ABSTRACT
Genetic exchange between different Leishmania strains in the sand fly vector has been experimentally demonstrated and is supported by population genetic studies. In nature, opportunities for Leishmania interstrain mating are restricted to flies biting multiply infected hosts or through multiple bites of different hosts. In contrast, self-mating could occur in any infected sand fly. By crossing two recombinant lines derived from the same Leishmania major strain, each expressing a different drug-resistance marker, self-hybridization in L. major was confirmed in a natural sand fly vector, Phlebotomus duboscqi, and in frequencies comparable to interstrain crosses. We provide the first high resolution, whole-genome sequencing analysis of large numbers of selfing progeny, their parents, and parental subclones. Genetic exchange consistent with classical meiosis is supported by the biallelic inheritance of the rare homozygous single nucleotide polymorphisms (SNPs) that arose by mutation during the generation of the parental clones. In contrast, heterozygous SNPs largely failed to be transmitted in Mendelian ratios for reasons not understood. SNPs that were heterozygous in both parents, however, recombined to produce homozygous alleles in some hybrids. For trisomic chromosomes present in both parents, transmittal to the progeny was only altered by self-hybridization, involving a gain or loss of somy in frequencies predicted by a meiotic process. Whole-genome polyploidization was also observed in the selfing progeny. Thus, self-hybridization in Leishmania, with its potential to occur in any infected sand fly, may be an important source of karyotype variation, loss of heterozygosity, and functional diversity.
KEYWORDS: Leishmania, hybridization, meiosis, sand fly
INTRODUCTION
Leishmaniases are sand fly-transmitted diseases of humans and domestic animals caused by kinetoplastid protozoan parasites of the genus Leishmania. The severity of the clinical outcomes in humans range from localized, self-limiting cutaneous lesions to mucosal involvement, to visceral disease that is fatal in the absence of treatment. These clinical types typically have distinct parasite species associations, with over 20 species described to date (1). Leishmania have a dimorphic life cycle consisting of extracellular promastigotes that multiply asexually within the digestive tract of the female sand fly vector, and intracellular amastigotes that multiply asexually within phagocytic cells, mainly macrophages, of their vertebrate hosts. Leishmania are largely diploid, although most if not all Leishmania strains show various degrees of aneuploidy (2). Frequent copy number variations are observed at the level of whole chromosomes or individual genes that are associated with drug resistance or tissue tropism (3–8). Amplification of specific chromosomes has also been linked to the fitness gain of Leishmania promastigotes during their growth in vitro (9).
In addition to cycles of clonal expansion, experimental studies have revealed that the extracellular promastigote stage(s) of Leishmania can hybridize in the sand fly vector (10–13) and in axenic culture (14, 15). Using pairwise combinations of parental lines expressing distinct drug resistant markers, double-drug-resistant (DDR) lines could be recovered from sand flies or cultures coinoculated with different strains and even different species of Leishmania. Whole-genome sequence (WGS) analyses of a large number of experimental hybrids generated in sand flies revealed parental chromosome contributions that were consistent with a meiotic process, either classical meiosis involving the generation and fusion of haploid gametic cells, or a tetraploid meiotic cycle involving intermediates formed by fusion of diploid cells. Thus, the progeny clones were found to be heterozygous at virtually all marker loci that were homozygous and different between the parents, with balanced contributions from each parent (16). Backcross progeny clones revealed genome-wide patterns of recombination, demonstrating that classical crossing over occurs at meiosis (16). Occasional “3n” and “4n” hybrid genotypes were also observed, while kinetoplast maxicircle DNA was always inherited from only one parent. These experimental findings supported what had long been argued based on the many examples of intra- and interspecies hybrid genotypes detected in field isolates (17–32), that sex constitutes a natural reproductive mode in the genus.
Sexual reproduction can itself be highly variable, with mating systems predominated by outcrossing, selfing, or both (33). Selfing refers to mating between haploid cells or nuclei derived from the same diploid individual, and is a particular case of inbreeding, which more generally refers to mating between genetically highly homogeneous cells or nuclei whether derived from the same diploid individual or not. In Leishmania, extreme inbreeding has been inferred from population genetic studies of Leishmania braziliensis strains based on the large deficiency in the heterozygosity of microsatellite loci analyzed (34), which is inconsistent with strictly clonal reproduction that will tend to accumulate divergence between alleles within the same loci (35). Whole-genome sequencing of Leishmania infantum isolates from Turkey or Leishmania tropica isolates from the Middle East revealed variable and patchy distribution of heterozygosity consistent with outcrossing with subsequent selfing or inbreeding via sexual or parasexual mating (27, 30).
Selfing or intraclonal mating has been experimentally demonstrated in the related kinetoplastid pathogen, Trypanosoma brucei, during its cyclical development in the tsetse fly vector (36, 37). In Leishmania, the direct demonstration that selfing can occur in the vector is confined to a single report in which two hybrid clones were recovered from sand flies coinfected with two recombinant lines derived from the same strain of L. infantum but bearing different drug resistance and fluorescence genes (38). More recently, two recombinant lines derived from the same strain of L. tropica were used in coculture experiments to generate 3 clones that were shown to have hybridized their drug resistance and fluorescence markers in vitro (39). In the present studies, we report that L. major is capable of self-hybridization, and provide the first high resolution, whole-genome sequencing analysis of large numbers of selfing progeny. By assessing allele frequencies and chromosome copy number in the parents, their subclones and their hybrid progeny, we could compare the extent to which self-hybridization contributes to genome alterations in Leishmania.
RESULTS
Generation of selfing hybrids in L. major.
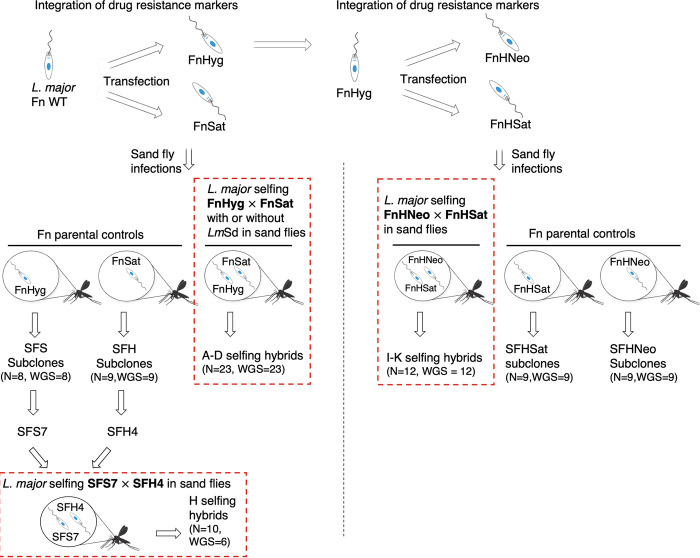
We carried out a total of 8 independent self-mating experiments in Phlebotomus duboscqi sand flies infected with 3 different mating pairs of recombinant clonal lines of the L. major Fn strain (LmFn) (Table 1). Fig. 1 summarizes these experiments with respect to the different conditions and recombinant parental clones used in our attempts to increase the efficiency of self-hybridization, and/or to minimize the genomic differences between the parents that arose during generation of the recombinant lines.
TABLE 1.
Summary of self-mating experiments and hybrid recovery
| Experiment | Cross | No. of clean gutsa | No. of hybrids | Hybrid recovery (%) | Hybrid labels |
|---|---|---|---|---|---|
| 1 | Fn Sat × Fn Hyg × Sd Sat | 100 | 7 (Self) | 7 | A1–8 |
| Fn Sat × Fn Hyg × Sd Hyg | 43 | 1 (Self) | 2.3 | ||
| 2 | Fn Sat × Fn Hyg × Sd Sat | 118 | 0 | 0 | B1–2 |
| Fn Sat × Fn Hyg | 121 | 2 | 1.7 | ||
| 3 | Fn Sat × Fn Hyg × Sd Sat | 118 | 1 (Self) | 0.84 | C1 |
| Fn Sat × Fn Hyg | 122 | 0 | 0 | ||
| 4 | Fn Sat × Fn Hyg × Sd Sat | 126 | 6 (Self) | 4.8 | D1–4;6−13 |
| Fn Sat × Fn Hyg | 124 | 6 | 4.8 | ||
| 5 | SFS7 × SFH4 | 76 | 10 | 15.6 | H1–10 |
| 6 | Fn HygNeo × Fn HygSat | 72 | 2 | 2.7 | I1–2 |
| 7 | Fn HygNeo × Fn HygSat | 47 | 3 | 6.4 | J1–3 |
| 8 | Fn HygNeo × Fn HygSat | 52 | 7 | 13.5 | K1–7 |
| Total | 1,119 | 45 | 4.0 |
Clean guts refers to the absence of fungal or bacterial contamination in the promastigote selection media containing the midgut homogenate.
FIG 1.
Schematic diagram summarizing the different self-hybridization experiments performed in this work. L. major Fn cells were transfected to generate parental clones carrying different drug resistance markers that were used in sand fly coinfection for self-mating, selection and cloning of double-drug-resistant hybrids. Self-hybridization experiments involving 3 different sets of parental clones were performed, highlighted by red dashed boxes. Each parental clone was also used for single infection of sand flies, recovery from individual flies, and subsequent isolation of parental subclones in culture. In total, 45 selfing hybrid clones and 35 parental subclones were generated, of which 41 of the selfing hybrids, and all 35 of the subclones were analyzed by WGS along with their respective parental clones. L. major Sd (LmSd) was included in the initial crosses between FnSat × FnHyg to evaluate the impact of, including a heterologous strain on the selfing frequencies.
Altogether, we generated a total of 45 selfing hybrids from 1,119 infected sand flies (4.0%, Table 1), a frequency within the range seen in interstrain crosses previously described (12, 16). Selfing efficiency was not significantly affected by inclusion of a heterologous strain or use of more fly-adapted parental clones. We performed a total DNA analysis and WGS on 41 hybrid clones, including for comparison a total of 35 subclones of the parents, in each case generated from promastigotes recovered from an individual midgut singly infected with one of the parental clones (Fig. 1). Sand fly midgut colonization and subsequent in vitro selection and cloning represent important bottlenecks during experimental mating. We therefore compared findings between selfing hybrids and the 35 parental subclones manipulated in a similar manner.
