formation.19,21 Deletion of matAB in S. coelicolor M1152 significantly resulted in a dispersed growth phenotype (data not shown) and led to better productivity. The more PNAG is produced the larger the pellets, and thus the technology may be widely applicable.
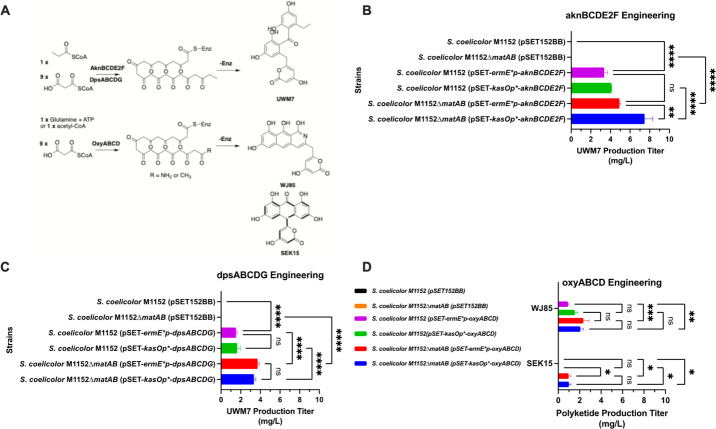
Engineering of the aknBCDE2F and dpsABCDG operons in S. coelicolor M1152ΔmatAB and S. coelicolor M1152 resulted in the expected production of UWM7 as the major metabolite at production titers of 1–7.5 mg/L in SG shake flask experiments (Figure 4). UWM7 exhibited a retention time of 8.42 min with a UV maximum of 290 nm and the expected mass in ESI+ mode [M + H]+ = 399 m/z, as previously described (Supporting Information, Figure S13).38 In each case, the corresponding S. coelicolor M1152ΔmatAB resulted in a statistically significant increase in UWM7 production titers as compared to S. coelicolor M1152 (p ≤ 0.01), and the aknBCDE2F operon was twice as productive as the dpsABCDG operon. The best construct featured the kasOp* promoter fused to the aknBCDE2F operon (7.5 mg/L UWM7), which resulted in 33% greater UWM7 as compared to the ermE*p promoter fused to the aknBCDE2F operon (p ≤ 0.05) (Figure 4).
Figure 4.
Engineering of UWM7 and WJ85 minimal aromatic polyketides. (A) Biosynthesis scheme for the synthesis of UWM7 from the DpsABCDG/AknBCDE2F minPKS and WJ85 from the OxyABCD minPKS. (B) Production titers of UWM7 resulting from the engineering of pSET-ermE*p-aknBCDE2F and pSET-kasOp*-aknBCDE2F in S. coelicolor M1152 and M1152ΔmatAB. (C) Production titers of UWM7 resulting from the engineering of pSET-ermE*p-dpsABCDG and pSET-kasOp*-dpsABCDG in S. coelicolor M1152 and M1152ΔmatAB. (D) Production titers of SEK15 and WJ85 resulting from the engineering of pSET-ermE*p-oxyABCD and pSET-kasOp*-oxyABCD in S. coelicolor M1152 and M1152ΔmatAB. ANOVA was carried out to determine the statistical significance between strains. The statistically significant comparisons are reflected with asterisks. The statistical significance of observed results was established with a p < 0.05. * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001.
Engineering of oxyABCD into the host strains resulted in the production of WJ85 as determined via HPLC-MS analysis (Supporting Information, Figure S14). WJ85 exhibited a retention time of 9.54 min with a UV maximum at 290 nm, and it exhibited a parental mass of [M + H]+ = 368 m/z, as previously described.37 The ermE*p-oxyABCD construct resulted in the production of 2.3 mg/L WJ85 in S. coelicolor M1152ΔmatAB, as compared to the kasOp*-oxyABCD construct, which resulted in the production of 2.0 mg/L WJ85, although the difference between these two promoters was not statistically significant in this case (Figure 4). Due to the deletion of the ΔmatAB operon, the S. coelicolor M1152ΔmatAB strain accumulated 1 mg/L SEK15, which was not detected in S. coelicolor M1152. This result was interpreted to mean that S. coelicolor M1152ΔmatAB strain exhibited improved production characteristics for minimal aromatic polyketides. The differences in production titer of WJ85 and SEK15 between M1152 and M1152ΔmatAB were determined to be statistically significant based on ANOVA statistical analysis (Figure 4). Due to the low production yields of the oxyABCD operon, we did not pursue its use any further. In summation, we chose S. coelicolor M1152ΔmatAB for further experiments to build out anthracyclinone pathways.
Engineering of Ketoreductases, Aromatases, and Cyclases
We next engineered reducing ketoreductase (KR), aromatase (ARO), and cyclase (CYC) gene cassettes into S. coelicolor M1152ΔmatAB to generate tricyclic anthracyclinones (Figure 5). Starting from the enzyme-tethered poly-β-ketothioester synthesized by Snoa123, the 3-oxoacyl-[acyl carrier protein] ketoreductase (SnoaD/AknA) reduces the poly-β-ketothioester at 9-position, followed by C7–C12 first ring cyclization and aromatization (SnoaE/AknE1), C5–C14 s ring cyclization and C3–C16 third ring cyclization by the second-third ring cyclase (SnoaM/AknW), and C-12 oxidation by the anthrone oxygenase (SnoaB/AknX) (Figure 1). Genetic cassettes for aknAE1W, aknAE1WX, snoaDEM, and snoaDEMB were codon-optimized and cloned as described above under the control of the strong p15 promoter from Streptomyces albus and cloned into a separate integrating vector pENSV1 for two plasmid coexpression or spliced into the pSET152BB-kasOp*-snoa123 construct for expression of the entire cassette on a single plasmid.24,42 Heterologous expression of snoa123+snoaDEM, snoa123+snoaDEMB, snoa123+aknAE1W, and snoa123+aknAE1WX on two plasmids resulted in the production of SEK15, and C7–C12 cyclized metabolites SEK43 and SEK43b, C7–C12 and C9–C14 cyclized metabolite S2502, and tricyclic anthracyclinones nogalonic acid and its rearrangement product decarboxy-nogalonic acid, as expected (Figure 5, Supporting Information, Figure S10). These metabolites were confirmed based on a comparison to authenticated biosynthetic standards from S. lividans TK24/pSY21c.11 Percent conversion was calculated based on the integration of the peak areas at λ = 290 nm since the polyketides exhibit similar molar absorptivity coefficients at this wavelength (Figure 5).11 This finding supports the role of AknA as a ketoreductase, AknE1 as an ARO/CYC, AknW as a second and third-ring cyclase, and AknX as an anthrone oxygenase. Previously, Chung et al. demonstrated that the AknX anthrone oxygenase enhances the oxidation of emodin anthrone to emodin anthraquinone, although C-12 oxidation can also occur spontaneously.10 Expression of the C-12 anthraquinol oxidase increased the formation of correctly cyclized tricyclic anthracyclinone intermediate, which confirms its essential role in the oxidation of nogalonic acid. Coexpression of the snoa123 minPKS genes with the KR/ARO/CYC cassettes on one plasmid resulted in higher metabolic flux toward nogalonic acid (Figure 5). One possible explanation for this was offered by Yang et al., who observed that expression of type II PKS genes on the same plasmid in E. coli resulted in higher production of carminic acid.43 Yang et al. postulated that colocalization of the minPKS genes and cyclase genes enhances the formation of the transient polyketide synthase complex, enhancing the production titer. Interestingly, the coexpression of snoa123+aknAE1WX on the same plasmid construct resulted in approximately 15% conversion to nogalonic acid. Expression of the snoa123+snoaDEMB construct resulted in a 50% conversion to nogalonic acid (Figure 5). These results demonstrate that the systematic testing of different orthologous gene combinations can provide additional insight into the compatibility of heterologous polyketide synthase components from related pathways. In addition, this approach can be used to provide biochemical evidence for gene products from uncharacterized pathways or for which only bioinformatic description is provided (e.g., AknA, AknE1, AknW).
Figure 5.
Engineering of KR/ARO/CYC cassettes with the snoa123 minPKS. (A) Chromatograms of metabolites from two-plasmid snoa123 and KR/ARO/CYC heterologous expression experiments. (B) Disposition of metabolites from two-plasmid snoa123 and KR/ARO/CYC heterologous expression experiments. (C) Chromatograms of metabolites from one-plasmid snoa123 and KR/ARO/CYC heterologous expression experiments. (D) Disposition of metabolites from one-plasmid snoa123 and KR/ARO/CYC heterologous expression experiments.
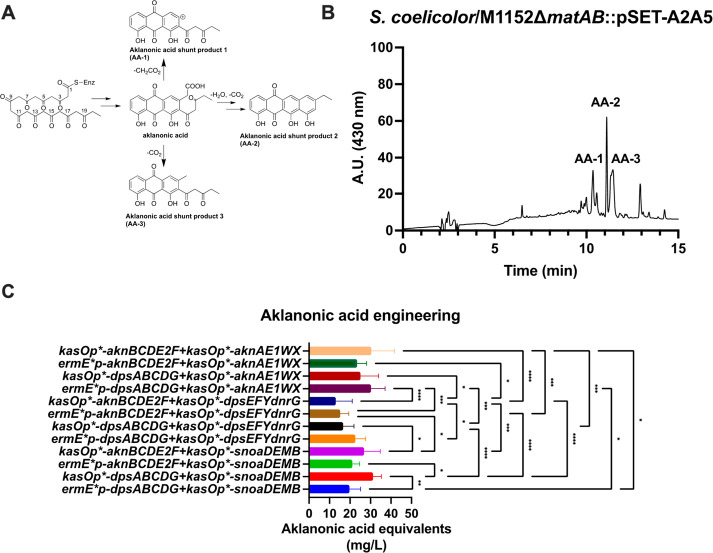
We next decided to test the potential of BIOPOLYMER for combinatorial biosynthesis by carrying out a full factorial experiment for the biosynthesis of aklanonic acid. We designed this experiment by cloning combinations of the aknBCDE2F and dpsABCDG minPKS operons with the aknAE1WX, snoaDEMB, and dpsEFY+dnrG KR/ARO/CYC/OXY operons. The KR/ARO/CYC/OXY operons were all cloned under the control of the strong kasOp* promoter since the strong transcriptional regulation of tailoring genes correlates with improved cyclic product formation and diminished shunt product accumulation in the actinorhodin pathway.44 Expression of aknBCDE2F and dpsABCDG with KR/ARO/CYC/OXY genes resulted in the accumulation of three aklanonic acid-derived shunt products (AA-1, AA-2, and AA3), as previously described (Figure 6).4 Mass spectroscopy confirmed the production of the three degradation products: AA-1, [M – H]− = 337 m/z; AA-2, [M – H]− = 333 m/z; AA-3, [M – H]− = 351 m/z (Supporting Information, Figure S85). All combinations assessed resulted in the production of aklanonic acid, which indicates that there is a significant biosynthetic collaboration between minPKS enzymes and ketoreductase/cyclase enzymes between the nogalamycin, daunorubicin, and aclacinomycin biosynthetic pathways (Figure 6). The production titers of aklanonic acid varied between 20 mg/L to 35 mg/L with the best combinations consisting of recombinant PKS systems dpsABCDG+snoaDEMB and dpsABCDG+aknAE1WX and the native aknBCDE2F+aknAE1WX pathway (Figure 6). We performed an analysis of variance (ANOVA) of all 12 strains to determine if the differences in production titer between the different strains were statistically significant (Figure 6). The ANOVA indicated that the observed results between many of the comparisons were statistically significant, which provides good evidence that the combinatorial biosynthesis of the minPKS and cyclase gene cassettes was responsible for the observed differences in production titer.
