Abstract
Strains of Staphylococcus aureus were transformed with plasmid DNA containing a Photorhabdus luminescens lux operon (luxABCDE) that was genetically modified to be functional in both gram-positive and gram-negative bacteria. S. aureus cells containing this novel lux construct, downstream of an appropriate promoter sequence, are highly bioluminescent, allowing the detection of fewer than 100 CFU in vitro (direct detection of exponentially dividing cells in liquid culture). Furthermore, these bacteria produce light stably at 37°C and do not require exogenous aldehyde substrate, thus allowing S. aureus infections in living animals to be monitored by bioluminescence. Two strains of S. aureus 8325-4 that produce high levels of constitutive bioluminescence were injected into the thigh muscles of mice, and the animals were then either treated with the antibiotic amoxicillin or left untreated. Bioluminescence from bacteria present in the thighs of the mice was monitored in vivo over a period of 24 h. The effectiveness of the antibiotic in the treated animals could be measured by a decrease in the light signal. At 8 h, the infection in both groups of treated animals had begun to clear, as judged by a decrease in bioluminescence, and by 24 h no light signal could be detected. In contrast, both groups of untreated mice had strong bioluminescent signals at 24 h. Quantification of CFU from bacteria extracted from the thigh muscles of the mice correlated well with the bioluminescence data. This paper shows for the first time that bioluminescence offers a method for monitoring S. aureus infections in vivo that is sensitive and noninvasive and requires fewer animals than conventional methodologies.
Many strains of Staphylococcus aureus, which cause a wide variety of diseases ranging from pyoderma to toxic shock syndrome, are methicillin and gentamicin resistant, with some strains also showing limited resistance to vancomycin. Such bacteria are a major problem in nosocomial, or hospital-acquired, infections (3, 9, 18). Without the development of new antibiotics it is possible that, given time, such bacteria will be untreatable by conventional means. In order to combat such bacterial infections, novel and effective drugs will be needed. In addition, new approaches for screening candidate antibiotics both in vitro and in vivo are essential to accelerate the development of new antiinfectives. To this end, bioluminescence offers a method that is sensitive and innocuous and allows only live, or viable, cells to be detected. Furthermore, in vivo detection of bioluminescent bacteria is noninvasive, allowing rapid monitoring of the infective state of eukaryotic cells both in culture and in animals (4, 7). Such a method of monitoring in vivo bioluminescent organisms in living animals has been described for gram-negative bacteria and it was demonstrated to correspond to bacterial CFU data with a correlation coefficient of 0.98 (H. L. Rocchetta, J. W. Foley, C. J. Boylan, P. W. Iverson, P. R. Contag, D. E. Jenkins, and T. R. Parr, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 797, 1999). However, until now, in vivo monitoring has not been achieved for gram-positive bacterial infections.
Synthesis of light in naturally occurring bioluminescent bacteria is encoded by five essential genes that are organized in an operon such as luxCDABE. Blue-green light is emitted from these bacteria with a peak at 490 nm as a result of a heterodimeric luciferase (encoded by luxAB) catalyzing the oxidation of reduced flavin mononucleotide (FMNH2) and a long-chain fatty aldehyde (synthesized by a fatty acid reductase complex encoded by luxCDE). Although a number of additional lux genes in bioluminescent bacteria have been identified, only luxA-E are essential for the biosynthesis of light (10). The lux operon can be readily moved into a variety of gram-negative bacteria to confer a bioluminescent phenotype. However, because all identified species of naturally occurring marine and terrestrial bioluminescent bacteria are gram negative, the transformation of gram-positive bacteria to a light phenotype has been somewhat limited due to the differing genetics of these two bacterial groups.
Bioluminescent strains of S. aureus have been constructed (5, 13). However, these bacteria contain the firefly luciferase gene (luc) or variations of isolated bacterial luxAB luciferase genes, each of which require the addition of an exogenous substrate to allow bioluminescence. Moreover, most bioluminescent gram-positive bacteria have been generated using bacterial luciferase genes that encode enzymes that are unstable at temperatures above 30°C (8). Although such bacteria are useful for environmental studies (e.g., the assessment of food products for contamination by such bacteria), luxAB constructs that only permit bioluminescence to occur below 30°C are of limited use for experimentation on pathogenicity carried out at 35°C and above in vivo.
In this report, we describe the construction of a novel Photorhabdus luminescens lux operon, where each gene (luxA, -B, -C, -D, and -E) has been modified by the introduction of a gram-positive ribosome binding site (RBS). This novel operon, preceded by an appropriate promoter sequence, allowed several highly bioluminescent S. aureus strains to be generated. These labeled bacteria could be detected in vivo noninvasively, allowing antibiotic efficacy to be efficiently assessed in real time.
MATERIALS AND METHODS
Incorporation of gram-positive RBSs upstream of luxA, -B, -C, -D, and -E by PCR.