Heterologous strains are not required to initiate genetic exchange in L. major.
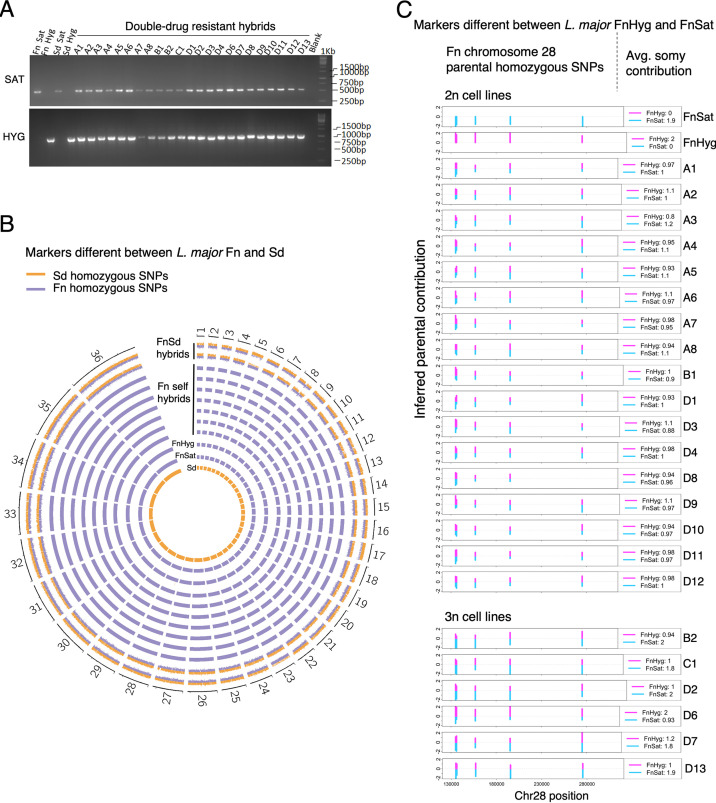
In the first series of experiments (experiments1 to 4), we coinfected sand flies with 2 recombinant clones resistant to either Hygromycin B (Hyg) or Nourseothricin (Sat), referred to as FnHyg or FnSat, respectively, and selected for DDR promastigotes. After an initial coinfection experiment with these parents failed to yield any hybrids, a third L. major clone was included in the sand fly coinfections. In different fungi such as Candida albicans and Cryptococcus neoformans, the presence of an opposite mating type (a or ɑ) promotes same sex or self-mating (40, 41). To test whether a similar phenomenon might occur in L. major, the infections included a third L. major clone, derived from the LmSd strain from Senegal, and transfected with a resistance gene to either Hyg or Sat (SdHyg or SdSat). Hybridization between LmFn and LmSd was demonstrated previously in sand flies (12) and thus potentially could contain hypothetical opposite “mating types.” While DDR hybrids were recovered from the flies infected with all 3 lines, in the experiments where the coinfections with just FnSat and FnHyg were run in parallel (experiments 2 to 4), the same frequency of hybrid recovery was obtained (1.9% and 2.2%, respectively).
All 23 DDR lines were cloned and genotyped by PCR, with a single clone from each line submitted to further analysis. All 23 clones (hybrid clones A to D) were PCR positive for both the Hyg and Sat resistance markers on chromosome 27 (Fig. 2A), confirming their hybrid genotypes. To determine whether the 16 DDR clones recovered from midguts that included the heterologous strain resulted from hybridization between self-strains, FnSat and FnHyg, or from interstrain hybridization with LmSd, we performed genome-wide single nucleotide polymorphism (SNP) analysis (Fig. 2B, see Table S1A in the supplemental material). Whole-genome sequencing reads of the parental and the 23 DDR clones, and 2 LmFn x LmSd hybrids (FnSd1 and FnSd2) generated previously (12), were aligned to the L. major Fn FV1 genome obtained from TritrypDB v50 (http://tritrypdb.org) and yielded an average coverage per sample of 46.85 (SD = 31.33; Table S1B). The mapped reads were processed to obtain total read depth, reference, and alternate allele frequencies using PAINT software (42). The number of heterozygous SNPs across the genomes of the FnSd1 and FnSd2 hybrids was 34,714 and 34,496, respectively (Table S1A, Fn reference genome). These SNPs resulted from biparental allelic inheritance of the SNPs that were homozygous and different between the LmFn and the LmSd parents (Fig. 2B). In the 23 self-DDR clones, the number of heterozygous SNPs was approximately 50 times lower, ranging from 626 to 825 (Fn reference genome) and more closely resembles the number of heterozygous SNPs in the FnSat and FnHyg parents (1310 and 1124, respectively). Moreover, we found no evidence for inheritance of LmSd alleles in any of the DDR clones (Fig. 2B). Thus, all the DDR parasites generated in the presence of LmSd resulted from hybridization between FnSat and FnHyg, and the presence of LmSd was not required to obtain self-hybrids. Indeed, there was a clear preference for self-mating, although it is possible that the Fn parents had a growth advantage over the Sd parent in the flies. In any event, these experiments did not inform as to the presence or absence of mating types in L. major.
FIG 2.
Genotyping of selfing hybrids. (A) PCR confirmation of the selectable parental markers Sat and Hyg in parental and hybrid progeny clones. Selfing hybrids shown were generated from crosses between FnSat and FnHyg in the presence or absence of LmSd to evaluate the impact of including a heterologous strain on the selfing frequencies; 1 kb: DNA ladder. (B) Circos plots of the frequencies of parental SNP markers that are homozygous and distinct between Sd (orange bars) and Fn lines (blue bars). Genome-wide heterozygosity of the parental markers was observed in the FnSd hybrids generated previously (12) (FnSd1 and FnSd2). Only Fn markers were detected in the DDR hybrids, allowing us to conclude that they are selfing hybrids. Data from seven representative selfing hybrids are shown (A1, A2, A3, A4, C1, D1, and D7). The frequencies of the parental alleles are represented as histogram bars for each genome position. Chromosome labels are shown on the outer circle. (C) Bottlebrush plots showing biparental inheritance of homozygous SNPs that are different between FnHyg and FnSat on chromosome 28 (chr28). Blue and magenta bars indicate SNPs mapping to the FnSat and FnHyg parents, respectively. The vertical distances correspond to the inferred allelic depth that was normalized across the genome which was assigned an average ploidy of 2n or 3n (selfing hybrids B2, C1, D2, D6, D7 and D13). The average somy contribution of the respective Fn parents is shown on the right inset of each plot. The data for one of the allelic markers on self-hybrid D8 was lost due to filtering.
Whole-genome sequencing reads of parental and hybrid clones. (A) Summary of homozygous and heterozygous SNPs in hybrids generated in the presence of L. major Sd. (B) Summary statistics of sequencing reads and alignment quality. Download Table S1, DOCX file, 0.03 MB (27.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Selfing between subclones of L. major Fn parental lines.
To test whether recent passage through sand flies might increase the frequency of self-hybrid formation, we singly infected flies with either FnSat or FnHyg, and generated subclones of each parent from the promastigotes recovered from individual flies. We carried out an additional selfing experiment using a subclone of FnSat, SFS7, crossed with a subclone of FnHyg, SFH4 (Fig. 1). Ten DDR lines were recovered from 10 different midguts out of a total of 76 flies (15.6%) (Table 1, experiment 5), each of which were cloned (hybrid clones H) and confirmed by PCR to have both the Sat and Hyg resistance markers (Fig. S1A). Of note, the high hybridization efficiency of the SFS7 and SFH4 subclones was not reproducible in a subsequent cross, suggesting that the variability was related more to differences in the fly populations used, as these experiments involved adults released from different generations of the colonized flies.
PCR confirmation of the selectable parental markers in parental and hybrid progeny clones. (A) PCR for the drug resistance markers Sat and Hyg in the parents and progeny from crosses between SFS7 and SFH4. (B) PCR for the selectable parental markers Neo and Sat in the parents and progeny from crosses between FnHNeo and FnHSat. 1kb: DNA ladder. Primers used amplify the specified integrated constructs on chromosome 27. Download FIG S1, TIF file, 1.0 MB (1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A final set of selfing hybrids was generated using a new pair of recombinant L. major Fn parental lines that were more closely coordinated in the number of mitotic generations removed from their common clone of origin, and thus might better reflect the conditions of natural intraclonal mating in the vector. Starting with the FnHyg clone, which we chose because it is the more euploid of the clones derived from LmFn that we have sequenced to date, we introduced an additional resistance marker, either to Sat or Neomycin (Neo), and derived antibiotic resistant clones referred to as FnHSat or FnHNeo (Fig. 1). Following coinfection of sand flies, midgut promastigotes were selected for growth in both Sat and Neo. In 3 independent experiments, a total of 12 DDR promastigote lines were recovered from a total of 171 flies dissected at day 10 postinfection (7.0%) (Table 1, experiments 5 to 8). The DDR lines were cloned (hybrid clones I to K) and genotyped by PCR to confirm that they were positive for both the Sat and Neo resistance markers on chromosome 27 (Fig. S1B).
Selfing hybrids show 2n and 3n DNA contents.
To determine the total DNA content of the selfing hybrids, parasites were fixed, digested with RNase, stained with propidium iodide, and analyzed by flow cytometry, as described (10, 12). While 23 of the 29 hybrids generated in crosses involving the FnSat × FnHyg or SFS7 × SFH4 showed an approximate 2n DNA content, 6 were close to 3n (B2, C1, D2, D6, D7, and D13) (Fig. S2A). This observation is in line with our previous analyses of experimental interstrain hybrids that showed triploid frequencies of 0 to 33% in different pairwise combinations of L. major strains (10, 12). The total DNA content analysis of the 12 hybrids generated between FnHSat and FnHNeo revealed that they were all close to 2n (Fig. S3A). An increase in ploidy was not observed in any of the 17 parental subclones derived from either FnSat or FnHyg (Fig. S2B), or in the 18 subclones derived from either FnHSat or FnHNeo (Fig. S3B).