Figure 6.
Engineering of aklanonic acid via combinatorial biosynthesis. (A) Scheme depicting degradation of aklanonic acid to its three detected shunt products (AA-1, AA-2, and AA-3). (B) Chromatogram of aklanonic acid degradation metabolites produced by a representative strain. (C) Aklanonic acid production titers resulting from full-factorial combinatorial biosynthesis of minPKS and KR/ARO/CYC/OXY genes. Error bars depict standard deviation. ANOVA was carried out to determine the statistical significance between strains. The statistically significant comparisons are reflected with asterisks. The statistical significance of observed results was established with a p < 0.05. * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001.
Engineering of Nogalamycinone and Structural Characterization of Anthracyclinones
We next decided to complete the aglycone engineering pathway by introducing the nogalonic acid methyltransferase, fourth-ring cyclase, and 7-ketoreductase from the nogalamycin biosynthetic pathway (Figure 1). The resulting construct, pSET152BB-ermE*p-snoa123+kasOp*-aknAE1WX+sp44-snoaLCF was transformed into S. coelicolor M1152ΔmatAB and assessed for production of nogalamycinone. The order of genes in the snoaLCF operon was determined to be important for the complete conversion of nogalonic acid to nogalamycinone. Initially, cloning of the snoaC before snoaL and subsequent expression of the resulting plasmid yielded incomplete conversion from nogalonic acid to nogalamycinone. The resulting strains produced six compounds that could be detected via HPLC-MS, including nogalamycinone (1) which could be confirmed via comparison to an authentic HPLC standard (Figure S18). Interestingly, a new early eluting product was also detected that corresponded to a glucosylated derivative of nogalamycinone, as determined by mass spectrometric analysis in ESI– mode ([M – H]− = 559 m/z). The resulting strain was scaled up in a 5 L SG shake flask fermentation, extracted with 3 × 5 L of ethyl acetate, and fractionated via SiO2 column chromatography in chloroform:methanol systems on a Teledyne Combiflash 100 auto purification system. A second 2 L fermentation of the strain was carried out to isolate the unknown glucosylated nogalamycinone derivative. Compounds 1–6 were purified from additional prep-HPLC experiments.
The physicochemical properties of compounds 1–6 have been summarized in the experimental section (Supporting Information, Figures S16–S82, Tables S3 and S4). The chemical structures of the known anthracyclines nogalamycinone (1), auramycinone (2), 7-deoxy-nogalamycinone (3), and 7-deoxyauramycinone (4) were established by 1H NMR, 13C NMR, 1H,1H-COSY, 1H,13C-HMBC, and 1H,1H-NOESY NMR spectroscopic measurements (Supporting Information, Tables S3 and S4, Figures S16 and S17). Unexpectedly, both nogalamycinone and auramycinone were produced, which result from fourth ring cyclization reactions that generate the 9(S) and 9(R) products.13,45 Compounds 1 and 2 were produced in an approximately 2:1 ratio, as were their shunt products 3 and 4, which indicates that nogalamycinone and 7-deoxynogalamycinone were the preferred enzymatic products in the strain.
It is generally accepted that the codon-optimization of genes is known to improve translational efficiency by substituting rare codons in an mRNA sequence for preferred codons with abundant tRNAs, which leads to an increase in the concentration of the protein.46 What is less understood is whether codon-optimization disrupts information that is present in wild-type mRNAs (e.g., rare codons that slow down the translation or that influence translation via binding to rRNA), which could impact protein conformation and function.47 Hu et al. verified 342 synonymous codon variants of the scFv antibody and observed widely varying production titers, solubility, and binding affinity (e.g., ranging from no binding affinity to 10–8 M), while all proteins encoded the same original amino acid sequence.48 Sander et al. demonstrated that synonymous codon changes in a fluorescent protein impacted the fluorescence and can alter the folded structure due to differences in the rate of translation and folding of the N- and C-termini.49 It could be hypothesized that the production of 3 and 4 by codon optimized SnoaL could be due to protein conformational differences from the wild-type SnoaL which impact the binding of NAME in the active site, but this must be examined further.
The formation of 3, 4, and 5 requires the elimination of the 7-hydroxyl group, possibly due to the presence of a promiscuous CytA-like enzyme that is present in S. coelicolor. Gui et al. have characterized CytA as a promiscuous anthracycline-inactivating enzyme that reduces the C-7 position for a broad variety of anthracyclines in the cytorhodin pathway according to an NADH-dependent mechanism.50
Compound 5 was obtained as a yellow solid and displayed UV–vis characteristics like metabolites 1–4. The molecular formula of 5 was established as C21H20O7 based on (+)- and (−)-HRESIMS indicative of two additional protons as compared to compounds 3 and 4 (Supporting Information, Figure S64 and S65). Comparison of the 1H and 13C NMR of 5 and 3/4 (Supporting Information, Tables S3 and S4) revealed 5 to lack the C-9/C-10 bond connection and the methine proton signal at C-10 position (CH-10) in compounds 3–4 was replaced by CH2 at δH 3.83 (CH2-10). In addition, an additional oxygenated methine signal was observed at δH 3.80 (m, CH-9) in compound 5. Furthermore, the singlet methyl signals in compounds 3/4 were replaced by a doublet methyl signal in 5 at δH 1.22 (d, 6.2). This was consistent with the presence of 1H–1H COSY correlations CH3-13/H-9, H-9/CH2-8, CH2-8/CH2-7 for 5 (Supporting Information, Figure S16). All the remaining 2D-NMR (1H,1H-COSY, HMBC, and NOESY) correlations are in full agreement with structure 5 (Supporting Information, Figures S63–S72). Based on the cumulative spectroscopic data, 5 differed solely from those of compounds 3-4 via their C-9/C-10 bond connection. Consistent with the cumulative 1D and 2D NMR data analysis, structure 5 was established as depicted in Figure 1 and named 9,10-seco-7-deoxy-nogalamycinone (5).
Compound 6 was isolated as a yellow solid and displayed UV–vis characteristics like anthracyclinones 1–4. The molecular formula of 6 was established as C27H28O13 based on (+) and (−)-HRESIMS, with Δm/z = 162 higher than those of 1, indicative of the presence of an extra hexose moiety in 6 (Supporting Information, Figures S74 and S75). Based on the 1H NMR, 13C NMR, NOESY, and 1H,1H-COSY data, the carbohydrate was unambiguously assigned as d-glucose (Supporting Information, Table S5 and Figure S17). Compared to 1, the 13C/1H/HSQC NMR of 6 (Supporting Information, Table S1–S3) highlighted the presence of additional signals for the O-glycoside moiety in compound 6. Based on the 1H NMR, 13C NMR, NOESY, and 1H,1H-COSY data, the carbohydrate was unambiguously assigned as d-glucose (Supporting Information, Table S5, Figure S17 and Figures S76−S82). The connection of the sugar moiety at the 4-position was established based on the observed critical HMBC correlation from H-1′ (δH 5.12, d, J = 7.7 Hz) to C-4 (δC 160.2). All the remaining 2D-NMR (1H,1H-COSY, HMBC, and NOESY) correlations are in full agreement with structure 6 (Supporting Information, Figure S17 and Figures S76–S82). As a new natural product and closely related to 1, compound 6 was designated as 4-β-d-glucosyl-nogalamycinone (6).
Full Pathway Engineering of Anthracyclinones
We decided to complete the pathway engineering of other anthracyclinones, including auramycinone (2), aklavinone (7), and 9-epi-aklavinone (8). Fujiwara et al. isolated auramycinone as a C-20 anthracyclinone from S. galilaeus OBB-111, and aklavinone is a C-21 anthracyclinone that serves as the backbone for doxorubicin and aclacinomycin A.51,52 9-epi-Aklavinone was reported as a hybrid anthracyclinone resulting from the heterologous expression of snoaL from the nogalamycin biosynthetic pathway in a mutant strain of S. peucetius M18.5 Using the strains S. coelicolor M1152ΔmatAB::pS2S5 (nogalonic acid producer) and S. coelicolor M1152ΔmatAB::pA2A5 (aklanonic acid producer) as hosts, we cloned different combinations of O-methyltransferase genes (aknG, dnrC, snoaC), fourth-ring cyclases (dnrD, kyc34, aknH), and 7-ketoreductases (snoaF, dnrE, aknU) under the control of the strong sp44 promoter and cloned them into the TG1-actinophage integrating vector pENTG1.24 In brief, pENTG1 is a vector that encodes bla and vph for ampicillin and viomycin selection, respectively, and a codon-optimized version of the TG1 integrase and corresponding attP site for integration into the S. coelicolor TG1 attB chromosomal locus at a single copy.53,54 The resulting vectors were expected to result in production of 9(S)-configured anthracyclinones (e.g., pENTG1-sp44-snoaC+kyc34+snoaF, pENTG1-sp44-snoCLF) or 9(R)-configured anthracyclinones (e.g., pENTG1-sp44-dnrCDE and pENTG1-sp44-aknGHU). Coexpression of these vectors in either of the two strains resulted in the production of the expected anthracyclinones (Figure 7). To produce nogalamycinone, snoaLCF and snoaC+kyc34+snoaF both resulted in the complete conversion of nogalonic acid. This result also provides experimental proof that kyc34 from the keyicin biosynthetic pathway encodes a NAME cyclase, like snoaL.55 Similarly, coexpression of aknGHU in S. coelicolor M1152ΔmatAB::pS2S5 resulted in the complete conversion of nogalonic acid to 2. For the production of 7, when S. coelicolor M1152ΔmatAB::pA2A5 was complemented with aknGHU, complete conversion of aklanonic acid was observed. Lastly, the complementation of the aklanonic acid producer with either snoaLCF or snoaC+kyc34+snoaF resulted in approximately 80% conversion to 9-epi-aklavinone. This experiment demonstrates that the BIOPOLYMER system can be used to produce all the naturally occurring reduced anthracyclinone analogues.