The five genes of the P. luminescens lux operon, luxA, -B, -C, -D, and E, were amplified using PCR. The primers were designed to replace the gram-negative RBS AGG with the gram-positive RBS AGGAGG, ensuring that this site was at least 7 nucleotides upstream of the start codon (ATG) of each gene. PCR was performed with a Geneamp PCR system 9700 automated thermocycler (Perkin-Elmer Applied Biosystems, Norwalk, Conn.) in 200-μl thin-walled PCR tubes (Molecular BioProducts, San Diego, Calif.). Reactions were carried out in 50-μl volumes containing 5 μl of 10× PCR buffer (supplied with Taq DNA polymerase; Boehringer Mannheim, Indianapolis, Ind.), 2.0 mM MgCl2, 50 pmol of each oligonucleotide primer (Table 1), a 0.2 mM concentration of each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP; Amersham Pharmacia Biotech, Piscataway, N.J.), 1 U of Taq DNA polymerase (Boehringer Mannheim) and 10 ng of plasmid DNA containing the P. luminescens luxCDABE cassette (pSB417) (17). Amplification of each gene was achieved with 30 cycles at 95°C for 15 s, 50°C for 30 s, and 72°C for 60 s, followed by a final extension step at 72°C for 2 min. PCR-amplified DNA was purified prior to cloning procedures by passing complete reaction volumes through spin columns (PCR purification kit; Qiagen, Valencia, Calif.).
TABLE 1.
Oligonucleotide sequences used to amplify the P. luminescens genes from pSB417a
| Gene | Primer | Sequence | Restriction endonucleases |
|---|---|---|---|
| luxA | XAF3 | CCCCGGATCCTGCAGATGAAGCAAGAGGAGGACTCTCTATG .... | BamHI, PstlI |
| XAR | GGCGGATCCGTCGACTTAATATAATAGCGAACGTTG .... | BamHI, SalI | |
| luxB | XBF | GGGAATTCTCGAGGAGGAGAGAAAGAAATGAAATTTGGA .... | EcoRI, XhoI |
| XBR | GGCGGATCCGTCGACTTAGGTATATTCCATGTGGTAC .... | BamHI, SalI | |
| luxC | XCF | GGGAATTCTCGAGGAGGATGGCAAATATGACTAA .... | EcoRI, XhoI |
| XCR | GGCGGATCCGTCGACTTATGGGACAAATACAAGGAAC .... | BamHI, SalI | |
| luxD | XDF | GGGAATTCTCGAGGAGGAGTAAAAGTATGGAAAATGA .... | EcoRI, XhoI |
| XDR | GGCGGATCCGTCGACTTAAGACAGAGAAATTGCTTGA .... | BamHI, SalI | |
| luxE | XEF | GGGAATTCTCGAGGAGGAAAACAGGTATGACTTCATATG .... | EcoRI, XhoI |
| XER | GGCGGATCCGTCGACTTAACTATCAAACGCTTCGGTTA .... | BamHI, SalI |
Oligonucleotide sequences used to amplify the P. luminescens luxA, B, C, D, and E genes from pSB417 by PCR (see main text for PCR conditions). Nucleotides highlighted in bold show the positions and sequences of the different restriction sites (identified in far-right column) incorporated to facilitate cloning. Gram-positive RBSs sites are underscored by solid lines. Start and stop codons are underscored by broken lines.
Assembly of a modified luxABCDE cassette on pBluescript.
The luciferase (luxAB) and fatty acid reductase genes (luxCDE) were assembled on separate pBluescript SK(−) vectors (Stratagene, La Jolla, Calif.) using restriction enzymes sites introduced by PCR. luxA PCR-amplified DNA was inserted into pBluescript SK(−) at the BamHI and SalI sites (directionally orientated downstream of the lacZ promoter), and the plasmid was electroporated into Escherichia coli strain DH5α. Positive luxA clones, grown overnight at 37°C on Luria-Bertani (LB) medium containing 100 μg of ampicillin per ml (as were all subsequent clonings in E. coli) were selected by PCR (conditions as described above) using M13 forward (5′ - GTA AAA CGA CGG CCA GT - 3′) and reverse (5′-GGA AAC AGC TAT GAC CAT G - 3′) primers, whose sequences flank the pBluescript SK(−) polylinker. Templates in these reactions were added as whole cells, and lysis occurred at the first 95°C incubation. Plasmid DNA from selected clones and all clones subsequently deemed positive was prepared using standard alkaline lysis procedures (Plasmid Spin Miniprep kit; Qiagen). luxB PCR-amplified DNA was then cut with SalI/XhoI and inserted at the SalI site downstream of the cloned luxA gene. This ligation was again electroporated into E. coli DH5α, and luxAB clones and selected by screening for bioluminescence in the presence of exogenously added n-decyl aldehyde (Sigma, St. Louis, Mo.). Bioluminescence was detected using a photon-counting intensified charge-coupled device (ICCD) camera (model 2400-32; Hamamatsu Photonics, Bridgewater, N.J.).
Assembly of a separate luxCDE cassette in pBluescript SK(−) was achieved by the sequential cloning of luxC, -D, and -E using the compatible enzymes SalI and XhoI. Each step of the cloning procedure was confirmed by screening potential E. coli transformants by PCR using M13 forward and reverse primers (as described above). The fidelity of the final luxCDE cassette was confirmed by inserting this sequence, cut with SalI/XhoI, at the SalI site downstream of the luciferase genes in the above pBluescript SK(−) luxAB vector and screening for bioluminescent colonies using the ICCD camera. The modified lux operon containing the rearranged lux genes preceded by gram-positive RBS was designated luxABCDE, to distinguish it from the unmodified native operon luxCDABE.
Transformation of Staphylococcus aureus to a bioluminescent phenotype.
