ABSTRACT
Pheromone-inducible conjugation in the Enterococcus faecalis pCF10 system is regulated by the PrgX transcription factor through binding interactions at two operator binding sites (XBS1 and XBS2) upstream of the transcription start site of the prgQ operon encoding the conjugation machinery. Repression of transcription requires the interaction of a PrgX tetramer with both XBSs via formation of a DNA loop. The ability of PrgX to regulate prgQ transcription is modulated by its interaction with two antagonistic regulatory peptides, ICF10 (I) and cCF10 (C); the former peptide inhibits prgQ transcription, while the latter peptide enhances prgQ transcription. In this report, we used electrophoretic mobility shift assays (EMSAs) and DNase footprinting to examine binding interactions between the XBS operator sites and various forms of PrgX (Apo-X, PrgX/I, and PrgX/C). Whereas a previous model based on high-resolution structures of PrgX proposed that the functional differences between PrgX/C and PrgX/I resulted from differences in PrgX oligomerization state, the current results show that specific differences in XBS2 occupancy by bound tetramers account for the differential regulatory properties of the two peptide/PrgX complexes and for the effects of XBS mutations on regulation. The results also confirmed a DNA looping model of PrgX function.
IMPORTANCE Peptide pheromones regulate antibiotic resistance transfer in Enterococcus faecalis. Here, we present new data showing that pheromone-dependent regulation of transfer genes is mediated via effects on the structures of complexes between peptides, the intracellular peptide receptor, and operator sites on the target DNA.
KEYWORDS: antibiotic resistance, cell-cell signaling, gene transfer, sex pheromone
INTRODUCTION
In Gram-positive bacteria, quorum sensing and other forms of intercellular communication commonly employ peptides as signaling molecules, although the mechanisms for production and for sensing of these molecules are diverse (1–4). Expression of transfer functions encoded by many conjugative plasmids in Enterococcus faecalis is activated via peptide sex pheromones secreted by plasmid-free recipient cells and sensed by plasmid-carrying donors (5–7). It is noteworthy that the initial identification of a specific bacterial signaling peptide and the first high-resolution structure of a signaling peptide receptor were both first reported in studies of enterococcal pheromone-responsive plasmid systems (8–11). For all pheromone plasmids analyzed thus far, the inducing peptides are produced by proteolytic processing of the cleaved signal sequences of chromosomally encoded, secreted lipoproteins (10). To help control spurious autoinduction of donor cells by endogenous pheromone, these plasmids also encode a competitive inhibitor peptide produced from a small gene adjacent to the locus encoding the pheromone receptor peptide, but in the opposite orientation (12–14).
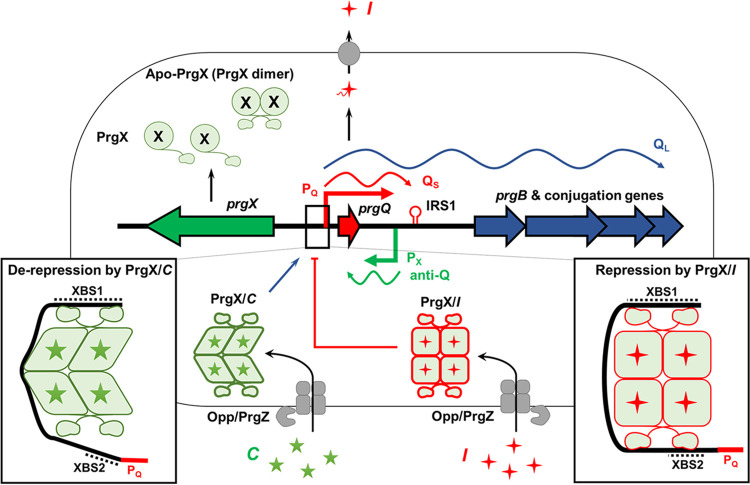
The pheromone-responsive plasmid pCF10 has been extensively studied as a model system for pheromone-induced conjugation (Fig. 1) (15, 16) and is the focus of the current report. In the pCF10 system, the pheromone peptide C (for clumping induction of mating cells; sequence LVTLVFV) is produced from the signal sequence of the secreted CcfA lipoprotein encoded by the chromosome (10, 11). Via the concerted activity of the secreted PrgZ binding protein and the chromosomal oligopeptide permease system, extracellular C is imported by donor cells (17–19), where it binds to the intracellular master regulator PrgX, abolishing repression of transcription of the pheromone-inducible prgQ operon encoding the pCF10 conjugation machinery (Fig. 1). PrgX is the founding member of the RPPR family of transcription factors; these proteins share similar structures but little amino acid sequence homology, and they are functionally related in that their regulatory activities are modulated by interaction with signaling peptides (1, 4). The first gene of the conjugation operon, prgQ, encodes a 22-amino-acid peptide whose C terminus is processed to form the inhibitor peptide I (sequence AITLIFI) (12). The C and I peptides compete for binding to both PrgZ and PrgX; ultimately, the induction state within a donor cell is determined by the relative intracellular concentrations of these two peptides (20). The prgX gene is adjacent to prgQ but in the opposite orientation, consistent with the conserved genetic organization of the peptide-sensing machinery noted above. Furthermore, the promoters and transcription start sites (TSSs) for these two genes are each within the opposing operon in an antisense orientation (21–26). The transcription start site for prgQ (see Fig. S1 in the supplemental material) was mapped physically using an RNA ligase-mediated reverse transcription method, which served as the basis for subsequent methods of distinguishing 5′ ends of RNAs representing transcription starts from processing products, with predicted −10 and −35 regions deduced from the determined TSS resemble typical microbial promoters (27). The complementary 5′ ends of prgX and prgQ transcripts (anti-Q and Qs) (Fig. 1) result from a 220-bp region where both strands of the DNA template are transcribed, resulting in multiple levels of antagonistic posttranscriptional regulation by antisense and transcription interference mechanisms (14, 22, 23, 26); mathematical modeling and experimental analyses of the system suggest that these multiple mechanisms cause the system to function as a bistable genetic switch (25) and impart stochastic variation in the levels of induction among individual cells in a population exposed to C (20).
FIG 1.