Figure 7.
Full pathway engineering of anthracyclinone polyketide synthases. Engineering of methyltransferase, fourth-ring cyclase, and ketoreductase genes furnished anthracyclinone pathways in S. coelicolor M1152ΔmatAB. (A) S. coelicolor M1152ΔmatAB::pS2S5 was complemented with different constructs to biosynthesize different amounts of 1 and 2. (B) S. coelicolor M1152ΔmatAB::pA2A5 was complemented with different constructs to biosynthesize different amounts of 7 and 8. (C) Production titer of anthracyclinones in strains expressing the entire pathway on two plasmids. Strains were grown in SG-TES liquid media for 5 days. Error bars reflect the SD of six replicates. Experimental groups were compared using a one-way ANOVA test to determine statistical significance. The statistical significance of observed results was established with a p < 0.05. * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001. (D) HPLC chromatogram traces of different strains at 430 nm with major metabolites highlighted with an asterisk (*): (i) S. coelicolor M1152ΔmatAB::pSET152; (ii) S. coelicolor M1152ΔmatAB::A2A5 (aklanonic acid); (iii) S. coelicolor M1152ΔmatAB::S2S5 (nogalonic acid); (iv) S. coelicolor M1152ΔmatAB::S2S5A6 (auramycinone); (v) S. coelicolor M1152ΔmatAB::S2S5S6 (nogalamycinone); (vi) S. coelicolor M1152ΔmatAB::A2A5A6 (aklavinone); (vii) S. coelicolor M1152ΔmatAB::A2A5S6 (9-epi-aklavinone).
Next, we cloned the aklavinone, 9-epi-aklavinone, auramycinone, and nogalamycinone pathways onto a single integrating plasmid. To accomplish this, a new version of pSET152 was cloned in which the strong synthetic tt-sbi-A transcriptional terminator from Mycobacterium tuberculosis and the strong fd phage terminator were inserted 5′ and 3′ of the BioBricks cloning sites,56−58 which was designated as pSET154BB (Supporting Information, Table S1). The resulting plasmids encoding aklavinone (e.g., pSET154BB-kasOp*-aknBCDE2F+kasOp*-aknAE1WX+sp44-aknGHU), auramycinone (pSET154BB-kasOp*-snoa123+kasOp*-snoaDEMB+sp44-aknGHU), and nogalamycinone (pSET154BB-kasOp*-snoa123+kasOp*-snoaDEMB+sp44-snoaC+kyc34+snoaF) were transformed into S. coelicolor M1152ΔmatAB to quantify the production titers of the intended anthracyclinones. In addition, we also transformed in the nogalamycinone pathway plasmid pSET152BB-kasOp*-snoa123+kasOp*-aknAE1WX + sp44-snoaCLF, which was used to isolate compounds 1–6 for structure elucidation. The strains were fermented in SG-TES media and E1 media, which are two media used in our laboratories to produce polyketides. In the case of the first four plasmids for aklavinone, 9-epi-aklavinone, auramycinone, and nogalamycinone, the yields of anthracyclinones were lower than those obtained with the two-plasmid expression system (Figure 8). S. coelicolor M1152ΔmatAB harboring these constructs produced a mean production titer of 2.22 mg/L aklavinone in SG-TES media and 0.67 mg/L in E1 media, 1.35 mg/L 9-epi-aklavinone in SG-TES media and 0.98 mg/L 9-epi-aklavinone in E1 media, 0.94 mg/L auramycinone in SG-TES media and 0.39 mg/L auramycinone in E1 media, and 3.22 mg/L nogalamycinone in SG-TES media and 1.32 mg/L nogalamycinone in E1 media. In contrast, the fully codon-optimized construct pSET152BB-kasOp*-snoa123+kasOp*-aknAE1WX+sp44-snoaCLF resulted in the production of approximately 33-fold higher levels of nogalamycinone at 103 mg/L in SG-TES media and 45 mg/L in E1 media E1 media. The yields of anthracyclinones indicate that the metabolic flux is controlled by the expression of the minimal polyketide synthase. This is especially highlighted by the greatly increased production titer of nogalamycinone resulting from the fully codon-optimized nogalamycinone construct, which could be explained by the enhanced translation of the Snoa123 minPKS complex. This result indicates that additional optimization of the aknBCDE2F minPKS cassette is still required. Several strategies could be pursued, including codon-optimization, additional promoter engineering, multiplexed site-specific genome engineering (MSGE) to increase the number of ϕC31 attB sites in the chromosome, and even engineering of the endogenous housekeeping sigma factor σhrdB to establish a self-sustaining production system (StSS).59−61 As determined by Student’s t-test, SG-TES media was better than E1 media to produce all four anthracyclinones in this strain (p ≤ 0.05).
Figure 8.

Production titers resulting from the expression of anthracyclinone pathways on a single plasmid. S. coelicolor M1152ΔmatAB was transformed with vectors (A) pSET154BB-kasOp*-aknBCDE2F+kasOp*-aknAE1WX+sp44-aknGHU (pEN10001, aklavinone pathway), (B) pSET154BB-kasOp*-aknBCDE2F+kasOp*-aknAE1WX+sp44-snoaC+kyc34+snoaF (pEN10002, 9-epi-aklavinone pathway), (C) pSET154BB-kasOp*-snoa123+kasOp*-snoaDEMB+sp44-aknGHU (pEN10003, auramycinone pathway), or (D) pSET154BB-kasOp*-snoa123+kasOp*-snoaDEMB+sp44-snoaC+kyc34+snoaF (pEN10004, wild-type nogalamycinone pathway) or pSET152BB-kasOp*-snoa123+kasOp*-aknAE1WX + sp44-snoaCLF (pRW10000, codon-optimized nogalamycinone pathway) and grown in shake flasks of SG-TES or E1 liquid media for 5 days. Error bars reflect the standard deviation of six replicates. Post hoc Student’s t tests were performed to determine statistical significance. The statistical significance of observed results was established with a p < 0.05. * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001.
Human Cancer Cell Viability Assays
Next, we assessed the antiproliferative activity of anthracyclinones 1–7 and compared the activity to anthracycline compounds previously isolated in our lab: nogalamycin A (9), nogalamycin KO (10), nogalamycin R (11), 3′,4′-demethoxy-nogalose-1-hydroxy-nogalamycinone (12), and aclacinomycin T (13).33,62,63 First, the compounds were assessed in a single dose (80 μM) viability assay against a representative set of human cancer cell lines [A549 (non-small-cell lung epithelial carcinoma), PC3 (prostate adenocarcinoma), and MKL1 (polyomavirus-positive Merkel carcinoma) and MCC26 (virus-negative Merkel carcinoma) (Figure 9)]. The most active compounds were anthracyclines nogalamycin A (9), nogalamycin R (11), 3′,4′-demethoxy-nogalose-1-hydroxy-nogalamycinone (12), and aclacinomycin T (13). This result was expected since the glycoside moiety of the anthracyclines is essential for binding to DNA and inhibition of DNA topoisomerases.64 Compounds 6 and 10 exhibited slight antiproliferative activity (6: 75–85% T/C; 10: 50–75% T/C). In the single dose experiment 1, 2, and 7 exhibited antiproliferative activity (<10% T/C) comparable to previously reported compounds 9, 11, 12, and 13. Compounds 3–5 did not exhibit any activity in the assay.
Figure 9.
% Viability of A549 (non-small-cell lung) and PC3 (prostate) human cancer cell lines, and Merkel cells (MKL1 and MCC26) (after 72 h) at 80 μM concentration of compounds 1–7 and 9–13.
The dose–response relationships of 1, 2, 6, 7, and 9–13 were established for A549 and PC3 cells and Merkel cells MKL1 and MCC26 (Supporting Information, Figure S83 and S84). The half-maximal inhibitory concentration values (IC50) were determined for the entire cancer cell panel (Table 3). Previously reported compound 9 was the most cytotoxic among the set tested, with IC50 values ranging from 21 to 95 nM. In contrast, compound 7 was the most active among the analogues derived from the current study, with IC50 values ranging from 8 to 26 μM. This is consistent with previous studies highlighting the contribution of anthracycline glycosylation to potency, selectivity, and ADMET.65
Table 3. Cytotoxic Activities of Compounds 1–13a.
| IC50 μM |
IC50 μM |
|||
|---|---|---|---|---|
| compounds | A549 | PC3 | MKL1 | MCC26 |
| Nogalamycinone (1) | 30.30 ± 2.30 | 18.60 ± 3.61 | 29.30 ± 10.30 | 36.50 ± 5.34 |
| Auramycinone (2) | 31.00 ± 0.96 | 13.90 ± 3.66 | 26.10 ± 1.16 | 30.40 ± 2.01 |
| 7-Deoxy-nogalamycinone (3) | >80 | >80 | >80 | >80 |
| 7-Deoxyauramycinone (4) | >80 | >80 | >80 | >80 |
| 9,10-seco-7-Deoxy-nogalamycinone (5) | >80 | >80 | >80 | >80 |
| 4-β-d-Glucosyl-nogalamycinone (6) | >80 | >80 | >80 | >80 |
| Aklavinone (7) | 17.80 ± 6.24 | 7.72 ± 0.57 | 22.70 ± 1.79 | 26.10 ± 0.66 |
| Nogalamycin A (9) | 0.056 ± 0.011 | 0.021 ± 0.002* | 0.062 ± 0.035* | 0.095 ± 0.044* |
| Nogalamycin KO (10) | >80 | >80 | >80 | >80 |
| Nogalamycin R (11) | 32.30 ± 4.09 | 28.30 ± 3.10 | 18.50 ± 2.64 | 35.60 ± 2.73 |
| 3′,4′-Demethoxy-nogalose-1-hydroxy-nogalamycinone (12) | 2.88 ± 0.23 | 2.88 ± 0.79 | 1.07 ± 0.011 | 3.50 ± 0.36 |
| Aclacinomycin T (13) | 3.62 ± 0.087 | 2.00 ± 0.15 | 2.35 ± 0.78 | 10.70 ± 4.32 |
Cytotoxicity IC50 values were obtained after 72 h incubation. Actinomycin D and H2O2 [A549 (non-small-cell lung), PC3 (prostate) human cancer cell lines, and Merkel cells MKL1 and MCC26] were used as a positive control at 20 μM and 1 mM concentration, respectively (0% viable cells, n = 3, but *(n = 6)).