Total cellular DNA content (ploidy) in FnHyg × FnSat crosses. Total DNA contents measured by flow cytometry on propidium iodide-stained parasites for the selfing hybrids in (A) and for the parental subclones in (B), as described in Materials and Methods. Data are presented as dark blue lines for each sample. 2n (red and gray histograms) and 3n (light blue histogram) reference samples were included in each plot for comparison. Triploid hybrids are B2, C1, D2, D6, D7 and D13. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Total cellular DNA content (ploidy) in FnHNeo × FnHSat crosses. Total DNA contents measured by flow cytometry on propidium iodide-stained parasites for the selfing hybrids in (A) and for the parental subclones in (B), as described in Materials and Methods. Download FIG S3, TIF file, 1.0 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Segregation of homozygous and heterozygous SNPs.
Whole-genome sequencing analysis of the parents and their hybrid progeny was used to identify homozygous and heterozygous SNPs that were different between the parents and the Fn reference genome, or between the parents, and to evaluate how the chromosomes marked by these SNPs were transmitted to the progeny. In previous studies of interstrain hybrids (12, 13), DDR parasites were confirmed to be full genomic hybrids by the appearance of new heterozygous SNPs across the nuclear genome, in each case acquired by the biallelic inheritance of the SNPs that were homozygous and different between the parents. Providing comparable evidence in the selfing hybrids was more challenging since the parents showed far fewer homozygous SNPs, differentiating strains or species. WGS nonetheless revealed 5 homozygous SNPs that were different between FnSat and FnHyg, all of which were close to the 5′ end of chromosome 28 (Fig. 2C). Importantly, all these markers appeared as heterozygous SNPs in each of the selfing progeny, with the 2n hybrids showing a roughly equal contribution from each parent. For the 3n hybrids, the extra chromosome appears to have been contributed by the FnSat parent in 5 of the 6 progeny clones (selfing hybrids B2, C1, D2, D6, D7, and D13). These markers were maintained as homozygous SNPs in each of 17 subclones of the parental lines (8 from FnSat and 9 from FnHyg) (Fig. S4). Thus, the hybridization extended to loci that were unlinked to the drug resistance markers on chromosome 27, and for chromosome 28 at least, the allotments were consistent with Mendelian ratios.
Homozygous parental SNP markers in FnSat and FnHyg subclones. Bottlebrush plots showing homozygous SNPs that are different between FnHyg and FnSat on chromosome 28 (chr 28). SNPs mapping to the FnSat and FnHyg parents are represented as blue and magenta bars, respectively. The vertical distances correspond to the inferred allelic depth that was normalized across the genome after assigning an average ploidy of 2n. Download FIG S4, TIF file, 0.5 MB (478.4KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
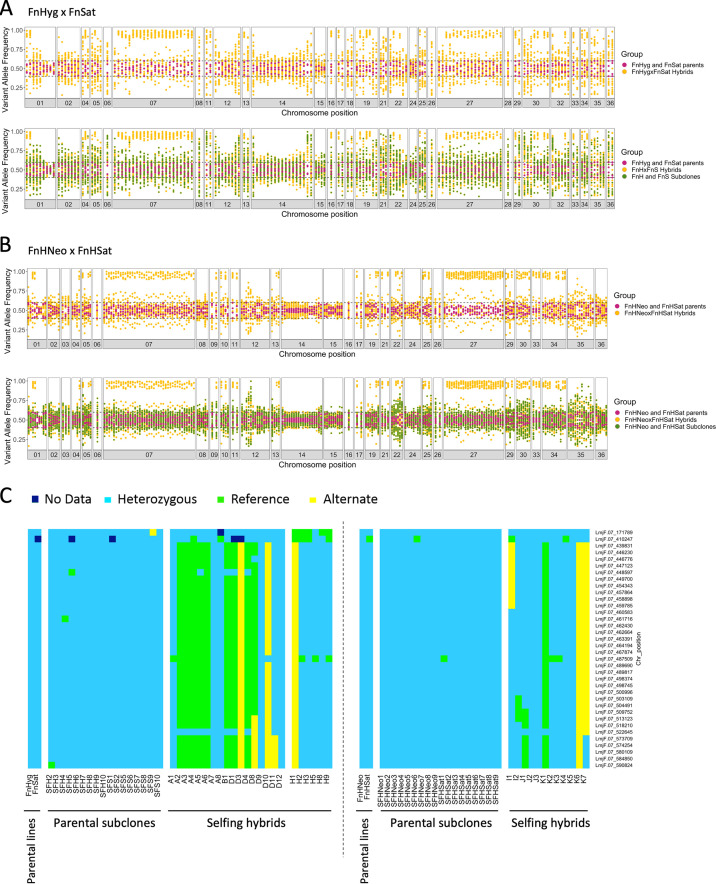
Testing for segregation of heterozygous alleles was another approach used to provide evidence that the DDR clones were the products of self-mating. While generation of the parental lines also resulted in heterozygous SNPs that were different between the parents (e.g., 648 alleles that were homozygous in FnHyg and heterozygous in FnSat), we focused our analysis on the SNPs that were heterozygous in both parents relative to the Fn reference genome, as the segregation of alleles that are identical in the parents would seem more relevant to the consequences of self-hybridization that would naturally occur. Under a Mendelian system, 50% of the progeny clones should remain heterozygous at these loci, while 25% should be homozygous for the reference alleles, and 25% homozygous for the alternative alleles. The results of the 2n selfing hybrids from FnHyg × FnSat (Fig. 3A) and FnHNeo × FnHSat (Fig. 3B) indicate that for most of these markers there was a significant deviation between expected and observed frequencies. For some chromosomes, little or no loss of heterozygosity (LOH) involving any of the markers was observed (e.g., chromosomes 2, 12, 14, 15, and 24), while for other chromosomes, the LOH was confined to only some of the markers on the same chromosome (e.g., chromosomes 1, 5, and 19). In contrast, chromosomes that combined their markers into either homozygous or heterozygous genotypes in expected frequencies were also observed (e.g., chromosomes 7, 27, and 30) (Fig. 3A and B). Fig. 3C shows the inheritance patterns for individual 2n hybrids of the heterozygous SNPs on chromosome 7, which followed Mendelian ratios, including evidence of crossing over in hybrids D8, D11, I1, I2, J1, and K7. Importantly, for each of the markers for which LOH was observed in some of the hybrid progeny, all were retained as heterozygous SNPs in each of the parental subclones (Fig. 3A to C). Overall, homozygous conversion was confined to the hybrid progeny and was observed for markers on 18 different chromosomes, even if in many cases the frequencies did not follow Mendelian ratios.
FIG 3.
Loss of heterozygosity in selfing hybrids. Segregation pattern of SNPs that are heterozygous in both parents is shown by the frequency of variant alleles identified on different chromosomes for 17 FnHyg x FnSat 2n hybrids (A) and 12 FnHNeo × FnHSat 2n hybrids (B). Parental SNP frequencies are shown in magenta, the corresponding allele on the same position are shown as yellow dots for the 2n hybrid progeny. The corresponding allele frequencies of the 2n parental subclones are represented by green dots in the bottom panels; 9 subclones of FnHyg and 8 subclones of FnSat in A, 9 subclones of FnHNeo and 9 subclones FnHSat in B. SNPs were considered heterozygous if their frequencies were between 0.4 and 0.6 on that genomic position, and homozygous if ≥ 0.85. Allele frequencies of 1.0 were subtracted by a random number between 0 and 0.05 before plotting to transform them into frequencies of 1.0 to 0.95 for a clearer visualization of data points. (C) Inheritance of parental heterozygous SNPs in high SNP density locus on chromosome 7 (position 439831 to 590824). Individual SNPs are indicated on the y axis on the right side of the panel and the type of SNP (heterozygous, homozygous matching L. major reference genome sequence and homozygous matching the alternate allele) is indicated by color. SFH and SFS refer to subclones derived from FnHyg and FnSat, respectively, and the 2n hybrid progeny are labeled A1 to H9. No significant difference between observed and expected frequencies: Chi square value, 0.7115; degrees of freedom, 2; P = 0.7007. SFHNeo and SFHSat refer to subclones derived from FnHNeo and FnHSat, respectively, and the 2n hybrid progeny are labeled I1 to K7. The segregation pattern of all homozygous and heterozygous SNPs in both parents is shown in Fig. S6.
Analysis of normalized coverage values at the loci carrying heterozygous SNPs revealed a coverage bias higher than 2-fold the chromosome average, in regions associated with apparent segregation distortion (Fig. S5A). For those alleles segregating as either heterozygous or homozygous SNPs in the hybrids, sequence coverage was comparable to the chromosome average (less than 2-fold; Fig. S5B). Thus, the discrepancies between expected and observed frequencies likely arise, at least in part, from marker loci with a high density of sequence repeats or copy number variation.
Chromosome loci with sequence coverage bias are associated with segregation distortion of heterozygous SNPs. Coverage plots showing the single base read depth normalized to the individual chromosome mean values in representative parental FnSat line (black lines). (A) Heterozygous parental SNPs (red points) that did not segregate in Mendelian ratios are found within genomic regions with elevated coverage (normalized coverage > 2) across chromosomes (chromosomes 12, 14 and 15 in the upper, middle and lower panels, respectively). (B) Heterozygous parental SNPs (red points) that segregated to generate homozygous alleles in hybrids are found outside regions of elevated coverage (normalized coverage < 2) across the chromosomes shown (chromosomes 7, 27, and 30 in the upper, middle and lower panel, respectively). Blue arrows highlight heterozygous SNPs on chromosomes 27 and 30 within regions with coverage bias and whose segregation does not result in homozygous hybrid alleles (see Fig. 3A). Dashed grey lines highlight base read depth values equal to the chromosome mean (Y = 1). Y axis scale is limited to between 0 and 4 for closer inspection of values within such range. Download FIG S5, TIF file, 1.3 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Somy inheritance associated with self-hybridization in L. major.