Overview of peptide-dependent regulation of pCF10 conjugation by PrgX. Transcription of conjugation genes from pCF10 is regulated by the interaction of PrgX and peptides. Donor cells carrying pCF10 secrete basal levels of inhibitor peptide (I, red stars) from pCF10 and reimport I via the Opp/PrgZ system (17). The inhibitor forms a tetramer with PrgX, which stably binds to the prgQ promoter (PQ) and represses transcription. When a recipient cell is near a donor, the donor imports pheromone peptide (C, green stars) produced by the recipient via Opp/PrgZ. Both peptides cause PrgX to form a tetramer, which binds to pCF10 DNA. However, the structure of the PrgX/C tetramer is distorted relative to PrgX/I (8, 9) with a predicted decrease in stability in solution (the experimentally determined structural differences between PrgX/I and PrgX/C tetramer are indicated by the differences in the shapes of the monomers in this and subsequent figures). The structural alterations in PrgX/C tetramers also weaken the interaction of XBS2 with PrgX/C, leading to dissociation from the DNA near the promoter, allowing access to RNA polymerase. Also, antisense interactions between small RNAs (QS and anti-Q) modulate expression of conjugation-related genes posttranscriptionally (15).
PrgX is essential for regulation of the pheromone response and acts primarily as a negative regulator of prgQ operon transcription (8, 22). Extensive genetic, biochemical, and structural studies of the molecular mechanisms by which the interactions of the C and I peptides with PrgX modulate initiation of transcription from the prgQ promoter, PQ, have been carried out, as reviewed in reference 28. PrgX molecules have a predicted DNA binding domain at the N terminus and a central region mediating homodimer formation, which is essential for function (8, 9). Initial DNase I footprinting and genetic studies of Apo-PrgX (23) demonstrated the existence of two PrgX binding sites upstream from prgQ TSS. XBS1 is a high-affinity binding site about 110 bp upstream of the transcription start site, and XBS2 is a lower-affinity binding site extending from approximately −13 to −27 relative to the prgQ TSS (Fig. S1), placing it between the −10 and −35 regions of PQ. The 214-bp segment of pCF10 upstream from the TSS shown in Fig. S1 is both sufficient and necessary for active, PrgX-regulated prgQ transcription (29). While footprinting studies of the interactions of RNA polymerase with PQ have not been carried out, competition between PrgX and RNA polymerase for binding to the XBS2 region has been demonstrated (2). Interestingly, the two XBS regions are on the same face of the DNA helix, and there is substantial evidence for formation of a DNA loop via protein-protein interactions between pairs of PrgX dimers bound to the two XBSs (Fig. 1) (2, 8, 9); the major outcome resulting from these cooperative interactions is increased PrgX occupancy at XBS2. In vitro, Apo-PrgX can act as a repressor of PQ in runoff transcription assays, and point mutations in the XBS regions reduced PrgX transcription repression in vitro (29). In vivo studies demonstrated that I enhances PrgX-mediated PQ repression, while C has the opposite effect (8, 14). Two publications (8, 9) reporting high-resolution structures of Apo-PrgX and PrgX complexed with either C or I led to the first working model for the mechanism of PrgX control of PQ transcription and how PrgX regulation is modulated by the two antagonistic peptides. This original model suggested that replacement of I by C in PrgX complexes would disrupt DNA-bound PrgX tetramers in vivo, destabilizing XBS2 binding, and resulting in increased transcription initiation from PQ (2, 8). Figure S2 depicts this model, where 3 pathways for control of PQ activity in donor cells are shown. As indicated in this illustration, Apo-PrgX exists as a dimer in solution, and the Apo protein can bind both XBS operator sites at high protein concentrations, resulting in repression of prgQ transcription in vitro. Based on structural data (8, 9), it was believed that tetramers of PrgX/I would be stable in solution, strengthening the repressing looped DNA complex in vivo, while stable PrgX/C tetramers would be unlikely to exist in vivo at significant concentrations, destabilizing the loop, reducing XBS occupancy, and increasing prgQ transcription.
A recent biochemical analysis of effects of the C and I peptides on oligomerization of PrgX and on PrgX/XBS binding (2), as well as the new information reported here, enable refinement of the original model, both in terms of molecular mechanistic details (based on in vitro studies) and in relation to how the system may function in the cytoplasm of pCF10-containing cells in response to changes in the relative concentrations of the C and I peptides, as depicted in Fig. 1. The necessity for refining the working model was first suggested by the results reported by Chen et al. (2), showing that binding of either C or I to PrgX dimers shifted the predominant oligomeric state to a tetramer, both peptide-bound forms of PrgX showed increased DNA binding, as determined by electrophoretic mobility shift assays (EMSAs), and both peptides bound PrgX with similar extremely high affinities. These results suggested that in vivo, conversion from repressed bound complex to an induced bound complex probably resulted from replacement of a DNA-bound I-tetramer or an Apo-tetramer with a C-tetramer rather than by replacement of I peptide with C peptide in preformed complexes. These new results raised the possibility that peptide-dependent differences in the structures of PrgX-DNA complexes near the XBS2 site (rather than changes in PrgX oligomerization) could account for the variable induction states associated with PrgX/C versus PrgX/I complexes. The main objective of the current study was to examine these predicted structural differences directly. To accomplish this goal, we used DNase I footprinting to examine PrgX/XBS interactions at both operator sites; we compared the results obtained with wild-type probes to those observed with probes containing either XBS point mutations or insertions of additional DNA between the XBSs. EMSA analyses were also carried out to independently confirm the formation of the expected protein/DNA complexes. Our experiments revealed the differential effects of C and I on these interactions. The results reported here generally support the predicted structural differences between the protein-DNA complexes at XBS2 in the induced versus uninduced conformations, as shown in Fig. 1; they also show that a point mutation reducing PrgX repression in vitro strongly altered protein-DNA interactions at XBS2. The data also demonstrate enhancement of binding by both peptide-PrgX complexes to XBS1 and confirm the requirement of a PrgX tetramer-stabilized DNA loop for functional regulation of conjugation gene expression.
RESULTS
Differential biological activities of C and I peptides in donor cells correlate with the structures of PrgX-peptide-operator DNA complexes.