Conclusions
The “mixing and matching” of PKS components for the reconstitution of non-native aromatic polyketide synthases has been the subject of numerous in vivo studies, which provided the foundational basis for the biosynthetic logic of these enzymes.66−70 However, these experiments often utilized PKS enzymes from disparate pathways, which resulted in recombinant PKS systems that produced low yields of the expected novel polyketides, or that failed.71 This resulted in the observation that the failure of polyketide combinatorial biosynthesis derives from the inflexibility of downstream enzymes to recognize novel substrates or lack of enzyme solubility.72 In contrast, focusing on enzymes from evolutionarily related pathways might provide more fertile ground for combinatorial biosynthesis efforts and improved substrate turnover to produce novel polyketides. More recently, the use of cell-free biosynthesis (also known as “combinatorial biosynthetic enzymology”) has demonstrated the success of this “mix-and-match” approach via the use of PKS enzymes from closely related pathways for the synthesis of defucogilvocarcin M and steffimycinone in vitro.73,74
A “Design-Build-Test-Learn” (DBTL) approach has been suggested to uncover the logic of recombinant PKS systems.75 This is the approach that we have currently undertaken with the development of the BIOPOLYMER toolbox, via the identification of PKS gene orthologs that interact positively and result in the production of expected polyketides in S. coelicolor M1152ΔmatAB. In this work, the “design phase” resulted in the selection of different S. coelicolor hosts, strong promoters, vector combinations, and gene orthologs. The “build phase” was facilitated by the use of the BioBricks-[RFC 10] synthetic biology standard and straightforward transformation of Streptomyces spp. using intergeneric conjugation. The “test phase” carried out several different comparisons of anthracyclinone genes to identify those with the best conversion percentage and production yield of the expected polyketide. Lastly, the “learn phase” resulted in the identification of an optimal production host, S. coelicolor M1152ΔmatAB, promoter and gene combinations, and the structure elucidation of several anthracyclinones (1–4) and new compounds 9,10-seco-7-deoxy-nogalamycinone (5) and 4-O-β-d-glucosyl-nogalamycinone (6). Furthermore, the production platform was used to generate newly engineered metabolites for testing in mammalian cancer cell viability assays. These assays confirmed that the 7-O-glycoside is important for the anticancer activity of anthracyclines, including nogalamycin A (9), 3′,4′-demethoxy-nogalose-1-hydroxy-nogalamycinone (12), and aclacinomycin T (13). Nevertheless, the observation that nogalamycinone, auramycinone, and aklavinone exhibited moderate cytotoxicity is encouraging for further efforts to generate diverse anthracycline analogues incorporating these pharmacophores.
The pathway engineering studies described here resulted in the unexpected generation of 7-deoxygenated metabolites, 3 and 4, and two new anthracycline metabolites, 9,10-seco-7-deoxy-nogalamycinone (5) and 4-O-β-d-glucosyl-nogalamycinone (6). Despite being a genome minimalized “superhost”, the genome S. coelicolor M1152ΔmatAB still encodes hypothetical proteins that can interact with heterologous pathways in an unanticipated fashion. Compounds 3, 4, and 5 are thought to derive from the dehydration of the 7-hydroxyl group via an ancillary CytA-like enzyme.50 CytA has been shown to catalyze the 7-reduction of a wide variety of anthracycline saccharide chains within the cytorhodin pathway. CytA could be part of a larger family of promiscuous enzymes encoded within actinomycete genomes, perhaps as an evolutionary self-defense strategy against xenobiotics. Studies are ongoing to identify this reductase within S. coelicolor M1152.
Compound 6 is thought to derive from a hypothetical glucosyltransferase that catalyzes the transfer of NDP-d-glucose to the 4-position of nogalamycinone. Studies are ongoing to identify the glucosyltransferase responsible for the transfer of NDP-d-glucose to nogalamycinone. The macrolide glucosyltransferase OleD has been mutagenized via directed evolution to afford one of the most substrate-promiscuous glycosyltransferase catalysts for the glycorandomization of a variety of diverse natural products.76−79 Notably, 6 has a similar 4-O-glycosylation pattern to the previously discovered mutactimycin PR, andicoquinones A–D, komodoquinone A, and histomodulin.80 In the cancer cell line cytotoxicity assays, the 4-β-d-glucose substitution was deleterious to the topoisomerase II inhibition of 6 since this compound was only slightly cytotoxic (Figure 9). Instead, the closely related 4-O-β-d-glucopyranuronosyl-ε-rhodomycinone (histomodulin) has been shown to exhibit upregulation of major histocompatibility class-I molecules on the surface of T-cells.81 This class of 4-O-glucosides could be further investigated for unique immunomodulatory activities as potential new anticancer or anti-infective agents.
In summation, BIOPOLYMER is a flexible synthetic biology platform for the rational programming of S. coelicolor to produce aromatic polyketide natural products. We anticipate that BIOPOLYMER will be useful for providing access to known and new anthracycline natural products for antiproliferative activity studies. We also expect that BIOPOLYMER could be useful for studying cryptic type II PKS pathways and for providing a robust toolset for unraveling their biosynthetic logic.
Methods
Bacterial Strains and Growth Conditions
Escherichia coli JM109 and E. coli ET12567 were grown in LB broth or LB agar at 37 °C as previously described.82E. coli JM109 was used for plasmid propagation and subcloning, while E. coli ET12567/pUZ8002 was used as the conjugation donor host for mobilizing expression vectors into S. coelicolor and S. lividans expression hosts as previously described.83 When appropriate, ampicillin (100 μg mL–1), kanamycin (25 μg mL–1), apramycin (25 μg mL–1), viomycin (25 μg mL–1), and nalidixic acid (30 μg mL–1) were supplemented to media to select for recombinant microorganisms.
S. lividans and S. coelicolor derivative strains were routinely maintained on Soya-Mannitol Flour (SFM) agar supplemented with 10 mM MgCl2 and International Streptomyces Project medium #4 (ISP4) (BD Difco) at 30 °C as described previously.83S. lividans K4–114 was a gift from Prof. Dr. Lou Charkoudian (Haverford College, PA). S. coelicolor M1146 and S. coelicolor M1152 were gifts from Prof. Dr. Mervyn Bibb’s laboratory (John Innes Centre, Norwich, UK). S. coelicolor M1152ΔmatAB was generated via replacement of the sco2963-sco2962 matAB locus via PCR-ReDirect mutagenesis as previously described (Supporting Information, Method 1).19 For liquid culturing, S. coelicolor derivative strains were grown in TSB media (3 mL) to ferment the seed culture and then grown in a modified 50 mL SG-TES liquid medium (soytone 10 g, glucose 20 g, yeast extract 5 g, TES free acid 5.73 g, CoCl2 1 mg, per liter) or 50 mL E1 medium for production for 7 days.84 All media and reagents were purchased from Thermo-Fisher Scientific.
General Manipulations
Routine genetic cloning and plasmid manipulation were carried out in E. coli JM109 (New England Biolabs). E. coli ET12567/pUZ8002 was used as the host for intergeneric conjugation with S. coelicolor as previously described.83E. coli chemically competent cells were prepared using the Mix and Go! E. coli Transformation Kit (Zymo Research). E. coli was transformed with plasmid DNA via chemically competent heat-shock transformation as described previously. Plasmid DNA was isolated via the Wizard Plus SV Minipreps DNA Purification System by following the manufacturer’s protocols (Promega). All molecular biology reagents and enzymes used for plasmid construction were purchased from New England Biolabs. The conjugation donor host E. coli ET12567/pUZ8002 was transformed with constructs for mobilization into S. coelicolor strains, as previously described. For each transformation, 9–12 independent exconjugants were plated to DNA plates supplemented with antibiotics and grown for 4–5 days until the formation of vegetative mycelium.
