Abstract
Background
Genome-wide association studies (GWAS) have identified some variants associated with subclinical atherosclerosis (SCA) in general population but lacking sufficient validation. Besides traditional risk factors, whether and how would genetic variants associate with SCA among people with HIV (PWH) remains to be elucidated.
Method
A large original GWAS and gene-environment interaction analysis of SCA were conducted among Chinese PWH (n = 2850) and age/sex-matched HIV-negative controls (n = 5410). Subgroup analyses by age and functional annotations of variants were also performed.
Results
Different from HIV-negative counterparts, host genome had a greater impact on young PWH rather than the elders: one genome-wide significant variant (rs77741796, P = 2.20 × 10−9) and eight suggestively significant variants (P < 1 × 10−6) were identified to be specifically associated with SCA among PWH younger than 45 years. Seven genomic loci and 15 genes were mapped to play a potential role on SCA among young PWH, which were enriched in the biological processes of atrial cardiac muscle cell membrane repolarization and molecular function of protein kinase A subunit binding. Furthermore, genome-wide interaction analyses revealed significant HIV-gene interactions overall as well as gene-environment interactions with alcohol consumption, tobacco use and obesity among PWH. The identified gene-environment interaction on SCA among PWH might be useful for discovering high-risk individuals for the prevention of SCA, particularly among those with tobacco use and alcohol consumption.
Conclusion
The present study provides new clues for the genetic contribution of SCA among young PWH and is the starting point of precision intervention targeting HIV-related atherosclerosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-022-03817-6.
Keywords: Subclinical atherosclerosis, HIV, GWAS, Interaction, Chinese
Background
Cardiovascular diseases (CVDs) have been identified as a major cause of death among people with HIV (PWH) in the antiretroviral therapy (ART) era [1]. Most forms of CVDs originate from atherosclerosis, a chronic inflammatory disease of blood vessels among elderly population [2]. Of note, there is an increase in incidental atherosclerosis among PWH [3], and HIV infection appears to increase the risk of carotid plaque [4, 5]. Atherosclerosis starts early in life and progresses silently [2] and thus it is necessary to identify atherosclerosis from early subclinical stages. We previously observed a disproportionally higher risk and earlier onset of subclinical atherosclerosis (SCA) among young PWH than HIV-negative counterparts in the Comparative HIV and Aging Research in Taizhou (CHART) cohort [6]. This age-specific association between HIV and SCA is independent of traditional risk factors of CVDs and suggestive of unrecognized unique mechanisms linking HIV infection with SCA [6].
Atherosclerosis is a complex disease with the involvement of multiple factors such as smoking, alcohol use and genetics [7]. It has been reported that genetics plays a vital role in atherosclerosis development [8], accounting for 30–50% of the variance in SCA [9]. Genome-wide association studies (GWAS) and meta-analyses have identified a number of genetic variants that contribute to the risk of SCA in the general population [10–12]. However, whether and how would the genetic variants associate with SCA differentially among PWH remains to be elucidated, especially in Asian people. The contributing effect of HIV infection could involve different sets of genes and biological pathways in SCA development [9]. A GWAS study conducted in 2010 reported two SNPs (rs2229116 and rs7177922) in tight linkage disequilibrium (LD) in the RYR3 gene associated with SCA in 171 White HIV-infected men, which was also the only GWAS in relation to SCA among PWH [13].
Therefore, in the present study, we conducted a large GWAS of SCA among Chinese PWH and HIV-negative counterparts based on the CHART cohort in an attempt to compare the differences of genetic associations with SCA between these two groups. The possible underlying mechanism of earlier onset of SCA among PWH as previously revealed [6] was explored by age-specific stratified analyses. Furthermore, genome-wide gene-environment interaction analyses of SCA that incorporate HIV infection, alcohol consumption, tobacco use and obesity were also performed.
Methods
Study design and participants
Participants were enrolled from the CHART cohort, which is an ongoing prospective cohort study specifically designed to facilitate epidemiological and pathophysiological understandings of aging-related comorbidities among Chinese PWH and comparative HIV-negative individuals [14]. The present cross-sectional study was based on the baseline survey of CHART conducted in 2017–2020. Details about the CHART cohort have been described elsewhere [6].
As of Jan. 2020, an aggregate of 8260 including 2850 PWH and 5410 HIV-negative individuals were enrolled. Eventually included in the analyses were 7904 (95.7%) without missing data on cIMT and after genotyping quality control. Written informed consent was obtained from all study participants. The study was approved by the Institutional Review Board of Fudan University School of Public Health, Shanghai, China.
Data collection and measurements
Questionnaire interview and physical examination
A standardized structured questionnaire was administered face-to-face by trained health staffs to collect information on age, sex, tobacco and regular alcohol use, physical activities and history of non-communicable diseases (NCDs). Regular alcohol use was defined as alcohol use at least 3 times per week. Smoking status was classified as “never”, “previous” or “current”, with current smoking defined as having smoked at least one cigarette in the past 30 days. Physical examinations of waist circumference, hip circumference, height, weight and blood pressures (BP) were carried out. Body mass index (BMI) was calculated and general obesity was defined as BMI ≥ 24 kg/m2. The cutoff of waist to hip ratio (WHR) for abdominal obesity was defined as 0.90 for men and 0.85 for women [14].
Hypertension was defined as systolic BP ≥ 140 mmHg or diastolic BP ≥ 90 mmHg, or prior clinical diagnosis of hypertension [15]. Diabetes was defined as HbA1c ≥ 6.5% or a prior clinical diagnosis. Metabolic syndrome (MS) was defined according to standardized protocol [16]. Dyslipidemia was defined as TC ≥ 6.2 mmol/L, LDL ≥ 4.1 mmol/L, HDL < 1.0 mmol/L or TG ≥ 2.3 mmol/L [17]. HIV-related variables were extracted from the national HIV/AIDS Comprehensive Response Information Management System (CRIMS) [18]. Nadir CD4 count was defined as the lowest CD4 count as recorded.
SCA measurements and outcome definition
One of the reliable and valid measure of SCA is carotid intima-media thickness (cIMT) [6, 19]. Intima-media thickness (IMT) of the left common carotid artery was measured by trained sonographers using a high-resolution B-mode ultrasound imager (LOGIQ P5 pro, GE, Indianapolis, USA), in accordance with standard procedures. Briefly, an IMT image was obtained on about 10 mm of the longitudinal carotid length which is free of plaque with an identified double-line pattern.
Subclinical carotid atherosclerosis was defined as a cIMT of 780 μm or more, according to our previous published study [6]. The average cIMT values were also categorized into < 780, 780–1000 and > 1000 μm [20]. We assigned two SCA-related phenotypes: one quantitative, using continuous cIMT values and the other categorical, termed “binary-cIMT” with the cutoff of 780 μm.
Genotyping and quality control
Genomic DNA was extracted from whole peripheral blood samples using a commercial DNA extraction kit (Qiagen) and was quantified using PicoGreen reagent (Invitrogen). We genotyped study samples for 664,165 SNPs on the Infinium™ Chinese Genotyping Array-24 v1.0 BeadChip. We then performed quality control using PLINK 1.9 [21] at sample level and at SNP level according to the following criteria: (1) individual level: call rate < 95%, gender discrepancies checking, heterozygosity rate outliers (> 6 sd.), and unexpected duplicates; (2) SNP level: missing data > 5%, minor allele frequencies (MAF) < 0.05, and deviated from Hardy–Weinberg equilibrium (HWE) (P < 10–6). Principal component analysis (PCA) was done in PLINK 1.9 for the remaining 372,728 SNPs and the first five principal components (PCs) were extracted and employed in further association analyses.
Genome-wide association (GWA) analyses
GWA analyses were performed under additive genetic effects assumption. For continuous phenotypes, linear mixed model (LMM) was applied; for dichotomous phenotypes, generalized linear mixed model (GLMM) was used. LMM-based methods are usually preferred over linear regression-based methods largely because they can account for population stratification [22, 23] and relatedness without the need to remove related individuals [24]. LMM was conducted through fastGWA model which is an extremely resource-efficient approach implemented in the GCTA software package [25, 26]. GLMM was conducted through fastGWA-GLMM which is a resource-efficient tool for GLMM based GWAS analysis for binary traits in biobank-scale data such as the UK Biobank [24]. For all analyses, we adjusted for following parameters as covariates: age (continuous variable), sex, regular alcohol use, current smoking status, BMI, and the first five PCs.
We also created quantile–quantile (QQ) plot and Manhattan plot using the R package “CMplot”. A QQ plot was used to evaluate the overall significance of the GWAS, and the deviation of the observed versus the expected distribution of the P values was represented by the inflation factor (λGC). We further performed age-specific stratified analyses both in PWH and HIV-negative counterparts. The genome-wide significance threshold was considered at P value less than 5 × 10–8, and P value less than 1 × 10–6 indicated a suggestive significance threshold [27, 28]. Plots of representative SNPs were generated using LocusZoom online software [29].
Genome-wide interaction analyses
In order to test the interaction between environmental factors and genetic variants, we conducted a genome-wide interaction analysis by including a two-way interaction parameter based on the equation: . Here, Y is the vector of the observed cIMT measurement, β0 is a constant, β1 and β2 are the main effects of SNP and environmental factors, respectively, β4 is the main effects of other covariates and β3 is the interaction term to be tested. Environmental factors included HIV infection, alcohol consumption, tobacco use and obesity, respectively.
Age-specific interaction effects of HIV infection and genetic variants on SCA were measured based on the same equation among participants under or above 45 years old (at and above 45 years old).
Statistical analyses
Comparisons of baseline characteristics, stratified by HIV serostatus, were performed using Student’s t test and analysis of variance (ANOVA) for normally distributed continuous variables, Mann Whitney U test for continuous variables with skewed distributions, and chi-square test for categorical variables. Distribution of cIMT was also analyzed. Logistic regressions were conducted to examine the association of baseline characteristics and SCA.