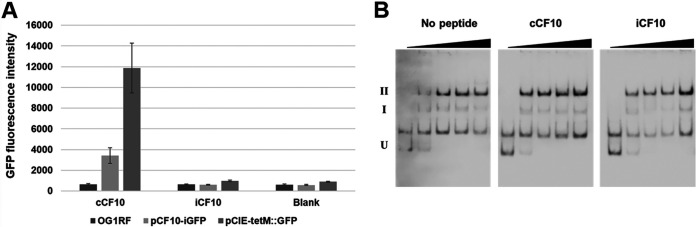
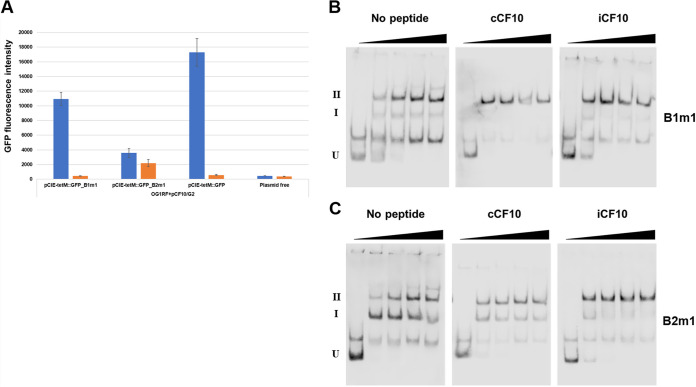
To examine the relationship between regulation of prgQ operon expression by PrgX in vivo and PrgX-XBS interactions, we used green fluorescent protein (GFP) transcription-reporter fusion constructs, EMSAs, and DNase I footprinting assays. Two different reporter constructs were used. In pCF10-iGFP, the fusion was made in the context of pCF10, whereas in pCIE-tetM::GFP, the same reporter was transcriptionally fused to PQ in the context of a multicopy cloning vector to facilitate subsequent manipulation of the XBS regulatory region (see Materials and Methods for details). As shown in Fig. 2A, both fusion strains were strongly induced by addition of C, with no GFP expression above the background in cells treated with I. The higher level of C-induced GFP expression with the pCIE-tetM::GFP reporter likely relates to a higher copy number of the vector; however, there was no significant background expression in I-treated cells carrying this plasmid.
FIG 2.
Biological activity of C and I peptides in donor cells versus peptide effects on DNA binding to the 5′ regulatory region of the prgQ operon. (A) Inducing activity of various peptides was determined using E. faecalis carrying pheromone-inducible plasmids. The gfp gene is located downstream of the regulatory region of both pCF10-iGFP and pCIE-tetM::GFP. GFP expression was induced by pheromone activity. Values plotted for this figure and all subsequent reporter assays are the means of 3 replicates with standard deviations indicated by the thin lines. (B) PrgX-peptide complexes bind to PrgX binding sites (XBSs). Electrophoretic mobility shift assays were performed using 30 fmol of digoxigenin-labeled LT DNA probes and PrgX complexes with various peptides. The concentrations of PrgX-peptide complexes are indicated by right triangles, and 0, 20, 50, 100, and 200 nM are shown in order from the lowest concentration. For this and all subsequent EMSA figures, U indicates unbound DNA probe, band I is a shifted species likely representing a PrgX dimer bound to XBS1, and band II represents a looped tetramer where each XBS is occupied by a dimer. EMSAs shown here and in subsequent figures are representative of a minimum of 3 replicates that produced similar results. The band between U and I represents a contaminating amplicon that does not bind PrgX.
Next, we examined the effect of C and I on PrgX binding to XBSs by using EMSAs with a double-stranded digoxigenin (DIG)-labeled DNA probe (WT, probe with wild type pCF10 sequence; see Fig. S1 in the supplemental material). When the PrgX complexes were reacted with a DIG-labeled WT probe containing both binding sites, two shifted bands (I and II) were observed (Fig. 2B). As previously noted, these likely represent DNA bound to PrgX dimers and tetramers, respectively. In the reactions with either PrgX/C or PrgX/I, a strong “supershift” to species II was noted at the lowest PrgX concentration (20 nM); with Apo-PrgX, comparable amounts of shifted species were noted at positions I and II. At higher concentrations of Apo-PrgX, increased amounts of species II were observed, suggesting the PrgX dimer could form a bound tetramer at high concentrations. These results are similar to those reported by Chen et al. (2). Note that we observed a spurious band migrating between band U (representing unbound probe DNA) and band I (representing shifted probe due to binding of XBS1 by a dimer) at various concentrations in EMSAs (similar bands were also observed experiments using probes with mutations presented below). There was no PrgX concentration-dependent shift in the migration of these bands in any experiments; they likely represent contaminating amplicons from the pGEM-T Easy plasmid templates used to generate probes (Materials and Methods), which do not interact specifically with PrgX.
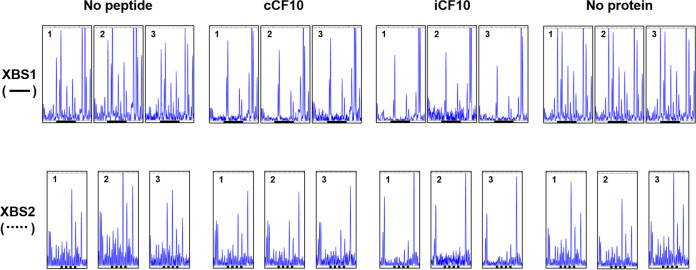
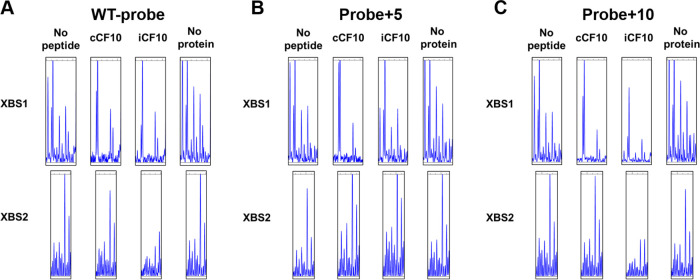
DNase I footprint assays using fluorescent (FAM or HEX)-labeled probes were performed to examine interactions between each binding site and PrgX-peptide complexes (Materials and Methods). DNase I footprints (Fig. 3 and Fig. S3) showed a clear difference in the binding interactions of different PrgX species for XBS1 versus XBS2. In the absence of PrgX, there are several predominant cleavage sites in both XBS1 and XBS2, which appear as peaks in the elution profiles from sequencing reactions. PrgX/XBS binding should protect the DNA from cleavage, reducing these peaks. Figure S3 shows the complete gene analyzer elution plots from a representative experiment involving a DNase-digestion WT probe in the absence (“no protein”) or presence of different forms of PrgX. All of these reactions showed a similar overall pattern of peaks, with most major differences localized to the previously identified XBS1 and XBS2 regions; strong similarity of all DNase cleavage products outside the XBS regions was observed in all footprinting assays presented here, as well as additional replicates not shown. For both XBS regions, the observed footprints produced by either PrgX/C or PrgX/I tended to extend leftward into regions flanking the original XBSs; since footprinting had never been carried out with either peptide and EMSAs (Fig. 2) showed that both peptides increased overall DNA binding, this result is not surprising. Figure 3 depicts XBS1, XBS2, and flanking region cleavage products of WT probes digested in the presence or absence of various forms of PrgX from 3 replicate experiments; the XBSs, as defined in the previous study (23), are indicated by solid or broken lines below the sequences. The overall cleavage patterns were highly reproducible. As expected, the protection of the XBS1 region for all PrgX complexes was observed, but the protection by Apo-PrgX was only increased modestly relative to the no-protein control, while strong XBS1 protection was observed for the reaction mixtures containing either PrgX/C or PrgX/I. Differences in protection of XBS2 by PrgX/I and PrgX/C versus Apo-PrgX were also observed. PrgX/I mediated the highest level of protection, as would be predicted for the decreased activity of PQ in cells grown in high levels of I. Even though Apo-PrgX has been shown to bind DNA containing XBSs (Fig. 2) (2, 8, 23, 30) and repress transcription from XBS-containing templates in vitro (29), it provided little protection in the XBS2 region in our assays (compare “no peptide” and “no protein” XBS2 region lanes in Fig. 3 and Fig. S3). PrgX/C complexes produced XBS2 region footprints similar in appearance to those produced by PrgX/I, but with somewhat reduced protection. The cumulative results suggest that both peptides can enhance PrgX/XBS binding, but stronger PrgX/I complexes at XBS2 could account for the differential effects of the peptides on regulation. To further examine the regulatory significance of differences in XBS2 binding by different forms of PrgX, we carried out additional experiments using DNA probes and reporter plasmids carrying mutations known to affect PrgX control of prgQ expression.