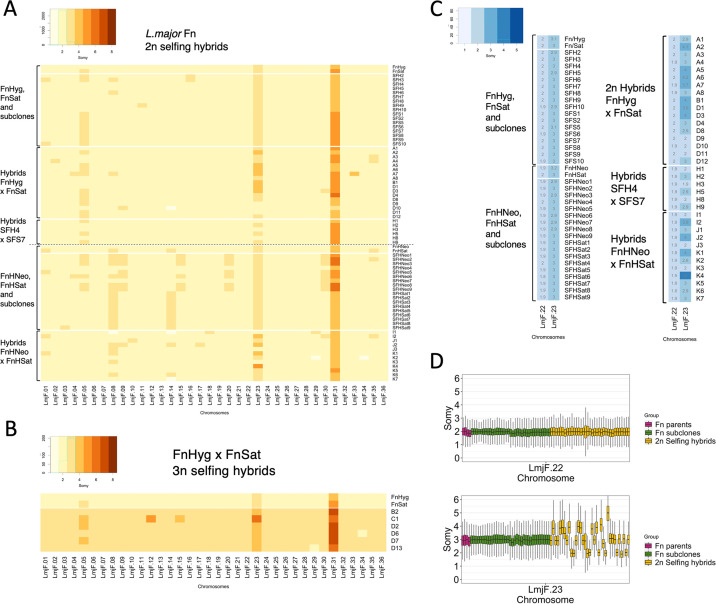
The WGS analysis of the hybrid clones revealed the copy number of the individual chromosomes. For somy quantification, the mean coverage for each chromosome was scaled to the ploidy of the cells. The somy profiles of the parents and hybrid clones, rounded off to the nearest integer value, are depicted as a heatmap in Fig. 4, with the 2n and 3n hybrids displayed separately (Fig. 4A and B, respectively). The numeric values for each somy, rounded to the nearest 0.1, are overlaid on the heatmaps in Fig. S7A and B. The noninteger values for many of the somies are most likely due to mosaic aneuploidy, which refers to somy variations that are present within clonal populations (2). The bulk analysis of somy at the population level, as was done here, will compute average values. To report the dispersion of somy scores along the length of the different chromosomes, we also performed a similar analysis using the GIP software (43) with sequence bins of 300 bp normalizing the mean coverage values by the median coverage of all bins (Fig. S8 and S9).
FIG 4.
Chromosome copy number variations generated by self-hybridization versus clonal growth. (A) Heatmap indicating copy number of all 36 chromosomes, rounded off to the nearest integer value, in the parental clones and 2n hybrids and subclones derived from each parent. (B) Heatmap of integer somies in the parental clones and 3n hybrids. (C) Heatmap of integer somies in the parental clones and all 2n hybrids and subclones for chromosomes 22 and 23. Overlaid are the somy values rounded to the nearest 0.1. (D) Boxplots of the somy scores for chromosomes 22 and 23 in Fn parental lines (magenta), 2n selfing hybrids (yellow), and Fn parental subclones (green) from all crosses pooled. The frequencies of selfing hybrids showing a mean chromosome 23 somy value of 2, 3, or 4 were 28.6%, 42.8%, and 25.8%, respectively. No significant difference between observed and expected frequencies: Chi square value, 0.8227; degrees of freedom, 2; P = 0.6628. GIP and giptools were used to calculate somy score distributions with genomic sequence coverage bins of 300 bp (gip.readthedocs.io/en/latest/giptools/karyotype.html). Somy score distributions for all chromosomes in 2n samples analyzed can be found in Fig. S8 and S9 for FnHyg × FnSat and FnHNeo × FnHSat crosses, respectively.
Segregation pattern of all homozygous and heterozygous SNPs in both parents. Plots represent the frequency of all variant alleles identified on different chromosomes for FnHyg x FnSat crosses in (A) and FnHNeo x FnHSat crosses in (B), regardless of whether SNPs were identical or distinct between the parents. Parental SNP frequencies are shown in magenta, the corresponding allele on the same position are shown as yellow dots for the 2n hybrid progeny, and as green dots for all the respective parental subclones analyzed. Frequencies of heterozygous alleles in chromosomes that have a somy higher than 2 fall outside the0.4 to 0.6 range (e.g., chromosomes 31 and 23). Download FIG S6, TIF file, 2.5 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Aneuploidy comparisons between hybrids and subclones. Heatmaps showing the chromosome somy values for parental lines and hybrids with 2n DNA content in (A) and 3n in (B). Color gradient represents somies rounded up to the nearest integer, with values within each cell corresponding to each somy rounded to the nearest 0.1. The same heatmaps are presented in Fig. 4 without the somy values. (C and D) Chromosome somy levels are presented on the Y axis, shown in (C) for FnHyg and FnSat parental lines (dark pink circles), or FnHNeo and FnHSat parental lines (dark green triangles) compared with their corresponding subclones (respective symbols in light colors); or shown in (D) for parental lines compared with their 2n selfing hybrids (respective symbols in light colors). Download FIG S7, TIF file, 2.7 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distribution of somy scores in FnHyg × FnSat crosses. Somy values are shown for each chromosome as boxplots generated using GIP and giptools with genomic sequence coverage bins of 300bp (gip.readthedocs.io/en/latest/giptools/karyotype.html). Parental lines are presented in magenta, parental subclones in green and 2n selfing hybrids in yellow. Download FIG S8, TIF file, 1.3 MB (1.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distribution of somy scores in FnHNeo × FnHSat crosses. Somy values are shown for each chromosome as boxplots generated using GIP and giptools with genomic sequence coverage bins of 300bp (gip.readthedocs.io/en/latest/giptools/karyotype.html). Parental lines are presented in magenta, parental subclones in green and 2n selfing hybrids in yellow. Download FIG S9, TIF file, 1.1 MB (1.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Despite originating from the same strain, differences in chromosome copy numbers were found between the parental clones that arose during the selection and cloning procedures. For the crosses involving FnSat and FnHyg, the parental somies were in each case close to 2n, except for chromosome 23 which had 3 copies in each parent, chromosome 31 which had 5 copies in FnSat and 4 copies in FnHyg, and chromosome 5 which was present in 3 copies in FnSat and 2 copies in FnHyg. For the 23 selfing hybrids that were close to 2n, 98.6% of the total of 759 chromosomes that were disomic in both parents, were transmitted as disomic chromosomes to the hybrids (Fig. 4A and Fig. S7C and D). For chromosome 5 that was trisomic in the FnSat parent, the hybrids were disomic 44% of the time and trisomic 56% of the time, close to expectations in which FnSat will have a roughly equal chance of transmitting 1 or 2 copies while the FnHyg parent will always transmit 1 copy. For chromosome 31, the hybrid somies ranged from 4 to 5, consistent with an expected contribution of 2 from the FnHyg parent, and of either 2 or 3 from the FnSat parent. Exceptions to the expected somy inheritance patterns were found in 8 of the 24 diploid progeny (1.4% of chromosomes), in which one, and sometimes multiple changes in chromosome copy number were observed. Most of the changes involved acquisition of an extra copy, typically from 2 to 3, including chromosomes 4 and 8 in hybrid A7, chromosome 35 in hybrids A2, A3 and A4, and chromosomes 2, 9 and 30 in hybrids A4, D10, and D3, respectively. Self-mating was also associated with a loss of somy, where chromosome 14 in hybrid D10 was present in only 1 copy despite being disomic in each parent.
Somy analysis of the FnHSat and FnHNeo parental clones revealed that they had also acquired different karyotypes. While the somy of the FnHNeo parent remained unchanged from FnHyg, the FnHSat parent acquired an extra copy for 4 chromosomes (chromosomes 1, 8, 14, and 35) that were disomic in the originating FnHyg clone (Fig. 4A). Of the chromosomes that remained disomic in both parents, 98% were transmitted in 2 copies to the 12 hybrids. For the chromosomes that were trisomic only in the FnHSat parent, chromosome 8 was inherited in close to expected frequencies, with 6 hybrids having 3 copies, and 5 having 2 copies. Unexpected aneuploidies in the hybrids involved 6 chromosomes that were disomic in each parent but were transmitted with an extra copy: chromosomes 18 and 29 in I1, chromosome 30 in I2, chromosome 10 in J1, and chromosomes 9 and 17 in J2. Monosomy in chromosomes 8, 14 (same as detected in hybrid D10), 29 and 34 was also observed.
For the six sequenced hybrids that were close to 3n in the DNA content analysis, the average coverage for each chromosome was scaled to a ploidy of 3. By this analysis, 96% of the chromosomes that were disomic in both parents were present in 3 copies in the hybrids. Somies varying from the expected number arising from whole-genome polyploidization included chromosomes 12 and 15 in hybrid C1 which were present in 5 and 4 copies instead of 3. A somy deficit for chromosomes 34 and 29 in hybrids D6 and D13, respectively, returned these chromosomes to a disomic state (Fig. 4B), which are the same chromosomes that showed monosomy in the 2n hybrid K2. The data are generally consistent with one parent having contributed an extra full genome, with a further gain or loss of somy involving a few chromosomes that occurred during meiosis or subsequent mitotic divisions.
Karyotype changes associated with clonal growth.
WGS of the parental subclones following their single passage through sand flies allowed us to track copy number variations in individual chromosomes that were associated with clonal growth, either as promastigote stages in the vector and/or during their subsequent growth in vitro during the cloning procedure. Of the 8 subclones of FnSat that were generated following passage through individual flies, only SFS10 demonstrated any change in somy, having acquired an extra copy of chromosome 1. Somy changes were more frequent in the 9 subclones generated from FnHyg, with 4 clones (SFH 2, 3, 6, and 9) having acquired an extra copy of at least one chromosome that was disomic in the originating clone (chromosomes 1, 5, 8, 11, and 16). No loss of somy was observed in any of the subclones. The subclones of each of the FnHSat and FnHNeo parents acquired a surprising karyotype diversity. Interestingly, 6 of the 9 subclones of FnHNeo displayed new aneuploidies that were identical across multiple chromosomes, each having acquired an extra copy of chromosomes 5, 8, 15, 20, and 30. Only the SFHNeo4 subclone remained unchanged from the originating FnHNeo clone. The karyotypes of the FnHSat subclones were largely identical to one another, though altered from the originating clone in having lost the extra copies of chromosomes 1 and 35, while retaining the extra copies of chromosomes 8 and 14. Altogether, the average somy was found to be present in extra copy in the subclones compared to their respective parent in 3.9% of the 1,260 chromosomes in the 35 2n subclones, while in the 35 2n hybrids this frequency was 1.1%, discounting the changes in somy that could be accounted for by meiotic segregation (discussed further below). Thus, we cannot conclude that self-hybridization itself contributed to these karyotype changes, since the hybrids will have also experienced clonal growth in the fly and in culture. In contrast, the loss of somy-producing chromosomes in single copy, while rare, was confined to the selfing hybrids (6 chromosomes, 0.5%; Fig. 4A and Fig. S7D).