We also calculated unweighted and weighted genetic risk scores (GRS) of selected risk variants (P < 1 × 10–6) for SCA. To calculate GRS for the ith subject from the selected risk variants, the following formula was used [30]:
Here is the number of risk alleles for the jth SNP in the ith subject () and is the weight or coefficient of the jth SNP. Unweighted genetic risk scores simply counted the number of alleles associated with SCA an individual carried across all potential risk variants, thus giving an equal weight to all risk alleles (=1). Weighted genetic risk scores were calculated likewise, with the associated beta estimates as for each selected SNP allele count. Weighting normally results in higher specificity of the GRS by assigning more weights to variants with stronger effects. A P value less than 0.05 served as statistical significance. Data were analyzed with SAS 9.4 software (SAS Institute, Cary, NC, USA).
Functional annotation, gene mapping and gene set analysis
Functional annotation was performed with Functional Mapping and Annotation (FUMA) [31], an online platform for the functional mapping of genetic variants. We first defined ‘independent significant SNPs’ as those surpassing a predefined threshold P value (1 × 10–6) and showing moderate to low LD (r2 < 0.6). We further defined ‘lead SNPs’ as the subset of independent SNPs (r2 < 0.1). In addition, we defined genomic risk loci by merging LD blocks of independent significant SNPs that have close physical position (< 250 kb).
SNPs in genomic risk loci were mapped to genes in FUMA using three strategies: position mapping, expression quantitative trait loci (eQTL) mapping and chromatin interaction mapping. Genes implicated by mapping of GWAS SNPs were further investigated using the GENE2FUNC procedure in FUMA, which provides enrichment of the list of mapped genes in MSigDB gene sets, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Geno Oncology (GO). Details are presented in Additional files 2, 3, 4, 5, 6 and 7.
Results
Demographic characteristics and risk factors of SCA
Finally included in the analyses were 2583 PWH and 5321 HIV-negative individuals. Of them, 74.2% were male and 52.4% (4139/7904) aged less than 45 years old. The cIMT phenotype subordinated an approximately normal distribution. Demographic characteristics of participants by HIV serostatus and SCA were summarized in Table 1. PWH had a higher prevalence of SCA than HIV negative counterparts in different categorial groups. PWH who had an older age, general/abdominal obesity, regular alcohol use, current/previous smoking status, hypertension, diabetes or MS, had a higher prevalence of SCA (all P < 0.05).
Table 1.
Sociodemographic characteristics and prevalence of subclinical atherosclerosis among study participants
| Characteristics | PWH (n = 2583) | HIV-negative counterparts (n = 5321) | P valuec | ||||
|---|---|---|---|---|---|---|---|
| SCA+ | SCA− | P valuea | SCA+ | SCA− | P valueb | ||
| Overall | 940 (36.4) | 1643 (63.6) | 1516 (28.5) | 3805 (71.5) | < 0.001d | ||
| Age (years) | < 0.001 | < 0.001 | < 0.001 | ||||
| 18–29 | 71 (14.1) | 431 (85.9) | 36 (3.6) | 971 (96.4) | |||
| 30–44 | 195 (23.1) | 651 (76.9) | 238 (13.3) | 1546 (86.7) | |||
| 45–59 | 332 (43.9) | 424 (56.1) | 557 (36.2) | 980 (63.8) | |||
| 60–89 | 342 (71.4) | 137 (28.6) | 685 (69.0) | 308 (31.0) | |||
| Male | 748 (37.1) | 1269 (62.9) | 0.167 | 1214 (31.5) | 2636 (68.5) | < 0.001 | 0.762 |
| BMI (kg/m3) (n = 7900) | 0.017 | < 0.001 | < 0.001 | ||||
| < 18.5 | 87 (33.7) | 171 (66.3) | 41 (14.2) | 247 (85.8) | |||
| 18.5–24.0 | 594 (35.0) | 1104 (65.0) | 573 (23.8) | 1830 (76.2) | |||
| > 24 | 256 (41.1) | 367 (58.9) | 902 (34.3) | 1728 (65.7) | |||
| Smoking status | < 0.001 | < 0.001 | < 0.001 | ||||
| Never | 513 (32.8) | 1049 (67.2) | 688 (22.9) | 2316 (77.1) | |||
| Previous | 144 (46.3) | 167 (53.7) | 272 (52.2) | 249 (47.8) | |||
| Current | 283 (39.9) | 427 (60.1) | 556 (31.0) | 1240 (69.0) | |||
| Regular alcohol use (n = 7888) | < 0.001 | < 0.001 | < 0.001 | ||||
| Yes | 66 (55.0) | 54 (45.0) | 316 (44.1) | 400 (55.9) | |||
| No | 874 (35.5) | 1589 (64.5) | 1196 (26.2) | 3393 (73.9) | |||
| Dyslipidemia | 0.095 | < 0.001 | 0.487 | ||||
| Yes | 565 (37.8) | 931 (62.2) | 894 (32.6) | 1846 (67.4) | |||
| No | 370 (34.6) | 701 (65.4) | 621 (24.1) | 1958 (75.9) | |||
| Total cholesterol (mmol/L, mean ± SD) | 4.80 ± 1.10 | 4.65 ± 1.00 | < 0.001 | 5.19 ± 1.03 | 5.03 ± 0.98 | < 0.001 | < 0.001 |
| LDL cholesterol (mmol/L, mean ± SD) | 2.52 ± 0.81 | 2.44 ± 0.71 | 0.018 | 3.00 ± 0.86 | 2.86 ± 0.81 | < 0.001 | < 0.001 |
| HDL cholesterol (mmol/L, median, IQR) | 1.0 (0.9, 1.3) | 1.0 (0.9, 1.2) | 0.905 | 1.1 (0.9, 1.3) | 1.1 (1.0, 1.4) | < 0.001 | < 0.001 |
| Triglycerides (mmol/L, median, IQR) | 1.7 (1.2, 2.5) | 1.6 (1.1, 2.4) | 0.004 | 2.0 (1.3, 2.9) | 1.7 (1.1, 2.6) | < 0.001 | < 0.001 |
| Abdominal obesity, measured as WHR (n = 7900) | 439 (41.7) | 614 (58.3) | < 0.001 | 974 (38.0) | 1589 (62.0) | < 0.001 | < 0.001 |
| Hypertension | < 0.001 | < 0.001 | < 0.001 | ||||
| Yes | 307 (53.9) | 262 (46.1) | 821 (46.8) | 934 (53.2) | |||
| No | 633 (31.4) | 1381 (68.6) | 695 (19.5) | 2871 (80.5) | |||
| Diabetes | < 0.001 | < 0.001 | < 0.001 | ||||
| Yes | 109 (56.5) | 84 (43.5) | 307 (53.7) | 265 (46.3) | |||
| No | 831 (34.8) | 1559 (65.2) | 1209 (25.5) | 3540 (74.5) | |||
| Metabolic syndrome | < 0.001 | < 0.001 | < 0.001 | ||||
| Yes | 337 (45.5) | 403 (54.5) | 796 (41.0) | 1147 (59.0) | |||
| No | 600 (32.6) | 1239 (67.4) | 716 (21.3) | 2646 (78.7) | |||
| Baseline CD4 (cells/μl) (n = 2563) | 0.024 | ||||||
| ≤ 200 | 177 (38.2) | 286 (61.8) | |||||
| 201–350 | 255 (40.0) | 382 (60.0) | |||||
| > 350 | 500 (34.2) | 963 (65.8) | |||||
| Nadir CD4 < 200 (cells/μl) (n = 2578) | 436 (37.8) | 719 (62.2) | 0.182 | ||||
| Years since HIV diagnosis (median, IQR) | 1.2 (0.2, 4.4) | 1.6 (0.3, 4.5) | 0.059 | ||||
aCompared between SCA+ and SCA− among PWH, assessed by chi-square test, student t test and Mann Whitney U test in appropriate
bCompared between SCA+ and SCA− among HIV-negative counterparts, assessed by chi-square test, student t test and Mann Whitney U test in appropriate
cCompared between PWH and HIV-negative counterparts among those with SCA, assessed by chi-square test, student t test and Mann Whitney U test in appropriate
dCompared the prevalence of SCA between PWH and HIV negative counterparts, assessed by chi-square test
PWH: people with HIV; BMI: body mass index; LDL: low-density lipoprotein; HDL: high-density lipoprotein; SCA: subclinical atherosclerosis
The prevalence of SCA was significantly higher among PWH than HIV-negative counterparts (36.39% vs. 28.49%, P < 0.001). Compared with the 18–29 age group, the age groups of 30–44 years (aOR = 2.55, 95% CI 2.03–3.20, P < 0.001), 45–59 years (aOR = 8.22, 95% CI 6.58–10.26, P < 0.001) and 60–89 years (aOR = 29.83, 95% CI 23.57–37.75, P < 0.001) were significantly associated with SCA which was also positively correlated to general obesity (aOR = 1.38, 95% CI 1.22–1.55, P < 0.001), previous (aOR = 1.25, 95% CI 1.04–1.50, P = 0.020) or current smoking status (aOR = 1.17, 95% CI 1.02–1.35, P = 0.024) and HIV infection (aOR = 1.77, 95% CI 1.56–2.00, P < 0.001), according to multiple logistic regression analysis (Additional file 1: Table S1).
Genetic variants associated with SCA
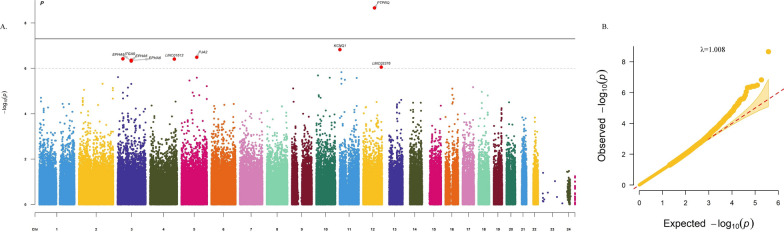
A total of 7904 participants and 372,728 SNPs were subject to final association analyses (Fig. 1). The association analyses with SCA were conducted for all participants, PWH and HIV-negative individuals, respectively. Manhattan plots and QQ plots were shown in Additional file 1: Figs. S1–6.