FIG 3.
Effects of C and I peptides on DNA binding to XBS regions assessed by DNase footprinting. The plots shown represent data from 3 replicate assays using the WT DNA probe (see Fig. S1 in the supplemental material) at 20 nM; the elution profiles of DNase-digested probe fragments as determined by analysis using the Applied Biosystems 3730 gene analyzer (Materials and Methods) in the regions representing the XBSs with flanking DNA sequence are shown. The solid heavy lines below the plots correspond to XBS1, and the broken heavy lines correspond to XBS2 as determined in initial footprinting studies of Apo-PrgX by Bae et al. (23). No peptide, reaction mixtures containing Apo-PrgX; cCF10, reaction mixtures containing PrgX/C; iCF10, reaction mixtures containing PrgX/I; and no protein, reaction mixtures containing neither PrgX or peptides. The concentration of PrgX-peptide complexes was 200 nM. See Materials and Methods for more details of nuclease protection protocols.
An essential requirement for a DNA loop connecting the two operator PrgX binding sites is confirmed by in vitro DNA binding assays.
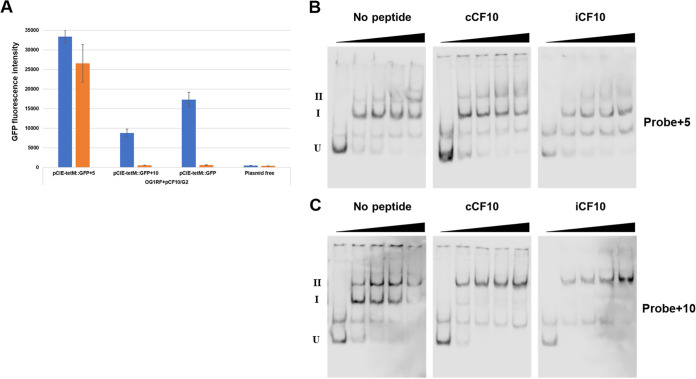
We found previously that a 5-bp insertion between the XBS sites abolished transcriptional regulation by PrgX and that a 10-bp insertion in the same region (see Fig. S1 for the sequences) retained wild-type regulation (2, 31). This likely results from differential effects of the insertions on the positions of the XBSs on the face of the DNA helix, supporting a model where repressing complexes contained a DNA loop between the XBSs (Fig. 1), stabilized by protein-protein interactions between dimers bound to each XBS (2, 31). To determine the effect of the insertion between XBSs on PQ expression in vivo using the GFP reporter system described above, pCIE-tetM::GFP with 5- or 10-bp insertions between the XBS sites were transferred into OG1RF(pCF10/G2), and the GFP expression was measured (Fig. 4A). With OG1RF carrying pCIE-tetM:GFP (wild-type regulatory region), GFP expression was induced by C but not I. However, with 5 bp inserted between the XBSs in an isogenic reporter, a high level of GFP expression was observed with or without C induction. When 10 bp were inserted into the same position, C-dependent expression of GFP was also observed, similar to the wild-type reporter construct. These results agreed with previous studies using a different reporter of prgQ transcription (2, 31), which also supported a DNA looping model for PQ regulation by PrgX.
FIG 4.
Effects of 5-bp and 10-bp insertions (shown in Fig. S3 in the supplemental material) between XBS1 and XBS2 on prgQ regulation in vivo and on PrgX binding to the XBSs. (A) Analysis of prgQ expression using GFP reporter fusion assays. The plasmids with insertions were prepared based on pCIE-tetM::GFP. GFP expression was induced for 1 h as described in Materials and Methods. Blue indicates pheromone induced, and orange indicates uninduced; means of 3 replicates with standard deviation. (B and C) Effect of insertions between XBSs on binding to Apo-PrgX, PrgX/C, and PrgX/I. EMSAs were performed as described in Fig. 2B. The concentrations of PrgX-peptide complexes are indicated by right triangles, and 0, 20, 50, 100, and 200 nM are shown in order from the left to right. (B) EMSA analysis of PrgX/DNA binding to a DNA probe containing the pCF10 regulatory region with a 5-bp insertion between XBS1 and XBS2. (C) EMSA analysis of PrgX/DNA binding to a DNA probe containing the pCF10 regulatory region with a 10-bp insertion between XBS1 and XBS2.
Next, the effect of insertions on the interaction between PrgX and XBSs in vitro was assessed with EMSA (Fig. 4B and C). Using the probe +5, the supershifted bands (position II) were observed at reduced levels relative to the wild-type probe (Fig. 2B), while the relative amount of the band at position I was increased in all PrgX complexes (Fig. 4B and Fig. S4). We believe that the small amounts of supershifted +5 bp species observed with all forms of PrgX probably represent tetramers bound to only XBS1 (Fig. S4). With the probe containing the 10-bp insert, the pattern of the shifted bands was similar to that of the wild-type probe (Fig. 2B, Fig. 4C, and Fig. S4). This suggested that probe +5 could bind tightly to XBS1 but poorly to both sites simultaneously, in contrast to the other two probes where the XBSs are predicted to be on the same face of the DNA helix (Fig. S4).