Karyotype changes unique to selfing hybrids.
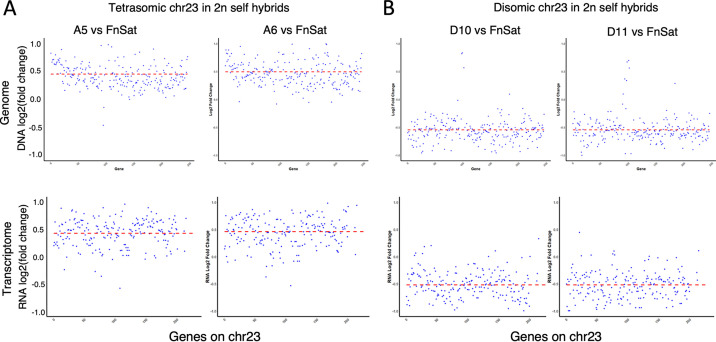
Despite the genomic plasticity associated with clonal growth, chromosome 23, which is trisomic in each of the parental lines, was stably transmitted as a trisomic chromosome to all 35 parental subclones. In contrast, 19 of the 35 2n hybrid clones were either tetrasomic (28.6%), or disomic (25.8%) for chromosome 23, in each case close to expectations under a meiotic process in which each parent has a 50% chance of transmitting either one or two copies. The somy of chromosome 23 is summarized as a heatmap and a boxplot in Fig. 4C and D, respectively, for all the parents, subclones, and 2n hybrids. Chromosome 22, which is disomic in each of the parents and was transmitted in 2 copies to all the hybrids and subclones, is shown for comparison (Fig. 4C and D). By RNA-seq analyses, we could confirm that the variations in copy numbers of chromosome 23 had a direct effect on the mRNA abundance of genes present on this chromosome. Hybrids A5 and A6, which each had a chromosome 23 somy close to 4, showed a 1.37-fold increase (0.46 log2 fold increase) relative to the FnSat parent in their average transcript abundance across this chromosome (Fig. 5A). For hybrids D10 and D11, which each had a chromosome 23 somy close to 2, their transcripts levels averaged 0.71-fold the levels in the FnSat parent (0.48 log2 fold decrease) (Fig. 5B). These results are consistent with previous studies showing the importance of gene dosage in the regulation of gene expression in Leishmania (9, 22, 44).
FIG 5.
Gene dosage effect due to chromosome copy numbers in chromosome 23 (chr23). The x axis represents the base pair position along chromosome 23. The y axis represents the DNA (upper panel) and RNA (lower panel) read counts in hybrids A5 and A6 (tetrasomic chr 23) relative to the FnSat parent (A), and in hybrids D10 and D11 (disomic chr23) relative to the FnSat parent (B).
DISCUSSION
Our findings extend to L. major the direct demonstration, previously shown for two clones of L. infantum (38), that members of the genus can undergo self-hybridization in a natural vector. More critically, our data provide the first high-resolution genomic analysis of large numbers of selfing hybrids in Leishmania. By singly introducing two different drug resistance markers into the same L. major strain, we could select for DDR hybrids in coinfected flies. Overall, hybrids were recovered from 3.7% of coinfected flies, within the range of recovery of interstrain hybrids previously described (10, 12, 13, 16). The presence of a heterologous strain, or recent passage of the parental lines through sand flies, did not reproducibly improve the frequency of self-hybridization. Genetic exchange consistent with a meiotic process is supported by the biallelic inheritance of the rare homozygous SNPs that arose by mutation during the generation of the parental lines. We could also assign a meiotic process to the close to predicted frequencies of selfing hybrids that inherited either 2, 3, or 4 copies of chromosomes that were trisomic in one or both parents. In contrast, and with the exception of a few chromosomes containing clusters of heterozygous SNPs, the inheritance of most heterozygous SNPs did not fit Mendelian expectations. Thus, while we can conclude that self-hybridization extends to loci unlinked to the resistance markers, we lack consistent evidence that the hybridization is driven by a meiotic-like process, which should operate genome wide.
In our previous studies of interstrain hybrids, we observed highly predictable, genome-wide patterns of chromosome inheritance when they were tracked using SNPs that were homozygous and different between the parents (16). Since the parental lines generated to select for self-hybridizing genomes contained only a single block of homozygous SNPs, we relied on the higher frequency and wider distribution of heterozygous SNPs in the parents to assess chromosomal inheritance. Overall, the disconnect between the behavior of the homozygous and heterozygous SNPs is difficult to explain. However, the observation that several parental heterozygous SNPs with apparent segregation bias were often associated with genomic regions with higher sequence coverage than the chromosome average (Fig. S5A), suggests that local copy number variations (CNVs) and/or paralogous sequence variants (PSVs) could be factors in this discrepancy. Correct assignment of sequence reads to multiple copy gene families represents a persisting challenge in genome reference mapping, particularly in Leishmania where gene copy number variability is a characteristic feature of their genomes. Even high quality Leishmania reference genomes may carry potentially misassembled regions due to the large density of repetitive sequences.
Another mechanism that might explain some of the anomalies in chromosome inheritance is parasexual mating, involving a tetraploid intermediate followed by random loss of chromosomes during mitotic divisions. This “random” process, however, would need to account for the fact that the chromosomes with only few exceptions were returned to a close to disomic state, and that for many of the disomic chromosomes (e.g., chromosomes 2, 12, 14, 15, and 24), heterozygosity was uniformly retained in every hybrid. Another explanation involves a potential loss of fitness induced by homozygosity in some of the genomic loci with heterozygous SNPs. Such a mechanism would require a strong haplotype selective pressure associated with sand fly colonization and/or in vitro growth, since these loci frequently remained heterozygous in all the genomes analyzed.
Assessing somy inheritance in Leishmania can also be confounded by the karyotype changes that accompany clonal growth. Copy number variations involving different chromosomes are commonly observed in cultured promastigotes, with specific karyotypes associated with a selective growth advantage of the cultured cells (9, 44–46). The contribution of mitotic versus meiotic divisions to aneuploid changes in Leishmania is not known. In our prior WGS analyses of interstrain hybrids generated in sand flies, somy values in the progeny were highly predictable based on a meiotic-like process (16). Unexpected somy inheritance patterns were nonetheless observed in roughly 2% of the hybrid chromosomes (16). Of particular relevance to the selfing hybrids described here, same sex mating in the pathogenic yeast Cryptococcus neoformans generated aneuploids that were not observed during asexual reproduction (47). By comparing the somy changes in the selfing hybrids, which also experienced clonal growth in the sand fly and in culture, with the somy changes in the parental subclones, which only experienced clonal growth, we could assess the possible contribution of hybridization to the karyotype plasticity observed. Because of mosaic aneuploidy, it is likely that the presence of “new” aneuploidies in the subclones reflects the selective recovery of aneuploidies that were already present at subintegral levels in the parent. In comparison of the 35 near diploid hybrids and the 35 near diploid subclones, neither the frequency of new somies (discounting the new somies predicted by meiotic segregation, discussed for chromosome 23 below), nor the frequency of hybrids with at least one somy difference from the parents, was greater in the selfing progeny. The new karyotypes that emerged during the generation of the parental clones and their subclones in most cases were defined by an increase of somy from 2 to 3 involving a subset of chromosomes, many of which have been observed in increased copy number in the bulk sequencing of cultured promastigotes of Leishmania donovani, L. infantum, Leishmania mexicana, or L. braziliensis (3, 9, 29, 48–50), and in the recent single-cell genome sequencing that elegantly revealed the evolution of mosaic aneuploidy in L. donovani (46). Together, these findings support the hypothesis that the increase in the dosage of genes present on particular chromosomes confers a fitness advantage to the cultured cells (9). For the chromosomes that were already present in extra copy in the parental clones, some chromosomes, e.g., chromosome 5, 8, 14, and 23, were stably transmitted in extra copy to each of the subclones, evidence that once acquired, the extra copies of these chromosomes were preserved under the selective conditions of axenic culture.
Despite the genome plasticity which can confound the analysis of allele and somy inheritance, WGS analysis of hybrids and parental subclones identified genomic changes that were unique to the selfing hybrids. These changes arose from the unique patterns of segregation and reassortment of chromosomes that were identical in both parents, and that may better reflect the consequences of self-hybridization in a sand fly infected with a single clonal lineage, as opposed to the allele and somy differences that arose between the parents during generation of the recombinant lines. Thus, chromosome 23, which is trisomic in both parents and was transmitted in 3 copies to all 35 subclones, was transmitted in either 2, 3, or 4 copies to the 35 hybrids in frequencies predicted by a meiotic process. Only genome hybridization was able to return this chromosome to a disomic state or to further amplify its copy number. These changes in average chromosome copy number were shown to be directly related to the average number of transcripts encoded by the genes present on this chromosome. Since selfing in the vector would be expected to similarly impact the heritable frequencies of any aneuploids that might arise during vegetative growth, self-hybridization should be considered as a potentially important source of karyotype diversity. Similarly, the combination of chromosomes from each parent carrying identical heterozygous alleles produced hybrid progeny that were homozygous at these loci for either the reference or alternative allele, and thus distinct from either parent or their subclones. This is the first direct evidence that self-hybridization can result in homozygous conversion of heterozygous alleles, and it lends support to the argument that the heterozygosity deficit in many Leishmania strains surveyed is the likely consequence of selfing or inbreeding that will produce offspring with homozygosity across the genome (34).