Fig. 1.
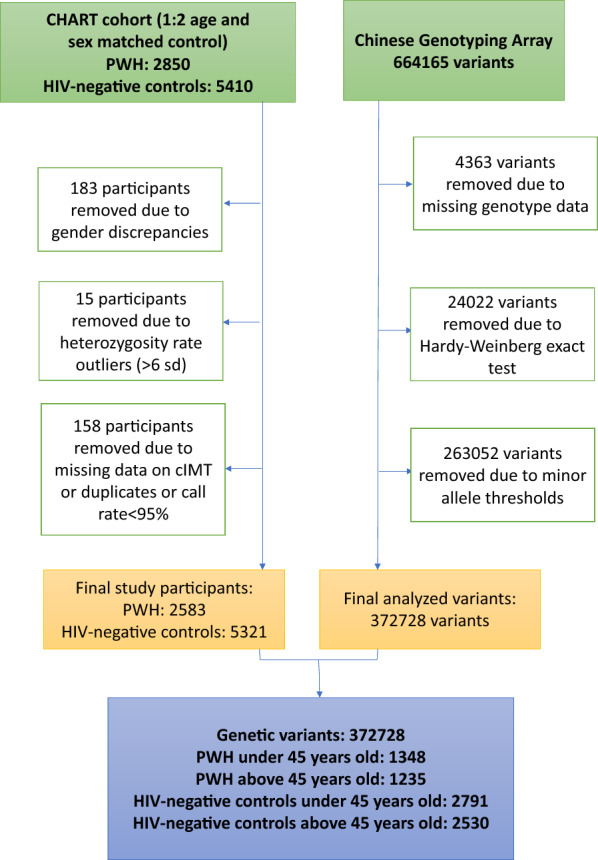
Flow chart of quality control. PWH: people with HIV
Two variants at EMC3 (rs3732968, P = 1.39 × 10–7; rs6786636, P = 7.64 × 10–7) and one variant at CRELD1 (rs2302786, P = 7.21 × 10–7) were potentially associated with cIMT among all participants (Additional file 1: Table S2). Among HIV-negative individuals, three variants at three genes reached the genome-wide association threshold for cIMT (rs2302786, P = 3.65 × 10–8, CRELD1; rs13096737, P = 4.73 × 10–8, EMC1; rs13146599, P = 4.80 × 10–8, CCSER1) and six variants at four genes reached suggestive evidence for cIMT (rs3774207, P = 1.33 × 10–7, CRELD1; rs6786636, P = 1.40 × 10–7, EMC3-AS1; rs3732968, P = 2.43 × 10–7, EMC3; rs8058808, P = 2.58 × 10–7, SLC7A5; rs3755783, P = 4.15 × 10–7, EMC3; rs10856885, P = 5.00 × 10–7, CCSER1) (Additional file 1: Table S2). Among PWH, no variant met the significant level and the most significant variant was rs6772280 (P = 3.66 × 10–6) located in the gene region of KCNAB1 on chromosome 3 (Table 2). Another genetic variant, located at the same gene, was also found among the top 4 most significant variants (rs78012168, P = 7.98 × 10–6) (Table 2).
Table 2.
Association of SNPs with cIMT among PWH in different groups
| SNP | CHR | Position (GRCh37) | Gene | Location | MAF | Minor allele | Major allele | β valuea | Adjusted P valuea |
|---|---|---|---|---|---|---|---|---|---|
| All PWH | |||||||||
| rs6772280 | 3 | 156,162,567 | KCNAB1 | Intronic | 0.109 | G | A | 0.07 | 3.66E−06 |
| rs77741796 | 12 | 80,815,650 | PTPRQ | Intergenic | 0.054 | T | C | 0.09 | 5.83E−06 |
| rs77595573 | 7 | 32,927,011 | KBTBD2 | Intronic | 0.115 | A | C | 0.07 | 6.19E−06 |
| rs78012168 | 3 | 156,150,321 | KCNAB1 | Intronic | 0.094 | A | G | 0.07 | 7.98E−06 |
| PWH under 45 years old | |||||||||
| rs77741796 | 12 | 80,815,650 | PTPRQ | Intergenic | 0.054 | T | C | 0.14 | 2.20E−09 |
| rs111815403 | 11 | 2,553,341 | KCNQ1 | Intronic | 0.061 | A | G | 0.11 | 1.50E−07 |
| rs35812497 | 5 | 108,563,812 | FER;PJA2 | Intergenic | 0.089 | A | G | 0.09 | 3.27E−07 |
| rs2507941 | 3 | 37,536,056 | ITGA9 | Synonymous | 0.064 | T | C | 0.11 | 3.85E−07 |
| rs148420952 | 4 | 171,482,271 | LINC01612 | Intergenic | 0.051 | T | C | 0.12 | 3.94E−07 |
| rs6762348 | 3 | 96,683,649 | EPHA6 | Intronic | 0.058 | G | A | 0.11 | 4.42E−07 |
| rs62263680 | 3 | 96,627,491 | EPHA6 | Intronic | 0.057 | C | T | 0.12 | 4.50E−07 |
| rs62262941 | 3 | 96,570,987 | EPHA6 | Intronic | 0.057 | C | T | 0.11 | 4.80E−07 |
| rs10847321 | 12 | 127,782,099 | LINC02376 | Intergenic | 0.056 | A | C | 0.11 | 8.91E−07 |
| PWH above 45 years old | |||||||||
| rs11948504 | 5 | 166,997,147 | TENM2 | intronic | 0.068 | G | T | 0.16 | 1.11E−06 |
| rs9851984 | 3 | 193,090,806 | ATP13A5 | intronic | 0.382 | G | A | − 0.07 | 6.51E−06 |
| rs36130341 | 7 | 343,194 | FAM20C | Intergenic | 0.273 | A | G | 0.07 | 7.70E−06 |
| rs35129955 | 4 | 181,960,818 | LINC00290 | Intergenic | 0.054 | C | T | 0.15 | 7.90E−06 |
| rs2430722 | 12 | 15,122,111 | PDE6H | Intergenic | 0.255 | C | T | − 0.08 | 1.14E−05 |
| rs6858162 | 4 | 40,555,674 | RBM47 | Intronic | 0.050 | T | C | 0.15 | 1.35E−05 |
aAssessed by linear mixed model, adjusted for age, sex, smoking status, regular alcohol use, BMI and the first five principal components of PCA
For binary cIMT, no variant reached the potential significance level among all participants, PWH and HIV negative participants (Additional file 1: Table S3).
Age-specific genetic variants associated with SCA
We further conducted stratified genetic association analyses among PWH and HIV-negative counterparts under or above 45 years old.
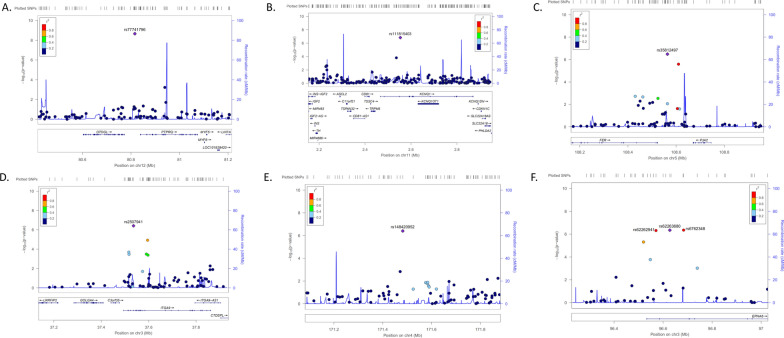
Manhattan plot and QQ plot in Fig. 2 showed the genetic association with cIMT among PWH under 45 years old. As shown in Table 2, among PWH under 45 years old, one variant at chromosome 12 (rs77741796, P = 2.20 × 10–9) reached genome-wide significance level with cIMT (Fig. 3A). Eight suggestively significant variants to cIMT were also identified, including variants located at KCNQ1 of chromosome 11 (rs111815403, P = 1.50 × 10–7) (Fig. 3B), FER/PJA2 of chromosome 5 (rs35812497, P = 3.27 × 10–7) (Fig. 3C), ITGA9 genes of chromosome 3 (rs2507941, P = 3.85 × 10–7) (Fig. 3D), and two variants located at chromosome 4 and 12, respectively (rs148420952, P = 3.94 × 10–7; rs10847321, P = 8.91 × 10–7) (Fig. 3E). Three of the eight variants were located at the EPHA6 of chromosome 3 (rs6762348, P = 4.42 × 10–7; rs62263680, P = 4.50 × 10–7; rs62262941, P = 4.80 × 10–7) (Fig. 3F). However, there was no significant or potentially significant variant associated with cIMT among PWH above 45 years old (Table 2).
Fig. 2.
Manhattan plot (A) and qqplot (B) of SNPs associated with cIMT among PWH under 45 years old. Manhattan plot showing the −log10 transformed two-tailed P value of each SNP from the GWAS on the y axis and base-pair positions along the chromosomes on the x axis. The red dots indicate SNPs surpassing Bonferroni-corrected suggestive significance threshold (P < 1 × 10–6). (n = 1348 individuals)
Fig. 3.