We used DNase I footprint assays with the same set of probes to further analyze the binding of PrgX to each XBS (Fig. 5). With the wild-type probe, the pattern of peaks was similar to Fig. 2C; protection near XBS1 was increased by all PrgX complexes, with both peptides increasing protection relative to that of the Apo protein (Fig. 5A). Using the probe with a 5-bp insert, the XBS1 protection was similar to the WT probe, but no significant protection of XBS2 was seen, suggesting PrgX/I complexes could not bind to XBS2 because they were shifted to the opposite face of the DNA helix insertion of 5 bp (Fig. 5B). On the other hand, for the 10-bp insertion, the protected regions near XBS2 were very similar to the WT probe (Fig. 5C). These results strongly support the DNA looping model, and they also indicate that the regulatory effects of the insertions are manifested by PrgX interactions at XBS2.
FIG 5.
DNase I footprinting analysis of XBS occupancy by PrgX in probes containing the 5-bp or 10-bp insertion mutations. DNase I footprint assay conditions and fragment analysis were performed as described in Fig. 3 and Materials and Methods. XBS1, TTAGGTATTGAATACGACACT; XBS2, TATAATACTTAGATG. (A) XBS occupancy using the WT probe. (B) XBS occupancy using the +5 probe. (C) XBS occupancy using the +10 probe.
Analysis of in vivo regulatory phenotypes and of PrgX-peptide-DNA complexes in vitro demonstrate a critical role of operator site XBS2 in regulation of pCF10 conjugation.
We examined the effect of point mutations in XBS1 or XBS2 on transcriptional regulation by PrgX in vivo and the XBS binding affinity of PrgX complexes. XBS1 contains a palindromic sequence (GTATTGAATAC), whereas only part of the palindrome is retained in XBS2 (AATAC), which probably accounts for the weaker affinity of the latter site for binding PrgX (23). EMSAs and in vitro transcription assays of the interactions of Apo-PrgX with these binding sites by Caserta et al. (29) showed that point mutations in XBS1 modestly reduced PrgX repression in vitro, while XBS2 point mutations strongly inhibited PrgX repression. In the present study, we used two of the point mutations (Fig. S2) examined in previous in vitro transcription studies to examine their effects on PrgX binding and regulation in vivo; the mutations involved changing two adjacent nucleotides (one pair for each site) to the complementary nucleotides. PQ expression by the strain carrying the XBS1 mutation (B1m1) was comparable to wild type but with a slight reduction in induced expression levels. In contrast, the XBS2 mutation (B2m1) nearly eliminated the differences in induced versus uninduced expression, resulting in a much higher basal level, as well as reduced induction relative to wild type (Fig. 6A).
FIG 6.
Effects of point mutations in XBS1 and XBS2 on prgQ regulation in vivo and on PrgX binding to the XBSs using EMSA. (A) The effect of XBS mutations on prgQ regulation was confirmed using E. faecalis carrying a coresident pheromone-inducible plasmid along with the reporter. The plasmids with XBS mutations were prepared based on pCIE-tetM::GFP. GFP expression was induced for 1 h as described in Materials and Methods. Blue indicates pheromone induced, and orange indicates uninduced. (B and C) EMSA analysis of PrgX DNA binding to probes with XBS mutations. Effect of XBS mutations of probe for binding to Apo-PrgX (PrgX dimer), PrgX/C, and PrgX/I. EMSAs were performed as described in Fig. 2B. The concentrations of PrgX-peptide complexes are indicated by right triangles, and 0, 20, 50, 100, and 200 nM are shown in order from the left to right.
In EMSAs, the shifting pattern of the B1m1 probe in the presence of Apo-PrgX was similar to that observed for the wild-type probe (compare Fig. 6B to Fig. 2B), and the shifts observed when the same probe bound either PrgX/C or PrgX/I were also similar to those observed with the wild-type probe, with an increased level of supershifted band II observed relative to Apo-PrgX for both peptide complexes. The B2m1 mutation, which had stronger regulatory effects in vivo (Fig. 6A), also reduced the ratio of band II/band I in all reactions (compare Fig. 2B to Fig. 6C). Since supershifts were still observed in all reaction mixtures containing PrgX, we speculate that the protein-DNA interactions at XBS2 could still be different from WT probe, e.g., the complexes might represent tetramers bound tightly to XBS1, but more weakly to XBS2 with the B2m1 probe. This question was further investigated by DNase I footprinting.
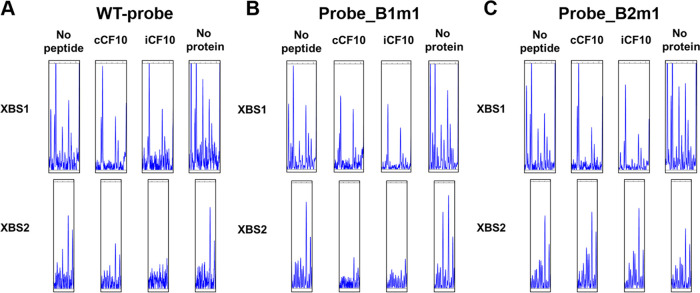
As depicted in Fig. 7, the XBS1 mutation had little or no effect on the protection of the XBS1 or XBS2 regions relative to the wild-type probe with any of the PrgX preparations. In fact, this mutation gave the appearance of increasing XBS1 protection very slightly, but since the cleavage patterns for the two probes in the absence of PrgX also showed subtle differences (compare “no protein” lanes in Fig. 7A and B), we think the biological significance of these differences is low. The XBS1 mutation also did not substantially alter the protection pattern in the XBS2 region (Fig. 7B). Stronger effects of the XBS2 mutation were apparent from the data shown in Fig. 7C. While this mutation did not substantially alter protection of the XBS1 region, it had a marked effect on the protection pattern of the XBS2 region. As can be seen from comparing Fig. 7A and C, the mutation essentially eliminated protection of this region by all 3 forms of PrgX, consistent with the dramatic deregulation of PQ expression observed in vivo (Fig. 6A). Figure S5 illustrates the predicted complexes and more closely compares the shifted mobilities observed with mutant probes.
FIG 7.
Effects of XBS point mutations on XBS1 and XBS2 occupancy assayed by DNase I footprinting analysis. Fragment analysis was performed as described in Fig. 3. The concentration of PrgX-peptide complexes was 200 nM. WT sequence and mutant sequences are shown in Fig. S1 in the supplemental material. (A) WT probe. (B) Probe with the XBS1 mutation. (C) Probe with the XBS2 mutation.