General Experimental Procedures
Ultraviolet–visible (UV–vis) spectra were taken directly from analytical HPLC runs and show relative intensities. All NMR spectra were recorded at 600 MHz (14.1 T) using a Bruker Avance Neo console equipped with a TCI 5 mm cryoprobe (all spectra were processed in Bruker Topspin 4.1.3 version, and 2D spectra were apodized with QSINE or SINE window functions and zero-filled to 256 × 1024 points), and spectra were analyzed and plotted using Mnova [where δ-values were referenced to respective solvent signals (CD3OD, δH 3.31 ppm, δC 49.15 ppm; CDCl3, δH 7.24 ppm, δC 77.23 ppm)] (Bruker BioSpin Corporation, Billerica, MA). High-resolution electrospray ionization (HRESI) mass spectra were recorded on AB SCIEX Triple TOF 5600 system. HPLC-UV/MS analyses were accomplished with an Agilent InfinityLab LC/MSD mass spectrometer (MS Model G6125B; Agilent Technologies, Santa Clara, CA, USA) equipped with an Agilent 1260 Infinity II Series Quaternary LC system and a Phenomenex NX-C18 column (250 × 4.6 mm, 5 μm) [Method: solvent A: H2O/0.1% formic acid, solvent B: CH3CN; flow rate: 0.5 mL min–1; 0–30 min, 5–100% B (linear gradient); 30–35 min, 100% B; 35–36 min, 100%–5% B; 36–40 min, 5% B]. HPLC-UV analyses were carried out in a Agilent 1260 system equipped with a photodiode array detector (PDA) and a Phenomenex C18 column (Phenomenex, Torrance, CA; 250 × 4.6 mm, 5 μm; solvent A: H2O/0.1% TFA, solvent B: CH3CN; flow rate: 1.0 mL min–1; 0–30 min, 5–100% B; 30–35 min, 100% B; 35–36 min, 100%–5% B; 36–40 min, 5% B). Semipreparative HPLC were carried out in a Agilent 1260 Infinity II (Prep HPLC) system equipped with a Diode Array Detector (DAD) and a Gemini 5 μm C18 110 Å, LC column 250 × 10 mm (Phenomenex, Torrance, CA) [Method A: H2O/0.025% TFA; solvent B: CH3CN; flow rate: 5.0 mL min–1; 0–2 min, 15% B; 2–31 min, 15–100% B; 31–33 min, 100% B; 33–34 min, 100–15% B; 34–36 min, 10% B; Method B: solvent A: H2O/0.025% TFA; solvent B: CH3CN; flow rate: 5.0 mL min–1; 0–2 min, 15% B; 2–28 min, 15–75% B; 28–30 min, 75–100% B; 30–32 min, 100% B; 32–34 min, 100–15% B; 34–36 min, 10% B; Method C: solvent A: H2O/0.025% TFA; solvent B: CH3CN; flow rate: 5.0 mL min–1; 0–2 min, 25% B; 2–28 min, 25–85% B; 28–30 min, 85–100% B; 30–32 min, 100% B; 32–34 min, 100–25% B; 34–36 min, 25% B; Method D: solvent A: H2O/0.025% TFA; solvent B: CH3CN; flow rate: 5.0 mL min–1; 0–2 min, 10% B; 2–22 min, 10–65% B; 22–25 min, 65–100% B; 25–27 min, 100% B; 27–28 min, 100–10% B; 28–30 min, 10% B]. All solvents used were of ACS grade and purchased from Pharmco-AAPER (Brookfield, CT). Size exclusion chromatography was performed on Sephadex LH-20 (25–100 μm; GE Healthcare, Piscataway, NJ). A549 and PC3 cells were obtained from ATCC (Manassas, VA). All other reagents used were reagent grade and purchased from Sigma-Aldrich (Saint Louis, MO).
Statistical Analyses
The statistical significance of the impact of genetic manipulations and combinatorially assessed variables on production was assessed via post hoc analysis. One-way ANOVA, two-way ANOVA, and Student’s t test analyses were performed using GraphPad Prism version 9.4.1 for Mac OS X, GraphPad Software, San Diego, CA, USA, www.graphpad.com.
Cancer Cell Line Viability Assay
A549 and PC3 cells were obtained from ATCC (Manassas, VA). Merkel cells MKL1 and MCC26 were a gift from Dr. Isaac Brownell’s laboratory (NIH/NIAMS, Bethesda, MD). All other reagents used were reagent grade and purchased from Sigma-Aldrich (Saint Louis, MO). Human cell line cytotoxicity [A549 (non-small-cell lung) and PC3 (prostate) human cancer cell lines, Merkel cells MKL1 and MCC26] assays were accomplished in triplicate following our previously reported protocols.85−88 Vehicle (DMSO) was used as the negative control, and actinomycin D and H2O2 (A549 and PC3; MKL1 and MCC26) were used as positive control at 20 μM and 1 mM concentration, respectively.
Isolation of Compounds 1–6
S. coelicolor M1152ΔmatAB::pRW10000 was grown on a soya mannitol flour (SFM) agar plate until well-sporulated for 4–5 days. Spores from the SFM agar plate were used to inoculate a seed culture of 50 mL tryptic soy broth in a 250 mL Erlenmeyer shake flask and were grown for 48 h in an orbital shaker at 30 °C at 220 rpm. One milliliter of seed culture was used to inoculate each of the 40 × 100 mL SG liquid media shake flasks (1% v/v inoculum) which were fermented for 5 days. The 4 L culture was extracted 4 × 4 L of ethyl acetate +0.1% formic acid and was dried in vacuo on a rotary evaporator. The resulting 2.3 g crude extract was dissolved in 9:1 chloroform–methanol and was loaded onto a 25 g SiO2 solid load cartridge and chromatographed on a 24 g RediSep Gold Normal Phase Silica cartridge (Teledyne-ISCO). The extract was fractionated using the gradient setting and chloroform–methanol systems [Method E: chloroform; solvent B: methanol; flow rate: 40.0 mL min–1; 0–15 min, 0–10% B.] Compounds 1–5 eluted in combined fraction at a retention time of 3.0 min. The compounds were individually resolved and purified via preparatory HPLC.
The same strain was separately grown in 2 L of E1 media to produce substantive amounts of compound 6. The 2 L fermentation was centrifuged to separate the cell pellet from the fermentation broth. The fermentation broth was extracted with 40 g Amberlite XAD-7 resin for 16 h, washed with 2 L of water, eluted with 1 L of methanol, and dried in vacuo on a rotary evaporator. Compound 6 was isolated via preparatory HPLC.
Physicochemical Properties of Compounds 1–6
Nogalamycinone (1)
C21H18O8 (398); yellow solid; HPLC-Rt = 22.72 min (Supporting Information, Figures S18 – S28); UV–vis λmax 198, 228, 258, 290 (sh), 430 nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz), see Tables S3 and S4; (−)-ESI-MS: m/z 397 [M – H]−; (+)-ESI-MS: m/z 381 [(M-H2O) + H]+, 363 [(M-2H2O) + H]+; (−)-HRESI-MS: m/z 397.0929 [M – H]− (calcd for C21H17O8, 397.0929); (+)-HRESI-MS: m/z 363.0860 [(M-2H2O) + H]+ (calcd. for C21H15O6, 363.0863), 421.0865 [M + Na]+ (calcd for C21H18O8Na, 421.0893), 819.1865 [2 M + Na]+ (calcd for C42H36O16Na, 819.1895).
Auramycinone (9-epi-Nogalamycinone) (2)
C21H18O8 (398); yellow solid; HPLC-Rt = 22.68 min (Supporting Information, Figures S29 – S39); UV–vis λmax 196, 228, 260, 290 (sh), 430 nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz), see Tables S3 and S4; (−)-ESI-MS: m/z 397 [M – H]−; (+)-ESI-MS: m/z 381 [(M-H2O) + H]+, 363 [(M-2H2O) + H]+; (−)-HRESI-MS: m/z 397.0930 [M – H]− (calcd for C21H17O8, 397.0929); (+)-HRESI-MS: m/z 363.0863 [(M-2H2O) + H]+ (calcd for C21H15O6, 363.0863), 421.0890 [M + Na]+ (calcd for C21H18O8Na, 421.0893), 819.1913 [2 M + Na]+ (calcd for C42H36O16Na, 819.1895).
7-Deoxy-nogalamycinone (3)
C21H18O7 (382); yellow solid; HPLC-Rt = 28.35 min (Supporting Information, Figures S40 – S50); UV–vis λmax 200, 228, 260, 290 (sh), 432 nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz), see Tables S3 and S4; (−)-ESI-MS: m/z 381 [M – H]−; (+)-ESI-MS: m/z 383 [M + H]+, 365 [(M-H2O) + H]+; (−)-HRESI-MS: m/z 381.0971 [M – H]− (calcd for C21H17O7, 381.0979); (+)-HRESI-MS: m/z 365.1241 [(M-H2O) + H]+ (calcd for C21H17O6, 365.1019), 383.1355 [M + H]+ (calcd for C21H19O7, 383.1126), 405.1187 [M + Na]+ (calcd for C21H18O7Na, 405.09447), 787.2493 [2 M + Na]+ (calcd for C42H36O14Na, 787.1997).
7-Deoxyauramycinone (9-epi-7-deoxy-nogalamycinone) (4)
C21H18O7 (382); yellow solid; HPLC-Rt = 28.02 min (Supporting Information, Figures S51 – S61); UV–vis λmax 200, 228, 260, 290 (sh), 432 nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz), see Tables S3 and S4; (−)-ESI-MS: m/z 381 [M – H]−; (+)-ESI-MS: m/z 405 [M + Na]+, 383 [M + H]+, 365 [(M-H2O) + H]+; (−)-HRESI-MS: m/z 381.0971 [M – H]− (calcd for C21H17O7, 381.0979); (+)-HRESI-MS: m/z 383.1188 [M + H]+ (calcd for C21H19O7, 383.1126).
9,10-seco-7-Deoxy-nogalamycinone (5)
C21H20O7 (384); yellow solid; HPLC-Rt = 27.95 min (Supporting Information, Figures S62 – S72); UV–vis λmax 200, 228, 260, 290 (sh), 434 nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz), see Tables S3 and S4; (−)-ESI-MS: m/z 383 [M – H]−; (+)-ESI-MS: m/z 367 [(M-H2O) + H]+; (−)-HRESI-MS: m/z 383.1129 [M – H]− (calcd for C21H19O7, 383.1136); (+)-HRESI-MS: m/z 367.1066 [(M-H2O) + H]+ (calcd for C21H19O6, 367.1176), 407.0976 [M + Na]+ (calcd for C21H20O7Na, 407.1101), 791.2076 [2 M + Na]+ (calcd for C42H40O14Na, 791.2310).
4-β-d-Glucosyl-nogalamycinone (6)
C27H28O13 (560); yellow solid; HPLC-Rt = 15.54 min (Supporting Information, Figures S73 – S82); UV–vis λmax 196, 226, 260, 286 (sh), 412 nm; 1H NMR (CD3OD, 600 MHz) and 13C NMR (CD3OD, 150 MHz), see Table S5; (−)-ESI-MS: m/z 559 [M – H]−; (+)-ESI-MS: m/z 363 [(M-glucose-2H2O) + H]+; (−)-HRESI-MS: m/z 559.1459 [M – H]− (calcd for C27H27O13, 559.1457); (+)-HRESI-MS: m/z 363.0860 [(M-glucose-2H2O) + H]+ (calcd for C21H15O6, 363.0863).