Regional plots of the association results for the significant susceptibility loci associated with cIMT among PWH under 45 years old. A rs77741796; B rs111815403; C rs35812497; D rs2507941; E rs148420952; F rs62263680, rs62262941, rs6762348. SNPs shown in red, orange, green, light blue and blue have r2 ≥ 0.8, r2 ≥ 0.6, r2 ≥ 0.4, r2 ≥ 0.2 and r2 < 0.2 with the tag SNP, respectively. Purple diamonds represent associations of tag SNPs identified in the GWAS stage
Among HIV-negative individuals under 45 years old, three variants were significantly associated with cIMT, two located at GRIP2 (rs34527568, P = 2.81 × 10–9; rs9863287, P = 7.04 × 10–9) and another at RBFOX1 (rs9932976, P = 4.41 × 10–8). Five variants were potentially related to cIMT (rs9938274, P = 7.13 × 10–8; rs10255973, P = 6.57 × 10–7; rs9385488, P = 9.71 × 10–7; rs118148069, P = 4.50 × 10–7; rs4485261, P = 4.77 × 10–7). Among HIV negative individuals above 45 years old, three variants (rs62487045, P = 4.53 × 10–9; rs8058808, P = 2.97 × 10–8; rs4661575, P = 3.63 × 10–8) reached genome-wide significance level with cIMT and six variants reached potential significance level (rs16891400, P = 7.43 × 10–8; rs587741, P = 1.83 × 10–7; rs28412203, P = 1.88 × 10–7; rs76462900, P = 3.65 × 10–7; rs1788783, P = 6.08 × 10–7; rs11625012, P = 8.84 × 10–7) (Additional file 1: Table S2).
For binary cIMT, no variant reached the potential significance level among PWH and HIV negative participants under or above 45 years old (Additional file 1: Table S3).
HIV-Gene interaction on SCA
Genome-wide interaction analyses were conducted among all participants and the most significant interaction effect was observed for rs78012168 at KCNAB1 (β = 0.09, Pinteract = 7.49 × 10–6)) (Table 3). Negative interactions with HIV infection were observed for the potential risk variants of cIMT to HIV-negative individuals (for three variants reaching the genome-wide association threshold: rs2302786, β = − 0.05, Pinteract = 0.001; rs13096737, β = − 0.07, Pinteract = 0.001; rs13146599, β = − 0.04, Pinteract = 0.022; and for six variants reaching suggestive association threshold: rs3774207, β = − 0.04, Pinteract = 0.002; rs6786636, β = − 0.04, Pinteract = 0.006; rs3732968, β = − 0.04, Pinteract = 0.017; rs8058808, β = − 0.05, Pinteract = 0.035; rs3755783, β = − 0.04, Pinteract = 0.006; rs10856885, β = − 0.03, Pinteract = 0.043) (Table 3). Both HIV infection and genetic variants remained significantly associated with cIMT in interaction analyses (all P < 0.001).
Table 3.
Association of SNP × HIV infection with cIMT among different groups
| SNP | CHR | Position (GRCh37) | MAF | Gene | Location | Minor allele | SNP × HIV | SNP | HIV | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β valuea | Adjusted P valuea | β valuea | Adjusted P valuea | β valuea | Adjusted P valuea | |||||||
| Among all participants | ||||||||||||
| rs78012168 | 3 | 156,150,321 | 0.098 | KCNAB1 | Intronic | A | 0.09 | 7.49E−06 | − 0.1 | 1.69E−04 | 0.04 | 3.48E−05 |
| rs1194716 | 10 | 54,196,773 | 0.199 | DKK1 | Intergenic | T | 0.06 | 1.17E−05 | − 0.08 | 4.74E−05 | 0.03 | 4.10E−03 |
| rs4814734 | 20 | 1,889,477 | 0.333 | SIRPA | Intronic | T | 0.05 | 1.33E−05 | − 0.06 | 1.98E−04 | 0.02 | 8.50E−02 |
| rs13096737 | 3 | 10,016,911 | 0.073 | EMC3 | Intronic | C | − 0.07 | 9.95E−04 | 0.14 | 5.38E−06 | 0.07 | 5.58E−13 |
| rs2302786 | 3 | 9,979,660 | 0.199 | CRELD1 | Intronic | G | − 0.05 | 1.26E−03 | 0.09 | 8.71E−06 | 0.07 | 9.41E−13 |
| rs3774207 | 3 | 9,985,656 | 0.200 | CRELD1 | Synonymous | T | − 0.04 | 2.20E−03 | 0.08 | 2.45E−05 | 0.07 | 1.91E−12 |
| rs3755783 | 3 | 10,029,289 | 0.167 | EMC3 | Intronic | G | − 0.04 | 5.90E−03 | 0.09 | 6.36E−05 | 0.07 | 3.60E−12 |
| rs6786636 | 3 | 10,045,703 | 0.170 | EMC3 | 3′-UTR | C | − 0.04 | 6.44E−03 | 0.09 | 4.89E−05 | 0.07 | 2.67E−12 |
| rs3732968 | 3 | 10,013,273 | 0.149 | EMC3 | Intronic | G | − 0.04 | 1.72E−02 | 0.09 | 2.35E−04 | 0.06 | 1.63E−11 |
| rs13146599 | 4 | 92,348,857 | 0.114 | CCSER1 | Intronic | A | − 0.04 | 2.21E−02 | 0.09 | 2.35E−04 | 0.06 | 1.63E−11 |
| rs8058808 | 16 | 87,839,943 | 0.052 | SLC7A5 | Intergenic | A | − 0.05 | 3.45E−02 | 0.12 | 4.17E−04 | 0.06 | 1.94E−11 |
| rs10856885 | 4 | 92,309,543 | 0.130 | CCSER1 | Intronic | C | − 0.03 | 4.29E−02 | 0.08 | 1.06E−03 | 0.06 | 5.64E−11 |
| Among participants under 45 years old | ||||||||||||
| rs9932976 | 16 | 6,505,665 | 0.119 | RBFOX1 | Intronic | C | − 0.09 | 3.47E−07 | 0.14 | 1.20E−08 | 0.1 | 1.38E−27 |
| rs9938274 | 16 | 6,505,208 | 0.118 | RBFOX1 | Intronic | T | − 0.09 | 4.00E−07 | 0.14 | 1.67E−08 | 0.1 | 9.96E−28 |
| rs1345439 | 16 | 48,751,012 | 0.133 | CBLN1 | Intergenic | A | 0.08 | 6.33E−07 | − 0.1 | 1.94E−05 | 0.06 | 6.29E−10 |
| rs2507941 | 3 | 37,536,056 | 0.064 | ITGA9 | Synonymous | T | 0.12 | 8.42E−07 | − 0.14 | 7.37E−05 | 0.07 | 7.07E−15 |
| rs6762348 | 3 | 96,683,649 | 0.058 | EPHA6 | Intronic | G | 0.12 | 1.93E−06 | − 0.12 | 2.80E−04 | 0.07 | 3.48E−15 |
| rs62263680 | 3 | 96,627,491 | 0.057 | EPHA6 | Intronic | C | 0.12 | 2.45E−06 | − 0.12 | 3.71E−04 | 0.07 | 2.98E−15 |
| rs62262941 | 3 | 96,570,987 | 0.057 | EPHA6 | Intronic | C | 0.12 | 3.13E−06 | − 0.12 | 4.77E−04 | 0.07 | 2.15E−15 |
| rs77741796 | 12 | 80,815,650 | 0.054 | PTPRQ | Intergenic | T | 0.11 | 8.89E−06 | − 0.09 | 1.18E−02 | 0.07 | 2.67E−16 |
| rs148420952 | 4 | 171,482,271 | 0.051 | LINC01612 | Intergenic | T | 0.11 | 4.05E−05 | − 0.09 | 1.21E−02 | 0.07 | 1.53E−16 |
| rs35812497 | 5 | 108,563,812 | 0.089 | FER, PJA2 | Intergenic | A | 0.08 | 1.14E−04 | − 0.07 | 1.53E−02 | 0.07 | 1.04E−13 |
| rs10847321 | 12 | 127,782,099 | 0.056 | LINC02376 | Intergenic | A | 0.09 | 1.69E−04 | − 0.07 | 3.06E−02 | 0.07 | 4.96E−16 |
| rs111815403 | 11 | 2,553,341 | 0.061 | KCNQ1 | Intronic | A | 0.07 | 2.11E−03 | − 0.04 | 2.65E−01 | 0.07 | 1.65E−16 |
| Among participants above 45 years old | ||||||||||||
| rs11948504 | 5 | 166,997,147 | 0.068 | TENM2 | Intronic | G | 0.19 | 7.16E−06 | − 0.21 | 2.72E−04 | − 0.01 | 9.34E−01 |
| rs2430722 | 12 | 15,122,111 | 0.255 | PDE6H | Intergenic | C | − 0.07 | 3.53E−03 | 0.08 | 1.37E−02 | 0.06 | 4.32E−03 |
| rs35129955 | 4 | 181,960,818 | 0.054 | LINC00290 | Intergenic | C | 0.12 | 6.55E−03 | − 0.11 | 6.05E−02 | 0.01 | 6.22E−01 |
| rs36130341 | 7 | 343,194 | 0.273 | FAM20C | Intergenic | A | 0.05 | 1.60E−02 | − 0.06 | 6.79E−02 | − 0.01 | 6.80E−01 |
| rs6858162 | 4 | 40,555,674 | 0.050 | RBM47 | Intronic | T | 0.11 | 1.76E−02 | − 0.09 | 1.72E−01 | 0.01 | 5.00E−01 |
| rs9851984 | 3 | 193,090,806 | 0.382 | ATP13A5 | Intronic | G | − 0.04 | 5.62E−02 | 0.03 | 3.18E−01 | 0.05 | 2.23E−02 |
aAssessed by generalized linear model, adjusted for age, sex, smoking status, regular alcohol use, BMI
Age-specific HIV-gene interactions on SCA
We then conducted genome-wide interaction analyses among participants under or above 45 years old. Among participants under 45 years old, four variants had a suggestive significant interaction effect with HIV infection (rs9932976, β = − 0.09, Pinteract = 3.47 × 10–7, RBFOX1; rs9938274, β = − 0.09, Pinteract = 4.00 × 10–7, RBFOX1; rs1345439, β = 0.08, Pinteract = 6.33 × 10–7, CBLN1; rs2507941, β = 0.12, Pinteract = 8.42 × 10–7, ITGA9) (Table 3).