In addition to the unique patterns of allele and somy inheritance associated with self-hybridization, only the selfing hybrids showed evidence of whole-genome polyploidization, with 6 of the 41 hybrids presenting an approximate 3n DNA content. The two selfing clones of L. infantum previously described were also close to 3n (38), as were all the L. tropica selfing clones generated in vitro (39). Polyploidy has been a common feature of interstrain hybrids generated in sand flies (10, 12, 13), and especially those generated in vitro (14, 15). In contrast, whole-genome polyploidization has not been described in the bulk sequencing analyses of cultured promastigotes, although triploid cells were detected by single-cell genome sequencing as a rare component of the mosaic aneuploidy observed in a clone of L. donovani (46). As suggested by the authors, these triploid cells could have arisen by self-hybridization in vitro.
The coexistence of asexual reproduction and selfing involving a largely homozygous lineage is intriguing because the offspring will have highly homologous genotypes. Apart from the homozygous conversion of the limited number of heterozygous alleles, and the karyotype changes driven by meiotic segregation of polysomic chromosomes, other advantages of sex have been proposed, mainly in fungi, that may be relevant to Leishmania. These include eliminating DNA and RNA viruses (51), promoting DNA structural repair (52–54), and resetting epigenetic signatures (55, 56). More critically, recombinations driven by self-mating may help solve the problem posed by “Muller’s ratchet,” in which deleterious mutations will tend to accumulate during asexual reproduction.
In summary, the present work documents the self-hybridization capability of L. major in a natural sand fly vector and provides insights into the role of this process in generating genotype changes that impact heterozygosity, chromosome copy number, and transcript abundance. The occurrence of self-hybridization in Leishmania suggests that these processes and other potential benefits of sex can occur in every infected sand fly, whether populated by a mix of genetically different parasites, or more commonly by a single clonal lineage.
MATERIALS AND METHODS
Parasites.
Self-mating hybrids were generated between two recombinant clones of L. major that were derived from a strain isolated from a patient with cutaneous leishmaniasis acquired in the Jordan Valley (MHOM/IL/80/Friedlin) (10). The two parental clones were generated by integrating either nourseothricin resistance (FnSat) or hygromycin B resistance (FnHyg) into one allele of the 18S rRNA cistrons located on chromosome 27, as was previously described (12). Two recombinant clones of L. major Sd were used as an outcross in this study. This strain was isolated from a patient with cutaneous lesions acquired in Senegal (MHOM/SN/74/SD) (58). SdHyg is heterozygous for an allelic replacement of the LPG5A locus on chromosome 24 by a hygromycin B resistance cassette (10). L. major SdSat was generated by integration of the nourseothricin resistance marker into one allele of the 18S rRNA cistrons (59). To generate an additional group of selfing hybrids using FnHyg, this parental line was transfected with either SwaI-digested pA2-GFP-Neo (60) or SwaI-digested pLEXSY-sat2 (Jena Bioscience, Germany). Linear fragments were integrated into the 18S rRNA locus, and the cloned recombinants were referred to as either FnHNeo or FnHSat.
All parasites were grown at 26°C in complete medium 199 (CM199) supplemented with 20% heat-inactivated Fetal calf serum (FCS), 100 U/mL penicillin, 100 mg/mL streptomycin, 2 mM l-glutamine, 40 mM HEPES, 0.1 mM adenine (in 50 mM HEPES), 5 mg/mL hemin (in 50% triethanolamine), and 1 mg/mL 6-biotin, and containing either 25 μg/mL hygromycin B (EMD Biosciences, San Diego, CA),100 μg/mL nourseothricin (Jena Bioscience, Germany), 20 μg/mL Geneticin (neomycin analogue; Thermo Fisher, Waltham, MA), or a combination of these antibiotics as necessary.
Sand fly infections and recovery of genetic hybrids.
Hybrid parasites were generated as previously described (10, 12). Briefly, two or more parasite clones, harboring different antibiotic resistance genes, were used to coinfect Phlebotomus duboscqi sand flies, obtained from a colony initiated from field specimens collected in Mali, by seeding heparinized mouse blood with promastigotes from logarithmic phase cultures, and feeding the flies artificially through chick skin. All animals used in this research were used under a study protocol approved by the NIAID Animal Care and Use Committee (protocol number LPD 68E). All aspects of the use of animals in this research were monitored for compliance with the Animal Welfare Act, the PHS Policy, the U.S. Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training, and the NIH Guide for the Care and Use of Laboratory Animals.
Flies were dissected 10 days postinfection, and double-drug-resistant promastigotes were selected for growth in CM199 with the respective antibiotics. Double-drug-resistant parasites were cloned as previously described (10). Each hybrid clone described in this report was singly derived from an individual sand fly gut. For genotype comparisons with parental subclones that had not been selected for hybrids, flies were singly infected with either FnSat, FnHyg, FnHNeo, or FnHSat, and midgut promastigotes recovered after 10 days were cloned from individual flies.
Ploidy analysis.
Ploidy was determined by flow cytometry as was previously described (6). Briefly, log-phase procyclic promastigotes were fixed in 0.4% paraformaldehyde and permeabilized in 100% methanol for 15 min. Fixed parasites were treated with 10 μg/mL RNase A and stained with 20 μg/mL propidium iodide for 30 min. Samples were loaded on a BD FACS CANTOII and the data were analyzed using Flowjo software (Becton, Dickinson, San Jose, CA).
DNA extraction, PCR, and genotyping.
DNA was extracted from the parental lines, subclones of the parental lines, and hybrid progeny clones, using Qiagen DNeasy blood and tissue kit. Antibiotic resistance genes were PCR-amplified using Bioline Mytaq mix, as described previously (12).
Whole-genome DNA sequencing, SNP, and somy analyses.
One hundred nanograms of DNA was prepared for sequencing using the TruSeq Nano DNA library preparation kit (Illumina, San Diego, CA). Primer dimers in the libraries were removed by an additional AMPure Bead XP (Beckman Coulter, Indianapolis, IN) purification using the beads at 0.9× to sample volume. The dual-indexed libraries were quantified by qPCR using the Kapa Library quantification kit for Illumina sequencing (Roche, Basel, Switzerland) and normalized to 2.0 nM stocks. The samples were pooled equitably for paired-end 200-bp sequencing on the Illumina HiSeq 2500 for FnHyg × FnSat selfies and subclones, or a single paired-end 150-bp sequencing run on an Illumina HiSeq X Ten performed by Psomagen (Rockville, MD), for the FnHNeo × FnHSat selfies and subclones. The short reads from the Illumina Hiseq sequencer were aligned to the L. major FV1 genome version 50 using bwa aligner at default parameters. The SNPs were determined using SAMtools software and by providing the chromosome somies as one of the inputs (61), using our in-house PAINT package (42). The output SNP file was filtered to remove SNPs with coverage <5 and minor allele frequency <0.15. Alleles were considered heterozygous if the variant base called had a frequency between 0.4 and 0.6 in that genomic position, and homozygous if ≥0.85. The homozygous and heterozygous SNPs were counted on every genomic position and depicted as line plots in circos software. The markers where the parents were homozygous but different from each other or where both parents were heterozygous were queried for allele information in the hybrids. The frequencies of alleles inherited from either parent are drawn as bottle brush plots (12) (Fig. 2C).
RNA extraction and RNA-seq.
RNA was extracted from purified metacyclic promastigotes, as described previously (57). Each one of the clones was prepared in four different biological replicates, originating from four different cryopreserved culture vials. Three hundred nanograms of extracted total RNA was prepared for sequencing using the TruSeq Stranded mRNA-Seq HT library preparation kit (Illumina, San Diego, CA). The dual-indexed libraries were quantified by qPCR using the Kapa Library quantification kit for Illumina sequencing (Roche, Basel, Switzerland) and normalized to 2.0 nM stocks. Samples were pooled equitably for two paired-end 100-bp sequencing runs on an Illumina HiSeq 2500 at Rocky Mountain Laboratories (RML, NIAID, Hamilton, MT) for the FnHyg × FnSat selfies and subclones. The reads from the Illumina HiSeq sequencer in fastq format were verified for quality control using the Fastqc software package (62). The low-quality segments of the read were trimmed and/or filtered with Trimmomatic (63) using seed mismatches-3, palindrome clip threshold-50, simple clip threshold-10, sliding window-4, required quality-20, minimum length after trimming-40, and maximum read length to error rate of 50:0.8. The quality filtered reads were mapped to the L. major Friedlin reference genome v50 (TritrypDB) using the RSEM package (64), using default parameters and the STAR aligner. Abundance normalization was performed using RSEM expectation-maximization algorithm. The expected RSEM counts were rounded to the nearest integer value and the transcripts with zero counts across all the samples were filtered out.
DNA versus RNA coverage.
GFOLD (65) was used to calculate DNA-seq log2 fold change values between each hybrid strain and parental strain for each chromosome. The GFOLD fold change values were plotted and compared to the DESeq2-generated log2 fold change values for the RNA-seq data.
Data availability.
The raw sequence data were deposited in the BioProject SRA database (PRJNA900217). The data that support the findings of this study are available from the corresponding author upon reasonable request.
ACKNOWLEDGMENTS
S.M.B. and D.E.D. received grant support from the National Institutes of Health (NIH R01 AI031078 and R01 AI029646). This work was supported in part by the Intramural Research Program of the NIAID, NIH.
E.I., T.R.F., J.S., S.M.B., and D.S. contributed to the conceptualization and design of the studies. E.I., T.R.F., J.S., B.M.J., K.G., D.E.D., S.M.B., and D.S. contributed to the acquisition and analysis of the data. E.I., T.R.F., J.S., S.M.B., and D.S. contributed to the drafting of the manuscript.
We declare no competing interests.
Footnotes
This article is a direct contribution from David L. Sacks, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Gerald Spaeth, Institut Pasteur, and Tim Downing, Dublin City University.
Contributor Information
David Sacks, Email: dsacks@nih.gov.
Joseph Heitman, Duke University.