The above luxABCDE construct was inserted into the gram-positive or -negative shuttle vector pMK4 (14) and tested for bioluminescence in different S. aureus strains using random host DNA fragments as promoters. The luxABCDE cassette was cut from pBluescript SK(−) using the restriction enzymes BamHI and SalI and ligated with similarly cut pMK4 plasmid DNA (oriented so that no promoter lay upstream of this cassette) (Fig. 1). This ligation was electroporated into E. coli DH5α and luxABCDE clones, grown overnight at 37°C on LB medium containing 100 μg of ampicillin (the gram-negative selectable marker found on pMK4) per ml, selected by screening for colonies emitting low levels of bioluminescence (light barely detectable using the Hamamatsu ICCD camera). Genomic DNA from a clinical isolate of methicillin-resistant S. aureus (MRSA) was cut with Sau3A in a partial digest (1) and ligated into pMK4 luxABCDE plasmid DNA cut with BamHI (BamHI enzyme left active to reduce background from religation of the empty vector DNA). This ligation mix was then electroporated into E. coli DH5α, and the resulting transformants were plated on LB medium containing 100 μg of ampicillin per ml. The colonies were scraped from the plates and pooled, and their plasmid DNA was extracted (Plasmid Spin Miniprep kit; Qiagen). This plasmid DNA was electroporated into competent S. aureus RN4220 cells, and the resulting transformants were selected on LB medium containing 5 μg of chloramphenicol (the gram-positive selectable marker found on pMK4) per ml. Highly bioluminescent colonies were then selected using the ICCD camera.
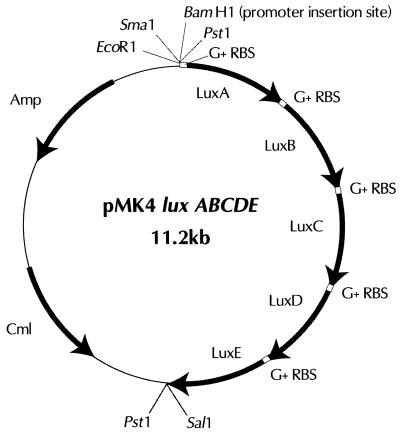
FIG. 1.
Schematic diagram of the plasmid pMK4 luxABCDE. Nucleotide sequences of the lux genes, ordered as shown, are as given in GenBank (accession number M90093) flanked by the relevant sequences shown in Table 1. This plasmid can be used to generate constitutive bioluminescent strains by ligating genomic DNA (partially digested by 4-base cutter) at the unique BamHI or SmaI site and selecting for light in the gram-positive bacterium from which the DNA was derived.
Comparing bioluminescence from S. aureus RN4220 strains containing the modified luxABCDE versus the native luxCDABE.
Plasmid DNA was prepared from several highly bioluminescent strains of S. aureus RN4220 containing pMK4 luxABCDE with different promoter fragments upstream of luxA (see above). The DNA was isolated using alkaline lysis (Plasmid Spin Miniprep kit; Qiagen) subsequent to treatment of the S. aureus cells with lysostaphin (Sigma) for 30 min at 37°C. A luxA backprimer (5′ - CCA CAC TCC TCA GAG ATG CG - 3′) was then used to sequence the S. aureus promoter fragments lying upstream of the lux operon in each plasmid. These promoter sequences were termed P1 through P73, and the plasmids were designated pMK4 luxABCDE Pn (where n is a number in the range of 1 to 73) Sequencing of the strain S. aureus RN4220 pMK4 luxABCDE P1 (data not shown) showed that this plasmid still contained an active BamHI site between the promoter sequence and luxA. This enabled the directional replacement of the modified luxABCDE with the native luxCDABE cassette cut from pSB417 (17) by using the restriction enzymes BamHI and SalI. The latter plasmid, pMK4 luxCDABE P1, was constructed in E. coli DH5α and then moved into S. aureus RN4220 using cloning and transformation procedures similar to those described above.
Bioluminescence from E. coli DH5α and S. aureus RN4220 containing either pMK4 luxABCDE P1 or pMK4 luxCDABE P1 was compared. Cultures of each of the latter bacterial strains, grown to exponential phase at 37°C in LB medium containing 100 μg of ampicillin per ml or 15 μg of chloramphenicol per ml, were diluted in 100-μl volumes across black 96-well microtiter plates in doubling dilutions (−0.3 log) and monitored for light over a period of 30 min at 37°C using the ICCD camera. The content of each well was then plated on LB agar to allow the number of CFU, ranging from approximately 101 to 107 CFU/ml, to be compared to levels of bioluminescence (in relative light units [RLU]).
Determining the temperature stability of bioluminescence in S. aureus.
Exponential cultures of the bacterial strains were grown to a concentration of approximately 107 CFU/ml at 30°C in LB medium containing 100 μg of ampicillin per ml or 15 μg of chloramphenicol per ml, and 1-ml volumes of each were placed in heating blocks set at 31, 33, 35, 37, 39, 41, 43, 45, and 47°C. After allowing the bacteria to acclimate and grow at each temperature for a period of 1 h, the nine heating blocks were sequentially placed inside a dark chamber, and light from each of the three cultures was recorded for a period of 1 min using the ICCD camera. To eliminate errors in the number of RLU arising from variations in bacterial numbers, each culture was plated on LB agar to allow the number of CFU to be recorded and the bioluminescence data to be normalized to the viable-cell number.
Estimating the minimum number of bioluminescent S. aureus CFU detectable in liquid culture.
Exponential cultures of bioluminescent S. aureus RN4220 pMK4 luxABCDE P1 were monitored using a highly sensitive liquid nitrogen-cooled integrating charge-coupled device (CCD) camera (model LN/CCD 1340-1300-EB/1; Princeton Instruments, Tucson, Ariz.) to determine the minimum number of CFU of this strain of bacteria detectable at 37°C. Volumes (100 μl) of exponentially growing cells were diluted across black 96-well microtiter plates from approximately 103 to 101 CFU/ml in doubling dilutions (−0.3 log) and monitored for light over a period of 10 min. The content of each well was then plated on LB agar to allow the number of CFU to be compared to levels of bioluminescence (in RLU).