Moreover, for potential risk variants of cIMT to young PWH, positive interactions of those variants with HIV infection were also observed among participants under 45 years old (rs2507941, β = 0.12, Pinteract = 8.42 × 10–7; rs6762348, β = 0.12, Pinteract = 1.93 × 10–6; rs62263680, β = 0.12, Pinteract = 2.45 × 10–6; rs62262941, β = 0.12, Pinteract = 3.13 × 10–6; rs77741796, β = 0.12, Pinteract = 8.89 × 10–6; rs148420952, β = 0.11, Pinteract = 4.05 × 10–5; rs35812497, β = 0.08, Pinteract = 1.14 × 10–4; rs10847321, β = 0.09, Pinteract = 1.69 × 10–4; rs111815403, β = 0.07, Pinteract = 0.002) (Table 3).
Among participants above 45 years old, no interaction term reached the suggestive significant level and the most significant interaction effect was observed in rs11948504 (β = 0.19, Pinteract = 7.16 × 10–6) (Table 3).
Gene interaction with potential risk factors of SCA among PWH
We also measured the interaction effect of genetic variants and traditional risk factors of SCA including alcohol consumption, tobacco use and increasing BMI category on cIMT among PWH, respectively. One variant at NKAIN2 (rs9375288, β = 0.09, Pinteract = 2.31 × 10–8) and six variants (rs817856, β = 0.39, Pinteract = 1.03 × 10–9; rs78096022, β = 0.41, Pinteract = 3.70 × 10–9; rs6513469, β = 0.50, Pinteract = 9.53 × 10–9; rs17392147, β = 0.83, Pinteract = 1.89 × 10–8; rs186333, β = 0.43, Pinteract = 2.70 × 10–8; rs78752139, β = 0.40, Pinteract = 4.28 × 10–8) had genome-wide significant interaction with alcohol consumption and tobacco use to cIMT among PWH, respectively (Table 4).
Table 4.
Association of SNP × traditional risk factors with cIMT among PWH
| CHR | SNP | Position (GRCh37) | MAF | Gene | Location | Minor allele | SNP × covariate | SNP | Covariate | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β valuea | P interaction | β valuea | P value | β valuea | P value | |||||||
| SNP × alcohol interaction | ||||||||||||
| 6 | rs9375288 | 124,194,087 | 0.123 | NKAIN2 | Intronic | G | 0.09 | 2.31E−08 | 0.03 | 8.10E−02 | 0.01 | 7.70E−01 |
| 12 | rs77741796 | 80,815,650 | 0.054 | PTPRQ | Intergenic | T | 0.10 | 6.45E−06 | 0.03 | 2.31E−01 | − 0.01 | 7.68E−01 |
| 3 | rs6762348 | 96,683,649 | 0.058 | EPHA6 | Intronic | G | 0.10 | 2.23E−05 | − 0.01 | 7.37E−01 | − 0.01 | 8.61E–01 |
| 3 | rs62262941 | 96,570,987 | 0.057 | EPHA6 | Intronic | C | 0.10 | 3.40E−05 | − 0.01 | 8.26E−01 | − 0.01 | 8.90E−01 |
| 3 | rs62263680 | 96,627,491 | 0.057 | EPHA6 | Intronic | C | 0.10 | 3.83E−05 | − 0.01 | 8.22E−01 | − 0.01 | 8.81E−01 |
| 4 | rs148420952 | 171,482,271 | 0.051 | LINC01612 | Intergenic | T | 0.09 | 1.74E−04 | 0.01 | 9.23E−01 | − 0.01 | 8.86E−01 |
| 12 | rs10847321 | 127,782,099 | 0.056 | LINC02376 | Intergenic | A | 0.05 | 1.97E−02 | 0.01 | 9.53E−01 | 0.01 | 8.51E−01 |
| SNP × tobacco interaction | ||||||||||||
| 9 | rs817856 | 110,118,754 | 0.115 | RAD23B | Intergenic | G | 0.39 | 1.03E−09 | − 0.02 | 2.15E−01 | − 0.04 | 1.81E−01 |
| 8 | rs78096022 | 132,359,092 | 0.107 | ADCY8 | Intergenic | T | 0.41 | 3.70E−09 | − 0.03 | 4.55E−02 | − 0.04 | 2.58E−01 |
| 20 | rs6513469 | 58,588,895 | 0.059 | CDH26 | 3′-UTR | C | 0.50 | 9.53E−09 | 0.01 | 7.63E−01 | − 0.03 | 3.73E−01 |
| 1 | rs17392147 | 9,060,210 | 0.054 | SLC2A7 | Intronic | C | 0.83 | 1.89E−08 | 0.02 | 4.43E−01 | 0.01 | 9.39E−01 |
| 5 | rs186333 | 23,202,461 | 0.074 | CDH12 | Intergenic | G | 0.43 | 2.70E−08 | − 0.01 | 6.68E−01 | − 0.02 | 5.47E−01 |
| 5 | rs78752139 | 103,593,227 | 0.092 | NUDT12 | Intergenic | T | 0.40 | 4.28E−08 | − 0.01 | 9.25E−01 | − 0.04 | 2.88E−01 |
| 11 | rs111815403 | 2,553,341 | 0.061 | KCNQ1 | Intronic | A | 0.22 | 3.67E−03 | 0.04 | 2.64E−02 | 0.01 | 8.74E−01 |
| 4 | rs148420952 | 171,482,271 | 0.051 | LINC01612 | Intergenic | T | − 0.24 | 1.39E−02 | 0.08 | 1.64E−04 | 0.06 | 4.87E−02 |
| 3 | rs2507941 | 37,536,056 | 0.064 | ITGA9 | Synonymous | T | 0.30 | 1.79E−02 | 0.05 | 7.73E−03 | 0.03 | 4.29E−01 |
| SNP × BMI category interaction | ||||||||||||
| 3 | rs2507941 | 37,536,056 | 0.064 | ITGA9 | Synonymous | T | 0.12 | 1.87E−04 | − 0.21 | 5.03E−03 | 0.01 | 5.19E−01 |
| 12 | rs10847321 | 127,782,099 | 0.056 | LINC02376 | Intergenic | A | 0.11 | 1.48E−03 | − 0.20 | 1.01E−02 | 0.01 | 3.62E−01 |
| 11 | rs111815403 | 2,553,341 | 0.061 | KCNQ1 | Intronic | A | 0.10 | 2.91E−03 | − 0.16 | 3.41E−02 | 0.01 | 3.77E−01 |
| 4 | rs148420952 | 171,482,271 | 0.051 | LINC01612 | Intergenic | T | 0.08 | 1.69E−02 | − 0.11 | 1.50E−01 | 0.01 | 2.86E−01 |
| 5 | rs35812497 | 108,563,812 | 0.089 | FER, PJA2 | Intergenic | A | 0.06 | 3.58E−02 | − 0.09 | 1.46E−01 | 0.01 | 3.63E−01 |
| 3 | rs6762348 | 96,683,649 | 0.058 | EPHA6 | Intronic | G | 0.06 | 4.18E−02 | − 0.09 | 2.20E−01 | 0.01 | 2.24E−01 |
aAssessed by generalized linear model, adjusted for age, sex, smoking status, regular alcohol use, BMI category
For the risk variants of cIMT to young PWH, six of them had a positive interaction effect with alcohol consumption (rs77741796, β = 0.10, Pinteract = 6.45 × 10–6; rs6762348, β = 0.10, Pinteract = 2.23 × 10–5; rs62262941, β = 0.10, Pinteract = 3.40 × 10–5; rs62263680, β = 0.10, Pinteract = 3.83 × 10–5; rs148420952, β = 0.09, Pinteract = 1.74 × 10–4; rs10847321, β = 0.05, Pinteract = 1.97 × 10–2); three of them had a significant interaction with tobacco use (rs111815403, β = 0.22, Pinteract = 3.67 × 10–3; rs148420952, β = -0.24, Pinteract = 1.39 × 10–2; rs2507941, β = 0.30, Pinteract = 1.79 × 10–2) and six of them had a positive interaction effect with increasing BMI category (rs2507941, β = 0.12, Pinteract = 1.87 × 10–4; rs10847321, β = 0.11, Pinteract = 1.48 × 10–3; rs111815403, β = 0.10, Pinteract = 2.91 × 10–3; rs148420952, β = 0.08, Pinteract = 1.69 × 10–2; rs35812497, β = 0.06, Pinteract = 3.58 × 10–2; rs6762348, β = 0.06, Pinteract = 4.18 × 10–2) (Table 4).
Summary statistics results of GWA analyses can be seen in Additional files 2, 3, 4, 5, 6 and 7.
Association of GRS with SCA
Genetic variants potentially associated with cIMT among PWH under 45 years old were selected to calculated for the unweighted and weighted GRS, and associations with cIMT and binary-cIMT were tested by GLM and logistic regression models, respectively. Both univariable and multivariable regression models were fitted adjusting for age, sex, regular alcohol use, current smoking status and BMI.
The unweighted GRS was significantly associated with cIMT level (β = 0.06, P < 2 × 10–16) and binary-cIMT (OR = 1.15, 95%CI: 1.04–1.25, P = 0.006) among PWH under 45 years old after adjustment (Additional file 1: Table S4). The weighted GRS demonstrated greater specificity for the cIMT level (β = 0.51, P = 2 × 10–16) and binary-cIMT (OR = 2.72, 95%CI: 1.15–6.42, P = 0.023) with greater evidence for association between genetic variants and SCA risk among PWH under 45 years old (Additional file 1: Table S5).
Functional annotation, gene mapping and gene set analysis
Using three gene mapping strategies in FUMA, we identified 7 genomic risk loci and 15 mapped genes associated with cIMT among PWH under 45 years old (Additional file 1: Tables S6, 7), 6 genomic risk loci and 11 mapped genes among HIV-negative counterparts under 45 years old (Additional file 1: Tables S8, 9).