Acknowledgments
Research reported in this publication was supported by the National Science Foundation under Grant No. ENG-2015951 (S.E.N.), by the National Cancer Institute of the National Institutes of Health under Award No. R15CA252830 (S.E.N.), the NIGMS-supported Center of Biomedical Research Excellence (COBRE) in Pharmaceutical Research and Innovation (CPRI, NIH P20 GM130456), the University of Kentucky College of Pharmacy, the National Center for Advancing Translational Sciences (UL1TR000117 and UL1TR001998) and a National Institutes of Health shared instrumentation Grant (S10OD28690), Novo Nordisk Foundation Grant No. NNF19OC0057511 (to M.M.-K) and Academy of Finland Grant No. 340013 (to M.M.-K.). We also acknowledge a grant from the Turku University Foundation and support from the China Scholarship Council 202107960010 (R.W.). G.v.W. was funded by ERC Advanced grant 101055020 from the European Research Council. We thank the College of Pharmacy NMR Center (University of Kentucky) for NMR support. We thank Dr. Lou Charkoudian for the gift of strain S. lividans K4-114. We thank Prof. Dr. Mervyn Bibb for the gift of strains S. coelicolor M1146 and M1152. We thank Dr. Isaac Brownell (Dermatology Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA) for the gift of Merkel cells MKL1 and MCC26.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00498.
Experimental procedure for generation of S. coelicolor M1152ΔmatAB strain, plasmid maps for vectors, sequences of BioBricks parts and coding sequences, high-resolution mass spectrometry data, NMR tables, NMR spectra, HPLC-UV–vis chromatograms, and dose–response data for human cancer cell lines (PDF)
Author Contributions
¶ R.W. and J.N. contributed equally to this work.
The authors declare the following competing financial interest(s): Material published in this report is covered under U.S. Patent Application No. 16/015,821 to Ferris State University.
Supplementary Material
References
- Hulst M. B.; Grocholski T.; Neefjes J. J. C.; van Wezel G. P.; Metsä-Ketelä M. Anthracyclines: Biosynthesis, Engineering and Clinical Applications. Nat. Prod. Rep. 2022, 39, 814. 10.1039/D1NP00059D. [DOI] [PubMed] [Google Scholar]
- Pang B.; Qiao X.; Janssen L.; Velds A.; Groothuis T.; Kerkhoven R.; Nieuwland M.; Ovaa H.; Rottenberg S.; van Tellingen O.; Janssen J.; Huijgens P.; Zwart W.; Neefjes J. Drug-Induced Histone Eviction from Open Chromatin Contributes to the Chemotherapeutic Effects of Doxorubicin. Nat. Commun. 2013, 4 (1), 1–13. 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgarhia V. B.; Strohl W. R. Minimal Streptomyces Sp. Strain C5 Daunorubicin Polyketide Biosynthesis Genes Required for Aklanonic Acid Biosynthesis. J. Bacteriol. 1997, 179 (8), 2690–2696. 10.1128/jb.179.8.2690-2696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm A.; Madduri K.; Ali A.; Hutchinson C. R. Characterization of the Streptomyces peucetius ATCC 29050 Genes Encoding Doxorubicin Polyketide Synthase. Gene 1994, 151 (1–2), 1–10. 10.1016/0378-1119(94)90625-4. [DOI] [PubMed] [Google Scholar]
- Torkkell S.; Kunnari T.; Palmu K.; Mäntsälä P.; Hakala J.; Ylihonko K. The Entire Nogalamycin Biosynthetic Gene Cluster of Streptomyces nogalater: Characterization of a 20-Kb DNA Region and Generation of Hybrid Structures. Molecular Genetics and Genomics 2001, 266 (2), 276–288. 10.1007/s004380100554. [DOI] [PubMed] [Google Scholar]
- Ylihonko K.; Tuikkanen J.; Jussila S.; Cong L.; Mäntsälä P. A Gene Cluster Involved in Nogalamycin Biosynthesis from Streptomyces nogalater: Sequence Analysis and Complementation of Early-Block Mutations in the Anthracycline Pathway. Molecular and General Genetics 1996, 251, 113–120. 10.1007/s004380050147. [DOI] [PubMed] [Google Scholar]
- Räty K.; Kantola J.; Hautala A.; Hakala J.; Ylihonko K.; Mäntsälä P. Cloning and Characterization of Streptomyces galilaeus Aclacinomycins Polyketide Synthase (PKS) Cluster. Gene 2002, 293 (1–2), 115–122. 10.1016/S0378-1119(02)00699-6. [DOI] [PubMed] [Google Scholar]
- Bao W.; Sheldon P. J.; Wendt-Pienkowski E.; Hutchinson C. R. The Streptomyces peucetius DpsC Gene Determines the Choice of Starter Unit in Biosynthesis of the Daunorubicin Polyketide. J. Bacteriol. 1999, 181 (15), 4690–4695. 10.1128/JB.181.15.4690-4695.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautala A.; Torkkell S.; Räty K.; Kunnari T.; Kantola J.; Mantsälä P.; Hakala J.; Ylihonko K. Studies on a Second and Third Ring Cyclization in Anthracycline Biosynthesis. J. Antibiot (Tokyo) 2003, 56 (2), 143–153. 10.7164/antibiotics.56.143. [DOI] [PubMed] [Google Scholar]
- Chung J.-y.; Fujii I.; Harada S.; Sankawa U.; Ebizuka Y. Expression, Purification, and Characterization of AknX Anthrone Oxygenase, Which Is Involved in Aklavinone Biosynthesis in Streptomyces galilaeus. J. Bacteriol. 2002, 184 (22), 6115–6122. 10.1128/JB.184.22.6115-6122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantola J.; Kunnari T.; Hautala A.; Hakala J.; Ylihonko K.; Mäntsälä P. Elucidation of Anthracyclinone Biosynthesis by Stepwise Cloning of Genes for Anthracyclines from Three Different Streptomyces Spp. Microbiology (Reading) 2000, 146 (1), 155–163. 10.1099/00221287-146-1-155. [DOI] [PubMed] [Google Scholar]
- Connors N. C.; Bartel P. L.; Strohl W. R. Biosynthesis of Anthracyclines: Enzymic Conversion of Aklanonic Acid to Aklavinone and ϵ-Rhodomycinone by Anthracycline-Producing Streptomycetes. J. Gen. Microbiol. 1990, 136, 1887. 10.1099/00221287-136-9-1887. [DOI] [Google Scholar]
- Sultana A.; Kallio P.; Jansson A.; Wang J. S.; Niemi J.; Mäntsälä P.; Schneider G. Structure of the Polyketide Cyclase SnoaL Reveals a Novel Mechanism for Enzymatic Aldol Condensation. EMBO J. 2004, 23 (9), 1911–1921. 10.1038/sj.emboj.7600201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkkell S.; Kunnari T.; Palmu K.; Hakala J.; Mantsala P.; Ylihonko K. Identification of a Cyclase Gene Dictating the C-9 Stereochemistry of Anthracyclines from Streptomyces nogalater. Antimicrob. Agents Chemother. 2000, 44 (2), 396–399. 10.1128/AAC.44.2.396-399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio P.; Sultana A.; Niemi J.; Mäntsälä P.; Schneider G. Crystal Structure of the Polyketide Cyclase AknH with Bound Substrate and Product Analogue: Implications for Catalytic Mechanism and Product Stereoselectivity. J. Mol. Biol. 2006, 357 (1), 210–220. 10.1016/j.jmb.2005.12.064. [DOI] [PubMed] [Google Scholar]
- Heinemann M.; Panke S. Synthetic Biology--Putting Engineering into Biology. Bioinformatics 2006, 22 (22), 2790–2799. 10.1093/bioinformatics/btl469. [DOI] [PubMed] [Google Scholar]
- Tippmann S.; Chen Y.; Siewers V.; Nielsen J. From Flavors and Pharmaceuticals to Advanced Biofuels: Production of Isoprenoids in Saccharomyces cerevisiae. Biotechnol J. 2013, 8 (12), 1435–1444. 10.1002/biot.201300028. [DOI] [PubMed] [Google Scholar]
- Yuzawa S.; Backman T. W. H.; Keasling J. D.; Katz L. Synthetic Biology of Polyketide Synthases. J. Ind. Microbiol. Biotechnol. 2018, 45 (7), 621–633. 10.1007/s10295-018-2021-9. [DOI] [PubMed] [Google Scholar]
- van Dissel D.; Claessen D.; Roth M.; van Wezel G. P. A Novel Locus for Mycelial Aggregation Forms a Gateway to Improved Streptomyces Cell Factories. Microb Cell Fact 2015, 14 (1), 44. 10.1186/s12934-015-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H.; Breitling R.; Takano E. Synthetic Biology in Streptomyces Bacteria. Methods Enzymol. 2011, 497, 485–502. 10.1016/B978-0-12-385075-1.00021-4. [DOI] [PubMed] [Google Scholar]
- van Dissel D.; Willemse J.; Zacchetti B.; Claessen D.; Pier G. B.; van Wezel G. P. Production of Poly-β-1,6-N-Acetylglucosamine by MatAB Is Required for Hyphal Aggregation and Hydrophilic Surface Adhesion by Streptomyces. Microbial Cell 2018, 5 (6), 269–279. 10.15698/mic2018.06.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylihonko K.; Hakala J.; Kunnari T.; Mantsalala P. Production of Hybrid Anthracycline Antibiotics by Heterologous Expression of Streptomycesnogalater Nogalamycin Biosynthesis Genes. Microbiology (N Y) 1996, 142 (8), 1965–1972. 10.1099/13500872-142-8-1965. [DOI] [PubMed] [Google Scholar]