Monitoring bioluminescent S. aureus 8325-4 infections in mice.
Plasmid DNA was isolated from S. aureus RN4220 pMK4 luxABCDE P1 and S. aureus RN4220 pMK4 luxABCDE P2 (as described above) and used to transform the pathogenic strain S. aureus 8325-4 (M. Smeltzer, University of Arkansas, Little Rock). Bioluminescent transformants of S. aureus 8325-4 were then tested in vivo in mice. Exponential cultures of S. aureus 8325-4 pMK4 luxABCDE P1 and S. aureus 8325-4 pMK4 luxABCDE P2, grown at 37°C in LB medium containing 15 μg of chloramphenicol per ml, were pelleted and then resuspended in phosphate-buffered saline (PBS). Bacterial concentrations were estimated spectrophotometrically by determining the absorbance at 600 nm, and concentrations were adjusted to approximately 5 × 107 CFU/ml by dilution with PBS. These cells were then held on ice for a short period until the mice were ready to be inoculated. Cell numbers were verified by plating dilutions of inoculum onto LB agar.
Mice were anesthetized with ketamine (100 mg/ml) and xylazine (20 mg/ml), mixed at a 4:1 ratio (vol/vol) just before use. A ketamine-xylazine mixture (100 mg/kg of body weight [dose based on ketamine concentration]) was injected intramuscularly into the right hamstring muscle. After anesthesia was established, the mice were injected in the left anterior tibialis with approximately 5 × 106 CFU in a total volume of 100 μl of PBS (twelve mice were injected with S. aureus 8325-4 pMK4 luxABCDE P1 and twelve mice were injected with S. aureus 8325-4 pMK4 luxABCDE P2). Six mice from each group were treated with a single oral dose of amoxicillin (10 mg/kg) immediately prior to anesthesia, with the remaining six mice from each group serving as untreated controls. Mice were imaged at 0, 4, 8, and 24 h postinfection using the Hamamatsu ICCD camera. At each time point, both dorsal and ventral images were taken. All images were collected for 5 min.
Quantification of bioluminescence data from mice.
Total photon emission from selected and defined areas within the images of each mouse was quantified with the LivingImage software package (Xenogen Corporation, Alameda, Calif.). The photon signal from the anterior tibialis muscle was quantified from the dorsal and ventral images of each mouse, and a dorsal-ventral average was calculated. This averaging corrects for light scattering differences due to mouse-to-mouse variation in the tissue depth of the bacteria.
Extraction and quantification of bacteria from the thigh muscle of mice.
After the 24-h imaging time point, the mice were sacrificed and the infected thigh muscles (both the anterior tibialis and the quadriceps muscle) were surgically removed. The muscle tissue was homogenized in 500 μl of PBS with a loose Dounce homogenizer. The tissue and bacterial suspension was then diluted in PBS by 105, 106, and 107 to a final volume of 1 ml for each dilution. A 100-μl volume of each dilution was plated out in duplicate onto LB agar plates and incubated overnight at 37°C. The following morning colonies were counted and the numbers of CFU for each tissue sample were estimated.
RESULTS
Gram-positive RBSs allow the P. luminescens lux operon to function in S. aureus.
Vallanoweth and Rabinowitz (17) have reported that gram-positive ribosomes are unable to translate mRNAs containing weak gram-negative RBSs. In addition, they stated that the spacing between the RBS and the start codon (most commonly ATG) on gram-positive mRNAs should ideally be 7 to 10 nucleotides. In the present study we used PCR to alter the RBS of each gene in the P. luminescens lux operon from AGG to AGGAGG, ensuring that this site was a minimum of 7 bases upstream of the start codons of each gene (Table 1). Interestingly, in E. coli, bioluminescence from clones containing the modified luxABCDE operon (that containing a gram-positive RBS) was found to be significantly greater than that from those containing the unmodified luxCDABE operon (that containing a gram-negative RBS), but only when both constructs lay downstream of a weak uninduced pBluescript lacZ promoter. Increasing transcription of both lux operons on pBluescript in E. coli by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium (solid or liquid culture) resulted in significantly more light from the unmodified luxCDABE operon but in a decrease in light from the modified luxABCDE operon. Low levels of bioluminescence could also be seen from E. coli clones containing a promoterless luxABCDE operon, greatly aiding the cloning of this cassette into the shuttle vector pMK4 lacking a recognized promoter (Fig. 1).
Random fragments of genomic DNA from S. aureus were ligated into pMK4 luxABCDE, such that efficiently expressed constitutive promoters could be identified using the modified lux operon as a reporter. Electrotransformation of S. aureus RN4220 with the mixture of plasmid DNAs in this ligation reaction mixture resulted in approximately 20,000 transformants, with between 5 and 10% of these colonies showing some degree of bioluminescence. From the 2,000 or so bioluminescent colonies, 73 moderately to highly bioluminescent transformants were isolated and further characterized for constitutive bioluminescence. The level of bioluminescence from the majority of these strains was comparable to that seen from gram-negative bacteria, such as E. coli, carrying an expressed luxCDABE operon (data not shown). Bioluminescence from pure cultures of many of these S. aureus strains was at a level that could be observed by the naked eye in a darkroom after less than a minute of acclimatization.