Among PWH under 45 years old, positional gene mapping aligned SNPs to 5 genes by genomic location, eQTL gene mapping matched SNPs to 8 genes by expression levels they influence, and chromatin interaction mapping annotated SNPs to 4 genes on the basis of 3D DNA-DNA interactions (Additional file 1: Table S7, S10–11). Of note, the variant rs2507941 was also mapped to SCN5A gene through chromatin interaction mapping (Additional file 1: Table S7). Eleven genes were notable as they were linked via eQTL mapping or chromatin interactions between two independent genomic risk loci (Additional file 1: Figs. S7–9). Circos plot of chromatin interactions among HIV negative counterparts under 45 years old can be seen in Additional file 1: Figs. S10–13.
Gene-set based analysis was performed to further evaluate the underlying disease mechanisms responsible for the genetic signals. The 2 significant GO biological processes and 6 significant GO molecular functions were identified among PWH under 45 years old (Table 5). Among those gene sets, there were 2 GO gene sets involved in pathogenesis of SCA, including regulation of atrial cardiac muscle cell membrane repolarization (FDR = 0.034) and molecular function of protein kinase A (PKA) subunit binding (FDR = 0.018). Gene-set results among HIV-negative individuals under 45 years old were also shown in Table 5.
Table 5.
The significant gene-set analyses for cIMT among participants under 45 years old
| Category | GeneSet | N | n | P-value | Adjusted P | Genes |
|---|---|---|---|---|---|---|
| PWH under 45 years old | ||||||
| GO_bp | regulation of atrial cardiac muscle cell membrane repolarization | 8 | 2 | 4.75E−06 | 0.034 | KCNQ1, SCN5A |
| GO_bp | atrial cardiac muscle cell membrane repolarization | 11 | 2 | 9.33E−06 | 0.034 | KCNQ1, SCN5A |
| GO_mf | protein kinase A catalytic subunit binding | 13 | 2 | 1.32E−05 | 0.018 | KCNQ1, PJA2 |
| GO_mf | protein kinase A regulatory subunit binding | 20 | 2 | 3.22E−05 | 0.018 | KCNQ1, PJA2 |
| GO_mf | protein phosphatase 1 binding | 20 | 2 | 3.22E−05 | 0.018 | KCNQ1, FER |
| GO_mf | protein kinase activity | 588 | 4 | 9.15E−05 | 0.038 | DCLK3, ACVR2B, EPHA6, FER |
| HIV-negative participants under 45 years old | ||||||
| GWAS catalog reported genes | Hyperopia | 6 | 2 | 1.09E−06 | 1.98E−03 | RBFOX1, LAMA2 |
| GWAS catalog reported genes | Urinary albumin-to-creatinine ratio in non-diabetics | 19 | 2 | 1.24E−05 | 6.44E−03 | SOGA3, SOGA3, C6orf58 |
| GWAS catalog reported genes | Spherical equivalent (joint analysis main effects and education interaction) | 20 | 2 | 1.38E−05 | 6.44E−03 | RBFOX1, LAMA2 |
| GWAS catalog reported genes | Spherical equivalent or myopia (age of diagnosis) | 175 | 3 | 1.42E−05 | 6.44E−03 | RBFOX1, LAMA2, DPP6 |
| GWAS catalog reported genes | Refractive error | 29 | 2 | 2.95E−05 | 1.07E−02 | RBFOX1, LAMA2 |
| GWAS catalog reported genes | Waist-to-hip ratio adjusted for BMI | 403 | 3 | 1.69E−04 | 4.30E−02 | ECHDC1, SOGA3, SOGA3, C6orf58 |
| GWAS catalog reported genes | Intracranial aneurysm | 72 | 2 | 1.84E−04 | 4.30E−02 | RBFOX1, CARHSP1 |
| GWAS catalog reported genes | Myopia | 73 | 2 | 1.89E−04 | 4.30E−02 | RBFOX1, LAMA2 |
Results of gene-set analyses for cIMT among PWH and HIV-negative counterparts under 45 years old. Gene-set analysis used the results from genes mapped in SNP-based analysis as input. N: Genes in Gene Set; n: Genes in Overlap.GO: gene ontology
Discussion
This study for the first time comprehensively compared and evaluated the genome-wide associated variants and gene-environment interaction in relation to SCA among PWH and HIV-negative individuals in Chinese population, indicating that the host genome had a greater impact on SCA among young PWH than the elder PWH. Nine novel genetic variants, seven genomic loci and 15 mapped genes were identified to be associated with SCA among PWH under 45 years old. Genetic variants had a significant interaction with HIV infection, tobacco use, alcohol use and obesity on the development of SCA. Aggregations of the identified genetic variants were highly associated with SCA among young PWH, as predicted by GRS. Using gene-set analyses, we demonstrated that genetic variants of SCA among PWH under 45 years old pointed towards a role of genes enriched in the biological process of cardiac muscle cell repolarization and molecular function of PKA subunit binding.
We previously reported that SCA could occur early in young HIV-infected adults in the CHART cohort [6]. Based on the same cohort, we found in the present study that one significant variant rs77741796 near PTPRQ gene and eight suggestive significant variants at KCNQ1/FER/PJA2/ITGA9/EPHA6 genes were associated with SCA among PWH under 45 years old (Table 2). There was no significant variant associated with SCA among PWH above 45 years old but significant variants can be found among HIV-negative individuals both under and above 45 years old. These results indicated that genetic predisposition may play a crucial role in the development of SCA among young HIV-infected adults instead of old PWH. Elderly PWH population usually have a higher prevalence of multimorbidity and traditional risk factors of CVDs, such as hypertension and metabolic syndrome [6, 32], and thus the role of genetics may be overshadowed. On the contrary, among young PWH with less traditional risk factors and accordingly lower prevalence of CVDs, the role of genetics may become prominent in the debut and progression of atherosclerosis. To what extent and how will the genetic variants impact on the development of SCA among PWH remains to be addressed in longitudinal prospective cohort studies.
In genome-wide interaction analyses among participants under 45 years old, we identified four genetic variants at RBFOX1/CBLN1/ITGA9 that had a suggestively significant interaction with HIV infection, indicating an age-specific interaction effect of HIV infection and genetic variant on the development of SCA. rs2507941 at ITGA9 was associated with cIMT both in GWA among young PWH and genome-wide interaction analyses. The protein that ITGA9 encodes can improve cell migration and regulate various cellular biological functions [33]. It was also reported that human ITGA9 was associated with blood pressure and linked to cardiovascular phenotypes [34]. The protein that RBFOX1 encodes is a muscle-specific isoform of an RNA splicing regulator and previous study identified that regulation of RNA splicing by RBFOX1 played a crucial role in transcriptome reprogramming during heart failure [35]. CBLN1 encodes a cerebellum-specific precursor protein that establishes parallel fiber-Purkinje cell synapses [36] but its role in SCA development was firstly reported.
For risk variants of cIMT to young PWH, a positive interaction effect with HIV infection was also identified among all young participants. This might be partially owing to a mixture of accelerated aging due to HIV infection and host genomic effects in the HIV-infected youngsters who had less traditional risk factors for SCA. In addition, risk variants of cIMT to HIV-negative individuals also had a negative interaction with HIV infection among all participants. The significant variants related to SCA among PWH and HIV-negative counterparts under 45 years old were also different. The underlying mechanism might be attributable to the integration of HIV proviral DNA into host genome, which could affect expression of host genes, influence basal and inducible transcription [37, 38], and thus manifest differential associations of genetic variants with SCA between comparable PWH and HIV-negative counterparts.
Genome-wide interaction analyses with traditional risk factors of SCA were also performed among PWH. These analyses identified variants at NKAIN2/RAD23B/ADCY8/CDH26/SLC2A7/CDH12/NUDT12 had a genome-wide significant interaction with alcohol consumption or tobacco use (Table 4). Previous study also revealed the strong association of NKAIN2 with alcohol dependence [39] and nicotine dependence [40]. The protein that RAD23B encodes is shown to elevate the nucleotide excision activity of 3-methyladenine-DNA glycosylase and plays a role in DNA damage recognition in base excision repair [41], the latter of which was usually caused by tobacco usage [42]. It was also reported that overexpression of a neuronal ADCY8 in sinoatrial node markedly impacted on heart rate and rhythm [43]. Cadherins (CDHs) formed adherens junctions and were known stabilizers of atherosclerotic plaques [44]. Overexpression of CDH12 and CDH26 might be related to myocardial infarction and progression of atherosclerosis [44]. SLC2A7 encodes a protein that catalyzes the uptake of sugars [45] through facilitated diffusion while NUDT12 regulates the concentrations of individual nucleotides [46], but their links to SCA were first reported in our study. Potential risk variants of cIMT to young PWH also had an interaction with alcohol consumption, tobacco use and obesity. These results strongly highlight the importance of controlling traditional risk factors of SCA, such as reducing alcohol use, smoking cessation and maintaining a good weight among PWH carrying high-risk alleles in an attempt to reduce SCA risk.
Using functional annotation of associated genetic variants, we found variants at KCNQ1 and SCN5A were associated with SCA among PWH under 45 years old. The KCNQ1 gene encodes a voltage-gated potassium channel required for repolarization phase of the cardiac action potential [47]. A cohort study in Japan has reported that SNPs at KCNQ1 were significantly associated with coronary epicardial endothelial dysfunction [48]. Animal experiment has confirmed that an imprinted antisense IncRNA in the KCNQ1 gene promotes macrophage lipid accumulation and accelerates the development of atherosclerosis through the miR-452-3p/HDAC3/ABCA1 pathway [8]. Protein encoded by SCN5A was primarily found in cardiac muscle and defects in this gene have been associated with atrial fibrillation (AF) and cardiomyopathy [49]. Previous study also indicated variants at SCN5A were related to increased AF risk and PR interval [50] but its relation to SCA was firstly reported.