DISCUSSION
This report proposes a new model (Fig. 1) for peptide-mediated modulation of the ability of PrgX to control PQ transcription based on structural, genetic, biochemical, and physiological analyses. In the original model (see Fig. S2 in the supplemental material) based on PrgX structures (2), the differential regulatory activities of PrgX/I versus PrgX/C were proposed to result from different effects of the bound peptides on PrgX oligomerization. It was suggested that PrgX/I complexes existed as stable tetramers in donor cells, while PrgX/C complexes were in the form of dimers; PrgX/I tetramers could interact tightly with both XBSs with the protein bound to XBS2 mediating repression of transcription. The predominant PrgX dimers formed in the presence of C would not bind tightly to XBS2, allowing RNA polymerase to access the promoter. The publication by Chen et al. (2) raised some questions about the oligomerization model when it was discovered that both peptides actually increased tetramer formation, suggesting the possibility that structural differences in the protein-DNA interactions at XBS2 mediated by PrgX/C versus PrgX/I tetramers might account for the distinct regulatory functions of the two peptide-protein complexes as illustrated in Fig. 1. To interrogate this model, we analyzed PrgX/XBS interactions using both EMSAs and DNase I footprinting. The results (Fig. 2C) strongly supported that PrgX oligomerization does not account for differential effects between C and I. Most importantly, the DNase footprinting assays suggest that differences in the protein-DNA interactions at XBS2 account for the different regulatory functions of PrgX/C versus PrgX/I. The model also suggests that Apo-PrgX dimers might bind independently to XBS1 and XBS2 prior to DNA loop formation, allowing for repression in vivo at high levels of Apo-PrgX. However, our DNase I footprinting data demonstrated that Apo-PrgX showed similar XBS occupancy to PrgX/C, with low affinity for XBS2. This suggests to us that endogenous levels of I production in uninduced donor cells might produce basal levels of PrgX/I complexes that keep the system off in the absence of exogenous C. It is also well established that the pCF10-endoded negative regulator PrgY acts to reduce endogenous C production by donor cells (32), allowing a very low concentration of PrgX/I to maintain repression.
Further support for the new model came from a comparison of PrgX-XBS interactions of DNA probes containing the wild-type sequence in the XBS sites and intervening region with probes containing mutations (see Fig. S1 for sequences). With probes containing a 5-bp insertion between XBSs, the XBSs are shifted to opposite faces of the DNA helix, while a 10-bp insertion restores the sites to the same face. In terms of both regulation in vivo (Fig. 3A) and DNA binding in vitro (Fig. 3B and C and Fig. 4). The constructs containing 10-bp insertions between the XBSs showed very similar behavior to the wild type, whereas the +5 constructs were unregulated in vivo, and EMSAs showed no supershifted bands indicative of a tetramer bound to the probe via a DNA loop (Fig. 3B). As shown in Fig. 4, the DNA probe containing the +5 insertion showed no protection of XBS2 even with PrgX/I complexes, whereas the +10 protection patterns were similar to wild-type probe. The cumulative data from these experiments provide a compelling confirmation of the DNA looping model and also show that the different regulatory phenotypes of +10 versus +5 constructs related to PrgX interactions with XBS2.
We then examined the effect of mutations on XBSs in EMSA by using probes with mutations in either XBS1 (Fig. S1, B1m1 probe) or XBS2 (Fig. S1, B2m1 probe) (Fig. 5 and 6). The alterations in regulatory phenotypes and DNA binding associated with the XBS1 mutation were relatively minor, but the XBS2 mutation largely abolished normal regulation (Fig. 5A) and protection of XBS2 by PrgX/I (Fig. 6). Interestingly, some supershifted probe was still observed with the XBS2 mutation in EMSAs (Fig. 5C); we suspect that the cooperativity between the various binding interactions could partially stabilize bound tetramer complexes in the B2m1 probe in spite of reduced XBS2 occupancy. In footprinting analysis (Fig. 7), the B2m1 mutation caused a substantial reduction of protection of the XBS2 region, even for PrgX/I, likely accounting for its effect on regulation.
The original analysis of PrgX interactions with pCF10 utilized only Apo-X and were carried out by analysis of DNase protection using Maxim-Gilbert sequencing (23). In spite of different experimental protocols and a long time period between studies, the protected sites identified in the present study generally agree with the previous results (23). The role of XBS1 and XBS2 mutations on PrgX regulation has also been studied using in vitro transcription assays, albeit using only Apo-X (29). These results also support the overall model of regulation proposed in Fig. 1. Further refinement of the details of the model might be possible via the use of both peptide variants of C and I (30) and PrgX variants containing point mutations with interesting regulatory phenotypes (33). Finally, a detailed analysis of high-resolution structures of PrgX-peptide-pCF10 operator DNA complexes using cryo-electron microscopy will likely reveal important insights into this complex regulatory system.
MATERIALS AND METHODS
Strains and plasmids.
Enterococcus faecalis strain OG1RF was grown statically at 37°C in M9-YE (34) or brain heart infusion (BHI; BD Franklin Lakes, NJ) broth. Escherichia coli strains were grown in Luria-broth (LB; Invitrogen) or on LB with 15 mg/mL agar (LB-agar). The following concentrations of antibiotics were used for selection: tetracycline (Tet), 10 μg/mL; chloramphenicol (Cm), 20 μg/mL for E. faecalis and E. coli; gentamicin (Gen), 1,000 μg/mL; rifampicin (Rif), 200 μg/mL; fusidic acid (Fus), 25 μg/mL for E. faecalis; carbenicillin (Carb), 50 μg/mL for E. coli; and kanamycin (Km), 30 μg/mL for E. coli. Peptide cCF10 or iCF10 (C or I) stock solutions were prepared in dimethylformamide (DMF) at 0.5 mg/mL and diluted into growth medium to the desired final concentration; synthetic peptides were obtained from Mimotopes (www.mimotopes.com).
Preparation of PrgX-peptide complexes.