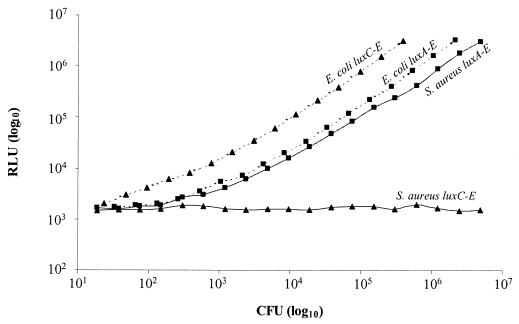
Plasmid DNA from the six brightest strains, designated S. aureus RN4220 pMK4 luxABCDE P1 through P6, was purified, and the flanking 500 bp upstream of luxA were sequenced. All six sequences were different (suggesting that different promoters had been selected), and each showed a high level of homology to gram-positive sequences (especially to S. aureus and Bacillus subtilis sequences) in GenBank (data not shown). Fortuitously, pMK4 luxABCDE P1 was found to still contain an active BamHI site between the promoter sequence and luxA. This enabled the directional replacement of luxABCDE with a luxCDABE cassette cut from pSB417 (18). By interchanging the lux cassettes so exactly, discrepancies in light due to promoter activity could essentially be eliminated, allowing a true comparison of the activity of the two operons to be made. Figure 2 shows that both luxCDABE and luxABCDE function similarly in E. coli (approximately fourfold difference), but only luxABCDE gives significant bioluminescence in S. aureus. The minimum number of S. aureus RN4220 pMK4 luxABCDE P1 CFU detectable at 37°C using a photon-counting ICCD camera was approximately 400 CFU. However, using a more sensitive liquid nitrogen-cooled integrating CCD camera we were able to detect as few as 80 CFU at 37°C.
FIG. 2.
Comparison of bioluminescence from S. aureus and E. coli containing the native luxCDABE operon versus that in strains containing the modified luxABCDE operon. Exponential cultures of S. aureus RN4220 pMK4 luxABCDE P1 (-■-), S. aureus RN4220 pMK4 luxCDABE P1 (-▴-), E. coli DH5α pMK4 luxABCDE P1 (‥■‥) and E. coli DH5α pMK4 luxCDABE P1 (‥▴‥) were diluted across black 96-well microtiter plates in doubling dilutions (−0.3 log) and monitored for light over a period of 30 min using the ICCD camera. The content of each well was then plated to allow the number of CFU to be compared to the level of bioluminescence (RLU).
Bioluminescence from S. aureus RN4220 pMK4 luxABCDE P1 is optimal at 37°C.
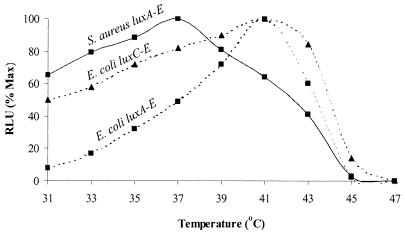
To monitor bioluminescence emitted from pathogenic bacteria within animals (4), it is essential that both the lux genes and Lux proteins function adequately at body temperature (i.e., around 37°C). Thus, in order to determine whether modification of the lux genes had altered the temperature range over which optimal bioluminescence occurs in bacterial cells, light arising from the modified luxABCDE cassette was compared to that from the native luxCDABE operon in both gram-negative and gram-positive bacteria between 31 and 47°C. Since S. aureus RN4220 pMK4 luxCDABE P1 was previously shown to be nonbioluminescent (Fig. 2), only S. aureus RN4220 pMK4 luxABCDE P1, E. coli DH5α pMK4 luxABCDE P1, and E. coli DH5α pMK4 luxCDABE P1 were tested over the above-mentioned temperature range. Figure 3 shows that the maximum light to be recorded from a culture of S. aureus RN4220 pMK4 luxABCDE P1 was at 37°C. Furthermore, between 31 and 41°C the light emission from this strain remained above 60% maximum, and it continued to do so for more than 4 h (data not shown), indicating that the Lux enzymes are stable over this temperature range. In contrast, both E. coli DH5α pMK4 luxABCDE P1 and E. coli DH5α pMK4 luxCDABE P1 gave maximum light at 41°C. Comparison of the light profiles from these two E. coli strains indicates that the luxABCDE construct functions less efficiently at lower temperatures than luxCDABE. Whether this is an accurate reflection of the functioning of this modified lux construct or of the P1 promoter is not known. However, since this construct was designed to work optimally in gram-positive bacteria, optimization of bioluminescence in gram-negative bacteria can still be performed using the native luxCDABE cassette.
FIG. 3.
Temperature stability of the modified luxABCDE operon. Exponential cultures of S. aureus RN4220 pMK4 luxABCDE P1 (-■-), E. coli DH5α pMK4 luxABCDE P1 (‥■‥), and E. coli DH5α pMK4 luxCDABE P1 (‥▴‥) were grown to approximately 1 × 107 CFU/ml at 30°C and 1-ml volumes of each were placed in heating blocks set at 31, 33, 35, 37, 39, 41, 43, 45, and 47°C. After 1 h at each of these elevated temperatures, the nine heating blocks were sequentially placed inside a dark chamber and light from each of the three cultures was recorded for a period of 1 min using the ICCD camera. Shown are the number of RLU at each of the temperatures, with this data expressed as a percentage of the maximum (Max) bioluminescence attained and adjusted for variation in the number of CFU.