- Elowitz M. B.; Leibler S. A Synthetic Oscillatory Network of Transcriptional Regulators. Nature 2000, 403 (6767), 335–338. 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Nguyen J. T.; Riebschleger K. K.; Brown K. v.; Gorgijevska N. M.; Nybo S. E. A BioBricks Toolbox for Metabolic Engineering of the Tetracenomycin Pathway. Biotechnol J. 2022, 17, 2100371 10.1002/biot.202100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty R. P.; Endy D.; Knight T. F. Engineering BioBrick Vectors from BioBrick Parts. J. Biol. Eng. 2008, 2 (1), 5. 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman M.; Logan R.; O’Brien K.; Seno E. T.; Nagaraja Rao R.; Schoner B. E. Plasmid Cloning Vectors for the Conjugal Transfer of DNA from Escherichia coli to Streptomyces Spp. Gene 1992, 116 (1), 43–49. 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Flett F.; Mersinias V.; Smith C. P.; Flett’ F.; Mersinias V.; Smith C. P. High Efficiency Intergeneric Conjugal Transfer of Plasmid DNA from Escherichia coli to Methyl DNA-Restricting Streptomycetes. FEMS Microbiol Lett. 1997, 155 (2), 223–229. 10.1016/S0378-1097(97)00392-3. [DOI] [PubMed] [Google Scholar]
- Wang W.; Li X.; Wang J.; Xiang S.; Feng X.; Yang K. An Engineered Strong Promoter for Streptomycetes. Appl. Environ. Microbiol. 2013, 79 (14), 4484–4492. 10.1128/AEM.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C.; Zhang Y.; Zhao X.; Hu Y.; Xiang S.; Miao J.; Lou C.; Zhang L.; Demain A. L. Exploiting a Precise Design of Universal Synthetic Modular Regulatory Elements to Unlock the Microbial Natural Products in Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (39), 12181–12186. 10.1073/pnas.1511027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziermann R.; Betlach M. C. Recombinant Polyketide Synthesis in Streptomyces: Engineering of Improved Host Strains. Biotechniques 1999, 26 (1), 106–110. 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P.; Bibb M. J. Engineering Streptomyces coelicolor for Heterologous Expression of Secondary Metabolite Gene Clusters. Microb Biotechnol 2011, 4 (2), 207–215. 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry C.; Pernodet J. L.; Lautru S. Modular and Integrative Vectors for Synthetic Biology Applications in Streptomyces Spp. Appl. Environ. Microbiol. 2019, 10.1128/AEM.00485-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siitonen V.; Claesson M.; Patrikainen P.; Aromaa M.; Mäntsälä P.; Schneider G.; Metsä-Ketelä M. Identification of Late-Stage Glycosylation Steps in the Biosynthetic Pathway of the Anthracycline Nogalamycin. ChemBioChem. 2012, 13 (1), 120–128. 10.1002/cbic.201100637. [DOI] [PubMed] [Google Scholar]
- MacNeil D. J.; Gewain K. M.; Ruby C. L.; Dezeny G.; Gibbons P. H.; MacNeil T. Analysis of Streptomycesavermitilis Genes Required for Avermectin Biosynthesis Utilizing a Novel Integration Vector. Gene 1992, 111 (1), 61–68. 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- Liu X.; Hua K.; Liu D.; Wu Z. L.; Wang Y.; Zhang H.; Deng Z.; Pfeifer B. A.; Jiang M. Heterologous Biosynthesis of Type II Polyketide Products Using E. coli. ACS Chem. Biol. 2020, 15 (5), 1177–1183. 10.1021/acschembio.9b00827. [DOI] [PubMed] [Google Scholar]
- Knight T. Idempotent Vector Design for Standard Assembly of Biobricks Idempotent Vector Design for Standard Assembly of Biobricks. Structure 2003, 1–11.12517331 [Google Scholar]
- Zhang W.; Watanabe K.; Wang C. C. C.; Tang Y. Heterologous Biosynthesis of Amidated Polyketides with Novel Cyclization Regioselectivity from Oxytetracycline Polyketide Synthase. J. Nat. Prod 2006, 69 (11), 1633–1636. 10.1021/np060290i. [DOI] [PubMed] [Google Scholar]
- Wohlert S. E.; Wendt-Pienkowski E.; Bao W.; Hutchinson C. R. Production of Aromatic Minimal Polyketides by the Daunorubicin Polyketide Synthase Genes Reveals the Incompatibility of the Heterologous DpsY and JadI Cyclases. J. Nat. Prod 2001, 64 (8), 1077–1080. 10.1021/np010067f. [DOI] [PubMed] [Google Scholar]
- Bibb M. J.; Janssen G. R.; Ward J. M. Cloning and Analysis of the Promoter Region of the Erythromycin-Resistance Gene (ErmE) of Streptomyces erythraeus. Gene 1986, 41 (2–3), E357. 10.1016/0378-1119(86)90122-8. [DOI] [PubMed] [Google Scholar]
- Nybo S. E.; Saunders J.; McCormick S. P. Metabolic Engineering of Escherichia coli for Production of Valerenadiene. J. Biotechnol. 2017, 262, 60–66. 10.1016/j.jbiotec.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Wardell J. N.; Stocks S. M.; Thomas C. R.; Bushell M.E. Decreasing the Hyphal Branching Rate of Saccharopolyspora Erythraea NRRL 2338 Leads to Increased Resistance to Breakage and Increased Antibiotic Production. Biotechnol. Bioeng. 2002, 78 (2), 141–146. 10.1002/bit.10210. [DOI] [PubMed] [Google Scholar]
- van Dissel D.; Claessen D.; van Wezel G. P. Morphogenesis of Streptomyces in Submerged Cultures.. Adv. Appl. Microbiol. 2014, 89, 1–45. 10.1016/B978-0-12-800259-9.00001-9. [DOI] [PubMed] [Google Scholar]
- van Wezel G. P.; Krabben P.; Traag B.ør. A.; Keijser B. J. F.; Kerste R.; Vijgenboom E.; Heijnen J. J.; Kraal B. Unlocking Streptomyces Spp. for Use as Sustainable Industrial Production Platforms by Morphological Engineering.. Appl. Environ. Microbiol. 2006, 72 (8), 5283–5288. 10.1128/AEM.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.; Zhang L.; Barton K. W.; Zhao H. Systematic Identification of a Panel of Strong Constitutive Promoters from Streptomyces albus. ACS Synth. Biol. 2015, 4 (9), 1001–1010. 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- Yang D.; Jang D.; Lee S. Y. Production of Carminic Acid by Metabolically Engineered Escherichia coli. Cite This: J. Am. Chem. Soc. 2021, 143, 5364–5377. 10.1021/jacs.0c12406. [DOI] [PubMed] [Google Scholar]
- Ji C. H.; Kim J. P.; Kang H. S. Library of Synthetic Streptomyces Regulatory Sequences for Use in Promoter Engineering of Natural Product Biosynthetic Gene Clusters. ACS Synth. Biol. 2018, 7, 1946–1955. 10.1021/acssynbio.8b00175. [DOI] [PubMed] [Google Scholar]
- Kendrew S. G.; Katayama K.; Deutsch E.; Madduri K.; Hutchinson C. R. DnrD Cyclase Involved in the Biosynthesis of Doxorubicin: Purification and Characterization of the Recombinant Enzyme †. Biochemistry 1999, 38 (15), 4794–4799. 10.1021/bi9827924. [DOI] [PubMed] [Google Scholar]
- Gaspar P.; Oliveira J. L.; Frommlet J.; Santos M. A. S.; Moura G. EuGene: Maximizing Synthetic Gene Design for Heterologous Expression. Bioinformatics 2012, 28 (20), 2683–2684. 10.1093/bioinformatics/bts465. [DOI] [PubMed] [Google Scholar]
- Al-Hawash A. B.; Zhang X.; Ma F. Strategies of Codon Optimization for High-Level Heterologous Protein Expression in Microbial Expression Systems. Gene Rep 2017, 9, 46–53. 10.1016/j.genrep.2017.08.006. [DOI] [Google Scholar]
- Hu S.; Wang M.; Cai G.; He M. Genetic Code-Guided Protein Synthesis and Folding in Escherichia coli. J. Biol. Chem. 2013, 288 (43), 30855–30861. 10.1074/jbc.M113.467977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander I. M.; Chaney J. L.; Clark P. L. Expanding Anfinsen’s Principle: Contributions of Synonymous Codon Selection to Rational Protein Design. J. Am. Chem. Soc. 2014, 136 (3), 858–861. 10.1021/ja411302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui C.; Chen J.; Xie Q.; Mo X.; Zhang S.; Zhang H.; Ma J.; Li Q.; Gu Y. C.; Ju J. CytA, a Reductase in the Cytorhodin Biosynthesis Pathway, Inactivates Anthracycline Drugs in Streptomyces. Commun. Biol. 2019, 10.1038/s42003-019-0699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S. K. Structure of Aklavinone, a DNA Binding Anthracycline Antibiotic. J. Antibiot (Tokyo) 1985, 38 (12), 1788–1791. 10.7164/antibiotics.38.1788. [DOI] [PubMed] [Google Scholar]
- Fujiwara A.; Hoshino T.; Tazoe M.; Fujiwara M. Auramycins and Sulfurmycins, New Anthracycline Antibiotics: Characterization of Aglycones, Auramycinone and Sulfurmycinone. J. Antibiot (Tokyo) 1981, 34 (5), 608–610. 10.7164/antibiotics.34.608. [DOI] [PubMed] [Google Scholar]
- Muroi T.; Kokuzawa T.; Kihara Y.; Kobayashi R.; Hirano N.; Takahashi H.; Haruki M. TG1 Integrase-Based System for Site-Specific Gene Integration into Bacterial Genomes. Appl. Microbiol. Biotechnol. 2013, 97 (9), 4039–4048. 10.1007/s00253-012-4491-4. [DOI] [PubMed] [Google Scholar]
- Morita K.; Yamamoto T.; Fusada N.; Komatsu M.; Ikeda H.; Hirano N.; Takahashi H. The Site-Specific Recombination System of Actinophage TG1. FEMS Microbiol Lett. 2009, 297 (2), 234–240. 10.1111/j.1574-6968.2009.01683.x. [DOI] [PubMed] [Google Scholar]