Moreover, three SNPs-rs6762348, rs62263680 and rs62262941 located at EPHA6 on chromosome 3 were identified to be associated with SCA among PWH under 45 years old. EPHA6 gene is predicted to enable transmembrane-ephrin receptor activity and is found to be associated with insulin signaling [51] and blood pressure phenotype [52], which are the known risk factors of atherosclerosis. We also identified that genetic variants at ITGA9, FER, PJA2, PTPRQ genes were significantly associated with SCA among PWH under 45 years old. Variants near PTPRQ reached genome-wide significance to cIMT among young PWH; this gene encodes a member of the type III receptor-like protein-tyrosine phosphatase family, playing roles in cellular proliferation and differentiation [53], which might have a link to cardiovascular disease [54]. FER regulated cell–cell adhesion and absence of FER protein tyrosine kinase could induce epithelial barrier dysfunction [55] which was regarded as a hallmark of many human panvascular diseases, including atherosclerosis, hypertension and diabetes [56]. One study demonstrated the association of PJA2 with atherosclerosis through protein–protein interaction network analysis [57]. The unweighted and weighted GRSs were significantly associated with SCA among PWH under 45 years old, which might be used as the predictive biomarker panel of SCA among young HIV-infected adults.
The gene set analyses revealed that genes related to SCA among PWH under 45 years old were enriched in regulation of atrial cardiac muscle cell membrane repolarization and molecular function of protein kinase A (PKA) catalytic subunit binding. KCNQ1 and SCN5A participated in the regulation of atrial cardiac muscle cell membrane repolarization which was involved in the process that modulates the establishment or extent of a membrane potential in the polarizing direction towards the resting potential in an atrial cardiomyocyte [58]. Dysregulation of atrial cardiac muscle cell membrane repolarization is related to long QT syndrome, sudden cardiac death, cardiac death and death from any cause [59–61]. KCNQ1 and PJA2 were involved in the catalytic subunit binding of PKA which is one of the master regulatory molecules in the heart. It has been reported that persistent activation of PKA signaling was linked to pathological hypertrophy and the progression to heart failure [62].
To our knowledge, this is the largest GWAS of SCA among comparative PWH and HIV-negative counterparts in Asia, and is the first that measured the genome-wide interaction effect of environmental factors and genetic variants on SCA. Nevertheless, our study has several limitations. First, replication study was not conducted, which may reduce the robustness of our results to some extent. However, using the stringent P-value could reduce the false discovery rate and candidate SNPs were presented for future validation. Second, since all genetic data were available within one cohort and were obtained using a single chip, no imputation of SNP genotypes was performed. Results of imputation analyses will also be reported in future work. Last, sample size for PWH under 45 years old was relatively small, although genome-wide significant variants were still identified. Future studies with a larger sample size are needed to validate these results.
Conclusion
In summary, the present GWAS indicated a greater impact of host genome on SCA among young Chinese PWH, as well as the interaction effects between genetic variants and environmental factors on HIV-related SCA development. Nine genetic variants, seven genomic loci and 15 mapped genes were identified to be associated with SCA among PWH under 45 years old. Pathways related to biological processes of atrial cardiac muscle cell membrane repolarization and molecular function of PKA subunit binding were implicated in pathogenesis of SCA in HIV-infected youngsters. Furthermore, the identified gene-environment interaction on SCA among PWH might be useful for discovering high-risk individuals for the prevention of SCA, particularly among those with tobacco use and alcohol consumption. The current study provides new clues for the causal mechanism of SCA among young Chinese HIV-infected adults, and is the starting point of precision intervention targeting HIV-related atherosclerosis.
Supplementary Information
Additional file 1: Supplemental material.
Additional file 2: Summary statistics of GWAS in relation to cIMT among all HIV negative control.
Additional file 3: Summary statistics of GWAS in relation to cIMT among all PWH.
Additional file 4: Summary statistics of GWAS in relation to cIMT among HIV negative control above 45 years old.
Additional file 5: Summary statistics of GWAS in relation to cIMT among HIV negative control under 45 years old.
Additional file 6: Summary statistics of GWAS in relation to cIMT among PWH above 45 years old.
Additional file 7: Summary statistics of GWAS in relation to cIMT among PWH under 45 years old.
Acknowledgements
Not applicable.
Role of the funding source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Abbreviations
- GWAS
Genome-wide association study
- SCA
Subclinical atherosclerosis
- PWH
People with HIV
- CVDs
Cardiovascular diseases
- ART
Antiretroviral therapy
- SNP
Single nucleotide polymorphism
- LD
Linkage disequilibrium
- NCDs
Non-communicable diseases
- BP
Blood pressures
- BMI
Body mass index
- WHR
Waist to hip ratio
- SBP
Systolic blood pressure
- DBP
Diastolic blood pressure
- TC
Total cholesterol
- TG
Triglyceride
- HDL
High density lipoprotein
- LDL
Low density lipoprotein
- MS
Metabolic syndrome
- cIMT
Carotid intima-media thickness
- PCA
Principal component analysis
- PCs
Principal components
- LMM
Linear mixed model
- GLMM
Generalized linear mixed model
- QQ plot
Quantile–quantile plot
- GRS
Genetic risk scores
- FUMA
Functional mapping and annotation
- eQTL
Expression quantitative trait loci
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Geno oncology
- aOR
Adjusted odds ratio
- CI
Confidence interval
Author contributions
NH proposed the research question and generally supervised the study. JH contributed to laboratory work, data analysis and manuscript drafting. HL and YD supervised subject enrollment, sample collection and data management. XL contributed to genomic data collection, management and analysis. KX advised on data analysis and manuscript drafting. XC, WS, SZ, MW and JX contributed to data collection and performing experiments. JH, HL, YD, XL and NH had full access to all the data. All authors critically reviewed and edited the manuscript and consented to final publication.
Funding
This study was supported by Yiwu Research Institute (Grant No. KCF201512 to NH) and Original Research Initiative Program (Grant No. IDF201021/001 to XL) of Fudan University, the National Natural Science Foundation of China (Grant No. 82173579 to NH, 81872671 to YD, 81803291 to XL), Taizhou City Foundation for Talents (Grant No. TZ2022-2 to HL), and Shanghai Municipal Health Commission (Grant No. GWV-10.1-XK16 to NH).
Availability of data and material
All the data in the paper or in the supplementary materials are free to obtain. Raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Written informed consent was obtained from all study participants. The study was approved by the Institutional Review Board of Fudan University School of Public Health, Shanghai, China.
Consent for publication
Not applicable.
Competing interests
No competing interests are declared.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiayu He and Haijiang Lin contributed equally
References
- 1.Feinstein MJ, et al. Characteristics, prevention, and management of cardiovascular disease in people living with HIV: a scientific statement from the American Heart Association. Circulation. 2019;140:e98–e124. doi: 10.1161/CIR.0000000000000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibanez B, et al. Progression of Early Subclinical Atherosclerosis (PESA) Study: JACC focus seminar 7/8. J Am Coll Cardiol. 2021;78:156–179. doi: 10.1016/j.jacc.2021.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Janjua SA, et al. Presence, characteristics, and prognostic associations of carotid plaque among people living With HIV. Circ Cardiovasc Imaging. 2017;10:e005777. doi: 10.1161/CIRCIMAGING.116.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrestha S, Irvin MR, Grunfeld C, Arnett DK. HIV, inflammation, and calcium in atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34:244–250. doi: 10.1161/ATVBAHA.113.302191. [DOI] [PubMed] [Google Scholar]
- 5.Zhi D, et al. Deep sequencing of RYR3 gene identifies rare and common variants associated with increased carotid intima-media thickness (cIMT) in HIV-infected individuals. J Hum Genet. 2015;60:63–67. doi: 10.1038/jhg.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H, et al. Age-specific associations between HIV infection and carotid artery intima-media thickness in China: a cross-sectional evaluation of baseline data from the CHART cohort. Lancet HIV. 2019;6:e860–e868. doi: 10.1016/S2352-3018(19)30263-2. [DOI] [PubMed] [Google Scholar]
- 7.Piepoli MF, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts) developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR) Eur Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu XH, et al. LncRNA kcnq1ot1 promotes lipid accumulation and accelerates atherosclerosis via functioning as a ceRNA through the miR-452-3p/HDAC3/ABCA1 axis. Cell Death Dis. 2020;11:1043. doi: 10.1038/s41419-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shendre A, et al. RYR3 gene variants in subclinical atherosclerosis among HIV-infected women in the Women's Interagency HIV Study (WIHS) Atherosclerosis. 2014;233:666–672. doi: 10.1016/j.atherosclerosis.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivapalaratnam S, et al. Genome-wide association studies in atherosclerosis. Curr Atheroscler Rep. 2011;13:225–232. doi: 10.1007/s11883-011-0173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strawbridge RJ, et al. Carotid intima-media thickness: novel loci, sex-specific effects, and genetic correlations with obesity and glucometabolic traits in UK Biobank. Arterioscler Thromb Vasc Biol. 2020;40:446–461. doi: 10.1161/ATVBAHA.119.313226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung MW, et al. Twenty-five novel loci for carotid intima-media thickness: a genome-wide association study in >45 000 individuals and meta-analysis of >100 000 individuals. Arterioscler Thromb Vasc Biol. 2022;42:484–501. doi: 10.1161/ATVBAHA.121.317007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha S, et al. A genome-wide association study of carotid atherosclerosis in HIV-infected men. AIDS. 2010;24:583–592. doi: 10.1097/QAD.0b013e3283353c9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan S, et al. Interaction of declined handgrip strength and HIV infection on neurocognitive impairment. J Neurovirol. 2021;28:217. doi: 10.1007/s13365-021-01036-1. [DOI] [PubMed] [Google Scholar]
- 15.Revision JCG. 2018 Chinese guidelines for prevention and treatment of hypertension-a report of the Revision Committee of Chinese guidelines for prevention and treatment of hypertension. J Geriatr Cardiol. 2019;16:182–241. doi: 10.11909/j.issn.1671-5411.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberti KG, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 17.Chinese Medical Association, C.M.J.P.H., Chinese Society of General Practice. Guideline for primary care of dyslipidemias (2019). Chin J General Pract. 2019; 18: 4.6–416 (2019).