Bioluminescent S. aureus allows effective antibiotic treatment to be monitored in living animals.
A number of pathogenic strains of S. aureus were transformed with the plasmids pMK4 luxABCDE P1 and pMK4 luxABCDE P2, including RN6390, 8325-4, and a clinical isolate of MRSA. Interestingly, RN6390 and 8325-4 were both found to produce similar levels of bioluminescence to RN4220 when transformed with each of the above-mentioned plasmids, whereas the transformed strain of MRSA was significantly brighter (two- to fourfold). Whether this increase in light was due to increased transcription or translation (the promoter sequences being derived from this MRSA strain) or was simply a factor of higher metabolic activity in this strain was not determined. However, the fact that fewer MRSA cells could be detected with these strains, both in culture and in vivo, than could be detected with other transformed S. aureus strains (data not shown), indicated that increased screening for more highly bioluminescent constructs may increase the sensitivity of detection still further.
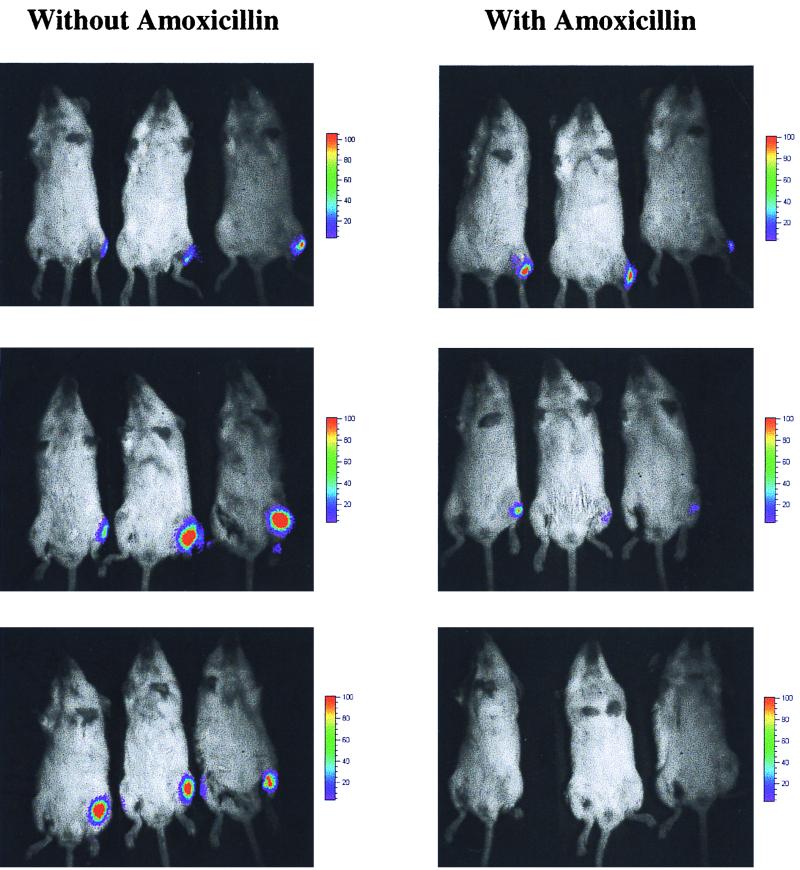
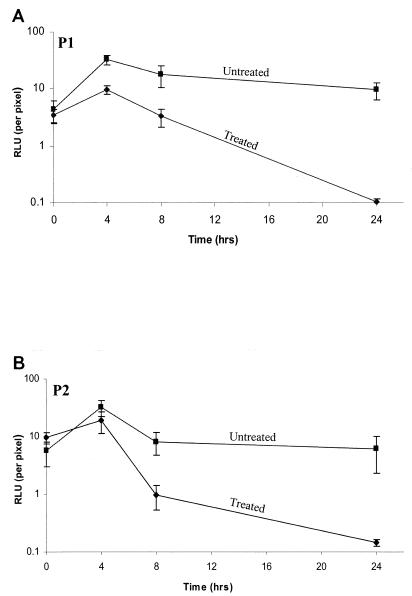
Figure 4 shows how bioluminescent S. aureus can be effectively monitored in living animals in real time and can be used to determine the efficacy of antibiotic therapy. Both S. aureus 8325-4 pMK4 luxABCDE P1 and S. aureus 8325-4 pMK4 luxABCDE P2 produced significant bioluminescent signals in mice, allowing treatment with amoxicillin to be effectively assessed (only P1 ventral images shown). Cumulative bioluminescence data collected from both the dorsal and ventral sides of each animal is shown in Fig. 5. At 8 h, the infection in both groups of treated animals had begun to clear, as judged by a decrease in bioluminescence, and by 24 h no signal could be detected. In comparison, both groups of untreated mice had strong bioluminescent signals at 24 h, corresponding to an apparent poor state of health as indicated by ruffled fur and dragging of the hindquarters during movement.
FIG. 4.
Monitoring the effects of amoxicillin on bioluminescent S. aureus in mice. Exponential cultures of the bioluminescent bacteria were injected intramuscularly into the left anterior tibialis (8 × 106 CFU) of 24 anesthetized mice, 12 mice with S. aureus 8325-4 pMK4 luxABCDE P1 and 12 mice with S. aureus 8325-4 pMK4 luxABCDE P2. Six mice from each group were treated with a single oral dose of amoxicillin (10 mg/kg) immediately prior to anesthesia, with the remaining six mice from each group serving as untreated controls. Shown are three treated and three untreated S. aureus 8325-4 pMK4 luxABCDE P1-infected mice, imaged ventrally for 5 min at 0, 8, and 24 h postinfection using the ICCD camera.