- Adnani N.; Chevrette M. G.; Adibhatla S. N.; Zhang F.; Yu Q.; Braun D. R.; Nelson J.; Simpkins S. W.; McDonald B. R.; Myers C. L.; Piotrowski J. S.; Thompson C. J.; Currie C. R.; Li L.; Rajski S. R.; Bugni T. S. Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chem. Biol. 2017, 12, 3093 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R.; Langner A.; Chang A. C. Y.; Cohen S. N.; Bujard H. Cloning and Analysis of Strong Promoters Is Made Possible by the Downstream Placement of a RNA Termination Signal. Proc. Natl. Acad. Sci. U. S. A. 1981, 78 (8), 4936–4940. 10.1073/pnas.78.8.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka J.; Kunisawa T. Characteristic Base Sequence Patterns of Promoter and Terminator Sites in ΦX174 and Fd Phage DNAs. J. Theor. Biol. 1982, 97 (3), 415–436. 10.1016/0022-5193(82)90374-5. [DOI] [PubMed] [Google Scholar]
- Huff J.; Czyz A.; Landick R.; Niederweis M. Taking Phage Integration to the next Level as a Genetic Tool for Mycobacteria. Gene 2010, 468 (1–2), 8–19. 10.1016/j.gene.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.; Wang M.; Wang S.; Xiong L.; Gao B.; Liu M.; Tao X.; Wang F.-Q.; Wei D. A Self-Sustained System Spanning the Primary and Secondary Metabolism Stages to Boost the Productivity of Streptomyces. ACS Synth. Biol. 2022, 11, 353-365. 10.1021/acssynbio.1c00473. [DOI] [PubMed] [Google Scholar]
- Li L.; Zheng G.; Chen J.; Ge M.; Jiang W.; Lu Y. Multiplexed Site-Specific Genome Engineering for Overproducing Bioactive Secondary Metabolites in Actinomycetes. Metab Eng. 2017, 40, 80–92. 10.1016/j.ymben.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Siegl T.; Tokovenko B.; Myronovskyi M.; Luzhetskyy A. Design, Construction and Characterisation of a Synthetic Promoter Library for Fine-Tuned Gene Expression in Actinomycetes. Metab. Eng. 2013, 19, 98-106. 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Siitonen V.; Selvaraj B.; Niiranen L.; Lindqvist Y.; Schneider G.; Metsä-Ketelä M. Divergent Non-Heme Iron Enzymes in the Nogalamycin Biosynthetic Pathway. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (19), 5251. 10.1073/pnas.1525034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocholski T.; Yamada K.; Sinkkonen J.; Tirkkonen H.; Niemi J.; Metsä-Ketelä M. Evolutionary Trajectories for the Functional Diversification of Anthracycline Methyltransferases. ACS Chem. Biol. 2019, 14 (5), 850–856. 10.1021/acschembio.9b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. V.; Wandi B. N.; Metsa-Ketela M.; Nybo S. E. Pathway Engineering of Anthracyclines: Blazing Trails in Natural Product Glycodiversification. J. Org. Chem. 2020, 85 (19), 12012–12023. 10.1021/acs.joc.0c01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng F.; Savkur R.; Fokt I.; Przewloka T.; Priebe W.; Chaires J. B. Base Specific and Regioselective Chemical Cross-Linking of Daunorubicin to DNA. J. Am. Chem. Soc. 1996, 118 (20), 4731–4738. 10.1021/ja9542606. [DOI] [Google Scholar]
- Fu H.; McDaniel R.; Hopwood D. A.; Khosla C. Engineered Biosynthesis of Novel Polyketides: Stereochemical Course of Two Reactions Catalyzed by a Polyketide Synthase. Biochemistry 1994, 33 (31), 9321–9326. 10.1021/bi00197a036. [DOI] [PubMed] [Google Scholar]
- McDaniel R.; Ebert-Khosla S.; Hopwood D. A.; Khosla C. Engineered Biosynthesis of Novel Polyketides: Manipulation and Analysis of an Aromatic Polyketide Synthase with Unproven Catalytic Specificities. J. Am. Chem. Soc. 1993, 115 (25), 11671–11675. 10.1021/ja00078a002. [DOI] [Google Scholar]
- McDaniel R.; Ebert-Khosla S.; Fu H.; Hopwood D. A.; Khosla C. Engineered Biosynthesis of Novel Polyketides: Influence of a Downstream Enzyme on the Catalytic Specificity of a Minimal Aromatic Polyketide Synthase. Proc. Natl. Acad. Sci. U. S. A. 1994, 91 (24), 11542–11546. 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel R.; Ebert-Khosla S.; Hopwood D. A.; Khosla C. Engineered Biosynthesis of Novel Polyketides. Science 1993, 262 (5139), 1546–1550. 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- McDaniel R.; Khosla C.; Hutchinson C. R. Engineered Biosynthesis of Novel Polyketides: Analysis of TcmN Function in Tetracenomycin Biosynthesis. J. Am. Chem. Soc. 1995, 117 (26), 6805–6810. 10.1021/ja00131a001. [DOI] [Google Scholar]
- Weissman K. J.; Leadlay P. F. Combinatorial Biosynthesis of Reduced Polyketides. Nat. Rev. Microbiol. 2005, 3, 925–936. 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- Weissman K. J. Genetic Engineering of Modular PKSs: From Combinatorial Biosynthesis to Synthetic Biology. Nat. Prod Rep 2016, 33 (2), 203–230. 10.1039/C5NP00109A. [DOI] [PubMed] [Google Scholar]
- Pahari P.; Kharel M. K.; Shepherd M. D.; van Lanen S. G.; Rohr J. Enzymatic Total Synthesis of Defucogilvocarcin M and Its Implications for Gilvocarcin Biosynthesis. Angew. Chem., Int. Ed. Engl. 2012, 51 (5), 1216–1220. 10.1002/anie.201105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Chen J.; Zhu H.; Rohr J. One-Pot Enzymatic Total Synthesis of Presteffimycinone, an Early Intermediate of the Anthracycline Antibiotic Steffimycin Biosynthesis. Org. Lett. 2017, 19 (3), 540–543. 10.1021/acs.orglett.6b03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poust S.; Hagen A.; Katz L.; Keasling J. D. Narrowing the Gap between the Promise and Reality of Polyketide Synthases as a Synthetic Biology Platform. Current Opinion in Biotechnology. 2014, 30, 32–39. 10.1016/j.copbio.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Blanchard S.; Thorson J. S. Enzymatic Tools for Engineering Natural Product Glycosylation. Curr. Opin Chem. Biol. 2006, 10 (3), 263–271. 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Williams G. J.; Zhang C.; Thorson J. S. Expanding the Promiscuity of a Natural-Product Glycosyltransferase by Directed Evolution. Nat. Chem. Biol. 2007, 3 (10), 657–662. 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- Williams G. J.; Goff R. D.; Zhang C.; Thorson J. S. Optimizing Glycosyltransferase Specificity via “Hot Spot” Saturation Mutagenesis Presents a Catalyst for Novobiocin Glycorandomization. Chem. Biol. 2008, 15 (4), 393–401. 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt R. W.; Peltier-Pain P.; Singh S.; Zhou M.; Thorson J. S. Broadening the Scope of Glycosyltransferase-Catalyzed Sugar Nucleotide Synthesis. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (19), 7648–7653. 10.1073/pnas.1220220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulst M. B.; Grocholski T.; Neefjes J. J. C.; van Wezel G. P.; Metsä-Ketelä M. Anthracyclines: Biosynthesis, Engineering and Clinical Applications. Nat. Prod. Rep. 2022, 39, 814. 10.1039/D1NP00059D. [DOI] [PubMed] [Google Scholar]
- Komatsu Y.; Takahashi O.; Hayashi H. Identification of the Anthracycline Antibiotic 4-O-(β-D-Glucopyranuronosyl)-ε-Rhodomycinone, Produced by Streptomyces ruber JCM3131, as an Up-Regulator of MHC Class-I Molecules in B16/BL6 Cells. J. Antibiot (Tokyo) 1998, 51 (1), 85–88. 10.7164/antibiotics.51.85. [DOI] [PubMed] [Google Scholar]
- Sambrook J.; W Russell D.. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2001; p 999. [Google Scholar]
- Kieser T.; Bibb M. J.; Buttner M. J.; Chater K. F.; Hopwood D. A.. Practical Streptomyces Genetics; John Innes Centre Ltd., 2000; p 529. 10.4016/28481.01 [DOI] [Google Scholar]
- Ylihonko K.; Hakala J.; Niemi J.; Lundell J.; Mantsala P. Isolation and Characterization of Aclacinomycin A-Non-Producing Streptomyces galilaeus (ATCC 31615) Mutants. Microbiology (N Y) 1994, 140 (6), 1359–1365. 10.1099/00221287-140-6-1359. [DOI] [PubMed] [Google Scholar]
- Savi D. C.; Shaaban K. A.; Gos F. M. W. R.; Ponomareva L. V.; Thorson J. S.; Glienke C.; Rohr J. Phaeophleospora Vochysiae Savi & Glienke Sp. Nov. Isolated from Vochysia divergens Found in the Pantanal, Brazil, Produces Bioactive Secondary Metabolites OPEN. Sci. Rep. 2018, 8, 3122. 10.1038/s41598-018-21400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban K. A.; Elshahawi S. I.; Wang X.; Horn J.; Kharel M. K.; Leggas M.; Thorson J. S. Cytotoxic Indolocarbazoles from Actinomadura melliaura ATCC 39691. J. Nat. Prod. 2015, 78 (7), 1723–1729. 10.1021/acs.jnatprod.5b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Shaaban K. A.; Elshahawi S. I.; Ponomareva L. v; Sunkara M.; Zhang Y.; Copley G. C.; Hower J. C.; Morris A. J.; Kharel M. K.; Thorson J. S. Frenolicins C–G, Pyranonaphthoquinones from Streptomyces Sp. RM-4-15. J. Nat. Prod. 2013, 76 (8), 1441-1447. 10.1021/np400231r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban K. A.; Wang X.; Elshahawi S. I.; Ponomareva L. v.; Sunkara M.; Copley G. C.; Hower J. C.; Morris A. J.; Kharel M. K.; Thorson J. S. Herbimycins D-F, Ansamycin Analogues from Streptomyces Sp. RM-7–15. J. Nat. Prod. 2013, 76 (9), 1619–1626. 10.1021/np400308w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.