For EMSA and DNase I footprint, His-tagged PrgX (His-PrgX) was purified following a previous protocol (23) with modifications. E. coli BL21(DE3) cells carrying pET28-PrgX were grown at 37°C until the culture reached the optical density at 600 nm (OD600) of 0.6, followed by inducing with 1 mM IPTG (isopropyl-β-d-thiogalactoside) for 4 h at 30°C. The induced cells were collected by centrifugation at 6,000 rpm at 4°C for 20 min. The pellet was resuspended in 15 mL of Ni column binding buffer (20 mM Tris-HCl [pH 8], 5 mM imidazole, and 0.5 M NaCl) supplemented with 15 mL of bacterial protein extraction reagent (B-PER) (Thermo Fisher) and 200 μL of 30 mg/mL lysozyme. After incubation at 37°C for 20 min, cells were sonicated on ice using QSonica Q125 with the following conditions (amplitude, 40%; probe, 4,420; 3 × 30 s on; 20 s off). PrgX-peptide complexes were obtained by incubating nickel columns containing bound Apo-PrgX with 1 ml of 1mg/ml peptide for 20 min at room temperature, and washing the column with 30 ml wash buffer prior to elution as described for Apo-PrgX. The column was washed with 30 mL of the binding buffer and 30 mL wash buffer (60 mM imidazole, 20 mM Tris-HCl [pH 8], and 0.5 M NaCl). The Apo-PrgX was eluted with buffer (500 mM imidazole, 20 mM Tris-HCl [pH 8], and 0.5 M NaCl). PrgX-peptide complexes were obtained by adding 1 mL of 1 mg/mL C or I in DMF were added to columns containing bound PrgX and incubated at room temperature (RT) for 20 min. The column was washed with 30 mL wash buffer, followed by elution. The eluted fractions containing His-PrgX were purified as previously described (2); because previous surface plasmon resonance binding studies (2) had shown that both C and I bind to PrgX with extremely high affinity (equilibrium dissociation constant (KD), ~10−13 mol L−1), our procedure yielded pure and stable PrgX-peptide complexes for DNA binding studies.
EMSAs.
Note that all of the DNA probes used for EMSA and for footprinting assays (described below) contain both XBS regions and flanking DNA, but the EMSA probes were shorter to optimize resolution of unshifted and shifted probe species on gels; see Fig. S1 in the supplemental material for complete sequences of the probes and the primers used to generate them. A DNA segment containing XBS1 and XBS2 sites (7,964 to 8,266 nucleotides [nt] of pCF10 [GenBank accession number AY855841]) was amplified from pCF10 using primers PrgX_pCIE_BS_F, ACAGAACCTATCTTAAACATTTTC, and PrgQ_pCIE_BS_R, CTATCAGATAAATATTAAGGTTATTG, and cloned into pGEM-T Easy. The WT DNA probe was amplified from the cloned plasmid using primers prgQ-5EM, CGATAAACTACAAAAATTTGTTAA, and 3′-Qp-RBS-EcoRI, CCCTCCTATAAAAACATCTTAACATCTAAGTATT, and purified with QIAquick PCR purification kit (Qiagen). To prepare DNA probes with 5-bp (GTACC) or 10-bp (GTACCTTCTA) insertions between XBS1 and XBS2 sites, fragments were amplified from pBK2+5 or pBK2+10 and cloned into pGEM-T Easy. For probes with the XBS1 mutation (B1m1), a template was generated by overlap extension PCR. In the first step, two fragments were amplified from pCF10 by using the primers PrgX_pCIE_BS_F and XmR, CACATCTTCGAGTGTCGTATTGTATACCTAAAATTAAAATATTAAC; XmF, GTTAATATTTTAATTTTAGGTATACAATACGACACTCGAAGATGTG; and PrgQ_pCIE_BS_R. Next, the fragments were annealed together, and PCR was performed using the outer primers (PrgX_pCIE_BS_F and PrgQ_pCIE_BS_R). The product was cloned into pGEM-T Easy. For probe with XBS2 mutation (B2m1), template was generated by QuikChange using primers B2m1_qc_F, CATATTTTAGTTGAAAATATAATTGTTAGATGTTAAG, and B2m1_qc_R, ATTATATTTTCAACTAAAATATGTATTTTTTTT. The probes were amplified from the plasmids and purified as well as the LT probe.
DNA probes were labeled with DIG-1-dUTP using the DIG gel shift kit, 2nd generation (Roche), according to the manufacturer’s instructions. EMSAs were performed as described by Chen et al. (2).
DNase I DNA footprint assay.
DNase I DNA footprint assays were modified from the procedure described in reference 35. The DNA probes were amplified from the same plasmids as EMSA using a 5-HEX-labeled forward primer and a 6-FAM-labeled reverse primer. PCR products were purified with QIAquick PCR purification kit (Qiagen) and used as DNA probe for footprint assay. We incubated 20 nM of the DNA probe with different concentrations (20, 50, 100, and 200 nM) of PrgX complexes for 15 min at 25°C in a buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 250 μg/mL heparin, 100 mM dithiothreitol (DTT), 1 μg of poly(dA-dT), and 0.1 μg of poly-l-lysine. After incubation, 5 μL DNase I (New England Biolabs) was added and the mixture was digested for 15 min at 25°C, followed by stopping the reaction by adding 25 μL of 0.5 M EDTA (pH 8.0). The digestion products were recovered using MinElute PCR purification kit (Qiagen), and samples were eluted with 15 μL Elution Buffer supplied with the kit. Samples were added to 1 μL of GeneScan 500 LIZ dye size standards (Applied Biosystems) and submitted to the University of Minnesota Genomics Center for detecting the fragments using a 3730 DNA analyzer (Applied Biosystems). The results were analyzed using Peak Scanner v1.0 (Applied Biosystems). The figures in this paper depicting footprint assays are all from reactions with 200 nM PrgX.
Reporter gene assay.
To confirm the effect of mutations or insertions in the binding region of PrgX on the transcriptional regulation in vivo, various plasmids were generated based on pCIE-tetM::GFP. pCIE-tetM::GFP with 5- or 10-bp insertion was generated by subcloning from pBK2+5 or pBK2+10. pCIE-tetM::GFP, pBK2 pBK2+5, and pBK2+10 were digested with the restriction enzymes XhoI and XbaI and ligated into pCIE-tetM::GFP. pCIE-tetM::GFP with XBS mutations was generated by QuikChange using the same primers as EMSA. The peptide-mediated induction by C was measured by reporter gene assay according to Segawa et al. with modifications (17). OG1RF-carrying plasmids were grown in M9-YE at 37°C for 15 h. The cultured strains were diluted to an OD600 of 0.1 in M9-YE and incubated at 37°C for 1 h, followed by addition of C (final concentration, 50 ng/mL) to induce the expression of GFP at 37°C for 1 h. The induced cells were centrifuged at 4,000 × rpm for 10 min and resuspended in 100 μL potassium phosphate-buffered saline (KPBS). The GFP fluorescence intensity was measured by using Synergy H1 Hybrid multimode microplate reader (BioTek). The results of 3 replicate assays for each strain/induction condition combination are presented as bar graphs with standard deviations. We note that the insertion of DNA between the two XBS sites appears to reduce PrgX protein levels (this region of the plasmid also encodes prgX mRNA transcribed from the opposite strand). To ensure that the level of PrgX in responder cells was not limiting, we inserted a coresident pCF10 derivative in the reporter strains for the results shown in Fig. 3A and Fig. 5A; this served to eliminate significant PQ expression in uninduced cells as indicated by comparing Fig. 3A and Fig. 5A to Fig. S3.