FIG. 5.
Bioluminescence data recorded from S. aureus 8325-4 pMK4 luxABCDE P1 (A)- and S. aureus 8325-4 pMK4 luxABCDE P2 (B)-infected mice. Each data set represents the mean number of RLU from six mice either untreated (■) or treated (⧫) with amoxicillin (10 mg/kg), imaged both dorsally and ventrally for 5 minutes at 0, 4, 8, and 24 h postinfection using the ICCD camera. Error bars, standard errors of the means.
Extraction of bacteria from the thigh muscles of each of the mice confirmed the bioluminescence data to be extremely accurate at predicting the number of viable bacteria in the tissue. For both strains of bacteria, S. aureus 8325-4 pMK4 luxABCDE P1 and S. aureus 8325-4 pMK4 luxABCDE P2, the number of CFU recovered from the thigh muscles of the untreated mice at 24 h postinfection was found to have increased approximately 25-fold (2 × 109 CFU/ml) over the inoculating dose (8 × 107 CFU/ml). In comparison, the number of CFU (P1 and P2) in the thigh muscles of the antibiotic-treated mice varied between 1.1 × 103 and 6.8 × 104 CFU/ml at 24 h postinfection, representing a decrease of at least 1,000-fold from the number of CFU used to inoculate the mice. Moreover, by plating all of the recovered bacteria on media containing no antibiotics, the plasmid stability of both bioluminescent S. aureus strains could be assessed. S. aureus 8325-4 pMK4 luxABCDE P2 was slightly more stable than S. aureus 8325-4 pMK4 luxABCDE P1, with 95% of P2 CFU retaining a high level of bioluminescence, as opposed to 85% of P1 CFU when plated in the absence of selection. However, since each CFU recovered from a mouse thigh muscle consists of approximately 105 to 106 cells derived from a single cell in the absence of selective pressure to maintain the lux plasmid, the percentage of bacteria still carrying plasmid DNA upon extraction from the animal was probably significantly higher.
DISCUSSION
Among the lux operons from bioluminescent bacteria, that from P. luminescens (2, 12) appears to be ideally suited for use in mammalian animal models, given that mammalian body temperatures lie within the temperature optimum for the bioluminescent enzymes from this bacterium (Fig. 2) (6, 15, 19). This is in contrast to the low optimum temperature for beetle luciferases (Luc) and other characterized bacterial luciferases, such as those from species of Vibrio (8, 10). In vivo monitoring of luciferase was initially pioneered with a gastrointestinal infectious disease model using bioluminescent gram-negative Salmonella enterica serovar Typhimurium (4). This study indicated not only that in vivo bioluminescent imaging is feasible but also that it can be quantitative and its use, as predicted, results in more information in less time than conventional assays. Although a large variety of gram-negative bacterial species can be labeled with the P. luminescens lux operon, the introduction of these or other bacterial lux operons into gram-positive bacteria results in little or no light, regardless of the strength of the promoter preceding this operon (our unpublished data). Data presented in this paper show that for the first time it is possible to generate bioluminescent gram-positive bacteria capable of producing high levels of light at 37°C without the addition of exogenous substrate (Fig. 2). The consequence of this finding is far reaching, especially in the development of real-time in vivo assays for antibiotic efficacy studies designed to monitor gram-positive infections in animal models, as is demonstrated in this study with bioluminescent S. aureus (Fig. 4).
In recent years there has been a dramatic increase in the number of antibiotic-resistant strains of gram-positive bacteria causing serious, life-threatening diseases in humans. This is especially true for strains of S. aureus, Streptococcus pneumoniae, Mycobacterium tuberculosis, and enterococci (3, 11), with a large number of these bacteria being resistant to a wide range of or, in some cases, all conventional antibiotics (e.g., MRSA and vancomycin-resistant enterococci). The occurrence of such multidrug-resistant bacteria is of increasing concern, since they represent a disease threat of epidemic proportions. Conventional methodologies for monitoring these gram-positive pathogens in vivo are not only cumbersome, but they also usually include biological assessment regimens that represent late or end stages of the disease process and often use death as an endpoint. The late-stage animal protocols are also considered more stressful to the animal in these models and less likely to give important information about the events early in disease. Alternatively, ex vivo assays can be employed to reveal disease processes. These assays require removal of tissue and, usually, sacrifice of the animal. As a consequence there is the loss of the contextual influences of the living animal. Ex vivo assays can be extraordinarily sensitive (e.g., PCR, with single-molecule sensitivity). However, such assays are typically plagued with various artifacts such as sampling limitations, where only a small amount of tissue or limited numbers of cells can be analyzed. The use of bioluminescent noninvasive imaging strategies to reveal the real-time effects of potential therapeutic agents on gram-positive bacterial infections in animal models would greatly accelerate the analyses of compounds under development. The spatial and temporal differences in the bioluminescent signals between treated and untreated animals (Fig. 4) can be used to assess the effects of compounds on specific biological processes in vivo.