REFERENCES
- 1.Pace D. 2014. Leishmaniasis. J Infect 69:S10–8. doi: 10.1016/j.jinf.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Sterkers Y, Lachaud L, Bourgeois N, Crobu L, Bastien P, Pages M. 2012. Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania. Mol Microbiol 86:15–23. doi: 10.1111/j.1365-2958.2012.08185.x. [DOI] [PubMed] [Google Scholar]
- 3.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, Quail MA, Rijal S, Sanders M, Schonian G, Stark O, Sundar S, Vanaerschot M, Hertz-Fowler C, Dujardin JC, Berriman M. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leprohon P, Legare D, Raymond F, Madore E, Hardiman G, Corbeil J, Ouellette M. 2009. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res 37:1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang WW, Ramasamy G, McCall LI, Haydock A, Ranasinghe S, Abeygunasekara P, Sirimanna G, Wickremasinghe R, Myler P, Matlashewski G. 2014. Genetic analysis of Leishmania donovani tropism using a naturally attenuated cutaneous strain. PLoS Pathog 10:e1004244. doi: 10.1371/journal.ppat.1004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz AK, Titus R, Beverley SM. 1993. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc Natl Acad Sci USA 90:1599–1603. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beverley SM. 1991. Gene amplification in Leishmania. Annu Rev Microbiol 45:417–444. doi: 10.1146/annurev.mi.45.100191.002221. [DOI] [PubMed] [Google Scholar]
- 8.Ubeda JM, Legare D, Raymond F, Ouameur AA, Boisvert S, Rigault P, Corbeil J, Tremblay MJ, Olivier M, Papadopoulou B, Ouellette M. 2008. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol 9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prieto Barja P, Pescher P, Bussotti G, Dumetz F, Imamura H, Kedra D, Domagalska M, Chaumeau V, Himmelbauer H, Pages M, Sterkers Y, Dujardin JC, Notredame C, Spath GF. 2017. Haplotype selection as an adaptive mechanism in the protozoan pathogen Leishmania donovani. Nat Ecol Evol 1:1961–1969. doi: 10.1038/s41559-017-0361-x. [DOI] [PubMed] [Google Scholar]
- 10.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL. 2009. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadlova J, Yeo M, Seblova V, Lewis MD, Mauricio I, Volf P, Miles MA. 2011. Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One 6:e19851. doi: 10.1371/journal.pone.0019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DE, Kauffmann F, Barhoumi M, Grigg M, Owens K, Fay M, Dobson DE, Shaik J, Beverley SM, Sacks D. 2013. The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672. doi: 10.1371/journal.pgen.1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romano A, Inbar E, Debrabant A, Charmoy M, Lawyer P, Ribeiro-Gomes F, Barhoumi M, Grigg M, Shaik J, Dobson D, Beverley SM, Sacks DL. 2014. Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc Natl Acad Sci USA 111:16808–16813. doi: 10.1073/pnas.1415109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louradour I, Ferreira TR, Duge E, Karunaweera N, Paun A, Sacks D. 2022. Stress conditions promote Leishmania hybridization in vitro marked by expression of the ancestral gamete fusogen HAP2 as revealed by single-cell RNA-seq. Elife 11. doi: 10.7554/eLife.73488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louradour I, Ferreira TR, Ghosh K, Shaik J, Sacks D. 2020. In Vitro Generation of Leishmania Hybrids. Cell Rep 31:107507. doi: 10.1016/j.celrep.2020.03.071. [DOI] [PubMed] [Google Scholar]
- 16.Inbar E, Shaik J, Iantorno SA, Romano A, Nzelu CO, Owens K, Sanders MJ, Dobson D, Cotton JA, Grigg ME, Beverley SM, Sacks D. 2019. Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania. PLoS Genet 15:e1008042. doi: 10.1371/journal.pgen.1008042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banuls AL, Guerrini F, Le Pont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. 1997. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol 44:408–411. doi: 10.1111/j.1550-7408.1997.tb05716.x. [DOI] [PubMed] [Google Scholar]
- 18.Belli AA, Miles MA, Kelly JM. 1994. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology 109:435–442. doi: 10.1017/S0031182000080689. [DOI] [PubMed] [Google Scholar]
- 19.Chargui N, Amro A, Haouas N, Schonian G, Babba H, Schmidt S, Ravel C, Lefebvre M, Bastien P, Chaker E, Aoun K, Zribi M, Kuhls K. 2009. Population structure of Tunisian Leishmania infantum and evidence for the existence of hybrids and gene flow between genetically different populations. Int J Parasitol 39:801–811. doi: 10.1016/j.ijpara.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Dujardin JC, Banuls AL, Llanos-Cuentas A, Alvarez E, DeDoncker S, Jacquet D, Le Ray D, Arevalo J, Tibayrenc M. 1995. Putative Leishmania hybrids in the Eastern Andean valley of Huanuco, Peru. Acta Trop 59:293–307. doi: 10.1016/0001-706x(95)00094-u. [DOI] [PubMed] [Google Scholar]
- 21.Evans DA, Kennedy WP, Elbihari S, Chapman CJ, Smith V, Peters W. 1987. Hybrid formation within the genus Leishmania? Parassitologia 29:165–173. [PubMed] [Google Scholar]
- 22.Iantorno SA, Durrant C, Khan A, Sanders MJ, Beverley SM, Warren WC, Berriman M, Sacks DL, Cotton JA, Grigg ME. 2017. Gene expression in Leishmania is regulated predominantly by gene dosage. mBio 8. doi: 10.1128/mBio.01393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly JM, Law JM, Chapman CJ, Van Eys GJ, Evans DA. 1991. Evidence of genetic recombination in Leishmania. Mol Biochem Parasitol 46:253–263. doi: 10.1016/0166-6851(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 24.Nolder D, Roncal N, Davies CR, Llanos-Cuentas A, Miles MA. 2007. Multiple hybrid genotypes of Leishmania (viannia) in a focus of mucocutaneous Leishmaniasis. Am J Trop Med Hyg 76:573–578. doi: 10.4269/ajtmh.2007.76.573. [DOI] [PubMed] [Google Scholar]
- 25.Odiwuor S, De Doncker S, Maes I, Dujardin JC, Van der Auwera G. 2011. Natural Leishmania donovani/Leishmania aethiopica hybrids identified from Ethiopia. Infect Genet Evol 11:2113–2118. doi: 10.1016/j.meegid.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Ravel C, Cortes S, Pratlong F, Morio F, Dedet JP, Campino L. 2006. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int J Parasitol 36:1383–1388. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA, Smith DF. 2014. Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092. doi: 10.1371/journal.pgen.1004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torrico MC, De Doncker S, Arevalo J, Le Ray D, Dujardin JC. 1999. In vitro promastigote fitness of putative Leishmania (Viannia) braziliensis/Leishmania (Viannia) peruviana hybrids. Acta Trop 72:99–110. doi: 10.1016/s0001-706x(98)00076-x. [DOI] [PubMed] [Google Scholar]
- 29.Van den Broeck F, Savill NJ, Imamura H, Sanders M, Maes I, Cooper S, Mateus D, Jara M, Adaui V, Arevalo J, Llanos-Cuentas A, Garcia L, Cupolillo E, Miles M, Berriman M, Schnaufer A, Cotton JA, Dujardin JC. 2020. Ecological divergence and hybridization of Neotropical Leishmania parasites. Proc Natl Acad Sci USA 117:25159–25168. doi: 10.1073/pnas.1920136117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glans H, Lind Karlberg M, Advani R, Bradley M, Alm E, Andersson B, Downing T. 2021. High genome plasticity and frequent genetic exchange in Leishmania tropica isolates from Afghanistan, Iran and Syria. PLoS Negl Trop Dis 15:e0010110. doi: 10.1371/journal.pntd.0010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rougeron V, De Meeus T, Banuls AL. 2017. Reproduction in Leishmania: a focus on genetic exchange. Infect Genet Evol 50:128–132. doi: 10.1016/j.meegid.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Tibayrenc M, Ayala FJ. 2013. How clonal are Trypanosoma and Leishmania? Trends Parasitol 29:264–269. doi: 10.1016/j.pt.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Billiard S, Lopez-Villavicencio M, Hood ME, Giraud T. 2012. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol 25:1020–1038. doi: 10.1111/j.1420-9101.2012.02495.x. [DOI] [PubMed] [Google Scholar]
- 34.Rougeron V, De Meeus T, Hide M, Waleckx E, Bermudez H, Arevalo J, Llanos-Cuentas A, Dujardin JC, De Doncker S, Le Ray D, Ayala FJ, Banuls AL. 2009. Extreme inbreeding in Leishmania braziliensis. Proc Natl Acad Sci USA 106:10224–10229. doi: 10.1073/pnas.0904420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mark Welch DB, Meselson MS. 2001. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc Natl Acad Sci USA 98:6720–6724. doi: 10.1073/pnas.111144598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peacock L, Ferris V, Bailey M, Gibson W. 2009. Intraclonal mating occurs during tsetse transmission of Trypanosoma brucei. Parasit Vectors 2:43. doi: 10.1186/1756-3305-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tait A, Buchanan N, Hide G, Turner CM. 1996. Self-fertilisation in Trypanosoma brucei. Mol Biochem Parasitol 76:31–42. doi: 10.1016/0166-6851(95)02528-6. [DOI] [PubMed] [Google Scholar]
- 38.Calvo-Alvarez E, Alvarez-Velilla R, Jimenez M, Molina R, Perez-Pertejo Y, Balana-Fouce R, Reguera RM. 2014. First evidence of intraclonal genetic exchange in trypanosomatids using two Leishmania infantum fluorescent transgenic clones. PLoS Negl Trop Dis 8:e3075. doi: 10.1371/journal.pntd.0003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez-Corbo C, Dominguez-Asenjo B, Perez-Pertejo Y, Garcia-Estrada C, Bello FJ, Balana-Fouce R, Reguera RM. 2022. Axenic interspecies and intraclonal hybrid formation in Leishmania: successful crossings between visceral and cutaneous strains. PLoS Negl Trop Dis 16:e0010170. doi: 10.1371/journal.pntd.0010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alby K, Schaefer D, Bennett RJ. 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 42.Shaik JS, Dobson DE, Sacks DL, Beverley SM. 2021. Leishmania sexual reproductive strategies as resolved through computational methods designed for aneuploid genomes. Genes (Basel) 12:167. doi: 10.3390/genes12020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spath GF, Bussotti G. 2022. GIP: an open-source computational pipeline for mapping genomic instability from protists to cancer cells. Nucleic Acids Res 50:e36. doi: 10.1093/nar/gkab1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumetz F, Imamura H, Sanders M, Seblova V, Myskova J, Pescher P, Vanaerschot M, Meehan CJ, Cuypers B, De Muylder G, Spath GF, Bussotti G, Vermeesch JR, Berriman M, Cotton JA, Volf P, Dujardin JC, Domagalska MA. 2017. Modulation of aneuploidy in Leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. mBio 8. doi: 10.1128/mBio.00599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mannaert A, Downing T, Imamura H, Dujardin JC. 2012. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol 28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Negreira GH, Monsieurs P, Imamura H, Maes I, Kuk N, Yagoubat A, Van den Broeck F, Sterkers Y, Dujardin JC, Domagalska MA. 2022. High throughput single-cell genome sequencing gives insights into the generation and evolution of mosaic aneuploidy in Leishmania donovani. Nucleic Acids Res 50:293–305. doi: 10.1093/nar/gkab1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni M, Feretzaki M, Li WJ, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franssen SU, Durrant C, Stark O, Moser B, Downing T, Imamura H, Dujardin JC, Sanders MJ, Mauricio I, Miles MA, Schnur LF, Jaffe CL, Nasereddin A, Schallig H, Yeo M, Bhattacharyya T, Alam MZ, Berriman M, Wirth T, Schonian G, Cotton JA. 2020. Global genome diversity of the Leishmania donovani complex. Elife 9. doi: 10.7554/eLife.51243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imamura H, Downing T, Van den Broeck F, Sanders MJ, Rijal S, Sundar S, Mannaert A, Vanaerschot M, Berg M, De Muylder G, Dumetz F, Cuypers B, Maes I, Domagalska M, Decuypere S, Rai K, Uranw S, Bhattarai NR, Khanal B, Prajapati VK, Sharma S, Stark O, Schonian G, De Koning HP, Settimo L, Vanhollebeke B, Roy S, Ostyn B, Boelaert M, Maes L, Berriman M, Dujardin JC, Cotton JA. 2016. Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. Elife 5. doi: 10.7554/eLife.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, Harris D, Her Y, Herzyk P, Imamura H, Otto TD, Sanders M, Seeger K, Dujardin JC, Berriman M, Smith DF, Hertz-Fowler C, Mottram JC. 2011. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coenen A, Kevei F, Hoekstra RF. 1997. Factors affecting the spread of double-stranded RNA viruses in Aspergillus nidulans. Genet Res 69:1–10. doi: 10.1017/s001667239600256x. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein H, Byerly HC, Hopf FA, Michod RE. 1985. Genetic damage, mutation, and the evolution of sex. Science 229:1277–1281. doi: 10.1126/science.3898363. [DOI] [PubMed] [Google Scholar]
- 53.Gorelick R, Carpinone J. 2009. Origin and maintenance of sex: the evolutionary joys of self sex. Biological J the Linnean Society 98:707–728. doi: 10.1111/j.1095-8312.2009.01334.x. [DOI] [Google Scholar]
- 54.Silliker ME, Liotta MR, Cummings DJ. 1996. Elimination of mitochondrial mutations by sexual reproduction: two Podospora anserina mitochondrial mutants yield only wild-type progeny when mated. Curr Genet 30:318–324. doi: 10.1007/s002940050139. [DOI] [PubMed] [Google Scholar]
- 55.Wasson JA, Ruppersburg CC, Katz DJ. 2013. Restoring totipotency through epigenetic reprogramming. Brief Funct Genomics 12:118–128. doi: 10.1093/bfgp/els042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spence PJ, Brugat T, Langhorne J. 2015. Mosquitoes reset malaria parasites. PLoS Pathog 11:e1004987. doi: 10.1371/journal.ppat.1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inbar E, Hughitt VK, Dillon LA, Ghosh K, El-Sayed NM, Sacks DL. 2017. The transcriptome of Leishmania major developmental stages in their natural sand fly vector. mBio 8. doi: 10.1128/mBio.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Most M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Vainola R, Wolf JB, Zinner D. 2013. Hybridization and speciation. J Evol Biol 26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 59.Ahmed S, Colmenares M, Soong L, Goldsmith-Pestana K, Munstermann L, Molina R, McMahon-Pratt D. 2003. Intradermal infection model for pathogenesis and vaccine studies of murine visceral leishmaniasis. Infect Immun 71:401–410. doi: 10.1128/IAI.71.1.401-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chagas AC, Oliveira F, Debrabant A, Valenzuela JG, Ribeiro JM, Calvo E. 2014. Lundep, a sand fly salivary endonuclease increases Leishmania parasite survival in neutrophils and inhibits XIIa contact activation in human plasma. PLoS Pathog 10:e1003923. doi: 10.1371/journal.ppat.1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews S. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 63.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. 2012. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28:2782–2788. doi: 10.1093/bioinformatics/bts515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole-genome sequencing reads of parental and hybrid clones. (A) Summary of homozygous and heterozygous SNPs in hybrids generated in the presence of L. major Sd. (B) Summary statistics of sequencing reads and alignment quality. Download Table S1, DOCX file, 0.03 MB (27.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
PCR confirmation of the selectable parental markers in parental and hybrid progeny clones. (A) PCR for the drug resistance markers Sat and Hyg in the parents and progeny from crosses between SFS7 and SFH4. (B) PCR for the selectable parental markers Neo and Sat in the parents and progeny from crosses between FnHNeo and FnHSat. 1kb: DNA ladder. Primers used amplify the specified integrated constructs on chromosome 27. Download FIG S1, TIF file, 1.0 MB (1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Total cellular DNA content (ploidy) in FnHyg × FnSat crosses. Total DNA contents measured by flow cytometry on propidium iodide-stained parasites for the selfing hybrids in (A) and for the parental subclones in (B), as described in Materials and Methods. Data are presented as dark blue lines for each sample. 2n (red and gray histograms) and 3n (light blue histogram) reference samples were included in each plot for comparison. Triploid hybrids are B2, C1, D2, D6, D7 and D13. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Total cellular DNA content (ploidy) in FnHNeo × FnHSat crosses. Total DNA contents measured by flow cytometry on propidium iodide-stained parasites for the selfing hybrids in (A) and for the parental subclones in (B), as described in Materials and Methods. Download FIG S3, TIF file, 1.0 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Homozygous parental SNP markers in FnSat and FnHyg subclones. Bottlebrush plots showing homozygous SNPs that are different between FnHyg and FnSat on chromosome 28 (chr 28). SNPs mapping to the FnSat and FnHyg parents are represented as blue and magenta bars, respectively. The vertical distances correspond to the inferred allelic depth that was normalized across the genome after assigning an average ploidy of 2n. Download FIG S4, TIF file, 0.5 MB (478.4KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chromosome loci with sequence coverage bias are associated with segregation distortion of heterozygous SNPs. Coverage plots showing the single base read depth normalized to the individual chromosome mean values in representative parental FnSat line (black lines). (A) Heterozygous parental SNPs (red points) that did not segregate in Mendelian ratios are found within genomic regions with elevated coverage (normalized coverage > 2) across chromosomes (chromosomes 12, 14 and 15 in the upper, middle and lower panels, respectively). (B) Heterozygous parental SNPs (red points) that segregated to generate homozygous alleles in hybrids are found outside regions of elevated coverage (normalized coverage < 2) across the chromosomes shown (chromosomes 7, 27, and 30 in the upper, middle and lower panel, respectively). Blue arrows highlight heterozygous SNPs on chromosomes 27 and 30 within regions with coverage bias and whose segregation does not result in homozygous hybrid alleles (see Fig. 3A). Dashed grey lines highlight base read depth values equal to the chromosome mean (Y = 1). Y axis scale is limited to between 0 and 4 for closer inspection of values within such range. Download FIG S5, TIF file, 1.3 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Segregation pattern of all homozygous and heterozygous SNPs in both parents. Plots represent the frequency of all variant alleles identified on different chromosomes for FnHyg x FnSat crosses in (A) and FnHNeo x FnHSat crosses in (B), regardless of whether SNPs were identical or distinct between the parents. Parental SNP frequencies are shown in magenta, the corresponding allele on the same position are shown as yellow dots for the 2n hybrid progeny, and as green dots for all the respective parental subclones analyzed. Frequencies of heterozygous alleles in chromosomes that have a somy higher than 2 fall outside the0.4 to 0.6 range (e.g., chromosomes 31 and 23). Download FIG S6, TIF file, 2.5 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Aneuploidy comparisons between hybrids and subclones. Heatmaps showing the chromosome somy values for parental lines and hybrids with 2n DNA content in (A) and 3n in (B). Color gradient represents somies rounded up to the nearest integer, with values within each cell corresponding to each somy rounded to the nearest 0.1. The same heatmaps are presented in Fig. 4 without the somy values. (C and D) Chromosome somy levels are presented on the Y axis, shown in (C) for FnHyg and FnSat parental lines (dark pink circles), or FnHNeo and FnHSat parental lines (dark green triangles) compared with their corresponding subclones (respective symbols in light colors); or shown in (D) for parental lines compared with their 2n selfing hybrids (respective symbols in light colors). Download FIG S7, TIF file, 2.7 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distribution of somy scores in FnHyg × FnSat crosses. Somy values are shown for each chromosome as boxplots generated using GIP and giptools with genomic sequence coverage bins of 300bp (gip.readthedocs.io/en/latest/giptools/karyotype.html). Parental lines are presented in magenta, parental subclones in green and 2n selfing hybrids in yellow. Download FIG S8, TIF file, 1.3 MB (1.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distribution of somy scores in FnHNeo × FnHSat crosses. Somy values are shown for each chromosome as boxplots generated using GIP and giptools with genomic sequence coverage bins of 300bp (gip.readthedocs.io/en/latest/giptools/karyotype.html). Parental lines are presented in magenta, parental subclones in green and 2n selfing hybrids in yellow. Download FIG S9, TIF file, 1.1 MB (1.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The raw sequence data were deposited in the BioProject SRA database (PRJNA900217). The data that support the findings of this study are available from the corresponding author upon reasonable request.