ACKNOWLEDGMENTS
We thank Yuqing Chen for assistance with protein and reagent preparation.
This research was supported by USPHS grant 5R35GM118079 to G.M.D. T.S. was a recipient of a fellowship from the Uehara Memorial Foundation.
Footnotes
Supplemental material is available online only.
Contributor Information
Gary M. Dunny, Email: dunny001@umn.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Perez-Pascual D, Monnet V, Gardan R. 2016. Bacterial cell-cell communication in the host via RRNPP peptide-binding regulators. Front Microbiol 7:706. 10.3389/fmicb.2016.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Bandyopadhyay A, Kozlowicz BK, Haemig HAH, Tai A, Hu WS, Dunny GM. 2017. Mechanisms of peptide sex pheromone regulation of conjugation in Enterococcus faecalis. Microbiologyopen 6:e00492-13. 10.1002/mbo3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler V, Keller W, Grohmann E. 2019. Regulation of Gram-positive conjugation. Front Microbiol 10:1134. 10.3389/fmicb.2019.01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neiditch MB, Capodagli GC, Prehna G, Federle MJ. 2017. Genetic and structural analyses of RRNPP intercellular peptide signaling of Gram-positive bacteria. Annu Rev Genet 51:311–333. 10.1146/annurev-genet-120116-023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA 75:3479–3483. 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler JR, Dunny GM. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 25:1377–1388. 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Fujimoto S, Clewell DB. 1998. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc Natl Acad Sci USA 95:6430–6435. 10.1073/pnas.95.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozlowicz BK, Shi K, Gu ZY, Ohlendorf DH, Earhart CA, Dunny GM. 2006. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol 62:958–969. 10.1111/j.1365-2958.2006.05434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi K, Brown CK, Gu Z-Y, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. 2005. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci USA 102:18596–18601. 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antiporta MH, Dunny GM. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol 184:1155–1162. 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori M, Sakagami Y, Ishii Y, Isogai A, Kitada C, Fujino M, Adsit JC, Dunny GM, Suzuki A. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J Biol Chem 263:14574–14578. 10.1016/S0021-9258(18)68258-4. [DOI] [PubMed] [Google Scholar]
- 12.Ruhfel RE, Manias DA, Dunny GM. 1993. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J Bacteriol 175:5253–5259. 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol 176:7405–7408. 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, Cook LCC, Shu CC, Chen Y, Manias DA, Ramkrishna D, Dunny GM, Hu WS. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci USA 110:7086–7090. 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breuer RJ, Hirt H, Dunny GM. 2018. Mechanistic features of the enterococcal pCF10 sex pheromone response and the biology of Enterococcus faecalis in its natural habitat. J Bacteriol 200:e00733-17. 10.1128/JB.00733-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol 187:1044–1054. 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segawa T, Johnson CM, Berntsson RP, Dunny GM. 2021. Two ABC transport systems carry out peptide uptake in Enterococcus faecalis: their roles in growth and in uptake of sex pheromones. Mol Microbiol 116:459–469. 10.1111/mmi.14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard BA, Podbielski A, Hedberg PJ, Dunny GM. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA 93:260–264. 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berntsson RPA, Schuurman-Wolters GK, Dunny G, Slotboom DJ, Poolman B. 2012. Structure and mode of peptide binding of pheromone receptor PrgZ. J Biol Chem 287:37165–37170. 10.1074/jbc.M112.386334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breuer RJ, Bandyopadhyay A, O'Brien SA, Barnes AMT, Hunter RC, Hu W-S, Dunny GM. 2017. Stochasticity in the enterococcal sex pheromone response revealed by quantitative analysis of transcription in single cells. PLoS Genet 13:e1006878. 10.1371/journal.pgen.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CM. 2011. Reciprocal regulation between the prgQ and prgX operons of pCF10, a conjugative plasmid of Enterococcus faecalis. University of Minnesota, Minneapolis, MN. [Google Scholar]
- 22.Bae T, Clerc-Bardin S, Dunny GM. 2000. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol 297:861–875. 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- 23.Bae T, Kozlowicz B, Dunny GM. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J Mol Biol 315:995–1007. 10.1006/jmbi.2001.5294. [DOI] [PubMed] [Google Scholar]
- 24.Bae T, Dunny GM. 2001. Dominant-negative mutants of prgX: evidence for a role for PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol Microbiol 39:1307–1320. 10.1111/j.1365-2958.2001.02319.x. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, Dunny GM, Hu WS. 2011. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci USA 108:9721–9726. 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson CM, Haemig HHA, Chatterjee A, Wei-Shou H, Weaver KE, Dunny GM. 2011. RNA-mediated reciprocal regulation between two bacterial operons is RNase III dependent. mBio 2:e00189-11. 10.1128/mBio.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bensing BA, Meyer BJ, Dunny GM. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci USA 93:7794–7799. 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 29.Caserta E, Haemig HAH, Manias DA, Tomsic J, Grundy FJ, Henkin TM, Dunny GM. 2012. In vivo and in vitro analyses of regulation of the pheromone-responsive prgQ promoter by the PrgX pheromone receptor protein. J Bacteriol 194:3386–3394. 10.1128/JB.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fixen KR, Chandler JR, Le T, Kozlowicz BK, Manias DA, Dunny GM. 2007. Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery. J Bacteriol 189:1399–1406. 10.1128/JB.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozlowicz BK. 2005. The molecular mechanism and peptide signaling response of PrgX used to control pheromone-induced conjugative transfer of pCF10. University of Minnesota, Minneapolis, MN. [Google Scholar]
- 32.Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol 190:1172–1183. 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozlowicz BK, Bae T, Dunny GM. 2004. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol Microbiol 54:520–532. 10.1111/j.1365-2958.2004.04286.x. [DOI] [PubMed] [Google Scholar]
- 34.Dunny GM, Clewell DB. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J Bacteriol 124:784–790. 10.1128/jb.124.2.784-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaparian RR, Olney SG, Hustmyer CM, Rowe-Magnus DA, van Kessel JC. 2016. Integration host factor and LuxR synergistically bind DNA to coactivate quorum-sensing genes in Vibrio harveyi. Mol Microbiol 101:823–840. 10.1111/mmi.13425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6. Download jb.00298-22-s0001.pdf, PDF file, 0.6 MB (572.1KB, pdf)