Although this study only describes bioluminescent S. aureus, we have used the modified luxABCDE operon to successfully transform a wide range of gram-positive bacteria, including strains of S. pneumoniae and Listeria monocytogenes (data not shown). In each case, the gram-positive bacteria carrying this modified lux operon are highly bioluminescent and can be monitored in vivo in animals. Plasmid loss in the absence of antibiotic selection has been shown to be minimal from each of the latter bacteria over a period of 24 to 48 h in vivo (>80% plasmid retention), with no observable genetic structural instability. However, since plasmids are of limited use for monitoring regulation due to artifacts involving, among other things, copy number and supercoiling, we have recently constructed a number of luxABCDE transposons. These have been successfully used to stably transform and monitor gene regulation in a number of gram-positive bacteria (our unpublished results). The ability to monitor gene regulation in gram-positive bacteria using bioluminescence should also prove extremely valuable for in vitro uses, such as environmental monitoring and for applications in the food industry.
The noninvasive approach described here allows for experimental protocols that are significantly faster. Furthermore, because biostatistics are improved through collection of multiple data points in the same animal, the overall number of animals required is reduced. These two benefits should improve the capacity to relieve the bottleneck of animal studies that occurs in drug development as high-throughput technologies increase the number of potential lead compounds that require animal evaluation. We believe that this technology also increases the quality of animal model data to provide more information for selecting the drug candidates that will be clinically successful.
ACKNOWLEDGMENTS
We thank B. Nelson and M. Cable (Bioimaging Section, Xenogen Corporation) for their assistance with experimental design and data collection using the two CCD cameras, C. Safieddine (Animal Husbandry Section, Xenogen Corporation) and R. Gungon (Infectious Disease Section, Xenogen Corporation) for assisting with the animal work, and D. Jenkins (Oncology Section, Xenogen Corporation) and C. Contag (University of Stanford, Stanford, California) for help with preparing the manuscript. We also thank M. Winson (University of Wales, Aberystwyth, United Kingdom) for kindly supplying us with the P. luminescens lux plasmid, pSB417, and M. Smeltzer (University of Arkansas) for the S. aureus strain 8325-4.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 2.Boemare N E, Akhhurst R J, Mourant R G. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol. 1993;43:249–255. [Google Scholar]
- 3.Carbon C. Costs of treating infections caused by methicillin-resistant staphylococci and vancomycin-resistant enterococci. J Antimicrob Chemother. 1999;44:31–36. doi: 10.1093/jac/44.suppl_1.31. [DOI] [PubMed] [Google Scholar]
- 4.Contag C H, Contag P R, Mullins J I, Spilman S D, Stevenson D K, Benaron D A. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 5.Corbisier P, Ji G, Nuyts G, Mergeay M, Silver S. luxAB gene fusions with the arsenic and cadmium resistance operons of Staphylococcus aureus plasmid pI258. FEMS Microbiol Lett. 1993;110:231–238. doi: 10.1111/j.1574-6968.1993.tb06325.x. [DOI] [PubMed] [Google Scholar]
- 6.Frackman S, Anhalt M, Nealson K H. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J Bacteriol. 1990;172:5767–5773. doi: 10.1128/jb.172.10.5767-5773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis K P, Gallagher M P. Light emission from a Mudlux transcriptional fusion in Salmonella typhimurium is stimulated by hydrogen peroxide and by interaction with the mouse macrophage cell line J774.2. Infect Immun. 1993;61:640–649. doi: 10.1128/iai.61.2.640-649.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill P J, Rees C E, Winson M K, Stewart G S. The application of lux genes. Biotechnol Appl Biochem. 1993;17:3–14. [PubMed] [Google Scholar]
- 9.Howe R A, Wootton M, Walsh T R, Bennett P M, MacGowan A P. Expression and detection of hetero-vancomycin resistance in Staphylococcus aureus. J Antimicrob Chemother. 1999;44:675–678. doi: 10.1093/jac/44.5.675. [DOI] [PubMed] [Google Scholar]
- 10.Meighen E A. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 11.Petrini B, Hoffner S. Drug-resistant and multidrug-resistant tubercle bacilli. Int J Antimicrob Agents. 1999;13:93–97. doi: 10.1016/s0924-8579(99)00111-9. [DOI] [PubMed] [Google Scholar]
- 12.Rainey F, Ehlers R-U, Stackebrandt E. Inability of the polyphasic approach to systematics to determine the relatedness of the genera Xenorhabdus and Photorhabdus. Int J Syst Bacteriol. 1995;45:379–381. doi: 10.1099/00207713-45-2-379. [DOI] [PubMed] [Google Scholar]
- 13.Steidler L, Yu W, Fiers W, Remaut E. The expression of the Photinus pyralis luciferase gene in Staphylococcus aureus Cowan I allows the development of a live amplifiable tool for immunodetection. Appl Environ Microbiol. 1996;62:2356–2359. doi: 10.1128/aem.62.7.2356-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan M A, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 15.Szittner R, Meighen E. Nucleotide sequence, expression and properties of luciferase coded by lux genes from terrestrial bacterium. J Biol Chem. 1990;265:16581–16587. [PubMed] [Google Scholar]
- 16.Vellanoweth R L, Rabinowitz J C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 17.Winson M K, Swift S, Hill P J, Sims C M, Griesmayr G, Bycroft B W, Williams P, Stewart G S. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]
- 18.Wong S S, Ho P L, Woo P C, Yuen K Y. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin Infect Dis. 1999;29:760–767. doi: 10.1086/520429. [DOI] [PubMed] [Google Scholar]
- 19.Xi L, Cho K W, Tu S C. Cloning and nucleotide sequences of lux genes and characterization of luciferase of Xenorhabdus luminescens from a human wound. J Bacteriol. 1991;173:1399–1405. doi: 10.1128/jb.173.4.1399-1405.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]