- 18.Ma Y, et al. Cohort profile: the Chinese national free antiretroviral treatment cohort. Int J Epidemiol. 2010;39:973–979. doi: 10.1093/ije/dyp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krikke M, Arends JE, Van Lelyveld S, Hoepelman A, Visseren F. Greater carotid intima media thickness at a younger age in HIV-infected patients compared with reference values for an uninfected cohort. HIV Med. 2017;18:275–283. doi: 10.1111/hiv.12428. [DOI] [PubMed] [Google Scholar]
- 20.Ssinabulya I, et al. Subclinical atherosclerosis among HIV-infected adults attending HIV/AIDS care at two large ambulatory HIV clinics in Uganda. PLoS ONE. 2014;9:e89537. doi: 10.1371/journal.pone.0089537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsdottir J, McPeek MS. MASTOR: mixed-model association mapping of quantitative traits in samples with related individuals. Am J Hum Genet. 2013;92:652–666. doi: 10.1016/j.ajhg.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53:1616–1621. doi: 10.1038/s41588-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, et al. A resource-efficient tool for mixed model association analysis of large-scale data. Nat Genet. 2019;51:1749–1755. doi: 10.1038/s41588-019-0530-8. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek EJ, et al. Genome-wide interaction study of late-onset asthma with seven environmental factors using a structured linear mixed model in Europeans. Front Genet. 2022;13:765502. doi: 10.3389/fgene.2022.765502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond, R.K. et al. Biological constraints on GWAS SNPs at suggestive significance thresholds reveal additional BMI loci. Elife10(2021). [DOI] [PMC free article] [PubMed]
- 29.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machiela MJ, et al. Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: a report from the female lung cancer consortium in Asia. Int J Cancer. 2015;137:311–319. doi: 10.1002/ijc.29393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masenga SK, et al. Hypertension and metabolic syndrome in persons with HIV. Curr Hypertens Rep. 2020;22:78. doi: 10.1007/s11906-020-01089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, et al. ITGA9: potential biomarkers and therapeutic targets in different tumors. Curr Pharm Des. 2022;28:1412–1418. doi: 10.2174/1381612828666220501165644. [DOI] [PubMed] [Google Scholar]
- 34.Sun G, et al. A bioinformatics perspective on the links between tetraspanin-enriched microdomains and cardiovascular pathophysiology. Front Cardiovasc Med. 2021;8:630471. doi: 10.3389/fcvm.2021.630471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao C, et al. RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure. J Clin Invest. 2016;126:195–206. doi: 10.1172/JCI84015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito-Ishida A, Okabe S, Yuzaki M. The role of Cbln1 on Purkinje cell synapse formation. Neurosci Res. 2014;83:64–68. doi: 10.1016/j.neures.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Symons J, Cameron PU, Lewin SR. HIV integration sites and implications for maintenance of the reservoir. Curr Opin HIV AIDS. 2018;13:152–159. doi: 10.1097/COH.0000000000000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohn LB, Chomont N, Deeks SG. The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe. 2020;27:519–530. doi: 10.1016/j.chom.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang KS, et al. Family-based association analysis of alcohol dependence in the COGA sample and replication in the Australian twin-family study. J Neural Transm (Vienna) 2011;118:1293–1299. doi: 10.1007/s00702-011-0628-3. [DOI] [PubMed] [Google Scholar]
- 40.Lind PA, et al. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, et al. Cytoplasmic RAD23B interacts with CORO1C to synergistically promote colorectal cancer progression and metastasis. Cancer Lett. 2021;516:13–27. doi: 10.1016/j.canlet.2021.05.033. [DOI] [PubMed] [Google Scholar]
- 42.Tang MS, et al. DNA damage, DNA repair and carcinogenicity: tobacco smoke versus electronic cigarette aerosol. Mutat Res Rev Mutat Res. 2022;789:108409. doi: 10.1016/j.mrrev.2021.108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moen JM, et al. Overexpression of a neuronal type adenylyl cyclase (Type 8) in sinoatrial node markedly impacts heart rate and rhythm. Front Neurosci. 2019;13:615. doi: 10.3389/fnins.2019.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derda AA, et al. Gene expression profile analysis of aortic vascular smooth muscle cells reveals upregulation of cadherin genes in myocardial infarction patients. Physiol Genomics. 2018;50:648–657. doi: 10.1152/physiolgenomics.00042.2017. [DOI] [PubMed] [Google Scholar]
- 45.Cheeseman C. GLUT7: a new intestinal facilitated hexose transporter. Am J Physiol Endocrinol Metab. 2008;295:E238–E241. doi: 10.1152/ajpendo.90394.2008. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, et al. Decapping enzyme NUDT12 partners with BLMH for cytoplasmic surveillance of NAD-Capped RNAs. Cell Rep. 2019;29:4422–4434.e13. doi: 10.1016/j.celrep.2019.11.108. [DOI] [PubMed] [Google Scholar]
- 47.Dotzler SM, et al. Suppression-replacement KCNQ1 gene therapy for type 1 Long QT syndrome. Circulation. 2021;143:1411–1425. doi: 10.1161/CIRCULATIONAHA.120.051836. [DOI] [PubMed] [Google Scholar]
- 48.Yoshino S, et al. Sex-specific genetic variants are associated with coronary endothelial dysfunction. J Am Heart Assoc. 2016;5:e002544. doi: 10.1161/JAHA.115.002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soh MS, et al. Rare sudden unexpected death in epilepsy SCN5A variants cause changes in channel function implicating cardiac arrhythmia as a cause of death. Epilepsia. 2022;63:e57–e62. doi: 10.1111/epi.17254. [DOI] [PubMed] [Google Scholar]
- 50.Ilkhanoff L, et al. A common SCN5A variant is associated with PR interval and atrial fibrillation among African Americans. J Cardiovasc Electrophysiol. 2014;25:1150–1157. doi: 10.1111/jce.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balakrishnan A, Guruprasad KP, Satyamoorthy K, Joshi MB. Interleukin-6 determines protein stabilization of DNA methyltransferases and alters DNA promoter methylation of genes associated with insulin signaling and angiogenesis. Lab Invest. 2018;98:1143–1158. doi: 10.1038/s41374-018-0079-7. [DOI] [PubMed] [Google Scholar]
- 52.Li C, et al. Genome-wide gene-sodium interaction analyses on blood pressure: the genetic epidemiology network of salt-sensitivity study. Hypertension. 2016;68:348–355. doi: 10.1161/HYPERTENSIONAHA.115.06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, et al. Protein tyrosine phosphatase receptor-type Q: structure, activity, and implications in human disease. Protein Pept Lett. 2022;29:567–573. doi: 10.2174/0929866529666220511141826. [DOI] [PubMed] [Google Scholar]
- 54.Heyde A, et al. Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell. 2021;184:1348–1361.e22. doi: 10.1016/j.cell.2021.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi W, Ebbert KV, Craig AW, Greer PA, McCafferty DM. Absence of Fer protein tyrosine kinase exacerbates endotoxin induced intestinal epithelial barrier dysfunction in vivo. Gut. 2005;54:1091–1097. doi: 10.1136/gut.2004.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu S, et al. Endothelial dysfunction in atherosclerotic cardiovascular diseases and beyond: from mechanism to pharmacotherapies. Pharmacol Rev. 2021;73:924–967. doi: 10.1124/pharmrev.120.000096. [DOI] [PubMed] [Google Scholar]
- 57.Lu Y, Zhang X, Hu W, Yang Q. The identification of candidate biomarkers and pathways in atherosclerosis by integrated bioinformatics analysis. Comput Math Methods Med. 2021;2021:6276480. doi: 10.1155/2021/6276480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.https://www.ebi.ac.uk/QuickGO/term/GO:0060372. GO:0060372-regulation of atrial cardiac muscle cell membrane repolarization-Biological Process. (2022/6/8).
- 59.Tikkanen JT, et al. Long-term outcome associated with early repolarization on electrocardiography. N Engl J Med. 2009;361:2529–2537. doi: 10.1056/NEJMoa0907589. [DOI] [PubMed] [Google Scholar]
- 60.Pargaonkar VS, et al. Long-term prognosis of early repolarization with J-wave and QRS slur patterns on the resting electrocardiogram: a cohort study. Ann Intern Med. 2015;163:747–755. doi: 10.7326/M15-0598. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz PJ, et al. Mutation location and IKs regulation in the arrhythmic risk of long QT syndrome type 1: the importance of the KCNQ1 S6 region. Eur Heart J. 2021;42:4743–4755. doi: 10.1093/eurheartj/ehab582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saad NS, Elnakish MT, Ahmed AAE, Janssen PML. Protein kinase a as a promising target for heart failure drug development. Arch Med Res. 2018;49:530–537. doi: 10.1016/j.arcmed.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental material.
Additional file 2: Summary statistics of GWAS in relation to cIMT among all HIV negative control.
Additional file 3: Summary statistics of GWAS in relation to cIMT among all PWH.
Additional file 4: Summary statistics of GWAS in relation to cIMT among HIV negative control above 45 years old.
Additional file 5: Summary statistics of GWAS in relation to cIMT among HIV negative control under 45 years old.
Additional file 6: Summary statistics of GWAS in relation to cIMT among PWH above 45 years old.
Additional file 7: Summary statistics of GWAS in relation to cIMT among PWH under 45 years old.
Data Availability Statement
All the data in the paper or in the supplementary materials are free to obtain. Raw data that support the findings of this study are available from the corresponding author upon reasonable request.